Notes
Article history
The research reported in this issue of the journal was funded by the EME programme as award number 13/122/01. The contractual start date was in February 2015. The draft manuscript began editorial review in October 2020 and was accepted for publication in July 2023. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The EME editors and production house have tried to ensure the accuracy of the authors’ manuscript and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this article.
Permissions
Copyright statement
Copyright © 2024 Henriksen et al. This work was produced by Henriksen et al. under the terms of a commissioning contract issued by the Secretary of State for Health and Social Care. This is an Open Access publication distributed under the terms of the Creative Commons Attribution CC BY 4.0 licence, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. See: https://creativecommons.org/licenses/by/4.0/. For attribution the title, original author(s), the publication source – NIHR Journals Library, and the DOI of the publication must be cited.
2024 Henriksen et al.
Chapter 1 Introduction
The use of medical imaging has steadily and rapidly increased, becoming a central pillar in the management of patients, particularly in the setting of cancer care. With increasing use of complex modalities such as computed tomography (CT) and magnetic resonance imaging (MRI) for diagnosis, treatment planning and clinical studies, it has become desirable to use image-vision methods, such as machine-learning (ML) methods, to assist radiological experts in clinical diagnosis, quantification tasks and treatment planning. 1 In recent years, there have been rapid developments in both machine-learning methods as well as MRI techniques. For example, less than 10 years ago the use of whole-body magnetic resonance imaging (WB-MRI) protocols was uncommon due to many limitations, such as the forbidding acquisition times and limited availability. This past decade has shown substantial progress in WB-MRI protocols. This very promising technique has now started to move from the research setting to becoming more commonly used in clinical practice. It is currently recommended by the National Institute for Health and Care Excellence (NICE) for the detection of lesions in the bone marrow in myeloma and is used for detection of metastatic bone disease in prostate and breast cancer. 2–4 This clinical use has been supported by recent technological developments and validation of WB-MRI by multiple studies and consensus papers. 5 The STREAMLINE study of WB-MRI, in patients with newly diagnosed lung or colorectal cancer, reported similar staging accuracy using WB-MRI but with a reduced number of tests needed to reach the final determination of stage of disease and with a reduction in time to staging and cost. 6–8 A single WB-MRI was also preferred by patients. 9,10 As a result, WB-MRI is progressively proposed by radiologists as an efficient examination for an expanding range of indications. 11 Multi-modality WB-MRI is emerging as a new imaging standard for many diseases requiring detection and monitoring of skeletal and soft-tissue involvement in cancer. 12 WB-MRI has been successfully used in several cancers for the detection of bone, lymph nodes and visceral metastases and their monitoring under treatment. 13 Among these, metastatic cancers to bone and hematological malignancies, mainly multiple myeloma, benefit from WB-MRI. The potential benefits of using WB-MRI are considered to be a reduction in ionising radiation exposure, increased health outcomes due to reduced waiting periods and decreased patient anxiety caused by waiting periods for multiple staging investigations.
However, the radiological interpretation of WB-MRI scans requires a high level of expertise and training. Interpretation of WB-MRI requires integration of a large number of image data from multiple sequences. As a result, the reading process can become rather time-consuming for inexperienced readers, with increased risk of misinterpretations. 14 This has hindered the translation of this technique into widespread use and the routine use of WB-MRI is currently limited to a relatively small number of expert centres.
The principal challenge for translating WB-MRI in clinical routine comes from the technical skills for image acquisition and the large number of data to be reviewed. Computer-aided image analysis may alleviate the workflow. Such automatic algorithms could ultimately facilitate the process of reading WB scans by reducing the reading time (RT) and improving the diagnostic accuracy of WB-MRI.
The advent of deep learning has pushed medical image analysis to new levels, rapidly replacing more traditional ML and computer vision pipelines. Detection of anatomical tissue of interest is crucial for quantitative analysis of WB images. Many image detection and segmentation algorithms have been proposed and some machine-learning algorithms can now perform image analysis tasks with performance equal, or even superior, to the one achieved by human experts. 15,16 Automatically derived measurements and visual guides, obtained with machine-learning techniques, may serve as a valuable aid in many clinical tasks and are highly likely to transform the ways we see and use medical imaging. Reliable machine-learning algorithms are required for the accurate delineation of anatomical structures and/or lesions from different modalities of medical images.
In this project, our aim was to develop machine-learning methods for improving the diagnostic performance and reducing the RT of WB-MRI in colon and lung cancer in order to support the translation of WB-MRI into routine clinical care for the benefit of patients. Given the pragmatic setting of MAchine Learning In whole Body Oncology (MALIBO), we believe that the methodological steps and challenges described here can be of invaluable assistance, and can serve as a guide, to groups who would like to apply similar studies in the future, not only for MRI, but in radiology generally. 17
Background
Whole-body MRI, including diffusion-weighted magnetic resonance imaging (DW-MRI), is currently an active research interest in oncology imaging as a non-invasive technique for the detection of metastatic disease, as well as a potential biomarker for clinical use and drug development. 18 Meta-analyses support further development of WB-MRI in clinical practice, in view of the promising sensitivities and specificities for detection of metastases (pooled sensitivity 0.92 and pooled specificity of 0.9319,20). DW-MRI is now standard in WB imaging. DW-MRI allows quantification of water diffusivity in tissues and has been found to be sensitive for detecting tumour sites in organs and bones, with visible changes in the MRI signal intensity due to a reduction in water diffusivity associated with the highly cellular nature of tumour tissue. 21 The characteristic appearances of the bone marrow have been studied in relatively small numbers of patients without metastatic disease, and in patients with breast cancer, myeloma and prostate cancer. 22–24
Other than the requirement for extensive training and potentially slow nature of manual reads, one of the main issues when using WB-MRI for staging of patients with cancer is the potential number of false interpretations. Many ‘normal’ anatomical structures (such as lymph nodes) may reflect similar diffusion properties compared to pathological regions. The possibility of using computer-assisted reading or machine-learning (ML) techniques has been considered in aiding interpretation of complex MRI data sets. One group evaluated the topography of whole- body adipose tissue and proposed an algorithm that enables reliable and completely automatic profiles of adipose distribution from the WB data set, reducing the examination and analysis time to less than half an hour. 25 Another group has developed a parametric modelling approach for computer-aided detection of vertebral column metastases in WB-MRI. 26 ML techniques have previously been developed to differentiate benign (86 cases) from malignant (49 cases) in soft-tissue tumours using a large MRI database of multicentre, multimachine MRI images, but without using diffusion-weighted imaging (DWI). 27 Co-investigators at Imperial College London have previously developed methods for organ localisation in WB DIXON MRI and accurate semantic segmentation on CT. 28–32
Machine learning for image segmentation
Medical image segmentation aims to identify regions of interests (ROIs) from the image volume that are relevant to diagnosis or image interpretation. 33 Numerous researchers have proposed various automated segmentation systems, including, but not limited to, active contour, graph cut and clustering. These segmentation algorithms are built on traditional methods such as edge detection filters and mathematical methods. However, due to developments in neural networks in the past decade, convolutional neural networks (CNNs) dictate the state of the art in biomedical image segmentation. 34,35 One of the notable network architectures is based on encoder–decoder method for semantic segmentation, including fully convolutional networks (FCNs) and U-Net. 36,37 A successful three dimensional (3D) neural network for brain tumour segmentation was DeepMedic, introduced by Kamnitsas et al. 15 Kamnitsas et al. later enhanced an ensemble by combining three different network architectures, namely 3D FCN, 3D U-Net and DeepMedic, trained with different loss functions (Dice loss and cross-entropy) and different normalisation schemes. 38
Whole-body magnetic resonance imaging for oncology
Simultaneously, MRI techniques have experienced rapid development, allowing the use of WB-MRI in evaluating for cancer or vascular disease,12 which was not possible in the last decade. Recently, a meta-analysis was conducted to evaluate the diagnostic performance of WB-DWI technique in detection of primary and metastatic malignancies compared with that of whole-body positron emission tomography/CT (WB-PET/CT). 39 It was found that WB-DWI has similar, good diagnostic performance for the detection of primary and metastatic malignancies compared with WB-PET/CT [area under curve (AUC) of WB-MRI 0.966 vs. AUC of WB-PET/CT 0.984]. This suggests that WB-MRI can be used to replace WB-PET/CT in certain clinical settings, such as some cancer studies.
Cancer is a leading cause of death worldwide, accounting for an estimated 9.6 million deaths in 2018 (www.who.int/news-room/fact-sheets/detail/cancer). Colorectal and lung cancer are the third and fourth most common cancers in the UK, accounting for 13% and 12% of all new cancers, respectively, and they are the leading causes of cancer-related deaths in the UK. In both cancers, detection of metastatic disease is fundamental to treatment strategy. 8 Although a range of imaging tests are available for diagnosis and staging, including CT and PET-CT, there is growing interests in using WB-MRI as an alternative to multimodality staging pathways. This is because WB-MRI does not impart diagnostic ionising radiation to patients, and promising data support its ability to stage. 8,40 The recently reported National Institute for Health and Care Research (NIHR)-funded STREAMLINE study40 supports the use of WB-MRI for lung and colorectal cancer staging and this study was based on the data sourced from the STREAMLINE study. Apart from 51 WB-(DW)-MRI data set that have already been acquired,41 as part of whole-body protocol optimisation study, the trial used the WB-MRI data predominantly from the NIHR-funded STREAMLINE-C and STREAMLINE-L studies. 8 Additional cases were obtained from the CRUK (Cancer Research United Kingdom) funded MELT study (Whole-Body Functional and Anatomical MRI: Accuracy in Staging and Treatment Response Monitoring in Adolescent Hodgkin’s Lymphoma Compared to Conventional Multi-modality Imaging: NCT01459224). 42 Also, data from the MASTER study [MRI Accuracy in STaging and Evaluation of Treatment Response in Cancer (Lymphoma and Prostate-MASTER L and MASTER P)] were employed (12/LO/0428). 43 These data sets demonstrated additional cases of nodal disease and bone metastases, thereby ensuring a variation in the distribution of disease used to develop the ML algorithms. However, due to extensive heterogeneity in the data from the different studies, the STREAMLINE studies provided the final cohort for the MALIBO study.
Existing literature using machine learning for lesion detection from whole-body magnetic resonance imaging study
We searched PubMed for articles with medical subject headings (MeSH) and full-text searched for ‘ML and MRI’, ‘WB-MRI and lesion detection’, ‘WB-MRI segmentation’ and ‘ML and WB-MRI’. We did not set the beginning of the publish time from PubMed, but our ending publish time was until 1 October 2020.
With ‘ML and MRI’ as keyword, we found there were 3325 papers from PubMed. As we can see, the number of publications in ML and MRI is increasing constantly. There are also number of available reviews. 44–50 This covers a wide range of research works from neuronal networks to deep learning and from image segmentation to disease prediction.
A total of 817 papers were found from PubMed using ‘WB-MRI and lesion detection’ as keywords. Many of these are for detection of bone lesions (using ‘WB-MRI and lesion detection and bone’ as keywords there are 356 papers, i.e. 356/817 = 0.44), suggesting there is great interest in applying WB-MRI in general and WB-DWI for bone study in particular.
If we used ‘WB-MRI segmentation’ as keywords from PubMed, there were fewer articles, which include prostate,51 skeleton,52 blood vessel segmentation based on magnetic resonance angiography (MRA)53 and manual tumour segmentation. 54 When limiting the keywords to ‘ML and WB-MRI’, besides our previous studies14,17 and a study for small-animal organ segmentation from WB-MRI using multiple support vector machine (SVM)-kNN (k-nearest neighbour) classifiers,55 there were only the following articles that are described below:
Firstly, a SVM method was used to segment prostate from WB-MRI scans. The method employed 3D neighbourhood information to build classification vectors from automatically generated features and randomly selected 16 MRI examinations for validation. 56 The result suggested that the SVM for prostate segmentation can segment the prostate in WB-MRI scans with good segmentation quality.
Secondly, a combined segmentation method which included image thresholds, Dixon fat-water segmentation and component analysis were adopted to detect the lungs. MRI images are segmented into five tissue classes (not including bone), and each class is assigned a default linear attenuation value. The method was assessed using WB-MRI. 57
Thirdly, a fully automated algorithm for extraction of the 3D-arterial tree and labelling the tree segments from WB-MRA sequences was presented. The algorithm developed consists of two core parts: (1) 3D-volume reconstruction from different stations with simultaneous correction of different types of intensity inhomogeneity, and (2) extraction of the arterial tree and subsequent labelling of the pruned extracted tree. A subjective visual validation of the method, with respect to the extracted tree, was performed. The results indicated clinical potential of the approach in enabling fully automated and accurate analysis of the entire arterial tree. This was the first study that not only automatically extracts the WB-MRA arterial tree, but also labels the vessel tree segments. 58
Thus, there are few studies in the field of using ML for human WB-MRI evaluation. The major reasons for this may include challenges related to segmenting and labelling anatomical regions due to appearance variations, the frequent presence of imaging artefacts, and a paucity and variability of annotated data. In addition, ML, particularly, with deep NN (neural network), has only been more widely developed in the last decade. As a result, there were not many described ML methods for human WB-MRI studies. Furthermore, WB-MRI itself is a relatively new technique which has only been established in the clinical setting in the last decade. 11 Although there were only a few related studies in this field, the application of ML methods to WB-MRI is thought to be of potential value in oncological imaging, to support the radiologist reading a complex imaging study that requires integration of fairly extensive information.
Clinical studies that this study relates to
The MALIBO study proposed to use WB-MRI data predominantly from the NIHR-funded STREAMLINE L and C studies. These are multicentre prospective cohort studies that evaluated WB-MRI in newly diagnosed non-small cell lung cancer (250 patients; STREAMLINE-L; ISRCTN50436483) and colorectal cancer (322 patients; STREAMLINE-C; ISRCTN43958015). 40 The studies initially defined WB-MRI acquisition, quality assurance and analysis protocols applicable to daily NHS practice. The objectives of both studies are the same. The primary objective was to evaluate whether early WB-MRI increases detection rate for metastases compared to standard NICE-approved diagnostic pathways for each of the tumour types studied (a full description of the diagnostic pathways is available). 6–8,40 Secondary objectives included assessing the influence of WB-MRI on time to and nature of first major treatment decision following definitive staging. At 12-month patient follow-up, a multidisciplinary consensus panel defined the reference standard for tumour stage considering all clinical, pathological, post-mortem and imaging follow-up. Accuracy was defined per lesion, per organ and per patient.
The STREAMLINE-C study recruited patients from 16 UK hospitals between March 2013 and August 2016 with a final number of evaluable patients of 299, 68 (23%) of whom had metastasis at baseline. The STREAMLINE-L study recruited patients from 16 UK hospitals between March 2013 and September 2016 with a final number of evaluable patients of 187, 52 (28%) of whom had metastasis at baseline. The ISRCTN for STREAMLINE-L is ISRCTN50436483 and for STREAMLINE-C, it is ISRCTN43958015. 40
Additional cases were obtained from the CRUK-funded MELT study (Whole-Body Functional and Anatomical MRI: Accuracy in Staging and Treatment Response Monitoring in Adolescent Hodgkin’s Lymphoma Compared to Conventional Multi-modality Imaging, NCT01459224)59 and the University College London Hospital (UCLH)-sponsored MASTER study, including cases with lymphoma and prostate cancer was also used. 42,43 The original justification for using these data sets was that they would demonstrate additional cases of nodal disease and sclerotic bone metastases, thereby ensuring a variation in the distribution of disease used to develop the ML algorithm, as the cases from STREAMLINE studies are likely to have more non-nodal metastatic sites, such as liver (LVR) and lytic bone metastases. The purpose of the MELT study was to compare staging accuracy as well as response assessment using WB-MRI with standard investigations in patients with newly diagnosed Hodgkin’s lymphoma. It was a prospective observational cohort study. The primary outcome measures were: per-site sensitivity and specificity of MRI for nodal and extra-nodal sites and concordance in final disease stage with the multimodality reference standard (at staging). The reference standard for the MELT study was contemporaneous multidisciplinary tumour board (MDT) with all other staging, for example, PET-CT and CT at the time of diagnosis and initial staging. Secondary outcome measures included: (1) interobserver agreement for MRI radiologists, and (2) evaluation of different MRI sequences on diagnostic accuracy; simulated effect of MRI on clinical management.
Rationale for the study
In order to make WB-DW-MRI a useful and clinically relevant tool within the NHS, a method that could assist the radiologist in improving diagnostic accuracy while reducing RT would be beneficial to deliver better accuracy, productivity and cost-effectiveness. An important aspect in the development of diagnostic support systems is semantic understanding of input data. In the case of WB-DW-MRI, it is essential to ‘teach’ the computer to automatically detect and localise different anatomical structures and discriminate normal and pathological appearances. A computer system that is able understand what is shown in an image can be effectively used to implement an intelligent radiology inspection tool. Such a tool may greatly support the radiologist when reading the large amount of MRI data, with integration of different MRI sequences. Guided navigation to ROI, automatic adjustment of organ and tissue specific visualisation parameters, and quantification of volume and extent of suspicious regions are some of the features that such a system would provide and thus, potentially reduce the time needed for an expert to perform diagnostic tasks. Previous ML methods (described in Image pre-processing for whole-body-magnetic resonance imaging: correction of fat-water swaps in Dixon magnetic resonance imaging) can be adapted to WB-DW-MRI to allow automatic vertebrae localisation,26 to automatically exclude false-positive detections in suspicious regions and to discriminate malignant from benign structures28–32). These methods have yielded promising results for their respective tasks. They are all based on a particular concept of ML called supervised learning. In supervised learning the assumption is that some annotated training data are available that can be used to train a predictor model. Here, the annotations reflect the output value that one would like to infer for new patient images. The training data can be defined as a set T = {(X!, Y!)} of pairs of input data Y!, here a WB-DW-MRI, and some desired output Y!, for example, a point-wise probability map that indicates the likelihood for each image point to be malignant. Using the training data, the aim of an employed learning procedure is then to estimate the conditional probability distribution (Y|X). Having a good estimate of this distribution allows prediction of output Y for any new input data X. In the context of WB-MRI for staging, the automatically obtained predictions for a new patient image can be integrated in a radiology inspection tool, for example to automatically navigate to or highlight suspicious region.
Objectives of this study
As this is a new area for oncology imaging research, there were no previous ML or CNN developed tools to apply to WB-MRI for tumour staging nor for lesion detection. To the best of our knowledge, the applicability of this CNNs method for lesion segmentation from WB-MRI had not been investigated in a clinical setting at the time the study was commenced. Allowing for this, the main purpose of our project was to study the possibility to apply state-of-the-art CNN methods for lesion detection on human WB-MRI, particularly, for detecting lung and colon cancer and any metastatic lesions. Our plan was to develop, compare and select the most appropriate ML methods to achieve these goals.
Primary and secondary objectives
The primary objective of Phases 1 and 2 of the study was to develop a ML method (or modify an available method) for the detection of cancer lesions on WB-MRI.
The primary objective of Phase 3 of the study was to compare the diagnostic test accuracy of WB-MRI, as read by independent, experienced readers in patients being staged for cancer, with and without the aid of ML support, against the reference standard from the source studies.
Secondary objectives
The planned secondary objectives of this study were:
-
To compare the RT of WB-MRI scans, as recorded by experienced readers, with and without the assistance of ML methods.
-
To determine the interobserver variability of WB-MRI diagnosis of primary and metastatic lesions by different radiologists, with and without the assistance of ML methods.
-
To evaluate the diagnostic accuracy of WB-(DW)-MRI, as reported by non-experienced readers, with and without the assistance of ML methods.
-
To estimate the diagnostic performance of WB-(DW)-MRI when using different combinations of acquired MRI sequences, with and without the support of ML methods.
-
To determine the number of potential, additional staging tests that would be redundant if ML methods were applied in WB-(DW)-MRI, by means of a simple cost-effectiveness analysis.
Most of the study objectives were achieved, step by step, in the three phases of this study.
Study design
This is an observational study (study limited to working with data), using three different patient cohorts, being evaluated in series during three consecutive phases.
Phase 1: segmentation of normal organs
Development and optimisation of a ML pipeline to automatically identify anatomical structures of interest in WB-MRI in healthy volunteers. For automatically labelling anatomical structures of interest, we extended previous work that automatically segments abdominal organs from CT data. 30 More specifically, we used a hierarchical weighting approach in which the anatomical atlases were constructed first at participant level and then followed by atlas construction at organ level and finally at voxel level. This approach has been shown to accommodate the significant body anatomical variability across different participants. By combining this with patch-based segmentation we were able to accurately and robustly annotate anatomical structures of interest. In order to construct the anatomical atlases, whole body MRI data sets from 50 healthy volunteers were used; these were collected under a separate ethics approval [Imperial College Research Ethics Committee (ICREC) 08/H0707/58]. The initial ML output was the ability to automatically segment the normal organs.
Phase 2: ‘training and validation set’ in cancer lesions
In the second phase, we planned to develop the ML pipeline for the automatic detection and identification of cancer lesions. For this we learnt shape and appearance models that were specific to the anatomical regions identified in Phase 1. These models allowed the probabilistic interpretation of the images in terms of a generative model. Classification was carried out using advanced ML techniques based on ensemble classifiers such as random forests (RFs). 60 WB-MRI scans from the STREAMLINE L and C, MELT and MASTER studies with established disease stage (main study reference standard, described above) were used to train ML detection of metastases. During the course of the study, it became apparent that the differences in sequences and acquisition parameters of the studies were too different to allow appropriate ML training and the study went forward with only the STREAMLINE C (STC) and L (STL) data sets, which had similar MRI protocols. Despite restricting Phases 2 and 3 to the STREAMLINE data, significant challenges were encountered due to the variation in scans acquired at the 16 recruitment centres. We undertook a power calculation to determine the number of cases with and without metastases that would be needed for the Phase 3 study, based on a hypothetical diagnostic test accuracy improvement. All of the remaining eligible data was then used for training. Allocation of cases to Phases 2 and 3 was undertaken by the study statistician in order to allow appropriate cases for training and ‘held-back’ cases for validation. A total of 245 WB-MR scans were allocated to Phase 2 for model development (the original protocol planned for a minimum of 150 scans), of which 19 could not be used due to technical failure, with a final number of 226 WB-MRI available for development. All lesions were segmented on T2 and DWI volumes and checked by an expert (accredited radiologist) using the reference standard to ensure that ground truth (GT) was as accurate as possible. Initial radiology outputs were reviewed by expert radiologist and ML team to identify areas for improvement and the ML development focused on accurate detection of the GT segmented sites of disease. The algorithm was gradually improved and was then tested on the internal validation set to ensure sensitivity of detection was met, before progressing to the clinical testing. A validation step in phase 2 was undertaken on 45 cases that were allocated to Phase 2 training but withheld from model development. Sensitivity for detection of metastatic lesions by the algorithm was evaluated using a Dice coefficient metric, with sufficient overlap and probability thresholds achieved. The threshold was determined as we developed the algorithm in the early stages of Phase 2. Our initially planned reader study, at the end of Phase 2, was not undertaken and the described computational method was employed. Cases used for the sensitivity check at the end of Phase 2 were not used for any ML training or read by radiologists in Phase 3.
Phase 3: ‘clinical validation set’
The allocated Phase 3 data set for clinical validation was comprised of 193 WB-MRI (217 were originally planned in the protocol) from the STREAMLINE C and L studies that had not been used for Phase 2 training. These scans were to be read by experienced readers with the fixed final ML support, which was provided as a probability map (a heatmap) which could be overlayed on the T2-weighted stitched volumes of the WB-MRI and this was used as the index test. The WB-MRI reads by experienced radiologists without ML support was the comparator test. The per-patient specificity and sensitivity of WB-MRI assessment, with and without ML support, was determined using the established reference standard from the main study. An interim analysis of the first 50–70 consecutive cases was planned but was not undertaken due to late running of the study. A scribe recorded the reader findings on the study case report form (CRF) and the RT was recorded. Substudies included: (1) Reads by new (inexperienced) WB-MRI readers; (2) Repeat reads (in random order and at time interval) with and without ML support to measure RT and interobserver variation. This ensured parity in computer setup between the reads, as there may have been variation in the original main study reads related to use of either picture archiving and communication system (PACS), Biotronics3D platform or other software, in addition to differences in internet speeds when reads were performed online for the main study.
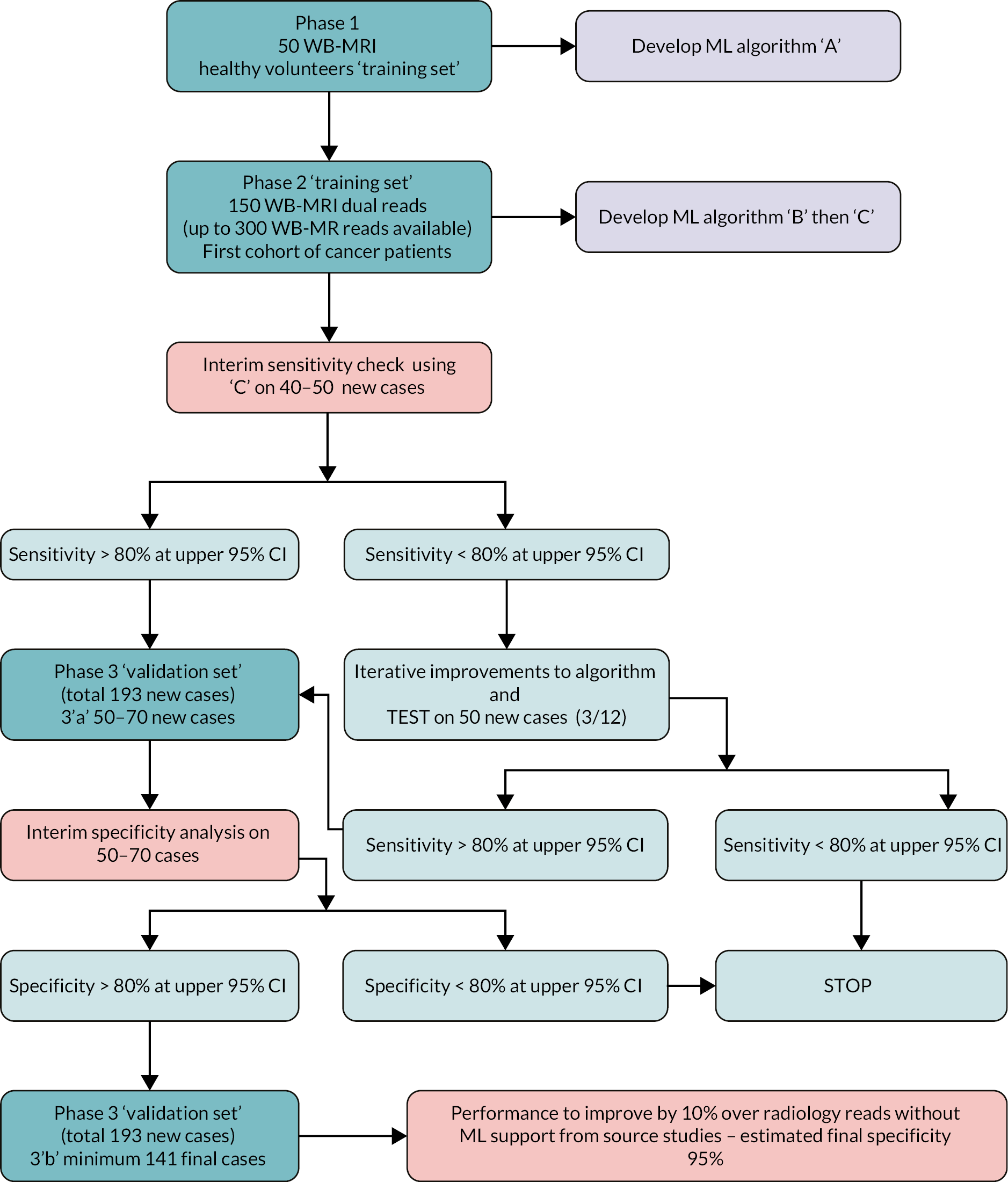
Figure 1 shows the study design flow diagram for this project.
FIGURE 1.
Planned study design flow diagram from the study protocol.
Patient public involvement
A patient representative took part in the development of the grant application, with all study materials being developed during the course of the study planning. The patient representative also was part of the trial steering committee. Discussions concerning use of patient data in de-identified format were constructive and the idea of using ML techniques to potentially improve accuracy was seen as a positive area of research. This study did not recruit patients directly and so there were no patient-facing materials for review. However, the patient representative has been kept abreast of the developments through the course of the study.
Chapter 2 Phase 1: healthy volunteer data collection and pre-processing fat-water swap artefact
This chapter includes material previously published by the authors in references. 41,61 The MRI protocol used for the healthy volunteer WB-MRI acquisition was first published in reference 41 and permission to reproduce the MRI protocol was given by the American Journal of Roentgenology. The development of a methodology for correction of fat-water swap was published in reference 61 as open access article distributed in accordance with the terms of the Creative Commons Attribution (CC BY 4.0) license, permits others to distribute, remix, adapt and build upon this work, for commercial use, provided the original work is properly cited. See: http://creativecommons.org/licenses/by/4.0/. The text below includes minor additions and formatting changes to the original text.
Volunteers and magnetic resonance imaging scans
The source study for the healthy volunteer data was approved by the local ethics committee (ICREC 08/H0707/58 for optimisation of MRI protocols used in clinical practice and translational research) and written consent was obtained from the participants. Fifty-one healthy volunteers [24 men (mean age = 37 years, age range = 23–67 years) and 27 women (mean age = 39 years, range = 23–68 years)] were scanned with WB-MRI from February 2012 to May 2014 at a single institution and scanned on the same machine.
Details about the volunteers’ population and scan protocol can be found in the reference. 41 The inclusion and exclusion criteria were the following:
Inclusion criteria:
-
male or female, healthy volunteers were aged 18–100 years;
-
written, informed consent was provided.
Exclusion criteria:
-
any co-existing medical illness;
-
contraindications to MRI (e.g. patients with pacemakers, metal surgical implants and aneurism clips, patients suffering from claustrophobia).
Magnetic resonance imaging protocol for healthy volunteers
Whole-body magnetic resonance imaging was performed on a moving-table 1.5-T system (Avanto with Syngo MR B17, Siemens Healthcare) using the body coil for transmission and the neck and body phased-array coils as receive coils. Four different imaging stations were used to achieve full body coverage, from the top of the neck to mid-thighs. Axial slices were acquired during free-breathing for DWI, whereas breath-holds were used for the three first stations for anatomic imaging. The acquisition time was approximately 45–50 minutes depending on the breath-holding ability of the examined participant. DWI slice-matched T1-weighted imaging with Dixon and T2-weighted imaging was performed to assist with the delineation of abdominal organs and bone marrow. The MRI protocol is provided in Table 1. 41
| DW-MRI | T1-W MRI | T2-W MRI | |
|---|---|---|---|
| Sequence type | SS SE EPI | VIBE with DIXON | HASTE |
| FOV (mm) | 450 × 366 | 450 × 351 | 450 × 366 |
| Matrix size | 128 × 128 interpolated |
320 × 202 | 256 × 256 |
| No of slices/thickness/distance (mm) | 50/5/0% | 56/5/20% | 50/5/0% |
| TR (ms) | 9000 | 7.54 | 767 |
| TE (ms) | 72 | 2.38/4.76 | 92 |
| Bandwidth (Hz/pixel) | 2056 | 300 | 399 |
| Flip angle | 90 | 10 | 130 |
| N A | 4 | 1 | 2 |
| Fat suppression | STIR (TI = 180 ms) |
N/A | N/A |
| b-values (s/mm2) | 0, 150, 400, 750, 1000 | N/A | N/A |
| Parallel acquisition | GRAPPA 2 | GRAPPA 2 | GRAPPA 2 |
| No stations | 4, free-breathing | 4, (3 with breath-holds) | 4, (3 with breath-holds) |
| TA (min)/station | 8.17 | 0.15 | 1.18 |
Representative slices showing ROIs used to calculate apparent diffusion coefficient (ADC) values are given in Figure 23 of Appendix 1. Also, scatterplots of ADCALL with age are displayed in Figure 24 of Appendix 1, and ADC values calculated from perfusion-sensitive WB-DWI protocol (ADCALL) vary with fat fraction (FF), shown in Figure 25 of Appendix 1.
Image pre-processing for whole-body magnetic resonance imaging: correction of fat-water swaps in Dixon magnetic resonance imaging61
In Phase 1 of the MALIBO project, we collected WB-MR images in 51 healthy volunteers with a Dixon sequence (Table 1). The Dixon method is a MRI sequence based on chemical shift and designed to achieve uniform fat suppression. However, fat- and water-only swap artefact may occur in up to 10% of scans, impacting on subsequent analysis.
We developed a new method to correct fat-water swap, based on regressing fat- and water-only images from in- and out-of-phase images by learning the conditional distribution of image appearance. We demonstrated the effectiveness of our approach on WB-MRI with various types of fat-water swaps. 61
Chapter 3 Phase 1: fully automatic, multi-organ segmentation in normal whole-body magnetic resonance imaging, using classification forests, convolutional neural networks and a multi-atlas approach14
Parts of this chapter are reproduced from Lavdas et al. 14 under licence agreement with John Wiley and Sons, number 5176490914331. Phase 1 of the MALIBO study involved the development and optimisation of a ML pipeline to automatically identify and segment anatomical structures of interest in normal WB-(DW)-MRI. In order to construct the anatomical atlases, WB-MRI data sets from 51 healthy volunteers were used (see Chapter 2 for description of data set or Lavdas et al. 41). These had already been collected under a separate ethics approval (ICREC 08/H0707/58). During this phase, we developed shape and appearance models that were specific to the normal anatomical regions as identified in WB-MRI. Classification was carried out using advanced supervised ML techniques based on ensemble classifiers, such as RFs,62 deep-learning algorithms such as CNNs-(15) or a multi-atlas (MA) approach. 63 In this phase, three different algorithms for automatic segmentation of the healthy organs and bones were completed. This phase has been successfully completed and published. 14
Machine-learning methods for image segmentation
Imaging protocol
Please see Chapter 2 for the full description of the MRI protocol that was used for scanning the 51 healthy volunteers. The full imaging protocol is shown in Table 1.
Machine-learning pipeline
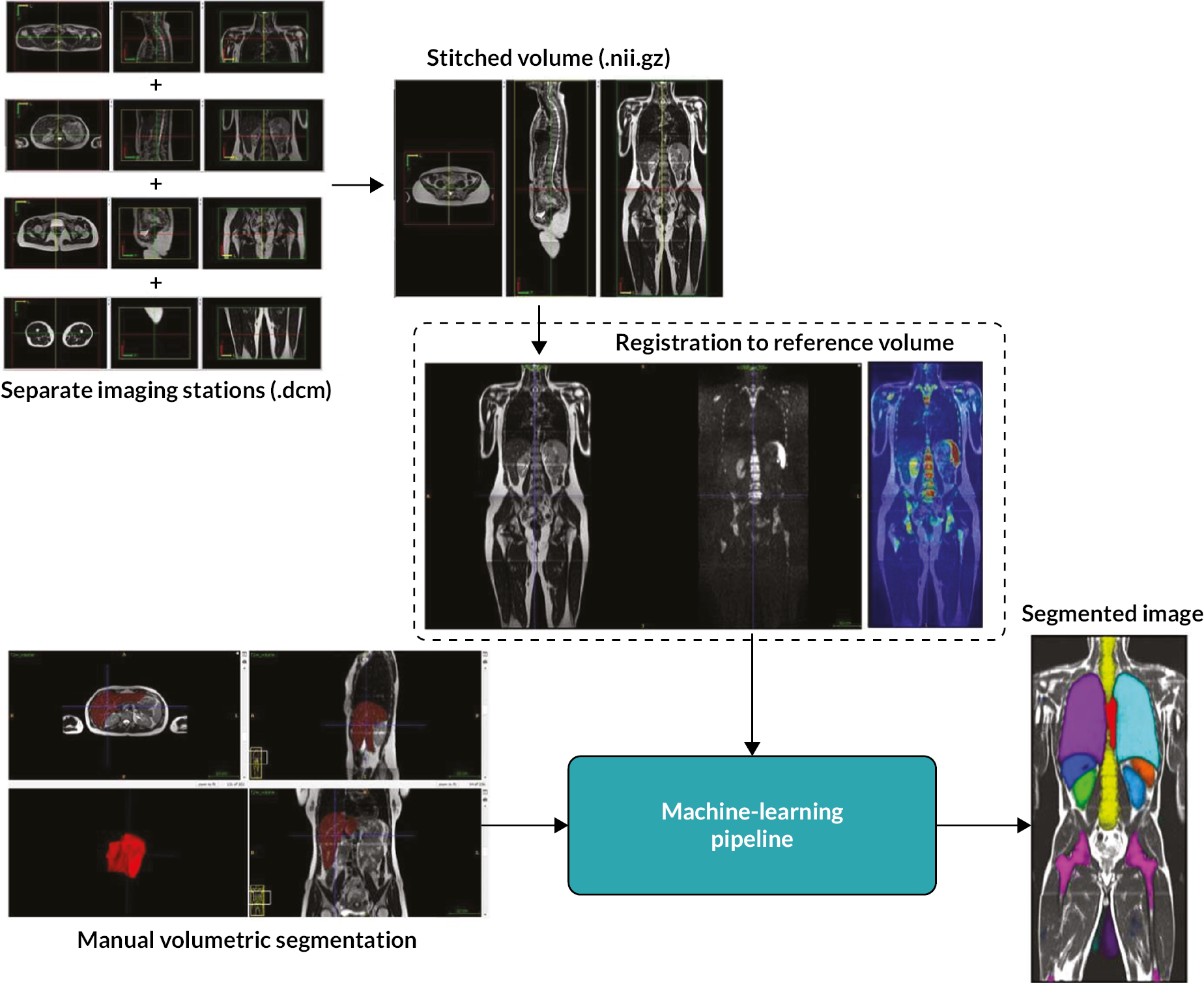
Digital Imaging and Communications in Medicine (DICOM) data from individual imaging stations were stitched into single Neuroimaging Informatics Technology Initiative (NIfTI) volumes (https://nifti.nimh.nih.gov/). The stitching method was performed in the following: First, MRI images were loaded from the individual stations (or sub-volumes), typically about six but this may vary between participants. Next, the first station that is provided in the list of stations was used as a reference for the in-plane resolution and number of voxels (because these can vary between stations). Using this reference standard, we resampled each station to the same in-plane resolution as the reference station. After that, there is a read out of the physical location of each station from the DICOM header information [this is typically stored in a DICOM tag called ‘Image Position (Patient)’]. Finally, using the physical location of each station, it calculates the output volume where all stations are ‘stitched’ together to form a continuous volume. Adjacent stations often have a small overlapping region. The information of the station that lies cranially (from a head-to-toe direction) was simply used.
The stitched MRI volume data were used as input data to the algorithms, while training was based on manual annotation of the anatomic ROI on the T2-weighted volumes, first segmented by two radiology trainees and then checked by a MRI expert. The expert checked the segmentations, which were adjusted, if needed, and agreed in consensus. When multimodal MRI data were used as input to classification forests (CFs) and CNNs (e.g. T2w + T1w + DWI data, where T1w refers to T1w in- and opposed-phase images from the DIXON acquisitions, and DWI refers to b = 1000 s/mm2 images and ADC maps), an extra registration step was attempted, to add to the data preparation pipeline. However, in the final pipeline, no registration was undertaken. During this step, T1w and DWI volumes were registered to the T2w volumes using an affine transformation. A schematic overview of the data preparation process, including the registration step, is given in Figure 2. Data from different imaging stations are stitched to single volumes and then intrapatient registration is performed (when using multimodal MRI data as input to the algorithms). Manual segmentation and annotation of the anatomic ROI was also performed to generate training data for the ML algorithms.
FIGURE 2.
Magnetic resonance imaging image data preparation pipeline. Diagrammatic flowchart of the data preparation process for the ML pipelines. CFs algorithm. Reproduced from Lavdas et al. 14
Classification forests are powerful, multilabel classifiers that facilitate simultaneous segmentation of multiple organs. They have very good generalisation properties, meaning that the algorithm can be effectively trained using a relatively small number of annotated example data, a particularly important advantage in the clinical setting. CFs are a supervised, discriminative learning technique, which is based on RFs; an ensemble of weak classifiers called decision trees. Each decision tree is constructed in a way that it produces a partitioning of the training data, for example, image points that carry organ label information, in a way that training data with same labels are grouped together. This is achieved by building the trees from the root node down to the leaf nodes. Internal nodes, so called split nodes, separate the incoming data into two sets. Leaf nodes then correspond to small clusters of training data from which label statistics are computed and are used for predictions at testing time. Data splitting in the trees is based on an objective function, which maximises the information gained over empirical label distributions. The goal is to select discriminative features at split nodes that are best for partitioning the data. Different trees are built by injecting randomness for both feature selection and training data subsampling. This ensures decorrelation between trees and has proven to yield good generalisation properties. During testing, image points from a new image are ‘pushed’ through each tree until a leaf node is reached. The label statistics over training data that are stored in the leaf nodes are aggregated over different trees by simple averaging, and a final decision on the most likely label is made based on this aggregation. Intuitively, image points will fall into leaf nodes that contain similar image points from the training data with respect to the features that are evaluated along the path from root to leaf node. An attractive property of CFs is their ability to automatically select the right image features for a given task, from a potentially very large and high-dimensional pool of possible features. This is important because preselecting or handcrafting image features beforehand can be very difficult, as it is not known in advance which features are discriminative for the task at hand. In CFs, the user only has to provide weak guidance on the ranges of parameters that are used to randomly generate potential features. In this work, we make use of the popular offset box features, which have been shown to provide effective means of capturing local and contextual information. 64–66 Box features are very efficient to compute, which is beneficial for training and testing. In box features, intensity averages are calculated within randomly sized and displaced 3D boxes. Two types of features are computed: single-box and two-box features. Single-box features simply correspond to the average intensity of all voxels from a particular MRI sequence that fall into a 3D box. Two-box features return the difference between the averages computed for each of the two boxes and generalise intensity gradient features. Here, each box can be taken from a different MRI sequence and, thus, yield cross-sequence information.
Tuning parameters for our algorithm have been set accordingly to knowledge from previous applications, such as vertebra localisation in whole-body CT scans. We have used CFs extensively for related tasks for which cross-validation has been used to optimise hyperparameters such as tree depth. In this work, we used 50 trees with a maximum tree depth of 30. The stopping criterion for growing trees is if either the objective function (information gain) cannot be further improved or the number of training samples in a leaf fall below a threshold of four samples. We found that neither increasing the number of trees nor the tree depth increases the segmentation accuracy of the CFs.
Convolutional neural networks algorithm
Convolutional neural networks are feed-forward artificial neural networks, which have recently emerged as powerful ML methods for image analysis tasks, such as segmentation. CNNs are capable of learning complex, nonlinear data associations between the input images and segmentation labels through layers of feature extractors. Each layer performs multiple convolutional filter operations on the data coming in from the previous layer and outputs feature responses, which are then processed by the next layer. The last layer in the network combines all the outputs to make a prediction about the most likely class label for each voxel in an image. The parameters of the convolutions and weights for combining feature responses are learnt during the training stage, using an algorithm called back-propagation. The layered architecture enables CNNs to learn complex features automatically without any need for guidance from the user. The features correspond to a sequence of filter kernels learnt in consecutive layers of the neural network. A final feature that is used for classification thus can correspond to a nonlinear combination of individual features that are extracted hierarchically. This is also called features-of-features, as filter kernels in deeper layers are applied to the feature responses of earlier layers. This is different to CFs, where the user has to define a pool of potential features beforehand from which the most discriminative ones are then selected during CF training. However, CNNs come with an increased computational cost during training, and they have multiple meta-parameters that need to be highly tuned to achieve optimal performance, a process which can be challenging for less experienced users. In addition, defining the right network architecture is a challenge on its own and a field of active research.
In this project, we made use of a recently published CNN approach that we developed originally for the task of brain lesion segmentation in multiparametric MRI. The approach, called DeepMedic,15 uses a dual pathway CNN that processes an image at different levels of resolution simultaneously. This has the advantage that features are based on both local and contextual information, something that can be particularly appealing in the case of whole-body multiorgan segmentation. For example, the left and right kidneys (LKDN and RKDN) might look very similar locally and share similar features at small scale, but the contextual features that cover larger regions of the images allow the discrimination between the left and right body parts.
The CNN configuration used here follows largely the default configuration that has been previously used for brain lesion segmentation. To accommodate for larger context in the case of organ segmentation, the receptive field for the low-resolution pathway has been increased by using an image downsampling factor of 3. We use a dual pathway (two resolutions), 11-layer deep CNN, where the last two layers correspond to fully connected layers, which combine the features extracted on the two resolution pathways. We employ 50–70 feature maps (FMs) (i.e. different kernels) for each layer. The network architecture is fully convolutional and there are no max-pooling layers, which we find to increase segmentation accuracy. The CNN architecture is a balance between model capacity, training efficiency and memory demands.
Multi-atlas algorithm
A MA label propagation approach was also employed in this study. 67 MA segmentation uses a set of atlases (images with corresponding segmentations) that represent the interparticipant variability of the anatomy to be segmented. Each atlas is registered to the new image to be segmented using a deformable image registration. The MA approach accounts for anatomical shape variability and is more robust than single-atlas propagation methods in that at any errors associated with propagation, are averaged out when combining multiple atlases. The approach employed here makes use of efficient 3D intensity-based image registration68 with free-form deformations as the transformation model and correlation coefficient as the similarity measure. Majority voting is used to derive the final tissue label at each voxel.
Implementation, training and validation procedure
Training of CFs and CNNs is a demanding process computationally and in our case took up to 12 hours for CFs and 30 hours for CNNs for a single fold with 27 images, when using a quad-core Intel Xeon 3.5 GHz workstation with 32 GB RAM and a NVIDIA Titan X graphics processor unit (GPU). Our CFs implementation uses all available central processor units (CPUs), while the CNN implementation runs mostly on the GPU. Training only needs to be performed once. Testing of new data points to obtain the full segmentation of an image is a particularly efficient process and takes about a minute for CFs and CNNs. Note that the MA algorithm does not require any training, but has considerably longer running time during testing which scales linearly with the number of atlases. To segment a single image using 27 atlases takes about 15 minutes on CPU.
We ran five-fold cross-validation experiments on 34 artefact-free data sets to assess the agreement of segmentations between the ones from the developed algorithms and the ones from the clinical experts. All data sets were inspected by an expert radiologist before being selected for validation. Data sets with severe motion artefacts or DWI data sets with severe distortion artefacts, and therefore severe misalignment, were excluded from validation.
We report six metrics (three overlap and three surface distance-based measures) to assess the agreement between automatic segmentation results from our algorithms and the manual segmentations performed by the clinical experts. The Dice similarity coefficient (DSC) quantifies the match between the two segmentations (1 = complete overlap, 0 = no overlap). Recall (RE) can be expressed in terms of sensitivity (1 = no misses) and precision (PR) can be expressed in terms of specificity (1 = no false positives). The average surface distance (ASD) is the average of all the distances from points on the boundary of the automatic segmentation to the boundary of the manual segmentation (0 = perfect match), the root-mean-square surface distance (RMSSD) is calculated in the same way as the ASD, except that the distances are now squared (0 = perfect match). Finally, the Hausdorff distance (HD) or maximum surface distance is the maximal distance from a point in the first segmentation to a nearest point in manual segmentation (0 = perfect match). 69 The three surface distance metrics are expressed in millimetres and are unbounded.
We measured the above metrics for the right and left lungs (RLNG and LLNG), LVR, gallbladder (GBLD), RKDN and LKDN, spleen (SPLN), pancreas (PNCR), bladder (BLD), spine (SPN) and pelvic bones, including the femurs [pelvis (PLVS)] for all three algorithms, when using T2w volumes as inputs. Then, we did the same when using all imaging combinations (T2w + T1w + DWI) as inputs to CFs and CNNs.
Statistical analysis
One-way analysis of variances (ANOVA) was used to compare the mean metrics for all the examined structures between the three algorithms. Post hoc analysis (multiple comparisons) was performed with a Tukey test. In cases where the homogeneity of variances was violated, a Kruskal–Wallis test was used. A Mann–Whitney U-test was used to compare the performance between CFs and CNNs when using T2w volumes as input to the algorithms and when using all imaging combinations (T2w + T1w + DWI). ANOVA and Mann–Whitney U-tests were similarly used to compare the DSC of individual anatomical labels between the three algorithms and between CFs and CNNs when using different imaging inputs. Finally, a Mann–Whitney U-test was used to compare the DSC between CFs with all imaging combinations (T2w + T1w + DWI) as input and CNNs with T2w images as input only, for each anatomical label. A significance level of 0.05 was used for all tests. Statistical analysis was performed in SPSS 21.0 for Windows (SPSS, Chicago, IL, USA).
Results
It is noteworthy that an ‘at a glance’ qualitative assessment reveals that CNNs outperform CFs and the MA algorithm in DSC, RE and PR (Figure 3), while the MA algorithm seems to perform best in terms of surface distance metrics, namely ASD, RMSSD and HD.
FIGURE 3.
Examples of segmentation results from three algorithms. T2w representative coronal (top row) and axial slices (bottom row), manual and automatic segmentations of major organs (lungs, heart, kidneys, LVR, and SPLN) and bones (SPN and femurs) from the three algorithms.
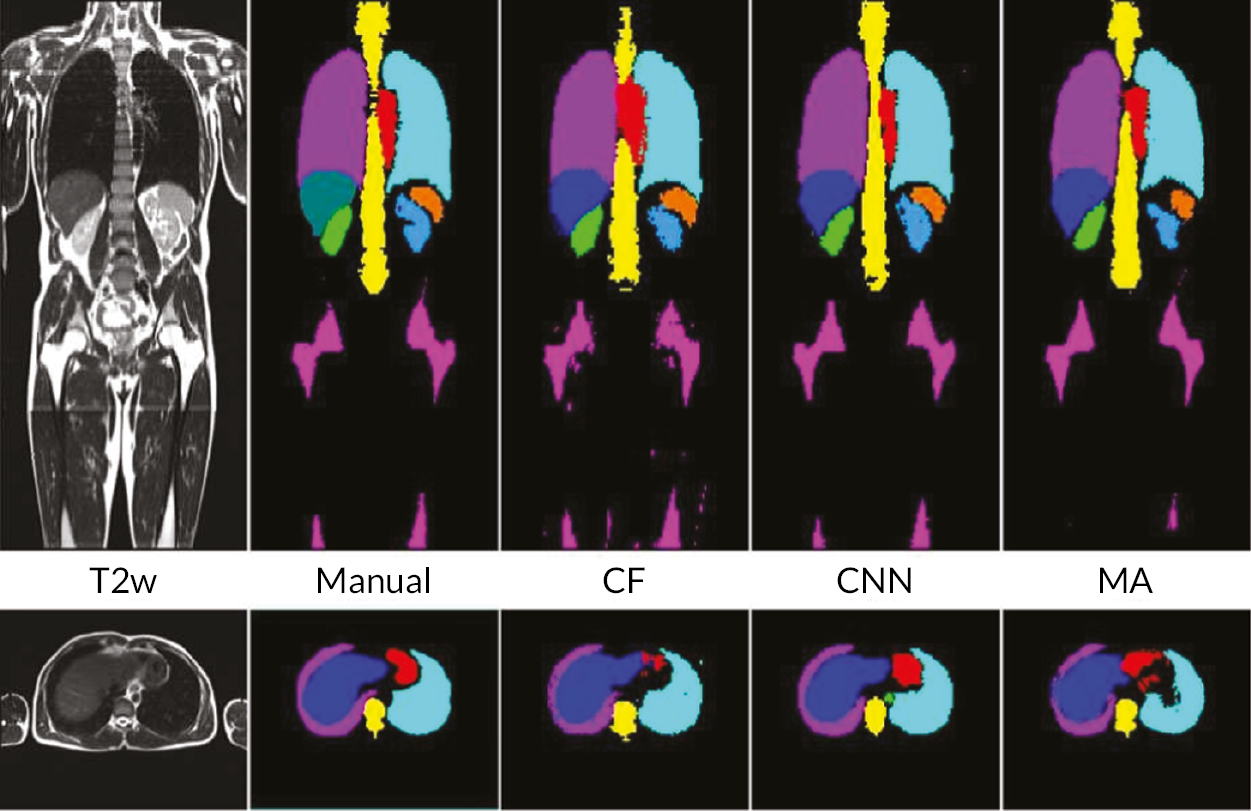
A visual example of automatic segmentation results from the three algorithms in the coronal and axial plane is shown in Figure 3.
A bar chart that provides a pictorial representation of the mean metrics (DSC, RE, PR, ASD, RMSSD and HD) for the segmented organs when using T2w volumes as input to all three algorithms is shown in Appendix 1, Figure 26.
Table 2 shows the pooled mean metrics ± standard deviation (SD) from all the segmented structures for the three algorithms. It also shows the p-values from the ANOVA when comparing the metrics between the three algorithms. It is seen that CNNs provide the highest mean DSC (0.81 ± 0.13), RE (0.83 ± 0.14) and PR (0.82 ± 0.10) compared to CFs and the MA algorithm, but not statistically significant (p = 0.271, 0.294 and 0.185, respectively). On the contrary, the MA algorithm returns the lowest ASD (4.22 ± 2.42 mm), RMSSD (6.13 ± 2.55 mm), and HD (38.9 ± 28.9 mm) when compared to CFs and CNNs, which is statistically significant (p = 0.005, 0.004 and 0.001, respectively).
| DSC | RE | PR | ASD (mm) | RMSSD (mm) | HD (mm) | |
|---|---|---|---|---|---|---|
| CFs | 0.70 ± 0.18 | 0.73 ± 0.18 | 0.71 ± 0.14 | 13.5 ± 11.3 | 34.6 ± 37.6 | 185.7 ± 194.0 |
| CNNs | 0.81 ± 0.13 | 0.83 ± 0.14 | 0.82 ± 0.10 | 5.48 ± 4.84 | 17.0 ± 13.3 | 199.0 ± 101.2 |
| MA | 0.71 ± 0.22 | 0.70 ± 0.24 | 0.77 ± 0.15 | 4.22 ± 2.42 | 6.13 ± 2.55 | 38.9 ± 28.9 |
| p | 0.271 | 0.294 | 0.185 | 0.005 | 0.004 | 0.001 |
Table 3 reports the DSC, the most commonly used metric to assess agreement between manual and automatic segmentations, for individual anatomical structures (labels) when the three algorithms (CFs, CNNs and MA) are using the T2w images as inputs only. It also shows the p-values from the ANOVA, when comparing the DSC between the three algorithms for each anatomical label.
| DSC | p-value | |||
|---|---|---|---|---|
| CFs | CNNs | MA | ||
| RLNG | 0.92 ± 0.03 | 0.95 ± 0.01 | 0.93 ± 0.01 | < 0.001 |
| LLNG | 0.92 ± 0.03 | 0.95 ± 0.01 | 0.93 ± 0.01 | < 0.001 |
| LVR | 0.85 ± 0.03 | 0.93 ± 0.01 | 0.86 ± 0.04 | < 0.001 |
| GBLD | 0.38 ± 0.26 | 0.56 ± 0.19 | 0.24 ± 0.26 | < 0.001 |
| RKDN | 0.75 ± 0.09 | 0.87 ± 0.03 | 0.77 ± 0.07 | < 0.001 |
| LKDN | 0.65 ± 0.19 | 0.84 ± 0.11 | 0.72 ± 0.13 | < 0.001 |
| SPLN | 0.57 ± 0.18 | 0.79 ± 0.11 | 0.58 ± 0.14 | < 0.001 |
| PNCR | 0.47 ± 0.13 | 0.62 ± 0.09 | 0.40 ± 0.14 | < 0.001 |
| BLD | 0.65 ± 0.22 | 0.75 ± 0.21 | 0.69 ± 0.23 | 0.162 |
| SPN | 0.80 ± 0.04 | 0.87 ± 0.01 | 0.87 ± 0.02 | < 0.001 |
| PLVS | 0.73 ± 0.05 | 0.81 ± 0.03 | 0.79 ± 0.06 | < 0.001 |
It is worth noting that CNNs performed significantly better (p < 0.001) than CFs and the MA algorithm in segmenting all the anatomies of interest, except for the BLD (p = 0.162).
A bar chart that provides a pictorial representation of the mean metrics (DSC, RE, PR, ASD, RMSSD and HD) for the segmented organs when using T2w volumes and all imaging combinations (T2w + T1w + DWI) as input to CFs and CNNs, is shown in Figure 26 in Appendix 1.
Pooled mean metrics ± SD are presented in Table 31 in Appendix 1. The bar plot of the mean measure metrics is plotted in Figures 26 and 27 of Appendix 1.
It is confirmed that the performance of CFs is improved when all imaging combinations are used (T2w + T1w + DWI) as input, when compared to using T2w volumes only. This is reflected in all metrics (DSC = 0.74 ± 0.16 vs. 0.70 ± 0.17, RE = 0.78 ± 0.16 vs. 0.73 ± 0.18, PR = 0.74 ± 0.13 vs. 0.71 ± 0.14, ASD = 7.89 ± 7.55 mm vs. 13.5 ± 11.2 mm, RMSSD = 20.9 ± 27.1 mm vs. 34.6 ± 37.6 mm and HD = 170.7 ± 194.0 mm vs. 185.7 ± 194.0 mm). On the contrary, the performance of CNNs is better when using T2w volumes only as input, rather than using all imaging combinations (T2w + T1w + DWI). This is again reflected in all metrics (DSC = 0.81 ± 0.12 vs. 0.77 ± 0.14, RE = 0.82 ± 0.14 vs. 0.79 ± 0.15, PR = 0.82 ± 0.10 vs. 0.79 ± 0.11, ASD = 5.48 ± 4.84 mm vs. 9.23 ± 8.04 mm, RMSSD = 17.0 ± 13.3 mm vs. 25.2 ± 19.1 mm and HD = 199.0 ± 101.2 mm vs. 215.9 ± 98.6 mm). No significant differences were found in the performance of CFs and CNNs when using different T2w only and all imaging combinations (T2w + T12w + DWI) as inputs.
Table 31 (in Appendix 1) shows the pooled mean metrics T SD from all the segmented structures for CFs and CNNs, when using T2w only volumes and all imaging combinations (T2w + T1w + DWI) as inputs. It also shows the p-values from the Mann–Whitney U-test when comparing the two-input cases for CFs and CNNs.
Table 4 shows the DSC for all the anatomical labels, when CFs and CNNs are being used with T2w images only (CFs_T2w and CNNs_T2w) as inputs and when using all imaging combinations (T2w + T1w + DWI) as input to the two algorithms (CFs_all and CNNs_all). It also shows the p-values from the Mann–Whitney U-tests when comparing the DSC between CFs and CNNs used with different imaging inputs.
| DSC | p-value | DSC | p-value | |||
|---|---|---|---|---|---|---|
| CFs_T2w | CFs_all | CNNs_T2w | CNNs_all | |||
| RLNG | 0.92 ± 0.03 | 0.92 ± 0.02 | 0.564 | 0.95 ± 0.01 | 0.94 ± 0.01 | 0.001 |
| LLNG | 0.92 ± 0.03 | 0.92 ± 0.02 | 0.500 | 0.95 ± 0.01 | 0.93 ± 0.03 | 0.003 |
| LVR | 0.85 ± 0.03 | 0.90 ± 0.02 | < 0.001 | 0.93 ± 0.01 | 0.91 ± 0.03 | < 0.001 |
| GBLD | 0.38 ± 0.26 | 0.38 ± 0.25 | 0.976 | 0.56 ± 0.19 | 0.49 ± 0.18 | 0.079 |
| RKDN | 0.75 ± 0.09 | 0.79 ± 0.06 | 0.093 | 0.87 ± 0.03 | 0.84 ± 0.05 | < 0.001 |
| LKDN | 0.65 ± 0.19 | 0.73 ± 0.13 | 0.023 | 0.84 ± 0.11 | 0.78 ± 0.13 | < 0.001 |
| SPLN | 0.57 ± 0.18 | 0.67 ± 0.15 | < 0.001 | 0.79 ± 0.11 | 0.69 ± 0.13 | < 0.001 |
| PNCR | 0.47 ± 0.13 | 0.55 ± 0.11 | 0.017 | 0.62 ± 0.09 | 0.57 ± 0.11 | 0.051 |
| BLD | 0.65 ± 0.22 | 0.74 ± 0.18 | 0.046 | 0.75 ± 0.21 | 0.74 ± 0.16 | 0.411 |
| SPN | 0.80 ± 0.04 | 0.83 ± 0.03 | < 0.001 | 0.87 ± 0.01 | 0.85 ± 0.05 | 0.044 |
| PLVS | 0.73 ± 0.05 | 0.74 ± 0.05 | 0.135 | 0.81 ± 0.03 | 0.78 ± 0.06 | 0.069 |
It is seen that the addition of extra imaging modalities (T1w + DWI) as input to CFs_T2w, significantly improves the segmentation performance (p = 0.046) for many anatomical structures (LVR, LKDN, SPLN, PNCR, BLD and SPN). By contrast, the addition of T1w + DWI to CNNs_T2w, significantly deteriorates the DSC (p = 0.044) for most the examined anatomies of interest (RLNG, LLNG, LVR, RKDN, LKDN, SPLN and SPN). Finally, Table 5 shows and compares the DSC from all anatomical labels, when segmented by the two algorithms with the best DSC performance as reported above, namely CFs_all and CNNs_T2w. It also shows the p-values from the Mann–Whitney U-tests to compare the DSC between the two algorithms for all the examined structures.
| DSC | p-value | ||
|---|---|---|---|
| CFs all | CNNs T2w | ||
| RLNG | 0.92 ± 0.02 | 0.95 ± 0.01 | < 0.001 |
| LLNG | 0.92 ± 0.02 | 0.95 ± 0.01 | < 0.001 |
| LVR | 0.90 ± 0.02 | 0.93 ± 0.01 | < 0.001 |
| GBLD | 0.38 ± 0.25 | 0.56 ± 0.19 | 0.002 |
| RKDN | 0.79 ± 0.06 | 0.87 ± 0.03 | < 0.001 |
| LKDN | 0.73 ± 0.13 | 0.84 ± 0.11 | < 0.001 |
| SPLN | 0.67 ± 0.15 | 0.79 ± 0.11 | < 0.001 |
| PNCR | 0.55 ± 0.11 | 0.62 ± 0.09 | 0.008 |
| BLD | 0.74 ± 0.18 | 0.75 ± 0.21 | 0.384 |
| SPN | 0.83 ± 0.03 | 0.87 ± 0.01 | < 0.001 |
| PLVS | 0.74 ± 0.05 | 0.81 ± 0.03 | < 0.001 |
It is striking that CNNs_T2w scored significantly better DSCs than CFs in all the examined organs (p < 0.008), aside from the BLD (p = 0.384). The segmentation performance was notably improved when using CNNs_T2w, even for organs with great variability in appearance, such as the GBLD (0.38 ± 0.25 for CNNs_T2w vs. 0.56 ± 0.19 for CFs_all, p = 0.002).
Discussion
All the algorithms tested in this study permitted automatic, multiorgan segmentation in whole-body MRI of healthy volunteers with very good agreement to the segmentations, performed manually by clinical experts. Accurate, multiorgan automatic segmentation in whole-body MRI is the first step in training ML algorithms to recognise normality. This will lead the way to developing automatic identification and segmentation algorithms for lesions, such as primary or metastatic tumours, with increased sensitivity and specificity. These algorithms could ultimately facilitate the process of reading whole-body scans in cancer patients by reducing the RT, and possibly, improving the diagnostic accuracy of WB-MRI. These algorithms may also assist in quantifying the extent of normal tissues such as muscle or fat.
Our analysis showed that CNNs outperformed CFs and the MA algorithm when T2w volumes were used as input to the algorithms and when using pooled overlap-evaluation metrics (DSC, RE and PR) to assess the accuracy of segmentation. When the performance of the algorithms was assessed with pooled surface distance metrics (ASD, RMSSD and HD), it was the MA algorithm that performed best. Single misinterpreted voxels in CFs and CNNs can greatly elevate ASD, RMSSD and HD; these metrics are particularly sensitive to outliers.
We then assessed the pooled metrics performance of CFs and CNNs when using all imaging combinations (T2w + T1w + DWI) as input, arguing that maximisation of training information to the algorithms might improve the performance of segmentation. We found that the performance of CFs was improved, however not significantly, when using all imaging combinations as input for training. The opposite was observed for CNNs.
The findings for the pooled metrics analysis, described above, were corroborated by a ‘per-organ’ quantitative analysis of the commonly used DSC, to assess the performance of our segmentation algorithms. This analysis confirmed that for all individual anatomical structures (except for the BLD), the algorithm that returned the greatest DSC was CNNs with T2w images only used as input.
Because our structural scans were acquired using breath-holds and the DWI ones with free breathing, we found that there was significant displacement between soft tissues in anatomical areas adjacent to the diaphragm between these types of scans. As the employed affine registration method cannot fully compensate for nonlinear motions caused by breathing, we assume that misregistration could be the reason why the performance of CNNs, despite performing better than the other two algorithms when using T2w volumes as input only, was degraded when using all imaging combinations as input for training. A more robust, nonlinear registration method could improve the accuracy of CNNs and further improve the performance of CFs, so we are currently looking into methods to address this issue. Alternatively, we could have generated training data by manually segmenting the structures of interest on each sequence type separately, but this would be a rather strenuous and time-consuming approach. Further work would need to address the performance limitations of our algorithms when segmenting organs with big variability in appearance (e.g. the GBLD or the PNCR).
Conclusion
In conclusion, in this phase of the MALIBO study we have developed and evaluated three state-of-the-art algorithms that automatically segment healthy organs and bones in whole-body MRI with accuracy comparable to the one achieved manually by clinical experts. An algorithm based on CNNs and trained using T2w only images as input performs favourably when compared to CFs or a MA algorithm, trained with either T2w only images or a combination of imaging inputs (T2w + T1w + DWI). Using multimodal MRI data as input for training, the developed algorithms did not improve the segmentation performance in this work, but it is anticipated to improve the segmentation performance if more effective WB registration between the various imaging modalities can be performed. This investigation is the first step towards developing robust algorithms for the automatic detection and segmentation of benign and malignant lesions in whole-body MRI scans for staging of cancer patients.
Chapter 4 Reverse classification accuracy and domain adaptation
This chapter is based on material previously published by the authors in Valindria et al. 70,71 with minor modification. This material is reproduced in line with Creative Commons licence BY 4.0.
Reverse classification accuracy: predicting segmentation performance in the absence of ground truth70
When integrating computational tools such as automatic segmentation into clinical practice, it is of utmost importance to be able to assess the level of accuracy on new data, and in particular, to detect when an automatic method fails. However, this is difficult to achieve due to absence of GT. Segmentation accuracy on clinical data might be different from what is found through cross-validation because validation data are often used during incremental method development, which can lead to overfitting and unrealistic performance expectations. Before deployment, performance is quantified using different metrics, for which the predicted segmentation is compared to a reference segmentation, often obtained manually by an expert. However, little is known about the real performance after deployment when a reference is unavailable. In this paper, we introduce the concept of reverse classification accuracy (RCA)70 as a framework for predicting the performance of a segmentation method on new data. In RCA, we take the predicted segmentation from a new image to train a reverse classifier which is evaluated on a set of reference images with available GT. The hypothesis is that if the predicted segmentation is of good quality, then the reverse classifier will perform well on at least some of the reference images. We validate our approach on multiorgan segmentation with different classifiers and segmentation methods. Our results indicate that it is indeed possible to predict the quality of individual segmentations, in the absence of GT. Thus, RCA is ideal for integration into automatic processing pipelines in clinical routine and as part of large-scale image analysis studies.
Introduction
Segmentation is an essential component in many image analysis pipelines that aim to extract clinically useful information from medical images to inform clinical decisions in diagnosis, treatment planning, or monitoring of disease progression. A multitude of approaches have been proposed for solving segmentation problems, with popular techniques based on graph cuts,72 MA label propagation,63 statistical models73 and supervised classification. 74 Traditionally, performance of a segmentation method is evaluated on an annotated database using various evaluation metrics in a cross-validation setting. These metrics reflect the performance in terms of agreement75 of a predicted segmentation compared to a reference ‘GT.’ (For simplicity, we use the term GT to refer to the best-known reference, which is typically a manual expert segmentation.) Commonly used metrics include DSC76 and other overlap-based measures,77 but also metrics based on volume differences, surface distances, and others. 78–80 A detailed analysis of common metrics and their suitability for segmentation evaluation can be found in Konukoglu et al. 81
Once a segmentation method is deployed in routine, little is known about its real performance on new data. Due to the absence of GT, it is not possible to assess performance using traditional evaluation measures. However, it is critical to be able to assess the level of accuracy on clinical data,82 and in particular, it is important to detect when an automatic segmentation method fails. Especially when the segmentation is an intermediate step within a larger automated processing pipeline where no visual quality control of the segmentation results is feasible. This is of high importance in large-scale studies such as the UK Biobank Imaging Study83 where automated methods are applied to large cohorts of several thousand images, and the segmentation is to be used for further statistical population analysis. In this study, we are asking the question whether it is possible to assess segmentation performance and detect failure cases when there is no GT available to compare with. One possible approach to monitor the segmentation performance is to occasionally select a random data set, obtain a manual expert segmentation and compare it to the automatic one. While this can merely provide a rough estimate about the average performance of the employed segmentation method, in clinical routine we are interested in the per-case performance and want to detect when the automated method fails. The problem is that the performance of a method might be substantially different on clinical data and is usually lower than what is found through cross-validation on annotated data carried out beforehand due to several reasons. Firstly, the annotated database is normally used during incremental method development for training, model selection and FT of hyper-parameters. This can lead to overfitting84 which is a potential cause for lower performance on new data. Secondly, the clinical data might be different due to varying imaging protocols or artefacts caused by pathology. To this end, we propose a general framework for predicting the real performance of deployed segmentation methods on a per-case basis in the absence of GT.
Related work
Retrieving an objective performance evaluation without GT has been an issue in many domains, from remote sensing,85 graphics,86 to marketing strategies. 87 In computer vision, several works evaluate the segmentation performance by looking at contextual properties,88 by separating the perceptual salient structures89 or by automatically generating semantic GT. 90,91 However, these methods cannot be applied to a more general task, such as an image with many different class labels to be segmented. An attempt to compute objective metrics, such as PR and RE with missing GT, is proposed by Lamiroy and Sun92 but it cannot be used for data sets with partial GT since it applies a probabilistic model under the same assumptions. Another stand-alone method to consider is the meta-evaluation framework, where image features are used in a ML setting to provide a ranking of different methods,93 but this does not allow the estimation of segmentation performance on an individual image level.
Meanwhile, unsupervised methods94,95 aim to estimate the segmentation accuracy directly from the images and label maps using, for example, information-theoretic and geometrical features. While unsupervised methods can be applied to scenarios where the main purpose of segmentation is to yield visually consistent results that are meaningful to a human observer, the application in medical settings is unclear.
When there are multiple reference segmentations available, a similarity measure index can be obtained by comparing an automatic segmentation with the set of references. 96 In medical imaging, the problem of performance analysis with multiple references which may suffer from intrarater and inter-rater variability has been addressed. 97,98 The simultaneous truth and performance level estimation (STAPLE) approach97 has led to the work of Bouix et al. 99 that proposed techniques for comparing the relative performance of different methods without the need of GT. Here, the different segmentation results are treated as plausible references, thus can be evaluated through STAPLE and the concept of common agreement. Another work by Sikka and Deserno100 has quantitatively evaluated the performance of several segmentation algorithms by region correlation matrix. The limitation of this work is that it cannot evaluate the segmentation performance of a particular method on a particular image independently.
Recent work has explored the idea of learning a regressor to directly predict segmentation accuracy from a set of features that are related to various segmentation energy terms. 101 Here, the assumption is that those features are well suited to characterise segmentation quality. In an extension for a security application, the same features as in Kohlberger et al. 101 are extracted and used to learn a generative model of good segmentations that can be used to detect outliers. 102 Similarly, the work of Frounchi et al. 103 considers training of a classifier that is able to discriminate between consistent and inconsistent segmentations. However, the approaches101,103 can only be applied when a training database with good and bad segmentations is available from which a mapping from features to segmentation accuracy can be learnt. Examples of bad segmentations can be generated by altering parameters of automatic methods, but it is unclear whether those examples resemble realistic cases of segmentation failure. The generative model approach in Grady et al. 102 is appealing as it only requires a database of good segmentations. However, there is still the difficulty of choosing appropriate thresholds on the probabilities that indicate bad or failed segmentations. Such an approach cannot not be used to directly predict segmentation scores such as DSC, but can be useful to inform automatic quality control or to automatically select the best segmentation from a set of candidates. In the general ML domain, the lack-of-label problem has been tackled by exploiting transfer learning104 using a reverse validation to perform cross-validation when the number of labelled data are limited. The basic idea of reverse validation104 is based on reverse testing, where a new classifier is trained on predictions on the test data and evaluated again on the training data. This idea of reverse testing is closely related to our approach as we will discuss in the following section.
Contribution
The main contribution of this study is the introduction of the concept of RCA to assess the segmentation quality of an individual image in the absence of GT. RCA can be applied to evaluate the performance of any segmentation method on a per-case basis. To this end, a classifier is trained using a single image with its predicted segmentation acting as pseudo GT. The resulting reverse classifier (or RCA classifier) is then evaluated on images from a reference database for which GT is available. It should be noted that the reference database can be (but does not have to be) the training database that has been used to train, cross-validate and fine-tune the original segmentation method. The assumption is that in ML approaches, such a database is usually already available, but it could also be specifically constructed for the purpose of RCA. Our hypothesis is that if the segmentation quality for a new image is high, then the RCA classifier trained on the predicted segmentation used as pseudo GT will perform well at least on some of the images in the reference database, and similarly, if the segmentation quality is poor, the classifier is likely to perform poorly on the reference images. For the segmentations obtained on the reference images through the RCA classifier, we can quantify the accuracy, for example, using DSC, since reference GT is available. It is expected that the maximum DSC score over all reference images correlates well with the real DSC that one would get on the new image if GT were available. Although the idea of RCA is similar to reverse validation104 and reverse testing,105 the important difference is that in our approach we train a reverse classifier on every single instance while the approaches in references104,105 train single classifiers over the whole test set and its predictions jointly to find out what the best original predictor is. RCA has the advantage of allowing to predict the accuracy for each individual case, while at the same time aggregating over such accuracy predictions allows drawing conclusions for the overall performance of a particular segmentation method.
In the following, we will first present the details of RCA and then evaluate its applicability to a multi-organ segmentation task by exploring the prediction quality of different segmentation metrics for different combinations of segmentation methods and RCA classifiers. Our results indicate that, at least to some extent, it is indeed possible to predict the performance level of a segmentation method on each individual case, in the absence of GT. Thus, RCA is ideal for integration into automatic processing pipelines in clinical routine and as part of large-scale image analysis studies.
Reverse classification accuracy
The RCA framework is based on the idea of training reverse classifiers on individual images utilising their predicted segmentation as pseudo GT. In this work, we employ three different methods for realising the RCA classifier and evaluate each in different combinations with three state-of-the-art image segmentation methods. Details about the different RCA classifiers are provided in the following sections.
Learning reverse classifiers
Given an image I and its predicted segmentation SI, we aim to learn a function fI,SI(x): Rn 7 → C that acts as a classifier by mapping feature vectors x Є Rn extracted for individual image points to class labels c Є C. In theory, any classification approach could be utilised within the RCA framework for learning the function fI,SI. We experiment with three different methods reflecting state-of-the-art ML approaches for voxel-wise classification and atlas-based label propagation.
Atlas forests (AFs): The first approach we consider for learning a RCA classifier is based on the recently introduced concept of AFs106 which demonstrates the feasibility of using RFs62 to encode individual atlases, that is images with corresponding segmentations. RFs have become popular for general image segmentation tasks as they naturally handle multiclass problems and are computationally efficient. Since they operate as voxel-wise classifiers, they do not (necessarily) require preregistration of the images neither at training nor testing time. Although in Zikic et al. 106 spatial priors have been incorporated by means of registering location probability maps to each atlas and new image, this is not a general requirement for using AFs to encode atlases. In fact, the way we employ AFs within our RCA framework does not require any image registration. The forest-based RCA classifiers in this study are all trained with the same set of parameters of maximum depth 30 and 50 trees. As we follow a very standard approach for RFs, we refer to previous works106,107 for more details. It is worth noting that, similar to previous work, we employ simple box features which can be efficiently evaluated using integral images. This has the advantage that feature responses do not need to be precomputed. Instead, we randomly generate a large pool of potential features (typically around 10,000) by drawing values randomly for the feature parameters such as box sizes and offsets from predefined ranges. At each split node we then evaluate on the fly a few hundred box features with a brute force search for optimal thresholds over the range of feature responses to greedily find the most discriminative feature/threshold pair. This strategy has proven successful in a number of works using RFs for various tasks.
Deep learning: The second approach was to experiment with CNNs as RCA classifiers. Here, we utilise DeepMedic, a 3D CNN architecture for automatic segmentation. 15 The architecture is computationally efficient as it can handle large image context by using a dual pathway for multiscale processing. CNNs have shown to be able to learn highly complex and discriminative data associations between input data and target output. The architecture of the network is defined by the number of layers and the number of activation functions in each layer. In CNNs, each activation function corresponds to a learnt convolutional filter, and each filter produces a FM by convolving the outputs of the previous layer. Through the sequential application of many convolutions, highly complex features are learnt that are then used to produce voxel-wise predictions at the final, fully connected layer. CNNs are a type of deep-learning approach which normally requires large amounts of training data in order to perform well due to the thousands (or millions) of parameters corresponding to the weights of the filters. To be able to act as a RCA classifier, that is trained on a single image, we require a specialised architecture. Here, we reduce the number of FMs in each layer by one third compared to the default setting of DeepMedic. We also cut the FMs in the last fully connected layers, from 150 to 45. By reducing the FMs without changing the architecture, in terms of number of layers the network preserves, its capability to see large image context as the size of the receptive field remains unchanged. With a smaller number of filters, the number of parameters is substantially decreased, which leads to faster computations, but, more importantly, reduces overfitting when trained on a single image. Training is performed in a patch-wise manner where the original input image is divided into 3D patches that are then sampled during training using backpropagation and batch normalisation. For details about the training procedure and further analysis of DeepMedic, we refer to Kamnistas et al. 15
Atlas-based label propagation: The third approach we consider is atlas-based label propagation. Label propagation, using multiple atlases, have been shown to yield state-of-the-art results on many segmentation problems. 63 A common procedure in MA methods is to use non-rigid registration to align the atlases with the image to be segmented and then perform label fusion strategies to obtain predictions for each image point. Although MA methods based on registration are not strictly voxel-wise classifiers as they operate on the whole image during registration, the final stage of label fusion can be considered as a voxel-wise classification step. Here, we make use of an approach that has been originally developed in the context of segmentation of cardiac MRI67. For the purpose of RCA, however, there is only a single atlas and, thus, no label fusion is required. Using single atlas-label propagation then boils down to making use of an efficient non-rigid registration technique as the one described in Bai et al. 67 For RCA, the single atlas then corresponds to the image and its predicted segmentation for which we want to estimate the segmentation quality. We use the same configuration for image registration as in Bai et al. 67 and refer to this study for further details.
Predicting segmentation accuracy
For the purpose of assessing the quality of an individual segmentation, we train a RCA classifier fI,SI on a single image I that has been segmented by any segmentation method, where SI denotes the predicted segmentation that acts here as pseudo GT during classifier training. Our objective is to estimate the quality of SI in the absence of GT. To this end, we define the segmentation function FI,SI(J) = SJ that applies the trained RCA classifier fI,SI to all voxels (or more precisely to the features extracted at each voxel) of another image J which produces a segmentation SJ. Assuming that for the image J a reference GT segmentation SJGT is available, we can now compute any segmentation evaluation metric on the pair (SJ, SJGT). The underlying hypothesis in our RCA framework is that there is a correlation between the values computed on (SJ, SJGT) and the values one would get for the pair (SI, SIGT), where SIGT is the reference GT of image I which in practice, however, is unavailable.
It is unlikely that this assumption of correlation holds for an arbitrary reference image J. In fact, the RCA classifier fI,SI is assumed to work best on images that are somewhat similar to I. Therefore, we further assume that a suitable reference database is available that contains multiple segmented images (or atlases) Τ={(Jk,SJkGT)}k=1m that capture the expected variability. Such a database is commonly available in the context of ML and MA-based segmentation approaches but could also be generated specifically for the purpose of RCA. If already available, we can re-use existing training databases that might have been previously used during method development and/or cross-validation and parameter tuning. When testing the RCA classifier on all of the available m reference images, we expect that the RCA classifier performs well on at least some of these, if and only if the predicted segmentation SI is of good quality. If SI is of bad quality, we expect the RCA classifier to perform poorly on all reference images. This leads to our definition of a proxy measure for predicting the segmentation accuracy as:
where ρ is any evaluation metric, such as DSC, assuming higher values correspond to higher quality segmentations. (For metrics where a lower value indicates better quality, such as surface distance, we can simply replace the max with a min operator.) Here, we only look for the maximum value that is found across all reference images, as this seems to be a good indicator of the quality of the segmentation SI. Other statistics could be considered, such as the average of the top three scores, but we found that the maximum score works best as a proxy. Note, that the mean or median scores are not very useful measures as we do not expect the RCA classifier to work well on the majority of the reference images. Afterall, the RCA classifier does overfit to the single image and will not generalise to perform well on dissimilar images. Nonetheless, as we will demonstrate in the experiments, ρ¯ indeed provides accurate estimates for the segmentation quality in a wide range of settings.
Summary
The following provides a summary of the required steps for using RCA in practice within a processing pipeline for automatic image segmentation. Given an image I to be segmented:
-
Run the automated image segmentation method to obtain predicted segmentation SI.
-
Train a RCA classifier on image I and its predicted segmentation SI to obtain an image segmenter FI,SI.
-
Evaluate the RCA classifier on a reference database with images for which GT is available Τ= {(Jk, SJkGT)}k=1m to obtain segmentations ∀ k FI,SI(Jk) = SJk.
-
Compute the segmentation quality of SI using a proxy measure ρ¯(SI) according to Equation 1 ρ¯S1= max1≤k≤m ρ(FI,SI(Jk), SJkGT).
Depending on the application, a threshold may be defined on ρ¯ to flag images with poor segmentation quality that need manual inspection, or to automatically identify high-quality segmentations suitable for further analysis.
Reverse classification accuracy experimental validation
In order to test the effectiveness of the RCA framework, we explore a comprehensive multiorgan segmentation task on WB-MRI. In this application, we evaluate the prediction accuracy of RCA in the context of three different state-of-the-art segmentation methods, a RF approach,74 a deep-learning approach using 3D CNNs,15 and a probabilistic MA label propagation approach. 67 The data set used to validate our framework is from our MALIBO study. We collected WB, multi-sequence MRI (T1w Dixon and T2w images) of 35 healthy volunteers. Detailed manual segmentations of 15 anatomical structures, including abdominal organs (heart, LLNG/RLNG, LVR, adrenal gland, GBLD, LKDN/RKDN, SPLN, PNCR, BLD) and bones (SPN, left/right clavicle, PLVS), have been generated by clinical experts as part of the study. These manual segmentations will serve as GT in the quantitative evaluation.
Experimental setting
We use threefold cross-validation to automatically segment all 525 structures (15 organs × 35 participants) with each of the three different segmentation methods, namely RFs, CNNs and MA. In each fold, we use the RCA framework with three different methods for realising the RCA classifier, namely AFs, constrained CNNs, and single-atlas, as described above. Using the RCA classifiers that are trained on each image for which we want to assess segmentation quality, we obtain segmentations on all reference images which are then compared to their manual reference GT. Since the GT is available for all 35 cases, we can compare the predicted versus the real segmentation accuracy for all cases and all organs under various settings with 9 different combinations of segmentation methods and RCA classifiers.
Quantifying prediction accuracy
The DSC is the most widely used measure for evaluating segmentation performance (despite some well-known shortcomings of DSC as discussed in Konukoglu et al. 81) and in our main results we focus on evaluating how well DSC can be predicted using our RCA framework. In order to quantify prediction accuracy, we consider three different measures, namely the correlation between predicted and real DSC, the mean absolute error (MAE) and a classification accuracy. Arguably, the most important measure for direct evaluation of how well RCA works is the MAE, as it directly tells us how close the predicted DSC is to the real one. Correlation is interesting, as it tells us something about the relation between predicted and real scores. We expect high correlation in order for RCA to be useful, but we might not always have an identity relation, as there could be a bias in the predictions. For example, if the predicted score is consistently lower than the real score, this can still be useful in practice, and will be indicated by high correlation but might not yield low MAEs. In such a case, a calibration might be considered as we will discuss later on. We also explore whether the predictions can be used to categorise segmentations according to their quality. We argue that for many clinical applications it is already of great value to be able to discriminate between good-, bad- and possibly medium-quality segmentations and that the absolute segmentation scores are of less importance. For proof of principle, we consider a three-category classification by grouping segmentations within DSC ranges [0.0, 0.6] for ‘bad’, [0.6, 0.8] for ‘medium’ and [0.8, 1.0] for ‘good’ cases. Note, that those ranges are somewhat arbitrary, in particular, as the quality of absolute DSC values is highly depending on the structure of interest. So in practice, those ranges would need to be adjusted specifically to the application at hand.
Results for predicting Dice similarity coefficients
Our main results are summarised in Table 6 where we report the quantitative analysis of the predicted accuracy for nine different settings consisting of three different segmentation methods and three different ways of realising the RCA classifier. In Figure 4 we provide the scatterplots of real versus predicted DSC for all 9 settings with 525 data points each.
| Segmentation method | RCA classifier | Correlation | MAE | Accuracy 3-categories | |||
|---|---|---|---|---|---|---|---|
| All | No zeros | All | No zeros | All | No zeros | ||
| RFs | AFs | 0.881 | 0.867 | 0.120 | 0.130 | 0.783 | 0.776 |
| CNNs | AFs | 0.828 | 0.630 | 0.166 | 0.245 | 0.623 | 0.500 |
| MA | AFs | 0.863 | 0.877 | 0.168 | 0.177 | 0.749 | 0.726 |
| RFs | Constrained CNNs | 0.721 | 0.718 | 0.252 | 0.271 | 0.653 | 0.631 |
| CNNs | Constrained CNNs | 0.756 | 0.662 | 0.225 | 0.292 | 0.592 | 0.472 |
| MA | Constrained CNNs | 0.773 | 0.686 | 0.209 | 0.237 | 0.693 | 0.642 |
| RFs | Single-atlas | 0.955 | 0.946 | 0.051 | 0.052 | 0.888 | 0.880 |
| CNNs | Single-atlas | 0.973 | 0.892 | 0.052 | 0.065 | 0.811 | 0.756 |
| MA | Single-atlas | 0.962 | 0.947 | 0.067 | 0.072 | 0.822 | 0.798 |
FIGURE 4.
Scatterplots using three different RCA classifiers.
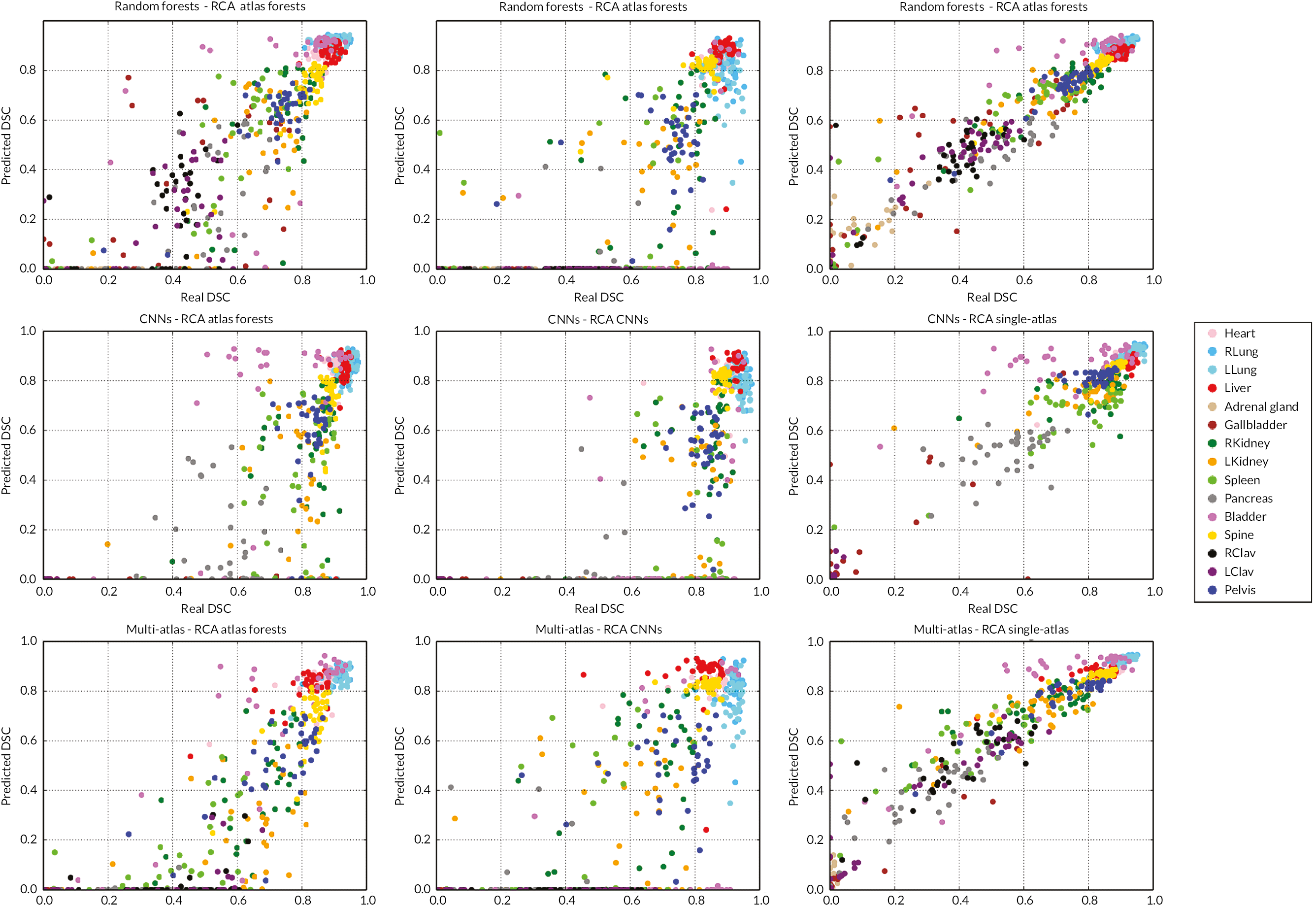
Overall, we observe high correlation between predicted and real DSC for both AFs and single-atlas when used as RCA classifiers, with single-atlas showing correlations above 0.95 for all three segmentation methods. The single-atlas approach also yields the lowest MAEs between 0.05 and 0.07, and good three-category classification accuracies between 81% and 89%. This is visually confirmed by the scatterplots in the right column of Figure 17 which show good linear relation close to the diagonal between predicted and real scores for most structures in the case where RFs or MA are used as the original segmentation method. When using AFs for RCA, we still observe good correlation but the relationship between predicted and real scores is off-diagonal with larger spread towards lower quality segmentation. The correlation is still good and above 0.82, MAEs are between 0.12 and 0.17 with classification accuracy going down to 0.62%, 0.75% and 0.78% depending on the original segmentation method. For the case of the constrained CNNs, we observe that the prediction quality is lowest confirmed by the scatterplots and all quantitative measures, with correlations below 0.78 and MAEs above 0.2. The constrained CNNs seem to only work for predicting segmentation accuracy in case of major organs such as LVR, lungs, and the SPN but clearly struggle with smaller structures leading to many zero predictions even when the real DSC is rather high. This is most likely caused by the difficulty of training the CNNs with single images and small structures which does not provide sufficient amounts of training data.
Figure 28 in Appendix 1 shows an example for predicting the accuracy of a LVR segmentation. Next to a slice from a T2w MRI volume we show the GT manual segmentation together with the result from a RF. Underneath, we show the 24 segmentations obtained on the reference database when using the single-atlas RCA approach. The bar plot in the same figure shows the variation of the 24 DSC scores. Similarly, the bar plots in Figure 27 (in Appendix 1) of two more examples illustrate the distribution of DSC scores when predicting a good-quality segmentation on the left, and a poor-quality segmentation on the right. The three examples support the hypothesis that selecting the maximum score across the reference database according to equation (ρ¯S1= max1≤k≤m ρ(FI,SI(Jk), SJkGT)) is a good proxy for predicting segmentation quality.
Scatterplots of predicted and real DSC of multiple structures for three different segmentation methods (rows) using three different RCA classifiers (columns) (see Figure 4). High correlation and low prediction errors are obtained when employing the single-atlas label propagation as RCA classifier (right column). There is also good correlation with predictions in case of AFs (left) with larger spread towards lower quality segmentations. The constrained CNNs (middle column) are less suitable for RCA which is likely due to the difficulty of training on single images. Both AFs and constrained CNNs work best for larger organs such as LVR, lungs, and PLVS while leading to many zero predictions for smaller structures such as adrenal gland and clavicles. The single-atlas label propagation makes accurate predictions of segmentation quality across all 15 anatomical structures. A summary of the plots is given in Table 6.
Some of the original segmentation methods have problems segmenting structures such as the adrenal gland and clavicles. The CNNs, in particular, failed to segment adrenal glands in most cases. Because the real DSC for these is zero with no voxels labelled in the segmentation map, the RCA predictions are always correct as there are no labels for the RCA classifier for this structure. In order to investigate the effect of those zero predictions on the quantitative results, we also report in Table 9 under the columns ‘No zeros’ the correlations, MAEs and classification accuracies when structures with a real DSC of zero are excluded. We observe that the zero predictions have mostly an impact on CNNs, either employed as original segmentation method or as RCA classifier. For AFs and single-atlas the effect on the accuracies is very little, confirming that those both are well suited within the RCA framework, independent of the original segmentation method.
| Segmentation method | RCA classifier | Correlation | MAE | Accuracy 3-categories | |||
|---|---|---|---|---|---|---|---|
| Direct | Calibrated | Direct | Calibrated | Direct | Calibrated | ||
| All | No zero | All | No zero | All | No zero | ||
| RFs | AFs | 0.881 | 0.833 | 0.120 | 0.105 | 0.783 | 0.809 |
| CNNs | AFs | 0.828 | 0.929 | 0.166 | 0.079 | 0.623 | 0.783 |
| MA | AFs | 0.863 | 0.939 | 0.168 | 0.065 | 0.749 | 0.745 |
| RFs | Constrained CNNs | 0.721 | 0.826 | 0.252 | 0.104 | 0.653 | 0.768 |
| CNNs | Constrained CNNs | 0.756 | 0.961 | 0.225 | 0.049 | 0.592 | 0.830 |
| MA | Constrained CNNs | 0.773 | 0.874 | 0.209 | 0.100 | 0.693 | 0.787 |
| RFs | Single-atlas | 0.955 | 0.872 | 0.051 | 0.089 | 0.888 | 0.815 |
| CNNs | Single-atlas | 0.973 | 0.967 | 0.052 | 0.048 | 0.811 | 0.811 |
| MA | Single-atlas | 0.962 | 0.918 | 0.067 | 0.080 | 0.822 | 0.825 |
The bar plots (Figure 5) show two examples for predicting the real DSC (red) in case of a good-quality (left) and bad-quality segmentation (right) using a database with 24 reference images with available GT. The predicted DSC (green bar) selected according to Eq. (ρ¯S1= max1≤k≤m ρ(FI,SI(Jk), SJkGT)) matches well the real DSC.
FIGURE 5.
Dice similarity coefficient plots from good-quality (left) and bad-quality segmentation (right).
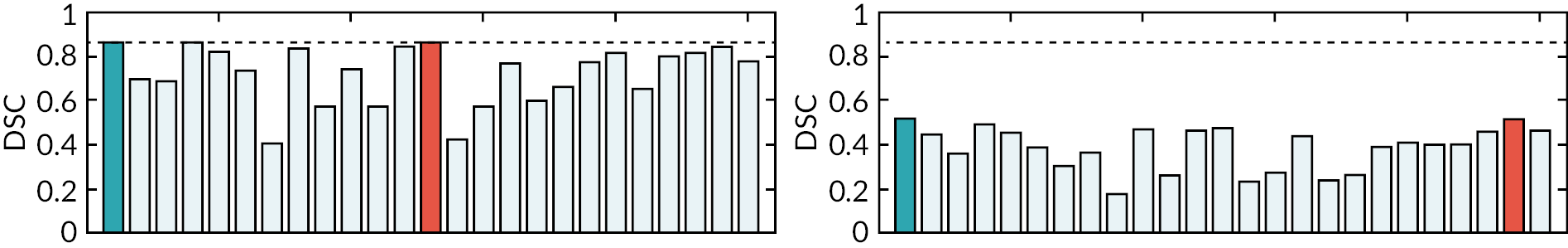
Scatterplots for the experiment of detecting segmentation failure when using degraded RFs with limited depth as the segmentation method. AFs (left) and single-atlas label propagation (right) make highly accurate predictions in the low DSC ranges and thus are able to correctly detect such failed segmentations, with the exception of the BLD. Constrained CNNs are again less suitable for RCA with many zero predictions.
Detecting segmentation failure
In clinical routine it is of great importance to be able to detect when an automated method fails. We conducted a dedicated experiment to investigate how well RCA can predict segmentation failure. From the scatterplots in Figure 27 in Appendix 1 we can see that all three segmentation methods perform reasonably well on most major organs with no failure cases among structures such as LVR, heart, and lungs. In order to further demonstrate that RCA can predict failure cases in these structures, we utilise degraded RFs by limiting the tree depth at test time to 8. This leads to much worse segmentation results for most structures which is confirmed in the corresponding scatterplots shown in Figure 6. Again, we evaluate the performance of the three different RCA classifiers, AFs, constrained CNNs and single-atlas. The results are summarised in Table 7. The constrained CNNs are again suffering from many zero predictions and less suitable for making accurate predictions. AFs and single-atlas, however, result in high correlations, low MAEs and very good classification accuracies. Low real DSC scores are correctly predicted and failed segmentations are identified. The only exception here is the BLD. This might be explained by the unique appearance of the BLD in the multispectral MRI with hyper-intensities in the T2w image, and its largely varying shape between participants. It appears that even a badly segmented BLD can be sufficient for the RCA classifier to learn its appearance and segment the BLD well on at least one of the reference images. Overall, the experiment suggests that RCA with AFs and single-atlas can be employed in automatic quality control, for example, in large-scale studies where it is important to be able to detect failed segmentations which should be excluded from subsequent analyses.
| RCA classifier | Correlation | MAE | Accuracy 3-Categories |
|---|---|---|---|
| AFs | 0.853 | 0.096 | 0.884 |
| Constrained CNNs | 0.237 | 0.139 | 0.924 |
| Single-atlas | 0.875 | 0.097 | 0.928 |
FIGURE 6.
Scatterplots for the experiment for detecting segmentation failure.
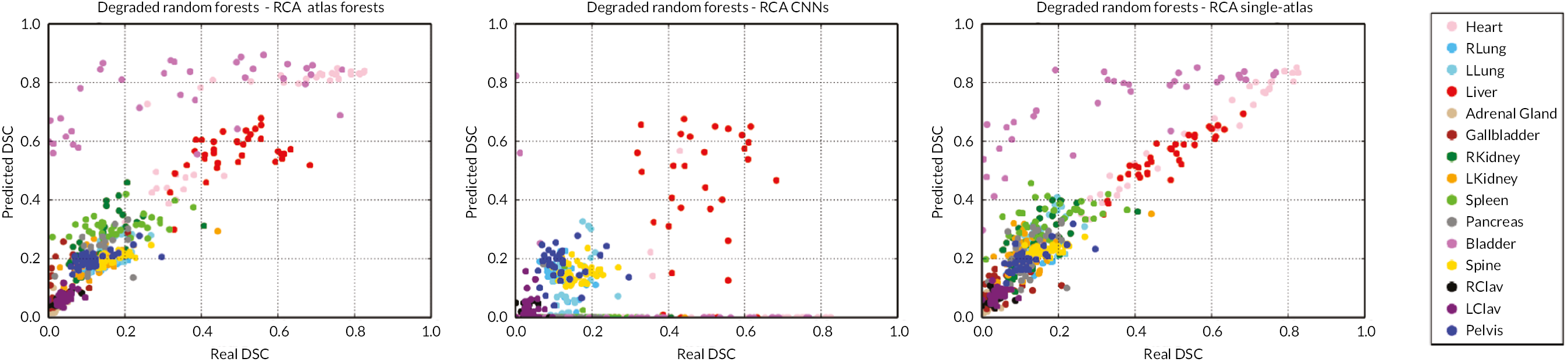
Results for predicting different segmentation metrics
We further explore the ability to predict other evaluation metrics rather than DSC. We consider the following metrics: Jaccard index (JI), PR, RE, ASD, HD and relative volume difference (RVD). For this experiment, we use RFs as segmentation method, and AFs for RCA. The results are summarised in Table 8.
| Metric | Correlation | MAE | Accuracy 3-Categories |
|---|---|---|---|
| DSC (0–1) | 0.881 | 0.120 | 0.783 |
| JI (0–1) | 0.899 | 0.110 | 0.749 |
| PR (0–) | 0.572 | 0.242 | 0.638 |
| RE (0–1) | 0.833 | 0.170 | 0.663 |
| HD (0–150) | 0.189 | 41.38 | 0.387 |
| ASD (0–10) | 0.022 | 4.120 | 0.344 |
| RVD (0–1) | 0.474 | 0.051 | 0.678 |
Good correlation is obtained between predicted and real overlap-based scores, with low MAEs, and high accuracies. Since Jaccard is directly related to DSC [JI = DSC/(2 − DSC)], it is expected that the predictions are of similar quality. Prediction accuracy for PR is lower than for RE. The two metrics capture different parts of segmentation error; under-segmentation is not reflected in PR, while over-segmentation is not captured in RE [DSC = 2 · PR · RE/(PR + RE)]. Distance-based errors are unbounded, so we define thresholds for HD and ASD, and errors above are clipped to the threshold value, which is set to 150 mm for HD and 10 mm for ASD. This also allows us to define ranges for the error categorisation. For HD, we use the ranges [0, 10], [10, 60] and [60, 150] for good-, medium- and bad-quality segmentations. For ASD we divide the range into [0, 2] for good, [2, 5] for medium and [5, 10] for bad segmentation quality. Compared to overlap-based metrics, the RCA predictions for HD and ASD are not convincing with low correlation, high MAE and low classification accuracy. RVD is the ratio of the absolute difference between the reference and predicted segmentation volume and the reference volume. Perfect segmentation will result in a value of zero. As RVD is also unbounded, we use a threshold of one to indicate maximum error. The predictions for RVD are good, with high classification accuracy of 0.68%, similar to the overlap-based scores. In conclusion, it seems RCA works very well for overlap-based measures and for RVD to some extent, while distance-based metrics cannot be accurately predicted with the current setting and would require further investigation.
Reverse classification accuracy discussion and conclusion
The experimental validation of the RCA framework has shown that it is indeed possible to accurately predict the quality of segmentations in the absence of GT, with some limitations. We have explored different methods for realising the RCA classifier and could demonstrate that AFs and in particular, single-atlas label propagation yield accurate predictions for different segmentation methods. As the RCA framework is generic, other methods can be considered and it might be necessary to select the most appropriate one for the application at hand. We have also experimented with a constrained CNN trained on single images, which only works well for major organs such as LVR, lungs and SPN. There might be other more appropriate architectures for the purpose of RCA, which will be explored as part of future work.
An appealing property of the proposed framework is that unlike the supervised methods in Frounchi et al. 103 and Kohlberger et al. 101 no training data are required that captures examples of good and bad segmentations. Instead, in RCA we simply rely on the availability of a reference database with available GT segmentations. The drawback, however, is that we assume a linear relationship between predicted and real scores which should be close to an identity mapping, something we only found in the case of using single-atlas label propagation (cf. right column of Figure 27 in Appendix 1). In the case of off-diagonal correlation, as, for example, found for AFs, an extension to RCA could be considered where the predictions are calibrated. This, however, requires training data from which a regression function could be learnt, similar to Kohlberger et al. 101 In order to demonstrate the potential of such an approach, we perform a simple experiment on the data that we used for conducting the main evaluation. After obtaining all predicted DSC scores, we run a leave-one-participant-out validation where in each fold we use RF regression to calibrate the predictions. The results are summarised in Table 9 where we compare the quantitative measures before and after calibration. Both the MAEs and classification accuracies improve significantly for the case of AFs and constrained CNNs. For single-atlas, however, the results remain similar due to the already close to identify relationship between predicted and real scores before calibration.
In our experiments we have found that best predictions are obtained for overlap-based measures such as DSC and JI. Whether those measures are sufficient to fully capture segmentation quality is debatable. Still, DSC is the most widely considered measure and being able to accurately predict DSC in the absence of GT has high practical value. Besides being useful for clinical applications where the goal is to identify failed segmentations after deployment of a segmentation method, we see an important application of RCA in large-scale imaging studies and analyses. In settings where thousands of images are automatically processed for the purpose of deriving population statistics, it is not feasible to employ manual quality control with visually inspection of the segmentation results. Here, RCA can be an effective tool to automatically extract the subset of high-quality segmentations which can be used for subsequent analysis. We are currently exploring this in the context of population imaging on the UK Biobank imaging data where image data of more than 10,000 participants are available which will be subsequently increased to 100,000 over the next couple of years. The UK Biobank data will enable the discovery of imaging biomarkers that correlate with non-imaging information such as lifestyle, demographics, and medical records. In the context of such large-scale analysis, automatic quality control is a necessity and we believe the RCA framework makes an important contribution in this emerging area of biomedical research. In future work, we will further explore the use of RCA for other image analysis and segmentation tasks. To facilitate the wide application of RCA and use by other researchers, the implementations of all employed methods are made publicly available on the website of the Biomedical Image Analysis group (https://biomedia.doc.ic.ac.uk/software/).
Domain adaptation for magnetic resonance angiography organ segmentation using reverse classification accuracy71
The variations in multicentre data in medical imaging studies have brought the necessity of domain adaptation. Despite the advancement of ML in automatic segmentation, performance often degrades when algorithms are applied on new data acquired from different scanners or sequences than the training data. Manual annotation is costly and time-consuming if it has to be carried out for every new target domain. In this study, we investigate automatic selection of suitable participants to be annotated for supervised domain adaptation using the concept of RCA. RCA predicts the performance of a trained model on data, from the new domain and different strategies of selecting participants to be included in the adaptation via transfer learning, are evaluated. We perform experiments on a two-centre MRI database for the task of organ segmentation. We show that participant selection via RCA can reduce the burden of annotation of new data for the target domain.
Introduction
Machine learning has led to significant advances in medical imaging, particularly with big improvements in medical image segmentation. Performance, however, depends on the availability of sufficient amounts of labelled samples for supervised learning, and also whether the test data are coming from the same domain as the training data. In clinical practice, the source domain (S) on which the classifier is trained might be different from the target domain (T) with clinical data. The images from these domains are samples from different appearance distributions. The mismatch of distributions is caused by various factors such as the use of different scanners, types of sequences, or biases in patient cohorts – often causing a trained algorithm to perform poorly on new data. In a scenario where the tasks are the same, but the source and target domains are different, domain adaptation is usually performed to address the domain disparity problem. 108
Domain adaptation can be categorised into three settings, supervised, semi-supervised and unsupervised. Our work focuses on supervised domain adaptation methods, which uses labelled data from the target domain. In the context of CNNs, supervised domain adaptation can be approached by training from scratch or fine-tuning (FT) a network pre-trained on the source domain. 109
An approach called domain adaptation for supervised learning from sparsely annotated MRI110 explored a supervised domain adaptation by introducing a weighting scheme in RFs and SVMs for segmentation. This approach preserves the segmentation quality even though only sparse annotated data are used. Another approach tackles the segmentation difficulty of images from different scanners and imaging protocols by weighting different SVM-based classifiers for transfer learning. 111 More recent work has tried to explore transfer learning in CNNs in medical imaging,112,113 which confirmed the potential of fine-tuned and fully trained CNNs.
However, the importance of participant selection in transfer learning has not been widely studied yet. Most works114 attempt to integrate active learning with domain adaptation. Intuitively, active learning is chosen since it develops a criterion to determine the ‘value’ of a candidate for annotation.
In active domain adaptation, the classifier selects among the limited labels on target data by combining hybrid oracles. However, obtaining the oracles is costly. Instead of selecting instances to be added to the training set,115 selects a bag of instances by self-training. However, this approach is more effective when the source and target domains differ substantially. An appealing work is a recent application of active learning in domain adaptation for biomedical images. 114 Active learning chooses the candidates with higher entropy and higher diversity, which are expected to improve the current performance. In Shin et al. ,114 a pre-trained CNN is further fine-tuned continuously by incorporating newly annotated samples in each iteration to enhance the performance incrementally. Although they can reduce the annotation cost, the iterative scheme can be time-consuming, especially if applied to volumetric data. Previous work on participant selection with active learning requires iterations and only deals with classification of 2D images.
Instead of using iterative active learning, we propose a framework to select the ‘most valuable’ samples from the unlabelled target domain to be annotated – using RCA70 (or see previous section). We address the question whether RCA can be employed to select fewer participants to reduce the cost of annotation in supervised domain adaptation. To answer this question, we systematically conducted several experiments. Our contributions are: (1) demonstrating the effective use of RCA as a selector for n-participants in target domain to be incorporated into the training data set, (2) comparing different strategies for supervised domain adaptation with the RCA selection, (3) studying how the training size and the combination of target samples affect segmentation performance.
Data sets
Source domain (S): The data set is obtained from our Phase 1 of MALIBO study and includes abdominal T1-weighted MRI Dixon images of 35 healthy participants. We consider this data set as the source domain used to train the initial classifier in a supervised manner as manual organ annotations are available for all participants. The images have size of (256 × 208 × 202) and resolution (1.64 × 1.64 × 5) mm.
Target domain (T): Data for the target domain are obtained from the UK Biobank (UK Biobank Application ID 12579, ‘ML for Abnormality Detection in Imaging Studies with Application to Auto-QC and identification of Pathology-specific Outliers through Correlation with Non-Image Data’, PI Dr Ben Glocker, approval date 1 March 2016). We use 45 participants with manually annotated T1-weighted Dixon MRI images which have been acquired with a similar protocol as the MALIBO data. The main obvious differences are the image size (224 × 168 × 366) and resolution (2.23 × 2.23 × 3) mm.
The source and target data set are acquired at different centres. UK Biobank data are acquired with a Siemens 1.5T MAGNETOM Aera scanner while in MALIBO a Siemens 1.5T MAGNETOM Avanto was used (see Image pre-processing for whole-body-magnetic resonance imaging: correction of fat-water swaps in Dixon magnetic resonance imaging). For this study, we use the T1-weighted in-phase images from the Dixon protocol. We focus on organ segmentation within WB scans. As a pre-processing, image intensities in both data sets are normalised to zero-mean and unit-variance. Images are resampled to the same size and physical resolution. Visual examples from the source and target database are depicted in Figure 7. Despite the similarity of the scanning protocol and same scanner manufacturer, the drop-in segmentation accuracy when applying a model trained on MALIBO and tested on UK Biobank data is striking, as we will show in our experiments. The images seem to encode a significant bias in their appearance which is not obvious upon visual inspection.
FIGURE 7.
Examples of WB scans from source (MALIBO) and target (UK Biobank) database.
Supervised domain adaptation
We apply RCA with the single-atlas registration classifier as described in previous section or see. 70 The test image together with its predicted segmentation is registered to a set of reference images such that the predicted segmentation can be quantitatively compared to the manual segmentations of the reference images by computing DSC. It is expected that the maximum DSC score over all reference images correlates well with the real DSC. RCA acts as a selector for picking up participants with high and low confidence in segmentation accuracy. Our hypothesis is that transfer learning with specific n-participant selection is better than picking-up random participants from the target domain, and thus fewer manually labelled participants are needed from the target domain. In the following, we call this ‘domain adaptation using RCA’ or DARCA.
In our experiments, we employ DeepMedic15 as the base network for 3D organ segmentation. We use the default 11-layer-deep, multiscale, parallel convolutional pathways architecture.
The main approaches of supervised domain adaptation with CNNs are either training from scratch or FT. 113 Using RCA as a selector, n-participants from the target domain are added to the training data. With the new training data containing source and n-target (S + T) participants, we train the network from scratch. Meanwhile, we can transfer the parameters from a pre-trained network and fine-tune on another database. To build the model, we fine-tune the pre-trained network (from S-only training), with the n-target participants selected by RCA. Based on the results in Ghafoorian et al. 116 FT the last layer achieves the best performance compared to using more convolutional layers for FT. We fine-tuned only the last layer of the pre-trained networks and used the same optimisation but with fewer epochs. Based on our experiments FT all the layers are shown to have lower accuracy and more training time is needed.
Domain adaptation experiments and results
We present results for using different strategies to investigate the effect of RCA-based participant selection for domain adaptation, as shown in Figure 8. We use threefold cross-validation with the same random splits in all experiments. As the baseline, we trained the network with all S data and tested it to segment the T images. We predict the DSCs of all target segmentations using RCA. After we sort their DSCs (lowest to highest confidence), we select n-participants from T domain to be included with their corresponding manual annotations in the training set mimicking an active learning approach.
FIGURE 8.
Overview of the strategies we tried for n-participant selection in DARCA: 1. Training from scratch. 2. FT. 3. Iterative DARCA.
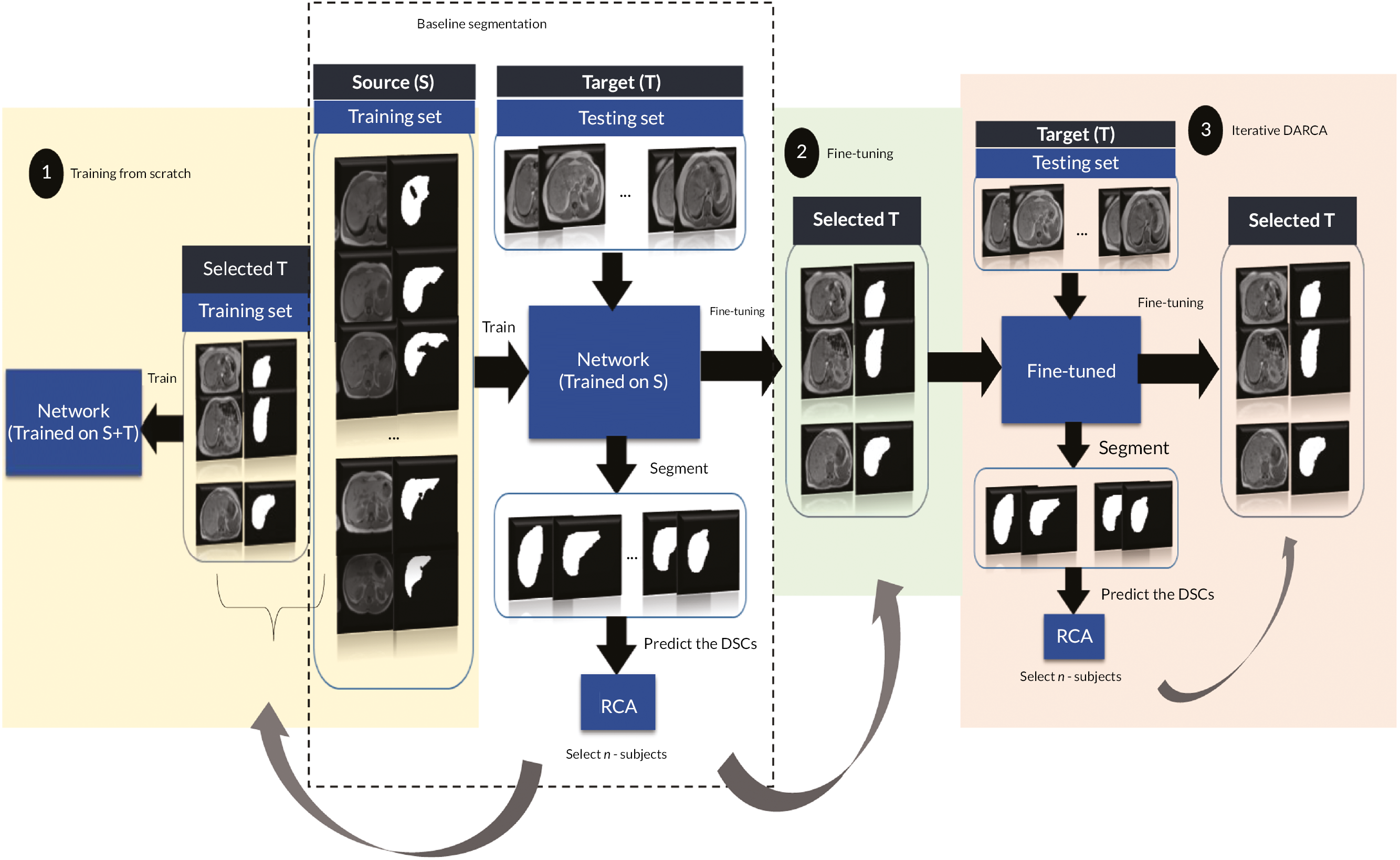
Training from scratch
We set our baseline as the segmentation of T with an S-only trained network, whereas the upper-bound is the segmentation of T with T-only trained network on LVR segmentation, as shown in Table 10.
| Training | DSC [mean (stdv)] |
|---|---|
| Train on S (baseline) | 0.639 (0.149) |
| Train on T (upper-bound) | 0.873 (0.046) |
Figure 8 shows that RCA selects n-participants from T domain, to be manually labelled and incorporated into training data set. For this experiment, we compared best-/worst-5 participants selected by RCA and the real best-/worst-5 (real DSCs from the target GT). Besides, we also run random-5 participant selection (repeated with three different random combinations and taking the average) to be trained from scratch.
In the training-from-scratch strategy, we train S + T data simultaneously with the same optimisation, update-rule, number of epochs, loss function, and regularisation techniques as in the baseline. From Table 11 column one, we can see that picking up five participants from target domain has already improved the accuracy, compared to the baseline. Table 11 shows that the segmentation accuracy using the best-5 or worst-5 participant selection outperformed the random selection. Moreover, RCA selection gives a relatively similar segmentation accuracy to the selection using real DSC.
| Strategies of sample selection | Training from scratch | FT | Iterative |
|---|---|---|---|
| Baseline | 0.639 (0.149) | 0.639 (0.149) | 0.639 (0.149) |
| All T | 0.831 (0.074) | 0.830 (0.066) | N/A |
| Random 5 | 0.720 (0.103) | 0.710 (0.172) | N/A |
| Worst-5 (real) | 0.797 (0.051) | 0.619 (0.256) | N/A |
| Worst-5 (RCA) | 0.799 (0.048) | 0.771 (0.156) | 0.828 (0.072) |
| Best-5 (real) | 0.747 (0.152) | 0.723 (0.173) | N/A |
| Best-5 (RCA) | 0.755 (0.148) | 0.687 (0.191) | 0.777 (0.107) |
| Best-5 and worst-5 (real) | 0.823 (0.058) | 0.842 (0.050) | N/A |
| Best-5 and worst-5 (RCA) | 0.831 (0.063) | 0.835 (0.065) | N/A |
As we increase the number of annotated target images to be incorporated in training from scratch, the accuracy does improve, as shown in Table 32 in Appendix 1. From Table 11, T segmentation with lowest confidence also gives a good contribution to the current CNN, therefore we combine five target participants with highest confidence and five participants with lowest confidence into the training. The result shows that this combination achieves similar accuracy to the one that incorporated all annotated T participants. Hence, we reduce the ‘less valuable’ samples for training by RCA selection. Unfortunately, training from scratch requires more time since the networks must learn from the beginning.
Fine-tuning a pre-trained network
In Figure 8(2), we fine-tune the pre-trained network (from S), with n-participants selected by RCA. FT requires less time than training from scratch. Here, we fine-tune with three different selections (random, real DSC, and RCA) at different set size (2, 5, 10, 15 and all of the T data). Table 32 in Appendix 1 shows the results when we fine-tune using all of the T data, which are very similar (DSC: 0.830) to when we train from scratch (DSC: 0.831).
However, RCA seems worse to predict the real segmentation accuracy of FT. There is a gap between best-/worst-5 real selection and best-/worst-5 RCA accuracy (see Table 4). One of the reasons could be due to the under-estimation of RCA prediction on the baseline segmentation accuracies on T, with 0.88 correlation and 0.15 MAE. As explained in the experiments of multiple-organs segmentation,70 the RCA prediction for organs with the real DSCs between (0.6–0.8) is not as accurate as in organs with real DSCs above 0.8. In our case, the average real DSCs of baseline LVR segmentation is 0.639, but RCA under-estimated the mean of predicted DSCs to be 0.497.
As we did in Training from scratch, we also combined best-5 and worst-5 in T domain to be annotated and incorporated in FT. These 10 combined participants give much better accuracy than when we choose only the best-10 participants (see Table 32 in Appendix 1). Similarly, picking up the worst sample in increasing number, results in lower accuracy than the ‘best 5 and worst 5’. Best-5 and worst-5 annotated samples with real selection shows the best results (DSC: 0.842), and RCA selection also gives similar results (DSC: 0.835). Additionally, this best-5 and worst-5 combination (real and RCA selection) performs better than when we use all of the annotated participants from T (DSC: 0.830). Hence, we cut the cost of annotation by 67%, using only 10 selected participants instead of 30 participants and achieve a higher accuracy.
Pseudo-labels for fine-tuning
In this experiment we investigate the use of pseudo GT labels in a semi-supervised way. In Fine-tuning a pre-trained network, we incorporated the n-participants by FT with their GT. What if, instead of using the real annotations, we use the predicted labels as pseudo GT, which are the baseline segmentation results – for training. In the previous work by Lee,117 pseudo-labels (PL) are used for semi-supervised learning with pre-trained and FT scheme. PL are defined as labels that have maximum predicted probability and seen as equivalent to entropy regularisation, which encourages low density separation between classes.
However, from Table 32 in Appendix 1, it is clear that using PL cannot improve the segmentation performance on the target domain. FT with all of the pseudo-labelled participants in T gives the worst result amongst all. The noisy labels negatively impact the segmentation performance. Hence, training using PL seems not suitable for domain adaptation in our application, since it assumes the baseline classifier to be of good quality, while by default it should be considered to be severely suboptimal.
Iterative domain adaptation using reverse classification accuracy
Different from the previous strategies, here, we wish to mimic the active learning domain adaptation,118 where at each iteration, RCA chooses n-participants from the target domain to fine-tune the baseline networks (see Figures 8 and 9). At the first iteration, we fine-tune the baseline network with best-5 participants selected by RCA. This new network is used to segment the test images from target domain for which accuracy is again predicted using RCA. At the second iteration, we fine-tune the network again with the best-5 and worst-5 selected participants.
FIGURE 9.
Plot for different n-selection training size in different strategies FT, FT with PL, and training from scratch (S + T). Similar trends are shown between real and RCA selection on FT and (S + T) with different size.
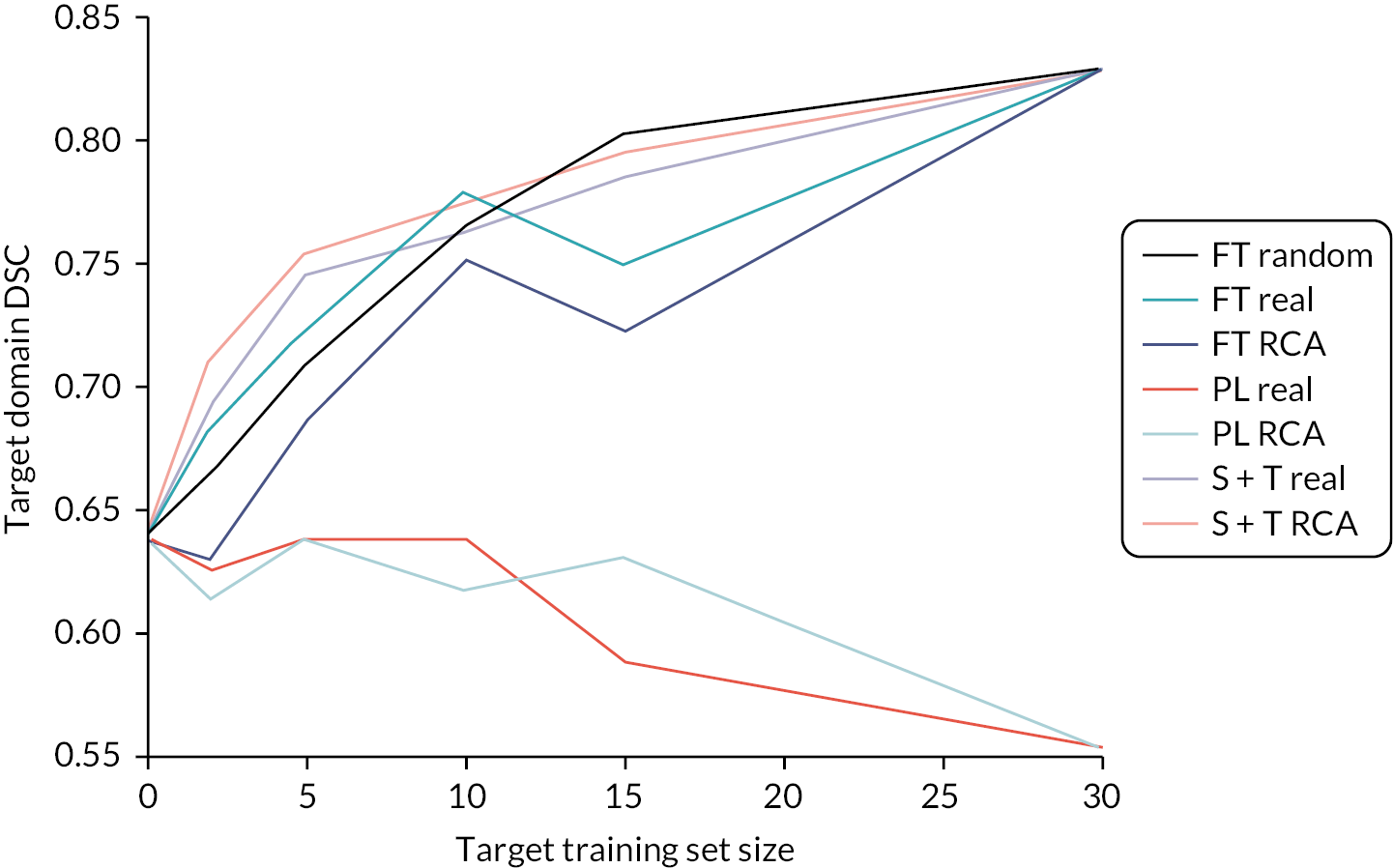
The results in Table 32 in Appendix 1 show that the second iteration with worst-5 participants gives higher accuracies than FT with best-5 RCA. The combination of best-5 (1st iteration), and worst-5 (2nd iteration) by RCA performs almost the same (DSC: 0.828) as FT using all of the labelled target data (DSC: 0.830). Additionally, the 2nd iteration with worst-5 RCA selection generally improves the 1st iteration (by best-5 RCA selection) accuracies. Hence, with fewer labelled data we can save time with similar results.
Fine-tuning in right kidney segmentation
From the three different strategies of DARCA in LVR segmentation, we can see that FT with DARCA gives better results, less time-consuming (compared to training from scratch), and no iterative scheme needed. Also, from the results in LVR segmentation (see Table 11), combination of best-5 and worst-5 participants always gives better or similar results than using all of the participants from domain T, in all strategies. To validate these results, we also explore DARCA-FT in a different task: RKDN segmentation.
Similarly, the best results of RKDN segmentation with FT are achieved when we combine best-5 and worst-5 participant selection (Table 12). The result (DSC: 0.716 with RCA selection) is better than when FT with all of the participants from T (DSC: 0.658), and similar to when we train from scratch using all target participants. This means we could cut the processing and annotation time. Figure 10 and Figure 11 depict some examples on how DARCA-FT with combination of best-5 and worst-5 participant selection improves the baseline and gives the best segmentation accuracies.
FIGURE 10.
Domain adaptation using reverse classification accuracy FT in LVR segmentation. Combination of lowest and highest RCA prediction can give a better result than FT with random selection and with all of the target participants.
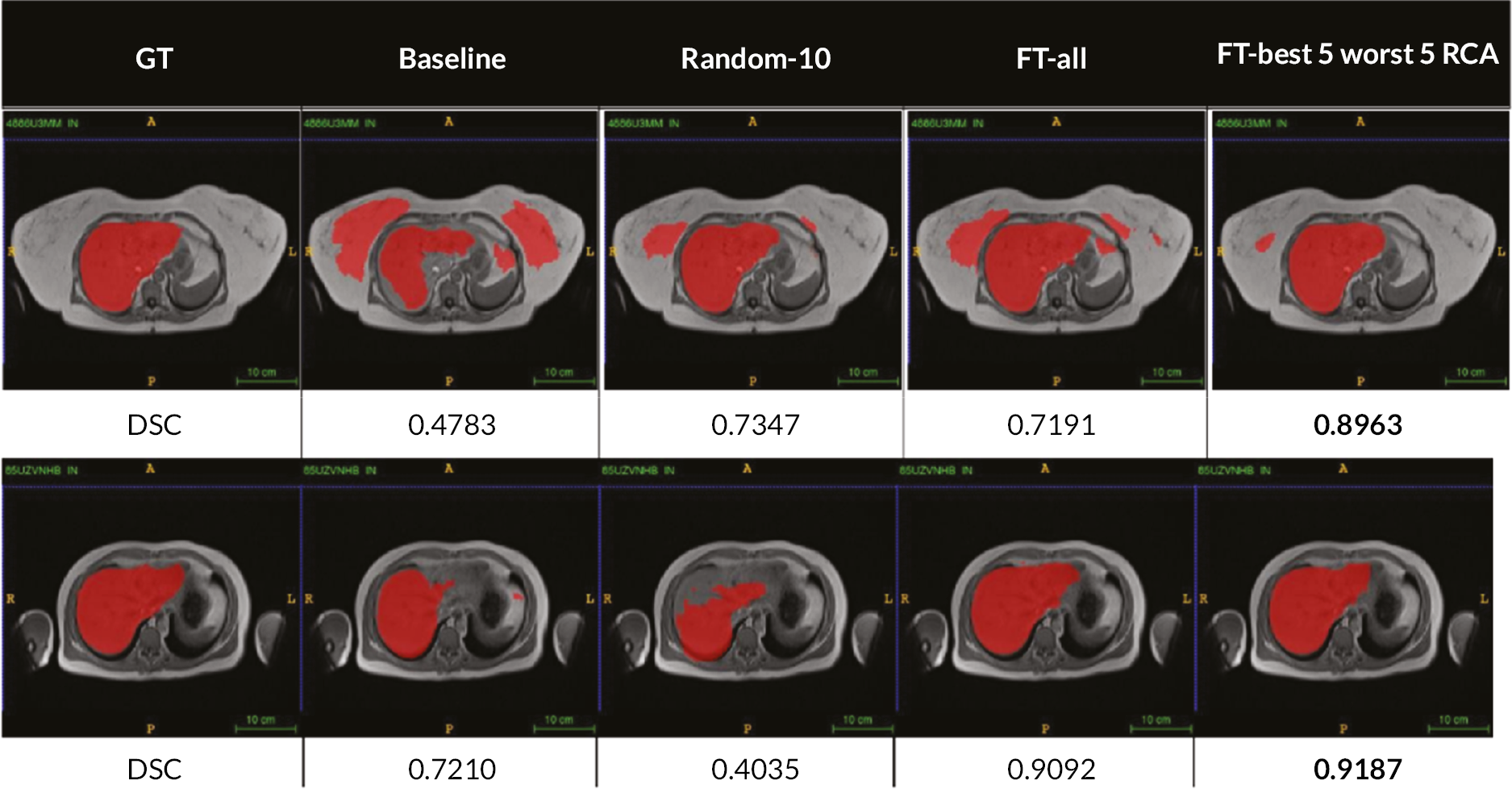
FIGURE 11.
Domain adaptation using reverse classification accuracy FT in RKDN segmentation. Combination of lowest and highest RCA prediction can give a better result than FT with all of the target participants.
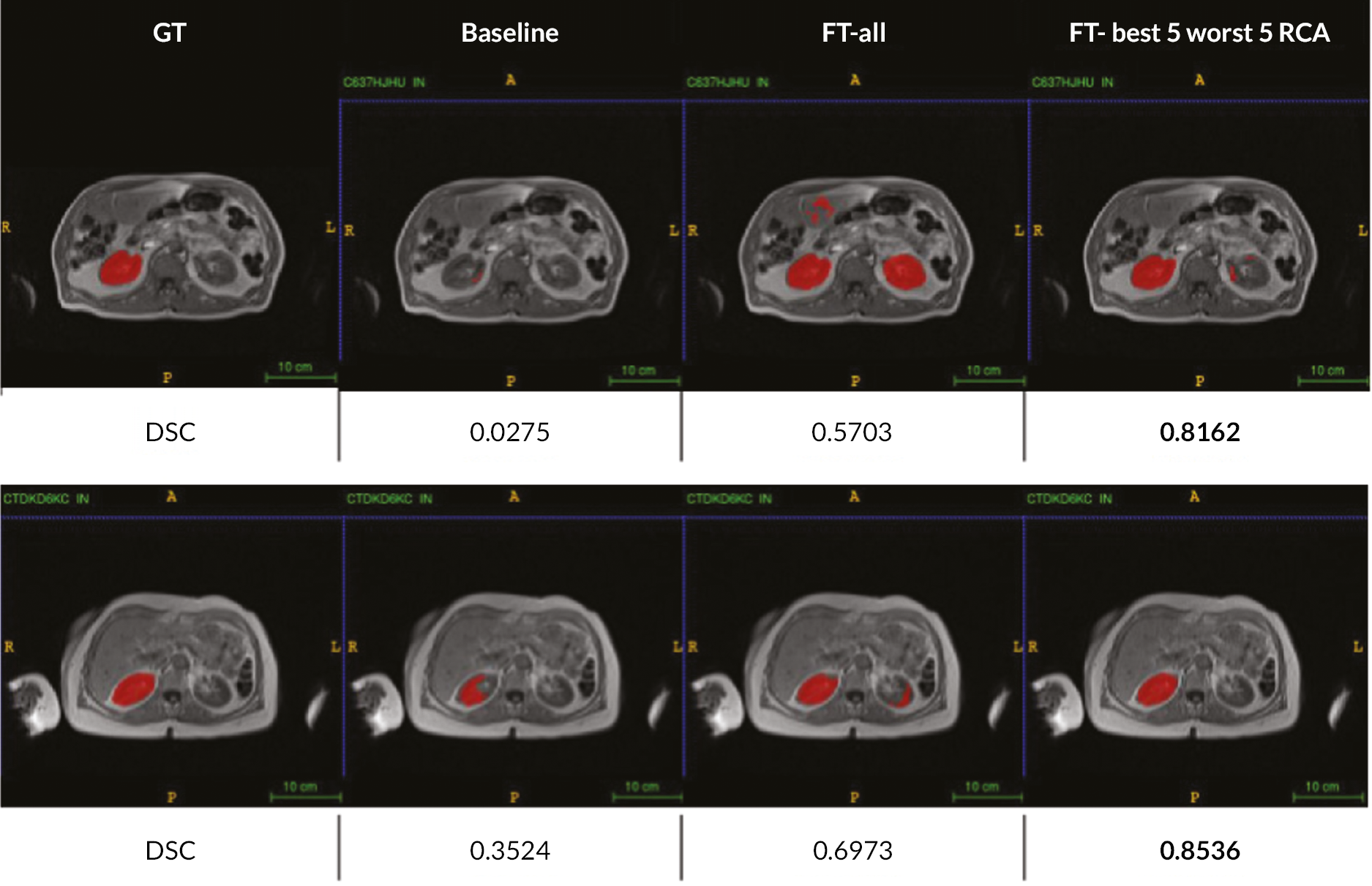
| Strategies | FT [mean (stdv)] |
|---|---|
| Lower bound | 0.417 (0.263) |
| Training from scratch (all T) | 0.719 (0.106) |
| FT all T | 0.658 (0.114) |
| FT random 5 | 0.506 (0.278) |
| FT worst-5 (real) | 0.416 (0.254) |
| FT worst-5 (RCA) | 0.358 (0.274) |
| FT best-5 (real) | 0.500 (0.293) |
| FT best-5 (RCA) | 0.421 (0.319) |
| Best-5 and worst-5 (real) | 0.726 (0.126) |
| Best-5 and worst-5 (RCA) | 0.716 (0.122) |
Discussion and conclusion
Set size and participant selection are important in domain adaptation, where usually labels are not available in one of the domains. Thus, we explored whether it will be useful to select only the ‘valuable’ participants by RCA to be annotated. PL, which normally are used in semi-supervised learning and regularisation, seem not to be useful in supervised DARCA. As observed in Figure 9 the performance drops as we increase the number of PL in FT. PL will introduce more noise confusing the training of the networks and make them incapable to be applied to the new domain.
All of our strategies in DARCA (training from scratch, FT and iterative) show a consistent result, combination of best-5 and worst-5 participant selection yields best results. RCA selection of those combined participants also results in a similar accuracy to the real selection, compared to a bigger gap between RCA and real selection when FT with only best or worst participant selection.
In this scheme, DARCA shows its potential to leverage the highest and lowest confident participants, to be incorporated in the domain adaptation process. We demonstrated that DARCA with few labelled data can perform similarly and/or better to full-size labelled target data. In the examples of Figures 24 and 25, DARCA with best-5 and worst-5 participants show consistent results across different tasks (LVR and kidney segmentation).
In the case of real DSCs between (0.6, 0.8), RCA underestimates the predictions. 70 This led to a different participant selection. In future, an improvement of RCA prediction for medium level DSCs (0.6–0.8) needs to be investigated so that it can work more accurately. The study only focuses at a predefined number of selected participants, and a more thorough exploration needs to be done in future work. Traditional active learning may have more flexibility regarding the number of ‘valuable’ samples to be included, but it requires iterations, which is time-consuming. DARCA only needs RCA to predict the DSCs from the baseline segmentation where we can select the highest and lowest predicted participants to be incorporated in a quick FT procedure. Therefore, we can conclude that DARCA could save processing time (no iterations needed) and annotation time with promising results avoiding the need for a large annotated target database.
Chapter summary
In this chapter, we only employed MALIBO data from healthy volunteers. Based on the data, we have developed RCA and domain method for medical image segmentation. These methods are useful for image segmentation if there is no GT available. We use threefold cross-validation to automatically segment all 15 organs from 35 participants with each of the 3 different segmentation methods, that is RFs, CNNs and MA. Overall, we found high correlation between predicted and real DSC for both AFs and single-atlas when used as RCA classifiers, with the single-atlas showing correlations above 0.95 for all three segmentation methods. In the future, it will be interesting to use these methods for lesion segmentation from patients’ data.
Chapter 5 Developing machine-learning method for clinical whole-body magnetic resonance imaging study: Phase 2 training and validation methods and model selection
Parts of this chapter are reproduced from Lavdas et al. 17 with permission from Clinical Radiology. The aim of Phase 2 was to develop an algorithm to detect and highlight possible cancer lesions on clinical WB-MRI. The primary output of the algorithm is a lesion probability map that can be visualised concurrently with the original MRI images during radiological reading, with the aim of providing valuable information to the reader that would increase the accuracy of lesion detection and speed up the reading process.
As this study uses a ‘human-in-the-loop’ approach, particular focus was ultimately given on an algorithm with high sensitivity to detect cancer lesions to make sure no lesion is being missed while there was less concern about false positives as these could be ruled out during radiological reading.
Whole-body magnetic resonance imaging data
In Phases 2 and 3 of the MALIBO project, the study used WB-MRI data from the NIHR-funded STREAMLINE-C and STREAMLINE-L studies. 6–8 These were multicentre, prospective cohort studies that evaluate WB-MRI in newly diagnosed colorectal cancer patients (STREAMLINE-C: ISRCTN43958015) and lung cancer patients (STREAMLINE-L: ISRCTN50436483), recruited from 16 NHS centres in England. The STREAMLINE studies evaluated the potential role of WB-MRI as in single investigation for staging patients with lung or colon cancer when compared to the current standard of care pathways. A full description of the STREAMLINE-C and STREAMLINE-L studies can be found in the references. 6–8,40
Ethical approval and consent
Ethical approval for retrospective use of previously acquired patient data was obtained (ICREC Reference 15IC2647). The MALIBO study did not directly collect patient imaging data, but relied on data from previous NIHR- and CRUK-funded trials (herein referred to as ‘contributing studies’40). The ethical approval for Phase 1 of the trial was in place (ICREC 08/H0707/58). Ethical approvals for Phases 2 and 3 (contributing studies) were also in place as per their individual protocols. 40 There were no material ethical concerns related to the MALIBO study with no perceived risk or benefit to individual patients. However, there was a significant interest in improving patient care, as indicated in section 60 of the Health and Social Care Act (2001). All patients gave written informed consent prior to participation in any of the contributing studies. Consent for the use of scans in future research was also obtained in the case of participants in the contributing studies. The need to re-consent participants for the use of the patient data was waived by the ethics committee. All patient data were de-identified and held in a secure central imaging server 3Dnet™ (www.3dnetmedical.com/public/), provided by Biotronics3D (London, UK). The data are also held on password-protected Imperial College London university computers for the purposes of the ML algorithms’ development.
Inclusion/exclusion criteria for evaluated cases
No patients were directly recruited into the MALIBO study. Recruitment and scanning of patients had taken place under separate studies (contributing studies), using their own ethical approval. The STREAMLINE-C study recruited patients from 16 hospitals between March 2013 and August 2016 with a final number of evaluable patients of 299, 68 (23%) of whom had metastasis at baseline. The STREAMLINE-L study recruited patients from 16 UK hospitals between March 2013 and September 2016 with a final number of evaluable patients of 187, 52 (28%) of whom had metastasis at baseline (see Figure 26). A total of 438 patient scans were available to the MALIBO team for evaluation and the remaining 48 scans were not to be available on the image repository (reasons not known).
Additional data from the CRUK funded MELT study (Whole-Body Functional and Anatomical MRI: Accuracy in Staging and Treatment Response Monitoring in Adolescent Hodgkin’s Lymphoma Compared to Conventional Multi-modality Imaging: NCT01459224)59 and the MASTER study [MRI Accuracy in STaging and Evaluation of Treatment Response in Cancer (Lymphoma and Prostate-MASTER L and MASTER P; 12/LO/0428)]43,59 were considered but then excluded due to significant differences in the images protocol and it was not felt possible to train the model which such variable data.
The following inclusion/exclusion criteria summarise the patient population for the MALIBO study Phases 2 and 3.
Inclusion and exclusion criteria
Inclusion criteria:
-
patient eligible for and consented to take part in one of the contributing studies: STREAMLINE C or L, MELT, MASTER
-
patient completed the study imaging assessments successfully
-
image DICOM data available on the image repository
-
consensus reference standard from the source study available.
Exclusion criteria:
-
patient that consented to contributing studies but did not complete the WB-MRI scan
-
scan could not be adequately completed due to, for example technical reasons
-
ML algorithm failed to produce results due to technical problems with the scan (missing sections, corrupted data or in the case of training data, poor quality of ADC/DWI images or extreme artefacts)
-
MELT and MASTER data due to incompatibility of imaging protocols.
Inclusion/exclusion criteria for the contributing studies can be found in previous works. 8,42,43,59
Reference standard for sites of disease: STREAMLINE study
The reference standard for the site of the primary tumour and presence and site of metastatic lesions was established as part of the STREAMLINE study: At 12-month patient follow-up, a multidisciplinary consensus panel defined the reference standard for tumour stage considering all clinical, pathological, post-mortem and imaging follow-up. Accuracy was defined per lesion, per organ and per patient.
Allocation of cases to Phases 2 and 3
Allocation of patients for Phase 2 and 3 testing was based on the available 438 STREAMLINE-C and STREAMLINE-L scans (Figure 12). Of these reads, 97 were pre-allocated to Phase 2. This initial allocation had been undertaken due to delays in the completion of the source study and a decision was made to provide a number of scans to the MALIBO team to allow the start of the time-consuming manual segmentations, while awaiting the STREAMLINE studies to complete. This initial allocation was random and consecutive, as to when the scans became available on the image sharing platform from the sponsor. To ensure that the training set (Phase 2) and validation set (Phase 3) contained a similar array of reads, 97 cases were subsequently assigned to Phase 3 by the study statistician to ensure that the proportion of study type (lung or colon), study site (hospital) and presence of metastases (LVR, bone or nodal sites) matched. When an equal proportion could not be reached exactly, study site was subsequently removed from the matching criteria.
FIGURE 12.
Phases 2 and 3 CONSORT diagram.
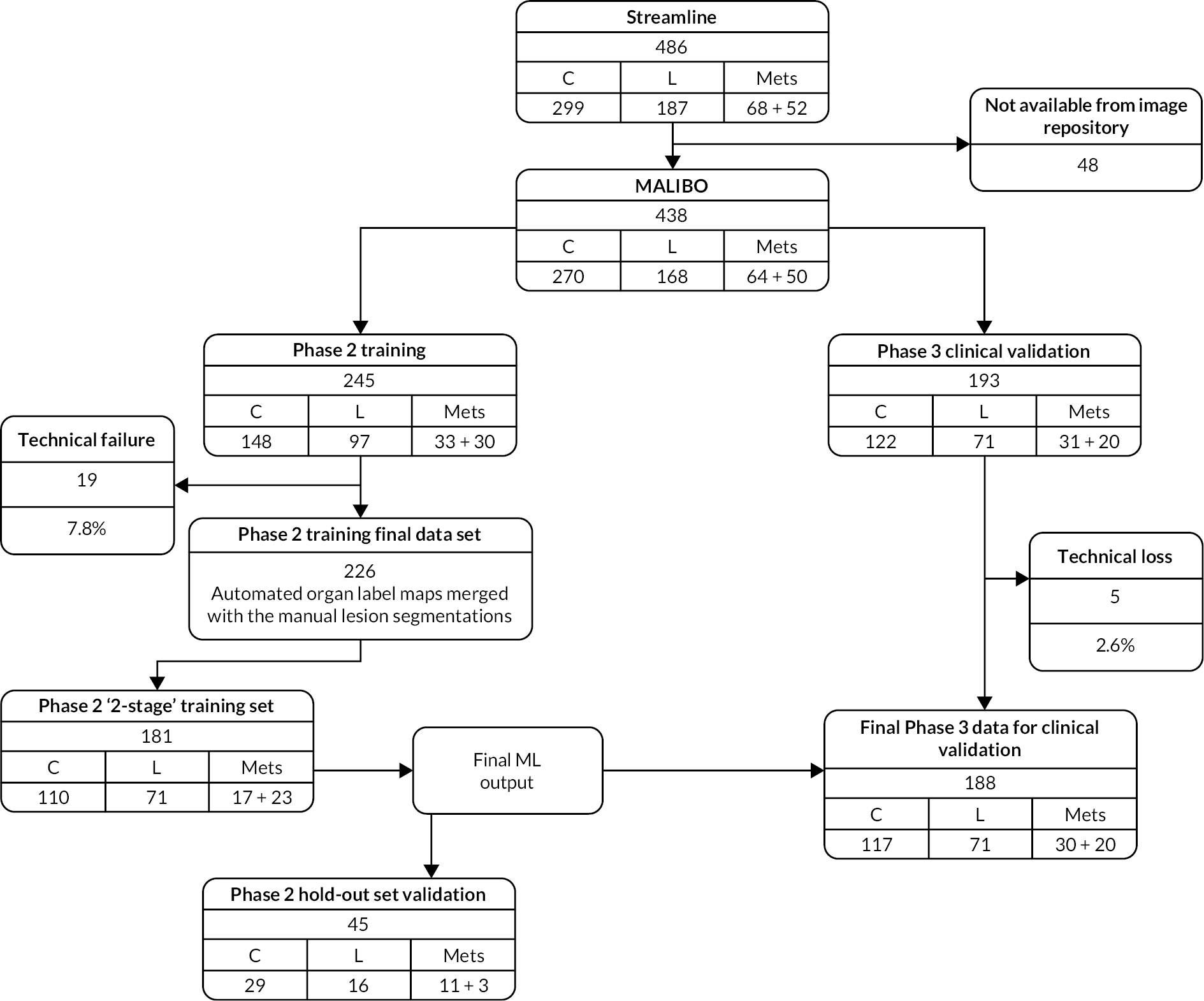
To adequately power the primary analysis, the remaining unallocated cases were then allocated at random between Phases 2 and 3 while ensuring that 141 reads with no recorded metastatic tumours were allocated to Phase 3. As before, cases were allocated such that the two phases had a similar proportion of cases based on study type (lung or colon), study site (hospital) and presence of metastases (LVR, bone and nodal).
Following the allocation process, 245 reads were assigned to Phase 2 and 193 reads were assigned to Phase 3. Frequency tables were run on both sets of data to validate the proportions between the two data sets matched for the variables described above.
Image preparation
There are many challenges for applying ML methods to clinical studies using WB-MRI, and this was particularly so for this study as the source data were obtained from 16 different centres. We encountered a range of data quality challenges (artefacts) in the data sets used in MALIBO. For example, in some participants, there were missing slices in the data; it was not uncommon to collect data with radio frequency interference; for some participants, there were motion artefacts; image artefacts were also present due to RF field inhomogeneities with resulting dielectric shading.
Whole-body magnetic resonance imaging scans were provided by the source study in their raw, unstitched format, with typically four stations for each of the axial WB-MRI sequences. Each station of the axial T2-weighted sequence, high b-value diffusion sequence and the accompanying ADC map were stitched together into single volumes, as described in Chapter 3.
Image quality
The versatility of MRI is the modality’s ‘blessing and curse’. A WB-MRI protocol puts both the machine and the patient through a stressful test, which usually is not tolerable for repetition. Therefore, a WB-MRI examination with DWI, will be extremely prone to artefacts. It is very common that through the rapid clinical workflow, the acquired images are not checked thoroughly during acquisition and imaging data sets of compromised quality can be ‘passed through the sieve’ of the clinical workflow.
It should be stressed, however, that the quality of the data sets might have been suitable for the objectives of the clinical study and not all of the issues were externally triggered [e.g. distortions in echo-planar imaging (EPI) DWI acquisitions are unavoidable], but they may have caused challenges to the ML algorithms with a subsequent detrimental effects to their performance.
This indicates the importance of having imaging data of readiness level of ‘Band A’, appropriate for the task at hand, as described by Lawrence119 for ML studies. It is acknowledged, however, that when multi-centre data are collected, the scenario above is unrealistic, so loss of some cases that are not usable for ML, as happened in MALIBO, might be unavoidable, although this selective removal of cases was only undertaken in the data set allocated to model training. We lost 19 of the 245 (7.8%) WB-MRI scans allocated to the model development as they were not suited for ML purposes and had to be discarded (see CONSORT diagram Figure 26). For the data set allocated to the final Phase 3 clinical validation, all cases were included with no selection, although some cases were lost due to technical failure (see below, 5 of 193 allocated cases could not be used, 2.6%).
Image registration
The use of multimodality MRI data (‘multichannel’ data as commonly referred to in computer science terminology) has shown to improve algorithmic performance in tasks like brain lesion segmentation. However, using multichannel inputs for algorithm training requires registered imaging data sets between modalities, so that annotated data from a single modality can be used in the interest of time-efficiency when generating training data. Matched data sets from different modalities, is a task which can be performed efficiently enough in the brain, where no gross motion or anatomical deformation is expected between acquisitions, with a rigid registration algorithm. In body imaging, where there might be significant organ motion and deformation between acquisitions, a rigid registration might not suffice. The task proved to be even more challenging with whole-body MRI data. Furthermore, when we attempted to register DWI volumes to anatomical volumes, we encountered the extra challenge from the significant geometric distortion of the EPI-acquired, high b-value DW volumes. We qualitatively assessed registration between DWI and anatomical volumes, when using a 12 degrees-of-freedom affine registration, but with mixed results. A non-rigid registration using free-form deformations was also tested, but the time required to apply on the tens of WB data sets used in MALIBO was unacceptably long. In the first phase of MALIBO project, we simply used slice-matched acquisitions, resampled to match the spatial resolution of the reference volumes. For the Phase 2 and Phase 3 data from the STREAMLINE-C and STREAMLINE-L studies we decided to avoid registration between diffusion and structure scans as much as possible, as the main cause of discrepancy between the modalities was caused by patient breathing. This affected anatomical areas in and surrounding the lungs but was deemed acceptable compared to running an affine or nonrigid registration which might have introduced misalignment in other areas, rather making it worse. In order to match the modalities in terms of number of voxels and physical voxel size, a simple resampling algorithm was used with linear interpolation. Scans were visually checked, and the large majority of cases did not show any concerning misalignment between structural T1w, T2w and diffusion scans.
Training data for machine learning
Generating training data for ML algorithms is one of the most important, but also laborious and time-consuming processes. Manual, volumetric segmentations performed by clinical experts, were used to ensure reliable and accurate information for model training. These labelled data were also used as the GT to compare with, when evaluating algorithmic performance in the Phase 2 hold-out set. In MALIBO, we used ITK-SNAP120 to manually generate annotated WB images (see Appendices 2 and 3 for the usage of the software for this project; and for the manual segmentation method). Labelling of healthy structures (23 anatomical structures, including organs and bones) occupied a significant proportion of Phase 1 of the project, but this work was of paramount importance as in Phase 2 where we used a two-stage approach, to identify cancer lesions as will be discussed in Two-stage approach. For Phase 2, the manual annotations concerned all cancer lesions, including primary tumours and metastatic lesions that were visible to the expert annotator. All primary tumours and metastases were manually segmented in both diffusion and structural T2w scans, using the final reference standard for sites of disease from the STREAMLINE consensus reference standard. These primary and metastatic lesion segmentations were then fused into a single lesion segmentation map for each participant and these fused lesion maps were used for training within the two-stage approach as discussed below.
Machine-learning pipeline
The choice of the exact machine algorithm is difficult to make beforehand, and often different variants and alternatives need to be considered during development. This was the case in Phase 1 where we evaluated three different approaches for the organ segmentation, namely MA, CFs and CNNs. Based on this previous phase and our experience from other studies, such as brain tumour segmentation, we decided to employ and validate two approaches for cancer lesion detection in Phase 2, namely CFs and CNNs. CFs are powerful, multilabel classifiers, which facilitate the simultaneous segmentation of multiple structures. They have good generalisation properties, which means they can effectively be trained using a limited number of data sets. Both traits were desirable in MALIBO. Our CNNs implementation was based on DeepMedic, an approach which has been shown to perform very well in brain lesion segmentation with multiparametric MRI data. The details of the hyperparameters used for the CFs and network architecture for the CNNs, can be found elsewhere. CNNs performed consistently better in healthy organ segmentation in Phase 1 of MALIBO, so it was the algorithm of choice for Phase 2 of the project (lesion detection). However, we decided to also give the CFs a try as ultimately the task in Phase 2 is less of a segmentation task where we want to get exact boundaries around cancer lesions, but more of a lesion detection task where a heatmap should flag up suspicious regions to the human reader.
Feature crafting for supervised learning requires the definition of set of potentially useful features that are quickly and efficiently computed and provide adequate information for the algorithm to successfully perform the task at hand. In MALIBO, we have used two types of ‘box features’ for our CF algorithm. Box features are intensity-based features that are computed ‘on the fly’ and provide both local and contextual information extracted from the images. The CNNs are capable of learning highly complex features on their own during training and therefore, do not require any feature crafting.
One-stage approach
We initially experimented with a simple one-stage approach by training CFs and CNNs on the annotated Phase 2 data. This resulted in binary classification methods that were assigning a probability to each voxel in a multichannel WB-MRI whether it is believed to be part of a cancer lesion or not. Higher probabilities indicated that a voxel is more likely to show signs of cancer. Due to the massive imbalance between normal and cancer voxels, this approach resulted in algorithms with low sensitivity, and was deemed insufficient for the task.
Two-stage approach
Ultimately, we opted for a two-stage approach leveraging the training data and algorithm for normal structure segmentation from Phase 1. Running the Phase 1 multiorgan CNN segmentation on all Phase 2 data provided automatic organ maps for all patient scans. This required an intermediate step of registering Phase 2 data with a rigid registration algorithm to a template participant from the Phase 1 data (Figure 13). This was to compensate for the different fields-of-view of Phase 1 and Phase 2. While the healthy volunteer Phase 1 data were covering the body from shoulders to knees, the Phase 2 patient data included the head which affects the performance of the Phase 1 algorithm. The registration is automatic and fast, and only required to obtain the organ masks. These are then mapped back with the inverse transformation to the original Phase 2 data. For Phase 2, there is no reference segmentation of organs to compare with, so we assessed the quality of these segmentations visually and they appeared to be sufficient for the following purpose.
FIGURE 13.
Data generation process for the two-stage approach in Phase 2. (a) An example of a T2w WB-MRI scans from a participant in Phase 2 data. (b) After registration to a template scan of Phase 1 data. (c) Output of the organ segmentation algorithm developed in Phase 1. (d) After mapping the organ segmentations back to the original Phase 2 scan. (e) The manual lesion segmentation overlaid on the T2w scan. (f) Merged organ segmentations and cancer lesion segmentation overlaid on the T2w scan which is used for training the final Phase 2 multiclass segmentation algorithm.
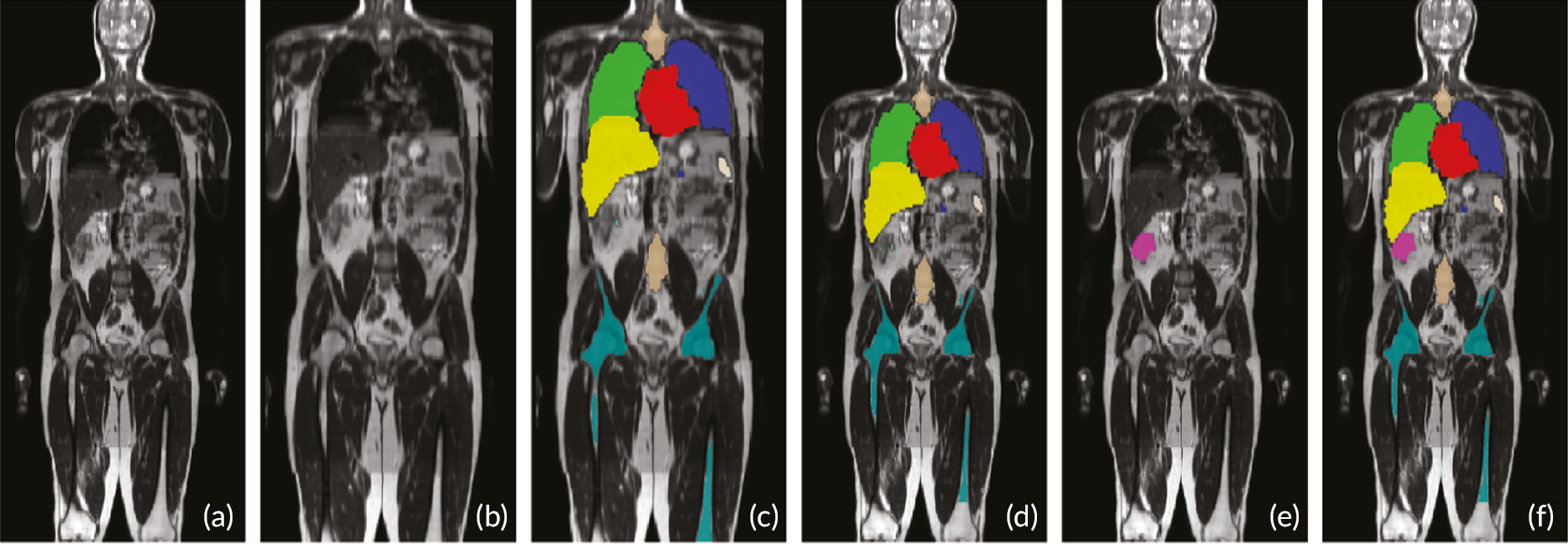
We merged the automatically generated organ maps with the manually segmented cancer lesions for Phase 2 data, by replacing the labels for all voxels that were marked as cancer with a new additional label indicating the presence of a cancer lesion. This was implemented by adding a new label to the list of organ labels. This resulted in all 226 scans from Phase 2 having multiclass segmentation maps where the organ labels were generated automatically using the CNN algorithm from Phase 1, while the cancer lesions were labelled manually. We then used the training set of 181 scans for training the two multi-class algorithms (CFs and CNNs based on DeepMedic) which are both capable of predicting the organ labels and cancer lesions jointly. This multiclass approach results in a much better distribution of voxels over class labels, and the ML algorithm has an easier task to learn class-specific features, rather than the binary task where all normal structures are merged into a single class. We confirmed this on the 45 scans in the Phase 2 validation set which showed a much higher sensitivity for detection of cancer lesions than the initial one-stage approach.
Post-processing to generate final lesion detection maps
This final two-stage approach could then be readily applied directly to Phase 3 data without any further pre-processing requirements on the input MRI, with the output being a probability map for each of the structures including the normal organs as defined in Phase 1 and the cancer lesions as defined in Phase 2 (Figure 14). The probability map for the cancer lesions is output of interest for the reader study in Phase 3. We found that the raw probability maps could be significantly improved for the final use with a customised post-processing pipeline. The probability maps produced by the CNN were smoothed with a Gaussian filter with a kernel size of 5 mm, then normalised to the range [0, 1], and thresholded to reduce false-positive detections. We also tested a larger kernel size of 10 mm, but 5 mm was found to be better based on visual assessment on the 45 validation cases.
FIGURE 14.
Cancer lesion detection. (a) Input T2w scan. (b) Diffusion scan. (c) Manual reference lesion segmentation overlaid on diffusion scan. (d) Post-processed lesion probability map from the CNN algorithm. (e) Post-processed lesion probability map from the CF algorithm.
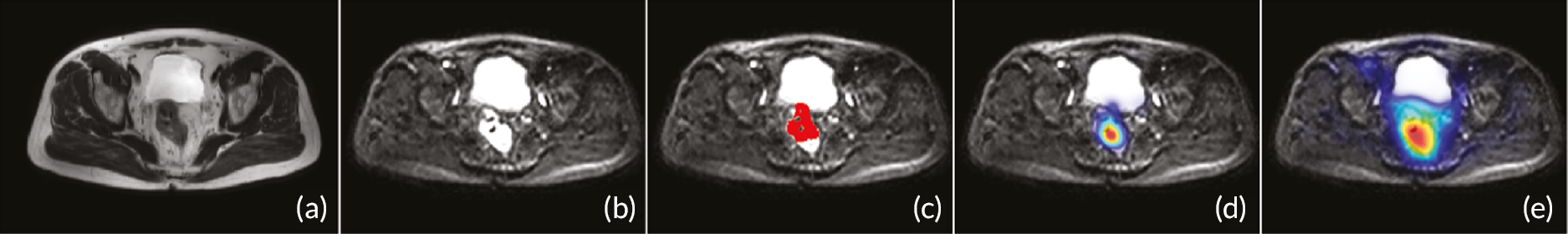
Results
We summarised the quantitative results over the 45 Phase 2 validation cases by plotting RE, PR and Dice curves (Figures 15–17).
FIGURE 15.
DeepMedic_multiclass curves.
FIGURE 16.
RF_multiclass curve.
FIGURE 17.
DeepMedic_binary curve.
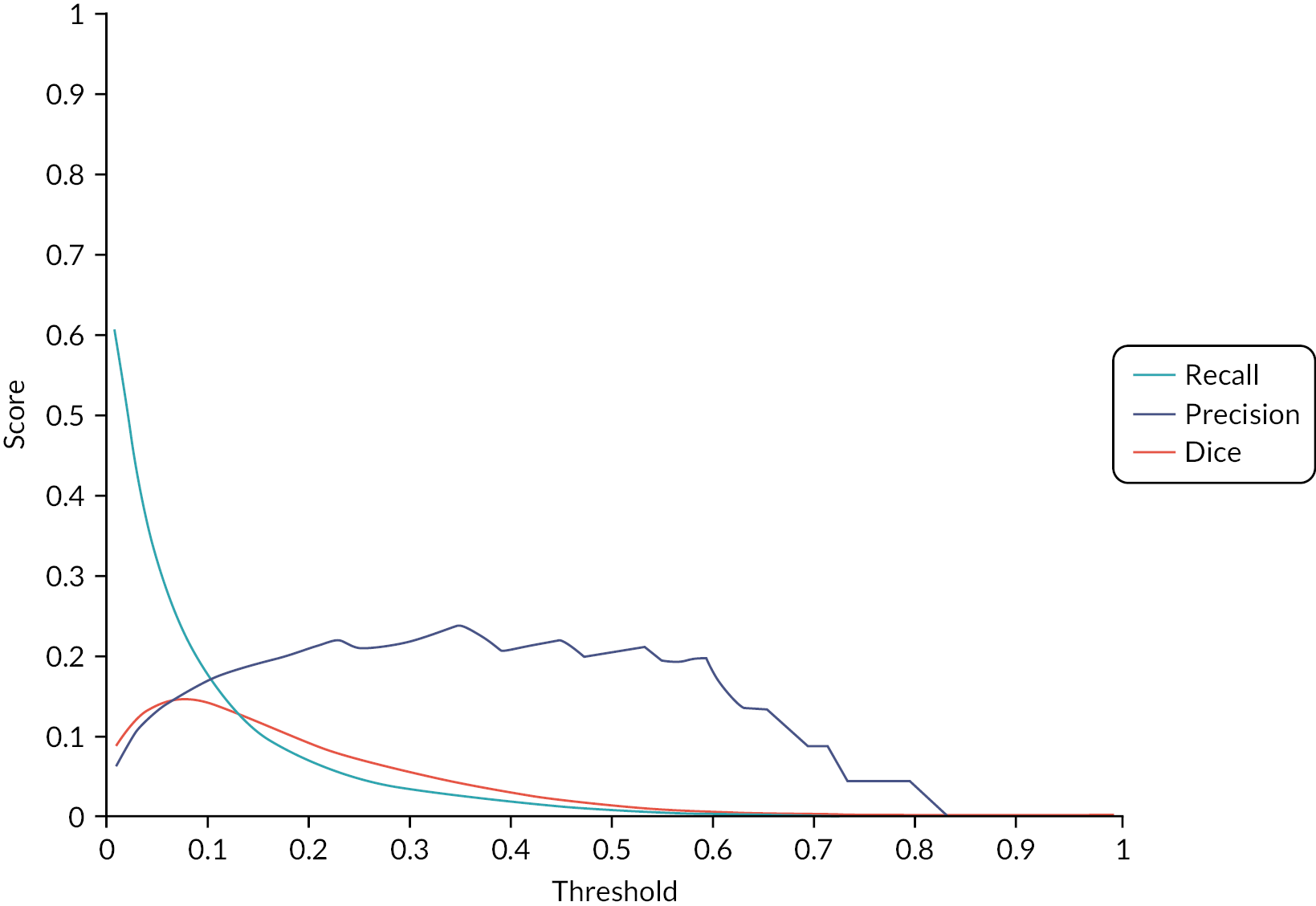
Figure 15 shows the multiclass DeepMedic CNN that we favoured overall as the best method. Multi-class here means that the algorithm predicts simultaneously labels for healthy organs and lesions.
Figure 16 shows the index curves for multi-class approach using RFs instead of CNNs.
Figure 17 displays a DeepMedic CNN that does not know about healthy organs, but is only trained to predict lesions versus non-lesions. Comparing this plot with the dm_multiclass nicely backs this up.
All three plots (see Figures 15–17) have three curves each for PR, RE and Dice. The important one is Dice, so whatever plot shows the highest Dice values is the best method. As we can see, in terms of Dice score, dm_multiclass > rf_multiclass > dm_binary.
It should be pointed out that these curves were plotted over different thresholds that one can place on the probability map. This is not relevant for the reader study (Phase 3 study), as the human sees recalibrated maps. It is also important to note that PR and RE here are on a voxel-level, so not to be confused with sensitivity/PR on a lesion level (as assessed in Phase 3).
Validation
Whether the task at hand is organ or lesion classification, segmentation or detection the core of the pipeline will most commonly be an accurate and robust classifier. In MALIBO Phase 2, we were interested in lesion detection and localisation rather than segmentation. We therefore had to employ a scheme to evaluate the CF and CNN-based voxelwise classification algorithms, but now in terms of detection task. A specific automatic evaluation procedure was implemented to calculate detection accuracy. This takes the manual reference segmentation and post-processed lesion probability from the ML algorithm as an input, and calculates the true positive rate, positive predictive value and F1 score, based on a connected component analysis and a user defined threshold (in millimetres) on the distance between true and prediction lesion location. We found the CNN-based algorithm to produce significantly fewer false positives. However, we kept the probability maps from the CFs for further evaluation in Phase 3, as false positives might not necessarily be a problem for a human-in-the-loop reader study. Thus, two final algorithms were available to radiologists in clinical validation Phase 3, each with two different kernel levels (rf kernel 5 and 10 and dm kernel 5 and 10). The ML output dm kernel 5 was selected to go forward but the rf kernel 5 was also provided as an alternative if the reader wished to use it.
Chapter 6 Machine-learning clinical validation: Phase 3 methods and results for performance evaluations
During Phase 3, the clinical validation ‘hold out’ set of WB-MRI data from the STREAMLINE study were read by experienced readers with the final ML algorithm support (termed algorithm ‘C’ in the original study protocol). The per-patient specificity and sensitivity of WB-MRI assessment, with and without ML support, were determined using the established reference standard from the contributing study. The RT was also recorded at this stage. Experienced and inexperienced readers undertook radiology reads and inter-rater agreement was evaluated.
Background
The algorithms taken forward into clinical validation both used a ‘two-stage’ approach for lesion detection on WB-MRI. In the first stage, the information from Phase 1 healthy organs/bones was used to identify the location of organs; in the second stage, the lesions are detected (as described in Phase 2 of MALIBO, Chapter 5). In addition, we refined and improved the computer algorithm to reach a substantial sensitivity for lesion detection, allowing for the fact that the final reading study was to be undertaken with experienced readers who could choose to ignore false-positive sites on the probability maps.
Verification of the clinical performance and utility of ML tools for radiology involves more than the demonstration of their technical soundness. 121 In the current study and final Phase 3 of the MALIBO project, we aimed to develop a robust methodology to verify and evaluate the ML lesion detection tool on WB-MRI in a near-real clinical setting. We compared the diagnostic test accuracy of the index test (WB-MRI with ML support, WB-MRI-ML) with the comparator test (WB-MRI standard, WB-MRI-SD) by a number of independent radiologists. We also measured RT using an independent scribe.
Reading platform
In clinical settings, PACS is used for hosting medical images and associated reader’s reports. In MALIBO, we have used a secure cloud-based central imaging server (3Dnet™), provided by Biotronics3D, to ensure that images and related ML output, are hosted in an secure environment where the research team could control the work-lists for each reader, ensuring blinding between reading rounds and strict allocation of cases with and without ML support.
Readers were trained online and were shown how to use a hanging protocol in Biotronics3D, so that stitched volumes from different imaging modalities, alongside the ML output, could be opened and browsed simultaneously, as shown in Figure 19 (or see Appendix 4). This setting also allowed for anatomical co-localisation using cross-hairs and also fusion between the colour-mapped ML output (probability map or ‘heat’ map) and any of the MRI modalities.
Data preparation for reading platform
Briefly, DICOM data from individual imaging stations had been previously stitched into a single NIfTI (https://nifti.nimh.nih.gov/) volume according to slice location to form whole-body volumes. 14,17 The original DICOM data were retained for ‘gluing’ back to the converted ‘header less’ images for uploading to the reading platforms 3Dnet™. The WB-MRI data of T2w, DWI and ADC map were registered for ML analysis and for viewing by the radiologists. To assist the radiologists in finding the necessary series, the series were all re-numbered in a uniform way so that they would appear in the same order in the series list on PACS, as all the cases came from multiple different hospitals and had different series orders. To ensure the required level of blinding, the secure central imaging server 3Dnet™ was employed to store and display WB-MRI and ML outputs. All the WB-MRI data sets from the contributing studies were anonymised (by the source study sites) before upload to the 3Dnet™ which enables rapid and simple upload of complex imaging data sets via a standard internet connection. The disadvantage of this system is that it can only process images with DICOM format; thus the NIfTI image volumes could not be used.
One hundred and ninety-three patient cases (122 colon cases and 71 lung cases), from the contributing studies, were initially allocated to Phase 3 of the study. One colon case (STC042) was removed prior to allocating Phase 3 cases to readers due to a technical failure to convert and upload image files which was recognised prior to the final allocation of cases to readers. Post allocation, four additional colon cases were excluded from the analysis: two of them were because of missing ADC maps (STC144 and STC151), one (STC062) had the corrupted DW image which leads to no-ML output and the other (STC223) was removed due to the failure to convert and upload image files (the same problem as STC042 but this was only recognised after the case has been allocated to a reader). Overall, 188 cases with ML output were adopted in the evaluation (see Figure 26).
Data conversion and viewing system
Four of the final ML algorithms were run on all 188 (rf kernel 5 and 10 and dm kernel 5 and 10). The ML output dm kernel 5 was selected to go forward into the clinical validation and in addition the rf kernel 5 output was available on PACS for additional use at reader’s discretion. The ML output was available for each WB-MRI-ML case as an additional series on PACS. The ML outputs which take the form of NIfTI were converted to DICOM format as the imaging server 3Dnet™ can only store and display DICOM image. A Python (www.python.org/) script was created to convert NIfTI images in an Ubuntu 18.04 Linux system (https://ubuntu.com/). In addition, MRI sequences were re-ordered for the central imaging server 3Dnet™ with T2w, ADC, DWI show on the top of the list on the server, followed by ML output. This was achieved by modifying the DICOM tags of the MRI images. After that, the converted images alongside with the original images were uploaded in 3Dnet™ system for radiologists to assess. As requested by 3Dnet™, Google Chrome (www.google.com/chrome/) and Mozilla Firefox (www.mozilla.org/en-GB/) (only one radiologist used this browser) browsers were used to view WB-MRI images. Each WB-MRI scan was copied and one copy was given a new unique identifier (UID) in order to allow one data set with ML output and the other as standard on PACS. Thus, each original case from the contributing STREAMLINE study had two versions on the MALIBO study folder in Biotronics3D: for example, STC001-SD or STC001-ML would be the STREAMLINE-C patient 001 with one standard version (SD) and one version with available ML output (ML).
Experimental design for the reads
Study design (allocation of reads)
One hundred and ninety-three Phase 3 cases were allocated at random by the study statistician to 18 experienced radiologists, defined as consultant radiologists that regularly reported reading WB-MRI in their standard clinical practice. Each reader was assigned 11 or 10 reads that would be assessed exclusively by them for the purpose of specificity and sensitivity testing. The randomisation was stratified by read type (colon or lung), presence of metastases (yes or no) and by original recruitment site to ensure that each reader had a similar set of reads. The allocation also ensured that no reader could assess read packages created at their home clinic/site. Allocated cases were then selected at random at a 1 : 1 ratio as to whether they would be presented with or without the additional ML documentation at round 1.
To allow for inter-rater assessments four or five additional reads were assigned to each reader in a way that would ensure that each read was assessed by two different readers. The same stratification method as described above was used in this process.
To allow for intrarater assessments, any reader that made themselves available for an additional third round of reads would have 10 reads (6 STC, 4 STL) selected from their original allocation (either primary or inter-rater) at random and then assigned whether to assess these with or without ML (at 1 : 1 ratio).
For 12 additional radiologists that were deemed inexperienced in reading WB-MRI or were experienced, 10 or 14 cases of the 193 Phase 3 cases were assigned at random using the same methodology as described above. Six or 10 cases were assigned for the purpose of establishing specificity and sensitivity with the other four cases used for inter-rater assessments. So each experienced reader was allocated 16 reads per session and each inexperienced reader was allocated 10–14 (three readers have 14 reads and four readers have 10 reads) reads per session.
The three rounds for the radiologist readers took place between 8 November 2019 and 6 March 2020. All readers took part in the first two reading rounds with paired reads. To reduce the possibility of RE bias, there were at least four weeks interval between reading rounds. Figure 18 shows the diagram for the 1st and 2nd round pipeline for experienced readers.
FIGURE 18.
Diagram showing experimental design for clinical evaluation of the ML method. Sixteen cases allocation for each experienced radiologist. The allocated reads include a selection of cases for inter-rater agreement (masked to the reader).
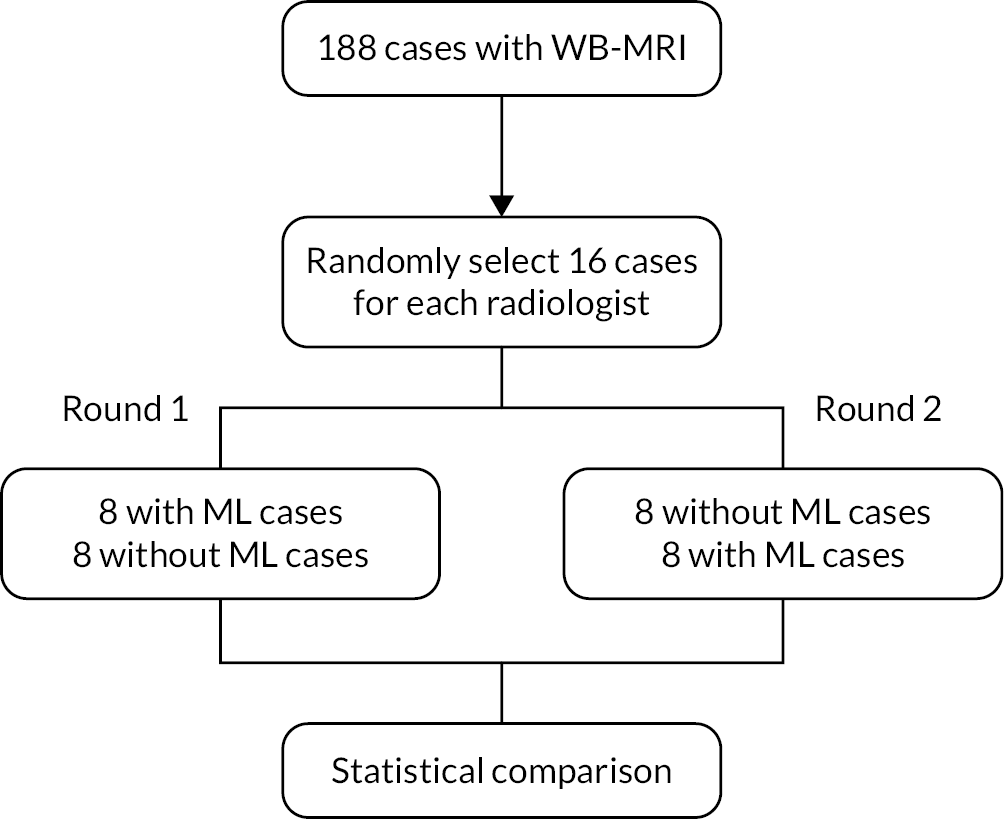
Independent radiologists training
Several weeks before the 1st round read, an instruction sheet (see Appendix 4) for using the 3Dnet™ system was sent to all the radiologists who took part in the study. The instructions listed the suggested optimal methods to view MRI images. Radiologists were able to login to the system and practice using the system with a set of data sets from Phase 2 of the MALIBO project. This practice demonstration only used the data which were not included for the Phase 3 study and had the option of viewing the cases with or without ML support, from the 45 cases in the hold-out Phase 2 validation. There were several live on-line training events provided by an applications specialist from 3Dnet™ medical. In addition, on the day of the 1st round read, a computer scientist or a scribe was assigned to set up the 3Dnet™ system and refresh the readers on how to use the system to display and overlay the ML images as a colour-encoded heatmap onto the T2w image. The computer scientist also showed the radiologists how to find the useful image sequences, allocate multiple images together on the screen, adjust image contrast and measure tumour size. Once radiologists were sure how to use the system, the reads and ML output evaluation were carried out. This demonstration only used the data which were not included for Phase 3 study.
Six- or eight-view windows (see Figure 19) was suggested to use the display MRI images depending on the total number of image sequence availability. Radiologists were advised to put the images in the order of T2w, DWI and ADC on the top row (see Figure 19) and lay ML image (if this is the case with ML support), dedicated LVR and dedicated brain scans at the bottom row. Radiologists could allocate more windows if they felt it was necessary.
FIGURE 19.
An example for read window layout (case number STC010-ML). (a) T2w image; (b) DWI image; (c) ADC map; (d) RF detection result overlaid on T2w image; (e) dedicated LVR image; (f) dedicated brain image.
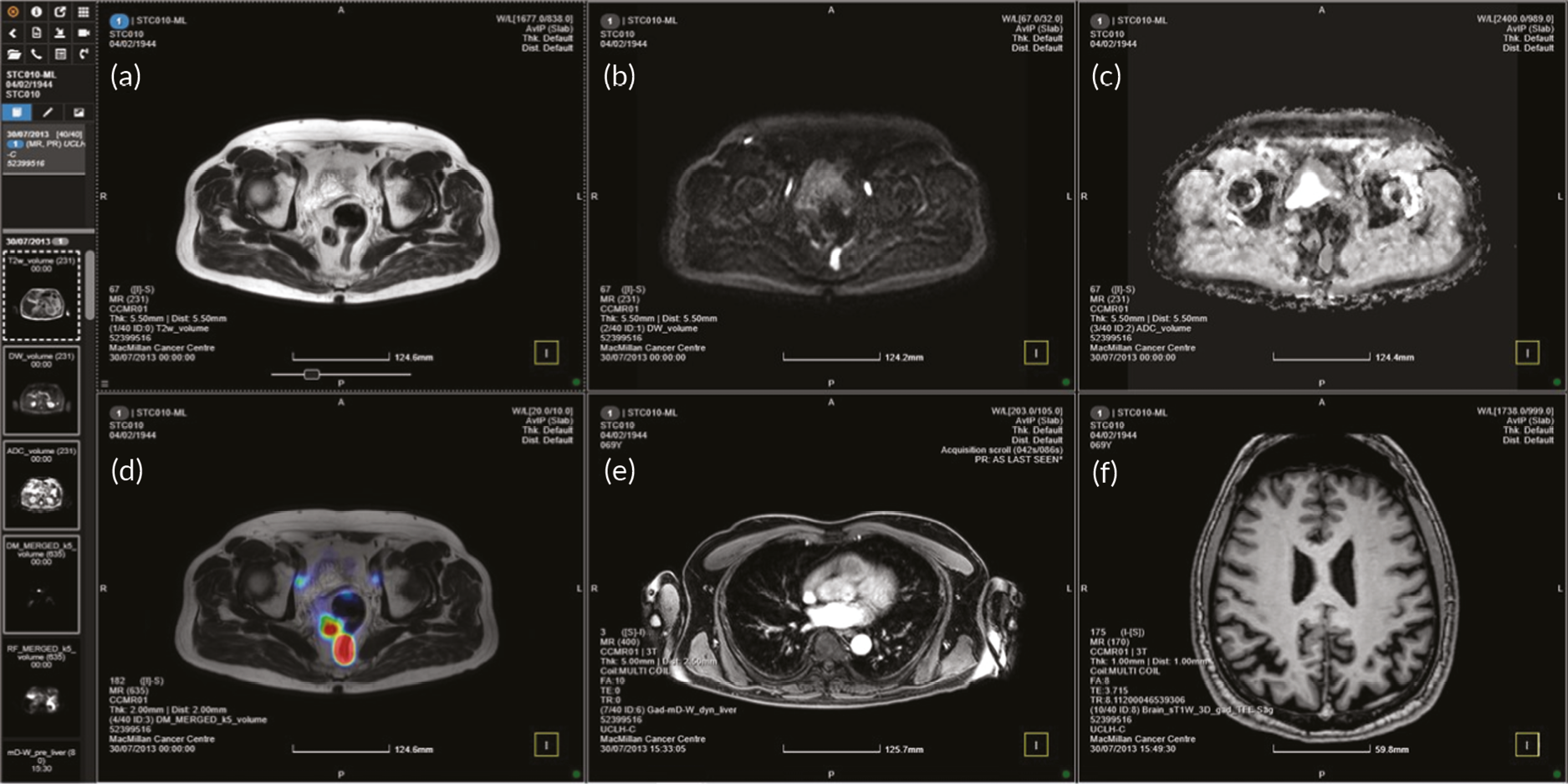
The ML output images were overlaid onto the WB-MRI T2w scans in the form of a threshold, coloured probability map or heatmap (see Figure 19D). The overlay threshold (suggested 65%) was decided by the radiologists and computing scientists in consensus and this reflected the extent of the underlying T2w imaging and the extent of the ML heatmap; this could be easily adjusted by the reader at any time. Following this decision, it was at the radiologists’ discretion to make a call on a lesion’s true or false appearance and characterisation. Window settings could be adjusted for all sequences, including the ML output volume.
To achieve correct sequential allocation of cases and secure blinding of the reader, we created two folders on 3Dnet™ system for each case and these two folders were identical except the ML folder has one additional ML result to support the radiologists for the report. Radiologists were able to access only the cases for a specific reading round. No other cases could be accessed, thus ensuring that all the reads in Phase 3 were blinded.
Reading method
Whole-body magnetic resonance imaging scans were reported by 25 independent radiologists (including 18 WB-experienced radiologists and 7 inexperienced in WB-MRI) who were blinded to the results of the other imaging and clinical information apart from know whether the primary tumour was lung or colon, as was the case in the source study. All radiologists took part in the 1st and 2nd round reads, but only eight (six WB-experienced and two WB-inexperienced radiologists) participated in the 3rd round (intrarater) read.
The detection of primary and metastatic lesions, incorporating ML support and radiologist’s expertise, was recorded using a proforma similar to the one used in the STREAMLINE studies,8 but adapted for the MALIBO study to account for the incorporation of ML (see Appendices 5 and 6).
After MRI images were set up on the 3Dnet™ system as shown in Figure 19 (or Appendix 4), beginning and ending RT and tumour stage and were recorded. T2w, DWI and ADC image qualities were evaluated. Readers were asked to detect a primary tumour based on all available information, including ML output if available. If the primary tumour was found, then four confidence level, from low (1) to high (4) was asked to be given by the reader to decide the confidence of the tumour detection. There after the maximum dimension of the tumour was measured and recorded. Cancer stage was also evaluated by the reader. For lung cases, regional nodal status was recorded. The detection of non-skeletal and skeletal metastatic sites was documented and incidental findings were also recorded in the case report form (see Appendix 5 for STC case and Appendix 6 for STL case).
In order to ensure parity in the time of reads, for the cases in which ML heatmaps were available, the readers were asked to undertake their clinical read together with the heatmap and record the sites of disease. Once this was completed the timer was stopped. The reader was then asked to review the sites of ML detection and assign one of four levels of detection related purely to the ML output. Four levels of probability were recorded by visual inspection by readers, from low (1) to high (4), to indicate the perceived probability of the ML for lesion detection by the reader – although the reader may have over-ruled the ML output in the clinical read.
A scribe was assigned to assist the radiologists for the report and completion of the CRF forms for all reading rounds. The scribe ensured appropriate knowledge of the PACS system prior to the worklist beginning, independently timed the clinical read and ensured that CRF forms were fully completed. The completed CRF forms were copied, scanned and sent to University College London (UCL) Cancer Trial Unit for input the data into the study database.
Statistical analysis
Encrypted data were provided to the study statistician by the UCL Cancer Trial Unit over a secure network and were imported into Statistical Analysis System v9.4 for analysis.
Primary analysis to investigate for a difference in specificity rates between reads with ML assistance compared to those without was carried out using McNemar’s test for paired proportions. Results are presented as a difference in proportions between the two arms with the corresponding 95% confidence interval (CI). Due to the nature of the results a two-sided test had to be carried out instead of the one-sided test anticipated. Significance testing was based on the binomial distribution of the discordant pairs between the two assessment groups (ML and no-ML).
Full details of this methodology, including formulae, can be seen in section A7 of the statistical analysis plan (SAP, Appendix 7). In some cases, the analysis performed differed slightly than originally specified:
-
Following primary analysis, it was clear that a two-sided test rather than a one-sided test was required throughout. The SAP does allow for this eventuality but for a sacrifice of power in the design.
-
The SAP stated that frequencies of confidence scores will be visualised using a bar charts. This will no longer be the case as the medium is not suitable for displaying the outcome data. The corresponding 4 × 4 frequency tables will be included in the results.
Results
Review of data
One hundred and ninety-three patient scans were initially assigned to Phase 3 of the MALIBO study (see Figure 20). Of these, one was removed due to technical problems prior to allocation to readers. Of 192 allocated to readers, 188 were included for the primary analysis, as 4 further scans could not be evaluated due to a lack of assessable ML images. This left 138 sets with negative reference standards (no presence of detectable metastases) and 50 with positive reference standards (presence of detectable metastases) (Figure 20).
FIGURE 20.
Flowchart of reference standard read data allocated for Phase 3 testing.
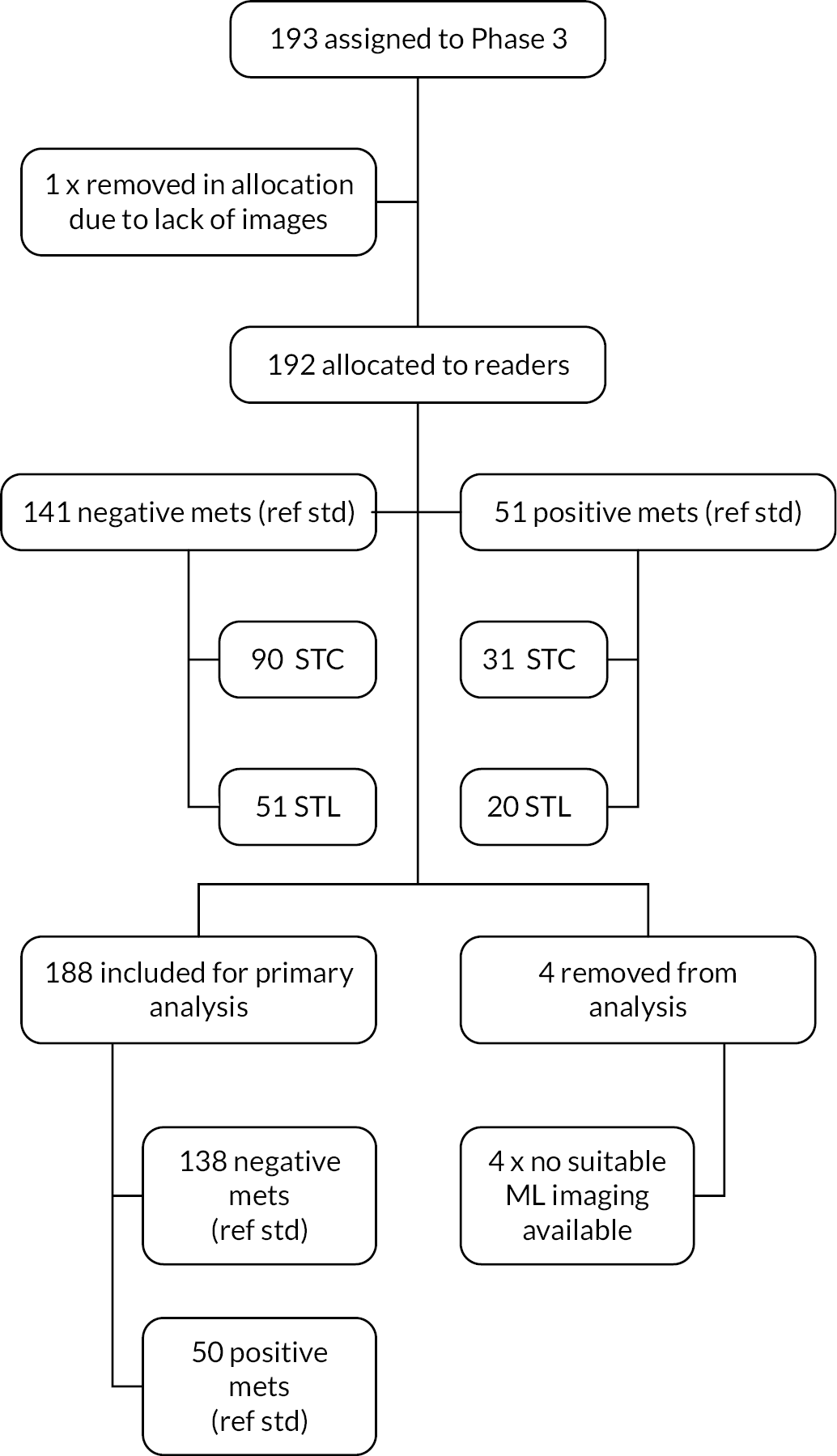
Primary analysis: specificity per patient
The primary outcome measure for this trial was the per-patient specificity amongst experienced readers of WB-MRI with ML algorithm support, compared to the standard radiology read (WB-MRI without ML algorithm support) against the reference standard established in the STREAMLINE study. Specificity is defined as the proportion of cases with negative reference standard, which has been correctly classified as negative by the reading radiologist, based on WB-MRI with or without ML algorithm support.
Table 13 displays the overall frequency of whether a patient had at least one metastatic tumour detected in MALIBO reads in comparison to the reference standard amongst experienced readers and was repeated both with and without the assistance of ML, respectively. The corresponding data detailing negative reference standards were then carried over to Table 14 in order to investigate specificity. Using ML, readers were not able to detect metastatic tumours in 119 of the 138 patients with negative reference standards. This translates to a specificity rate of 86.2%. Without ML, 121 of the 138 patients with negative reference standards were unable to have metastatic tumours detected, a specificity rate of 87.7%. The corresponding difference in proportions when using ML is −1.5% (95% CI −6.4% to 3.5%) with the derived p-value (0.387) indicating that there is no evidence to reject a null hypothesis of no difference in specificity rates between arms.
| Reference standard | MALIBO | ||
|---|---|---|---|
| Frequency: n (%) | Negative | Positive | Total |
| (a) Without ML | |||
| Negative | 121 (64.4) | 17 (9.0) | 138 (73.4) |
| Positive | 15 (8.0) | 35 (18.6) | 50 (26.6) |
| Total | 136 (72.3) | 52 (27.7) | 188 (100.0) |
| (b) With ML | |||
| Negative | 119 (63.3) | 19 (10.1) | 138 (73.4) |
| Positive | 17 (9.0) | 33 (17.6) | 50 (26.6) |
| Total | 136 (72.3) | 52 (27.7) | 188 (100.0) |
| no-ML | ML | ||
|---|---|---|---|
| Frequency: n (%) | Negative | Positive | Total |
| Negative | 114 (82.6) | 7 (5.1) | 121 (87.7) |
| Positive | 5 (3.6) | 12 (8.7) | 17 (12.3) |
| Total | 119 (86.2) | 19 (13.8) | 138 (100.0) |
Secondary analysis: sensitivity per patient
Table 15 shows whether metastatic tumours were detected in patients with positive reference standards when using ML or no-ML. Of the 50 patients with metastases present within the reference standard, 33 were able to be detected by readers when using ML. This translates to a sensitivity rate of 66.0%. Without ML, 35 patients were established to have metastatic tumours, providing a sensitivity rate of 70.0%. The corresponding difference in proportions when using ML is −4.0% (95% CI −13.5% to 5.5%) with the derived p-value (0.344), indicating that there is no evidence to reject a null hypothesis of no difference in sensitivity rates between arms.
| no-ML | ML | ||
|---|---|---|---|
| Frequency: n (%) | Negative | Positive | Total |
| Negative | 13 (26.0) | 2 (4.0) | 15 (30.0) |
| Positive | 4 (8.0) | 31 (62.0) | 35 (70.0) |
| Total | 17 (34.0) | 33 (66.0) | 50 (100.0) |
Secondary analysis: specificity and sensitivity per site
A breakdown of the specificity and sensitivity rates per site of lesion can be found in Tables 16 and 17, respectively. Generally, specificity was not affected based on usage of the ML algorithm with difference in proportions ranging from 1.6% down to −0.5%. In all cases, per-site specificity remained above 95%.
| Site | n | Specificity | Difference in proportions | |||
|---|---|---|---|---|---|---|
| ML (%) | no-ML (%) | ∆ (%) | LCI | UCI | ||
| LVR | 165 | 98.2 | 98.2 | 0.0 | −2.4 | 2.4 |
| Lung | 178 | 95.5 | 95.5 | 0.0 | −2.7 | 2.7 |
| Adrenal | 184 | 98.4 | 96.7 | 1.6 | −0.2 | 3.5 |
| Kidney | 187 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Brain | 182 | 98.9 | 98.9 | 0.0 | 0.0 | 0.0 |
| Pleura | 187 | 97.3 | 97.9 | −0.5 | −2.9 | 1.8 |
| SPLN | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PNCR | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Peritoneum | 185 | 97.8 | 98.4 | −0.5 | −1.6 | 0.5 |
| Bowel | 188 | 99.5 | 99.5 | 0.0 | −1.5 | 1.5 |
| Chest | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PLVS (non-skeletal) | 186 | 99.5 | 100.0 | −0.5 | −1.6 | 0.5 |
| Skull | 187 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Cervix | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Thorax | 184 | 99.5 | 100.0 | −0.5 | −1.6 | 0.5 |
| Lumbar | 184 | 99.5 | 98.9 | 0.5 | −0.5 | 1.6 |
| Sternum | 187 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PLVS (skeletal) | 186 | 99.5 | 100.0 | −0.5 | −1.6 | 0.5 |
| Clavicle | N/A | N/A | N/A | |||
| Ribs | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Other skeletal | 188 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Site | n | Sensitivity | Difference in proportions | |||
|---|---|---|---|---|---|---|
| ML (%) | no-ML (%) | ∆ (%) | LCI | UCI | ||
| LVR | 23 | 60.9 | 69.6 | −8.7 | −20.2 | 2.8 |
| Lung | 10 | 10.0 | 0.0 | 10.0 | −8.6 | 28.6 |
| Adrenal | 4 | 50.0 | 50.0 | 0.0 | 0.0 | 0.0 |
| Kidney | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Brain | 6 | 66.7 | 50.0 | 16.7 | −13.2 | 46.5 |
| Pleura | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Peritoneum | 3 | 0.0 | 33.3 | −33.3 | −86.7 | 20.0 |
| PLVS | 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Skull | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Thorax | 4 | 25.0 | 0.0 | 25.0 | −17.4 | 67.4 |
| Lumbar | 4 | 25.0 | 0.0 | 25.0 | −17.4 | 67.4 |
| Sternum | 1 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PLVS skeletal | 2 | 0.0 | 50.0 | −50.0 | −119 | 19.3 |
Investigating per-site sensitivity was hindered by the lack of positive cases within the reference standards with only two sites (LVR and lung) having 10 cases or more to relate to. With 23 positive cases (see Table 17), LVR produced a difference of −8.7% (95% CI −20.2 to 2.8) in metastatic tumour detection when using ML. The results for lung cancer provided very low sensitivity rates with 10.0% (95% CI 0.5 to 45.9) in the machine-learning arm and 0% (95% CI 0.0 to 34.5) without ML. It should be noted that these figures are based on a small sample of positive lung cases such that the upper CIs are hitting more moderate values of 45.9% (ML) and 34.5% (No-ML), respectively.
Secondary analysis: analysis for inexperienced readers
Investigating per-patient and per-site specificity and sensitivity for less experienced readers produced similar results. Fifty-three reads were assessed amongst 7 readers; 38 with negative reference standards, 15 with positive standards. For specificity, readers when using both ML and no-ML were able to not detect metastases in 29 of the 38 read sets with negative reference standards. This translates to a specificity rate of 76.3% (95% CI 59.4% to 88.0%) with a difference of 0.0% (95% CI −15.0% to 15.0%) (Table 18). For sensitivity, readers using ML were able to detect metastases in 11 of the 15 read sets with positive reference standards, a sensitivity rate of 73.3% (95% CI 44.8% to 91.1%). Without ML this figure decreases to 9 out of 15, a sensitivity rate of 60.0% (95% CI 32.9% to 82.5%). The corresponding difference in sensitivity rates when using ML is 13.3% (95% CI −7.9% to 34.5%) (Table 19).
| no-ML | ML | ||
|---|---|---|---|
| Frequency: n (%) | Negative | Positive | Total |
| Negative | 23 (60.5) | 6 (15.8) | 29 (76.3) |
| Positive | 6 (15.8) | 3 (7.9) | 9 (23.7) |
| Total | 29 (76.3) | 9 (23.7) | 38 (100.0) |
| no-ML | ML | ||
|---|---|---|---|
| Frequency: n (%) | Negative | Positive | Total |
| Negative | 3 (20.0) | 3 (20.0) | 6 (40.0) |
| Positive | 1 (6.7) | 8 (53.3) | 9 (60.0) |
| Total | 4 (26.7) | 11 (73.3) | 15 (100.0) |
A breakdown of the specificity and sensitivity rates per site of lesion for inexperienced readers can be found in Tables 20 and 21. The output is again generally similar to that found in the corresponding table for experienced readers (see Tables 16 and 17) with ML and no-ML producing similar results with small proportions of difference. The specificity values are slightly smaller in some cases with values heading down towards 90% (in comparison the lowest specificity in the experienced readers was above 95%). It should be noted that due to the low numbers of positive reference standards available in this subset of data that no realistic inferences can be made regarding difference in proportions in sensitivity.
| Site | n | Specificity | Difference in proportions | |||
|---|---|---|---|---|---|---|
| ML | no-ML | ∆ | LCI | UCI | ||
| LVR | 46 | 95.7 | 95.7 | 0.0 | −8.5 | 8.5 |
| Lung | 51 | 96.1 | 90.2 | 5.9 | −2.6 | 14.3 |
| Adrenal | 52 | 98.1 | 100.0 | −1.9 | −5.7 | 1.8 |
| Kidney | 52 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Brain | 50 | 98.0 | 100.0 | −2.0 | −5.9 | 1.9 |
| Pleura | 52 | 98.1 | 94.2 | 3.8 | −3.6 | 11.3 |
| SPLN | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PNCR | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Peritoneum | 52 | 98.1 | 100.0 | −1.9 | −5.7 | 1.8 |
| Bowel | 53 | 96.2 | 100.0 | −3.8 | −8.9 | 1.4 |
| Chest | 53 | 98.1 | 100.0 | −1.9 | −5.6 | 1.8 |
| PLVS non-skeletal | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Skull | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Cervix | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Thorax | 53 | 100.0 | 96.2 | 3.8 | −1.4 | 8.9 |
| Lumbar | 53 | 100.0 | 98.1 | 1.9 | −1.8 | 5.6 |
| Sternum | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| PLVS skeletal | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Clavicle | N/A | N/A | N/A | |||
| Ribs | 53 | 98.1 | 100.0 | −1.9 | −5.6 | 1.8 |
| Other skeletal | 53 | 100.0 | 100.0 | 0.0 | 0.0 | 0.0 |
| Site | n | Specificity | Difference in proportions | |||
|---|---|---|---|---|---|---|
| ML | no-ML | ∆ | LCI | UCI | ||
| LVR | 7 | 71.4 | 71.4 | 0.0 | 0.0 | 0.0 |
| Brain | 3 | 66.7 | 100.0 | −33.3 | −86.7 | 20.0 |
| Lung | 2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Adrenal | 1 | 100.0 | 0.0 | 100.0 | 100 | 100 |
| Kidney | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Pleura | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Peritoneum | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Secondary analysis: time to complete reads
Combining experienced and inexperienced reads across both rounds 1 and 2, the overall mean (SD) RT of WB-MRI with ML algorithm support is 560 (260) seconds. Without using ML algorithm support, the time increases slightly to 595 (610) seconds (Table 22). Thus, using ML mean RT is lowered by an average of 35 seconds (95% CI −60 to 140), an average percentage reduction of 6.2% (95% CI −10.0% to 22.8%). Table 22 contains additional summary statistics and also breaks these results down by reader ability (experienced/inexperienced) and read order (rounds 1 and 2). Round 2 read times also dropped, regardless of ML assistance, read type (Table 23 and 24), or reader experience by an average of 226 seconds (95% CI 147 to 304) or 31.8%, (95% CI 20.8% to 42.8%).
Comparing experienced read times to inexperienced read times we see that experienced readers complete their reads approximately 2 minutes faster than their inexperienced counterparts, however, the difference in time deduction depends on the read round. On average, experienced readers completed their reads 100 seconds (95% CI −75 to 274) faster (or 12.6%, 95% CI −9.6% to 34.8%) than their inexperienced counterparts for round 1 reads, and 81 seconds (95% CI 10 to 152) faster (or 14.8%, 95% CI 1.2% to 27.8%) for round 2.
| Read round | Without ML | With ML | |||||
|---|---|---|---|---|---|---|---|
| n | Mean (SD) | Median (IQR) | n | Mean (SD) | Median (IQR) | ||
| Experienced readers | All reads | 188 | 595 (610) | 480 (300–720) | 188 | 560 (260) | 540 (360–720) |
| Round 1 | 92 | 715 (824) | 600 (360–780) | 96 | 663 (259) | 600 (450–810) | |
| Round 2 | 96 | 481 (236) | 420 (300–600) | 92 | 453 (216) | 390 (300–570) | |
| Round 3 | 21 | 454 (206) | 420 (300–540) | 20 | 411 (156) | 390 (300–510) | |
| Inexperienced readers | All reads | 53 | 691 (412) | 600 (420–900) | 53 | 645 (329) | 600 (360–840) |
| Round 1 | 26 | 842 (476) | 630 (540–1020) | 27 | 736 (382) | 660 (360–840) | |
| Round 2 | 27 | 544 (275) | 540 (300–660) | 26 | 552 (235) | 480 (360–720) | |
| Round 3 | 7 | 351 (112) | 360 (240–480) | 6 | 410 (158) | 360 (300–420) | |
| Colon | Without ML | With ML | |||||
|---|---|---|---|---|---|---|---|
| Read round | n | Mean (SD) | Median (IQR) | n | Mean (SD) | Median (IQR) | |
| Experienced readers | All reads | 117 | 597 (357) | 540 (360–780) | 117 | 560 (257) | 540 (360–720) |
| Round 1 | 58 | 703 (417) | 600 (360–840) | 59 | 655 (249) | 600 (420–840) | |
| Round 2 | 59 | 492 (249) | 480 (300–600) | 58 | 463 (228) | 390 (300–660) | |
| Round 3 | 12 | 455 (161) | 450 (330–570) | 10 | 432 (162) | 420 (300–600) | |
| Inexperienced readers | All reads | 33 | 758 (481) | 600 (360–1020) | 33 | 705 (368) | 660 (360–840) |
| Round 1 | 13 | 1057 (559) | 960 (600–1320) | 20 | 756 (423) | 600 (420–840) | |
| Round 2 | 20 | 564 (303) | 540 (300–780) | 13 | 626 (259) | 660 (420–720) | |
| Round 3 | 5 | 396 (100) | 420 (360–480) | 3 | 460 (227) | 360 (300–720) | |
| Lung | Without ML | With ML | |||||
|---|---|---|---|---|---|---|---|
| Read round | n | Mean (SD) | Median (IQR) | n | Mean (SD) | Median (IQR) | |
| Experienced readers | All reads | 71 | 593 (885) | 420 (300–720) | 71 | 559 (268) | 540 (360–720) |
| Round 1 | 34 | 736 (1253) | 540 (360–720) | 37 | 675 (276) | 660 (480–780) | |
| Round 2 | 37 | 462 (213) | 420 (300–660) | 34 | 434 (195) | 390 (300–540) | |
| Round 3 | 9 | 453 (265) | 420 (300–480) | 10 | 390 (156) | 330 (300–420) | |
| Inexperienced readers | All reads | 20 | 579 (229) | 570 (450–630) | 20 | 546 (228) | 510 (360–660) |
| Round 1 | 13 | 628 (243) | 600 (480–660) | 7 | 677 (249) | 600 (540–840) | |
| Round 2 | 7 | 489 (181) | 540 (360–600) | 13 | 475 (189) | 420 (360–600) | |
| Round 3 | 2 | 240 (0) | 240 (240–240) | 3 | 360 (60) | 360 (300–420) | |
Model investigating difference in ML and non-ML RT in seconds and percentage adjusted for fixed-effects read round (when ML is applied, 1st or 2nd round) and Read package (lung or colon cancer). Additional clustering effect applied for reader experience.
Value estimates relate to the overall estimated effect adjusted for all other covariates within the model. Intercept refers to value at reference standards round 1 and lung packages.
To investigate ML versus non-ML difference in read time a regression analysis was carried out using paired data comparing ML against their respecting non-ML read. The regression model was adjusted for fixed-effect co-variates; read tumour type (lung and colon) and read round (whether ML was used in the 1st or 2nd round of reading). A clustering effect for reader experience was also included. Assumptions for regression modelling held and residuals were found to be normally distributed. Table 25 contains regression estimates (in seconds and as a percentage) for estimated effects of read round and read package when investigating paired ML versus non-ML difference in read time. While package type was not found to influence difference in read time, the output indicated read round to have a significant (p = 0.0281) effect. The estimated effect on ML/non-ML difference between rounds 1 and 2 is −486 seconds (95% CI −760 to −213). Post hoc testing to estimate the subsequent effect on read time when using ML at round 2 is −286 (95% CI −370 to −201) seconds. Similar post hoc testing of percentage difference estimated ML to reduce round 2 read times by −11% (95% CI −61% to 26%).
| Effect | Value | Effect estimate (95% CI) | |
|---|---|---|---|
| Secs | Intercept | 226 (−250 to 702) | |
| ReadRound | Round 2 | −486 (−760 to 213)** | |
| Package | Colon | −51 (−626 to 524) | |
| %age | Intercept | 64% (−32% to 160%)* | |
| ReadRound | Round 2 | −77% (−113% to −41%)** | |
| Package | Colon | −9% (−43% to 25%) |
Secondary analysis: confidence in reads
Tables 26–29 indicate the frequencies each confidence score provided by the reader for ML and non-ML. For experienced readers without ML assistance, 77.1% (815) of primary tumour detection checks resulted in no tumour being found with high confidence (score of 1). For metastatic site checks, this increases to 95.5% (4852). Adding ML assistance, 77.5% (819) of primary tumour detection checks resulted in no tumour being found with high confidence and 95.5% (4851) in relation to metastatic site checks. When looking at tumour detection with high confidence (score of 4), primary tumour detection checks without ML assistance accounted for 14.9% (157) of read scores with 0.7% (34) in metastatic sites. With ML assistance primary tumour detection checks with a score of 4 were recorded 15.6% (165) times with 0.6% (31) in metastatic sites. Looking at experienced readers we can see that 87.4% (924) and 93.9% (4774) confidence scores remained the same in both ML and non-ML reads in primary tumour sites (see Table 26) and metastatic tumour sites (see Table 27), respectively. In comparison, for non-experienced readers 82.9% (247) and 95.2% (1363) confidence scores remained the same in both ML and non-ML reads in primary tumour sites (see Table 28) and metastatic tumour sites respectively (see Table 29).
| Confidence score | With ML | ||||||
|---|---|---|---|---|---|---|---|
| 1 (%) | 2 (%) | 3 (%) | 4 (%) | 999 (%) | Total (%) | ||
| Without ML | 1 (%) | 771 (72.9) | 17 (1.6) | 1 (0.1) | 16 (1.5) | 10 (0.9) | 815 (77.1) |
| 2 (%) | 29 (2.7) | 13 (1.2) | 3 (0.3) | 1 (0.1) | 0 (0.0) | 46 (4.4) | |
| 3 (%) | 7 (0.7) | 4 (0.4) | 10 (0.9) | 18 (1.7) | 0 (0.0) | 39 (3.7) | |
| 4 (%) | 12 (1.1) | 0 (0.0) | 13 (1.2) | 130 (12.3) | 2 (0.2) | 157 (14.9) | |
| Total (%) | 819 (77.5) | 34 (3.2) | 27 (3.6) | 165 (15.6) | 12 (1.1) | 1057 (100.0) | |
| Confidence score | With ML | ||||||
|---|---|---|---|---|---|---|---|
| 1 (%) | 2 (%) | 3 (%) | 4 (%) | 999 (%) | Total (%) | ||
| Without ML | 1 (%) | 4709 (97.7) | 108 (2.1) | 14 (0.3) | 5 (0.1) | 15 (0.4) | 4852 (95.5) |
| 2 (%) | 104 (2.0) | 36 (0.7) | 3 (0.1) | 1 (0.0) | 0 (0.0) | 146 (2.8) | |
| 3 (%) | 10 (0.2) | 6 (0.1) | 8 (0.2) | 4 (0.1) | 0 (0.0) | 28 (0.6) | |
| 4 (%) | 3 (0.1) | 1 (0.0) | 9 (0.2) | 21 (0.4) | 0 (0.0) | 34 (0.7) | |
| 999 (%) | 24 (0.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 24 (0.5) | |
| Total (%) | 4851 (95.5) | 151 (3.0) | 34 (0.7) | 31 (0.6) | 15 (0.3) | 5084 (100.0) | |
| Confidence score | With ML | |||||
|---|---|---|---|---|---|---|
| 1 (%) | 2 (%) | 3 (%) | 4 (%) | Total (%) | ||
| Without ML | 1 (%) | 213 (71.5) | 9 (3.0) | 8 (2.7) | 9 (3.0) | 239 (80.2) |
| 2 (%) | 4 (1.3) | 2 (0.7) | 0 (0.0) | 0 (0.0) | 6 (2.0) | |
| 3 (%) | 6 (2.0) | 0 (0.0) | 1 (0.3) | 6 (2.0) | 13 (4.4) | |
| 4 (%) | 6 (2.0) | 0 (0.0) | 2 (0.7) | 31 (10.4) | 39 (13.1) | |
| 999 (%) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | |
| Total (%) | 230 (77.2) | 11 (3.7) | 11 (3.7) | 46 (15.4) | 298 (100.0) | |
| Confidence score | With ML | |||||
|---|---|---|---|---|---|---|
| 1 (%) | 2 (%) | 3 (%) | 4 (%) | Total (%) | ||
| Without ML | 1 (%) | 1353 (94.5) | 18 (1.3) | 11 (0.8) | 1 (0.1) | 1383 (96.6) |
| 2 (%) | 17 (1.2) | 0 (0.0) | 2 (0.1) | 1 (0.1) | 20 (1.4) | |
| 3 (%) | 8 (0.6) | 3 (0.2) | 1 (0.1) | 1 (0.1) | 13 (0.9) | |
| 4 (%) | 3 (0.2) | 1 (0.1) | 1 (0.1) | 9 (0.6) | 14 (1.0) | |
| 999 (%) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | |
| Total (%) | 1382 (96.6) | 22 (1.5) | 15 (1.0) | 12 (0.8) | 1431 (100.0) | |
The extent at which confidence levels differ between ML and non-ML reads seems to differ when looking at primary tumour sites in comparison to metastatic tumour sites. Twenty-eight (2.6%) and 15 (5.0%) experienced and non-experienced reads respectively jump from high confidence in detection to non-detection (scores of 1 and 4) and vice versa. The corresponding jump from scores representing low confidence in detection and non-detection (scores of 2 and 3) are lower with values of 7 (0.7%) and 0 (0%). In metastatic tumour sites, changes in high confidence (1 and 4) and low confidence scores (2 and 3) are a lot closer. Experienced readers have 8 (0.2%) sets of scores alternating between 1 and 4, and 9 (0.2%) alternating between 2 and 3. Non-experienced readers have 4 (0.3%) sets of scores alternating between 1 and 4, and 5 (0.3%) alternating between 2 and 3.
Secondary analysis: size of tumours
Figure 21 shows scatterplots of the MALIBO recorded tumour size against the reference standard for both colon and lung data sets (Plots A and B). Both plots demonstrate a fairly strong positive correlation along the line where the MALIBO measurement matches the reference measurement (X = Y) indicating that the measurements taken within the MALIBO reads correspond fairly well with those provided in the reference standards.
FIGURE 21.
Tumour measurements (mm) in relation to the reference standard for experienced readers. Plot A: Scatterplot of recorder size in mm against reference standard for colon data. Plot B: Scatterplot of recorder size in mm against reference standard for lung data.
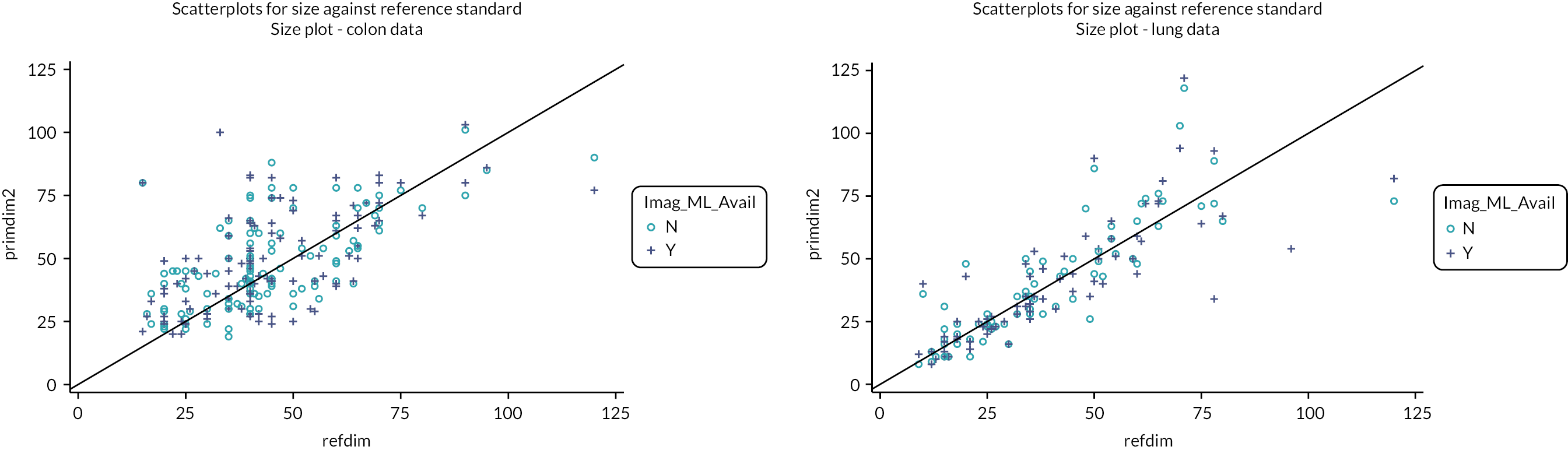
Looking at Figure 21 in more detail we see that in plot A, for both ML and non-ML-assisted reads, a greater proportion of the data points lie above the reference line indicating a mean read difference greater than zero. Plots A and B in Figure 22 reinforce this with the distribution curve of both plots (representing distribution of all the differences in measurement found within in ML and non ML) peaking above zero. We can estimate the mean (SD) difference to be + 3.0 mm (16.3) with ML assistance and + 3.0 mm (14.6) without. Likewise, 95% of the size differences (in mm) with ML assistance were estimated to be in the range (−29.6, 35.5) and (−26.1, 32.2) without. This represents appreciable variability.
FIGURE 22.
Histogram plot. Plot A: Histogram w/distribution curve for difference in mm against reference standard under non-ML. Plot B: Histogram w/distribution curve for difference in mm against reference standard with ML.
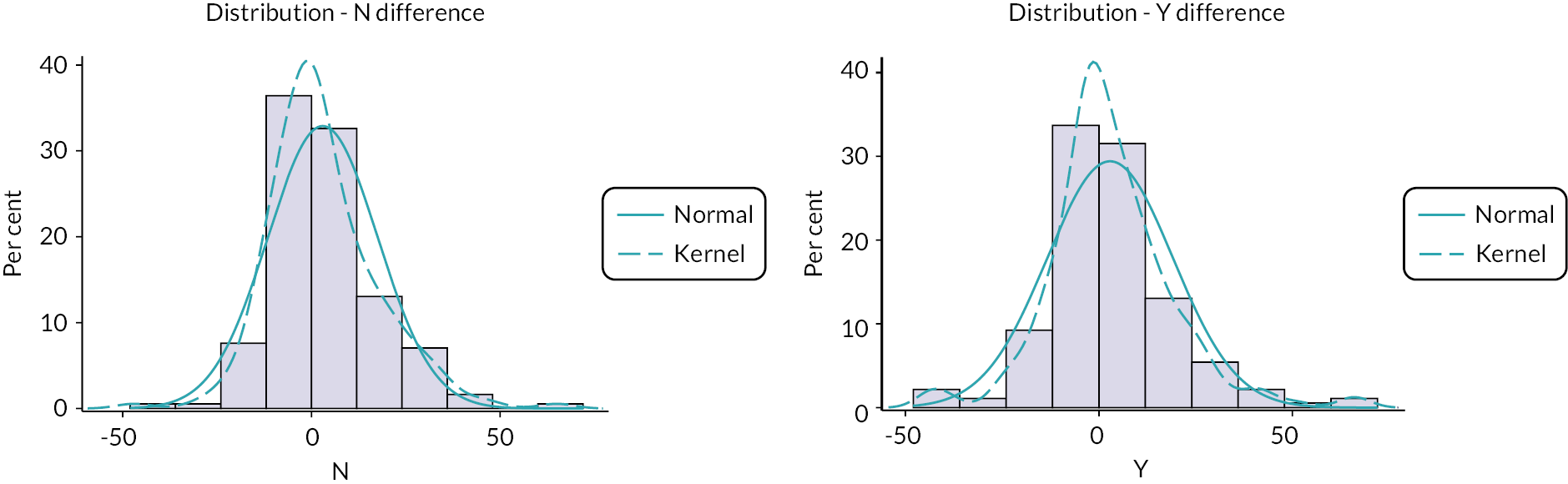
Looking at the size of primary tumours, when investigating mean difference in millimetres in comparison to the reference standard, the mean difference in sizes when using ML is on average 3.0 mm greater than the reference standard (95% CI 0.6 to 5.3). Without ML the mean difference stays at 3.0 mm (95% CI 0.9 to 5.2). Comparing the two groups we see that using ML reduces the mean difference on average by 0.1 mm (95% CI −3.1 to 3.2).
For non-experienced readers the mean (SD) difference in read sizes when compared to the reference standard is +6.8 mm (20.7) with ML assistance and +5.1 mm (19.2) without. Likewise, 95% of the size differences (in mm) with ML assistance were estimated to be in the range (−34.7, 48.2) and (−33.4, 43.5) without. Subsequently, the mean difference in sizes when using ML is on average 6.8 mm greater than the reference standard (95% CI 1.0 to 12.5). Without ML this difference reduces to 5.1 mm (95% CI −0.4 to 10.5). Comparing the two groups we see that using ML increases the mean difference on average by 1.7 mm (95% CI −6.1 to 9.5).
Similar results were found looking at the size difference as a percentage value in relation to the reference standard. For experienced readers, with ML assistance the mean (SD) percentage difference estimated that tumours were measured on average 14.4% (53.9) larger than the reference standard. Without ML this average reduced marginally to 14.3% (51.4). Ninety-five per cent of the size differences (as a percentage) with ML assistance were estimated to be in the range (−93.4, 112.3) and (−88.5, 117.2) without. When comparing the two arms, using ML output resulted in a primary tumour size on average 14.4% greater than the value provided in the reference standard (95% CI 6.6 to 22.3). The corresponding figure without ML assistance was 14.3% (95% CI 6.8 to 21.8). Subsequently, we see that using ML assistance in comparison to not using ML assistance produced an average percentage increase in tumour measurement (in relation to the reference standard) by 0.1% (95% CI −10.7 to 11.0) (Table 30).
| Reader level | Arm | n | Mean | Lower CIa | Upper CIa | SD |
|---|---|---|---|---|---|---|
| (a) Summary statistics of tumour difference values against reference standard (mm) | ||||||
| Experienced | no-ML | 183 | 3.0 | 0.9 | 5.2 | 14.6 |
| ML | 183 | 3.0 | 0.6 | 5.3 | 16.3 | |
| Inexperienced | no-ML | 51 | 5.1 | −0.4 | 10.5 | 19.2 |
| ML | 52 | 6.8 | 1 | 12.5 | 20.7 | |
| (b) Summary statistics of tumour difference values against reference standard (%b) | ||||||
| Experienced | no-ML | 183 | 14.3 | 6.8 | 21.8 | 51.4 |
| ML | 183 | 14.4 | 6.6 | 22.3 | 53.9 | |
| Inexperienced | no-ML | 51 | 23.1 | 4.8 | 41.5 | 65.2 |
| ML | 52 | 28.7 | 8.6 | 48.7 | 72 | |
For non-experienced readers, with ML assistance the mean (SD) percentage difference estimated that tumours were measured on average 28.7% (72.0) larger than the reference standard. Without ML this average reduced to 23.1% (65.2). Ninety-five per cent of the size differences (as a percentage) with ML assistance were estimated to be in the range (−115.3, 172.6) and (−107.3, 153.5) without. When comparing the two arms, using ML output on average increased the primary tumour size by 28.7% (95% CI 8.6 to 48.7) while the corresponding figure without was 23.1% (95% CI 4.8 to 41.5). Subsequently we see that using ML assistance increases the average percentage increase in measurement (compared to the reference standard) by, on average 5.5% (95% CI −21.3 to 32.4) (Table 30).
Secondary analysis: inter- and intrareader analysis
Using the methodology described in the SAP (Appendix 6, Secondary outcome analysis) and based on pairing 93 reads amongst 18 readers; Cohen’s kappa for the interobserver variance amongst experienced readers when using ML was derived as 0.64 (95% CI 0.47 to 0.81). Without machine-learning assistance the interobserver variance was unaffected with a kappa statistic of 0.66 (95% CI 0.47 to 0.81). This can be interpreted as moderate agreement between readers. 122 When investigating the smaller cohort of inexperienced readers (24 reads paired between 7 readers) the kappa statistic when using ML was derived as 0.27 (95% CI −0.08 to 0.64) and as 0.12 (95% CI −0.18 to 0.46) without machine-learning assistance. This can be interpreted as low agreement with ML, but no agreement without.
For intrarater reads, based on a sample of 30 tests, the corresponding kappa statistics when comparing round 3 experienced reads with their counterpart in round 1/2 is 0.61 (95% CI 0.13 to 1.00) with ML and 0.46 (95% CI 0.10 to 0.74) without. With only a sample of 10 tests amongst inexperienced readers, ML had 9 out of 10 round 3 tests match their round 1/2 counterpart. This produced a kappa value of 0.62 (95% CI 0.29 to 1.00). All 10 tests without ML produced the same result in round 3 testing. This provides a kappa value of 1.00 (95% CI N/A). For experienced readers we could potentially see that using ML provides a greater consistency in results but would need more data to confirm. The sample for inexperienced readers was too small to provide a valid comparison but ultimately shows the volatility of testing within a small subsample with a difference in one test result resulting in a kappa reduction of 0.38.
Chapter 7 Discussion
Whole-body magnetic resonance imaging is accurate, efficient and cost-effective for cancer staging. 6,7 However, it is not widely used as a diagnostic or staging tool and this could be due to the perceived or real difficulties faced by the radiologist in reading such complex studies of the whole body, with the need to integrate many different MRI sequences. We proposed to develop a ML algorithm to support radiologists in reading WB-MRI in patients with cancer by automatically detecting and demonstrating suspected cancer lesions using a heat map. The over-arching vision was to develop a ML method to assist radiologists reporting WB-MRI, by providing an efficient and accurate tool for identification of metastatic lesions for concurrent use by radiologists, as a human–AI interaction application, as this could assist clinical translation. Our starting point was to train for detection of normal organs using a healthy volunteer data set. We then trained a model for cancer lesion detection. Finally, we tested the algorithm in an environment that was close to a real clinical environment.
Phase 1: healthy volunteer anatomic labelling study
Phase 1 of the study allowed an exploration of techniques to train an algorithm to identify various normal organs and bones on WB-MRI and to automatically segment the normal anatomy. 14,41 We achieved reasonable Dice overlap for many organs, such as the lungs and LVR, but certain organs had poor Dice overlap, for example the PNCR. This finding was similar to other groups who have worked on MRI organ segmentation. 123,124
Many aspects of Phase 1 ran smoothly, as the DICOM data were very homogeneous in nature, having been acquired on a single scanner at a single centre. In addition, many of the participants were young and able to tolerate the scan with ease, thus ensuring high-quality images in most cases. None of the healthy volunteer scans needed to be omitted due to technical failure. Data preparation was also relatively straightforward in terms of stitching the stations to form complete WB-MRI volumes for the T2w, DWI and ADC sequences, although some challenges will be discussed. Many additional exploratory studies were undertaken with the Phase 1 data, including the development of the Dixon-Fix software for fat-water swap and the work on reverse classification in the absence of GT and domain adaptation. 71 These areas lend themselves to wider use in areas beyond WB-MRI. In addition, some detailed work on the bone marrow was undertaken using the Phase 1 DICOM data, important basic work in understanding differences in the bone marrow appearances by gender and age which will be important in future work on bone marrow disease, such as myeloma or in metastatic bone disease. 41
One of the main challenges that we faced, initially in Phase 1, was unexpected. Registration between the T2w and the DWI sequences was poor. T2w images are acquired in breath-hold, whereas DWI are acquired with gentle breathing. This resulted in significant challenges for registration that, despite many methods being applied and many attempts, we could not overcome. We also identified registration issues between the DWI and ADC maps, which should not be the case as the ADC maps are calculated using the DWI sequence. However, the stitching process can result in some differences between volumes and the registration process following station stitching is not straightforward. Ultimately, for both Phase 1 healthy volunteers and in the patient studies, Phases 2 and 3, we were unable to register the different sequences. This leaves room for some improvement: registration of areas of concern could allow a more nuanced identification of disease. For example, the ML was very good at picking up sites of high signal intensity on the diffusion sequence, but could not always differentiate the areas with T2 shine through, which are bright on ADC map against areas of restricted diffusion, which are dark on the ADC map. Being able to capture the combined signal intensity appearances from multiple sequences is highly likely to reduce the number of incorrect (false-positive) lesion identification.
Phase 2: training for detection of cancer lesions
Following allocation of STREAMLINE cases to Phase 2 (training) and Phase 3 (clinical validation), we trained a model for the detection of cancer lesions. Training was based on expert manual segmentations of disease sites that were confirmed on the consensus reference standard of the STREAMLINE study. This was a time-consuming task, requiring multiple annotators. Occasionally, we found what appeared to be a discrepancy between a site of disease on the images and the consensus reference standard, resulting in uncertainty of the reference standard. However, we complied with the reference standard as the GT, although a review panel was used to evaluate these lesions in order to ensure there were no significant GT errors.
We developed a two-stage method, first using the organ-segmentation model from Phase 1 followed by lesion detection model developed during Phase 2 of the study. This two-stage method allowed us to overcome some limitations that we encountered when using conventional CNN methods alone and we verified the final method on multimodality MRI images from a multicentre relatively large data set. These data were collected from a representative range of district general and teaching hospitals, with all imaging performed and interpreted according to usual local protocols, to increase the generalisability and robustness of our methods for clinical application. However, many challenges were faced with the image data suitability for training (and testing) an ML algorithm. Many of the cases had significant issues, such as lacking part of the DWI, which prevented use in the study. In some cases, the acquisition resulted in some gaps in the coverage of the body. Those cases that were allocated to model training had to be removed as these fundamental problems would have interfered with the training. However, in the clinical test set (allocated as 193), all cases were included except five in which we were unable to successfully convert between DICOM and NIfTI formats and upload to the PACS system failed or in those cases in which ML output failed, due to lack of a complete DWI sequence. These problems could potentially have been overcome if time allowed. However, due to delays in receipt of the DICOM data from the contributing study, we had severe time limitations for the fine-checking of the data prior to statistical allocation, and thus, cases that were incomplete were included in the allocation.
Phase 2 training was brief due to the affected timelines. Despite this, two models were developed with reasonable sensitivity. The quantitative results for the model outputs were tested on the 45 Phase 2 validation cases by plotting RE, PR and Dice curves and the 2 best performing models went forward for clinical validation.
Phase 3: clinical validation of machine-learning support tool during radiology reads
We developed a method to evaluate the diagnostic performance of concurrent ML support for radiologists reading WB-MRI in a clinical setting using a retrospective data set. We assessed the diagnostic performance for the detection of metastatic disease based on standard reads (without ML support) and with concurrent ML support (a combined human–AI interpretation).
Overall, the diagnostic accuracy was only moderate for the detection of metastatic lesions. For experienced readers, per-patient specificity for detection of metastases was 86.2% (WB-MRI + ML) and 87.7% (WB-MRI + SD), (difference −1.5%, 95% CI −6.4% to 3.5%; p = 0.387); per-patient sensitivity was 66.0% (WB-MRI + ML) and 70.0% (WB-MRI + SD) (difference −4.0%, 95% CI −13.5% to 5.5%; p = 0.344). For inexperienced readers (53 reads, 15 with metastases), per-patient specificity was 76.3% in both groups with sensitivities of 73.3% (WB-MRI + ML) and 60.0% (WB-MRI + SD). Per-site specificity remained high within all sites: above 95% (experienced) or 90% (inexperienced). Per-site sensitivity was highly variable due to low number of lesions in each site.
While the results fail to satisfy our initial hypothesis that ML can improve specificity in detecting per-patient metastatic tumours, the data indicate that the ML heatmaps did not hinder the read process and could potentially make reads slightly quicker.
The ML output was tested on a relatively heterogeneous WB-MRI data set (multi-institution and multi-vendor) making the ML output vendor neutral and therefore hopefully more generalisable. The outputs had to be available to the radiologist in a format that was practical and meaningful, and we managed this using the Biotronics3D cloud-based PACS. We organised the scans and ML probability heatmaps on a system that was a familiar platform for radiologists. For cases being read with ML support, radiologists were able to view the heatmaps alongside the native WB-MRI volumes or they could overlay the heatmap on top of the T2 or diffusion sequence. We had the facility to fully blind each read without ML support from the those reads with ML support.
In Phase 3, we evaluated the ML method in an approximation of a real clinical setting with several independent radiologists from NHS hospitals, including many that were experienced in reading WB-MRI as well as a number of radiologists that were inexperienced in reading WB-MRI. The reads all took place in a large NHS radiology reporting room.
The statistical methodologies used for analysis of Phase 3 were traditional diagnostic test accuracy methods with robust evaluation of the index test against the comparator.
The analysis did produce some interesting results that may not have been expected. Investigating per-site sensitivity for lung metastases, of the 10 available reference reads, only 1 case with lung metastases was picked up by the reader using ML with 0 detected without (a sensitivity rate of 10% and 0%, respectively). The failure to detect lung metastases can most likely be attributed to the underlying imaging technique which is not sensitive for detection of small lung nodules. However, detection of the primary lung masses was very high.
One surprising result was the lower intrarater agreement compared to the inter-rater agreement. While we could attribute some of this to the comparatively smaller sample of data it does not explain why the value of kappa is lower in both ML and non-ML. It is likely that the 3rd round of reads incurred a study-specific effect where readers were just keen to complete the reads at a potential cost of accuracy.
The overall mean times of reads were also interesting. These were lower than anticipated for WB-MRI reads. We can speculate on the possible reasons for this: it may be that the preparation of the case with the scribe, who also asks focused and directed questions to the reader concerning particular aspects of the scan, results in an efficient system for reading the scan; the stitched volumes for T2w, DWI and ADC may have meant that some other unstitched sequences were not fully reviewed, as the available number of unprocessed sequences in many cases was very large and cumbersome to find any particular desired sequence for review, with each contributing site having slightly different sequence naming conventions and ordering of the sequences. The presence of the scribe may have engendered a consideration to work quickly, particular as the readers knew that they were being timed. In addition, the knowledge that the report was not going to affect patient care may also have an effect. The speed of reporting may have had an effect on the diagnostic accuracy, but it is not possible to test for this. However, the readers were not ‘rushed’ by the time available for reads, as each reader was booked for reading sessions for either a full day for a complete reading round (16 cases for experienced readers) or a half day for a half ready round (8 cases for experienced readers), allowing for 30 minutes per read. As each read was significantly shorter than the planned allowed time, the available time for the reads was clearly not a factor in the relatively short time used for each read.
We found slightly shorter mean RTs using ML of approximately 6% for both experienced and inexperienced readers. The limited time reduction is unlikely to affect daily practice. However, importantly, the use of the ML output did not make RTs longer. This was the first time that all readers included in this study had used ML output in WB-MRI and the need to check appearances on the ML heatmaps might have slowed down the process of radiology reading.
Machine learning in whole-body oncology results in context
Machine-learning methods have been extensively applied in radiology for lesion segmentation and classification, patient risk stratification or patient outcome prediction based on radiological images with different modalities. However, fewer studies have undertaken clinical evaluation of ML in WB-MRI. ML has been applied to compare segments of WB-MRI in chronic non-bacterial osteitis pre and post treatment,125 with good sensitivity but low specificity of 33% for detection of treatment response. A study in evolutive lymphoma and residual masses in WB-MRI included methods for automatic segmentation of the lesions, their localisation, their enumeration and the generation of the parametric ADC map. It was found that combining functional, anatomical and morphological features, using a deep learning CNN method, resulted in a very high accuracy for classifying residual masses of 98.5%. No publication has been identified concerning ML for detection of metastases in WB-MRI.
There are few publications related to improving registration for multimodal WB-MRI. Ceranka et al. evaluated four different strategies for alignment of DWI and T1w images. 126 The best method was found to be a two-step process with groupwise mosaicking of ADC stations followed by registration to the T1w image. However, this remains a relatively unexplored area, perhaps due to many challenges.
Change in RT when using ML or artificial intelligence (AI) outputs has been evaluated in other studies, although not to our knowledge in reading WB-MRI. A decrease in RT of 11% has been reported in the detection of lung nodules, using a deep learning-based computer-aided diagnosis (DL-CAD) system in patients with suspected lung cancer. 127 In a recent study evaluating AI support in breast tomosynthesis, the use of DL-CAD reading support in breast tomosynthesis resulted in a mean decrease in RT from 41 to 36 seconds,128 a mean decrease of 11%. 129 Other studies in breast tomosynthesis have reported decreases of between 14% and 52.7%. 130,131 A DL-CAD system in prostate MRI reduced RT by 21%, from a median of 103 to 81 seconds. 132 ML methods in the detection of intracranial haemorrhage on CT have demonstrated improved diagnostic performance with a reduction in RT from 68 to 43 seconds.
Strengths and limitations
There are several strengths in this study.
-
We developed and employed state-of-the-art ML methods to attempt to improve the diagnostic performance of WB-MRI. To the best of our knowledge, this is the first to apply ML methods for the detection of lung or colon cancer using WB-MRI. The WB-MRI scans were obtained from a prospective study with a confirmed reference standard for sites of disease. The scans were acquired from multiple sites and manufacturers, with multimodality images and different number of image scan sequences.
-
A relatively large number of independent radiologists took part in the study, this makes this study robust comparing with the study if there is only a small number of radiologists, although had time allowed, it would have been beneficial for each reader to have read a larger portion of the scans. The use of a scribe for CRF filling ensured some homogeneity of reading methods and ensured that readers would have technical support for the PACS tool; the scribe also ensured an external way of measuring RT that was done in the same way for each reader.
-
We adopted an efficient reading platform which is cloud-based. This has several advantages: we were able to efficiently transfer the scans from the STREAMLINE study to the dedicated MALIBO server; within this we could allocate cases into separate worklists which included cases for Phase 2 reviews of ML outputs, which were also used for online training of the readers, who could join from anywhere with an internet connection. We were able to create two copies of each scan, one with and one without ML output, and could create dedicated worklists for individual readers to ensure masking of cases. Although readers could have undertaken the reads at any location, we decided to do this within a normal radiology reporting room, in an attempt at making the environment as similar as possible to daily clinical work.
-
The design and statistical methods employed for the final clinical study were based on classical diagnostic test accuracy studies to try to ensure robust testing methodologies. The statistical plan for the model development was more difficult to plan as we were uncertain at the time of writing the protocol how the model development would progress. However, a computational evaluation of the model output on the Phase 2 hold-out validation set was used to determine the best eventual model candidate to take forward to clinical testing.
There were many obstacles to overcome in the study.
-
Data availability
Data availability for Phases 2 and 3 from the STREAMLINE study was delayed, due to unexpected delays in completing the study, resulting in significant problems with the MALIBO study timelines. In hindsight, it is now clear that it is best not to plan a ML model development study until all the data are fully available for a particular project because without the data it is impossible to proceed. In the face of this difficulty, the team worked on other developments using the Phase 1 data and had a small portion of the STREAMLINE data to commence work on segmentation and work on image registration tasks. Due to time constraints, we were unable to spend as much time as wished to further develop and test the model at Phase 2. There were regular false-positive detections of normal anatomy (e.g. the uterus, prostate gland – which may have been mis-identified as many lesions were rectal cancers) and these are areas for future improvements.
-
Data preparedness
One of the biggest challenges was the wide variability of the WB-MRI scans, which were acquired from 16 centres as part of a multicentre prospective study. There was a variation of scan vendor at the 16 sites with different software versions and slight differences in acquisition parameters. These variations in scan acquisition and protocol resulted in significant challenges for ML development. However, the challenge for ML development with this heterogeneous input data may also be considered a strength of the study, in that the method developed is working towards a generic and pragmatic ML output. The range of WB-MRI appearances was also a challenge for individual radiology readers, as they had a number of cases from different scanner types and slightly different protocols at each reading round, unlike a typical reporting session whereby scans are typically from a single centre with much more homogeneous acquisition parameters which become familiar to the site radiologist. The MRI images obtained from 16 different hospitals demonstrate significant variation in image quality. Some had poor ADC maps, while others have fat water swap artefacts. It is challenging to correct these artefacts as it requires robust algorithms to adapt to different images. Poor quality of DWI and ADC maps can lead to incorrect decision for both human reader and ML methods and in some of these cases, ML outputs were not successful. There were also significant challenges to register T2w, ADC and DWI images. We have tested various image registration methods, including both linear and non-linear methods and we found it is difficult to realise robust image registration with reasonable accuracy. Ultimately, we did not succeed in registration of the different sequences.
-
Reference standard
The reference standard for metastatic lesions was based on an expert consensus panel at 1 year following patient recruitment into the source study. This is an accepted form of reference standard but does leave room for opinion, in the absence of histology for every site of suspected disease. However, this particular limitation is not one that can be readily overcome as it is not possible nor ethical to biopsy every site of suspected metastatic disease for reference standard confirmation.
-
Reader-associated factors
As there were 18 experienced readers, there existed a risk that variance within reader interpretation could inadvertently effect results. To counter this, all readers were randomised a similar proportion of reads based on type (lung and colon), presence of patient metastases (negative and positive reference values) and site of read. Likewise, the paired analysis to derive Cohen’s kappa for inter-reader agreement was stratified in the same way. Due to time and resource constraints it was not feasible to establish viable estimates of agreement between all 18 readers (this would result in 153 separate pairs); however, the inter-rater analysis was able to establish an overall consensus amongst experienced readers. The inter-reader agreement statistic of > 0.6 for both ML and no-ML suggests that despite the large cohort of readers used, the consistency of assessment is fairly strong throughout, thus validating the results produced in the main cohort of experienced readers.
Extending this to the inexperienced reader cohort the two main issues are the number of readers (and reads) in this subgroup and the lack of overall agreement in assessments. To an extent this was expected due to the smaller sample of data and the greater likelihood of variance in the range of abilities in the less experienced inexperienced readers.
The intrarater agreement was also hard to assess. Again, due to time and resource constraints it was originally not planned to incorporate a third round of reads in order to allow for intrarater testing. While a third round of data was obtained, only five ML and five non-ML reads were able to be re-assessed by two readers, resulting in a very small subset of data.
We were ambitious with regards to the ability of ML algorithm to train to high standard on the limited data planned. Specificity on a per-patient basis did not reach the high specificity of the source STREAMLINE study and some speculative reasons for this could be:
-
Readers were allocated scans from a multitude of different hospitals – they were not used to the appearance of the all the various sequences from different sites/different scanners. Whereas in the STREAMLINE study, radiologists read WB-MRI from their own site, although masked to the other staging investigations.
-
The availability of stitched volumes of the T2, DWI, ADC but only unstitched Dixon T1w images, in addition to the large number of unstitched sequences available to the readers for each case on PACS, as provided by the STREAMLINE study, may have resulted in readers relying on the few stitched volumes as these are much easier to read – and not checking all the other sequences – perhaps reflected in the short RTs.
-
There were many additional sites of ‘suspected disease’ highlighted by the ML heatmap, most of which could be ignored or over-looked by the readers and this may have affected the reader specificity. However, the specificity achieved in MALIBO was lower than STREAMLINE in the both standard WB-MRI reads as well as the ML reads, so this is unlikely to account for the lower specificity.
-
MALIBO reads were entirely retrospective, not affecting patient care, whereas STREAMLINE WB-MRI reads were revealed in the multidisciplinary team meeting at the time of patient treatment planning. Thus, there may have been a difference in the level of concern for the effect on patient care, which may have resulted in this difference in reading accuracy.
-
-
Completion of study objectives
We were unable to fulfil two of our secondary objectives, due to time limitations related to delay in receipt of data. We were unable to undertake a study to evaluate different combinations of acquired MRI sequences with and without ML support. The COVID pandemic and UK national lockdown took place just as we finished the radiology reads that have been reported and it was not possible to undertake any further reading rounds with different combinations MRI sequences. In addition, we did not undertake a simple health economic analysis. However, as there was a very limited difference in RT with ML support and no difference in diagnostic performance, an inference can be made that it is unlikely that health economic analysis would demonstrate any benefit to using ML support in the current study.
Clinical and public acceptance is an important consideration in ML-related imaging studies. The validation of the developed ML tools needs to stand up to scrutiny and the methods used for testing the tools need to be clear to clinical radiologists. In MALIBO, we have devised a viewing framework that is widely used by radiologists and incorporates the ML tools into a typical clinical environment for robust testing. This field of work is relatively new and further developments will be needed in order to identify whether ML support will ultimately improve the interpretation of complex scans, such as WB-MRI. However, the steps taken in this current study represent a part of a discovery process for building AI-supported imaging interpretation.
Conclusions
-
Phase 1 demonstrated that an ML algorithm could be developed for the accurate segmentation of most healthy organs on WB-MRI.
-
Phase 2 developed a ML output using a two-phase approach, with a first step being the organ segmentation followed by a second step for lesion detection.
-
Phase 3 clinical validation of the ML output demonstrated equivalent performance for the detection of metastatic lesions in patients with lung or colon cancer on WB-MRI when ML methods were applied to assist clinical radiology reads. There was a slight decrease in RT when using ML.
Chapter 8 Implications for practice and future research
The use of ML methods that automatically identify normal anatomical structures and subsequently detect abnormal lesions has the potential to improve the diagnostic accuracy and reduce the RT of WB-MRI scans obtained from patients having cancer staging. ML methods may support radiologists in complex reading tasks, particularly where significant expertise and training are required but not always available in an overstretched workforce. This ambitious project took steps in the direction of clinical translation. However, the work is not ready yet for deployment into the clinical arena and areas for further work have been identified.
Recommendations for future research
Further study within the current work.
With respect to the current study, future work should include:
-
Analysis of the cases in detail with respect to false-positive and false-negative lesion detection in order to evaluate the cause of failure/error and work on improving model training. Review the ML output together with the reference standard would allow to identify the sites that were missed or disregarded by readers but actually correctly identified by the ML output.
-
The work could be expanded to allow not only detection of metastatic lesions but also detection of the primary tumour mass.
-
Automated segmentation of lesions for lesion characterisation, for example with radiomics analysis.
-
The ML output on Phase 3 cases should be tested against manual expert segmentations, in order to evaluate the detection of lesions by the algorithm, independent of radiologist interaction.
Further research in this field of study.
With respect to the wider radiology and ML community, the following research needs were identified:
-
Harmonise WB-MRI protocols for different applications, in order to be able to harness the potential of ML model development. There is a clear need to improve standardisation of image acquisition across vendors as well as sites. Variation in acquisition may hamper widely generalisability ML tools. Homogeneity of acquisition may help the field overall. Attempts at protocol harmonisation for WB-MRI should be related to specific applications (bone marrow/soft tissue or PET/MRI).
-
Image post-processing is an area that needs further development, including work in stitching of imaging stations to create whole-body image volumes, in the hope of being able to allow accurate image registration, particularly with respect to sequences acquired in breath-hold or non-breath-hold.
-
There is scope for further work in the ML analysis of the bone and bone marrow appearances by gender and age in order to increase knowledge of normal marrow, allowing detection of marrow abnormalities as well as recognition and quantification of osteoporosis.
-
Future research work could include investigating the application of the developed methods for lesion detection on FDG-PET/CT and CT images. It may also be possible to combine PET/CT with WB-MRI for cancer detection. This may increase the accuracy of the detection as there are more features available from different modalities of the image.
Additional information
Contributions of authors
Andrea Rockall (https://orcid.org/0000-0001-8270-5597) (Professor of Radiology) was the Chief Investigator, conceived the study design, contributed to the protocol writing and study management as member of the Trial Management Group, interpreted trial imaging and performed drafting and final editing of the report.
Xingfeng Li (https://orcid.org/0000-0002-6640-1048) (Research Associate) co-ordinated Phase 2 and Phase 3 imaging data, integration of data with Biotronics3D, training radiologists on use of Biotronics3D, scribing of radiology reads, performed drafting and editing of the report.
Nicholas Johnson (https://orcid.org/0000-0002-3702-5530) (Research Associate) contributed to the protocol writing, co-ordinated Phase 1 of the study, development of the data processing pipeline for Phases 1 and 2, initial data management and annotation of Phase 2 data.
Ioannis Lavdas (https://orcid.org/0000-0003-3680-5954) (Research Associate) contributed to the protocol writing, co-ordinated Phase 1 of the study, development of the data processing pipeline for Phases 1 and 2, initial data management and annotation of Phase 2 data.
Shalini Santhakumaran (https://orcid.org/0000-0003-0988-9339) (Statistician) wrote the statistical analysis plan and performed the stratified randomisation of cases for Phases 2 and 3.
A Toby Prevost (https://orcid.org/0000-0003-1723-0796) (Senior Statistician) contributed to statistical analysis plan, statistical analysis of Phase 3 and editing of the report.
Dow-Mu Koh (https://orcid.org/0000-0001-7654-8011) (Radiologist) contributed to the study design, interpreted trial imaging, collected trial data and contributed to the final report.
Shonit Punwani (https://orcid.org/0000-0002-1014-0870) (Radiologist) contributed to the study design, interpreted trial imaging, collected trial data and contributed to the final report.
Vicky Goh (https://orcid.org/0000-0002-2321-8091) (Radiologist) contributed to the study design, interpreted trial imaging, collected trial data and contributed to the final report.
Nishat Bharwani (https://orcid.org/0000-0002-6236-1480) (Radiologist), contributed to the study design, interpreted trial imaging, collected trial data and contributed to the final report.
Amandeep Sandhu (https://orcid.org/0000-0001-9790-5102) (Radiologist), contributed to the study data management and annotation, interpreted trial imaging, collected trial data and contributed to the final report.
Harbir Sidhu (https://orcid.org/0000-0003-3564-3383) (Radiologist), contributed to the study data management and annotation, interpreted trial imaging, collected trial data and reviewed the final report.
Andrew Plumb (https://orcid.org/0000-0003-1322-5113) (Radiologist), contributed to the study data management and annotation, interpreted trial imaging, collected trial data and contributed to the final report.
James Burn (https://orcid.org/0000-0002-4151-3377) (Radiologist), contributed to the study data management and annotation, interpreted trial imaging, collected trial data and reviewed the final report.
Aisling Fagan (https://orcid.org/0000-0003-3455-4397) (Radiologist), contributed to the study data management and annotation, interpreted trial imaging, collected trial data and reviewed the final report.
Alf Oliver (https://orcid.org/0000-0002-7380-1074) (patient representative), contributed to the study design, study management as member of the TMG, and reviewed the final report.
Georg J Wengert (https://orcid.org/0000-0002-1854-9128) (Radiologist), developed training manual for Phase 3 reads, trained readers on Biotronics3D, interpreted trial imaging, collected trial data and reviewed the final report.
Daniel Rueckert (https://orcid.org/0000-0002-5683-5889) (Senior Computer Scientist), contributed to the study design, study management as member of the TMG, interpreted trial imaging outputs and reviewed the final report.
Eric Aboagye (https://orcid.org/0000-0003-2276-6771) (Senior Imaging Scientist), contributed to the study design, study management as member of the TMG and reviewed the final report.
Stuart Taylor (https://orcid.org/0000-0002-6765-8806) (Radiologist), helped conceive the study design, contributed to the protocol writing and study management as member of the TMG, interpreted trial imaging and contributed to editing of the report.
Ben Glocker (https://orcid.org/0000-0002-4897-9356) (Senior Computer Scientist) contributed to the study design, contributed to the protocol writing, study management as member of the TMG, interpreted trial imaging, developed machine learning methodology for all phases of the study and contributed to the final report.
Acknowledgements
The investigators thank the healthy volunteers and patients that participated in the contributing studies. We thank the sponsors and funders of the contributing studies for data-sharing. We thank the following for their support of the study:
Hanna Nicholas, Cancer Research UK Imperial Centre: Clinical Trials Section/Imperial Clinical Trials Unit.
Bhavesh Pratap, Imperial College London.
Harry Hatzakis, Biotronics3D.
Tarekur Chowdhury, Biotronics3D.
Giacomo Falcone, Biotronics3D.
MALIBO Investigators as Group authorship
Phase 3 radiology reader contributors
Tara D Barwick, Imperial College Healthcare NHS Trust and Department of Surgery and Cancer, Faculty of Medicine, Imperial College London, experienced WB-MRI reader.
Peter Boavida, Homerton University Hospital NHS Trust, experienced WB-MRI reader.
Katja N DePaepe, Cambridge University Hospitals NHS Foundation Trust, Cambridge, experienced WB-MRI reader.
Edward W Johnston, Royal Marsden Hospital, experienced WB-MRI reader.
Christian Kelly-Morland, Guy’s and St. Thomas’ NHS Foundation Trust, Department of Cancer Imaging King’s College London School of Biomedical Engineering and Imaging Sciences, experienced WB-MRI reader.
Amish Lakhani, Paul Strickland Scanner Centre, Mount Vernon Cancer Centre and Imperial College Healthcare NHS Trust, Imperial College London, experienced WB-MRI reader.
Christina Messiou, Royal Marsden Hospital and The Institute of Cancer Research, experienced WB-MRI reader.
James Russell, Imperial College Healthcare NHS Trust, inexperienced WB-MRI reader.
Miriam Salib, Imperial College Healthcare NHS Trust, inexperienced WB-MRI reader.
Heminder Sokhi, The Hillingdon Hospitals NHS Foundation Trust and Paul Strickland Scanner Centre, Mount Vernon Hospital, experienced WB-MRI reader.
Neil Soneji, Imperial College Healthcare NHS Trust, London, UK and Royal Marsden Hospital, experienced WB-MRI reader.
Nina Tunariu, Royal Marsden Hospital and The Institute of Cancer Research, experienced WB-MRI reader.
Sarah Vinnicombe, Thirlestaine Breast Centre, Gloucestershire NHS Foundation Trust, University of Dundee, inexperienced WB-MRI reader.
Kathryn Wallitt, Imperial College Healthcare NHS Trust, experienced WB-MRI reader.
Machine learning contributors
Qi Dou, Faculty of Engineering, Dept of Computing, Imperial College London, model development, Phase 2 machine learning.
Vanya Valindria, Imperial College London, Computing Science, Phase 1.
Konstantin Kamnitsas, Faculty of Engineering, Dept of Computing, Imperial College London, Phase 1 machine learning.
Wenjai Bai, Faculty of Engineering, Dept of Computing, Imperial College London, Phase 1 machine learning.
Ender Konukoglu, Faculty of Engineering, Dept of Computing, Imperial College London, Phase 1 machine learning.
Henrietta Mair, Chelsea and Westminster Hospital NHS Trust, image annotator.
Ala Haqiqi, Imperial College Healthcare NHS Trust, image annotator.
Statistics, trials unit and imaging research support contributors
Deborah Ashby, Imperial Clinical Trials Unit, statistical methodology.
Jane Warwick, Imperial Clinical Trials Unit, statistical methodology.
Xinxue Liu, Imperial Clinical Trials Unit, statistical methodology.
Lesley Honeyfield, Imperial College Healthcare NHS Trust, Phase 1 and STREAMLINE data curation.
Krystyna Reczko, University College London Clinical Trials Unit.
Laura White, University College London Clinical Trials Unit.
Trial Steering Committee
David Lomas (chair), Bradley J Erickson, Dr Shah-Jalal Sarker, Alf Oliver (Public and Patient representative).
The project is supported by researchers at the Imperial College London NIHR Biomedical Research Centre, CRUK Imperial Centre, NIHR University College London Hospitals Biomedical Research Centre, NIHR Biomedical Research Centre and the NIHR Clinical Research Facilities and the Royal Marsden Hospital and Institute of Cancer Research. This research has been conducted using the UK Biobank Resource.
Patient data statement
This work uses data provided by patients and collected by the NHS as part of their care and support. Using patient data is vital to improve health and care for everyone. There is huge potential to make better use of information from people’s patient records, to understand more about disease, develop new treatments, monitor safety and plan NHS services. Patient data should be kept safe and secure, to protect everyone’s privacy, and it’s important that there are safeguards to make sure that they are stored and used responsibly. Everyone should be able to find out about how patient data are used. #datasaveslives You can find out more about the background to this citation here: https://understandingpatientdata.org.uk/data-citation.
Data-sharing statement
All data requests should be submitted to the corresponding author for consideration. Access to available anonymised data may be granted following review.
Ethics statement
Ethical approval for retrospective use of previously acquired patient data was obtained (ICREC Reference 15IC2647). The MALIBO study did not directly collect patient imaging data, but relied on data from previous contributing studies. The ethical approval for Phase 1 of the trial was in place (ICREC 08/H0707/58). Ethical approvals for Phases 2 and 3 (contributing studies) were also in place as per their individual protocols. There were no material ethical concerns related to the MALIBO study with no perceived risk or benefit to individual patients. However, there was a significant interest in improving patient care, as indicated in section 60 of the Health and Social Care Act (2001). All patients gave written informed consent prior to participation in any of the contributing studies. Consent for the use of scans in future research was also obtained in the case of participants in the contributing studies. The need to re-consent participants for the use of the patient data was waived by the ethics committee. All patient data were de-identified and held in a secure central imaging server 3DnetTM (www.3dnetmedical.com/public/), provided by Biotronics3D (London, UK). The data are also held on password-protected Imperial College London university computers for the purposes of the ML algorithms’ development.
Information governance statement
Imperial College London is committed to handling all personal information in line with the UK Data Protection Act (2018) and the General Data Protection Regulation (EU GDPR) 2016/679.
Under the Data Protection legislation, Imperial College London is the Data Controller, and you can find out more about how we handle personal data, including how to exercise your individual rights and the contact details for our Data Protection Officer. https://www.imperial.ac.uk/clinical-trials-unit/dataprotection/
Disclosure of interests
Full disclosure of interests: Completed ICMJE forms for all authors, including all related interests, are available in the toolkit on the NIHR Journals Library report publication page at https://doi.org/10.3310/KPWQ4208.
Primary conflicts of interest: Andrea Rockall: EME grant to institution; RoClub Advisory Board, virtual share options; European Society of Radiology Board of Directors, no payment. Vicky Goh: Siemens Healthineers to institution; travel support payment to self; Academic Committee, Royal College of Radiologists, unpaid, Workforce Group, NIHR Imaging Group, unpaid. Stuart A Taylor: EME funding committee. Shonit Punwani: Cancer research UK; CLIMATE clinical trial funding; The Urology Foundation, LIMIT clinical trial funding; NVISION consulting for hyperpolarised MRI and shares; patient application Luminal water imaging, PCT/GB2021/050911. A Toby Prevost: NIHR Public Health Research Funding Committee member 2014–2020; NIHR Covid-19 Recovery and Learning Funding Committee 2020; NIHR Public Health Interventions Responsive Studies Team PHIRST-CONNECT Independent Advisory Board Member 2020–present no payment. Nicholas Johnson: Note: No participation as DSMB/IDMC member for any trials within oncology or machine learning. Current membership of one DSMB as independent statistician but in an entirely different field (respiratory). Daniel Rueckert: Grants EPSRC (Heartflow), Wellcome Trust (Alexpander von Humboldt Foundation); Innovate UK; NIHR; consulting fees to self from IXICO and Heartflow; Ben Glocker: EU Commission Grant No. 757173, ERC-2017-STG; Innovate UK UKRI London Medical Imaging & Artificial Intelligence Centre for Value Based Healthcare; Heartflow grant, industry collaboration; Kheiron Medical Technologies and Heartflow, part-time employee with stock options as part of compensation package; Eric Aboagye: AstraZeneca paid to institution; Advisory Board, Radiopharm theranostics paid to self and to institution.
Publications
Lavdas I, Rockall AG, Castelli F, Sandhu RS, Papadaki A, Honeyfield L, et al. Apparent diffusion coefficient of normal abdominal organs and bone marrow from whole-body DWI at 1.5 T: the effect of sex and age. AJR Am J Roentgenol 2015;205(2):242–50. https://doi.org/10.2214/AJR.14.13964
Glocker B, Konukoglu E, Lavdas I, Iglesias JE, Aboagye EO, Rockall AG, et al. Correction of fat-water swaps in Dixon MRI. In Medical Image Computing and Computer-assisted Intervention – MICCAI 2016. Cham: Springer International Publishing; 2016.
Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Sandhu A, et al. Fully automatic, multiorgan segmentation in normal whole-body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Med Phys 2017;44(10):5210–20.
Valindria VV, Lavdas I, Bai W, Kamnitsas K, Oboagye EO, Rockall AG, et al. Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans Med Imaging 2017;36(8):1597–606.
Valindria VV, Lavdas I, Bai W, Kamnitsas K, Oboagye EO, Rockall AG, et al. Domain adaptation for MRI organ segmentation using reverse classification accuracy. arXiv preprint arXiv:1806.00363, 2018.
Lavdas I, Glocker B, Rueckert D, Taylor SA, Oboagye EO, Rockall AG. Machine learning in whole-body MRI: experiences and challenges from an applied study using multicentre data. Clin Radiol 2019;74(5):346–56.
Disclaimers
This article presents independent research. The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, the MRC, the EME programme or the Department of Health and Social Care. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, the EME programme or the Department of Health and Social Care.
References
- Sharma N, Aggarwal LM. Automated medical image segmentation techniques. J Med Phys 2010;35:3-14.
- Team N . Myeloma: Diagnosis and Management 2018.
- Petralia G, Padhani AR, Pricolo P, Zugni F, Martinetti M, Summers PE, et al. Whole-body magnetic resonance imaging (WB-MRI) in oncology: recommendations and key uses. La Radiologia Medica 2019;124:218-33.
- Jacobs MA, Macura KJ, Zaheer A, Antonarakis ES, Stearns V, Wolff AC, et al. Multiparametric whole-body MRI with diffusion-weighted imaging and ADC mapping for the identification of visceral and osseous metastases from solid tumors. Acad Radiol 2018;25:1405-14.
- Barnes A, Alonzi R, Blackledge M, Charles-Edwards G, Collins DJ, Cook G, et al. UK quantitative WB-DWI technical workgroup: consensus meeting recommendations on optimisation, quality control, processing and analysis of quantitative whole-body diffusion-weighted imaging for cancer. Br J Radiol 2018;91.
- Taylor SA, Mallett S, Ball S, Beare S, Bhatnagar G, Bhowmik A, et al. Streamline Investigators . Diagnostic accuracy of whole-body MRI versus standard imaging pathways for metastatic disease in newly diagnosed non-small-cell lung cancer: the prospective STREAMLINE L trial. Lancet Respir Med 2019;7:523-32.
- Taylor SA, Mallett S, Beare S, Bhatnagar G, Blunt D, Boavida P, et al. STREAMLINE Investigators . Diagnostic accuracy of whole-body MRI versus standard imaging pathways for metastatic disease in newly diagnosed colorectal cancer: the prospective STREAMLINE C trial. Lancet Gastroenterol Hepatol 2019;4:529-37.
- Taylor SA, Mallett S, Miles A, Beare S, Bhatnagar G, Bridgewater J, et al. Streamlining staging of lung and colorectal cancer with whole body MRI; study protocols for two multicentre, non-randomised, single-arm, prospective diagnostic accuracy studies (STREAMLINE C and STREAMLINE L). BMC Cancer 2017;17.
- Miles A, Evans RE, Halligan S, Beare S, Bridgewater J, Goh V, et al. STREAMLINE Investigators . Predictors of patient preference for either whole-body magnetic resonance imaging (WB-MRI) or CT/PET-CT for staging colorectal or lung cancer. J Med Imaging Radiat Oncol 2020;64:537-45.
- Miles A, Taylor SA, Evans REC, Halligan S, Beare S, Bridgewater J, et al. STREAMLINE Investigators . Patient preferences for whole-body MRI or conventional staging pathways in lung and colorectal cancer: a discrete choice experiment. Eur Radiol 2019;29:3889-900.
- Pasoglou V, Michoux N, Larbi A, Van Nieuwenhove S, Lecouvet F. Whole body MRI and oncology: recent major advances. Br J Radiol 2018;91.
- Lauenstein TC, Semelka RC. Emerging techniques: whole-body screening and staging with MRI. J Magn Reson Imaging 2006;24:489-98.
- Ceranka J, Verga S, Kvasnytsia M, Lecouvet F, Michoux N, de Mey J, et al. Multi-atlas segmentation of the skeleton from whole-body MRI-Impact of iterative background masking. Magn Reson Med 2020;83:1851-62.
- Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Sandhu A, et al. Fully automatic, multiorgan segmentation in normal whole-body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Med Phys 2017;44:5210-20.
- Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61-78.
- Hesamian MH, Jia W, He X, Kennedy P. Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging 2019;32:582-96.
- Lavdas I, Glocker B, Rueckert D, Taylor SA, Aboagye EO, Rockall AG. Machine learning in whole-body MRI: experiences and challenges from an applied study using multicentre data. Clin Radiol 2019;74:346-56.
- Padhani AR, Koh DM, Collins DJ. Whole-body diffusion-weighted MR imaging in cancer: current status and research directions. Radiology 2011;261:700-18.
- Machado Medeiros T, Altmayer S, Watte G, Zanon M, Basso Dias A, Henz Concatto N, et al. 18F-FDG PET/CT and whole-body MRI diagnostic performance in M staging for non–small cell lung cancer: a systematic review and meta-analysis. Eur Radiol 2020;30:3641-9.
- Wu LM, Gu HY, Zheng J, Xu X, Lin LH, Deng X, et al. Diagnostic value of whole-body magnetic resonance imaging for bone metastases: a systematic review and meta-analysis. J Magn Reson Imaging 2011;34:128-35.
- Koh DM, Collins DJ. Diffusion-weighted MRI in the body: applications and challenges in oncology. AJR Am J Roentgenol 2007;188:1622-35.
- Messiou C, Collins DJ, Morgan VA, deSouza NM. Optimising diffusion weighted MRI for imaging metastatic and myeloma bone disease and assessing reproducibility. Eur Radiol 2011;21:1713-8.
- Padhani AR, van Ree K, Collins DJ, D’Sa S, Makris A. Assessing the relation between bone marrow signal intensity and apparent diffusion coefficient in diffusion-weighted MRI. AJR Am J Roentgenol 2013;200:163-70.
- Eschmann SM, Pfannenberg AC, Rieger A, Aschoff P, Müller M, Paulsen F, et al. Comparison of 11C-choline-PET/CT and whole body-MRI for staging of prostate cancer. Nuklearmedizin 2007;46:161-8; quiz N47.
- Würslin C, Machann J, Rempp H, Claussen C, Yang B, Schick F. Topography mapping of whole body adipose tissue using a fully automated and standardized procedure. J Magn Reson Imaging 2010;31:430-9.
- Jerebko AK, Schmidt GP, Zhou X, Bi J, Anand V, Liu J, et al. Robust parametric modeling approach based on domain knowledge for computer aided detection of vertebrae column metastases in MRI. Inf Process Med Imaging 2007;20:713-24.
- Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 2010;31:680-9.
- Medical Image Computing and Computer-assisted Intervention – MICCAI 2011. Berlin; Heidelberg: Springer; 2011.
- Joint Classification-Regression Forests for Spatially Structured Multi-object Segmentation. Computer Vision – ECCV 2012. Berlin; Heidelberg: Springer; 2012.
- Medical Image Computing and Computer-assisted Intervention – MICCAI 2012. Berlin; Heidelberg: Springer; 2012.
- Medical Image Computing and Computer-assisted Intervention – MICCAI 2013. Berlin; Heidelberg: Springer; 2013.
- Medical Image Computing and Computer-assisted Intervention – MICCAI 2012. Berlin; Heidelberg: Springer; 2012.
- Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annu Rev Biomed Eng 2000;2:315-37.
- International Conference on Medical Image Computing and Computer-assisted Intervention. Cham: Springer; 2016.
- Medical Image Computing and Computer-assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015.
- Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2018.
- No New-net. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2019.
- Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2018.
- Li B, Li Q, Nie W, Liu S. Diagnostic value of whole-body diffusion-weighted magnetic resonance imaging for detection of primary and metastatic malignancies: a meta-analysis. Eur J Radiol 2014;83:338-44.
- Taylor SA, Mallet S, Miles A, Morris S, Quinn L, Clarke CS, et al. Whole-body MRI compared with standard pathways for staging metastatic disease in lung and colorectal cancer: the Streamline diagnostic accuracy studies. Health Technol Assess 2019;23:1-270.
- Lavdas I, Rockall AG, Castelli F, Sandhu RS, Papadaki A, Honeyfield L, et al. Apparent diffusion coefficient of normal abdominal organs and bone marrow from whole-body DWI at 1.5 T: the effect of sex and age. Am J Roentgenol 2015;205:242-50.
- Latifoltojar A, Hall-Craggs M, Bainbridge A, Rabin N, Popat R, Rismani A, et al. Whole-body MRI quantitative biomarkers are associated significantly with treatment response in patients with newly diagnosed symptomatic multiple myeloma following bortezomib induction. Eur Radiol 2017;27:5325-36.
- Johnston EW, Latifoltojar A, Sidhu HS, Ramachandran N, Sokolska M, Bainbridge A, et al. Multiparametric whole-body 3.0-T MRI in newly diagnosed intermediate- and high-risk prostate cancer: diagnostic accuracy and interobserver agreement for nodal and metastatic staging. Eur Radiol 2019;29:3159-69.
- Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol 2018;15:504-8.
- Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol 2019;16:391-403.
- Giger ML. Machine learning in medical imaging. J Am Coll Radiol 2018;15:512-20.
- Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol 2017;10:257-73.
- Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics 2017;37:505-15.
- Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology 2018;288:318-28.
- Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik 2019;29:102-27.
- Habes M, Schiller T, Rosenberg C, Burchardt M, Hoffmann W. Automated prostate segmentation in whole-body MRI scans for epidemiological studies. Phys Med Biol 2013;58:5899-915.
- Ishiguchi H, Ito S, Kato K, Sakurai Y, Kawai H, Fujita N, et al. Diagnostic performance of 18F-FDG PET/CT and whole-body diffusion-weighted imaging with background body suppression (DWIBS) in detection of lymph node and bone metastases from pediatric neuroblastoma. Ann Nucl Med 2018;32:348-62.
- Laible M, Schoenberg SO, Weckbach S, Lettau M, Winnik E, Bischof J, et al. Whole-body MRI and MRA for evaluation of the prevalence of atherosclerosis in a cohort of subjectively healthy individuals. Insights Imaging 2012;3:485-93.
- Heffler MA, Le LQ, Xi Y, Chhabra A. Tumor segmentation of whole-body magnetic resonance imaging in neurofibromatosis type 1 patients: tumor burden correlates. Skeletal Radiol 2017;46:93-9.
- Akselrod-Ballin A, Dafni H, Addadi Y, Biton I, Avni R, Brenner Y, et al. Multimodal correlative preclinical whole body imaging and segmentation. Sci Rep 2016;6.
- Habes M, Schiller T, Rosenberg C, Burchardt M, Hoffmann W. Automated prostate segmentation in whole-body MRI scans for epidemiological studies. Phys Med Biol 2013;58:5899-915.
- Hofmann M, Bezrukov I, Mantlik F, Aschoff P, Steinke F, Beyer T, et al. MRI-based attenuation correction for whole-body PET/MRI: quantitative evaluation of segmentation- and atlas-based methods. J Nucl Med 2011;52:1392-9.
- Shahzad R, Dzyubachyk O, Staring M, Kullberg J, Johansson L, Ahlström H, et al. Automated extraction and labelling of the arterial tree from whole-body MRA data. Med Image Anal 2015;24:28-40.
- Latifoltojar A, Punwani S, Lopes A, Humphries PD, Klusmann M, Menezes LJ, et al. Whole-body MRI for staging and interim response monitoring in paediatric and adolescent Hodgkin’s lymphoma: a comparison with multi-modality reference standard including 18F-FDG-PET-CT. Eur Radiol 2019;29:202-12.
- Antonio C, Jamie S, Ender K. Decision Forests: A Unified Framework for Classification, Regression, Density Estimation, Manifold Learning and Semi-supervised Learning: Now. 2012.
- Medical Image Computing and Computer-assisted Intervention - MICCAI 2016. Cham: Springer International Publishing; 2016.
- Breiman L. Random forests. Mach Learn 2001;45:5-32.
- Iglesias JE, Sabuncu MR. Multi-atlas segmentation of biomedical images: a survey. Med Image Anal 2015;24:205-19.
- Cuingnet R, Prevost R, Lesage D, Cohen LD, Mory B, Ardon R. Medical Image Computing and Computer-assisted Intervention – MICCAI 2012: 15th International Conference, Nice, France, October 1–5, 2012, Proceedings, Part III. Berlin; Heidelberg: Springer; 2012.
- Glocker B, Pauly O, Konukoglu E, Criminisi A. Computer Vision – ECCV 2012: 12th European Conference on Computer Vision, Florence, Italy, October 7–13, 2012, Proceedings, Part IV. Berlin; Heidelberg: Springer; 2012.
- Glocker B, Konukoglu E, Haynor DR. Medical Recognition, Segmentation and Parsing. London: Academic Press, Elsevier; 2015.
- Bai W, Shi W, O’Regan DP, Tong T, Wang H, Jamil-Copley S, et al. A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: application to cardiac MR images. IEEE Trans Med Imaging 2013;32:1302-15.
- Glocker B, Komodakis N, Tziritas G, Navab N, Paragios N. Dense image registration through MRFs and efficient linear programming. Med Image Anal 2008;12:731-41.
- Heimann T, van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 2009;28:1251-65.
- Valindria VV, Lavdas I, Bai W, Kamnitsas K, Aboagye EO, Rockall AG, et al. Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans Med Imaging 2017;36:1597-606.
- Valindria VV, Lavdas I, Bai W, Kamnitsas K, Aboagye EO, Rockall AG, et al. Domain adaptation for MRI organ segmentation using reverse classification accuracy 2018.
- Boykov Y, Funka-Lea G. Graph cuts and efficient ND image segmentation. Int J Comput Vis 2006;70:109-31.
- Heimann T, Meinzer H-P. Statistical shape models for 3D medical image segmentation: a review. Med Image Anal 2009;13:543-63.
- Geremia E, Zikic D, Clatz O, Menze B, Glocker B, Konukoglu E, et al. Decision Forests for Computer Vision and Medical Image Analysis. Springer; 2013.
- Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, et al. Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Acad Radiol 2004;11:178-89.
- Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297-302.
- Crum WR, Camara O, Hill DL. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans Med Imaging 2006;25:1451-61.
- Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15.
- International Conference on Medical Image Computing and Computer-assisted Intervention. Springer; 2007.
- Patch-based Evaluation of Image Segmentation 2014.
- Konukoglu E, Glocker B, Ye DH, Criminisi A, Pohl KM. Discriminative segmentation-based evaluation through shape dissimilarity. IEEE Trans Med Imaging 2012;31:2278-89.
- MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge. 2007.
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med 2015;12.
- Dwork C, Feldman V, Hardt M, Pitassi T, Reingold O, Roth A. The reusable holdout: Preserving validity in adaptive data analysis. Science 2015;349:636-8.
- Baraldi A, Bruzzone L, Blonda P. Quality assessment of classification and cluster maps without ground truth knowledge. IEEE Trans Geosci Remote Sens 2005;43:857-73.
- Liu Y, Wang J, Cho S, Finkelstein A, Rusinkiewicz S. A no-reference metric for evaluating the quality of motion deblurring. ACM Trans Graph 2013;32:1-12.
- Classification of Proxy Labeled Examples for Marketing Segment Generation. 2011.
- Correia PL, Pereira F. Stand-alone objective segmentation quality evaluation. EURASIP J Adv Signal Process 2002;2002.
- Ge F, Wang S, Liu T. New benchmark for image segmentation evaluation. J Electron Imaging 2007;16.
- International Conference on Advanced Concepts for Intelligent Vision Systems. Berlin; Heidelberg: Springer; 2008.
- 2013 IEEE International Conference on Multimedia and Expo (ICME). San Jose, CA: IEEE; 2013.
- International Workshop on Graphics Recognition. Berlin; Heidelberg: Springer; 2011.
- 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06). New York, NY: IEEE; 2006.
- Chabrier S, Emile B, Rosenberger C, Laurent H. Unsupervised performance evaluation of image segmentation. EURASIP J Adv Signal Process 2006;2006.
- Zhang H, Fritts JE, Goldman SA. Image segmentation evaluation: a survey of unsupervised methods. Comput Vis Image Underst 2008;110:260-80.
- Unnikrishnan R, Pantofaru C, Hebert M. Toward objective evaluation of image segmentation algorithms. IEEE Trans Pattern Anal Mach Intell 2007;29:929-44.
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 2004;23:903-21.
- 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; 2011.
- Bouix S, Martin-Fernandez M, Ungar L, Nakamura M, Koo M-S, McCarley RW, et al. On evaluating brain tissue classifiers without a ground truth. Neuroimage 2007;36:1207-24.
- Medical Imaging 2010: Image Perception, Observer Performance, and Technology Assessment. International Society for Optics and Photonics; 2010.
- International Conference on Medical Image Computing and Computer-assisted Intervention. Berlin; Heidelberg: Springer; 2012.
- European Conference on Computer Vision. Berlin; Heidelberg: Springer; 2012.
- Frounchi K, Briand LC, Grady L, Labiche Y, Subramanyan R. Automating image segmentation verification and validation by learning test oracles. Inf Softw Technol 2011;53:1337-48.
- Joint European Conference on Machine Learning and Knowledge Discovery in Databases. Berlin; Heidelberg: Springer; 2010.
- Reverse Testing: An Efficient Framework to Select Amongst Classifiers under Sample Selection Bias. 2006.
- Zikic D, Glocker B, Criminisi A. Encoding atlases by randomized classification forests for efficient multi-atlas label propagation. Med Image Anal 2014;18:1262-73.
- Criminisi A, Shotton J, Konukoglu E. Decision forests: a unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Found Trends Comput Graph Vis 2012;7:81-227.
- Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010;22:1345-59.
- CNN Features Off-the-shelf: an Astounding Baseline for Recognition. 2014.
- Goetz M, Weber C, Binczyk F, Polanska J, Tarnawski R, Bobek-Billewicz B, et al. DALSA: Domain Adaptation for Supervised Learning From Sparsely Annotated MR images. IEEE Trans Med Imaging 2016;35:184-96.
- Opbroek A, Vernooij MW, Ikram MA, Bruijne M. Weighting training images by maximizing distribution similarity for supervised segmentation across scanners. Med Image Anal 2015;24:245-54.
- Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285-98.
- Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning?. IEEE Trans Med Imaging 2016;35:1299-312.
- 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 21–26 July 2017. 2017.
- Ardehaly EM, Culotta A. Proceedings of the Twenty-fifth International Joint Conference on Artificial Intelligence. New York, NY: AAAI Press; 2016.
- International Conference on Medical Image Computing and Computer Assisted Intervention. n.d.
- The Simple and Efficient Semi-supervised Learning Method for Deep Neural Networks. 2013.
- Active Supervised Domain Adaptation. Berlin; Heidelberg: Springer; 2011.
- Lawrence ND. Data readiness levels 2017.
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31:1116-28.
- Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018;286:800-9.
- McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276-82.
- Kart T, Fischer M, Küstner T, Hepp T, Bamberg F, Winzeck S, et al. Deep learning-based automated abdominal organ segmentation in the UK biobank and German National Cohort Magnetic Resonance Imaging Studies. Invest Radiol 2021;56:401-8.
- Zheng H, Chen Y, Yue X, Ma C, Liu X, Yang P, et al. Deep pancreas segmentation with uncertain regions of shadowed sets. Magn Reson Imaging 2020;68:45-52.
- Bhat CS, Chopra M, Andronikou S, Paul S, Wener-Fligner Z, Merkoulovitch A, et al. Artificial intelligence for interpretation of segments of whole body MRI in CNO: pilot study comparing radiologists versus machine learning algorithm. Pediatr Rheumatol Online J 2020;18.
- Ceranka J, Polfliet M, Lecouvet F, Michoux N, de Mey J, Vandemeulebroucke J. Registration strategies for multi-modal whole-body MRI mosaicing. Magn Reson Med 2018;79:1684-95.
- Kozuka T, Matsukubo Y, Kadoba T, Oda T, Suzuki A, Hyodo T, et al. Efficiency of a computer-aided diagnosis (CAD) system with deep learning in detection of pulmonary nodules on 1-mm-thick images of computed tomography. Jpn J Radiol 2020;38:1052-61.
- Pinto MC, Rodriguez-Ruiz A, Pedersen K, Hofvind S, Wicklein J, Kappler S, et al. Impact of artificial intelligence decision support using deep learning on breast cancer screening interpretation with single-view wide-angle digital breast tomosynthesis. Radiology 2021;300:529-36.
- van Winkel SL, Rodríguez-Ruiz A, Appelman L, Gubern-Mérida A, Karssemeijer N, Teuwen J, et al. Impact of artificial intelligence support on accuracy and reading time in breast tomosynthesis image interpretation: a multi-reader multi-case study. Eur Radiol 2021;31:8682-91.
- Conant EF, Toledano AY, Periaswamy S, Fotin SV, Go J, Boatsman JE, et al. Improving accuracy and efficiency with concurrent use of artificial intelligence for digital breast tomosynthesis. Radiol Artif Intell 2019;1.
- Chae EY, Kim HH, Jeong JW, Chae SH, Lee S, Choi YW. Decrease in interpretation time for both novice and experienced readers using a concurrent computer-aided detection system for digital breast tomosynthesis. Eur Radiol 2019;29:2518-25.
- Winkel DJ, Tong A, Lou B, Kamen A, Comaniciu D, Disselhorst JA, et al. A novel deep learning based computer-aided diagnosis system improves the accuracy and efficiency of radiologists in reading biparametric magnetic resonance images of the prostate: results of a multireader, multicase study. Invest Radiol 2021;56:605-13.
- Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20:37-46.
- Landis JR, Koch GG. A one-way components of variance model for categorical data. Biometrics 1977;33:671-9.
- Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6.
- McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947;12:153-7.
- Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull 1945;1:80-3.
- Gwet KL. Testing the difference of correlated agreement coefficients for statistical significance. Educ Psychol Meas 2016;76:609-37.
- Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 1927;22:209-12.
Appendix 1 Supplementary tables and figures
FIGURE 23.
Apparent diffusion coefficient maps. Representative slices show ROIs used to calculate ADC values for red bone marrow and yellow bone marrow. (a–d) ADC maps show ROIs (outlined areas) used to calculate ADC values for red bone marrow in first thoracic vertebra (T1) (a), second thoracic vertebra (T2) (b), and third thoracic vertebra (T3) (c) and in yellow bone marrow in left and right femoral heads (d).
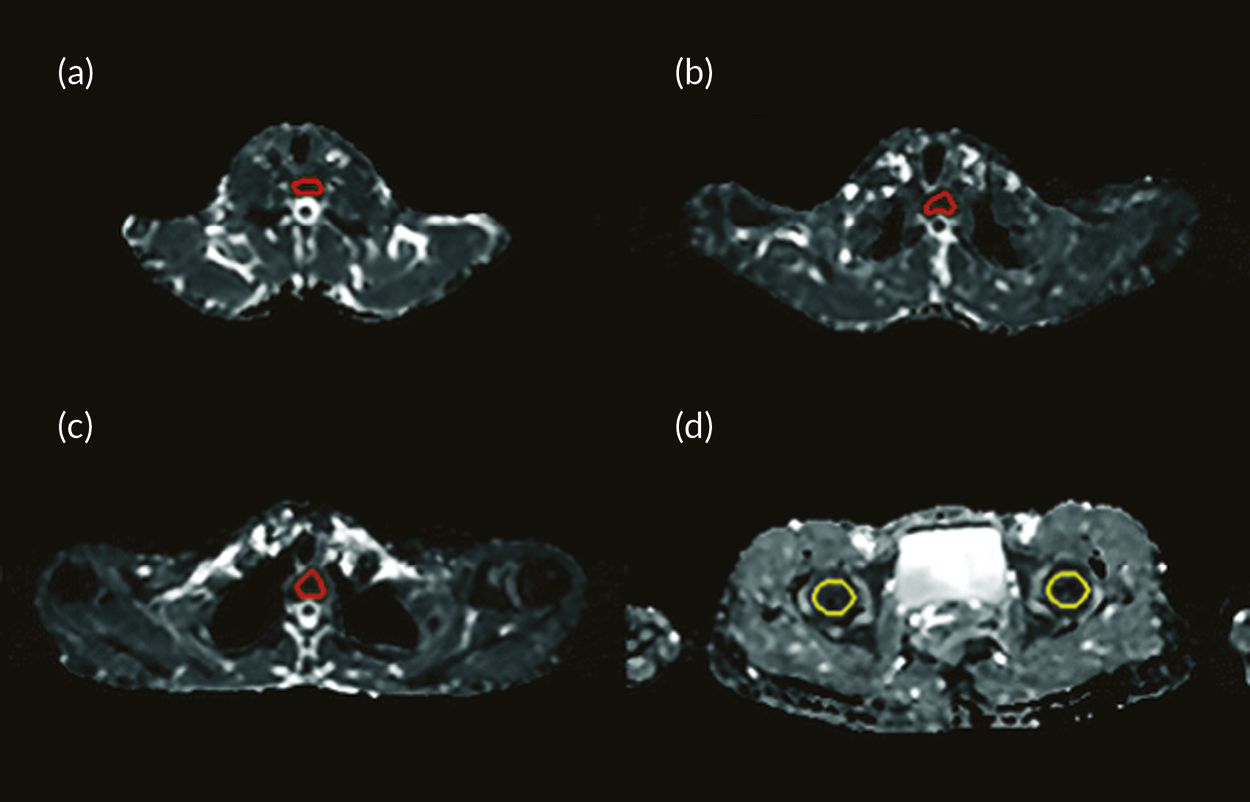
FIGURE 24.
Scatterplots showing the variation of ADCALL with age. Scatterplots showing the variation of ADCALL with age in the (a) LVR parenchyma (r = −0.37/p = 0.008 for all volunteers, r = −0.36/p = 0.11 for male volunteers and r = −0.49/p = 0.01 for female volunteers) and (b) yellow bone marrow (r = −0.35/p = 0.013 for all volunteers, r = −0.41/p = 0.046 for male volunteers and r = −0.53/p = 0.004 for female volunteers).
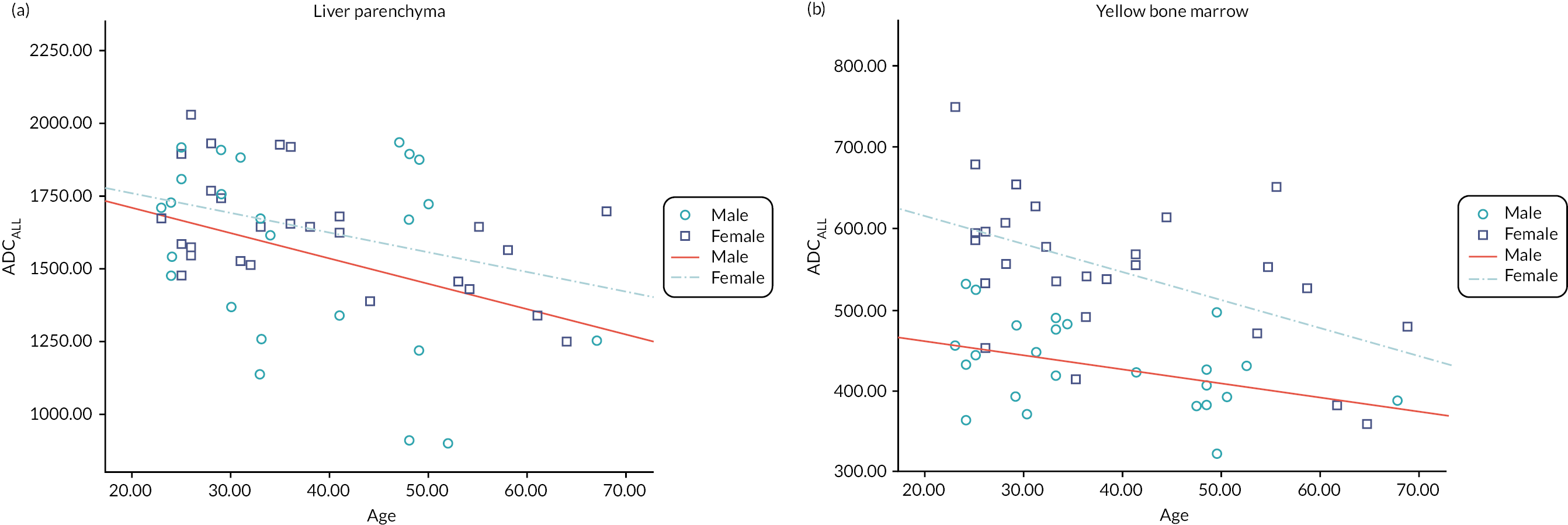
FIGURE 25.
Scatterplots show ADC values calculated from perfusion-sensitive WB-DWI protocol (ADCALL) vary with FF. (a) Scatterplot shows ADCALL values in red bone marrow vary with FF (r = –0.12, p = 0.41 for all volunteers; r = 0.17, p = 0.48 for male volunteers; r = –0.46, p = 0.02 for female volunteers). (b) Scatterplot shows ADCALL values in yellow bone marrow vary with FF (r = –0.49, p = 0.001 for all volunteers; r = –0.21, p = 0.37 for male volunteers; r = –0.41, p = 0.04 for female volunteers).
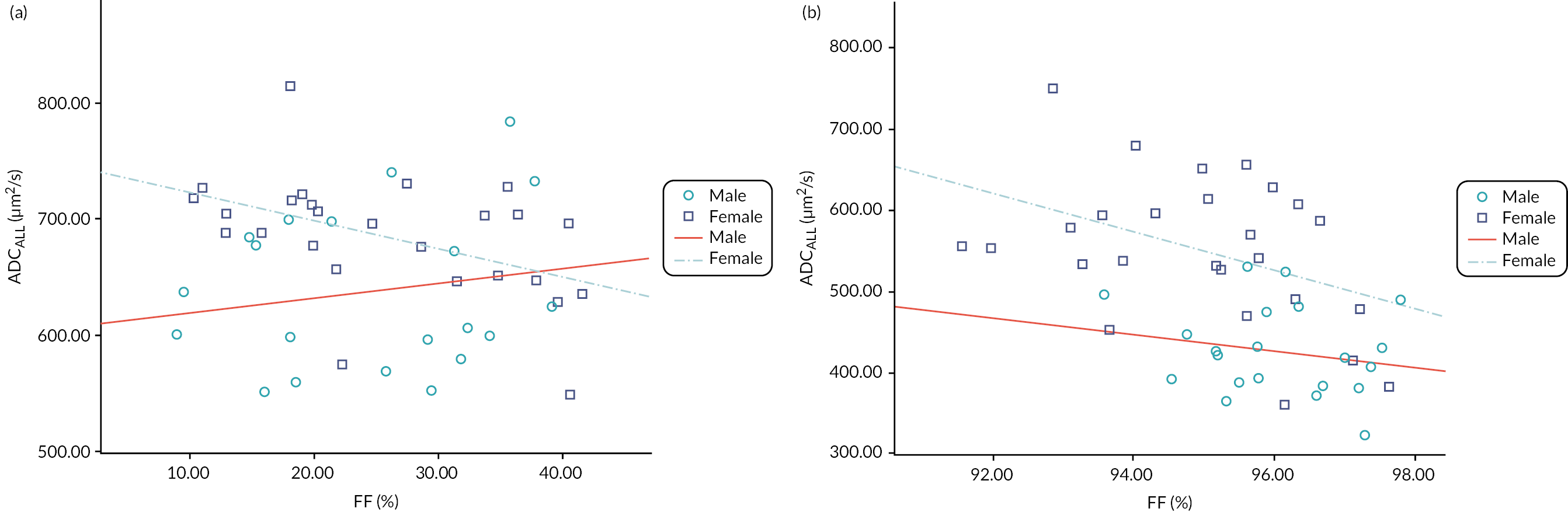
| DSC | RE | PR | ASD (mm) | RMSSD (mm) | HD (mm) | |
|---|---|---|---|---|---|---|
| CFs_T2w | 0.70 ± 0.17 | 0.73 ± 0.18 | 0.71 ± 0.14 | 13.5 ± 11.2 | 34.6 ± 37.6 | 185.7 ± 194.0 |
| CFs_all | 0.74 ± 0.16 | 0.78 ± 0.16 | 0.74 ± 0.13 | 7.89 ± 7.55 | 20.9 ± 27.1 | 170.7 ± 194.0 |
| p-value | 0.491 | 0.412 | 0.533 | 0.039 | 0.309 | 0.974 |
| CNNs_T2w | 0.81 ± 0.12 | 0.82 ± 0.14 | 0.82 ± 0.10 | 5.48 ± 4.84 | 17.0 ± 13.3 | 199.0 ± 101.2 |
| CNNs_all | 0.77 ± 0.14 | 0.79 ± 0.15 | 0.79 ± 0.11 | 9.23 ± 8.04 | 25.2 ± 19.1 | 215.9 ± 98.6 |
| p-value | 0.412 | 0.450 | 0.450 | 0.178 | 0.224 | 0.224 |
FIGURE 26.
Bar chart showing the mean measured metrics. DSC (a), RE (b), PR (c), ASD (d), RMSSD (e) and HD (f) for the segmented organs (RLNG and LLNG, LVR, GBLD, RKDN and LKDN, SPLN, PNCR and BLD) and bones (SPN and PLVS) for the three algorithms (CFs), (CNNs) and (MA).
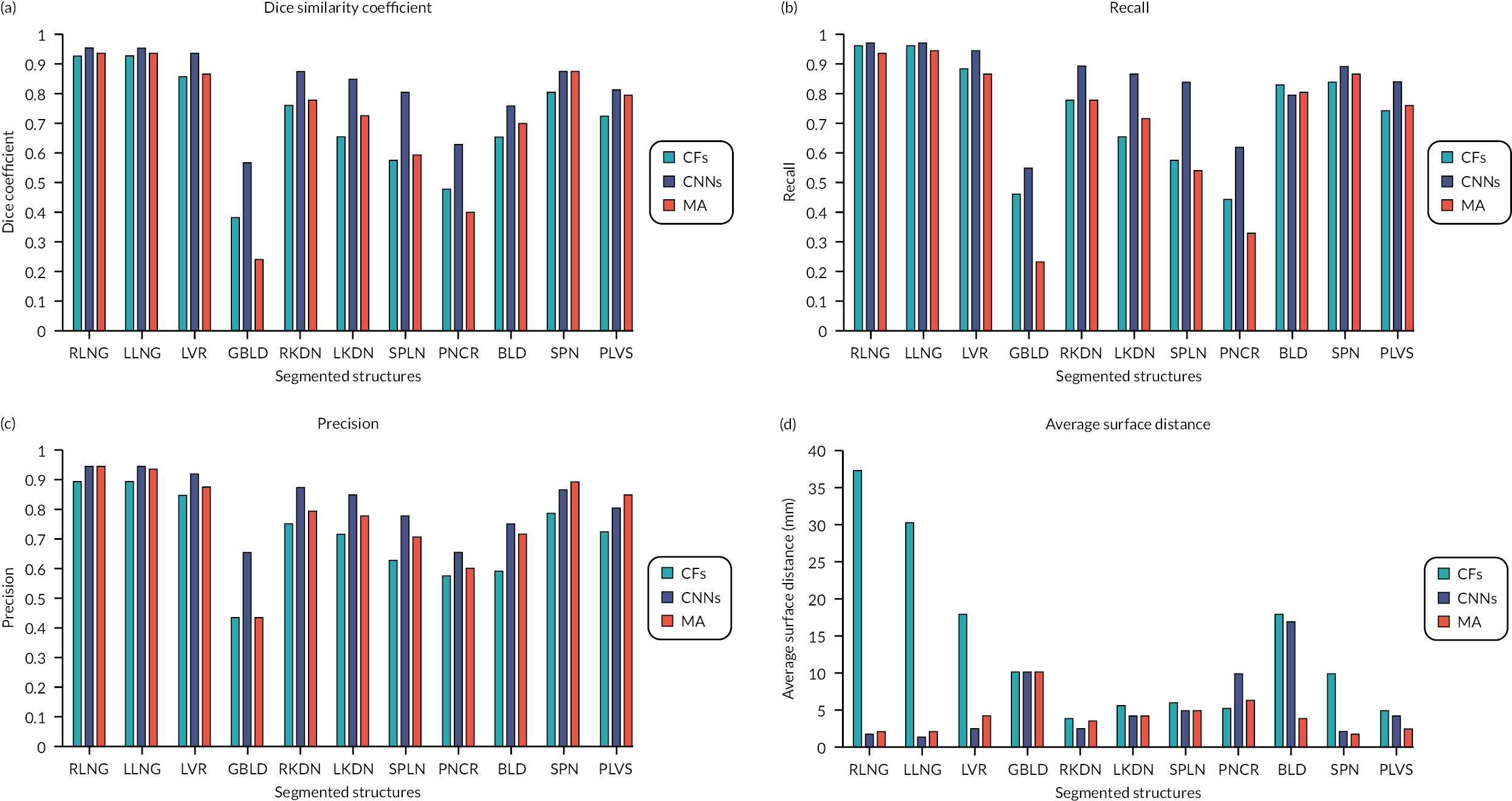
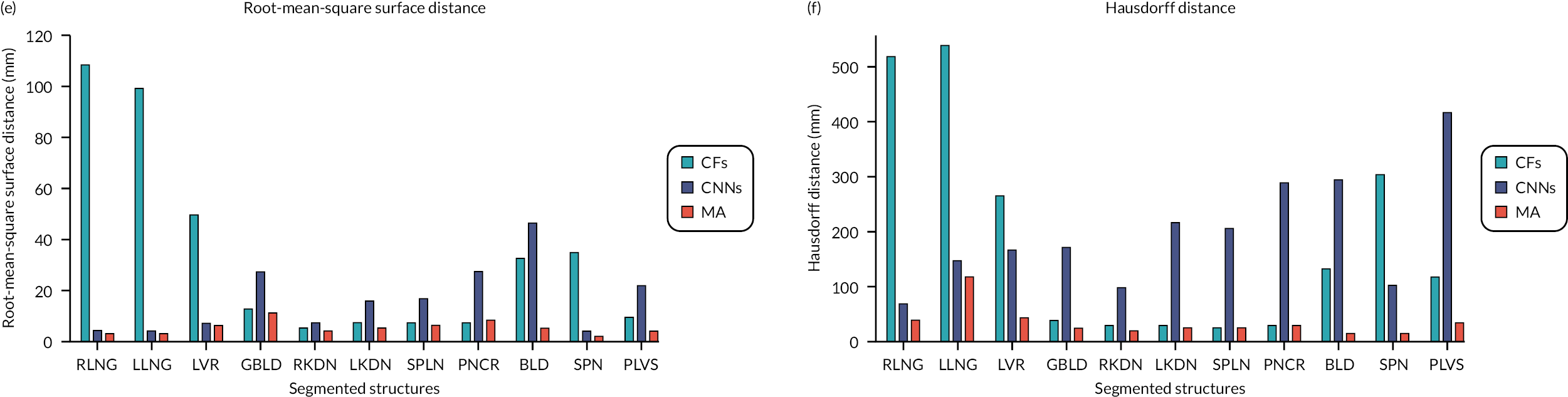
FIGURE 27.
Bar chart comparing the mean measured metrics. DSC (a), RE (b), PR (c), ASD (d), RMSSD (e) and HD (f) for the segmented organs (RLNG and LLNG: right and left lungs, GBLD, RKDN and LKDN, SPLN, PNCR and BLD) and bones (SPN and PLVS), when using T2w volumes and all imaging combinations (T2w + T1w + DWI) as inputs to CFs and B CNNs.
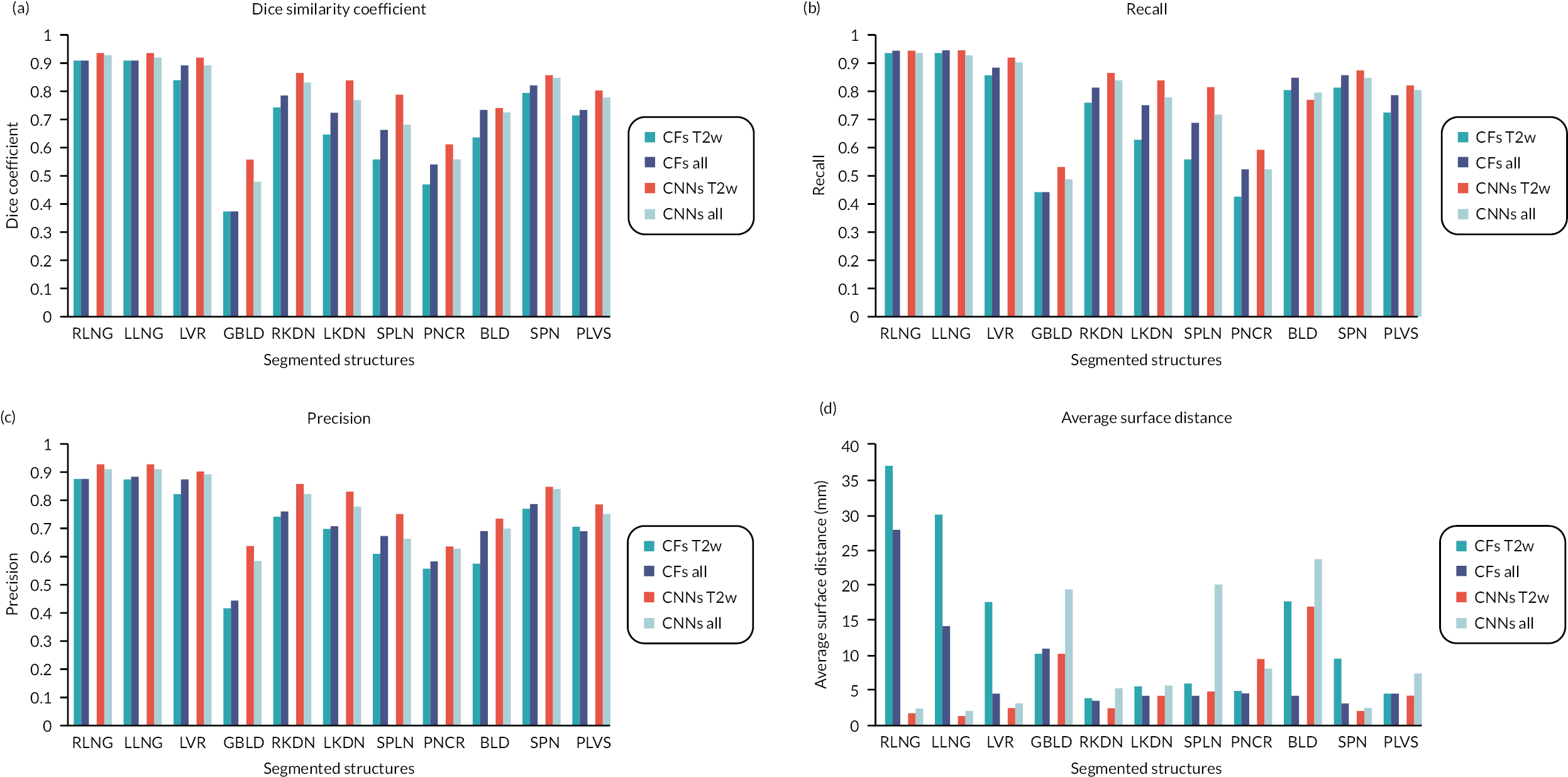
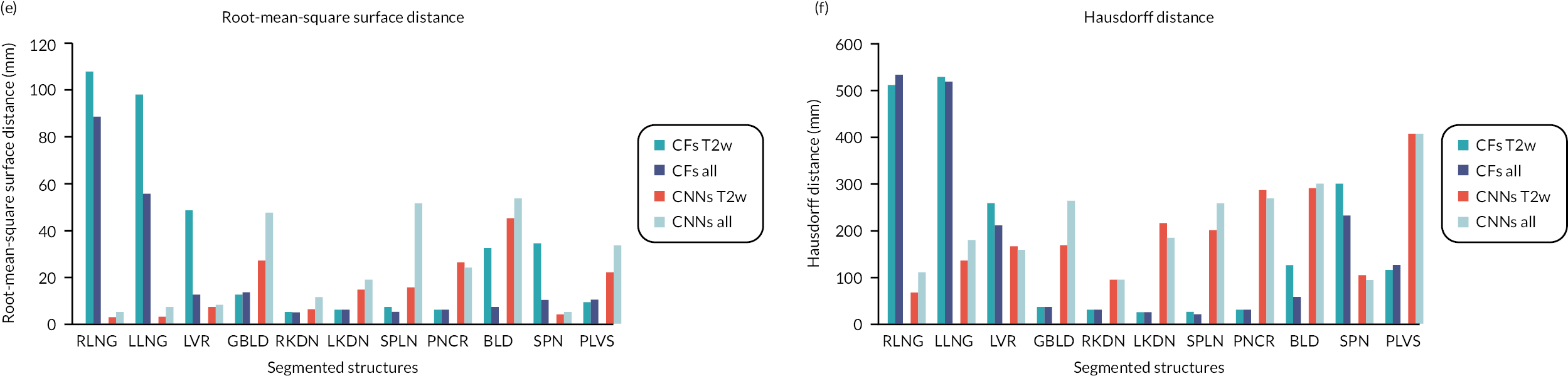
FIGURE 28.
Examples for LVR segmentation. Visual examples for predicting the segmentation quality of the LVR for a new image (slice from a T2w volume) shown on the top. Its GT segmentation (red coloured) is unknown in practice, and we want to estimate the quality of the predicted, automatic segmentation shown on the most right (green coloured). By taking the predicted segmentation as pseudo GT and training a RCA classifier we can obtain segmentations on a reference database with 24 images with available GT. The bar plot shows the real DSC in red and the different DSC values obtained for the reference images shown below. The green bar corresponds to the maximum DSC and is selected as predicted DSC for the new image according to Equation 1 which matches well the real DSC.
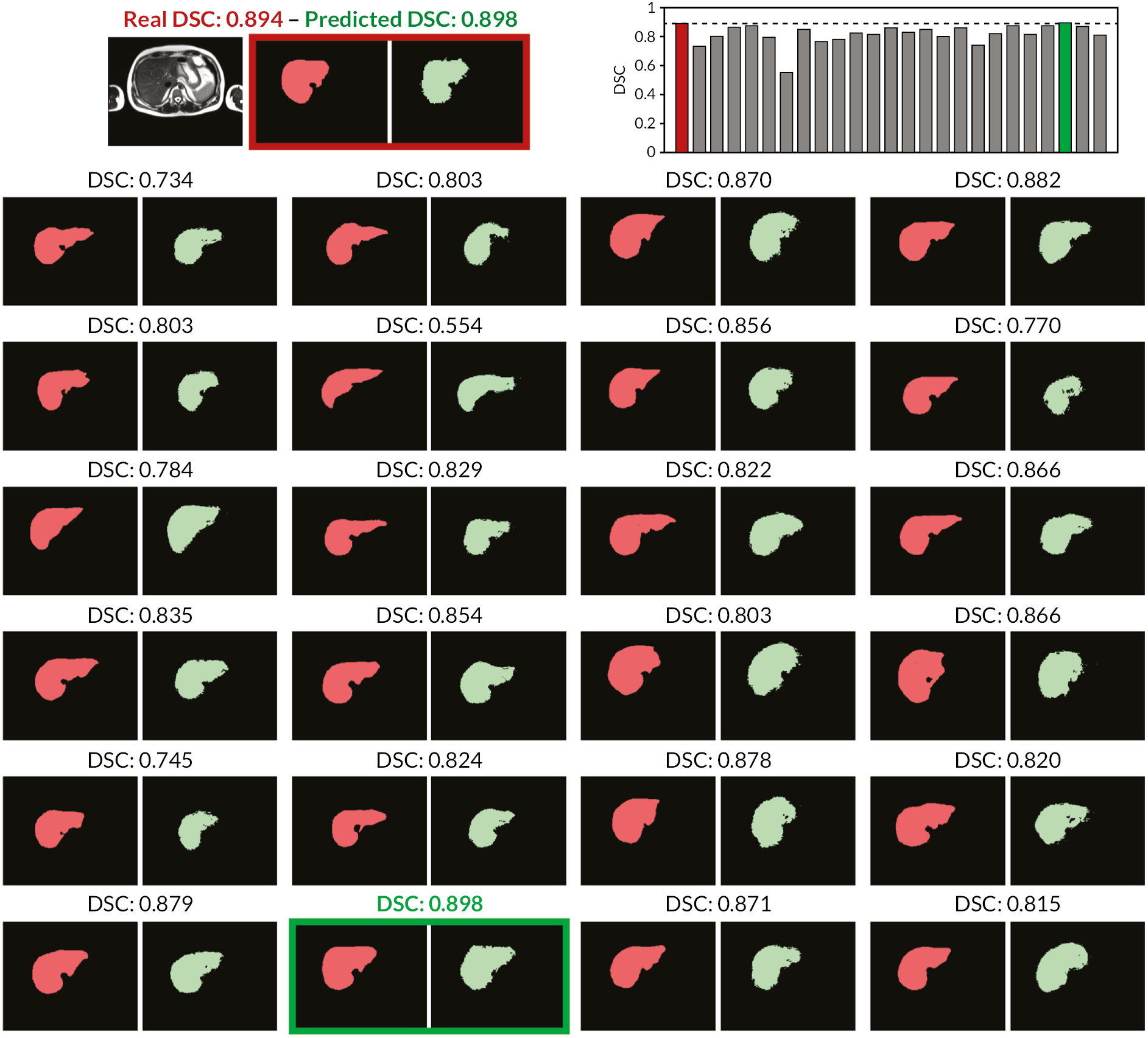
| Strategies | 0 | 2 | 5 | 10 | 15 | 30 (all) |
|---|---|---|---|---|---|---|
| FT random-n | 0.639 (0.149) | 0.665 (0.245) | 0.710 (0.172) | 0.765 (0.157) | 0.803 (0.086) | 0.830 (0.066) |
| FT best-n (real) | 0.639 (0.149) | 0.684 (0.225) | 0.723 (0.173) | 0.780 (0.176) | 0.750 (0.178) | 0.830 (0.066) |
| FT best-n (RCA) | 0.639 (0.149) | 0.631 (0.234) | 0.687 (0.191) | 0.753 (0.166) | 0.722 (0.229) | 0.830 (0.066) |
| PL best-n (real) | 0.639 (0.149) | 0.625 (0.162) | 0.639 (0.123) | 0.640 (0.131) | 0.589 (0.196) | 0.553 (0.145) |
| PL best-n (RCA) | 0.639 (0.149) | 0.614 (0.146) | 0.640 (0.125) | 0.619 (0.580) | 0.632 (0.139) | 0.553 (0.145) |
| S + T best-n (real) | 0.639 (0.149) | 0.692 (0.164) | 0.747 (0.152) | 0.763 (0.151) | 0.786 (0.282) | 0.831 (0.063) |
| S + T best-n (RCA) | 0.639 (0.149) | 0.711 (0.160) | 0.755 (0.148) | 0.776 (0.282) | 0.797 (0.277) | 0.831 (0.063) |
Appendix 2 Using ITK-SNAP for checking segmentation
-
Download the free software from the web – either for mac or pc.
-
Once downloaded, click on ‘file’ then ‘open main image’.
-
Browse to find the dicom file for your first case – wherever you saved these from the download file. I usually check the T2 first so I will take you through my steps.
-
Select T2 volume (note: it will open zipped files so you do not have to unzip). You will need to do a few clicks through some questions but then T2 should open in the viewer.
-
I like to also have the diffusion and ADC opened for reference so I can check against these. So next I click on ‘file’ then ‘add another image’ then I browse to open the Diffusion, then I repeat for the ADC.
-
You may need to increase the image size, by clicking on the magnifying glass icon. To do this, on mac, I click on the ctrl button and adjust with my mouse. However, you can also use the standard options just below the icons 1 × 2 × 4×.
-
I then fix the image contrast settings if needed. To do this click on ‘tools’ then ‘image contrast’. A scary graph pops up but it is actually really easy to use. You will see a list of the images open on the left of the pop up box. Choose the T2 then play with the round dots on the graph to make the image as you wish it to be. Then you can click on DW and do the same thing and then ADC if needed. You can always go back to image contrast and adjust at any time. This gets very quick once you are used to it.
-
Next you need to open the segmentation – in this case for T2 as this is the ‘main image’. To open the segmentation, go to ‘segmentation’ then ‘open segmentation’.
-
If you have also added extra images to help you (I always open the main image and then I add the other two sequences) then the segmentation will overlay on those but will likely not match – that’s fine.
-
The segmentation will then hopefully match the tumour – either primary, regional nodes, or mets – you need to check the position of these against the reference standard on the spreadsheet. To remove the segmentation push the S key, to make for opaque, push the A key, to make darker, push the D key.
-
If any need changing or adjusting, then click on the paintbrush. You can adjust the size of the paint brush on the sliding tool just below the paint brush. To add you just paint over the area you want to add. To remove you hold right click down and erase over the area you want to remove.
-
When happy, click ‘segmentation’ then ‘save segmentation as’ and add your initials to the end of the standard stem, for example _AR for me, just ahead of.nii.gz which is needed.
-
Then ‘unload the segmentation’ under the segmentation tab and upload the next one, for example, If you have done the primary, then upload the nodes or mets, until all done for T2.
-
If you need to add a new segmentation according to the consensus reference, then after you unload the previous segmentation, choose the paintbrush and colour in the appropriate item, for example Regional nodes. Then save as: STC-XXX_T2w_REGND_MET_initials or if LVR met the STC-XXX_DW_LVR_MET_initials etc.
-
Repeat the process for DW.
-
Then move onto the next case.
-
When you have done a set, please upload the new segmentations, with your initials, to the drop box under the appropriate segmentation folder, STC or STL.
Appendix 3 Phase 2 segmentation checking methods
MALIBO_Phase2_2019 (main folder shared with you in box).
Folder structure and where to find what you need:
There are three main folders:
-
Data:
-
This contains the DICOM data and is divided into folders for STL and STC. Within each are the folders for each patient containing the DICOM data T2, DWI and ADC. If there are multiple volumes, always use the one called ADCfinal_volume, regDWfinal_volume, (reDWvolume) and T2w_volume. These are the volumes that apply to the segmentations. I am sorry there are a number of options but I could not easily sort this out.
-
Please download the cases allocated to you and save in a folder on your computer. Please remember the data are shared confidentially with us by the sponsor of STREAMLINE study and we do not have permission to use the scans for any other purpose. Please delete the scans from your computer when you have finished checking the segmentations.
-
-
Segmentations:
-
The segmentations are divided up into DW and T2 segmentation folders. Once you have checked please save the checked segmentation into the folder with your name on it called ‘checked by (your name)’. Can you possibly add your initials at the end of the checked cases so in other words if I checked it, I would add (standard stem_AR).
-
To check the segmentations (see full explanation below): The segmentations will load up onto the image volume that you load into ITK-snap and should match the open ‘main image’ such that if you open the T2 volume as the ‘main image’, then open the T2 segmentation for it to match.
-
-
Documents (to download the reference standard):
-
MALIBO STC or STL DATA EXTRACTS 26.10.2018 are the documents with the reference standard. You can see the increasing degree of information at the lower tabs. So the first tab gives the main position and size of the tumour and stage. Then other tabs give the presence of nodal mets/skeletal mets/non skeletal mets and the details of these according to the STREAMLINE reference standard. You need to check against this to confirm that you agree with the segmentations.
-
Appendix 4 User manual for using 3D Biotronics platform
User Manual for training radiologists to use 3D Biotronics platform
Go to Google Chrome and open www.3dnetmedical.com/portal.
Login with your username and password (make sure this is the username that you have been given that links you to the MALIBO study folder), go then to PACS (upper row on the left side).
------- ----------- ------------ --------- ----------- --------- ---------- ------- -------- ----------
A window with 80 training cases will open.
For individual sear, please use the search bar. We suggest:
-
STL-039 -070, -103, -211.
-
STC-007, -089 (though overly does not work on this one), -096, -274.
By double clicking a STL/STC case will open.
Chose the second icon from the left (Series) to open all given series
-------- ------- -------- ------- --------- ---------- ----------- ---------- --------- -------
Select the appropriate images stated on the CRF sheet (T2 ax stack, DW ax stack, ADC ax stack, ML output if available), as well as other appropriate series (T1 dynamic LVR, lung, head/neck) by checking the icon box.
Afterwards click View to open.
-------- --------- --------- ------------- -------------- ------------ ------------- -----------
After a new window has opened, you can click F11 for full screen mode.
Use the indicated icon on the sidebar to open a dedicated PACS interface.
Choose sequences from the hanging protocol on the left sidebar by drag and drop.
The following image illustrates the recommended user layout for the readings:
Once you have established your own preferred layout, you can connect all sequences to scroll them together via:
Position all images at the same anatomic level then you must keep ‘Ctrl + Shift’ down and select each chosen sequence by clicking them. Each selected window will be marked with blue outlines (choose perhaps only the volumes/stacks and ML).
Choose right mouse button and select the icon on the open window to link them.
You have to hold Ctrl±Shift down until the link is complete!
Now you can scroll through the images.
--------- -------- ---------- ---------- ---------- ------------ ---------- ------------- -------------
For windowing, use right mouse button or select W/L in the right upper corner. This will open a new window, where the appropriate settings can be done.
To merge the T2 and the ML sequences together keep again Ctrl + Shift down and select both sequences by clicking them.
Afterwards, click with the right mouse button over the T2 image (!) and select the icon as indicated.
------------ ------------- ------------- ------------- ---------- ---------- --------- -------------
Once you have created the merged image you can further adjust the balance to a certain side by selecting the icon again and adjust the threshold button on the right side. We recommend a 65% threshold selection.
In case of improper windowing of the anatomic sequence (T2), as in this particular example.
Please load another T2 sequence into a different window.

As all 80 training cases from the Phase 2 trial have inconsistent sequences, there are some with four ML series! You can select and use all four in your training. Some cases have no-ML output.
The DM5 should be merged together to the T2 sequence and used primarily for the reading as it has the highest specificity.
The RF5 should be used secondarily and only to check for small lesions as it has high sensitivity, but low specificity.
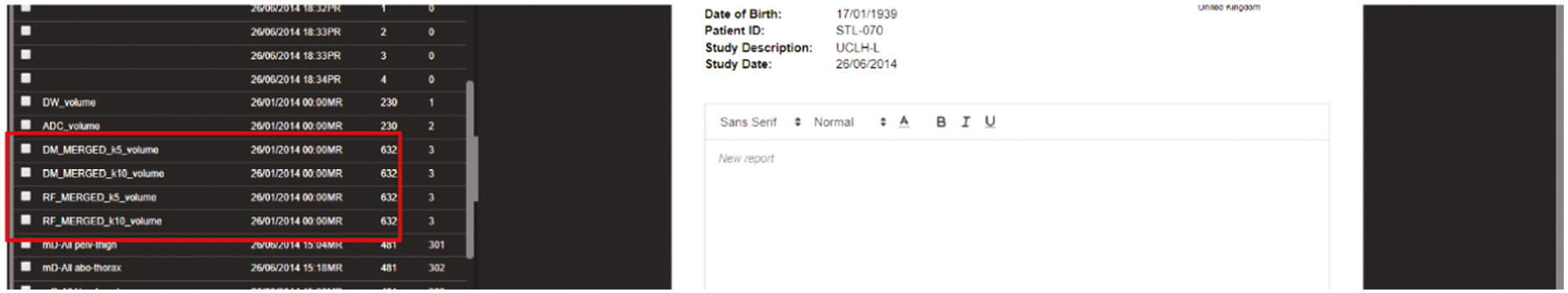
Scoring the ML output.
During your Phase 3 read, you will be asked to give ‘your opinion’ in relation to the primary tumour position and stage, nodal stage and presence and position of mets. Once ‘your opinion’ is recorded, you will be asked to go back and record what the ML output was like – on a score of 1 (no colour at all – no probability of cancer lesion), 2 (blue green colour – low probability of cancer lesion), 3 (orange, moderate probability of cancer lesion), 4 (red, high probability of cancer lesion). See examples below, score 2 for bowel, score 3 for bowel and score 4 for posterior mediastinum.
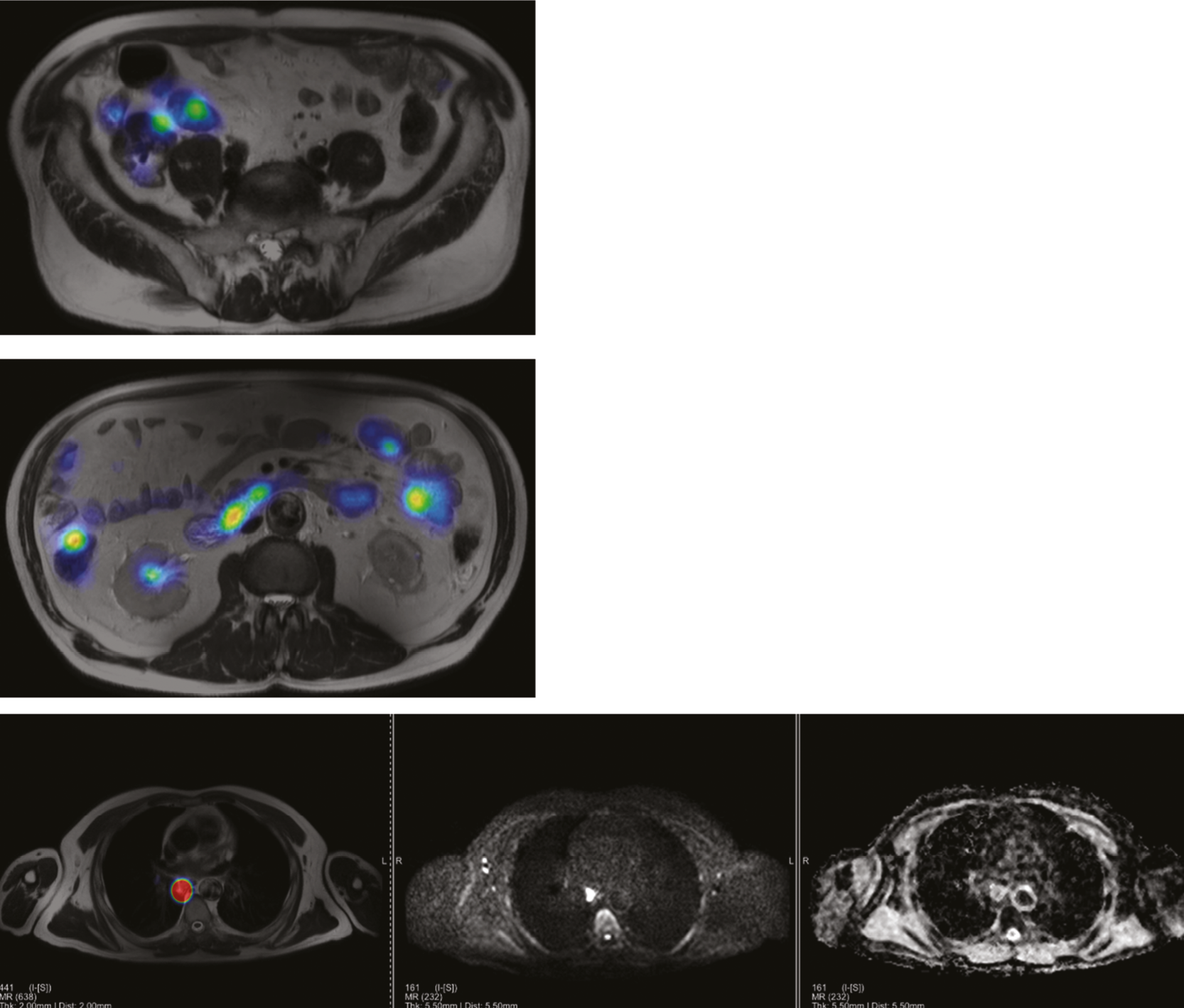
Appendix 5 MALIBO STC CRF

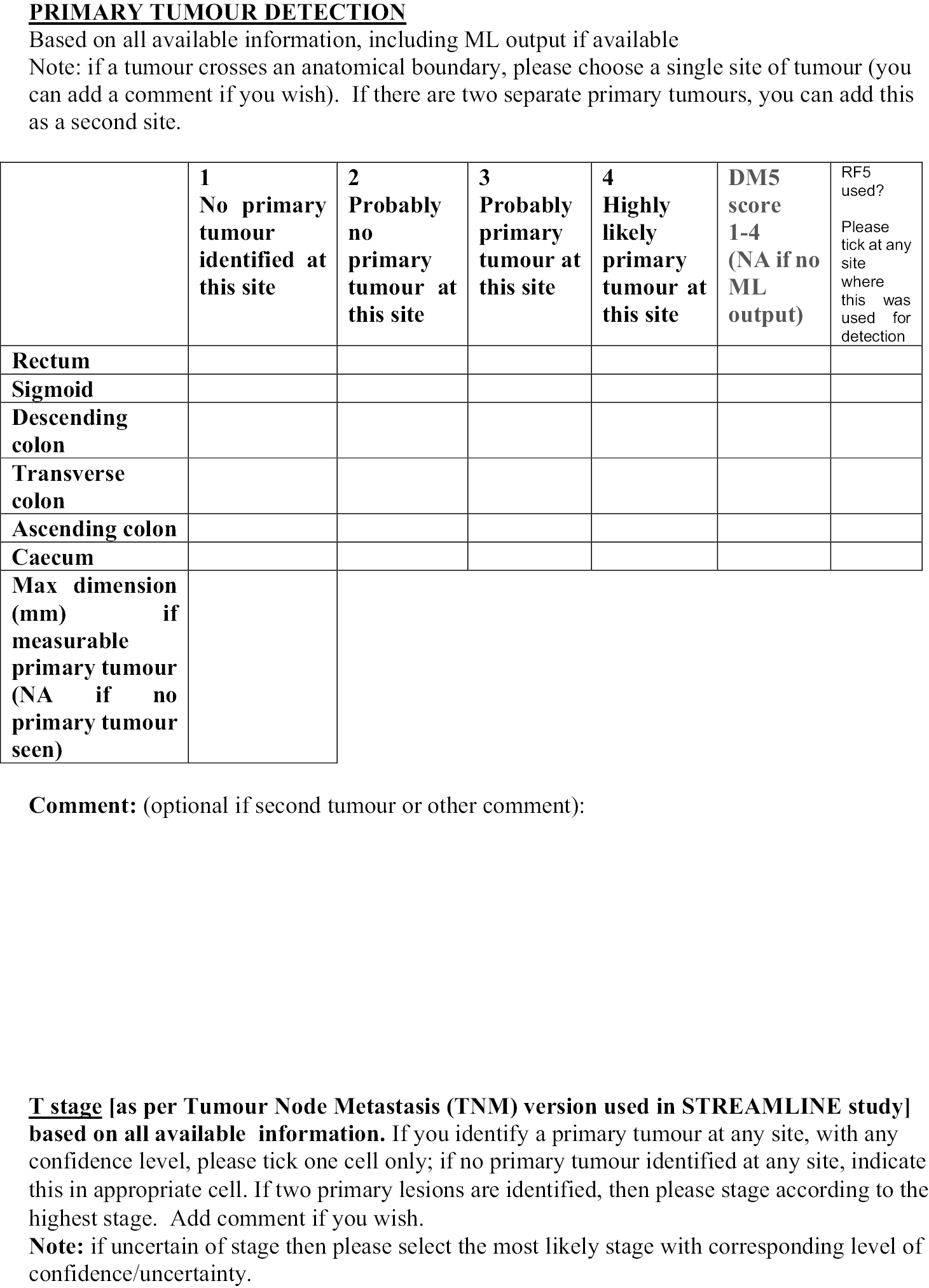
Appendix 6 MALIBO STL CRF

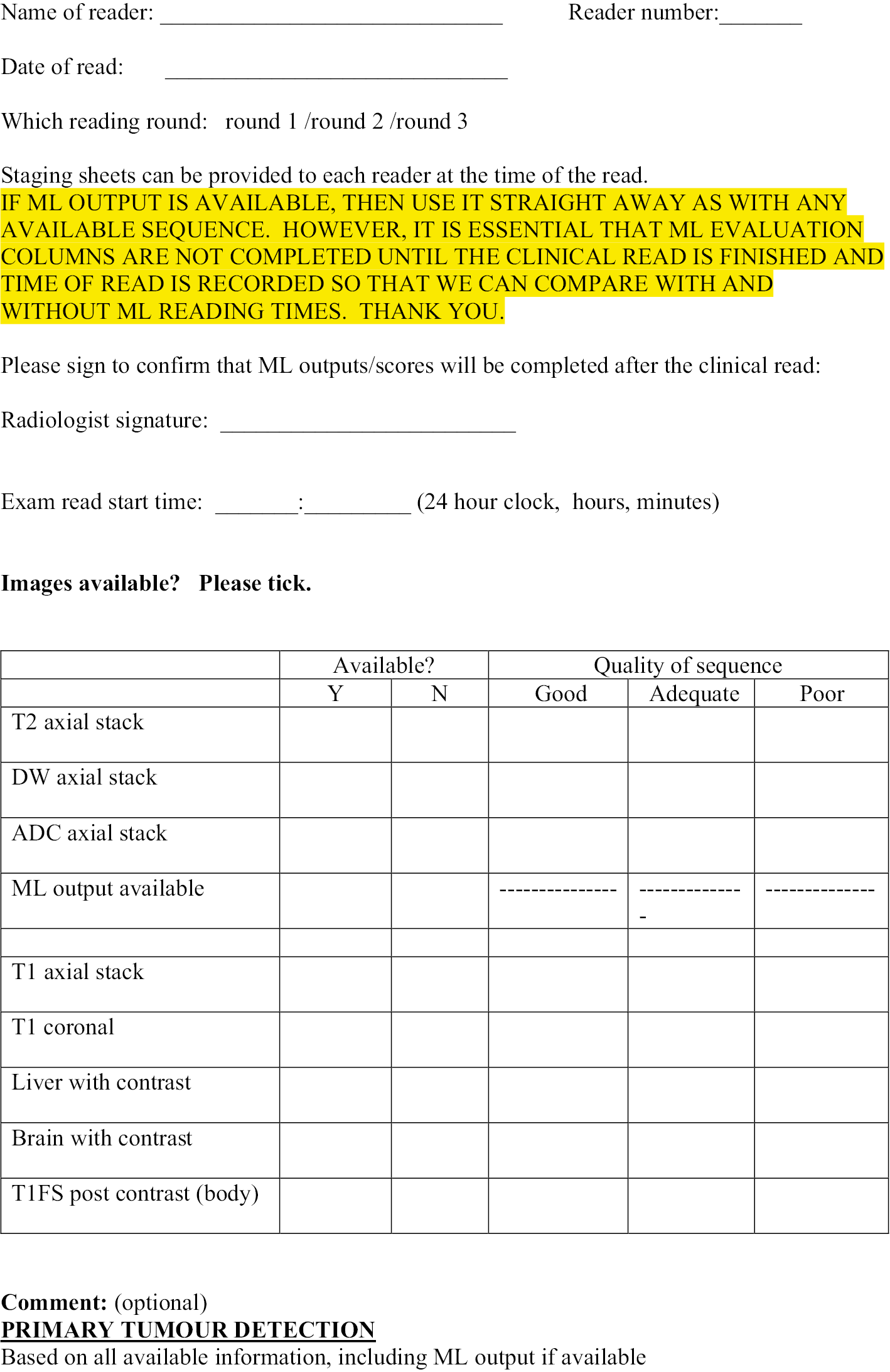
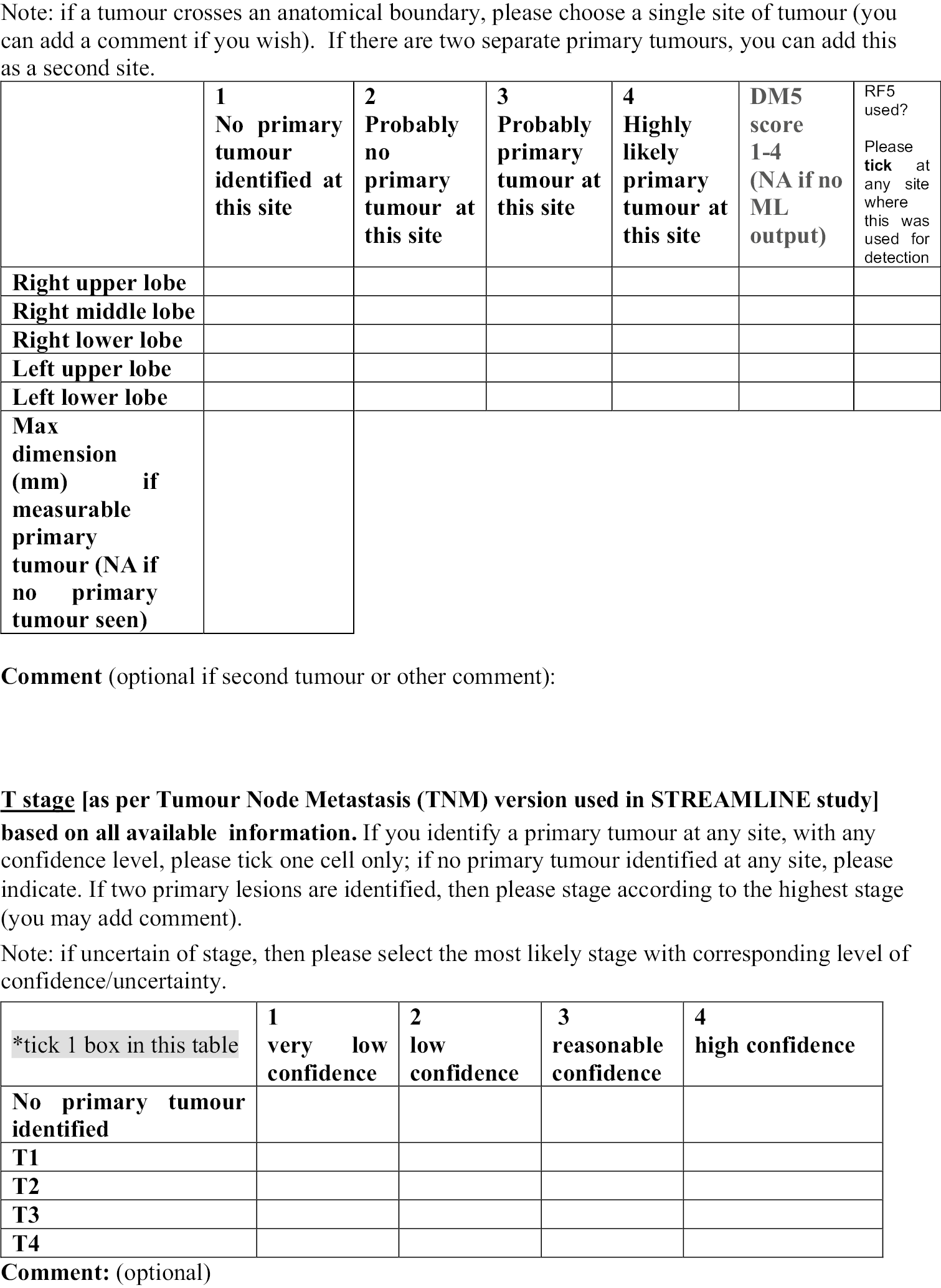
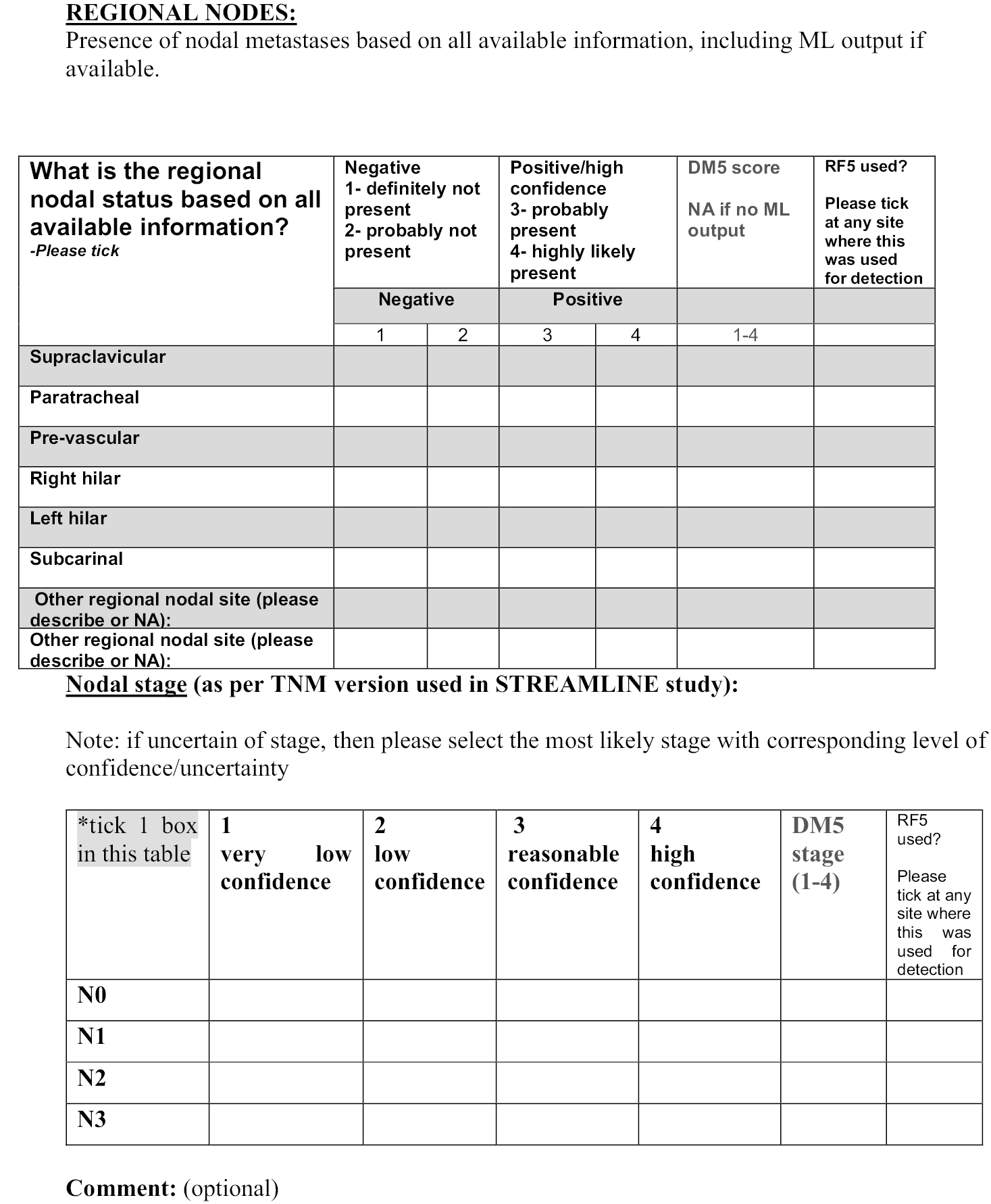
Appendix 7 Statistical analysis plan for machine learning in whole-body oncology project (version 1.1; 24 January 2020)
Statistical analysis plan background
The use of WB-MRI for the detection of metastatic disease is an active area of research in oncology imaging. In particular DW-MRI, which quantifies water diffusivity, can detect tumour sites in organs and bones. Limitations of WB-DW-MRI are the risk of false positives (as many ‘normal’ anatomical structures can look similar to pathological tissue) and the long reading times due to the large number of complex images. ML techniques have previously been used to assist in reading MRI data by developing algorithms to differentiate between benign and malignant cases, though not using DWI. In this study ML methods using WB-DW-MRI will be developed and evaluated for staging of patients with cancer.
Research questions
The primary research question is as follows. Is the specificity of WB-DW-MRI scans, in patients being staged for cancer, significantly improved with no subsequent loss of sensitivity when ML methods are applied? The secondary research question includes:
-
Can the RT of WB-DW-MRI scans be reduced, with a reduction of associated radiology costs, when ML techniques are employed to assist experienced radiologists?
-
Can interobserver variability be reduced by the use of ML methods in experienced or new WB-MRI radiologists?
-
Can the application of ML methods in WB-MRI increase the diagnostic accuracy delivered by less experienced radiologists?
-
Can intraobserver variability be reduced by the use of ML methods in experienced WB-MRI radiologists (exploratory)?
Study objective
The primary objective of this project is to compare the diagnostic accuracy of WB-DW-MRI, as read by experienced radiologists, in patients being staged for cancer, with and without the aid of ML methods against the reference standard of full clinical diagnosis at 12-month follow-up period.
The secondary objectives are in the following:
-
To compare the RT of WB-MRI scans.
-
To assess interobserver variability.
-
To test the diagnostic accuracy of non-experienced radiologists.
-
To achieve these objectives, we need to design statistical analysis experiments systematically.
Design
Patient data
This is an observational study using patient data from the STREAMLINE study. If deemed compatible with the developed algorithm, data from the MELT and MASTER studies may also be used.
The primary data source is STREAMLINE,8 a pair of multicentre prospective cohort studies that evaluate WB-MRI in newly diagnosed non-small cell lung cancer (250 patients; STREAMLINE-L; ISRCTN50436483) and colorectal cancer (322 patients; STREAMLINE-C; ISRCTN43958015). The primary objective for both studies was to evaluate whether early WB-MRI increases detection rate for metastases compared to standard NICE (www.nice.org.uk)-approved diagnostic pathways. Secondary objectives included assessing influence of WB-MRI on time to and nature of first major treatment decision following definitive staging. At 12-month patient follow-up, a multidisciplinary consensus panel defined the reference standard for tumour stage considering all clinical, pathological, post-mortem and imaging follow-up.
Cases from STREAMLINE are likely to have more non-nodal metastatic sites, such as LVR and lytic bone metastases. Therefore, additional cases of nodal disease and sclerotic bone metastases will be acquired from two other studies to ensure variation in the distribution of disease used to develop the ML algorithm. The MELT study (100 patients; NCT01459224) is a prospective observational cohort study to compare staging accuracy using WB-MRI with standard investigations in patients with newly diagnosed Hodgkin’s lymphoma. Data from the MASTER study, including cases with lymphoma and prostate cancer, will also be used (the myeloma cases from MASTER are unlikely to be used).
In addition, WB-MRI data sets from 51 healthy volunteers will be used; these have been collected under a separate ethics approval (ICREC 08/H0707/58). 14
Study design
The statistical study design is mainly in the second and third phases of this project. The anatomical atlases from Phase 1 were used in Phase 2 for anatomic mapping of healthy organs in the study scans.
In Phase 2, the ML produced a probabilistic map indicating the likelihood that the tissue is malignant. The scans and disease segmentations, based on the source study reference standard, were used to inform the algorithm using relatively sparse data. Training of the algorithm requires sufficient data from the three source studies to allow identification for different sites of disease, while holding back the data to be used in testing. The ML algorithm was refined in successive iterations until a final algorithm is obtained. An analysis of per-lesion sensitivity was performed at this stage using approximately 40–50 new patient data sets (to allow for sufficient positive cases). If the upper 95% CI of the sensitivity by algorithm ‘C’ is less than 80%, then further work on the algorithm will need to be undertaken prior to proceeding to Phase 3.
The assessment of study outcomes was carried out in the third phase of this project. A second set of WB-MRI data relating to 191 participants from the STREAMLINE, MELT and MASTER studies was read by expert radiologists, both with and without ML support, in a similar way to a cross-over design. The timing and order of the reads were randomised. Study outcomes were recorded for each read. For the diagnostic accuracy measures, the ‘gold standard’ was regarded as the reference standard from the main study. A subset of the scans was re-read (both with and without ML support) by another radiologist to assess interobserver variability. A further subset was read by non-experienced radiologists to estimate diagnostic accuracy in this group.
Groups for comparison
The groups for comparison were read with the aid of ML compared to reads without the aid of ML. The groups are paired as each scan will be read by the same radiologist with and without the aid of ML. The primary comparison was amongst experienced radiologists and a separate, secondary comparison was amongst inexperienced radiologists for a subset of 30 patients.
Study population
No patients were directly recruited into this study. Recruitment and scanning of patients have taken place under the separate contributing studies.
Study population for contributing studies
STREAMLINE L and C study inclusion criteria: histopathologically confirmed or suspected lung cancer or colorectal cancer being staged for initial treatment planning; written informed consent. Exclusion criteria include any contraindication to MRI scanning.
MELT (if applicable) study inclusion criteria: aged 6–18 years with participant/guardian informed consent, histologically confirmed Hodgkin’s lymphoma, treated with the Euronet chemotherapy regime. Exclusion criteria: contraindications to MRI, previous other malignancy or pregnancy/nursing.
MASTER (if applicable) study inclusion criteria: diagnosis of prostate cancer, lymphoma or myeloma.
Additional inclusion and exclusion criteria for this study
Phase 1 inclusion criteria: healthy volunteers aged 18–100 years, written informed consent.
Exclusion criteria: any co-existing medical illness, contraindications to MRI.
Phase 2 inclusion criteria: patient eligible for and consented to take part in one of the contributing studies (STREAMLINE C/L plus, if applicable, MELT and MASTER), patient completed the study imaging assessments and the reference standard from the source study is available.
Exclusion criteria: patients that consented to contributing study but did not undergo the scan, or the scan could not be adequately completed. In addition, cases will be excluded if the scan data is significantly corrupted during data transfer or contains significant artefacts, with marked obscuration of the images, such that the scan could not be reasonably processed for ML. This evaluation is made by the study MRI physicist and a record of each case will be maintained. Cases will also be excluded if the source study reference standard is not available.
Phase 3 inclusion criteria: any patients that were eligible for Phase 2 are automatically eligible for Phase 3.
Exclusion criteria: any patients whose reads were used in Phase 2 for the development of the ML algorithm are not considered for Phase 3.
Blinding
The study cannot be blinded to the addition of ML. The readers evaluating the sensitivity and specificity of WB-MRI with or without ML support will be blinded to the reference standard, including the original primary diagnosis and the stage of disease. To prevent any RE bias occurring in the Phase 3 analysis, readers will not be assigned scans originating from their home practice/site as part of the source studies.
Sample size
The sample size required for Phase 3 based on patients with no metastases is 141. This is based on McNemar’s test of paired proportions (McNemar, 1947), assuming ML support will improve specificity by 10%, from 86% to 96%, with a type 1 error of 0.05 (one-sided) and 90% power. Further, as per section 9.1 of the protocol, it is expected that 193 cases from the STREAMLINE study are available for use. Amongst the 193 patients from STREAMLINE studies, 51 are expected to have metastatic disease. Based on the above, we are anticipating an improvement in specificity of 10% from 86%. Regarding sensitivity, on the assumption that the sensitivity of WB-MR with ML support is no less than that of WB-MR alone (88%), a sample size of 51 metastases will provide an expected 95% CI for the sensitivity of WB-MR with ML support of 79–97%.
Randomisation
This study is not a randomised controlled trial; however, stratified randomisation will be employed to allocate scans between Phases 2 and 3 to reduce the risk of a difference in the scans analysed in these phases. Randomisation will be stratified on: (1) type of primary tumour (colon, lung), (2) presence of metastatic disease: LVR, bone, nodal. Randomisation will be undertaken by the study statistician when complete reference standard data are available and the cases that could not be processed for ML have been determined by the physicist.
Randomisation will also be used to assign and order the cases. First, cases will be randomly assigned to readers, maintaining an equal number of cases per reader as far as possible, based on the strata of type of primary tumour, presence of metastatic disease and site such that each radiologist with have a similar proportion of each ‘type’. Once assigned to each reader, a random selection of half the cases will be chosen to be assigned ML first, and the remainder non-ML first. These scans will comprise the first set received by the reader. The reverse set will be provided once the first set has been completed and a month has elapsed.
In order to allow for the inter-reader analysis, each radiologist will be assigned 10 reads that will be read only by the assigned radiologist. The remainder will be read by two radiologists, randomly assigned such that the proportions of primary tumour type and presence of metastatic disease remain even amongst all experienced radiologists.
In order to assess the effect of ML amongst non-experienced radiologists, the same randomisation procedure as detailed above for experienced radiologists will be utilised.
Analysis sets
The analysis population was included all WB-MRI scans where both the unassisted read and the ML-assisted read were completed by a radiologist.
Variables of analysis
Primary outcome
The primary outcome measure was the per-patient specificity of WB-MRI against the reference standard established in the main study. The observed data are summarised in Table 35.
| (a) | Patient classification without ML | (b) | Patient classification with ML | |||||
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Total | Negative | Positive | Total | |||
| Reference standard | Negative | a 1 | b 1 | a1 + b1 = n− | a 2 | b 2 | a2 + b2 = n− | |
| Positive | c 1 | d 1 | c1 + d1 = n+ | c 2 | d 2 | c2 + d2 = n+ | ||
| Total | a1 + c1 | b1 + d1 | a1 + b1 + c1 + d1 = N | a2 + c2 | b2 + d2 | a2 + b2 + c2 + d2 = N | ||
Specificity is defined as the proportion of patients with negative reference standard (metastasis not present anywhere) which have been correctly classified as negative by the radiologist, that is a1/(a1 + b1) without ML and a2/(a2 + b2) with ML.
Radiologist classification
Whole-body diffusion-weighted magnetic resonance imaging will be assessed for the presence of disease, using an imaging volume from the brain to mid-thighs. Reads will proceed using all sequences from the source study, with experienced and non-experienced WB-MRI readers. The sites of disease to be described will include the primary tumour site, presence and site of metastatic lesions and any significant incidental findings.
Reference standard
The reference standards for this study are taken from the contributing studies. For the STREAMLINE data, at 12-month patient follow-up, a multidisciplinary consensus panel defined the reference standard for tumour site and stage considering all clinical, pathological, post-mortem and imaging follow-up. The reference standard for the MELT study is contemporaneous MDT with all other staging, for example PET-CT and CT, at the time of diagnosis and initial staging.
Secondary outcomes
Per-patient sensitivity of WB-MRI against the reference standard established in the main study. Sensitivity is defined as the proportion of patients with positive reference standard (at least one metastatic deposit) which have been correctly classified as positive by the radiologist, that is d1/(c1 + d1) without ML and d2/(c2 + d2) with ML.
Per-lesion specificity of WB-MRI against the reference standard established in the main study. Specificity is defined as the proportion of lesions with negative reference standard which have been correctly classified as negative by the radiologist. A similar table to Table 1 can be constructed on a per-lesion basis.
Per-lesion sensitivity of WB-MRI against the reference standard established in the main study. Sensitivity is defined as the proportion of lesions with positive reference standard which have been correctly classified as positive by the radiologist.
Confidence of the tumour detection diagnosis at site ‘x’ by the radiologist reading the WB-MRI:
Tumour size: Size of the largest organ deposit, the second largest organ deposits, the number of additional deposits ≥ 6 mm, the number of additional deposits < 6 mm.
Reading time: The total time taken by the radiologist to read and report the WB-MRI in minutes, not including time taken to complete the CRF.
Interobserver variability: Interobserver variability in a subset of scans with reads by two different radiologists will be measured by the kappa coefficient, generalised to non-unique raters. 133,134 Both ML and non-ML reads will be repeated. For each participant i, i = 1, …, N, let xi be the total number of positive diagnoses (0, 1 or 2) from the two radiologists, and ρ¯ be the overall proportion of positive ratings. Then the between-participant mean square error is approximated by:
and the within-participant mean square error is:
and kappa is defined as:
Kappa is calculated separately for reads with and without ML assistance.
Intraobserver variability will be measured and compared using the kappa coefficient in the same method as above. Instead of comparing ‘Reader 1’ and ‘Reader 2’, ‘Period 1’ and ‘Period 2’ will be used instead to represent the two different times the same read was assessed. Again, Kappa is calculated separately for reads with and without ML assistance and compared.
Cost of radiology RT measured as per hour staff costs in Great British pounds for consultant radiologists.
Other variables
The following variables are recorded both as part of the reference standard and the radiologist assessment in Phase 3:
-
site of tumour;
-
tumour stage (N-stage; M-stage).
Confidence of the N/M-stage diagnosis by the radiologist reading the WB-MRI:

-
size of largest and second largest deposits at staging;
-
number of additional deposits < 6 mm;
-
number of additional deposits ≥ 6 mm;
-
radiologist ID;
-
radiologist level (experienced or non-experienced);
-
date of read.
Statistical methodology
General methodology
This SAP does not describe the ML aspect of the analysis. For clarity, the analysis described by this SAP is as follows:
-
interim analysis of the per-lesion sensitivity in Phase 2;
-
analysis of outcomes in Phase 3.
Statistical significance is set at p = 0.05 throughout and CIs. The primary outcome analysis comparing the per-patient specificities with and without ML will be a one-sided test and CI for the difference; all other tests and CIs are two-sided. The only hypothesis test and p-value to be presented concerns the primary outcome and, as such, there will be no adjustment for multiple testing.
Results will be presented according to the STARD guidelines135 where possible and checklist items have been noted in this SAP.
Missing data (STARD item 16)
This study will use scans and follow-up data already collected within the STREAMLINE studies and also MELT and MASTER studies (if deemed compatible). Patients with missing or indeterminate reference standard data, or with missing or inadequate scan data from the source studies, will not be eligible for this study, but numbers will be reported in the patient flow diagram. In Phase 3, any missing data from the radiology reads will be queried with the reader and attempts made to complete the data. Any missing data remaining will be reported but excluded from the analysis. If only part of a patient’s report is missing, then the remainder of the data will be used where possible. Inconclusive diagnoses in Phase 3 will be reported but excluded from the analysis.
As it is expected that missing data will be at a minimum, no data will be imputed for the purpose of the primary or secondary analysis.
Baseline characteristics (STARD items 20 and 21)
The following information (from the reference standard) will be described for all cases, with those used in Phases 2 and 3 shown separately:
-
location of primary tumour (colon, lung);
-
maximum dimension of primary tumour (cm, median and interquartile range);
-
location of metastatic disease (LVR, bone, nodal);
-
N-stage (N0, N1, N2, N3);
-
M-stage (M0, M1a, M1b).
Primary outcome analysis (STARD items 23 and 24)
The per-patient specificities of WB-MRI with and without ML, for experienced radiologists, against reference standard were presented with 95% CI calculated using the Wilson method. The normal approximation is unsuitable as the proportions are likely to be close to 1. The proportions were compared using McNemar’s test for paired proportions. 136 Using the same notation for the negative reference standard cases as used in Table 33 (a1, b1, a2, b2, n−), we can construct a 2 × 2 table to compare the (paired) proportions of patients classified as negative using the reference standard that are correctly identified as negative by the radiologist with and without ML (see Table 36):
| With ML | ||||
|---|---|---|---|---|
| Negative | Positive | Total | ||
| Without ML | Negative | I | J | a1 |
| Positive | K | L | b1 | |
| Total | a2 | b2 | n − | |
The null hypothesis is that the two specificities (the marginal probabilities in Table 2) are the same, and the alternative hypothesis is that the specificity is higher with ML:
McNemar’s test statistic is:
Under the null hypothesis, T follows a χ2 distribution on 1 degree of freedom. The p-value from a one-sided test will be reported.
Results will be expressed as an absolute difference in proportions:
As a one-sided test is being used, assuming the point estimate of the absolute difference in proportions is positive, the one-sided 95% CI is:
where:
A one-sided test is being used as it is not expected that specificity could be worsened by the addition of ML assistance. In the event that the point estimate is negative, a two-sided test and CI will be presented, acknowledging the loss in power to demonstrate a difference.
Should the number of discordant pairs (j + k) be small, an exact test will be performed instead. The p-value is calculated from the binomial distribution as:
Secondary outcome analysis
Per-patient sensitivity, per-lesion sensitivity and per-lesion specificity of WB-MRI with and without ML, for experienced radiologists, against the reference standard were reported with 95% CIs. The difference in sensitivity/specificity and 95% CI was calculated similarly to the primary outcome though no hypothesis test was performed.
Per-patient sensitivities and specificities of WB-MRI with and without ML, for inexperienced radiologists, against reference standard was reported with 95% CIs. The difference in sensitivity/specificity and 95% CI was calculated similarly to the primary outcome though no hypothesis test will be performed.
| Confidence with ML | ||||
|---|---|---|---|---|
| 1 | … | 4 | ||
| Confidence without ML | 1 | |||
| … | ||||
| 4 | ||||
| Without ML | With ML | Absolute difference % (95% CI)a |
|
|---|---|---|---|
| Per-patient specificity, n/N % (95% CI) | |||
| Per-patient sensitivity, n/N % (95% CI) | |||
| Per-lesion specificity, n/N % (95% CI) | |||
| Per-lesion sensitivity, n/N % (95% CI) |
| Experienced radiologists | Inexperienced radiologists | |||
|---|---|---|---|---|
| Without ML | With ML | Without ML | With ML | |
| Confidence, median (IQR) | ||||
| Reference positive | ||||
| Reference negative | ||||
| RT, median (IQR) | ||||
| Interobserver variance, κ (95% CI) | ||||
| Intraobserver variance, κ (95% CI) | ||||
Confidence: The agreement in confidence (on a scale of 1–4) between WB-MRI with and without the ML support will be described using a 4 × 4 table, as below, and visualised using a bar charts. This will be done overall and by reference standard diagnosis.
Tumour size as assessed with and without ML will be compared using scatterplots. The size of the largest deposit as measured with ML will be plotted against the size of the largest deposit as measured without ML. A reference line of x = y will be added to indicate the same size measured by both methods, and colours used to indicate the reference standard diagnosis. This will be repeated for the size of second largest deposit, and the number of additional deposits.
Reading time will be compared between WB-MRI with and without the ML support by calculating the paired difference (RT with ML – RT without ML) for each scan, as they are read by the same radiologist. The paired differences will be analysed using regression, adjusting the standard errors for clustering at the radiologist level and including covariates: order of reads (ML first/second) and type of primary tumour (colon, lung). The estimated mean difference in RT (with 95% CI) at the mean level of covariates will be obtained from the intercept term in the regression. The associations with read order and type of tumour will also be reported. A 1-month gap between reading sessions under the provision that the gap is sufficient enough to remove any RE bias. In the event where times between reading sessions vary between readers, an additional covariate may be added and its interaction with other covariates to explore dilution of their effects due to RE (time between reads in days).
A transformation of the dependent variable may be required if the regression assumptions are not met. If the assumptions are still not met after transformation, a Wilcoxon signed rank test137 was used to test for an unadjusted difference in RT.
Interobserver variance: Summary statistics of the proportions of concordant and discordant diagnosis between two experienced radiologists will be reported for both methods. Interobserver variance will be measured by kappa (κ) coefficient as described in Reading platform. A 95% CIs for kappa will be calculated using bootstrapping (bias-corrected method). We will assess whether the interobserver variance is reduced using ML by comparing the estimated values of kappa with and without ML, using the method of Gwet138 which is similar to the paired t-test.
Intraobserver variance: this will be calculated and compared using the same methodology as above replacing the reads from two different experienced radiologists with tow reads from the same radiologist, taken at a different time.
Costs of radiology RT: Estimated cost savings per WB-MRI will be calculated by multiplying any reduction in RT in hours (as per the above RT analysis) by the associated hourly radiologist reading costs. If appropriate, this will be performed separately for cases with and without metastases.
Analysis for inexperienced readers
In addition to the above analyses described in Primary outcome analysis (STARD items 23 and 24) and Secondary outcome analysis, to satisfy the third secondary objective, a duplicate set of analyses will be carried out based on reads carried out by a cohort of approximately 7–8 non-experienced readers. Likewise, difference in ML-effect sizes between experienced and non-experienced cohorts will be assessed to investigate whether any effect derived from using ML output is affected by the experience of reader.
The non-experienced reader cohort may potentially have a variance in abilities (e.g. some may be at consultant level with experience of reporting cancer while some are at the trainee level). In the event where this occurs, an additional set of sensitivity analyses will be run allowing for the inclusion of a stratification variable for ‘ability’ (consultants/trainees/etc.).
Subgroup analysis
The following subgroup analysis was performed for diagnostic accuracy outcomes, with subgroups defined by the reference standard:
-
size of lesion (above or below median);
-
location of primary tumour (colon, lung);
-
location of metastatic disease (LVR, bone, nodal);
-
N-stage;
-
M-stage.
Interim analysis
Interim analysis, concerning per-lesion sensitivity, will be undertaken as part of Phase 2. It will be carried out using 40–50 new patient data sets. The per-lesion sensitivity will be calculated as the proportion of metastatic lesions which are correctly identified by the ML algorithm. Correct identification is defined as achieving a particular threshold for the Dice coefficient, which quantifies the overlap of the areas identified as lesions by the ML algorithm with the true lesion area, as defined by the clinical expert. A suitable threshold for the Dice coefficient will be defined as part of the ML process, prior to the interim analysis. A 95% CI for the sensitivity will be calculated using the Wilson method. 139 We will require the upper 95% CI of the sensitivity no less than 80%. If this is not met, then further work on the algorithm will be required. This is not a formal stopping rule, but rather the check to prevent proceeding to Phase 3 if the algorithm is not identifying lesions at all. The cases used for the first interim analysis will not have been used for ML training or read by radiologists. Therefore, if the algorithm is sufficiently sensitive to proceed to Phase 3, the cases used in the first interim analysis can be part of the validation set. They will therefore be selected from those allocated to Phase 3.
A proposed interim analysis for per-patient specificity, for reads assisted by the ML algorithm in Phase 3, will no longer be performed due to time constraints.
Sensitivity analysis
Any difference in specificity seen in the primary analysis may be affected by the order or timing of the scans. It is also possible that the statistical significance is overstated if outcomes for the same radiologist are correlated. Conditional logistic regression will be used to obtain an odds ratio comparing the specificity with and without ML adjusting for the scan order (ML first/second), time between scans (if regression assumptions in section Secondary outcome analysis do not hold), and using robust standard errors to allow for clustering by radiologist.
To investigate whether the quality of sequence has an effect on the study results the primary analysis will be re-run using just reads where either the T2 axial stack or the DW axial stack has been deemed ‘good’ quality. The same set of reads will be used to re-run secondary analyses for per-patient specificity and sensitivity. To investigate the effect sequence quality on read time, the regression model in section Secondary outcome analysis will include covariates for T2 stack quality and DW stack quality.
As per Analysis for inexperienced readers, in the event where the non-experienced reader cohort has varying grades of ability/experience an additional sensitivity analysis will be run within the non-experienced cohort, including an additional variable to consider any effect this ability difference may have.
List of abbreviations
- 3D
- three-dimensional
- ADC
- apparent diffusion coefficient
- AFs
- atlas forests
- ANOVA
- one-way analysis of variances
- ASD
- average surface distance
- AUC
- area under curve
- BLD
- bladder
- CNNs
- convolutional neural networks
- CPU
- central processor unit
- CRF
- case report form
- CRUK
- Cancer Research United Kingdom
- CT
- computed tomography
- DARCA
- domain adaptation using RCA
- DSC
- Dice similarity coefficient
- DWI
- diffusion weighted imaging
- EPI
- echo-planar imaging
- FCN
- fully convolutional network
- FM
- feature map
- GBLD
- gallbladder
- GPU
- graphics processor unit
- GT
- ground truth
- HD
- Hausdorff distance
- ICREC
- Imperial College Research Ethics Committee
- JI
- Jaccard index
- LKDN
- left kidney
- LLNG
- left lung
- MA
- multi-atlas
- MAE
- mean absolute error
- MALIBO
- MAchine Learning In whole Body Oncology
- MDT
- Multidisciplinary Tumour Board
- MeSH
- medical subject headings
- ML
- machine learning
- MRI
- magnetic resonance imaging
- NICE
- National Institute for Health and Care Excellence
- NIfTI
- Neuroimaging Informatics Technology Initiative
- NIHR
- National Institute for Health and Care Research
- PACS
- picture archiving and communications system
- PET-CT
- positron emission tomography–computed tomography
- PLVS
- pelvis
- PNCR
- pancreas
- PR
- precision
- RCA
- reverse classification accuracy
- RE
- recall
- RF
- random forest
- RKDN
- right kidney
- RLNG
- right lung
- RMSSD
- root-mean-square surface distance
- ROI
- regions of interest
- RT
- reading time
- RVD
- relative volume difference
- SAP
- statistical analysis plan
- SPLN
- spleen
- SPN
- spine
- STAPLE
- simultaneous truth and performance level estimation
- SVM
- support vector machine
- UCL
- University College London
- WB-MRA
- whole-body magnetic resonance angiography
- WB-MRI
- whole-body magnetic resonance imaging
Notes
Supplementary material can be found on the NIHR Journals Library report page (https://doi.org/10.3310/KPWQ4208).
Supplementary material has been provided by the authors to support the report and any files provided at submission will have been seen by peer reviewers, but not extensively reviewed. Any supplementary material provided at a later stage in the process may not have been peer reviewed.
