Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 12/34/01. The protocol was agreed in July 2012. The assessment report began editorial review in February 2013 and was accepted for publication in August 2013. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors:
none
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2014. This work was produced by Westwood et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Objective
The overall objective of this project is to summarise the evidence on the clinical effectiveness and cost-effectiveness of commercial or UK in-house epidermal growth factor receptor tyrosine kinase (EGFR-TK) mutation (hereafter to be referred to as EGFR mutation) tests to identify those previously untreated adults with locally advanced or metastatic non-small cell lung cancer (NSCLC) who may benefit from first-line treatment with EGFR-TK inhibitors (EGFR-TKIs; gefitinib or erlotinib). In order to address the clinical effectiveness, data on the analytical validity of the different EGFR mutation tests (sensitivity/specificity for detection of mutations known to be linked to treatment effectiveness) are required. Because methods of testing EGFR mutation status differ both in terms of the mutations targeted and limit of detection (the lowest proportion of tumour cells with a mutation that can be detected), the definition of EGFR mutation positive varies according to which test is used. All testing methods are essentially reference standard methods for classifying mutation status, as defined by the specific test characteristics, and it is therefore not useful to select any particular test as the reference standard. In addition, the relationship between the effectiveness of EGFR-TKIs and the presence of specific mutations or combinations of mutations, as well as the relationship between the effectiveness of EGFR-TKIs and the level of mutation present, is uncertain. Therefore, the following research questions were formulated to address the review objectives:
-
What is the technical performance of the different EGFR mutation tests (e.g. proportion tumour cells needed, failures, costs, turnaround time)?
-
What is the accuracy (clinical validity) of EGFR mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors (TKIs)? If individual patient data (IPD) are available, what are the associations between individual mutations detected and patient outcome?
-
How do clinical outcomes from treatment with EGFR-TKIs vary according to which test is used to select patients for treatment?
-
What is the cost-effectiveness of the use of the different EGFR mutation tests to decide between standard chemotherapy or anti-EGFR-TKIs?
Chapter 2 Background and definition of the decision problem(s)
Population
The indication for this assessment is the detection of mutations in the EGFR-TK oncogene in previously untreated adults with locally advanced or metastatic NSCLC. The presence of EGFR mutations can affect the response of tumours to standard chemotherapy and oral EGFR-TKIs, and mutation status is thus used to select the most appropriate course of treatment. 1,2
The 2010 age-standardised incidence rate for lung cancer in England was 55.9 per 100,000 in men and 37.9 per 100,000 in women. Since 2001 the incidence rate has declined by 15% for men and increased by 10.8% for women. 3 In 2009 there were 35,406 new cases of lung cancer recorded in England and Wales, and in 2010 there were 29,914 deaths from lung cancer. 4 The National Lung Cancer Audit (NLCA) data for 2010 included 32,347 new cases for England and Wales, of which 19,379 (71.9%) were histologically confirmed NSCLC and 5932 (18%) were stage IIIB or IV NSCLC. 5 The prevalence of EGFR mutations in NSCLC varies widely with population ethnicity. Estimates from observational studies ranged from 4.5% in a study conducted in Italy6 to approximately 40% in two studies conducted in Japan and Taiwan. 7,8 The great majority of EGFR mutations occur in adenocarcinomas; from three studies,6–8 with a total of 1238 participants (189 with EGFR mutation-positive tumours), only one mutation occurred in a patient with tumour cytology other than adenocarcinoma. The prevalence of EGFR mutations in NSCLC (adenocarcinoma) therefore ranged from 10.4% in the Italian study6 to 50% and 39% in the Japanese and Taiwanese studies, respectively. 7,8
Lung cancer incidence and mortality rates are strongly age related. In the UK between 2007 and 2009 three-quarters of new cases were diagnosed in people aged > 65 years, and between 2008 and 2010 around 78% of lung cancer deaths were in people aged > 65 years. In the UK, lung cancer incidence and lung cancer mortality rates in men have been declining since the early 1970s but both continue to increase in women. Gender-specific time trends in lung cancer reflect patterns in past smoking behaviour. 4 Lung cancer incidence and mortality rates are also related to socioeconomic factors. Age-standardised incidence rates are twice as high and age-standardised mortality rates are around three times higher in the most deprived wards of England and Wales compared with the least deprived wards. 4,9
Lung cancer survival rates are generally low because a substantial proportion of patients present at an advanced stage, when curative treatment is no longer possible. 4,10 The latest cancer survival statistics for England and Wales for patients diagnosed in the period 2005–9 and followed up to 2010 show 1-year age-standardised survival rates of 27% in men and 30% in women; 5-year age-standardised survival rates were 7% and 9% in men and women, respectively. 11
Intervention technologies
There are a variety of tests available for EGFR mutation testing; Table 1 summarises the methods currently used in UK NHS laboratories participating in the UK National External Quality Assurance Scheme (NEQAS) pilot scheme for EGFR mutation testing, which responded to a request to provide information to the National Institute for Health and Care Excellence (NICE). The tests used can be broadly classified into two subgroups: mutation screening and targeted mutation detection. Mutation screening tests screen samples for all EGFR mutations (known and novel), whereas targeted tests analyse samples for specific known mutations. Successful mutation analysis is dependent on a sufficient quantity of tumour tissue in the sample. The limit of detection varies between different assay methods, with some studies reporting mutation detection when the proportion of tumour cells in a sample is < 10% and Sanger sequencing requiring up to 25% of tumour cells (see Table 1). 12,13 There is some evidence that EGFR mutations can be accurately detected in plasma;14 however, biopsy tissue or cytology samples remain the gold standard. Clinical opinion, provided by specialist advisors during scoping, suggested that plasma testing is currently a ‘research-only’ application, which should not be included in this assessment. Further, clinical opinion also stated that cytology samples should be considered equivalent to biopsy. In 2009, a European multidisciplinary workshop ‘EGFR Testing in NSCLC: From Biology to Clinical Practice’ was held by the International Association for the Study of Lung Cancer and the European Thoracic Oncology Platform. This workshop included 122 molecular biologists, pathologists, chest physicians, surgeons and medical oncologists, and produced consensus recommendations for the implementation of EGFR mutation testing in Europe. 12 Although there was no consensus on which laboratory test should be used, emphasis was placed upon the importance of standardisation and validation, and a recommendation was made that EGFR mutation testing should be undertaken only in a quality assured, accredited setting. 12 Participants also agreed that the decision to request EGFR mutation testing should be made by the treating physician and that results should be reported within 7 working days of request. 12
| Sequencing method | Targeted (mutations targeted)/screening test | Methodology |
|---|---|---|
| Commercial tests | ||
| Therascreen® Kit/ARMS (Qiagen, Venlo, the Netherlands) Therascreen® Pyro kit (Qiagen, Venlo, the Netherlands) |
Targeted (version 1–28 mutations, version 2–29 mutations) Targeted (28 mutations) |
Real-time PCR Pyrosequencing |
| Roche cobas® EGFR Mutation Testing Kit (Roche Molecular Systems, Inc., Branchburg, NJ, USA) | Targeted (41 mutations) | Real-time PCR |
| In-house tests | ||
| Sanger sequencing | All mutations | Usually PCR but variation in detail |
| Fragment length analysis | Varies | PCR followed by fluorescence to determine fragment size |
| Pyrosequencing | Varies | PCR followed by pyrosequencing reaction |
| TaqMan/real-time PCR/EntroGen | Targeted (details unclear) | Real-time PCR |
| HRM analysis | All mutations | PCR followed by HRM |
| Single-strand conformation analysis | Screening (> 98% of all mutations) | PCR followed by electrophoresis |
| SNaPshot/RFLP/other | Targeted (details unclear) | PCR RFLP |
| Mass spectrometry | Targeted (details unclear) | Mass spectrometry |
| Next-generation sequencing | Screening | DNA first fragments into small segments that can be sequenced in parallel reactions |
Targeted mutation detection tests
The different targeted tests look for different numbers and combinations of EGFR mutations and are able to detect different levels of mutation. For example, a sample may contain a high proportion of tumour cells, but only a low proportion of these may harbour mutations, and a low proportion of mutation, although detectable by some tests, may not be clinically significant. Thus tests may differ in their ability to accurately select patients who are likely to benefit from chemotherapy with TKIs. EGFR mutations are known to be restricted to four exons (18–21), with deletions in exon 19 and point mutations in exon 21 accounting for > 90%. 6,7,13 Observational studies have linked deletions in exon 19, point mutations at codons 858 and 861 of exon 21, and point mutations at codon 719 of exon 18 to tumours that are responsive to treatment with gefitinib. 13,15
The licensed indication for the TKIs gefitinib and erlotinib is treatment of locally advanced or metastatic NSCLC in patients who are previously untreated and whose tumours test positive for EGFR mutations. NICE Technology Appraisal 192 recommends gefitinib as an option for the first-line treatment of people with locally advanced or metastatic NSCLC if they test positive for an EGFR mutation. 1 The mutation test used in the trial that informed NICE Technology Appraisal 192 was version 1 of the Therascreen® EGFR polymerase chain reaction (PCR) Kit (Qiagen, Venlo, the Netherlands); it should be noted that this version is no longer being marketed and has been superseded by version 2, the Therascreen® EGFR RGQ PCR Kit (Qiagen, Venlo, the Netherlands). NICE Technology Appraisal 258 recommends erlotinib as an option for the first-line treatment of people with locally advanced or metastatic NSCLC if they test positive for an EGFR mutation. 2 Trials used in this assessment were conducted only in patients whose tumours were EGFR mutation positive, and used a direct sequencing approach to select patients with exon 19 deletions or exon 21 L858R point mutations for inclusion. 2,16
The Therascreen EGFR RGQ PCR Kit is a molecular diagnostic kit for detection of the 29 most common EGFR mutations against a background of wild-type genomic deoxyribonucleic acid (DNA). It uses real-time PCR on the Rotor-Gene Q 5plex HRM Instrument (a real-time PCR cycler). All versions of the Therascreen EGFR PCR Kit and the Therascreen® EGFR Pyro Kit (Qiagen, Venlo, the Netherlands) will be included in the assessment. The mutations detected by the currently available Therascreen EGFR RGQ PCR Kit include 19 deletions in exon 19, T790M, L858R, L861Q, G719X (Therascreen detects the presence of these mutations but does not distinguish between them), S768I, and three insertions in exon 20; version 1 of the Therascreen EGFR PCR Kit, as used in the studies included in this assessment but no longer available, detected the same mutations. A version of the Therascreen EGFR PCR Kit that did not detect the resistance mutation T790M was previously marketed by Qiagen but this version is no longer available and was not used in any of the studies included in this review. Versions 1 and 2 of the Therascreen EGFR PCR Kit, referred to in this assessment, may therefore be considered equivalent. The Therascreen EGFR RGQ PCR kit includes all reagents needed to perform a PCR-based assay, where specific areas of DNA containing mutations are targeted by amplification refractory mutation system (ARMS) primers and Scorpions technology is used to detect amplifications of those specific areas of DNA. The test uses DNA isolated from formalin-fixed and paraffin-embedded (FFPE) tissue obtained from lung biopsy. The Therascreen EGFR RGQ PCR Kit uses a two-step procedure. The first step is performance of the control assay to assess the total DNA in a sample. The second step is to complete the mutation assay for the presence or absence of mutated DNA.
The cobas® EGFR Mutation Testing Kit (Roche Molecular Systems, Inc., Branchburg, NJ, USA) is a CE-marked real-time PCR test for the detection of 41 EGFR mutations [G719X (G719S/G719A/G719C) in exon 18, 29 deletions and complex mutations in exon 19, T790M in exon 20, S768I in exon 20, five insertions in exon 20, L858R point mutation in exon 21]. The first step is to process the tumour tissue using the cobas® DNA Sample Preparation Kit. The second step is PCR amplification and detection of EGFR mutations using complementary primer pairs and fluorescently labelled probes. The PCR is run using the cobas® z 480 analyser (Roche Molecular Systems, Inc., Branchburg, NJ, USA), which automates amplification and detection. cobas® 4800 software (Roche Molecular Systems, Inc., Branchburg, NJ, USA) provides automated test result reporting.
Pyrosequencing methods are usually set up to detect specific EGFR-TK mutations and are sometimes used to look for point mutations alongside fragment length analysis to detect deletions and insertions. The process involves first extracting DNA from the sample and amplifying it using PCR. The PCR product is then cleaned up before the pyrosequencing reaction. The reaction involves the sequential addition of nucleotides to the mixture. A series of enzymes incorporate nucleotides into the complementary DNA strand, generate light proportional to the number of nucleotides added and degrade unincorporated nucleotides. The DNA sequence is determined from the resulting pyrogram trace.
Fragment length analysis can be used to detect deletions in exon 19 and insertions in exon 20. DNA is first extracted from the sample and then amplified and labelled with fluorescent dye using PCR. Amplified DNA is mixed with size standards and is analysed using capillary electrophoresis. The fluorescence intensity is monitored as a function of time, and analysis software can determine the size of the fragments. The presence or absence of a deletion/insertion can then be reported.
Mutation screening tests
Direct sequencing is used to screen for all EGFR mutations (known and novel) in exons 18–21. This process is known as ‘comprehensive testing’ and has been considered as the routine method for detecting EGFR mutations; however, it requires larger tumour samples than other methods. Randomised controlled trials (RCTs) comparing the effectiveness of erlotinib with standard chemotherapy, in participants whose tumours were EGFR mutation positive, selected participants using direct sequencing to identify mutations in exon 19 or 21. A comparison of version 1 of the Therascreen EGFR PCR Kit with direct sequencing reported that Therascreen was ‘more sensitive’, i.e. some EGFR mutations were detected, which were not identified by direct sequencing. This was ascribed to low density of tumour cells in the sample. 17 Other mutation screening methods include single-strand confirmation polymorphism, high-resolution melt (HRM) analysis and next-generation sequencing.
For single-strand conformation polymorphism, DNA is first extracted from the sample and amplified using PCR. The PCR product is then prepared for analysis by heat denaturing and analysed using capillary electrophoresis under non-denaturing conditions. Sequence variations (single point mutations and other small changes) are detected through electrophoretic mobility differences.
High-resolution melt analysis detects all mutations, known and novel. The DNA is first extracted from the sample and amplified using PCR. The HRM reaction is then performed. This involves a precise warming of the DNA, during which the two strands of DNA ‘melt’ apart. Fluorescent dye that binds only to double-stranded DNA is used to monitor the process. A region of DNA with a mutation will ‘melt’ at a different temperature to the same region of DNA without a mutation. These changes are documented as melt curves and the presence or absence of a mutation can be reported.
Next-generation sequencing can also be used to identify all mutations. As with Sanger sequencing, there is much variation in the methodology used. The concept is similar to Sanger sequencing; however, the sample DNA is first fragmented into a library of small segments that can be sequenced in parallel reactions.
Care pathway
Diagnosis and staging of lung cancer
Guidance from NICE on the diagnosis and treatment of lung cancer was updated in 2011. 18 Patients referred for suspected lung cancer should initially undergo urgent chest radiography. If the chest radiograph is suggestive of lung cancer a contrast-enhanced computed tomography (CT) scan of the thorax, upper abdomen and lower neck is performed. Patients can then undergo a variety of diagnostic and staging investigations, which should be selected to provide the most information with the least risk to the patient. Most pathways in the diagnostic algorithm include biopsy for histological confirmation and tissue typing (e.g. to confirm if NSCLC is adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma or large cell carcinoma). The mediastinal lymph nodes are assessed for malignancy using positron emission tomography (PET)-CT, or endobronchial ultrasound (EBUS)-guided transbronchial needle aspiration (TBNA), or endoscopic ultrasound (EUS)-guided fine-needle aspiration (FNA), or non-ultrasound-guided TBNA. Patients with clinical and/or radiological features of advanced/metastatic disease may undergo further imaging (e.g. PET/CT or MRI) with possible biopsy of the most accessible site. 18
Where biopsy is undertaken, DNA extraction and mutation analysis may be carried out on the biopsy tissue, after pathological examination, to determine whether the tumour is EGFR mutation positive or negative. NICE clinical guidance recommends that adequate samples are taken without unacceptable risk to the patient to permit tumour subtyping and measurement of predictive markers. 18 For the 32,347 cases of lung cancer recorded in the 2010 NLCA data, the median [interquartile range (IQR)] percentage of patients receiving a histological–cytological diagnosis was 76.0% (70.5–83.6%) across NHS Trusts in England and Wales. NLCA data for 2010 reported a median of 20.0% (IQR 13.1–28.9%) of patients with NSCLC with unspecified histology for NHS trusts in England and Wales. 5 This assessment will assume that, in line with current clinical guidance, biopsy is undertaken in all patients for whom it is considered possible and clinically appropriate. However, the proportion of patients in whom the biopsy sample is inadequate is an important consideration for this assessment, as it represents a requirement for additional mutation testing, possible additional invasive procedures (in order to obtain an adequate sample) and associated additional costs.
Treatment of non-small cell lung cancer
Once NSCLC has been confirmed, NICE clinical guidance recommends that chemotherapy should be offered to people with stage III or IV (locally and regionally advanced or metastatic) NSCLC and a good performance status (PS) [World Health Organization (WHO) 0, 1 or Karnofsky score 80–100] with the aim of improving survival, disease control (DC) and quality of life. Treatment with curative intent is not possible for these patients. First-line chemotherapy should be a combination of a single third-generation drug (docetaxel, gemcitabine, paclitaxel or vinorelbine) and a platinum drug (carboplatin or cisplatin). People who are unable to tolerate a platinum combination may be offered single-agent chemotherapy with a third-generation drug. 18 Pemetrexed in combination with cisplatin is recommended as a first-line treatment for patients with locally advanced or metastatic NSCLC if the histology of the tumour has been confirmed as adenocarcinoma or large cell tumour. 19 The most recent data for England and Wales (NLCA 2011) suggest that the median proportion of patients with stage III or IV NSCLC receiving chemotherapy was 51.5% (IQR 48.2–64%); however, the case ascertainment rate for this measure was < 50%. 5
The NICE Technology Appraisal 192 recommends the EGFR-TKI gefitinib as an option for the first-line treatment of people with locally advanced or metastatic NSCLC who test positive for EGFR mutation. 1 NICE Technology Appraisal 258 recommends erlotinib as an option for the first-line treatment of people with locally advanced or metastatic NSCLC if they test positive for an EGFR mutation. 2 NICE guidance does not currently include any recommendations on the type of diagnostic tests used to identify EGFR mutations, and there is no consensus on which testing method should be preferred for clinical decision-making. 12
Measuring response to treatment
In 1979 the WHO and the International Union Against Cancer introduced criteria for the classification of the response of solid tumours to treatment. 20 These criteria were an early attempt to standardise reporting of response outcomes and were widely adopted. However, some problems with their use have subsequently developed: there has been variation in the methods used for incorporating into response assessments the change in size of measurable lesions, as defined by WHO; the minimum lesion size and number of lesions to be recorded have also varied; the definitions of progressive disease (PD) have sometimes been related to change in a single lesion and sometimes to change in overall tumour load (sum of the measurements of all lesions); and there has been confusion around how to use three-dimensional measures from new technologies, such as CT and magnetic resonance imaging (MRI), in the context of WHO criteria. 21 The Response Evaluation Criteria in Solid Tumours (RECIST) group is a collaborative initiative that was initiated to review the WHO criteria. The RECIST criteria use the same categories [complete response (CR), partial response (PR), stable disease (SD) and PD]. 21 RECIST guidance states that ‘CT and MRI are the best currently available and most reproducible methods for measuring target lesions selected for response assessment’ and that imaging-based evaluation is generally preferable to clinical examination. It is suggested that follow-up assessments every 6–8 weeks is a ‘reasonable norm’. 21 Taking into account the longest diameter for only all target lesions, the RECIST criteria, as they are applicable to this assessment, can be summarised as follows:21
-
CR Disappearance of all target lesions and no new lesions.
-
PR At least 30% decrease in the sum of the longest diameter of target lesions, taking the sum of the baseline diameters as the reference, and no new lesions.
-
PD At least a 20% increase in the sum of the longest diameter of target lesions, taking the smallest sum of the longest diameters recorded since treatment started as the reference, or appearance of one or more new lesions.
-
SD Neither sufficient shrinkage to be classified as PR or sufficient increase to be classified as PD, taking the smallest sum of the longest diameters recorded since treatment started as the reference, and no new lesions.
Best overall response is defined as the best response recorded from the start of treatment to disease progression. 21
This assessment compares the performance and cost-effectiveness of EGFR mutation testing options currently available in the NHS in England and Wales to identify previously untreated adults with locally advanced or metastatic NSCLC who may benefit from first-line treatment with EGFR inhibitors (gefitinib or erlotinib).
Chapter 3 Assessment of clinical effectiveness
A systematic review was conducted to summarise the evidence on the clinical effectiveness of the different EGFR mutation testing options currently available in the NHS in England and Wales for the identification of previously untreated adults with locally advanced or metastatic NSCLC who may benefit from first-line treatment with EGFR-TKIs (gefitinib or erlotinib). Systematic review methods followed the principles outlined in the Centre for Reviews and Dissemination (CRD) guidance for undertaking reviews in health care,22 the NICE Diagnostic Assessment Programme interim methods statement23 and the Cochrane Handbook for diagnostic test accuracy reviews. 24
Systematic review methods
Search strategy
Search strategies were based on target condition and intervention, as recommended in the CRD guidance for undertaking reviews in health care and the Cochrane Handbook for diagnostic test accuracy reviews. 22,25
Candidate search terms were identified from target references, browsing database thesauri (e.g. MEDLINE MeSH and EMBASE Emtree), existing reviews identified during the rapid appraisal process and initial scoping searches. These scoping searches were used to generate test sets of target references, which informed text mining analysis of high-frequency subject indexing terms using EndNote X4 reference management software (Thomson Reuters, CA, USA). Strategy development involved an iterative approach testing candidate text and indexing terms across a sample of bibliographic databases and aimed to reach a satisfactory balance of sensitivity and specificity.
The following databases were searched for relevant studies from 2000 to August 2012:
-
MEDLINE (OvidSP) (2000 to July 2012 week 1)
-
MEDLINE In-Process & Other Non-Indexed Citations and Daily Update (OvidSP) (up to 17 July 2012)
-
EMBASE (OvidSP) (2000 to 2012 week 28)
-
Cochrane Database of Systematic Reviews (CDSR) (internet) (2000 to 2012/Issue 7)
-
Cochrane Central Register of Controlled Trials (CENTRAL) (internet) (2000 to 2012/Issue 7)
-
Database of Abstracts of Reviews of Effects (DARE) (via The Cochrane Library) (2000 to 2012/Issue 3)
-
Health Technology Assessment database (HTA) (via The Cochrane Library) (2000 to 2012/Issue 3)
-
Science Citation Index (SCI) (Web of Science) (2000 to 18 July 2012)
-
Latin American and Caribbean Health Sciences Literature (LILACS) (internet) (2000 to 6 July 2012) http://regional.bvsalud.org/php/index.php?lang=en
-
BIOSIS Previews (Web of Knowledge) (2000 to 24 August 2012)
-
NIHR Health Technology Assessment programme (internet) (2000 to 18 July 2012)
-
PROSPERO (International Prospective Register of Systematic Reviews) (internet) (up to 19 July 2012) www.crd.york.ac.uk/prospero/
Completed and ongoing trials were identified by searches of the following resources:
-
National Institutes of Health (NIH) ClinicalTrials.gov (2000 to 19 July 2012) (internet) www.clinicaltrials.gov/
-
Current Controlled Trials (2000 to 30 August 2012) (internet) www.controlled-trials.com/
-
WHO International Clinical Trials Registry Platform (ICTRP) (2000 to 30 August 2012) (internet) www.who.int/ictrp/en/
Searches were undertaken to identify studies of EGFR-TK mutation testing in NSCLC. The main EMBASE strategy for each set of searches was independently peer reviewed by a second information specialist, using the Peer Review of Electronic Search Strategies Evidence-Based Checklist (PRESS-EBC). 26 Search strategies were developed specifically for each database and the keywords associated with NSCLC were adapted according to the configuration of each database. Searches took into account generic and other product names for the intervention. No restrictions on language or publication status were applied. Limits were applied to remove animal studies. Full search strategies are reported in Appendix 1.
Electronic searches were undertaken for the following conference abstracts:
-
American Society of Clinical Oncology (ASCO) conference proceedings (2007 to 2012) (internet) www.asco.org/ASCOv2/Meetings/Abstracts
-
European Society of Medical Oncology (ESMO) conference proceedings (2007 to 2012) (internet) www.esmo.org/no_cache/education/abstracts-and-virtual-meetings.html
-
2008 33rd ESMO Congress, Stockholm – http://annonc.oxfordjournals.org/content/vol19/suppl_8/
-
2009 European Cancer Congress (ECCO) 15 and 34th ESMO Multidisciplinary Congress – www.ejcancer.info
-
2010 35th ESMO Congress, Milan – http://annonc.oxfordjournals.org/content/21/suppl_8
-
2011 ECCO 16 and 36th ESMO Multidisciplinary Congress, Brussels – www.ejcancer.info/issues
-
2012 37th ESMO Congress, Vienna – http://annonc.oxfordjournals.org/content/23/suppl_9
-
-
World Conference on Lung Cancer (International Association for the Study of Lung Cancer) (2007 to 2012) (internet) http://iaslc.org/
-
14th World Conference on Lung Cancer – http://journals.lww.com/jto/toc/2011/06001
-
13th World Conference on Lung Cancer – http://journals.lww.com/jto/Citation/2009/09001/Abstracts.1.aspx
-
12th World Conference on Lung Cancer – http://journals.lww.com/jto/toc/2007/08001
-
Identified references were downloaded in EndNote X4 software for further assessment and handling.
References in retrieved articles were checked for additional studies. The final list of included papers was also checked on PubMed for retractions, errata and related citations. 27–29
Inclusion and exclusion criteria
Separate inclusion criteria were developed for each of the three clinical effectiveness questions; these are summarised in Table 2.
| Question | What is the technical performance of the different EGFR mutation tests? | What is the accuracy of EGFR mutation testing, using any test, for predicting response to treatment with TKIs? | How do outcomes from treatment with EGFR-TKIs vary according to which test is used to select patients for treatment? |
|---|---|---|---|
| Participants | Adult patients (≥ 18 years) with treatment naive, locally and regionally advanced or metastatic (stage IIIB or IV) NSCLC | Adult patients (≥ 18 years) with treatment naive, locally and regionally advanced or metastatic (stage IIIB or IV) NSCLC | Adult patients (≥ 18 years) with treatment naive, locally and regionally advanced or metastatic (stage IIIB or IV) NSCLC Patients who test positive on any EGFR mutation test |
| Setting | Secondary or tertiary care | ||
| Interventions (index test) | Any commercial or in-house EGFR mutation test | Any commercial or in-house EGFR mutation test | EGFR-TKIs |
| Comparators | NA | NA | Standard care |
| Reference standard | NA | Response to treatment with TKIs (e.g. PFS) | NA |
| Outcomes | Proportion tumour cells needed, failures, turnaround time, costs, expertise/logistics of test | OS or PFS in patients whose tumours are EGFR positive vs. EGFR negative. Test accuracy – the number of TP, FN, FP and TN. IPD if available | Overall survival or PFS |
| Study design | Survey of NHS laboratories participating in the UK NEQAS pilot scheme for EGFR mutation testing | RCTs, CCTs and cohort studies | RCTs (CCTs and cohort studies where no RCTs were identified) |
Inclusion screening and data extraction
Two reviewers (MW and PW) independently screened the titles and abstracts of all reports identified by searches and any discrepancies were discussed and resolved by consensus. Full copies of all studies that were deemed potentially relevant were obtained and the same two reviewers independently assessed these for inclusion; any disagreements were resolved by consensus. Details of studies excluded at the full paper screening stage are presented in Appendix 5.
Studies provided by the manufacturers of Therascreen (Qiagen) and cobas EGFR Mutation Testing Kit were first checked against the project reference database in EndNote X4; any studies not already identified by our searches were screened for inclusion following the process described above.
Data were extracted on the following: study design/details, participant details (e.g. tumour stage, histological diagnosis, PS, smoking status, ethnicity), EGFR mutation test(s) and mutations targeted, clinical outcomes, test performance outcome measures (against treatment response as reference standard), details of specific mutations identified by outcome measure (where reported) and test failure rates. Data were extracted by one reviewer, using a piloted, standard data extraction form, and checked by a second reviewer (MW and PW); any disagreements were resolved by consensus. Full data extraction tables are provided in Appendix 2.
Quality assessment
The risk of bias in included RCTs was assessed using the Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. 30 Studies used to derive accuracy data, for the ability of EGFR mutation tests to predict treatment response, were assessed using QUADAS-2. 31 The version of QUADAS-2 used in this report did not include assessment of applicability because both the index test and study population were tightly defined by our inclusion criteria, and clinical outcome measures were treated as the reference standard. Studies that provided both accuracy data and data on the effectiveness of treatment with TKIs following testing were assessed using both tools. Risk of bias assessments were undertaken by one reviewer and checked by a second reviewer (MW and PW), and any disagreements were resolved by consensus.
The results of the risk of bias assessments were summarised and presented in tables and graphs in the results of the systematic review, and are presented in full, by study, in Appendix 3.
Survey of laboratories providing epidermal growth factor receptor mutation testing
We conducted a web-based survey (October 2012) to gather data on the technical performance characteristics of EGFR mutation tests. We sent an e-mail invitation to NHS laboratories participating in the UK NEQAS pilot scheme for EGFR mutation testing, which had responded to a request to provide information to NICE at the start of this assessment. We used the SurveyMonkey (SurveyMonkey, Palo Alto, CA, USA) online software to run the survey. We structured the survey into sections on:
-
laboratory details
-
EGFR testing methods
-
logistics
-
technical methods
-
costs.
Where possible we used multiple choice options with tick boxes to make the survey quick and easy to complete. A copy of the survey is provided in Appendix 4.
Methods of analysis/synthesis
The results of studies included in this review were summarised by research question (see Chapter 1), i.e. studies providing technical information on EGFR mutation testing in NHS laboratories in England and Wales (see What are the technical performance characteristics of the different epidermal growth factor receptor mutation tests?, below), studies providing information on the accuracy of EGFR mutation tests for predicting response to TKI treatment (see What is the accuracy of epidermal growth factor receptor mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors?, below), and studies reporting information on how clinical outcomes may vary according to which test is used to select patients for TKI treatment (see How do outcomes from treatment with epidermal growth factor receptor inhibitors vary according to which test is used to select patients for treatment?, below). We planned to use a bivariate/hierarchical summary receiver operating characteristic (SROC) random-effects model to generate summary estimates and an SROC curve for test accuracy data,32–34 and a DerSimonian and Laird random-effects model to generate summary estimates of treatment effects. However, because the review identified a relatively small number of studies with between-study variation in participant characteristics, methods used to test for EGFR mutations and mutations targeted, we did not consider meta-analyses to be appropriate and have provided a structured narrative synthesis.
For all studies that provided data on accuracy for the prediction of response to treatment with TKIs, the absolute numbers of true-positive (TP), false-negative (FN), false-positive (FP) and true-negative (TN) test results, as well as sensitivity and specificity values, with 95% confidence intervals (CIs) are presented in results tables, for each reference standard response [e.g. objective response (OR), DC] reported. Where reported, data on the numbers of failed EGFR mutation tests and reasons for failure were also included in the results tables. The results of individual studies were plotted in the receiver operating characteristic (ROC) plane to illustrate the trade-off between sensitivity and specificity, and for ease of comparison between test methods; separate plots were provided for each reference standard response. For RCTs providing information on how clinical outcomes may vary according to which test is used to select patients for TKI treatment, hazard ratios (HRs), with 95% CIs, are provided for survival outcome measures [progression-free survival (PFS), overall survival (OS)] and relative risks (RRs), with 95% CIs, are reported for tumour response outcomes (OR and DC). The results of individual studies were illustrated in forest plots. Between-study clinical heterogeneity was assessed qualitatively. There were insufficient studies to assess heterogeneity statistically, such as the chi-squared test and I2-statistic. 35
This report contains reference to confidential information provided as part of the NICE appraisal process. This information has been removed from the report and the results, discussions and conclusions of the report do not include the confidential information. These sections are clearly marked in the report.
Results of the assessment of clinical effectiveness
The literature searches of bibliographic databases identified 6932 references. After initial screening of titles and abstracts, 152 were considered to be potentially relevant and ordered for full-paper screening. No additional papers were ordered based on screening of papers provided by test manufacturers. One conference abstract,36 which was provided as part of the submission from Roche Molecular Systems, was included in the review; all other studies submitted cited in industry submissions had already been identified by bibliographic database searches. No additional studies were identified from searches of clinical trials registries. One study considered to be potentially relevant and ordered for full-paper screening was published in Japanese and no translation could be obtained. 37 Figure 1 shows the flow of studies through the review process, and Appendix 5 provides details, with reasons for exclusions, of all publications excluded at the full-paper screening stage.
FIGURE 1.
Flow of studies through the review process.
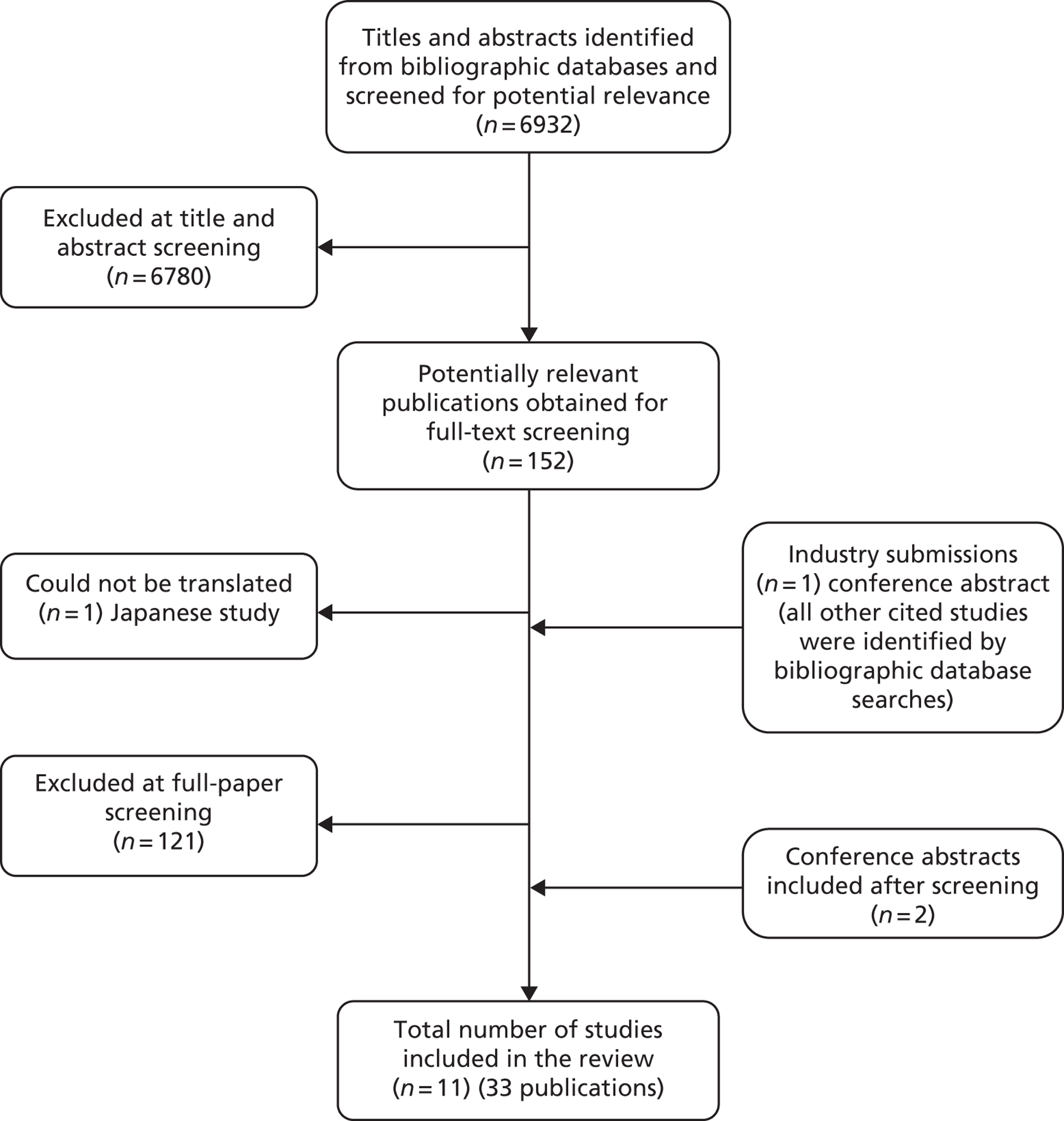
Based on the searches and inclusion screening described above, 31 publications of 11 studies were included in the review. Hand-searching of conference proceedings resulted in the identification of two additional publications38,39 for two previously identified trials. 40,41 A total of 11 studies in 33 publications were therefore included in the review.
One study was included only for information on the technical performance characteristics of an EGFR mutation test from a UK NHS laboratory. 42 Four studies reported data on tumour response following treatment with TKIs in a group of patients tested for EGFR mutations; all patients in the group were treated, regardless of mutation status. 43–46 These studies provide information on the accuracy of various EGFR mutation tests for the prediction of response to treatment with TKIs. Three RCTs compared the effectiveness of TKIs with that of standard chemotherapy in patients whose tumours were positive for EGFR mutations. 16,40,47 A further study36 reported a re-analysis of subset samples from the EURTAC trial40 using the cobas EGFR Mutation Test. Because the method used to determine mutation status varied between trials, these RCTs provide information on how clinical outcomes may vary according to which test is used to select patients for TKI treatment. The remaining two studies, the IRESSA Pan-Asia Study (IPASS) and the First-SIGNAL study, could be analysed to provide both accuracy and clinical effectiveness data. 41,48,49 These studies were RCTs that compared TKIs with standard chemotherapy in patients with NSCLC who were not initially tested for EGFR mutations; subgroup analyses were reported for patients in whom EGFR-TK mutation status was determined. The IPASS study was reported in two full-paper publications. Throughout this report it is cited either as both publications,48,49 or the specific publication from which the reported data were extracted. Multiple publications of other studies did not provide additional data and are listed in the data extraction tables in Appendix 2. For the remainder of the report, these studies are cited using the primary publication, as given above.
All included studies were published in 2006 or later and all RCTs were published in 2010 or later. Of the studies providing information on test accuracy, two were conducted in Europe,43,45 one in the USA,44 and three in East Asia. 41,46,49 With the exception of one European trial, EURTAC,40 all RCTs were conducted in East Asia. With one exception – the North East Japan Study Group (NEJSG) trial47 – all RCTs were funded by the manufacturers of TKIs (Hoffmann-La Roche Ltd or AstraZeneca); the re-analysis of samples from the EURTAC trial36 was funded by Roche Molecular Systems.
Full details of the characteristics of study participants, study inclusion and exclusion criteria, EGFR mutation test used and mutations targeted, TKI intervention and (where applicable) standard chemotherapy comparator are reported in the data in the extraction tables presented in Appendix 2. For studies providing test accuracy data, full details of the EGFR mutation testing process are reported as part of the QUADAS-2 risk of bias assessment (see Appendix 3).
What are the technical performance characteristics of the different epidermal growth factor receptor mutation tests?
Literature review
One study that evaluated the technical performance of EGFR mutation tests was included in the review. The study was conducted in the Department of Molecular Diagnostics at the Royal Marsden Hospital and the Institute of Cancer Research; this laboratory also contributed to our survey. The study reported data for 2 years of EGFR testing from January 2009 to January 2011. During year 1 of the testing period, version 1 of the Therascreen EGFR PCR Kit was used; during year 2 a combination of Therascreen EGFR PCR, fragment analysis (for exon 19 deletions and exon 20 insertions) and direct sequencing (for the rarer exon 19 or exon 21 mutations) was used. A total of 121 patients (152 samples) were tested during year 1 and 755 patients during year 2. The mean turnaround time for the Therascreen EGFR PCR test alone during year 1 was 4.9 business days (95% CI 4.5 to 5.5 days). However, the actual time from the test request to the result was 17.8 days (95% CI 16.4 to 19.4 days). The test failure rate was 19% (29/152 samples) but this improved over time from 33% during the first 3 months to 13% during the last 3 months of year-1 testing. The failure rate was lower in year 2, at only 5%.
Laboratory survey results
There were 24 UK laboratories participating in the 2012–13 UK NEQAS pilot scheme for EGFR mutation testing; 14 of these had responded to a request to provide information to NICE at the start of this assessment and were invited to participate in the survey. Thirteen of the 14 laboratories invited to participate in the survey completed our online questionnaire (response rate 93%). Three laboratories used more than one EGFR testing method and so completed the questionnaire more than once.
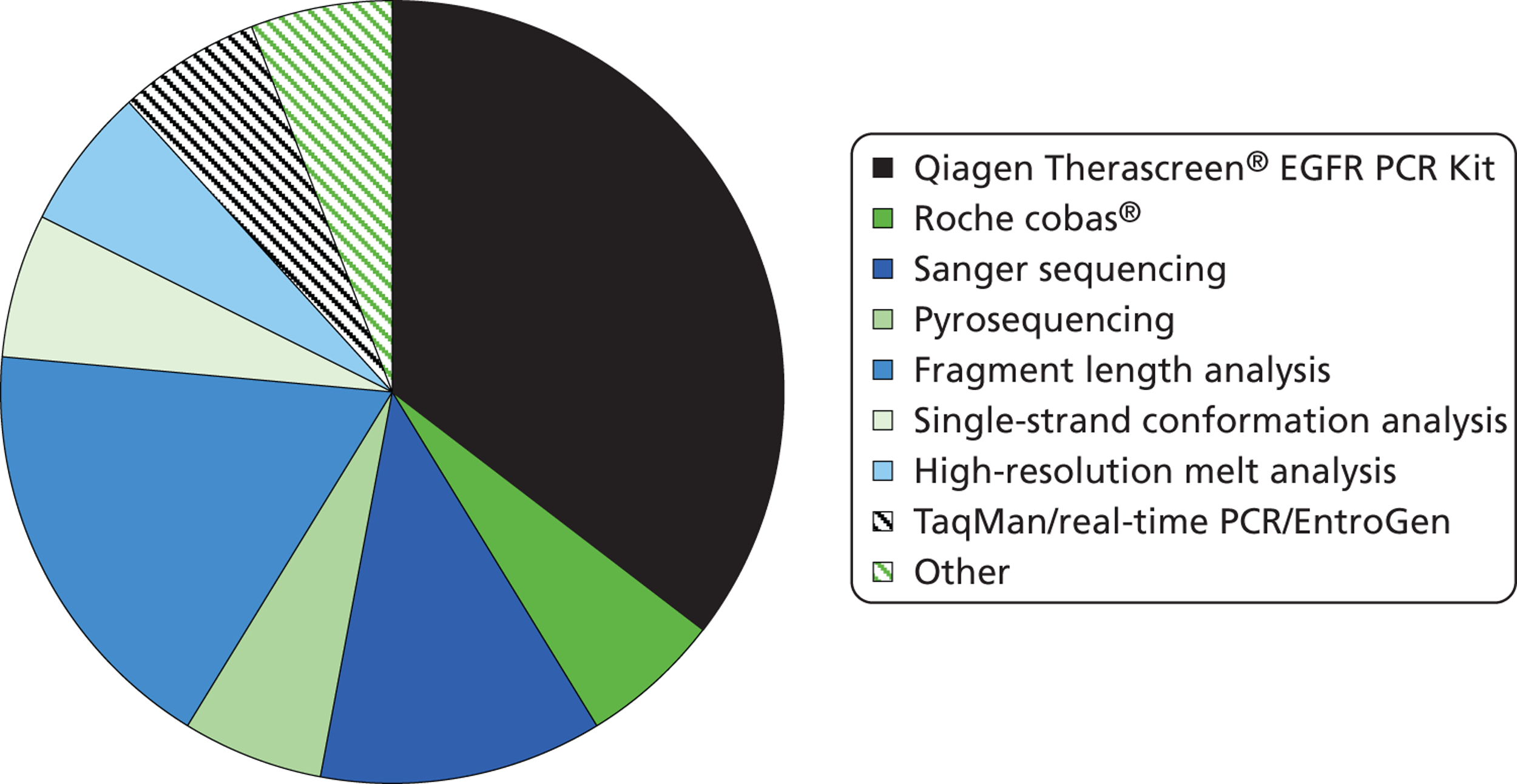
Epidermal growth factor receptor mutation test methods (see Figure 2 and Table 3)
The Therascreen EGFR PCR Kit was the most commonly used EGFR mutation test (Figure 2 and Table 3), with six laboratories using this test. A combination of fragment length analysis and pyrosequencing was used in three laboratories, and Sanger sequencing in two; other tests were each used in single laboratories. Most laboratories that used the Therascreen EGFR PCR Kit cited ease of use (n = 5) and/or proportion of tumour cells required (n = 5) as their reasons for choosing this method; three laboratories also cited mutation coverage and two cited cost. All laboratories that used fragment length analysis cited cost as a reason for their choice of this method; one also cited proportion of tumour cells required, mutation coverage and flexibility of method; another also cited ease of use; and the third claimed that accuracy was high. The two laboratories that used Sanger sequencing both cited mutation coverage as a reason for choice, and one also cited cost and ease of use; both used a second testing option for samples with insufficient tumour cells or for verification of mutations. Although only three laboratories completed the questionnaire separately for more than one test, 11 laboratories answered the question on reason for using more than one EGFR testing method. Reasons for this included insufficient tumour cells (n = 3), verification of mutations (n = 5), validating a new method (n = 1), ‘back up technique in case kits are made unavailable’, ‘methods are complementary and detect different mutations’ and ‘coverage of mutations and simplicity, cost’. Of the laboratories that completed the questionnaire more than once, one used the Therascreen EGFR PCR test but is also developing and validating a new next-generation sequencing method, which it thinks may be cheaper and target more mutations. The second used Sanger sequencing and Roche cobas, and cites verification of mutations and insufficient tumour cell as its reason for using multiple tests. The third used Sanger sequencing, TaqMan/real-time PCR/EntroGen and fragment length analysis, and also cites verification of mutations and insufficient tumour cell as its reason for using multiple tests. Two further laboratories indicated that they use a combination of pyrosequencing and fragment length analysis as complementary tests that detect different mutations; laboratories using fragment length analysis always do so as part of a strategy that involves more than one test.
FIGURE 2.
Epidermal growth factor receptor mutation test used in NHS laboratories in England and Wales participating in the UK NEQAS pilot scheme for EGFR mutation testing.
| EGFR mutation test used | Reasons for choosing test | Mutations targeted |
|---|---|---|
| Qiagen Therascreen EGFR PCR Kit | Ease of use | 28/29 mutations in Therascreen Kit |
| Proportion of tumour cells required; ease of use; ‘We had a trainee project comparing several different methods. Qiagen picked up more mutations than Sanger (more sensitive), and was very easy to use’ | ||
| Proportion of tumour cells required; mutation coverage | ||
| Proportion of tumour cells required; mutation coverage; ease of use | ||
| Cost; proportion of tumour cells required; ease of use | ||
| Cost; proportion of tumour cells required; ease of use; mutation coverage | ||
| Fragment length analysis and pyrosequencing | Cost; proportion of tumour cells required; mutation coverage; not a black box method so easily modified if required | All exon 18–21 mutations |
| Cost; ‘Sensitivity is greater than Sanger and specificity is good. Equipment for pyrosequencing is in house and is a platform used reliable for many molecular pathology investigations’ | Exon 19 deletions Insertions in exon 20 Exon 21 – L858R mutation Targeted exon 18–21 mutations; 12 mutations in total but other mutations may be detected if they are within the same region |
|
| Sanger sequencing and/or fragment length analysis/TaqMan/real-time PCR [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]a | Sanger sequencing: cost; ease of use; mutation coverage; fits in with laboratory high throughput sequencing pipeline so samples will be processed quickly Fragment length analysis: cost; ease of use TaqMan/real-time PCR: cost; ease of use |
Sanger sequencing: all exon 18–21 mutations Fragment length analysis: exon 19 deletions TaqMan/real-time PCR: Exon 21 – L858R mutation |
| Sanger sequencing and/or Roche cobas (used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)b | Sanger sequencing: mutation coverage Roche cobas: proportion of tumour cells required |
Sanger sequencing: all exon 18–21 mutations Roche cobas: 41 mutations in cobas kit |
| Next-generation sequencing, stated ‘in process of developing and validation’ | Cost; proportion of tumour cells required; mutation coverage; capacity to test multiple genes/samples/patients | Potentially all |
| HRM analysis | Mutation coverage; ease of use | All exon 18–21 mutations |
| Single-strand conformation analysis | Cost; ease of use; the vast majority of cases (90%) are EGFR wild type, therefore an easy method that reliably detects wild-type cases with ease of analysis seems cost-effective | All exon 18–21 mutations |
| Pyrosequencing | Cost; mutation coverage | Exon 19 deletions Insertions in exon 20 Exon 21 – L858R mutation |
Epidermal growth factor receptor mutation test logistics (see Figure 3 and Table 4)
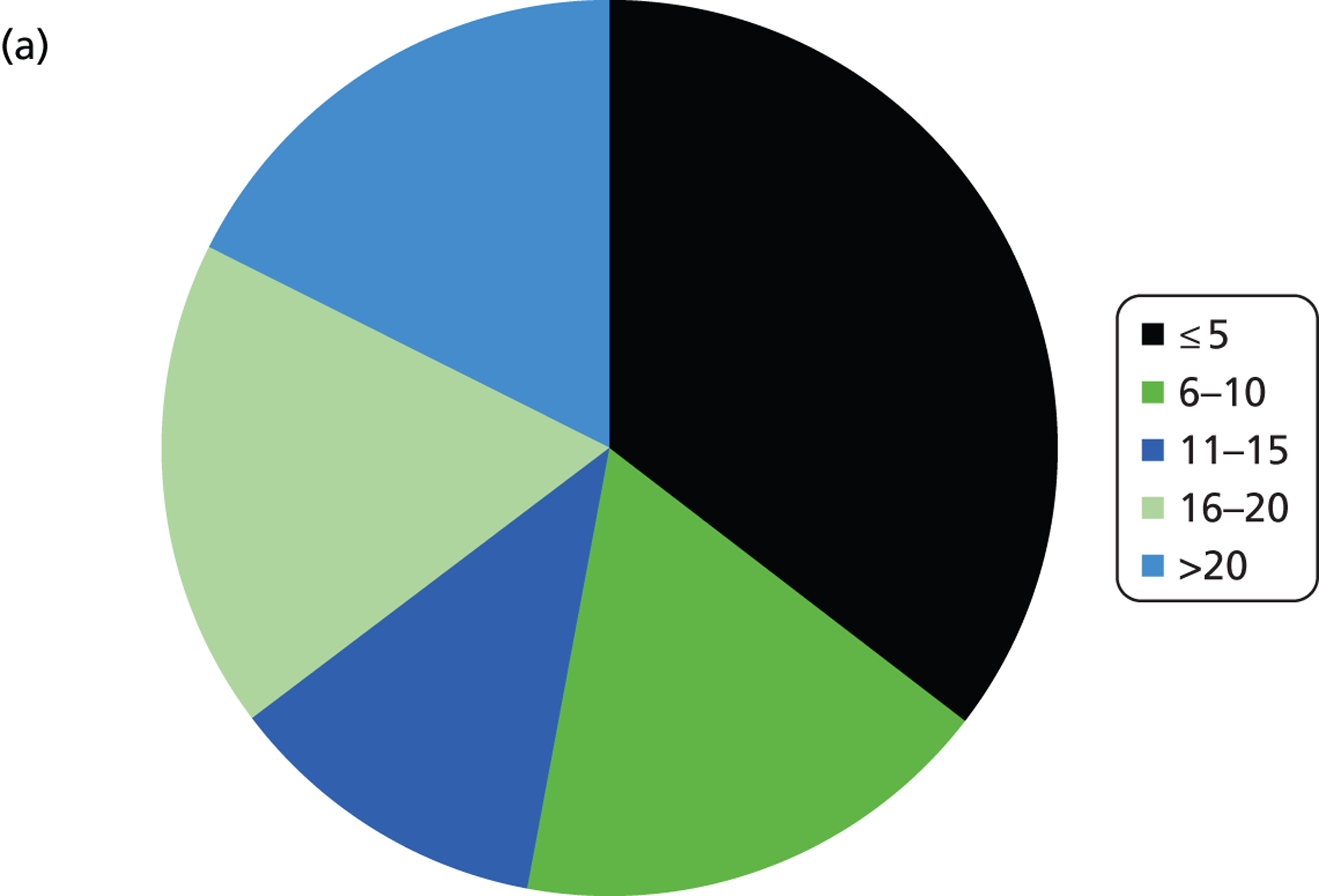
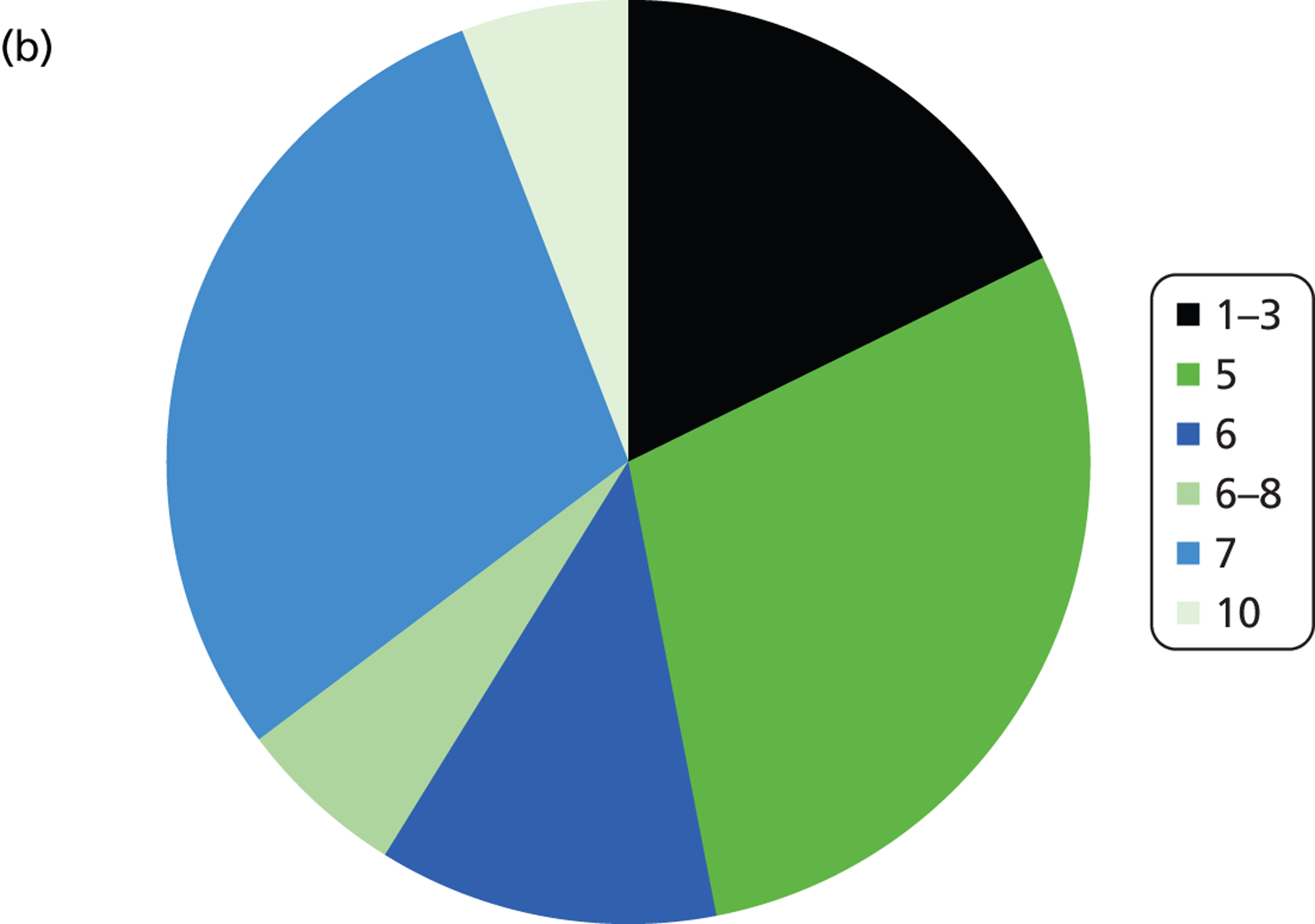
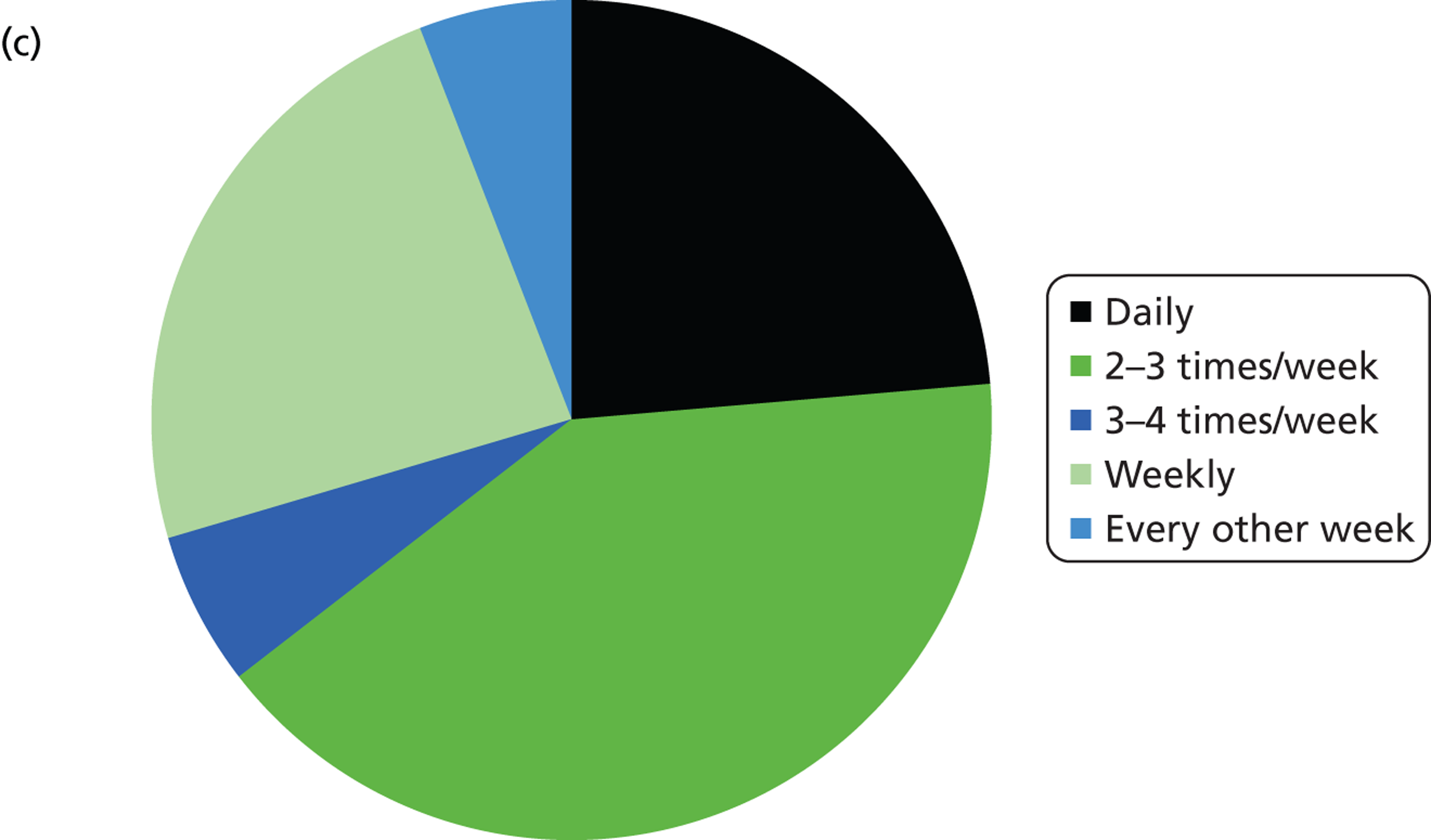
The number of samples screened for EGFR mutations in a typical week varied by laboratory from less than five (six laboratories) to > 20 (three laboratories). The batch size ranged from less than 3 to 10 samples (Figure 3 and Table 4). Only laboratories with five or fewer samples screened per week ran batches of three or fewer. Only one laboratory had a batch size of 10 and this laboratory screened > 20 samples per week; all other laboratories had batch sizes of between five and eight. For the Therascreen EGFR PCR test, all batch sizes were five or seven. The frequency at which the laboratories ran the test ranged from daily to every other week, although the laboratory that ran the test every other week stated that they would match demand. Three laboratories stated that they waited for a minimum batch size (five to seven samples), although one of these stated that they would match demand.
FIGURE 3.
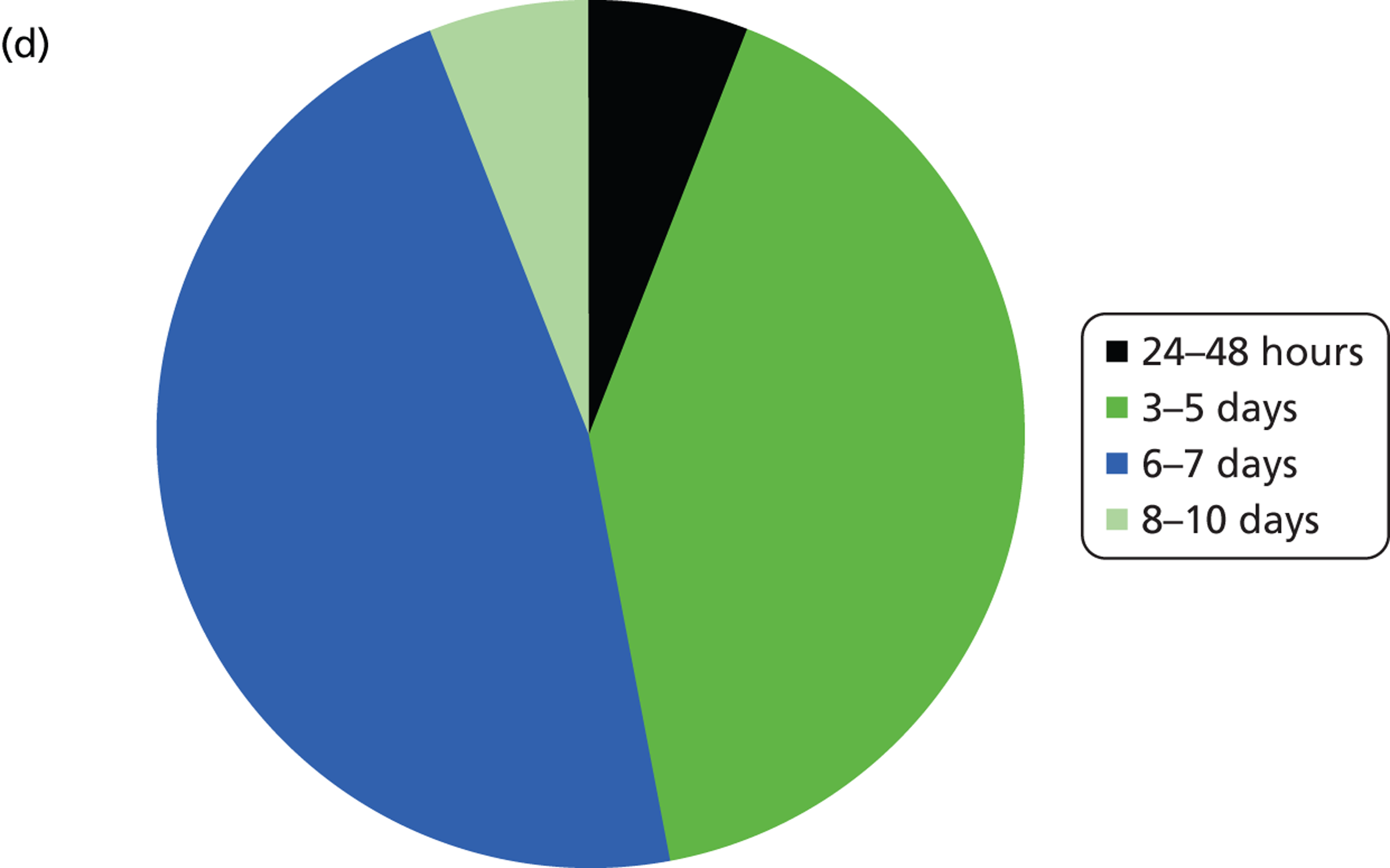
Summary of logistic information. (a) In a typical week, how many samples do you screen for EGFR mutations?; (b) what is you average batch size (number of samples)?; (c) how often do you run the EGFR mutation test?; (d) on average, how long does it take from receiving a sample at the laboratory to sending a result back to the clinician?
| EGFR mutation test | Samples per week | Batch size | Frequency of test | Wait for batch size? | Time from receiving test to returning result to clinician |
|---|---|---|---|---|---|
| Qiagen Therascreen EGFR PCR Kit | > 20 | 7 | Daily | No | 3–5 days |
| > 20 | 7 | Three to four times per week | Yes | 3–5 days | |
| 11–15 | 7 | Weekly | No | 8–10 days | |
| 6–10 | 7 | Weekly plus further run when required | No | 3–5 days | |
| ≤ 5 | 5 | Weekly | No | 24–48 hours | |
| ≤ 5 | 5 | Every other week | Yes, but will match demand | 6–7 days | |
| Fragment length analysis and pyrosequencing | 6–10 | 5 | Two to three times per week | No | 6–7 days |
| 6–10 | 5 | Two to three times per week | No | 6–7 days | |
| Sanger sequencing and/or fragment length analysis/TaqMan/real-time PCR [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]a | ≤ 5 | 1–3 | Daily | No | 6–7 days |
| Sanger sequencing and/or Roche cobas [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]b | 16–20 | 6 | Two to three times per week | Yes | 6–7 days |
| 16–20 | 6 | Two to three times per week | No | 3–5 days | |
| HRM analysis | 11–15 | 7 | Two to three times per week | No | 3–5 days |
| Next-generation sequencing | ≤ 5 | 5 | Weekly | No | 3–5 days |
| Pyrosequencing | 16–20 | 6–8 | Two to three times per week | No | 6–7 days |
| Single-strand conformation analysis | > 20 | 10 | Two to three times per week | No | 3–5 days |
The majority of laboratories had a turnaround time from receiving the sample to reporting the result to the clinician of 3–5 or 6–7 days with only one laboratory having a time of 24–48 hours and one having a time of 8–10 days. The laboratory with the shortest turnaround time was one that used the Therascreen EGFR PCR test, and which tested fewer than five samples per week. The laboratory with the longest turnaround time was also a laboratory that used Therascreen EGFR PCR, but had a higher throughput of 11–15 samples per week. Neither of these two laboratories waited for a minimum batch size before running the test.
Epidermal growth factor receptor mutation test technical performance
The minimum reported percentage of tumour cell required varied between laboratories, even for those using the same EGFR mutation test (Table 5). For the Therascreen EGFR PCR test, two laboratories reported that < 1% of tumour cells were required, three laboratories reported that 1–5% of tumour cells were required, and one reported that 6–10% of tumour cells were required. The two laboratories that used fragment length analysis and pyrosequencing both reported that a minimum of 1–5% of tumour cells were required. Sanger sequencing needed the greatest percentage of tumour cells, with a requirement of > 30%. HRM analysis and Roche cobas required 6–10%; all other methods were reported to require 1–5% of tumour cells. One laboratory, which used a combination of either fragment length analysis, Sanger sequencing or TaqMan/real-time PCR/EntroGen indicated on the questionnaire that the minimum percentage of tumour cells required was 30%, but stated that they had no failed samples and that ‘we always get a result out even if using only one of the three methods’.
The estimated total number of failed samples ranged from 0% to 10% with the number of failed samples due to insufficient tumour cells ranging from 0% to 5%. The most common reasons for failed tests were insufficient tumour cell count and poor-quality DNA/DNA degradation.
| Test | Minimum percentage of tumour cells required | Estimate of total failed samples (%) | Estimate of failures due to insufficient tumour cells (%) | Reasons for failed tests |
|---|---|---|---|---|
| Qiagen Therascreen EGFR PCR Kit | ≤ 1% | 0 | 0 | All met assay quality control criteria |
| ≤ 1% | 10 | NR | Large number of original failures related to samples not validated for kit (bone, cerebrospinal fluid, etc.). Most other failures due to inhibition (i.e. require a dilution factor) | |
| 1–5 | 5 | NR; not included in 5 | Unknown reason in most cases; decalcification for bone specimens is a classical cause of failure; for others it is assumed to be due to DNA degradation owing to delay in formalin fixation | |
| 1–5 | 1 | 1 | Low levels of amplifiable DNA | |
| 1–5 | 2 | 0 | DNA degradation or scanty material | |
| 6–10 | 5 | 5 | NR | |
| Fragment length analysis and pyrosequencing | 1–5 | 5 | NR | Poor-quality DNA – we do not test the tumour load but rely on information from the referring pathologist; if they do not supply this information then we add a caveat. We rarely fail samples but may be reporting on non-tumour DNA if incorrect samples are sent |
| 1–5 | 5 | 2 | Insufficient sample mainly | |
| Sanger sequencing and/or fragment length analysis/TaqMan/real-time PCR [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]a | > 30 | 0 | 0 | We always get a result out even if using only one of the three methods [55 fails on sequencing; 6/77 (7.8%) fluorescent PCR fails; 7/74 (9.55%) L858R real-time PCR fails] Reasons for failed tests usually insufficient quantity of tissue and DNA quality |
| Sanger sequencing and/or Roche cobas [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]b | > 30 | 4 | 3 | Poor DNA quality and low tumour cell count |
| 6–10 | 5 | 4 | Insufficient tumour cell count and poor samples that are degraded | |
| Pyrosequencing | 1–5 | 5 | 2 | Poor quality DNA, generally due to inadequate fixation |
| HRM analysis | 6–10 | 0.2 | 0.2 | Lack of good PCR amplification |
| Single-strand conformation analysis | 1–5 | 10 | 2 | Degraded DNA (70%), low DNA quantity (25%), technical errors (5%) |
| Next-generation sequencing | 1–5 | NR | NR | NR – state that in the process of validation |
Epidermal growth factor receptor mutation test costs
The cost of the EGFR mutation tests ranged from £110 to £190 and the price charged by the laboratories ranged from £120 to £200 (Table 6). Most laboratories reported that the cost of the test was the same as the price charged for the test; where there was a difference this ranged from £10 to £37.50 per test. The variation in the cost of the test was similar within tests to what it was between tests, with no single test appearing more or less expensive than any of the other tests despite most laboratories citing cost of test as their reason for selecting a particular EGFR mutation testing method. Costs were similar for laboratories using single tests and those using strategies involving multiple tests. The cost and price charged for the Therascreen EGFR PCR test ranged from £120 to £190.
| Test | What is the cost (£) of the test (including purchase costs, personnel, material and overheads)? | What is the price (£) that you charge for the test? |
|---|---|---|
| Qiagen Therascreen EGFR PCR Kit | 190 | 190 |
| 180 | 180 | |
| Approximately 160 | 160 | |
| Approximately 120 | 157.50 | |
| Real cost unknown | 120 | |
| 120 | 120 | |
| Fragment length analysis and pyrosequencing | 175, excluding overheads | 200 |
| 150 | 175 | |
| Sanger sequencing and/or fragment length analysis/TaqMan/real-time PCR [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]a | NR | 140 |
| Sanger sequencing and/or Roche cobas [used for verification of mutations, or where sample contains insufficient tumour cells for Sanger sequencing (< 30%)]b | NR | 120 |
| NR | 140 | |
| Pyrosequencing | ≈ 175 | 175 |
| HRM analysis | 140 | 150 |
| Single-strand conformation analysis | 110 | 140 |
| Next-generation sequencing | NR | NR |
What is the accuracy of epidermal growth factor receptor mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors?
Six studies – two RCTs41,48,49 and four cohort studies43–46 – provided data on the accuracy of EGFR mutation testing for predicting response to treatment in patients with stage IIIB or IV NSCLC when they are treated with TKIs. Three studies were conducted in patients treated with gefitinib,41,46,48 and three were conducted in patients treated with erlotinib. 43–45 These studies are particularly useful as they provide full information on the extent to which EGFR mutation tests are able to discriminate between patients who will benefit from TKI treatment and those who will not. We defined TPs as those patients with an EGFR mutation who have a positive response to TKI treatment. Where presence or absence of OR was the reference standard, a positive response was defined as best observed response = CR or PR. Where presence or absence of DC was the reference standard, a positive response was defined as best observed response = CR, PR or SD. FPs were defined as those patients with an EGFR mutation who did not have a positive response to TKI treatment (SD or PD) for the reference standard OR, or disease progression for the reference standard DC; FNs were defined as those without an EGFR mutation who had a positive response to TKI treatment; and TNs were defined as those without EGFR mutation who did not have a positive response to TKI treatment. Full definitions of CR, PR, SD and PD are provided in Chapter 2 (see Measuring response to treatment).
Study details
Participant characteristics varied across studies. Four studies did not report any details of the ethnicity of participants,41,43,45,46 one study included mainly white participants,44 and one study included almost entirely (> 99%) East Asian participants. 48 All studies reported a high (> 75%) proportion of participants with stage IV disease. Most study participants had a histological diagnosis of adenocarcinoma, but the proportion varied (range 45% to 100%). Only two studies specifically reported the inclusion of any patients with squamous cell carcinoma (9%44 and 15%43); neither study reported separate data for these patients. Three studies included mainly (92%),48 or only participants who had never smoked;41,45 one study included mainly (71%) patients who had never smoked; and the remaining two studies included mainly (70%43 and 90%44) current and former smokers. Full details of study participants are reported in Appendix 2.
Five studies evaluated direct sequencing methods for the identification of any EGFR mutation: three assessed exons 18–21,43,45,46 one assessed exons 19–2141 and one assessed exons 18–24. 44 In one study two patients, one with the exon 20 resistance mutation T790M and one with a previously undescribed exon 20 mutation V802I, were classified as test negative,43 and in one study two patients with a non-sensitising mutation G863S were classified as test negative. 45 One study assessed version 1 of the Therascreen EGFR PCR Kit, which detects 19 exon 19 deletions (does not distinguish between individual deletions), exon 21 point mutations L858R and L861Q, the exon 20 mutations S768I and T790M, exon 18 mutations G719X (does not distinguish between G719S, G719A and G719C) and three exon 20 insertions. 48
All but one study used the RECIST criteria21 to evaluate response to TKI treatment and response was defined as the best response to TKI treatment observed during treatment. In the other study, criteria used were not clearly defined. 41 Tumour response was assessed every 6 weeks,43,44,49 every 8 weeks,45,46 or every 9 weeks41 during treatment. Three studies did not report the duration of TKI treatment, i.e. the response evaluation period, and this could not be assumed to be the same as the follow-up period for the study, as all studies allowed further therapies after disease progression. 43,44,46 The remaining three studies reported similar median treatment durations of 5.4 to 5.7 months. 41,45,49 All studies reported data for OR (best observed response was PR or CR) and all but one41 also reported data for DC (best observed response was PR, CR or SD).
Epidermal growth factor receptor mutation test accuracy
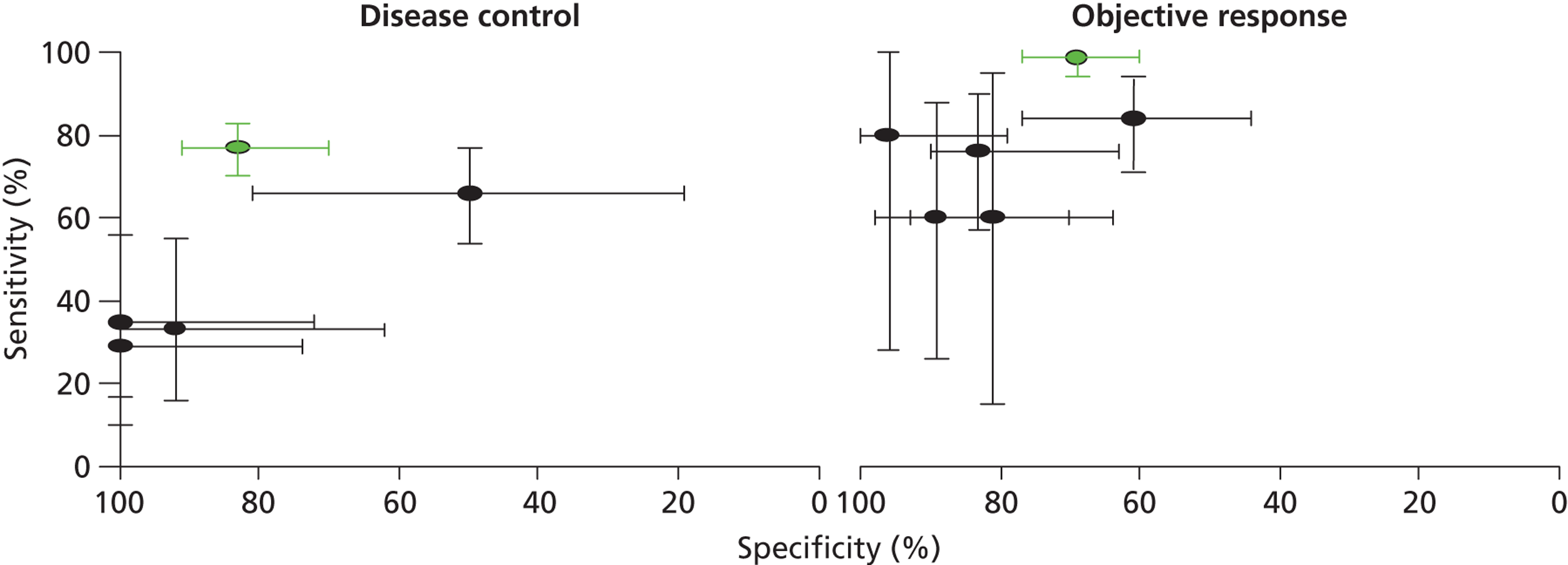
The Therascreen EGFR PCR Kit appeared to have the best overall performance for discriminating between patients who are likely to benefit from TKI treatment and those who are not. The sensitivity and specificity estimates for OR were 99% (95% CI 94% to 100%) and 69% (95% CI 60% to 77%), respectively. 49 As might be expected, the specificity was higher when a lower threshold (DC) was used to define response to treatment and, conversely, sensitivity was higher when a higher threshold (OR) was used to define response to treatment (see Table 7). It should be noted that although initial examination may appear to indicate a better performance for the Therascreen EGFR PCR Kit, only one data set was available for this test and no studies reported direct comparisons between tests conducted in the same population. Figure 4 illustrates the results for all studies reporting accuracy data with the Therascreen EGFR PCR Kit study (IPASS) indicated in green. Four of the five studies that used direct sequencing methods to identify EGFR mutations reported high estimates of specificity (> 80%) for OR, and specificities ranged from 60% to 80%. 41,43–45 Three of these studies also assessed DC; specificities remained high (> 90%), whereas sensitivity estimates were very low (≤ 35%). 43–45 The remaining direct sequencing study reported low sensitivity (66%) and specificity (50%) for DC, and low specificity (61%) with high sensitivity (84%) for OR. All direct sequencing studies had small sample sizes, reflected in the wide CIs around sensitivity and specificity estimates. There were no clear common participant characteristics across studies that reported similar sensitivity or specificity estimates for DC or OR. All test accuracy results are summarised in Table 7. It is possible that the lower specificity values observed in two studies46,49 may be explained, at least in part, by the classification of resistance mutations as a positive result for EGFR mutation testing. The four direct sequencing studies that reported high specificity estimates for DC and/or OR41,43–45 either stated that patients whose tumours showed resistance or non-sensitising mutations were classified as EGFR mutation negative, or did not identify any patients whose tumours showed these types of mutation (Table 8). Although the number of resistance mutations identified was generally small, their potential effect on specificity estimates was magnified by the very small sample size in most studies. Data relating best response to individual mutations appeared to indicate that there may be a less favourable response to TKIs in patients with T790M or other exon 20 mutations (see Table 8); [Commercial-in-confidence (CiC) information has been removed]. The most commonly observed mutations were exon 19 deletions and the exon 21 point mutation L858R, and most patients with these mutations achieved a minimum response of SD. Two studies did not report sufficient information to derive best response data by mutation type and both of these studies identified only exon 19 deletions and exon 21 point mutation L858R. 41,45 One study reported a CR in three patients whose tumours were positive for EGFR mutations and no CRs in patients whose tumours were negative for EGFR mutations;49 none of the other studies reported any CRs.
| Study | EGFR test and mutations targeted | Non-evaluable samples | DC | OR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | Sensitivity (95% CI) | Specificity (95% CI) | TP | FP | FN | TN | Sensitivity (95% CI) | Specificity (95% CI) | |||
| Fukuoka (IPASS) (2011)48,49 | Therascreen EGFR PCR Kit (version 1) | 386/609 unknown mutation status (number with insufficient sample quality NR) | 121 | 10 | 36 | 47 | 77 (70 to 83)a | 83 (70 to 91)a | 94 | 37 | 1 | 82 | 99 (94 to 100)a | 69 (60 to 77) |
| Giaccone (2006)43 | Direct sequencing (nested PCR) of all exon 18–21 mutations | 24/53 no sample available, no samples of insufficient quality reported | 5 | 0 | 12 | 12 | 29 (10 to 56)a | 100 (74 to 100)a | 4 | 1 | 1 | 23 | 80 (28 to 100)a | 96 (79 to 100)a |
| Han (First-SIGNAL) (2012)41 | Direct sequencing (PCR) of all exon 19–21 mutations | 53/159 unknown mutation status (number with insufficient sample quality NR) | NR | NR | NR | NR | NR | NR | 22 | 4 | 7 | 20 | 76 (57 to 90)a | 83 (63 to 95)a |
| Jackman (2007)44 | Direct sequencing (34 samples), or WAVE-HS (Transgenomic, Omaha, NE, USA) (nine samples) for inadequate samples (< 50% of tumour cells) of all exon 18–24 mutations | 4/80 no sample available, 26/80 samples of insufficient quality | 9 | 0 | 17 | 11 | 35 (15 to 56)a | 100 (72 to 100)a | 3 | 6 | 2 | 26 | 60 (15 to 95)a | 81 (64 to 93)a |
| Pallis (2012)45 | Direct sequencing (PCR) of all exon 18–21 mutations | 13/49 no sample available, no samples of insufficient quality reported | 8 | 1 | 16 | 11 | 33 (16 to 55)a | 92 (62 to 100)a | 6 | 3 | 4 | 23 | 60 (26 to 88)a | 89 (70 to 98) |
| Yang (2008)46 | Direct sequencing (PCR) of all exon 18–21 mutations | 16/106 EGFR mutation status not successfully determined, no details reported | 47 | 5 | 24 | 5 | 66 (54 to 71)a | 50 (19 to 81)a | 38 | 14 | 7 | 22 | 84 (71 to 94)a | 61 (44 to 77)a |
FIGURE 4.
Receiver operating characteristic plane plots comparing EGFR mutation testing methods for the prediction of response to treatment with TKIs. Plots show sensitivity and specificity estimates with 95% CIs.
| Study | EGFR mutation | n | Best response | |||
|---|---|---|---|---|---|---|
| CR | PR | SD | PD | |||
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | CiC information has been removed | |
| Giaccone (2006)43 | Exon 19 deletion only | 5 | 0 | 4 | 1 | 0 |
| Jackman (2007)44 | Exon 19 deletion only | 3 | 0 | 2 | 1 | 0 |
| Exon 21 L858R only | 5 | 0 | 1 | 4 | 0 | |
| Exon 19 deletion and exon 21 L861Q | 1 | 0 | 0 | 1 | 0 | |
| Yang (2008)46 | Exon 19 deletion only | 20 | 0 | 19 | 0 | 1 |
| Exon 21 L858R only | 22 | 0 | 17 | 5 | 0 | |
| Exon 21 L861R | 1 | 0 | 0 | 1 | 0 | |
| Exon 21 L858R and H850D | 2 | 0 | 0 | 1 | 1 | |
| Exon 21 L861Q and R831H | 1 | 0 | 0 | 1 | 0 | |
| Exon 20 SVD 786–770 insertions | 3 | 0 | 1 | 0 | 2 | |
| Exon 21 L858R and exon 20 S768I | 1 | 0 | 1 | 0 | 0 | |
| Exon 21 L858R and exon 20 T790M | 1 | 0 | 0 | 0 | 1 | |
| Exon 21 L861Q and exon 20 R776H | 1 | 0 | 0 | 1 | 0 | |
The IPASS trial, which used version 1 of the Therascreen EGFR PCR Kit, reported the minimum quantity of DNA required to detect 1% for each mutation targeted (1.5 ng for all mutations, except insertions that required 3.0 ng). 49 No direct sequencing study reported information on the limit of detection of the EGFR mutation test method used. Two studies specified a minimum proportion of tumour cells as a sample quality prerequisite for testing: these were 50% of tumour cells44 and 80% of tumour cells,45 respectively. Details of non-evaluable samples were generally poorly reported; any information reported is presented in Table 7.
QUADAS-2 assessments
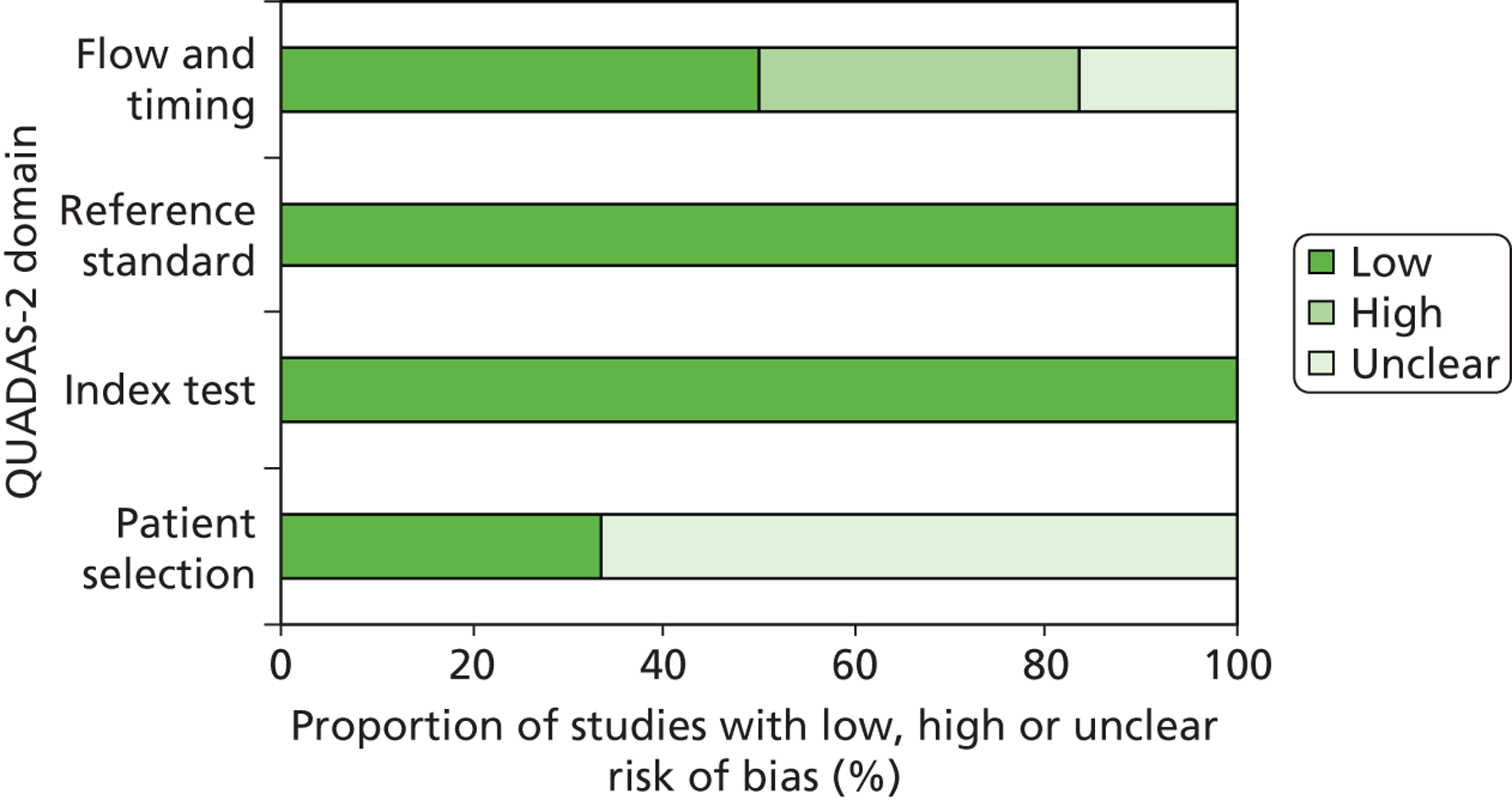
All studies in this section were rated as ‘low’ risk of bias for the ‘index test’ and ‘reference standard’ domains of the quality assessment tool. 41,43–46,49,50 The two RCTs – IPASS48,49 and First-SIGNAL41 – were rated at ‘low’ risk of bias for participant selection; none of the other studies reported details of participant selection and consequently all were rated as ‘unclear’ risk of bias for this domain. Three studies had a ‘high’ risk of bias rating for any domain. 41,44,46 All of these were for the ‘flow and timing’ domain. For two cohorts the ‘high’ risk of bias rating arose because patients who were not evaluable for response were excluded from the analysis and these patients were judged to represent a significant proportion of the study population. 44,46 One RCT was rated as at ‘high’ risk of bias for the ‘flow and timing’ domain because only a small proportion of trial participants were assessed for tumour EGFR mutation status, no reasons were reported for why participants were not assessed, and no information was available to assess possible differences between those with known mutation status and those without. The results of QUADAS-2 assessments are summarised in Table 9 and Figure 5, and full QUADAS-2 assessments for each study are provided in Appendix 3.
| Study | Risk of bias | |||
|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | |
| Fukuoka (IPASS) (2011)48,49 | ☺ | ☺ | ☺ | ☺ |
| Giaccone (2006)43 | ? | ☺ | ☺ | ☺ |
| Han (First-SIGNAL) (2012)41 | ☺ | ☺ | ☺ | ? |
| Jackman (2007)44 | ? | ☺ | ☺ | ☹ |
| Pallis (2012)45 | ? | ☺ | ☺ | ☺ |
| Yang (2008)46 | ? | ☺ | ☺ | ☹ |
FIGURE 5.
Summary of QUADAS-2 results.
How do outcomes from treatment with epidermal growth factor receptor inhibitors vary according to which test is used to select patients for treatment?
Five RCTs provided data on the clinical effectiveness of TKIs compared with standard chemotherapy in patients with stage IIIB or IV NSCLC, whose tumours tested positive for EGFR mutations,16,40,41,47,49 and one additional study36 reported data for a subgroup of patients from the EURTAC trial,40 whose samples had been re-analysed using a different EGFR mutation testing method (cobas EGFR Mutation Test). The trials compared the TKIs gefitinib or erlotinib with various single agent or combination standard chemotherapy regimens (Table 10). Three of the trials included only patients with EGFR mutation-positive tumours,16,40,47 and the remaining two trials (IPASS and First-SIGNAL) included chemotherapy-naive patients with stage IIIB or IV NSCLC, and reported a subgroup analysis for patients who had received EGFR mutation testing. 41,48
| Study | EGFR test and mutations targeted | Total number of participants (n), non-evaluable samples | Intervention | Comparator | Outcome | Effect estimate (95% CI) |
|---|---|---|---|---|---|---|
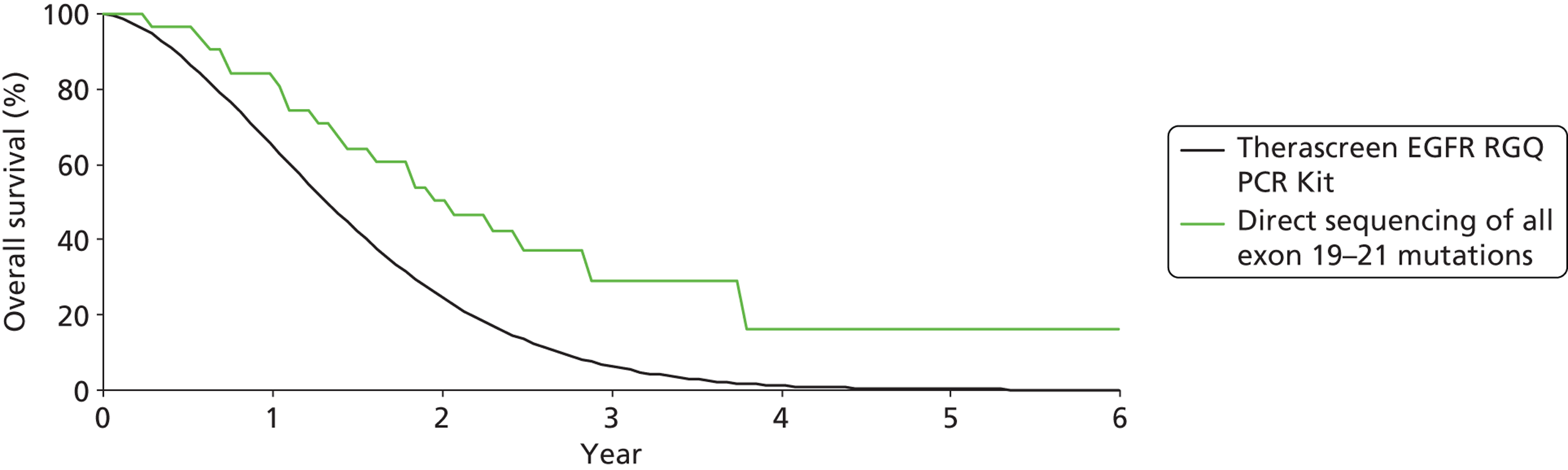
| Benlloch (EURTAC) (2012)36 | cobas EGFR Mutation Testing Kit | n = 135 37 no tumour block available, and two with insufficient tumour material |
Erlotinib | Cisplatin plus docetaxel or gemcitabine | PFS | HR 0.35 (0.21 to 0.58) |
| Fukuoka (IPASS) (2011)48,49 | Therascreen EGFR PCR Kit (version 1) | n = 261, mutation-positive subgroup Whole trial (n = 1217): 437 samples evaluable, 534 samples unavailable, 118 cytology samples excluded as the biomarker kit used was not validated for these samples, and 128 histology samples inadequate for testing |
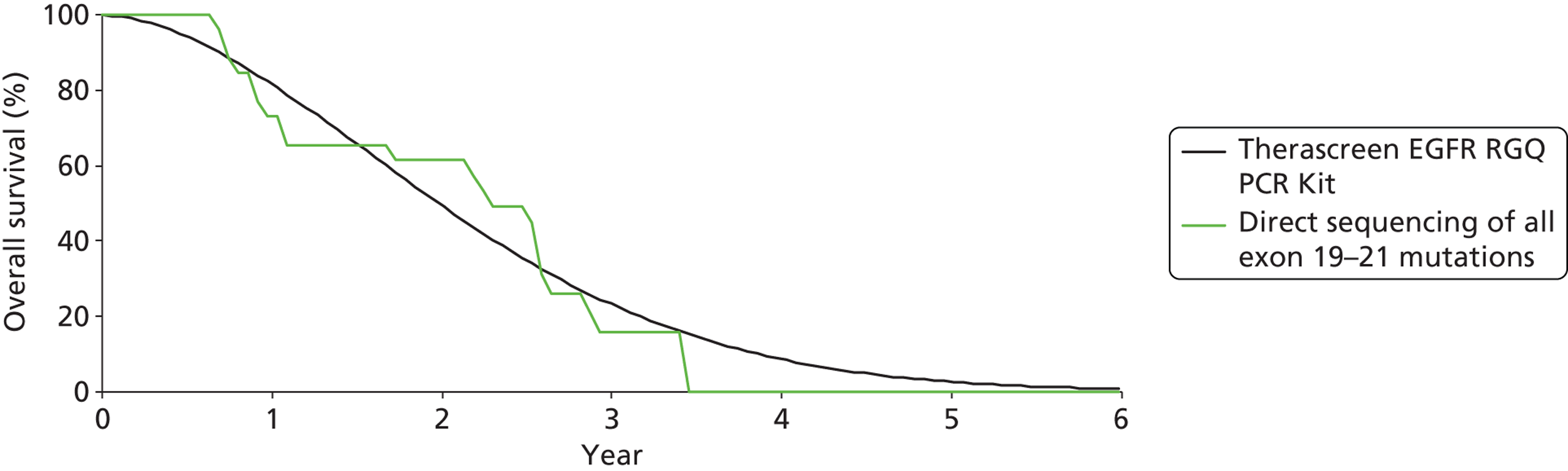
Gefitinib | Carboplatin plus paclitaxel | PFS | HR 0.48 (0.36 to 0.64) |
| OS | HR 1.00 (0.76 to 1.33) | |||||
| DC | RR 1.05 (0.96 to 1.15) | |||||
| OR | RR 1.51 (1.23 to 1.88) | |||||
| Han (First-SIGNAL) (2012)41 | Direct sequencing (PCR) of all exon 19–21 mutations | n = 42 mutation-positive subgroup Whole trial (n = 313): 217 patients were not assessable for tumour EGFR mutation status (reasons not reported) |
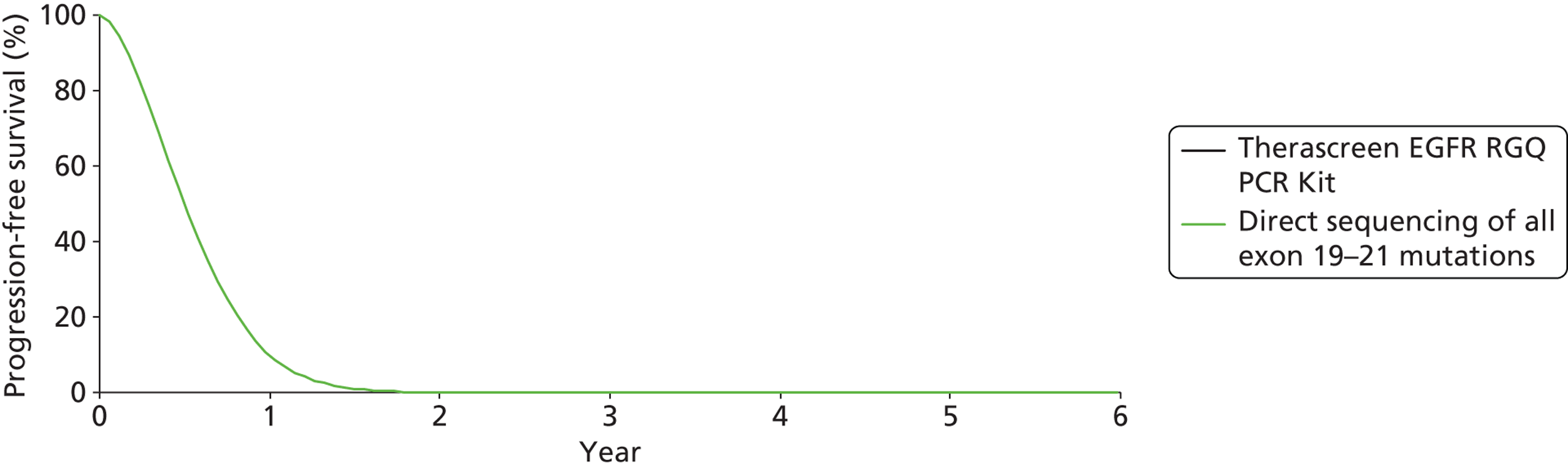
Gefitinib | Gemcitabine plus cisplatin | PFS | HR 0.54 (0.27 to 1.10) |
| OS | HR 1.04 (0.50 to 2.18) | |||||
| OR | RR 2.26 (1.31 to 4.65) | |||||
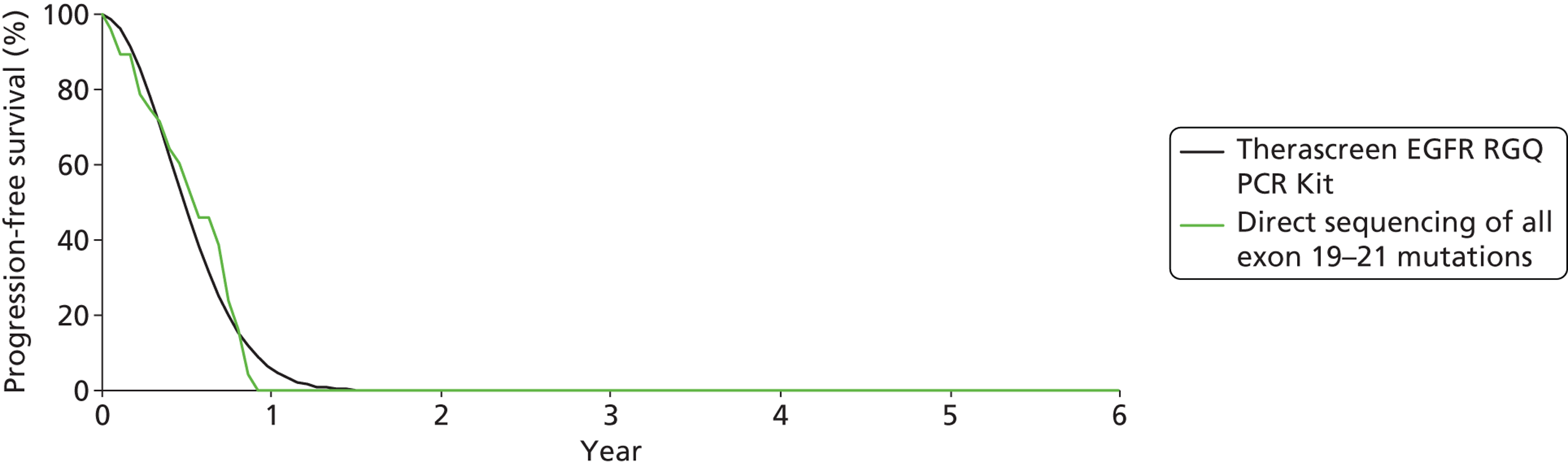
| Maemondo (NEJSG) (2010)47 | Fragment length analysis; exon 19 deletions, exon 21 point mutations (L858R, L861Q), exon 18 point mutations (G719A, G719C, G719S), exon 20 point mutation (T790M) | n = 227 None reported |
Gefitinib | Carboplatin plus paclitaxel | PFS | HR 0.30 (0.22 to 0.41) |
| OS | HR 0.89 (0.63 to 1.24) | |||||
| DC | RR 1.12 (1.00 to 1.27) | |||||
| OR | RR 2.40 (1.81 to 3.26) | |||||
| Rosell (EURTAC) (2012)40 | Sanger sequencing; exon 19 deletions and exon mutation 21 L868R | n = 150 None reported |
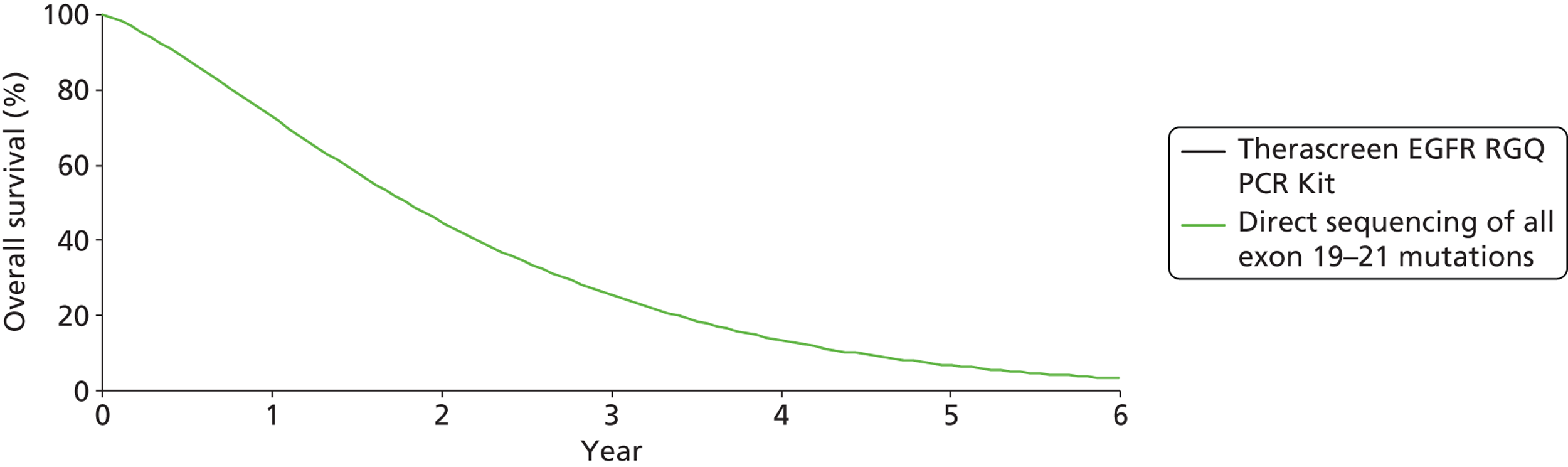
Erlotinib | Cisplatin plus docetaxel or gemcitabine | PFS | HR 0.37 (0.25 to 0.54) |
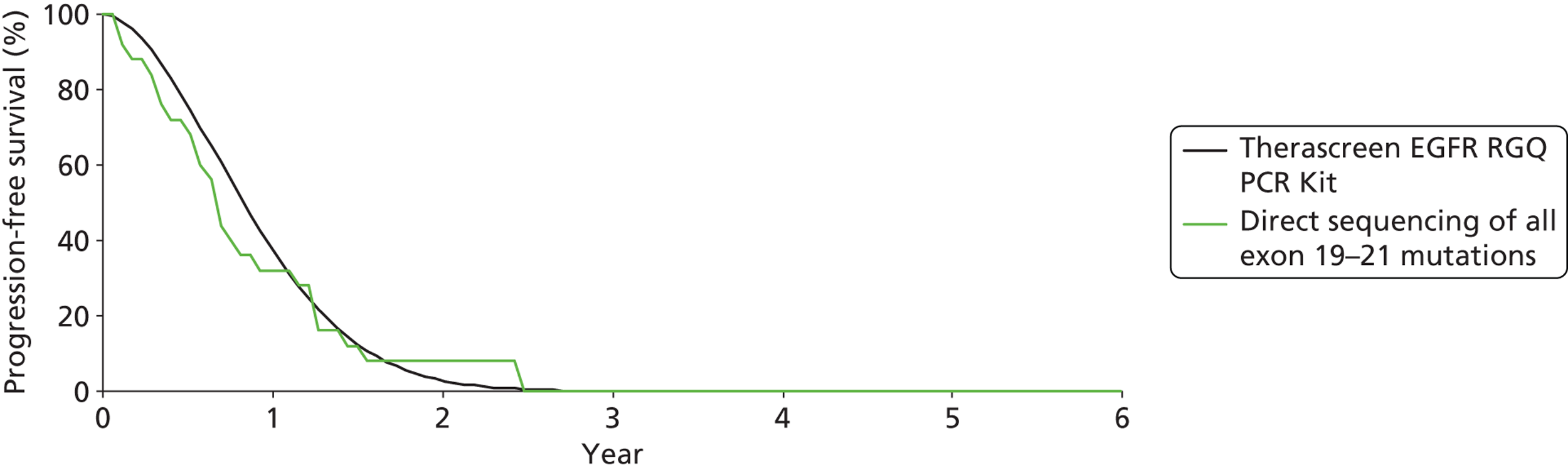
| OS | HR 1.04 (0.65 to 1.68) | |||||
| DC | RR 1.21 (1.00 to 1.47) | |||||
| OR | RR 3.89 (2.34 to 6.68) | |||||
| Zhou (OPTIMAL) (2011)16 | Direct sequencing (PCR based); exon 19 deletions and exon mutation 21 L868R | n = 154 None reported |
Erlotinib | Carboplatin plus gemcitabine | PFS | HR 0.16 (0.10 to 0.26) |
| DC | RR 1.18 (1.06 to 1.35) | |||||
| OR | RR 2.30 (1.70 to 3.23) |
Study details
Participant characteristics varied across studies. Four studies were conducted in East Asia: one reported that it included > 99% East Asian participants,48 and three other studies did not report details of participant ethnicity but were conducted entirely in Japan,47 China16 and South Korea. 41 The remaining study was conducted in multiple centres across Spain and France and included almost entirely (> 99%) white patients. 40 One study included only participants who had never smoked,41 one study included mainly (94%) participants who had never smoked48 and the remaining studies included similar proportions of participants who had never smoked (range 62–71%). 16,40,47 One study included only participants with adenocarcinoma,41 and in the remaining studies approximately 90% of participants had a histological diagnosis of adenocarcinoma. Two studies reported the inclusion of very small numbers of participants with squamous cell carcinoma (n = 547 and n = 140). The majority of participants (> 75%) in all studies had stage IV disease. Full details of study participants are reported in Appendix 2.
The included trials used various methods to assess EGFR mutation status. Two studies, the EURTAC40 and OPTIMAL16 trials, used direct sequencing methods; however, both limited the definition of positive EGFR mutation status to the presence of an ‘activating mutation’ (exon 19 deletions or exon 21 mutation L858R). One additional study reported the results of a re-analysis of samples from the EURTAC study using the cobas EGFR Mutation Test, which can detect 41 EGFR mutations [G719X (G719S/G719A/G719C) in exon 18; 29 deletions and complex mutations in exon 19; T790M in exon 20; S768I in exon 20; five insertions in exon 20; and L858R point mutation in exon 21]. 36 The remaining three studies also used EGFR mutation tests that targeted a wider range of mutations. The IPASS trial used version 1 of the Therascreen EGFR PCR Kit, which detects 19 exon 19 deletions (does not distinguish between individual deletions); exon 21 point mutations L858R and L861Q; the exon 20 resistance mutation T790M; exon 20 mutation S768I; exon 18 mutations G719X (does not distinguish between G719S, G719A and G719C); and three exon 20 deletions. 48 The NEJSG trial used fragment length analysis, targeting exon 19 deletions, exon 21 point mutations (L858R, L861Q), exon 18 point mutations (G719A, G719C, G719S) and exon 20 point mutation (T790M). 47 The First-SIGNAL trial used direct sequencing of exons 19–21. 41
The primary outcome measure, reported by all studies, was PFS, defined as the time from date of randomisation to when progression was first observed or death. Three studies reported intention-to-treat (ITT) analyses of PFS,36,40,49 and three studies excluded withdrawals and patients who did not receive study treatments (four patients,47 four patients41 and 21 patients16); full details of withdrawals are reported as part of the risk of bias assessment (see Appendix 3). With the exception of the re-analysis of samples from the EURTAC trial,36 studies also reported response to treatment outcomes (DC and/or OR). All but one trial used the RECIST criteria21 to evaluate best observed response to treatment during the study period. The First-SIGNAL trial reported that response was evaluated according to the WHO criteria20 but provided no further details. Tumour response was assessed every 6 weeks,16,40,49 every 9 weeks,41 or every 2 months47 until progression. Some limited data were also reported for CR and OS.
Clinical outcomes in patients with epidermal growth factor receptor mutation-positive tumours who were treated with tyrosine kinase inhibitors compared with those treated with standard chemotherapy
All studies in this section reported improvements in OR and improvements or trends towards improvement in PFS for patients with EGFR mutation-positive tumours who were treated with TKIs compared with those treated with standard chemotherapy. There were no clear differences in treatment effect, regardless of which EGFR mutation test (selective for activating mutations exon 19 deletions and exon 21 L858R, or targeting a wider range of mutations) was used to select patients (Figures 6 and 7). Based on subgroup analyses conducted within the trials, three trials reported no significant difference in the HR for PFS between patients with exon 19 deletions and those with the exon 21 mutation L858R. 40,47,48 However, the IPASS study also noted that, although the OR rate was higher in patients with exon 19 deletions who were treated with gefitinib (84.8%) than in those who were treated with standard chemotherapy (43.2%), there was no significant difference between the two treatment groups for patients with the exon 21 mutation L858R (OR rates were 60.9% and 53.2% for the gefitinib and standard chemotherapy groups, respectively). 48 One trial also reported that HRs for PFS did not differ significantly between patients with and without previous surgery, radiotherapy or adjuvant/neoadjuvant chemotherapy, by age, gender or PS; subgroup analyses by smoking status indicated that the treatment effect in favour of gefitinib was significant only in patients who had never smoked [HR 0.24 (95% CI 0.15 to 0.39)]. 40 One further trial noted that HRs for PFS appeared similar across all clinical subgroups (age, gender, PS, disease stage, histology and smoking status). 48 However, the authors noted that the trial was not powered to detect differences between subgroups. Where reported, the median PFS for participants with EGFR mutation-positive tumours in the TKI group was 9.7 months (95% CI 8.4 to 12.3 months),40 10.8 months47 and 13.1 months (95% CI 10.6 to 16.5 months). 16 The corresponding PFS values in the standard chemotherapy groups were 5.2 (95% CI 4.3 to 5.8) months,40 5.4 months47 and 4.6 months (95% CI 4.2 to 5.4 months). 16 The OR rates for participants with EGFR mutation-positive tumours in the TKI groups were 71% (94/132),49 58% (50/86),40 74% (84/114)47 and 83% (68/82). 16 The corresponding OR rates in the standard chemotherapy groups were 47% (61/129),49 15% (13/87),40 31% (35/114)47 and 36% (26/72). 16 Where DC was used as the outcome measure, the observed benefits of TKI treatment were generally more marginal, but there were no clear differences between studies using different EGFR mutation testing methods (Figure 8). Three studies reported OS40,47,48 but none found a significant difference between patients treated with TKIs and those treated with standard chemotherapy (see Table 10). Four studies reported data on the number of patients with CR as the best observed response; the numbers of CR were small in all cases (two,40 two,16 three48 and five47 patients in the TKI groups, and one49 patient in one standard chemotherapy group).
FIGURE 6.
Progression-free survival in patients with EGFR mutation-positive tumours who were treated with TKIs compared with those treated with standard chemotherapy.
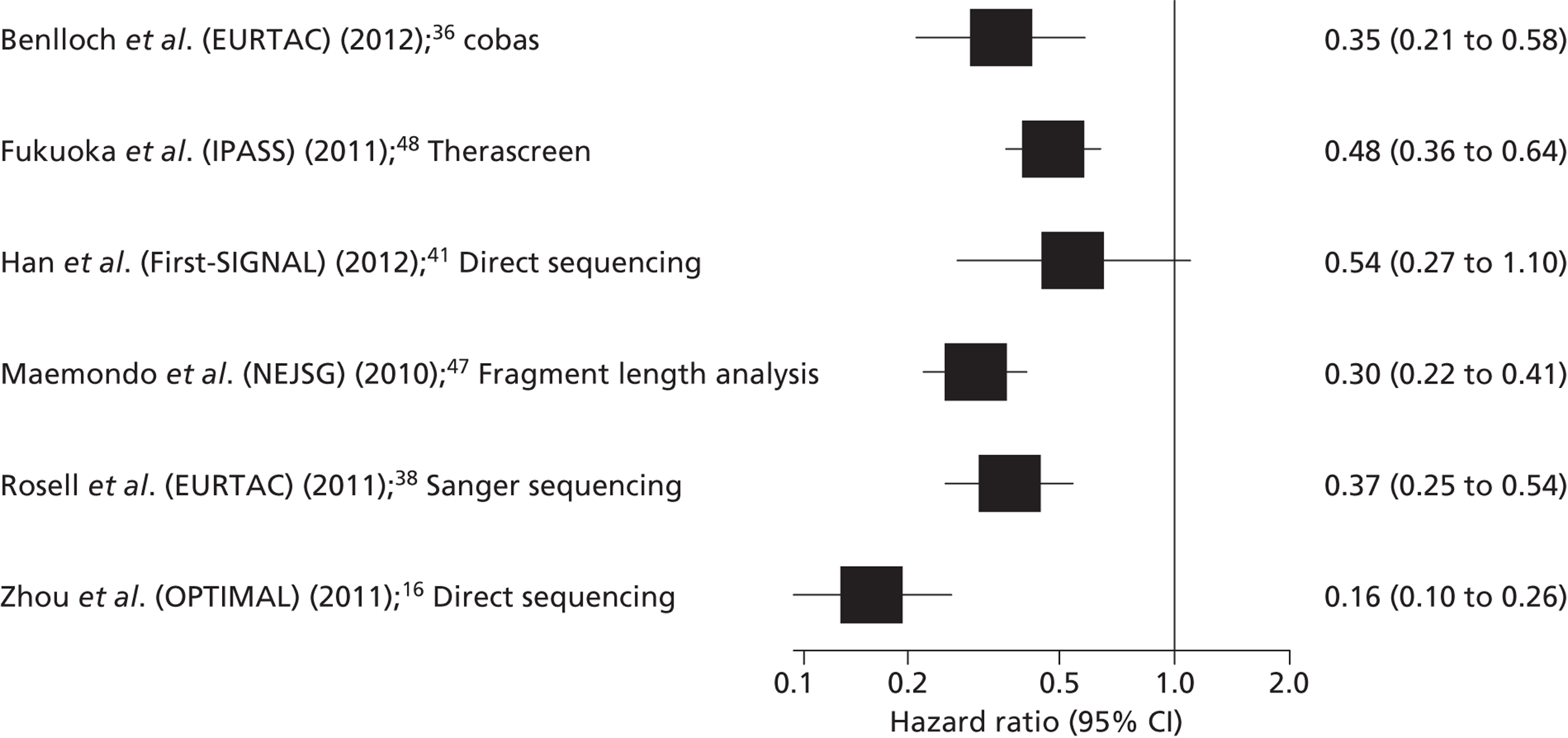
FIGURE 7.
Objective response in patients with EGFR mutation-positive tumours who were treated with TKIs compared with those treated with standard chemotherapy.
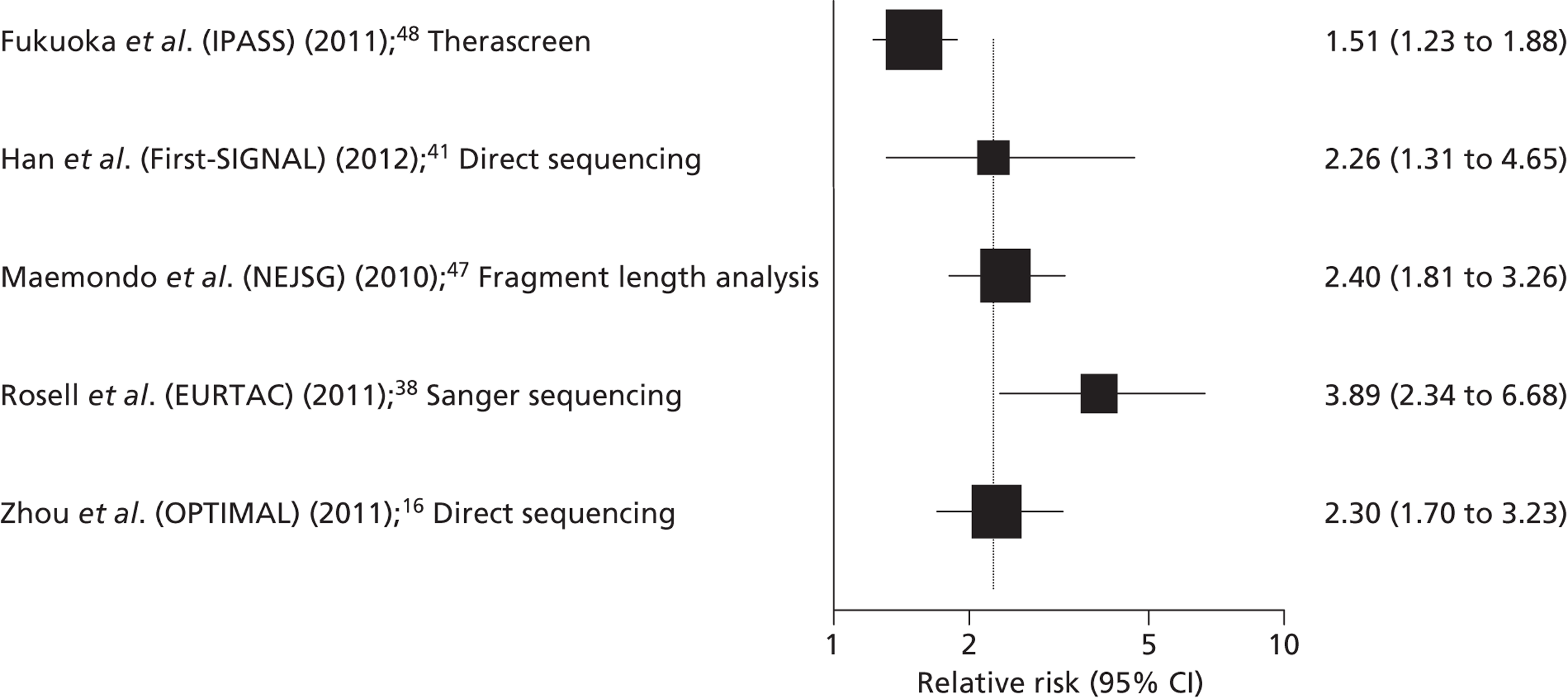
FIGURE 8.
Disease control in patients with EGFR mutation-positive tumours who were treated with TKIs compared with those treated with standard chemotherapy.
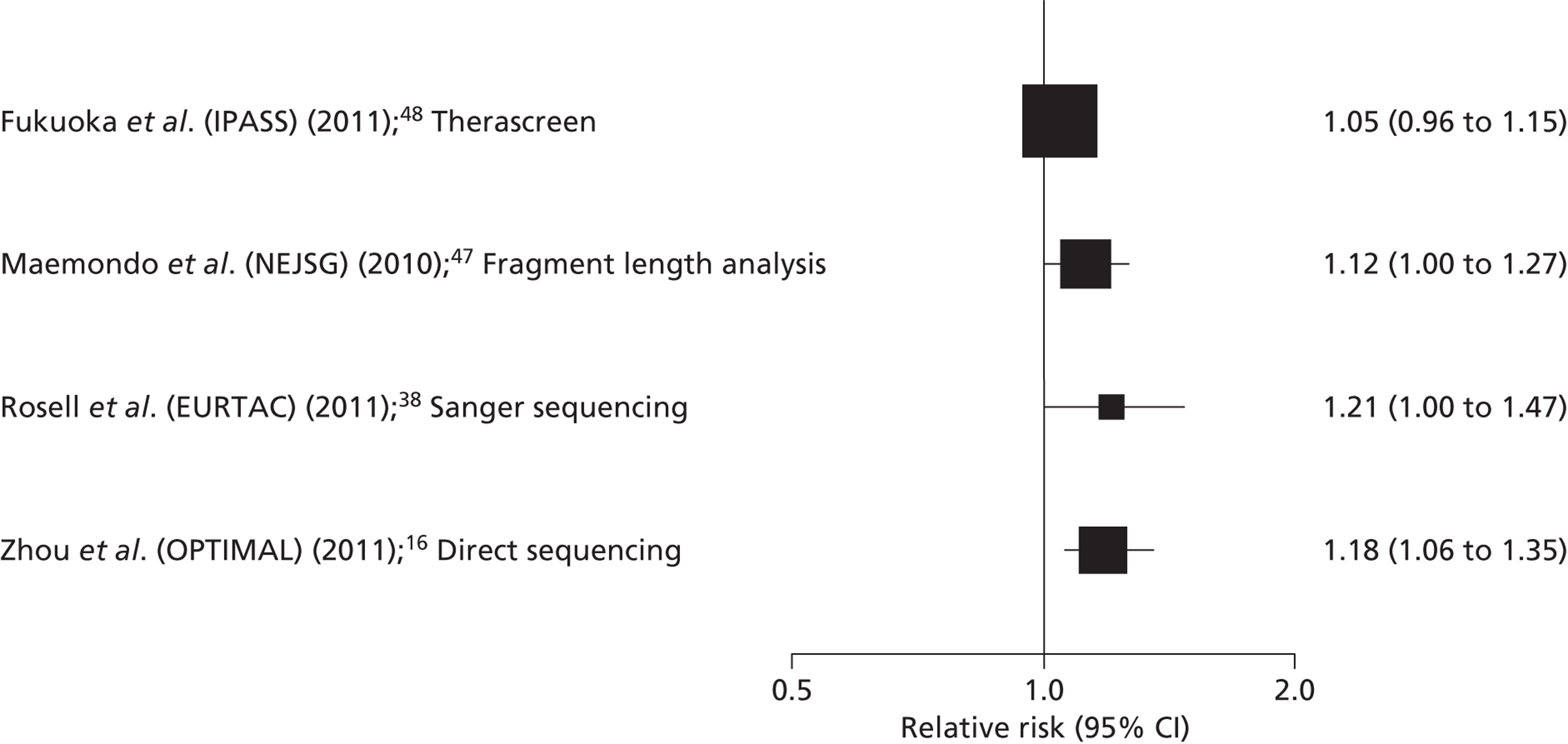
Minimum sample requirements
The IPASS trial, which used version 1 of the Therascreen EGFR PCR Kit, reported the minimum quantity of DNA required to detect 1% for each mutation targeted was 1.5 ng for all mutations except insertions that required 3.0 ng. 48 The study36 that reported data for a subgroup of patients from the EURTAC trial,40 whose samples had been re-analysed using cobas EGFR Mutation Test, reported that the cobas EGFR Mutation Test had a lower ‘invalid rate’ (8.8%) than Sanger sequencing (15.5%), and noted that the cobas EGFR Mutation Test requires a total DNA input of 150 ng. No other trial reported information on the limit of detection of the EGFR mutation test method used. Details of non-evaluable samples were generally poorly reported; any information reported is presented in Table 10.
Clinical outcome for studies that provided data for patients according to tyrosine kinase inhibitor mutation test status
The results of the IPASS subgroup analyses indicated that PFS was significantly longer for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-positive subgroup [HR 0.48 (95% CI 0.36 to 0.64)], and significantly shorter for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-negative subgroup [HR 2.85 (95% CI 2.05 to 3.98)],49 whereas results in the subgroup with unknown mutation status were similar to those observed in the whole study population [HR 0.68 (95% CI 0.58 to 0.81) and HR 0.75 (95% CI 0.65 to 0.85), respectively. The results of the First-SIGNAL subgroup analyses showed a trend towards longer PFS for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-positive subgroup [HR 0.54 (95% CI 0.27 to 1.10)] and a trend towards significantly shorter PFS for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-negative subgroup [HR 1.42 (95% CI 0.82 to 2.47)]; the small size of the EGFR mutation tested subgroup in this study is reflected in the wide CIs around these estimates. 41
In the IPASS trial, the OR rates for mutation-negative participants were 1% (1/91) for the TKI group and 24% (20/85) for the standard chemotherapy group, and for participants whose mutation status was unknown the OR rates were 43% (167/386) for the TKI group and 29% (115/394) for the standard chemotherapy group. The First-SIGNAL trial reported similar data on OR rates for participants whose tumours tested negative for EGFR mutations [26% (7/27) for the TKI group and 52 (14/27) for the standard chemotherapy group]. 41
Risk of bias
All of the studies in this section were rated as ‘low’ or ‘unclear’ risk of bias for randomisation, allocation concealment and selective outcome reporting. All studies were rated as ‘high’ risk of bias for blinding of study participants and personnel; blinding of study participants and personnel was not possible in these trials because of the different routes of administration used for the treatment and comparator arms [oral TKI vs. intravenous (i.v.) standard chemotherapy]. However, only one study was rated as ‘high’ risk of bias for blinding of outcome assessors;16 three studies reported independent outcome assessment40,41,47 and the remaining study did not report details of outcome assessor blinding. 48,49 With the exception of the OPTIMAL16 and First-SIGNAL41 trials, all studies were rated as ‘low’ risk of bias for incomplete reporting of outcome data; all other studies either reported ITT analyses40,48 or very small numbers of withdrawals (< 2% of the total study population). 47 For risk of bias assessment, EURTAC trial40 and the re-analysis of the EURTAC trial were treated as one study. The results of risk of bias assessments are summarised in Table 11 and Figure 9, and full risk of bias assessments for each study are provided in Appendix 3.
| Study | Risk of bias | |||||
|---|---|---|---|---|---|---|
| Randomisation | Allocation concealment | Participant and personnel blinding | Outcome assessor blinding | Incomplete outcome data | Selective outcome reporting | |
| Fukuoka (IPASS) (2011)48,49 | ? | ? | ☹ | ? | ☺ | ☺ |
| Han (First-SIGNAL) (2012)41 | ? | ? | ☹ | ☺ | ☹ | ☺ |
| Maemondo (NEJSG) (2010)47 | ? | ? | ☹ | ☺ | ☺ | ☺ |
| Rosell (EURTAC) (2012)40/Benlloch (2012)36 | ☺ | ? | ☹ | ☺ | ☺ | ☺ |
| Zhou (OPTIMAL) (2011)16 | ☺ | ☺ | ☹ | ☹ | ☹ | ☺ |
FIGURE 9.
Summary of risk of bias assessments.
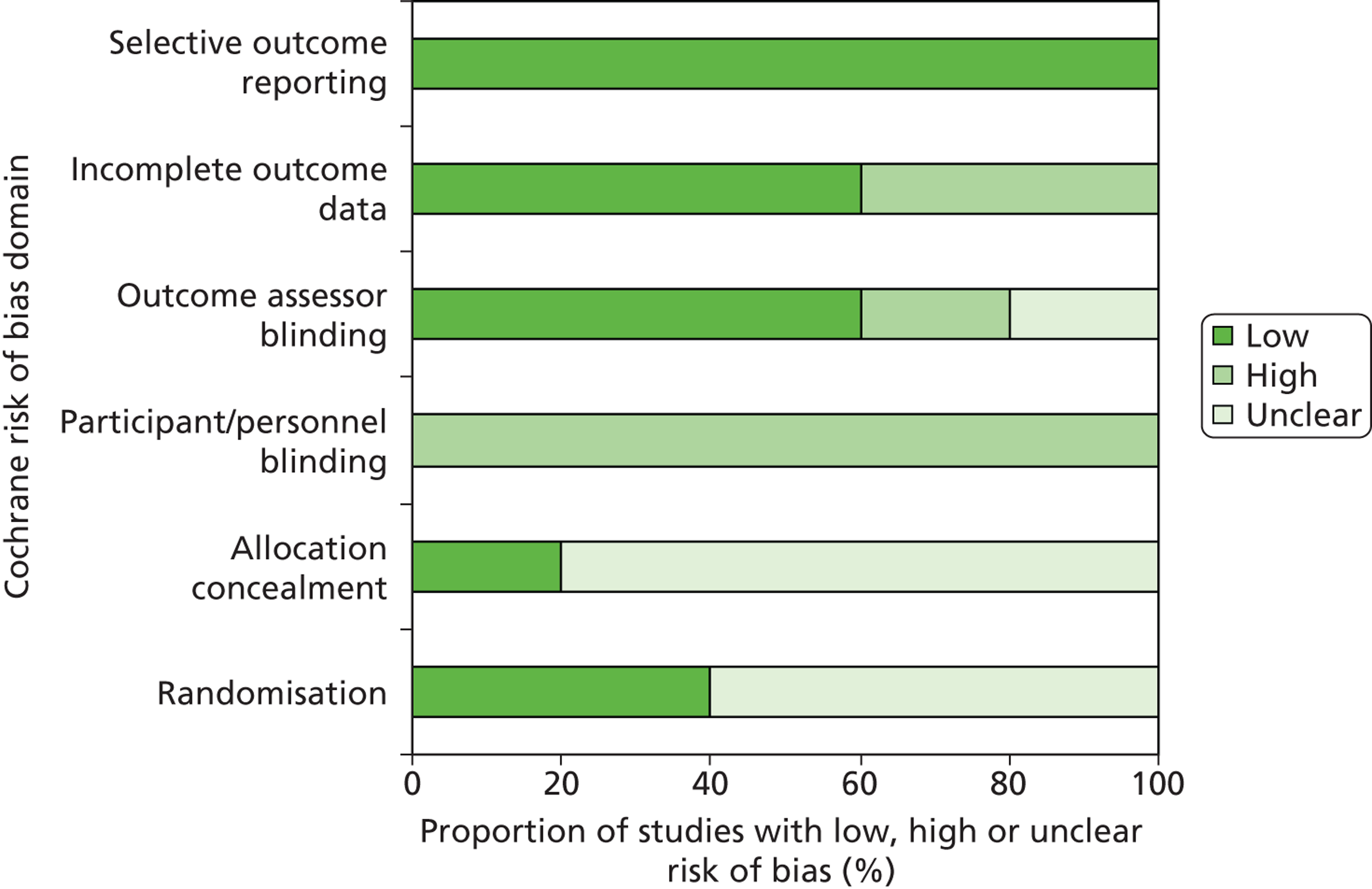
Chapter 4 Assessment of cost-effectiveness
This chapter explores the cost-effectiveness of the use of different EGFR mutation tests to decide between standard chemotherapy and EGFR-TKIs in patients with previously untreated locally advanced or metastatic NSCLC.
Review of economic analyses of tyrosine kinase inhibitor mutation testing
Search strategy
Searches were undertaken to identify cost-effectiveness studies of EGFR-TK testing in NSCLC. As with the clinical effectiveness searching, the main EMBASE strategy for each set of searches was independently peer reviewed by a second information specialist, using the PRESS-EBC checklist. 26 Search strategies were developed specifically for each database, and searches took into account generic and other product names for the intervention. All search strategies are reported in Appendix 1.
The following databases were searched for relevant studies from 2000 to present:
-
MEDLINE (OvidSP) (2000 to September 2012 week 4)
-
MEDLINE In-Process Citations and Daily Update (OvidSP) (2000 to 29 August 2012)
-
EMBASE (OvidSP) (2000–2012 week 34)
-
NHS Economic Evaluation Database (NHS EED) (via The Cochrane Library) (2000–2012/Issue 3)
-
Health Economic Evaluation Database (HEED) (Wiley) (2000 to 30 August 2012)
-
Science Citation Index (SCI) (Web of Science) (2000 to 29 August 2012)
Additional searches were undertaken to update the Resource Utilisation searches in the manufacturer’s submission for STA 192. 51 For this work, the following resources were searched:
-
MEDLINE (OvidSP) (2000 to September 2012 week 4)
-
Medline In-Process & Other Non-Indexed Citations and Daily Update (OvidSP) (2000 to 29 August 2012)
-
EMBASE (OvidSP) (2000–2012 week 40)
-
NHS Economic Evaluation Database (NHS EED) (Internet) (2009 to 30 August 2012)
-
Cumulative Index to Nursing and Allied Health Literature (CINAHL) (EBSCOhost) (2009 to 24 August 2012).
Identified references were downloaded in EndNote X4 software for further assessment and handling.
References in retrieved articles were checked for additional studies.
Inclusion criteria
Studies reporting a full economic analysis that related explicitly to the test–treat combination of EGFR mutation testing and treatment with EGFR-TKIs were eligible for inclusion. Specifically, one of the comparators included EGFR mutation testing and for this comparator the treatment decision was guided by the test result; patients whose tumour was EGFR mutation negative were also included in the treatment pathway.
Results
The search retrieved 606 references. Studies were independently assessed for inclusion by two health economists (BR and AvA) and any disagreements were resolved by discussion. After initial screening of titles and abstracts four studies remained, all of which were published as conference abstracts only. During the course of the assessment we identified two additional studies: one published as a conference abstract only and one published as a full paper and a conference abstract. In total, six studies were included, of which only one was published as a full paper. A summary of the full paper by Borget et al. 52 is provided in Table 12, with a quality checklist based on Drummond et al. 53 shown in Table 13. A condensed summary of the conference abstracts is provided in Table 14.
| Study details | Borget (2012)52 |
|---|---|
| Population | Patients with advanced NSCLC in whom at least one platinum-based chemotherapy regimen had failed and who were eligible for erlotinib or chemotherapy |
| Time horizon | 30 months |
| Objective | To compare the cost-effectiveness ratios of three hypothetical strategies for NSCLC |
| Source of effectiveness information |
|
| Comparators |
|
| Unit costs | Source unclear, probably French health-care payer? |
| Measure of benefit | QALYs |
| Study type | Cost–utility analysis: Markov model |
| Model assumptions | Patients who progressed were assumed to receive palliative care until death |
| Perspective | French health-care payer |
| Discount rate | 3% for costs only |
| Uncertainty around cost-effectiveness ratio expressed | Yes, in numbers for one-way sensitivity analyses, incremental cost-effectiveness planes and cost-effectiveness acceptability curves for PSA |
| Sensitivity analysis | One-way sensitivity analyses (selection criteria for second strategy, prevalence of EGFR mutation, biological testing cost, post progression cost, erlotinib tariff) and PSA |
| Outcome (cost and Lys/QALYs) per comparator | No selection: 0.478 QALYs €21,025 Clinically guided: 0.558 QALYs €16,005 Biologically guided: 0.559 QALYs €15,210 |
| Summary of incremental analysis | The biologically and clinically guided strategies were dominant, but the biological strategy was slightly less expensive than the clinical strategy |
| Borget (2012)52 | |
|---|---|
| Study design | |
| The research question is stated | ✓ |
| The economic importance of the research question is stated | ✓ |
| The viewpoint(s) of the analysis is clearly stated and justified | ✓ |
| The rationale for choosing alternative programmes or interventions compared is stated | ✓ |
| The alternatives being compared are clearly described | ✓ |
| The form of economic evaluation used is stated | ✓ |
| The choice of form of economic evaluation is justified in relation to the questions addressed | ✓ |
| Data collection | |
| The source(s) of effectiveness estimates used is stated | ✓ |
| Details of the design and results of effectiveness study are given (if based on a single study) | ✓ |
| Details of the methods of synthesis or meta-analysis of estimates are given (if based on a synthesis of a number of effectiveness studies) | NA |
| The primary outcome measure(s) for the economic evaluation is clearly stated | ✓ |
| Methods to value benefits are stated | ✓ |
| Details of the subjects from whom valuations were obtained were given | ✓ |
| Productivity changes (if included) are reported separately | NA |
| The relevance of productivity changes to the study question is discussed | ✗ |
| Quantities of resource use are reported separately from their unit costs | ✗ |
| Methods for the estimation of quantities and unit costs are described | ✓ |
| Currency and price data are recorded | ✗ |
| Details of currency of price adjustments for inflation or currency conversion are given | ✗ |
| Details of any model used are given | ✓ |
| The choice of model used and the key parameters on which it is based are justified | ✓ |
| Analysis and interpretation of results | |
| Time horizon of costs and benefits is stated | ✓ |
| The discount rate(s) is stated | ✓ |
| The choice of discount rate(s) is justified | ✗ |
| An explanation is given if costs and benefits are not discounted | NA |
| Details of statistical tests and CIs are given for stochastic data | ✓ |
| The approach to sensitivity analysis is given | ✓ |
| The choice of variables for sensitivity analysis is justified | ✓ |
| The ranges over which the variables are varied are justified | ✓ |
| Relevant alternatives are compared | ✓ |
| Incremental analysis is reported | ✓ |
| Major outcomes are presented in a disaggregated as well as aggregated form | ✗ |
| The answer to the study question is given | ✓ |
| Conclusions follow from the data reported | ✓ |
| Conclusions are accompanied by the appropriate caveats | ✓ |
| Study details | Arrieta (2010)54 | Chen (2011)55 | Jacob (2011)56 | Lopes (2011)57 | Shiroiwa (2012)58 |
|---|---|---|---|---|---|
| Population | Patients with advanced NSCLC | Patients with advanced NSCLC in Ontario | Patients with NSCLC in Sweden | Patients with advanced NSCLC | Patients with NSCLC in Japan |
| Objective | Assess cost-effectiveness of EGFR mutation testing | Assess the cost-effectiveness of EGFR mutation testing to guide first-line gefitinib treatment | Evaluate the cost-effectiveness of a treatment strategy with gefitinib, based on data from the IPASS trial49 | Determine the cost-effectiveness of EGFR mutation testing and first-line treatment with gefitinib for patients with EGFR positive tumours | Not stated |
| Comparators |
|
|
|
|
|
| Method of analysis | Discrete event simulation/Markov model | Decision-analytic model | Markov model | Markov model | Not stated |
| Measure of benefit | Progression-free months | LYs, QALYs | QALYs | QALYs | LYs |
| Outcome (cost and LYs/QALYs) | Progression-free months 7.57 in testing strategy, 7.11 in no testing strategy; costs not stated | Not specified | Not specified | Not specified | Not specified |
| Summary of incremental analysis | ICER of testing vs. no testing: US$1379.49 per progression-free month gained | ICER for testing vs. no testing US$46,021 per LY and US$81,071 per QALY gained | Test and treat strategy associated with a QALY gain of 0.0116 at an IC of €300; ICER for test and treat strategy was €25,900 | EGFR testing and first-line treatment with gefitinib was found to be dominant compared with standard care | ICER of (1) vs. (3) was US$12,000 ICER of (2) vs. (3) was US$46,500a |
Borget et al. 52 developed a Markov model to compare three hypothetical strategies for second-line treatment with erlotinib in patients with NSCLC for whom at least one platinum-based chemotherapy regimen had failed and who were eligible for erlotinib or chemotherapy.
The three hypothetical strategies were:
-
no patient selection, all patients receive erlotinib
-
clinically guided, patients with favourable clinical features (female, never smokers, with adenocarcinoma) receive erlotinib, others receive docetaxel
-
biologically guided, patients with known EGFR mutations received erlotinib, others receive docetaxel.
Clinical inputs were derived from IPD in the ERMETIC study59 and the GFPC0506 study. 60 Utilities were derived from population-based studies of advanced NSCLC performed in the UK. 61 Total costs included the following categories: chemotherapy drugs, erlotinib, supportive treatments (including treatment for adverse events), transfusion and hospitalisation for any reason, costs after progression and palliative care.
Total quality-adjusted life-years (QALYs) were 0.478, 0.558 and 0.559 for the no selection, clinically guided and biologically guided strategies, respectively. The respective total costs were €21,025, €16,005 and €15,210. The no selection strategy was both the least effective and the most expensive. The biologically and clinically guided strategies had comparable effectiveness, but the biologically guided strategy was slightly less expensive. Results were robust in the sensitivity analyses.
Although this study was of good quality, it does not match our decision problem, as it concerns second-line use of EGFR-TKIs, whereas this assessment concerns first-line treatment with TKIs. The conference abstracts identified all concern the first-line use of TKIs, but do not provide sufficient information to be of use. However, as all were relatively recent, more informative full publications may follow.
Model structure and methodology
Epidermal growth factor receptor tyrosine kinase mutation tests considered in the model
In the health-economic analysis, the cost-effectiveness of different methods for EGFR-TK mutation testing to decide between standard chemotherapy and anti-EGFR-TKIs in patients with locally advanced or metastatic NSCLC was assessed. A range of methods for EGFR-TK mutation testing are currently used in NHS laboratories in England and Wales.
Ideally, the performance of these tests would be assessed against an objective measure of the true presence/absence of a clinically relevant EGFR-TK mutation (the ‘reference standard’). Comparative effectiveness of treatment (TKI vs. chemotherapy) conditional upon the true or false presence/absence of the EGFR-TK mutation could then be determined. However, each different testing method targets a different range of mutations and has different limits of detection (lowest proportion of mutation detectable in tumour cells) and the exact combination of mutation type and level that will provide optimal treatment selection remains unclear. For this reason, assessment of test performance based on comparison with a conventional ‘reference standard’ is currently not possible. In this situation, an alternative way to determine the relative value of diagnostic methods for EGFR-TK mutation testing is to use studies that report on the comparative treatment effect in patients with different EGFR mutation status (positive, negative or unknown) as defined using different EGFR mutation tests. As outlined in the previous chapter, information on comparative effectiveness (PFS and OS) of TKI and chemotherapy in patients with mutation-positive, mutation-negative and mutation-unknown tumours was available for only the Therascreen EGFR PCR Kit48,49 and in patients with mutation-positive and mutation-negative tumours for one type of direct sequencing (direct sequencing of all exon 19–21 mutations). 41 A major assumption underlying the use of these data in the health-economic modelling, however, is that the difference in comparative treatment effect between the two treatments (e.g. TKI vs. chemotherapy) is solely attributable to the use of different mutation tests. Although direct sequencing of all exon 19–21 mutations is not listed in the scope, it was included in the analyses because of a lack of effectiveness and/or survival data on other direct sequencing methods.
In the absence of evidence on the comparative treatment effect in patients with different EGFR mutation status as defined using different EGFR mutation tests, one could consider the accuracy of different EGFR mutation tests for the prediction of response to treatment with TKIs; in this case, response to treatment with TKIs serves as a clinical reference standard. This type of accuracy data was available for two other direct sequencing tests (direct sequencing of all exon 18–21 mutations46 and direct sequencing or WAVE-HS (Transgenomic, Omaha, NE, USA) for inadequate samples (< 50% of tumour cells of all exon 18–24 mutations44). These studies provided no data on the relative PFS and/or OS separately for patients with mutation-positive and mutation-negative tumours. Therefore, evidence available on the relative PFS and OS for mutation positives and mutation negatives, as observed for direct sequencing of all exon 19–21 mutations, was ‘linked’ to direct sequencing of all exon 18–21 mutations46 and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells). Again, although the test strategy direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) is not listed in the scope, it was included in the analysis because it was the only test for which information on the proportion of patients with unknown mutations status was available.
For the remaining EGFR mutation tests listed in the scope, no accuracy data or information to predict (relative) treatment response, PFS or OS in mutation-positive patients (after treatment with TKIs), mutation-negative patients or patients with unknown mutation status (after treatment with doublet chemotherapy) were available. As a result, for the remaining tests, it was only possible to make a comparison based on differences in technical performance and test costs retrieved from the online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different epidermal growth factor receptor mutation tests?), while assuming equal prognostic value across tests. The latter assumption was not based on evidence of equality but rather absence of any reliable evidence to model a difference in prognostic value for these tests.
Based on the information available to us, three analyses were performed:
-
‘Evidence on comparative effectiveness available’ analysis Therascreen EGFR PCR Kit compared with direct sequencing of all exon 19–21 mutations in order to estimate cost and QALYs using the observed response to treatment and relative PFS and OS data. Information on relative (HR of TKI vs. chemotherapy) PFS and OS in patients with mutation-positive tumours and patients with mutation-negative tumours is not available for other tests. Therefore, in this analysis direct sequencing of all exon 19–21 mutations was used as the closest approximation available to the comparator listed in the scope (direct sequencing of all exon 18–21 mutations).
-
‘Linked evidence’ analysis In this analysis, besides Therascreen EGFR PCR Kit compared with direct sequencing of all exon 19–21 mutations, two other direct sequencing tests [direct sequencing of all exon 18–21 mutations and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells)] for which accuracy data to predict response to treatment were available were included. This was based on the assumption that for the last two direct sequencing methods, the relative PFS and OS for mutation positives and mutation negatives correlate perfectly with relative PFS and OS, as observed for direct sequencing of all exon 19–21 mutations.
-
‘Assumption of equal prognostic value’ analysis For all tests for which information on cost and/or technical performance was available from the online survey. This includes the tests for which neither comparative effectiveness nor response data were available. In this analysis we assessed whether the tests were likely to be cost-effective given an assumption of equal prognostic value and test-specific information on cost and failure rate only. The equal prognostic value assigned was based on data for the Therascreen EGFR PCR Kit, as this was the only test for which prognostic data were available on patients with positive, negative and unknown mutation status. In addition, other tests used in NHS laboratories in England and Wales were considered to have technical characteristics (low limit of detection and similar proportion of tumour cells required for analysis), which were more similar to this test than to direct sequencing methods and would therefore be more likely to have similar prognostic value to the Therascreen EGFR PCR Kit than to direct sequencing. The following tests were included in this analysis:
-
Therascreen EGFR PCR Kit
-
Direct sequencing of exons 19–21
-
Direct sequencing or WAVE-HS for samples with insufficient tumour cells
-
Direct sequencing of exons 18–21
-
Fragment length analysis combined with pyrosequencing
-
Sanger sequencing and fragment length analysis/PCR of negative samples
-
Roche cobas test
-
HRM analysis
-
single-strand conformation analysis
-
Sanger sequencing or Roche cobas for samples with insufficient tumour cells
-
Sanger sequencing or Therascreen for samples with insufficient tumour cells
-
next-generation sequencing
-
Therascreen and Pyrosequencing Kit.
-
Direct sequencing of all exon 18–21 mutations was taken as the comparator in the ‘linked evidence’ and ‘assumption of equal prognostic value’ analyses.
Consistency with related assessments
This assessment does not update the appraisal of gefitinib for the first-line treatment of locally advanced or metastatic NSCLC. 1 In order to ensure consistency between the modelling approach used in Technology Appraisal 192 and the assessment of the cost-effectiveness of different methods for EGFR-TK mutation testing in this report, the assessment group received the health-economic model submitted by AstraZeneca for Technology Appraisal 192. This model calculates the expected cost-effectiveness of gefitinib compared with doublet chemotherapy for the first-line treatment of patients with locally advanced or metastatic NSCLC with a positive EGFR mutation test based on Therascreen EGFR PCR Kit. This model, together with the amendments suggested and made by the External Review Group (ERG), was used to inform the development of a de novo model in which the long-term consequences of using different EGFR mutation tests were assessed not only in patients with a positive EGFR mutation test, but also in patients with a negative test result, or an unknown test result. The assessment group tested the consistency between the de novo model, the AstraZeneca model, and the amendments made by the ERG. We compared the results of patients with a positive EGFR mutation test using Therascreen EGFR PCR Kit with the initial manufacturer’s submission. Subsequently, the ERG amendments were incorporated and ICERs from the de novo model were compared with ICERs, as reported in the final appraisal determination of Technology Assessment 1921 (see Appendix 6 for results). Furthermore, the health-economic analysis did not assess any differences between different TKIs.
Model structure
In the health-economic model the mean expected costs, life-years (LYs) and QALYs were calculated for each alternative.
The health-economic analysis considers the long-term consequences of technical performance and accuracy of the different tests/test combinations followed by treatment with either standard chemotherapy or a TKI in patients with NSCLC. For this purpose a decision tree and a Markov model were developed. The decision tree was used to model the test result (positive, negative or unknown) and the treatment decision. Patients with a positive test result receive an anti-EGFR-TKI. It is assumed that patients with a negative test result or unknown EGFR mutation status will receive doublet chemotherapy (pemetrexed and cisplatin), as the negative consequences of treatment with TKIs in FPs are greater than the negative consequences of treatment with doublet chemotherapy in FNs. 49 The decision tree is shown in Figure 10.
FIGURE 10.
Decision tree structure. CTX, chemotherapy.
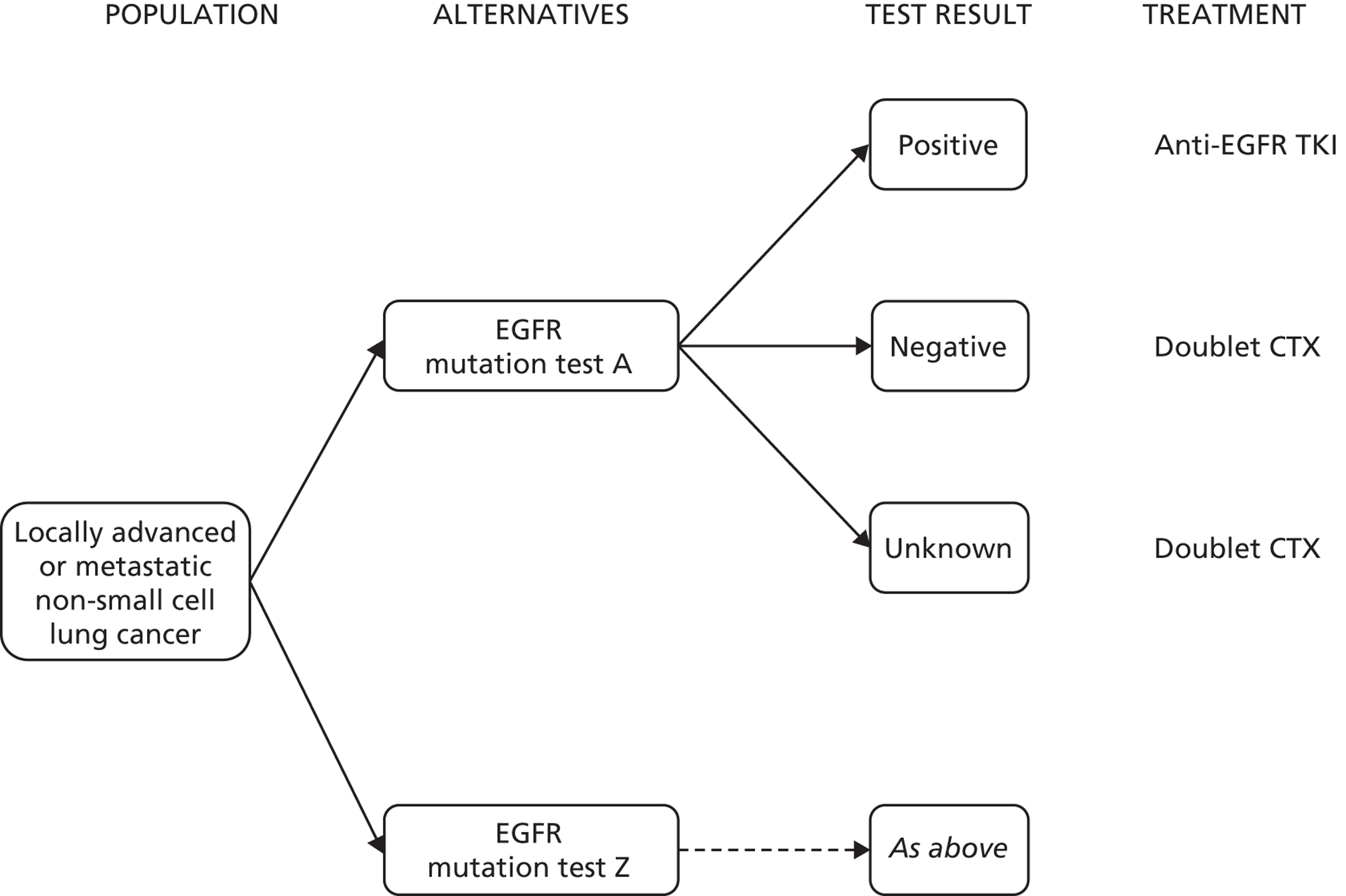
The long-term consequences in terms of costs and QALYs were estimated using a Markov model with a cycle time of 21 days (resembling the duration of one cycle of chemotherapy) and a time horizon of 6 years. Health states in the Markov model are progression free (subdivided into ‘response’ and ‘SD’), disease progression and death. In the progression-free state, patients are on treatment (either TKI or doublet chemotherapy). In each cycle these patients are subdivided over the ‘SD’ and ‘response’ states, based on the OR rate, in order to account for a difference in quality of life between those states. In addition, disutilities and costs associated with treatment related characteristics (i.v. or oral therapy) are modelled. For adverse events of treatment, disutilities and costs were applied for a single cycle in the model. The Markov model structure is shown in Figure 11. The model is described in more detail in NICE Technology Appraisal 192. 51
FIGURE 11.
Markov model structure.
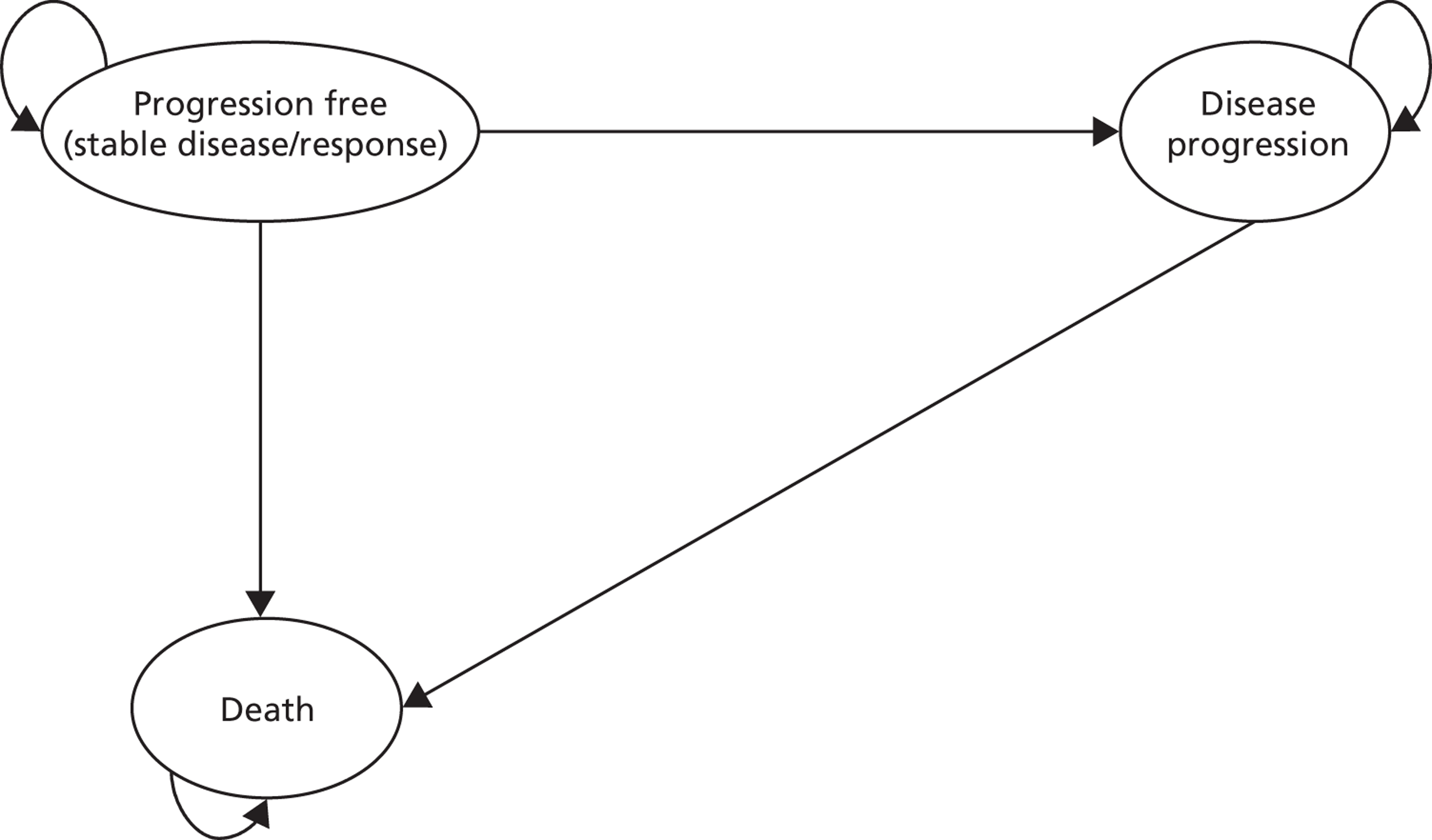
Model parameters
Estimates for model input parameters were retrieved from the industry submission for NICE Technology Appraisal 192,51 the assessment of the clinical effectiveness of different EGFR mutation tests (see Chapter 3, What is the accuracy of epidermal growth factor receptor mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors? and How do outcomes from treatment with epidermal growth factor receptor inhibitors vary according to which test is used to select patients for treatment?), an online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different EGFR mutation tests?) and the Personal Social Services Research Unit (PSSRU). 62
Test result
The proportions of test failures for the EGFR mutation tests were based on the online survey of NHS laboratories in England and Wales. The proportions of positive and negative test results were based on the estimated proportions of EGFR mutation-positive patients in England and Wales [16.6%, standard error (SE) 0.8%],63 the test accuracy (sensitivity and specificity with OR to TKI as reference standard, see Table 7) and the proportion of patients with an unknown test result. The proportions of patients with an unknown test result were based on the proportions of patients without mutation status relative to the number of patients for whom a tissue sample was available in the trials. As the trials do not represent clinical practice, this might be an overestimation of the proportion of patients with an unknown test result in clinical practice. One possible reason for this is that, in the trials, the tissue samples were not generally taken for the purpose of EGFR mutation testing, and may therefore have been inadequate more often than would be the case in current clinical practice. In contrast, the results of the online survey of reference laboratory in England and Wales are likely to provide an underestimation of the total proportion of patients with an unknown test result, as the reference laboratories are not likely to have insight into the total proportion of pre-test failures (samples considered inadequate by the pathologist and therefore not sent to the laboratory). In the base-case analysis the proportion of patients with an unknown test result was based on the literature, whereas in a sensitivity analysis the results of the online survey were used.
The proportion of TP, TN, FN and FP test results were calculated by:
Subsequently, the proportions of patients with a mutation-positive (TP + FP), mutation-negative (TN + FN) test result were calculated. The results are listed in Tables 15 and 16. As is apparent from Table 16, there are substantial differences in the proportions of positive and unknown test results between the various tests. This is a result of the reported numbers of unknowns in the trials and the test accuracy estimates calculated from the trials (see Table 7). As noted previously, the number of unknowns derived in this way may not reflect true clinical practice and is influenced by how the trials were designed and performed. The accuracy estimates also have important limitations, which are described in Chapter 3 (see What is the accuracy of epidermal growth factor receptor mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors?) and Chapter 5 (see Clinical effectiveness).
| Input parameter [estimated value (SE)] | Distribution | Source | ||
|---|---|---|---|---|
| Proportion of EGFR mutation-positive patients in England and Wales | ||||
| Proportion of mutation positives | 16.6% (0.8%) | Beta | Rosell (2009)63 | |
| Test accuracy | Sensitivity | Specificity | ||
| Therascreen | 98.9% (1.0%) | 68.9% (4.2%) | Beta | Mok (2009)49 |
| Direct sequencing of all exon 19–21 mutations | 75.9% (7.8%) | 83.3% (7.5%) | Beta | Han (2012)41 |
| Direct sequencing of all exon 18–21 mutations | 84.4% (5.3%) | 61.1% (8.0%) | Beta | Yang (2008)46 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | 60.0% (20.0%) | 81.3% (6.8%) | Beta | Jackman (2007)44 |
| Probability of unknown test result | ||||
| Therascreen | 22.7% (1.8%) | Beta | Mok (2009)49 | |
| Direct sequencing of all exon 19–21 mutations | Assumed equal to direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | |||
| Direct sequencing of all exon 18–21 mutations | Assumed equal to direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | |||
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | 37.7% (5.8%) | Beta | Jackman (2007)44 | |
| Mutation test | Probability (SE) of test resulta | ||
|---|---|---|---|
| Positive | Unknown | Negative | |
| Therascreen | 32.8% (2.9%) | 22.7% (1.8%) | 44.6% (3.0%) |
| Direct sequencing of all exon 19–21 mutations | 16.5% (4.2%) | 37.7% (5.2%) | 45.8% (5.5%) |
| Direct sequencing of all exon 18–21 mutations | 29.0% (4.6%) | 37.7% (4.2%) | 33.4% (4.8%) |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | 16.0% (4.4%) | 37.7% (5.8%) | 46.4% (6.0%) |
In the third analysis (‘assumption of equal prognostic value’), the probability of positive, unknown and negative test results were assumed to be equal to the Therascreen EGFR PCR Kit for all tests. This assumption was relaxed in a sensitivity analysis.
Response to treatment
Patients who are in the progression-free state are subdivided into the ‘SD’ and ‘response’ states based on the objective tumour response rate. For patients with positive test results, the objective tumour response rate after treatment with TKIs (Table 17) was used and the objective tumour response rate after treatment with doublet chemotherapy was used for the remaining patients (negative or unknown test results).
| Mutation test | OR rate (SE)a,b | Source | ||
|---|---|---|---|---|
| Positive | Unknown | Negative | ||
| Therascreen EGFR PCR Kit | 0.712 (0.039) | 0.292 (0.023) | 0.235 (0.046) | Mok (2009)49 |
| Direct sequencing of all exon 19–21 mutations | 0.846 (0.069) | As for Therascreen EGFR PCR Kit | 0.484 (0.098)c | Han (2012)41 |
| Direct sequencing of all exon 18–21 mutations | 0.731 (0.061) | As for Therascreen EGFR PCR Kit | As direct sequencing of all exon 19–21 mutations | Yang (2008)46 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | 0.333 (0.149) | As for Therascreen EGFR PCR Kit | As direct sequencing of all exon 19–21 mutations | Jackman (2007)44 |
Survival
As was the case in NICE Technology Appraisal 192, two separate Weibull models were used to estimate cycle-dependent transitions for PFS and OS while on doublet chemotherapy for positive, negative and unknown mutation status. Figure 12 provides a schematic representation of the modelling approach.
FIGURE 12.
Modelling of overall and PFS.
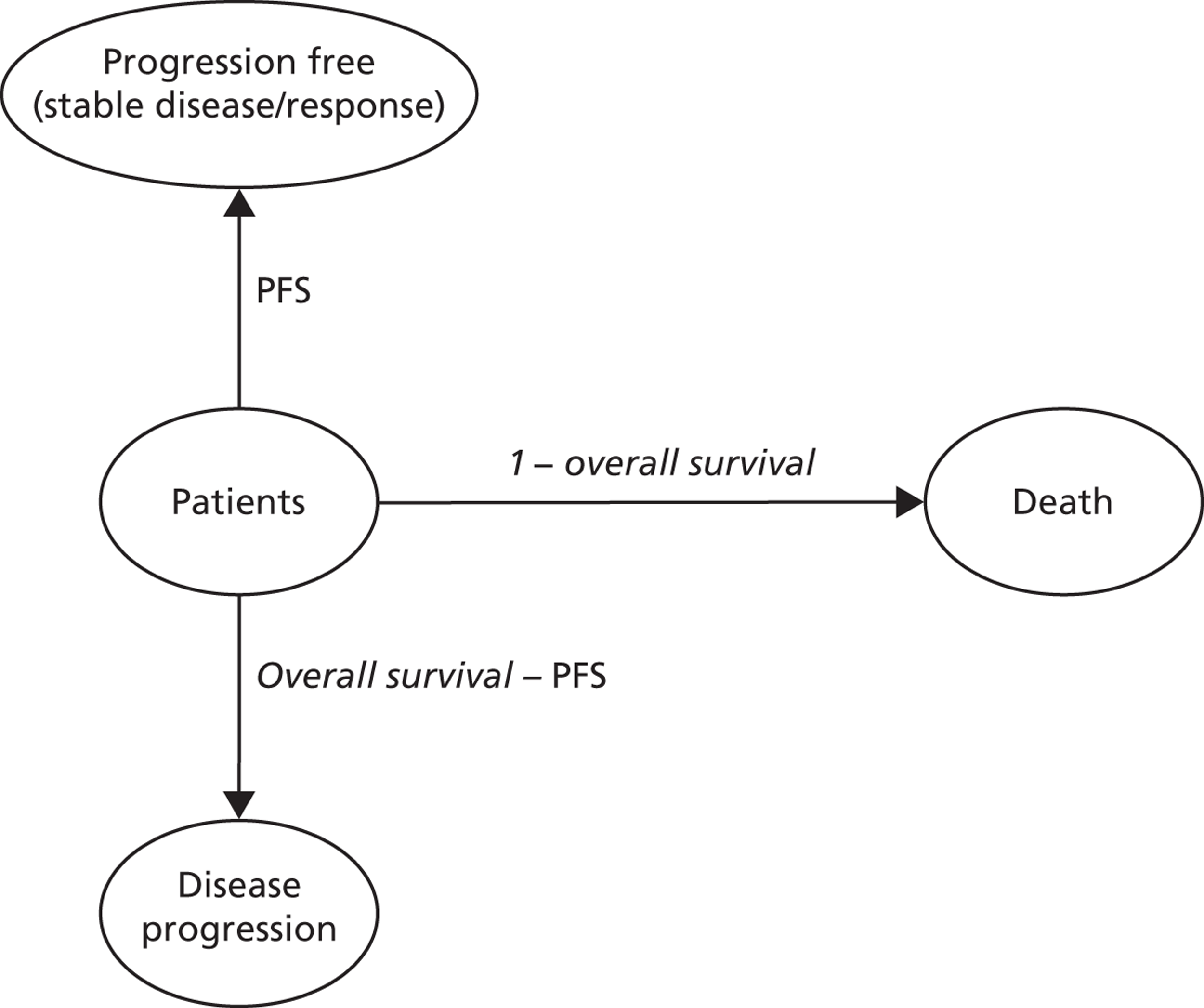
For testing using the Therascreen EGFR PCR Kit, PFS and OS were modelled using the Weibull regression models based on the IPASS study49 and a HR for TKI (based on a meta-analysis and mixed-treatment comparison) used in NICE Technology Appraisal 192. 51 The Weibull regression models have separate lambda and alpha parameters for patients with mutation-positive, -unknown and -negative tumours, and are based on treatment with doublet chemotherapy (Table 18).
To estimate PFS and OS for patients treated with TKIs after a positive test result using the Therascreen EGFR PCR Kit, a HR of 0.43 (95% CI 0.34 to 0.53) was applied to the Weibull function for mutation positives. This HR was modelled using a log-normal distribution.
For direct sequencing of all exon 19–21 mutations, PFS and OS for mutation positives after EGFR-TKI and negatives after doublet chemotherapy were modelled using Kaplan–Meier curves extracted from the First-SIGNAL trial. 41 The corresponding SEs were calculated using the Peto method. 64 In the First-SIGNAL trial, mutation-negative patients were treated with gemcitabine and cisplatin. 41 The PFS and OS estimates obtained for these mutation-negative patients were adjusted (HR = 1.087 for PFS, and HR = 1.087 for OS) to correspond with treatment with paclitaxel and carboplatin. 51 PFS and OS for patients with tumours of unknown mutation status were based on the IPASS Weibull model for unknown mutations, as these were not reported in the First-SIGNAL trial.
Consistent with the use of pemetrexed and cisplatin as doublet chemotherapy, the HRs reported in Table 19 were used to recalculate PFS and OS for both comparators. Accordingly, OR rate presented in Table 17 was recalculated to correspond with pemetrexed and cisplatin. These HRs and odds ratios were retrieved from the updated mixed-treatment comparison from NICE Technology Appraisal 192.
| Estimate | Lower 95% CI | Upper 95% CI | Distribution | |
|---|---|---|---|---|
| HRs progression free and OS | ||||
| PFS | 0.88 | 0.74 | 1.05 | Log-normal |
| OS | 0.78 | 0.65 | 0.93 | Log-normal |
| Odds ratios | ||||
| OR rate | 1.64 | 1.15 | 2.27 | Log-normal |
| Neutropenia | 0.46 | 0.07 | 1.62 | Log-normal |
| Febrile neutropenia | 0.19 | 0.01 | 0.84 | Log-normal |
| Fatigue | 2.62 | 1.30 | 4.65 | Log-normal |
| Nausea and vomiting | 10.92 | 1.11 | 41.94 | Log-normal |
| Diarrhoea | 1.00 | – | – | Fixed |
| Hair loss (grade 2) | 1.00 | – | – | Fixed |
| Anaemia | 1.62 | 0.54 | 3.75 | Log-normal |
The PFS and OS curves for patients tested with the Therascreen EGFR PCR Kit and with direct sequencing of all exon 19–21 mutations for the ‘evidence on comparative effectiveness available’ analysis are presented in Figures 13 and 14.
In the ‘linked evidence’ analysis, PFS and OS for patients tested with direct sequencing of all exon 18–21 mutations and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) were assumed equal to the PFS and OS as described above for direct sequencing of all exon 19–21 mutations. PFS and OS for patients tested with the Therascreen EGFR PCR Kit and with direct sequencing of all exon 19–21 mutations in the ‘linked evidence’ analysis was equal to the estimates used in the ‘evidence on comparative effectiveness available’ analysis.
Adverse events
The occurrence of adverse events was assumed to be dependent on treatment and independent of EGFR mutation status, i.e. adverse events for patients with mutation-negative and mutation-unknown tumours were assumed to be equal after chemotherapy. The occurrence of adverse events is presented in Table 20.
| Adverse event per treatment | Probability | SE | Distribution |
|---|---|---|---|
| Neutropenia | 0.0% | – | Fixed |
| Febrile neutropenia | 0.0% | – | Fixed |
| Fatigue | 0.0% | – | Fixed |
| Nausea and/or vomiting | 0.0% | – | Fixed |
| Diarrhoea | 5.3% | – | Fixed |
| Hair loss (grade 2) | 1.2% | – | Fixed |
| Rash | 2.3% | – | Fixed |
| Anaemia | 1.5% | – | Fixed |
| Neutropenia | 33.3% | – | Fixed |
| Febrile neutropenia | 3.9% | – | Fixed |
| Fatigue | 2.3% | – | Fixed |
| Nausea and/or vomiting | 4.7% | – | Fixed |
| Diarrhoea | 0.8% | – | Fixed |
| Hair loss (grade 2) | 31.6% | – | Fixed |
| Rash | 0.0% | – | Fixed |
| Anaemia | 9.3% | – | Fixed |
As for PFS and OS, the occurrence of adverse events after doublet chemotherapy (as presented in Table 20) is adjusted using the odds ratios in Table 19 to correspond to treatment with pemetrexed and cisplatin. The odds ratios for diarrhoea and hair loss were assumed to be 1.00 (resulting in an equal occurrence of toxicity as paclitaxel and carboplatin), as no data were available to calculate these odds ratios. 51
Health-state utilities
Utility values were in line with those used in the industry submission for NICE Technology Appraisal 19251 and based on the study by Nafees et al. 61 Utility estimates in the manufacturer’s model were adopted from a single UK study in which utility values were derived from a survey of 105 members of the general public who were asked to value descriptions of health states of second-line chemotherapy for patients with NSCLC. This study did not provide utility estimates associated with the mode of delivery of treatment (oral vs. i.v.), so the manufacturer used utility values previously applied in NICE Technology Appraisal guidance 162 (Erlotinib for the treatment of relapsed NSCLC), which examined second-line chemotherapy for patients with NSCLC and included utilities related to oral (erlotinib) and i.v. treatment. 66 Utilities for health states and adverse events were calculated using a baseline utility for SD with no adverse events of 0.653 (SE 0.022). This baseline utility was increased in case of treatment response and/or decreased using adverse events and/or treatment related disutilities (Table 21).
| Estimate | SE | Distribution | Source | |
|---|---|---|---|---|
| Health-state utilities | ||||
| Baseline utility (progression-free, SD) | 0.653 | 0.022 | Beta | Nafees (2009)61 |
| Disease progression (disutility) | 0.180 | 0.022 | Beta | Nafees (2009)61 |
| Progression-free response (utility increment) | 0.019 | 0.007 | Beta | Nafees (2009)61 |
| Disutilities related to adverse events (grade 3 or 4)a | ||||
| Neutropenia | 0.090 | 0.015 | Beta | Nafees (2009)61 |
| Febrile neutropenia | 0.090 | 0.016 | Beta | Nafees (2009)61 |
| Fatigue | 0.073 | 0.018 | Beta | Nafees (2009)61 |
| Nausea and/or vomiting | 0.048 | 0.016 | Beta | Nafees (2009)61 |
| Diarrhoea | 0.047 | 0.016 | Beta | Nafees (2009)61 |
| Hair loss (grade 2) | 0.045 | 0.015 | Beta | Nafees (2009)61 |
| Skin and subcutaneous tissue disorders | 0.032 | 0.012 | Beta | Nafees (2009)61 |
| Anaemia | 0.073 | 0.018 | Beta | Lilly (2008)67 |
| Disutilities related to treatment | ||||
| Intravenous therapy | 0.043 | 0.020 | Beta | Roche (2006)68 |
| Oral therapy | 0.014 | 0.012 | Beta | Roche (2006)68 |
If the mutation tests were to differ substantially in turnaround time, there could be a difference in process disutility associated with waiting for a test result, or even health outcome owing to delayed start of treatment. To investigate this, an item on turnaround time was included in the online survey. The results (see Chapter 3, What are the technical performance characteristics of the different EGFR mutation tests?) showed that the tests were very similar. In most laboratories, the turnaround times were generally between 3 and 7 days. One laboratory (using the Therascreen EGFR PCR Kit) had a turnaround time of 1 to 2 days and one laboratory (also using the Therascreen EGFR PCR Kit) had a turnaround time of between 8 and 10 days. Based on these results, it was assumed in the health-economic analysis that the turnaround times were not test driven, and therefore the tests did not differ with respect to process disutility or health outcomes associated as a result of waiting for the test results.
Resource use and costs
Resource use and costs were taken from NICE Technology Appraisal 192,51 with the exception of the EGFR mutation test costs. These costs were based on the online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different EGFR mutation tests?).
Test costs
For patients with a positive or negative test result, the full test costs as reported in Table 22 were accounted for. For this purpose, the charged prices from the online survey of NHS laboratories in England and Wales (see Chapter 3, What are the technical performance characteristics of the different EGFR mutation tests?) were used. These costs were either the same as or did not differ substantially from the actual test costs; the charged prices were reported for more tests than the actual test costs, and the incremental test costs are similar (see Table 22). To calculate test costs for patients with an unknown mutation status, it is necessary to differentiate between patients with an unknown mutation because the sample was considered inadequate by the pathologist before sending the specimen to the laboratory (pre-laboratory clinical failure), and patients with a sample that was considered adequate by the pathologist but which results in a failure once inside the laboratory (technical failures within the laboratory). In the case of an unknown mutation status owing to a pre-laboratory clinical failure, no test costs were taken into account. In the case of an unknown mutation status due to a technical failure within the laboratory, full test costs were taken into account. This proportion was calculated based on the proportion of patients with an unknown mutation status as taken from the literature (see Tables 15 and 16) and the total proportion of technical failures in the laboratories as reported in the online survey (see Table 5), using the following formula:
The results of the calculations of the proportion of patients with unknown test results for which test costs are included are presented in Table 23.
| Test | Test costs | Charged price | Distribution | Source | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean (SE)a | Range | n | Mean (SE)a | Range | |||||||||
| Therascreen EGFR PCR Kit | 5 | 154.00 | (14.70) | 120.00 | – | 190.00 | 7 | 154.58 | (12.01) | 120.00 | – | 190.00 | Gamma | Online survey |
| Direct sequencing of exons 19–21b | 0 | 175.00 | (14.70) | 175.00 | – | 175.00 | 0 | 147.50 | (27.50) | 120.00 | – | 175.00 | Gamma | Online survey |
| Direct sequencing or WAVE-HS for samples with insufficient tumour cellsb | 0 | 175.00 | (14.70) | 175.00 | – | 175.00 | 0 | 147.50 | (27.50) | 120.00 | – | 175.00 | Gamma | Online survey |
| Direct sequencing of exons 18–21b | 0 | 175.00 | (14.70) | 175.00 | – | 175.00 | 0 | 147.50 | (27.50) | 120.00 | – | 175.00 | Gamma | Online survey |
| Fragment length analysis combined with pyrosequencing | 2 | 162.50 | (12.50) | 150.00 | – | 175.00 | 2 | 187.50 | (12.50) | 175.00 | – | 200.00 | Gamma | Online survey |
| Sanger sequencing and fragment length analysis/PCR of negative samplesc | 0 | NR | – | 1 | 140.00 | (27.50) | 140.00 | – | 140.00 | Gamma | Online survey | |||
| Roche cobas test | 0 | NR | – | 1 | 140.00 | (27.50) | 140.00 | – | 140.00 | Gamma | Online survey | |||
| HRM analysis | 1 | 140.00 | (14.70) | 140.00 | – | 140.00 | 1 | 150.00 | (27.50) | 150.00 | – | 150.00 | Gamma | Online survey |
| Single-strand conformation analysis | 1 | 110.00 | (14.70) | 110.00 | – | 110.00 | 1 | 140.00 | (27.50) | 140.00 | – | 140.00 | Gamma | Online survey |
| Sanger sequencing or Roche cobas for samples with insufficient tumour cellsd | 0 | NR | – | – | 0 | 130.00 | (19.34)e | 120.00 | – | 140.00 | Gamma | Online survey | ||
| Sanger sequencing or Therascreen for samples with insufficient tumour cellsf | 0 | 154.00 | (14.70) | 120.00 | – | 190.00 | 0 | 137.30 | (14.88)e | 120.00 | – | 190.00 | Gamma | Online survey |
| Next-generation sequencingg | 0 | NR | – | – | 0 | NR | – | – | – | |||||
| Therascreen and Pyrosequencing Kitf | 0 | NR | – | – | 0 | NR | – | – | – | |||||
| Test | Total proportion of patients with unknown test result (SE)a | Distribution | Source | Proportion of technical failures in laboratory (SE)b | No. of reporting laboratories | Distribution | Proportion of patients with an unknown mutation due to a technical failure (full test costs) |
|---|---|---|---|---|---|---|---|
| Analyses 1 and 2b | |||||||
| Therascreen EGFR PCR Kit | 22.7% (1.8%) | Beta | Mok (2009)49 | 3.8% (1.5%) | 6 | Beta | 3.1% |
| Direct sequencing of exons 19–21c | As for direct sequencing or WAVE-HS | 4.5% (0.5%) | 0 | Beta | 2.9% | ||
| Direct sequencing or WAVE-HS for samples with insufficient tumour cellsc | 37.7% (4.2%) | Beta | Jackman (2007)44 | 4.5% (0.5%) | 0 | Beta | 2.9% |
| Direct sequencing of exons 18–21c | As for direct sequencing or WAVE-HS | 4.5% (0.5%) | 0 | Beta | 2.9% | ||
| Analysis 3b | |||||||
| Therascreen EGFR PCR Kit | 22.7% (1.8%) | Beta | Mok (2009)49 | 3.8% (1.5%) | 6 | Beta | 3.1% |
| Direct sequencing of exons 19–21c | As for Therascreen EGFR PCR Kit | 4.5% (0.5%) | 0 | Beta | 3.6% | ||
| Direct sequencing or WAVE-HS for samples with insufficient tumour cellsc | As for Therascreen EGFR PCR Kit | 4.5% (0.5%) | 0 | Beta | 3.6% | ||
| Direct sequencing of exons 18–21c | As for Therascreen EGFR PCR Kit | 4.5% (0.5%) | 0 | Beta | 3.6% | ||
| Fragment length analysis combined with pyrosequencing | As for Therascreen EGFR PCR Kit | 5.0% (1.5%) | 2 | Beta | 4.1% | ||
| Sanger sequencing and fragment length analysis/PCR of negative samplesd | As for Therascreen EGFR PCR Kit | 0.1% (1.5%) | 1 | Beta | 0.1% | ||
| Roche cobas test | As for Therascreen EGFR PCR Kit | 5.0% (1.5%) | 1 | Beta | 4.1% | ||
| HRM analysis | As for Therascreen EGFR PCR Kit | 0.2% (1.5%) | 1 | Beta | 0.2% | ||
| Single-strand conformation analysis | As for Therascreen EGFR PCR Kit | 10.0% (1.5%) | 1 | Beta | 8.6% | ||
| Sanger sequencing or Roche cobas for samples with insufficient tumour cellse | As for Therascreen EGFR PCR Kit | 4.5% (1.0%)g | 0 | Beta | 3.6% | ||
| Sanger sequencing or Therascreen for samples with insufficient tumour cellsf | As for Therascreen EGFR PCR Kit | 3.9% (1.1%)g | 0 | Beta | 3.2% | ||
| Next-generation sequencingh | As for Therascreen EGFR PCR Kit | NR | 0 | – | – | ||
| Therascreen and Pyrosequencing Kith | As for Therascreen EGFR PCR Kit | NR | 0 | – | – | ||
| Type of costs | Costs | SE | Distribution | Source |
|---|---|---|---|---|
| Treatment costs | ||||
| TKIa | CiC information has been removed | – | Fixed | NICE Technology Appraisal 19251 |
| Resource use | ||||
| No. of chemotherapy cycles | 4.0 | – | Fixed | ERG65 |
| Costs per chemotherapy cycleb | ||||
| Pemetrexed and cisplatin | £1536.30 | – | Fixed | ERG65 |
| Chemotherapy administration | £307.00 | £80.61 | Gamma | ERG65 |
| Transport | £28.00 | £3.57 | Gamma | NICE Technology Appraisal 19251 |
| Adverse event costs (grade 3 or 4)c | ||||
| Neutropenia | £92.80 | – | Fixed | NICE Technology Appraisal 19251 |
| Febrile neutropenia | £2286.00 | – | Fixed | NICE Technology Appraisal 19251 |
| Fatigue | £38.90 | – | Fixed | NICE Technology Appraisal 19251 |
| Nausea and vomiting | £700.79 | – | Fixed | NICE Technology Appraisal 19251 |
| Diarrhoea | £867.12 | – | Fixed | NICE Technology Appraisal 19251 |
| Hair loss (grade 2) | £0.00 | – | Fixed | NICE Technology Appraisal 19251 |
| Skin and subcutaneous tissue disorders | £116.82 | – | Fixed | NICE Technology Appraisal 19251 |
| Anaemia | £615.04 | – | Fixed | NICE Technology Appraisal 19251 |
| Other | ||||
| Patient monitoring (per cycle) | CiC information has been removed | CiC information has been removed | Gamma | NICE Technology Appraisal 19251 |
| Second-line therapy following disease progression (per cycle) | £1022.05 | – | Fixed | NICE Technology Appraisal 19251 |
| Probability of second-line therapy following disease progression | 61.0% | 4.3% | NICE Technology Appraisal 19251 | |
| Best supportive care (per cycle)d | £599.69 | – | Fixed | NICE Technology Appraisal 19251 |
Model analyses
Expected mean costs, LYs and QALYs were estimated for all EGFR mutation tests. Long-term costs, LYs and QALYs were discounted using the UK discount rates of 3.5% for both costs and effects. Based on the estimated outcomes (probabilistic), the incremental cost-effectiveness ratio (ICER) was calculated by dividing the incremental cost (ICs) by the incremental QALYs. The ICER represents the costs of an additional QALY gained and was used to estimate the cost-effectiveness of a strategy opposed to (1) direct sequencing of all exon 18–21 mutations and (2) the next best alternative. All outcomes are based on probabilistic sensitivity analyses with 5000 simulations using parameter distributions as presented in this section.
Overview of main model assumptions
The main assumptions in the health-economic analyses were:
-
The differences in relative treatment response, PFS and OS reported in the First-SIGNAL trial41 and those reported for the IPASS trial49 are attributable solely to the different tests used (Therascreen EGFR PCR Kit and direct sequencing of all exon 19–21 mutations, respectively) to distinguish between patients whose tumours are EGFR mutation positive (and receive TKI treatment) and patients whose tumours are EGFR mutation negative (and receive doublet chemotherapy) (‘evidence of comparative effectiveness available’ and ‘linked evidence’ analyses).
-
To calculate the sensitivity and specificity of the tests, required to calculate the proportion of positive and negative test results (see Table 15), patients who tested positive were categorised as FP if no treatment response was observed after TKI, whereas patients were categorised as TP if treatment response was observed TKI. Similarly, patients who tested negative were categorised as FN if treatment response was observed after TKI, whereas patients were categorised as TN if no treatment response was observed after TKI (all analyses).
-
The proportion of patients with unknown mutation status relative to the number of patients for whom a tissue sample was available in the trials44,49 provides a realistic approximation of the proportion of patients with an unknown test result in clinical practice (all analyses).
-
The OR rate, PFS and OS in patients with an unknown test result as reported in the IPASS trial49 is generalisable to direct sequencing methods (‘evidence of comparative effectiveness available’ and ‘linked evidence’ analyses).
-
The probability of an unknown test result as reported in the study by Jackman et al. 44 [direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) is generalisable to other direct sequencing methods (‘linked evidence’ analysis)].
-
The OR rate in patients with a negative test result as reported in the First-SIGNAL trial41 is generalisable to other direct sequencing methods (‘linked evidence’ analysis).
-
PFS and OS in patients with a positive or negative test result reported in the First-SIGNAL trial41 (direct sequencing of exons 19–21) are generalisable to other direct sequencing methods (exons 18–21) (‘linked evidence’ analysis). In other words, we assumed there is no clinical significance in testing exon 18 mutations.
Sensitivity analyses
For analyses 1 and 2, in a sensitivity analysis the costs reported in Table 24 were updated. For all three analyses, in a sensitivity analysis the proportion of unknown patients was based on the results of the online survey instead of the literature (see Table 5).
Sensitivity analysis using updated costs
In this sensitivity analysis, the costs reported in Table 23 were updated based on price indices and 2012 reference costs (Table 25), with the exception of EGFR-TKI treatment costs.
| Type of costs | Costs | SE | Distribution | Source |
|---|---|---|---|---|
| Treatment costs | ||||
| Costs per chemotherapy cycle | ||||
| Chemotherapy administration | £333.67 | £83.01 | Reference costs 201269 | |
| Transporta | £30.07 | STA 19251 and PSSRU62 | ||
| Adverse event costs (grade 3 or 4)a | ||||
| Neutropenia | £99.66 | – | Fixed | STA 19251 and PSSRU62 |
| Febrile neutropenia | £2455.00 | – | Fixed | STA 19251 and PSSRU62 |
| Fatigue | £41.78 | – | Fixed | STA 19251 and PSSRU62 |
| Nausea and vomiting | £752.60 | – | Fixed | STA 19251 and PSSRU62 |
| Diarrhoea | £931.23 | – | Fixed | STA 19251 and PSSRU62 |
| Hair loss (grade 2) | £0.00 | – | Fixed | STA 19251 and PSSRU62 |
| Skin and subcutaneous tissue disorders | £125.46 | – | Fixed | STA 19251 and PSSRU62 |
| Anaemia | £660.51 | – | Fixed | STA 19251 and PSSRU62 |
| Other | ||||
| Patient monitoring (per cycle) | £113.00 | £28.26 | Gamma | Reference costs 201269 |
| Second-line therapy following disease progression (per cycle)a | £1098.00 | – | Fixed | STA 19251 and PSSRU62 |
| Best supportive care (per cycle)a | £644.32 | – | Fixed | STA 19251 and PSSRU62 |
Sensitivity analysis using the proportion of patients with unknown mutation status based on online survey results
This sensitivity was performed for all three analyses. The proportion of patients with unknown mutation status was based on the survey results, as reported in Table 23, instead of the trials.
Results of cost-effectiveness analyses
This section reports the results of the ‘evidence on comparative effectiveness available’ analysis, the ‘linked evidence’ analysis and the ‘assumption of equal prognostic value’ analysis. In the tables the strategies are ranked by costs from least to most expensive. For the ‘evidence on comparative effectiveness’ analysis, the comparator from the scope (direct sequencing of all exon 18–21 mutations) could not be included. Therefore, direct sequencing of all exon 19–21 mutations was used as comparator in the ‘evidence on comparative effectiveness’ analyses. In the ‘linked evidence’ and ‘assumption of equal prognostic value’ analyses, direct sequencing of exons 18–21 was included and hence was used as the comparator. For all analyses the results are presented in two ways: first, compared with the comparator (direct sequencing of all exon 18–21 mutations or of all exon 19–21 mutations) and, second, compared with the next most cost-effective strategy.
‘Evidence on comparative effectiveness available’ analysis
The probabilistic results of the ‘evidence on comparative effectiveness available’ analysis are shown in Table 26. It should be noted that this analysis is based on a number of assumptions outlined above (see Model analyses), of which the following two are particularly problematic:
-
The proportion of patients with a positive or negative test result after the use of these tests in the NHS population was estimated based on the proportion of EGFR mutation-positive patients in England and Wales, the proportion of patients with an unknown test result, and test accuracy for the prediction of treatment response derived from two separate trials. 41,49
-
The differences in relative treatment response, PFS and OS between the results of First-SIGNAL41 that were used to model EGFR mutation testing with direct sequencing of all exon 19–21 mutations and the results of the IPASS trial48,49 that were used to model EGFR mutation testing with the Therascreen EGFR PCR Kit, are assumed to be solely attributable to the different tests used to distinguish between patients who are EGFR mutation positive (and receive TKI treatment) and patients who are EGFR mutation negative (and receive doublet chemotherapy).
| Strategy | Costs | QALYs | Compared with direct sequencing (exons 19–21) | ||
|---|---|---|---|---|---|
| Incremental costs | Incremental QALYs | ICER | |||
| Base case | |||||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.902 | –0.207 | £32,167 | |
| Direct sequencing of all exon 19–21 mutations a | CiC information has been removed | 1.109 | |||
| Sensitivity analysis: updated costs | |||||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.874 | –£9194 | –0.286 | £32,196 |
| Direct sequencing of all exon 19–21 mutations a | CiC information has been removed | 1.160 | |||
| Sensitivity analysis: unknowns from survey | |||||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.905 | –£7130 | –0.206 | £34,555 |
| Direct sequencing of all exon 19–21 mutations a | CiC information has been removed | 1.111 | |||
In this analysis, the Therascreen EGFR PCR Kit was both less effective and less costly than direct sequencing of all exons 19–21 at an ICER of £32,167. The lower costs and QALYs for the Therascreen EGFR PCR Kit can be explained by the fact that patients whose tumours are mutation negative do worse on OS in the IPASS trial48,49 than in First-SIGNAL,41 whereas for mutation-positive patients the outcome is similar, and for unknowns it is the same (by assumption) – see Figures 13 and 14. Therefore, on average, with the Therascreen EGFR PCR Kit strategy patients have shorter survival, and therefore fewer QALYs than testing with direct sequencing of all exons 19–21. The apparent shorter survival also reduces costs. The cost-effectiveness acceptability curve (Figure 15) shows that at a threshold value of £32,500 direct sequencing of all exons 19–21 becomes the preferred strategy.
FIGURE 15.
Cost-effectiveness acceptability curve for ‘evidence on comparable effectiveness available’ analysis, base case.
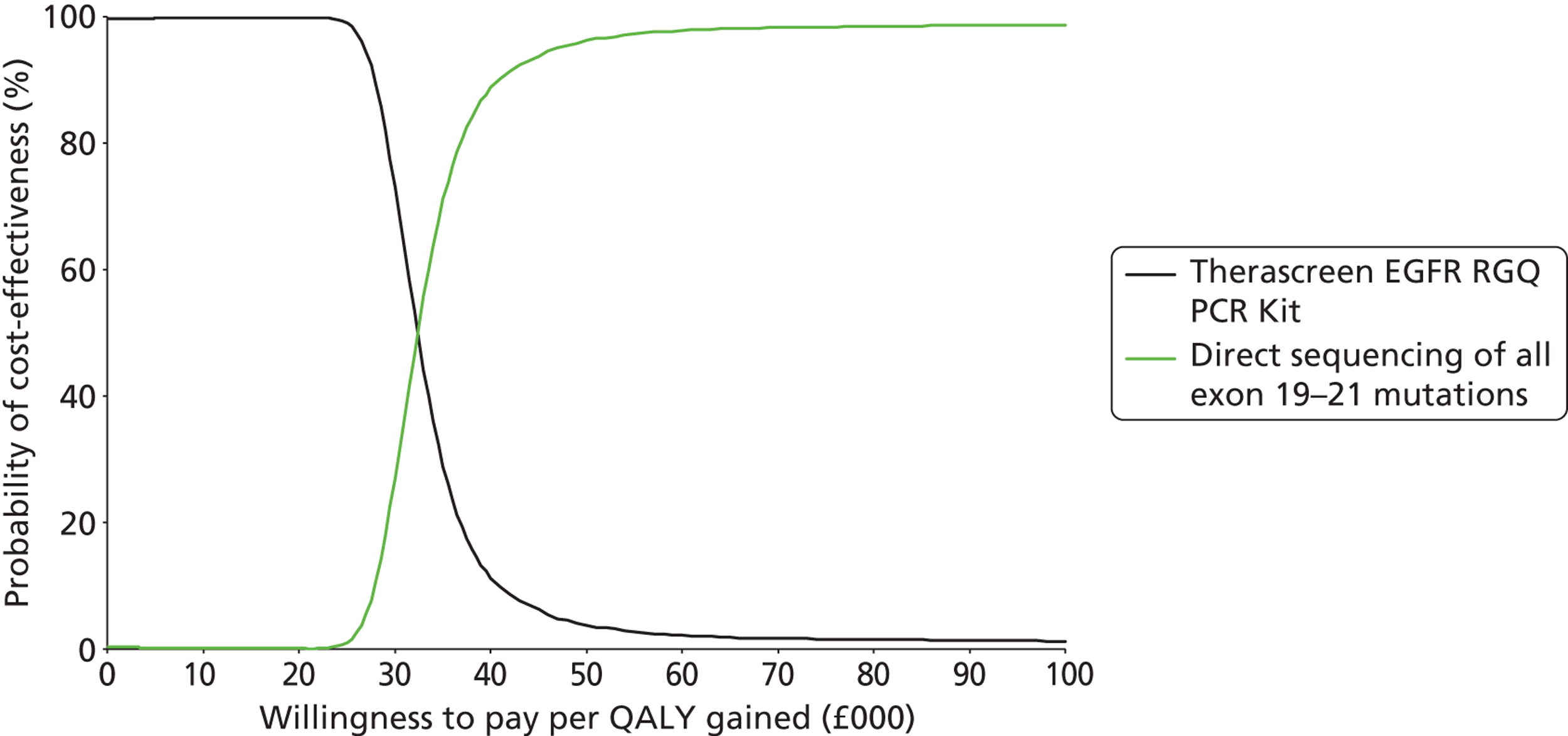
Results were robust for changed assumptions in the sensitivity analyses, in the sense that testing with Therascreen EGFR PCR Kit was always less effective and less expensive. The ICERs amounted to £34,555 (unknowns from survey) and £32,196 (updated costs). The cost-effectiveness acceptability curves for the sensitivity analyses are presented in Appendix 7.
‘Linked evidence’ analysis
The ‘linked evidence’ analysis includes four tests, i.e. all tests for which either evidence on relative effectiveness or accuracy was available. Table 27 shows the probabilistic results of this analysis.
| Strategy | Costs | QALYs | Compared with direct sequencing (exons 18–21) | ||
|---|---|---|---|---|---|
| Incremental costs | Incremental QALYs | ICER | |||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.902 | –£6040 | –0.190 | £31,849 |
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.092 | |||
| Direct sequencing of all exon 19–21 mutations a | CiC information has been removed | 1.109 | £619 | 0.017 | £35,634 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) a | CiC information has been removed | 1.109 | £658 | 0.017 | £38,251 |
| Strategy | Costs | QALYs | Comparator | Compared with next cost-effective strategy | ||
|---|---|---|---|---|---|---|
| Incremental costs | Incremental QALYs | ICER | ||||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.902 | ||||
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.092 | Therascreen EGFR PCR Kit | £6040 | 0.190 | £31,849 |
| Direct sequencing of all exon 19–21 mutations a | CiC information has been removed | 1.109 | Direct sequencing (exons 19–21) | £619 | 0.017 | £35,634 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) a | CiC information has been removed | 1.109 | Direct sequencing (exons 19–21) | £39 | 0.000 | Dominated |
This analysis was also based on a number of assumptions, including those described above (see Model analyses and ‘Evidence on comparative effectiveness available’ analysis). The following additional assumption should be particularly noted:
-
For direct sequencing of all exon 18–21 mutations and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells), the relative PFS and OS for mutation positives and mutation negatives correlate perfectly with relative PFS and OS as observed for direct sequencing of all exon 19–21 mutations in the First-SIGNAL trial. 41
In the base-case analysis, compared with direct sequencing of all exon 18–21 mutations, the Therascreen EGFR PCR Kit was less costly and less effective at an ICER of £31,849 per QALY lost. Direct sequencing of all exon 19–21 mutations and direct sequencing or WAVE-HS for inadequate samples were both more expensive and more effective than the comparator. For thresholds of < £33,500, testing with the Therascreen EGFR PCR Kit is the preferred strategy, then direct sequencing of all exon 18–21 mutations is preferred up to a threshold of £39,000, at which direct sequencing of all exon 19–21 mutations has the highest probability of being cost-effective (Figure 16). The sensitivity analyses (see Appendix 7) show that these findings are quite robust in the sense that compared with direct sequencing of all exon 18–21 mutations the Therascreen EGFR PCR Kit is always less expensive and less effective, and the remaining two tests are more effective and more expensive.
FIGURE 16.
Cost-effectiveness acceptability curve for ‘linked evidence’ analysis.
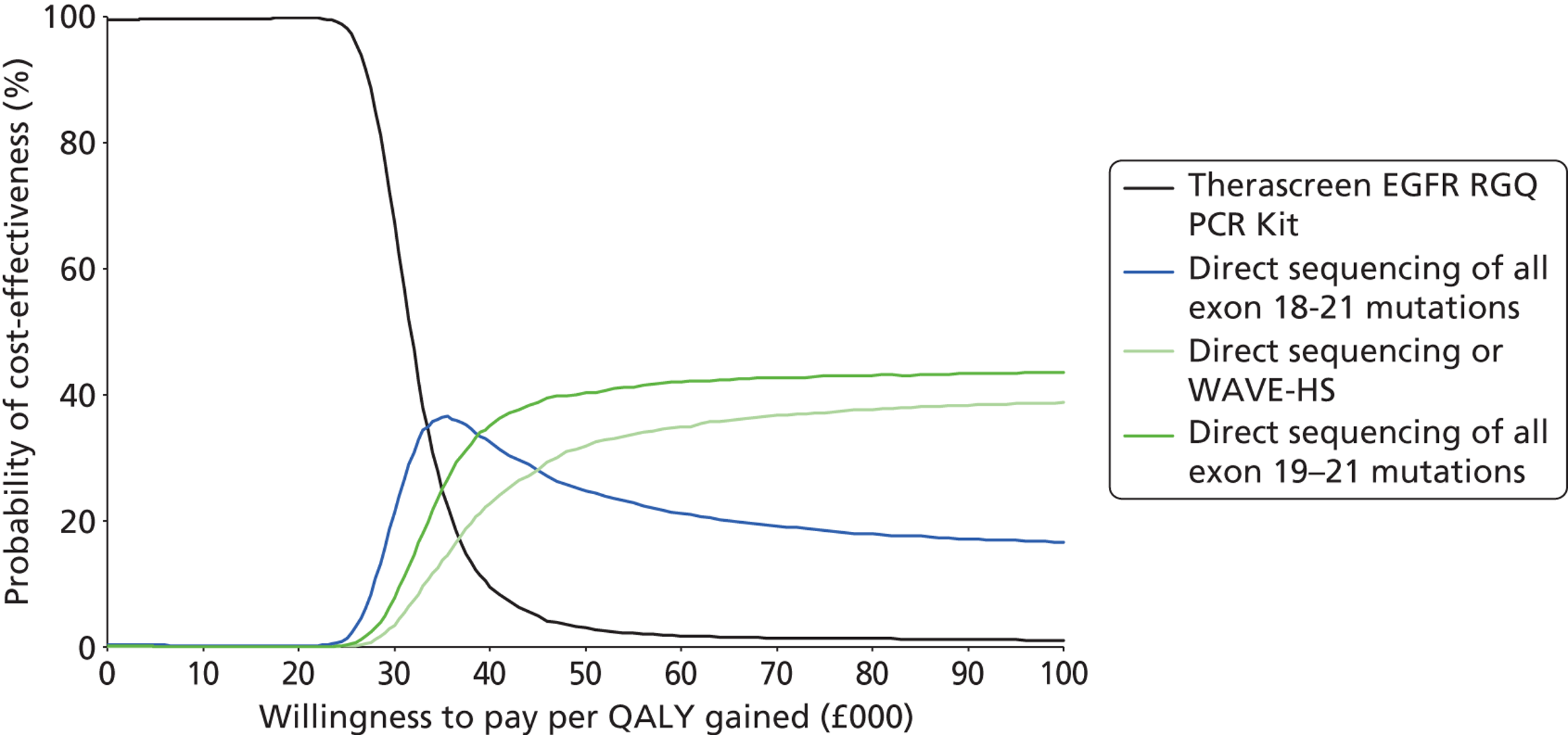
‘Assumption of equal prognostic value’ analysis
The ‘assumption of equal prognostic value’ analysis included all tests for which information on cost and/or technical performance was available from the online survey of NHS laboratories in England and Wales. This includes the tests for which neither comparative effectiveness nor response data were available. Therefore, this analysis assessed whether the tests were likely to be cost-effective given an assumption of equal prognostic value (based on the prognostic value of testing with the Therascreen EGFR PCR Kit, as this was the only test for which prognostic data were available on patients with positive, negative and unknown tumour EGFR mutation status) and test-specific information on cost only. As a result, the strategies differ only with respect to costs. As shown in Table 28, Sanger sequencing or Roche cobas for samples with insufficient tumour cells is the least expensive strategy, and fragment length analysis combined with pyrosequencing is the most expensive strategy. However, the difference between the costs of these strategies amounts to only £477 (< 1% of total strategy costs).
| Strategy | Costs (95% CI) | Incremental costs compared with direct sequencing (exons 18–21) |
|---|---|---|
| Sanger sequencing or Roche cobas for samples with insufficient tumour cells | CiC information has been removed | –£15 |
| Sanger sequencing and fragment length analysis/PCR of negative samples | CiC information has been removed | –£11 |
| Sanger sequencing or Therascreen EGFR PCR Kit for samples with insufficient tumour cells | CiC information has been removed | –£9 |
| Roche cobas | CiC information has been removed | –£9 |
| HRM analysis | CiC information has been removed | –£3 |
| Direct sequencing of exons 19–21 a | CiC information has been removed | £0 |
| Direct sequencing of exons 18–21 | CiC information has been removed | |
| Single-strand conformation analysis | CiC information has been removed | £1 |
| Direct sequencing or WAVE-HS a | CiC information has been removed | £1 |
| Therascreen EGFR PCR Kit | CiC information has been removed | £5 |
| Fragment length analysis combined with pyrosequencing | CiC information has been removed | £33 |
In a sensitivity analysis the proportion of patients with tumours of unknown mutation status were taken from the online survey of NHS laboratories in England and Wales, rather than based on the literature. As a result, in this sensitivity analysis a difference in health outcomes (QALYs) is modelled. The results in Table 29 show that this assumption has some impact on the relative costs and effects of the strategies, in the sense that single-strand conformation analysis is now the most costly. This is caused by the fact that the percentage of failures as reported in the survey is the highest for single-strand conformation analysis (10%, n = 1), whereas for Sanger sequencing and fragment length analysis/PCR it is 0% (n = 1). A higher failure rate will, in its turn, lead to a lower proportion of patients with mutation-positive and mutation-negative tumours and, therefore, on average, to higher costs. This is because patients with an unknown mutation status are more costly than the average of the patients with a known (positive or negative) mutation status. The cost-effectiveness acceptability curve is presented in Figure 17.
| Strategy | Costs | QALYs | Compared with direct sequencing of all exon 18–21 mutations | Compared with next best strategy | |||||
|---|---|---|---|---|---|---|---|---|---|
| Incremental costs | Incremental QALYs | ICER | Comparator | Incremental costs | Incremental QALYs | ICER | |||
| Sanger sequencing and fragment length analysis/PCR of negative samples | CiC information has been removed | 0.871 | –£226 | –0.007 | £33,437 | ||||
| HRM analysis | CiC information has been removed | 0.871 | –£211 | –0.007 | £31,848 | Sanger sequencing and fragment length analysis/PCR of negative samples | £14 | 0.000 | Extended dominance |
| Sanger sequencing or Therascreen EGFR PCR Kit for samples with insufficient tumour cells | CiC information has been removed | 0.877 | –£40 | –0.001 | £45,629 | Sanger sequencing and fragment length analysis/PCR of negative samples | £186 | 0.006 | Extended dominance |
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.877 | –£26 | –0.001 | £24,977 | Sanger sequencing and fragment length analysis/PCR of negative samples | £200 | 0.006 | Extended dominance |
| Sanger Sequencing or Roche cobas for samples with insufficient tumour cells | CiC information has been removed | 0.878 | –£18 | 0.000 | Dominated | Sanger sequencing and fragment length analysis/PCR of negative samples | £207 | 0.007 | £30,602 |
| Direct sequencing or WAVE-HS a | CiC information has been removed | 0.878 | £0 | 0.000 | Dominated | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £18 | 0.000 | Dominated |
| Direct sequencing of exons 18–21 | CiC information has been removed | 0.878 | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £18 | 0.000 | Dominated | |||
| Direct sequencing of exons 19–21 a | CiC information has been removed | 0.878 | £0 | 0.000 | £615,549 | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £19 | 0.000 | Dominated |
| Roche cobas | CiC information has been removed | 0.879 | £15 | 0.001 | £19,501 | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £33 | 0.001 | Extended dominance |
| Fragment length analysis combined with pyrosequencing | CiC information has been removed | 0.879 | £62 | 0.001 | £79,807 | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £81 | 0.001 | Extended dominance |
| Single-strand conformation analysis | CiC information has been removed | 0.886 | £264 | 0.008 | £31,080 | Sanger sequencing or Roche cobas for samples with insufficient tumour cells | £283 | 0.008 | £33,338 |
FIGURE 17.
Cost-effectiveness acceptability curve for ‘assumption of equal prognostic value’ analysis, sensitivity analysis: unknown based on survey.
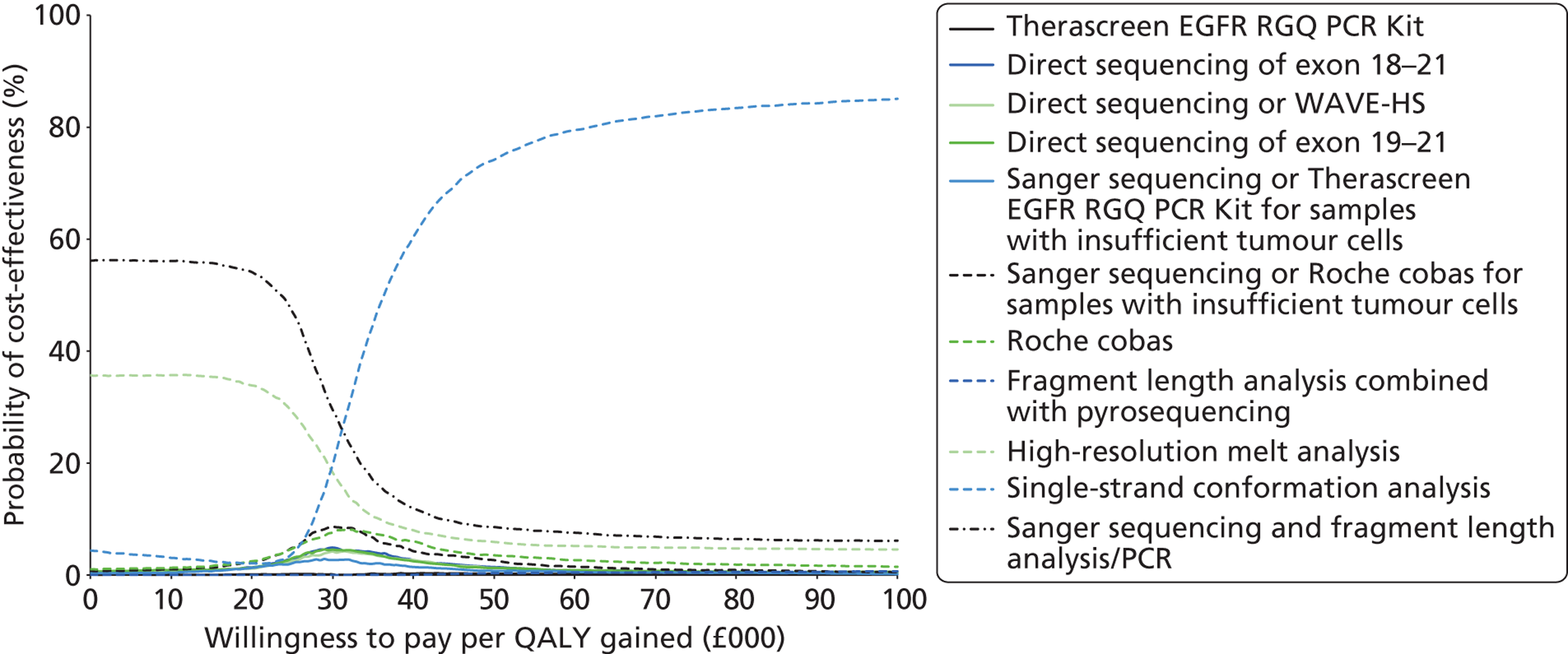
Chapter 5 Discussion
Statement of principal findings
Clinical effectiveness
There was no strong evidence that any one EGFR mutation test had greater accuracy than any other test, although there was a suggestion that Therascreen EGFR PCR Kit may be more accurate than direct sequencing for predicting response to treatment with TKIs. Eleven studies were included in the review; these evaluated the Therascreen EGFR PCR Kit (version 1), direct sequencing, cobas EGFR Mutation Testing Kit, fragment length analysis, and Sanger sequencing. Six studies (two RCTs and four cohort studies) provided data on the accuracy of EGFR mutation testing for predicting response to treatment with TKIs in patients with stage IIIB or IV NSCLC. Five RCTs, including two that also provided accuracy data, reported data on the clinical effectiveness of TKIs compared with standard chemotherapy in patients with stage IIIB or IV NSCLC with EGFR mutation-positive tumours; one additional study reported data for a subgroup of patients from one of these RCTs, whose biopsy samples had been re-analysed using a different EGFR mutation testing method. The remaining study was included as a supplement to the survey of laboratories in England and Wales that currently provide EGFR mutation testing, and did not report any data on clinical outcomes.
The survey of laboratories providing EGFR mutation testing indicated that the Therascreen EGFR PCR Kit was the single most commonly used method (6 out of 13 respondents). Reasons cited by respondents for their choice of the Therascreen EGFR PCR Kit were proportion of tumour cells required; ease of use; cost; and mutations covered. There was no clear indication that choice of test method was related to volume of throughput. Most respondents reported turnaround times – from receipt of sample to reporting to the clinician – of between 3 and 7 days. The only laboratory to report a turnaround time of < 3 days (24–48 hours) used the Therascreen EGFR PCR Kit. All respondents reported turnaround times of less than the 10 working day maximum recommended by the European EGFR Workshop Group. 12 With the exception of those whose testing strategy included direct sequencing methods, all respondents reported a minimum requirement for testing at or < 10% of tumour cells, with some of the laboratories that used the Therascreen EGFR PCR Kit reporting minimum requirements as low as 1%. Although most respondents included costs in their reasons for choosing a particular test, it is worth noting that a relatively narrow range of costs was reported across all tests (£110–190), with a similar level of variation apparent within a single test, Therascreen EGFR PCR Kit (£120–190). When contacted by NICE, UK NEQAS stated:
Error rates are not always method related and it is not always possible to obtain data from all the labs committing critical genotyping errors. Therefore, any data which could be provided would be skewed with processing and reporting issues rather than being method related. There has been no correlation between any method used for EGFR testing and errors since we started providing the scheme in 2010.
Studies that provided data on test accuracy assessed the Therascreen EGFR PCR Kit (version 1) or direct sequencing methods (exons 18 or 19 to exons 21 or 24). No studies were identified that reported accuracy data for any other EGFR mutation testing method. The Therascreen EGFR PCR Kit appeared to have the best overall performance for discriminating between patients who are likely to benefit from TKI treatment and those who are not. The sensitivity and specificity estimates for OR were 99% (95% CI 94% to 100%) and 69% (95% CI 60% to 77%), respectively, with specificity increasing and sensitivity decreasing where a lower response threshold (DC) was used. 49 Four of the five direct sequencing studies reported high estimates of specificity (> 80%) for OR, with sensitivities ranging from 60% to 80%. 41,43–45 Three of these studies also assessed DC and reported high specificities (> 90%) and very low sensitivities (≤ 35%). 43–45 The remaining direct sequencing study reported low sensitivity (66%) and specificity (50%) for DC and low specificity (61%) with high sensitivity (84%) for OR.
There were no clear common participant characteristics, across studies, which reported similar sensitivity or specificity estimates for DC or OR. Specificity estimates may have been affected by the way in which resistance mutations were classified; the three direct sequencing studies that reported high specificity estimates either stated that patients whose tumours showed resistance or non-sensitising mutations were classified as EGFR mutation negative or did not identify any patients with tumours showing these types of mutation. Although the number of resistance mutations identified was generally small, their potential effect on specificity estimates was magnified by the very small sample size in most studies. The most commonly observed mutations were exon 19 deletions and the exon 21 point mutation L858R; most patients in the included studies who had these mutations achieved a minimum response of SD when treated with TKIs. Large database studies provide some support for the idea that mutations in exon 20, and in particular the mutation T790M, may be associated with a lack of response to TKIs (see Clinical effectiveness, below). A second possible explanation may be that the Therascreen EGFR PCR Kit has a lower limit of detection, i.e. it is able to detect EGFR mutations at a lower abundance (fewer cancer cells carrying the mutation) than direct sequencing methods. A lower limit of detection would be beneficial only if it could be shown that patients whose tumours have a lower abundance of EGFR mutation benefit from treatment with TKIs, and the apparent improved diagnostic performance of the Therascreen EGFR PCR Kit, compared with direct sequencing methods, indicates that this may be the case. However, none of the studies identified by this review reported data on the relationship between abundance of EGFR mutation and response to first-line TKI treatment in patients with stage IIIB or IV NSCLC.
The five RCTs included in this review compared the TKIs gefitinib or erlotinib with various single agent or combination standard chemotherapy regimens and reported data on PFS. Three of the trials included only patients with EGFR mutation-positive tumours,40,47 and the remaining two trials (IPASS and First-SIGNAL) included chemotherapy naive patients with stage IIIB or IV NSCLC and reported a subgroup analysis for patients who had received EGFR mutation testing using the Therascreen EGFR PCR Kit (version 1)48,49 or direct sequencing. 41 Though derived from a subgroup analysis of tested patients, data from these trials were most the most complete available, in that they provided information on the effectiveness of TKIs compared with standard chemotherapy in both test-positive and test-negative patients. The results of the IPASS subgroup analyses indicated that PFS was significantly longer for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-positive subgroup [HR 0.48 (95% CI 0.36 to 0.64)] and significantly shorter for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-negative subgroup [HR 2.85 (95% CI 2.05 to 3.98)]. This trial formed the basis of the technology appraisal that informed NICE guidance TA 192 on gefitinib for the first-line treatment of locally advanced or metastatic NSCLC. 1 The results of the First-SIGNAL trial indicated a trend towards longer PFS for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-positive subgroup [HR 0.54 (95% CI 0.27 to 1.10)] and a trend towards significantly shorter PFS for patients receiving gefitinib than for those receiving standard chemotherapy in the EGFR mutation-negative subgroup [HR 1.42 (95% CI 0.82 to 2.47)]. 41 The remaining trials provided information on only the effectiveness of TKIs compared with standard chemotherapy in patients with EGFR mutation-positive tumours; HRs for PFS ranged from 0.48 (95% CI 0.36 to 0.64) to 0.16 (95% CI 0.10 to 0.26). The included trials used various methods to assess EGFR mutation status. Two trials used direct sequencing methods, but limited the definition of positive EGFR mutation status to the presence of an ‘activating mutation’ (exon 19 deletions or exon 21 mutation L858R). These two trials were included in the technology appraisal that informed NICE guidance TA 258 on erlotinib for the first-line treatment of locally advanced or metastatic EGFR-TK mutation-positive NSCLC. 2 The re-analysis of samples from one of these trials and the two remaining trials used EGFR mutation tests that targeted a wider range of mutations, including resistance mutations. Overall, there were no clear differences in any measure of TKI treatment effect (PFS, OR or DC), regardless of which EGFR mutation test (selective for activating mutations exon 19 deletions and exon 21 L858R, or targeting a wider range of mutations) was used to select patients. No study reported a significant difference in TKI treatment effect between patients with exon 19 deletions and those with the exon 20 mutation L858R. One additional trial, the Western Japan Oncology Group study, was included in TA 258 but did not meet the inclusion criteria for our review, as it focused on the treatment of patients with postoperative recurrence with or without postoperative adjuvant chemotherapy; patients with stage IIIB or IV NSCLC were also included but no separate data were reported for them. 70 EGFR mutation testing in this study also targeted exon 19 deletions and the exon 21 mutation L858R, and used a combination of fragment analysis and direct sequencing methods; the reported treatment effect of TKI (gefitinib) compared with standard chemotherapy (cisplatin plus docetaxel) was similar to that seen in the trials included in our review [PFS HR 0.49 (95% CI 0.34 to 0.71)]. 70
The estimates of the effectiveness of first-line treatment with TKIs, compared with standard chemotherapy, in patients with advanced NSCLC whose tumours tested positive for an EGFR mutation reported by studies included in this review, were consistent with pooled estimates reported in recent systematic reviews. Three systematic reviews had inclusion criteria that matched ours in terms of population intervention and comparator but which did not specify reporting of EGFR testing methods. All three reviews reported pooled HRs, which indicated increased PFS in patients with EGFR mutation-positive tumours who were treated with TKIs compared with those treated with standard chemotherapy [HR 0.43 (95% CI 0.32 to 0.58),71 HR 0.37 (95% CI 0.27 to 0.52)72 and HR 0.45 (95% CI 0.36 to 0.58)73]. Two reviews also reported significantly higher OR rates [RR 5.68 (95% CI 3.17 to 10.18)]72 and HR 2.08 (95% CI 1.75 to 2.46)73 for patients treated with TKIs, and no significant difference in OS between the two treatment groups. 72,73
Cost-effectiveness
The review of economic analyses of different methods for EGFR TK mutation testing to decide between standard chemotherapy or EGFR-TKIs for first-line treatment of patients with locally advanced or metastatic NSCLC found one full paper52 and five conference abstracts. 54–58 The full paper did not fit the decision problem, as it concerned second-line use of anti-EGFR-TKIs. Although the conference abstracts were all about first-line use of TKIs, they did not provide enough specific information to be of use; future full publications may provide more information.
In the health-economic analysis, the cost-effectiveness of different methods for EGFR-TK mutation testing to decide between standard chemotherapy or EGFR-TKIs for first-line treatment of patients with locally advanced or metastatic NSCLC was assessed. In light of the scarce evidence that was available, three analyses were performed: ‘evidence on comparative effectiveness available’, ‘linked evidence’, and ‘assumption of equal prognostic value’. Direct sequencing of all exon 18–21 mutations, the comparator, could only be included in the last two analyses.
In the ‘evidence on comparative effectiveness available’ analysis, testing with the Therascreen EGFR PCR Kit was compared with direct sequencing of all exon 19–21 mutations in order to estimate lifetime cost and QALYs using the observed response to treatment and the available relative PFS and OS data. The results of this analysis suggested that direct sequencing of all exon 19–21 mutations was both more effective and more costly than testing with the Therascreen EGFR PCR Kit at an ICER of £32,167 per QALY gained. The sensitivity analyses all resulted in similar outcomes. The key drivers behind this result were the differences in the proportion of patients with EGFR mutation positive, unknown mutation and mutation-negative tumours, and differences in OR, PFS and OS. In particular, the predicted OS for mutation-negative patients differed substantially between the studies using the Therascreen EGFR PCR Kit48,49 and the study which used direct sequencing of all exon 19–2141 (see Figure 13). OS for mutation negatives after testing using the Therascreen EGFR PCR Kit was substantially lower than for testing using direct sequencing of all exon 19–21 mutations, whereas PFS was similar. As a result, testing using the Therascreen EGFR PCR Kit appeared less effective in terms of QALYs but was also less costly as the gained LYs for direct sequencing of all exon 19–21 mutations were mainly spent in the relatively expensive disease progression health state.
It should be noted that this analysis was based on a number of assumptions, of which the following two are particularly problematic:
-
The proportion of patients with a positive or negative test result after the use of these tests in the NHS population was estimated based on the proportion of EGFR mutation-positive patients in England and Wales, the proportion of patients with an unknown test result and test accuracy for the prediction of treatment response derived from two separate trials. 41,49
-
The differences in relative treatment response, PFS and OS between the results of the First-SIGNAL trial,41 that were used to model direct sequencing of all exon 19–21 mutations, and the results of the IPASS trial,48,49 that were used to model testing using the Therascreen EGFR PCR Kit, are solely attributable to the different tests used to distinguish between patients whose tumours are EGFR mutation positive (and who receive TKI treatment) and patients whose tumours are EGFR mutation negative (and who receive doublet chemotherapy).
The results of the ‘evidence on comparative effectiveness available’ analysis should therefore be interpreted on the condition that these assumptions hold. Moreover, the uncertainty presented surrounding the results is an underestimation of the true uncertainty, as the uncertainty associated with the assumptions was not parameterised and is therefore not reflected in the probabilistic sensitivity analyses.
In the ‘linked evidence’ analysis, two other direct sequencing tests [direct sequencing of all exon 18–21 mutations and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells)] for which accuracy data to predict response to treatment with TKIs were available were also included in the analysis. The results of this analysis showed that, compared with direct sequencing of all exons 18–21 mutations, the Therascreen EGFR PCR Kit was less effective and less costly (ICER £31,849), whereas the other tests were more effective and more expensive (ICERs £35,634 and £38,251). Sensitivity analyses did not show any substantial changes to these results. However, it should be noted that this analysis is also based on a number of substantive assumptions, including those described for the ‘evidence on comparative effectiveness available’ analysis. The following additional assumption should also be noted:
-
For direct sequencing of all exon 18–21 mutations and direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells), the relative PFS and OS for mutation positives and mutation negatives correlates perfectly with relative PFS and OS as observed for direct sequencing of all exon 19–21 mutations in the First-SIGNAL trial. 41
The same caveat for the interpretation of the results and surrounding uncertainty as explained above for the ‘evidence on comparative effectiveness available’ analysis applies to the interpretation of the results of the ‘linked evidence’ analysis.
The ‘assumption of equal prognostic value’ analysis included all tests for which information on cost and/or technical performance were available from the online survey of NHS laboratories in England and Wales. This included the tests for which neither comparative effectiveness nor response data were available. Therefore, in this analysis, the costs of the tests were assessed given an assumption of equal prognostic value and test-specific information on costs only. For this purpose, the prognostic value of all tests was based on the Therascreen EGFR PCR Kit, as this was the only test for which prognostic data were available on patients with positive, negative and unknown tumour mutation status. In addition, other tests used in NHS laboratories in England and Wales were considered to have technical characteristics (low limit of detection and similar proportion of tumour cells required for analysis) that were more similar to this test than to direct sequencing methods and would therefore be more likely to have similar prognostic value to the Therascreen EGFR PCR Kit than to direct sequencing. The results of the ‘assumption of equal prognostic value’ analysis indicated that the strategies were almost equal, i.e. the lowest total strategy cost was (CiC information has been removed) (Sanger sequencing or Roche cobas) compared with (CiC information has been removed) for the most expensive strategy (fragment length analysis combined with pyrosequencing). The sensitivity analysis, where the number of unknowns was based on results from the online survey of NHS laboratories in England and Wales, instead of being assumed equal based on literature, showed a slightly larger range of costs (CiC information has been removed) and a small range in QALYs (0.871 to 0.886) for the included mutation tests.
Strengths and limitations of assessment
Clinical effectiveness
Extensive literature searches were conducted in an attempt to maximise retrieval of relevant studies. These included electronic searches of a variety of bibliographic databases, as well as screening of clinical trials registers and conference abstracts to identify unpublished studies. Because of the known difficulties in identifying test accuracy studies using study design-related search terms,74 and potential need to include non-RCTs, search strategies were developed to maximise sensitivity at the expense of reduced specificity. Thus, large numbers of citations were identified and screened, many of which did not meet the inclusion criteria of the review.
The possibility of publication bias remains a potential problem for all systematic reviews. Considerations may differ for systematic reviews of test accuracy studies. It is relatively simple to define a positive result for studies of treatment, for example a significant difference which favours treatment between the treatment and control groups. This is not the case for test accuracy studies, which measure agreement between index test and reference standard. It would seem likely that studies finding greater agreement (high estimates of sensitivity and specificity) will be published more often. This distinction may be less applicable to studies in this review which provided accuracy data, as in all cases these studies aimed to assess the effectiveness of treatment with TKIs in different patient groups rather than being primarily focused upon test performance. Our review included small numbers of clinically heterogeneous studies, both for the accuracy of EGFR mutation testing to predict response to treatment with TKIs and for the relative effectiveness of TKIs in populations selected using different EGFR mutation test methods. We were therefore unable to undertake any meta-analyses or formal assessment of publication bias. However, our search strategy included a variety of routes to identify unpublished studies and resulted in the inclusion of a number of conference abstracts.
Clear inclusion criteria were specified in the protocol for this review and the one protocol modification that occurred during the assessment has been highlighted in the protocol. The eligibility of studies for inclusion is therefore transparent. In addition, we have provided specific reasons for excluding all of the studies considered potentially relevant at initial citation screening (see Appendix 5). The review process followed recommended methods to minimise the potential for error and/or bias;22 studies were independently screened for inclusion by two reviewers and data extraction and quality assessment were carried out by one reviewer and checked by a second (MW and PW). Any disagreements were resolved by consensus.
Studies included in this review were assessed for risk of bias using published tools appropriate to study design and/or the type of data extracted. Studies that provided data on the accuracy of EGFR mutation testing to predict response to treatment with TKIs were assessed using a modification of the QUADAS-2 tool. 31 QUADAS-2 is structured into four key domains covering participant selection, index test, reference standard and the flow of patients through the study (including timing of tests). Each domain is rated for risk of bias (low, high or unclear); the participant selection, index test and reference standard domain are also separately rated for concerns regarding the applicability of the study to the review question (low, high or unclear). The version of QUADAS-2 used in this report did not include assessment of applicability because both the index test and study population were tightly defined by our inclusion criteria and clinical outcome measures were treated as the reference standard. Studies that provided data on the effectiveness of treatment with TKIs, compared with standard chemotherapy, in patients with EGFR mutation-positive tumours were all RCTs or subgroup analyses from RCTs. These studies were therefore assessed using the Collaboration’s tool for assessing risk of bias in randomised trials. 25,30 The results of the risk of bias assessment are reported, in full, for all included studies (see Appendix 3) and in summary in the results (see Chapter 3, What is the accuracy of epidermal growth factor receptor mutation testing, using any test, for predicting response to treatment with tyrosine kinase inhibitors? and How do outcomes from treatment with epidermal growth factor receptor inhibitors vary according to which test is used to select patients for treatment?). The main potential sources of bias identified were exclusion of withdrawals from the analyses (for both studies providing data on the accuracy of EGFR mutation tests to predict response to TKIs and RCTs of TKIs in patients with EGFR mutation-positive tumours) and blinding of participants and personnel in treatment trials, which was not possible owing to the different delivery modes of intervention and comparator drugs.
All of the studies included in this review have some limitations in respect of their ability to address the overall aim of comparing the clinical effectiveness of different EGFR mutation tests to determine which patients may benefit from treatment with TKIs and which should receive standard chemotherapy. The IPASS48,49 and First-SIGNAL41 trials represent the closest approximation to the ideal study in that they provide full information on the comparative treatment effect (TKI vs. standard chemotherapy) for both patients with EGFR mutation-positive and EGFR mutation-negative tumours, for which mutation status was defined using the Therascreen EGFR PCR Kit (version 1) and direct sequencing, respectively. However, data were derived from subgroup analyses of patients included in the original trial who had received EGFR mutation testing and, in the case of the First-SIGNAL study, this subgroup included a small number of participants and was poorly described. 41 Because methods of testing EGFR mutation status differ both in terms of the mutations targeted and limit of detection (the lowest proportion of tumour cells with a mutation that can be detected), the definition of EGFR mutation positive varies according to which test is used. All testing methods are essentially reference standard methods for classifying mutation status, as defined by the specific test characteristics. It is therefore not useful to compare tests solely in terms of their ability to detect particular combinations of mutations. The essential clinical question is ‘which testing method is best at classifying patients, such that the maximum treatment effect is achieved both for mutation-positive patients who receive TKIs and mutation-negative patients who receive standard chemotherapy?’ To fully address this question, IPASS-type data would be required for patients with mutation-positive tumours and patients with mutation-negative tumours, as defined by each proposed classification method (i.e. each different EGFR test). Following the IPASS trial and subsequent NICE recommendations,1,75 obtaining these data may be problematic, as it could be argued that a trial for which patients are randomised to TKI or standard chemotherapy regardless of tumour EGFR mutation status would be unethical. Additionally, once the principle had been established that TKIs are more effective in EGFR mutation-positive patients, subsequent trials have tended to focus on assessing the effectiveness of various TKIs in populations with EGFR mutation-positive tumours; trials are not primarily concerned with the method used to establish mutation status. An alternative approach to this problem is provided by studies that report sufficient data to calculate the accuracy of different EGFR mutation tests for predicting response to treatment with TKIs. These studies provide information on the extent to which different EGFR mutation tests are able to discriminate between patients who will respond to TKI treatment and those who will not; treatment response data are reported for patients with EGFR mutation-positive and EGFR mutation-negative tumours. However, we were able to identify only four studies of this type: all used direct sequencing methods, three pre-dated the IPASS trial, and three had very small sample sizes, which were reflected in the wide CIs around sensitivity and specificity estimates. In addition, no study reported data for more than one EGFR mutation test, hence any apparent differences in test performance observed between studies may have arisen as a result of differences in study populations. Trials that compared the effectiveness of TKIs with that of standard chemotherapy in patients with advanced NSCLC whose tumours tested positive for EGFR mutations were also included in this review. These trials were included with the aim of providing some indication on how the favourable TKI treatment effect seen in patients with mutation-positive tumours in the IPASS trial may vary according to how these patients are selected (which EGFR mutation test is used). However, it should be noted that differences between these studies, other than the way in which positive EGFR mutation status is defined, particularly in relation to the baseline participant characteristics, may contribute to any differences in treatment effects observed. In addition, these trials can provide no information about the relative effectiveness of TKIs and standard chemotherapy in patients whose tumours are classified as EGFR mutation negative by tests other than the Therascreen EGFR PCR Kit. Some trials reported the results of subgroup analyses to assess possible variation in treatment effect (e.g. smoking history, tumour histology); however, trials were generally not powered to detect any difference in treatment effect between subgroups.
This assessment assumes equivalent treatment effects for the two TKIs (gefitinib and erlotinib), which are recommended by NICE as first-line treatments for patients with advanced, EGFR mutation-positive NSCLC. 1,75 This assumption is supported by the conclusion of the appraisal committee in NICE guidance 258 that ‘there was insufficient evidence to suggest a difference in clinical effectiveness between erlotinib and gefitinib’. 75 No RCTs directly comparing gefitinib and erlotinib have been identified and the results of indirect treatment comparisons vary. 75,76 Our review identified one retrospective Taiwanese study comparing gefitinib and erlotinib, which did not meet our inclusion criteria. This study included 224 patients, with known tumour EGFR mutation status, who had received TKI treatment (124 gefitinib and 100 erlotinib) but was not restricted to first-line treatment; no significant difference between the two treatments was observed for either PFs or OR rate. 77
Cost-effectiveness
A de novo probabilistic model was developed to assess the cost-effectiveness of different methods for EGFR-TK mutation testing to decide between standard chemotherapy or EGFR-TKIs for first-line treatment of patients with locally advanced or metastatic NSCLC. In order to be consistent with related assessments/appraisals, we first ensured that the results for patients with an EGFR positive mutation tumour using the Therascreen EGFR PCR Kit in the de novo model were similar to the results of these patients in the initial manufacturer’s model used in NICE Technology Appraisal 192. 51 Subsequently, the ERG amendments were incorporated and ICERs from the de novo model were compared with ICERs as reported in the final appraisal determination of STA 19251 (see Appendix 6 for results).
Test failures and costs were based on information obtained from the online survey of NHS laboratories in England and Wales. These real-life data provided an important source of information, which is likely to be representative of clinical practice.
In the assessment of economic value of different tests, a link has to be established between test accuracy, clinical value (e.g. treatment response, PFS, OS) and relative cost-effectiveness. Ideally, the performance of EGFR mutation tests would be assessed against an objective measure of the true presence/absence of a clinically relevant EGFR-TK mutation (the ‘reference standard’), and comparative effectiveness of treatment (TKI vs. chemotherapy) conditional upon the true or false presence/absence of the EGFR-TK mutation would be determined. However, each different testing method targets a different range of mutations and has different limits of detection (lowest proportion of mutation detectable in tumour cells) and the exact combination of mutation type and level that will provide optimal treatment selection remains unclear. For this reason, assessment of test performance based on comparison with a conventional ‘reference standard’ is not currently possible. In this situation, an alternative way to determine the relative value of diagnostic methods for EGFR-TK mutation testing is to use studies that report on the comparative treatment effect in patients with different EGFR mutation status (positive, negative, or unknown) as defined using different EGFR mutation tests. Thus, OR on anti-EGFR-TKIs was assumed to correlate perfectly with the ‘true’ presence/absence of the EGFR-TK mutation. The use of alternative measures of EGFR-TK mutation in the assessment of cost-effectiveness might impact the proportion of mutation positives and negatives (see Tables 15 and 16) and thus might substantially impact the assessment of cost-effectiveness (in either direction) as this is one of the key drivers of cost-effectiveness. In the absence of an objective measure of the ‘true’ presence/absence of a clinically significant EGFR-TK mutation (i.e. which mutations, present at what levels, as defined by which testing method, will result in differential treatment effects for TKIs vs. standard chemotherapy), the current cost-effectiveness assessment is, at best, an approximation of the ‘true’ cost-effectiveness of test-guided treatments.
Evidence on the comparative treatment effect in patients with different tumour EGFR mutation status (positive, negative or unknown) as defined using different tests was available for only two tests (the Therascreen EGFR PCR Kit and direct sequencing of all exon 19–21 mutations). A major assumption underpinning our analyses was that the differences in OR, PFS and OS observed in the two included studies from which these data were derived41,48,49 can be solely ascribed to differences in test performance. In practice this assumption would seem unlikely to hold true. These differences could also be caused by differences in participant characteristics, differences in the standard chemotherapy regimen or differences in treatment strategies following progression that may affect OS, all of which were apparent between these two studies.
It was not part of the scope of this assessment to update the appraisal of gefitinib for the first-line treatment of locally advanced or metastatic NSCLC (NICE Technology Appraisal 192). 1 However, the ERG’s report for NICE Technology Appraisal 19265 noted that the cost-effectiveness of the ‘EGFR mutation test + TKI treatment if positive and doublet chemotherapy if negative’ strategy compared with the ‘doublet chemotherapy without EGFR mutation testing’ strategy is conditional upon the accuracy of the mutation test used to distinguish between patients who receive TKI treatment and patients who receive doublet chemotherapy. This is a simplification of the issue because, as described previously, each EGFR mutation testing method identifies a subtly different combination of type and level of EGFR mutation, and the clinical significance of these different combinations is largely unknown. It is particularly problematic if a test defines positive mutation status for a type and/or level of mutation that is not clinically significant (associated with response to treatment with TKIs), as the patients thus ‘falsely’ identified as having mutation-positive tumours will experience a loss of survival time and quality of life due to not receiving the most effective treatment option for them, while still experiencing treatment related adverse events; the costs of treatment are also considerably increased. The effects of this might even outweigh the relative gains of TKI treatment compared with doublet chemotherapy for those patients correctly selected for TKI treatment. Therefore, the economic evaluation of TKI treatment should not be seen as an assessment of the relative value of the drug in isolation from the mutation test used to select eligible patients, but as an assessment of a specific ‘mutation test’–’treatment’ combination, which may not be valid if other methods for mutation testing are used. For this assessment, this means that the results described are partial in the sense that the ‘doublet chemotherapy without EGFR mutation testing’ strategy was not taken into account.
Uncertainties
Clinical effectiveness
As discussed above (see Strengths and limitations of assessment, Clinical effectiveness), one key consideration when selecting an EGFR mutation testing method is the variation between tests in limit of detection (i.e. the minimum percentage of mutation in tumour cells required to produce a positive result). A lower limit of detection can enhance the ability of laboratories to produce results from poor-quality samples. Similarly, methods of specimen handling, for example laser-capture microdissection, may affect the ability of the EGFR mutation testing method to detect mutations present at a low level. However, it should not be assumed that a lower limit of detection will necessarily result in a more clinically effective test, as it is possible that TKIs may be less effective in patients with a low proportion of tumour cells harbouring mutation. Discussions with clinical experts suggest that there is ongoing uncertainty around this issue as quantitative results of EGFR mutation testing are not routinely reported. None of the studies that met the inclusion criteria for this review reported any data on variation in treatment effect with the proportion of tumour cells having EGFR mutations. A Chinese study, which did not meet our inclusion criteria, assessed tissue bank tumour samples from NSCLC patients who had been treated with gefitinib at any stage during the course of their disease. 78 This study analysed samples using both direct DNA sequencing and the Therascreen EGFR PCR Kit: samples that were positive by both methods were classified as having a high abundance of EGFR mutations; samples that were positive using the Therascreen EGFR PCR Kit and negative on direct sequencing were classified as having a low abundance of EGFR mutations; and samples that were negative on both tests were classified as wild type. The results of this study were mixed: median PFS was significantly longer in both the high abundance [11.3 months (95% CI 7.4 to 15.2 months)] and low abundance [6.9 months (95% CI 5.5 to 8.4 months)] groups compared with wild type [2.1 months (95% CI 1.0 to 3.2 months)]; however, for other outcome measures (OR rate and OS) benefits were limited to the high-abundance group. 78 It should also be noted that this study provides no information on the relative effectiveness of standard chemotherapy in these patient groups.
A further area of uncertainty concerns the clinical value of detecting rare mutations and possible resistance mutations. The majority of the evidence on the effectiveness of first-line treatment with TKIs in patients with EGFR mutation-positive NSCLC has been derived from patients with exon 19 deletions or the exon 21 mutation L858R. This is unsurprising, as these account for > 90% of all EGFR mutations. 6,7,13 The additional clinical value of using tests that target a wider range of mutations remains uncertain, as the low frequency of most EGFR mutations makes it very difficult to adequately assess treatment effects in patients with mutations other than exon 19 deletions or L858R. Some of the studies in our review that provided data on the accuracy of EGFR testing in predicting response to treatment with TKIs reported response data by individual mutation; these data appeared to indicate that there may be a less favourable response to TKIs in patients with T790M or other exon 20 mutations (see Table 8); however, these data were very limited. There are a number of registry studies that did not meet our inclusion criteria, but which have reported some information of clinical response in patients with different EGFR mutations. Murray et al. compiled a database of 202 articles, which provided data on 2,548 NSCLC patients (disease stage and previous treatment not specified) who had been treated with TKIs. 79 This study reported an OR rate of 86% for patients with a mutation in exon 19 compared with 33% for those with a mutation in exon 20; subgroup analysis indicated that a mutation in exon 20, in the absence of T790M, was associated with an OR rate of 68% (similar to that for mutations in exon 18 or 21). 79 Of the 115 different mutations for which response data were available, only 13 demonstrated PD as a response, of which eight were located in exon 20. However, as noted by the authors, some caution is required in interpreting these data, as the two most common mutations account for > 90%, with T790M occurring in only around 2% of patients. 79 An observational study conducted in 15 of 28 French National Cancer Institute laboratories identified 1048 EGFR mutations from 10,117 patients with NSCLC who were tested. 80 Of these, 108 were rare mutations (48 in exon 18 and 60 in exon 20); 36 of these patients received a TKI and were evaluable for response. The best response was progression in 18 patients, stabilisation in 11 patients and PR in seven patients. 80 REASON, a large registry study of > 4000 patients at 151 centres in Germany, aims to generate data on EGFR mutation status and clinical response to TKIs in patients with stage IIIB or stage IV data; however, to date, this study has been published only as a conference abstract, with no data for specific mutations. 81 A similar programme, EGFR FASTnet, exists in Italy, although again we have not been able to identify any publication that reports mutation-specific response data. 82,83 Both programmes are supported by AstraZeneca.
The clinical significance of rare mutations and the possible increased risk of ‘false-positives’ associated with the use of EGFR mutation tests that are able to detect very low levels of mutation were both highlighted as areas requiring further research by the European EGFR Workshop Group in a 2009 multidisciplinary consensus meeting on the implementation of EGFR mutation testing. 12
As with the issue of rare mutations, there is uncertainty regarding the clinical effectiveness of identifying EGFR mutations in non-adenocarcinoma NSCLC. The majority of the evidence on the effectiveness of first-line treatment with TKIs in patients with EGFR mutation-positive NSCLC has been derived from patients with adenocarcinomas. All but one41 of the studies included in our review included small numbers of patients with other histological diagnoses, but none reported separate data for these patients. We identified one retrospective analysis of patients with advanced NSCLC and known EGFR mutation status (determined by direct sequencing), which did not meet our inclusion criteria but which reported comparative data on 12 patients with non-adenocarcinoma and 269 with EGFR mutation-positive adenocarcinoma who were treated with TKIs. 84 OR and DC rates were lower in patients with non-adenocarcinoma than in those with adenocarcinoma (50% vs. 78% and 75% vs. 89%, respectively), and PFS was also significantly longer in the adenocarcinoma group [11.27 months (95% CI 9.87 to 12.67 months) vs. 3.67 months (95% CI 1.34 to 5.99) months]. 84 Similar results were reported for a systematic review which compared data for 33 EGFR mutation-positive non-adenocarcinoma NSCLC patients treated with gefitinib from 15 studies with adenocarcinoma patients from the same studies. 85 Although it appears that patients with non-adenocarcinoma NSCLC, which is positive for EGFR mutations, may derive less benefit from treatment with TKIs than those with adenocarcinomas, it should be noted that this question was outside the scope of our review and the studies discussed above do not provide any information on the relative effectiveness of TKIs and standard chemotherapy regimens in this group of patients.
A wide variety of EGFR mutation test methods are currently used by accredited NHS laboratories in England and Wales; however, for the majority of these methods, no studies were identified that could provide data linking the results of EGFR testing to the effectiveness treatment. Therefore, the potential clinical effects of using different EGFR tests to make decisions on first-line treatment in patients with stage IIIB or IV remains uncertain. The available data were for version 1 of the Therascreen EGFR PCR Kit and for direct sequencing methods targeting various mutations. Version 1 of the Therascreen kit is no longer being actively marketed by Qiagen and equivalent data are not available for its replacement – the Therascreen EGFR RGQ PCR Kit – or for the Therascreen EGFR Pyro Kit. However, it may be reasonable to assume equivalent diagnostic performance for all three products, as both versions of the Therascreen EGFR PCR Kit target the same mutations and the Therascreen EGFR Pyro Kit targets a similar set of mutations, with the addition of one further exon 19 deletion and the exon 21 mutation L861R and the loss of three exon 20 insertions. 86,87 All three methods have a low limit of detection (≤ 5%). 86,87 The Therascreen EGFR Pyro Kit can also produce quantitative results. 87 No data are currently available for next-generation sequencing; a method for this is currently being developed and validated by one NHS laboratory but next-generation sequencing is not yet in routine clinical use in any of NHS laboratories in England and Wales that responded to our survey.
Cost-effectiveness
Major assumptions were made in order to be able to model the relative cost-effectiveness of different EGFR mutation tests. It was assumed that the differences in relative treatment response, PFS and OS between the results of First-SIGNAL trial41 and the results of the IPASS trial48,49 were solely attributable to the different mutation tests used (the Therascreen EGFR PCR Kit and direct sequencing of all exons 19–21, respectively) to distinguish between patients whose tumours are EGFR mutation positive and those whose tumours are EGFR mutation negative (‘evidence of comparative effectiveness’ and ‘linked evidence’ analyses). As described in the previous section, it is highly questionable whether this assumption would hold. Furthermore, in order to calculate the proportion of patients with a positive and negative test result, patients who tested positive were categorised as FP if no treatment response was observed after TKI, whereas patients were categorised as TP if treatment response was observed after TKI. Similarly, patients who tested negative were categorised as FN if treatment response was observed after TKI, whereas patients were categorised as TN if no treatment response was observed after TKI. Ideally, the categorisation of true/false positives/negatives should be based on an objective measure of the true presence/absence of a clinically relevant EGFR-TK mutation. However, as previously described, the uncertainty around the exact definition of a clinically relevant mutation is such that this is not currently possible. It was also assumed that the proportion of patients with unknown mutation status relative to the number of patients for whom a tissue sample was available, as reported in the trials included in the systematic review,44,49 provides a realistic approximation of the proportion of patients with an unknown test result in clinical practice. Outcomes in patients with unknown tumour mutation status were only reported in the IPASS trial. 48,49 These results were used to model the outcomes in patients with an unknown test result for the other testing methods considered in this assessment, assuming that the OR rate, PFS and OS in patients with an unknown test result after use of the Therascreen EGFR PCR Kit, as reported in the IPASS trial,48,49 were generalisable to the direct sequencing methods. In the ‘linked evidence’ analysis, information from Jackman et al. 44 [direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells)] and the First-SIGNAL trial41 were used to model the other direct sequencing methods if information was missing, thus assuming this information was generalisable to the other direct sequencing methods. The extent to which these results are actually generalisable to testing methods other than the Therascreen EGFR PCR Kit is unknown.
Moreover, as this model was partially based on the evidence and model structure used in the appraisal of gefitinib for the first-line treatment of locally advanced or metastatic NSCLC (NICE Technology Appraisal 192),1,51 the assumptions underlying that appraisal also apply to this assessment; for instance, assumptions regarding the applicability of the findings in the trials to the population in England and Wales.
Finally, it should be emphasised that the uncertainty resulting from the above mentioned assumptions was not parameterised in the model and is therefore not reflected in the probabilistic sensitivity analyses and hence cost-effectiveness acceptability curves.
Chapter 6 Conclusions
Implications for service provision
There was no strong evidence that any one EGFR mutation test had greater accuracy than any other test. There was a suggestion that Therascreen EGFR PCR Kit may be more accurate than direct sequencing for predicting response to treatment with TKIs, although it should be noted that only one data set was available for this test and no studies reported direct comparisons between the Therascreen EGFR PCR Kit and other tests, conducted in the same population. The clinical effectiveness of TKIs, in patients whose tumours are positive for EGFR, did not appear to vary according to which test was used to determine EGFR mutation status.
The results of the ‘evidence on comparative effectiveness available’ analysis and the ‘linked evidence’ analysis both indicated that the Therascreen EGFR PCR Kit was less effective and less expensive than direct sequencing (all exon 19–21 mutations and all 18–21 mutations, respectively) at £31,000–35,000 per QALY lost. The lower QALYs for the Therascreen EGFR PCR Kit seem counterintuitive, as the accuracy data show a higher accuracy for Therascreen EGFR PCR Kit. This contradiction possibly results from the problematic and substantial assumptions made to arrive at the economic results. In particular, the assumption that the differences in treatment response and survival between tests, as observed between the different studies, are solely attributable to the different tests used. This ignores all other factors that can explain variations in outcomes between the studies. Therefore, these outcomes of the assessment of cost-effectiveness should be interpreted with extreme caution.
The results of the ‘assumption of equal prognostic value’ analysis (including all tests for which information on cost and/or technical performance was available from the online survey of NHS laboratories in England and Wales) showed that the costs of the EGFR mutation tests were very similar [range from (CiC information has been removed) for Sanger sequencing or Roche cobas for samples with insufficient tumour cells to (CiC information has been removed) for fragment length analysis combined with pyrosequencing].
There are no data on the clinical or cost-effectiveness of Therascreen EGFR Pyro Kit or next-generation sequencing. No published studies were identified for either of these two methods and neither method is currently in routine clinical use in any of NHS laboratories in England and Wales that responded to our survey; one laboratory is currently developing and validating a next-generation sequencing method.
Suggested research priorities
The available data have limitations in respect of their ability to address the overall aim of this assessment, to compare the clinical effectiveness of different EGFR mutation tests to determine which patients may benefit from treatment with TKIs and which should receive standard chemotherapy. Because each different testing method potentially selects a subtly different population, based on the targeting of a different range of mutations and different limits of detection, the most informative studies are those that provide full information on the comparative treatment effect (TKI vs. standard chemotherapy) for both patients with EGFR mutation-positive and EGFR mutation-negative tumours. Studies of this type are available for only two testing methods, direct sequencing and the Therascreen EGFR PCR Kit (version 1), and further similar trials are unlikely as randomisation of patients to TKIs or standard chemotherapy, regardless of EGFR mutation status, would be against current clinical guidance and would almost certainly be considered unethical. One possible solution to this problem would be to re-test stored samples from previous studies, where patient outcomes are already known, using those EGFR mutation testing methods for which adequate data are currently unavailable. This approach could provide a ‘black box’ answer, whereby the relative effectiveness of TKIs and standard chemotherapy in patients with EGFR mutation-positive and negative tumours could be determined for each test. However, it would not provide any information on the underlying reason for any observed differences between tests. As they are likely to represent the most practical approach to obtaining informative data, retrospective, comparative accuracy studies, using stored samples for which the patient outcome is already known, should be given priority.
Newer methods of EGFR mutation testing, for example the Therascreen EGFR Pyro Kit, can provide quantitative results. Should quantitative testing become part of routine practice, longitudinal follow-up studies relating the level of mutation and/or the presence or rarer mutations to patient outcomes would become possible. Studies of this type could help to assess which features of EGFR mutation tests are likely to be important in determining their clinical effectiveness and should be considered going forward.
Building upon information gained from the two study types described above, preliminary research to develop a multifactorial prediction model should be considered. Initially, research of this type is likely to be exploratory in nature; however, models developed could form the basis of tools that will eventually help determine more accurately which patients are likely to benefit from treatment with EGFR-TKIs.
As the uncertainties associated with clinical effectiveness forced the major assumptions in the economic evaluation, this type of research would also facilitate economic analyses of EGFR mutation testing.
Acknowledgements
The authors acknowledge the clinical advice and expert opinion provided by Mr Paul Roberts, Consultant Cytogeneticist, Leeds Teaching Hospitals NHS Trust; Dr Phillipe Taniere, Consultant Histopathologist, University Hospitals Birmingham NHS Foundation Trust; Professor Ian Cree, Professor of Pathology, Warwick Medical School, University Hospitals Coventry and Warwickshire; Dr Mark Slade, Consultant Respiratory Physician and Clinical Director, Papworth Hospital NHS Foundation Trust; Dr Fiona Blackhall, Consultant Medical Oncologist, The Christie NHS Foundation Trust; and Mrs Mani Elliott, Chemotherapy Nurse specialist, Hull and East Yorkshire Hospitals NHS Trust. The authors would like to thank all of the NHS laboratories – providing EGFR mutation testing – that kindly completed our on-line survey. The authors would also like to thank AstraZeneca UK Ltd for providing access to the cost-effectiveness model used in NICE Technology Appraisal 192. Finally, the authors would like to thank the lay members of the NICE Diagnostics Advisory Committee and Assessment Sub-group for providing input on the patients’ perspective at key stages of the assessment process.
Contributions of authors
Marie Westwood and Penny Whiting planned and performed the systematic review and interpretation of evidence.
Manuela Joore, Thea van Asselt and Bram Ramaekers planned and performed the cost-effectiveness analyses and interpreted results.
Nigel Armstrong contributed to planning and interpretation of cost-effectiveness analyses, and acquisition of input data for modelling.
Kate Misso devised and performed the literature searches and provided information support to the project.
Jos Kleijnen and Johan Severens provided senior advice and support to the systematic review and cost-effectiveness analyses, respectively.
All parties were involved in drafting and/or commenting on the report.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Gefitinib for the First-Line Treatment of Locally Advanced or Metastatic Non-Small-Cell Lung Cancer. London: NICE; 2010.
- Erlotinib for the First-Line Treatment of Locally Advanced or Metastatic EGFR-TK Mutation-Positive Non-Small Cell Lung Cancer. London: NICE; 2012.
- Cancer Registrations in England, 2010: Number of New Cases Diagnosed by Site. London: ONS; 2012.
- Lung Cancer: UK Incidence Statistics. London: Cancer Research UK; 2012.
- National Lung Cancer Audit Report 2011. Report for the Audit Period 2010. Leeds: The NHS Information Centre; 2011.
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 2005;23:857-65. http://dx.doi.org/10.1200/JCO.2005.08.043.
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919-23. http://dx.doi.org/10.1158/0008-5472.CAN-04-2818.
- Huang S-F, Liu H-P, Li L-H, Ku Y-C, Fu Y-N, Tsai H-Y, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 2004;10:8195-203. http://dx.doi.org/10.1158/1078-0432.CCR-04-1245.
- Romeri E, Baker A, Griffiths C. Mortality by deprivation and cause of death in England and Wales, 1999–2003. Health Stat Q 2006;32:19-34.
- National Lung Cancer Audit Report. Key Findings about the Quality of Care for People with Lung Cancer in England Incorporating Headline and Completeness Data from Wales. Report for the Audit Period 2005. Leeds: The NHS Information Centre; 2006.
- Office for National Statistics (ONS) . Cancer Registrations in England, 2010 2012. www.ons.gov.uk/ons/dcp171778_263537.pdf (accessed 10 July 2012).
- Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol 2010;5:1706-13. http://dx.doi.org/10.1097/JTO.0b013e3181f1c8de.
- Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T. A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn 2006;8:335-41. http://dx.doi.org/10.2353/jmoldx.2006.050104.
- Yung TKF, Chan KCA, Mok TSK, Tong J, To K-F, Lo YMD. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. http://dx.doi.org/10.1158/1078-0432.CCR-08-2622.
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. http://dx.doi.org/10.1056/NEJMoa040938.
- Zhou C, Wu Y-L, Chen G, Feng J, Liu X-Q, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. http://dx.doi.org/10.1016/S1470-2045(11)70184-X.
- Horiike A, Kimura H, Nishio K, Ohyanagi F, Satoh Y, Okumura S, et al. Detection of epidermal growth factor receptor mutation in transbronchial needle aspirates of non-small cell lung cancer. Chest 2007;131:1628-34. http://dx.doi.org/10.1378/chest.06-1673.
- Lung Cancer: the Diagnosis and Treatment of Lung Cancer (CG121). London: NICE; 2011.
- Lung Cancer (Non-Small-Cell, First Line Treatment) – Pemetrexed (TA181). London: NICE; 2009.
- WHO Handbook for Reporting Results of Cancer Treatment. WHO Offset Publication No. 48. Geneva: WHO; 1979.
- Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-16.
- Systematic Reviews: CRD’s Guidance for Undertaking Reviews in Health Care. York: University of York; 2009.
- Diagnostics Assessment Programme Manual. Manchester: NICE; 2011.
- Cochrane Diagnostic Test Accuracy Working Group . Handbook for DTA Reviews: The Cochrane Collaboration 2009. http://srdta.cochrane.org/handbook-dta-reviews (accessed 23 March 2011).
- Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. The Cochrane Collaboration; 2011.
- McGowan J, Sampson M, Lefebvre C. An evidence based checklist for the peer review of electronic search strategies (PRESS EBC). Evid Based Libr Inf Pract 2010;5:1-6.
- Royle P, Waugh N. Should systematic reviews include searches for published errata?. Health Info Libr J 2004;21:14-20. http://dx.doi.org/10.1111/j.1471-1842.2004.00459.x.
- Wright K, McDaid C. Madrid: Cochrane Colloquium, Cochrane Collaboration; 2011.
- Wright K, McDaid C. Reporting of article retractions in bibliographic databases and online journals. J Med Libr Assoc 2011;99:164-7. http://dx.doi.org/10.3163/1536-5050.99.2.010.
- Higgins JP, Altman DG, Gotzsche PC, Juni P, Moher D, Oxman AD, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011;343.
- Whiting PF, Rutjes AWS, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. http://dx.doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. http://dx.doi.org/10.1016/j.jclinepi.2005.02.022.
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics 2007;8:239-51. http://dx.doi.org/10.1093/biostatistics/kxl004.
- Harbord RM, Whiting P, Sterne JA, Egger M, Deeks JJ, Shang A, et al. An empirical comparison of methods for meta-analysis of diagnostic accuracy showed hierarchical models are necessary. J Clin Epidemiol 2008;61:1095-103. http://dx.doi.org/10.1016/j.jclinepi.2007.09.013.
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. http://dx.doi.org/10.1002/sim.1186.
- Benlloch S, Taron M, Botero ML, Bertran-Alamillo J, Mayo C, Gimenez Capitan A, et al. Retrospective EGFR mutation testing of clinical specimens from the EURTAC trial of erlotinib in non-small cell lung cancer (NSCLC) using a novel allele-specific PCR (AS-PCR) assay n.d.
- Sugio K, Uramoto H, Takenoyama M, Hanagiri T, Yasumoto K. Molecular targeted therapy and tailor-made therapy for lung cancer. Kyobu Geka 2008;61:37-42.
- Rosell R, Gervais R, Vergnenegre A, Massuti B, Felip E, Cardenal F, et al. Erlotinib versus chemotherapy (CT) in advanced non-small cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) mutations: interim results of the European Erlotinib Versus Chemotherapy (EURTAC) phase III randomized trial. Paper presented at ASCO Annual Meeting 2011; 3–7 June 2011 ; Chicago, Illinois. J Clin Oncol 2011;29.
- Lee JS, Park K, Kim SW, Lee DH, Kim HT, Han JT, et al. A randomized phase III study of gefitinib (IRESSA) versus standard chemotherapy (gemcitabine plus cisplatin) as first-line treatment for never smokers with advanced or metastatic adenocarcinoma of the lung. J Thorac Oncol 2009;4.
- Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. http://dx.doi.org/10.1016/S1470-2045(11)70393-X.
- Han JY, Park K, Kim SW, Lee DH, Kim HY, Kim HT, et al. First-SIGNAL: first-line single-agent IRESSA versus gemcitabine and cisplatin trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol 2012;30:1122-8. http://dx.doi.org/10.1200/JCO.2011.36.8456.
- Leary AF, de Castro DG, Nicholson AG, Ashley S, Wotherspoon A, O’Brien MER, et al. Establishing an EGFR mutation screening service for non-small cell lung cancer: sample quality criteria and candidate histological predictors. Eur J Cancer 2012;48:61-7. http://dx.doi.org/10.1016/j.ejca.2011.09.022.
- Giaccone G, Gallegos Ruiz M, Le Chevalier T, Thatcher N, Smit E, Rodriguez JA, et al. Erlotinib for frontline treatment of advanced non-small cell lung cancer: a phase II study. Clin Cancer Res 2006;12:6049-55. http://dx.doi.org/10.1158/1078-0432.CCR-06-0260.
- Jackman DM, Yeap BY, Lindeman NI, Fidias P, Rabin MS, Temel J, et al. Phase II clinical trial of chemotherapy-naive patients ≥ 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol 2007;25:760-6. http://dx.doi.org/10.1200/JCO.2006.07.5754.
- Pallis AG, Voutsina A, Kentepozidis N, Giassas S, Papakotoulas P, Agelaki S, et al. A phase II trial of erlotinib as front-line treatment in clinically selected patients with non-small-cell lung cancer. Clin Lung Cancer 2012;13:129-35. http://dx.doi.org/10.1016/j.cllc.2011.08.004.
- Yang C-H, Yu C-J, Shih J-Y, Chang Y-C, Hu F-C, Tsai M-C, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol 2008;26:2745-53. http://dx.doi.org/10.1200/JCO.2007.15.6695.
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. http://dx.doi.org/10.1056/NEJMoa0909530.
- Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. http://dx.doi.org/10.1200/JCO.2010.33.4235.
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. http://dx.doi.org/10.1056/NEJMoa0810699.
- Ohe Y, Ishikura S, Yokoyama A, Atagi S, Shibata T, Tamura T, et al. Safety and efficacy trial of cisplatin (P) and vinorelbine (V) followed by gefitinib (G) and concurrent thoracic radiotherapy (TRT) for unresectable locally advanced non-small cell lung cancer (LA-NSCLC). 4th Asia Pacific Lung Cancer Conference, APLCC, Seoul, 2–4 December 2010. J Thorac Oncol 2010;5:401-2.
- Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd; 2010.
- Borget I, Cadranel J, Pignon JP, Quoix E, Coudert B, Westeel V, et al. Cost-effectiveness of three strategies for second-line erlotinib initiation in nonsmall-cell lung cancer: the ERMETIC study part 3. Eur Respir J 2012;39:172-9. http://dx.doi.org/10.1183/09031936.00201210.
- Drummond MF, Jefferson TO. Guidelines for authors and peer reviewers of economic submissions to the BMJ. BMJ 1996;313:275-83. http://dx.doi.org/10.1136/bmj.313.7052.275.
- Arrieta O, Anaya P, Lopez RJ, Polanco AC. Cost-effectiveness analysis of EGFR mutation testing in patients with advanced non small-cell lung cancer (ANSCLC) treated with gefitinib or carboplatin-paclitaxel. Paper presented at 15th Annual International Meeting of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR 2010); 15–19 May 2010; Atlanta, GA. Value Health 2010;13:A37-8. http://dx.doi.org/10.1016/S1098-3015(10)72164-1.
- Chen W, Ellis P, Levin L, Krahn M. Cost-effectiveness of epidermal growth factor receptor gene mutation testing in the selection of first-line therapy for patients with advanced non-small cell lung cancer in Ontario. Paper presented at 16th Annual International Meeting of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR); 21–25 May 2011; Baltimore, MD. Value Health 2011;14. http://dx.doi.org/10.1016/j.jval.2011.02.459.
- Jacob J, Henriksson M, Brattström D. Cost-effectiveness of gefitinib versus doublet chemotherapy in first-line treatment of non-small cell lung cancer (NSCLC) in Sweden. Paper presented at ISPOR 13th Annual European Congress; 6–9 November 2010; Prague. Value Health 2010;13. http://dx.doi.org/10.1016/S1098-3015(11)71995-7.
- Lopes GD, Segel J, Tan D, Do Y, Mok T, Finkelstein E. Cost-effectiveness of epidermal growth-factor receptor mutation testing and first-line treatment with gefitinib for advanced non-small-cell lung cancer. EJC Suppl 2011;9.
- Shiroiwa T, Miyoshi Y, Tsutani K. Cost-effectiveness analysis of EGFR testing and gefitinib for nonsmall-cell lung cancer (NSCLC) in Japan. Paper presented at 17th Annual International Meeting of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR); 2–6 June 2012; Washington, DC. Value Health 2012;15.
- Cadranel J, Coudert B, Mauguen A, Friard S, Quoix E, Madelaine J, et al. Clinical and biological predictors of progression-free survival (PFS) and overall survival (OS) in patients (pts) with advanced non-small-cell lung cancer (NSCLC) treated by erlotinib in the ERMETIC cohort. Eur Respir J 2010.
- Vergnenegre A, Corre R, Berard H, Paillotin D, Dujon C, Robinet G, et al. Cost-effectiveness of second-line chemotherapy for non-small cell lung cancer: an economic, randomized, prospective, multicenter phase III trial comparing docetaxel and pemetrexed: the GFPC 05–06 study. J Thorac Oncol 2011;6:161-8. http://dx.doi.org/10.1097/JTO.0b013e318200f4c1.
- Nafees B, Stafford M, Gavriel S, Bhalla S, Watkins J. Health state utilities for non-small cell lung cancer. Health Qual Life Outcomes 2008;6. http://dx.doi.org/10.1186/1477-7525-6-84.
- Curtis L. Unit Costs of Health and Social Care 2011. www.pssru.ac.uk/pdf/uc/uc2011/uc2011.pdf (accessed 18 December 2012).
- Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. http://dx.doi.org/10.1056/NEJMoa0904554.
- Cantor AB. Projecting the standard error of the Kaplan–Meier estimator. Stat Med 2001;20:2091-7. http://dx.doi.org/10.1002/sim.856.
- Brown T, Boland A, Bagust A, Oyee J, Hockenhull J, Dundar Y, et al. Gefitinib for the first-line treatment of locally advanced or metastatic non-small cell lung cancer. Health Technol Assess 2010;14:71-9.
- Erlotinib for the Treatment of Non-Small-Cell Lung Cancer (TA162). London: NICE; 2008.
- Single Technology Appraisal (STA) for Pemetrexed for the First Line Treatment of Non-Small Cell Lung Cancer. Eli Lilly and Company Ltd; 2008.
- Achieving Clinical Excellence in the Treatment of Relapsed Non-Small Cell Lung Cancer. Roche Products Ltd; 2006.
- NHS Reference Costs 2011–2012. London: DoH; 2012.
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. http://dx.doi.org/10.1016/S1470-2045(09)70364-X.
- Wang F, Wang LD, Li B, Sheng ZX. Gefitinib compared with systemic chemotherapy as first-line treatment for chemotherapy-naive patients with advanced non-small cell lung cancer: a meta-analysis of randomised controlled trials. Clin Oncol 2012;24:396-401. http://dx.doi.org/10.1016/j.clon.2011.09.013.
- Gao G, Ren S, Li A, Xu J, Xu Q, Su C, et al. Epidermal growth factor receptor-tyrosine kinase inhibitor therapy is effective as first-line treatment of advanced non-small-cell lung cancer with mutated EGFR: a meta-analysis from six phase III randomized controlled trials. Int J Cancer 2012;131:E822-9. http://dx.doi.org/10.1002/ijc.27396.
- Bria E, Milella M, Cuppone F, Novello S, Ceribelli A, Vaccaro V, et al. Outcome of advanced NSCLC patients harboring sensitizing EGFR mutations randomized to EGFR tyrosine kinase inhibitors or chemotherapy as first-line treatment: a meta-analysis. Ann Oncol 2011;22:2277-85. http://dx.doi.org/10.1093/annonc/mdq742.
- Whiting P, Westwood M, Beynon R, Burke M, Sterne JA, Glanville J. Inclusion of methodological filters in searches for diagnostic test accuracy studies misses relevant studies. J Clin Epidemiol 2011;64:602-7. http://dx.doi.org/10.1016/j.jclinepi.2010.07.006.
- National Institute for Health and Clinical Excellence (NICE) . Erlotinib Monotherapy for Maintenance Treatment of Non-Small-Cell Lung Cancer. 2011. http://guidance.nice.org.uk/TA227 (accessed 10 July 2012).
- Schwander B, De Castro Carpeno J, Heigener DF, Wright E, Bischoff HG, Walzer S. Comparative effectiveness assessment of erlotinib versus gefitinib in first-line EGFR activating mutation positive non-small cell lung cancer. Paper presented at 16th Annual International Meeting of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR); 21–25 May 2011; Baltimore, MD. Value Health 2011;14. http://dx.doi.org/10.1016/j.jval.2011.02.865.
- Wu WS, Chen YM, Tsai CM, Shih JF, Chiu CH, Chou KT, et al. Erlotinib has better efficacy than gefitinib in adenocarcinoma patients without EGFR-activating mutations, but similar efficacy in patients with EGFR-activating mutations. Exp Ther Med 2012;3:207-13.
- Zhou Q, Zhang X-C, Chen Z-H, Yin X-L, Yang J-J, Xu C-R, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. http://dx.doi.org/10.1200/JCO.2010.33.3757.
- Murray S, Dahabreh IJ, Linardou H, Manoloukos M, Bafaloukos D, Kosmidis P. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol 2008;3:832-9. http://dx.doi.org/10.1097/JTO.0b013e31818071f3.
- Beau-Faller M, Prim N, Ruppert A-M, Nanni I, Besse B, Escande F, et al. Rare epidermal growth factor receptor (EGFR) mutations in 10,117 patients with non-small cell lung cancer (NSCLC) evaluated by the French ERMETIC IFCT network: clinical, molecular, and survival data. Paper presented at ASCO Annual Meeting 2012; 1–5 June 2012; Chicago, IL. J Clin Oncol 2012;30.
- Schuette W, Eberhardt WEE, von der Schulenburg J-MG, Dietel M, Schirmacher P, Zaun S, et al. EGFR mutation analysis from REASON: a registry for the epidemiologic and scientific evaluation of EGFR mutation status in newly diagnosed NSCLC patients stage IIIB/IV. Paper presented at ASCO Annual Meeting 2012; 1–5 June 2012; Chicago, IL. J Clin Oncol 2012;30.
- Normanno N, Pinto C, Taddei GL, Mari E, Troncone G, Graziano P, et al. EGFR FASTnet: the Italian network for epidermal growth factor receptors (EGFR) mutation analysis in non-small cell lung cancer (NSCLC). Paper presented at ASCO Annual Meeting; 3–7 June 2011; Chicago, IL. J Clin Oncol 2011;29.
- Normanno N, Pinto C, Taddei GL, Troncone G, Graziano P, De Maglio G, et al. Frequency and clinical correlations of epidermal growth factor receptor (EGFR) mutations in a large cohort of Italian non-small cell lung cancer (NSCLC) patients (pts) within the EGFR FASTnet program. Paper presented at ASCO Annual Meeting 2012; 1–5 June 2012; Chicago, IL. J Clin Oncol 2012;30.
- Cho SH, Park LC, Ji JH, Park S, Hwang DW, Lee JY, et al. Efficacy of EGFR tyrosine kinase inhibitors for non-adenocarcinoma NSCLC patients with EGFR mutation. Cancer Chemother Pharmacol 2012;70:315-20. http://dx.doi.org/10.1007/s00280-012-1876-0.
- Shukuya T, Takahashi T, Kaira R, Ono A, Nakamura Y, Tsuya A, et al. Efficacy of gefitinib for non-adenocarcinoma non-small-cell lung cancer patients harboring epidermal growth factor receptor mutations: a pooled analysis of published reports. Cancer Sci 2011;102:1032-7. http://dx.doi.org/10.1111/j.1349-7006.2011.01887.x.
- Limburg, the Netherlands: Qiagen; 2012.
- Limburg, the Netherlands: Qiagen; 2011.
- Chen Y-M, Tsai C-M, Fan W-C, Shih J-F, Liu S-H, Wu C-H, et al. Phase II randomized trial of erlotinib or vinorelbine in chemo-naive, advanced, non-small cell lung cancer patients aged 70 years or older. J Thorac Oncol 2012;7:412-18.
- Oizumi S, Kobayashi K, Inoue A, Maemondo M, Sugawara S, Yoshizawa H, et al. Quality of life with gefitinib in patients with EGFR-mutated non-small cell lung cancer: quality of life analysis of North East Japan Study Group 002 Trial. Oncologist 2012;17:863-70. http://dx.doi.org/10.1634/theoncologist.2011-0426.
- Yang CH, Fukuoka M, Mok TS, Wu YL, Thongprasert S, Saijo N, et al. Final overall survival (OS) results from a phase III, randomised, open-label, first-line study of gefitinib (G) v carboplatin/paclitaxel (C/P) in clinically selected patients with advanced nonsmall cell lung cancer (NSCLC) in Asia (IPASS). Paper presented at 35th ESMO Congress; 8–12 October 2010; Milan. Ann Oncol 2010;21:viii1-2.
- Ohe Y, Ichinose Y, Nishiwaki Y, Yamamoto N, Negoro S, Duffield E, et al. Phase III, randomized, open-label, first-line study of gefitinib (G) versus carboplatin/paclitaxel (C/P) in selected patients (pts) with advanced non-small cell lung cancer (NSCLC) (IPASS): evaluation of recruits in Japan. Paper presented at 2009 Annual Meeting of the American Society of Clinical Oncology (ASCO); 29 May to 2 June 2009; Orlando, FL. J Clin Oncol 2009;27.
- Wu Y, Mok T, Chu D, Han B, Liu X, Zhang L, et al. Evaluation of clinically selected patients (pts) with advanced non-small cell lung cancer (NSCLC) recruited in China in a phase III, randomized, open-label, first-line study in Asia of gefitinib (G) versus carboplatin/paclitaxel (C/P) (IPASS). Paper presented at Annual Meeting of the American Society of Clinical Oncology (ASCO); 29 May to 2 June 2009; Orlando, FL. J Clin Oncol 2009;27.
- Fukuoka M, Wu Y, Thongprasert S, Yang C, Chu D, Saijo N, et al. Biomarker analyses from a phase III, randomized, open-label, first-line study of gefitinib (G) versus carboplatin/paclitaxel (C/P) in clinically selected patients (pts) with advanced non-small cell lung cancer (NSCLC) in Asia (IPASS). Paper presented at 2009 Annual Meeting of the American Society of Clinical Oncology (ASCO); 29 May to 2 June 2009; Orlando, FL. J Clin Oncol 2009;27.
- Mok T, Wu YL, Thongprasert S, Yang CH, Chu D, Saijo N, et al. Phase III, randomised, open-label, first-line study of gefitinib (G) vs carboplatin/paclitaxel (C/P) in clinically selected patients (PTS) with advanced non-small-cell lung cancer (NSCLC) (IPASS). Paper presented at 34th Congress of the European Society for Medical Oncology (ESMO); 12–16 September 2008; Stockholm. Ann Oncol 2008;19.
- Inoue A, Kobayashi K, Maemondo M, Sugawara S, Oizumi S, Isobe H, et al. Updated overall survival results from a randomized phase III trial comparing gefitinib with carboplatin-paclitaxel for chemo-naive non-small cell lung cancer with sensitive EGFR gene mutations (NEJ002). Ann Oncol 2013;24:54-9. http://dx.doi.org/10.1093/annonc/mds214.
- Minegishi Y, Inoue A, Kobayashi K, Gemma A, Maemondo M, Oizumi S, et al. First line gefitinib versus first line chemotherapy by carboplatin plus paclitaxel in non-small cell lung cancer patients with EGFR mutations: a phase III study (002) by North East Japan Gefitinib Study Group. EJC Suppl 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)72190-1.
- Kinoshita I, Inoue A, Kobayashi K, Maemondo M, Sugawara S, Oizumi S, et al. Phase III study of gefitinib versus chemotherapy by carboplatin (CBDCA) plus paclitaxel (TXL) as first-line therapy for non-small cell lung cancer (NSCLC) with EGFR mutations: North East Japan Gefitinib Study group trial 002 (NEJ002). Paper presented at 14th Congress of the APSR and 3rd Joint Congress of the APSR/ACCP; 14–18 November 2009; Seoul. Respirology 2009;14.
- Kobayashi K, Inoue A, Maemondo M, Sugawara S, Isobe H, Oizumi S, et al. First-line gefitinib versus first-line chemotherapy by carboplatin (CBDCA) plus paclitaxel (TXL) in non-small cell lung cancer (NSCLC) patients (pts) with EGFR mutations: a phase III study (002) by North East Japan Gefitinib Study Group. J Clin Oncol 2009;27.
- Inoue A, Kobayashi K, Maemondo M, Sugawara S, Oizumi S, Saijo Y, et al. A randomized phase III study comparing gefitinib with carboplatin (CBDCA) plus paclitaxel (TXL) for the first-line treatment of non-small cell lung cancer (NSCLC) with sensitive EGFR mutations: NEJ002 study. Paper presented at Joint ECCO 15–34th ESMO Multidisciplinary Congress; 20–24 September 2009; Berlin. Eur J Cancer Suppl 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)72038-5.
- De Marinis F, Rosell R, Vergnenegre A, Massuti B, Felip E, Gervais R, et al. Erlotinib vs chemotherapy (CT) in advanced non-small-cell lung cancer (NSCLC) patients (P) with epidermal growth factor receptor (EGFR) activating mutations: the EURTAC phase II randomized trial interim results. Paper presented at 2011 European Multidisciplinary Cancer Congress; 23–27 September 2011; Stockholm. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)72328-0.
- Zhou C, Wu YL, Chen G, Feng JF, Liu X, Wang C, et al. Updated efficacy and quality-of-life (QoL) analyses in OPTIMAL, a phase III, randomized, open-label study of first-line erlotinib versus gemcitabine/carboplatin in patients with EGFR-activating mutation-positive (EGFR Act Mut+) advanced non-small cell lung cancer (NSCLC). Paper presented at ASCO Annual Meeting; 3–7 June 2011; Chicago, IL. J Clin Oncol 2011;29.
- Wang CL, Zhou C, Wu YL, Lu S, Zhang L, Zhang Y, et al. First-line erlotinib vs gemcitabine/carboplatin (GC) in activating EGFR mutation-positive advanced non-small cell lung cancer (NSCLC): efficacy and safety results from phase III optimal study in Chinese patients (pts). Paper presented at 4th Asia Pacific Lung Cancer Conference (APLCC); 2–4 December 2010; Seoul. J Thorac Oncol 2010;5:S373-4.
- Liu X, Wu Y, Zhou C, Lu S, Zhang L, Zhi X, et al. Biomarker analyses from the phase III, randomized, openlabel, OPTIMAL study of first-line erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM) in Chinese patients (pts) with advanced non-small cell lung cancer (NSCLC) whose tumors harbor activating EGFR mutations. Paper presented at Chicago Multidisciplinary Symposium in Thoracic Oncology; 9–11 December 2010; Chicago, IL. J Thorac Oncol 2010;5:514-15.
- Wang J, Zhou C, Wu Y, Lu S, Zhang L, Zhang Y, et al. Optimal: phase III study of first-line erlotinib versus gemcitabine plus carboplatin (GC) in Chinese patients (pts) with activating EGFR mutation-positive advanced non-small cell lung cancer (NSCLC). Paper presented at Chicago Multidisciplinary Symposium in Thoracic Oncology; 9–11 December 2010; Chicago, IL. J Thorac Oncol 2010;5:507-8.
- Wu YL, Zhou C, Chen G, Feng J, Liu X, Wang C, et al. First biomarker analyses from a phase III, randomised, open-label, first-line study of erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM) in Chinese patients (PTS) with advanced non-small-cell lung cancer (NSCLC) with EGFR activating mutations (optimal, CTONG-0802). Paper presented at 35th ESMO Congress; 8–12 October 2010; Milan. Ann Oncol 2010;21.
- Zhou C, Wu YL, Chen G, Feng J, Liu X, Wang C, et al. Efficacy results from the randomised phase III optimal (CTONG 0802) study comparing first-line erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM), in Chinese advanced non-small-cell lung cancer (NSCLC) patients (PTS) with EGFR activating mutations. Paper presented at 35th ESMO Congress; 8–12 October 2010; Milan. Ann Oncol 2010;21.
- Zhou C, Wu Y, Chen G, Feng J, Liu X, Wang C, et al. Preliminary results of randomized phase III study comparing efficacy and safety of first-line erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM) in Chinese advanced non-small cell lung cancer (NSCLC) patients (pts) with EGFR-activating mutations (OPTIMAL). Paper presented at Annual Meeting of the American Society of Clinical Oncology (ASCO); 4–8 June 2010; Chicago, IL. J Clin Oncol 2010;28.
- Teck LW, Huat TB, Ravindran K, Balram C. EGFR mutational status and clinical outcome to gefitinib monotherapy in chemo-naive non-small-cell lung cancer patients. Pharmacogenomics 2008;9.
- First-Line Treatment for Adenocarcinoma Patients with Epidermal Growth Factor Receptor (EGFR) Mutation. NCT00344773. Bethesda, MD: National Library of Medicine; 2010.
- A Chinese Randomized Crossover Study of Erlotinib Versus Docetaxel/Cisplatin in Previously Untreated Stage IIIB/IV Lung Adenocarcinoma with EGFR Mutations. NCT01131429. Bethesda, MD: National Library of Medicine; 2010.
- Ahn M-J, Park B-B, Ahn JS, Kim SW, Kim H-T, Lee JS, et al. Are there any ethnic differences in molecular predictors of erlotinib efficacy in advanced non-small cell lung cancer?. Clin Cancer Res 2008;14:3860-6. http://dx.doi.org/10.1158/1078-0432.CCR-07-4608.
- Aydiner A. Efficacy of gefitinib in patients with epidermal growth factor receptor mutation positive advanced non-small-cell lung cancer: a meta-analysis of randomized controlled trials. Paper presented at 2011 European Multidisciplinary Cancer Congress; 23–27 September 2011; Stockholm. Eur J Cancer 2011;47.
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005;97:643-55. http://dx.doi.org/10.1093/jnci/dji112.
- Carlson JJ, Garrison LP, Ramsey SD, Veenstra DL. Epidermal growth factor receptor genomic variation in NSCLC patients receiving tyrosine kinase inhibitor therapy: a systematic review and meta-analysis. J Cancer Res Clin Oncol 2009;135:1483-93. http://dx.doi.org/10.1007/s00432-009-0595-3.
- Chang C-H, Chen K-Y, Young-Xu Y, Kurth T, Orav EJ, Yang P-C, et al. The safety and efficacy of gefitinib versus platinum-based doublets chemotherapy as the first-line treatment for advanced non-small-cell lung cancer patients in East Asia: a meta-analysis. Lung Cancer 2008;62:242-52. http://dx.doi.org/10.1016/j.lungcan.2008.03.001.
- Chen P, Wang L, Liu B, Zhang HZ, Liu HC, Zou Z. EGFR-targeted therapies combined with chemotherapy for treating advanced non-small-cell lung cancer: a meta-analysis. Eur J Clin Pharmacol 2011;67:235-43. http://dx.doi.org/10.1007/s00228-010-0965-4.
- Chou T-Y, Chiu C-H, Li L-H, Hsiao C-Y, Tzen C-Y, Chang K-T, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res 2005;11:3750-7. http://dx.doi.org/10.1158/1078-0432.CCR-04-1981.
- Chung KP, Wu SG, Wu JY, Yang JH, Yu CJ, Wei PF, et al. Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res 2012;18:3470-7. http://dx.doi.org/10.1158/1078-0432.CCR-11-2353.
- Cohen V, Agulnik JS, Ang C, Kasymjanova G, Batist G, Small D, et al. Epidermal growth factor receptor mutations detected by denaturing high-performance liquid chromatography in nonsmall cell lung cancer: impact on response to therapy with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer 2010;116:4309-17. http://dx.doi.org/10.1002/cncr.25214.
- Cohen V, Agulnik JS, Jarry J, Batist G, Small D, Kreisman H, et al. Evaluation of denaturing high-performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in nonsmall-cell lung cancer. Cancer 2006;107:2858-65. http://dx.doi.org/10.1002/cncr.22331.
- Cohen VA, Jarry JS, Batist J, Small G, Kreisman D, Tejada H, et al. Evaluation of denaturing high-performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in non-small-cell lung cancer. Cancer 2006;107:2858-65.
- Crosby C, Smye-Rumsby T, Plummeridge M, Braybrooke J, Dangoor A, De Winton E, et al. EGFR mutation testing in non-small cell lung cancer: the Bristol experience. J Med Genet 2011;48.
- Dahabreh IJ, Linardou H, Siannis F, Kosmidis P, Bafaloukos D, Murray S. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2010;16:291-303. http://dx.doi.org/10.1158/1078-0432.CCR-09-1660.
- de Braud F, Noberasco C, Curigliano G, De Pas M, Dodaro L, Manzoni S, et al. Correlation between epidermal growth factor receptor expression and tumour response in patients with advanced non-small-cell lung cancer treated with gefitinib (‘Iressa’, ZD1839). Br J Cancer 2003;89.
- De Greve J, Van Meerbeeck JP, Vansteenkiste JF, Teugels E, Geers C, Meert A, et al. First-line erlotinib in advanced non-small cell lung cancer (NSCLC) carrying an activating EGFR mutation: a multicenter academic phase II study in Caucasian patients (pts) (NCT00339586)-FIELT study group. Paper presented at ASCO Annual Meeting 2011; 3–7 June 2011; Chicago, IL. J Clin Oncol 2011;29.
- De Pas T, Toffalorio F, Manzotti M, Fumagalli C, Spitaleri G, Catania C, et al. Activity of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with non-small cell lung cancer harboring rare epidermal growth factor receptor mutations. J Thorac Oncol 2011;6:1895-901. http://dx.doi.org/10.1097/JTO.0b013e318227e8c6.
- Dickson R, Bagust A, Boland A, Blundell M, Davis H, Dundar Y, et al. Erlotinib monotherapy for the maintenance treatment of non-small cell lung cancer after previous platinum-containing chemotherapy: NICE single technology appraisal. Pharmacoeconomics 2011;29:1051-62. http://dx.doi.org/10.2165/11591600-000000000-00000.
- Eaton H, Dutton L, Wallace AJ, Howard E, Gaunt L, Blackhall F, et al. Epidermal Growth Factor Receptor (EGFR) mutation testing in non-small cell lung cancer (NSCLC) patients. J Med Genet 2011;48.
- Eberhardt W, Thomas M, von der Schulenburg JMG, Dietel M, Schirmacher P, Gutendorf B, et al. EGFR mutation testing and first line treatment of patients with advanced NSCLC and positive EGFR mutation status: results from a German registry. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)72456-X.
- Edwards SJ, Welton N, Borrill J. Tolerability of first-line treatments of locally advanced or metastatic non-small-cell lung cancer (NSCLC): a systematic review and adjusted indirect comparison. Paper presented at ISPOR 13th Annual European Congress; 6–9 November 2011; Prague. Value Health 2010;13. http://dx.doi.org/10.1016/S1098-3015(11)71898-8.
- Edwards SJ, Welton N, Borrill J. Gefitinib compared with doublet chemotherapy for first-line treatment of non-small-cell lung cancer (NSCLC): a systematic review and adjusted indirect comparison. Paper presented at ISPOR 13th Annual European Congress; 6–9 November 2010; Prague. Value Health 2010;13:A252-3. http://dx.doi.org/10.1016/S1098-3015(11)71908-8.
- Enting D, Januszewski A, Benepal T. EGFR-mutation analysis, treatment and outcomes in newly diagnosed patients with non-small cell lung cancer (NSCLC): a single-institution retrospective review. Lung Cancer 2012;75. http://dx.doi.org/10.1016/S0169-5002(12)70014-4.
- Feld R, Sridhar SS, Shepherd FA, Mackay JA, Evans WK. Use of the epidermal growth factor receptor inhibitors gefitinib and erlotinib in the treatment of non-small cell lung cancer: a systematic review. J Thorac Oncol 2006;1:367-76. http://dx.doi.org/10.1097/01243894-200605000-00018.
- Feng JF, Wu YL, Zhou CC, Lu S, Zhang L, Zhi XY, et al. Additional biomarker analyses from the phase III OPTIMAL study of first-line erlotinib versus gemcitabine/carboplatin (GC) in Chinese patients (pts) with advanced activating EGFR mutation-positive non-small cell lung cancer (NSCLC). J Thorac Oncol 2010;5.
- Gao H, Ding X, Wei D, Cheng P, Su X, Liu H, et al. Efficacy of erlotinib in patients with advanced non-small cell lung cancer: a pooled analysis of randomized trials. Anticancer Drugs 2011;22:842-52. http://dx.doi.org/10.1097/CAD.0b013e328349c303.
- Goss G, Ferry D, Wierzbicki R, Laurie SA, Thompson J, Biesma B, et al. Randomized phase II study of gefitinib compared with placebo in chemotherapy-naive patients with advanced non-small-cell lung cancer and poor performance status. J Clin Oncol 2009;27:2253-60. http://dx.doi.org/10.1200/JCO.2008.18.4408.
- Gracia M, Majem M, Aguillo MTM, Banaclocha NM, Provencio M, Arriola E, et al. Effect of sample type and turnaround time (TAT) on the feasibility of non-small cell lung cancer (NSCLC) epidermal growth factor receptor (EGFR) mutation testing in routine clinical practice: results from the Spanish REASON study. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)71185-6.
- Gupta R, Dastane AM, McKenna R, Marchevsky AM. The predictive value of epidermal growth factor receptor tests in patients with pulmonary adenocarcinoma: review of current ‘best evidence’ with meta-analysis. Hum Pathol 2009;40:356-65.
- Han SW, Jeong S, Choi IS, Kim DN, Chung DH, Im SA, et al. EGFR and K-ras mutations as determinants of gefitinib sensitivity in non-small-cell lung cancer (NSCLC). J Clin Oncol 2005;23.
- Han S-W, Kim T-Y, Hwang PG, Jeong S, Kim J, Choi IS, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol 2005;23:2493-501. http://dx.doi.org/10.1200/JCO.2005.01.388.
- Han S-W, Kim T-Y, Lee K-H, Hwang PG, Jeon YK, Oh D-Y, et al. Clinical predictors versus epidermal growth factor receptor mutation in gefitinib-treated non-small-cell lung cancer patients. Lung Cancer 2006;54:201-7. http://dx.doi.org/10.1016/j.lungcan.2006.07.007.
- Han Y, Xu J-m, Duan H-q, Song S-t, Liu X-q, Zhang Y, et al. EGFR mutation predicts response and prognosis in IRESSA-treated advanced-stage non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi 2007;29:278-83.
- Hata A, Katakami N, Kunimasa K, Yoshioka H, Fujita S, Kaji R, et al. How sensitive are epidermal growth factor receptor tyrosine kinase inhibitors for squamous cell carcinoma of the lung harboring epidermal growth factor receptor gene-sensitive mutations? Paper presented at ASCO Annual Meeting 2011; 3–7 June 2011; Chicago, IL. J Clin Oncol 2011;29.
- Hou M-M, Huang S-F, Kuo H-P, Yang C-T, Tsai Y-H, Yu C-T, et al. Erlotinib treatment in patients with advanced lung adenocarcinoma with CISH-positive and CISH-negative EGFR gene alterations. Anticancer Res 2012;32:1107-12.
- Hsieh M-H, Fang Y-F, Chang W-C, Kuo H-P, Lin S-Y, Liu H-P, et al. Complex mutation patterns of epidermal growth factor receptor gene associated with variable responses to gefitinib treatment in patients with non-small cell lung cancer. Lung Cancer 2006;53:311-22. http://dx.doi.org/10.1016/j.lungcan.2006.06.005.
- Ibrahim EM. Frontline gefitinib in advanced non-small cell lung cancer: meta-analysis of published randomized trials. Ann Thorac Med 2010;5:153-60. http://dx.doi.org/10.4103/1817-1737.65047.
- Inoue A, Kobayashi K, Usui K, Maemondo M, Ando M, Gemma A, et al. Gefitinib provides an beneficial treatment to poor PS or super-elderly patients with EGFR mutation-positive NSCLC (North-East Japan Gefitinib Study Group). Ann Oncol 2008;19.
- Inoue A, Minegishi Y, Maemondo M, Okinaga S, Morikawa N, Kobayashi K, et al. A final results of a phase II study of first-line gefitinib for elderly patients (PTS) with advanced non-small cell lung cancer (NSCLC) harboring epidermal growth factor receptor (EGFR) mutations; NEJ 003 study. Paper presented at 35th ESMO Congress; 8–12 October 2010; Milan. Ann Oncol 2010;21:141-2.
- Jackman DM, Miller VA, Cioffredi L-A, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res 2009;15:5267-73. http://dx.doi.org/10.1158/1078-0432.CCR-09-0888.
- Johnson BE, Lucca J, Rabin MS, Lynch TJ, Ostler P, Skarin AT, et al. Preliminary results from a phase II study of the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in patients > 70 years of age with previously untreated advanced non-small cell lung carcinoma. J Clin Oncol 2004;22.
- Kasahara K, Sone T, Kimura H, Nishio K, Tamura T, Shibata K, et al. Correlation between epidermal growth factor receptor gene status and clinical outcome of gefitinib in Japanese patients with non-small cell lung cancer. J Clin Oncol 2006;24.
- Kashii T, Okamoto I, Urata Y, Hirashima T, Kudoh S, Ichinose Y, et al. EGFR mutation-based phase II multicenter trial of gefitinib in advanced non-small cell lung cancer (NSCLC) patients (PTS): results of West Japan Thoracic Oncology Group Trial (WJTOG0403). Ann Oncol 2006;17.
- Kim YH, Masago K, Togashi Y, Sakamori Y, Okuda C, Mio T, et al. EGFR mutation: significance as a stratification factor in the era of molecular-targeted therapy. Oncol Lett 2011;2:383-7. http://dx.doi.org/10.3892/ol.2011.240.
- Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006;12:3915-21. http://dx.doi.org/10.1158/1078-0432.CCR-05-2324.
- Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. http://dx.doi.org/10.1038/sj.bjc.6603949.
- Kris M, Mok T, Kim E, Douillard JY, Fukuoka M, Thatcher N. Response and progression-free survival in 1006 patients with known EGFR mutation status in phase III randomized trials of gefitinib in individuals with non-small cell lung cancer. Paper presented at Joint ECCO 15–34th ESMO Multidisciplinary Congress; 20–24 September 2009; Berlin. Eur J Cancer Suppl 2009;7:505-6. http://dx.doi.org/10.1016/S1359-6349(09)71716-1.
- Ku GY, Haaland BA, de Lima Lopes G. Gefitinib vs. chemotherapy as first-line therapy in advanced non-small cell lung cancer: meta-analysis of phase III trials. Lung Cancer 2011;74:469-73. http://dx.doi.org/10.1016/j.lungcan.2011.04.008.
- Kunimasa K, Katakami N, Masago K, Yoshioka H, Tomii K, Kaneda T, et al. Customized chemotherapy on the basis of EGFR mutation status for elderly patients with advanced non-small-cell lung cancer. Paper presented at European Multidisciplinary Cancer Congress; 23–27 September 2011; Stockholm: Sweden. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)71267-9.
- Lee DH, Kim S-W, Suh C, Han YH, Lee J-S. Phase II study of erlotinib for chemotherapy-naive patients with advanced or metastatic non-small cell lung cancer who are ineligible for platinum doublets. Cancer Chemother Pharmacol 2011;67:35-9. http://dx.doi.org/10.1007/s00280-010-1280-6.
- Lee DH, Song HJ, Lee YM, Choi YH, Kim SW, Suh C, et al. Prospective study of erlotinib comparing chemotherapy-naive non-small cell lung cancer patients having an activating mutation in EGFR gene with those having wild-type EGFR gene. EJC Suppl 2008;6. http://dx.doi.org/10.1016/S1359-6349(08)72147-5.
- Lilenbaum R, Axelrod R, Thomas S, Dowlati A, Seigel L, Albert D, et al. Randomized phase II trial of erlotinib or standard chemotherapy in patients with advanced non-small-cell lung cancer and a performance status of 2. J Clin Oncol 2008;26:863-9. http://dx.doi.org/10.1200/JCO.2007.13.2720.
- Liu S, Wang D, Chen B, Wang Y, Zhao W, Wu J. The safety and efficacy of EGFR TKIs monotherapy versus single-agent chemotherapy using third-generation cytotoxics as the first-line treatment for patients with advanced non-small cell lung cancer and poor performance status. Lung Cancer 2011;73:203-10. http://dx.doi.org/10.1016/j.lungcan.2010.12.006.
- Massuti B, Moran T, Porta R, Queralt C, Cardenal F, Mayo C, et al. Multicenter prospective trial of customized erlotinib for advanced non-small cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) mutations: final results of the Spanish Lung Cancer Group (SLCG) trial. Paper presented at Annual Meeting of the American Society of Clinical Oncology (ASCO); 2 May – 2 June 2009; Orlando, FL. J Clin Oncol 2009;27.
- Massuti B, Porta R, Paz-Ares L, Cardenal F, Lopez-Vivanco G, Provencio M, et al. First-line erlotinib in stage IV non-small cell lung cancer (NSCLC) patients (pts) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR). Lung Cancer 2006;52:S25-6.
- Massuti B, Reguart N, Vivanco GL, Provencio M, Isla D, Camps C, et al. First-line erlotinib in stage IV non-small-cell lung cancer (NSCLC) patients (P) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR). Ann Oncol 2006;17.
- Meert AP, Martin B, Delmotte P, Berghmans T, Lafitte JJ, Mascaux C, et al. The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J 2002;20:975-81. http://dx.doi.org/10.1183/09031936.02.00296502.
- Miller VA, Zakowski M, Riely GJ, Pao W, Hussain S, Ladanyi M, et al. EGFR mutation, immumohistochemistry (IHC) and chromogenic in situ hybridization (CISH) as predictors of sensitivity to erlotinib and gefitinib in patients (pts) with NSCLC. J Clin Oncol 2005;23.
- Minegishi Y, Maemondo M, Okinaga S, Morikawa N, Inoue A, Kobayashi K, et al. First-line gefitinib therapy for elder advanced non-small cell lung cancer patients with epidermal growth factor receptor mutations: multicenter phase II trial (NEJ 003 study). Paper presented at Annual Meeting of the American Society of Clinical Oncology (ASCO); 4–8 June 2010; Chicago, IL. J Clin Oncol 2010;28.
- Morita S, Okamoto I, Kobayashi K, Yamazaki K, Asahina H, Inoue A, et al. Combined survival analysis of prospective clinical trials of gefitinib for non-small cell lung cancer with EGFR mutations. Clin Cancer Res 2009;15:4493-8. http://dx.doi.org/10.1158/1078-0432.CCR-09-0391.
- Murray S, Evangelou E, Linardou H, Dahabreh IJ, Kosmidis P, Bafaloukos D, et al. Predictive significance of EGFR somatic mutations and gene copy number in NSCLC patients treated with single agent tyrosine kinase inhibitors: a systematic review and meta-analysis. Paper presented at 2nd European Lung Cancer Conference; 28 April to 1 May 2010; Geneva. J Thorac Oncol 2010;5:55-6.
- Murray S, Evangelou E, Linardou H, Kosmidis P, Bafaloukos D, Ioannidis J. Predictive and prognostic significance of somatic mutations in the tyrosine kinase (TK) domain of EGFR in NSCLC patients treated with single agent gefitinib: a systematic review and meta-analysis. Paper presented at 34th Congress of the European Society for Medical Oncology (ESMO); 23–16 September 2008, Stockholm. Ann Oncol 2008;19.
- Na II, Rho JK, Choi YJ, Kim CH, Koh JS, Ryoo B-Y, et al. Clinical features reflect exon sites of EGFR mutations in patients with resected non-small-cell lung cancer. J Korean Med Sci 2007;22:393-9. http://dx.doi.org/10.3346/jkms.2007.22.3.393.
- Naoki K, Okamoto H, Soejima K, Nakachi I, Fujii T, Hida N, et al. New sensitive methods to detect EGFR gene mutations in lung cancer specimens, structure-specific flap endonuclease based method; comparison with direct sequencing. Paper presented at 34th Congress of the European Society for Medical Oncology (ESMO); 23–16 September 2008; Stockholm. Ann Oncol 2008;19.
- Naoki K, Soejima K, Okamoto H, Hamamoto J, Hida N, Nakachi I, et al. The PCR-invader method (structure-specific 5′ nuclease-based method), a sensitive method for detecting EGFR gene mutations in lung cancer specimens; comparison with direct sequencing. Int J Clin Oncol 2011;16:335-44.
- Okamoto I, Kashii T, Urata Y, Hirashima T, Kudoh S, Ichinose Y, et al. EGFR mutation-based phase II multicenter trial of gefitinib in advanced non-small cell lung cancer (NSCLC) patients (pts): results of West Japan Thoracic Oncology Group trial (WJTOG0403). J Clin Oncol 2006;24.
- Pallis AG, Voutsina A, Kalikaki A, Souglakos J, Briasoulis E, Murray S, et al. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer 2007;97:1560-6. http://dx.doi.org/10.1038/sj.bjc.6604068.
- Park SH, Ha SY, Lee J-I, Lee H, Sim H, Kim YS, et al. Epidermal growth factor receptor mutations and the clinical outcome in male smokers with squamous cell carcinoma of lung. J Korean Med Sci 2009;24:448-52. http://dx.doi.org/10.3346/jkms.2009.24.3.448.
- Paz-Ares L, Soulieres D, Klughammer B, Melezinek I, Moecks J, Keil L, et al. Clinical outcomes in patients with EGFR mutations: pooled analysis of NSCLC patients treated with either an EGFR TKI or chemotherapy. Paper presented at Joint ECCO 15–34th ESMO Multidisciplinary Congress; 20–24 September 2009; Berlin. Eur J Cancer Suppl 2009;7:551-2.
- Paz-Ares L, Soulieres D, Melezinek I, Moecks J, Keil L, Mok T, et al. Clinical outcomes in non-small-cell lung cancer patients with EGFR mutations: pooled analysis. J Cell Mol Med 2010;14:51-69. http://dx.doi.org/10.1111/j.1582-4934.2009.00991.x.
- Paz-Ares L, Sanchez JM, Garcia-Velasco A, Massuti B, Lopez-Vivanco G, Provencio M, et al. A prospective phase II trial of erlotinib in advanced non-small cell lung cancer (NSCLC) patients (p) with mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR). J Clin Oncol 2006;24.
- Pesek M, Benesova L, Belsanova B, Mukensnabl P, Bruha F, Minarik M. Dominance of EGFR and insignificant KRAS mutations in prediction of tyrosine-kinase therapy for NSCLC patients stratified by tumor subtype and smoking status. Anticancer Res 2009;29:2767-73.
- Petrelli F, Borgonovo K, Cabiddu M, Barni S. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small-cell lung cancer: a meta-analysis of 13 randomized trials. Clin Lung Cancer 2012;13:107-14. http://dx.doi.org/10.1016/j.cllc.2011.08.005.
- Petruzelka L. Erlotinib as first-line treatment of European non-small-cell lung cancer (NSCLC) patients (pts.) with epidermal growth factor receptor (EGFR) activating mutations (mut plus). Lung Cancer 2012;77. http://dx.doi.org/10.1016/j.lungcan.2012.05.042.
- Plant R, Chetiyawardana S, Thompson J, Williams S, Ghafoor Q. Use of Gefitinib (Iressa) in patients with EGFR positive NSCLC in the Pan Birmingham Cancer Network. Paper presented at 10th Annual British Thoracic Oncology Group Conference (BTOG); 25–27 January 2012; Dublin. Lung Cancer 2012;75. http://dx.doi.org/10.1016/S0169-5002(12)70018-1.
- Reck M, Heller A, Foernzler D, Moecks J, Ward C, De Rosa F, et al. Molecular markers such as EGFR and kRAS mutations as predictors of sensitivity to erlotinib in patients (pts) with NSCLC: exploratory subanalyses of TALENT, a phase III trial. Lung Cancer 2005;49.
- Ricciardi S, Tedesco B, Migliorino MR, Graziano P, Leone A, Di Salvia M, et al. EGFR mutations in patients with non-small cell lung cancer (NSCLC) and correlation with sensitivity to erlotinib. EJC Suppl 2008;6. http://dx.doi.org/10.1016/j.ejcsup.2008.06.065.
- Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:839-44. http://dx.doi.org/10.1158/1078-0432.CCR-05-1846.
- Rizvi N, Pao W, Miller V, Rusch V, Heelan R, Ladanyi M, et al. A prospective study to correlate EGFR mutations with gefitinib response in early stage NSCLC. Lung Cancer 2005;49. http://dx.doi.org/10.1016/S0169-5002(05)80257-0.
- Satouchi M, Mitsudomi T, Ebi N, Saka H, Sawa Y, Sasaki J, et al. A phase III trial of gefitinib versus cisplatin/docetaxel for NSCLC patients with activating EGFR mutation; WJTOG 3405. Ann Oncol 2010;21.
- Schneider C-P, Heigener D, Schott-von-Romer K, Gutz S, Laack E, Digel W, et al. Epidermal growth factor receptor-related tumor markers and clinical outcomes with erlotinib in non-small cell lung cancer: an analysis of patients from German centers in the TRUST study. J Thorac Oncol 2008;3:1446-53. http://dx.doi.org/10.1097/JTO.0b013e31818ddcaa.
- Shih J-Y, Gow C-H, Yu C-J, Yang C-H, Chang Y-L, Tsai M-F, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer 2006;118:963-9. http://dx.doi.org/10.1002/ijc.21458.
- Shukuya T, Takahashi T, Ono A, Nakamura Y, Tsuya A, Kenmotsu H, et al. The efficacy of gefitinib for non-adeno non-small-cell lung cancer patients harboring EGFR mutations: a pooled analysis. J Thorac Oncol 2010;5.
- Sone T, Kasahara K, Kimura H, Nishio K, Mizuguchi M, Nakatsumi Y, et al. Comparative analysis of epidermal growth factor receptor mutations and gene amplification as predictors of gefitinib efficacy in Japanese patients with non-small cell lung cancer. Cancer 2007;109:1836-44.
- Sun JM, Won YW, Kim ST, Kim JH, Choi YL, Lee J, et al. The different efficacy of gefitinib or erlotinib according to epidermal growth factor receptor exon 19 and exon 21 mutations in Korean non-small cell lung cancer patients. J Cancer Res Clin Oncol 2011;137:687-94. http://dx.doi.org/10.1007/s00432-010-0928-2.
- Sunaga N, Yanagitani N, Kaira K, Tomizawa Y, Iijima H, Otani Y, et al. Phase II study of the efficacy of gefitinib in patients with non-small cell lung cancer with the EGFR mutations. J Clin Oncol 2006;24.
- Sutani A, Nagai Y, Udagawa K, Uchida Y, Murayama Y, Tanaka T, et al. Phase II study of gefitinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) gene mutations detected by PNA-LNA PCR clamp. J Clin Oncol 2006;24.
- Takano T, Ohe Y, Furuta K, Tsuta K, Nomoto K, Matsuno Y, et al. EGFR mutations detected by high-resolution melting analysis (HRMA) as a predictor of response and survival in non-small cell lung cancer (NSCLC) patients treated with gefitinib. J Clin Oncol 2006;24.
- Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol 2005;23:6829-37. http://dx.doi.org/10.1200/JCO.2005.01.0793.
- Takano T, Ohe Y, Tsuta K, Fukui T, Sakamoto H, Yoshida T, et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res 2007;13:5385-90. http://dx.doi.org/10.1158/1078-0432.CCR-07-0627.
- Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res 2005;11:1167-73.
- Tsai CM, Chiu CH, Chou TY, Li LH, Hsiao CY, Chen M, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. J Clin Oncol 2005;23.
- Tsurutani J, Mitsudomi T, Mori S, Okamoto I, Kaname N, Tada H, et al. A phase III, first-line trial of gefitinib versus cisplatin plusdocetaxel for patients with advanced or recurrent non-small cell lung cancer (NSCLC) harboring activating mutation of the epidermal growth factor receptor (EGFR) gene: a preliminary results of WJTOG 3405. Paper presented at Joint ECCO 15–34th ESMO Multidisciplinary Congress; 20–24 September 2009; Berlin. Eur J Cancer Suppl 2009;7. http://dx.doi.org/10.1016/S1359-6349(09)71715-X.
- Tyagi P. Updated data from the Iressa survival in lung cancer trial. Clin Lung Cancer 2005;6:340-2. http://dx.doi.org/10.1016/S1525-7304(11)70364-3.
- van Zandwijk N, Mathy A, Boerrigter L, Ruijter H, Tielen I, de Jong D, et al. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: Retro- and prospective observations in non-small-cell lung cancer. Ann Oncol 2007;18:99-103. http://dx.doi.org/10.1093/annonc/mdl323.
- Villaflor VM, Buckingham L, Gale M, Coon J, Mauer AM, Muzzafar T, et al. EGFR mutations and pAKT expression as potential predictors of gefitinib efficacy in non-small cell lung cancer (NSCLC) patients (pts). J Clin Oncol 2005;23.
- Wang T, Ma L. Pooled analysis of the trials of erlotinib monotherapy for epidermal growth factor receptor (EGFR)-mutant advanced non-small cell lung cancer. Zhongguo Fei Ai Za Zhi 2009;12:1237-41.
- Webb S, Thomas M, Metcalf C, Segal A, Nowak AK, Bentel J, et al. EGFR mutation testing in NSCLC: patterns of care and outcomes in Western Australia. Asia Pac J Clin Oncol 2009;5:66-71. http://dx.doi.org/10.1111/j.1743-7563.2009.01192.x.
- Won Y-W, Han J-Y, Lee GK, Park S-Y, Lim KY, Yoon K-A, et al. Comparison of clinical outcome of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations. J Clin Pathol 2011;64:947-52. http://dx.doi.org/10.1136/jclinpath-2011-200169.
- Wu J-Y, Shih J-Y, Chen K-Y, Yang C-H, Yu C-J, Yang P-C. Gefitinib therapy in patients with advanced non-small cell lung cancer with or without testing for epidermal growth factor receptor (EGFR) mutations. Medicine 2011;90:159-67. http://dx.doi.org/10.1097/MD.0b013e31821a16f4.
- Wu J-Y, Wu S-G, Yang C-H, Chang Y-L, Chang Y-C, Hsu Y-C, et al. Comparison of gefitinib and erlotinib in advanced NSCLC and the effect of EGFR mutations. Lung Cancer 2011;72:205-12. http://dx.doi.org/10.1016/j.lungcan.2010.08.013.
- Wu J-Y, Yu C-J, Chang Y-C, Yang C-H, Shih J-Y, Yang P-C. Effectiveness of tyrosine kinase inhibitors on ‘uncommon’ epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 2011;17:3812-21. http://dx.doi.org/10.1158/1078-0432.CCR-10-3408.
- Wu J-Y, Yu C-J, Yang C-H, Wu S-G, Chiu Y-H, Gow C-H, et al. First- or second-line therapy with gefitinib produces equal survival in non-small cell lung cancer. Am J Respir Crit Care Med 2008;178:847-53. http://dx.doi.org/10.1164/rccm.200803-389OC.
- Wu Y-L, Zhong W-Z, Li L-Y, Zhang X-T, Zhang L, Zhou C-C, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: a meta-analysis based on updated individual patient data from six medical centers in mainland China. J Thorac Oncol 2007;2:430-9. http://dx.doi.org/10.1097/01.JTO.0000268677.87496.4c.
- Wu Y, Zhong W, Li L, Zhang L, Zhou C, Liu W, et al. EGFR mutations and the correlation with gefitinib therapy in Chinese NSCLC: a systematic review based on individual patient data from 5 medical centers in China. J Clin Oncol 2006;24.
- Xu C, Zhou Q, Wu YL. Meta-analysis of the chemotherapy versus EGFR-TKI in different selections of patients. Paper presented at ASCO Annual Meeting; 3–7 June 2011; Chicago, IL. J Clin Oncol 2011;29.
- Yang J, Wu YL, Saijo N, Thongprasert S, Chu DT, Chen YM, et al. Efficacy outcomes in first-line treatment of advanced NSCLC with gefitinib (G) vs carboplatin/paclitaxel (C/P) by epidermal growth factor receptor (EGFR) gene-copy number score and by most common EGFR mutation subtypes. Exploratory data from IPASS. Paper presented at 2011 European Multidisciplinary Cancer Congress; 23–27 September 2011; Stockholm. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)72444-3.
- Yoshida K, Okamoto I, Kobayashi K, Sunaga N, Sugio K, Inoue A, et al. Comparison of the efficacy between chemotherapy and gefitinib as 1st line setting in patients with EGFR mutation positive NSCLC. Paper presented at 34th Congress of the European Society for Medical Oncology (ESMO); 12–16 September 2008; Stockholm: Sweden. Ann Oncol 2008;19.
- Yoshida K, Yatabe Y, Park J, Ogawa S, Park JY, Shimizu J, et al. Clinical outcomes of advanced non-small cell lung cancer patients screened for epidermal growth factor receptor gene mutations. J Cancer Res Clin Oncol 2010;136:527-35. http://dx.doi.org/10.1007/s00432-009-0685-2.
- Zhang XH, Li C, Dai CF, Zhou BS. Epidermal growth factor receptor mutations and their correlation with epidermal growth factor receptor-tyrosine kinase inhibitor therapy and association with the characteristics of patients with non-small-cell lung cancer: a meta-analysis. Thoracic Cancer 2011;2:101-8. http://dx.doi.org/10.1111/j.1759-7714.2011.00051.x.
- Zhang X, Xu L, Wang H, Zhu Y, Liu Z, Yue W, et al. The relationship between EGFR mutations and response and prognosis of tyrosine kinase inhibitors in advanced non-small-cell lung cancer. Zhongguo Fei Ai Za Zhi 2008;11:206-13.
- Zhong W, Xia Y, Wang MZ, Chen MJ, Zhang L, Zhao J, et al. EGFR gene mutations in advanced non-small cell lung cancer and its influence on effect of gefitinib. Respirology 2011;16.
Appendix 1 Literature search strategies
Clinical effectiveness search strategies
EMBASE (OvidSP): 2000 to 2012 week 28
Searched 18 July 2012.
-
erlotinib/ or (Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (11,968)
-
gefitinib/ or (Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (13,035)
-
or/1-2 (18,405)
-
lung non small cell cancer/ (45,170)
-
(nsclc or nsclcs).ti,ab,ot,hw. (22,339)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (9347)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (35,098)
-
(lclc or lclcs).ti,ab,ot,hw. (56)
-
or/4-8 (59,672)
-
Receptor, Epidermal Growth Factor/ (34,579)
-
(epidermal growth factor receptor$ or epidermis growth factor receptor$ or transforming growth factor alpha receptor$).ti,ab,ot. (24,964)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (183)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (1421)
-
(EGFR or EGFRTK).ti,ab,ot. (30,350)
-
EGF receptor$.ti,ab,ot. (8985)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (46)
-
or/10-18 (56,110)
-
3 and 9 and 19 (4768)
-
lung non small cell cancer/di [Diagnosis] (5261)
-
diagnostic test/ (53,292)
-
diagnosis/ (875,184)
-
differential diagnosis/ (295,658)
-
laboratory diagnosis/ (40,591)
-
laboratory test/ (100,888)
-
diagnos$.ti,ab,ot. (1,925,228)
-
(test or tests or testing or tested).ti,ab,ot. (2,207,310)
-
((lab or labs or laborator$) adj2 (procedure$ or exam$)).ti,ab,ot. (15,288)
-
or/21-29 (4,581,699)
-
9 and 19 and 30 (2035)
-
animal/ or animal experiment/ (3,398,728)
-
(rat or rats or mouse or mice or murine or rodent or rodents or hamster or hamsters or pig or pigs or porcine or rabbit or rabbits or animal or animals or dogs or dog or cats or cow or bovine or sheep or ovine or monkey or monkeys).mp. (5,489,895)
-
or/32-33 (5,489,895)
-
exp human/ or human experiment/ (13,717,180)
-
34 not (34 and 35) (4,418,831)
-
20 or 31 (5626)
-
37 not 36 (5547)
-
limit 38 to yr=“2000 -Current” (5500)
-
limit 39 to embase (4910)
-
remove duplicates from 40 (4897)
MEDLINE (OvidSP): 2000 to July 2012 week 1
Searched 18 July 2012.
-
Quinazolines/ (11,462)
-
(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (2563)
-
(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (3588)
-
or/1-3 (12,590)
-
Carcinoma, Non-Small-Cell Lung/ (26,828)
-
(nsclc or nsclcs).ti,ab,ot,hw. (14,105)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (6982)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (24,330)
-
(lclc or lclcs).ti,ab,ot,hw. (42)
-
or/5-9 (37,809)
-
Receptor, Epidermal Growth Factor/ (25,521)
-
epidermal growth factor receptor$.ti,ab,ot. (20,251)
-
epidermis growth factor receptor$.ti,ab,ot. (0)
-
transforming growth factor alpha receptor$.ti,ab,ot. (10)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (243)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (1223)
-
EGFR.ti,ab,ot. (19,756)
-
EGFRTK.ti,ab,ot. (10)
-
EGF receptor$.ti,ab,ot. (8278)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (13)
-
or/11-22 (38,939)
-
4 and 10 and 23 (2059)
-
Carcinoma, Non-Small-Cell Lung/di [Diagnosis] (1966)
-
“diagnostic techniques and procedures”/ or diagnostic tests, routine/ (7996)
-
clinical laboratory techniques/ or molecular diagnostic techniques/ (19,825)
-
Diagnosis/ (16,321)
-
Diagnosis, Differential/ (355,501)
-
diagnos$.ti,ab,ot. (1,397,222)
-
(test or tests or testing or tested).ti,ab,ot. (1,683,480)
-
((lab or labs or laborator$) adj2 (procedure$ or exam$)).ti,ab,ot. (10,488)
-
or/25-32 (3,069,443)
-
10 and 23 and 33 (887)
-
24 or 34 (2529)
-
animals/ not (animals/ and humans/) (3,660,877)
-
35 not 36 (2499)
-
limit 37 to yr=“2000 -Current” (2463)
-
remove duplicates from 38 (2318)
MEDLINE In-Process Citations (OvidSP): 2000 to 17 July 2012
MEDLINE Daily Update (OvidSP): 2000 to 17 July 2012
Searched 18 July 2012.
-
Quinazolines/ (31)
-
(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (278)
-
(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (233)
-
or/1-3 (432)
-
Carcinoma, Non-Small-Cell Lung/ (78)
-
(nsclc or nsclcs).ti,ab,ot,hw. (1275)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (468)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (1905)
-
(lclc or lclcs).ti,ab,ot,hw. (6)
-
or/5-9 (2407)
-
Receptor, Epidermal Growth Factor/ (57)
-
epidermal growth factor receptor$.ti,ab,ot. (1221)
-
epidermis growth factor receptor$.ti,ab,ot. (0)
-
transforming growth factor alpha receptor$.ti,ab,ot. (0)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (5)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (48)
-
EGFR.ti,ab,ot. (1697)
-
EGFRTK.ti,ab,ot. (1)
-
EGF receptor$.ti,ab,ot. (195)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (2)
-
or/11-22 (2307)
-
4 and 10 and 23 (163)
-
Carcinoma, Non-Small-Cell Lung/di [Diagnosis] (7)
-
“diagnostic techniques and procedures”/ or diagnostic tests, routine/ (23)
-
clinical laboratory techniques/ or molecular diagnostic techniques/ (86)
-
Diagnosis/ (1)
-
Diagnosis, Differential/ (316)
-
diagnos$.ti,ab,ot. (71,695)
-
(test or tests or testing or tested).ti,ab,ot. (101,066)
-
((lab or labs or laborator$) adj2 (procedure$ or exam$)).ti,ab,ot. (550)
-
or/25-32 (160,664)
-
10 and 23 and 33 (103)
-
24 or 34 (219)
-
animals/ not (animals/ and humans/) (3555)
-
35 not 36 (219)
-
limit 37 to yr=“2000 -Current” (219)
-
remove duplicates from 38 (215)
Cochrane Database of Systematic Reviews (CDSR) (Wiley): Issue 7, 2012
Cochrane Central Register of Controlled Trials (CENTRAL) (Wiley): Issue 7, 2012
Database of Abstracts of Reviews of Effects (DARE) (Wiley): Issue 3, 2012
Health Technology Assessment Database (HTA) (Wiley): Issue 3, 2012
Search limited to 2000 to 2012.
Searched 18 July 2012.
#1 MeSH descriptor Quinazolines, this term only (612)
#2 (Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9):ti,ab,kw (130)
#3 (Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2):ti,ab,kw (171)
#4 (#1 OR #2 OR #3) (738)
#5 MeSH descriptor Carcinoma, Non-Small-Cell Lung, this term only (1952)
#6 (nsclc or nsclcs or lclc or lclcs):ti,ab (2101)
#7 (lung* NEAR/3 (adeno-carcinoma* or adenocarcinom*)):ti,ab,kw (73)
#8 ((non-small NEXT cell) NEAR/3 lung*):ti,ab,kw (3584)
#9 ((large NEXT cell) NEAR/3 lung*):ti,ab,kw (4)
#10 (#5 OR #6 OR #7 OR #8 OR #9) (3812)
#11 MeSH descriptor Receptor, Epidermal Growth Factor, this term only (264)
#12 (epidermal NEXT growth NEXT factor NEXT receptor*):ti,ab,kw (405)
#13 (epidermis NEXT growth NEXT factor NEXT receptor*):ti,ab,kw (0)
#14 (transforming NEXT growth NEXT factor NEXT alpha NEXT receptor*):ti,ab,kw (0)
#15 (tgf-alpha NEAR/2 receptor*):ti,ab,kw (1)
#16 (urogastrone NEAR/2 receptor*):ti,ab,kw (0)
#17 ((erbB1 or erbB-1 or erbB) NEAR/2 (protein* or receptor*)):ti,ab,kw (292)
#18 (EGFR or EGFRTK):ti,ab,kw (446)
#19 (EGF NEXT receptor*):ti,ab,kw (23)
#20 (Cobas NEAR/3 EGFR) (0)
#21 (Cobas NEAR/3 (epidermal NEXT growth NEXT factor)) (0)
#22 (thera-screen* or therascreen*) (0)
#23 (#11 OR #12 OR #13 OR #14 OR #15 OR #16 OR #17 OR #18 OR #19 OR #20 OR #21 OR #22) 921
#24 (#4 AND #10 AND #23) (103)
#25 MeSH descriptor Carcinoma, Non-Small-Cell Lung, this term only with qualifier: DI (72)
#26 MeSH descriptor Diagnostic Techniques and Procedures, this term only (95)
#27 MeSH descriptor Diagnostic Tests, Routine, this term only (251)
#28 MeSH descriptor Clinical Laboratory Techniques, this term only (111)
#29 MeSH descriptor Molecular Diagnostic Techniques, this term only (33)
#30 MeSH descriptor Diagnosis, this term only (73)
#31 MeSH descriptor Diagnosis, Differential, this term only (1330)
#32 diagnos*:ti,ab,kw (70,823)
#33 (test or tests or testing or tested):ti,ab,kw (127,012)
#34 ((lab or labs or laborator*) NEAR/2 (procedure* or exam*)):ti,ab,kw (605)
#35 (#25 OR #26 OR #27 OR #28 OR #29 OR #30 OR #31 OR #32 OR #33 OR #34) (174,193)
#36 (#10 AND #23 AND #35) (38)
#37 (#24 OR #36), from 2000 to 2012 (116)
CDSR search retrieved 0 references.
CENTRAL search retrieved 96 references.
DARE search retrieved 7 references.
HTA search retrieved 11 references.
PROSPERO (International Prospective Register of Systematic Reviews) (Internet): up to 19 July 2012
Searched 19 July 2012.
Displayed all records (n = 650) and browsed the titles for the following terms:
| Terms | Records |
|---|---|
| lung | 0/7 |
| Small cell | 0/2 |
| Nsclc | 0/1 |
| Therascreen | 0 |
| Thera-screen | 0 |
| Cobas | 0 |
| EGF | 0 |
| erbb | 0 |
| urogastrone | 0 |
| tgf | 0 |
| Growth factor | 0 |
| Erlotinib | 0 |
| gefitinib | 0 |
| Total | 0 |
Latin American and Caribbean Health Sciences (LILACS): 2000 to 6 July 2012
http://regional.bvsalud.org/php/index.php?lang=en
Searched 19 July 2012.
| Terms (date limits applied in EndNote) | Records |
|---|---|
| (“Quinazolinas” or MH:D03.438.786 or Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2 or Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319-69-9) AND (lung$ or Pulmón or Pulmão or Pulmonar or nsclc or nsclcs or lclc or lclcs or MH:C04.588.894.797.520.109.220.249 or MH:C08.381.540.140.500 or MH:C08.785.520.100.220.500) | 13 |
| (lung$ or Pulmon or Pulmao or Pulmonar or nsclc or nsclcs or lclc or lclcs or MH:C04.588.894.797.520.109.220.249 or MH:C08.381.540.140.500 or MH:C08.785.520.100.220.500) AND (“Receptor, Epidermal Growth Factor” or “Receptor del Factor de Crecimiento Epidermico” or “Receptor do Fator de Crescimento Epidermico” or MH:D08.811.913.696.620.682.725.400.100 or MH:D12.776.543.750.060.249 or MH:D12.776.543.750.750.360.300 or MH:D12.776.543.750.750.400.340 or thera-screen$ or therascreen$ or “EGF receptor” or EGFR or EGFRTK or erbB1 or erbB-1 or erbB or urogastrone or tgf-alpha or “transforming growth factor” or “epidermis growth factor receptor” or “epidermal growth factor receptor”) | 14/25 |
| Total | 27 |
Spanish and Portuguese translations of MeSH terms identified using the DECS (Health Sciences Descriptors) thesaurus: http://decs.bvs.br/I/homepagei.htm
Date limit applied within EndNote Library.
Clinicaltrials.gov (Internet)
http://clinicaltrials.gov/ct2/search/advanced
Limited 1 January 2000 to 19 July 2012.
Searched 19 July 2012.
Advanced search option – search terms box:
| Search terms | Condition | Intervention | Records |
|---|---|---|---|
| (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR (epidermal growth factor*) OR erbb OR ERBB1 OR urogastrone) | (lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS) | (Erlotinib OR Nsc-718781 OR nsc718781 OR osi-774 OR osi774 OR r-1415 OR r1415 OR tarceva OR cp-358774 OR cp358774 OR 183321-74-6 OR 183319-69-9 OR Gefitinib OR Geftinat OR Geftib OR iressa OR zd-1839 OR zd1839 OR 184475-35-2) | 180 |
| (diagnos* OR test OR tests OR testing OR tested OR (lab procedure*) OR (lab exam*) OR (labs procedure*) OR (labs exam*) OR (laborator* procedure*) OR (laborator* exam*)) | (lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS) | (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR (epidermal growth factor*) OR erbb OR ERBB1 OR urogastrone) | 54 |
| Total | 234 |
metaRegister of Controlled Trials (mRCT) (Internet)
Up to 30 August 2012.
Searched 30 August 2012.
| Search terms | Results |
|---|---|
| (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR (epidermal growth factor*) OR erbb OR ERBB1 OR urogastrone) AND (lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS) | 302 |
| (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR erbb OR ERBB1 OR urogastrone) AND (Erlotinib OR r1415 OR tarceva OR 183321-74-6 OR 183319-69-9 OR Gefitinib OR Geftinat OR Geftib OR iressa OR zd1839 OR 184475-35-2) | 195 |
| (epidermal growth factor*) AND (Erlotinib OR r1415 OR tarceva OR 183321-74-6 OR 183319-69-9 OR Gefitinib OR Geftinat OR Geftib OR iressa OR zd1839 OR 184475-35-2) | 100 |
| Total | 597 |
WHO International Clinical Trials Registry Platform (ICTRP) (Internet)
Limited to 1 January 2000 to 30 August 2012.
Searched 30 August 2012.
Advanced search option:
| Title | Condition | Intervention | Records |
|---|---|---|---|
| (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR (epidermal growth factor*) OR erbb OR ERBB1 OR urogastrone) | (lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS) | (Erlotinib OR Nsc-718781 OR nsc718781 OR osi-774 OR osi774 OR r-1415 OR r1415 OR tarceva OR cp-358774 OR cp358774 OR 183321-74-6 OR 183319-69-9 OR Gefitinib OR Geftinat OR Geftib OR iressa OR zd-1839 OR zd1839 OR 184475-35-2) | 62 |
| (lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS) | (Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR (epidermal growth factor*) OR erbb OR ERBB1 OR urogastrone) | 82 | |
| Total | 144 |
BIOSIS Previews (Web of Knowledge): 2000 to 24 August 2012
Searched 30 July 2012.
Advanced search (Lemmatization off):
# 30 44 #28 not #29
Databases=BIOSIS Previews Timespan=2000–2012
# 29 5,773,678 TS=(cat or cats or dog or dogs or animal or animals or rat or rats or hamster or hamster or feline or ovine or canine or bovine or sheep OR macaque* OR monkey*)
# 28 1954 #21 or #27
# 27 889 #7 and #20 and #26
# 26 1933,480 #22 or #23 or #24 or #25
# 25 45,681 TS=((laborator* NEAR procedure*) or (laborator* NEAR exam*))
# 24 99 TS=((labs NEAR procedure*) or (labs NEAR exam*))
# 23 685 TS=((lab NEAR procedure*) or (lab NEAR exam*))
# 22 1,913,268 TS=(diagnos* OR test or tests or testing or tested)
# 21 1411 #3 and #7 and #20
# 20 34,254 #8 or #9 or #10 or #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18 or #19
# 19 13 TS=(thera-screen* or therascreen*)
# 18 0 TS=(Cobas NEAR epidermal NEAR growth NEAR factor)
# 17 0 TS=(Cobas NEAR EGFR)
# 16 7196 TS=(EGF NEAR receptor*)
# 15 20,895 TS=(EGFR or EGFRTK)
# 14 3256 TS=((erbB1 or erbB-1 or erbB) NEAR (protein* or receptor*))
# 13 0 TS=(urogastrone NEAR receptor*)
# 12 700 TS=(tgf-alpha NEAR receptor*)
# 11 1 TS=(transform NEAR growth NEAR factor NEAR alpha NEAR receptor*)
# 10 1962 TS=(transforming NEAR growth NEAR factor NEAR alpha NEAR receptor*)
# 9 62 TS=(epidermis NEAR growth NEAR factor NEAR receptor*)
# 8 21,660 TS=(epidermal NEAR growth NEAR factor NEAR receptor*)
# 7 27,387 #4 or #5 or #6
# 6 19,225 TS=((non-small NEAR cell NEAR lung*) or (large NEAR cell NEAR lung*))
# 5 10,230 TS=((lung* NEAR adeno-carcinoma*) OR (lung NEAR adenocarcinom*))
# 4 8560 TS=(nsclc or nsclcs or lclc or lclcs)
# 3 4669 #1 or #2
# 2 3230 TS=(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2)
# 1 2401 TS=(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319-69-9)
American Society of Clinical Oncology (ASCO) Conference Proceedings: 2007 to 2012
www.asco.org/ASCOv2/Meetings/Abstracts
Searched 26 October 2012.
Searched 2007–12 Annual Meetings:
| Keywords | Search for keyword in title | Search for keyword in Abstract | Total |
|---|---|---|---|
| Therascreen | 0 | 13 | 13 |
| Thera-screen | 0 | 0/13 | 0 |
| Cobas | 2 | 14/16 | 16 |
| EGFR-TK | 1 | 18/19 | 19 |
| Epidermal growth factor mutation | 19 | 19 | |
| Epidermal growth factor mutations | 38 | 38 | |
| EGFR mutation | 78 | 78 | |
| EGFR mutations | 109/116 | 109 | |
| Total | 292 |
ESMO Conference Proceedings (European Society of Medical Oncology): 2007 to 2012
www.esmo.org/no_cache/education/abstracts-and-virtual-meetings.html
Searched 31 October 2012.
-
2008 33rd ESMO Congress, Stockholm: http://annonc.oxfordjournals.org/content/vol19/suppl_8/
-
2009 ECCO 15 and 34th ESMO Multidisciplinary Congress: www.ejcancer.info
-
2010 35th ESMO Congress, Milan: http://annonc.oxfordjournals.org/content/21/suppl_8
-
2011 ECCO 16 and 36th ESMO Multidisciplinary Congress, Brussels: www.ejcancer.info/issues
-
2012 37th ESMO Congress, Vienna - http://annonc.oxfordjournals.org/content/23/suppl_9
| Intervention | 2008 | 2009 | 2010 | 2011 | 2012 |
|---|---|---|---|---|---|
| Therascreen | 0 | 0 | 0 | 4 | 3 |
| Thera-screen | 0 | 0 | 0 | 0 | 0 |
| Cobas | 0 | 0 | 0 | 0 | 4 |
| EGFR-TK | 0 | 0 | 1 | 0 | 1 |
| EGFR TK | 24 | 0 | 23 | 0 | 34 |
| Epidermal growth factor mutation | 40 | 0 | 38 | 0 | 62 |
| Epidermal growth factor mutations | 40 | 0 | 38 | 0 | 62 |
| EGFR mutation | 35 | 0 | 31 | 27 | 50 |
| EGFR mutations | 35 | 0 | 31 | 29 | 50 |
| Total | 174 | 2 | 162 | 63 | 266 |
| Total after deduplication | 41 | 2 | 38 | 51 | 65 |
World Conference on Lung Cancer (International Association for the Study of Lung Cancer): 2007 to 2012
Searched 30 October 2012.
-
14th World Conference on Lung Cancer: http://journals.lww.com/jto/toc/2011/06001
-
13th World Conference on Lung Cancer: http://journals.lww.com/jto/Citation/2009/09001/Abstracts.1.aspx
-
12th World Conference on Lung Cancer: http://journals.lww.com/jto/toc/2007/08001
| Intervention | 2007 | 2009 | 2011 |
|---|---|---|---|
| Therascreen | 0 | 1 | 1 |
| Thera-screen | 0 | 0 | 0 |
| Cobas | 0 | 0 | 0 |
| EGFR-TK | 20 | 44 | 25 |
| Epidermal growth factor mutation | 0 | 0 | 1 |
| Epidermal growth factor mutations | 0 | 0 | 0/1 |
| Total | 20 | 45 | 27 |
PubMed Related Citations search undertaken for included studies
Results sorted by Link Ranking.
Searched 24 October 2012.
Of 30 included studies, 12 references were indexed on PubMed. For each reference, the first 20 related citations were retrieved by carrying out a Related Citations search using PubMed’s similarity matching algorithm. These records were downloaded for screening. All related citations were checked against the EndNote Library to remove duplicate, and only new unique references were imported and screened.
| Reference | PMID | Result retrieved |
|---|---|---|
| #5011. Chen88 | 22157367 | 20/151 |
| #1591. Fukuoka48 | 21670455 | 20/141 |
| #6550. Giaccone43 | 17062680 | 20/424 |
| #6471. Jackman44 | 17228019 | 20/220 |
| #5109. Leary42 | 22036089 | 20/111 |
| #5637. Maemondo47 | 20573926 | 20/447 |
| #7377. Mok49 | 19692680 | 20/275 |
| #7220. Oizumi89 | 22581822 | 20/97 |
| #4980. Pallis45 | 22000696 | 20/208 |
| #1295. Rosell40 | 22285168 | 20/999 |
| #6145. Yang46 | 18509184 | 20/579 |
| #7352. Zhou16 | 21783417 | 20/787 |
| Total | 240/4438 | |
| Following duplicate removal, number of records screened | 26 | |
Cost-effectiveness search strategies
Review of cost-effectiveness literature
NHS Economic Evaluation Database (NHS EED) (Wiley): 2000 to 2012, Issue 3
Searched 30 August 2012.
#1 MeSH descriptor Quinazolines, this term only (613)
#2 (Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9):ti,ab,kw (131)
#3 (Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2):ti,ab,kw (171)
#4 (#1 OR #2 OR #3) (739)
#5 MeSH descriptor Carcinoma, Non-Small-Cell Lung, this term only (1953)
#6 (nsclc or nsclcs or lclc or lclcs):ti,ab (2101)
#7 (lung* NEAR/3 (adeno-carcinoma* or adenocarcinom*)):ti,ab,kw (73)
#8 ((non-small NEXT cell) NEAR/3 lung*):ti,ab,kw (3585)
#9 ((large NEXT cell) NEAR/3 lung*):ti,ab,kw (4)
#10 (#5 OR #6 OR #7 OR #8 OR #9) (3813)
#11 MeSH descriptor Receptor, Epidermal Growth Factor, this term only (265)
#12 (epidermal NEXT growth NEXT factor NEXT receptor*):ti,ab,kw (406)
#13 (epidermis NEXT growth NEXT factor NEXT receptor*):ti,ab,kw (0)
#14 (transforming NEXT growth NEXT factor NEXT alpha NEXT receptor*):ti,ab,kw (0)
#15 (tgf-alpha NEAR/2 receptor*):ti,ab,kw (1)
#16 ( urogastrone NEAR/2 receptor*):ti,ab,kw (0)
#17 ((erbB1 or erbB-1 or erbB) NEAR/2 (protein* or receptor*)):ti,ab,kw (292)
#18 (EGFR or EGFRTK):ti,ab,kw (449)
#19 (EGF NEXT receptor*):ti,ab,kw (23)
#20 (Cobas NEAR/3 EGFR) (0)
#21 (Cobas NEAR/3 (epidermal NEXT growth NEXT factor)) (0)
#22 (thera-screen* or therascreen*) (0)
#23 (#11 OR #12 OR #13 OR #14 OR #15 OR #16 OR #17 OR #18 OR #19 OR #20 OR #21 OR #22) (924)
#24 (#4 OR #23) (1476)
#25 (#10 AND #24), from 2000 to 2012 (8)
EMBASE (OvidSP): 2000 to 2012 week 28
Searched 30 August 2012.
-
health-economics/ (31,839)
-
exp economic-evaluation/ (188,273)
-
exp health-care-cost/ (180,330)
-
exp pharmacoeconomics/ (156,985)
-
or/1-4 (433,309)
-
(econom$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (521,624)
-
(expenditure$ not energy).ti,ab. (20,859)
-
(value adj2 money).ti,ab. (1141)
-
budget$.ti,ab. (21,476)
-
or/6-9 (543,342)
-
5 or 10 (796,287)
-
letter.pt. (796,544)
-
editorial.pt. (414,244)
-
note.pt. (527,749)
-
or/12-14 (1,738,537)
-
11 not 15 (716,763)
-
(metabolic adj cost).ti,ab. (768)
-
((energy or oxygen) adj cost).ti,ab. (2933)
-
((energy or oxygen) adj expenditure).ti,ab. (17,921)
-
or/17-19 (20,863)
-
16 not 20 (712,137)
-
exp animal/ (1,796,019)
-
exp animal-experiment/ (1,636,900)
-
nonhuman/ (3,899,172)
-
(rat or rats or mouse or mice or hamster or hamsters or animal or animals or dog or dogs or cat or cats or bovine or sheep).ti,ab,sh. (4,729,791)
-
or/22-25 (6,695,652)
-
exp human/ (13,830,628)
-
exp human-experiment/ (303,941)
-
27 or 28 (13,832,063)
-
26 not (26 and 29) (5,273,705)
-
21 not 30 (661,610)
-
erlotinib/ or (Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (12,196)
-
gefitinib/ or (Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (13,183)
-
or/32-33 (18,694)
-
lung non small cell cancer/ (45,891)
-
(nsclc or nsclcs).ti,ab,ot,hw. (22,788)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (9502)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (35,647)
-
(lclc or lclcs).ti,ab,ot,hw. (56)
-
or/35-39 (60,634)
-
Receptor, Epidermal Growth Factor/ (35,039)
-
(epidermal growth factor receptor$ or epidermis growth factor receptor$ or transforming growth factor alpha receptor$).ti,ab,ot. (25,326)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (183)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (1443)
-
(EGFR or EGFRTK).ti,ab,ot. (31,087)
-
EGF receptor$.ti,ab,ot. (9045)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (50)
-
or/41-49 (57,139)
-
34 or 50 (66,682)
-
31 and 40 and 51 (743)
-
limit 52 to yr=“2000 -Current” (743)
-
remove duplicates from 53 (736)
-
limit 54 to embase (703)
Costs filter:
Centre for Reviews and Dissemination. NHS EED Economics Filter: EMBASE (Ovid) weekly search [Internet]. York: Centre for Reviews and Dissemination; 2010 (cited 17 March 2011). Available from: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html
MEDLINE (OvidSP): 2000 to August 2012 week 4
Searched 30 August 2012:
-
economics/ (26,369)
-
exp “costs and cost analysis”/ (167,172)
-
economics, dental/ (1844)
-
exp “economics, hospital”/ (18,137)
-
economics, medical/ (8482)
-
economics, nursing/ (3868)
-
economics, pharmaceutical/ (2362)
-
(economic$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (369,952)
-
(expenditure$ not energy).ti,ab. (15,358)
-
(value adj1 money).ti,ab. (18)
-
budget$.ti,ab. (15,574)
-
or/1-11 (487,655)
-
((energy or oxygen) adj cost).ti,ab. (2460)
-
(metabolic adj cost).ti,ab. (652)
-
((energy or oxygen) adj expenditure).ti,ab. (14,385)
-
or/13-15 (16,851)
-
12 not 16 (483,875)
-
letter.pt. (757,777)
-
editorial.pt. (305,167)
-
historical article.pt. (285,776)
-
or/18-20 (1,335,091)
-
17 not 21 (457,810)
-
animals/ not (animals/ and humans/) (3,680,958)
-
22 not 23 (430,529)
-
Quinazolines/ (11,624)
-
(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (2631)
-
(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (3648)
-
or/25-27 (12,777)
-
Carcinoma, Non-Small-Cell Lung/ (27,182)
-
(nsclc or nsclcs).ti,ab,ot,hw. (14,335)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (7087)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (24,664)
-
(lclc or lclcs).ti,ab,ot,hw. (42)
-
or/29-33 (38,312)
-
Receptor, Epidermal Growth Factor/ (25,843)
-
epidermal growth factor receptor$.ti,ab,ot. (20,568)
-
epidermis growth factor receptor$.ti,ab,ot. (0)
-
transforming growth factor alpha receptor$.ti,ab,ot. (10)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (243)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (1239)
-
EGFR.ti,ab,ot. (20,234)
-
EGFRTK.ti,ab,ot. (10)
-
EGF receptor$.ti,ab,ot. (8326)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (14)
-
or/35-46 (39,620)
-
28 or 47 (47,729)
-
24 and 34 and 48 (90)
-
limit 49 to yr=“2000 -Current” (90)
-
remove duplicates from 50 (87)
Costs filter:
Centre for Reviews and Dissemination. NHS EED Economics Filter: MEDLINE (Ovid) monthly search [Internet]. York: Centre for Reviews and Dissemination; 2010 (cited 28 September 2010]). Available from: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html
MEDLINE In-Process Citations (OvidSP): 2000 to 29 August 2012
MEDLINE Daily Update (OvidSP): 2000 to 29 August 2012
Searched 30 August 2012:
-
economics/ (1)
-
exp “costs and cost analysis”/ (111)
-
economics, dental/ (0)
-
exp “economics, hospital”/ (5)
-
economics, medical/ (1)
-
economics, nursing/ (0)
-
economics, pharmaceutical/ (0)
-
(economic$ or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic$).ti,ab. (29,272)
-
(expenditure$ not energy).ti,ab. (821)
-
(value adj1 money).ti,ab. (2)
-
budget$.ti,ab. (1501)
-
or/1-11 (30,841)
-
((energy or oxygen) adj cost).ti,ab. (155)
-
(metabolic adj cost).ti,ab. (53)
-
((energy or oxygen) adj expenditure).ti,ab. (652)
-
or/13-15 (838)
-
12 not 16 (30,593)
-
letter.pt. (17,056)
-
editorial.pt. (10,671)
-
historical article.pt. (96)
-
or/18-20 (27,818)
-
17 not 21 (30,243)
-
animals/ not (animals/ and humans/) (1498)
-
22 not 23 (30,207)
-
Quinazolines/ (16)
-
(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319 69 9).ti,ab,ot,hw,rn. (259)
-
(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2).ti,ab,ot,hw,rn. (223)
-
or/25-27 (402)
-
Carcinoma, Non-Small-Cell Lung/ (30)
-
(nsclc or nsclcs).ti,ab,ot,hw. (1282)
-
(lung$ adj3 (adeno-carcinoma$ or adenocarcinom$)).ti,ab,ot. (455)
-
((non-small cell or large cell) adj3 lung$).ti,ab,ot. (1908)
-
(lclc or lclcs).ti,ab,ot,hw. (6)
-
or/29-33 (2376)
-
Receptor, Epidermal Growth Factor/ (23)
-
epidermal growth factor receptor$.ti,ab,ot. (1197)
-
epidermis growth factor receptor$.ti,ab,ot. (0)
-
transforming growth factor alpha receptor$.ti,ab,ot. (0)
-
((tgf-alpha or urogastrone) adj2 receptor$).ti,ab,ot. (6)
-
((erbB1 or erbB-1 or erbB) adj1 (protein$ or receptor$)).ti,ab,ot. (44)
-
EGFR.ti,ab,ot. (1666)
-
EGFRTK.ti,ab,ot. (1)
-
EGF receptor$.ti,ab,ot. (189)
-
(Cobas adj3 EGFR).af. (0)
-
(Cobas adj3 epidermal growth factor).ti,ab,ot. (0)
-
(thera?screen$ or therascreen$).af. (2)
-
or/35-46 (2255)
-
28 or 47 (2406)
-
24 and 34 and 48 (12)
-
limit 49 to yr=“2000 -Current” (12)
-
remove duplicates from 50 (12)
Costs filter:
Centre for Reviews and Dissemination. NHS EED Economics Filter: MEDLINE (Ovid) monthly search [Internet]. York: Centre for Reviews and Dissemination; 2010 [cited 28.9.10]. Available from: www.york.ac.uk/inst/crd/intertasc/nhs_eed_strategies.html
Health Economic Evaluation Database (HEED) (Internet): up to 30 August 2012
http://onlinelibrary.wiley.com/book/10.1002/9780470510933
Searched 30 August 2012:
Compound search, (all data), unable to limit by date:
Erlotinib OR Nsc-718781 OR nsc718781 OR osi-774 OR osi774 OR r-1415 OR r1415 OR tarceva OR cp-358774 OR cp358774 OR 183321-74-6 OR 183319 69 9 OR Gefitinib OR Geftinat OR Geftib OR iressa OR zd-1839 OR zd1839 OR 184475-35-2
AND
lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS
N=41
(Therascreen OR Thera-screen OR Cobas OR EGF OR EGFR OR EGFRTK OR TGF OR epidermal OR erbb OR ERBB1 OR urogastrone)
AND
(lung* OR NSCLC OR NSCLCS OR LCLC OR LCLCS)
N=8
HEED search retrieved 49 records.
Science Citation Index (SCI) (Web of Knowledge): 2000 to 29 August 2012
Search limited to 2000 to 2012.
Searched 30 August 2012.
Advanced search (Lemmatization off):
# 32 146 #11 and #15 and #31
# 31 40,025 #30 OR #29 OR #28
# 30 6378 TS=(Gefitinib or Geftinat or Geftib or iressa or zd-1839 or zd1839 or 184475-35-2)
# 29 3959 TS=(Erlotinib or Nsc-718781 or nsc718781 or osi-774 or osi774 or r-1415 or r1415 or tarceva or cp-358774 or cp358774 or 183321-74-6 or 183319-69-9)
# 28 36,741 #16 or #17 or #18 or #19 or #20 or #21 or #22 or #23 or #24 or #25 or #26 or #27
# 27 13 TS=(thera-screen* or therascreen*)
# 26 0 TS=(Cobas NEAR epidermal NEAR growth NEAR factor)
# 25 0 TS=(Cobas NEAR EGFR)
# 24 8720 TS=(EGF NEAR receptor*)
# 23 21,367 TS=(EGFR or EGFRTK)
# 22 3781 TS=((erbB1 or erbB-1 or erbB) NEAR (protein* or receptor*))
# 21 13 TS=(urogastrone NEAR receptor*)
# 20 702 TS=(tgf-alpha NEAR receptor*)
# 19 0 TS=(transform NEAR growth NEAR factor NEAR alpha NEAR receptor*)
# 18 819 TS=(transforming NEAR growth NEAR factor NEAR alpha NEAR receptor*)
# 17 68 TS=(epidermis NEAR growth NEAR factor NEAR receptor*)
# 16 20,767 TS=(epidermal NEAR growth NEAR factor NEAR receptor*)
# 15 34,161 #12 or #13 or #14
# 14 24,966 TS=((non-small NEAR cell NEAR lung*) or (large NEAR cell NEAR lung*))
# 13 8029 TS=((lung* NEAR adeno-carcinoma*) OR (lung NEAR adenocarcinom*))
# 12 15,691 TS=(nsclc or nsclcs or lclc or lclcs)
# 11 484,626 #9 not #10
# 10 1,160,972 TS=(cat or cats or dog or dogs or animal or animals or rat or rats or hamster or hamster or feline or ovine or canine or bovine or sheep OR macaque* OR monkey*)
# 9 508,156 #4 not #8
# 8 26,623 #5 or #6 or #7
# 7 14,802 TS=((energy or oxygen) NEAR expenditure)
# 6 1295 TS=(metabolic NEAR cost)
# 5 11,720 TS=((energy or oxygen) NEAR cost)
# 4 521,849 #1 or #2 or #3
# 3 796 TS=(value NEAR money)
# 2 10,358 TS=(expenditure* not energy)
# 1 517,471 TS=(economic* or cost or costs or costly or costing or price or prices or pricing or pharmacoeconomic* or budget*)
Update of manufacturer’s search in gefitinib STA 192 (appendix 10.4: resource utilisation)51
EMBASE (OvidSP): January 2009 to August 2012 week 34
Searched 30 August 2012:
-
Socioeconomics/ (102,445)
-
Cost benefit analysis/ (61,757)
-
Cost-effectiveness analysis/ (82,283)
-
Cost of illness/ (13,206)
-
Cost control/ (42,633)
-
Economic aspect/ (99,388)
-
Financial management/ (96,915)
-
Health care cost/ (111,972)
-
Health care financing/ (10,847)
-
Health economics/ (31,839)
-
Hospital cost/ (12,121)
-
(fiscal or financial or finance or funding).tw. (88,152)
-
Cost minimization analysis/ (2109)
-
(cost adj estimate$).mp. (1709)
-
(cost adj variable$).mp. (135)
-
(unit adj cost$).mp. (1997)
-
or/1-16 (603,136)
-
Carboplatin/ or carboplatin.mp. or Paraplatin.mp. (38,879)
-
Cisplatin/ or Cisplatin.mp. or Platinol.mp. (115,027)
-
Paclitaxel/ or Paclitaxel.mp. or Taxol.mp. (58,541)
-
Topotecan/ or Topotecan.mp. or Hycamtin.mp. (7943)
-
(irinotecan or Campto).mp. (21,091)
-
(docetax?l or Taxotere).mp. (27,947)
-
(vinorelbine or Navelbine).mp. (11,861)
-
(gemcitabine or Gemzar).mp. (27,119)
-
(zactima or ZD6474).mp. (615)
-
(bevacizumab or Avastin).mp. (23,392)
-
(pemetrexed or Alimta).mp. (4688)
-
(erlotinib or Tarceva).mp. (12,217)
-
(bortezomib or Velcade).mp. (11,513)
-
(vinflunine or Javlor).mp. (456)
-
(cetuximab or Erbitux).mp. (12,952)
-
(gefitinib or Iressa).mp. (13,204)
-
Vatalanib.mp. (2098)
-
Panitumumab.mp. (3458)
-
platinum compounds/ or platinum.mp. (40,273)
-
Taxoids/ or taxoid$.mp. (3083)
-
exp antineoplastic protocols/ (61,147)
-
Antineoplastic Agent/ (204,229)
-
Angiogenesis Inhibitor/ (12,049)
-
Antimetabolite/ (5559)
-
antineoplastic agents, alkylating/ (12,712)
-
antineoplastic agents, phytogenic/ (204,229)
-
or/18-43 (479,713)
-
Lung non Small Cell Cancer/ (45,891)
-
(non small cell or non-small-cell or nonsmall cell or nsclc).mp. (53,758)
-
(lung$ or pulmon$).mp. (1,175,884)
-
(cancer or tumor$ or tumour$ or carcino$ or blastom$ or squamous or neoplas$ or sarcom$ or lymphom$ or adenocarcinom$).mp. (3,339,510)
-
46 and 47 and 48 (53,042)
-
45 or 49 (53,042)
-
17 and 44 and 50 (861)
-
limit 51 to yr=“2006 -Current” (586)
-
(2009$ or 201$).em. (4,309,768)
-
52 and 53 (419)
-
remove duplicates from 54 (416)
-
limit 55 to embase (405)
Update of search strategy from appendix 10.4:
AstraZeneca UK Ltd. Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd, 2010 (cited 18 July 2012). 233pp.
MEDLINE (OvidSP): 2009 to August 2012 week 4
Searched 30 August 2012:
-
Economics/ (26,369)
-
“costs and cost analysis”/ (40,051)
-
Cost allocation/ (1921)
-
Cost-benefit analysis/ (54,902)
-
Cost control/ (19,311)
-
Cost savings/ (7775)
-
Cost of illness/ (15,410)
-
Cost sharing/ (1769)
-
“deductibles and coinsurance”/ (1348)
-
Medical savings accounts/ (462)
-
Health care costs/ (23,671)
-
Direct service costs/ (974)
-
Drug costs/ (11,210)
-
Employer health costs/ (1044)
-
Hospital costs/ (6965)
-
Health expenditures/ (12,574)
-
Capital expenditures/ (1914)
-
Value of life/ (5232)
-
exp economics, hospital/ (18,137)
-
exp economics, medical/ (13,308)
-
Economics, nursing/ (3868)
-
Economics, pharmaceutical/ (2362)
-
exp “fees and charges”/ (26,011)
-
exp budgets/ (11,515)
-
(low adj cost).mp. (16,966)
-
(high adj cost).mp. (6653)
-
(health?care adj cost$).mp. (3110)
-
(fiscal or funding or financial or finance).tw. (66,638)
-
(cost adj estimate$).mp. (1193)
-
(cost adj variable).mp. (27)
-
(unit adj cost$).mp. (1276)
-
(economic$ or pharmacoeconomic$ or price$ or pricing).tw. (141,586)
-
or/1-32 (403,794)
-
Carboplatin/ or carboplatin.mp. or Paraplatin.mp. (11,002)
-
Cisplatin/ or Cisplatin.mp. or Platinol.mp. (47,742)
-
Paclitaxel/ or Paclitaxel.mp. or Taxol.mp. (22,534)
-
Topotecan/ or Topotecan.mp. or Hycamtin.mp. (2326)
-
(irinotecan or Campto).mp. (6288)
-
(docetax?l or Taxotere).mp. (7860)
-
(vinorelbine or Navelbine).mp. (2936)
-
(gemcitabine or Gemzar).mp. (8196)
-
(zactima or ZD6474).mp. (170)
-
(bevacizumab or Avastin).mp. (6308)
-
(pemetrexed or Alimta).mp. (1286)
-
(erlotinib or Tarceva).mp. (2611)
-
(bortezomib or Velcade).mp. (3482)
-
(vinflunine or Javlor).mp. (148)
-
(cetuximab or Erbitux).mp. (2956)
-
(gefitinib or Iressa).mp. (3609)
-
Vatalanib.mp. (249)
-
Panitumumab.mp. (586)
-
platinum compounds/ or platinum.mp. (22,109)
-
Taxoids/ or taxoid$.mp. (7866)
-
exp antineoplastic protocols/ (95,249)
-
Antineoplastic Agents/ (171,356)
-
Angiogenesis Inhibitors/ (13,184)
-
Antimetabolites/ (7082)
-
antineoplastic agents, alkylating/ (7002)
-
antineoplastic agents, phytogenic/ (22,154)
-
or/34-59 (336,727)
-
lung neoplasms/ (148,687)
-
carcinoma, non-small-cell lung/ (27,182)
-
(non small cell or non-small-cell or nonsmall cell or nsclc).mp. (32,947)
-
(lung$ or pulmon$).mp. (849,108)
-
(cancer or tumor$ or tumour$ or carcino$ or blastom$ or squamous or neoplas$ or sarcom$ or lymphom$ or adenocarcinom$).mp. (2,650,656)
-
63 and 64 and 65 (32,779)
-
61 or 62 or 66 (151,633)
-
33 and 60 and 67 (367)
-
limit 68 to yr=“2006 -Current” (204)
-
(2009$ or 201$).ed. (2,869,749)
-
69 and 70 (142)
-
remove duplicates from 71 (129)
Update of search strategy from appendix 10.4:
AstraZeneca UK Ltd. Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd, 2010 (cited 18 July 2012). 233pp.
MEDLINE In-Process Citations (OvidSP): 2009 to 29 August 2012
MEDLINE Daily Update (OvidSP): 2009 to 29 August 2012
Searched 30 August 2012.
-
Economics/ (1)
-
“costs and cost analysis”/ (14)
-
Cost allocation/ (0)
-
Cost-benefit analysis/ (32)
-
Cost control/ (3)
-
Cost savings/ (7)
-
Cost of illness/ (11)
-
Cost sharing/ (2)
-
“deductibles and coinsurance”/ (0)
-
Medical savings accounts/ (1)
-
Health care costs/ (38)
-
Direct service costs/ (2)
-
Drug costs/ (17)
-
Employer health costs/ (0)
-
Hospital costs/ (3)
-
Health expenditures/ (11)
-
Capital expenditures/ (2)
-
Value of life/ (0)
-
exp economics, hospital/ (5)
-
exp economics, medical/ (2)
-
Economics, nursing/ (0)
-
Economics, pharmaceutical/ (0)
-
exp “fees and charges”/ (7)
-
exp budgets/ (4)
-
(low adj cost).mp. (2925)
-
(high adj cost).mp. (456)
-
(health?care adj cost$).mp. (296)
-
(fiscal or funding or financial or finance).tw. (4322)
-
(cost adj estimate$).mp. (64)
-
(cost adj variable).mp. (3)
-
(unit adj cost$).mp. (75)
-
(economic$ or pharmacoeconomic$ or price$ or pricing).tw. (10,972)
-
or/1-32 (18,244)
-
Carboplatin/ or carboplatin.mp. or Paraplatin.mp. (451)
-
Cisplatin/ or Cisplatin.mp. or Platinol.mp. (1599)
-
Paclitaxel/ or Paclitaxel.mp. or Taxol.mp. (927)
-
Topotecan/ or Topotecan.mp. or Hycamtin.mp. (71)
-
(irinotecan or Campto).mp. (297)
-
(docetax?l or Taxotere).mp. (501)
-
(vinorelbine or Navelbine).mp. (127)
-
(gemcitabine or Gemzar).mp. (524)
-
(zactima or ZD6474).mp. (10)
-
(bevacizumab or Avastin).mp. (695)
-
(pemetrexed or Alimta).mp. (96)
-
(erlotinib or Tarceva).mp. (257)
-
(bortezomib or Velcade).mp. (313)
-
(vinflunine or Javlor).mp. (12)
-
(cetuximab or Erbitux).mp. (245)
-
(gefitinib or Iressa).mp. (219)
-
Vatalanib.mp. (5)
-
Panitumumab.mp. (57)
-
platinum compounds/ or platinum.mp. (4025)
-
Taxoids/ or taxoid$.mp. (27)
-
exp antineoplastic protocols/ (47)
-
Antineoplastic Agents/ (185)
-
Angiogenesis Inhibitors/ (18)
-
Antimetabolites/ (2)
-
antineoplastic agents, alkylating/ (4)
-
antineoplastic agents, phytogenic/ (15)
-
or/34-59 (8676)
-
lung neoplasms/ (77)
-
carcinoma, non-small-cell lung/ (30)
-
(non small cell or non-small-cell or nonsmall cell or nsclc).mp. (2057)
-
(lung$ or pulmon$).mp. (24,317)
-
(cancer or tumor$ or tumour$ or carcino$ or blastom$ or squamous or neoplas$ or sarcom$ or lymphom$ or adenocarcinom$).mp. (86,846)
-
63 and 64 and 65 (2012)
-
61 or 62 or 66 (2060)
-
33 and 60 and 67 (13)
-
limit 68 to yr=“2006 -Current” (13)
-
(2009$ or 201$).ed. (556,714)
-
69 and 70 (4)
-
remove duplicates from 71 (4)
Update of search strategy from appendix 10.4:
AstraZeneca UK Ltd. Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd, 2010 (cited 18 July 2012). 233pp.
NHS Economic Evaluation Database (NHS EED) (Internet): 1 January 2009 to 30 August 2012
www.york.ac.uk/inst/crd/index_databases.htm
Searched 23 August 2012.
-
(carboplatin or Paraplatin) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (10)
-
(Cisplatin or Platinol) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (19)
-
(Paclitaxel or Taxol) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (32)
-
(Topotecan or Hycamtin) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (2)
-
((irinotecan or Campto)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (8)
-
((docetaxal or docetaxel or Taxotere)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (37)
-
((vinorelbine or Navelbine)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (11)
-
((gemcitabine or Gemzar)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (8)
-
((zactima or ZD6474)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (0)
-
((bevacizumab or Avastin)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (18)
-
((pemetrexed or Alimta)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (14)
-
((erlotinib or Tarceva)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (9)
-
((bortezomib or Velcade)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (3)
-
((vinflunine or Javlor)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (0)
-
((cetuximab or Erbitux)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (8)
-
((gefitinib or Iressa)) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (3)
-
(Vatalanib) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (0)
-
(Panitumumab) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (1)
-
(Advanced non-small cell lung cancer) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (10)
-
(NSCLC) IN NHSEED WHERE PD FROM 01/01/2009 TO 30/08/2012 (9)
-
#1 OR #2 OR #3 OR #4 OR #5 OR #6 OR #7 OR #8 OR #9 OR #10 OR #11 OR #12 OR #13 OR #14 OR #15 OR #16 OR #17 OR #18 (104)
-
#19 OR #20 (16)
-
#21 AND #22 (14)
Update of search strategy from appendix 10.4:
AstraZeneca UK Ltd. Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd, 2010 (cited 18 July 2012). 233pp.
CINAHL (EBSCOhost): January 2009 to 24 August 2012
Searched 30 August 2012.
-
S1 (MH “Financial Management”) OR (MH “Financial Support”) OR (MH “Financing, Organized”) OR (MH “Business”) (17,359)
-
S2 (MH “Economics”) (5503)
-
S3 S2 not S1 (4967)
-
S4 (MH “Health Resource Allocation”) OR (MH “Health Resource Utilization”) (12,749)
-
S5 TX cost or costs or economic* or pharmacoeconomic* or price* or pricing* (164,114)
-
S6 S3 or S4 or S5 (172,311)
-
S7 PT ( Editorial or Letter or News ) OR MH Animal studies OR SO Cochrane library OR AU Anonymous (279,784)
-
S8 S6 not S7 (159,174)
-
S9 MH Carboplatin (607)
-
S10 MH Cisplatin (1427)
-
S11 MH Paclitaxel (1551)
-
S12 MH Antineoplastic Agents (12,767)
-
S13 MH Antimetabolites (106)
-
S14 MH Antimetabolites, antineoplastic (608)
-
S15 MH Angiogenesis Inhibitors (1370)
-
S16 S9 or S10 or S11 or S12 or S13 or S14 or S15 (16,663)
-
S17 TX carboplatin or paraplatin or cisplatin or Platinol or Paclitaxel or Taxol or Topotecan or Hycamtin or irinotecan or Campto or docetax?l or Taxotere or vinorelbine or Navelbine or gemcitabine or Gemzar or zactima or ZD6474 or bevacizumab or Avastin or pemetrexed or Alimta or erlotinib or Tarceva or bortezomib or Velcade or vinflunine or Javlor or cetuximab or Erbitux or gefitinib or Iressa or Vatalanib or Panitumumab or platinum or taxoid* (7964)
-
S18 TX carboplatin or paraplatin or cisplatin or Platinol or Paclitaxel or Taxol or Topotecan or Hycamtin or irinotecan or Campto or docetax?l or Taxotere or vinorelbine or Navelbine or gemcitabine or Gemzar or zactima or ZD6474 or bevacizumab or Avastin or pemetrexed or Alimta or erlotinib or Tarceva or bortezomib or Velcade or vinflunine or Javlor or cetuximab or Erbitux or gefitinib or Iressa or Vatalanib or Panitumumab or platinum or taxoid* (7964)
-
S19 MH lung neoplasms (8574)
-
S20 MH carcinoma, non-small-cell lung (2144)
-
S21 TX ( non small cell or non-small-cell or nonsmall cell or nsclc ) OR TX ( lung* or pulmon* ) OR TX ( cancer or tumor* or tumour* or carcino* or blastom* or squamous or neoplasm* or sarcom* or lymphoma* or adenocarcinom* ) (254,889)
-
S22 S18 or S19 or S20 (15,916)
-
S23 S16 or S17 (19,714)
-
S24 S22 and S21 and S8 Limiters - Published Date from: 20060101-20121231 (382)
-
Entry date limit (2009-2012) applied in EndNote. Final results = 241
Update of search strategy from appendix 10.4:
AstraZeneca UK Ltd. Single Technology Appraisal (STA) for Gefitinib for the first line treatment of locally advanced or metastatic non-small lung cancer (submission to National Institute for Health and Clinical Excellence) [Word document provided by AstraZeneca]. Luton, UK: AstraZeneca UK Ltd, 2010 (cited 18 July 2012). 233pp.
Appendix 2 Data extraction tables
Studies that provided information on the accuracy epidermal growth factor receptor mutation testing for predicting response to treatment with tyrosine kinase inhibitors
| Study details | Selection criteria | Population | Intervention | EGFR mutation test details |
|---|---|---|---|---|
| Study details Fukuoka (IPASS) (2011)48,49,90–94 Countries China, Hong Kong, Taiwan, Japan, Indonesia Study design RCT Funding AstraZeneca Recruitment March 2006 to October 2007 No. treated with gefitinib for whom EGFR mutation test results were available 223 |
Inclusion criteria Adults (minimum 18 years) with stage IIIB or IV NSCLC (histological or cytological diagnosis) with histological features of adenocarcinoma. Non-smokers (< 100 cigarettes over lifetime) or former light smokers (stopped smoking at least 15 years previously and had a maximum of 10 pack-years of smoking). No previous chemotherapy, biological, or immunotherapy. WHO PS 0–2, measurable disease (RECIST) with at least one measurable lesion not previously irradiated, adjuvant chemotherapy permitted if not platinum based and completed > 6 months previously, neutrophil count > 2000 μl, adequate liver function Exclusion criteria None reported |
Age < 65 years: 170 |
Intervention Gefitinib Dose Oral 250 mg Frequency Daily Duration 5.6 months (range 0.1 to 22.8) |
EGFR mutation test Therascreen EGFR PCR Kit (version 1) Manufacturer DxS, Manchester, UK Mutations targeted 29 mutations in Therascreen kit |
| No. male 48 |
||||
| Ethnicity > 99% East Asian |
||||
| Smoking status Never smoked: 206 Current/former smoker: 17 |
||||
| Histological features NR |
||||
| Disease stage at entry IIIB: 40 IV: 183 Not stated: 0 |
||||
| PS: WHO 0 or 1: 204 2: 19 |
||||
| Previous treatments None reported |
| Study details | Selection criteria | Population | Intervention | EGFR mutation test details |
|---|---|---|---|---|
| Study details Giaccone (2006)43 Countries The Netherlands, France Study design Cohort Funding Hofmann-La Roche Ltd, AstraZeneca, Genentech Recruitment January 2004 to July 2004 No. enrolled: 54 No. treated: 53 |
Inclusion criteria Adults (≥ 18 years), NSCLC (histological or cytological diagnosis), not amenable to radical surgery or radiotherapy. No prior chemotherapy or other systemic treatment, measurable disease (RECIST), PS 0–2, life expectancy ≥ 12 weeks, time since prior surgery or radiotherapy ≥ 4 weeks, granulocyte count ≥ 1500 μl, platelet count ≥ 100,000 μl, bilirubin and transaminases ≤ 1.5 × upper limit of normal, creatinine clearance ≥ 60 ml/minute, negative pregnancy test in females of childbearing age. Patients with brain metastases were included if there was no evidence of progression in the brain and in the absence of corticosteroid treatment Exclusion criteria Unstable systemic disease (active infection, uncontrolled hypertension, unstable angina, congestive heart failure, myocardial infarction in the previous year, serious cardiac arrhythmia requiring medication), any other malignancy in previous 5 years (except carcinoma in situ of the cervix or squamous cell skin cancer), significant eye disorders (severe dry eye syndrome, Sjögren syndrome, severe exposure keratinitis, any other disorder likely to increase the risk of corneal epithelial lesions) |
Age Median: 60 years Range: 30–80 years |
Intervention Erlotinib Dose 150 mg Frequency Daily |
EGFR mutation test Direct sequencing (nested PCR) Manufacturer Not specified Analysis Software Sequencing of PCR products was done with the ABI PRISM 310 Genetic analyzer (Applied Biosystems) Mutations targeted All exon 18–21 mutations |
| No. male 22 |
||||
| Ethnicity Not stated |
||||
| Smoking status Never smoked: 16 Former smoker: 0 Current smoker: 0 Current/former smoker: 37 |
||||
| Histological features Adenocarcinoma: 24 Bronchoalveolar: 6 Squamous: 8 Not stated or other: 15 |
||||
| Disease stage at entry IIIB: 11 IV: 42 |
||||
| PS: scale not stated 0: 13 1: 32 2: 8 |
||||
| Previous treatments Surgery 8, radiotherapy 5, surgery and radiotherapy 3, none 37 |
||||
| Study details Han (First-SIGNAL) (2012)39,41 Country South Korea Study design RCT Funding AstraZeneca Recruitment October 2005 to November 2007 No. treated: Treated with gefitinib, for whom EGFR test results were available: 53 |
Inclusion criteria Adults (> 18 years), chemotherapy naive, never smokers, stage IIIB/IV adenocarcinoma NSCLC with measurable or non-measurable disease, PS 0–2, adequate bone marrow, liver and renal function Exclusion criteria Known severe hypersensitivity to gefitinib or any constituents, any evidence of clinically active interstitial lung disease, severe or uncontrolled systemic disease, concomitant use of phenytoin, carbamazepine, riampin, barbiturate or St John’s wort, and unstable brain metastases |
Baseline characteristics not reported for EGFR tested subgroups; see table in following section for whole trial patient characteristics | Intervention Gefitinib Dose 250 mg Frequency Daily |
EGFR mutation test Direct sequencing PCR Manufacturer Not specified Analysis software Not specified Mutations targeted Exons 19–21 |
| Study details Jackman (2007)44 Country USA Study design Cohort Funding National Institutes of Health, National Cancer Institute Specialised Program of Research Excellence in Lung Cancer, Doris and William Krupp Research Fund in Thoracic Oncology, Genentech Inc. Recruitment March 2003 to May 2005 No. enrolled 82 No. treated 80 |
Inclusion criteria Stage IIIB/IV NSCLC (histological or cytological diagnosis), age ≥ 70 years. ECOG PS 0–2, WBC ≥ 3000 μl, haemoglobin ≥ 9.0 g/dl, platelet count ≥ 100,000 μl, total bilirubin ≤ 1.5 mg/dl, AST ≤ 2 × upper limit of normal, creatinine ≤ 1.5 mg/dl, measurable or assessable lesions (RECIST), life expectancy ≥ 8 weeks. Patients with stable brain metastases after surgical resection and/or cranial radiation were eligible Exclusion criteria Prior chemotherapy or treatment with any ErbB1- or ErbB2-targeted agent, major surgery or radiation therapy in the previous 21 days, any malignancy in the previous 5 years (except non-melanoma skin cancers or definitively treated cervical cancer), any active gastrointestinal disorder that alters motility or absorption, severe and unstable comorbidities |
Age Median: 75 years Range: 70–91 years |
Intervention Erlotinib Dose 150 mg; dose reductions allowed for toxicity Frequency Daily |
EGFR mutation test Direct sequencing (34 samples) or WAVE-HS (nine samples) for inadequate samples (< 50% of tumour cells) Manufacturer Transgenomic Inc., Omaha, NE, USA (for WAVE-HS) Mutations targeted All exon 18–24 mutations |
| No. male 40 |
||||
| Ethnicity White: 76 Black: 3 Asian: 1 |
||||
| Smoking status Never smoked: 8 Former smoker: 67 Current smoker: 5 |
||||
| Histological features Adenocarcinoma: 47 Bronchoalveolar: 4 Squamous: 7 Not stated or other: 22 |
||||
| Disease stage at entry IIIB: 12 IV: 68 |
||||
| PS: ECOG 0: 13 1: 59 2: 8 |
||||
| Previous treatments Surgery 13, radiotherapy 12, surgery and radiotherapy 9, none 46 |
| Study details | Selection criteria | Population | Intervention | EGFR mutation test details |
|---|---|---|---|---|
| Study details Pallis (2012)45 Country Greece Study design Cohort Funding Cretan Association for Biomedical Research Recruitment December 2004 to October 2008 No. enrolled: 49 No. treated: 49 |
Inclusion criteria Adult (≥ 18 years) chemotherapy naive non-smokers (< 100 cigarettes over lifetime), with inoperable stage IIIB or IV NSCLC (histological or cytological diagnosis), with histological features of adenocarcinoma. At least one measurable lesion (RECIST), ECOG PS 0–2, life expectancy ≥ 3 months, adequate organ function (serum bilirubin ≤ 1.5 × upper limit of normal, AST and ALT ≤ 2.5 × upper limit of normal in the absence of perceptible liver metastases or ≤ 5 × upper limit of normal in presence of liver metastases, serum creatinine ≤ 1.5 × upper limit of normal, neutrophil count ≥ 1500 μl, platelet count ≥ 100,000 μl). Patients with central nervous system metastases were eligible, provided they had been irradiated and were clinically and radiologically stable Exclusion criteria Active infection, history of significant cardiac disease (unstable angina, congestive heart failure, myocardial infarction in the previous 6 months, ventricular arrhythmias) |
Age Median: 68 years Range: 36–81 years |
Intervention Erlotinib Dose 150 mg Frequency Daily Duration Median 5.67 months |
EGFR mutation test Direct sequencing (PCR) Manufacturer Not specified Mutations targeted All exon 18–21 mutations All specimens contained at least 80% of tumour cells |
| No. male: 17 | ||||
| Ethnicity Not stated |
||||
| Smoking status Never smoked: 49 Former smoker: 0 Current smoker: 0 |
||||
| Histological features Adenocarcinoma: 46 Bronchoalveolar: 3 |
||||
| Disease stage at entry IIIB: 7 IV: 42 |
||||
| PS: ECOG 0: 11 1: 35 2: 3 |
||||
| Previous treatments None reported |
| Study details | Selection criteria | Population | Intervention | EGFR mutation test details |
|---|---|---|---|---|
| Study details Yang (2008)46 Country Taiwan Study design Cohort Funding Taiwan National Science Council and Department of Health, AstraZeneca Taiwan Recruitment May 2005 to April 2006 No. enrolled: 106 No. treated: 106 |
Inclusion criteria NSCLC (histological or cytological diagnosis) stage IIIB or IV, not amenable to curative treatment. Tumour measurable on imaging, ECOG PS 0–2, adequate liver function (bilirubin ≤ 2.0 mg/dl, transaminases < 2.5 × upper limit of normal, ALP < 5 × upper limit of normal), adequate renal function (serum creatinine < 2 mg/dl), adequate bone marrow function (haemoglobin > 10 g/dl, neutrophil count > 2000 μl, platelet count > 100,000 μl), no prior systemic anti-cancer treatment, no immediate need for palliative radiotherapy, candidacy for cisplatin–based combination chemotherapy, life expectancy > 6 months. Patients with central nervous system metastases were eligible if they were clinically stable 6 weeks after radiotherapy Exclusion criteria Secondary malignancies and major systemic diseases. Central nervous system metastases |
Age Median: 67 years Range: 32–86 years |
Intervention Gefitinib Dose 250 mg Frequency Daily |
EGFR mutation test Direct sequencing (PCR) Manufacturer Not specified Mutations targeted All exon 18–21 mutations |
| No. male: 38 | ||||
| Ethnicity Not stated |
||||
| Smoking status Never smoked: 75 Former smoker: 19 Current smoker: 12 |
||||
| Histological features Adenocarcinoma: 97 Not stated or other: 9 |
||||
| Disease stage at entry IIIB: 10 IV: 96 |
||||
| PS: ECOG 0: 0 1: 98 2: 8 |
||||
| Previous treatments None reported |
Studies that provided information on how clinical outcomes may vary according to which test is used to select patients for tyrosine kinase inhibitor treatment
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Benlloch (2012)36 Countries France, Spain and Italy Study design RCT Funding Hoffmann-La Roche Recruitment February 2007 to January 2011 No. enrolled: 173 No. treated: 150 No. with cobas EGFR test: 135 |
Inclusion criteria Adults (> 18 years) with histologically confirmed NSCLC [stage IIIB (with pleural effusion) or stage IV], measurable or evaluable disease, and the presence of activating EGFR mutations (exon 19 deletions or exon 21 mutation L858R). No history of chemotherapy for metastatic disease (neoadjuvant or adjuvant chemotherapy was allowed if it ended at least 6 months before study entry). Patients with asymptomatic, stable brain metastases were eligible for inclusion Exclusion criteria None stated Re-analysis of a subset of EURTAC using a different test. Of 174 patients in the EURTAC trial 39 were excluded from this study (37 no tumour block available and two insufficient tumour material) |
Age | Intervention (dose) Erlotinib (150 mg daily) Duration Median 8.2 months (range 0.3 to 32.9) No. of participants 77 |
Intervention (dose) i.v. cisplatin (75 mg/m2) plus docetaxel (75 mg/m2) administered on day 1 of a 3-week cycle or i.v. cisplatin (75 mg/m2) plus gemcitabine 1.25 g/m2) with cisplatin administered on day 1 and gemcitabine on days 1 and 8 of a 3-week cycle Median 4 cycles (range 1–6 cycles, 2–4 cycles) Patients who were ineligible for cisplatin received i.v. carboplatin Duration Median 2.8 months (range 0.7–5.1 months, 1.0–2.6 months) No. of participants 73 |
EGFR mutation test cobas® EGFR mutation test Manufacturer Hoffmann-la Roche, Basel, Switzerland Mutations targeted L858R (exon 21) and 29 exon 19 deletions |
||
| Mean (SD) | 63 (11) | 64 (9) | |||||
| Age range | NR | NR | |||||
| No. male | 28 | 19 | |||||
| Ethnicity All but two patients were white |
|||||||
| Smoking status | |||||||
| Never smoked | 57 | 63 | |||||
| Former smoker | 22 | 12 | |||||
| Current smoker | 7 | 12 | |||||
| Histological features | |||||||
| Adenocarcinoma | 82 | 78 | |||||
| Bronchoalveolar | 0 | 2 | |||||
| Squamous | 1 | 0 | |||||
| NS/other | 3 | 7 | |||||
| Disease stage at entry | |||||||
| IIIB | 6 | 5 | |||||
| IV | 78 | 82 | |||||
| NS | 2 | 0 | |||||
| PS: ECOG | |||||||
| 0 | 27 | 30 | |||||
| 1 | 47 | 45 | |||||
| 2 | 12 | 12 | |||||
| Previous treatments | NR | NR | |||||
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Fukuoka (IPASS) (2011)48,49,90–94 Countries China, Hong Kong, Taiwan, Japan, Indonesia Study design RCT Funding AstraZeneca Recruitment March 2006 to October 2007 No. enrolled: 1217 No. treated: 1196 No. in EGFR mutation-positive subgroup: 261 |
Inclusion criteria Adults (minimum 18 years) with stage IIIB or IV NSCLC (histological or cytological diagnosis) with histological features of adenocarcinoma. Non-smokers (< 100 cigarettes over lifetime) or former light smokers (stopped smoking at least 15 years previously and had a maximum of 10 pack-years of smoking). No previous chemotherapy, biological, or immunotherapy. WHO PS 0–2, measurable disease (RECIST) with at least one measurable lesion not previously irradiated, adjuvant chemotherapy permitted if not platinum-based and completed > 6 months previously, neutrophil count > 2000 μl, adequate liver function Exclusion criteria None reported Comments Data were extracted for the EGFR-positive subgroup (132 treated with gefitinib and 129 treated with SC). Full separate baseline data were not available for these patients |
Age | Intervention (dose) Gefitinib (250 mg daily) Duration 5.6 months (range 0.1 to 22.8) No. of participants 607 (132 in EGFR mutation-positive subgroup) |
Intervention (dose) Carboplatin (variable), paclitaxel (200 mg per m2) Administered on day 1 of 3-week cycle Median six cycles 3.4 (range 0.7 to 5.8) No. of participants 589 (129 in mutation-positive subgroup) |
EGFR mutation test Therascreen EGFR PCR Kit (version 1) Manufacturer Qiagen Mutations targeted 29 mutations in Therascreen Kit |
||
| Median (range) | 57 (25–84) | 57 (25–84) | |||||
| Subgroup (< 65 years) | 95 | 90 | |||||
| No. male | 125 | 127 | |||||
| Ethnicity | |||||||
| Chinese | 314 | 304 | |||||
| Japanese | 114 | 119 | |||||
| Other East Asian | 179 | 184 | |||||
| Other | 2 | 1 | |||||
| Smoking status | |||||||
| Never smoked | 571 | 569 | |||||
| Former smoker | 38 | 39 | |||||
| Subgroup (never smoked) | 124 | 122 | |||||
| Histological features | |||||||
| Adenocarcinoma | 581 | 591 | |||||
| Bronchoalveolar | 27 | 15 | |||||
| NS/other | 1 | 2 | |||||
| Disease stage at entry | |||||||
| IIIB | 150 | 144 | |||||
| IV | 459 | 463 | |||||
| NS | 0 | 1 | |||||
| Subgroup (IIIB) | 19 | 29 | |||||
| PS | |||||||
| 0 | 157 | 161 | |||||
| 1 | 391 | 382 | |||||
| 2 | 61 | 65 | |||||
| Subgroup (0 or 1) | 119 | 122 | |||||
| Previous treatments | NR | NR | |||||
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Han (First-SIGNAL) (2012)39,41 Country South Korea Study design RCT Funding AstraZeneca Recruitment October 2005 to November 2007 No. enrolled 313 No. treated 309 No. in EGFR mutation-positive subgroup 42 |
Inclusion criteria Adults (> 18 years), chemotherapy naive, never smokers, stage IIIB/IV adenocarcinoma NSCLC with measurable or non-measurable disease, PS 0–2, adequate bone marrow, liver and renal function Exclusion criteria Known severe hypersensitivity to gefitinib or any constituents, any evidence of clinically active interstitial lung disease, severe or uncontrolled systemic disease, concomitant use of phenytoin, carbamazepine, riampin, barbiturate or St John’s wort, and unstable brain metastases Comments Data were extracted for the EGFR-positive subgroup (26 treated with gefitinib and 16 treated with SC). Separate baseline data were not available for these patients |
Age | Intervention (dose) Gefitinib (250 mg daily) Duration Median 163 days (range 11–885) No. of participants 159 (26 in EGFR mutation positive subgroup) |
Intervention (dose) i.v. gemcitabine (1250 mg/m2) on day 1 and day 8 and cisplatin (75 mg/m2) on day 1 of 3 week cycles Duration Median number of cycles 6 (range 1 to 9) No. of participants 150 (16 in EGFR mutation-positive subgroup) |
EGFR mutation test Direct sequencing PCR Manufacturer Not specified Analysis software Not specified Mutations targeted Exons 19–21 |
||
| Median | 52 | 57 | |||||
| Age range | 32–74 | 19–74 | |||||
| No. male | 19 | 16 | |||||
| Ethnicity Not stated |
|||||||
| Smoking status | |||||||
| Never smoked | 159 | 150 | |||||
| Histological features | |||||||
| Adenocarcinoma | 159 | 150 | |||||
| Disease stage at entry | |||||||
| IIIB | 17 | 14 | |||||
| IV | 142 | 136 | |||||
| PS: ECOG | |||||||
| 0 | 41 | 31 | |||||
| 1 | 104 | 105 | |||||
| 2 | 14 | 14 | |||||
| Previous treatments | NR | NR | |||||
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Maemondo (NEJSG) (2010)47,89,95–99 Country Japan Study design RCT Funding Japan Society for Promotion of Science, the Japanese Foundation for the Multidisciplinary Treatment of Cancer, and the Co-operative Oncology Group Recruitment March 2006 to May 2009 No. enrolled 228 No. treated 227 |
Inclusion criteria Chemotherapy naive, aged 20–75 years, histologically or cytologically confirmed stage IIIB or IV NSCLC, or recurrent disease after surgery, no indication for further surgery or curative radiotherapy. Patients who had received palliative radiation therapy for brain or bone metastases > 2 weeks previously were eligible. Confirmed presence of sensitive EGFR mutations. Lesions evaluable by RECIST. ECOG PS 0 or 1. Normal bone marrow function (WBC count ≥ 4000/μl, platelet count ≥ 100,000/μl, haemoglobin ≥ 9.0 g/dl), normal liver function (AST and ALT ≤ 2 × upper limit of normal, total serum bilirubin ≤ 1.5 mg/dl). Normal renal function (creatinine clearance ≥ 40). Prognosis > 3 months Exclusion criteria Interstitial pneumonia or pulmonary fibrosis. Positive for resistant EGRF mutation T790M. Radiation therapy for primary lesions. Severe complications (uncontrolled heart, lung, liver, or kidney disease, or diabetes mellitus), pregnant or lactating women, severe malabsorption syndrome, diseases affecting digestive function, receipt of systemic steroids for ≥ 4 weeks, pleural effusion, pericardial effusion and/or peritoneal effusion requiring tube drainage, unless stable for at least 2 weeks after drainage, contraindications for gefitinib, carboplatin or paclitaxel, active double cancers (intramucosal tumours were not considered to be independent cancers), patients judged inappropriate for enrolment by attending physicians |
Age | Intervention (dose) Gefitinib (250 mg daily) Duration Median 308 days (range 14–1219) No. of participants 114 |
Intervention (dose) i.v. paclitaxel (220 mg/m2) and carboplatin (variable) Administered on day 1 of 3-week cycle for median of 4 cycles (range 1–7) No. of participants 113 |
EGFR mutation test Fragment length analysis Manufacturer NS Mutations targeted Exon 19 deletions, exon 21 point mutations (L858R, L861Q), exon 18 point mutations (G719A, G719C, G719S), exon 20 point mutation (T790M) |
||
| Mean (SD) | 64 (8) | 63 (9) | |||||
| Age range | 43–75 | 35–75 | |||||
| No. male | 42 | 41 | |||||
| Ethnicity Not stated |
|||||||
| Smoking status | |||||||
| Never smoked | 75 | 66 | |||||
| Current/former | 39 | 48 | |||||
| Histological features | |||||||
| Adenocarcinoma | 103 | 110 | |||||
| Squamous | 3 | 2 | |||||
| NS/other | 8 | 2 | |||||
| Disease stage at entry | |||||||
| IIIB | 15 | 21 | |||||
| IV | 88 | 84 | |||||
| NS/other | 11 | 9 | |||||
| PS: ECOG | |||||||
| 0 | 54 | 57 | |||||
| 1 | 59 | 55 | |||||
| 2 | 1 | 2 | |||||
| Previous treatments | NR | NR | |||||
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test details | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Rosell (EURTAC) (2012)38,40,100 Countries France, Spain and Italy Study design RCT Funding Hoffmann-la Roche and Red Tematica de Investigacion Cooperativa en Cancer grant Recruitment February 2007 to January 2011 No. enrolled 173 No. treated 150 |
Inclusion criteria Adults (> 18 years) with histologically confirmed NSCLC [stage IIIB (with pleural effusion), or stage IV], measurable or evaluable disease, and the presence of activating EGFR mutations (exon 19 deletions or exon 21 mutation L858R). No history of chemotherapy for metastatic disease (neoadjuvant or adjuvant chemotherapy was allowed if it ended at least 6 months before study entry). Patients with asymptomatic, stable brain metastases were eligible for inclusion Exclusion criteria None stated |
Age | Intervention (dose) Erlotinib (150 mg daily) Duration Median 8.2 months (range 0.3 to 32.9) No. of participants 77 |
Intervention (dose) i.v. cisplatin (75 mg/m2) plus docetaxel (75 mg/m2) administered on day 1 of a 3-week cycle or i.v. cisplatin (75 mg/m2) plus gemcitabine 1.25 g/m2) with cisplatin administered on day 1 and gemcitabine on days 1 and 8 of a 3-week cycle Median 4 cycles (range 1–6, 2–4) Patients who were ineligible for cisplatin received i.v. carboplatin Duration Median 2.8 months (range 0.7–5.1, 1.0–2.6) No. of participants 73 |
EGFR mutation test Sanger sequencing. All mutations were independently confirmed with PCR fragment length analysis for exon 19 deletions and TaqMan® assay (Applied Biosystems) for exon 21 point mutation L858R Manufacturer Not specified Mutations targeted Exon 19 and 21 mutations |
||
| Mean (SD) | 63 (11) | 64 (9) | |||||
| Age range | NR | NR | |||||
| No. male | 28 | 19 | |||||
| Ethnicity All but two patients were white |
|||||||
| Smoking status | |||||||
| Never smoked | 57 | 63 | |||||
| Former smoker | 22 | 12 | |||||
| Current smoker | 7 | 12 | |||||
| Histological features | |||||||
| Adenocarcinoma | 82 | 78 | |||||
| Bronchoalveolar | 0 | 2 | |||||
| Squamous | 1 | 0 | |||||
| NS/other | 3 | 7 | |||||
| Disease stage at entry | |||||||
| IIIB | 6 | 5 | |||||
| IV | 78 | 82 | |||||
| NS | 2 | 0 | |||||
| PS: ECOG | |||||||
| 0 | 27 | 30 | |||||
| 1 | 47 | 45 | |||||
| 2 | 12 | 12 | |||||
| Previous treatments | NR | NR | |||||
| Study details | Selection criteria | Participant details | Intervention details | EGFR mutation test details | |||
|---|---|---|---|---|---|---|---|
| Criteria | EGFR-TKI | SC | EGFR-TKI | SC | |||
| Study details Zhou (OPTIMAL) (2011)16,101–107 Country China Study design RCT Funding Hoffmann-la Roche and the Science and technology Commission of Shanghai Municipality Recruitment August 2008 to July 2009 No. enrolled 165 No. treated 154 |
Inclusion criteria Adults (> 18 years) with histologically confirmed advanced or recurrent stage IIIB or stage IV NSCLC and a confirmed activating EGFR mutation (exon 19 deletions or exon 21 mutation L858R). Measurable disease according to RECIST. ECOG PS 0–2. Adequate haematological, biochemical and organ function Exclusion criteria Patients with uncontrolled brain metastases, or who had received previous systemic anticancer therapy for advanced disease (adjuvant or neoadjuvant therapy allowed for non-metastatic disease in which relapse had occurred ≥ 6 months after final treatment) |
Age | Intervention (dose) Erlotinib (150 mg daily) Duration Median 55.5 weeks (range 3.1 to 93) No. of participants 82 |
Intervention (dose) i.v. gemcitabine (1 g/m2) administered on days 1 and 8 and i.v. carboplatin (variable) administered on day 1 of a 3-week cycle Median 4 (range 1–6) cycles Duration Median 10.4 weeks (range 1.0 to 18.9) No. of participants 72 |
EGFR mutation test Direct sequencing (PCR-based). Test confirmation methods were applied at the same time: agarose gel electrophoresis for exon 19 deletions; Cycleave® real-time PCR for exon 21 L858R point mutations Manufacturer Not specified Mutations targeted Exons 19 and 21 |
||
| Median, years | 57 | 59 | |||||
| Age range, years | 31–74 | 36–78 | |||||
| No. male | 34 | 29 | |||||
| Ethnicity Not stated |
|||||||
| Smoking status | |||||||
| Never smoked | 59 | 50 | |||||
| Current/former | 23 | 22 | |||||
| Histological features | |||||||
| Adenocarcinoma | 72 | 62 | |||||
| NS/other | 10 | 10 | |||||
| Disease stage at entry | |||||||
| IIIB | 11 | 5 | |||||
| IV | 71 | 67 | |||||
| PS: ECOG | |||||||
| 0–1 | 75 | 69 | |||||
| 2 | 7 | 3 | |||||
| Previous treatments | NS | NS | |||||
Appendix 3 Risk of bias assessments
QUADAS-2 assessments
Study: Fukuoka (2011)48,49
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: RCT, only patients treated with gefitinib included for accuracy evaluation |
|
| Was a consecutive or random sample of patients enrolled? | Yes |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? | RISK: low |
| DOMAIN 2: INDEX TEST(S) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Biomarker status was determined by analysing paraffin-embedded archival tumour tissue. Scientists were blinded to clinical outcome and treatment. Samples underwent central histopathological review; only those considered suitable for downstream biomarker analysis were progressed (on the basis of quality, sample source and tumour content). If a patient provided more than one sample, the appropriate section was selected before database lock and analysed on the basis of sample quality and largest area of tumour tissue. EGFR mutations were detected by using an amplification mutation refractory system with an EGFR mutation detection kit (DxS, Manchester, UK). Tumours were considered EGFR mutation positive if at least one of 29 EGFR mutations was detected. Additional validation for samples with T790M mutations was performed by using three methods: DNA sequencing, multithreaded electronic PCR sequencing and an alternative amplification mutation refractory system assay |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? | RISK: low |
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by RECIST criteria21) acted as reference standard. Tumour response was assessed every 6 weeks until disease progression |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct or its interpretation have introduced bias? | RISK: low |
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: A total of 386 patients had unknown mutation status as they declined consent for biomarker analysis, had no available tumour sample or had samples of insufficient quality for EGFR mutation analysis. All cytology samples were excluded as biomarker kit used was not validated for these samples. A further 9 patients were not evaluated for tumour response. Describe the time interval and any interventions between index test(s) and reference standard: Follow-up continued for over 2 years |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | No |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: low |
Study: Giaccone (2006)43
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: No details on how patients were enrolled other than inclusion criteria |
|
| Was a consecutive or random sample of patients enrolled? | Unclear |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? |
RISK: unclear
|
| DOMAIN 2: INDEX TEST(S) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Paraffin embedded tumour material was cut in 4-μm-thick sections and placed on to glass slides, stained with haematoxylin and eosin and the presence of tumour cells was verified by a pathologist. Tumour cells were microdissected and genomic DNA was isolated using the QIAamp DNA Micro Kit (Qiagen, Venlo, the Netherlands). Nested PCRs were carried out using primers to amplify exons 18–21 of EGFR. To facilitate sequencing, internal primers incorporated an M13 Tag. Sequencing of PCR products was done with the ABI PRISM 310 Genetic analyser (Applied Biosystems, Foster City, CA, USA). Mutations were confirmed by sequencing independent PCR products. Because of concerns about the sensitivity of direct sequencing, DNA from 22 independent samples were analysed by other institutions in a blinded fashion |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? |
RISK: low
|
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by RECIST criteria21) acted as reference standard. Tumour response was assessed at six weeks and subsequent assessment frequency was unclear. |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct or its interpretation have introduced bias? |
RISK: low
|
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2×2 table: Histological material was not available for 24 patients and so EGFR mutation analysis could not be performed Describe the time interval and any interventions between index test(s) and reference standard: The median duration of response was 333 days |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | Yes |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: low |
Study: Han (First-SIGNAL) (2012)41
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: RCT, only patients treated with gefitinib included for accuracy evaluation |
|
| Was a consecutive or random sample of patients enrolled? | Yes |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? |
RISK: low
|
| DOMAIN 2: INDEX TEST(S) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Genomic DNA was extracted from paraffin-embedded tissue blocks or cells blocks of cytology specimens, whichever were available by using the QIAamp DNA mini kit (Qiagen, Valencia, CA). To detect somatic mutations of EGFR genes, exons 19, 20 and 21 were amplified by PCR and directly sequenced according to the method previously reported. All PCR direct sequencing reactions were repeated twice to confirm the results |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? |
RISK: low
|
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by WHO criteria20) acted as reference standard. Tumour response was assessed every 9 weeks during treatment. |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct, or its interpretation have introduced bias? |
RISK: low
|
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: A total of 217 patients were not assessed for mutation status; reasons were not given and no information on differences between those with and without known mutation status; 43 received standard chemotherapy so did not contribute to accuracy data. Of the 159 patients who received gefitinib, 53 were assessed for tumour EGFR mutation status Describe the time interval and any interventions between index test(s) and reference standard: Follow-up continued for over 4 years |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | Yes |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: Unclear |
Study: Jackman (2007)44
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: No details on how patients were enrolled other than inclusion criteria |
|
| Was a consecutive or random sample of patients enrolled? | Unclear |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? |
RISK: Unclear
|
| DOMAIN 2: INDEX TEST(S) If more than one index test was used, please complete for each test |
|
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Tumour specimens (frozen or paraffin embedded) were collected from previous diagnostic or surgical procedures. No specific requirements were prospectively mandated for the type of tumour specimen. For patients with sufficient tissue for direct DNA sequencing, tumour cells were isolated by microdissection. Exons 18 through 24 of the EGFR were amplified and sequenced according to previously described methods. For tumour samples deemed inadequate by a molecular pathologist for direct sequencing based on a high percentage of normal cells (< 50% of tumour cells) mutation analysis was performed with the WAVE-HS platform using previously published methods. All detected mutations were confirmed by repeat analysis |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear |
| Could the conduct or interpretation of the index test have introduced bias? |
RISK: low
|
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by RECIST criteria21) acted as reference standard. Tumour response was assessed every 6 weeks during treatment. |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Unclear |
| Could the reference standard, its conduct or its interpretation have introduced bias? | RISK: low |
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: Four samples could not be obtained from other hospitals, seven patients had not consented to EGFR testing, 26 samples judged inadequate for testing. Response was not assessable in six patients with EGFR mutation-negative tumours Describe the time interval and any interventions between index test(s) and reference standard: Median time to progression was 3.5 months (95% CI 2 to 5.5 months). Median survival was 10.9 months, follow-up continued for over 2 years |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | No |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: high |
Study: Pallis (2012)45
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: No details on how patients were enrolled other than inclusion criteria |
|
| Was a consecutive or random sample of patients enrolled? | Unclear |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? |
RISK: unclear
|
| DOMAIN 2: INDEX TEST(S) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Tumour samples obtained from formalin-fixed paraffin embedded tissue blocks made on initial diagnosis. Microdissection was used to ensure that specimens contained at least 80% of tumour cells. DNA sequence of exons 18–21 of EGFR were determined by direct forward and reverse sequencing of the PCR product from nested PCR reactions as described previously |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? |
RISK: low
|
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by RECIST criteria21) acted as reference standard. Tumour response was assessed every 8 weeks during treatment |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Yes |
| Could the reference standard, its conduct or its interpretation have introduced bias? |
RISK: low
|
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: All patients had data on outcome (reference standard). Thirteen patients did not have data on mutation status, because samples were not available Describe the time interval and any interventions between index test(s) and reference standard: Median duration of response 10.2 months (95% CI 7.4 to 12.9 months), median follow-up time was 18.9 months (range 0.6 to 50.7 months) |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | Yes |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: low |
Study: Yang (2008)46
| DOMAIN 1: PATIENT SELECTION | |
|---|---|
| A. Risk of bias | |
| Describe methods of patient selection: No details on how patients were enrolled other than inclusion criteria |
|
| Was a consecutive or random sample of patients enrolled? | Unclear |
| Was a case–control design avoided? | Yes |
| Did the study avoid inappropriate exclusions? | Yes |
| Could the selection of patients have introduced bias? |
RISK: unclear
|
| DOMAIN 2: INDEX TEST(S) | |
| A. Risk of bias | |
| Describe the index test and how it was conducted and interpreted: Most tumour samples were obtained from paraffin-embedded blocks made on initial diagnosis. Alternatively, DNA extracted from pleural fluid-derived cancer cells were also used for analysis. DNA sequence of exons 18–21 were determined by direct forward and reverse sequencing of the PCR product from nested PCR reactions as described previously |
|
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes |
| Could the conduct or interpretation of the index test have introduced bias? |
RISK: low
|
| DOMAIN 3: REFERENCE STANDARD | |
| A. Risk of bias | |
| Describe the reference standard and how it was conducted and interpreted: Best overall response to treatment (as defined by RECIST criteria21) acted as reference standard. Tumour response was assessed every 8 weeks during treatment |
|
| Is the reference standard likely to correctly classify the target condition? | Yes |
| Were the reference standard results interpreted without knowledge of the results of the index test? | Yes |
| Could the reference standard, its conduct or its interpretation have introduced bias? |
RISK: low
|
| DOMAIN 4: FLOW AND TIMING | |
| A. Risk of bias | |
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table: EGFR mutation status was not successfully determined in 16 patients, and nine patients did not have data on outcome, reasons were not given Describe the time interval and any interventions between index test(s) and reference standard: Median time to treatment failure was 5.5 months. Duration of follow-up was a minimum of 12 months |
|
| Was there an appropriate interval between index test(s) and reference standard? | Yes |
| Did all patients receive a reference standard? | No |
| Did patients receive the same reference standard? | Yes |
| Were all patients included in the analysis? | No |
| Could the patient flow have introduced bias? | RISK: high |
Risk of bias assessments
Studies: Fukuoka (IPASS) (2011)48 and Mok (2009)49
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | No details reported | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | Open label | High |
| Outcome assessor blinding | No details reported | Unclear |
| Incomplete outcome data | 246 withdrawals in gefitinib arm: 223 died, 19 withdrew consent, 5 lost to follow-up 276 withdrawals in carboplatin–paclitaxel arm: 227 died, 46 withdrew consent, 2 lost to follow-up, 1 did not meet eligibility criteria 386 patients in the gefitinib arm and 394 patients in the carboplatin–paclitaxel arm had unknown mutation status as they declined consent for biomarker analysis, had no available tumour sample, or had samples of insufficient quality for EGFR mutation analysis. All cytology samples were excluded as biomarker kit used was not validated for these samples. Baseline data similar to overall population and between intervention groups for subgroup with known mutation status Subgroup analysis was reported with data available for all subgroups |
Low |
| Selective outcome reporting | Details on main outcomes reported | Low |
Study: Han (First-SIGNAL) (2012)41
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | No details reported | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | Open label | High |
| Outcome assessor blinding | All measurable and non-measurable lesions were independently assessed by a referee radiologist who was blinded to treatment assignment | Low |
| Incomplete outcome data | Four patients withdrew consent before treatment in SC group; no other withdrawals; 43 were assessable for EGFR mutation status in SC group, 53 in gefitinib group were assessable for EGFR mutation status. Only 23 mutation positive in gefitinib and 16 in SC. Reasons for not assessing mutation status in other patients not stated | High |
| Selective outcome reporting | Details on main outcomes reported | Low |
Study: Maemondo (NEJSG) (2010)47
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | No details reported | Unclear |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | No details reported, but one treatment is oral and the other i.v. | High |
| Outcome assessor blinding | Treatment response and PFS were determined by external review of CT films by experts who were not aware of treatment assignments | Low |
| Incomplete outcome data | Three patients in the standard chemotherapy group were not evaluated in the PFS population (one had a severe allergic reaction to paclitaxel and two withdrew consent) | Low |
| Selective outcome reporting | Details on main outcomes reported | Low |
Studies: Rosell (EURTAC) (2012)40 and Benlloch (EURTAC) (2012)36
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | Central randomisation by an independent clinical research organisation using a computer-generated system, patients registered via fax, stratified randomisation (mutation type and ECOG PS) | Low |
| Allocation concealment | No details reported | Unclear |
| Participant/personnel blinding | States that this was not possible owing to different drug administration routes (oral vs. i.v.) | High |
| Outcome assessor blinding | PFS and treatment responses were confirmed by external review of CT scans by a central review board | Low |
| Incomplete outcome data | Nine patients from the erlotinib group and 10 from the standard chemotherapy group could not be assessed for response (non-measurable disease at baseline or time of response assessment) | Low |
| Selective outcome reporting | Details on main outcomes reported | Low |
Study: Zhou (OPTIMAL) (2011)16
| Support for judgement | Risk of bias | |
|---|---|---|
| Random sequence generation | Central computerised randomisation by an independent clinical research organisation | Low |
| Allocation concealment | Allocation communicated via e-mail or telephone | Low |
| Participant/personnel blinding | States that participants and clinicians were not masked owing to the nature of the treatments | High |
| Outcome assessor blinding | Independent review was not done | High |
| Incomplete outcome data | One patient did not receive erlotinib (no target lesion), 10 patients did not receive standard chemotherapy (nine withdrew consent and one did not start treatment) For treated patients, one patient in the erlotinib group withdrew consent and one was lost to follow-up. In the standard chemotherapy group there were four protocol violations (treated with erlotinib) and four patients were lost to follow-up |
High |
| Selective outcome reporting | Details on main outcomes reported | Low |
Appendix 4 Survey of NHS laboratories in England and Wales participating in the UK NEQAS pilot scheme for epidermal growth factor receptor mutation testing
| LABORATORY DETAILS | ||
|---|---|---|
| This questionnaire has been designed to collect information to inform a NICE diagnostic assessment review on EGFR testing. 1. At which laboratory are you based?
|
||
| EGFR MUTATION TESTING METHODS | ||
| If you use more than one method to test for EGFR mutations in your laboratory, please complete this questionnaire separately for each EGFR mutation test used. | ||
2. Which EGFR sequencing method do you currently use in your laboratory? NB If you use more than one method, please just select one method and then complete the questionnaire again for any other methods
|
||
3. Why have you chosen the EGFR mutation testing method(s) that you have (please select all that apply):
|
||
4. If you use more than one EGFR mutation testing method, what is the reason for using more than one method:
|
||
5. Which mutations does your EGFR mutation testing method aim to detect?
|
||
| LOGISTICS | ||
6. In a typical week, how many samples do you screen for EGFR mutations?
8. How often do you run the EGFR mutation test?
|
||
10. On average, how long (in actual days i.e. including working and non-working days) does it take from receiving a sample at the lab to sending a result back to the clinician?
|
||
| TECHNICAL PERFORMANCE | ||
11. What is the limit of detection of the EGFR mutation test in terms of the % of tumour cells?
|
||
| Total number of samples screened (type 1000 if providing an estimate): | ||
| 13. Total number of failed samples: | ||
| 14. No. of failures due to insufficient tumour cells in sample: | ||
| 15. What are the reasons for failed tests? | ||
| COSTS | ||
| 16. What is the cost of the test (including purchase costs, personnel, material and overheads)? | ||
| 17. If you do not have this information, please provide any information on cost that you have available | ||
| 18. What is the price that you charge for the test? | ||
| 19. Do you have any final comments? | ||
| Thank you for taking the time to complete the survey. If you use more than one EGFR mutation testing method in your laboratory please could you complete the survey again for the other testing methods. | ||
Appendix 5 Table of excluded studies with rationale
| Study | Reason for exclusion (once a study had failed on one criterion it was not assessed further) | |||
|---|---|---|---|---|
| 1. Not a primary study | 2. Did not include adults with chemotherapy naive, locally/regionally advanced/metastatic (IIIB or IV) NSCLC | 3. EGFR mutation test not performed and/or test and mutation not specified or deducible | 4. Study did not report on response to treatment, survival or PFS | |
| Ahn (2008)111 | ✗ | ✓ | ||
| AstraZeneca (2010)109 | ✗ | ✗ | ✗ | ✓ |
| Aydiner (2011)112 | ✓ | |||
| Bria (2011)73 | ✓ | |||
| Cappuzzo (2005)113 | ✗ | ✓ | ||
| Carlson (2009)114 | ✓ | |||
| Chang (2008)115 | ✓ | |||
| Chen (2011)116 | ✓ | |||
| Chen (2012)88 | ✗ | ✗ | ✗ | ✓ |
| Chinese PLA General Hospital (2010)110 | ✗ | ✗ | ✗ | ✓ |
| Chou (2005)117 | ✗ | ✓ | ||
| Chung (2012)118 | ✗ | ✓ | ||
| Cohen (2010)119 | ✓ | |||
| Cohen (2006)120 | ✗ | ✓ | ||
| Cohen (2006)121 | ✗ | ✓ | ||
| Crosby (2011)122 | ✗ | ✓ | ||
| Dahabreh (2010)123 | ✓ | |||
| de Braud (2003)124 | ✗ | ✓ | ||
| De Greve (2011)125 | ✗ | ✓ | ||
| De Pas (2011)126 | ✗ | ✓ | ||
| Dickson (2011)127 | ✗ | ✓ | ||
| Eaton (2011)128 | ✗ | ✓ | ||
| Eberhardt (2011)129 | ✗ | ✗ | ✓ | |
| Edwards (2010)130 | ✓ | |||
| Edwards (2010)131 | ✓ | |||
| Enting (2012)132 | ✗ | ✗ | ✗ | ✓ |
| Feld (2006)133 | ✓ | |||
| Feng (2010)134 | ✗ | ✗ | ✓ | |
| Gao (2012)72 | ✓ | |||
| Gao (2011)135 | ✓ | |||
| Goss (2009)136 | ✗ | ✗ | ✓ | |
| Gracia (2011)137 | ✗ | ✓ | ||
| Gupta (2009)138 | ✓ | |||
| Han (2005)139 | ✗ | ✓ | ||
| Han (2005)140 | ✗ | ✓ | ||
| Han (2006)141 | ✗ | ✓ | ||
| Han (2007)142 | ✗ | ✓ | ||
| Hata (2011)143 | ✗ | ✓ | ||
| Hou (2012)144 | ✗ | ✗ | ✓ | |
| Hsieh (2006)145 | ✗ | ✓ | ||
| Ibrahim (2010)146 | ✓ | |||
| Inoue (2008)147 | ✗ | ✗ | ✗ | ✓ |
| Inoue (2010)148 | ✗ | ✗ | ✓ | |
| Jackman (2009)149 | ✓ | |||
| Johnson (2004)150 | ✗ | ✗ | ✓ | |
| Kasahara (2006)151 | ✗ | ✓ | ||
| Kashii (2006)152 | ✗ | ✓ | ||
| Kim (2011)153 | ✗ | ✓ | ||
| Kimura (2006)154 | ✗ | ✓ | ||
| Kimura (2007)155 | ✗ | ✗ | ✓ | |
| Kris (2009)156 | ✓ | |||
| Ku (2011)157 | ✓ | |||
| Kunimasa (2011)158 | ✗ | ✗ | ✓ | |
| Lee (2011)159 | ✗ | ✗ | ✓ | |
| Lee (2008)160 | ✗ | ✗ | ✓ | |
| Lilenbaum (2008)161 | ✗ | ✗ | ✓ | |
| Liu (2011)162 | ✓ | |||
| Massuti (2009)163 | ✗ | ✓ | ||
| Massuti (2006)164 | ✗ | ✗ | ✗ | ✓ |
| Massuti (2006)165 | ✗ | ✓ | ||
| Meert (2002)166 | ✓ | |||
| Miller (2005)167 | ✗ | ✓ | ||
| Minegishi (2010)168 | ✗ | ✗ | ✗ | ✓ |
| Mitsudomi (2010)70 | ✓ | |||
| Morita (2009)169 | ✓ | |||
| Murray (2010)170 | ✓ | |||
| Murray (2008)171 | ✓ | |||
| Na (2007)172 | ✗ | ✓ | ||
| Naoki (2008)173 | ✗ | ✓ | ||
| Naoki (2011)174 | ✗ | ✓ | ||
| Okamoto (2006)175 | ✗ | ✓ | ||
| Pallis (2007)176 | ✗ | ✓ | ||
| Park (2009)177 | ✗ | ✓ | ||
| Paz-Ares (2009)178 | ✓ | |||
| Paz-Ares (2010)179 | ✓ | |||
| Paz-Ares (2006)180 | ✗ | ✗ | ✗ | ✓ |
| Pesek (2009)181 | ✗ | ✓ | ||
| Petrelli (2012)182 | ✓ | |||
| Petruzelka (2012)183 | ✓ | |||
| Plant (2012)184 | ✗ | ✓ | ||
| Reck (2005)185 | ✗ | ✗ | ✓ | |
| Ricciardi (2008)186 | ✗ | ✓ | ||
| Riely (2006)187 | ✗ | ✓ | ||
| Rizvi (2005)188 | ✗ | ✓ | ||
| Rosell (2009)63 | ✗ | ✓ | ||
| Satouchi (2010)189 | ✗ | ✓ | ||
| Schneider (2008)190 | ✗ | ✓ | ||
| Shih (2006)191 | ✗ | ✓ | ||
| Shukuya (2010)192 | ✓ | |||
| Shukuya (2011)85 | ✓ | |||
| Sone (2007)193 | ✗ | ✓ | ||
| Sun (2011)194 | ✗ | ✓ | ||
| Sunaga (2006)195 | ✗ | ✓ | ||
| Sutani (2006)196 | ✗ | ✓ | ||
| Takano (2006)197 | ✗ | ✓ | ||
| Takano (2005)198 | ✗ | ✓ | ||
| Takano (2007)199 | ✗ | ✓ | ||
| Teck (2008)108 | ✓ | |||
| Tokumo (2005)200 | ✗ | ✓ | ||
| Tsai (2005)201 | ✗ | ✓ | ||
| Tsurutani (2009)202 | ✗ | ✗ | ✓ | |
| Tyagi (2005)203 | ✗ | ✓ | ||
| van Zandwijk (2007)204 | ✗ | ✓ | ||
| Villaflor (2005)205 | ✗ | ✓ | ||
| Wang (2012)71 | ✓ | |||
| Wang (2009)206 | ✗ | ✓ | ||
| Webb (2009)207 | ✗ | ✓ | ||
| Won (2011)208 | ✗ | ✓ | ||
| Wu (2011)209 | ✗ | ✓ | ||
| Wu (2011)210 | ✗ | ✓ | ||
| Wu (2011)211 | ✗ | ✓ | ||
| Wu (2008)212 | ✗ | ✗ | ✗ | ✓ |
| Wu (2007)213 | ✗ | ✓ | ||
| Wu (2006)214 | ✓ | |||
| Xu (2011)215 | ✓ | |||
| Yang (2011)216 | ✓ | |||
| Yoshida (2008)217 | ✓ | |||
| Yoshida (2010)218 | ✗ | ✓ | ||
| Zhang (2011)219 | ✓ | |||
| Zhang (2008)220 | ✗ | ✓ | ||
| Zhong (2011)221 | ✗ | ✓ | ||
Appendix 6 Consistency check with the model used in Single Technology Appraisal 192
Deterministic outcomes for patients with epidermal growth factor receptor mutation-positive tumours as tested with Therascreen EGFR PCR Kit and treated with gefitinib
| Model used | Costs (£)a | QALYsb | LYsc |
|---|---|---|---|
| De novo model | CiC information has been removed | CiC information has been removed | CiC information has been removed |
| Manufacturer model in STA 19251 | CiC information has been removed | 1.111 | CiC information has been removed |
| De novo model with ERG amendmentsd | CiC information has been removed | 1.111 | CiC information has been removed |
Appendix 7 Cost-effectiveness acceptability curves and results for sensitivity analyses
Cost-effectiveness acceptability curve for ‘evidence on comparative effectiveness’ analysis, sensitivity analysis: updated costs
Cost-effectiveness acceptability curve for ‘evidence on comparative effectiveness’ analysis, sensitivity analysis: unknown based on survey
Probabilistic results for ‘linked evidence’ analysis, sensitivity analysis: updated costs
| Strategy | Costs | QALYs | Compared with direct sequencing of all exon 18–21 mutations | ||
|---|---|---|---|---|---|
| Incremental costs (£) | Incremental QALYs | ICER | |||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.905 | –6444 | –0.189 | 34,169 |
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.094 | |||
| Direct sequencing of all exon 19–21 mutations | CiC information has been removed | 1.111 | 685 | 0.018 | 38,659 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | (CiC information has been removed.) | 1.111 | 723 | 0.018 | 41,156 |
| Strategy | Costs | QALYs | Compared with next cost-effective strategy | |||
|---|---|---|---|---|---|---|
| Comparator | Incremental costs (£) | Incremental QALYs | ICER | |||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.905 | ||||
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.094 | Therascreen | 6444 | 0.189 | 34,169 |
| Direct sequencing of all exon 19–21 mutations | CiC information has been removed | 1.111 | Therascreen | 7130 | 0.206 | 34,555 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | CiC information has been removed | 1.111 | Therascreen | 7168 | 0.206 | 34,765 |
Cost-effectiveness acceptability curve for ‘linked evidence’ analysis, sensitivity analysis: updated costs
Probabilistic results for ‘linked evidence’ analysis, sensitivity analysis: unknown based on survey
| Strategy | Costs | QALYs | Compared with direct sequencing of all exon 18–21 mutations | ||
|---|---|---|---|---|---|
| Incremental costs (£) | Incremental QALYs | ICER | |||
| Therascreen EGFR PCR Kit | CiC information has been removed | 0.874 | –8220 | –0.258 | 31,880 |
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.132 | |||
| Direct sequencing of all exon 19–21 mutations | CiC information has been removed | 1.160 | 973 | 0.028 | 35,138 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | (CiC information has been removed.) | 1.159 | 1014 | 0.027 | 37,452 |
| Strategy | Costs | QALYs | Compared with next cost-effective strategy | |||
|---|---|---|---|---|---|---|
| Comparator | Incremental costs (£) | Incremental QALYs | ICER | |||
| Therascreen | CiC information has been removed | 0.874 | ||||
| Direct sequencing of all exon 18–21 mutations | CiC information has been removed | 1.132 | Therascreen | 8220 | 0.258 | 31,880 |
| Direct sequencing of all exon 19–21 mutations | CiC information has been removed | 1.160 | Therascreen | 9194 | 0.286 | 32,196 |
| Direct sequencing or WAVE-HS for inadequate samples (< 50% of tumour cells) | CiC information has been removed | 1.159 | Therascreen | 9234 | 0.285 | 32,409 |
Cost-effectiveness acceptability curve for ‘linked evidence’ analysis, sensitivity analysis: unknown based on survey
Appendix 8 National Institute for Health and Care Excellence guidance relevant to epidermal growth factor receptor mutation testing and the first-line treatment of locally advanced or metastatic non-small cell lung cancer
Clinical guidelines
National Institute for Health and Clinical Excellence. Lung Cancer: the Diagnosis and Treatment of Lung Cancer (CG121). London: NICE; April 2011. 40pp. URL: http://guidance.nice.org.uk/CG121 (accessed 20 June 2012). Date for review: 2014.
Technology appraisals: first-line treatment
National Institute for Health and Clinical Excellence. Pemetrexed for the First-Line Treatment of Non-small-cell Lung Cancer. NICE technology appraisal guidance 181. London: NICE; 2009. 32pp. URL: http://guidance.nice.org.uk/TA181 (accessed 18 December 2012). Date for review: January 2010.
National Institute for Health and Clinical Excellence. Gefitinib for the first-line treatment of locally advanced or metastatic non-small-cell lung cancer. NICE technology appraisal guidance 192. London: NICE; 2010. 45pp. URL: http://guidance.nice.org.uk/TA192 (accessed 20 June 2012). Date for review: April 2013.
Technology appraisals: second-line treatment
National Institute for Health and Clinical Excellence. Erlotinib for the Treatment of Non-small-cell Lung Cancer. NICE technology appraisal guidance 162 [internet]. London: NICE, 2008. 29pp. URL: http://guidance.nice.org.uk/TA162 (accessed 18 December 2012). Date for review: June 2010.
National Institute for Health and Clinical Excellence. Pemetrexed for the Treatment of Non-small-cell Lung Cancer. NICE technology appraisal guidance 124 [internet]. London: NICE, 2007. 20pp. URL: http://guidance.nice.org.uk/TA124 (accessed 18 December 2012). Date for review: January 2010.
National Institute for Health and Clinical Excellence. Erlotinib for the First-line Treatment of Locally Advanced or Metastatic EGFR-TK Mutation-positive Non-small cell Lung Cancer. NICE technology appraisal guidance 258 [internet]. London: NICE, 2012. 43pp. URL: http://guidance.nice.org.uk/TA258 (accessed 18 December 2012).
Technology Appraisals: maintenance treatment
National Institute for Health and Clinical Excellence. Pemetrexed for the Maintenance Treatment of Non-small-cell Lung Cancer. NICE technology appraisal guidance 190. London: NICE; 2010. 29pp. URL: http://guidance.nice.org.uk/TA190 (accessed 18 December 2012). Date for review: November 2012.
National Institute for Health and Clinical Excellence. Erlotinib Monotherapy for Maintenance Treatment of Non-small-cell Lung Cancer. NICE technology appraisal guidance 227. London: NICE; 2011. 52pp. URL: http://guidance.nice.org.uk/TA227 (accessed 10 July 2012). Date for review: April 2013.
Under development
National Institute for Health and Care Excellence (NICE). Crizotinib for Previously Treated Non-small-cell Lung Cancer Associated with an Anaplastic Lymphoma Kinase Fusion Gene. NICE technology appraisal guidance 296. London: NICE, 2013. URL: http://publications.nice.org.uk/crizotinib-for-previously-treated-non-small-cell-lung-cancer-associated-with-an-anaplastic-lymphoma-ta296 (accessed 27 February 2014).
Appendix 9 PRISMA checklist
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| TITLE | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both | Title page, p. i |
| ABSTRACT | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number | Executive summary, pp. xvii–xxi PROSPERO registration, p. xxi |
| INTRODUCTION | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known | Chapter 2, Background, pp. 3–8 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS) | Chapter 1, Objective, p. 1 |
| METHODS | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g., Web address), and, if available, provide registration information including registration number | PROSPERO registration: CRD42012002828 URL: www.crd.york.ac.uk/prospero/display_record.asp?ID=CRD42012002828 |
| Eligibility criteria | 6 | Specify study characteristics (e.g. PICOS, length of follow-up) and report characteristics (e.g. years considered, language, publication status) used as criteria for eligibility, giving rationale | Table 2, p. 11 |
| Information sources | 7 | Describe all information sources (e.g. databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Search strategy, pp. 9–10 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated | Appendix 1 |
| Study selection | 9 | State the process for selecting studies (i.e. screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis) | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Inclusion screening and data extraction, p. 10 |
| Data collection process | 10 | Describe method of data extraction from reports (e.g. piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Inclusion screening and data extraction, pp. 10–11 |
| Data items | 11 | List and define all variables for which data were sought (e.g. PICOS, funding sources) and any assumptions and simplifications made | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Inclusion screening and data extraction, pp. 10–11 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Quality assessment, pp. 11–12 |
| Summary measures | 13 | State the principal summary measures (e.g. risk ratio, difference in means) | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Methods of analysis/synthesis, p. 12 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (e.g. I2) for each meta-analysis | Chapter 3, Assessment of clinical effectiveness, Systematic review methods, Methods of analysis/synthesis, p. 12 |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (e.g. publication bias, selective reporting within studies) | NA |
| Additional analyses | 16 | Describe methods of additional analyses (e.g., sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified | NA |
| RESULTS | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram | Chapter 3, Assessment of clinical effectiveness, Results of the assessment of clinical effectiveness, pp.13–14 Figure 1, p. 13 |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (e.g. study size, PICOS, follow-up period) and provide the citations | Table 7, p. 24 Table 10, p. 28 Appendix 2 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12) | Table 9, p. 27 Figure 5, p. 27 Table 11, p. 32 Figure 9, p. 33 Appendix 3 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (a) simple summary data for each intervention group (b) effect estimates and CIs, ideally with a forest plot | Table 7, p. 24 Figure 4, p. 23 Table 10, p. 28 Figures 6–8, p. 30–31 |
| Synthesis of results | 21 | Present results of each meta-analysis done, including CIs and measures of consistency. | NA |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see Item 15) | NA |
| Additional analysis | 23 | Give results of additional analyses, if done [e.g. sensitivity or subgroup analyses, meta-regression (see Item 16)] | NA |
| DISCUSSION | |||
| Summary of evidence | 24 | Summarise the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (e.g. health-care providers, users, and policy-makers | Chapter 5, Discussion, Statement of principal findings, pp. 63–67 |
| Limitations | 25 | Discuss limitations at study and outcome level (e.g. risk of bias), and at review-level (e.g. incomplete retrieval of identified research, reporting bias) | Chapter 5, Discussion, Strengths and limitations of the assessment, Uncertainties, pp. 67–73 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research | Chapter 6, Conclusions, pp. 75–76 |
| FUNDING | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (e.g. supply of data); role of funders for the systematic review | p. ii |
Glossary
- Cost-effectiveness analysis
- An economic analysis that converts effects into health terms and describes the costs for additional health gain.
- Decision modelling
- A theoretical construct that allows the comparison of the relationship between costs and outcomes of alternative health-care interventions.
- False negative
- Incorrect negative test result – number of diseased persons with a negative test result.
- False positive
- Incorrect positive test result – number of non-diseased persons with a positive test result.
- Incremental cost-effectiveness ratio
- The difference in the mean costs of two interventions in the population of interest divided by the difference in the mean outcomes in the population of interest.
- Index test
- The test whose performance is being evaluated.
- Markov model
- An analytic method particularly suited to modelling repeated events, or the progression of a chronic disease over time.
- Meta-analysis
- Statistical techniques used to combine the results of two or more studies and obtain a combined estimate of effect.
- Meta-regression
- Statistical technique used to explore the relationship between study characteristics and study results.
- Metastasis
- The spread of a disease from one organ or part to another non-adjacent organ or part.
- Opportunity costs
- The cost of foregone outcomes that could have been achieved through alternative investments.
- Publication bias
- Bias arising from the preferential publication of studies with statistically significant results.
- Quality-adjusted life-year
- A measure of health gain, used in economic evaluations, in which survival duration is weighted or adjusted by the patient’s quality of life during the survival period.
- Quality of life
- An individual’s emotional, social and physical well-being and their ability to perform the ordinary tasks of living.
- Receiver operating characteristic curve
- A graph that illustrates the trade-offs between sensitivity and specificity which result from varying the diagnostic threshold.
- Reference standard
- The best currently available diagnostic test, against which the index test is compared.
- Sensitivity
- Proportion of people with the target disorder who have a positive test result.
- Specificity
- Proportion of people without the target disorder who have a negative test result.
- True negative
- Correct negative test result – number of non-diseased persons with a negative test result.
- True positive
- Correct positive test result – number of diseased persons with a positive test result.
List of abbreviations
- ARMS
- amplification refractory mutation system
- ASCO
- American Society of Clinical Oncology
- CI
- confidence interval
- CiC
- commercial-in-confidence
- CR
- complete response
- CRD
- Centre for Reviews and Dissemination
- CT
- computed tomography
- DC
- disease control
- DNA
- deoxyribonucleic acid
- EBUS
- endobronchial ultrasound
- ECCO
- European Cancer Congress
- EGFR
- epidermal growth factor receptor
- EGFR-TK
- epidermal growth factor receptor tyrosine kinase
- EGFR-TKI
- epidermal growth factor receptor tyrosine kinase inhibitor
- ERG
- External Review Group
- ESMO
- European Society of Medical Oncology
- EUS
- endoscopic ultrasound
- FFPE
- formalin-fixed and paraffin-embedded
- FN
- false negative
- FNA
- fine-needle aspiration
- FP
- false positive
- HR
- hazard ratio
- HRM
- high-resolution melt
- IC
- incremental cost
- ICER
- incremental cost-effectiveness ratio
- IPD
- individual patient data
- IQR
- interquartile range
- ITT
- intention to treat
- i.v.
- intravenous
- LY
- life-year
- MRI
- magnetic resonance imaging
- NEQAS
- National External Quality Assurance Scheme
- NICE
- National Institute for Health and Care Excellence
- NLCA
- National Lung Cancer Audit
- NSCLC
- non-small cell lung cancer
- OR
- objective response
- OS
- overall survival
- PCR
- polymerase chain reaction
- PD
- progressive disease
- PET
- positron emission tomography
- PFS
- progression-free survival
- PR
- partial response
- PRESS-EBC
- Peer Review of Electronic Search Strategies Evidence-Based Checklist
- PS
- performance status
- PSSRU
- Personal Social Services Research Unit
- QALY
- quality-adjusted life-year
- RCT
- randomised controlled trial
- RECIST
- Response Evaluation Criteria in Solid Tumours
- ROC
- receiver operating characteristic
- RR
- relative risk
- SD
- stable disease
- SE
- standard error
- SROC
- summary receiver operating characteristic
- TBNA
- transbronchial needle aspiration
- TKI
- tyrosine kinase inhibitor
- TN
- true negative
- TP
- true positive
- WHO
- World Health Organization
Note
This monograph is based on the Technology Assessment Report produced for NICE. The full report contained a considerable number of data that were deemed commercial-in-confidence. The full report was used by the Appraisal Committee at NICE in their deliberations. The full report with each piece of commercial-in-confidence data removed and replaced by the statement ‘commercial-in-confidence information (or data) removed’ is available on the NICE website: www.nice.org.uk. The present monograph presents as full a version of the report as is possible while retaining readability, but some sections, sentences, tables and figures have been removed. Readers should bear in mind that the discussion, conclusions and implications for practice and research are based on all the data considered in the original full NICE report.