Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 12/14/01. The protocol was agreed in July 2012. The assessment report began editorial review in March 2013 and was accepted for publication in October 2013. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
none
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2015. This work was produced by Huxley et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Background
Nature of the disease
Breast cancer affects the breast tissue of either women or men, although breast cancer in men is relatively uncommon. In breast cancer, cells making up the breast begin to divide in an uncontrolled manner forming tumours. Initially they are confined within the structures of the breast where the cells would normally occur and the tumour is referred to as ductal carcinoma in situ (DCIS). Later the cells become locally invasive extending into parts of the breast and surrounding tissue where they would not normally occur. The breast cancer cells also enter the lymph drainage system, which leads to breast cancer cells lodging and growing in lymph nodes (LNs), particularly those in the armpit – the axillary lymph nodes (ALNs). These are called regional metastases. ALNs containing local metastases can vary from a near-normal size of ≤ 0.5 cm in length to a greatly enlarged, 4 cm in length.
In addition, breast cancer cells can enter the bloodstream from where they can spread to distant parts of the body such as the lungs, bones and the liver. These are called distant metastases. Without treatment the proliferation of the breast cancer cells and their spread around the body leads to death. The reasons why breast cancer develops are not completely understood, although genetic predisposition has been increasingly recognised as a contributing factor in a minority of women. 1
Breast cancer is a very important challenge to health. Each year in England2 and Wales3 approximately 40,000 individuals develop breast cancer. As illustrated in Table 1, the majority of these are women aged > 40 years. Breast cancer is responsible for >10,000 deaths each year in England and Wales,4 although the mortality rate has been declining from a peak in the mid-1980s (Figure 1). Breast cancer is the most commonly occurring female cancer and the second most common cause of cancer death after lung cancer, accounting for one in six of all female cancer deaths. Approximately one in nine women will develop breast cancer at some stage of their life.
| Population and country | Age (years) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10–39 | 40–44 | 45–49 | 50–54 | 55–59 | 60–64 | 65–69 | 70–74 | 75–79 | 80–84 | 85+ | Total | |
| Women, Walesa | 90 | 133 | 241 | 278 | 280 | 358 | 357 | 219 | 262 | 184 | 223 | 2625 |
| Women, Englandb | 1798 | 2528 | 4164 | 4637 | 4005 | 5618 | 5009 | 3430 | 3492 | 3073 | 3505 | 41,259 |
| Men, Walesa | 0 | 0 | 0 | 0 | 4 | 1 | 2 | 5 | 0 | 2 | 1 | 15 |
| Men, Englandb | 7 | 11 | 11 | 19 | 30 | 37 | 47 | 59 | 47 | 48 | 37 | 353 |
| Total | 1895 | 2672 | 4416 | 4934 | 4319 | 6014 | 5415 | 3713 | 3801 | 3307 | 3766 | 44,252 |
FIGURE 1.
Age-standardised rates for female breast cancer mortality in the UK, 1971–2010. 4 Reproduced with permission from Cancer Research UK. URL: www.cancerresearchuk.org/cancer-info/cancerstats/types/breast/mortality/uk-breast-cancer-mortality-statistics#trends (accessed 5 August 2014).

Staging of breast cancer
There is a well-defined system for recording the severity and extent of the spread of breast cancer. This is called the TNM (tumour, node, metastasis) classification and is based on tumour size, spread to LNs and presence of distant metastases5–8 (Tables 2 and 3).
| Stage | Description |
|---|---|
| T: tumour stage | |
| Tx | Primary tumour cannot be assessed |
| T0 | No evidence of primary tumour |
| Tis | Carcinoma in situ |
| T1 | Tumour ≤ 2 cm across |
| T2 | Tumour > 2 cm to 5 cm across |
| T3 | Tumour > 5 cm across |
| T4 | Tumour of any size with direct extension to skin or chest wall, or inflammatory breast cancer |
| N: LN stage | |
| Nx | Nodal stage cannot be assessed |
| N0 | No metastases to any ipsilateral LNs |
| N1 | Metastases to one to three axillary nodes or axillary nodes that are mobile |
| N2 | Metastases to four to nine axillary nodes or axillary nodes that are fixed to one another or other structures or clinically apparent metastases to internal mammary nodes |
| N3 | Metastasis to nodes above or below the collarbone (supraclavicular/infraclavicular) or to both axillary and internal mammary nodes, or to 10+ axillary nodes |
| M: metastasis stage | |
| Mx | Presence of metastases cannot be assessed |
| M0 | No distant metastases |
| M1 | Distant metastases |
| Stage | T | N | M |
|---|---|---|---|
| 0 (DCIS/LCIS) | Tis | N0 | M0 |
| I | T1 | N0 | M0 |
| IIA | T0–1 | N1 | M0 |
| T2 | N0 | M0 | |
| IIB | T2 | N1 | M0 |
| T3 | N0 | M0 | |
| IIIA | T0–2 | N2 | M0 |
| T3 | N1–2 | M0 | |
| IIIB | T4 | N0–2 | M0 |
| IIIC | T (any) | N3 | M0 |
| IV | T (any) | N (any) | M1 |
Increasing size of the main tumour and spread to ALNs are two of the key features denoting worsened stage. The presence of distant metastases denotes the worst stage IV, irrespective of the size of the main tumour or spread to regional LNs. The majority of patients present with stage I or stage II cancer, as illustrated in Table 4. 10 These figures, combined with information on how tumour size inter-relates with nodal status, suggest that in the UK 33% of patients present with spread to LNs. 11 This agrees with a systematic review12 including 13 studies undertaken to inform the National Institute for Health and Care Excellence (NICE) clinical guideline13 on early and locally advanced breast cancer, which reported a mean prevalence of LN-positive status of 31.4%, with a range of 18–59%. Other sources have suggested a higher value of 41% for the percentage of patients presenting with spread to LNs. 9
| Stage | n | % among all patients | % among patients with known stage |
|---|---|---|---|
| I | 6788 | 38 | 41 |
| II | 7361 | 41 | 45 |
| III | 1490 | 8 | 9 |
| IV | 821 | 5 | 5 |
| Unknown | 1376 | 8 | N/A |
| All (excluding unknown) | 17,836 (16,460) |
By definition, early breast cancer is cancer that has not spread beyond the breast or the ALNs on the same side as the tumour, that is, stages I (any), II (any) or IIIA.
Recent modifications to the staging system for breast cancer also recognise the size of the local metastases in the LNs. Macrometastases are defined as tumour deposits in which one dimension is > 2 mm. Micrometastases, deposits that are only discernible microscopically, measure > 0.2 mm with no dimension being > 2 mm. Isolated tumour cells (ITCs) are also recognised as part of the N0 category and by definition no dimension of any collection of ITCs must exceed 0.2 mm.
Prognosis
With early identification and treatment the outlook for patients with breast cancer is good. The overall 5-year survival rate is approximately 80%. 14 The survival rate varies with age, with patients aged > 80 years having survival rates of < 80%, as shown in Table 5.
| Age (years) | |||||||
|---|---|---|---|---|---|---|---|
| 15–39 | 40–49 | 50–59 | 60–69 | 70–79 | 80–99 | All ages | |
| 5-year survival rate (%) | 84 | 89 | 90 | 90 | 81 | 69 | 85 |
Stage also influences survival, as shown in Table 6. In women diagnosed with breast cancer in the West Midlands between 1985 and 1989 and followed up until 1999, the 5-year survival rate was 88% for stage I, 69% for stage II, 43% for stage III and 12% for stage IV. 15 This pattern is maintained in data from the National Cancer Data Base in the USA for patients who were diagnosed in 2001 and 2002. 16
| Data source | Stage of disease | ||||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | ||||
| IIA | IIB | IIIA | IIIB | IIIC | |||
| UKa | 88 | 69 | 43 | 12 | |||
| USAb | 88 | 81 | 74 | 67 | 41 | 49 | 15 |
In addition to age, tumour size and spread, prognosis is also related to tumour grade and receptor status. With respect to the latter, positive oestrogen receptor status and positive progesterone receptor status denote better prognosis and overexpression of human epidermal growth factor 2 denotes poorer prognosis. These additional factors are incorporated into tools to estimate prognosis, such as the Nottingham Prognostic Index17 and Adjuvant! Online,18 which are used to guide treatment, particularly the use of adjuvant chemotherapy in early breast cancer. The effectiveness and cost-effectiveness of gene expression profiling and expanded immunochemistry tests to enhance the Nottingham Prognostic Index17 and Adjuvant! Online18 are currently under consideration by the NICE Diagnostic Assessment Committee. 19
The implications for the prognosis of micrometastases compared with macrometastases are currently unclear, but micrometastases are counted as local metastasis and are treated in a similar manner to macrometastases (see Clinical pathway for staging of breast cancer and subsequent surgery to the axilla and Other approaches to treating the spread of breast cancer tumour cells beyond the sentinel lymph nodes). In contrast, ITCs are not currently counted as local metastasis and they are assumed to have a similar prognosis to no LN metastases (N0). 20
Management of disease
General clinical pathway for suspected breast cancer
The tests and treatment advised for patients with suspected breast cancer are outlined in the NICE clinical guideline on early and locally advanced breast cancer. 13 This is summarised as an algorithm and reproduced in Figure 2.
FIGURE 2.
Clinical pathway for breast cancer. a, Following the publication of the Cancer Reform Strategy,21 by December 2009 all patients presenting with breast problems and referred by their general practitioner to a specialist should be seen within 2 weeks in England. b, Include repeat core biopsy/open biopsy/magnetic resonance imaging, etc. c, Not all patients will require staging. 22 d, For elderly or unfit patients, surgery may not be appropriate. For locally advanced but not metastatic cancer, primary systemic therapy precedes therapeutic surgery in order to reduce the size of the tumour. e, Could include breast conservation (wide local incision), mastectomy and axillary staging (SLNB, sampling or clearance). FNAC, fine-needle aspiration cytology; MDT, multidisciplinary team. Source: National Institute for Health and Care Excellence. 13

In the initial assessment, anyone suspected of breast cancer, whether through primary care or through the breast screening programme, is referred to an assessment clinic where the clinical examination is repeated, mammography and ultrasound are undertaken and a biopsy of the lesion(s) is performed (usually a core biopsy). Ultrasound of the axilla is performed and further biopsies of suspicious LNs are taken (fine-needle aspirations or core). A multidisciplinary team (MDT) meeting considers the results from these investigations and makes a definitive recommendation regarding proposed management to discuss with the patient.
If breast cancer is detected, surgery is usually performed to remove the cancer unless neoadjuvant therapy is considered appropriate. Staging of the axilla is performed in cases of invasive breast cancer in the manner described in the next section. All findings, including the pathology of the removed tumour and the results of any staging procedures, are then discussed at a further MDT meeting where decisions are made about further surgery and whether adjuvant hormone therapy, chemotherapy, immunotherapy or radiotherapy is required.
Clinical pathway for staging of breast cancer and subsequent surgery to the axilla
The detailed steps for investigating if a suspected breast cancer has spread to the axilla and the degree of that spread are also outlined in the NICE guideline. 13 These are summarised in Figure 3. Further information on key steps in this pathway is given in the following sections.
FIGURE 3.
Clinical pathway for staging of breast cancer and subsequent surgery to the axilla. –, negative; +, positive.

Four different scenarios are recognised depending on the findings of the initial assessment:
-
Likely DCIS. Provided axillary ultrasound reveals no abnormalities and the patient is not considered high risk, no further investigations of the axilla are undertaken.
-
Likely invasive breast cancer with negative axillary ultrasound (no abnormal LNs identified). In these patients, at the time of the surgical removal of the main breast tumour, a sentinel lymph node biopsy (SLNB) is undertaken. In a SLNB a weakly radioactive solution and a blue dye are injected into the breast before surgery to identify the first LN(s) to which the breast drains lymph in a particular individual. The sentinel lymph nodes (SLNs) become blue and/or can be detected using a radioactivity counter. They are most frequently found in the axilla of the same side. These SLNs are then removed to determine if the cancer has spread from the original site. This is carried out by histopathology, which involves cutting very thin slices of the SLNs, staining them and then a medically qualified specialist carefully examining them under a microscope. Histopathology takes several days and sometimes further investigations are required for a definitive diagnosis, for example immunohistochemistry. If no breast cancer cells meeting the criteria for metastasis or single ITC are found in the SLN(s) no further action needs to be taken. However, if the breast cancer has spread to the SLNs, all of the relevant LNs in the axilla need to be removed in a further operation called axillary lymph node dissection (ALND). This provides treatment by removing all of the tumour cell-bearing LNs and others within a defined anatomical boundary and it provides detailed staging information by allowing the number of LNs with metastases to be precisely quantified, as the LNs removed are subjected to further histopathological examination. Occasionally, a SLNB cannot be undertaken. In this case four-node sampling, in which four LNs are removed and examined without specific evidence that they are the SLNs, may be performed instead to provide further information.
-
Likely invasive breast cancer with positive axillary ultrasound but normal ultrasound-guided fine-needle aspiration cytology (FNAC). At the time of the surgical removal of the main breast cancer a SLNB is undertaken, followed by the same actions as in (b) if cancer is not found or is confirmed to have spread to the SLNs.
-
Likely invasive breast cancer with positive axillary ultrasound and confirmed abnormality on ultrasound-guided FNAC. In this case the patient would proceed directly to ALND without undergoing a SLNB.
The preference in NICE guidance for the use of a strategy employing ultrasound and FNAC to triage the need for a SLNB was underpinned by a model-based cost-effectiveness analysis undertaken as part of the preparation of the NICE clinical guideline on early and locally advanced breast cancer. 13
Accuracy of clinical examination
Clinical examination of the axilla involving palpation is subject to error as a method of detecting the spread of breast cancer to the ALNs. The sensitivity of the technique has been estimated as 46% based on pooling of a number of studies. 24–29 The fact that over half of axillary metastases are not detected by palpation is the reason why additional investigations are required. The main reason why palpation is unsuccessful is often because the presence of metastases does not always lead to a change in size or texture of the ALNs, coupled with the fact that the ALNs are not always easy to examine.
Accuracy of ultrasound-guided fine-needle aspiration cytology
Like palpation, ultrasound and FNAC are imperfect techniques. Their accuracy was considered in detail as part of the NICE clinical guideline on early and locally advanced breast cancer (guidance 1.1.3 on preoperative staging of the axilla13 and section 2.2, pages 77–8, of the evidence review report12).
The key facts identified were:
-
LNs can be visualised by ultrasound in 81% of cases, although there is considerable variation.
-
Using LNs that were suspicious on ultrasound based on their size (>5mm) and configuration as the diagnostic criteria, sensitivity was 69% and specificity was 75% when patients with palpable and non-palpable ALNs were combined. The accuracy was improved when cases with palpable LNs only were included and was worse when cases with non-palpable LNs were included.
-
The staging performance of ultrasound-guided FNAC was a sensitivity of 43% and a specificity of 100%, with an accompanying positive predictive value (PPV) of 99% and a negative predictive value (NPV) of 72%.
Accuracy and adverse effects of axillary lymph node dissection
Axillary lymph node dissection, described earlier, has been considered the ‘gold standard’ procedure for staging the axilla. It is very accurate in establishing the presence of axillary disease and has the therapeutic advantage of being associated with a high long-term local disease control rate. 9
However, ALND is associated with significant complications, including a 21% incidence of arm lymphoedema (general swelling),29–31 a 22% incidence of seromas (pockets of fluid under the skin)29,32 and a 14% infection rate. 29,33 In addition, insertion of a surgical drain during surgery is commonplace (79%) and usually necessitates prolongation of hospital stay. 33 Pain, limited mobility, numbness and sensory loss are also common. In total, 80% of women are claimed to suffer some adverse event. 34 It is for these reasons that there has been a focus on performing ALND for its therapeutic effects, applicable in patients with the spread of breast cancer to the ALNs, rather than as a more widely applied diagnostic tool.
Accuracy and adverse effects of sentinel lymph node biopsy
Sentinel lymph node biopsy, described earlier, although still a surgical procedure of the axilla, is much simpler than ALND and is associated with a much lower rate of side effects. Thus, the incidence of lymphoedema falls to 7%,35 seroma to 7%29,32 surgical drain requirement to 2%33,36 and infection incidence to 2%. 29,33,36
Sentinel lymph node biopsy was considered in detail as part of the NICE clinical guideline on early and locally advanced breast cancer (guidance 1.4.1–1.4.4 on surgery to the axilla, invasive breast cancer,13 and section 3.3, pages 308–10, of the evidence review report12). The key features identified were:
-
The overall SLN localisation rate was 96.4%.
-
The pooled estimate of the false-negative rate was 7%, that is, sensitivity is 93%.
-
The mean proportion of patients with positive SLNs was 42%.
-
Patients treated by SLNB do not appear to have poorer rates of disease-free survival or overall survival, or of axillary recurrence in the short term, than patients treated by axillary clearance employing ALND.
Challenge to measuring accuracy: tissue allocation bias
Accuracy indicates the degree to which a test of interest correctly identifies if a patient/sample has the target disease (its sensitivity) or correctly identifies that the patient/sample does not have the disease (its specificity), the true disease state being identified by applying a reference standard to all patients/samples.
A challenge for measuring the accuracy of tests aiming to identify if breast cancer has spread to ALNs is that the tumour cells are not evenly distributed throughout the LN. Further, the tests of interest often consume the LN so that once it has been used for one test it cannot be used by another, necessitating that the sample be partitioned if multiple tests are to be carried out. Thus, apparent errors in accuracy may be introduced not just because a new test truly fails to identify tumour cells that are present, but also because the portion of the LN used in the test was not the portion that contained the tumour cells. This is referred to as tissue allocation bias (TAB) or sampling bias, which can lead to underestimation of sensitivity when the portion allocated to the new test does not contain the tumour cells. This is illustrated in Figure 4. Underestimation of specificity can also occur if the new test is allocated a portion of the LN containing tumour cells and the reference standard is allocated the portion without the tumour cells.
FIGURE 4.
Diagram illustrating the effect of TAB or sampling bias.

The impact of TAB needs careful consideration both in the context of the particular new technologies under consideration and because histopathology is a commonly used reference test and it is itself affected by TAB as the portion of the LN submitted to histopathology may not contain the tumour cells. This is further complicated by the fact that histopathology examines only a finite number of the slices in the portion of the LN allocated to it, which, even though likely to be equally spaced throughout the LN portion, do not represent all of it.
The implications of TAB are revisited later in this chapter and throughout the report.
Other approaches to treating the spread of breast cancer tumour cells beyond the sentinel lymph nodes
The current management indicated above, particularly that micrometastases and macrometastases in the SLNs should be followed by ALND, is based on the consensus at the time the current NICE guideline on the subject was published in 2009.
Pre-eminently this has been precipitated by the results of a randomised controlled trial comparing ALND with no ALND in women with invasive breast cancer and SLN spread – the American College of Surgeons Oncology Group Z0011 trial. 37 The participants were adult women with histologically confirmed invasive breast cancer with a clinical size of ≤ 5 cm, no palpable adenopathy and a SLN metastatic breast cancer documented by frozen section, touch preparation or haematoxylin–eosin staining on permanent section. They were ineligible if they had three or more positive SLNs, matted nodes or gross extranodal disease or if they had received neoadjuvant hormonal therapy or chemotherapy. In total, 445 women were randomly allocated to ALND following SLNB and 446 were randomly allocated to no ALND following SLNB, although this number of participants was well below the target for recruitment (total of 1900). At a median follow-up of 6.3 years, 5-year overall survival was 91.8% with ALND and 92.5% without ALND. This is equivalent to an adjusted hazard ratio (HR) of 0.87 [95% confidence interval (CI) 0.62 to 1.23]. At the same follow-up, 5-year disease-free survival was 82.2% with ALND and 83.9% without ALND. This is equivalent to an adjusted HR of 0.88 (95% CI 0.65 to 1.25). The conclusion was that, among patients with limited SLN metastatic breast cancer treated with breast conservation and systemic therapy, the use of SLNB alone compared with SLNB and ALND did not result in inferior survival. Although the lack of power of the study and the limited radiotherapy quality assurance are important provisos, it does explain why there is growing caution about the use of ALND when the amount of spread to the SLNs is limited and women are also likely to receive adjuvant therapy.
In addition to possible changes in the use of ALND, there is also debate about the role of axillary irradiation as an alternative to ALND when SLNB is positive. Currently there are no prospective data to guide indications for axillary irradiation in the absence of ALND. The results of the ongoing AMAROS (After Mapping of the Axilla, Radiotherapy or Surgery?) trial38 comparing axillary irradiation with ALND after SLNB are awaited. In the absence of level 1 evidence, pragmatic recommendations for locoregional irradiation have been suggested. 39 It may be appropriate, for example, to consider axillary irradiation after SLNB for patients with low-volume macrometastases (one to two positive nodes) or high-risk micrometastases. It should be noted that there are no data on the role of axillary irradiation after a positive sentinel node analysed by one-step nucleic acid amplification (OSNA) or equivalent technologies.
Description of the technologies under assessment
Rationale
One of the problems with current practice with respect to investigating whether breast cancer has spread to the ALNs is the need to wait for the histopathology results from the SLNB indicating whether or not spread has occurred. Thus, if spread to the ALNs has occurred, there is inevitably a delay before performing ALND relative to a situation in which excision of the suspected breast cancer and ALND could be carried out in one operation. Also, the individual must be admitted to hospital, receive an anaesthetic and be operated on for a second time. The ALND itself may be more difficult because of the recent previous operation and, as a result, complication rates may be higher than if the ALND immediately followed the SLNB. Adjuvant treatments, if required, may also be delayed.
Two new methods of examining LNs removed in SLNB intraoperatively, similar in approach, have been claimed as ways to achieve SLNB followed immediately by ALND when required, avoiding any delays and a second operation. However, the new methods may limit the opportunity for a MDT to consider the appropriateness of ALND, taking account of all of the information that could potentially be available at the time of the second operation under current practice, unless each possible outcome has been discussed preoperatively.
One-step nucleic acid amplification
The RD-100i OSNA system (henceforth referred to as OSNA) (Sysmex, Norderstedt, Germany) is an automated molecular test that uses OSNA technology. The test analyses and amplifies genetic material (messenger ribonucleic acid, mRNA) from solubilised biopsy samples of SLN tissue and detects the presence of the cytokeratin-19 (CK19) gene, a biological marker associated with breast cancer and not normally present in LN tissue. It is claimed that OSNA will provide a result within a short time and can therefore be used during breast surgery to determine whether other LNs should be removed at the same time as the initial tumour.
One-step nucleic acid amplification does not require the mRNA to be extracted from the tissue and purified before being analysed. The expression level of CK19 mRNA correlates with the size of the LN tumour cell foci. As the foci may not be evenly distributed throughout the node, the system provides more accurate results if more of the node is analysed because there is less risk of TAB (sample bias). The result is most accurate if the entire node is used but in this case no follow-up histopathology is possible. The system can be used with half of the LN (one piece or alternate slices), allowing for the possibility of follow-up histopathology but potentially decreasing the accuracy of the results because of the increased risk of TAB. The time to receive results is dependent on the number of LNs analysed, but the test takes approximately 30–45 minutes. This includes the time to prepare the LNs, dissect them out and trim away fat, solubilise them and run the test. The OSNA test result is expressed both quantitatively and qualitatively; – for LN-negative test results, + for LNs with a micrometastatic tumour burden (> 250 copies of CK19 mRNA/µl) and ++ for LNs with a macrometastatic tumour burden (> 5000 copies of CK19 mRNA/µl). Thus, 250 copies of CK19 mRNA/µl is the threshold or cut-off level defining the tumour load in the SLNs, above which further treatment with ALND is triggered.
The analyser amplifies and detects the CK19 mRNA by using six different primers that have been specifically designed to avoid the amplification of CK19 pseudogenes or their transcripts; amplification of these would lead to false-positive results. Undesired amplification of genomic deoxyribonucleic acid (DNA) is avoided by precipitation of DNA at low pH during sample preparation and by using an isothermal reaction temperature of 65°C.
The manufacturer estimates that 1% of breast tumours do not express CK19 mRNA and therefore, if cancer spreads to the LNs from these tumours, CK19 mRNA will not be detected even though the LNs are metastatic. Prescreening of tumour biopsies for CK19 expression could be carried out before using the OSNA test to reduce the small risk of false-negative results for SLNs with actual tumour cell foci.
A (Conformitée Européenne) CE mark has been obtained for this technology.
Metasin
The Metasin test is an intraoperative molecular test developed within the NHS at the Princess Alexandra Hospital in Harlow, Essex. The claims for its effect on the management of patients with breast cancer are similar to those for OSNA. The test has similarities to a discontinued commercial test (GeneSearch BLN assay; Veridex, Warren, NJ, USA) and uses the technique of quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) to detect two predictive markers of metastases, CK19 and mammaglobin. Mammaglobin is expressed mainly by breast epithelial cells and high levels are associated with breast cancer. A reference gene, porphobilinogen deaminase (PBGD), is used to confirm the validity of the mRNA used in the test and two other controls, positive and negative, are also included. The test uses reagents that can be purchased from Roche and QIAGEN and can be used on any platform (PCR machine). This in-house test differs from the discontinued commercial test by using distinctly different and unique primer–probe combinations to detect the CK19 and mammaglobin genes. It is reported to take 6–10 minutes to extract and purify mRNA from the tissue followed by 26 minutes to obtain the results of the test.
Although somewhat unclear, Metasin appears to be semiquantitative according to Sundaresan. 40 The thresholds are calculated by crossing-point (Cp) values, the points at which the fluorescence from the DNA-associated fluorescent probes increase above background during amplification. The values for micrometastasis are quoted as Cp > 25 and < 32 for CK19 and Cp > 25.9 and < 32 for mammaglobin. Presumably, the values for macrometastasis are above these ranges.
Prescreening of tumour biopsies for CK19 and mammaglobin mRNA expression could be carried out before using the Metasin test because, like the CK19 biomarker, mammaglobin is not expressed in all breast tumours. The proportion of breast cancer tumours that do not express mammaglobin mRNA is not known.
A CE mark has very recently been obtained for this technology.
Proposed clinical pathway
Either OSNA or Metasin could be used as a replacement for current normal practice, in which case the SLNs are used in their entirety. However, OSNA or Metasin could also be used adjunctively, in which half of each LN (one piece or alternate slices) is used for OSNA or Metasin intraoperative testing and the remaining half is examined using standard postoperative histopathology. Where the new tests have been introduced in practice the first model is the most commonly followed as it maximises the claimed benefits of the new technology.
The proposed clinical pathways are illustrated in Figures 5 and 6. This emphasises that changes to the clinical pathway occur only in patients whose ultrasound is negative or in patients whose ultrasound is positive but whose FNAC is negative.
FIGURE 5.
Proposed clinical pathway for staging of breast cancer and subsequent surgery to the axilla: OSNA or Metasin used as a replacement for SLNB. a, In cases in which the OSNA/Metasin test does not provide a result or provides a result that is uninterpretable, the OSNA/Metasin test would first be repeated using the remaining solubilised LN. Failing this, four-node sampling could be carried out, followed by delayed ALND in the case of positive LNs being identified in the sample or immediate ALND with the patient’s prior informed consent for contingency.
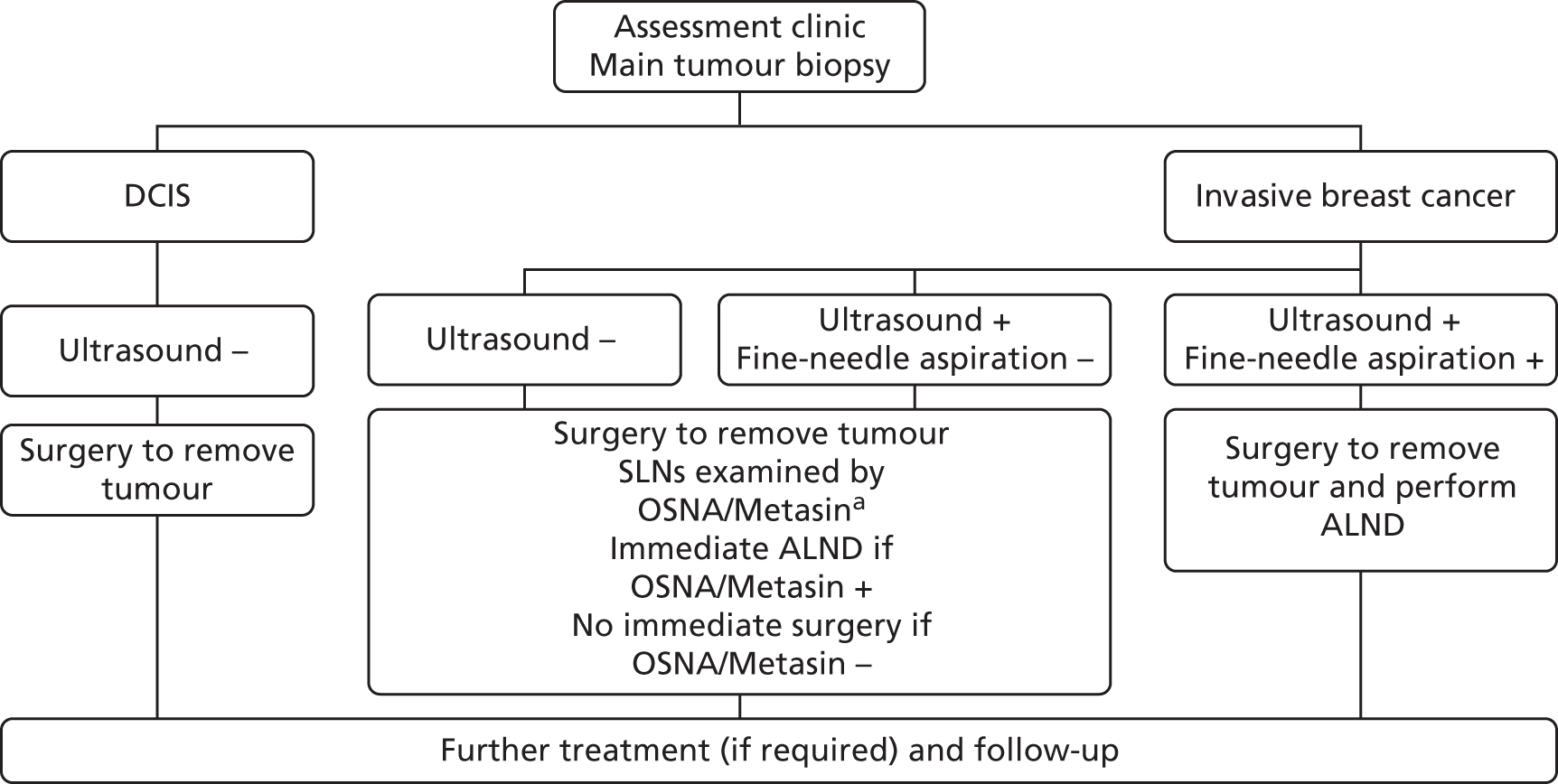
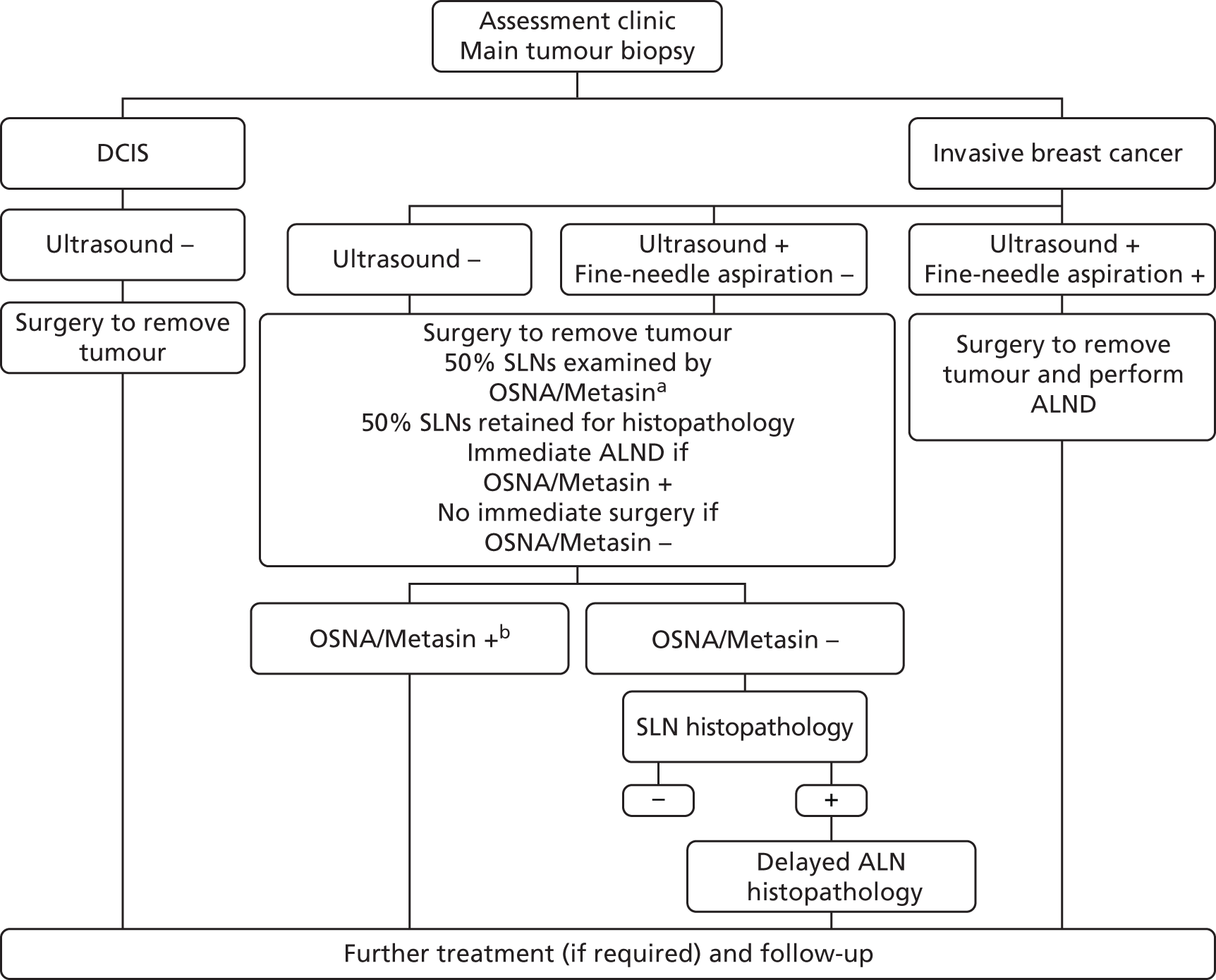
FIGURE 6.
Proposed clinical pathway for staging of breast cancer and subsequent surgery to the axilla: OSNA or Metasin used adjunctively. a, In cases in which OSNA/Metasin does not provide a result or provides a result that is uninterpretable, the OSNA/Metasin test would first be repeated using the remaining solubilised LN. Failing this, histopathology is performed on the remaining 50% of the SLN. b, No histopathology is performed on the SLN if the OSNA/Metasin test is positive. The ALND sample is still examined histopathologically as normal. –, negative; +, positive.
Other technologies
There are two other pathological methods that can be used intraoperatively, frozen section and touch imprint cytology. Frozen section involves a section of the LN being snap frozen, stained and sliced before being viewed by a consultant histopathologist. Touch imprint cytology involves the LN being sliced followed by the cut surface of the node being imprinted on to a slide, which is then stained and viewed by a consultant histopathologist. Both intraoperative pathological methods can be used to determine if ALND needs to be performed at the same time as the first surgery, and postoperative histopathology analysis is usually carried out to reduce the risk of a false-negative result. However, in practice, these intraoperative methods are rarely used because they have low accuracy and pathology resources are very limited within the NHS. 41
Measuring the accuracy of one-step nucleic acid amplification and Metasin
As already introduced earlier, one aspect of the evaluation of new tests is measuring their accuracy by calculating their sensitivity and specificity. This requires specification of the best available method of identifying the target condition of interest, the reference standard. In the case of the new technologies in question the target condition is the true presence of breast cancer cells in the SLNs. The ideal reference standard is thus histopathological examination of the SLNs in their entirety. However, practically this is impossible because tissue is required by both the test of interest and the reference standard. This leads to a compromise reference standard, with LNs split into several sections, often four, and alternate sections being allocated to either the test whose accuracy is being evaluated or the reference standard. However, as described earlier, this means that tissue allocation or sampling bias will operate, which needs to be carefully considered when interpreting the results of test accuracy studies. This can include a careful analysis of discrepant results to try to identify if sampling is a potential explanation for apparent false-negative or false-positive results, recognising that there are other reasons for these. The limitations of histopathology as a reference standard, given that all of the LN sample can never be examined because of the finite number of slices that can be taken, are important among these.
Further, it is known that histopathological examination of SLNs has a false-negative rate relative to examining all LNs removed in an ALND. Accepting the use of histopathological examination of SLNs as the reference standard for OSNA and Metasin implies that these error rates of SLNB histopathology are also suffered by the new tests of interest. Whether or not these errors could be avoided by the new tests could theoretically be investigated by using ALND findings as the reference standard, but the ethical considerations of exposing all OSNA-/Metasin-negative patients to the side effects of ALND greatly reduce the acceptability of such an approach.
Implications for comparing the effectiveness and cost-effectiveness of one-step nucleic acid amplification and Metasin with that of current practice
The following report needs to extend its assessment beyond the accuracy of the new tests of interest to their impact on patients and the health service. In the situation in which there is no direct and rigorous research evidence on whether introducing OSNA or Metasin will lead to improved patient outcomes, a linked evidence approach using economic modelling is likely to be required. In this, the likely consequences of errors in diagnosis are translated into outcomes. The estimates of sensitivity and specificity from accuracy studies are generally used to capture the difference in error rates between the new tests and current practice, so it is important that the use of the new tests in the accuracy studies is similar to the use of the tests in practice and that the reference standard used in the accuracy studies is as close as possible to that used in current diagnostic practice. The issues raised in the preceding section suggest that the similarity between current histopathological practice and the reference standard in the accuracy studies may require close attention. An immediate difficulty, however, is that what constitutes average current practice with regard to histopathological examination of SLNs is difficult to define. As a consequence, it is assumed that histopathology as typically performed in current practice is of the same standard as that used by diagnostic research studies reviewed for the purpose of this report.
Main potential consequences of using one-step nucleic acid amplification and Metasin compared with current practice
Although based on claims that require substantiation (one of the main purposes of this study), the previous background information suggests that the main anticipated effects of introducing intraoperative OSNA or Metasin for those undergoing SLNB are:
-
ALND will be performed as a single operation following immediately after primary tumour removal, rather than as a separate second operation as in current practice.
-
This in turn may lead to reduced anxiety in patients who no longer have to wait to find out if they need a second operation and to a reduced time to adjuvant treatment, if this is required.
-
The reduced time to ALND may, however, complicate the decision-making process of the MDT.
-
There may be fewer adverse effects of ALND when it is performed immediately after primary tumour removal than when it is performed later as a second operation.
-
One rather than two operations may also lead to reduced hospital costs.
-
There will be increased costs associated with OSNA or Metasin, which will be offset by reduced histopathology costs when OSNA or Metasin are used as replacement tests.
-
Any potential benefits of OSNA and Metasin will be offset if they introduce diagnostic errors, indicated by either their sensitivity or their specificity being < 100%.
-
If false negatives are introduced, women with macro- or micrometastases will be misidentified as SLNB negative and will not undergo ALND, which may compromise their outcome with respect to breast cancer.
-
If false positives are introduced, women who are SLNB negative will be misidentified as having macro- or micrometastases, with any resulting side effects but without any outcome benefits with respect to breast cancer.
The economic model will need to attempt to capture all of the above potential consequences.
Chapter 2 Definition of the decision problem
The question addressed by this health technology assessment is as set out in the final scope published by NICE and reproduced here for reader convenience.
A protocol was developed a priori by the authors to address the decision problem. The aspects of this involving systematic review were registered on the PROSPERO website (registration number CRD42012002889).
The methods used to address specific aspects of the decision problem are detailed at the beginning of each of the relevant chapters that follow.
Decision question
Are the RD-100i OSNA system and any alternative technologies identified during scoping clinically effective and cost-effective if used in the NHS in England?
Population
Individuals with invasive breast cancer who undergo a SLNB.
Intervention
-
The RD-100i OSNA system using a whole-node sample.
-
The RD-100i OSNA system using a half-node sample with postoperative histopathology confirmation.
Alternative diagnostic technologies
-
The Metasin test using a whole-node sample (intraoperative in-house molecular test developed at Princess Alexandra Hospital, Harlow, Essex).
-
The Metasin test using a half-node sample with postoperative histopathology confirmation.
Comparators
Postoperative standard histopathology alone.
Health-care setting
Secondary and tertiary care settings.
Health outcomes
Clinical considerations
The intermediate measures for consideration include:
-
diagnostic test accuracy (DTA)
-
test failure rate
-
discordant test results
-
time to test result
-
duration of anaesthesia.
The clinical outcomes for consideration include:
-
patient anxiety associated with waiting time for results and not knowing the extent of surgery before the operation
-
the number of repeat operations (except for re-excision of positive margins)
-
time in operating theatre
-
time to start of and nature of adjuvant therapy
-
morbidity and mortality from biopsies, axillary dissections, first and second operations and treatment of cancer
-
adverse events from false test results including patient distress and sequelae.
Data on these outcomes are likely to be used along with clinical utility scores to estimate quality-adjusted life-years (QALYs).
Cost considerations
The cost analysis will be based on the UK NHS setting and will comprise both NHS and Personal Social Services costs.
The costs for consideration include:
-
the costs of equipment, any additional tests (pre-screening), reagents and consumables
-
staff and staff training costs
-
equipment maintenance costs
-
costs associated with surgeon time and the management of operating theatre time
-
medical costs arising from ongoing care following test results, including those associated with surgery, time spent in hospital and treatment of cancer
-
medical costs arising from adverse events, including those associated with biopsies, surgery, cancer treatment and false test results.
The cost of the hardware for the RD-100i OSNA system is approximately £70,000 [excluding value-added tax (VAT)] The cost of consumables is approximately £150–250 per patient (excluding VAT). This consumable cost is dependent on the number of tests performed per theatre day and the number of patient samples tested. The maintenance cost is £6180 per annum (excluding VAT) following the expiry of the 1-year warranty.
Chapter 3 Assessment of clinical effectiveness
Methods for reviewing clinical effectiveness
The diagnostic accuracy of the OSNA and Metasin tests was assessed by a systematic review of research evidence. The review was undertaken following the principles published by the NHS Centre for Reviews and Dissemination. 42
Identification of studies
The following bibliographic databases were searched: MEDLINE, MEDLINE-In Process & Other Non-Indexed Citations and EMBASE (all via Ovid), Web of Science (including conference proceedings, via ISI), The Cochrane Library (all) and the NHS Economic Evaluations Database (EED) (via The Cochrane Collaboration). The searches did not use any form of limit (e.g. date) and covered up to 2012, weeks 29–30 (see Appendix 1 for details).
The following clinical trials registries were also searched: ClinicalTrials.gov, Current Controlled Trials, the World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP) and the EU Clinical Trials Register. The Google search engine (Google Inc., Menlo Park, CA, USA) was also used to identify grey literature and conference publications.
Items included after full-text screening were forward citation chased using Web of Science (Thompson Reuters).
Searches were deduplicated and managed using EndNote X5 (Thomson Reuters, CA, USA).
Relevant studies were identified in two stages. Titles and abstracts returned by the search strategy were examined independently by two researchers (TJH and HC) and screened for possible inclusion. Disagreements were resolved by discussion. Full texts of the identified studies were obtained. Two researchers (TJH and HC) examined these independently for inclusion or exclusion and disagreements were again resolved by discussion.
Inclusion and exclusion criteria
Population
Studies of individuals with invasive breast cancer who underwent a (sentinel) LN biopsy during the primary operation to excise a suspected breast cancer were included.
Interventions and comparators
Studies of OSNA or Metasin as used at the thresholds recommended by the manufacturer or designer were included. The reference standard was postoperative histopathology, performed on fresh sections of tissue. Frozen section and touch imprint cytology were excluded as comparators as they were not felt to be sufficiently feasible for widespread implementation (or intervention).
Outcomes
No study was excluded on the basis of outcomes, provided that it appeared relevant to those listed in the decision problem:
-
test failure rate
-
DTA
-
discordant test results
-
time to test result
-
duration of anaesthesia/time in operating theatre
-
number of repeat operations (except for re-excision of positive margins)
-
time to start of, and nature of, adjuvant therapy.
The clinical outcomes for consideration include:
-
patient anxiety associated with waiting for results and not knowing the extent of surgery before the operation
-
adverse events from false test results including patient distress and sequelae
-
morbidity and mortality from biopsies, axillary dissections, first and second operations and treatment of cancer.
Study design
For the review of test accuracy, the protocol made provision for all study designs unless evidence on the intervention and outcome of interest was already available from designs less open to bias as judged with reference to standard hierarchies of evidence. 42
Systematic reviews were used as a source for finding further studies and to compare with our systematic review. For the purpose of this review, a systematic review was defined as including:
-
a focused research question
-
explicit search criteria that are available to review, either in the document or on application
-
explicit inclusion/exclusion criteria defining the population(s), intervention(s), comparator(s) and outcome(s) of interest
-
a critical appraisal of included studies, including consideration of internal and external validity of the research
-
a synthesis of the included evidence, whether narrative or quantitative.
Studies were excluded if they did not match the inclusion criteria and, in particular, were:
-
preclinical and animal studies
-
reviews, editorials and opinion pieces
-
case reports
-
studies with < 10 participants.
Data extraction strategy
Data were extracted by one reviewer (TJH) using a standardised data extraction form in Microsoft Access 2010 (Microsoft Corporation, Redmond, WA, USA) and were checked by a second reviewer (HC). Disagreements were resolved by discussion, with involvement of a third reviewer if necessary. Data extraction forms for each included study can be found in Appendix 2.
Critical appraisal strategy
The methodological quality of the studies was assessed, when applicable to the design of the study, according to criteria specified by The Cochrane Collaboration’s tool for assessing risk of bias. 42 The Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool was used for test accuracy studies. 43
Quality was assessed by one reviewer and judgements were checked by a second. Any disagreement was resolved by discussion, with involvement of a third reviewer as necessary. The two instruments are summarised in the following section. The results were tabulated and the relevant aspects described in the data extraction forms.
Internal validity
The instruments sought to assess the following considerations.
The Cochrane Collaboration’s tool for assessing risk of bias
-
Was the allocation sequence adequately generated?
-
Was allocation adequately concealed?
-
Was knowledge of the allocated intervention adequately prevented during the study?
-
Were incomplete outcome data adequately addressed?
-
Are reports of the study free of suggestion of selective outcome reporting?
-
Was the study apparently free of other problems that could put it at high risk of bias?
Quality Assessment of Diagnostic Accuracy Studies tool
-
Description of patient selection:
-
Was a consecutive or random sample of patients enrolled?
-
Was a case–control design avoided?
-
Did the study avoid inappropriate exclusions?
-
Could the selection of patients have introduced bias?
-
Are there concerns that the included patients do not match the review question?
-
-
Description of index and reference tests:
-
Was the index test assessor blind to the results of the reference standard and vice versa?
-
Was a threshold prespecified?
-
Could the conduct or interpretation of the index test or reference standard have introduced bias?
-
Are there concerns that the conduct or interpretation of the question have introduced bias for the index test or reference standard?
-
Is the reference standard likely to classify the target condition?
-
-
Description of patient flow and timing:
-
Did all patients receive a reference standard and was it the same test for each?
-
Were all patients included in the analysis?
-
Could the patient flow have introduced bias?
-
External validity
External validity was judged according to the ability of a reader to consider the applicability of findings to a patient group and service setting. Study findings can be generalisable only if they provide enough information to conclude that a cohort is representative of the affected population at large. Therefore, studies whose samples appeared to be typical of the UK breast cancer population were judged to be externally valid.
Methods of data synthesis
Details of the extracted data and quality assessment for each individual study are presented in structured tables and as a narrative description. Any possible effects of study quality on the effectiveness data are discussed. Data on test accuracy are presented as sensitivity, specificity and concordance, when available.
Meta-analysis
Meta-analysis of DTA was performed using the bivariate method44 implemented in Stata/SE 12.1 (StataCorp LP, College Station, TX, USA) using the command metandi. 45 Studies were included in the meta-analysis only if the numbers of true positives, true negatives, false negatives and false positives were all reported in the text or could be unambiguously inferred from other figures in the text. Meta-analysis using the full bivariate method was not performed when there were fewer than four included studies as the model cannot generally be estimated with fewer than four studies. When the full bivariate model could not be estimated (either because of too few studies or because of other convergence errors) we reduced the complexity of the model (as, for example, in Pennant et al. 46) by setting the correlation parameter to zero (effectively reducing the model to two independent univariate random-effects analyses) and performing the analysis directly using the Stata command xtmelogit.
The bivariate method, when calculated using maximum likelihood estimation and without covariates, is equivalent to the hierarchical summary receiver operating characteristic (HSROC) model47–49 and this can be used to provide a summary receiver operating characteristic (SROC) curve and prediction region as well as a summary estimate and confidence region of sensitivity and specificity.
The SROC curve is designed to show how sensitivity and specificity are traded off against each other in different studies, through variation of the positivity threshold. If, and only if, there is reason to believe that the positivity threshold might vary between studies, we provide a SROC curve and prediction region.
Interpreting the results from the diagnostic studies
Test accuracy
In most of the studies, the accuracy of the interventions has been evaluated against the reference (gold) standard of postoperative histopathology. The results are generally reported as follows:
-
Sensitivity = true positives/(true positives + false negatives). This is the probability of detecting the presence of metastases in someone with metastases.
-
Specificity = true negatives/(false positives + true negatives). This is the probability of not detecting metastases in someone without metastases. In this instance a high specificity is required to avoid unnecessary ALND.
-
PPV = true positives/(true positives + false positives). This is the probability of someone with a positive result actually having metastases.
-
NPV = true negatives/(true negatives + false negatives). This is the probability of someone with a negative test result actually not having metastases.
-
Accuracy or concordance with reference standard = (true positives + true negatives)/(true positives + true negatives + false positives + false negatives). This is the percentage of test results correctly identified by the test, that is, the rate of agreement with the reference standard.
-
Discordance = cases of disagreement between the reference standard and the index test
Discordant case analysis
Many studies used a single-gate design whereby, in a single sample of individuals with unknown metastatic status, portions of the same node were allocated to either a molecular assay or histopathology. However, because of the spatial distribution of metastasis within a LN, and the use of different parts of the LN for different diagnostic tests, TAB may occur (i.e. discordant results between tests may result from genuine differences between the tissue samples). Unfortunately, it is not possible to use the same tissue for both tests; the tissue used for the molecular assays cannot be used for histology, and the formalin-fixed and paraffin-embedded tissue required for permanent sections is not suitable for quantitative mRNA measurements.
Some studies have attempted to address this issue by performing extensive histopathological and molecular analysis on discordant cases to ascertain whether the test results are true and occur because of differences in the allocated tissue. The results are then adjusted (discordant cases resulting from TAB are removed).
Results of test accuracy
The results of the assessment of clinical effectiveness will be presented as follows:
-
an overview of the quantity and quality of available evidence together with a table summarising all included trials (see Table 8), a table of patient characteristics (see Table 9) and a summary table of key quality indicators (see Table 10)
-
a critical review of the available evidence for each of the stated research questions including the quantity and quality of available evidence; a summary table of the study characteristics; a summary table of the population characteristics; study results in terms of sensitivity, specificity and discordant case analysis presented in narrative and tabular form; a comparison of the results in terms of time to analysis; and a summary table of abstracts identified but not included in the review.
Quantity and quality of research available
Number of studies identified
The electronic searches retrieved a total of 665 titles and abstracts. Fifty-nine additional papers were found by searching the bibliographies of included studies and by forward chasing. A total of 589 papers were excluded, based on screening titles and abstracts. The full texts of the remaining 135 papers were requested for more in-depth screening, of which 16 published papers50–65 (two reporting the same study) and two unpublished papers40,66 were included in the review. The process of study selection is shown in Figure 7.
FIGURE 7.
Summary of study selection.
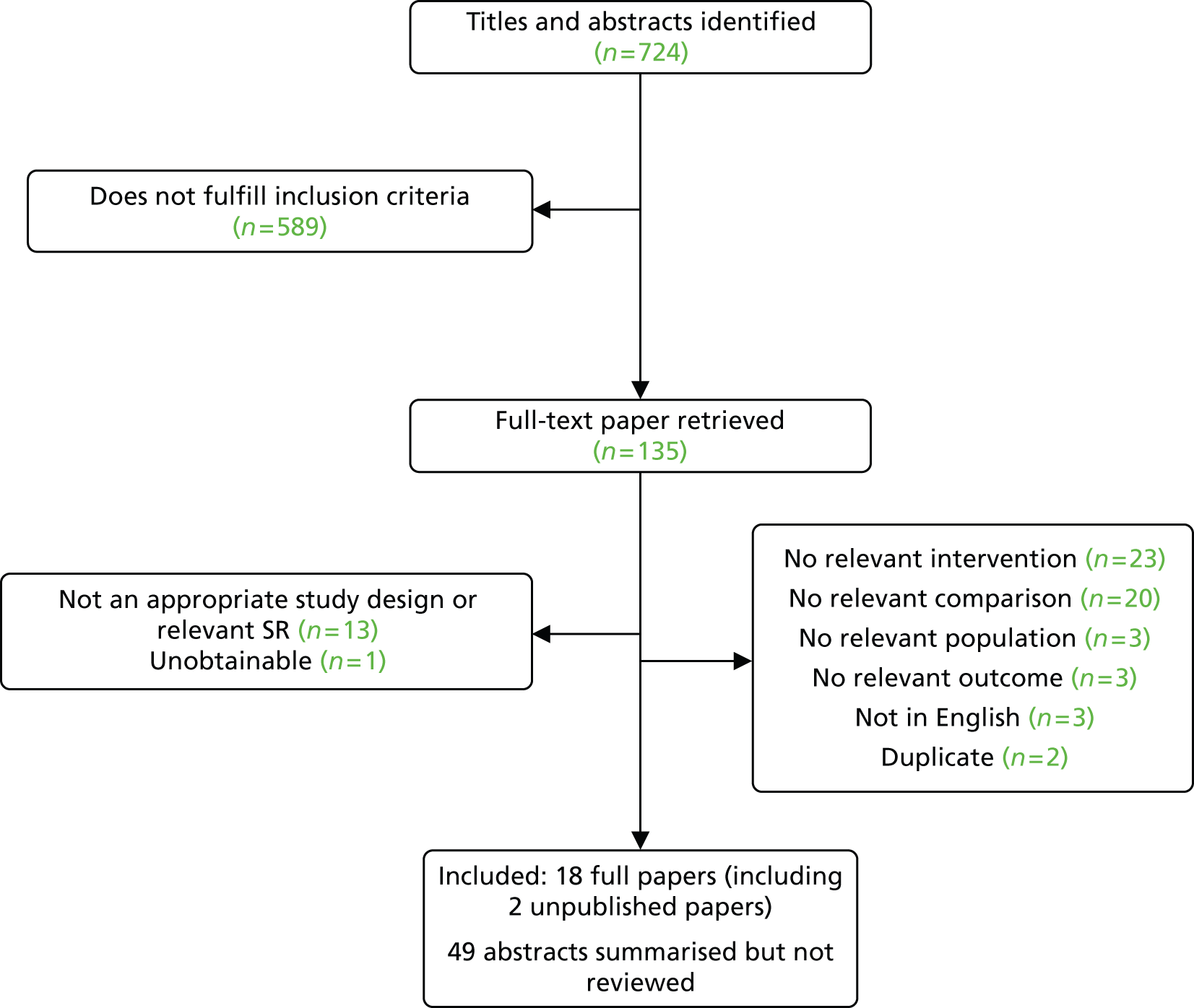
Number of studies excluded
Papers were excluded for at least one of the following reasons: duplicate publication, narrative review and publication (systematic reviews and individual studies) not considering relevant intervention, population, comparison or outcomes. The bibliographic details of studies retrieved as full papers and subsequently excluded, along with the reasons for their exclusion, are detailed in Appendix 3.
Number and description of included studies
Sixteen test accuracy papers were included for OSNA,50–65 with two papers reporting the same study. Two unpublished papers40,66 for Metasin were identified and assessed using the Standards for the Reporting of Diagnostic Accuracy Studies (STARD) in Table 7.
| Section and topic | STARD criteria | Sundaresan40 | McDowell66 |
|---|---|---|---|
| Title/abstract/keywords | 1. Identify the article as a study of diagnostic accuracy | ✗ | Academic-in-confidence information has been removed |
| Introduction | 2. State the research questions or study aims, such as estimating diagnostic accuracy or comparing accuracy between tests or across participant groups | ✓ | Academic-in-confidence information has been removed |
| Methods | |||
| Participants | 3. Describe the study population: the inclusion and exclusion criteria, setting and locations where the data were collected | ✗ | Academic-in-confidence information has been removed |
| 4. Describe participant recruitment: was recruitment based on presenting symptoms, results from previous tests or the fact that the participants had received the (evaluated) index tests or the (gold) reference standard? | ✗ | Academic-in-confidence information has been removed | |
| 5. Describe participant sampling: was the study population a consecutive series of participants defined by the selection criteria in items 3 and 4? If not, specify how participants were further selected | ✗ | Academic-in-confidence information has been removed | |
| 6. Describe data collection: was data collection planned before the index test and reference standard were performed (prospective study) or after (retrospective study)? | ✓ | Academic-in-confidence information has been removed | |
| Test methods | 7. Describe the reference standard and its rationale | ✓ | Academic-in-confidence information has been removed |
| 8. Describe technical specifications of material and methods involved including how and when measurements were taken, and/or cite references for index tests and reference standard | ✗ | Academic-in-confidence information has been removed | |
| 9. Describe definition of and rationale for the units, cut-offs and/or categories of the results of the index tests and the reference standard | ✗ | Academic-in-confidence information has been removed | |
| 10. Describe the number, training and expertise of the persons executing and reading the index tests and the reference standard | ✗ | Academic-in-confidence information has been removed | |
| 11. Describe whether or not the readers of the index tests and reference standard were blind (masked) to the results of the other test and describe any other clinical information available to the readers | ✗ | Academic-in-confidence information has been removed | |
| Statistical methods | 12. Describe methods for calculating or comparing measures of diagnostic accuracy, and the statistical methods used to quantify uncertainty (e.g. 95% CIs) | ✓ | Academic-in-confidence information has been removed |
| 13. Describe methods for calculating test reproducibility, if done | ✗ | Academic-in-confidence information has been removed | |
| Results | |||
| Participants | 14. Report when study was done, including beginning and ending dates of recruitment | ✗ | Academic-in-confidence information has been removed |
| 15. Report clinical and demographic characteristics of the study population (e.g. age, sex, spectrum of presenting symptoms, comorbidity, current treatments, recruitment centres) | ✗ | Academic-in-confidence information has been removed | |
| 16. Report the number of participants satisfying the criteria for inclusion that did or did not undergo the index tests and/or the reference standard; describe why participants failed to receive either test (a flow diagram is strongly recommended) | ✗ | Academic-in-confidence information has been removed | |
| Test results | 17. Report time interval from the index tests to the reference standard, and any treatment administered between | NA | Academic-in-confidence information has been removed |
| 18. Report distribution of severity of disease (define criteria) in those with the target condition; other diagnoses in participants without the target condition | ✗ | Academic-in-confidence information has been removed | |
| 19. Report a cross-tabulation of the results of the index tests (including indeterminate and missing results) by the results of the reference standard; for continuous results, the distribution of the test results by the results of the reference standard | ✓ | Academic-in-confidence information has been removed | |
| 20. Report any adverse events from performing the index tests or the reference standard | ✗ | Academic-in-confidence information has been removed | |
| Estimates | 21. Report estimates of diagnostic accuracy and measures of statistical uncertainty (e.g. 95% CI) | ✓ (no CIs) | Academic-in-confidence information has been removed |
| 22. Report how indeterminate results, missing responses and outliers of the index tests were handled | ✗ | Academic-in-confidence information has been removed | |
| 23. Report estimates of variability of diagnostic accuracy between subgroups of participants, readers or centres, if done | ✗ | Academic-in-confidence information has been removed | |
| 24. Report estimates of test reproducibility, if done | ✗ | Academic-in-confidence information has been removed | |
| Discussion | 25. Discuss the clinical applicability of the study findings | ✓ | Academic-in-confidence information has been removed |
The search also identified 49 abstracts,67–115 some of which repeated the data in the full papers, some of which provided supplementary information and some of which were not associated with a full paper. The data in the abstracts are presented in Appendix 4. However, because of the lack of accompanying details, no quality assessment has been performed.
Summary information for the 18 test accuracy studies included in the review is provided in Table 8.
| Study | Year | Patients, n | SLN or ALN, n | Centre | Designa | Outcomes |
|---|---|---|---|---|---|---|
| Metasin | ||||||
| McDowell66 | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) |
| Sundaresan40 | Unpublished | 1265 | 2279 SLNs | Multicentre, UK | Single gate | Test accuracy, time to analysis |
| OSNA | ||||||
| Bernet50 | 2011 | NR | 55 SLNs | Multicentre, Spain | Observation | Time to analysis |
| Bernet Vegue51 | 2012 | 55 | 567 non-SLNs | Multicentre, Spain | Single gate | Test accuracy |
| Castellano52 | 2012 | 279 | Unclear | Turin, Italy | Cohort | Test accuracy, non-SLN involvement |
| Choi53 | 2010 | 199 | 284 SLNs | Seoul, Korea | Single gate | Test accuracy |
| Feldman54 | 2011 | 496 | 1044 SLNs | Multicentre, USA | Single gate | Test accuracy |
| Godey55 | 2012 | 723 | Unclear | Rennes, France | Cohort | Test accuracy |
| Guillen-Paredes56 | 2011 | 80 | 114 SLNs | Murcia, Spain | Cohort | Operating time, days in hospital, costs |
| Khaddage57 | 2011 | 46 | 80 SLNs | Saint-Etienne, France | Single gate | Test accuracy |
| Le Frère-Belda58 | 2012 | 233 | 503 SLNs | Multicentre, France | Single gate | Time to analysis, test accuracy |
| Osako59 | 2011 | 183 | Non-SLNs | Cancer Institute Hospital, Tokyo, Japan | Cohort | Test accuracy |
| Schem60 | 2009 | 93 | 343 ALNs | University Clinic of Schleswig-Holstein, Albertinen Hospital, Germany | Single gate | Test accuracy |
| Snook61 | 2011 | 204 | 395 SLNs | Multicentre, UK | Single gate | Test accuracy, time to analysis |
| Tamaki62 | 2009 | 198 | 674 ALNs + SLNs | Multicentre, Japan | Single gate | Test accuracy |
| Tamaki63 | 2012 | 417 | 775 SLNs | Multicentre, Japan | Single gate | Test accuracy |
| Tsujimoto64 | 2007 | 101 | 325 ALNs + SLNs | Multicentre, Japan | Single gate | Test accuracy |
| Visser65 | 2008 | 32 | 346 ALNs | Alkmaar and Amsterdam | Single gate | Test accuracy |
Study characteristics
One-step nucleic acid amplification and Metasin have standard procedures and thresholds; therefore, unless otherwise stated, the included studies have complied with the manufacturers’ instructions. Both methods are semiquantitative and differentiate between micro- and macrometastases, (academic-in-confidence information has been removed).
In contrast to the molecular methods, there is some heterogeneity with regard to the reference standard, particularly in the number of levels examined, for example one-level analysis will involve analysis of one section of node, whereas five-level analysis will examine five sections. As such, for one-level histopathology it is likely that macrometastases will be identified but micrometastases may be missed. A micrometastasis is considered to be > 0.2 mm and < 2 mm and a macrometastasis is considered to be > 2 mm. As there is no way to analyse the whole node, this method cannot be 100% sensitive.
Some studies report cases of ITCs with histopathological analysis. These will fall below the threshold for OSNA and Metasin and are generally considered LN negative as their clinical significance is unknown. 60
The majority of included studies comply with a single-gate design, in which a single sample of individuals with unknown metastatic status is assessed by both the diagnostic test under scrutiny and the reference standard. No studies utilising a two-gate design, in which the test under scrutiny is performed on a sample that includes individuals with known metastatic status (using the reference standard), were identified. However, three cohort studies have been included in which different patient populations were utilised for each test. The inclusion of cohort studies enabled the identification of data based on whole-node analysis, something that would not be possible for OSNA or Metasin using classic DTA study designs, such as the single-gate or the two-gate design.
A general issue for all of the studies is that a portion of node tissue is allocated to the index test and a portion is allocated to the reference standard. As such, the tests are analysing different samples of tissue, which cannot be reused between tests. As metastases may be distributed unevenly, TAB may occur, that is, metastases may exist in the tissue sample provided for one test but not in the tissue sample provided for the other test.
It should also be noted that studies examining ALNs as well as, or instead of, SLNs were included.
Metasin
There is currently no published evidence for the test accuracy of Metasin; however, we have received draft reports of two single-gate studies.
Sundaresan40 reports on a postoperative evaluation of Metasin using 2279 nodes from 1265 patients with breast cancer in which 608 of the 1265 cases were from patients already assessed using the GeneSearch BLN assay. Six centres contributed tissue homogenates, although two centres were able to provide frozen ribonucleic acid (RNA) only. Three-level histology, that is, examination of three sections, was performed although the author states that all laboratories followed their own standard operating procedures for analysis of slides. The author also mentions that there was no uniformity in the protocols followed for sentinel and axillary clearance.
(Academic-in-confidence information has been removed.)
One-step nucleic acid amplification
An assessment of the reliability of OSNA as a single test for SLNs was reported by Castellano et al. 52 This was a single-centre cohort study with 279 patients. Histology was performed on fresh SLNs, initially sliced at 2 mm after which further step sections at 100 µm were performed. Positive rates for both cohorts were presented.
A cohort study reported by Godey et al. 55 compares the positivity rate of OSNA performed in a routine clinical setting with historical postoperative histology results for 723 patients. Histology was performed on 250-µm sections of the node until no tissue was left.
Guillen-Paredes et al. 56 present a retrospective cost–benefit analysis. This cohort study aimed to analyse the economic costs of the intraoperative OSNA assay compared with the costs of the conventional postoperative histological and immunohistochemical assay. Histology was performed on 4-mm sections of the node. Results include operative time, days in hospital, costs and postoperative complications.
Osako et al. 59 report on a retrospective cohort study of 183 patients at a single centre. Intraoperative OSNA was compared with one-level histology (examination of one slice) performed on five to seven non-SLNs from the same patient. Positive rates were described for both methods.
The aim of a multicentre study presented by Bernet et al. 50 was to compare OSNA with histology and evaluate its feasibility intraoperatively. Fifty-five SLNs were investigated using a single-gate study design; however, the results appear to include touch imprint and frozen section analysis supported by postoperative histopathology and therefore these results are not included in the review. Relevant outcomes that are included in the review are times of extraction, dissection, preparation and analysis.
A paper by Bernet Vegue et al. 51 reported on the Breast Complete Lymphadenectomy OSNA Study for Enhanced Review-I (B-CLOSER-I). Eight hospitals were involved in this single-gate study comparing histopathology and OSNA for the pathological staging of ALNs after identification of positive SLNs in patients with primary breast cancer. Fifty-five patients were recruited consecutively, providing 567 non-SLNs for analysis. Both OSNA and histopathology were performed postoperatively. Tissue used for OSNA analysis was stored at –80°C. Two phases are reported, both utilising a single-gate design: the validation phase and routine use. For the validation phase, histopathology was five level (examination of five slices) whereas for routine use a central 1-mm slice was used for histopathology (one level), with the remainder being used for OSNA. For both phases discordant cases were investigated using additional molecular analysis, performed by qRT-PCR. Discussed outcomes include discordance during the validation phase.
The sensitivity, specificity and accuracy of intraoperative OSNA compared with that of three-level histology are described in a study by Choi et al. ,53 in which 199 patients had 284 SLNs analysed. With regard to discordance, clinical information, status of non-SLNs and expression of CK19 protein in LN metastasis foci were evaluated on a patient basis.
A multicentre single-gate study reported by Feldman et al. 54 recruited 496 patients, providing 1044 SLNs for analysis. The node was sectioned into six with alternate slices used for histopathology and OSNA. The slices for histopathology were then dissected at 200-µm intervals for haematoxylin–eosin staining and pan-CK immunohistochemistry. The results were evaluated by blinded pathologists. For discordant case analysis, blank histopathology was checked and OSNA was retested using Western blot analysis and qRT-PCR. This study used the RD-110i system with a reagent kit that was different from that used in the other included studies.
Khaddage et al. 57 reported on a multicentre, single-gate study based in France, with concordance, sensitivity and specificity as outcomes. The study included a validation phase with 46 patients and a routine phase with 197 patients. Histopathology for the validation phase was five level and for the routine phase was one level. Both node- and patient-level analyses are presented. The results of the OSNA investigation were not known to the histopathologist and vice versa. Discordant case analysis of the validation phase was performed by qRT-PCR.
Le Frère-Belda et al. 58 reported a study assessing the intraoperative diagnostic performance of OSNA compared with extensive histological evaluation. This was a multicentre single-gate study involving eight clinical centres in France. Alternate slices of dissected SLNs were used, along with five-level histology. It should be noted that two centres reused frozen section samples, which may impair integrity for final histology. In cases of discordance, with positive OSNA and negative histology, 200-µm skip spaces for all slices were analysed. Samples were also sent to Sysmex for blind molecular analysis by qRT-PCR.
A study by Schem et al. 60 considered the performance of OSNA compared with that of five-level histology. This two-centre study used a blinded, single-gate, experimental design and analysed 343 ALNs. OSNA samples were stored frozen at –80°C. Discordant samples were analysed by further levels of histology, Western blot analysis and qRT-PCR. In addition, 120 histopathologically negative samples were cut into further levels for determining specificity. Sensitivity and discordance were also reported.
Snook et al. 61 reported on a UK single-gate, prospective, multicentre evaluation of OSNA involving four centres. In total, 204 patients were recruited, providing 395 SLNs, although there are no further details on recruitment. OSNA slices were snap frozen at –80°C. Five-level histology was performed and molecular analysis for discordant samples was carried out by qRT-PCR. Sensitivity, specificity, accuracy, PPV, NPV and discordance were reported.
A Japanese multicentre study including two single-gate trials is reported by Tamaki et al. 62 This study examined the validity of OSNA for clinical use. Seven centres were involved, two in trial 1, three in trial 2 and two in both. Trial 1 compared intraoperative OSNA with detailed histology for the detection of metastases in 124 ALNs. Alternate slices of the node were allocated to each technology; for histology, the tissue was dissected into 0.2-mm sections. Trial 2 was designed to replicate routine use and so only one-level histology was performed. For discordant cases, however, histology was performed as in trial 1, alongside Western blot analysis. Sensitivity, specificity and discordance were reported.
The same trial was reported by Tsujimoto et al. ,64 who present data from six centres for 101 patients and 325 ALNs. Intraoperative OSNA was compared with three-level histology. The results were examined by three third-party (blinded) pathologists. The authors state that calculations for sensitivity and specificity were not appropriate because separate tissue was used for both tests; therefore, the results were evaluated as concordance with histology. Discordant samples were analysed using qRT-PCR and Western blotting.
A subsequent Japanese multicentre study by Tamaki et al. 63 investigated the clinical use of OSNA compared with one-level histology. This study used a single-gate design and involved 198 patients and 674 ALNs and SLNs.
Finally, Visser et al. 65 reported on a single-gate study that tested OSNA on 346 ALNs compared with five-level histology. To investigate if the results were influenced by sampling bias, the histological work-up was extended to all levels in the first 120 histologically negative LN samples (and also to paraffin blocks of discordant cases). In addition, the homogenised LN lysates of samples were subjected to qRT-PCR and Western blot analysis.
Population characteristics
In general, patient characteristics were poorly reported, as were inclusion and exclusion criteria. Not all studies provided the age range of participants and, often, only information on tumour staging was provided. Patient characteristics are presented in Table 9.
| Characteristic | Bernet 201150 | Bernet Vegue 201251 | Castellano 201252 | Choi 201053 | Feldman 201154 | Godey 201255 | Guillen-Parades 201156 | Khaddage 201157 | Le Frère-Belda 201158 | Osako 201159 | Schem 200960 | Snook 201161 | Tamaki 200962 | Tamaki 201263 | Tsujimoto 200764 | Visser 200865 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intervention | O + H | O + H | O | H | O + H | O + H | O | H | O + H | H | O + H clinical | O + H routine | O + H | O | H | O + H | O + H | O + H trial 1 | O + H trial 2 | O + H | O + H | O + H |
| n | 55 | 55 | 110 | 169 | 199 | 496 | 258 | 355 | 233 | 45 | 46 | 197 | 233 | 119 | 64 | 93 | 204 | 36 | 185 | 439 | 101 | 32 |
| Age (years), median (range) | 59 (23–87) | 66.7 (38–82) | 61.2 (23–86) | 44.5a | 58.8b (28–88) | 56.8b | 56.9b | 58 (30–93) | 61.89b | 58 (30–93) | 53 (27–86) | 56 (39–81) | 55.9b | 54.7b | 56.1b (25–90) | |||||||
| Clinical stage, n (%) | ||||||||||||||||||||||
| 0 | 11 (5.5) | 41 (17.7) | 2 (6) | 14 (9) | 5 | 0 | ||||||||||||||||
| I | 21 (38.2) | 132 (66.3) | 175 (75.4) | 8 (24) | 51 (31) | 183 (43.9) | 41 | 8 | ||||||||||||||
| II | 22 (40.0) | 54 (27.1) | 13 (5.6) | 14 (41) | 64 (40) | 110 (26.4) | 49 | 15 | ||||||||||||||
| III | 12 (21.8) | 2 (1.0) | 2 (0.9) | 5 (15) | 7 (4) | 70 (16.8) | 5 | 7 | ||||||||||||||
| IV | 11 (5.5) | 1 (0.4) | 0 | 0 | 1 | 2 | ||||||||||||||||
| Unknown | 2 (6) | 0 | 54 (12.9) | 5 | ||||||||||||||||||
| Tumour size, n (%)c | ||||||||||||||||||||||
| ≤ 1 cm | 33 (30) | 41 (24) | ||||||||||||||||||||
| 1.1–1.5 cm | 19 (17) | 45 (27) | ||||||||||||||||||||
| > 1.5 cm | 58 (53) | 83 (49) | ||||||||||||||||||||
| Clinical tumour classification, n (%) | ||||||||||||||||||||||
| T0 | 7 | 1 | ||||||||||||||||||||
| Tis | 1 (1.8) | 8 (4.0) | 21 (4.2) | 2 | 3 | 0 | 21 | 50 (12) | ||||||||||||||
| T1 | 32 (58.2) | 129 (64.8) | 327 (65.9) | 16 | 13 | 34 | 141 | 46 | 133 | 254 (60.9) | ||||||||||||
| T2 | 21 (38.2) | 56 (28.1) | 124 (25) | 17 | 29 | 2 | 30 | 36 | 60 | 111 (26.6) | ||||||||||||
| T3 | 1 (1.8) | 2 (1.0) | 5 (1) | 2 | 1 | 4 | 5 | 2 (0.5) | ||||||||||||||
| T4 | 1 | 1 | 7 | |||||||||||||||||||
| Tx | 4 (2.0) | 19 (3.8) | ||||||||||||||||||||
| Nodal status, n (%) | ||||||||||||||||||||||
| pN0 | 153 (76.9) | 387 (78) | 225 (97.0) | 46 | 60 | 14 | ||||||||||||||||
| pN1 | 37 (18.6) | 84 (16.9) | 7 (3.0) | 115 (96.6) | 62 (96.9) | 27 | 35 | 10 | ||||||||||||||
| pN2 | 5 (2.5) | 14 (2.8) | 3 (3.4) | 2 (3.1) | 13 | 2 | 6 | |||||||||||||||
| pN3 | 4 (2.0) | 4 (0.8) | 7 | 4 | 2 | |||||||||||||||||
| Histopathological type, n (%) | ||||||||||||||||||||||
| IDC | 44 (80.0) | 81 (74) | 109 (64) | 165 (82.9) | 348 (70.2) | 212 | 313 | 31 | 37 | 36 | 148 | 164 (70.4) | 110 (92.4) | 57 (89.1) | 68 | 160 (78.8) | 32 (94) | 130 (79) | 305 (73.1) | 87 | 30 | |
| ILC | 7 (12.7) | 16 (14) | 29 (17) | 9 (4.5) | 40 (8.1) | 46 | 42 | 2 | 5 | 5 | 16 | 34 (14.6) | 4 (3.4) | 2 (3.1) | 21 | 22 (10.8) | 1 (3) | 7 (4) | 24 (5.8) | 4 | 2 | |
| DCIS | 1 (1.8) | 9 (4.5) | 1 | 2 | 5 | 21 | 23 (9.9) | 0 | 18 (11) | 53 (12.7) | 5 | |||||||||||
| Other | 3 (5.5) | 13 (12) | 31 (18) | 16 (8.1) | 109 (21.7) | 1 | 1 | 0 | 12 | 12 (5.2) | 4 | 16 (7.9) | 1 (3) | 9 (5) | 35 (8.4) | 5 | ||||||
| HER2, n (%) | ||||||||||||||||||||||
| Negative | 49 (89.1) | 108 (98) | 144 (85) | 106 (89.1) | 55 (85.9) | 334 (87.8) | ||||||||||||||||
| Positive | 6 (10.9) | 2 (2) | 25 (15) | 13 (6.3) | 13 (10.9) | 9 (14.1) | 51 (12.2) | |||||||||||||||
Metasin
(Academic-in-confidence information has been removed.) Sundaresan et al. 40 provide no details on patient characteristics.
One-step nucleic acid amplification
Two52,59 of the cohort studies reveal reasonably homogeneous patient populations with regard to age and clinical status. A relatively small sample size was used for each patient group in the study reported by Guillen-Paredes,56 with a difference of ∼7 years for mean age. The histology group had proportionally more participants with a T2 tumour classification than the OSNA group.
Across many of the single-gate studies, minimal information is given regarding population characteristics. In general, the populations are heterogeneous, with studies including patients across the spectrum of tumour and nodal staging, although some studies have included patients with cancer of only one or two stages. It should also be noted that, for some studies, the number of recruited patients is small whereas the number of nodes analysed is comparatively large.
Assessment of study quality
A summary of the quality assessment of studies included in the review is provided in Table 10; this is followed by a narrative summary. The main issue within the included studies is TAB, which occurs when tissue cannot be reused between the index test and the reference test. In short, because of an uneven distribution of metastases, the test results may be discordant but accurate for the tissue received. Many studies attempted to address this, either by more extensive histology or by additional molecular analyses, such as qRT-PCR and Western blotting for CK19 protein. The results should then be recalculated after adjustment for TAB, generally with an increase in OSNA sensitivity and specificity.
| QUADAS-2 domain | Study | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OSNA | Metasin | |||||||||||||||||
| aBernet 201150 | Bernet Vegue 201251 | Castellano 201252 | Choi 201053 | Feldman 201154 | Godey 201255 | Guillen-Paredes 201156 | Khaddage 201157 | Le Frère Belda 201258 | Osako 201159 | Schem 200960 | Snook 201161 | Tamaki 200962 | Tamaki 201263 | Tsujimoto 200764 | Visser 200865 | Academic-in-confidence information has been removed | Sundaresan40 | |
| Patient selection | ||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | N | Y | U | U | U | U | U | U | U | Y | U | U | U | U | U | U | b | U |
| Was a cohort study design avoided? (Y/N/U)c | NA | Y | N | Y | Y | N | N | Y | Y | N | Y | Y | Y | Y | Y | Y | b | Y |
| Did the study avoid inappropriate exclusions? (Y/N/U) | U | Y | Y | Y | Y | N | Y | Y | Y | Y | U | Y | Y | Y | U | Y | b | U |
| Could the selection of patients have introduced bias? (H/L/U) | U | L | U | U | U | U | U | U | U | L | U | U | U | U | U | U | b | U |
| Concerns that the included patients do not match the review question? (H/L/U) | L | L | L | L | L | L | L | L | L | L | U | L | L | L | L | L | b | L |
| Index test | ||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | U | NA | U | Y | NA | Y | Y | U | U | Y | Y | Y | U | Y | U | b | U |
| If a threshold was used, was it prespecified? (Y/N/U) | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | b | Y |
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U)d | L | U | L | L | L | U | L | L | L | L | L | L | L | L | H | L | b | U |
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | L | L | L | L | U | L | L | L | L | L | L | L | L | L | L | b | L |
| Reference standard | ||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | b | Y |
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | NA | U | NA | U | Y | U | NA | Y | Y | U | U | Y | Y | U | U | U | b | U |
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U)e | L | U | U | U | L | U | U | L | U | L | U | U | L | U | L | U | b | U |
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | L | L | L | L | U | L | L | L | L | L | L | L | L | L | L | b | L |
| Flow and timingf | ||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | NA | Y | N | Y | Y | N | NA | Y | Y | N | Y | Y | Y | Y | Y | Y | b | Y |
| Did all patients receive the same reference standard? (Y/N/U) | NA | Y | Y | Y | Y | N | NA | Y | Y | Y | Y | Y | Y | U | Y | Y | b | Y |
| Were all samples (that should have been) included in the analysis? (Y/N/U) | U | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | b | Y |
| Could the patient flow have introduced bias? (H/L/U) | U | L | U | U | U | U | U | L | L | L | U | U | L | L | U | U | b | U |
| Additional items | ||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U)g | NA | N | NA | N | Y | NA | NA | Y | Y | NA | Y | Y | Y | N | N | Y | b | N |
| Are there concerns about selective reporting of outcomes? (H/L/U) | U | L | L | L | L | H | L | L | L | L | L | L | L | L | U | L | b | L |
Other concerns included lack of detail on patient recruitment, minimal information on patient characteristics and unclear sampling methods, that is, no evidence was given on sample replicates and reproducibility for molecular analysis. Furthermore, unless otherwise mentioned, test failures, such as operator or instrument error, were not reported. It was also often not mentioned whether or not the outcome assessors were blinded. For example, it is not inconceivable that the pathologist performing the intraoperative molecular analysis also performed the more subjective postoperative histopathology. Papers were also often unclear on the robustness of the histopathology, for example if results were checked by a second party.
A potential conflict of interest also features heavily, (academic-in-confidence information has been removed) and the majority of the OSNA studies were financially supported by Sysmex.
Metasin
(Academic-in-confidence information has been removed.)
The second draft paper was presented by Sundaresan,40 who created the Metasin test. This study design is experimental, with six centres contributing tissue homogenates that were then compared with historical three-level histology. As such, no patient characteristics are included. Two centres were able to provide only frozen homogenates, which may impact on the quality of the RNA. Technical details for the Metasin test are not provided. No cases were excluded because of suspected TAB and no retrospective discordant case analysis was performed. However, when possible, RNA was reanalysed using an independent panel of markers. It is not clear if the initial analysis was performed by blinded pathologists. The failure rate for assays was reported (1.2%). According to the authors this was possibly because there was insufficient RNA in the samples.
One-step nucleic acid amplification
Castellano et al. 52 provide a comprehensive flow chart. Patients were not selected randomly, although patient groups were compared and no significant differences were found for the characteristics reported. Histology involved 2-mm slices of the whole node, which were then sectioned at 100 µm.
Godey et al. 55 compare a historical cohort using a 1-mm central slice sectioned every 250 µm. A flow diagram documents the pathway of patients through the study. However, reporting of information on patient characteristics for the whole study is minimal. The authors mention that for three patients there were technical problems with OSNA, but this was not elaborated on.
Guillen-Paredes et al. 56 provide a retrospective cost–benefit analysis using two relatively small patient groups. Inclusion criteria are detailed but few patient characteristics are presented. Histology involved preparing 4-mm sections of the LN, subsectioned into 15 × 4-µm slices for examination.
In the final cohort study, reported by Osako et al. ,59 OSNA is compared with historical single-section histology. Detailed exclusion criteria and comprehensive patient characteristics are included, with no significant heterogeneity being found between the groups. Some of the nodes were frozen at –80°C before OSNA, which may affect the quality of the mRNA. Laboratory consumables were funded by Sysmex.
Bernet et al. 50 report two trials within one paper. For trial 1 it was unclear whether fresh or frozen sections were used as the comparator; therefore, the results from this trial are not included in the review. Trial 2 investigates the timing of the procedural steps for OSNA for 55 cases and was therefore included in the review.
The study by Bernet Vegue et al. 51 reports on a single-gate study using only one-level histopathology, with the remaining tissue used for OSNA, which is likely to result in substantial TAB. The authors claim that this approach was chosen to replicate routine use. Patients were recruited consecutively across eight hospitals, although the achieved sample size is relatively small at 55 patients. Reasonably comprehensive reporting of patient characteristics was provided. No details were given on blinding of pathologists and there was no further investigation into discordant cases, although further details such as copies/µl were provided. There may be a conflict of interest as the study was funded by Sysmex.
The study reported by Choi et al. 53 does not provide any information on recruitment or a diagram of patient flow. However, analysis on all appropriate samples appears to have been performed and clinicopathological characteristics of patients are given. Of note, macrometastasis or micrometastasis was confirmed by both or either of intraoperative histopathological examination of frozen section specimens and postoperative histopathological (three-level) examination of permanent tissue specimens. Although it appears from this that some of the reference standard data may therefore have been based on frozen section specimens alone, closer examination of the entirety of the data has led us to believe that this is not the case. In particular, data are presented on discordant frozen section and final histopathology results and these suggest that there were no cases for which frozen section results were positive and final histology was negative. As such, it appears that a positive histology result was based on two scenarios: first, both the frozen section and the final histology were positive and, second, the frozen section results were negative and the final histology was positive. Therefore, all positive histology cases would have had a positive final histology result.
The study is unclear on whether or not the pathologists were blinded and provides no detail on test failures or replicates for OSNA. Discordance was evaluated using clinical information, status of non-SLNs and CK19 protein expression in metastatic foci of LNs. It is unclear how the CK19 protein expression was determined. This study was also supported by Sysmex.
Feldman et al. 54 report a study funded by Sysmex comparing OSNA with ‘extensive’ histology of 200-µm intervals of approximately 3 × 1 mm slices. Patient recruitment is not mentioned although patient characteristics are comprehensively reported. No patient flow diagram is included and the number of SLNs after discordance (n = 1018) does not comply with the numbers before discordance (n = 1044) minus the resolved cases. The population is somewhat heterogeneous, including all classifications of tumour and LN status. Histopathologists were blinded and discordant case analysis was performed using Western blotting and qRT-PCR, although no details of this are given.
A single-gate study reported by Le Frère-Belda et al. 58 provides a flow chart of the SLN samples as well as patient characteristics. This study investigates a reasonably heterogeneous population, although the majority had pN0 node status. No information was given on recruitment other than inclusion criteria. Duplicate samples were used for OSNA. Five-level histology was performed, although it should be noted that in five centres frozen sections were reused for this, which may have resulted in degradation of the sample. Discordant case analysis was performed by extensive histopathology and blind molecular analysis (SLN homogenates were shipped to Sysmex for this analysis). It is unclear if the original analysis was blinded. One sample was excluded because of a manipulation error. Laboratory consumables were funded by Sysmex.
The study reported by Khaddage et al. 57 compared OSNA with five-level histology for clinical evaluation and with one-level histology for routine use. Although replicates are not mentioned, positive and negative controls are confirmed. No patient flow diagram is provided and there are no details on recruitment other than minimal inclusion/exclusion criteria. However, clinical characteristics of the patient population are given and all samples appear to have been analysed. Pathologists were blinded to both test results. Discordant case analysis was performed only on the clinical study using only qRT-PCR (two cases), with TAB likely to be less of an issue in this study than in the routine study (17 cases). No details of test failures, other than one case of a false positive as a result of an invalid control, were given. The study was funded by Sysmex.
The study reported by Schem et al. 60 provides minimal information on patient characteristics or recruitment. Five-level histology was used as the reference standard and the outcome assessors were blinded. The samples for OSNA were frozen at –80°C. Specificity was calculated on the first 120 negative nodes (out of 343) using extended histology. Histology was also extended on discordant cases with Western blotting and qRT-PCR performed on the homogenates. Comprehensive details are provided on all of these techniques other than the replicates and test failures. This study was also supported by Sysmex.
Snook et al. 61 report a prospective study comparing OSNA with five-level histology. No patient flow diagram is included, although numbers appear accurate, and minimal patient characteristics are provided. It is unclear if recruitment was consecutive. Outcome assessors were blinded to the OSNA results. Discordant case analysis was performed by Western blotting and qRT-PCR. No financial contribution was received from any organisation; however, support in the form of training and advice and compensation for the additional workload as a result of the study protocol and all necessary material for OSNA analysis were provided by Sysmex.
Tamaki et al. 62 present one of two papers on a Japanese multicentre study. A validity and a routine use trial are reported. For the validity trial, histopathology was extensive, with node sections taken at 0.2-mm intervals. For routine use, a more standard three-level method was used. Brief patient characteristics are presented for each trial with no details on recruitment. No patient flow diagram is provided but the numbers of nodes extracted and analysed comply. Outcome assessors were blinded and discordant cases were analysed by Western blotting and qRT-PCR, although technical details are not provided. This study was supported by Sysmex.
Tsujimoto et al. 64 appear to report on the same trial as Tamaki et al. ,62 with OSNA compared with three-level histology. Minimal details are provided on patient characteristics and there is no description of recruitment. LNs were stored at –80°C until needed. Samples were analysed in duplicate. It is unclear from this report if pathologists were blinded, although if this is in fact the same trial as described in Tamaki et al. 62 then it is reported that they were. Histopathological samples were examined by three third-party pathologists. As in Tamaki et al. ,62 discordant case analysis was performed by Western blotting and qRT-PCR, although it is reported here that these tests were not performed on all samples. Sysmex are acknowledged but there is no explicit mention of financial support.
A second paper by Tamaki et al. 63 reports another multicentre study based in Japan. Comprehensive patient clinical characteristics are given with minimal details provided on recruitment. The description of the OSNA assay is sparse and only one-level histology is employed. No discordant case analysis was performed.
The final study by Visser et al. 65 employed three-level histology on ALNs, which, if negative, underwent a further four-level investigation. The number of patients included was relatively low at 32, although the total number of nodes analysed was 346. No details were provided on patient recruitment and patient characteristics were minimally reported. There was no evidence of replicates for OSNA, but for discordant case analysis qRT-PCR was performed in duplicate. Frozen samples were used for OSNA (the first 120 histologically negative samples). The pathologists do not appear to have been blinded.
Assessment of test accuracy
The assessment of test accuracy for both OSNA and Metasin is hindered by TAB and by comparison with an inconsistent reference standard. When discordant samples are further investigated (usually using either more extensive histopathology or molecular analysis by qRT-PCR or Western blotting) and attributed to TAB, analyses can be adjusted by excluding such cases; however, adjustment cannot be made for heterogeneity between studies with regard to the reference standard; histopathology may be performed extensively across five levels of a LN or more perfunctorily using only one level of a central slice.
It should be noted that ITCs which are found by immunohistochemical staining and not detected by OSNA are considered LN negative as their clinical significance is unknown. 65
Individual results are described alongside a narrative description in the following sections. Summarised results, which have been stratified, are presented in Tables 27–32.
Metasin
(Academic-in-confidence information has been removed.)
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | |
|---|---|---|
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | |
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
The second paper on Metasin, by Sundaresan,40 which like the first is unpublished and therefore has not undergone formal peer review, reports a sensitivity of 92%, a specificity of 97% and an accuracy of 96%. (Academic-in-confidence information has been removed.) Sundaresan40 used three-level histology as the reference standard. In total, 20 cases (1.6%) were reported as failed assays, which the authors consider may have been because of insufficient RNA in the material submitted for molecular analysis. The authors report 56 patients to be discordant and, although they do not present evidence, they consider TAB to be largely responsible (Table 12).
| Metasin | Three-level histopathology (number of cases) | |
|---|---|---|
| Positive | Negative | |
| Positive | 249 | 36 |
| Negative | 20 | 940 |
One-step nucleic acid amplification
Cohort studies
In the study by Castellano et al. 52 the rate of negative cases determined by standard histology (73%) was highly comparable with the rate of negative SLNs determined by OSNA (71%), with no adjustment for TAB. The rate of micrometastases detected by OSNA was significantly higher (18%) than that determined by the standard procedure (8%; p < 0.01), whereas the rate of macrometastases detected by OSNA was lower (11% vs. 20%). The authors mention that this may be a reflection of different patient populations rather than the different techniques. Nevertheless, the positive rates were similar between the two techniques (Table 13).
| Test | Castellano 201252 | Godey 201255 | Osako 201159 |
|---|---|---|---|
| Histology | 28 | 24.8 | 20.3 (95% CI 11.7 to 32.6) |
| OSNA | 29 | 24.4 | 55.5 (95% CI 46.1 to 64.5) |
The authors hypothesise that the morphological evaluation of the diameter of metastases does not perfectly correlate with the tumour load as evaluated by mRNA copies of CK19 and that there could be an overestimation of macrometastases (> 2 mm) compared with micrometastases by histology (> 0.2 mm and < 2 mm) or an underestimation of macrometastases compared with micrometastases by OSNA.
The study by Godey et al. 55 reports a positive rate of 24.4% for OSNA, which is comparable to the rate reported for OSNA by Castellano et al. 52 (29%) (see Table 13). The time required to obtain the final results of the OSNA assay (including transport to the laboratory) depended on the number of SLNs studied. The mean time to receive the final results was 32.9 minutes [standard deviation (SD) 4.9 minutes] for one SLN (n = 94), 36.4 minutes (SD 4.5 minutes) for two SLNs (n = 144), 41.6 minutes (SD 5.2 minutes) for three SLNs (n = 87) and 48.5 minutes (SD 8.7 minutes) for four SLNs (n = 39).
Guillen-Paredes et al. 56 present a cost–benefit analysis. They report a shorter total operative time for OSNA patients, with the mean total time difference between histopathology [mean (SD) 78 (48.02) minutes] and OSNA [mean (SD) 62.14 (21.93) minutes] being statistically significant (p < 0.005). However, when considering only the time of the first operation, the time for histology is shorter (histology 57.11 minutes vs. OSNA 62.14 minutes), although this difference was not statistically significant (p = 0.15).
In the histology group the mean hospital stay after the first operation was 1.8 days (range 1–13 days, SD 2.04 days) and after the second operation was 2.41 days (range 1–6 days, SD 1.29 days, n = 12 patients), resulting in a total hospital stay of 2.44 days, compared with a mean of 1.54 days for OSNA (range 1–4 days, SD 0.78 days); this difference was statistically significant (p < 0.001). In the analysis of complications (minor, major and no complications), there was a statistically significant difference between the histology group 1 (57 patients with complications, considering the first and second operations) and the OSNA group38 (p = 0.015).
The final cohort study is presented by Osako et al. 59 The entire node was used in the OSNA group whereas only a single section of the node was used in the histology group. No significant difference was seen between the positive rate of histological macrometastases and the positive rate of OSNA (++) (18.8%, 95% CI 10.5% to 30.8% vs. 25.2%, 95% CI 17.9% to 34.2%; p = 0.420). However, a significant difference was seen between the positive rate of histological micrometastases and the positive rate of OSNA (+) (1.6%, 95% CI 0.1% to 9.5% vs. 30.3%, 95% CI 22.3% to 39.5%; p < 0.001).
Single-gate studies
The study by Bernet et al. 50 has been included in the review only for the data on the time taken for OSNA analysis (see Assessment of analysis time).
The study by Bernet Vegue et al. 51 investigated only a 1-mm central node slice by histopathology, with the remainder allocated to OSNA (therefore, a high rate of discordance was predicted). Values for sensitivity and specificity were not provided in the paper. A statistically significant overall discrepancy was shown between OSNA and histopathology (p < 0.001).
Discordant results were observed in 51 (9%) nodes, although the discordant case analysis table shows discordant results for only 47 nodes (Table 14). Of the histopathology-negative nodes, 14 were identified as ++ (2.7% of total) 25 as + (4.9% of total) and 8 as low expression (1.6% of total) by OSNA (the authors reported the respective results as 2.5%, 4.5% and 1.4%). The authors suggest that OSNA is of particular relevance for the identification of low-volume metastases.
| OSNA | One-level histopathology (number of cases) | ||
|---|---|---|---|
| Macrometastasis | Micrometastasis | Negative | |
| ++ | 1 | 4 | 14 |
| + | 0 | 1 | 25 |
| + i (low expression) | 0 | 0 | 8 |
| – | 0 | 0 | 514 |
When individual nodes were considered, there were no false negatives for OSNA. In contrast, > 80% of the discordant nodes found to be positive for metastases were not identified by conventional one-level histology. The authors state that one-level histology was chosen to reflect the minimum standard used in many laboratories. 51
The study by Choi et al. 53 reported a sensitivity of 77.8% (95% CI 60% to 90%) for OSNA, which is comparatively low. The authors indicate that this is likely to be the result of TAB; however, the dissection of the node was carried out using a similar method to that used in other studies. The authors report a specificity of 96.3% (95% CI 92% to 99%) and an accuracy of 93% (95% CI 88% to 96%).
Fourteen cases of discordance occurred (Table 15). In one false-positive case (OSNA positive, histology negative), the histopathological result of ITC detection as well as the existence of metastasis in other SLNs (not used for this study) was attributed to the localisation of metastatic foci in the LN. For three more discordant cases the OSNA result was close to the cut-off value, indicating the existence of weak positivity in the sample. For another, even though the OSNA assay provided a ++ judgement, the authors were unable to find any other evidence to support this judgement. They hypothesised that this result was due to the localisation of metastatic foci in the LN as the probability of OSNA false positives is considered to be very low based on the results of previous studies. For another OSNA-positive, histology-negative case, the OSNA assay provided positive results for each of two LNs from this patient. This was also considered to be due to the localisation of metastatic foci in the LNs.
| OSNA | Three-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| ++ | 19 | 2 | 1 | 1 |
| + | 3 | 3 | 0 | 4 |
| + i (low expression) | 1 | 0 | 0 | 0 |
| – | 4 | 4 | 3 | 154 |
The false-negative cases (OSNA–, histology+) were investigated immunohistochemically to confirm the protein level of CK19 in tumours. Four cases showed expression of CK19 protein in < 10% of the tumour. The cause of these discordant results may have been the inability of the OSNA assay to detect metastasis of breast cancers with low CK19 expression. In two other cases, metastases were histopathologically detected by the postoperative examination of permanent specimens. CK19 immunohistochemical analysis of two cases was not performed because of insufficient tissue and these results remain inconclusive.
Following adjustment for TAB, the study by Feldman et al. 54 reported a sensitivity of 82.7%, a specificity of 97.7% and an accuracy of 95.8% for OSNA compared with reference pathology.
Seventy-one cases were considered discordant, including two pathology assessment reversals because of TAB. (Table 16). Although OSNA failed to detect nine macrometastases and 22 micrometastases that were identified by reference pathology, it detected nine macrometastases and 29 micrometastases that were identified as negative or as ITCs by reference pathology. All of these nine macrometastases were identified as true misses by reference pathology, and recalculation of the assay performance for macrometastasis yielded a PPV of 100% for an OSNA (++) result.
| OSNA | Three-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| ++ | 77 | 9 | 1 | 8 |
| + | 9 | 12 | 0 | 29 |
| – | 9 | 22 | 14 | 854 |
For macrometastases, all nine discordant OSNA (++) results were identified as true positives, and nine of 29 discordant OSNA (+) results were also identified as true positives on discordant case analysis. Although 20 of the 29 discordant OSNA (+) results could not be confirmed, the median CK19 copy number in these samples was close to the assay cut-off, suggesting that these cases involved very small micrometastases, and this did not rule out the possibility of TAB.
The sensitivity and specificity of OSNA were lower than the values reported in previous smaller studies, possibly because this study evaluated solely SLNs; slicing was at 1-mm intervals rather than at 2-mm intervals; and there were a large number of micrometastases.
The study by Le Frère-Belda et al. 58 reported a sensitivity of 91.1% (95% CI 80.3% to 97.1%) and a specificity of 97.2% (95% CI 95.1% to 98.6%) for OSNA compared with five-level histology after adjustment for TAB.
The authors report 39 cases of discordance (Table 17) and mention that, as different parts of the node were used for each method, because each technique required a different method of tissue preparation, discrepancies between the OSNA results and the histological results were expected. 58 Twenty-seven cases were histology negative/OSNA positive. All 27 samples remained histologically negative after further examination of histological slices, whereas 15 samples had OSNA-positive lysates after further molecular analysis by qRT-PCR; this was suggested by the authors to be due to TAB. Twelve histology-positive/OSNA-negative samples were found. One was not investigated by further molecular analysis and seven were found to be negative on further molecular analysis.
| OSNA | Five-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| n = 503 SLNs | ||||
| ++ | 37 | 6 | 0 | 3 |
| + | 5 | 3 | 1 | 23 |
| – | 3 | 9 | 27 | 386 |
| n = 233 patients | ||||
| ++ | 22 | 6 | 0 | 3 |
| + | 2 | 3 | 3 | 17 |
| – | 2 | 7 | 17 | 151 |
For the study presented by Godey et al. ,55 a positive rate of 24.4% is reported for OSNA and a positive rate of 24.8% is reported for histology; the rate of 24.4% for OSNA is comparable to the rate of 29% for OSNA reported by Castellano et al. 52
Khaddage et al. 57 reported on OSNA compared with both five-level histology and routine one-level histology. For the extensive histology after adjustment for TAB, presented in Table 18, the sensitivity, specificity and accuracy of OSNA were 100%, 98.4% and 98.7% respectively.
| OSNA | Five-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasisa | ITCs | Negative | |
| n = 80 SLNs (validation study) | ||||
| ++ | 11 | 2 | – | 0 |
| + | 2 | 0 | – | 1 |
| – | 0 | 0 (2) | 2 | 60 |
| n = 46 patients (validation study) | ||||
| ++ | 6 | 2 | 0 | 0 |
| + | 0 | 0 | 0 | 1 |
| – | 0 | 0 (2) | 2 | 33 |
| One-level histopathology | ||||
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| n = 197 patients (routine use) | ||||
| ++ | 9 | 1 | 1 | 2 |
| + | 8 | 7 | 1 | 13 |
| – | 0 | 0 | 1 | 154 |
Only three results appear to be discordant for five-level histology. In the two cases in which micrometastases were detected by histology but the OSNA sample was negative, further molecular analysis by qRT-PCR of CK19 and two breast cancer-specific markers displayed negative results; therefore, TAB was assumed to have occurred. For the one case with a positive result for OSNA but not histopathology, the sample was very close to the cut-off level, indicating very small tumour deposits. Seventeen cases were discordant for routine use histology, all histology negative and OSNA positive, three of which had non-SLN involvement and fourteen of which indicated micrometastases and the likelihood of TAB.
The study by Schem et al. 60 reported a sensitivity of 98.1% and a specificity of 91.7% for OSNA compared with five-level histopathology. After adjustment for TAB, sensitivity increased to 100% and specificity to 96.5%. A total of 28 cases were considered discordant (Table 19).
| OSNA | Five-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| ++ | 90 | 7 | 0 | 9 |
| + | 7 | – | 1 | 16 |
| – | 0 | 2 | 2 | 209 |
Ribonucleic acid and proteins were extracted from the lysates of the 28 discordant cases, followed by analysis for CK19 by qRT-PCR and Western blotting. If the data obtained by the additional analyses were consistent with the results obtained by OSNA, TAB was considered likely and these samples were excluded from the sample cohort.
In both of the OSNA-negative/histology-positive samples and in 11 out of the 26 OSNA-positive/histology-negative samples, discordant case analysis revealed equivalent results to those obtained with the OSNA assay. As such, it cannot be fully excluded that an even higher proportion of discordant results was due to TAB because the homogenates were exposed to long storage and transport times, which might have lowered the concentration and quality of the RNA and proteins. A further six OSNA-positive/histology-negative samples had a CK19 mRNA copy number of < 1000/µl. The authors suggest that, with a cut-off level of 250 copies/µl, these positive OSNA results very likely indicate a low tumour burden in the LNs, which was probably absent in the tissue sections used for histological investigation. For samples close to the cut-off level, long storage and travel times can affect the concentration and quality of the RNA and proteins.
Snook et al. 61 compared OSNA with five-level histology. OSNA was reported to have a sensitivity of 91.7%, a specificity of 96.9% and an accuracy of 96.0%, after adjustment for TAB.
A total of 33 cases were considered to be discordant, with 17 reported to be due to TAB following qRT-PCR and Western blot analysis. Sixteen samples were reported as truly discordant, 10 false positives and six false negatives (Table 20). The OSNA false positives with a low copy number were attributed to the possible presence of small micrometastases that were not apparent on histology, although the authors also mention that there is a possibility of contamination as CK19 is expressed on the surface of cells of epithelial origin, benign or malignant, such as breast, colonic or gastric cells. 116 Ideally, the cut-off value for the OSNA test reduces the likelihood of this.
| OSNA | Five-level histopathology (number of cases) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITCs | Negative | |
| n = 395 SLNs | ||||
| ++ | 48 | 1 | 0 | 0 |
| + | 8 | 9 | 0 | 10 |
| – | 4 | 2 | 20 | 293 |
| n = 194 patients | ||||
| ++ | 33 | 1 | 0 | 0 |
| + | 5 | 5 | 1 | 7 |
| – | 4 | 1 | 11 | 126 |
As histopathology is prone to sampling bias, that is, the number of levels of histopathology can influence the result, the authors also question if histopathology is appropriate as a gold standard. With regard to further molecular analysis, the authors consider the possibility of a reduction in RNA concentration in the frozen lysates of SLN samples because of the freeze–thaw effect, which could account for some of the discordant cases, despite repeat molecular testing.
Tamaki et al. 62 present results for two trials, as reported in Table 21, which are also reported on by Tsujimoto et al. 64 Trial 1 compared the specificity of OSNA with that of detailed histology (0.2-mm sections), to confirm the validity of the index test. Trial 2 was intended to replicate routine use and only three-level histology took place.
| OSNA | 0.2-mm section histopathology, number of cases (n = 124 ALNs, trial 1) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITC | Negative | |
| Positive | 16 | 3 | NR | 3 |
| Negative | 0 | 1 | NR | 101 |
| OSNA | Three-level histopathology (n = 551 ALNs, trial 2) | |||
| Macrometastasis | Micrometastasis | ITC | Negative | |
| Positive | 64 | 6 | NR | 22 |
| Negative | 4 | 6 | NR | 348 |
Sensitivity and specificity for trial 1 were reported as 95% and 97%, respectively, whereas sensitivity and specificity for trial 2 were reported as 87.5% and 94%, respectively, before adjustment for TAB and 87.7% and 94.3%, respectively, after adjustment for TAB.
In trial 2, 32 nodes displayed discordant results (7.1%). For eight of the 10 false-negative nodes, not all of the serial sections contained metastasised foci and therefore they may not have been in the OSNA samples. Two nodes from different patients among the false-negative cases displayed very weak expression of CK19 mRNA. The primary tumours of these patients also stained negative for CK19 on immunohistochemistry.
Isolated tumour cells were found in the remaining specimens of five of the 22 false-positive nodes after additional sectioning, which had not been detected by routine pathological examination using 2-mm interval sections. In another eight false-positive nodes, no tumour cells were found in the pieces remaining after pathological examination, but lymphatic vascular invasions were observed in the main tumours. In addition, the lysates of two of the nodes preserved for the OSNA assay contained a significant amount of CK19 protein.
As for the pathologically positive LNs (diameter of metastasis > 0.2 mm), 87.7% could be detected using the OSNA assay, whereas 94.1% of the macrometastases (> 2 mm) were assessed as positive.
The second paper by Tamaki et al. 63 compared OSNA with one-level histology (Table 22).
| OSNA | One-level histopathology, number of cases [n = 417 patients (SLNs)] | |
|---|---|---|
| Positive | Negative | |
| Positive | 58 | 36 |
| Negative | 8 | 315 |
Forty-four cases were discordant, considered to be inevitable by the authors because of the method of tissue sampling used. In eight cases OSNA was negative but postoperative pathological examination identified metastasis. Of these, seven patients had micrometastases and discordance was suspected to be due to uneven allocation of minuscule metastases in a SLN. However, in the remaining patient with macrometastasis, low expression of the CK19 protein in the main tumour was confirmed, leading to a negative OSNA result, although, the authors state that it is not necessarily the case that low expression of CK19 protein is reflected in low expression of CK19 mRNA.
There were also 36 OSNA-positive, histology-negative discordant cases, including two with ITCs in the SLNs as assessed by pathology. Of these, non-SLN metastases were subsequently identified in seven patients; therefore, the OSNA results in these patients were accurate. Microinvasion was suspected in a core-needle biopsy specimen from one patient. Two patients had widespread DCIS that measured > 6 cm and another had multiple lesions. A further two patients had high-grade DCIS.
The authors suggest that, despite the cut-off, OSNA can detect metastases with high sensitivity even in tumours diagnosed pathologically as DCIS and that such findings may result in an upgrade of the clinical stage of such tumours.
Tsujimoto et al. 64 report on the same trial as Tamaki et al. 62 Compared with three-level histology (n = 325 SLNs and ALNs), OSNA sensitivity was reported as 91.1% and accuracy was reported as 98.2% (Table 23).
| OSNA | Three-level histopathology, number of cases (n = 325 SLNs and ALNs) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITC | Negative | |
| ++ | 34 | 0 | 0 | 0 |
| + | 6 | 3 | 0 | 4 |
| – | 0 | 2 | 13 | 263 |
| OSNA | Three-level histopathology, number of cases (n = 81 SLNs) | |||
| Macrometastasis | Micrometastasis | ITC | Negative | |
| ++ | 11 | 0 | 0 | 0 |
| + | 1 | 2 | 0 | 1 |
| – | 0 | 2 | 3 | 61 |
| OSNA | 0.2 mm-interval histopathology, number of cases (n = 144 SLNs from pN0 patients) | |||
| Macrometastasis | Micrometastasis | ITC | Negative | |
| ++ | 0 | 0 | 0 | 0 |
| + | 0 | 0 | 0 | 0 |
| – | 0 | 0 | 3 | 141 |
Six discordant cases were observed (for the trial with 325 nodes) between the OSNA assay and histopathology. Western blot analysis of two discordant cases (OSNA positive/histology negative) showed the presence of an amount of CK19 protein equivalent to micrometastasis. The authors considered that, although the possible presence of benign epithelial cells such as glandular inclusions in the LNs cannot be eliminated, the results may be better explained by the presence of metastatic foci in the LNs in light of the results of the specificity study and the amount of CK19 protein expression. This is further supported by the lack of false positives reported for the pN0 patients. Two other cases were negative according to the OSNA assay but were judged positive for micrometastasis according to three-level histopathology. The two remaining cases were not tested.
The final single-gate study included in this review was presented by Visser et al. ,65 who compared OSNA with five-level histology (Table 24). After adjustment for TAB, a sensitivity of 95.3%, specificity of 97.1% and accuracy of 96.8% were reported.
| OSNA | Five-level histopathology, number of cases (n = 346 ALNs) | |||
|---|---|---|---|---|
| Macrometastasis | Micrometastasis | ITC | Negative | |
| ++ | 50 | 4 | 0 | 2 |
| + | 2 | 5 | 0 | 13 |
| – | 1 | 2 | 3 | 264 |
To establish the level of discordance as a result of TAB, the first 120 histologically negative LNs, as determined by five-level histology, were cut into further levels at intervals of 250 µm.
Seven discordant cases, considered to be due to TAB, are not included in Table 24. A further 18 unresolved cases were reported. In eight cases, only Western blot results for CK19 protein could be obtained because poor-quality RNA did not allow qRT-PCR analysis to be performed. One of three histology-positive/OSNA-negative samples yielded negative results. In the lysates of the other two samples, CK19 protein levels were slightly above the cut-off value, suggesting the presence of small tumour deposits. In 11 histologically negative/OSNA-positive samples, low CK19 mRNA copy numbers (250–750/µL) were found with OSNA. Six of these could be further analysed by qRT-PCR, whereas the remaining five samples suffered from poor RNA quality. The same was true for one sample with a high CK19 mRNA copy number.
Discordance
Studies show a range of discordance, from 1.8% to 8.6% (Table 25); further details are provided in Table 26. The greatest discordance was generally for OSNA false positives. This is a concern as it would potentially lead to unnecessary complete axillary dissection in patients. This may be due to epithelial contamination although, for OSNA, the CK19 copy number cut-off corresponds to the presence of 5000 tumour cells. Therefore, it is unlikely that positive results would occur because of epithelial displacement or illegitimate transcription, which involves only 500–1000 non-tumour cells. 65 By contrast, false-negative tests may result in a lack of treatment in the early stages of cancer, leading to a worse prognosis. The patient may also be required to undergo a second operation for ALND.
| Study | Total cases (%) | Attributed to TAB (%) | H+/O– or H+/M– (%) | H–/O+ or H–/M+ (%) |
|---|---|---|---|---|
| OSNA | ||||
| Bernet Vegue 201251 | 8.3 | NR | 0 | 8.3 |
| Choi 201053 | 7 | NR | 4 | 3 |
| Feldman 201154 | 6.6 | 2.7 | 3.0 | 3.6 |
| Khaddage 201157 (study phase) | 3.7 | 0 | 2.5 | 1.2 |
| Khaddage 201157 (routine use) | 8.6 | NR | 0 | 8.6 |
| Le Frère-Belda 201258 | 7.7 | 4.3 | 2.3 | 5.4 |
| Schem 200960 | 8.2 | 3.8 | 0.6 | 7.6 |
| Snook 201161 | 8.3 | 4.3 | 3.0 | 5.3 |
| Tamaki 200962 (trial 2) | 7.1 | NR | 2.2 | 4.9 |
| Tamaki 201263 | 5.7 | NR | 1.0 | 4.6 |
| Tsujimoto 200764 | 1.8 | NR | 0.6 | 1.2 |
| Visser 200865 | 5.2 | 2 | 0.9 | 4.3 |
| Metasin | ||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
| Sunderasan40 | 4.2 | NR | 1.5 | 2.6 |
| Study | n | Discordant analysis | Unadjusted 2 × 2 | Adjusted 2 × 2 | Numbers reclassified | Comments on nature and impact of discordant case analysis | |||
|---|---|---|---|---|---|---|---|---|---|
| Bernet Vegue 201251 | |||||||||
| Axillary node level | 567 | No further analysis performed | TP | 6 | TP | – | FP to TN | – | Results not adjusted. In total, 17 FPs had < 1000 copies/µl; relatively close to cut-off for micrometastases |
| FP | 39 | FP | – | FP to TP | – | ||||
| FN | 0 | FN | – | FP to FN | – | ||||
| TN | 522 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Choi 201053 | |||||||||
| Patient level | 199 | In discordant cases, clinical information, status of non-SLNs and expression of CK19 protein in LN metastatic foci were evaluated on a patient basis | TP | 27 | TP | – | FP to TN | – | Results not adjusted. For one FP and one FN, discordance was attributed to the location of metastases. Three FPs had a low copy number and weak positivity. One FP had metastases in other SLNs and the result was considered to be the result of TAB. Four FNs displayed < 10% CK19 by immunohistochemistry and were therefore not detected by OSNA. For one FP and three FNs the discordance was unresolved |
| FP | 6 | FP | – | FP to TP | – | ||||
| FN | 9 | FN | – | FP to FN | – | ||||
| TN | 157 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Feldman 201154 | |||||||||
| SLN level | 1044 | Blank tissue sections were stained with CK19-specific antibody; back-up samples were retested with OSNA to check for operator errors; Western blot analysis of CK19 and qRT-PCR | TP | 107 | TP | 125 | FP to TN | – | There were two pathology assessment reversals after assessment of TAB; unclear whether this affects TP or TN result. In total, 28 discordant results were resolved but specific results were not given other than 18 FPs were found to be TPs by histology (assumed to be because of TAB) and one FN was confirmed by molecular analysis. In total, 20 FPs had small deposits close to the cut-off level. A total of 1018 nodes were used for the adjusted analysis but it is unclear which results have been changed as the numbers do not agree |
| FP | 38 | FP | 20 | FP to TP | – | ||||
| FN | 31 | FN | – | FP to FN | – | ||||
| TN | 868 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Khaddage 201157 | |||||||||
| Patient level (study phase) | 46 | Discordant case analysis consisted of qRT-PCR | TP | 8 | TP | 8 | FP to TN | 0 | Two FNs – in one case micrometastases were confined to two of five-level histology; in the other case, one slice was metastasis free for histology. Further molecular analysis was negative indicating TAB. One FP – close to the cut-off level indicating very small tumour deposits |
| FP | 1 | FP | 1 | FP to TP | 0 | ||||
| FN | 2 | FN | 0 | FP to FN | 0 | ||||
| TN | 35 | TN | 35 | FPs excluded | 0 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 2 | ||||||||
| Other | 0 | ||||||||
| SLN level (study phase) | 80 | TP | 15 | TP | 15 | FP to TN | 0 | ||
| FP | 1 | FP | 1 | FP to TP | 0 | ||||
| FN | 2 | FN | 0 | FP to FN | 0 | ||||
| TN | 62 | TN | 62 | FPs excluded | 0 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 2 | ||||||||
| Other | 0 | ||||||||
| Patient level (clinical phase) | 197 | TP | 25 | TP | – | FP to TN | 0 | ||
| FP | 17 | FP | – | FP to TP | 0 | ||||
| FN | 0 | FN | – | FP to FN | 0 | ||||
| TN | 155 | TN | – | FPs excluded | 0 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 0 | ||||||||
| Other | 0 | ||||||||
| Le Frère-Belda 201258 | |||||||||
| Patient level | 233 | FPs (O+, H–): slides made for all the portions of the half-node allocated to histology (b and d) that were not used initially FPs (O+, H–) and FNs (O+, H–): remaining homogenate from the portions of the half-node allocated to OSNA (a and c) shipped to Sysmex for blind molecular analysis |
TP | 33 | TP | 32 | FP to TN | 0 | Study assumes that TAB is the only reason why there may be discordance. If initial histology was confirmed or initial OSNA+ or OSNA– results were confirmed, TAB was assumed to have occurred and the patient affected was excluded from the analysis |
| FP | 23 | FP | 12 | FP to TP | 0 | ||||
| FN | 9 | FN | 3 | FP to FN | 0 | ||||
| TN | 168 | TN | 168 | FPs excluded | 11 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 6 | ||||||||
| Other | 1 TP excluded | ||||||||
| SLN level | 503 | TP | 51 | TP | 51 | FP to TN | 0 | ||
| FP | 27 | FP | 12 | FP to TP | 0 | ||||
| FN | 12 | FN | 5 | FP to FN | 0 | ||||
| TN | 413 | TN | 413 | FPs excluded | 15 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 7 | ||||||||
| Other | 0 | ||||||||
| Academic-in-confidence information has been removed | |||||||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to TN | – | |
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to TP | – | ||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to FN | – | ||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to TN | – | Academic-in-confidence information has been removed |
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to TP | – | ||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FP to FN | – | ||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Schem 200960 | |||||||||
| ALN level | 343 | For discordant results, histological work-up was extended until no tissue remained in the paraffin blocks. The homogenates were also analysed by Western blotting and qRT-PCR | TP | 104 | TP | 104 | FP to TN | 0 | Provided that the supplemental analyses gave the same result as OSNA (either histology or molecular), the samples were excluded as an uneven distribution of the metastases (TAB) was likely to be the cause. The two FNs also gave negative results on Western blotting and qRT-PCR. For 11 FPs analyses indicated the presence of tumour deposits in the slices allocated to OSNA |
| FP | 26 | FP | 15 | FP to TP | 0 | ||||
| FN | 2 | FN | 0 | FP to FN | 0 | ||||
| TN | 211 | TN | 211 | FPs excluded | 11 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 2 | ||||||||
| Other | 0 | ||||||||
| Snook 201161 | |||||||||
| Patient level | 194 | Homogenates were analysed using qRT-PCR and Western blot analysis | TP | – | TP | 44 | FP to TN | – | After discordant case analysis and the exclusion of samples affected by TAB, nine patients were excluded from the final analysis. No further details were given |
| FP | – | FP | 8 | FP to TP | – | ||||
| FN | – | FN | 5 | FP to FN | – | ||||
| TN | – | TN | 137 | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| SLN level | 395 | TP | – | TP | 66 | FP to TN | 0 | There were 33 discordant nodes. In total, 17 nodes considered to be affected by TAB (11 FPs, six FNs) were excluded from the final analysis. The remaining 16 cases were unresolved (10 FPs, six FNs). Four FPs with a low copy number were assumed to have small micrometastases. Of the six FNs, three had metastases in ‘internal’ levels and unlikely to be present in OSNA slices. A further two suggest metastases only on one or two levels | |
| FP | – | FP | 10 | FP to TP | 0 | ||||
| FN | – | FN | 6 | FP to FN | 0 | ||||
| TN | – | TN | 313 | FPs excluded | 11 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 6 | ||||||||
| Other | – | ||||||||
| Sundaresan40 | |||||||||
| Patient level | 1245 | Not described – deeper-level histology | TP | 249 | TP | 251 | FP to TN | 0 | One FN was changed to ITCs. Deeper levels of histology revealed metastases in two cases |
| FP | 36 | FP | 34 | FP to TP | 2 | ||||
| FN | 20 | FN | 19 | FP to FN | 0 | ||||
| TN | 940 | TN | 941 | FPs excluded | 0 | ||||
| FN to TN | 1 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 0 | ||||||||
| Other | 0 | ||||||||
| SLN level | 2206 | Not described | TP | 341 | TP | 343 | FP to TN | 0 | |
| FP | 60 | FP | 58 | FP to TP | 2 | ||||
| FN | 35 | FN | 34 | FP to FN | 0 | ||||
| TN | 1770 | TN | 1771 | FPs excluded | 0 | ||||
| FN to TN | 1 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 0 | ||||||||
| Other | 0 | ||||||||
| Tamaki 200962 (trial 2) | |||||||||
| SLN level | 450 | For discordant nodes, the remaining pathological specimen blocks were sectioned at 0.2-mm intervals and examined for CK19. The lysate was examined for CK19 protein expression by Western blotting | TP | 70 | TP | 71 | FP to TN | 0 | For eight FNs, uneven localisation of tumour cells was found in remnants of the nodes. Two other FNs showed faint expression of CK19 by immunohistochemistry. In five of 22 FPs, some foci of tumour cells were detected in node remnants. For eight FPs, there were no tumour cells in remnants but there were lymphatic vascular invasions in the main tumours, with two containing significant CK19 protein in the lysates. Further analysis of the nine remaining nodes showed no signs of metastasis. The results were adjusted for only one node, with a FP confirmed by reanalyisis of histology to be a TP; therefore, the two techniques agreed |
| FP | 22 | FP | 21 | FP to TP | 1 | ||||
| FN | 10 | FN | 10 | FP to FN | 0 | ||||
| TN | 348 | TN | 348 | FPs excluded | 0 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 0 | ||||||||
| Other | 0 | ||||||||
| Tamaki 201263 | |||||||||
| Patient level | 417 | No additional analyses mentioned in the methods section | TP | 58 | TP | – | FP to TN | – | No adjustments made; however, seven FNs were assessed to have micrometastases by histopathology. Macrometastasis was present in one FN but immunohistochemistry revealed low CK19 protein expression.There were ITCs in SLNs from two FPs and non-SLN metastases in seven FPs. Therefore, nine FPs harboured cancer cells in the ALNs |
| FP | 36 | FP | – | FP to TP | – | ||||
| FN | 8 | FN | – | FP to FN | – | ||||
| TN | 315 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Tsujimoto 200764 | |||||||||
| Patient level (ALNs and SLNs) | 325 | qRT-PCR and CK19 Western blot analysis of the lysates were carried out. Histopathology was repeated, examined and evaluated by third-party pathologists | TP | 43 | TP | – | FP to TN | – | Adjustment of results was not performed. There were six discordant cases. In two FPs, micrometastasis was observed on Western blot analysis. For the other two FPs, Western blot analysis was not performed. The reasons for the discordance for the FNs are unclear |
| FP | 4 | FP | – | FP to TP | – | ||||
| FN | 2 | FN | – | FP to FN | – | ||||
| TN | 276 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| SLN level | 81 | TP | 14 | TP | – | FP to TN | – | ||
| FP | 1 | FP | – | FP to TP | – | ||||
| FN | 2 | FN | – | FP to FN | – | ||||
| TN | 64 | TN | – | FPs excluded | – | ||||
| FN to TN | – | ||||||||
| FN to TP | – | ||||||||
| FN to FP | – | ||||||||
| FNs excluded | – | ||||||||
| Other | – | ||||||||
| Visser 200865 | |||||||||
| ALN level | 346 | For discordant samples, histology was extended to all levels. The lysates were analysed by qRT-PCR and Western blot analysis | TP | 61 | TP | 61 | FP to TN | 0 | If further analysis yielded a result compatible with a positive OSNA result, these samples were excluded from the final analysis because of the indication of sampling bias. Seven FPs were considered to be the result of sampling bias, leaving 11 discordant cases unresolved. In eight cases, poor-quality RNA prevented analysis by qRT-PCR. One FN case gave a negative result. The two other FNs had values close to the cut-off level. In 11 FP samples the copy number was also low |
| FP | 15 | FP | 8 | FP to TP | 0 | ||||
| FN | 3 | FN | 3 | FP to FN | 0 | ||||
| TN | 267 | TN | 267 | FPs excluded | 7 | ||||
| FN to TN | 0 | ||||||||
| FN to TP | 0 | ||||||||
| FN to FP | 0 | ||||||||
| FNs excluded | 0 | ||||||||
| Other | 0 | ||||||||
Although the true number of discordant cases and the number that result from TAB cannot be fully differentiated, the studies that have adjusted their results display increased specificity and sensitivity for OSNA. Interestingly, the studies that analyse a full node with extensive histology showed no significant difference in positive rates between OSNA and histology,52,55 whereas the study using one-level histology demonstrated a significantly higher positive rate for OSNA. 59
Summary of test accuracy
The results are summarised and stratified according to SLNs, ALNs, patients, before TAB and after TAB in Tables 27–32. An overall summary is provided in Table 33.
With regard to Metasin, the results must be used with caution as they have been taken from unpublished, non-peer-reviewed papers. These papers are also very much in draft form and therefore lacking in detail. That said, with regard to quality, many of the issues (such as lack of information about replicate measurements and therefore no estimate of the reproducibility and the robustness of the test) apply to both papers reporting OSNA and (academic-in-confidence information has been removed).
One-step nucleic acid amplification detects only CK19 mRNA expression. It has been reported that 98.2% of breast cancer tumours express CK19 protein, leaving 1.8% of patients for whom this technique may be invalid. 60 However, Tamaki et al. 62 suggest that it is not necessarily the case that low expression of CK19 protein is reflected in low expression of CK19 mRNA. In contrast, Metasin, which also identifies mammaglobin, (academic-in-confidence information has been removed) compared with that of OSNA, which ranges from 77.8% to 80%. 40
As the reference standard, immunohistochemistry is capable of detecting ITCs, unlike OSNA (and presumably Metasin). However, as the American Society of Clinical Oncology (ASCO)117 and the NICE clinical guideline for early breast cancer13 indicate that ITCs have an unknown clinical significance, and there are insufficient data to recommend appropriate treatment, ITCs were considered histologically negative throughout this review.
Although the aim of OSNA and Metasin is to perform intraoperative molecular analysis, some studies employed a more experimental approach and used frozen samples of RNA, either initially or in discordant case analysis. Both storage and the freeze–thaw effect have been known to adversely affect RNA, effectively leading to a reduction in concentration.
Histopathology will always be limited by sampling bias as only a certain number of slices are taken. This can be increased or reduced according to requirements but will have cost implications and there will always be unanalysed tissue. Furthermore, there is the possibility of observer subjectivity. In contrast, for OSNA or Metasin the whole node can be used and the semiquantitative test is objective.
There is also the issue of TAB. Unfortunately, because of the nature of the OSNA/Metasin tests and of histopathology, it is not possible to use the same tissue sample for both tests and thus TAB is likely to occur. There is no DTA study design that can resolve this issue and, although cohort studies can provide us with some data on whole-node analysis, the tissue being analysed by each test is still, of course, different. Studies have attempted to mitigate the issue of TAB by investigating discordant cases. However, there is a lack of consistency between studies with regard to the methods used to investigate discordant cases, and there will always be uncertainty attached to decisions about whether TAB has occurred.
With regard to the overall results, the range for specificity is lower for results presented by patient rather than by node, before and after adjustment for TAB, as shown in Tables 27–33. The results produced by meta-analysis agree with this (Table 34). Sensitivity before adjustment for TAB is similar for both patients and nodes, whereas, after TAB adjustment, sensitivity is slightly greater for nodes. This may be because some studies use a larger number of nodes from a small number of patients.
| Study | Patients, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Metasin | ||||||||
| (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | NR | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) |
| Sundaresan40 | 1265a | Three | 249 | 940 | 20 | 36 | 92 (89 to 95)b | 97 (95 to 97)b |
| OSNA | ||||||||
| Choi 201053 (SLNs) | 199 | Three | 27 | 157 | 9 | 6 | 77.8 (60 to 90) | 96.3 (92 to 99) |
| Khaddage 201157 (SLNs) | 46 | Five | 8 | 35 | 2 | 1 | 80.0 (44.4 to 97.5)b | 97.2 (85.5 to 99.9)b |
| Khaddage 201157 (SLNs) | 197 | One | 25 | 155 | 0 | 17 | 100 (88.7 to 100)c | 90.1 (84.6 to 94.1)c |
| Le Frère-Belda 201258 (SLNs) | 233 | Five | 33 | 168 | 9 | 23 | 78.6 (63.1 to 89.7) | 88.0 (82.4 to 92.3) |
| Tamaki 201263 (SLNs) | 417 | One | 58 | 315 | 8 | 36 | 87.9 (77.5 to 94.6)c | 89.7 (86.1 to 92.7)c |
| Bernet Vegue 201251 (non-SLNs) | 55 | One | 6 | 26 | 0 | 23 | 100 (41.4 to 100)c | 53.1 (38.3 to 67.5)c |
| Study | Patients, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| OSNA | ||||||||
| Khaddage 201157 | 46 | Five | NR | NR | NR | NR | 100a | 97.2a |
| Le Frère-Belda 201258 | 215 | Five | 32 | 168 | 3 | 12 | 91.4 (76.9 to 98.2) | 93.3 (88.6 to 96.6) |
| Snook 201161 | 194 | Five | 44 | 137 | 5 | 8 | 89.8 (77.8 to 96.6)b | 94.5 (89.4 to 97.6)a |
| Study | Sample, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Metasin | ||||||||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
| Sundaresan40 | 2279a | Three | 341 | 1770 | 35 | 60 | 93 (87.3 to 93.4)b,c | 97 (95.8 to 97.5)b |
| OSNA | ||||||||
| Feldman 201154 | 1044 | Three | 107 | 868 | 31 | 38 | 77.5 (69.7 to 84.2) | 95.8 (94.3 to 97.0) |
| Khaddage 201157 | 80 | Five | 15 | 62 | 2 | 1 | 88.2 (63.6 to 98.5)b | 98.4 (91.5 to 100)b |
| Le Frère-Belda 201258 | 503 | Five | 51 | 413 | 12 | 27 | 80.9 (69.0 to 89.8) | 93.9 (91.2 to 96.0) |
| Tsujimoto 200764 | 81 | Three | 14 | 64 | 2 | 1 | 87.5 (61.7 to 98.4)d | 98.5 (91.7 to 100)d |
| Study | Sample, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| OSNA | ||||||||
| Feldman 201154 | 1018 | Three | NR | NR | NR | NR | 82.7a | 97.7a |
| Khaddage 201157 | 78 | Five | NR | NR | NR | NR | 100a | 98.4a |
| Le Frère-Belda 201258 | 481 | Five | 51 | 413 | 5 | 12 | 91.1 (80.3 to 97.1) | 97.2 (95.1 to 98.6) |
| Snook 201161 | 395 | Five | 66 | 313 | 6 | 10 | 91.7 (82.7 to 96.9)b | 96.9 (94.4 to 98.5)b |
| Study | Sample, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| OSNA | ||||||||
| Bernet 201150 | 567 ALNs | One | 6 | 522 | 0 | 39 | 100 (60.7 to 100)a | 93.0 (90.6 to 95.0)a |
| Schem 200960 | 343 ALNs | Five | 104 | 211 | 2 | 26 | 98.1 (93.4 to 99.8)b | 91.7 (84.3 to 92.7)c |
| Tamaki 200962 | 124 ALNs | Sectioned at 0.2-mm intervals | 19 | 101 | 1 | 3 | 95 (75.1 to 99.9) | 97.1 (91.8 to 99.4) |
| Tamaki 200962 | 450 ALNs | Three | 70 | 348 | 10 | 22 | 87.5 (78.2 to 93.8) | 94.1 (91.0 to 96.3) |
| Tsujimoto 200764 | 325 ALNs/SLNs | Three | 43 | 276 | 2 | 4 | 95.6 (84.9 to 99.5)a | 98.6 (96.4 to 99.6)a |
| Visser 200865 | 346 ALNs | Three | 61 | 267 | 3 | 15 | 95.3 (84.9 to 99.5)b | 94.7 (96.4 to 99.6)b |
| Study | Sample, n | Histology level | H+/O+ | H–/O– | H+/O– | H–/O+ | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|---|---|---|
| OSNA | ||||||||
| Schem 200960 | 330 ALNs | Five | NR | NR | NR | NR | 100a | 95.6a |
| Tamaki 200962 | 450 ALNs | Sectioned at 0.2-mm intervals | 71 | 348 | 10 | 21 | 87.7 (78.5 to 93.9) | 94.3 (95.3 to 98.8) |
| Visser 200865 | 339 ALNs | Three | NR | NR | NR | NR | 95.3a | 97.1a |
| Sample type | Sensitivity (%) | Specificity (%) |
|---|---|---|
| Patients before adjustment for TAB (Metasin) | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
| Patients before adjustment for TAB (OSNA – SLNs) | 77.8–80.0 | 88.0–97.2 |
| Patients after adjustment for TAB (OSNA – SLNs) | 89.8–100 | 93.3–97.2 |
| SLNs before adjustment for TAB (OSNA) | 77.5–88.2 | 93.9–98.4 |
| SLNs after adjustment for TAB (OSNA) | 82.7–100 | 96.9–98.4 |
| ALNs before adjustment for TAB (OSNA) | 87.5–98.1 | 91.7–97.1 |
| ALNs after adjustment for TAB (OSNA) | 87.7–100 | 94.3–97.1 |
With the exception of the study by Feldman et al. ,54 the ranges for sensitivity and specificity for ALNs and SLNs appear to be similar. The results of the meta-analysis for OSNA are shown in Table 34.
| Sample type | Adjustment for TAB | Number of studies | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|
| Patient | No | 5 | 84.5 (74.7 to 91.0) | 91.8 (87.8 to 94.6) |
| Patient | Yes | 3 | 91.3 (83.6 to 95.6) | 94.2 (91.2 to 96.2) |
| SLN | No | 4 | 79.9 (74.2 to 84.6) | 95.5 (94.1 to 96.5) |
| SLN | Yes | 5 | 89.0 (82.1 to 93.4) | 97.5 (96.6 to 98.2) |
| ALN | No | 6 | 95.1 (90.0 to 97.6) | 94.9 (91.2 to 96.9) |
| ALN | Yes | 4 | 96.5 (87.3 to 99.1) | 96.2 (93.4 to 97.8) |
For the meta-analysis of test accuracy based on analysis of SLNs without adjustment for TAB, we were not aware of a compelling reason to believe that the positivity threshold might vary between studies and so for consistency with meta-analyses for the other subgroups (see below) we do not include a HSROC curve or prediction region. Figure 8 shows the results of this meta-analysis.
FIGURE 8.
(a) Meta-analysis and (b) forest plot of test accuracy of OSNA for patients (based on analysis of SLNs) without adjustment for TAB. a, results of the ‘routine use’ patient sample reported by Khaddage et al. ;57 b, results of the ‘validation’ patient sample reported by Khaddage et al. 57 (see Table 18). FN, false negative; FP, false positive; TN, true negative; TP, true positive.
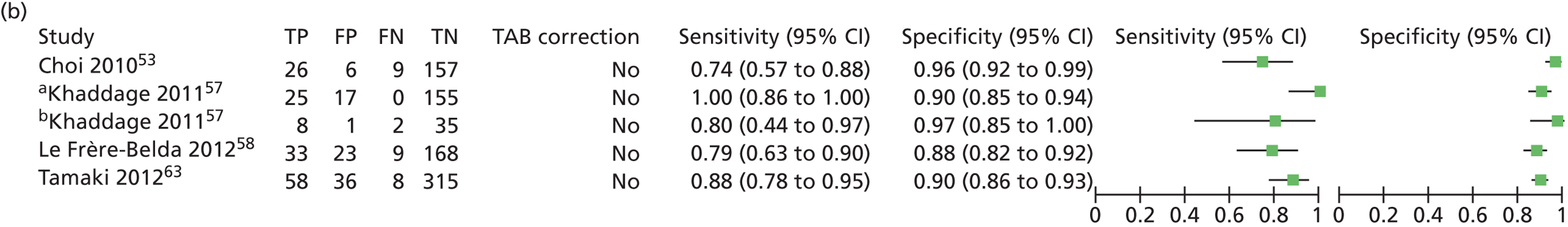
As there were only three studies available to estimate the test accuracy of OSNA for patients after adjustment for TAB, we set the correlation parameter to zero as described in Assessment of test accuracy. The results are displayed in Figure 9.
FIGURE 9.
(a) Meta-analysis and (b) forest plot of the test accuracy of OSNA for patients (based on analysis of SLNs) with adjustment for TAB. a, Results of the ‘validation’ patient sample reported by Khaddage et al. 57 (see Table 18); b, adjusted results of the patient sample reported by Le Frère-Belda et al. 58 (see Table 17, bottom), which includes 18 discordant cases that could not be evaluated for TAB. FN, false negative; FP, false positive; TN, true negative; TP, true positive.
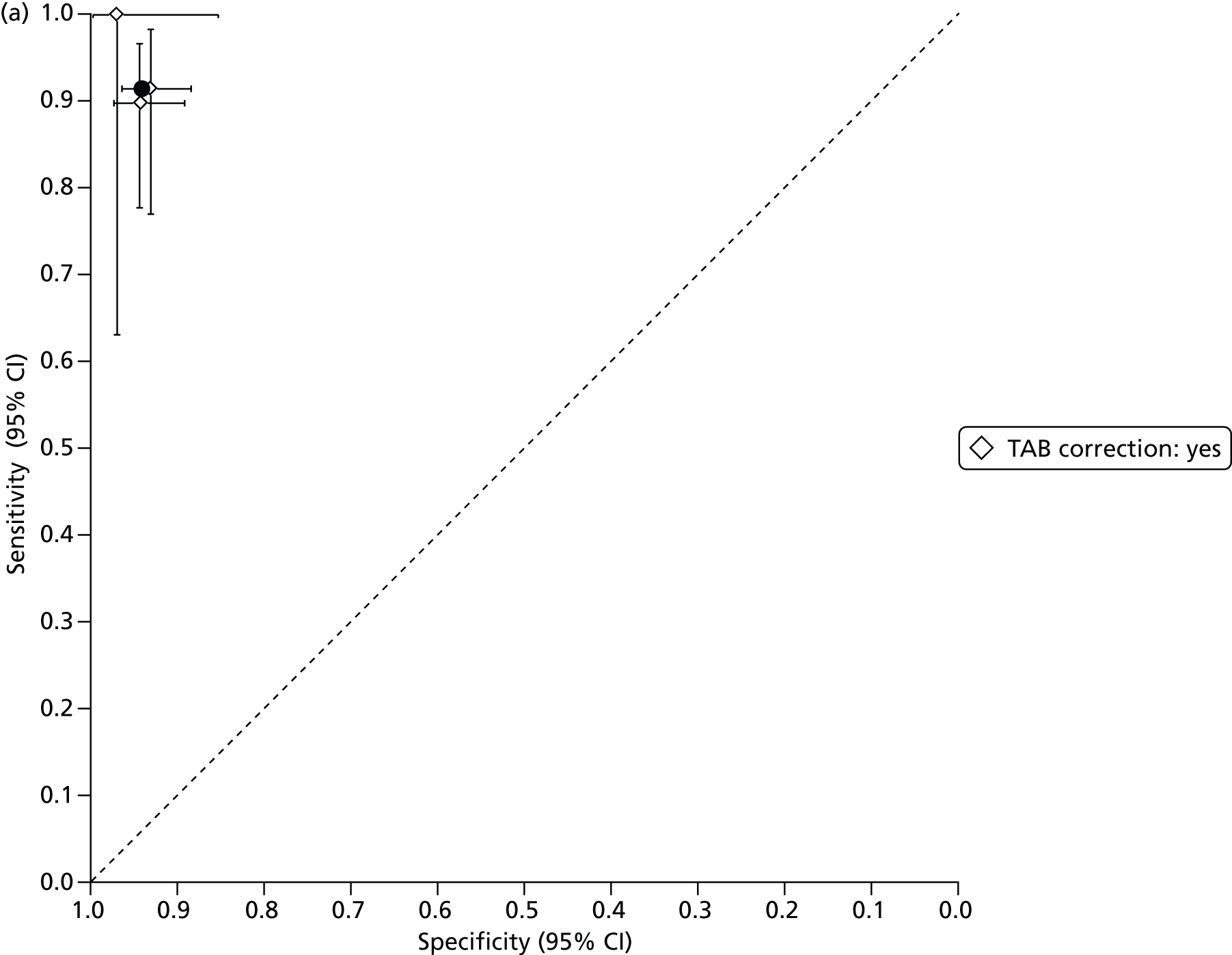

For the meta-analyses of the test accuracy of OSNA for SLNs and ALNs we do not present SROC curves or prediction regions because the studies report the same copies/µl thresholds of 250 and 5000 for the OSNA results of + (micrometastases) and ++ (macrometastases) respectively.
We attempted to perform meta-analyses of the test accuracy of OSNA for SLNs with and without adjustment for TAB using the full bivariate method. In both of these meta-analyses we encountered atypical results in the output such that the between-study correlation parameter (σAB/σA2σB2 in Reitsma et al. 44) is estimated as –1 with no estimate of standard error (SE) or CI. This phenomenon has been recognised before and Riley et al. 118 conclude that it is likely to occur when there are few studies or the within-study variation is large (i.e. the studies themselves are small) and is due to sensible bounds being placed on the model to avoid the maximum likelihood estimation including a correlation outside the range [–1,1]. Riley et al. 118 also conclude that there is no systematic bias in the summary estimates of sensitivity and specificity introduced by this phenomenon and that CIs of the summary points are conservative. However, as described in Assessment of test accuracy, we set the correlation parameter to zero and repeated the analysis, the results of which are shown in Figures 10 and 11. Computed summary points and CIs differed only at the third decimal point.
FIGURE 10.
(a) Meta-analysis and (b) forest plot of the test accuracy of OSNA in SLNs without adjustment for TAB. FN, false negative; FP, false positive; TN, true negative; TP, true positive.

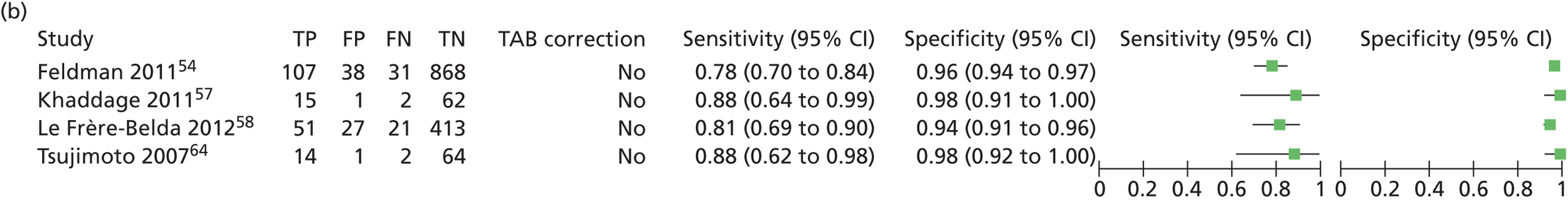
FIGURE 11.
(a) Meta-analysis and (b) forest plot of the test accuracy of OSNA in SLNs with adjustment for TAB. a, Adjusted results of the SLN sample reported by Feldman et al. 54 (see Table 16), which includes 26 cases that could not be evaluated for TAB; b, adjusted results of the ‘validation’ SLN sample reported by Khaddage et al. 57 (see Table 18), which includes two discordant cases that could not be evaluated for TAB; c, adjusted results of the SLN sample reported by Le Frère-Belda et al. 58 (see Table 17, top), which includes 22 discordant cases that could not be evaluated for TAB; d, adjusted results of the SLN sample reported by Tsujimoto et al. 64 (see Table 23, middle), which includes one discordant case that could not be evaluated for TAB. FN, false negative; FP, false positive; TN, true negative; TP, true positive.


Figures 12 and 13 show the results of the meta-analyses of the test accuracy of OSNA in ALNs before and after adjustment for TAB respectively. The bivariate method converged with no abnormal results. Note that there were six studies to inform the meta-analysis before adjustment for TAB but only four studies to inform the meta-analysis after adjustment and we believe that this accounts for the significantly larger confidence region shown in Figure 13 as removing two studies from the subgroup without adjustment for TAB gives a confidence region of a comparable size.
FIGURE 12.
(a) Meta-analysis and (b) forest plot of the test accuracy of OSNA in ALNs without adjustment for TAB. a, Results of the trial 1 ALN sample reported by Tamaki et al. 62 (see Table 21); b, results of the trial 2 ALN sample reported by Tamaki et al. 62 (see Table 21). FN, false negative; FP, false positive; TN, true negative; TP, true positive.


FIGURE 13.
(a) Meta-analysis and (b) forest plot of the test accuracy of OSNA in ALNs with adjustment for TAB. a, Adjusted results of the ALN sample reported by Schem et al. 60 (see Table 19), which includes 13 discordant cases that could not be evaluated for TAB; b, results of the trial 2 ALN sample reported by Tamaki et al. 62 (see Table 21); c, adjusted results of the ALN and SLN sample reported by Tsujimoto et al. 64 (see Table 23, top); d, adjusted results of the ALN sample reported by Visser et al. 65 (see Table 24), which includes seven discordant cases that could not be evaluated for TAB. FN, false negative; FP, false positive; TN, true negative; TP, true positive.
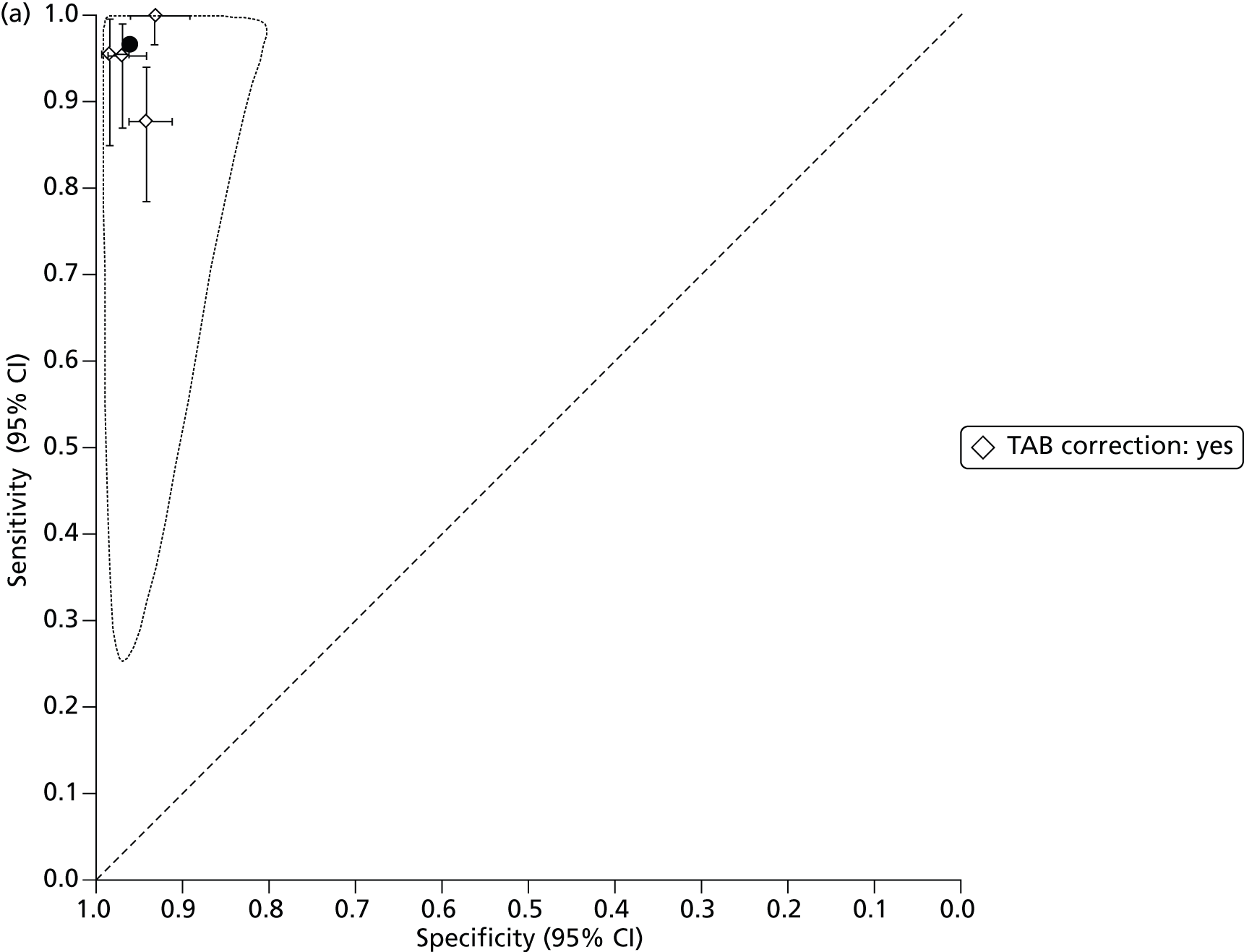

Assessment of analysis time
The time taken for analysis of LNs in the different studies is displayed in Table 35. It is unclear if the timings are directly comparable as the description of exactly which procedures were being monitored was sometimes ambiguous. That said, the results seem fairly consistent for both methods: between < 30 and 39.6 minutes for one node, increasing by approximately 5–10 minutes per additional node analysed.
| Nodes, n | Median time to analysis (minutes) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OSNA | Metasin | ||||||||
| Bernet 201150a,b | Choi 201053a,c | Feldman 201154d | Godey 201255a,e | Khaddage 201157f | Le Frère-Belda 201258g | Snook 201161h | Tsujimoto 200764i | Sundaresan40 | |
| 1 | 39.6 (range 26–70) | 35.2 | 33.0 | 32.9 (SD 4.9) | 33 | 32 (range 22–97) | < 30 | 36 | |
| 2 | 44.8 | 39.6 | 36.4 (SD 4.5) | 37 | 40 | 42 (range 30–73) | 42 | ||
| 3 | 50.4 | 45.2 | 41.6 (SD 5.2) | 48 | 51 (range 38–73) | 46 | |||
| 4 | 50.0 | 48.5 (SD 8.7) | 54 | 62 (range 46–90) | |||||
It was also noted by Bernet et al. 50 that the longest and most variable time period corresponded to the stage when the node was transported from the operating room to the pathology department. The time taken for macroscopic processing of the samples could also fluctuate significantly depending on the training level of the pathologist involved. The least variable time period corresponded to the homogenisation of the tissue, preparation of the diluted sample and amplification using the amplification equipment.
The study be Guillen-Paredes et al. 56 did not provide a time to analysis but compared operative time, days in hospital and complications between histology and OSNA, as shown in Tables 36 and 37.
| Test | Mean (SD) intervention time (minutes) | Mean (SD) days in hospital | ||||
|---|---|---|---|---|---|---|
| First operation | Second operation | Total | First admission | Second admission | Total | |
| Histology | 57.11 (23.93) | 78.33 (NR) | 78 (48.02) | 1.8 (2.04) | 2.41 (1.29) | 2.44 (3.00) |
| OSNA | 62.14 (21.93) | NA | 62.14 (21.93) | 1.54 (0.78) | NA | 1.54 (0.78) |
| Test | Absolute no. intervention time (minutes) | Absolute no. days in hospital | ||||
| First operation | Second operation | Total | First admission | Second admission | Total | |
| Histology | 2570 | 940 | 3510 | 81 | 29 | 110 |
| OSNA | 2175 | NA | 2175 | 54 | NA | 54 |
| Test | Complications in first intervention | Complications in second intervention | ||||
|---|---|---|---|---|---|---|
| None | Minor | Major | None | Minor | Major | |
| Histology | 28 | 17 | 0 | 4 | 8 | 0 |
| OSNA | 24 | 10 | 1 | NA | NA | NA |
Table 37 indicates that, overall, patients undergoing OSNA experienced fewer complications, although the one major complication occurred in the OSNA group.
Chapter 4 Assessment of cost-effectiveness: systematic review
Systematic review of existing cost-effectiveness evidence
Search strategy
The main objective of this review was to identify and evaluate published studies that examine the cost-effectiveness of intraoperative tests in the diagnosis of ALN metastases. As a secondary objective, we also used the studies to inform the design of the model used in our independent economic assessment.
We reviewed published economic evaluations of intraoperative molecular assessment for metastasis in early breast cancer to identify evidence relevant to current NHS practice. In addition to the electronic databases searched in the effectiveness review, EconLit and the bibliographies of relevant studies were searched for cost, cost-effectiveness and cost–utility studies. Forward citations of identified studies were searched for any relevant publications published after the initial search.
Relevant studies were then identified in two stages. Titles and abstracts returned by the search strategy were examined independently by two researchers (NH and RM) and screened for possible inclusion. Disagreements were resolved by discussion. Full texts of the potentially relevant studies were obtained. Two researchers (NH and RM) examined these independently for inclusion or exclusion and disagreements were resolved by discussion.
Description of the included studies
The initial search identified a total of 13 abstracts, seven82,87,110,111,114,119,120 of which were conference presentations and six9,56,121–124 of which represented four individual studies. One study measured the costs of intraoperative options that are not relevant to the UK (touch imprint and frozen section)121 and one study analysed the diagnostic pathway leading to SLN biopsy or ALND. 9,122 The remaining two studies56,123 were identified as relevant to our review. A second report of the study by Cutress et al. 123 was available from the NHS Technology Adoption Centre124 and was used to complement the information from Cutress et al. 123 See Appendix 5 for the quality assessment of all available studies reported in journal articles and Appendix 6 for details of the excluded studies.
The characteristics of the studies are summarised in Table 38. Both were single-centre observational studies that compared an intraoperative test with histopathology as the gold standard for assessing SLNB. The study by Cutress et al. 123 was set in the UK and assessed the GeneSearch assay whereas the study by Guillen-Paredes et al. 56 was conducted in Spain and evaluated the OSNA assay. Both found their respective intraoperative test to be cost-effective compared with histopathology, with both assays being cost-saving while reducing theatre time and length of hospital stay. The UK study also considered a strategy in which axillary clearance was performed on all patients instead of assessing them using SLNB, but this practice is no longer recommended by NICE13 and the study did not find it to be cost-effective. Guillen-Paredes et al. 56 also measured the benefits of each strategy by assessing the incidence of minor and major complications during surgery. They found significantly fewer complications in the OSNA group, although this group was also the only one to have a major complication (no details were given). Neither study looked at outcomes beyond the diagnostic phase.
| Study | Setting, perspective | Population | Study purpose | Study approach | Diagnostic comparators | Outcomes measured | Base results |
|---|---|---|---|---|---|---|---|
| Cutress 2010123 | UK health-care setting, hospital trusts | Patients diagnosed with breast cancer eligible for SLNB | Cost–benefit analysis of the use of the intraoperative test GeneSearch to diagnose SLN metastases | Prospective, single-centre, observational study; costs estimated retrospectively | Model 1: No SLNB, axillary clearance performed Model 2: SLNB analysed by histopathology – node negative, no further treatment; node positive, axillary clearance in second operation Model 3: Half-node SLNB analysed with GeneSearch – node negative, no further treatment; node positive, axillary clearance in same operation. Rest of the node analysed with histopathology |
Costs, theatre time, length of hospital stay | Cost-effectiveness improves with intraoperative testing – 28% avoid a second operation and cost per case reduced from £2891 to £2833 |
| Guillen-Paredes 201156 | Spain, hospital setting | Patients with early-stage breast cancer who are ultrasound negative and who underwent a SLNB | Cost–benefit analysis of the use of OSNA to diagnose SLN metastases | Retrospective, single-centre study | Group 1: Histopathology – metastases, axillary clearance in second operation Group 2: OSNA – metastases, axillary clearance in first operation |
Costs, theatre time, length of hospital stay | Overall, group 2 had a cost saving of €319.99 per patient per intervention, a shorter theatre time and length of hospital stay and fewer complications |
It must be noted that the study by Cutress et al. ,123 although providing a unique source of evidence on resource use and costs of intraoperative molecular testing in the UK, provides limited, if any, evidence on the economic outcomes of a specific intraoperative test as it refers to a test that has been withdrawn from the market (GeneSearch). Metasin, one of the intraoperative testing technologies being evaluated in this assessment, uses the same markers as GeneSearch, CK19 and mammaglobin, but different primer–probe combinations and it is therefore expected to perform differently in routine practice from GeneSearch.
Quality appraisal
A quality appraisal was carried out on the two studies using the checklist of Drummond and Jefferson. 125 A summary of the results is provided in Table 39.
| Criteria | Cutress 2010123 | Guillen-Paredes 201156 |
|---|---|---|
| Study design | ||
| The research question is stated | ✓ | ✓ |
| The economic importance of the research question is stated | ✓ | ✓ |
| The viewpoint(s) of the analysis are clearly stated and justified | ||
| The rationale for choosing alternative programmes or interventions compared is stated | Partial | ✓ |
| The alternatives being compared are clearly described | ✓ | ✓ |
| The form of economic evaluation used is stated | ✓ | ✓ |
| The choice of form of economic evaluation is justified in relation to the question addressed | ✗ | ✗ |
| Data collection | ||
| The source(s) of effectiveness estimates used are stated | ✓ | ✓ |
| Details of the design and results of the effectiveness study are given (if based on a single study) | ✓ | ✓ |
| Details of the methods of synthesis or meta-analysis of estimates are given (if based on a synthesis of a number of effectiveness studies) | NA | NA |
| The primary outcome measure(s) for the economic evaluation are clearly stated | Partial | ✓ |
| Methods to value benefits are stated | ✗ | ✗ |
| Details of the subjects from whom valuations were obtained are given | NA | NA |
| Productivity changes (if included) are reported separately | ✗ | ✗ |
| The relevance of productivity changes to the study question is discussed | NA | NA |
| Quantities of resource use are reported separately from their unit costs | ✓ | ✓ |
| Methods for the estimation of quantities and unit costs are described | ||
| Currency and price date are recorded | ✓ | Partial |
| Details of currency of price adjustments for inflation or currency conversion are given | ✗ | ✗ |
| Details of any model used are given | NA | NA |
| The choice of model used and the key parameters on which it is based are justified | NA | NA |
| Analysis and interpretation of results | ||
| Time horizon of costs and benefits is stated | ✓ | ✓ |
| The discount rate(s) are stated | NA | NA |
| The choice of discount rate(s) is justified | NA | NA |
Study design
Both studies were observational and are therefore open to bias and confounding. They both stated their research question and the approach to economic evaluation but no justification was given by either study for the economic evaluation study design used in relation to the research question. The viewpoint of both analyses was implicitly justified by the public health systems in which the studies were conducted and the local practice at the respective centres, and both studies acknowledged the limited generalisability of their findings because of the availability of different intraoperative testing technologies in other centres.
Data
Details of the methods of patient recruitment were given. Although both studies were based on single-gate designs and are thus subject to TAB, neither adjusted for it in the analysis of accuracy. Both studies reported the methods of collecting health-care resource use data and applying unit costs to them, but only the Spanish study56 explicitly stated the primary outcome measures of its evaluation in terms of the health benefits of intraoperative diagnosis. Both studies reported unit costs and quantities separately but they provided explanations only for the estimation of unit costs not quantities. The Spanish study56 did not state the date for the unit costs used and neither study provided details on whether any price and currency conversion adjustments were made. Neither study valued health benefits nor examined changes in productivity or its associated costs.
Analysis and interpretation of the results
Neither study analysed outcomes beyond the end of the diagnostic phase and therefore the studies did not require the use of a discount rate. As no sensitivity analyses were provided, the degree to which the reported cost differences may have been the result of chance alone cannot be established.
In summary, only one study was found for OSNA56 and none for Metasin. One study was found on GeneSearch,123 which served as the basis for the development of Metasin (see Chapter 1, Metasin) but may not produce the same outcomes. Although the OSNA study56 is likely to reflect the study centre’s practice in Spain, the GeneSearch study123 is no longer relevant as the technology has been withdrawn from the market. The results in both studies are likely to be biased because of the inherent limitations of observational studies, which commonly lack control for the effect of confounders on outcomes. The validity of these studies’ findings is made more uncertain by their lack of reporting of the methods used to measure diagnostic accuracy outcomes. Further, the degree of uncertainty in the estimates, and the extent to which the observed differences may be explained by chance, cannot be established. Nor can it be known to what extent the results may be generalisable across countries or indeed jurisdictions within the UK. This suggests that the existing evidence on economic outcomes is unlikely to serve to inform medical decision-making in the context studied here.
Submissions from sponsoring companies
No economic studies of Metasin were submitted by its sponsor. For this technology, only one conference abstract114 was found that included any information on costs; it reported only the cost per assay (£35.00; a different estimate, based on more detailed information obtained from the sponsor, was used in the model; see Model parameter values, Costs). As for OSNA, only a file with illustrations of spreadsheets from a decision model of its costs and accuracy, developed for Belgium by Professor Lieven Annemanns (Ghent University, Belgium), was made available, as unpublished and academically confidential work (NICE technical management team technical analyst Sarah Byron, 1 July 2012, personal communication of information provided by the manufacturer). (Academic-in-confidence information has been removed.)
The model considered only the cost differences between OSNA and the status quo for diagnostic testing of metastatic early breast cancer. The status quo was mixed: 90% of patients underwent intraoperative testing by touch imprint cytology or frozen section, with confirmatory postoperative histopathology for patients with negative test results, and the remaining 10% of patients were subject to postoperative histopathology testing only. The analysis assumed a SLN-positive rate of 39%. In addition to the costs of the diagnostic tests, the analysis included the costs of further investigations after a negative intraoperative test result, the initial surgery without LN dissection, the extended surgery with intraoperative testing and ALND, and the cost of the second surgery including the hospital stay. The model analysed the costs accruing over a 1-year period following primary surgery for breast cancer. The authors’ rationale for this choice was that ‘it is anticipated that the systematic application of OSNA will lead to equivalent test results and long term patient management as current postoperative testing’ (NICE technical management team technical analyst Sarah Byron, 1 July 2012, personal communication of information provided by the manufacturer).
Using values of 95.6% for sensitivity and 96.7% for specificity, the mean per patient costs were estimated to be €771 (£603; at gross domestic product purchasing power parity126) for the status quo and €604 (£473) for OSNA, resulting in a mean per patient cost saving with the latter of €167 (£131). The authors argue that their analysis ‘would be equally applicable to the UK but should show a greater economic benefit given that most sentinel lymph node analysis in breast cancer patients in the UK market is performed post-operatively so savings in second surgery would be more pronounced’ (NICE technical management team technical analyst Sarah Byron, 1 July 2012, personal communication of information provided by the manufacturer).
The results of this analysis are of limited value because of the lack of information made available to the Evidence Review Group (ERG) on the methods behind the analysis. The analysis was based on aggregate measures of costs and so quantities of resource use were not discernible. The analysis claims to account for costs up to 1 year after the initial operation but from the available detail it is unclear if any costs other than those associated with the diagnostic pathway were included. The analysis did not attempt to account for the costs of adverse events nor the effects in terms of health benefits and quality of life. No account was made of the degree of uncertainty in the model estimates, thus preventing an assessment of the likelihood that chance alone may explain the reported cost effects of OSNA.
Independent Evidence Review Group assessment
Objective of the analysis
The main analysis compares OSNA and Metasin with histopathology from the perspective of the NHS. The evaluation is presented for the outcomes occurring up to staging of the axilla. In addition, a separate analysis evaluated the long-term outcomes of intraoperative testing options in terms of QALYs and costs. The long-term analysis is intended as an illustration of the relative size of the benefits of intraoperative diagnosis and the effect of uncertainty on its expected lasting impact.
Description of the model
The model is split into two separate sections (diagnostic and management) to encompass both immediate and long-term outcomes. As the technology is expected to have a larger impact on the short-term outcomes, it is this section that we particularly focus on.
Diagnostic pathway
This section of the model is used to both inform the rest of the model and provide the intermediate outcomes described in the definition of the decision problem (see Chapter 2, Decision question).
The Evidence Review Group diagnostic pathway
Patients enter the model having had a SLNB performed during their initial tumour removal. The model then splits into three different strategies to encompass each of the possible combinations of diagnostic tests: intraoperative only (OSNA or Metasin, not in combination), histopathology only or a combination of an intraoperative assay and follow-up histopathology. These three strategies are shown in Figure 14. In the following descriptions, positive test results refer to results that indicate metastases in the SLN. The three modelled pathways are therefore:
-
Current practice. The SLNB is analysed using histopathology of the full node. If positive for metastases a second surgery is performed in which ALND occurs.
-
Add-in strategy. Half of the SLN from the SLNB is analysed during removal of the tumour using one of the intraoperative tests (OSNA or Metasin). Those with a positive result receive ALND during that surgery. For those with a negative result, the other half of the SLN is kept to be analysed by histopathology. Patients whose metastases were not detected at the intraoperative stage but whose histopathology is positive receive ALND as a second operation.
-
Replacement strategy. The full SLN is assessed using the intraoperative test, with no histopathology. Those with a positive result will receive ALND during their tumour removal.
FIGURE 14.
Diagnostic pathway to test for axillary metastases.
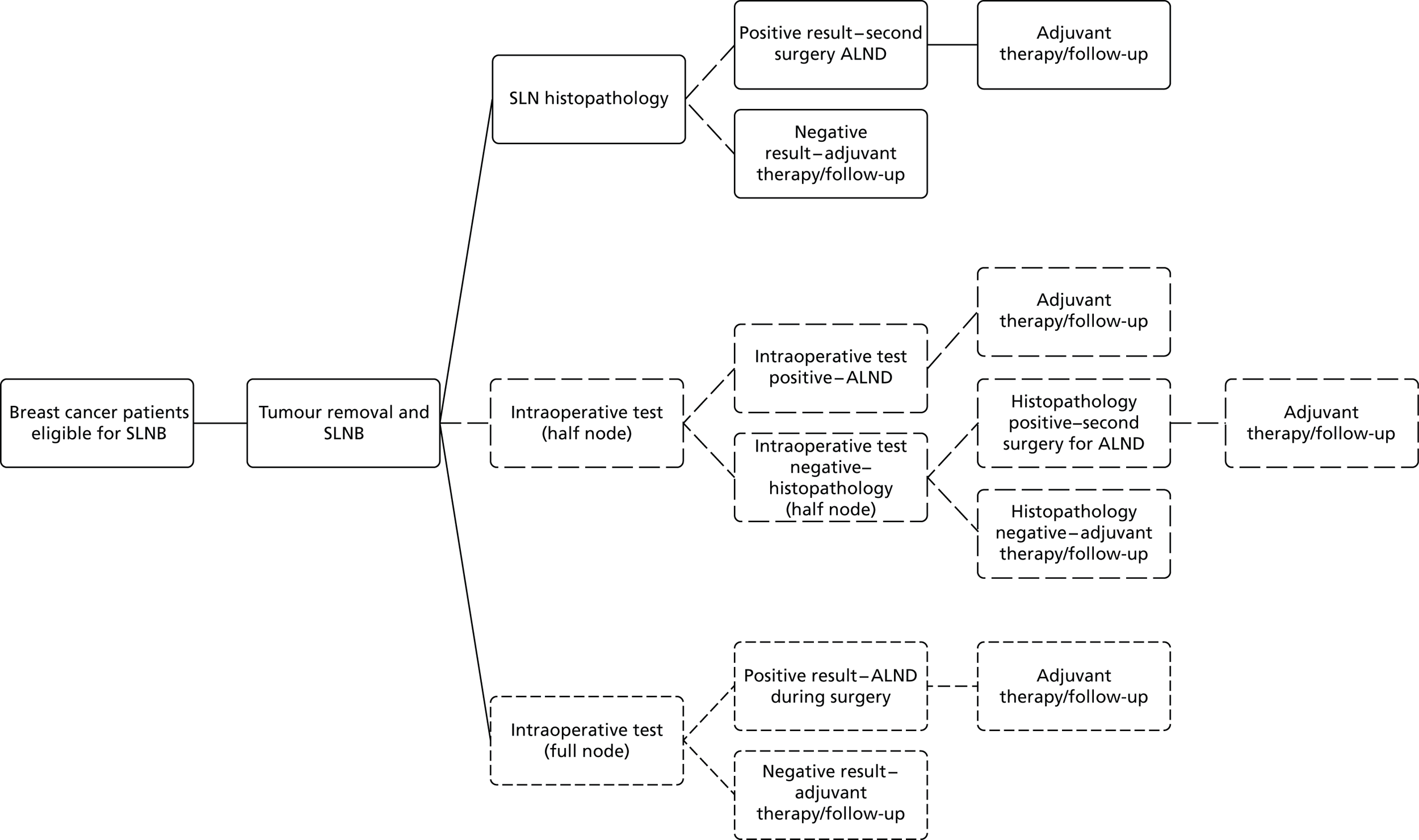
At this stage we calculate the intermediate costs and patient outcomes. This analysis was developed in Microsoft Excel 2010 (version 14.0.7128.5000, Microsoft Corporation, Redmond, WA, USA).
Assumptions
In this section of the model the following assumptions are made:
-
Compliance to all procedures offered to the patient is 100%. This includes compliance to ALND and therefore all patients who test positive for SLN metastases receive ALND. This assumption was adopted on the basis of the opinion elicited from clinical experts advising the ERG on this topic (see Appendix 7 for a list of the clinical experts).
-
The failure rate of SLNB, discussed in Chapter 1 (see Accuracy and adverse effects of sentinel lymph node biopsy), is treated as 0. In reality, SLNB has a failure rate of < 5% (the consensus from clinical experts advising the ERG on this topic). As this is relatively low and there is no evidence on a difference in impact when comparing the use of SLNB in intraoperative testing with the use of SLNB in histopathology, we have to assume that the impact is the same and treat it as 0.
-
It is unclear from the clinical effectiveness systematic review what a failed intraoperative test would be, as it is not something that is discussed in the studies. The number of reports that provide the number of failed tests is low, which suggests that the failure rates are low; in addition, our experts were not aware of failure rates, suggesting that the concepts of a failed intraoperative test and the rate of failure are not yet well established. As the understanding of failure for OSNA is unclear, it is impossible to model successfully the impact of knowing when a test has failed in the model. Therefore, for the purposes of the model we assume the known failure rate to be 0. Instances in which the failure status of intraoperative testing is unknown directly impact on the sensitivity and specificity of the test and therefore can be implicitly incorporated into the model using sensitivity analysis of the sensitivity and specificity parameter values.
-
In the absence of relevant empirical data from studies using full-node analysis, half-node analysis data are used for all test accuracy data for all pathways. We expect this to underestimate the accuracy of these tests in a full-node situation and thus serve as a conservative analysis of their benefits.
-
Similarly, based on the data, histopathology is assumed to have a sensitivity and specificity of 1 as it is used as the gold standard in the evaluative studies. This may also slightly underestimate the relative accuracy of the intraoperative tests, as the accuracy of histopathology will not actually be 1.
Post diagnosis (management pathway)
After the diagnostic pathway our cohort splits into various management subgroups:
-
Patients with SLN metastases who test positive for metastases and who receive ALND (in either their first surgery or as a separate second surgery for ALND) and who also test positive for additional axillary metastases when the material from their ALND is tested. The proportion of patients assumed to have additional axillary metastases after a false-positive SLN metastases diagnosis is assumed to be small enough that it can be modelled as 0%. The proportion of patients assumed to have additional axillary metastases after a true-positive SLN metastases diagnosis is taken from the Z0011 trial37 and is set to 27.3%.
-
Patients who test positive for metastases and who receive ALND (in either their first surgery or as a separate second surgery for ALND) but who do not test positive for additional axillary metastases when the material from their ALND is tested. These patients include those who were correctly identified as having SLN metastases and those who were incorrectly identified as having SLN metastases.
-
Patients without SLN metastases who test negative for metastases and do not receive ALND.
-
Patients with SLN metastases who test negative for metastases and do not receive ALND.
A visual representation of these subgroups is provided in Figure 15.
FIGURE 15.
Post-diagnosis subgroups. This diagram shows the possible pathways of patients immediately post diagnosis and their true SLN metastases status.

Once these subgroups are established, the model moves into its second section: the management pathway. This section of the model calculates the long-term outcomes, most notably the costs and QALYs. We model each of the groups from this point using as a basis the discrete event simulation (DES) model previously utilised by the School of Health and Related Research Technology Assessment Group (ScHARR-TAG) team,9,122 with updated parameters.
The School of Health and Related Research model structure
The ScHARR uses a DES model built in Simul8 2009 (Simul8 Corporation, Boston, MA, USA) for its economic evaluation of diagnostic imaging leading to SLNB or ALND without SLNB (without intraoperative testing) for the diagnosis of metastasis in early breast cancer. A DES models individual patients with individual attributes, such as the patient’s diagnostic and disease history. Unlike Markov state transition models, in which the transition probabilities are evaluated during fixed time intervals, the transit of patients through states in a DES may occur continuously according to a statistical distribution function. If it is possible for a patient to make a transition from the current state to multiple states then the model samples the time to each possible destination and compares them. The destination state with the shortest sampled time is the one to which the simulated patient moves to. 9 DES models are more flexible than Markov models but, as they are usually more complex, they take longer to build and run and, more importantly, are more demanding in terms of evidence to populate their parameters. 127 As the existing ScHARR model had already been subject to peer review,122 and because of the high element of uncertainty involved in extrapolating outcomes beyond the diagnostic phase in this evaluation, the benefits of building a de novo model did not justify the costs.
A basic layout of the ScHARR model is provided in Figure 16. Once the process of testing for SLN metastases, surgery for tumour removal and possible ALND has been completed, patients enter a state of adjuvant therapy. This involves chemotherapy and hormonal therapy (when appropriate) for those diagnosed with metastases and hormonal therapy alone (when appropriate) in patients diagnosed without SLN metastases. After adjuvant therapy patients may move into a disease-free state (post-adjuvant therapy state) and potentially stay there for the rest of their lifetime. During or after adjuvant therapy, some patients may have a locoregional relapse (see Glossary). Patients in the post-adjuvant therapy state may experience locoregional or metastatic relapse. Patients in the locoregional relapse state may go into remission or advance to metastatic relapse. Patients in remission may remain in that state until death or enter a metastatic relapse state (for the purposes of the model there cannot be a further locoregional relapse). The model does not consider metastatic disease curable and therefore patients in this state may move to a death state only, either from breast cancer or other causes. Patients in all states can die from other causes.
FIGURE 16.
The ScHARR model for post diagnosis of axillary metastases.

The authors of the ScHARR model claimed that there were problems in deciding whether ALN metastases identified during follow-up are the result of recurrence or a previous misdiagnosis (a previous false-negative test). As this problem was beyond the scope of their project, they did not explicitly model it. In practice, it is difficult to obtain conclusive evidence that a given recurrence results from secondary metastatic spread (from some subclinical distant site), and recurrence in the axilla would be presumed to be disease that already existed at presentation but which was not eradicated by standard therapy, that is, in a LN that was not removed by ALND. The same argument applies to extra-LN recurrences, which represent cancer cells not removed by surgery or eradicated by systemic therapy. This issue is similarly beyond the scope of our assessment and therefore we adopt the model’s approach as reasonable for our purpose.
The following are the ScHARR model assumptions:
-
Lymphoedema is the only long-term adverse event considered and is classified as either mild/moderate or severe, based on the research data available. It affects both costs and quality of life for the rest of the patient’s life. We also model only lymphoedema as no relevant papers on other long-term adverse events were found. To incorporate the effect of lymphoedema on patients we populated the respective parameters using evidence from studies of patient-reported outcomes for lymphoedema.
-
Adjuvant therapy occurs for a maximum 5-year period, consistent with the recommended follow-up period stated in the NICE guidelines for early breast cancer,13 but the model allows for patients to move to another state before the end of this 5-year period. In this state, patients who test positive for additional axillary metastases, confirmed on ALND, receive chemotherapy for 6 months followed by hormonal therapy for 4.5 years. Patients who test negative for SLN metastases or who test positive for SLN metastases only and not for further axillary metastases on ALND receive hormonal therapy for 5 years.
-
After a locoregional relapse further locoregional relapse cannot occur; only metastatic relapse can occur. This is a simplification of clinical practice but was retained so that the model could be kept simple and because this was not the main focus of our assessment.
-
Death rates for non-breast cancer causes are based on UK mortality statistics and are applied across all health states. These are not adjusted to exclude breast cancer mortality and so they may overestimate the risk of dying from non-breast cancer causes. In our model we use mortality statistics for England as this was the population stated in the objective in our protocol. As in the ScHARR model we use the statistics for women only as a very small proportion of breast cancer cases (< 1%) occur in men. 14
Because we relied on the treatment phase of the ScHARR model with limited modifications, we quality assessed the model and found that it met the standard quality criteria required for health economic evaluation models. 125 The ScHARR model depicts a very similar problem to that analysed in this study, but does not consider intraoperative testing approaches. Although this is presumably because intraoperative tests were outside its remit, the authors do not state this. However, this has a limited impact as it affects only the diagnostic stage of the model, which we do not use. The model does not value productivity changes, which are beyond our remit too. Overall, the ScHARR model was well reported and transparent and we therefore felt comfortable adapting it to our purposes. During the course of our review process we have worked with two of the ScHARR model authors in adapting and updating their model for our purposes; one (YM) has become a co-author of this report and the support of the other (KC) has been acknowledged.
Source of the model parameter values
Diagnostic pathway
Test accuracy probabilities
In our analyses histopathology is assumed to be the gold standard and is given an accuracy of 100% (Table 40), as this generally is how the sensitivities and specificities of the intraoperative tests have been assessed in the literature (see Chapter 3, Assessment of test accuracy).
| Test | Sensitivity (%) | Specificity (%) | Source |
|---|---|---|---|
| Histopathology | 100.0 | 100.0 | Assumed (used as the gold standard in studies) |
| OSNAa | 84.5 | 91.8 | Meta-analysis results (see Chapter 3, Assessment of test accuracy) |
| OSNAb | 91.4 | 93.3 | Le Frère-Belda et al.58 |
| 100.0 | 97.2 | Khaddage et al.57 | |
| 89.8 | 94.5 | Snook et al.61 | |
| Metasina | 92.6 | 96.3 | Sundaresan40 |
Accuracies for Metasin have been provided by authors of unpublished papers40,66 and accuracies for OSNA were provided by the patient-level results of the clinical systematic review. Sufficient studies on OSNA that did not adjust for TAB were found to allow us to carry out a meta-analysis for the sensitivity and specificity of the test. As there were too few studies on OSNA that adjusted for TAB and only two studies on Metasin, we used the meta-analysed accuracy values for OSNA without adjustment for TAB and the Sundaresan report40 on Metasin (which did not adjust for TAB) for our base-case analysis and used the other studies57,58,61 individually in our sensitivity analyses. For neither histopathology nor intraoperative testing have we accounted for the false-negative rate associated with SLNB. This rate is normally < 10%128 (e.g. Cooper et al. 9 model it as 7%) and occurs either as a result of metastases being in a node other than the SLNB being examined or as a result of the preparation procedure for histopathology. As there are no data on what proportion of the failure rate would specifically affect histopathology (or intraoperative testing), we have not modelled it. It is therefore important to note that the model may underestimate the accuracy of the intraoperative tests.
The node-positive prevalence is set at 20% in the base case, which is consistent with the values found in the systematic review of accuracy studies.
Adverse event probabilities
We found no new relevant studies in the population of interest that served to update the probabilities of adverse events in the ScHARR model. For the short-term adverse event probabilities we used the original model’s values. 9 We examined the original studies that ScHARR used to populate both long- and short-term outcomes to verify that the probability values for events in its long-term model applied to our intermediate outcomes. For lymphoedema we confirmed ScHARR’s values by returning to the same original sources as ScHARR and similarly splitting patients into those with mild/moderate lymphoedema and those with severe lymphoedema (Table 41). Patients were divided into these groups either on the basis of their response30 or on the swelling measurement (> 5 cm was severe in the study by McLaughlin et al. 31). Patients were also split by the treatment to the LNs: either SLNB or ALND. In all of the studies ALND followed SLNB and therefore the reported probabilities included the risk level of SLNB. The costs, disutility and probabilities for lymphoedema are incorporated in the treatment phase of the model as it is an adverse event with long-term implications.
| Event | Probability | Source |
|---|---|---|
| Short-term adverse events | ||
| Infection | ||
| SLNB | 0.072 | Cooper et al.9 |
| ALND | 0.221 | |
| Seroma | ||
| SLNB | 0.020 | Cooper et al.9 |
| ALND | 0.792 | |
| Surgical drain | ||
| SLNB | 0.021 | Cooper et al.9 |
| ALND | 0.142 | |
| Long-term adverse event: lymphoedema | ||
| SLNB | 0.068 | Crane-Okada et al.30 |
| Mild/moderate lymphoedema | 0.045 | Adjusted using Mak et al.129 |
| Severe lymphoedema | 0.023 | |
| ALND | 0.214 | Crane-Okada et al.,30 McLaughlin et al.,31 Blanchard et al.29 |
| Mild/moderate lymphoedema | 0.142 | Adjusted using Mak et al.129 |
| Severe lymphoedema | 0.072 | Adjusted using Mak et al.129 |
Costs
Costs for tests were taken directly from manufacturer/academic submissions. When only a range of costs was provided [e.g. the costs for OSNA were given as £300–400 (Nice technical management team technical analyst, Sarah Byron, 1 July 2012 personal communication)], we took the midpoint of that range as our base cost (£350 in the case of OSNA). The costs of Metasin provided by the sponsor of the technology were used. After the analysis had been completed the sponsor provided an indicative update of the costs (£60–80 for kit reagents, for up to four nodes, and a reagents and consumables cost of £20 per node), which were not used to revise the analysis because of time constraints and because the results were not likely to be significantly altered. (However, with a private company having taken over the commercial development of the test, the cost-effectiveness analysis may be updated if and when a finalised list price for the CE-marked Metasin test is received.) Histopathology costs were based on data provided by the NHS Technology Adoption Centre, which in turn were based on evidence from a microcosting study conducted at the Queen Alexandra Hospital123 (reflated to 2010 prices) and reported in technical detail by researchers at the York Health Economics Consortium (YHEC),124 and include the cost of confirmation by immunohistochemistry (Table 42).
| Test or surgery | Cost (£) | Source |
|---|---|---|
| Histopathology | 472 | Cutress et al.123 |
| OSNA | 350 | Manufacturer information submitted to NICE |
| Metasin | 74 | Manufacturer (Matt Hayday, Head of Corporate Governance, Princess Alexandra Hospital NHS Trust, Harlow, 19 November 2012, personal communication) |
| Breast surgery with SLNB | 584 | NHS reference costs 2010–2011130 updated using the same method as ScHARR using the results of Pandharipande et al.131 |
| With intraoperative test | 12 | Additional time cost calculated: 3 minutes for 54% of patients;67 extra cost124 updated to 2010 costs132 |
| Breast surgery with ALND | 3855 | Breast surgery cost adjusted using the technique of Cooper et al.9 |
| Secondary operation for ALND | 3569 | NHS reference costs 2010–2011130 |
| Additional hospital stay for surgery with ALND | 106 | Burke and Patton,124 updated to 2010 costs |
| Additional hospital stay for second surgery for ALND | 512 | Burke and Patton,124 updated to 2010 costs |
| Short-term adverse event | 333 | Jeruss et al.,133 updated to 2010 prices |
| Mild lymphoedema | 71 | Cooper et al.,9 updated to 2010 prices |
| Moderate/severe lymphoedema | 1269 | Cooper et al.,9 updated to 2010 prices |
For surgery costs we relied mainly on NHS reference costs. 130 An overall average value for breast surgery was calculated using costs for lumpectomy and mastectomy [Healthcare Research Group (HRG) codes JA07D, JA07E, JA07F, JA07G and JA07H for mastectomy and HRG codes JA09E, JA09F, JA09G and JA09H for lumpectomy, chosen on advice from ScHARR]. 130 These costs were weighted by two-thirds for lumpectomy and one-third for mastectomy to represent the split between lumpectomy and mastectomy cases according to the opinion from our specialist advisors. The cost of a single surgery for ALND used the reference cost for procedures on the lymphatic system (HRG code WA24Z). There were no reference costs that included extra costs for SLNB or ALND procedures undertaken during breast surgery. We therefore used the same procedure as employed by Cooper et al. 9 to incorporate these additional costs. Using the results reported by Pandharipande et al. 131 we calculated the ratio between the cost of breast surgery with SLNB and the cost of breast surgery alone ($7537/$5264) and the ratio between breast surgery with SLNB and ALNB and breast surgery with SLNB alone ($11,244/$7537). The base cost of breast surgery (£1804.90 from the NHS reference costs130) is then multiplied by these ratios to give the costs for breast surgery with SLNB and breast surgery with SLNB and ALND.
The costs of short-term adverse events were taken from the same source as the ScHARR model133 as no other papers were identified. No papers were identified for updating the annual lymphoedema costs and as these were originally provided to ScHARR by the Sheffield Lymphoedema Service we considered this an appropriate source and updated the costs to 2010.
Most studies report only the time that it takes to run an intraoperative test and not the impact that it has on the length of surgery. The exception is a recent abstract by Ng et al. ,67 which stated that in 54% of cases breast surgery was complete before the results for OSNA were received. The median time over this surgery was 3 minutes. Applying this to an entire cohort gives an average waiting time of 1.62 minutes. To cost this we applied this time delay to costs from the microcosting study from the YHEC,124 which reports unit costs for various surgical procedures. These costs were updated to 2010 costs using the cost convertor developed by Shemilt et al. 132 Using the costs for a surgeon (£159.74 per hour), theatre staff (£2.60 per minute) and anaesthetist staff (£124.24 per hour) we calculated that the extra cost of waiting for intraoperative results was £11.88. We used this value for all intraoperative tests, even though it was calculated using only OSNA data, as there were no equivalent data for Metasin and none of the reported Metasin processing times were contrary to any of the assumptions made.
A consequence of an ALND procedure, either in the first breast surgery or in a separate surgery, is extra days spent in hospital by the patient. The cost of this extended stay was calculated using the results of the YHEC microcosting study,124 subtracting the length of stay for a standard surgery (2.1 days) from the length of stay for surgery that includes ALND (2.7 days) and from the combined length of stay for the first and second surgeries (5 days) for those who have ALND as a separate surgery. These were multiplied by the updated 2010 ward costs (£176.44 per day124) to give the costs of this extra hospital stay. The cost of short-term adverse events was adjusted to 2010 prices because, as reported previously, no more recent reports were found. 9,133
In univariate sensitivity analyses, the unit costs of diagnostic testing in primary breast surgery were varied according to the range of values reported in the literature for its most important cost elements. The recently published paper by the YHEC provides unit costs and resource use and is widely known. 124 We therefore examined costs based on this information. To ensure that it would directly compare with our approach using NHS reference costs, we examined the length of stay per operation both in our reference case and using the YHEC data. In our base case the average length of stay for a single breast surgery was approximately 2 days; this compared with a length of stay in the paper by the YHEC of 2.1 days124 (Table 43). The average length of stay for surgery on the lymphatic system (our ALND-alone procedure) was approximately 2.65 days, which is not dissimilar from the YHEC average of 2.9 days. 124
| Resource | Surgery with SLNB | Second surgery | Surgery with axillary clearance |
|---|---|---|---|
| Procedure time (minutes) | 54 | 60 | |
| Procedure time – biopsy and analysis (minutes) | 53 | ||
| Procedure time – operation (minutes) | 40 | ||
| Anaesthetic preparation/recovery (minutes) | 20 | 20 | 20 |
| Theatre turnaround time (minutes) | 20 | 20 | 20 |
| Length of stay (days) | 2.1 | 2.9 | 2.7 |
| Hospital sterilisation and disinfection unit (trays) | 1 | 1 | 1 |
| Physiotherapist (minutes) | 10 | 10 | |
| MDT appointment (minutes) | 10 | 10 | |
| MDT meeting (minutes) | 3 | 3 |
We therefore calculated surgery costs based on the YHEC information, with unit costs updated to 2010 costs (Table 44). As in our base-case analysis we assumed that the ward stay would be the same for all patients without axillary clearance, regardless of how the SLNB was analysed. The YHEC reported intraoperative analysis times that were shorter than the surgery times and so no additional time was factored in in this case. On the advice of our experts, the time taken by the MDT is the same for all diagnostic testing procedures and therefore the second operation for ALND does not incur an additional MDT cost as it does in the YHEC report.
| Resource | Unit cost (£) |
|---|---|
| Direct costs | |
| Surgeon (per hour) | 159.74 |
| Ward costs (per day) | 176.44 |
| Indirect costs | |
| Theatre staff (per minute) | 2.60 |
| Anaesthetist staff (per hour) | 124.24 |
| Theatre stock consumables (by case) | 64.73 |
| Anaesthetic administration and junior doctors (by case) | 68.91 |
| Hospital sterilisation and disinfection unit (by case) | 17.75 |
| Anaesthetic gases (by case) | 25.06 |
| Physiotherapist (per hour) | 25.06 |
| MDT appointment (per hour) | 81.43 |
| MDT meeting (per hour) | 644.16 |
Total surgery costs, given in Table 45, are calculated by multiplying the resource use by the relevant unit cost. For basic surgery this includes the cost of theatre time for each member of the surgical team, the theatre stock cost, the ward cost for the length of stay and the cost of the MDT meetings. As additional ward costs for surgery with ALND and ALND alone are already calculated separately as part of the base-case analysis, these are not included in those particular operation costs. However, these two surgeries do incur the cost of a physiotherapist, because of the ALND.
| Surgery | Cost (£) |
|---|---|
| Operation with SLNB (with ward costs, MDT meeting) | 1282 |
| Operation with SLNB and ALND | 1580 |
| ALND only | 1284 |
As Burke and Patton124 did not present values for the uncertainty around these estimates, in sensitivity analysis we used a separate report by Burke and Setters134 to obtain alternative values of resource items that might have the largest effect on the cost difference between diagnostic strategies. The alternative values that applied were a shortening in the time of surgery with ALND by 10 minutes and changes in the MDT meeting length (range 10–20 minutes). By varying the surgery costs by ±10% we were able to incorporate the variations that these changes caused.
Utilities
Patients who undergo testing by histopathology have to wait approximately 2 weeks for the results. We surmised that this would incur some level of disutility to patients because of the associated anxiety of waiting, and imputed it using the health state valuation equation provided by Dolan. 135 Dolan estimated the health state values of the European Quality of Life-5 Dimensions (EQ-5D) classification system, which measures health-related quality of life in terms of five dimensions: mobility, self-care, ability to perform usual activities, pain/discomfort and anxiety/depression. In turn, each of these dimensions is measured on three levels: no problem, moderate problems and severe problems. The utility weights for these levels were estimated from responses to relative valuation questions in a survey of the UK general public. For our purposes, only the utility decrement due to anxiety/depression was relevant and we adopted the decrement specific to the severe level. The equation for this takes the form:
where α is the constant (0.081) applied to any level of disutility, AD is the constant (0.071) applied to any level of disutility associated with anxiety or depression, A2 is the constant (0.094) applied to severe levels of anxiety or depression and N3 is the constant (0.269) applied when any of the five dimensions of the EQ-5D are recorded as severe. Our patients were assumed to already have a utility of < 1 (meaning that we did not need to apply the α value) and to be moving from a state of no anxiety/depression to a state of severe anxiety/depression and it was assumed that this anxiety/depression would be the only dimension of the EQ-5D that was graded as severe. This gave us a decrement of –0.236 – 0.269 = –0.505. Once we had used the formula to calculate a value for Y, we adjusted it to apply for only 2 weeks. This gave us a reduction in undiscounted QALYs of 0.019 for the 2 weeks spent waiting for histopathology results.
We also assumed that patients who undergo a second operation would have a disutility. To account for this we used utilities from the literature. 136 None was specific but a value of 0.62 was provided for the 2 months following breast cancer surgery. We subtracted this from the highest utility in the adjuvant therapy states (0.82) and adjusted for the 2 months to give a disutility of 0.03.
Treatment phase
Health state transition probabilities
No studies were found that allowed us to update all of the health state transition probability values appropriately and therefore we returned to the papers used by ScHARR and kept all of the parameters and SEs used in the ScHARR model9 (Table 46). The papers used included previous cost-effectiveness models131,137,139 and a prospective cohort study. 138 The cost-effectiveness models used data from the Early Breast Cancer Trialists’ Collaborative Group,137,140 Adjuvant! Online18,131 and a retrospective US study. 139
| Transition | Probability | SE | Distribution | Source |
|---|---|---|---|---|
| Annual probability of locoregional recurrence: no SLN metastases/no additional axillary metastases | 0.03 | 0.0017 | Beta | Cooper et al.,9 Orr et al.137 |
| Annual probability of locoregional recurrence: SLN metastases and additional axillary metastases | 0.09 | 0.0052 | Beta | Cooper et al.,9 Orr et al.137 |
| Annual probability of locoregional recurrence: SLN metastases, tested negative | 0.14 | 0.0082 | Beta | Cooper et al.,9 Orr et al.137 |
| Annual probability of metastatic recurrence: true negative, false positive | 0.0023 | 0.00014 | Beta | Cooper et al.,9 Pandharipande et al.131 |
| Annual probability of metastatic recurrence: true positive | 0.0052 | 0.00030 | Beta | Cooper et al.,9 Pandharipande et al.131 |
| Annual probability of metastatic recurrence: false negative | 0.0094 | 0.00054 | Beta | Cooper et al.,9 Pandharipande et al.131 |
| Annual probability of metastatic relapse from locoregional recurrence | 0.18 | 0.010 | Beta | Cooper et al.,9 Kamby and Sengelov138 |
| Annual probability of death from locoregional recurrence | 0.30 | 0.017 | Beta | Cooper et al.,9 Orr et al.137 |
| Annual probability of metastatic relapse from remission | 0.13 | 0.0075 | Beta | Cooper et al.,9 Kamby and Sengelov138 |
| Annual probability of death from metastatic relapse | 0.37 | 0.021 | Beta | Cooper et al.,9 Ward et al.139 |
When patients enter the model, their maximum life expectancy is set using life tables. 141 It is possible for a patient to die before this time in the model, depending on his or her transitions through the health states. Each time a patient enters a state, the time to each of the next plausible states is sampled from an exponential distribution using the annual transition probabilities. The state with the shortest time delay is the state that the patient then moves to following that delay and the process is then repeated. This analysis simulates the individual experiences of a cohort of 5000 patients, to calculate patient-level results (first-order Monte Carlo simulation) and additionally randomly samples from the independent model parameter distributions (second-order Monte Carlo simulation). There is therefore a probabilistic sensitivity analysis built into the model.
Health state costs
Relevant studies and NHS costs were used to calculate the health state costs (Table 47). The cost of adjuvant therapy was calculated using methods reported elsewhere. 9 This assumes that patients with test-negative SLN status (true or false) and patients who test positive for SLN metastases but who have no further metastases found on ALND will receive hormonal therapy for 5 years when appropriate. Patients who were found to have SLN metastases and additional axillary metastases receive chemotherapy for 6 months using regimens commonly used in the UK at corresponding prices. 9 They also then receive hormonal therapy for 4.5 years. To calculate an average cost per patient, assumptions are made about the hormonal therapy provided. It is assumed that 81% of patients are oestrogen receptor positive and will therefore respond to hormonal therapy. 139 As in the ScHARR model, we assume that, of these, 90% will receive some form of aromatase inhibitor and the other 10% will receive tamoxifen. 9 Regardless of hormonal therapy status, each patient will also have a yearly outpatient follow-up appointment and a mammogram. The combined average annual cost per patient for hormonal adjuvant therapy and follow-up is calculated to be £1086.75. The cost of the post-adjuvant therapy state is assumed to be £0 and the costs of locoregional and metastatic relapse and remission and death are taken from the study by Karnon et al. ,143 which was a UK cost–utility analysis that estimated these costs based on a cost study of 199 women in Edinburgh.
| State | Cost (£) | Parameter | Source |
|---|---|---|---|
| Cost of adjuvant therapy: TN, FP, FN | 1087 | Annual cost | NHS reference costs 2010–2011130 for outpatient follow-up + NHS reference costs 2002–2003142 for mammogram, cost updated + Ward et al.139 cost for hormone therapy, cost updated |
| Cost of adjuvant therapy: TP | 9447 first year | Cost for 6 months | Cooper et al.,9 cost updated |
| 1087 after first year | Annual cost after first 6 months | NHS reference costs 2010–2011130 for outpatient follow-up + NHS reference costs 2002–2003142 for mammogram, cost updated + Ward et al.139 cost for hormone therapy, cost updated | |
| Cost post adjuvant therapy | 0 | Annual cost | Cooper et al.,9 assumption |
| Cost of locoregional recurrence | 13,745 | Annual cost | Karnon et al.;143 2005 prices reflated to 2010 using the Hospital and Community Health Services Index144 |
| Cost of remission | 108 | Annual cost | Karnon et al.;143 2005 prices reflated to 2010 using the Hospital and Community Health Services Index144 |
| Cost of metastatic relapse | 10,443 | Annual cost | Karnon et al.;143 2005 prices reflated to 2010 using the Hospital and Community Health Services Index144 |
| Cost of death | 5713 | Cost of event | Karnon et al.;143 2005 prices reflated to 2010 using the Hospital and Community Health Services Index144 |
All costs are updated to prices for the year 2010 as required and, as we had no data on the uncertainty around these estimates, values were sampled from a uniform distribution of ±10% around the mean estimate for sensitivity analysis.
Health state utilities
All health state utilities were derived from the Tengs and Wallace study136 using the same parameters and SEs as ScHARR (Table 48). The Tengs and Wallace study136 was a comprehensive study and no new data were available for these particular variables. Each parameter was given a beta distribution.
| State | Utility | SE | Distribution | Source |
|---|---|---|---|---|
| Adjuvant therapy: TN, FP, FN | 0.82 | 0.18 | Beta | Tengs and Wallace136 (adjuvant therapy) |
| Adjuvant therapy: TP | 0.74 | 0.26 | Beta | Tengs and Wallace136 (chemotherapy) |
| Post therapy | 0.94 | 0.11 | Beta | Tengs and Wallace136 |
| Locoregional recurrence | 0.70 | 0.19 | Beta | Tengs and Wallace136 |
| Remission | 0.85 | 0.19 | Beta | Tengs and Wallace136 (after first recurrence) |
| Metastatic relapse | 0.40 | 0.19 | Beta | Tengs and Wallace136 |
| Death | 0.00 | By definition |
We also accounted for the general population change in utility that occurs with age, using the same formula as in the original ScHARR analysis, which adjusted a previously reported formula. 145 The utilities for the health states are adjusted using the following formula at the end of each year in the model:
In our model, the starting age is 56 years, the mode across studies in our systematic review (see Chapter 3, Population characteristics). Although the formula was not specifically estimated for women, analysis of Canadian longitudinal population survey data suggests that the evolution of utility with age does not significantly differ by gender. 146
Other utility values, including lifetime utility decrements for lymphoedema into the treatment phase of 9.9% for mild/moderate lymphoedema and 12.3% for severe lymphoedema, were obtained from the same source129 used in the ScHARR model. 9 These were estimated based on quality-of-life data obtained using the FACT-B+4 questionnaire (Functional Assessment of Cancer Therapy – Breast, adding a four-item arm subscale) from breast cancer patients who suffer from different degrees of lymphoedema and therefore they do not necessarily represent the true utility decrements for lymphoedema. However, when ScHARR performed a sensitivity analysis around these utilities it was found to not significantly affect the results. 9,122
Implementation of sensitivity analyses
In terms of uncertain parameters of the diagnostic phase of the model we focused mainly on the cost drivers and accuracy as part of sensitivity analyses. We looked at the impact of altering the costs of the tests and surgeries, particularly the cost of a second surgery, by setting ranges appropriate to the evidence that we had obtained from the literature. Although there was no additional information on the variation in costs, we looked at cost ranges of ±10%. As well as looking at the differences in accuracy between the studies that did not account for TAB and the studies that did, we performed a threshold analysis of sensitivity in which the specificity is held constant and the sensitivity is increased in steps of 5% from a minimum of 70% (the lower end of the CI produced in the meta-analysis). This process was repeated for specificity, holding sensitivity constant. The impact of accuracy and costs was assessed in both the short-term and long-term results.
For the purposes of assessing the effect of uncertainty in the long-term outcomes on the results, we ran supplementary univariate sensitivity analysis of the costs of adjuvant therapy, adjusting the mean by ±10% in two different simulations. Adjuvant therapy is the cost parameter that varies the most in direct relation to the outcome of the diagnostic tests and therefore is the most likely to impact on the model results.
We also considered the impact of the disutility associated with waiting for histopathology results and a second operation for ALND. For the disutility associated with anxiety, we examined the scenarios in which the disutility was 0, in which there was some disutility (0.006, using Dolan’s135 moderate anxiety value of 0.071, adjusted for 2 weeks) and in which there was extreme disutility (0.023), which used Dolan’s formula but assumed that there was nothing else wrong with the patient. For the disutility associated with a second operation, we looked at the scenarios in which the disutility was adjusted by ±20% and in which the disutility was 0.
For the base-case parameters, a total of 900 simulations were run according to the parameter distributions (see Tables 46 and 47 and Health state costs) and with other parameters fixed at base-case values and presented the mean incremental costs and incremental QALYs. One-way and two-way sensitivity analyses were performed using a total of 500 simulations and, as for the base case, were presented as the mean incremental cost, mean incremental QALYs and the ratio of these two estimates across model simulations. Despite the limited evidence on DTA parameters, we also conducted a probabilistic analysis based on base-case parameter values, accounting for the sampling uncertainty in the sensitivity and specificity of OSNA from the three study reports that adjusted for TAB (see Table 40), using 900 simulations, assuming independent variation in diagnostic test and other parameters, but imposing the negative correlation between OSNA sensitivity and specificity as described by Cooper et al. 9 The results are presented as the probability of OSNA being cost-effective as a function of the cost-effectiveness threshold.
Discount rate
Costs and utilities are discounted at a rate of 3.5% in accordance with NICE guidelines. 147
Measure used to synthesise costs and benefits
Intermediate results
The following measures were used to synthesise costs and health benefits in the diagnostic phase:
-
incremental cost per patient
-
incremental cost per case correctly diagnosed
-
incremental cost per additional positive case detected
-
incremental cost per additional negative case detected.
Although we had set out to present, in addition to these measures, estimates of the cost per second operation avoided, this measure turned out to be of no additional informational value and these results are therefore not presented.
Long-term analysis
The following measures were used to synthesise costs and health benefits over the lifetime of patients:
-
incremental costs
-
incremental QALYs
-
incremental cost per QALY gained.
Because the status quo, histopathology, was assumed to be the gold standard in the clinical effectiveness studies, it tended to be the option with the highest benefits (only one study reported evidence that diagnostic accuracy with OSNA was sufficiently close to that of histopathology to generate more QALYs with OSNA than with histopathology). To avoid the complication in interpretation that arises with the incremental cost-effectiveness ratio (ICER) of the new technology, intraoperative testing, which has lower costs than the status quo, the results are presented for the more relative to the less effective among the three options, of half-node intraoperative diagnosis, full-node intraoperative diagnosis and histopathology, according to total QALYs.
Results
Base case
Diagnostic phase
The studies used to inform the economic analyses are presented in Table 40 (see Chapter 3, Assessment of study quality, for a discussion on the strengths and weakness of the selected studies). For the purpose of the economic analyses the results are presented by test option and whether estimates of test accuracy were controlled for TAB.
Ranking the diagnostic strategies by diagnostic accuracy and comparing the incremental costs and yields between increasingly accurate diagnostic strategies allowed the calculation of incremental costs per additional patient correctly diagnosed ratios and the identification of dominated options, that is, those that had higher costs and lower yields than the alternative. Table 49 summarises the cost–accuracy analysis for accuracy estimates unadjusted for TAB. Under the NHS reference costs130 costing system, half-node OSNA dominated histopathology for each additional node-positive case detected, but histopathology dominated half-node OSNA for each additional node-negative case detected and had an incremental cost per additional negative case detected of £8994 relative to full-node OSNA, which is lower than the conventional cost-effectiveness threshold. When YHEC cost values124 were used, histopathology dominated half-node OSNA for all measures of accuracy and consistently had an ICER of < £13,000 compared with full-node OSNA.
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | ||
| Half node | Full node | ||||
| Accuracya | 1.0000 | 0.9344 | 0.9034 | 0.0310 | 0.0656 |
| Sensitivity × prevalenceb | 0.2000 | 0.2000 | 0.1690 | 0.0310 | 0 |
| Specificity × (1 – prevalence)b | 0.8000 | 0.7344 | 0.7344 | 0 | 0.0656 |
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (£) | 3987 | 3897 | 3397 | 500 | 90 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node OSNA extendedly dominated | 6108c | |||
| Incremental cost per additional node-positive case detected (£) | 16,123 | Histopathology dominated | |||
| Incremental cost per additional node-negative case detected (£) | Half-node OSNA dominated | 8994c | |||
| Analysis using costs based on the YHEC model124 | |||||
| Cost per patient (£) | 2228 | 2284 | 1855 | 429 | –56 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node OSNA extendedly dominated | 3866c | |||
| Incremental cost per additional node-positive case detected (£) | Half-node OSNA extendedly dominated | 12,046c | |||
| Incremental cost per additional node-negative case detected (£) | Half-node OSNA dominated | 5693c | |||
The discrepancy between the YHEC cost results and the NHS reference cost results occurred for a couple of reasons. First, YHEC costs for surgery were significantly lower than the NHS reference costs for surgery, giving greater weight to other costs, such as adverse event costs, but there was also a difference in the ratios between the different types of surgery (breast surgery with SLNB alone, breast surgery with SLNB and ALND, breast surgery for ALND alone). In particular, the cost ratio between histopathological and intraoperative test-positive patients using YHEC costs was significantly lower than that obtained using the NHS reference costs. This meant that the cost impact of a second surgery for patients diagnosed by histopathology was greatly reduced and costs such as the additional cost of histopathology in OSNA test-negative patients and the cost of short-term adverse events in the half-node strategy had a greater influence. This made the half-node strategy more expensive than histopathology alone. This gave rise to the seemingly inconsistent results between the NHS reference cost analysis, in which half-node OSNA was not dominated by histopathology, and the YHEC cost analysis, in which half-node OSNA was dominated by histopathology.
This analysis was repeated for the three individual OSNA studies that adjusted for TAB and the incremental results are reported in Table 50. Similar results were found for the studies by Le Frère-Belda et al. 58 and Snook et al. ,61 but not for the best-case scenario provided by Khaddage et al. ,57 in which sensitivity was 100% and specificity was 97.2%. Here, full-node OSNA had the same accuracy as half-node OSNA and so histopathology was compared directly with full-node OSNA. Full-node OSNA dominated histopathology for a node-positive diagnosis and any additional node-negative patients detected by histopathology cost £27,300 per case identified under the NHS reference costs. This reduced to £17,100 under YHEC costing.
| Measure | Incremental results | |||||
|---|---|---|---|---|---|---|
| Le Frère-Belda 201258 | Snook 201161 | Khaddage 201157 | ||||
| Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | |
| Accuracya | 0.0172 | 0.0536 | 0.0204 | 0.0440 | 0 | 0.0224 |
| Sensitivity × prevalenceb | 0.0172 | 0 | 0.0204 | 0 | 0 | 0 |
| Specificity × (1 – prevalence)b | 0 | 0.0536 | 0 | 0.0440 | 0 | 0.0224 |
| Analysis using NHS reference costs130 for ALND | ||||||
| Cost per patient (£) | 437 | 150 | 458 | 152 | 367 | 244 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node OSNA extendedly dominated | 8289c | Half-node OSNA extendedly dominated | 9463c | Half-node OSNA dominated | 27,300c |
| Incremental cost per additional node-positive case detected (£) | 25,426 | Histopathology dominated | 22,435 | Histopathology dominated | Half-node OSNA dominated | Histopathology dominated |
| Incremental cost per additional node-negative case detected (£) | Half-node OSNA dominated | 10,949c | Half-node OSNA dominated | 13,850c | Half-node OSNA dominated | 27,300c |
| Analysis using costs based on the YHEC model124 | ||||||
| Cost per patient (£) | 398 | –26 | 411 | –29 | 367 | 16 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node OSNA extendedly dominated | 5254c | Half-node OSNA extendedly dominated | 5933c | Half-node OSNA dominated | 17,100c |
| Incremental cost per additional node-positive case detected (£) | Half-node OSNA extendedly dominated | 21,629c | Half-node OSNA extendedly dominated | 18,731c | Half-node OSNA dominated | Histopathology dominated |
| Incremental cost per additional node-negative case detected (£) | Half-node OSNA dominated | 6941c | Half-node OSNA dominated | 8684c | Half-node OSNA dominated | 17,100c |
Part of the cost-effectiveness assessment of the short-term outcomes (i.e. of the diagnostic phase) examined the disutility of second operations and waiting for histopathology results. The diagnostic costs and utility per patient are reported in Table 51. For strategies that do not involve histopathology, this utility was 1 as there was neither the need to wait for test results nor a second surgery directly resulting from the intraoperative test. Using the NHS reference cost scenario, full-node OSNA dominated half-node OSNA and histopathology as its increased QALY gains, measured until the end of the diagnostic phase, were less costly. Under the YHEC costing strategy, half-node OSNA provided a small QALY gain over histopathology, with an ICER of £4832 per QALY gained, but full-node OSNA continued to dominate half-node analysis and histopathology. It is estimated that 4.1% of the 76.5% of patients who have to wait for confirmatory postoperative histopathology results under the half-node testing strategy end up with a positive diagnosis as opposed to the 20% expected positivity rate under the postoperative histopathology-only strategy.
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | OSNA | Difference half-node OSNA vs. histopathology | Difference full-node OSNA vs. half-node OSNA | ||
| Half node | Full node | ||||
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (£) | 3987 | 3897 | 3397 | –90 | –500 |
| Utility | 0.9739 | 0.9854 | 1 | 0.0115 | 0.0146 |
| Incremental cost per QALY gained | Half-node OSNA dominated | Histopathology dominateda | |||
| Analysis using costs based on the YHEC model124 | |||||
| Cost per patient (£) | 2228 | 2284 | 1855 | 56 | –429 |
| Utility | 0.9739 | 0.9854 | 1 | 0.0115 | 0.0146 |
| Incremental cost per QALY gained | Half-node OSNA extendedly dominated | Histopathology dominateda | |||
According to these short-term costs and benefits, full-node OSNA is the most cost-effective test for short-term utility. Using the meta-analysis accuracy values for OSNA (sensitivity 84.5%, specificity 91.8%), histopathology appears to be cost-effective in terms of short-term accuracy, although this begins to look questionable when the effect of TAB is accounted for and OSNA has a higher sensitivity and specificity. In all scenarios OSNA is less costly than histopathology.
Long-term analysis
Thus far, the analysis has considered only the short-term costs and consequences of the different diagnostic options, effectively assuming that no utility benefits accrue from increased diagnostic accuracy. In the long-term scenario we examined costs and QALYs, which are presented in Table 52 for the studies with no adjustment for TAB, and account for all costs and the benefits of accurate diagnosis through improved patient management net of the disutility of anxiety from waiting for the postoperative diagnosis and undergoing a second operation. The strategies were then ordered by increasing number of QALYs, with full-node OSNA having the fewest QALYs (9.222) and histopathology having the most (9.321). As this demonstrates, the QALY difference was approximately 0.10 (i.e. the equivalent of 5 weeks of life in full health). This QALY difference occurs as a direct consequence of the accuracy of the tests, as a higher accuracy leads to a greater number of correct diagnoses. As the purpose of diagnosis is to inform management strategies, an increase in the number of correct diagnoses leads to an increase in the correct management of patients and therefore patients gain the most QALYs possible. The QALY gain from the higher accuracy of histopathology is small, but it may be seen as cost-effective; under the NHS reference costing and ignoring half-node OSNA, as this is a strategy that is extendedly dominated and unlikely to continue once OSNA has been validated, histopathology had an ICER of £4324 per QALY gained compared with full-node OSNA. Under YHEC costing, histopathology dominated half-node OSNA, that is, OSNA had higher costs and produced fewer QALYs. In this scenario the ICER comparing histopathology with full-node OSNA was £2150 per QALY gained.
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | Half-node OSNA | Full-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | |
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (discounted) (£)a | 20,530 | 20,523 | 20,099 | 424 | 7 |
| QALYs (discounted)a | 9.321 | 9.307 | 9.222 | 0.085 | 0.015 |
| Incremental cost per QALY gained (£)a | Half-node OSNA extendedly dominated | 4324b | |||
| Analysis using costs based on the YHEC model124 | |||||
| Costs per patient (discounted) (£)a | 18,771 | 18,910 | 18,556 | 353 | –139 |
| Incremental cost per QALY gained (£) | Half-node OSNA extendedly dominated | 2150b | |||
For the TAB-adjusted studies (presented in Table 53), half-node OSNA remained extendedly dominated using the Le Frère-Belda58 values. In this case, histopathology compared with full-node OSNA had ICERs of £9493 per QALY gained using NHS reference costs130 and £5215 using YHEC costs. 124 Using the values of Snook et al. ,61 half-node OSNA was extendedly dominated using YHEC costs (and histopathology had an ICER compared with full-node OSNA of < £4850 per QALY gained) but was no longer extendedly dominated using NHS reference costs, and had an ICER compared with full-node OSNA of £8063 per QALY gained. Comparing histopathology with half-node OSNA, the ICER was £14,967 per QALY gained. More importantly, Khaddage et al. 57 suggested a much better result for OSNA, with full-node OSNA dominating half-node OSNA and histopathology (full-node OSNA had a higher QALY gain and lower costs). This was because of the influence of the short-term disutility for anxiety and a second operation applied (when applicable) to patients undergoing histopathology testing in the histopathology and OSNA half-node arms. Without this disutility the results using the Khaddage et al. 57 values for sensitivity and specificity altered slightly so that, although in the long term half-node OSNA was still dominated by full-node OSNA, this was now because they had the same QALY gain but full-node OSNA was less expensive. The ICER between histopathology and full-node OSNA was £68,432 per QALY gained using NHS reference costs and £41,619 per QALY gained using YHEC costs, suggesting that histopathology was not that cost-effective compared with full-node OSNA.
| Measure | Incremental results | |||||
|---|---|---|---|---|---|---|
| Le Frère-Belda 201258 | Snook 201161 | Khaddage 201157 | ||||
| Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. histopathologya | Difference full-node OSNA vs. half-node OSNAa | |
| Analysis using NHS reference costs130 for ALND | ||||||
| Cost per patient (discounted) (£)b | 395 | 82 | 408 | 96 | –216 | –367 |
| QALYs (discounted)b | 0.041 | 0.010 | 0.051 | 0.006 | 0.0025 | 0.0151 |
| Incremental cost per QALY gained (£) | Half-node OSNA extendedly dominated | 9493c | 8063 | 14,967 | Histopathology dominated | Half-node OSNA dominated |
| Analysis using costs based on the YHEC model124 | ||||||
| Cost per patient (discounted) (£)b | 356 | –94 | 361 | –85 | –367 | –13 |
| Incremental cost per QALY gained (£) | Half-node OSNA extendedly dominated | 5215c | Half-node OSNA extendedly dominated | 4850c | Histopathology dominated | Half-node OSNA extendedly dominated |
Sensitivity analysis
For the purposes of sensitivity analysis we chose to report only the findings of the NHS reference costing strategy.
Accuracy
As the TAB-adjusted results have already indicated, test accuracy has a direct impact on the cost-effectiveness of the tests. A threshold analysis was therefore conducted to investigate sensitivity and specificity separately. In the case of threshold analysis for sensitivity, specificity was held constant and sensitivity was increased in steps of 5% over a range of 70–100%. The opposite was then performed for specificity. This sensitivity analysis was conducted on the full-node OSNA results, for which the change in accuracy would be most notable (the overall sensitivity for half-node OSNA was fixed at 100%). Short-term utility results are not reported as the utility for OSNA was not affected by the accuracy of the test.
The short-term results of the threshold analysis for sensitivity are presented in Table 54. Table 54 demonstrates that, when specificity is held at 91.8%, the proportion of node-positive cases detected increased by 1% each time the sensitivity increased by 5%. The short-term costs of OSNA also increased by approximately £18 for each step increase in sensitivity (therefore decreasing the difference in costs between OSNA and histopathology) and the combination of both increased the cost–accuracy ICERs between histopathology and OSNA. The ICER for cost per case correctly identified increased from £5104 per additional case correctly identified by histopathology when OSNA had 70% sensitivity to £8162 per additional case correctly identified by histopathology when OSNA had 100% sensitivity. The ICERs for cost per additional node-positive case detected were consistently larger, ranging from £10,685 per additional node-positive case detected when OSNA had 70% sensitivity to OSNA dominating histopathology when OSNA had 100% sensitivity. However, in this case, the ICER increased much faster as the sensitivity of OSNA neared 100%, as Figure 17 shows.
| Measure | Incremental outcomes of histopathology vs. OSNA of varying sensitivitya | |||||||
|---|---|---|---|---|---|---|---|---|
| Base case: 0.845 | 0.70 | 0.75 | 0.80 | 0.85 | 0.90 | 0.95 | 1.00 | |
| Accuracyb | 0.0966 | 0.1256 | 0.1156 | 0.1056 | 0.0956 | 0.0856 | 0.0756 | 0.0656 |
| Sensitivity × prevalencec | 0.031 | 0.06 | 0.05 | 0.04 | 0.03 | 0.02 | 0.01 | 0 |
| Analysis using NHS reference costs130 for ALND | ||||||||
| Cost per patient (£) | 590 | 641 | 623 | 606 | 588 | 571 | 553 | 535 |
| Incremental cost per additional patient correctly diagnosed (£) | 6108 | 5104 | 5394 | 5737 | 6153 | 6666 | 7315 | 8162 |
| Incremental cost per additional node-positive case detected (£) | 19,033 | 10,685 | 12,470 | 15,147 | 19,609 | 28,532 | 55,303 | Histopathology dominated |
FIGURE 17.
Comparison of cost–accuracy results for threshold analysis of OSNA sensitivity.
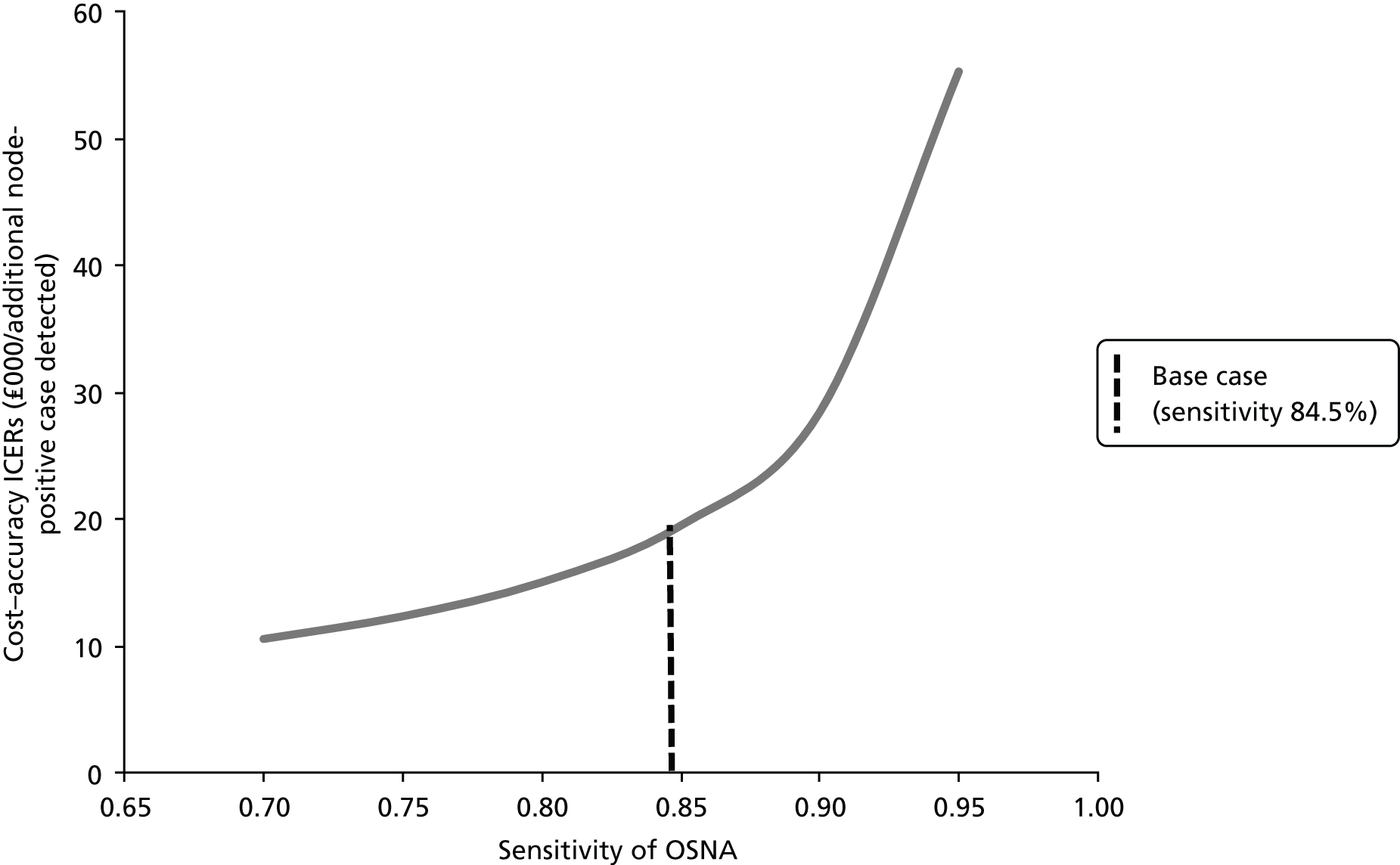
The long-term results, presented in Table 55, show a similar finding to the accuracy results: as the sensitivity of OSNA increased, so did the ICER for the cost per QALY gained by histopathology. This ranged from £2119 per QALY gained when OSNA had a sensitivity 70% to £14,193 per QALY gained when OSNA had a sensitivity of 95%. At 100% sensitivity OSNA dominated histopathology, having more QALYs at a lower cost. Unlike the accuracy results, the cost difference between histopathology and OSNA also increased each time the sensitivity increased. Again, as the sensitivity of OSNA neared 100% the ICER began to increase much faster, as demonstrated in Figure 18.
| Measure | Incremental outcomes of histopathology vs. OSNA of varying sensitivitya | |||||||
|---|---|---|---|---|---|---|---|---|
| Base case: 0.845 | 0.70 | 0.75 | 0.80 | 0.85 | 0.90 | 0.95 | 1.00 | |
| QALYs (discounted)b | 0.0997 | 0.1939 | 0.1614 | 0.1289 | 0.0964 | 0.0639 | 0.0314 | –0.0011 |
| Analysis using NHS reference costs130 for ALND | ||||||||
| Cost per patient (discounted)b (£) | 431 | 411 | 418 | 425 | 431 | 438 | 445 | 452 |
| Incremental cost per QALY (£) | 4324 | 2119 | 2588 | 3294 | 4476 | 6862 | 14,193 | Histopathology dominated |
FIGURE 18.
Comparison of long-term cost-effectiveness for threshold analysis of OSNA sensitivity.

The results of the threshold analysis for specificity are presented in Tables 56 and 57. When sensitivity is held at 84.5%, the proportion of node-negative cases detected by OSNA increases by 4% each time the specificity is increased by 5% and the cost decreases by approximately £70. As with the threshold analysis for sensitivity, the accuracy ICERs for histopathology increased with increasing specificity of OSNA. In terms of the cases correctly identified, the ICERs ranged from £1043 per additional case correctly diagnosed by histopathology when OSNA had a specificity of 70% to £22,761 per additional case correctly diagnosed when OSNA had a specificity of 100%. For the node-negative cases, the ICERs ranged from £1178 per node-negative case detected when OSNA had a specificity of 70% to OSNA dominating histopathology when OSNA had a specificity of 100%. This time both cost–accuracy measures had ICERs that increased more quickly as the specificity of OSNA approached 100%. Figure 19 illustrates this pattern for detecting node-negative cases. In this figure, the ICER for histopathology increases as the specificity of OSNA increases. The node-negative cases curve appears truncated because at the maximum specificity (100%) OSNA dominates histopathology (same diagnostic yield, lower costs), which is not represented on the graph.
| Measure | Incremental outcomes of histopathology vs. OSNA of varying specificitya | |||||||
|---|---|---|---|---|---|---|---|---|
| Base case: 0.918 | 0.70 | 0.75 | 0.80 | 0.85 | 0.90 | 0.95 | 1.00 | |
| Accuracyb | 0.0966 | 0.2710 | 0.2310 | 0.1910 | 0.1510 | 0.1110 | 0.0710 | 0.0310 |
| Specificity × (1 – prevalence)c | 0.0656 | 0.24 | 0.20 | 0.16 | 0.12 | 0.08 | 0.04 | 0.00 |
| Analysis using NHS reference costs130 for ALND | ||||||||
| Cost per patient (£) | 590 | 283 | 353 | 424 | 494 | 565 | 635 | 706 |
| Incremental cost per additional patient correctly diagnosed (£) | 6108 | 1043 | 1529 | 2218 | 3272 | 5087 | 8945 | 22,761 |
| Incremental cost per additional node-negative case detected (£) | 8994 | 1178 | 1766 | 2648 | 4118 | 7058 | 15,878 | Histopathology dominated |
| Measure | Incremental outcomes of histopathology vs. OSNA of varying specificitya | |||||||
|---|---|---|---|---|---|---|---|---|
| Base case: 0.918 | 0.70 | 0.75 | 0.80 | 0.85 | 0.90 | 0.95 | 1.00 | |
| QALYs (discounted)b | 0.0097 | 0.1660 | 0.1508 | 0.1356 | 0.1203 | 0.1051 | 0.0899 | 0.0747 |
| Analysis using NHS reference costs130 for ALND | ||||||||
| Cost per patient (discounted) (£)b | 431 | –98 | 24 | 145 | 266 | 387 | 508 | 630 |
| Incremental cost per QALY (£) | 4324 | OSNA dominated | 156 | 1068 | 2210 | 3683 | 5655 | 8430 |
FIGURE 19.
Comparison of cost–accuracy results for threshold analysis of OSNA specificity.

The long-term cost-effectiveness results, presented in Table 57 and Figure 20, show that the long-term costs of OSNA decreased as the specificity and QALY gain increased. This meant that for the lowest specificity of 70%, OSNA was dominated by histopathology as it was both more expensive and produced fewer QALYs. The largest ICER for histopathology, when OSNA had 100% specificity, was £8430 per QALY gained. As with the previous results, the ICER increase was more pronounced as the cost difference between histopathology and OSNA increased (i.e. as the specificity of OSNA increased), but this increase was not as severe as it was for the short-term outcomes in the specificity threshold analysis or for the long-term cost-effectiveness outcomes in the sensitivity threshold analysis.
FIGURE 20.
Comparison of long-term cost-effectiveness for threshold analysis of OSNA.
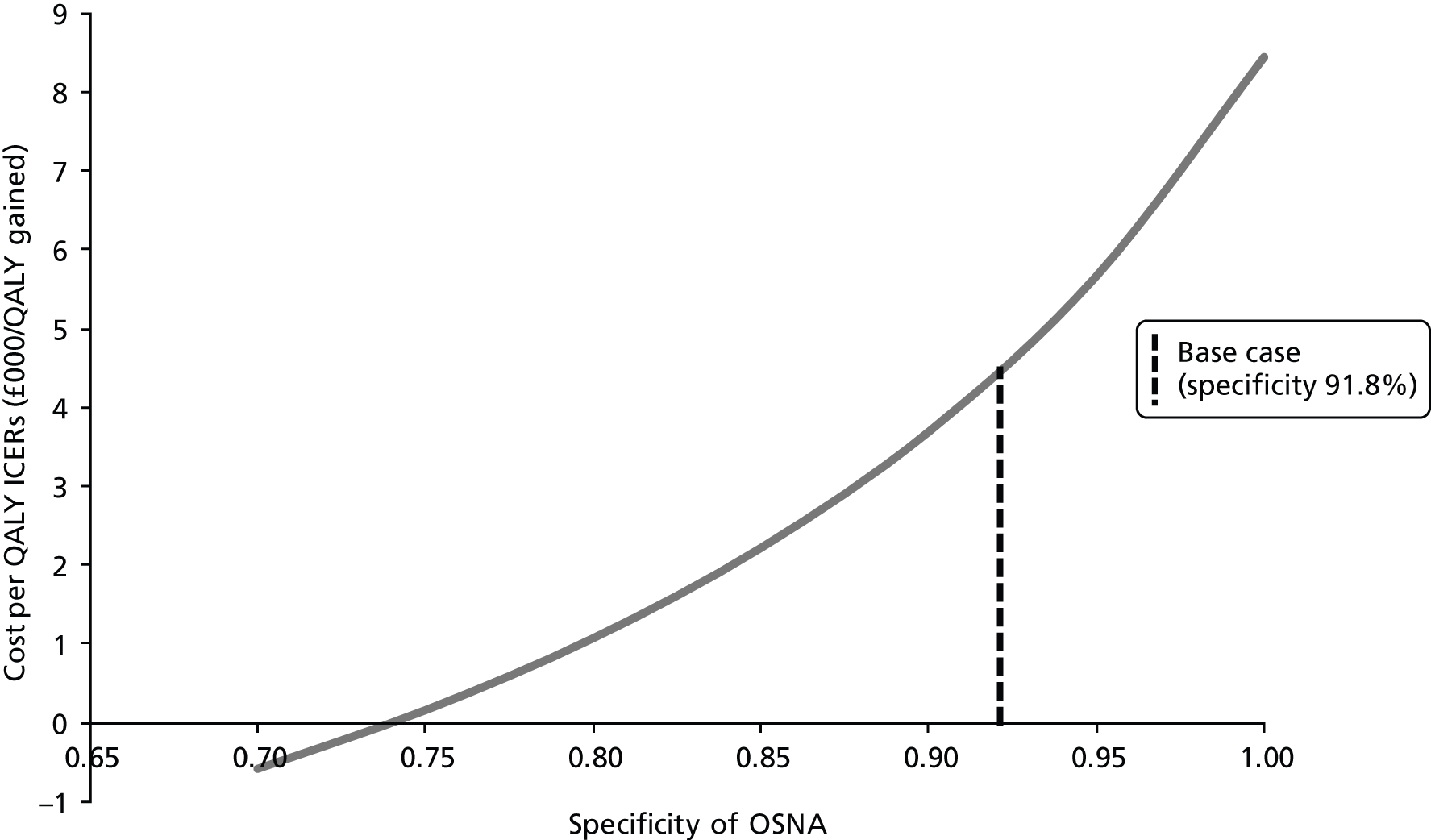
These threshold analyses demonstrate no obvious significant differences from the base-case results, although all ICERs increased as either the sensitivity or the specificity increased. Starting at about 90%, the increases in sensitivity or specificity resulted in exponential increases in the ICERs. Individually, the findings from the base case are repeated, with specificity seemingly having a larger impact on the short-term outcome of cost–accuracy and sensitivity having a larger impact on the long-term cost-effectiveness.
Overall, these threshold analyses suggest that, if the true values of sensitivity and specificity for OSNA lie within the range of 90–100%, close to the values for the sensitivity and specificity of histopathology, the cost-effectiveness of OSNA may increase greatly. Furthermore, sensitivity and specificity are not independent and therefore altering one would alter the other. For simplicity, this analysis does not analyse the relationship between specificity and sensitivity and therefore it may not adequately represent the uncertainty in the results. However, as the TAB-adjusted results from Khaddage et al. 57 demonstrate, when both specificity and sensitivity are sufficiently high, OSNA becomes much more cost-effective.
The uncertainty in the results, accounting for the dependency of sampling variation in sensitivity and specificity, is measured by the probability at which full OSNA is cost-effective relative to postoperative histopathology. Figure 21 presents this probability as a function of the value of a QALY to NHS decision-makers. One curve is shown for each of the three studies on the diagnostic performance of OSNA that adjust for TAB and a fourth curve presents the results corresponding to accuracy values (sensitivity 0.914, specificity 0.968) obtained by combining all data on patients from the three studies, with the sensitivity and specificity being the weighted averages of the estimates of the individual studies, with the respective weights given in proportion to the relative sample sizes of the studies. Although adopting the values reported by Khaddage et al. 57 leads to the opposite result to that obtained using sensitivity–specificity values from the other two studies (i.e. at the £20,000 threshold OSNA has a higher probability of being cost-effective than histopathology), the study weight in the analysis using combined accuracy data is limited by its small size relative to the other two studies. On the basis of all available data, at a cost-effectiveness threshold of £20,000 per QALY, OSNA has a less than one in three chance of being cost-effective.
FIGURE 21.
Probability of full-node OSNA being cost-effective by cost-effectiveness threshold.
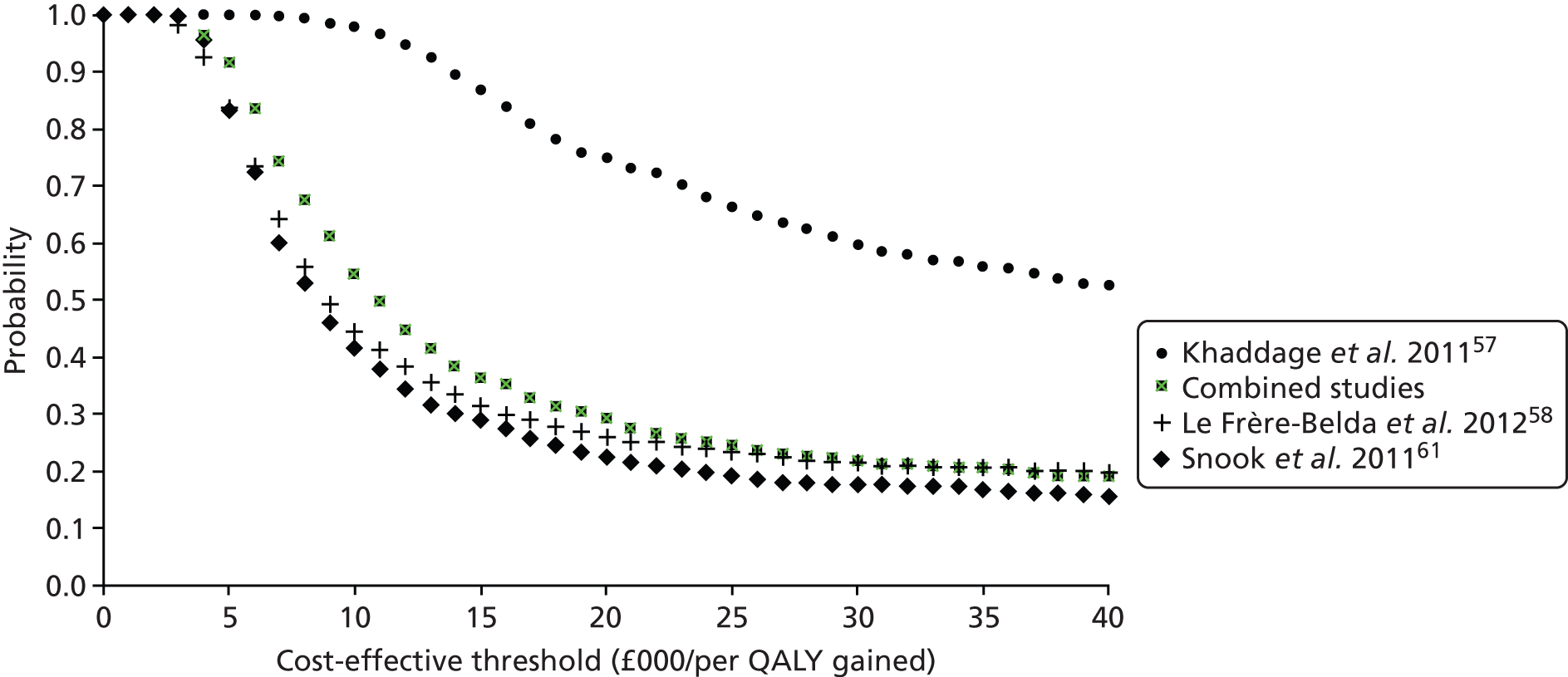
Prevalence
The prevalence of LN metastases in the population directly affects the accuracy of the tests and their outcomes. To investigate the effect fully, we considered the effect of prevalence when our base-case value of 20% was halved to 10% and doubled to 40%.
When prevalence was 10%, histopathology dominated half-node OSNA and had an ICER of < £20,000 per additional case correctly identified compared with full-node OSNA, both overall and when split into node-negative and node-positive patients. Short-term cost–utility results remained the same, with full-node OSNA dominating the rest. In the long term, histopathology dominated half-node OSNA and the ICER for histopathology compared with full-node OSNA was £2626 per QALY gained.
When the prevalence was increased to 40%, the accuracy results were similar to those of the base case, with histopathology being cost-effective for additional cases correctly identified, half-node OSNA dominating histopathology for detecting additional node-positive patients and full-node OSNA dominating half-node OSNA for additional node-negative patients detected. The ICER for histopathology compared with full-node OSNA for detecting node-negatives cases increased to £24,680 per additional case detected. OSNA once again dominated the short-term utility analyses. In the long term half-node OSNA dominated histopathology as it now had a slightly higher QALY gain (0.0001). Half-node OSNA had an ICER of £2208 per QALY gained compared with full-node OSNA.
Short-term costs
In the short term the costs of the tests were altered by ±10%. Altering the cost of histopathology did not affect the overall outcomes, simply increasing or reducing individual ICERs accordingly. Decreasing the cost of OSNA to £315 similarly did not greatly affect the results but increasing it to £385 resulted in histopathology dominating half-node OSNA in the long term, with an ICER of £3972 for histopathology compared with full-node OSNA.
The cost of separate ALND surgery was altered using values provided in the NHS reference costs. 130 A low second surgery cost (£1608) meant that, in the short term, histopathology dominated half-node OSNA for all accuracy ICERs and, compared with full-node OSNA, all ICERs were < £7000 per additional case detected. For short-term utility, full-node OSNA still dominated half-node OSNA and histopathology. In the long term histopathology dominated half-node OSNA and, compared with full-node OSNA, histopathology had an ICER of £388 per QALY gained. A high second surgery cost (£4871) had no significant effect on short-term outcomes. Long-term ICERs increased to £15,384 per QALY gained for histopathology compared with half-node OSNA and £5469 per QALY gained for half-node OSNA compared with full-node OSNA.
Long-term costs
Adjuvant therapy costs were altered by ±10%. This affected only the long-term results and did not greatly influence them. A high cost for patients undergoing hormonal adjuvant therapy (£1195) increased the ICERs slightly, with histopathology having an ICER of £4353 compared with full-node OSNA. A low cost for patients undergoing chemotherapy (£8502) resulted in the smallest ICERs, reducing the ICER for histopathology compared with full-node OSNA to £4237 per QALY gained.
The sensitivity analyses performed are presented in Tables 58 and 59. These include sensitivity analyses performed on the disutility associated with anxiety from waiting for results and a second operation and the costs of the first breast surgery (with or without ALND). There were no significant differences in the results. Overall, we found that the most influential parameters were the sensitivity and specificity of OSNA, which means that it is very important to have good-quality data on these values. Additional two-way sensitivity analyses (see Appendix 8) showed that, to be cost-effective, full-node OSNA has to have sensitivity and specificity estimates of ≥ 95%.
| Parameter | Base case | Sensitivity analysis | Cost–accuracy ICERs using NHS reference costs130 | |||||
|---|---|---|---|---|---|---|---|---|
| Incremental cost per additional patient correctly diagnosed (£) | Incremental cost per additional node-positive case detected (£) | Incremental cost per additional node-negative case detected (£) | ||||||
| Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | |||
| Base case | NA | NA | Half-node OSNA extendedly dominated | 6108a | 16,123 | Histopathology dominated | Half-node OSNA dominated | 8994a |
| Prevalence (%) | 20 | 10 | Half-node OSNA extendedly dominated | 3112a | Half-node OSNA extendedly dominated | 17,930a | Half-node OSNA dominated | 3766a |
| 40 | Half-node OSNA extendedly dominated | 10,919a | 9132 | Histopathology dominated | Half-node OSNA dominated | 24,680a | ||
| Cost of histopathology (£) | 472 | 425 | Half-node OSNA extendedly dominated | 5619a | 14,957 | Histopathology dominated | Half-node OSNA dominated | 8274a |
| 519 | Half-node OSNA extendedly dominated | 6597a | 17,289 | Histopathology dominated | Half-node OSNA dominated | 9714a | ||
| Cost of OSNA (£) | 350 | 315 | Half-node OSNA extendedly dominated | 6470a | 16,123 | Histopathology dominated | Half-node OSNA dominated | 9528a |
| 385 | Half-node OSNA extendedly dominated | 5746a | 16,123 | Histopathology dominated | Half-node OSNA dominated | 8461a | ||
| Cost of second surgery (£) | 3569 | 1608 | Half-node OSNA extendedly dominated | 2048a | Half-node OSNA extendedly dominated | 6383a | Half-node OSNA dominated | 3016a |
| 4871 | Half-node OSNA extendedly dominated | 8805a | 17,425 | Histopathology dominated | Half-node OSNA dominated | 12,966a | ||
| Parameter | Base case | Sensitivity analysis | Short-term and long-term cost-effectiveness ICERs using NHS reference costs130 | |||
|---|---|---|---|---|---|---|
| Short-term incremental cost per QALY gained (£) | Long-term incremental cost per QALY gained (cost and QALYs discounted) (£)b | |||||
| Difference half-node OSNA vs. histopathology | Difference full-node OSNA vs. half-node OSNA | Difference half-node OSNA vs. full-node OSNA | Difference histopathology vs. half-node OSNA | |||
| Base case | NA | NA | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4324c |
| Prevalence (%) | 20 | 10 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 2626c |
| 40 | Half-node OSNA extendedly dominated | Histopathology dominatedc | 2208 | Histopathology dominated | ||
| Cost of histopathology (£) | 472 | 425 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 3850c |
| 519 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4797c | ||
| Cost of OSNA (£) | 350 | 315 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4675c |
| 385 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 3972c | ||
| Cost of second surgery (£) | 3569 | 1608 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 388c |
| 4871 | Half-node OSNA extendedly dominated | Histopathology dominatedc | 5469 | 15,384 | ||
| Annual cost of adjuvant therapy (hormone therapy and follow-up) (£) | 1087 | 978 | NA | NA | Half-node OSNA extendedly dominated | 4311c |
| 1195 | NA | NA | Half-node OSNA extendedly dominated | 4353c | ||
| 6-month cost of adjuvant therapy (chemotherapy and follow-up) (£) | 9447 | 8502 | NA | NA | Half-node OSNA extendedly dominated | 4237c |
| 10,392 | NA | NA | Half-node OSNA extendedly dominated | 4402c | ||
| Disutility of anxiety from waiting for histopathology results | 0.019 | None | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 3618c |
| 0.006 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 3805c | ||
| 0.023 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4463c | ||
| Disutility of second surgery for ALND | 0.033 | None | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4052c |
| 0.027 | Half-node OSNA extendedly dominated | Histopathology dominatedc | Half-node OSNA extendedly dominated | 4266c | ||
| 0.04 | Half-node OSNA extended dominated | Histopathology dominatedc | Half-node OSNA extended dominated | 4382c | ||
Metasin results
As an alternative index test we compared Metasin with histopathology. This was performed as an illustrative exercise as the accuracy values for Metasin come from a draft paper (Sundaresan40) that had not yet been peer reviewed at the time of this review. Furthermore, the only cost information for Metasin was provided by personal correspondence with the Princess Alexandra Hospital on behalf of the author of the draft paper (Matt Hayday, The Princess Alexandra Hospital NHS Trust, 19 November 2012, personal communication). This value reflected the cost of the non-CE-marked Metasin test as no list price for the test is yet available even though Metasin has received a CE mark. The cost-effectiveness analyses may be updated if and when a finalised list price for the CE-marked Metasin test is received. Updated cost estimates were made available after the modelling analyses were completed but as these did not seem significantly different from those used in the analysis the changes were not implemented.
The cost–accuracy results are reported in Table 60 and show that half-node Metasin is extendedly dominated by histopathology, which, when compared with full-node OSNA, results in £20,302 per additional case detected using NHS reference costs130 and £14,990 using YHEC costing. 124 As with OSNA, half-node Metasin dominated histopathology for additional node-positive case detected as it had the same accuracy but was less expensive than histopathology. Furthermore, full-node Metasin dominated half-node Metasin for detecting node-negative cases. The ICER for node-negative case detected for histopathology compared with full-node Metasin was £30,453 using NHS reference costs and £22,484 using YHEC costs. The cost-effectiveness of histopathology compared with Metasin therefore depends on the costing strategy and threshold used.
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | Metasin | Difference half-node Metasin vs. full-node Metasin | Difference histopathology vs. half-node Metasin | ||
| Half node | Full node | ||||
| Accuracya | 1.0000 | 0.9704 | 0.9556 | 0.0148 | 0.0296 |
| Sensitivity × prevalenceb | 0.2000 | 0.2000 | 0.1852 | 0.0148 | 0 |
| Specificity × (1 – prevalence)b | 0.8000 | 0.7704 | 0.7704 | 0 | 0.0296 |
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (£) | 3987 | 3523 | 3086 | 437 | 465 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node Metasin extendedly dominated | 20,302c | |||
| Incremental cost per additional node-positive case detected (£) | 29,515 | Histopathology dominated | |||
| Incremental cost per additional node-negative case detected (£) | Half-node Metasin dominated | 30,453c | |||
| Analysis using costs based on the YHEC model124 | |||||
| Cost per patient (£) | 2228 | 1966 | 1562 | 403 | 263 |
| Incremental cost per additional patient correctly diagnosed (£) | Half-node Metasin extendedly dominated | 14,990c | |||
| Incremental cost per additional node-positive case detected (£) | 27,230 | Histopathology dominated | |||
| Incremental cost per additional node-negative case detected (£) | Half-node Metasin dominated | 22,484c | |||
Short-term utility results for Metasin are reported in Table 61. Regardless of costing strategy Metasin dominated histopathology, having a lower cost and producing more QALYs. It is estimated that of the 78.5% of half-node Metasin patients with a negative result, who have to wait for histopathology test results, 1.9% would go on to have a positive diagnosis, in contrast to the 20% of expected positive diagnoses with postoperative histopathology. In the long term, histopathology compared with half-node Metasin had an ICER of > £460,000 per QALY gained using NHS reference costs and an ICER of > £240,000 per QALY gained using YHEC costs (Table 62). The ICER between half-node and full-node Metasin remained < £13,000 for both costing strategies.
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | Metasin | Difference half-node Metasin vs. histopathology | Difference full-node Metasin vs. half-node Metasin | ||
| Half node | Full node | ||||
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (£) | 3987 | 3523 | 3086 | –465 | –437 |
| Utility | 0.9739 | 0.9849 | 1 | 0.0110 | 0.0151 |
| Incremental cost per QALY gained (£) | Half-node Metasin extendedly dominated | Histopathology dominateda | |||
| Analysis using costs based on the YHEC model124 | |||||
| Cost per patient (£) | 2228 | 1966 | 1562 | –263 | –403 |
| Incremental cost per QALY gained (£) | Half-node Metasin extendedly dominated | Histopathology dominateda | |||
| Measure | Mean estimates | Incremental results | |||
|---|---|---|---|---|---|
| Histopathology | Half-node Metasin | Full-node Metasin | Difference half-node Metasin vs. full-node Metasin | Difference histopathology vs. half-node Metasin | |
| Analysis using NHS reference costs130 for ALND | |||||
| Cost per patient (£) | 20,530 | 20,103 | 19,702 | 401 | 427 |
| QALYs | 9.321 | 9.320 | 9.288 | 0.032 | 0.001 |
| Incremental cost per QALY gained (£) | 12,374 | 467,113 | |||
| Analysis using costs based on the YHEC model124 | |||||
| Cost per patient (£) | 18,771 | 18,546 | 18,179 | 367 | 225 |
| Incremental cost per QALY gained (£) | 11,329 | 246,089 | |||
Chapter 5 Discussion
Statement of principal findings
Clinical effectiveness
Seventeen studies were included that investigated the performance of either OSNA or Metasin for detecting metastases in the sentinel or axillary LNs of breast cancer patients. Two of the included studies reported on Metasin; both were unpublished and were reported in draft form. The remaining 15 studies reported on OSNA, with two papers reporting the same study.
The studies were assessed against the Cochrane risk of bias tool148 and the QUADAS-2 tool. 43 The majority of studies were considered to be at low risk of bias, although many were unclear regarding their method of patient recruitment and lacked detail on patient characteristics. Often, no evidence was given of sample replicates and reproducibility for molecular analysis. Furthermore, test failures were not reported for most studies. Reported outcomes were also limited, for example no data were found for clinical outcomes, such as patient anxiety and number of repeat operations. Only one study56 provided evidence on time in the operating theatre. A potential conflict of interest also features heavily as one of the two unpublished Metasin studies was performed at the institution in which the technology was developed and the majority of the OSNA studies were financially supported by Sysmex.
With regard to test accuracy, there are two main issues. The first is the strong assumption that the reference standard is the most accurate measure of the target disorder. In this case, the reference standard (i.e. histopathology), although plausible, has been performed with varying levels of analysis and, as such, it may not be a true indicator of the target condition. The second concern is TAB, which occurs when different portions of tissue are allocated to the index and reference tests and cannot then be reused between them. Studies have dealt with this in a variety of ways, some reanalysing both the histopathology and the molecular samples, some choosing to reanalyse just one technology and some doing neither. The majority of studies that have adjusted for TAB have taken a conservative approach by excluding affected samples, which we consider to be a reasonable practice.
A summary of the results is presented in Table 63. As there were only two studies for Metasin, a meta-analysis was not performed. The displayed data for this test were taken from draft papers, before peer review. Therefore, the results, (academic-in-confidence information has been removed), must be used with caution.
| Measure | Sample type | Adjustment for TAB | No. of studies | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) |
|---|---|---|---|---|---|
| (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) | (Academic-in-confidence information has been removed) |
| Pooled OSNA | Patient | No | 5 | 84.5 (74.7 to 91.0) | 91.8 (87.8 to 94.6) |
| Pooled OSNA | Patient | Yes | 3 | 91.3 (83.6 to 95.6) | 94.2 (91.2 to 96.2) |
| Pooled OSNA | SLN | No | 4 | 79.9 (74.2 to 84.6) | 95.5 (94.1 to 96.5) |
| Pooled OSNA | SLN | Yes | 5 | 89.0 (82.1 to 93.4) | 97.5 (96.6 to 98.2) |
| Pooled OSNA | ALN | No | 6 | 95.1 (90.0 to 97.6) | 94.9 (91.2 to 96.9) |
| Pooled OSNA | ALN | Yes | 4 | 96.5 (87.3 to 99.1) | 96.2 (93.4 to 97.8) |
It should be noted that more than one SLN may be removed from a patient, which, as shown in Table 63, may have implications for sensitivity and specificity data when grouped either by patient or by node. Adjustment for TAB clearly improves test accuracy, increasing sensitivity of OSNA from 79.9% to 89.0% and increasing specificity of OSNA from 95.5% to 97.5% at the node level, with similar increases at patient level.
Studies that analysed ALNs were also included in this review. An increased sensitivity (ALN 95.1% vs. SLN 79.9%) and similar specificity (ALN 94.9% vs. SLN 95.5%) compared with the values found in the analysis of SLNs were seen before adjustment for TAB. Subsequent TAB adjustment increases the SLN sensitivity but has little effect on ALN data.
With regard to the time taken to perform the OSNA test, despite the lack of detail in the studies explaining which aspects of the procedure were monitored, the times reported ranged from < 30 minutes to 39.6 minutes for one node. This increases by approximately 5–10 minutes per additional node analysed.
Overall, and despite the concerns raised with regard to the reference standard and TAB, we feel that the studies were well performed and produced consistent results.
Cost-effectiveness
The short-term results, up to the diagnostic phase, suggest that OSNA is less accurate than the reference standard of histopathology but that it saves costs and results in less quality-of-life losses in patients from anxiety about a delayed diagnosis and second operation. With regard to Metasin the evidence base is currently only illustrative and requires validation as it is limited to two unpublished studies and may therefore be neither of the same quality nor as generalisable as the evidence on OSNA, which has accumulated over several studies performed in a wider variety of settings. It is possible that there is more confidence in the accuracy of OSNA and this should be taken into account when assessing the technologies, as OSNA still has the potential for cost-savings compared with current practice.
Limiting our analysis to the evidence on OSNA compared with histopathology revealed that there may be a trade-off between histopathology and intraoperative testing but that this depends on the estimates of costs adopted. Valuing resource use according to the current national reference costs, and judged on the ability to identify node-positive patients, histopathology was found to be inferior to OSNA. Whether the higher costs of using OSNA adjunctively are justified by the additional sensitivity of the test strategy, however, depends on whether decision-makers are prepared to pay £16,000 per additional case detected. On the other hand, when estimates derived from a microcosting study developed by YHEC in a study commissioned by the NHS Technology Adoption Centre are used to value resource use, histopathology may be cost-effective as long as the cost-effectiveness threshold is > £12,000 per node-positive case detected. In contrast, when the disutility of anxiety and a second operation is taken into account and the diagnostic options are compared in terms of costs and such disutility, OSNA emerges as the clearly preferred option with lower costs and less harm (disutility) inflicted on patients.
These results would be sufficient for medical decision-making if one entertained the idea that a more accurate diagnosis does not have significant health benefits for patients (i.e. QALY gains). Despite the inherent uncertainty of extrapolating benefits from the diagnostic phase into the remaining lifetime of patients undergoing SLNB, the present analysis has adapted an existing model of early breast cancer patient management developed by ScHARR, to gain insight into the potential significance of benefits from diagnostic accuracy gains in this clinical area. After accounting for the disutility of anxiety from waiting for postoperative diagnostic test results and a second operation, the long-term analysis using the adopted ScHARR model suggests that histopathology may produce large enough benefits from improved patient management compared with intraoperative testing approaches to offset its short-term disadvantage attributable to the costs and disutility associated with a delayed diagnosis and second operation. Indeed, the results suggest that as long as the NHS is willing to pay > £4000 per QALY gained histopathology should be retained as the optimal diagnostic approach for metastatic diagnosis in early breast cancer.
The base-case result that histopathology is cost-effective is, however, not robust to adjustment for TAB. Of the three studies that provided good-quality evidence on sensitivity and specificity, two supported histopathology as the cost-effective option at the conventional £20,000 per QALY threshold whereas the third, by Khaddage et al. ,57 results in full-node OSNA being unambiguously dominant.
A comment is also warranted regarding the relevant cost-effectiveness threshold for the present analyses. Typically in cost-effectiveness analyses, the status quo is less effective and less costly than the new technologies against which it is being compared. In the present case the reverse applies as histopathology, the current standard, is both more effective (i.e. produces higher QALYs) and more costly than the new intraoperative approaches. To facilitate interpretation we have presented the results of the three-arm comparison (intraoperative test with half node vs. intraoperative test with full node vs. histopathology) by ranking the three options by increasing level of effectiveness and calculating incremental costs, benefits and ICERs between adjacent options. That way we can retain the standard interpretation of ICERs for decision-making. However, it is argued by some economists that the threshold for cost-effectiveness should be higher for new technologies that involve foregoing some benefit in exchange for cost-savings, as in the case of OSNA or Metasin relative to histopathology in our present analysis. If this is the case then histopathology may well have an ICER relative to OSNA that is larger than the conventional £20,000 per QALY threshold and still be considered cost-effective. The problem is that no conventional thresholds for new technologies with lower costs and lower benefits exist.
Strengths and limitations of the assessment
Clinical effectiveness
The strengths of this systematic review are that it was conducted by an independent research team using the latest evidence in line with a prespecified protocol. The search strategy did not restrict by study design and also included forward chasing. The studies were independently screened by two reviewers, with data extraction and quality appraisal performed by one reviewer and checked by a second. Any disagreements were resolved by consensus. A large number of abstracts were identified that were not included in the review as quality appraisal could not be performed. However, a table of results has been compiled in Appendix 4 for interest.
The assessment of Metasin should be treated with caution. Only two studies were included, both of which were unpublished and therefore had not been peer reviewed.
For all studies there was a wide variance in the histopathology performed, for example one level, three level or five level. It is questionable whether the use of one-level histopathology is appropriate as a reference standard. The methods employed to analyse discordant cases and subsequently adjust for TAB were not consistent across studies. However, for the majority of studies, a conservative approach of excluding discordant cases was followed. Studies reported patient-level data, node data, ALN data or SLN data or a combination of these. Often, for the ALN analysis, the recruited population was relatively small but a large number of nodes was analysed. On some occasions it was unclear whether fresh or frozen samples had been used for histopathology.
Cost-effectiveness
The scope of our analysis was limited by some assumptions adopted to address data limitations. We have not accounted for the possibility of a second operation because of involved or close margins of the primary tumour. Indeed, a second operation rate of 9% for OSNA and 39% for histopathology, including 5% and 13%, respectively, for margin insufficiency without ALND resection has been reported. 149 No previous analysis has looked at this complex issue in terms of its costs, let alone its long-term health implications. Our current analysis does not look at how the cost and health benefit balance of intraoperative testing relative to histopathology and between intraoperative testing options may be affected by the current controversy about the effectiveness of ALND, particularly in relation to micrometastatic disease. 150 If part of the claimed advantages of OSNA, the detection of micrometastatic involvement, has no associated survival benefits, its long-term cost-effectiveness may be overestimated in our present analysis. Undertaking objective analysis of this issue is complicated, however, as the threshold definitions of micrometastasis for histopathology and OSNA differ and because of the current uncertainty associated with the emerging evidence on the benefits of ALND in small tumours found in the SLN.
Another limitation that originated from the lack of evidence to inform our analysis pertained to the potential loss of valuable information for patient management that full node use by intraoperative testing incurs relative to histopathology. 151 The extent to which this may affect our results is open to highly speculative judgement.
We have not looked at the cost-effectiveness implications of possible options that have been considered for increasing the sensitivity of the diagnostic strategies considered here. For example, OSNA reliance on the CK19 marker for detecting metastasis may lead to the test missing a small proportion of patients with metastatic breast cancer whose tumours do not express the marker. It has been suggested that preoperative testing of the primary tumour may identify the subgroup of patients whose tumours do not express CK19 and who require an alternative approach for metastatic diagnosis. The same approach could be adopted for Metasin, to exclude the possibility that those testing negative may be false-negative cases resulting from the lack of expression of CK19 and mammaglobin, the markers used by the test to identify positive cases.
Uncertainties
Clinical effectiveness
The primary concern in the studies in which a portion of the node was provided for molecular analysis and a portion provided for histopathology was TAB. Many studies attempted to address this; however, whether the discordant results were truly due to TAB or whether a proportion of cases had been missed is unclear.
The histopathology methods were sometimes ambiguously reported and the variety of methods compared is likely to be reflected in the heterogeneous data.
A significant proportion of the studies was funded by the manufacturers and there may therefore be a potential conflict of interest.
From the perspective of the patient, the case for intraoperative testing is made in terms of reducing anxiety about diagnosis and the need for a second operation. Although having a preference for one as opposed to two operations seems logical a priori, it is by no means obvious that patients will always give more weight to reduced anxiety from not having a second operation than to the uncertainty of occurrence of ALND during the first operation. Because there is no available evidence on patient views about this decision, we adopted the claim at face value in our base-case analysis, but from sensitivity analyses noted that our conclusion that intraoperative testing is not cost-effective would be strengthened by weakening the effect on anxiety. Moreover, for patients needing further surgery because of close/involved margins, the benefits of intraoperative SLNB would be reduced relative to the base case. In contrast to the negative effects of waiting for the results of SLNB with histopathology, which we accounted for, our modelled analysis could not account for the benefits in terms of patient management outcomes of additional information obtainable from postoperative histopathology.
Cost-effectiveness
Throughout our analyses we have assumed that histopathology is the gold standard. There are, however, indications that histopathologists may miss low-volume metastases. 152–154 Because of the low frequency of this omission relative to the occurrence of TAB, our analysis may have adopted a slightly conservative stance.
Our analysis was based on the estimated rate of axillary node positivity in patients with a positive SLNB result in a randomised trial (Z0011 study;37 see Chapter 1, Other approaches to treating the spread of breast cancer tumour cells beyond the sentinel lymph nodes), who were selected from among those with a better prognosis. Although the positive axillary node rate may therefore be underestimated, the associated bias in the cost-effectiveness results is attenuated by the fact that the axillary node positivity parameter affects cost and health outcomes of both intraoperative and histopathology strategies. In any case the bias makes our results conservative, as it reduces the importance of the sensitivity advantage of postoperative histopathology relative to OSNA.
The model on which our analysis is based did not account for the adverse effect of ALND in terms of altered shoulder function. 38 This has previously been found to occur post surgery in up to 39% of cases and have health-related quality of life effects as important as those of lymphoedema,155,156 which were accounted for in the analysis. We found no studies that evaluated the long-term health-related utility impact of altered shoulder function in our population. By omitting the impact on shoulder function our analysis is likely to overestimate the benefits of a correct positive diagnosis and underestimate the benefits of a correct negative test result for any given diagnostic option. However, this omission, as shown by sensitivity analysis of lymphoedema, does not matter for our results, depending as they do on differences in costs and benefits between options, given the small difference in ALND use between the intraoperative and the postoperative diagnostic arms.
The decision model assumed that adjuvant therapy is given for 5 years, as originally specified by SCHARR. 9 This may not reflect current practice because, if patients receive hormonal therapy, it is for at least 5 years after completion of chemotherapy; extended adjuvant letrozole is currently licensed for 5 years after 5 years of tamoxifen, and new evidence suggesting that 10 years of tamoxifen is better than 5 years157 implies that there will be a group of patients who will receive endocrine therapy for 10 years. Although the treatment course specified in the model may not apply to actual practice, the bias arising would affect both arms of the analysis and would have a small impact on the difference in costs and benefits and consequent cost-effectiveness results. Assuming that the extended course of adjuvant therapy is likely to result in an increase in health benefits by a factor that is higher than the factor for the increase in costs relative to the ScHARR model’s adjuvant treatment specification, the bias in the treatment pathway would overestimate the ICER of histopathology relative to OSNA and our results could therefore be seen as conservative, strengthening the conclusion that OSNA is not cost-effective.
We present our results under two costing approaches, which have been used by other assessment groups previously. They have significant implications for decision-making because, as discussed in relation to the short-term comparison between OSNA and histopathology, the two costing approaches have contrasting results, that is, the status quo, histopathology, is inferior to intraoperative testing in one case and a cost-effectiveness prospect in the other.
Our probabilistic analysis of uncertainty shows that the results are robust to parameter uncertainty. However, structural uncertainty may be very important but could not be addressed within the scope of our review. This means only that our ability to model the patient’s life course is limited, and so we cannot estimate with precision what the final outcome will be, and suggests that a shorter time frame may be meaningful for decision-making. In essence, this may be summarised by assuming that differences in outcomes last only a few years (e.g. 5 years), a period within which patient management and outcomes may be predicted with precision.
Chapter 6 Conclusions
Implications for service provision
The OSNA and Metasin tests may provide some advantages over histopathology. They are automated and standardised, with the capacity to be used intraoperatively. However, information gained by histology, such as the location of metastatic foci and morphological evaluation, is not provided by these intraoperative tests. OSNA displays a higher rate of false positives, possibly because of contamination from epithelial cells. In contrast, false negatives may occur when the breast cancer does not express CK19 protein.
(Academic-in-confidence information has been removed.)
It is not clear that the adoption of OSNA or Metasin will bring significant benefits to patients and cost-savings to the NHS. There remain important questions regarding whether, ultimately, the reduction in NHS costs with intraoperative testing is worth the reduction in diagnostic accuracy and potentially inferior long-term health outcomes for patients. Although the long-term uncertainty of breast cancer patient management and outcomes may determine long-term patient benefits, thus limiting the effects of the reduced diagnostic accuracy of the intraoperative tests on survival and quality of life, it is likely that a decision-maker would err on the side of caution and wait for more evidence before abandoning the current standard, postoperative histopathology. Some centres may not feel at ease performing ALND without the information that is currently available at MDT meetings through postoperative histopathology. Ultimately, in view of the current uncertainty in the evidence base on the benefits of performing ALND and the impact of this uncertainty on clinicians’ views, there is a possibility that intraoperative testing for SLNs will not be routine practice in the future.
Suggested research priorities
The uncertainty in decision-making because of a lack of data on, and variation in, available estimates of diagnostic accuracy and a lack of data on short-term implications of diagnostic approaches and long-term patient management may be reduced by undertaking further research. Priority should be placed on documenting the performance of intraoperative tests under different protocols. There is limited information about the performance of such tests under routine established practice and many of the existing studies are likely to apply to an early experience with these technologies. Peer-reviewed and formally published research on Metasin is also essential as the presumed low cost of this test makes it a potentially attractive option for the NHS.
Greater clarity on the true costs of the alternative tests, and the variation in resource utilisation at the level of the patient, would help to identify the significance and the distribution of costs and benefits of intraoperative diagnosis. Observational studies should be conducted to verify and quantify the hypothesised effects of introducing intraoperative SLNB, namely a reduction in the number of operations and costs to hospitals and the amelioration of anxiety and its impact on patients’ and their families’ quality of life by discarding postoperative diagnosis and second operations. Although much uncertainty is likely to remain over the magnitude of the long-term benefits of increased diagnostic accuracy, a natural target for great returns on investment in research may be found in documenting what the impact on patient-reported outcomes is of uncertainty associated with a delayed diagnosis and a second possible operation. Indeed, we may expect greater prominence of this aspect of the decision-making problem in populations with a greater prevalence of node-positive SLNs.
However, improving on the accuracy of estimates for OSNA in particular and overcoming concerns about TAB and the validity of currently available reference standards may be challenging. A test–treat randomised trial may be the only way to truly resolve whether the introduction of intraoperative testing in SLNB would be effective and thus cost-effective. The outcome would need to be the locoregional recurrence rate or even survival to capture the trade-off between the potential short-term gains associated with a single operation for achieving ALND and the longer-term disbenefits arising from the occurrence of false-negative and false-positive cases with intraoperative testing.
Also, a strong assumption in this report is that ALND is the usual best treatment if micro- and macrometastases, or their equivalents, are identified in a SLNB. Evidence on this is evolving and needs to be followed closely as it could impact on decision-making around intraoperative testing in SLNB in the future.
Acknowledgements
We would like to thank Katie Cooper, Research Fellow in Health Economics, ScHARR.
We would also like to acknowledge Sue Whiffin and Jenny Lowe for their administrative support throughout the project.
Contributions of authors
Nicola Huxley developed the short-term model and adapted the long-term model, executed the economic model and wrote the sections on the design and results of the economic model.
Tracey Jones-Hughes assessed abstracts and titles for inclusion, led the systematic review of clinical effectiveness and contributed to the writing and editing of the report.
Helen Coelho assessed abstracts and titles for inclusion and contributed to the writing and editing of the report.
Tristan Snowsill performed the statistical analysis and contributed to the writing of the report.
Chris Cooper designed and carried out the literature searches for the systematic reviews and identification of model parameters and contributed to the writing and editing of the report.
Yang Meng co-authored the original long-term economic model, advised on its adaptation to the analysis and contributed to writing the report.
Chris Hyde developed the protocol and contributed to the systematic review, design of the model and the writing and editing of the report. He is the director of the Technology Assessment Review Group at the Peninsula Technology Assessment Group (PenTAG).
Rubén Mújica-Mota contributed to the development of the protocol, led the systematic review of economic evaluations, contributed to the design of the analysis and writing and editing of the report and was the overall lead for the project and final report and guarantor of the report.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Dumitrescu RG, Cotarla I. Understanding breast cancer risk – where do we stand in 2005?. J Cell Mol Med 2005;9:208-21. http://dx.doi.org/10.1111/j.1582-4934.2005.tb00350.x.
- Office for National Statistics . Cancer Statistics: Registrations, England (Series MB1), No. 41, 2010 n.d. www.ons.gov.uk/ons/publications/re-reference-tables.html?edition=tcm%3A77-262496 (accessed 6 October 2014).
- Welsh Cancer Intelligence and Surveillance Unit . Cancer Incidence in Wales 2003–2007 2009. www.wales.nhs.uk/sites3/Documents/242/Cancer%20Incidence%20in%20Wales%202003–2007.pdf (accessed 1 July 2012).
- Cancer Research UK . Breast Cancer Mortality Statistics n.d. www.cancerresearchuk.org/cancer-info/cancerstats/types/breast/mortality/#sex (accessed 4 January 2013).
- Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin 2006;56:37-4. http://dx.doi.org/10.3322/canjclin.56.1.37.
- Cancer Research UK . TNM Breast Cancer Staging n.d. www.cancerhelp.org.uk/help/default (accessed 29 November 2010).
- American Cancer Society . How Is Breast Cancer Staged? n.d. www.cancer.org/docroot/CRI/content/CRI_2_4_3X_How_is_breast_cancer_staged_5.asp (accessed 29 November 2010).
- Cancer Research UK . Number Stages of Breast Cancer n.d. www.cancerhelp.org.uk/help/default.asp?page=3315 (accessed 29 November 2010).
- Cooper KL, Meng Y, Harnan S, Ward SE, Fitzgerald F, Papaioannou D, et al. Positron emission tomography (PET) and magnetic resonance imaging (MRI) for the assessment of axillary lymph node metastases in early breast cancer: systematic review and economic evaluation. Health Technol Assess 2011;15. http://dx.doi.org/10.3310/hta15040.
- Lyratzopoulos G, Abel GA, Barbiere JM, Brown CH, Rous BA, Greenberg DC. Variation in advanced stage at diagnosis of lung and female breast cancer in an English region 2006–2009. Br J Cancer 2012;106:1068-75. http://dx.doi.org/10.1038/bjc.2012.30.
- Carter C, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989;63:181-7. http://dx.doi.org/10.1002/1097-0142(19890101)63:1<181::AID-CNCR2820630129>3.0.CO;2-H.
- Breast Cancer (Early and Locally Advanced): Diagnosis and Treatment – Evidence Review. London: National Collaborating Centre for Cancer; 2009.
- Breast Cancer (Early and Locally Advanced): Diagnosis and Treatment. London: NICE; 2009.
- Cancer Survival Rates. Survival Rates in England, Patients Diagnosed 2001–2006 Followed Up to 2007. London: ONS; 2009.
- 0–10 Year Relative Survival for Cases of Breast Cancer by Stage Diagnosed in the West Midlands 1985–1989 Followed Up to the End of 1999, as at January 2002. Birmingham: WMCI; 2002.
- American Cancer Society . Breast Cancer Survival Rates by Stage n.d. www.cancer.org/cancer/breastcancer/detailedguide/breast-cancer-survival-by-stage (accessed 5 August 2012).
- Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 1992;22:207-19. http://dx.doi.org/10.1007/BF01840834.
- Adjuvant! Online . Welcome to Adjuvant! Online n.d. www.adjuvantonline.com/index.jsp (accessed 29 November 2012).
- National Institute for Health and Care Excellence . Gene Expression Profiling and Expanded Immunohistochemistry Tests to Guide the Use of Adjuvant Chemotherapy in Early Breast Cancer Management: MammaPrint, Oncotype DX, IHC4 and Mammostrat n.d. http://guidance.nice.org.uk/DT/4 (accessed 9 November 2012).
- Pazaiti A, Fentiman IS. Which patients need an axillary clearance after sentinel node biopsy. Int J Breast Cancer 2011;2011:1-9. http://dx.doi.org/10.4061/2011/195892.
- Cancer Reform Strategy. London: Department of Health; 2007.
- Management of Breast Cancer in Women: A National Clinical Guideline. Edinburgh: SIGN; 2005.
- Kvistad KA, Rydland J, Smethurst HB, Lundgren S, Fjosne HE, Haraldseth O. Axillary lymph node metastases in breast cancer: preoperative detection with dynamic contrast-enhanced MRI. Eur Radiol 2000;10:1464-71. http://dx.doi.org/10.1007/s003300000370.
- Mumtaz H, Hall-Craggs MA, Davidson T, Walmsley K, Thurell W, Kissin MW, et al. Staging of symptomatic primary breast cancer with MR imaging. AJR Am J Roentgenol 1997;169:417-27. http://dx.doi.org/10.2214/ajr.169.2.9242745.
- Smith IC, Welch AE, Chilcott F, Heys SD, Sharp P, Eremin O. Gamma emission imaging in the management of breast disorders. Eur J Surg Oncol 1998;24:320-9. http://dx.doi.org/10.1016/S0748-7983(98)80016-4.
- Wahl RL, Siegel BA, Coleman RE, Gatsonis CG. PET Study Group . Prospective multicenter study of axillary nodal staging by positron emission tomography in breast cancer: a report of the staging breast cancer with PET Study Group. J Clin Oncol 2004;22:227-85.
- Wallace IW, Champton HR. Axillary nodes in breast cancer. Lancet 1972;1:217-18. http://dx.doi.org/10.1016/S0140-6736(72)90619-8.
- Fisher B, Wolmark N, Bauer M, Redmond C, Gebhardt M. The accuracy of clinical nodal staging and of limited axillary dissection as a determinant of histologic nodal status in carcinoma of the breast. Surg Gynecol Obstet 1981;152:765-72.
- Blanchard DK, Donohue JH, Reynolds C, Grant CS. Relapse and morbidity in patients undergoing sentinel lymph node biopsy alone or with axillary dissection for breast cancer. Arch Surg 2003;138:482-7. http://dx.doi.org/10.1001/archsurg.138.5.482.
- Crane-Okada R, Wascher RA, Elashoff D, Giuliano AE. Long-term morbidity of sentinel node biopsy versus complete axillary dissection for unilateral breast cancer. Ann Surg Oncol 2008;15:1996-2005. http://dx.doi.org/10.1245/s10434-008-9909-y.
- McLaughlin SA, Wright MJ, Morris KT, Giron GL, Sampson MR, Brockway JP, et al. Prevalence of lymphedema in women with breast cancer 5 years after sentinel lymph node biopsy or axillary dissection: objective measurements. J Clin Oncol 2008;26:5213-19. http://dx.doi.org/10.1200/JCO.2008.16.3725.
- Purushotham AD, Upponi S, Klevesath MB, Bobrow L, Millar K, Myles JP, et al. Morbidity after sentinel lymph node biopsy in primary breast cancer: results from a randomised controlled trial. J Clin Oncol 2005;23:4312-21. http://dx.doi.org/10.1200/JCO.2005.03.228.
- Mansel RE, Fallowfield L, Kissin M, Goyal A, Newcombe RG, Dixon JM, et al. Randomized multicenter trial of sentinel node biopsy versus standard axillary treatment in operable breast cancer: the ALMANAC trial. J Natl Cancer Inst 2006;98:566-609. http://dx.doi.org/10.1093/jnci/djj158.
- D’Angelo-Donovan D, Dickson-Witmer D, Petrelli NJ. Sentinel lymph node biopsy in breast cancer: a history and current clinical recommendations. Surg Oncol 2012;21:196-200. http://dx.doi.org/10.1016/j.suronc.2011.12.005.
- Liu CQ, Guo Y, Shi JY, Sheng Y. Late morbidity associated with a tumour-negative sentinel lymph node biopsy in primary breast cancer patients: a systematic review. Eur J Cancer 2009;45:1560-8. http://dx.doi.org/10.1016/j.ejca.2009.02.012.
- Wilke LG, McCall LM, Posther KE, Whitworth PW, Reintgen DS, Leitch AM, et al. Surgical complications associated with sentinel lymph node biopsy: results from a prospective international cooperative group trial. Ann Surg Oncol 2006;13:491-500. http://dx.doi.org/10.1245/ASO.2006.05.013.
- Giuliano AE Hunt KK, Ballman KV, Beitsch PD, Whitworth PW, Blumencranz PW, . Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis. A randomized clinical trial. JAMA 2011;305:569-75. http://dx.doi.org/10.1001/jama.2011.90.
- Straver ME, Meijnen P, van Tienhoven G, van de Velde CJ, Mansel RE, Bogaerts J, et al. Role of axillary axillary clearance after a tumor-positive sentinel node in the administration of adjuvant therapy in early breast cancer. J Clin Oncol 2010;28:731-7. http://dx.doi.org/10.1200/JCO.2008.21.7554.
- Haffty BG, Hunt KK, Harris JR, Buccholz TA. Positive sentinel nodes without axillary dissection: implications for the radiation oncologist. J Clin Oncol 2011;29:4479-81. http://dx.doi.org/10.1200/JCO.2011.36.1667.
- Sundaresan V. Metasin-BLNA, the NHS solution for the intra-operative assessment of sentinel lymph nodes from breast cancer patients: the multi-centre validation of 1265 cases. J Pathol 2013;229.
- Intraoperative Tests (RD-100i OSNA System and Metasin Test) for Detecting Sentinel Lymph Node Metastases in Breast Cancer. Final Scope. London: NICE; 2012.
- Undertaking Systematic Reviews of Research on Effectiveness. York: NHS Centre for Reviews and Dissemination; 2001.
- Whiting PF, Westwood ME, Rutjes AW, Reitsma JB, Bossuyt PN, Kleijnen J. Evaluation of QUADAS, a tool for the quality assessment of diagnostic accuracy studies. BMC Med Res Methodol 2006;6. http://dx.doi.org/10.1186/1471-2288-6-9.
- Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Clin Epidemiol 2005;58:982-90. http://dx.doi.org/10.1016/j.jclinepi.2005.02.022.
- Harbord RM, Whiting PF. Metandi: meta-analysis of diagnostic accuracy using hierarchical logistic regression. Stata J 2009;9:211-29.
- Pennant M, Takwoingi Y, Pennant L, Davenport C, Fry-Smith A, Eisinga A, et al. A systematic review of positron emission tomography (PET) and positron emission tomography/computed tomography (PET/CT) for the diagnosis of breast cancer recurrence. Health Technol Assess 2010;14. http://dx.doi.org/10.3310/hta14500.
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med 2001;20:2865-84. http://dx.doi.org/10.1002/sim.942.
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JAC. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics 2007;8:239-51. http://dx.doi.org/10.1093/biostatistics/kxl004.
- Arends LR, Hamza TH, van Houwelingen JC, Heijenbrok-Kal MH, Hunink MGM, Stijnen T. Bivariate random effects meta-analysis of ROC curves. Med Decis Making 2008;28:621-38. http://dx.doi.org/10.1177/0272989X08319957.
- Bernet L, Cano R, Martinez M, Duenas B, Matias-Guiu X, Morell L, et al. Diagnosis of the sentinel lymph node in breast cancer: a reproducible molecular method: a multicentric Spanish study. Histopathology 2011;58:863-9. http://dx.doi.org/10.1111/j.1365-2559.2011.03836.x.
- Bernet Vegue L, Rojo F, Hardisson D, Iturriagagoitia AC, Panades MJ, Velasco A, et al. Comparison of molecular analysis and histopathology for axillary lymph node staging in primary breast cancer: results of the B-CLOSER-I study. Diagn Mol Pathol 2012;21:69-76. http://dx.doi.org/10.1097/PDM.0b013e318241117b.
- Castellano I, Macri L, Deambrogio C, Balmativola D, Bussone R, Ala A, et al. Reliability of whole sentinel lymph node analysis by one-step nucleic acid amplification for intraoperative diagnosis of breast cancer metastases. Ann Surg 2012;255:334-42. http://dx.doi.org/10.1097/SLA.0b013e31823000ed.
- Choi YL, Ahn SK, Bae YK, Park IA, Min JW, Lee KW, et al. One-step nucleic acid amplification (OSNA): intraoperative rapid molecular diagnostic method for the detection of sentinel lymph node metastases in breast cancer patients in Korean cohort. J Breast Cancer 2010;13:366-74. http://dx.doi.org/10.4048/jbc.2010.13.4.366.
- Feldman S, Krishnamurthy S, Gillanders W, Gittleman M, Beitsch PD, Young PR, et al. A novel automated assay for the rapid identification of metastatic breast carcinoma in sentinel lymph nodes. Cancer 2011;117:2599-607. http://dx.doi.org/10.1002/cncr.25822.
- Godey F, Leveque J, Tas P, Gandon G, Poree P, Mesbah H, et al. Sentinel lymph node analysis in breast cancer: contribution of one-step nucleic acid amplification (OSNA). Breast Cancer Res Treat 2012;131:509-16. http://dx.doi.org/10.1007/s10549-011-1808-4.
- Guillen-Paredes MP, Carrasco-Gonzalez L, Chaves-Benito A, Campillo-Soto A, Carrillo A, Aguayo-Albasini JL. One-step nucleic acid amplification (OSNA) assay for sentinel lymph node metastases as an alternative to conventional postoperative histology in breast cancer: a cost–benefit analysis. Cir Esp 2011;89:456-62. http://dx.doi.org/10.1016/j.ciresp.2011.04.013.
- Khaddage A, Berremila SA, Forest F, Clemenson A, Bouteille C, Seffert P, et al. Implementation of molecular intra-operative assessment of sentinel lymph node in breast cancer. Anticancer Res 2011;31:585-90.
- Le Frère-Belda MA, Bats AS, Gillaizeau F, Poulet B, Clough KB, Nos C, et al. Diagnostic performance of one-step nucleic acid amplification for intraoperative sentinel node metastasis detection in breast cancer patients. Int J Cancer 2012;130:2377-86. http://dx.doi.org/10.1002/ijc.26291.
- Osako T, Iwase T, Kimura K, Yamashita K, Horii R, Akiyama F. Accurate staging of axillary lymph nodes from breast cancer patients using a novel molecular method. Br J Cancer 2011;105:1197-202. http://dx.doi.org/10.1038/bjc.2011.350.
- Schem C, Maass N, Bauerschlag DO, Carstensen MH, Loning T, Roder C, et al. One-step nucleic acid amplification – a molecular method for the detection of lymph node metastases in breast cancer patients; results of the German study group. Virchows Arch 2009;454:203-10. http://dx.doi.org/10.1007/s00428-008-0703-9.
- Snook KL, Layer GT, Jackson PA, de Vries CS, Shousha S, Sinnett HD, et al. Multicentre evaluation of intraoperative molecular analysis of sentinel lymph nodes in breast carcinoma. Br J Surg 2011;98:527-35. http://dx.doi.org/10.1002/bjs.7347.
- Tamaki Y, Akiyama F, Iwase T, Kaneko T, Tsuda H, Sato K, et al. Molecular detection of lymph node metastases in breast cancer patients: results of a multicenter trial using the one-step nucleic acid amplification assay. Clin Cancer Res 2009;15:2879-84. http://dx.doi.org/10.1158/1078-0432.CCR-08-1881.
- Tamaki Y, Sato N, Homma K, Takabatake D, Nishimura R, Tsujimoto M, et al. Routine clinical use of the one-step nucleic acid amplification assay for detection of sentinel lymph node metastases in breast cancer patients. Results of a multicenter study in Japan. Cancer 2012;118:3477-83. http://dx.doi.org/10.1002/cncr.26683.
- Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, Akiyama F, et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res 2007;13:4807-16. http://dx.doi.org/10.1158/1078-0432.CCR-06-2512.
- Visser M, Jiwa M, Horstman A, Brink A, Pol RP, van Diest P, et al. Intra-operative rapid diagnostic method based on CK19 mRNA expression for the detection of lymph node metastases in breast cancer. Int J Cancer 2008;122:2562-7. http://dx.doi.org/10.1002/ijc.23451.
- McDowell A, Yiangou C, Wise M, Agrawal A, Gabriel FG, Sundaresan V, et al. Intra-operative sentinel node assessment by QRT-PCR: experience from a single centre n.d.
- Ng VV, Charlton FI, Cunnick GH. The use of intra-operative one step nucleic acid amplification (OSNA) to analyse sentinel lymph nodes in patients with breast cancer – does it impact on operating times?. Br J Surg 2011;98.
- Garcia-Estepa R, Martinez-Ferez IM. Efficacy of a molecular method for detection of lymph node metastases in early breast cancer. Eur J Cancer Suppl 2010;8. http://dx.doi.org/10.1016/S1359-6349(10)72271-0.
- Di Filippo F, Mottolese M, Botti C, Marandino F, Psaila A, Perri P, et al. A prospective clinical study for molecular intra-operative detection of lymph node metastasis in breast cancer patients by one step nucleic acid amplification (OSNA) in comparison to intensive histological investigation. J Clin Oncol 2009;27.
- Buglioni S, Del Chierico F, Conti S, Visca P, Perri P, Di Filippo F, et al. A prospective clinical study for molecular intra-operative detection of lymph node metastasis in breast cancer patients by ‘one step nucleic acid amplification (OSNA)’ in comparison with intensive histological investigation. Breast 2009;18. http://dx.doi.org/10.1016/S0960-9776(09)70134-1.
- Buglioni S, Casini B, Marino M, Perri P, Terrenato I, Di Filippo F, et al. One Step nucleic acid amplification (OSNA) assay for molecular detection of sentinel lymph node metastases in early breast cancer classified according to molecular subtypes: an observational prospective study. Eur J Cancer Suppl 2010;8. http://dx.doi.org/10.1016/S1359-6349(10)70365-7.
- Kaneko T, Akiyama F, Tsujimoto M, Noguchi S, Inaji H, Nakamura S, et al. Rapid detection of lymph node metastasis in breast cancer patients by ‘one-step nucleic acid amplification (OSNA)’: results from a multi-institutional clinical study. Breast Cancer Res Treat 2007;106.
- Masuda N, Akiyama F, Tsuda H, Noguchi S, Tsujimoto M, Inaji H, et al. A new one-step nucleic acid amplification (OSNA) with cytokeratin (CK)19 mRNA for intra-operative detection of lymph node (LN) metastasis in breast cancer patients: results from the Multi-institutional Clinical Study. Breast 2007;16. http://dx.doi.org/10.1016/S0960-9776(07)70094-2.
- Tsuda H, Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, et al. A multicenter clinical study for intraoperative detection of lymph node metastasis in breast cancer patients by the one-step nucleic acid amplification method. Breast Cancer Res Treat 2006;100.
- Sato N, Honma K, Noguchi S, Tamaki Y, Tsuda H, Kinoshita T, et al. Multi-institutional evaluation of sentinel lymph node (SLN) examination by one-step nucleic acid amplification (OSNA) assay in breast cancer: performance of metastases detection and prediction of additional non-sentinel lymph node (non-SLN) involvement. J Clin Oncol 2011;29.
- Takabatake D, Noguchi S, Akiyama F, Sato N, Tsujimoto M, Tsuda H, et al. Evaluation of clinical utility of sentinel lymph node (SLN) examination by one-step nucleic acid amplification (OSNA) assay in breast cancer. Eur J Cancer 2011;47. http://dx.doi.org/10.1016/S0959-8049(11)71587-8.
- Peston D, Shousha S, Sinnett D. Intra-operative assessment of sentinel node biopsy for breast carcinoma using OSNA technique. The Charing Cross experience. Virchows Arch 2009;455:S75-6.
- Snook KL, Layer GT, Jackson P, Al-Hashimi S, Woodland J, de Vries CS, et al. OSNA((R)) for rapid molecular analysis of breast cancer lymph nodes: the Guildford experience. Eur J Cancer Suppl 2007;5:13-4. http://dx.doi.org/10.1016/S1359-6349(07)71732-9.
- Snook K, Layer G, Jackson P, de Vries C, Shousha S, Sinnett H, et al. Sentinel node OSNA (R) analysis in the UK hospital setting: how long does it take?. Ann Surg Oncol 2008;15:40-1.
- Snook KL, Kissin MW, Layer GT, Jackson P, de Vries C, Shousha S, et al. ‘One step nucleic acid amplification’ for rapid molecular analysis of breast cancer lymph nodes: the way towards one stop sentinel node surgery?. Breast Cancer Res Treat 2007;106.
- Kissin MW, Snook KL, Layer GT, Jackson P, de Vries CS, Shousha S, et al. Intraoperative molecular sentinel lymph node analysis with OSNA: multicentre prospective UK evaluation. Cancer Res 2009;69. http://dx.doi.org/10.1158/0008-5472.SABCS-1003.
- Nizar S, Shrotria S, Shambayati B, Albertini F. Intraoperative sentinel node analysis in breast cancer – a pilot study of OSNA and imprint cytology. Eur J Surg Oncol 2010;36. http://dx.doi.org/10.1016/j.ejso.2010.08.028.
- Chaudhry A, Massey E, Cook J, Jenkins M, Winters Z, Rayter Z. One step nucleic acid amplification (OSNA) of sentinel lymph nodes in breast cancer. Eur J Surg Oncol 2011;37. http://dx.doi.org/10.1016/j.ejso.2011.08.046.
- Massey E, Chaudhry A, Cook J, Jenkins M, Winters Z, Rayter Z. The impact on operating times of using OSNA in sentinel node surgery. Eur J Surg Oncol 2011;37. http://dx.doi.org/10.1016/j.ejso.2011.08.003.
- Beitsch P, Taylor W, Jordan J, Garcia M, Kocian L. Rapid detection of sentinel lymph node metastasis in breast cancer by OSNA assay. Breast Cancer Res Treat 2007;106:S131-2.
- Tomlins A, Starczynski J, Paterson I. Validation study of one-step nucleic acid amplification (OSNA) analysis of axillary sentinel lymph nodes in breast cancer. Eur J Surg Oncol 2011;37.
- Iqbal M, Mecci AJ, Whisker L, Parkes M, Smith A, Sandhu F, et al. Implementation of one step nucleic acid amplification (OSNA) for intra-operative assessment of sentinel lymph nodes in a DGH. Eur J Surg Oncol 2012;38:439-40. http://dx.doi.org/10.1016/j.ejso.2012.02.110.
- Ng VV, Charlton F, Cunnick G. Analysing sentinel lymph nodes with intra-operative one step nucleic acid amplification (OSNA) – does it impact on operating times?. Eur J Surg Oncol 2011;37. http://dx.doi.org/10.1016/j.ejso.2011.03.033.
- Ng VV, Charlton F, Chia Y, Cunnick G. A comparison of nodal positivity between ‘one step nucleic acid amplification’ (OSNA) and routine pathology of sentinel lymph nodes. Eur J Surg Oncol 2011;37. http://dx.doi.org/10.1016/j.ejso.2011.03.010.
- Ng VV, Charlton FI, Chia Y, Cunnick GH. Comparing nodal positivity between ‘one step nucleic acid amplification’ (OSNA) and routine pathology of sentinel lymph nodes – is OSNA more accurate in detecting metastases in breast cancer?. Br J Surg 2011;98.
- Remoundos DD, Wilson HA, Ng VV, Ahmed F, Chia Y, Cunnick GH. Detection rates of micrometastasis in sentinel nodes: a comparison of intraoperative one-step nucleic-acid amplification (OSNA) versus routine histopathology. Eur J Cancer 2012;48.
- Bilous M, E-Elder E, French J, Pathmanathan N, Salisbury E, Mahajan H, et al. Optimising intraoperative assessment of sentinel lymph nodes in breast cancer – one step nucleic acid amplification assay compared with imprint cytology. Eur J Cancer 2012;48. http://dx.doi.org/10.1016/S0959-8049(12)70476-8.
- Godey F, Bendavid CA, Gandon G, Tas P, Rouquette S, Blanchot J, et al. Breast sentinel lymph node analysis intraoperative with OSNA (one step nucleic acid amplification) to avoid second surgery for axillary lymph node dissection. Breast 2011;20:S56-7. http://dx.doi.org/10.1016/S0960-9776(11)70184-9.
- Peoch M, Boutheille C, Seffert P, Kaddhage A. Molecular intra-operative assessment of sentinel lymph node in breast cancer: a report about routine use. Virchows Arch 2011;459.
- Khaddage A, Bouteille C, Seffert R, Peoc’h M. Intra-operative sentinel node metastasis detection in breast cancer by one-step nucleic acid amplification (OSNA): from validation to routine use. Breast 2009;18. http://dx.doi.org/10.1016/S0960-9776(09)70125-0.
- Godey F, Khaddage A, Bendavid-Athias C, Bouteille C, Tas P, Foucher F, et al. Intra-operative sentinel node metastasis detection in breast cancer by one-step nucleic acid amplification (OSNA): the Saint-Etienne Hospital and Rennes Cancer Institute experience. Ann Oncol 2009;20.
- Godey F, Bendavid C, Tas P, Gandon G, Foucher F, Rouquette S, et al. Intra operative sentinel node metastasis detection in breast cancer by one step nucleic acid amplification: Rennes Cancer Institute experience. Eur J Cancer Suppl 2010;8:151-2. http://dx.doi.org/10.1016/S1359-6349(10)70348-7.
- Levine E, Krishnamurthy S, Gittleman M, Young P, Streck C, Boolbol S, et al. Prospective comparison of the OSNA breast cancer system to imprint cytology for the intraoperative analysis of sentinel lymph nodes from cancer of the breast. Ann Surg Oncol 2010;17.
- US OSNA Breast Cancer Sentinel Lymph Node Study Group . Prospective multicenter study of a novel fully automated molecular test for identification of metastatic carcinoma in axillary sentinel lymph nodes in breast cancer – the US OSNA Breast Cancer Sentinel Lymph Node Study Group. Mod Pathol 2010;23.
- Schem C, Maass N, Do B, Donat W, Carstensen M, Tiemann K. One-step nucleic acid amplification for intra-operative detection of lymph node metastases in breast cancer patients. Breast Cancer Res Treat 2007;106.
- Schem C, Mathiak M, Hilpert F, Mundhenke C, Bauerschlag D, Jonat W. One step nucleic acid amplification (OSNA) as an intra-operative diagnostic tool for the assessment of the sentinel lymph node status in breast cancer patients. Eur J Cancer Suppl 2010;8. http://dx.doi.org/10.1016/S1359-6349(10)70346-3.
- Jimbo K, Kinoshita T, Hojo T, Asaga S, Suzuki J. A new development in sentinel lymph node biopsy in breast cancer using a combination of molecular and histological methods. Eur J Cancer 2012;48. http://dx.doi.org/10.1016/S0959-8049(12)70604-4.
- Suzuki M, Nanjo H, Sugiyama T. OSNA is suitable for intraoperative analysis of sentinel lymph node metastasis in breast cancer. Eur J Cancer 2011;47:S373-4. http://dx.doi.org/10.1016/S0959-8049(11)71588-X.
- Rai Y, Ohi Y, Kaikura M, Tamada S, Baba S, Yotsumoto D, et al. Intraoperative one-step nucleic acid amplification assay (OSNA) to detect sentinel lymph node (SLN) metastasis in breast cancer – an evaluation of 703 cases in a single institution. Eur J Cancer 2012;48. http://dx.doi.org/10.1016/S0959-8049(12)70584-1.
- Wahab T, Davidson T, Banerjee S, Ullah Md Z, Byron N, Davison S, et al. Results of a ‘belt and braces’ approach to using OSNA: what to do when there is discordance with histology. Eur J Surg Oncol 2012;38. http://dx.doi.org/10.1016/j.ejso.2012.02.139.
- Siso C, Morales S, Iglesias E, Gonzalez S, Canosa C, Pon M. Could axillary dissection be avoided after neoadjuvant chemotherapy in patients with prior positive axillary sentinel lymph node by a RT-PCR method?. Eur J Cancer 2012;48. http://dx.doi.org/10.1016/S0959-8049(12)70668-8.
- Krishnamurthy S, Tang S, Wang X, Sahin AA, Pantoja LM, Lucci A, et al. Utility of a one-step nucleic acid amplification (OSNA) assay for comprehensive examination of axillary lymph nodes in breast cancer. Cancer Res 2009;69. http://dx.doi.org/10.1158/0008-5472.SABCS-09-1012.
- Mizoo T, Shien T, Nogami T, Iwamoto T, Motoki T, Taira N, et al. Efficacy of one-step nucleic acid amplification (OSNA) for intraoperative diagnosis of breast cancer metastases. Eur J Cancer 2012;48. http://dx.doi.org/10.1016/S0959-8049(12)70174-0.
- Al-Ramadhani S, Balaraman P, McKenzie J, Jader S, Morgan M, Sundaresan V. Metasin – a rapid and robust PCR assay for intra-operative sentinel node diagnosis. Eur J Surg Oncol 2010;36:1105-6. http://dx.doi.org/10.1016/j.ejso.2010.08.125.
- Johns R, Dabbas N, McDowell A, Gabriel G, Agrawal A, Cree I, et al. Breast cancer sentinel node intraoperative molecular diagnosis: GeneSearch BLN assay vs. Metasin assay. Eur J Surg Oncol 2011;37:S6-7. http://dx.doi.org/10.1016/j.ejso.2011.03.024.
- Johns R, Dabbas N, McDowell A, Cree I, Wise M. Breast cancer sentinel node intra-operative molecular diagnosis: implementation of the Metasin assay. Br J Surg 2011;98.
- Simoes T, Yiangou C, McDowell A. Comparison of Metasin versus GeneSearch (Veridex) in intraoperative analysis (IOA) of sentinel lymph nodes (SLN) in breast cancer (BC). Eur J Surg Oncol 2012;38. http://dx.doi.org/10.1016/j.ejso.2012.02.020.
- Sundaresan V, Brown S, George D, Sai Giridhar P, Al-Ramadhani S, Bustin SA, et al. The Metasin-effusion assay: a quantitative multiplexed real time qPCR assay for the detection of metastatic carcinoma in biopsy material and cells from cytological preparations; potential for results in under 30 mins (a POCT?). J Pathol 2012;226.
- Sundaresan V, Al-Ramadhani S, Balaraman P, Holt S, Mansel R, Cree I, et al. Metasin-BLNA: the NHS solution cost effective-rapid intraoperative molecular assessment of sentinel lymph nodes from breast cancer patients. Eur J Surg Oncol 2011;37. http://dx.doi.org/10.1016/j.ejso.2011.03.039.
- Holt FS, Al-Ramadhani S, Bustin S, Al-sam S, Sundaresan V, Sai-Gridar P, et al. The Metasin-BLNA, the NHS solution, qPCR real time assay for the detection of metastatic cancer in sentinel lymph nodes from patients with breast cancer: running live at Harlow and Portsmouth: the 1000 case milestone reached. J Pathol 2012;226.
- Chu PG, Weiss LM. Keratin expression in human tissues and neoplasms. Clin Cancer Res 2002;13:4807-16.
- Lyman GH, Giuliano AE, Somerfield MR, Benson AB III, Bodurka HJ, Burstein H, et al. American Society of Clinical Oncology guideline recommendations for sentinel lymph node biopsy in early-stage breast cancer. J Clin Oncol 2005;23:7703-20. http://dx.doi.org/10.1200/JCO.2005.08.001.
- Riley RD, Abrams KR, Sutton AJ, Lambert PC, Thompson JR. Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Med Res Methodol 2007;7. http://dx.doi.org/10.1186/1471-2288-7-3.
- Al-Ramadhani S, Balaraman P, George D, Morgan M, Jader S, McDowell A, et al. Metasin: a novel rapid RT-PCR assay for the analysis of sentinel lymph nodes from patients with breast cancer, in the intra-operative setting: analysis of the first 1000 cases. J Pathol 2011;224.
- Nishimura K, Sugiyama D, Moronobu A, Kawano S, Kumagai S. Cost utility analysis on one-step nucleic acid amplification method for the detection of axillary metastasis in breast cancer. Cancer Res 2009;69. http://dx.doi.org/10.1158/0008-5472.SABCS-1014.
- Classe JM, Baffert S, Sigal-Zafrani B, Fall M, Rousseau C, Alran S, et al. Cost comparison of axillary sentinel lymph node detection and axillary lymphadenectomy in early breast cancer. A national study based on a prospective multi-institutional series of 985 patients ‘on behalf of the Group of Surgeons from the French Unicancer Federation’. Ann Oncol 2012;23:1170-7. http://dx.doi.org/10.1093/annonc/mdr355.
- Meng Y, Ward S, Cooper K, Harnan S, Wyld L. Cost-effectiveness of MRI and PET imaging for the evaluation of axillary lymph node metastases in early stage breast cancer. Eur J Surg Oncol 2011;37:40-6. http://dx.doi.org/10.1016/j.ejso.2010.10.001.
- Cutress RI, McDowell A, Gabriel FG, Gill J, Jeffrey MJ, Agrawal A, et al. Observational and cost analysis of the implementation of breast cancer sentinel node intraoperative molecular diagnosis. J Clin Pathol 2010;63:522-9. http://dx.doi.org/10.1136/jcp.2009.072942.
- Burke M, Patton T. The Cost Impact of Implementing Intra-Operative Testing for the Diagnosis of Patients with Metastatic Breast Cancer in England. York: York Health Economics Consortium NHS Technology Adoption Centre, University of York; 2010.
- Drummond MF, Jefferson TO. Guidelines for authors and peer reviewers of economic submissions to the BMJ. BMJ 1996;313:275-83. http://dx.doi.org/10.1136/bmj.313.7052.275.
- Organisation for Economic Co-operation and Development . PPPs and Exchange Rates n.d. http://stats.oecd.org/Index.aspx?datasetcode=SNA_TABLE4 (accessed 1 November 2012).
- Karnon J. Alternative decision modelling techniques for the evaluation of health care technologies: Markov processes versus discrete event simulation. Health Econ 2003;12:837-48. http://dx.doi.org/10.1002/hec.770.
- Cody HS. Clinicopathologic factors associated with false-negative sentinel lymph-node biopsy in breast cancer. Ann Surg 2006;244. http://dx.doi.org/10.1097/01.sla.0000230027.27680.97.
- Mak SS, Mo KF, Suen JJS, Chan SL, Ma WL, Yeo W. Lymphedema and quality of life in Chinese women after treatment for breast cancer. Eur J Oncol Nurs 2009;13:110-15. http://dx.doi.org/10.1016/j.ejon.2009.01.005.
- Department of Health . NHS Reference Costs 2010–2011 n.d. www.dh.gov.uk/en/Publicationsandstatistics/Publications/PublicationsPolicyAndGuidance/DH_131140 (accessed 5 November 2011).
- Pandharipande PV, Harisinghani MG, Ozanne EM, Specht MC, Hur C, Lee JM, et al. Staging MR lymphangiography of the axilla for early breast cancer: cost-effectiveness analysis. AJR Am J Roentgenol 2008;191:1308-19. http://dx.doi.org/10.2214/AJR.07.3861.
- Shemilt I, Thomas J, Morciano M. A web-based tool for adjusting costs to a specific target currency and price year. Evid Policy 2010;6:51-9. http://dx.doi.org/10.1332/174426410X482999.
- Jeruss JS, Hunt KK, Xing Y, Krishnamurthy S, Meric-Bernstam F, Cantor SB, et al. Is intraoperative touch imprint cytology of sentinel lymph nodes in patients with breast cancer cost effective?. Cancer 2006;107:2328-36. http://dx.doi.org/10.1002/cncr.22275.
- Burke M, Setters J. Breast Lymph Node Assay. York: York Health Economics Consortium NHS Technology Adoption Centre, University of York; 2010.
- Dolan P. Modeling valuations for EuroQol health states. Med Care 1997;35:1095-108. http://dx.doi.org/10.1097/00005650-199711000-00002.
- Tengs TO, Wallace A. One thousand health-related quality-of-life estimates. Med Care 2000;38:583-637. http://dx.doi.org/10.1097/00005650-200006000-00004.
- Orr RK, Col NF, Kuntz KM. A cost-effectiveness analysis of axillary node dissection in postmenopausal women with estrogen receptor-positive breast cancer and clinically negative axillary nodes. Surgery 1999;126:568-76. http://dx.doi.org/10.1016/S0039-6060(99)70100-5.
- Kamby C, Sengelov L. Pattern of dissemination and survival following isolated locoregional recurrence of breast cancer – a prospective study with more than 10 years of follow up. Breast Cancer Res Treat 1997;45:181-92. http://dx.doi.org/10.1023/A:1005845100512.
- Ward S, Simpson E, Davis S, Hind D, Rees A, Wilkinson A. Taxanes for the adjuvant treatment of early breast cancer: systematic review and economic evaluation. Health Technol Assess 2007;11. http://dx.doi.org/10.3310/hta11400.
- Lancet. 1992.
- National Life Tables, 2007–2009. London: ONS; 2010.
- Department of Health . NHS Reference Costs 2002–2003 n.d. http://collections.europarchive.org/tna/20100509080731/http://dh.gov.uk/en/Publicationsandstatistics/Publications/PublicationsPolicyAndGuidance/DH_4070195 (accessed 12 November 2012).
- Karnon J, Delea T, Barghout V. Cost utility analysis of early adjuvant letrozole or anastrozole versus tamoxifen in postmenopausal women with early invasive breast cancer: the UK perspective. Eur J Health Econ 2008;9:171-83. http://dx.doi.org/10.1007/s10198-007-0058-1.
- Curtis L. Unit Costs of Health & Social Care 2011. Canterbury: PSSRU, University of Kent; 2011.
- Ara R, Brazier JE. Populating an economic model with health state utility values: moving toward better practice. Value Health 2010;13:509-18. http://dx.doi.org/10.1111/j.1524-4733.2010.00700.x.
- Asakawa K, Senthilselvan A, Feeny D, Johnson J, Rolfson D. Trajectories of health-related quality of life differ by age among adults: results from an eight-year longitudinal study. J Health Econ 2012;31:207-18. http://dx.doi.org/10.1016/j.jhealeco.2011.10.002.
- Guide to the Methods of Technology Appraisal. London: NICE; 2008.
- Higgins JPT, Altman DG, Sterne JAC, Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0 [updated March 2011]. The Cochrane Collaboration; 2011.
- Klingler S, Marchal F, Rauch P, Kenouchi O, Chrétien A, Genin P, et al. Intraoperative detection of lymph node metastasis using one-step nucleic acid amplification (OSNA) in breast cancer patients: effect on second surgery rate and delay for adjuvant therapy. J Clin Oncol 2012;30.
- Giuliano AE, McCall L, Beitsch P, Whitworth PW, Blumencranz P, Leitch AM, et al. Locoregional recurrence after sentinel lymph node dissection with or without axillary dissection in patients with sentinel lymph node metastases. Ann Surg 2010;252:426-33.
- Cserni G. Intraoperative analysis of sentinel lymph nodes in breast cancer by one-step nucleic acid amplification. J Clin Pathol 2012;65:193-9. http://dx.doi.org/10.1136/jclinpath-2011-200301.
- Mesker WE, Torrenga H, Sloos WCR, Vrolijk H, Tollenaar RAEM, de Bruin PC, et al. Supervised automated microscopy increases sensitivity and efficiency of detection of sentinel node micrometastases in patients with breast cancer. J Clin Pathol 2004;57:960-4. http://dx.doi.org/10.1136/jcp.2004.017368.
- Weaver DL, Krag DN, Manna EA, Ashikaga T, Waters BL, Harlow SP, et al. Detection of occult sentinel lymph node micrometastases by immunohistochemistry in breast cancer – an NSABP protocol B-32 quality assurance study. Cancer 2006;107:661-7. http://dx.doi.org/10.1002/cncr.22074.
- Cserni G, Bianchi S, Vezzosi V, Peterse H, Sapino A, Arisio R, et al. The value of cytokeratin immunohistochemistry in the evaluation of axillary sentinel lymph nodes in patients with lobular breast carcinoma. J Clin Pathol 2006;59:518-22. http://dx.doi.org/10.1136/jcp.2005.029991.
- Nagel P, Bruggink E, Wobbes T, Strobbe L. Arm morbidity after complete axillary lymph node dissection for breast cancer. Acta Chir Belg 2003;103:212-16.
- Land SR, Kopec JA, Julian TB, Brown AM, Anderson SJ, Krag DN, et al. Patient-reported outcomes in sentinel node-negative adjuvant breast cancer patients receiving sentinel-node biopsy or axillary dissection: National Surgical Adjuvant Breast and Bowel Project Phase III Protocol B-32. J Clin Oncol 2010;28:3929-36. http://dx.doi.org/10.1200/JCO.2010.28.2491.
- Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V, et al. Long term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 2013;381:805-16. http://dx.doi.org/10.1016/S0140-6736(12)61963-1.
- Le Frère-Belda MA, Charon Barra C, Crouet H, Houvenaeghel G, Khaddage A, Leroux A, et al. Intra-operative sentinel lymph node metastasis detection in breast cancer by ‘one-step nucleic acid amplification (OSNA)’ – results of the French multicentre prospective study. Eur J Cancer Suppl 2008;6. http://dx.doi.org/10.1016/S1359-6349(08)70331-8.
- Al-Ramadhani S, Sai-Giridhar P, George D, Gopinath P, Arkoumani E, Jader S, et al. Metasin – an intra-operative RT-qPCR assay to detect metastatic breast cancer in sentinel lymph nodes. Int J Mol Sci 2013;14:12931-52. http://dx.doi.org/10.3390/ijms140712931.
Appendix 1 Literature search strategy
The search strategy focuses on the interventions under consideration for this review in the context of the specific area in which the tests are applied: the LNs. Also, independently of the interventions, the search draws in literature on the biological markers CK19 and mammaglobin (in the context of the test area), which aims to help serve any modelling that may relate to this project. The search was not limited by language or methodology or to humans exclusively. The search was run from database inception.
Database search results
Table 64 details the databases searched. The Web of Science search included the Conference Proceedings Citation Index. Records were downloaded and managed in EndNote X5 (Thompson Reuters, Philadelphia, PA, USA).
| Database | Results |
|---|---|
| MEDLINE via Ovid | 197 |
| Medline In-Process & Other Non-Indexed Citations via Ovid | 15 |
| EMBASE via Ovid | 624 |
| Web of Science via ISI | 93 |
| Cochrane Database of Systematic Reviews, Cochrane Central Register of Controlled Trials, Cochrane Methodology Register, Database of Abstracts of Reviews of Effects, Health Technology Assessment Database and NHS Economic Evaluation Database via The Cochrane Library | 18 |
| NHS Economic Evaluation Database (EED) via The Cochrane Library | 4 |
| Total | 951 |
| EndNote deduplication | –286 |
| Unique records to screen | 665 |
Bibliographic search annex
Ovid MEDLINE(R)
Host: Ovid.
Data parameters: 1946 to July week 3 2012.
Date searched: 1 August 2012.
Search strategy: Table 65.
Hits: 197.
| # | Searches | Results |
|---|---|---|
| 1 | Sysmex.mp. | 464 |
| 2 | (RD100i or RD-100i or (RD and 100i) or OSNA or One-step nucleic acid amplification).mp. | 23 |
| 3 | 1 or 2 | 486 |
| 4 | Metasin.mp. | 0 |
| 5 | “98/79/EC”.tw. | 16 |
| 6 | 3 or 4 or 5 | 502 |
| 7 | Cytokeratin 19.mp. | 1217 |
| 8 | (CK19 adj5 (gene or lymph)).mp. | 42 |
| 9 | Mammaglobin B/ or Mammaglobin A/ | 179 |
| 10 | mammaglobin.mp. | 242 |
| 11 | 7 or 8 or 9 or 10 | 1441 |
| 12 | 6 or 11 | 1933 |
| 13 | Sentinel Lymph Node Biopsy/ | 6859 |
| 14 | exp Lymph Nodes/ | 65,568 |
| 15 | (lymph$ adj3 node$).mp. | 169,538 |
| 16 | 13 or 14 or 15 | 171,356 |
| 17 | 12 and 16 | 197 |
Ovid MEDLINE(R) In-Process & Other Non-Indexed Citations
Host: OVID.
Data parameters: from inception to 31 July 2012.
Date searched: 1 August 2012.
Search strategy: Table 66.
Hits: 15.
| # | Searches | Results |
|---|---|---|
| 1 | Sysmex.mp. | 26 |
| 2 | (RD100i or RD-100i or (RD adj1 100i) or OSNA or One-step nucleic acid amplification).mp. | 5 |
| 3 | 1 or 2 | 30 |
| 4 | Metasin.mp. | 0 |
| 5 | “98/79/EC”.tw. | 0 |
| 6 | 3 or 4 or 5 | 30 |
| 7 | Cytokeratin 19.mp. | 61 |
| 8 | (CK19 adj5 (gene or lymph)).mp. | 5 |
| 9 | Mammaglobin B/ or Mammaglobin A/ | 0 |
| 10 | mammaglobin.mp. | 5 |
| 11 | 7 or 8 or 9 or 10 | 69 |
| 12 | 6 or 11 | 97 |
| 13 | Sentinel Lymph Node Biopsy/ | 0 |
| 14 | exp Lymph Nodes/ | 0 |
| 15 | (lymph$ adj3 node$).mp. | 4941 |
| 16 | 13 or 14 or 15 | 4941 |
| 17 | 12 and 16 | 15 |
EMBASE
Host: Ovid.
Data parameters: 1974 to 2012 week 30.
Date searched: 1 August 2012.
Search strategy: Table 67.
Hits: 624.
| # | Searches | Results |
|---|---|---|
| 1 | Sysmex.mp. | 1135 |
| 2 | (RD100i or RD-100i or (RD and 100i) or OSNA or “One-step nucleic acid amplification”).mp. | 98 |
| 3 | 1 or 2 | 1225 |
| 4 | Metasin.mp. | 11 |
| 5 | “98/79/EC”.tw. | 32 |
| 6 | 3 or 4 or 5 | 1268 |
| 7 | Cytokeratin 19.mp. | 3691 |
| 8 | (CK19 adj5 (gene or lymph)).mp. | 79 |
| 9 | Mammaglobin B/ or Mammaglobin A/ | 44 |
| 10 | mammaglobin.mp. | 425 |
| 11 | 7 or 8 or 9 or 10 | 4053 |
| 12 | 6 or 11 | 5266 |
| 13 | Sentinel Lymph Node Biopsy/ | 7986 |
| 14 | exp lymph node/ | 96,163 |
| 15 | (lymph$ adj3 node$).mp. | 239,855 |
| 16 | 13 or 14 or 15 | 241,216 |
| 17 | 12 and 16 | 624 |
Web of Science (Science Citation Index Expanded, Social Sciences Citation Index, Arts and Humanities Citation Index, Conference Proceedings Citation Index – Science, Conference Proceedings Citation Index – Social Science & Humanities, Book Citation Index – Science, Book Citation Index – Social Science and Humanities)
Host: ISI.
Data parameters: 1899–2012.
Date searched: 1 August 2012.
Search strategy: Table 68.
Hits: 93.
| # | Searches | Results |
|---|---|---|
| 1 | Topic=((“RD100i” or “RD-100i” or (RD NEAR/1 100i) or “OSNA” or “One-step nucleic acid amplification”)) | – |
| 2 | Topic=(“Metasin”) | – |
| 3 | 1 or 2 | 93 |
The Cochrane Library
Host: www.thecochranelibrary.com/view/0/index.html
Data parameters: Issue 7 of 12, July 2012.
Date searched: 1 August 2012.
Search strategy: Table 69.
Hits: Cochrane Database of Systematic Reviews = 4; Cochrane Central Register of Controlled Trials = 13 and NHS Economic Evaluation Database = 1 (total = 18).
| # | Searches | Results |
|---|---|---|
| 1 | (Sysmex):ti,ab,kw | 10 |
| 2 | (RD100i or RD-100i or (RD and 100i) or OSNA or (One-step nucleic acid amplification)) | 8 |
| 3 | Metasin | 0 |
| 4 | (#1 OR #2 OR #3) | 18 |
NHS Economic Evaluations Database (EED)
Host: via The Cochrane Collaboration.
Data parameters: Issue 7 of 12, July 2012.
Date searched: 1 August 2012.
Search strategy: Table 70.
| # | Searches | Results |
|---|---|---|
| 1 | All Data: Sysmex | – |
| 2 | All Data: (RD100i or RD-100i or (RD and 100i) or OSNA or (One-step nucleic acid amplification)) | – |
| 3 | All Data: Metasin | 4 |
Trial registries
Table 71 shows the trial registries that were searched.
| Registry | Results |
|---|---|
| ClinicalTrials.gov | 3 |
| Current Controlled Trials | 0 |
| WHO ICTRP | 4 |
| EU Clinical Trials Register | 0 |
| Total | 7 |
ClinicalTrials.gov
Searched: 1 August 2012.
Results: 3 (Table 72).
| Search | Results |
|---|---|
| OSNA | 3 |
| One-step nucleic acid amplification | 0 |
| Metasin | 0 |
-
Clinical evaluation of OSNA breast cancer system to extensive frozen section histopathology (www.clinicaltrials.gov/ct2/show/NCT01368744?term=OSNA&rank=1).
-
Clinical evaluation of OSNA breast cancer system in breast cancer patients receiving neoadjuvant therapy (www.clinicaltrials.gov/ct2/show/NCT01140776?term=OSNA&rank=2).
-
Clinical evaluation of OSNA breast cancer system to test sentinel lymph nodes from patients with breast cancer (www.clinicaltrials.gov/ct2/show/NCT01136369?term=OSNA&rank=3).
Current Controlled Trials
URL: www.controlled-trials.com/
Searched: 1 August 2012.
Results: 0 (Table 73).
| Search | Results |
|---|---|
| OSNA | 0 |
| One-step nucleic acid amplification | 0 |
| Metasin | 0 |
World Health Organization International Clinical Trials Registry Platform
Searched: 1 August 2012.
Results: 5 (Table 74).
| Search | Results |
|---|---|
| OSNA | 4 |
| One-step nucleic acid amplification | 1 |
| Metasin | 0 |
-
Clinical evaluation of OSNA breast cancer system to extensive frozen section histopathology (http://apps.who.int/trialsearch/Trial.aspx?TrialID=NCT01368744).
-
Clinical evaluation of molecular detection for sentinel lymph node examination in breast cancer patients (http://apps.who.int/trialsearch/Trial.aspx?TrialID=JPRN-UMIN000005321).
-
Clinical evaluation of OSNA breast cancer system in breast cancer patients receiving neoadjuvant therapy (http://apps.who.int/trialsearch/Trial.aspx?TrialID=NCT01140776).
-
Clinical evaluation of OSNA breast cancer system to test sentinel lymph nodes from patients with breast cancer (http://apps.who.int/trialsearch/Trial.aspx?TrialID=NCT01136369).
-
A clinical study of intraoperative diagnosis of sentinel lymph node metastasis in head and neck cancer patients using bimolecular methods (http://apps.who.int/trialsearch/Trial.aspx?TrialID=JPRN-UMIN000006508).
EU Clinical Trials Register
URL: www.clinicaltrialsregister.eu/
Searched: 1 August 2012.
Results: 0 (Table 75).
| Search | Results |
|---|---|
| OSNA | 0 |
| One-step nucleic acid amplification | 0 |
| Metasin | 0 |
Google searches
Searched: 1 August 2012.
Search term: OSNA
-
All of the searches below were conducted using the advanced search function to restrict identified reports to those in PDF format.
www.sysmex-lifescience.com/files/lifescience_patients_en.pdf
www.sysmex-lifescience.com/files/lifescience_en.pdf
www.sysmex-lifescience.com/files/OSNA%20Produktflyer_EN_150.pdf
www.sysmex-lifescience.com/files/poster_san_antonio_breast_cancer_meeting_2007_german_osna_study.pdf
www.osnaelectronics.net/safety_light/interfaces-process-automation.pdf
www.translational-medicine.com/content/pdf/1479-5876-8-83.pdf
www.sysmex-lifescience.com/files/sysmex_OSNA_breastcancer_en.pdf
http://pannonia-pathology.com/sites/default/files/presentations/anna_sapino.pdf
-
All of the searches below were conducted using the advanced search function without limit or filter.
www.translational-medicine.com/content/8/1/83
www.asco.org/ascov2/Meetings/Abstracts?&vmview=abst_detail_view&confID=58&abstractID=40334
Search term: Metasin
-
All of the searches below were conducted using the advanced search function to restrict identified reports to those in PDF format.
www.pathsoc.org/files/meetings/winter2010/05.01.106552ProgMAINv10(web).pdf
-
All of the searches below were conducted using the advanced search function, without limit or filter.
No hits.
Forward citation chasing
Review of clinical effectiveness
Database: Web of Science.
Host: Thomson Reuters.
Date searched: 15 October 2012.
Search by: Jenny Lowe.
Results: Table 76.
| Study | Article title | Results |
|---|---|---|
| Bernet Vegue 201251 | Comparison of molecular analysis and histopathology for axillary lymph node staging in primary breast cancer: results of the B-CLOSER-I study | 0 |
| Castellano 201252 | Reliability of whole sentinel lymph node analysis by one-step nucleic acid amplification for intraoperative diagnosis of breast cancer metastases | 2 |
| Choi 201053 | One-step nucleic acid amplification (OSNA): intraoperative rapid molecular diagnostic method for the detection of sentinel lymph node metastases in breast cancer patients in Korean cohort | 0 |
| Feldman 201154 | A novel automated assay for the rapid identification of metastatic breast carcinoma in sentinel lymph nodes | 9 |
| Godey 201255 | Sentinel lymph node analysis in breast cancer: contribution of one-step nucleic acid amplification (OSNA) | 0 |
| Guillen-Paredes 201156 | One-step nucleic acid amplification (OSNA) assay for sentinel lymph node metastases as an alternative to conventional postoperative histology in breast cancer: a cost–benefit analysis | 2 |
| Khaddage 201157 | Implementation of molecular intra-operative assessment of sentinel lymph node in breast cancer | 4 |
| Le Frére-Belda 2008157 | Intra-operative sentinel lymph node metastasis detection in breast cancer by ‘one-step nucleic acid amplification (OSNA)’ – results of the French multicentre prospective study | 0 |
| Osako 201159 | Accurate staging of axillary lymph nodes from breast cancer patients using a novel molecular method | 1 |
| Schem 200960 | One-step nucleic acid amplification – a molecular method for the detection of lymph node metastases in breast cancer patients; results of the German study group | 25 |
| Snook 201161 | Multicentre evaluation of intraoperative molecular analysis of sentinel lymph nodes in breast carcinoma | 11 |
| Tamaki 200962 | Molecular detection of lymph node metastases in breast cancer patients: results of a multicenter trial using the one-step nucleic acid amplification assay | 44 |
| Tamaki 201263 | Routine clinical use of the one-step nucleic acid amplification assay for detection of sentinel lymph node metastases in breast cancer patients. Results of a multicenter study in Japan | 0 |
| Tusjimoto 200764 | One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients | 71 |
| Total | 169 | |
| Deleting duplicate records | –76 | |
| Unique items to screen | 93 | |
| Deleting duplicate records against the master search | –35 | |
| Unique items to screen | 58 |
Review of cost-effectiveness
Database: Web of Science.
Host: Thomson Reuters.
Date searched: 9 October 2012.
Search by: Chris Cooper.
Results: Table 77.
| Study | Article title | Results |
|---|---|---|
| Cutress 2010123 | Observational and cost analysis of the implementation of breast cancer sentinel node intraoperative molecular diagnosis | 9 |
| Guillen-Paredes 201156 | One-step nucleic acid amplification (OSNA) assay for sentinel lymph node metastases as an alternative to conventional postoperative histology in breast cancer: a cost–benefit analysis | 2 |
| Iqbal 201287 | Implementation of one step nucleic acid amplification (OSNA) for intra-operative assessment of sentinel lymph nodes in a DGH | 0 |
| Total | 11 |
Appendix 2 Clinical effectiveness: quality appraisal and data extraction forms
Castellano et al.52
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to assess the reliability of OSNA as a single test on whole SLNs as a method of intraoperative diagnosis and staging of SLNs in breast cancer Study design: cohort Country: Italy Number of centres: 1 Funding: unknown Notes: OSNA SLNs were also analysed using imprint cytology and the two results were compared (almost like a single-gate study embedded within the parallel-group study), but that comparison is not relevant for this review |
Number of participants: 110 OSNA, 169 histology Number of SLNs or ALNs: unclear Recruitment procedure: unclear Inclusion criteria: patients who did not have suspicious ALNs after ultrasound or positive cytological smears. For the OSNA cohort the primary tumour had to express CK19 in > 80% of the tumour cells Exclusion criteria: NR Sample attrition/dropout: 13 patients transferred to histology because of the lack of CK19 expression |
Index test (technical details): the minimum weight for one OSNA reaction was 50 mg and the maximum was 600 mg. SLNs were homogenised using lysis buffer for 90 seconds on ice. CK19 mRNA was determined on a RD100i system according to the standard curve: micrometastases (+) was defined as 250–5000 CK19 mRNA copies/µl and macrometastases (++) was defined as > 5000 CK19 mRNA copies/µl; < 250 CK19 copies/µl indicated a negative result Reference standard (technical details): histopathology; four slices were placed in bioboxes, formalin fixed and paraffin embedded. Slices were step sectioned at 100-µm intervals until extinction. The first two consecutive sections for each step were used for haematoxylin–eosin staining and immunohistochemistry. Metastatic deposits were measured in two dimensions and categorised as follows: pN0(i+), malignant cells < 0.2 mm, single tumour cells or a cluster of < 200 cells; pN1mi, micrometastases > 0.2 mm and/or > 200 cells; pN1a, metastases in one to three ALNs or at least one metastasis > 2.0 mm Details of SLN detection: SLNs were identified using a combination of blue dye and radioactive isotopes. Blue-stained nodes and nodes with high radioactive counts were considered to be SLNs Extraction and division of SLNs: SLNs were cleared from fat tissue, weighed and cut along the short axis. Four slices were step sectioned at 100-µm intervals until extinction Discordance analysis: NA Outcome assessor: NR Blinding: NA |
Accuracy outcomes: positive and negative rates Process outcomes: NR Clinical outcomes: NR Other: NR Unit of analysis: patient Discordant case analysis: NA Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||
| NA, not applicable; NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNAHistologyPatients, n110169Age (years), median (range)66.7 (38–82)61.2 (23–86)Tumour size, n (%) < 10 mm33 (30)41 (24) 1.1–1.5 cm19 (17)45 (27) > 1.5 cm58 (53)83 (49)Histopathological type, n (%) Invasive ductal carcinoma81 (74)109 (64) Invasive lobular carcinoma16 (14)29 (17) DCIS Other13 (12)31 (18)HER2, n (%) Negative108 (98)144 (85) Positive2 (2)25 (15)HER2, human epidermal growth factor receptor 2. | Characteristic | OSNA | Histology | Patients, n | 110 | 169 | Age (years), median (range) | 66.7 (38–82) | 61.2 (23–86) | Tumour size, n (%) | < 10 mm | 33 (30) | 41 (24) | 1.1–1.5 cm | 19 (17) | 45 (27) | > 1.5 cm | 58 (53) | 83 (49) | Histopathological type, n (%) | Invasive ductal carcinoma | 81 (74) | 109 (64) | Invasive lobular carcinoma | 16 (14) | 29 (17) | DCIS | Other | 13 (12) | 31 (18) | HER2, n (%) | Negative | 108 (98) | 144 (85) | Positive | 2 (2) | 25 (15) | HER2, human epidermal growth factor receptor 2. | |||||||||||||
| Characteristic | OSNA | Histology | |||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 110 | 169 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 66.7 (38–82) | 61.2 (23–86) | |||||||||||||||||||||||||||||||||||||||||||||||||
| Tumour size, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||
| < 10 mm | 33 (30) | 41 (24) | |||||||||||||||||||||||||||||||||||||||||||||||||
| 1.1–1.5 cm | 19 (17) | 45 (27) | |||||||||||||||||||||||||||||||||||||||||||||||||
| > 1.5 cm | 58 (53) | 83 (49) | |||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 81 (74) | 109 (64) | |||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 16 (14) | 29 (17) | |||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 13 (12) | 31 (18) | |||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 108 (98) | 144 (85) | |||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 2 (2) | 25 (15) | |||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Resuts | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Total casesNegative, n (%)ITCs (%)Micrometastases, n (%)Macrometastases, n (%)OSNA11078 (71)–20 (18)12 (11)Histology169112 (66)11 (7)13 (8)33 (20) | Total cases | Negative, n (%) | ITCs (%) | Micrometastases, n (%) | Macrometastases, n (%) | OSNA | 110 | 78 (71) | – | 20 (18) | 12 (11) | Histology | 169 | 112 (66) | 11 (7) | 13 (8) | 33 (20) | ||||||||||||||||||||||||||||||||||
| Total cases | Negative, n (%) | ITCs (%) | Micrometastases, n (%) | Macrometastases, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | 110 | 78 (71) | – | 20 (18) | 12 (11) | ||||||||||||||||||||||||||||||||||||||||||||||
| Histology | 169 | 112 (66) | 11 (7) | 13 (8) | 33 (20) | ||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Recruitment: unclear Replicates: unclear whether replicate samples were analysed Outcome assessment: unclear whether the histology was checked by more than one independent pathologist |
|||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
Le Frère-Belda et al.58
| Design | Participants | Tests | Outcomes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to assess OSNA for intraoperative SLN metastasis detection in breast cancer patients, using final histology as the reference standard Study design: single gate Country: France Number of centres: 8 Funding: laboratory consumables funded by Sysmex |
Number of participants: 233 Number of SLNs or ALNs: 503 samples from 456 SLNs Recruitment procedure: NR Inclusion criteria: all breast cancer patients scheduled for surgery with SLN biopsy were considered for enrolment Exclusion criteria: patients who had other types of cancer with metastatic spread, patients given neoadjuvant therapy or patients aged < 18 years Sample attrition/dropout: NR |
Index test (technical details): the assay was performed in duplicate on a pure sample and on a diluted sample (1/10). Homogenates were then stored at –80 °C. Results were automatically characterised by the CK19 mRNA copy number/µl of the original tissue homogenate. A (++) positive result (CK19 mRNA copy number > 5000/µl) was associated with macrometastasis, a (+) positive result (copy numbers between 250 and 5000/µl) was associated with micrometastasis and a negative result (copy numbers no greater than 250/µl) was associated with either ITCs or no tumour Reference standard (technical details): five ribbons were cut with a 200-µm skip space. From each ribbon three sections were prepared, one for haematoxylin–eosin staining and two for immunohistochemistry. Macrometastasis was defined as a tumour deposit > 2 mm and micrometastasis as a tumour deposit > 0.2 mm and ≤ 2 mm. Tumour deposits ≤ 0.2 mm were categorised as ITCs and recorded as histologically negative pN0(i+) Details of SLN detection: NR Extraction and division of SLNs: the excised SLNs were cut into four equal slices. Two alternate slices were prepared for OSNA and the other two slices were fixed in 4% buffered formaldehyde and embedded in a paraffin blockDiscordance analysis: when OSNA was positive and histology negative, consecutive ribbons with a 200-µm skip space were cut until depletion of the remaining paraffin-embedded SLN slices. The sections were stained with haematoxylin–eosin and immunostained with CK19 and AE1/AE3 antibodies (a mixture of two different clones of anticytokeratin monoclonal antibodies AE1 and AE3). In all cases of discrepancy, the SLN homogenates were subjected to further blind molecular analysis. qRT-PCR was performed for CK19 and the breast tissue-specific markers SPDEF (SAM pointed domain containing ETS transcription factor) and FOXA1 (forkhead box A1). CK19 protein expression was assessed using Western blotting analysis. If OSNA and the intensive molecular investigation showed the same results (both negative or both positive) this was taken to indicate TAB, that is, the presence of tumour deposit in either of the two slices used for histology or the two slices used for OSNA Outcome assessor: NR Blinding: yes |
Accuracy outcomes: sensitivity, specificity, PPV, NPV, positive likelihood ratio (LR+), negative likelihood ratio (LR–) Process outcome: median time for OSNA testing Clinical outcomes: NR Other: NR Unit of analysis: patient and node Discordant case analysis: yes Test failures: yes |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNAPatients, n233Age (years), median (range)58 (30–93)Clinical stage, n (%) 041 (17.7) I175 (75.4) II13 (5.6) III2 (0.9) IV1 (0.4)Nodal status, n (%) pN0225 (97.0) pN17 (3.0) pN2 pN3Histopathological type, n (%) Invasive ductal carcinoma164 (70.4) Invasive lobular carcinoma34 (14.6) DCIS23 (9.9) Other12 (5.2)HER2, n (%) Negative Positive13 (6.3)HER2, human epidermal growth factor receptor 2. | Characteristic | OSNA | Patients, n | 233 | Age (years), median (range) | 58 (30–93) | Clinical stage, n (%) | 0 | 41 (17.7) | I | 175 (75.4) | II | 13 (5.6) | III | 2 (0.9) | IV | 1 (0.4) | Nodal status, n (%) | pN0 | 225 (97.0) | pN1 | 7 (3.0) | pN2 | pN3 | Histopathological type, n (%) | Invasive ductal carcinoma | 164 (70.4) | Invasive lobular carcinoma | 34 (14.6) | DCIS | 23 (9.9) | Other | 12 (5.2) | HER2, n (%) | Negative | Positive | 13 (6.3) | HER2, human epidermal growth factor receptor 2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Characteristic | OSNA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 233 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 58 (30–93) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | 41 (17.7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I | 175 (75.4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| II | 13 (5.6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| III | 2 (0.9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IV | 1 (0.4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN0 | 225 (97.0) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1 | 7 (3.0) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 164 (70.4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 34 (14.6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 23 (9.9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 12 (5.2) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, n (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 13 (6.3) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| After TAB exclusion: OSNAFive-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 503 SLNs++37603+53123–3927386n = 233 patients++22603+23317–2717151 Before TAB exclusion per sample: Sensitivity (%) (95% CI)Specificity (%) (95% CI)PPV (%) (95% CI)NPV (%) (95% CI)OSNA LR+OSNA LR–Discordance (%)80.9 (69.0 to 89.8)93.9 (91.2 to 96.0)65.4 (53.7 to 75.8)97.2 (95.1 to 98.6)13.20.207.7 Before TAB exclusion per patient: Sensitivity (%) (95% CI)Specificity (%) (95% CI)PPV (%) (95% CI)NPV (%) (95% CI)OSNA LR+OSNA LR–Discordance (%)78.6 (63.1 to 89.1)88.0 (82.4 to 92.3)58.9 (44.9 to 71.9)94.9 (90.5 to 97.7)6.50.207.5 Nodes, nMedian time to analysis (minutes)133240348454 Test failures – one sample excluded because of a manipulation error |
OSNA | Five-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 503 SLNs | ++ | 37 | 6 | 0 | 3 | + | 5 | 3 | 1 | 23 | – | 3 | 9 | 27 | 386 | n = 233 patients | ++ | 22 | 6 | 0 | 3 | + | 2 | 3 | 3 | 17 | – | 2 | 7 | 17 | 151 | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | OSNA LR+ | OSNA LR– | Discordance (%) | 80.9 (69.0 to 89.8) | 93.9 (91.2 to 96.0) | 65.4 (53.7 to 75.8) | 97.2 (95.1 to 98.6) | 13.2 | 0.20 | 7.7 | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | OSNA LR+ | OSNA LR– | Discordance (%) | 78.6 (63.1 to 89.1) | 88.0 (82.4 to 92.3) | 58.9 (44.9 to 71.9) | 94.9 (90.5 to 97.7) | 6.5 | 0.20 | 7.5 | Nodes, n | Median time to analysis (minutes) | 1 | 33 | 2 | 40 | 3 | 48 | 4 | 54 | ||||||||||||||
| OSNA | Five-level histopathology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 503 SLNs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 37 | 6 | 0 | 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 5 | 3 | 1 | 23 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 3 | 9 | 27 | 386 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 233 patients | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 22 | 6 | 0 | 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 2 | 3 | 3 | 17 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 2 | 7 | 17 | 151 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | OSNA LR+ | OSNA LR– | Discordance (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 80.9 (69.0 to 89.8) | 93.9 (91.2 to 96.0) | 65.4 (53.7 to 75.8) | 97.2 (95.1 to 98.6) | 13.2 | 0.20 | 7.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | OSNA LR+ | OSNA LR– | Discordance (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 78.6 (63.1 to 89.1) | 88.0 (82.4 to 92.3) | 58.9 (44.9 to 71.9) | 94.9 (90.5 to 97.7) | 6.5 | 0.20 | 7.5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodes, n | Median time to analysis (minutes) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 33 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 40 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 48 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 54 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Recruitment: unclear whether patients were recruited consecutively or randomly Analysis: five centres reused frozen section samples, which may impair their integrity for final histology Outcome assessment: unclear whether histology was assessed by more than one independent pathologist Conflict of interest: laboratory consumables were purchased by Sysmex |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Choi et al.53
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to assess the clinical utility and applicability of the OSNA assay in breast cancer treatment in Korea by comparing it with histopathological examination Study design: single gate Country: Republic of Korea Number of centres: 1 Funding: Sysmex |
Number of participants: 199 (after exclusions – see below) Number of SLNs or ALNs: 284 SLNs Recruitment procedure: NR Inclusion criteria: Included patients were suspected as being negative for LN metastasis from initial clinical assessment, and were scheduled for SLNB Exclusion criteria: patients receiving neoadjuvant therapy before undergoing SLNB and those who had already undergone SLNB were excluded from the study Sample attrition/dropout: one patient was excluded because she was finally diagnosed as not having breast cancer but large B-cell lymphoma |
Index test (technical details): each LN was homogenised in glycine buffer. The solutions (10 times diluted and 100 times diluted solutions) were mixed with six different CK19 primers, four deoxynucleoside triphosphates, reverse transcriptase, DNA synthetase and magnesium sulphate. The resulting solution was reacted at a constant temperature of 65 °C. Complementary deoxyribonucleic acid (cDNA) was synthesised from CK19 mRNA in the LN-homogenised solution using reverse transcriptase. The degree of DNA amplified product was calculated using a calibration curve, with standards of known CK19 mRNA concentration. A negative result occurred when the CK19 mRNA concentration of both the 10 times diluted solution and the 100 times diluted solution was < 250 copies/µl. In a positive (++) result the CK19 mRNA concentration in the 10 times diluted solution was ≥ 5000 copies/µl; in a positive (+) result the CK19 mRNA concentration in the 10 times diluted solution was < 5000 and ≥ 250 copies/µl; and in a positive (+I, i.e. positive with reaction inhibited result) the CK19 mRNA concentration in the 10 times diluted solution was < 250 copies/µl and the CK19 mRNA concentration in the 100 times diluted solution was ≥ 250 copies/µl Reference standard (technical details): each SLN was cut along its longitudinal axis into sections of 1.5- to 2.0-mm thickness. For the postoperative histopathological examination, three-level sections were prepared at 200-µm intervals. Three sections were obtained at each level for haematoxylin–eosin staining, anticytokeratin antibody (AE1/AE3) immunohistochemical staining and unstaining. The presence/absence of metastases was determined from haematoxylin–eosin staining and AE1/AE3 staining. Based on the TNM classification of the American Joint Committee on Cancer (7th edition), metastatic deposits were recorded as ITCs if their largest diameter was < 0.2 mm, as micrometastases if they were > 0.2 mm but ≤ 2 mm and as macrometastases if they were > 2 mm. LN samples were regarded as positive if at least one micrometastasis or macrometastasis was found. ITC results were considered as negative. Macrometastasis or micrometastasis was confirmed by both or either of intraoperative histopathological examination of frozen section specimens and postoperative histopathological examination of permanent tissue specimens Details of SLN detection: for the detection of sentinel nodes, both radioisotope and blue dye were used in most patients and radioisotope only was used in some patients. SLNs were defined as any blue-stained nodes or any nodes with radioactive counts of ≥ 10% Extraction and division of SLNs: resected LNs were equally sectioned into blocks along their long axis at 2-mm intervals. Two alternate blocks were subjected to the OSNA assay and the remaining two blocks were subjected to intraoperative and postoperative histopathological examination. If LNs were < 4 mm in the short axis they were cut in half. One half was subjected to the OSNA assay and the other half to histopathological examination. Each LN was subjected to the OSNA assay and histopathological examination Outcome assessor: NR Blinding: unclear Discordant case analysis: clinical information, status of non-SLNs and expression of CK19 protein in LN metastatic foci were evaluated on a patient-level basis |
Accuracy outcomes: sensitivity, specificity, PPV, NPV, concordance rate Process outcomes: the rapidity of the OSNA assay was investigated by measuring the turnaround time (i.e. the time between starting homogenisation and obtaining the results of the assay) Clinical outcomes: NR Other: none reported Unit of analysis: patient Discordant case analysis: yes Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n199Age (years), median (range)40–49Clinical stage, n (%) 011 (5.5) I132 (66.3) II54 (27.1) III2 (1.0) IV11 (5.5)Clinical tumour classification, n (%) T0 Tis8 (4.0) T1129 (64.8) T256 (28.1) T32 (1.0) T4 Tx4 (2.0)Nodal status, n (%) pN0153 (76.9) pN137 (18.6) pN25 (2.5) pN34 (2.0)Histopathological type, n (%) Invasive ductal carcinoma165 (82.9) Invasive lobular carcinoma9 (4.5) DCIS9 (4.5) Other16 (8.1) | Characteristic | OSNA plus histology | Patients, n | 199 | Age (years), median (range) | 40–49 | Clinical stage, n (%) | 0 | 11 (5.5) | I | 132 (66.3) | II | 54 (27.1) | III | 2 (1.0) | IV | 11 (5.5) | Clinical tumour classification, n (%) | T0 | Tis | 8 (4.0) | T1 | 129 (64.8) | T2 | 56 (28.1) | T3 | 2 (1.0) | T4 | Tx | 4 (2.0) | Nodal status, n (%) | pN0 | 153 (76.9) | pN1 | 37 (18.6) | pN2 | 5 (2.5) | pN3 | 4 (2.0) | Histopathological type, n (%) | Invasive ductal carcinoma | 165 (82.9) | Invasive lobular carcinoma | 9 (4.5) | DCIS | 9 (4.5) | Other | 16 (8.1) | |||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 199 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 40–49 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | 11 (5.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I | 132 (66.3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| II | 54 (27.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| III | 2 (1.0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IV | 11 (5.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | 8 (4.0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 129 (64.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 56 (28.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | 2 (1.0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tx | 4 (2.0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN0 | 153 (76.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1 | 37 (18.6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2 | 5 (2.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN3 | 4 (2.0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 165 (82.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 9 (4.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 9 (4.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 16 (8.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNAThree-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 199 patients++19211+3304+i1000–443154 Sensitivity (%) (95% CI)Specificity (%) (95% CI)Discordance (%)n = 199 patients77.8 (0.60 to 0.90)96.3 (0.92 to 0.99)7 Nodes, nMean time to analysis (minutes)135.2244.8350.4450.0 Overall, 39.0 minutes |
OSNA | Three-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 199 patients | ++ | 19 | 2 | 1 | 1 | + | 3 | 3 | 0 | 4 | +i | 1 | 0 | 0 | 0 | – | 4 | 4 | 3 | 154 | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | n = 199 patients | 77.8 (0.60 to 0.90) | 96.3 (0.92 to 0.99) | 7 | Nodes, n | Mean time to analysis (minutes) | 1 | 35.2 | 2 | 44.8 | 3 | 50.4 | 4 | 50.0 | |||||||||||||
| OSNA | Three-level histopathology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 199 patients | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 19 | 2 | 1 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 3 | 3 | 0 | 4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| +i | 1 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 4 | 4 | 3 | 154 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 199 patients | 77.8 (0.60 to 0.90) | 96.3 (0.92 to 0.99) | 7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodes, n | Mean time to analysis (minutes) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 35.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 44.8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 50.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 50.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Recruitment: unclear Replicates: unclear whether replicate samples were analysed Outcome assessment: unclear whether the histology was checked by more than one independent pathologist Conflict of interest: the study was funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Feldman et al.54
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to compare the performance of the OSNA system with that of a detailed histopathological examination of the LN and with immunohistochemistry for the detection of metastatic carcinoma in axillary SLNs in patients with early-stage breast cancer Study design: single gate Country: USA Number of centres: 11 Funding: Sysmex |
Number of participants: 496 Number of SLNs or ALNs: 1044 SLNs Recruitment procedure: NR Inclusion criteria: patients aged > 18 years with a clinical Tis, T1 or T2 primary breast cancer who were awaiting lymphatic mapping and SLN biopsy were eligible for enrolment Exclusion criteria: locally advanced breast cancer (tumours classified as T3 or T4), DCIS in patients who were undergoing breast-conserving surgery, clinically palpable suspicious ALNs, previous diagnosis of another type of carcinoma, previous breast or axillary surgery and preoperative neoadjuvant therapy Sample attrition/dropout: NR |
Index test (technical details): the SLN slices were homogenised in 4 ml of OSNA lysis buffer and centrifuged according to the manufacturer’s instructions. The device was calibrated to designate samples that contained ≥ 250 copies/µl of CK19 mRNA as positive for metastatic tumour. Cut-off values, system calibration and calculation of the CK19 mRNA level of the sample from the calibration curve followed those of Tsujimoto et al.64 Reference standard (technical details): the SLNs allocated to histopathology were fixed in formalin and embedded in paraffin. Pathologists at the individual clinical sites evaluated the SLNs according to their local standard protocols for clinical management. Paraffin blocks of the SLNs were cut at 200-µm intervals (levels) until all tissue was depleted. At each level, three 5-µm sections were cut; the first section for each level was stained with haematoxylin–eosin and the third section from the third level was stained immunohistochemically using pan-CK antibodies. The remaining sections were used for additional staining, as required. All slides were sent to a central reference pathology laboratory for independent, blinded pathologist evaluation. Tumour deposits in the SLNs were classified according to American Joint Committee on Cancer guidelines Details of SLN detection: blue dye used in 34 patients (6.9%), technetium-99m sulphur colloid (radiocolloid) used in 107 patients (21.6%) and both used in 355 patients (71.6%) Extraction and division of SLNs: SLNs were included only if they were 4–20 mm in size along the long axis with a thickness ranging from 4 mm to 10 mm. SLNs were sectioned into an average of six pieces along the long axis into 1-mm slices. Alternate slices of the LN were subjected either to analysis with the OSNA system or to detailed histopathological examination Outcome assessor: two independent pathologists Blinding: yes Discordant case analysis: performed by Western blotting and qRT-PCR |
Accuracy outcomes: sensitivity and specificity, agreement, NPV and PPV Process outcome: time to analysis Clinical outcomes: NR Other: none Unit of analysis: SLN Discordant case analysis: yes Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n496Age (years), median (range)58.8 (28–88)Clinical tumour classification, n (%) T0 Tis21 (4.2) T1327 (65.9) T2124 (25) T35 (1) T4 Tx19 (3.8)Nodal status, n (%) pN0387 (78) pN184 (16.9) pN214 (2.8) pN34 (0.8) pNx7 (1.4)Histopathological type, n (%) Invasive ductal carcinoma348 (70.2) Invasive lobular carcinoma40 (8.1) DCIS Other109 (21.7)HER2, n (%) Negative PositiveHER2, human epidermal growth factor receptor 2. | Characteristic | OSNA plus histology | Patients, n | 496 | Age (years), median (range) | 58.8 (28–88) | Clinical tumour classification, n (%) | T0 | Tis | 21 (4.2) | T1 | 327 (65.9) | T2 | 124 (25) | T3 | 5 (1) | T4 | Tx | 19 (3.8) | Nodal status, n (%) | pN0 | 387 (78) | pN1 | 84 (16.9) | pN2 | 14 (2.8) | pN3 | 4 (0.8) | pNx | 7 (1.4) | Histopathological type, n (%) | Invasive ductal carcinoma | 348 (70.2) | Invasive lobular carcinoma | 40 (8.1) | DCIS | Other | 109 (21.7) | HER2, n (%) | Negative | Positive | HER2, human epidermal growth factor receptor 2. | |||||||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 496 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 58.8 (28–88) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | 21 (4.2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 327 (65.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 124 (25) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | 5 (1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tx | 19 (3.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN0 | 387 (78) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1 | 84 (16.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2 | 14 (2.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN3 | 4 (0.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pNx | 7 (1.4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 348 (70.2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 40 (8.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 109 (21.7) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNAThree-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 1044 SLNs++77918+912029–92214854 Sensitivity (%) (95% CI)Specificity (%) (95% CI)Discordance (%)n = 1044 SLNs77.5 (69.7 to 84.2)95.8 (94.3 to 97.0)6.8 Nodes, nInterquartile mean time to analysis (minutes)133.0239.6345.2 |
OSNA | Three-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 1044 SLNs | ++ | 77 | 9 | 1 | 8 | + | 9 | 12 | 0 | 29 | – | 9 | 22 | 14 | 854 | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | n = 1044 SLNs | 77.5 (69.7 to 84.2) | 95.8 (94.3 to 97.0) | 6.8 | Nodes, n | Interquartile mean time to analysis (minutes) | 1 | 33.0 | 2 | 39.6 | 3 | 45.2 | ||||||||||||||||||
| OSNA | Three-level histopathology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 1044 SLNs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 77 | 9 | 1 | 8 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 9 | 12 | 0 | 29 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 9 | 22 | 14 | 854 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 1044 SLNs | 77.5 (69.7 to 84.2) | 95.8 (94.3 to 97.0) | 6.8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodes, n | Interquartile mean time to analysis (minutes) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 33.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 39.6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 45.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Recruitment: unclear whether recruitment was consecutive or randomised Patient flow: the number of SLNs after discordance (1018) does not comply with the numbers before discordance (1044) minus the resolved cases (28) Replicates: unclear whether replicate samples were analysed Conflict of interest: the study was funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Bernet Vegue et al.51
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: description of the results of the B-CLOSER-I study with regard to staging Study design: single gate Country: Spain Number of centres: 8 Funding: Sysmex |
Number of participants: 55 after exclusions Number of SLNs or ALNs: 567 ALNs Recruitment procedure: consecutive Inclusion criteria: in all cases, tumours were confirmed as CK19 positive by immunohistochemistry before SLNB. All patients had undergone ALND after positive SLNB diagnosed by OSNA Exclusion criteria: patients were excluded if they had metastatic disease, had received neoadjuvant therapy or were judged unsuitable because of concomitant disease, and if < 10 ALNs were obtained by ALND Sample attrition/dropout: two patients with < 10 ALNs excluded |
Index test (technical details): the LN tissue was homogenised. CK19 mRNA was then amplified by RT-LAMP (reverse transcription loop-mediated isothermal amplification). Results were classified according to the following cut-off values for CK19 mRNA copy number: < 100 copies/µl, negative; 100–250 copies/ml, negative (low expression); 250 to 5000 copies/µl, micrometastasis; and > 5000 copies/µl, macrometastasis Reference standard (technical details): the central tissue slice was fixed and embedded in paraffin for histopathological analysis and the remaining tissue was stored at –80 °C before analysis by OSNA assay. A 5-mm paraffin section was obtained from each central slice and stained with haematoxylin–eosin. Macrometastases, micrometastases and ITCs were defined by American Joint Committee on Cancer TNM criteria. When ITCs were identified by histopathology, serial sections of the remainder of the block were analysed for the presence of micrometastases or macrometastases Details of SLN detection: NR Extraction and division of SLNs: in LNs obtained by ALND weighing (without surrounding fat) > 50 mg (the cut-off for validity using the OSNA method), a central longitudinal 1-mm slice was taken from each node and allocated to histology, with the rest of the node assigned to OSNA Outcome assessor: NR Blinding: NR Discordant case analysis: no further analysis |
Accuracy outcome: concordance Process outcomes: NR Clinical outcomes: NR Other: NR Unit of analysis: patient and ALN Discordant case analysis: cases reported but no further analysis Test failures: NR |
||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n55Age (years), median (range)59 (23–87)Clinical stage, n (%) 0 I21 (38.2) II22 (40) III12 (21.8) IV UnknownHistopathological type, n (%) Invasive ductal carcinoma44 (80.0) Invasive lobular carcinoma7 (12.7) DCIS1 (1.8) Other3 (5.5)HER2, n (%) Negative49 (89.1) Positive6 (10.9)HER2, human epidermal growth factor receptor 2. | Characteristic | OSNA plus histology | Patients, n | 55 | Age (years), median (range) | 59 (23–87) | Clinical stage, n (%) | 0 | I | 21 (38.2) | II | 22 (40) | III | 12 (21.8) | IV | Unknown | Histopathological type, n (%) | Invasive ductal carcinoma | 44 (80.0) | Invasive lobular carcinoma | 7 (12.7) | DCIS | 1 (1.8) | Other | 3 (5.5) | HER2, n (%) | Negative | 49 (89.1) | Positive | 6 (10.9) | HER2, human epidermal growth factor receptor 2. | ||||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||
| Patients, n | 55 | ||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 59 (23–87) | ||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n (%) | |||||||||||||||||||||||||||||||||||||||||
| 0 | |||||||||||||||||||||||||||||||||||||||||
| I | 21 (38.2) | ||||||||||||||||||||||||||||||||||||||||
| II | 22 (40) | ||||||||||||||||||||||||||||||||||||||||
| III | 12 (21.8) | ||||||||||||||||||||||||||||||||||||||||
| IV | |||||||||||||||||||||||||||||||||||||||||
| Unknown | |||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 44 (80.0) | ||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 7 (12.7) | ||||||||||||||||||||||||||||||||||||||||
| DCIS | 1 (1.8) | ||||||||||||||||||||||||||||||||||||||||
| Other | 3 (5.5) | ||||||||||||||||||||||||||||||||||||||||
| HER2, n (%) | |||||||||||||||||||||||||||||||||||||||||
| Negative | 49 (89.1) | ||||||||||||||||||||||||||||||||||||||||
| Positive | 6 (10.9) | ||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | |||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||
| OSNAOne-level histopathologyMacrometastasisMicrometastasisNegativen = 567 non-SLNs++1414+0125+i (low expression)008–00514 | OSNA | One-level histopathology | Macrometastasis | Micrometastasis | Negative | n = 567 non-SLNs | ++ | 1 | 4 | 14 | + | 0 | 1 | 25 | +i (low expression) | 0 | 0 | 8 | – | 0 | 0 | 514 | |||||||||||||||||||
| OSNA | One-level histopathology | ||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | Negative | |||||||||||||||||||||||||||||||||||||||
| n = 567 non-SLNs | |||||||||||||||||||||||||||||||||||||||||
| ++ | 1 | 4 | 14 | ||||||||||||||||||||||||||||||||||||||
| + | 0 | 1 | 25 | ||||||||||||||||||||||||||||||||||||||
| +i (low expression) | 0 | 0 | 8 | ||||||||||||||||||||||||||||||||||||||
| – | 0 | 0 | 514 | ||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||
| Recruitment: small sample size with relatively large number of ALNs Replicates: unclear whether replicate samples were analysed Outcome assessment: unclear whether the histology was checked by more than one independent pathologist Conflict of interest: the study was funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | L | ||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||
Godey et al.55
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to present OSNA results in a routine clinical setting compared with histology Study design: single gate embedded in cohort Country: France Number of centres: unclear Funding: NR |
Number of participants: 722 Number of SLNs or ALNs: 810 SLNs Recruitment procedure: NR Inclusion criteria: clinically node-negative early-stage breast cancer undergoing axillary SLN procedure Exclusion criteria: NR Sample attrition/dropout: 108 patients with in situ carcinoma, tumour size > T1c or invasive carcinoma other than ductal or lobular forms were excluded from the OSNA cohort group in the analysis to avoid bias, because these categories of patients were not represented in the (historical) histology cohort group |
Index test (technical details): SLNs cleared from fat and extranodal tissue were homogenised. SLNs weighing > 600 mg were cut and analysed separately with two or more molecular analyses. OSNA analysis was carried out in duplicate with a pure and a diluted sample (1/10) of SLN lysates. The CK19 mRNA copy number/µl of lysate was classified as follows: < 250/µl, no metastasis; 250–5000/µl, micrometastasis; > 5000/µl, macrometastasis. The OSNA assay was not calibrated to detect ITCs. If the copy number was > 250/µl in the diluted preparation only, the OSNA result was classified as positive with inhibition of the amplification reaction; Patients with at least one SLN macrometastasis were classified as macrometastatic, those with at least one SLN micrometastasis were classed as micrometastatic and those with at least one metastasis with inhibition were classed as metastatic Reference standard (technical details): histological analysis was performed on SLN tissue sections embedded in paraffin blocks and sectioned every 250 µm until the block was completely cut. Each level was initially stained with standard haematoxylin–eosin. If no metastases were revealed by conventional staining, then immunohistochemical labelling of all levels was carried out. Final examination of axillary non-SLNs was investigated by permanent histology (each 2-mm section of the LN was analysed with haematoxylin–eosin staining) in both the OSNA and the historical cohort Details of SLN detection: localisation of the sentinel node was identified with technetium-99m-labelled colloid injected the day before surgery and 3 hours after axillary lymphoscintigraphy, followed by subcutaneous injection of 2 ml of patent blue dye on the day of the procedure. SLNs were cut by the pathologist and touch imprints were performed intraoperatively Extraction and division of SLNs: a 1-mm thick central slice was stained for postoperative histology. The remaining portion of the node was used for OSNA analysis intraoperatively Outcome assessor: NR Blinding: NR Discordant case analysis: NR |
Accuracy outcome: positivity rate Process outcome: time for analysis Clinical outcomes: NR Other: NR Unit of analysis: patient Discordant case analysis: NA Test failures: issues with three samples for OSNA; no further details |
||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNAHistologyPatients, n258355Age (years), median56.856.9Clinical tumour classification, n T0 Tis T1a, b or c19, 93, 14616, 125, 214 T2 T3 T4 TxHistopathological type, n Invasive ductal carcinoma212313 Invasive lobular carcinoma4642 DCIS Other | Characteristic | OSNA | Histology | Patients, n | 258 | 355 | Age (years), median | 56.8 | 56.9 | Clinical tumour classification, n | T0 | Tis | T1a, b or c | 19, 93, 146 | 16, 125, 214 | T2 | T3 | T4 | Tx | Histopathological type, n | Invasive ductal carcinoma | 212 | 313 | Invasive lobular carcinoma | 46 | 42 | DCIS | Other | |||||||||||||||||||||||
| Characteristic | OSNA | Histology | |||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 258 | 355 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median | 56.8 | 56.9 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n | |||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | |||||||||||||||||||||||||||||||||||||||||||||||||||
| T1a, b or c | 19, 93, 146 | 16, 125, 214 | |||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Tx | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 212 | 313 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 46 | 42 | |||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA-positive rate 24.4%, histology-positive rate 24.8% Technical problems with OSNA for three patients; no further details Nodes, nMean (SD) time to analysis (minutes)132.9 (4.9)236.4 (4.5)341.6 (5.2)448.5 (8.7) |
Nodes, n | Mean (SD) time to analysis (minutes) | 1 | 32.9 (4.9) | 2 | 36.4 (4.5) | 3 | 41.6 (5.2) | 4 | 48.5 (8.7) | |||||||||||||||||||||||||||||||||||||||||
| Nodes, n | Mean (SD) time to analysis (minutes) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 32.9 (4.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 36.4 (4.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 41.6 (5.2) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 48.5 (8.7) | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Outcome assessment: unclear whether the histology was checked by more than one independent pathologist |
|||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U)a | H | ||||||||||||||||||||||||||||||||||||||||||||||||||
Bernet et al.50
| Design | Participants | Tests | Outcomes | ||||
|---|---|---|---|---|---|---|---|
| Objective: to compare OSNA with conventional histology and evaluate the feasibility of OSNA for intraoperative evaluation of SLNs in breast cancer surgery Study design: observation Country: Spain Number of centres: 1 (for trial 2) Funding: NR Notes: trial 1 was not included in this review because the comparator was excluded. Trial 2 was included for process outcomes |
Number of participants: 55 Number of SLNs or ALNs: unclear Recruitment procedure: NR Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: NR |
Index test (technical details): the OSNA protocol consisted of homogenisation of tissue and subsequent isothermal (65 °C) amplification of CK19. Tissue homogenates from each LN were kept frozen at –80 °C as a backup for possible future studies Reference standard (technical details): NA Details of SLN detection: NR Extraction and division of SLNs: the entire node was submitted to the OSNA assay in all cases, except in nine cases in which alternate slices were studied by both methods Outcome assessor: NA Blinding: NA Discordant case analysis: NA |
Accuracy outcomes: NA Process outcome: time from receipt of node to analytical report Unit of analysis: node Discordant case analysis: NA Test failures: NR |
||||
| NA, not applicable; NR, not reported. | |||||||
| Participant characteristics | |||||||
| NR | |||||||
| Results | |||||||
| Nodes, nMean (range) time to analysis (minutes)139.6 (26–70) | Nodes, n | Mean (range) time to analysis (minutes) | 1 | 39.6 (26–70) | |||
| Nodes, n | Mean (range) time to analysis (minutes) | ||||||
| 1 | 39.6 (26–70) | ||||||
| Methodological issues | |||||||
| Replicates: unclear whether replicate samples were analysed | |||||||
| Quality appraisal | |||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | N | ||||||
| Was a cohort study design avoided? (Y/N/U) | NA | ||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | U | ||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | ||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||
| Did all patients receive a reference standard? (Y/N/U) | N | ||||||
| Did all patients receive the same reference standard? (Y/N/U) | NA | ||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | U | ||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | NA | ||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | U | ||||||
Guillen-Paredes et al.56
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to analyse the economic costs of intraoperative OSNA compared with those of conventional deferred histological and immumohistochemical assay carried out in hospital Study design: cohort Country: Spain Number of centres: 1 Funding: Foundation for Healthcare Research and Training of the Murcia Region (FFIS) |
Number of participants: Histology 45; OSNA 35 Number of SLNs or ALNs: 114 SLNs Recruitment procedure: patients were recruited from an administrative database that recorded all SLNBs reported by the pathology department since the implementation of this technique in hospital in 2002 for the study of SLNs in breast cancer patients Inclusion criteria: patients with breast cancer stages pT1/2 N0 M05 with clinically negative and ultrasound-negative ALNs who underwent SNLB along with appropriate breast cancer surgery in the same intervention at the breast unit of the hospital between 15 October 2008 and 15 December 2009 Exclusion criteria: patients who had received neoadjuvant treatment, those who refused to sign the informed consent, those who could not undergo the planned surgery because of a high anaesthetic risk, those who had undergone previous extensive breast surgery, those who had undergone a SLNB with local anaesthesia before the definitive breast surgery (as they were candidates for immediate reconstruction or for receiving neoadjuvant chemotherapy with clinical N0 to reduce tumour size), pregnant women and men Sample attrition/dropout: NR |
Index test (technical details): the SLN was sent fresh to the pathology department (if more than one LN was available all were sent at the same time). The fat was separated from the LN and the LN was sectioned if it weighed > 600 mg. The samples were then lysed and centrifuged. Results were classified according to the number of CK19 mRNA copies per tumour cell: no metastasis, < 250 copies/µl; micrometastasis, from 250 to 5000 copies/µl; macrometastasis, > 5000 copies/µl Reference standard (technical details): after initial preparation of the LN using 4-mm sections fixed in formalin and embedded in paraffin, 15 × 4 µm-thick serial sections were cut and stained with haematoxylin–eosin and immunohistochemically for cytokeratins. A pathologist examined the samples by microscope and classified them as negative (no metastatic cells), ITCs (focus of malignant cells < 0.2 mm), micrometastasis (> 0.2 mm and ≤ 2 mm) and macrometastasis (> 2 mm) Details of SLN detection: the SLN was defined as that which had an activity > 10% of the maximum activity detected Extraction and division of SLNs: details as above Outcome assessor: pathologist using a conventional optical microscope Blinding: NA Discordant case analysis: NA |
Accuracy outcomes: NR Process outcomes: operative time, days in hospital, hospital costs Clinical outcomes: complications, lymphadenectomy Unit of analysis: patient Discordant case analysis: NA Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NA, not applicable; NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ClassificationOSNAHistologyPatients, n3545Age (years), median55.5461.89Clinical tumour classification, n T0 Tis23 T11613 T21729 T3 T4 TxHistopathological type, n Invasive ductal carcinoma3137 Invasive lobular carcinoma25 DCIS12 Other11 | Classification | OSNA | Histology | Patients, n | 35 | 45 | Age (years), median | 55.54 | 61.89 | Clinical tumour classification, n | T0 | Tis | 2 | 3 | T1 | 16 | 13 | T2 | 17 | 29 | T3 | T4 | Tx | Histopathological type, n | Invasive ductal carcinoma | 31 | 37 | Invasive lobular carcinoma | 2 | 5 | DCIS | 1 | 2 | Other | 1 | 1 | |||||||||||||||||||||||||||||||||||
| Classification | OSNA | Histology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 35 | 45 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median | 55.54 | 61.89 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | 2 | 3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 16 | 13 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 17 | 29 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tx | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 31 | 37 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 2 | 5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 1 | 2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 1 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AnalysisMean (SD) intervention time (minutes)Mean (SD) days in hospitalFirst operationSecond operationTotalFirst admissionSecond admissionTotalHistology57.11 (23.93)78.33 (NR)78 (48.02)1.8 (2.04)2.41 (1.09)2.44 (0.78)OSNA62.14 (48.02)NA62.14 (21.93)1.54 (0.78)NA1.54 (0.78)NA, not applicable; NR, not reported. TestComplications in first interventionComplications in second interventionNoneMinorMajorNoneMinorMajorHistology28170480OSNA24101NANANANA, not applicable. |
Analysis | Mean (SD) intervention time (minutes) | Mean (SD) days in hospital | First operation | Second operation | Total | First admission | Second admission | Total | Histology | 57.11 (23.93) | 78.33 (NR) | 78 (48.02) | 1.8 (2.04) | 2.41 (1.09) | 2.44 (0.78) | OSNA | 62.14 (48.02) | NA | 62.14 (21.93) | 1.54 (0.78) | NA | 1.54 (0.78) | NA, not applicable; NR, not reported. | Test | Complications in first intervention | Complications in second intervention | None | Minor | Major | None | Minor | Major | Histology | 28 | 17 | 0 | 4 | 8 | 0 | OSNA | 24 | 10 | 1 | NA | NA | NA | NA, not applicable. | |||||||||||||||||||||||
| Analysis | Mean (SD) intervention time (minutes) | Mean (SD) days in hospital | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| First operation | Second operation | Total | First admission | Second admission | Total | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histology | 57.11 (23.93) | 78.33 (NR) | 78 (48.02) | 1.8 (2.04) | 2.41 (1.09) | 2.44 (0.78) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | 62.14 (48.02) | NA | 62.14 (21.93) | 1.54 (0.78) | NA | 1.54 (0.78) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NA, not applicable; NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Test | Complications in first intervention | Complications in second intervention | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| None | Minor | Major | None | Minor | Major | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histology | 28 | 17 | 0 | 4 | 8 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | 24 | 10 | 1 | NA | NA | NA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NA, not applicable. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Recruitment: patients do not appear to have been recruited consecutively or randomly Analysis: unclear whether histopathology results were checked by an independent pathologist |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Khaddage et al.57
| Design | Participants | Tests | Outcomes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to evaluate the intraoperative performance of OSNA in comparison to postoperative histology and then to introduce the technique into routine practice Study design: single gate Country: France Number of centres: multicentre; number NR Funding: Sysmex |
Number of participants: validation study 46; routine study 197 Number of SLNs or ALNs: validation study 80 SLNs; routine study unclear Recruitment procedure: NR Inclusion criteria: for both patient cohorts inclusion criteria were a minimum age of 18 years and assignment for SLNB Exclusion criteria: neoadjuvant treatment and the presence of metastatic disease other than breast carcinoma Sample attrition/dropout: NR |
Index test (technical details): the SLN slices were homogenised. The results were presented on the RD-100i in qualitative categories (++, +, –) and further specified by CK19 mRNA copy number/µl: 0–249 copies/µl (–), 250–5000 copies/µl (+), and > 5000 copies/µl (++). A result indicating a (+) was comparable to the presence of a micrometastasis and a result indicating a (++) was comparable to a macrometastasis Reference standard (technical details): two alternate slices were embedded in paraffin and postoperatively cut at 200-µm intervals (five levels). Each level was subjected to haematoxylin–eosin and immunohistochemical staining for CK19 protein as well as immunohistochemistry with AE1/AE3 as a pan-CK marker. For routine use, a central slide of 1 mm from each SLN was analysed by one level of haematoxylin–eosin staining and one level of immunohistochemistry (AE1/AE3). Non-SLNs were cut into 2-mm slices and one level of haematoxylin–eosin staining was performed for each slice. Tumour deposits were classified according to the TNM classification of the Union for International Cancer Control (sixth edition) and the American Joint Committee on Cancer (sixth edition). ITCs or a tumour-free SLN were defined as a negative histological result Details of SLN detection: NR Extraction and division of SLNs: during the clinical study, nodes were cleared from fat after SLNB and intraoperatively cut into four equal slices of 1–2 mm thickness. Two alternate slices were analysed by OSNA and the remaining two slices were subjected to histology Outcome assessor: NR Blinding: the results of OSNA were not known to the investigator of histology and vice versa Discordant case analysis: consisted of qRT-PCR for CK19 mRNA and two breast cancer-specific markers [SAM pointed domain containing ETS transcription factor, (SPDEF) and forkhead box A1 (FOXA1)] as well as beta-actin for RNA control. RNA was extracted from 200 µl of the homogenate. The cut-off levels for each marker were determined according to the qRT-PCR results of a series of histologically positive and negative LNs from breast cancer patients |
Accuracy outcomes: Concordance, sensitivity, specificity Process outcome: time to analysis Unit of analysis: node and patient Discordant case analysis: yes Test failures: NR |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histology validationOSNA plus histology routinePatients, n46197Age (years), median (range)(≥ 18)(≥ 18)Clinical tumour classification, n T071 Tis021 T134141 T2230 T321 T411 TxHistopathological type, n Invasive ductal carcinoma36148 Invasive lobular carcinoma516 DCIS521 Other012 | Characteristic | OSNA plus histology validation | OSNA plus histology routine | Patients, n | 46 | 197 | Age (years), median (range) | (≥ 18) | (≥ 18) | Clinical tumour classification, n | T0 | 7 | 1 | Tis | 0 | 21 | T1 | 34 | 141 | T2 | 2 | 30 | T3 | 2 | 1 | T4 | 1 | 1 | Tx | Histopathological type, n | Invasive ductal carcinoma | 36 | 148 | Invasive lobular carcinoma | 5 | 16 | DCIS | 5 | 21 | Other | 0 | 12 | ||||||||||||||||||||||||||||||||||||||||||||
| Characteristic | OSNA plus histology validation | OSNA plus histology routine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 46 | 197 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | (≥ 18) | (≥ 18) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | 7 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | 0 | 21 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 34 | 141 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 2 | 30 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | 2 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tx | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 36 | 148 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 5 | 16 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 5 | 21 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 0 | 12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%)Specificity (%)Discordance (%)Validation studyn = 46 patients before TAB80.097.23.7n = 46 patients after TAB10097.2n = 80 SLNs before TAB88.298.4n = 80 SLNs after TAB10098.4 | Sensitivity (%) | Specificity (%) | Discordance (%) | Validation study | n = 46 patients before TAB | 80.0 | 97.2 | 3.7 | n = 46 patients after TAB | 100 | 97.2 | n = 80 SLNs before TAB | 88.2 | 98.4 | n = 80 SLNs after TAB | 100 | 98.4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Validation study | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 46 patients before TAB | 80.0 | 97.2 | 3.7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 46 patients after TAB | 100 | 97.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 80 SLNs before TAB | 88.2 | 98.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 80 SLNs after TAB | 100 | 98.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNAFive-level histopathology – validation studyMacrometastasisMicrometastasisITCsNegativen = 80 SLNs++112–0+20–1–00 (2)260 (62)n = 46 patients++6200+0001–00 (2)233 (35)OSNAOne-level histopathology – routine useMacrometastasisMicrometastasisITCsNegativen = 197 patients++91–3+87–14–00–155Values in parentheses indicate case numbers before discordant case investigation. Median time to analysis for two nodes: 37 minutes |
OSNA | Five-level histopathology – validation study | Macrometastasis | Micrometastasis | ITCs | Negative | n = 80 SLNs | ++ | 11 | 2 | – | 0 | + | 2 | 0 | – | 1 | – | 0 | 0 (2) | 2 | 60 (62) | n = 46 patients | ++ | 6 | 2 | 0 | 0 | + | 0 | 0 | 0 | 1 | – | 0 | 0 (2) | 2 | 33 (35) | OSNA | One-level histopathology – routine use | Macrometastasis | Micrometastasis | ITCs | Negative | n = 197 patients | ++ | 9 | 1 | – | 3 | + | 8 | 7 | – | 14 | – | 0 | 0 | – | 155 | Values in parentheses indicate case numbers before discordant case investigation. | |||||||||||||||||||||||||
| OSNA | Five-level histopathology – validation study | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 80 SLNs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 11 | 2 | – | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 2 | 0 | – | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 0 (2) | 2 | 60 (62) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 46 patients | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 6 | 2 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 0 | 0 | 0 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 0 (2) | 2 | 33 (35) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | One-level histopathology – routine use | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 197 patients | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 9 | 1 | – | 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 8 | 7 | – | 14 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 0 | – | 155 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Values in parentheses indicate case numbers before discordant case investigation. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Recruitment: unclear whether patients have been recruited consecutively or randomly Analysis: unclear whether histopathology results were checked by an independent pathologist |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
McDowell (unpublished)
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
|---|---|---|---|
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | Academic-in-confidence information has been removed |
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | |||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
| Academic-in-confidence information has been removed | Academic-in-confidence information has been removed | ||
Osako et al.59
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to determine the performance of the OSNA assay as an accurate nodal staging tool compared with routine histological examination Study design: cohort Country: Japan Number of centres: 1 Funding: Sysmex contributed to funding of laboratory consumables |
Number of participants: 183 Number of SLNs or ALNs: NR Recruitment procedure: consecutive Inclusion criteria: patients with clinically and ultrasonographically node-negative pT1–2 breast cancer who had undergone ALND after a positive SNLB with OSNA between April 2009 and September 2010 Exclusion criteria: patients with three or more positive SLNs; SLN identification without using the radioisotope tracer; previous excision of primary tumour; heterochronous ipsilateral breast cancer recurrence; neoadjuvant drug therapy Sample attrition/dropout: NR |
Index test (technical details): after removal of the extranodal tissue, whole LNs were homogenised. A standard positive control containing 5000 copies/µl of CK19 mRNA and a negative control with no CK19 mRNA were used for quality assurance in each run. LNs that exceeded the specified maximum weight of 600 mg were cut into two or more pieces and processed as separate nodes. The number of CK19 mRNA copies/µl was calculated and the result assessed in accordance with the following cut-off classification levels: ≥ 5000: positive (++); 250 to < 5000: positive (+); < 250 and ≥ 250 in diluted (as opposed to measurement) sample: positive with reaction inhibited (+i); < 250: negative. All SLNs and a small number of non-SLNs were assessed intraoperatively. Almost all non-SLNs in ALND specimens were assessed postoperatively after freezing at –80 °C. The frozen non-SLNs were assessed in the same manner as fresh nodes at a later date Reference standard (technical details): all non-SLNs were sliced in half along the long axis after formalin fixation. One of the cut surfaces was examined after haematoxylin–eosin staining. Five to seven nodes were embedded in paraffin in one cassette. Immunohistochemistry was not used for evaluation of non-SLNs. The non-SLN specimens were classified into three categories according to the 7th American Joint Committee on Cancer staging manual: positive, micrometastasis, negative, ITCs (< 0.2 mm) or no tumour cells Details of SLN detection: the radioisotope tracer used was 1.5 mCi/ml of technetium-99m phytate. A day before surgery, the tracer was injected in the area of the tumour and the retrotumoural space. In addition, 2–3 ml of vital dye, indigocarmine, was injected into the peritumoral space or areola at the time of surgery Extraction and division of SLNs: all non-SLNs were sliced in half along the long axis after formalin fixation. Discussion refers to three-level histology, although this is not clear Outcome assessor: NR Blinding: NA Discordant case analysis: NA |
Accuracy outcome: positive rate Unit of analysis: patient Discordant case analysis: NA Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NA, not applicable; NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNAHistologyPatients, n11964Age (years), median (range)53 (27–86)56 (39–81)Nodal status, n (%) pN0 pN1115 (96.6)62 (96.9) pN23 (3.4)2 (3.1) pN3Histopathological type, n (%) Invasive ductal carcinoma110 (92.4)57 (89.1) Invasive lobular carcinoma4 (3.4)2 (3.1) DCIS Other/special type5 (4.2)5 (7.8)HER2, n (%) Negative106 (89.1)55 (85.9) Positive13 (10.9)9 (14.1)HER2, human epidermal growth factor receptor 2. | Characteristic | OSNA | Histology | Patients, n | 119 | 64 | Age (years), median (range) | 53 (27–86) | 56 (39–81) | Nodal status, n (%) | pN0 | pN1 | 115 (96.6) | 62 (96.9) | pN2 | 3 (3.4) | 2 (3.1) | pN3 | Histopathological type, n (%) | Invasive ductal carcinoma | 110 (92.4) | 57 (89.1) | Invasive lobular carcinoma | 4 (3.4) | 2 (3.1) | DCIS | Other/special type | 5 (4.2) | 5 (7.8) | HER2, n (%) | Negative | 106 (89.1) | 55 (85.9) | Positive | 13 (10.9) | 9 (14.1) | HER2, human epidermal growth factor receptor 2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Characteristic | OSNA | Histology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 119 | 64 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | 53 (27–86) | 56 (39–81) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1 | 115 (96.6) | 62 (96.9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2 | 3 (3.4) | 2 (3.1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 110 (92.4) | 57 (89.1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 4 (3.4) | 2 (3.1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other/special type | 5 (4.2) | 5 (7.8) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 106 (89.1) | 55 (85.9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 13 (10.9) | 9 (14.1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TestPositive rate for non-SLNs (%) (95% CI)Histology20.3 (11.7 to 32.6)OSNA55.5 (46.1 to 64.5) SLN stageOverall axillary stage for histologypN1mipN1apN2apN3aUpstaging rate (%)nn%n%n%n%pN1mi (sn)211990.514.814.8009.5pN1a (sn)41––3482.949.837.317.1pN2a (sn)2––––2100000All641929.73554.7710.934.714.1mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. SLN stageOverall axillary stage for OSNApN1mipN1apN2apN3aUpstaging rate (%)nn%n%n%n%pN1mi (sn)504386.0612.00012.014.0pN1a (sn)65––5483.1913.823.116.9pN2a (sn)4––––250.0250.050.0All1194336.16050.4119.254.216.8mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. |
Test | Positive rate for non-SLNs (%) (95% CI) | Histology | 20.3 (11.7 to 32.6) | OSNA | 55.5 (46.1 to 64.5) | SLN stage | Overall axillary stage for histology | pN1mi | pN1a | pN2a | pN3a | Upstaging rate (%) | n | n | % | n | % | n | % | n | % | pN1mi (sn) | 21 | 19 | 90.5 | 1 | 4.8 | 1 | 4.8 | 0 | 0 | 9.5 | pN1a (sn) | 41 | – | – | 34 | 82.9 | 4 | 9.8 | 3 | 7.3 | 17.1 | pN2a (sn) | 2 | – | – | – | – | 2 | 100 | 0 | 0 | 0 | All | 64 | 19 | 29.7 | 35 | 54.7 | 7 | 10.9 | 3 | 4.7 | 14.1 | mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. | SLN stage | Overall axillary stage for OSNA | pN1mi | pN1a | pN2a | pN3a | Upstaging rate (%) | n | n | % | n | % | n | % | n | % | pN1mi (sn) | 50 | 43 | 86.0 | 6 | 12.0 | 0 | 0 | 1 | 2.0 | 14.0 | pN1a (sn) | 65 | – | – | 54 | 83.1 | 9 | 13.8 | 2 | 3.1 | 16.9 | pN2a (sn) | 4 | – | – | – | – | 2 | 50.0 | 2 | 50.0 | 50.0 | All | 119 | 43 | 36.1 | 60 | 50.4 | 11 | 9.2 | 5 | 4.2 | 16.8 | mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Test | Positive rate for non-SLNs (%) (95% CI) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histology | 20.3 (11.7 to 32.6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | 55.5 (46.1 to 64.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SLN stage | Overall axillary stage for histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1mi | pN1a | pN2a | pN3a | Upstaging rate (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n | n | % | n | % | n | % | n | % | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1mi (sn) | 21 | 19 | 90.5 | 1 | 4.8 | 1 | 4.8 | 0 | 0 | 9.5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1a (sn) | 41 | – | – | 34 | 82.9 | 4 | 9.8 | 3 | 7.3 | 17.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2a (sn) | 2 | – | – | – | – | 2 | 100 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All | 64 | 19 | 29.7 | 35 | 54.7 | 7 | 10.9 | 3 | 4.7 | 14.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SLN stage | Overall axillary stage for OSNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1mi | pN1a | pN2a | pN3a | Upstaging rate (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n | n | % | n | % | n | % | n | % | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1mi (sn) | 50 | 43 | 86.0 | 6 | 12.0 | 0 | 0 | 1 | 2.0 | 14.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1a (sn) | 65 | – | – | 54 | 83.1 | 9 | 13.8 | 2 | 3.1 | 16.9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2a (sn) | 4 | – | – | – | – | 2 | 50.0 | 2 | 50.0 | 50.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All | 119 | 43 | 36.1 | 60 | 50.4 | 11 | 9.2 | 5 | 4.2 | 16.8 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| mi, micrometastases (> 0.2 mm and/or > 200 cells but none > 2.0 mm); sn, sentinel node evaluation performed after treatment. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: unclear whether histopathology results were checked by an independent pathologist Conflict of interest: consumables funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | NA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Schem et al.60
| Design | Participants | Tests | Outcomes | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to evaluate the performance of OSNA compared with that of histology Study design: single gate Country: Germany Number of centres: 2 Funding: Sysmex |
Number of participants: 93 Number of SLNs or ALNs: 343 ALNs Recruitment procedure: NR Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: NR |
Index test (technical details): two alternate LN slices out of four were homogenised together. The LN lysates were stored at −80 °C until further use. A CK19 mRNA copy number of < 250/µl was regarded as −; copy numbers between 250 and 5000/µl were regarded as +; and copy numbers > 5000/µl were regarded as ++ Reference standard (technical details): the two remaining LN slices were fixed with neutral buffered formaldehyde and embedded in the same paraffin block. Each slice was identified by colour coding. Two initial haematoxylin–eosin sections (representing frozen sections of the SLN), one initial level and four additional levels with a 0.1-mm skip space were cut from the 343 blocks. Each level consisted of four 4-µm sections: one was used for haematoxylin–eosin staining, one for immunohistochemistry with the pan-anti-CK antibody LU5, one for CK19 immunohistochemistry and one was a spare section. For the specificity study, the paraffin blocks of 120 histologically negative samples, analysed by five-level histology, were cut into further levels until depletion. Immunohistochemistry was then performed and deparaffinised sections were cooked in tris–ethylenediaminetetraacetic acid–sodium citrate buffer (pH 7.8) for 4 minutes. After blocking, incubation with the primary antibody was performed for 40 minutes and with the secondary antibody for 30 minutes. Visualisation was achieved with diaminobenzidine tetrahydrochloride and staining employed the LU5 antibody. Metastatic deposits were recorded, according to the TNM classification of the Union for International Cancer Control (sixth edition) and the American Joint Committee on Cancer (sixth edition), as ITCs if their largest diameter was < 0.2 mm, as micrometastases if they were > 0.2 mm and ≤ 2 mm in diameter and as macrometastases if they were > 2 mm in diameter. LNs with ITCs were considered as negative Details of SLN detection: NR Extraction and division of SLNs: 343 LN samples were longitudinally cut into four nearly equal slices with a special cutting tool consisting of three blades either 1 or 2 mm apart. ALNs were categorised according to their size: ALNs with a minor axis < 0.4 cm were excluded from the analysis; ALNs with a minor axis between 0.4 and 0.6 cm (group 1) were centrally cut into four slices with the 1-mm cutting tool; ALNs between 0.6 and 1.0 cm (group 2) were centrally cut into four slices with a 2-mm cutting tool; ALNs with a minor axis > 1.0 cm (group 3) were either halved or cut into several pieces, and each piece, depending on its size, was treated in a similar fashion as described for groups 1 and 2. Alternate slices were allocated to the OSNA method and to histology at five levels. The slices used for OSNA were shock frozen in liquid nitrogen and stored at −80 °C before the analysis Outcome assessor: NR Blinding: Yes Discordant case analysis: cases with discordant results between the OSNA assay and five-level histological examination were subjected to extended histological work-up that involved slicing the remaining tissue in the paraffin blocks until depletion. The homogenates of these discordant cases were also analysed by Western blotting and qRT-PCR. When these supplemental analyses gave the same result as the OSNA assay, the related samples were excluded from the analysis as TAB was likely to be the cause of the discrepant results |
Accuracy outcomes: concordance, sensitivity, specificity Unit of analysis: node Discordant case analysis: yes Test failures: NR |
|||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | ||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | ||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n93Clinical tumour classification, n T0 Tis T16 T236 T34 T41Nodal status, n pN046 pN127 pN213 pN37Histopathological type, n Invasive ductal carcinoma68 Invasive lobular carcinoma21 DCIS Other (mixed)4 | Characteristic | OSNA plus histology | Patients, n | 93 | Clinical tumour classification, n | T0 | Tis | T1 | 6 | T2 | 36 | T3 | 4 | T4 | 1 | Nodal status, n | pN0 | 46 | pN1 | 27 | pN2 | 13 | pN3 | 7 | Histopathological type, n | Invasive ductal carcinoma | 68 | Invasive lobular carcinoma | 21 | DCIS | Other (mixed) | 4 | ||||||||||||
| Characteristic | OSNA plus histology | |||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 93 | |||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n | ||||||||||||||||||||||||||||||||||||||||||||
| T0 | ||||||||||||||||||||||||||||||||||||||||||||
| Tis | ||||||||||||||||||||||||||||||||||||||||||||
| T1 | 6 | |||||||||||||||||||||||||||||||||||||||||||
| T2 | 36 | |||||||||||||||||||||||||||||||||||||||||||
| T3 | 4 | |||||||||||||||||||||||||||||||||||||||||||
| T4 | 1 | |||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n | ||||||||||||||||||||||||||||||||||||||||||||
| pN0 | 46 | |||||||||||||||||||||||||||||||||||||||||||
| pN1 | 27 | |||||||||||||||||||||||||||||||||||||||||||
| pN2 | 13 | |||||||||||||||||||||||||||||||||||||||||||
| pN3 | 7 | |||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | ||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 68 | |||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 21 | |||||||||||||||||||||||||||||||||||||||||||
| DCIS | ||||||||||||||||||||||||||||||||||||||||||||
| Other (mixed) | 4 | |||||||||||||||||||||||||||||||||||||||||||
| Results | ||||||||||||||||||||||||||||||||||||||||||||
| Before TAB exclusion OSNAFive-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 343 ALNs++90709+7–116–022209 Sensitivity (%)Specificity (%)Discordance (%)n = 343 ALNs before exclusion for TAB98.191.78.2n = 330 ALNs after exclusion for TAB10095.64.5 |
OSNA | Five-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 343 ALNs | ++ | 90 | 7 | 0 | 9 | + | 7 | – | 1 | 16 | – | 0 | 2 | 2 | 209 | Sensitivity (%) | Specificity (%) | Discordance (%) | n = 343 ALNs before exclusion for TAB | 98.1 | 91.7 | 8.2 | n = 330 ALNs after exclusion for TAB | 100 | 95.6 | 4.5 | |||||||||||
| OSNA | Five-level histopathology | |||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||
| n = 343 ALNs | ||||||||||||||||||||||||||||||||||||||||||||
| ++ | 90 | 7 | 0 | 9 | ||||||||||||||||||||||||||||||||||||||||
| + | 7 | – | 1 | 16 | ||||||||||||||||||||||||||||||||||||||||
| – | 0 | 2 | 2 | 209 | ||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | ||||||||||||||||||||||||||||||||||||||||||
| n = 343 ALNs before exclusion for TAB | 98.1 | 91.7 | 8.2 | |||||||||||||||||||||||||||||||||||||||||
| n = 330 ALNs after exclusion for TAB | 100 | 95.6 | 4.5 | |||||||||||||||||||||||||||||||||||||||||
| Methodological issues | ||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: unclear whether histopathology results were checked by an independent pathologist Conflict of interest: consumables funded by Sysmex |
||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | ||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||
Snook et al.61
| Design | Participants | Tests | Outcomes | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to evaluate OSNA as a potential intraoperative diagnostic tool via a multicentre prospective study that was undertaken to reassess the accuracy of OSNA diagnosis compared with intensive histopathological examination and to investigate the feasibility of intraoperative use of OSNA to diagnose LN metastases Study design: single gate Country: UK Number of centres: 4 Funding: The Juniper Trust and BUFFER (Breast Unit Fund for Education and Research), both registered charities, funded the salary of a clinical research fellow, who was registered with the University of Surrey during the period of her MD research. There was no financial contribution from any commercial organisation Notes: the study was undertaken in two phases. The technical performance phase was designed to familiarise each site with the molecular biological test. The technical performance and accuracy of the OSNA method of diagnosing breast cancer LN metastasis was compared with histology using both sentinel and non-sentinel axillary nodes for analysis. LN specimens for OSNA analysis were snap frozen at −80 °C for analysis at a time suitable for the laboratory. Each site had to achieve a concordance of at least 80% (94% concordance was achieved across the four sites following discordant case analysis) in a minimum of 40 LNs before starting the second phase. This next phase, the clinical equivalence study, was designed to investigate the feasibility of the intraoperative use of OSNA. Only SLNs were used for this phase. LN specimens were analysed by OSNA immediately on arrival at the histopathology laboratory to simulate the intraoperative scenario |
Number of participants: 204 Number of SLNs or ALNs: 393 LNs, dissected to 417 samples Recruitment procedure: NR Inclusion criteria: SLNs from patients with a preoperative diagnosis of breast carcinoma, undergoing mastectomy or breast-conserving surgery, were identified and removed surgically using the standard technique employed at each study site Exclusion criteria: patients who had undergone neoadjuvant chemotherapy and those with a previous diagnosis of a potentially metastatic malignancy were excluded from the study Sample attrition/dropout: NR |
Index test (technical details): lysates of homogenised LN samples were prepared manually before amplification. OSNA provided a qualitative result (++, + or –) and a quantitative result (copy number of CK19 mRNA): (++)/copy number > 5000/µl (macrometastasis, > 2 mm); (+)/copy number 250–5000/µl (micrometastasis, > 0.2 mm to ≤ 2 mm); (−)/copy number 0–250/µl (negative). Simultaneous positive and negative controls were performed with the specimen analysis to ensure quality control Reference standard (technical details): one initial ribbon and additional ribbons with a 0.25-mm skip space were cut from two out of four alternate slices. From the initial ribbon and from the subsequent four ribbons (giving a total of five levels, equating to 10 levels of analysis in total as two separate slices of node were analysed), three 3-µm sections were used for haematoxylin–eosin staining, standard immunohistochemistry with pan-CK clone AE1/AE3 and CK19 immunohistochemistry. In the event of discordance, further levels were taken from the remaining ribbons, using the same protocol, until the entire paraffin block had been examined. Histopathology slides were examined by pathologists masked to the results of OSNA Details of SLN detection: a combination of injected radioisotope, scintigraphy, hand-held gamma probe and blue dye injection was used for the identification of SLNs. SLNs were identified according to the New Start criteria (hot, blue, a combination of both, palpable and suspicious) Extraction and division of SLNs: on arrival in the histopathology laboratory, each node was categorised into one of four groups according to its size and sliced longitudinally with a purpose-designed cutting instrument into four 1- or 2-mm slices. The total weight of each node for slicing did not exceed 1.2 g; nodes weighing > 1.2 g were divided into portions (giving two or more node samples) before slicing. Two alternate slices were colour coded, immediately fixed with neutral buffered formaldehyde and processed to paraffin blocks for histopathological analysis. The remaining two slices were allocated to OSNA analysis and snap frozen at −80°C (technical performance phase) or analysed immediately for the clinical equivalence study Outcome assessor: NR Blinding: yes Discordant case analysis: stored homogenate was analysed further by qRT–PCR. If the discordant case investigation supported the OSNA result, it was concluded that TAB had occurred and the respective samples were excluded from the analysis. Total RNA was extracted from the homogenates of discordant samples. qRT-PCR for breast cancer-specific markers was then carried out with CK19, SPDEF (SAM pointed domain containing Ets transcription factor) and FOXA1 (forkhead box A1). In addition, Western blot analysis for CK19 using 20 µl of lysate was performed. At least one marker in addition to beta-actin had to be positive to classify a result as truly discordant |
Accuracy outcomes: concordance, sensitivity, specificity Process outcome: time to test Unit of analysis: patient and node Discordant case analysis: yes Test failures: yes |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyClinical tumour classification, n T0 Tis T1133 T260 T35 T4Histopathological type, n (%) Invasive ductal carcinoma160 (78.8) Invasive lobular carcinoma22 (10.8) DCIS Other16 (7.9) | Characteristic | OSNA plus histology | Clinical tumour classification, n | T0 | Tis | T1 | 133 | T2 | 60 | T3 | 5 | T4 | Histopathological type, n (%) | Invasive ductal carcinoma | 160 (78.8) | Invasive lobular carcinoma | 22 (10.8) | DCIS | Other | 16 (7.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Characteristic | OSNA plus histology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 133 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 60 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | 5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 160 (78.8) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 22 (10.8) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 16 (7.9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| After TAB exclusion OSNAFive-level histopathologyMacrometastasisMicrometastasisITCNegativen = 395 SLNs++48100+89010–4220293n = 194 patients++33100+5517–4111126 Sensitivity (%)Specificity (%)Discordance (%)n = 417n = 194 patients after exclusion for TAB89.894.5n = 395 SLNs after exclusion for TAB91.796.9 Nodes, nMedian (range) time to analysis (minutes)132 (22–97)242 (30–73)351 (38–73)462 (46–90) Test failures: six technical errors reported |
OSNA | Five-level histopathology | Macrometastasis | Micrometastasis | ITC | Negative | n = 395 SLNs | ++ | 48 | 1 | 0 | 0 | + | 8 | 9 | 0 | 10 | – | 4 | 2 | 20 | 293 | n = 194 patients | ++ | 33 | 1 | 0 | 0 | + | 5 | 5 | 1 | 7 | – | 4 | 1 | 11 | 126 | Sensitivity (%) | Specificity (%) | Discordance (%) | n = 417 | n = 194 patients after exclusion for TAB | 89.8 | 94.5 | n = 395 SLNs after exclusion for TAB | 91.7 | 96.9 | Nodes, n | Median (range) time to analysis (minutes) | 1 | 32 (22–97) | 2 | 42 (30–73) | 3 | 51 (38–73) | 4 | 62 (46–90) | ||||||||||||||||||||
| OSNA | Five-level histopathology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITC | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 395 SLNs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 48 | 1 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 8 | 9 | 0 | 10 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 4 | 2 | 20 | 293 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 194 patients | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 33 | 1 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 5 | 5 | 1 | 7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 4 | 1 | 11 | 126 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 417 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 194 patients after exclusion for TAB | 89.8 | 94.5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 395 SLNs after exclusion for TAB | 91.7 | 96.9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodes, n | Median (range) time to analysis (minutes) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | 32 (22–97) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 42 (30–73) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 51 (38–73) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 62 (46–90) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: unclear whether histopathology results were checked by an independent pathologist |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sundaresan (unpublished)
| Design | Participants | Tests | Outcomes | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: NR Study design: to describe the validation of Metasin, a novel real time PCR assay for the detection of metastatic cancer in SLNs from breast cancer patients Country: UK Number of centres: 6 centres Funding: NR |
Number of participants: 1265 Number of SLNs or ALNs: 2279 SLNs Recruitment procedure: NR Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: NR |
Index (technical details): determination of the cut-off values was carried out relative to Veridex data and morphology, for verification of the thresholds for macrometastasis (> 2 mm) and micrometastasis (< 2 mm and > 0.2 mm) categories. The Cp values were determined for CK19 (Cp values < 25) and mammaglobin (< 25.9). CK19 thresholds for micrometastasis (CK19 > 25 and < 32) and mammoglobin (Cp > 25.9 and < 32) were similarly identified. For RNA extraction and quantification, the research protocol was adopted from the GeneSearch assay. Further details have been published since this review was completed159 Reference standard (technical details): SLNs were sectioned at three levels/steps of 150 µm. Nodal micrometastatic (< 2 mm and > 0.2 mm) and macrometastatic (> 2 mm) disease were interpreted as positive Details of SLN detection: SLNs were identified by a combination of the use of blue dye and radiation, according to an established conventional protocol following the New Start programme Extraction and division of SLN: six centres contributed tissue homogenates and RNA from patients treated for breast cancer. Two centres provided only frozen RNA. The remaining institutions contributed lymph node homogenates stored at –80 °C. LNs were serially sliced in the longitudinal plane into an even number of approximately 2-mm slices. Alternate slices were submitted for conventional histopathological analysis and for homogenisation and RNA preparation Discordance analysis: cases were followed up by examination of the block by extra levels and selectively examining with MNF116 immunostaining. Cases in which the molecular assay was positive but histology was negative and for which homogenates were available for analysis were subject to RNA re-extraction and examination with an independent panel of markers. Retrospective discordant case analysis could not be uniformly followed Outcome assessor: NR Blinding: NR |
Accuracy outcomes: sensitivity, specificity and concordance Clinical outcomes: NR Other: NR Unit of analysis: patient Discordant case analysis: yes Test failures: 1.2% – insufficient RNA |
|||||||||||||||||||||||||||||||||||||
| NR, not reported. | ||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | ||||||||||||||||||||||||||||||||||||||||
| Not reported | ||||||||||||||||||||||||||||||||||||||||
| Results | ||||||||||||||||||||||||||||||||||||||||
| MetasinThree-level histopathologyPositiveNegativen = 1265 patientsaPositive24936Negative20940a 20 tests failed for 20 patients and were discarded. Sensitivity (%)Specificity (%)Discordance (%)n = 1245 patientsa92.696.34.5a Excludes 20 patients with failed tests. Nodes (n)Median time to analysis (minutes)136242346 Test failure – 1.2% because of insufficient mRNA in the sample |
Metasin | Three-level histopathology | Positive | Negative | n = 1265 patientsa | Positive | 249 | 36 | Negative | 20 | 940 | a 20 tests failed for 20 patients and were discarded. | Sensitivity (%) | Specificity (%) | Discordance (%) | n = 1245 patientsa | 92.6 | 96.3 | 4.5 | a Excludes 20 patients with failed tests. | Nodes (n) | Median time to analysis (minutes) | 1 | 36 | 2 | 42 | 3 | 46 | ||||||||||||
| Metasin | Three-level histopathology | |||||||||||||||||||||||||||||||||||||||
| Positive | Negative | |||||||||||||||||||||||||||||||||||||||
| n = 1265 patientsa | ||||||||||||||||||||||||||||||||||||||||
| Positive | 249 | 36 | ||||||||||||||||||||||||||||||||||||||
| Negative | 20 | 940 | ||||||||||||||||||||||||||||||||||||||
| a 20 tests failed for 20 patients and were discarded. | ||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | ||||||||||||||||||||||||||||||||||||||
| n = 1245 patientsa | 92.6 | 96.3 | 4.5 | |||||||||||||||||||||||||||||||||||||
| a Excludes 20 patients with failed tests. | ||||||||||||||||||||||||||||||||||||||||
| Nodes (n) | Median time to analysis (minutes) | |||||||||||||||||||||||||||||||||||||||
| 1 | 36 | |||||||||||||||||||||||||||||||||||||||
| 2 | 42 | |||||||||||||||||||||||||||||||||||||||
| 3 | 46 | |||||||||||||||||||||||||||||||||||||||
| Methodological issues | ||||||||||||||||||||||||||||||||||||||||
| See STARD table (Chapter 3, Results of test accuracy, Table 7) | ||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | ||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | |||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | |||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | |||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (H/L/U) | N | |||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | |||||||||||||||||||||||||||||||||||||||
Tamaki et al.63
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to determine the usefulness of the OSNA assay for clinical use in SLNB of breast cancer Study design: single gate Country: Japan Number of centres: 11 Funding: Nakatani Foundation of Electronic Measuring Technology Advancement |
Number of participants: 439 Number of SLNs or ALNs: 775 Recruitment procedure: NR Inclusion criteria: patients with Tis to T2, clinically LN-negative primary breast cancer who underwent SLNB between August 2009 and December 2010 at one of the participating hospitals. Patients who had a preoperative diagnosis of DCIS were enrolled in the study when a surgeon judged that SLNB was needed. Patients who underwent SLNB before receiving preoperative systemic chemotherapy were also eligible for the analysis of sensitivity of the OSNA assay Exclusion criteria: those who received chemotherapy or hormone therapy before SLNB were excluded from the study. Men were also excluded Sample attrition/dropout: 21 of the originally enrolled patients were excluded from the analysis because of significant violations against the study protocol, including eight patients who received preoperative systemic chemotherapy before SLNB, 10 patients who were not examined with the OSNA assay, two patients whose central sections of the SLN did not undergo pathological examination as a permanent specimen for haematoxylin–eosin staining and one patient who was a man. Two patients who had benign intraductal papilloma confirmed after surgery, one who had a clinical T4 tumour and two who had clinically evident ALN metastases were also excluded because they did not meet the general criteria for SLNB candidates. Conversely, two patients who had T3 tumours that finally were diagnosed as DCIS and T1 invasive cancer were included. The final total enrolment was 413 patients who had 417 SLNBs eligible for analysis |
Index test (technical details): a SLN was assessed with the OSNA assay according to the cut-off levels of calculated CK19 mRNA copy number/µl determined by Tsujimoto et al.64 and the results were classified as negative (< 250 copies/µl); (+) positive (> 250 and < 5000 copies/µl); (++) positive (5000 copies/µl); or (+i) positive (inhibited in the regular sample and > 250 copies/µl in the diluted sample) Reference standard (technical details): a 1-mm slice was cut from the longitudinal central part of the SLN, fixed as a permanent section for staining with H&E and examined postoperatively by a pathologist Details of SLN detection: SLNs were detected using both radiocolloids and blue dye, radiocolloids only or blue dye only Extraction and division of SLNs: removed SLNs were assessed immediately with OSNA. Patients had ALND recommended according to OSNA and/or other clinicopathological factors. Non-SLNs underwent routine pathological examination using H&E staining. Fat tissue surrounding the SLN was trimmed off. A 1-mm thick slice was then cut out from the longitudinal central part of the SLN, fixed as a permanent section for staining with haematoxylin–eosin and examined postoperatively by a pathologist. The remaining part of the LN was immediately examined with the OSNA assay Outcome assessor: NR Blinding: yes Discordant case analysis: discussed but not analysed |
Accuracy outcomes: positive rate, concordance Unit of analysis: patient Discordant case analysis: discussed but not analysed Test failures: unclear – although 98.3% examined successfully with OSNA |
||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n439Age (years), mean (range)56.1 (25–90)Clinical stage, n (%) 0 I183 (43.9) II110 (26.4) III70 (16.8) IV Unknown54 (12.9)Clinical tumour classification, n (%) T0 Tis50 (12) T1254 (60.9) T2111 (26.6) T32 (0.5) T4Histopathological type, n (%) Invasive ductal carcinoma305 (73.1) Invasive lobular carcinoma24 (5.8) DCIS53 (12.7) Other35 (8.4)HER2 , n (%) Positive51 (12.2) Negative334 (87.8)HER2, human epidermal growth factor receptor 2. | Characteristic | OSNA plus histology | Patients, n | 439 | Age (years), mean (range) | 56.1 (25–90) | Clinical stage, n (%) | 0 | I | 183 (43.9) | II | 110 (26.4) | III | 70 (16.8) | IV | Unknown | 54 (12.9) | Clinical tumour classification, n (%) | T0 | Tis | 50 (12) | T1 | 254 (60.9) | T2 | 111 (26.6) | T3 | 2 (0.5) | T4 | Histopathological type, n (%) | Invasive ductal carcinoma | 305 (73.1) | Invasive lobular carcinoma | 24 (5.8) | DCIS | 53 (12.7) | Other | 35 (8.4) | HER2 , n (%) | Positive | 51 (12.2) | Negative | 334 (87.8) | HER2, human epidermal growth factor receptor 2. | ||||||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 439 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), mean (range) | 56.1 (25–90) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I | 183 (43.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| II | 110 (26.4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| III | 70 (16.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IV | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unknown | 54 (12.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical tumour classification, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tis | 50 (12) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T1 | 254 (60.9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T2 | 111 (26.6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T3 | 2 (0.5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| T4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 305 (73.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 24 (5.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 53 (12.7) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 35 (8.4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2 , n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 51 (12.2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 334 (87.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HER2, human epidermal growth factor receptor 2. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNAOne-level histopathologyPositiveNegativen = 417 patients (SLN)Positive5836Negative8315 Discordance (%)n = 417 patients5.7 |
OSNA | One-level histopathology | Positive | Negative | n = 417 patients (SLN) | Positive | 58 | 36 | Negative | 8 | 315 | Discordance (%) | n = 417 patients | 5.7 | |||||||||||||||||||||||||||||||||||||||||
| OSNA | One-level histopathology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 417 patients (SLN) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 58 | 36 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 8 | 315 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Discordance (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 417 patients | 5.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: unclear whether histopathology results were checked by an independent pathologist Recruitment: unclear if recruitment was consecutive or random |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tamaki et al.62
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to develop a more efficient method for intraoperative detection of LN metastasis Study design: single gate Country: Japan Number of centres: 6 Funding: not known – Sysmex had some involvement in the study Notes: trial 1 – full histology; trial 2 – frozen section then full histology |
Number of participants: two trials: trial 1: n = 36 patients, n = 149 nodes; trial 2: n = 185 patients, n = 551 nodes Recruitment procedure: unknown Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: trial 1: five nodes, one patient withdrew; 19 nodes – lack of lymphatic tissue; one node – technical error. Trial 2: eight nodes, three patients withdrew; 26 nodes – six patients had neoadjuvant chemotherapy; 36 nodes – lack of lymphatic tissue; 31 nodes did not meet the study specification |
Index test (technical details): pieces obtained from ALNs were homogenised. A LN was assessed as negative when there were < 250 copies/µl of CK19 mRNA and positive when there were ≥ 250 copies/µl Reference standard (technical details): in the case of LNs from pN0 patients, two out of four alternate blocks were further sliced at 0.2-mm intervals, followed by examination of each alternate slice with haematoxylin–eosin and CK19 immunohistochemistry. A total of 144 LNs in which neither micrometastases nor macrometastases were observed were used for the study of false-positive results with OSNA Details of SLN detection: NR Extraction and division of SLNs: a fresh LN with a short axis of 4–12 mm was divided into four blocks at 1- or 2-mm intervals. Two alternate blocks were used for OSNA. Two slices were cut from each of the three cutting surfaces and used for the permanent three-level histopathological examination with haematoxylin–eosin and CK19 immunohistochemistry Outcome assessor: histology was checked by independent pathologists Blinding: NR Discordant case analysis (trial 2 only): discordant results between OSNA and three-level histopathology led to repeated histopathological analysis of blocks assigned to histopathology. All slides were examined and evaluated by three independent pathologists. The final analysis of all results of the histopathological examinations was performed by a study group comprised of representatives from the different facilities |
Accuracy outcomes: concordance, sensitivity, specificity Unit of analysis: node Discordant case analysis: yes Test failures: one technical error |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyTrial 1Trial 2Patients, n36185Age (years), mean55.954.7Clinical stage, n (%) 02 (6)14 (9) I8 (24)51 (31) IIA14 (41)64 (40) IIB3 (9)28 (17) III5 (15)7 (4) IV00 Unknown2 (6)0Histopathological type, n (%) Invasive ductal carcinoma32 (94)130 (79) Invasive lobular carcinoma1 (3)7 (4) DCIS018 (11) Other1 (3)9 (5) | Characteristic | OSNA plus histology | Trial 1 | Trial 2 | Patients, n | 36 | 185 | Age (years), mean | 55.9 | 54.7 | Clinical stage, n (%) | 0 | 2 (6) | 14 (9) | I | 8 (24) | 51 (31) | IIA | 14 (41) | 64 (40) | IIB | 3 (9) | 28 (17) | III | 5 (15) | 7 (4) | IV | 0 | 0 | Unknown | 2 (6) | 0 | Histopathological type, n (%) | Invasive ductal carcinoma | 32 (94) | 130 (79) | Invasive lobular carcinoma | 1 (3) | 7 (4) | DCIS | 0 | 18 (11) | Other | 1 (3) | 9 (5) | ||||||||||||||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Trial 1 | Trial 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 36 | 185 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), mean | 55.9 | 54.7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | 2 (6) | 14 (9) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I | 8 (24) | 51 (31) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IIA | 14 (41) | 64 (40) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IIB | 3 (9) | 28 (17) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| III | 5 (15) | 7 (4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IV | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unknown | 2 (6) | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 32 (94) | 130 (79) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 1 (3) | 7 (4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 0 | 18 (11) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other | 1 (3) | 9 (5) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Before TAB exclusions 0.2-mm section histopathologyOSNAMacrometastasisMicrometastasisITCsNegativeTrial 1: n = 124 ALNsPositive1633Negative01101Three-level histopathologyOSNAMacrometastasisMicrometastasisITCsNegativeTrial 2: n = 450 ALNsPositive64622Negative46348 Sensitivity (%) (95% CI)Specificity (%) (95% CI)Discordance (%)Trial 1: n = 124 ALNs95 (75.1–99.9)97.1 (91.8–99.4)3.2Trial 2: n = 450 ALNs87.5 (78.2–93.8)94.1 (91.0–96.3)7.1 |
0.2-mm section histopathology | OSNA | Macrometastasis | Micrometastasis | ITCs | Negative | Trial 1: n = 124 ALNs | Positive | 16 | 3 | 3 | Negative | 0 | 1 | 101 | Three-level histopathology | OSNA | Macrometastasis | Micrometastasis | ITCs | Negative | Trial 2: n = 450 ALNs | Positive | 64 | 6 | 22 | Negative | 4 | 6 | 348 | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | Trial 1: n = 124 ALNs | 95 (75.1–99.9) | 97.1 (91.8–99.4) | 3.2 | Trial 2: n = 450 ALNs | 87.5 (78.2–93.8) | 94.1 (91.0–96.3) | 7.1 | ||||||||||||||||||||||||
| 0.2-mm section histopathology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Trial 1: n = 124 ALNs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 16 | 3 | 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 0 | 1 | 101 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Three-level histopathology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNA | Macrometastasis | Micrometastasis | ITCs | Negative | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Trial 2: n = 450 ALNs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Positive | 64 | 6 | 22 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Negative | 4 | 6 | 348 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Discordance (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Trial 1: n = 124 ALNs | 95 (75.1–99.9) | 97.1 (91.8–99.4) | 3.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Trial 2: n = 450 ALNs | 87.5 (78.2–93.8) | 94.1 (91.0–96.3) | 7.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: unclear whether histopathology results were checked by an independent pathologist Recruitment: unclear if recruitment was consecutive or random Conflict of interest: the study was funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tsujimoto et al.64
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to develop a more efficient method for intraoperative detection of LN metastasis Study design: single gate Country: Japan Number of centres: 6 Funding: NR |
Number of participants: 101 patients (81 SLNs from 49 patients) Number of SLNs or ALNs: 325 SLNs and ALNs, 81 SLNs Recruitment procedure: NR Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: NR |
Index test (technical details): a histopathologically negative LN (≤ 600 mg) was homogenised and centrifuged at 10,000 g for 1 minute at room temperature. A 20-µl sample of supernatant was subject to RT-LAMP (reverse transcription loop-mediated isothermal amplification) in a gene amplification detector (RD-100i) Reference standard (technical details): two slices were cut from each of the three cutting surfaces and used for permanent three-level histology with haematoxylin–eosin and CK19 immunohistochemistry. Macrometastasis and micrometastasis were defined according to the TNM classification of the Union for International Cancer Control (sixth edition) and the American Joint Committee on Cancer (sixth edition). All samples for histopathology were examined by independent pathologists Details of SLN detection: NR Extraction and division of SLNs: a fresh LN with a short axis of 4–12 mm was divided into four blocks at 1- or 2-mm intervals. Two out of four alternate blocks were used for OSNA. Two slices were cut from each of the three cutting surfaces and used for the permanent three-level histopathological examination with haematoxylin–eosin and CK19 immunohistochemistry Outcome assessor: three independent pathologists. Conflicting results were settled consensually Blinding: unclear, although blinded in paper by Tamaki et al.,62 which reports on the same trial Discordant case analysis: in cases with discordant results between OSNA and three-level histopathology occurred, a histopathological analysis of the two blocks assigned to histopathology was repeated. All slides were examined and evaluated by three independent pathologists. The final analysis of all results of the histopathological examinations was performed by a study group comprised of representatives from the different facilities. In the analysis of discordant cases, qRT-PCR and CK19 Western blot analysis of the lysates from discordant cases were carried out (further details of this process are provided in the paper). A cut-off value for CK19 protein expression between histopathologically positive and negative LNs was determined by Western blot analysis. The cut-off value was determined by statistical analysis of the amount of CK19 measured by Western blot analysis of 37 histopathologically negative LNs from 16 pN0 patients |
Accuracy outcomes: concordance, sensitivity, specificity Process outcome: time to analysis Unit of analysis: node Discordant case analysis: yes Test failures: NR |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n101Age (years), median (range)NRClinical stage, n 05 I41 II49 III5 IV1Nodal status, n pN060 pN135 pN22 pN34Histopathological type, n Invasive ductal carcinoma87 Invasive lobular carcinoma4 DCIS5 Others5 | Characteristic | OSNA plus histology | Patients, n | 101 | Age (years), median (range) | NR | Clinical stage, n | 0 | 5 | I | 41 | II | 49 | III | 5 | IV | 1 | Nodal status, n | pN0 | 60 | pN1 | 35 | pN2 | 2 | pN3 | 4 | Histopathological type, n | Invasive ductal carcinoma | 87 | Invasive lobular carcinoma | 4 | DCIS | 5 | Others | 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 101 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | NR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0 | 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I | 41 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| II | 49 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| III | 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IV | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN0 | 60 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN1 | 35 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN2 | 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| pN3 | 4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 87 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DCIS | 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Others | 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OSNAThree-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 325 SLNs and ALNs++34000+6304–0213263n = 81 SLNs++11000+1201–023610.2-mm interval histopathologyMacrometastasisMicrometastasisITCsNegativen = 144 SLNs from pN0 patients++0000+0000–003141 Sensitivity (%)Specificity (%)Discordance (%)325 nodes (ALNs or SLNs)91.1NR7.4 Time to analysis: < 30 minutes |
OSNA | Three-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 325 SLNs and ALNs | ++ | 34 | 0 | 0 | 0 | + | 6 | 3 | 0 | 4 | – | 0 | 2 | 13 | 263 | n = 81 SLNs | ++ | 11 | 0 | 0 | 0 | + | 1 | 2 | 0 | 1 | – | 0 | 2 | 3 | 61 | 0.2-mm interval histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 144 SLNs from pN0 patients | ++ | 0 | 0 | 0 | 0 | + | 0 | 0 | 0 | 0 | – | 0 | 0 | 3 | 141 | Sensitivity (%) | Specificity (%) | Discordance (%) | 325 nodes (ALNs or SLNs) | 91.1 | NR | 7.4 | |||||||||||||||||||||||
| OSNA | Three-level histopathology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 325 SLNs and ALNs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 34 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 6 | 3 | 0 | 4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 2 | 13 | 263 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 81 SLNs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 11 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 1 | 2 | 0 | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 2 | 3 | 61 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 0.2-mm interval histopathology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| n = 144 SLNs from pN0 patients | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ++ | 0 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| + | 0 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| – | 0 | 0 | 3 | 141 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 325 nodes (ALNs or SLNs) | 91.1 | NR | 7.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Analysis: histopathology results were checked by an independent pathologist Recruitment: unclear if recruitment was consecutive or random |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Visser et al.65
| Design | Participants | Tests | Outcomes | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Objective: to test the suitability of OSNA for intraoperative SLN analysis Study design: single gate Country: the Netherlands Number of centres: 2 Funding: Sysmex |
Number of participants: 32 Number of SLNs or ALNs: 346 ALNs and SLNs Recruitment procedure: NR Inclusion criteria: NR Exclusion criteria: NR Sample attrition/dropout: NR |
Index test (technical details): NR Reference standard (technical details): LNs were cut with the blades of the cutting device 1 mm apart for LNs with a minor axis of 4–6 mm and 2 mm apart for LNs with a minor axis of 6–10 mm. LNs with a minor axis > 10 mm were halved and the resulting pieces were then cut either in 1-mm or the 2-mm slices depending on the size of the pieces. From two alternate slices out of four, initially three 4 µm-thick sections were stained with haematoxylin–eosin, CAM5.2 (CK8) and an anti-CK19 antibody respectively. If the initial sections were tumour positive no further sections were cut. Otherwise, additional sections (n = 3) at further levels and at an interval of 250 µm (usually four) were cut and analysed. Separate sections containing non-neoplastic epithelial cells were included in each staining procedure as a positive control for both antibodies. The size of a metastasis was determined by measuring its largest diameter and the metastasis was categorised as ITCs (< 0.2 mm), micrometastasis (> 0.2 mm and < 2.0 mm) or macrometastasis (≥ 2.0 mm). LNs containing ITCs were recorded as LN negative and designated as N0(i+) according to the Union for International Cancer Control (sixth edition) TNM classification Details of SLN detection: NR Extraction and division of SLNs: LN samples were cut in four equal slices. Two of these slices, alternately chosen, were snap frozen in liquid nitrogen and stored at –80 °C until OSNA analysis was performed. The remaining two slices were fixed in 4% buffered formaldehyde and embedded in a single paraffin block for histological examination at five levels Outcome assessor: microscopic evaluation was carried out by two pathologists masked to the results of the OSNA analysis Blinding: no Discordant case analysis: the histological work-up was extended to all levels in the first 120 histologically negative LN samples and for paraffin blocks of discordant cases. The homogenised LN lysates of samples with discordant OSNA vs. histology results were subjected to qRT-PCR and Western blot analysis. In case these investigations yielded a result compatible with a positive OSNA result these samples were excluded from the final analysis as they were likely affected by sampling bias |
Accuracy outcomes: concordance, sensitivity, specificity Process outcomes: NR Clinical outcomes: NR Unit of analysis: ALN Discordant case analysis: yes Test failures: NR |
||||||||||||||||||||||||||||||||||||||||
| NR, not reported. | |||||||||||||||||||||||||||||||||||||||||||
| Participant characteristics | |||||||||||||||||||||||||||||||||||||||||||
| CharacteristicOSNA plus histologyPatients, n32Age (years), median (range)NRClinical stage, n 00 I8 II15 III7 IV2 UnknownNodal status, n pN014 pN110 pN26 pN32Histopathological type, n Invasive ductal carcinoma30 Invasive lobular carcinoma2 DCIS Other | Characteristic | OSNA plus histology | Patients, n | 32 | Age (years), median (range) | NR | Clinical stage, n | 0 | 0 | I | 8 | II | 15 | III | 7 | IV | 2 | Unknown | Nodal status, n | pN0 | 14 | pN1 | 10 | pN2 | 6 | pN3 | 2 | Histopathological type, n | Invasive ductal carcinoma | 30 | Invasive lobular carcinoma | 2 | DCIS | Other | |||||||||
| Characteristic | OSNA plus histology | ||||||||||||||||||||||||||||||||||||||||||
| Patients, n | 32 | ||||||||||||||||||||||||||||||||||||||||||
| Age (years), median (range) | NR | ||||||||||||||||||||||||||||||||||||||||||
| Clinical stage, n | |||||||||||||||||||||||||||||||||||||||||||
| 0 | 0 | ||||||||||||||||||||||||||||||||||||||||||
| I | 8 | ||||||||||||||||||||||||||||||||||||||||||
| II | 15 | ||||||||||||||||||||||||||||||||||||||||||
| III | 7 | ||||||||||||||||||||||||||||||||||||||||||
| IV | 2 | ||||||||||||||||||||||||||||||||||||||||||
| Unknown | |||||||||||||||||||||||||||||||||||||||||||
| Nodal status, n | |||||||||||||||||||||||||||||||||||||||||||
| pN0 | 14 | ||||||||||||||||||||||||||||||||||||||||||
| pN1 | 10 | ||||||||||||||||||||||||||||||||||||||||||
| pN2 | 6 | ||||||||||||||||||||||||||||||||||||||||||
| pN3 | 2 | ||||||||||||||||||||||||||||||||||||||||||
| Histopathological type, n | |||||||||||||||||||||||||||||||||||||||||||
| Invasive ductal carcinoma | 30 | ||||||||||||||||||||||||||||||||||||||||||
| Invasive lobular carcinoma | 2 | ||||||||||||||||||||||||||||||||||||||||||
| DCIS | |||||||||||||||||||||||||||||||||||||||||||
| Other | |||||||||||||||||||||||||||||||||||||||||||
| Results | |||||||||||||||||||||||||||||||||||||||||||
| OSNAFive-level histopathologyMacrometastasisMicrometastasisITCsNegativen = 346 ALNs++50402+25013–123264 | OSNA | Five-level histopathology | Macrometastasis | Micrometastasis | ITCs | Negative | n = 346 ALNs | ++ | 50 | 4 | 0 | 2 | + | 2 | 5 | 0 | 13 | – | 1 | 2 | 3 | 264 | |||||||||||||||||||||
| OSNA | Five-level histopathology | ||||||||||||||||||||||||||||||||||||||||||
| Macrometastasis | Micrometastasis | ITCs | Negative | ||||||||||||||||||||||||||||||||||||||||
| n = 346 ALNs | |||||||||||||||||||||||||||||||||||||||||||
| ++ | 50 | 4 | 0 | 2 | |||||||||||||||||||||||||||||||||||||||
| + | 2 | 5 | 0 | 13 | |||||||||||||||||||||||||||||||||||||||
| – | 1 | 2 | 3 | 264 | |||||||||||||||||||||||||||||||||||||||
| Sensitivity (%)Specificity (%)Discordance (%)n = 346 ALNs before adjustment for TAB95.394.75.2n = 339 ALNs after adjustment for TAB95.397.13.2 | Sensitivity (%) | Specificity (%) | Discordance (%) | n = 346 ALNs before adjustment for TAB | 95.3 | 94.7 | 5.2 | n = 339 ALNs after adjustment for TAB | 95.3 | 97.1 | 3.2 | ||||||||||||||||||||||||||||||||
| Sensitivity (%) | Specificity (%) | Discordance (%) | |||||||||||||||||||||||||||||||||||||||||
| n = 346 ALNs before adjustment for TAB | 95.3 | 94.7 | 5.2 | ||||||||||||||||||||||||||||||||||||||||
| n = 339 ALNs after adjustment for TAB | 95.3 | 97.1 | 3.2 | ||||||||||||||||||||||||||||||||||||||||
| Methodological issues | |||||||||||||||||||||||||||||||||||||||||||
| Replicates: unclear whether replicate samples were analysed Recruitment: unclear if recruitment was consecutive or random Conflict of interest: consumables funded by Sysmex |
|||||||||||||||||||||||||||||||||||||||||||
| Quality appraisal | |||||||||||||||||||||||||||||||||||||||||||
| Was a consecutive or random sample of patients enrolled? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| Was a cohort study design avoided? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Did the study avoid inappropriate exclusions? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Could the selection of patients have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| Concerns that the included patients do not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||
| Were the index test results interpreted without knowledge of the results of the reference standard? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| If a threshold was used, was it prespecified? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Could the conduct or interpretation of the index test have introduced bias? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the index test, its conduct or its interpretation differ from the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||
| Is the reference standard likely to correctly classify the target condition? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Were the reference standard results interpreted without knowledge of the results of the index test? (Y/N/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| Could the reference standard, its conduct or its interpretation have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| Are there concerns that the target condition as defined by the reference standard does not match the review question? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive a reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Did all patients receive the same reference standard? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Were all samples (that should have been) included in the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Could the patient flow have introduced bias? (H/L/U) | U | ||||||||||||||||||||||||||||||||||||||||||
| Were samples suspected of TAB excluded from the analysis? (Y/N/U) | Y | ||||||||||||||||||||||||||||||||||||||||||
| Are there concerns about selective reporting of outcomes? (H/L/U) | L | ||||||||||||||||||||||||||||||||||||||||||
Appendix 3 Clinical effectiveness review: excluded studies
| Papers excluded | Reason for exclusion |
|---|---|
| 1. Abdul-Rasool S, Kidson SH, Panieri E, Dent D, Pillay K, Hanekom GS. An evaluation of molecular markers for improved detection of breast cancer metastases in sentinel nodes. J Clin Pathol 2006;59:289–97 | Exclude on intervention |
| 2. Aihara T, Fujiwara Y, Ooka M, Sakita I, Tamaki Y, Monden M. Mammaglobin B as a novel marker for detection of breast cancer micrometastases in axillary lymph nodes by reverse transcription-polymerase chain reaction. Breast Cancer Res Treat 1999;58:137–40 | Exclude on population |
| 3. Allende D, Denninghoff V, Avagnina A, Elsner B. Sentinel lymph nodes study: how to do it right? The Argentinean experience. Breast J 2008;14:216–17 | Exclude on study design |
| 4. Al-Ramadhani S, Balaraman P, George D, Jader S, McKenzie J, Al-Sam S, et al. Molecular markers for the detection of metastatic breast cancer in sentinel lymph nodes: an adjunct to the metasin BLNA? 6th Joint Meeting of the British Division of the International Academy of Pathology and the Pathological Society of Great Britain and Ireland, Ghent Pathology 2011, 10–13 May 2011, p. S4 | Exclude on study design |
| 5. Anonymous. Sentinel lymph node biopsy for patients with breast cancer. Oncol Forum 2003;6:8–10 | Unobtainable |
| 6. Anonymous. 6th Joint Meeting of the British Division of the International Academy of Pathology and the Pathological Society of Great Britain and Ireland, Ghent Pathology 2011. Journal of Pathology Conference | Exclude on population |
| 7. Babar M, Madani R, Thwaites L, Jackson P, Chakravorty A, Irvine T, et al. Intraoperative molecular detection of lymph node metastases and micro-metastases – results of the first UK centre using the one step nucleic acid amplification assay. Eur J Cancer 2011;47(Suppl. 1):S333 | Exclude on study design |
| 8. Backus J, Laughlin T, Wang Y, Belly R, White R, Baden J, et al. Identification and characterization of optimal gene expression markers for detection of breast cancer metastasis. J Mol Diagn 2005;7:327–36 | Exclude on intervention |
| 9. Basu NN, Nash T, Doddi S, Desai A. An assessment of the impact of OSNA (one step nuclear acid amplification) analysis on the rates of axillary clearance in breast cancer patients. Eur J Cancer 2012;48:S213–14 | Exclude on outcomes |
| 10. Bedrosian I. Commentary on ‘Long-term follow-up study of a prospective multicenter sentinel node trial: molecular detection of breast cancer sentinel node metastases’: Verbanac KM, for the ECU/AAMC Sentinel Node Study Group (the Brody School of Medicine at East Carolina Univ, Greenville, NC; et al). Ann Surg Oncol 2010;17:S368–77. Breast Dis 2011;22:392–3 | Exclude on study design |
| 11. Belda M, Barra CC, Crouet H, Houvenaeghel G, Khaddage A, Leroux A, et al. Intra-operative sentinel lymph node metastasis detection in breast cancer by ‘one-step nucleic acid amplification (OSNA)’ – results of the French multicentre prospective study. Eur J Cancer Suppl 2008;6:54 | Exclude on comparator |
| 12. Berger J, Mueller-Holzner E, Fiegl H, Marth C, Daxenbichler G. Evaluation of three mRNA markers for the detection of lymph node metastases. Anticancer Res 2006;26:3855–60 | Exclude on intervention |
| 13. Bernet L, Martinez-Benaclocha M, Chica FS, Munoz RC, Medrano J, Gonzalez JP. Optimization of the intraoperative sentinel node procedure in breast cancer with one-step nucleic acid amplification (OSNA) method. Eur J Cancer Suppl. 2010;8:159–60 | Exclude on comparator |
| 14. Bernet L, Cano R, Martinez M, Duenas B, Matias-Guiu X, Morell L, et al. Diagnosis of the sentinel lymph node in breast cancer: a reproducible molecular method: a multicentric Spanish study. Histopathology 2011;58:863–9 | Exclude on comparator |
| 15. Blumencranz P, Pieretti M, Asad H, Zanchi A. Quantitative intra-operative RT-PCR assay predicts size of sentinel node metastases. Cancer Res 2009;69(2 Suppl. 1):1002 | Exclude on intervention |
| 16. Branagan G, Hughes D, Jeffrey M, Crane-Robinson C, Perry PM. Detection of micrometastases in lymph nodes from patients with breast cancer. Br J Surg 2002;89:86–9 | Exclude on intervention |
| 17. Campbell J, A-Ramadhani S, Brown S, Bustin S, Jader S, Sai Giridhar P. The PaXgene reagent, the pathologist’s answer to have your cake & eat it: formalin-free fixation & processing of tissues fit for qPCR using the metasin-BLNA & diagnostic pathology. J Pathol 2012;226:S13 | Exclude on study design |
| 18. Cannone M, Oliveri C, Roz E, Rispoli F, Ferrarese S, Alexiadis S, et al. Molecular markers of breast cancer cells identified in fine needle aspiration samples from resected sentinel lymph nodes. Acta Cytol 2006;50:271–6 | Exclude on comparator |
| 19. Cano Munanoz R, Bernet L, Martinez-Benaclocha M, Sevilla Chica F, Medrano J, Gonzalez JP. Sentinel lymph node diagnosis in breast cancer: comparison between two different molecular methods. Eur J Cancer Suppl. 2010;8:147 | Exclude on comparator |
| 20. Cepedello Boiso I, Urena Lara M, Duenas Rodriguez B, Ramirez Tortosa C, Martinez Ferrol J. Sentinel lymph node biopsy after neoadjuvant chemotherapy in breast cancer. Eur J Nucl Med Mol Imaging 2011;38(2 Suppl.):S419–20 | Exclude on comparator |
| 21. Cepedello Boiso I, Urena Lara M, Ramirez Tortosa C, Duenas Rodriguez B, Martinez Ferrol J. Sentinel lymph node biopsy in breast cancer: routine histological investigation versus OSNA. Eur J Nucl Med Mol Imaging 2011;38(2 Suppl.):S126 | Exclude on comparator |
| 22. Choi YL, Ahn SK, Bae YK, Park IA, Min JW, Lee KW, et al. One-step nucleic acid amplification (OSNA): intraoperative rapid molecular diagnostic method for the detection of sentinel lymph node metastases in breast cancer patients in Korean cohort. J Breast Cancer 2010;13:366–74 | Exclude on intervention |
| 23. Cserni G. Overinterpretation of the role of cytokeratin 19 RT-PCR of sentinel nodes in breast carcinoma. Surgery 2003;133:124 | Exclude on study design |
| 24. Cutress RI, McDowell A, Gabriel FG, Gill J, Jeffrey MJ, Agrawal A, et al. Observational and cost analysis of the implementation of breast cancer sentinel node intraoperative molecular diagnosis. J Clin Pathol 2010;63:522–9 | Exclude on intervention |
| 25. Daniele L, Annaratone L, Allia E, Mariani S, Armando E, Bosco M, et al. Technical limits of comparison of step-sectioning, immunohistochemistry and RT-PCR on breast cancer sentinel nodes: a study on methacarn-fixed tissue. J Cell Mol Med 2009;13:4042–5 | Exclude on intervention |
| 26. Dauplat MM, Penault-Llorca F. Sentinel lymph node biopsy intraoperative evaluation: state of art and emerging molecular assays. Med Nucl Imagerie Fonctionnelle Metab 2010;34:23–6 | Exclude on language |
| 27. Dell’Orto P, Biasi MO, Del Curto B, Zurrida S, Galimberti V, Viale G. Assessing the status of axillary sentinel lymph nodes of breast carcinoma patients by a real-time quantitative RT-PCR assay for mammaglobin 1 mRNA. Breast Cancer Res Treat 2006;98:185–90 | Exclude on comparator |
| 28. Denninghoff V, Allende D, Paesani F, Garcia A, Avagnina A, Perazzo F, et al. Sentinel lymph node molecular pathology in breast carcinoma. Diagn Mol Pathol 2008;17:214–19 | Exclude on intervention |
| 29. Fischer S, Asa SL. Application of immunohistochemistry to thyroid neoplasms. Arch Pathol Lab Med 2008;132:359–72 | Exclude on intervention |
| 30. Ghaffari SR, Sabokbar T, Tahmasebi S, Dastan J, Shorakae S, Moradi A, et al. Combining mammaglobin and carcinoembryonic mRNA markers for early detection of micrometastases from breast cancers – a molecular study of 59 patients. Asian Pac J Cancer Prev 2006;7:396–8 | Exclude on intervention |
| 31. Gillanders WE, Mikhitarian K, Hebert R, Mauldin PD, Palesch Y, Walters C, et al. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Ann Surg 2004;239:828–40 | Exclude on intervention |
| 32. Gimbergues P, Dauplat MM, Cayre A, Durando X, Le Bouedec G, Finat-Duclos F, et al. Correlation between molecular metastases in sentinel lymph nodes of breast cancer patients and St Gallen risk category. Eur J Surg Oncol 2007;33:16–22 | Exclude on intervention |
| 33. Görgens DR, Tulusan AH, Sopov I. Intra-operative detection of sentinel lymph node metastasis in breast cancer by OSNA (one step nucleic acid amplification). Breast 2011;20(Suppl. 1):S58 | Exclude on comparator |
| 34. Guillen-Paredes MP, Carrasco-Gonzalez L, Chavez-Benito A, Aguayo-Albasini JL. Application of the OSNA technique for intraoperative analysis of sentinel lymph node in breast cancer. Cir Esp 2011;89:261–2 | Exclude on language |
| 35. Hasui Y, Yamasaki M, Amano C, Yoshimoto M, Funakoshi K, Fujimoto K. Development of quality control material for OSNA assay. Clin Chem 2008;54:A158 | Exclude on study design |
| 36. Inokuchi M, Ninomiya I, Tsugawa K, Terada I, Miwa K. Quantitative evaluation of metastases in axillary lymph nodes of breast cancer. Br J Cancer 2003;89:1750–6 | Exclude on intervention |
| 37. Jackson P, Irvine T, Layer G, Kissin M. The intraoperative molecular analysis of sentinel lymph node metastases and micro-metastases in breast cancer patients using one step nucleic acid amplification on whole nodes. Eur J Surg Oncol 2012;38:455 | Exclude on comparator |
| 38. Laia BV, Marcos MB, Refael CM, Francisco SC, Jose T, Blai BS, et al. Molecular diagnosis of sentinel lymph nodes for breast cancer: one step ahead for standardization. Diagn Mol Pathol 2011;20:18–21 | Exclude on comparator |
| 39. Le Frère-Belda MA, Lecuru F, Bats AS, Clough KB, Charon-Barra C, Crouet H, et al. Preoperative detection of sentinel node metastasis in breast cancer by the ‘OSNA’ technique (one-step nucleic acid amplification): results of a multicentric French study. Bull Cancer 2008;95:576 | Exclude on language |
| 40. Le Frère-Belda MA, Bats AS, Gillaizeau F, Poulet B, Clough KB, Nos C, et al. Diagnostic performance of one-step nucleic acid amplification for intraoperative sentinel node metastasis detection in breast cancer patients. Int J Cancer 2011;130:2377–86 | Exclude on comparator |
| 41. Madani R, Jafferbhoy S, Thwaites L, Jackson P, Layer GT, Irvine TE, et al. One-step nucleic acid amplification in detection of lymph node metastases in breast cancer patients: are patients being over treated? Eur J Cancer Suppl 2010;8:23 | Exclude on comparator |
| 42. Madani R, Jafferbho S, Thwaites L, Jackson P, Layer G, Irvine T, et al. One-step nucleic acid amplification: an intraoperative test for detection of lymph node metastases in breast cancer patients. results of the first UK centre. Eur J Surg Oncol 2011;36:1111 | Exclude on study design |
| 43. Mansel R. An update of sentinel node biopsy in breast cancer. Eur J Cancer 2009;45(Suppl. 1):447–8 | Exclude on study design |
| 44. Manzotti M, Dell’Orto P, Maisonneuve P, Zurrida S, Mazzarol G, Viale G. Reverse transcription-polymerase chain reaction assay for multiple mRNA markers in the detection of breast cancer metastases in sentinel lymph nodes. Int J Cancer 2001;95:307–12 | Exclude on intervention |
| 45. Nishimura K, Sugiyama D, Moronobu A, Kawano S, Kumagai S. Cost utility analysis on one-step nucleic acid amplification method for the detection of axillary metastasis in breast cancer. Cancer Res 2009;69:110 | Exclude on outcomes |
| 46. Nissan A, Jager D, Roystacher M, Prus D, Peretz T, Eisenberg I, et al. Multimarker RT-PCR assay for the detection of minimal residual disease in sentinel lymph nodes of breast cancer patients. Br J Cancer 2006;94:681–5 | Exclude on intervention |
| 47. Osako T, Iwase T, Kimura K, Yamashita K, Horii R, Yanagisawa A, et al. Intraoperative molecular assay for sentinel lymph node metastases in early stage breast cancer: a comparative analysis between one-step nucleic acid amplification whole node assay and routine frozen section histology. Cancer 2011;117:4365–74 | Exclude on comparator |
| 48. Pinero A. Present and future of sentinel lymph node biopsy in breast cancer staging. Clin Trans Oncol 2010;12:457–8 | Exclude on study design |
| 49. Rebollo-Aguirre AC, Gallego-Peinado M, Menjon-Beltran S, Garcia-Garcia J, Pastor-Pons E, Chamorro-Santos CE, et al. Sentinel lymph node biopsy in patients with operable breast cancer treated with neoadjuvant chemotherapy. Rev Esp Med Nucl 2012;31:117–23 | Exclude on comparator |
| 50. Remoundos DD, Wilson HA, Ng VV, Ahmed F, Chia Y, Cunnick GH. Sentinel node biopsy analysis using intraoperative one-step nucleic-acid amplification (OSNA): are we really saving patients a second operation? Eur J Cancer 2012;48:S224–5 | Exclude on outcomes |
| 51. Rothe M, Wingfield S, Barranco P, Charache S. Robotics in the hematology laboratory. An evaluation of the productivity of the Sysmex HS-330. Am J Clin Pathol 1995;103:154–8 | Exclude on population |
| 52. Saha S, Sirop S, Chakravarty B, Wiese D, Soni M, Singla A, et al. Comparison of a novel molecular assay with touch imprint and permanent histology of sentinel lymph nodes in early-stage breast cancer. J Clin Oncol 2010;28(15 Suppl. 1):642 | Exclude on intervention |
| 53. Sanchez-Mendez J, Lopez-Rodriguez MJ, Escabias Del Pozo C, Oliver Goldaracena JM, Redondo Sanchez A, Belinchon Olmeda B, et al. Effect of one step molecular intraoperative method in detection of micrometastasis in sentinel node and management of them. Eur J Cancer 2012;48:S207–8 | Exclude on comparator |
| 54. Sansano I, Espinosa M, Iglesias C, Aizpurua M, Sancho M, Garcia C, et al. Multicentric comparative study between one-step nucleic acid amplification (OSNA) whole node assay and standard histology for breast sentinel lymph node: molecular assay can avoid secondary surgeries and predict no other node involvement. Mod Pathol 2012;25:65A | Duplicate |
| 55. Sapino A, Macri L, Lupo R, Marazzi A, Pecchioni C, De Rosa P, et al. Intraoperative assessment of sentinel node in breast cancer performing one step nucleic acid amplification (OSNA) assay on the whole lymph node. Breast 2011;20:S59 | Exclude on comparator |
| 56. Schimanski S, Gorgens D, Tulusan A. Intra-operative detection of sentinel lymph node metastasis in breast cancer by one step nucleic acid amplification (OSNA). Eur J Cancer 2012;48:S75 | Exclude on comparator |
| 57. Schroder CP, Ruiters MH, de Jong S, Tiebosch AT, Wesseling J, Veenstra R, et al. Detection of micrometastatic breast cancer by means of real time quantitative RT-PCR and immunostaining in perioperative blood samples and sentinel nodes. Int J Cancer 2003;106:611–18 | Exclude on intervention |
| 58. Span PN, Van Laarhoven HW, Sweep FC. Mammaglobin RT-qPCR for breast cancer metastasis detection in sentinel lymph nodes. Cancer Sci 2010;101:2501 | Exclude on study design |
| 59. Sua LF, Silva NM, Vidaurreta M, de la Orden V, Veganzones S, Rafael S, et al. Rapid detection of sentinel lymph node metastases in different techniques and comparison in low-grade breast carcinomas. Colomb Med 2012;43:28–37 | Exclude on intervention |
| 60. US OSNA Breast Cancer Sentinel Lymph Node Study Group. Prospective multicenter study of a novel fully automated molecular test for identification of metastatic carcinoma in axillary sentinel lymph nodes in breast cancer – the US OSNA Breast Cancer Sentinel Lymph Node Study Group. Lab Invest 2010;90(Suppl. 1s):75A | Duplicate |
| 61. Velasco A, Morales S, Panades MJ, Gonzalez S, Vilardell F, Canosa C, et al. May axillary dissection be avoided after micro metastases in sentinel lymph node analyzed by OSNA method? J Clin Oncol 2011;29(27 Suppl. 1): abstract 18 | Exclude on study design |
| 62. Verbanac KM, Min CJ, Mannie AE, Lu J, O’Brien KF, Rosman M, et al. Long-term follow-up study of a prospective multicenter sentinel node trial: molecular detection of breast cancer sentinel node metastases. Ann Surg Oncol 2010;17(Suppl. 3):368–77 | Exclude on intervention |
| 63. Vieites B, Mendoza E, Robles MJ, Palacios J. Histologic sections, immunohistochemistry and OSNA-CK19 in sentinel lymph node: a comparative study. Eur J Cancer Suppl 2010;8:154 | Exclude on comparator |
| 64. Vilardell F, Novell A, Martin J, Santacana M, Velasco A, Diez-Castro MJ, et al. Importance of assessing CK19 immunostaining in core biopsies in patients subjected to sentinel node study by OSNA. Virchows Arch 2012;460:569–75 | Exclude on comparator |
| 65. Wallwiener CW, Wallwiener M, Kurth RR, Rohm C, Neubauer H, Banys MJ, et al. Molecular detection of breast cancer metastasis in sentinel lymph nodes by reverse transcriptase polymerase chain reaction (RT-PCR): identifying, evaluating and establishing multi-marker panels. Breast Cancer Res Treat 2011;130:833–44 | Exclude on intervention |
| 66. Wang YS, Ouyang T, Wu J, Liu YH, Cao XC, Sun X, et al. A comparative study of one-step nucleic acid amplification (OSNA), frozen section and touch imprint cytology for intra-operative assessment of breast cancer sentinel lymph node – China multicenter study CBCSG-001c. Eur J Cancer 2012;48:S214 | Exclude on intervention |
| 67. Whisker L, Mecci AJ, Stanton D, Kneeland C, Iqbal M, Clarke D. Can the Memorial Sloan Kettering (MSK) nomogram be used to improve theatre planning when undertaking intra-operative assessment of sentinel lymph nodes (SLN)? Eur J Surg Oncol 2012;38:440 | Exclude on intervention |
| 68. Woelfl S, Bogner ST, Broinger G, Bergmayr A, Puhringer K, Schrenk P, et al. Intra-operative use of one step nucleic acid amplification (OSNA) for whole sentinel lymph node analysis in breast cancer patients. Eur J Cancer 2012;48:S89 | Exclude on study design |
Appendix 4 Table of abstracts
| Study | Design | SLNs | Outcomes |
|---|---|---|---|
| Garcia-Estepa 201068 | Systematic review | Concordance 91.7–98.2%, sensitivity 87.5%–98.1%, specificity 89–98.5% | |
| Di Filippo 200969 | Single gate | 247 SLNs | Concordance 96.7%, specificity 96.8%, sensitivity 96.4% |
| Buglioni 200970 | 228 SLNs | Concordance 96.9%, specificity 97.2%, sensitivity 96.1% | |
| Buglioni 201071 | 416 SLNs | Concordance 95%, specificity 95%, sensitivity 94% | |
| Kaneko 200772 | Single gate | 141 ALNs | Specificity 96.9% (95% CI 92.3% to 99.3%) |
| 469 ALNs | Concordance 93.0% (95% CI 90.3% to 95.1%) | ||
| Masuda 200773 | 178 ALNs | Specificity 97.5% (95% CI 93.5% to 99.3%), positivity rate 17.9%, concordance 94% (95% CI 91.9% to 95.8%) | |
| Tsuda 200674 | 144 SLNs | Concordance with three-level histology 98%, specificity 100% | |
| Sato 201175 | Single gate | 415 patients | PPV of OSNA (++) for non-SLN metastases 44.0%, PPV of OSNA (+) for non-SLN metastases 17.6% (p = 0.01) |
| Takabatake 201176 | 417 patients | PPV of OSNA (++) for non-SLN metastases 44.0%, PPV of OSNA (+) for non-SLN metastases 17.6% (p = 0.01) | |
| Peston 200977 | Single gate | 100 SLNs | Concordance 96%, sensitivity 92%, specificity 97%. OSNA results achieved, on average, within 30 minutes for two nodes |
| DTA | |||
| Snook 200778 | 45 ALNs | Concordance 93.3%, sensitivity 100%, specificity 91.2% | |
| Snook 200879 | Median time for analysis of one SLN 32 minutes | ||
| Snook 200780 | 87 ALNs | Concordance 90.8%, sensitivity 90%, specificity 91% | |
| Kissin 200981 | 396 SLNs | Concordance 96.2%, sensitivity 91.5%, specificity 97.2, PPV 87.7%, NPV 98.1%. Minimum time to reach a result on a single node was 22 minutes | |
| Nizar 201082 | Single gate | 31 patients | Specificity 82%, sensitivity 50%. OSNA requires investment of nearly £60,000, with regular servicing and replenishment of reagents costing nearly £1500 per month |
| Chaudhry 201183 | Single gate | 251 SLNs | Sensitivity 93%, specificity 89%, increased to 94% if accounting for TAB |
| Massey 201184 | Cohort | Mean time 40.5, 51.8, 54.0 and 61.5 minutes for one, two, three and four nodes respectively. Operation time was changed by –48 to +65 minutes (median +20 minutes) | |
| Beitsch 200785 | Single gate | 58 SLNs | Concordance 94.8% |
| Tomlins 201186 | Single gate | 62 SLNs | Concordance 95% |
| Iqbal 201287 | Observation | 99 SLNs | Mean time for SLN analysis 49.7 minutes (range 37–94 minutes). A second operation was avoided for 33% of all patients studied |
| Ng 201188 | Observation | 100 patients | Median time for SLNB to be performed 12 minutes (range 2–57 minutes). Median time for telephone result 44 minutes (range 28–75 minutes). In 54% the operation had finished before receiving the results; median waiting time 3 minutes |
| Ng 201189 | Cohort | 200 patients | OSNA-positive rate 39%, histology-positive rate 19% |
| Ng 201190 | Observation | 100 patients | Median time for SLNB to be performed 12 minutes (range 2–57 minutes). Median time for telephone result 44 minutes (28–75 minutes). In 54% the operation had finished before receiving the results; median waiting time 3 minutes |
| Ng 201167 | Cohort | 200 patients | OSNA-positive rate 39%, histology-positive rate 19% |
| Remoundos 201291 | Cohort | 602 SLNs | OSNA-positive rate: macrometastases 19%, micrometastases 21%; histology-positive rate: macrometastases 19%, micrometastases 2% |
| Bilous 201292 | Single gate | 211 SLNs | Specificity 96.3%, sensitivity 95.8% |
| Godey 201193 | Cohort | 344 SLNs | OSNA-positive rate 21.3%, histology-positive rate 25%, concordance 138/160 patients. Median time to analysis 35 minutes for two SLNs and ∼5 additional minutes per additional node |
| Cohort | 367 patients | OSNA-positive rate 24.32%, histology-positive rate 24.79% | |
| Peoch 201194 | Single gate | 197 patients | OSNA-positive rate 21.3%. Median time for analysis for two SLNs 37 minutes |
| Khaddage 200995 | Single gate | 80 SLNs | Validation study (adjusted for TAB): sensitivity 100%, concordance 97.7%, specificity 97.1%; routine practice study (unadjusted for TAB): sensitivity 100%, concordance 94.9%, specificity 92.9%. Median time taken to analyse two SLNs 35–37 minutes |
| Godey 200996 | Single gate | 175 SLNs | OSNA-positive rate 18%, 7/91 cases discordant. Median time for analysis for two SLNs 35–37 minutes |
| Godey 201097 | Single gate | DTA, time to analysis | |
| Levine 201098 | Single gate | DTA | |
| US OSNA Breast Cancer Sentinel Lymph Node Study Group 201099 | Single gate | DTA | |
| Schem 2007100 | Single gate | 188 LNs | DTA |
| Schem 2010101 | Single gate | 110 patients | Sensitivity 93.3% |
| Jimbo 2012102 | Single gate | Concordance 91.5%, sensitivity 90.3%, specificity 93.3% | |
| Suzuki 2011103 | Single gate | Concordance 95.1%, specificity 96.9% | |
| Rai 2012104 | Observation | Average time to result 36 minutes. Of 703 patients, 581 (83%) were OSNA negative, 56 (8.0%) were OSNA-positive micrometastasis and 66 (9.4%) were OSNA-positive macrometastasis. PPV OSNA (++) 57.6%, PPV OSNA (+) 17.9% | |
| Wahab 2012105 | Single gate | 196 SLNs | Sensitivity 94%, specificity 96.6%, concordance 96% |
| Siso 2012106 | Observation | 49 patients | 53.1% OSNA positivity rate; no comparison with histopathology; results were contrasted with ALND |
| Krishmamurthy 2009107 | Single gate | 279 ALNs | Kappa coefficient between histology and OSNA was 0.87 (95% CI 0.72 to 1.00) |
| Mizoo 2012108 | Single gate | 36 SLNs | Compared with intraoperative frozen section. Mean time to analysis 38.9 minutes (34.9 minutes for one node, 46.4 minutes for two nodes, 55 minutes for three to four nodes) |
| Al-Ramadhani 2010109 | Single gate | 122 patients, 275 nodes | Sensitivity and specificity rates of > 95% |
| Johns 2011110 | Single gate | 127 patients, 202 SLNs | 204/978 cases were histology and Metasin positive (21%). In total, 21 cases (2.1%) were histology-only positive and 27 cases (2.7%) were Metasin-only positive: discordance of 5% of the total |
| Johns 2011111 | Single gate | 127 patients, 202 SLNs | The Metasin assay displays the same diagnostic accuracy as GeneSearch BLN with a considerable cost reduction |
| Simoes 2012112 | Single gate’ | 1167 patients, 2018 SLNs | The false-negative rate (from sampling error) was 0.68% for GeneSearch and 0.92% for Metasin |
| Sundaresan 2012113 | Single gate | > 700 cases | The Metasin-BLNA assay is cheap (£35) for two nodes; fast result in < 46 minutes (total assay time from receipt in laboratory) in > 75% of cases |
| Sundaresan 2011114 | Single gate | > 700 cases | Discordance < 4% (with histology and GeneSearch) |
| Holt 2012115 | Single gate | 485 cases, 493 cases post-August 2010 | Discordance 3.8% against histopathology. After August 2010, discordance 5% against histopathology |
Appendix 5 Cost-effectiveness: quality appraisal of studies using the checklist of Drummond and Jefferson125
| Criteria | Cutress 2010123 | Guillen-Paredes 201156 | Burke 2010124 | Cooper 2011,9 Meng 2011122 | Classe 2012121 |
|---|---|---|---|---|---|
| Study design | |||||
| The research question is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The economic importance of the research question is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The viewpoint(s) of the analysis are clearly stated and justified | ✓ | ✓ | ✓ | ✓ | ✓ |
| The rationale for choosing alternative programmes or interventions compared is stated | Partial | ✓ | ✓ | ✗ | ✗ |
| The alternatives being compared are clearly described | ✓ | ✓ | ✓ | ✓ | ✓ |
| The form of economic evaluation used is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The choice of form of economic evaluation is justified in relation to the question addressed | ✗ | ✗ | ✓ | ✓ | ✗ |
| Data collection | |||||
| The source(s) of effectiveness estimates used are stated | ✓ | ✓ | ✓ | ✓ | ✗ |
| Details of the design and results of the effectiveness study are given (if based on a single study) | ✓ | ✓ | ✓ | NA | ✓ |
| Details of the methods of synthesis or meta-analysis of estimates are given (if based on a synthesis of a number of effectiveness studies) | NA | NA | NA | ✓ | NA |
| The primary outcome measure(s) for the economic evaluation are clearly stated | Partial | ✓ | Partial | ✓ | ✓ |
| Methods to value benefits are stated | ✗ | ✗ | ✗ | ✗ | ✗ |
| Details of the subjects from whom valuations were obtained are given | NA | NA | NA | NA | NA |
| Productivity changes (if included) are reported separately | ✗ | ✗ | ✗ | ✗ | ✗ Attempted |
| The relevance of productivity changes to the study question is discussed | NA | NA | NA | NA | NA |
| Quantities of resource use are reported separately from their unit costs | ✓ | ✓ | ✓ | ✓ | ✓ |
| Methods for the estimation of quantities and unit costs are described | Partial | Partial | Partial | ✓ | ✓ |
| Currency and price date are recorded | ✓ | Partial | ✓ | ✓ | ✓ |
| Details of currency of price adjustments for inflation or currency conversion are given | ✗ | ✗ | ✗ | Partial | ✗ |
| Details of any model used are given | NA | NA | ✓ | ✓ | NA |
| The choice of model used and the key parameters on which it is based are justified | NA | NA | ✓ | ✓ | NA |
| Analysis and interpretation of results | |||||
| Time horizon of costs and benefits is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The discount rate(s) is stated | NA | NA | NA | ✓ | NA |
| The choice of discount rate(s) is justified | NA | NA | NA | ✓ | NA |
| An explanation is given if costs and benefits are not discounted | NA | NA | ✓ | NA | NA |
| Details of statistical tests and confidence intervals are given for stochastic data | ✗ | ✓ | ✗ | ✓ | ✓ |
| The approach to sensitivity analysis is given | NA | NA | ✓ | ✓ | ✓ |
| The choice of variables for sensitivity analysis is justified | NA | NA | ✓ | Partial | ✗ |
| The ranges over which the variables are varied are justified | NA | NA | ✗ | ✓ | ✗ |
| Relevant alternatives are compared | ✓ | ✓ | ✓ | ✓ | ✓ |
| Incremental analysis is reported | ✗ | ✗ | ✗ | ✓ | ✓ |
| Major outcomes are presented in a disaggregated as well as an aggregated form | ✓ | ✓ | ✗ | ✓ | ✓ |
| The answer to the study question is given | ✓ | ✓ | Partial | ✓ | ✓ |
| Conclusions follow from the data reported | ✓ | ✓ | ✓ | ✓ | ✓ |
| Conclusions are accompanied by the appropriate caveats | ✗ | ✗ | ✓ | ✓ | ✗ |
Appendix 6 Cost-effectiveness review: excluded studies
Appendix 7 Expert advisors
| Title | Name | Specialty | Affiliation |
|---|---|---|---|
| Mr | Simon Pain | Consultant Breast and Endocrine Surgeon | Norfolk and Norwich University Hospital |
| Mr | Zenon Rayter | Consultant Surgeon | Bristol Royal Infirmary |
| Professor | Ian Kunkler | Consultant and Honorary Professor in Clinical Oncology | Edinburgh Cancer Centre |
| Professor | Graham Layer | Consultant Surgeon and Director of Professional Standards | Royal Infirmary of Edinburgh |
| Dr | Abeer Shaaban | Consultant Pathologist | St James’s University Hospital, Leeds |
| Dr | Deirdre Ryan | Consultant Pathologist | Barts Health NHS Trust, London |
Appendix 8 Two-way threshold analysis of one-step nucleic acid amplification sensitivity and specificity
The two-way sensitivity analyses examine the effect of altering both specificity and sensitivity while fixing other parameters at their base-case values. Here, we present the long-term ICERs comparing histopathology with full-node OSNA, as this analysis accounts for all costs and benefits of the tests. As a guide, we chose a cost-effectiveness threshold of £30,000 per QALY gained.
First, we present the results (Table 78) using the range of sensitivity and specificity values that were used in the one-way threshold analyses (70–100% for both sensitivity and specificity; see Tables 54–57). This demonstrates that histopathology was cost-effective at a threshold of £30,000 per QALY gained for most values in the analysis. However, when the specificity was 90% and the sensitivity was 100%, the ICER increased to > £94,000 per QALY gained. Furthermore, when the sensitivity was 95% and the specificity was 100%, the ICER increased to > £100,000. When the sensitivity was 100% and the specificity was ≥ 95%, OSNA dominated histopathology, having lower costs and larger QALY gains.
| OSNA specificity (%) | Incremental cost (£) per QALY gained for histopathology vs. full-node OSNA | |||||||
|---|---|---|---|---|---|---|---|---|
| OSNA sensitivity (%) | ||||||||
| Base case: 84.5 | 70 | 75 | 80 | 85 | 90 | 95 | 100 | |
| Base case: 91.8 | 4324 | 2119 | 2588 | 3294 | 4476 | 6862 | 14,193 | OSNA dominated histopathology |
| 70 | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA | Histopathology dominates OSNA |
| 75 | 156 | 15 | 49 | 96 | 164 | 270 | 460 | 898 |
| 80 | 1068 | 543 | 667 | 841 | 1099 | 1526 | 2366 | 4776 |
| 85 | 2210 | 1146 | 1389 | 1736 | 2277 | 3234 | 5386 | 14,691 |
| 90 | 3683 | 1842 | 2242 | 2835 | 3807 | 5691 | 10,899 | 94,097 |
| 95 | 5655 | 2652 | 3266 | 4214 | 5875 | 9529 | 24,166 | OSNA dominates histopathology |
| 100 | 8430 | 3608 | 4518 | 5997 | 8823 | 16,367 | 100,308 | OSNA dominates histopathology |
Table 79 narrows down the range of sensitivity and specificity values at which the ICER crossed the £30,000 threshold. The increments in specificity are shown in steps of 2%, to keep the table concise. As the table shows, if the sensitivity of OSNA was 100%, the specificity had to be > 88% for the ICER to be > £30,000 per QALY gained (at 87% specificity the ICER was £24,928 per QALY gained). When the specificity was ≥ 92% and the sensitivity was 100%, OSNA dominated histopathology, having higher QALY gains and lower costs. If the specificity was 100% the sensitivity of OSNA had to be at least 93% to have an ICER > £30,000 per QALY gained (at 92% sensitivity the ICER was £24,683). In this case the sensitivity had to reach 96% before OSNA dominated histopathology. As sensitivity and specificity were decreased below 100%, the ICERs decreased. When we looked at reducing both sensitivity and specificity but keeping the ICER > £30,000 per QALY gained, the lowest values that they could take were 95% for specificity and 96% for sensitivity (the ICER was £34,639 per QALY gained compared with histopathology).
| OSNA specificity (%) | Incremental cost (£) per QALY gained for histopathology vs. full-node OSNA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| OSNA sensitivity (%) | |||||||||
| Base case: 84.5 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | |
| Base case: 91.8 | 4324 | 9972 | 11,720 | 14,193 | 17,958 | 24,389 | 37,867 | 84,008 | OSNA dominates histopathology |
| 88 | 3046 | 6264 | 7115 | 8225 | 9730 | 11,890 | 15,248 | 21,185 | 34,527 |
| 90 | 3683 | 8001 | 9232 | 10,899 | 13,279 | 16,957 | 23,393 | 37,547 | 94,097 |
| 92 | 4399 | 10,221 | 12,041 | 14,631 | 18,608 | 25,498 | 40,345 | 95,774 | OSNA dominates histopathology |
| 94 | 5209 | 13,159 | 15,947 | 20,204 | 27,506 | 42,943 | 97,181 | OSNA dominates histopathology | OSNA dominates histopathology |
| 96 | 6132 | 17,228 | 21,746 | 29,425 | 45,362 | 98,379 | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology |
| 98 | 7194 | 23,238 | 31,259 | 47,620 | 99,410 | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology |
| 100 | 8430 | 33,015 | 49,733 | 100,308 | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology | OSNA dominates histopathology |
Although these results suggest that there are values for sensitivity and specificity at which OSNA may be considered cost-effective, there is little evidence in the clinical effectiveness review to suggest that these values are the true sensitivity and specificity of OSNA.
Glossary
- Axillary lymph node
- Lymph node located in the axilla or armpit.
- Axillary lymph node dissection
- Removal of some or all of the lymph nodes within the axilla.
- Cost-effectiveness analysis
- An economic analysis that converts effects into health terms and describes the costs of additional health gain.
- Cost-effectiveness threshold
- The maximum incremental cost-effectiveness ratio that a technology may have if it is to be considered cost-effective. This may be interpreted as the maximum cost that the NHS is willing to pay to produce one unit of benefit (e.g. quality-adjusted life-year or patient correctly diagnosed) with the technology.
- Crossing point
- The point during a quantitative polymerase chain reaction at which the level of fluorescence is above background.
- Decision modelling
- A theoretical construct that allows a comparison of the relationship between costs and outcomes of alternative health-care interventions.
- Distant metastases
- Cancer that has spread from the original (primary) tumour to distant organs or distant lymph nodes.
- Dominant
- A diagnostic option that costs less and results in more benefits (i.e. quality-adjusted life-years or patients correctly diagnosed) than the alternative.
- Dominated
- A diagnostic option that costs more and results in fewer benefits (i.e. quality-adjusted life-years or patients correctly diagnosed) than the alternative.
- Extendedly dominant
- A diagnostic option with a lower incremental cost-effectiveness ratio (relative to a third option) and more benefits than the alternative.
- Extendedly dominated
- A diagnostic option with a higher incremental cost-effectiveness ratio (relative to a third option) and fewer benefits than the alternative.
- False negative
- Incorrect negative test result – number of diseased persons with a negative test result.
- False positive
- Incorrect positive test result – number of non-diseased persons with a positive test result.
- Incremental cost-effectiveness ratio
- The difference in the mean costs of two interventions in the population of interest divided by the difference in the mean outcomes in the population of interest. This may be interpreted as the cost of producing one unit of benefit (e.g. quality-adjusted life-year or patient correctly diagnosed) with NHS resources.
- Index test
- The test whose performance is being evaluated.
- Locoregional metastases
- Metastasis (spread) of a cancer only within the region in which it arose.
- Lymph node
- An organ of the immune system that filters foreign particles and bacteria from lymph fluid.
- Meta-analysis
- Statistical techniques used to combine the results of two or more studies and obtain a combined estimate of effect.
- Metastatic disease
- The spread of cancer from one organ or body part to another organ or body part.
- Polymerase chain reaction
- A technology used for amplifying deoxyribonucleic acid sequences.
- Primary tumour
- A tumour growing at the anatomical site where tumour progression began.
- Quality-adjusted life-year
- A measure of health gain, used in economic evaluations, in which survival duration is weighted or adjusted by the patient’s quality of life during the survival period.
- Quantitative reverse transcriptase-polymerase chain reaction
- The cloning of genes by reverse transcribing the ribonucleic acid of interest into its deoxyribonucleic acid (DNA) complement through the use of reverse transcriptase. Subsequently, the newly synthesised complementary DNA is amplified using traditional polymerase chain reaction techniques and may be quantitatively measured using fluorescent probes.
- Receiver operating characteristic curve
- A graph which illustrates the trade-offs between sensitivity and specificity that result from varying the diagnostic threshold.
- Reference standard
- The best currently available diagnostic test against which the index test is compared.
- Regional metastases
- The spread of cancer beyond the initial site to regional lymph nodes.
- Reverse transcription loop-mediated isothermal amplification
- A technique to amplify messenger ribonucleic acid directly from tissue lysates.
- Sensitivity
- The proportion of people with the target disorder who have a positive test result.
- Sentinel lymph node
- The first lymph node(s) to which cancer cells are likely to spread from a primary tumour.
- Specificity
- The proportion of people without the target disorder who have a negative test result.
- Summary receiver operating characteristic plot or curve
- A diagram used to plot the results of studies included in a systematic review of test accuracy. If a meta-analysis is performed it may include a summary point or a summary curve or both.
- Tissue allocation bias
- This occurs when tumour deposits exist in different portions of tissue, which are separately allocated to the index and reference test.
- True negative
- Correct negative test result – number of non-diseased persons with a negative test result.
- True positive
- Correct positive test result – number of diseased persons with a positive test result.
List of abbreviations
- ALN
- axillary lymph node
- ALND
- axillary lymph node dissection
- B-CLOSER-I
- Breast Complete Lymphadenectomy OSNA Study for Enhanced Review-I
- CE
- Conformitée Européenne
- CI
- confidence interval
- CK19
- cytokeratin-19
- Cp
- crossing point
- DCIS
- ductal carcinoma in situ
- DES
- discrete event simulation
- DNA
- deoxyribonucleic acid
- DTA
- diagnostic test accuracy
- EQ-5D
- European Quality of Life-5 Dimensions
- ERG
- Evidence Review Group
- FNAC
- fine-needle aspiration cytology
- HEED
- Health Economic Evaluations Database
- HR
- hazard ratio
- HRG
- Healthcare Research Group
- HSROC
- hierarchical summary receiver operating characteristic
- ICER
- incremental cost-effectiveness ratio
- ICTRP
- International Clinical Trials Registry Platform
- ITC
- isolated tumour cell
- LN
- lymph node
- MDT
- multidisciplinary team
- mRNA
- messenger ribonucleic acid
- NICE
- National Institute for Health and Care Excellence
- NPV
- negative predictive value
- OSNA
- one-step nucleic acid amplification
- PPV
- positive predictive value
- QALY
- quality-adjusted life-year
- qRT-PCR
- quantitative reverse transcriptase-polymerase chain reaction
- QUADAS
- Quality Assessment of Diagnostic Accuracy Studies
- RNA
- ribonucleic acid
- ScHARR
- School of Health and Related Research
- SD
- standard deviation
- SE
- standard error
- SLN
- sentinel lymph node
- SLNB
- sentinel lymph node biopsy
- SROC
- summary receiver operating characteristic
- STARD
- Standards for the Reporting of Diagnostic Accuracy Studies
- TAB
- tissue allocation bias
- TNM
- tumour, node, metastasis
- VAT
- value added tax
- WHO
- World Health Organization
- YHEC
- York Health Economics Consortium
This monograph is based on the Technology Assessment Report produced for the National Institute for Health and Care Excellence (NICE). The full report contained a considerable number of data that were deemed academic-in-confidence. The full report was used by the Appraisal Committee at NICE in their deliberations. The full report with each piece of academic-in-confidence information (or data) removed and replaced by the statement ‘academic-in-confidence information has been removed’ is available on the NICE website: www.nice.org.uk.
The present monograph presents as full a version of the report as possible while retaining readability, but some sections, sentences, tables and figures have been removed. Readers should bear in mind that the discussion, conclusions and implications for practice and research are all based on the data considered in the original full NICE report.