Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 14/69/04. The protocol was agreed in May 2015. The assessment report began editorial review in August 2015 and was accepted for publication in November 2015. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Gavin D Perkins is a member of the Health Services and Delivery Research (researcher-led) panel. Ronan McMullan reports grant funding (2012–17) as a member of the Innovate UK-funded consortium, with industry partner (Randox Ltd), to develop and validate a novel point-of-care diagnostic test for sepsis. Paul Dark reports grants from Innovate UK (Technology Strategy Board) during the conduct of the study.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2016. This work was produced by Stevenson et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Background and definition of the decision problem
Background to sepsis and bloodstream infection
Sepsis is a condition characterised by the body’s inflammatory response to an infection. Sepsis is diagnosed where there is evidence of systemic inflammation, in addition to a documented or presumed bloodstream infection. Systemic illness often occurs when bacteria or fungi invade normally sterile parts of the body. One example of this is the invasion of bacteria or fungi into the bloodstream, a process that often causes an inflammatory immune response. A pictorial representation of the relationship between systemic inflammatory response syndrome (SIRS), infection, sepsis and severe sepsis is provided in Figure 1.
FIGURE 1.
The relationship between SIRS, infection, sepsis and severe sepsis. 1 Reproduced with permission from the Royal College of Physicians.

If sepsis is not treated with antibiotics it can progress to severe sepsis or septic shock and can lead to multiple organ failure and death. Severe sepsis occurs when the body’s response to infection interferes with the functioning of vital organs, such as the heart, kidneys, lungs or liver. Severe sepsis has historically been defined as infection and the presence of at least two SIRS criteria;2 however, a recent paper suggests that the need for two or more SIRS criteria excludes one in eight patients with infection, organ failure and substantially increases mortality risk. 3 SIRS criteria are a fever of > 38 °C or < 36 °C; heart rate of > 90 beats per minute; respiratory rate of > 20 breaths per minute or arterial carbon dioxide tension of < 32 mmHg; or an abnormal white blood cell count [> 12,000 cells per µl or < 4000 cells per µl, or > 10% immature neutrophils (bands)]. 4
Septic shock occurs in severe cases of sepsis and is defined as persistent sepsis-induced hypotension (low blood pressure) despite adequate fluid resuscitation. Septic shock prevents organs from receiving enough oxygenated blood. Complications of septic shock can include respiratory failure, heart failure, kidney injury or failure, and abnormal blood clotting. Severe sepsis is a time-critical condition and delays in recognition and the subsequent administration of appropriate treatment can adversely impact on outcomes. It has been reported that the survival rate of untreated patients with sepsis decreases by the hour. 5
The cost implications of sepsis are considerable. The consequences, in terms of mortality and morbidity, are severe, with Levy et al. 6 reporting a mortality rate of 46% for septic patients with both hypotension and a blood lactate concentration ≥ 4 mmol/l. However, compliance with the 2004 Surviving Sepsis Guidelines7 appears to reduce both mortality and length of stay outcomes; Levy et al. 8 report that mortality was lower (29.0%) in those with high compliance with the resuscitation bundle than in those with low compliance (38.6%). Hospital mortality rates dropped 0.7% for every 3 months of participation with the campaign, and hospital and intensive care unit (ICU) length of stay decreased 4% for every 10% increase in site compliance – all of these reductions were statistically significant. An estimate of mortality in patients with early septic shock was 29% at 90 days. 9 Lower estimates of mortality have been provided in a recent study of patients with hospital-acquired infection with 13% mortality at 28 days,10 and in data from Australia and New Zealand which report in-hospital mortality as approximately 10% for SIRS-positive sepsis and 20% for SIRS-negative sepsis. 3
Severe sepsis is one of the most common reasons for admission to a critical care unit, accounting for almost one-third of all admissions. In the UK, sepsis is estimated to be responsible for 100,000 hospital admissions and 37,000 deaths per year. 11
Bacterial infections are the most common cause of bloodstream infection; however, they can also be caused by viral and fungal infections. The most common sites of infection leading to sepsis are the lungs, urinary tract, abdomen and pelvis. Other sources of infection leading to sepsis include skin infections (such as cellulitis), post-surgical infections and infections of the nervous system (such as meningitis or encephalitis). Bacteria can be categorised into three groups: Gram-positive bacteria, Gram-negative bacteria and, very rarely, Gram-indeterminate bacteria.
Patients who are currently, or have recently been, hospitalised are at risk of acquiring a health-care-associated infection and are, therefore, at increased risk of sepsis and bloodstream infection. It is thought that the increasing number of invasive procedures (such as catheterisation), immunosuppressive therapy, antibiotic therapy and life support measures has resulted in an increase in health-care-associated bloodstream infections. 12 In 2011, an estimated 6.4% [95% confidence interval (CI) 4.7% to 8.7%] of patients in acute care hospitals were diagnosed with a health-care-associated infection, with the largest proportion (23.4%) within the ICU. 13 Of patients with a health-care-associated infection, it was estimated that 7.6% had a bloodstream infection. 13 Septic shock is most commonly associated with Gram-negative bacterial bloodstream infections, but shock can also be associated with bloodstream infections caused by Gram-positive bacteria, particularly with fulminant pneumococcal, Lancefield group A streptococcal and Staphylococcus aureus infections. 14 Community-acquired bloodstream infections occur in people who have not had recent contact with health-care services. The spectrum of pathogens isolated from these people may differ from those associated with health-care-acquired bloodstream infection. 12
Bloodstream infection is also a risk for people who are immunocompromised, particularly among people with neutropenia (abnormally low neutrophil levels in the blood), who are at risk of developing neutropenic sepsis. People who are immunocompromised have a higher incidence of infections caused by pathogens that pose low risk to those whose immune system is not impaired, such as Pseudomonas species, Listeria monocytogenes, Corynebacterium species, Candida species, coagulase-negative staphylococci, enterococci and viridans streptococci. Polymicrobial infections are also more common among people who are immunocompromised. 12
The bacteria most commonly associated with bloodstream infection in adults in England, Wales and Northern Ireland are outlined in Table 1.
| Name of bacteria | Percentage of all bacteria associated with bloodstream infection | Group of bacteria |
|---|---|---|
| Escherichia coli | 36 | – |
| Staphylococcus aureus (MSSA) | 9.7 | + |
| Klebsiella spp. | 7.8 | – |
| Non-pyogenic streptococci | 7.1 | + |
| Other Gram negative | 6.4 | – |
| Enterococcus spp. | 6.3 | + |
| Pseudomonas spp. | 4.3 | – |
| Streptococcus pneumoniae | 4.2 | + |
| Other Gram positive | 4.2 | + |
| Proteus spp. | 3.1 | – |
| Enterobacter spp. | 2.2 | – |
| Staphylococcus aureus (MRSA) | 1.6 | + |
| Bacteroides spp. | 1.5 | – |
| Group B streptococci | 1.4 | + |
| Group A streptococci | 1.4 | + |
| Diphtheroids | 1.2 | + |
| Serratia spp. | 1.0 | – |
| Acinetobacter spp. | 0.7 | – |
The types of pathogens causing bloodstream infection in children can also be slightly different from those isolated from adults with bloodstream infection. Pathogens particularly associated with community-acquired bloodstream infection in children include Streptococcus pneumoniae, Neisseria meningitidis, S. aureus and Escherichia coli. The profile of pathogens associated with health-care-associated infections in children is thought to be similar to that associated with health-care-associated infections in adults; however, polymicrobial infection and anaerobic bacteraemia are thought to occur less frequently among children. 12
Diagnosis of sepsis
Diagnostic criteria for sepsis are listed in the Surviving Sepsis Campaign guidelines16 (adapted from Levy et al. 17). In summary, regular observations of all vital signs should be taken and recorded, kidney and liver function tests should be performed, and inflammatory biomarkers and serum lactate concentration should be measured. These guidelines state that a diagnosis of sepsis should be based on infection, documented or suspected, in conjunction with hyperthermia or hypothermia, tachycardia and at least one indication of altered organ function (see below). The diagnostic criteria for sepsis include the following variables:
-
General variables – temperature of > 38.3 °C or < 36 °C, heart rate > 90 beats per minute, rapid breathing, altered mental status, significant oedema or high blood sugar concentration in the absence of diabetes.
-
Inflammatory variables – low or high white blood cell count or more than 10% of immature forms, raised plasma C-reactive protein or raised plasma procalcitonin.
-
Haemodynamic and tissue perfusion variables – low blood pressure or raised blood lactate (a concentration of ≥ 4 mmol/l suggests tissue hypoperfusion).
-
Organ dysfunction variables – low blood oxygen, reduced urine output, increased creatinine levels (indicating impaired kidney function), coagulation abnormalities, absent bowel sounds, reduced platelet count or raised plasma bilirubin levels.
Current standard of care for patients with suspected bloodstream infections or sepsis
The diagnostic work-up of sepsis and bloodstream infection is described in several guidelines:
-
the National Institute for Health and Care Excellence (NICE) Clinical Guideline 151: Prevention and Management of Neutropenic Sepsis in Cancer Patients18
-
the Royal College of Obstetricians and Gynaecologists: Green-Top Guideline 64a. Bacterial Sepsis in Pregnancy19
-
the Royal College of Obstetricians and Gynaecologists: Green-Top Guideline 64b. Bacterial Sepsis following Pregnancy20
-
Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock. 16
In addition, a NICE Clinical Guideline, Sepsis: The Recognition, Diagnosis and Management of Severe Sepsis, is currently in development with an estimated publication date of July 2016. 21 Furthermore, the Commissioning for Quality and Innovation, which is currently developing a payment framework, has announced new sepsis mandates to monitor adherence to the sepsis care pathway across the NHS. 22
The Surviving Sepsis Campaign guidelines make the following specific recommendations relating to the detection of localised and bloodstream infection:16
-
At least two sets of blood cultures should be collected (aerobic and anaerobic) before antimicrobial therapy is initiated, provided this does not significantly delay (> 45 minutes) the start of antimicrobial administration. At least one sample should be drawn percutaneously and one drawn through each vascular access device, unless the device was recently (< 48 hours) inserted. The blood cultures can be drawn at the same time if they are obtained from different sites. Cultures of other fluids, such as urine, cerebrospinal fluid, wound exudate, respiratory secretions or other bodily fluids, which may be the source of infection should be obtained before initiation of antimicrobial therapy, as long as doing so does not significantly delay the start of antimicrobial administration.
-
Imaging studies such as computerised tomography or radiography should be performed in order to confirm a potential source of infection.
-
Assays to diagnose systemic fungal infection should be used if available, and invasive candidiasis is suspected.
The Surviving Sepsis Campaign guidelines recommend care ‘bundles’ which should be initiated during the diagnostic work-up of a patient. The 3-hour bundle should be completed within 3 hours of a patient developing symptoms that are indicative of sepsis:
-
measure blood lactate levels to identify tissue hypoperfusion
-
obtain blood cultures prior to administration of antibiotics
-
administer broad-spectrum antibiotics
-
administer 30 ml/kg of crystalloid for hypotension or lactate ≥ 4 mmol/l.
The 6-hour bundle should be completed within 6 hours of presentation in the emergency department or recording of symptoms if in hospital when sepsis starts:
-
apply vasopressors (for hypotension that does not respond to initial fluid resuscitation) to maintain a mean arterial pressure of ≥ 65 mmHg
-
in the event of persistent arterial hypotension despite volume resuscitation (septic shock) or initial lactate concentration of ≥ 4 mmol/l:
-
– measure central venous pressure
-
– measure central venous oxygen saturation
-
-
remeasure lactate concentration, if initial lactate was elevated.
The treatment of sepsis varies based on the initial infection, the organs affected and the extent of tissue damage. The management of severe sepsis and septic shock is described by the Surviving Sepsis Campaign in its International Guidelines for the Management of Severe Sepsis and Septic Shock. 16 All patients with severe sepsis or septic shock will require initial resuscitation, antimicrobial therapy, source control (where appropriate) and fluid therapy. Some patients may require additional treatment with vasopressors, inotropic therapy, corticosteroids or other supportive therapy.
It is recommended that intravenous empiric antimicrobials should be administered within the first hour of recognition of septic shock and severe sepsis. The initial antimicrobial therapy should include one or more drugs that have activity against all likely pathogens (bacterial and/or fungal or viral) and that penetrate in adequate concentrations into the tissues presumed to be the source of sepsis. 16 Such treatment is typically referred to as broad spectrum. Frequently used broad-spectrum antibiotics for more serious infections include beta-lactams and aminoglycosides. Carbapenems are often the last option in patients with hard-to-treat infections. 23
The choice of empirical antimicrobial therapy is often based on:
-
the patient’s history, including drug intolerances
-
recent treatment with antibiotics
-
underlying disease
-
the clinical syndrome
-
susceptibility patterns of pathogens in the local community and hospital
-
microbiology reports identifying pathogens that have previously colonised or infected the patient.
Clinicians should also consider whether or not a fungus is a likely causative pathogen when selecting initial therapy and administer, when appropriate, empirical antifungal therapy.
The use of antimicrobials varies between hospitals, as prescribing choices are influenced by local resistance and susceptibility patterns. The choice of antimicrobials is also influenced by the suspected source of the infection and local prescribing protocols may be developed for:
-
urinary tract infections
-
upper respiratory tract infections
-
lower respiratory tract infections
-
soft-tissue infections
-
central nervous system infections
-
gastrointestinal infections and genital tract infections
-
bloodstream infections
-
eye, ear, nose and throat infections
-
sepsis of unknown origin.
Current practice for detecting pathogens in those with suspected bloodstream infection or sepsis
The current practice for detecting pathogens in those with suspected bloodstream infection or sepsis consists of clinical assessment in conjunction with blood culture. However, within the NICE scope for this project,24 an additional comparator of clinical assessment in conjunction with blood culture and matrix-absorbed laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (MS) was included in recognition of the fact that some hospitals are incorporating MALDI-TOF MS within their standard practice. MALDI-TOF MS has an advantage of shortening the time required for identifying the causative pathogen when a blood culture becomes positive.
Blood culture
Blood culture is required for the detection and subsequent identification of bloodstream bacteria and fungi, and to provide potential definitive antimicrobial susceptibility data. Standards for the investigation of blood cultures are available from Public Health England. 12 A blood culture set for the diagnosis of bloodstream infection is defined as one aerobic and one anaerobic bottle. 12 For adult patients it is recommended that 20–30 ml of blood be cultured per set, and that two consecutive blood culture sets from two separate sites should be collected during any 24-hour period for each septic episode. The first set should be taken prior to the administration of antimicrobial treatment as the presence of antibiotics or antifungals may inhibit the growth of pathogens in the blood culture. 12 Blood culture bottles should be incubated within 4 hours of the blood sample being taken, with many laboratories now using automated culture systems such as the BACTEC (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD, USA) or BacT/ALERT (Oganon Teknika Corp., Durham, NC, USA) systems, which alert laboratory staff once growth has been detected.
The time taken for a blood culture bottle to show positivity is variable and can depend on the individual pathogen, the volume of cultured blood, the concentration of organisms in the sample, whether or not there are multiple pathogens and whether or not the patient had recently received antibiotics prior to the blood being sampled. 25,26 A median time to positivity of approximately 15 hours has been reported, but with a wide range for individual samples. 25,26
When a blood culture bottle has been detected as positive, it is recommended that:
-
Gram staining and rapid antigen testing should be performed within 2 hours.
-
Direct or automated isolate identification should be performed within 24 hours (extending to 48 hours if traditional microbiology techniques, such as morphological identification are used). Rapid species identification may be done following blood culture using techniques such as MALDI-TOF MS.
-
Identification should be followed by susceptibility testing to determine to which antimicrobials the identified pathogen is susceptible. A preliminary report should be made within 24 hours.
-
A preliminary positive report is made within 2 hours of identification and susceptibility testing, and a final positive report should be made within 5 days of the sample arriving in the laboratory. 12
The first target is not typically met by laboratories because if the blood culture is detected as positive during the night, Gram staining would not occur until the laboratory opened in the morning.
If a blood culture is not positive within 48 hours of sample receipt in the laboratory it is recommended that a preliminary negative report is provided with a final negative report issued within 5 days, unless extended culture is being undertaken, for example if fungi or unusual, fastidious or slow-growing organisms are suspected. 12
Blood culture results may not detect pathogens within an individual’s bloodstream because of the transient nature of bloodstream infections and a low number of organisms present in a blood sample; there can often be fewer than 1 × 103 colony-forming units (cfus) per litre in adults with a bloodstream infection. 12 The presence of antibiotic treatment prior to the blood being sampled can also result in pathogens not being detected. Conversely, blood culture results may identify a pathogen that is not in an individual’s bloodstream as pathogens transferred from the skin during the drawing of blood can contaminate the culture. To reduce the incidence of such false-positive results, current standards recommended that contamination rates are no higher than 3%. 12 In addition, several criteria may be used to differentiate between contamination and true bloodstream infection, which include the identity and clinical significance of the pathogen, the number of positive blood culture sets and positive culture bottles, and the quantity of growth detected.
Blood culture sample collection differs for infants and neonates, for whom a single aerobic bottle or low-volume blood culture bottle may be requested. 12 Criteria for calculating total blood culture volumes in neonates and children are based on weight rather than age, and relate to total patient blood volume. It has been suggested that the volume of blood drawn should be no more than 1% of the patient’s total blood volume. 12 The magnitude of bacteraemia is usually higher in infants and children than that in adults and, therefore, the sensitivity of detection is not believed to be significantly reduced by lower blood-to-medium ratios. 12
Although blood culture is considered the gold standard, a number of limitations regarding its use were identified; for example, it has been estimated that only 30–60% of blood cultures taken from patients with sepsis are positive. 27 This may indicate poor sensitivity, which may be attributed to commencement of antimicrobial therapy prior to sample collection, low pathogen levels in blood and inadequate blood sampling. Additionally, blood culture does not always pick up fungal pathogens. 28
Matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
Following a blood culture becoming positive, it is possible to use MALDI-TOF MS to provide an identification of the pathogen more quickly than by standard phenotypic techniques alone. Details on MALDI-TOF MS have been provided by Schubert et al. ,29 where pathogens were identified from an agar plate. Recently, however, Sepsityper (Bruker, Billerica, MA, USA), a preparation method prior to MALDI-TOF MS, has been developed, allowing MALDI-TOF MS to be used directly on a positive blood culture bottles without the need for growing pathogens on an agar plate. The use of Sepsityper can thus provide a result more quickly than standard culture-based identification techniques or MALDI-TOF MS used in conjunction with agar plates. Morgenthaler and Kostrzewa30 report that ‘the use of the Sepsityper sample preparation kit leads to a reduction in overall time to results from 8 to > 48 hours (in some studies > 100 hours), depending on the microorganism growth rate on solid phase culture plates’. The level of Sepsityper use in England is currently unknown.
A recently completed National Institute for Health Research-funded study RAPIDO (A Prospective Randomised, Multicentre Trial to Assess the Impact of Laboratory based Rapid Diagnosis on Outcome in patients with Blood Stream Infections) has compared MALDI-TOF MS with standard practice having recruited 4536 patients from the UK. 31 However, at the time of writing, the data analysis had not been fully conducted. The primary outcome measure within the randomised controlled trial (RCT) is the 28-day all-cause mortality between the two arms. Following personal communication with Dr Leeming (Dr John Leeming, North Bristol NHS Trust, 2015, personal communication), it was identified that Sepsityper had been used in the MALDI-TOF MS arm in all centres bar Newcastle upon Tyne, where the centre used its own method.
The risk of antimicrobial resistance
Broad-spectrum antibiotics administered to patients with suspected sepsis are a mainstay of treatment; however, these interventions cannot be used indiscriminately without risking unwanted consequences. Antimicrobial resistance describes the development of resistance to existing antimicrobial medications (including antibiotics, antifungals and antivirals) among bacteria, viruses and fungi. As existing antimicrobial medications are becoming less effective, strategies such as the UK Five Year Antimicrobial Resistance Strategy23 have been introduced to help conserve the effectiveness of existing treatments. One of the key priorities outlined in the UK Five Year Antimicrobial Resistance Strategy is the introduction of antimicrobial stewardship programmes which aim to promote the rational prescribing of antimicrobial medications, and the use of existing and new rapid diagnostic tests.
Recent surveillance data for England suggest that rates of meticillin-resistant S. aureus infection have fallen, while there is an increase in the incidence of bloodstream infections caused by resistant Enterobacteriaceae (Gram-negative bacteria) such as Klebsiella spp. and E. coli. Of particular concern in some regions of England, such as the north-west and Greater London, is the increasing resistance to carbapenem antibiotics which are often used as a last resort for treating severe infections.
Clinicians prescribing antimicrobial therapy should take into account the Department of Health’s guidance on antimicrobial stewardship, which is based on the ‘start smart then focus’ strategy. 32 The guidance recommends that, when antimicrobials are administered empirically, the patient is reviewed after 48–72 hours to allow an ‘antimicrobial prescribing decision’ to be made. This decision should take into account available microbiology results to determine whether therapy can be stopped or changed, that is, the de-escalation, substitution or addition of antimicrobial agents to the treatment plan. 32 Narrowing the spectrum of antimicrobial coverage and reducing the duration of therapy is thought to be associated with a reduction in the risk of a patient developing a superinfection, a reduction in the selection of resistant organisms and a reduction in the treatment of related side effects. Adverse events associated with the use of broad-spectrum antimicrobials may include diarrhoea, nausea, vomiting, hearing loss, damage to the kidneys and an increased risk of developing a superinfection with Clostridium difficile.
Narrowing the spectrum of antimicrobial coverage may also be associated with an increase in treatment efficacy, as certain broad-spectrum antibiotics may not be as effective as related narrow-spectrum antibiotics against certain pathogens. 32 In addition, a reduction in agents may result in cost savings.
The National Institute for Health and Care Excellence recently issued a draft clinical guideline on antimicrobial stewardship which discussed the evidence for de-escalation of antimicrobials. 33 A conclusion of this draft guideline was that five RCTs had assessed the impacts of de-escalation (although only three are explicitly referenced34–36), four of which were set in ICUs, the exception being hospital based, and only one of which, by Leone et al. ,35 was in patients with sepsis. The Guideline Development Group found no evidence from these RCTs that de-escalation between 48 and 72 hours increased patient mortality. The Guideline Development Group found little evidence of increased length of ICU or hospital stay but noted the exception of Leone et al. ,35 which was classified as a low-quality RCT, who recruited 116 patients with severe sepsis who were randomised to de-escalation or continuation of empirical antimicrobial treatment. Leone et al. 35 reported statistically significantly greater rates of superinfection in the de-escalation group (27% vs. 11%; p = 0.03) and in the mean number of antibiotic days (9 vs. 7.5 days; p = 0.03), although the increase in median duration of ICU stay (9 vs. 8 days) was not statistically significant (p = 0.71). The Guideline Development Group noted that it identified no health economic evidence regarding which interventions, systems and processes are effective or cost-effective in reducing antimicrobial resistance without causing harm to patients, nor did it identify any health economic evaluations that included outcomes of antimicrobial resistance.
We have used the term ICU throughout the report as this is the term often used in the published literature, although we recognise that care can also be provided in other critical care settings. We have assumed that such settings are encompassed by the ICU categorisation.
The External Assessment Group note that clinical advice received during the scoping process stated that a barrier to de-escalation in practice could be the resistance of family members to change the treatment in a patient who was clearly improving and, thus, the extent to which de-escalation would occur in clinical practice is unclear.
The potential benefits and possible harms of a test that could provide earlier information on pathogen
The individual characteristics of the three tests evaluated in this report [LightCycler SeptiFast Test MGRADE® (Roche Diagnostics, Risch-Rotkreuz, Switzerland); SepsiTest™ (Molzym Molecular Diagnostics, Bremen, Germany); and IRIDICA BAC BSI assay (Abbott Diagnostics, Lake Forest, IL, USA)] are detailed in the following section. The aim of this section is to explain the benefits that could be provided by tests that report information on the type of bacteria earlier than standard blood culture methods, with or without MALDI-TOF MS, which can be used with or without Sepsityper. Were a rapid test to have a sensitivity of 100% and a specificity of 100% in identifying the pathogen(s), caused by bloodstream infection, that is, the test was perfect, management strategies could be quickly altered dependent on whether or not there was presence of a pathogen. Were a pathogen to be identified, then treatment could be tailored to that pathogen alongside de-escalation of antimicrobial treatment by removing the components of broad-spectrum treatment to which either the pathogen was not sensitive, or to which a targeted treatment was more effective. Were a pathogen not identified, then treatment could be de-escalated or removed entirely. Owing to the rapid identification by the test, these benefits would be achieved more quickly than through standard techniques.
The advantages of earlier appropriate treatment have been reported in the published literature. A Spanish, retrospective, matched, cohort study37 attempted to determine the attributable mortality and excess length of stay associated with inadequate empirical antimicrobial therapy between 1997 and 2006. Therapy was considered inadequate when no effective drug against the isolated pathogen(s) was included in the empirical antibiotic treatment within the first 24 hours of admission to the ICU, or the doses and pattern of administration were not in accordance with current medical standards. From 87 matched pairs, 59 (67.8%) died in the inadequate group compared with 25 (28.7%) in the control group. Removing pairs with nosocomial infection still showed 31.4% excess in mortality (65.7% in the inadequate group vs. 34.3% in the control group). In those without a nosocomial infection there was a significant reduction in the length of stay in ICU associated with adequate treatment (7 days in the inadequate group vs. 9 days in the control group; p = 0.02).
Using a generalised linear model, adjusted for confounders, Zilberberg et al. 38 estimated that the excess length of hospitalisation was 7.7 days (95% CI 0.6 to 13.5 days) and excess costs were US$13,398 (95% CI US$1060 to US$26,736) when a patient had inadequate antifungal treatment. Inadequate antifungal treatment was defined as a treatment delay of ≥ 24 hours from candidaemia onset or inadequate dose of an antifungal agent active against the pathogen.
Arnold et al. 39 attempted to estimate the costs of inappropriate treatment of candidaemia from 167 consecutive patients, which was defined as delayed antifungal therapy > 24 hours from culture collection. Twenty-two patients had appropriate therapy; 145 did not. Length of stay was shorter in the appropriately treated group than in the non-appropriately treated group (7 vs. 10.4 days, respectively; p = 0.037) and the costs were lower (US$15,832 vs. US$33,021, respectively; p < 0.001).
Morrell et al. 40 retrospectively analysed 157 consecutive patients who, over a 4-year period, developed a Candida bloodstream infection, of whom 50 (32%) died during hospitalisation. The number of patients in whom antifungal treatment was not delayed (> 12 hours) was nine, while treatment was delayed in 148 patients. The adjusted odds ratio associated with delay in antifungal treatment was 2.09 (95% CI 1.53 to 2.84). Delays in antifungal treatment were also associated with a longer stay in ICU (9.4 days, compared with 0.4 days for those in whom treatment was not delayed; p = 0.019).
It is unlikely that the tests evaluated would be 100% sensitive and 100% specific, meaning that the consequences of misdiagnoses would also need to be considered. These take the form of false positives (a pathogen that is not present is identified) and false negatives (a pathogen that is present in the blood culture is not identified). The consequences of these misdiagnoses are likely to differ. In the case of false positives, there is the risk of overtreatment, which would incur cost and could increase the risk of antimicrobial resistance; in the case of false negatives, withdrawal of treatment could put the patient at increased risk of morbidity and mortality.
However, it is known that diagnostic inaccuracy is not confined to the new tests and can occur in standard techniques and, therefore, that standard techniques provide an inaccurate gold standard that may result in biased evaluation of the interventions. This is believed most likely where the correct identification of a pathogen could be classed as a false positive if it was not detected by blood culture. As detailed in this report, some clinical experts believe that such results would provide valuable information in the patient treatment decision, despite adversely affecting the specificity of the test against blood culture.
Description of the technologies under assessment
Our research aims to evaluate the clinical effectiveness and cost-effectiveness of three tests that potentially allow the rapid detection and identification of bacterial and fungal deoxyribonucleic acid (DNA) in the bloodstream of patients suspected of having sepsis. These tests are the SeptiFast, SepsiTest and IRIDICA BAC BSI assays, which will be compared with blood culture, with or without, MALDI-TOF MS. Each test is intended to be run directly on whole-blood samples without prior incubation or preculture steps, allowing an earlier initial assessment of the patient. It is anticipated that blood cultures and clinical judgement would be required in conjunction with each test to provide additional, potentially more definitive, data on the most effective antimicrobial to use, as data on this provided by the interventions are very limited. This section details the three technologies; the comparators have been described in Current practice for detecting pathogens in those with suspected bloodstream infection or sepsis. For brevity, where the test name alone is provided it should be assumed that this denotes its use in conjunction with blood cultures and clinical judgement. Similarly, any reference to blood culture, with or without MALDI-TOF MS, also denotes these being used in conjunction with clinical judgement.
LightCycler SeptiFast Test MGRADE
The LightCycler SeptiFast Test MGRADE – henceforth referred to as SeptiFast – is a Conformité Européenne (CE)-marked in vitro diagnostic real-time polymerase chain reaction (PCR) test that simultaneously detects and identifies bacterial and fungal DNA. The test requires 1.5 ml of ethylenediaminetetraacetic acid (EDTA)-treated whole blood, which can be processed without prior incubation or culturing. SeptiFast involves three distinct processes: specimen preparation by mechanical lysis and purification of DNA; real-time PCR amplification of target DNA in three parallel reactions (Gram-positive bacteria, Gram-negative bacteria and fungi); and detection using fluorescence-labelled probes specific to the target DNA. The test takes around 6 hours in optimal conditions, but could take longer depending on laboratory workflow.
The SeptiFast Identification Software set v2.0 (Roche Diagnostics, Risch-Rotkreuz, Switzerland) analyses the samples and generates a report including relevant laboratory data and details of the identified species. The software also includes a crossing point cut-off rule that is intended to reduce the positive rate for coagulase-negative staphylococci and Streptococcus spp. based on the assumption that they are contaminants and not causal agents when the crossing point value is < 20.
Where S. aureus is identified in a sample, an aliquot of the SeptiFast Test MGRADE eluate can be further tested for the presence of the mecA gene using the LightCycler SeptiFast MecA Test MGRADE. The test is intended to determine the likely meticillin resistance of S. aureus through PCR using the LightCycler 2.0 instrument.
The bacterial and fungal species that can be detected by SeptiFast are shown in Table 2.
| Bacteria | Fungi | |
|---|---|---|
| Gram negative | Gram positive | |
| E. coli | S. aureus | Candida albicans |
| Klebsiella (pneumoniae/oxytoca) | Coagulase-negative staphylococci (including Staphylococcus epidermidis and Staphylococcus haemolyticus) | Candida tropicalis |
| Serratia marcescens | S. pneumoniae | Candida parapsilosis |
| Enterobacter (cloacae/aerogenes) | Streptococcus spp. (including Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus mitis) | Candida krusei |
| Proteus mirabilis | Enterococcus faecium | Candida glabrata |
| Pseudomonas aeruginosa | Enterococcus faecalis | Aspergillus fumigatus |
| Acinetobacter baumannii | ||
| Stenotrophomonas maltophilia | ||
The test has an analytical sensitivity of 100 cfu/ml for coagulase-negative staphylococci, Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus pneumoniae and Streptococcus mitis. The minimum analytical sensitivity for all other pathogens detected by SeptiFast is 30 cfu/ml.
SepsiTest
SepsiTest is a CE-marked PCR test for detecting bacterial and fungal DNA in 1 ml of K-EDTA- (potassium-EDTA) or citrate-treated whole blood. The test is able to identify species from more than 200 genera of bacteria and 65 genera of fungi. The manufacturer states that SepsiTest can identify Candida krusei, although this organism has not been found in any study to date.
SepsiTest involves three distinct processes: extracting and purifying microbial DNA using centrifugation, universal PCR and Sanger sequencing. The PCR result is available after 4 hours in optimal conditions, although it could take longer depending on laboratory workflow, and indicates whether bacteria or fungi are present in the sample. Amplicons from positive samples are then sequenced to confirm the PCR result and to determine which bacterial or fungal species are present. Where readable sequences are available from sequence analysis, bacteria and fungi can be identified using the SepsiTest-BLAST online tool (www.sepsitest-blast.de/en/index.html). Sequencing results are typically available in 3–4 hours in optimal conditions, depending on the analyser used, equating to a time of 8 hours from drawing blood, but could take longer based on laboratory workflow.
The analytical sensitivity of SepsiTest ranges from 10 to 80 cfu/ml, depending on the target species.
Shortly before the submission of this report, Molzym Molecular Diagnostics, on 22 July 2015, informed NICE that it had updated SepsiTest to version 4.0 (date of change 1 July 2015). The changes reported by the company include the implementation of an internal extraction control to validate the extraction of DNA; the removal of the internal control from the kits; and the fact that processing of duplicate samples is no longer recommended. In consultation with NICE, a decision was taken to exclude the updated version of SepsiTest from the analyses described in this report, primarily because no data on the diagnostic accuracy associated with this version were provided. Given the potentially large change compared with the previous version regarding the removal of the duplicate sample, it could not be assumed, without supportive evidence, that the results from previous studies were applicable to the latest version of SepsiTest.
IRIDICA BAC BSI
The IRIDICA BAC BSI assay – henceforth referred to as IRIDICA – is a CE-marked in vitro diagnostic test for detecting and identifying bacterial and candidal DNA in 5 ml of EDTA-treated whole blood. The test can also detect the mecA (Staphylococcus-specific meticillin resistance), vanA and vanB (Enterococcus-specific vancomycin resistance) and Klebsiella pneumoniae carbapenemase-producing (Gram-negative-associated carbapenem resistance) genes, which are associated with antibiotic resistance. The test is designed for use with the IRIDICA system, which combines broad-range PCR with electrospray ionisation time-of-flight mass spectrometry to amplify and detect pathogens. The IRIDICA system includes a proprietary database and software that identifies the organism present in the sample by comparing the sequence of the sample with a library of known sequences. The IRIDICA system was developed incrementally from a previous test called PLEX-ID (Abbott Diagnostics, Lake Forest, IL, USA), although the final IRIDICA system has key differences from PLEX-ID as it uses a greater volume of whole blood (5 vs. 1.5 ml, respectively) and has different desalter and mass spectrometry modules. The company supplied confidential data regarding the equivalency of IRIDICA and PLEX-ID, which the company declared demonstrated that the limits of detection of four core organisms were comparable in IRIDICA and PLEX-ID. Based on these data, the External Assessment Group was comfortable with including data from studies that used IRIDICA–PLEX-ID hybrid systems.
The IRIDICA assay is able to detect over 780 bacterial and candidal species. The mean limit of detection for the assay is 39 cfu/ml, with a range of 0.25–128 cfu/ml depending on the target species. The estimated time to result is 5 hours and 55 minutes in optimal conditions, although it may take longer depending on laboratory workflow.
The decision problem
This report aims to evaluate the clinical effectiveness and cost-effectiveness of the three interventions in comparison with blood culture, with or without MALDI-TOF MS. As detailed in The potential benefits and possible harms of a test that could provide earlier information on pathogen, there are reasons to believe that a quicker identification of pathogens can produce health benefits. The quickest time at which clinically important information would be available for each test is provided in An estimation of the time to clinically important information associated with each intervention and comparator.
It is anticipated that good compliance with the guidelines described in Current standard of care for patients with suspected bloodstream infections or suspected sepsis and Current practice for detecting pathogens in those with suspected bloodstream infection or sepsis will be associated with better patient outcomes. As such, the potential gains associated with the three tests have likely been reduced since the introduction of the guidelines. Given the early use of broad-spectrum antibiotics, it is anticipated that there will be much greater clinical utility in accurately determining the specific pathogen causing the infection rather than in determining whether or not the patient has sepsis.
An estimation of the time to clinically important information associated with each intervention and comparator
Table 3 denotes estimations of time to clinically relevant events in the detection of pathogens associated with bloodstream infections. It is noted that for the interventions it has been assumed that workflow is optimal; that is, that the test result will be reported back in a timely manner and not delayed because of staff hours, waiting for additional blood to be gathered which will be tested simultaneously or transport times. For the comparators, the time of day has been included in the estimates to produce a range of possible time-to-event data. As such, the timings presented in Table 3 are favourable to the interventions.
| Test | Time to indication of whether bacteria or fungi are present (hours) for SepsiTesta or time to indication of Gram stain positive or Gram stain negative in positive cultures | Time to preliminary identification of type of organism | Time to preliminary antimicrobial sensitivity data | Time to earliest possible identification of precise bacteria or fungia,b |
|---|---|---|---|---|
| Interventions | ||||
| SeptiFast | 6 hours | |||
| IRIDICA | 6 hours | |||
| SepsiTest | 4 hours (denoted as x) | (x) + 3–4 hours (range 7–8 hours) | ||
| Comparators | ||||
| Blood culture | 15 hours (denoted as y) (range 12–48 hours)c | (y) + 12–24 hours (denoted as z)d | (z) | (z) + 12–18 hourse (range 36–90 hours) |
| Blood culture with MALDI-TOF MS | (y) | (z) | (z) | (z) (range 24–72 hours) |
| Blood culture with MALDI-TOF MS and Sepsityper | (y) | (y) + 1–13 hoursd | (z) | (z) (range 24–72 hours) |
Chapter 2 Assessment of clinical effectiveness
A systematic review of the literature and meta-analysis (where appropriate) was undertaken to evaluate the clinical effectiveness of the SeptiFast, SepsiTest and IRIDICA assays in conjunction with clinical assessment for rapidly identifying bloodstream bacteria and fungi.
A review and meta-analysis was undertaken in accordance with the guidelines published by the Centre for Reviews and Dissemination for undertaking systematic reviews41 and the Cochrane Diagnostic Test Accuracy Working Group on the meta-analysis of diagnostic tests. 42,43
This report contains reference to confidential information provided as part of the NICE appraisal process. This information has been removed from the report and the results, discussions and conclusions of the report do not include the confidential information. These sections are clearly marked in the report.
Methods for reviewing effectiveness
Identification of studies
Electronic databases
Studies were identified by searching the following electronic databases and research registers from January 2006 to May 2015:
-
MEDLINE In-Process & Other Non-Indexed Citations and MEDLINE (via OvidSP)
-
EMBASE (via OvidSP)
-
Cochrane Database of Systematic Reviews (via Wiley Online Library)
-
Cochrane Central Register of Controlled Trials (via Wiley Online Library)
-
Health Technology Assessment Database (via Wiley Online Library)
-
Database of Abstracts of Review of Effects (via Wiley Online Library)
-
Science Citation Index Expanded (via the Web of Science)
-
Conference Proceedings Index-Science (via the Web of Science)
-
World Health Organization’s International Clinical Trials Registry Platform
-
Current Controlled Trials
-
National Institutes of Health ClinicalTrials.gov
-
Manufacturer and User Facility Device.
Sensitive keyword strategies using free text and, where available, thesaurus terms using Boolean operators and database-specific syntax were developed to search the electronic databases. Synonyms relating to the condition (e.g. sepsis) were combined with terms for the test (i.e. SeptiFast, SepsiTest and IRIDICA). No language restrictions were used on any database; however, the clinical effectiveness searches were date restricted. To date, all included rapid molecular tests (SeptiFast, SepsiTest and IRIDICA assay) have received a CE mark for use on whole-blood samples. For the SeptiFast test, clinical studies on whole-blood samples were first published in abstract form by Raglio et al. 44 in 2006 with subsequent full-text peer-reviewed publications by Mancini et al. 45 and Louie et al. 46 in 2008. The SeptiFast test gained its CE mark in 2006. For the SepsiTest assay, studies evaluating the use of SepsiTest on whole-blood samples in the clinical setting were first published in abstract form by Disqué et al. 47 in 2008, with a subsequent full-text peer-reviewed publication by Wellinghausen et al. 48 in 2009. SepsiTest received a CE mark in 2008. For the IRIDICA assay, studies evaluating the use of IRIDICA on whole-blood samples in the clinical setting were first published by Bacconi et al. ,49 in 2014, who used an IRIDICA–PLEX-ID hybrid system. The final version of the IRIDICA platform received a CE mark in 2014 and has been available for purchase by the NHS since 16 November 2014. Based on these data, the clinical effectiveness searches were limited by date from 2006 to May 2015. The search strategy of the current review updated the search strategy of an existing review on SeptiFast50 and amended it within the scope of the current review (i.e. the search strategy was amended to include generic, trademark or other product names of all the relevant index tests, other bacterial or fungal gene terms were added and were combined with PCR and population terms, and a limit to exclude all animal-only studies was introduced). An example of the MEDLINE search strategy is provided in Appendix 1.
Other resources
To identify additional published, unpublished and ongoing studies, the reference lists of all relevant studies were checked and a citation search of relevant articles (using the Web of Science Citation Index Expanded and Conference Proceedings Citation Index – Science) was undertaken to identify articles that cite the relevant articles. In addition, systematic keyword searches of the World Wide Web were undertaken using the Google search engine (Google Inc., Mountain View, CA, USA), key experts in the field were contacted and company submissions were screened for published or unpublished data additional to those identified in studies retrieved from the literature search.
All identified citations from the electronic searches and other resources were imported into, and managed using, the Reference Manager bibliographic software (version 12.0; Thomson ResearchSoft, San Francisco, CA, USA).
Inclusion and exclusion criteria
The inclusion of potentially relevant articles was undertaken using a three-step process. First, all titles were examined for inclusion by one reviewer (LU). Any citations that clearly did not meet the inclusion criteria (e.g. non-human, unrelated to sepsis) were excluded. Second, all abstracts were examined independently by two reviewers (LU and AP) and the full manuscript of all potentially eligible articles that were considered relevant was obtained, where possible. Third, two reviewers independently assessed the full-text articles (n = 177) for inclusion (LU and AP). All potential studies for inclusion (n = 87) were then adjudicated by three clinical experts independently (GDP, PD and RM). Any disagreements in the selection process were resolved through discussion and included by consensus between the two reviewers and three clinicians. The relevance of each article for the systematic review was assessed according to the criteria below.
Study design
All clinical diagnostic accuracy studies that evaluated the index test with standard culture results (with or without MALDI-TOF MS) on patients’ whole-blood samples during the management of suspected sepsis were included. In reviews of test accuracy the ‘index test’ (the test of which the performance is being evaluated) can be viewed as the intervention.
Reviews of primary studies were not included in the analysis but were retained for discussion and identification of additional studies. Moreover, the following publication types were excluded from the review: animal models; biological studies; narrative reviews, editorials and opinions; case reports; non-English-language papers; and reports published as meeting abstracts only when insufficient methodological details are reported to allow critical appraisal of study quality.
Population
All studies of adults and children (of any age) with suspected bloodstream infections in secondary care (i.e. departments and wards providing care for acutely unwell patients and/or critical care units) who required blood cultures were included. Potential subgroups of interest included people with a suspected health-care-associated infection, people with a suspected community-acquired infection, children and neonates, people who are immunocompromised and people exposed to antibiotics prior to blood sample collection. Following clinical advice, people with febrile neutropenia were also considered as potential subgroup of interest. This group of patients usually undergo blood culture testing as their ability to show the classical signs of sepsis are impaired and failing to treat an underlying infection can result in mortality. This practice is supported by a recent large, retrospective study by Kaukonen et al. ,3 which found that a significant number of poor outcomes from severe systemic infection occurs in the absence of SIRS criteria at inception.
Target conditions
Suspected sepsis, including severe sepsis and septic shock as defined by Levy et al. 17
Interventions (index test)
The following tests (in conjunction with clinical assessment) performed on whole-blood samples for the detection of bloodstream bacterial and fungal pathogens were included:
-
SeptiFast
-
SepsiTest
-
IRIDICA assay (extended to include preceding versions of the test if the authors believed that the data were likely to be generalisable to the IRIDICA assay).
Comparator test (reference standard)
The reference tests included current standard care to define the target condition, which included blood culture (in conjunction with clinical assessment) for the identification of bloodstream bacterial and fungal pathogens with or without MALDI-TOF MS. Where studies were identified that included more than one intervention, these would also form comparators for each intervention.
Outcomes
The outcomes of the review included a range of intermediate measures (such as diagnostic accuracy, discordant results with blood culture, time to result, time to treatment, test failure rates, duration of ICU and/or hospital stay, duration of broad- and narrow-spectrum antimicrobial therapy, readmission rate and change in antimicrobial treatment plan) and clinical outcome measures {such as side effects associated with broad-spectrum antimicrobial use, morbidity and mortality, severity of disease [as measured by scoring systems such as the Acute Physiology and Chronic Health Evaluation (APACHE) II, Simplified Acute Physiology Score (SAPS) II and the Sequential Organ Failure Assessment (SOFA)], rates of superinfection (including C. difficile), rates of resistant infections and health-related quality of life}, where available.
Data abstraction strategy
Data abstraction was performed by one of three reviewers into a standardised data extraction form and independently checked for accuracy by a second reviewer (AP, LU or MMJ). Discrepancies were resolved by discussion between the two reviewers and, if agreement could not be reached, a third reviewer was consulted. When multiple publications of the same study were identified, data were extracted and reported as a single study. Moreover, as this review of three rapid molecular tests incorporated an update of the most recent review of SeptiFast by Dark et al. ,50 all relevant data were extracted from the systematic review in the first instance, but were cross-checked for accuracy with the original papers. When necessary, additional data were extracted from the original papers. For the review of SepsiTest and IRIDICA, all data were extracted from the original papers. Unpublished study data from the company (which were received during the review process) that met the inclusion criteria were also extracted and quality assessed in accordance with the procedures outlined in this chapter.
The following information was extracted for all studies when reported: study characteristics (e.g. author, year of publication, country, study design, setting, funding), participant details (e.g. age, sex, inclusion and exclusion criteria), test details, reference standard details and outcomes (including definitions).
Quality assessment strategy
The methodological quality of each included study was assessed by one of three reviewers and independently checked by a second reviewer (AP, LU or MMJ). Disagreements were resolved by discussion between the two reviewers and, if agreement could not be reached, a third reviewer was consulted. The study quality characteristics were assessed according to (adapted) criteria based on those proposed by Whiting et al. 51 [Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2)]. Further details are provided in Appendix 2.
Methods of data synthesis
The extracted data and quality assessment variables were presented for each study, both in structured tables and as a narrative description. The analysis comprised a narrative synthesis and pairwise meta-analysis.
Meta-analysis
Where sufficient data existed, a meta-analysis was undertaken to generate pooled estimates of diagnostic parameters. The number of true positives, false negatives, false positives and true negatives from each study was meta-analysed to estimate sensitivity and specificity under the assumption that blood culture was 100% sensitive and specific. In brief, a bivariate normal model was used to model the population logit sensitivities and specificities in each study to account for correlation between sensitivity and specificity within studies. 52 We assumed that the observed number of true positives in study i, TPi, was binomially distributed, with parameter, πAi, representing the study-specific sensitivity given the total number of positives on the reference test such that:
Similarly, we assumed that the observed number of true negatives in study i, TNi, was binomially distributed with parameter, πBi, representing the study-specific specificity given the total number of negatives on the reference test such that:
We transform the parameters to the real line using the logit transformation such that:
Sensitivity and specificity are correlated within each study such that higher values for sensitivity tend to be associated with lower values for specificity, and vice versa. We model this by assuming that the study-specific logits for sensitivity and specificity arise from a bivariate normal distribution with population logits for sensitivity and specificity, (µA,µB)T, respectively, and variance–covariance matrix, ΣAB, such that:
σA2 represents the variability in the logit sensitivities between studies, σB2 represents the variability in the logit specificities between studies and σAB represents the covariance of the logit sensitivity and logit specificity.
The model was completed by giving the uncertain parameters the following prior distributions:
IW represents the inverse Wishart distribution on υ degrees of freedom.
This prior distribution has a between-study standard deviation (SD) of 1.5 [95% credible interval (CrI) 0.4 to 32.4].
Where there were relatively few studies to estimate the variance–covariance matrix, ΣAB, a weakly informative prior distribution was used such that:
This prior distribution has a between-study SD of 0.5 (95% CrI 0.3 to 1.4).
Reasons for the heterogeneity in sensitivity and specificity between studies were explored using metaregression. Models with and without covariates were compared using the deviance information criterion, which provides a relative measure of goodness of fit that penalises complexity and can be used to compare different models for the same likelihood and data. 53
All parameters were estimated using Markov chain Monte Carlo simulation implemented using the WinBUGS software package (version 1.4.3; MRC Biostatistics Unit, Cambridge, UK). 54 Analyses were conducted in R using the R2WinBUGS interface package (version 3.1.2; R Foundation for Statistical Computing, Vienna, Austria). 55 Convergence was assessed using the Gelman–Rubin convergence statistic. Convergence was achieved relatively quickly and generally within 5000 iterations; in practice, a burn-in of 10,000 iterations was used. There was no evidence of high autocorrelation between successive samples of the Markov chains. Results were displayed as forest plots and summary receiver operating curve plots with 95% CrIs and 95% prediction intervals for sensitivity and specificity.
Narrative synthesis
A meta-analysis was not conducted on a range of intermediate measures (i.e. time to result, time to treatment, test failure rates, duration of ICU and/or hospital stay, duration of broad- and narrow-spectrum antimicrobial therapy, readmission rate and change in antimicrobial treatment plan) and clinical outcome measures (e.g. side effects associated with broad-spectrum antimicrobial use, morbidity and mortality, severity of disease, rates of superinfection, rates of resistant infections and health-related quality of life), as the necessary data were not available or it was inappropriate to statistically pool studies because of their variability in reporting outcome data. Therefore, as suggested by the guidance produced by the Cochrane Collaboration56 and the Centre for Reviews and Dissemination for undertaking systematic reviews,41,57 a narrative synthesis of included studies (grouped by outcome) was undertaken.
Clinical effectiveness results
Quantity and quality of research available
Number of studies identified/included
The literature searches identified 2892 citations. Of these, 66 studies met the inclusion criteria. A flow chart describing the process of identifying relevant literature can be found in Figure 2.
FIGURE 2.
Study flow chart (adapted from Moher et al. 58): clinical effectiveness review. a, Two studies included both SeptiFast and SepsiTest and are counted as individual studies in each test comparison (meta-analysis) with the reference standard.

Number and type of studies excluded
A total of 111 full-text articles were excluded, as they did not meet all the prespecified inclusion criteria. The majority of the articles were excluded primarily on the basis of having insufficient information to allow calculation of a diagnostic 2 × 2 metrics table (which includes data for true positives, false negatives, false positives and true negatives), incorrect population or interventions, or data reported in abstract form that were replaced by published full-text papers. A full list of excluded studies with reasons for exclusion is presented in Appendix 3.
Assessment of effectiveness
Description of included studies (design and patient characteristics)
Study design characteristics
The design characteristics of the 66 included studies that evaluated the effectiveness of the SeptiFast, SepsiTest and IRIDICA in patients with suspected sepsis are summarised in Table 4 (further details are provided in Appendix 4).
| Author (year) | Country | Clinical setting | Study designa | Total number of patients (paired blood tests) | Outcomes (unit of analysis) | Commercially funded |
|---|---|---|---|---|---|---|
| Single index test studies: SeptiFast compared with blood culture | ||||||
| Raglio et al. (2006)44 (abstract) | NR | NR | Single gate, NR | 74 (114) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Bingold et al. (2007)59 (abstract) | Germany | Intensive/critical care | Single gate, NR | 21 (134) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Klemm et al. (2007)60 (abstract) | Germany | Intensive/critical care | Single gate, NR | 44 (56) | Test accuracy (patient), other intermediary/clinical outcomes | NR |
| Lodes et al. (2008)61 (abstract) | Germany | Intensive/critical care | Single gate, NR | 137 (358) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Louie et al. (2008)46 | USA | Emergency department, in hospital and intensive/critical care | Single gate, prospective | 200 (200) | Test accuracy (patient), other intermediary/clinical outcomes | Roche Diagnostics |
| Mancini et al. (2008)45 | Italy | In hospital and unclear if intensive/critical care | Single gate, NR | 34 (103) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics |
| Vince et al. (2008)62 (correspondence) | Croatia | In hospital and intensive/critical care | Single gate, NR | 36 (39) | Test accuracy (sample) | NR |
| Dark et al. (2009)63 (correspondence) | UK | Intensive/critical care | Single gate, NR | 50 (90) | Test accuracy (pathogen) | NR |
| Dierkes et al. (2009)64 | Germany | Intensive/critical care | Single gate, retrospective | 77 (99) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Gimeno et al. (2009)65 (abstract) | Spain | NR | Single gate, prospective | 19 (45) | Test accuracy (sample) | NR |
| Lehmann et al. (2009)66 | Germany | Emergency department, in hospital and intensive/critical care | Single gate, retrospective | 436 (NR) | Intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Lodes et al. (2009)67 | Germany | Intensive/critical care | Single gate, prospective | 52 (258) | Test accuracy (sample) | NR |
| Palomares et al. (2009)68 (abstract) | Spain | Intensive/critical care | Single gate, NR | 73 (76) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Paolucci et al. (2009)69 (correspondence) | Italy | NR | Single gate, retrospective | 34 (NR) | Test accuracy (patient), other intermediary/clinical outcomes | NR |
| Varani et al. (2009)70 | Italy | In hospital and unclear if intensive/critical care | Single gate, NR | 100 (130) | Test accuracy (episode) | NR |
| von Lilienfeld-Toal et al. (2009)71 | Germany | In hospital | Single gate, prospective | 70 (784) | Test accuracy (pathogen) | Roche Diagnostics (partly) |
| Westh et al. (2009)72 | Germany | NR | Single gate, NR | 359 (558) | Test accuracy (pathogen), other intermediary/clinical outcomes | Roche Diagnostics |
| Berger et al. (2010)73 (abstract) | Austria | Neonatal unit | Single gate, NR | 38 (38) | Test accuracy (patient) | NR |
| Bloos et al. (2010)74 | Germany and France | Intensive/critical care | Single gate, prospective | 142 (236) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics |
| Lamoth et al. (2010)75 | Switzerland | In hospital | Single gate, prospective | 86 (237) | Test accuracy (episode) | Roche Diagnostics |
| Lehmann et al. (2010)76 | Germany | Intensive/critical care | Single gate, prospective | 108 (453) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics |
| Maubon et al. (2010)77 | France | In hospital and unclear if intensive/critical care | Single gate, prospective | 110 (110) | Test accuracy (patient), other intermediary/clinical outcomes | Roche Diagnostics |
| Regueiro et al. (2010)78 | Spain | In hospital and intensive/critical care | Single gate, NR | 72 (106) | Test accuracy (sample), other intermediary/clinical outcomes | No |
| Soki et al. (2010)79 (abstract) | Hungary | In hospital and intensive/critical care | Single gate, NR | 159 (162) | Test accuracy (sample) | NR |
| Tsalik et al. (2010)80 | USA | Emergency department | Single gate, NR | 306 (306) | Test accuracy (patient) other intermediary/clinical outcomes | No |
| Wallet et al. (2010)81 | France | Intensive/critical care | Single gate, prospective | 72 (102) | Test accuracy (pathogen) other intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Yanagihara et al. (2010)82 | Japan | In hospital and emergency department | Single gate, prospective | 212 (400) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics |
| Bravo et al. (2011)83 | Spain | In hospital and intensive/critical care | Single gate, NR | 53 (53) | Test accuracy (episode) | NR |
| Hettwer et al. (2011)84 | Germany | Emergency department | Single gate, prospective | 113 (113) | Test accuracy (patient) | Roche Diagnostics |
| Josefson et al. (2011)85 | Sweden | In hospital | Single gate, prospective | 1093 (1141) | Test accuracy (patient), other intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Lucignano et al. (2011)86 | Italy | In hospital and intensive/critical care | Single gate, retrospective | 803 (1553) | Test accuracy (sample) | NR |
| Obara et al. (2011)87 | Japan | Emergency department, in hospital and intensive/critical care | Single gate, NR | 54 (78) | Test accuracy (sample) | Roche Diagnostics (partly) |
| Vrioni et al. (2011)88 (abstract) | Greece | NR | Single gate, NR | 33 (33) | Test accuracy (patient), other intermediary/clinical outcomes | NR |
| Alvarez et al. (2012)89 | Spain | Intensive/critical care | Single gate, retrospective | 102 (NR) | Intermediary/clinical outcomes | NR |
| Grif et al. (2012)90 | Austria | In hospital and intensive/critical care | Single gate, prospective | 61 (71) | Test accuracy (sample), other intermediary/clinical outcomes | Pfizer |
| Guido et al. (2012)91 | Italy | In hospital and unclear if intensive/critical care | Single gate, NR | 166 (166) | Test accuracy (sample) | NR |
| Lodes et al. (2012)92 | Germany | Intensive/critical care | Single gate, NR | 104 (148) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Mauro et al. (2012)93 | Italy | In hospital and unclear if intensive/critical care | Single gate, NR | 79 (79) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Pasqualini et al. (2012)94 | Italy | In hospital and unclear if intensive/critical care | Single gate, NR | 391 (391) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Rath et al. (2012)95 | Germany | Intensive/critical care | Single gate, prospective | 170 (225) | Test accuracy (sample) | NR |
| Tschiedel et al. (2012)96 | Germany | In hospital and intensive/critical care | Single gate, retrospective | 75 (110) | Test accuracy (sample) other intermediary/clinical outcomes | NR |
| Herne et al. (2013)97 | Estonia | In hospital and intensive/critical care | Single gate, retrospective | 144 (160) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Kasper and Altiok (2013)98 | Austria | NR | Single gate, NR | 46 (NR) | Test accuracy (patient) | Roche Diagnostics (partly) |
| Paolucci et al. (2013)99 | Italy | In hospital | Single gate, prospective | 201 (437) | Test accuracy (episode), other intermediary/clinical outcomes | No |
| Rodrigues et al. (2013)100 (abstract) | Brazil | NR | Single-gate, prospective RCT | 46 (NR) | Intermediary/clinical outcomes | NR |
| Avolio et al. (2014)101 | Italy | Emergency department and intensive/critical care | Single gate, prospective | 525 (525) | Test accuracy (pathogen), other intermediary/clinical outcomes | NR |
| Burdino et al. (2014)102 | Italy | In hospital and intensive/critical care | Single gate, NR | 1024 (1186) | Test accuracy (sample) | NR |
| Mancini et al. (2014)103 | Italy | In hospital | Single gate, retrospective and prospective data | 228 (NR) | Intermediary/clinical outcomes | Roche Diagnostics |
| Markota et al. (2014)104 | Slovenia | Intensive/critical care | Single gate, prospective | 57 (63) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Ozkaya-Parlakay et al. (2014)105 | Turkey | NR | Single gate, prospective | 69 (79) | Test accuracy (sample), other intermediary/clinical outcomes | NR |
| Schaub et al. (2014)106 | Switzerland | Emergency department | Single gate, prospective | 110 (205) | Test accuracy (patient), other intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Sitnik et al. (2014)107 | Brazil | Intensive/critical care (and oncology patients) | Single gate, prospective | 114 (114) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Diagnostics (partly) |
| Barbanti et al. (2015)108 (abstract) | Italy | In hospital | Single gate, NR | 491 (1837) | Test accuracy (sample) | NR |
| Calitri et al. (2015)109 | Italy | In hospital and intensive/critical care | Single gate, retrospective | 289 (NR) | Test accuracy (episode) | No |
| Idelevich et al. (2015)110 | Germany | NR | Single-gate, prospective RCT | 150 (253) | Test accuracy (pathogen), other intermediary/clinical outcomes | Roche Diagnostics and Pfizer (partly) |
| Warhurst et al. (2015)111 | UK | Intensive/critical care | Single gate, prospective | 795 (NR) | Test accuracy (pathogen) other intermediary/clinical outcomes | No |
| Single index test studies: SeptiFast compared with blood culture plus MALDI-TOF MS | ||||||
| Tafelski et al. (2015)112 | Germany | Intensive/critical care | Single-gate, prospective RCT | 78 (78) | Test accuracy (sample), other intermediary/clinical outcomes | Roche Deutschland GmbH |
| Single index test studies: SepsiTest compared with blood culture | ||||||
| Wellinghausen et al. (200948) | Germany | Intensive/critical care | Single gate, prospective | 187 (342) | Test accuracy (sample), other intermediary/clinical outcomes | No |
| Nieman et al. (2015)113 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Single index test studies: SepsiTest compared with blood culture plus MALDI-TOF MS | ||||||
| Loonen et al. (2014)114 | The Netherlands | Emergency department | Single gate, retrospective | 125 (NR) | Test accuracy (sample), other intermediary/clinical outcomes | Molzym GmbH (partly) |
| Single index test studies: IRIDICA compared with blood culture | ||||||
| Bacconi et al. (2014)49 | USA | Emergency department | Single gate, prospective | 331 (331) | Test accuracy (sample) | NR but majority of authors are employees of Ibis Biosciences (an Abbott company) |
| Delco-Volante et al. (2015)115 (conference presentation) | NR | NR | Single gate, prospective | NR (81) | Test accuracy (sample) | Abbott |
| Vincent et al. (2015)116 | Belgium, UK, Switzerland, France, Poland and Germany | Intensive/critical care | Single gate, prospective | 529 (NR) | Test accuracy (sample), other intermediary/clinical outcomes | Ibis Biosciences, Inc., Abbott |
| Metzgar et al. (2015)117 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Two index test studies: SeptiFast and SepsiTest compared with blood culture | ||||||
| Leitner et al. (2013)118 | Austria | NR | Single gate, NR | 57 (75) | Test accuracy (sample) | No |
| Schreiber and Nierhaus (2013)119 | Germany | Intensive/critical care | Single gate, prospective | 50 (NR) | Test accuracy (patient), other intermediary/clinical outcomes | Molzym GmbH and Roche Diagnostics (partly) |
In total, 56 single index test studies compared SeptiFast with blood culture,44–46,59–111 and one study112 evaluated SeptiFast with blood culture and MALDI-TOF MS. All SeptiFast studies were single gate in design (i.e. same patient characteristics for both reference standard and index test). With the exception of three RCTs,100,110,112 all SeptiFast studies were diagnostic cohort studies. Two single index test studies48,113 compared SepsiTest with blood culture, and one study evaluated SepsiTest with blood culture and MALDI-TOF MS. 114 Two three-arm studies118,119 compared both SeptiFast and SepsiTest with blood culture. Four single index test studies compared IRIDICA with blood culture,49,115–117 two of which employed IRIDICA–PLEX-ID hybrid systems49,116 (confidential data suggest that the IRIDICA CE-certified systems is equivalent to the hybrid systems). All SepsiTest and IRIDICA studies were single-gate diagnostic cohort studies.
Two SeptiFast studies46,80 and one IRIDICA study49 were conducted in North America. One IRIDICA study did not report the country. 115 Two SeptiFast studies100,107 were conducted in Brazil, two were undertaken in Japan82,87 and one was undertaken in Turkey. 105 Two SeptiFast studies63,111 were undertaken in the UK. (Confidential information has been removed.)
Twenty-four of the SeptiFast studies,46,65,67,71,74–77,81,82,84,85,90,95,99–101,104–107,110–112 one multitest SeptiFast and SepsiTest study,119 (confidential information has been removed) had data collected prospectively.
Eight of the SeptiFast studies64,66,69,86,89,96,97,109 and one SepsiTest study114 had a retrospective design. One SeptiFast study103 evaluated samples collected both retrospectively and prospectively. (Confidential information has been removed.)
Where reported, the sampling period ranged from 2 months45 to 66 months. 80
The clinical setting (e.g. community, emergency department, hospital, intensive/critical care, general/specialist) was not reported for nine SeptiFast studies,44,65,69,72,88,98,100,105,110 or for one multitest SeptiFast and SepsiTest study118 (confidential information has been removed). The setting across the remaining studies varied (see Table 4). Explicit inclusion and exclusion criteria for patients were reported for seven SeptiFast studies,74,80,83,106,110–112 (confidential information has been removed) and one IRIDICA study. 116
With the exception of the SeptiFast study by Warhurst et al. ,111 which reported on health-care-associated bloodstream infections, and the SeptiFast study by Josefson et al. ,85 which reported on community-acquired bloodstream infections, reporting of whether infection was community or hospital acquired was unclear in the remainder of the included studies.
Patient characteristics of included studies
The patient characteristics of the included studies are summarised in Table 5 (further details are provided in Appendix 4). Twenty-four of the SeptiFast studies,44,59–63,65,68–70,72,73,79,81,82,84,86,88,93,98,99,101,102,108 one SepsiTest study,48 and one multitest SeptiFast and SepsiTest study118 (confidential information has been removed) did not report on the mean or median age of patients. Six of the SeptiFast studies70,85,93,96,99,111 included both adults and children, two included children and neonates86,109 and one included children and infants. 105 Three SeptiFast studies69,73,98 and one IRIDICA study115 included neonates and infants, and one SepsiTest study included adults and children. 48 (Confidential information has been removed.)
| Author (year) | Population | Mean age, years (median) | % male | % suspected community-/hospital-acquired infection | % immunocompromised patients | % antibiotics at time of blood sample collection |
|---|---|---|---|---|---|---|
| Single index test studies: SeptiFast (without MALDI-TOF) | ||||||
| Raglio et al. (2006)44 (abstract) | Patients with suspected sepsis | NR | NR | NR | NR | NR |
| Bingold et al. (2007)59 (abstract) | Surgical patients with severe sepsis and septic shock | NR | NR | NR | NR | NR |
| Klemm et al. (2007)60 (abstract) | Patients in intensive care with suspected sepsis | NR | NR | NR | NR | NR |
| Lodes et al. (2008)61 (abstract) | Surgical intensive care patients with suspected sepsis | NR | NR | NR | NR | NR |
| Louie et al. (2008)46 | Adults with suspected sepsis | (46.5 average median) | 61 | NR | 4 | NR |
| Mancini et al. (2008)45 | Adult neutropenic patients (with haematological malignancies) with suspected sepsis | 47 | 67.6 | NR | 44.1 | NR |
| Vince et al. (2008)62 (correspondence) | Patients with suspected sepsis | NR | NR | NR | NR | 100 |
| Dark et al. (2009)63 (correspondence) | Adults with suspected sepsis | NR | NR | NR | NR | NR |
| Dierkes et al. (2009)64 | Adults with suspected sepsis | (55) | 63.6 | NR | 45 | 79.2 |
| Gimeno et al. (2009)65 | Patients (oncohaematological) with febrile neutropenia | NR | NR | NR | NR | 100 |
| Lehmann et al. (2009)66 | Adults with suspected sepsis | 54.8 | 61.5 | NR | NR | NR |
| Lodes et al. (2009)67 | Adults with suspected sepsis | 60.5 | 57.7 | NR | NR | NR |
| Palomares et al. (2009)68 (abstract) | Adults with suspected sepsis | NR | NR | NR | NR | 93.2 |
| Paolucci et al. (2009)69 (correspondence) | Neonates with suspected sepsis | NR | NR | NR | NR | NR |
| Varani et al. (2009)70 | Adults and children (immunocompromised) with suspected sepsis | NR | NR | NR | 100 | 100 |
| von Lilienfeld-Toal et al. (2009)71 | Adults (haematological) with febrile neutropenia | (60) | 54 | NR | NR | 0 |
| Westh et al. (2009)72 | Patients with suspected sepsis | NR | NR | NR | NR | NR |
| Berger et al. (2010)73 (abstract) | Very low birthweight infants (neonates) with suspected sepsis | NR | NR | NR | NR | NR |
| Bloos et al. (2010)74 | Adults with severe sepsis or septic shock | 66 | 68.5 | NR | NR | 95.8 on antibiotics (unclear if prior to blood sampling) |
| Lamoth et al. (2010)75 | Adults (haematological) with febrile neutropenia | (54) | 62 | NR | NR | NR |
| Lehmann et al. (2010)76 | Adults with suspected sepsis | 58.4 | 66.7 | NR | NR | NR |
| Maubon et al. (2010)77 | Patients with malignancies and suspected sepsis | 56.3 | 60.9 | NR | NR | 88.2 |
| Regueiro et al. (2010)78 | Adults with suspected sepsis | 64 | 73.6 | NR | NR | NR |
| Soki et al. (2010)79 (abstract) | Septic patients in intensive care or with haematological malignancies | NR | NR | NR | NR | NR |
| Tsalik et al. (2010)80 | Adults with suspected sepsis | 54.1 | 54.9 | NR | NR | 22.5 |
| Wallet et al. (2010)81 | Adults with suspected sepsis | NR | NR | NR | NR | NR |
| Yanagihara et al. (2010)82 | Patients with suspected sepsis | NR | 64.6 | NR | NR | NR |
| Bravo et al. (2011)83 | Adult ICU patients who were critically ill with suspected sepsis | (65.5) | 62.3 | NR | NR | 0 |
| Hettwer et al. (2011)84 | Adults with suspected sepsis | NR | NR | NR | NR | NR |
| Josefson et al. (2011)85 | Adults and children with suspected sepsis | (67) | 55.5 | 100 (community acquired) | NR | NR |
| Lucignano et al. (2011)86 | Neonates and children with suspected sepsis | NR | NR | NR | NR | NR |
| Obara et al. (2011)87 | Adults with suspected sepsis | 61.6 | 64.8 | NR | NR | NR |
| Vrioni et al. (2011)88 (abstract) | Patients with suspected sepsis | NR | NR | NR | NR | NR |
| Alvarez et al. (2012)89 | Adults with severe sepsis or septic shock | 64.9 | 78.4 | NR | NR | NR |
| Grif et al. (2012)90 | Adults with suspected sepsis | 55.6 | 68.9 | NR | NR | 91.8 |
| Guido et al. (2012)91 | Adult neutropenic patients (with haematological malignancies) with suspected sepsis | (66.1) | 62 | NR | NR | 0 |
| Lodes et al. (2012)92 | Adults with suspected sepsis | 63.1 | 71.1 | NR | NR | 79.7% of samples under antibiotic therapy and 41.9% under antifungal |
| Mauro et al. (2012)93 | Adults and children, immunocompromised with suspected sepsis | NR | 51.9 | NR | 100 | 5 |
| Pasqualini et al. (2012)94 | Adults with suspected sepsis | (73) | 55 | NR | 4.3 | 48.8 |
| Rath et al. (2012)95 | Adults (who have undergone liver transplantation or other major abdominal surgery) with suspected sepsis | 56.4 | 56.5 | NR | NR | NR |
| Tschiedel et al. (2012)96 | Adults and children with suspected sepsis | (6) | 49.3 | NR | NR | NR |
| Herne et al. (2013)97 | Adults with suspected sepsis | 58 | 42.4 | NR | NR | 99.3 |
| Kasper and Altiok (2013)98 | Very low-birthweight premature infants with suspected sepsis | NR | NR | NR | NR | 0 |
| Paolucci et al. (2013)99 | Adults and children (haematological) with severe febrile neutropenia | NR | NR | NR | NR | 0 |
| Rodrigues et al. (2013)100 (abstract) | Adults with suspected sepsis | 64.5 | 67.4 | NR | NR | 0 |
| Avolio et al. (2014)101 | Adults with suspected sepsis | NR | NR | NR | NR | NR |
| Burdino et al. (2014)102 | Adults with suspected sepsis | NR | NR | NR | 10.5 | 89 |
| Mancini et al. (2014)103 | Adults (haematological patients) with suspected sepsis | 48.6 | 66.7 | NR | NR | NR |
| Markota et al. (2014)104 | Adults with severe sepsis or septic shock | 59.5 | 66.7 | NR | NR | 61.9 |
| Ozkaya-Parlakay et al. (2014)105 | Children with severe sepsis or septic shock | 2.7 | 62.3 | NR | 0 | NR |
| Schaub et al. (2014)106 | Adults with suspected sepsis | (64) | 51 | NR | 13.6 | 14.5 |
| Sitnik et al. (2014)107 | Adults with suspected sepsis | 49.7 | 64.9 | NR | NR | NR |
| Barbanti et al. (2015)108 | Patients (haematological) with febrile neutropenia and suspected sepsis | NR | NR | NR | NR | NR |
| Calitri et al. (2015)109 | Children and neonates with suspected sepsis, febrile neutropenia, fever without focus or localised infective focus | (6.8) | 63.3 | NR | NR | NR |
| Idelevich et al. (2015)110 | Adults with febrile neutropenia or afebrile neutropenia with sepsis | 52.4 | 59.3 | NR | NR | 100 |
| Warhurst et al. (2015)111 | Adults (over 16 years) with suspected sepsis | (58) | 60 | 100 (health-care acquired) | NR | 85.7 |
| Single index test studies: SeptiFast (with MALDI-TOF) | ||||||
| Tafelski et al. (2015)112 | Adults with suspected sepsis | (63, average median) | 64.1 | NR | 15.4 | NR |
| Single index test studies: SepsiTest (without MALDI-TOF) | ||||||
| Wellinghausen et al. (2009)48 | Adults and children with SIRS, sepsis (79.1%) or haematological patients with neutropenic fever (20.9%) | NR | NR | NR | NR | NR |
| Nieman et al. (2015)113 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Single index test studies: SepsiTest (with MALDI-TOF) | ||||||
| Loonen et al. (2014)114 | Adults with suspected sepsis | 64.7 | 59.2 | NR | NR | NR |
| Single index test studies: IRIDICA | ||||||
| Bacconi et al. (2014)49 | Adults with suspected sepsis | NR | NR | NR | NR | NR |
| Delco-Volante et al. (2015)115 (conference presentation) | Neonates with suspected sepsis | (0) | NR | NR | NR | 0 |
| Vincent et al. (2015)116 | Adults with suspected or proven sepsis or severe infection | 61 | 64.6 | NR | 16.6 | NR |
| Metzgar et al. (2015)117 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Two index test studies: SeptiFast and SepsiTest | ||||||
| Leitner et al. (2013)118 | Critically ill patients with suspected sepsis | NR | NR | NR | NR | NR |
| Schreiber and Nierhaus (2013)119 | Adults with suspected sepsis | (64) | 80 | NR | NR | 72 |
Twenty-two SeptiFast studies,44,45,62,63,65,68,70–72,75,78,79,81,83,85,88,96,98–100,105,108 one multitest SeptiFast and SepsiTest study,118 (confidential information has been removed) did not report details or a reference to a guideline for defining sepsis. The remaining studies provided a description or a reference to a guideline for defining sepsis for included patients; however, these definitions and descriptions varied across studies and were sometimes not explicitly clear (see Appendix 4).
Ten SeptiFast studies45,46,64,70,93,94,102,105,106,112 and one IRIDICA study116 reported on the proportion of the included patients who were immunocompromised. In addition, Paolucci et al. 69 reported that one patient was affected by primary congenital immunodeficiency; however, it was unclear if other immunocompromised patients were included in this study.
Twenty-three SeptiFast studies62,64,65,68,70,71,77,80,83,90–94,97–100,102,104,106,110,111 reported on the proportion of patients receiving antimicrobial therapy at the time of blood sampling. In addition, it was unclear in one SeptiFast study74 if patients received antimicrobial therapy (98%) prior to blood sampling. Of the 23 SeptiFast studies, six71,83,91,98–100 reported that none of the included patients had received antimicrobial therapy at the time of blood sampling. Similarly, in one IRIDICA study115 none of the included patients received antimicrobial therapy at the time of blood sampling. In one multitest SeptiFast and SepsiTest study,119 the majority of patients (72%) received antimicrobial therapy at recruitment. (Confidential information has been removed.)
The SeptiFast studies by Alvarez et al. ,89 Bloos et al. ,74 Bingold et al. 59 and Markota et al. 104 reported that all included participants had severe sepsis or septic shock. Bloos et al. 74 also reported a mean SOFA score of 10 and SAPS II score of 49 for the entire cohort. Markota et al. 104 reported a mean admission APACHE score for the cohort of 25 (SD ± 7.6). The SeptiFast studies by Herne et al. 97 and Lehmann et al. 76 reported that all included patients had severe sepsis. Seven SeptiFast studies46,77,80,83,100,106,112 and the one multitest SeptiFast and SepsiTest study119 reported mixed samples of patients with sepsis, severe sepsis and septic shock in varying proportions. The SeptiFast RCT by Rodrigues et al. 100 reported a mean APACHE II score of 17 for the SeptiFast group and 17 for the blood culture group, but was unclear whether or not this was at study entry or following testing. The SeptiFast RCT by Tafelski et al. 112 reported a median SAPS II on admission for the SeptiFast group of 40 (interquartile range 32–50) and the blood culture group of 47 (interquartile range 34–65). Schreiber and Nierhaus119 also reported a median SAPS II score of 41 (interquartile range 33 to 49) for the entire cohort. The IRIDICA study by Vincent et al. 116 reported a mean SOFA score at baseline of 7.6 (SD 4.2) indicating a 15–20% risk in mortality in ICU, but did not report proportions of the patients with sepsis, severe sepsis or septic shock. The remainder of the included studies did not report on the proportion of patients with sepsis, severe sepsis or septic shock, or disease severity.
Across the included studies, the number of patients analysed ranged from 19 (45 paired blood samples) (SeptiFast – Gimeno et al. 65) to 1093 (1114 paired blood samples) (SeptiFast – Josefson et al. 85).
Details of index and reference tests, blood sampling methods and Conformité Européenne approval
A detailed summary of the index and reference tests, blood samples taken and interval between the index and reference test, CE approval of the blood volume used for testing, definition of a true positive, laboratory working times and the unit of analysis (pathogen/sample/patient/episode) is presented in Appendix 4.
Thirty-four of the SeptiFast studies reported on the blood volume used for the SeptiFast test. 45,69,71–78,80–83,85–87,90–94,96–99,101–103,107,109–112,114 Of these, nine studies reported blood volumes that did not comply with CE approval: Lehmann et al. ,76 Lodes et al. 92 and von Lilienfeld-Toal et al. 71 all reported using 1 ml in adults; Bloos et al. ,74 Lamoth et al. ,75 Paolucci et al. 99 and Sitnik et al. 107 all reported using 3 ml in adults; Berger et al. 73 reported using 0.1 ml in neonates and infants; and Kasper and Altiok98 reported using 0.1–0.7 ml in neonates and infants. The remainder of the SeptiFast studies did not report the blood volume used for the test.
Thirty-eight SeptiFast studies reported that blood drawn for SeptiFast and for blood culture were drawn at the same time. 45,60,64,65,68,70–72,74–76,78,81–87,90–99,101,102,104–107,110–112 Of these, one SeptiFast study reported that blood drawn for SeptiFast and for blood culture were drawn within 1 hour,102 and another study reported that blood drawn for SeptiFast and for blood culture were drawn within 12 hours of each other. 97 The remainder of the SeptiFast studies did not report on when blood samples were drawn.
Across the studies evaluating SeptiFast, the studies by Lehmann et al. ,66 Westh et al. ,72 Tsalik et al. ,80 Wallet et al. 81 and Yanagihara et al. 82 all reported that either the BACTEC or BacT/ALERT blood systems were used and was dependant on the testing site performing the assay. Across the remaining SeptiFast studies, 19 reported using the BACTEC system. 60,64,70,71,75,76,79,83,85–87,92–96,107,110,112 Of these, one used the BACTEC system with MALDI-TOF MS. 112 Seventeen studies reported using the BacT/ALERT system. 45,78,84,90,91,97–104,106,108,109,111 The remaining studies did not report the method used.
Details of the laboratory working times or when assays were carried out for the index test were reported by 13 studies evaluating SeptiFast. 45,64,76,90,96,97,99,101–104,110,112 Working times were 7 days per week for four SeptiFast studies,45,64,97,101 6 days per week for one SeptiFast study104 and 5 days per week for six SeptiFast studies. 90,96,99,103,110,112 For the remainder of the studies reporting on working times, it was unclear how many days of the week laboratories were working.
Definition of a true positive was reported by 12 SeptiFast studies. 74,76,83,85,86,94,97,101,106,112,118,119 Definition of a true positive varied across these studies (Table 6).
| Study | Risk of bias | Applicability concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| Raglio et al. (2006)44 (abstract) | U | U | U | U | U | U | U |
| Bingold et al. (2007)59 (abstract) | U | U | U | U | U | U | U |
| Klemm et al. (2007)60 (abstract) | U | U | U | U | U | U | U |
| Lodes et al. (2008)61 (abstract) | U | U | U | U | U | U | U |
| Louie et al. (2008)46 | U | U | U | U | N | U | U |
| Mancini et al. (2008)45 | U | U | U | L | N | N | U |
| Vince et al. (2008)62 (correspondence) | U | U | U | U | U | U | U |
| Dark et al. (2009)63 (correspondence) | U | L | U | U | N | U | U |
| Dierkes et al. (2009)64 | U | U | U | U | Y | U | U |
| Gimeno et al. (2009)65 (abstract) | U | U | U | U | U | U | U |
| Lehmann et al. (2009)66 | U | U | U | H | N | U | U |
| Lodes et al. (2009)67 | U | U | U | U | U | U | U |
| Palomares et al. (2009)68 (abstract) | U | U | U | U | U | U | U |
| Paolucci et al. (2009)69 (correspondence) | U | U | U | U | N | N | U |
| Varani et al. (2009)70 | U | U | U | L | N | U | U |
| Von Lilienfeld-Toal et al. (2009)71 | U | U | U | U | U | Y | U |
| Wellinghausen et al. (2009)48 | U | U | U | L | U | N | U |
| Westh et al. (2009)72 | U | U | U | H | U | N | U |
| Berger et al. (2010)73 (abstract) | U | U | U | U | U | Y | U |
| Bloos et al. (2010)74 | U | L | U | U | N | Y | U |
| Lamoth et al. (2010)75 | U | U | U | U | U | Y | U |
| Lehmann et al. (2010)76 | U | U | U | L | N | Y | U |
| Maubon et al. (2010)77 | U | U | U | U | N | N | U |
| Regueiro et al. (2010)78 | U | U | U | L | N | N | U |
| Soki et al. (2010)79 (abstract) | U | U | U | U | U | U | U |
| Tsalik et al. (2010)80 | U | U | U | H | N | N | U |
| Wallet et al. (2010)81 | U | U | U | H | U | N | U |
| Yanagihara et al. (2010)82 | U | U | U | H | N | N | U |
| Bravo et al. (2011)83 | H | L | U | U | Y | N | U |
| Hettwer et al. (2011)84 | U | U | U | H | N | U | U |
| Josefson et al. (2011)85 | U | U | U | H | U | N | U |
| Lucignano et al. (2011)86 | U | U | U | H | U | N | U |
| Obara et al. (2011)87 | U | U | U | L | N | N | U |
| Vrioni et al. (2011)88 (abstract) | U | U | U | U | U | U | U |
| Alvarez et al. (2012)89 | U | H | U | U | N | U | U |
| Grif et al. (2012)90 | U | U | U | L | N | N | U |
| Guido et al. (2012)91 | U | U | U | L | U | N | U |
| Lodes et al. (2012)92 | U | U | U | L | N | Y | U |
| Mauro et al. (2012)93 | U | U | U | L | N | N | U |
| Pasqualini et al. (2012)94 | U | U | U | L | U | N | U |
| Rath et al. (2012)95 | U | U | U | L | Y | U | U |
| Tschiedel et al. (2012)96 | U | U | U | L | U | N | U |
| Herne et al. (2013)97 | U | U | U | H | N | N | U |
| Kasper and Altiok (2013)98 | U | U | U | L | U | Y | U |
| Leitner et al. (2013)118 | U | U | U | U | U | U | U |
| Paolucci et al. (2013)99 | U | U | U | U | U | Y | U |
| Rodrigues et al. (2013)100 (abstract) | U | U | U | U | U | U | U |
| Schreiber and Nierhaus (2013)119 | U | L | U | U | N | U | U |
| Avolio et al. (2014)101 | U | U | U | H | Y | N | U |
| Bacconi et al. (2014)49 | U | U | U | U | U | N | U |
| Burdino et al. (2014)102 | U | U | U | L | U | N | U |
| Loonen et al. (2014)114 | U | U | U | H | U | N | U |
| Mancini et al. (2014)103 | U | U | U | U | N | N | U |
| Markota et al. (2014)104 | U | U | U | U | N | U | U |
| Ozkaya-Parlakay et al. (2014)105 | U | U | U | U | N | U | U |
| Schaub et al. (2014)106 | U | U | U | U | N | U | U |
| Sitnik et al. (2014)107 | U | U | U | U | N | Y | U |
| Barbanti et al. (2015)108 | U | U | U | U | U | U | U |
| Calitri et al. (2015)109 | U | U | U | U | Y | N | U |
| Delco-Volante et al. (2015)115 (conference presentation) | U | U | U | U | U | Y | U |
| Idelevich et al. (2015)110 | U | U | U | U | U | N | U |
| Tafelski et al. (2015)112 | H | L | L | L | N | N | U |
| Warhurst et al. (2015)111 | L | L | L | L | N | N | U |
| Vincent et al. (2015)116 | N | N | U | U | N | N | U |
| Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
A range of metrics (units of analyses) was used to assess the diagnostic accuracy of SeptiFast. The unit of analyses was ‘patients’ in 11 studies,46,60,69,73,77,80,84,85,88,98,106 pathogens in seven studies63,71,72,81,101,110,111 and episodes in five studies. 70,75,83,99,109 For the remainder of the studies evaluating SeptiFast against blood culture (with or without MALDI-TOF MS) the unit of analysis was samples. Although the heterogeneity in the metrics has the potential to introduce some bias, the impact on the results was believed to be modest.
Thirty studies evaluating SeptiFast against blood culture included contaminants in the diagnostic test accuracy analysis in this assessment report. 44,45,64,67–70,73,76,78,81–85,87,91–98,105–107,109,110,120 and eight studies reported that contaminants were excluded. 46,72,80,86,99,101,102,111 For the remainder of the SeptiFast studies it was unclear if contaminants were included or excluded.
(Confidential information has been removed.) One study of SepsiTest did not report when bloods were drawn. 114 One study performed blood culture using a BACTEC system,48 (confidential information has been removed) and one study reported using BacT/ALERT with MALDI-TOF MS. 114 (Confidential information has been removed.) Definition of a true positive was reported by one SepsiTest study. 48 (Confidential information has been removed.)
(Confidential information has been removed.) The study by Delco-Volante et al. 115 reported using 0.5 ml of blood in neonates and infants. (Confidential information has been removed.)
Neither of the studies evaluating both SeptiFast and SepsiTest against blood culture reported the volume of blood used for the index test assay. 118,119 Leitner et al. 118 reported that the reference standard and index tests were performed on blood samples drawn at the same time. Schreiber and Nierhaus119 did not report if blood samples for the index tests and blood culture assay were drawn at the same time. Both studies reported that blood culture was undertaken using the BACTEC system. Both studies reported a definition of a true positive. Neither study reported on laboratory working times. The unit of analysis for the study by Leitner et al. 118 was samples and the unit of analysis for Schreiber and Nierhaus119 was patients. Both studies included contaminants in the diagnostic test accuracy analysis.
Quality characteristics
The QUADAS-2 tool,51 designed to evaluate the methodological quality of diagnostic accuracy studies, comprises four key domains: patient selection, index test, reference standard, and flow and timing. Using a set of signalling questions, each domain is assessed in terms of risk of bias [low, high or unclear risk (in the event of insufficient data in the publication to answer the corresponding question)] and the first three domains are also assessed in terms of applicability (no, yes or unclear concerns).
The overall methodological quality of the 66 included studies is summarised in Figure 3 and Table 7. The methodological quality of the included studies, as assessed using the QUADAS-2 tool, was variable. With the exception of Warhurst et al. ,111 all other studies were considered to be at risk of bias and to have concerns regarding applicability. 121
FIGURE 3.
Risk of bias and applicability concerns graph: review authors’ judgements about each domain presented as percentages across included studies.
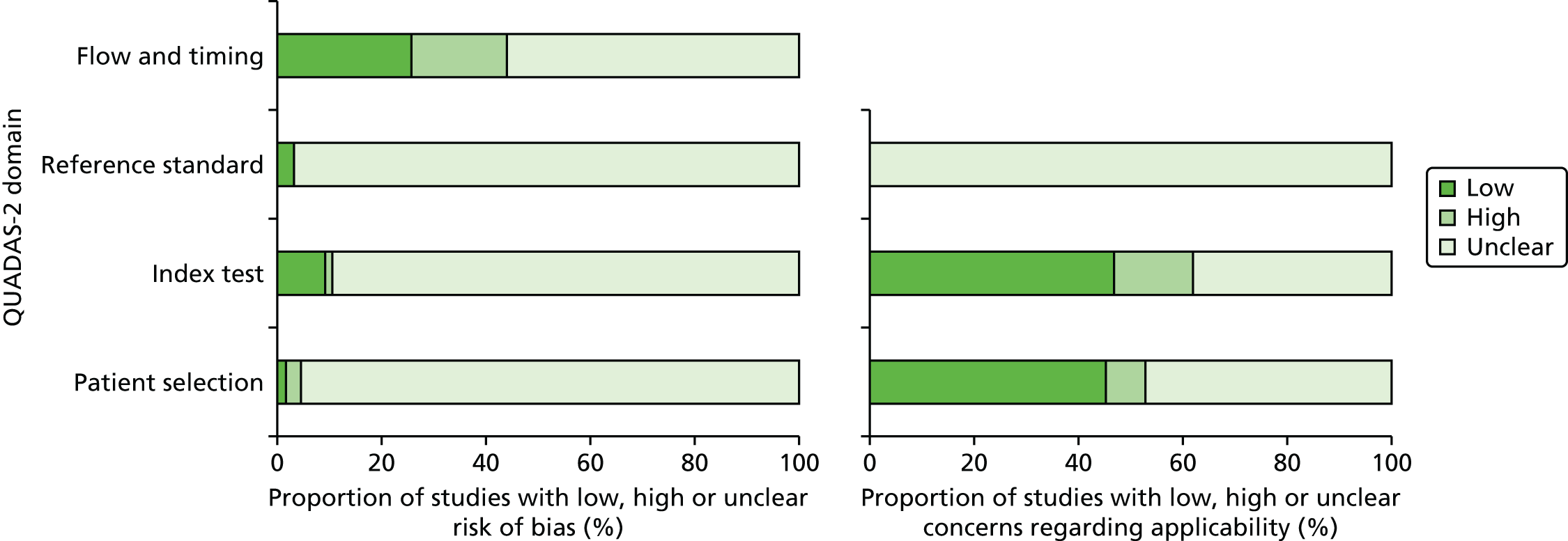
| Author (year) | Time to pathogen identification: index test | Time to treatment | Test failure rates | Mortality | Duration of ICU and/or hospital stay | Duration of antibiotic therapy | Reported changes in antimicrobial treatment plan |
|---|---|---|---|---|---|---|---|
| Single index test studies: SeptiFast | |||||||
| Raglio et al. (2006)44 | ✓ | ||||||
| Bingold et al. (2007)59 | ✓ | ||||||
| Klemm et al. (2007)60 | ✓ | ||||||
| Louie et al. (2008)46 | ✓ | ||||||
| Mancini et al. (2008)45 | ✓ | ||||||
| Dierkes et al. (2009)64 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Lehmann et al. (2009)66 | ✓ | ✓ | ✓ | ||||
| Palomares et al. (2009)68 | ✓ | ||||||
| Paolucci et al. (2009)69 | ✓ | ||||||
| Westh et al. (2009)72 | ✓ | ||||||
| Bloos et al. (2010)74 | ✓ | ✓ | |||||
| Lehmann et al. (2010)76 | ✓ | ||||||
| Maubon et al. (2010)77 | ✓ | ✓ | |||||
| Regueiro et al. (2010)78 | ✓ | ||||||
| Tsalik et al. (2010)80 | ✓ | ✓ | ✓ | ||||
| Wallet et al. (2010)81 | ✓ | ✓ | |||||
| Hettwer et al. (2011)84 | ✓ | ||||||
| Josefson et al. (2011)85 | ✓ | ✓ | |||||
| Vrioni et al. (2011)88 | ✓ | ✓ | |||||
| Alvarez et al. (2012)89 | ✓ | ✓ | |||||
| Grif et al. (2012)90 | ✓ | ✓ | |||||
| Lodes et al. (2012)92 | ✓ | ✓ | |||||
| Mauro et al. (2012)93 | ✓ | ||||||
| Pasqualini et al. (2012)94 | ✓ | ||||||
| Tschiedel et al. (2012)96 | ✓ | ✓ | |||||
| Herne et al. (2013)97 | ✓ | ✓ | |||||
| Paolucci et al. (2013)99 | ✓ | ||||||
| Rodrigues et al. (2013)100 | ✓ | ✓ | ✓ | ✓ | |||
| Avolio et al. (2014)101 | ✓ | ||||||
| Mancini et al. (2014)103 | ✓ | ✓ | ✓ | ||||
| Markota et al. (2014)104 | ✓ | ✓ | ✓ | ||||
| Ozkaya-Parlakay et al. (2014)105 | ✓ | ✓ | |||||
| Schaub et al. (2014)106 | ✓ | ✓ | ✓ | ||||
| Sitnik et al. (2014)107 | ✓ | ||||||
| Idelevich et al. (2015)110 | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Tafelski et al. (2015)112 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Warhurst et al. (2015)111 | ✓ | ✓ | ✓ | ||||
| Single index test studies: SepsiTest | |||||||
| Loonen et al. (2014)114 | ✓ | ||||||
| Single index test studies: IRIDICA | |||||||
| Confidential information has been removed | Confidential information has been removed | ||||||
| Vincent et al. (2015)116 | ✓ | ✓ | |||||
| Two index test studies: SeptiFast and SepsiTest | |||||||
| Schreiber and Nierhaus (2013)119 | ✓ | ||||||
(Confidential information has been removed.)
(Confidential information has been removed.)
Effectiveness of the interventions
This section presents the results of the following separately:
-
an assessment of diagnostic test accuracy (meta-analysis, where applicable) of each diagnostic tests (i.e. SeptiFast, SepsiTest and IRIDICA in conjunction with clinical assessment) for rapidly identifying bloodstream bacteria and fungi
-
an assessment of each diagnostic test on a range of other intermediate and clinical outcome measures (narrative synthesis).
Analyses were undertaken to assess the sensitivity of the results to alternative priors but these made little difference and thus only the results using the priors detailed in Meta-analysis have been presented.
Diagnostic test accuracy
A total of 62 studies contributed to the meta-analysis of sensitivity and specificity, including two studies118,119 that were three-arm (two index tests) studies. For simplicity, the correlation between tests was ignored in the analyses.
In total, 54 studies44–46,59–65,67–88,90–99,101,102,104–111,118,119 evaluated SeptiFast compared with blood culture, four studies48,113,118,119 evaluated SepsiTest compared with blood culture and four studies49,115–117 evaluated IRIDICA compared with blood culture. Separate meta-analyses are presented for each of these three tests in SeptiFast test, SepsiTest and IRIDICA assay compared with blood culture. In addition, one study112 evaluated SeptiFast compared with blood culture plus MALDI-TOF MS and one study114 evaluated SepsiTest compared with blood culture plus MALDI-TOF MS. As there was only one study for each of these comparisons, no meta-analysis was conducted and the data were summarised narratively.
The pooled sensitivity and specificity of SeptiFast compared with blood culture (54 studies) were 0.65 (95% CrI 0.60 to 0.71) and 0.86 (95% CrI 0.84 to 0.89), respectively (Figure 4). The 95% prediction intervals of 0.29 to 0.90 (sensitivity) and 0.62 to 0.96 (specificity) suggest considerable uncertainty in predicting the sensitivity and specificity of a new study. The between-study SDs for logit sensitivity and specificity were estimated to be 0.76 (95% CrI 0.57 to 1.01) and 0.66 (95% CrI 0.53 to 0.85), respectively, with a correlation of –0.05 (95% CrI –0.38 to 0.28). Figure 5 presents the joint distribution for sensitivity and specificity. The proportion of discordant results with blood culture (i.e. cases of disagreement between the reference standard and the index test) varied across studies from 6% to 46%, with a median of 17%.
FIGURE 4.
Sensitivity and specificity of SeptiFast compared with blood culture. FN, false negative; FP, false positive; PrI, prediction interval; TN, true negative; TP, true positive.
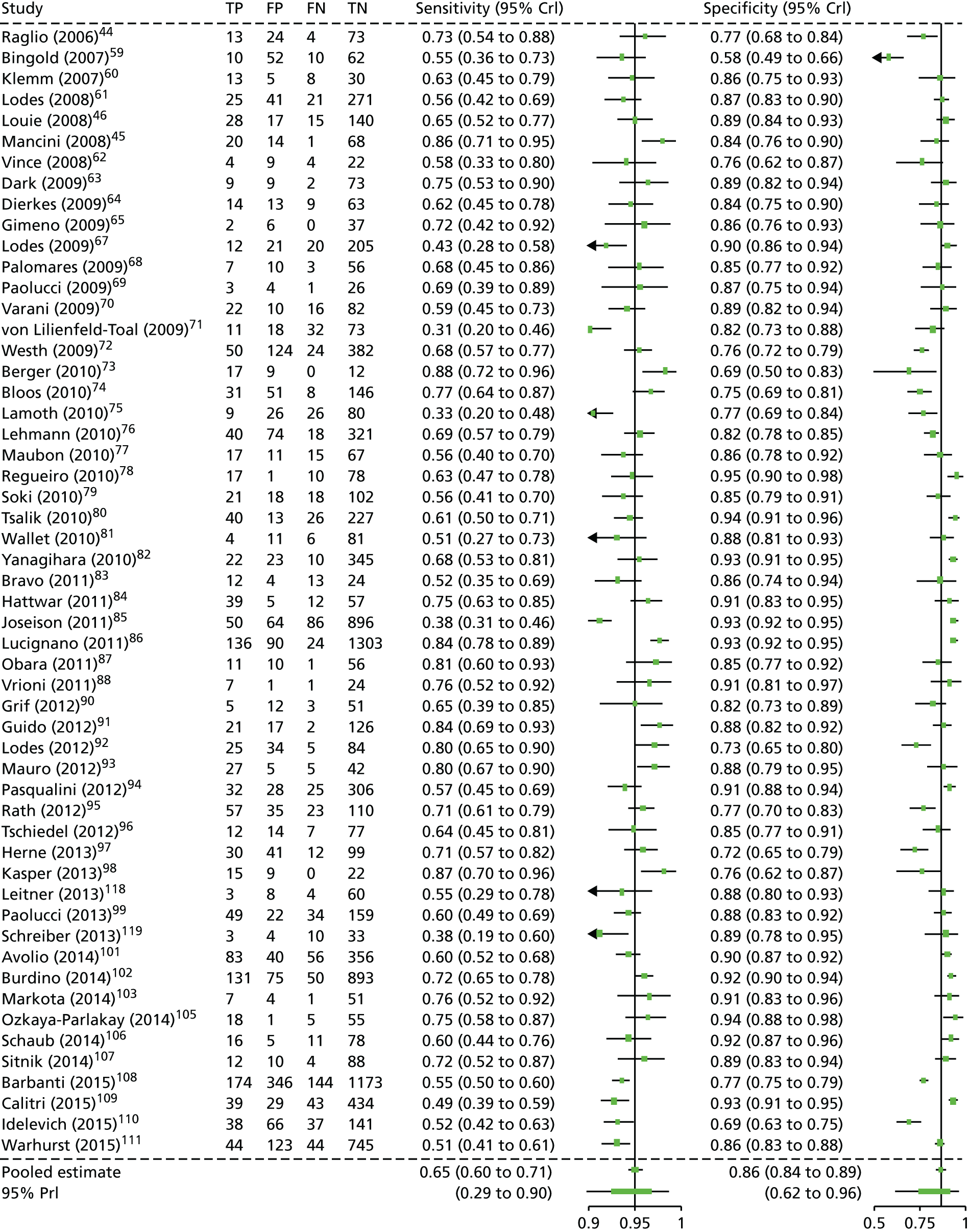
FIGURE 5.
Summary receiver operating curve plot of SeptiFast compared with blood culture (all studies). The 95% prediction interval indicates the extent of heterogeneity between studies. The green circles represent the sensitivity and specificity estimates from each study, with the circle size reflecting the study sample size.
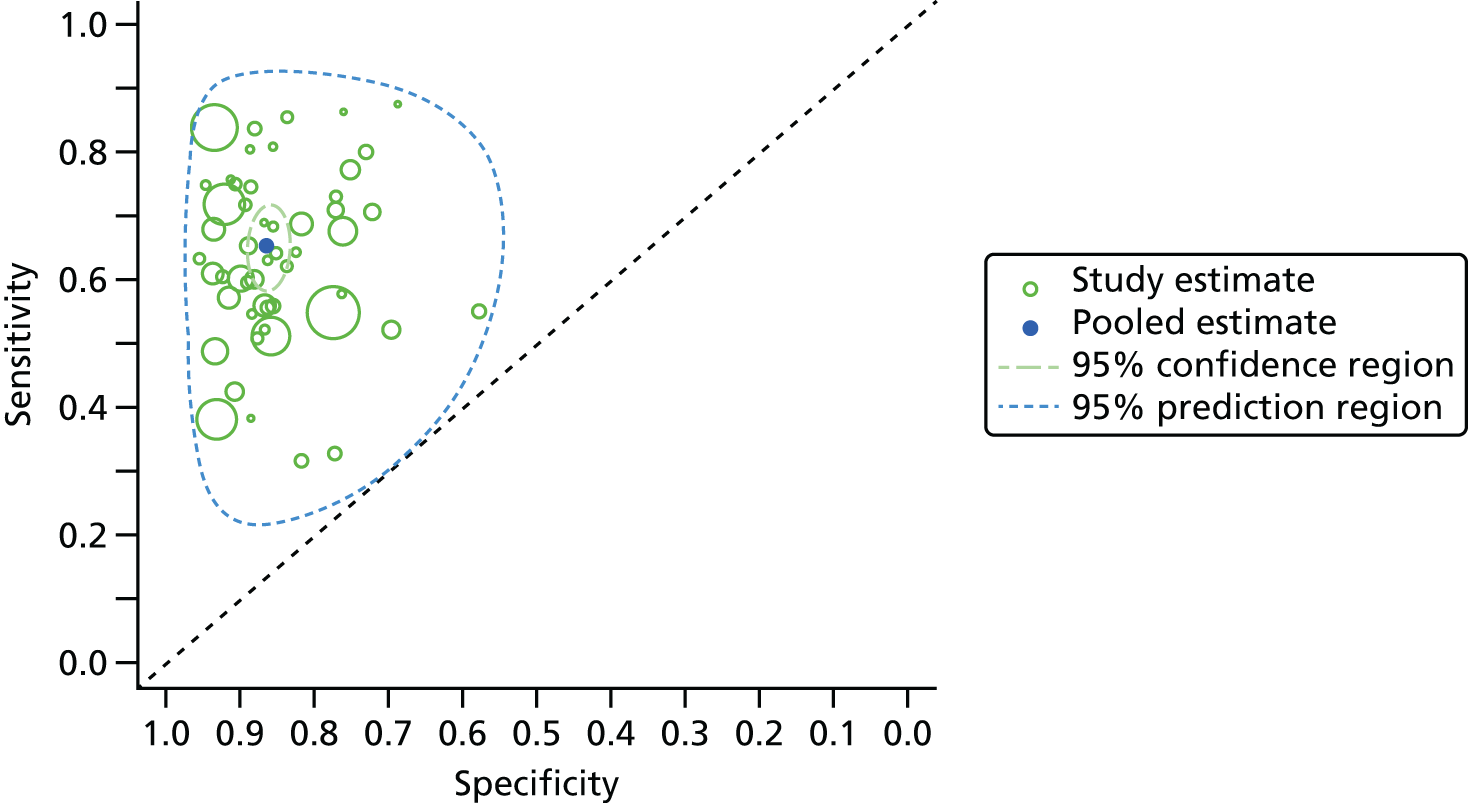
Additional analyses were undertaken for the following subgroups only: neonates and children, antibiotic use prior to blood sample collection, suspected community- or health-acquired infection, patients with febrile neutropenia and studies that included/excluded contaminants in the data analysis. There was insufficient information on studies at low risk of bias (see Quality characteristics) and people who were immunocompromised to allow a meaningful estimate of test accuracy. None of the subgroups analysed was shown to significantly affect the results.
-
Neonates and children Six studies provided data on children and neonates. Of these, three studies included neonates only,69,73,98 one included children only aged 1 month to 17 years105 and two included both neonates and children. 86,109 Of the remaining studies, six were conducted in adults and children,70,85,93,96,99,111 28 were conducted in adults45,46,63,64,67,68,71,74–76,78,80,81,83,84,87,90–92,94,95,97,101,102,104,106,110,119 and 14 did not report the age of participants. 44,59–62,65,72,77,79,82,88,107,108,118 Based on comparison of models with and without covariates for an age category, there was no evidence that sensitivity and specificity was affected by the age of the subjects (see Appendix 5).
-
People exposed to antibiotics prior to blood sample collection The proportion of patients receiving antibiotics prior to blood draw was recorded in 24 studies24,62,64,65,68,70,71,74,77,80,83,90–94,97–99,104,106,110,111,119 and ranged from 0% to 100% with a median of 72%. The remaining studies either did not report prior exposure to antibiotics or provided only limited information and were, therefore, excluded from the analysis. There was no evidence that exposure to antibiotics prior to blood sample collection affected the estimates of sensitivity and specificity (see Appendix 5).
-
People with suspected community- or health-acquired infection Clinical setting was used as a proxy for suspected community- or health-acquired infection. Studies were grouped according to whether infection was diagnosed in hospital (38 studies45,59–64,67,68,70,71,73–79,81,83,85,86,90–97,99,102,104,107–109,111,119), emergency department (three studies80,84,106), mixed setting of emergency or other hospital department (four studies46,82,87,101) or not recorded (nine studies44,65,69,72,88,98,105,110,118). Based on comparison of models with and without covariates for the clinical setting, there was no evidence that this affected sensitivity and specificity (see Appendix 5).
-
People with febrile neutropenia In total, eight studies provided data on patients with febrile neutropenia. 45,65,71,75,91,99,108,110 Of these, six studies included patients (100%) with febrile neutropenia only. 65,71,75,91,99,108 Studies by Mancini et al. 45 and Idelevich et al. 110 reported that 92% and 98% of patients had febrile neutropenia, respectively. Based on a comparison of models with and without covariates for the presence of patients with febrile neutropenia, there was no evidence that this affected sensitivity and specificity (see Appendix 5).
-
Studies with inclusion/exclusion of contaminants In total, 32 studies44,45,64,67–70,73,76,78,81–85,87,91–98,105–110,118,119 included contaminants in the reported results, eight studies46,72,80,86,99,101,102,111 excluded contaminants and 14 did not report on handling of contaminants. 59–63,65,71,74,75,77,79,88,90,104 Based on a comparison of models with and without covariates for the inclusion/exclusion of contaminants, there was no evidence that this affected sensitivity and specificity (see Appendix 5).
Only one study,112 which compared the SeptiFast test with MALDI-TOF MS, provided data on diagnostic test accuracy. This study reported a sensitivity and specificity of 0.58 (95% CI 0.30 to 0.86) and 0.74 (95% CI 0.64 to 0.85), respectively.
The pooled sensitivity and specificity of SepsiTest compared with blood culture was 0.48 (95% CrI 0.21 to 0.74) and 0.86 (95% CrI 0.78 to 0.92), respectively (Figure 6). The 95% prediction intervals of 0.07 to 0.90 (sensitivity) and 0.66 to 0.95 (specificity) suggest considerable uncertainty in predicting the sensitivity and specificity of a new study. The between-study SDs for logit sensitivity and specificity were estimated to be 0.90 (95% CrI 0.50 to 1.92) and 0.45 (95% CrI 0.27 to 0.90), with a correlation of –0.03 (95% CrI –0.73 to 0.68). Figure 7 presents the joint distribution for sensitivity and specificity, and highlights the extent of the heterogeneity between studies (as indicated by the 95% prediction interval). Owing to insufficient information provided in the included studies, planned subgroup analyses were not conducted and there were insufficient studies to conduct meaningful analyses.
FIGURE 6.
Confidential information has been removed.
FIGURE 7.
Confidential information has been removed.
Only one study114 that compared the SepsiTest assay with MALDI-TOF MS provided data on diagnostic test accuracy. This study reported a sensitivity and specificity of 0.11 (95% CI 0.00 to 0.23) and 0.96 (95% CI 0.92 to 1.00), respectively.
The pooled sensitivity and specificity of IRIDICA compared with blood culture was 0.81 (95% CrI 0.69 to 0.90) and 0.84 (95% CrI 0.71 to 0.92), respectively (Figure 8). The 95% prediction intervals of 0.55 to 0.94 (sensitivity) and 0.50 to 0.96 (specificity) suggest considerable uncertainty in predicting the sensitivity and specificity of a new study. The between-study SDs for logit sensitivity and specificity were estimated to be 0.46 (95% CrI 0.28 to 0.93) and 0.65 (95% CrI 0.39 to 1.27), with a correlation of 0.06 (95% CrI –0.71 to 0.75). Figure 9 presents the joint distribution for sensitivity and specificity and highlights the extent of the heterogeneity between studies (as indicated by the 95% prediction interval). Owing to insufficient information provided in the included studies, planned subgroup analyses were not conducted and there were insufficient studies to conduct meaningful analyses.
FIGURE 8.
Confidential information has been removed.
FIGURE 9.
Confidential information has been removed.
Other intermediate measures and clinical outcomes
A total of 41 studies provided data on one or more intermediate and/or clinical outcome measures: 37 SeptiFast studies,44–46,59,60,64,66,68,69,72,74,76–78,80,81,84,85,88–90,92–94,96,97,99–101,103–107,110–112 one SepsiTest study,114 two IRIDICA studies116,117 and one study evaluating both SeptiFast and SepsiTest. 119 A brief summary of the studies reporting data on each of the intermediate and clinical outcomes measure is presented in Table 7.
Across the studies reporting intermediate and/or clinical outcomes, the majority of studies reported data for the whole patient cohort, as opposed to comparative data for the index and reference test. Furthermore, for some outcomes, for example mortality, it was often unclear at what point the outcome was assessed. These limitations in reporting prohibited any statistical analysis to pool any intermediate and/or clinical outcome across included studies. None of the included studies provided data on readmission rates, adverse events associated with broad-spectrum antimicrobial use, morbidity, changes in disease severity over time, rates of superinfection, rates of resistant infection or health-related quality of life.
A summary of the studies reporting the times to pathogen identification of the index and reference test is presented in Table 8. Twenty-one SeptiFast studies reported data on the time to availability of results/pathogen identification. 44–46,59,60,64,68,69,77,80,81,85,88,93,96,97,101,106,107,110,112 However, for the majority of these studies, it was unclear if the value was the mean or median, and variance estimates or ranges were not reported. Across these studies, the reported time to pathogen identification with SeptiFast ranged from 4 hours68 to a median of 26.25 hours [range 6.75–79 hours (for samples collected at beginning of a weekend)]. 64 In contrast, the time to pathogen identification using blood cultures (with or without MALDI-TOF MS) ranged from 24 hours (minimum) to a median of 80 hours. Although the majority of studies that reported data on time to pathogen identification suggest that results are obtained sooner using SeptiFast than using blood culture, laboratory working times (e.g. laboratories operating on weekdays only) might delay the time to pathogen identification in real clinical practice. Time to pathogen identification was not reported by any of the studies evaluating SepsiTest or IRIDICA.
| Author (year) | Time to pathogen identification: index test | Time to pathogen identification: reference test |
|---|---|---|
| SeptiFast studies (non-comparative) | ||
| Raglio et al. (2006)44 | 16–30 hours | 5–7 days |
| Bingold et al. (2007)59 | 6 hours | 24–48 hours |
| Klemm et al. (2007)60 | 6.5 hours (minimum) | 2 days |
| Louie et al. (2008)46 | 6.54 hours (mean) | 65 hours (median) (range 24–214 hours) |
| Mancini et al. (2008)45 | NR | Detection with blood culture (lowest to highest range of mean number of hours): 10 hours for E.coli to 22.2 hours for coagulase-negative staphylococci. Definitive identification (lowest to highest range of mean number of hours): 44.2 hours for coagulase-negative staphylococci to 56.6 hours for E. faecalis |
| Dierkes et al. (2009)64 | 18 hours (median): twice daily analysis (range 6.75–74 hours for samples collected at beginning of weekend). 26.25 hours (median): once-daily analysis (range 6.75–79 hours for samples collected at beginning of weekend) | NR |
| Palomares et al. (2009)68 | 4 hours | 6.5 hours |
| Paolucci et al. (2009)69 | Information on antimicrobial susceptibility or microorganism viability ≈ 8 hours | 48–72 hours |
| Maubon et al. (2010)77 | 6.5 hours | NR |
| Tsalik et al. (2010)80 | 6.5 hours (approximately) | NR |
| Wallet et al. (2010)81 | 7–15 hours | 24–72 hours |
| Josefson et al. (2011)85 | 6 hours | NR |
| Vrioni et al. (2011)88 | 7–15 hours | 24–72 hours |
| Mauro et al. (2012)93 | 6 hours (approximately) | NR |
| Tschiedel et al. (2012)96 | 17 hours (range 6–17 hours) | 48 hours (range 48–120 hours, median 120 hours) |
| Herne et al. (2013)97 | NR (range 5–22 hours) | NR |
| Avolio et al. (2014)101 | Mean 16.6 hours (95% CI 14.9 to 18.2 hours) or median 15 hours (range 13–17 hours) (excludes SeptiFast and blood culture negative results) | Mean 84.2 hours (95% CI 82 to 86.4 hours) or median 80 hours (range 79–84 hours) (excludes SeptiFast and blood culture-negative results) |
| Schaub et al. (2014)106 | 6 hours | Median 16 hours (range 6–44 hours) |
| Sitnik et al. (2014)107 | < 8 hours (mean) | 3.5 days (mean) for blood culture-positive results. 5 days for blood culture-negative results |
| SeptiFast studies (comparative: RCTs) | ||
| Idelevich et al. (2015)110 | 20.3 hours (mean) | 58.3 hours (mean) |
| Tafelski et al. (2015)112 | Mean 15.9 hours (SD ± 5.9 hours) (95% CI 14.1 to 17.7 hours, assuming n = 41)a | Mean 38.1 hours (SD ± 11.6 hours) (95% CI 34.4 to 41.8 hours, assuming n = 37)a |
Time to treatment was reported by three SeptiFast studies, one of which was the RCT by Tafelski et al. 112 comparing SeptiFast with blood culture and MALDI-TOF MS, and two of which were the RCTs by Rodrigues et al. 100 and Idelevich et al. 110 comparing SeptiFast with blood culture. The time to treatment modification reported by all three RCTs was shorter in the SeptiFast group than in the blood culture group. Tafelski et al. 112 reported that the mean (SD) time from initially drawing blood to adaptation of empirical antimicrobial treatment was 18.8 (SD 5.6) hours in the intervention group (SeptiFast, n = 4), compared with 38.3 (SD 14.5) hours in the control group (n = 5) (p-value for difference not reported). The number of patients with therapy modification based on a positive diagnostic test was 4 out of 41 (9.8%) in the SeptiFast group and 5 out of 37 (13.5%) in the blood culture group. Rodrigues et al. 100 reported a mean time to change in therapy of 580 minutes (9.7 hours) with SeptiFast, compared with 3007 minutes (50.1 hours) (p-value for the between-group difference = 0.004) for blood culture. The number of patients in whom an adjustment of treatment was performed was 6 out of 17 (35%) in the SeptiFast group and 7 out of 29 (24%) in the blood culture group. Idelevich et al. 110 reported that the median time to the first change to a targeted antimicrobial therapy was significantly shorter with SeptiFast (21.4 hours, range 16.2–46.3 hours) than with blood culture (47.5 hours, range 7.3–59.2 hours) (p-value for the between-group difference = 0.018). There were 14 and 12 changes in antimicrobial therapy as a result of microbiological findings in the study group and the control group, respectively.
Seven SeptiFast studies64,72,84,99,106,111,112 reported information relating to failure rates. In the SeptiFast studies, test failure rates ranged from 1.5%106 to 24.2%. 84 A summary of the failure rates reported by the SeptiFast studies is presented in Table 9. No data on failure rates associated with SepsiTest were available.
| Author (year) | Reported test failure rate details |
|---|---|
| SeptiFast studies | |
| Dierkes et al. (2009)64 | One failure was attributed to technical problems during the analysis; however, no further details were provided |
| Westh et al. (2009)72 | 70/558 (12.5% of episodes) |
| Hettwer et al. (2011)84 | 38/157 (24.2%) |
| Paolucci et al. (2013)99 | 100/437 (22.9% of samples corresponding to 75 febrile episodes) |
| Schaub et al. (2014)106 | 3/205 (1.5% of samples had technical failure where the internal control was not detected) |
| Tafelski et al. (2015)112 | 4/37 (10.8%) |
| Warhurst et al. (2015)111 | 69/1006 (6.9% of episodes) [SeptiFast assay failure: reagent control (n = 6), internal control (n = 56) and other reasons (n = 7)] |
(Confidential information has been removed.)
The clinical significance of failure rates is that no additional information will be provided by the tests. As such, it is expected that the patient will be treated as would be the case without the test being available.
Thirteen of the included studies,64,74,76,80,89,100,103–106,110–112 all of which evaluated SeptiFast compared with blood culture, reported details of ICU and/or hospital stay. Details of these studies and the length of stay are reported in Table 10. Across the RCTs, Idelevich et al. 110 reported no statistically significant between-group difference in either ICU or hospital length of stay (p = 0.815 and p = 0.235, respectively); Tafelski et al. 112 also reported no significant between-group differences in either ICU or hospital length of stay (p ≥ 0.05 for both comparisons); and Rodrigues et al. 100 reported no statistically significant between-group difference in hospital stay (p = 0.632). Across the other studies reporting this outcome, data were often reported as characteristics of the included participants and it was often unclear if the length of stay was up to, including and/or after blood sampling.
| Study author (year) | Duration of ICU and/or hospital stay |
|---|---|
| SeptiFast studies (non-comparative) | |
| Dierkes et al. (2009)64 | Hospital stay: 35 days (median) |
| Bloos et al. (2010)74 | ICU stay: 13 days (median) Hospital stay: 34 days (median) |
| Lehmann et al. (2010)76 | ICU stay true negatives: 17 days (range 1–89 days) ICU stay true positives: 36 days (range 8–87 days) Hospital stay true negatives: 23 days (range 1–93 days) Hospital stay true negatives: 38 days (range 8–90 days) |
| Tsalik et al. (2010)80 | Hospital stay: 6.3 days (mean) |
| Alvarez et al. (2012)89 | ICU stay, SeptiFast: 22.9 days (mean) (SD ± 29.9 days) ICU stay, blood culture: 31.0 days (mean) (SD ± 19.4 days) Hospital stay, SeptiFast: 18.3 days (mean) (SD ± 21.4 days) Hospital stay, blood culture: 21.3 days (mean) (SD ± 23.4 days) Between-group difference ICU and hospital: p < 0.05 |
| Mancini et al. (2014)103 | Hospital stay: no between-group differences were observed (no data reported) |
| Markota et al. (2014)104 | Hospital stay: 27 days (mean) (SD ± 28.9 days) |
| Ozkaya-Parlakay et al. (2014)105 | ICU stay: 15.3 days (mean) (SD ± 23.8 days) |
| Schaub et al. (2014)106 | Hospital stay: 11 days (median) |
| Warhurst et al. (2015)111 | ICU stay: 16 days (IQR 9–30 days) |
| SeptiFast studies (comparative: RCTs) | |
| Rodrigues et al. (2013)100 | Hospital stay, SeptiFast: 32 days (mean) Hospital stay, blood culture: 31 days (mean) Between-group difference: p = 0.632 |
| Idelevich et al. (2015)110 | ICU stay, SeptiFast: 0.8 days (mean) (SD ± 4.0 days) ICU stay, blood culture: 0.9 days (mean) (SD ± 3.4 days) Hospital stay, SeptiFast: 40.4 days (mean) (SD ± 25.3 days) Hospital stay, blood culture: 42.9 days (mean) (SD ± 22.0 days) Between-group difference: hospital, p = 0.235; ICU, p = 0.815 |
| Tafelski et al. (2015)112 | ICU stay, SeptiFast: 34 days (range 13–65 days) ICU stay, blood culture: 32 days (range 16–57 days) Hospital stay, SeptiFast: 53 days (range 33–79 days) Hospital stay, blood culture: 37 days (range 20–76 days) |
Details of change in antimicrobial treatment plan were reported by 14 SeptiFast studies64,66,77,81,88,90,92,96,97,100,103,104,110,112 and one IRIDICA study. 116 Details of these studies and the reported changes are presented in Table 11.
| Author (year) | Reported changes in antimicrobial treatment plan |
|---|---|
| SeptiFast studies (non-comparative) | |
| Dierkes et al. (2009)64 | SeptiFast: from pathogens identified by SeptiFast only, five (7.7%) patients had an adjustment of antimicrobial therapy |
| Lehmann et al. (2009)66 | Blood culture: in 49 out of 467 (9.5%) episodes, antimicrobial treatment was changed |
| Maubon et al. (2010)77 | SeptiFast: results would have significantly improved treatment in 11 (10%) patients, and prompted immediate antimicrobial therapy not given initially in three patients |
| Wallet et al. (2010)81 | SeptiFast: on the basis of results, 8 out of 72 (11.1%) patients had an adjustment of antimicrobial therapy |
| Vrioni et al. (2011)88 | SeptiFast: on the basis of results, 5 out of 33 (15.2%) patients had an adjustment of antimicrobial therapy |
| Grif et al. (2012)90 | SeptiFast and concordant results from blood culture: 3 out of 33 (9.1%) patients had an adjustment of antimicrobial therapy SeptiFast and concordant results from samples from body sites: 5 out of 33 (15.2%) patients had an adjustment of antimicrobial therapy |
| Lodes et al. (2012)92 | SeptiFast: on the basis of results, 25 out of 148 (16.9%) samples had an adjustment of antimicrobial therapy |
| Tschiedel et al. (2012)96 | Patients with positive SeptiFast: 35 out of 75 (46%) had an adjustment of antimicrobial therapy Patients with negative SeptiFast: 5 out of 75 (6%) had an adjustment of antimicrobial therapy |
| Herne et al. (2013)97 | SeptiFast: on the basis of results, 21 out of 54 (39%) positive cases had an adjustment of antimicrobial therapy |
| Mancini et al. (2014)103 | Reports no between-group differences were observed in changes in management (propensity matching) |
| Markota et al. (2014)104 | SeptiFast: on the basis of results, four (6.3%) samples had an adjustment of antimicrobial therapy |
| SeptiFast studies (comparative: RCTs) | |
| Rodrigues et al. (2013)100 | SeptiFast: on the basis of results, 6 out of 17 (35%) patients had an adjustment of antimicrobial therapy Blood culture: on the basis of results, 7 out of 29 (21%) patients had an adjustment of antimicrobial therapy Between-group difference not reported |
| Idelevich et al. (2015)110 | SeptiFast: on the basis of results, 7 out of 74 (9.5%) patients had an adjustment of antimicrobial therapy Blood culture: on the basis of results, 8 out of 76 (10.5%) patients had an adjustment of antimicrobial therapy Between-group difference not reported |
| Tafelski et al. (2015)112 | SeptiFast and blood culture with MALDI-TOF MS: on the basis of results, 4 out of 41 (9.8%) patients had an adjustment of antimicrobial therapy Blood culture with MALDI-TOF MS: on the basis of results, 5 out of 37 (13.5%) patients had an adjustment of antimicrobial therapy Between-group difference not statistically significant: p ≥ 0.05 |
| IRIDICA studies | |
| Vincent et al. (2015)116 | A panel of three independent experts (randomly selected from a pool of seven) would have recommended a change in management, including initiation of therapy, altered antimicrobial spectrum and/or change in duration of therapy, based on the IRIDICA results. The panel recommended a change in management in 41% of patients. Where IRIDICA tests were positive and blood culture results were negative, a change in management was recommended in 57% of patients; no values were provided for other scenarios |
Nine of the SeptiFast studies reported on changes in antimicrobial therapy based on the SeptiFast results. 64,77,81,88,90,92,96,97,104 These studies did not report on changes based on blood culture results. One SeptiFast study reported on changes based on the blood results only. 66 The SeptiFast RCT by Rodrigues et al. 100 reported that 6 out of 17 (35%) patients in the SeptiFast group and 7 out of 29 (21%) patients in the blood culture group had an adjustment of antimicrobial therapy. The corresponding numbers for the RCT by Idelevich et al. 110 were 7 out of 74 (9.5%) patients in the SeptiFast group and 8 out of 76 (10.5%) patients in the blood culture group. The RCT by Tafelski et al. 112 reported that 4 out of 41 (9.8%) patients in the SeptiFast and blood culture with MALDI-TOF MS group and 5 out of 37 (13.5%) patients in the blood culture with MALDI-TOF MS group had an adjustment of antimicrobial therapy. A p-value for the between-group difference was not reported by any of these RCTs.
Seventeen of the SeptiFast studies,64,66,74,78,80,85,89,90,92,94,100,103–105,110–112 one SepsiTest study,114 one IRIDICA study116 and one study evaluating both SeptiFast and SepsiTest119 reported data on mortality. The majority of studies reported data for the whole patient cohort, as opposed to comparative data for the index and reference test. A summary of the mortality rates across all included studies is presented in Table 12. Across the RCTs, the SeptiFast RCT by Tafelski et al. 112 reported ICU mortality of 7 out of 41 (17%) participants in the SeptiFast group and 8 out of 37 (22%) participants in the blood culture group. The between-group difference was not statistically different (p ≥ 0.05); the SeptiFast RCT by Idelevich et al. 110 reported that five (6.6%) participants in the blood culture group and three (4.1%) participants in the SeptiFast group died, but did not report when this occurred. The SeptiFast RCT by Rodrigues et al. 100 reported that the between-group difference in 28-day mortality was not statistically significant.
| Author (year) | Reported mortality details |
|---|---|
| Single index test studies: SeptiFast (non-comparative) | |
| Dierkes et al. (2009)64 | In-hospital mortality: 33% |
| Lehmann et al. (2009)66 | 30-day mortality: 33.8% of 467 episodes |
| Bloos et al. (2010)74 | 29.9% – location/time period NR |
| Regueiro et al. (2010)78 | 37.5% – location/time period NRa |
| Tsalik et al. (2010)80 | 2.6% – location/time period NR |
| Josefson et al. (2011)85 | 30-day mortality: 4% |
| Alvarez et al. (2012)89 | 28-day mortality SeptiFast: 29% 28-day mortality blood culture: 24% 6-month mortality SeptiFast: 41.6% 6-month mortality blood culture: 37% Between-group difference: p = NS |
| Grif et al. (2012)90 | 24-hour mortality: 61% |
| Lodes et al. (2012)92 | 43.2% – location/time period NR |
| Pasqualini et al. (2012)94 | In-hospital: 12% |
| Mancini et al. (2014)103 | The mortality difference in the original propensity score matching was not significant: 8.24% (prospective cohort) vs. 13.48% (retrospective cohort) (p = 0.39). However, in a more stringently matched group, SeptiFast was reported to be associated with lower mortality rates [3.13% (n = 2 deaths) in the prospective cohort and 14.71% (n = 10 deaths) in the retrospective cohort (p = 0.04)] |
| Markota et al. (2014)104 | In-hospital mortality: 52.6% |
| Ozkaya-Parlakay et al. (2014)105 | 25.3% – location/time period NR |
| Idelevich et al. (2015)110 | SeptiFast: 4.1% – location/time period NR Blood culture: 6.6% – location/time period NR Between-group difference: p = 0.719 |
| Single index test studies: SeptiFast (comparative: RCTs) | |
| Rodrigues et al. (2013)100 | 28-day mortality SeptiFast: 53% 28-day mortality blood culture: 59% Between-group difference: p = 0.765 |
| Tafelski et al. (2015)112 | ICU mortality, SeptiFast and blood culture with MALDI-TOF MS: 17% ICU mortality, blood culture with MALDI-TOF MS: 22% Between-group difference: not statistically significant (p ≥ 0.05) |
| Warhurst et al. (2015)111 | 28-day mortality: 14% |
| Single index test studies: SepsiTest | |
| Loonen et al. (2014)114 | 3.2% – location/time period NR |
| Single index test studies: IRIDICA | |
| Vincent et al. (2015)116 | 29% – location/time period NR |
| Two index test studies: SeptiFast and SepsiTest | |
| Schreiber and Nierhaus (2013)119 | ICU mortality: 16% 28-day mortality: 24% |
In summary, the current evidence for the impact any of the index tests evaluated against blood culture on intermediary and clinical outcomes, such as mortality and reduced length of stay in critical care units, is limited. A small number of RCTs of low methodological quality have not shown SeptiFast to produce a statistically significant improvement compared with blood culture and there are presently no RCTs comparing the other index tests (SepsiTest or IRIDICA) of interest with blood culture, or any RCTs comparing any of the index tests of interest in a head-to-head manner.
Additional information on matrix-absorbed laser desorption time-of-flight mass spectrometry
Although not an intervention, and therefore omitted from the systematic review of clinical effectiveness, information on the diagnostic accuracy and in the potential benefits associated with MALDI-TOF MS was required. Two recent systematic reviews have been published:30,122 one focusing on the time taken to identify microbial organisms from positive blood cultures122 and one reviewing the performance of Sepsityper kit in conjunction with MALDI-TOF MS. 30
Dixon et al. 122 identified 10 studies which provided evidence that MALDI-TOF MS is associated with faster identification of pathogens, usually 24 hours sooner than blood culture alone. Where data were reported, MALDI-TOF MS was associated with a reduction in hospital costs and length of stay. However, the authors state that ‘all the included studies were observational and their findings have a relatively high risk of bias’ and that ‘MALDI-TOF MS has the potential to reduce length of stay and costs while improving patient outcomes, but more and better evidence, including that on cost-effectiveness, is required’.
Morgenthaler and Kostrzewa30 summarise data from 21 reports to assess the reliability of the Sepsityper kit in the rapid identification of bloodstream infection. It was reported that ‘no relevant misidentification on the genus level was reported at a log (score) cut-off of 1.6’, whereas time to a result was reduced by several hours or days.
In addition to these reviews, papers known to the authors, submitted by the company or identified in the sifting related to the review of economic evaluations of the interventions were read to provide additional information regarding MALDI-TOF MS. Citation searching was performed to identify further information. It is noted that often MALDI-TOF MS was introduced in conjunction with another change, such as the establishment of an antimicrobial stewardship team, and, therefore, the exact gain attributable to MALDI-TOF MS was unknown.
Perez et al. 123 report the implementation of an evidence-based intervention that integrated MALDI-TOF MS, rapid antimicrobial susceptibility testing and near-real-time antimicrobial stewardship practices. A comparison of the results before and after testing was made. The mean hospital length of stay for survivors (n = 100) after bloodstream infection onset was 9.9 days in the pre-intervention group, compared with 8.1 days in the intervention group (n = 101; p = 0.01). Within a multivariate model, receiving active antibiotic therapy at 48 hours was associated with a hazard ratio for discharge of 2.90 (95% CI 1.15 to 7.33; p = 0.02) and the intervention was associated with a hazard ratio for discharge of 1.38 (95% CI 1.01 to 1.88; p = 0.04). Total hospitalisation costs were US$45,709 in the pre-intervention cohort and US$26,162 in the intervention cohort.
A further paper124 reported a pre–post quasi-experimental study that analysed the impact of MALDI-TOF MS with an antimicrobial stewardship team. The intervention (n = 256) decreased time to organism identification compared with previous treatment prior to MALDI-TOF with an antimicrobial stewardship team (n = 245) (55.9 vs. 84.0 hours; p < 0.001) and improved time to effective antibiotic therapy (20.4 vs. 30.1 hours; p = 0.021), optimal antibiotic therapy (47.3 vs. 90.3 hours; p < 0.001) and length of ICU stay (8.3 vs. 14.9 days; p = 0.014). The 30-day all-cause mortality was lower in the intervention arm than in the pre-intervention arm (12.7% vs. 20.3%; p = 0.021), as was length of hospitalisation (14.2 vs. 11.4 days; p = 0.066).
A study in Texas (USA) compared the outcomes of antibiotic-resistant Gram-negative bacteraemia in 112 patients admitted during January 2009 to November 2011 and 157 patients admitted during February 2012 to June 2013 following the introduction of an intervention (MALDI-TOF MS and antimicrobial stewardship). 125 Time to initiation of active treatment was 90 hours in the first group and 32 hours in the second group (p < 0.001). There were 33 (21%) deaths from any cause in the pre-intervention cohort and 10 (9%) in the intervention cohort. In multivariate logistic regression, the intervention was a significant predictor of survival (odds ratio 0.28, 95% CI 0.12 to 0.71; p = 0.008). A significant reduction in average total hospital costs was observed, from US$78,991 to US$52,693.
A quasi-experimental study126 was conducted to evaluate the effect of introducing MALDI-TOF MS plus antimicrobial stewardship team review on the treatment of hospitalised patients in whom blood samples tested positive for coagulase-negative staphylococci (n = 324). Before the introduction of the intervention, 117 positive cultures (72%) were deemed to be contaminated and 46 (28%) were from patients with bacteraemia. The corresponding figures after the introduction of the intervention were 129 (80%) and 32 (20%), respectively. Following the introduction of MALDI-TOF MS plus antimicrobial stewardship team review, patients with bacteraemia received optimal therapy sooner (34.4 vs. 58.7 hours; p = 0.032) and exhibited a lower mortality rate than patients treated before the introduction of the intervention (3.1% vs. 21.7%; p = 0.023). In addition, following introduction of the intervention, patients whose blood samples were contaminated experienced a shorter duration of unnecessary antibiotic therapy (1.31 vs. 3.89 days; p = 0.032) and underwent fewer vancomycin trough assays (0.88 vs. 1.95; p < 0.001). However, rates of mortality, duration of hospitalisation stay, recurrent bloodstream infections and 30-day hospital readmissions were not significantly different.
A paper by Martiny and Debaugnies127 reports that the use of MALDI-TOF MS resulted in the modification of treatment in 21 out of 157 adults and 1 out of 40 children.
The potential benefits associated with MALDI-TOF MS, albeit not from RCTs, is believed to be the motivation for the RAPIDO study,31 which will report the required data for assessing the clinical effectiveness of MALDI-TOF MS, typically in conjunction with Sepsityper and blood culture compared with blood culture alone within clinical practice. It is envisaged that the results from this study will make the preceding data in this section largely redundant.
Chapter 3 Assessment of cost-effectiveness
Systematic review of existing economic evidence
This section of the report describes a review of the existing published evidence on the economic impact of the SeptiFast, SepsiTest and IRIDICA tests to rapidly detect and identify bacterial and fungal DNA, which may be present in the bloodstream of people who are suspected of having sepsis. As previously stated, earlier versions of the IRIDICA BAC BSI assay were assumed by the authors to provide generalisable data, and these have been included in the review, with explicit reference made to the version of IRIDICA.
Methods
Electronic resources
A systematic search was undertaken of the existing published literature evaluating the economic impact of the SeptiFast, SepsiTest and IRIDICA tests to rapidly detect and identify bacterial and fungal DNA that may be present in the bloodstream in people who are suspected of having sepsis.
Studies were identified by searching the following electronic databases and research registers:
-
MEDLINE In-Process & Other Non-Indexed Citations and MEDLINE (via OvidSP), searched from 1948 to May 2015
-
EMBASE (via OvidSP), searched from 1980 to May 2015
-
Cochrane Database of Systematic Reviews (via Wiley Online Library), searched from 1996 to May 2015
-
Cochrane Central Register of Controlled Trials (via Wiley Online Library), searched from 1898 to May 2015
-
Health Technology Assessment Database (via Wiley Online Library), searched from 1995 to May 2015
-
Database of Abstracts of Review of Effects (via Wiley Online Library), searched from 1995 to May 2015
-
NHS Economic Evaluation Database (via Wiley Online Library), searched from 1995 to May 2015
-
Science Citation Index Expanded (via the Web of Science), searched from 1899 to May 2015
-
Conference Proceedings Index-Science (via the Web of Science), searched from 1990 to May 2015
-
World Health Organization’s International Clinical Trials Registry Platform, searched from 2007 to May 2015
-
Current Controlled Trials, searched from 2000 to May 2015
-
NIH ClinicalTrials.gov, searched from 2000 to May 2015
-
Manufacturer and User Facility Device, searched from 1991 to May 2015
-
MEDION database.
Sensitive keyword strategies using free text and, where available, thesaurus terms using Boolean operators and database-specific syntaxes were developed to search the electronic databases. Synonyms relating to the condition (e.g. sepsis) and the test (i.e. SeptiFast, SepsiTest and IRIDICA) were combined with a search filter aimed at restricting results to economic and cost-related studies (used in the searches of MEDLINE and EMBASE). No language restrictions were used on any database; however, the searches were restricted by date (see Chapter 2, Methods for reviewing effectiveness, for further details). In brief, CE approval for the oldest rapid molecular test (i.e. SeptiFast) was obtained in 2006. As a result, no relevant economic evaluations were expected to be published prior to this date. An example of the MEDLINE search strategy is provided in Appendix 6.
Other resources
To identify additional published, unpublished and ongoing studies, the reference lists of all relevant studies were checked and a citation search of relevant articles (using the Web of Science Citation Index Expanded and Conference Proceedings Citation Index – Science) was undertaken to identify articles that cited the relevant articles. In addition, systematic keyword searches of the World Wide Web were undertaken using the Google search engine, key experts in the field were contacted and company submissions were screened for published or unpublished data additional to those identified in studies retrieved from the literature search.
All identified citations from the electronic searches and other resources were imported into, and managed using, the Reference Manager bibliographic software.
Studies were selected for inclusion according to predetermined inclusion and exclusion criteria. A summary of these criteria is provided in Table 13.
| Criteria | Included | Excluded |
|---|---|---|
| Countries | All | No restriction |
| Settings | All | No restriction |
| Study design | Economic evaluations (model or study based) comparing one of the interventions listed below with an appropriate comparator, including other interventions if applicable | Non-economic evaluation Cost study of one test only (comparison of costs of different reagents or techniques) |
| Population | Adults and children (of any age) with suspected bloodstream infections in secondary care (i.e. departments and wards providing care for acutely unwell patients and/or critical care units) who required blood cultures | Those people not meeting the inclusion criteria |
| Target condition | People with suspected sepsis | People who do not have suspected sepsis |
| Comparator test | Blood culture with or without MALDI-TOF MS | Other tests done in house |
| Interventions (index test) | SeptiFast, SepsiTest and IRIDICA | Economic evaluations that do not investigate one of the interventions of interest in at least one of the arms |
| Outcomes | Cost minimisation, cost-effectiveness, cost–utility analysis | Other forms of economic evaluations |
Studies were selected for inclusion through a two-stage process:
-
Level 1 screening: titles and abstracts were independently examined for inclusion by two reviewers (RR and MS). Any disagreements in the selection process were resolved through discussion.
-
Level 2 screening: full manuscripts of selected citations were then retrieved and assessed by one reviewer (RR). A second reviewer (MS) performed an independent quality check to ensure that the inclusion criteria were applied correctly. Any disagreements in the selection process were resolved through discussion.
No formal quality assessment was conducted. When assessing the methodological quality of the economic literature, a number of checklists are available; however, quality assessment checklists for assessing economic evaluations of diagnostic tests are limited. Similarly, the majority of checklists focus on the quality of reporting rather than the methodological quality of a study. Owing to these limitations the relevance of each study to the decision problem is discussed within Descriptive summary of the study included in the review, Critique of the studies included in the review and Relevance of existing economic evaluations for National Institute for Health and Care Excellence decision-making.
Results
Identified studies
A total of 89 citations were retrieved. Of these, 77 citations were identified via database searching and an additional 12 citations were retrieved through other sources (Figure 10).
Eighty references were excluded at title and abstract stage. Nine references related to eight studies were examined at full-text level and four studies (corresponding to four references) were identified as meeting the inclusion criteria of the systematic review of economic evaluations. 89,103,128,129 These included an economic evaluation of the IRIDICA–PLEX-ID hybrid assay reported in a poster presentation (submitted by the company). 128 It was highlighted that the system evaluated in the poster presentation was not the final IRIDICA BAC BSI assay but was an earlier version that used components of PLEX-ID and is assumed to be equivalent (see Chapter 1, IRIDICA BAC BSI).
Five papers were excluded after retrieval of the full papers,130–134 the rationale being that the results were published in full elsewhere,130 they were other interventions131–133 and there was an absence of an economic evaluation and inappropriate intervention. 134
Descriptive summary of the study included in the review
A tabulated summary of the key characteristics of the studies included in the economic review, as determined by the authors of this report, is presented in Table 14. It was not possible for the External Assessment Group to check the economic models, as only the publications were available in the public domain.
| Parameter | Alvarez et al. (2012)89 | Bilkovski et al. (2014)128 | Lehmann et al. (2010)129 | Mancini et al. (2014)103 |
|---|---|---|---|---|
| Country | Spain | Unclear | Unclear | Italy |
| Study type | Within-study (observational retrospective non-matched study) economic evaluation | Mathematical model [based on the RADICAL (Rapid Diagnosis of the Critically Ill)119 study and data from two MALDI-TOF MS studies] | Mathematical model (evidence from different sources combined) | Within-study (observational propensity score-matched study) economic evaluation |
| Economic evaluation | Cost minimisation | Cost minimisation | Cost-effectiveness/cost–utility analysis Cost per incremental survivor Cost per QALY gained |
Cost minimisation |
| Rationale for the approach used | No difference in observed mortality | Not provided | NA | Not provided |
| Intervention | SeptiFast | IRIDICA–PLEX-ID hybrid | SeptiFast | SeptiFast |
| Comparator | BC | BC | BC | BC |
| Funder of the study | Unclear | Abbott Diagnostics | Roche Diagnostics | Roche Diagnostics |
| Target population and condition | Patients diagnosed with severe sepsis and septic shock | Critically ill patients with suspected BSI | Patients with a sepsis episode. Predominantly hospital-acquired infection | Haematological patients with signs of systemic inflammatory response syndrome with suspected sepsis |
| Setting | ICU | Majority ICU | Post-surgical and ICU patients | Haematology and bone marrow transplant units |
| Age | ≈ 65 ± 14 years | 60.4 ± 18.8 years | > 60 years | ≈ 50 ± 14 years |
| Source used for the impact of the test on treatment modification | NA – within-study economic evaluation | RADICAL study116 | Evidence collected prospectively from five hospitals (two German, one Italian, one Spanish and one US hospital)66 | NA – within-study economic evaluation |
| Source used for the impact of the test on clinical outcomes (mortality, length of stay) | NA – within-study economic evaluation | Two studies of MALDI-TOF MS, in addition to an antimicrobial stewardship programme124,125 | Pooled data on the impact of inadequate treatment on outcomes from two previously published studies135,136 conducted in the USA | NA – within-study economic evaluation |
| Study perspective | Health-care perspective | Health-care perspective | Health-care perspective | Health-care perspective |
| Discounting | NA | NA | Not stated | NA |
| Time horizon | NA | NA | Lifetime | NA |
| Cost categories included in the economic evaluation | Antibiotic treatment ICU stay Ward stay SeptiFast test |
ICU stay Ward stay IRIDICA–PLEX-ID hybrid |
SeptiFast test | Diagnostic and laboratory assays (including SeptiFast test) Instrumental diagnostic procedures Administered therapeutic agents (empiric and pathogen-targeted therapy) Non-anti-infectious drugs to manage SIRSS-SS-related complications |
| Cost of the intervention | €183 (based on seven patients) per patient including reagent cost, personnel cost and imputable structural costs | US$250 | €300 | €178.75 per sample (average of four samples plus positive and negative controls per each run, including both reagents and personnel costs) |
| Unit costs for other resource use | Stay in ward: €273 Stay in ICU: €1058 Cost of antibiotics taken directly from hospital – values not reported |
Non-ICU stay per day: US$2122 ICU stay per day: US$3500 |
NA | Taken directly from hospital – values not reported |
| Measurement of benefits | NA | NA | Mortality Morbidity (associated with sepsis) – QALYs |
NA |
| Utility values | NA | NA | Sepsis: 0.68 | NA |
| Results | Total costs (test vs. BC) per patient: €32,228 vs. €42,198; p = 0.05 (saving of €9970) 96.3% probability of cost savings |
Total costs (test vs. BC): US$19,375,716 vs. US$20,499,088 per 422 tests | €11,44 (95% CI €9321 to €14,977) per incremental survivor €3107 (95% CI €2523 to €4055) per QALY gained |
Total costs (test vs. BC) per patient: €1579.80 (median €1075.47) vs. €2010.53 (median €1105.18); p = 0.05 (saving of €430.73) |
| Breakdown of clinical results | ICU length of stay (control vs. test) (31.0 ± 19.4 vs. 22.9 ± 29.9 days) Hospital length of stay (21.3 ± 23.4 vs. 18.3 ± 21.4 days) ICU length of stay (survivors) (24.1 ± 21.9 vs. 18.3 ± 11.4 days) Number of antibiotics used per patient (5.1 ± 3.1 vs. 4.2 ± 2.2) |
4.2 days saved in hospital stay in patients with a positive test (1.6 days in all patients) 1.8 days saved in ICU stay in patients with a positive test (0.7 in all patients) |
80.5 days (95% CI 48 to 113 days) potential earlier treatment Absolute reduction in mortality of 2.6% |
NA |
| Breakdown of cost results | Antibiotic treatment costs (test vs. control): €2812 vs. €3,576; p < 0.05 ICU costs: €24,246 vs. €32,798; p < 0.05 Ward costs: €5988 vs. €5824; p < 0.05 |
Hospital cost (per 422 tests) (test vs. control): US$19,185,816 vs. US$20,414,688 | Classical diagnostic and instrumental procedures assays (test vs. BC): €652.79 vs. €625.66; p = 0.68 Medication costs: €927.01 vs. €1384; p = 0.02 |
Of the four identified economic studies (corresponding to three full texts89,103,129 and one poster presentation128), three economic evaluations (full text) compared the addition of SeptiFast to blood culture with blood culture alone89,103,129 and one compared the addition of the IRIDICA–PLEX-ID hybrid assay to blood culture with blood culture alone (poster presentation). 128 No economic evaluations of SepsiTest were identified. None of the four published economic evaluations was conducted in a UK setting. However, the RADICAL study,116 used for the impact of the IRIDICA–PLEX-ID hybrid assay on treatment modification, included two UK (out of nine) sites. 116
Two103,129 out of the three SeptiFast studies were funded by Roche Diagnostics; it was unclear whether or not the third study89 was funded by the company. The IRIDICA–PLEX-ID hybrid cost minimisation study was funded by Abbott Diagnostics.
The target population, condition and setting varied between the four identified economic studies. Mancini et al. 103 included haematological patients with signs of systemic inflammatory response syndrome with suspected sepsis (SIRS-SS). Alvarez et al. 89 included patients diagnosed with severe sepsis and septic shock. Lehmann et al. 129 included all post-surgical and ICU patients with a sepsis episode (predominantly hospital-acquired infection), while Bilkovski et al. 128 included critically ill patients with suspected bloodstream infection.
Three studies were cost-minimisation studies. 89,103,128 Two were conducted within studies: a non-matched retrospective study evaluating SeptiFast89 and a propensity score-matched study evaluating SeptiFast against blood culture. 103 The Alvarez et al. study89 justified the use of cost minimisation given the absence of mortality data associated with the use of SeptiFast. The third cost minimisation study was undertaken using a decision tree model128 and evaluated the IRIDICA–PLEX-ID hybrid assay by combining evidence from the RADICAL study116 on the impact of the test in terms of treatment decision, and evidence from MALDI-TOF MS studies123,124 on the impact of rapid identification on the reduction in hospital and ICU length of stay. The main assumptions within the model were that (a) all patients start on empiric antimicrobial therapy and (b) only patients testing positive using the IRIDICA–PLEX-ID hybrid assay experience a reduction in length of stay.
Only one study was a cost-effectiveness (cost–utility) analysis and estimated the ‘cost per incremental survivor’ and the cost per quality-adjusted life-year (QALY) gained of introducing SeptiFast. 129 An algebraic model was constructed, which independently estimated the potential cost impacts and clinical outcomes associated with a change in treatment plan as a result of the earlier identification of inadequate treatment through the use of SeptiFast. Only positive SeptiFast results were considered to provide sufficient evidence to allow a treatment change, with the authors concluding that ‘withdrawal of antimicrobial treatment upon a PCR negative result is not recommended’. 129 Cost savings and increased health associated with quicker adequate treatment were estimated assuming a relationship between a reduction of 1 day in inadequate treatment and changes in both length of stay and mortality. This study used evidence collected prospectively from five hospitals to inform the change in treatment decision: two based in Germany, one in Spain, one in Italy and one in the USA. 66 In addition, the modelling uses pooled data on the impact of inadequate treatment on outcomes from two previously published studies135,136 conducted in the USA.
Although not clearly stated by the authors, it is believed by the authors of this report that Mancini et al. 103 and Alvarez et al. 89 report results from an Italian and Spanish setting, respectively. In the case of the studies by Lehmann et al. 129 and Bilkovski et al.,128 the country on which the analysis was based is unclear, as the data came from multicentre studies.
The economic evaluation was conducted in patients in the ICU in three studies. 89,128,129 All studies appear to use a hospital (health-care payer) perspective, although this was not explicitly stated in two studies. 103,128 Uncertainty was examined in two studies. 89,129 Only one study used QALYs as a measure of benefit. 129
The cost of performing SeptiFast varied between studies and ranged from €178.75103 (£128.70 assuming an exchange rate of €1 to £0.72137) to €300129 (£216). One study evaluated the IRIDICA–PLEX-ID hybrid assay and assumed a cost of US$250 per test128 (£160.67 assuming an exchange rate of US$1 to £0.64). 137
Two studies89,103 considered antibiotics costs, and both reported a reduction in antibiotics costs associated with the use of SeptiFast. One study included the savings in classical diagnostic assays103 with the use of SeptiFast. Three of the four studies considered the costs associated with ICU/hospital stay, and all reported a reduction in hospital/ICU stay with the use of the test. 76,89,128 None of the studies identified considered the impact on costs associated with the potential reduction in antibiotic resistance.
Overall, all three cost-minimisation studies reported a reduction in total costs, with the additional cost of SeptiFast and the IRIDICA–PLEX-ID hybrid assay being outweighed by the savings in antibiotics and/or hospital costs. 89,103,128 Mancini et al. 103 reported an overall saving of €430.73 per patient receiving a SeptiFast test and blood culture compared with those diagnosed using only blood culture (€1579.80 vs. €2010.53), considering savings in diagnostic and instrumental assays, and medications (including antibiotics, antimycotics, antiviral agents and other drugs). Alvarez et al. 89 reported a saving of €9970 per SeptiFast test, with the majority of savings achieved based on a reduction in ICU length of stay. Bilkovski et al. 128 reported a total saving of US$1,123,372 over 422 patients tested (equating to a saving of £2662 per patient tested), with the IRIDICA–PLEX-ID hybrid assay associated with a reduction in hospital or ICU length of stay. Finally, Lehmann et al. 129 reported that the cost of the SeptiFast test could be recovered if the daily medical costs were above €717 and suggested that this was likely to be the case. 129 The authors reported the cost per incremental survivor and cost per QALY gained to be €11,477 (95% CI €9321 to €14,977) and €3107 (95% CI €2523 to €4055), respectively.
Critique of the studies included in the review
The Lehmann et al. 129 economic evaluation, which compared the use of SeptiFast and conventional blood culture, appears to be a reasonably well-conducted cost-effectiveness analysis based on the description provided by the authors. However, this study has a number of limitations that may restrict the generalisability to UK practice, not only because none of the hospitals contributing to the study was located in the UK. The data used in the Lehmann et al. 129 cost-effectiveness model on the potential impact of the test in terms of treatment modification are relatively out of date and were collected prior to recent guidelines on the diagnosis and treatment of sepsis. 16,18–20 Therefore, the extent to which the inadequate treatment observed in Lehmann et al. 66 is generalisable to current practice in the UK is not known. Furthermore, the studies on how earlier adequate treatment translates into reduced morbidity and mortality were dated and cohort based and, as acknowledged by Lehmann et al. ,129 could be potentially confounded. Lehmann et al. 129 used a relative risk of non-survival between immediate and delayed adequate antimicrobial treatment of 2.32, but report in their discussion section that a value of 1.8 estimated in a clinical trial of immunomodulating therapy for severe sepsis138 would have been a better estimate of the relative risk of non-survival associated with inadequate treatment. Using this value led to an increase in the cost per incremental survivor from €11,477 to €14,670 (an alternative cost per QALY gained value was not reported). It should be noted that the relative risk is still relatively dated as it was published in 2003. 138 The relative risk of mortality was pooled from three studies, rather than the more appropriate method of meta-analysing. The same limitations of the relationship between inadequate treatment and mortality also apply to the relationship between inadequate treatment and length of hospitalisation, which uses data from the two old studies. It is noted that, in the original Lehmann et al. 66 paper, data were collected from two separate sites of attendance: ICU or surgical ward, and emergency room or other. The estimated gainable days of adequate treatment per 100 SeptiFast tests were 36.4 days for the ICU or surgical ward group and 10.6 days for the remaining attendance method. It is unclear if the evaluation by site was preplanned or if the analysis on ICU patients alone could be viewed as data dredging; ideally a replication of the study within the ICU would provide more conclusive data. The failure rate of the SeptiFast test was also not considered. In the model, patients receiving SeptiFast are assumed to experience a mortality benefit associated with rapid identification. Although this may be plausible, so far, evidence on the impact of SeptiFast on clinical outcomes of mortality and hospital length of stay is inconclusive from five comparative studies. 89,103,110,112,139
The cost-minimisation study conducted by Alvarez et al. 89 is based on a retrospective non-matched comparison of the costs in a cohort prior to the introduction of SeptiFast and a cohort following the use of SeptiFast. Alvarez et al. 89 reported a non-significant increase in mortality in the cohort receiving the intervention at 28 days (29% vs. 24%, non-significant; p-value not reported), and at 6 months (41.6% vs. 37%, non-significant; p-value not reported). In contrast, the authors reported a significant reduction in ICU length of stay for survivors (31.0 ± 19.4 vs. 22.9 ± 29.9 days; p < 0.05), hospital length of stay (21.3 ± 23.4 vs. 18.3 ± 21.4 days; p < 0.05) and ICU length of stay (24.1 ± 21.9 vs. 18.3 ± 11.4 days; p < 0.05). The authors calculated the total cost to be €42,198 for the control group and €32,228 for the intervention group, corresponding to a saving of €9970 (with more than 85% of savings attributable to a reduction in ICU length of stay). This study has serious limitations that need further consideration. The results need to be interpreted with caution because of the retrospective nature of this study, the small sample size of each arm (48 patients in the intervention group and 54 patients in the control group) and the imbalances between patient characteristics, as shown in Table 15. It is also unclear whether patients were randomly selected to receive SeptiFast or whether these patients were selected based on their characteristics. Although the authors reported no differences in terms of patients’ ages, sex distribution or risk indices, it is unclear from the paper how these risk indices were calculated. There appear to be large imbalances in the initial diagnoses that could impact on ICU length of stay (main source of savings). It should be noted that no difference in ICU or hospital length of stay was observed in other comparative studies (RCT or propensity-matched score studies). 103,110,112,139 The authors report a saving in antibiotics (excluding cost of the test) of €764 per patient [as a result of a reduction in the number of antibiotics used per patient of 0.9 (from 5.1 to 4.2)]. The authors do not say which antibiotics were used in the study, or for how long. It should be noted that, in England, antibiotics are typically given as a 5- to 7-day course, with a daily cost around £50. The failure rate of the SeptiFast test was not considered.
| Baseline characteristics | Group | |
|---|---|---|
| Control (n = 54) | Intervention (n = 48) | |
| Age (years), mean ±SD | 65 ± 14.7 | 54 ± 12.9 |
| Sex, % male | 83.30% | 72.90% |
| Initial diagnoses, % | ||
| Emergency abdominal surgery | 20.37 | 18.75 |
| Elective abdominal surgery | 3.70 | 4.17 |
| Pneumonia | 7.41 | – |
| Pancreatitis | 1.85 | 14.58 |
| CNS lesions | 16.67 | 10.42 |
| Polytrauma/head injuries | 7.41 | 41.67 |
| Heart surgery | 37.04 | 4.17 |
| Major vascular surgery | 5.56 | 2.08 |
| Pneumonectomy | – | 2.08 |
The second cost-minimisation study, by Mancini et al. 103 evaluated the use of SeptiFast with blood culture and is based on a propensity score-matched approach with 101 matched SIRS-SS episodes. The authors reported a trend (although not significant) towards a reduction in SIRS-SS-related mortality associated with SeptiFast. The authors reported that the differences in mortality reached statistical significance when the tolerance (calliper) used for the propensity matching was reduced to only 5%, corresponding to 77 matched episodes (64 patients in the intervention group and 68 in the control group). Although the use of a lower calliper improved the quality of matching, it reduced the precision by decreasing the sample size and could also introduce bias, as the estimate is no longer the effect of treatment in the treated subjects, but the effect of treatment in those treated subjects for whom a control was found. 140 Using the tight calliper of 0.05, the authors found a mortality rate of 3.13% in the intervention group and 14.71% in the control group (i.e. a difference in the mortality rate of 11.58%). Based on a pilot study, the authors reported that 278 pairs of SIRS events were needed to demonstrate a 10.6% difference in mortality (9.7% in the intervention group vs. 20.3% in the control group), which is greater than the numbers analysed in Mancini et al. 103 The results from this study therefore need to be interpreted with caution. It should also be noted that the PCR test was implemented under optimal conditions. The authors found no reduction in SIRS-SS episode length, even under a tight strict matching criterion. In this study, the SeptiFast test was estimated to lead to a reduction in medication. Based on information provided in a supplementary appendix associated with the paper, the cost of antibiotics was higher in patients receiving SeptiFast (€11.78). In contrast, the costs of antimycotics (commonly called antifungals in England) (€376.62), antiviral agents (€1.65) and other drugs (€91.39) were reduced. Although the two cohorts were propensity matched, it is unclear whether or not the suspected organisms in the two cohorts were matched, which may affect the antibiotics prescribed. In the control arm, antimycotics represented approximately 53% of total medication costs. This figure appears relatively high compared with that expected in England based on Warhurst et al. 10 It is also unclear how the cost of antibiotics and antifungals differ between England and Italy. The failure rate of the SeptiFast test was not considered, nor was it clear whether patients were randomly selected to receive SeptiFast or were selected based on their characteristics.
Finally, the Bilkovski et al. study128 is a decision tree using evidence from the RADICAL study116 on the impact of the IRIDICA–PLEX-ID hybrid assay (referred to by the authors as the PCR/ESI-MS) in terms of treatment decision and studies on MALDI-TOF MS123,124 for the impact of the test on clinical outcomes. This study was available only as a poster presentation and, therefore, it is difficult to judge its quality. However, the External Assessment Group notes some inconsistencies. In the RADICAL study, Vincent et al. 116 reported that, among the 529 patients included, the overall mortality rate was 29%, suggesting that about 375 patients survived hospital discharge. However, the economic evaluation conducted by Bilkovski et al. 128 is based on 422 patients and includes only 290 patients surviving to hospital discharge. Patients who tested positive using the IRIDICA–PLEX-ID hybrid were assumed to experience a reduction in hospital and ICU length of stay because of the earlier availability of results and potential therapy changes based on data from two previous studies of MALDI-TOF MS. 123,124 The validity of this assumption is uncertain as MALDI-TOF MS and the IRIDICA–PLEX-ID hybrid assay are used differently and provide different information. Notably, studies of MALDI-TOF MS are conducted in patients with a positive blood culture only. Therefore, the impact of treatment changes when blood culture is negative is unknown and there may be unintended consequences. Importantly, studies on MALDI-TOF MS123,124 used in the economic evaluation evaluated the impact of MALDI-TOF MS in combination with an antimicrobial stewardship programme compared with blood culture prior to the programme. Hence, the impacts of MALDI-TOF MS in these studies are highly likely to be confounded by the simultaneous introduction of an antimicrobial stewardship programme. These studies were also non-randomised in nature and, therefore, subject to potential biases. Bilkovski et al. 128 predicted that the use of the IRIDICA–PLEX-ID hybrid system would lead to a reduction in hospital and ICU length of stay of 1.6 days and 0.7 days, respectively, in all patients and of 4.2 days and 1.8 days, respectively, in patients with positive IRIDICA-PLEX-ID results. It is unclear how the model estimate relates to the impact of the test in practice and whether or not such savings would be observed. As previously stated, evidence from five comparative studies of SeptiFast on the impact of a similar test in terms of mortality or ICU and hospital length of stay is inconclusive. 89,100,103,110,112 Test failure was not considered in the analysis.
Relevance of existing economic evaluations for National Institute for Health and Care Excellence decision-making
Overall, the existing economic evidence has limited relevance to the current UK setting. To date, only two of the three tests (SeptiFast and IRIDICA–PLEX-ID hybrid) have supporting published economic evidence. However, a number of limitations are noted.
There were a number of issues in the evaluations that require further consideration:
-
It is unclear if results are generalisable to the UK. Notably, the current standard of care, the type of antibiotics used and costs may differ between countries.
-
All economic evaluations compared the use of either SeptiFast or the IRIDICA–PLEX-ID hybrid test with blood culture. No comparison is provided against MALDI-TOF MS, which is an increasing part of current practice in some units in the UK.
-
Two studies included economic evaluations. 89,103 However, there are limitations because of the retrospective nature of these studies and potential biases associated with patient selection. Notably, the study by Alvarez et al. 89 is believed to be highly confounded, with large imbalances between groups in terms of initial diagnosis: in the control group (n = 54), 20 patients had had heart surgery and four patients had polytrauma/head injuries whereas, in the SeptiFast group (n = 48), only two patients had had heart surgery but 20 suffered from polytrauma/head injuries.
-
Two studies used modelling approaches to assess the impact of SeptiFast129 or the IRIDICA–PLEX-ID hybrid assay,128 compared with blood culture, on treatment modification and the resulting effect in terms of inadequate therapy or rapid identification. The results produced contradict the evidence of the impact of the tests on clinical outcomes. The evidence so far is inconclusive, despite five comparative studies comparing the impact of SeptiFast with that of blood culture, with or without MALDI-TOF MS, on mortality and hospital/ICU length of stay. 89,100,103,110,112 In the SeptiFast modelling study129 the impact of the test on treatment change and on clinical outcomes was based on relatively old evidence that was collected prior to recent guidelines on the diagnosis and treatment of sepsis. 16,18–20 Bilkovski et al. 128 based their model of the IRIDICA-PLEX-ID hybrid assay on the results of two earlier MALDI-TOF MS studies. 123,124 Both of these studies128,129 suffer from limitations: not only were they non-randomised, and therefore subject to biases, but they evaluated the use of MALDI-TOF MS in combination with an antimicrobial stewardship programme only in patients with positive blood cultures. Furthermore, the potential impact of the test on treatment modification was estimated retrospectively, which may have introduced a further source of bias.
-
The cost of SeptiFast differed among the three studies89,103,129 and does not necessarily reflect the most likely cost estimated by the External Assessment Group. Similarly, the cost of the IRIDICA–PLEX-ID hybrid assay128 does not necessarily reflect the most likely cost estimated by the External Assessment Group.
-
The range of costs included varied between studies. With the exception of Mancini et al. ,103 most savings are attributable to a reduction in length of hospital stay. However, as previously mentioned, there is no robust evidence on the impact of any of the interventions on hospital or ICU length of stay. 89,100,103,110,112
-
Mancini et al. 103 reported a reduction in the costs associated with ‘classical diagnostic assays and instrumental procedures’ following the introduction of SeptiFast and a large reduction in the costs of empirical therapy predominantly associated with antifungals. It is unclear whether or not these reductions are generalisable to the NHS in England.
-
The identified economic evaluations focused on the positive impact of the test, for example a potential reduction in antibiotics and/or length of stay. It is noted that tests may be associated with unintended detrimental effects, such as a greater incidence of superinfection, as reported by Leone et al. ,35 who evaluated the effects of de-escalation in patients with sepsis. Furthermore, any detrimental effects that may occur as a result of the wrong decision being made have been ignored.
-
The results are also likely to be optimistic as the interventions appear to be implemented under optimal conditions in the majority of identified economic evaluations103,129 rather than considering delays that may occur under current practice conditions.
-
The benefits associated with better antibiotic stewardship are not included in the economic evaluations; however, the External Assessment Group acknowledges the difficulty of robustly quantifying such benefits.
-
It is also unclear whether or not patients were preselected in the studies.
-
Finally, the failure rates within interventions were not considered. A recent UK study conducted by Warhurst et al. 10 suggests a test failure rate of 7% for SeptiFast. An alternative study reported failure rates of 22.9%. 99
Independent economic assessment: conceptual model and methods
The conceptual model developed by the External Assessment Group was relatively simplistic because of the lack of appropriate data. A decision tree approach was adopted with a lifetime horizon and discounting undertaken at 3.5% per annum.
The NICE diagnostic reference case141 requests that cost-effectiveness is presented in terms of cost per QALY gained. This has been adhered to although the authors highlight the considerable uncertainty in any estimate because of the lack of robust data on key components of the calculation.
The cost per QALY can be divided into the incremental costs incurred and the incremental QALYs gained.
The incremental costs should consider:
-
the cost of each test/comparator
-
the net effect on hospital length of stay for both ICU and non-ICU patients, noting that rapid tests could be detrimental to the patient as well as beneficial
-
the net effect on the costs of antimicrobial treatment
-
any net cost impact associated with the potential impact on antimicrobial resistance.
The incremental QALYs would ideally consider:
-
the impact on sepsis-related mortality
-
the impact of net effect on hospital length of stay for both ICU and non-ICU patients, noting that rapid tests could be detrimental to the patient as well as beneficial
-
any net QALY impact associated with the potential impact on antimicrobial resistance.
Although the costs of the tests and comparators can be estimated relatively well from current data, there are no conclusive data on any other parameter listed in the bullet points above that were identified in the External Assessment Group’s review. Therefore, a scenario analysis was undertaken in which these values were estimated by clinical experts.
Within the model it was assumed that the rapid identification of a pathogen could result in changes in four key outcomes: 30-day mortality rates; the length of stay in an ICU; the length of stay in the hospital; and the costs associated with antimicrobial treatment. Of these, changes in the mortality rate were assumed to affect QALYs only, with the remaining categories assumed to affect costs only. This is a simplification in that, for example, additional time in an ICU may be associated with slightly lower QALYs, but the impact of such omissions was assumed not to affect the overall conclusions. In all scenarios the potential impact of better antimicrobial stewardship in terms of drug resistance was not evaluated because of both the complexity of such a task and the absence of information on how the tests would reduce antimicrobial use.
It was assumed that negative tests would not impact on any of the four key outcomes. This assumption was supported by the clinical experts to the External Assessment Group. The decision to ignore negative tests was because of the potential fatal consequences if treatment was withdrawn from a patient with sepsis. Acknowledged reasons for a false-negative result include the test being unable to detect the pathogen or the quantity of the pathogen being below the test’s limit of detection. Similarly, tests that would be denoted as failures were assumed to have no impact on the four key outcomes. Both negative tests and failures would, however, be associated with the cost of the test.
A pictorial representation of the conceptual model is provided in Figure 11. The net cost impact and the net QALY impact of rapid identification were used to estimate a cost per QALY gained ratio.
FIGURE 11.
The components within the conceptual model.
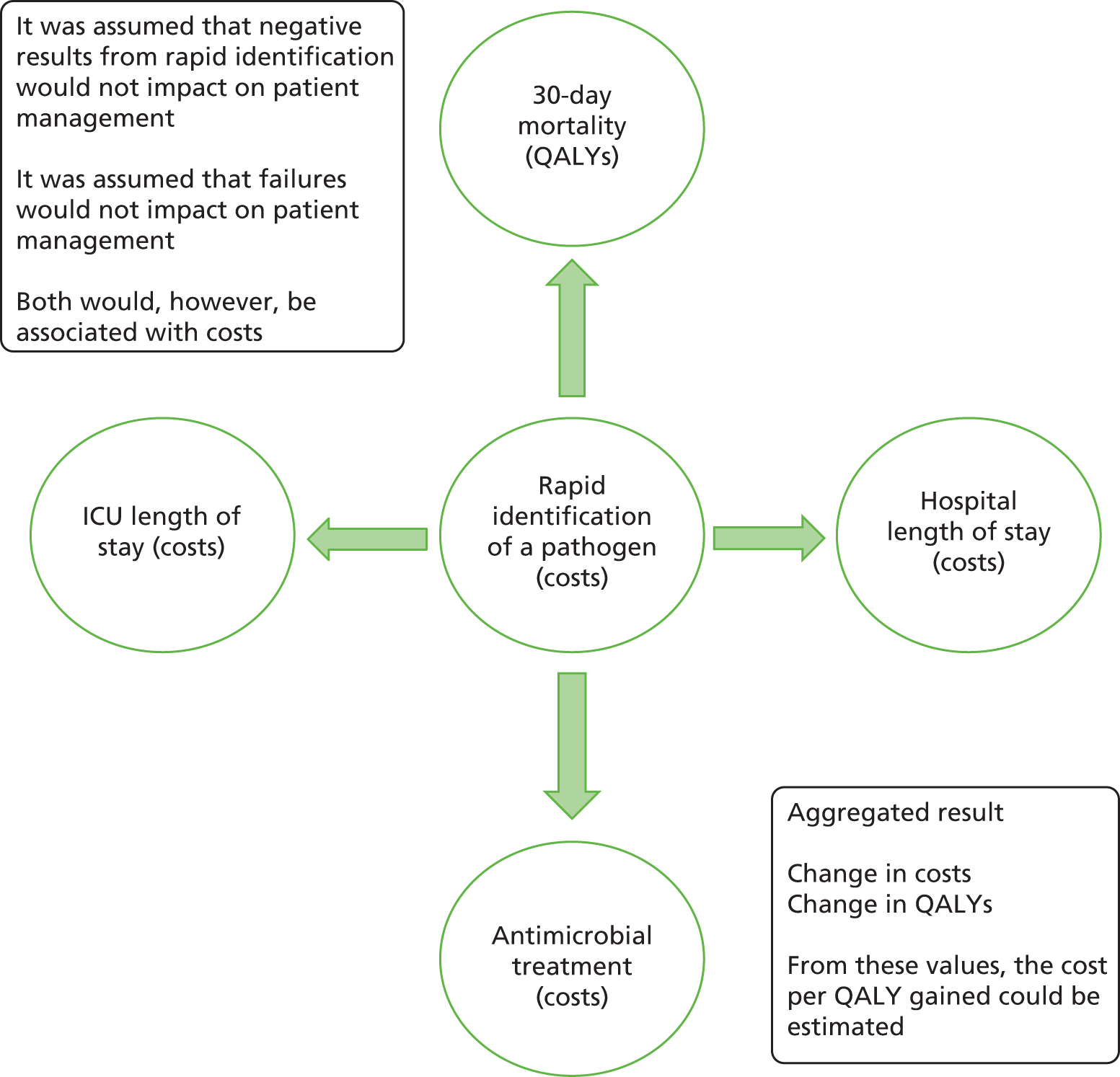
The evaluations presented by the External Assessment Group have been divided into five categories.
-
Base case 1: an analysis based on currently published evidence.
-
Base case 2: an analysis in which parameter values were populated by estimates from clinical experts in order to estimate the cost-effectiveness of each test. This has a benefit in that if, in base case 1, the rapid tests offered little or no benefit compared with the comparators, based on the absence, or lack of statistical significance, of the required data, then clinical beliefs could be incorporated.
-
Threshold analyses were undertaken to guide decision-makers on the likelihood of the interventions having cost per QALY gained values of £20,000 or lower and of £30,000 or lower, as it was assumed that experts in the field would be more confident in providing an indication of whether the value of a parameter was greater than, or less than, a threshold value than in estimating a value in the absence of data (as was requested in base case 2). The variables assessed within the threshold analysis in the threshold base case were the number of mortalities within 30 days that were prevented and the reduction in days in ICU. For simplicity, and to allow thresholds to be presented purely in terms of net 30-day mortality or net cost, it was assumed that no additional QALY gain was associated with a reduction in ICU duration of stay. The results are presented allowing for a mixture of both net mortalities and of net reduced ICU stay. In an alternative analysis, the thresholds of both the reduction in the net number of ICU days and the net reduced costs of antimicrobial treatment are also presented.
-
Analyses comparing the interventions with MALDI-TOF MS based on published literature.
-
Analyses of data taken from studies in which more than one intervention were compared directly.
Given the large divergence in results produced by base cases 1 and 2, the External Assessment Group decided that probabilistic sensitivity analyses would provide spurious accuracy with respect to the decision being undertaken. As such, only deterministic answers have been provided. If robust data are produced in relation to the efficacy of the interventions on key patient outcomes, then probabilistic sensitivity analyses should be conducted.
The lack of evidence for heterogeneous diagnostic accuracy among subgroups resulted in the External Assessment Group providing only an overall measurement of cost-effectiveness rather than by subgroup. Although the cost-effectiveness may differ among subgroups – for example, a neonate would be expected to accrue more QALYs than an adult – these do not affect the fundamental uncertainty of whether or not the interventions would be associated with any key patient outcome.
For all but the threshold analyses, the incremental cost per test has been calculated accounting for the net effect on ICU and hospital length of stay, and changes in the costs of antimicrobial treatment. The rate of positivity for each test must also be known, as it has been assumed that only positive intervention tests would result in a change in management.
As an illustrative example, assuming that the cost of a test was £400, that each positive test was associated with a 0.1-day reduction in ICU length of stay, a 0.3-day reduction in hospital stay and a £50 reduction in antimicrobial treatment, and a 20% rate of positivity, the incremental costs would be estimated to be:
Note that these values are for illustrative purposes and are not necessarily those used in the modelling exercises that are detailed in later sections.
The incremental QALYs have been calculated assuming 11.32 discounted QALYs per 30-day mortality avoided (see The quality-adjusted life-year gains associated with preventing a 30-day mortality). Thus, if an intervention was assumed to reduce 0.01 deaths per test, then the discounted QALYs gained would be 0.1132. To calculate the numbers of deaths avoided, data are required on the assumed underlying mortality rate at 30 days, the estimated reduction in the rate of 30-day mortality associated with each test and the rate of positivity. As an example, if it was assumed that the underlying mortality rate was 13%, the reduction following a positive test was 5% and the rate of positivity was 20%, then the estimated number of deaths prevented would be 13% × 5% × 20% per test, which equals 0.0013.
The principles outlined above in calculating incremental costs and QALYs have been maintained throughout the analyses undertaken in this report. For simplicity, the example provided above did not distinguish between the assumed impacts of the interventions when a subsequent blood culture was either negative or positive although, as detailed in Model parameters assumed for base case 1, separate values for these were provided by the clinical experts.
Independent economic assessment: populating the model
General model parameters
The number of blood samples that need analysing per day
Three broad scenarios were undertaken regarding the number of blood samples that need to be analysed each day: based on the number observed in a recent clinical study10 assuming an increase for community-acquired infection (2.4 blood samples); assuming 17 blood samples; and assuming 68 blood samples. These values were thought to provide a wide range of plausible values that could be undertaken within units of different sizes.
Assuming the numbers of samples that need analysing based on the number observed in a recent clinical study
These data were calculated from data reported in Warhurst et al. ,10 as summarised in Table 16. This source was chosen as it was a recent, large, high-quality study set in England. In this study there was a central hub, in Salford, with a SeptiFast machine that supplied results to four sites. A monthly rate per site was calculated and then these were added to estimate a monthly throughput. This was assumed to be plausibly representative of the use of a centrally located machine relevant to an intervention serving a number of satellite hospitals. Note that the daily value does not equal the average across the Warhurst et al. 10 study because of the different lengths of enrolment in the study by site. It is not reported why the monthly rate in site 4 was considerably lower than in the remaining sites.
| Site number | Number of months in study by Warhurst et al.10 | Number of tests sent from the site for analysis | Estimated tests per month |
|---|---|---|---|
| 1 | 30 | 481 | 16.03 |
| 2 | 30 | 343 | 11.43 |
| 3 | 21 | 170 | 8.10 |
| 4 | 13 | 12 | 0.92 |
| Total | 1006 | 36.48 |
The sites’ requirements sum to 36.48 tests per month, although results were provided for only 922 out of 1006 samples: the SeptiFast had a failure rate of 69 out of 1006 (7%) and a further 15 samples were lost for clinical reasons. The Warhurst et al. study10 included only health-care-acquired sepsis. Based on clinical opinion, it was assumed that 50% of sepsis cases are health care acquired and 50% are community acquired, and thus, to include both types of sepsis, the number of tests required was doubled to 72.96 each month, or 2.40 tests each day. It was assumed that two tests would be analysed on 60% of days and three tests would be analysed on 40% of days. However, on clinical advice, it was further assumed that testing would be carried out on only 5 days a week; therefore, the numbers of tests were divided so that there were three times the expected number of tests on Monday compared with Tuesday to Friday. It was assumed that all tests sent for processing would be tested and that no allowance would be made for the possibility of rejecting requests for tests on blood that was sampled on a Friday evening but which would not be processed until Monday morning. This assumption was supported by our clinical advisors, who deemed that this would be how the system would work in practice, although clearly the gain in speed of identification is reduced for those samples collected on Friday evening and over the weekend. It was assumed that there would be two runs on a Monday, with an initial run analysing those samples collected after staff had finished working on a Friday evening and a second run analysing those samples collected on the Monday itself.
Assuming that the number of samples that need to be analysed each day is 17
In this example, the value was set to approximately seven times that estimated from the Warhurst et al. study. 10 This value is also the maximum number of blood samples that one IRIDICA system can process in 1 day, assuming three runs per day, although it is noted that SeptiFast could process 21 tests over three runs and, thus, the selected number may favour IRIDICA. In this scenario it is assumed that practice has been changed to accommodate the interventions being used 7 days a week, and that each intervention could be run three times daily. The costs of these changes in standard practice have not been incorporated, although a statement of the magnitude of this compared with any potential savings has been made.
Assuming that the number of samples that need to be analysed each day is 68
In this example, the value was set to approximately 28 times that estimated from the Warhurst et al. study. 10 This scenario assumes that four SeptiFast machines and four IRIDICA machines would be required to process the requested blood samples. In this scenario it is assumed that practice has been changed to accommodate the interventions being used 7 days a week and that each intervention could be run three times daily. The costs of these changes in standard practice have not been incorporated, although a statement of the magnitude of this compared with any potential savings has been made.
The estimated costs of the interventions and matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
For all tests there is a fixed cost and a variable cost that is determined by the numbers of tests undertaken. Following advice obtained from our clinical advisors, we assumed that the purchase cost of a machine could be equally divided over an assumed 7-year period of use. Annual costs associated with the maintenance of the machines were incorporated.
For each intervention, the cost will be dependent on whether or not the equipment required for the specific test (SeptiFast, SepsiTest, IRIDICA and MALDI-TOF MS) is already available within the microbiology laboratory. SeptiFast and IRIDICA have their own bespoke PCR machines, whereas SepsiTest can be run on generic PCR machines. This led to two scenarios being evaluated for each intervention. These, in order of cost, are (1) purchasing of the required machine for the specific test or a generic PCR machine; and (2) no additional machinery needing to be purchased. In both cases consumable costs would be incurred.
The estimation of the cost of an average test is far from robust, relying on uncertain assumptions in the number of tests per day and machinery costs, and therefore adds additional uncertainty.
Note that, in accordance with the NICE reference case for diagnostic evaluations, value-added tax (VAT) is not included in economic evaluations. 141 Based on advice from our clinical experts, it was assumed that no additional staff costs or room costs would be incurred if any of the interventions were purchased, although this is less likely to be the case should it be assumed that 17 or 68 samples need to be analysed per day. For simplicity, neither transport costs associated with sample testing nor additional staff training was included.
No discounts associated with bulk purchasing of equipment has been assumed for any intervention. It is possible that these exist in reality; if the costs of the test are lower than those assumed in the analyses, then the incremental cost-effectiveness ratio (ICER) would be lower than those presented with all other variables held constant.
The costs associated with SeptiFast
Data provided by Roche Diagnostics on the list prices of the required Roche instruments to run a SeptiFast test are shown in Table 17. The sum of these individual items is £26,397, excluding VAT.
| Instrument | Category number | List price (£) |
|---|---|---|
| LightCycler 2.0 Instrument with Software 4.1 | 03 531 414 001 | 18,000.00 |
| LC Carousel Centrifuge 2.0 | 03 709 582 001 | 3060.00 |
| MagNA Lyser Instrument (230 V) | 03 358 976 001 | 4500.00 |
| Multi-Colour Compensation Set | 4484355001 | 536.76 |
| SeptiFast Cooling Block | 4555864001 | 300.45 |
Additionally, Roche Diagnostics reports an annual service charge of £3000, excluding VAT, that covers any repairs required (including a replacement system if necessary) and an annual calibration check.
The key consumables required to run a test, along with their list price, are shown in Table 18.
| Product name | Category number | List price (£) | Number per item |
|---|---|---|---|
| LightCycler M Grade Capillaries | 3612066001 | 883.93 | 768 |
| LightCycler SeptiFast Kit, CE | 4469046001 | 1422.62 | 54 |
| LightCycler SeptiFast mecA Kit, CE | 4488814001 | 624.44 | 10 |
| SeptiFast Lys Kit, CE | 4404432001 | 745.95 | 200 |
| SeptiFast Prep Kit, CE | 4404459001 | 320.73 | 10 |
Roche Diagnostics estimated the cost per test based on the number of tests per run, assuming one daily run and a 5-day working week. Running more tests per run reduces the average reagent cost per sample because of the requirement of having two control samples for each run. The costs estimated by Roche Diagnostics, excluding VAT, for reagent costs are replicated in Table 19. From Table 19 it can be calculated that the marginal cost of one additional sample in a run is £122.00.
| Tests per run | Reagent cost per reportable result (£) |
|---|---|
| 1 | 333.99 |
| 2 | 228.00 |
| 3 | 192.68 |
| 4 | 175.01 |
| 5 | 164.41 |
| 6 | 157.35 |
| 7 | 152.30 |
| Scenario | Average cost per test assuming samples per day calculated from Warhurst et al.10 (£) | Average cost per test assuming 17 samples per day (£) | Average cost per test assuming 68 samples per day (£) |
|---|---|---|---|
| Assuming that SeptiFast machinery needs to be purchaseda | 205.54 | 160.52 | 154.28 |
| Assuming no additional machinery is needed | 201.23 | 159.91 | 153.67 |
The estimated average costs per SeptiFast test are provided in Table 20 for the three scenarios related to the number of blood samples to be analysed (2.39, 17 and 68) and for the two purchasing scenarios (where the SeptiFast platform does or does not need to be purchased). In both purchasing scenarios, consumable costs will be incurred.
| Price (€) | |
|---|---|
| Apparatus | |
| Thermomixer (24 × 2.0-ml tubes, adjust at 37 °C, 56 °C and 70 °C) | ≈2000 |
| Cooling racks for 1.5-ml tubes (adjust at 4 °C and –20 °C) | ≈50 |
| Vortexer (e.g. VWR, Germany) | ≈170 |
| Bench-top microcentrifuge (≥ 13.000 rpm, ≥ 12.000 × g) | ≈1100 |
| Clear work places | |
|
≈3000 |
|
≈8000 |
| One set of precision pipettes: up to 10 µl, up to 20 µl, up to 200 µl and up to 1000 µl (e.g. Eppendorf, Germany) | ≈400 |
| Thermocycler | |
|
≈3000 |
|
≈4000 |
|
≈15,000 |
| Sequencing analysis as service (GATC, Germany) | ≈5 per sequencing |
| ABI 310 (Applied Biosystems, CA, USA) refurbished | ≈30,000 |
| Plastic ware | |
| Pipette tips with aerosol filter (e.g. Biosphere®, Sarstedt AG & Co., Germany) | |
| PCR tubes | |
| Chemicals | |
| DNA decontamination: DNA Exitus®, Applichem, Germany | |
| Agarose gel (2%) for standard PCR | |
| A container for plastic waste (pipette tips, vials, tubes) and another for liquid waste, autoclavable | |
| Others | |
| Sterile disposables | |
| Gloves (e.g. Kimberly-Clark, Germany) | |
| Sleeves (e.g. Cardinal Health, Ireland) | |
| Bouffant covers (e.g. VWR, Germany) | |
| Overshoes (e.g. hygi, Germany) | |
The costs associated with SepsiTest
As previously detailed (see Chapter 1, SepsiTest), an updated version of SepsiTest was recently released. This version does not require duplicate samples and thus the cost of the reaction per sample is halved. Owing to the lack of diagnostic accuracy data on the updated version of the test, the analysis conducted in this report is based on the previous version of SepsiTest.
The company (Molzym Molecular Diagnostics) that manufactures SepsiTest reports that the list price for 48 reactions for use in SepsiTest is £3000, including VAT, which equates to £2500, excluding VAT, and a cost per reaction of £52.08, excluding VAT. Each test is assumed to require two slots of a 96-well PCR machine, and that two controls (one negative and one positive) are required for each run. Thus, a maximum of 47 SepsiTest analyses can be performed in one run. The costs of the controls were not provided by Molzym Molecular Diagnostics.; these were estimated to be £104.17 for the pair, which is the cost of two reactions. This is likely to underestimate the cost as additional costs associated with spiking blood to produce the positive control have not been included, although this omission is unlikely to affect the conclusions of the report.
The cost of additional sequencing following a positive SepsiTest was assumed to be €11 for bacteria and €5.50 for fungi based on information supplied by Molzym Molecular Diagnostics. These costs include VAT at 19% and are equivalent to €9.24 and €4.62, respectively, once VAT is deducted. Assuming an exchange rate of €1 to £0.72,137 these values were calculated as £6.66 and £3.33, respectively.
The costs of the machinery required to undertake PCR testing and Sanger sequencing were estimated from data provided by Molzym Molecular Diagnostics. These costs are reproduced in Table 21. The data were assessed by a clinical expert advising the External Assessment Group and were deemed appropriate. Therefore, it was considered not unreasonable to assume a cost of €60,000 (a high estimate based on the values in Table 21) to purchase the equipment needed to undertake PCR and to cover any maintenance required. This is equivalent to £43,200, assuming an exchange rate of €1 to £0.72. 137
In order that the profile of bacteria and fungi was representative of that observed in England, an assumption was made that the proportion of positive results for bacteria and fungi identified by SeptiFast would be applicable to SepsiTest. This proportion, calculated from table 14 of Warhurst et al. ,10 was 18 fungi from 167 positive SeptiFast tests (10.8%), which was used to estimate a weighted cost of additional sequencing of £6.30. Based on the data synthesis conducted (see Figure 6), it was assumed that SepsiTest had a sensitivity of 0.48 and a specificity of 0.86, which, when combined with an assumed blood culture-positive rate of 8.7% calculated from Warhurst et al. ,10 equated to a positivity rate of 17% for SepsiTest. The company manufacturing SepsiTest confirmed that the cost per SepsiTest assay is not dependent on throughput.
The estimated average costs per SepsiTest assay are provided in Table 22. These costs assume the sensitivity and specificity of SepsiTest reported in the data synthesis. In alternative analyses, when specific trial data are used, these costs will change slightly as the number of sequencing tests needed will differ owing to assumed alternative diagnostic accuracy data.
| Scenario | Average cost per test assuming samples per day calculated from Warhurst et al.10 (£) | Average cost per test assuming 17 samples per day (£) | Average cost per test assuming 68 samples per day (£) |
|---|---|---|---|
| Assuming a generic PCR machine and sequencer needs to be purchaseda | 149.53 | 112.29 | 108.55 |
| Assuming no additional machinery is needed | 142.48 | 111.36 | 108.30 |
The costs associated with an IRIDICA test
Abbott Diagnostics reports that the cost of the IRIDICA analyser is £268,000, excluding VAT, and that the cost of annual maintenance is £30,150, excluding VAT. The cost of an IRIDICA test is reported to be £174, excluding VAT. Following clarification, the manufacturer provided costs in relation to the number of tests per day, as the control test needs to be changed every 24 hours. These costs are summarised in Table 23. According to Abbott Diagnostics, it is possible to analyse 23 samples per day, assuming that one control is used at the start of the day along with five samples and that subsequent runs do not need a control and therefore six samples can be analysed simultaneously. The External Assessment Group comments that, although it is technically possible to conduct four runs, each taking slightly < 6 hours, in a 24-hour period, this is unlikely to be possible in practice. Therefore, the External Assessment Group assumed that 17 tests per day represents the limit of an IRIDICA machine. The estimated average costsper IRIDICA test are provided in Table 24.
| Number of samples to be analysed | Total cost per sample (£) |
|---|---|
| 1 | 362.04 |
| 2 | 273.29 |
| 3 | 242.53 |
| 4 | 226.28 |
| 5 | 215.82 |
| 6 | 211.78 |
| 7 | 208.38 |
| 8 | 205.40 |
| 9 | 202.69 |
| 10 | 200.18 |
| 11 | 197.80 |
| 12 | 197.28 |
| 13 | 196.56 |
| 14 | 195.71 |
| 15 | 194.73 |
| 16 | 193.65 |
| 17 | 192.49 |
| 18 | 192.44 |
| 19 | 192.21 |
| 20 | 191.93 |
| 21 | 191.31 |
| 22 | 190.68 |
| 23 | 189.96 |
| Scenario | Average cost per test assuming samples per day calculated from Warhurst et al.10 (£) | Average cost per test assuming 17 samples per day (£) | Average cost per test assuming 68 samples per day (£) |
|---|---|---|---|
| Assuming the IRIDICA analyser needs to be purchaseda | 314.61 | 203.52 | 203.52 |
| Assuming no additional machinery is needed | 270.89 | 197.35 | 197.35 |
The costs associated with a matrix-absorbed laser desorption time-of-flight mass spectrometry system
The costs associated with MALDI-TOF MS were provided by Bruker UK Limited (Coventry, UK) (Erika Tranfield, Bruker UK, 2015, personal communication). It was assumed that these costs would be generalisable to other MALDI-TOF MS systems such as bioMérieux’s VITEK® MS system (Marcy-l'Étoile, France). The cost of the MALDI-TOF MS machine was assumed to be £125,000. A further technology, the Sepsityper kit, is available at a cost of approximately £3 per test. The Sepsityper kit is a sample preparation method that involves the lysis of blood cells, followed by centrifugation and washing steps to produce a pellet of bacteria or fungi, which is further processed by standard MALDI-TOF MS methods. Given that Sepsityper was in widespread use in the RAPIDO trial,31 a large UK study comparing MALDI-TOF MS and standard practice with standard practice alone, this will be used for the purposes of this economic evaluation. On clinical advice, no further costs for MALDI-TOF MS were assumed in addition to that for the Sepsityper kit (assumed to be £3), as these are relatively inexpensive. The costs for preventative maintenance are dependent on the number of maintenances performed per year: for two maintenances per year the cost is £13,985, whereas the cost for 3–5 maintenances per year is £17,000. It was assumed that the higher number of preventative maintenances applies.
It is noted that MALDI-TOF MS can be used for many investigations other than just the use of pathogen identification in those with suspected sepsis. Based on clinical advice it was assumed that only 50% of the purchase and maintenance cost would be attributable to sepsis investigations. Our clinical advisors did not advocate attributing the costs of any of the interventions to non-sepsis-related disease areas.
Data on the number of blood culture bottles that are flagged as positive were taken from Warhurst et al. 10 and assumed generalisable to England. Data from table 6 of Warhurst et al. 10 show that, of 922 episodes, 80 were blood culture positive (8.7%), and it was assumed that these samples would undergo further analysis via MALDI-TOF MS. As such, it was estimated that the MALDI-TOF MS machine would process 8.7% of the daily throughput of 2.40 tests (see Assuming the numbers of samples that need analysing based on the number observed in a recent clinical study), which is 0.21 tests per day. When the assumed number of blood samples that need to be analysed was increased to 17 and 68 per day, the number of samples analysed using MALDI-TOF MS was increased to 1.48 and 5.92 per day, respectively. The estimated costs of MALDI-TOF MS are provided in Table 25.
| Scenario | Average cost per test assuming samples per day calculated from Warhurst et al.10 (£) | Average cost per test assuming 17 samples per day (£) | Average cost per test assuming 68 samples per day (£) |
|---|---|---|---|
| Assuming a MALDI-TOF MS machine needs to be purchaseda | 232.39 | 35.35 | 11.09 |
| Assuming no additional machinery is needed | 114.88 | 18.78 | 6.94 |
Care should be taken not to directly compare the costs per test between the interventions and MALDI-TOF MS as all samples would be processed by the interventions, while only those where the blood culture had tested positive would be analysed by MALDI-TOF MS.
The costs associated with blood culture
Given that blood culture would be used alongside all interventions and alongside MALDI-TOF MS, the costs would have no bearing on the incremental costs associated with the intervention tests and MALDI-TOF MS. For this reason no resources were spent in trying to ascertain the costs per blood sample and the cost was assumed to be £0 in all analyses.
The assumed failure rates of the interventions
Both SeptiFast and IRIDICA use internal controls that could be subject to failure, as could the controls on a PCR machine used by SepsiTest. For SeptiFast, a 6.9% failure rate was reported in Warhurst et al. ,10 although a greater value of 22.9% was reported in Paolucci et al. 99 Data on the failure rate of IRIDICA have been reported in Metzgar et al,117 and indicate a rate of (confidential information has been removed). No data on the failure rate of SepsiTest were identified.
In the base case it has been assumed that SeptiFast has a failure rate of 6.9%. In sensitivity analyses a failure rate of 11.7% was assumed for SeptiFast based on a naive pooling of failure results from Warhurst et al. 10 (69 failures) and from Paolucci et al. 99 (100 failures) divided by the numbers of samples analysed in all of the SeptiFast versus blood culture trials combined (11,659 samples), assuming no failures in any SeptiFast study. This results in an estimated failure rate for SeptiFast of 1.4%. For the base case for IRIDICA, a failure rate of (confidential information has been removed) was assumed based on a naive pooling of data from all of the studies assuming no failures in any study but Metzgar et al. 117
Although Warhurst et al. ,10 Paolucci et al. 99 and Metzgar et al. 117 explicitly stated that any failures were excluded from analyses of diagnostic accuracy, it is not clear if failures occurred in the remaining identified studies but were not reported as such and were treated as a negative result. If failures had been excluded from the analysis of diagnostic accuracy but not reported, then this would be beneficial to the relevant intervention.
No data were identified on the failure rate of SepsiTest. Rather than assign an arbitrary value to this parameter, the value was set to zero with the acknowledgement that this was likely to be favourable to SepsiTest.
The quality-adjusted life-year gains associated with preventing 30-day mortality
It was assumed that each 30-day mortality avoided is associated with a gain of 11.32 discounted QALYs. This value was calculated based on (a) the estimated number of discounted life-years for a typical patient and (b) the estimated quality of life after a sepsis episode to account for the possible reduced quality of life in sepsis survivors. It should be noted that the discounted QALY gains for neonates and children would be greater than those for adults because of their longer life expectancy, although the exact increase is uncertain.
Although there is evidence that the survival after a sepsis episode may be lower than in the general population,142–144 for simplicity we assumed that survival of patients with sepsis was comparable with that of the general population. National life tables for England and Wales for the period 2011–13145 were used to estimate the life expectancy of a typical patient, assuming an age of 58 years and a sex split of 60%/40% (male/female) as reported in Warhurst et al. 10
Patients were assumed to have a utility value of 0.68 based on the European Quality of Life-5 Dimensions score reported by Cuthbertson et al. 142 at 5 years after a severe sepsis episode, which is similar to the value reported by Drabinski et al. 146 If the utility predicted for the general population for an age and sex profile147 was lower than 0.68, this value was used instead. A discount rate of 3.5% per annum was used, as recommended in the NICE reference case. 141
The assumed cost of 1 day’s treatment in an intensive care unit
This value was calculated from NHS reference costs. 148 Service code CCU03 [medical adult patients (unspecified specialty)] was assumed to be representative of treatment for sepsis patients. This service code is subdivided by the number of organs supported, ranging from 0 to ≥ 6, and an average of the reported average unit costs weighted by activity levels was calculated. This resulted in a cost of ICU care of £1057 per day. This is slightly more than the weighted average for service code CCU002 [surgical adult patients (unspecified specialty)] selected, which was £987 per day.
The assumed cost of 1 day’s treatment in a standard hospital ward
This value was calculated from NHS reference costs,148 assuming that the average excess bed-day cost per non-elective impatient of £275 was appropriate.
The assumed cost of typical empirical antimicrobial treatment for sepsis
Based on the advice of our clinical advisors, it was assumed that 7 days’ treatment with either 18 g of piperacillin/tazobactam per day or 3 g of meropenem per day was an appropriate empirical treatment for typical sepsis patients. Using British National Formulary costs,149 these prices were estimated to be £51.60 per day (assuming 4.5 g every 6 hours) for piperacillin/tazobactam and £48.00 (assuming 1 g every 8 hours) for meropenem. Given the uncertain proportion of the drugs used in England, it was assumed that a cost per day of £50, equating to a cost for a course of treatment of £350, was not unreasonable. However, an expert clinician on the Diagnostic Appraisal Committee commented that these costs may be high for adults admitted to regular hospital wards or for children. If this was the case, then the value used for a course of treatment would be favourable to the interventions.
The assumed 30-day mortality rate for those with suspected sepsis
It was assumed that the 28-day mortality rate reported in a recent health technology assessment,111 set in England, could be generalised to a 30-day mortality rate. This value was 13% (95% CI 11% to 16%),10 with the rate of hospital mortality being 21% (95% CI 17% to 23%). This value has some support from data provided by Kaukonen et al. ,3 which analysed hospital mortality rates in patients with severe sepsis in Australia and New Zealand and showed a decrease across time, with values of approximately 10% for SIRS-positive sepsis and 20% for SIRS-negative sepsis.
An alternative value of 29%, as reported in Mouncey et al. 9 (albeit for 90-day mortality), in patients with early septic shock was tested in sensitivity analyses. This value was supported by data from Levy et al. ,6 who reported hospital mortality rates of 29% when there was high compliance with a resuscitation bundle, although the patients included in this study were those with severe sepsis and septic shock and would be likely to have a worse prognosis.
Model parameters assumed for base case 1
In base case 1, only data from the published literature related to patient outcomes were included in an economic evaluation to estimate the cost-effectiveness of each test.
Based on the literature identified by the External Assessment Group, no data were found that provided a conclusive and non-confounded indication that any of the interventions provided benefits in terms of 30-day mortality (see Chapter 2, Mortality), length of stay in ICU or length of stay in hospital (see Chapter 2, Duration of intensive care unit and/or hospital stay). One study of a propensity score-matched design was identified103 and indicated a significant reduction in the costs of empirical therapy, but this study had a number of limitations. First, the study population was of haematological patients who were prescribed empirical antifungal drugs, which is not typical for a suspected sepsis patient. Second, the cost savings predominantly came from the reduction in relatively expensive empiric antifungals; however, according to our clinical experts the most widely used antifungal treatment in England is fluconazole, which is now relatively cheap, with a cost of £1.83 per day for a dose of 400 mg. 149
Data were found showing that the time to therapy modification was much shorter following the introduction of SeptiFast (18.8 hours compared with 38.3 hours),112 but no data were provided on the change in costs of modified therapy. Changes in costs were also not provided for the studies identified in Table 9. Therefore, it was assumed that the costs of antimicrobials were unchanged in the base case.
Thus, in base case 1, it was assumed that the results did not differ depending on whether an intervention was used or was not used. An analysis was undertaken to estimate the reduction in antimicrobial costs needed in order for an intervention to be cost neutral.
Model parameters assumed for base case 2
In base case 2, parameter values were populated using estimates from clinical experts in order to estimate the cost-effectiveness of each test if the benefits anticipated were realised. A document (reproduced in Appendix 7) was sent to the clinicians on the Diagnostic Assessment Committee and to the clinical experts who are authors of this report with a request to estimate key parameters – supporting information identified by the External Assessment Group was also supplied. Seven clinical experts responded, with a wide variation in the answers provided. Although the clinicians were asked for ranges in their answers, six of the seven clinicians provided point estimates only. Typically the clinicians reported that the task was difficult to complete, and the majority assumed the same values for SeptiFast, IRIDICA and SepsiTest. The values for MALDI-TOF MS were typically less favourable than for the three interventions as information on the pathogen would be provided at a later time point than for the interventions. The average values from the clinical experts shown in Table 26 relate to the situation that an intervention is positive and the subsequent blood culture is positive; those in Table 27 relate to the situation that an intervention is positive and the subsequent blood culture is negative. Note that monetary savings in antimicrobial costs were transformed into a percentage reduction, assuming a typical course of treatment cost of £350.
| Parameter | SeptiFast | SepsiTest | IRIDICA | MALDI-TOF MS |
|---|---|---|---|---|
| Average net effect on ICU length of stay (days) | –0.607 | –0.671 | –0.736 | –0.175 |
| Average net effect on hospital length of stay (days) | –1.050 | –1.214 | –1.329 | –0.758 |
| Average net effect on the cost of antimicrobials | –17.78% | –21.63% | –25.92% | –14.26% |
| Net effect on 30-day mortality | –3.16% | –3.87% | –4.59% | –3.00% |
| Parameter | SeptiFast | SepsiTest | IRIDICA |
|---|---|---|---|
| Average net effect on ICU length of stay (days) | –0.571 | –0.629 | –0.700 |
| Average net effect on hospital length of stay (days) | –1.307 | –1.471 | –1.586 |
| Average net effect on the cost of antimicrobials | –28.98% | –31.84% | –37.12% |
| Net effect on 30-day mortality | –3.93% | –4.64% | –5.36% |
The clinicians predicted comparable gains when the blood culture was negative with when the blood culture was positive. This is believed to be because the clinical experts trusted the intervention result rather than the blood culture result in this scenario. This contrasts with the majority of diagnostic accuracy studies, in which blood culture is assumed to be the gold standard, and casts uncertainty over meta-analyses in which this assumption is made.
Consideration was given to attempting to construct a distribution to represent the values provided by the clinicians. However, we decided against this as it was not clear that it would provide significantly better data than an aggregate value. In addition, it would necessitate analysis at the individual clinician level as well as further assumption regarding the distribution type and the range surrounding the mid-point answers provided.
Data from individual clinicians were used in sensitivity analyses. These values are shown in Tables 28–31 for the three interventions and MALDI-TOF MS when the blood cultures were subsequently positive, and in Tables 32–34 for the three tests when the subsequent blood cultures were negative.
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –0.2 | –0.5 | –5% | –3% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | –0.1 | –0.2 | –£40 (–11%) | –0.1% |
| 4 | –2.5 (range –4 to –1) | –1.5 (range –3 to –0) | Not known (assumed to be £0) | –1.0% (range –2% to 0%) |
| 5 | –1 | –3 | –£175 (–50%) | –2% |
| 6 | –0.001 | 0 | –18% | 0% |
| 7 | –0.45 | –1.15 | –25% | –15% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobial drugs | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –0.2 | –0.5 | –5% | –3% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | –0.1 | –0.2 | –£40 (–11%) | –0.1% |
| 4 | –2.5 (range –4 to –1) | –1.5 (range –3 to –0) | Not known (assumed to be £0) | –1.0% (range –2% to –0%) |
| 5 | –1 | –3 | –£175 (–50%) | –2% |
| 6 | 0 | 0 | 0% | 0% |
| 7 | –0.9 | –2.3 | –70% | –20% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –0.2 | –0.5 | –5% | –3% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | –0.1 | –0.2 | –£40 (–11%) | –0.1% |
| 4 | –2.5 (range –4 to –1) | –1.5 (range –3 to –0) | Not known (assumed to be £0) | –1.0 (range –2 to –0) |
| 5 | –1 | –3 | –£175 (–50%) | –2% |
| 6 | –0.001 | 0 | –20% | 0% |
| 7 | –1.35 | –3.1 | –80% | –25% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –0.1 | –0.2 | –2% | –1% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | No answers provided | |||
| 4 | 0 | 0 | 0 | 0 |
| 5 | –0.5 | –1.5 | –£100 (–29%) | –1% |
| 6 | 0 | 0 | –10% | 0 |
| 7 | –0.45 | –1.85 | –30% | –15% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –1 | –1 | –15% | –8% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | 0 | –1.5 | –£80 (–23%) | 0% |
| 4 | –2.5 (range –4 to –1) | –3.5 (range –5 to –2) | Not known (assumed to be £0) | –3.5% (range –5% to –2%) |
| 5 | 0 | –1 | –£700 (–100%a) | –0% |
| 6 | –0.05 | 0.0 | –25% | 0% |
| 7 | –0.45 | –1.15 | –25% | –15% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –1 | –1 | –15% | –8% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | 0 | –1.5 | –£80 (–23%) | 0% |
| 4 | –2.5 (range –4 to –1) | –3.5 (range –5 to –2) | Not known (assumed to be £0) | –3.5% (range –5 to –2) |
| 5 | 0 | –1 | –£700 (–100%a) | –0% |
| 6 | 0 | 0 | 0% | 0% |
| 7 | –0.9 | –2.3 | –70% | –20% |
| Clinician | Average net effect on ICU length of stay (days) | Average net effect on hospital length of stay (days) | Average net effect on the cost of antimicrobials | Net effect on 30-day mortality |
|---|---|---|---|---|
| 1 | –1 | –1 | –15% | –8% |
| 2 | 0 | –1 | –15% | –1% |
| 3 | 0 | –1.5 | –£80 (–23%) | 0% |
| 4 | –2.5 (range –4 to –1) | –3.5 (range –5 to –2) | Not known (assumed to be £0) | –3.5 (range –5 to –2) |
| 5 | 0 | –1 | –£700 (–100%a) | –0% |
| 6 | 0.05 | 0 | –27% | 0% |
| 7 | –1.35 | –3.1 | –80% | –25% |
Independent economic model: results
As previously stated, the results will be presented in three broad categories: (1) base case 1, in which only data from the published literature considered appropriate are included in an economic evaluation to estimate the cost-effectiveness of each test; (2) base case 2, in which parameters were populated from estimates by clinical experts in order to estimate the cost-effectiveness of each test; and (3) a series of threshold analyses. For clarity, threshold analyses indicate the value of a parameter at which the decision is likely to change. In the results presented in this report, these values indicate the level above which the cost per QALY reduces to an assumed value (either £20,000 or £30,000 per QALY). In addition, supplementary analyses using studies deemed to provide additional information on head-to-head comparisons between interventions, or of an intervention with MALDI-TOF MS, have been undertaken. Given the results of the data synthesis, which did not show a difference in diagnostic accuracy by subgroup, only one set of analyses is presented. It is acknowledged that this is likely to underestimate the gains in mortality prevented associated with neonates, but it was deemed that this would not affect the conclusion, which would remain uncertain.
Results from base case 1
The estimated costs and estimated QALYs when the machinery required for the intervention needs to be purchased are shown in Table 35 and when no additional machinery needs to be purchased are provided in Table 36. For brevity, these results are presented only for the cost per test estimated using the number of blood samples per day from the Warhurst et al. 10 study; the conclusions remain the same at lower cost of tests owing to the assumed lack of QALY gain. The conclusion is that all the tests are dominated, in that they are associated with an additional cost but are assumed to provide no additional QALYs.
| Test | Incremental cost per test compared with blood culture (£) | Incremental QALYs gained per test compared with blood culture | Cost per QALY gained compared with blood culture |
|---|---|---|---|
| SeptiFast | 205.54 | 0.00 | Dominated |
| SepsiTest | 149.53 | 0.00 | Dominated |
| IRIDICA | 314.61 | 0.00 | Dominated |
| Test | Incremental cost per test compared with blood culture (£) | Incremental QALYs gained per test compared with blood culture | Cost per QALY gained compared with blood culture |
|---|---|---|---|
| SeptiFast | 201.23 | 0.00 | Dominated |
| SepsiTest | 142.48 | 0.00 | Dominated |
| IRIDICA | 270.89 | 0.00 | Dominated |
An analysis was undertaken to estimate the reduction in antimicrobial costs needed per test in order that the introduction of the intervention would be cost neutral. These results are shown in Table 37 when machinery needs to be purchased and in Table 38 when no additional machinery is required.
| Test | Required reduction in antimicrobial treatment costs assuming samples per day, calculated from Warhurst et al.10 (%) | Required reduction in antimicrobial treatment costs, assuming 17 samples per day (%) | Required reduction in antimicrobial treatment costs, assuming 68 samples per day (%) |
|---|---|---|---|
| SeptiFast | 59 | 46 | 44 |
| SepsiTest | 43 | 32 | 31 |
| IRIDICA | 90 | 58 | 58 |
| Test | Required reduction in antimicrobial treatment costs assuming samples per day, calculated from Warhurst et al.10 (%) | Required reduction in antimicrobial treatment costs, assuming 17 samples per day (%) | Required reduction in antimicrobial treatment costs, assuming 68 samples per day (%) |
|---|---|---|---|
| SeptiFast | 57 | 46 | 44 |
| SepsiTest | 41 | 32 | 31 |
| IRIDICA | 77 | 56 | 56 |
If it is assumed that a reduction in antibiotic costs will be achieved only in the event of a positive test, then, assuming the positivity rates calculated for each test based on their estimated sensitivity and specificity values, the costs of the tests cannot be recouped from reduced antimicrobial treatment costs alone.
Results from base case 2
Results are presented separately using the average value from all clinician responses and by individual clinician. The results are also differentiated based on the assumed mortality rate associated with suspected sepsis. As clinicians provided estimates for the potential benefit of MALDI-TOF MS in addition to the interventions, this technique has also been included in the tables. These results are divided into those assuming a mortality rate of 13% (from Warhurst et al. 10) and those assuming a mortality rate of 29% (from Mouncey et al. 9). For SeptiFast, the results from Warhurst et al. 10 have been given primacy as this is an English study10 and was assessed as high quality; however, sensitivity analyses have been undertaken using the evidence from the data synthesis undertaken in this report. For SepsiTest and IRIDICA, the results presented use the evidence from the data synthesis.
The machinery-purchasing requirements vary according to the machinery already available to process the blood samples. In order to facilitate an estimation of the relative cost-effectiveness of each intervention and MALDI-TOF MS across these combinations, the results have been summarised in terms of net monetary benefit (NMB)150 compared with blood culture. Comparison of NMB is simple, and the strategy with the highest NMB is estimated to be the most cost-effective. Figures 12–23 present findings that are combinations of the assumed mortality rate (either 13% or 29%), the maximum acceptable incremental cost-effectiveness ratio (MAICER) (either £20,000 or £30,000 per QALY gained) and the number of blood samples that need to be analysed per day (2.4, 17 or 68). It is likely that the mortality rate is inversely correlated with the number of tests per day, in that high throughput may be associated with a greater proportion of relatively minor investigations. This is noted as a limitation, but has not been formally investigated. Following this summary, the results for each intervention in each scenario are presented reporting incremental QALYs, incremental costs and ICERs compared with blood culture. All results are presented assuming a timescale of testing of 1 year, although discounted QALYs accrued in future years are included.
FIGURE 12.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 13% and 2.4 blood samples to be analysed per day.

FIGURE 13.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 13% and 2.4 blood samples to be analysed per day.

FIGURE 14.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 29% and 2.4 blood samples to be analysed per day.

FIGURE 15.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 29% and 2.4 blood samples to be analysed per day.
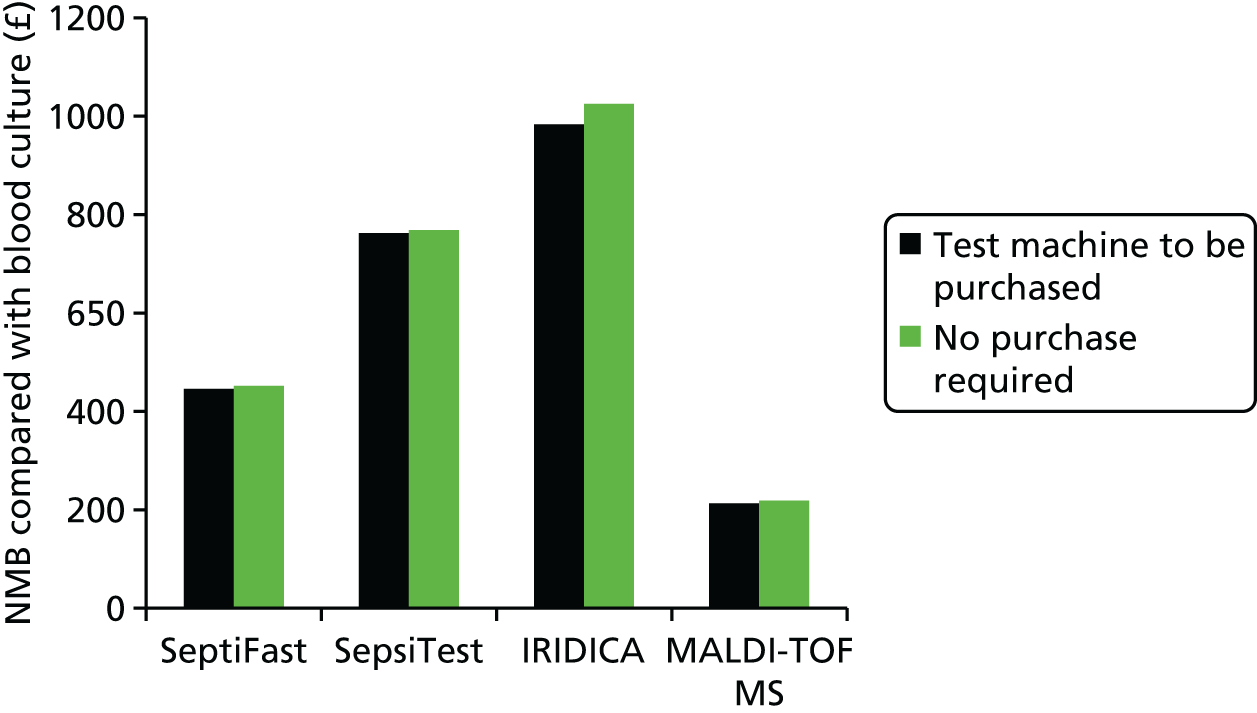
FIGURE 16.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 13% and 17 blood samples to be analysed per day.

FIGURE 17.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 13% and 17 blood samples to be analysed per day.

FIGURE 18.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 29% and 17 blood samples to be analysed per day.

FIGURE 19.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 29% and 17 blood samples to be analysed per day.
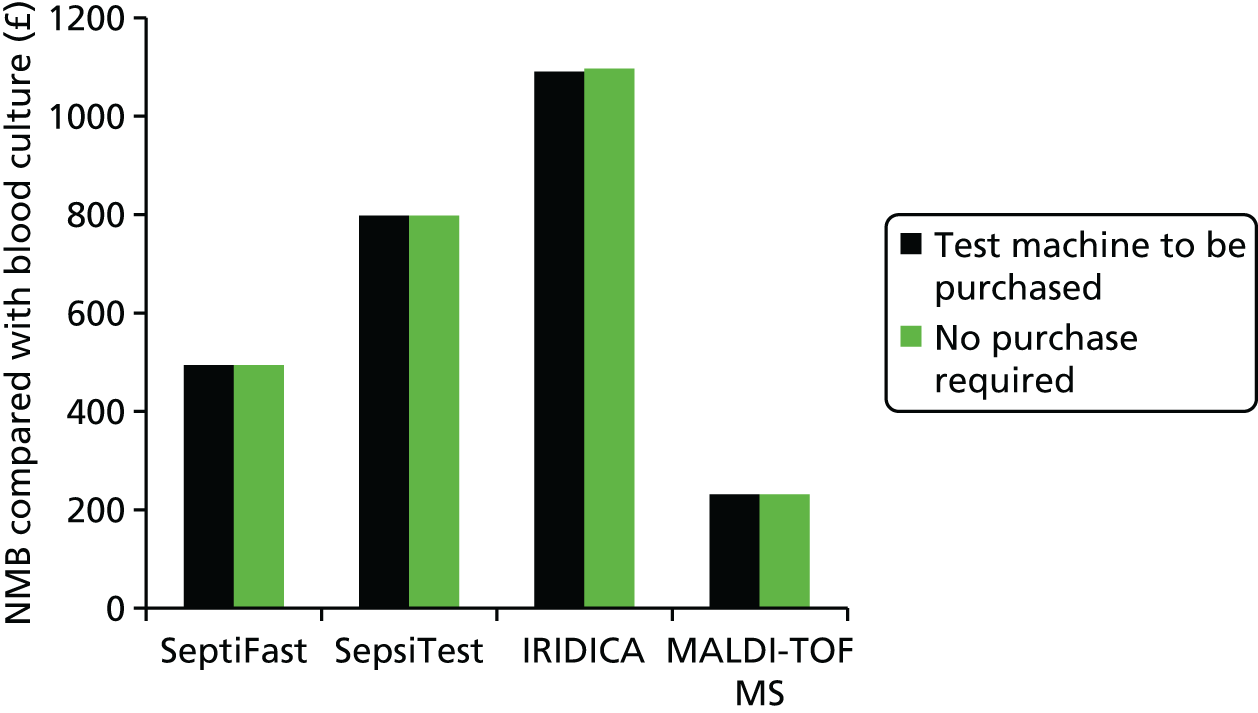
FIGURE 20.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 13% and 68 blood samples to be analysed per day.
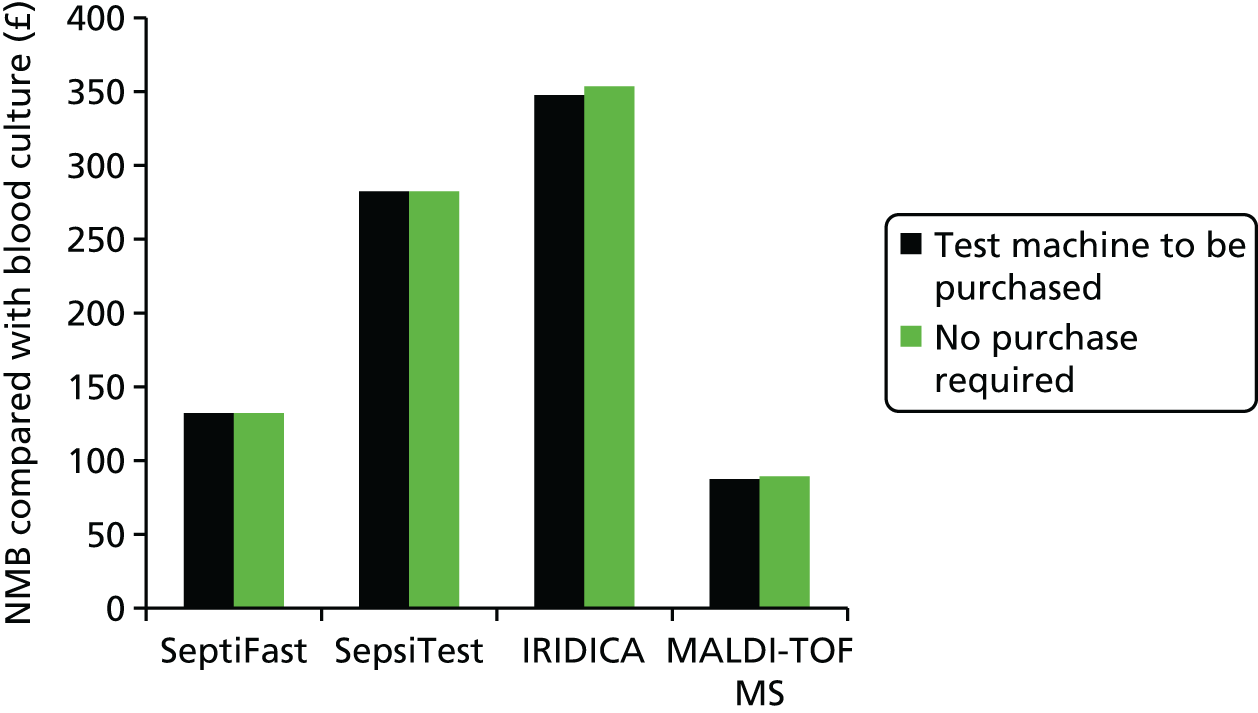
FIGURE 21.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 13% and 68 blood samples to be analysed per day.
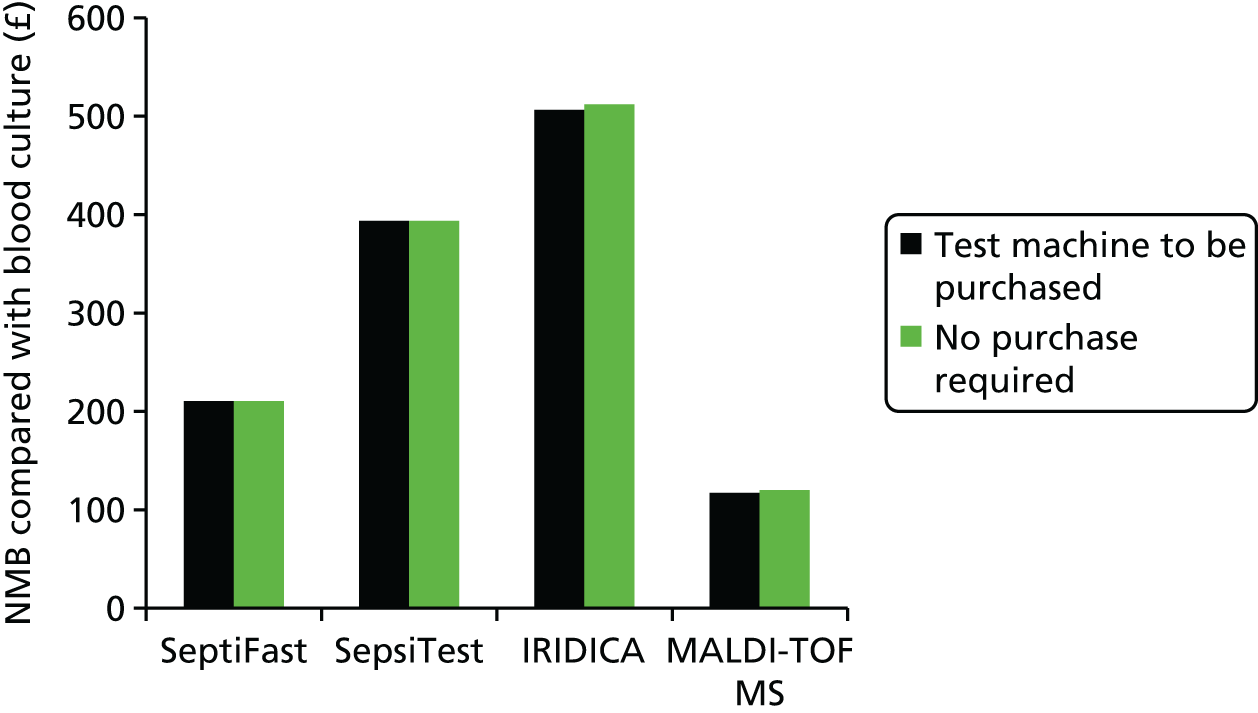
FIGURE 22.
Net monetary benefit assuming a MAICER of £20,000, a mortality rate of 29% and 68 blood samples to be analysed per day.

FIGURE 23.
Net monetary benefit assuming a MAICER of £30,000, a mortality rate of 29% and 68 blood samples to be analysed per day.

It is seen that, regardless of the scenario, all three interventions and MALDI-TOF MS produced a positive net benefit compared with blood culture. This conclusion was not affected regardless of whether or not the acquisition cost of the machine was incorporated in the calculation.
In the scenario in which the mortality rate is assumed to be 13%, the MAICER £20,000 per QALY and the number of blood samples per day 2.4, SepsiTest or IRIDICA has the highest estimated NMB depending on the assumption regarding machine purchase. For all other scenarios, IRIDICA is estimated to have the highest NMB. However, these results are highly uncertain and it remains plausible that each (or none) of the interventions is most cost-effective.
However, as will be detailed later, these results are highly dependent on the individual clinician questioned and thus there is large uncertainty in these estimates.
Results from base case 2 using the average clinician values and assuming a 30-day mortality rate of 13%
The estimated costs and estimated QALYs for each intervention and MALDI-TOF MS compared with blood culture when it is assumed that the machinery required for the intervention needs to be purchased are shown in Table 39. The results when no additional machinery needs to be purchased are provided in Table 40.
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | 67,878 | 6.88 | 9862 |
| SepsiTest | –15,963 | 9.72 | Dominating |
| IRIDICA | 73,501 | 13.96 | 5264 |
| MALDI-TOF MS | –548 | 0.23 | Dominating |
| Assuming 17 blood samples per day | |||
| SeptiFast | 201,782 | 48.81 | 4134 |
| SepsiTest | –343,990 | 68.96 | Dominating |
| IRIDICA | –168,633 | 99.01 | Dominating |
| MALDI-TOF MS | –13,094 | 1.65 | Dominating |
| Assuming 68 blood samples per day | |||
| SeptiFast | 652,257 | 195.22 | 3341 |
| SepsiTest | –1,470,568 | 275.82 | Dominating |
| IRIDICA | –674,533 | 396.06 | Dominating |
| MALDI-TOF MS | –56,914 | 6.59 | Dominating |
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | 64,107 | 6.88 | 9314 |
| SepsiTest | –22,134 | 9.72 | Dominating |
| IRIDICA | 35,215 | 13.96 | 2522 |
| MALDI-TOF MS | –1322 | 0.23 | Dominating |
| Assuming 17 blood samples per day | |||
| SeptiFast | 198,011 | 48.81 | 4057 |
| SepsiTest | –350,161 | 68.96 | Dominating |
| IRIDICA | –206,919 | 99.01 | Dominating |
| MALDI-TOF MS | –13,869 | 1.65 | Dominating |
| Assuming 68 blood samples per day | |||
| SeptiFast | 637,173 | 195.22 | 3264 |
| SepsiTest | –1,476,739 | 275.82 | Dominating |
| IRIDICA | –827,676 | 396.06 | Dominating |
| MALDI-TOF MS | –57,688 | 6.59 | Dominating |
It is seen that the cost per QALY values are relatively low for all interventions. It is assumed that any costs associated with allowing machines to be run on a 7 days per week, 24 hours per day basis could be subsumed into the intervention costs while still producing ICERs that are below £20,000 per QALY gained. To illustrate this, if it was assumed that there were additional costs of £100,000 to operate SeptiFast continuously, then the ICER, assuming 17 samples to be analysed per day and machine purchase, would be calculated as:
Incremental cost (see Table 39): £201,782 + £100,000 = £301,782.
Incremental QALYs (see Table 39): 48.81.
ICER: £301,782/£48.81 = £6183.
Results from base case 2 using the average values from the clinician survey and assuming a 30-day mortality rate of 29%
The estimated costs and estimated QALYs for each intervention test compared with blood culture, when it is assumed that the machinery required for the intervention needs to be purchased, are shown in Table 41. The results when no additional machinery needs to be purchased are provided in Table 42.
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | 67,878 | 15.35 | 4421 |
| SepsiTest | –15,963 | 21.69 | Dominating |
| IRIDICA | 73,501 | 31.15 | 2360 |
| MALDI-TOF MS | –548 | 0.52 | Dominating |
| Assuming 17 blood samples per day | |||
| SeptiFast | 201,782 | 108.88 | 1853 |
| SepsiTest | –343,990 | 153.82 | Dominating |
| IRIDICA | –168,427 | 220.88 | Dominating |
| MALDI-TOF MS | –13,094 | 3.67 | Dominating |
| Assuming 68 blood samples per day | |||
| SeptiFast | 652,257 | 435.50 | 1498 |
| SepsiTest | –1,470,568 | 615.30 | Dominating |
| IRIDICA | –674,533 | 883.51 | Dominating |
| MALDI-TOF MS | –56,914 | 14.69 | Dominating |
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | 64,107 | 15.35 | 4175 |
| SepsiTest | –22,134 | 21.69 | Dominating |
| IRIDICA | 35,215 | 31.15 | 1131 |
| MALDI-TOF MS | –15,240 | 0.52 | Dominating |
| Assuming 17 blood samples per day | |||
| SeptiFast | 198,011 | 108.88 | 1819 |
| SepsiTest | –350,161 | 153.82 | Dominating |
| IRIDICA | –206,919 | 220.88 | Dominating |
| MALDI-TOF MS | –159,840 | 3.67 | Dominating |
| Assuming 68 blood samples per day | |||
| SeptiFast | 637,173 | 435.50 | 1463 |
| SepsiTest | –1,476,739 | 615.30 | Dominating |
| IRIDICA | –827,676 | 883.51 | Dominating |
| MALDI-TOF MS | –664,860 | 14.69 | Dominating |
Results from base case 2 using the average values from the clinical survey, using the results from the data synthesis for SeptiFast
In the base case, for SeptiFast, it is assumed that data from Warhurst et al. 10 are the most appropriate, as these data were taken from a large study (853 patients) in an English population that was believed to be the study with the highest quality (see Table 6). The only study with a similar level of quality was that by Tafelski et al. 112 but, as that was a small study (88 patients) set in Germany, the results are believed to be not as generalisable as those from Warhurst et al. 10
In an alternative scenario, the impact of using the estimated results from the synthesis of diagnostic accuracy data (see Figure 3) was explored. The mid-point values of 65% sensitivity and 86% specificity were used, with the prevalence of sepsis identified by blood culture by episode assumed to be that reported in table 6 of Warhurst et al. ,10 of 80 out of 922 (8.7%). These data result in an estimated positivity rate of 18.4% for SeptiFast with 30.6% of subsequent blood cultures also being positive and 67% being negative. In this analysis, a failure rate of 1.4% was assumed for SeptiFast, based on a naive pooling of failure results from Warhurst et al. 10 (69 failed samples) and from Paolucci et al. 99 (100 failed samples) divided by the number of samples analysed in the SeptiFast versus blood culture trials (11,659 samples).
The estimated cost per QALY values for SeptiFast, obtained using the results of the synthesis of diagnostic accuracy, are shown in Table 43 (assuming a 13% mortality rate) and Table 44 (assuming a 29% mortality rate).
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| Assuming SeptiFast machine purchase required | 39,631 | 8.64 | 4588 |
| Assuming no purchase required | 35,860 | 8.64 | 4152 |
| Assuming 17 blood samples per day | |||
| Assuming SeptiFast machine purchase required | 1477 | 61.25 | 24 |
| Assuming no purchase required | –2294 | 61.25 | Dominating |
| Assuming 68 blood samples per day | |||
| Assuming SeptiFast machine purchase required | –148,963 | 245.00 | Dominating |
| Assuming no purchase required | –164,047 | 245.00 | Dominating |
| Scenario and test | Incremental cost per annum compared with blood culture (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with blood culturea | Cost per QALY gained compared with blood culture (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| Assuming SeptiFast machine purchase required | 39,631 | 19.27 | 2057 |
| Assuming no purchase required | 35,860 | 19.27 | 1861 |
| Assuming 17 blood samples per day | |||
| Assuming SeptiFast machine purchase required | 1477 | 136.63 | 11 |
| Assuming no purchase required | –2294 | 136.63 | Dominating |
| Assuming 68 blood samples per day | |||
| Assuming SeptiFast machine purchase required | –148,963 | 546.53 | Dominating |
| Assuming no purchase required | –164,047 | 546.53 | Dominating |
It is seen that, using the pooled results for SeptiFast, rather than the results of Warhurst et al. 10 study, would be more favourable to SeptiFast, with estimated ICERs below £10,000 in all scenarios analysed regardless of the diagnostic accuracy data used.
Results from base case 2 using the individual clinician values
Cost-effectiveness results produced by individual clinicians are provided in Table 45 for SeptiFast, Table 46 for SepsiTest, Table 47 for IRIDICA and Table 48 for MALDI-TOF MS. For concision, only the cost per QALY values are presented, with incremental costs and incremental QALY values omitted. Only the results assuming 2.4 tests a day have been documented, as the purpose was to show the concordance between individual clinicians, which is largely unaffected by the numbers of blood samples assumed to be analysed per day.
| Clinician | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 13% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 13% (£) | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 29% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 29% (£) |
|---|---|---|---|---|
| 1 | 6036 | 5720 | 2706 | 2564 |
| 2 | 73,884 | 71,870 | 33,120 | 32,217 |
| 3 | 2,137,101 | 2,075,380 | 958,011 | 930,343 |
| 4 | Dominating | Dominating | Dominating | Dominating |
| 5 | 42,886 | 39,800 | 19,225 | 17,841 |
| 6 | Dominated | Dominated | Dominated | Dominated |
| 7 | 2985 | 2851 | 1338 | 1278 |
| Clinician | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 13% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 13% (£) | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 29% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 29% (£) |
|---|---|---|---|---|
| 1 | Dominating | Dominating | Dominating | Dominating |
| 2 | 37,700 | 34,874 | 16,900 | 15,633 |
| 3 | 1,294,903 | 1,179,881 | 580,474 | 528,912 |
| 4 | Dominating | Dominating | Dominating | Dominating |
| 5 | Dominating | Dominating | Dominating | Dominating |
| 6 | Dominated | Dominated | Dominated | Dominated |
| 7 | Dominating | Dominating | Dominating | Dominating |
| Clinician | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 13% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 13% (£) | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 29% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 29% (£) |
|---|---|---|---|---|
| 1 | 6737 | 4541 | 3020 | 2036 |
| 2 | 78,491 | 64,489 | 35,186 | 28,909 |
| 3 | 2,288,469 | 1,857,349 | 1,025,865 | 832,605 |
| 4 | Dominating | Dominating | Dominating | Dominating |
| 5 | 50,386 | 28,830 | 22,587 | 12,924 |
| 6 | Dominated | Dominated | Dominated | Dominated |
| 7 | Dominating | Dominating | Dominating | Dominating |
| Clinician | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 13% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 13% (£) | Only the machinery required for the intervention needs to be purchased. Mortality rate assumed to be 29% (£) | No additional machinery needs to be purchased. Mortality rate assumed to be 29% (£) |
|---|---|---|---|---|
| 1 | 10,265 | 255 | 4601 | 114 |
| 2 | Dominating | Dominating | Dominating | Dominating |
| 3 | Did not answer this question | |||
| 4 | Dominated | Dominated | Dominated | Dominated |
| 5 | Dominating | Dominating | Dominating | Dominating |
| 6 | Dominated | Dominated | Dominated | Dominated |
| 7 | Dominating | Dominating | Dominating | Dominating |
For all tests the answers were highly discordant between clinicians, with answers ranging from dominated (higher cost and the same or fewer QALYs) to dominating (lower cost the same or more QALYs), indicating high levels of uncertainty in the assumed effectiveness of the interventions and MALDI-TOF MS.
Results from the threshold analyses
Threshold analyses are presented for each intervention in comparison with blood culture and with MALDI-TOF MS. It is assumed that, where a comparison with MALDI-TOF MS is made, the unit already had a MALDI-TOF MS machine in place. The analyses have been undertaken assuming that an intervention needs to be purchased – it is assumed that if a laboratory already had one of the interventions in place then this would be routinely used.
Thresholds are reported for net reduction in mortality and net reduction in ICU length of stay combined, and for net reduction in antimicrobial costs and net reduction in ICU length of stay combined. All threshold results are presented per 100 positive tests and for 100 tests, irrespective of the test result. For the net reduction in antimicrobial costs and net reduction in ICU length of stay combined, there is no separate curve based on the MAICER as it is assumed that both factors affect cost only and the decision regarding cost-effectiveness reduces to one of cost minimisation.
Owing to the number of figures presented, the threshold analyses are contained in Appendix 8. Note that, in all analyses, the diagnostic accuracy for SeptiFast has been taken from Warhurst et al. ,10 as this was assumed to be a more representative study of English practice and was graded as a higher-quality study than the remaining studies (see Table 6).
In summary, relatively small mortality gains would be required for the interventions to achieve a cost per QALY gained of £20,000. The threshold levels compared with blood culture, assuming that 2.4 samples per day need to be analysed, are shown in Table 49. The values assuming that the comparator is MALDI-TOF MS are shown in Table 50.
| Test | Per 100 tests | Per 100 positive tests | ||||
|---|---|---|---|---|---|---|
| Reduction in 30-day mortality (lives) | Reduction in ICU length of stay (days) | Reduction in antimicrobial treatment costs (£) | Reduction in 30-day mortality (lives) | Reduction in ICU length of stay (days) | Reduction in antimicrobial treatment costs (£) | |
| SeptiFast | 0.09 | 19.45 | 205.54 | 0.62 | 133.82 | 1414.50 |
| SepsiTest | 0.07 | 14.15 | 149.53 | 0.39 | 83.46 | 882.15 |
| IRIDICA | 0.14 | 29.76 | 314.61 | 0.65 | 140.23 | 1482.28 |
| Test | Per 100 tests | Per 100 positive tests | ||||
|---|---|---|---|---|---|---|
| Reduction in 30-day mortality (lives) | Reduction in ICU length of stay (days) | Reduction in antimicrobial treatment costs (£) | Reduction in 30-day mortality (lives) | Reduction in ICU length of stay (days) | Reduction in antimicrobial treatment costs (£) | |
| SeptiFast | 0.09 | 18.50 | 195.57 | 0.59 | 127.33 | 1345.90 |
| SepsiTest | 0.06 | 13.20 | 139.56 | 0.36 | 77.89 | 823.34 |
| IRIDICA | 0.13 | 28.82 | 304.65 | 0.63 | 135.79 | 1435.32 |
These values assume no change in either of the two remaining parameters. All other scenarios require lower threshold values to attain a cost per QALY gained of £20,000.
Results from the studies comparing SeptiFast with matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry and comparing SepsiTest with matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
Two studies had a comparator of MALDI-TOF MS in addition to blood culture. These were the studies by Tafelski et al. ,112 in which the index test was SeptiFast, and by Loonen et al. ,114 in which the index text was SepsiTest. The cost-effectiveness of SeptiFast and SepsiTest was estimated using the data from the trials with MALDI-TOF MS as a comparator and assuming the average benefits estimated by the clinicians. These results were then compared with the results produced by the indirect comparisons of SeptiFast and MALDI-TOF MS, and of SepsiTest and MALDI-TOF MS, generated through the evidence provided by the clinical experts relative to blood culture to see if they were concordant.
The benefits associated with the interventions were amended to account for any benefit associated with a positive MALDI-TOF MS. Thus, for example, if (as an illustrative example) SeptiFast was associated with an estimated reduction in ICU stay of 0.607 per positive test and MALDI-TOF MS with a reduction of 0.175 day per positive test, then, assuming that MALDI-TOF MS had a sensitivity of 0.798 compared with blood culture, which was the reliable identification value at species level from Morgenthaler and Kostrzewa,30 the benefit of a positive SeptiFast test when the accompanying blood culture was positive in terms of ICU length of stay reduction would be calculated as (0.607 – 0.175) × 0.798, which equals 0.468 days.
The costs of MALDI-TOF MS were subtracted from the costs of the tests based on the estimated number of tests performed. The results are provided in Table 51 (assuming a mortality rate of 13%) and Table 52 (assuming a mortality rate of 29%). Given the relatively low ICERs, for concision, evaluations in which machinery was not required to be purchased to use SeptiFast or SepsiTest are not reported. As no failures were mentioned by Tafelski et al. 112 or Loonen et al. ,114 it was assumed that there were none for the analyses presented.
| Scenario and test | Incremental cost per annum compared with MALDI-TOF MS (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with MALDI-TOF MSa | Cost per QALY gained compared with MALDI-TOF MS (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | –38,345 | 12.40 | Dominating |
| SepsiTest | 84,087 | 2.41 | 34,848 |
| Assuming 17 blood samples per day | |||
| SeptiFast | –499,681 | 87.95 | Dominating |
| SepsiTest | 417,246 | 17.11 | 24,385 |
| Assuming 68 blood samples per day | |||
| SeptiFast | –2,128,094 | 351.79 | Dominating |
| SepsiTest | 1,599,875 | 68.44 | 23,375 |
| Scenario and test | Incremental cost per annum compared with MALDI-TOF MS (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 year compared with MALDI-TOF MSa | Cost per QALY gained compared with MALDI-TOF MS (£)b |
|---|---|---|---|
| Assuming 2.4 blood samples per day | |||
| SeptiFast | –38,345 | 37.67 | Dominating |
| SepsiTest | 84,087 | 5.38 | 15,621 |
| Assuming 17 blood samples per day | |||
| SeptiFast | –499,681 | 196.19 | Dominating |
| SepsiTest | 417,246 | 38.17 | 10,931 |
| Assuming 68 blood samples per day | |||
| SeptiFast | –2,128,094 | 784.76 | Dominating |
| SepsiTest | 1,599,875 | 152.68 | 10,479 |
The results indicate that SeptiFast appears to be more cost-effective than MALDI-TOF MS when aggregated clinicians values are used. The results for SepsiTest are less conclusive, with estimated values of > £20,000 per QALY gained when a mortality rate of 13% is assumed, and with values below this when a mortality rate of 29% is assumed.
These results differ from those of the main analyses, in which SepsiTest appeared considerably more cost-effective than both MALDI-TOF MS and SeptiFast (see Figures 12–23).
Results from studies comparing SeptiFast and SepsiTest simultaneously with blood culture
Two studies evaluated both SeptiFast and SepsiTest against blood culture: those by Schreiber and Nierhaus119 and Leitner et al. 118 The cost-effectiveness of these studies was estimated to see if it was concordant with the results produced by the indirect comparisons of SeptiFast and SepsiTest generated through the evidence provided by the clinical experts relative to blood culture. Neither study mentioned the SeptiFast failure rate and, thus, it was assumed to be zero. It was further assumed that machinery would need to be purchased for both tests. The ICERs for SeptiFast compared with SepsiTest are provided in Table 53 (mortality rate assumed to be 13%) and in Table 54 (mortality rate assumed to be 29%). It is seen that in all of the scenarios the ICER for SeptiFast compared with SepsiTest is > £30,000 per QALY gained. This conclusion is concordant with those of the main analyses as shown in Figures 12–23.
| Study | Test | Incremental cost per annum (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 yeara | Cost per QALY gained (£)b | Cost per QALY of SeptiFast compared with SepsiTest (£)c |
|---|---|---|---|---|---|
| Assuming 2.4 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 87,022 | 5.81 | 14,975 | 90,855 |
| SepsiTest | 49,147 | 5.39 | 9111 | ||
| Leitner et al.118 | SeptiFast | 67,686 | 6.95 | 9739 | Dominated |
| SepsiTest | –7910 | 9.27 | Dominating | ||
| Assuming 17 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 337,533 | 41.21 | 8191 | 74,363 |
| SepsiTest | 117,708 | 38.25 | 3077 | ||
| Leitner et al.118 | SeptiFast | 200,421 | 49.28 | 4067 | Dominated |
| SepsiTest | –286,890 | 65.73 | Dominating | ||
| Assuming 68 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 1,195,262 | 164.83 | 7252 | 69,267 |
| SepsiTest | 376,224 | 153.00 | 2459 | ||
| Leitner et al.118 | SeptiFast | 646,815 | 197.13 | 3281 | Dominated |
| SepsiTest | –1,242,168 | 262.91 | Dominating | ||
| Study | Test | Incremental cost per annum (£) | Incremental QALYs gained through number of avoided 30-day mortalities within 1 yeara | Cost per QALY gained (£)b | Cost per QALY of SeptiFast compared with SepsiTest (£)c |
|---|---|---|---|---|---|
| Assuming 2.4 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 87,022 | 12.96 | 6713 | 40,728 |
| SepsiTest | 49,147 | 12.03 | 4084 | ||
| Leitner et al.118 | SeptiFast | 67,686 | 15.50 | 4366 | Dominated |
| SepsiTest | –7910 | 20.68 | Dominating | ||
| Assuming 17 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 337,533 | 91.92 | 3672 | 33,335 |
| SepsiTest | 117,708 | 85.33 | 1379 | ||
| Leitner et al.118 | SeptiFast | 200,421 | 109.94 | 1823 | Dominated |
| SepsiTest | –286,890 | 146.62 | Dominating | ||
| Assuming 68 blood samples per day | |||||
| Schreiber and Nierhaus119 | SeptiFast | 1,195,262 | 367.69 | 3251 | 31,051 |
| SepsiTest | 376,224 | 341.31 | 1102 | ||
| Leitner et al.118 | SeptiFast | 646,815 | 439.75 | 1471 | Dominated |
| SepsiTest | –1,242,168 | 586.50 | Dominating | ||
Interpretation of the independent economic model results
Forming conclusions based on the economic modelling is very difficult because of the lack of high-quality evidence regarding the impact of the interventions on hard patient outcomes, such as sepsis-related mortality, length of stay in the ICU and on changes in the costs of antimicrobial therapy.
In base case 1, which includes only data from the published literature on patient-related outcomes, all interventions were estimated to be dominated as there was no evidence that any knowledge gained translated into a benefit for the patient and all interventions were associated with additional cost.
In base case 2, parameter values were populated from estimates provided by clinical experts. The results contrasted with those in base case 1. Using the average values provided by the clinical experts, the estimated cost per QALY for all interventions was below a threshold of £20,000 per QALY in all scenarios. However, when the results were broken down by individual clinician estimates, there was a wide variation in the cost per QALY. In the scenario of a 13% mortality rate and 2.4 samples analysed per day, costs per QALY estimates were > £20,000 for four out of seven clinicians for SeptiFast, for two out of seven clinicians for SepsiTest, for four out of seven clinicians for IRIDICA and for two out of six clinicians for MALDI-TOF MS. The clinical experts commented on the difficulty of the task of populating the model parameters and thus all results should be treated with caution.
The External Assessment Group also caution against forming conclusions from a direct comparison of the interventions, although, for completeness, these results have been presented along with a description of the key assumptions and data driving these comparative results. IRIDICA is estimated to have much better sensitivity than either SeptiFast or SepsiTest, and this results in an increase in QALYs and reduced costs resulting from reductions in ICU and hospital lengths of stay and changes in antimicrobial costs. The specificity of the interventions is similar, although the specificity of IRIDICA is marginally lower than that of SeptiFast or SepsiTest; this will result in QALY gains and cost savings as the clinical experts believed false positives to be associated with an imperfect reference standard rather than an inaccurate test. Additionally, the data provided by the expert clinicians indicated that a positive IRIDICA test would be more beneficial than the other tests. The QALY gains and cost savings associated with IRIDICA were sufficient to offset its higher costs and to confer on IRIDICA the highest NMB. The second highest NMB was typically for SepsiTest, not because of its inherent accuracy, as it has lower sensitivity than SeptiFast, but because the assumed cost per test is lower and the benefit per test estimated by the clinicians is higher. MALDI-TOF MS was presumed to be better than blood culture, but as the number of samples tested using MALDI-TOF MS was much smaller, with only those that were blood culture positive being tested, the NMB was lower than for any of the interventions.
Additional analyses undertaken using the results from multitest studies of SeptiFast, SepsiTest and blood culture and the data provided by clinicians, were concordant with base case 2, in that SeptiFast has an estimated cost per QALY gained value of > £20,000 compared with SepsiTest. However, the indirect results produced when using studies directly comparing MALDI-TOF MS were contradictory, with SeptiFast estimated to dominate SepsiTest.
Given the discordant results between base case 1 and base case 2, the External Assessment Group cannot confidently suggest any cost per QALY gained value for the interventions. It is clear that the majority of clinicians questioned believe that the interventions are likely to be cost-effective, yet there are no conclusive data to show that the tests provide a benefit to patients.
Threshold analyses were carried out, as these may be helpful to decision-makers in formulating guidance. It was seen that relatively small mortality gains would be required for the interventions to achieve a cost per QALY gained of £20,000 compared with standard practice.
The External Assessment Group comments that studies comparing the use of an intervention with standard practice, with the results of the tests fed into a treatment management plan, are urgently needed to produce more definitive estimates of the cost per QALY gained. The RAPIDO study31 is undertaking this for MALDI-TOF MS in addition to blood culture and clinical judgement. Although this study was recently completed, data analysis had not been fully conducted at the time of writing. When the results of the clinical effectiveness and cost-effectiveness of the addition of MALDI-TOF MS are known, the best choice for standard practice in any future trial should be more certain.
Chapter 4 Discussion
Statement of principal findings
Clinical effectiveness
A comprehensive systematic review and meta-analysis (where applicable) were undertaken to evaluate the clinical effectiveness of three interventions (SeptiFast, SepsiTest and IRIDICA) in conjunction with clinical assessment for rapidly identifying bloodstream bacteria and fungi in people with suspected sepsis.
For the review of diagnostic test accuracy, 62 studies44–46,48,49–65,67–88,90–99,101,102,104–119 of varying methodological quality were included. Most of these studies were considered to be at risk of bias and there are concerns about their applicability. Pooled effects for sensitivity and specificity across 54 studies44–46,59–65,67–88,90–102,104–111,118,119 (comprising 10,010 patients) comparing SeptiFast with blood culture found that the specificity of SeptiFast (0.86, 95% CrI 0.84 to 0.89) is higher than its sensitivity (0.65, 95% CrI 0.60 to 0.71). Similarly, one study112 that compared SeptiFast with blood culture plus MALDI-TOF MS found that SeptiFast had higher specificity (0.74, 95% CI 0.64 to 0.85) than sensitivity (0.58, 95% CI 0.30 to 0.86). However, because of the deficiencies in the quality of the included studies, these data may not be reliable and should be treated with caution. Moreover, the prediction intervals of the pooled estimates indicate a substantial amount of heterogeneity between studies, particularly for sensitivity. Reasons for the observed heterogeneity in sensitivity and specificity between studies were explored using metaregression for several potentially relevant characteristics including age category (adults, and children and neonates), antibiotic use at the time of blood sampling, community- or health-acquired infection, inclusion/exclusion of contaminants and patients with febrile neutropenia. There was no evidence to suggest that the pooled sensitivity and specificity were affected by these subgroups.
Pooled effects for sensitivity and specificity across four studies48,113,118,119 (comprising 460 patients) comparing SepsiTest with blood culture suggest that SepsiTest has a higher specificity (0.86, 95% CrI 0.78 to 0.92) than sensitivity (0.48, 95% CrI 0.21 to 0.74). Although the pooled estimate indicates low sensitivity, the associated CrI is large. Comparison with blood culture plus MALDI-TOF MS in a single study114 also showed higher specificity than sensitivity (0.96, 95% CrI 0.92 to 1.00; and 0.11, 95% CrI 0.00 to 0.23, respectively). Despite substantial amounts of heterogeneity between studies, analyses for potential causes of this heterogeneity could not be explored because of the small number of studies included. Owing to the deficiencies in the quality of the included studies, the sensitivity and specificity data for SepsiTest may not be reliable and should be treated with caution.
The pooled effects for sensitivity and specificity across four studies49,115–117 [comprising 860 patients across two studies115,117 (data not reported for other studies)] comparing IRIDICA with blood culture suggest that IRIDICA has a higher specificity (0.84, 95% CrI 0.71 to 0.92) than sensitivity (0.81, 95% CrI 0.69 to 0.90), although the difference between sensitivity and specificity is small. Despite substantial amounts of heterogeneity between studies, analyses for potential causes of this heterogeneity could not be explored owing to the small number of studies included. Owing to the deficiencies in the quality of the included studies, the sensitivity and specificity data for IRIDICA may not be reliable and should be treated with caution.
Although 41 studies44–46,59,60,64,66,68,69,72,74,76–78,80,81,84,85,88–90,92–94,96,97,99–101,103–107,110–112,114,116,117,119 across the three interventions reported data on one or more intermediate (e.g. time to pathogen identification, time to treatment, test failure rates, duration of stay in hospital or critical care units, and change in antimicrobial treatment plan) and/or clinical outcome measures (e.g. mortality), the majority of studies reported data for the whole patient cohort, as opposed to comparative data for the index and reference test. Few clinical trials have been conducted on the likely impact and safety of acting on the results of the real-time PCR assays in patients in any setting, although three RCTs,100,110,112 all of SeptiFast, were identified. One did not investigate patient outcomes112 and one was predominantly in febrile neutropenia patients and reported no significant difference in mortality, length of stay (ICU or hospital) or fever duration. 110 The third RCT was published in abstract form only and did not report a difference in mortality. 100
Given the potentially fatal consequences of removing treatment from patients with sepsis, it is not anticipated that negative tests in isolation would be acted on in clinical practice were an intervention introduced. This is because none of the three tests has the very high sensitivity needed to be used as a ‘rule-out’ test, which would reassure clinicians that a negative result would be associated with a very low probability of a patient having sepsis. Therefore, the main advantage of the tests in patients who are receiving broad-spectrum antibiotics would be in providing an earlier indication of the pathogen than would be achieved by standard methods, and in the potential focusing of antibiotic treatments. If negative tests are not acted upon, then the numbers of false-positive tests are reduced in populations with a high incidence of sepsis, such as patients in the ICU. It may be that the benefits of the tests are greater in high-incidence populations.
In addition, the three interventions provide very limited data regarding antimicrobial sensitivity. Definitive data on this are needed, to be obtained if possible, using standard culture methods undertaken in parallel with the interventions.
Cost-effectiveness
A systematic review of the literature was undertaken to identify cost-effectiveness analyses relating to the interventions. Two of these were within-study analyses, one using propensity scoring to match patients103 and one not. 89 The remaining two presented results from modelling studies, with one evaluating SeptiFast66 and one evaluating an IRIDICA–PLEX-ID hybrid. 128 The External Assessment Group noted limitations of all four studies and constructed a de novo mathematical model, and reported results under a number of scenarios. In base case 1, only documented statistically significant benefits associated with the tests were included, resulting in an estimation that all of the interventions provided no benefit. In order to investigate alternative scenarios, clinicians from the Diagnostic Appraisal Committee and clinicians who are authors of this report were asked, for each intervention, to provide estimates of the benefits associated with a positive test; this formed base case 2. At the aggregate level, all of the interventions were estimated to have cost per QALY gained values < £20,000. However, these results must be taken with caution as the clinicians noted the difficulty of the task and there was a wide divergence of opinion among the individual clinicians.
Additional analyses, using the data provided by clinicians, were undertaken to assess whether or not the estimated results were altered by analysing individual studies that assessed an intervention versus MALDI-TOF MS, or where two interventions were compared simultaneously within a study. The results from the studies against MALDI-TOF MS were concordant with those produced in base case 2 for individual interventions; however, indirectly, SeptiFast appeared to dominate SepsiTest, which did not occur in base case 2. For trials that assessed SeptiFast and SepsiTest simultaneously, the ICER for SeptiFast compared with SepsiTest was consistently > £30,000 per QALY, which was concordant with base case 2. It is commented that the results of the evaluation of SeptiFast with SepsiTest are driven by the relative costs of each test rather than the diagnostic accuracy, and also the assumed benefits assigned to each test by the expert clinicians. The External Assessment Group notes that the specificities of the tests are comparable, but the sensitivity of SeptiFast is estimated to be greater than that for SepsiTest.
To provide potentially useful information to the Diagnostic Appraisal Committee, threshold analyses were undertaken in relation to 30-day mortalities prevented, reduction in the number of days in the ICU and reduced antimicrobial costs.
Strengths and limitations of the assessment
Clinical effectiveness
The strengths of this systematic review are that it was conducted using robust methods, including the development of a prespecified protocol, comprehensive searching of published and unpublished evidence (including contact with clinical experts in the field and checking evidence submitted by the companies that manufacture the tests), study selection (including adjudication by three independent clinical experts), and data extraction by a minimum of two independent reviewers and a formal assessment of methodological quality. Statistical evaluation of diagnostic test accuracy was undertaken using statistically rigorous methods, allowing for the correlation between sensitivity and specificity, and potential between-study heterogeneity. Reasons for the heterogeneity in sensitivity and specificity between studies were explored using metaregression and parameter estimates were produced using Markov chain Monte Carlo simulation.
The assessment of methodological quality was generally hampered by the poor quality of reporting in the included SeptiFast, SepsiTest and IRIDICA studies, with the majority of studies being classified as being at unclear risk of bias on most assessment domains. Although a number of abstracts were included in the current systematic review, differences often occur between data reported in conference abstracts and their corresponding full reports; however, differences in results are usually not very large. 41
The pooled estimates of sensitivity and specificity for SeptiFast, SepsiTest and IRIDICA were estimated assuming that the reference standard was 100% sensitive and specific; however, this is unlikely to be the case. In practice, a wide range of factors is known to influence the diagnostic accuracy of blood cultures. For example, this may include antimicrobial treatment prior to blood sampling, low blood sample volumes, lack of replicate blood culture sets, delays in incubation and contamination during sampling. 10,12 As a result, the reported estimates of sensitivity and specificity are likely to be biased (underestimated) compared with those that would be obtained using a perfect reference standard. In addition, diagnostic metrics in the included studies were measured using different units: patients, sample episodes or species/pathogen level. Such analyses create a ‘unit of analyses’ error and may have contributed to the heterogeneity in the results.
Although no other systematic reviews or meta-analyses were identified for SepsiTest or IRIDICA, the present overall findings and conclusion for SeptiFast compared with blood culture were consistent with the review and meta-analysis by Dark et al. ,50 with pooled effects for sensitivity and specificity (across 41 SeptiFast studies, which were also included in the current review) of 0.68 (95% CI 0.63 to 0.73) and 0.86 (95% CI 0.84 to 0.89), respectively. An earlier systematic review of SeptiFast by Chang et al. 151 observed similar specificities (0.92, 95% CI 0.90 to 0.95), but higher sensitivities (0.75, 95% CI 0.65 to 0.83) across 34 SeptiFast studies. That review included a number of studies that were excluded from the present review because of publication type (foreign language, n = 2) or because they did not meet our inclusion criterion of ‘suspected sepsis’ (n = 2). In addition, Chang et al. 151 pooled studies comparing SeptiFast results against various reference standards to produce composite overall diagnostic accuracy metrics. These factors may have contributed to the the fact that diagnostic performance metrics were higher than those found in the present review and by Dark et al. 50
Cost-effectiveness
A systematic review of the cost-effectiveness literature associated with the interventions was undertaken. The External Assessment Group noted limitations with the identified evidence and therefore constructed a de novo model. A strength of the modelling work undertaken is that a framework for modelling interventions that provide rapid information on bloodstream bacteria and fungi has been established. The framework allows for there to be a benefit associated with false-positive tests and thus explicitly incorporates the fact that blood culture, with or without MALDI-TOF MS, is an imperfect reference test.
A fundamental limitation is that there are few robust data to populate the mathematical model. The External Assessment Group attempted to reduce this limitation by asking clinical experts to provide data to be used in an evaluation. The robustness of any conclusions is severely limited given the feedback from clinicians regarding the difficulty of the task and also because of the large heterogeneity of results produced from individual clinicians, which range from the interventions dominating to the interventions being dominated.
Further limitations are acknowledged in the model, which was simplistic, although none is expected to influence the conclusion that until further research is performed no robust assessment of cost-effectiveness can be made. The limitations of the model include the lack of modelling regarding antimicrobial stewardship benefits; the cost implications of any service reconfiguration required to move to a 24 hours a day, 7 days a week, service; any training costs required; any utility differential in survivors with and without any intervention; the possibility that only a sequencer need be purchased to run SepsiTest; that the estimates for the sensitivity of MALDI-TOF MS have been used at species level; and that any discounts associated with undertaking large quantities of tests have been omitted.
Uncertainties
Clinical effectiveness
All of the included studies compared the index test with a reference standard (blood culture with or without MALDI-TOF MS). No studies were identified that compared all of the index tests of interest directly with each other (end-to-end studies). In addition, there are very few robust data at present that report the impact of interventions on hard clinical outcomes such as mortality and reduced length of stay in critical care or in hospital.
Cost-effectiveness
The key uncertainty relates to the estimated cost-effectiveness of each intervention. The results produced by the External Assessment Group indicate that at an aggregate level clinicians believe the interventions to provide information that, if acted on, would improve key patient outcomes of mortality or ICU length of stay. However, there are no data currently available to support these views and no definitive conclusions can be provided until further research is undertaken.
Chapter 5 Conclusion
Implications for service provision
Given the considerable uncertainty in the cost-effectiveness results produced for each intervention, it is uncertain what the implications in the NHS would entail. Were the interventions deemed to be a cost-effective use of resources, it is likely that reconfigurations of working practice, such as moving to a 7-day working schedule, would be required in order that the interventions could provide results more quickly than under the present system.
Research recommendations
Despite the growing evidence base for all three interventions, a number of key issues need to be addressed. First, all future clinical studies incorporating SeptiFast, SepsiTest and IRIDICA need to be better reported, in accordance to the standards for the reporting of diagnostic accuracy studies statement. 152 Once robust diagnostic accuracy data have been established, there is a need for a pragmatic trial in which the results from the interventions are allowed to change patient management and for these results to be compared with standard practice in order to allow robust estimates of the clinical effectiveness and cost-effectiveness of an intervention to be estimated. A process evaluation, running alongside such a pragmatic trial, may also be of value to understand how the tests are used in the NHS. At present, there are very few data that report the impact of interventions on hard clinical outcomes such as mortality and reduced length of stay in critical care units. Any such trials should wait until the results from the RAPIDO trial31 are published in order that key information on the clinical utility of MALDI-TOF MS compared with blood culture is known. Finally, research into logistical issues, such as the numbers of hospitals serviced by the machine and the number of days that the machine operates to determine the optimal use of the interventions in England, is required.
Conclusions
Clinical effectiveness
SeptiFast, SepsiTest and IRIDICA appear to have higher specificity values than sensitivity values. However, because of the deficiencies in study quality in the included studies, these data may not be reliable and should be treated with caution. Moreover, there are no head-to-head comparisons of all these tests and there are few robust data that report the impact of interventions on hard clinical outcomes, such as mortality and reduced length of stay in critical care units; the data that do exist have not shown any intervention to produce a statistically significant improvement. In order to produce a definitive conclusion on the clinical effectiveness of the interventions appropriate studies need to be conducted (see Research recommendations).
Cost-effectiveness
There is considerable uncertainty associated with all analyses within this assessment and a definitive estimate of the cost-effectiveness of each intervention cannot be provided. This is largely because of the limitations of the evidence base. The studies recommended in Research recommendations would reduce this uncertainty. Threshold analyses have been provided that may allow decision-makers to estimate whether or not the interventions are likely to meet a level at which the decision-makers would consider the interventions to be cost-effective.
Acknowledgements
The authors wish to thank Andrea Shippam [Programme Manager, School of Health and Related Research (ScHARR)] for providing administrative support and in preparing and formatting the report, and Mark Clowes (Information Specialist, ScHARR) for assisting with the literature searching.
Contributions of authors
Matt Stevenson (Professor of Health Technology Assessment) and Rachid Rafia (Research Fellow) were responsible for the acquisition of data, analysis and interpretation of data, model construction (for the health economic evaluations), and drafting and revising of the final report.
Abdullah Pandor (Senior Research Fellow), Marrissa Martyn-St James (Research Fellow) and Lesley Uttley (Research Fellow) co-ordinated the review and were responsible for the acquisition of data, analysis and interpretation of data (for the systematic review), and drafting and revising the final report.
John Stevens (Reader in Decision Science) and Jean Sanderson (Research Associate) were responsible for the statistical analyses, interpretation of data, and drafting and revising the final report.
Ruth Wong (Information Specialist) was responsible for developing and undertaking the electronic literature searches.
Gavin D Perkins (Professor of Critical Care Medicine), Ronan McMullan (Senior Lecturer and Consultant Microbiologist) and Paul Dark (Reader and Honorary Consultant Intensivist) were responsible for providing expert clinical advice throughout the project, and drafting and revising the final report.
Data sharing statement
Data can be obtained from the corresponding author, subject to their being non-confidential.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Acute Care Toolkit 9: Sepsis. London: Royal College of Physicians; 2015.
- Anon . American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med 1992;20:864-74. http://dx.doi.org/10.1097/00003246-199206000-00025.
- Kaukonen K-M, Bailey M, Pilcher D, Cooper DJ, Bellomo R. Systemic inflammatory response syndrome criteria in defining severe sepsis. N Engl J Med 2015;372:1629-38. http://dx.doi.org/10.1056/NEJMoa1415236.
- Bone R, Balk R, Cerra F, Dellinger R, Fein A, Knaus W, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644-55. http://dx.doi.org/10.1378/chest.101.6.1644.
- Kumar A, Roberts D, Wood K, Light B, Parrillo J, Sharma S, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2005;34:1589-96. http://dx.doi.org/10.1097/01.CCM.0000217961.75225.E9.
- Levy M, Dellinger R, Townsend S, Linde-Zwirble WT, Marshall JC, Bion J, et al. The surviving sepsis campaign: results of an international guideline-based performance improvement program targeting severe sepsis. Crit Care Med 2010;38:367-74. http://dx.doi.org/10.1097/CCM.0b013e3181cb0cdc.
- Dellinger R, Carlet J, Masur H, Gerlach H, Calandra T, Cohen J, et al. The surviving sepsis campaign guidelines for management of severe sepsis and septic shock. Crit Care Med 2004;32:858-73. http://dx.doi.org/10.1097/01.CCM.0000117317.18092.E4.
- Levy M, Rhodes A, Phillips G, Townsend S, Schorr C, Beale R, et al. Surviving sepsis campaign: association between performance metrics and outcomes in a 7.5-year study. Int Care Med 2014;40:1623-33. http://dx.doi.org/10.1007/s00134-014-3496-0.
- Mouncey PR, Osborn TM, Power GS, Harrison DA, Sadique MZ, Grieve RD, et al. Trial of early, goal-directed resuscitation for septic shock. N Engl J Med 2015;372:1301-11. http://dx.doi.org/10.1056/NEJMoa1500896.
- Warhurst G, Dunn G, Chadwick P, Blackwood B, McAuley D, Perkins GD, et al. Rapid detection of health-care-associated bloodstream infection in critical care using multipathogen real-time polymerase chain reaction technology: a diagnostic accuracy study and systematic review. Health Technol Assess 2015;19. http://dx.doi.org/10.3310/hta19350.
- Daniels R. Surviving the first hours in sepsis: getting the basics right (an intensivist’s perspective). J Antimicrob Chemother 2011;66:11-23. http://dx.doi.org/10.1093/jac/dkq515.
- UK Standards for Microbiology Investigations: Investigation of Blood Cultures (for Organisms other than Mycobacterium Species). London: Public Health England; 2014.
- English National Point Prevalence Survey on Healthcare Associated Infections and Antimicrobial Use, 2011: Preliminary Data. London: Health Protection Agency; 2012.
- English Surveillance Programme for Antimicrobial Utilisation and Resistance (ESPAUR). London: Health Protection Agency; 2014.
- Davies S. Annual Report of the Chief Medical Officer, Volume Two, 2011, Infections and the Rise of Antimicrobial Resistance. London: Department of Health; 2013.
- Dellinger R, Levy M, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 2013;41:580-637. http://dx.doi.org/10.1097/CCM.0b013e31827e83af.
- Levy M, Fink M, Marshall J, Abrahm E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International sepsis definitions conference. Crit Care Med 2003;31:1250-6.
- Neutropenic Sepsis: Prevention and Management of Neutropenic Sepsis in Cancer Patients. NICE Guidelines [CG151]. London: NICE; 2012.
- Bacterial Sepsis in Pregnancy. Green-top Guideline No. 64a. London: Royal College of Obstetricians and Gynaecologists; 2012.
- Bacterial Sepsis following Pregnancy Green-top Guideline No. 64b. London: Royal College of Obstetricians and Gynaecologists; 2012.
- Sepsis: The Recognition, Diagnosis and Management of Severe Sepsis. London: NICE; 2015.
- Commissioning for Quality and Innovation (CQUIN) Guidance for 2015/16. Redditch: NHS England; 2015.
- UK Five Year Antimicrobial Resistance Strategy 2013 to 2018. London: Department of Health; 2013.
- Sepsis: The LightCycler SeptiFast test MGRADE, SepsiTest and IRIDICA BAC BSI Assay for Rapidly Identifying Bloodstream Bacteria and Fungi Final Scope. London: NICE; 2015.
- Peralta G, Rodriguez-Lera M, Garrido J, Andorena L, Roiz M. Time to positivity in blood cultures of adults with Streptococcus pneumoniae bacteremia. BMC Infect Dis 2006;6. http://dx.doi.org/10.1186/1471-2334-6-79.
- Afshari A, Schrenzel J, Ieven M, Harbarth S. Bench-to-bedside review: rapid molecular diagnostics for bloodstream infection – a new frontier?. Crit Care 2012;16. http://dx.doi.org/10.1186/cc11202.
- Raad I, Hanna H, Maki D. Intravascular catheter-related infections: advances in diagnosis, prevention, and management. Lancet Infect Dis 2007;7:645-57. http://dx.doi.org/10.1016/S1473-3099(07)70235-9.
- Vincent J-L, Sakr Y, Sprung C, Ranieri VM, Reinhart K, Gerlach H, et al. Sepsis in European intensive care units: results of the SOAP study. Crit Care Med 2006;34:344-53. http://dx.doi.org/10.1097/01.CCM.0000194725.48928.3A.
- Schubert S, Weinert K, Wagner C, Gunzl B, Wieser A, Maier T, et al. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J Mol Diagn 2011;13:701-6. http://dx.doi.org/10.1016/j.jmoldx.2011.07.004.
- Morgenthaler N, Kostrzewa M. Rapid identification of pathogens in positive blood culture of patients with sepsis: review and meta-analysis of the performance of the sepsityper kit. Int J Microbiol 2015;2015. http://dx.doi.org/10.1155/2015/827416.
- ISRCTN Registry . A Prospective Randomised, Multicentre Trial to Assess the Impact of Laboratory Based Rapid Diagnosis on Outcome in Patients With Blood Stream Infections 2015. www.isrctn.com/ISRCTN97107018 (accessed April 2016).
- Advisory Committee on Antimicrobial Resistance and Healthcare Associated Infection - Antimicrobial Stewardship ‘Start Smart then Focus’: Guidance for Antimicrobial Stewardship in Hospitals (England). London: Department of Health; 2011.
- Antimicrobial Stewardship. Antimicrobial Stewardship: Systems and Processes for Effective Antimicrobial Medicine Use. Medicines Practice Guideline. Methods, evidence and recommendations. London: NICE; 2015.
- Kim J, Chung J, Choi S, Jang H, Hong S, Lim C, et al. Early use of imipenem/cilastatin and vancomycin followed by de-escalation versus conventional antimicrobials without de-escalation for patients with hospital-acquired pneumonia in a medical ICU: a randomized clinical trial. Crit Care 2012;16. http://dx.doi.org/10.1186/cc11197.
- Leone M, Bechis C, Baumstarck K, Lefrant J-Y, Albanese J, Jaber S, et al. De-escalation versus continuation of empirical antimicrobial treatment in severe sepsis: a multicenter non-blinded randomized noninferiority trial. Intens Care Med 2014;40:1399-408. http://dx.doi.org/10.1007/s00134-014-3411-8.
- Oosterheert J, Bonten M, Schneider M, Buskens E, Lammers J-W, Hustinx W, et al. Effectiveness of early switch from intravenous to oral antibiotics in severe community acquired pneumonia: multicentre randomised trial. BMJ 2006;333. http://dx.doi.org/10.1136/bmj.38993.560984.BE.
- Garnacho-Montero J, Ortiz-Leyba C, Herrera-Melero I, Aldabó-Pallás T, Cayuela-Dominguez A, Marquez-Vacaro JA, et al. Mortality and morbidity attributable to inadequate empirical antimicrobial therapy in patients admitted to the ICU with sepsis: a matched cohort study. J Antimicrob Chemother 2008;61:436-41. http://dx.doi.org/10.1093/jac/dkm460.
- Zilberberg M, Kollef M, Arnold H, Labelle A, Micek S, Kothari S, et al. Inappropriate empiric antifungal therapy for candidemia in the ICU and hospital resource utilization: a retrospective cohort study. BMC Infect Dis 2010;10. http://dx.doi.org/10.1186/1471-2334-10-150.
- Arnold HM, Micek ST, Shorr AF, Zilberberg MD, Labelle AJ, Kothari S, et al. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy 2010;30:361-8. http://dx.doi.org/10.1592/phco.30.4.361.
- Morrell M, Fraser V, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 2005;49:3640-5. http://dx.doi.org/10.1128/AAC.49.9.3640-3645.2005.
- Systematic Reviews: CRD’s Guidance for Undertaking Reviews in Health Care. York: Centre for Reviews and Dissemination; 2009.
- Leeflang M, Deeks JJ, Gatsonis C, Bossuyt P. Cochrane diagnostic test accuracy working group. Systematic reviews of diagnostic test accuracy. Ann Intern Med 2008;149:889-97. http://dx.doi.org/10.7326/0003-4819-149-12-200812160-00008.
- The Cochrane Collaboration . Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy. Version 1.0.0 2013. http://methods.cochrane.org/sdt/handbook-dta-reviews (accessed April 2016).
- Raglio A, Rizzi M, Amer M, Mangia M, Luca M, Goglio A. Sepsis Diagnosis by Real-Time PCR (SeptiFast Kit, Roche Diagnostics): Preliminary Results and Possible Applications n.d.
- Mancini N, Clerici D, Diotti R, Perotti M, Ghidoli N, De Marco D, et al. Molecular diagnosis of sepsis in neutropenic patients with haematological malignancies. J Med Microbiol 2008;57:601-4. http://dx.doi.org/10.1099/jmm.0.47732-0.
- Louie RF, Tang Z, Albertson T, Cohen S, Tran N, Kost GJ. Multiplex polymerase chain reaction detection enhancement of bacteremia and fungemia. Crit Care Med 2008;36:1487-92. http://dx.doi.org/10.1097/CCM.0b013e31816f487c.
- Disqué C, Kochem AJ, Mühl H, Lorenz MG, Sakka SG. PCR detection of sepsis-inducing pathogens in blood using SepsiTest™ blood. Int J Med Microbiol 2008;298.
- Wellinghausen N, Kochem A, Disqué C, Mühl H, Gebert S, Winter J, et al. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol 2009;47:2759-65. http://dx.doi.org/10.1128/JCM.00567-09.
- Bacconi A, Richmond G, Baroldi M, Laffler T, Blyn LB, Carolan H, et al. Improved sensitivity for molecular detection of bacterial and Candida infections in blood. J Clin Microbiol 2014;52:3164-74. http://dx.doi.org/10.1128/JCM.00801-14.
- Dark P, Blackwood B, Gates S, McAuley D, Perkins G. Accuracy of LightCycler® SeptiFast for the detection and identification of pathogens in the blood of patients with suspected sepsis: a systematic review and meta-analysis. Intens Care Med 2015;41:21-33. http://dx.doi.org/10.1007/s00134-014-3553-8.
- Whiting PF, Rutjes A, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. http://dx.doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Reitsma JB, Glas AS, Rutjes AWS, Scholten RJPM, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90. http://dx.doi.org/10.1016/j.jclinepi.2005.02.022.
- Spiegelhalter DJ, Best NG, Carlin BR, van der Linde A. Bayesian measures of model complexity and fit. J R Stat Soc B Met 2002;64:583-616. http://dx.doi.org/10.1111/1467-9868.00353.
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Stat Comput 2000;10:325-37. http://dx.doi.org/10.1023/A:1008929526011.
- Sturtz S, Ligges U, Gelman A. R2WinBUGS: a package for running WinBUGS from R. J Stat Softw 2005;12:1-16. http://dx.doi.org/10.18637/jss.v012.i03.
- Higgins JPT, Green SE. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 2011. www.cochrane-handbook.org (accessed April 2016).
- Khan K, Kunz R, Kleijnen J, Antes G. Systematic Reviews to Support Evidence-Based Medicine. Boca Raton, FL: CRC Press; 2011.
- Moher D, Liberati A, Tetzlaff J, Altman D. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLOS Med 2009;6. http://dx.doi.org/10.1371/journal.pmed.1000097.
- Bingold T, Hunfeld K, Scheller B, Ronneberg T, Sartorius S, Wahrmann M, et al. Clinical utility of a new PCR-based assay for rapid pathogen detection (SeptiFast) in patients with clinical sepsis compared to standard blood culture. Infection 2007;35.
- Klemm M, Prinz M, Nowak A, Morgenstern T, Meisner M, Rothe K, et al. Clinical application of SeptiFast, a PCR method for the detection of bacteraemia, in intensive care patients. Infection 2007;35.
- Lodes U, Bohmeier B, Meyer F, Koenig B, Lippert H. W1043 microbiologic diagnostic with the novel lightcycler Septifast® test during the early phase of surgical sepsis mainly caused by peritonitis is quicker and more sensitive than conventional microbiological culture. Gastroenterology 2008;134. http://dx.doi.org/10.1016/S0016-5085(08)62901-7.
- Vince A, Lepej S, Barsic B, Dusek D, Mitrovic Z, Serventi-Seiwerth R, et al. LightCycler SeptiFast assay as a tool for the rapid diagnosis of sepsis in patients during antimicrobial therapy. J Med Microbiol 2008;57:1306-7. http://dx.doi.org/10.1099/jmm.0.47797-0.
- Dark P, Chadwick P, Warhurst G. Detecting sepsis-associated bloodstream infection acquired in intensive care using multi-pathogen real-time PCR. J Infect 2009;59:296-8. http://dx.doi.org/10.1016/j.jinf.2009.08.002.
- Dierkes C, Ehrenstein B, Siebig S, Linde H, Reischl U, Salzberger B. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis 2009;9. http://dx.doi.org/10.1186/1471-2334-9-126.
- Gimeno E, Sorli ML, Abella E, Alvarez A, Pablo JPJ, Gimenez MT, et al. Molecular diagnosis of bacteremia in neutropenic febrile oncohematological patients. Preliminary results. Haematologica 2009;94:415-16.
- Lehmann LE, Alvarez J, Hunfeld K-P, Goglio A, Kost G. Potential clinical utility of polymerase chain reaction in microbiological testing for sepsis. Crit Care Med 2009;37:3085-90. http://dx.doi.org/10.1097/CCM.0b013e3181b033d7.
- Lodes U, Meyer F, Konig B, Lippert H. Microbiological sepsis screening in surgical ICU patients with the ‘lightCycler’ Septifast test – a pilot study. Zentralbl Chir 2009;134:249-53. http://dx.doi.org/10.1055/s-0028-1098776.
- Palomares J, Puche B, Martos A, Lucena F, Marin E, Martin-Mazuelos E. Rapid Molecular Diagnosis Of Severe Sepsis In Patients With Symptoms Of Severe Sepsis n.d.
- Paolucci M, Capretti M, Dal Monte P, Corvaglia L, Landini M, Varani S, et al. Laboratory diagnosis of late-onset sepsis in newborns by multiplex real-time PCR. J Med Microbiol 2009;58:533-4. http://dx.doi.org/10.1099/jmm.0.003848-0.
- Varani S, Stanzani M, Paolucci M, Melchionda F, Castellani G, Nardi L, et al. Diagnosis of bloodstream infections in immunocompromised patients by real-time PCR. J Infect 2009;58:346-51. http://dx.doi.org/10.1016/j.jinf.2009.03.001.
- von Lilienfeld-Toal M, Lehmann LE, Raadts A. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J Clin Microbiol 2009;47:2405-10. http://dx.doi.org/10.1128/JCM.00491-09.
- Westh H, Lisby G, Breysse F, Boddinghaus B, Chomarat M, Gant V, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect 2009;15:544-51. http://dx.doi.org/10.1111/j.1469-0691.2009.02736.x.
- Berger A, Altiok I, Mechtler T, Langgartner M, Böhm J, Herkner K, et al. Adaption of the Roche Septifast system for the early detection of neonatal sepsis in very low birthweight infants. Klin Padiatr 2010;222. http://dx.doi.org/10.1055/s-0030-1261288.
- Bloos F, Hinder F, Becker K, Sachse S, Mekontso Dessap A, Straube E, et al. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intens Care Med 2010;36:241-7. http://dx.doi.org/10.1007/s00134-009-1705-z.
- Lamoth F, Jaton K, Prod’hom G, Senn L, Bille J, Calandra T, et al. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol 2010;48:3510-16. http://dx.doi.org/10.1128/JCM.00147-10.
- Lehmann LE, Hunfeld K, Steinbrucker M, Brade V, Book M, Seifert H, et al. Improved detection of blood stream pathogens by real-time PCR in severe sepsis. Intens Care Med 2010;36:49-56. http://dx.doi.org/10.1007/s00134-009-1608-z.
- Maubon D, Hamidfar-Roy R, Courby S, Vesin A, Maurin M, Pavese P, et al. Therapeutic impact and diagnostic performance of multiplex PCR in patients with malignancies and suspected sepsis. J Infect 2010;61:335-42. http://dx.doi.org/10.1016/j.jinf.2010.07.004.
- Regueiro B, Varela-Ledo E, Martinez-Lamas L, Rodriguez-Calvino J, Aguilera A, Santos A, et al. Automated extraction improves multiplex molecular detection of infection in septic patients. PLOS ONE 2010;5. http://dx.doi.org/10.1371/journal.pone.0013387.
- Soki J, Szabo E, Hajdu E, Lazar A, Nagy E. Use of a Commercially Available Multiplex Real-Time PCR Assay (SeptiFast) to Detect Bacterial and Fungal Pathogens in Septic Patients n.d.
- Tsalik EL, Jones D, Nicholson B, Waring L, Liesenfeld O, Park L, et al. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. J Clin Microbiol 2010;48:26-33. http://dx.doi.org/10.1128/JCM.01447-09.
- Wallet F, Nseir S, Baumann L, Herwegh S, Sendid B, Boulo M, et al. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin Microbiol Infect 2010;16:774-9. http://dx.doi.org/10.1111/j.1469-0691.2009.02940.x.
- Yanagihara K, Kitagawa Y, Tomonaga M, Tsukasaki K, Kohno S, Seki M, et al. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care 2010;14. http://dx.doi.org/10.1186/cc9234.
- Bravo D, Blanquer J, Tormo M, Aguilar G, Borras R, Solano C, et al. Diagnostic accuracy and potential clinical value of the LightCycler SeptiFast assay in the management of bloodstream infections occurring in neutropenic and critically ill patients. Int J Infect Dis 2011;15:e326-31. http://dx.doi.org/10.1016/j.ijid.2011.01.003.
- Hettwer S, Wilhelm J, Schürmann M, Ebelt H, Hammer D, Amoury M, et al. Microbial diagnostics in patients with presumed severe infection in the emergency department. Klin Intensivmed Notfmed 2011;107:53-62. http://dx.doi.org/10.1007/s00390-011-0287-5.
- Josefson P, Strålin K, Ohlin A, Ennefors T, Dragsten B, Andersson L, et al. Evaluation of a commercial multiplex PCR test (SeptiFast) in the etiological diagnosis of community-onset bloodstream infections. Eur J Clin Microbiol Infect Dis 2011;30:1127-34. http://dx.doi.org/10.1007/s10096-011-1201-6.
- Lucignano B, Ranno S, Liesenfeld O, Pizzorno B, Putignani L, Bernaschi P, et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. J Clin Microbiol 2011;49:2252-8. http://dx.doi.org/10.1128/JCM.02460-10.
- Obara H, Aikawa N, Hasegawa N, Hori S, Ikeda Y, Kobayashi Y, et al. The role of a real-time PCR technology for rapid detection and identification of bacterial and fungal pathogens in whole-blood samples. J Infect Chemother 2011;17:327-33. http://dx.doi.org/10.1007/s10156-010-0168-z.
- Vrioni G, Daniil I, Mamali V, Kimouli M, Mylona-Petropoulou D, Themeli-Digalaki K, et al. Preliminary Clinical Study Using a Multiplex Blood PCR for Rapid Detection of Bacterial and Fungal Pathogens in ICU Patients With Presumed Sepsis n.d.
- Alvarez J, Mar J, Varela-Ledo E, Garea M, Matinez-Lamas L, Rodriguez J, et al. Cost analysis of real-time polymerase chain reaction microbiological diagnosis in patients with septic shock. Anaesth Intens Care 2012;40:958-63.
- Grif K, Fille M, Wurzner R, Weiss G, Lorenz I, Gruber G, et al. Rapid detection of bloodstream pathogens by real-time PCR in patients with sepsis. Wien Klin Wochenschr 2012;124:266-70. http://dx.doi.org/10.1007/s00508-012-0159-4.
- Guido M, Quattrocchi M, Zizza A, Pasanisi G, Pavone V, Lobreglio G, et al. Molecular approaches in the diagnosis of sepsis in neutropenic patients with haematological malignances. J Prev Med Hyg 2012;53:104-8.
- Lodes U, Bohmeier B, Lippert H, Konig B, Meyer F. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch Surg 2012;397:447-55. http://dx.doi.org/10.1007/s00423-011-0870-z.
- Mauro MV, Cavalcanti P, Perugini D, Noto A, Sperli D, Giraldi C. Diagnostic utility of LightCycler SeptiFast and procalcitonin assays in the diagnosis of bloodstream infection in immunocompromised patients. Diagn Microbiol Infect Dis 2012;73:308-11. http://dx.doi.org/10.1016/j.diagmicrobio.2012.04.006.
- Pasqualini L, Mencacci A, Leli C, Montagna P, Cardaccia A, Cenci E, et al. Diagnostic performance of a multiple real-time PCR assay in patients with suspected sepsis hospitalized in an internal medicine ward. J Clin Microbiol 2012;50:1285-8. http://dx.doi.org/10.1128/JCM.06793-11.
- Rath P, Saner F, Paul A, Lehmann N, Steinmann E, Buer J, et al. Multiplex PCR for rapid and improved diagnosis of bloodstream infections in liver transplant recipients. J Clin Microbiol 2012;50:2069-71. http://dx.doi.org/10.1128/JCM.00745-12.
- Tschiedel E, Steinmann J, Buer J, Onnebrink J, Felderhoff-Muser U, Rath P, et al. Results and relevance of molecular detection of pathogens by SeptiFast – a retrospective analysis in 75 critically ill children. Klin Padiatr 2012;224:12-6. http://dx.doi.org/10.1055/s-0031-1285878.
- Herne V, Nelovkov A, Kutt M, Ivanova M. Diagnostic performance and therapeutic impact of LightCycler SeptiFast assay in patients with suspected sepsis. Eur J Microbiol Immunol 2013;3:68-76. http://dx.doi.org/10.1556/EuJMI.3.2013.1.10.
- Kasper DC, Altiok I. Molecular detection of late-onset neonatal sepsis in premature infants using small blood volumes: proof-of-concept. Neonatology 2013;103:268-73. http://dx.doi.org/10.1159/000346365.
- Paolucci M, Stanzani M, Melchionda F, Tolomelli G, Castellani G, Landini MP, et al. Routine use of a real-time polymerase chain reaction method for detection of bloodstream infections in neutropaenic patients. Diagn Microbiol Infect Dis 2013;75:130-4. http://dx.doi.org/10.1016/j.diagmicrobio.2012.10.012.
- Rodrigues C, Dos Santos MS, Filho HHC, Charbel CE, da Silva LCS, Rossi F, et al. Rapid molecular test (SeptiFast) reduced time for adjustment of antibiotic treatment in comparison with conventional blood cultures in critically ill sepsis patients: a randomized controlled clinical trial (preliminary results). Crit Care 2013;17. http://dx.doi.org/10.1186/cc12926.
- Avolio M, Diamante P, Modolo M, De Rosa R, Stano P, Camporese A. Direct molecular detection of pathogens in blood as specific rule-in diagnostic biomarker in patients with presumed sepsis: our experience on a heterogeneous cohort of patients with signs of infective systemic inflammatory response syndrome. Shock 2014;42:86-92. http://dx.doi.org/10.1097/SHK.0000000000000191.
- Burdino E, Ruggiero T, Allice T, Milia M, Gregori G, Milano R, et al. Combination of conventional blood cultures and the SeptiFast molecular test in patients with suspected sepsis for the identification of bloodstream pathogens. Diagn Microbiol Infect Dis 2014;79:287-92. http://dx.doi.org/10.1016/j.diagmicrobio.2014.03.018.
- Mancini N, Sambri V, Corti C, Ghidoli N, Tolomelli G, Paolucci M, et al. Cost-effectiveness of blood culture and a multiplex real-time PCR in hematological patients with suspected sepsis: an observational propensity score-matched study. Expert Rev Mol Diagn 2014;14:623-32. http://dx.doi.org/10.1586/14737159.2014.916212.
- Markota A, Seme K, Golle A, Poljak M, Sinkovič A. SeptiFast real-time PCR for detection of bloodborne pathogens in patients with severe sepsis or septic shock. Collegium Antropol 2014;38:829-33.
- Ozkaya-Parlakay A, Cengiz AB, Ceyhan M, Hascelik G, Kara A, Celik M, et al. Evaluation of multiplex real time polymerase chain reaction and procalcitonin in the diagnosis of sepsis. Clin Lab 2014;60:1075-81.
- Schaub N, Boldanova T, Noveanu M, Arenja N, Hermann H, Twerenbold R, et al. Incremental value of multiplex real-time PCR for the early diagnosis of sepsis in the emergency department. Swiss Med Wkly 2014;144. http://dx.doi.org/10.4414/smw.2014.13911.
- Sitnik R, Marra AR, Petroni RC, Ramos OP, Martino MD, Pasternak J, et al. SeptiFast for diagnosis of sepsis in severely ill patients from a Brazilian hospital. Einstein (Sao Paulo) 2014;12:191-6. http://dx.doi.org/10.1590/S1679-45082014AO2932.
- Barbanti MC, Greco R, Mancini N, Orsini A, Crucitti L, Forcina A, et al. Improving the diagnostic algorithm for sepsis: adjuvant role of SeptiFast in 491 consecutive hematological patients. Bone Marrow Transpl 2015;50.
- Calitri C, Denina M, Scolfaro C, Garazzino S, Licciardi F, Burdino E, et al. Etiological diagnosis of bloodstream infections through a multiplex real-time polymerase chain reaction test in pediatric patients: a case series from a tertiary Italian hospital. Infect Dis (Lond) 2015;47:73-9. http://dx.doi.org/10.3109/00365548.2014.967291.
- Idelevich EA, Silling G, Niederbracht Y, Penner H, Sauerland MC, Tafelski S, et al. Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: a randomized controlled study. Med Microbiol Immunol 2015;204:585-92. http://dx.doi.org/10.1007/s00430-014-0385-7.
- Warhurst G, Maddi S, Dunn G, Ghrew M, Chadwick P, Alexander P, et al. Diagnostic accuracy of SeptiFast multi-pathogen real-time PCR in the setting of suspected healthcare-associated bloodstream infection. Intens Care Med 2015;41:86-93. http://dx.doi.org/10.1007/s00134-014-3551-x.
- Tafelski S, Nachtigall I, Adam T, Bereswill S, Faust J, Tamarkin A, et al. Randomized controlled clinical trial evaluating multiplex polymerase chain reaction for pathogen identification and therapy adaptation in critical care patients with pulmonary or abdominal sepsis. J Int Med Res 2015;43:364-77. http://dx.doi.org/10.1177/0300060514561135.
- Nieman A, Schade R, Savelkoul P, Beishuizen A, Henrich B, Lamik Wolters B, et al. A Prospective Multicenter Evaluation of Direct Molecular Detection of Blood Stream Infection from a Clinical Perspective 2015.
- Loonen AJ, de Jager CP, Tosserams J, Kusters R, Hilbink M, Wever PC, et al. Biomarkers and molecular analysis to improve bloodstream infection diagnostics in an emergency care unit. PLOS ONE 2014;9. http://dx.doi.org/10.1371/journal.pone.0087315.
- Delco-Volante C, Karam O, Posfay-Barbe K. Detection of Neonate Bloodstream Infections by PCR ESI-MS: A Proof-of-Concept Study (Oral Presentation #I038) 2015.
- Vincent J-L, Brealey D, Libert N, Abidi N, O’Dwyer M, Zacharowski K, et al. Rapid diagnosis of infection in the critically ill (RADICAL), a multicenter study of molecular detection in bloodstream infections, pneumonia and sterile site infections. Crit Care Med 2015;43:2283-91. http://dx.doi.org/10.1097/CCM.0000000000001249.
- Metzgar D, Frinder M, Rothman R, Peterson S, Carroll KC, Zhang S, et al. A Rapid and Sensitive Assay for Direct Identification of Bacteria and Candida in Blood 2015.
- Leitner E, Kessler H, Spindelboeck W, Hoenigl M, Putz-Bankuti C, Stadlbauer-Köllner V, et al. Comparison of two molecular assays with conventional blood culture for diagnosis of sepsis. J Microbiol Methods 2013;92:253-5. http://dx.doi.org/10.1016/j.mimet.2012.12.012.
- Schreiber J, Nierhaus A. Comparison of three different commercial PCR assays for the detection of pathogens in critically ill sepsis patients. Med Klinik Intensivmed Notfmed 2013;108:311-18. http://dx.doi.org/10.1007/s00063-013-0227-1.
- Czaja J, Jachymek W, Niedziela T, Lugowski C, Aldova E, Kenne L. Structural studies of the O-specific polysaccharide from Plesiomonas shigelloides strain CNCTC 113/92. Eur J Biochem 2000;267:1672-9. http://dx.doi.org/10.1046/j.1432-1327.2000.01161.x.
- Tests for Rapidly Identifying Bloodstream Bacteria and Fungi (LightCycler SeptiFast Test MGRADE SepsiTest and IRIDICA BAC BSI Assay): Outcomes. London: NICE; 2016.
- Dixon P, Davies P, Hollingworth W, Stoddart M, MacGowan A. A systematic review of matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry compared to routine microbiological methods for the time taken to identify microbial organisms from positive blood cultures. Eur J Clin Microbiol 2015;34:863-76. http://dx.doi.org/10.1007/s10096-015-2322-0.
- Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Land GA, et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 2013;137:1247-54. http://dx.doi.org/10.5858/arpa.2012-0651-OA.
- Huang AM, Newton D, Kunapuli A, Gandhi T. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013;57:1237-45. http://dx.doi.org/10.1093/cid/cit498.
- Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Peterson LE, et al. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J Infection 2014;69:216-25. http://dx.doi.org/10.1016/j.jinf.2014.05.005.
- Nagel JL, Huang AM, Kunapuli A, Gandhi TN, Washer LL, Lassiter J, et al. Impact of antimicrobial stewardship intervention on coagulase-negative Staphylococcus blood cultures in conjunction with rapid diagnostic testing. J Clin Microbiol 2014;52:2849-54. http://dx.doi.org/10.1128/JCM.00682-14.
- Martiny D, Debaugnies F. Impact of rapid microbial identification directly from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on patient management. Clin Microbiol Infect 2013;19:E568-81. http://dx.doi.org/10.1111/1469-0691.12282.
- Bilkovski R, Sampath R, Huiras M, Durtschi A, Ecker D, Chalfin D, et al. The Potential Clinical and Economic Value of Rapid Diagnosis of Suspected Bloodstream Infections Using PCR ESI-MS n.d.
- Lehmann LE, Herpichboehm B, Kost G, Kollef M, Stüber F. Cost and mortality prediction using polymerase chain reaction pathogen detection in sepsis: evidence from three observational trials. Crit Care 2010;14. http://dx.doi.org/10.1186/cc9294.
- Zaniolo O, Povero M, Pradelli L, Pizzorno B, Mancini N. An observational propensity score-matched study to evaluate cost-effectiveness of a real-time PCR-based assay in patients with suspected Sepsisk. Value Health 2014;17:A672-3. http://dx.doi.org/10.1016/j.jval.2014.08.2493.
- Wong JR, Bauer KA, Mangino JE, Goff DA, Bauer KA. Antimicrobial stewardship pharmacist interventions for coagulase-negative staphylococci positive blood cultures using rapid polymerase chain reaction. Ann Pharmacother 2012;46:1484-90. http://dx.doi.org/10.1345/aph.1R439.
- Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program’s impact. Clin Infect Dis 2010;51:1074-80. http://dx.doi.org/10.1086/656623.
- Hermsen ED, Shull SS, Klepser DG, Iwen PC, Armbrust A, Garrett J, et al. Pharmacoeconomic analysis of microbiologic techniques for differentiating staphylococci directly from blood culture bottles. J Clin Microbiol 2008;46:2924-9. http://dx.doi.org/10.1128/JCM.00623-08.
- Beyda ND, Eiland EH. Rapid molecular testing in bloodstream infections: improving patient outcomes, reducing health care costs, and aiding clinical pharmacists in decision making. US Pharm 2009;34:HS9-13.
- Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000;118:146-55. http://dx.doi.org/10.1378/chest.118.1.146.
- Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 1999;115:462-74. http://dx.doi.org/10.1378/chest.115.2.462.
- Google . Google Exchange Rate 2015. www.google.co.uk/search?q=exchange+rate+euro+pound&ie=utf-8&oe=utf-8&gws_rd=cr&ei=PnR9VbiPI8bl7gaC_IGgBg (accessed June 2015).
- Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med 2003;115:529-35. http://dx.doi.org/10.1016/j.amjmed.2003.07.005.
- Rodríguez-Sánchez B, Sánchez-Carrillo C, Ruiz A, Marín M, Cercenado E, Rodríguez-Créixems M, et al. Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin Microbiol Infect 2014;20:O421-7. http://dx.doi.org/10.1111/1469-0691.12455.
- Lunt M. Selecting an appropriate caliper can be essential for achieving good balance with propensity score matching. Am J Epidemiol 2014;179:226-35. http://dx.doi.org/10.1093/aje/kwt212.
- Diagnostic Assessment Programme Manual. London: NICE; 2011.
- Cuthbertson BH, Elders A, Hall S, Taylor J, MacLennan G, Mackirdy F, et al. Mortality and quality of life in the five years after severe sepsis. Crit Care 2013;17. http://dx.doi.org/10.1186/cc12616.
- Davis JS, He V, Anstey NM, Condon JR. Long term outcomes following hospital admission for sepsis using relative survival analysis: a prospective cohort study of 1,092 patients with 5 year follow up. PLOS ONE 2014;9. http://dx.doi.org/10.1371/journal.pone.0112224.
- Wang HE, Szychowski JM, Griffin R, Safford MM, Shapiro NI, Howard G. Long-term mortality after community-acquired sepsis: a longitudinal population-based cohort study. BMJ Open 2014;4. http://dx.doi.org/10.1136/bmjopen-2013-004283.
- Office For National Statistics . National Life Tables, United Kingdom, 1980–82 to 2011–2013 2015. www.ons.gov.uk/ons/publications/re-reference-tables.html?edition=tcm%3A77-365199 (accessed June 2015).
- Drabinski A, Williams G, Formica C. Observational evaluation of health state utilities among a cohort of sepsis patients. Value Health 2001;4:128-9. http://dx.doi.org/10.1046/j.1524-4733.2001.40202-165.x.
- Ara R, Brazier J. Populating an economic model with health state utility values: moving toward better practice. Value Health 2010;13:509-18. http://dx.doi.org/10.1111/j.1524-4733.2010.00700.x.
- Reference Cost Collection: National Schedule of Reference Costs – Year 2013–14 – NHS Trusts and NHS Foundation Trusts. London: Department of Health; 2015.
- British National Formulary. London: BMJ Group and Pharmaceutical Press; n.d.
- Stinnett AA, Mullahy J. Net health benefits: a new framework for the analysis of uncertainty in cost-effectiveness analysis. Med Decis Making 1998;18:S68-80.
- Chang S, Hsieh W, Liu T, Lee S, Wang C, Chou H, et al. Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis – a systemic review and meta-analysis. PLOS ONE 2013;8. http://dx.doi.org/10.1371/journal.pone.0062323.
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clin Chem 2003;49:1-6. http://dx.doi.org/10.1373/49.1.1.
- Reinhart K, Brunkhorst F, Bone H, Bardutzky L, Dempfle C-E, Forst H, et al. Prävention, Diagnose, Therapie und Nachsorge der Sepsis Erste Revision der S2k-Leitlinien der Deutschen Sepsis-Gesellschaft e.V. (DSG) und der Deutschen Interdisziplinären Vereinigung für Intensiv- und Notfallmedizin (DIVI). Anaesthesist 2010;59:347-70. http://dx.doi.org/10.1007/s00101-010-1719-5.
- Dellinger R, Levy M, Carlet J, Bion J, Parker M, Jaeschke R, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock. Intens Care Med 2008;34:17-60. http://dx.doi.org/10.1007/s00134-007-0934-2.
- Levy M, Fink M, Marshall J, Abrahm E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International sepsis definitions conference. Intens Care Med 2003;29:530-8. http://dx.doi.org/10.1007/s00134-003-1662-x.
- Goldstein B, Giroir B, Randolph A. Members of the International Consensus on Pediatric Sepsis . International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 2005;6:2-8. http://dx.doi.org/10.1097/01.PCC.0000149131.72248.E6.
Appendix 1 Literature search strategies for the review of clinical effectiveness: a MEDLINE example
| MEDLINE search strategy | Details |
|---|---|
| Database searched | Ovid MEDLINE In-Process & Other Non-Indexed Citations and Ovid MEDLINE |
| Platform or provider used | Ovid SP |
| Date of coverage | 1948 to May 2015 |
| Search undertaken | Initial search February 2015 |
| Updated search | May 2015 |
Search strategy
-
exp Sepsis/
-
sepsis.mp.
-
septic?emia.mp.
-
Shock, Septic/
-
((septic or endotoxic or toxic) adj shock).tw.
-
Bacteremia/
-
bacter?emia.mp.
-
Fungemia/
-
fung?emia.mp.
-
Systemic Inflammatory Response Syndrome/
-
sirs.mp.
-
blood$ infection$.tw.
-
blood poison$.tw.
-
or/1-13
-
septifast.mp.
-
lightcycler.mp.
-
15 or 16
-
14 and 17
-
sepsitest.mp.
-
iridica.mp.
-
(plex id or plex-id).mp.
-
or/19-21
-
exp Polymerase Chain Reaction/
-
polymerase chain reaction$.tw.
-
pcr$.mp.
-
Gene Amplification/
-
Nucleic Acid Amplification Techniques/
-
or/23-27
-
Genes, Bacterial/ or Genes, Fungal/
-
(exp bacteria/ or exp Fungi/) and exp Nucleic Acids/
-
((bacteri$ or fung$) adj3 (dna or gene$ or nucleic acid$)).tw.
-
blood culture$.tw.
-
or/29-32
-
14 and 28 and 33
-
18 or 22 or 34
-
Animals/ not (Humans/ and Animals/)
-
35 not 36
-
limit 37 to yr=“2006 –Current”
Appendix 2 The QUADAS-2 tool (adapted) for the methodological assessment of diagnostic studies51
| Quality domain | Scoring | Summary judgement |
|---|---|---|
| Risk of bias | ||
| Patient selection | ||
| Was a consecutive or random sample of patients enrolled? | ‘Yes’ if states consecutive or random ‘No’ if states another method of patient sampling/selection ‘Unclear’ if unclear or not reported |
Could the selection of patients have introduced bias? ‘Low risk’ if all domains are ‘yes’ ‘High risk’ if one or more domain is ‘no’ ‘Unclear risk’ anything in between |
| Was a case–control design avoided? | ‘Yes’ ‘No’ ‘Unclear’ if insufficient information provided |
|
| Did the study avoid inappropriate exclusions? | ‘Yes’ if the study provides explicit exclusion criteria and appropriately select participants that are typical of patients with bloodstream infection/suspected sepsis ‘No’ if the study has made inappropriate exclusions from the group it set out to select (i.e. unrepresentative of people with bloodstream infection/suspected sepsis) ‘Unclear’ if insufficient information provided |
|
| Index test | ||
| Were the index test results interpreted without knowledge of the results of the reference standard? | ‘Yes’ if index test was interpreted without knowledge (blind) of the results of the reference standard or the index test was clearly interpreted before the reference standard was known ‘No’ if results of reference standard were already known ‘Unclear’ if insufficient details are provided |
Could the conduct or interpretation of the index test have introduced bias? ‘Low risk’ if all domains are ‘yes’ ‘High risk’ if one or more domain is ‘no’ ‘Unclear risk’ anything in between |
| Reference standard | ||
| Is the reference standard likely to correctly classify the target condition? | ‘Yes’ if clinical standard described and is consistent with published standard operating procedures ‘No’ if reference standard falls short of standard operating procedures ‘Unclear’ if insufficient information provided |
Could the conduct or interpretation of the reference standard have introduced bias? ‘Low risk’ if all domains are ‘yes’ ‘High risk’ if one or more domain is ‘no’ ‘Unclear risk’ anything in between |
| Were the reference standard results interpreted without knowledge of the results of the index test? | ‘Yes’ if the reference standard was interpreted blind to the index test or the reference standard was clearly interpreted before the index test was known ‘No’ if the results of the index test were known ‘Unclear’ if insufficient information is provided |
|
| Flow and timing | ||
| Was there an appropriate interval between index test(s) and reference standard? | ‘Yes’ if reference standard and index tests performed on blood samples drawn at the same time ‘No’ if reference standard and index tests not performed on blood samples drawn at different times ‘Unclear’ if insufficient information is provided |
Could the patient flow have introduced bias? ‘Low risk’ if all domains are ‘yes’ ‘High risk’ if one or more domain is ‘no’ ‘Unclear risk’ anything in between |
| Did all patients receive a reference standard? | ‘Yes’ if all participants who received the index test also verified using the reference test ‘No’ if not all (or some) of the participants who received the index test also underwent the reference test (partially verified). If all participants did not receive the reference test, how many did not (of the total) ‘Unclear’ if insufficient information is provided |
|
| Did patients receive the same reference standard? | ‘Yes’ if the same reference test was used regardless of the index test results ‘No’ if different reference tests are used depending on results of the index tests. If different reference tests are used, what were the reasons and how many participants were involved? ‘Unclear’ if insufficient information is provided |
|
| Were all patients included in the analysis? | ‘Yes’ if all patients who were recruited/enrolled into the study were included in the analysis or if sufficient explanation is provided for any discrepancy ‘No’ if there are participants excluded from the analysis and no/insufficient explanation is given for any discrepancy ‘Unclear’ if insufficient information is given to assess whether or not any patients were excluded from the analysis |
|
| Applicability | ||
| Patients | ||
| Are there concerns that the included patients and settings do not match the review question? | Scored in relation to the description of included patients | ‘Yes’ if the sample is unrepresentative of people with bloodstream infection/suspected sepsis ‘No’ if characteristics of participants are well described and typical of patients with bloodstream infection/suspected sepsis ‘Unclear’ if characteristics are not well described |
| Index test | ||
| Is there concern that the index test, its conduct, or interpretation, differ from the review question (i.e. CE protocol followed)? | Scored in relation to the CE mark protocol for SeptiFast, SepsiTest and IRIDICA | ‘Yes’ if CE mark protocol for SeptiFast, SepsiTest and IRIDICA is not followed ‘No’ if CE mark protocol for SeptiFast, SepsiTest and IRIDICA is followed ‘Unclear’ if insufficient details provided |
| Reference standard | ||
| Is there concern that the target condition, as defined by the reference standard, does not match the review question? | Scored in relation to description of the reference standard | ‘Yes’ if full details of reference standard are not provided, for example the reference standard may be free of bias but the target condition that it defines may differ from the target condition specified in the review question ‘No’ if full details are provided ‘Unclear’ if insufficient details provided |
Appendix 3 Clinical effectiveness review: table of excluded studies with rationale
| Reference details | Reason for exclusion |
|---|---|
| Clinicaltrials.gov. Benefit of SeptiFast Multiplex PCR in the Etiologic Diagnosis and Therapeutic Approach for Onco-Hematology Patients Presenting Sepsis. 2010. URL: http://ClinicalTrials.gov/show/NCT00561639 (accessed April 2016) | Trial record with no study results |
| Clinicaltrials.gov. Diagnosis of Septicaemia by Detection of Microbial DNA in Blood in Severe Infections. 2011. URL: http://ClinicalTrials.gov/show/NCT00709358 (accessed April 2016) | Protocol: SeptiFast EVAMICA trial |
| Clinicaltrials.gov. Value of the LightCycler© SeptiFast Test MGRADE for the Pathogen Detection in Neutropenic Hematological Patients. 2012. URL: http://ClinicalTrials.gov/show/NCT01114165 (accessed April 2016) | Trial record of Idelevich et al.a |
| Clinicaltrials.gov. Evaluation in the Treatment of Nosocomial Sepsis Comparing Polymerase Chain Reaction with Conventional Blood Culture. 2013. URL: http://ClinicalTrials.gov/show/NCT01450358 (accessed April 2016) | Replaced (protocol) by full-text paper reported by Rodrigues et al.b |
| Clinicaltrials.gov. Optimal Antibiotic Treatment of Moderate to Severe Bacterial Infections. 2014. URL: http://ClinicalTrials.gov/show/NCT01338116 (accessed April 2016) | Ongoing SeptiFast trial – estimated completion January 2017 |
| Abbott Molecular Inc. IRIDICA BAC BSI Assay – Package Insert Ref: 08N22-010. Des Plaines, IL: Abbott Molecular Inc.; 2014 | Replaced (package insert) by full-text paper reported by Metzgar et al.c |
| Afsharpaiman S, Mamishi S, Pourakbari B, Siyadati A, Tabatabaee P, Khotaee G. Diagnosis of bacteremia using universal PCR in febrile ill children. Acta Med Iranica 2007;45:131–8 | Cultured samples (positive) |
| Al-Zahrani AKH, Ghonaim MM, Hussein YM, Eed EM, Khalifa AS, Dorgham LS, et al. Evaluation of recent methods versus conventional methods for diagnosis of early-onset neonatal sepsis. J Infect Develop Countries 2015;9:388–93 | Not intervention (test) of interest |
| Arabestani MR, Fazzeli H, Nasr Esfahani B. Identification of the most common pathogenic bacteria in patients with suspected sepsis by multiplex PCR. J Infect Develop Countries 2014;8:461–8 | Cultured samples (positive) |
| Avolio M, Diamante P, Zamparo S, Modolo M, Grosso S, Zigante P, et al. Molecular identification of bloodstream pathogens in patients presenting to the emergency department with suspected sepsis. Shock 2010;34:27–30 | Diagnostic metrics data included in Avolio et al.d (confirmed by authors) |
| Avolio M, Diamante P, Zamparo S, Modolo ML, Grosso S, Zigante P, et al. Evaluation of molecular detection of bloodstream pathogens in 144 patients arriving in the emergency room with clinical signs of sepsis. Clin Microbiol Infect 2010;16:S533–4 | Replaced (abstract) by full-text paper reported by Avolio et al.e |
| Baraki H, Al AA, Schilling T, Pichlmaier M, Martens A, Haverich A, et al. Are universal rRNA gene PCR and sequencing tests an alternative to conventional culture analysis for infected alloplastic implants? Thorac Cardiovasc Surg 2012;60:114 | Specimens from tissue samples only |
| Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program's impact. Clin Infec Dis 2010;51:1074–80 | Cultured samples (positive) |
| Bernaschi P, Ranno S, Lucignano B, Pizzorno B, Liesenfeld O, Menichella D. Value of multiplex-PCR (SeptiFast) for the diagnosis of bacterial and fungal pathogens in newborns and children with suspected sepsis. Clin Microbiol Infect 2010;16:S541 | Replaced (abstract) by full-text paper reported by Lucignano et al.f |
| Bilkovski R, Sampath R, Huiras M, Durtschi A, Ecker D, Chalfin D, et al. The Potential Clinical and Economic Value of Rapid Diagnosis of Suspected Bloodstream Infections using PCR/ESI-MS. 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 5–9 September 2014 | Replaced (abstract/poster) by full-text paper reported by Vincent et al.g |
| Bingold T, Just L, Roenneberg T, Sartorius S, Hunfeld K, Wissing H. Septifast allows more rapid detection of liver transplant patients being at risk of sepsis than blood culture or inflammatory parameters. Infection 2009;37:40 | Not target population (patients scheduled for orthotopic liver transplant) |
| Brealey D, Libert N, Pugin J, Chalfin D, Sampath R, Ecker D, et al. RADICAL Study: Rapid Diagnosis of Suspected Bloodstream Infections using PCR/ESI-MS. 54th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 5–9 September 2014 | Replaced (abstract/poster) by full-text paper reported by Vincent et al.g |
| Burdino E, Milia M, Milano R, Gregori G, Allice T, Ruggiero T, et al. Evaluation of the SEPTIFAST real-time PCR for rapid identification of blood pathogens in patients with suspected sepsis: an experience in a northwestern Italy hospital. Clin Microbiol Infect 2012;18:505 | Replaced (abstract) by full-text paper reported by Burdino et al.h |
| Cambau E, Courcol R, Veerabudun V, Bretagne S, Durand-Zaleski I, Bastuji-Garin S, et al. Does Performing DNA Detection in Blood Improves the Microbial Diagnosis of Severe Bloodstream Infections? First Results of the EVAMICA study. Proceedings of the 23rd European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 27–30 April 2013 | Insufficient information to allow calculation of diagnostic 2 × 2 table (SeptiFast EVAMICA trial) |
| Casalta JP, Gouriet F, Roux V, Thuny F, Habib G, Raoult D. Evaluation of the LightCycler SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur J Clin Microbiol 2009;28:569–73 | Not target population (patients with infective endocarditis) |
| Chaidaroglou A, Manoli E, Gkouziouta A, Gourzi P, Pantou M, Saroglou G, et al. Clinical impact of a multiplex real-time PCR assay (SeptiFast) for the rapid detection of pathogens in patients with end-stage heart failure bridged to heart transplantation with ventricular assist devices. Crit Care 2010;14:S13 | Not target population (implantable ventricular-assist device patients suspected of infection) |
| Chaidaroglou A, Manoli E, Gkouziouta A, Aggelaki D, Pantou M, Saroglou G, et al. The use of a multiplex real-time PCR assay for the detection of bacterial and fungal bloodstream infections in thoracic allograft recipients. J Heart Lung Transpl 2011;30(Suppl. 1):S146 | Not target population (thoracic allograft recipients) |
| Chaidaroglou A, Manoli E, Gkouziouta A, Kolovou V, Gourzi P, Pandou M, et al. The contribution of a multiplex real-time PCR assay (SeptiFast) for the rapid detection of bacterial and fungal bloodstream infections in patients with end-stage heart failure bridged to heart transplantation with ventricular assist devices. Transplantation 2012;94:945 | Not target population (implantable ventricular-assist device patients suspected of infection) |
| Chaidaroglou A, Manoli E, Gkouziouta A, Gourzi P, Pantou M, Kolovou V, et al. The contribution of a multiplex real-time PCR to detect bacterial and fungal bloodstream infections in a cohort of thoracic allograft recipients. Transplantation 2012;94:77 | Not target population (thoracic allograft recipients) |
| Chaidaroglou A, Manoli E, Marathias E, Gkouziouta A, Saroglou G, Alivizatos P, et al. Use of a multiplex polymerase chain reaction system for enhanced bloodstream pathogen detection in thoracic transplantation. J Heart Lung Transpl 2013;32:707–13 | Not target population (thoracic allograft recipients) |
| Chan KYY, Lam HS, Cheung HM, Chan AKC, Li K, Fok TF, et al. Rapid identification and differentiation of Gram-negative and Gram-positive bacterial bloodstream infections by quantitative polymerase chain reaction in preterm infants. Crit Care Med 2009;37:2441–7 | Cultured samples (positive/negative) |
| Clerici D, Mancini N, Forno B, Cappelli B, Mastaglio S, Messina C, et al. Molecular diagnosis by lightcycler? SeptiFast in the preemptive treatment of invasive fungal infections: a 5 cases report. Haematologica 2009;94:415 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Conen A, Schaub N, Achermann Y, Battegay M, Frei R, Trampuz A. Is multiplex PCR (SeptiFast) useful for diagnosis of infectious endocarditis? Clin Microbiol Infect 2009;15:S531–2 | Not target population (patients with suspected native or prosthetic valve infective endocarditis) |
| Dark P, Dunn G, Chadwick P, Young D, Bentley A, Carlson G, et al. The clinical diagnostic accuracy of rapid detection of healthcare-associated bloodstream infection in intensive care using multipathogen real-time PCR technology. BMJ Open 2011;1:e000181 | Replaced (protocol) by full-text paper reported by Warhurst et al.i |
| Diamante P, Avolio M, Zamparo S, Grosso S, Tosoni N, Zigante P, et al. [Molecular diagnosis of sepsis: the experience at the Pordenone hospital.] Riv Ital Medi Lab 2010;6:205–10 | Foreign language (Italian) |
| Disqué C, Kochem AJ, Mühl H, Lorenz MG, Sakka SG. PCR detection of sepsis-inducing pathogens in blood using SepsiTest™. Int J Med Microbiol 2008;298:7 | No outcome data |
| Disqué C, Sakka S, Wellinghausen N. Klinische Evaluation von SepsiTest™ zum universellen PCR-Nachweis von bakteriellen Erregern in Vollblut. Der Mikrobiologe Heft 1 2010;20:13–16 | Foreign language (German) |
| Disqué C, Mühl H, Gebert S, Winter J, Matten J, Wellinghausen N. Microbe detection in whole blood without blood culture. J Mol Diagn 2010;12:882 | Replaced (abstract) by full-text paper reported by Wellinghausen et al.j |
| Disqué C, Gebert S, Kochem AJ, Mühl H, Matten J, Sakka SG, et al. A Multicentre Study of Bacteraemia using a New Commercial Universal 16S rDNA PCR Test. 8th World Congress on Trauma, Shock, Inflammation and Sepsis, Munich, Germany, 9–13 March 2010 | Replaced (abstract) by full-text paper reported by Wellinghausen et al.j |
| Disqué C, Mühl H, Keim S, Lorenz MG. Automated extraction of microbial DNA from whole blood for the universal PCR detection of pathogens. Infection 2011;39:S115–16 | No comparator (study investigating the influence of blood volume) |
| Disqué C, Linow M, Murphy N. Broad-range microbial DNA isolation from clinical specimens for universal PCR diagnosis. J Mol Diagn 2012;14:687 | Specimens (liquid and tissue) from different body sites and mixed population |
| Disqué C, Keim S, Mühl H, Lorenz MG. DNA extraction from broad range of micro-organisms for molecular diagnosis. Clin Microbiol Infect 2012;18:775 | Specimens (liquid and tissue) from different body sites and mixed population) |
| Draz NI, Taha SE, Abou Shady NM, Abdel Ghany YS. Comparison of broad range 16S rDNA PCR to conventional blood culture for diagnosis of sepsis in the newborn. Egypt J Med Hum Genet 2013;14:403–11 | Not intervention (test) of interest |
| Dubská L, Vyskočilová M, Minaríková D, Jelínek P, Tejkalová R, et al. LightCycler SeptiFast technology in patients with solid malignancies: clinical utility for rapid etiologic diagnosis of sepsis. Crit Care 2012;16:404 | Not target population (patients with solid malignancy) |
| Elwan AE, Zarouk WA. Diagnosis of neonatal bacterial sepsis by polymerase chain reaction. J Biol Sci 2009;9:533–40 | Not intervention (test) of interest |
| Enomoto M, Morioka I, Morisawa T, Yokoyama N, Matsuo M. A novel diagnostic tool for detecting neonatal infections using multiplex polymerase chain reaction. Neonatology 2009;96:102–8 | Not intervention (test) of interest |
| Gosiewski T, Jurkiewicz-Badacz D, Sroka A, Brzychczy-Wloch M, Bulanda M. A novel, nested, multiplex, real-time PCR for detection of bacteria and fungi in blood. BMC Microbiol 2014;14:144 | Not intervention (test) of interest |
| Greco R, Clerici D, Mancini N, Clementi M, Lorentino F, Crucitti L, et al. Multiplex PCR-based assay (SeptiFast) for rapid detection of pathogens in febrile neutropenia: results in 273 consecutive patients. Bone Marrow Transplant 2012;47:S77–8 | Replaced (abstract) by full-text paper reported by Barbanti et al.k |
| Greco R, Mancini N, Lorentino F, Crucitti L, Barbanti C, Forcina A, et al. Rapid molecular detection of pathogens in 1941 blood samples from 516 consecutive patients with febrile neutropenia. Bone Marrow Transplant 2014;49:S334 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Grif K, Heller I. Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis. J Clin Microbiol 2012;50:2250–4 | Specimens (liquid and tissue) from different body sites |
| Haag H, Locher F, Nolte O. Molecular diagnosis of microbial aetiologies using SepsiTest™ in the daily routine of a diagnostic laboratory. Diagn Microbiol Infect Dis 2013;76:413–18 | Specimens (liquid and tissue) from different body sites |
| Halasz E, Petro M, Bojtos I, Myszoglad R, Simon J. Rapid diagnosis of sepsis with molecular biological methods. Clin Chem Lab Med 2012;50:eA32–3 | Insufficient information to allow calculation of diagnostic 2 × 2 table (not a diagnostic study) |
| Halliday CL, Sorrell TC, Chen SCA. Detection of multiple fungal species in blood samples by real-time PCR: an interpretative challenge. J Clin Microbiol 2014;52:3515–16 | Letter/comment with no details of intervention |
| Hettwer S, Wilhelm J, Hammer D, Schürmann M, Amoury M, Scheubel S, et al. Sepsis in the emergency department: pathogen identification by blood cultures and PCR. Crit Care 2009;13:S154 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Holmes C, Kirk-Granger H, Perera N. The Clinical Usefulness of Multiplex PCR for the Detection of Bacteria and Fungi in the Blood of Patients with Haematological Malignancies. Proceedings of the 24th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 10–13 May 2014 | Not target population (haematological malignancies) |
| Horvath A, Peto Z, Urban E, Vagvolgyi C, Somogyvari F. A novel, multiplex, real-time PCR-based approach for the detection of the commonly occurring pathogenic fungi and bacteria. BMC Microbiol 2013;13:300 | Not intervention (test) of interest |
| Idelevich EA, Niederbracht Y, Tafelski S, Nachtigall I, Berdel WE, Peters G, et al. Clinical value of the SeptiFast Multiplex PCR test in hematologic patients with neutropenic fever or sepsis: interim study results. Int J Med Microbiol 2011;301:7 | Replaced (abstract) by full-text paper reported by Idelevich et al.l |
| Irwin AD, Barton T, Grant A, Williams R, Carrol ED. SepsiTest molecular diagnosis of bacteraemia in febrile paediatric patients. Clin Microbiol Infect 2012;18:505 | Not target population (patients with increased levels of C-reactive protein) |
| Jordan JA, Durso MB, Butchko AR, Jones JG, Brozanski BS, Durso MB, et al. Evaluating the near-term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing. J Mol Diagn 2006;8:357–63 | Not intervention (test) of interest |
| Jordana-Lluch E, Carolan HE, Giménez M, Sampath R, Ecker DJ, Quesada D, et al. Rapid diagnosis of bloodstream infections with PCR followed by mass spectrometry. PLOS ONE 2013;8:e62108 | Not intervention (test) of interest (used older extraction method on PLEX-ID system, thus not comparable to the current IRIDICA platform) |
| Josefson P, Stralin K, Ohlin A, Ennefors T, Dragsten B, Andersson L, et al. Evaluation of a commercial multiplex PCR (SeptiFast) in the aetiological diagnosis of community-acquired bloodstream infections. Clin Microbiol Infect 2010;16:S541–2 | Replaced (abstract) by full-text paper reported by Josefson et al.m |
| Kalenka A, Timm J, Schmid S, Beck G. Value of LightCycler SeptiFast in detection of ventilator-associated pneumonia. Intens Care Med 2009;35:S197 | Not target population (patients with suspected ventilator associated pneumonia) |
| Kalenka A, Schmid S, Timm J, Beck G. Lightcycler SeptiFast as a tool to enhance the detection of bacteremia and fungemia in patients with intraabdominal infection during antimicrobiological therapy. Intens Care Med 2009;35:S196 | No comparator (i.e. not vs. blood culture) |
| Kaleta EJ, Clark AE, Johnson DR, Gamage DC, Wysocki VH, Cherkaoui A, et al. Use of PCR coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J Clin Microbiol 2011;49:345–53 | Cultured samples (positive) |
| Karam El Din ZA, Mokhtar E, El-Shereef A, Abd El KA, Al-Tounisy A. Rapid diagnosis of neonatal sepsis caused by yeast infection. Mycoses 2012;55:75 | Not intervention (test) of interest |
| Kim B, Park S, Kim T, Kim J, Rim D, Choi T, et al. [Clinical efficacy evaluation of multi-parameter realtime polymerase chain reaction for the central venous catheter-related blood stream infection.] Infect Chemother 2011;43:240–4 | Foreign language (Korean) |
| Kuhn C, Disqué C, Mühl H, Orszag P, Stiesch M, Haverich A. Molecular diagnosis of the etiological agents of infectious endocarditis using commercial universal rRNA gene PCR plus sequencing tests. Infection 2011;39:S124–25 | Replaced (abstract) by full-text paper reported by Kuhn et al.n |
| Kuhn C, Disqué C, Mühl H, Orszag P, Stiesch M, Haverich A. Evaluation of commercial universal rRNA gene PCR plus sequencing tests for identification of bacteria and fungi associated with infectious endocarditis. J Clin Microbiol 2011;49:2919–23 | Not target population (patients with suspected infectious endocarditis and used valvular and blood samples for analysis) |
| Lefort A, Chartier L, Sendid B, Wolff M, Mainardi J, Podglajen I, et al. Diagnosis, management and outcome of Candida endocarditis. Clin Microbiol Infect 2012;18:E99–109 | Not target population (patients with Candida endocarditis) |
| Lehmann LE, Alvarez J, Hunfeld KP, Goglio AP, Kost GJ, Louie RF, et al. Model analysis of clinical utility of PCR in microbiological testing for sepsis. Infection 2009;37:44 | Not intervention (test) of interest |
| Leli C, Cardaccia A, D'Alo F, Ferri C, Bistoni F, Mencacci A. A prediction model for real-time PCR results in blood samples from febrile patients with suspected sepsis. J Med Microbiol 2014;63:649–58 | Study aim to develop prediction model from positive SepsitFast results only |
| Liberto MC, Puccio R. Applications of LightCycler Staphylococcus M-GRADE assay to detect Staphylococcus aureus and coagulase-negative staphylococci in clinical blood samples and in blood culture bottles. Infezioni in Medicina 2006;14:71–6 | Not intervention (test) of interest |
| Liu CL, Ai HW, Wang WP, Chen L, Hu HB, Ye T, et al. Comparison of 16S rRNA gene PCR and blood culture for diagnosis of neonatal sepsis. Arch Pediatrie 2014;21:162–9 | Not intervention (test) of interest |
| Lodes U, Lippert H. [Molecular biological sepsis diagnostic using multiplex PCR in surgical intensive care as suitable alternative to conventional microbial culture – a representative overview.] Zentralbl Chir 2011;136:135–42 | Foreign language (German) |
| Markota A, Golle A, Sinkovic A. Polymerase chain reaction analysis in patients with sepsis. Intens Care Med 2013;39:S453 | Diagnostics metrics data (from patients and samples) included in a full-text study by Markota et al.o |
| Martinez MDM, Arredondo AR, Alvarez EM, Prieto AMP, Caballero MP, Mari JMN. Lightcycler septifast trading system in molecular microbiological diagnosis of neonatal sepsis. J Matern-Fetal Neonat Med 2010;23(Suppl. 1):156 | Coagulase-negative staphylococci detection only |
| Mencacci A, Leli C, Montagna P, Cardaccia A, Meucci M, Bietolini C, et al. Diagnosis of infective endocarditis: comparison of the LightCycler SeptiFast real-time PCR with blood culture. J Med Microbiol 2012;61:881–3 | Not target population (patients with suspected infective endocarditis) |
| Mencacci A, Leli C, Cardaccia A, Meucci M, Moretti A, D'Alo F, et al. Procalcitonin predicts real-time PCR results in blood samples from patients with suspected sepsis. PLOS ONE 2012;7:e53279 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Merisescu M, Luminos M, Jugulete G, Florea D, Streinu-Cercel A. Molecular diagnosis of severe bacterial sepsis in children. Crit Care 2012;16:67 | Cultured samples (blood and/or fluids) |
| Merisescu M, Jugulete G, Streinu-Cercel A, Luminos M. Plex Id role in the diagnosis of acute bacterial sepsis with E-coli in children. Pediatr Crit Care Med 2014;15(Suppl. 1):150–1 | Klebsiella pneumoniae detection only (no details on sample type, comparator methods or useable outcome data) |
| Meyer T, Franke G, Polywka S, Lutgehetmann M, Gbadamosi J, Magnus T, et al. Improved detection of bacterial central nervous system infections by use of a broad-range PCR assay. J Clin Microbiol 2014;52:1751–3 | Specimens from cerebrospinal fluid samples |
| Molina JM, Cordoba J, Ramirez P, Gobernado M. [Automatic detection of bacterial and fungal infections in blood.] Enferm Infect Microbiol Clin 2008;26(Suppl. 9):75–80 | Foreign language (Spanish) |
| Mongelli G, Romeo MA, Denaro C, Gennaro M, Fraggetta F, Stefani S. An added value of multi-pathogen probe-based real-time PCR SeptiFast in the rapid diagnosis of bloodstream infections in patients with bacteremia. J Med Microbiol 2015;64:670–5 | Not target population (febrile patients with suspected bacteraemia) |
| Moore MS, McCann CD, McCarroll M, May L, Jordan JA. Direct detection of bacteria from blood of ED and ICU patients being evaluated for bloodstream infection. J Mol Diagn 2014;16:727 | Not intervention (test) of interest |
| Mundy L, Hiller JE. Rapid Molecular Assays for the Diagnosis of Sepsis and Identification of Sepsis Causing Pathogens. Adelaide, SA: Adelaide Health Technology Assessment (AHTA). Horizon Scanning Prioritising Summary. 2010. URL: www.horizonscanning.gov.au/internet/horizon/publishing.nsf/Content/C8A5BA60BD01A93ECA257757000A2015/$File/Volume_27_June_2010_sepsis.pdf (accessed April 2016) | Review |
| Niederbracht Y, Idelevich EA, Penner H, Berdel WE, Peters G, Silling G, et al. Applicability of a commercial multiplex PCR test for identification of true blood stream infections with coagulase-negative staphylococci in neutropenic hematological patients. Int J Med Microbiol 2013;303:13 | Coagulase-negative staphylococci detection only |
| Nieman A, Rozemeijer W, de Jong E, Beishuizen B, van Agtmael M, Savelkoul P, et al. Molecular detection of bacterial bloodstream infections: validation of the SepsiTest™ assay. Ned Tijdschr Med Microbiol 2010;18:109 | Preclinical validation study |
| Nieman A, De Jong E, Beishuizen B, Koek A, Savelkoul PH, Schade RP. Molecular detection of bacterial bloodstream infections: the SepsiTest assay. Clin Microbiol Infect 2011;17:S729 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Novak-Frazer L, Rautemaa-Richardson R, Denning D, Bowyer P. Multiplex PCR for the rapid diagnosis of skin/soft tissue and bloodstream infections. Clin Microbiol Infect 2012;18:122–3 | Specimens from whole blood and wound swabs (and insufficient information to allow calculation of diagnostic 2 × 2 table) |
| Ohlin A, Backman A, Ewald U, Schollin J, Bjorkqvist M. Diagnosis of neonatal sepsis by broad-range 16S real-time polymerase chain reaction. Neonatology 2012;101:241–6 | Not intervention (test) of interest |
| Orszag P, Disqué C, Keim S, Lorenz M, Wiesner O, Hadem J, et al. Monitoring of patients supported by extracorporeal membrane oxygenation for systemic infections by broad-range rRNA gene PCR amplification and sequence analysis. J Clin Microbiol 2013;52:307–11 | Not target population (patients supported by extracorporeal membrane oxygenation) |
| Ortiz Ibarra F, Reyna J, Trevino P, Fernandez L, Lara G, Valenzuela E, et al. A standardized protocol for the multiplex PCR technique Septifast Roche for neonatal samples with suspected sepsis. Crit Care 2012;16:79 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Palomares JC, Puche B, Martos A, Lucena F, Marin M, Martin-Mazuelos E. Rapid molecular diagnosis of severe sepsis in patients with SIRS. Clin Microbiol Infect 2009;15:S529–30 | Replaced (abstract) by full-text paper reported by Palomares et al.p |
| Pleskova T, Greplova K, Bacikova L, Dubska L. What is the added value of molecular microbiology in sepsis? Clin Chem Lab Med 2011;49:S610 | Cultured samples (positive) and unclear if cancer patients have sepsis |
| Popov DA, Vostrikova TI. [The first experience of application of PCR techniques in real-time mode to diagnose bacteriemia during postoperational period in cardiosurgery patients.] Klin Labor Diagn 2011;8:49–52 | Foreign language (Russian) |
| Raineri SM, Canzio D, Sarno C, Cascio ND, Mineo G, Chiaramonte R, et al. LightCycler SeptiFast in early diagnosis of sepsis: our experience. Crit Care 2009;13:S153 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Ratanarat R, Cazzavillan S, Ricci Z, Rassu M, Segala C, de Cal M, et al. Usefulness of a molecular strategy for the detection of bacterial DNA in patients with severe sepsis undergoing continuous renal replacement therapy. Blood Purificat 2007;25:106–11 | Not intervention (test) of interest |
| Reier-Nilsen T, Farstad T, Nakstad B, Lauvrak V, Steinbakk M. Comparison of broad range 16S rDNA PCR and conventional blood culture for diagnosis of sepsis in the newborn: a case control study. BMC Pediatr 2009;9:5 | Not intervention (test) of interest |
| Rogina P, Skvarc MM, Stubljar DD. Diagnostic utility of broad range bacterial 16S rRNA gene PCR with degradation of human and free bacterial DNA in bloodstream infection is more sensitive than an in-house developed PCR without degradation of human and free bacterial DNA. Mediators Inflamm 2014;2014:108592 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Sahre H, Vogel S, Park JW, Weber S. [Sepsis-diagnostics with help of SeptiFast®-systems: experience on a internal intensive care unit.] Infection 2007;35:9–10 | Foreign language (German) |
| Sakka S, Wellinghausen N, Kochem AJ, Disqué C, Muehl H, Gebert S, et al. A multicentre study of bacteraemia using a new commercial universal 16S rDNA PCR test. Inflamm Res 2010;59:s137 | Replaced (abstract) by full-text paper reported by Wellinghausen et al.j |
| Sampath R. Advanced techniques for detection and identification of viral contaminants using the Ibis PLEX-ID universal biosensor. Pda J Pharm Sci Tech 2011;65:690 | Conference abstract not available |
| Santolaya ME, Farfan MJ, De La Maza, Farfan MJ. Diagnosis of bacteremia in febrile neutropenic episodes in children with cancer: microbiologic and molecular approach. Pediatr Infect Dis J 2011;30:957–61 | Not intervention (test) of interest |
| Schaub N, Boldanova T, Noveanu M, Arenja N, Hermann H, Twerenbold R, et al. Incremental value of multiplex real-time PCR for the early diagnosis of sepsis in the emergency department. Swiss Med Wkly 2014;144:w13911 | Diagnostic metrics data included in Schaub et al.q |
| Shaat SS, El Shazly SA, Badr Eldin MM, Barakat SS, Hashish MH. Role of polymerase chain reaction as an early diagnostic tool for neonatal bacterial sepsis. J Egypt Public Health Assoc 2013;88:160–4 | Not intervention (test) of interest |
| Sitnik R, Marra A, Petroni R, Ramos O, Martino M, Pasternak J, et al. SeptiFast for diagnosis of sepsis in severely ill patients from a Brazilian Hospital. J Med Diagn 2011;13:736 | Replaced (abstract) by full-text paper reported by Sitnik et al.r |
| Skvarc M, Stublar D, Rogina P. Broad-range 16S rRNA gene PCR using SepsiTest in conjunction with valid clinical data and sepsis biomarkers improve sepsis diagnostics. Int J Med Microbiol 2012;302:6 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Steinmann J, Buer J, Paul A, Saner F, Rath PM. Diagnostic performance of multiplex PCR for diagnosis of bloodstream infections in liver transplant recipients with suspected sepsis. Int J Med Microbiol 2011;301:51 | Replaced (abstract) by full-text paper reported by Rath et al.s |
| Stubljar D, Rogina P, Skvarc M, Pavlovic A. Diagnostic Accuracy of sCD14 (Presepsin) is Comparable to Procalcitonin (PCT) for the Diagnosis of Bacterial Infections in Critically Ill Patients. Proceedings of the 23rd European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 27–30 April 2013 | Not intervention (test) of interest |
| Tafelski S, Nachtigall I, Idelevich E, Silling G, Becker K, Faust J, et al. Impact of Septi-Fast for Pathogen Detection in Critical-Care Patients with Sepsis: Results from a Randomized Controlled Clinical Trial. Proceedings of the 23rd European Congress of Clinical Microbiology and Infectious Diseases, Berlin, Germany, 27–30 April 2013 | Replaced (abstract) by full-text paper reported by Tafelski et al.t |
| Torres-Martos E, Pérez-Ruiz M, Pedrosa-Corral I, Peña-Caballero M, Jiménez-Valera MM, Pérez-Ramirez MD, et al. [Evaluation of the LightCycler SeptiFast test in newborns and infants with clinical suspicion of sepsis.] Enferm Infecc Microbiol Clin 2013;31:375–9 | Foreign language (Spanish) |
| Tsalik EL, Jones D, Nicholson B, Caram LB, Liesenfeld O, Fowler VG, et al. Detection of bacterial and fungal pathogens associated with sepsis in patients presenting to the emergency room. Intens Care Med 2009;35:S196 | Insufficient information to allow calculation of diagnostic 2 × 2 table |
| Vrsajkov V, Pejakovic J, Dragic Z, Radanovic B. Pathogen identification in septic patients: clinical importance of PCR and blood cultures. Intens Care Med 2014;40(Suppl. 1):259 | Not intervention (test) of interest |
| Warhurst G, Dunn G, Chadwick P, Blackwood B, McAuley D, Perkins GD, et al. Rapid detection of health-care-associated bloodstream infection in critical care using multipathogen real-time polymerase chain reaction technology: a diagnostic accuracy study and systematic review. Health Technol Assess 2015;19(35) | Replaced (Health Technology Assessment monograph) by full-text journal paper reported by Warhurst et al.i |
| Zerweck A, Raepple D, Finck A, Finke J, Bertz H. PCR-based molecular diagnostic tool (Septifast) in septic patients undergoing induction therapy or stem cell transplantation in haematological malignancies. Bone Marrow Transplant 2010;45:S228 | Not target condition (patients undergoing induction therapy and or stem cell transplantation) |
| Ziegler I, Josefson P, Olcen P, Molling P, Stralin K. Quantitative data from the SeptiFast real-time PCR is associated with disease severity in patients with sepsis. BMC Infect Dis 2014;14:155 | Diagnostic metrics data (secondary analysis) included in Josefson et al.m |
Appendix 4 Study and population characteristics of the included studies
| Author (year); country; source of funding; manuscript type | Study design; study type; sampling method; sampling period | Department and wards providing care for acutely unwell patients or critical care units | Clinical setting | Inclusion criteria (illness aetiology) | Exclusion criteria |
|---|---|---|---|---|---|
| Single index test studies: SeptiFast | |||||
| Dierkes et al. (2009)64 Germany Sponsor/funding: NR. However, SeptiFast provided free of charge by Roche Full text |
Single gate Retrospective study Sampling method: NR 8 months (July 2006–March 2007) |
Surgical and medical wards | Intensive/critical care | Inclusion criteria: NR (chart review) | NR |
| Raglio et al. (2006)44 European multicentre Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
NR | NR | Reported that patients fulfilling SIRS criteria were included | NR |
| Bingold et al. (2007)59 Germany Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Anaesthesiological/surgical ICU | Intensive/critical care | Reported that significant elevated inflammatory parameters (PCT, IL-6, LBP) at study entry; informed consent were patients included | NR |
| Klemm et al. (2007)60 Germany Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Intensive care | Intensive/critical care | Reported that clinical suspicion for sepsis [fever (> 38 °C) or hypothermia (< 36 °C) and additionally leucocytosis (> 12.000 µl), leucopenia (< 4.000 µl), leucocyte left shift (> 10%), tachycardia (> 90 beats per minute), tachypnoea (> 20 breaths per minute) or hyperventilation (PaCO2 < 33 mmHg)] were patients included | NR |
| Lodes et al. (2008)61 Germany Sponsor/funding: NR Abstract |
Single gate Study type: NR Consecutive sample Sampling period: NR |
Surgical intensive care unit | Intensive/critical care | Reported that surgical patients with SIRS on intensive care were included | NR |
| Louie et al. (2008)46 USA Roche Diagnostics and a grant from the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health Full text |
Single gate Prospective study Sampling method: NR 34 months |
University medical centre | Emergency department, in hospital and intensive/critical care | Inclusion criteria: adults from emergency department, intensive care unit and general medicine with suspected bloodstream infection and at least two SIRS criteria | NR |
| Mancini et al. (2008)45 Italy Roche Full text |
Single gate Study type: NR Consecutive sample 2 months |
Haematology unit | In hospital and unclear if intensive/critical care | Reported that 73 (70.9%) samples were drawn from heavily neutropenic patients | NR |
| Vince et al. (2008)62 Croatia, two centres Partly supported by Ministry of Science, Education and Sports of the Republic of Croatia Correspondence |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
ICU (n = 17), outside ICU (n = 9) and Department of Haematology following bone marrow or peripheral blood stem cell transplantation (n = 10) | In hospital and intensive/critical care | Reported that patients with a clinical diagnosis of sepsis who were treated with antimicrobial therapy were included | NR |
| Dark et al. (2009)63 UK Sponsor/funding: NR Correspondence |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Samples from neuro-injured, general surgical and general internal medicine patients | Intensive/critical care | Reported that patients with new episodes of suspected bloodstream infection were included | NR |
| Gimeno et al. (2009)65 Spain, single centre Sponsor/funding: NR Abstract |
Single gate Prospective Sampling method: NR 5 months (dates NR) |
NR | NR | Haematological patients with febrile neutropenia | NR |
| Lehmann et al. (2009)66 Germany, five centres Supported, in part, by Roche Diagnostics Full text |
Single gate Retrospective study Sampling consecutive Sampling period: NR |
ICU, emergency room, medical and surgical wards | Emergency department, in hospital and intensive/critical care | Included patients were aged ≥ 18 years, suspected sepsis, a blood culture drawn and subsequent antibiotic treatment initiation or change | NR |
| Lodes et al. (2009)67 Germany Sponsor/funding: NR Full text |
Single gate Prospective study Consecutive sample 4 months (May–August 2006) |
Surgical ICU | Intensive/critical care | Surgical patients with SIRS and subsequent need of intensive care were included | NR |
| Palomares et al. (2009)68 Spain, single centre Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
ICU | Intensive/critical care | Reported that patients with suspected bloodstream infection and two SIRS were included | NR |
| Paolucci et al. (2009)69 Italy Sponsor/funding: NR Correspondence |
Single gate Retrospective study Sampling method: NR Sampling period: NR |
NR | NR | Reported that patients included were newborns aged > 3 days with late-onset sepsis | NR |
| Varani et al. (2009)70 Italy, three centres Sponsor/funding: NR Full text |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Paediatric oncology and haematology unit | In hospital and unclear if intensive/critical care | Reported that immunocompromised patients with haematological malignancies in whom sepsis was suspected were included | NR |
| von Lilienfeld-Toal et al. (2009)71 Germany, single centre Roche Molecular Diagnostics (all reagents, instruments and disposables were obtained from Roche Molecular Diagnostics) Full text |
Single gate Prospective Sampling method: NR 16 months (September 2001–February 2002; April 2003–January 2004) |
Tertiary care hospital haematology ward | In hospital | NR (included patients with febrile neutropenia after chemotherapy for haematological malignancies) | NR |
| Westh et al. (2009)72 Germany, multicentre (n = 6) Research funding from Roche Diagnostics Full text |
Single gate Study type: NR Sampling method: NR 5 months (June–October 2004) |
NR | NR | Specific inclusion criteria NR (all patients included were clinically suspected to have bacterial or fungal sepsis) | NR |
| Berger et al. (2010)73 Austria Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Neonatology | Neonatal unit | Very low-birthweight infants | NR |
| Bloos et al. (2010)74 Germany, France Roche Full text |
Single gate Prospective study Sampling method: NR December 2005–April 2007 |
Intensive care | Intensive/critical care | Presence of severe sepsis or septic shock according to the ACCP/SCCM consensus criteria | Exclusion criteria: aged < 18 years or previous enrolment in this trial |
| Lamoth et al. (2010)75 Switzerland, single centre Roche Diagnostics Full text |
Single gate Prospective Consecutive 14 months (September 2006–November 2007) |
University hospital isolation ward | In hospital | NR (included febrile neutropenic adult haematological patients undergoing induction or consolidation chemotherapy for acute leukaemia or autologous haematopoietic stem cell transplantation) | NR |
| Lehmann et al. (2010)76 Germany, two centres Research funding, reagents, and equipment from Roche Diagnostics Full text |
Single gate Prospective study Sampling method: NR Sampling period: NR |
Surgical ICUs | Intensive/critical care | Reported that all adult patients who were clinically suspected of suffering from severe sepsis of bacterial or fungal origin were included. Inclusion followed after independent decision of the physician in charge to call for a blood culture | NR |
| Maubon et al. (2010)77 France, single centre Roche Full text |
Single gate Prospective study Consecutive sample 12 months |
Teaching hospital | In hospital and unclear if intensive/critical care | Reported that patients with solid or haematological malignancies were admitted for suspected infection with at least one sign of sepsis, with or without organ dysfunction were included | NR |
| Regueiro et al. (2010)78 Spain, single centre Partly supported by the Spanish Ministerio de Ciencia e Innovacion Full text |
Single gate Study type: NR Sampling method: NR 13 months (May 2007–May 2008) |
Intensive care and anaesthesiology services | In hospital and intensive/critical care | Reported that patients who all met criteria for SIRS and suspected sepsis on admission were included | NR |
| Soki et al. (2010)79 Hungary, single centre Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
ICU and haematology department | In hospital and intensive/critical care | Reported that patients displaying symptoms of sepsis with or without antibiotic therapy were included | NR |
| Tsalik et al. (2010)80 USA, two centres National Institutes of Health grant and Roche Full text |
Single gate Study type: NR Sampling method: NR 66 months |
University medical centre emergency department (trauma centre) and Veterans Affairs Medical Centre | Emergency department | Patients were considered for inclusion in the study if they had a known or suspected infection on the basis of clinical data at the time of screening, and if they exhibited two or more signs of SIRS within a 24-hour period | Patients were excluded if they were aged < 18 years, if they had an imminently terminal comorbid condition or if they were participating in an ongoing clinical trial. Only subjects admitted to the hospital and for whom blood culture results were available were included in this analysis |
| Wallet et al. (2010)81 France, single centre Sponsor/funding: NR Roche Molecular Diagnostics provided materials to perform the study Full text |
Single gate Consecutive study Sampling method: NR 6 months |
ICU | Intensive/critical care | All patients with fever (≥ 38 °C) or hypothermia (≤ 36 °C) were eligible | NR |
| Yanagihara et al. (2010)82 Japan, multicentre (n = 3) Research funding, reagents and equipment from Roche Diagnostics Full text |
Single gate Prospective study Sampling method: NR 1 year (May 2007–April 2008) |
Departments of Surgery, Haematology, Emergency, Cardiopulmonary and ICU | In hospital and emergency department | Reported that patients (treated or untreated) with SIRS caused by bacterial or fungal infection, and for whom blood culture was considered to be required for identification of the causative pathogens were included | NR |
| Bravo et al. (2011)83 Spain Sponsor/funding: NR Full text |
Single gate Study type: NR Non-consecutive sample 8 months (February–September 2009) |
Medical and surgical ICU | In hospital and intensive/critical care | Development of a febrile episode in neutropenic or ICU patients that required hospital admission, or occurred during hospital stay | Receipt of empirical antibiotic treatment prior to blood sampling for analysis |
| Hettwer et al. (2011)84 Germany Roche Full text |
Single gate Prospective study Sampling method: NR August 2006–March 2009 |
Emergency department | Emergency department | Reported that patients aged > 18 years and admitted with clinical signs prompting physician to draw blood culture were included | NR |
| Josefson et al. (2011)85 Sweden, single centre Research grant: County Council of Örebro. Reagents and technical assistance: Roche Diagnostics Full text |
Single gate Prospective study Consecutive sample 1 year (October 2007–September 2008) |
Department of Infectious Diseases | In hospital | Reported that all patients who were subjected to blood culture at the department and gave their informed consent were included | Patients with HIV and with hepatitis B and C infections |
| Lucignano et al. (2011)86 Italy, single centre Sponsor/funding: NR Full text |
Single gate Retrospective study Sampling method: NR 26 months |
ICUs and surgery; oncology, haematology and neonatology; emergency department and paediatrics | In hospital and intensive/critical care | Reported that patients with clinical suggestion of SIRS with suspected bacterial or fungal infection, availability of a filled-out questionnaire with demographic, clinical and laboratory information, and collection of paired blood samples for SeptiFast and two blood samples for cultures from a peripheral vein or a central venous line were included | NR |
| Obara et al. (2011)87 Japan Partly funded by Roche Full text |
Single gate Study type: NR Consecutive sample 6 months (September 2004–March 2005) |
University hospital | Emergency department, in hospital and intensive/critical care | Reported that patients with suspected bacterial/fungal infection and at least two criteria of SIRS were included | NR |
| Vrioni et al. (2011)88 Greece, two centres Sponsor/funding: NR Abstract |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
NR | NR | Reported that patients with presumed sepsis in ICU were included | NR |
| Alvarez et al. (2012)89 Spain Sponsor/funding: NR Full text |
Single gate Retrospective study Sampling method: NR (cost-minimisation study) 12 months |
ICU | ICU | Patients with a diagnosis of severe sepsis or septic shock were included | NR |
| Grif et al. (2012)90 Austria Pfizer Full text |
Single gate Prospective study Consecutive sample 3 months (January 2009–March 2009) |
ICU and general wards | In hospital and intensive/critical care | Reported that patients with presumed sepsis (> 2 SIRS criteria) were included | NR |
| Guido et al. (2012)91 Italy Sponsor/funding: NR Full text |
Single gate Study type: NR Consecutive sample 12 months (January 2010–December 2010) |
Haematology department | In hospital and unclear if intensive/critical care | Reported that patients with febrile neutropenia (temperature > 38.0 °C) and neutrophil count < 0.5 × 109/L in presence of acute and/or chronic blood disorders or bone marrow transplant were included | NR |
| Lodes et al. (2012)92 Germany, single centre Sponsor/funding: NR Full text |
Single gate Study type: NR Consecutive sample 20 months |
ICU, Department of Surgery | Intensive/critical care | Reported that patients in ICU with clinical diagnosis of sepsis within last 24 hours at risk of abdominal sepsis with non-threatening SIRS were included | NR |
| Mauro et al. (2012)93 Italy, single centre Sponsor/funding: NR Full text |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
Department of Paediatric Oncology and Department of Internal Medicine | In hospital and unclear if intensive/critical care | Reported that immunocompromised patients, which was defined as patients with any of the following: neutropenia (neutrophil count <1 × 103/µl), exposure to immunosuppressive agents, haematological malignancy, or solid tumour, were included | NR |
| Pasqualini et al. (2012)94 Italy, single centre Sponsor/funding: NR Full text |
Single gate Study type: NR Consecutive 5 months |
Department of Internal Medicine | In hospital and unclear if intensive/critical care | Reported that patients suspected of having SIRS caused by bacterial or fungal infection, and for whom blood culture was performed for causative pathogen identification were included | NR |
| Rath et al. (2012)95 Germany, single centre Sponsor/funding: NR Full text |
Single gate Prospective study Sampling method: NR 24 months (May 2009–April 2011) |
Department of General, Visceral and Transplant Surgery | Intensive/critical care | Reported that ICU patients with suspected sepsis according to the criteria of the ACCP/SCCM were included | NR |
| Tschiedel et al. (2012)96 Germany, multicentre Sponsor/funding: NR Full text |
Single gate Retrospective study Sampling method: NR 19 months (May 2009–December 2010) |
Different hospitals | In hospital and intensive/critical care | Reported that critically ill patients with symptoms of systemic infection were included | NR |
| Herne et al. (2013)97 Estonia Sponsor/funding: NR Full text |
Single gate Retrospective study Sampling method: NR March 2007–July 2011 |
Acute and intensive care | In hospital and intensive/critical care | Reported that patients with severe infection from intensive care or other parts of hospital were included. Patients with clinically proven sepsis or septic shock, or severe infection without known etiologic agent, were included. Patients with only blood culture or SeptiFast collected samples were included | NR |
| Kasper and Altiok (2013)98 Austria, Roche Molecular Diagnostics provided materials to perform the study Full text |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
NR | NR | Specific inclusion criteria: NR (very low-birthweight infants when infection was suspected after 72 hours of life were included) | NR |
| Paolucci et al. (2013)99 Italy, single centre The University of Bologna, the Italian Ministry of Education, University and Research (MIUR) Full text |
Single gate Prospective Consecutive 22 months (June 2008–March 2010) |
Haematology and the Paediatric Oncology and Haematology Unit, St Orsola-Malpighi University Hospital, Bologna, Italy | In hospital | NR (included severely neutropenic with haematological malignancies) | NR |
| Rodrigues et al. (2013)100 Brazil Sponsor/funding: NR Abstract |
Single gate Prospective RCT Sampling method: NR 7 months |
NR (patients from cardiology hospital) | NR | Included adult patients aged > 18 years staying > 48 hours in hospital, with clinical suspicion of sepsis | NR |
| Avolio et al. (2014)101 Italy, single centre Sponsor/funding: NR Full text |
Single gate Prospective study Consecutive sample 3 years (September 2008–December 2011) |
Emergency and ICU | Emergency department and intensive/critical care | Patients aged > 18 years, admitted to the emergency department of the Santa Maria degli Angeli Hospital (Pordenone, Italy) with suspected bloodstream infections and at least two criteria of SIRS, were included | NR |
| Burdino et al. (2014)102 Italy Sponsor/funding: NR Full text |
Single gate Study type: NR Sampling method: NR October 2008–December 2012 |
Samples from infectious disease, ICU, cardiology, internal and surgical departments | In hospital and intensive/critical care | Reported that patients who had signs and symptoms of sepsis as defined by SIRS with suspected bacterial or fungal infections were included | NR |
| Mancini et al. (2014)103 Italy, two centres Roche Diagnostics Full text |
Single gate Retrospective data compared with prospective data Sampling method: NR (propensity score study) Sampling period: 34 months, retrospective; 24 months prospective |
Haematology and Bone Marrow Transplant Unit | In hospital | Haematological patients with suspected sepsis were included | NR |
| Markota et al. (2014)104 Slovenia, single centre Sponsor/funding: NR Full text |
Single gate Prospective study Sampling method: NR 13 months (September 2011–September 2012) |
Medical ICU | Intensive/critical care | Specific inclusion criteria NR (adults who fulfilled the criteria for severe sepsis or septic shock were included) | NR |
| Ozkaya-Parlakay et al. (2014)105 Turkey, single centre Sponsor/funding: NR Full text |
Single gate Prospective study Consecutive sample 30 months (September 2009–February 2012) |
Paediatrics department, university hospital | NR | Reported that patients included were aged between 1 month and 17 years without immune deficiency | NR |
| Schaub et al. (2014)106 Switzerland, single centre Research grants from the University of Basel, the Department of Internal Medicine and Roche Full text |
Single gate Prospective study Consecutive sample 20 months (June 2007–January 2009) |
University hospital | Emergency department | Patients presenting to the emergency department with suspected sepsis were included. Both patients with and without prior antimicrobial therapy were included | Patients aged < 18 years |
| Sitnik et al. (2014)107 Brazil, multicentre (n = 2) Roche Diagnostics donated all material multiplex PCR testing and training on test workflow Full text |
Single gate Prospective study Consecutive sample 11 months (December 2008–October 2009) |
ICU, emergency room and oncology patients | Intensive/critical care (and oncology patients) | Reported that all patients with infection plus two or more SIRS were included | NR |
| Barbanti et al. (2015)108 Italy, single centre Sponsor/funding: NR Abstract |
Single gate NR Consecutive Months NR (2009–13) |
Haematology and bone marrow transplant unit | In hospital | Haematological patients with febrile neutropenia were included | NR |
| Calitri et al. (2015)109 Italy, single centre None Full text |
Single gate Retrospective Sampling method: NR 37 months (September 2009–September 2012) |
Wards (various), ICU | In hospital and intensive/critical care | Paediatric patients with suspected sepsis, febrile neutropenia, fever without focus or localised infective focus were included | NR |
| Idelevich et al. (2015)110 Germany, single centre Partly funded by Roche Diagnostics and Pfizer Full text |
Single gate Prospective RCT Sampling method: NR 28 months (May 2010–September 2012) |
University Hospital Münster | NR | Patients who developed febrile neutropenia according to IDSA criteria were included. Afebrile neutropenic patients fulfilling sepsis criteria were also eligible to participate. Patient inclusion took place from Sunday afternoon until noon on Friday | Patients with of non-infectious causes of fever were excluded |
| Tafelski et al. (2015)112 Germany, multicentre Roche Deutschland Full text |
RCT Randomised, double-blind, parallel-group trial Consecutive sample 20 months (August 2010–March 2012) |
ICUs | Intensive/critical care | Patients were eligible for study inclusion when they presented with signs of sepsis of suspected abdominal or pulmonary origin, caused by an unknown pathogen when blood culture diagnostics were indicated. Sepsis was defined as suspected or proven infection causing systemic inflammation with at least two of the following:
|
Exclusion criteria were patients aged < 18 years, pregnant, in police custody, missing or withdrawn informed consent, or participating in another prospective clinical study |
| Warhurst et al. (2015)111 UK, multicentre (n = 4) UK National Institute for Health Research Health Technology Assessment programme Full text |
Single gate Prospective study Consecutive sample 2 years 6 months (30 July 2010–31 January 2013) |
Critical care | Intensive/critical care | Patients (aged ≥ 16 years) in a critical care setting and identified by the treating clinician as having clinical suspicion of developing a suspected bloodstream infection after ≥ 48 hours of hospital admission or recent exposure to hospital care were included. Suspicion of bloodstream infection was based on the development of two or more SIRS criteria. Patient inclusion was then based entirely on the clinical decision to perform urgent blood culture investigations | Patients already recruited into the study, except where a subsequent, new episode of suspected health-care-associated bloodstream infection had developed, and/or patients placed on an end-of-life care pathway were excluded |
| Single index test studies: SepsiTest | |||||
| Wellinghausen et al. (2009)48 Germany, multicentre Bundesministerium für Wirtschaft und Technologie Full text |
Single gate Prospective Sampling method: NR 11 months |
Departments of medicine, paediatrics, and surgical intensive care | In hospital and intensive/critical care | ICU patients with SIRS or sepsis, haematology/oncology patients with fever and neutropenia (one site), or patients with other forms of hereditary or acquired immunodeficiency and fever (one site) were included | NR |
| Loonen et al. (2014)114 The Netherlands, single centre Sponsor/funding: NR Full text |
Single gate Retrospective study – data acquired retrospectively from the laboratory information system Sampling method: NR 5 months (November–December 2011 and October–December 2012) |
Hospital emergency department | Emergency department | Reported that patients with two or more SIRS criteria and clinical signs of infection presenting at the emergency department were included | NR |
| Nieman et al. (2015)113 Confidential information has been removed |
Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Single index test studies – IRIDICA | |||||
| Bacconi et al. (2014)49 USA, single centre Sponsor/funding: NR but majority authors are employees of Ibis Biosciences (an Abbott company) Full text |
Single gate Prospective study Sampling method: NR 4 months (January–April 2012) |
Hospital emergency department | Emergency department | Subjects were considered eligible if they were aged ≥ 18 years, were having blood cultures drawn as part of clinical care and were able to provide informed consent | NR |
| Delco-Volante et al. (2015)115 Country: NR Abbott Conference presentation |
Single gate Prospective study Consecutive sample 17 months (August 2013–December 2014) |
NR | NR | NR [patients included neonates (aged < 28 days) if the treating physicians diagnosed a suspected sepsis and intended to treat them with antibiotics] | NR |
| Vincent et al. (2015)116 Belgium, UK, Switzerland, France, Poland, Germany Ibis Biosciences Inc., Abbott Full text |
Single gate Study type: NR Sampling method: NR 8 months (October 2013–June 2014) |
ICU | Intensive/critical care | Patients were considered for inclusion if they had (1) suspected or proven severe infection or sepsis; or (2) suspected or proven health-care-associated pneumonia, ventilator-associated pneumonia or severe community-acquired pneumonia | Patients were excluded if the treating clinician expected the patient to be discharged from the ICU on the day of evaluation or the following day, the treatment intent was palliative, the clinician was not committed to aggressive treatment or death was deemed imminent and inevitable |
| Metzgar et al. (2015)117 Confidential information has been removed |
Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Two index test studies: SeptiFast and SepsiTest | |||||
| Leitner et al. (2013)118 Austria Institute of Hygiene, Microbiology and Environmental Medicine, Medical University of Graz Full text |
Single gate Study type: NR Sampling method: NR Sampling period: NR |
NR | NR | Reported that critically ill patients were included | NR |
| Schreiber and Nierhaus (2013)119 Germany, single centre Molzym GmbH & Co. KG, Sirs-Lab GmbH and Roche Diagnostics GmbH provided materials Full text |
Single gate Prospective study Sampling method: NR 4 months (April–July 2009) |
Department of intensive care medicine | Intensive/critical care | Inclusion criteria – minimum age of 18 years, as well as clinical symptoms and signs consistent with the diagnosis of sepsis according to the sepsis criteria of the German Sepsis Competence Network | NR |
| Author (year) | Adults/children/neonates; mean (SD) age, n/N (%) males; number immunocompromised; number exposed to antibiotics prior to blood sample collection; previous antibiotic exposure (history) | Sepsis – n (%) suspected, severe, shock; severity of disease | Definition of sepsis | Sites of infection, other symptoms | Number of patients recruited/included, not followed up, analysed; number of paired blood samples |
|---|---|---|---|---|---|
| Single index test studies: SeptiFast | |||||
| Dierkes et al. (2009)64 | Adults; aged 55 years (median) 49/77 (64%) male 35 (45%) were immunocompromised Concomitant antibiotic therapy at the time of specimen collection had been administered in 61 patients, with 83 samples (81%) studied Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Patients with presumed sepsis – not defined | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 77 patients analysed 99 paired samples |
| Raglio et al. (2006)44 | Adults/children/neonates: NR; age, NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 74 patients analysed 114 paired samples |
| Bingold et al. (2007)59 | Adults/children/neonates: not stated; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
100% severe or septic shock | Severe sepsis and septic shock according to the S2 guidelines of the German Society of Sepsis153 | Site of infection: NR Other symptoms: NR |
21 patients included Number of patients not followed up: NR Samples from 21 patients analysed 134 paired samples |
| Klemm et al. (2007)60 | Adults/children/neonates: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Patients enrolled in this study had evidence for a new focus of infection and a clinical suspicion for sepsis – not defined | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 44 patients analysed 56 paired samples |
| Lodes et al. (2008)61 | Adults/children/neonates: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was classified according to the ACCP/SCCM consensus conference criteria2 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 137 patients analysed 358 paired samples |
| Louie et al. (2008)46 | Adults; aged > 18 years [male, median 47 (range 18–80) years; female, median 46 (range 18–91) years] 122/200 (61%) male AIDS, immunosuppressant, 8 (4%) Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Septic shock/MODS, 2 (1%) | Suspected bloodstream infection and at least two criteria of the SIRS criteria | Site of infection: NR Respiratory/pneumonia, 44 (22%); trauma/abscess, 34 (17%); cancer/neutropenic fever, 29 (14.5%); line infection, 23 (11.5%); cellulitis, 17 (8.5%); urinary/pyelonephritis, 14 (7%); endocarditis/cardiovascular, 12 (6%); AIDS, immunosuppressant, 8 (4%); gastrointestinal, 6 (3%); postoperative fever, 5 (2.5%); septic shock/MODS, 2 (1%); other, 6 (3%) |
200 patients included Number of patients not followed up: NR Samples from 200 patients analysed 200 paired samples |
| Mancini et al. (2008)45 | Adults; aged 47 (range 21–69) years 23/34 (67.6%) male Number immunocompromised: NR (all haematological malignancies) Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Underlying disease, n (%): AML, 14 (41.2%); ALL, 5 (14.7%); HD, 5 (14.7%); NHD 3, (8.8%); MDS, 2 (5.9%); CLL, 2 (5.9%); MM, 2 (5.9%); Biph. AL, 1 (2.9%) |
34 patients included Number of patients not followed up: NR Samples from 34 patients analysed 103 paired samples |
| Vince et al. (2008)62 | Adults/children/neonates: NR; age: NR Number male: NR Haematology patients following bone marrow or peripheral blood stem cell transplantation (10/39, 25.6%) 36/36 (100%) patients (empirical antimicrobial therapy) Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 36 patients analysed 39 paired samples |
| Dark et al. (2009)63 | Adults; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
50 patients included Number of patients not followed up: NR Samples from 50 patients analysed 90 paired samples |
| Gimeno et al. (2009)65 | Adults/children: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample 19/19 (100%). All patients received antibiotics and/or antifungal prophylaxis Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR 19 patients analysed 45 paired blood samples |
| Lehmann et al. (2009)66 | Adults; mean age 54.8 (range 18–92) years 268/436 (61%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was defined according to the SCCM/ACCP consensus conference guidelines of 19924 | Intra-abdominal sepsis, 136; nosocomial pneumonia, 112; community-acquired pneumonia, 19; multiorgan dysfunction syndrome, 13; catheter-related sepsis, 61; neutropenic fever, 47; pyelonephritis, 24; genitourinary infection, 13; wound infection, 10; bone/joint infection, 14; other, 102 Other symptoms: NR |
Number of patients recruited: NR 436 patients with 467 episodes of antimicrobial treatment were included in the study in total Paired blood samples: NR |
| Lodes et al. (2009)67 | Adults (range: NR); ages 60.5 (14.7) years 30/52 (58%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was classified according to the ACCP/SCCM consensus conference criteria2 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 52 patients analysed 258 paired samples |
| Palomares et al. (2009)68 | Adults; age: NR Number male: NR Number immunocompromised: NR 68/73 patients (93%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 73 patients analysed 76 paired samples |
| Paolucci et al. (2009)69 | Neonates aged > 3 days; mean age NR (however, newborns were aged > 3 days) Number male: NR One patient was affected by primary congenital immunodeficiency Antibiotics prior to blood sample: NR (however, three blood cultures – newborns received antibiotics prior to blood sampling) Previous antibiotic exposure: NR |
Clinical suspicion of sepsis | Sepsis was based on the presence of at least one clinical sign suggestive of clinical sepsis, and elevated C-reactive protein > 2.0 mg | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 34 patients analysed Number of paired blood samples NR |
| Varani et al. (2009)70 | Adults and children; age: NR (85 adults and 15 children) Number male: NR All immunocompromised All patients received antibiotics and antifungal prophylaxis Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR (patients with neutropenia or fever with signs and symptoms of infection) | Site of infection: NR 50, AML; 17, ALL; 15, lymphoma; 7, MM; 3, chronic myeloproliferative disorders; 6, solid tumours; 1, autoimmune thrombocytopenia; and 1, haemophagocytic lymphohistiocytosis |
100 patients included Number of patients not followed up: NR Samples from 100 patients analysed 130 paired samples |
| von Lilienfeld-Toal et al. (2009)71 | Adults; median age 60 (IQR 49–66) years 38/70 (54%) male Number immunocompromised: NR Antibiotics prior to blood sample: none Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | AML, 50 (71%); ALL, 8 (11%); NHD, 4 (6%); MM, 2 (3%); MDS, 2 (3%); aplastic anaemia, 2 (3%); metastatic carcinoma, 2 (3%) | Number of patients recruited: NR Number of patients not followed up: NR 70 patients analysed (119 episodes) 784 paired blood samples |
| Westh et al. (2009)72 | Adults/children/neonates: NR; age: NR Number male: NR Number immunocompromised: NR Limited details provided regarding patients receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 359 patients analysed 558 paired samples |
| Berger et al. (2010)73 | Neonates; age: NR Number male: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Clinical sepsis suspicion – sepsis not defined | Site of infection: NR Other symptoms: NR |
38 patients included Number of patients not followed up: NR Samples from 38 patients analysed 38 paired samples |
| Bloos et al. (2010)74 | Adults; aged 66 years 68.5% male None immunocompromised 95.8% on antibiotics (unclear if prior to blood sampling) Previous antibiotic exposure: NR |
100% severe sepsis or septic shock Severity of disease SOFA score of 10 for entire sepsis cohort |
Severe sepsis or septic shock according to ACCP/SCCM consensus criteria2 | Lung (40%), abdomen (16.9%), bloodstream (9.3%), catheter-related (9.3%) Mechanical ventilation (81.7%) |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 142 patients analysed 236 paired samples |
| Lamoth et al. (2010)75 | Adults; median age 54 (range 17–71) years 53/86 (62%) male Number immunocompromised: NR Antibiotics prior to blood sample 144 (61%) samples drawn under AB therapy Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | AML, 37 (43%); ALL, 9 (10%); lymphoma, 12 (14%); MM, 22 (26%); other haematological malignancies, 6 (7%); chemotherapy induction/consolidation for acute leukaemia, 45 (52%); autologous stem cell transplant, 37 (43%); other chemotherapy, 4 (5%) | Number of patients recruited: NR Number of patients not followed up: NR 86 patients analysed 237 paired blood samples |
| Lehmann et al. (2010)76 | Adults (range 18–84); age 58.37 years 72/108 (67%) male Number immunocompromised: NR [18.5% (20/108) had neoplasms] Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
100% severe sepsis | Severe sepsis was classified according to the ACCP/SCCM consensus conference criteria2 | Abdominal sepsis (n = 35); sepsis following CV surgery (n = 28); pneumonia/ARDS (n = 23); tissue infection following trauma (n = 15); osteomyelitis (n = 2); genitourinary infection (n = 2); mediastinitis (n = 2); catheter related (n = 1) Chronic comorbidities (some had multiple): neoplasms (18.5%); liver failure (16.7%); NYHA III/IV heart failure (50.9%); chronic renal failure (45.4%); diabetes (19.4%) |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 108 patients analysed 453 paired samples |
| Maubon et al. (2010)77 | Adults/children/neonates: NR; age: 56.3 (13.7) years 57/110 (60.9%) male Number immunocompromised: NR (all haemato-oncology study cohorts) 97/110 (88.2%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
64/110 (58%) had severe sepsis and 27/110 (25%) had septic shock | Suspected infection with at least one sign of sepsis, with or without organ dysfunction, as defined by the SIRS criteria | Site of infection: NR Acute leukaemia, 48 (43.6%); lymphoma, 28 (25.5%); myeloma, 10 (9.1%); other haematological malignancy, 9 (8.2%); solid tumour 15 (13.6%) |
110 patients included Reports on pathogens identified in 50 patients with documented sepsis not followed up Samples from 110 patients analysed 110 paired samples |
| Regueiro et al. (2010)78 | Adults, 21–92; aged 64 (range 21–92) years 53/72 (73.6%) male Number immunocompromised: NR [6/72 (8.3%) oncology] Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease entire cohort APACHE II scores: 0–4, n = 0; 5–9, n = 2; 10–14, n = 9; 15–19, n = 15; 20–24, n = 12; 25–29; n = 15; 30–34, n = 11; > 34, n = 8 |
NR | Site of infection: NR Basal disease: respiratory, 18; cardiovascular, 18; alcoholism, 10; oncologic, 6; digestive, 6; psychiatric, 4; neurological, 2; various causes, 6; other cause, 2 |
72 patients included Number of patients not followed up: NR Samples from 72 patients analysed 106 paired samples |
| Soki et al. (2010)79 | Adults/children/neonates: NR; age: NR Number male: NR Patients with haematology malignancies with fever (126/162), no further details provided Limited details provided regarding patients receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 159 patients analysed 162 paired samples |
| Tsalik et al. (2010)80 | Adults, 18–97; aged 54.1 (range 18–97) years 168/306 (54.9%) male Number immunocompromised: NR There were 69 subjects (22.5% of 306) who received at least one dose of antibiotic before blood was collected for culture or PCR Previous antibiotic exposure: NR |
Non-infected SIRS positive, 43 (14.1%); sepsis, 184 (60.1%); severe sepsis, 42 (13.7%); septic shock, 37 (12.1%) | Sepsis was defined as SIRS with evidence of infection but no evidence of end-organ damage. Severe sepsis occurred in the presence of end-organ damage, which included metabolic damage, haematological damage, pulmonary damage or renal damage. Sepsis in the presence of hypotension, despite fluid challenge, or a blood lactate concentration of 4 mmol/l was defined as septic shock | Lung, 55 (18.0%); urinary tract, 46 (15.0%); skin, 41 (13.4%); intra-abdominal, 25 (8.2%); intravascular catheter, 16 (5.2%); other, 32 (10.5%); unknown, 91 (29.7%) Other symptoms: NR |
306 patients included Number of patients not followed up: NR Samples from 306 patients analysed 306 paired samples |
| Wallet et al. (2010)81 | Adults; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR (antimicrobial prescription was prospectively recorded on the day of blood culture sampling) Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR (patient status was defined according to SIRS criteria) | The most frequent suspected site of infection was the respiratory tract: 66% (59/90) of samples with negative blood culture results and 70% (7/10) of samples with positive blood culture results Other symptoms: NR |
72 patients included Number of patients not followed up: NR Samples from 72 patients analysed 102 paired samples |
| Yanagihara et al. (2010)82 | Adults/children/neonates: NR; age: NR 137/212 (65%) male Varied including immune deficiency (33/407 samples), tumour (51/407 samples) Of the pathogens detected by SeptiFast or blood culture, 40 were from patients who had been administered antibiotics and 32 of these 40 samples were from patients that had been administered antibiotics that matched the spectra of the antibiotics Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was classified according to the ACCP/SCCM consensus conference criteria2 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 212 patients analysed 400 paired samples |
| Bravo et al. (2011)83 | Adult; aged 65.5 median (range 23–86) years 33/53 (62%) male Number immunocompromised: NR None receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Severe sepsis or septic shock n = 15 | NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 53 patients analysed 53 paired samples |
| Hettwer et al. (2011)84 | Adults; aged 62.3 (± 18.1) years 94 (61%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
53.7% had severe sepsis or shock | At least 2/4 SIRS criteria. Patients also stratified according to procalcitonin and assessed against APACHE II and SOFA | Medical non-pneumonogen, n = 55/153 (37%); medical pneumonogen, n = 63 (42%); urogenital, n = 17 (11%); other, n = 14 (9%) Procalcitonin (ng/ml): 14.4 ± 42.3 |
211 patients recruited (153 with sepsis) PCR and blood culture available for 113 patients 113 paired samples |
| Josefson et al. (2011)85 | Adults and children (range 14–98 years); aged 67 years (median) 607/1093 (56%) male Number immunocompromised: NR 36/200 pathogen detections in presence of antibiotics Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
1540 patients included 447 patients did not have results from at least one blood culture/PCR set not followed up Samples from 1093 patients analysed 1141 paired samples |
| Lucignano et al. (2011)86 | Neonates and children; age: NR Number male: NR Number immunocompromised: NR [272/803 (33.9%) from oncology, haematology, neonatology] Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
The condition of sepsis was defined when a SIRS was in the presence of or a result of suspected or proven infection | Site of infection: NR Clinical wards: heart surgery, n = 323; haematology, n = 272; cardiology, n = 208. Others, other symptoms: NR |
811 patients recruited 8 not followed up Samples from 803 patients analysed 1553 paired samples |
| Obara et al. (2011)87 | Adults (30–86); aged 61.6 (range 30–86) years 35/54 (64.8%) male Number immunocompromised: NR [21/54 (38.9%) haematology] Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
At least two criteria of the SIRS | Site of infection: NR Other symptoms: NR |
54 patients included Number of patients not followed up: NR Samples from 54 patients analysed 78 paired samples |
| Vrioni et al. (2011)88 | Adults/children/neonates: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 33 patients analysed 33 paired samples |
| Alvarez et al. (2012)89 | Adults; mean age 64.9 years 55 (54.5%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
All had severe sepsis or septic shock | Dellinger et al.154 and by Levy et al.155 | Emergency abdominal surgery: BC = 9, SF = 11; elective abdominal surgery, BC = 2, SF = 2; pneumonia, BC = 0, SF = 4; pancreatitis, BC = 7, SF = 1; CNS lesion, BC = 5, SF = 9; polytrauma/head, BC = 20, SF = 4; heart surgery, BC = 2, SF = 20; vascular surgery, BC = 1, SF = 1; pneumonectomy, BC = 1, SF = 0 Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 102 patients analysed Number of paired blood samples: NR |
| Grif et al. (2012)90 | Adults/children/neonates: NR; age 55.6 years 42/61 (69%) male Number immunocompromised: NR 56/61 (91.8%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
At least two SIRS criteria – sepsis not defined | Site of infection: NR Other symptoms: NR |
61 patients included Number of patients not followed up: NR Samples from 61 patients analysed 71 paired samples |
| Guido et al. (2012)91 | Adults; age 66.1 median (range 23–82) years 103/166 (62%) male None immunocompromised None receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Febrile neutropenia – sepsis not defined | Site of infection: NR Other symptoms: NR |
166 patients included Number of patients not followed up: NR Samples from 166 patients analysed 166 paired samples |
| Lodes et al. (2012)92 | Adults (20–88 years); age 63.1 (14.1) years 74/104 (71.1%) male Number immunocompromised: NR [malignant neoplasm 38 (36.5%)] 79.7% of all blood samples were taken under antibiotic therapy and 41.9% were taken under antifungal therapy Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was classified according to the ACCP/SCCM consensus conference criteria2 | All abdominal Main diagnosis by surgery: malignant neoplasm, 38 (36.5%); peritonitis, 23 (22.1%); hepatorenal syndrome, liver failure, liver cirrhosis, 14 (13.5%); haemorrhage, 7 (6.7%); ischaemia, 6 (5.8%); pancreatitis, 5 (4.8%); urosepsis, 2 (1.9%); gall bladder perforation, choledocholithiasis, 2 (1.9%); hernia incarceration, 2 (1.9%); others, 5 (4.8%) |
104 patients included Number of patients not followed up: NR Samples from 104 patients analysed 148 paired samples |
| Mauro et al. (2012)93 | Adults and children; aged 5–68 years 41/79 (51.9%) male All immunocompromised All but 4 patients had blood culture drawn before starting antimicrobial therapy Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Suspected bloodstream infections and at least 2 criteria for SIRS | Site of infection: NR 21, ALL; 2, Wilms’ tumour; 3, hepatocellular carcinoma; 2, allogeneic stem cell transplantations; 18, NHD; 4, colon cancer under chemotherapy; 2, ovarian cancer; 1, leishmania visceral; and 32, exposure to glucocorticoids for autoimmune disease |
79 patients included Number of patients not followed up: NR Samples from 79 patients analysed 79 paired samples |
| Pasqualini et al. (2012)94 | Adults; aged 73 median (range 20–99) years 215/391 (55%) male 17 (4%) immune deficiency 191/391 (48.8%) had been receiving antibiotic treatment for ≥ 24 hours Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Suspected sepsis, suspected of having SIRS caused by bacterial or fungal infection | Site of infection: NR History of CV disease, 88 (22%); malignancy, 70 (18%); dementia, 46 (12%); chronic lung disease, 44 (11%); diabetes, 41 (10%); chronic renal failure, 28 (7%); immune deficiency, 17 (4%); chronic liver disease, 16 (5%); gangrene, 3 (1%) |
391 patients included Number of patients not followed up: NR Samples from 391 patients analysed 391 paired samples |
| Rath et al. (2012)95 | Adults; aged 52.6 (10.9) (range 27–70) years 72 (64.6%) male Number immunocompromised: NR (all liver transplant patients) Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Patients with suspected sepsis according to the criteria of the ACCP/SCCM17 | Site of infection: NR Cirrhosis [alcoholic, 21 (26.5%); infectious (hepatitis B/C), 17 (21.5%); NASH, 4 (5.0%); other (autoimmune, unknown), 11 (13.9%)]; hepatocellular carcinoma, 12 (15.1%); primary sclerosing cholangitis, 7 (8.8%); acute liver failure, 4 (5.0%); liver cysts, 3 (3.7%); malignancy, 48 (60.7%); abdominal infection, 27 (29.6%); abdominal organ perforation, 12 (13.1%); colon ischaemia, 4 (4.3%) |
170 patients included Number of patients not followed up: NR Samples from 170 patients analysed 225 paired samples |
| Tschiedel et al. (2012)96 | Adults and children; median age 6 (range 0–24) years 37/75 (49%) male 64 samples were drawn from immunosuppressed patients (58%) 97 samples were drawn from patients (88%) under antibiotic treatment at time of sample taking Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR 87% of patients suffered from severe underlying disease such as: organ transplantation, malignant illnesses, cystic fibrosis, pulmonary hypertension, cardiac vitium, spinal muscular atrophy, and renal insufficiency with dialysis. 14 samples (13%) were taken from previously healthy patients |
75 patients included Number of patients not followed up: NR Samples from 75 patients analysed 110 paired samples |
| Herne et al. (2013)97 | Adults; aged 58 (range 20–81) years 61/144 (42%) male Number immunocompromised: NR (haemato-oncology study cohorts) 143/144 (99%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
100% severe sepsis | Clinically suspected sepsis or septic shock or severe infection without known etiologic agent – sepsis not defined | Acute pneumonia (43%), central venous catheter-associated bloodstream infection (11%), acute peritonitis (12%), septic endocarditis (10%), acute pancreatitis (10%), and acute urinary tract infection (4%) A total of 108 (75%) patients had multifocal infection with concurrent diagnoses and polymicrobial aetiology |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 144 patients analysed 160 paired samples |
| Kasper and Altiok (2013)98 | Neonates of very low birthweight; age: NR (however, for blood culture-positive sepsis and blood culture-negative clinical sepsis group, age range 23.3–30.1 weeks) Number male: NR Number immunocompromised: NR [neonates mean (assumed) birthweight 818 ± 242 g] None receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
46 patients included Number of patients not followed up: NR Samples from 46 patients analysed Number of paired blood samples: NR |
| Paolucci et al. (2013)99 | Adults and children; age: NR (23 children, 178 adults) Number male: NR Number immunocompromised: NR (included two cases of autoimmune thrombocytopenia) Antibiotics prior to blood sample: none Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Haematological malignancies (105 AML, 23 ALL, 34 lymphoma, 15 MM and 8 chronic myeloproliferative disorders), severe aplastic anaemia (4 patients), solid tumours (9 patients) or other disorders (2 cases of autoimmune thrombocytopenia, 1 case of haemophagocytic lymphohistiocytosis) | Number of patients recruited: NR Number of patients not followed up: NR 201 patients analysed (339 episodes) 437 paired blood samples |
| Rodrigues et al. (2013)100 | Adults; age SF: 63 (46–75) years, BC: 66 (39–85) years 31/46 (67%) Male Number immunocompromised: NR Antibiotics prior to blood sample: none Previous antibiotic exposure: none |
Septic shock SF: 9/17 (53), BC: 16/29 (55) | NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR 46 patients analysed Number of paired blood samples: NR |
| Avolio et al. (2014)101 | Adults; age: NR Number male: NR None immunocompromised Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Suspected BSIs and at least two SIRS criteria | Site of infection: NR Other symptoms: NR |
830 patients included 305 cases did not have a BC assay (not requested by clinician if patients under antibiotic treatment at time of blood sampling, or previous blood culture negative such as long-term critically ill patients) Samples from 525 patients analysed 525 paired samples |
| Burdino et al. (2014)102 | Adults; age NR Number male: NR Number immunocompromised: NR (10.5% HIV infection) 89% receiving empirical antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis as defined by SIRS with suspected bacterial or fungal infections | Site of infection: NR Other symptoms: NR |
1024 patients included Number of patients not followed up: NR Samples from 1024 patients analysed 1186 paired samples |
| Mancini et al. (2014)103 | Adults; mean age 48.6 years 152/228 (67%) male Number immunocompromised: NR [100% haematological (55.7% with AML)] Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was defined according to Dellinger et al.16 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Six episodes in the prospective cohort and four in the retrospective were excluded for incomplete compilation of the study records Retrospective cohort, 134 episodes in 115 patients; prospective cohort, 131 episodes in 113 patients analysed Number of paired blood samples: NR |
| Markota et al. (2014)104 | Adults; aged 59.5 (14.8) years 38/57 (66.7%) male Number immunocompromised: NR 39/57 (61.9%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
All had severe sepsis or septic shock Severity of disease: entire cohort mean admission APACHE score 25 (± 7.6) |
Sepsis was defined according to Dellinger et al.154 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 57 patients analysed 63 paired samples |
| Ozkaya-Parlakay et al. (2014)105 | Children; aged 1 month to 17 years, age 2.71 (4.11) years (73.4% < 2 years) 43/69 (62.3%) male None (no immune deficiency) Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR (patients with two signs of SIRS included) | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 69 patients analysed 79 paired samples |
| Schaub et al. (2014)106 | Adults; aged ≥ 18 years, median 64 years 56/110 (60%) male 15/110 (14%) immunosuppressed (drug induced or HIV) Antibiotic therapy prior to presentation at ED had been started in 16 (15%) patients Previous antibiotic exposure: NR |
Sepsis without organ dysfunction in 61 patients (77%), severe sepsis in 13 patients (17%) and septic shock in five patients (6%) | Sepsis and its severity were defined according to the 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference.155 Patients were only considered to have sepsis if they had both SIRS and infection | Pulmonary, 38 (35%); urogenital, 19 (17%); abdominal, 8 (7%); musculoskeletal, 3 (3%); skin 7 (6%); ear, nose and throat, 5 (5%); other 3 (3%); systemic, 5 (5%); no focus found, 3 (3%) Diabetes, 26 (24%); renal impairment, 22 (20%); immunosuppression, 15 (14%) |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 110 patients analysed 205 paired samples |
| Sitnik et al. (2014)107 | Adults; aged 49.7 (24.8) years 74/114 (64.9%) male Oncology patients (38/114), no further details provided Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was classified according to the ACCP/SCCM consensus conference criteria4 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 114 patients analysed 114 paired samples |
| Barbanti et al. (2015)108 | Adults/children: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR 491 patients analysed Number of paired blood samples: NR |
| Calitri et al. (2015)109 | Children and neonates (8 preterm newborns); median age 6.8 (IQR 2.7–13.1) years 183/289 (63.3%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR (however, high rate of patients received antibiotic and/or antifungal treatment at time of sampling) Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
International Paediatric Sepis Consensus156 | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR 289 patients analysed (545 episodes) Number of paired blood samples: NR |
| Idelevich et al. (2015)110 | Adults; mean age 52.4 years [SF, 50.4 (14.4) years; BC, 54.4 (15.2) years] 89/150 (59.3%) male [SF, 45/74 (60.8%); BC, 44/76 (57.9%)] Number immunocompromised: NR Antibiotics prior to blood sample 150/150 (100%) Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Sepsis was defined according to the SCCM/ACCP consensus conference guidelines of 19924 | AML SF = 33 (44.6), BC = 42 (55.3); ALL SF = 8 (10.8), BC = 12 (15.8); MM SF = 12 (16.2), BC = 11 (14.5); NHD SF = 18 (24.3), BC = 6 (7.9); chronic myeloid leukaemia SF = 1 (1.4), BC = 3 (3.9); MDS SF = 1 (1.4), BC = 1 (1.3); others SF = 1 (1.4), BC = 1 (1.3) | Number of patients recruited: NR Number of patients not followed up: NR 150 patients analysed 253 paired blood samples |
| Tafelski et al. (2015)112 | Adults (range 47–74 years); age BC + SF: 67 years (median); BC: 59 years (median) BC + SF: 26 (63%) male; BC: 24 (65%) male BC + SF: 6/41 (15%); BC: 6/37 (16%) immunocompromised Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Septic shock BC + SF: 20/41 (49%) BC: 25/37 (68%) Median SAPS II on admission: BC + SF 40 (IQR 32–50); BC: 47 (IQR 34–65) |
Sepsis defined as suspected or proven infection causing systemic inflammation with at least two of the following: (1) leucocyte count < 4 or > 12/nl; (2) body temperature < 36 °C or fever > 38 °C; (3) tachypnoea > 20 breaths per minute or hyperventilation (PaCO2 < 32 mmHg); (4) tachycardia > 90 beats per minute | Abdominal BC + SF: 15/41(37%); BC: 8/37 (22%); pulmonary BC + SF: 26/41(63%); BC: 29/37 (78%) Other symptoms: NR |
100 patients included 22 (unable to provide informed consent) not followed up Samples from 78 BC + SF: 41; BC: 37 patients analysed 78 paired samples |
| Warhurst et al. (2015)111 | Adults and children; aged ≥ 16 years, median 58 (44 to 68) years 553/795 (60%) male Number immunocompromised: NR 788/795 (85%) receiving antibiotic treatment at time of blood collection Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Suspicion of bloodstream infection was based on the development of two or more SIRS criteria as defined by Levy et al.155 | Site of infection: NR Other symptoms: NR |
795 patients included Number of patients not followed up: NR Samples from 795 (922 episodes) patients analysed Number of paired blood samples: NR |
| Single index test studies – SepsiTest | |||||
| Wellinghausen et al. (2009)48 | 173 adults and 14 children < 18 years; age: NR Number male: NR Number immunocompromised: none 8 of the 13 patients received broad-spectrum antimicrobials before sampling Previous antibiotic exposure: NR |
148 patients (79.1%) were ICU patients fulfilling the criteria for SIRS or sepsis, and 39 patients (20.9%) were haematological patients with neutropenic fever | NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 187 patients analysed 342 paired samples |
| Loonen et al. (2014)114 | Adults; positive blood culture aged 68.9 (17.3) years; negative blood culture aged 60.4 (18.0) years Positive blood culture, 17/125 (13.6%) male; negative blood culture, 57/125 (45.6%) male Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
140 patients included 15 – alternative diagnosis without infection not followed up Samples from 125 patients analysed Number of paired blood samples: NR |
| Nieman et al. (2015)113 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Single index test studies: IRIDICA | |||||
| Bacconi et al. (2014)49 | Adults; aged ≥ 18 years Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
Patients with suspected sepsis enrolled | Site of infection: NR Other symptoms: NR |
331 patients included Number of patients not followed up: NR Samples from 331 patients analysed 331 paired samples |
| Delco-Volante et al. (2015)115 | Neonates (< 28 days old); age: NR Number male: NR Number immunocompromised: NR (neonates) None – before the initiation of antibiotics Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Number of patients analysed: NR 81 paired samples |
| Vincent et al. (2015)116 | Adults; aged 61 (17.9) years 64.6% male Immunosuppressed, 90/543 (16.6%) Within 30 days prior to hospitalisation, 12.7%; initiated during hospitalisation, 62.7%; initiated at or after enrolment: 22.1% Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease entire cohort: mean SOFA score at baseline: 7.6 (4.2) indicating 15–20% risk in mortality in ICU |
NR | Site of infection: NR Other symptoms: NR |
543 patients included 14 did not have matching PCR/ESI-MS or standard-of-care microbiology results not followed up Samples from 529 patients analysed Number of paired blood samples: NR |
| Metzgar (2015)117 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Two index test studies: SeptiFast and SepsiTest | |||||
| Leitner et al. (2013)118 | Adults/children/neonates: NR; age: NR Number male: NR Number immunocompromised: NR Antibiotics prior to blood sample: NR Previous antibiotic exposure: NR |
Suspected, severe, shock: NR Severity of disease: NR |
NR | Site of infection: NR Other symptoms: NR |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 57 patients analysed 75 paired samples |
| Schreiber and Nierhaus (2013)119 | Adults; age median 64 (IQR 51–70) years 10/50 (20%) male Number immunocompromised: NR (2/50 (1%) had bone marrow transplant) Antibiotics prior to blood sample: NR 36/50 (72%) had received antibiotic treatment at recruitment |
Sepsis, 10 (20%); severe sepsis, 13 (26%); septic shock, 27 (54%) Severity of disease entire cohort: SAPS II, median 41 (IQR 33–49) |
Diagnosis of sepsis according to the sepsis criteria of the German Sepsis Competence Network153 | Site of infection: NR Reasons for admission: surgical 20 (40%) (abdominal, 7; chest, 6; trauma, 7); medical, 24 (48%) (pneumonia, 15; pancreatitis/cholangitis, 2; bone marrow transplant, 2; unknown focus, 5) |
Number of patients recruited: NR Number of patients not followed up: NR Samples from 50 patients analysed Number of paired blood samples: NR |
| Author (year) | Name of test, sampling method, interval between index test and reference standard | CE-approved blood volume | Laboratory test performed on same day as sampling method | Name of reference standard, sampling method, culture methods | Definition of true positive: index test/reference test | Laboratory working times for analysis | Unit of analysis (metric) | Contaminants included? |
|---|---|---|---|---|---|---|---|---|
| Single index test studies: SeptiFast | ||||||||
| Raglio et al. (2006)44 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Sample | Included |
| Bingold et al. (2007)59 | LightCycler SeptiFast Test (MGRADE NR) 3 ml of kEDTA venous or arterial blood When samples drawn: NR |
Unclear | NR | Blood culture with swabs from suspicious sites for microbiological diagnostics Sampling method: NR Culture method: NR |
NR | NR | Samples | NR |
| Klemm et al. (2007)60 | LightCycler SeptiFast Test (MGRADE NR) 2 × 3.5 ml of EDTA–blood Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | Same day | Blood culture and procalcitonin as a clinical marker of sepsis 2 × 10 ml of blood BACTEC BD (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) |
NR | NR | Patients | NR (four samples contaminated with dermal Streptococci deliberately not reported by SeptiFast interpretive software) |
| Lodes et al. (2008)61 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Sample | NR |
| Louie et al. (2008)46 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: 3 ml of EDTA for PCR testing When samples drawn: NR |
Unclear | NR | Blood culture plus clinical chart review Sample: whole blood Site: NR Volume: NR (the PCR sample was collected after a sample was drawn for blood culture) Culture method: NR |
NR | NR | Patients | Excluded |
| Mancini et al. (2008)45 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: at least 1.5 ml of K-EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: at least 20 ml of blood (average of 25 ml inoculated into aerobic and anaerobic bottles) BacT/ALERT 3D automated blood culture system (bioMérieux, Marcy-l’Etoile, France), with monitoring of carbon dioxide production within each bottle every 10 minutes 24 hours per day. All bottles signalled as positive were removed from the instrument, and an aliquot was taken for Gram stain and culture on solid media. Sensitivity to antibiotics were performed with the VITEK 2 system (bioMérieux, Marcy-l’Etoile, France) |
NR | 7 days per week (from 8 a.m. to 7 p.m.) | Samples | Included |
| Vince et al. (2008)62 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Samples | NR |
| Dark et al. (2009)63 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture with input from results of other cultures Sampling method: NR Culture method: NR |
NR | NR | Pathogens | NR (three detected in blood culture but not in SeptiFast, assigned as true negatives) |
| Dierkes et al. (2009)64 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: NR Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture concordance. Clinical data were extracted by chart review and by the data provided for the test application Sample: whole blood Site: NR Volume: 2 × 10-ml bottles BACTEC 9240. Both aerobic and anaerobic blood culture bottles were inoculated directly with 10 ml of blood each and delivered to the microbiology department together with the aliquot for analysis with the SeptiFast. Blood cultures were incubated for 7 days |
NR | 7 days per week (Monday to Friday from 8 a.m. to 7 p.m.; Saturday, Sunday and holidays from 9 a.m. to 4 p.m.) | Samples | Included |
| Gimeno et al. (2009)65 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venous or catheter Volume: 3 ml Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Traditional blood cultures (aerobic and anaerobic) | NR | NR | Samples | NR |
| Lehmann et al. (2009)66 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | The study was not designed for method comparison, therefore multiple blood culture tests per episode were allowed. Blood culture was performed using BACTEC or BacT/ALERT | NR | NR | Incomplete diagnostic data reported | Incomplete diagnostic data reported |
| Lodes et al. (2009)67 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Samples | Included |
| Palomares et al. (2009)68 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: 3 ml in EDTA bottles Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Sample | Included |
| Paolucci et al. (2009)69 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: peripheral venous Volume: 1.5 ml When samples drawn: NR |
Yes | NR | Blood culture plus clinical. Bloodstream infection was confirmed by the presence of clinical signs of infection or additional microbiological data Sample: whole blood Site: peripheral venous Volume: 1.0 ml of blood Blood cultures were performed according to the CLSI protocol. No further detail reported |
NR | NR | Patients | Included |
| Varani et al. (2009)70 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: adults, peripheral veins; children, central venous catheter Volume: K-EDTA 3 ml was sampled and processed for patients who weighed 45 kg and 1.5 ml for those who weighed < 45 kg Reference standard and index tests performed on blood samples drawn at the same time |
Unclear (mixed based on weight – immunocompromised patients) | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system and processed according to the CLSI |
NR | NR | Febrile episodes | Included |
| von Lilienfeld-Toal et al. (2009)71 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous or catheter Volume: 1.5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
No [1 ml (adults)] | NR | Blood culture was performed using the BACTEC system | NR | NR | Pathogen | NR |
| Westh et al. (2009)72 | SeptiFast lys kit, the SeptiFast prep kit, and the LightCycler SeptiFast kit, all MGRADE Sample: whole blood Site: venous Volume: used 1.5 ml for assay (5 ml drawn) Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture in conjunction with clinical adjudication. Identification of micro-organisms from a suspected infectious focus within 48 hours of the episode was used to resolve discrepancies in the results Sample: whole blood Site: venous Volume: 8–10 ml of blood for each aerobic and anaerobic bottle for each system Blood culture was performed using the BACTEC or BacT/ALERT system. Each blood culture was performed in a pair of aerobic/anaerobic bottles. Blood for one or two additional blood culture sets was collected from each patient within a 24-hour period and included in episode evaluation |
NR | NR | Pathogen | Excluded |
| Berger et al. (2010)73 | LightCycler SeptiFast Test (MGRADE NR) Modified DNA extraction for very low birthweight infants protocol to decrease blood volume requirements to 1.0 ml When samples drawn: NR |
No [0.1 ml (neonates)] | NR | Blood culture with clinical and laboratory signs of infection Volume: 0.1 ml of blood Sampling method: NR |
NR | NR | Patients | Included |
| Bloos et al. (2010)74 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venous Volume: 10 ml, 3 ml from 10 ml EDTA treated Reference standard and index tests performed on blood samples drawn at the same time |
No [3 ml (adults)] | Same day as severe sepsis suspected | Blood culture and clinical/laboratory confirmation Whole blood (20 ml) for conventional cultures Pair of blood cultures incubated at 37 °C and monitored for 8 days. Isolated micro-organisms and their susceptibilities were determined by standard methods and criteria |
Clinical adjudicators reviewed the patient data and corresponding microbiological culture results of the presumed site of infection | NR | Samples | NR |
| Lamoth et al. (2010)75 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venous Volume: 1 ml Reference standard and index tests performed on blood samples drawn at the same time |
No [3 ml (adults)] | For SeptiFast assays, DNA was extracted from the EDTA whole-blood tubes within 48–72 hours after sampling | Blood culture was performed using the BACTEC system | NR | NR | Episodes | NR |
| Lehmann et al. (2010)76 | SeptiFast Prep Kit and the LightCycler (MGRADE NR) Sample: whole blood Site: NR Volume: used 1 ml EDTA whole-blood sample (9 ml was drawn for further PCR analysis) Reference standard and index tests performed on blood samples drawn at the same time |
No [1 ml (adults)] | NR | Blood culture and clinical/laboratory confirmation A pair of aerobic/anaerobic blood culture bottles. Also provides analysis for a constructed gold standard including blood culture and other microbiological tests (sensitivity 0.83 and specificity 0.93) Sample: whole blood Site: NR Volume: 20 µl for pair of aerobic and anaerobic blood culture bottles Blood culture was performed using the BACTEC system. All blood cultures were processed using semi-automated blood culture systems according to the manufacturer’s instructions. The blood culture system and the local laboratory software automatically registered time to positive blood culture |
A bloodstream infection was defined as a positive blood culture result, obtained and analysed as set forth by the current DGHM procedures. Whether micro-organisms identified by PCR represented true infection or contamination was evaluated retrospectively by taking into account the identity of the micro-organism detected and by comparing PCR results with corresponding blood culture findings | NR (however laboratories were not 24 hours a day, 7 days per week) | Samples | Included |
| Maubon et al. (2010)77 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 1.5 ml EDTA When samples drawn: NR |
Yes | NR | Blood culture plus clinical: leucocyte count, C-reactive protein and procalcitonin measurement, two sets of bacterial and fungal blood cultures, urine culture, chest radiograph and, when appropriate, specific viral and fungal tests Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Patients | Unclear |
| Regueiro et al. (2010)78 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous or arterial draw Volume: 1.5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venous or arterial draw Volume: 10 ml Blood culture was performed using the BacT/ALERT system. Once flagged by the instrument for detectable growth, fluid was withdrawn for Gram stain and appropriate agar-based culture plates. Isolated colonies were analysed either by an automated identification system VITEK II |
NR | NR | Samples | Included |
| Soki et al. (2010)79 | SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture: NR if with clinical confirmation or not Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system. Blood culture sets (one aerobic and one anaerobic bottle) were used |
NR | NR | Sample | NR |
| Tsalik et al. (2010)80 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 1.5 ml When samples drawn: NR |
Yes | NR | Blood culture and clinical confirmation Sample: whole blood Site: NR Volume: the volume inoculated was not monitored Blood culture was performed using the BacT/ALERT system plus BACTEC |
NR | NR | Patients | Excluded |
| Wallet et al. (2010)81 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venepuncture, site NR Volume: EDTA 5 ml; volume for DNA preparation was 1.5 ml Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venepuncture, site NR Volume: 10 ml Blood culture was performed using the BacT/ALERT system plus BACTEC |
NR | NR | Pathogens | Included |
| Yanagihara et al. (2010)82 | SeptiFast-Lys and Prep MGRADE kits Sample: whole blood Site: NR Volume: used 1.5 ml for assay (10 ml drawn) Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Blood for DNA detection kit was stored at –20 °C for up to 72 hours before testing | Blood culture in conjunction with clinical adjudication. When the result of blood culture analysis was positive, the sample was identified using each site’s identification system Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC and BacT/ALERT system. Blood culture bottles in which results were positive were sent to one commercial laboratory to confirm valid identification of micro-organisms |
NR | NR | Sample | Included |
| Bravo et al. (2011)83 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous Volume: 1.5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venous Volume: paired 10-ml bottles of blood BACTEC 9420 blood culture. A pair of bottles for aerobic and anaerobic bacteria and an additional bottle for fungal recovery. The blood cultures were incubated for a maximum of 7 days. Each bottle was inoculated with 10 ml of blood. The different sets of blood cultures were obtained at intervals of 30 minutes. Direct smear examination (Gram staining) was performed from positive blood cultures as soon as detected, obtained at 30-minute intervals |
The significance of either the isolation of a potentially contaminating micro-organism in a single set of blood cultures or the detection of CoNS DNA in blood by the SeptiFast assay was judged on the basis of clinical grounds | NR | Episodes | Included |
| Hettwer et al. (2011)84 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venous Volume: 10 ml EDTA Reference standard and index text performed on blood samples drawn at the same time |
Unclear | While blood culture/SeptiFast samples collected at same time, SeptiFast measurements obtained 5 months after all blood culture results were available | Blood culture with microbiological data and clinical outcome Sample: whole blood Volume: 2 × 10-ml bottles of blood BacT/ALERT analysed at the Institute of Medical Microbiology. When an aerobic and/or anaerobic bottle returned a positive result, Gram stain procedure and assay culture were performed according to standardised procedures |
NR | NR | Patients | Included |
| Josefson et al. (2011)85 | SeptiFast Lys Kit MGRADE and the MagNA Lyser Sample: whole blood Site: venous (for all samples) Volume: 1.5 ml EDTA whole-blood sample Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Whole blood was stored for a maximum of 4 hours at room temperature, or up to 3 days at +4 °C, or 3 months at −70 °C prior to DNA preparation | Blood culture and clinical/laboratory confirmation One BACTEC Plus Aerobic/F bottle and one BACTEC Plus Anaerobic/F bottle Sample: whole blood Site: venous (for all samples) Volume: 8–10 ml for each aerobic and anaerobic bottle After transport at room temperature, the bottles were placed in a BACTEC 9240 incubator, with monitoring for pH changes every 10 minutes for 6 days. All signalling bottles were opened and an aliquot was taken for microscopy after Gram staining, culture on solid media, and further analyses for species designation |
A positive PCR result was considered to be fully supported when an identical micro-organism was isolated in the blood culture of the same blood culture/PCR set | NR | Patients | Included |
| Lucignano et al. (2011)86 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: peripheral or central venous line Volume: ≥ 1.5 ml paired blood samples for SeptiFast Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture plus clinical (vital signs) and laboratory variables (leucocyte count, C-reactive protein, microbiological evidence of infection focus) Sample: whole blood Site: peripheral or central venous line Volume: 0.5–10 ml of blood depending on whether aerobic or anaerobic bottle Blood culture was performed using the BACTEC system. The bottles were then incubated at 35 °C in BACTEC 9240/9120 blood culture system cabinets for 8 days. In case of positivity, Gram staining and culture on solid medium were performed; definitive organism identification and antibiotic susceptibility were determined with accredited routine laboratory methods [VITEK 2 system (bioMérieux, Durham, NC, USA) or Phoenix (BD Diagnostics) system] |
The condition of sepsis was defined when SIRS was in the presence, or a result, of suspected or proven infection, ascertained by the microbiology routine team, which addressed the final interpretation of the results (contaminants vs. pathogens) on the basis of type of microbe, TTP, number of positive blood cultures for the same microbe and patient data provided by clinicians | NR | Samples | Excluded |
| Obara et al. (2011)87 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: 1.5 ml K-EDTA for the molecular method Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture: NR if with clinical confirmation or not Sample: whole blood Site: NR Volume: 5 ml of blood Blood culture was performed using the BACTEC system. After cultivation, the strain was identified by WalkAway 96 SI system and auto scan-4 (Dade Behring GmbH, Germany) |
NR | NR | Samples | Included |
| Vrioni et al. (2011)88 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | NR | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Patients | NR |
| Alvarez et al. (2012)89 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture, tracheal aspirate, urine, surgical wounds, intravenous catheters and other sources. No further detail reported | NR | NR | No diagnostic data reported | No diagnostic data reported |
| Grif et al. (2012)90 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous Volume: 1.5 ml processed from 5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Samples collected every day except weekends and testing done immediately | Blood culture alone and blood culture with other microbiological cultures Sample: whole blood Site: venous Volume: 20 ml BacT/ALERT 3D, two bottles incubated for maximum 5 days at 37 °C. From all bottles signalled as positive, micro-organisms were isolated and identified according to standard laboratory methods |
NR | NR (but samples collected every day except on weekends and testing done immediately with both assays) | Samples | NR |
| Guido et al. (2012)91 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous Volume: 1.5 ml K-EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venous Volume: 10 ml BacT/ALERT 3D 2 bottles. All bottles signalled as positive were removed from the instrument and an aliquot was taken for Gram stain and culture on solid media for subsequent analysis. Identification and determination of sensitivity to antibiotics were performed with the VITEK 2 system |
NR | NR (but positive blood culture bottles from automated blood culture system removed between 8 a.m. and 7 p.m. for further analysis) | Samples | Included |
| Lodes et al. (2012)92 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: NR Volume: 1 ml EDTA for assay (a pair of aerobic/anaerobic blood culture bottles – 9 ml whole blood collected by EDTA for further PCR analysis) Reference standard and index tests performed on blood samples drawn at the same time |
No [1 ml (adults)] | NR | Blood culture plus clinical diagnosis (text suggests clinical diagnosis) Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system |
NR | NR | Samples | Included |
| Mauro et al. (2012)93 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 1.5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR BACTEC 9240 – one set of blood cultures (aerobic/anaerobic and fungal). When the blood culture gave a positive signal, Gram staining was carried out. An aliquot of positive blood culture was plated onto solid media and incubated for 24/48 hours, and identification was carried out with a VITEK 2 system |
NR | NR | Samples | Included |
| Pasqualini et al. (2012)94 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 1.5 ml K-EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system. All bottles flagged positive were removed from the instrument, and an aliquot was taken for Gram stain and culture on solid media for subsequent analysis. Identification and determination of sensitivity to antibiotics were performed with conventional methods and with the Phoenix automatic system |
Micro-organisms detected by SeptiFast were considered to be pathogens if the results of the DNA kit coincided with the results of the blood culture analysis | NR | Samples | Included |
| Rath et al. (2012)95 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venepuncture or central venous catheter Volume: 3 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venepuncture or central venous catheter Volume: 8–10 ml of blood per bottle Blood culture was performed using the BACTEC system |
NR | NR | Samples | Included |
| Tschiedel et al. (2012)96 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 1.5 ml Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system |
NR | NR (however, laboratories open until 11 p.m. Monday to Friday) | Samples | Included |
| Herne et al. (2013)97 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous Volume: 1.5 ml from 3 ml K2EDTA All blood culture/SeptiFast samples collected within 12 hours of each other |
Yes | Perform SeptiFast from 8 a.m. to 4 p.m. only, 7 days of the week | Blood culture alone and blood culture with clinical and other microbiological cultures Sample: whole blood Site: venous Volume: > 2 × 10 ml bottles BacT/ALERT 3D. At least two sets of 10-ml paired bottles 0.5–1 hour apart during the acute febrile episode. Blood cultures for aerobes and anaerobes were incubated up to 7 days, those for fungi for up to 11 days. In the event of positive blood cultures, micro-organisms were identified according to standard laboratory procedures:
|
Defined all cases in which both methods gave positive results as well as all cases in which a positive result in either SeptiFast or blood culture was considered clinically relevant as true positive results | 7 days per week (SeptiFast assay performed from 8 a.m. to 4 p.m. only) | Samples | Included |
| Kasper and Altiok (2013)98 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 0.1–0.7 ml (maximum) blood sample into 0.8 ml K3EDTA vacutainers Reference standard and index tests performed on blood samples drawn at the same time |
No (0.1–0.7 ml, neonates) | Samples analysed immediately or stored at –20 °C and processed next day | Blood culture plus clinical diagnosis Sample: whole blood Site: NR Volume: 0.5–1.0 ml of blood Blood culture was performed using the BacT/ALERT system. Only aerobic paediatric bottle was inoculated and incubated for 7 days (because of limited blood volumes) by standard microbiological procedures |
NR | NR | Patients | Included |
| Paolucci et al. (2013)99 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR Reference standard and index tests performed on blood samples drawn at the same time |
No (3 ml, adults) | Blood samples intended for PCR analyses arriving to the laboratory after 12 p.m. were stored at 4 °C until the next PCR session was programmed | Blood culture was performed using the BacT/ALERT system | NR | Blood cultures were accepted for automated incubation from 8 a.m. to 7 p.m. (Monday to Saturday). PCR was performed once per day from Monday to Friday (samples were received by 12 p.m.). Blood samples intended for PCR analyses arriving to the laboratory after 12 p.m. were stored at 4 °C until the next PCR session was programmed (time limit for storage was 72 hours, as suggested by the manufacturer) | Episodes | Excluded |
| Rodrigues et al. (2013)100 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture was performed using the BacT/ALERT system | NR | NR | Incomplete diagnostic data reported | Incomplete diagnostic data reported |
| Avolio et al. (2014)101 | LightCycler SeptiFast Test MGRADE 1.5 ml of whole blood was collected in sterile EDTA-KE Reference standard and index tests performed on blood samples drawn at the same time |
Yes | SeptiFast assays were performed once daily for samples collected in the previous 24 hours | Blood culture and clinical/laboratory confirmation A blood culture bottle of 20 ml of blood either by venepuncture or from an intravenous access device BacT/ALERT 3D automated system (bioMérieux). All instances in which a maximum of three sets (six bottles) of blood cultures for patient, obtained during a 24-hour period and arrived simultaneously at laboratory was included. When aerobic/anaerobic bottles gave a positive signal, Gram staining was carried out |
Micro-organisms detected by SeptiFast were considered to be pathogens if results coincided with those of blood culture and/or in accordance of the American College of Clinical Pharmacy/Society of Critical Care Medicine Conference Committee definition of infection | 7 days per week (Monday to Friday from 7:30 a.m. to 5 p.m.; Saturday and Sunday from 7:30 a.m. to 12:30 p.m.) | Pathogen | Excluded |
| Burdino et al. (2014)102 | LightCycler SeptiFast Test MGRADE Sample: whole blood Sire: venous Volume: 1.5 ml EDTA Reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | Blood culture with clinical and laboratory signs of infection. Clinical evidences, laboratory findings, and microbiological data combined with the identification of the same bacteria from other body sites as defined by the Weinstein algorithm were used to confirm pathogens vs. irrelevant and/or contaminants for both blood culture and SeptiFast Sample: whole blood Site: venous Volume: 8–10 ml of blood bioMérieux blood culture bottles incubated at least 24–72 hours (for positive blood cultures) to 5 days for negative results |
NR | NR (but assays performed daily with one or more runs with dedicated personnel) | Samples | Excluded |
| Mancini et al. (2014)103 | LightCycler SeptiFast Test (MGRADE NR) Sample: uncultured blood Site: NR Volume: 1.5 ml Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Yes | NR | BacT/ALERT 3D blood culture system | NR | The molecular assay in the prospective cohort was organised assuring two daily sessions from Monday to Friday | Incomplete diagnostic data reported | Incomplete diagnostic data reported |
| Markota et al. (2014)104 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: peripheral veins Volume: NR but EDTA 5 ml drawn Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | On same day if sampled before 6 p.m., next day if sampled after 6 p.m. | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: peripheral veins Volume: 20 ml (10 ml inoculated in each aerobic/anaerobic bottle) Blood culture was performed using the BacT/ALERT system. All blood culture bottles signalled as positive were processed according to standard microbiology laboratory procedures |
NR | SeptiFast assays were performed from 8 a.m. to 4 p.m. on weekdays and from 8 a.m. to 1 p.m. on Saturdays | Sample | NR |
| Ozkaya-Parlakay et al. (2014)105 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venepunctures, site NR Volume: NR Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | If possible, PCR was evaluated immediately, if not, stored at –20 °C | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venepunctures, site NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Sample | Included |
| Schaub et al. (2014)106 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venepunctures, site NR Volume: NR, but collected in 2.7-ml EDTA tubes Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | The turnaround time of MRT-PCR results was calculated on the assumption that the MRT-PCR could be performed 24 hours a day, 7 days a week and using the reported turnaround time of the assay of approximately 6 hours | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venepunctures, site NR Volume: 25 ml inoculated into an aerobic and anaerobic bottle Blood culture was performed using the BacT/ALERT system |
Patients were categorised as ‘true positive’ or ‘true negative’ for bacterial sepsis on the basis of the expanded reference standard, for example conventional microbiological methods such as culture of blood/urine/sputum, rapid antigen testing in throat swabs (streptococcal rapid antigen test) or urine (Streptococcus pneumoniae, Legionella pneumophila) | NR (however, positive blood culture bottles removed from automated system from 8 a.m. to 5 p.m.) | Patients | Included |
| Sitnik et al. (2014)107 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: NR Volume: 5 ml collected from each patient in sterile EDTA tube (mechanical lysis on 3 ml) Reference standard and index tests performed on blood samples drawn at the same time |
No [3 ml adults)] | Blood samples stored at –20 °C and multiplex PCR testing done twice per week | Blood culture in conjunction with clinical adjudication Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system. When a positive signal was obtained, Gram staining of the blood culture medium in the bottles was performed. Samples were plated onto blood agar, chromogenic agar and anaerobic blood agar. Identification of bacterial or fungal species as well as antibiotic sensitivity tests were then carried out using the VITEK 2 system and API 32 °C for yeast |
NR | NR | Sample | Included |
| Barbanti et al. (2015)108 | LightCycler SeptiFast Test (MGRADE NR) Sample: NR Site: NR Volume: NR Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Unclear | NR | Blood culture was performed using the BacT/ALERT system | NR | NR | Samples | Included |
| Calitri et al. (2015)109 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: venous or catheter Volume: 1.5 ml EDTA Unclear if reference standard and index tests performed on blood samples drawn at the same time |
Unclear | Samples were processed within a few hours of collection for the SeptiFast test; blood culture collected on same day or if unavailable the nearest blood culture performed was recorded (± 48 hours maximum from SeptiFast collection) | Blood culture was performed using the BacT/ALERT system. Each blood culture consisted of one bottle aerobic or anaerobic cultures, each inoculated with minimum blood required according to paediatric age group | NR | NR | Episodes | Included |
| Idelevich et al. (2015)110 | LightCycler SeptiFast Test MGRADE Sample: whole blood Site: peripheral veins in adults central venous catheter in children Volume: 3 ml ≤ 45 kg, 1.5 ml > 45 kg Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Samples for PCR analyses that arrived at the laboratory after 12 p.m. were stored at 4 °C and processed on the next day. The median time to the arrival of blood culture samples at the microbiological laboratory was 13.3 hours (12.9 hours in the study group and 13.6 hours in the control group; p = 0.14). It took 14.8 hours for whole-blood samples of the study group patients to arrive at the PCR department. This includes the arrival time at the microbiological laboratory and forwarding samples to the PCR department | Blood culture was performed using the BACTEC system. The blood culture was incubated for at maximum 7 days | NR | Blood culture Monday to Friday 7:30 a.m. to 6:00 p.m., Saturday 7:30 a.m. to 1:30 p.m.; PCR Monday to Friday 7:30 a.m. to 6:00 p.m. | Pathogen | Included |
| Tafelski et al. (2015)112 | LightCycler SeptiFast Test (MGRADE NR) Sample: whole blood Site: venous or arterial site Volume: 1.5 ml for SeptiFast assay Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Samples were processed immediately for the SeptiFast test; however, in the control group, samples were stored at –80 °C for later analysis | Blood culture plus MALDI-TOF MS with clinical adjudication Sample: whole blood Site: peripheral venous blood Volume: 10 ml each for blood culture (anaerobic or aerobic bottle) Blood culture was performed using the BACTEC system. Positive bottles were streaked onto a set of agar plates and subjected to direct Gram staining. Pathogens identified using MALDI-TOF MS or biochemical identification test (VITEK 2). Commercial procedures used for antimicrobial susceptibility testing (VITEK 2 or Etest) |
For analysis of the test performance of the LightCycler SeptiFast PCR test, blood culture results were used as the diagnostic gold standard for detecting bacteraemia | 5 days a week (6 a.m. to 6 p.m.) | Samples | NR |
| Warhurst et al. (2015)111 | SeptiFast Test (MGRADE NR) Sample: whole blood Site: Two separate sites (including one peripheral site) Volume: used 1.5 ml for assay (20 ml drawn) Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Blood for DNA detection kit was stored at 4 °C for up to 72 hours before PCR analysis | Blood culture in conjunction with clinical adjudication. Blood cultures entered the standard clinical pathway, and the results were returned directly to the clinical service at each centre Sample: whole blood Site: two separate site (including one peripheral site) Volume: two blood samples of 20 ml from two separate sites Blood culture was performed using BacT/ALERT system. Blood cultures entered the standard clinical pathway |
NR | NR | Pathogens | Excluded |
| Single index test studies – SepsiTest | ||||||||
| Wellinghausen et al. (2009)48 | SepsiTest Sample: whole blood Site: NR Volume: 1 ml in duplicate for adults and 1 ml in single for children Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Samples were sent for PCR from the local laboratory to the central study laboratory in Bremen within 2 days. Blood cultures were incubated at the local laboratories in automated BACTEC 9240 systems for up to 7 days | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: 20 ml for adults, 3–5 ml for children Blood culture was performed using the BACTEC system in adults and BACTEC PED system in children |
Probable to true bacteraemia was assigned if:
|
NR | Samples | Included |
| Loonen et al. (2014)114 | SepsiTest Sample: remnant whole blood 1 ml When samples drawn: NR |
Yes | NR | Blood culture plus MALDI-TOF MS and clinical/laboratory confirmation Remnant whole blood 1 ml Blood culture was performed using the BacT/ALERT system. Two pairs of aerobic and anaerobic bottles were obtained and incubated for at least 5 days and a maximum of 7 days |
NR | NR | Samples | Included |
| Nieman et al. (2015)113 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Single index test studies – IRIDICA | ||||||||
| Bacconi et al. (2014)49 | IRIDICA–PLEX-ID Hybrid Sample: whole blood Site: venepuncture, arm Volume: 5–15 ml Reference standard and index tests performed on blood samples drawn within 30 minutes |
Yes | Kept at 4 °C within 30-minute collection | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: venepuncture, arm Volume: NR Blood culture was performed using the BACTEC system. The bottles were incubated and monitored for 5 days before being called negative. Positive blood cultures were removed immediately from the instrument, and a Gram stain was performed |
When PCR/ESI-MS-positive but culture-negative specimens were confirmed by repeat PCR/ESI-MS testing of additional replicate specimens and the confirmed detections were considered true positives | NR | Samples | NR |
| Delco-Volante et al. 2015115 | IRIDICA Sample: whole blood Site: venepuncture Volume: 0.5 ml Reference standard and index tests performed on blood samples drawn at the same time |
No [0.5 ml (neonates)] | NR | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
NR | NR | Samples | Included |
| Vincent et al. (2015)116 | IRIDICA–PLEX-ID Hybrid Sample: whole blood Site: NR Volume: minimum 5 ml Reference standard and index tests performed on blood samples drawn at the same time |
Yes | Specimens were cooled to 4 °C within 30 minutes of collection and maintained at 4 °C or frozen at –20 °C until analysis | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Sampling method: NR Culture method: NR |
The simple kappa statistic was used to estimate the agreement between culture and PCR/ESI-MS, but it under-represents this agreement because of the requirement that the PCR/ESI-MS assay and blood culture results matched in terms of organism identity in order to be considered a true positive in the contingency table | NR | Samples | Excluded |
| Metzgar et al. (2015)117 | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed | Confidential information has been removed |
| Two index test studies: SeptiFast and SepsiTest | ||||||||
| Leitner et al. (2013)118 | SeptiFast Lys Kit and the SeptiFast Prep Kit MGRADE: NR Sample: whole blood Site: venous or indwelling peripheral or central catheter Volume: 6 ml in EDTA tubes Reference standard and index tests performed on blood samples drawn at the same time SepsiTest Sample: whole blood Site: venous or indwelling peripheral or central catheter Volume: NR (but collected in 6 ml EDTA tubes) Reference standard and index tests performed on blood samples drawn at the same time |
Unclear | Samples were tested in parallel with blood culture | Blood culture and clinical/laboratory confirmation Blood culture sets (blood cultures) consisting of three pairs of aerobic/anaerobic blood culture bottles. If the instrument reported a blood culture bottle positive, conventional biochemical identification methods and susceptibility testing were done Sample: whole blood Site: Venous or indwelling peripheral or central catheter Volume: NR Blood culture was performed using the BACTEC system. The blood culture was incubated for a maximum 7 days |
Bacterial pathogens were considered true positive if growing in at least one blood culture bottle. Potential skin contaminants were considered true positive only if the identical organism was growing in two or more blood culture bottles | NR | Samples | Included |
| Schreiber and Nierhaus (2013)119 | LightCycler SeptiFast Test MGRADE: NR Sample: whole blood Site: NR Volume: NR When samples drawn: NR SepsiTest Sample: whole blood Site: NR Volume: NR When samples drawn: NR |
Unclear | Immediately after sampling, the containers for blood cultures were transported to the Institute of Medical Microbiology for cultivation at 37 °C. The EDTA blood samples were stored at 4–10 °C and the NA was extracted within 24 hours, in accordance with the manufacturers’ specifications | Blood culture and clinical/laboratory confirmation Sample: whole blood Site: NR Volume: NR Blood culture was performed using the BACTEC system |
The blood culture and PCR results were compared with the final clinical diagnosis and categorised as true positive or false positive and true negative or false negative | NR | Patients | Included |
Appendix 5 Diagnostic test accuracy, additional information
| Model | DICa |
|---|---|
| Covariate adjustment | |
| Standard model | 630.10 |
| Age categories | 624.55 |
| Febrile neutropenia | 630.78 |
| Clinical setting | 630.26 |
| Inclusion/exclusion of contaminants | 631.43 |
| Model parameter | Regression coefficient, median (95% CrI), logit scale |
|---|---|
| Sensitivity | –0.17 (–1.16 to 0.78) |
| Specificity | –0.58 (–1.24 to 0.10) |
Appendix 6 Literature search strategies for the review of cost-effectiveness: a MEDLINE example
| MEDLINE search strategy | Details |
|---|---|
| Database searched | Ovid MEDLINE In-Process & Other Non-Indexed Citations and Ovid MEDLINE |
| Platform or provider used | Ovid SP |
| Date of coverage | 1948 to May 2015 |
| Search undertaken | Initial search February 2015 |
| Updated search | May 2015 |
Search strategy
-
exp Sepsis/
-
sepsis.mp.
-
septic?emia.mp.
-
Shock, Septic/
-
((septic or endotoxic or toxic) adj shock).tw.
-
Bacteremia/
-
bacter?emia.mp.
-
Fungemia/
-
fung?emia.mp.
-
Systemic Inflammatory Response Syndrome/
-
sirs.mp.
-
blood$ infection$.tw.
-
blood poison$.tw.
-
or/1-13
-
septifast.mp.
-
lightcycler.mp.
-
15 or 16
-
14 and 17
-
sepsitest.mp.
-
iridica.mp.
-
(plex id or plex-id).mp.
-
or/19-21
-
exp Polymerase Chain Reaction/
-
polymerase chain reaction$.tw.
-
pcr$.mp.
-
Gene Amplification/
-
Nucleic Acid Amplification Techniques/
-
or/23-27
-
Genes, Bacterial/ or Genes, Fungal/
-
(exp bacteria/ or exp Fungi/) and exp Nucleic Acids/
-
((bacteri$ or fung$) adj3 (dna or gene$ or nucleic acid$)).tw.
-
blood culture$.tw.
-
or/29-32
-
14 and 28 and 33
-
18 or 22 or 34
-
Animals/ not (Humans/ and Animals/)
-
35 not 36
-
limit 37 to yr=“2006 -Current”
-
exp “Costs and Cost Analysis”/
-
Economics/ (26570)
-
exp Economics, Hospital/
-
exp Economics, Medical/
-
Economics, Nursing/
-
exp models, economic/
-
Economics, Pharmaceutical/
-
exp “Fees and Charges"/
-
exp Budgets/
-
budget$.tw.
-
ec.fs.
-
cost$.ti.
-
(cost$ adj2 (effective$ or utilit$ or benefit$ or minimi$)).ab.
-
(economic$ or pharmacoeconomic$ or pharmaco-economic$).ti.
-
(price$ or pricing$).tw.
-
(financial or finance or finances or financed).tw.
-
(fee or fees).tw.
-
(value adj2 (money or monetary)).tw.
-
quality-adjusted life years/
-
(qaly or qalys).af.
-
(quality adjusted life year or quality adjusted life years).af.
-
or/39-59
-
38 and 60
-
38 not 61
Appendix 7 Population of key parameters by clinical estimates: reproduction of the correspondence sent to the clinical experts
Appendix 8 Results from the threshold analyses
The threshold analyses are divided into three categories based on the number of samples assumed to require analysing per day (2.4, 17 or 68). In each category each intervention is compared with both blood culture and MALDI-TOF MS. In these analyses it is assumed that the comparator has already been purchased and that the intervention will require purchasing.
Assuming 2.4 samples a day require analysing
Threshold analyses for SeptiFast versus blood culture
FIGURE 24.
Threshold analyses for SeptiFast vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 2.4 samples need to be analysed per day.

FIGURE 25.
Threshold analyses for SeptiFast vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 2.4 samples need to be analysed per day.
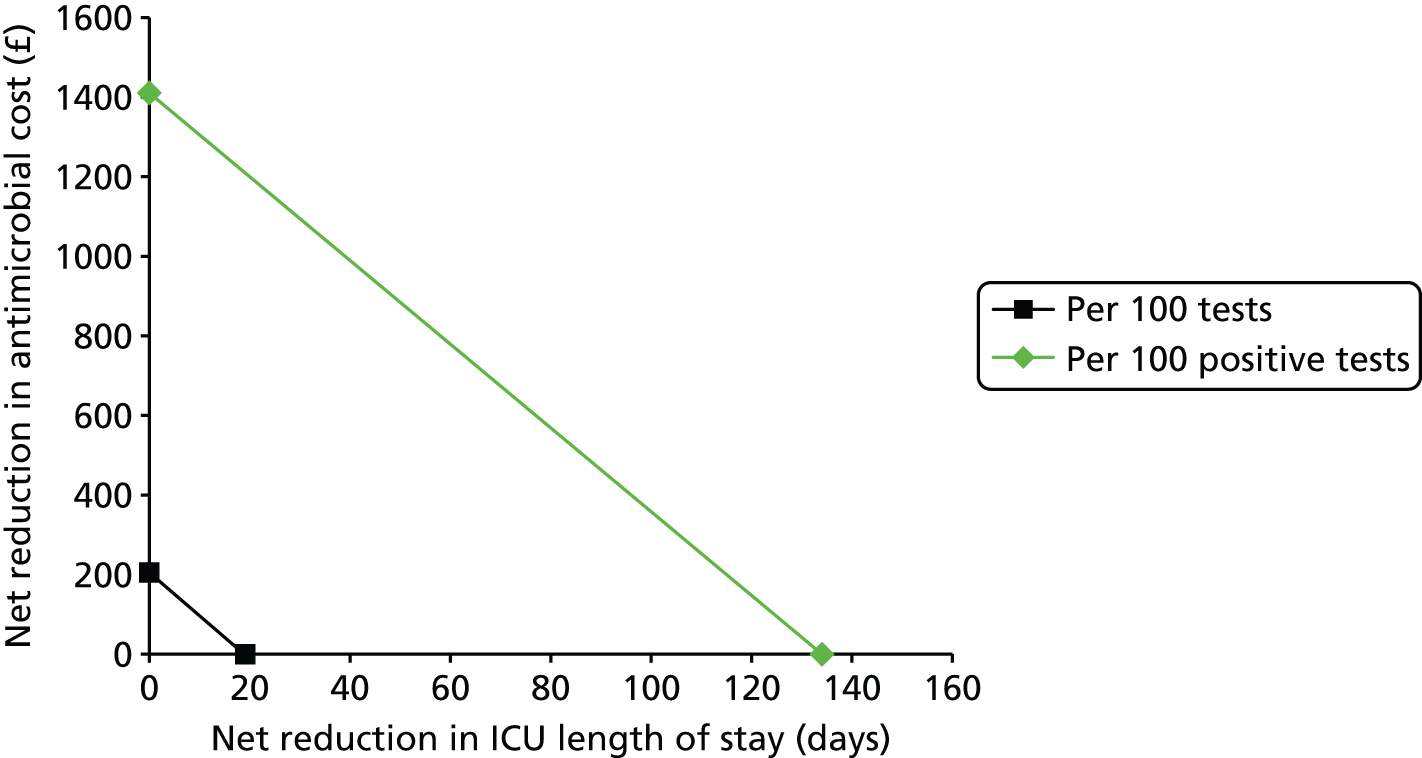
Threshold analyses for SeptiFast versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 26.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 2.4 samples need to be analysed per day.
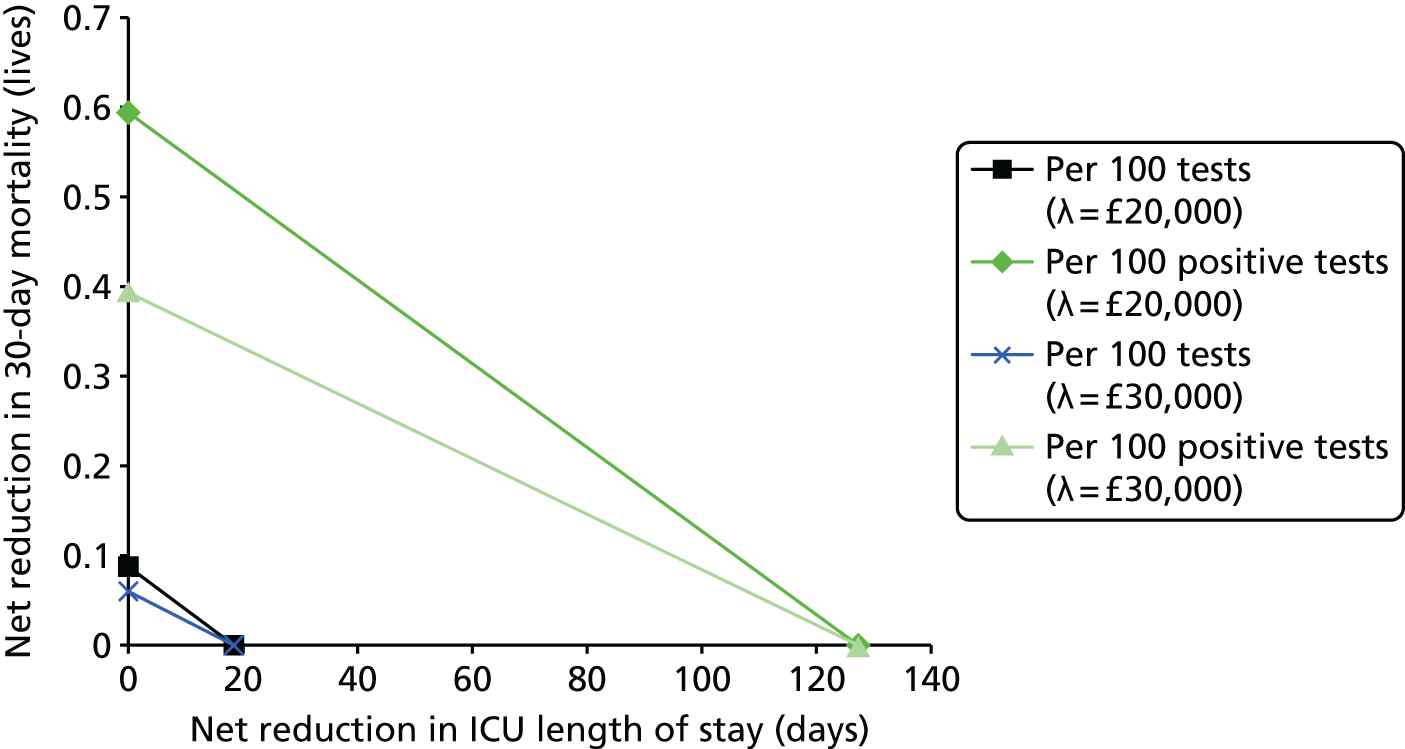
FIGURE 27.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 2.4 samples need to be analysed per day.

Threshold analyses for SepsiTest versus blood culture
FIGURE 28.
Threshold analyses for SepsiTest vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 2.4 samples need to be analysed per day.

FIGURE 29.
Threshold analyses for SepsiTest vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 2.4 samples need to be analysed per day.

Threshold analyses for SepsiTest versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 30.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 2.4 samples need to be analysed per day.

FIGURE 31.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 2.4 samples need to be analysed per day.
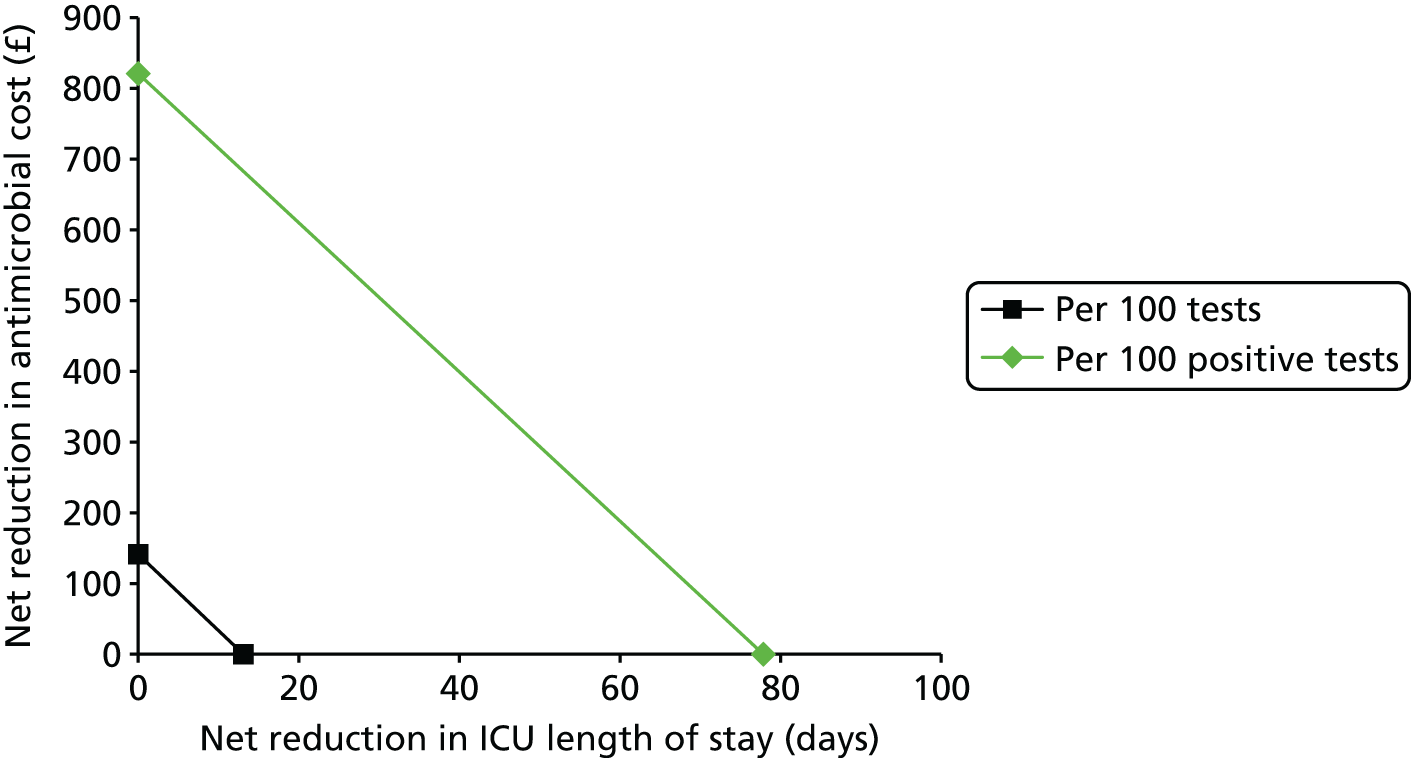
Threshold analyses for IRIDICA versus blood culture
FIGURE 32.
Threshold analyses for IRIDICA vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 2.4 samples need to be analysed per day.

FIGURE 33.
Threshold analyses for IRIDICA vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 2.4 samples need to be analysed per day.

Threshold analyses for IRIDICA versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 34.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 2.4 samples need to be analysed per day.
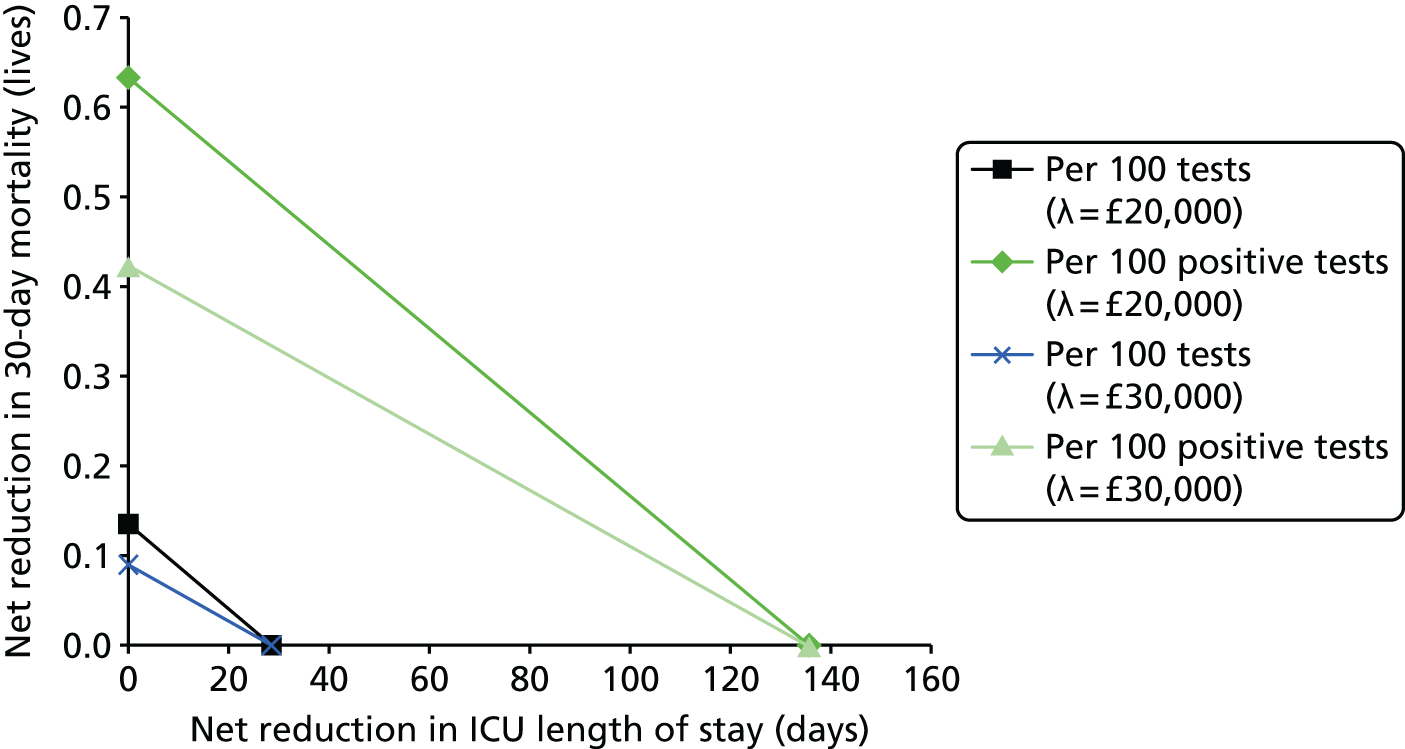
FIGURE 35.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 2.4 samples need to be analysed per day.

Assuming 17 samples a day require analysing
Threshold analyses for SeptiFast versus blood culture
FIGURE 36.
Threshold analyses for SeptiFast vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 17 samples need to be analysed per day.

FIGURE 37.
Threshold analyses for SeptiFast vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 17 samples need to be analysed per day.
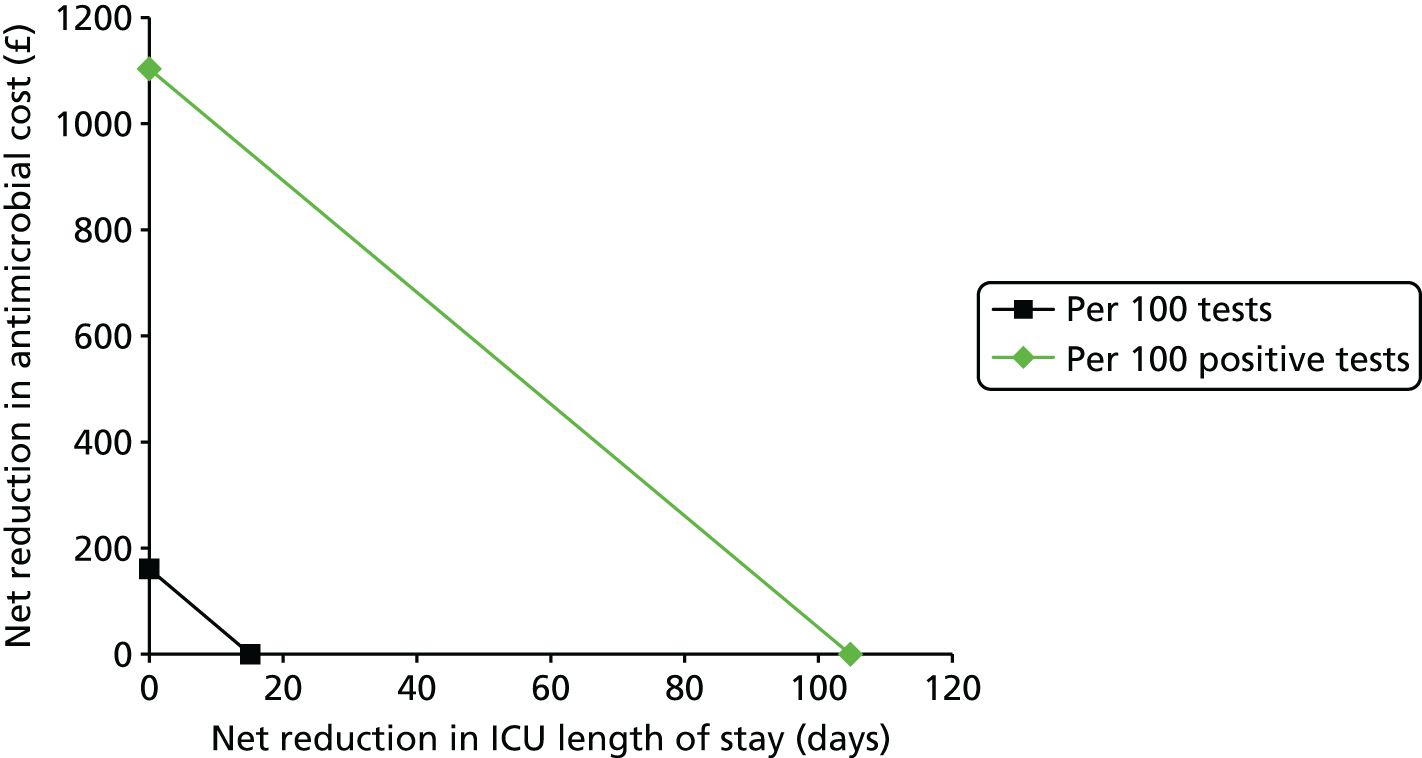
Threshold analyses for SeptiFast versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 38.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 17 samples need to be analysed per day.
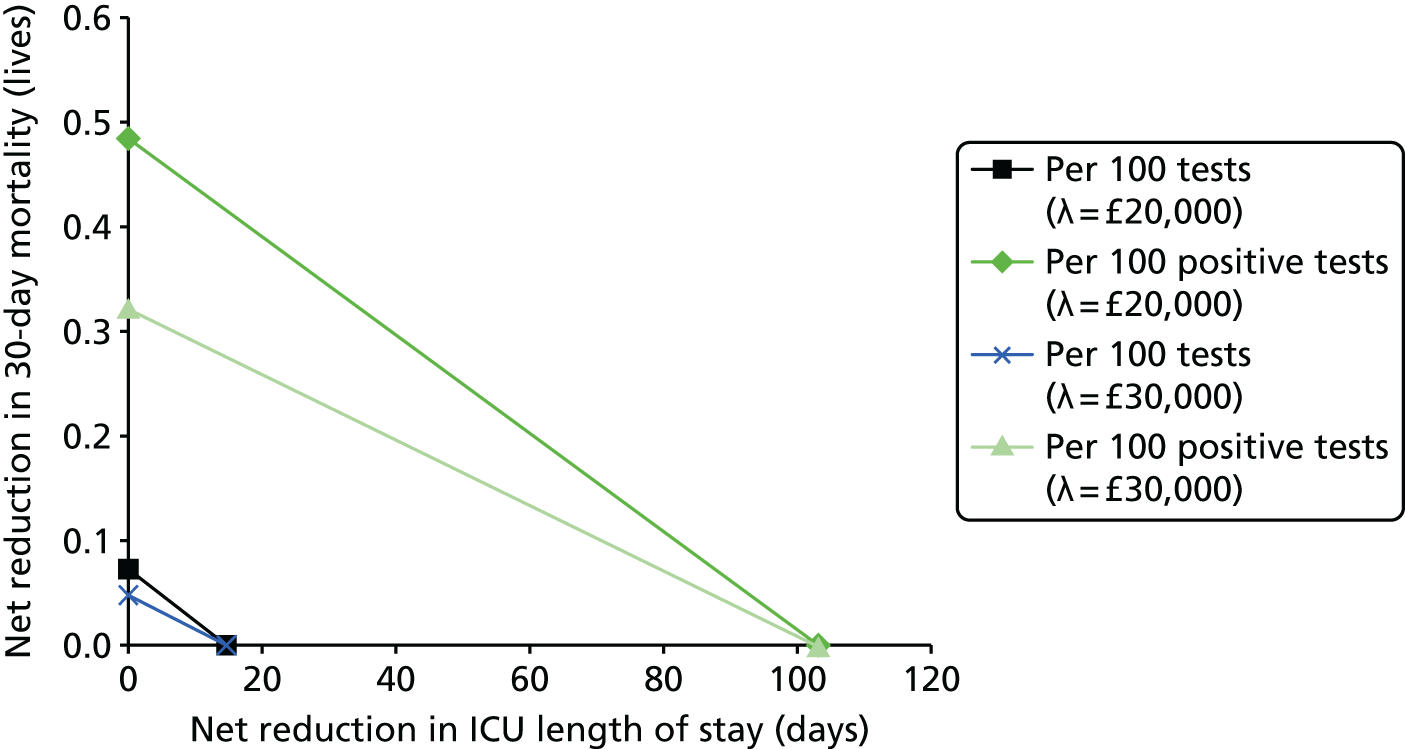
FIGURE 39.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 17 samples need to be analysed per day.

Threshold analyses for SepsiTest versus blood culture
FIGURE 40.
Threshold analyses for SepsiTest vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 17 samples need to be analysed per day.
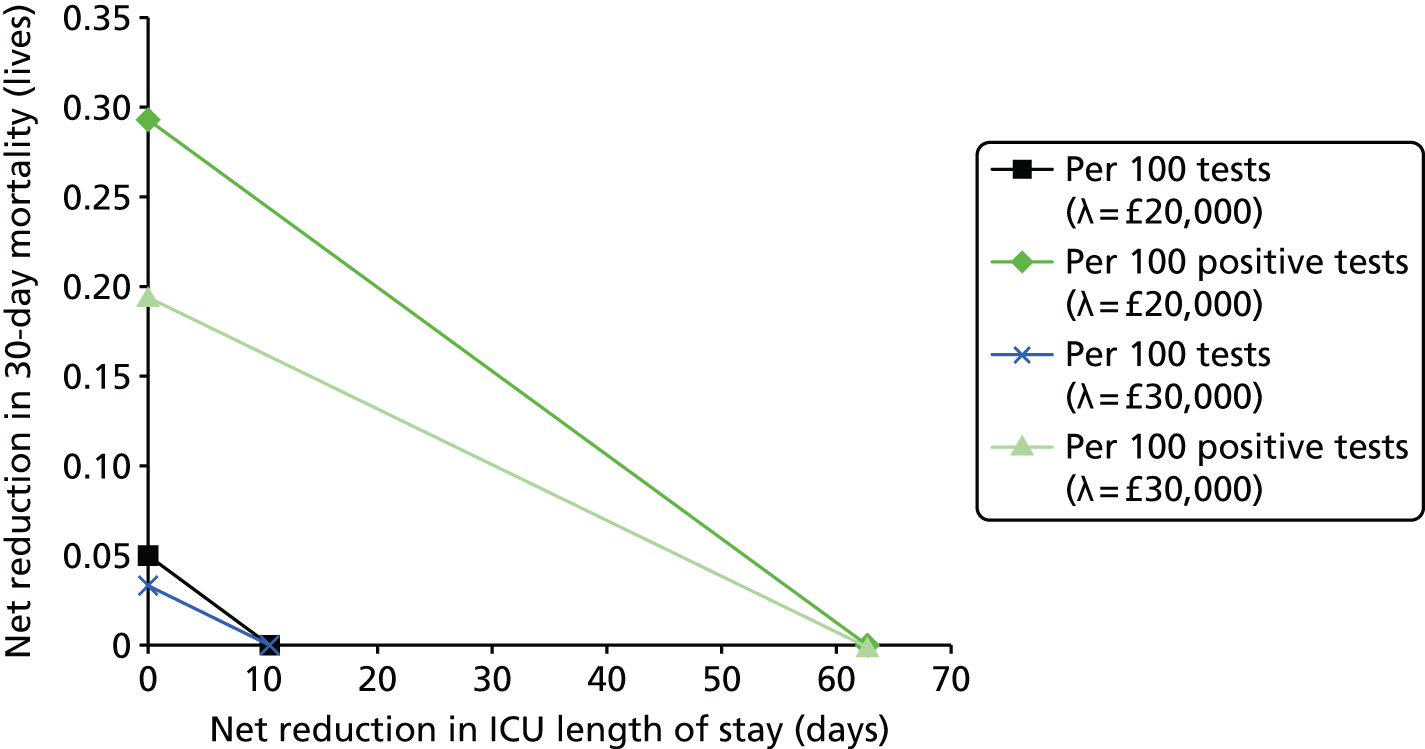
FIGURE 41.
Threshold analyses for SepsiTest vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 17 samples need to be analysed per day.

Threshold analyses for SepsiTest versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 42.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 17 samples need to be analysed per day.
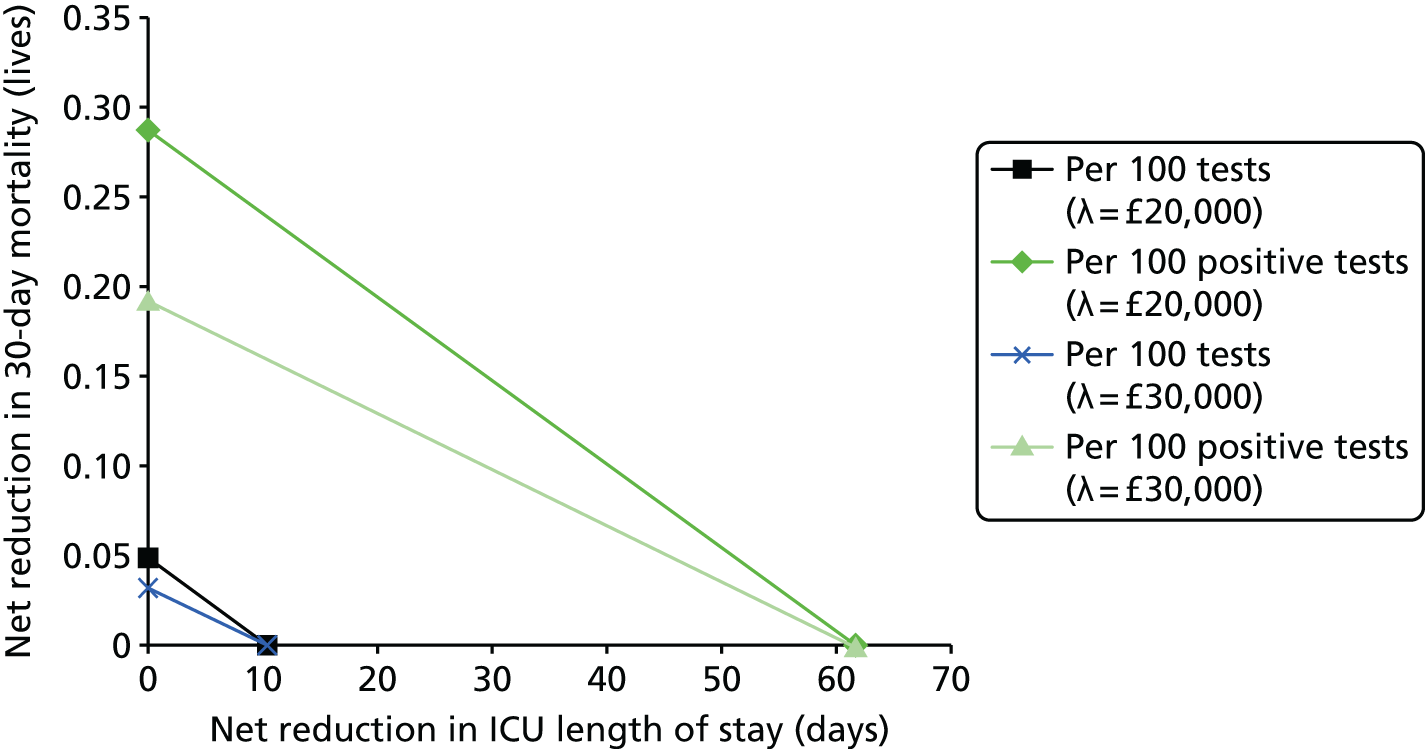
FIGURE 43.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 17 samples need to be analysed per day.
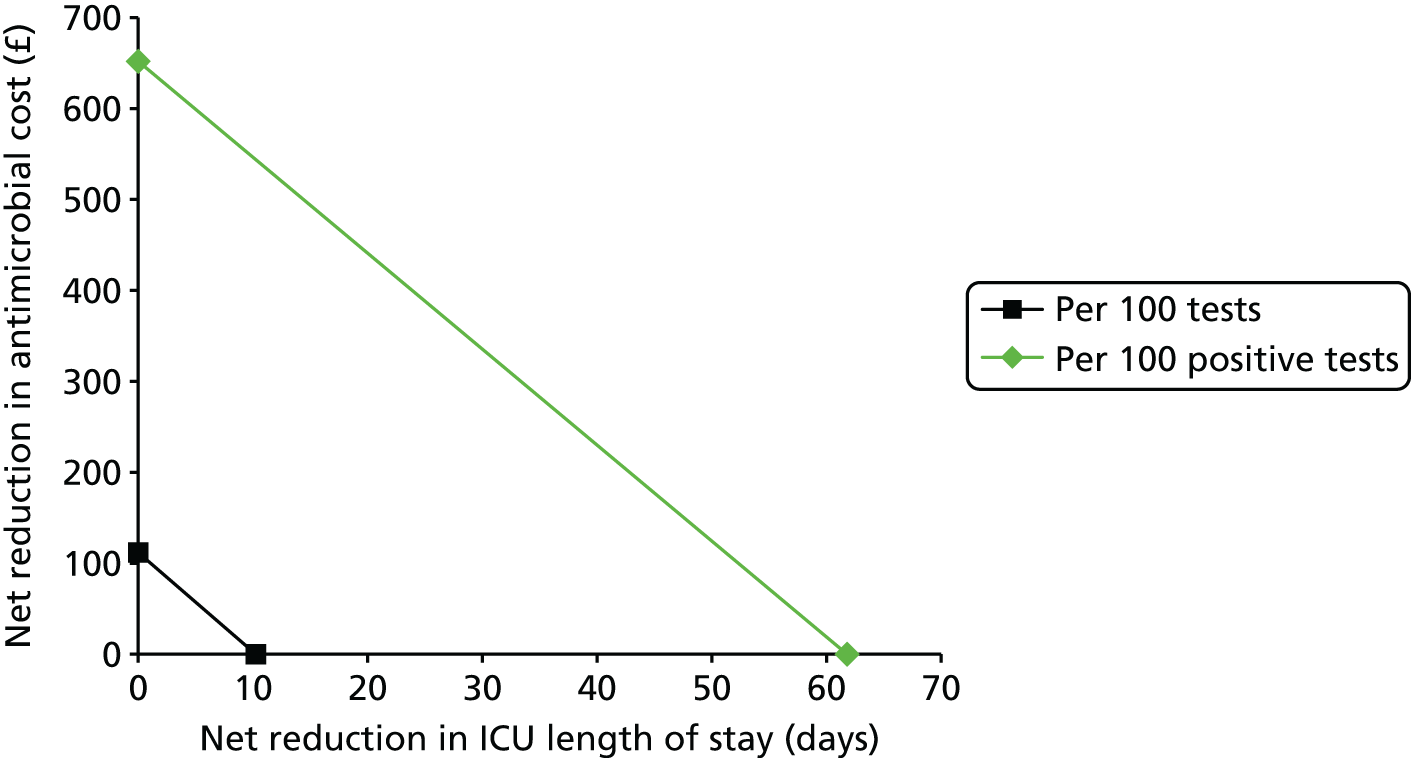
Threshold analyses for IRIDICA versus blood culture
FIGURE 44.
Threshold analyses for IRIDICA vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 17 samples need to be analysed per day.

FIGURE 45.
Threshold analyses for IRIDICA vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 17 samples need to be analysed per day.

Threshold analyses for IRIDICA versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 46.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 17 samples need to be analysed per day.
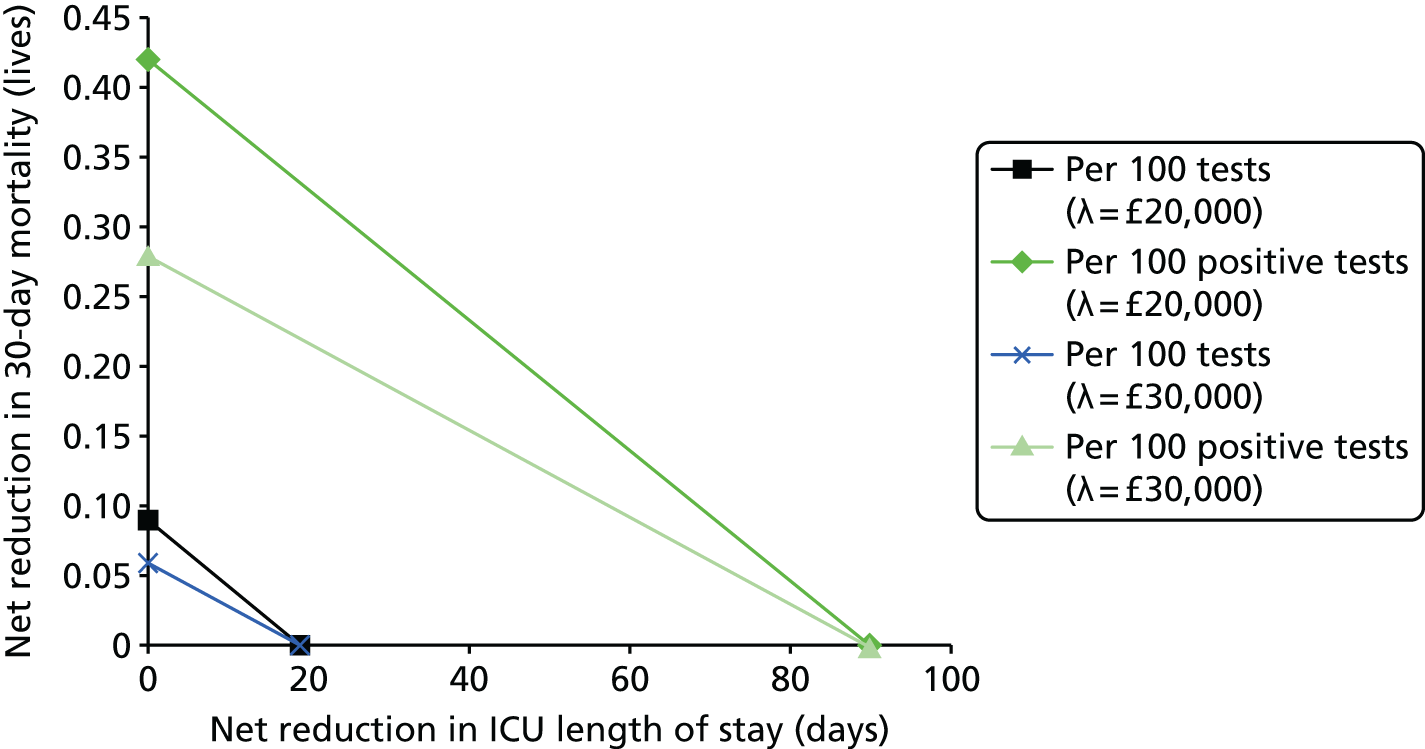
FIGURE 47.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 17 samples need to be analysed per day.

Assuming 68 samples a day require analysing
Threshold analyses for SeptiFast versus blood culture
FIGURE 48.
Threshold analyses for SeptiFast vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 68 samples need to be analysed per day.
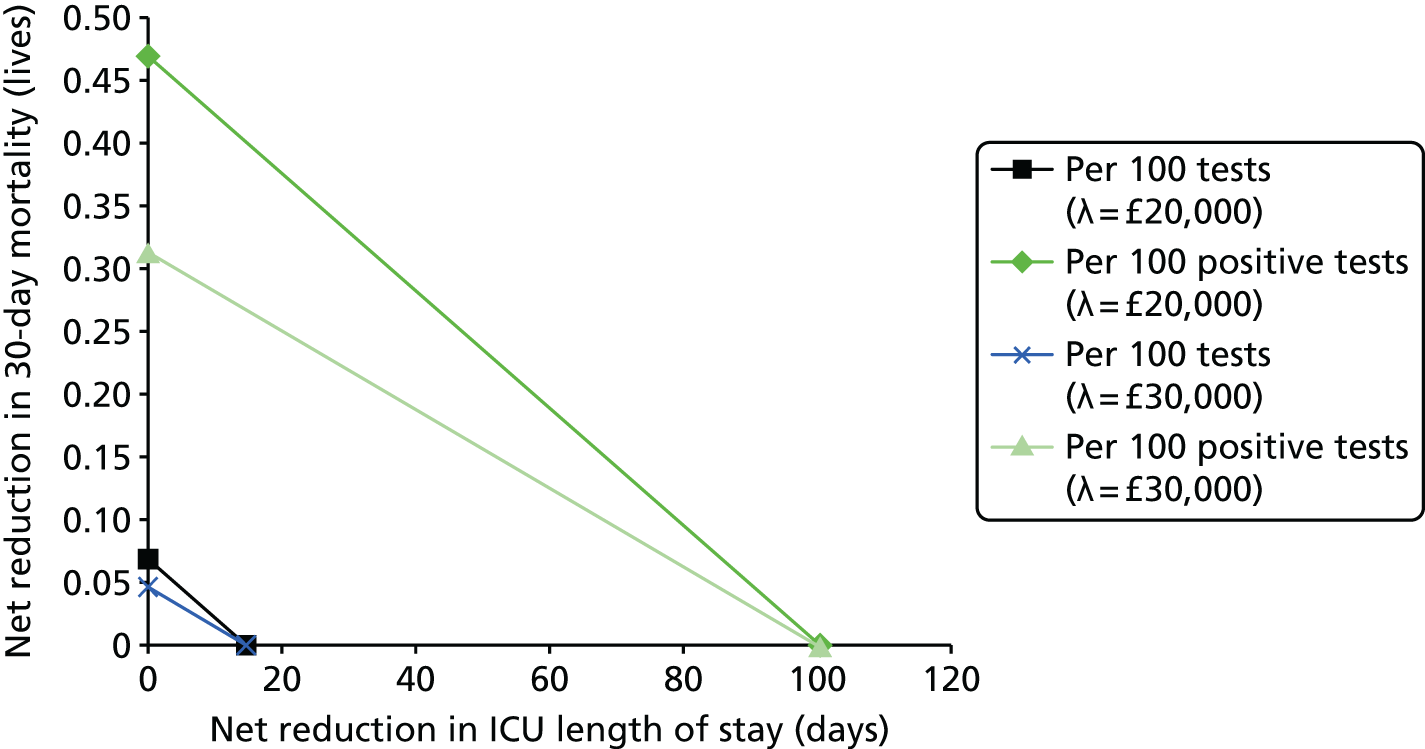
FIGURE 49.
Threshold analyses for SeptiFast vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 68 samples need to be analysed per day.

Threshold analyses for SeptiFast versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 50.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 68 samples need to be analysed per day.

FIGURE 51.
Threshold analyses for SeptiFast vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the SeptiFast machine needs to be purchased and 68 samples need to be analysed per day.

Threshold analyses for SepsiTest versus blood culture
FIGURE 52.
Threshold analyses for SepsiTest vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 68 samples need to be analysed per day.
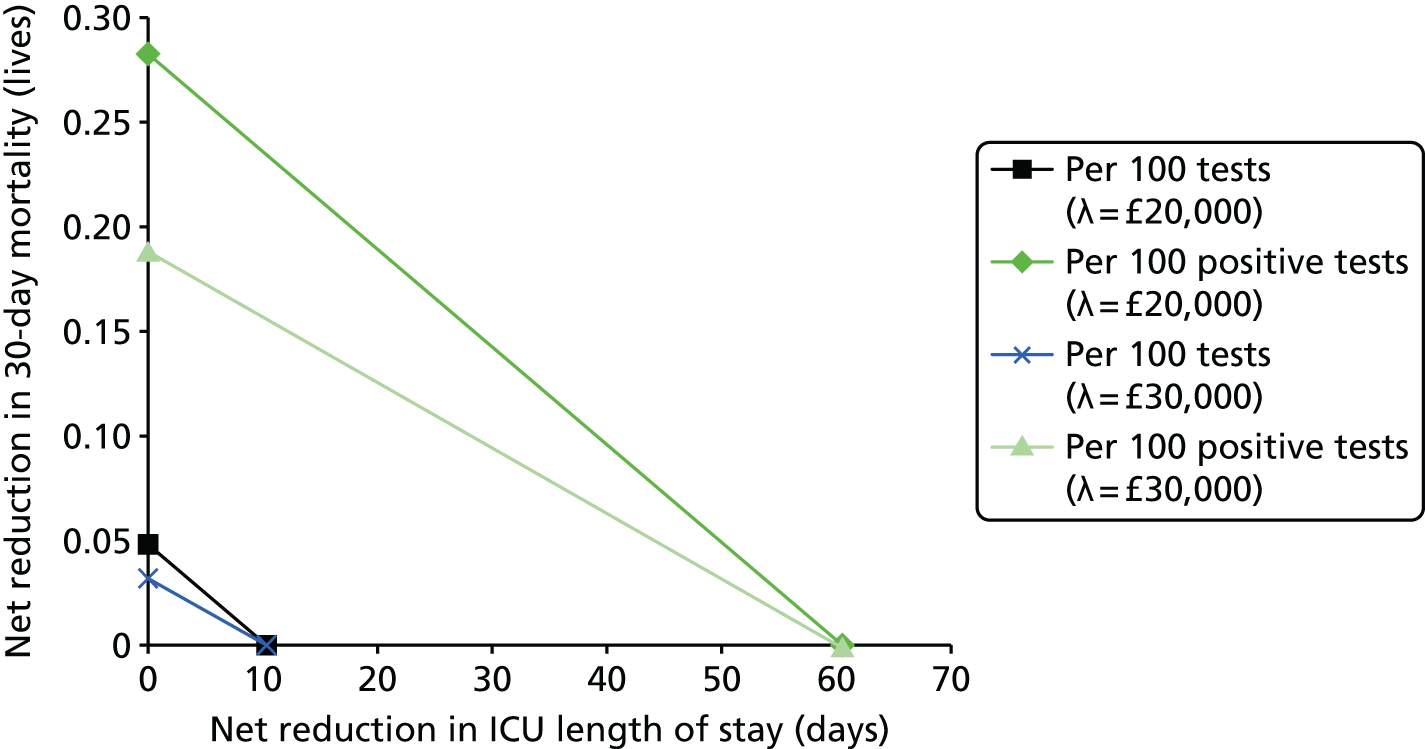
FIGURE 53.
Threshold analyses for SepsiTest vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 68 samples need to be analysed per day.
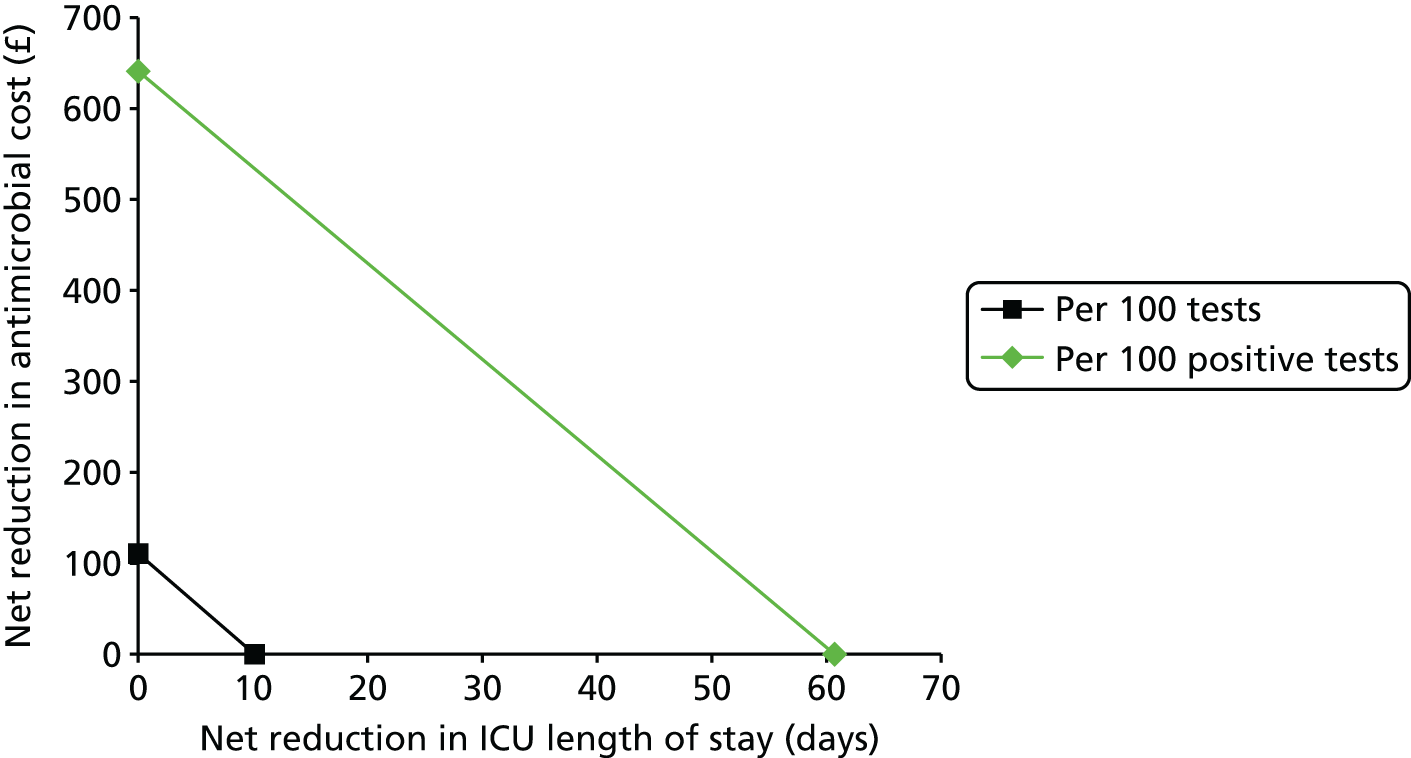
Threshold analyses for SepsiTest versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 54.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the machinery related to SepsiTest needs to be purchased and 68 samples need to be analysed per day.

FIGURE 55.
Threshold analyses for SepsiTest vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that machinery related to SepsiTest needs to be purchased and 68 samples need to be analysed per day.

Threshold analyses for IRIDICA versus blood culture
FIGURE 56.
Threshold analyses for IRIDICA vs. blood culture using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 68 samples need to be analysed per day.

FIGURE 57.
Threshold analyses for IRIDICA vs. blood culture using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 68 samples need to be analysed per day.

Threshold analyses for IRIDICA versus matrix-absorbed laser desorption/ionisation time-of-flight mass spectrometry
FIGURE 58.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in mortality and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 68 samples need to be analysed per day.

FIGURE 59.
Threshold analyses for IRIDICA vs. MALDI-TOF MS using net reduction in antimicrobial treatment costs and net reduction in ICU length of stay combined. Assuming that the IRIDICA machine needs to be purchased and 68 samples need to be analysed per day.

Glossary
- Aliquot
- A portion of a total amount of a solution.
- Amplicon
- A piece of deoxyribonucleic acid or ribonucleic acid that is the source and/or product of natural or artificial amplification or replication events.
- Antigen
- Any structural substance that serves as a target for the receptors of an adaptive immune response.
- Antimicrobial medications
- Drugs used to treat infections, such as antibiotics and antifungal and antiviral drugs.
- Bacteraemia
- The presence of bacteria in the blood.
- Bloodstream infection
- The presence of bacteria in the blood.
- Broad-spectrum antibiotic
- An antibiotic that acts against a wide range of disease-causing bacteria.
- Colony-forming unit
- A unit used to estimate the number of viable bacteria or fungal cells in a sample.
- Conformité Européenne mark
- A manufacturer’s declaration that the product meets the requirements of the applicable Electronic Commerce directives.
- Dominated
- Of an intervention, more expensive and providing the same or fewer additional quality-adjusted life-years than another intervention, or equally expensive and providing fewer quality-adjusted life-years than another intervention.
- Dominating
- Of an intervention, less expensive and providing the same or more quality-adjusted life-years than another intervention, or equally expensive and providing more quality-adjusted life-years than another intervention.
- Empiric treatment
- A therapy based on clinical experience in the absence of complete information.
- Gram-indeterminate bacteria
- Bacteria that do not respond predictably to Gram staining and, therefore, cannot be determined as either Gram positive or Gram negative.
- Gram-negative bacteria
- Bacteria that give a negative result in the Gram stain test.
- Gram-positive bacteria
- Bacteria that give a positive result in the Gram stain test.
- Gram staining
- Differentiates bacteria by the chemical and physical properties of their cell walls by detecting peptidoglycan, which is present in a thick layer in Gram-positive bacteria.
- Incremental cost-effectiveness ratio
- The additional cost per unit increase in effectiveness (often measured in quality-adjusted life-years).
- Incremental net monetary benefit
- A measure of the cost-effectiveness of a test at a given cost per quality-adjusted life-year gained threshold. The greater the incremental net monetary benefit, the more cost-effective the test.
- Index test
- The test of which the performance is being evaluated.
- Lysis
- The breaking down of the membrane of a cell, often by viral, enzymic or osmotic mechanisms that compromise its integrity.
- Maximum acceptable incremental cost-effectiveness ratio
- The largest value that society is assumed to be willing to spend to purchase a one-unit increase in effectiveness.
- Narrow-spectrum antibiotic
- An antibiotic effective against specific families of bacteria.
- Nosocomial infection
- Hospital-acquired infection.
- Polymerase chain reaction
- A technology in molecular biology used to amplify a single copy or a few copies of a piece of deoxyribonucleic acid across several orders of magnitude, generating thousands to millions of copies of a particular deoxyribonucleic acid sequence.
- Propensity score matching
- A statistical matching technique that attempts to estimate the effect of a treatment, policy or other intervention by accounting for the covariates that predict receiving the treatment.
- Quality-adjusted life-year
- A measure of both the longevity and quality of life. The higher the quality-adjusted life-years, the longer a person is likely to live for and/or the better the quality of life the person is predicted to have.
- Reference standard
- The best test currently available.
- Sanger sequencing
- A method of deoxyribonucleic acid sequencing.
- Sensitivity
- The proportion of people with the target condition that receive a positive test result. It is not uncommon for the true status of the patient to be determined by the reference standard even if that is an imperfect test.
- Sepsis
- A condition characterised by the body’s inflammatory response to an infection.
- Septic shock
- Persistent sepsis-induced hypotension (low blood pressure) despite adequate fluid resuscitation.
- Severe sepsis
- Occurs when the body’s response to infection interferes with the functioning of vital organs, such as the heart, kidneys, lungs or liver.
- Single gate
- A study design in which only patients with the target condition are recruited.
- Specificity
- The proportion of people without the target condition who receive a negative test result. It is not uncommon for the true status of the patient to be determined by the reference standard, even if that is an imperfect test.
- Superinfection
- A second infection superimposed on an earlier one, especially by a different microbial agent of exogenous or endogenous origin, which is resistant to the treatment used against the first infection.
List of abbreviations
- APACHE
- Acute Physiology And Chronic Health Evaluation
- CE
- Conformité Européenne
- cfu
- colony-forming unit
- CI
- confidence interval
- CrI
- credible interval
- DNA
- deoxyribonucleic acid
- EDTA
- ethylenediaminetetraacetic acid
- ICER
- incremental cost-effectiveness ratio
- ICU
- intensive care unit
- MAICER
- maximum acceptable incremental cost-effectiveness ratio
- MALDI-TOF
- matrix-absorbed laser desorption/ionisation time of flight
- MS
- mass spectrometry
- NICE
- National Institute for Health and Care Excellence
- NMB
- net monetary benefit
- PCR
- polymerase chain reaction
- QALY
- quality-adjusted life-year
- QUADAS
- Quality Assessment of Diagnostic Accuracy Studies
- RCT
- randomised controlled trial
- SAPS
- Simplified Acute Physiology Score
- SD
- standard deviation
- SIRS
- systemic inflammatory response syndrome
- SIRS-SS
- systemic inflammatory response syndrome with suspected sepsis
- SOFA
- Sequential Organ Failure Assessment
- VAT
- value-added tax
This monograph is based on the Technology Assessment Report produced for NICE. The full report contained a considerable number of data that were deemed confidential. The full report was used by the Appraisal Committee at NICE in their deliberations. The full report with each piece of confidential data removed and replaced by the statement ‘confidential (or data) removed’ is available on the NICE website: www.nice.org.uk.
The present monograph presents as full a version of the report as is possible while retaining readability, but some sections, sentences, tables and figures have been removed. Readers should bear in mind that the discussion, conclusions and implications for practice and research are based on all the data considered in the original full NICE report.
