Notes
Article history
The research reported in this issue of the journal was funded by the HTA programme as project number 09/75/01. The contractual start date was in April 2011. The draft report began editorial review in February 2015 and was accepted for publication in July 2015. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Professor E Andrea Nelson was a member of the Health Technology Assessment Commissioning Board.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2016. This work was produced by Nelson et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Background
Diabetes: prevalence and complications
Worldwide prevalence of diabetes mellitus was estimated at around 2.8% in 20001 and this is predicted to increase to 7.7% (affecting 439 million adults) by 2030,2 largely as a result of the obesity epidemic. 3,4 In the USA, the prevalence of diabetes was 8.3% in 2012, which is a sharp increase (more than doubling) compared with the prevalence in 1990, which was 3.5%. 5 Estimates from the USA predict that Americans born in 2000 will have a one in three lifetime risk of developing type 2 diabetes. 6 In the UK, the prevalence of diabetes is 6.0% (3.2 million people),7 and Diabetes UK estimates that there are around 630,000 people with diabetes who have not yet been diagnosed. 8 Treatment of diabetes in the UK cost approximately £23.7B in 2010/11, accounting for approximately 10% of the total health resource expenditure.
Both type 1 and type 2 diabetes can lead to serious health problems. 9 Complications of diabetes, especially in patients with poorly controlled blood sugar levels, include damage to the eyes, kidneys, nerves and arteries. In the feet, diabetes-related peripheral neuropathy leads to changes to foot architecture (hence increasing pressure on plantar surfaces, including those unaccustomed to load-bearing),10,11 reduced sweating (hence dry, cracking skin), poor sensation (hence susceptibility to trauma) and accelerated atherosclerotic disease, which leads to reduced circulation, with consequent problems with healing wounds and fighting infection. These peripheral neuropathic and vascular changes, either alone or in combination, predispose the foot to ulceration and its sequalae. 12,13
Diabetic ulcer infection: epidemiology and aetiology
It is estimated that the proportion of people with diabetes in the UK who have ever had a foot ulcer is around 6%,14 and that lifetime risk of ulceration is 15–25%. 15,16 Diabetic foot ulcers (DFUs) can take many weeks or often months to heal and have a negative impact on patients’ functional ability and quality of life. Foot ulceration in diabetic individuals also has a wider societal impact, such as reduced work productivity, high health-care costs and financial loss. 17–21 An open wound related to foot ulceration, combined with various immunological perturbations caused by diabetes, frequently results in infection. Prospective studies have found that about half of recent-onset DFUs are clinically infected at presentation. 22 Diabetic foot infection is thought to be the most common cause of diabetes-related hospital admissions and precedes approximately 80% of non-traumatic lower limb amputations. 15,23–27
Foot infections in people with diabetes can be hard to manage because of the associated impaired arterial supply to the legs, as well as impaired function of the immune system (especially those related to defects in polymorphonuclear leucocytes). This leads to an increased risk of progression of infection with contiguous spread to deeper tissues (including bone), and proximal extension up the foot and leg, as well as systemic spread into the blood stream. Therefore, many diabetic foot infections require some level of lower extremity amputation as a limb-sacrificing, but potentially life-saving, measure. 14,28,29
The incidence of lower extremity amputations is 10–30% higher in people with diabetes than in the general population,28,29 and about 85% of these amputations are preceded by a foot ulcer infection. 23,24,26,30–33 Limb amputation is associated with major consequences, as it dramatically reduces health-related quality of life, is expensive for both the patient and the health-care system and is associated with a 5-year mortality of over 50%. 15 To reduce the risk of foot ulceration, accelerate the healing of open ulcers and identify and treat infection promptly, many health-care systems have deployed multidisciplinary foot teams to co-ordinate foot care. The prevention of foot ulceration and amputation involves optimising glycaemic control and foot care. This may include supplying pressure-relieving shoes or insoles, undertaking surgical interventions promptly and optimally treating any infection. Providing this care involves input from the specialties of general practice, diabetology, nursing, dietetics, podiatry, orthotics, vascular surgery and infectious diseases/clinical microbiology.
Wound infection: definition, identification and characterisation
All chronic wounds, including DFUs, have bacteria on their surface that may originate from the surrounding normal skin flora, as well as opportunistic bacteria, such as gut flora. Therefore, the presence of bacteria in a wound does not indicate the presence of infection. When the host tissues show no inflammatory response or incur no damage associated with the bacterial growth, then the wound is described as ‘colonised,’ rather than infected. At this stage, there is typically thought to be a ‘balance’ between the growth of the several species of bacteria and no single organism usually dominates. When a critical density or high virulence of colonising organisms causes damage to host tissues, the wound is deemed to be ‘infected’. Therefore, infection of chronic wounds is usually a clinical diagnosis based on signs and symptoms of host tissue inflammation, such as pyrexia, purulent secretions, pain or tenderness, erythema, warmth and induration. 34–38 Although some investigators and clinicians also describe a quantitative diagnostic criterion for the presence of infection (e.g. a bacterial load of > 105 colony-forming units per gram of tissue), there is no agreement on this. 39 In chronic wounds, a single cut-off point for bacteria has been found to be insufficient for defining infection; other factors, such as the number or virulence of the bacterial groups and the presence of biofilm, are also important. 40
When a wound infection is diagnosed, the therapeutic approach depends on the whole clinical situation. Because in infected diabetic foot wounds the consequences of delayed antibiotic treatment can be profound, empiric antibiotic therapy should usually be initiated immediately. The antimicrobial regimen is usually selected in accordance with departmental protocols that are based on the probable causative organisms and their susceptibility patterns. Concurrently, samples for microbiological analysis are taken to identify the infecting organisms within the wound and their susceptibility to a range of antimicrobials. The resulting microbiological information guides subsequent modifications of the empiric antibiotic therapy required should the infection not improve and resistant organisms be isolated. 34–38,41,42 The culture and sensitivity results also allow a change from broad- to narrow-spectrum antibiotic agents, thus following the principles of antibiotic stewardship. 34–38,41
The microbiological analysis of specimens from the ulcer is useful only if the specimen is properly collected and processed and reported accurately and promptly. The aim is to acquire a wound sample that identifies all pathogens while avoiding colonising flora. First, the ulcer must be cleaned, which may involve debridement to remove necrotic material or callus and undermining tissues. Second, a specimen is taken from the site of infection, using one of a number of specimen-acquisition techniques, such as wound swabbing, fluid sampling using a fine-needle aspiration, or tissue sampling (by biopsy or curettage). 36,37,42 Taking a tissue sample either uses a tool to extract a ‘punch biopsy’ or scrapes the base of a wound with a sharp-edged dermal curette or scalpel blade to obtain ulcer tissue from the debrided ulcer bed. 43
It is important that the culture of the sample obtained reflects an accurate profile of the bacterial environment in the ulcer. Either failing to identify a true pathogen or identifying a coloniser as a pathogen can lead to inappropriate treatment of an infected wound. Therefore, it is important that health-care staff use a technique that will give a specimen that provides an accurate account of the bacteria present, including their number and sensitivity to antibiotics. Most published guidelines recommend obtaining a tissue specimen rather than a swab, in order to increase the likelihood of accurately reflecting the organisms associated with clinical infection at initial presentation. 34,36,37,42
In clinical practice, however, samples from wounds are often taken with a cotton swab. 44–47 The advantages of a wound swab include the almost universal availability of the equipment, the relative ease of the technique, the low cost of the swab and the fact that little training is needed to perform this correctly, which means that it can be done by non-clinician staff. 45 Furthermore, there is little risk of harm using a swab to collect a tissue sample. The disadvantages of a swab include the concerns that it may not collect those bacteria responsible for the infection deep within the tissues (e.g. as happens if an appropriate technique is used), that it will collect the colonising bacteria on the wound surface, or that it will fail to provide an environment conducive to growth of obligate anaerobes and other fastidious organisms (i.e. those that may be present in the wound but die in a swab device that does not provide an adequate medium for their survival). To counter these problems, advocates of wound swabbing have specified how to prepare the ulcer bed (i.e. removing dead tissue that may contain non-pathogenic bacterial groups) and how to obtain a sample from deep in the ulcer (by pressing to collect fluid from deep in the subcutaneous tissues, as described by Levine et al. 48 in 1976), as well as the optimal storage and transport procedures (use of charcoal swab, transport medium and swift delivery to the laboratory to maintain the viability of fastidious organisms). 48
In contrast, the reported advantage of tissue sampling is that the specimen is likely to contain the pathogens responsible for tissue destruction and infection. However, tissue-sampling techniques require disruption or cutting of the ulcer bed to obtain a specimen and this may lead to bleeding or pain (although most DFUs are complicated by neuropathy, which reduces the ability to perceive pain). Some clinical staff may need additional training to be able to take these samples safely and they also require some basic equipment: sharp sterile blades (scalpel), dermal curettes or a biopsy cutter. Using appropriate storage and transport procedures (transport medium and swift delivery to laboratory to maintain the viability of fastidious organisms) is still required.
Processing method
Accurate characterisation of the bacterial flora depends on both the sample collection method and the processing method. Standard culture and plating techniques involve the multiplication of the bacteria in a medium, by growing them on various types of culture plates, identifying the organisms and assessing their sensitivity to antimicrobial agents. It is thought that some organisms do not survive collection and transport and, hence, a swab (or occasionally tissue sample) does not fully reflect the organisms causing the wound infection. These ‘fastidious’ organisms remain undetected in the laboratory but may be important pathogens. 49 As these uncultured organisms cannot be identified by standard microbiological methods, appropriate antibiotic selection is problematic. This may partly account for the fact that approximately 10–20% of diabetic foot wounds fail to respond to initial antibiotic treatment. There is, therefore, some question over whether or not alternative techniques to identify bacteria within a sample, either instead of or in addition to sample plating and culture, may provide a more accurate picture of the wound flora. Modern molecular (or genotypic) techniques, such as polymerase chain reaction (PCR), have been proposed for this as the equipment for these tests become more readily available in hospitals. 50 It is not yet clear, however, how the results of these molecular tests, which generally identify more pathogens, should be interpreted. 51
The full report of culture results can take 4 or 5 days to be returned to the clinician. This delay in reporting, combined with the effects of antibiotic treatment given in the intervening period, means that the laboratory result may be out of date and that the wound flora may have changed. Therefore, a clinician reviewing an ulcer that has not improved with treatment cannot presume that the bacteria described in the microbiologists report are the same pathogens responsible for an infected ulcer 5 days later. Quicker techniques for microbiological analyses, such as genetic fingerprinting, that take 1 day or less, may help to address this delay. 52
These newer microbiological analysis techniques multiply the genetic material of the bacteria rather than grow them in culture. Genetic fingerprint techniques are then used to identify the bacteria group from its deoxyribonucleic acid (DNA)/ribonucleic acid (RNA) signature. 52 Culture-based methodology may not identify minor, although possibly important, components of a mixed bacterial population, whereas genetic fingerprinting techniques can. 53 Therefore, we also conducted a small substudy to compare identification of ulcer pathogens using conventional culture versus PCR techniques. This enabled us to determine the agreement between analysis techniques, that is, how does the quicker molecular technique reflect the bacterial load captured by swabs and tissues samples in the foot ulcer compared with swab and curettage specimens (e.g. for those organism not identified via plating and culture).
Diabetic foot ulcer guideline recommendations for infection (diagnosis/identification and characterisation and treatment)
Several guidelines and consensus documents aimed at improving the care for people with DFUs have been published over the past decade. 9,15,34–37,41,42,54,55 In this report, we have focused on three guidelines: (1) the UK National Institute for Health and Care Excellence (NICE) guidance on inpatient management of diabetic foot problems;37 (2) the Infectious Diseases Society of America (IDSA) guidelines for the diagnosis and treatment of diabetic foot infections;1,2,36 and (3) the International Working Group on the Diabetic Foot (IWGDF) guidelines on the management and prevention of the diabetic foot. 42,56 IDSA guidelines were first published in 200434,35 and are widely used. The IWGDF guidelines were published in 200842 and the latest NICE guidance in 2011. 37 The IDSA guidelines have recently been updated and provide details on the strength of the recommendations and the quality of the supporting evidence,36 making them the most current and comprehensive guidelines for the diagnosis and management of DFUs.
National Institute for Health and Care Excellence guidance37 recommends that clinicians should evaluate a diabetic patient presenting with a foot wound at three levels: the patient as a whole, the affected foot or limb and the infected wound. For infected wounds, an appropriately obtained specimen for culture is recommended prior to starting empiric antibiotic therapy, if possible. NICE guidance37 recommends sending a specimen for culture that is from deep tissue, obtained by biopsy or curettage and after the wound has been cleansed and debrided. The guidance advises against taking swab specimens, especially of inadequately debrided wounds, as they are likely to provide less accurate results. The IWGDF guidelines have the same message about obtaining the specimen but also mention the value of obtaining a Gram-stained smear of the wound in addition to culture. 42 For infected wounds, the IDSA guidelines34–36 recommend sending a specimen for culture that is from deep tissue, obtained by biopsy or curettage after the wound has been cleansed and debrided. The guidelines also advise against obtaining for culture by swabbing the wound or wound drainage. 36 In summary, all the clinical guidelines34–37,42 agree on their preference of tissue sample (obtained by biopsy, curettage or aspiration) to wound swab specimens.
The need for research
Although clinicians commonly use swab samples to provide information on the bacteria in a clinically infected wound, the current major guidelines all recommend tissue specimens over swab samples. 34–38 This is mainly because swabs can be contaminated with colonising flora, can miss deep pathogens and may be less likely to grow anaerobic and some fastidious aerobic organisms. However, the strength of this recommendation was specified only in IDSA guidelines,36 where it was ‘moderate’ (i.e. further definitive research is likely to have an important impact on future recommendations). 57
Three primary studies of culture techniques informing the guidelines were those conducted by Pellizzer et al. ,58 Slater et al. 59 and Bill et al. 60 Pellizzer et al. 58 assessed the reliability of results of ulcer swabbing versus deep tissue biopsy in 29 diabetic patients with a limb-threatening foot infection, who were neither recently treated with antibiotics nor hospitalised. This selected population does not reflect many of the patients with foot infections seen in outpatient clinics, who have often had recent antibiotic therapy. The study did not report on the agreement between swab and tissue samples, but, rather, simply on the number of bacterial colonies in each. Their conclusion that tissue samples are better than swab samples was based on a comparison of the numbers of isolates in only 21 participants remaining in the study at 30 days. Their finding may be due to chance as they performed 20 comparisons without adjustment for multiple testing. Furthermore, a method that identifies more colonies may be collecting more colonising bacteria and, therefore, is not necessarily ‘better’. The unpaired analysis presented means that we cannot readily compare the two techniques using appropriate statistical methods.
Slater et al. 59 aimed to evaluate the accuracy of swab compared with deep tissue (obtained via needle aspiration) cultures in diabetic wounds of varying depth and severity. Their study, however, included only 30 people with ulcers (in a sample of 60, in which the other patients had deep abscesses, etc.) and it is not clear if the results were heterogeneous across types of wounds or apply to tissue samples collected using scalpel or curette. In 62% of the samples, there was a similar profile of organisms isolated from the swab and the deep tissue sample, whereas in 20% of samples the swab identified more organisms and in 18% the deeper tissue sample picked up more organisms. These data were not stratified by the presence or absence of an ulcer or by ulcer type (i.e. neuropathic or ischaemic). This study identified that there can be two forms of disagreement between swabbing and sampling, with swabs identifying more organisms or tissue samples identifying more organisms; hence, they did not consider either technique to be a gold standard.
In a 2006 systematic review of the diagnosis and management of infection in DFUs,61 only one study that evaluated sample acquisition and reported agreement in sufficient detail to allow appropriate analysis was identified. This study by Bill et al. 60 included 18 patients with a pressure ulcer, 10 with a DFU, 5 with a venous leg ulcer, and 5 with an arterial ulcer. In this study, quantitative analysis of bacterial growth from a punch biopsy taken from the centre of the wound was compared with that of a wound swab. Using a definition of infection of a bacterial load of > 106 bacteria per gram of tissue in the punch biopsy, the authors reported a sensitivity for wound swabbing of 79%, meaning that the swab failed to detect approximately one in five wound infections as defined by punch biopsy. The derived likelihood ratios suggested that the wound swab was not a useful method of identifying infection in chronic wounds. Interpretation of this study’s findings is impeded by its small size and heterogeneity in the ulcer population. We cannot be sure that these data are directly transferable to the population of interest here, namely people with a DFU and a clinically diagnosed ulcer infection (there is no reason to sample uninfected ulcers and inclusion of people with uninfected foot ulcers may reduce the external validity of the study). In addition, there were potential sources of bias, such as no description of blind test verification and lack of clarity over whether or not the same clinical data were available when test results were interpreted as would be available when the test is used in practice.
Two studies62,63 have been published since the IDSA, NICE and IWGDF guidelines. Mutluoglu et al. 62 assessed the reliability of cultures of superficial swabs by comparing them with cultures of concomitantly obtained deep tissue specimens in patients with DFUs. They retrospectively reviewed the notes from 54 patients from whom there were 89 pairs of samples, one a superficial swab and the other deep tissue. The results showed a 73% concordance between swab cultures and deep tissue biopsies, which dropped to 69.2% when sterile pairs of cultures were excluded. Compared with deep tissue specimens, in 11.2% of cases swabs detected additional species, in 9.0% of cases swabs detected fewer species and in 6.7% the two techniques identified totally different organisms. The study concluded that superficial swab cultures are not sufficiently accurate to identify the causative organisms in patients with an infected DFU. They described three forms of disagreement: swabs identified more organisms, tissue samples identified more organisms and the techniques found different organisms.
Demetriou et al. 63 assessed the diagnostic performance of swabs versus tissue cultures in 50 consecutive diabetic patients with a foot ulcer, 28 of which were neuropathic and 22 of which were neuroischaemic. The authors stated that 36 (72%) wounds were infected, based on ‘the presence of at least 2 of the following criteria: local swelling or induration, erythema greater than 0.5 cm in any direction around the ulcer, local tenderness or pain, local increase of temperature, and purulent discharge’. Overall, the results showed that swabs reported significantly more isolates than tissue cultures, this difference being more evident in neuropathic than in neuroischaemic ulcers. They defined the tissue sample as the ‘gold standard’ for the diagnosis of infection, and reported swab culture sensitivity of 100% and negative predictive value of 100%, but the specificity was only 14.3% in neuropathic and 18.2% in neuroischaemic ulcers. They concluded that swabs are useful only to rule out infection. Given that guidelines do not recommend sampling/swabbing uninfected ulcers, the inclusion of 14 people in this study with uninfected ulcers reduces its external validity.
In summary, we concluded that the existing evidence regarding the results of cultures of specimens obtained by swabbing versus tissue sampling was derived from small, heterogeneous and, often, methodologically poor studies. Thus, there is a lack of robust evidence on the most appropriate method to use in routine clinical practice.
The question addressed in this study was not how to diagnose infection in a DFU, but rather what was the best way to collect a sample to characterise the bacterial flora. We therefore set out to describe the patterns of agreement and disagreement between swab and tissue samples. To help advise clinicians on the best technique to identify pathogens and to avoid colonising organisms in DFUs, we conducted a series of studies. The first was the ‘main study’, followed by three substudies:
-
main study: cross-sectional study to determine the patterns of agreement between culture results of contemporaneously collected swab versus tissue samples
-
substudy 1: independent ‘virtual’ clinical review of the appropriateness of empirical antimicrobial therapy based on the results of swabs compared with tissue samples to describe the potential clinical relevance of any differences in sampling results from swabs and tissue
-
substudy 2: a pilot comparative study of results of standard plating and culture techniques versus the molecular technique of PCR
-
substudy 3: a study of the prognosis of diabetic foot infection.
The main study was the first large, cross-sectional, multicentre study to examine agreement and disagreement of culture results between swab and tissue sampling techniques taken at the same time in a large group of patients with a clinically infected DFU. 64 Each of the studies is described in detail in Chapters 2–5.
Study aims
The primary aim of the main study (patterns of agreement between swab sampling and tissue sampling) was to evaluate concordance between culture results from wound swabs and tissue samples from the same patient (see Chapter 2).
The aim of the clinical panel review study was to evaluate whether or not any differences in bacterial profiles from specimens obtained from swabs and tissue samples are clinically relevant. This was done by ascertaining from a panel of clinicians whether or not the reports from a swab or tissue sample would have resulted in a change in clinical management (see Chapter 3).
The aim of the pilot comparative study of standard plating and culture techniques versus PCR was to assess the concordance between results from specimens taken by conventional culture techniques and by molecular techniques (see Chapter 4).
The aim of the prognosis of foot infection study was to determine the outcome of patients with an infected DFU at 12 months post registration and to explore prognostic factors that may be related to time to wound healing (see Chapter 5).
Patient and public involvement
In this study, patient and public involvement was achieved by using our links with diabetes organisations at the national (Diabetes UK), regional (North West Diabetes Local Research Network) and local (School of Healthcare Service User Group) levels.
As this work was commissioned by the NHS Health Technology Assessment (HTA) programme, then there had been patient and public involvement engagement at the prioritisation stage, and this informed the commissioners as regards the importance and relevance of the clinical question.
During the study we were fortunate to recruit a patient representative, Mrs Christine Thomas, as a member of the Study Steering Committee (SSC). She played a key part in the SSC meetings and advised the study team at different stages, including at the writing of patient and public-facing information. She also had an important role in shaping all aspects of the communications with patients as regards consent, particularly when moving to verbal consent. Furthermore, Mrs Thomas advised the study team about the dissemination of the initial results to participants at the end of the study and reviewed draft communications.
Chapter 2 Patterns of agreement between swab sampling and tissue sampling
Introduction
For infected DFUs, the accurate identification of pathogens, rather than colonising bacteria, is a prerequisite for selecting targeted antibiotic therapy to ensure optimal patient outcomes and avoid the acquisition of antibiotic resistance. Currently available evidence from the main diabetic foot infection guidelines (NICE,37 IDSA1,2,36 and the IWGDF42,56) and other studies62,63 is not sufficiently robust to advise clinicians on the best technique to identify pathogens in DFUs.
Objectives
The primary objective of the COncordance in DIabetic Foot Infection (CODIFI) main agreement study was to assess the level of agreement and patterns of disagreement between culture results from specimens taken by both surface swabs and tissue sampling from DFUs with suspected infection. We were interested in comparing three major microbiological parameters:
-
reported presence of isolates likely to be pathogens
-
the number of bacterial pathogens reported
-
the presence of antimicrobial resistance among likely pathogens.
Secondary objectives of the main agreement study were to compare rates of sampling-related adverse events (AEs) and the costs of sampling using each of the two techniques.
Methods
Study design
A multicentre, cross-sectional study involving 400 patients with a DFU with suspected infection requiring antibiotic therapy was conducted (Figure 1). Consenting patients had both a swab and tissue sample taken from their suspected infected DFU for conventional plating and culture.
FIGURE 1.
Study flow diagram. Adapted from Nelson EA, Backhouse MR, Bhogal MS, Wright-Hughes A, Lipsky BA, Nixon J, et al. Concordance in diabetic foot ulcer infection. BMJ Open 2013;3:e002370. Used under Creative Commons Attribution-Non Commercial 2.0 licence. Co-I, co-ordinator; PI, principal investigator.

Eligibility
All patients at least 18 years of age with a DFU that the attending clinician suspected was either a new case of infection or a chronic infection were screened for enrolment against the eligibility criteria below.
A DFU was considered to be any open wound on the foot (below the malleoli/ankle) in a patient with a diagnosis of diabetes mellitus. Each patient underwent an eligibility screen by a member of the research team, prior to entry, and an anonymised log was used to capture patient demographics along with reasons for not entering the study.
Inclusion criteria
-
Patient had a diagnosis of diabetes mellitus (type 1 or type 2).
-
Patient had a suspected foot ulcer infection, with or without bone involvement, based on clinical signs and symptoms using IDSA/IWGDF36,42 criteria and the judgement of the investigator.
-
The clinical plan was to treat the patient with antibiotics for their infected ulcer.
-
Patient was at least 18 years of age at the time of signing the consent form.
Exclusion criteria
-
The clinician deemed it inappropriate to take a tissue sample or a swab sample for any reason.
-
The patient had already been recruited to the study.
Recruitment and registration
The study was approved by the Sheffield Research Ethics Committee (reference 11/YH/0078) and had central and local NHS permissions at each participating centre prior to data collection.
Patients were recruited from multidisciplinary primary and secondary care-based foot ulcer/diabetic clinics and hospital wards, by a member of the research team (usually a clinical research nurse). Potential patients were provided with a patient information leaflet outlining all aspects of the study and given the chance to read it and to ask any questions they may have about the study. Written informed consent was documented by the patient and member of the local team. Informed written consent was obtained from all patients prior to entering the study.
Patients were registered via a 24-hour automated telephone registration system that automatically sent confirmation of successful registration through to the site.
Assessments
Sample acquisition
Clinicians in the participating sites participated in a study information session to instruct them on techniques for swab and tissue sample acquisition. An e-learning package was also developed and issued to all sites, detailing study procedures, including video footage of the correct use of both sampling techniques.
After wound cleansing (using sterile saline and gauze) and debridement (removal of necrotic tissue, foreign material, callus, undermining, usually with sharp instruments), a physician or podiatrist obtained specimens from the wound for cultures in one of the following ways:
-
Rubbing a sterile, cotton-tipped swab over the wound surface to sample superficial wound fluid and tissue debris. The swab was pressed with sufficient pressure on the wound bed to capture expressed wound fluid and was positioned deep in the ulcer to collect from likely infected areas. This is the wound swabbing technique described by Levine et al. 48
-
Immediately after the cotton swab had been collected, a tissue sample was removed from the same area of the ulcer bed. This procedure was performed using sterile equipment and aseptic technique, involved removal of a small piece of wound tissue at the base of the wound by scraping or scooping using a dermal curette or sterile scalpel blade.
Sample transport and processing
Each sample was placed individually in the standard transport medium used at the site and delivered to the local medical microbiology laboratory in accordance with routine clinical practice. A UK national standard method was used for collecting and processing samples. 65,66 Both samples from each patient were processed in the same local laboratory as routine clinical samples. Neither sample was labelled as having been taken as part of a clinical study. Our goal was to ensure that, as far as possible, the reports reflected current sample processing methods in each laboratory, rather than these samples having received special attention or processing.
Clinical assessments
A member of the research team used a case report form (see Appendix 5) to record patient demographics, diabetes status and foot health history, including current or proposed antibiotic treatment and wound dressings. Details of the index ulcer were recorded using each of the Perfusion, Extent/Size, Depth/Tissue loss, Infection, Sensation (PEDIS),67 Wagner grade,68 and Clinical Signs and Symptoms Classification for Infection69 schemes. The research team also filled out other study-related documentation, which was forwarded to the study co-ordinating centre at the University of Leeds.
Centre differences questionnaire
A ‘centre differences questionnaire’ aimed to capture, from each centre and laboratory processing samples, details relating to the clinical acquisition of samples, specimen transport, sample analysis, methods of reporting results of samples by the laboratory and local antibiotic protocols for infected DFUs.
End points
Coprimary end points
In order to assess agreement and patterns of disagreement between results from the swab and tissue samples, three coprimary end points were defined.
Reported presence of likely pathogens
The first coprimary end point was originally defined as the reported presence or not of the following likely pathogens, identified by the UK Health Protection Agency (HPA) as likely pathogens from limb-threatening DFUs:65,66
-
Staphylococcus aureus Rosenbach 1884 (categorised by the presence or absence of meticillin resistance)
-
Streptococcus species Rosenbach 1884
-
Enterobacter aerogenes Hormaeche and Edwards 1960
-
Escherichia coli (Migula 1895) Castellani and Chalmers 1919
-
Pseudomonas species Migula 1984
-
Corynebacterium species Lehmann and Neumann 1896
-
anaerobic cocci (i.e. mixed anaerobes)
-
Fusobacterium species Knorr 1922
-
Bacteroides fragilis (Veillon and Zuber 1898) Castellani and Chalmers 1919
-
Prevotella bivia (Holdeman and Johnson 1977) Shah and Collins 1990.
A revised definition was implemented to include the most prevalent pathogens, defined as those reported in ≥ 10% of patients (in either swab or tissue samples). This overall prevalence rate was determined based on statistical justification of the sample size calculation; we also used clinical discretion to determine whether or not the end point would include pathogens with an overall prevalence below 10%.
An overall summary of pathogens reported59 allowed for the comparison of all pathogens reported within each sample and an assessment of whether or not agreement was influenced by any of a number of covariates.
Antimicrobial resistance
Presence or absence of resistance to antibiotics to which the specific species is ordinarily susceptible among likely pathogens, as reported by standard techniques for:
-
meticillin-resistant S. aureus (MRSA)
-
meticillin-resistant coagulase-negative staphylococci (CNS)
-
vancomycin-resistant Enterococcus species.
Number of pathogens
Number of pathogens reported per specimen.
Secondary end points
Adverse events
The secondary end point relating to AEs was the number of patients with a study-related event categorised as an expected AE, defined as bleeding of concern attributable to the sampling method or patient-reported pain before and after each sampling technique, or as a related unexpected serious adverse event (RUSAE).
Costs
A full economic evaluation was beyond the scope of this study. The cost data collected were the laboratory costs, including all components used in processing and reporting of swab and tissue samples. Costs of these procedures were requested from the microbiologists at study centres.
Derivation involving reported pathogens
Microbiology laboratories reported pathogens at a range of taxonomic levels (species, genus, family and group); therefore, the end points for the prevalence, overall summary and number of pathogens required derivation in order to allow for comparison of pathogens reported within each sample at a meaningful level.
Staphylococcus aureus is used in reference to non-MRSA, whereas MRSA is used to describe S. aureus that is meticillin resistant.
The majority of pathogens were included at the genus level, with the exception of S. aureus (identified at the species level) and vancomycin-resistant and non-resistant Enterococcus spp. (included separately by vancomycin resistance). The following groups of pathogens were also included as part of the first coprimary end point for reported presence or absence: Gram-positive cocci, Gram-negative cocci, Gram-positive bacilli, Gram-negative bacilli, anaerobes (where possible as anaerobic cocci or anaerobic rods), CNS and Enterobactereaceae (including coliforms).
Furthermore, the following isolate designations were considered unlikely to represent pathogenic organisms in a sample from a DFU and were not included in the end points: yeasts, skin flora, normal flora, mixed flora, skin organisms, bacterial flora, enteric flora and faecal flora.
Statistical methods
Sample size
The sample size calculation was based on the primary outcome of reported ‘presence or absence of a pathogen’ for the whole sample overall.
To be confident that swabs adequately sampled wound flora, it was assumed that the chance corrected agreement between swab and tissue samples needed to be at least ‘good’ (usually defined as a κ-statistic > 0.6). 70 Of course, the κ-statistic alone does not convey the distribution of disagreement between swabs and tissue samples. Good overall agreement, with balanced disagreement, would be clinically important if tests were to be regarded as interchangeable. Therefore, the total sample size was based on there being good agreement and reasonably balanced disagreement for clinically important and less prevalent pathogens.
Using a two-sided McNemar’s test at the 5% level of significance, a sample size of 399 patients would provide 80% power to detect a difference of ≥ 3% in the reported presence of a pathogen, assuming an overall prevalence of the pathogen of 10% and 5% disagreement between the swab and tissue samples. This amount of agreement would also result in a κ-statistic of ≈ 0.7, and the calculation was based on the expected prevalence of less common pathogens, such as Pseudomonas (present in 10% of samples in Pellizer et al. 58). It was, therefore, planned that a total of 400 patients would be recruited. Further details of the sample size calculation are provided in Appendix 1.
Analysis methods
All data analyses and summaries were performed using SAS® version 9.2 (SAS Institute Inc., Cary, NC, USA),71 with the exception of exact confidence intervals (CIs) only, which were calculated within R version 3.0 (The R Foundation for Statistical Computing, Vienna, Austria). 72 All significance tests were two-sided and conducted at the 5% level of significance, with p-values and 95% CI provided where appropriate.
Patient populations
The full analysis set consisted of all patients registered and consented to take part in the study, regardless of their adherence to the study protocol or eligibility violation.
The evaluable population consisted of all registered and consented patients with evaluable swab and tissue samples. Patients for whom the swab or tissue samples were not successfully collected or were lost, or for whom the sample results were lost, were excluded from this evaluable population.
The per-protocol (PP) population consisted of all registered and consented patients for whom there were no protocol violations. Patients who did not satisfy the eligibility criteria, or those for whom a protocol deviation in the collection or processing of either sample had occurred, were excluded from the PP population.
Coprimary end point analysis
Reported presence of likely pathogens
The first coprimary end point was defined for each patient as the reported presence or absence of each likely pathogen reported from the result of culture of the swab and tissue sample. Patients for whom either the swab or tissue sample result was not available were excluded from the primary end point, with analysis conducted on the evaluable population.
For each likely pathogen, cross-tabulations of reported presence were generated to investigate agreement and the pattern of disagreement. For each pathogen, the following statistics are presented:
-
overall percentage prevalence, calculated as the proportion of patients with the pathogen reported from either the swab or tissue sample results
-
the percentage of patients for whom the swab and tissue sample results agreed or disagreed in the reported presence of the pathogen
-
the unadjusted κ-statistic (with 95% CIs) and the prevalence- and bias-adjusted kappa (PABAK), which evaluates chance corrected agreement between swab and tissue samples. Strength of agreement according to the κ-statistic was categorised as shown in Table 1
-
the difference in percentage prevalence between the swab and tissue samples (tissue percentage minus swab percentage) with 95% CIs (accounting for paired samples)
-
McNemar’s test of the difference between the swab and tissue samples in the percentage of samples reporting the pathogen accounting for paired samples.
| Strength of agreement | Value of kappa (κ) |
|---|---|
| Poor | < 0.0 |
| Slight | 0.0–0.2 |
| Fair | 0.21–0.4 |
| Moderate | 0.41–0.6 |
| Substantial | 0.61–0.8 |
| Almost perfect | 0.81–1.0 |
When the proportion of disagreement was low, leading to cell counts in the cross-tabulations of < 5, a small-sample binomial version of McNemar’s test was used to provide an exact p-value73 and the exact 95% CI for the difference in percentage prevalence was calculated using an inductive method. 74
Cross-tabulations on the semiquantitative extent of bacterial growth (none, + to +++) and weighted κ-statistics by type of DFU (neuropathic, ischaemic, without inferential statistics) were also generated.
An overall summary of the pathogens reported was further generated,59 with each patient’s pair of results (swab and tissue sample) coded as follows: swab and tissue sample report the same pathogens; swab reports same pathogens as tissue sample plus extra pathogens; tissue sample reports same pathogens as swab plus extra pathogens; both tissue sample and swab report different pathogens (with or without overlap in pathogen). Multinomial logistic regression analysis was used to model the proportion of patients in each category, compared with the reference category ‘swab and tissue sample report the same pathogens’ on pre-specified baseline factors to investigate their relevance in determining agreement between sample results. These factors included type of ulcer (ischaemic or neuroischaemic vs. neuropathic); Wagner ulcer grade (1 to 5); recent (on the day of sampling) systemic or topical antimicrobial therapy; or dressing and wound duration (< 56 days vs. ≥ 56 days, and continuous on the log-scale). The centre from which the patient was enrolled was included in each model as a random effect in order to allow for additional variability in outcome by centre and estimates of the effect of baseline factors without directly requiring the estimation of individual centre effects. Estimates of odds ratios for each covariate are presented along with 95% CIs and p-values (based on the change in –2 log-likelihood).
Reported presence of antimicrobial resistance among likely pathogens
Meticillin-resistant S. aureus, meticillin-resistant CNS and vancomycin-resistant Enterococcus were the three antimicrobial-resistant pathogens identified for exploration. For each of these resistant pathogens, cross-tabulations were created (reported presence or absence of resistant pathogen) and the following statistics presented: PABAK, unadjusted κ-statistic and overall percentage agreement. McNemar’s test was used to test for a difference between swab and tissue sampling techniques in the proportion of samples in which the specified resistant pathogen was reported.
For each resistant pathogen the following codes were also created: resistant pathogen reported by swab not tissue sample; resistant pathogen reported by tissue sample not swab; swab and tissue sample results agree. To determine if agreement is influenced by specified covariates, multinomial regression modelling was planned to model these categories on type of ulcer (predominantly neuropathic or ischaemic), ulcer grade, pre-sampling antibiotic therapy, pre-sampling antimicrobial dressing, wound duration and centre.
Number of pathogens reported
Summaries (including cross-tabulations) on the number of pathogens reported per specimen were generated for swab versus tissue samples. Samples were further coded as follows: tissue sample had two or more extra pathogens reported; tissue sample had one extra pathogen reported; tissue sample and swab had the same number of pathogens reported; swab had one extra pathogen reported; swab had two or more extra pathogens reported.
Ordinal logistic regression analysis, based on the proportional odds model,75 was used to model the number of pathogens reported per specimen on pre-specified baseline factors to investigate their relevance in determining agreement between sample results. These factors included type of ulcer (ischaemic or neuroischaemic vs. neuropathic); Wagner ulcer grade (1–5); recent systemic or topical antimicrobial therapy or dressing; and wound duration (< 56 days vs. ≥ 56 days, and continuous on the log-scale). Centre was included in each model as a random effect in order to allow for additional variability in outcome by centre and estimates of the effect of baseline factors without directly requiring the estimation of individual centre effects. Estimates of odds ratios for each covariate are presented along with 95% CIs and p-values (based on the change in –2 log-likelihood).
Analysis population
Patients for whom both swab and tissue sample results were available were included in the coprimary end points, with analysis conducted on the evaluable population.
Missing data
As part of the study design, efforts were made to collect complete data; however, where data remained missing, this was assumed to be missing at random, and multiple imputation (MI)76 was used to impute missing baseline covariates, thereby allowing inclusion of the 28 (7.1%) patients with missing data for at least one candidate baseline factor. The pattern and prevalence of missing data among covariates considered within the regression analysis of the coprimary end points are presented in Appendix 1, Table 81.
The outcome and all baseline covariates (including type of ulcer, Wagner ulcer grade, recent systemic or topical antimicrobial therapy or dressing, wound duration) to be considered in each regression analyses were included in the MI models alongside centre. A total of 10 imputations were conducted using the Markov chain Monte Carlo (MCMC) method77 with multiple chains, initial values from the expectation–maximisation (EM) algorithm, 200 burn-in iterations, and the assumption of normality for factors with missing data (thus, imputations were made on a continuous scale). 71 For dichotomous factors, imputations were not restricted for ‘implausible values’ and thus continuous imputations were rounded to plausible values for the dichotomous factor (with a small proportion of missing data the bias introduced as a result of this method is minimal). 78 This method was used as the pattern of missing data was arbitrary and non-monotone.
For the 10 imputed data sets, the odds ratios generated through the regression analyses were combined using Rubin’s rules;79 therefore, reported estimates reflect the average of estimates across the imputed data sets, and estimated standard errors include variability across the imputed sets as well as the usual uncertainty in parameter estimates. The mean change in –2 log-likelihood was used to calculate the overall p-value.
Derivation
A number of common scales were used to quantify the extent of growth of a pathogen, specific to each recruiting site. In order of severity of growth within a scale, these were: +/++/+++; +/++/+++/++++; scanty/light/moderate/heavy; scanty/+/++/+++; light, moderate, heavy. The reported growth for each pathogen was derived onto one 3-point scale reported as +/++/+++ (Table 2).
| Scale | Derived level of growth | ||
|---|---|---|---|
| 1: + | 2: ++ | 3: +++ | |
| Scanty/light/moderate/heavy | Scanty/light | Moderate | Heavy |
| +/++/+++/++++ | + | ++ | +++/++++ |
| +/++/+++ | + | ++ | +++ |
| Scanty/+/++/+++ | Scanty/+ | ++ | +++ |
| Light/moderate/heavy | Light | Moderate | Heavy |
κ-statistic weights were selected to reflect the ordinal nature of extent of growth, in which the difference between a sample with an extent of growth of + and ++ is far smaller than the difference between ++ and +++, owing to the increase in dilution factors used to determine the extent of growth (10-fold increase). To account for this relationship, while allowing greater differentiation between the highest level of growth (+++), the following exponential values were assigned to each level of growth, from which linear Cicchetti–Allison agreement weights were derived:80 pathogen not reported (= 1), + (= 2.7), ++ (= 7.4), +++ (= 20.1).
As the choice of values for each level of growth was somewhat arbitrary, a sensitivity analysis assessed the impact of these weights, in which levels of growth were assigned the following linear values: pathogen not reported – 0, + – 1, ++ – 2, +++ – 3.
To account for pathogens reported at various taxonomic ranks and to determine whether or not swab and tissue results reported the same pathogens, pathogens were compared according to pre-defined groups set out in Appendix 1 [i.e. largely at the genus level, and at the higher group level where further detail was not reported from the laboratory result (e.g. Gram-positive cocci rather than S. aureus)]. For example, where a pathogen was reported at the species level it was compared with the corresponding alternative sample at the genus level (e.g. E. coli belongs to the Escherichia genus). If, however, one sample reported the pathogen at a taxonomic rank higher than the genus level, such as ‘Gram-negative bacilli’ with the corresponding alternative sample reporting the pathogen in more detail (in this scenario ‘Escherichia’), then we did not class the patient’s results as reporting the same pathogens. This was based on clinical relevance of pathogens and overcame discrepancies in the level of reporting.
The summary and number of pathogens reported per specimen was calculated independently for both the swab and tissue samples.
Samples were identified where more than one strain or species of pathogen (in which we were interested in the genus level or higher) was reported. In these samples, a single pathogen at the level of interest was retained for comparison with the corresponding swab or tissue sample in the summary of pathogens and with the count of the number of pathogens within the sample.
Samples from which results of a Gram-stained smear had been reported in addition to those from the culture were identified by the reporting of the following groups of pathogens: Gram-positive bacilli, Gram-negative bacilli, Gram-positive cocci, Gram-positive cocco-bacillus and Gram-negative cocci. Gram stain results were then compared with pathogens reported within the corresponding culture result, and pathogens belonging to the group of pathogens reported by the Gram stain were further identified. Where a pathogen belonged to the same group as that reported by the Gram stain, it was considered likely that both referred to the same pathogen and the corresponding Gram stain result was excluded from the summary and number of pathogens reported from the swab or tissue sample. For example, where both Gram-positive cocci and S. aureus were reported, because S. aureus is a type of Gram-positive cocci, only S. aureus was retained in the summary and number of pathogens. Conversely, where the results of a Gram stain were provided and no pathogens identified by the culture belonged to the group identified by the Gram stain, all pathogens were included. Details of all samples for which this derivation was applied are in Appendix 1.
Secondary end point analysis
Adverse events
Safety analyses presents summaries of all expected AEs (bleeding of concern that is attributable to either of the sampling methods, pain as reported by the patient before and immediately after acquisition of each sample) and RUSAEs. The number of events and number of patients with events are also summarised.
Results
Sample size
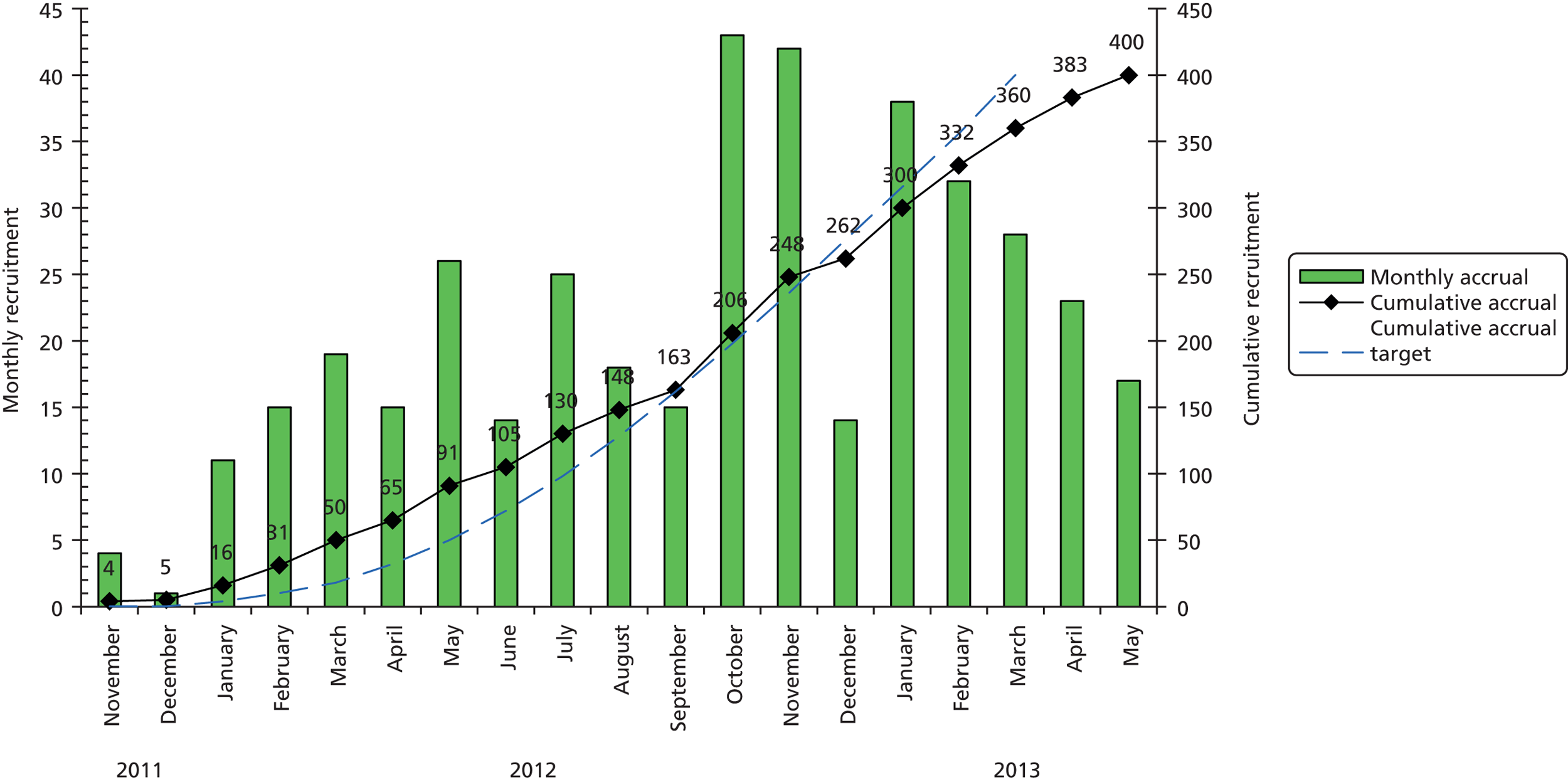
In total, 680 patients were screened for recruitment into CODIFI and 401 patients were enrolled between November 2011 and May 2013. One patient was excluded as the written informed consent was lost. Of 27 centres, 25 recruited patients into the study; Figure 2 shows the number of patients recruited and screened per centre, and Figure 3 shows the overall, monthly and cumulative recruitment of patients to the study together with our original target. See Appendix 1 for full centre names.
FIGURE 2.
Screening and recruitment by centre. SJUH, St James’s University Hospital.

FIGURE 3.
Monthly and cumulative recruitment.
Analysis populations
The numbers of patients recruited to CODIFI and included in the full analysis set, the evaluable population and the PP population are each summarised in Table 3.
| Analysis population | Total (N = 400), n (%) |
|---|---|
| Full analysis set | 400 (100.0%) |
| Evaluable population | 395 (98.8%) |
| PP population | 386 (96.5%) |
Full analysis set
No patient withdrew consent for the samples to be used for research purposes and hence only the one patient without informed consent was excluded from the full analysis set.
Evaluable population
The evaluable population consisted of the 395 (98.8%) patients who had both swab and tissue sample results available. Patients with a protocol deviation involving the loss of one or both samples or results were excluded. The number of patients excluded from the evaluable population and the reasons are summarised in Table 4.
| Exclusions and reasons for exclusion | Total (N = 400), n (%) |
|---|---|
| Excluded from the evaluable population | 5 (1.3) |
| Protocol deviation: swab not processed by laboratory | 2 (40.0) |
| Swab sample used for other purpose: MRSA screen | 1 (20.0) |
| Swab and curettage samples were lost | 1 (20.0) |
| Swab sample was lost | 1 (20.0) |
| Excluded from the PP population | 14 (3.5) |
| Eligibility violation: clinical plan not to treat with antibiotics | 3 (21.4) |
| Clinical plan not to treat with antibiotics and no suspected infection | 2 (14.3) |
| Protocol deviation: tissue sample taken before the swab sample | 3 (21.4) |
| Swab not processed by laboratory | 2 (14.3) |
| Swab sample used for other purpose: MRSA screen | 1 (7.1) |
| Swab and tissue sample was lost | 1 (7.1) |
| Swab sample was lost | 1 (7.1) |
| Second swab sample taken after the tissue sample | 1 (7.1) |
Per-protocol population
The PP population consisted of the 386 (96.5%) patients without an eligibility violation or protocol deviation. The number of patients excluded from the PP population and the reasons are summarised in Table 4. Given that only an additional nine patients were excluded from the PP population compared with the evaluable population, no analyses were repeated for the PP population.
Study conduct
Figure 4 presents a flow diagram depicting the study conduct and analysis population.
FIGURE 4.
Patient flow diagram.

Baseline characteristics
Tables 5–13 summarise the baseline characteristics, including patient demographics, information about diabetes, clinical details, ulcer characteristics, PEDIS classification, clinical signs and symptoms, and antibiotic regimens immediately pre and post sampling. Because the evaluable population was very similar to the full analysis set with respect to baseline characteristics, characteristics of the full sample only are detailed below.
| Patient demographics | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Age (years) | ||
| Mean (SD) | 63.1 (13.3) | 63.1 (13.4) |
| Median (range) | 63.0 (26–99) | 63.0 (26–99) |
| Missing | 0 | 0 |
| Sex, n (%) | ||
| Male | 316 (79.0) | 311 (78.7) |
| Female | 84 (21.0) | 84 (21.3) |
| Ethnicity, n (%) | ||
| White | 377 (94.3) | 372 (94.2) |
| Other mixed background | 1 (0.3) | 1 (0.3) |
| Asian: Indian | 3 (0.8) | 3 (0.8) |
| Asian: Pakistani | 11 (2.8) | 11 (2.8) |
| Other Asian background | 2 (0.5) | 2 (0.5) |
| Black: Caribbean | 3 (0.8) | 3 (0.8) |
| Black: African | 1 (0.3) | 1 (0.3) |
| Other ethnic group | 2 (0.5) | 2 (0.5) |
| Site of recruitment, n (%) | ||
| Hospital ward | 53 (13.3) | 53 (13.4) |
| Outpatient clinic | 319 (79.8) | 314 (79.5) |
| Community clinic | 28 (7.0) | 28 (7.1) |
| Diabetes details | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Diabetes type, n (%) | ||
| Type 1 | 58 (14.5) | 58 (14.7) |
| Type 2 | 342 (85.5) | 337 (85.3) |
| Duration of diabetes (years) | ||
| Mean (SD) | 16.8 (11.0) | 16.9 (11.0) |
| Median (range) | 15.0 (0.04–57) | 15.0 (0.04–57) |
| Missing | 3 | 3 |
| HbA1C (%) | ||
| Mean (SD) | 8.72 (2.29) | 8.71 (2.29) |
| Median (range) | 8.10 (4.6–17.2) | 8.10 (4.6–17.2) |
| Missing | 6 | 6 |
| Current diabetes treatment, n (%) | ||
| Yes | 385 (96.3) | 381 (96.5) |
| No | 15 (3.8) | 14 (3.5) |
| Diabetes treatment details, n (%) | ||
| Oral hypoglycaemic agent | 107 (27.8) | 106 (27.8) |
| Insulin | 168 (43.6) | 166 (43.6) |
| Both oral hypoglycaemic agent and insulin | 109 (28.3) | 108 (28.3) |
| Oral hypoglycaemic agent and exenatide | 1 (0.3) | 1 (0.3) |
| Ulcer characteristics | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Location of ulcer(s), n (%) | ||
| Ulcers on both right and left foot | 60 (15.0) | 59 (14.9) |
| Ulcer(s) on right foot only | 173 (43.3) | 169 (42.8) |
| Ulcer(s) on left foot only | 167 (41.8) | 167 (42.3) |
| Total number of ulcers, n (%) | ||
| 1 | 268 (67.0) | 264 (66.8) |
| 2 | 78 (19.5) | 78 (19.7) |
| 3 | 43 (10.8) | 43 (10.9) |
| 4 | 6 (1.5) | 6 (1.5) |
| 5 | 1 (0.3) | 1 (0.3) |
| 6 | 3 (0.8) | 2 (0.5) |
| 7 | 1 (0.3) | 1 (0.3) |
| Mean (SD) | 1.5 (0.9) | 1.5 (0.9) |
| Median (range) | 1.0 (1–7) | 1.0 (1–7) |
| Missing | 0 | 0 |
| Index ulcer characteristics | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Foot containing index ulcer, n (%) | ||
| Right foot | 205 (51.3) | 201 (50.9) |
| Left foot | 195 (48.8) | 194 (49.1) |
| Index ulcer location, n (%)a | ||
| Apex | 47 (11.8) | 45 (11.4) |
| Interdigital | 25 (6.3) | 25 (6.3) |
| Plantar | 172 (43.0) | 170 (43.0) |
| Dorsum | 56 (14.0) | 56 (14.2) |
| Digital | 90 (22.5) | 89 (22.5) |
| Other | 8 (2.0) | 8 (2.0) |
| Missing | 2 (0.5) | 2 (0.5) |
| Duration of index ulcer (months) | ||
| Mean (SD) | 5.58 (12.28) | 5.52 (12.17) |
| Median (range) | 1.84 (0.1–144.0) | 1.84 (0.1–144.0) |
| Missing | 4 | 4 |
| First or recurrent index ulcer, n (%) | ||
| Incident | 288 (72.0) | 283 (71.6) |
| Recurrent | 110 (27.5) | 110 (27.8) |
| Missing | 2 (0.5) | 2 (0.5) |
| Aetiology of index ulcer, n (%) | ||
| Ischaemic | 14 (3.5) | 14 (3.5) |
| Neuropathic | 202 (50.5) | 199 (50.4) |
| Both ischaemic and neuropathic | 182 (45.5) | 180 (45.6) |
| Missing | 2 (0.5) | 2 (0.5) |
| Antimicrobial dressing on the ulcer, n (%) | ||
| Yes | 241 (60.3) | 238 (60.3) |
| No | 154 (38.5) | 152 (38.5) |
| Missing | 5 (1.3) | 5 (1.3) |
| PEDIS classification and ulcer debridement | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Perfusion, n (%) | ||
| Grade 1: no symptoms/signs of PAD | 200 (50.0) | 197 (49.9) |
| Grade 2: symptoms or signs of PAD, but no CLI | 192 (48.0) | 190 (48.1) |
| Grade 3: CLI | 8 (2.0) | 8 (2.0) |
| aExtent/size: estimated index ulcer area, cm2 | ||
| Mean (SD) | 6.76 (15.16) | 6.60 (14.85) |
| Median (range) | 1.77 (0.01–138.2) | 1.77 (0.01–138.2) |
| Missing | 3 | 3 |
| Depth/tissue loss, n (%) | ||
| Grade 1: superficial full-thickness ulcer | 131 (32.8) | 130 (32.9) |
| Grade 2: ulcer penetrating below dermis to skin structures | 134 (33.5) | 132 (33.4) |
| Grade 3: all subsequent layers of foot, including bone/joint | 135 (33.8) | 133 (33.7) |
| Infection, n (%) | ||
| Grade 1: no symptoms/signs of inflammation | 2 (0.5) | 2 (0.5) |
| Grade 2: inflammation of skin/subcutaneous tissue only | 149 (37.3) | 148 (37.5) |
| Grade 3: extensive erythema deeper than skin/subcutaneous tissue | 237 (59.3) | 234 (59.2) |
| Grade 4: systemic inflammatory response syndrome | 12 (3.0) | 11 (2.8) |
| Sensation, n (%) | ||
| Grade 1: no loss of protective sensation | 27 (6.8) | 27 (6.8) |
| Grade 2: loss of protective sensation | 373 (93.3) | 368 (93.2) |
| Ulcer debridement undertaken, n (%) | ||
| Yes | 351 (87.8) | 347 (87.8) |
| No | 49 (12.3) | 48 (12.2) |
| Presence of clinical signs and symptoms | Full analysis set (N = 400), n (%) | Evaluable population (N = 395), n (%) |
|---|---|---|
| Wound odour | 127 (31.8) | 126 (31.9) |
| Pocketing in wound | 170 (42.5) | 168 (42.5) |
| Discoloured granulation tissue | 225 (56.3) | 220 (55.7) |
| Friable granulation tissue | 204 (51.0) | 202 (51.1) |
| Recent increase in paina | 125 (31.3) | 123 (31.1) |
| Recent decrease in paina | 9 (2.3) | 9 (2.3) |
| Recent increase in wound sizea | 246 (61.5) | 241 (61.0) |
| Breakdown of epitheliuma | 126 (31.5) | 124 (31.4) |
| Wagner grade | Full analysis set (N = 400), n (%) | Evaluable population (N = 395), n (%) |
|---|---|---|
| Grade 1: superficial diabetic ulcer (partial or full thickness) | 136 (34.0) | 135 (34.2) |
| Grade 2: ulcer extension ligament, tendon, joint capsule or deep fascia without abscess or osteomyelitis | 134 (33.5) | 132 (33.4) |
| Grade 3: deep ulcer with abscess, osteomyelitis or joint sepsis | 122 (30.5) | 120 (30.4) |
| Grade 4: gangrene localised to portion of forefoot or heel | 7 (1.8) | 7 (1.8) |
| Grade 5: extensive gangrenous involvement of the entire foot | 1 (0.3) | 1 (0.3) |
| Antibiotic therapy | Full analysis set (N = 400) | Evaluable population (N = 395) |
|---|---|---|
| Patient on a pre-sampling antibiotic therapy regimen, n (%) | ||
| Yes | 187 (46.8) | 186 (47.1) |
| None prescribed | 194 (48.5) | 190 (48.1) |
| Missing | 19 (4.8) | 19 (4.8) |
| Days spent on pre-sampling antibiotic therapy | ||
| Mean (SD) | 14.6 (21.9) | 14.7 (21.9) |
| Median (range) | 7.0 (1–145) | 7.0 (1–145) |
| Missing | 1 | 1 |
| Change to antibiotic therapy: immediately post sampling, n (%) | ||
| Yes | 248 (62.0) | 244 (61.8) |
| No | 133 (33.3) | 132 (33.4) |
| Missing | 19 (4.8) | 19 (4.8) |
| Antibiotic regimen | Full analysis set (N = 400), n (%) | Evaluable population (N = 395), n (%) |
|---|---|---|
| No pre-sampling antibiotic but initiation immediately post sampling | 168 (42.0) | 164 (41.5) |
| No pre-sampling antibiotic and no initiation immediately post sampling | 26 (6.5) | 26 (6.6) |
| On a pre-sampling antibiotic with or without a change immediately post sampling | 187 (46.8) | 186 (47.1) |
| Unknown whether or not there was a pre-sampling antibiotic but initiation/change immediately post sampling | 19 (4.8) | 19 (4.8) |
Tables 5 and 6 summarise patient demographics and diabetes details, respectively. The median age of patients was 63 years (range 26–99 years); 79% of patients were male; and the majority of patients (94.3%) were of white ethnic origin. Recruitment of patients was from outpatient clinics for 79.8% of patients, hospital wards for 13.3% and community clinics for 7%. The median duration of diabetes in enrolled patients was 15 years (range 2 weeks–57 years); 14.5% and 85.5% of patients had type 1 or type 2 diabetes, respectively; and the vast majority of patients (96.3%) were receiving treatment for their diabetes.
Tables 7 and 8 summarise patients’ ulcer characteristics. The total number of DFUs ranged from one to seven per patient, with one ulcer observed for 67.0% of patients, two ulcers for 19.5%, and three or more ulcers for 13.6% of patients. The anatomic site of the index ulcer, from which both the swab and tissue samples were obtained, was most commonly the plantar surface (43.0%), the digital surface (22.5%), the dorsum (14.0%) or the apex (i.e. tip of toe, 11.8%). The duration of the index ulcer varied to a large degree, with a median of 1.84 months (range 3 days–12 years). A total of 72.0% of patients’ index ulcers were incident as opposed to recurrent. Only 3.5% of ulcers were solely ischaemic, 50.5% of ulcers were neuropathic only, and 45.5% of ulcers were both ischaemic and neuropathic.
Tables 9–11 summarise ulcer characterisation according to the PEDIS criteria, clinical signs and symptoms, and Wagner scale. Almost all patients (98%) had a grade 1 or 2 perfusion rating (no critical limb ischaemia); approximately equal proportions of patients had a grade 1–3 depth/tissue loss rating; the majority of patients (93.3%) had grade 2 sensation (loss of protective sensation); and the majority of patients had an infection of either grade 2 (inflammation of skin/subcutaneous tissue only, 37.3%) or grade 3 (extensive erythema deeper than skin/subcutaneous tissue, 59.3%). The majority of patients had an ulcer debridement undertaken at the baseline visit (87.8%), with the median area measuring 1.77 cm2 (range 0.01–138.2 cm2). The clinical signs and symptoms classification of patients’ index ulcers revealed that 31.8% of patients had a foul wound odour; 42.5% had pocketing in the wound; 56.3% had discoloured granulation tissue; 51.0% had friable granulation tissue; 31.3% had a recent increase in pain, as opposed to the 2.3% who had a recent decrease in pain; 61.5% had a recent increase in wound size; and 31.5% had a breakdown of epithelium. Furthermore, of all index ulcers, 34.0% were classified as grade 1 (superficial diabetic ulcer); 33.5% were classified as grade 2 (ulcer extension to ligament, tendon, joint capsule or deep fascia without abscess or osteomyelitis); 30.5% were classified as grade 3 (deep ulcer with abscess, osteomyelitis or joint sepsis); 1.8% were classified as grade 4 (gangrene localised to a portion of forefoot or heel); and 0.3% were classified as grade 5 (extensive gangrenous involvement of the entire foot).
Tables 12 and 13 and Figure 5 summarise the antibiotic regimens patients were prescribed immediately pre and post sampling. Prior to sampling, 60.3% of patients had been treated with an antimicrobial dressing or agent on the infected ulcer. Furthermore, 46.8% of patients were on a systemic antibiotic regimen, with the most frequently prescribed antibiotics being flucloxacillin (31.1%), clindamycin (18.3%), co-amoxiclav (13.1%), ciprofloxacin (13.1%) and metronidazole (7.2%). The patient’s antibiotic regimen was changed following clinical assessment and specimen sampling, but before microbiology results were available, in 62.0% of patients. Among the 42.0% of patients who were not on an antibiotic regimen prior to sampling, treatment was initiated immediately post sampling. Finally, 6.5% of patients were not on an antibiotic regimen prior to sampling and did not have an antibiotic regimen initiated immediately post sampling.
FIGURE 5.
Prescribed antibiotics pre and post sampling (antibiotics are not mutually exclusive).

Microbiology results
Microbiology reports of culture results for swab and tissue samples produced a total of 79 different microbial isolates from the 395 evaluable patients.
Table 14 presents the number of patients with at least one pathogen reported. At least one pathogenic isolate was reported from swab results in 277 (70.1%) patients and from tissue results in 340 (86.1%) patients. On swab sample results, only isolates not likely to be pathogenic (defined as mixed skin flora, normal flora, enteric flora, yeast, faecal flora) were reported for 39 (9.9%) patients, and no isolates were reported at all for 79 (20.0%) patients. Based on tissue results, only isolates not likely to be pathogenic were reported for 15 (3.8%) patients and no isolates were reported at all for 40 (10.1%) patients.
| Reporting of pathogens | Specimen type | |
|---|---|---|
| Swab (N = 395), n (%) | Tissue (N = 395), n (%) | |
| No pathogens reported | 118 (29.9) | 55 (13.9) |
| No isolates reported at all | 79 (20.0) | 40 (10.1) |
| Only isolates not likely to be pathogenic reporteda | 39 (9.9) | 15 (3.8) |
| At least one pathogen reported | 277 (70.1) | 340 (86.1) |
Table 15 presents the pathogens most frequently reported, following their grouping at a range of taxonomic levels. The most frequently reported groups of pathogens from at least one of the patient’s swab or tissue sample were Gram-positive cocci (70.6%), Gram-negative bacilli (36.7%), Enterobacteriaceae including coliforms (26.6%), anaerobes (23.8%) and Gram-positive bacilli (11.1%). The most frequently reported genus- and species-level pathogens were S. aureus (35.7%), Streptococcus (16.7%), Enterococcus (14.9%), CNS (12.2%), Corynebacterium (9.4%), Pseudomonas (8.6%) and MRSA (8.1%). The prevalence of additional genus- and species-level pathogens were all < 6%.
| Pathogensa | Swab (N = 395) | Tissue (N = 395) | Overall (N = 395) |
|---|---|---|---|
| Groups of pathogens, n (%) | |||
| Gram-positive cocci | 211 (53.4) | 265 (67.1) | 279 (70.6) |
| Gram-negative bacilli | 96 (24.3) | 133 (33.7) | 145 (36.7) |
| Enterobacteriaceae (including coliforms) | 68 (17.2) | 91 (23.0) | 105 (26.6) |
| Overall anaerobes | 48 (12.2) | 75 (19.0) | 94 (23.8) |
| Anaerobes (type not reported) | 42 (10.6) | 64 (16.2) | 83 (21.0) |
| Anaerobic cocci | 3 (0.8) | 6 (1.5) | 6 (1.5) |
| Anaerobic rods | 3 (0.8) | 5 (1.3) | 5 (1.3) |
| Gram-positive bacilli | 4 (1.0) | 43 (10.9) | 44 (11.1) |
| Gram-negative cocci | 0 (0.0) | 1 (0.3) | 1 (0.3) |
| Genus-level pathogens, n (%) | |||
| Streptococcus | 48 (12.2) | 61 (15.4) | 66 (16.7) |
| Enterococcus (excluding vancomycin resistant) | 25 (6.3) | 53 (13.4) | 59 (14.9) |
| CNS | 9 (2.3) | 47 (11.9) | 48 (12.2) |
| Corynebacterium | 4 (1.0) | 36 (9.1) | 37 (9.4) |
| Pseudomonas | 26 (6.6) | 26 (6.6) | 34 (8.6) |
| Proteus | 14 (3.5) | 20 (5.1) | 23 (5.8) |
| Enterobacter | 4 (1.0) | 11 (2.8) | 11 (2.8) |
| Klebsiella | 3 (0.8) | 8 (2.0) | 10 (2.5) |
| Candida | 5 (1.3) | 5 (1.3) | 9 (2.3) |
| Acinetobacter | 3 (0.8) | 5 (1.3) | 6 (1.5) |
| Citrobacter | 2 (0.5) | 4 (1.0) | 4 (1.0) |
| Bacteroides | 1 (0.3) | 2 (0.5) | 2 (0.5) |
| Prevotella | 1 (0.3) | 2 (0.5) | 2 (0.5) |
| Enterococcus (vancomycin resistant) | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| Species-level pathogens, n (%) | |||
| S. aureus | 125 (31.6) | 125 (31.6) | 141 (35.7) |
| MRSA | 27 (6.8) | 31 (7.8) | 32 (8.1) |
| E. coli | 6 (1.5) | 13 (3.3) | 15 (3.8) |
| Morganella morganii | 2 (0.5) | 5 (1.3) | 6 (1.5) |
| Serratia marcescens | 1 (0.3) | 5 (1.3) | 5 (1.3) |
| Stenotrophomonas maltophilia | 1 (0.3) | 3 (0.8) | 3 (0.8) |
| Peptoniphilus asaccharolyticus | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| Raoultella planticola | 1 (0.3) | 1 (0.3) | 1 (0.3) |
Coprimary end points
Coprimary end point: reported presence of likely pathogens
Most prevalent pathogens
Table 16 presents full cross-tabulations of the reported presence of the most prevalent pathogens (those with prevalence > 8%), Figure 6 depicts this information and Table 17 presents statistics relating to the agreement and differences in reporting of these pathogens.
| Pathogen (overall prevalence) | Swab results, n (%) | Tissue results, n (%) | |||
|---|---|---|---|---|---|
| At least one pathogen (88.1%) | Not reported | Reported | Total | ||
| Swab | Not reported | 47 (11.9) | 71 (18.0) | 118 (29.9) | |
| Reported | 8 (2.0) | 269 (68.1) | 277 (70.1) | ||
| Total | 55 (13.9) | 340 (86.1) | 395 (100.0) | ||
| Gram-positive cocci (70.6%) | Not reported | Reported | Total | ||
| Swab | Not reported | 116 (29.4) | 68 (17.2) | 184 (46.6) | |
| Reported | 14 (3.5) | 197 (49.9) | 211 (53.4) | ||
| Total | 130 (32.9) | 265 (67.1) | 395 (100.0) | ||
| Gram-negative bacilli (36.7%) | Not reported | Reported | Total | ||
| Swab | Not reported | 250 (63.3) | 49 (12.4) | 299 (75.7) | |
| Reported | 12 (3.0) | 84 (21.3) | 96 (24.3) | ||
| Total | 262 (63.3) | 133 (33.7) | 395 (100.0) | ||
| Enterobacteriaceae (26.6%) | Not reported | Reported | Total | ||
| Swab | Not reported | 290 (73.4) | 37 (9.4) | 327 (82.8) | |
| Reported | 14 (3.5) | 54 (13.7) | 68 (17.2) | ||
| Total | 304 (77.0) | 91 (23.0) | 395 (100.0) | ||
| Overall anaerobes (23.8%) | Not reported | Reported | Total | ||
| Swab | Not reported | 301 (76.2) | 46 (11.6) | 347 (87.8) | |
| Reported | 19 (4.8) | 29 (7.3) | 48 (12.2) | ||
| Total | 320 (81.0) | 75 (19.0) | 395 (100.0) | ||
| Gram-positive bacilli (11.1%) | Not reported | Reported | Total | ||
| Swab | Not Reported | 351 (88.9) | 40 (10.1) | 391 (99.0) | |
| Reported | 1 (0.3) | 3 (0.8) | 4 (1.0) | ||
| Total | 352 (89.1) | 43 (10.9) | 395 (100.0) | ||
| Streptococcus (16.7%) | Not reported | Reported | Total | ||
| Swab | Not reported | 329 (83.3) | 18 (4.6) | 347 (87.8) | |
| Reported | 5 (1.3) | 43 (10.9) | 48 (12.2) | ||
| Total | 334 (84.6) | 61 (15.4) | 395 (100.0) | ||
| Enterococcus (excluding vancomycin resistant) (14.9%) | Not reported | Reported | Total | ||
| Swab | Not reported | 336 (85.1) | 34 (8.6) | 370 (93.7) | |
| Reported | 6 (1.5) | 19 (4.8) | 25 (6.3) | ||
| Total | 342 (86.6) | 53 (13.4) | 395 (100.0) | ||
| CNS (12.2%) | Not reported | Reported | Total | ||
| Swab | Not reported | 347 (87.8) | 39 (9.9) | 386 (97.7) | |
| Reported | 1 (0.3) | 8 (2.0) | 9 (2.3) | ||
| Total | 348 (88.1) | 47 (11.9) | 395 (100.0) | ||
| Corynebacterium (9.4%) | Not reported | Reported | Total | ||
| Swab | Not reported | 358 (90.6) | 33 (8.4) | 391 (99.0) | |
| Reported | 1 (0.3) | 3 (0.8) | 4 (1.0) | ||
| Total | 359 (90.9) | 36 (9.1) | 395 (100.0) | ||
| Pseudomonas (8.6%) | Not reported | Reported | Total | ||
| Swab | Not reported | 361 (91.4) | 8 (2.0) | 369 (93.4) | |
| Reported | 8 (2.0) | 18 (4.6) | 26 (6.6) | ||
| Total | 369 (93.4) | 26 (6.6) | 395 (100.0) | ||
| S. aureus (35.7%) | Not reported | Reported | Total | ||
| Swab | Not reported | 254 (64.3) | 16 (4.1) | 270 (68.4) | |
| Reported | 16 (4.1) | 109 (27.6) | 125 (31.6) | ||
| Total | 270 (68.4) | 125 (31.6) | 395 (100.0) | ||
| MRSA (8.1%) | Not reported | Reported | Total | ||
| Swab | Not reported | 363 (91.9) | 5 (1.3) | 368 (93.2) | |
| Reported | 1 (0.3) | 26 (6.6) | 27 (6.8) | ||
| Total | 364 (92.2) | 31 (7.8) | 395 (100.0) | ||
FIGURE 6.
Reported presence of at least one pathogen and most prevalent pathogens. VRE, vanconycin-resistant Enterococcus.
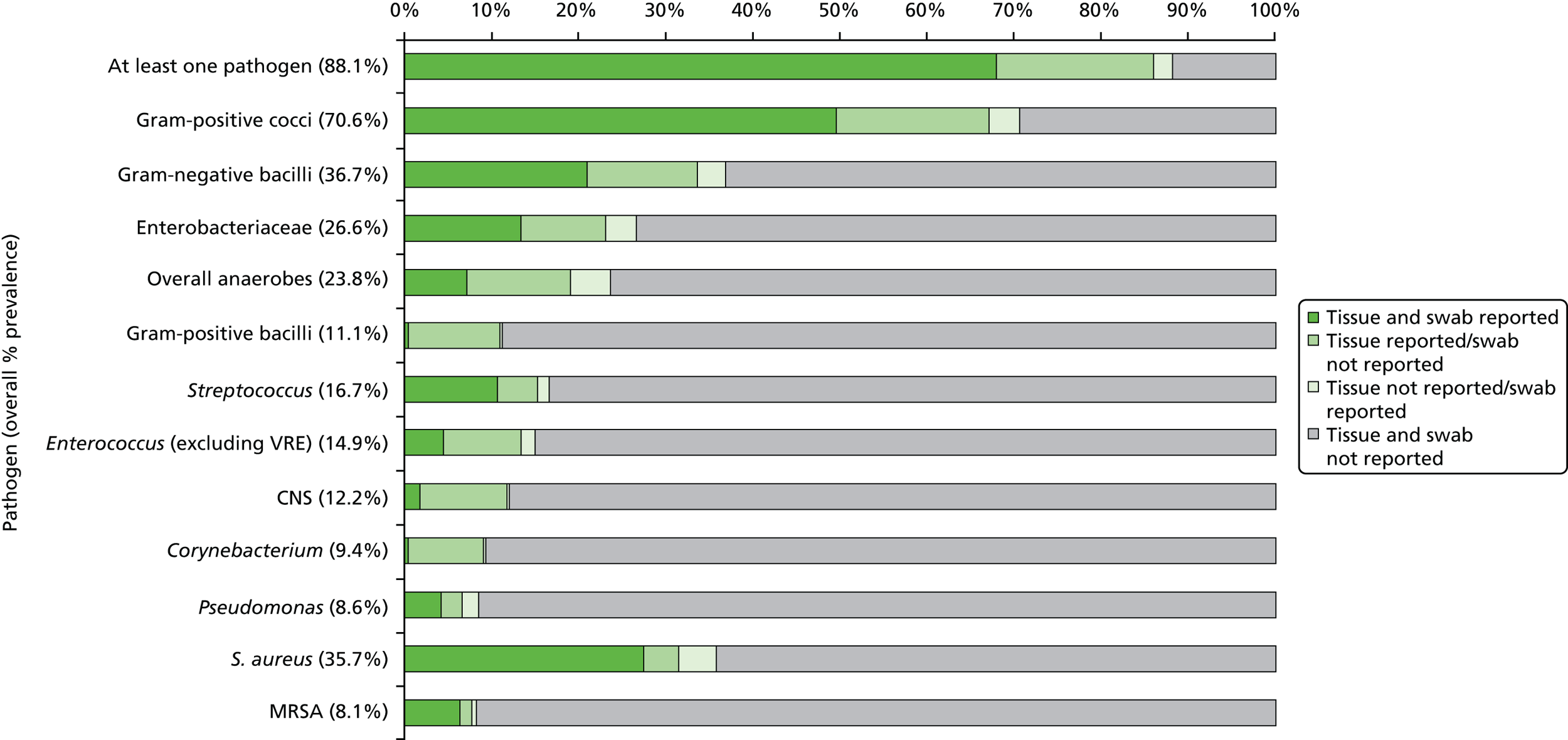
| Pathogens | Overall prevalence, % | Disagreement, % | Difference, % (95% CI)a | McNemar’s p-value | Agreement, % | κ (95% CI) | PABAK |
|---|---|---|---|---|---|---|---|
| ≥ 1 pathogen | 88.1 | 20.0 | 15.9 (11.8 to 20.1) | < 0.0001 | 80.0 | 0.44 (0.34 to 0.53) | 0.60 |
| Gram-positive cocci | 70.6 | 20.8 | 13.7 (9.4 to 18.0) | < 0.0001 | 79.2 | 0.57 (0.50 to 0.65) | 0.58 |
| Gram-negative bacilli | 36.7 | 15.4 | 9.4 (5.6 to 13.1) | < 0.0001 | 84.6 | 0.63 (0.55 to 0.71) | 0.69 |
| Enterobactereaceae (including coliforms) | 26.6 | 12.9 | 5.8 (2.3 to 9.3) | 0.0013 | 87.1 | 0.60 (0.50 to 0.70) | 0.74 |
| Overall anaerobes | 23.8 | 16.5 | 6.8 (2.9 to 10.8) | 0.0008 | 83.5 | 0.38 (0.26 to 0.50) | 0.67 |
| Gram-positive bacilli | 11.1 | 10.4 | 9.9 (6.9 to 13.5) | < 0.0001b | 89.6 | 0.11 (–0.01 to 0.23) | 0.79 |
| Streptococcus | 16.7 | 5.8 | 3.3 (0.9 to 5.6) | 0.0067 | 94.2 | 0.76 (0.66 to 0.85) | 0.88 |
| Enterococcus (excluding vancomycin resistant) | 14.9 | 10.1 | 7.1 (4.0 to 10.1) | < 0.0001 | 89.9 | 0.44 (0.30 to 0.58) | 0.80 |
| CNS | 12.2 | 10.1 | 9.6 (6.7 to 12.9) | < 0.0001b | 89.9 | 0.26 (0.11 to 0.41) | 0.80 |
| Corynebacterium | 9.4 | 8.6 | 8.1 (5.4 to 11.2) | < 0.0001b | 91.4 | 0.13 (–0.01 to 0.28) | 0.83 |
| Pseudomonas | 8.6 | 4.1 | 0.0 (–2.0 to 2.0) | 1.0000 | 95.9 | 0.67 (0.52 to 0.82) | 0.92 |
| S. aureus | 35.7 | 8.1 | 0.0 (–2.8 to 2.8) | 1.0000 | 91.9 | 0.81 (0.75 to 0.87) | 0.84 |
| MRSA | 8.1 | 1.5 | 1.0 (–0.2 to 2.8) | 0.2188b | 98.5 | 0.89 (0.80 to 0.98) | 0.97 |
Overall, there was evidence of a significant difference [15.9% (95% CI 11.8% to 20.1%); p-value < 0.0001] between the swab and tissue samples in the percentage reporting at least one pathogen (86.1% of patients with tissue sample vs. 70.1% of patients with swab sample) (see Table 17).
Among the most prevalent pathogens, overall agreement between swab and tissue sample results was at least 79%. The κ-values for the chance corrected agreement suggested:
-
almost perfect agreement for MRSA [κ = 0.89 (95% CI 0.80 to 0.98)] and S. aureus [κ = 0.81 (95% CI 0.75 to 0.87)]
-
substantial agreement for Streptococcus [κ = 0.76 (95% CI 0.66 to 0.85)], Pseudomonas [κ = 0.67 (95% CI 0.52 to 0.82)] and Gram-negative bacilli [κ = 0.63 (95% CI 0.55 to 0.71)]
-
moderate agreement for Enterobactereaceae (including coliforms) [κ = 0.60 (95% CI 0.50 to 0.70)], Gram-positive cocci [κ = 0.57 (95% CI 0.50 to 0.65)] and Enterococcus [κ = 0.44 (95% CI 0.30 to 0.58)]
-
fair agreement for overall anaerobes [κ = 0.38 (95% CI 0.26 to 0.50)] and CNS [κ = 0.26 (95% CI 0.11 to 0.41)]
-
slight agreement for Corynebacterium [κ = 0.13 (95% CI –0.01 to 0.28)] and Gram-positive bacilli [κ = 0.11 (95% CI –0.01 to 0.23)].
The PABAK for the majority of pathogens showed a considerably higher estimate of agreement after accounting for the low prevalence of the majority of pathogens.
For the majority of pathogens, there was evidence of a significant difference (McNemar’s p-value < 0.01), with reported prevalence higher in the tissue sample results than the swab results, with the exception for S. aureus, MRSA and Pseudomonas. Symmetrical disagreement was observed for S. aureus and Pseudomonas, with the pathogen reported in one sample but not the other an equal number of times for the two samples. The reported prevalence of MRSA was non-statistically higher in tissue samples than swab samples.
Semi-quantitative extent of bacterial growth
Table 18 presents cross-tabulations of the level of growth of each of the most prevalent pathogens, by swab and tissue samples, and the associated κ-values. The overall κ-value does not account for the ordinal levels of growth, whereas the weighted κ-value quantifies the relative difference between levels of growth. κ-values were calculated after excluding patients with a missing level of growth for either the swab or tissue sample.
| Tissue results: level of growth | Total, n (%) | κ-valuea | κ | 95% CI | |||||
|---|---|---|---|---|---|---|---|---|---|
| Not reported | Reported: no growth | + | ++ | +++ | |||||
| Gram-positive cocci | |||||||||
| Swab results, n (%) | (n = 340) | ||||||||
| Not reported | 116 (29.4) | 13 (3.3) | 25 (6.3) | 14 (3.5) | 16 (4.1) | 184 (46.6) | Overall | 0.49 | 0.42 to 0.55 |
| Reported: no growth | 3 (0.8) | 33 (8.4) | 1 (0.3) | 0 (0.0) | 1 (0.3) | 38 (9.6) | Weighted 1 | 0.58 | 0.51 to 0.66 |
| + | 4 (1.0) | 1 (0.3) | 22 (5.6) | 3 (0.8) | 0 (0.0) | 30 (7.6) | Weighted 2 | 0.60 | 0.54 to 0.67 |
| ++ | 3 (0.8) | 0 (0.0) | 13 (3.3) | 15 (3.8) | 10 (2.5) | 41 (10.4) | |||
| +++ | 4 (1.0) | 3 (0.8) | 9 (2.3) | 21 (5.3) | 65 (16.5) | 102 (25.8) | |||
| Total | 130 (32.9) | 50 (12.7) | 70 (17.7) | 53 (13.4) | 92 (23.3) | n = 395 | |||
| Gram-negative bacilli | |||||||||
| Swab results, n (%) | (n = 354) | ||||||||
| Not reported | 250 (63.3) | 5 (1.3) | 14 (3.5) | 12 (3.0) | 18 (4.6) | 299 (75.7) | Overall | 0.48 | 0.39 to 0.57 |
| Reported: no growth | 4 (1.0) | 18 (4.6) | 2 (0.5) | 3 (0.8) | 5 (1.3) | 32 (8.1) | Weighted 1 | 0.49 | 0.38 to 0.61 |
| + | 2 (0.5) | 1 (0.3) | 6 (1.5) | 0 (0.0) | 2 (0.5) | 11 (2.8) | Weighted 2 | 0.53 | 0.43 to 0.63 |
| ++ | 3 (0.8) | 2 (0.5) | 4 (1.0) | 10 (2.5) | 2 (0.5) | 21 (5.3) | |||
| +++ | 3 (0.8) | 1 (0.3) | 5 (1.3) | 5 (1.3) | 18 (4.6) | 32 (8.1) | |||
| Total | 262 (66.3) | 27 (6.8) | 31 (7.8) | 30 (7.6) | 45 (11.4) | n = 395 | |||
| Enterobacteriaceae (including coliforms) | |||||||||
| Swab results, n (%) | (n = 359) | ||||||||
| Not reported | 290 (73.4) | 6 (1.5) | 12 (3.0) | 5 (1.3) | 14 (3.5) | 327 (82.8) | Overall | 0.45 | 0.33 to 0.56 |
| Reported: no growth | 4 (1.0) | 17 (4.3) | 2 (0.5) | 1 (0.3) | 3 (0.8) | 27 (6.8) | Weighted 1 | 0.50 | 0.35 to 0.64 |
| + | 2 (0.5) | 1 (0.3) | 2 (0.5) | 0 (0.0) | 0 (0.0) | 5 (1.3) | Weighted 2 | 0.51 | 0.38 to 0.64 |
| ++ | 3 (0.8) | 1 (0.3) | 3 (0.8) | 5 (1.3) | 0 (0.0) | 12 (3.0) | |||
| +++ | 5 (1.3) | 1 (0.3) | 2 (0.5) | 3 (0.8) | 13 (3.3) | 24 (6.1) | |||
| Total | 304 (77.0) | 26 (6.6) | 21 (5.3) | 14 (3.5) | 30 (7.6) | n = 395 | |||
| Overall anaerobes | |||||||||
| Swab results, n (%) | (n = 377) | ||||||||
| Not reported | 301 (76.2) | 9 (2.3) | 12 (3.0) | 14 (3.5) | 11 (2.8) | 347 (87.8) | Overall | 0.32 | 0.21 to 0.43 |
| Reported: no growth | 4 (1.0) | 4 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 (2.0) | Weighted 1 | 0.43 | 0.29 to 0.57 |
| + | 7 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 8 (2.0) | Weighted 2 | 0.44 | 0.31 to 0.56 |
| ++ | 1 (0.3) | 1 (0.3) | 1 (0.3) | 4 (1.0) | 6 (1.5) | 13 (3.3) | |||
| +++ | 7 (1.8) | 0 (0.0) | 0 (0.0) | 2 (0.5) | 10 (2.5) | 19 (4.8) | |||
| Total | 320 (81.0) | 14 (3.5) | 13 (3.3) | 20 (5.1) | 28 (7.1) | n = 395 | |||
| Gram-positive bacilli | |||||||||
| Swab results, n (%) | (n = 389) | ||||||||
| Not reported | 351 (88.9) | 6 (1.5) | 15 (3.8) | 10 (2.5) | 9 (2.3) | 391 (99.0) | Overall | 0.06 | –0.00 to 0.13 |
| + | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | Weighted 1 | 0.07 | –0.01 to 0.16 |
| ++ | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 2 (0.5) | Weighted 2 | 0.11 | –0.01 to 0.24 |
| +++ | 0 (0.0) | 0 (0.0) | 1 (0.3) | 1 (0.3) | 0 (0.0) | 2 (0.5) | |||
| Total | 352 (89.1) | 6 (1.5) | 16 (4.1) | 11 (2.8) | 10 (2.5) | n = 395 | |||
| Streptococcus | |||||||||
| Swab results, n (%) | (n = 384) | ||||||||
| Not reported | 329 (83.3) | 6 (1.5) | 5 (1.3) | 1 (0.3) | 6 (1.5) | 347 (87.8) | Overall | 0.65 | 0.55 to 0.75 |
| Reported: no growth | 0 (0.0) | 4 (1.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 5 (1.3) | Weighted 1 | 0.68 | 0.56 to 0.80 |
| + | 2 (0.5) | 0 (0.0) | 5 (1.3) | 1 (0.3) | 0 (0.0) | 8 (2.0) | Weighted 2 | 0.74 | 0.65 to 0.84 |
| ++ | 1 (0.3) | 0 (0.0) | 1 (0.3) | 4 (1.0) | 2 (0.5) | 8 (2.0) | |||
| +++ | 2 (0.5) | 0 (0.0) | 2 (0.5) | 7 (1.8) | 16 (4.1) | 27 (6.8) | |||
| Total | 334 (84.6) | 10 (2.5) | 13 (3.3) | 13 (3.3) | 25 (6.3) | n = 395 | |||
| Enterococcus (excluding vancomycin resistant) | |||||||||
| Swab results, n (%) | (n = 384) | ||||||||
| Not reported | 336 (85.1) | 6 (1.5) | 9 (2.3) | 12 (3.0) | 7 (1.8) | 370 (93.7) | Overall | 0.39 | 0.24 to 0.54 |
| Reported – no growth | 0 (0.0) | 4 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (1.0) | Weighted 1 | 0.52 | 0.34 to 0.70 |
| + | 2 (0.5) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 3 (0.8) | Weighted 2 | 0.47 | 0.31 to 0.64 |
| ++ | 1 (0.3) | 1 (0.3) | 0 (0.0) | 3 (0.8) | 1 (0.3) | 6 (1.5) | |||
| +++ | 3 (0.8) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 8 (2.0) | 12 (3.0) | |||
| Total | 342 (86.6) | 11 (2.8) | 10 (2.5) | 16 (4.1) | 16 (4.1) | n = 395 | |||
| CNS | |||||||||
| Swab results, n (%) | (n = 389) | ||||||||
| Not reported | 347 (87.8) | 5 (1.3) | 22 (5.6) | 7 (1.8) | 5 (1.3) | 386 (97.7) | Overall | 0.23 | 0.08 to 0.37 |
| + | 0 (0.0) | 0 (0.0) | 2 (0.5) | 0 (0.0) | 0 (0.0) | 2 (0.5) | Weighted 1 | 0.34 | 0.11 to 0.57 |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 2 (0.5) | 3 (0.8) | Weighted 2 | 0.31 | 0.13 to 0.50 |
| +++ | 1 (0.3) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 2 (0.5) | 4 (1.0) | |||
| Total | 348 (88.1) | 6 (1.5) | 24 (6.1) | 8 (2.0) | 9 (2.3) | n = 395 | |||
| Corynebacterium | |||||||||
| Swab results, n (%) | (n = 390) | ||||||||
| Not reported | 358 (90.6) | 5 (1.3) | 10 (2.5) | 10 (2.5) | 8 (2.0) | 391 (99.0) | Overall | 0.07 | –0.00 to 0.15 |
| ++ | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 2 (0.5) | Weighted 1 | 0.08 | –0.01 to 0.17 |
| +++ | 0 (0.0) | 0 (0.0) | 1 (0.3) | 1 (0.3) | 0 (0.0) | 2 (0.5) | Weighted 2 | 0.13 | –0.01 to 0.27 |
| Total | 359 (90.9) | 5 (1.3) | 11 (2.8) | 11 (2.8) | 9 (2.3) | n = 395 | |||
| Pseudomonas | |||||||||
| Swab results, n (%) | (n = 391) | ||||||||
| Not reported | 361 (91.4) | 1 (0.3) | 1 (0.3) | 3 (0.8) | 3 (0.8) | 369 (93.4) | Overall | 0.58 | 0.42 to 0.75 |
| Reported: no growth | 0 (0.0) | 2 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 3 (0.8) | Weighted 1 | 0.61 | 0.40 to 0.82 |
| + | 4 (1.0) | 0 (0.0) | 3 (0.8) | 0 (0.0) | 1 (0.3) | 8 (2.0) | Weighted 2 | 0.61 | 0.44 to 0.79 |
| ++ | 2 (0.5) | 0 (0.0) | 1 (0.3) | 3 (0.8) | 0 (0.0) | 6 (1.5) | |||
| +++ | 2 (0.5) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 6 (1.5) | 9 (2.3) | |||
| Total | 369 (93.4) | 3 (0.8) | 6 (1.5) | 6 (1.5) | 11 (2.8) | n = 395 | |||
| S. aureus | |||||||||
| Swab results, n (%) | (n = 363) | ||||||||
| Not reported | 254 (64.3) | 1 (0.3) | 11 (2.8) | 3 (0.8) | 1 (0.3) | 270 (68.4) | Overall | 0.64 | 0.57 to 0.71 |
| Reported: no growth | 4 (1.0) | 25 (6.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 29 (7.3) | Weighted 1 | 0.7 | 0.62 to 0.79 |
| + | 3 (0.8) | 1 (0.3) | 14 (3.5) | 1 (0.3) | 0 (0.0) | 19 (4.8) | Weighted 2 | 0.74 | 0.68 to 0.81 |
| ++ | 6 (1.5) | 0 (0.0) | 8 (2.0) | 8 (2.0) | 2 (0.5) | 24 (6.1) | |||
| +++ | 3 (0.8) | 1 (0.3) | 9 (2.3) | 10 (2.5) | 30 (7.6) | 53 (13.4) | |||
| Total | 270 (68.4) | 28 (7.1) | 42 (10.6) | 22 (5.6) | 33 (8.4) | n = 395 | |||
| MRSA | |||||||||
| Swab results, n (%) | (n = 391) | ||||||||
| Not reported | 363 (91.9) | 0 (0.0) | 1 (0.3) | 2 (0.5) | 2 (0.5) | 368 (93.2) | Overall | 0.73 | 0.61 to 0.85 |
| Reported: no growth | 0 (0.0) | 4 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (1.0) | Weighted 1 | 0.79 | 0.67 to 0.91 |
| + | 1 (0.3) | 0 (0.0) | 3 (0.8) | 0 (0.0) | 0 (0.0) | 4 (1.0) | Weighted 2 | 0.83 | 0.73 to 0.93 |
| ++ | 0 (0.0) | 0 (0.0) | 2 (0.5) | 1 (0.3) | 2 (0.5) | 5 (1.3) | |||
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (0.8) | 11 (2.8) | 14 (3.5) | |||
| Total | 364 (92.2) | 4 (1.0) | 6 (1.5) | 6 (1.5) | 15 (3.8) | n = 395 | |||
Agreement on the level of growth (according to the primary weighting) was somewhat skewed owing to the prevalence of each pathogen and the proportion of patients with discordant results where a pathogen was reported in one sample and not the other. Therefore, the level of growth in one sample was often in comparison with the lack of presence of the pathogen rather than a corresponding level of growth. The range for agreement was substantial for Streptococcus, Pseudomonas, S. aureus and MRSA; moderate for Gram-positive cocci, Gram-negative bacilli, Enterobacteriaceae and Enterococcus; fair for CNS and Corynebacterium; and slight for Gram-positive bacilli.
Summary of pathogens
Table 19 presents the overall summary of all pathogens reported by specimen type for the evaluable population and by baseline characteristics. Figure 7 presents the overall summary by centre. Overall, there is a difference in the pathogens reported by the two techniques for 58.0% of patients. Findings among the 395 patient pairs of results were swab and tissue results reported the same pathogens in 42.0% of patients; swab results reported additional pathogens to those in the tissue in 8.1% of patients; tissue reported additional pathogens to those in the swab in 36.7% of patients; and the tissue sample and swab specimens reported different pathogens, with or without overlap, in 13.2% of patients.
| Baseline characteristics | Swab and tissue report the same pathogens, n (%) | Swab reports additional pathogens to the tissue, n (%) | Tissue reports additional pathogens to the swab, n (%) | Swab and tissue report different pathogens,d n (%) |
|---|---|---|---|---|
| Total (n = 395) | 166 (42.0)a | 32 (8.1)b | 145 (36.7)c | 52 (13.2) |
| Type of ulcer | ||||
| Any ischaemia (± neuropathy) (n = 194) | 87 (44.8) | 17 (8.8) | 68 (35.1) | 22 (11.3) |
| Neuropathic only (n = 199) | 79 (39.7) | 15 (7.5) | 76 (38.2) | 29 (14.6) |
| Missing (n = 2) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 1 (50.0) |
| Grade of ulcer | ||||
| Grade 1 (n = 135) | 60 (44.4) | 12 (8.9) | 47 (34.8) | 16 (11.9) |
| Grade 2 (n = 132) | 59 (44.7) | 8 (6.1) | 48 (36.4) | 17 (12.9) |
| Grade 3, 4 or 5 (n = 128) | 47 (36.7) | 12 (9.4) | 50 (39.1) | 19 (14.8) |
| Pre-sampling antibiotic therapy | ||||
| Yes (n = 186) | 78 (41.9) | 12 (6.5) | 71 (38.2) | 25 (13.4) |
| No (n = 190) | 80 (42.1) | 15 (7.9) | 70 (36.8) | 25 (13.2) |
| Missing (n = 19) | 8 (42.1) | 5 (26.3) | 4 (21.1) | 2 (10.5) |
| Presence of antimicrobial dressing or agent | ||||
| Yes (n = 238) | 101 (42.4) | 20 (8.4) | 82 (34.5) | 35 (14.7) |
| No (n = 152) | 61 (40.1) | 11 (7.2) | 63 (41.4) | 17 (11.2) |
| Missing (n = 5) | 4 (80.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) |
| Wound duration (by median) | ||||
| < 56 days (n = 189) | 72 (38.1) | 13 (6.9) | 81 (42.9) | 23 (12.2) |
| ≥ 56 days (n = 202) | 94 (46.5) | 18 (8.9) | 62 (30.7) | 28 (13.9) |
| Missing (n = 4) | 0 (0.0) | 1 (25.0) | 2 (50.0) | 1 (25.0) |
FIGURE 7.
Overall summary of all pathogens reported by centre. SJUH, St James’s University Hospital.
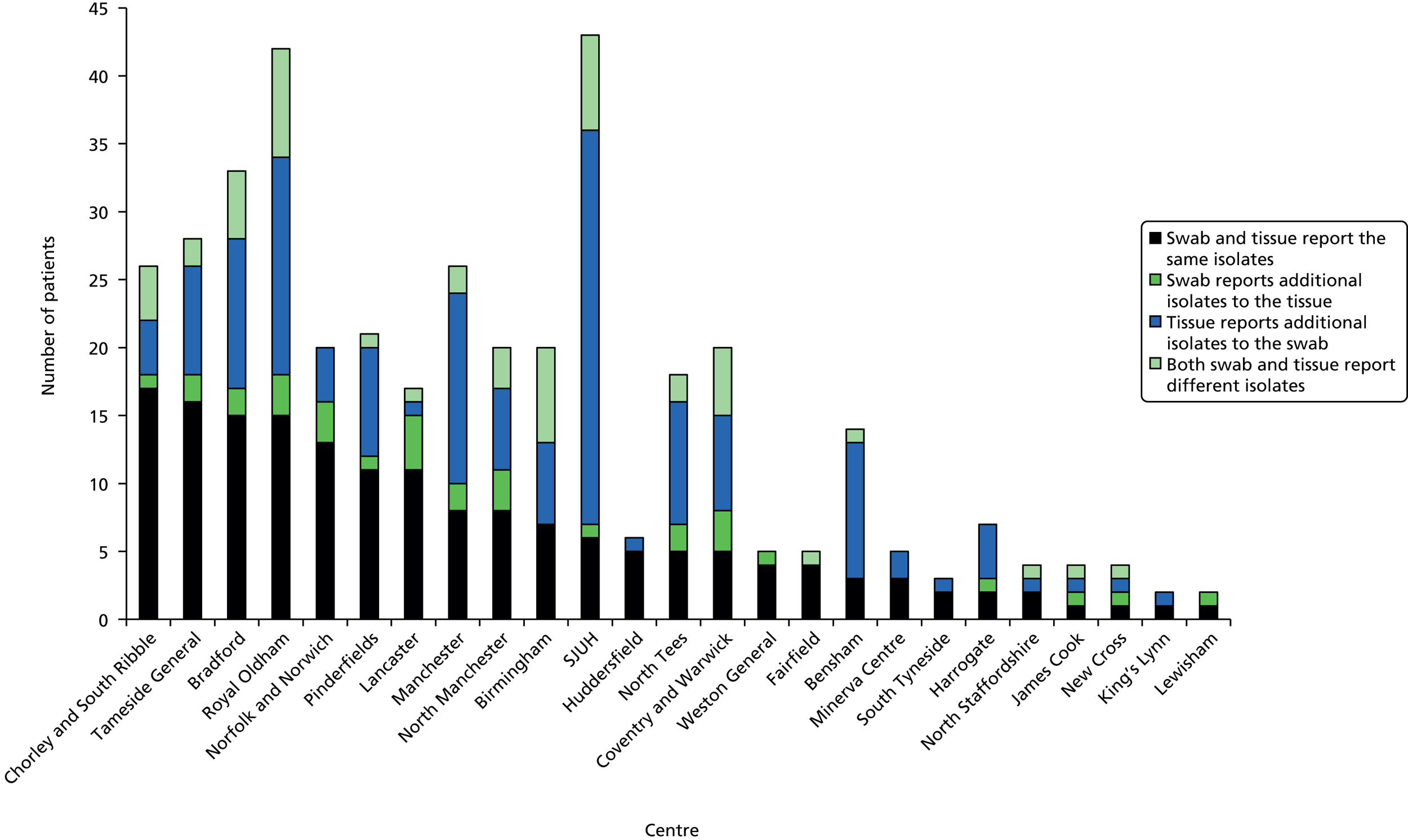
Multinomial regression analyses
Multinomial regression modelling with a random effect for centre (and MIs to allow for missing data) was used to assess whether or not agreement, based on the overall summary of pathogens, was influenced by the pre-specified baseline covariates (Table 20).
| Multinomial regressionanalyses | Summary of pathogens (reference: both swab and tissue report the same pathogens) | Odds ratioa (95% CI) | AICb | df | p-valuec |
|---|---|---|---|---|---|
| Null model | 941.29 | ||||
| Ulcer type: any ischaemia (± neuropathy) vs. neuropathic only | 945.72 | 3 | 0.6663 | ||
| Swab reports additional pathogens to the tissue | 1.03 (0.48 to 2.20) | ||||
| Tissue reports additional pathogens to the swab | 0.86 (0.53 to 1.40) | ||||
| Both swab and tissue report different pathogens | 0.68 (0.35 to 1.31) | ||||
| Ulcer grade | 949.16 | 6 | 0.6598 | ||
| Grade 2 vs. grade 1 | Swab reports additional pathogens to the tissue | 0.68 (0.26 to 1.78) | |||
| Tissue reports additional pathogens to the swab | 1.08 (0.60 to 1.93) | ||||
| Both swab and tissue report different pathogens | 1.14 (0.51 to 2.54) | ||||
| Grade 3/4/5 vs. grade 1 | Swab reports additional pathogens to the tissue | 1.28 (0.52 to 3.11) | |||
| Tissue reports additional pathogens to the swab | 1.60 (0.87 to 2.95) | ||||
| Both swab and tissue report different pathogens | 1.55 (0.69 to 3.45) | ||||
| Pre-sampling antibiotic therapy: yes vs. no | 946.28 | 3 | 0.8001 | ||
| Swab reports additional pathogens to the tissue | 0.80 (0.36 to 1.80) | ||||
| Tissue reports additional pathogens to the swab | 1.14 (0.69 to 1.89) | ||||
| Both swab and tissue report different pathogens | 1.10 (0.56 to 2.16) | ||||
| Antimicrobial dressing: yes vs. no | 943.44 | 3 | 0.2782 | ||
| Swab reports additional pathogens to the tissue | 1.13 (0.51 to 2.51) | ||||
| Tissue reports additional pathogens to the swab | 0.69 (0.40 to 1.19) | ||||
| Both swab and tissue report different pathogens | 1.38 (0.66 to 2.89) | ||||
| Wound duration (median split): ≥ 56 days vs. < 56 days | 941.48 | 3 | 0.1216 | ||
| Swab reports additional pathogens to the tissue | 1.06 (0.49 to 2.32) | ||||
| Tissue reports additional pathogens to the swab | 0.57 (0.35 to 0.93) | ||||
| Both swab and tissue report different pathogens | 0.88 (0.46 to 1.70) | ||||
| Log-wound duration (continuous) | 944.97 | 3 | 0.5091 | ||
| Swab reports additional pathogens to the tissue | 0.95 (0.72 to 1.25) | ||||
| Tissue reports additional pathogens to the swab | 0.88 (0.74 to 1.04) | ||||
| Both swab and tissue report different pathogens | 0.93 (0.74 to 1.18) | ||||
None of the baseline factors [ulcer type (any ischaemia/neuropathic only), ulcer grade (Wagner grade 1/grade 2/grade 3, 4 or 5), pre-sampling antibiotic therapy (yes/no), antimicrobial dressing or agent (yes/no), wound duration (considered dichotomously as < 56 days/≥ 56 days and continuously on the log-scale)] was found to have a significant overall impact on agreement. However, comparison of the individual outcomes did suggest that patients with a wound duration ≥ 56 days had significantly reduced odds of their tissue sample reporting additional pathogens to the swab sample, as opposed to their swab and tissue reporting the same pathogens, with an odds ratio of 0.57 (95% CI 0.35 to 0.93). This finding was not, however, supported on the continuous scale for wound duration.
Coprimary end point 2: reported presence of antimicrobial resistance among likely pathogens
Likely pathogens
Of the three pathogens of interest, no meticillin-resistant CNS was reported and vancomycin-resistant Enterococcus was reported for just one patient in both their swab and tissue sample results (Table 21).
| Antimicrobial resistance | Swab (N = 395), n (%) | Tissue (N = 395), n (%) | Overall prevalence (N = 395), n (%) |
|---|---|---|---|
| MRSA | 27 (6.8) | 31 (7.8) | 32 (8.1) |
| Vancomycin-resistantEnterococcus | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| Meticillin-resistant CNS | 0 (0) | 0 (0) | 0 (0) |
Meticillin-resistant S. aureus was reported in 32 (8.1%) patients overall, with overall agreement of 98.5% between swab and tissue samples. In 5 (1.3%) patients, the pathogen was reported in the tissue results but not the swab, and in 1 (0.3%) patient, the pathogen was reported in the swab but not the tissue results (see Table 16). As such, a difference of 1.0% (exact 95% CI –0.2% to 2.8%) was reported, with McNemar’s test suggesting that this was not a significant difference (exact p-value = 0.2188) (see Table 17).
To evaluate whether or not agreement was influenced by pre-specified covariates, multinomial regression modelling had been proposed based on the outcomes of reported MRSA: by swab not tissue, by tissue not swab, swab and tissue results agree. However, given the small number of patients whose swab and tissue sample results did not agree [6 (1.6%)], this analysis was not appropriate and was not performed.
Additional sensitivities and resistances
In addition to the three pathogens of interest, resistance and sensitivity to antibiotics were collected where reported for all pathogens within a patient’s swab or tissue sample.
Patients’ swab or tissue sample results were reported to contain pathogens with a resistance to a maximum of eight different antibiotics and sensitivity to a maximum of 10 different antibiotic agents, of any of the antibiotics for which samples were tested (Table 22). There were 123 (31.1%) patients whose swab sample reported pathogen(s) with resistance to at least one antibiotic agent, whereas 165 (41.8%) patients’ tissue samples reported pathogen(s) with resistance to at least one resistant antibiotic agent. Overall, from either the swab or tissue sample results there were 185 (46.8%) patients for whom resistance to at least one antibiotic agent was reported. A greater proportion of patients’ sample results reported at least one antibiotic to which pathogens were sensitive. There were 221 (55.9%) patients whose swab samples reported pathogen(s) with sensitivity to at least one antibiotic agent, 268 (67.8%) patients whose tissue sample reported pathogen(s) with sensitivity to at least one antibiotic agent and 284 (71.9%) patients for whom the swab or tissue sample reported pathogen(s) with sensitivity to at least one antibiotic agent.
| Number of antibiotics | Antibiotic resistance | Antibiotic sensitivity | ||||
|---|---|---|---|---|---|---|
| Swab (N = 395) | Tissue (N = 395) | Overall (N = 395) | Swab (N = 395) | Tissue (N = 395) | Overall (N = 395) | |
| Number of antibiotics | ||||||
| Mean (SD) | 0.6 (1.16) | 0.9 (1.35) | 1.0 (1.45) | 1.6 (1.80) | 2.1 (1.98) | 2.5 (2.14) |
| Median (range) | 0.0 (0–6) | 0.0 (0–8) | 0.0 (0–8) | 1.0 (0–9) | 2.0 (0–9) | 2.0 (0–10) |
| IQR | (0.0–1.0) | (0.0–1.0) | (0.0–2.0) | (0.0–3.0) | (0.0–3.0) | (0.0–4.0) |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 |
| Number of antibiotics, n (%) | ||||||
| 0 | 272 (68.9) | 230 (58.2) | 210 (53.2) | 174 (44.1) | 127 (32.2) | 111 (28.1) |
| 1 | 62 (15.7) | 77 (19.5) | 81 (20.5) | 38 (9.6) | 39 (9.9) | 27 (6.8) |
| 2 | 31 (7.8) | 41 (10.4) | 51 (12.9) | 63 (15.9) | 69 (17.5) | 64 (16.2) |
| 3 | 10 (2.5) | 21 (5.3) | 19 (4.8) | 56 (14.2) | 68 (17.2) | 68 (17.2) |
| 4 | 14 (3.5) | 17 (4.3) | 21 (5.3) | 35 (8.9) | 50 (12.7) | 58 (14.7) |
| 5 | 3 (0.8) | 4 (1.0) | 6 (1.5) | 16 (4.1) | 14 (3.5) | 28 (7.1) |
| 6 | 3 (0.8) | 4 (1.0) | 5 (1.3) | 10 (2.5) | 18 (4.6) | 23 (5.8) |
| 7 | 0 (0.0) | 0 (0.0) | 1 (0.3) | 2 (0.5) | 5 (1.3) | 10 (2.5) |
| 8 | 0 (0.0) | 1 (0.3) | 1 (0.3) | 0 (0.0) | 4 (1.0) | 4 (1.0) |
| 9 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| 10 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) |
The most frequently reported antibiotic agent to which at least one pathogen isolated from a patient’s swab or tissue sample was resistant was penicillin, with a resistance observed in either the swab or tissue sample results for 72 (18.2%) patients; that is, 47 (11.9%) patients’ swab samples and 63 (15.9%) patients’ tissue samples. Figure 8 presents all reported antibiotics to which pathogens were found to be resistant within patients’ samples. A similar pattern is observed across all reported antibiotic agents with each reported more often in the tissue sample than in the swab.
FIGURE 8.
Reported antibiotic resistances for pathogens within patients swab and tissue sample results.

The most frequently reported antibiotic agent to which at least one pathogen isolated from a patient’s swab or tissue sample was sensitive was flucloxacillin, with a sensitivity observed in either the swab or tissue sample results for 144 (36.5%) patients; that is, 126 (31.9%) patients’ swab samples and 126 (31.9%) patients’ tissue samples. Figure 9 presents all reported antibiotics to which pathogens were found to be resistant within patients’ samples, with a similar pattern observed across the majority of reported antibiotic agents, with all agents but erythromycin reported in the same or a greater percentage of patients in the tissue sample than the swab sample.
FIGURE 9.
Reported antibiotic sensitivities for pathogens within patients swab and tissue sample results.

Coprimary end point 3: number of pathogens reported per specimen
The third coprimary end point evaluated agreement between the two specimen collection methods for microbiological characterisation determined by the number of pathogens reported per specimen.
Tables 23 and 24 present the cross-tabulation and summary statistics of the number of pathogens reported from each sample. A median of 1 pathogen was reported in both samples, and the mean number of pathogens reported in the swab and tissue samples was 1 and 1.5, respectively, with a slightly higher level of variation observed in the tissue samples. The number of pathogens ranged from 0 to 4 in the swab sample and 0 to 6 in the tissue sample. A greater proportion of swab results reported no pathogens compared with tissue results (29.9% vs. 13.9%), whereas similar proportions of samples reported just one pathogen (45.1% and 40.8%). Where more than one pathogen was reported, there was consistently a greater frequency of patients with more pathogens in the tissue sample than the swab sample results.
| Swab results | Tissue results, n (%) | Total | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 or more | ||
| 0 | 47 (11.9) | 44 (11.1) | 16 (4.1) | 10 (2.5) | 1 (0.3) | 118 (29.9) |
| 1 | 7 (1.8) | 96 (24.3) | 50 (12.7) | 17 (4.3) | 8 (2.0) | 178 (45.1) |
| 2 | 1 (0.3) | 20 (5.1) | 43 (10.9) | 13 (3.3) | 4 (1.0) | 81 (20.5) |
| 3 | 0 (0.0) | 1 (0.3) | 6 (1.5) | 8 (2.0) | 1 (0.3) | 16 (4.1) |
| 4 or more | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.5) | 2 (0.5) |
| Total | 55 (13.9) | 161 (40.8) | 115 (29.1) | 48 (12.2) | 16 (4.1) | 395 (100.0) |
| Number of pathogens reported per specimen | Swab (N = 395) | Tissue (N = 395) |
|---|---|---|
| Mean (SD) | 1.0 (0.84) | 1.5 (1.04) |
| Median (range) | 1.0 (0–4) | 1.0 (0–6) |
| Range | 0–4 | 0–6 |
| Number of pathogens: frequency, n (%) | ||
| 0 | 118 (29.9) | 55 (13.9) |
| 1 | 178 (45.1) | 161 (40.8) |
| 2 | 81 (20.5) | 115 (29.1) |
| 3 | 16 (4.1) | 48 (12.2) |
| 4 | 2 (0.5) | 13 (3.3) |
| 5 | 0 (0.0) | 2 (0.5) |
| 6 | 0 (0.0) | 1 (0.3) |
A summary of the number of pathogens by baseline characteristics, presented in Table 25, shows that for approximately half (49.6%) of all patients the same number of pathogens were reported for the tissue and swab sample; for 41.5% of patients the tissue sample reported at least one more pathogen than the swab; and for 8.9% of patients the swab sample reported at least one more pathogen than the tissue. Figure 10 presents the summary of the number of pathogens by centre.
| Baseline characteristics | 1: swab sampling had ≥ 2 extra pathogens reported | 2: swab sampling had 1 extra pathogen reported | 3: tissue and swab sampling had the same number of pathogens reported | 4: tissue sampling had 1 extra pathogen reported | 5: tissue sampling had ≥ 2 extra pathogens reported |
|---|---|---|---|---|---|
| Total (N = 395), n (%) | 2 (0.5) | 33 (8.4) | 196 (49.6) | 108 (27.3) | 56 (14.2) |
| Type of ulcer, n (%) | |||||
| Any ischaemia (± neuropathy) (n = 194) | 0 (0.0) | 18 (9.3) | 101 (52.1) | 50 (25.8) | 25 (12.9) |
| Neuropathic only (n = 199) | 2 (1.0) | 15 (7.5) | 94 (47.2) | 57 (28.6) | 31 (15.6) |
| Missing (n = 2) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 1 (50.0) | 0 (0.0) |
| Grade of ulcer, n (%) | |||||
| Grade 1 (n = 135) | 1 (0.7) | 13 (9.6) | 69 (51.1) | 33 (24.4) | 19 (14.1) |
| Grade 2 (n = 132) | 0 (0.0) | 8 (6.1) | 68 (51.5) | 35 (26.5) | 21 (15.9) |
| Grade 3, 4 or 5 (n = 128) | 1 (0.8) | 12 (9.4) | 59 (46.1) | 40 (31.3) | 16 (12.5) |
| Pre-sampling antibiotic therapy, n (%) | |||||
| Yes (n = 186) | 0 (0.0) | 15 (8.1) | 92 (49.5) | 51 (27.4) | 28 (15.1) |
| No (n = 190) | 2 (1.1) | 13 (6.8) | 94 (49.5) | 54 (28.4) | 27 (14.2) |
| Missing (n = 19) | 0 (0.0) | 5 (26.3) | 10 (52.6) | 3 (15.8) | 1 (5.3) |
| Presence of antimicrobial dressing or agent, n (%) | |||||
| Yes (n = 238) | 0 (0.0) | 23 (9.7) | 123 (51.7) | 58 (24.4) | 34 (14.3) |
| No (n = 152) | 2 (1.3) | 9 (5.9) | 69 (45.4) | 50 (32.9) | 22 (14.5) |
| Missing (n = 5) | 0 (0.0) | 1 (20.0) | 4 (80.0) | 0 (0.0) | 0 (0.0) |
| Wound duration, n (%) | |||||
| < 56 days (n = 189) | 1 (0.5) | 13 (6.9) | 85 (45.0) | 57 (30.2) | 33 (17.5) |
| ≥ 56 days (n = 202) | 1 (0.5) | 19 (9.4) | 110 (54.5) | 49 (24.3) | 23 (11.4) |
| Missing (n = 4) | 0 (0.0) | 1 (25.0) | 1 (25.0) | 2 (50.0) | 0 (0.0) |
FIGURE 10.
Summary of the number of pathogens reported by centre. SJUH, St James’s University Hospital.

Ordinal regression analyses
Ordinal regression modelling with a random effect for centre (and MIs to allow for missing data) was used to assess whether or not agreement, based on the summary of the number of pathogens, was influenced by the pre-specified baseline covariates. The results are presented in Table 26.
| Baseline characteristics | Odds ratioa (95% CI) | AICb | df | p-valuec |
|---|---|---|---|---|
| Null model | 917.72 | |||
| Ulcer type: any ischaemia (± neuropathy) vs. neuropathic only | 0.90 (0.61 to 1.33) | 919.45 | 1 | 0.6030 |
| Ulcer grade | 920.16 | 2 | 0.4587 | |
| Grade 2 vs. grade 1 | 1.33 (0.82 to 2.15) | |||
| Grade 3, 4 or 5 vs. grade 1 | 1.27 (0.78 to 2.07) | |||
| Pre-sampling antibiotic therapy: yes vs. no | 1.25 (0.81 to 1.91) | 918.56 | 1 | 0.2828 |
| Antimicrobial dressing: yes vs. no | 0.76 (0.49 to 1.18) | 918.16 | 1 | 0.2127 |
| Wound duration (median split): ≥ 56 days vs. < 56 days | 0.64 (0.43 to 0.95) | 914.62 | 1 | 0.0240d |
| Log-wound duration (continuous) | 0.92 (0.80 to 1.05) | 918.15 | 1 | 0.2101 |
Of the baseline factors [ulcer type (any ischaemia/neuropathic only), ulcer grade (Wagner grade 1/grade 2/grade 3, 4 or 5), pre-sampling antibiotic therapy (yes/no), antimicrobial dressing or agent (yes/no), wound duration (considered dichotomously as < 56 days/≥ 56 days and continuously on the log-scale)], only wound duration (< 56 days/≥ 56 days) was found to have a statistically significant association (p-value = 0.0240). The associated odds ratio of 0.64 (95% CI 0.43 to 0.95) suggests that patients whose ulcer has been present for 56 days or more had significantly reduced odds of having a higher outcome (i.e. in the direction that the tissue sampling had two or more extra pathogens) than those whose ulcer has been present for fewer than 56 days.
Owing to the significance of wound duration, a forward selection model building approach was used to determine if further covariates had an influential effect on outcome in the model containing wound duration and centre random effect. Table 27 presents the results of the model building. There was no further significant improvement in the fit of the model on the addition of any additional baseline factors, and so the final model contained wound duration and random-centre effect and is presented in Table 28 and Figure 11.
| Additional baseline characteristic | Reduction in df | AICa | Reduction in –2log-likelihood | p-valueb |
|---|---|---|---|---|
| Wound duration (median split) | 914.62 | |||
| + Ulcer type | 1 | 916.53 | 0.086 | 0.7688 |
| + Ulcer grade | 2 | 916.69 | 1.925 | 0.3820 |
| + Pre-sampling antibiotic therapy | 1 | 915.94 | 0.674 | 0.4115 |
| + Antimicrobial dressing | 1 | 915.60 | 1.019 | 0.3128 |
| Fixed effect | Odds ratio (95% CI) | Estimates of fixed effect | Random-centre effect | ||||
|---|---|---|---|---|---|---|---|
| Parameter estimate | Standard error | p-value | Parameter estimate | Standard error | Test of H0: random intercept variance = 0 | ||
| Wound duration: ≥ 56 days vs. < 56 days | 0.64 (0.43 to 0.95) | –0.4501 | 0.2008 | 0.0250 | 0.4689 | 0.2065 | < 0.0001 |
FIGURE 11.
Plot of predicted random-centre effect in the model with fixed effect for wound duration (median split). SJUH, St James’s University Hospital.
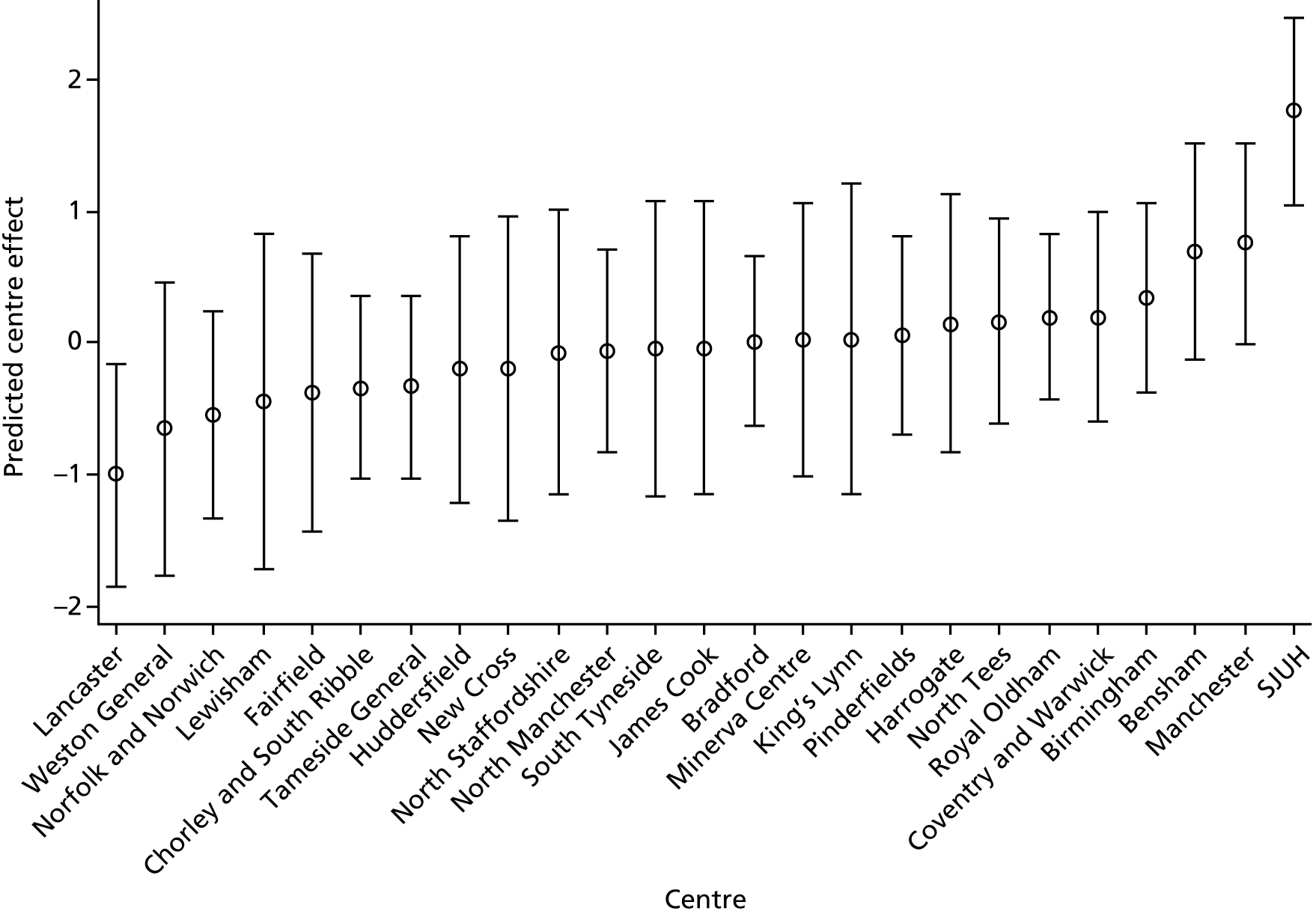
Graphical plots were used to assess the proportional odds assumption for each baseline factor and can be found in Appendix 1. The proportional odds assumption was supported for all factors with the exception of centre, which was, however, fitted as a random effect negating the need for proportional odds.
The figure presents the ranked predicted random-centre effect on the parameter estimate of wound duration of –0.4501 (see Table 28). The parameter estimate relates to the odds of a higher outcome (i.e. tissue sample finds two or more pathogens), with a negative value reducing these odds, and a positive increasing the odds. The presented predicted centre effects vary considerably, across both positive and negative values, and therefore impact on the likely odds of a higher outcome and variability around the estimate across centres.
Secondary end points
Adverse events
During the collection of swab and tissue samples, AEs consisting of bleeding of concern were reported for 30 (7.5%) patients: for 3 (0.8%) patients this was attributable to swab sampling; for 24 (6.0%) patients it was attributable to tissue sampling; and for 3 (0.8%) patients it was attributable to both swab and tissue sampling (Table 29).
| Swab sampling | Tissue sampling, n (%) | ||
|---|---|---|---|
| Yes | No | Total | |
| Yes | 3 (0.8) | 3 (0.8) | 6 (1.5) |
| No | 24 (6.0) | 370 (92.5) | 394 (98.5) |
| Total | 27 (6.8) | 373 (93.3) | 400 (100.0) |
Patient-reported pain, collected before sampling and immediately following both swab and tissue sampling, is summarised in Tables 30 and 31. At baseline, prior to sampling, 74% of patients reported no pain, 15% reported mild pain, 8% reported moderate pain and 3% reported severe pain. Comparing pain ratings after swab and tissue sampling, 5 (1.3%) patients reported an increased pain score immediately after swab sampling compared with tissue sampling, 37 (9.3%) patients reported an increased pain score immediately after tissue sampling compared with swab sampling, and 358 (89.5%) patients reported the same pain score immediately after swab and tissue sampling. Patient-rated pain is also presented according to patients’ type of ulcer: ischaemic, neuropathic or neuroischaemic ulcers (Table 32).
| Verbal rating scale pain score, n (%) | Before sampling (n = 400) | Immediately after swab sampling (n = 400) | Immediately after tissue sampling (n = 400) |
|---|---|---|---|
| No pain | 296 (74.0) | 299 (74.8) | 281 (70.3) |
| Mild pain | 60 (15.0) | 50 (12.5) | 55 (13.8) |
| Moderate pain | 32 (8.0) | 42 (10.5) | 51 (12.8) |
| Severe pain | 12 (3.0) | 9 (2.3) | 13 (3.3) |
| Pain score after swab sampling | Pain score after tissue sampling, n (%) | Total | |||
|---|---|---|---|---|---|
| No pain | Mild pain | Moderate pain | Severe pain | ||
| No pain | 279 (69.8) | 17 (4.3) | 2 (0.5) | 1 (0.3) | 299 (74.8) |
| Mild pain | 1 (0.3) | 35 (8.8) | 13 (3.3) | 1 (0.3) | 50 (12.5) |
| Moderate pain | 1 (0.3) | 2 (0.5) | 36 (9.0) | 3 (0.8) | 42 (10.5) |
| Severe pain | 0 (0.0) | 1 (0.3) | 0 (0.0) | 8 (2.0) | 9 (2.3) |
| Total | 281 (70.3) | 55 (13.8) | 51 (12.8) | 13 (3.3) | 400 (100.0) |
| Ulcer aetiologya | Verbal rating scale pain score, n (%) | ||
|---|---|---|---|
| Before sampling | Immediately after swab sampling | Immediately after tissue sampling | |
| Ischaemic (n = 14) | |||
| No pain | 5 (35.7) | 4 (28.6) | 3 (21.4) |
| Mild pain | 2 (14.3) | 2 (14.3) | 2 (14.3) |
| Moderate pain | 5 (35.7) | 5 (35.7) | 5 (35.7) |
| Severe pain | 2 (14.3) | 3 (21.4) | 4 (28.6) |
| Neuropathic (n = 202) | |||
| No pain | 154 (76.2) | 156 (77.2) | 145 (71.8) |
| Mild pain | 29 (14.4) | 24 (11.9) | 28 (13.9) |
| Moderate pain | 14 (6.9) | 18 (8.9) | 22 (10.9) |
| Severe pain | 5 (2.5) | 4 (2.0) | 7 (3.5) |
| Both ischaemic and neuropathic (n = 182) | |||
| No pain | 136 (74.7) | 138 (75.8) | 132 (72.5) |
| Mild pain | 29 (15.9) | 24 (13.2) | 25 (13.7) |
| Moderate pain | 12 (6.6) | 18 (9.9) | 23 (12.6) |
| Severe pain | 5 (2.7) | 2 (1.1) | 2 (1.1) |
No unexpected serious AEs related to the specimen collections were reported.
Sampling costs
Sampling costs were provided by only one CODIFI study site, but information was also provided from an additional non-study site by a microbiologist. At the study site, the quoted swab cost was £15.55, whereas the cost for a tissue sample was £16.53. At the non-study site, the swab cost was quoted as £3.91 and the tissue sample as £5.85. These costs do not include sampling equipment, transport or staff costs. It was not possible to obtain full economic costs within the confines of the study. Many sites considered this information as commercially sensitive.
Centre differences
Completed site difference questionnaires were received from 22 of the 25 participating sites. For full details and tables summarising the responses, see Appendix 3.
Tissue samples were collected using a scalpel at 20 of these sites and a dermal curette and one site.
There were no differences in the time taken for swab and tissue samples to reach the laboratory. The majority of laboratories reported no clear differences in the time taken from receipt of swab and tissue samples to commencement of processing, with just 4 of 17 reporting slightly more urgent/quicker time to processing for tissue samples. There were, however, clear differences in the transport media used for the two sampling techniques. Swabs were all transported with an Amies nutritional growth medium, whereas the vast majority of tissue samples were either transported in a dry container (11/17) or a dry container with saline (3/17). The remaining three tissue samples were transported using nutritional media (Amies = 2, Stuarts = 1).
Further differences were identified in the analysis and reporting of samples. Only 3 out of 19 laboratories reported performing a Gram-stained smear on both swab and tissue samples, whereas 9 out of 19 laboratories performed these on tissue samples only, the remaining 6 out of 19 never performed one, with 1 out of 19 performing them only on request.
A variety of systems were used to report amount of bacterial growth, with 8 out of 18 using combinations of scanty/light/moderate/heavy, 4 out of 18 using combinations of +/++/+++/++++, and 4 out of 18 not reporting amount of growth.
Isolates were reported to a variety of taxonomic ranks, ranging from species, genus and other. It is reported that 16 out of 18 laboratories report to the same level for swab and tissue samples, whereas 1 out of 18 reported that all tissue isolates are provided to the species level and only significant organisms are provided in such detail for the swab. However, differences are more apparent when considering whether or not all recovered isolates are reported to the clinician. Only 8 out of 18 laboratories reported that the same isolates are reported from swab and tissue samples. In contrast, the remaining 10 out of 18 laboratories report that all are reported from a tissue sample, whereas reporting of those from a swab sample depends on a mix of clinical details, clinical significance, whether or not there is heavy pure growth and whether or not they are clinically significant pathogens; those that are not are reported as enteric or skin flora. In 16 out of 19 laboratories it was reported that their standard procedures allow identification of the same isolates; however, 3 out of 16 laboratories said that their standard procedures would not allow this, and one of these reported that the tissue samples are also put into a broth.
A total of 12 out of 13 laboratories reported that the same antibacterial agents were tested in swab and tissue samples, with one laboratory reporting additional agents for the tissue sample.
Discussion
This study is a cross-sectional multicentre study to examine agreement and disagreement between swab and tissue sampling techniques in patients with a suspected DFU infection. The conclusions drawn from this study will help to determine if the extra effort and cost of sampling tissue is potentially worthwhile. Furthermore, if there is disagreement, we aimed to determine whether one method provided additional information or different information.
The key results were that a significant proportion of wounds suspected to be infected had microbiology reports that indicated no growth, and a further proportion indicated no pathogens. There was a higher proportion of swab samples than tissue samples that had no reported pathogens. There are a number of possible explanations why the culture results may report no pathogens, such as the clinical diagnosis being incorrect (e.g. inflammation was mistaken for infection). Given the lack of validated tools for diagnosing the presence of wound infection and the acknowledged risk of missing infection in diabetic foot ulceration, it is understandable that clinical diagnosis might prioritise sensitivity over specificity (accepting practice that misclassifies people as having an infected ulcer when it is not, but not missing anyone with an infected ulcer). Furthermore, it may be that sampling technique as currently practised in these centres may not have adequately captured wound flora. For example, if there was inadequate wound ulcer debridement, then swabbing or taking a tissue sample from a sloughy ulcer area will be likely to collect surface contaminants, with or without wound tissue bacteria. Last, the lack of any organisms identified from a sample may be attributable to them not having survived the transport to the microbiology laboratory. Our information from sites indicated a range of transport media, and in some centres dry tissue samples were transported, which may not adequately support fastidious organisms or anaerobes. Although the sites were practising to HPA standards, there was considerable variation in collection and microbiology practice and this heterogeneity may be important. As part of this study, we attempted to ensure that all centres practised appropriate sample collection by developing and delivering to them a training package in person or remotely, and we updated staff when turnover occurred.
In almost two-thirds of patients (58%), there was a difference in the described biome from the microbiology culture results depending on whether swab or tissue sampling was used. Furthermore, in half of the patients (50.4%), there was a difference in the number of pathogens reported. However, this was not invariably that tissue samples had a greater yield (i.e. they reported the information contained in the paired swab sample) and additional organisms (although this was the case for 36.7% of patients). In a minority of cases (8.1%), the swab sample had a greater yield than the tissue sample, that is, it reported additional information over the tissue samples. This variation in yield may be attributable to variation in the bacterial profile across a wound surface. Therefore, a swab taken from an area of a tissue subsequently removed for tissue sampling would be a closer comparison, able to account of the spread of bacteria, although the area of tissue removal would preclude this practice in many wounds, as it would potentially impact healing.
A greater proportion of tissue sample results detailed at least one antibiotic to which pathogens within the sample were resistant (41.8% vs. 31.1%) and sensitive (67.8% vs. 55.9%); however, it was not always the case that the tissue samples reported all the information contained in the paired swab sample.
The fact that a tissue sample is able to collect bacteria from deep within the wound bed and that a swab relies on the capture of bacteria from the fluid expressed by pressing the wound (as per Levine et al. ’s48 technique) means that one might expect a tissue sample to collect additional deep bacteria. We found that tissue samples provide information on more pathogens, but it is not clear from this study if the added information might have an effect on clinical decision-making. Wound microbiology results are only one aspect of the clinical assessment, with direct assessment of the wound progress during treatment likely to be an important cue for determining progress or deterioration in a wound and guiding treatment. If clinicians currently default to swabbing wounds, then it is uncertain if a move to tissue sampling is warranted.
It is clear that more patients experienced a higher degree of pain with tissue sampling than swabbing, regardless of the ulcer type. In addition, post-sampling bleeding was noted more often after tissue sampling than swabbing – bleeding of concern was attributable to tissue sampling in 24 (6.0%) patients, attributable to swab sampling in 3 (0.8%) patients and attributable to both swab and tissue sampling in 3 (0.8%) patients. The limited information indicates that there is a small difference in costs for tissue sampling and swab sampling and, hence, the question remains as to the added clinical value of tissue sampling over swabbing.
One of the strengths of our study is that we recruited a large study population with a single aetiology, all clinically suspected to be infected and intended to be treated on antibiotics. As the question we sought to address (i.e. is the selection of sampling by swab or tissue collection best) arises in the management of infected foot ulcers, we have studied this group rather than a consecutive series of ulcers or a unselected sample. This is because bacterial sampling in chronic wounds is not used to diagnose infection; if it were, then a study sample that included both infected and uninfected wounds would be essential. One should not usually collect microbiology from a clinically uninfected wound and, therefore, only patients with clinically infected wounds were recruited. The study was pragmatic in that it allowed clinicians to diagnose infection according to their current clinical practice. This means that our study is relevant to contemporary practice in a range of settings and not just specialist centres. The processing of samples using current NHS laboratory practice is also a strength, as results are thus applicable to regular clinical settings.
Given the lack of a gold standard, our study did not consider diagnostic accuracy, but rather agreement; hence, we used appropriate statistical methods to summarise and analyse our data. Most previous studies asking a similar question have presumed that a tissue sample was the criterion standard against which swab specimens are judged as providing true- and false-positive results. We believe that this is not appropriate, as it presupposes that there is a criterion standard assumed to be the tissue sample, whereas there is evidence from studies that have found swabs to report additional isolates in 11% (Mutluoglu et al. 62) and 8.1% of samples (CODIFI), and different isolates in 6.7% (Mutluoglu et al. 62) and 13.2% (CODIFI).
Our study was also significantly larger than previous investigations in the area.
Overall, the results indicate that the results of tissue and swab cultures are different in a substantial minority of cases, with tissue sampling usually providing reports with higher numbers of pathogens. This is potentially attributable to the less detailed reporting by some microbiology laboratories for the swab specimens. These factors favouring tissue samples must be weighed against the slightly more complex process of collecting tissue specimens, the slightly greater pain and bleeding, and the possibly slightly higher cost.
Chapter 3 Independent clinical review of the appropriateness of empirical antimicrobial therapy based on swab and tissue sampling information
Introduction
In the main study (see Chapter 2), the extent of agreement and pattern of disagreement on presence and number of pathogens reported was investigated, and we concluded that tissue samples more often reported additional information than swab samples. At the outset of the study, however, we were aware that if one method of sampling did provide more information as assessed by the number of pathogens identified, this did not necessarily mean that the additional microbiology information would be considered ‘more informative’ by clinicians. We therefore wished to investigate the clinician’s perspective on the microbiology reports. In order to determine whether or not a microbiology report from tissue sampling was ‘more informative’ than a swab report, we presented paired reports (a swab report and a tissue report from the same patient) to clinicians to assess the microbiology information within a patient vignette, and we analysed how the clinician would have responded to the available information. Therefore, this clinical panel review study investigated the clinical usefulness of the information provided by tissue versus swab samples, using an expert clinical panel blinded to the type of specimen to interpret the results. We assumed that a more informative culture report would allow a more appropriate decision regarding the choice of an empiric antibiotic regimen.
The best available test of the clinical utility of whether either swab or tissue information was better would be a trial in which clinicians were provided with results from one or the other sample, but this was not possible within the remit of this commissioned study.
If the method of sampling makes no difference to ongoing treatment decisions, then the sampling method that is the quickest, cheapest and best tolerated by patients would be the recommended method for clinical practice. However, if the choice of sampling method does affect subsequent treatment choice, then there is a trade-off between clinical usefulness and cost, specimen collection-related pain or bleeding and clinician time and skill in taking the sample.
Objectives
The main objective of this substudy was to compare the proportion of patients for whom the empirical antibiotic regimen (initially prescribed by the attending medical team immediately following swab and tissue sampling) was ‘appropriate,’ based on culture and sensitivity results of swab or tissue samples. This was assessed by a review of the microbiology data by a blinded clinical panel (with a record of empirical antimicrobial regimen prescribed).
A further objective of this substudy was to assess whether or not the appropriateness of the selected empirical antibiotic regimen based on the swab versus the tissue culture results was influenced by the clinical characteristics of the patient and of the infected ulcer.
A further exploratory objective was to evaluate the inter- and intrarater reliability for reviewers involved in the clinical review.
Methods
Study design
Main substudy
Appropriateness of the empirical antibiotic regimen was assessed by a blinded ‘virtual’ clinical panel, conducted on a subsample of 250 patients recruited to the main CODIFI study whose sample results were sent for review. Further subsamples of 30 patients from these 250 patients were additionally assessed by reviewers to allow us to ascertain the degree of agreement between different reviewers (inter-rater reliability) as well as consistency of reviewers from one time point to another (intrarater reliability).
Reviewers were provided with anonymised patient vignettes comprising (1) key baseline clinical information including, age, sex and baseline clinical assessment (PEDIS); (2) the microbiology laboratory results (including antibiotic sensitivities and resistance of pathogens identified); and (3) the patient’s empirical antibiotic regimen [none, or name(s) of antibiotic(s)], that is, the antibiotic regimen patients were initially prescribed immediately following swab and tissue sampling based only on clinical (not microbiological) knowledge acquired during the visit. Vignettes were provided with only a reference code to blind reviewers to the source of the vignette for both patient study number and sample type (swab or tissue).
To conduct the review, patient vignettes were randomly assigned to reviewers. Each reviewer received both the swab and tissue vignette (i.e. paired vignettes) for a number of patients; however, reviewers were not informed that they would receive paired vignettes. Paired vignettes were provided in separate rounds to avoid bias via the matching of vignettes based on patients’ baseline clinical and demographic information. Each round contained a mixture of results from swab and tissue samples, and the second round of vignettes (containing vignettes based on the same patients as in the first round but for the corresponding swab or tissue sample) were sent only when the first had been completed and returned, approximately 2 weeks later. Reviews from both the first and second round of vignettes were required to ensure that the same reviewer reviewed the vignettes generated from the same patient’s swab and tissue sample results.
This process was repeated in two batches. The first batch consisted of patient vignettes for the first 200 patients recruited to the study. Following the review of vignettes from the first batch, a second batch of vignettes were generated and sent for review to increase the number of patients included in the clinical review to 250 and to allow the assessment of inter- and intrarater reliability.
Inter- and intrarater reliability substudy
To assess the inter-rater reliability for reviewers involved in the clinical review, vignettes from 30 patients were randomly selected from batch 1 across reviewers who had returned their vignettes at that time (reviewer 1). These vignettes were then sent to multiple reviewers, selected randomly from those who had agreed to receive a second batch at that time, in batch 2 (reviewer 2). Similarly, to assess the intrarater reliability for the reviewers, vignettes from a further 30 patients were randomly selected from batch 1 (first review). These vignettes were then sent to the same reviewer in batch 2 (second review); the substudy was designed so that each reviewer considered the same vignettes twice.
Assessments and outcomes
Reviewers were asked to consider each vignette and comment as to (1) whether or not they considered there to be pathogens identified in the laboratory report that were not covered by the empirical antimicrobial regimen (where ‘regimen’ includes no prescription or name of prescribed antibiotic); and (2) if so, whether or not knowing this information would have led them to prescribe an alternative antibiotic regimen. The following questions were put to reviewers:
-
Q1: ‘Are there any pathogens identified in the laboratory report that are not covered by the prescribed antimicrobial regimen? (Y/N)’
-
Q2: ‘If you answered ‘yes’ to question 1, would knowing this information lead you to prescribe an alternative antibiotic regimen for this patient? (Y/N)’.
The clinical panel’s judgement of how a patient’s prescribed empirical antibiotic regimen would change (i.e. either initiation of antibiotic therapy or change to existing antibiotic regimen after review of the microbiology results) were derived as:
-
no change required to regimen (including initiation of therapy) (either Q1: N, or Q1: Y and Q2: N)
-
change required to regimen (Q1: Y and Q2: Y, or Q1: N and Q2: Y).
When pathogens that were not covered by the prescribed empirical antibiotic regimen were identified, sample pairs (swab and tissue samples) were further coded as:
-
swab but not tissue sample indicates pathogens that are not covered by the prescribed empirical antimicrobial regimen
-
tissue but not swab sample indicates pathogens that are not covered by the prescribed empirical antimicrobial regimen
-
swab and tissue sample in agreement on whether or not pathogens are covered by the prescribed empirical antimicrobial regimen.
A summary of the requirement for a change in antibiotic therapy (including initiation of antibiotics) was coded as:
-
swab but not tissue sample indicates a need for change
-
tissue sample but not swab indicates a need for change
-
swab and tissue sample in agreement on a need for change.
Sample size
The a priori sample size of 250 patients provided the following scenario of the power available to detect a difference in the proportion of samples in which results indicate an inappropriate empirical antibiotic regimen:
-
80% power to detect a difference of 5.5%, with a discordance of 10% (overall proportion of sample pairs whose results lead to a differing decision). For example, this assumed that the proportion of results from one sample (swab or tissue) indicating an inappropriate empirical antibiotic regimen is 2.25%, versus 7.75% in the corresponding paired sample.
Two further subsamples of 30 patients (based on feasibility) were randomly selected from the main substudy to provide data to assess the inter- and intrarater reliability of the reviewers in an exploratory manner. A sample size was selected based on the general rule of thumb that it takes at least 30 patients to estimate a parameter (the κ-statistic). 82,83 This number of samples also prevented overburdening reviewers with additional vignettes to those in the main clinical review.
Patient population
The clinical review population consisted of the sample of patients whose antibiotic regimen and microbiology results underwent review by the clinical panel. The clinical panel were all principal investigators on the CODIFI study and all had prescribing rights.
Analysis
Main substudy
Summaries were generated for each sample on whether pathogens that were or were not covered by the empirical antimicrobial regimen were identified and on whether or not a change/initiation in therapy was required. McNemar’s test was used to identify if one sample identified significantly more patients requiring a change/initiation in therapy.
Multinomial regression analysis was conducted (in which baseline factors were included as a single fixed effect) to evaluate the association between baseline factors on the requirement for a change/initiation of therapy (i.e. swab and tissue sample in agreement on change/initiation in therapy, etc.). Baseline factors to determine whether or not agreement on the requirement for a change in therapy (including initiation of antibiotic therapy) was influenced by any of the specified covariates were type of ulcer (ischaemic, neuroischaemic/neuropathic); Wagner ulcer grade (1–5); recent receipt of systemic or topical antimicrobial therapy or dressing; and wound duration. Owing to the wound duration being highly positively skewed, this factor was assessed in two ways: (1) logarithm of wound duration on the continuous scale; and (2) dichotomised at the median of 56 days (8 weeks).
A term for reviewer was included in the model as a random effect (regardless of significance) to allow for additional variability in outcome by reviewer and for estimates of the effect of baseline factors without directly requiring the estimation of individual reviewer effects. The impact of centre on agreement was also investigated in the model; however, it was not included in the model alongside reviewer owing to the large number of degrees of freedom each category required.
Multiple imputation was used to handle missing data,76 thereby allowing inclusion of the 21 (8.5%) patients with missing data for at least one candidate baseline factor. (For the pattern and extent of missing data, see Tables 81 and 82. ) The outcome and all baseline covariates (including type of ulcer, Wagner ulcer grade, recent systemic or topical antimicrobial therapy or dressing, wound duration) to be considered in the regression analyses were included in the MI models alongside centre. A total of 10 imputations were made using the MCMC method77 with multiple chains, initial values from the EM algorithm, 200 burn-in iterations and the assumption of normality for baseline factors with missing data (thus, imputations were made on a continuous scale). 71 For dichotomous factors, imputations were not restricted for ‘implausible values’ and, therefore, continuous imputations were rounded to plausible values for the dichotomous factor. 78 This method was used as the pattern of missing data was arbitrary and non-monotone.
Model fit statistics were compared between models with and without each baseline factor as a fixed effect. A chi-squared test (with degrees of freedom equal to the reduction in the degrees of freedom between each model) was used to test whether or not the reduction in the –2 log-likelihood between each model suggested a significant improvement in model fit.
For the 10 imputed data sets, the odds ratios generated through the regression analyses were combined using Rubin’s rules;79 therefore, reported estimates reflect the average of estimates across the imputed data sets and estimated standard errors include variability across the imputed sets as well as the usual uncertainty in parameter estimates. The mean change in –2 log-likelihood was used to calculate the overall p-value.
Inter- and intrarater reliability substudy
Summaries were generated to compare the results of sample reviews by different reviewers, and from one time point to another, to assess inter- and intrarater reliability.
As each of the inter-rater reliability validation samples were not reviewed by the same two reviewers, the κ-statistic is not appropriate. Instead, samples were each reviewed by two reviewers from a set of multiple reviewers and Krippendorff’s alpha reliability estimate84 is reported (with 95% CIs produced through bootstrapping with 2000 samples), which is applicable to any number of reviewers and where there are incomplete data (samples were not reviewed by all reviewers). Krippendorff’s alpha ranges from 0.00 to 1.00 and, for interpretation, Krippendorff states that ‘It is customary to require α ≥ 0.800. Where tentative conclusions are still acceptable, α ≥ 0.667 is the lowest conceivable limit’. 84,85 The κ-statistic is reported as a sensitivity analysis and to provide continuity with results from the main review and intrarater test–retest validation.
Similarly, the intrarater test–retest validation was not undertaken by a single reviewer; however, we were not interested in the differences between reviewers here, but rather we wished to obtain an estimate of within-reviewer reliability and so the κ-statistic is reported.
Sample pairs were further coded as (1) reviewers agree on requirement for a change in therapy for both swab and tissue sample; (2) reviewers disagree on requirement for a change in therapy for both swab and tissue sample; (3) reviewers disagree on requirement for a change in therapy for swab but not tissue sample; and (4) reviewers disagree on requirement for a change in therapy for tissue but not swab sample. The proportion of pairs within each group was reported.
Results
The clinical panel review involved a total of 13 reviewers and sample results from 250 patients.
Of the 250 patients whose microbiology results were included in the clinical review, 30 were also used as an inter-rater ‘validation’ sample to assess the reliability between reviewers, and another 30 were used as an intrarater ‘validation’ sample to assess reliability within reviewers (i.e. comparing their responses on the same data on two different occasions). This corresponded to a total of 310 paired patient vignettes reviewed (310 swab sample and 310 tissue samples). Three patients were excluded from the evaluable clinical review population: one from batch 1, as the patient was subsequently found to have a protocol deviation for which their swab sample was not sent to the laboratory; and two from batch 2, as a result of missing responses from reviewers. Therefore, a total of 247 patients were included in the evaluable clinical review population, corresponding to 307 paired patient vignettes across the main study and the inter- and intrarater reliability substudies. Figure 12 presents a diagram of the process, the number of patient vignettes and the number of reviewers involved in the clinical review. Patterns of missing data and samples of patient vignettes are presented in Appendix 2.
FIGURE 12.
Diagram of the clinical review process, population and reviewers.

Reviewers were each sent between 13 and 31 paired patient vignettes, provided in two batches, each consisting of two rounds (in order to separate vignettes from patients swab and tissue samples).
Table 33 presents a summary of all the reviewers involved in the clinical review and the number of patients whose vignettes (pairs of vignettes) were reviewed. This includes those forming the main clinical review sample used to address the comparison of the appropriateness of the empirical antibiotic regimen between swab and tissue samples and also the additional inter- and intrarater validation samples.
| Reviewer | Batch 1 | Batch 2 | Overall | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical reviewa (inter-rater reviewer 1) | Total | Clinical reviewa | Inter-rater reviewer 2 | Intrarater | Total | Clinical review (inter-rater reviewer 1) | Inter-rater reviewer 2 | Intrarater | Total | |
| Ab | 13 (4) | 13 | 13 (4) | 13 | ||||||
| B | 13 (2) | 13 | 9 | 3 | 5 | 17 | 22 (2) | 3 | 5 | 30 |
| Cb | 13 (5) | 13 | 13 (5) | 13 | ||||||
| Dc | 13 | 4 | 17 | 13 | 4 | 17 | ||||
| Eb | 14 | 14 | 14 | 14 | ||||||
| F | 13 | 13 | 9 | 3 | 4 | 16 | 22 | 3 | 4 | 29 |
| G | 14 (4) | 14 | 10 | 3 | 4 | 17 | 24 (4) | 3 | 4 | 31 |
| H | 13 (2) | 13 | 8d | 4 | 4 | 16 | 21 (2) | 4 | 4 | 29 |
| I | 13 (3) | 13 | 10 | 3 | 4 | 17 | 23 (3) | 3 | 4 | 30 |
| J | 13 (4) | 13 | 9 | 3 | 5 | 17 | 22 (4) | 3 | 5 | 30 |
| Kb | 13 (3) | 13 | 13 (3) | 13 | ||||||
| L | 13 (3)d | 13 | 11d | 4 | 15 | 24 (3) | 4 | 28 | ||
| M | 13 | 13 | 10 | 3 | 4 | 17 | 23 | 3 | 4 | 30 |
| Total | 158 | 158 | 89 | 30 | 30 | 149 | 247 | 30 | 30 | 307 |
Summaries of the appropriateness of empirical antibiotic regimen
Pathogens identified in the laboratory report that are not covered by the prescribed antimicrobial regimen
As presented in Table 34, reviewers concluded that for 103 (41.7%) patients, the empirical antibiotic regimen would not cover the pathogens reported from the swab sample results and that for 131 (53%) patients the empirical antibiotic regimen would not cover the pathogens reported from the tissue sample. Note that this includes 24 patients for whom no empirical antibiotic therapy had been prescribed; in these cases, clinicians had (presumably) elected to wait for results from the swab and/or tissue sample before prescribing an antibiotic regimen. The overall agreement for whether the swab or tissue sample results suggested that the empirical antibiotic regimen was sufficient to cover the range of pathogens reported within each sample and their corresponding antibiotic sensitivities was 71.7%. Discordance was observed for 49 (19.8%) patients, for whom the tissue sample results suggested that the empirical antibiotic regimen did not cover the pathogens reported, whereas the swab sample results did. Furthermore, in 21 (8.5%) patients, the swab sample results suggested that the empirical antibiotic regimen did not cover the pathogens reported, whereas the tissue sample results did.
| Swab results | Tissue results, n (%) | Total, n (%) | |
|---|---|---|---|
| Covered | Not covered | ||
| Covered | 95 (38.5) | 49 (19.8) | 144 (58.3) |
| Not covered | 21 (8.5) | 82 (33.2) | 103 (41.7) |
| Total | 116 (47.0) | 131 (53.0) | 247 (100.0) |
Table 35 summarises the presence of pathogens in the laboratory report that were or were not covered by the empirical antimicrobial regimen depending on whether or not there was a prescribed antimicrobial regimen.
| Patients on an empirical antimicrobial regimen | Patients not on an empirical antimicrobial regimen | |||||
|---|---|---|---|---|---|---|
| Tissue results, n (%) | Total, n (%) | Tissue results, n (%) | Total, n (%) | |||
| Swab results | Covered | Not covered | Covered | Not covered | ||
| Covered | 93 (41.7) | 47 (21.1) | 140 (62.8) | 2 (8.3) | 2 (8.3) | 4 (16.7) |
| Not covered | 20 (9.0) | 63 (28.3) | 83 (37.2) | 1 (4.2) | 19 (79.2) | 20 (83.3) |
| Total | 113 (50.7) | 110 (49.3) | 223 (100.0) | 3 (12.5) | 21 (87.5) | 24 (100.0) |
Change/initiation in therapy required?
Reviewers’ responses in relation to the need for a change/initiation in therapy depending on results from swab or tissue sample are cross-tabulated in Table 36. Reviewers concluded that for 132 (53.4%) patients, results from the swab or tissue sample suggested that a change/initiation in therapy was required. For 110 (44.5%) patients, a change from the empirical antibiotic regimen would be required based on the results of the tissue sample, whereas in 88 (35.6%) patients, a change from the empirical antibiotic regimen would be required based on the results of the swab sample. There was, therefore, a discordance for 66 (26.7%) patients over whether the swab or tissue sample results suggested the requirement for a change in patients’ empirically prescribed antibiotic regimen. For 22 (8.9%) patients, the tissue sample results suggested no change, whereas the swab sample results suggested that a change was required, and there were 44 (17.8%) patients for whom the swab sample results suggested no change, whereas the tissue sample results suggested that a change/initiation was required.
| Swab results | Tissue results, n (%) | Total, n (%) | |
|---|---|---|---|
| Change/initiation to therapy required | No change/initiation to therapy required | ||
| Change/initiation to therapy required | 66 (26.7) | 22 (8.9) | 88 (35.6) |
| No change/initiation to therapy required | 44 (17.8) | 115 (46.6) | 159 (64.4) |
| Total | 110 (44.5) | 137 (55.5) | 247 (100.0) |
Table 37 summarises the requirement for a change/initiation in therapy in accordance with whether or not there was a prescribed empirical antimicrobial regimen.
| Swab results | Patients on an empirical antimicrobial regimen | Patients NOT on an empirical antimicrobial regimen | ||||
|---|---|---|---|---|---|---|
| Tissue results, n (%) | Total, n (%) | Tissue results, n (%) | Total, n (%) | |||
| Change/initiation to therapy required | No change/initiation to therapy required | Change/initiation to therapy required | No change/initiation to therapy required | |||
| Change/initiation to therapy required | 50 (22.4) | 21 (9.4) | 71 (31.8) | 16 (66.7) | 1 (4.2) | 17 (70.8) |
| No change/initiation to therapy required | 39 (17.5) | 113 (50.7) | 152 (68.2) | 5 (20.8) | 2 (8.3) | 7 (29.2) |
| Total | 89 (39.9) | 134 (60.1) | 223 (100.0) | 21 (87.5) | 3 (12.5) | 24 (100.0) |
Based on the requirement for a change/initiation in therapy, Table 38 presents associated statistics and McNemar’s test for a difference between samples.
| Statistic | Overall | Patients on an empirical antimicrobial regimen | Patients not on an empirical antimicrobial regimen |
|---|---|---|---|
| Overall prevalence of required change/initiation, % | 53.4 | 49.3 | 91.7 |
| Overall agreement, % | 73.3 | 73.1 | 75.0 |
| Unadjusted kappa | |||
| Value (95% CI) | 0.45 (0.34 to 0.56) | 0.42 (0.30 to 0.54) | 0.27 (–0.13 to 0.68) |
| Asymptotic standard error | 0.06 | 0.06 | 0.21 |
| PABAK | 0.47 | 0.46 | 0.50 |
| Difference in percentage of patients requiring a change/initiation in therapy (tissue – swab) | |||
| Value (95% CI) | 8.9% (2.6% to 15.3%) | 8.1% (1.3% to 14.8%) | 16.7% (–2.2% to 35.5%) |
| McNemar’s testa | |||
| Degrees of freedom | 1 | 1 | 1 |
| Asymptotic p-value | 0.0068 | 0.0201 | 0.1025 |
| Exact p-value | 0.0092 | 0.0273 | 0.2188 |
There was 73.3% overall agreement on the requirement for a change/initiation in therapy between swab and tissue samples, with a kappa value of 0.45 (95% CI 0.34 to 0.56), which represents moderate agreement. 70 The PABAK of 0.47 similarly represents moderate agreement after adjusting the kappa for imbalances caused by differences in the prevalence [the 132 (53.4%) patients for whom swab or tissue sample results suggested a change/initiation in therapy] and bias.
There was significant evidence of a difference in the proportion of patients with a required change/initiation in therapy in the two samples (p-value = 0.0068), with the requirement for change reported in 8.9% more tissue samples than swab samples (95% CI 2.6% to 15.3%).
Overall summary of the requirement for a change/initiation in therapy
Table 39 presents a summary of the requirement for a change/initiation in therapy by specimen type for each patient, based on the outcomes for the multinomial logistic regression modelling. For almost three-quarters (73.3%) of patients, the reviewer agreed on the need for a change/initiation in therapy or not, based on the results of the swab and tissue sample. However, for 17.8% of patients, the reviewer found that the tissue sample results suggested the need for a change to patients’ empirical antibiotic regimen, whereas the swab sample results did not. Conversely, in 8.9% of patients, the reviewer found that the swab sample results suggested the need for a change to patients’ empirical antibiotic regimen, whereas the tissue sample results did not.
| Requirement for a change/initiation in therapy | Total, n (%) |
|---|---|
| 1: Swab but not tissue indicates change/initiation in therapy | 22 (8.9) |
| 2: Tissue but not swab indicates change/initiation in therapy | 44 (17.8) |
| 3: Swab and tissue agree on change/initiation in therapy | 181 (73.3) |
| Total | 247 (100) |
Table 40 presents the summary of the requirement for a change/initiation in therapy by whether the patient was on an empirical prescribed antibiotic regimen or not, type of ulcer, ulcer grade, previous antibiotic therapy (prior to initiation of empirical antibiotic regimen), presence of antimicrobial dressing or agent, wound duration and reviewer. The clinical review panel was twice as likely to conclude that a change in therapy was required based on tissue samples (vs. swabs) rather than on swab samples (vs. tissue samples).
| Patient characteristics | Swab but not tissue indicates change/initiation in therapy (N = 22) | Tissue but not swab indicates change/initiation in therapy (N = 44) | Swab and tissue agree on change/initiation in therapy (N = 181) | Total (N = 247) |
|---|---|---|---|---|
| On an empirical antibiotic regimen, n (%) | ||||
| Yes | 21 (9.4) | 39 (17.5) | 163 (73.1) | 223 (90.3) |
| No | 1 (4.2) | 5 (20.8) | 18 (75.0) | 24 (53.9) |
| Type of ulcer, n (%) | ||||
| Any ischaemia (± neuropathy) | 13 (10.5) | 23 (18.5) | 88 (71.0) | 124 (50.2) |
| Neuropathic only | 9 (7.3) | 21 (17.1) | 93 (75.6) | 123 (49.8) |
| Ulcer grade, n (%) | ||||
| Grade 1 | 4 (4.7) | 14 (16.5) | 67 (78.8) | 85 (34.4) |
| Grade 2 | 11 (12.9) | 14 (16.5) | 60 (70.6) | 85 (34.4) |
| Grade 3, 4 or 5 | 7 (9.1) | 16 (20.8) | 54 (70.1) | 77 (31.2) |
| Wound duration, n (%) | ||||
| < 56 days | 8 (7.3) | 18 (16.5) | 83 (76.1) | 109 (44.1) |
| ≥ 56 days | 14 (10.2) | 25 (18.2) | 98 (71.5) | 137 (55.5) |
| Missing | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (0.4) |
| Previous antibiotic therapy, n (%) | ||||
| Yes | 11 (9.3) | 22 (18.6) | 85 (72.0) | 118 (47.8) |
| No | 10 (8.8) | 19 (16.7) | 85 (74.6) | 114 (46.2) |
| Missing | 1 (6.7) | 3 (20.0) | 11 (73.3) | 15 (6.1) |
| Antimicrobial dressing, n (%) | ||||
| Yes | 14 (8.5) | 32 (19.5) | 118 (72.0) | 164 (66.4) |
| No | 8 (10.3) | 12 (15.4) | 58 (74.4) | 78 (31.6) |
| Missing | 0 (0.0) | 0 (0.0) | 5 (100.0) | 5 (2.0) |
Association between baseline factors on extent of agreement on the requirement for a change/initiation in therapy
Multinomial regression modelling with a random effect for reviewers (and MI to allow for missing data) was used to assess whether extent of agreement, based on the overall summary of the requirement for a change or initial in therapy, was influenced by the pre-specified baseline covariates (Table 41).
| Baseline characteristics | ‘Swab and tissue agree on requirement for change/initiation in therapy’ vs. a change/initiation in therapy indicated by | Odds ratioa (95% CI) | AICb | p-valuec |
|---|---|---|---|---|
| Null model | 376.64 | |||
| Patient on an empirical antimicrobial regimen: yes vs. no | 379.68 | 0.6203 | ||
| 1: Swab but not tissue | 2.32 (0.29 to 18.48) | |||
| 2: Tissue but not swab | 0.86 (0.30 to 2.50) | |||
| Ulcer type: any ischaemia (± neuropathy) vs. neuropathic only | 379.68 | 0.6205 | ||
| 1: Swab but not tissue | 1.53 (0.62 to 3.77) | |||
| 2: Tissue but not swab | 1.16 (0.59 to 2.25) | |||
| Ulcer grade | 380.12 | 0.3408 | ||
| Grade 2 vs. grade 1 | 1: Swab but not tissue | 3.07 (0.92 to 10.22) | ||
| 2: Tissue but not swab | 1.11 (0.48 to 2.54) | |||
| Grade 3/4/5 vs. grade 1 | 1: Swab but not tissue | 2.17 (0.60 to 7.86) | ||
| 2: Tissue but not swab | 1.43 (0.63 to 3.22) | |||
| Pre-sampling antibiotic therapy: yes vs. nod | 380.47 | 0.9218 | ||
| 1: Swab but not tissue | 1.07 (0.43 to 2.65) | |||
| 2: Tissue but not swab | 1.08 (0.54 to 2.15) | |||
| Antimicrobial dressing: yes vs. nod | 379.97 | 0.7171 | ||
| 1: Swab but not tissue | 0.85 (0.34 to 2.15) | |||
| 2: Tissue but not swab | 1.29 (0.62 to 2.71) | |||
| Wound duration: < 56 days vs. ≥ 56 daysd | 379.69 | 0.6211 | ||
| 1: Swab but not tissue | 0.67 (0.27 to 1.69) | |||
| 2: Tissue but not swab | 0.82 (0.41 to 1.64) | |||
| Log-wound duration (continuous)d | 377.93 | 0.2576 | ||
| 1: Swab but not tissue | 1.23 (0.90 to 1.66) | |||
| 2: Tissue but not swab | 1.15 (0.91 to 1.45) | |||
None of the baseline factors was found to be significant at the 10% level. Therefore, there was no evidence of an association between any of baseline factors on the extent of agreement on the requirement for a change/initiation in therapy between swab and tissue samples.
The overall test of the covariance parameter based on the change in likelihood for the model with and without the random intercept for reviewer was not statistically significant (p-value = 0.5937). However, it remains important to account for the variation in the data attributable to the reviewer, and, therefore, the random effect for reviewer remained in the null model during testing of covariate effects. Figure 13 displays the ranked predicted random reviewer effect for the null model, for outcome ‘2 – Tissue but not Swab indicates change/initiation in therapy’ compared with the reference ‘3 – Swab and Tissue agree on change/initiation in therapy’.
FIGURE 13.
Plot of predicted random reviewer effect in the null model for outcome: tissue but not swab indicates change/initiation in therapy.

The figure presents the ranked predicted random reviewer effect for the null model. Note that the magnitude of the predicted reviewer effects are comparable to the natural log of the odds presented in Table 41. For example, for the tissue but not the swab indicating a change in therapy versus the swab and tissue agreeing on the requirement to change therapy for patients on an empirical antibiotic regimen against those who are not, the odds ratio is 0.86, with a log-odds of –0.15.
Validation of reviewers’ assessments of appropriateness of empirical antibiotic regimen: inter-rater reliability
A total of 12 of the 13 reviewers were involved in the review of patients’ samples contributing to the inter-rater reliability validation at varying levels, providing at least one of the following: a review of initial patient vignettes for the main clinical review and inter-rater reliability by another reviewer only (three reviewers); a review of a copy of initial patient vignettes for inter-rater reliability only (three reviewers) or both (six reviewers) (see Table 33).
Table 42 presents a cross-tabulation of reviewers’ agreement on whether or not there were pathogens present in swab or tissue results that were not covered by the empirically prescribed antibiotic regimen. Within swab samples, reviewers agreed for 21 (70%) patients, and reviewers agreed for 24 (80%) patients based on their tissue results.
| Swab, n (%) | Tissue, n (%) | |||||
|---|---|---|---|---|---|---|
| Reviewer 2 | Reviewer 2 | |||||
| Reviewer 1 | Covered | Not covered | Total | Covered | Not covered | Total |
| Covered | 13 (43.3) | 4 (13.3) | 17 (56.7) | 11 (36.7) | 3 (10.0) | 14 (46.7) |
| Not covered | 5 (16.7) | 8 (26.7) | 13 (43.3) | 3 (10.0) | 13 (43.3) | 16 (53.3) |
| Total | 18 (60.0) | 12 (40.0) | 30 (100.0) | 14 (46.7) | 16 (53.3) | 30 (100.0) |
Cross-tabulations for reviewers’ responses in relation to the requirement for a change/initiation in antibiotic regimen based on swab and tissue sample results are presented in Table 43 and corresponding agreement statistics are presented in Table 44. Based on patients’ swab samples, reviewers agreed for 21 (70%) patients, and based on patients’ tissue sample results, reviewers agreed for 25 (83.3%) patients. Krippendorff’s alpha reliability estimate of 0.35 (95% CI 0.16 to 0.55) for reviewers’ agreement for swab samples represents fair agreement;70 however, this falls well below the conceivable limit of 0.667 proposed by Krippendorff. For reviewers’ agreement for tissue samples, the kappa value of 0.66 (95% CI 0.51 to 0.80) represents substantial agreement70 and is in line with Krippendorff’s conceivable limit.
| Swab, n (%) | Tissue, n (%) | |||||
|---|---|---|---|---|---|---|
| Reviewer 2 | Reviewer 2 | |||||
| Reviewer 1 | Change/initiation to therapy required | No change/initiation to therapy required | Total | Change/initiation to therapy required | No change/initiation to therapy required | Total |
| Change/initiation to therapy required | 6 (20.0) | 6 (20.0) | 12 (40.0) | 10 (33.3) | 3 (10.0) | 13 (43.3) |
| No change/initiation to therapy required | 3 (10.0) | 15 (50.0) | 18 (60.0) | 2 (6.7) | 15 (50.0) | 17 (56.7) |
| Total | 9 (30.0) | 21 (70.0) | 30 (100.0) | 12 (40.0) | 18 (60.0) | 30 (100.0) |
| Sensitivity analysis | |||||
|---|---|---|---|---|---|
| Overall agreement | Krippendorff’s alpha reliability estimate (95% CI) | Unadjusted kappa (95% CI) | Unadjusted kappa ASE | PABAK | |
| Swab samples (n = 30) | 70.0% | 0.35 (0.16 to 0.55) | 0.35 (0.01 to 0.69) | 0.17 | 0.40 |
| Tissue samples (n = 30) | 83.3% | 0.66 (0.51 to 0.80) | 0.66 (0.38 to 0.93) | 0.14 | 0.67 |
Table 45 further compares reviewers’ opinions on the requirement for a change/initiation in therapy by linking reviewers’ responses for swab and tissue results from the same patients. For the largest proportion of patients, 19 (63.3%) patients, reviewers agreed on the requirement for a change/initiation in therapy for both swab and tissue sample results. However, for three (10%) patients, reviewers disagreed on the requirement for a change/initiation in therapy for both swab and tissue sample results; in six (20%) patients, reviewers disagreed on the requirement for a change/initiation in therapy for swab but agreed for tissue sample results; and in two (6.7%) patients, reviewers disagreed on the requirement for a change/initiation in therapy for tissue but agreed for swab sample results.
| Reviewers’ agreement | Total, n (%) |
|---|---|
| Reviewers agree on requirement for a change/initiation in therapy for both swab and tissue sample | 19 (63.3) |
| Reviewers disagree on requirement for a change/initiation in therapy for both swab and tissue sample | 3 (10.0) |
| Reviewers disagree on requirement for a change/initiation in therapy for swab but not tissue sample | 6 (20.0) |
| Reviewers disagree on requirement for a change/initiation in therapy for tissue but not swab sample | 2 (6.7) |
| Total | 30 (100) |
Validation of reviewers’ assessment of appropriateness of empirical antibiotic regimen: intrarater reliability
A total of 7 of the 13 reviewers were involved in the review of patients’ samples that contributed to the intrarater reliability validation, each re-reviewing an additional four to five patient vignettes during the second batch of reviews.
Table 46 presents a cross-tabulation of reviewers’ agreement on whether or not there were pathogens present on the swab or tissue results that were not covered by the empirical antibiotic regimen given to the patient. For the swab samples, reviewers agreed on re-review in 26 (86.7%) patients; similarly, they agreed on re-review for the tissue samples in 26 (86.7%) patients.
| First review | Swab, n (%) | Tissue, n (%) | ||||
|---|---|---|---|---|---|---|
| Second review | Total | Second review | Total | |||
| Covered | Not covered | Covered | Not covered | |||
| Covered | 17 (56.7) | 1 (3.3) | 18 (60.0) | 14 (46.7) | 1 (3.3) | 15 (50.0) |
| Not covered | 3 (10.0) | 9 (30.0) | 12 (40.0) | 3 (10.0) | 12 (40.0) | 15 (50.0) |
| Total | 20 (66.7) | 10 (33.3) | 30 (100.0) | 17 (56.7) | 13 (43.3) | 30 (100.0) |
Cross-tabulations for reviewers’ responses in relation to the requirement for a change/initiation in antibiotic regimen based on swab and tissue sample results are presented in Table 47 and corresponding agreement statistics are presented in Table 48. Based on patients’ swab samples, reviewers agreed on re-review for 27 (90%) patients, and based on patients’ tissue sample results, reviewers agreed on re-review for 24 (80%) patients. The κ-statistic of 0.77 (95% CI 0.53 to 1.00) for reviewers’ agreement on re-review for swab samples represents substantial agreement, as does the PABAK of 0.8 after adjusting the kappa for imbalances caused by differences in prevalence and bias. 70 For reviewers’ agreement for tissue samples on re-review, the κ-statistic of 0.59 (95% CI 0.29 to 0.88) represents moderate agreement,70 as does the PABAK of 0.60.
| First review | Swab, n (%) | Tissue, n (%) | ||||
|---|---|---|---|---|---|---|
| Second review | Total | Second review | Total | |||
| Change/initiation to therapy required | No change/initiation to therapy required | Change/initiation to therapy required | No change/initiation to therapy required | |||
| Change/initiation to therapy required | 8 (26.7) | 3 (10.0) | 11 (36.7) | 9 (30.0) | 4 (13.3) | 13 (43.3) |
| No change/initiation to therapy required | 0 (0.0) | 19 (63.3) | 19 (63.3) | 2 (6.7) | 15 (50.0) | 17 (56.7) |
| Total | 8 (26.7) | 22 (73.3) | 30 (100.0) | 11 (36.7) | 19 (63.3) | 30 (100.0) |
| Label | Overall agreement, % | Unadjusted κ-statistics (95% CI) | Unadjusted κ-statistic ASE | PABAK |
|---|---|---|---|---|
| Swab samples (n = 30) | 90.0 | 0.77 (0.53 to 1.00) | 0.12 | 0.80 |
| Tissue samples (n = 30) | 80.0 | 0.59 (0.29 to 0.88) | 0.15 | 0.60 |
Table 49 further compares reviewers’ re-reviews on the requirement for a change/initiation in therapy by linking responses for swab and tissue results from the same patients. For 21 (70.0%) patients (the largest proportion), the reviewer agreed on re-review on the requirement for a change/initiation in therapy for both swab and tissue sample results. However, for three (10%) patients, the reviewer disagreed on the requirement for a change/initiation in therapy for both swab and tissue sample results on re-review; and in six (20%) patients, the reviewer disagreed on the requirement for a change/initiation in therapy for swab but agreed for tissue sample results on re-review.
| Summary of reviewers’ agreement | Total, n (%) |
|---|---|
| Reviewer agreed on requirement for a change/initiation in therapy for both swab and tissue sample | 21 (70.0) |
| Reviewer disagreed on requirement for a change/initiation in therapy for swab but not tissue sample | 3 (10.0) |
| Reviewer disagreed on requirement for a change/initiation in therapy for tissue but not swab sample | 6 (20.0) |
| Total | 30 (100) |
Discussion
This substudy set out to investigate the potential clinical impact of providing either a swab result or a tissue result to a clinician who was tasked with making a decision on antibiotic therapy for a patient with an infected DFU. The previous chapter discussed the finding that tissue samples usually identified more pathogens than swabs, but we wanted to determine if this equated to providing more clinically useful information. We therefore assessed if there was a difference in a swab sample compared with a tissue sample for providing information indicating that empiric antimicrobial therapy was adequate and if the reports from swab or tissues indicated whether or not a change in antimicrobial therapy was needed (including the need to initiate antibiotic). In order to understand the reliability of the data collected through the panel review, we also assessed the inter- and intrarater reliability for the clinical assessors making these judgements. We are not aware of any previous studies that have attempted to understand the potential clinical impact of differences in microbiology reports by using clinical vignettes in this way.
We found that one in five patients had swab results that indicated that all the pathogens were covered, but the tissue results indicated that all pathogens were not covered by the empirical antibiotic regimen. Correspondingly, clinicians reported that in nearly one in five patients (17.8%), the swab results indicated that no change in therapy was required, whereas the tissue results did suggest that a change in therapy was required. In a smaller number of cases (8.9%), the swab results indicated that a change in therapy was required when the tissue samples did not. These recommended changes to antibiotic regimen (including patients who had received no antibiotics at the outset) were based only on clinical vignette information and hence did not represent the full set of information that should be available to clinicians when reviewing the appropriateness of antibiotic therapy, such as change in ulcer state and changes in local and systemic findings of infection, for example.
Overall, we observed that 9% more tissue samples than swab samples indicated that a change in therapy was required (17.8% vs. 8.9%). Therefore, if the treating clinician had the tissue sample results rather than the swab results they would probably change the antibiotic regimen for 1 in every 11 patients. This is a potential ‘number needed to treat differently’ of 11. If the additional cost, skill and/or discomfort associated with tissue compared with swab sampling is perceived as modest, then for every 11 people in whom a tissue sample is taken (rather than a swab), a different treatment decision may be taken.
The inter-rater reliability results suggested slight to modest overall agreement between different reviewers (Krippendorff’s alpha of 0.35 and 0.66 for swab and tissue samples, respectively), indicating substantial variability between reviewers. This emphasises the importance, therefore, of ensuring that the same reviewer reviewed swab and tissue samples in the main clinical review.
There are a number of limitations to this work. As mentioned, we provided the reviewing clinicians with a limited data set which did not include all the information usually available in clinical practice to evaluate whether or not a change in antibiotic therapy was needed. We note, however, that the study also asked reviewing clinicians to make a judgement on whether or not empiric antibiotics were appropriate, and this judgement did not require additional clinical cues, such as ulcer status.
We provided blinded, paired information to clinicians and attempted to mask them to whether the result came from tissue or swab samples, to prevent bias. We do not, however, know if our blinding was successful. As tissue samples reported more pathogens, the reviewing clinicians might have guessed that longer reports (with more isolates) came from tissue samples and shorter reports from swab samples.
Given the increased attention on providing appropriate antibiotic therapy, these results may indicate a potentially higher rate of change in therapy (one would hope, but cannot guarantee, from broad-spectrum to targeted narrow-spectrum antimicrobials) following collection of a tissue result. It is not known, however, if a clinician would always initiate the change from broad- to narrow-spectrum antibiotics if an ulcer is improving, or what factors would prompt such a change (such as audit of prevalence of broad- and narrow-spectrum regimens). Identifying additional pathogens might lead to an increased used of broad-spectrum antimicrobials, thus threatening best practice antibiotic stewardship.
The current data also do not tell us anything about whether or not resampling after initiation of empiric antibiotics is needed to tailor the antibiotic regimen post receipt of the microbiology report. If a change/initiation in therapy is indicated or required, then should a patient’s ulcer be recultured when microbiology results are returned?
The fact that there were 9% of patients in whom swab results indicated a change/initiation in therapy when tissue results did not means that choosing either approach will potentially miss microbiology information that might lead to a change/initiation in therapy. Therefore, each method provides complementary information and, in certain circumstances, it might be appropriate to use both methods, for example if empiric therapy has not resulted in improvement.
It also needs to be borne in mind that tissue sampling may not be appropriate in all clinical cases or settings. In some patients, for example those with clotting disorders at high risk of bleeding, then the risk/benefit of tissue sampling over swabbing will differ from other patients.
Further work would be needed to determine whether clinical review of the swab and tissue results in these patients actually led to a real change in antibiotic regimen, rather than these ‘virtual panel’ results.
Chapter 4 A pilot comparative study of plating and culture techniques versus polymerase chain reaction
Introduction
Traditional methods of pathogen detection, such as microscopy of stained smears, plating specimens for culture and performing biochemical tests to identify the isolated organism, have major limitations. 86–89 These include the inability to culture some pathogens, a low sensitivity of pathogen detection,86–88 a lack of diagnostic specificity89 and the length of time and cost for processing. 89 Furthermore, in patients receiving antibiotic therapy, cultures may be falsely negative. Recent years have seen the advent of novel molecular (genotypic) methods, such as PCR, which have transformed the characterisation of organisms and diagnosis. 52 Traditional (phenotypic) culture methods may not identify minor, although possibly important, components of a mixed bacterial population; molecular methods provide an alternative with increased sensitivity (partly related to their ability to retrieve information on organisms that do not survive transport to the laboratory or are difficult to culture) and reduced time to identify the pathogens. 52,53,86,87,89
A study by Davies et al. 86 compared bacterial microflora of 18 (8 healing and 10 non-healing) chronic venous leg ulcers identified by using plating/culture methodologies and by PCR techniques. Although culture-based methods revealed that the majority of both wound types carried the aerobes Staphylococcus and Pseudomonas spp. (89% and 80% in healing and non-healing ulcers, respectively), PCR also identified strains not detected by plating/culture methods. The results of this86 study are consistent with a previous study87 in which the molecular approaches demonstrated significantly greater bacterial diversity of the wound microflora than that revealed by plating/culture methods. Thomsen et al. 90 also compared the plating/culture and PCR techniques in 14 chronic venous ulcers. They reported that PCR identified flora not found by traditional methods. Importantly, they found variation in the flora within the wound, such that a single sample may not represent the biome adequately. Similarly, other studies have consistently demonstrated the conflicting results of plating/culture methods and molecular techniques, the latter being more sensitive than the former. 91–93 More importantly, application of the molecular methods in guiding treatment enables a targeted approach to wound care, which has the potential to improve patient outcomes. 94–96
Ideally, wounds would be treated with antibiotics only after receiving results from microbial analysis, in order to limit overprescription and to ensure that narrow-spectrum antibiotics are used when possible. This is, however, only possible with rapid techniques, such as PCR. Furthermore, traditional culturing methods may be biased as a diagnostic tool as they select for easily cultured organisms, such as S. aureus, and select against difficult to culture bacteria, such as obligate anaerobes. 97 The main disadvantage of DNA-based PCR techniques is the inability to distinguish viable DNA sequences from inactive or dead organisms,98–100 unless supplementary methods are used. 101,102
This small substudy examined the identification of pathogens present in suspected infected DFUs using both conventional plating and culture-based techniques and PCR. This enabled us to investigate the agreement between the two analysis techniques (i.e. whether or not organisms identified by PCR reflected the bacterial load captured by plating/culture techniques). Moreover, we were able to obtain further detailed information on the agreement in identification of organisms between the two analysis techniques.
Aims and objectives
The aim of the CODIFI PCR substudy was to examine the agreement between organisms identified by traditional analytical techniques (plating/culture) compared with state-of-the-art-PCR molecular methods in suspected infected DFUs.
The objectives were to:
-
compare the pathogens reported by conventional plating and culture for both swab and tissue samples against those identified by PCR
-
compare the pathogens reported from swab and tissue samples based on PCR (alongside the culture results as reference).
Methods
For patients included in the substudy, a second swab sample was taken and the tissue sample was cut in two so that samples obtained using either sampling technique could be analysed using both PCR and conventional plating/culture.
Eligibility and consent
All patients registered into the main CODIFI study were eligible and able to consent to the substudy.
Sample collection and transportation
One swab sample and half of the tissue samples taken for molecular PCR analysis, identified by study number, patients’ date of birth and date taken, were sent by first class post at ambient temperature to Micropathology Ltd (Coventry, UK). Upon receipt, samples were stored at –70°C. Batches were defrosted before being processed. Further details on the methods for the PCR analysis were developed into a study standard operating procedure.
Polymerase chain reaction analysis
A semiquantitative PCR analysis was conducted by Micropathology Ltd. This method included a reference standard in each PCR test. The level of amplified DNA in each sample was expressed as a ratio of the reference standard. This method enabled comparison of species prevalence across a variety of samples. PCR analysis results provided by Micropathology Ltd contained details of pathogens detected from each of the swab and tissue samples for those pathogens comprising at least 5% of the total microbial load within a sample. This cut-off point was chosen to enable potentially clinical relevant pathogens to be analysed, rather than the potentially large number of pathogens with very low prevalence that would be identified by PCR and sequencing but not necessarily by routine culture. Reported pathogens were detailed at a mix of the species and genus levels, and the within-sample percentage of the pathogen relative to the total microbial load within a sample was also provided.
End points
Results were compared using conventional plating and culture versus PCR techniques for:
-
the number and presence of pathogens reported via conventional plating and molecular PCR techniques from swab samples
-
the number and presence of pathogens reported via conventional plating and molecular PCR techniques from tissue samples.
Swab versus tissue sampling:
-
the number and presence of pathogens reported via molecular PCR techniques from the swab and tissue sample.
Statistical methods
Sample size
The pilot substudy planned to collect samples from approximately 20 patients, based on feasibility, to allow an evaluation of the level of agreement and inform a powered, definitive study.
Patient population
The evaluable microbiology substudy population consisted of patients for whom evaluable swab and tissue samples were available, by both plating/culture and PCR techniques, and where PCR results were also obtainable; and written informed consent has been received, with no withdrawal of consent for the use of their samples for research purposes.
Analysis methods
Conventional plating and culture versus molecular polymerase chain reaction techniques
An overall summary of pathogens reported using plating/culture and PCR was generated independently for both the swab and tissue samples. Each pair of results (‘PCR’ and ‘cultured’) for both swab and tissue samples were coded as:
-
PCR and culture reports the same pathogens
-
PCR reports same pathogens as culture plus extra pathogens
-
culture reports same pathogens as PCR plus extra pathogens
-
both culture and PCR report different pathogens (with or without overlap in pathogens found; for example, culture reports coliforms and S. aureus and PCR reports S. aureus and E. coli).
Summaries (including cross-tabulations) on the number of pathogens reported were produced.
Swab versus tissue sampling
An overall summary of pathogens reported using PCR techniques by swab and tissue sample was produced. Each pair of results (swab and tissue samples) was coded as follows, with corresponding codes for the plating/culture results provided as a reference:
-
swab and tissue sampling report the same pathogens
-
swab reports same pathogens as tissue sampling plus extra pathogens
-
tissue sampling reports same pathogens as swab plus extra pathogens
-
both tissue and swab sampling report different pathogens (with or without overlap in pathogens found).
Summaries (including cross-tabulations) on the number of pathogens reported were produced.
Derivation
Derivation followed that of the main study, as detailed below.
Isolates from the plating and culture results that were considered not likely to represent pathogenic organisms (yeasts, skin flora, normal flora, mixed flora, skin organisms, bacterial flora, enteric flora and faecal flora) were excluded.
Given the range of taxonomic levels for reported pathogens, pathogens were primarily summarised and included at the genus level, with the exception of S. aureus, for which the interest lay at the species level. Pathogens reported at a taxonomic rank higher than the genus level were retained for the analysis and included at the level reported.
The summary and number of pathogens reported per specimen were calculated independently for both plating and culture and PCR for swab and tissue samples. Where more than one strain or species of a pathogen (in which we were interested in the genus level) was reported, a single pathogen at the level of interest was retained for comparison in the summary of pathogens and the count of the number of pathogens within the sample from the specific technique.
Results
Substudy population
A total of 14 patients from four centres were involved in the substudy; however, the evaluable microbiology substudy population consisted of 12 patients with evaluable swab and tissue samples, by both plating/culture and PCR techniques (Table 50). Two patients were excluded: one because both swab and tissue samples provided to Micropathology Ltd were insufficient and a subsequent review of the culture result for both swab and tissue sample found that no isolates had been reported in these either; and the other because the swab sample was insufficient and subsequent review of the culture results for both swab and tissue sample found only ‘mixed skin/normal flora’ reported from each sample, whereas for the evaluable tissue sample sent to Micropathology Ltd PCR reported Providencia alcalifaciens.
| Availability of PCR results by centre | Total (N = 14) |
|---|---|
| Yes, n (%) | 12 (85.7) |
| N00006: James Cook University Hospital | 1 (8.3) |
| N00036: Norfolk and Norwich University Hospital | 4 (33.3) |
| N00040: Pinderfields General Hospital | 1 (8.3) |
| N00488: Royal Oldham Hospital | 6 (50.0) |
| No, n (%) | 2 (14.3) |
| N00488: Royal Oldham Hospital | 2 (100.0) |
Furthermore, there was one patient included within the evaluable microbiology substudy population, whose second swab sample was taken after collection of the tissue sample.
Reported pathogens
Table 51 presents the groups of pathogens reported in order of overall prevalence, with pathogens reported via PCR comprising those that made up at least 5% of the total microbial load within the sample (reported in any one of swab or tissue sample by PCR or culture techniques).
| Pathogens | Swab, n (%) | Tissue, n (%) | Overall prevalence (N = 12), n (%) | ||
|---|---|---|---|---|---|
| PCR (N = 12) | Culture (N = 12) | PCR (N = 12) | Culture (N = 12) | ||
| S. aureus | 5 (41.7) | 5 (41.7) | 4 (33.3) | 5 (41.7) | 6 (50.0) |
| Enterococcus | 2 (16.7) | 0 (0.0) | 0 (0.0) | 4 (33.3) | 4 (33.3) |
| Finegoldia | 4 (33.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (33.3) |
| Prevotella | 2 (16.7) | 0 (0.0) | 3 (25.0) | 0 (0.0) | 4 (33.3) |
| Anaerobes | 0 (0.0) | 1 (8.3) | 0 (0.0) | 2 (16.7) | 3 (25.0) |
| Anaerococcus | 3 (25.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 3 (25.0) |
| Proteus | 2 (16.7) | 0 (0.0) | 2 (16.7) | 1 (8.3) | 3 (25.0) |
| Enterobacter | 2 (16.7) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 2 (16.7) |
| Enterobacteriaceae | 2 (16.7) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 2 (16.7) |
| Lactobacillus | 2 (16.7) | 0 (0.0) | 2 (16.7) | 0 (0.0) | 2 (16.7) |
| Streptococcus | 1 (8.3) | 1 (8.3) | 2 (16.7) | 2 (16.7) | 2 (16.7) |
| Acinetobacter | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Arthrobacter | 0 (0.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Candida | 0 (0.0) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 1 (8.3) |
| Coliform | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) | 1 (8.3) |
| Corynebacterium | 0 (0.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Fusobacterium | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Helcococcus | 1 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) |
| Klebsiella | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (8.3) | 1 (8.3) |
| Peptoniphilus | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Providencia | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Pseudomonas | 0 (0.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
Overall, the most prevalent pathogen was S. aureus, reported in at least one sample and for at least one technique in six (50%) patients; Enterococcus, Finegoldia and Prevotella were reported in four (33.3%) patients; anaerobes, Anaerococcus and Proteus in three (25%) patients; Enterobacter, Enterobacteriaceae, Lactobacillus and Streptococcus in two (16.7%) patients; and Acinetobacter, Arthrobacter, Candida, coliforms Corynebacterium, Fusobacterium, Helcococcus, Klebsiella, Peptoniphilus, Providencia and Pseudomonas in one (8.3%) patient.
Conventional plating/culture versus polymerase chain reaction techniques
Table 52 presents an overall summary of pathogens reported using plating/culture and PCR. Each pair of results (PCR and culture) for both swab and tissue samples was compared to assess if PCR and culture report the same pathogens, if additional pathogens were reported via one technique but not the other, or if different pathogens were reported with or without overlap. The same distribution of this response was observed for both swab and tissue samples; however, it should be noted that these do not all correspond to the same patient samples finding the same response for swab and tissue samples.
| Summary of pathogens reporteda | Swab (N = 12), n (%) | Tissue (N = 12), n (%) |
|---|---|---|
| PCR and culture report the same pathogens | 4 (33.3) | 4 (33.3) |
| PCR reports additional pathogens to the culture | 6 (50.0) | 6 (50.0) |
| Culture reports additional pathogens to PCR | 0 (0.0) | 0 (0.0) |
| Different pathogens reported (with or without overlap) | 2 (16.7) | 2 (16.7) |
For both swab and tissue samples, the PCR and culture resulted in the reporting of the same pathogens for four (33.3%) patients, whereas for six (50%) patients, PCR resulted in the reporting of additional pathogens compared with the culture results. In the remaining two (16.7%) patients, different isolates were reported via PCR and culture reports (with or without overlap), and there were no samples in which additional pathogens were reported from the culture results.
Where PCR resulted in the reporting of additional pathogens compared with the culture results, for swab samples this was attributable to the reporting of no pathogens in the culture results for five (42%) patients, and for the tissue samples there were two (17%) patients for whom the culture results reported no pathogens. These proportions are slightly higher in this subpopulation than in the full evaluable population, at 29.9% and 13.9%, respectively.
Number of pathogens
Tables 53 and 54 present cross-tabulations of the number of pathogens reported using PCR and plating/culture for swab and tissue samples, respectively, and Table 55 presents summary statistics for the number of pathogens for each type of sample and technique.
| PCR results | Culture results, n (%) | Total, n (%) | ||
|---|---|---|---|---|
| 0 | 1 | 2 | ||
| 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 1 | 0 (0.0) | 4 (33.3) | 0 (0.0) | 4 (33.3) |
| 2 | 1 (8.3) | 1 (8.3) | 0 (0.0) | 2 (16.7) |
| 3 | 1 (8.3) | 1 (8.3) | 1 (8.3) | 3 (25.0) |
| 4 | 2 (16.7) | 0 (0.0) | 0 (0.0) | 2 (16.7) |
| 5 | 1 (8.3) | 0 (0.0) | 0 (0.0) | 1 (8.3) |
| Total | 5 (41.7) | 6 (50.0) | 1 (8.3) | 12 (100.0) |
| PCR results | Culture results, n (%) | Total, n (%) | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| 0 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 1 | 1 (8.3) | 4 (33.3) | 1 (8.3) | 0 (0.0) | 6 (50.0) |
| 2 | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) | 2 (16.7) |
| 3 | 1 (8.3) | 0 (0.0) | 2 (16.7) | 0 (0.0) | 3 (25.0) |
| 4 | 0 (0.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Total | 2 (16.7) | 5 (41.7) | 4 (33.3) | 1 (8.3) | 12 (100.0) |
| Number of pathogens | Swab | Tissue | ||
|---|---|---|---|---|
| PCR | Culture | PCR | Culture | |
| (N = 12) | (N = 12) | (N = 12) | (N = 12) | |
| Mean (SD) | 2.5 (1.38) | 0.7 (0.65) | 1.9 (1.08) | 1.3 (0.89) |
| Median (range) | 2.5 (1–5) | 1.0 (0–2) | 1.5 (1–4) | 1.0 (0–3) |
| IQR | (1.0–3.5) | (0.0–1.0) | (1.0–3.0) | (1.0–2.0) |
| Number of pathogens: frequency, n (%) | ||||
| 0 | 0 (0.0) | 5 (41.7) | 0 (0.0) | 2 (16.7) |
| 1 | 4 (33.3) | 6 (50.0) | 6 (50.0) | 5 (41.7) |
| 2 | 2 (16.7) | 1 (8.3) | 2 (16.7) | 4 (33.3) |
| 3 | 3 (25.0) | 0 (0.0) | 3 (25.0) | 1 (8.3) |
| 4 | 2 (16.7) | 0 (0.0) | 1 (8.3) | 0 (0.0) |
| 5 | 1 (8.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Comparison of swab and tissue samples based on polymerase chain reaction results
An overall summary of pathogens reported using PCR by swab and tissue sample is presented in Table 56, with the corresponding culture results also presented to provide reference. Table 57 presents a cross-tabulation of the number of pathogens reported from swab and tissue samples using PCR analysis.
| Summary of pathogens | PCR (N = 12), n (%) | Culture (N = 12) , n (%) |
|---|---|---|
| Swab and tissue report the same pathogens | 3 (25.0) | 6 (50.0) |
| Swab reports additional pathogens to the tissue | 4 (33.3) | 0 (0.0) |
| Tissue reports additional pathogens to the swab | 2 (16.7) | 4 (33.3) |
| Different pathogens reported (with or without overlap) | 3 (25.0) | 2 (16.7) |
| Swab results | Tissue results, n (%) | Total, n (%) | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 1 | 3 (25.0) | 1 (8.3) | 0 (0.0) | 0 (0.0) | 4 (33.3) |
| 2 | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 2 (16.7) |
| 3 | 1 (8.3) | 1 (8.3) | 0 (0.0) | 1 (8.3) | 3 (25.0) |
| 4 | 1 (8.3) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 2 (16.7) |
| 5 | 0 (0.0) | 0 (0.0) | 1 (8.3) | 0 (0.0) | 1 (8.3) |
| Total | 6 (50.0) | 2 (16.7) | 3 (25.0) | 1 (8.3) | 12 (100.0) |
Based on the results of the PCR analysis, overall it appears that greater numbers and types of pathogens were reported within swab samples compared with tissue samples. However, it may be that swab samples resulted in more polymorphous samples with a lower total microbial load (more reported pathogens but each at a lower within-sample prevalence relative to total microbial load and with a lower total microbial load overall) compared with tissue samples.
Discussion
This study has found that PCR techniques, when used to analyse either swab or tissue samples, yield a higher number of reported pathogens than conventional plating and culture, with the number of pathogens reported by PCR dependent on the applied percentage cut-off of total microbial load within a sample, selected to be 5% in our study. Whether the higher number of pathogens is meaningful clinically is not clear, as the yield in PCR may be of persistent microbial DNA that is not associated with any current infection but that remains in the wound.
In our study, we used a single laboratory to analyse wound flora with PCR techniques. Although for this study the results were not available for clinical decision-making as they were batch processed, the results from such a laboratory can be returned within 24 hours. The arguments for greater use of PCR techniques (over culture and plating) are twofold: they make available increased amounts of information (often including identification of new bacterial species); and they allow for quicker analysis. This, however, needs to be set against the patchy availability and cost (which may change as technology advances) of such techniques. Furthermore, the impact of a higher yield from PCR on the complexity of laboratory results may affect clinical decision-making, particularly because clinicians have been accustomed to microbiology reports in which there has been selection of organisms for reporting (as identified in a small number of cases from our survey of practice) rather than reporting of the wound biome overall. The identification of a large number of species at low frequency provides clinical teams with increased data, this then requires selection of the meaningful information on likely pathogens associated with the current clinical picture, and hence ability to identify contaminants.
Although PCR techniques are often represented as a gold-standard, they too are vulnerable to contamination and (as per plating and culture) require standardised techniques and validated processes. There is a potential for false-positive results associated with the very high sensitivity. 103,104 Fenollar et al. 104 reported a comparison of plating and culture techniques in sampling for bone and joint infections (525 patients) and identified 50 discordant results: seven cases where different organisms were identified by the two analysis techniques, 21 where PCR only was positive (culture was negative), and 22 cases where the culture was positive (13 culture contaminants and nine ‘false-negative’ PCR results) and the PCR negative. They identified that in 475 out of 525 cases, the results were in agreement but their baseline event rate was much lower than in this study, as their patients did not have to be infected to enter the study. The utility of comparing PCR and plating/culture techniques in uninfected wounds is not clear given that these techniques are, currently, not used to determine the presence, or otherwise, of infection.
Chapter 5 Prognosis of infected diabetic foot ulcers
Introduction
Understanding the natural history of DFUs is an important pre-requisite to the planning of clinical trials to examine new interventions in the management of DFUs. It enables an informed approach when incorporating various design parameters, such as sample size estimates, follow-up period, frequency of visits and important outcomes.
The natural history of a condition can be reported from inception cohorts, but these are not commonly available in the DFU literature. The largest relevant prospective cohort study is the Eurodiale (European Study Group on Diabetes and the Lower Extremity) study,105 which recruited 1232 patients from 14 European countries. The study collected data on the characteristics of patients with DFUs, diagnostic and management procedures, health-care organisation, quality of life measurement and resource use. Patients with a new DFU were followed up monthly until healing of the foot, lower-leg amputation, death or non-healing after 1 year. 106 They reported that 58% of ulcers were infected at first presentation, with the prevalence of infection varying markedly between centres (range 28–74%); this may be due to referral patterns, standards of care or sensitivity of infection diagnosis. Data are reported on healing for 998 patients, of whom 77% healed within the follow-up period (12 months). This high rate of healing requires further confirmation, as the Eurodiale study reported higher healing and lower adverse outcome rates than earlier studies: only 5% of patients underwent a major amputation and 6% died. 106 This may reflect the potential reporting bias associated with a large proportion of missing data.
As well as overall prognosis, we also wanted to understand the relationship between patient and wound characteristics and outcomes. In the UK, Ince et al. 107 explored the relationships between time to healing of DFUs and baseline characteristics of the patients and their ulcers in a large prospective cohort study. They reported on 449 participants referred to a specialist clinic and found four variables independently associated with median time to healing: (1) area of the ulcer; (2) severity of peripheral arterial disease (PAD); (3) ulcer site; and (4) duration of diabetes.
In our previous systematic review,61 we identified that there were no data available specifically exploring the influence of infection status on the time to healing. Trials designed to investigate agents for treating DFUs, such as topical agents/antibiotics, may recruit populations that are homogeneous in respect of their infection status (infected ulcers or uninfected ulcers only). It would be useful to know the clinical outcomes in infected ulcers, as these are at highest risk of adverse sequelae. Characterising this patient population should allow both event rate estimation for trial planning, as well as providing data on prognosis that would be useful for the patients, their families and health-care service planning. It is essential that future trials commissioned in the area of DFUs are informed by accurate data on natural history, in the context of modern diabetes management protocols.
Some studies have reported that factors such as ulcer area, diabetes duration, severity of PAD and ulcer site provide prognostic information. However, the prognostic value of the newest tools, such as the PEDIS classification scheme, has not been assessed in this way. When Schaper et al. 108 examined the relationship between infection, neuropathy and ischaemia, they concluded that infection itself had no effect on healing in people with neuropathy, but it did have an effect on the healing of those with ischaemia (healing rate of 77% in people with ischaemia and no infection and 64% in those with ischaemia and infection). This suggests that treatment of infection in people with peripheral neuropathy may restore the potential for healing, whereas in people with lower extremity ischaemia, usual care for infection in the absence of addressing their ischaemia may be insufficient. This could be due to poor antibiotic penetration when administered systemically in those with PAD.
Schaper et al. 108 concluded that the profile of people with DFUs is changing, with a move away from the simple neuropathic foot to a more complex clinical presentation. This is in the context of an ageing population in which more people may have concurrent arterial disease. They note that more emphasis should be placed on developing strategies to improve outcome for neuroischaemic foot ulcers and, in particular, those with PAD and infection.
Aims and objectives
The aim of the CODIFI prognosis of foot infection study was to determine the medium-term outcome of patients with a suspected infected DFU at 12 months post registration and to explore various prognostic factors that may be related to time to wound healing.
The objectives were to:
-
describe the medium-term clinical outcome of patients with a suspected infected DFU, including wound healing, need for surgical treatment (amputation/revascularisation) or death during follow-up
-
conduct an exploratory analysis of the prognostic factors related to time to wound healing in patients with a suspected infected DFU.
Methods
The CODIFI prognosis of foot infection follow-up study was an extension to the original multicentre cross-sectional CODIFI study design involving 400 patients with a DFU with suspected infection requiring antibiotic therapy64 (see Chapter 2).
The prognosis study involved the addition of a case-note review to allow identification of clinical outcomes 12 months after baseline swab and tissue sampling.
The prognostic substudy was submitted as a substantial amendment (version 4.0) for ethical approval on 19 October 2012. Approval was granted on 29 November 2012. By the time of this approval, 248 participants had been recruited into CODIFI. We therefore needed to obtain both prospective consent for subsequent participants for follow-up at 12 months (via a revised protocol sent for ethical review), as well as obtain retrospective consent for those recruited under the initial protocols (again via an application to the Research Ethics Committee).
Eligibility and consent
All patients registered into the main CODIFI study were eligible to consent to the prognosis study.
The CODIFI main study patient information leaflet was updated (version 4.0) to incorporate additional information on the prognostic 12-month case-note review substudy (see Appendix 4) and to request permission to access patient notes 12 months post registration. The patient information leaflet was used not only to gain consent for new patients recruited to the main study but also to retrospectively gain the consent of patients already in the study for the 12-month case-note review.
As the rate of return of retrospective consent forms was low, with no improvement over time, a new short ‘addendum consent’ form was designed to improve return. This simplified two-page consent form asked patients to give consent for the case-note review only rather than for the full CODIFI study; however, this had little impact on the rates of consent (18 consents over a 2-month period). Following consultation with sites and the study’s public patient involvement officer, the study team contacted the National Research Ethics Service Sheffield committee to explore the possibility of obtaining verbal consent for the 12-month case-note review over the telephone, and a verbal consent ‘script’ was produced and submitted to ethics. Following its approval and implementation, the impact of this approach was significant (95 consents obtained over a 2-month period) and proved an invaluable tool to obtain retrospective consent for case-note review.
Case-note review/assessments
A detailed case-note review, conducted by research nurses and podiatrists at each centre, captured information regarding patient status at the time of review and clinical outcomes relating to the patients’ index ulcer in the 12-month period after CODIFI baseline swab and tissue sampling, referred to herewith as 12 months post sampling. This information included healing, reoccurrence, revascularisation surgery, surgical amputation and other events considered to be clinically relevant.
End points
Medium-term outcomes of patients with an infected diabetic foot ulcer
Medium-term outcomes were defined as clinically important events relating to the index ulcer within 12 months of collection of the wound samples and included wound healing, reoccurrence, lower extremity amputation, lower extremity revascularisation and death. The index ulcer was the ulcer from which swab and tissue samples were collected in the main CODIFI study. We did not collect information about new infection or the date on which infection was said to have resolved.
Prognostic factors relating to healing
Pre-specified baseline patient and wound characteristic factors were identified to explore the relationship with time to healing of the index ulcer, and to determine whether or not differential rates of healing were indicated.
Patients’ clinical characteristics from the detailed baseline assessment and microbiological data from the swabs and tissue samples were included. The full list of baseline assessment candidate factors include:
-
age (continuous and median split)
-
type of ulcer (ischaemic or neuroischaemic/neuropathic)
-
ulcer grade (Wagner grade: 1/2/≥ 3)
-
PEDIS:
-
PAD: perfusion (grade 1/≥ 2)
-
ulcer area: extent (continuous)
-
depth (grade 1/2/3)
-
infection (grade ≤ 2/3/4)
-
sensation (grade 1/2)
-
-
ulcer site (apex/interdigital/digital/plantar/dorsum/calcaneal/other)
-
incident or recurrent ulcer
-
diabetes duration (continuous and median split)
-
glycated haemoglobin (HbA1C) (continuous)
-
patient receiving insulin therapy (yes/no)
-
type of diabetes (type 1/type 2)
-
wound duration (< 56 days, ≥ 56 days)
-
prior antibiotic therapy (at the time of baseline sampling: yes/no)
-
prior antimicrobial dressing on ulcer (at the time of baseline sampling: yes/no).
The full list of baseline microbiology factors include:
-
any reported pathogens (no pathogens reported/at least one pathogen reported)
-
most prevalent pathogens, reported in either swab or tissue sample (yes/no):
-
anaerobes
-
MRSA
-
Gram-positive cocci
-
Gram-negative bacilli
-
Enterobacteriaceae
-
Gram-positive bacilli
-
S. aureus
-
Streptococcus
-
Enterococcus excluding vancomycin-resistant species
-
CNS
-
Corynebacterium
-
Pseudomonas.
-
Statistical methods
Sample size
As the prognosis study was an extension of the original CODIFI design, at the time of the funding request it was anticipated that the follow-up population would contain at least 200 patients and up to a maximum of 400 patients should all patients be willing to reconsent.
Medium-term outcomes
Considering the healing rate at 12 months, we estimated the accuracy with which we would be able to estimate the healing rate for varying numbers of patients and rates of healing, with accuracy based on ‘precision’ corresponding to the half width of the 95% CI. Assuming a healing rate of 50%, the minimum expected sample size of 200 patients would provide ± 6.9% precision and the maximum sample size of 400 patients would provide ± 4.9% precision, with improved precision as the healing rate departs from 50% (Table 58).
| Number of patients | Healing rate, % | Precision (half width of 95% CI), % |
|---|---|---|
| 200 | 40 or 60 | ±6.8 |
| 50 | ±6.9 | |
| 300 | 40 or 60 | ±5.5 |
| 50 | ±5.7 | |
| 350 | 40 or 60 | ±5.1 |
| 50 | ±5.2 | |
| 400 | 40 or 60 | ±4.8 |
| 50 | ±4.9 |
Prognostic factors relating to healing
The number of parameter estimates that could be included in the exploratory prognostic model of time to healing was also considered for varying numbers of participants and healing rates. 105 Assuming a healing rate of 50%, the minimum expected sample size of 200 patients would allow for a maximum of 10 parameter estimates in the model, and the maximum sample size of 400 patients would allow for 20 parameter estimates, with a reduction in the number of estimates if the healing rate should be < 50% (Table 59).
| Number of patients | Healing rate, % | Number of parameter estimatesa |
|---|---|---|
| 200 | 40 | 8 |
| 50 | 10 | |
| 300 | 40 | 12 |
| 50 | 15 | |
| 350 | 40 | 14 |
| 50 | 17 | |
| 400 | 40 | 16 |
| 50 | 20 |
Analysis methods
Patient populations
The follow-up population consisted of the sample of all registered and consented CODIFI patients for whom written or verbal informed consent for the 12-month case review had been obtained (or the patient had died) and the 12-month case-note review had been conducted. If either one of these criteria was not met, the patient was excluded from the follow-up population.
Medium-term outcomes
The numbers of patients with a healing, reoccurrence, amputation, revascularisation or other event for their index ulcer and time to event were summarised overall and by patients’ healing status.
A competing risk analysis using cumulative incidence functions was conducted in order to estimate the cumulative incidence of healing at 12 months, adjusted for lower extremity amputation and death. Lower extremity amputation or death were considered to be competing risks, as the occurrence of either event made it impossible to subsequently observe the event of interest, namely healing. Therefore, in patients with a lower extremity amputation or in patients who had died, healing was unobserved. Cumulative incidence functions were therefore used to adjust for these competing events, as the standard Kaplan–Meier method would lead to a biased estimate of the healing rate, as it assumes that patients with a competing event could still heal if observed for a longer period of time, which as detailed is not the case for such competing risks.
Under the competing risk analysis, patients who were alive, without a lower extremity amputation and without healing of their index ulcer at 12 months were censored at the earliest of the dates of their case-note review or 12 months after sampling. As death or amputation meant that healing of the index ulcer could no longer be observed, where an ulcer was not reported to have healed and death or amputation were reported, they were considered to be competing risks. As such, patients whose index ulcer had not healed and who had an amputation or who had died prior to the end of the 12 months’ follow-up were considered to have a competing event at the date of amputation or at their date of death, respectively. The use of cumulative incidence functions to account for the competing risks means that participants were removed from the at-risk set at the time of their competing event (i.e. those whose ulcers have the opportunity to heal).
Prognostic factors relating to healing
An exploratory analysis was conducted to model the relationship of baseline factors with the cumulative incidence of healing, using the proportional subdistribution hazards model109 for competing risks data.
Pre-specified baseline factors were individually fitted in a univariate analysis to explore the association with time to healing, and all factors were subsequently fitted in the same model in a preliminary multivariable analysis to examine the independent effects of baseline factors with time to healing. Factors found to be significant at the 10% level in the preliminary multivariable analyses, based on the presented p-value associated with the Wald test, were selected for inclusion in a final multivariable model. Healing estimates at 12 months were calculated as predicted from the univariate analysis, in which each factor was considered individually and independent of other baseline factors.
Cross-tabulations were generated for all baseline factors found to be significant at the 10% level in the univariate analysis, and the chi-squared test statistic was calculated to examine associations between factors.
In order to test the assumption of proportional hazards, competing risks were treated as censored events in order to generate log-cumulative hazards plots by the selected factor. Under the proportional hazards assumption, the lines were expected to be parallel and not to cross, for each level of the factor. The plots presented were from the first imputed data set. To further investigate departures from the proportional hazards assumption, a time-dependent covariate was included in the univariate model for two level factors.
Missing data
As part of the study design, efforts were made to collect complete follow-up data for patients in the follow-up population; however, where data remained missing this was assumed to be missing at random and MI76 was used to impute the time of healing for patients whose index ulcer was known to have healed but date of healing was unknown, and for patients for whom at least one baseline covariate was missing. This allowed for the inclusion of 43 (14.4%) patients with missing data for at least one missing baseline covariate or date of healing; for details of missing data items and imputed healing times, see Appendix 3.
The outcome (healing) and all covariates to be considered in the prognostic model were included in the MI model alongside centre and indicators for the occurrence of other outcomes (reoccurrence, revascularisation, amputation, death). MI was conducted by patients’ healed status; thus, separate imputation analyses were performed for patients in whom the index ulcer had healed and for patients in whom it had not.
A total of 10 imputations were conducted using the MCMC method77 with multiple chains, initial values from the EM79 algorithm, 200 burn-in iterations and the assumption of normality for factors with missing data (thus, imputations were made on a continuous scale). For continuous variables with missing data (healing time, extent of ulcer, diabetes duration and HbA1C), minimum imputation values of zero were specified, whereas imputations for dichotomous factors were not restricted for ‘implausible values’ and, thus, continuous imputations were rounded to plausible values for the dichotomous factor78 (owing to the small proportion of missing data, the bias introduced as a result of this method is anticipated to be minimal).
In order to present graphically the cumulative incidence of healing (in the presence of competing risks amputation and death), the mean imputed healing time for each patient with missing data was used; to present the number left at each month, the mean number left across all 10 imputed data sets was used.
Estimated healing rates at 6 and 12 months reflect the average healing rate across the imputed data sets, and estimated CIs include variability across the imputed sets as well as the usual uncertainty in parameter estimates. For the 10 imputed data sets, estimated healing rates (accounting for competing risks) were combined using Rubin’s rules79 and to present the number healed and left at 12 months, the mean across the 10 imputed data sets was used.
Results from the 10 imputed data sets for the proportional subdistribution hazards model109 were also combined using Rubin’s rules;79 therefore, reported estimates reflect the average of estimates across the imputed data sets, and estimated standard errors include variability across the imputed sets as well as the usual uncertainty in parameter estimates. For categorical factors with more than two levels (ulcer site, Wagner grade and PEDIS depth), the overall type-3 chi-squared statistics were pooled and p-values were calculated via the procedure proposed by Li et al. 110
Missing data were not imputed for patients recruited into CODIFI who were not part of the follow-up population (owing to lack of reconsent) for whom no follow-up data were obtained, owing to the strong assumptions required in order to impute multiple outcomes (healing, amputation, death) and the timing of these outcomes in relation to baseline sampling. In order to assess the generalisability of our results, that is, whether or not the follow-up population (n = 299) was representative of all patients including those not followed up (n = 101), the characteristics of these patients are presented in Appendix 3.
Results
Follow-up population
Of the 400 patients who were part of the CODIFI cross-sectional study, the long-term follow-up analysis population consisted of 299 (74.8%) patients with a completed case-note review at 12 months post sampling and baseline swab and (or) tissue sampling from their index ulcer. Baseline characteristics of patients in the follow-up population and those not included in the population can be found in Appendix 3. Baseline characteristics are similar across both groups of patients, and observed differences appear to occur by chance (owing to the number of variables compared) as differences do not consistently suggest a ‘better’ or ‘worse’ population represented by either group. Table 60 summarises the number of patients included and excluded from the follow-up population, the type of consent attained and the reasons for exclusion.
| Included in the follow-up population | Full analysis set (N = 400), n (%) |
|---|---|
| Yes | 299 (74.8) |
| Full consent (at enrolment main study) | 13 (4.3) |
| Full reconsent | 124 (41.5) |
| Addendum reconsent | 18 (6.0) |
| Verbal reconsent | 95 (31.8) |
| Patient died: reconsent not collected | 49 (16.4) |
| No | 101 (25.3) |
| Patient did not respond to request for consent by site | 55 (54.5) |
| Patient lost to follow-up | 23 (22.8) |
| Consent was not attained, reason unknown | 11 (10.9) |
| Patient had died and case-note review not completed | 7 (6.9) |
| Patient unable to consent (dementia)/lacked capacity | 2 (2.0) |
| Patient provided incomplete consent | 1 (1.0) |
| Patient refused consent | 1 (1.0) |
| Patient consented but case-note review not completed | 1 (1.0) |
| Total | 400 (100) |
As described in Methods, only 13 patients consented to the review of their case notes at 12 months post sampling as part of their initial consent into the CODIFI study. The majority of patients 124 (41.5%) consented in the subsequent CODIFI reconsent process; 18 (6%) consented by the addendum consent and a further 95 (31.8%) consented via verbal consent over the telephone. At the time of obtaining consent, 49 (16.4%) patients in the follow-up population were found to have died. We considered how to ensure that our data did not exclude those patients who died within 12 months of the study or between consent and follow-up. We concluded that these data were crucial for a complete picture of the prognosis for people with infected foot ulcers and identified that consent was not necessary for the collection of the routine data we sought if a patient was deceased. The records of these patients were therefore included in the follow-up review.
The most frequent reasons for patients not being part of the follow-up population were that the patient was known to be alive but did not respond to the enrolment site’s consent request (54.5%) or the patient was lost to follow-up (22.8%). Investigation of the time at which patients entered the study in accordance with whether or not a case-note review had been conducted found no associated pattern of missing data, such that patients were not more likely to be missing the case-note review if recruited earlier into the trial (i.e. when reconsent would have been requested after a far longer period of time since their last involvement in the study). Missing case-note reviews were also spread across centres rather than occurring as a result of the lack of reviews in single centres.
Follow-up summary
The case-note review was undertaken 12 months or more after baseline sample collection for the majority of patients; however, the review took place prior to the anniversary of sample collection for 17 (5.7%) patients reported to have been alive, resulting in a minimum follow-up length of 10.6 months (Table 61). Events reported beyond the 12-month period were removed; however, three events (two healing and one reoccurrence) reported just outside the 12-month follow-up period (within 13 months) are included in the analysis for completeness.
| Length of follow-up | Total (N = 299) |
|---|---|
| Follow-up to at least 12 months or death, n (%) | |
| Yes | 282 (94.3) |
| No | 17 (5.7) |
| Length of follow-up (months) for patients alive with follow-up < 12 months | |
| N | 17 |
| Mean (SD) | 11.5 (0.53) |
| Median (range) | 11.9 (10.58–11.96) |
Medium-term outcomes
Cross-tabulations of patients’ healing status, against the other outcomes of death, amputation, revascularisation surgery and reoccurrence, are presented in Table 62. The index ulcer was reported to have healed in 136 (45.5%) patients; however, of these, 13 (9.6%) patients’ index ulcers then reoccurred within the follow-up period. A total of 45 (15.1%) patients died within the 12-month follow-up period, 52 (17.4%) had an amputation of the same limb on which the index ulcer was found (or on the same limb) and 18 (6.0%) had revascularisation surgery.
| Index ulcer healed? | Total | ||
|---|---|---|---|
| Yes | No | ||
| Patient died, n (%) | |||
| Yes | 8 (2.7) | 37 (12.4) | 45 (15.1) |
| No | 128 (42.8) | 126 (42.1) | 254 (84.9) |
| Total | 136 (45.5) | 163 (54.5) | 299 (100.0) |
| Amputation (of or on the limb of the index ulcer), n (%) | |||
| Yes | 12 (4.0) | 40 (13.4) | 52 (17.4) |
| No | 124 (41.5) | 123 (41.1) | 247 (82.6) |
| Total | 136 (45.5) | 163 (54.5) | 299 (100.0) |
| Revascularisation surgery, n (%) | |||
| Yes | 8 (2.7) | 10 (3.3) | 18 (6.0) |
| No | 128 (42.8) | 153 (51.2) | 281 (94.0) |
| Total | 136 (45.5) | 163 (54.5) | 299 (100.0) |
| Index ulcer reoccurred, n (%) | |||
| Yes | 13 (9.6) | N/A | 13 (4.3) |
| No | 123 (90.4) | N/A | 123 (90.4) |
| Total | 136 (100.0) | N/A | 136 (100.0) |
There were 12 (4.0%) patients whose index ulcer was reported to have healed and for whom an amputation of, or on, the same limb as the index ulcer was also reported; for two of these patients, the amputation occurred after the reported healing of the index ulcer, and for 10 patients the amputation occurred prior to reported healing of the index ulcer (Table 63). Although it may appear counterintuitive for there to be an amputation and later healing of the index ulcer, the area of amputation in relation to the index ulcer suggests a different amputation site to that of the index ulcer. Amputations were predominantly performed on one or more digits, confirming that healing was in reference to the index ulcer rather than the amputation site.
| Summary | Total (N = 12), n (%) |
|---|---|
| Amputation after index ulcer healed without reoccurrence of index ulcer | 2 (16.7) |
| Amputation before index ulcer healed | 10 (83.3) |
The time to each outcome by 12 months is presented in Table 64. Of the 136 patients whose index ulcer healed (excluding 12 patients whose date of healing was missing), the median time to healing was 4.5 months (range 0.5–12.9 months). Of the 12 patients whose ulcers were reported to have reoccurred after initially healing (excluding one patient with missing reoccurrence date), the median time to reoccurrence from healing was 1.7 months (range 0.3–10.7 months). The median time to death for the 45 patients was 5.6 months (range 0.6–11.5 months) and the median time to amputation of the index ulcer/limb for 52 patients was 2 months (range 0.0–10.6 months). Finally, of the 16 patients who had revascularisation surgery (excluding 2 patients whose date of surgery was missing), the median time to surgery was 3.0 months (range 0.1–9.5 months).
| Time to outcomea | Total |
|---|---|
| Time to first healing (n = 136) | |
| Mean (SD) | 5.5 (3.47) |
| Median (range) | 4.5 (0.5–12.9) |
| Missing | 12 |
| Time from healing to reoccurrence (n = 13) | |
| Mean (SD) | 4.1 (4.01) |
| Median (range) | 1.7 (0.3–10.7) |
| Missing | 1 |
| Time to death (n = 45) | |
| Mean (SD) | 5.9 (3.23) |
| Median (range) | 5.6 (0.6–11.5) |
| Missing | 0 |
| Time to amputation (n = 52) | |
| Mean (SD) | 3.0 (3.05) |
| Median (range) | 2.0 (0.0–10.6) |
| Missing | 0 |
| Time to revascularisation surgery (n = 18) | |
| Mean (SD) | 3.4 (2.85) |
| Median (range) | 3.0 (0.1–9.5) |
| Missing | 2 |
Table 65 presents the number and types of ‘other’ events as reported by the local team (podiatrists or research nurses). A total of 45 ‘other’ events were reported in 37 (12.4%) patients with substantial variability in the type of other events reported.
| Other event occurred | Total (n = 299) |
|---|---|
| Yesa | 37 (12.4%) |
| No | 262 (87.6%) |
| Type of other event (non-mutually exclusive categories) | |
| Charcot | 2 |
| Further amputation | 2 |
| Further healing following reoccurrence | 2 |
| Further ulcer(s) | 11 |
| Further ulcer(s) and healing | 3 |
| Incision and drainage | 1 |
| Kidney and pancreas transplant | 1 |
| Necrobiosis lipoidica to legs | 1 |
| Osteomyelitis | 1 |
| Skin graft and negative pressure therapy | 1 |
| Surgical correction of deformity | 1 |
| Debridement (surgical and non-surgical) | 7 |
| Contralateral amputation | 4 |
| Contralateral angioplasty | 1 |
| Contralateral gangrene | 1 |
| Contralateral healing of ulcer | 1 |
| Contralateral osteomyelitis | 1 |
| Contralateral revascularisation | 1 |
| Contralateral ulceration | 3 |
A summary of patients’ first occurring outcome (healing, revascularisation, surgery, amputation, death) is presented in Table 66 and provides further detail of the outcomes reported subsequent to the first.
| Clinical outcome sequence | Total (N = 299), n (%) |
|---|---|
| Index ulcer healed | 122 (40.8) |
| Healed only | 110 (90.2) |
| Healed then revascularisation surgery | 1 (0.8) |
| Healed and revascularisation (order unknown missing) | 1 (0.8) |
| Healed then amputation | 2 (1.6) |
| Healed then patient died | 8 (6.6) |
| Revascularisation surgery | 10 (3.3) |
| Revascularisation surgery only | 1 (10.0) |
| Revascularisation surgery then healed | 4 (40.0) |
| Revascularisation surgery then amputation | 3 (30.0) |
| Revascularisation surgery then patient died | 1 (10.0) |
| Revascularisation surgery then amputation then healed | 1 (10.0) |
| Amputation (of or on the limb of the index ulcer) | 46 (15.4) |
| Amputation only | 26 (56.5) |
| Amputation then healed | 8 (17.4) |
| Amputation then revascularisation surgery | 4 (8.7) |
| Amputation then patient died | 6 (13.0) |
| Amputation then revascularisation surgery then healed | 1 (2.2) |
| Amputation and revascularisation surgery then patient died | 1 (2.2) |
| Patient died (without prior events) | 29 (9.7) |
| Healing, amputation or death not reported | 92 (30.8) |
Cumulative incidence of healing in the presence of competing risks: death and amputation
Table 67 presents the occurrence of amputation and death in patients whose index ulcer had not healed. Of the 163 (54.5%) patients whose index ulcer had not healed, 93 (57.1%) were known to be alive and without amputation at 12 months and were censored at the earliest of their case-note review or at 12 months post sampling; 33 (20.2%) were known to be alive with amputation before 12 months and seven (4.3%) were known to have died with amputation before 12 months. These patients therefore had a competing event at their date of amputation and 30 (18.4%) patients died without amputation before 12 months and therefore had a competing event at their date of death.
| Healing of the index ulcer | Total (N = 299) |
|---|---|
| Number of patients with index ulcer healed, n (%) | |
| Yes | 136 (45.5) |
| No | 163 (54.5) |
| Index ulcer not healed, n (%) | |
| Patient alive with amputation before 12 months | 33 (20.2) |
| Patient alive without amputation at 12 months | 93 (57.1) |
| Patient died with amputation before 12 months | 7 (4.3) |
| Patient died without amputation before 12 months | 30 (18.4) |
Figure 14 shows the estimated cumulative incidence curves of the time to healing or the competing risks of death or amputation. Corresponding healing estimates at 6 and 12 months are presented in Table 68. The estimated 6 months’ post sampling healing rate was 27.5% (95% CI 22.4% to 32.5%) and the 12 months’ post sampling healing rate was 44.5% (95% CI 38.9% to 50.1%). The median time to healing for all patients in the follow-up population was not reached and is not estimated.
FIGURE 14.
Cumulative incidence functions of healing in the presence of competing risks: death and amputation.

| Month | Healing estimate (95% CI) | Number healed | Number with competing amputation event | Number with competing death event | Number censored | Number left |
|---|---|---|---|---|---|---|
| 6 | 27.5% (22.4% to 32.5%) | 82 | 32 | 17 | 0 | 168 |
| 12 | 44.5% (38.9% to 50.1%) | 133 | 40 | 30 | 93 | 3a |
A total of 43 (14.4%) patients had at least one missing baseline covariate or date of healing (details of missing data items and imputed healing times are provided in Appendix 3).
Prognostic factors relating to healing: exploratory analysis
Tables 69 and 70 present summary tables of patients’ healed status by baseline factors included in the prognostic modelling.
| Baseline characteristics | Healed, N = 136 (45.5%) | Not healed, N = 163 (54.5%) |
|---|---|---|
| Age (median split), n (%) | ||
| ≤ 63 years (n = 146) | 58 (39.7) | 88 (60.3) |
| > 63 years (n = 153) | 78 (51.0) | 75 (49.0) |
| Ulcer type, n (%) | ||
| Any ischaemia (± neuropathy) (n = 142) | 49 (34.5) | 93 (65.5) |
| Neuropathic only (n = 155) | 85 (54.8) | 70 (45.2) |
| Missing (n = 2) | 2 (100.0) | 0 (0.0) |
| Wagner ulcer grade, n (%) | ||
| Grade 1 (n = 104) | 56 (53.8) | 48 (46.2) |
| Grade 2 (n = 93) | 41 (44.1) | 52 (55.9) |
| Grade 3, 4 or 5 (n = 102) | 39 (38.2) | 63 (61.8) |
| PEDIS (perfusion: PAD), n (%) | ||
| Grade 1 (n = 147) | 85 (57.8) | 62 (42.2) |
| Grade ≥ 2 (n = 152) | 51 (33.6) | 101 (66.4) |
| PEDIS (depth), n (%) | ||
| Grade 1 (n = 96) | 51 (53.1) | 45 (46.9) |
| Grade 2 (n = 100) | 47 (47.0) | 53 (53.0) |
| Grade 3 (n = 103) | 38 (36.9) | 65 (63.1) |
| PEDIS (infection), n (%) | ||
| Grade 2 (n = 104) | 55 (52.9) | 49 (47.1) |
| Grade 3 (n = 185) | 78 (42.2) | 107 (57.8) |
| Grade 4 (n = 10) | 3 (30.0) | 7 (70.0) |
| PEDIS (sensation), n (%) | ||
| Grade 1 (n = 20) | 12 (60.0) | 8 (40.0) |
| Grade 2 (n = 279) | 124 (44.4) | 155 (55.6) |
| Ulcer site, n (%) | ||
| Apex, interdigital, digital (n = 119) | 56 (47.1) | 63 (52.9) |
| Plantar (n = 133) | 62 (46.6) | 71 (53.4) |
| Dorsum (n = 38) | 15 (39.5) | 23 (60.5) |
| Other/missing (n = 9) | 3 (33.3) | 6 (66.7) |
| Incident or recurrent, n (%) | ||
| Incident (n = 206) | 88 (42.7) | 118 (57.3) |
| Recurrent (n = 91) | 46 (50.5) | 45 (49.5) |
| Missing (n = 2) | 2 (100.0) | 0 (0.0) |
| Diabetes duration (median split), n (%) | ||
| < 15 years (n = 136) | 65 (47.8) | 71 (52.2) |
| ≥ 15 years (n = 161) | 71 (44.1) | 90 (55.9) |
| Missing (n = 2) | 0 (0.0) | 2 (100.0) |
| Patient receiving insulin therapy, n (%) | ||
| Yes (n = 211) | 89 (42.2) | 122 (57.8) |
| No (n = 88) | 47 (53.4) | 41 (46.6) |
| Type of diabetes, n (%) | ||
| Type 1 (n = 40) | 17 (42.5) | 23 (57.5) |
| Type 2 (n = 259) | 119 (45.9) | 140 (54.1) |
| Wound duration (median split), n (%) | ||
| < 56 days (n = 143) | 77 (53.8) | 66 (46.2) |
| ≥ 56 days (n = 152) | 56 (36.8) | 96 (63.2) |
| Missing (n = 4) | 3 (75.0) | 1 (25.0) |
| Prior antibiotic use (at the time of baseline sampling), n (%) | ||
| Yes (n = 139) | 60 (43.2) | 79 (56.8) |
| No (n = 145) | 71 (49.0) | 74 (51.0) |
| Missing (n = 15) | 5 (33.3) | 10 (66.7) |
| Prior antimicrobial dressing on ulcer (at the time of baseline sampling), n (%) | ||
| Yes (n = 175) | 69 (39.4) | 106 (60.6) |
| No (n = 119) | 64 (53.8) | 55 (46.2) |
| Missing (n = 5) | 3 (60.0) | 2 (40.0) |
| Only ulcer on index foot, n (%) | ||
| Single index ulcer on index foot (n = 222) | 113 (50.9) | 109 (49.1) |
| > 1 ulcer on index foot (n = 77) | 23 (29.9) | 54 (70.1) |
| Baseline microbiologya | Healed, N = 136 (45.5%) | Not healed, N = 163 (54.5%) |
|---|---|---|
| Any reported pathogens, n (%) | ||
| Yes (n = 263) | 118 (44.9) | 145 (55.1) |
| No (n = 36) | 18 (50.0) | 18 (50.0) |
| Overall anaerobes, n (%) | ||
| Yes (n = 69) | 28 (40.6) | 41 (59.4) |
| No (n = 230) | 108 (47.0) | 122 (53.0) |
| MRSA, n (%) | ||
| Yes (n = 27) | 8 (29.6) | 19 (70.4) |
| No (n = 272) | 128 (47.1) | 144 (52.9) |
| Gram-positive cocci, n (%) | ||
| Yes (n = 217) | 95 (43.8) | 122 (56.2) |
| No (n = 82) | 41 (50.0) | 41 (50.0) |
| Gram-negative bacilli, n (%) | ||
| Yes (n = 110) | 45 (40.9) | 65 (59.1) |
| No (n = 189) | 91 (48.1) | 98 (51.9) |
| Enterobacteriaceae | ||
| Yes (n = 79) | 31 (39.2) | 48 (60.8) |
| No (n = 220) | 105 (47.7) | 115 (52.3) |
| Gram-positive bacilli, n (%) | ||
| Yes (n = 31) | 15 (48.4) | 16 (51.6) |
| No (n = 268) | 121 (45.1) | 147 (54.9) |
| S. aureus, n (%) | ||
| Yes (n = 107) | 51 (47.7) | 56 (52.3) |
| No (n = 192) | 85 (44.3) | 107 (55.7) |
| Streptococcus, n (%) | ||
| Yes (n = 44) | 21 (47.7) | 23 (52.3) |
| No (n = 255) | 115 (45.1) | 140 (54.9) |
| Enterococcus (excluding vancomycin resistant), n (%) | ||
| Yes (n = 48) | 18 (37.5) | 30 (62.5) |
| No (n = 251) | 118 (47.0) | 133 (53.0) |
| CNS, n (%) | ||
| Yes (n = 38) | 24 (63.2) | 14 (36.8) |
| No (n = 261) | 112 (42.9) | 149 (57.1) |
| Corynebacterium, n (%) | ||
| Yes (n = 26) | 13 (50.0) | 13 (50.0) |
| No (n = 273) | 123 (45.1) | 150 (54.9) |
| Pseudomonas, n (%) | ||
| Yes (n = 24) | 10 (41.7) | 14 (58.3) |
| No (n = 275) | 126 (45.8) | 149 (54.2) |
Tables 71 and 72 summarise the results of the exploratory univariate and adjusted multivariable analysis investigating the relationship of baseline factors with the cumulative incidence of healing. A proportional subdistribution hazards model109 for competing risks data was used to adjust for each factor individually in the univariate analysis and adjusted for all factors simultaneously in the multivariable analysis (as detailed in the methods section).
| Baseline factors | Healing estimate at 12 months (95% CI) |
|---|---|
| Ulcer type | |
| Any ischaemia (± neuropathy) | 33.2% (25.4% to 41.0%) |
| Neuropathic only | 55.3% (47.6% to 63.0%) |
| Wagner ulcer grade | |
| Grade 1 | 56.1% (46.3% to 65.8%) |
| Grade 2 | 41.6% (32.5% to 50.8%) |
| Grade 3, 4 or 5 | 36.2% (27.3% to 45.1%) |
| PEDIS: perfusion | |
| Grade 1 | 58.0% (50.1% to 65.8%) |
| Grade ≥ 2 | 31.8% (24.9% to 38.8%) |
| PEDIS: depth | |
| Grade 1 | 55.8% (45.6% to 66.0%) |
| Grade 2 | 43.6% (35.0% to 52.3%) |
| Grade 3 | 35.5% (27.0% to 44.0%) |
| PEDIS: infection | |
| Grade 2 | 54.2% (45.2% to 63.2%) |
| Grade 3 and 4 | 39.6% (33.2% to 46.0%) |
| Ulcer on index foot only | |
| Single index ulcer on index foot | 49.7% (43.4% to 55.9%) |
| > 1 ulcer on index foot | 29.6% (19.5% to 39.7%) |
| Wound duration (median split) | |
| ≥ 56 days | 35.3% (28.4% to 42.3%) |
| < 56 days | 55.1% (46.5% to 63.7%) |
| Antimicrobial dressing | |
| Yes | 38.8% (32.0% to 45.6%) |
| No | 53.1% (44.9% to 61.4%) |
| MRSA | |
| Yes | 26.8% (11.5% to 42.2%) |
| No | 46.3% (40.6% to 52.1%) |
| CNS | |
| Yes | 60.4% (46.7% to 74.1%) |
| No | 42.1% (36.3% to 48.0%) |
| Baseline factors | df | Univariate | Preliminary multivariable | Final multivariable | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | ||
| Age (continuous) | 1 | 1.00 (0.99 to 1.02) | 0.5887 | 1.02 (1.00 to 1.04) | 0.0169* | 1.02 (1.01 to 1.04) | 0.0081* |
| Age: > 63 vs. ≤ 63 years | 1 | 1.39 (0.99 to 1.95) | 0.0543* | ||||
| Diabetes duration years (continuous) | 1 | 0.99 (0.98 to 1.01) | 0.5268 | 1.00 (0.98 to 1.02) | 0.7518 | ||
| Diabetes duration: ≥ 15 vs. < 15 years | 1 | 0.85 (0.61 to 1.20) | 0.3615 | ||||
| Diabetes type: type 2 vs. type 1 | 1 | 1.11 (0.67 to 1.84) | 0.6915 | 0.78 (0.41 to 1.49) | 0.4524 | ||
| Patient receiving insulin therapy: yes vs. no | 1 | 0.80 (0.57 to 1.13) | 0.2038 | 0.84 (0.54 to 1.32) | 0.4578 | ||
| HbA1C (continuous) | 1 | 0.96 (0.89 to 1.03) | 0.2518 | 0.99 (0.89 to 1.09) | 0.8237 | ||
| Ulcer type: any ischaemia vs. neuropathic only | 1 | 0.50 (0.35 to 0.71) | < 0.0001* | 1.09 (0.59 to 2.02) | 0.7837 | ||
| Ulcer grade | 2 | 0.0113* | 0.3343 | ||||
| Grade 2 vs. grade 1 | 0.65 (0.44 to 0.98) | 0.0397 | 0.56 (0.25 to 1.23) | 0.1477 | |||
| Grade ≥ 3 vs. grade 1 | 0.55 (0.36 to 0.82) | 0.0038 | 0.59 (0.25 to 1.37) | 0.2159 | |||
| PEDIS perfusion: grade ≥ 2 vs. grade 1 | 1 | 0.44 (0.31 to 0.62) | < 0.0001* | 0.43 (0.22 to 0.83) | 0.0113* | 0.37 (0.25 to 0.55) | < 0.0001* |
| Ulcer extent (continuous) | 1 | 0.99 (0.97 to 1.01) | 0.2766 | 1.00 (0.99 to 1.01) | 0.8500 | ||
| PEDIS: depth | 2 | 0.0172* | 0.3298 | ||||
| Grade 2 vs. grade 1 | 0.70 (0.47 to 1.05) | 0.0843 | 1.61 (0.74 to 3.49) | 0.2311 | |||
| Grade 3 vs. grade 1 | 0.54 (0.35 to 0.83) | 0.0046 | 1.16 (0.47 to 2.91) | 0.7451 | |||
| PEDIS: infection – grade 3 and 4 vs. grade 2 | 1 | 0.65 (0.46 to 0.91) | 0.0135* | 0.91 (0.59 to 1.41) | 0.6831 | ||
| PEDIS: sensation – grade 2 vs. grade 1 | 1 | 0.70 (0.40 to 1.23) | 0.2139 | 0.60 (0.33 to 1.11) | 0.1036 | ||
| Incident or recurrent ulcer: recurrent vs. incident | 1 | 1.11 (0.79 to 1.56) | 0.5603 | 1.27 (0.85 to 1.91) | 0.2487 | ||
| Only ulcer on index foot: yes vs. no | 1 | 1.96 (1.25 to 3.07) | 0.0034* | 1.91 (1.15 to 3.17) | 0.0122* | 1.90 (1.18 to 3.06) | 0.0081* |
| Ulcer location | 3 | 0.6810 | 0.9856 | ||||
| Dorsum vs. apex/interdigital/digital | 0.72 (0.41 to 1.27) | 0.2572 | 1.01 (0.52 to 1.94) | 0.9828 | |||
| Other/missing vs. apex/interdigital/digital | 0.68 (0.20 to 2.33) | 0.5371 | 1.08 (0.25 to 4.74) | 0.9141 | |||
| Plantar vs. apex/interdigital/digital | 0.92 (0.64 to 1.32) | 0.6451 | 1.09 (0.70 to 1.70) | 0.7007 | |||
| Wound duration: ≥ 56 vs. < 56 days | 1 | 0.54 (0.39 to 0.76) | 0.0004* | 0.46 (0.30 to 0.70) | 0.0003* | 0.55 (0.39 to 0.77) | 0.0005* |
| Previous antibiotic therapy: yes vs. no | 1 | 0.86 (0.61 to 1.21) | 0.3791 | 0.91 (0.61 to 1.36) | 0.6475 | ||
| Antimicrobial dressing: yes vs. no | 0.65 (0.46 to 0.91) | 0.0123* | 0.77 (0.52 to 1.14) | 0.1934 | |||
| Any reported pathogens: yes vs. no | 1 | 0.82 (0.50 to 1.36) | 0.4487 | 0.79 (0.36 to 1.70) | 0.5414 | ||
| Overall anaerobes: yes vs. no | 1 | 0.83 (0.55 to 1.26) | 0.3884 | 0.78 (0.48 to 1.27) | 0.3146 | ||
| MRSA: yes vs. no | 1 | 0.50 (0.26 to 0.97) | 0.0419* | 0.67 (0.28 to 1.63) | 0.3802 | ||
| Gram-positive cocci: yes vs. no | 1 | 0.83 (0.58 to 1.19) | 0.3039 | 0.82 (0.39 to 1.69) | 0.5841 | ||
| Gram-negative bacilli: yes vs. no | 1 | 0.77 (0.54 to 1.10) | 0.1474 | 1.34 (0.56 to 3.17) | 0.5071 | ||
| Enterobacteriaceae including coliforms: yes vs. no | 1 | 0.75 (0.51 to 1.11) | 0.1449 | 0.63 (0.27 to 1.49) | 0.2952 | ||
| Gram-positive bacilli: yes vs. no | 1 | 1.08 (0.64 to 1.81) | 0.7768 | 1.06 (0.26 to 4.38) | 0.9364 | ||
| MSSA: yes vs. no | 1 | 1.20 (0.84 to 1.70) | 0.3170 | 1.27 (0.79 to 2.05) | 0.3229 | ||
| Streptococcus: yes vs. no | 1 | 1.08 (0.68 to 1.72) | 0.7383 | 1.10 (0.65 to 1.83) | 0.7292 | ||
| Enterococcus excluding vancomycin resistant: yes vs. no | 1 | 0.72 (0.44 to 1.15) | 0.1702 | 0.86 (0.50 to 1.47) | 0.5713 | ||
| CNS: yes vs. no | 1 | 1.69 (1.11 to 2.59) | 0.0147* | 1.98 (1.08 to 3.61) | 0.0270* | 1.53 (0.98 to 2.40) | 0.0603* |
| Corynebacterium: yes vs. no | 1 | 1.17 (0.67 to 2.07) | 0.5794 | 0.89 (0.18 to 4.35) | 0.8894 | ||
| Pseudomonas: yes vs. no | 1 | 0.82 (0.45 to 1.48) | 0.5055 | 0.66 (0.27 to 1.66) | 0.3818 | ||
Univariate analysis of incidence of healing
Univariate analysis (see Table 72) showed a significant association (p-value < 0.1) between the incidence of healing and perfusion (p-value < 0.0001), ulcer type (p-value = < 0.0001), wound duration (p-value = 0.0004), presence of other ulcers (p-value = 0.0034), Wagner ulcer grade (p-value = 0.0113), presence of an antimicrobial dressing (p = 0.0123), infection (p-value = 0.0135), ulcer depth (p-value = 0.0172), CNS (p-value = 0.0147) and MRSA (p-value = 0.0419).
Significant associations were such that the incidence of healing was lower for patients with:
-
ischaemic ulcers compared with neuropathic ulcers, with a hazard ratio (HR) of 0.5 (95% CI 0.35 to 0.71)
-
a Wagner ulcer grade of 2 (ulcer extension without abscess or osteomyelitis) or ≥ 3 (at least a deep ulcer with abscess, osteomyelitis or sepsis) compared with a grade 1 ulcer (superficial), with HRs of 0.65 (95% CI 0.44 to 0.98) and 0.55 (95% CI 0.36 to 0.82), respectively
-
a perfusion grade ≥ 2 (indicating PAD), with a HR of 0.44 (95% CI 0.31 to 0.62)
-
a depth/tissue loss grade 2 (penetrating to skin structures) or grade 3 ulcer (penetrating all layers of foot) compared with a grade 1 ulcer (superficial), with HRs of 0.70 (95% CI 0.47 to 1.05) and 0.54 (95% CI 0.35 to 0.83), respectively
-
a ≥ grade 3 infection (extensive erythema or systemic inflammatory response) compared with a grade 2 infection (infection without involvement of deeper tissues or systemic signs), with a HR of 0.65 (95% CI 0.46 to 0.91)
-
older ulcers (≥ 56 days), with a HR of 0.54 (95% CI 0.39 to 0.76)
-
an antimicrobial dressing on their ulcer at baseline, with a HR of 0.65 (95% CI 0.46 to 0.91)
-
MRSA reported from their index ulcer at baseline, with a HR of 0.50 (95% CI 0.26 to 0.97).
The incidence of healing was higher for patients with:
-
a single ulcer on their index foot, with a HR of 1.96 (95% CI 1.25 to 3.07)
-
CNS reported from their index ulcer at baseline, with a HR of 1.69 (95% CI 1.11 to 2.59).
Age, categorised at the median, was also found to be associated with an increased incidence of healing for patients > 63 years of age; however, this result is somewhat counterintuitive and is not supported by the analysis performed using a continuous age parameter. This association was not supported by an association with healing for other categorisations of age, suggesting a spurious result for the significant association for age.
Table 71 presents 12-month healing estimates for factors found to have a significant relation to the incidence of healing in the univariate analysis (excluding age). Furthermore, plots of the cumulative incidence of healing, by each of these factors can be found in Appendix 3.
Preliminary multivariable analysis
Preliminary multivariable analysis included all potentially prognostic factors in a single proportional subdistribution hazards model, with the following showing a significant association with the incidence of healing: wound duration (p-value = 0.0003), perfusion (p-value = 0.0113), presence of other ulcers (p-value = 0.0122), age (continuous, p-value = 0.0169) and CNS (p-value = 0.0270) (see Table 72).
The slightly increased healing incidence for age should, however, be interpreted with some caution owing to both the proximity of the HR to 1 (with the lower limit of the CI including 1, which represents no difference) and the lack of consistency between the univariate and multivariable model in which there was no evidence of a difference in healing when age was considered independently of other factors on the continuous scale. Age was further investigated in terms of associations with other factors in order to identify if this result was attributable to an unusual case mix of patients, for example, whether or not older patients had less severe ulcers. The Cochran–Armitage test for trend between age, split according to quartiles, found significant trends (at the 10% level) with ulcer type, death, dressing, wound duration, PEDIS infection and perfusion; however, in each case the trend was such that older patients had the more severe ulcer in terms of each prognostic factor.
The assumption of proportional hazards was valid for the majority of factors (see Appendix 3). However, there was some evidence against the proportional hazards assumption for CNS, with an increased incidence of healing in patients without CNS prior to 1 month post sampling, but an increased and increasing incidence of healing in patients with CNS after 1 month (i.e. non-constant HR). The assumption is satisfied for the reference grade 1 of both the depth/tissue loss classification and Wagner ulcer grade; however, there is some suggestion of non-proportionality for grades 2 and 3+ (for the Wagner scale) owing to the reduced differences observed in the cumulative incidence for these levels, which may suggest that grouping grades > 1, for both the depth/tissue loss classification and Wagner ulcer grade, may not result in a substantial loss of information on the likely incidence of healing.
Final multivariable analysis
The final multivariable analysis included only those factors found to be significant in the preliminary multivariable model: perfusion (p-value = < .0001), wound duration (p-value = 0.0005), presence of other ulcers (p-value = 0.0081), age (continuous, p-value = 0.0081) and CNS (p-value = 0.0603) (see Table 72).
Similar to estimates for the previous models, these factors were such that the incidence of healing was lower for patients with:
-
perfusion grade ≥ 2 relative to patients with grade 1, with a HR of 0.37 (95% CI 0.25 to 0.55)
-
older ulcers (≥ 56 days), with a HR of 0.55 (95% CI 0.39 to 0.77).
The incidence of healing was higher for:
-
patients with a single ulcer on their index foot relative to patients with more than one ulcer, with a HR of 1.90 (95% CI 1.18 to 3.06)
-
older patients, increasing with each year of age, with a HR of 1.02 (95% CI 1.01 to 1.04); however, this is suspected to be a spurious relationship
-
patients with CNS reported from their ulcer at baseline, with a HR of 1.53 (95% CI 0.98 to 2.40).
Associations between factors
When all factors were included in the complete multivariable model, ulcer type, depth, infection, Wagner ulcer grade, antimicrobial dressing and MRSA were no longer significant at the 10% level owing to correlations with other baseline factors. The influence of these factors on the incidence of healing was therefore better explained by other associated factors, in this case the remaining significant factors: PEDIS perfusion, only ulcer, wound duration, CNS and age. Details of associations between factors with a significant association with healing in the univariate analysis (excluding age) can be found in Table 73.
| Ulcer type | Ulcer grade | PEDIS perfusion | PEDIS depth | PEDIS infection | Only ulcer | Wound duration | Dressing | MRSA | CNS | |
| < 0.0001 | < 0.0001 | < 0.0001 | 0.0036 | 0.0300 | 0.0219 | 0.0012 | 0.0042 | 0.0453 | Ulcer type | |
| < 0.0001 | < 0.0001 | < 0.0001 | Ulcer grade | |||||||
| < 0.0001 | 0.0002 | 0.0423 | < 0.0001 | PEDIS perfusion | ||||||
| < 0.0001 | PEDIS depth | |||||||||
| PEDIS infection | ||||||||||
| Only ulcer | ||||||||||
| 0.0201 | Wound duration | |||||||||
| Dressing | ||||||||||
| 0.0376 | MRSA | |||||||||
| CNS |
Ulcer type, perfusion, depth, infection and Wagner ulcer grade were all found to have significant pairwise associations; of these, perfusion was the only factor to remain significant in the complete multivariable model, with a higher perfusion grade associated with ischaemic ulcers, higher depth grade, higher infection grade and higher Wagner ulcer grade.
Ulcer type was also further associated with only ulcer, wound duration and CNS, which remained significant in the multivariable model, such that ischaemic ulcers were associated with having more than one ulcer, older ulcers and less presence of CNS.
The presence of an antimicrobial dressing, non-significant in the multivariable model, was associated with higher perfusion grades and older ulcers which remained significant in the multivariable model, suggesting that the association between antimicrobial dressing and a reduced incidence of healing may be explained by the antimicrobial dressing having been present on more severe ulcers.
Finally, MRSA was also found to be non-significant in the multivariable model, which may be explained by the association with ulcer types in which there was a greater presence of MRSA for ischaemic ulcers. There was also a negative association with CNS, in which MRSA and CNS were not reported from the same sample for any patients; therefore, MRSA was present only when CNS was not and vice versa, which may also help explain the greater incidence of healing when CNS was present (i.e. because MRSA was not).
Discussion
This substudy has reported on the prognosis associated with infected DFUs. This information is necessary for the planning of clinical trials of interventions for infected foot ulcers and, in our analysis, we have been able to identify relationships between factors associated with poor prognosis, as well as competing risks, which are important for trial planning.
For clinicians, these data confirm the poor prognosis associated with infected DFUs. The healing rate is lower than other published healing rates from non-selected cohorts. For example, in the largest study, Eurodiale, the 12-month outcome (in 1232 consecutive patients, of whom 58% had infection at first presentation) found that 77% of the patients healed (with or without a minor amputation), 5% underwent a major amputation and 6% died. 22 Although one may suggest that our results lend credence to the proposal that infection is associated with poor outcomes, Prompers et al. 22 did not find this within Eurodiale, and the authors observed no differences for major amputation or healing rate between neuropathic ulcers with and without infection. They did observe that infection was a risk factor for minor amputation and suggest that their result indicates that aggressive treatment of infection in the participating centres meant that the impact of infection on healing was minimised. 22
This study confirmed the prognostic value (using univariable analysis techniques) of ulcer perfusion grade (PAD vs. no PAD), ulcer type (ischaemic vs. non-ischaemic), wound duration (< 56 days, ≥ 56 days), presence of other ulcers, Wagner ulcer grade, presence of antimicrobial dressing, infection grade, depth grade, presence of CNS, presence of MRSA and age with the incidence of healing. We propose that the association of increased healing with higher age may be spurious as we are unaware of biological mechanisms that might explain increase healing at older age, and the association did not endure when age was modelled as a continuous factor rather than dichotomised one.
The association of CNS with an increased risk of healing may be in part attributable to a negative association between MRSA and CNS.
The observation of reduced healing for ulcers that had an antimicrobial dressing on at baseline was not expected. However, this is not an indication that antimicrobial dressings per se are associated with delayed healing (there is, in fact, very little robust evidence of the relationship between antimicrobial dressings and improved outcomes such as healing). It may be, in this population, a proxy for wound severity (i.e. the most severe ulcers were those that had an antimicrobial dressing applied).
There were correlations between many of the factors associated with healing. Subsequent multivariable modelling to account for this suggested that wound duration, ulcer perfusion grade and presence of other ulcers provide the best independent predictors of healing.
We found no association between diabetic control and outcomes, and this may represent low power to detect an association. This is in contrast to the recent work of Christman et al. 111 who found that HbA1C was associated with healing outcomes in a retrospective study of 187 Americans with diabetes (average 2.3 wounds per patient, the majority of which were foot ulcers). Diabetic control was a significant factor only in insensate neuropathic wounds rather than in the whole sample, however.
Unlike Ince et al. ,107 we found no association between duration of diabetes and outcomes. Margolis et al. 112 identified an association between wound area, wound depth, wound duration and healing (using multivariate analysis on a data set with > 31,000 patients); however, we found no association between these variables and healing in our population. This may be due to power or to the overwhelming effect of infection in determining healing outcomes for this group.
Of the domains within the PEDIS assessment tool, perfusion proved to be the strongest prognostic factor in our study. Depth and infection were both also shown to be associated with healing but were not, however, retained in the multivariate model including all factors, and sensation and area (extent) did not emerge from the model at all. All participants were clinically diagnosed as infected; therefore, infection compared ulcers with extensive erythema or systemic inflammatory response to those without involvement of deeper tissues or systemic signs. The lack of association between area and healing is also is different from other studies. Our analysis has investigated the association of single pathogens with healing and, in future, we may be able to investigate the associations for different spreads of pathogens. This would allow us to determine the impact of infection profiles on clinical outcomes.
One of the strengths of this study is the acknowledgement of and the approach to dealing with competing risks. This is the first study we are aware of that has dealt with patient death or amputation in this way when estimating time to healing and the prognostic value of different clinical factors. In using cumulative incidence functions to account for the competing risks of death or amputation, which prohibit the incidence of healing, rather than censoring patients at their date of death or amputation (i.e. Ince et al. 107), we have not overestimated the rate of healing. Indeed, had a conventional proportional hazards regression analyses been applied, the healing rate at 12 months would have been overestimated by almost 10%.
One of the limitations of this study is the incomplete patient follow-up. We are not able to determine whether or not those missing from the follow-up population were systematically different from those who we did include. We were able to determine that missing case-note reviews for the 101 patients without follow-up data were not found to be associated with the timing of patient entry into CODIFI, or by study centre with patients appearing to be missing a case-note review at random. Missing case-note review could indicate that a patient lost contact with a site (i.e. owing to healing and no need for contact) or equally that they were ill or still with ulcer and did not want to provide consent.
As the pattern of having both healing and amputation at different sites on the foot made for complex data collection and analysis, researchers in this area may wish to design studies to ensure completeness of data recording in terms of the relationship between multiple ulcers and how to reference the index ulcer > 12 months after initial sample collection. It would have been useful also to collect overall patient outcome (i.e. ulcer free, not just index ulcer).
In this follow-up study, we did not control recall to hospital for follow-up or frequency of patient appointments. Given that we undertook case-note review at 12-month follow-up, some patients would have been between appointments or would not have been seen for some length of time at 365 days.
Clinicians completing the follow-up data additionally provided data on a range of ‘other’ events. These ‘other’ reported events may be clinically important events to collect in future studies.
Chapter 6 Conclusions
Results from main study: agreement between swab and tissue sampling
We compared the reported presence of isolates likely to be pathogens using both techniques. Swab results of 395 patients with DFUs and suspected infection yielded no bacterial isolates at all in 20% of samples. Despite these wounds being clinically assessed as infected, 29.9% of swab reports noted no potential pathogens. For tissue sampling the rates were lower: 10% had no bacterial isolates reported and 13.9% of reports cited no pathogens (only contaminating/colonising flora). Given that wounds are not sterile, the high ‘no isolates’ rate suggests that sample collection and/or transport were less than optimal. In addition, the ‘no pathogens’ rate might indicate that clinicians have a low threshold for suspecting infection in DFUs given the clinical consequences of failing to detect and treat foot infection promptly. This may result in some wounds with suspected infection being merely colonised (i.e. yielding no pathogens). However, the high ‘no isolates’ rate suggests that another explanation is that collection and transport of samples were suboptimal and, hence, pathogens in the wound were either not collected or did not survive transport. Tissue samples were better than wound swabs in collecting and transporting to the laboratory live bacteria from DFUs, with half the chance of reporting no isolates compared with wound swabbing (10% vs. 20%). To improve the collection of microbiology samples from wounds with suspected infection, it may be necessary to improve collection and transport such that fastidious and anaerobic organisms are preserved for subsequent culture and analysis. We had specified that clinicians used the Levine et al. 48 technique for swab sample collection, noting that this was identified as superior to two other swab techniques by Gardner et al. ;113 however, the pragmatic nature of the study meant that we did not restrict sampling to a subset of clinical staff whose technique had been assessed as producing the sufficient wound surface pressure to express wound fluid from the ulcer bed, nor did our protocol require validation regarding the level of cleansing and debridement performed prior to sampling.
Overall, the most frequently reported pathogens were S. aureus (35.7%), Streptococcus (16.7%), Enterococcus (14.9%), CNS (12.2%), Corynebacterium (9.4%), Pseudomonas (8.6%) and MRSA (8.1%). Considering each of the potential pathogens separately allowed us to determine patterns of agreement across different pathogens. This was important, as combining pathogens would mask potentially important differences. It is possible to have disagreement in identification of pathogens when both methods report the same prevalence: swab reports 10% and tissue sample reports 10%, but there is symmetrical disagreement whereby some tissue results are positive and swab reports are negative for a particular pathogen and vice versa. We therefore reported both prevalence of identification of pathogens and agreement rates by pathogen.
Reported prevalence of pathogens was identical for S. aureus and Pseudomonas and very similar (1% different) for MRSA, whereas for all other groups of potential pathogens the prevalence was statistically significantly higher from tissue samples than from swab samples. In 80% of patients there was agreement in the parameter ‘at least one pathogen identified’, meaning that the disagreement rate was 20% for ‘≥ 1 pathogen’. This does not convey the extent of variation between pathogens, however, and the disagreement was low for MRSA (1.5%), Pseudomonas (4.1%) and S. aureus (8.1%). For anaerobes, by way of contrast, disagreement was 16.5%. Overall, therefore, the agreement between wound swabbing and tissue sampling is high for three of the most prevalent pathogens, S. aureus, MRSA and Pseudomonas. For other pathogens, the swab reported organisms significantly less often than tissue samples. This might indicate that wound swabbing and tissue sampling have comparable yields for these three organisms. When sampling is designed to provide information on the wound biome with no prior expectation of the infecting organisms, however, then using a tissue sample will more often report the presence of organisms than a swab sample. It is not possible from these data to determine whether the higher yield is associated with higher survivability of organisms or better collection.
As well as measuring disagreement (proportion of samples in which the tissue and swab samples did not agree regarding the presence/absence of a specified organism), the commissioning brief determined that we should also report the agreement using the κ-statistic. There is discussion among statisticians about the use of Cohen’s kappa in measuring agreement (which will not be rehearsed here), but, for completeness, we reported percentage disagreement as well as kappa and PABAK. Although the kappa and PABAK for these outcomes were at a level usually associated with ‘fair’ to ‘high’ agreement, researchers have argued that they are imperfect summary statistics for determining the ability to replace one assessment tool with another, as the consequences of disagreement (such as not identifying an aggressive pathogen with one method), and hence the clinical implications of delayed diagnosis and treatment need to be considered. Guidelines for reporting agreement studies note that some authors require high agreement levels when tools are used to make individual and important clinical decisions, for example, suggesting that a tool should have reliability levels of at least 0.9 when being used to make an individual assessment of pressure ulcer risk. 114
The number of different bacterial pathogens reported differed between tissue sampling and wound swabbing. Overall, tissue sampling identified more species than wound swabbing, with a median of 1.5 pathogens identified per tissue sample and 1 per wound swab. Given that the wound swab collected wound tissue from a greater area than the tissue sample, it might be expected that wound swabs would report a higher number of potential pathogens; therefore, this finding might be attributable to poorer pathogen survival in swab samples than tissue samples or insufficient pressure used during swab sample collection to allow deeper-wound-fluid-containing pathogens to be expressed.
We also reported the proportions of patients in whom the same bacteria were identified in each report, where additional bacteria were identified in the tissue sample over the swab sample (but all the swab bacteria were also identified), where additional bacteria were identified in the swab over the tissue sample (but all the tissue sample bacteria were reported in the swab sample) and where there was a difference in pathogens reported. Swab and tissue results reported the same pathogens in 42% of patients; a swab reported additional pathogens to those in the tissue sample in 8.1% of patients; tissue samples reported additional pathogens to those in the swab in 36.7% of patients; and, the tissue sample and swab specimens reported different pathogens, with or without overlap, in 13.2% of patients. These differences were unrelated to any clinical characteristics (type of ulcer, etc.) except potentially the wound duration: for wounds open for > 56 days, the odds of their tissue sample reporting additional pathogens (compared with swab sample) was lower than for young wounds, although this relationship was not present when duration was analysed as a continuous variable. This may be due to the change in biome as wounds age and become increasingly polymicrobial, for example, or may be a chance finding; therefore, it is worthy of further study in future research. The chance of identifying additional isolates (likely pathogens) with a tissue sample is therefore potentially clinically significant; however, if these isolates are present in low numbers then it may represent an increase in overall information about the wound biome with no impact on the clinical assessment regarding the likely cause of infection and the appropriate antibiotic regimen. For this reason, we added the substudy whereby a clinical panel review was undertaken to determine whether or not the (usually) higher amount of information from tissue samples translated into different analyses by clinicians reviewing the microbiology reports while blind to source (swab or tissue). Overall, the microbial diversity (number of species) was higher when wounds were sampled using tissue rather than swab.
The microbial load, assessed semiquantitatively, was compared between tissue and swab. Agreement between the two techniques for MRSA (overall κ = 0.73), Pseudomonas (overall κ = 0.58), Streptococcus (overall κ = 0.65) and S. aureus (overall κ = 0.64) was in the ‘moderate’ to ‘substantial’ range, but was lower for anaerobes (overall κ = 0.32). We were unable to conclude overall if the microbial load (as summarised in +/++/+++) was consistently higher for either sampling technique.
The presence of antimicrobial resistance among likely pathogens was low (33 patients overall), and there was disagreement about the presence of resistance in only 6 patients and, hence, detailed analysis was not undertaken.
The rates of sampling-related AEs were higher for tissue sampling than for wound swabbing, both in terms of the pain reported by patients and sampling-related bleeding (6% in cases related to tissue sampling alone, 0.8% cases related to swab sampling alone, and related to sampling of any form in 0.8% of other cases). This equates to a number needed to harm of 20: for every 20 patients undergoing tissue sampling rather than wound swabbing, there would be one additional case of clinically concerning bleeding. We did not have any reports of further sequelae of sampling; we did not collect data at the next visit, for example, but there were no reports of RUSAEs throughout the study.
One-quarter of patients reported pain before sampling, and we recorded sampling-associated pain after swabbing and again after tissue sampling (swabbing always preceded tissue sample collection). A total of 5 patients (1.3%) reported that pain increased (from baseline) on swabbing, and 37 patients (9.3%) reported that it increased after tissue sampling. This equates to a number needed to harm of 13, although overall (in 89.5% of patients) ulcer pain remained the same after sampling. Despite the vast majority of these patients having neuropathy, we cannot assume that these wounds are painless, that patient comfort is not affected by sampling technique or that, overall, swab sampling is associated with a lower risk of sampling pain.
We sought to report the costs of sampling using each of the two techniques but found limited information on the real costs of techniques, with some centres considering such information commercially sensitive and hence being unwilling to provide it. The cost of sampling can include a number of elements, not all of which will have been included in the costs quoted by those centres providing information. For example, the clinical time involved in wound bed preparation (cleansing and debridement) may not be captured. The relative differences between swab costs and tissue sample costs was 4% in one site (£15.55 vs. £16.53) and 49% in another (£3.91/£5.85). These costs do not include sampling equipment, transport or clinician time. Further work is required to understand the economics of wound swabbing versus tissue sampling.
It appears, therefore, that if one wishes to identify the presence of S. aureus, MRSA or Pseudomonas in a clinically infected wound (diagnosed with regard to clinical appearance, signs and symptoms) then tissue sampling is, broadly speaking, comparable to swab sampling for these clinically important pathogens both in terms of identifying the presence of the pathogen and in determining the bacterial load in a semiquantitative manner. By taking a tissue sample instead of a swab, however, it is likely that a larger number of pathogens will be identified. It is not clear whether this difference in yield is due to poorer collection, survival in transport or lower reporting of the bacteria collected using wound swabs. Interestingly, the small number of swabs analysed in the substudy of molecular microbiology (PCR) techniques had a higher number of pathogens identified than either the cultured swab results, or the tissue samples (cultured or PCR), and as PCR is able to identify and multiply dead bacteria, this suggests that swabs collect more bacteria than tissue but that more of the species die in transport.
In a small proportion of cases, swab samples identify pathogens in addition to those identified from tissue sampling, which is likely to be attributable to the heterogeneity in location of pathogens across the wound surface and the fact that a wound swab gathers material from a larger area. This means that choosing one sample technique leads to a trade-off in information for 58% of patients. One approach might be to consider the two techniques as complementary, but performing both procedures would lead to increased costs and patient pain/bleeding complications. Choosing to take a tissue sample over a swab also leads to a small increase in the number of patients experiencing pain and bleeding of clinical concern.
Results from the substudies
Clinical review panel
A clinical panel review substudy was undertaken to identify if any differences identified in the main study in sampling yield were potentially clinically relevant. If one sampling technique provided more information but did not result in any therapeutic changes (compared with a reference method of sampling), then the additional information may not be clinically important. As all patients in this study had both samples taken and clinicians had access to both reports, we were not able to identify whether or not any clinical changes initiated were attributable to either swab or tissue sampling microbiology reports. We presented paired and blinded microbiology reports to clinicians to ask them if patients’ antimicrobial regimes ‘covered’ the pathogens reported, and also if a change in therapy was warranted, based solely on the microbiology results. The decision to change antimicrobial therapy would not, in practice, be taken on the basis of a laboratory test result alone, with clinical assessment of the patients and their wound playing a major part in clinical decision-making; however, as clinicians were presented with imperfect information (i.e. with no clinical data) for both swabs and tissue sample microbiology reports, we propose that this approach allows some analysis of the amount of clinically relevant information provided by the laboratory reports.
We assessed the inter- and intrarater reliability of these assessments and determined that the intrarater reliability for 7 of 13 assessors was ‘moderate to substantial’70 for a change in therapy (κ = 0.59 for tissue and κ = 0.77 for swab results), with inter-rater reliability lower at κ = 0.35 for swab and κ = 0.66 for tissue samples, meaning that there was at least ‘fair’70 agreement between the clinical reviewers for swab samples but substantial agreement for tissue samples. Having established adequate agreement between clinicians (and where agreement was low, this was overcome as each clinician reviewed both patients swab and tissue sample vignettes), we determined the proportion of patients for whom the prescribed antimicrobial regimen was deemed to be insufficient. For 1 in 5 patients (19.8%), a tissue sample report was assessed by clinicians as indicating that the empirical antibiotic regimen did not cover the pathogens reported (whereas the swab sample results did). Conversely, for 1 in 12 (8.5%) patients, the swab sample results suggested that the empirical antibiotic regimen did not cover the pathogens reported, whereas the tissue sample results did. A change of strategy from swab sampling to tissue sampling would result in 11.5% (1 in 9) of patients additionally being deemed to ‘not be covered’ by their current regimen. Any sampling regimen using a single method of ulcer sampling would be associated with potentially missing pathogens and the resultant assessment would be influenced by the source of the laboratory report.
The results for ‘change in therapy required’ were very similar, given the relationship between these two outcomes. For 1 in 12 (8.9%) patients, the tissue sample results suggested no change, whereas the swab sample results did suggest that a change was required. For 1 in 6 (17.8%) patients, the swab sample results suggested no change, whereas the tissue sample results did suggest that a change/initiation was required. A change of strategy from swab sampling to tissue sampling would result in 8.9% (1 in 11) of patients potentially having a change in antimicrobial regimen.
Although it is tempting to conclude that clinicians should use the sampling technique that result in the highest number of pathogens and the highest rate of potential therapeutic changes, the data from this study cannot determine whether or not tissue sampling is necessarily associated with better clinical outcomes. In managing infected DFUs, there may be a trade-off between tissue sampling and swabbing. Swabbing may be less expensive and require less training to perform, fewer patients may experience sampling pain or bleeding, and the reduced report of pathogens may be sufficient to guide therapy changes at clinical reassessment. Indeed, the survey of practice at sites indicated that microbiologists, working with clinicians in the clinics/wards, sometimes used their expertise to filter the content of the laboratory reports so that they included the material most relevant to the clinical condition (and omitting some low number isolates likely to be non-pathogenic) (see Appendix 3). Neither swab result nor tissue sample microbiology results produced by plating and culture arrive in the clinic in sufficient time to guide initiation of antimicrobials for the vast majority of patients and, hence, currently the role of wound sampling is to guide the tailoring of therapy at day 3 and beyond, when a clinical assessment will also be undertaken. Although plating and culture are the mainstay of microbiological analysis in wounds, there are a number of potential sampling scenarios, as outlined below:
-
Sampling to be undertaken upon first clinical assessment of infection using a swab, and any subsequent sampling to be undertaken by tissue sample. This would be, for example, should the clinical assessment or microbiology report determine that the empirical antimicrobial regimen is inadequate. This would expose only patients for whom additional information was needed to the potential harms of tissue sampling.
-
Initial sampling with both swab and tissue sample performed to capture the maximum amount of wound microbiology information. This would expose all patients to the harms of tissue sampling.
-
Sampling undertaken at initial assessment by taking a piece of tissue, knowing that this gives a better yield than swabbing, after determining that the risk of pain and bleeding is warranted. Some organisms are likely to be missed by tissue sampling alone, however.
-
Sampling undertaken at initial assessment by swabbing using Levine et al. ’s technique,48 as happens in some sites at present. This gives a lower yield than tissue sampling but reduces the risk of pain and bleeding. A large proportion of potential pathogens are likely to be missed by swabbing alone, however.
For other clinical scenarios (e.g. not DFUs) in which wound samples are taken with the goal of identifying the infecting organisms prior to antibiotic prescription, the trade-offs may be different.
Comparing culture and polymerase chain reaction techniques
For 14 patients, both swab and tissue samples were sent for analysis of bacterial RNA (dead and alive) and the results were compared with culture microbiology results. There was agreement between PCR and culture for one in three patients, but in half the patients, PCR analysis reported additional pathogens. PCR techniques more frequently reported the presence of a higher number of pathogens (in terms of bacterial diversity); for example, for the two swabs taken from each wound, the PCR analysed swab reported a median of 2.5 pathogens and the culture results a median of 1 pathogen. The difference was smaller for the tissue results, with PCR reporting a median of 1.5 pathogens and culture results a median of 1 pathogen.
In contrast to the results found using culture techniques, swab samples reported more pathogens than tissue samples (when both analysed by PCR). For the 12 samples with complete data, the culture results were that the swab and tissue agreed in 50% of patients and that tissue samples reported additional pathogens (i.e. over swab in 33%, with disagreement in 16.7%). Using PCR, however, the agreement rate was lower at 25%, and swabs identified more pathogens than tissue samples in 33% of patients, tissue identified additional pathogens in 16.7% of patients and there was disagreement in 25%. This indicates that both the choice of sampling technique and the method of analysis affects the yield from swabbing versus tissue sampling. The lower yield from swab samples and culture may be related to low viability of the bacteria sampled, secondary to sampling technique, transport media or delay in analysis owing to timing of collection from clinic/ward or at the laboratory. Any change in sampling technique (e.g. from swab to tissue) must be considered with reference to the sample analysis technique as the roll-out of PCR facilities in more hospitals may mean that wound samples can be analysed more quickly and that the differential yield identified using culture techniques is minimised when using PCR. This would influence the trade-off between swabbing and tissue sampling.
Prognosis of foot ulcer infection
We determined, at around 12 months, if patients had experienced ulcer healing, a revascularisation procedure, amputation, reulceration or had died. As there are no validated tools available to make a clinical diagnosis of resolution of infection, we did not collect data on the time to resolution. Over the 12-month period after ulcer infection, some patients experienced multiple events and, therefore, we used competing risks analyses to appropriately consider the risks of events. The relationship between these outcomes and baseline characteristics, including pathogens identified, were assessed.
The ulcer healing rate at around 12 months was 45.5%, indicating the poor prognosis associated with infected foot ulcers. Of those unhealed ulcers, 20% were not healed at 12 months despite an amputation, 57% of patients had an unhealed ulcer, and the remaining 22.7% of patients had died. Accounting for the competing risks of death or amputation, the estimated cumulative incidence of healing at 12 months post sampling was 44.5% (95% CI 38.9% to 50.1%).
With a univariate analysis we identified an association between reduced risk of healing and the following factors: ischaemic (vs. neuropathic) aetiology, increasing Wagner grade, increasing perfusion grade, increased ulcer depth/tissue loss, infection at grade 3 or above (extensive erythema or systemic inflammatory response), ulcer duration ≥ 56 days (vs. < 56 days), presence of an antimicrobial dressing and MRSA.
Factors associated with increased risk of healing included having a single foot ulcer, the presence of CNS and age (> 63 years vs. ≤ 63 years).
Following multivariable analyses, the remaining prognostic factors for healing were as follows (direction of association not specified unless not identified in previous studies): perfusion, wound duration, presence of other ulcers (a single ulcer more likely to heal), patient age, and the presence of CNS (ulcer more likely to heal in presence of this organism, probably because presence of CNS was inversely related to the presence of MRSA). The associations with ulcer duration and perfusion have been identified previously; we believe the higher risk of healing with increasing age to be a spurious association and that antimicrobial dressing may not be independent of the ulcer status, rather than it having a direct effect of delaying healing.
Scope of the study
This study did not set out to determine whether swabbing or tissue sampling were more accurate at identifying infection. We understood that in the UK the normal practice is to make a diagnosis of wound infection in DFU by considering signs and symptoms and not by using a laboratory test (such as swab or tissue sample). This is due to the time delay associated with getting a result, the potential for a false-negative result (e.g. owing to organism death during transport/media not supporting fastidious anaerobes) and the need for rapid (if not immediate) initiation of antibiotics. The lack of an agreed and validated definition of chronic wound infection means that we were not able either to compare sampling results with a gold-standard diagnosis or to determine reliably when ulcer infection was resolved.
This study cannot identify the exact source of the difference between tissue sampling, for example did the two techniques collect organisms differentially, did organisms survive the trip to the laboratory differentially, or were they handled differently in the laboratory (plating, culture), or were the differences at report stage? To study this we would have needed to change current practice, and we sought to provide information to guide UK NHS practice (as per the HTA remit) rather than to undertake more explanatory analyses.
Strengths of this study
This is the largest comparison of the two main methods of sampling and the first study to report detailed data on paired ulcers for each pathogen and to examine the relationship between baseline characteristics and agreement using multivariable modelling. A strength of the study is its external validity – there were few exclusion criteria and patients were recruited in normal practice settings (specialist clinics and hospital wards) resulting in a generalisable study population. In addition, samples were taken by members of the attending clinical teams and processed by local laboratories, providing a ‘real world’ comparison of the two techniques.
All centres received training updates on swab and tissue sampling to minimise sample differences; the overall agreement between the isolates reported from the two types of specimens was relatively high but overall tissue sampling (and subsequent culture) reported more pathogens than swab sampling with culture.
Previous reports comparing swab to tissue specimens have been small, single-centre, studies but generally had findings similar to ours. Bill et al. 60 compared culture results for a variety of chronic wounds from a swab with a punch biopsy (the reference standard) and found the sensitivity to be 79%. Their study of only 38 patients with several sources of bias regarded quantitative culture of the biopsy as a gold-standard reference test for infection, rather than the alternative, namely clinical signs and symptoms. In a retrospective study of 89 concomitantly obtained pairs of samples from 54 patients with DFUs (87% clinically infected), Mutluoglu et al. 62 found that culture results of superficial swabs did not correlate well with those obtained from deep tissue. Compared with tissue specimens, swab cultures had a positive predictive value of 84.4%, negative predictive value of 44.0% and overall accuracy of 73.0%. The use of the term ‘predictive value’ presumes the presence of a gold-standard for diagnosis, which we have not done. In keeping with our study, 52% of their patients had received antibiotic therapy at presentation. In a study of 50 patients with an infected DFU (who had not recently received antibiotic therapy), Demetriou et al. 63 used a tissue specimen culture against which to compare a swab specimen culture. The results obtained from tissue cultures were the same as those obtained by swab in only 50% of patients. Whereas the sensitivity of swab compared with tissue cultures was 100%, the specificity was ≤ 40%, and because of the isolation of ‘contaminating’ flora it was < 20% for ‘true pathogens’. Similar to our results, they noted no important differences in results between patients with neuropathic compared with neuroischaemic ulcers.
The substantial proportion of samples in our study that reported no pathogens may reflect the difficulty in establishing a diagnosis threshold for clinical infection in the diabetic foot, which is related to the frequent presence of peripheral neuropathy and arterial disease. 115 Alternatively, it may be related to poor sampling technique, transport media that fail to maintain viability of pathogens, or a choice by the microbiologist to report only those pathogens, according to their professional judgement threshold, necessary for reporting (i.e. they may be assessing ‘necessity for reporting’ when faced with a modest number of mixed flora).
Implications for practice
A key issue is how much, and what type of, information on ulcer flora is most useful for clinicians managing patients with a DFU. Certainly, clinicians want to know which organisms are causing infection so they can optimally target antibiotic therapy. However, providing them with comprehensive microbiology reports listing many organisms in addition to the predominant pathogen, perhaps including unusual isolates present in low numbers, may not necessarily aid clinical decision-making. We do not know if treatment based on a more detailed microbiogram leads to more effective care in terms of the likelihood of, or time to resolution of, infection, or the prevention of treatment-associated adverse effects and antibiotic resistance.
Given the global emergency associated with antibiotic-resistance related to overuse of this precious and limited resource, we need to be cautious about recommending a technique that may lead to unnecessarily broad-spectrum prescribing. Furthermore, the bacterial flora present in the wound at the time of sampling presentation may differ from those present after initial empiric antibiotic therapy, when culture results are reported. Studies have found swabs reporting additional isolates in 11%62 and 8.1% of samples (CODIFI), and different isolates in 6.7%62 and 13.2% (CODIFI) of sample, hence the more invasive technique of tissue sampling cannot be relied on to identify all the organisms identified by the less invasive technique. The two techniques collect information from two different parts of the ulcer biome: deep collection in one small area (another part of the wound may have a different bacterial profile) or superficial collection from a slightly larger area (again noting that other parts of the wound may have a different profile), and the final report depends on not only sampling, but also transport and microbiologist practice.
Implications for research
Future studies should determine whether or not one of the methods of sampling would lead to improved patient outcomes and better antimicrobial stewardship.
Further work is needed to understand the value of sampling at presentation with infection (plus empiric antimicrobial therapy as per local protocol) in terms of both clinical outcomes and antimicrobial stewardship.
Further studies are needed to confirm the variation in difference in reporting according to the observed pathogens which we observed.
We do not understand the role that laboratory tests have in the assessment and tailoring of further therapy in people with infected DFUs. Future research should therefore determine (1) the merits of rapid diagnostic testing over plating and culture when managing infected DFUs and (2) the impact of sampling followed by rapid PCR in terms of definitive (rather than empiric) therapy at first presentation.
Acknowledgements
Contributions of authors
All authors have completed authorship forms and qualify for authorship according to the International Committee of Medical Journal guidance.
Professor E Andrea Nelson (Professor of Wound Healing) initiated the study, led the grant application development, led the study team, drafted the final report and is the principal investigator.
Miss Alexandra Wright-Hughes (Senior Statistician) was a member of the study team, drafted the statistical analysis plan, undertook the statistical analyses, drafted the statistical results and commented on the study report.
Miss Sarah Brown (Principal Statistician) the supervising statistician, was a coapplicant on the study grant, was a member of the study team, contributed to the drafting of the statistical analysis plan, oversaw the statistical analyses and the preparation of the statistical results and commented on the study report – she is the guarantor for the study data.
Professor Benjamin A Lipsky (Professor of Medicine) was a coapplicant on the study grant, was a member of the study team, contributed to the drafting of the study report and revised it for intellectual content.
Dr Michael Backhouse (National Institute for Health Research Fellow, Podiatrist) was the first clinical study co-ordinator and a member of the study team; he initiated sites for recruitment, contributed to the drafting of the study report and revised it for intellectual content.
Dr Moninder Bhogal (Senior Trial Co-ordinator) was the trials unit study co-ordinator and a member of the study team; he was responsible for ethics and governance applications and co-ordinated the study team, contributed to the drafting of the study report and revised it for intellectual content.
Dr Mwidimi Ndosi (Lecturer in Adult Nursing) was the second clinical study co-ordinator and a member of the study team; he oversaw the prognostic study, contributed to the drafting of the study report and revised it for intellectual content.
Miss Catherine Reynolds (Senior Data Manager) was the data manager and a member of the study team; she was responsible for data quality, contributed to the drafting of the study report and revised it for intellectual content
Mrs Gill Sykes (Clinical Lead, Podiatrist) was a coapplicant on the study grant, a member of the study team and reviewed the study report for intellectual content.
Professor Christopher Dowson (Professor in Microbiology) was a coapplicant on the study grant, a member of the study team and was responsible for the PCR analysis aspects of the study; he reviewed the study report for intellectual content.
Dr Michael Edmonds (Consultant Physician) was a coapplicant on the study grant, a member of the study team and reviewed the study report for intellectual content.
Professor Peter Vowden (Consultant Vascular Surgeon) was a coapplicant on the study grant, was a member of the study team, contributed patients to the study and reviewed the study report for intellectual content.
Dr Edward B Jude (Consultant Physician, Diabetologist and Endocrinologist) was a coapplicant on the study grant, was a member of the study team, contributed patients to the study and reviewed the study report for intellectual content.
Mr Tom Dickie (Foot Health Manager) was a coapplicant on the study grant, was a member of the study team, contributed patients to the study and reviewed the study report for intellectual content.
Professor Jane Nixon (Professor of Tissue Viability) was a coapplicant on the study grant, a member of the study team, the lead for the Clinical Trials Unit activity and responsible for these aspects; she contributed to the drafting of the study report and reviewed it for intellectual content.
Acknowledged contributors
The authors wish to thank the members of the SSC: Professor John Deeks, Professor Rose Cooper, Dr Roger Gadsby, Dr Anne-Maree Keenan and Mrs Christine Thomas, who was the patient representative. Dr Anne-Maree Keenan and Mrs Christine Thomas reviewed patient information and public-facing documents.
The authors also wish to thank the following research collaborators across 25 hospitals, and their teams: Professor Peter Vowden (Bradford Teaching Hospitals, Bradford), Dr Kilimangalam Narayanan (Queen Elizabeth Hospital, Gateshead), Professor Satyan Rajbhandari (Chorley and South Ribble Hospital, Chorley), Dr Cuong Dang (North Manchester General Hospital, Manchester), Dr Deirdre Maguire (Harrogate District Hospital, Harrogate), Dr Robert Moisey (Huddersfield Royal Infirmary, Huddersfield), Dr Simon Ashwell (James Cook University Hospital, Middlesbrough), Dr Frank Bowling (Manchester Royal Infirmary, Manchester), Dr Varadarajan Baskarn (New Cross Hospital, Wolverhampton), Dr Ketan Dhatariya (Norfolk and Norwich University Hospital, Norwich), Dr Ryan D’Costa (Pinderfields General Hospital, Wakefield), Dr Mujahid Saeed (Queen Elizabeth Hospital, Birmingham), Dr Sunil Sharma (Queen Elizabeth Hospital, King’s Lynn), Dr Paul Smith (Royal Lancaster Infirmary, Lancaster), Dr Deepak Bhatnagar (Royal Oldham Hospital, Oldham), Dr Khaled Dukhan (South Tyneside District Hospital, South Shields), Dr Jonathan Bodansky (St James’s University Hospital, Leeds), Dr Edward Jude (Tameside General Hospital, Lancashire), Dr Harpal Randeva (University Hospital Coventry, Coventry), Dr Yashica Nathan (University Hospital Lewisham, Lewisham), Dr Fahmy Hanna (University Hospital of North Staffordshire, Stoke-on-Trent), Dr Sony Anthony (University Hospital of North Tees, Stockton-on-Tees) and Dr Parag Singhal (Weston General Hospital, Weston-Super-Mare).
Publications
Nelson EA. University study on diabetic foot care. J Diabetes Nurs 2012;16:38.
Nelson EA. University study on diabetic foot care. Podiatry Now 2012;15:11.
Nelson EA, Backhouse MR, Bhogal MS, Wright-Hughes A, Lipsky BA, Nixon J. Concordance in diabetic foot ulcer infection (CODIFI): a study protocol. BMJ Open 2013:13; e002370.
Data sharing statement
Requests for data should be made to the corresponding author.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 2004;27:1047-53. http://dx.doi.org/10.2337/diacare.27.5.1047.
- Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pr 2010;87:4-14. http://dx.doi.org/10.1016/j.diabres.2009.10.007.
- Hauner H, Holt RIG, Cockram C, Flyvbjerg A, Goldstein BJ. Textbook of Diabetes. Oxford: Wiley-Blackwell; 2011.
- Hex N, Bartlett C, Wright D, Taylor M, Varley D. Estimating the current and future costs of type 1 and type 2 diabetes in the UK, including direct health costs and indirect societal and productivity costs. Diabet Med 2012;29:855-62. http://dx.doi.org/10.1111/j.1464-5491.2012.03698.x.
- Geiss LS, Wang J, Cheng YLJ, Thompson TJ, Barker L, Li YF, et al. Prevalence and incidence trends for diagnosed diabetes among adults aged 20 to 79 years, United States, 1980–2012. JAMA 2014;312:1218-26. http://dx.doi.org/10.1001/jama.2014.11494.
- Narayan KM, Boyle JP, Thompson TJ, Sorensen SW, Williamson DF. Lifetime risk for diabetes mellitus in the United States. JAMA 2003;290:1884-90. http://dx.doi.org/10.1001/jama.290.14.1884.
- Quality and Outcomes Framework: Achievement, Prevalence and Exceptions Data, 2012/13. London: Health and Social Care Information Centre; 2013.
- Diabetes UK . Diabetes in the UK 2012 n.d. www.diabetes.org.uk/diabetes-in-the-uk-2012 (accessed 17 December 2014).
- Frykberg RG, Zgonis T, Armstrong DG, Driver VR, Giurini JM, Kravitz SR, et al. Diabetic foot disorders: a clinical practice guideline (2006 revision). J Foot Ankle Surg 2006;45:S1-S66. http://dx.doi.org/10.1016/S1067-2516(07)60001-5.
- Sawacha Z, Spolaor F, Guarneri G, Contessa P, Carraro E, Venturin A, et al. Abnormal muscle activation during gait in diabetes patients with and without neuropathy. Gait Posture 2012;35:101-5. http://dx.doi.org/10.1016/j.gaitpost.2011.08.016.
- Abouaesha F, van Schie CHM, Griffths GD, Young RJ, Boulton AJM. Plantar tissue thickness is related to peak plantar pressure in the high-risk diabetic foot. Diabetes Care 2001;24. http://dx.doi.org/10.2337/diacare.24.7.1270.
- Vinik AI, Maser RE, Mitchell BD, Freeman R. Diabetic autonomic neuropathy. Diabetes Care 2003;26. http://dx.doi.org/10.2337/diacare.26.5.1553.
- Richard JL, Lavigne JP, Sotto A. Diabetes and foot infection: more than double trouble. Diabetes Metab Res Rev 2012;28:46-53. http://dx.doi.org/10.1002/dmrr.2234.
- Williams R, Airey M, Boulton A, Connor H, Cavanagh P. The Foot in Diabetes. Chichester: Wiley; 2000.
- Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA 2005;293:217-28. http://dx.doi.org/10.1001/jama.293.2.217.
- Lavery LA, Armstrong DG, Wunderlich RP, Tredwell J, Boulton AJ. Diabetic foot syndrome: evaluating the prevalence and incidence of foot pathology in Mexican Americans and non-Hispanic whites from a diabetes disease management cohort. Diabetes Care 2003;26:1435-8. http://dx.doi.org/10.2337/diacare.26.5.1435.
- Iversen MM, Tell GS, Riise T, Hanestad BR, Østbye T, Graue M, et al. History of foot ulcer increases mortality among individuals with diabetes ten-year follow-up of the Nord-Trøndelag health study, Norway. Diabetes Care 2009;32:2193-9. http://dx.doi.org/10.2337/dc09-0651.
- Vileikyte L. Diabetic foot ulcers: a quality of life issue. Diabetes Metab Res Rev 2001;17:246-9. http://dx.doi.org/10.1002/dmrr.216.
- Meijer JW, Trip J, Jaegers SM, Links TP, Smits AJ, Groothoff JW, et al. Quality of life in patients with diabetic foot ulcers. Disabil Rehabil 2001;23:336-40. http://dx.doi.org/10.1080/09638280010005585.
- Vileikyte L, Rubin RR, Leventhal H. Psychological aspects of diabetic neuropathic foot complications: an overview. Diabetes Metab Res Rev 2004;20:S13-18. http://dx.doi.org/10.1002/dmrr.437.
- Boulton AJ, Kirsner RS, Vileikyte L. Clinical practice. Neuropathic diabetic foot ulcers. N Eng J Med 2004;351:48-55. http://dx.doi.org/10.1056/NEJMcp032966.
- Prompers L, Huijberts M, Schaper N, Apelqvist J, Bakker K, Edmonds M, et al. Resource utilisation and costs associated with the treatment of diabetic foot ulcers. Prospective data from the Eurodiale Study. Diabetologia 2008;51:1826-34. http://dx.doi.org/10.1007/s00125-008-1089-6.
- Reiber GE. The epidemiology of diabetic foot problems. Diabet Med 1996;13:S6-11.
- Armstrong DG, Lavery LA, Quebedeaux TL, Walker SC. Surgical morbidity and the risk of amputation due to infected puncture wounds in diabetic versus nondiabetic adults. J Am Podiat Med Assn 1997;87:321-6. http://dx.doi.org/10.7547/87507315-87-7-321.
- Alpizar M, Eltayeb K, Gubanov N, Loche M, Laporte R, Pina E, et al. Comparing the incidence of lower-extremity amputations across the world – the Global Lower-Extremity Amputation Study. Diabet Med 1995;12:14-8. http://dx.doi.org/10.1111/j.1464-5491.1995.tb02055.x.
- Unwin N. Global Lower Extremity Amputation Study Group . Epidemiology of lower extremity amputation in centres in Europe, North America and East Asia. The Global Lower Extremity Amputation Study Group. Br J Surg 2000;87:328-37. http://dx.doi.org/10.1046/j.1365-2168.2000.01344.x.
- Gregg EW, Sorlie P, Paulose-Ram R, Gu Q, Eberhardt MS, Wolz M, et al. Prevalence of lower-extremity disease in the U.S. adult population ≥ 40 years of age with and without diabetes: 1999–2000 National Health and Nutrition Examination Survey. Diabetes Care 2004;27:1591-7. http://dx.doi.org/10.2337/diacare.27.7.1591.
- Trautner C, Haastert B, Giani G, Berger M. Incidence of lower limb amputations and diabetes. Diabetes Care 1996;19:1006-9. http://dx.doi.org/10.2337/diacare.19.9.1006.
- Siitonen OI, Niskanen LK, Laakso M, Siitonen JT, Pyorala K. Lower-extremity amputations in diabetic and nondiabetic patients. A population-based study in eastern Finland. Diabetes Care 1993;16:16-20. http://dx.doi.org/10.2337/diacare.16.1.16.
- Ramsey SD, Newton K, Blough D, McCulloch DK, Sandhu N, Reiber GE, et al. Incidence, outcomes, and cost of foot ulcers in patients with diabetes. Diabetes Care 1999;22:382-7. http://dx.doi.org/10.2337/diacare.22.3.382.
- Oyibo SO, Jude EB, Tarawneh I, Nguyen HC, Armstrong DG, Harkless LB, et al. The effects of ulcer size and site, patient’s age, sex and type and duration of diabetes on the outcome of diabetic foot ulcers. Diabet Med 2001;18:133-8. http://dx.doi.org/10.1046/j.1464-5491.2001.00422.x.
- Apelqvist J, Larsson J, Agardh CD. Long-term prognosis for diabetic patients with foot ulcers. J Intern Med 1993;233:485-91. http://dx.doi.org/10.1111/j.1365-2796.1993.tb01003.x.
- Armstrong DG, Lavery LA, Harkless LB. Validation of a diabetic wound classification system: the contribution of depth, infection, and ischemia to risk of amputation. Diabetes Care 1998;21:855-9. http://dx.doi.org/10.2337/diacare.21.5.855.
- Lipsky BA. International consensus group on diagnosing treating the infected diabetic foot . A report from the international consensus on diagnosing and treating the infected diabetic foot. Diabetes Metab Res Rev 2004;20:S68-77. http://dx.doi.org/10.1002/dmrr.453.
- Lipsky BA, Berendt AR, Deery HG, Embil JM, Joseph WS, Karchmer AW, et al. Diagnosis and treatment of diabetic foot infections. Clin Infect Dis 2004;39:885-910. http://dx.doi.org/10.1086/424846.
- Lipsky BA, Berendt AR, Cornia PB, Pile JC, Peters EJ, Armstrong DG, et al. 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis 2012;54:e132-73. http://dx.doi.org/10.1093/cid/cis346.
- Diabetic Foot Problems: Inpatient Management of Diabetic Foot Problems. Manchester: NICE; 2011.
- Lipsky B. Empirical therapy for diabetic foot infections: are there clinical clues to guide antibiotic selection?. Clin Microbiol Infec 2007;13:351-3. http://dx.doi.org/10.1111/j.1469-0691.2007.01697.x.
- Kallstrom G. Are quantitative bacterial wound cultures useful?. J Clin Microbiol 2014;52:2753-6. http://dx.doi.org/10.1128/JCM.00522-14.
- Trengove NJ, Stacey MC, McGechie DF, Mata S. Qualitative bacteriology and leg ulcer healing. J Wound Care 1996;5:277-80.
- Steed DL, Attinger C, Colaizzi T, Crossland M, Franz M, Harkless L, et al. Guidelines for the treatment of diabetic ulcers. Wound Repair Regen 2006;14:680-92. http://dx.doi.org/10.1111/j.1524-475X.2006.00176.x.
- Apelqvist J, Bakker K, van Houtum WH, Schaper NC. International Working Group on the Diabetic Foot Editorial B . Practical guidelines on the management and prevention of the diabetic foot: based upon the International Consensus on the Diabetic Foot (2007). Prepared by the International Working Group on the Diabetic Foot. Diabetes Metab Res Rev 2008;249:S181-7. http://dx.doi.org/10.1002/dmrr.848.
- Senior C. Assessment of infection in diabetic foot ulcers. J Wound Care 2000;9:313-17. http://dx.doi.org/10.12968/jowc.2000.9.7.25999.
- Kelly F. Infection control: validity and reliability in wound swabbing. Br J Nurs 2003;12:959-60. http://dx.doi.org/10.12968/bjon.2003.12.16.11437.
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14:244-69. http://dx.doi.org/10.1128/CMR.14.2.244-269.2001.
- Angel DE, Lloyd P, Carville K, Santamaria N. The clinical efficacy of two semi-quantitative wound-swabbing techniques in identifying the causative organism(s) in infected cutaneous wounds. Int Wound J 2011;8:176-85. http://dx.doi.org/10.1111/j.1742-481X.2010.00765.x.
- Dow G. Bacterial swabs and the chronic wound: when, how, and what do they mean?. Ostomy Wound Manage 2003;49:S8-13.
- Levine NS, Lindberg RB, Mason AD, Pruitt BA. The quantitative swab culture and smear: a quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma 1976;16:89-94. http://dx.doi.org/10.1097/00005373-197602000-00002.
- Sotto A, Richard J-L, Jourdan N, Combescure C, Bouziges N, Lavigne J-P. Miniaturized oligonucleotide arrays a new tool for discriminating colonization from infection due to Staphylococcus aureus in diabetic foot ulcers. Diabetes Care 2007;30:2051-6. http://dx.doi.org/10.2337/dc07-0461.
- Spichler A, Hurwitz B, Armstrong D, Lipsky B. Microbiology of diabetic foot infections: from Louis Pasteur to ‘crime scene investigation’. BMC Med 2015;13. http://dx.doi.org/10.1186/s12916-014-0232-0.
- Lipsky BA, Richard J-L, Lavigne J-P. Diabetic foot ulcer microbiome: one small step for molecular microbiology. . . One giant leap for understanding diabetic foot ulcers?. Diabetes 2013;62:679-81. http://dx.doi.org/10.2337/db12-1325.
- Gasser RB. Molecular tools--advances, opportunities and prospects. Vet Parasitol 2006;136:69-8. http://dx.doi.org/10.1016/j.vetpar.2005.12.002.
- Redkar R, Kalns J, Butler W, Krock L, McCleskey F, Salmen A, et al. Identification of bacteria from a non-healing diabetic foot wound by 16 S rDNA sequencing. Mol Cell Probe 2000;14:163-9. http://dx.doi.org/10.1006/mcpr.2000.0303.
- Armstrong D, Attinger C, Boulton A, Frykberg R, Kirsner R, Lavery L, et al. Guidelines regarding negative wound therapy (NPWT) in the diabetic foot. Ostomy Wound Manage 2004;50:S3-27.
- Pinzur MS, Slovenkai MP, Trepman E, Shields NN. Guidelines for diabetic foot care: recommendations endorsed by the Diabetes Committee of the American Orthopaedic Foot and Ankle Society. Foot and Ankle Int 2005;26:113-19.
- Bakker K, Schaper NC. International Working Group on the Diabetic Foot Editorial Board . The development of global consensus guidelines on the management and prevention of the diabetic foot 2011. Diabetes Metab Res Rev 2012;28:116-18. http://dx.doi.org/10.1002/dmrr.2254.
- Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008;336:924-6. http://dx.doi.org/10.1136/bmj.39489.470347.AD.
- Pellizzer G, Strazzabosco M, Presi S, Furlan F, Lora L, Benedetti P, et al. Deep tissue biopsy vs. superficial swab culture monitoring in the microbiological assessment of limb-threatening diabetic foot infection. Diabet Med 2001;18:822-7. http://dx.doi.org/10.1046/j.1464-5491.2001.00584.x.
- Slater R, Lazarovitch T, Boldur I, Ramot Y, Buchs A, Weiss M, et al. Swab cultures accurately identify bacterial pathogens in diabetic foot wounds not involving bone. Diabet Med 2004;21:705-9. http://dx.doi.org/10.1111/j.1464-5491.2004.01221.x.
- Bill TJ, Ratliff CR, Donovan AM, Knox LK, Morgan RF, Rodeheaver GT. Quantitative swab culture versus tissue biopsy: a comparison in chronic wounds. Ostomy Wound Manage 2001;47:34-7.
- Nelson EA, O’Meara S, Craig D, Iglesias C, Golder S, Dalton J, et al. A series of systematic reviews to inform a decision analysis for sampling and treating infected diabetic foot ulcers. Health Technol Assess 2006;10. http://dx.doi.org/10.3310/hta10120.
- Mutluoglu M, Uzun G, Turhan V, Gorenek L, Ay H, Lipsky BA. How reliable are cultures of specimens from superficial swabs compared with those of deep tissue in patients with diabetic foot ulcers?. J Diabetes Complicat 2012;26:225-9. http://dx.doi.org/10.1016/j.jdiacomp.2012.03.015.
- Demetriou M, Papanas N, Panopoulou M, Papatheodorou K, Bounovas A, Maltezos E. Tissue and swab culture in diabetic foot infections: neuropathic versus neuroischemic ulcers. Int J Low Extrem Wounds 2013;12:87-93. http://dx.doi.org/10.1177/1534734613481975.
- Nelson EA, Backhouse MR, Bhogal MS, Wright-Hughes A, Lipsky BA, Nixon J, et al. Concordance in diabetic foot ulcer infection. BMJ Open 2013;3. http://dx.doi.org/10.1136/bmjopen-2012-002370.
- Public Health England . Investigation of Skin, Superficial and Non-Surgical Wound Swabs n.d. www.gov.uk/government/publications/smi-b-11-investigation-of-skin-superficial-and-non-surgical-wound-swabs (accessed 17 December 2014).
- Public Health England . Investigation of Tissues and Biopsies n.d. https://www.gov.uk/government/publications/smi-b-17-investigation-of-tissues-and-biopsies (accessed 17 December 2014).
- Schaper N. Diabetic foot ulcer classification system for research purposes: a progress report on criteria for including patients in research studies. Diabetes Metab Res Rev 2004;20:S90-S95. http://dx.doi.org/10.1002/dmrr.464.
- Wagner F. The diabetic foot. Orthopedics 1987;10:163-72.
- Gardner SE, Frantz RA, Troia C, Eastman S, MacDonald M, Buresh K, et al. A tool to assess clinical signs and symptoms of localized infection in chronic wounds: development and reliability. Ostomy Wound Manage 2001;47:40-7.
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159-74. http://dx.doi.org/10.2307/2529310.
- The MI Procedure. SAS/STAT 9.22 User’s Guide. Cary, NC: SAS Institute Inc.; 2010.
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria 2013. www.R-project.org/ (accessed 17 March 2014).
- Agresti A. Categorical Data Analysis. New York, NY: Wiley; 2014.
- Wang WZ. An inductive order construction for the difference of two dependent proportions. Stat Probabil Lett 2012;82:1623-8. http://dx.doi.org/10.1016/j.spl.2012.03.035.
- Mccullagh P. Regression-models for ordinal data. J Roy Stat Soc B Met 1980;42:109-42.
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. New York, NY: Wiley; 2002.
- Brooks S, Gelman A, Jones G, Meng X-L. Handbook of Markov Chain Monte Carlo. Boca Raton, FL: Chapman & Hall/CRC Press; 2011.
- White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med 2011;30:377-99. http://dx.doi.org/10.1002/sim.4067.
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York, NY: Wiley; 1987.
- Cicchetti DV, Allison T. A new procedure for assessing reliability of scoring EEG sleep recordings. Am J EEG Technol 1971;11:101-10.
- Kundin JI. A new way to size up a wound. Am J Nurs 1989;89:206-7.
- Lancaster GA, Dodd S, Williamson PR. Design and analysis of pilot studies: recommendations for good practice. J Eval Clin Pract 2004;10:307-12. http://dx.doi.org/10.1111/j. 2002.384.doc.x.
- Browne RH. On the use of a pilot sample for sample-size determination. Stat Med 1995;14:1933-40. http://dx.doi.org/10.1002/sim.4780141709.
- Hayes AF, Krippendorff K. Answering the call for a standard reliability measure for coding data. Commun Methods Meas 2007;1:77-89. http://dx.doi.org/10.1080/19312450709336664.
- Krippendorff K. Content Analysis: An Introduction to Its Methodology. Thousand Oaks, CA: Sage; 2004.
- Davies CE, Hill KE, Wilson MJ, Stephens P, Hill CM, Harding KG, et al. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J Clin Microbiol 2004;42:3549-57. http://dx.doi.org/10.1128/JCM.42.8.3549-3557.2004.
- Hill KE, Davies CE, Wilson MJ, Stephens P, Harding KG, Thomas DW. Molecular analysis of the microflora in chronic venous leg ulceration. J Med Microbiol 2003;52:365-9. http://dx.doi.org/10.1099/jmm.0.05030-0.
- Fall A, Thompson RCA, Hobbs RP, Morgan-Ryan U. Morphology is not a reliable tool for delineating species within cryptosporidium. J Parasitol 2003;89:399-402. http://dx.doi.org/10.1645/0022-3395(2003)089[0399:MINART]2.0.CO;2.
- Jex AR, Smith HV, Monis PT, Campbell BE, Gasser RB. Cryptosporidium--biotechnological advances in the detection, diagnosis and analysis of genetic variation. Biotechnol Adv 2008;26:304-17. http://dx.doi.org/10.1016/j.biotechadv.2008.02.003.
- Thomsen TR, Aasholm MS, Rudkjøbing VB, Saunders AM, Bjarnsholt T, Givskov M, et al. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen 2010;18:38-49. http://dx.doi.org/10.1111/j.1524-475X.2009.00561.x.
- Rhoads DD, Wolcott RD, Sun Y, Dowd SE. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci 2012;13:2535-50. http://dx.doi.org/10.3390/ijms13032535.
- Melendez JH, Frankel YM, An AT, Williams L, Price LB, Wang NY, et al. Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin Microbiol Infec 2010;16:1762-9. http://dx.doi.org/10.1111/j.1469-0691.2010.03158.x.
- Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, et al. Community analysis of chronic wound bacteria using 16s rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLOS ONE 2009;4. http://dx.doi.org/10.1371/journal.pone.0006462.
- Dowd SE, Wolcott RD, Kennedy J, Jones C, Cox SB. Molecular diagnostics and personalised medicine in wound care: assessment of outcomes. J Wound Care 2011;20. http://dx.doi.org/10.12968/jowc.2011.20.5.232.
- Wolcott RD, Cox SB, Dowd SE. Healing and healing rates of chronic wounds in the age of molecular pathogen diagnostics. J Wound Care 2010;19:272-8. http://dx.doi.org/10.12968/jowc.2010.19.7.48898.
- Tuttle MS, Mostow E, Mukherjee P, Hu FZ, Melton-Kreft R, Ehrlich GD, et al. Characterization of bacterial communities in venous insufficiency wounds using conventional culture and molecular diagnostic methods. J Clin Microbiol 2011;49:3812-19. http://dx.doi.org/10.1128/JCM.00847-11.
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLOS ONE 2008;3. http://dx.doi.org/10.1371/journal.pone.0003326.
- Tong J, Liu C, Summanen P, Xu H, Finegold SM. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 2011;17:64-8. http://dx.doi.org/10.1016/j.anaerobe.2011.03.004.
- Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches. Microbial Ecol Fems Microbiol Ecol 2009;67:6-20. http://dx.doi.org/10.1111/j.1574-6941.2008.00629.x.
- Heininger A, Binder M, Ellinger A, Botzenhart K, Unertl K, Döring G. DNase pretreatment of master mix reagents improves the validity of universal 16S rRNA gene PCR results. J Clin Microbiol 2003;41:1763-5. http://dx.doi.org/10.1128/JCM.41.4.1763-1765.2003.
- Sharma JK, Gopalkrishna V, Das BC. A simple method for elimination of unspecific amplifications in polymerase chain reaction. Nucleic Acids Res 1992;20:6117-18. http://dx.doi.org/10.1093/nar/20.22.6117.
- Bae S, Wuertz S. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl Environ Microb 2009;75:2940-4. http://dx.doi.org/10.1128/AEM.01333-08.
- Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Ag 2007;30:S7-15. http://dx.doi.org/10.1016/j.ijantimicag.2007.06.024.
- Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 2006;44:1018-28. http://dx.doi.org/10.1128/JCM.44.3.1018-1028.2006.
- Harrell FE. Regression Modeling Strategies: with Applications to Linear Models, Logistic Regression, and Survival Analysis. Nashville, TN: Springer; 2001.
- Prompers L, Huijberts M, Apelqvist J, Jude E, Piaggesi A, Bakker K, et al. High prevalence of ischaemia, infection and serious comorbidity in patients with diabetic foot disease in Europe. Baseline results from the Eurodiale study. Diabetologia 2007;50:18-25. http://dx.doi.org/10.1007/s00125-006-0491-1.
- Ince P, Kendrick D, Game F, Jeffcoate W. The association between baseline characteristics and the outcome of foot lesions in a UK population with diabetes. Diabet Med 2007;24:977-81. http://dx.doi.org/10.1111/j.1464-5491.2007.02189.x.
- Schaper NC, Andros G, Apelqvist J, Bakker K, Lammer J, Lepantalo M, et al. Specific guidelines for the diagnosis and treatment of peripheral arterial disease in a patient with diabetes and ulceration of the foot 2011. Diabetes Metab Res Rev 2012;28:S236-7. http://dx.doi.org/10.1002/dmrr.2252.
- Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999;94:496-509. http://dx.doi.org/10.1080/01621459.1999.10474144.
- Li KH, Meng XL, Raghunathan TE, Rubin DB. Significance levels from repeated p-values with multiply-imputed data. Stat Sin 1991;1:65-92.
- Christman AL, Selvin E, Margolis DJ, Lazarus GS, Garza LA. Hemoglobin A1c predicts healing rate in diabetic wounds. J Invest Dermatol 2011;131:2121-7. http://dx.doi.org/10.1038/jid.2011.176.
- Margolis DJ, Allen-Taylor L, Hoffstad O, Berlin JA. Diabetic neuropathic foot ulcers: the association of wound size, wound duration, and wound grade on healing. Diabetes Care 2002;25:1835-9. http://dx.doi.org/10.2337/diacare.25.10.1835.
- Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, Scherubel M. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen 2006;14:548-57. http://dx.doi.org/10.1111/j.1743-6109.2006.00162.x.
- Kottner J, Audige L, Brorson S, Donner A, Gajewski BJ, Hrobjartsson A, et al. Guidelines for Reporting Reliability and Agreement Studies (GRRAS) were proposed. Int J Nurs Stud 2011;48:661-71. http://dx.doi.org/10.1016/j.ijnurstu.2011.01.016.
- Edmonds M, Foster A. The use of antibiotics in the diabetic foot. Am J Surg 2004;187:25S-28S. http://dx.doi.org/10.1016/S0002-9610(03)00300-3.
- Note 37944: Plots to Assess the Proportional Odds Assumption in an Ordinal Logistic Model. Cary, NC: SAS Institute Inc.; 2012.
Appendix 1 Supplementary information for Chapter 2
Main study sample size: patterns of agreement and disagreement
The sample size required depended on two key factors: the proportion of pairs for which the two samples disagree (discordance) and the clinically significant difference. The amount of discordance is dependent on the prevalence of the pathogen, with pathogens with low prevalence having much lower levels of discordance than pathogens with a high degree of prevalence. Therefore, the sample size had to cover both low and high levels of discordance to allow for pathogens with low and high prevalence. Assuming a lower prevalence level of 10% and 5% discordance, we assumed the following scenario, as summarised in Table 74.
| Sample | Tissue + | Tissue – | Total |
|---|---|---|---|
| Swab + | 7.5% | 1% | 8.5% |
| Swab – | 4% | 87.5% | 91.5% |
| Total | 11.5% | 88.5% | 100% |
This allowed us to formulate the sample size based on the McNemar’s test for a difference as follows (Figure 15):
-
significance: two-sided 5% level of significance
-
power, %: 80
-
discordance, %: 1 + 4 = 5
-
difference, %: 4 – 1 = 3
-
McNemar’s test: n = 399 pairs
-
kappa: rounding up to n = 400 provides κ = 0.7299.
FIGURE 15.
nQuery Advisor output for the CODIFI main sample size.

All participating centres are listed in Table 75.
| Centre | |
|---|---|
| Code | Name |
| N00003 | Queen Elizabeth Hospital, Birmingham |
| N00006 | James Cook University Hospital, Middlesbrough |
| N00034 | New Cross Hospital, Wolverhampton |
| N00036 | Norfolk and Norwich University Hospital, Norwich |
| N00040 | Pinderfields General Hospital, Wakefield |
| N00050 | St James’s University Hospital, Leeds |
| N00073 | South Tyneside District General, South Shields |
| N00075 | Bradford Royal Infirmary, Bradford |
| N00076 | Harrogate District Hospital, Harrogate |
| N00077 | Huddersfield Royal Infirmary, Huddersfield |
| N00080 | Manchester Royal Infirmary, Manchester |
| N00081 | Royal Lancaster Infirmary, Lancaster |
| N00163 | Queen Elizabeth Hospital, King’s Lynn |
| N00199 | Scarborough General Hospital, Scarborough |
| N00251 | University Hospital Lewisham, London |
| N00260 | North Manchester General Hospital, Manchester |
| N00294 | Weston General Hospital, Weston-super-Mare |
| N00390 | University Hospital of North Tees, Stockton-on-Tees |
| N00449 | Fairfield General Hospital, Bury |
| N00470 | Tameside General Hospital, Ashton-under-Lyne |
| N00488 | Royal Oldham Hospital, Oldham |
| N00511 | University Hospital, Coventry |
| N00522 | University Hospital of North Staffordshire, Stoke-on-Trent |
| N00936 | Bensham Hospital, Gateshead |
| N09829 | Chorley and South Ribble Hospital, Chorley |
| N15868 | Minerva Centre, Preston |
Pathogen derivations: group and summary of pathogens
To account for pathogens reported at various taxonomic ranks and to determine whether or not swab and tissue results reported the same pathogens, pathogens were compared according to predefined groups set out in Table 76 and in the details below [i.e. largely at the genus level and at the higher group level where further detail was not reported from the laboratory result (e.g. Gram-positive cocci rather than S. aureus)].
| Level grouping | Isolate/s | Isolates belonging to this group |
|---|---|---|
| Groups of isolates | Gram-positive cocci | Enterococcus, Gemella, Helcococcus, Peptoniphilus, Staphylococcus, Streptococcus, Pediococcus |
| Gram-negative cocci | Neisseria | |
| Gram-positive bacilli | Actinomyces, Bacillus, Clostridium, Corynebacterium, Propionibacterium | |
| Gram-negative bacilli | Achromobacter, Acinetobacter, Alcaligenes, Bacteroides, Citrobacter, Enterobacter, Escherichia, Fusobacterium, Klebsiella, Morganella, Prevotella, Proteus, Pseudomonas, Raoultella, Serratia, Stenotrophomonas, Moraxella, Pasteurella | |
| Enterobacteriaceae including coliforms (interest at the genus level) | Enterobacter, Citrobacter, Escherichia, Klebsiella, Morganella, Proteus, Raoultella, Serratia, coliform | |
| Overall anaerobes (interest at the cocci/rod level) | Anaerobes Anaerobic cocci: Peptococcus, Peptoniphilus, Peptostreptococcus, Veillonella Anaerobic rods: Actinomyces, Bacteroides, Clostridium, Fusobacterium, Lactobacillus, Prevotella, Propionibacterium |
|
| Genus-level isolates | Achromobacter | Achromobacter xylosoxidans |
| Acinetobacter | Acinetobacter baumannii | |
| Alcaligenes | Alcaligenes faecalis | |
| Bacteroides | Bacteroides fragilis | |
| Candida | Candida albicans, Candida guilliermondii, Candida parapsilosis | |
| Citrobacter | Citrobacter braakii, Citrobacter sedlakii, Citrobacter freundii, Citrobacter koseri | |
| CNS | Staphylococcus capitis, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus pettenkoferi, Staphylococcus simulans | |
| Corynebacterium | Corynebacterium amycolatum, Corynebacterium striatum, diptheroid | |
| Enterobacter | Enterobacter aerogenes, Enterobacter cloacae | |
| Enterococcus (excluding vancomycin resistant) | Enterococcus faecalis (excluding vancomycin resistant), Enterococcus raffinosus (excluding vancomycin resistant), group D Streptococcus (excluding vancomycin resistant) | |
| Enterococcus (vancomycin resistant) | E. faecalis (vancomycin resistant), E. raffinosus (vancomycin resistant), group D Streptococcus (vancomycin resistant) | |
| Escherichia | E. coli | |
| Gemella | Gemella morbillorum | |
| Helcococcus | Helcococcus kunzii | |
| Klebsiella | Klebsiella oxytoca, Klebsiella pneumoniae | |
| Morganella | Morganella morganii | |
| Neisseria | ||
| Pasteurella | Pasteurella dagmatis | |
| Pediococcus | ||
| Peptoniphilus | Peptoniphilus asaccharolyticus | |
| Prevotella | Prevotella bivia, Prevotella melaninogenica | |
| Proteus | Proteus mirabilis, Proteus vulgaris | |
| Pseudomonas | Pseudomonas aeruginosa | |
| Raoultella | Raoultella planticola | |
| Serratia | Serratia marcescens | |
| Stenotrophomonas | Stenotrophomonas maltophilia | |
| Streptococcus | Streptococcus agalactiae, Streptococcus anginosus, Streptococcus constellatus, Streptococcus dysgalactiae, Streptococcus gallolyticus, Streptococcus milleri, group B, C, G Streptococcus, Viridans, Streptococcus | |
| Species-level isolates | S. aureus (excluding MRSA) | |
| MRSA | ||
| Other | Coccobacillus, Gram-positive coccobacillus, Staphylococcus | |
| Isolates not likely to represent pathogenic organisms | Enteric flora, faecal flora, mixed skin/normal flora, pus, yeast | |
-
For the majority of pathogens, interest was at the genus level.
-
If, however, over all the sample results, only one species belonging to the genus is reported, the species is reported.
-
-
Interest was at the subgenus or species level for: CNS; S. aureus, by meticillin resistance; and at the subgenus level for Enterococcus, depending on vancomycin resistance.
-
Pathogens could also be reported at a level higher than genus, either at the family level or as per the below groups. In the comparison of pathogens within each sample, the groups of pathogens were compared as per:
-
Gram-positive cocci
-
Gram-negative cocci
-
Gram-positive bacilli
-
Gram-negative bacilli
-
anaerobes:
-
anaerobic cocci
-
anaerobic rods
-
-
family name.
-
-
If a pathogen was reported at the genus level in one sample but at the family or group level in another, samples were not considered to contain the same pathogens. Table 76 sets out a number of examples.
-
Enterobacteriaceae (including coliforms) is included in Table 77 (and in cross-tabulations within Chapter 2) as a group of pathogens in addition to the respective genus-level pathogens of interest comprising this group, owing to the reporting of ‘coliforms’ without detail of the specific genus or species.
| Outcome | Swab results | Tissue results |
|---|---|---|
| Swab and tissue sampling report the same pathogens | E. coli (Genus = Escherichia) | Escherichia |
| Corynebacterium | Diptheroid (used to represent corynebacteria) | |
| Coliforms (these are Enterobacteriaceae) | Enterobacteriaceae | |
| Swab sampling reports same pathogens as tissue sampling plus extra pathogens | Acinetobacter, Escherichia | Acinetobacter |
| Coliforms, S. aureus | Enterobacteriaceae | |
| Gram-positive bacilli, Corynebacterium | Gram-positive bacilli | |
| Tissue sampling reports same pathogens as swab plus extra pathogens | Anaerobic cocci | Anaerobic cocci, Peptococcus |
| Pseudomonas | Pseudomonas, Citrobacter | |
| Gram-negative bacilli | Gram-negative bacilli, Gram-negative cocci | |
| Both tissue and swab sampling report different pathogens (with or without overlap) | Gram-positive cocci (Enterococcus is a Gram-positive cocci, but insufficient information is available to determine if they refer to the same isolate) | Enterococcus |
| MRSA (interested at species level) | Staphylococcus | |
| CNS (interested in differentiating this group) | Staphylococcus | |
| Anaerobes (insufficient information to determine type), Acinetobacter | Anaerobic cocci, Acinetobacter |
Derivation for the summary and number of pathogens in the presence of multiple pathogens at the level of interest
As detailed in Chapter 2, Methods, where more than one strain or species of a pathogen was reported within a sample, a single pathogen at the level of interest was retained for comparison with the corresponding swab or tissue sample in the summary of pathogens and included in the count of the number of pathogens within the sample. Table 78 presents the data for which this derivation was applied.
| Sample | Trial number | Pathogen retained (at level of interest) | Pathogens reported |
|---|---|---|---|
| Swab | 234 | Streptococcus | Streptococcus and group B Streptococcus |
| 235 | Coliform | Coliform × 2 | |
| 298 | S. aureus (excluding MRSA) | S. aureus × 2 | |
| 386 | Coliform | Coliform × 2 | |
| Tissue | 167 | CNS | CNS × 2 |
| 212 | Streptococcus | Group B and group C Streptococcus | |
| 235 | Escherichia | E. coli × 2 | |
| 278 | Streptococcus | Streptococcus and Streptococcus dysgalactiae | |
| 294 | Corynebacterium | Corynebacterium × 2 and diptheroid | |
| 304 | Corynebacterium | Corynebacterium × 2 | |
| 344 | CNS | CNS and Staphylococcus pettenkoferi | |
| 386 | Coliform | Coliform × 2 |
Derivation for the summary and number of pathogens in the presence of results from a Gram stain
As detailed in Chapter 2, Methods, a derivation was applied for samples from which results of a Gram stain were reported in addition to those from the culture. As per Table 79, Gram stain results (in shaded boxes) were compared with pathogens reported within the corresponding culture result and pathogens belonging to the group of pathogens reported by the Gram stain were further identified (matched by colour in Table 79). Where a pathogen belonged to the same group as that reported by the Gram stain, it was considered likely that both referred to same pathogen and the corresponding Gram stain result was excluded (indicated by a strikethrough in Table 79) from the summary and number of pathogens reported from the swab or tissue sample. Conversely, where the results of a Gram stain were provided and no pathogens identified by the culture belonged to the group identified by the Gram stain, all pathogens were included
| Sample | Trial number | Pathogens | Derivation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Swab | 52 | CNS | Anaerobic GNB | No change | |||||
| 154 | Coliform | Candida | GPC | GNB | GNB removed as coliform in group | ||||
| 276 | S. aureus | GNB | No change | ||||||
| 279 | GNB | No change | |||||||
| Tissue | 9 | GPC | GNB | Coliform | Anaerobes | GNB removed as coliform in group | |||
| 12 | S. aureus | GPB | No change | ||||||
| 43 | Klebisella oxytoca | GPC | GNB | GNB removed as K. oxytoca in group | |||||
| 45 | S. aureus | GPB | No change | ||||||
| 52 | CNS | Enterococcus | Anaerobic GNB | No change | |||||
| 55 | GNB | Coliform | GNB removed as coliform in group | ||||||
| 82 | S. aureus | GPC | GPC removed as S. aureus in group | ||||||
| 96 | MRSA | GPC | GNB | GPB | GPC removed as MRSA in group | ||||
| 132 | GNB | Enterobacter | GNB removed as Enterobacter in group | ||||||
| 138 | GPC | No change | |||||||
| 147 | Anaerobic cocci | GPB | Group G Streptococcus | GPC | GPC removed as group G Streptococcus in group | ||||
| 148 | GPC | No change | |||||||
| 185 | Group G Streptococcus | GPC | GPC removed as group G Streptococcus in group | ||||||
| 204 | MRSA | GNB | Enterobacter cloacae | GNB removed as E. cloacae in group | |||||
| 207 | Pseudomonas | GPC | GNB | GNB removed as Pseudomonas in group | |||||
| 215 | GNB | Pseudomonas aeruginosa | GNB removed as P. aeruginosa in group | ||||||
| 239 | GPC | GNB | No change | ||||||
| 253 | S. aureus | G. morbillorum | GPB | Gram positive cocco-bacillus | Streptococcus constellatus | GPC | GNB | GPC removed as S. aureus, G. morbillorum and S. constellatus in group | |
| 254 | Streptococcus dysgalactiae | GPC | GPC removed as S. dysgalactiae in group | ||||||
| 270 | MRSA | GPC | GPC removed as MRSA belongs to group | ||||||
| 279 | GNB | GPC | No change | ||||||
| 292 | S. aureus | GPC | GPC removed as S. aureus in group | ||||||
| 293 | S. aureus | GPC | GPC removed as S. aureus in group | ||||||
| 299 | GPB | No change | |||||||
| 301 | GPC | GNB | No change | ||||||
| 302 | Diptheroid | Neisseria | Streptococcus dysgalactiae | GPC | GNB | GPC removed as S. dysgalactiae in group | |||
| 311 | S. aureus | Diptheroid | Candida guilliermondii | GPB | GPC | GPB removed as diptheroid in group/GPC removed as S. aureus in group | |||
| 322 | S. aureus | CNS | GPC | GPC removed as S. aureus and CNS in group | |||||
| 331 | S. aureus | GPC | GPC removed as S. aureus in group | ||||||
| 354 | S. aureus | Streptococcus | GNB | No change | |||||
| 356 | GPC | GPB | GNB | No change | |||||
| 369 | E. coli | GPC | GNB | Citrobacter koseri | Proteus mirabilis | GNB removed as E. coli, C. koseri and P. mirabilis in group | |||
Semiquantitative extent of bacterial growth by type of diabetic foot ulcer
Table 80 presents cross-tabulations for the extent of growth for the most prevalent pathogens by the type of ulcer (ischaemic/both ischaemic and neuropathic and neuropathic only).
| Tissue results: level of growth, n (%) | Total, n (%) | |||||
|---|---|---|---|---|---|---|
| Not reported | Reported: no growth | + | ++ | +++ | ||
| Gram-positive cocci: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 60 (30.9) | 5 (2.6) | 10 (5.2) | 8 (4.1) | 9 (4.6) | 92 (47.4) |
| Reported: no growth | 2 (1.0) | 14 (7.2) | 1 (0.5) | 0 (0.0) | 1 (0.5) | 18 (9.3) |
| + | 2 (1.0) | 0 (0.0) | 11 (5.7) | 1 (0.5) | 0 (0.0) | 14 (7.2) |
| ++ | 3 (1.5) | 0 (0.0) | 8 (4.1) | 11 (5.7) | 5 (2.6) | 27 (13.9) |
| +++ | 4 (2.1) | 2 (1.0) | 3 (1.5) | 6 (3.1) | 28 (14.4) | 43 (22.2) |
| Total | 71 (36.6) | 21 (10.8) | 33 (17.0) | 26 (13.4) | 43 (22.2) | n = 194 |
| Gram-positive cocci: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 56 (28.1) | 7 (3.5) | 15 (7.5) | 6 (3.0) | 7 (3.5) | 91 (45.7) |
| Reported: no growth | 1 (0.5) | 19 (9.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 20 (10.1) |
| + | 2 (1.0) | 1 (0.5) | 11 (5.5) | 2 (1.0) | 0 (0.0) | 16 (8.0) |
| ++ | 0 (0.0) | 0 (0.0) | 4 (2.0) | 4 (2.0) | 5 (2.5) | 13 (6.5) |
| +++ | 0 (0.0) | 1 (0.5) | 6 (3.0) | 15 (7.5) | 37 (18.6) | 59 (29.6) |
| Total | 59 (29.6) | 28 (14.1) | 36 (18.1) | 27 (13.6) | 49 (24.6) | n = 199 |
| Gram-negative bacilli: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 122 (62.9) | 2 (1.0) | 6 (3.1) | 6 (3.1) | 11 (5.7) | 147 (75.8) |
| Reported: no growth | 1 (0.5) | 12 (6.2) | 1 (0.5) | 1 (0.5) | 1 (0.5) | 16 (8.2) |
| + | 0 (0.0) | 0 (0.0) | 3 (1.5) | 0 (0.0) | 1 (0.5) | 4 (2.1) |
| ++ | 1 (0.5) | 1 (0.5) | 0 (0.0) | 4 (2.1) | 0 (0.0) | 6 (3.1) |
| +++ | 0 (0.0) | 0 (0.0) | 3 (1.5) | 2 (1.0) | 16 (8.2) | 21 (10.8) |
| Total | 124 (63.9) | 15 (7.7) | 13 (6.7) | 13 (6.7) | 29 (14.9) | n = 194 |
| Gram-negative bacilli: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 128 (64.3) | 3 (1.5) | 8 (4.0) | 6 (3.0) | 7 (3.5) | 152 (76.4) |
| Reported: no growth | 3 (1.5) | 5 (2.5) | 1 (0.5) | 2 (1.0) | 3 (1.5) | 14 (7.0) |
| + | 2 (1.0) | 1 (0.5) | 3 (1.5) | 0 (0.0) | 1 (0.5) | 7 (3.5) |
| ++ | 2 (1.0) | 1 (0.5) | 4 (2.0) | 6 (3.0) | 2 (1.0) | 15 (7.5) |
| +++ | 3 (1.5) | 1 (0.5) | 2 (1.0) | 3 (1.5) | 2 (1.0) | 11 (5.5) |
| Total | 138 (69.3) | 11 (5.5) | 18 (9.0) | 17 (8.5) | 15 (7.5) | n = 199 |
| Enterobacteriaceae (including coliforms): ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 142 (73.2) | 3 (1.5) | 5 (2.6) | 1 (0.5) | 9 (4.6) | 160 (82.5) |
| Reported: no growth | 1 (0.5) | 12 (6.2) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 15 (7.7) |
| + | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| ++ | 1 (0.5) | 1 (0.5) | 0 (0.0) | 2 (1.0) | 0 (0.0) | 4 (2.1) |
| +++ | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 12 (6.2) | 14 (7.2) |
| Total | 144 (74.2) | 16 (8.2) | 7 (3.6) | 5 (2.6) | 22 (11.3) | n = 194 |
| Enterobacteriaceae (including coliforms): neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 147 (73.9) | 3 (1.5) | 7 (3.5) | 4 (2.0) | 4 (2.0) | 165 (82.9) |
| Reported: no growth | 3 (1.5) | 5 (2.5) | 2 (1.0) | 0 (0.0) | 2 (1.0) | 12 (6.0) |
| + | 2 (1.0) | 1 (0.5) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 4 (2.0) |
| ++ | 2 (1.0) | 0 (0.0) | 3 (1.5) | 3 (1.5) | 0 (0.0) | 8 (4.0) |
| +++ | 5 (2.5) | 1 (0.5) | 1 (0.5) | 2 (1.0) | 1 (0.5) | 10 (5.0) |
| Total | 159 (79.9) | 10 (5.0) | 14 (7.0) | 9 (4.5) | 7 (3.5) | n = 199 |
| Overall anaerobes: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 150 (77.3) | 2 (1.0) | 6 (3.1) | 5 (2.6) | 6 (3.1) | 169 (87.1) |
| Reported: no growth | 3 (1.5) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (2.6) |
| + | 4 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 5 (2.6) |
| ++ | 0 (0.0) | 0 (0.0) | 1 (0.5) | 2 (1.0) | 3 (1.5) | 6 (3.1) |
| +++ | 3 (1.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (3.1) | 9 (4.6) |
| Total | 160 (82.5) | 4 (2.1) | 7 (3.6) | 7 (3.6) | 16 (8.2) | n = 194 |
| Overall anaerobes: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 149 (74.9) | 7 (3.5) | 6 (3.0) | 9 (4.5) | 5 (2.5) | 176 (88.4) |
| Reported: no growth | 1 (0.5) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (1.5) |
| + | 3 (1.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (1.5) |
| ++ | 1 (0.5) | 1 (0.5) | 0 (0.0) | 2 (1.0) | 3 (1.5) | 7 (3.5) |
| +++ | 4 (2.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 4 (2.0) | 10 (5.0) |
| Total | 158 (79.4) | 10 (5.0) | 6 (3.0) | 13 (6.5) | 12 (6.0) | n = 199 |
| Gram-positive bacilli: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 174 (89.7) | 4 (2.1) | 5 (2.6) | 4 (2.1) | 5 (2.6) | 192 (99.0) |
| ++ | 1 (0.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) |
| Total | 175 (90.2) | 4 (2.1) | 5 (2.6) | 5 (2.6) | 5 (2.6) | n = 194 |
| Gram-positive bacilli: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 175 (87.9) | 2 (1.0) | 10 (5.0) | 6 (3.0) | 4 (2.0) | 197 (99.0) |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) |
| +++ | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| Total | 175 (87.9) | 2 (1.0) | 11 (5.5) | 6 (3.0) | 5 (2.5) | n = 199 |
| Streptococcus: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 166 (85.6) | 3 (1.5) | 1 (0.5) | 0 (0.0) | 4 (2.1) | 174 (89.7) |
| Reported: no growth | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 3 (1.5) |
| + | 2 (1.0) | 0 (0.0) | 3 (1.5) | 0 (0.0) | 0 (0.0) | 5 (2.6) |
| ++ | 1 (0.5) | 0 (0.0) | 0 (0.0) | 3 (1.5) | 0 (0.0) | 4 (2.1) |
| +++ | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 6 (3.1) | 8 (4.1) |
| Total | 170 (87.6) | 5 (2.6) | 4 (2.1) | 4 (2.1) | 11 (5.7) | n = 194 |
| Streptococcus: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 161 (80.9) | 3 (1.5) | 4 (2.0) | 1 (0.5) | 2 (1.0) | 171 (85.9) |
| Reported: no growth | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) |
| + | 0 (0.0) | 0 (0.0) | 2 (1.0) | 1 (0.5) | 0 (0.0) | 3 (1.5) |
| ++ | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 2 (1.0) | 4 (2.0) |
| +++ | 1 (0.5) | 0 (0.0) | 2 (1.0) | 6 (3.0) | 10 (5.0) | 19 (9.5) |
| Total | 162 (81.4) | 5 (2.5) | 9 (4.5) | 9 (4.5) | 14 (7.0) | n = 199 |
| Enterococcus (excluding vancomycin resistant): ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 166 (85.6) | 2 (1.0) | 2 (1.0) | 7 (3.6) | 3 (1.5) | 180 (92.8) |
| Reported: no growth | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) |
| ++ | 1 (0.5) | 1 (0.5) | 0 (0.0) | 3 (1.5) | 1 (0.5) | 6 (3.1) |
| +++ | 3 (1.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (1.5) | 6 (3.1) |
| Total | 170 (87.6) | 5 (2.6) | 2 (1.0) | 10 (5.2) | 7 (3.6) | n = 194 |
| Enterococcus (excluding vancomycin resistant): neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 168 (84.4) | 4 (2.0) | 7 (3.5) | 5 (2.5) | 4 (2.0) | 188 (94.5) |
| Reported: no growth | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) |
| + | 2 (1.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 3 (1.5) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 5 (2.5) | 6 (3.0) |
| Total | 170 (85.4) | 6 (3.0) | 8 (4.0) | 6 (3.0) | 9 (4.5) | n = 199 |
| CNS: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 178 (91.8) | 1 (0.5) | 7 (3.6) | 2 (1.0) | 2 (1.0) | 190 (97.9) |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) |
| +++ | 1 (0.5) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 3 (1.5) |
| Total | 179 (92.3) | 2 (1.0) | 7 (3.6) | 3 (1.5) | 3 (1.5) | n = 194 |
| CNS: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 168 (84.4) | 4 (2.0) | 14 (7.0) | 5 (2.5) | 3 (1.5) | 194 (97.5) |
| + | 0 (0.0) | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 2 (1.0) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) |
| Total | 168 (84.4) | 4 (2.0) | 16 (8.0) | 5 (2.5) | 6 (3.0) | n = 199 |
| Corynebacterium: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 179 (92.3) | 3 (1.5) | 2 (1.0) | 4 (2.1) | 4 (2.1) | 192 (99.0) |
| ++ | 1 (0.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) |
| Total | 180 (92.8) | 3 (1.5) | 2 (1.0) | 5 (2.6) | 4 (2.1) | n = 194 |
| Corynebacterium: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 177 (88.9) | 2 (1.0) | 8 (4.0) | 6 (3.0) | 4 (2.0) | 197 (99.0) |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) |
| +++ | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| Total | 177 (88.9) | 2 (1.0) | 9 (4.5) | 6 (3.0) | 5 (2.5) | n = 199 |
| Pseudomonas: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 175 (90.2) | 1 (0.5) | 0 (0.0) | 2 (1.0) | 2 (1.0) | 180 (92.8) |
| Reported: no growth | 0 (0.0) | 2 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) |
| + | 1 (0.5) | 0 (0.0) | 2 (1.0) | 0 (0.0) | 1 (0.5) | 4 (2.1) |
| ++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) |
| +++ | 1 (0.5) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 5 (2.6) | 7 (3.6) |
| Total | 177 (91.2) | 3 (1.5) | 3 (1.5) | 3 (1.5) | 8 (4.1) | n = 194 |
| Pseudomonas: neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 184 (92.5) | 1 (0.5) | 1 (0.5) | 1 (0.5) | 187 (94.0) | |
| Reported: no growth | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | |
| + | 3 (1.5) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 4 (2.0) | |
| ++ | 2 (1.0) | 1 (0.5) | 2 (1.0) | 0 (0.0) | 5 (2.5) | |
| +++ | 1 (0.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 2 (1.0) | |
| Total | 190 (95.5) | 3 (1.5) | 3 (1.5) | 3 (1.5) | n = 199 | |
| S. aureus (excluding MRSA): ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 135 (69.6) | 0 (0.0) | 6 (3.1) | 2 (1.0) | 1 (0.5) | 144 (74.2) |
| Reported: no growth | 2 (1.0) | 8 (4.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (5.2) |
| + | 0 (0.0) | 0 (0.0) | 5 (2.6) | 1 (0.5) | 0 (0.0) | 6 (3.1) |
| ++ | 4 (2.1) | 0 (0.0) | 5 (2.6) | 4 (2.1) | 1 (0.5) | 14 (7.2) |
| +++ | 1 (0.5) | 0 (0.0) | 4 (2.1) | 5 (2.6) | 10 (5.2) | 20 (10.3) |
| Total | 142 (73.2) | 8 (4.1) | 20 (10.3) | 12 (6.2) | 12 (6.2) | n = 194 |
| S. aureus (excluding MRSA): neuropathic only | ||||||
| Swab results | ||||||
| Not reported | 118 (59.3) | 1 (0.5) | 5 (2.5) | 1 (0.5) | 0 (0.0) | 125 (62.8) |
| Reported: no growth | 2 (1.0) | 17 (8.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 19 (9.5) |
| + | 3 (1.5) | 1 (0.5) | 9 (4.5) | 0 (0.0) | 0 (0.0) | 13 (6.5) |
| ++ | 1 (0.5) | 0 (0.0) | 3 (1.5) | 4 (2.0) | 1 (0.5) | 9 (4.5) |
| +++ | 2 (1.0) | 1 (0.5) | 5 (2.5) | 5 (2.5) | 20 (10.1) | 33 (16.6) |
| Total | 126 (63.3) | 20 (10.1) | 22 (11.1) | 10 (5.0) | 21 (10.6) | n = 199 |
| MRSA: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 171 (88.1) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 1 (0.5) | 174 (89.7) |
| Reported: no growth | 0 (0.0) | 4 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (2.1) |
| + | 1 (0.5) | 0 (0.0) | 3 (1.5) | 0 (0.0) | 0 (0.0) | 4 (2.1) |
| ++ | 0 (0.0) | 0 (0.0) | 2 (1.0) | 1 (0.5) | 2 (1.0) | 5 (2.6) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 6 (3.1) | 7 (3.6) |
| Total | 172 (88.7) | 4 (2.1) | 6 (3.1) | 3 (1.5) | 9 (4.6) | n = 194 |
| MRSA: ischaemic/both ischaemic and neuropathic | ||||||
| Swab results | ||||||
| Not reported | 190 (95.5) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 1 (0.5) | 192 (96.5) |
| +++ | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 5 (2.5) | 7 (3.5) |
| Total | 190 (95.5) | 0 (0.0) | 0 (0.0) | 3 (1.5) | 6 (3.0) | n = 199 |
Missing data
Table 81 presents the pattern of missing baseline covariate data considered within the analysis models for the coprimary end points for the evaluable population in which an arbitrary missing data pattern was observed. There were no missing data on centre, 19 patients had missing ‘pre-sampling antibiotic therapy’ information, 5 patients had no information on use of antimicrobial wound dressing, 2 had wound duration information missing and 2 had both wound duration and ulcer-type information missing.
| Centre | Antimicrobial dressing | Pre-sampling antibiotic therapy | Ulcer type | Wound duration | Wagner ulcer grade | Total (N = 395), n (%) |
|---|---|---|---|---|---|---|
| ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | 367 (92.9) |
| ✗ | ✗ | ✗ | ✗ | Missing | ✗ | 2 (0.5) |
| ✗ | ✗ | ✗ | Missing | Missing | ✗ | 2 (0.5) |
| ✗ | ✗ | Missing | ✗ | ✗ | ✗ | 19 (4.8) |
| ✗ | Missing | ✗ | ✗ | ✗ | ✗ | 5 (1.3) |
Graphical plots to assess the proportional odds assumption for the coprimary end point: number of pathogens
Figures 16–21 present graphical plots of the empirical cumulative logit function for each covariate included in the ordinal regression analysis for the coprimary end point of number of pathogens.
FIGURE 16.
Plot to assess the proportional odds assumption for ulcer type (n = 393) (proportionality satisfied).
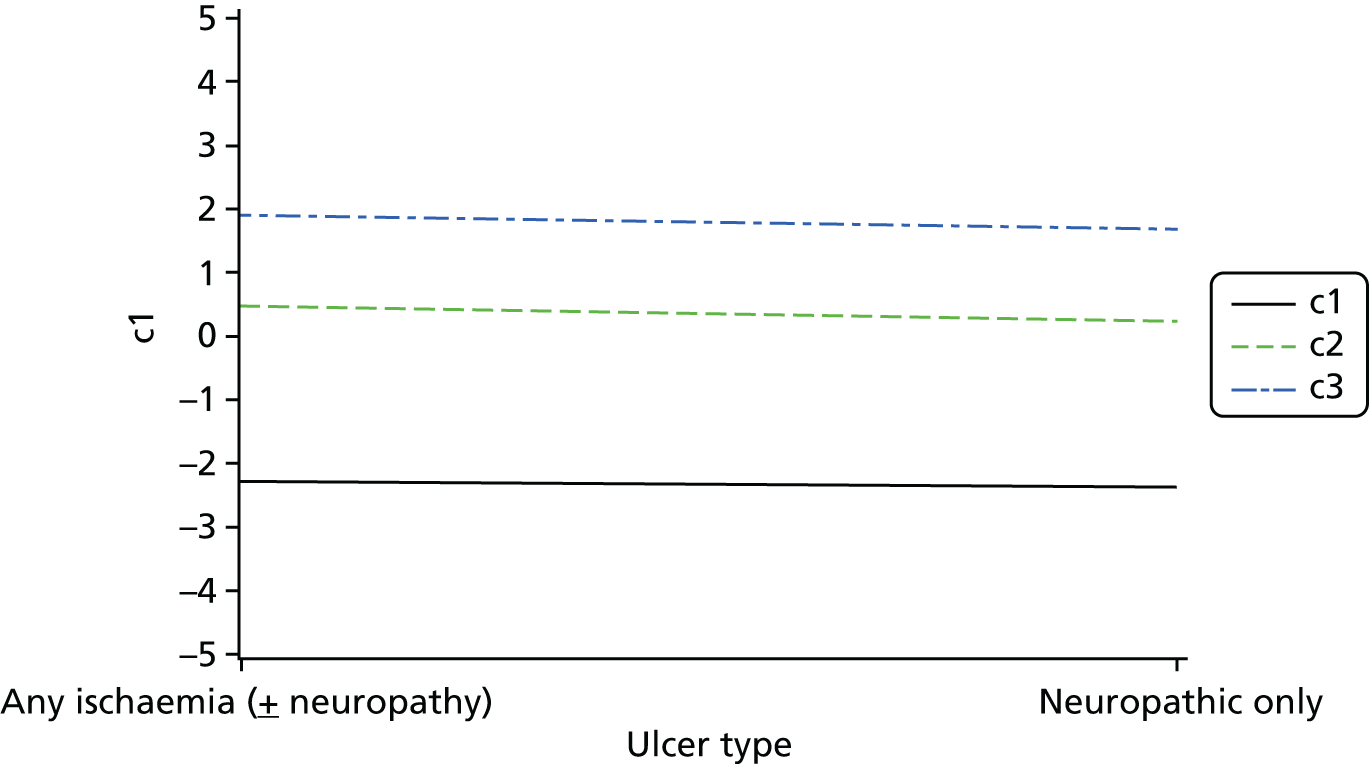
FIGURE 17.
Plot to assess the proportional odds assumption for ulcer grade (n = 395) (proportionality satisfied).

FIGURE 18.
Plot to assess the proportional odds assumption for previous antibiotic therapy (n = 376) (proportionality satisfied).

FIGURE 19.
Plot to assess the proportional odds assumption for antimicrobial dressing (n = 390) (proportionality satisfied).

FIGURE 20.
Plot to assess the proportional odds assumption for wound duration (n = 391) (proportionality satisfied).
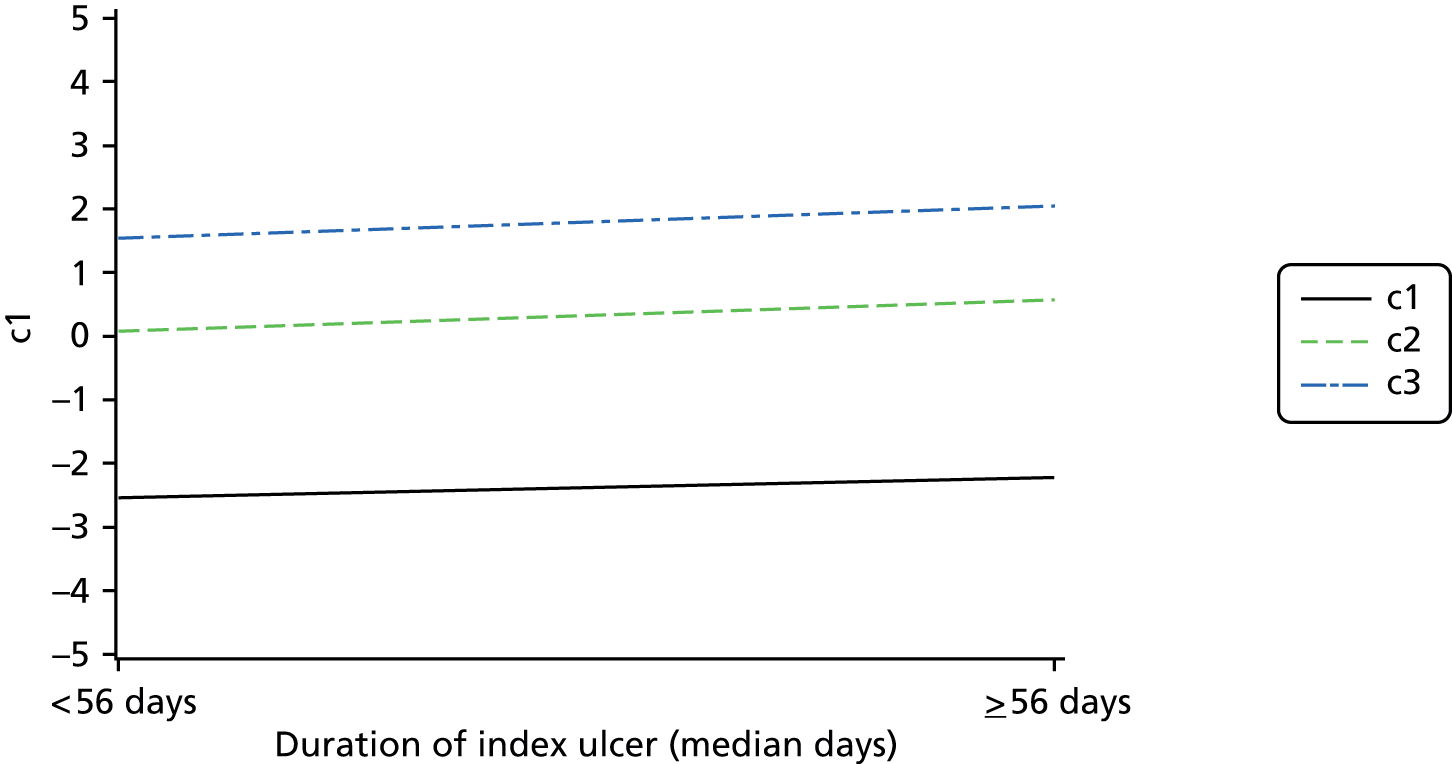
FIGURE 21.
Plot to assess the proportional odds assumption for centre (n = 395) (proportionality not satisfied: centre fitted in ordinal regression analysis as a random effect).

Given that there are four ordered response levels (outcome: swab sampling had one or more extra isolate reported; tissue and swab sampling had the same number of isolates reported; tissue sampling had one extra isolate reported; tissue sampling had two or more extra isolates reported), three cumulative logits were computed and plotted as per the SAS Note 37944. 116 To assess the proportional odds assumption for each covariate, if the empirical cumulative logits look approximately parallel, then this provides evidence that a proportional odds model is appropriate.
Each figure supports the proportional odds assumption with the exception of centre, which was instead fitted as a random effect in the regression model.
Appendix 2 Supplementary information for Chapter 3
Missing data
Table 82 presents the pattern of missing baseline covariate data considered within the analysis model for the evaluable clinical review population, in which an arbitrary missing data pattern is observed. There were no missing data on centre, 15 patients had missing ‘pre-sampling antibiotic therapy’ information, 5 had no information on use of antimicrobial wound dressing and 1 had missing wound duration information.
| Centre | Antimicrobial dressing | Pre-sampling antibiotic therapy | Ulcer type | Wound duration | Wagner ulcer grade | Total (N = 247), n (%) |
|---|---|---|---|---|---|---|
| ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | 226 (91.5) |
| ✗ | ✗ | ✗ | ✗ | Missing | ✗ | 1 (0.4) |
| ✗ | ✗ | Missing | ✗ | ✗ | ✗ | 15 (6.1) |
| ✗ | Missing | ✗ | ✗ | ✗ | ✗ | 5 (2.0) |
Patient vignette: 82766
A 75-year-old male with diabetes attended the clinic a few days ago because of a clinically infected foot ulcer. You are now asked to review the microbiology report and the antibiotics prescribed when the patient attended the clinic, and answer the following questions.
Prescribed antibiotic regimen: flucloxacillin
Microbiology laboratory report: S. aureus (growth: scanty; sensitive to: co-fluampicil, clarithromycin and erythromycin; resistant to: penicillin.)
Clinical assessment a few days ago indicated that the ulcer was classified as follows on the PEDIS scale:
Perfusion: grade 2
Extent: 2 cm × 2.5 cm
Depth: grade 1
Infection: grade 2
Sensation: grade 1
Question 1
Are there any pathogens identified in the laboratory report that are not covered by the prescribed antimicrobial regimen? Please consider both the pathogens and their sensitivity and resistance.
Yes □
No □
If ‘yes’ please list which pathogens are not covered by the existing antibiotic regimen:
__________________________________________________________________________________________________________________________________
Question 2
If you answered ‘yes’ to question 1, would knowing this information lead you to prescribe an alternative antibiotic regimen for this patient?
Yes □
No □
If ‘yes’ please describe the antibiotic regimen you would prescribe for this patient:
__________________________________________________________________________________________________________________________________
Signature. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . . . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .
Name
Date. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . . . . . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .
Patient vignette: 85277
A 75-year-old male with diabetes attended the clinic a few days ago because of a clinically infected foot ulcer. You are now asked to review the microbiology report and the antibiotics prescribed when the patient attended clinic, and answer the following questions.
Prescribed antibiotic regimen: flucloxacillin
Microbiology laboratory report: Staphylococcus aureus (Growth: ++; sensitive to: flucloxacillin; resistant to: no antibiotics recorded).
Clinical assessment a few days ago indicated that the ulcer was classified as follows on the PEDIS scale:
Perfusion: grade 2
Extent: 2 cm × 2.5 cm
Depth: grade 1
Infection: grade 2
Sensation: grade 1
Question 1
Are there any pathogens identified in the laboratory report that are not covered by the prescribed antimicrobial regimen? Please consider both the pathogens and their sensitivity and resistance.
Yes □
No □
If ‘yes’ please list which pathogens are not covered by the existing antibiotic regimen:
__________________________________________________________________________________________________________________________________
Question 2
If you answered ‘yes’ to question 1, would knowing this information lead you to prescribe an alternative antibiotic regimen for this patient?
Yes □
No □
If ‘yes’ please describe the antibiotic regimen you would prescribe for this patient:
__________________________________________________________________________________________________________________________________
Signature. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . . . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .
Name
Date. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . . . . . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .. . .
Appendix 3 Supplementary information for Chapter 5
Baseline characteristics for the follow-up population, compared with patients not in the follow-up population
Tables 83–85 present the baseline characteristics for patients in the follow-up population and those not in the follow-up population.
| Recruiting centre | In the follow-up population (N = 299), n (%) | Not in the follow-up population (N = 101), n (%) | Total: full analysis set (N = 400), n (%) |
|---|---|---|---|
| N00003: Queen Elizabeth Hospital Birmingham | 14 (4.7) | 7 (6.9) | 21 (5.3) |
| N00006: James Cook University Hospital | 4 (1.3) | 0 (0.0) | 4 (1.0) |
| N00034: New Cross Hospital Wolverhampton | 5 (1.7) | 0 (0.0) | 5 (1.3) |
| N00036: Norfolk and Norwich University Hospital | 19 (6.4) | 1 (1.0) | 20 (5.0) |
| N00040: Pinderfields General Hospital | 18 (6.0) | 3 (3.0) | 21 (5.3) |
| N00050: St James’s University Hospital Leeds | 29 (9.7) | 15 (14.9) | 44 (11.0) |
| N00073: South Tyneside District General | 1 (0.3) | 2 (2.0) | 3 (0.8) |
| N00075: Bradford Royal Infirmary | 28 (9.4) | 6 (5.9) | 34 (8.5) |
| N00076: Harrogate District Hospital | 7 (2.3) | 0 (0.0) | 7 (1.8) |
| N00077: Huddersfield Royal Infirmary | 5 (1.7) | 1 (1.0) | 6 (1.5) |
| N00080: Manchester Royal Infirmary | 19 (6.4) | 7 (6.9) | 26 (6.5) |
| N00081: Royal Lancaster Infirmary | 14 (4.7) | 3 (3.0) | 17 (4.3) |
| N00163: Queen Elizabeth Hospital King’s Lynn | 1 (0.3) | 1 (1.0) | 2 (0.5) |
| N00251: University Hospital Lewisham | 2 (0.7) | 0 (0.0) | 2 (0.5) |
| N00260: North Manchester General Hospital | 19 (6.4) | 2 (2.0) | 21 (5.3) |
| N00294: Weston General Hospital | 5 (1.7) | 0 (0.0) | 5 (1.3) |
| N00390: University Hospital of North Tees | 12 (4.0) | 6 (5.9) | 18 (4.5) |
| N00449: Fairfield Hospital | 2 (0.7) | 3 (3.0) | 5 (1.3) |
| N00470: Tameside General Hospital | 15 (5.0) | 13 (12.9) | 28 (7.0) |
| N00488: Royal Oldham Hospital | 27 (9.0) | 15 (14.9) | 42 (10.5) |
| N00511: University Hospitals Coventry and Warwick | 10 (3.3) | 10 (9.9) | 20 (5.0) |
| N00522: University Hospital of North Staffordshire | 4 (1.3) | 0 (0.0) | 4 (1.0) |
| N00936: Bensham Hospital | 13 (4.3) | 1 (1.0) | 14 (3.5) |
| N09829: Chorley and South Ribble hospital | 23 (7.7) | 3 (3.0) | 26 (6.5) |
| N15868: Minerva Centre, Preston | 3 (1.0) | 2 (2.0) | 5 (1.3) |
| Patient demographics | In the follow-up population (n = 299) | Not in the follow-up population (n = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Age, years | |||
| Mean (SD) | 64.3 (12.8) | 59.3 (14.2) | 63.1 (13.3) |
| Median (IQR) | 64.0 (56.0–74.0) | 58.0 (49.0–72.0) | 63.0 (54.0–73.0) |
| Range | 28–99 | 26–91 | 26–99 |
| Age at sampling (years, by median), n (%)a | |||
| ≤ 63 years | 146 (48.8) | 62 (61.4) | 208 (52.0) |
| > 63 years | 153 (51.2) | 39 (38.6) | 192 (48.0) |
| Sex, n (%) | |||
| Male | 233 (77.9) | 83 (82.2) | 316 (79.0) |
| Female | 66 (22.1) | 18 (17.8) | 84 (21.0) |
| Ethnicity, n (%) | |||
| White | 284 (95.0) | 93 (92.1) | 377 (94.3) |
| Other mixed background | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Asian: Indian | 2 (0.7) | 1 (1.0) | 3 (0.8) |
| Asian: Pakistani | 8 (2.7) | 3 (3.0) | 11 (2.8) |
| Other Asian background | 2 (0.7) | 0 (0.0) | 2 (0.5) |
| Black: Caribbean | 1 (0.3) | 2 (2.0) | 3 (0.8) |
| Black: African | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Other ethnic group | 0 (0.0) | 2 (2.0) | 2 (0.5) |
| Site of recruitment | |||
| Hospital ward | 38 (12.7) | 15 (14.9) | 53 (13.3) |
| Outpatient clinic | 241 (80.6) | 78 (77.2) | 319 (79.8) |
| Community clinic | 20 (6.7) | 8 (7.9) | 28 (7.0) |
| Diabetes details | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Diabetes type, n (%) | |||
| Type 1 | 40 (13.4) | 18 (17.8) | 58 (14.5) |
| Type 2 | 259 (86.6) | 83 (82.2) | 342 (85.5) |
| Duration of diabetes (years)a | |||
| Number of patients with missing data | 2 | 1 | 3 |
| Mean (SD) | 17.2 (11.1) | 15.5 (10.5) | 16.8 (11.0) |
| Median (IQR) | 15.0 (10.0–23.0) | 14.5 (8.0–21.5) | 15.0 (9.0–23.0) |
| Range | 0–57 | 0–44 | 0–57 |
| Duration of diabetes (split by median), n (%)b | |||
| < 15 years | 136 (45.5) | 50 (49.5) | 186 (46.5) |
| ≥ 15 years | 161 (53.8) | 50 (49.5) | 211 (52.8) |
| Missing | 2 (0.7) | 1 (1.0) | 3 (0.8) |
| Current diabetes treatment, n (%) | |||
| Yes | 289 (96.7) | 96 (95.0) | 385 (96.3) |
| No | 10 (3.3) | 5 (5.0) | 15 (3.8) |
| Diabetes treatment details, n (%)c | |||
| Oral hypoglycaemic agent | 77 (26.6) | 30 (31.3) | 107 (27.8) |
| Insulin | 126 (43.6) | 42 (43.8) | 168 (43.6) |
| Both, oral hypoglycaemic agent and insulin | 85 (29.4) | 24 (25.0) | 109 (28.3) |
| Other | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Total | 289 (100.0) | 96 (100.0) | 385 (100.0) |
| HbA1C (%) | |||
| Number of patients | 294 | 100 | 394 |
| Number of patients with missing data | 5 | 1 | 6 |
| Mean (SD) | 8.61 (2.24) | 9.04 (2.40) | 8.72 (2.29) |
| Median (IQR) | 8.10 (7.00–9.90) | 8.45 (6.95–10.70) | 8.10 (7.00–10.20) |
| Range | 4.6–17.2 | 5.4–17.1 | 4.6–17.2 |
Tables 86–93 present the clinical assessment, antibiotic regimen at baseline and ulcer characteristics and classification.
| Clinical assessment | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Temperature (°C) | |||
| Number of patients with missing data | 7 | 3 | 10 |
| Mean (SD) | 36.61 (0.62) | 36.58 (0.61) | 36.60 (0.62) |
| Median (IQR) | 36.60 (36.20–37.00) | 36.60 (36.20–37.00) | 36.60 (36.20–37.00) |
| Range | 35.0–39.5 | 35.1– 38.6 | 35.0–39.5 |
| Heart rate (beats/minute) | |||
| Number of patients with missing data | 3 | 1 | 4 |
| Mean (SD) | 80.8 (13.6) | 83.0 (13.5) | 81.4 (13.6) |
| Median (IQR) | 80.0 (72.0–90.0) | 81.5 (72.0–94.0) | 80.0 (72.0–90.5) |
| Range | 50–118 | 58–120 | 50–120 |
| Respiratory rate (breaths/minute) | |||
| Number of patients with missing data | 4 | 0 | 4 |
| Mean (SD) | 18.1 (4.5) | 17.9 (3.5) | 18.1 (4.3) |
| Median (IQR) | 18.0 (15.0–20.0) | 18.0 (15.0–20.0) | 18.0 (15.0–20.0) |
| Range | 9–40 | 10–28 | 9–40 |
| Ulcer characteristics | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Location of ulcer(s), n (%) | |||
| Ulcers on both right and left foot | 50 (16.7) | 10 (9.9) | 60 (15.0) |
| Ulcer(s) on right foot only | 125 (41.8) | 48 (47.5) | 173 (43.3) |
| Ulcer(s) on left foot only | 124 (41.5) | 43 (42.6) | 167 (41.8) |
| Total number of ulcers, n (%) | |||
| 1 | 194 (64.9) | 74 (73.3) | 268 (67.0) |
| 2 | 60 (20.1) | 18 (17.8) | 78 (19.5) |
| 3 | 36 (12.0) | 7 (6.9) | 43 (10.8) |
| 4 | 4 (1.3) | 2 (2.0) | 6 (1.5) |
| 5 | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| 6 | 3 (1.0) | 0 (0.0) | 3 (0.8) |
| 7 | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Total number of ulcers | |||
| Mean (SD) | 1.6 (1.0) | 1.4 (0.7) | 1.5 (0.9) |
| Median (IQR) | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) |
| Range | 1–7 | 1–4 | 1–7 |
| Index ulcer characteristics | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Foot containing index ulcer, n (%) | |||
| Right foot | 150 (50.2) | 55 (54.5) | 205 (51.3) |
| Left foot | 149 (49.8) | 46 (45.5) | 195 (48.8) |
| Other ulcers on index foot, n (%) | |||
| Single index ulcer on index foot | 222 (74.2) | 82 (81.2) | 304 (76.0) |
| > 1 ulcer on index foot | 77 (25.8) | 19 (18.8) | 96 (24.0) |
| Index ulcer location, n (%)a | |||
| Apex | 31 (10.4) | 16 (15.8) | 47 (11.8) |
| Interdigital | 18 (6.0) | 7 (6.9) | 25 (6.3) |
| Plantar | 133 (44.5) | 39 (38.6) | 172 (43.0) |
| Dorsum | 38 (12.7) | 18 (17.8) | 56 (14.0) |
| Digital | 70 (23.4) | 20 (19.8) | 90 (22.5) |
| Other | 7 (2.3) | 1 (1.0) | 8 (2.0) |
| Missing | 2 (0.7) | 0 (0.0) | 2 (0.5) |
| Duration of index ulcer (months) | |||
| Number of patients with missing data | 4 | 0 | 4 |
| Mean (SD) | 4.80 (8.28) | 7.86 (19.68) | 5.58 (12.28) |
| Median (IQR) | 1.84 (0.69–6.00) | 1.84 (0.69–4.60) | 1.84 (0.69–6.00) |
| Range | 0.1–75.0 | 0.2–144.0 | 0.1–144.0 |
| First or recurrent index ulcer, n (%) | |||
| Incident | 206 (68.9) | 82 (81.2) | 288 (72.0) |
| Recurrent | 91 (30.4) | 19 (18.8) | 110 (27.5) |
| Missing | 2 (0.7) | 0 (0.0) | 2 (0.5) |
| Aetiology of index ulcer, n (%) | |||
| Any ischaemia (± neuropathy) | 142 (47.5) | 54 (53.5) | 196 (49.0) |
| Neuropathic only | 155 (51.8) | 47 (46.5) | 202 (50.5) |
| Missing | 2 (0.7) | 0 (0.0) | 2 (0.5) |
| Antimicrobial dressing on the infected ulcer, n (%) | |||
| Yes | 175 (58.5) | 66 (65.3) | 241 (60.3) |
| No | 119 (39.8) | 35 (34.7) | 154 (38.5) |
| Missing | 5 (1.7) | 0 (0.0) | 5 (1.3) |
| Antibiotic regimen | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Currently on antibiotic regimen, n (%) | |||
| Yes | 139 (46.5) | 48 (47.5) | 187 (46.8) |
| No | 145 (48.5) | 49 (48.5) | 194 (48.5) |
| Missing | 15 (5.0) | 4 (4.0) | 19 (4.8) |
| Days spent on current antibiotic regimen | |||
| Number of patients | 138 | 48 | 186 |
| Number of patients with missing data | 1 | 0 | 1 |
| Mean (SD) | 15.2 (21.8) | 13.2 (22.2) | 14.6 (21.9) |
| Median (IQR) | 7.0 (4.0–16.0) | 5.5 (2.0–13.0) | 7.0 (3.0–14.0) |
| Range | 1–145 | 1–124 | 1–145 |
| Proposed new antibiotic regimen, n (%) | |||
| Yes | 198 (66.2) | 50 (49.5) | 248 (62.0) |
| No | 91 (30.4) | 42 (41.6) | 133 (33.3) |
| Missing | 10 (3.3) | 9 (8.9) | 19 (4.8) |
| Summary of patients pre and post sampling antibiotic regimen, n (%) | |||
| Not a pre-sampling antibiotic regimen with initiation immediately post sampling | 131 (43.8) | 37 (36.6) | 168 (42.0) |
| Not on a pre-sampling antibiotic regimen with no initiation immediately post sampling | 14 (4.7) | 12 (11.9) | 26 (6.5) |
| On a pre-sampling antibiotic regimen with or without a change immediately post sampling | 139 (46.5) | 48 (47.5) | 187 (46.8) |
| Unknown whether on a pre-sampling antibiotic regimen but initiation/change immediately post sampling | 15 (5.0) | 4 (4.0) | 19 (4.8) |
| PEDIS classification | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Perfusion (PAD), n (%) | |||
| Grade 1 | 147 (49.2) | 53 (52.5) | 200 (50.0) |
| Grade 2 | 146 (48.8) | 46 (45.5) | 192 (48.0) |
| Grade 3 | 6 (2.0) | 2 (2.0) | 8 (2.0) |
| Depth/tissue loss, n (%) | |||
| Grade 1 | 96 (32.1) | 35 (34.7) | 131 (32.8) |
| Grade 2 | 100 (33.4) | 34 (33.7) | 134 (33.5) |
| Grade 3 | 103 (34.4) | 32 (31.7) | 135 (33.8) |
| Infection, n (%) | |||
| Grade 1 | 0 (0.0) | 2 (2.0) | 2 (0.5) |
| Grade 2 | 104 (34.8) | 45 (44.6) | 149 (37.3) |
| Grade 3 skin/subcutaneous tissue | 185 (61.9) | 52 (51.5) | 237 (59.3) |
| Grade 4 | 10 (3.3) | 2 (2.0) | 12 (3.0) |
| Sensation, n (%) | |||
| Grade 1 | 20 (6.7) | 7 (6.9) | 27 (6.8) |
| Grade 2 | 279 (93.3) | 94 (93.1) | 373 (93.3) |
| PEDIS classification | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Ulcer debridement undertaken, n (%) | |||
| Yes | 261 (87.3) | 90 (89.1) | 351 (87.8) |
| No | 38 (12.7) | 11 (10.9) | 49 (12.3) |
| Estimated index ulcer area (cm)a | |||
| Number of patients | 297 | 100 | 397 |
| Number of patients with missing data | 2 | 1 | 3 |
| Mean (SD) | 6.71 (15.37) | 6.91 (14.57) | 6.76 (15.16) |
| Median (IQR) | 1.77 (0.63–5.50) | 1.57 (0.79–6.73) | 1.77 (0.63–6.15) |
| Range | 0.0–138.2 | 0.0–94.2 | 0.0–138.2 |
| Clinical signs and symptoms | In the follow-up population (N = 299) | Not in the follow-up population (N = 101) | Total: full analysis set (N = 400) |
|---|---|---|---|
| Wound odour, n (%) | |||
| Yes | 93 (31.1) | 34 (33.7) | 127 (31.8) |
| No | 206 (68.9) | 67 (66.3) | 273 (68.3) |
| Pocketing in wound, n (%) | |||
| Yes | 127 (42.5) | 43 (42.6) | 170 (42.5) |
| No | 172 (57.5) | 58 (57.4) | 230 (57.5) |
| Discoloured granulation tissue, n (%) | |||
| Yes | 162 (54.2) | 63 (62.4) | 225 (56.3) |
| No | 137 (45.8) | 38 (37.6) | 175 (43.8) |
| Friable granulation tissue, n (%) | |||
| Yes | 146 (48.8) | 58 (57.4) | 204 (51.0) |
| No | 153 (51.2) | 43 (42.6) | 196 (49.0) |
| Recent increase in pain, n (%) | |||
| Yes | 95 (31.8) | 30 (29.7) | 125 (31.3) |
| No | 203 (67.9) | 71 (70.3) | 274 (68.5) |
| Missing | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Recent decrease in pain, n (%) | |||
| Yes | 7 (2.3) | 2 (2.0) | 9 (2.3) |
| No | 289 (96.7) | 99 (98.0) | 388 (97.0) |
| Missing | 3 (1.0) | 0 (0.0) | 3 (0.8) |
| Recent increase in wound size, n (%) | |||
| Yes | 186 (62.2) | 60 (59.4) | 246 (61.5) |
| No | 113 (37.8) | 40 (39.6) | 153 (38.3) |
| Missing | 0 (0.0) | 1 (1.0) | 1 (0.3) |
| Breakdown of epithelium, n (%) | |||
| Yes | 94 (31.4) | 32 (31.7) | 126 (31.5) |
| No | 204 (68.2) | 69 (68.3) | 273 (68.3) |
| Missing | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Wagner grade | In the follow-up population (N = 299), n (%) | Not in the follow-up population (N = 101), n (%) | Total: full analysis set (N = 400), n (%) |
|---|---|---|---|
| Grade 1 | 104 (34.8) | 32 (31.7) | 136 (34.0) |
| Grade 2 | 93 (31.1) | 41 (40.6) | 134 (33.5) |
| Grade 3 sepsis | 96 (32.1) | 26 (25.7) | 122 (30.5) |
| Grade 4 | 5 (1.7) | 2 (2.0) | 7 (1.8) |
| Grade 5 foot | 1 (0.3) | 0 (0.0) | 1 (0.3) |
Missing data
Table 94 presents the level of missing data present for the long-term follow-up population, for the outcome time to healing and baseline covariates. Table 95 presents summary statistics of the 10 imputed healing times derived for each of the 12 patients with missing time to healing and overall. Compared with the time of healing in patients where the healing date was known, imputed times were on average higher (median 6.5 months).
| Missing dataa | Total (N = 299), n (%) |
|---|---|
| Yes | 43 (14.4) |
| Healing date | 12 (4.0) |
| Diabetes duration | 2 (0.7) |
| HbA1c | 5 (1.7) |
| Extent of ulcer | 2 (0.7) |
| Ulcer type | 2 (0.7) |
| Recurrent ulcer | 2 (0.7) |
| Ulcer duration | 4 (1.3) |
| Previous antibiotic therapy | 15 (5.0) |
| Antimicrobial dressing | 3 (1.0) |
| No | 256 (85.6) |
| Trial number | Number of imputations | Mean | SD | Median | Lower quartile | Upper quartile | Minimum | Maximum |
|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 7.0 | 2.60 | 6.2 | 5.02 | 9.07 | 4.1 | 11.3 |
| 43 | 10 | 8.8 | 2.39 | 8.8 | 6.49 | 10.10 | 6.4 | 13.9 |
| 57 | 10 | 9.3 | 2.94 | 8.8 | 6.77 | 10.86 | 5.6 | 15.2 |
| 82 | 10 | 2.8 | 2.53 | 2.1 | 0.72 | 4.61 | 0.5 | 7.7 |
| 104 | 10 | 4.4 | 2.73 | 4.5 | 3.07 | 5.20 | 0.3 | 9.5 |
| 108 | 10 | 5.4 | 4.04 | 4.4 | 1.85 | 8.48 | 0.7 | 13.0 |
| 113 | 10 | 4.8 | 1.91 | 4.6 | 3.09 | 6.03 | 2.2 | 7.8 |
| 117 | 10 | 7.8 | 3.43 | 8.8 | 5.31 | 10.30 | 2.1 | 12.7 |
| 158 | 10 | 5.0 | 3.37 | 4.4 | 1.59 | 6.66 | 1.2 | 11.2 |
| 222 | 10 | 5.7 | 2.39 | 5.0 | 4.14 | 7.33 | 3.0 | 10.3 |
| 331 | 10 | 10.1 | 3.53 | 10.0 | 7.87 | 12.91 | 5.4 | 16.2 |
| 386 | 10 | 8.3 | 2.20 | 8.1 | 6.67 | 9.55 | 4.9 | 12.4 |
| All 12 patients | 120 | 6.6 | 3.52 | 6.5 | 4.2 | 9.1 | 0.3 | 16.2 |
Cumulative incidence curves
Plots of the cumulative incidence of healing, with 95% CIs, in the presence of the competing risks of death and amputation by each of the factors found to be significant in the univariate analysis are presented in Figures 22–31. Note that the actual cumulative incidence curves were obtained via estimation of the cumulative incidence functions using non-parametric methods and are based on the first imputed data set only.
FIGURE 22.
Cumulative incidence of healing by perfusion grade.

FIGURE 23.
Cumulative incidence of healing by age group.

FIGURE 24.
Cumulative incidence of healing by the presence of more than one ulcer.
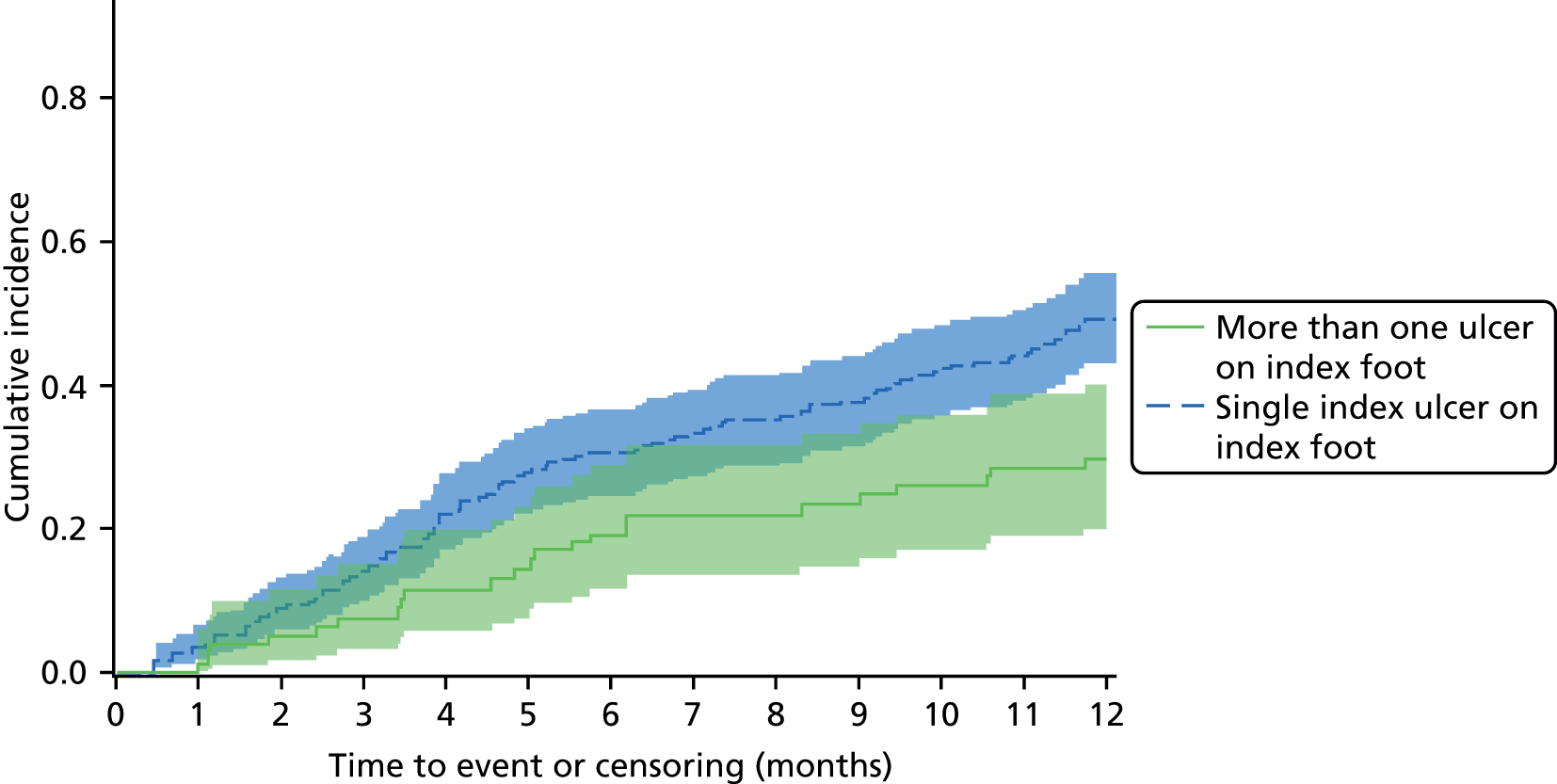
FIGURE 25.
Cumulative incidence of healing by presence of CNS.
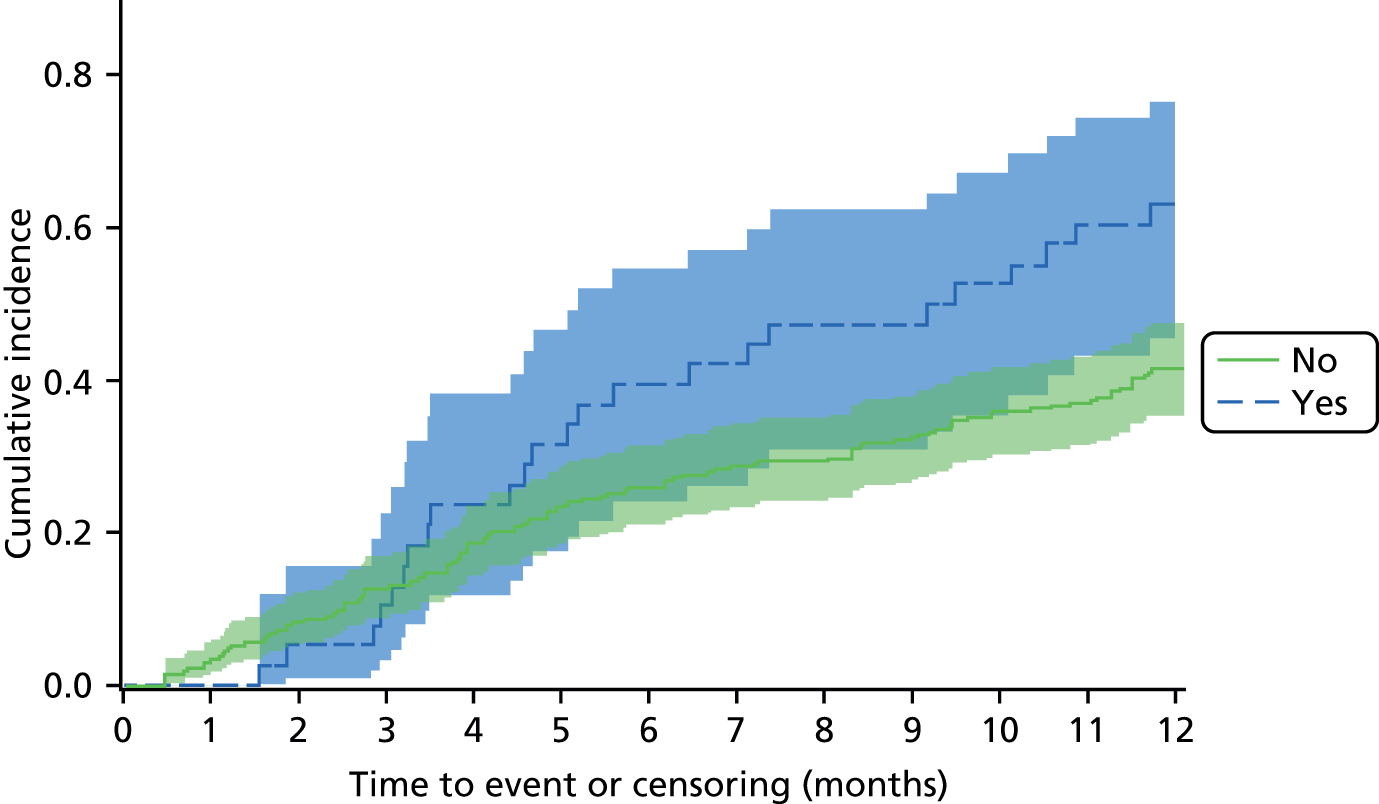
FIGURE 26.
Cumulative incidence of healing by wound duration.
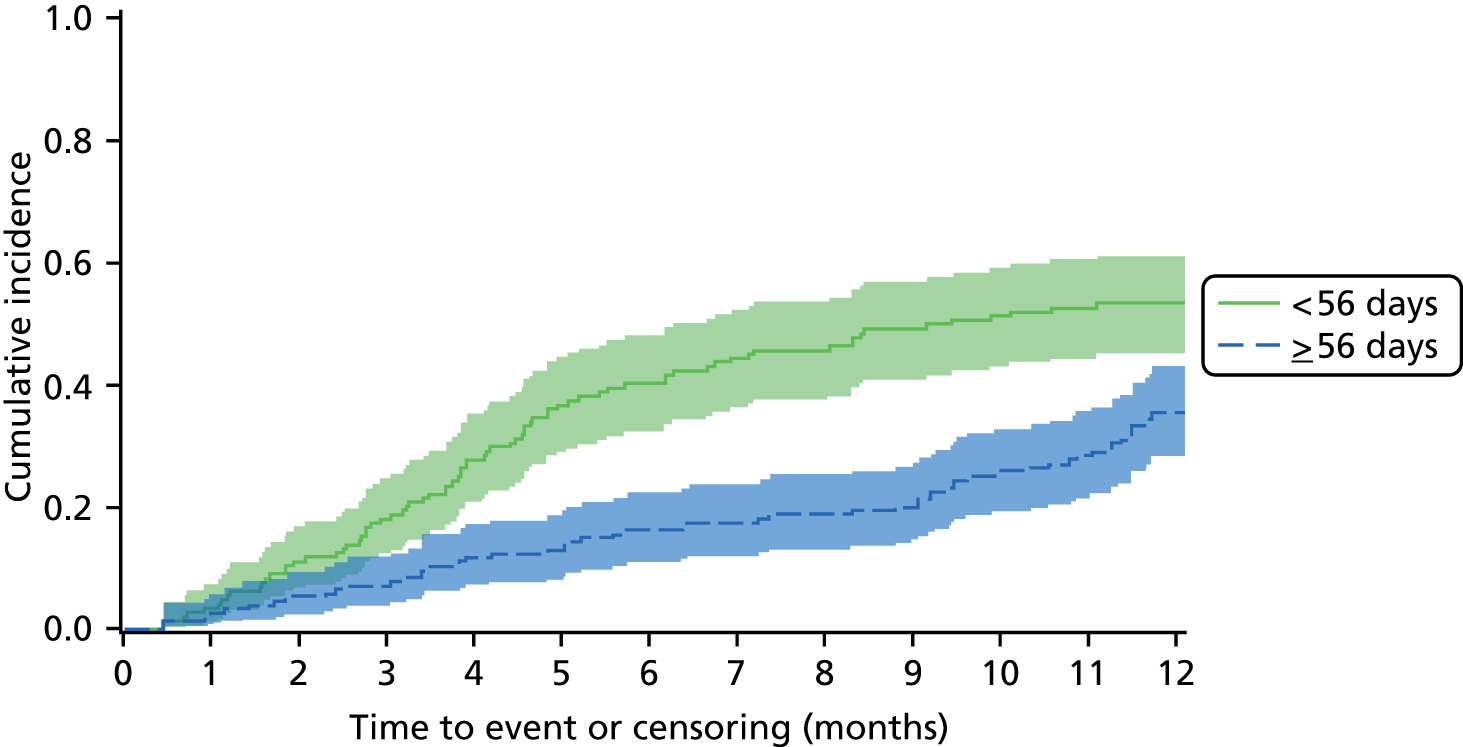
FIGURE 27.
Cumulative incidence of healing by PEDIS depth/tissue loss grade.

FIGURE 28.
Cumulative incidence of healing by PEDIS infection grade.
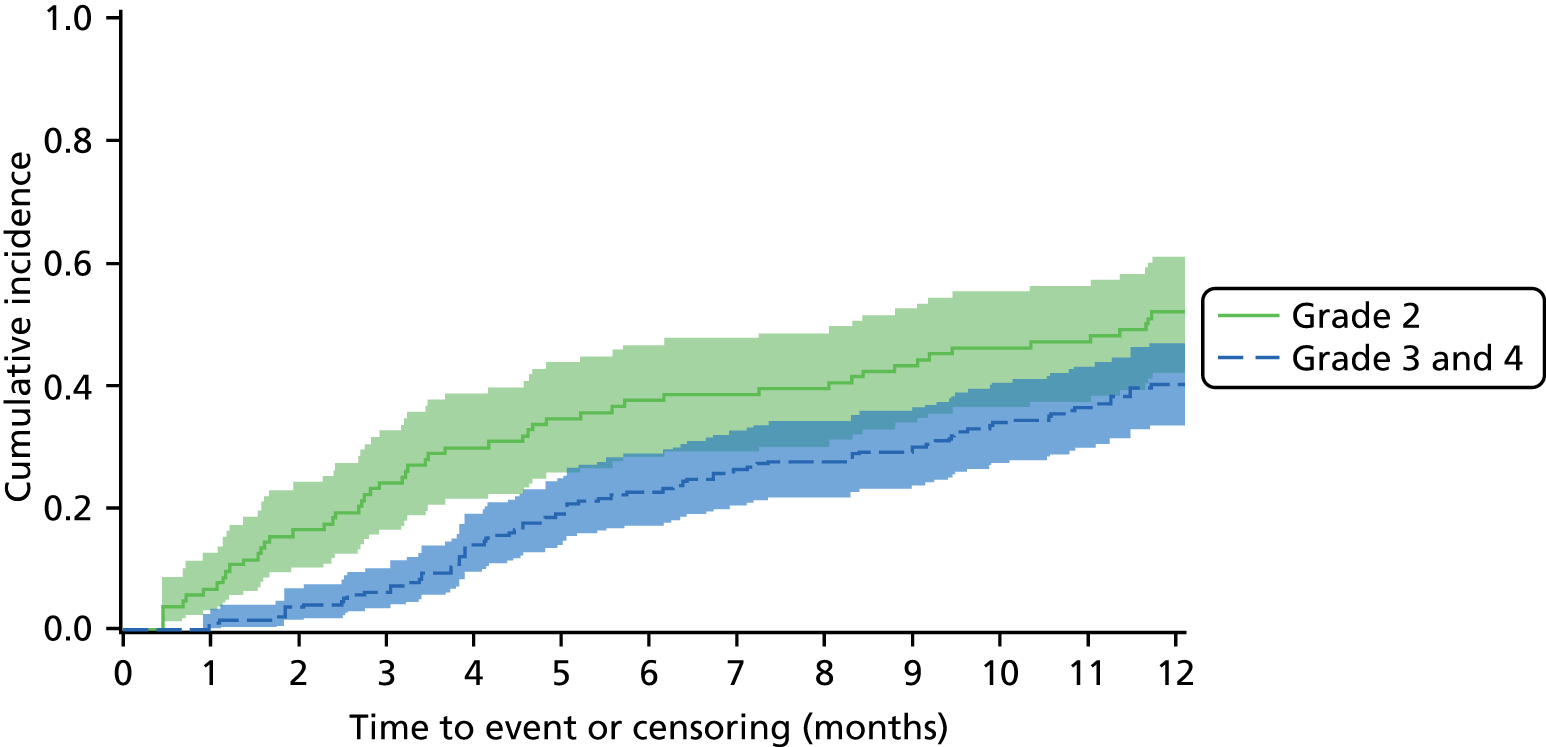
FIGURE 29.
Cumulative incidence of healing by Wagner ulcer grade.
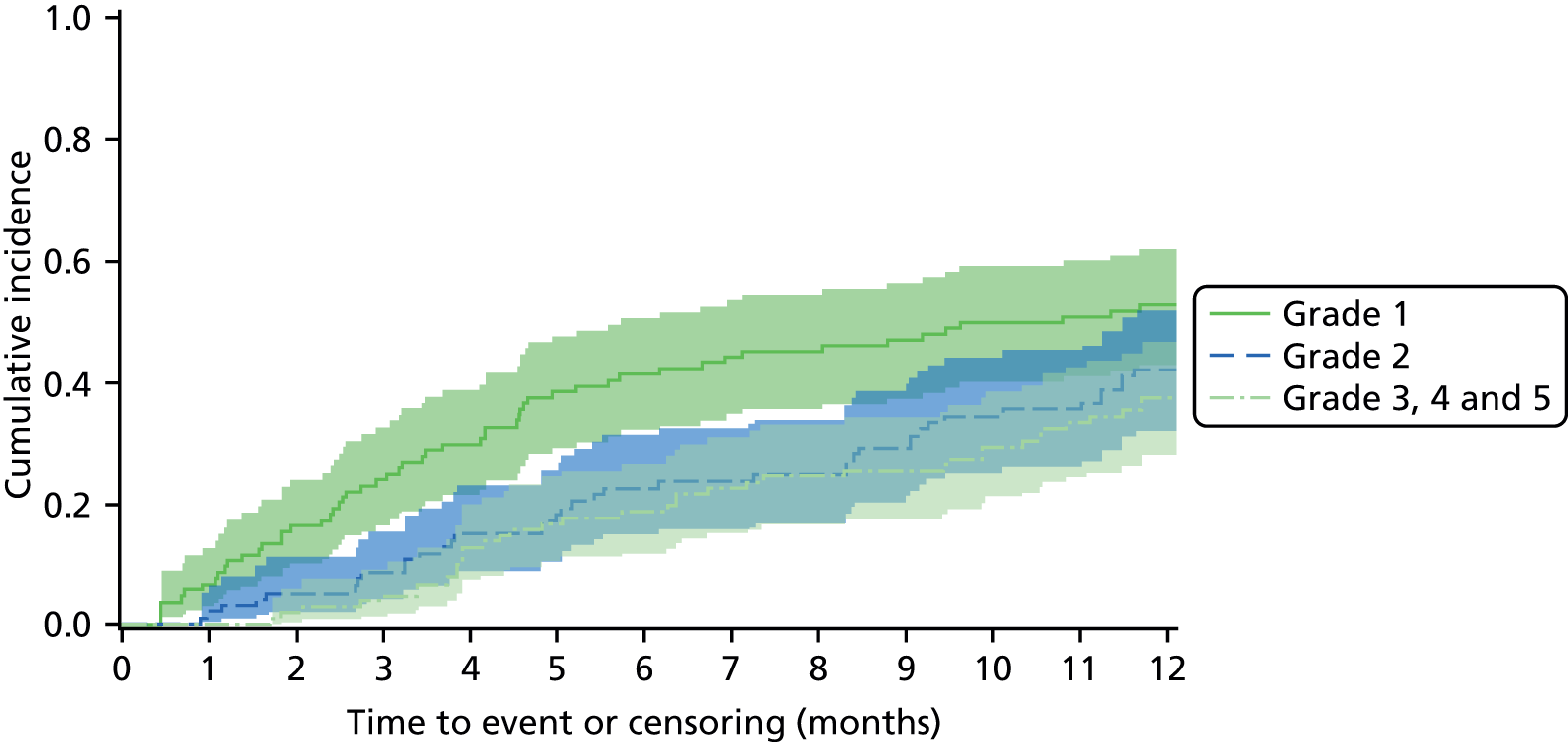
FIGURE 30.
Cumulative incidence of healing by the presence of an antimicrobial dressing.
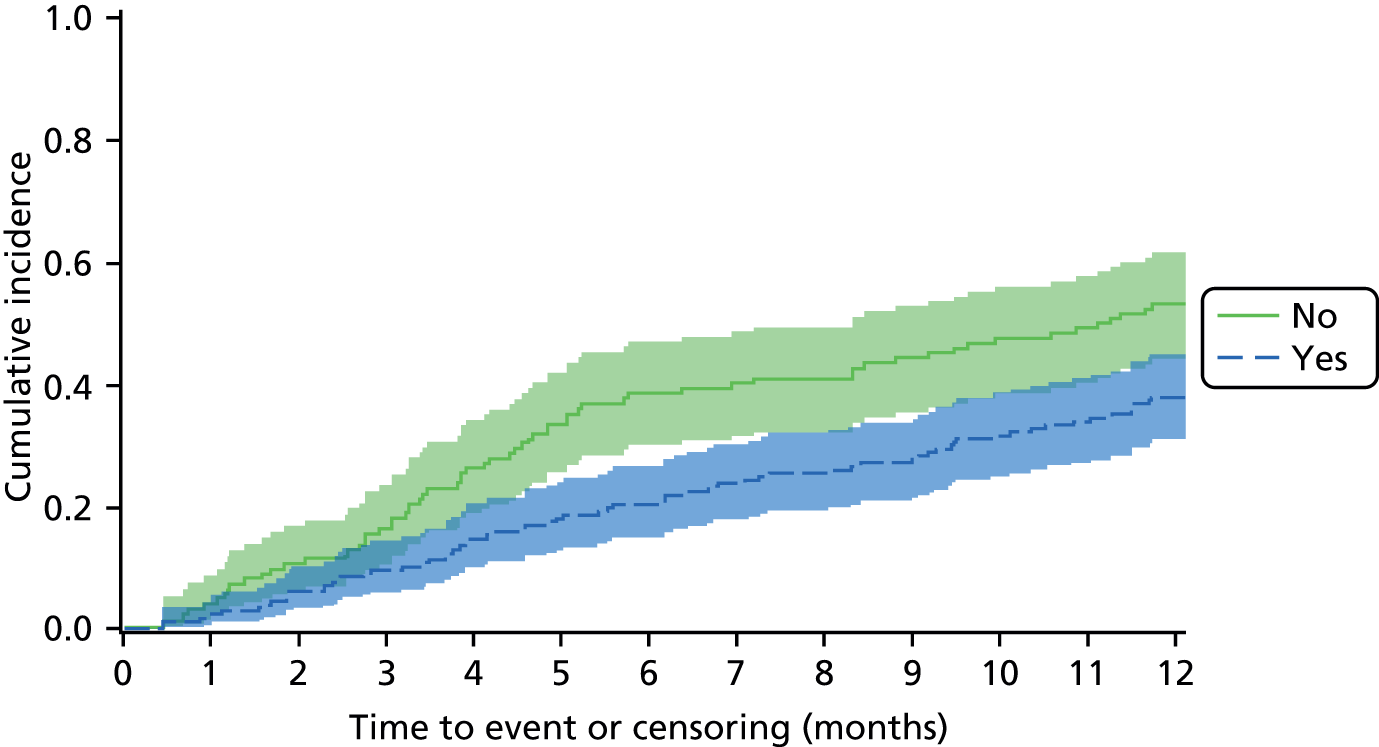
FIGURE 31.
Cumulative incidence of healing by the presence of MRSA.
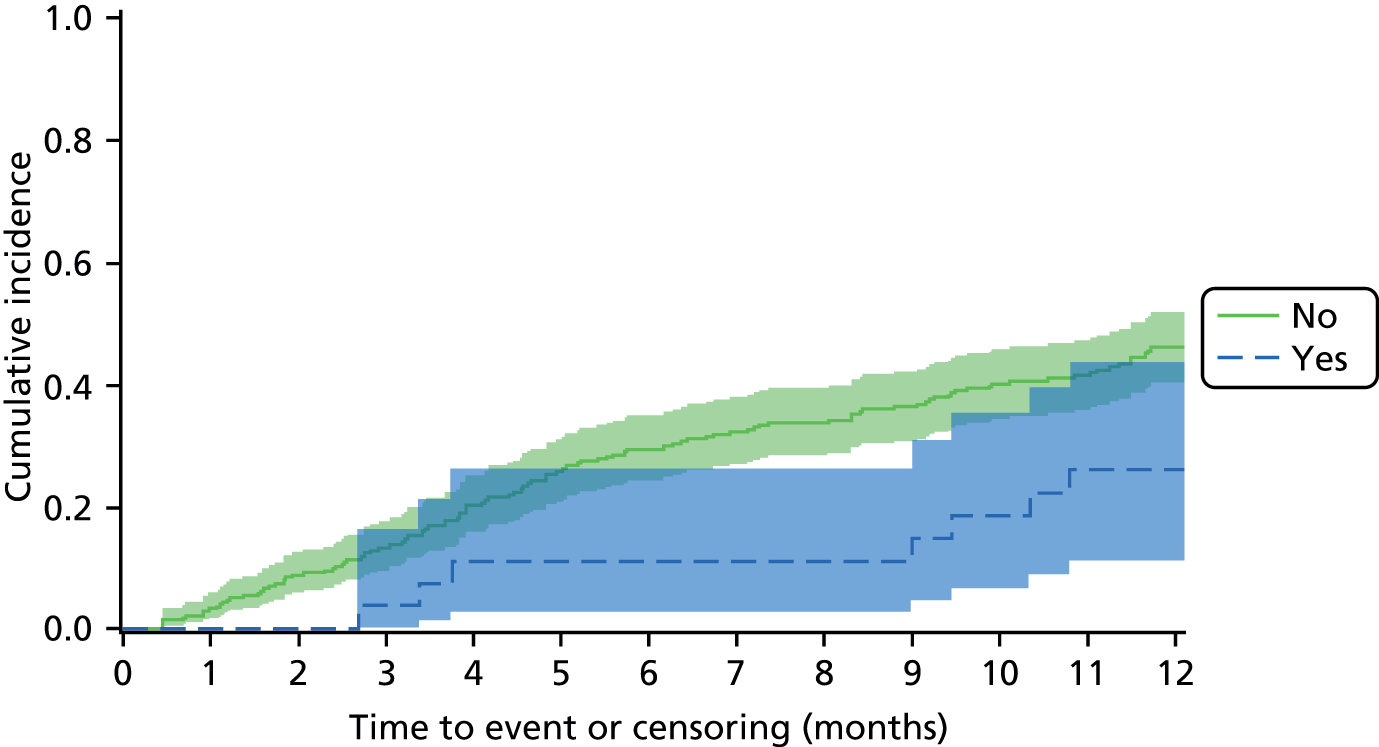
Proportional hazards assumption
In order to test the assumption of proportional hazards, competing risks were treated as censored events in order to generate log-cumulative hazards plots by the selected factor. Under the proportional hazards assumption we would expect the lines to be parallel and not to cross for each level of the factor. The plots presented were from the first imputed data set. To further investigate departures from the proportional hazards assumption, a time-dependent covariate was included in the univariate model for two level factors.
The plots suggest that for the majority of factors the proportional hazards assumption holds. However, there is some question over the assumption for CNS, although the violation of proportional hazards occurs at a point at which there are very few healing events. For this two-level factor, the assumption of proportional hazards was further investigated via the inclusion of a time-dependent covariate, which suggested further evidence against the proportional hazards assumption for CNS with an increased incidence of healing in patients without CNS prior to 1 month post sample, but an increased and increasing incidence of healing in patients with CNS after 1 month (i.e. non-constant HR).
Furthermore, the proportional hazards assumption appears to be satisfied for the reference grade 1 of both the depth/tissue loss classification and Wagner ulcer grade; however, lines do cross for grades 2 and 3 (Wagner grades 3/4/5) owing to the reduced differences observed in the cumulative incidence for these levels. This suggests that grouping grades > 1, for both the depth/tissue loss classification and Wagner ulcer grade, may not result in a substantial loss of information on the likely incidence of healing.
Log-cumulative hazard plots
Figures 32–36 present the log-cumulative hazard plots of time to healing by perfusion, age, presence of more than one ulcer, wound duration and CNS.
FIGURE 32.
Log-cumulative hazard plot of time to healing by perfusion. Proportional hazards assumption satisfied.
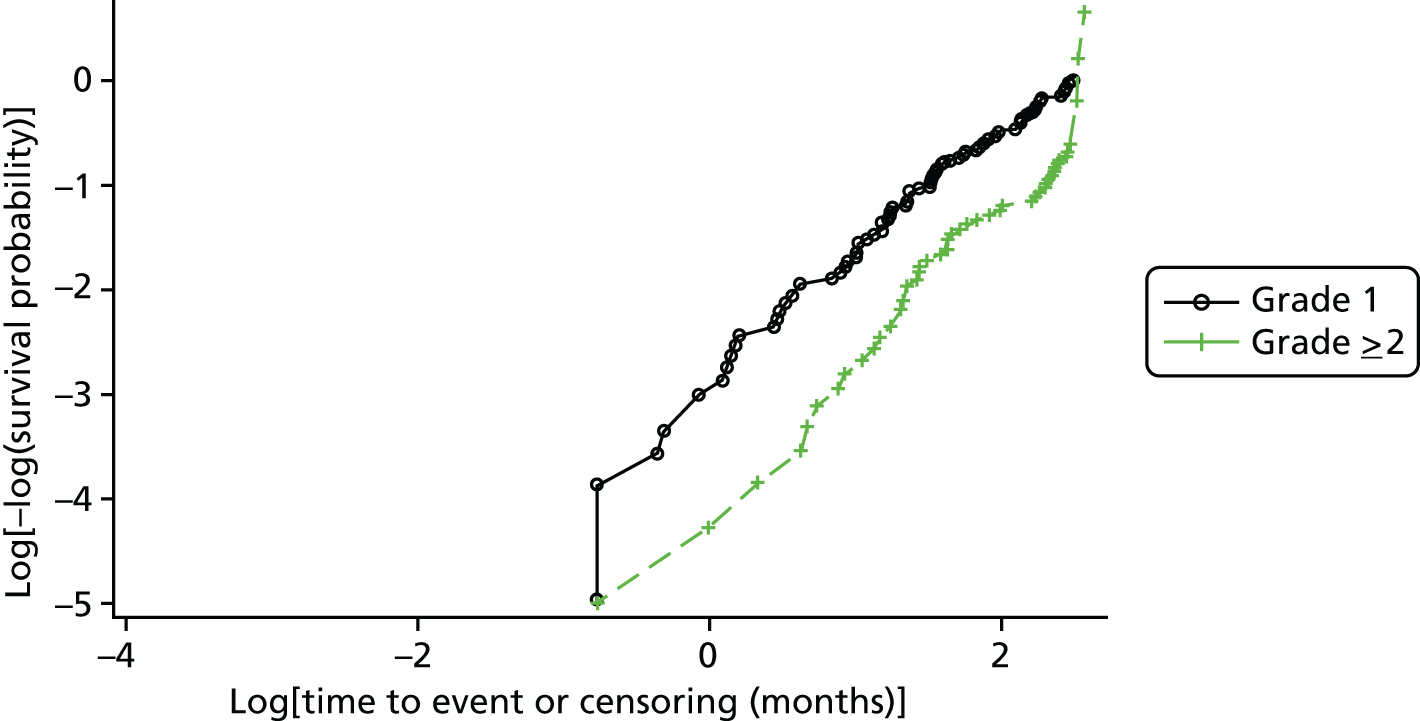
FIGURE 33.
Log-cumulative hazard plot of time to healing by age. Curves are very close and appear to cross early on; however, this is after only very few healing events, hence proportional hazards assumption is satisfied.

FIGURE 34.
Log-cumulative hazard plot of time to healing by presence of more than one ulcer. Proportional hazards assumption satisfied.
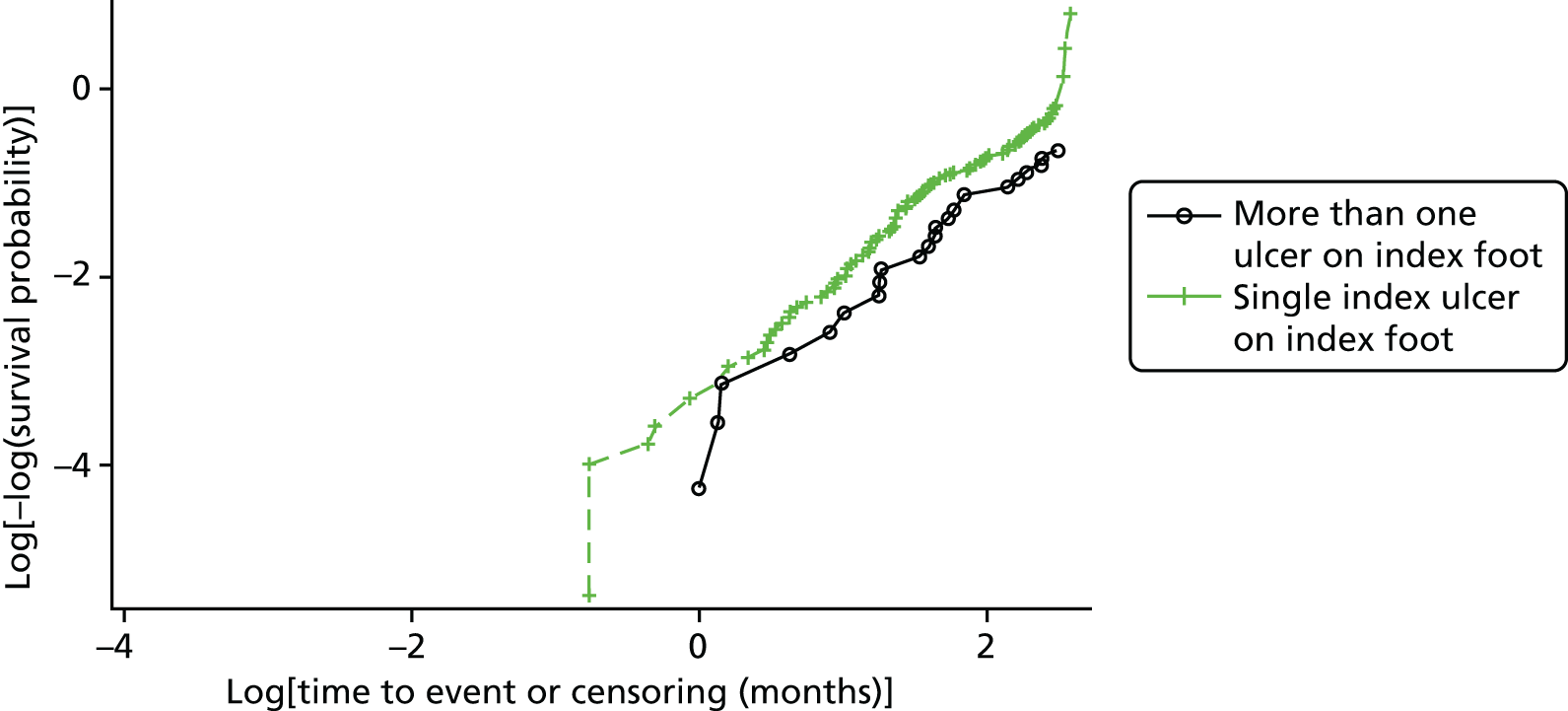
FIGURE 35.
Log-cumulative hazard plot of time to healing by wound duration. Proportional hazards assumption satisfied.
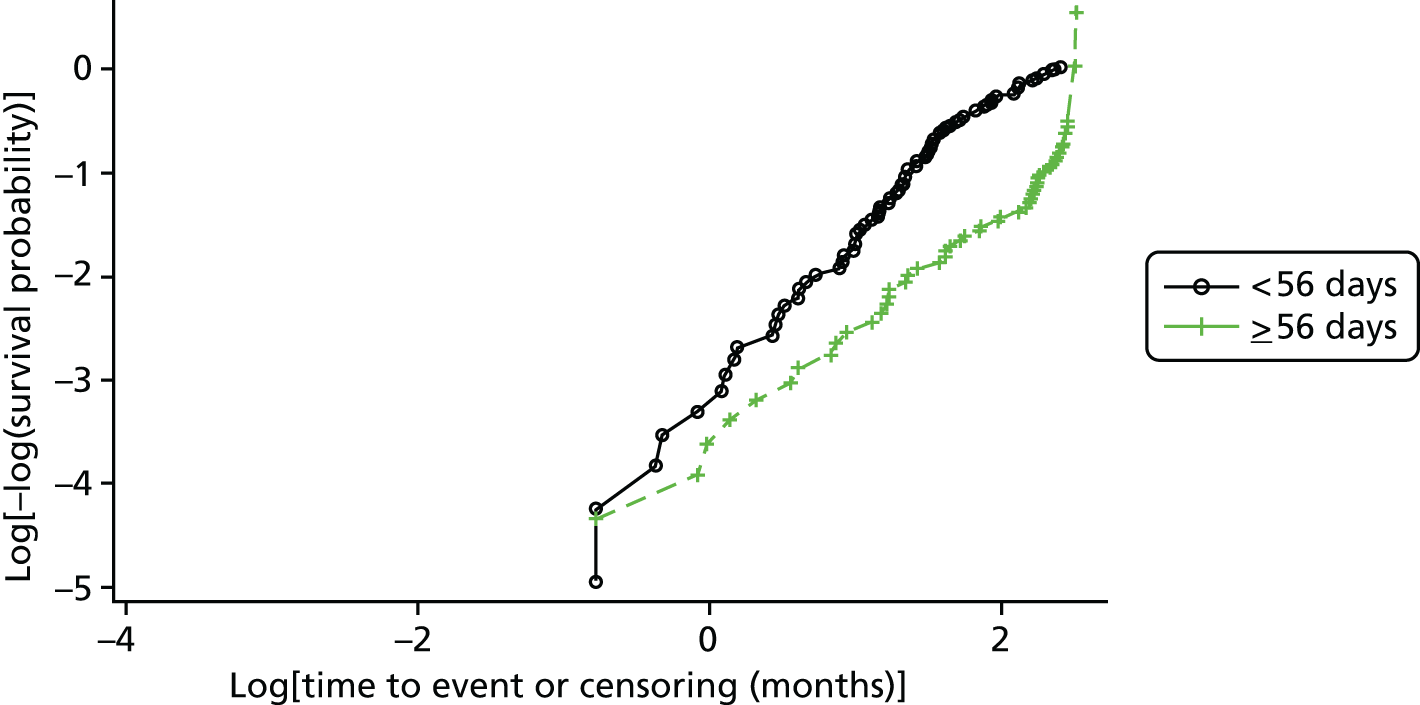
FIGURE 36.
Log-cumulative hazard plot of time to healing by presence of CNS. Curves cross and, hence, the proportional hazards assumption is questionable; however, there are few healing events in those with CNS, in particular prior to the point at which lines cross.

Figures 37–41 present the log-cumulative hazard plots of time to healing by depth/tissue loss, infection grade, ulcer grade, presence of antimicrobial dressing and MRSA.
FIGURE 37.
Log-cumulative hazard plot of time to healing by PEDIS depth/tissue loss grade. Proportional hazards assumption satisfied for grade 1; however, lines do cross for grades 2 and 3.
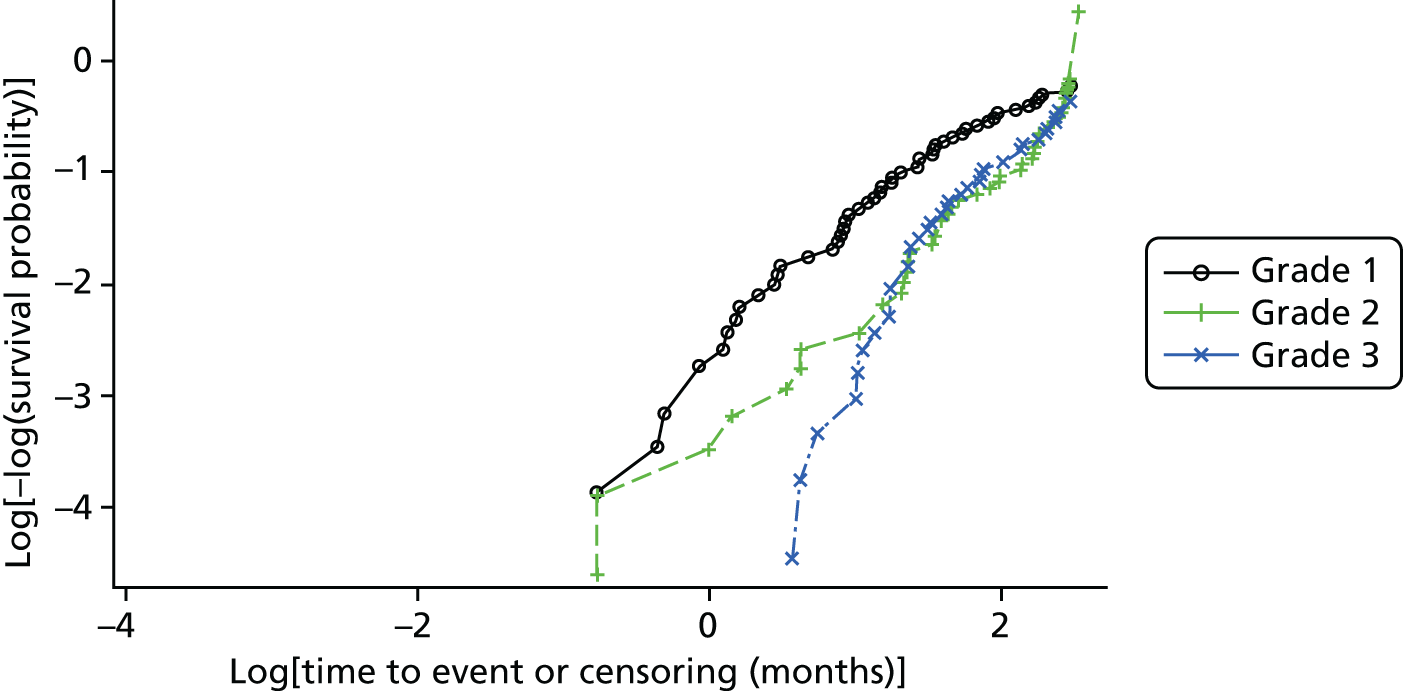
FIGURE 38.
Log-cumulative hazard plot of time to healing by PEDIS infection grade. Proportional hazards assumption satisfied.

FIGURE 39.
Log-cumulative hazard plot of time to healing by Wagner ulcer grade. Proportional hazards assumption satisfied for grade 1; however, lines do cross for grades 2 and 3, 4, 5.
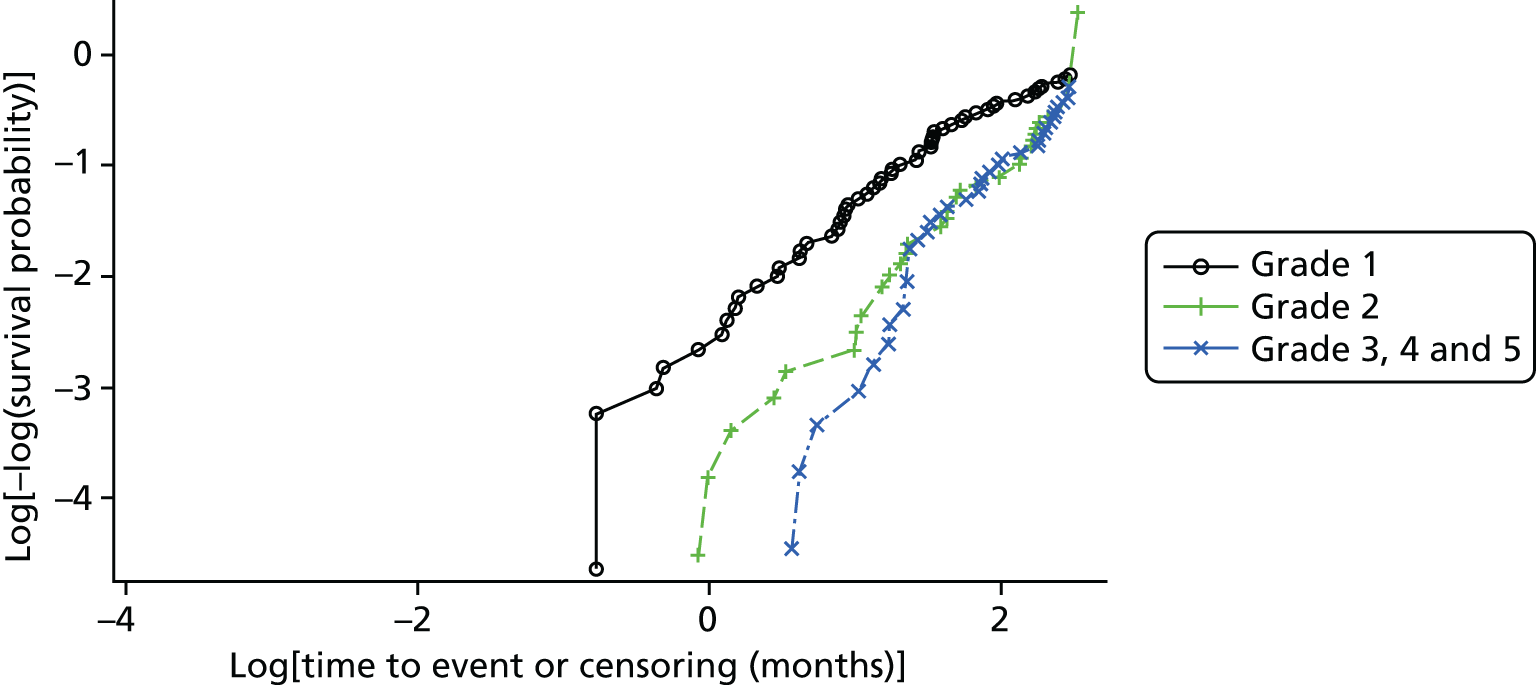
FIGURE 40.
Log-cumulative hazard plot of time to healing by presence of antimicrobial dressing. Proportional hazards assumption satisfied.

FIGURE 41.
Log-cumulative hazard plot of time to healing by presence of MRSA. Proportional hazards assumption satisfied.

Time-dependent covariate
Table 96 shows that the interaction of the presence of CNS against time (having applied a log function) is significant. The parameter estimate for the interaction term suggests that the likelihood of healing increases over time for patients with CNS reported at baseline. However, the negative parameter estimate for the presence of CNS suggests that prior to month 1, patients with no CNS reported were more likely to heal than those in which it was reported. This relationship is demonstrated in Figure 42 which presents the relative hazard of healing over time for a patient who had CNS reported at baseline relative to those who did not.
| Model | Parameter estimate | p-value | HR (95% CI) |
|---|---|---|---|
| CNS: With Time × CNS interaction | |||
| CNS: Yes vs. No | 0.204243 | 0.5833 | 1.23 (0.59 to 2.54) |
| Interaction: CNS × Time | 0.059799 | 0.2945 | 1.06 (0.95 to 1.19) |
| CNS: With log(Time) × CNS interaction | |||
| CNS: Yes vs. No | –0.180176 | 0.6739 | 0.84 (0.36 to 1.93) |
| Interaction: CNS × log(Time) | 0.479331 | 0.0567 | 1.61 (0.99 to 2.64) |
FIGURE 42.
Plot of the relative hazard of healing for a patient with CNS vs. a patient without, over time. RH, relative hazard.

Appendix 4 Centre differences
Questionnaire responses
| Responding sites | |
|---|---|
| Bensham | |
| Bradford | |
| Chorley and South Ribble | |
| Fairfields | |
| Huddersfield Royal Infirmary | |
| Manchester Diabetes Centre, CMFT NHS Trust | |
| Minerva Centre | |
| New Cross Hospital | |
| Norfolk and Norwich University Hospital NHS Foundation Trust | |
| North Manchester General Hospital | |
| North Tees and Hartlepool Foundation Trust | |
| Pinderfields Hospital | |
| Queen Elizabeth Hospital King’s Lynn | |
| Royal Oldham | |
| Scarborough Hospital | |
| St James’s University Hospital | |
| Tameside NHS Foundation Trust | |
| James Cook University Hospital | |
| University Hospitals Coventry and Warwickshire NHS Trust | |
| University Hospital Birmingham | |
| University Hospital North Staffordshire | |
| Weston General Hospital | |
| Grand total | n = 22 |
Clinical acquisition of samples
The majority of responding sites (20/21) used a scalpel to collect tissue samples, of which the majority (16/20), used a cutting motion, whereas a minority (4/20) used a scraping motion.
| Technique | Count |
|---|---|
| Dermal curette | 1 |
| Scalpel | 20 |
| Grand total | 21 |
| Technique | Count |
|---|---|
| Not applicable | 1 |
| Use a cutting motion | 16 |
| Use a scraping motion | 4 |
| Grand total | 21 |
Transport, analysis and reporting of samples by laboratory
Transport
There are no differences in the time it takes for swab and tissue samples to reach the laboratory. The majority of laboratories report no clear difference in the time it takes from receipt of swab and tissue samples to processing, with just 4 out of 17 reporting slightly more urgent/quicker time to processing for tissue samples.
| Laboratory location | Count |
|---|---|
| No | 9 |
| Path laboratory is at hospital site: clinics are held at both hospital site and community centres around the city. Therefore, samples arrive quicker from clinics held at the hospital | 1 |
| Yes | 11 |
| Yes and no: recruitment takes place at both hospitals | 1 |
| Grand total | 22 |
| Time for swab to arrive at laboratory | Count |
|---|---|
| ≤ 1 hour | 8 |
| 2–4 hours | 7 |
| If at North Tees within 1 hour of sampling at Hartlepool; if at Hartlepool within 2 hours | 1 |
| If sent from hospital site: immediately via airtube system in pods. 2 hours if sent from a community care clinic | 1 |
| ≤ 24 hours | 1 |
| Unknown | 2 |
| Grand total | 20 |
| Time differences | Count |
|---|---|
| Same | 19 |
| Grand total | 19 |
| Refrigeration of samples | Count |
|---|---|
| Samples are kept overnight but not refrigerated | 3 |
| Samples are never kept overnight before transit | 16 |
| Yes samples are refrigerated overnight before transit | 1 |
| Grand total | 20 |
| Differences in overnight refrigeration | Count |
|---|---|
| Same | 18 |
| Same: however, slight difference in frequency from rarely (swab) to never (tissue) | 1 |
| Same: however, slight difference in frequency from rarely (tissue) to never (swab) | 1 |
| Grand total | 20 |
| Time to swab processing | Count |
|---|---|
| ≤ 1 hour | 7 |
| 1–2 hours | 2 |
| 2–4 hours | 3 |
| 6–12 hours | 1 |
| Between 20 minutes and 18 hours depending on time of day | 1 |
| Depends on the time of arrival to the laboratory. Swabs are processed within 1 hour until 16.00 and then refrigerated overnight and processed within 16 hours | 1 |
| Same day | 1 |
| 2 hours unless urgent | 1 |
| Unknown: setups done throughout the day but will not be done after 23.00 until 08.00 the next day | 1 |
| Unknown | 1 |
| Grand total | 19 |
| Differences in time to swab processing | Count |
|---|---|
| Same | 13 |
| Same (but tissue sample usually with more urgency) | 1 |
| Swab 2 hours vs. tissue 1 hour | 1 |
| Swab 2–4 hours vs. tissue 2 hours | 1 |
| Tissue processing continues later into the day (20.00 vs. 16.00) and if refrigerated are processed within 12 hours vs. 16 hours | 1 |
| Grand total | 17 |
| Swab transport medium | Count |
|---|---|
| Amies (Amies Plain) | 4 |
| Amies CIR or Amies Charcoal | 12 |
| Amies CIR or Amies Charcoal/none received in sterile universal | 1 |
| Amies Liquid | 1 |
| Standard swab sample tube | 1 |
| Grand total | 19 |
| Tissue sample transport medium | Count |
|---|---|
| Amies (Amies Plain) | 1 |
| Amies CIR or Amies Charcoal | 1 |
| Dry pot (no medium) | 2 |
| Dry Universal with a few drops of sterile saline | 1 |
| In sterile sample pot (no medium) | 1 |
| No transport media used, sample placed in aseptic universal container with saline | 1 |
| None/no transport medium used | 2 |
| Plain sterile universal container | 6 |
| Sterile saline | 1 |
| Stuarts CIR or Stuarts Charcoal | 1 |
| Grand total | 17 |
Analysis
Just 3 out of 19 laboratories report performing a Gram stain on both swab and tissue samples, whereas 9 out of 19 laboratories perform these only on tissue samples; the remaining 6 never perform one, with 1 performing them only on request (Table 109).
| Gram staining | Count |
|---|---|
| Never performs a Gram stain | 6 |
| Only on request | 1 |
| Only performs a Gram stain on tissue samples | 9 |
| Performs a Gram stain on both swabs and tissue samples | 2 |
| Performs a Gram stain on both swabs (deep sites) and tissue samples | 1 |
| Grand total | 19 |
Reporting
A combination of systems are used to report growth, with a slight majority (8/18) using combinations of scanty/light/moderate/heavy, 4 out of 18 using combinations of +/++/+++/++++, and 4 out of 18 not reporting growth (Table 110).
| System for reporting bacterial growth | Count |
|---|---|
| +/++/+++ | 3 |
| +/++/+++ or +/++/+++/++++ | 1 |
| Both | 1 |
| Light/moderate/heavy | 5 |
| Scanty/light/moderate/heavy | 2 |
| Scanty/moderate/heavy | 1 |
| Scanty, profuse, mixed, a growth | 1 |
| My laboratory does not report amount bacterial growth | 3 |
| My laboratory does not report bacterial growth/+/++/+++ | 1 |
| Grand total | 18 |
Isolates are reported to a variety of taxonomic ranks ranging from species, genus and other. It is reported that 16 out of 18 laboratories report to the same level for swab and tissue samples, whereas 1 out of 18 reports that tissue isolates are provided to the species level and only significant organisms are provided in such detail for the swab. However, differences are more apparent when considering whether or not all recovered isolates are reported to the clinician, with only 8 out of 18 laboratories reporting that the same isolates are reported from swab and tissue samples. In contrast, the remaining 10 laboratories report that all isolated are reported from a tissue sample, whereas reporting of those from a swab sample depends on a mix of clinical details, clinical significance, whether or not there is heavy pure growth, and whether or not those pathogens that are not reported as enteric or skin flora are significant. In 16 out of 19 laboratories, it was reported that standard procedures allow identification of the same isolates; however, 3 out of 16 laboratories said that standard procedures would not allow this, of which 1 laboratory mentioned that the tissue samples are also put into a broth (Tables 110–117).
| Taxonomic rank | Count |
|---|---|
| Dependant on organism: we identify ‘significant pathogens’ (e.g. S. aureus, P. aeruginosa, etc.) | 1 |
| Genus | 2 |
| Genus/species | 2 |
| Genus/species/other | 1 |
| Other | 2 |
| Species | 10 |
| Grand total | 18 |
| Taxonomic rank | Count |
|---|---|
| Genus | 2 |
| Genus/species | 2 |
| Other | 3 |
| Species | 11 |
| Grand total | 18 |
| Differences in taxonomic rank isolates | Count |
|---|---|
| Insufficient tissue details but appears to reference subset of genus, whereas swab references genus/species/other | 1 |
| Only significant organisms reported in greater detail for swab. Tissue reported to species level | 1 |
| Same | 16 |
| Grand total | 18 |
| Reported/not reported (with reasons) | Count |
|---|---|
| No | 15 |
| Significant pathogens reported but others are grouped into enteric flora or skin flora, etc. | 1 |
| Biomedical science decides depending on clinical significance of Istalks | 2 |
| Clinically significant isolates are determined by national HPA guidelines or if indicated by the medical microbiologist | 1 |
| Depends on clinical details given. Common pathogens such as S. aureus | 1 |
| Depends on type of sample and clinical details | 1 |
| Heavy pure growth | 1 |
| No: may be reported as mixed growth with no obvious pathogen or interpretive comment | 1 |
| Only if clinically necessary | 1 |
| Pathogens are reported. Others may be reported as normal skin flora | 1 |
| Skin flora | 1 |
| Skin flora not reported as individual isolate | 1 |
| Target organisms as per the HPA SMI | 1 |
| We report significant pathogens. Mixed skin flora for example does not get reported to species level | 1 |
| Yes | 4 |
| Grand total | 19 |
| Reporting | Count |
|---|---|
| No | 4 |
| No: may be reported as mixed growth with no obvious pathogen other interpretive comment. Depends on number/amount of growth and species isolated in conjunction with clinical data provided | 1 |
| Pathogens are reported. Others may be reported as normal skin flora | 1 |
| Target organisms as per HPA SMI | 1 |
| Type of sample and clinical diagnosis | 1 |
| Yes | 14 |
| Generally we report everything, but we might add a significance comment to an isolate of CNS | 1 |
| Grand total | 18 |
| Reporting of isolates | Count |
|---|---|
| All reported tissue not swab | 1 |
| All reported tissue not swab (depends on clinical details) | 1 |
| All reported tissue not swab (only if clinically significant) | 5 |
| All reported tissue not swab (only if heavy pure growth) | 1 |
| All reported tissue not swab (significant pathogens reported but others grouped into enteric or skin flora) | 1 |
| All reported tissue not swab (skin flora) | 1 |
| Same | 8 |
| Grand total | 18 |
| Identification of same isolates across samples | Count |
|---|---|
| No | 3 |
| Not necessarily: the same pathogens would be identified but not skin flora | 1 |
| Tissue samples are put up in a broth culture as well | 1 |
| Yes | 16 |
| Grand total | 19 |
Reporting: antimicrobial resistance for S. aureus, coagulase-negative Staphylococcus and enterococci
None of the responding 19 laboratories reported that they tested the antimicrobial sensitivity of S. aureus to meticillin for swab or tissue samples. A total of 12 out of 13 laboratories reported that the same agents were tested in swab and tissue samples, with 1 laboratory reporting additional agents for the tissue sample.
None of the responding 19 laboratories reported that they tested the antimicrobial sensitivity of CNS to meticillin for swab or tissue samples; 4 out of 19 laboratories reported that they tested no agents for swab samples; and 1 out of 17 laboratories reported that they tested no agents for tissue samples. In 10 out of 12 laboratories, the same agents (or lack of) were reported to be tested for in swab and tissue samples; however, the remaining two laboratories do not test agents for swab samples but do for tissue samples.
When enterococci is isolated, antimicrobial sensitivity to vancomycin is reported to be tested for in 10 out of 19 laboratories for swab samples and 13 out of 17 laboratories for tissue samples. In 9 out of 12 laboratories, the same agents were reported to be tested for swab and tissue samples; however, the remaining 3 laboratories reported that no agents are tested for swab samples, whereas numerous agents are tested for in tissue samples (Tables 118–126).
| Agents | Count |
|---|---|
| Flucloxacillin, erythromycin, clindamycin, trimethoprim, doxycycline, clarithromycin, fusidin, cefoxitin | 1 |
| Benzylpenicillin, chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, fuscidic acid, nitrofurantoin, oxacillin, rifampicin, gentamicin, linezolid, mupirocin, teicoplanin, tetracycline, tgercycline, vancomycin, trimethoprim | 2 |
| Beta-lactams, macrolides | 1 |
| Cefoxitin as marker for flucloxacillin, erythromycin, clindamycin, doxycycline, co-trimoxazole, rifampicin, vancomycin, linezolid, mupirocin | 1 |
| Cefoxitin, oxacillin, vancomycin, erythromycin, fusidic acid, tetracycline, mupirocin | 1 |
| Chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, flucloxacillin, fusidic acid, gent, mupirocin, lizenzolid, pencillin, rifampicin, teicoplanin, tetracycline, tigecycline, trimethoprim, vancomycion (VITEK card) | 1 |
| Ciprofloxacin, tetracycline, gentamicin, erythromycin, flucloxacillin, fusidic acid | 1 |
| Clindamycin, gentamicin, mupirocin, oxacillin, tetracycline | 1 |
| E, doxycycline, W5, CN, FD, FOX, neomycin, C10 | 1 |
| Flucloxacillin, cefoxatin, erythromycin, pencillin, fusidic acid, chloramphenicol, ciprofloxacin, erythromycin, clindamycin, daptomycin, gentomycin, linezolid, muprirocin, teicoplanin, tetracycline, tigecycline, vancomycin | 1 |
| Flucloxacillin, erythromycin, fusidic acid, rifampicin, gentamicin, tetracycline, clindamycin, cefradine, chloramphenicol, daptomycin, linezolid, penicillin, tigecycline, vancomycion, teicoplanin, trimethoprim | 1 |
| Pencillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Penicillin, cefoxitin, erythromycin, clindamycin, tetracycline, vancomycin, ciprofloxacin, neomycin, fusidic acid, mupirocin, rifampacin, gentamycin | 1 |
| Penicillin, erythromycin, clindamycin, flucloxacillin, gentamicin, vancomycin, fusidic acid, rifampicin, ciprofloxacin, linezolid, daptomycin | 1 |
| Penicillin, erythromycin, clindamycin, tetracycline, rifampicin, cefotaxine, vancomycin, mupirocin | 1 |
| Penicillin, erythromycin, flucloxacillin, tetracycline, rifampicin, trimethoprim, linezolid, vancomycin, mupirocin, gentamicin, ciprofloxacin, fusidic acid | 1 |
| Sensitivities on these isolates are performed on Biomeriuex Vitek2 system with a P620 card which has 22 antibiotics | 1 |
| Grand total | 18 |
| Agents | Count |
|---|---|
| Beta-lactams, macrolides | 1 |
| Cefoxitin as marker for flucloxacillin, erythromycin, clindamycin, doxycycline, co-trimoxazole, rifampicin, vancomycin, linezolid, mupirocin | 1 |
| Cefoxitin, oxacillin, vancomycin, erythromycin, fusidic acid, tetracycline, mupirocin | 1 |
| Chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, flucloxacillin, fusidic acid, gentamicin, mupirocin, lizenzolid, penicillin, rifampicin, teicoplanin, tetracycline, tigecycline, trimethoprim, vancomycin (VITEK card) | 1 |
| Ciprofloxacin, tetracycline, gentamicin, erythromycin, flucloxacillin, fusidic acid | 1 |
| Clindamycin, gentamicin, mupirocin, oxacillin, tetracycline | 1 |
| E, doxycycline, W5, CN, FD, FOX, neomycin, C10 | 1 |
| Flucloxacillin, erythromycin, clindamycin, trimethoprim, doxycycline, clarithromycin, fusidin, rifampicin, cefoxitin, gentamicin, vancomycin, teicoplanin, linezolid, daptomycin | 1 |
| Flucloxacillin, cefoxatin, erythromycin, pencillin, fusidic acid, chloramphenicol, ciprofloxacin, erythromycin, clindamycin, daptomycin, gentomycin, linezolid, muprirocin, teicoplanin, tetracycline, tigecycline, vancomycin | 1 |
| Flucloxacillin, erythromycin, fusidic acid, rifampicin, gentamicin, tetracycline, clindamycin, cefradine, chloramphenicol, daptomycin, linezolid, penicillin, tigecycline, vancomycion, teicoplanin, trimethoprim | 1 |
| Penicillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Penicillin, cefoxitin, erythromycin, clindamycin, tetracycline, vancomycin, ciprofloxacin, neomycin, fusidic acid, mupirocin, rifampacin, gentamycin | 1 |
| Penicillin, erythromycin, clindamycin, tetracycline, rifampicin, cefotaxine, vancomycin, mupirocin | 1 |
| Penicillin, erythromycin, flucloxacillin, tetracycline, rifampicin, trimethoprim, linezolid, vancomycin, mupirocin, gentamicin, ciprofloxacin, fusidic acid | 1 |
| Penicillin, erythromycin, clindamycin, flucloxacillin, gentamicin, vancomycin, fusidic acid, rifampicin, ciprofloxacin, linezolid, daptomycin | 1 |
| Sensitivities on these isolates are performed on the Biomeriuex Vitek2 system with a P620 card which has 22 antibiotics | 1 |
| Grand total | 16 |
| Difference in agents tested against for S. aureus | Count |
|---|---|
| Same | 12 |
| Tissue tests additional (rifampicin, gentamicin, vancomycin, teicoplanin, linezolid, daptomycin) | 1 |
| Grand total | 13 |
| Agents | Count |
|---|---|
| As above | 1 |
| Benzylpenicillin, chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, fuscidic acid, nitrofurantoin, oxacillin, rifampicin, gentamicin, linezolid, mupirocin, teicoplanin, tetracycline, tigecycline, vancomycin, trimethoprim | 2 |
| Beta-lactams, macrolides | 1 |
| Cefoxitin, oxacillin, vancomycin, erythromycin, fusidic acid, tetracycline, mupirocin | 1 |
| Chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, flucloxacillin, fusidic acid, gentamicin, mupirocin, lizenzolid, penicillin, rifampicin, teicoplanin, tetracycline, tigecycline, trimethoprim, vancomycin (VITEK card) | 1 |
| Clindamycin, gentamicin, mupirocin, oxacillin, tetracycline | 1 |
| E, doxycycline, W5, CN, FD, FOX, neomycin, C11 | 1 |
| Flucloxacillin, erythromycin, fusidic acid, rifampicin, gentamicin, tetracycline, clindamycin, cefradine, chloramphenicol, daptomycin, linezolid, penicillin, tigecycline, vancomycion, teicoplanin, trimethoprim | 1 |
| N/A | 1 |
| Nil | 1 |
| None | 1 |
| Pencillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Penicillin, cefoxitin, erythromycin, clindamycin, tetracycline, vancomycin, ciprofloxacin, neomycin, fusidic acid, mupirocin, rifampacin, gentamycin | 1 |
| Penicillin, erythromycin, clindamycin, tetracycline, rifampicin, cefotaxine, vancomycin, mupirocin | 1 |
| Penicillin, erythromycin, flucloxacillin, tetracycline, rifampicin, trimethoprim, linezolid, vancomycin, mupirocin, gentamicin, ciprofloxacin, fusidic acid | 1 |
| Probably none | 1 |
| Sensitivities on these isolates are performed on the Biomeriuex Vitek2 system with P620 card which has 22 antibiotics | 1 |
| Ticked | 1 |
| Grand total | 19 |
| Agents | Count |
|---|---|
| As above | 1 |
| As for S. aureus | 1 |
| Beta-lactams, macrolides | 1 |
| Cefoxitin, oxacillin, vancomycin, erythromycin, fusidic acid, tetracycline, mupirocin | 1 |
| Chloramphenicol, ciprofloxacin, clindamycin, dapto, erythromycin, flucloxacillin, fusidic acid, gentamycin, mupirocin, lizenzolid, penicillin, rifampicin, teicoplanin, tetracycline, tigecycline, trimethoprim, vancomycion (VITEK card) | 1 |
| Clindamycin, gentamicin, oxacillin, tetracycline | 1 |
| E, doxycycline, W5, CN, FD, FOX, neomycin, C11 | 1 |
| Flucloxacillin, erythromycin, clindamycin, trimethoprim, doxycycline, clarithromycin, fusidin, rifampicin, cefoxitin, gentamicin, vancomycin, teicoplanin, linezolid, daptomycin | 1 |
| Flucloxacillin, erythromycin, fusidic acid, rifampicin, gentamicin, tetracycline, clindamycin, cefradine, chloramphenicol, daptomycin, linezolid, penicillin, tigecycline, vancomycion, teicoplanin, trimethoprim | 1 |
| N/A | 1 |
| Pencillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Penicillin, cefoxitin, erythromycin, clindamycin, tetracycline, vancomycin, ciprofloxacin, neomycin, fusidic acid, mupirocin, rifampacin, gentamycin | 1 |
| Penicillin, erythromycin, clindamycin, flucloxacillin, gentamicin, vancomycin, fusidic acid, rifampicin, ciprofloxacin, linezolid, daptomycin | 1 |
| Penicillin, erythromycin, clindamycin, tetracycline, rifampicin, cefotaxine, vancomycin, mupirocin | 1 |
| Penicillin, erythromycin, flucloxacillin, tetracycline, rifampicin, trimethoprim, linezolid, vancomycin, mupirocin, gentamicin, ciprofloxacin, fusidic acid | 1 |
| Sensitivities on these isolates are performed on the Biomeriuex Vitek2 system with P620 card which has 22 antibiotics | 1 |
| Ticked | 1 |
| Grand total | 17 |
| Difference in agents tested against | Count |
|---|---|
| Same | 9 |
| Same (N/A) | 1 |
| Tissue tests numerous, swab tests none | 1 |
| Tissue tests numerous, swab tests none | 1 |
| Grand total | 12 |
| Agents | Count |
|---|---|
| Amoxicillin, co-amoxicillin, imipenem, linezolid, vancomycin, teicoplanin, tetracycline | 1 |
| Amoxicillin, vancomycin, tetracycline | 1 |
| Ampicillin, linezolid, vancomycin, gentamicin, teicoplanin | 1 |
| Ampicillin, clindamycin, erythromycin, high-level gentomycin, linezolid, penicillin, quinupristin/dalfopristin, teicoplanin, tetracycline, tigecycline, vancomycin | 1 |
| Ampicillin, vancomycin | 1 |
| Chloramphenicol, ciprofloxacin, clindamycin, daptomycin, erythromycin, linezolid, teicoplanin, tetracycline, trimethoprim, vancomycin | 2 |
| Glycopeptides, beta-lactams | 1 |
| N/A | 1 |
| None | 2 |
| P, AML, TE, TEC, vancomycin, linezolid | 1 |
| Pencillin, ampicillin, vancomycin, teicoplanin, linezolid, dalfopristin/quinupristin, high-level gentamycin | 1 |
| Pencillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Probably none | 1 |
| Sensitivities on these isolates are performed on the Biomeriuex Vitek2 system with P607 card which has 20 antibiotics | 1 |
| Ticked | 1 |
| Vancomycin, amoxicillin | 1 |
| Vancomycin, amoxicillin, gentamycin | 1 |
| Grand total | 19 |
| Agents | Count |
|---|---|
| Amoxicillin, vancomycin, teicoplanin, gentamicin (high level), linezolid, syndercid | 1 |
| Amoxicillin, co-amoxicillin, imipenem, linezolid, vancomycin, teicoplanin, tetracycline | 1 |
| Amoxicillin, vancomycin, linezolid | 1 |
| Amoxicillin, vancomycin, tetracycline | 1 |
| Ampicillin, linezolid, vancomycin, gentamicin, teicoplanin | 1 |
| Ampicillin, clindamycin, erythromycin, high-level gentomycin, linezolid, penicillin, quinupristin/dalfopristin, teicoplanin, tetracycline, tigecycline, vancomycin | 1 |
| Ampicillin, vancomycin | 1 |
| Ampicillin, vancomycin, linezolid, co-trimoxazole | 1 |
| Glycopeptides, beta-lactams | 1 |
| Pencillin, amoxicillin, teicoplanin, tetracycline, vancomycin, linezolid | 1 |
| Pencillin, ampicillin, vancomycin, teicoplanin, linezolid, dalfopristin/quinupristin, high-level gentomycin | 1 |
| Pencillin, erythromycin, clindamycin, ciprofloxacin, co-amoxiclav, fusidic acid, gentamycin, rifampicin, mupricollin, linezolid, tetracycline | 1 |
| Sensitivities on these isolates are performed on the Biomeriuex Vitek2 system (BioMérieux, Inc., Durham, NC, USA) with P607 card which has 20 antibiotics | 1 |
| Ticked | 1 |
| Vancomycin, amoxicillin | 1 |
| Vancomycin, amoxicillin, gentamycin | 1 |
| When reported ampicillin, gentamicin, teicoplanin, vancomycin, linezolid | 1 |
| Grand total | 17 |
| Difference in agents tested against | Count |
|---|---|
| Same | 9 |
| Tissue tests numerous, swab tests none | 1 |
| Tissue tests numerous, swab tests none | 1 |
| Tissue tests numerous, swab tests none (N/A) | 1 |
| Grand total | 12 |
Local antibiotic protocols
Questions asked:
-
Local antibiotic protocol for infected DFUs, including specific antibiotics, dose, inclusions and contradictions, in:
-
outpatients
-
inpatients.
-
-
Local antibiotic protocol for patients with infected DFUs and osteomyelitis if different from the above.
-
Any other antibiotic protocols you have.
Responses to these questions were provided via free text, with a wide range of in-depth detail provided; for example, at least one site has attached their full protocol/documentation. As such quantitative analysis is not practical here, paper copies of each site’s responses are available for qualitative review if necessary.
Appendix 5 Case report forms
Original forms, questionnaires and trial documentation are included as separate files:
-
screening log
-
eligibility checklist
-
baseline assessment
-
sample collection
-
additional sample collection
-
registration
-
swab sample microbiology
-
swab sample microbiology continuation
-
curettage sample microbiology
-
curettage sample microbiology continuation
-
clinical review
-
RUSAE-related medical history
-
RUSAE
-
withdrawal
-
long-term follow-up
-
protocol violation.
List of abbreviations
- AE
- adverse event
- CI
- confidence interval
- CNS
- coagulase-negative staphylococci
- CODIFI
- COncordance in DIabetic Foot Infection
- DFU
- diabetic foot ulcer
- DNA
- deoxyribonucleic acid
- EM
- expectation–maximisation
- HbA1C
- glycated haemoglobin
- HPA
- Health Protection Agency
- HR
- hazard ratio
- HTA
- Health Technology Assessment
- IDSA
- Infectious Diseases Society of America
- IWGDF
- International Working Group on the Diabetic Foot
- MCMC
- Markov chain Monte Carlo
- MI
- multiple imputation
- MRSA
- meticillin-resistant Staphylococcus aureus
- NICE
- National Institute for Health and Care Excellence
- PABAK
- prevalence- and bias-adjusted kappa
- PAD
- peripheral arterial disease
- PCR
- polymerase chain reaction
- PEDIS
- Perfusion, Extent/Size, Depth/Tissue loss, Infection, Sensation
- PIL
- patient information leaflet
- PP
- per protocol
- RNA
- ribonucleic acid
- RUSAE
- related unexpected serious adverse event
- SSC
- Study Steering Committee