Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 15/17/03. The protocol was agreed in December 2015. The assessment report began editorial review in June 2016 and was accepted for publication in October 2016. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Sian Taylor-Phillips is partly supported by the National Institute for Health Research Collaboration for Leadership in Applied Health Research and Care West Midlands at the University Hospitals Birmingham NHS Foundation Trust.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2017. This work was produced by Freeman et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Introduction
Description of the health problem
Gastroenteritis is a common, transient disorder usually caused by infection with viruses, bacteria or parasites. It is estimated that around 25% of people in the UK have a gastrointestinal infection each year. 1 Gastroenteritis is characterised by acute onset of diarrhoea with or without vomiting. 2 Depending on the cause of the infection, the symptoms of gastroenteritis can take from a few hours to a few days to develop. Most cases resolve without treatment within days, although persistent or severe symptoms may lead sufferers to contact the health services. Patients may be managed in the community or admitted for observation, symptom management and diagnosis. Severe diarrhoea can quickly cause dehydration, which may be life-threatening. The most commonly identified pathogens in England in the Second Infectious Intestinal Disease in the Community Study were norovirus, sapovirus, Campylobacter and rotavirus. 1 Gastroenteritis can also occur in people who are currently taking, or who have recently taken, antibiotics. This is known as antibiotic-associated diarrhoea, which is frequently caused by Clostridium difficile. 3
Diarrhoea may have non-infectious causes, such as inflammatory bowel disease, and it is therefore sometimes desirable to be able to identify or rule out infectious causes of gastroenteritis in people who present to health services with diarrhoea or vomiting. Differential diagnoses for gastroenteritis include non-gastrointestinal infections (e.g. pneumonia, urinary tract infection or human immunodeficiency virus infection), irritable bowel syndrome, inflammatory bowel disease, coeliac disease, side effects of medications, endocrinopathy (e.g. diabetes or hyperthyroidism) and secretory tumours. 2
Patients who are in hospital and have suspected infectious diarrhoea may be nursed in an isolation bay or side room until infection has been ruled out or symptoms subside. Patients in isolation are asked not to enter other areas of the ward or hospital until members of a hospital infection prevention and control team have advised them otherwise, which can potentially result in a poor patient experience. In addition, isolation facilities may be limited and experience periods of overdemand; thus, it is not always possible to isolate all patients. Procedures such as endoscopy may also be cancelled if there is a risk of transmitting infection. The use of rapid gastrointestinal pathogen panel (GPP) tests might reduce the amount of time spent in isolation for some patients if the primary reason for isolation is infection risk rather than symptom management. In addition, for people presenting to primary care services who require faecal microbiology tests, the more rapid provision of test results may provide earlier information for people who are in regular contact with young children and older people, helping them to reduce the risk of transmission within their work and/or home environment. This could bring forward the timing of specific treatment for pathogens such as giardiasis or typhoid for which antimicrobial treatment is usual, as well as to guide the type of treatment provided for very ill or vulnerable patients for whom clinical assessment supports antimicrobial treatment. Early diagnosis may also accelerate exclusion advice to minimise the spread of infection.
The National Institute for Health and Care Excellence (NICE) Diagnostics Advisory Committee is tasked with providing guidance to the NHS about the use of the integrated multiplex tests for patients with acute diarrhoea with or without vomiting, thought to be as result of infectious gastroenteritis. To inform the Diagnostics Advisory Committee, the external assessment group has provided this assessment of the clinical accuracy and cost-effectiveness of selected multiplex panels as a replacement or adjunct for standard assessment procedures. The potential value of the multiplex tests is in rapidly determining the presence and nature of infection, which may be bacterial, viral or parasitic.
Clear definition of interventions
Three integrated multiplex tests have been evaluated as interventions: xTAG GPP (Luminex, Toronto, ON, Canada), FilmArray (BioFire Diagnostics, Salt Lake City, UT, USA) and Faecal Pathogens B (AusDiagnostics, Beaconsfield, NSW, Australia). Clinical judgement is used when interpreting multiplex test findings and may be further informed by current routine tests or other confirmatory testing. This assessment focuses on panels combining investigation of bacteria, viruses and parasites and does not consider partial panels that could be combined to cover all pathogen groups.
xTAG Gastrointestinal Pathogen Panel
The xTAG GPP is a Conformité Européenne (CE)-marked, qualitative, highly multiplexed polymerase chain reaction (PCR) test for the simultaneous detection and identification of nucleic acids from up to 15 gastroenteritis-causing viruses, parasites and bacteria (Box 1). It can analyse human stool samples that are fresh, frozen or in a holding medium. It is intended to be used in a laboratory setting.
Campylobacter.
C. difficile, toxin A/B.
Escherichia coli O157.
Enterotoxigenic E. coli LT/ST.
Shiga-like toxin-producing E. coli stx1/stx 2.
Salmonella.
Shigella.
Vibrio cholerae.
Yersinia enterocolitica.
VirusesAdenovirus 40/41.
Norovirus GI/GII (genogroup).
Rotavirus A.
ParasitesCryptosporidium.
Entamoeba histolytica.
Giardia.
The assay uses reverse transcription PCR and the procedure includes five phases.
Using the laboratory’s amplification platform:
-
pre-treatment of the sample
-
nucleic acid extraction and purification using an automated nucleic acid extraction system
-
broad-range PCR using a thermal cycler
-
bead hybridisation and detection using a thermal cycler.
Using the Luminex analyser:
-
data acquisition and analysis [using Luminex 100/200 (Luminex, Toronto, ON, Canada) or MAGPIX analyser (Luminex, Toronto, ON, Canada)].
A total of 10 µl of purified sample is required for the initial broad-range PCR reaction, which amplifies nucleic acids that are present in the sample. A total of 5 µl of the broad-range PCR product is then added to a hybridisation and detection reaction, in which target nucleic acids bind to species-specific tagged beads. When pathogen nucleic acid is present, fluorescence is emitted by a streptavidin and R-phycoerythrin conjugate, which is included in the reaction.
Fluorescence intensity is measured by either the Luminex 100/200 or MAGPIX analyser, determining which bacterial, viral or parasitic deoxyribonucleic acid (DNA) is present in the sample. Positive and negative controls should be included in each test run. The company recommends that three negative controls (RNase-free water) and at least one positive control (known positive sample) should be included in each run. The assay also contains an internal control, which is added to each sample prior to extraction and indicates whether or not the assay is functioning as intended.
The estimated turnaround time for the xTAG GPP is 5–6 hours, including sample preparation time. This turnaround time may increase if extraction products are transferred by hand to the PCR thermocycler and will therefore vary depending on the number of samples. Up to 96 samples (including controls) can be processed in one run, depending on the capacity of a laboratory’s PCR thermocyclers. The test does not provide any information on antimicrobial resistance genes or antimicrobial susceptibility.
FilmArray Gastrointestinal Panel
The FilmArray Gastrointestinal Panel is a CE-marked, qualitative, highly multiplexed PCR test that can simultaneously detect and identify up to 22 pathogens (Box 2) from stool samples in Cary–Blair transport media. It is intended for use within a clinical laboratory and should be used in conjunction with other clinical and laboratory findings.
Campylobacter (jejuni, coli and upsaliensis).
C. difficile (toxin A/B).
Plesiomonas shigelloides.
Salmonella.
Yersinia enterocolitica.
Vibrio (parahaemolyticus, vulnificus and cholerae).
Vibrio cholerae.
Enteroaggregative Escherichia coli.
Enteropathogenic E. coli.
Enterotoxigenic E. coli lt/st.
Shiga-like toxin-producing E. coli stx1/stx2.
E. coli O157.
Shigella/enteroinvasive E. coli.
VirusesAdenovirus F 40/41.
Astrovirus.
Norovirus GI/GII.
Rotavirus A.
Sapovirus (I, II IV and V).
ParasitesCryptosporidium.
Cyclospora cayetanensis.
E. histolytica.
Giardia lamblia.
The FilmArray GPP is intended for use with FilmArray and FilmArray 2.0 integrated systems, which include automated sample preparation. Each FilmArray unit can process one sample per hour and the FilmArray 2.0 system allows FilmArray units to be linked to process up to eight samples per hour depending on how many modules are purchased. All reagents required for sample preparation, reverse transcription, PCR and detection are provided freeze-dried in a single-use pouch. Before inserting the reagent pouch into the analyser, the sample is combined with sample buffer and is injected into the pouch along with a hydration solution. The system automatically processes a sample through the following stages once a pouch has been inserted:
-
nucleic acid purification
-
reverse transcription and multiplex PCR
-
second-stage ‘nested’ PCR with species-specific primers
-
detection with melting curve analysis.
The system extracts and purifies nucleic acids, which then undergo reverse transcription and are amplified in the first broad-range PCR reaction. A second nested PCR reaction containing species-specific primers is run to detect and identify pathogens present in the sample by fluorescence. Each single-use pouch also contains two internal controls, one ribonucleic acid process control assay and one control assay for the second-stage PCR. Both controls must be positive for the sample to be reported. Results are reported automatically using the FilmArray software (version 2, FilmArray, Salt Lake City, UT, USA).
Faecal Pathogens B (16Plex)
The Faecal Pathogens B assay is a CE-marked, highly multiplexed, qualitative PCR test, which can detect and identify up to 15 pathogens from nucleic acid extracted from fresh faecal samples. The pathogens detected by the assay are shown in Box 3. The assay is intended to be used in conjunction with the High-Plex Multiplex Tandem PCR system (AusDiagnostics, Beaconsfield, NSW, Australia) and Easy-Plex results software (version 1.3, AusDiagnostics, Beaconsfield, NSW, Australia). The assay procedure includes the following processes:
-
nucleic acid extraction and purification using the laboratory’s platform
-
broad-range PCR (using the High-Plex system)
-
real-time PCR with species-specific primers (using the High-Plex system)
-
detection with melting curve analysis
Salmonella spp.
Shigella spp. and Shigella enteroinvasive Escherichia coli.
Campylobacter spp.
C. difficile.
Shiga toxin 1 and 2.
E. coli O157.
VirusesRotavirus A.
Norovirus genogroup I and II.
Adenovirus group F and group G.
Sapovirus.
Astrovirus.
ParasitesGiardia lamblia (18S).
Cryptosporidium (parvus and hominis).
E. histolytica (not dispar).
In the first PCR step, broad-range primers are used and the product of this reaction is diluted and divided into a number of real-time PCR reactions, which use nested species-specific primers to detect and identify any pathogens present in the sample by fluorescence. Results are reported using the Easy-Plex results software. When multiple pathogens are present in a sample, the software provides a relative quantification of targets, providing guidance about the relative importance of each detected pathogen. Each tube used for the broad-range PCR includes an internal positive control (spike) and the company advises that both positive and negative (water) controls are included in each run. Up to 24 samples can be processed in one run. The estimated test turnaround time is 3–4 hours. The assay is intended to be used in conjunction with other clinical and laboratory findings.
Comparative technical overview of the three gastrointestinal pathogen panels
The differing configurations and capacities of the three panels are summarised in Table 1.
| Features of GPPs | xTAG (Luminex) | FilmArray (BioFire Diagnostics) | Faecal Pathogens B (AusDiagnostics) |
|---|---|---|---|
| Number of pathogens | 15 | 22 | 15 |
| Pre-treatment time (minutes) | 30–45 | 2 | 45–60 |
| Turnaround time (hours) | 5 | 1 | 3–4 |
| Throughput (samples/run) | 96 (including controls) | 1 (including controls), up to 8 units per computer | 24 (including controls) |
| Starting material | 100 µl of fresh or newly frozen stool (raw or in holding medium, e.g. Cary–Blair) | 200 µl of Cary–Blair stool | Fresh stool |
| Detection method | Bead hybridisation following PCR | Melting curve following nested PCR | Melting curve following nested PCR |
| System | Open (not an integrated system requiring DNA extraction before loading for analysis and post PCR handling) | Closed [fully integrated system including sample preparation (DNA extraction), PCR reactions and detection] | Open (DNA extraction required before loading for analysis) |
Management of infectious gastroenteritis in hospitals
As previously described (see Clear definition of interventions), patients admitted to hospital with suspected infectious diarrhoea may be nursed with or without isolation. Use of an isolation bay or side room is preferable until infection has been ruled out; however, it may not always be possible to isolate patients when isolation facilities are in full use. When infection prevention control measures are advised for a patient, some procedures, which may not be classed as urgent (e.g. endoscopy), may be postponed until the infection has resolved. Gastrointestinal infections may not be the only reason for isolation of patients, which may be required, for example, for other communicable diseases such as respiratory virus infections, meticillin-resistant Staphylococcus aureus and multidrug-resistant Gram-negative bacteria (e.g. extended-spectrum beta-lactamase producers). Patients carrying, or at risk of carrying, carbapenemase-producing Enterobacteriaceae are an increasingly important cohort who require single-room isolation. 4
Management of infectious gastroenteritis in the community
When infectious gastroenteritis is suspected in the community, people are often advised to absent themselves from work or, in the case of children, from schools and nursery. 5 Advice is also given on reducing the risk of transmission, particularly when highly transmissible pathogens such as norovirus and Shigella are suspected. Infectious gastroenteritis can have particular implications for people in certain professions, such as people who handle food and health-care workers. Food handlers are typically advised to remain away from work until 48 hours after symptoms have resolved; however, infections with certain pathogens, including Salmonella typhi or Salmonella paratyphi and Escherichia coli O157, may require negative microbiology results before the person is able to return to work. 6 In some cases, the detection of suspected food-borne pathogens may result in public health teams initiating an outbreak investigation.
Place of the intervention in the treatment pathway(s)
The main clinical feature of gastrointestinal infection is diarrhoea, but other symptoms can include nausea, sudden onset of vomiting, blood or mucus in stool, or systemic features such as fever or malaise. 2 Public Health England (PHE) advises that no infection-specific treatment is warranted in most patients. Management involves strategies to maintain hydration and steps to prevent cross-infection.
In acute cases, diagnostic investigations are needed to confirm that an infection is present or to determine the causative pathogen. It is recommended that stool samples for microbiological diagnosis are taken in the following situations:
-
There is persistent diarrhoea or malabsorption.
-
There is blood, mucus or pus in the stool.
-
There is a history of diarrhoea and/or vomiting, and the patient is systemically unwell.
-
There is a history of recent hospitalisation.
-
There is a history of antibiotic therapy.
When parasitic infections are suspected, it is recommended that three samples are sent 2–3 days apart as ova, cysts and parasites are shed intermittently. 2 In the case of hospital-acquired gastroenteritis and diarrhoea, laboratories may employ a 3-day rule when deciding whether or not to process stool samples from inpatients for routine bacterial culture (Salmonella, Shigella, Campylobacter and E. coli O157), although testing for C. difficile should be done as soon as infective diarrhoea is suspected. The rule suggests that, in the case of suspected hospital-acquired infection, stool samples should not be sent to microbiology unless one or more of the following criteria are met:
-
diarrhoea develops within 3 days of admission
-
in the case of adults with nosocomial diarrhoea, one of the following is present:
-
pre-existing disease causing permanently altered gut function in a patient aged ≥ 65 years
-
human immunodeficiency virus infection
-
neutropenia/immunocompromised
-
-
a nosocomial outbreak (e.g. Salmonella) is suspected
-
non-diarrhoeal manifestations of enteric infections are suspected.
The setting for multiplex testing will include patients from community and hospital settings, with microbiology laboratories receiving samples from primary and secondary care services. The use of findings will be in their clinical settings.
Relevant comparators
The comparator will be standard microbiology techniques, outlined in the PHE syndromic algorithm for routine testing in cases of gastroenteritis and diarrhoea (see Figure 1). People who have a history of recent travel (to areas other than Western Europe, North America, Australia or New Zealand) may have additional primary testing for Vibrio and Plesiomonas species by bacterial culture. A two-stage testing approach is currently recommended for C. difficile, which involves an initial testing step using either a nucleic acid amplification test or enzyme immunoassay for glutamate hydrogenase. When the initial test is positive, a sensitive toxin enzyme immunoassay should be performed to detect the presence of the toxins that cause illness. 7 The syndromic algorithm also notes that laboratories may opt to test for norovirus only during cooler months when peak incidence occurs (November–April). Blood cultures may also be taken if a patient is systemically unwell. 8
The current laboratory pathway for routine screening of stool samples for people with diarrhoea and vomiting is shown below in Figure 1. 8
FIGURE 1.
Current laboratory pathway for routine screening of bacterial pathogens for people with diarrhoea and vomiting. EIA, enzyme immunoassay; NAAT, nucleic acid amplification test. Adapted with permission from PHE. 8
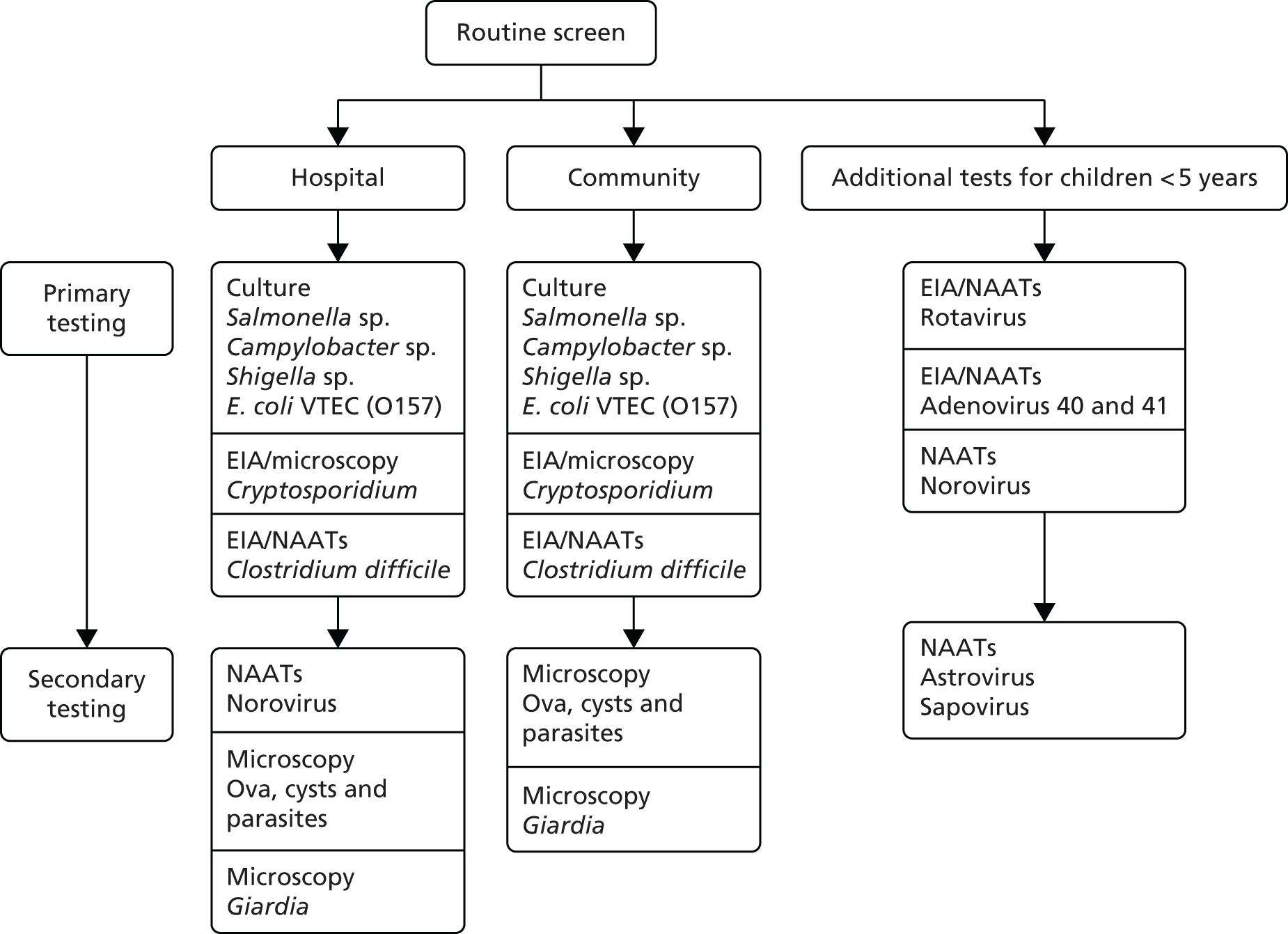
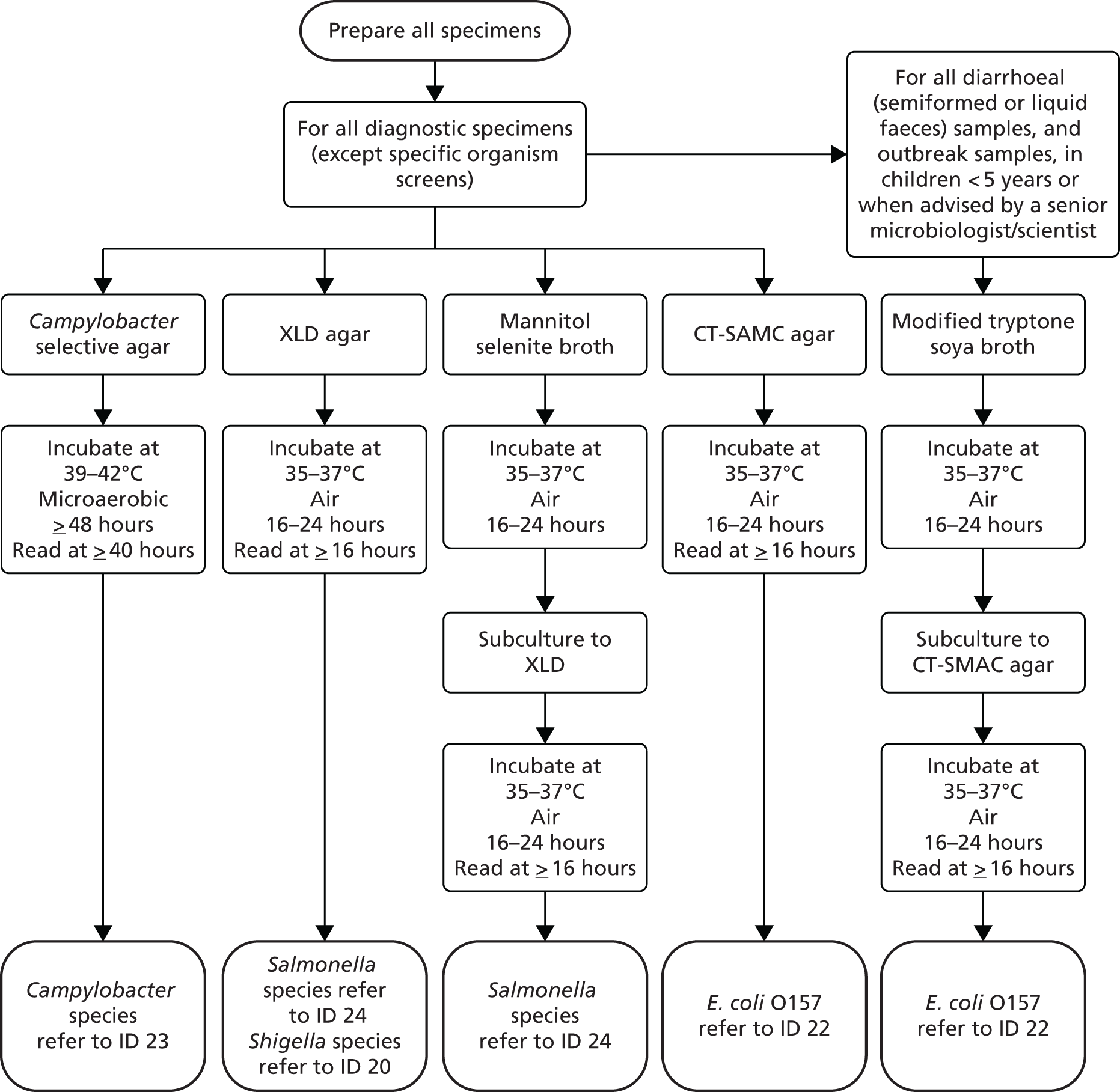
Multiple types of media may be required for bacterial culture and it may take up to 3 days for incubation and pathogen detection. The current bacterial culture protocols recommended by PHE for the investigation of faecal specimens for routine bacterial pathogens9 are shown in Figure 2. The standards also note that rapid diagnostic tests for the direct identification of bacteria directly from faeces, such as enzyme immunoassays and PCR, are available. These tests are thought to be highly accurate for Salmonella, Campylobacter and E. coli O157, but fewer data are available on their effectiveness for detecting toxin-producing bacteria such as Clostridium perfringens, Bacillus species and S. aureus. 9
FIGURE 2.
Overview of microbiological laboratory methods for bacterial pathogens according to Public Health recommendations. Reproduced with permission from PHE. 9
The need for an adequate reference standard
The standard algorithm (representing routine care) presented is an appropriate comparator for the multiplex panels. However, it is apparent that standard microbiology techniques, as described in the standard algorithm, cannot provide a reference standard to evaluate multiplex tests. First-line tests are not 100% accurate and may require confirmatory PCR assays in the case of diagnostic doubt. A range of PCR tests is used across the NHS, but these are used neither widely nor consistently. Multiplex tests may identify substantially higher levels of certain pathogens than standard microbiology. Whether these additional findings are either correct or clinically important (toxin-producing) needs to be determined by an external reference standard. Assessment of diagnostic accuracy depends on determining the performance of both index tests (the GPP panels) and comparator (conventional testing) against a common and adequate reference standard.
Comparison of pathogen coverage between Public Health England and gastrointestinal pathogen panels
A comparison of the pathogen coverage of the PHE algorithm and GPP panels is provided in Table 2.
| Pathogen | PHE | xTAG | FilmArray | Faecal Pathogens B |
|---|---|---|---|---|
| Adenovirus | ✗ | ✗ | ✗ | ✗ |
| Astrovirus | ✗ | – | ✗ | ✗ |
| C. difficile toxin A/B | ✗ | ✗ | ✗ | ✗ |
| Campylobacter | ✗ | ✗ | ✗ | ✗ |
| Cryptosporidium | ✗ | ✗ | ✗ | ✗ |
| Cyclospora cayetanensis | – | – | ✗ | – |
| E. coli O157 | ✗ | ✗ | ✗ | ✗ |
| Enteroaggregative E. coli | – | – | ✗ | – |
| Entamoeba histolytica | – | ✗ | ✗ | ✗ |
| Enteropathogenic E. coli | – | – | ✗ | – |
| Enterotoxigenic E. coli | – | ✗ | ✗ | ✗ |
| Giardia | ✗ | ✗ | ✗ | ✗ |
| Norovirus | ✗ | ✗ | ✗ | ✗ |
| Plesiomonas shigelloides | ✗ | – | ✗ | – |
| Rotavirus | ✗ | ✗ | ✗ | ✗ |
| Salmonella | ✗ | ✗ | ✗ | ✗ |
| Sapovirus | ✗ | – | ✗ | ✗ |
| Shigella/enteroinvasive E. coli | ✗ | ✗ | ✗ | ✗ |
| Shiga toxin-producing E. coli | – | ✗ | ✗ | ✗ |
| Vibrio (parahaemolyticus, vulnificus and cholerae) | – | – | ✗ | – |
| Vibrio cholerae | ✗ | ✗ | ✗ | – |
| Yersinia enterocolitica | – | ✗ | ✗ | – |
| Total | 14a | 15 | 22 | 15 |
Chapter 2 Definition of the decision problem
Decision question
This report, prepared for the NICE Diagnostics Assessment Programme, examines the clinical effectiveness and cost-effectiveness of GPPs, which include all three types of pathogen: bacteria, viruses and parasites. The report will help NICE when making recommendations about how well the tests work and whether or not the benefits are worth the cost of the tests when used in the NHS in England. The assessment considers both clinical improvement in patients’ symptoms and the cost of the tests using evidence identified through systematic reviews and information submitted to NICE during the evaluation process by the companies providing the GPP tests.
The decision question for this project is: what is the clinical effectiveness and cost-effectiveness of the xTAG, FilmArray and Faecal Pathogens B GPPs in the identification of gastrointestinal bacteria, viruses and parasites in patients with suspected gastroenteritis presenting in primary or secondary care, compared with conventional microbiological methods outlined in the PHE standard?8
Overall aim of the assessment
The overall aim of this report is to present evidence on the clinical effectiveness and cost-effectiveness of the xTAG, FilmArray and Faecal Pathogens B GPPs in the identification of gastrointestinal bacteria, viruses and parasites compared with conventional microbiological methods.
Objectives
To systematically review the evidence for the clinical effectiveness of the GPP tests (xTAG, FilmArray and Faecal Pathogens B), systematically review existing economic evaluations and develop a de novo economic model to assess the cost-effectiveness of GPP tests compared with the standard of care in England and Wales.
Chapter 3 Clinical effectiveness review
Methods
Identification and selection of studies
Search strategies for clinical effectiveness
The search strategy for the clinical effectiveness review is detailed in Appendix 1. Briefly, the search strategy included:
-
databases: Ovid MEDLINE 1946 to November week 3 2015; Ovid MEDLINE In-Process & Other Non-Indexed Citations 31 December 2015; Ovid EMBASE 1980 to 2015 week 52; Web of Science 1980 to 31 December 2015; Cochrane Database of Systematic Reviews issue 5 of 12, May 2016 – all sections. Weekly auto-alerts for emerging evidence were run in Ovid MEDLINE, Ovid EMBASE and PubMed from 1 January 2016 to 30 April 2016
-
reference lists of all reviews and included studies
-
websites of NICE, PHE, Food and Drug Administration (FDA) and the manufacturers of the multiplex PCR tests
-
ongoing studies of the following sources: National Institutes of Health ClinicalTrials.gov, Current Controlled Trials, World Health Organization International Clinical Trials Registry Platform and UK Clinical Trials Gateway.
Inclusion and exclusion of relevant studies
Inclusion criteria
Studies that satisfied the criteria outlined in Table 3 were included.
| Criterion | Detail |
|---|---|
| Population | Patients with acute diarrhoea with or without vomiting, thought to be a result of infective gastroenteritis, with test referrals from hospital and community. Subgroups evaluated include people in the community, people in hospital, children aged < 5 years, people with recent foreign travel and people who are immunocompromised |
| Intervention | xTAG, FilmArray or Faecal Pathogens B GPPs |
| Comparator | Standard microbiology techniques, outlined in the PHE syndromic algorithm for routine testing in cases of gastroenteritis and diarrhoea8 |
| Outcome | Outcomes of test performance – primary:
|
| Study design | Test–treat trials comparing clinically relevant outcomes (e.g. morbidity, mortality, length of stay and length of isolation) for patients randomised to either conventional testing or GPP |
| Clinical diagnostic test accuracy studies that compare the index tests (GPP) and the comparator (conventional methods) to an adequate reference standard (if an adequate reference standard exists or is reported) | |
| Studies that compare discrepant results between the index tests (GPP) and the comparator (conventional methods) using an unbiased umpire test (if an adequate reference standard does not exist or is not reported) | |
| Studies of agreement and disagreement between the index tests (GPP) and the comparator (conventional methods) without using an unbiased umpire test (neither an adequate reference standard nor an unbiased umpire test exists or is reported) | |
| Studies of head-to-head comparisons of different index tests (GPP) reporting agreement of tests | |
| Health-care setting | Clinical laboratory receiving samples from primary and secondary care |
Exclusion criteria
Studies that satisfied the criteria outlined in Table 4 were excluded.
| Criterion | Detail |
|---|---|
| Population | ‘Spiked’ samples, swab testing or non-representative populations |
| Intervention | Other modular or partial multiplex tests, index tests during outbreaks or for the routine management of chronic conditions |
| Study design | Reviews, biological studies, case reports, editorials and opinions, poster presentations without supporting abstracts, non-English-language reports, meeting abstracts without sufficient detail on test performance for 2 × 2 data table extraction per pathogen |
Spiked samples are most commonly samples from healthy volunteers to which pathogens have been added; therefore, the type and concentration of pathogen is known and the samples do not represent true clinical samples.
Using the information provided by the manufacturers
The information provided by Luminex, BioFire Diagnostics and AusDiagnostics (see Appendix 2 for an itemised list of documents received) was screened for three purposes:
-
to identify potential additional studies not identified by our searches
-
to identify unpublished test accuracy and clinical effectiveness data
-
to obtain information on the technical description of the three intervention assays.
Study selection strategy
All publications identified in searches from all sources were collated in EndNote (X7, Thomson Reuters, CA, USA) and de-duplicated. Two reviewers independently screened the titles and abstracts of all records identified by the searches and discrepancies were resolved through discussion. Full copies of all studies deemed potentially relevant were obtained and two reviewers independently assessed these for inclusion; any disagreements were resolved by consensus or discussion with a third reviewer. Records rejected at full-text stage and reasons for exclusion were documented. Three authors were contacted to confirm that the included study population was eligible. Eligible studies identified after 23 March 2016 were not included in the analysis but contributed to the discussion of the clinical effectiveness.
Data extraction strategy
Data were extracted by one reviewer using a piloted data extraction form. Completed data extraction forms are available from the authors. A second reviewer checked the extracted data. Any disagreements were resolved by consensus or discussion with a third reviewer. A sample data extraction form used in this review is available in Appendix 3.
Test results for GPP and comparator tests were extracted into 2 × 2 contingency tables following the format shown in Table 5. Ideally, studies provided a comparison for each sample of the full PHE algorithm and an index panel test. For studies in which each sample might receive only a selection of conventional methods (according to physician’s choice), the denominator was adjusted accordingly for each pathogen. Only samples that received (for each pathogen) both the index test and conventional methods were included in the 2 × 2 table. The four cells of the 2 × 2 table are subsequently referred to as a(+/+), b(−/+), c(+/−) and d(−/−).
| Test | Comparator + | Comparator – |
|---|---|---|
| GPP + | +/+ | –/+ |
| GPP – | +/– | –/– |
Quality assessment strategy
Quality assessment of eligible studies was undertaken with a highly tailored Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool. 10 A single reviewer determined the methodological quality of included studies and a second reviewer checked findings. Any disagreements were resolved by consensus or arbitration.
Quality assessment aimed to assess the risk of bias and applicability concerns of included studies on the pathogen level, where the GPP method was the index test, conventional methods were the comparator and any efforts to verify discordant results were assessed under the reference standard domain. The studies did not fit the conventional test accuracy study format, as there is no reference standard, and each pathogen investigated had to be considered individually in terms of test accuracy. Modifications to tailor the QUADAS-2 form to the research question in terms of the risk-of-bias assessment are detailed next (see Appendix 4 for the tailored QUADAS-2 form and guidance notes).
Patient selection domain
Addition of one signalling question: was only one sample per episode of diarrhoea included in the study?
It is important that studies do not include more than one sample per patient per episode of diarrhoea to avoid double counting. Many studies reported samples rather than patients, or studies reported greater numbers of samples included than numbers of patients, where it would be important to ascertain that samples are representative of one episode of diarrhoea. We assumed that a period of 1 month between samples was required to define separate episodes of diarrhoea.
Index test domain
The addition of one signalling question: was the index test undertaken as recommended by the manufacturer?
Bias may occur if the study deviates from the manufacturers’ recommendations in the conduct and interpretation of the GPP tests. Manufacturers provide clear package inserts for the handling of samples and equipment. Deviation in terms of type of sample (fresh, frozen, pure, in Cary–Blair medium), amount of sample and handling of sample if not tested immediately may result in systematic differences in the performance of the test.
Comparator domain
The addition of a domain for the comparator.
The eligible studies compared GPP testing with a comparator that consisted, broadly speaking, of a range of conventional microbiological tests that cannot be classed as the reference standard because GPP testing may be superior to conventional methods. Therefore, the comparator was assessed in addition to the index test and the reference standard. Similar signalling questions in terms of blinding and thresholds were considered for the comparator. In addition, one signalling question was added: was culture performed on fresh (not previously frozen) samples? Culture from frozen samples appear to be less reliable than culture from fresh samples, which will result in greater discrepancies between conventional and GPP methods for pathogens that are confirmed by culture. 11,12
Reference standard domain
The addition of one signalling question: was the reference standard independent and unbiased?
As there is no independent and reliable reference standard for the assessment, we looked for an independent and unbiased umpire test to be used in the studies to assess discordant results between index test and conventional methods [e.g. exposure, treatment effect (pathogen-specific treatment) or self-reported symptoms, previous antibiotic treatment, over-the-counter medicine].
Flow and timing domain
The adjustment of one signalling question: did all discordant samples receive a reference standard?
For a fair umpire test to be a valid verification method, all discordant results rather than a proportion should be tested.
The addition of one signalling question: did all samples receive the comparator methods for all pathogens considered in the study?
The risk of bias is high if patients received conventional tests only on the basis of prior assessments by physicians and according to symptoms.
Methods of analysis/synthesis
Following 2 × 2 data extraction, several approaches to data analysis were considered. In order to justify the approach, the limitations of the available data are explained, rendering more robust methods inappropriate.
Summary of strategies for analysis
Test–treat trials
The most pertinent study design would be a test–treat trial comparing clinically relevant outcomes such as morbidity, mortality, length of stay and length of isolation for patients randomised to either conventional testing or GPP. In the absence of an appropriate reference standard, and as the primary interest is the effect on clinically relevant outcomes, this would be the most informative study design. However, no test–treat trials investigating this question in the relevant population were identified.
Conventional test accuracy paradigm: calculation of sensitivity and specificity
Following the conventional diagnostic test accuracy paradigm, the ideal scenario would be to derive sensitivity and specificity as a measure of test performance of GPP testing. This would require testing all samples with a reference standard. The reference standard would need to be a highly accurate (and ideally independent) test, recognised as the best available. It is known that PCR can detect pathogen DNA at very low levels, including from non-viable organisms. Neither PCR nor culture can be used as a reference standard because we do not know which is more accurate and because it might introduce significant bias to define one or other test as the reference standard. There are no other appropriate tests to act as reference standard, therefore sensitivity and specificity cannot be calculated. 13
Discrepant analysis
Discrepant analysis14 involves retesting discordant results between the index test (GPP) and comparator test (conventional testing), using a resolving test with better discriminatory properties, to update the final 2 × 2 table. Sometimes this approach has been used in studies to calculate sensitivity and specificity of the tests using the assumption that when both tests agree (without resolving test) then they are both correct. This may produce inflated estimates of sensitivity and specificity. This was not an appropriate approach for this review15 because of the potential bias introduced and the lack of a suitable resolving test.
Fair umpire test
The fourth approach considered was not based on the conventional diagnostic test accuracy paradigm, in the absence of a reference standard. In this scenario, in addition to the 2 × 2 data of association between the GPP test and conventional methods, verification results for discordant outcomes using an unbiased, independent (but possibly imperfect) umpire test are considered, for example exposure levels to the pathogen of interest. The characteristics of discordant results in cell b of a 2 × 2 table (GPP positive, conventional test negative) are compared with those in cell c (GPP negative, conventional test positive). The characteristics to be compared must have a better than chance association with the condition or outcome of interest and, crucially, be unbiased towards one test or another. This can provide unbiased information on which is the most accurate test, but cannot be used to calculate sensitivity or specificity. 13 In anticipation of this approach, information on verification and characterisation of discordant results were sought, assessed for independence from the index test and comparator, and 2 × 2 tables updated with information on discordant outcomes to allow unbiased assessment of the direction of resolution of discordant results in favour of either the index test or the comparator. 13
This approach yielded no results: discordant results were verified only in a limited number of studies and only with PCR-based methods. These methods are not independent and unbiased, as PCR assays are not sufficiently different from GPP tests (i.e. the index test), which are also based on PCR methodology and should produce biased results in favour of GPP testing. No fair umpire test based using exposure or outcome variables was identified in any of the included studies.
Test agreement: kappa
Test agreement (Cohen’s kappa) was considered as a single measure of agreement between tests using the updated 2 × 2 tables. However, Cohen’s kappa is only really useful and interpretable for test–retest reliability, not for comparing different tests (conventional tests and GPP), when small deviations from a kappa of 1 could have clinically meaningful implications. Furthermore, Cohen’s kappa is unstable when individual cell counts are low in a 2 × 2 table, as they are for many pathogens in this review. 16 The use of a single measure of agreement, such as Cohen’s kappa in this instance, is, therefore, discouraged. 17
Methods of analysis
In the absence of any test–treat trials, a suitable reference standard or any fair umpire tests in any included studies, we followed the FDA recommendations on reporting results from studies evaluating diagnostic tests. 17 In the absence of a reference standard, the guidance discourages the reporting of sensitivity and specificity and recommends reporting measures of positive and negative test agreement.
We calculated positive agreement (a/a + c) and negative agreement (d/b + d) when benchmarked against the comparator, and positive agreement (a/a + b) and negative agreement (d/c + d) when benchmarked against GPP for each pathogen to determine the range of feasible outcomes of test agreement.
The resulting measures of agreement are not measures of performance, as it is not known which test is correct. Agreement could be poor because one test is more accurate than the other or because they are both poor. There is no statistical method to determine which scenario is correct. As recommended in the guidance,17 we used the original 2 × 2 table data without updating results from discrepant analyses because, when attempted in a minority of studies, the resolving test used was not a reference standard.
Positive and negative agreement benchmarked against conventional methods and positive and negative agreement benchmarked against GPP were then meta-analysed by pathogen if the denominator was ≥ 20 to achieve agreement estimates and explore heterogeneity.
We undertook a random-effects meta-analysis of proportions using the metaprop command in Stata SE 14.1 (StataCorp LLC, College Station, TX, USA). 18 The analysis was exploratory and no adjustment was made for the interdependence of positive and negative values. Exact binomial methods were used to estimate 95% confidence intervals (CIs) using the Freeman–Tukey transformation of proportions, and the I2-statistic of between-study heterogeneity was computed.
The analysis was undertaken for each pathogen at the sample level, assuming independence between samples within studies and between studies, that is, having one pathogen does not affect the likelihood of having another pathogen.
Definitions for the review
Definition of the Public Health England algorithm
The PHE algorithm8 consists of primary testing for Salmonella, Campylobacter, Shigella, E. coli O157, Cryptosporidium and C. difficile with additional tests for children aged < 5 years for rotavirus, adenovirus 40/41 and norovirus. Secondary testing in hospitalised adults is recommended for norovirus and ova, cysts and parasites including Giardia, whereas people in the community should not be tested for norovirus and children aged < 5 years should also be tested for astro- and sapovirus. Furthermore, our assessment included travellers as a subgroup of interest for which additional tests for Vibrio and Plesiomonas species are considered if they are returning from areas other than Western Europe, North America, Australia or New Zealand. Physicians following this algorithm use local tests that vary across the UK and may or may not request all tests detailed in the PHE algorithm, depending on symptoms and suspicion following patient assessment. In order to compare GPP testing with microbiology techniques, as outlined in the PHE syndromic algorithm for routine testing, a clear definition of the comparator is needed.
For the purpose of this review, no differentiation is being made between primary and secondary testing or differences in terms of population testing (children, adults, travellers) and seasonality (including norovirus testing in winter months only). For a sufficiently equivalent comparator, all pathogens mentioned in the syndromic PHE pathway8 for any of the populations of interest were included. A pragmatic judgement was required regarding whether or not the number of tested pathogens in the studies is sufficiently similar to the PHE algorithm [i.e. at least 75% (≥ 11 of the above pathogens) included in study].
In terms of tests, the following was assumed: testing is classed as sufficiently equivalent to the PHE algorithm if the comparator in the studies includes:
-
culture or PCR for bacteria (see final bullet point for C. difficile)
-
PCR or enzyme immunoassay for viruses
-
microscopy or enzyme immunoassay for parasites
-
PCR and/or enzyme immunoassay (plus toxin test) for C. difficile.
It is noteworthy that even if the type of method is equivalent, there are a huge number of different tests and testing kits currently used within the NHS for the various pathogens. Therefore, we have included studies using a wide range of conventional microbiology approaches, reflecting variation in health services practice.
Clinical effectiveness results
Search results
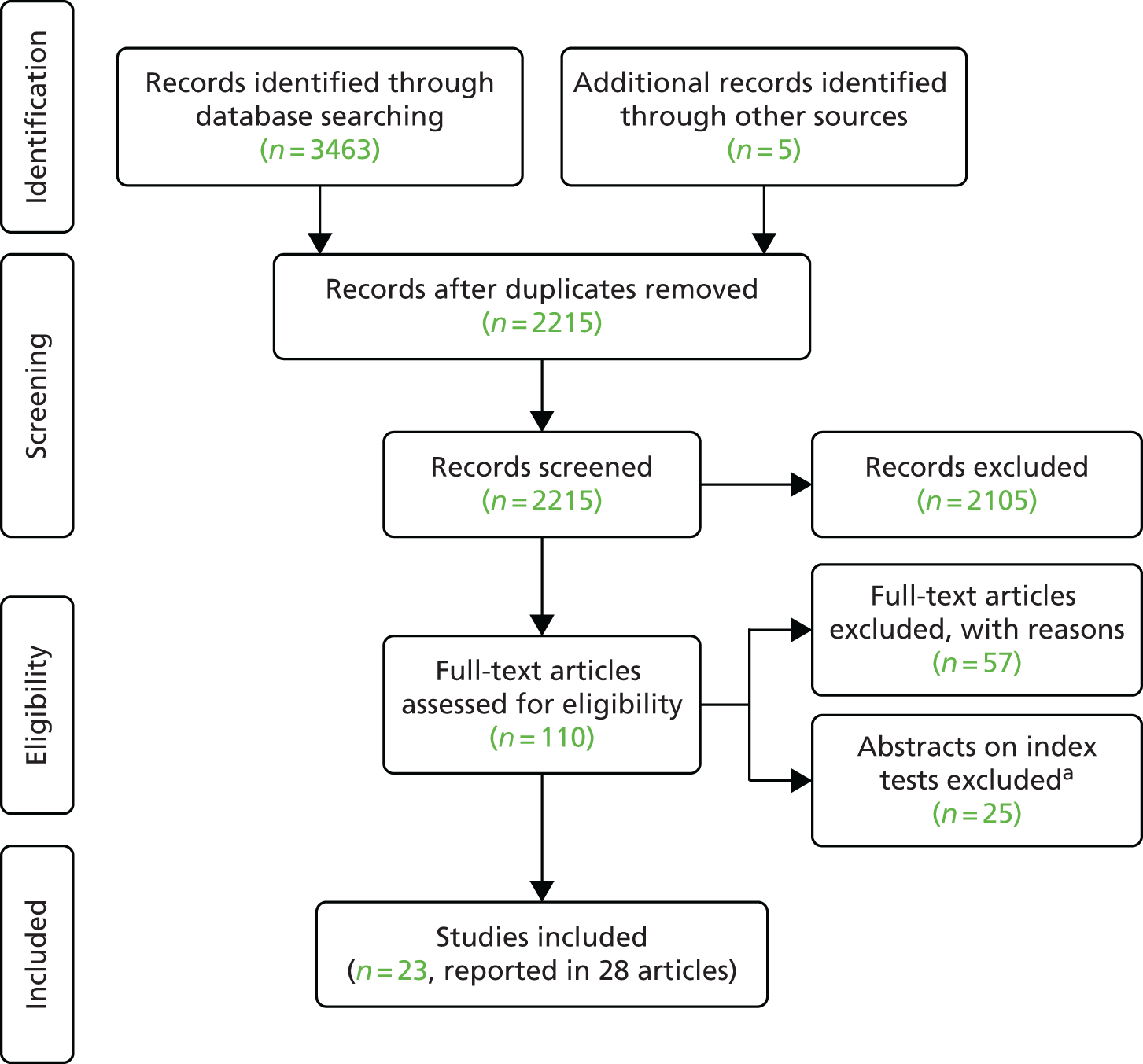
The process of study identification and selection for the clinical effectiveness review is illustrated in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) diagram (Figure 3). The search identified 3468 records. Following duplicate removal, we screened 2215 unique records, of which 110 were taken forward to full-text assessment. A total of 57 studies were subsequently excluded, and a further 25 abstracts that did not report sufficient data on the validation method, the comparator and/or the outcomes per pathogens tested also were excluded. The list of excluded full texts and abstracts with reasons can be found in Appendix 5. We included a total of 23 studies11,12,19–39 that were reported in 28 articles.
FIGURE 3.
The PRISMA flow diagram of study selection for the clinical effectiveness review. a, Without sufficient detail on test performance for 2 × 2 data table per pathogen.
The xTAG GPP assay trial for FDA approval tested 14 pathogens and was reported in two FDA documents,25,40 as well as one review by the company. 41 This study is referenced as FDA25 in this report. Similarly, the trial undertaken by BioFire for the approval of the FilmArray GPP test was reported variously as FDA report,42 as 510(k) submission to the FDA,43 as part of the instruction manual44 and as a published study. 20 This study is referenced as Buss et al. 20 in this report.
The search for ongoing trials identified one ongoing trial for which no outcomes were available for this review (see Appendix 6). Screening of the information provided by the companies did not identify any additional published or unpublished studies.
Study characteristics
The 23 studies11,12,19–39 included in the clinical effectiveness review are described in Table 6.
| Study reference; location; n | Study population, n (%) | GPP assay | Comparator PHE equivalent yes/no (justification) | Verification yes/no | 2 × 2 data (association between GPP and comparator) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Setting | IC | Travellers | Aged ≤ 5 years | |||||||
| Mixed | Hospital | Community | ||||||||
| Becker et al. 2015;19 Côte d’Ivoire; 68 | 0 | 0 | 68 (100) | NR | NR | 22 (32) | xTAG (15 pathogens) | No (only two PHE pathogens considered) | No | No (symptomatic and asymptomatic patients combined) |
| Beckmann et al. 2014;11 Switzerland; 296 | 120 (40) | NR | 176 (59) | NR | 176 (59) | 106 (36) | xTAG (15 pathogens) | No (only 5 PHE pathogens, children only EIA for viruses and travellers only microscopy for parasites) | Yes; xTAG positive samples and random sample of 72 negative samples | Incomplete |
| Buss et al. 2015;20 USA; 1556 | 164 (11) | NR | 1392 (89) | NR | NR | 539 (35) | FilmArray (23 pathogens, IUO version includes Aeromonas) | No (real-time PCR was used in conjunction with bidirectional sequencing) | Yes; discordant results | Complete |
| Claas et al. 2013;21 the Netherlands, USA, UK, Canada; 901 | 901 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | Yes (some laboratories used PCR for parasites; 11 PHE pathogens) | Yes; subset of samples (uninformative) | Complete |
| Coste et al. 2013;22 France; 54 samples from 49 patients | 0 | 54 (100) | 0 | 54 (100) | NR | NR | xTAG (number of pathogens considered, NR) | No (C. difficile tested with EIA and culture; 10 PHE pathogens) | Yes; stool samples positive for one of the enteric viruses or for Campylobacter | Complete |
| Deng et al. 2015;23 China; 290 | 70 (24) | NR | 220 (76) | NR | NR | NR | xTAG (15 pathogens) | No (culture confirmed by serotyping or sequencing) | Yes; discordant results | Complete |
| Duong et al. 2016;24 Vietnam; 479 | 479 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (only six PHE pathogens) | No | Complete |
| FDA 2012;25 USA, Canada; 1407 | 850 (60) | NR | 557 (40) | 493 (35) | NR | 26 (2) | xTAG (14 pathogens, excluding Yersinia) | No (conventional methods combined with PCR and sequencing for most pathogens) | Yes; discordant results | Complete |
| Gu et al. 2015;26 USA; 199 | 0 | 199 (100) | 0 | 199 (100) | NR | NR | xTAG (15 pathogens) and FilmArray (23 pathogens, IUO version with Aeromonas) | Yes (14 PHE pathogens tested) | No | Complete |
| Halligan et al. 2014;27 UK; 2187 | 0 | 1201 (55) | 986 (45) | NR | NR | NR | xTAG (15 pathogens) | Yes (11 PHE pathogens; Vibrio on request only) | No | Complete |
| Kahlau et al. 2013;28 Germany; 347 | 347 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (only five PHE pathogens included) | No | No |
| Khare et al. 2014;12 USA; 500 | 500 (100) | NR | NR | NR | NR | NR | xTAG (RUO version 15 pathogens) and FilmArray (IUO version 23 including Aeromonas) | Yes (11 PHE pathogens included) | Yes for most; discordant results | No (outcomes are associations between GPP and comparator + verification method) |
| Mengelle et al. 2013;29 France; 440 samples from 329 patients | 0 | 208 (63) | 121 (37) | 152 (46) | NR | NR | xTAG (15 pathogens) | Yes (11 PHE pathogens included) | Yes; only discordant adenovirus results | Complete |
| Pankhurst et al. 2014;30 UK; 839 | 839 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (only four PHE pathogens considered) | Yes; discordant results | Complete |
| Patel et al. 2014;31 USA; 167 | 167 (100) | NR | NR | NR | NR | NR | xTAG (11 pathogens) | No (only nine PHE pathogens considered, E. coli O157 followed up with 16S sequencing) | Yes; discordant results | No |
| Perry et al. 2014;32 UK; 1472 | 1472 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (11 PHE pathogens considered but all viruses only on request and Vibrio only for children aged < 2 years) | Yes; discordant results | No |
| Petterson et al. 2016;33 USA; 328 | 231 (70) | NR | 97 (30) | NR | NR | NR | xTAG (RUO version 15 pathogens) | No (only nine PHE pathogens considered) | Yes; discordant results and xTAG positive/conventional not tested samples | No |
| Rand et al. 2015;34 USA; 158 | 158 (100) | NR | NR | NR | NR | NR | FilmArray (IUO version with Aeromonas, 23 pathogens) | No (only two PHE pathogens considered: rotavirus and C. difficile if negative by conventional test) | Yes, for norovirus positive samples only | No (conventional positive samples not available) |
| Spina et al. 2015;35 Europe; 709 | 0 | 0 | 709 (100) | NR | NR | 108 (15) | FilmArray (22 pathogens) | Unclear (routine tests at local laboratories of 10 different countries) | No | No |
| Stockmann et al. 2015;36 USA; 339 | 339 (100) | NR | NR | NR | NR | 135 (40) | FilmArray (IUO version with Aeromonas, 23 pathogens) | Yes (11 PHE pathogens considered) | No | No |
| Vocale et al. 2015;37 Italy; 664 | 664 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (only 10 PHE pathogens considered) | No | No |
| Wessels et al. 2014;38 the Netherlands; 393 | 393 (100) | NR | NR | NR | NR | NR | xTAG (15 pathogens) | No (parasites and viruses detected by multiplex PCR, only 10 PHE pathogens) | Yes; discordant results and xTAG positive/conventional not tested samples | Incomplete (proportion for xTAG negative samples not reported) |
| Zboromyrska et al. 2014;39 Spain; 185 samples from 174 patients | 0 | 0 | 147 (100) | NR | 147 (100) | NR | xTAG (15 pathogens) | No (only seven PHE pathogens considered) | Yes; discordant results and xTAG positive/conventional not tested samples | Incomplete (proportion for xTAG negative samples not reported) |
Participants
The included patient population and the setting of studies were insufficiently characterised, possibly as result of the inclusion of samples rather than patients in the majority of studies. Only one study11 differentiated between hospital-based samples (from children admitted with suspected viral gastroenteritis and children presenting to the emergency room) and community-based samples [adult and child travellers whose samples were submitted by general practitioners (GPs)]; the study population consisted of children and travellers only. Seven studies19,22,26,27,29,35,39 reported the setting sufficiently to allow judgement on the origin of the infection (hospital vs. community). The majority of studies11,12,20,21,23–25,28,30–34,36–38 reported recruitment of hospitalised patients, for which a mixed population should be assumed (in terms of origin of infection) as it is unclear at what point in time during hospitalisation each infection occurred. Only one study was identified as a community study, set in Côte d’Ivoire. 19 Subgroups of interest were reported in 10 studies11,19,20,22,25,26,29,35,36,39 and, of these, one considered travellers only39 and two considered immunocompromised patients only. 22,26 Children aged < 5 years were not considered separately in any study, but the proportion of children aged < 5 years was reported in six studies11,19,20,25,35,36 and ranged from 2% to 40% of the total study population. These studies did not report outcomes by setting or subgroup at the pathogen level, which is why reported outcomes should be regarded as applicable to a mixed population. However, one study reported study-level results for hospital- and community-acquired infections. 27
Country of study
Eleven studies11,22,27–30,32,35,37–39 included participants from European countries, including three studies from the UK. 27,30,32 One study was multinational, covering North America and Europe,21 eight studies were from North America,12,20,25,26,31,33,34,36 two were from Asia23,24 and one was from Africa. 19 Therefore, the applicability of the study population in terms of prevalence of pathogens is questionable in 3 out of the 23 studies (see Quality considerations of included studies).
Index test
Overall, 17 studies11,19,21–25,27–33,37–39 evaluated xTAG and four studies20,34–36 evaluated FilmArray. Two studies12,26 compared both tests but no study was identified that assessed the Faecal Pathogens B assay. The majority of studies11,12,19,21,23,24,26–30,32,33,37–39 investigated xTAG for all 15 pathogens, of which two studies12,33 specified the research use only version (pre-FDA approval). However, two studies considered either 1131 (excluding adenovirus, Entamoeba histolytica, Vibrio cholerae and Yersinia) or 1425 pathogens (excluding Yersinia). Initial FDA approval of xTAG only covered 11 pathogens but was extended to 14 pathogens following an additional submission by Luminex requesting to extend reporting to a further three pathogens in 2014. 40 However, CE marking for xTAG covers all 15 pathogens. 48 Therefore, variation of the number of pathogens included in the studies exists. The assay methods are identical in the different versions. 40 Similarly, an investigational use only version of the FilmArray panel test included a test for Aeromonas. This pathogen is not included in the FDA-cleared version of the panel test. Therefore, the majority of FilmArray studies included 23 pathogens12,20,26,34,36 using the investigational use only version rather than the CE-marked 2235 pathogen panel.
Comparator
A methodological limitation of studies included in this review concerns the varying ways in which comparator tests are implemented between studies. Relatively few studies have used a mix of microbiological and PCR methods reflected in the PHE pathway, with an over-reliance on PCR methods and sequencing. A further difficulty is the inconsistent use of the full comparator method. In most studies the GPP system is compared with a subset of conventional pathogen tests rather than assessing the full PHE algorithm.
According to our definition of the PHE algorithm (see Definitions for the review), only six studies12,21,26,27,29,36 can be classed as having used a comparator sufficiently similar to the routine screening pathway recommended by PHE in terms of pathogens included (at least 11) and methods used. None of the studies tested patients or samples with all conventional tests. Pathogens were tested according to the physician’s request only. Two studies19,34 considered as few as two pathogens in common with the PHE algorithm, whereas two restricted conventional testing to only five pathogens28 and four pathogens. 30 Rand et al. 34 included samples negative for rotavirus and C. difficile only and Beckmann et al. 11 tested children only for viruses and travellers only for parasites with conventional methods. The two main clinical studies by the companies20,25 used a comparator that was significantly different (PCR and sequencing for the majority of pathogens) to typical conventional methods, whereas one study from the Netherlands38 also used methods different from the PHE methods, namely multiplexed PCR for parasites and viruses. Although multiplexing is emerging in UK laboratories for individual pathogens or types of pathogens, this is not specified in the PHE algorithm. 8 The assessment of the comparator in terms of equivalence to the algorithm was not possible for one multinational study35 in which conventional methods were described as routine tests undertaken at local laboratories in 10 different countries.
Outcomes
Ten studies20–27,29,30 reported results of association between the comparator and the GPP test in sufficient detail by pathogen to allow the construction of 2 × 2 tables by pathogen. These studies are further characterised in Table 7 and contributed data to the meta-analysis in Pathogen-level positive and negative agreement.
| Study reference; location; number of centres | GPP test | Population; sample size; characteristics | Study design | Agreement outcome reported |
|---|---|---|---|---|
| Buss et al. 2015;20 USA; 4 | FilmArray | n = 1556 | Study set up: culture was set up as part of routine clinical testing at four testing sites using their standard procedure, submitting physicians may have ordered testing in addition to stool culture; however, the results of such routine testing were not collected or utilised in this study for comparator analysis. FilmArray was performed at study sites Comparator: bacteria – specimens were tested using stool culture. Viruses/parasites/C. difficile toxins/STEC/ETEC/EPEC/EAEC – real-time PCR (two independent well-validated assays for each analyte different from the FilmArray GI Panel targets if possible) and sequence analysis were performed by BioFire personnel. Comparator was regarded positive if both tests were positive; if discrepant, both PCRs were repeated All samples received all comparator tests with two exceptions regarding interpretation of results: in order to follow the same algorithm as FilmArray utilises, EPEC results were classed as NA if they were STEC positive and E. coli O157 results were only considered if the STEC test was positive GPP: IUO version therefore including results for Aeromonas, all samples tested with FilmArray at study sites within 4 days of sampling, samples shipped frozen on a weekly basis Verification: discordant samples tested blinded by BioFire, PCR and sequencing using different targets to comparator method and FilmArray, or using enhanced methods (additional PCR cycles and replicate samples) or using bench top version of FilmArray (FilmArray primers in conventional real-time PCR) |
Positive per cent agreement and negative per cent agreement instead of sensitivity and specificity to indicate that a non-gold-standard assay (e.g. PCR) was used for the comparator analysis |
| Hospitalised: 164 (10.5%) | ||||
| ER: 42 (2.7%) | ||||
| Outpatients: 1350 (86.8%) | ||||
| Adults aged > 21 years: 584 (38%) | ||||
| Children aged 0–5 years: 539 (35%) Children and young people aged 6–21 years: 433 (27%) |
||||
| Claas et al. 2013;21 the Netherlands, USA, Canada, UK; 4 | xTAG | n = 901 | Study set up: each participating laboratory analysed specimens according to the routine diagnostic algorithm in place at that site and as ordered by the referring physician. All samples were shipped to Luminex for GPP testing Comparator: as ordered by referring physician at four study sites following their routine diagnostic algorithms. Bacteria – culture according to standard procedures at four sites. Bacterial toxins – EIA (North America). Parasites – microscopy or EIA (North America), microscopy or PCR (Europe). Viruses – PCR (Europe only) GPP: xTAG performed by Luminex Verification: a subset of samples was assessed by conventional PCR and bidirectional sequencing using validated primers targeting genomic regions distinct from those of the xTAG GPP, site of testing not reported |
Sensitivity and specificity |
| Coste et al. 2013;22 France; 1 | xTAG | 54 samples of 49 adult kidney transplant recipients | Study set up: stool samples were taken from each study participant at the time of the severe diarrhoea episode, parasitological tests were performed again on a second sample taken 72 hours after inclusion to deal with the shedding of intestinal protozoa. Routine microbiology testing at study site, samples stored and retrospectively tested by seven GPP assays at the study site Comparator: bacteria – culture. C. difficile – EIA and culture. Parasites – microscopy. Viruses – rapid antigen detection tests. Assumed that all patients received all mentioned conventional tests GPP: xTAG on stored frozen samples at study site Verification: Stool samples positive for one of the enteric viruses and for Campylobacter spp. as well as 23 negative samples by GPP were sent to a national reference centre for confirmation and typing |
Sensitivity, specificity, positive predictive value and negative predictive value for norovirus and Campylobacter |
| Median age: 51 years | ||||
| Range: 18–78 years | ||||
| Male: 30/49 (61%) | ||||
| Median post-transplantation term: 6.3 years (range 3 days to 24.2 years) | ||||
| Immunocompromised: 49/49 (100%) | ||||
| Deng et al. 2015;23 China; 1 | xTAG | 290 stool specimens of 290 diarrhoeal patients | Study set up: stool specimens prospectively collected and submitted to clinical laboratory. All stool samples were tested for all 17 pathogens using routine methods using standard procedures. All samples received single-plex PCR and sequencing for C. difficile. All samples received xTAG Comparator: Disregarding what the physician ordered, all samples were tested for 17 pathogens at a clinical laboratory. Bacteria – culture confirmed by gene sequencing, mass spectrometry or serotyping. Viruses – immunochromatography. Norovirus – real-time reverse-transcription PCR. Parasites – microscopy. C. difficile not tested GPP: xTAG performed at study site on all samples Verification: samples discordant between the routine tests and xTAG were tested by single-plex PCR and sequencing using primers from published literature, which were synthesised by Sangon Biotech (Shanghai, China) (assumed to be different from xTAG primers) |
Agreement using kappa coefficient test, sensitivity and specificity |
| Inpatients: 70/290 (24%) | ||||
| Outpatients: 220/290 (76%) | ||||
| Male: 186/290 (64%) | ||||
| Median age: 25 months | ||||
| Age range: 11 days to 83 years | ||||
| Duong et al. 2016;24 Vietnam; > 3 | xTAG | 479 patients hospitalised with diarrhoeal disease | Study set up: fresh stool was stored at 4 °C at sites and transported to the central study microbiology laboratory within 24 hours. The specimens were tested using microbiological culture and real-time PCR, then stored at –80 °C for xTAG testing at study laboratory. All samples were tested for Shigella, Salmonella and Campylobacter by culture and PCR, and all were tested for adenovirus, norovirus and rotavirus by PCR, culture and PCR were evaluated separately Comparator: All samples received all tests for pathogens considered in the study. Bacteria – culture and PCR. Viruses – PCR GPP: xTAG of all samples at study laboratory Verification: no verification undertaken, sensitivity and specificity were calculated for culture as gold standard and for PCR as gold standard |
Sensitivity and specificity |
| Adults age: median 50 years (IQR 33–64 years) | ||||
| Children age: 16.5 months (IQR 6.7–20 months) | ||||
| Adult male: 36/92 (39%) | ||||
| Children male: 221/387 (57%) | ||||
| FDA 2012;25 USA, Canada; 6 | xTAG | n = 1534 patients (including outbreak samples) | Study set up: prospective clinical specimens were submitted fresh to the sites and were processed according to their routine algorithm Comparator: conventional methods were ordered by the referring physician following routine methods at sites, but these were not included in analysis. Comparator methods for all prospective samples were undertaken in central reference laboratories. In the event that comparator results were not available for all targets on a given specimen, the specimen in question was excluded from performance calculations of xTAG Salmonella, Shigella, E. coli – culture. Campylobacter – culture and PCR/sequencing assay for Campylobacter-positive samples. STEC – broth enrichment and immunocard assay. ETEC – four PCR/sequencing assays. C. difficile – cytotoxicity assay. Cryptosporidium, Giardia – microscopy. Norovirus – real-time PCR and conventional PCR with bidirectional sequencing. Rotavirus – EIA and PCR/sequencing assay GPP: xTAG on all samples at six study sites. 14 pathogens (excluding Yersinia) Verification: discrepant results between the xTAG GPP and the reference methods were evaluated using analytically validated PCR/sequencing assays (bidirectional sequencing analysis) using primers not covered by the xTAG GPP kit primers or FDA-cleared molecular assays (i.e. for C. difficile toxin), central laboratories undertook verification |
Sensitivity and specificity (positive and negative per cent agreements) |
| Male: 632 (44.9%) | ||||
| Age: 12–21 years, n = 51 (3.6%); 21–65 years, n = 879 (62.5%); > 65 years, n = 426 (30.3%) | ||||
| Subject status: outpatients, n = 421 (29.9%); hospitalised n = 804 (57.1%); emergency department, n = 118 (8.4%); long-term care facility, n = 18 (1.3%); not determined, n = 46 (3.3%) | ||||
| Immune status: immunocompromised, n = 493 (35%); immunocompetent, n = 758 (54%); not determined, 156 (11.12%) | ||||
| Gu et al. 2015;26 USA; 1 | xTAG and FilmArray | 436 samples of 199 paediatric oncology patients | Study set up: after routine clinical testing, remnant samples were stored at –80 °C for 30–44 months before testing with GPP assays. All samples were tested with xTAG and FilmArray Comparator: standard-of-care testing at study site on clinical suspicion and subsequent ordering of routine test (172 samples). In addition to routine tests, astrovirus, norovirus and sapovirus PCR were tested on all samples, and standard care tests plus multiplex PCRs for three additional viruses made up the comparator. Viruses – multiplex PCR (adenovirus multiplex PCR detected all serotypes not limited to enteric adenovirus). Rotavirus – EIA. Bacteria – culture. C. difficile – PCR. Parasites – EIA GPP: all samples received xTAG and FilmArray at study site Verification: no verification undertaken, agreement was reported either out of 172 patients having received conventional methods or out of 199 patients having received comparator |
Sensitivity and specificity using in-house methods as reference standard |
| Halligan et al. 2014;27 UK; 1 | xTAG | n = 2187 | Study set up: clinicians were advised to investigate all cases of diarrhoea selecting tests from a menu including bacteria (Campylobacter, Salmonella, Shigella and E. coli O157 with Vibrio and Yersinia species available upon specific request), norovirus (samples received from children aged ≤ 5 years are automatically tested for rotavirus and faecal adenovirus in addition), parasites and C. difficile testing that is either performed on request or automatically on samples from patients aged > 65 years. Conventional testing was performed 7 days per week. Clinicians were advised not to send samples for bacterial culture if the onset of symptoms was > 3 days following hospital admission. Samples were stored at 4 °C until testing was complete. Clinicians were unable to request a GPP test directly; instead, whenever a request for conventional testing was received, a GPP request was included Comparator: testing according to physician’s request at study centre. Bacteria – culture. C. difficile – EIA followed by PCR. Viruses – EIA. Parasites – microscopy GPP: all samples received xTAG at study centre. Samples were batched for DNA extraction at 16.00 Monday–Thursday, further analysis commenced the following morning with results available at 15.00. Alternative run on Friday at 10.00 for late evening or Saturday reporting Verification: assumption that xTAG results are correct, no verification undertaken |
Agreement and kappa, sensitivity and specificity were not calculated because of the lack of a comparable reference standard or resolving assay |
| Hospital-associated cases: 1201/2187 (55%) | ||||
| Community-associated cases: 986/2187 (45%) | ||||
| Mengelle et al. 2013;29 France; 1 | xTAG | 440 samples of 329 diarrhoeic patients | Study set up: prospectively collected stool samples, C. difficile and E. coli pathovars tested in all samples from children, but only in those from certain adults: post-antibiotherapy diarrhoea and nosocomial outbreaks for toxigenic C. difficile or an epidemiological infection for STEC. Conventional and GPP undertaken at study site Comparator: all had at least one conventional test according to physician’s judgement. Bacteria – culture. C. difficile – immunochromatographic test. Viruses – rapid immunochromatographic test. Parasites – microscopy GPP: xTAG on all samples at study site Verification: samples showing discrepant adenovirus results were tested with an in-house real-time PCR |
Proportions of positives by conventional and GPP method and McNemar’s test |
| Immunosuppressed hospitalised patients: (1) 102 adult organ transplant recipients (mean age 50.6 years, median 56 years, range 17–75 years); (2) 50 immunocompromised children (mean age 5 years, median 7 years, range 0–14 years); (3) 56 children attending the neonatal unit (aged < 1 year); and (4) 121 children attending the emergency unit (mean age 2.80 years, median 9 years, range 0–16 years) were considered to be outpatients | ||||
| Pankhurst et al. 2014;30 UK; 1 | xTAG | n = 839 | Study set up: a retrospective study of fixed numbers of samples positive for C. difficile, Campylobacter spp., Salmonella spp. and norovirus, plus samples negative for all these pathogens. All samples collected were initially sent to the service microbiology laboratory for faecal culture and/or C. difficile toxin testing by hospital-based doctors or GPs as a result of a suspected enteric infection. xTAG was undertaken at the study site on all samples Comparator: initial diagnosis of the target faecal pathogens was performed in accordance with PHE guidelines in the service microbiology laboratory for faecal culture and/or C. difficile toxin testing requested by hospital-based doctors or GPs. All 839 patients had results for all four pathogens GPP: all samples were tested for all 15 pathogens, but only comparison conventional testing for four most common: C. difficile, Campylobacter spp., Salmonella spp. and norovirus, different laboratory to conventional testing. C. difficile – EIA testing for toxins A and B with subsequent serological and sensitivity testing. Salmonella and Campylobacter – culture. Norovirus: not reported Verification: unexpectedly positive or negative for target organisms were retested in duplicate using qPCR assays, same laboratory, not blinded samples. Positive on standard reference microbiology but negative on xTAG and confirmed negative on qPCR were considered negative, and samples negative on standard reference microbiology but positive on xTAG and confirmed positive on qPCR were considered positive |
Sensitivity and specificity |
An attempt to verify at least some of the discrepant results between GPP assay and conventional methods was undertaken in 15 studies. 11,12,20–23,25,29–34,38,39 However, only four studies reported outcomes of verification by pathogen. 20,23,25,30
Study design
Table 7 characterises the design heterogeneity among the studies reported, which should be noted when considering the pooled study outcomes. Studies retrospectively included samples with confirmed gastroenteritis and negative controls30 or prospectively included samples from patients with suspected gastroenteritis. 20–27,29 Comparator and GPP tests were both undertaken at the study site(s) in seven studies,22–27,29 whereas one study sent the samples to reference laboratories for conventional testing,30 one study sent the samples to Luminex for GPP testing21 and one study sent the samples to BioFire for comparator testing. 20 Verification was undertaken on all discrepant samples by four studies,20,23,25,30 on a subset of samples by Claas et al. ,21 on positive samples for enteric viruses and Campylobacter as well as 23 negative samples by Coste et al. ,22 and on samples with discrepant adenovirus results only by Mengelle et al. 29 The remaining three studies,24,26,27 which did not undertake verification, assumed either that the comparator methods are the reference standard to calculate sensitivity and specificity24,26 or assumed that the GPP assay is correct and did not calculate sensitivity and specificity because of the lack of a resolving assay. 27 The verification results are summarised in Analysis of discordant results.
Comparator assays varied considerably between studies. Testing included different pathogens and different assays according to the routine diagnostic algorithms in place at the study sites. Pankhurst et al. 30 only included the most common four pathogens for conventional testing by selecting fixed numbers of samples positive for C. difficile, Campylobacter, Salmonella and norovirus. The xTAG GPP Luminex trial25 did not evaluate test samples using traditional conventional methods within routine laboratory analysis, instead testing all samples at a reference laboratory; the FilmArray BioFire trial20 only considered conventional culture results as undertaken by a routine laboratory while the presence of other pathogens was tested at BioFire with various methods and a combination of methods. Gu et al. 26 added multiplex PCR testing to the in-house methods for four viruses that were not routinely tested for, creating a comparator of in-house methods consisting of conventional and multiplex PCR assays. In three studies21,27,29 participants received conventional testing according to the physician’s request not to provide results for all pathogens, for all included patients. Only Deng et al. 23 and Coste et al. 22 tested all patients with conventional methods, irrespective of whether or not they were requested by the physician. Two studies tested all samples with comparator methods, but these were not equivalent to conventional methods of the PHE algorithm. 20,25 In three studies,22,24,30 samples had received all conventional methods, but the studies only looked at a limited number of pathogens, and Gu et al. 26 constructed a comparator for which conventional methods were undertaken according to the physician’s judgement, but additional testing for astrovirus, norovirus and sapovirus was undertaken for all samples.
Use of GPP testing showed less variation across studies; 2 × 2 data were available from nine studies21–27,29,30 for xTAG and from two studies20,26 for FilmArray. Of interest is the batching method reported by Halligan et al. ,27 who batched samples for DNA extraction at 16.00 Monday–Thursday and subsequent analysis of samples the following morning with results available at 15.00. Alternative runs took place on Friday at 10.00 for late evening or Saturday reporting. This is the only study reporting details of batching of samples.
The majority of studies21–26,30 reported sensitivity and specificity using either GPP or conventional tests as a reference standard; these estimates were not used within this review, which instead used the 2 × 2 table data from the studies to estimate the positive and negative agreement between the GPP and conventional tests.
Quality considerations of included studies
Quality assessment for this review required significant adaptations to the QUADAS-2 tool to allow tailoring to the research question. This is because no adequate reference standard is available for the assessment. Furthermore, the comparator (the PHE algorithm) needed to be included in the quality assessment, in effect as a further index test. GPP assays identify more pathogens and may be superior to conventional microbiological methods; however, it is unlikely that GPP tests identify the same spectrum of information as conventional methods. Although conventional methods generally identify viable pathogens, the GPP assays detect the presence of microbial nucleic acids that may result from any non-infective colonisation, dead pathogen or disease-causing pathogen.
A further challenge for quality appraisal involved assessing study quality at the overall study level while the outcomes of the test accuracy studies were considered at the pathogen level. GPP testing systems evaluate a panel of pathogens each with their own test accuracy, threshold, prevalence and clinical importance. However, analyses at the pathogen level meant that varying inclusion within different versions of GPP panels (such as ‘investigational use only’ versions) did not present an applicability concern in this assessment; this only resulted in varying outcomes according to the number of pathogens evaluated.
The ideal study design of a test–treat trial is shown in Table 8. In the absence of a test–treat trial, the test accuracy study design that we consider feasible for the evaluation of GPP tests is detailed (Table 9). This study has been assumed to be a minimum requirement for a study of test accuracy to answer the research question, and the relevant retrieved studies were assessed against this ideal study design.
| Criterion | Details |
|---|---|
| Population | Consecutive series of diarrhoeal patients with suspected gastroenteritis who are eligible for routine laboratory testing |
| Setting | Microbiology laboratory centrally or within a hospital receiving samples from hospital and community patients |
| Index test | xTAG, FilmArray or Faecal Pathogens B |
| Comparator | Conventional tests according to the PHE standard |
| Study design | Prospective cohort randomised to either the index test or the comparator |
| Outcome | Morbidity, mortality, management (e.g. length of hospital stay and length of isolation), treatment |
| Criterion | Details |
|---|---|
| Population | Consecutive series of diarrhoeal patients with suspected gastroenteritis who are eligible for routine laboratory testing |
| Setting | Microbiology laboratory centrally or within a hospital receiving samples from hospital and community patients |
| Index test | xTAG, FilmArray or Faecal Pathogens B |
| Comparator | Conventional tests according to the PHE standard |
| Reference standard/fair umpire | Independent test relative to comparator and index test that may be imperfect but unbiased (fair umpire) to investigate discordant results |
| Study design | Prospective cohort receiving the comparator and at least one index test with follow-up of discordant results |
| Outcome | Comparison of characteristics of discordant results in cells b (GPP positive, conventional test negative) and c (GPP negative, conventional test positive) using a fair umpire test such as exposure (travellers, hospitalised patients, immunocompromised, small children), treatment effect (pathogen-specific treatment) or self-reported symptoms, previous antibiotic treatment and over-the-counter medicine, to allow unbiased assessment of the direction of resolution of discordant results in favour of either the index test or the comparator |
As there was no independent and reliable reference standard, we sought an independent and unbiased umpire test, used in the studies to assess discordant results between index test and conventional methods [e.g. exposure, treatment effect (pathogen-specific treatment) or self-reported symptoms, previous antibiotic treatment, over-the-counter medication].
Verification of discordant results by retesting, by optimising the index test and conventional methods, or by undertaking individual PCRs with primers different from the primers used in the GPP assay remain biased, as PCR assays are not sufficiently different from GPP tests, which are also based on PCR methodology and, therefore, should provide verification results favouring GPP findings.
In addition to the ideal study design, we defined a theoretical comparator test based on the PHE algorithm. 8 The definition considered for the quality appraisal is described in Definitions for the review.
Assessment of risk of bias and applicability of study outcomes to the research question
The assessment of risk of bias and applicability for the 23 studies included in the review using the QUADAS-2 tool are summarised in Table 10 and Figure 4.
| Study | Risk of bias | Applicability concerns | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Comparator | Reference standard | Flow and timing | Patient selection | Index test | Comparator | Reference standard | |
| Becker et al. 201519 | Unclear | High | High | NA | High | High | High | High | NA |
| Beckmann et al. 201411 | High | Low | Unclear | High | High | High | Low | High | High |
| Buss et al. 201520 | Unclear | Low | High | High | High | High | Low | High | High |
| Claas et al. 201321 | Unclear | Low | High | High | High | Low | Low | Low | High |
| Coste et al. 201322 | Low | Low | High | High | High | High | Low | High | High |
| Deng et al. 201523 | Unclear | Low | High | High | Low | High | Low | High | High |
| Duong et al. 201624 | Unclear | Low | High | NA | High | High | Low | High | NA |
| FDA 201225 | Unclear | Low | High | High | High | Low | Low | High | High |
| Gu et al. 201526 | High | Low | High | NA | High | High | Low | Low | NA |
| Halligan et al. 201427 | High | Low | High | NA | High | Low | Low | Low | NA |
| Kahlau et al. 201328 | Unclear | Low | High | NA | High | Low | Low | High | NA |
| Khare et al. 201412 | Unclear | Low | High | High | High | Low | Low | Low | High |
| Mengelle et al. 201329 | High | Unclear | High | High | High | Low | Unclear | Low | High |
| Pankhurst et al. 201430 | Unclear | Low | High | High | Low | Low | Low | High | High |
| Patel et al. 201431 | Unclear | Unclear | High | High | Unclear | Unclear | Unclear | High | High |
| Perry et al. 201432 | Unclear | High | High | High | High | Unclear | High | High | High |
| Petterson et al. 201633 | Low | Low | High | High | High | Low | Low | High | High |
| Rand et al. 201534 | Unclear | Low | High | High | High | High | Low | High | High |
| Spina et al. 201535 | Unclear | Unclear | High | NA | Unclear | Low | Unclear | Unclear | NA |
| Stockmann et al. 201536 | High | Unclear | High | NA | High | High | Unclear | Low | NA |
| Vocale et al. 201537 | Unclear | Low | High | NA | High | Low | Low | High | NA |
| Wessels et al. 201438 | High | Low | High | High | High | Low | Low | High | High |
| Zboromyrska et al. 201439 | Unclear | Low | High | High | High | High | Low | High | High |
FIGURE 4.
Concerns regarding (a) bias; and (b) applicability in included studies. NA, not applicable.

Risk of bias
In general, the methodological and reporting quality of the included studies was poor. Specifically, none of the studies used a reference standard against which the index tests and comparators (e.g. routine tests, PHE algorithm) could be reliably evaluated and compared. Instead, in most studies the index tests were compared with the routine tests/algorithms. Discrepant results between the two tests (GPP positive and routine test negative) were verified using confirmatory tests that were not adequately independent of index/routine tests. In many cases, the routine tests were not performed for all pathogens specified in the GPP and not all samples received the comparator (routine) tests for all pathogens. Some studies included more than one sample (or episode) per patient, which may have led to double counting within measures of agreement.
In up to 65% of the studies it was not clear how stool samples (or patients) were selected, and only 9% of the studies were rated as having a low risk of bias with regard to the patient selection domain (domain 1: patient selection). The selection process in the remaining 25% of the studies was rated as being at a high risk of bias. The use and interpretation of the index test (i.e. GPPs) was associated with a low risk of bias in about 75% of the studies (domain 2: index tests) because these tests were used on all samples independently (the GPP results were not influenced by the knowledge of routine test results), with a pre-specified threshold, and were implemented according to recommendations by the manufacturer. In contrast, almost all studies were rated being at a high risk of bias with regard to the use and interpretation of the comparator (i.e. routine) tests, mainly as a result of no pre-specified threshold (domain 3: comparator). In 65% of the studies, the use of verification tests (domain 4: reference standard) was associated with a high risk of bias. No verification tests were used in 8 (35%) studies (risk of bias not applicable). The between-test intervals and patient/sample flow for at least 80% of studies were rated as being at a high risk of bias (domain 5: flow and timing), with only two studies at a low risk of bias. 23,30
Applicability of study findings
The applicability of study findings was assessed with regard to four domains: patient selection, index test (i.e. GPPs), comparator (i.e. routine tests) test and reference standard (i.e. verification tests). In approximately 45% of the studies, the degree of applicability of the included patients was of low concern. The other 45% of the studies had a high concern for applicability. For the remaining two studies (9%), the applicability of included population could not be determined. 31,32 For at least 70% of studies, there was a low concern for the applicability of the index tests. Only two studies19,32 were rated as having high concern for the applicability with respect to the index tests (because of deviations from the manufacturer’s instructions: timing, storage or extraction). The applicability of a comparator was of low concern for six studies (26.1%)12,21,26,27,29,36 and of high concern for 16 studies (69.6%),11,19,20,22–25,28,30–34,37–39 in which the comparator methods were not consistent with those specified for the PHE algorithm. The applicability concern for a reference standard (i.e. verification tests) was high for all 15 studies using such tests. 11,12,20–23,25,29–34,38,39 The remaining eight studies did not use the verification tests (not applicable). 19,24,26–28,35–37
Agreement between gastrointestinal pathogen panel tests and conventional methods
Evidence retrieved approximated to a mixed population of acute and hospitalised patients, as the studies did not adequately report outcomes by setting or subgroup. No subgroup analysis could be undertaken as a result of a lack of data, more specifically for patients in the community, patients in hospital, children aged < 5 years, patients with recent foreign travel and patients who are immunocompromised. Sensitivity analyses limited to UK studies only, or studies judged as equivalent to the PHE algorithm, were not undertaken because, methodologically, all studies had substantial quality issues. The review identified only three studies from the UK: Halligan et al. 27 provided only conventional testing by clinician request, Perry et al. 32 did not report pathogen-level data and Pankhurst et al. 30 considered only four pathogens in the comparison of xTAG versus conventional methods. Assessing the equivalence of the conventional methods with the PHE algorithm was pragmatic and did not take into account that the standard is interpreted differently in different laboratories. These sensitivity analyses were not pre-planned and may lead to overanalysing weak evidence. Accepting the limitations of the studies, Halligan et al. 27 provides the largest UK study, which might be the most representative of the NHS setting. No one study could be found to adequately characterise the PHE pathway.
Pathogen-level positive and negative agreement
Pathogen-level meta-analysis of positive and negative agreement was undertaken. The 2 × 2 data of test agreement, by pathogen, for the studies eligible for inclusion in the meta-analysis are reported in Appendix 7. The meta-analytic outcomes reported in Agreement between xTAG and the comparator and Agreement between FilmArray and conventional methods are of an exploratory nature, summarising the available evidence and illustrating patterns in the data as well as heterogeneity. When considering the results, the following caveats apply.
-
The results do not reflect the performance of the tests, but the agreement between the GPP test and the comparator.
-
These findings are typically heterogeneous, probably reflecting both methodological and statistical heterogeneity and drawing from studies of variable quality.
-
Analyses are presented at the pathogen level, requiring independence assumptions both within and between pathogens (i.e. repeat samples of the same patient are not included and having one pathogen does not affect the likelihood of having another pathogen).
-
The comparator used in studies does not sufficiently align with the conventional methods described in the PHE algorithm8 in the majority of studies, as described in Definitions for the review.
-
Pooled summary estimates (across pathogens) have been included for information; however, these pooled estimates are not weighted by the prevalence of the different pathogens and include varying multiple usage of samples for which studies have tested samples with varying components of the conventional panel of tests, thus violating the independence assumption.
Agreement between xTAG and the comparator
Table 11 reports the positive (a/a + c) and negative (d/b + d) agreement between xTAG and conventional methods when conventional methods are the benchmark. When conventional testing provides the benchmark, virtually all positive cases found by xTAG are confirmed by conventional testing, leading to generally high levels of positive agreement findings. There are few additional positives identified by xTAG compared with the vast majority of specimens that are pathogen negative; thus, negative agreement remains high.
| RE | LCI | UCI | Number of studies contributing | Q | p-value | I 2 | |
|---|---|---|---|---|---|---|---|
| Positive agreement | |||||||
| C. difficile | 0.959 | 0.933 | 0.980 | 5 | 5.9 | 0.207 | 32% |
| Campylobacter | 0.959 | 0.924 | 0.985 | 6 | 8.0 | 0.157 | 37% |
| E. coli O157 | – | – | – | – | – | – | – |
| ETEC | – | – | – | – | – | – | – |
| STEC | – | – | – | – | – | – | – |
| Salmonella | 0.818 | 0.666 | 0.934 | 5 | 30.8 | 0.000 | 87% |
| Shigella | 0.989 | 0.949 | 1.000 | 3 | 3.6 | 0.164 | 45% |
| V. cholerae | – | – | – | – | – | – | – |
| Yersinia enterocolitica | – | – | – | – | – | – | – |
| Adenovirus | 0.558 | 0.413 | 0.699 | – | – | – | – |
| Norovirus | 0.927 | 0.893 | 0.956 | 7 | 10.9 | 0.093 | 45% |
| Rotavirus | 0.958 | 0.920 | 0.985 | 3 | 2.9 | 0.240 | 30% |
| Cryptosporidium | 0.914 | 0.794 | 0.989 | 1 | – | – | – |
| E. histolytica | – | – | – | – | – | – | – |
| Giardia | 1.000 | 0.935 | 1.000 | 1 | – | – | – |
| Negative agreement | |||||||
| C. difficile | 0.968 | 0.933 | 0.991 | 7 | 128.0 | 0.000 | 95% |
| Campylobacter | 0.968 | 0.950 | 0.982 | 10 | 83.7 | 0.000 | 89% |
| E. coli O157 | 0.995 | 0.990 | 0.998 | 6 | 10.8 | 0.055 | 54% |
| ETEC | 0.988 | 0.964 | 1.000 | 4 | 23.7 | 0.000 | 87% |
| STEC | 0.990 | 0.984 | 0.995 | 4 | 2.8 | 0.418 | 0% |
| Salmonella | 0.940 | 0.866 | 0.986 | 10 | 726.0 | 0.000 | 99% |
| Shigella | 0.985 | 0.965 | 0.997 | 8 | 120.0 | 0.000 | 94% |
| V. cholerae | 1.000 | 0.998 | 1.000 | 4 | 0.1 | 0.988 | 0% |
| Y. enterocolitica | 1.000 | 1.000 | 1.000 | 4 | 0.4 | 0.933 | 0% |
| Adenovirus | 0.990 | 0.983 | 0.996 | 2 | – | – | – |
| Norovirus | 0.969 | 0.944 | 0.987 | 12 | 239.0 | 0.000 | 95% |
| Rotavirus | 0.991 | 0.979 | 0.999 | 8 | 36.7 | 0.000 | 81% |
| Cryptosporidium | 0.989 | 0.954 | 1.000 | 5 | 77.4 | 0.000 | 95% |
| E. histolytica | 0.991 | 0.979 | 0.998 | 5 | 20.5 | 0.000 | 81% |
| Giardia | 0.989 | 0.970 | 0.999 | 7 | 46.4 | 0.000 | 87% |
| Overall agreement | |||||||
| Positive | 0.929 | 0.898 | 0.955 | 33 | 188.3 | 0.000 | 83% |
| Negative | 0.982 | 0.976 | 0.988 | 101 | 2080.8 | 0.000 | 95% |
Overall, more studies contributed to the calculation of negative than positive agreement as only studies with sufficient numbers (denominator ≥ 20) were included in the analysis. For a number of pathogens, E. coli O157, enterotoxigenic E. coli, shiga toxin-producing E. coli, V. cholerae, Yersinia enterocolitica and E. histolytica (rare pathogens or no test requested by physician and marked as empty rows in Tables 6 and 7), limited data were available and, therefore, no positive agreement was estimated. Overall, agreement is high (positive agreement 0.929 and negative agreement 0.982), with little variation between pathogens for both positive and negative agreement. Positive agreement for adenovirus was an exception, as positive agreement was considerably lower at 0.558. This is visualised in the forest plot in Figure 5. Gu et al. 26 reported that an additional 20 samples positive for adenovirus detected by comparator were a result of the use of multiplex PCR, which detected all serotypes, whereas xTAG only detected adenovirus 40/41, resulting in the poor agreement of tests for this virus. However, it is important to note that for enteric infections, adenovirus 40/41 is commonly implicated. In situations when a patient may have a systemic infection affecting, for example, the respiratory or urinary tracts, then all adenovirus serotypes should be looked for by multiplex PCR. Therefore, it is difficult to come to any firm conclusions about the positive agreement because of the different parameters (type 40/41 vs. multiplex) that were assessed. The outlying finding for Salmonella (Pankhurst et al. 30), which is caused by a worryingly high number of missed Salmonella infections by xTAG, is not as easily interpreted. Generally, both tests agreed in terms of absence of pathogens, masking the relatively small number of disagreements. A forest plot in Appendix 8 (see Figure 43) shows, however, that there are a few outliers when studies report a significantly higher number of positives for certain pathogens with xTAG than conventional methods, specifically Campylobacter and norovirus in a small study of 49 adult kidney transplant recipients22 and Salmonella in a study in which bacteria were tested by PCR as well as culture. 24
FIGURE 5.
Positive agreement: xTAG vs. conventional testing (benchmark). G2, genogroup 2; RE, random-effects estimate. Reproduced from Freeman et al. 2017. 49 © 2017 Freeman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
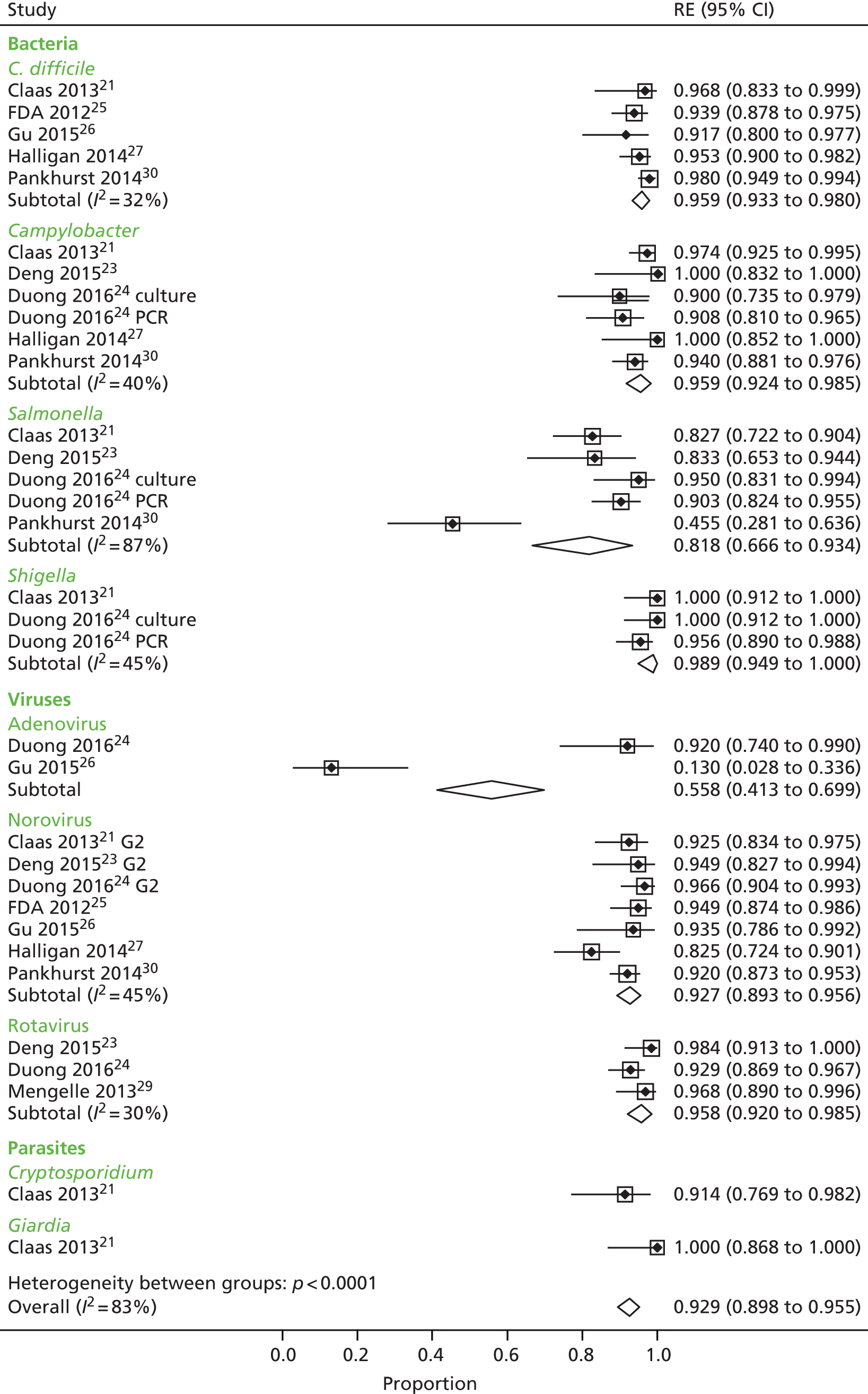
Used as a measure of overall heterogeneity of estimates, I2 is moderate (for positive agreement) to high (for negative agreement) at the pathogen level.
Table 12 shows the positive (a/a + b) and negative (d/c + d) agreement when xTAG is considered the benchmark. Strikingly, the positive agreement between xTAG and conventional methods is considerably smaller when xTAG provides the benchmark.
| RE | LCI | UCI | Number of studies contributing | Q | p-value | I 2 | |
|---|---|---|---|---|---|---|---|
| Positive agreement | |||||||
| C. difficile | 0.801 | 0.594 | 0.948 | 5 | 124.0 | 0.000 | 97% |
| Campylobacter | 0.639 | 0.398 | 0.849 | 7 | 167.0 | 0.000 | 96% |
| E. coli O157 | 0.750 | 0.534 | 0.920 | 1 | – | – | – |
| ETEC | – | – | – | – | – | – | – |
| STEC | – | – | – | – | – | – | – |
| Salmonella | 0.484 | 0.278 | 0.693 | 8 | 173.0 | 0.000 | 96% |
| Shigella | 0.734 | 0.381 | 0.971 | 3 | 61.6 | 0.000 | 97% |
| V. cholerae | – | – | – | – | – | – | – |
| Y. enterocolitica | – | – | – | – | – | – | – |
| Adenovirus | 0.570 | 0.425 | 0.710 | 2 | – | – | – |
| Norovirus | 0.774 | 0.584 | 0.920 | 8 | 215.0 | 0.000 | 97% |
| Rotavirus | 0.924 | 0.853 | 0.975 | 3 | 6.5 | 0.039 | 69% |
| Cryptosporidium | 0.508 | 0.407 | 0.608 | 2 | – | – | – |
| E. histolytica | – | – | – | – | – | – | – |
| Giardia | 0.337 | 0.237 | 0.444 | 2 | – | – | – |
| Negative agreement | |||||||
| C. difficile | 0.996 | 0.992 | 0.999 | 7 | 11.1 | 0.084 | 46% |
| Campylobacter | 0.998 | 0.994 | 1.000 | 10 | 37.4 | 0.000 | 76% |
| E. coli O157 | 1.000 | 1.000 | 1.000 | 6 | 3.4 | 0.637 | 0% |
| ETEC | 0.999 | 0.996 | 1.000 | 4 | 3.0 | 0.393 | 0% |
| STEC | 1.000 | 0.998 | 1.000 | 4 | 4.9 | 0.182 | 38% |
| Salmonella | 0.992 | 0.980 | 0.999 | 10 | 94.4 | 0.000 | 91% |
| Shigella | 1.000 | 0.999 | 1.000 | 8 | 12.0 | 0.099 | 42% |
| V. cholerae | 1.000 | 0.999 | 1.000 | 4 | 3.5 | 0.326 | 13% |
| Y. enterocolitica | 1.000 | 0.999 | 1.000 | 4 | 0.4 | 0.933 | 0% |
| Astrovirus | 0.989 | 0.971 | 0.999 | 7 | 68.7 | 0.000 | 91% |
| Norovirus | 0.995 | 0.990 | 0.998 | 12 | 34.9 | 0.000 | 69% |
| Rotavirus | 0.998 | 0.992 | 1.000 | 8 | 29.3 | 0.000 | 76% |
| Cryptosporidium | 1.000 | 0.999 | 1.000 | 5 | 2.0 | 0.743 | 0% |
| E. histolytica | 1.000 | 0.999 | 1.000 | 5 | 3.5 | 0.481 | 0% |
| Giardia | 1.000 | 1.000 | 1.000 | 7 | 1.8 | 0.941 | 0% |
| Overall agreement | |||||||
| Positive | 0.678 | 0.580 | 0.770 | 41 | 1340.5 | 0.000 | 97% |
| Negative | 0.998 | 0.997 | 0.999 | 101 | 429.2 | 0.000 | 77% |
The forest plot in Figure 6 visualises the inconsistency and variation in positive agreement between studies across all pathogens. When xTAG is the benchmark, the positive cases ‘missed’ by conventional testing have a considerable impact of the positive agreement findings.
FIGURE 6.
Positive agreement: conventional testing vs. xTAG (benchmark). G1, genogroup 1; G2, genogroup 2; RE, random-effects estimate. Reproduced from Freeman et al. 2017. 49 © 2017 Freeman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
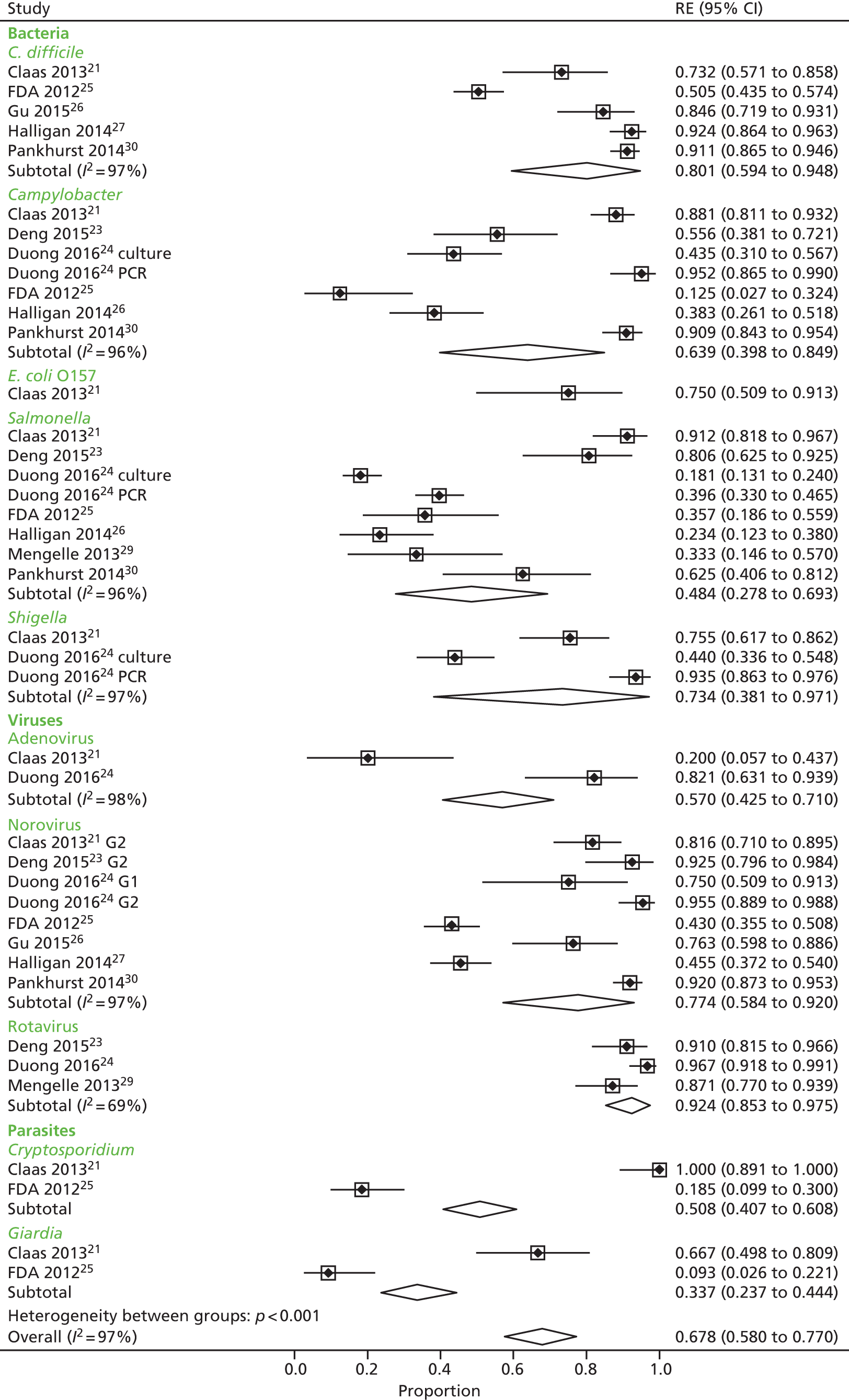
As the positive agreement overall is 0.678 (95% CI 0.580 to 0.770), inverting these figures means that xTAG finds about 1.5 times more pathology (95% CI 1.3 to 1.7), although this estimate does not reflect the prevalence of individual pathogens. Negative agreement was consistently very high across studies and pathogens (see Appendix 8, Figure 44) with the exception of Gu et al.,26 discussed previously. Heterogeneity is moderate to high when considering I2, and is higher for positive agreement than for negative agreement.
Agreement between FilmArray and conventional methods
Only two studies contributed to the evaluation of FilmArray; therefore, the agreement between FilmArray and conventional methods is based on one or two studies across the range of pathogens. Table 13 reports the positive (a/a + b) and negative (d/c + d) agreement between FilmArray and conventional methods when benchmarked against conventional methods. Positive agreement is very high for all pathogens, with the same study (Gu et al. 26) contributing an outlier value for adenovirus for FilmArray, as was the case for xTAG (Figure 7). Negative agreement is consistently high for all studies across all pathogens (see Appendix 8, Figure 45).
| RE | LCI | UCI | Number of studies contributing | Q | p-value | I 2 | |
|---|---|---|---|---|---|---|---|
| Positive agreement | |||||||
| C. difficile | 0.967 | 0.937 | 0.989 | 2 | – | – | – |
| Campylobacter | 0.971 | 0.881 | 1.000 | 1 | – | – | – |
| E. coli O157 | – | – | – | – | – | – | – |
| EAEC | 0.988 | 0.949 | 1.000 | 1 | – | – | – |
| EPEC | 0.991 | 0.976 | 0.999 | 1 | – | – | – |
| ETEC | 1.000 | 0.923 | 1.000 | 1 | – | – | – |
| Plesiomonas shigelloides | – | – | – | – | – | – | – |
| STEC | 1.000 | 0.949 | 1.000 | 1 | – | – | – |
| Salmonella | 1.000 | 0.945 | 1.000 | 1 | – | – | – |
| Shigella | 0.959 | 0.881 | 0.999 | 1 | – | – | – |
| Vibrio (parahaemolyticus, vulnificus and cholerae) | – | – | – | – | – | – | – |
| V. cholerae | – | – | – | – | – | – | – |
| Y. enterocolitica | – | – | – | – | – | – | – |
| Adenovirus | 0.722 | 0.606 | 0.826 | 2 | – | – | – |
| Astrovirus | – | – | – | – | – | – | – |
| Norovirus | 0.932 | 0.866 | 0.979 | 2 | – | – | – |
| Rotavirus | – | – | – | – | – | – | – |
| Sapovirus | 1.000 | 0.963 | 1.000 | 1 | – | – | – |
| Cyclospora cayetanensis | – | – | – | – | – | – | – |
| Cryptosporidium | – | – | – | – | – | – | – |
| E. histolytica | – | – | – | – | – | – | – |
| Giardia | 1.000 | 0.916 | 1.000 | 1 | – | – | – |
| Negative agreement | |||||||
| C. difficile | 0.971 | 0.962 | 0.979 | 2 | – | – | – |
| Campylobacter | 0.987 | 0.981 | 0.993 | 2 | – | – | – |
| E. coli O157 | 0.968 | 0.930 | 0.993 | 2 | – | – | – |
| EAEC | 0.982 | 0.974 | 0.988 | 1 | – | – | – |
| EPEC | 0.972 | 0.961 | 0.980 | 1 | – | – | – |
| ETEC | 0.994 | 0.990 | 0.997 | 1 | – | – | – |
| P. shigelloides | 0.990 | 0.985 | 0.995 | 1 | – | – | – |
| STEC | 0.997 | 0.993 | 0.999 | 1 | – | – | – |
| Salmonella | 0.998 | 0.994 | 1.000 | 2 | – | – | – |
| Shigella | 1.000 | 0.998 | 1.000 | 2 | – | – | – |
| Vibrio (parahaemolyticus, vulnificus and cholerae) | 0.999 | 0.997 | 1.000 | 1 | – | – | – |
| V. cholerae | 1.000 | 0.997 | 1.000 | 2 | – | – | – |
| Y. enterocolitica | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Adenovirus | 0.993 | 0.987 | 0.997 | 2 | – | – | – |
| Astrovirus | 1.000 | 0.998 | 1.000 | 2 | – | – | – |
| Norovirus | 0.991 | 0.985 | 0.995 | 2 | – | – | – |
| Rotavirus | 0.995 | 0.990 | 0.998 | 2 | – | – | – |
| Sapovirus | 0.994 | 0.989 | 0.997 | 2 | – | – | – |
| C. cayetanensis | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| Cryptosporidium | 1.000 | 0.997 | 1.000 | 2 | – | – | – |
| E. histolytica | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| Giardia | 1.000 | 0.997 | 1.000 | 2 | – | – | – |
| Overall agreement | |||||||
| Positive | 0.954 | 0.897 | 0.991 | 14 | 129.4 | 0.000 | 89% |
| Negative | 0.996 | 0.993 | 0.998 | 35 | 295.6 | 0.000 | 88% |
FIGURE 7.
Positive agreement: FilmArray vs. conventional testing (benchmark). EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; RE, random-effects estimate; STEC, shiga toxin-producing E. coli.

When benchmarked against FilmArray, positive agreement between FilmArray and conventional methods is lower (0.82 vs. 0.95) (Table 14) with greater variability across pathogens (Figure 8), whereas negative agreement (see Appendix 8, Figure 46) is consistently high, with the exception of Gu et al. 26 once again. Heterogeneity is not reported, as only one or two studies contributed data per pathogen.
| RE | LCI | UCI | Number of studies contributing | Q | p-value | I 2 | |
|---|---|---|---|---|---|---|---|
| Positive agreement | |||||||
| C. difficile | 0.817 | 0.766 | 0.864 | 2 | – | – | – |
| Campylobacter | 0.586 | 0.456 | 0.710 | 1 | – | – | – |
| E. coli O157 | – | – | – | – | – | – | – |
| EAEC | 0.752 | 0.667 | 0.829 | 1 | – | – | – |
| EPEC | 0.902 | 0.869 | 0.931 | 1 | – | – | – |
| ETEC | 0.710 | 0.536 | 0.858 | 1 | – | – | – |
| Plesiomonas shigelloides | – | – | – | – | – | – | – |
| STEC | 0.868 | 0.739 | 0.961 | 1 | – | – | – |
| Salmonella | 0.838 | 0.699 | 0.942 | 1 | – | – | – |
| Shigella | 0.959 | 0.881 | 0.999 | 1 | – | – | – |
| Vibrio (parahaemolyticus, vulnificus and cholerae) | – | – | – | – | – | – | – |
| V. cholerae | – | – | – | – | – | – | – |
| Y. enterocolitica | – | – | – | – | – | – | – |
| Adenovirus | 0.764 | 0.641 | 0.868 | 1 | – | – | – |
| Astrovirus | – | – | – | – | – | – | – |
| Norovirus | 0.851 | 0.771 | 0.917 | 2 | – | – | – |
| Rotavirus | – | – | – | – | – | – | – |
| Sapovirus | 0.780 | 0.664 | 0.877 | 1 | – | – | – |
| Cyclospora cayetanensis | – | – | – | – | – | – | – |
| Cryptosporidium | 0.750 | 0.555 | 0.906 | 1 | – | – | – |
| E. histolytica | – | – | – | – | – | – | – |
| Giardia | 0.741 | 0.557 | 0.891 | 1 | – | – | – |
| Negative agreement | |||||||
| C. difficile | 0.998 | 0.994 | 1.000 | 2 | – | – | – |
| Campylobacter | 1.000 | 0.999 | 1.000 | 2 | – | – | – |
| E. coli O157 | 1.000 | 0.989 | 1.000 | 2 | – | – | – |
| EAEC | 0.999 | 0.997 | 1.000 | 1 | – | – | – |
| EPEC | 0.997 | 0.994 | 1.000 | 1 | – | – | – |
| ETEC | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| P. shigelloides | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| STEC | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| Salmonella | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Shigella | 1.000 | 0.998 | 1.000 | 2 | – | – | – |
| Vibrio (parahaemolyticus, vulnificus and cholerae) | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| V. cholerae | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Y. enterocolitica | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Adenovirus | 0.997 | 0.993 | 0.999 | 2 | – | – | – |
| Astrovirus | 1.000 | 0.998 | 1.000 | 2 | – | – | – |
| Norovirus | 0.998 | 0.995 | 1.000 | 2 | – | – | – |
| Rotavirus | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Sapovirus | 1.000 | 0.999 | 1.000 | 2 | – | – | – |
| C. cayetanensis | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| Cryptosporidium | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| E. histolytica | 1.000 | 0.999 | 1.000 | 1 | – | – | – |
| Giardia | 1.000 | 1.000 | 1.000 | 2 | – | – | – |
| Overall agreement | |||||||
| Positive | 0.820 | 0.761 | 0.872 | 15 | 72.3 | 0.000 | 81% |
| Negative | 1.000 | 0.999 | 1.000 | 36 | 171.5 | 0.000 | 80% |
FIGURE 8.
Positive agreement: conventional testing vs. FilmArray (benchmark). EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; RE, random-effects estimate; STEC, shiga toxin-producing E. coli.
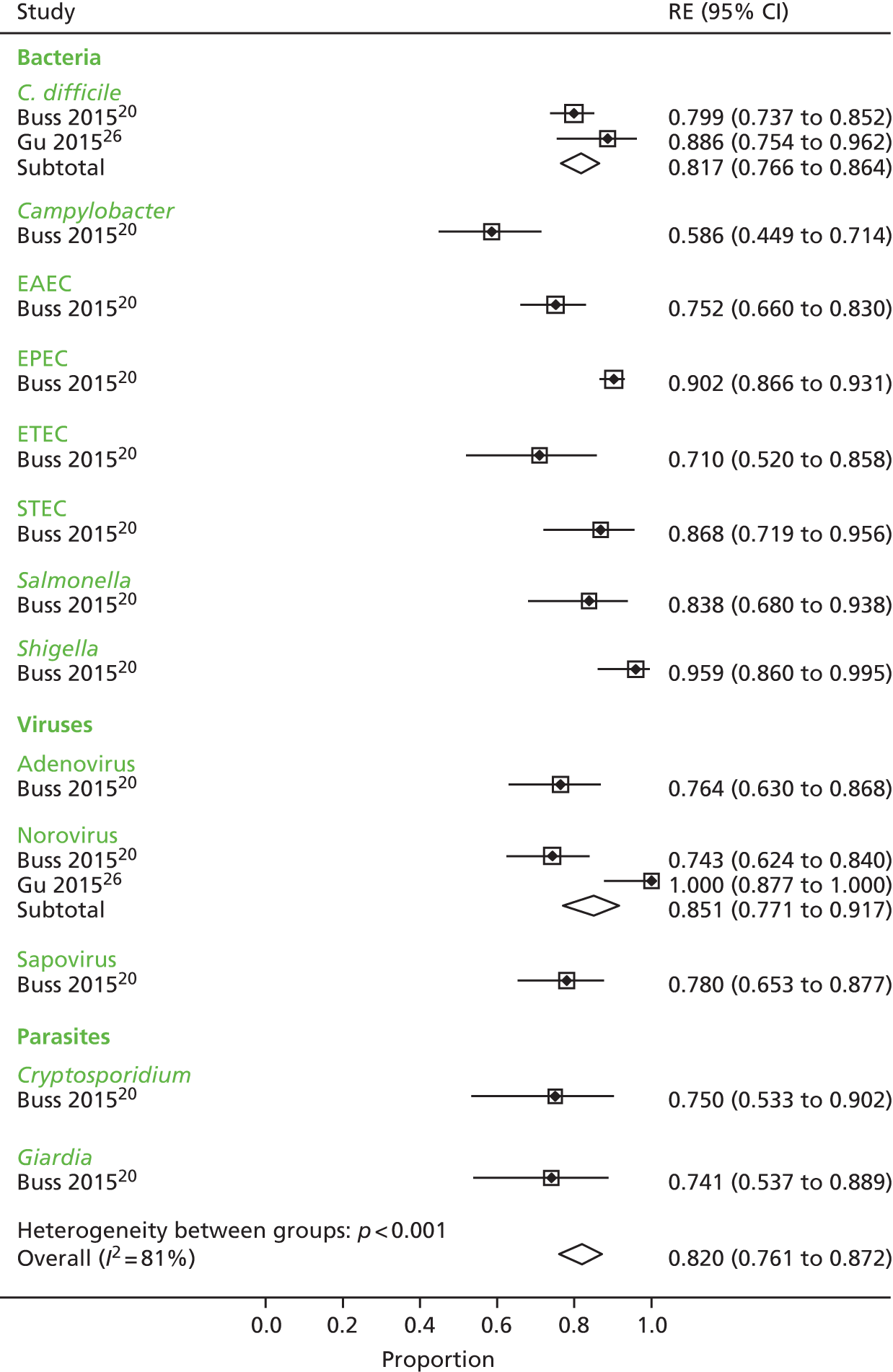
As positive agreement overall is 0.820 (95% CI 0.761 to 0.872), inverting these figures means that FilmArray finds about 1.2 times more pathology than conventional testing (95% CI 1.1 to 1.3), although this estimate does not reflect the prevalence of individual pathogens.
Aetiology of additional positive findings
Studies typically reported overall levels of detection of pathogens in addition to providing pathogen-level analysis (several studies only provided this overall analysis). Additional pathogens could arise in part because of a greater ‘sensitivity’ to detect specific pathogens, but also because of a greater coverage of pathogens within the GPP system than in the conventional method. Studies were reviewed to ascertain the extent of these two sources of additional positive findings, because simply reporting the total number of pathogens detected may mislead by confounding these issues.
Table 15 summarises the ascertainment of positive results. Studies reported this issue in a varying manner, but the agreement findings illustrate that GPP testing produces a greater number of pathogen-positive findings than conventional testing, and the extent to which greater overall pathology arises from the number of tests within the GPP system or a higher level of pathogen detection is inconclusive.
| Study reference | Samples positive with GPP (n/N) | Samples positive with conventional methods | Additional positives with conventional methods | Additional positives with GPP | Additional positives from GPP, for pathogens covered by conventional testing (improved ‘sensitivity’), n/N (%) | Additional positives from GPP, for pathogens not covered by conventional testing (greater coverage), n/N (%)a | Comments |
|---|---|---|---|---|---|---|---|
| Becker et al. 201519 | NR | NR | 54 (parasites not on xTAG) | NR | NR | 37 | 68 samples from Côte d’Ivoire were investigated for a range of parasites. Positives not reported by GPP/conventional method |
| Beckmann et al. 201411 | Children, 66/127; travellers, 21/185 | Children, 30/127; travellers, 63/185 | Children, 0; travellers, 62 | Children, 36; travellers, 20 | Children, 19/36 (53); travellers, 4/20 (20) | Children, 17/36 (47); travellers, 16/20 (80) | Microscopy on traveller’s samples identified a wider range of parasites than on GPP; children were tested for viruses only with conventional methods |
| Buss et al. 201520 | 832 (1180 pathogens)/1556 | 957/1556 | 14 | 237 | NR | NR | – |
| Claas et al. 201321 | NR | NR | 43 | 233 | 96/233 (41) | 137/233 (59) | 901 samples from paediatric and adult patients |
| Coste et al. 201322 | 36/54 | 13/54 | 7 | 30 | 30/30 (100) | 0/30 | Patients were immunocompromised kidney transplant patients; 6/7 additional samples with conventional test were EPEC positive (not in xTAG) |
| Deng et al. 201523 | 199/290 (pathogens in 159 positive stool samples) | 170/290 | 10 | 39 | NR | NR | – |
| Duong et al. 201624 | 404/479 | Culture, 105/479; PCR, 268/479 | Culture, 5; PCR, 35 | Culture, 258; PCR, 155 | NR | NR | xTAG was compared with culture and PCR |
| FDA 201225 | 486/1407 | 234/1407 | 19 | 413 | NR | NR | Conventional method here not in agreement with PHE algorithm as largely PCR based |
| Gu et al. 201526 | xTAG, 99/199; FilmArray, 94/199 | xTAG, 105/199; FilmArray, 122/199 | xTAG, 26; FilmArray, 40 | xTAG, 23; FilmArray, 11 | NR | NR | 20 samples positive for adenovirus by comparator, multiplex PCRs detected all serotypes, whereas GPP assays only detected adenovirus 40/41 |
| Halligan et al. 201427 | 483/2187 | 262/2187 | 21 | 215 | NR | NR | – |
| Kahlau et al. 201328 | 157/347 | NR | 7 | 27 | 8/27 (30) | 19/27 (70) | – |
| Khare et al. 201412 | xTAG, 69/230; FilmArray, 76/230 | xTAG, 19/230; FilmArray, 19/230 | NR | NR | NR | NR | – |
| Mengelle et al. 201329 | 176/440 | 85/440 | 10 | 96 | 66/96 (69) | 30/96 (31) | Patients were immunocompromised adults and children, children in neonatal unit and children in A&E |
| Pankhurst et al. 201430 | 561/839 | 544/839 | 46 | 104 | NR | NR | – |
| Patel et al. 201431 | Retrospective, 77/167; prospective, 86/211 | Retrospective, NR; prospective, 83/211 | Retrospective, NR; prospective, 3 | Retrospective, NR; prospective, 6 | NR | NR | Two laboratories tested same samples; joint confirmed detection reported |
| Perry et al. 201432 | Stored samples, 248/1000; concurrently tested samples, 155/472 | Stored samples, 90/1000; concurrently tested samples, 41/472 | Stored samples, 5; concurrently tested samples, NR | Stored samples, NR; concurrently tested samples, 114 | Stored samples, 73% of additional GPP calls; concurrently tested samples, 46/114 (40) | Stored samples, 27% of additional GPP calls; concurrently tested samples, 68/114 (60) | – |
| Petterson et al. 201633 | Centre 1 (tertiary care), 19/141; Centre 2 (general hospital), 47/187 | Centre 1 (tertiary care), 12/141; Centre 2 (general hospital), 31/187 | Centre 1 (tertiary care), 0; Centre 2 (general hospital), 5 | Centre 1 (tertiary care), NR; Centre 2 (general hospital), 22 | Centre 1 (tertiary care), NR; Centre 2 (general hospital), 5/22 (23) | Centre 1 (tertiary care), 6; Centre 2 (general hospital), 17/22 (77.3) | |
| Rand et al. 201534 | 35/158 | 0/158 | NA | 35 | 2/35 (6) | 33/35 (94) | Conventional only tested for C. difficile and rotavirus, and study only included samples that were negative on conventional testing |
| Spina et al. 201535 | 384/709 | 128/709 | NR | NR | NR | NR | – |
| Stockmann et al. 201536 | 244/378 | 175/378 | NR | NR | NR | NR | Children and young adults. Convenient sample was enriched for multiple conventional tests and positive samples |
| Vocale et al. 201537 | 356/664 | 301/664 | NR | NR | NR | NR | – |
| Wessels et al. 201438 | 76/393 (83 pathogens) | 95/393 | 55 | 43 | 12/43 (28) | 31/43 (72) | 51/55 additional by conventional testing were pathogens not on GPP panel |
| Zboromyrska et al. 201439 | 67/185 (86 pathogens) | 35/185 | 19 | 60 | NR | NR | Travellers |
Even in the studies in which the comparator was largely PCR based, GPP methods identified more pathogens. 25 Only three studies reported more additional samples with conventional testing than with GPP. 11,26,38 Gu et al. 26 reported that 20 of the additional samples with conventional testing were positive for adenovirus by multiplex PCR, which detected all serotypes, whereas GPP assays detected only adenovirus 40/41. Beckmann et al. 11 detected parasites in travellers using microscopy and detected additional parasites that were not available on the GPP test. Similarly, in Wessels et al. ,38 51 out of 55 additional positives by conventional methods were pathogens not present on the panel test.
Eight studies characterised the additional GPP positives by judging whether they were primarily a result of greater ‘sensitivity’ or greater coverage. 11,21,22,28,29,32,34,38 Proportions varied across studies and clearly depended on how well the conventional methods and GPP test agreed in terms of number and type of pathogens tested for in individual samples.
Using two methods for the comparator, Duong et al. 24 reported considerably more additional positives when GPP was compared with culture rather than PCR. Conducted in Vietnam, this study reported a coinfection rate of 58%, much higher than other studies (see Frequency and characterisation of multiple infections), resulting in a number of additional positives.
There is some suggestion from the meta-analyses that additional positive case findings (above conventional testing) may be greater with xTAG and FilmArray. However, this suggestion does not arise from a direct comparison: only one or two studies contribute to evidence for FilmArray, there is considerable heterogeneity between studies and the methods of pooling across pathogens do not reflect pathogen prevalence. Without validation with a reliable reference standard, it remains unknown whether additional positives are true or false positives; however, there is concern that at least some of the additional positives by panel testing are false positives because they identify non-viable pathogens.
Analysis of discordant results
In the absence of a reference standard to verify test results, a small number of studies attempted to verify samples that did not agree when tested with GPP and conventional methods (Table 16). In four studies,20,23,25,30 discordant samples were verified and results reported by pathogen, whereas one additional study only reported that verification produced similar results to GPP testing. 29 Verification methods were PCR based. Buss et al. 20 aimed to analyse discordant samples using PCR and sequencing assays that targeted DNA sequences different from both the FilmArray panel and the comparator method. Alternatively, if no distinct PCR assay was available, discordant samples were retested by the original molecular tests using enhanced methods (including up to 10 additional PCR cycles and up to 10 additional replicate samples) followed by sequencing. If still discrepant, samples were tested with a bench-top version of the FilmArray panel followed by sequencing. Deng et al. 23 sourced primers from the published literature for individual pathogens for single-plex PCR and followed up results by sequencing. Similarly, the xTAG study in the FDA report25 used validated primers that targeted genomic regions distinct from the xTAG GPP and followed up with bidirectional sequencing analysis to confirm pathogen identification. Mengelle et al. 29 used in-house real-time PCR to verify discordant results, and Pankhurst et al. 30 used single quantitative PCR for which the primers were probably different from xTAG.
| Study (GPP test) | Pathogen | Comparator | Verification method (judgement of bias) | Reported results of verificationa | |
|---|---|---|---|---|---|
| Buss et al. 201520 (FilmArray) | Adenovirus 40/41 | 2 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – |
| GPP + 42 | GPP + 13 (verified 11+d/0, 2 remain inconclusive) | ||||
| GPP – 2 (verified 1+e/0, 1 remains inconclusive) | GPP – 1499 | ||||
| C. difficile toxin A/B | 2 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 163 | GPP + 41 (verified 41+d/0) | ||||
| GPP – 2 (verified 1+e/0, 1 remains inconclusive) | GPP – 1350 | ||||
| Campylobacter | Culture | cadF and gyrA PCR (biased), BTFAb (biased) | Comparator + | Comparator – | |
| GPP + 34 | GPP + 24 (verified 19+d/0, 5 remain inconclusive) | ||||
| GPP – 1 (verified 1+e/0) | GPP – 1497 | ||||
| Cryptosporidium | 2 × PCR | ECP,b 18S rRNA gene PCR (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 18 | GPP + 6 (verified 6+d/0) | ||||
| GPP – 0 | GPP – 1532 | ||||
| E. coli O157 | Culture (STEC positive only) | rfbE PCR (biased) | Comparator + | Comparator – | |
| GPP + 3 | GPP + 1 (verified 1+d/0) | ||||
| GPP – 0 | GPP – 34 | ||||
| ETEC | 3 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 22 | GPP + 9 (verified 6+d/3–e) | ||||
| GPP – 0 | GPP – 1525 | ||||
| Giardia | 2 × PCR (one using published primers) | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 20 | GPP + 7 (verified 4+d/2–e, 2 remain inconclusive) | ||||
| GPP – 0 | GPP – 1529 | ||||
| Norovirus | PCR (using published primers) | PCR with multiple primer sets (biased), BTFAb,c (biased) | Comparator + | Comparator – | |
| GPP + 52 | GPP + 18 (verified 8+d/0, 10 remain inconclusive) | ||||
| GPP – 3 (verified 2+e/0, 1 remains inconclusive) | GPP – 1483 | ||||
| Plesiomonas shigelloides | Culture | hugA PCR (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 3 | GPP + 15 (verified 15+d/0) | ||||
| GPP – 0 | GPP – 1538 | ||||
| Rotavirus | 2 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 6 | GPP + 12 (verified 11+d/0, 1 remains inconclusive) | ||||
| GPP – 0 | GPP – 1538 | ||||
| Salmonella | Culture | stn PCR (biased) | Comparator + | Comparator – | |
| GPP + 31 | GPP + 6 (verified 6+d/0) | ||||
| GPP – 0 | GPP – 1519 | ||||
| Shigella | Culture | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 47 | GPP + 2 (2 remain inconclusive) | ||||
| GPP – 2 (2 remain inconclusive) | GPP – 1505 | ||||
| STEC | 2 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 33 | GPP + 5 (verified 5+d/0) | ||||
| GPP – 0 | GPP – 1518 | ||||
| V. cholerae | Culture | gyrB PCR (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 0 | GPP + 1 (verified 1+/0) | ||||
| GPP – 0 | GPP – 1555 | ||||
| Vibrio spp. | Culture | gyrB PCR (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 0 | GPP + 2 (verified 2+d/0) | ||||
| GPP – 0 | GPP – 1554 | ||||
| EAEC | 2 × PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 82 | GPP + 27 (verified 27+d/0) | ||||
| GPP – 1 (1 remains inconclusive) | GPP – 1446 | ||||
| EPEC | 2 × PCR (of samples that were STEC negative) | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 314 | GPP + 34 (verified 23+d/0, 11 remain inconclusive) | ||||
| GPP – 3 (3 remain inconclusive) | GPP – 1167 | ||||
| Astrovirus | PCR | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 7 | GPP + 1 (verified 1+d/0) | ||||
| GPP – 0 | GPP – 1548 | ||||
| Sapovirus | 2 × PCR (one with published primers and one with in-house primers) | ECPb (biased), BTFAc (biased) | Comparator + | Comparator – | |
| GPP + 46 | GPP + 13 (verified 12+d/0, 1 remains inconclusive) | ||||
| GPP – 0 | GPP – 1497 | ||||
| Deng et al. 201523 (xTAG) | Adenovirus 40/41 | Immunochromatography | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – |
| GPP + 3 | GPP + 0 | ||||
| GPP – 2 (verified 0/2–d) | GPP – 285 | ||||
| Campylobacter | Culture | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 20 | GPP + 16 (verified 16+d/0) | ||||
| GPP – 0 | GPP – 254 | ||||
| E. coli O157 | Culture confirmed by gene sequencing using published primers | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 1 | GPP + 2 (verified 2+d/0) | ||||
| GPP – 0 | GPP – 287 | ||||
| ETEC | Culture confirmed by gene sequencing using published primers | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 1 | GPP + 4 (verified 4+d/0) | ||||
| GPP – 0 | GPP – 285 | ||||
| Norovirus GII | Real-time reverse transcription PCR using PCR kit | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 37 | GPP + 3 (verified 0/3–e) | ||||
| GPP – 2 (verified 0/2+d) | GPP – 248 | ||||
| Rotavirus | Immunochromatography | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (unbiased) | Comparator + | Comparator – | |
| GPP + 61 | GPP + 6 (verified 6+d/0) | ||||
| GPP – 1 (verified 0/1–d) | GPP – 222 | ||||
| Salmonella | Culture confirmed by serotyping | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 25 | GPP + 6 (verified 3+d/3–e) | ||||
| GPP – 5 (verified 5+e/0) | GPP – 254 | ||||
| Shigella | Culture confirmed by serotyping | Single-plex PCR and sequencing with primers specifically for conserved regions of each target sourced from published literature (biased) | Comparator + | Comparator – | |
| GPP + 3 | GPP + 1 (verified 1+d/0) | ||||
| GPP – 0 | GPP – 286 | ||||
| FDA 201225 (xTAG) | Adenovirus | EIA and one PCR/sequencing assay | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – |
| GPP + 4 | GPP + 13 | ||||
| GPP – 1 (1+e/0) | GPP – 1154 | ||||
| C. difficile toxin A/B | Bartels cytotoxicity assay for C. difficile toxin | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased), or FDA cleared C. difficile toxin molecular assays (biased) | Comparator + | Comparator – | |
| GPP + 107 | GPP + 105 (verified 48+d/0, 57 remain inconclusive) | ||||
| GP – 7 | GPP – 922 | ||||
| Campylobacter | Culture (a PCR/sequencing assay was also performed directly on clinical specimens that were tested positive by culture for species identification only) | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 3 | GPP + 21 (verified 6+d/0, 15 remain inconclusive) | ||||
| GPP – 0 | GPP – 1155 | ||||
| Cryptosporidium | Microscopy | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 12 | GPP + 53 (verified 8+d/0, 45 remain inconclusive) | ||||
| GPP – 1 | GPP – 1131 | ||||
| E.coli O157 | Culture | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 2 | GPP + 9 (verified 4+d/0, 5 remain inconclusive) | ||||
| GPP – 0 | GPP – 1158 | ||||
| ETEC | Composite consisting of PCR/sequencing directly from clinical specimen using four PCR/sequencing assays | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 2 (verified 2+/0) | GPP + 4 | ||||
| GPP – 6 (verified 6+e/0) | 1156 | ||||
| Norovirus | Composite consisting of CDC real-time PCR and conventional PCR followed by bidirectional sequencing assays directly from clinical specimen | Validated PCR/sequencing assays (biased) | Comparator + | Comparator – | |
| GPP + 74 | GPP + 96 | ||||
| GPP – 4 (verified 0/4–d) | GPP – 1023 | ||||
| Salmonella | Culture | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 10 | GPP + 18 (verified 2+d/0,16 remain inconclusive) | ||||
| GPP – 0 | GPP – 1143 | ||||
| Shigella | Culture | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 2 | GPP + 17 (verified 2+d/0, 15 remain inconclusive) | ||||
| GPP – 0 | GPP – 1154 | ||||
| STEC | Broth enrichment followed by rapid immunoassay | Bidirectional sequencing analysis using analytically validated primers that targeted genomic regions distinct from the xTAG GPP (biased) | Comparator + | Comparator – | |
| GPP + 1 | GPP + 16 (verified 1+d/0, 15 remain inconclusive) | ||||
| GPP – 0 | GPP – 1153 | ||||
| Mengelle et al. 201329 (xTAG) | Adenovirus 40/41 | Rapid immunochromatographic tests | In-house real-time PCR (biased) | Verification gave results similar to those of xTAG | |
| Pankhurst et al. 201430 (xTAG) | Campylobacter | Culture | Single qPCR: primers probably different from xTAG (biased) | Comparator + | Comparator – |
| GPP + 110 | GPP + 11 (verified 11+d/0) | ||||
| GPP – 7 (verified 3+e/0, 4 remain inconclusive) | GPP – 711 | ||||
| C. difficile toxin A and B | EIA plus culture | Single qPCR: primers probably different from xTAG (biased) | Comparator + | Comparator – | |
| GPP + 195 | GPP + 19 (verified 15+d/0, 4 remain inconclusive) | ||||
| GPP – 4 (verified 2+e/0, 2 remain inconclusive) | GPP – 621 | ||||
| Norovirus | Quantitative PCR | Single qPCR (biased) | Comparator + | Comparator – | |
| GPP + 183 | GPP + 16 (verified 7+d/0, 9 remain inconclusive) | ||||
| GPP – 16 (verified 6+e/0, 10 remain inconclusive) | GPP – 624 | ||||
| Salmonella | Culture | Single qPCR: primers probably different from xTAG (biased) | Comparator + | Comparator – | |
| GPP + 15 | GPP + 9 (verified 0+/0, 9 remain inconclusive) | ||||
| GPP – 18 (verified 10+e/0, 8 remain inconclusive) | GPP – 797 | ||||
Even though the verification methods differed from conventional methods and used different molecular targets to GPP assays, they are not independent of the PCR methods used for either conventional methods or GPP assays, and they would be expected to resolve discordant results in favour of PCR-based methods, such as the GPP assays. Therefore, they are biased towards GPP testing. The findings are described briefly and are not considered further in the assessment of the GPP assays or economic modelling. Discordant analysis of GPP positive/conventional testing negative samples was generally in favour of GPP as anticipated; however, analysis of GPP negative/conventional positive outcomes more often favoured conventional testing (see Table 11). Discordant analysis failed to resolve all discordant samples and a considerable number of discordant results remained inconclusive following testing.
No particular pattern for any specific pathogen emerges from the discordant analysis. Finally, no study attempted independent verification of samples found to be pathogen positive by both methods.
Agreement between gastrointestinal pathogen panel tests in head-to-head comparisons
There are few comparative data of different GPP tests. Khare et al. 12 directly compared FilmArray with xTAG and concluded that both assays identify more pathogens than conventional methods and both have pathogens for which they do not appear to perform as well (FilmArray: Aeromonas, enteroaggregative E. coli and enteropathogenic E. coli; xTAG: Y. enterocolitica and norovirus). Similarly, Gu et al. 26 reported that both assays detected similar numbers of pathogens with differences between the two tests not reaching significance, considering either all pathogens on either panel (p = 0.34) or pathogens detectable only with both systems (p = 0.24). Gu et al. 26 reported differences in run failure rates when 10 out of 431 samples failed with xTAG, but none failed with FilmArray. A comparison of seven different multiplex systems provided no further information for the current review as, apart from xTAG, all other systems provided partial panel tests and the discrepancy in pathogens detected was significant. 22
In the absence of concordance analyses in these studies, whether or not tests identify the same pathogens in the same samples, no reliable assessment has been possible of the comparative performance of the different GPP tests.
Secondary outcomes
Studies reported the following secondary outcomes: GPP run failure (n = 8),20,25,26,29,31,33,35,38 multiple infections (n = 23),11,12,19–39 patient isolation (n = 2),27,34 turnaround times (n = 8),12,20,23,26–28,35,37 costs (n = 3)12,27,31 and other outcomes (n = 4). 22,27,31,34 No study reported on change in management by test outcome, health-related quality of life, morbidity or mortality. Reported secondary outcomes are summarised in this section.
Run failures/invalid test results
Gastrointestinal pathogen panel run failure results were reported in nine studies (Table 17). 20,25,26,29,31,33,35,38,39 In six studies,20,26,29,33,35,38 the run failure rate ranged from 0.8%20 to 7.8%. 29 Some of the reasons for these failures were ‘no call’, software error, loss of vacuum pressure in pouches or failed internal control. In one study,29 there was a repeated pattern of 34 failures (7.8%) for each pathogen, suggesting that these failures were test related rather than pathogen related. Likewise, reported failure rates were similar for each pathogen in the FDA report. 25 GPP failures appear much greater than for conventional testing when considering failures reported in the FDA report, which recorded failures only for C. difficile with conventional testing. Two other studies reported GPP run failure rates of ≥ 14%,25,31 and no explanations or reasons for these failures were provided. Only one study using two GPP tests (xTAG and FilmArray) reported comparative failure rates, with FilmArray producing no run failures whereas xTAG returned about 5%. 26
| Study reference (GPP assay) | Test failures for GPP samples, n/N (%) | Comments, resolution and retesting |
|---|---|---|
| Buss et al. 201520 (FilmArray) | 13/1557 (0.8) | One initial run aborted by user; three runs with software errors; nine runs with failed internal control; retest resulted in 12 valid results, one sample could not be retested within 4 days and was excluded |
| FDA 201225 (xTAG) | Campylobacter: 228/1407 (16.2) | No reasons for failure were given |
| C. difficile toxin A/B: 196/1407 (13.9) | ||
| Cryptosporidium: 210/1407 (14.9) | ||
| E. coli: 238/1407 (16.9) | ||
| ETEC: 239/1407 (16.9) | ||
| Giardia: 232/1407 (16.5) | ||
| Norovirus: 210/1407 (14.9) | ||
| Rotavirus: 241/1407 (17.12) | ||
| Salmonella: 236/1407 (16.8) | ||
| STEC: 237/1407 (16.8) | ||
| Shigella: 234/1407 (16.6) | ||
| Gu et al. 201526 (xTAG and FilmArray) | xTAG: 10/199 (5.0) | Failed samples underwent repeat testing, 7/10 were resolved |
| FilmArray: no failures | ||
| Mengelle et al. 201329 (xTAG) | 34/440 (7.8) for each pathogen (the repeated pattern of failures indicates that they were test related rather than pathogen related) | Uninterpretable because internal control was negative |
| Patel et al. 201431 retrospective (xTAG) | 19% and 14%, respectively, for two laboratories | 1 : 10 dilution of samples and retesting resulted in 3% total inhibition rate |
| Petterson et al. 201633 (XTAG) | Centre 1 (tertiary care): 4/145 (2.7) | Repeatedly ‘no call’ samples that were subsequently excluded from the analysis |
| Centre 2 (general hospital): 4/191 (2.1) | ||
| Spina et al. 201535 (FilmArray) | 36/709 (5.1) | Software error/loss of vacuum pressure in pouches |
| Wessels et al. 201438 (xTAG) | 30/393 (7.7) | ‘No call’ xTAG 10-fold dilution of samples reduced ‘no calls’ to 9/393 |
| Zboromyrska et al. 201439 (xTAG) | NR | The large number of initially inhibited results was addressed by adding an additional step to the GPP assay protocol: dilution of all extracted samples (1 : 10 in water) prior to amplification. Four of the inhibited samples were not resolved (negative by routine testing). No further inhibited results were obtained |
Test failures for conventional tests were reported for one study only,25 in which, for C. difficile toxin, the test failure rate was 6.8% (95/1407), whereas for the remaining pathogens no failure was recorded (Table 18).
| Study reference (GPP assay) | Test failures for conventional method samples, n/N (%) | Comments, resolution and retesting |
|---|---|---|
| FDA 201225 (xTAG) | Campylobacter: 0/1407 (0.0) | Inhibited results were presented as ‘invalid’ for each microbial target. No reasons for failure were given |
| C. difficile toxin A/B: 95/1407 (6.8) | ||
| Cryptosporidium: 0/1407 (0.0) | ||
| E. coli: 0/1407 (0.0) | ||
| ETEC: 0/1407 (0.0) | ||
| Giardia: 0/1407 (0.0) | ||
| Norovirus: 0/1407 (0.0) | ||
| Rotavirus: 0/1407 (0.0) | ||
| Salmonella: 0/1407 (0.0) | ||
| STEC: 0/1407 (0.0) | ||
| Shigella: 0/1407 (0.0) |
Frequency and characterisation of multiple infections
Table 19 summarises multiple infections as a proportion of positive GPP samples in the included studies. Multiple infections observed in the studies varied from 4%28,32 to 58%. 24 The studies with the greatest reported rate of multiple infections were from Vietnam (58%)24 and Côte d’Ivoire (54%). 19 Although the majority of multiple infections were double infections, Becker et al. 19 identified three samples with 10 pathogens, and the FDA study25 reported one sample with seven pathogens. Spina et al. 35 investigated stool samples from in- and outpatients and concluded that multiple infections were mainly identified in children aged < 5 years and outpatients. The three most frequent pathogens involved in multiple infections varied between studies, with no clear pattern emerging.
| Study reference, location (GPP/population) | Number of coinfections per positive GPP sample, n/N (%) | Three most commonly involved pathogens |
|---|---|---|
| Becker et al. 2015,19 Côte d’Ivoire (xTAG/community patients) | 37/68 (54)a
|
NR |
| Beckmann et al. 2014,11 Switzerland (xTAG/children and travellers) | 8/87 (9) (5 in children, 3 in travellers)
|
Rotavirus, ETEC and C. difficile |
| Buss et al. 2015,20 USA (FilmArray/routine stool samples for gastrointestinal testing | 262/832 (31)
|
Plesiomonas shigelloides, ETEC and adenovirus |
| Claas et al. 2013,21 the Netherlands, USA, UK, Canada (xTAG/paediatric and adult patients) | 86/NR
|
C. difficile, norovirus and Campylobacter |
| Coste et al. 2013,22 France (xTAG/adult kidney transplant recipients) | 7/36 (19)
|
Campylobacter, norovirus and either Giardia, STEC or rotavirus |
| Deng et al. 2015,23 China (xTAG/in- and outpatients, adults and children with diarrhoea) | 35/193 (18)
|
NR |
| Duong et al. 2016,24 Vietnam (xTAG/hospitalised patients with diarrhoea) | 233/404 (58)
|
Cryptosporidium, ETEC and norovirus |
| FDA 2012,25 USA, Canada (xTAG/mixed setting, immunocompromised and immunocompetent patients) | 91/486 (19)
|
Norovirus, C. difficile and Salmonella |
| Gu et al. 2015,26 USA (xTAG/immunocompromised paediatric patients) | 19/99 (19)
|
NR |
| Gu et al. 2015,26 USA (FilmArray/immunocompromised paediatric patients) | 14/94 (15)
|
NR |
| Halligan et al. 2014,27 UK (xTAG/hospitalised patients with diarrhoea) | 41/483 (8)
|
C. difficile, rotavirus and Salmonella |
| Kahlau et al. 2013,28 Germany (xTAG/hospitalised patients with diarrhoea) | 7/157 (4)
|
Norovirus, Salmonella and C. difficile |
| Khare et al. 2014,12 USA (FilmArray/routine stool samples for gastrointestinal testing | 86/318 (27)
|
EPEC, Yersinia and norovirus |
| Khare et al. 2014,12 USA (xTAG/routine stool samples for gastrointestinal testing) | 44/312 (14)
|
Norovirus, C. difficile and STEC |
| Mengelle et al. 2013,29 France (xTAG/immunosuppressed adults and children, children attending neonatal unit and children attending emergency unit) | 31/176 (18)
|
Rotavirus, norovirus and Salmonella |
| Pankhurst et al. 2014,30 UK (xTAG/hospitalised patients with diarrhoea) | 45/561 (8)
|
Norovirus, C. difficile and Campylobacter |
| Patel et al. 2014,31 USA (xTAG/paediatric and adult patients; retrospective) | 5/77 (6)
|
Norovirus, C. difficile and rotavirus |
| Patel et al. 2014,31 USA (xTAG/paediatric and adult patients; prospective) | 5/86 (6)
|
C. difficile, norovirus and Shigella |
| Perry et al. 2014,32 UK stored samples (xTAG/patients from hospital and community) | 10/248 (4)
|
C. difficile, norovirus and rotavirus |
| Perry et al. 2014,32 UK concurrently tested samples (xTAG/patients from hospital and community) | 6/155 (4)
|
C. difficile, norovirus and Shigella |
| Petterson et al. 2016,33 USA Centre 2 (general hospital) (xTAG/uninsured primary care patients) | 5/47 (11)
|
Norovirus, Giardia and Campylobacter |
| Petterson et al. 2016,33 USA Centre 1 (tertiary care) (xTAG/surgical, transplant and oncology patients) | 1/19 (5)
|
Norovirus and Giardia |
| Rand et al. 2015,34 USA (FilmArray/hospitalised patients with diarrhoea) | NR | EPEC, norovirus and EIEC/Shigella |
| Spina et al. 2015,35 Europe (FilmArray/community-acquired gastroenteritis in- or outpatients) | 64/384 (17)
|
EPEC, EAEC and Campylobacter |
| Stockmann et al. 2015,36 USA (FilmArray/children and young adults) | 77/244 (32)
|
NR |
| Vocale et al. 2015,37 Italy (xTAG/hospitalised patients) | 123/356 (35)
|
C. difficile, norovirus and Cryptosporidium |
| Wessels et al. 2014,38 the Netherlands (xTAG/patients with gastrointestinal complaints) | 6/76 (8)
|
NR |
| Zboromyrska et al. 2014,39 Spain (xTAG/travellers) | 20/70 (29)
|
Shigella, ETEC and EAEC |
Multiple infections in positive samples identified by the comparator were less frequently reported and the number of pathogens detected was generally lower. Spina et al. 35 reported that 5.4% (7/128) of positive samples were positive for two pathogens using conventional methods, Stockmann et al. 36 reported that 3.4% (6/175) of positive samples were positive for multiple infections with conventional methods and Gu et al. 26 reported that 2% (1/47) of samples tested positive for two pathogens using conventional methods. Deng et al. 23 reported a considerably higher rate of 16% (29/178) of multiple infections with conventional methods.
Patient isolation
Patient isolation as an outcome was reported in two studies (Table 20). 27,34 In the study by Halligan et al. ,27 a higher proportion of community-acquired inpatient cases versus hospital-acquired inpatient cases were isolated (69.0% vs. 52.1%), but subsequently removed from isolation (60.1% vs. 41.6%; p < 0.01). The same study found a tendency for the median number of days spent in isolation to be higher for patients with hospital-acquired infection (range 0–13.5 days) than for those with community-acquired infection (range 1–4 days). Rand et al. 34 reported that isolation could have been averted in the 25 patients (out of the 102) who tested negative on GPP (24.6%). The median isolation time for these patients was 3.8 days (interquartile range 1.8–11.0 days).
| Study reference, country | Patient isolation, n/N (%) | Time spent in isolation | ||
|---|---|---|---|---|
| Isolated | Removed from isolation | Not removed from isolation | ||
| Halligan et al. 2014,27 UK | Of the 409 CA cases, 282 (69.0%) were isolated and 127 (31%) were not isolated Of the 622 HA cases, 324 (52.1%) were isolated and 298 (47.9%) were not isolated |
CA cases: 246/409 (60.1) HA cases: 259/622 (41.6) |
CA cases: 36/409 (8.8%) HA cases: 65/622 (10.5%) Reason for not removing patients from isolation after negative GPP result:
|
Median isolation time (number of days) by pathogen CA cases:
|
| Rand et al. 2015,34 USA | GPP+/conventional– (n = 35):
|
NR | GPP–/conventional– (n = 102):
|
GPP–/conventional– (n = 25):
|
Turnaround times
The turnaround times for GPP testing were reported in seven studies (Table 21),12,20,23,26–28,35 of which two compared timings with conventional methods. 27,28 In two studies12,26 the results tended to demonstrate a longer turnaround time for xTAG than for FilmArray. However, turnaround times may apply to different throughputs, with FilmArray able to test up to eight samples (using eight units) at the same time and xTAG able to run up to 96 samples. One study found that median turnover time was significantly shorter for xTAG than for conventional methods (1 day vs. 3 days; p < 0.0005). 28 One study reported a wide variability in turnaround test times across different pathogens when using conventional methods. 27 This study reported separately the clinical and laboratory turnaround times for GPP testing, with clinical turnaround times being much longer (41.8 hours vs. 26.6 hours), but suggested that xTAG pragmatically may provide a next-day service with test outcomes, only available the following day, having allowed for batching. One study only reported time taken using conventional methods, reflecting the common experience of conventional testing taking up to 3 days to result and report. 37
| Study reference, country | Turnaround time (in days or hours) |
|---|---|
| Buss et al. 2015,20 USA | FilmArray: about 1 hour per specimen |
| Deng et al. 2015,23 China | xTAG: 5 hours |
| Gu et al. 2015,26 USA | FilmArray: total run time was approximately 1 hour with approximately 5 minutes of hands-on time |
| xTAG: total run time was approximately 6.5 hours with approximately 2.5 hours of hands-on time, including the sample processing and data analysis | |
| Halligan et al. 2014,27 UK | Median clinical turnaround times: xTAG: 41.8 hours for afternoon testing, compared with conventional methods (C. difficile, 17.3 hours; norovirus, 27.3 hours; adenovirus and rotavirus, 27 hours; bacterial culture, 66.5 hours; parasites, 66.5 hours); time includes sample collection and transport time |
| Laboratory turnaround time: xTAG 26.6 hours for afternoon run and 10.4 hours for morning run. In 205/2187 (9.4%) patients, xTAG was returned the day after collection and was significantly slower than the afternoon run | |
| Kahlau et al. 2013,28 Germany | Median turnover time (xTAG vs. conventional): 1 day (range 0.5–2 days) vs. 3 days; p = 0.0000021 |
| Khare et al. 2014,12 USA | Processing time per run: FilmArray (2 minutes), xTAG (45 minutes) |
| Time/run: FilmArray (about 1 hour), xTAG (about 3.5 hours) | |
| Throughput: FilmArray (1 specimen per run), xTAG (96 specimens per run) | |
| Separate extraction required: FilmArray (no), xTAG (yes; about 45 minutes) | |
| Spina et al. 2015,35 Austria, Ireland, Finland, France, Germany, Greece, Italy, Portugal, Romania and UK | FilmArray: The median number of days between sampling and testing at the central study laboratory was 11 (IQR: 7 days) |
| Vocale et al. 2015,37 Italy, the Netherlands | For conventional methods:
|
Costs
Three studies reported test-related costs (Table 22). 12,27,31 GPP testing was reported to be more costly than conventional tests (£150,641 for 2187 samples vs. £63,431 for 4467 samples). 27,31 Among GPPs, FilmArray was reported to be more costly than xTAG (£22,910 vs. £21,460 list price per instrument). 12 These costs reflect different fixed and variable cost assumptions and are reported for information only.
| Study reference, country | Costs (£) |
|---|---|
| Halligan et al. 2014,27 UK | Total cost for conventional test (n = 4467): 63,431 |
| Total cost for GPP xTAG (n = 2187): 150,641 | |
| Khare et al. 2014,12 USA | Per instrument:
|
List price reagent cost per specimen:
|
|
| Patel et al. 2014,31 USA | Per identification reagents (conventional vs. xTAG): 29 vs. 46.40 |
| Per specimen identification (conventional vs. xTAG): 145 vs. 36.25 |
Other outcomes
Other outcomes (e.g. clinical management change, user acceptance, length of stay, technical hands-on time) were reported for four studies and are summarised in Table 23. 22,27,31,34
| Study reference, country | Other outcomes |
|---|---|
| Coste et al. 2013,22 France | Management not changed as a result of GPP testing, but might be potentially |
| Intestinal endoscopy was performed in nine patients (18%). Endoscopy was performed if symptoms persisted for > 21 days with negative microbiological analyses and/or worsening symptoms (rectal haemorrhage, fever) | |
| Immunosuppressive therapy switching or dose reduction was observed in 13/54 episodes (24%) | |
| Halligan et al. 2014,27 UK | User acceptance: fast turnaround, enhanced infection and prevention control action |
| Conventional tests performed for every GPP test: 2 | |
| Patel et al. 2014,31 USA | Technician hands-on time (hours): conventional vs. xTAG (10 vs. 2.5) |
| Time to detection (hours): conventional vs. xTAG (72 vs. 5) | |
| Rand et al. 2015,34 USA | Length of stay, mean (SD): after the negative test for C. difficile and/or rotavirus length of stay in the hospital was 8.0 (± 9.1) days for those with an unsuspected hospital-acquired pathogen and was not statistically different from the 15.1 (± 22.2) days for those negative for all agents on the gastrointestinal panel |
Interpretation of the clinical effectiveness results and implications for the cost-effectiveness evaluation
Interpretation of findings
In the absence of an adequate reference standard or alternative methods of assessing test accuracy, positive and negative agreements have been produced against a benchmark, as recommended by FDA guidance. If conventional methods are an accurate determinant of important disease, then meta-analytic results (benchmarked against conventional testing) suggest that GPP testing is reliable and could replace current microbiological methods, although there would be an increase in false-positive reporting and potential overtreatment. However, if GPP testing is considered accurate (in the sense that current testing practices are missing clinically important pathology), then it would identify missed infections and could potentially result in more appropriate treatment. Currently, the clinical importance of the additional pathogens identified by GPP testing is uncertain, apart from, for example, enteroaggregative E. coli, which is not detected by routine culture.
A further uncertainty is the value of more rapid testing, achieved to a varying extent by each GPP system. In the vast majority of cases, hydration, hygiene and watchful waiting are required. Only for a select few pathogens is specific treatment recommended [C. difficile, some strains of Salmonella, Shigella, E. coli (non-shiga toxin-producing E. coli), Campylobacter and Giardia], although not all patients are treated (see Chapter 5, Treatment). If conventional methods do accurately determine important disease, then the introduction of GPP testing could potentially result in some earlier overtreatment of unnecessary cases, which might spontaneously resolve.
Agreement measures are not measures of test performance in a conventional sense, and only adequately designed research will resolve uncertainties about the introduction of GPP testing. It may not be possible to design a study with an adequate independent reference standard. Molecular methods may not be the best option to address the problem, as the presence of pathogen DNA does not necessarily answer the question of the clinical importance of the identified pathogens. A randomised controlled trial may be the best option, with patients randomised to conventional and GPP testing, and clinical effectiveness and cost-effectiveness outcomes used to determine the value of GPP testing. Currently, there is no adequate evidence of the impact of GPP testing on the management, treatment and outcome of patients as no test-and-treat trials have been conducted.
The NICE scope identified the PHE algorithm8 as the relevant comparator against which to consider GPP tests. However, the PHE algorithm was published in 2013 and may require updating to reflect progress in routine microbiological laboratories. For instance, the new PHE guidance for the interpretation of PCR assays for gastrointestinal pathogens states that a growing number of PHE and NHS virology laboratories use PCR assays for detection of viral gastrointestinal pathogens. 50 The same guidance recommends PCR as a first-line diagnostic test for the intestinal parasites Giardia lamblia, Cryptosporidium spp. and E. histolytica. This is not reflected in the national standard of practice used in this review. Similarly, this review has evaluated GPP systems according to their current specification, but it is anticipated that the coverage of these systems will continue to evolve in response to changing pathogen prevalence, hence the evaluation problem is a dynamic one.
Current evidence should be regarded as applicable to a mixed population of community and hospitalised patients, as the studies do not adequately report outcomes by setting or subgroup. Only one study from Switzerland11 reported receiving samples both from GP practices and hospitals (emergency room and admitted patients). However, the GP samples were from travellers returning from the tropics only and the hospital samples were paediatric samples with suspected viral gastroenteritis; the study sample may have low applicability to a routine clinical setting. One study27 reported that community-acquired infections show a much greater spectrum of pathogens present than patients with hospital-acquired infections. Thus, a goal for a future trial might include adequate stratification of different populations (e.g. community managed, community acquired and hospital managed, hospital acquired, children, travellers and immunocompromised patients).
A paucity of head-to-head comparative data of different GPP tests limits conclusions. Khare et al. 12 directly compared FilmArray with xTAG, and concluded that both assays identified more pathogens than conventional methods, but may not perform uniformly well against different pathogens. Gu et al. 26 reported that both assays detected similar numbers of pathogens, with differences between the two tests not reaching significance. Failure rates may vary between GPP systems, and further research is needed to inform this issue.
Discrepancies between GPP tests and conventional methods may result from differences not only in accuracy but also in their targets. For example, discrepancies reported by Gu et al. 26 for adenovirus may have been a result of the comparator PCR identifying all adenovirus strains, whereas the GPP system identified only adenovirus 40 and 41. Similarly, Pankhurst et al. 30 reported that xTAG has two targets for C. difficile (genes for toxins A and B) and includes two primers against norovirus GI and GII strains. However, the single quantitative PCRs used as comparators in the study targeted only the gene for toxin B or the GII strain. They classed a positive result on either target with GPP as indicating the presence of the organism. This misalignment problem may be exacerbated when comparing different GPP systems with their varying coverage.
Implications for health economic model
Uncertainties about how to interpret available evidence for the performance of GPP systems and the impact they may have on clinical care make any attempt to model the economic consequences of introducing GPP systems tentative. Consequently, an economic model has been developed to explore these uncertainties. In the model, conventional testing using the PHE algorithm is considered to be accurate, providing the benchmark for comparison. The consequence of replacing conventional testing with GPP systems is explored. Different populations and a range of assumptions are explored regarding the impact of adopting GPP systems on clinical care. These assumptions have been variously informed by the direct evidence, epidemiological sources and expert opinion.
Chapter 4 Cost-effectiveness review
Systematic review of existing cost-effectiveness evidence
Methods
Search strategy
A comprehensive review of the published literature including economic evaluations, economic models, cost studies and quality-of-life studies was performed. The systematic search included the following electronic databases on 29 January 2016:
-
Ovid MEDLINE – 1946 to January week 3 2016
-
Ovid MEDLINE In-Process & Other Non-Indexed Citations: 27 January 2016
-
EMBASE: 1974 to January 27 2016
-
Web of Science Core Collection.
The search terms included economic and quality-of-life terms combined with gastroenteritis and the different GPP tests. The search was limited to studies published in the English language and studies involving humans. The search strategy was developed, based on the clinical effectiveness review and with health economist input. Details of the search strategies are provided in Appendix 1.
Review strategy
Citations and abstracts from the electronic online databases were exported into a citation software package (EndNote X7) and duplicate citations were removed. Two reviewers independently reviewed titles and abstracts to identify potentially relevant papers for inclusion. Any discrepancies were resolved by discussion.
Inclusion criteria
Studies meeting the following criteria were included in the review:
-
study type – fully published economic evaluations (including any economic models)
-
population – patients with suspected infectious gastroenteritis
-
intervention – integrated multiplex PCR tests, which include xTAG, FilmArray and Faecal Pathogens B
-
comparator – standard microbiology techniques as outlined in the PHE pathway for routine testing in cases of gastroenteritis and diarrhoea
-
outcomes – cost–benefit, cost–consequences, cost-effectiveness or cost–utility studies reporting outcomes as clinical effectiveness measures or utility measures [utility, EuroQol-5 Dimensions (EQ-5D) or Short Form questionnaire-6 Dimensions score, or quality-adjusted life-years (QALYs)].
Exclusion criteria
Studies meeting the following criteria were excluded from the review:
-
non-English-language publications
-
studies not in humans
-
studies not in gastroenteritis and diarrhoea
-
studies that were not full economic evaluations (incremental costs and incremental benefits).
Studies that provided useful information for the economic model, such as resource use, costs, utilities and transition probabilities, were retained but were not included in this review.
Data extraction strategy
Data extraction was carried out by one reviewer using standardised data extraction sheets (www.journalslibrary.nihr.ac.uk/hta/hta19100) and was then checked by a second reviewer. Data extracted included the following information:
-
study details – study title, author names, source of publication, language and publication type
-
baseline characteristics – population (and subgroups), intervention, comparators, outcomes, study design, and setting and location
-
methods – study perspective, time horizon, discount rate, measurement of effectiveness, measurement and valuation of preference-based outcomes, resource use and costs, currency, price date and conversion, model type, assumptions and analytical methods
-
results – study parameters, incremental costs and outcomes, and reporting of uncertainty
-
discussion – study findings, limitations, generalisability and conclusions
-
other – sources of funding, conflicts of interest and any comments.
Quality assessment strategy
We critically appraised studies classed as full economic evaluations against the framework of quality assessment for economic evaluation studies developed by the Consolidated Health Economic Evaluation Reporting Standards (CHEERS) group. 51 The CHEERS framework comprises six dimensions (i.e. title and abstract, introduction, methods, results, discussion and other), including a series of questions check whether or not the criteria have been clearly reported. If the studies included any model-based economic evaluations, they were further critically appraised using the framework on quality assessment for economic modelling developed by Philips et al. 52 The framework assesses decision-analytic models used in economic evaluations under the dimensions of structure, data and consistency.
Results
Search results
The literature search identified 377 records through electronic database searches and other sources:
-
Ovid MEDLINE – 1946 to January week 3 2016 (n = 132)
-
Ovid MEDLINE In-Process & Other Non-Indexed Citations – 27 January 2016 (n = 12)
-
EMBASE – 1974 to January 27 2016 (n = 203)
-
Web of Science Core Collection (n = 30).
After removing duplicates, 222 records were screened for inclusion. On the basis of title and abstract sift only, 217 records were excluded. The remaining five records were included for full-text screening. 27,53–56 A further four articles were excluded at the full-text stage as these studies did not contain a full economic evaluation (see Appendix 9). 27,53–55
The literature search identified only one study,56 classed as a full economic evaluation, that simulated the GPP pathway of care and presented a cost–consequence type model (although not explicitly described) (Figure 9). Two further reports were identified in our search (and also by NICE) but were not included in our review: Abubakar et al. 57 did not look at the same interventions as outlined above in our inclusion criteria and Pankhurst et al. 30 did not provide a cost-effectiveness analysis. However, both studies contained some useful information for our economic model and so have been summarised in Additional information.
FIGURE 9.
A PRISMA flow diagram for the economic review.

Quality assessment
The quality of the reporting of the economic analysis provided by the Goldenberg et al. 56 study was assessed using the 25-point CHEERS checklist51 and is provided in Appendix 10.
The study was comprehensively completed, with 17 out of the 25 statements (68.0%) being completed, three statements (12.0%) partially completed, three statements (12.0%) not completed and two statements (8.0%) not applying. The quality of the reporting of the cost–consequence model, using the Philips et al. 52 checklist, is presented in Table 7. The article was not comprehensively reported, with just 11 out of the 57 statements (19.3%) completed, 7 statements (12.3%) partially completed, 11 statements (19.3%) not completed, 18 statements (31.6%) not applying and 10 statements unclear (17.5%). The poor quality of reporting was related to the authors conducting a partial rather than full economic evaluation.
Overview of included studies
Full economic evaluation
Goldenberg et al. 56 report an analysis based on a parallel testing study involving 800 patients and comparing the Luminex xTAG GPP with conventional laboratory testing (which included a combination of selective culture, microscopy and enzyme immunoassay). In this study, patients essentially received conventional testing and care according to clinicians’ usual clinical practice over an 8-month period. Conventional testing was performed 7 days per week and only one GPP test was performed per 5-day period so that GPP testing may have influenced the care of some patients, notably removal from isolation. Costs were estimated for 800 patients in a London hospital with suspected infectious gastroenteritis (diarrhoea and/or vomiting). The paper presented pathways for both the conventional (provided) and GPP (simulated) arms. To summarise the pathways: a patient could either be isolated or not isolated while they were waiting for test results, then when the test result arrived the infectious gastroenteritis agent might or might not be detected. If the agent was detected and the patient was infected with a communicable agent, then the patient was isolated and treated; however, if the agent was detected and was non-communicable, then the patient was treated without isolation. If the agent was not detected and symptoms persisted, then the patient was retested, and alternative causes were investigated; however, if the symptoms resolved then the patient was discharged. The authors assumed that all GPP tests were 100% accurate.
The findings were presented as a cost–benefit analysis from a NHS perspective. Although not explicitly stated, the time horizon for the analysis was the duration of the index episode; whether or not patients were readmitted is not reported. Resource use and costs included laboratory testing and patient isolation costs, although a comprehensive breakdown of these costs is not provided. Isolation costs for the conventional pathway were based on actual days observed and for the GPP pathway these days were estimated. Costs associated with false-positive and false-negative results and treatment costs were not included. Consequently, total costs presented may underestimate costs incurred within the NHS. The year for which resources were costed is not explicitly stated, limiting comparison with other studies. As the time frame of the study was under 1 year, no discounting of costs (and/or benefits) was performed by the authors.
The preferred outcome measure for NICE is the QALY, permitting comparisons with other diseases and interventions. Reported as a cost–benefit analysis, the value of outcomes (test accuracy, reduction of time in isolation) was expressed in Great British pounds (GBP). A common limitation of this form of analysis is that patient health outcomes are intangible and assumed proxied by tangible measures. Compared with the conventional pathway, the xTAG GPP pathway had a higher detection rate for isolated patients [37.2% (152/409) compared with 19.8% (81/409)], reduced isolation time from 2202 to 1447 days, but cost an additional £22,283 for the laboratory tests, resulting in total savings of £66,765. Overall, the total costs (isolation costs plus laboratory costs) for GPP testing were £44,482 lower than conventional testing. The authors also conducted sensitivity analyses to deal with uncertainty around key parameters used in the economic model. One key sensitivity analysis was around the time spent in isolation (time spent varied from 1 to 3 days), and in each case there was a net saving under the GPP testing pathway. The authors found that the break-even point associated with implementing the GPP pathway is a reduction of 252 isolation days. The Goldenberg et al. 56 study is summarised in Appendix 10.
The key methodological shortcomings of the Goldenberg et al. 56 paper were that the study simulated care following xTAG diagnosis (no real patient care was involved in this group), the xTAG GPP test was assumed to be 100% accurate, the days in isolation were estimated for the GPP pathway rather than observed and outcomes were not in the form of QALYs.
Additional information
The two further health technology assessment reports that were identified,30,57 as noted previously, have been summarised below.
Abubakar et al. 57 assessed the cost-effectiveness of rapid diagnostic tests (real-time PCR and enzyme-linked immunosorbent assay) compared with routine culture using a hypothetical decision tree model. The authors tested 10,000 faecal samples for enteric pathogens causing food-borne illnesses. A NHS perspective was adopted for the study and the time horizon was 1 year. Cost data came from hospitals and manufacturers; for example, microbiological investigations included average costs of capital and overheads of providing these services, test costs and staff costs. Treatment costs (antibiotic therapy or hospital bed-days) were excluded. Costs were presented in GBP in 2005 prices. As the study length was 1 year, no discounting was performed. Data from the clinical effectiveness review, which included data on isolation rates for each pathogen, were used as the measure of effectiveness in the study. Outcomes were presented as the cost per additional case detected. At baseline, testing one sample for Campylobacter, Salmonella and E. coli cost £18.85 with PCR, £15.66 with immunoassays and £15.01 by culture methods. The use of enzyme-linked immunosorbent assay test kits was a marginally less expensive strategy than PCR testing. In terms of characterising the uncertainty in the model, small changes in diagnostic accuracy and implementation cost estimates do not vary the findings. However, the most sensitive parameter was the isolation rate for each pathogen. The key limitations were the availability of the data for the model, no inconclusive test results were recorded and there were no repeat tests and, in addition, sensitivity and specificity for the comparator (bacterial culture) were assumed to be 1. The authors concluded that rapid testing in combination with routine culture is unlikely to be cost-effective. However, they believed that, if the cost of rapid technologies falls, it may become more cost-effective. In addition, they noted that, if multiple pathogens can be simultaneously detected, then these rapid tests may prove to be cost-effective.
Pankhurst et al. 30 aimed to assess the diagnostic accuracy of the Masscode gastric multiplex PCR test (Qiagen Masscode Technology, Hilden, Germany). The study was split into two phases. Phase 1 was a retrospective study assessing the sensitivity and specificity of the MassCode test against four major pathogens. In addition, the questionnaire survey examined current practice and the costs of managing infectious diarrhoea. Phase 2 was a prospective, real-time, parallel-group study estimating the cost-effectiveness of the MassCode test using a decision Markov model. However, because of analytical problems with the MassCode test, the project was stopped earlier than intended and the planned economic analyses were not carried out.
Summary
The cost-effectiveness search highlighted only one study that was classed as a full economic evaluation comparing an integrated multiplex PCR test (xTAG) with conventional care to rapidly identify gastrointestinal pathogens in people with suspected infectious gastroenteritis. However, the study had some key limitations, principally changes in isolation due to GPP were estimated not observed, and there was no explicit valuation of patient outcomes. In the next chapter, we build a de novo economic model comparing these integrated multiplex PCR tests with conventional care for people with suspected infectious gastroenteritis.
Chapter 5 Health economic methods and results
Objective
To develop a de novo model to explore the cost-effectiveness of GPP tests compared with standard care in England and Wales. This chapter describes the structure of the model, the model inputs, the assumptions made, the different scenarios that have been evaluated, the main results and the results from the sensitivity analyses.
Developing the model structure
Currently in the UK, patients with acute suspected gastroenteritis persisting for more than several days undergo routine faecal microbiology tests to investigate the cause of acute diarrhoea with or without vomiting. A decision tree model was constructed to see if the use of integrated multiplex PCR tests (xTAG, FilmArray and Faecal Pathogens B) is cost-effective compared with conventional care (the PHE pathway) for rapidly identifying gastrointestinal pathogens in people with suspected infectious gastroenteritis. In this section, the structure of the model is described. Conventional testing was chosen pragmatically as the benchmark for comparison within the model as it represents current practice.
Models developed
To assess the cost-effectiveness of integrated multiplex PCR tests compared with conventional care, a de novo decision tree model was developed in Microsoft Excel® (version 16, Microsoft Corporation, Redmond, WA, USA). A decision tree model was the most appropriate choice as gastroenteritis is a self-limiting illness and usually stops within 2 weeks (i.e. diarrhoea lasts for 5–7 days and stops within 2 weeks and vomiting lasts for 1–2 days and stops within 3 days). 58
The base economic model (model 1) includes hospitalised adult patients with suspected gastroenteritis (acute hospital admissions). The following subgroups of people with suspected gastroenteritis are explored in subsequent models: model 2, young children; model 3, people in the community; model 4, people who are immunocompromised; and model 5, people with a history of recent foreign travel (to areas other than Western Europe, North America, Australia or New Zealand).
Model 1: hospitalised adult patients with suspected gastroenteritis
The economic model was developed by determining the different clinical pathways for patients with suspected gastroenteritis. We have used information from the systematic review of clinical effectiveness, published literature and expert opinion to develop the different clinical pathways. The model structure is the same for both the GPP and conventional care arms (the main differences between the two arms are when test results are received and whether or not patients move from isolated to non-isolated care earlier, are treated earlier or are discharged earlier). Figure 10 shows the overall structure comparing the different treatment arms, Figure 11 shows the different pathways for patients who are isolated and Figure 12 shows the different pathways for patients who are not isolated.
FIGURE 10.
Overall model structure for patients with suspected gastroenteritis comparing the different treatment arms.
FIGURE 11.
The different pathways for isolated patients.

FIGURE 12.
The different pathways for non-isolated patients.
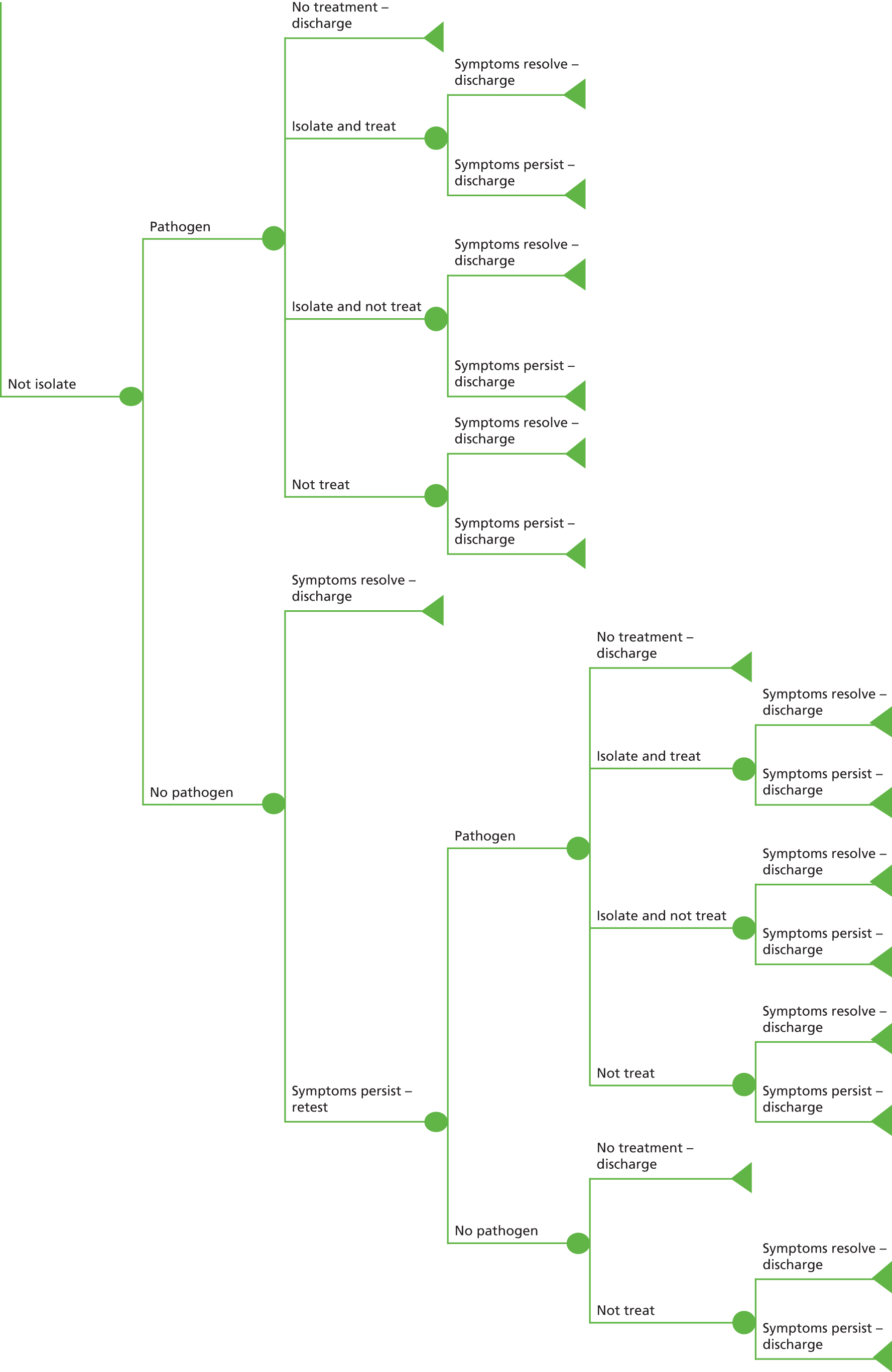
The pathway assumes that an adult patient is admitted to (or already in) hospital with suspected gastroenteritis (diarrhoea and/or vomiting). A decision is made by the medical staff regarding whether the patient goes to a side room (isolated) or stays on an open bay/ward (not isolated). This decision may depend on the availability of isolation rooms.
The patient provides a stool sample to test for suspected gastroenteritis. The stool sample is then sent to the laboratory (or transported in some instances when the laboratory is not on site) for testing. The model evaluates the consequences of using a multiplex integrated PCR test (xTAG, FilmArray or Faecal Pathogens B) or conventional testing. When results are returned, care is determined according to whether a pathogen(s) has or has not been detected. The presence of a pathogen is determined by conventional testing, as the benchmark test.
Isolated patients
In the following pathways, ‘treatment’ refers to pathogen-specific treatment over and above general care, hydration and hygiene management received by all patients. In the case of isolated patients in whom a pathogen has been detected, there are four options:
-
discharge the patient because symptoms have resolved
-
continue to isolate the patient and provide treatment
-
continue to isolate the patient and do not provide any treatment
-
deisolate the patient and do not provide any treatment.
With the final three options, symptoms can either resolve naturally, or can persist; in both cases, the patient is subsequently discharged when appropriate.
In the case of isolated patients in whom no pathogen has been detected, there are three options:
-
discharge the patient because symptoms have resolved naturally
-
keep the patient in isolation because symptoms persist
-
deisolate the patient and do not provide any treatment.
Patients in whom symptoms persist undergo retesting. (These patients remain in isolation.) The second test results either will or will not detect a pathogen. Patients in whom a pathogen is detected follow the same care pathway as isolated patients in whom a pathogen has been detected. In the case of patients in whom no pathogen is detected there are two options:
-
discharge the patient because symptoms have resolved
-
continue to isolate the patient and do not provide any treatment.
In the latter case, symptoms can either resolve naturally or persist; in both cases, the patient is subsequently discharged. Among patients who have been deisolated and who have received no treatment, symptoms can either resolve naturally or can persist; in both cases, the patient is subsequently discharged.
Non-isolated patients
For non-isolated patients in whom a pathogen has been detected, there are four options:
-
discharge the patient because symptoms have resolved
-
isolate the patient and provide treatment
-
isolate the patient and do not provide any treatment
-
do not provide any treatment.
With the final three options, symptoms can either resolve naturally, or can persist; in both cases, the patient is subsequently discharged when appropriate.
Among non-isolated patients in whom no pathogen has been detected, symptoms can resolve naturally or persist. Patients whose symptoms have resolved are discharged. Patients with persistent symptoms are retested. Those patients in whom a pathogen is detected follow the same care pathway as non-isolated patients in whom a pathogen has been detected. For patients in whom no pathogen is detected, there are two options:
-
discharge the patient because symptoms have resolved
-
do not provide any treatment.
In the case of the latter option, symptoms can either resolve naturally or persist; in both cases, the patient is subsequently discharged when appropriate.
Model assumptions and base-case analysis
As gastroenteritis is a self-limiting illness, most patients are treated symptomatically and there is no significant impact on quality of life for most patients beyond the symptomatic impact while the infection lasts. As a result of the lack of evidence with which to extrapolate benefits of multiplex testing, the model time frame is limited to the immediate index hospitalisation (approximately 2 weeks). The model does not take into account any sequelae or adverse events of treatment, subsequent readmission, persistent complications or mortality. In addition, the model does not consider transition costs (moving from one test to another or having to manage both in tandem); rather, a simple comparison of the two approaches to testing is made.
The base-case model is based on a hypothetical cohort of hospitalised adult patients with suspected gastroenteritis. The analysis is conducted from a NHS and Personal Social Services perspective. All costs are in GBP 2014/15 prices. Health outcomes were measured as QALYs gained. Results are expressed as an incremental cost per QALY gained. As the time frame is < 1 year, no discounting of costs or outcomes is performed. In the model, conventional testing provides the benchmark with which multiplex integrated PCR tests are compared.
Data required for the model
No clinical data were retrieved for Faecal Pathogens B, limiting subsequent consideration to its acquisition costs. Modelling of cost-effectiveness was possible for the xTAG GPP and FilmArray tests.
Prevalence and probabilities
Prevalence information for the model was provided from the clinical effectiveness meta-analysis review. Table 24 below shows the prevalence of pathogens and conditional agreement probabilities when conventional testing is the benchmark.
| Pathogen | Conventional testing | xTAG | FilmArray | ||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | True positive | False negative | True negative | False positive | ||
| Adenovirus | 0.01551 | 0.00865 | 0.00685 | 0.97465 | 0.00984 | 0.01120 | 0.00431 | 0.97760 | 0.00689 |
| C. difficile | 0.10342 | 0.09918 | 0.00424 | 0.86789 | 0.02869 | 0.10001 | 0.00341 | 0.87058 | 0.02600 |
| Campylobacter | 0.01162 | 0.01114 | 0.00048 | 0.95676 | 0.03163 | 0.01128 | 0.00034 | 0.97554 | 0.01285 |
| Cryptosporidium | 0.01125 | 0.01028 | 0.00097 | 0.97787 | 0.01088 | 0.01073 | 0.00052 | 0.98875 | 0.00000 |
| E. coli O157 | 0.00185 | 0.00171 | 0.00013 | 0.99316 | 0.00499 | 0.00176 | 0.00008 | 0.96621 | 0.03194 |
| ETEC | 0.01071 | 0.00995 | 0.00076 | 0.97742 | 0.01187 | 0.01071 | 0.00000 | 0.98336 | 0.00594 |
| Giardia | 0.00810 | 0.00810 | 0.00000 | 0.98099 | 0.01091 | 0.00810 | 0.00000 | 0.99190 | 0.00000 |
| Norovirus | 0.00472 | 0.04377 | 0.00345 | 0.92324 | 0.02954 | 0.04401 | 0.00000 | 0.94420 | 0.00858 |
| Rotavirus | 0.00285 | 0.00273 | 0.00012 | 0.98818 | 0.00897 | 0.00271 | 0.00013 | 0.99217 | 0.00499 |
| Salmonella | 0.01448 | 0.01184 | 0.00264 | 0.92639 | 0.05913 | 0.01448 | 0.00000 | 0.98355 | 0.00197 |
| Shigella | 0.01444 | 0.01428 | 0.00016 | 0.97078 | 0.01478 | 0.01385 | 0.00059 | 0.98556 | 0.00000 |
| Overall prevalence | 0.24143 | 0.22164 | 0.01979 | 0.53733 | 0.22124 | 0.22883 | 0.01260 | 0.65942 | 0.09915 |
Prevalences for conventional testing were obtained by pooling two large, representative studies (FDA evaluating xTAG,25 Buss et al. 20 evaluating FilmArray); thus, a common prevalence matrix was applied to both systems. The overall true pathogen detection rate, determined by conventional testing (assuming conventional testing is correct) is 24.1%, while the positive detection rate is 22.2% for xTAG GPP and 22.9% for FilmArray testing in the first round of testing; a second confirmatory test is required to identify false negatives. The true no-pathogen rate is 1 minus the prevalence. It is conservatively assumed that 25% of patients with false-positive results are retested, with 75% discharged without further management, as are all patients with true-negative results. Probabilities for positive and negative agreements for each pathogen were drawn from the clinical effectiveness review.
To estimate the proportion of patients who followed each pathway for the different tests requires knowledge of the proportion of patients who would be treated and not treated. These proportions were estimated by our clinical experts, as shown in Table 25.
| Pathogen | Treated and isolated | Not treated and isolated | Not treated and not isolated |
|---|---|---|---|
| Adenovirus | – | 1.00 | 0.00 |
| C. difficile | 0.95 | 0.05 | 0.00 |
| Campylobacter | 0.35 | 0.65 | 0.00 |
| Cryptosporidium | 0.00 | 1.00 | 0.00 |
| E. coli O157 | 0.00 | 1.00 | 0.00 |
| ETEC | 0.45 | 0.55 | 0.00 |
| Giardia | 0.95 | 0.025 | 0.025 |
| Norovirus | 0.00 | 1.00 | 0.00 |
| Rotavirus | 0.00 | 1.00 | 0.00 |
| Salmonella | 0.45 | 0.55 | 0.00 |
| Shigella | 0.45 | 0.55 | 0.00 |
These proportions (see Table 25), along with the prevalences (see Table 24), were used to estimate the proportions of patients who followed each pathway according to each pathogen (numbers are not shown here).
The overall probabilities (conditional) for patients in whom a pathogen was detected are shown in Table 26. When a test provides the benchmark, it is assumed that all pathogens are correctly identified and that second testing would identify no new pathology. When a test is being compared against the benchmark, second testing in the presence of persisting symptoms may detect missed pathology.
| Clinical pathway | Conventional testing as benchmark, first test | xTAG | FilmArray | ||
|---|---|---|---|---|---|
| First test | Second test | First test | Second test | ||
| Total treated and isolated | 0.5295 | 0.5506 | 0.2928 | 0.5428 | 0.2879 |
| Not treated and isolated | 0.1302 | 0.1317 | 0.1126 | 0.1345 | 0.0511 |
| Not treated and not isolated | 0.2933 | 0.2727 | 0.5239 | 0.2769 | 0.5898 |
| Not treated and discharged | 0.0470 | 0.0449 | 0.0707 | 0.0457 | 0.0712 |
The proportion of patients who were isolated and not isolated in each model was based on the study by Goldenberg et al. 56 (Table 27). Following advice from our clinical experts, we have assumed that the probability that a patient’s symptoms resolve naturally and that he or she is discharged is 0.75, and that the probability that patients’ symptoms persist and they are discharged is 0.25. These two probabilities do not influence the costs or QALYs within each of the different pathways. Again, on the advice of our clinical experts, among patients in whom neither the first nor the second test detects any pathogen, the probably of receiving no treatment and being discharged is 90%, while the probability of being isolated but receiving no pathogen-specific treatment is 10%. Table 28 shows the probabilities for the remaining pathways.
| Clinical pathway | Probability value | Source |
|---|---|---|
| Isolate | 0.5113 | Goldenberg et al.56 |
| Not isolate | 0.4888 | Goldenberg et al.56 |
| Symptoms resolve: discharge | 0.7500 | Clinical expert |
| Symptoms persist: discharge | 0.2500 | Clinical expert |
| Isolate/not isolate, no pathogen, symptoms persist: retest, no pathogen, no treatment – discharge | 0.9000 | Clinical expert |
| Isolate/not isolate, no pathogen, symptoms persist: retest, no pathogen, isolate and not treat | 0.1000 | Clinical expert |
| Probability value | Conventional | xTAG | FilmArray | Source |
|---|---|---|---|---|
| Isolate, no pathogen, symptoms resolve: discharge | 0.9573 | 0.9271 | 0.9673 | Goldenberg et al.56 |
| Isolate, no pathogen, symptoms persist: retest | 0.0213 | 0.0729 | 0.0327 | Clinical expert |
| Isolate, no pathogen, deisolate: not treat | 0.0213 | 0.0000 | 0.0000 | Clinical expert |
| Not isolate, no pathogen, symptoms resolve: discharge | 0.8868 | 0.9271 | 0.9673 | Goldenberg et al.56 |
| Not isolate, no pathogen, symptoms resolve: retest | 0.1132 | 0.0729 | 0.0327 | Goldenberg et al.56 |
Resource use and costs
The resource use and costs included in the economic model were those directly incurred by the NHS. Costs for the conventional and the GPP tests, bed-days, cleaning, any other tests or investigations, blood tests, medications and rehydration costs were all included in the analysis. All costs are presented in 2014/15 prices and any costs not in this financial year were adjusted using the Hospital and Community Health Service pay and price index. 59
Test costs
The test costs for the conventional arm were based on the study by Goldenberg et al. 56 The tests included routine culture methods, which can potentially detect Campylobacter, Salmonella, Shigella and E. coli O157; light or fluorescent microscopy, which can detect Giardia and Cryptosporidium; enzyme immunoassays [glutamate dehydrogenase (GDH)] or PCR for detecting C. difficile; PCR tests for detecting norovirus; and the combined immunochromatographic tests for detecting adenovirus and rotavirus. All test costs included consumables, labour and overheads. The costs for conventional tests are shown in Table 29.
| Resource use | Test cost (£) | Reference |
|---|---|---|
| Conventional tests | ||
| Culture tests | 11.30 | Goldenberg et al.56 |
| Light or fluorescent microscopy | 10.53 | Goldenberg et al.56 |
| Enzyme immunoassay (GDH) or PCR test for screening | 26.05 | Goldenberg et al.56 |
| PCR test | 11.30 | Clinical experta |
| Combined immunochromatographic tests | 7.00 | Goldenberg et al.56 |
| Total cost for conventional tests | 66.18 | – |
| Total costs (£) | Test cost per sample (£) | |
| Integrated PCR tests | ||
| xTAG GPP | ||
| Capital cost (total) | 25,000 | 0.34 |
| Maintenance cost (per annum) | 4190 | 0.48 |
| Nucleic extraction system (total) | 35,000 | 4.00 |
| Thermal cycler (total) | 7500 | 0.19 |
| Test kit for 96 wells | 1920 | 20.00 |
| Consumables/disposables | – | 8.66 |
| Staff costs (per hour) | 15.55 | 3.43 |
| Total costs | 37.10 | |
| FilmArray | ||
| Capital cost (total) | 203,000 | 5.13 |
| Maintenance cost (per annum) | 3400 | 0.39 |
| GI panel kit | 85.00 | 85.00 |
| Cary–Blair medium | 1.00 | 1.00 |
| Staff costs (per hour) | 15.55 | 2.01 |
| Total costs | 93.53 | |
| Faecal Pathogens B | ||
| Capital cost (total) | 28,320 | 0.72 |
| Maintenance cost (per annum) | 3172 | 0.36 |
| Step 1: tubes (per sample) | 7.60 | 7.60 |
| Step 2: plates (per sample) | 0.80 | 0.80 |
| Master mix (per sample) | 1.50 | 1.50 |
| Nucleic acid extraction (per sample) | 3.50 | 3.50 |
| Consumables/disposables | 0.66 | 0.66 |
| Staff costs (per hour) | 15.55 | 2.73 |
| Total costs | 17.86 | |
The base model assumes an average throughput by GPP of 24 samples per day, as representative of typical use, although some larger facilities may have considerably higher average throughput. The configurations chosen allow flexibility to manage peak loads within facilities.
To calculate staff costs per hour associated with processing the samples, we have used the cost of a medical laboratory assistant using Agenda for Change pay rates for 2015 band 4 mid-point (point 14: £20,844 plus 17.5% on-costs). 60 This was based on working an average of 42 weeks per annum and 37.5 hours per week.
For xTAG, to run 24 samples would take an average of 5–6 hours. Using the information above, the staff cost associated with each individual sample would be £3.43. The MAGPIX system (Luminex, Toronto, ON, Canada) costs £25,000 and, taking into account a 10-year lifespan and using a 3.5% discount rate,61 assuming that the system is used every day for 24 samples, the cost would be £0.34 per sample. Assuming that the annual maintenance cost is £4190, the cost of the nucleic acid extraction system is approximately £35,000 and the Thermal cycler costs £7500 (assuming for the final two products a 5-year lifespan and using a 3.5% discount rate), the total cost per sample would be £4.67. We have also included the cost per well, which equates to £20, and the cost of consumables of £8.66. In total, the cost per sample for xTAG is £37.10 (see Table 29).
For FilmArray, to run 24 samples would take an average of 3 hours (using FilmArray 2.0 with eight modules, which is run three times). Using the information above, the staff cost associated with each individual sample would be £2.01. The capital cost of the system is £28,000, including the costs of a further seven modules, taking the total capital cost up to £203,000. Taking into account a 5-year lifespan and using a 3.5% discount rate,61 assuming that the system is used every day for 24 samples, the cost would be £5.13 per sample. The annual maintenance cost is £3400 for one module, the cost for one gastrointestinal panel kit is £85.00 and the Cary–Blair medium cost is approximately £1. Using these costs, the total cost per sample would be £93.53 (see Table 29).
For Faecal Pathogens B, to run 24 samples would take an average of 2–3 hours. Using the information above, the staff cost associated with each individual sample would be £2.73. The capital cost of the system is £28,320, and taking into account a 5-year lifespan and using a 3.5% discount rate,61 assuming that the system is used every day for 24 samples, the cost would be £0.72 per sample. The annual maintenance cost is £3172; the costs for tubes (step 1), plates (step 2), plus the master mix is, in total, £10.26 per sample; and consumables and extraction come to £4.16 per sample. Using these costs, the total cost per sample would be £17.86 (see Table 29).
After GPP testing, for confirmatory purposes we have included the cost of enzyme immunoassay (GDH) or PCR test for screening (£26.05) when C. difficile has been detected and the cost of routine culture (£11.30) when E. coli O157, Salmonella and Shigella have been detected.
We have not added in the cost of waste disposal associated with culture of faecal samples. The cost of disposing of samples can be substantial; however, as confirmed by our clinical experts, this cost is common to both arms.
In addition, we have not added any further costs for samples that are then sent to PHE for subsequent testing. As confirmed by our clinical experts, sending these samples will not affect subsequent management for these patients.
Unit costs for bed-days, cleaning, any other tests or investigations, blood tests, medications and rehydration costs are presented in Table 30.
| Resource use | Base-case value (£) | Reference |
|---|---|---|
| Bed-days | ||
| General adult ward (cost per day) | 497.39 | Department of Health 201562 |
| Adult isolation ward (cost per day) | 593.89 | Department of Health 2015,62 HAI Task Force63 |
| Cleaning costs | ||
| Daily cleaning | 10.11 | Allen et al.64 |
| Disposables and staff time | 8.88 | Allen et al.64 |
| Spot cleaning and changing | 34.80 | Allen et al.64 |
| Blood tests | ||
| Full blood count | 7.00 | Department of Health 201562 |
| Routine chemistry and C-reactive protein test | 4.00 | Department of Health 201562 |
| Other tests/investigations | ||
| Abdominal X-ray | 161.00 | Department of Health 201562 |
| Flexible sigmoidoscopy | 781.89 | Department of Health 201562 |
| Medications (cost per pack) | ||
| Vancomycin (oral) | 132.47 | BNF65 |
| Metronidazole (oral) | 1.21 | BNF65 |
| Fidaxomicin (Dificlir®, Astellas) (oral) | 1350.00 | BNF65 |
| Erythromycin (oral) | 1.61 | BNF65 |
| Ciprofloxacin (oral) | 0.98 | BNF65 |
| Amoxicillin (oral) | 1.61 | BNF65 |
| Rehydration methods | ||
| Dioralyte® (Sanofi-Aventis) (adult) (cost per day) | 2.25 | BNF65 |
| Intravenous fluids (adult) (cost per day) | 2.31 | NICE 200966 |
| Administration of rehydration methods | ||
| Oral | 3.58 | Allen et al.64 |
| Intravenous fluids | 14.33 | Allen et al.64 |
Number of bed-days before obtaining test results
It has been assumed that the results of conventional testing will come back in 3 days and the results of GPP tests will come back more quickly: xTAG in 1 day and FilmArray in half a day. The use of bed-days takes into account the time taken to collect a stool sample, to send the stool sample to the laboratory, to process the test and obtain the results, to communicate the results and to decide, depending on the test results, if any action needs to be taken.
Patients in whom a pathogen was not detected in the first instance, and who are retested because of persisting symptoms, are assumed to consume the same number of bed-days as above before the second set of test results is obtained.
Number of bed-days after obtaining test results
The total number of bed-days spent in hospital by patients with suspected gastroenteritis was based on a weighted average obtained from the Admitted Patient Care spreadsheet within the Hospital Episodes Statistics data for 2014/15. 67 The primary diagnosis Healthcare Resource Group code for intestinal infectious diseases was A00–A09. During the time period 1 April 2014 to 31 March 2015, there were 201,141 finished consultant episodes relating to intestinal infectious diseases, and mean length of stay was 4 days (this is based on all patients regardless of age or sex). Further breakdown of codes A00–A09 enabled a calculation of the weighted average number of bed-days for the different pathogens (Table 31).
| Pathogen | Length of stay (days) |
|---|---|
| Campylobacter | 6 |
| C. difficile | 19 |
| E. coli | 6 |
| Parasites | 5 |
| Salmonella | 6 |
| Shigella | 5 |
| Viruses | 2 |
| Other | 4 |
Once test results have been obtained, whether by conventional testing or by one of the GPP tests, it has been assumed that the number of bed-days listed above will apply for the total number of bed-days (before and after test results) for both isolated and non-isolated patients, depending on the care pathway before the patient is discharged. However, when a pathogen was missed at the first test but identified at the second test, the patient stay would be increased by the time delay to the second test.
A number of further assumptions were also made with regard to bed-days. First, we have assumed that the total minimum number of bed-days for both arms would be 3 days (even though the results may come back earlier for GPP testing). This was so that we could be consistent with what happens with conventional care patients, as there is currently no evidence that patients would be discharged earlier. Some patients are already discharged earlier than the 3 days (as this is an average), presumably because of symptom resolution. Thus, the base model assumes that patients are essentially treated symptomatically (with simple hygiene and rehydration advice) independent of the method of testing. Second, we have assumed that, if patients were in isolation, they might remain in isolation without moving to an open ward/bay before being discharged. Third, for patients in whom C. difficile was detected, the number of bed-days after the test result was split evenly between an isolation room and an open ward/bay.
Costs for bed-days were obtained from NHS Reference Costs62 (see Table 30). For stay on an adult general ward, this cost was a weighted average based on patients with gastrointestinal infections without interventions from the elective inpatient spreadsheet. For patients who stayed in isolated wards, an additional cost of £96.50 was added. This cost was based on the additional cost of patients with meticillin-resistant S. aureus infection staying in single beds in single room isolation rather than open wards. 63
Daily cleaning and spot cleaning
For each day the patient spent in hospital, a cost for daily cleaning was included. This cost included time for domestic cleaning staff, specialist cleaning agents and special cleaning equipment as reported in a previous Health Technology Assessment publication64 looking at diarrhoea in older people who are admitted to hospital. We also assumed that, for every second day spent in hospital, patients would have ‘an accident’ relating to their diarrhoea and spot cleaning would need to take place. This cost, again, included time for domestic cleaning staff, specialist cleaning agents and special cleaning equipment, as well as spot cleaning and changing of beds, laundry and disposables such as gloves and aprons. 64 Unit costs for cleaning are shown in Table 30.
Blood tests and other investigations
We assumed that a full blood count will be carried out in every patient on admission, and that approximately 30% of patients will undergo routine blood chemistry and a C-reactive protein test. Unit costs for blood tests were obtained from the NHS Reference Costs from the Direct Access Pathology Services spreadsheet62 (see Table 30).
We also assumed that 1% of patients in whom a pathogen was detected would undergo flexible sigmoidoscopy, as would 10% of patients in whom no pathogen was detected. We estimated that abdominal radiography would be performed in 10% of patients in whom C. difficile was detected and in all patients in whom no pathogen was detected. It was assumed that 5% of patients in whom conventional testing detected any other pathogen would undergo abdominal radiography, as would 2% of patients in the GPP test arm, reflecting the impact of earlier microbiological diagnosis on other testing. Unit costs for the named tests above were obtained from NHS Reference Costs62 (see Table 30).
Treatment
For most gastrointestinal infections, antimicrobial therapy is not recommended except in certain situations, for example C. difficile infection or immunocompromise. Unit costs for drugs were obtained from the British National Formulary (BNF)65 (see Table 30) and we have assumed that patients complete each course of tablets/capsules prescribed either at hospital or at home. For patients in whom C. difficile infection is identified, either metronidazole (non-severe C. difficile) or vancomycin (severe C. difficile) is administered. For patients with severe C. difficile infection and multiple comorbidities, oral fidaxomicin (Dificlir®, Astellas) may be considered. 68 A typical course of metronidazole or vancomycin lasts for 10–14 days (and fidaxomicin lasts 10 days), and only 47% of patients complete antibiotic therapy in hospital. 30 In a study of C. difficile-associated diarrhoea in hospitalised patients in Northern Ireland, 55.2% of patients received vancomycin and 44.8% received metronidazole. 69 According to our clinical experts, approximately 4.5% of patients in hospital would be treated with fidaxomicin. The following proportions were applied in our economic model: 52.95% vancomycin, 42.55% metronidazole and 4.5% fidaxomicin. For C. difficile patients, two packs of vancomycin or metronidazole are needed for a 10- to 14-day course.
Certain other pathogens are sometimes treated in the NHS; patients infected with these pathogens are included in the isolate and treat arm of the economic model. For example, patients infected with Campylobacter may receive erythromycin and patients infected with Salmonella, Shigella or E. coli (non-shiga toxin producing) may receive either ciprofloxacin or amoxicillin (on the advice of our clinical experts we have assumed that 75% of patients would receive ciprofloxacin and 25% of patients would receive amoxicillin); and for patients with Giardia they may receive metronidazole. Treatment is generally not recommended for shiga toxin-producing E. coli O157.
In patients in whom Cryptosporidium is detected (this is a very rare occurrence), two drugs [paromomycin or nitazoxanide (Alina®, Lupin Pharmaceuticals Inc., MD, USA)] can be used. However, there is no standard approved treatment in the UK for Cryptosporidium;65 hence, we have not included any costs in the economic model for this pathogen.
We have not included any cost of administering the drugs, as this is included in the nursing costs, which are included in the cost of the bed-days.
Rehydration methods
Oral rehydration therapy is given only to patients who are clinically dehydrated; however, most hospitalised patients will receive some form of rehydration. We have assumed that, for each day in hospital, 60% of patients would receive oral rehydration and 30% of patients would receive intravenous fluids. We have assumed that each patient receiving oral rehydration would receive 200 ml of Dioralyte® (Sanofi-Aventis) every 4 hours; one pack of Dioralyte contains six sachets and costs £2.25. 65 We have assumed that patients receiving intravenous fluids would receive 2 l of sodium chloride 0.9% per day, at a cost of £2.31. 66
The cost of nursing staff time to administer rehydration solutions must also be included. These costs were based on the earlier study by Allen et al. ,64 who calculated the time spent administering oral medication as 5 minutes and time spent administering intravenous medications as 20 minutes. The cost of a hospital-based nurse (band 5 with qualifications) was obtained from the Unit Costs of Health and Social Care. 59 The unit costs for rehydration methods and the administering costs are shown in Table 30.
Utilities
A literature search was undertaken to identify utility values for patients with infective gastroenteritis. Most published values are used to assess the cost-effectiveness of rotavirus vaccination for children aged ≤ 5 years. 70–75 Although some of these studies differentiate utility values by severity levels, for example mild or severe gastroenteritis or hospitalisation,70,71,76–78 none of the studies provided utility values for adults. The most useful paper was by Minor et al. ,79 who estimated health-care costs associated with food-borne illnesses caused by known viruses, bacteria and parasites in the USA. They presented the information for different pathogens in terms of weighted quality-adjusted life-days (QALDs) lost. The authors suggested that the use of QALDs lost is a more helpful unit of analysis for acute illnesses than QALYs lost. The lost QALDs were estimated by taking the average quality weight of a food-borne illness (measured by the EQ-5D index) and the average US quality weight. They then multiplied the percentage QALD loss by the duration of the condition. To convert to QALYs we divided the QALDs values by 365. Table 32 shows the QALY loss by the different pathogens in our model.
| Pathogen | Mean QALY loss | 95% CI for PSA | Distribution for PSA | Source |
|---|---|---|---|---|
| Bacteria | ||||
| C. difficile | 0.0137 | – | Clinical expert estimate (NM) | |
| Campylobacter | 0.0057 | 0.0035 to 0.0080 | Beta | Minor et al.79 |
| Salmonella | 0.0049 | 0.0041 to 0.0070 | Beta | Minor et al.79 |
| Shigella | 0.0050 | 0.0033 to 0.0069 | Beta | Minor et al.79 |
| ETEC | 0.0044 | 0.0032 to 0.0055 | Beta | Minor et al.79 |
| E. coli O157 | 0.0126 | 0.0083 to 0.0169 | Beta | Minor et al.79 |
| E. coli (STEC + non-STEC) | 0.0085 | 0.0057 to 0.0112 | Beta | Minor et al.79 |
| Parasites | ||||
| Giardia | 0.0123 | 0.0045 to 0.0201 | Beta | Minor et al.79 |
| Cryptosporidium | 0.0039 | 0.0010 to 0.0068 | Beta | Minor et al.79 |
| Parasites (all) | 0.0081 | 0.0027 to 0.0134 | Beta | Minor et al.79 |
| Viruses | ||||
| Adenovirus | 0.0007 | 0.0005 to 0.0010 | Beta | Minor et al.79 |
| Norovirus | 0.0008 | 0.0006 to 0.0010 | Beta | Minor et al.79 |
| Rotavirus | 0.0032 | 0.0023 to 0.0041 | Beta | Minor et al.79 |
| Viruses (all) | 0.0008 | 0.0011 to 0.0020 | Beta | Minor et al.79 |
| Other | ||||
| Other (unspecified pathogen) | 0.0007 | 0.0005 to 0.0010 | Beta | Minor et al.79 |
Changes made to base-case model
Various changes were made to the base-case model for hospitalised adult patients (model 1) with suspected gastroenteritis in order to carry out analyses for the different subgroups, as noted below.
Model 2: young children with suspected gastroenteritis
The model structure for young children in hospital matches the adult hospital model.
Prevalence and probabilities
Table 33 shows the prevalence of pathogens and conditional agreement probabilities when conventional testing is the benchmark for young children, which was estimated by our clinical experts using the hospital prevalence for adults as a base case. The overall pathogen detection rate was 24.2% for conventional testing, 21.4% for xTAG and 22.3% for FilmArray testing.
| Pathogen | Conventional testing | xTAG | FilmArray | ||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | True positive | False negative | True negative | False positive | ||
| Adenovirus | 0.03461 | 0.01931 | 0.01530 | 0.95574 | 0.00965 | 0.02499 | 0.00962 | 0.95863 | 0.00676 |
| C. difficile | 0.00115 | 0.00111 | 0.00005 | 0.96688 | 0.03196 | 0.00112 | 0.00004 | 0.96988 | 0.02897 |
| Campylobacter | 0.04268 | 0.04093 | 0.00175 | 0.92668 | 0.03063 | 0.04145 | 0.00124 | 0.94487 | 0.01245 |
| Cryptosporidium | 0.01384 | 0.01265 | 0.00119 | 0.97531 | 0.01085 | 0.01321 | 0.00064 | 0.98616 | 0.00000 |
| E. coli O157 | 0.00461 | 0.00429 | 0.00033 | 0.99041 | 0.00498 | 0.00440 | 0.00021 | 0.96353 | 0.03185 |
| ETEC | 0.01071 | 0.00995 | 0.00076 | 0.97742 | 0.01187 | 0.01071 | 0.00000 | 0.98336 | 0.00594 |
| Giardia | 0.00473 | 0.00473 | 0.00000 | 0.98432 | 0.01095 | 0.00473 | 0.00000 | 0.99527 | 0.00000 |
| Norovirus | 0.05999 | 0.05561 | 0.00000 | 0.91087 | 0.02914 | 0.05591 | 0.00408 | 0.93155 | 0.00846 |
| Rotavirus | 0.04614 | 0.04421 | 0.00194 | 0.94527 | 0.00858 | 0.04402 | 0.00212 | 0.94909 | 0.00477 |
| Salmonella | 0.01154 | 0.00944 | 0.00210 | 0.92916 | 0.05931 | 0.01154 | 0.00000 | 0.98649 | 0.00198 |
| Shigella | 0.01154 | 0.01141 | 0.00013 | 0.97364 | 0.01483 | 0.01106 | 0.00047 | 0.98846 | 0.00000 |
| Overall prevalence | 0.24155 | 0.21364 | 0.02792 | 0.53569 | 0.22276 | 0.22313 | 0.01842 | 0.65728 | 0.10116 |
The proportions of young children treated in each scenario are assumed to be the same as reported in Table 25. The overall conditional probabilities for patients in whom a pathogen is detected are shown in Table 34.
| Clinical pathway | Conventional testing as benchmark (first test) | xTAG | FilmArray testing | ||
|---|---|---|---|---|---|
| First test | Second test | First test | Second test | ||
| Total treated and isolated | 0.1479 | 0.1579 | 0.0717 | 0.1571 | 0.0370 |
| Not treated and isolated | 0.1737 | 0.1847 | 0.0897 | 0.1837 | 0.0521 |
| Not treated and not isolated | 0.5932 | 0.5732 | 0.7458 | 0.5749 | 0.8146 |
| No treatment: discharged | 0.0852 | 0.0842 | 0.0928 | 0.0843 | 0.0963 |
All common probabilities, as shown in Table 27, remain the same. The other probabilities for model 2 are shown in Table 35.
| Clinical pathway | Conventional | xTAG | FilmArray | Source |
|---|---|---|---|---|
| Isolate, no pathogen, symptoms resolve: discharge | 0.9573 | 0.9266 | 0.9667 | Goldenberg et al.56 |
| Isolate, no pathogen, symptoms persist: retest | 0.0213 | 0.0734 | 0.0333 | Clinical expert assumption |
| Isolate, no pathogen, deisolate: not treat | 0.0213 | 0.0000 | 0.0000 | Clinical expert assumption |
| Not isolate, no pathogen, symptoms resolve: discharge | 0.8868 | 0.9266 | 0.9667 | Goldenberg et al.56 |
| Not isolate, no pathogen, symptoms resolve: retest | 0.1132 | 0.0734 | 0.0333 | Goldenberg et al.56 |
Resource use and costs
Conventional test costs and GPP costs, as shown in Table 29, remain the same.
We assumed that the number of days spent in hospital is the same for children as for adults infected with the same pathogen (obtained from Hospital Episodes Statistics67), as hospital stay did not differ by age or sex. The costs for bed-days were obtained from NHS Reference Costs62 based on a weighted average of paediatric, infectious and non-infectious gastroenteritis stay, and are shown in Table 36.
| Resource use | Base-case value (£) | Reference |
|---|---|---|
| Bed-days | ||
| Paediatric general ward (cost per day) | 765.43 | Department of Health 201562 |
| Paediatric isolation ward (cost per day) | 861.93 | Department of Health 2015;62 HAI Task Force63 |
| Medications (cost per pack) | ||
| Vancomycin (intravenous) | 12.99 | BNFC80 |
| Metronidazole (oral) | 1.21 | BNFC80 |
| Fidaxomicin (oral) | 1350.00 | BNF65 |
| Erythromycin (oral) | 1.61 | BNFC80 |
| Ciprofloxacin (oral) | 1.48 | BNFC80 |
| Amoxicillin (oral) | 1.37 | BNFC80 |
| Rehydration methods | ||
| Dioralyte (child) (cost per day) | 2.25 | BNF65 |
| Intravenous fluids (child) (cost per day) | 2.31 | NICE 200966 |
| Administration of rehydration methods | ||
| Oral | 3.58 | Allen et al.64 |
| Intravenous fluids | 14.33 | Allen et al.64 |
We have assumed that cleaning took place each day, as in the base model, and that spot cleaning took place every day, rather than every second day as in the base model.
We have assumed that the same percentage of patients as in the base model would undergo blood tests. We also assumed that 3% of patients in whom no pathogen was detected would undergo flexible sigmoidoscopy (patients in whom a pathogen was detected would not undergo flexible sigmoidoscopy). We estimated that 5% of patients in whom C. difficile was detected and all patients in whom no pathogen was detected would undergo abdominal radiography. It was assumed that 3% of patients in the conventional arm and 2% of patients in the GPP arm in whom any other pathogen was detected would undergo abdominal radiography.
The proportions of patients with C. difficile, Campylobacter, Salmonella, Shigella, E. coli (non-shiga toxin producing) or Giardia infection who received different treatments were assumed to be the same as in the base model. The costs of the drugs were taken from the BNF for Children80 (see Table 36). Patients infected with C. difficile require three vials of intravenous injections of vancomycin or two packs of metronidazole for the 10- to 14-day course.
We have assumed that, for each day in hospital, 75% of young children would receive oral rehydration and 20% of young children would receive intravenous fluids. The unit costs for the rehydration methods were obtained from the BNF for Children80 and are shown in Table 30 along with the administration costs.
Model 3: people in the community with suspected gastroenteritis
We have assumed that the model structure for people in the community is simplified (Figure 13).
FIGURE 13.
Overall model structure for patients with suspected gastroenteritis in the community.
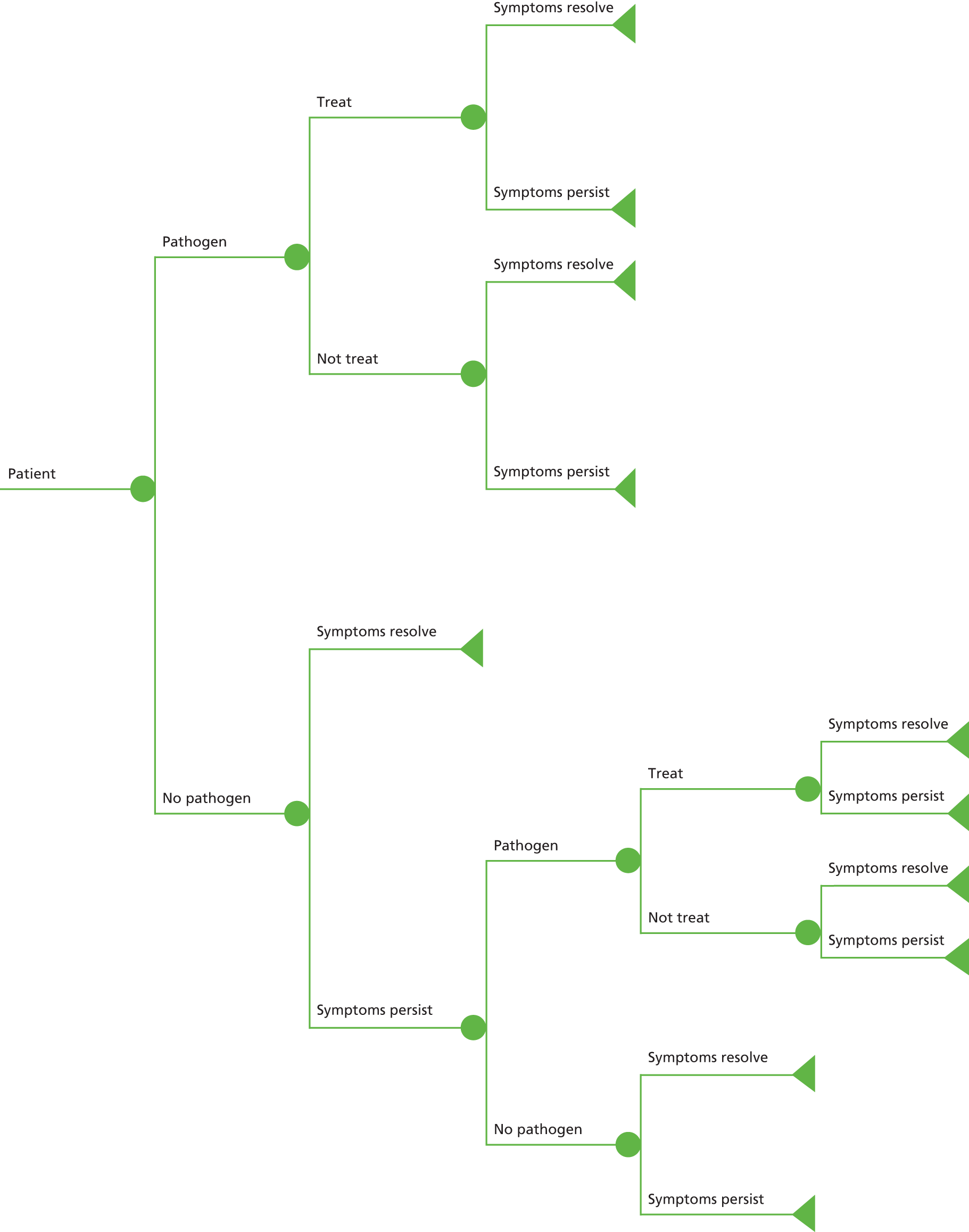
Prevalence and probabilities
Table 37 shows the prevalence of pathogens and conditional agreement probabilities when conventional testing is the benchmark for the community. Prevalences for conventional testing were obtained from the Second Study of Infectious Intestinal Disease in the Community. 1 The overall pathogen detection rate was 34.9% for conventional testing, 31.6% for xTAG and 32.5% for FilmArray testing. Probabilities for positive and negative agreements for each pathogen were drawn from the clinical effectiveness review.
| Pathogen | Conventional testing | xTAG | FilmArray | ||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | True positive | False negative | True negative | False positive | ||
| Adenovirus | 0.03432 | 0.01915 | 0.01517 | 0.95602 | 0.00966 | 0.02478 | 0.00954 | 0.95892 | 0.00676 |
| C. difficile | 0.00136 | 0.00130 | 0.00006 | 0.96668 | 0.03196 | 0.00131 | 0.00004 | 0.96968 | 0.02896 |
| Campylobacter | 0.07968 | 0.07641 | 0.00327 | 0.89087 | 0.02945 | 0.07737 | 0.00231 | 0.90836 | 0.01196 |
| Cryptosporidium | 0.01043 | 0.00953 | 0.00090 | 0.97869 | 0.01089 | 0.00995 | 0.00048 | 0.98957 | 0.00000 |
| E. coli O157 | 0.00115 | 0.00107 | 0.00008 | 0.99385 | 0.00499 | 0.00110 | 0.00005 | 0.96688 | 0.03196 |
| ETEC | 0.01071 | 0.00995 | 0.00076 | 0.97742 | 0.01187 | 0.01071 | 0.00000 | 0.98336 | 0.00594 |
| Giardia | 0.00695 | 0.00695 | 0.00000 | 0.98212 | 0.01092 | 0.00695 | 0.00000 | 0.99305 | 0.00000 |
| Norovirus | 0.12357 | 0.11455 | 0.00000 | 0.84926 | 0.02717 | 0.11517 | 0.00840 | 0.86854 | 0.00789 |
| Rotavirus | 0.07323 | 0.07015 | 0.00308 | 0.91843 | 0.00834 | 0.06986 | 0.00337 | 0.92214 | 0.00463 |
| Salmonella | 0.00808 | 0.00661 | 0.00147 | 0.93240 | 0.05952 | 0.00808 | 0.00000 | 0.98993 | 0.00198 |
| Shigella | 0.00000 | 0.00000 | 0.00000 | 0.98500 | 0.01500 | 0.00000 | 0.00000 | 1.00000 | 0.00000 |
| Overall prevalence | 0.34948 | 0.31568 | 0.03380 | 0.43075 | 0.21976 | 0.32528 | 0.02420 | 0.55043 | 0.10009 |
We have assumed that the proportion of patients treated and not treated, as shown in Table 25, would remain the same for the community. The overall conditional probabilities for patients in whom a pathogen is detected are shown in Table 38.
| Clinical pathway | Conventional testing as benchmark (first test) | xTAG | Film Array | ||
|---|---|---|---|---|---|
| First test | Second test | First test | Second test | ||
| Total treated | 0.1266 | 0.1332 | 0.0651 | 0.1334 | 0.0352 |
| Total not treated | 0.8734 | 0.8668 | 0.9349 | 0.8666 | 0.9648 |
Following advice from our clinical experts, we have assumed that the probability that a patient’s symptoms resolve naturally is 0.75 and the probability that a patient’s symptoms persist is 0.25. Other probabilities used in the model are shown in Table 39.
Resource use and costs
Conventional test costs and GPP test costs, as shown in Table 29, remain unchanged. The community model does not include costs incurred by the NHS, such as bed-days, cleaning, tests or investigations, or blood tests.
For both arms, each patient is assumed to visit a GP before the testing process takes place. As this cost is common to both arms, it has not been included. However, in the case of patients who need treatment, the model includes the cost of 5 minutes’ GP time to review results and prescribe treatment together with the cost of 10 minutes’ nurse time to contact the patient to explain the test result and ask the patient to come and collect the prescription. GP and nurse costs were obtained from the Unit Costs of Health and Social Care for 2015. 59
The proportions of patients with C. difficile, Campylobacter, Salmonella, Shigella, E. coli (non-shiga toxin producing) or Giardia infection who received different treatments were assumed to be the same as in the base model. However, on the advice of our clinical experts, we have assumed that 33.3% of C. difficile patients would receive vancomycin and 66.6% would receive metronidazole. The costs of the drugs were taken from the BNF65 (see Table 30).
We have assumed that 10% of patients in the community would receive oral rehydration. The unit costs for the different rehydration methods were obtained from the BNF65 (no administration costs have been included) (see Table 30).
Utilities
Utilities for the different pathogens remain the same as the base model.
Model 4: people who are immunocompromised with suspected gastroenteritis
We have assumed that the model structure for people who are immunocompromised in hospital is the same as the non-immunocompromised adult hospital model.
Prevalence and probabilities
Table 40 shows the prevalence of pathogens and conditional agreement probabilities when conventional testing is the benchmark for immunocompromised patients, which was estimated by our clinical experts using the hospital prevalence for adults as a base case. The overall pathogen detection rate was 31.1% for conventional testing, 28.7% for xTAG and 29.4% for FilmArray testing.
| Pathogen | Conventional testing | xTAG | FilmArray | ||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | True positive | False negative | True negative | False positive | ||
| Adenovirus | 0.01800 | 0.01004 | 0.00796 | 0.97218 | 0.00982 | 0.01300 | 0.00500 | 0.97513 | 0.00687 |
| C. difficile | 0.13500 | 0.12947 | 0.00554 | 0.83732 | 0.02768 | 0.13055 | 0.00446 | 0.83992 | 0.02509 |
| Campylobacter | 0.01800 | 0.01726 | 0.00074 | 0.95058 | 0.03142 | 0.01748 | 0.00052 | 0.96923 | 0.01277 |
| Cryptosporidium | 0.01500 | 0.01371 | 0.00129 | 0.97417 | 0.01084 | 0.01431 | 0.00069 | 0.98500 | 0.00000 |
| E. coli O157 | 0.00600 | 0.00557 | 0.00043 | 0.98903 | 0.00497 | 0.00572 | 0.00028 | 0.96219 | 0.03181 |
| ETEC | 0.01071 | 0.00995 | 0.00076 | 0.97742 | 0.01187 | 0.01071 | 0.00000 | 0.98336 | 0.00594 |
| Giardia | 0.00300 | 0.00300 | 0.00000 | 0.98603 | 0.01097 | 0.00300 | 0.00000 | 0.99700 | 0.00000 |
| Norovirus | 0.06900 | 0.06396 | 0.00504 | 0.90214 | 0.02886 | 0.06431 | 0.00469 | 0.92262 | 0.00838 |
| Rotavirus | 0.00300 | 0.00287 | 0.00013 | 0.98803 | 0.00897 | 0.00286 | 0.00014 | 0.99202 | 0.00499 |
| Salmonella | 0.00900 | 0.00736 | 0.00164 | 0.93154 | 0.05946 | 0.00900 | 0.00000 | 0.98902 | 0.00198 |
| Shigella | 0.02400 | 0.02374 | 0.00026 | 0.96136 | 0.01464 | 0.02302 | 0.00098 | 0.97600 | 0.00000 |
| Overall prevalence | 0.31071 | 0.28694 | 0.02377 | 0.46979 | 0.21950 | 0.29395 | 0.01676 | 0.59148 | 0.09781 |
Our clinical experts estimated that the proportions of patients who would be treated and not treated are as shown in Table 41.
| Pathogen | Treated and isolated | Not treated and isolated | Not treated and not isolated |
|---|---|---|---|
| Adenovirus | – | 1.00 | 0.00 |
| C. difficile | 0.95 | 0.05 | 0.00 |
| Campylobacter | 0.70 | 0.30 | 0.00 |
| Cryptosporidium | 0.00 | 1.00 | 0.00 |
| E. coli O157 | 0.00 | 1.00 | 0.00 |
| ETEC | 0.80 | 0.20 | 0.00 |
| Giardia | 0.95 | 0.025 | 0.025 |
| Norovirus | 0.00 | 1.00 | 0.00 |
| Rotavirus | 0.00 | 1.00 | 0.00 |
| Salmonella | 0.80 | 0.20 | 0.00 |
| Shigella | 0.80 | 0.20 | 0.00 |
The overall conditional probabilities for patients for whom a pathogen is detected are shown in Table 42.
| Clinical pathway | Conventional testing as benchmark (first test) | xTAG | Film Array | ||
|---|---|---|---|---|---|
| First test | Second test | First test | Second test | ||
| Total treated and isolated | 0.5750 | 0.5951 | 0.3325 | 0.5895 | 0.3213 |
| Not treated and isolated | 0.0609 | 0.0628 | 0.0390 | 0.0627 | 0.0309 |
| Not treated and not isolated | 0.3215 | 0.3016 | 0.5617 | 0.3068 | 0.5799 |
| No treatment: discharged | 0.0425 | 0.0405 | 0.0667 | 0.0410 | 0.0679 |
All common probabilities, as shown in Table 27, remain the same. The other probabilities for model 4 are shown in Table 43.
| Clinical pathway | Conventional | xTAG | FilmArray | Source |
|---|---|---|---|---|
| Isolate, no pathogen, symptoms resolve: discharge | 0.9573 | 0.9204 | 0.9645 | Goldenberg et al.56 |
| Isolate, no pathogen, symptoms persist: retest | 0.0213 | 0.0796 | 0.0355 | Clinical expert assumption |
| Isolate, no pathogen, deisolate: not treat | 0.0213 | 0.0000 | 0.0000 | Clinical expert assumption |
| Not isolate, no pathogen, symptoms resolve: discharge | 0.8868 | 0.9204 | 0.9645 | Goldenberg et al.56 |
| Not isolate, no pathogen, symptoms resolve: retest | 0.1132 | 0.0796 | 0.0355 | Goldenberg et al.56 |
Resource use and costs
Conventional test costs and GPP costs, as shown in Table 29, remain the same. We have assumed that only costs relating to the episode of diarrhoea have been included in this analysis; any additional costs associated with the hospital stay have not been included.
We assumed that the number of days spent in hospital is the same for immunocompromised as for non-immunocompromised adults infected with the same pathogen (obtained from Hospital Episodes Statistics67), as hospital stay did not differ by age or sex. The costs for bed-days were obtained from NHS Reference Costs62 and remain the same as model 1.
We have assumed that cleaning will take place each day and that spot cleaning will take place every second day, as in the base model.
We have assumed that every patient will undergo two full blood counts (one on admission and one after). The same percentage (30%) of patients as in the base model will undergo routine blood chemistry and a C-reactive protein test.
We also assumed that 20% of patients in whom a pathogen was not detected and only 1% of patients in whom a pathogen was detected would undergo flexible sigmoidoscopy test (the latter value remains the same as in the base model). We estimated that 10% of patients in whom C. difficile was detected and 20% of patients in whom no pathogen was detected would undergo abdominal radiography. It was assumed that 5% of patients in the conventional arm and 2% of patients in the GPP arm in whom any other pathogen was detected would undergo abdominal radiography.
The proportions of patients with C. difficile, Campylobacter, Salmonella, Shigella, E. coli (non-shiga toxin producing) or Giardia infection who received different treatments were assumed to be the same as in the base model. The costs of the drugs were taken from the BNF65 (see Table 30).
We have assumed that 60% of patients will receive oral rehydration and 30% will receive intravenous fluids, as in the base model.
Utilities
Utilities for the different pathogens remained the same as in the base model.
Model 5: people with a recent history of foreign travel with suspected gastroenteritis
We have assumed that the model structure for people with a recent history of foreign travel with suspected gastroenteritis is the same as for people in the community (see Figure 13).
Prevalence and probabilities
Table 44 shows the prevalence of pathogens and conditional agreement probabilities when conventional testing is the benchmark for recent travellers and prevalences were estimated by our clinical experts using the community prevalence for adults as a base case. The overall pathogen detection rate was 31.1% for conventional testing, 28.0% for xTAG and 29.2% for FilmArray testing.
| Pathogen | Conventional testing | xTAG | FilmArray | ||||||
|---|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | True positive | False negative | True negative | False positive | ||
| Adenovirus | 0.02700 | 0.01507 | 0.01193 | 0.96327 | 0.00973 | 0.01949 | 0.00751 | 0.96619 | 0.00681 |
| C. difficile | 0.00150 | 0.00144 | 0.00006 | 0.96655 | 0.03195 | 0.00145 | 0.00005 | 0.96954 | 0.02896 |
| Campylobacter | 0.10500 | 0.10070 | 0.00431 | 0.86636 | 0.02864 | 0.10196 | 0.00305 | 0.88337 | 0.01164 |
| Cryptosporidium | 0.01800 | 0.01645 | 0.00155 | 0.97120 | 0.01080 | 0.01717 | 0.00083 | 0.98200 | 0.00000 |
| E. coli O157 | 0.00300 | 0.00279 | 0.00021 | 0.99202 | 0.00499 | 0.00286 | 0.00014 | 0.96510 | 0.03190 |
| ETEC | 0.01200 | 0.01200 | 0.00000 | 0.97713 | 0.01087 | 0.01071 | 0.00000 | 0.98336 | 0.00594 |
| Giardia | 0.08550 | 0.07926 | 0.00624 | 0.88615 | 0.02835 | 0.01200 | 0.00000 | 0.98800 | 0.00000 |
| Norovirus | 0.00300 | 0.00287 | 0.00013 | 0.98803 | 0.00897 | 0.07969 | 0.00581 | 0.90627 | 0.00823 |
| Rotavirus | 0.02700 | 0.02209 | 0.00491 | 0.91462 | 0.05838 | 0.00286 | 0.00014 | 0.99202 | 0.00499 |
| Salmonella | 0.01800 | 0.01780 | 0.00020 | 0.96727 | 0.01473 | 0.02700 | 0.00000 | 0.97105 | 0.00195 |
| Shigella | 0.01071 | 0.00995 | 0.00076 | 0.97742 | 0.01187 | 0.01726 | 0.00074 | 0.98200 | 0.00000 |
| Overall prevalence | 0.31071 | 0.28041 | 0.03030 | 0.47001 | 0.21928 | 0.29245 | 0.01826 | 0.58889 | 0.10040 |
We have assumed that the proportion of patients treated and not treated, as shown in Table 25, will remain the same in the case of recent travellers. The overall conditional probabilities for patients in whom a pathogen is detected are shown in Table 45.
| Clinical pathway | Conventional testing as benchmark (first test) | xTAG | Film Array | ||
|---|---|---|---|---|---|
| First test | Second test | First test | Second test | ||
| Total treated | 0.2402 | 0.2512 | 0.1389 | 0.2503 | 0.0791 |
| Total not treated | 0.7598 | 0.7488 | 0.8611 | 0.7497 | 0.9209 |
Following advice from our clinical experts, we have assumed that the probability that a patient’s symptoms resolve naturally is 0.75 and the probability that symptoms persist is 0.25. Other probabilities used in the model are shown in Table 46.
Resource use and costs
Conventional test costs and GPP test costs, as shown in Table 29, remain the same. The recent travellers model does not include costs incurred by the NHS such as bed-days, cleaning, tests or investigations, or blood tests.
As with the community care model, for both testing methods each patient is assumed to visit a GP before the testing process takes place, at no net cost. Again, in the case of patients who need treatment, the model includes the cost of 5 minutes’ GP time to review results and prescribe treatment together with the cost of 10 minutes’ nurse time to contact the patient to explain the test result and ask the patient to come and collect the prescription. GP and nurse costs were obtained from the Unit Costs of Health and Social Care for 2015. 59
The proportions of patients with C. difficile, Campylobacter, Salmonella, Shigella, E. coli (non-shiga toxin producing) or Giardia infection who received different treatments were assumed to be the same as in the base model. The costs for the drugs were taken from the BNF65 (see Table 30).
We have assumed that 10% of recent travellers will receive oral rehydration. The unit costs for the rehydration methods were obtained from the BNF65 (no administration costs have been included) (see Table 30).
Utilities
Utilities for the different pathogens remained the same as in the base model.
Measuring cost-effectiveness
The models were constructed to compare the cost-effectiveness of integrated multiplex PCR tests and conventional testing in patients with suspected infectious gastroenteritis. The models estimated the mean total costs and total QALY losses avoided for each GPP test compared with conventional care for the initial index episode. The analysis was undertaken from a NHS and Personal Social Services perspective, and outcomes are reported as incremental cost-effectiveness ratios (ICERs), expressed in terms of cost per QALY gained.
To assess the uncertainty in parameter inputs used in the decision tree model and to check if the results obtained are robust, we present both deterministic and probabilistic results. For the deterministic analysis, we identify the key factors driving the cost-effectiveness. For the probabilistic sensitivity analysis, to reflect the amount and pattern of variation, the analysis was conducted by running the model with 1000 simulations and calculating the incremental costs and effects for each simulation. In each simulation, the value for each parameter was sampled from its probability distribution. We used gamma distributions for costs (except for test costs that we assumed were fixed) and beta distributions for utility values and probabilities. 81 For the costs and probabilities in the model, we assumed that the standard error was 0.1 of the mean value82,83 in order to calculate the alpha and beta values that are required for the probabilistic sensitivity analysis. These bootstrapped simulations were plotted on a cost-effectiveness plane and were used to present results as cost-effectiveness acceptability curves (CEACs), which indicated the probability of a GPP test being cost-effective compared with conventional testing using a willingness-to-pay threshold varying from £0 to £60,000.
Results of the cost-effectiveness modelling
The cost-effectiveness deterministic and probabilistic results for the GPP tests compared with conventional testing are presented in this section.
Model 1 results: hospitalised adult patients with suspected gastroenteritis
Table 47 shows the results when conventional testing is the benchmark for the adult hospital model (model 1). The deterministic model results show that both xTAG and FilmArray cost marginally less than conventional testing (£69 and £61 cheaper per patient, respectively) with marginally fewer QALYs lost (or more QALYs gained), and hence dominate conventional testing. The probabilistic findings are very similar, but show huge uncertainty in the changes in cost and QALYs gained, shown by the 95% CIs.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Net monetary benefit (£20,000/QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 3157 | 0.00309 | – | – | – | – |
| xTAG | 3089 | 0.00292 | –69 | 0.00018 | Dominant | 72 |
| Conventional | 3157 | 0.00309 | – | – | – | – |
| FilmArray | 3096 | 0.00297 | –61 | 0.00013 | Dominant | 64 |
| Probabilistic results | ||||||
| Conventional | 3159 | 0.00309 | – | – | – | – |
| xTAG | 3095 | 0.00293 | –63 (–929 to 791) | 0.00016 (–0.00071 to 0.00106) | Dominant | 67 (–802 to 947) |
| Conventional | 3159 | 0.00309 | – | – | – | – |
| FilmArray | 3107 | 0.00298 | –52 (–883 to 793) | 0.00012 (–0.00079 to 0.00104) | Dominant | 54 (–806 to 909) |
Figures 14 and 15 show the cost-effectiveness planes when conventional testing is the benchmark for model 1. In both figures, the bootstrap estimates are located in all four quadrants, with costs and outcomes negatively correlated. Figures 16 and 17 show the CEACs when conventional testing provides the benchmark. In both figures, GPP testing is of uncertain cost-effectiveness (approximately 57% probability for xTAG and 54% probability for FilmArray) at a willingness-to-pay threshold of £20,000 per QALY. The figures also highlight that willingness to pay has no effect and as a general rule, when estimating cost-effectiveness ratios with incremental QALYs close to zero, the ICER sign may switch.
FIGURE 14.
Cost-effectiveness plane: xTAG vs. conventional testing (benchmark) (model 1).

FIGURE 15.
Cost-effectiveness plane: FilmArray vs. conventional testing (benchmark) (model 1).
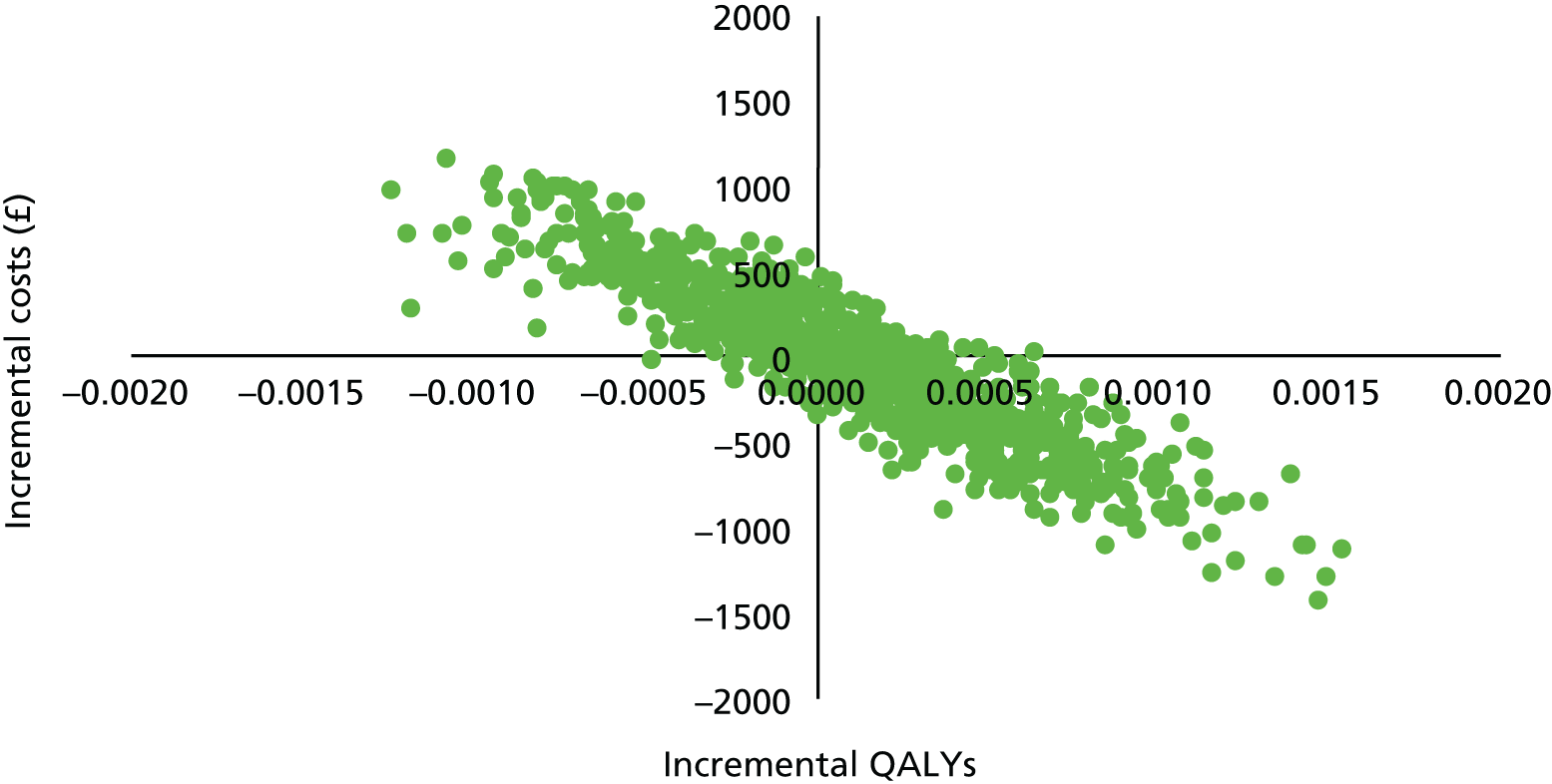
FIGURE 16.
Cost-effectiveness acceptability curve: xTAG vs. conventional testing (benchmark) (model 1).

FIGURE 17.
Cost-effectiveness acceptability curve: FilmArray vs. conventional testing (benchmark) (model 1).
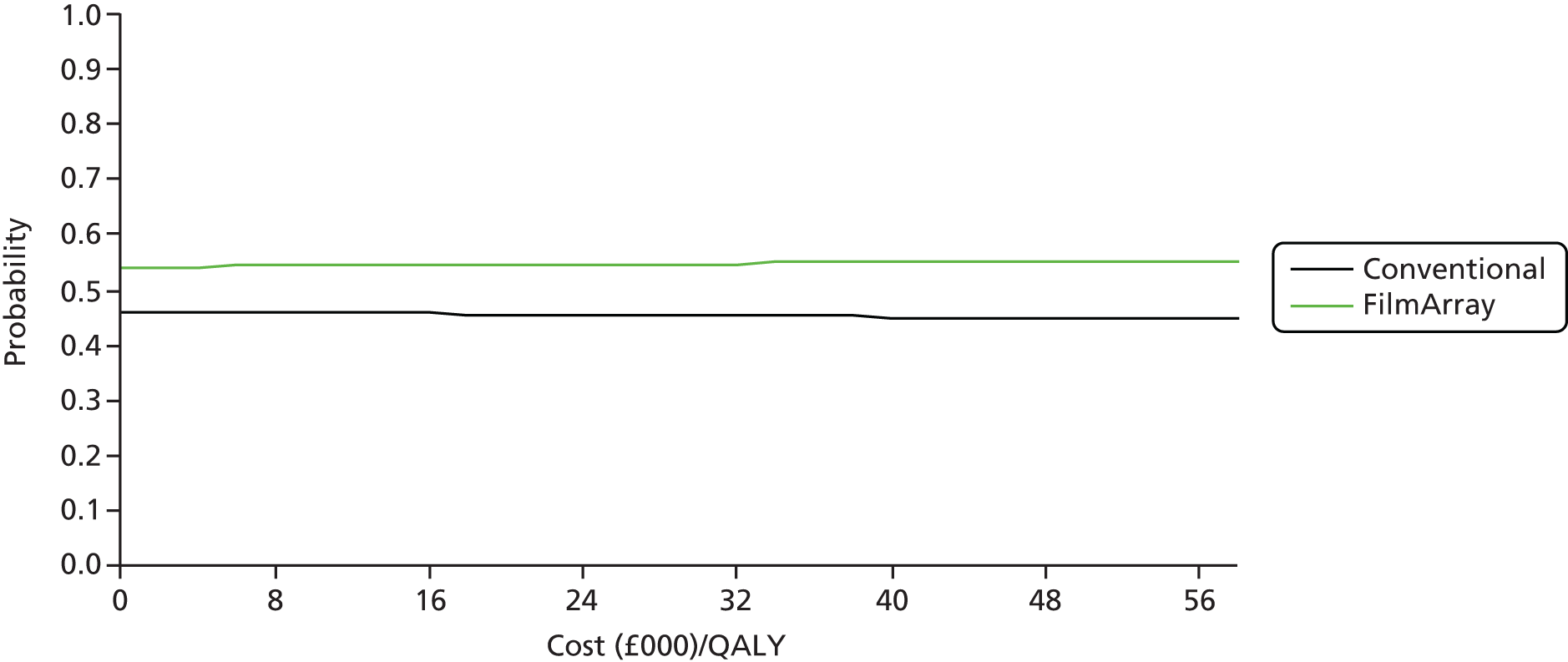
In the base-case analysis, we assume that the minimum total bed-days required with both methods of testing is 3 (even though the results would expect to be returned more quickly for GPP testing). In a scenario analysis we have tested the assumption that patients undergoing GPP testing leave hospital 1 day earlier than patients receiving conventional care, rather than after the same number of days (as in the base-case analysis). The results of this analysis are shown in Figures 18–20.
FIGURE 18.
Incremental mean costs (probabilistic results): model 1. (a) xTAG vs. conventional testing; and (b) FilmArray vs. conventional testing.

FIGURE 19.
Incremental mean QALYs (probabilistic results): model 1. (a) xTAG vs. conventional testing; and (b) FilmArray vs. conventional testing.
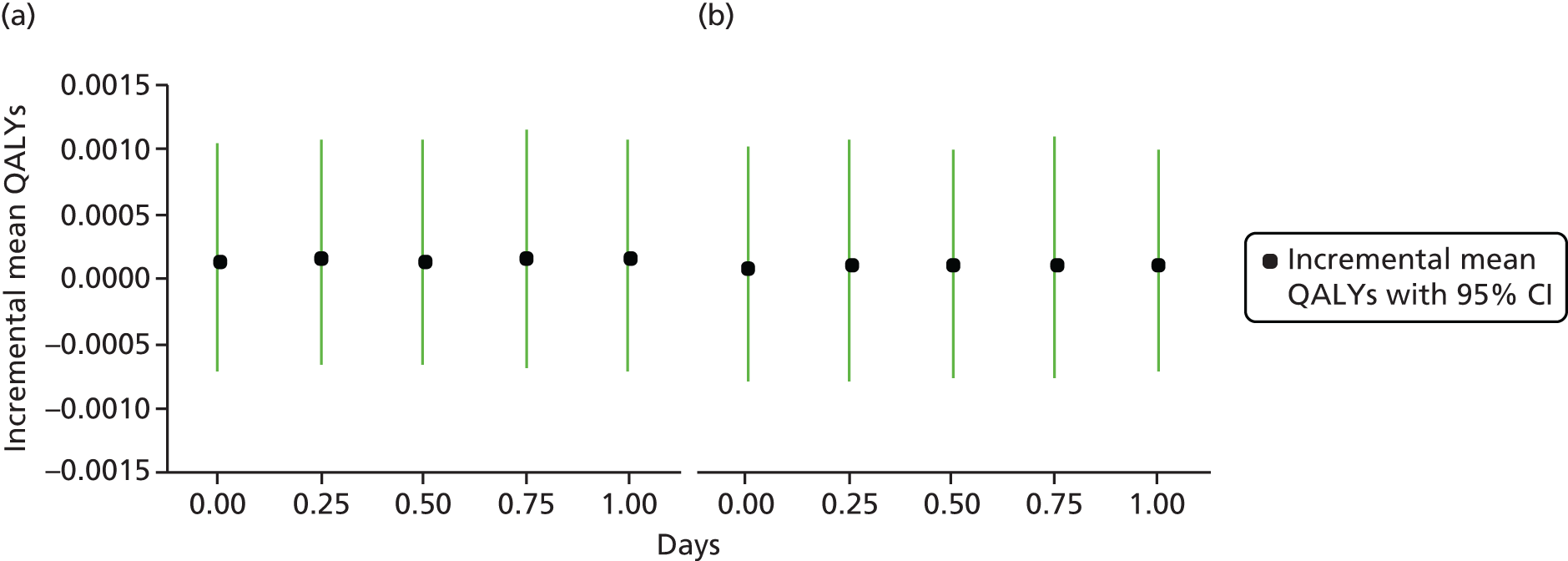
FIGURE 20.
Net monetary benefit (willingness-to-pay threshold = £20,000/QALY) (probabilistic results): model 1. (a) xTAG vs. conventional testing; and (b) FilmArray vs. conventional testing.
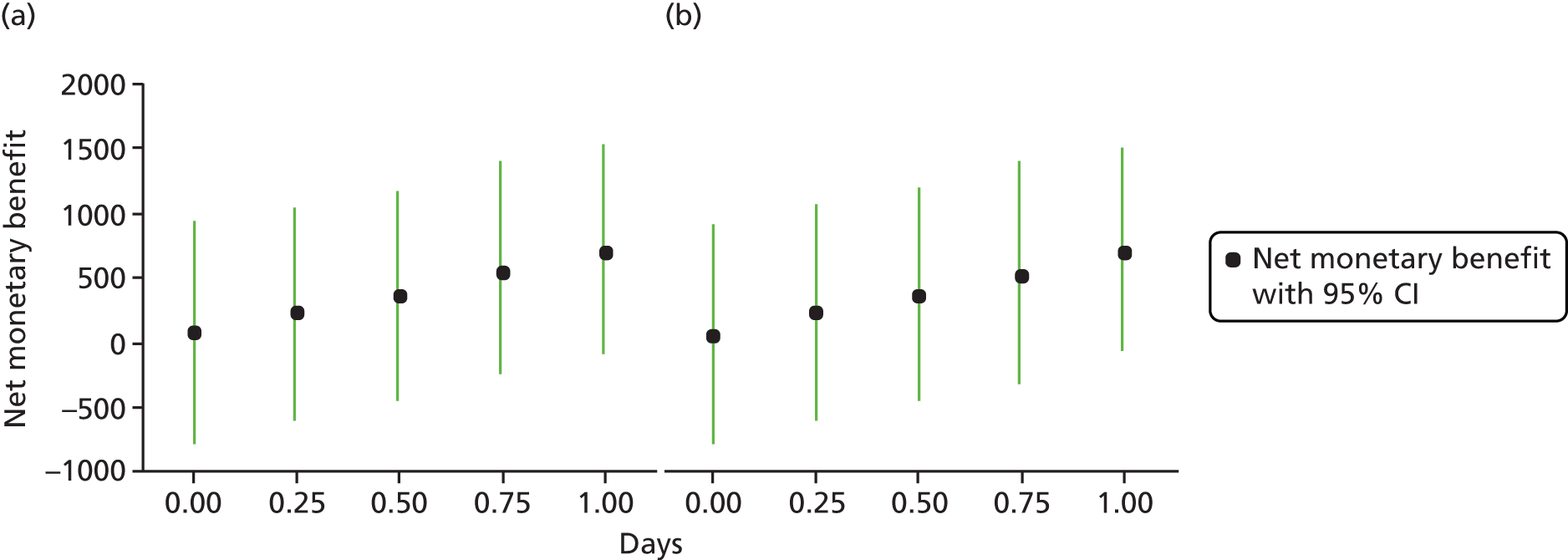
The incremental per-patient mean costs are £63 less for xTAG and £52 less for FilmArray testing than for conventional care when there is no difference in the overall length of stay (i.e. 3 days; see Figure 18). If patients are discharged, on average, 1 day earlier, the cost saving per patient rises to £679 for xTAG and to £677 for FilmArray.
Figure 19 shows the incremental mean QALYs gained when changing the length of stay for GPP testing. However, in the model no disutility is attached to being in hospital (the value to patients is unknown); thus, earlier discharge might result in a reduction of disutility further enhancing cost-effectiveness, if reductions in length of stay do occur. Figure 20 shows the net monetary benefit (which brings costs and QALYs with willingness to pay). With earlier discharge, the net monetary benefit of GPP testing increases, but the 95% CI does not exceed zero at a threshold of £20,000 per QALY.
Figures 21 and 22 show the one-way sensitivity analysis (tornado diagrams); a number of key variables were varied while holding others constant to understand what was influencing the net monetary benefit of GPP tests.
FIGURE 21.
Tornado diagram for net monetary benefit of conventional testing (benchmark) vs. xTAG: model 1. i.v., intravenous; WTP, willingness to pay.

FIGURE 22.
Tornado diagram for net monetary benefit of conventional testing (benchmark) vs. FilmArray: model 1. i.v., intravenous; WTP, willingness to pay.

Figure 21 is centred on the net monetary benefit ratio of £72 for xTAG compared with conventional testing. The most important variable is the number of bed-days, when the speed of reporting of findings is allowed to influence length of stay in patients without a pathogen. A variation of 50% in the number of bed-days for the GPP tests has a dramatic effect on net monetary benefit. For xTAG, by increasing the number of bed-days by 50% from 3 to 4.5 days the net monetary benefit ratio falls to –£575, whereas reducing the number of bed-days by 50% from 3 to 1.5 days increases the net monetary benefit ratio to £719. The potential value of earlier detection and discharge (if possible) would affect the majority of patients, and cost savings would dwarf any other influence in the model, such as assumptions about time in isolation or changes to isolation room use. The next most important variable is the false-positive rate, as an increase in the false-positive rate in the model gives rise to unnecessary further testing and care. Other influences include the costs of the tests themselves, the costs of bed-days and the probability of initial isolation. Results are similar for FilmArray and conventional testing (see Figure 22).
Model 2 results: young children with suspected gastroenteritis
Table 48 shows the results when conventional testing is the benchmark for the hospital admission model in young children (model 2). The deterministic results show that both xTAG and FilmArray are marginally less costly and marginally more effective (more QALYs gained), but again the probabilistic findings show that costs and QALY savings are highly uncertain.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Net monetary benefit (£20,000 per QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 3300 | 0.00267 | – | – | – | – |
| xTAG | 3237 | 0.00242 | –63 | 0.00025 | Dominant | 68 |
| Conventional | 3300 | 0.00267 | – | – | – | – |
| FilmArray | 3221 | 0.00249 | –79 | 0.00017 | Dominant | 82 |
| Probabilistic results | ||||||
| Conventional | 3313 | 0.00263 | – | – | – | – |
| xTAG | 3240 | 0.00239 | –73 (–955 to 849) | 0.00024 (–0.00049 to 0.00117) | Dominant | 78 (–856 to 977) |
| Conventional | 3313 | 0.00263 | – | – | – | – |
| FilmArray | 3231 | 0.00246 | –83 (–1094 to 858) | 0.00017 (–0.00061 to 0.00114) | Dominant | 86 (–869 to 1114) |
Figures 23 and 24 show the cost-effectiveness planes when conventional testing is the benchmark for model 2. Like model 1, the bootstrap estimates are located in all four quadrants, with costs and outcomes negatively correlated. Figures 25 and 26 show the CEACs when conventional testing provides the benchmark. In both figures, GPP testing is of uncertain cost-effectiveness (approximately 58% probability for xTAG and 57% probability for FilmArray) at a willingness-to-pay threshold of £20,000 per QALY. As mentioned in Model 1 results: hospitalised adult patients with suspected gastroenteritis, the figures highlight that willingness to pay has no effect and, as a general rule, when estimating cost-effectiveness ratios with incremental QALYs close to zero, the ICER sign may switch.
FIGURE 23.
Cost-effectiveness plane: xTAG vs. conventional testing (benchmark) (model 2).
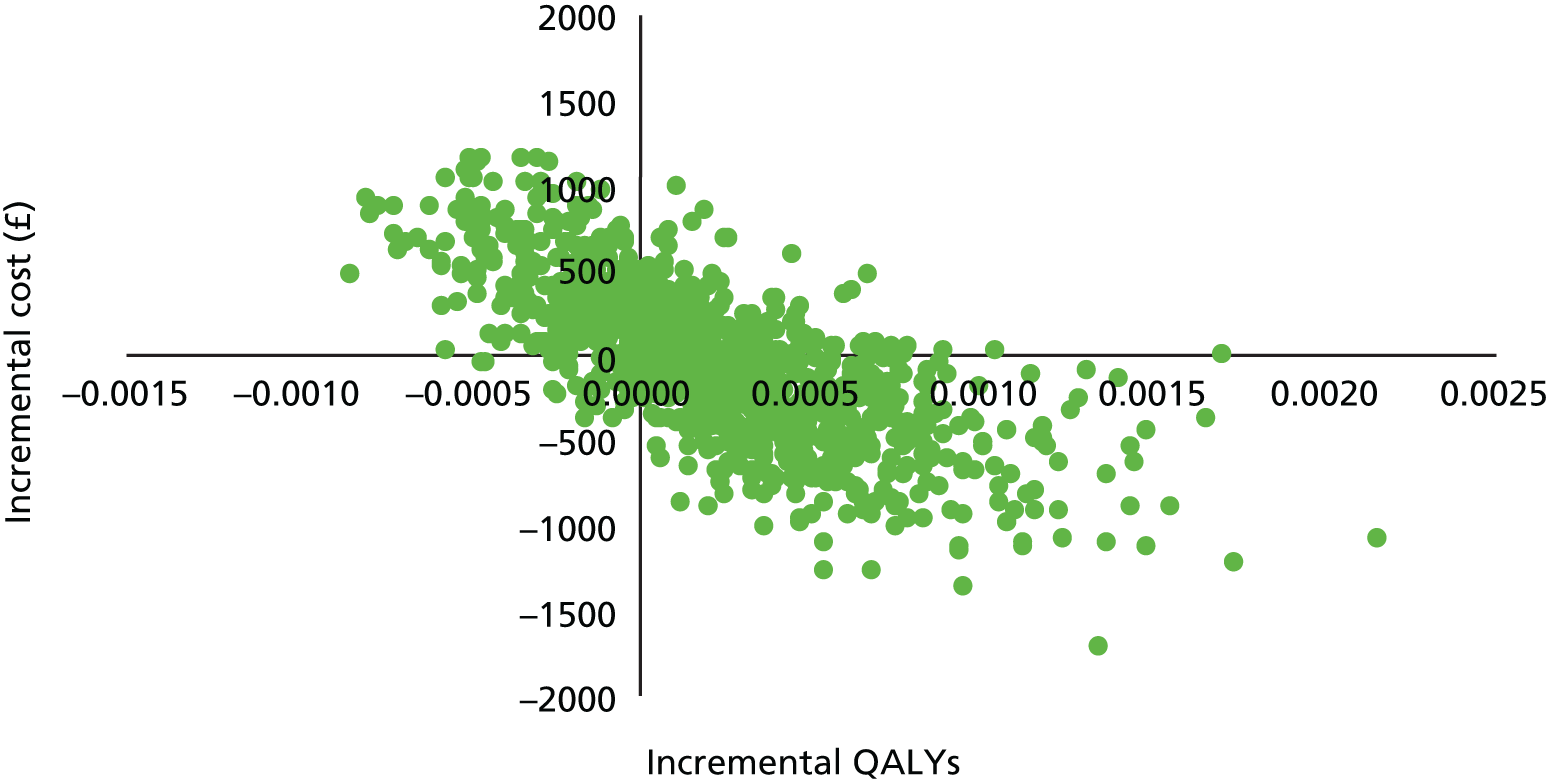
FIGURE 24.
Cost-effectiveness plane: FilmArray vs. conventional testing (benchmark) (model 2).
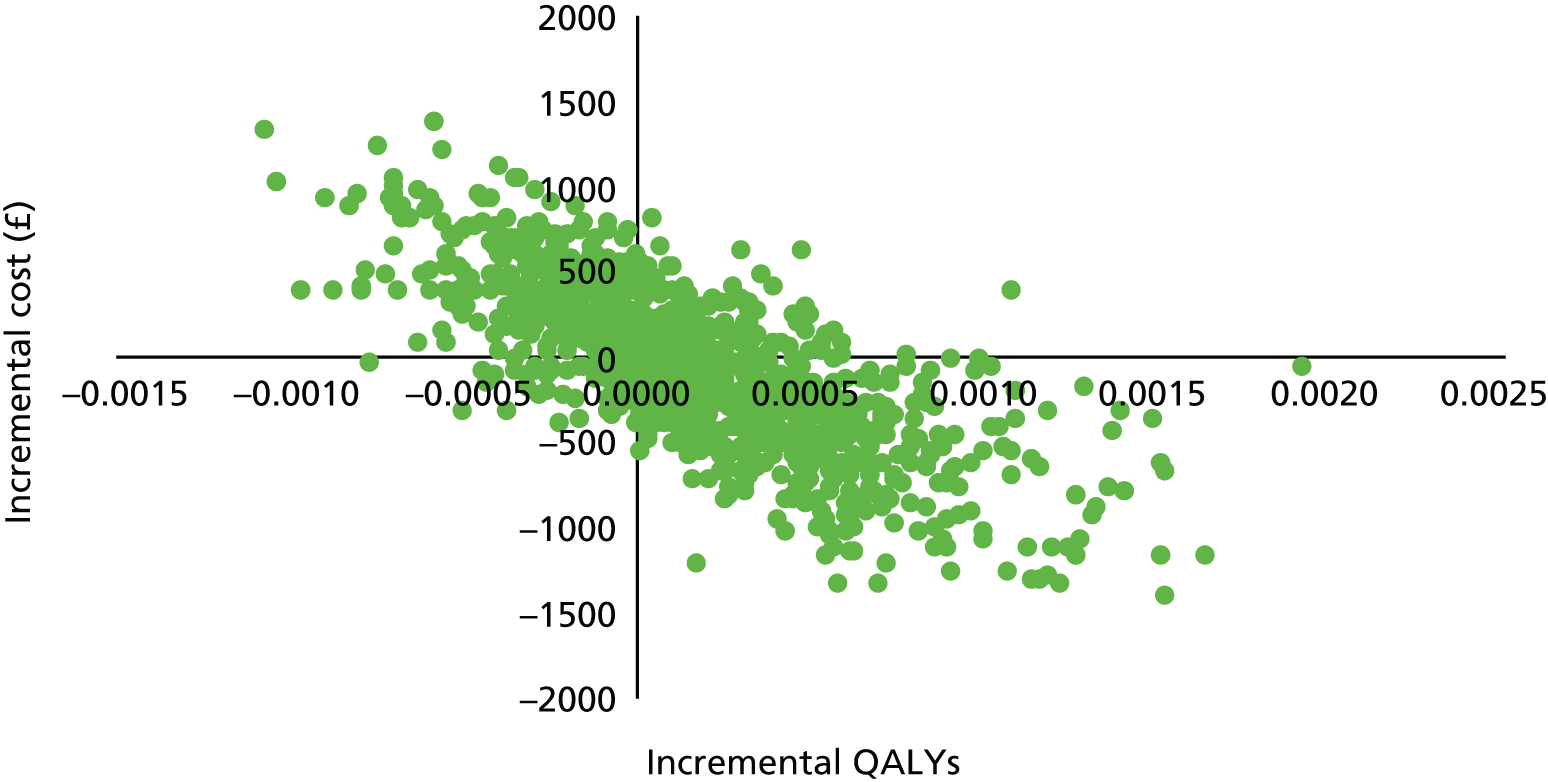
FIGURE 25.
Cost-effectiveness acceptability curve: xTAG vs. conventional testing (benchmark) (model 2).
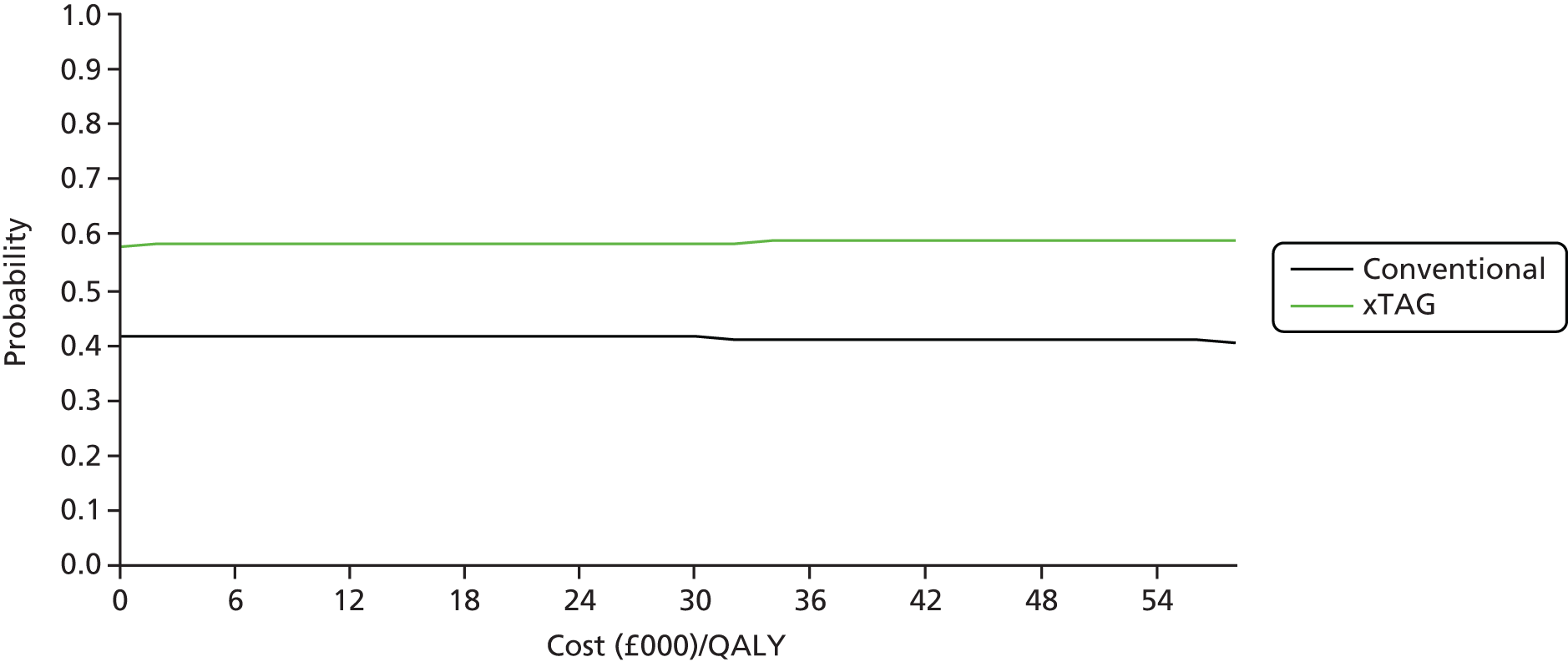
FIGURE 26.
Cost-effectiveness acceptability curve: FilmArray vs. conventional testing (benchmark) (model 2).

In a scenario analysis, we have tested the assumption that patients undergoing GPP testing leave hospital 1 day earlier than patients receiving conventional care, rather than after the same number of days (as in model 1). The incremental per-patient mean costs are £73 less for xTAG and £83 less for FilmArray testing than for conventional care when there is no difference in the overall length of stay (i.e. 3 days; Figure 27). If patients are discharged, on average, 1 day earlier, the cost saving per patient rises to £959 for xTAG and to £969 for FilmArray.
FIGURE 27.
Incremental mean costs (£) (probabilistic results): model 2. (a) xTAG vs. conventional testing; and (b) FilmArray vs. conventional testing.
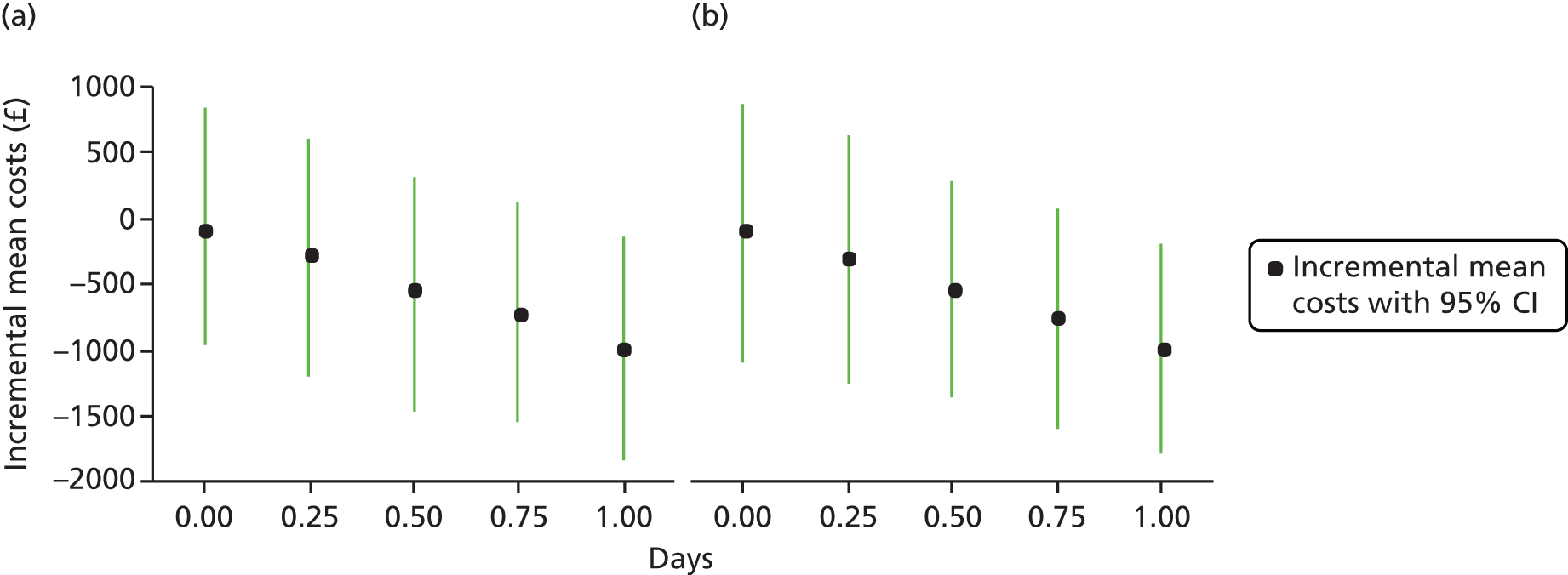
As with model 1, the incremental mean QALYs gained varied little when changing the length of stay for GPP testing, reflecting the fact that no disutility is attached to being in hospital. Figure 28 shows the net monetary benefit: with earlier discharge, the net monetary benefit of GPP testing increases and for this model with its assumptions. The 95% CI exceeds zero at a threshold of £20,000 per QALY, if GPP testing reduces length of stay by 1 day.
FIGURE 28.
Net monetary benefit (willingness-to-pay threshold = 20,000/QALY) (probabilistic results): model 2.
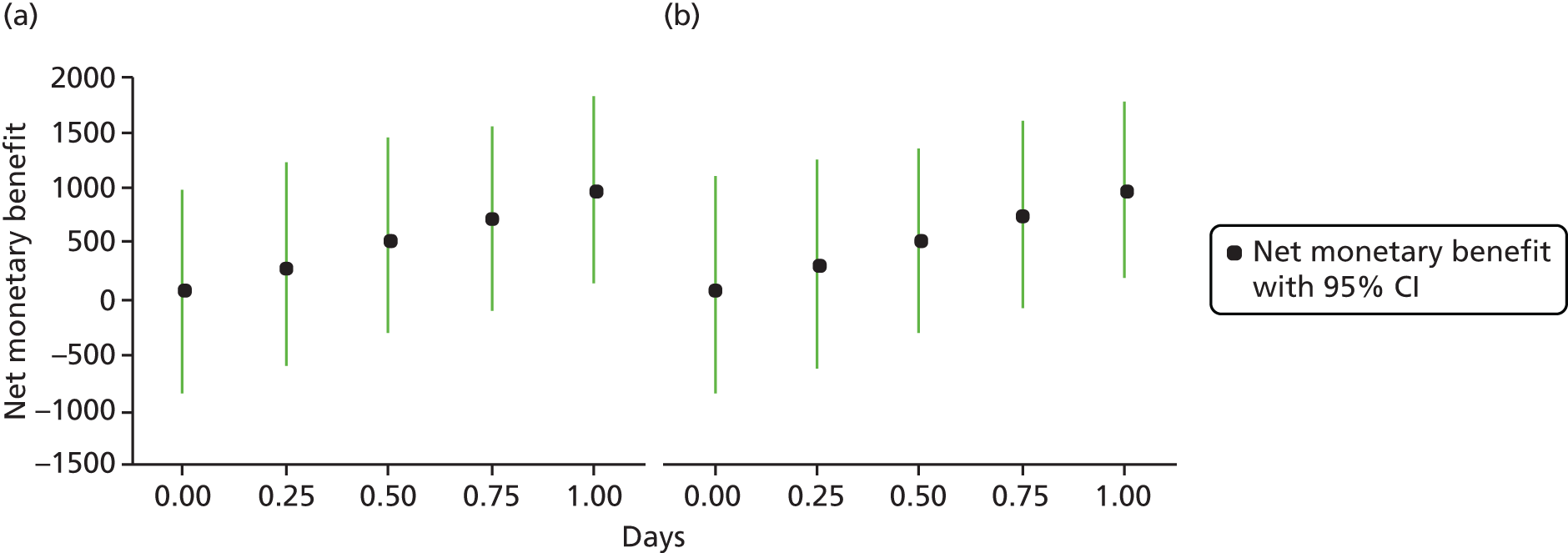
Model 3 results: people in the community with suspected gastroenteritis
Table 49 shows the results when conventional testing is the benchmark for people in the community with an episode of gastroenteritis (model 3). Use of xTAG resulted in a small net negative cost, whereas FilmArray marginally increased costs; both tests marginally improved quality of life. The probabilistic results showed that the differing costs were robust given the assumptions in the model. Consequently, xTAG appears cost-effective whereas FilmArray does not.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Net monetary benefit (£20,000/QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 72 | 0.00139 | – | – | – | – |
| xTAG | 40 | 0.00136 | –32 | 0.00003 | Dominant | 32 |
| Conventional | 72 | 0.00139 | – | – | – | – |
| FilmArray | 97 | 0.00138 | 25 | 0.00002 | 1,653,939 | –25 |
| Probabilistic results | ||||||
| Conventional | 72 | 0.00139 | – | – | – | – |
| xTAG | 40 | 0.00136 | –32 (–49 to –15) | 0.00003 (–0.00036 to 0.00045) | Dominant | 32 (9 to 56) |
| Conventional | 72 | 0.00139 | – | – | – | – |
| FilmArray | 97 | 0.00137 | 25 (0 to 48) | 0.00002 (–0.00036 to 0.00042) | 1,309,346 | –25 (–53 to 7) |
Figures 29 and 30 show the cost-effectiveness planes when conventional testing is the benchmark for model 3. Bootstrapped estimates for xTAG are primarily in the southern quadrants, denoting cost saving, whereas estimates for FilmArray are primarily in the northern quadrants, denoting increased cost. Figures 31 and 32 show the CEACs, which indicate that GPP testing probabilistically is nearly 100% cost-effective using xTAG but approximately 6% cost-effective using FilmArray, at a willingness-to-pay threshold of £20,000 per QALY.
FIGURE 29.
Cost-effectiveness plane: xTAG vs. conventional testing (benchmark) (model 3).

FIGURE 30.
Cost-effectiveness plane: FilmArray vs. conventional testing (benchmark) (model 3).

FIGURE 31.
Cost-effectiveness acceptability curve: xTAG vs. conventional testing (benchmark) (model 3).
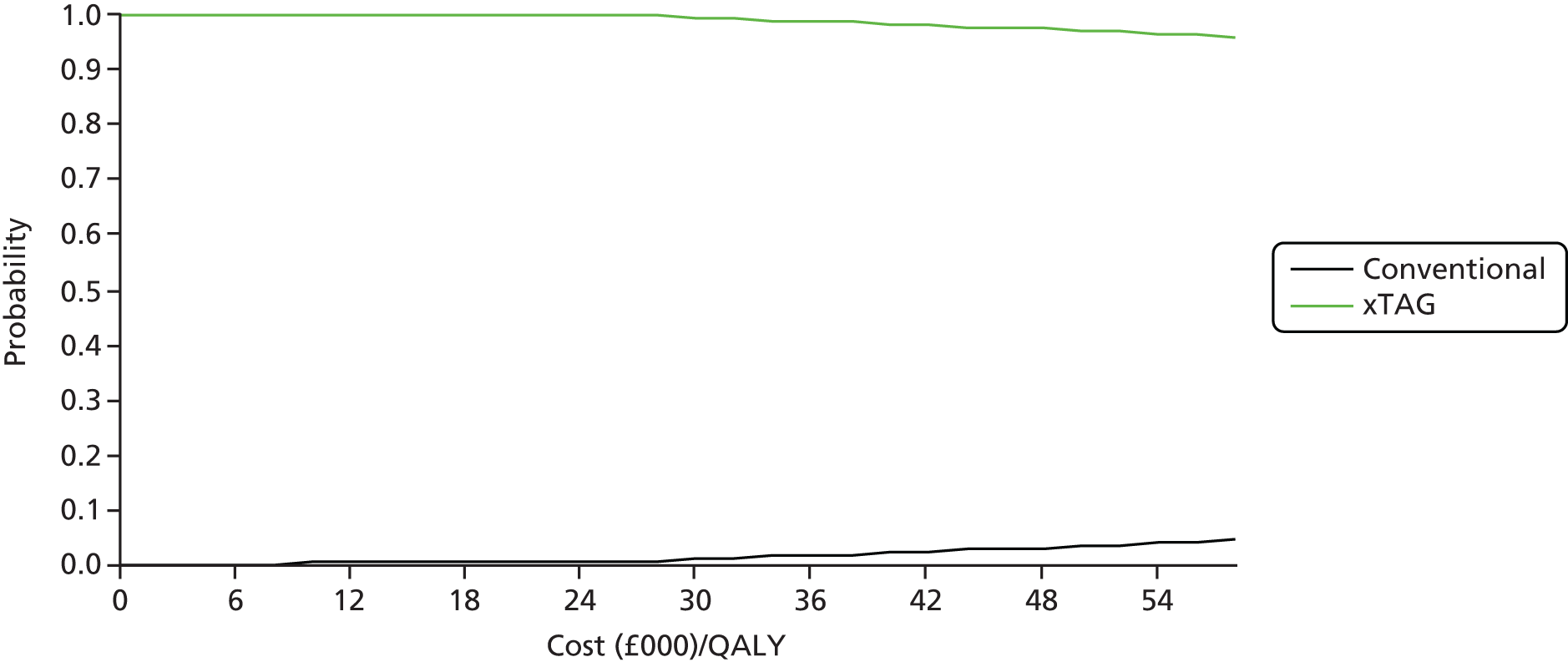
FIGURE 32.
Cost-effectiveness acceptability curve: FilmArray vs. conventional testing (benchmark) (model 3).
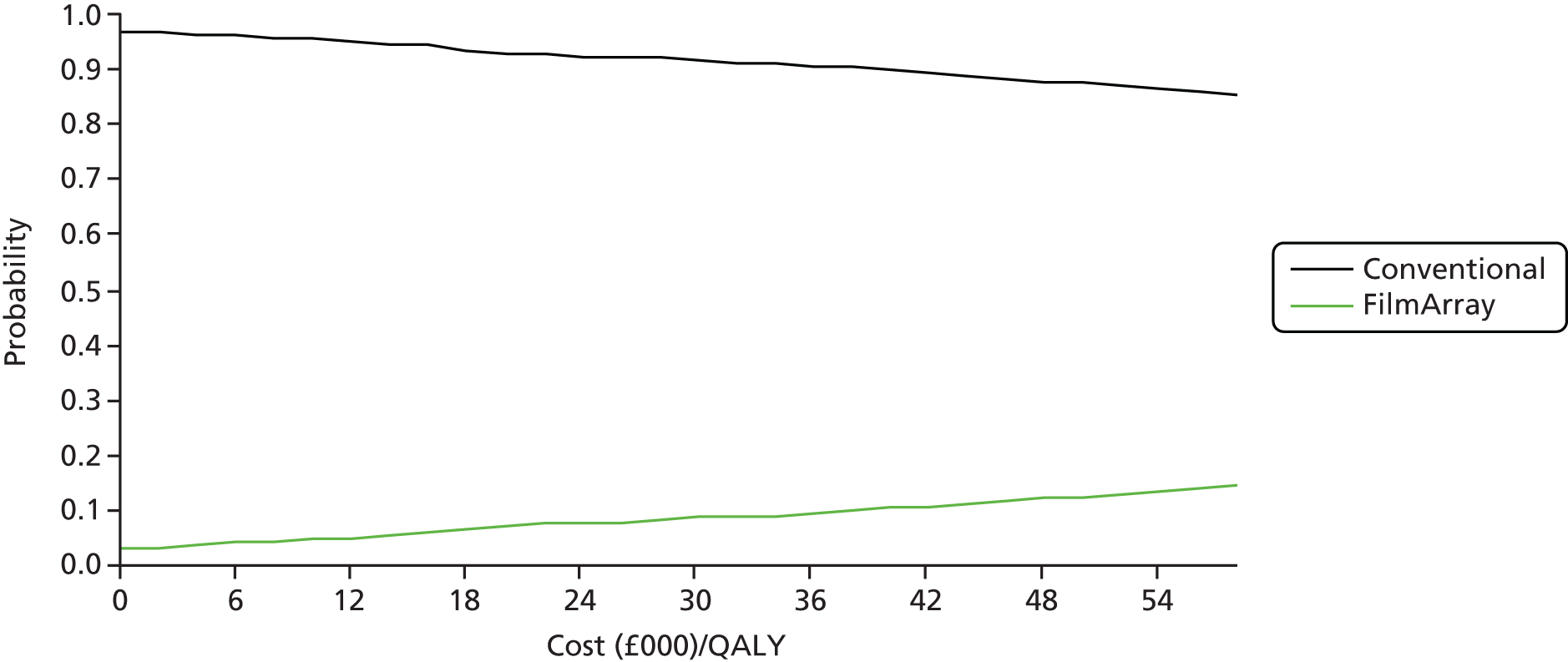
Figures 33 and 34 show one-way sensitivity analyses (tornado diagrams) of key variables. Each are varied in turn while holding others constant to understand what influences net monetary benefit, at a willingness-to-pay threshold of £20,000 per QALY.
FIGURE 33.
Tornado diagram for net monetary benefit conventional testing (benchmark) vs. xTAG: model 3. WTP, willingness to pay.

FIGURE 34.
Tornado diagram for net monetary benefit conventional testing (benchmark) vs. FilmArray: model 3. WTP, willingness to pay.

Net monetary benefit is centred around £32.30 for xTAG GPP when compared with conventional testing (see Figure 33). In this community model, all of the hospital costs are removed. The most important variables are assumptions about the costs of the conventional and GPP tests, as seen when these are varied by 50%. The finding is similar for FilmArray (see Figure 34), which is centred on a net monetary benefit ratio of –£24.90. Changing the cost of drugs, the cost of oral rehydration, the probability of detection (for the first test) and the utilities had little effect on the net monetary benefit.
Model 4 results: people immunocompromised with suspected gastroenteritis
Table 50 shows the results when conventional testing is the benchmark for patients who are immunocompromised (model 4). The deterministic results show that both xTAG and FilmArray are marginally less costly and more effective (more QALYs gained), but again the probabilistic findings show that cost and QALY savings are highly uncertain. Figures 35 and 36 show the cost-effectiveness planes when conventional testing is the benchmark for model 4. Like models 1 and 2, the bootstrap estimates are located in all four quadrants, with costs and outcomes negatively correlated. Figures 37 and 38 show the CEACs when conventional testing provides the benchmark. For both figures, GPP testing is of uncertain cost-effectiveness (approximately 55% probability for xTAG and 57% probability for FilmArray) at a willingness-to-pay threshold of £20,000 per QALY. As mentioned in Model 1 results: hospitalised adult patients with suspected gastroenteritis, Figures 37 and 38 highlight that willingness to pay has no effect and, as a general rule, when estimating cost-effectiveness ratios with incremental QALYs close to zero, the ICER sign may switch.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Net monetary benefit (£20,000/QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 3561 | 0.00384 | – | – | – | – |
| xTAG | 3486 | 0.00361 | –76 | 0.00023 | Dominant | 80 |
| Conventional | 3561 | 0.00384 | – | – | – | – |
| FilmArray | 3493 | 0.00367 | –68 | 0.00017 | Dominant | 72 |
| Probabilistic results | ||||||
| Conventional | 3563 | 0.00384 | – | – | – | – |
| xTAG | 3482 | 0.00361 | –81 (–1031 to 891) | 0.00023 (–0.00094 to 0.00134) | Dominant | 86 (–901 to 1056) |
| Conventional | 3563 | 0.00384 | – | – | – | – |
| FilmArray | 3486 | 0.00367 | –77 (–1024 to 871) | 0.00017 (–0.00088 to 0.00129) | Dominant | 80 (–883 to 1047) |
FIGURE 35.
Cost-effectiveness plane: xTAG vs. conventional testing (benchmark) (model 4).

FIGURE 36.
Cost-effectiveness plane: FilmArray vs. conventional testing (benchmark) (model 4).
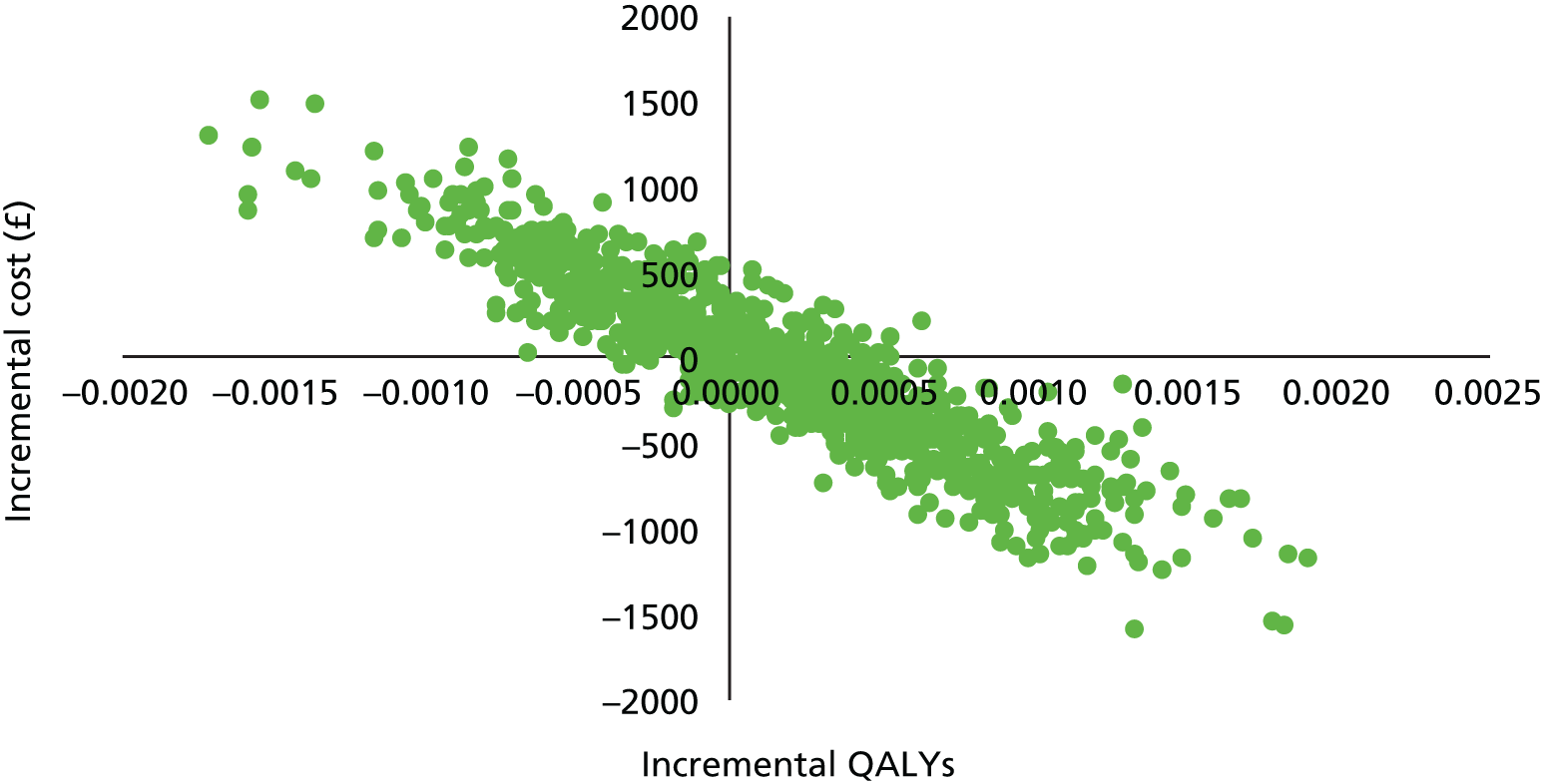
FIGURE 37.
Cost-effectiveness acceptability curve: xTAG vs. conventional testing (benchmark) (model 4).
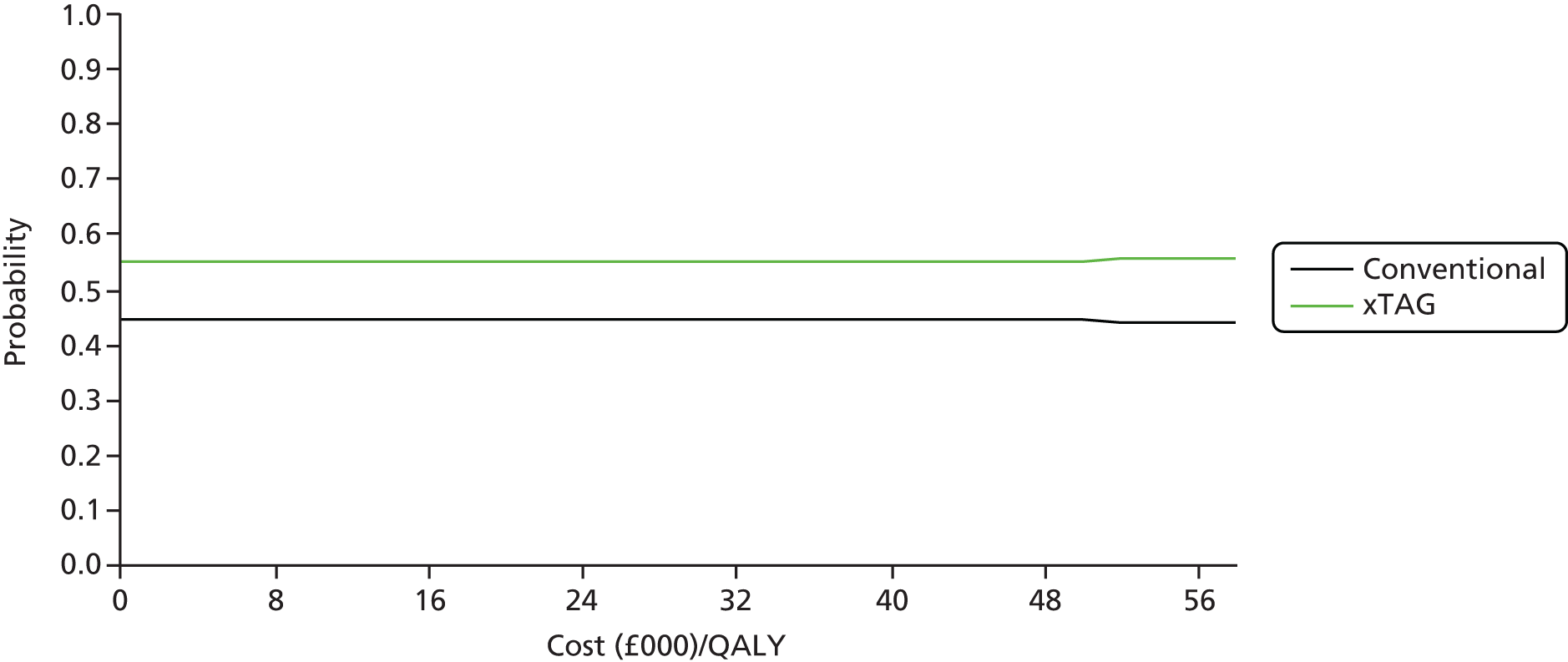
FIGURE 38.
Cost-effectiveness acceptability curve: FilmArray vs. conventional testing (benchmark) (model 4).

Model 5 results: people with a recent history of travel with suspected gastroenteritis
Table 51 shows the results when conventional testing is the benchmark for the recent travellers model (model 5). The deterministic results are very similar to the adult community model (model 3); xTAG dominates conventional testing, being cheaper (£32) and resulting in the loss of fewer QALYs. The probabilistic analysis points to small but significant cost savings and more uncertain QALY gains. This is shown graphically in the ICER plane (Figure 39) and CEAC (Figure 40), which find xTAG testing probabilistically to be approximately 99% likely to be cost-effective given the assumptions in the model.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Net monetary benefit (£20,000/QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 73 | 0.00180 | – | – | – | – |
| xTAG | 41 | 0.00174 | –32 | 0.00006 | Dominant | 33 |
| Conventional | 73 | 0.00180 | – | – | – | – |
| FilmArray | 98 | 0.00177 | 25 | 0.00002 | 1,020,674 | –25 |
| Probabilistic results | ||||||
| Conventional | 73 | 0.00181 | – | – | – | – |
| xTAG | 42 | 0.00175 | –31 (–48 to –16) | 0.00006 (–0.00044 to 0.00055) | Dominant | 32 (9 to 57) |
| Conventional | 73 | 0.00181 | – | – | – | – |
| FilmArray | 98 | 0.00176 | 25 (2 to 44) | 0.00004 (–0.00046 to 0.00055) | 560,220 | –24 (–52 to 7) |
FIGURE 39.
Cost-effectiveness plane: xTAG vs. conventional testing (benchmark) (model 5).
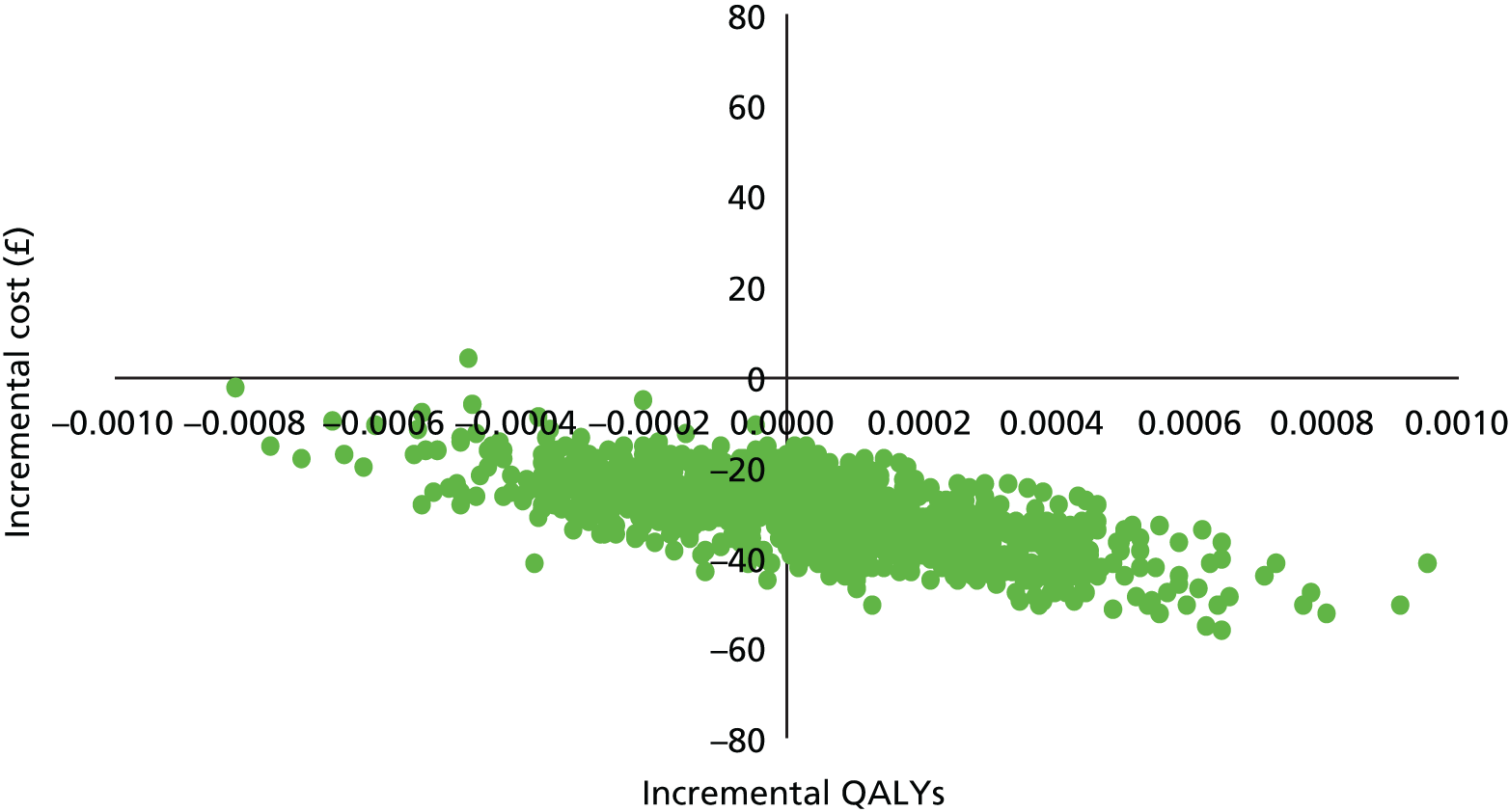
FIGURE 40.
Cost-effectiveness acceptability curve: xTAG vs. conventional testing (benchmark) (model 5).

Table 51 also shows the deterministic results for FilmArray vs. conventional care. Again, the results are similar to the community model, as shown in Model 3 results: people in the community with suspected gastroenteritis, in that the FilmArray test is slightly more expensive and effective than conventional care, although there is a considerable uncertainty about the QALYs gained. Probabilistic estimation is shown graphically in the cost-effectiveness plane (Figure 41) and CEAC (Figure 42); there is an approximately 6% probability that FilmArray is cost-effective at a willingness-to-pay threshold of £20,000 per QALY.
FIGURE 41.
Cost-effectiveness plane: FilmArray vs. conventional testing (benchmark) (model 5).
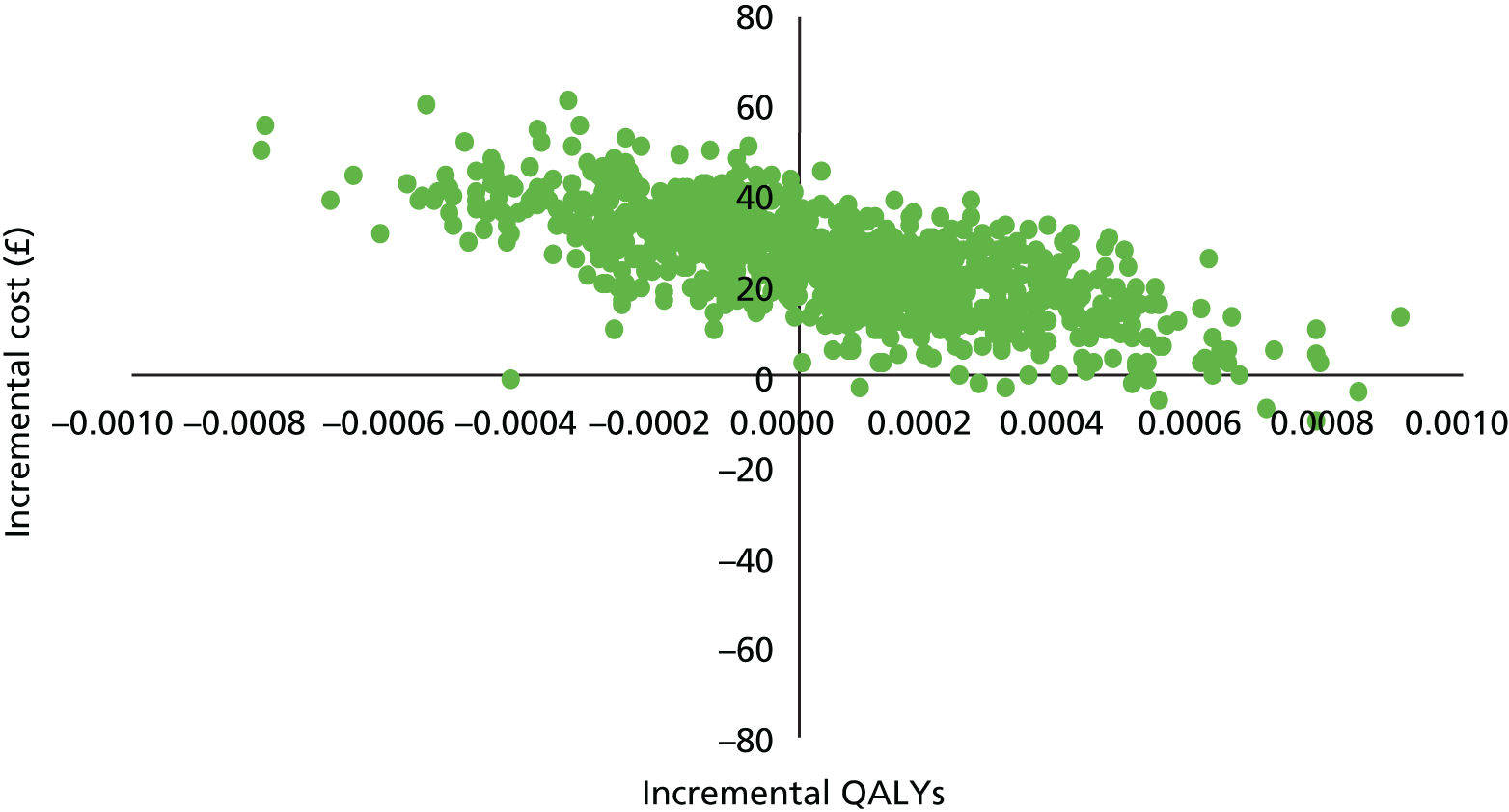
FIGURE 42.
Cost-effectiveness acceptability curve: FilmArray vs. conventional testing (benchmark) (model 5).
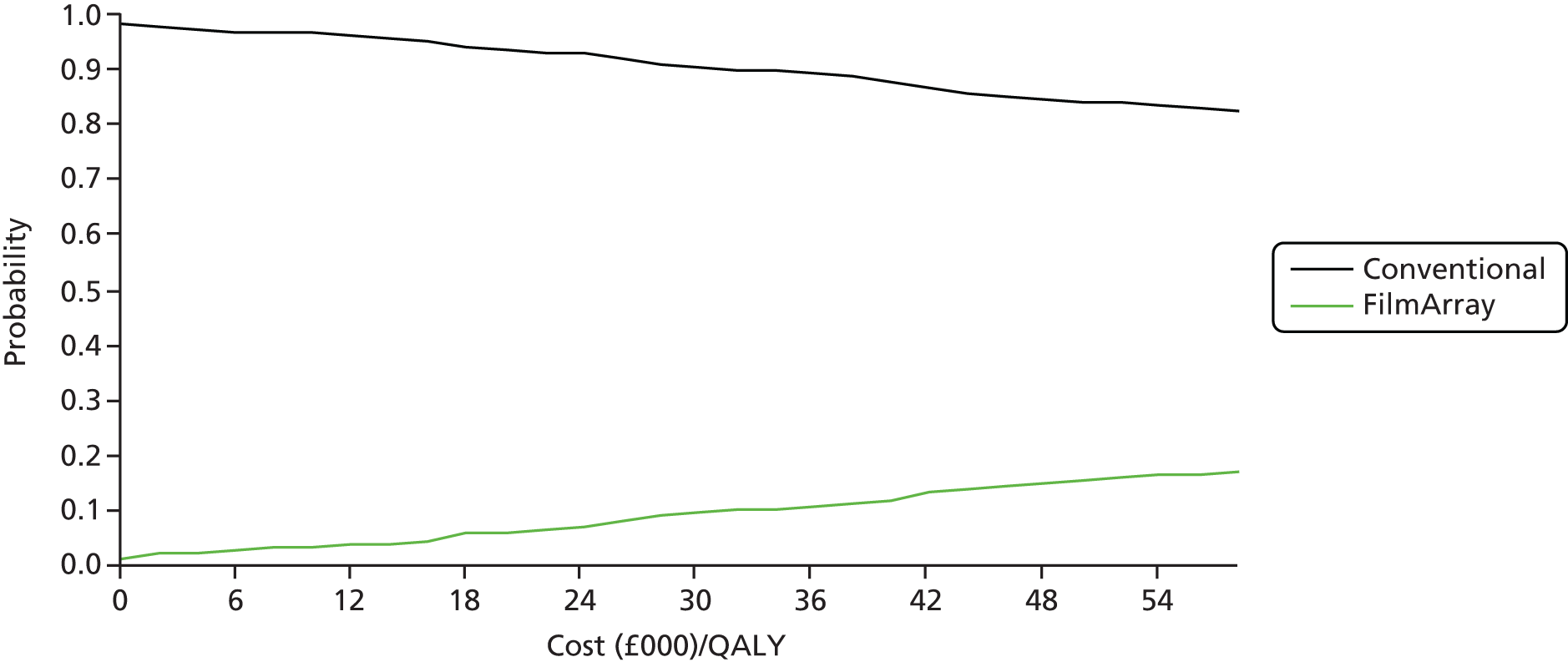
Additional analyses
Daily throughput of tests
We varied the average daily number of samples for each of the GPP tests. For all levels of throughput, one xTAG system is employed; for FilmArray, one system with four modules was employed for 12 samples per day, and eight modules for 24 and 48 samples per day. Table 52 provides the deterministic cost-effectiveness results when the number of samples is changed from 24 (base-case analysis) to 12 or 48 for model 1. There is very little difference in the incremental costs, and the overall results show that the GPP tests dominate conventional care.
| Test | Total mean costs (£) | Total mean QALY loss | Incremental costs (£) | Incremental QALYs gained | ICER (cost per QALY gained) | Net monetary benefit (£20,000/QALY) (£) |
|---|---|---|---|---|---|---|
| Deterministic results | ||||||
| Conventional | 3300 | 0.00267 | – | – | – | – |
| xTAG | 3237 | 0.00242 | –63 | 0.00025 | Dominant | 68 |
| Conventional | 3300 | 0.00267 | – | – | – | – |
| FilmArray | 3221 | 0.00249 | –79 | 0.00017 | Dominant | 82 |
| Deterministic results: 12 samples | ||||||
| Conventional | 3157 | 0.00309 | – | – | – | – |
| xTAG | 3094 | 0.00292 | –64 | 0.00018 | –359968 | 67 |
| Conventional | 3157 | 0.00309 | – | – | – | – |
| FilmArray | 3099 | 0.00297 | –59 | 0.00013 | –465006 | 61 |
| Deterministic results: 48 samples | ||||||
| Conventional | 3157 | 0.00309 | – | – | – | – |
| xTAG | 3086 | 0.00292 | –71 | 0.00018 | –399635 | 74 |
| Conventional | 3157 | 0.00309 | – | – | – | – |
| FilmArray | 3092 | 0.00297 | –65 | 0.00013 | –515654 | 68 |
Further additional analyses, as requested by the NICE committee, are shown in Appendix 11.
Discussion
For the base-case analysis, we modelled a hypothetical cohort of hospitalised adult patients with suspected gastroenteritis. We assumed that conventional testing provided a pragmatic benchmark with which to compare the GPP tests. The de novo economic model was a simple decision tree analysis, as gastroenteritis is a self-limiting illness and usually stops within 2 weeks. The analysis was conducted from a NHS and Personal Social Services perspective. Data for the model, including prevalence information, were obtained from a systematic clinical effectiveness review, published literature and expert opinion. Costs are reported in pounds sterling (£) in 2014/15 prices. Results are expressed as incremental cost per QALY gained. No discounting was performed as the time frame was < 1 year. We ran the model deterministically and probabilistically with 1000 bootstrapped iterations. We undertook various sensitivity analyses; the base-case model was adapted to look at various subgroups: young children, people in the community, immunocompromised patients and patients with a recent history of travel.
Methods and summary of findings
In the base model (adult hospitalised patients) and base-case assumptions, there was considerable uncertainty about the cost-effectiveness of GPP tests (Table 53). The pattern was similar for the two other hospital models involving young children and immunocompromised patients. These models show that varying pathogen prevalence does not significantly affect the cost-effectiveness of testing. Important variables are length of stay and the management of false-positive findings. In the community model, and likewise the recent travellers model, xTAG appeared cost-effective, whereas FilmArray did not. In community models, without the potential for hospital length-of-stay savings, cost-effectiveness is driven by the costs of the tests themselves and assumptions made when estimating these costs. All findings are sensitive to costs estimated for the two tests; findings would vary with different cost assumptions.
| Test | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Probability cost-effectivea | Net monetary benefit (£)a |
|---|---|---|---|---|---|
| Model 1: hospitalised adult patients | |||||
| xTAG | –63 (–929 to 791) | 0.00016 (–0.00071 to 0.00106) | Dominant | 0.566 | 67 (–802 to 947) |
| FilmArray | –52 (–883 to 793) | 0.00012 (–0.00079 to 0.00104) | Dominant | 0.544 | 54 (–806 to 909) |
| Model 2: hospitalised children | |||||
| xTAG | –73 (–955 to 849) | 0.00024 (–0.00049 to 0.00117) | Dominant | 0.583 | 78 (–856 to 977) |
| FilmArray | –83 (–1094 to 858) | 0.00017 (–0.00061 to 0.00114) | Dominant | 0.568 | 86 (–869 to 1114) |
| Model 3: people in the community | |||||
| xTAG | –32 (–49 to –15) | 0.00003 (–0.00036 to 0.00045) | Dominant | 0.997 | 32 (9 to 56) |
| FilmArray | 25 (0 to 48) | 0.00002 (–0.00036 to 0.00042) | 1,309,346 | 0.064 | –25 (–53 to 7) |
| Model 4: immunocompromised patients | |||||
| xTAG | –81 (–1031 to 891) | 0.00023 (–0.00094 to 0.00134) | Dominant | 0.553 | 86 (–901 to 1056) |
| FilmArray | –77 (–1024 to 871) | 0.00017 (–0.00088 to 0.00129) | Dominant | 0.570 | 80 (–883 to 1047) |
| Model 5: recent travellers | |||||
| xTAG | –31 (–48 to –16) | 0.00006 (–0.00044 to 0.00055) | Dominant | 0.994 | 32 (9 to 57) |
| FilmArray | 25 (2 to 44) | 0.00004 (–0.00046 to 0.00055) | 560,220 | 0.059 | 24 (–52 to 7) |
A sensitivity analysis identified the potential to reduce the number of bed-days as a key cost driver in the adult hospital. Although earlier discharge is plausible, it is by no means certain in the present hospital environment, in which patients are generally discharged as soon as possible and care may be symptom led rather than diagnosis led. In the absence of evidence, earlier discharge for GPP testing was explored in a scenario analysis, which indicated that if length of stay is reduced, then GPP testing is likely to be cost-effective.
Strengths and limitations
Although we undertook a thorough search of cost-effectiveness studies of the use of GPP tests compared with conventional testing for patients with suspected gastroenteritis, we could not identify any relevant economic models that could be adapted. Consequently, a de novo decision tree model was built to compare GPP testing with conventional care. A number of limitations apply to the economic model.
-
The model provides a representation of the clinical care pathway in the NHS, although practice and management will vary from site to site. We have compared ‘like with like’, that is, GPP tests are compared with a complete panel of conventional tests. Clinicians in some settings may order tests selectively and sequentially, as necessary, rather than as a complete panel of tests. As this strategy may not alter the pathway of care for most patients, conventional testing costs would be reduced. However, this strategy may result in a delayed diagnosis of treatable pathogens in a small minority of patients and needs to be formally evaluated – this was beyond the scope of the current work.
-
We could compare conventional testing with xTAG and FilmArray only; we could not retrieve any clinical data for Faecal Pathogens B.
-
We have assumed in the base-case model (adult hospital) that the minimum length of stay in hospital is 3 days; in practice, some patients may be discharged earlier, but this applies to both GPP and conventional testing.
-
The costs of the new GPP tests were based on the assumption that the machine would be running for 365 days per year, and an average of 24 samples would be run each day. We specified the capacity of GPP tests within the model to cope with variation on a daily basis and peak demand – theoretically, xTAG could be run twice a day in normal working hours and FilmArray units 8–10 times; thus, both configurations could cope with higher capacity. Although the economics change marginally in low-throughput settings, FilmArray appeared to be a comparatively expensive system.
-
Assumptions made about the costs of the GPP systems relative to conventional testing are important influences in the model.
-
We have not taken into account any broader societal costs such as lost productivity or time off work as a result of suspected gastroenteritis or length of stay in hospital. Similarly, no account has been taken of the costs of providing alternative care for children or other dependants of adults with gastroenteritis who are hospitalised or require care in the community. Costs may be underestimated and outcomes overestimated, as the model has not taken into account rare instances of mortality, recurrence or adverse events because of the lack of evidence. Thus, the model is limited to the initial index episode and does not consider readmissions. Buchanan et al. 84 stated that 10% of patients who have been discharged on antibiotics for infectious diarrhoea from hospital before a full course of treatment has been completed will be readmitted within 14 days of discharge.
-
We have not considered the management of outbreaks, as this was excluded from the scope of work.
-
We have not included costs for the treatment of Cryptosporidium, as there is no standard approved treatment in the UK for this infection.
-
We conducted a comprehensive literature search for disutility/utility values for gastroenteritis and found that most values in the literature were based around children with rotavirus – no adult utility values were identified for either hospital or community. Adult utilities used in the economic model for the different pathogens are based on food-borne illnesses from the USA, which used the EQ-5D.
-
In practice, each laboratory may receive a varying mix of hospital and community samples – the balance of this mix may affect the overall cost-effectiveness of GPP testing. We have not estimated weighted analyses based on variations in the mix of samples, as the five models presented are themselves tentative.
-
Finally, modelled changes in costs and QALYs are simulations and have not been observed. Findings should be verified through properly designed and conducted research.
Chapter 6 Discussion
Decision problem and objectives
The overall objective was to undertake a clinical effectiveness and cost-effectiveness analysis of the three gastrointestinal panel tests (xTAG, FilmArray and Faecal Pathogens B). The panels simultaneously test for common bacteria, viruses and parasites. The literature informing clinical effectiveness and cost-effectiveness was systematically reviewed and summarised. A de novo economic model was developed to assess the cost-effectiveness of GPP tests compared with current care in England and Wales.
Summary of methods and findings
Clinical effectiveness
We searched a number of databases including MEDLINE, EMBASE, Web of Science and The Cochrane Library. We found 2215 unique records, of which 23 were included. Of these studies, 10 reported sufficient data to assess agreement, by pathogen, comparing GPP assay and conventional methods using 2 × 2 tables. In general, the methodological quality of the included studies was poor. No adequate test–treat trials or diagnostic studies using a reference standard were retrieved, and no study conducted adequate discrepant analysis or applied a fair umpire approach. In the absence of adequate studies, positive and negative agreements were estimated, as recommended by FDA guidance, alternating conventional and GPP testing as the benchmark as an aid to explore differences in findings. Using conventional methods as a benchmark, GPP testing appears reliable and could replace current microbiological methods, although there would be an increase in additional positive reporting. Whether or not this would result in some overtreatment is uncertain; however, using GPP testing as the benchmark suggests that we might identify disease missed by current methods. Potentially, the consequence might be more appropriate treatment for pathology missed or not detected by conventional methods. Currently, the clinical importance of the additional pathogens identified by GPP testing is uncertain.
The review has not included data from 1847 patients from three recent studies that were identified after our deadline for considering studies for full review. 45–47 These three studies consisted of a case–control study of Ghanaian children aged < 6 years using xTAG,46 a study of 779 children aged < 18 years from the USA using FilmArray,45 and a Belgian study of 386 mixed patients that assessed FilmArray for the detection of shiga toxin-producing E. coli compared with an in-house culture-based PCR method. 47 This final study is the only one of the three that would have contributed 2 × 2 data for the meta-analysis; however, the study considered only 1 out of 23 pathogens detectable with FilmArray. Full inclusion of the three studies would therefore not have influenced or changed the conclusions of this review.
Current evidence is inadequate to compare the performance or value of different GPP systems. Direct comparison may not necessarily be appropriate when these systems are designed with different operational scale and throughput in mind, and provide overlapping but not identical coverage of pathogens. Further research is needed to understand failure rates because of the various GPP systems, or importance of multipathogen infections.
Cost-effectiveness
Given uncertainties about the performance of GPP systems and their impact on clinical care, any attempt at economic modelling is necessarily tentative. An economic model has been developed to explore these uncertainties. Pragmatically, as it reflects current practice, conventional testing serves as the benchmark in the model (i.e. it is assumed to correctly identify clinically important pathology). Exploring the impact of adopting GPP testing on clinical care requires multiple assumptions about the use of the tests and how the tests affect the care provided. These assumptions have been variously informed by the direct evidence, epidemiological sources and expert opinion.
These limitations accepted, the hospital admission models indicate that use of GPP systems might be cost-effective if they result in reduced length of stay. Community care use of GPP systems might be cost-effective, but findings are sensitive to assumptions about the cost of GPP tests. Current evidence precludes a robust conclusion, and so adoption should be further informed by findings from relevant and robust research.
Strengths and limitations
Assessment of clinical evidence has depended on measures of agreement, which are not measures of test accuracy in a conventional sense and should not be interpreted as such. For example, agreement analysis takes no consideration of whether or not both tests can agree and be wrong. The quality of available studies contributing to the clinical evidence review was generally poor.
Within the clinical studies identified, many pathogens were present only in very low numbers, and the context of studies included a mixture of different patient populations (e.g. children, immunocompromised patients, community), each with their own distribution of prevalence of pathogens.
As described, the economic model provides a representation of the clinical care pathway, and has populated a decision tree with probabilities and values from the clinical evidence review, epidemiological studies and clinical expert opinion. Although modelling methods permit an exploration of uncertainties, ultimately the reliability of the model depends on the belief that modelled changes would actually occur in clinical practice. It has not been possible to validate model findings with real-world data.
The scope of work has been limited to the management of patients in hospital and community settings who are tested for suspected infectious gastroenteritis. The public health implications of different patterns of testing or of special circumstances, such as infectious outbreaks, have not been considered.
Points for discussion
For the purpose of comparison within the economic model, the number of pathogens evaluated was constrained to match the current PHE algorithm. However, the prevalence and importance of pathogens varies over time and by context. Is the coverage within each of the panels adequate, or should this be explicitly considered (e.g. should detection of sapovirus be specified, one of the most commonly identified pathogens causing gastroenteritis in the community)? Sapovirus is included in the coverage of FilmArray and the Faecal Pathogens B assay but not xTAG. Changes in prevalence and virulence of pathogens, current coverage of GPPs and the ability of manufacturers to adapt coverage create a dynamic context for decision-making.
Assumptions in the hospital economic models about length of stay are key to findings. What is the opinion among committee members regarding whether length of stay is predominantly symptom driven or pathogen detection/exclusion driven? Is there scope with earlier detection for earlier discharge, or is this already efficiently managed in hospitals? Does the discharge destination further complicate matters given the need for further hydration, hygiene and watchful waiting, when provision may differ between home, care home and other institutional destinations?
The base-case model assumes an average throughput of 24 samples per day. Although we found no evidence that our findings were strongly influenced by throughput, the largest hospital trusts in England may currently process about 80 samples per day and the next 10–20 years may see a move towards superlaboratories, with just 10–20 laboratories in the country, implying still greater throughput. In parallel with these developments, GPP testing technology may further develop, making evidence requirements and timing of adoption points for discussion.
Chapter 7 Conclusions
The systematic review and cost-effectiveness model identify uncertainties about the adoption of GPP tests. GPP testing will generally correctly identify pathogens identified by conventional testing; however, these tests also generate considerable additional positive results of uncertain clinical importance with potential workplace implications for those diagnosed. Early diagnosis may change the management of patients positively or negatively, for example possibly with earlier discharge in the case of negative findings but possibly with unnecessary treatment in the case of positive findings when symptoms would spontaneously resolve with watchful waiting over the following few days. The economic model reflects one pattern of care for which patients are broadly tested in line with PHE guidance, although this practice is not universally followed. Different practices, such as sequential testing, would give rise to different patterns of cost and benefit. Model explorations suggest scope for GPP testing to be cost-effective, but considerable uncertainty remains.
Recommendations for further research
Further research is needed. It may be difficult to design a study with an adequate independent reference standard, although the clinical review suggests that such evidence may not be pivotal to an adoption decision. A randomised controlled test–treat trial, with patients randomised to conventional or GPP testing, recording total care received, and evaluating clinical effectiveness and cost-effectiveness outcomes, would determine whether or not GPP systems can realise potential cost savings and improve patient utility. Currently, there is no adequate evidence of the impact of GPP testing on the management, treatment and outcome of patients, as no test–treat trials have been conducted. In addition, further evaluation might consider the public health impact of different diagnostic approaches, as the results of testing are the basis for public health surveillance of these diseases.
Current evidence should be regarded applicable to a mixed population of community and hospitalised patients, as the studies do not adequately report outcomes by setting or subgroup. A goal for a future trial might be to include adequate stratification of different populations (e.g. community managed patients, community acquired and hospital managed patients, hospital acquired patients, children, travellers and immunocompromised patients).
The NICE scope identified the PHE algorithm8 as the relevant comparator against which to consider GPP tests. However, the PHE algorithm was published in 2013 and may need to be updated to reflect progress in the microbiological laboratories. For instance, the new PHE guidance for the interpretation of PCR assays for gastrointestinal pathogens states that a growing number of PHE and NHS virology laboratories use PCR assays for the detection of gastrointestinal viral pathogens. 50 The same guidance recommends PCR as a first-line diagnostic test for the intestinal parasites G. lamblia, Cryptosporidium spp. and E. histolytica. This is not reflected in the national standard of practice used in this review. Similarly, this review has evaluated GPP systems according to their current specification, but it is anticipated that the coverage of these systems will continue to evolve in response to changing pathogen prevalence, hence the evaluation problem is a dynamic one.
Acknowledgements
We would like to thank Karoline Munro for her help with the formatting and proofreading of the document.
Contributions of authors
Karoline Freeman (Research Fellow) was the senior clinical effectiveness reviewer, conducted the clinical effectiveness systematic review, and co-ordinated and drafted the report.
Hema Mistry (Assistant Professor) undertook the health economic review and modelling, and drafted the report.
Alexander Tsertsvadze (Senior Research Fellow) conducted the clinical effectiveness systematic review.
Pam Royle (Information Specialist) developed the search strategy and undertook searches.
Noel McCarthy (Professor of Evidence in Communicable Disease Epidemiology and Control) provided clinical expertise and extensive input into the model structure and model inputs.
Sian Taylor-Phillips (Assistant Professor) provided diagnostic and methodological expertise.
Rohini Manuel (Consultant Medical Microbiologist) provided clinical expertise and guidance.
James Mason (Professor of Health Economics) provided project management, conducted meta-analyses, supervised the economic analysis and drafted the report.
All authors were involved in revising the report.
Publication
Freeman K, Tsertsvadze A, Taylor-Phillips S, McCarthy N, Mistry H, Manuel R, Mason J. Agreement between gastrointestinal panel testing and standard microbiology methods for detecting pathogens in suspected infectious gastroenteritis: test evaluation and meta-analysis in the absence of a reference standard. PLOS ONE 2017;12:e0173196.
Data sharing statement
All available data are included in the appendices of this report.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Tam C, Viviani L, Adak B, Bolton E, Dodds J, Cowden JM, et al. The Second Study of Infectious Intestinal Disease in the Community (IID2 Study): Final Report 2011. www.food.gov.uk/science/research/foodborneillness/b14programme/b14projlist/b18021 (accessed 9 November 2015).
- NICE . NICE Clinical Knowledge Summaries: Gastroenteritis 2015. http://cks.nice.org.uk/gastroenteritis (accessed 1 November 2015).
- PHE . UK Standards for Microbiology Investigations: Processing of Faeces for Clostridium Difficile SMI B 10 2014. www.gov.uk/government/uploads/system/uploads/attachment_data/file/343912/B_10i1.5.pdf (accessed 1 November 2015).
- PHE . Guidance: Carbapenemase-Producing Enterobacteriaceae: Early Detection, Management and Control Toolkit for Acute Trusts 2014. www.gov.uk/government/publications/carbapenemase-producing-enterobacteriaceae-early-detection-management-and-control-toolkit-for-acute-trusts (accessed 9 June 2016).
- PHE . Guidance on Infection Control in Schools and Other Childcare Settings 2016. www.gov.uk/government/uploads/system/uploads/attachment_data/file/522337/Guidance_on_infection_control_in_schools.pdf (accessed 4 January 2017).
- Food Standards Agency . Food Handlers: Fitness to Work 2009. www.food.gov.uk/sites/default/files/multimedia/pdfs/publication/fitnesstoworkguide09v3.pdf (accessed 31 May 2016).
- Department of Health . Updated Guidance on the Diagnosis and Reporting of Clostridium Difficile 2012. www.gov.uk/government/uploads/system/uploads/attachment_data/file/215135/dh_133016.pdf (accessed 31 May 2016).
- PHE . UK Standards for Microbiology Investigations: Gastroenteritis and Diarrhoea: SMI S7 Issue 1 2013. www.gov.uk/government/uploads/system/uploads/attachment_data/file/344110/S_7i1.pdf (accessed 20 May 2016).
- PHE . UK Standards for Microbiology Investigations: Investigation of Faecal Specimens for Enteric Pathogens: SMI B 30 2014. www.gov.uk/government/uploads/system/uploads/attachment_data/file/343955/B_30i8.1.pdf (accessed 1 November 2015).
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. http://dx.doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Beckmann C, Heininger U, Marti H, Hirsch HH. Gastrointestinal pathogens detected by multiplex nucleic acid amplification testing in stools of pediatric patients and patients returning from the tropics. Infection 2014;42:961-70. http://dx.doi.org/10.1007/s15010-014-0656-7.
- Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 2014;52:3667-73. http://dx.doi.org/10.1128/JCM.01637-14.
- Glasziou P, Irwig L, Deeks JJ. When should a new test become the current reference standard?. Ann Intern Med 2008;149:816-22. https://doi.org/10.7326/0003-4819-149-11-200812020-00009.
- Rutjes AW, Reitsma JB, Coomarasamy A, Khan KS, Bossuyt PM. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol Assess 2007;11. https://doi.org/10.3310/hta11500.
- Cochrane Collaboration . Handbook for DTA Reviews: Chapter 6 2008. http://methods.cochrane.org/sdt/HANDBOOK-DTA-REVIEWS (accessed 10 May 2016).
- Feinstein AR, Cicchetti DV. High agreement but low kappa: I. The problems of two paradoxes. J Clin Epidemiol 1990;43:543-9. https://doi.org/10.1016/0895-4356(90)90158-L.
- FDA . Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests 2007. www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm (accessed 19 April 2016).
- Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Public Health 2014;72. http://dx.doi.org/10.1186/2049-3258-72-39.
- Becker SL, Chatigre JK, Gohou JP, Coulibaly JT, Leuppi R, Polman K, et al. Combined stool-based multiplex PCR and microscopy for enhanced pathogen detection in patients with persistent diarrhoea and asymptomatic controls from Côte d’Ivoire. Clin Microbiol Infect 2015;21:591.e1-10. http://dx.doi.org/10.1016/j.cmi.2015.02.016.
- Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, et al. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 2015;53:915-25. http://dx.doi.org/10.1128/JCM.02674-14.
- Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. Performance of the xTAG® gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 2013;23:1041-5. https://doi.org/10.4014/jmb.1212.12042.
- Coste JF, Vuiblet V, Moustapha B, Bouin A, Lavaud S, Toupance O, et al. Microbiological diagnosis of severe diarrhea in kidney transplant recipients by use of multiplex PCR assays. J Clin Microbiol 2013;51:1841-9. http://dx.doi.org/10.1128/JCM.03366-12.
- Deng J, Luo X, Wang R, Jiang L, Ding X, Hao W, et al. A comparison of Luminex xTAG® Gastrointestinal Pathogen Panel (xTAG GPP) and routine tests for the detection of enteropathogens circulating in Southern China. Diagn Microbiol Infect Dis 2015;83:325-30. http://dx.doi.org/10.1016/j.diagmicrobio.2015.07.024.
- Duong VT, Phat VV, Tuyen HT, Dung TT, Trung PD, Minh PV, et al. Evaluation of Luminex xTAG Gastrointestinal Pathogen Panel Assay for detection of multiple diarrheal pathogens in fecal samples in Vietnam. J Clin Microbiol 2016;54:1094-100. http://dx.doi.org/10.1128/JCM.03321-15.
- FDA . Evaluation Of Automatic Class III Designation (De Novo) For XTAG® Gastrointestinal Pathogen Panel (GPP) Decision Summary 2012. www.accessdata.fda.gov/cdrh_docs/reviews/K121454.pdf (accessed 30 October 2015).
- Gu Z, Zhu H, Rodriguez A, Mhaissen M, Schultz-Cherry S, Adderson E, et al. Comparative evaluation of broad-panel PCR assays for the detection of gastrointestinal pathogens in pediatric oncology patients. J Mol Diagn 2015;17:715-21. http://dx.doi.org/10.1016/j.jmoldx.2015.06.003.
- Halligan E, Edgeworth J, Bisnauthsing K, Bible J, Cliff P, Aarons E, et al. Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin Microbiol Infect 2014;20:O460-7. http://dx.doi.org/10.1111/1469-0691.12476.
- Kahlau P, Malecki M, Schildgen V, Schulz C, Winterfeld I, Messler S, et al. Utility of two novel multiplexing assays for the detection of gastrointestinal pathogens – a first experience. Springerplus 2013;2. http://dx.doi.org/10.1186/2193-1801-2-106.
- Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, et al. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin Microbiol Infect 2013;19:E458-65. http://dx.doi.org/10.1111/1469-0691.12255.
- Pankhurst L, Macfarlane-Smith L, Buchanan J, Anson L, Davies K, O’Connor L, et al. Can rapid integrated polymerase chain reaction-based diagnostics for gastrointestinal pathogens improve routine hospital infection control practice? A diagnostic study. Health Technol Assess 2014;18. https://doi.org/10.3310/hta18530.
- Patel A, Navidad J, Bhattacharyya S. Site-specific clinical evaluation of the Luminex xTAG gastrointestinal pathogen panel for detection of infectious gastroenteritis in fecal specimens. J Clin Microbiol 2014;52:3068-71. http://dx.doi.org/10.1128/JCM.01393-14.
- Perry MD, Corden SA, Howe RA. Evaluation of the Luminex xTAG Gastrointestinal Pathogen Panel and the Savyon Diagnostics Gastrointestinal Infection Panel for the detection of enteric pathogens in clinical samples. J Med Microbiol 2014;63:1419-26. http://dx.doi.org/10.1099/jmm.0.074773-0.
- Petterson J, York V, Ward P, Green N, She R. The value of a multiplexed gastrointestinal pathogen panel in 2 distinct patient populations. Diagn Microbiol Infect Dis 2016;85:105-8. http://dx.doi.org/10.1016/j.diagmicrobio.2015.12.020.
- Rand KH, Tremblay EE, Hoidal M, Fisher LB, Grau KR, Karst SM. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn Microbiol Infect Dis 2015;82:154-7. http://dx.doi.org/10.1016/j.diagmicrobio.2015.01.007.
- Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L, et al. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 2015;21:719-28. http://dx.doi.org/10.1016/j.cmi.2015.04.007.
- Stockmann C, Rogatcheva M, Harrel B, Vaughn M, Crisp R, Poritz M, et al. How well does physician selection of microbiologic tests identify Clostridium difficile and other pathogens in paediatric diarrhoea? Insights using multiplex PCR-based detection. Clin Microbiol Infect 2015;21:179.e9-15. http://dx.doi.org/10.1016/j.cmi.2014.07.011.
- Vocale C, Rimoldi SG, Pagani C, Grande R, Pedna F, Arghittu M, et al. Comparative evaluation of the new xTAG GPP multiplex assay in the laboratory diagnosis of acute gastroenteritis. Clinical assessment and potential application from a multicentre Italian study. Int J Infect Dis 2015;34:33-7.
- Wessels E, Rusman LG, van Bussel MJ, Claas EC. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG® GPP) testing in the diagnosis of infectious gastroenteritis. Clin Microbiol Infect 2014;20:O182-7. http://dx.doi.org/10.1111/1469-0691.12364.
- Zboromyrska Y, Hurtado JC, Salvador P, Alvarez-Martínez MJ, Valls ME, Mas J, et al. Aetiology of traveller’s diarrhoea: evaluation of a multiplex PCR tool to detect different enteropathogens. Clin Microbiol Infect 2014;20:O753-9. http://dx.doi.org/10.1111/1469-0691.12621.
- FDA . 510(K) Substantial Equivalence Determination Decision Summary: Number K140377 2014. www.accessdata.fda.gov/cdrh_docs/reviews/k140377.pdf (accessed 19 April 2016).
- Dunbar SA, Ritchie VB, Hoffmeyer MR, Rana GS, Zhang H. Luminex® multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods Mol Biol 2015;1225:1-27. http://dx.doi.org/10.1007/978-1-4939-1625-2_1.
- FDA . FilmArray® Gastrointestinal (GI) Panel Microorganism Multiplex Nucleic Acid-Based Assay: 510(K) Number: K140407 Substantial Equivalence Determination Decision Summary 2015. www.accessdata.fda.gov/cdrh_docs/reviews/k140407.pdf (accessed 20 May 2016).
- BioFire Diagnostics . 510(k) Summary BioFire Diagnostics, LLC – FilmArray Gastrointestinal (GI) Panel Kit 2014. www.accessdata.fda.gov/cdrh_docs/pdf14/K140407.pdf (accessed 12 May 2016).
- Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group . Preferred Reporting Items for Systematic reviews and Meta-Analyses: the PRISMA statement. BMJ 2009;339. http://dx.doi.org/10.1136/bmj.b2535.
- Stockmann C, Pavia AT, Graham B, Vaughn M, Crisp R, Poritz MA, et al. Detection of 23 gastrointestinal pathogens among children who present with diarrhea [published online ahead of print 4 May 2016]. J Pediatric Infect Dis Soc 2016. https://doi.org/10.1093/jpids/piw020.
- Eibach D, Krumkamp R, Hahn A, Sarpong N, Adu-Sarkodie Y, Leva A, et al. Application of a multiplex PCR assay for the detection of gastrointestinal pathogens in a rural African setting. BMC Infect Dis 2016;16. http://dx.doi.org/10.1186/s12879-016-1481-7.
- De Rauw K, Detemmerman L, Breynaert J, Piérard D. Detection of Shiga toxin-producing and other diarrheagenic Escherichia coli by the BioFire FilmArray® Gastrointestinal Panel in human fecal samples. Eur J Clin Microbiol Infect Dis 2016;35:1479-86. http://dx.doi.org/10.1007/s10096-016-2688-7.
- Dunbar SA, Das S. Luminex Corporation . XTAG Gastrointestinal Pathogen Panel – The State of the Art in Molecular GI Testing 2015. www.luminexcorp.com/eu/download/xtag-gastrointestinal-pathogen-panel-the-state-of-the-art-in-molecular-gi-testing-2/ (accessed 20 May 2016).
- Freeman K, Tsertsvadze A, Taylor-Phillips S, McCarthy N, Mistry H, Manuel R, et al. Agreement between gastrointestinal panel testing and standard microbiology methods for detecting pathogens in suspected infectious gastroenteritis: test evaluation and meta-analysis in the absence of a reference standard. PLOS ONE 2017;12. http://dx.doi.org/10.1371/journal.pone.0173196.
- PHE . Guidance for the Interpretation of PCR Assays for Gastrointestinal Pathogens 2013. www.gov.uk/government/publications/gastrointestinal-pathogens-interpreting-pcr-assays-guidance (accessed 12 May 2016).
- Husereau D, Drummond M, Petrou S, Carswell C, Moher D, Greenberg D, et al. Consolidated Health Economic Evaluation Reporting Standards (CHEERS) statement . BMJ 2013;346. http://dx.doi.org/10.1136/bmj.f1049.
- Philips Z, Ginnelly L, Sculpher M, Claxton K, Golder S, Riemsma R, et al. Review of guidelines for good practice in decision-analytic modelling in health technology assessment. Health Technol Assess 2004;8. https://doi.org/10.3310/hta8360.
- Bignardi GE, Settle C. Can the Luminex xTAG gastrointestinal pathogen panel really save money?. J Infect 2015;71:498-9. http://dx.doi.org/10.1016/j.jinf.2015.06.008.
- Binnicker MJ. Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness. J Clin Microbiol 2015;53:3723-8. https://doi.org/10.1128/JCM.02103-15.
- Freedman SB, Lee BE, Louie M, Pang XL, Ali S, Chuck A, et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 2015;15. http://dx.doi.org/10.1186/s12887-015-0407-7.
- Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD. A cost benefit analysis of the Luminex xTAG Gastrointestinal Pathogen Panel for detection of infectious gastroenteritis in hospitalised patients. J Infect 2015;70:504-11. http://dx.doi.org/10.1016/j.jinf.2014.11.009.
- Abubakar I, Irvine L, Aldus CF, Wyatt GM, Fordham R, Schelenz S, et al. A systematic review of the clinical, public health and cost-effectiveness of rapid diagnostic tests for the detection and identification of bacterial intestinal pathogens in faeces and food. Health Technol Assess 2007;11. https://doi.org/10.3310/hta11360.
- NICE . Diarrhoea and Vomiting in Children Overview: NICE Pathway 2015. http://pathways.nice.org.uk/pathways/diarrhoea-and-vomiting-in-children (accessed 1 November 2015).
- Curtis L, Burns A. Unit Costs of Health and Social Care 2015. www.pssru.ac.uk/project-pages/unit-costs/2015/ (accessed 20 May 2016).
- Health Careers . NHS Agenda for Change – Pay Rates 2016. www.healthcareers.nhs.uk/about/careers-nhs/nhs-pay-and-benefits/agenda-change-pay-rates (accessed 20 May 2016).
- Her Majesty’s Treasury . The Green Book: Appraisal and Evaluation in Central Government 2011. www.gov.uk/government/publications/the-green-book-appraisal-and-evaluation-in-central-governent (accessed 20 May 2016).
- Department of Health . NHS Reference Costs 2014 to 2015 2015. www.gov.uk/government/publications/nhs-reference-costs-2014-to-2015 (accessed 20 May 2016).
- HAI Task Force, Health Protection Scotland . NHS Scotland MRSA Screening Pathfinder Programme. Final Report Volume 2: An Assessment of the Economics, Implementation and Modelling of Universal MRSA Screening 2011. www.documents.hps.scot.nhs.uk/hai/mrsa-screening/pathfinder-programme/mrsa-pathfinder-vol2-2011-02-23.pdf (accessed 20 May 2016).
- Allen SJ, Wareham K, Wang D, Bradley C, Sewell B, Hutchings H, et al. A high-dose preparation of lactobacilli and bifidobacteria in the prevention of antibiotic-associated and Clostridium difficile diarrhoea in older people admitted to hospital: a multicentre, randomised, double-blind, placebo-controlled, parallel arm trial (PLACIDE). Health Technol Assess 2013;17. http://dx.doi.org/10.3310/hta17570.
- British National Formulary. London: BMJ Group, Pharmaceutical Press; 2014.
- NICE . Costing Statement: Diarrhoea and Vomiting in Children 2009. www.nice.org.uk/guidance/cg84/resources/costing-statement-242294941 (accessed 20 May 2016).
- Health and Social Care Information Centre . Hospital Episode Statistics, Admitted Patient Care – England, 2014–15 2015. www.hscic.gov.uk/catalogue/PUB19124 (accessed 20 May 2016).
- PHE . Updated Guidance on the Management and Treatment of Clostridium Difficile Infection 2013. www.gov.uk/government/publications/clostridium-difficile-infection-guidance-on-management-and-treatment (accessed 20 May 2016).
- Al-Eidan FA, McElnay JC, Scott MG, Kearney MP. Clostridium difficile-associated diarrhoea in hospitalised patients. J Clin Pharm Ther 2000;25:101-9. https://doi.org/10.1046/j.1365-2710.2000.00266.x.
- Atkins KE, Shim E, Carroll S, Quilici S, Galvani AP. The cost-effectiveness of pentavalent rotavirus vaccination in England and Wales. Vaccine 2012;30:6766-76. http://dx.doi.org/10.1016/j.vaccine.2012.09.025.
- Fisman DN, Chan CH, Lowcock E, Naus M, Lee V. Effectiveness and cost-effectiveness of pediatric rotavirus vaccination in British Columbia: a model-based evaluation. Vaccine 2012;30:7601-7. http://dx.doi.org/10.1016/j.vaccine.2012.10.034.
- Jit M, Edmunds WJ. Evaluating rotavirus vaccination in England and Wales. Part II. The potential cost-effectiveness of vaccination. Vaccine 2007;25:3971-9. https://doi.org/10.1016/j.vaccine.2007.02.070.
- Marlow R, Finn A, Trotter C. Quality of life impacts from rotavirus gastroenteritis on children and their families in the UK. Vaccine 2015;33:5212-6. http://dx.doi.org/10.1016/j.vaccine.2015.07.012.
- Rautenberg TA, Zerwes U, Foerster D, Aultman R. Evaluating the cost utility of racecadotril for the treatment of acute watery diarrhea in children: the RAWD model. Clinicoecon Outcomes Res 2012;4:109-16. http://dx.doi.org/10.2147/CEOR.S31238.
- Tilson L, Jit M, Schmitz S, Walsh C, Garvey P, McKeown P, et al. Cost-effectiveness of universal rotavirus vaccination in reducing rotavirus gastroenteritis in Ireland. Vaccine 2011;29:7463-73. http://dx.doi.org/10.1016/j.vaccine.2011.07.056.
- Martin A, Cottrell S, Standaert B. Estimating utility scores in young children with acute rotavirus gastroenteritis in the UK. J Med Econ 2008;11:471-84. http://dx.doi.org/10.3111/13696990802321047.
- Standaert B, Parez N, Tehard B, Colin X, Detournay B. Cost-effectiveness analysis of vaccination against rotavirus with RIX4414 in France. Appl Health Econ Health Policy 2008;6:199-216. http://dx.doi.org/10.2165/00148365-200806040-00003.
- Rozenbaum MH, Mangen MJ, Giaquinto C, Wilschut JC, Hak E, Postma MJ. Consensus Group on Dutch Rotavirus Vaccination (CoRoVa-Group) . Cost-effectiveness of rotavirus vaccination in the Netherlands; the results of a consensus model. BMC Public Health 2011;11. http://dx.doi.org/10.1186/1471-2458-11-462.
- Minor T, Lasher A, Klontz K, Brown B, Nardinelli C, Zorn D. The per case and total annual costs of foodborne illness in the United States. Risk Anal 2015;35:1125-39. http://dx.doi.org/10.1111/risa.12316.
- BNF for Children. London: BMJ Group, Pharmaceutical Press, and RCPCH Publications; 2015.
- Briggs A, Claxton K, Sculper M. Decision Modelling for Health Economic Evaluation. Oxford: Oxford University Press; 2006.
- Fox M, Mealing S, Anderson R, Dean J, Stein K, Price A, et al. The clinical effectiveness and cost-effectiveness of cardiac resynchronisation (biventricular pacing) for heart failure: systematic review and economic model. Health Technol Assess 2007;11. https://doi.org/10.3310/hta11470.
- Drummond M, McGuire A. Economic Evaluation in Health Care: Merging Theory with Practice. Oxford: Oxford University Press; 2001.
- Buchanan J, Wordsworth S, O’Connor L, Pike G, Walker AS, Wilcox MH, et al. Management of patients with suspected infectious diarrhoea in hospitals in England. J Hosp Infect 2015;90:199-207. http://dx.doi.org/10.1016/j.jhin.2014.12.021.
Appendix 1 Search strategy for clinical effectiveness and cost-effectiveness review
MEDLINE and EMBASE searches
-
exp Gastroenteritis/
-
exp *Diarrhea/
-
exp *Feces/
-
exp *Gastroenteritis/
-
exp *Gastrointestinal Diseases/
-
(gastrointestin* or stool* or enteric* or feces or faeces or diarrh?ea).tw.
-
1 or 2 or 3 or 4 or 5 or 6
-
Multiplex Polymerase Chain Reaction/
-
(xtag or Luminex or Filmarray or biofire).tw.
-
(“Faecal Pathogens B” or “Faecal Panel B” or ausdiagnostics).tw.
-
(multiplex* adj4 (PCR or polymerase chain reaction or assay* or panel* or test*)).tw.
-
(gastrointestinal pathogen panel or gastrointestinal infection panel).tw.
-
8 or 9 or 10 or 11 or 12
-
7 and 13
Ovid MEDLINE: 1946 to November Week 3 2015 – 1424 downloaded.
Ovid MEDLINE In-Process & Other Non-Indexed Citations: 31 December 2015 – 138 downloaded.
Ovid EMBASE: 1980 to 2015 Week 52 – 1803 downloaded.
| Number | Search terms |
|---|---|
| #4 | #3 OR #2 OR #1 DocType=All document types; Language=All languages; |
| #3 | TITLE: (xtag or Luminex or Filmarray or biofire or “Faecal Pathogens B” or “Faecal Panel B” or ausdiagnostics) AND TOPIC: (gastro*) DocType=All document types; Language=All languages; |
| #2 | TS=(“gastrointestinal pathogen panel” or “gastrointestinal infection panel”) DocType=All document types; Language=All languages; |
| #1 | TITLE: (multiplex* and (PCR or polymerase chain reaction or assay* or panel* or test*)) AND TOPIC: (gastrointestin*) DocType=All document types; Language=All languages; |
Downloaded: 98.
Auto-alerts
Weekly auto-alerts were run in Ovid MEDLINE, Ovid EMBASE and PubMed from 1 January 2016 to 31 April 2016 to check for any new studies added subsequent to the main searches.
Search strategies for cost-effectiveness studies
Ovid MEDLINE and Ovid MEDLINE In-Process & Other Non-Indexed Citations
Ovid MEDLINE was searched from 1946 to January week 3 2016; and Ovid MEDLINE In-Process & Other Non-Indexed Citations searched 27 January 2016.
-
exp Gastroenteritis/
-
exp *Diarrhea/
-
exp *Feces/
-
exp *Gastroenteritis/
-
exp *Gastrointestinal Diseases/
-
(gastrointestin* or stool* or enteric* or feces or faeces or diarrh?ea).tw.
-
1 or 2 or 3 or 4 or 5 or 6
-
Multiplex Polymerase Chain Reaction/
-
(xtag or Luminex or Filmarray or biofire).tw.
-
(“Faecal Pathogens B” or “Faecal Panel B” or ausdiagnostics).tw.
-
(multiplex* adj4 (PCR or polymerase chain reaction or assay* or panel* or test*)).tw.
-
(gastrointestinal pathogen panel or gastrointestinal infection panel).tw.
-
8 or 9 or 10 or 11 or 12
-
7 and 13
-
exp Economics/
-
exp “Costs and Cost Analysis”/
-
Health Status/
-
exp “Quality of Life”/
-
exp Quality-Adjusted Life Years/
-
(pharmacoeconomic* or pharmaco-economic* or economic* or cost*).tw.
-
(health state* or health status or “willingness to pay”).tw.
-
(qaly* or ICER* or utilit* or EQ5D or EQ-5D or euroqol or euro-qol or short form or SF-36 or SF36 or SF-6D or SF6D or SF-12 or SF12 or HUI).tw.
-
(markov or time trade off or TTO or standard gamble or hrql or hrqol or disabilit* or disutilit* or net-benefit* or contingent valuation).tw.
-
(quality adj2 life).tw.
-
(decision adj2 model).tw.
-
(visual analog* scale* or discrete choice experiment* or health* year* equivalen* or (willing* adj2 pay)).tw.
-
15 or 16 or 17 or 18 or 19 or 20 or 21 or 22 or 23 or 24 or 25 or 26
-
14 and 27
Downloaded: 144.
Ovid EMBASE
Searched 1974 to 27 January 2016.
-
exp Gastroenteritis/
-
exp *Diarrhea/
-
exp *Feces/
-
exp *Gastroenteritis/
-
exp *Gastrointestinal Diseases/
-
(gastrointestin* or stool* or enteric* or feces or faeces or diarrh?ea).tw.
-
1 or 2 or 3 or 4 or 5 or 6
-
Multiplex Polymerase Chain Reaction/
-
(xtag or Luminex or Filmarray or biofire).tw.
-
(“Faecal Pathogens B” or “Faecal Panel B” or ausdiagnostics).tw.
-
(multiplex* adj4 (PCR or polymerase chain reaction or assay* or panel* or test*)).tw.
-
(gastrointestinal pathogen panel or gastrointestinal infection panel).tw.
-
8 or 9 or 10 or 11 or 12
-
7 and 13
-
exp health economics/
-
exp health status/
-
exp “quality of life”/
-
exp quality adjusted life year/
-
(pharmacoeconomic* or pharmaco-economic* or economic* or cost*).tw.
-
(health state* or health status).tw.
-
(qaly* or ICER* or utilit* or EQ5D or EQ-5D or euroqol or euro-qol or short-form or SF-12 or SF12 or SF-36 or SF36 or SF-6D or SF6D or HUI).tw.
-
(markov or time trade off or TTO or standard gamble or hrql or hrqol or disabilit* or disutilit* or net benefit or contingent valuation).tw.
-
(quality adj2 life).tw.
-
(decision adj2 model).tw.
-
(visual analog* scale* or discrete choice experiment* or health* year* equivalen* or (willing* adj2 pay)).tw.
-
(resource* or quality of well-being or qwb).tw.
-
(utility* adj2 (value* or index* or health or measure* or estimate*)).tw.
-
15 or 16 or 17 or 18 or 19 or 20 or 21 or 22 or 23 or 24 or 25 or 26 or 27
-
14 and 28
Downloaded: 203.
| Number | Search terms |
|---|---|
| #6 | #5 AND #4 |
| #5 | TOPIC: (cost* or economic* or QALY* or quality of life or QoL) OR TOPIC: (health state* or EQ-5D or markov) |
| #4 | #3 OR #2 OR #1 |
| #3 | TOPIC: (xtag or Luminex or Filmarray or biofire or “Faecal Pathogens B” or “Faecal Panel B” or ausdiagnostics) AND TOPIC: (gastro*) |
| #2 | TOPIC: (“gastrointestinal pathogen panel”) OR TOPIC: (“gastrointestinal infection panel”) |
| #1 | TOPIC: (multiplex* and (PCR or polymerase chain reaction or assay* or panel* or test*)) AND TOPIC: (gastrointestin*) |
Downloaded: 30.
Other searches
-
Searched all sections of the Cochrane Database of Systematic Reviews (including the NHS Economic Evaluation Database: Issue 2 of 4, April 2015) – no additional studies found.
-
Cost-Effectiveness Analysis registry (https://research.tufts-nemc.org/cear4/): no additional studies found.
Databases searched for ongoing studies
-
National Institutes of Health ClinicalTrials.gov (www.clinicaltrials.gov/): 1 downloaded.
-
Current Controlled Trials (www.controlled-trials.com).
-
World Health Organization International Clinical Trials Registry Platform (www.who.int/ictrp/en/).
-
UK Clinical Trials Gateway (www.nihr.ac.uk/research/uk-clinical-trials-gateway.htm).
Appendix 2 Information submitted by companies
BioFire Diagnostics (FilmArray Gastrointestinal Panel)
-
FilmArray GI Panel Information Sheet.
-
FilmArray v1.5 user manual.
-
FilmArray v2.0 user manual.
-
FilmArray GI Laboratory Verification Advisory Notice.
-
Zeptometrix Panel for FilmArray GI Panel.
-
BioFire entericpathogens Final DR.
-
Selection of Publications.
-
WMMG abstract 26 May.
-
FilmArray GI Panel CE Declaration of Conformity.
-
IFU for FilmArray GI Panel.
-
FDA decision summary.
-
510K summary.
-
Summary for 2.0 and FAIV for GI.
-
GI Panel Publications.
-
FLM1-MKT-0126 FilmArray Gastrointestinal (GI) Panel Publications Summary.
-
Request for Information.
-
The Film Array GI Panel IMPACT Study.
-
FilmArray Powerpoint Pioneering Diagnostics.
AusDiagnostics (Faecal Pathogens B)
AusDiagnostics response
Content of request for information.
-
Response to Request for Information.
-
Appendix 1: Clinical Evidence Document: Clinical Evidence Report Faecal Pathogens High-Plex Kits.
-
Appendix 2: CE conformity and IFUs.
-
Instructions for Use High-Plex Faecal Pathogens Kit.
-
Instructions for Use High-Plex Diagnostic system Cat No 9150 Easy-Plex Processor and High Plex Analyser.
Luminex (xTAG® Gastrointestinal Pathogen Panel)
-
xTAG GPP Kit Package Insert.
-
Company Insight: Multiple Answers from a single sample.
-
Company Insight: Gastroenteritis: A serious medical and economic burden – a new approach.
-
Company Insight: Less pain with gastroenteritis.
-
NxTAG Respiratory Pathogen Panel Package Insert.
-
xTAG GPP (US) for use with MAGPIX.
-
Luminex Response to Request for information.
-
Declaration of Conformity.
-
xTAG Gastrointestinal Pathogen Panel – The state of the Art in Molecular GI Testing. Review of Literature and Summary of Publication works.
-
GPP consumables (pdf and Excel).
-
Gastroenteritis and “doing more with less” sponsored feature in hospitalhealthcare.com.
Appendix 3 Data extraction form
Data extraction form: clinical effectiveness of GPP for gastroenteritis.
First reviewer: Second reviewer:
| Study details | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study ID (EndNote reference) | |||||||||||||||||||||||||
| First author surname | |||||||||||||||||||||||||
| Year of publication | |||||||||||||||||||||||||
| Country | |||||||||||||||||||||||||
| Study setting | |||||||||||||||||||||||||
| Number of centres | |||||||||||||||||||||||||
| Duration of study | |||||||||||||||||||||||||
| Follow up (if applicable) | |||||||||||||||||||||||||
| Funding | |||||||||||||||||||||||||
| Aim of the study | |||||||||||||||||||||||||
| Inclusion/exclusion criteria for patients | |||||||||||||||||||||||||
| Inclusion criteria: | |||||||||||||||||||||||||
| Exclusion criteria: | |||||||||||||||||||||||||
| Participants | |||||||||||||||||||||||||
| Item | Hospital | Community | Total | ||||||||||||||||||||||
| Definition of subgroups | |||||||||||||||||||||||||
| Total number of patients (samples) at baseline | |||||||||||||||||||||||||
| Number followed up | |||||||||||||||||||||||||
| Description of study format (number of patients and reasons for receiving index test, validation test comparator test) | |||||||||||||||||||||||||
| Baseline characteristics of included patients | |||||||||||||||||||||||||
| Number included in analysis | |||||||||||||||||||||||||
| Adults (n) | |||||||||||||||||||||||||
| Children aged < 5 years (n) | |||||||||||||||||||||||||
| Travellers (n) | |||||||||||||||||||||||||
| Immunocompromised people (n) | |||||||||||||||||||||||||
| Index test | |||||||||||||||||||||||||
| Name of pathogen panel test(s) | |||||||||||||||||||||||||
| Proportion having received index test (n/N) | |||||||||||||||||||||||||
| Method of batching | |||||||||||||||||||||||||
| Pathogens tested: | |||||||||||||||||||||||||
| Adenovirus 40/41 (x/F/B) | |||||||||||||||||||||||||
| Astrovirus (F/B) | |||||||||||||||||||||||||
| C. difficile toxin A/B (x/F/B) | |||||||||||||||||||||||||
| Campylobacter (x/F/B) | |||||||||||||||||||||||||
| Cryptosporidium (x/F/B) | |||||||||||||||||||||||||
| Cyclospora cayetanensis (F) | |||||||||||||||||||||||||
| E. coli O157 (x/F/B) | |||||||||||||||||||||||||
| EAEC (F) | |||||||||||||||||||||||||
| E. histolytica (x/F/B) | |||||||||||||||||||||||||
| EPEC (F) | |||||||||||||||||||||||||
| ETEC (x/F) | |||||||||||||||||||||||||
| Giardia (x/F/B) | |||||||||||||||||||||||||
| Norovirus (x/F/B) | |||||||||||||||||||||||||
| Plesiomonas shigelloides (F) | |||||||||||||||||||||||||
| Rotavirus (x/F/B) | |||||||||||||||||||||||||
| Salmonella (x/F/B) | |||||||||||||||||||||||||
| Sapovirus (F/B | |||||||||||||||||||||||||
| Shigella (x/F/B) | |||||||||||||||||||||||||
| STEC (x/F/B) | |||||||||||||||||||||||||
| Vibrio (parahaemolyticus, vulnificus and cholerae) (F) | |||||||||||||||||||||||||
| V. cholerae (x/F) | |||||||||||||||||||||||||
| Y. enterocolitica (x/F) | |||||||||||||||||||||||||
| Conventional method (comparator) | |||||||||||||||||||||||||
| Did all patients receive at least one conventional test? | |||||||||||||||||||||||||
| Did all patients receive all available conventional tests? | |||||||||||||||||||||||||
| Methods and proportion having received each test: | |||||||||||||||||||||||||
| Adenovirus 40/41 | |||||||||||||||||||||||||
| Astrovirus | |||||||||||||||||||||||||
| C. difficile toxin A/B | |||||||||||||||||||||||||
| Campylobacter | |||||||||||||||||||||||||
| Cryptosporidium | |||||||||||||||||||||||||
| E. coli O157 | |||||||||||||||||||||||||
| Giardia | |||||||||||||||||||||||||
| Norovirus | |||||||||||||||||||||||||
| P. shigelloides | |||||||||||||||||||||||||
| Rotavirus | |||||||||||||||||||||||||
| Salmonella | |||||||||||||||||||||||||
| Sapovirus | |||||||||||||||||||||||||
| Shigella | |||||||||||||||||||||||||
| V. cholerae | |||||||||||||||||||||||||
| Other (not specified in PHE algorithm, please specify) | |||||||||||||||||||||||||
| Validation method (reference standard/fair umpire) | |||||||||||||||||||||||||
| Define patients who received validation method (e.g. GPP+/conventional– discordant only) | |||||||||||||||||||||||||
| Place where validation method was undertaken (same laboratory as index test?) | |||||||||||||||||||||||||
| Validation method (judgement biased/unbiased): | |||||||||||||||||||||||||
| Adenovirus 40/41 (x/F/B) | |||||||||||||||||||||||||
| Astrovirus (F/B) | |||||||||||||||||||||||||
| C. difficile toxin A/B (x/F/B) | |||||||||||||||||||||||||
| Campylobacter (x/F/B) | |||||||||||||||||||||||||
| Cryptosporidium (x/F/B) | |||||||||||||||||||||||||
| C. cayetanensis (F) | |||||||||||||||||||||||||
| E. coli O157 (x/F/B) | |||||||||||||||||||||||||
| EAEC (F) | |||||||||||||||||||||||||
| E. histolytica (x/F/B) | |||||||||||||||||||||||||
| EPEC (F) | |||||||||||||||||||||||||
| ETEC (x/F) | |||||||||||||||||||||||||
| Giardia (x/F/B) | |||||||||||||||||||||||||
| Norovirus (x/F/B) | |||||||||||||||||||||||||
| P. shigelloides (F) | |||||||||||||||||||||||||
| Rotavirus (x/F/B) | |||||||||||||||||||||||||
| Salmonella (x/F/B) | |||||||||||||||||||||||||
| Sapovirus (F/B) | |||||||||||||||||||||||||
| Shigella (x/F/B) | |||||||||||||||||||||||||
| STEC (x/F/B) | |||||||||||||||||||||||||
| Vibrio (parahaemolyticus, vulnificus and cholerae) (F) | |||||||||||||||||||||||||
| V. cholerae (x/F) | |||||||||||||||||||||||||
| Y. enterocolitica (x/F) | |||||||||||||||||||||||||
| Outcomes | |||||||||||||||||||||||||
| Specify measure(s) of test agreement reported (e.g. test accuracy, kappa, concordance, agreement) | |||||||||||||||||||||||||
| Record 3 × 2 table by pathogen reported in the following format: | Conventional method +Conventional method –Conventional not testedDescribe conventional methodDescribe validation methodGPP+a (verified +/–)b (verified +/–)# (verified +/–)GPP–c (verified +/–)d (verified +/–)# (verified +/–)–, negative; +, positive. | Conventional method + | Conventional method – | Conventional not tested | Describe conventional method | Describe validation method | GPP+ | a (verified +/–) | b (verified +/–) | # (verified +/–) | GPP– | c (verified +/–) | d (verified +/–) | # (verified +/–) | –, negative; +, positive. | ||||||||||
| Conventional method + | Conventional method – | Conventional not tested | Describe conventional method | Describe validation method | |||||||||||||||||||||
| GPP+ | a (verified +/–) | b (verified +/–) | # (verified +/–) | ||||||||||||||||||||||
| GPP– | c (verified +/–) | d (verified +/–) | # (verified +/–) | ||||||||||||||||||||||
| –, negative; +, positive. | |||||||||||||||||||||||||
| Adenovirus 40/41 (x/F/B) | |||||||||||||||||||||||||
| Astrovirus (F/B) | |||||||||||||||||||||||||
| C. difficile toxin A/B (x/F/B) | |||||||||||||||||||||||||
| Campylobacter (x/F/B) | |||||||||||||||||||||||||
| Cryptosporidium (x/F/B) | |||||||||||||||||||||||||
| C. cayetanensis (F) | |||||||||||||||||||||||||
| E. coli O157 (x/F/B) | |||||||||||||||||||||||||
| EAEC (F) | |||||||||||||||||||||||||
| E. histolytica (x/F/B) | |||||||||||||||||||||||||
| EPEC (F) | |||||||||||||||||||||||||
| ETEC (x/F) | |||||||||||||||||||||||||
| Giardia (x/F/B) | |||||||||||||||||||||||||
| Norovirus (x/F/B) | |||||||||||||||||||||||||
| P. shigelloides (F) | |||||||||||||||||||||||||
| Rotavirus (x/F/B) | |||||||||||||||||||||||||
| Salmonella (x/F/B) | |||||||||||||||||||||||||
| Sapovirus (F/B) | |||||||||||||||||||||||||
| Shigella (x/F/B) | |||||||||||||||||||||||||
| STEC (x/F/B) | |||||||||||||||||||||||||
| Vibrio (parahaemolyticus, vulnificus and cholerae) (F) | |||||||||||||||||||||||||
| V. cholerae (x/F) | |||||||||||||||||||||||||
| Y. enterocolitica (x/F) | |||||||||||||||||||||||||
| Overall 2 × 2 table results | |||||||||||||||||||||||||
| Patients with infections with index test (n) | |||||||||||||||||||||||||
| Patients with one pathogen (n) | |||||||||||||||||||||||||
| Patients with coinfections (n) | |||||||||||||||||||||||||
| With two pathogens (n) | |||||||||||||||||||||||||
| With three or more pathogens (n) | |||||||||||||||||||||||||
| Patients with infections with comparator (n) | |||||||||||||||||||||||||
| Patients with infections following validation (n) | |||||||||||||||||||||||||
| Change in treatment/management by test outcome | |||||||||||||||||||||||||
| Time spent in isolation | |||||||||||||||||||||||||
| Patients isolated | |||||||||||||||||||||||||
| Patients removed from isolation (n) | |||||||||||||||||||||||||
| Patients not removed from isolation | |||||||||||||||||||||||||
| Patients taken off/change in antibiotic treatment (n) | |||||||||||||||||||||||||
| Test failure rates | |||||||||||||||||||||||||
| Turnaround time | |||||||||||||||||||||||||
| Costs | |||||||||||||||||||||||||
| Morbidity | |||||||||||||||||||||||||
| Mortality | |||||||||||||||||||||||||
| HRQoL | |||||||||||||||||||||||||
| Other outcomes | |||||||||||||||||||||||||
| Authors’ conclusion | |||||||||||||||||||||||||
| Reviewer’s notes | |||||||||||||||||||||||||
Appendix 4 Quality Assessment of Diagnostic Accuracy Studies-2 quality appraisal tool and guidance notes
Guidance notes for study assessment using tailored Quality Assessment of Diagnostic Accuracy Studies-2
Risk of bias should be classed as low for each domain only if all questions could be answered with ‘yes’. If one or more signalling question is answered with ‘no’, the risk of bias should be classed as ‘high’ and, equally, if at least one question is answered with ‘unclear’, the risk of bias should be judged ‘unclear’.
Domain 1: patient selection
The study setting is primarily the microbiology laboratory, which is why samples are more frequently collected into the study than patients. Consecutive or random samples should be included in the study, patients should not have received prior testing and only one sample per episode should be considered in the study.
A. Risk of bias
Was a consecutive or random sample of patients enrolled?
This question should be answered with ‘yes’ only if the study clearly states that samples or patients were recruited consecutively or randomly (either retrospectively or prospectively).
Was a case–control design avoided?
This question can be answered with ‘yes’ if the study design is a prospective cohort. If the study is a case–control study this question should be answered with ‘no’.
Did the study avoid inappropriate exclusions?
If the study excludes > 10% of participants with or without specifying reasons, the exclusions should be considered as inappropriate and the question answered with ‘no’. This cut-off point has been determined pragmatically. The exclusion of samples that do not conform with the shape of the container is not classed as inappropriate exclusions as these would not be eligible patients for testing.
Was only one sample per episode included in the study?
If the sample size is equal to the number of patients enrolled in the study, this question can be answered with ‘yes’. If the study included only samples and did not specify whether or not there was one sample per episode per patient, the question should be answered with ‘unclear’. If the study included more samples than patients, the question should be answered with ‘no’ unless the study clearly states that only one sample was included per episode or specifies 1 month between samples (marking different episodes).
B. Concerns regarding applicability
If the setting is a microbiology laboratory and samples are from patients with diarrhoea, concerns regarding applicability are ‘low’. If there is uncertainty about the samples originating from patients with diarrhoea or samples from only test-negative (by conventional test) patients are included in the study, the study population is questionable and the concern of applicability should be classed as ‘unclear’ and ‘high’, respectively.
For studies undertaken in (developing) countries (including countries in Africa, Asia and South America) that are expected to have considerably different prevalences of pathogens, applicability of study results should be of ‘high’ concern.
If the study included a mixed population in terms of setting and subpopulations of interest (i.e. travellers, immunocompromised patients, children aged ≤ 5 years), the study population can be regarded as applicable to the population in which the test is likely to be undertaken (general clinical laboratory receiving samples from community, outpatient, accident and emergency and hospitalised patients). If the population is travellers or immunocompromised patients only, the study population is not applicable to the population in which the test is likely to be undertaken in, thus applicability concerns should be rated as ‘high’.
Domain 2: index test
The main sources of bias introduced by conducting and interpreting an index test are blinding and defining the threshold. The index test in this assessment is a laboratory testing kit that is analysed by computer software and results reported as ‘yes’ or ‘no’ following a pre-specified threshold to judge (1) elevated relative fluorescence (xTAG) and (2) fluorescent images of the DNA melt curve analysis of the PCR reactions (FilmArray). Blinding of personnel to the results of the comparator or the verification method is a ‘no bias’ issue.
A. Risk of bias
Were the index test results interpreted without knowledge of the results of the comparator and verification method?
This question can be answered with ‘yes’, as the test is completely objective.
Was a threshold explicitly pre-specified?
This question can be answered with ‘yes’, as the threshold is pre-specified within the GPP test and if the test or its interpretation has not been changed.
Add: did all samples receive the index test?
Answer the question with ‘yes’ if the study did not specify any selective testing with the GPP test.
Add: was the index test undertaken as recommended by the manufacturer?
At least one study reported deviation from manufacturer’s protocol because it used stool in Cary–Blair medium and not raw sample. However, the 510(k)Substantial Equivalence Determination Decision Summary40 submitted to the FDA added a claim for human stool in Cary–Blair medium. Therefore, stool samples in Cary–Blair is not a deviation from the recommended protocol. This question can be answered with ‘yes’ if the study clearly states that the protocol followed the manufacturer’s recommendations; it is ‘unclear’ if nothing is specified (or we shall assume that it was undertaken according to protocol) or if the GPP assay was undertaken by Luminex and BioFire, retrospectively. This question should be answered with ‘no’ if the recommended time period for GPP testing (within 3 days) was not adhered to or if the specimens were not frozen for longer time periods between sampling and testing with GPP (testing within 2–3 days or immediate freezing at –70 °C).
B. Concerns about applicability
If the GPP test is used per manufacturer’s recommendations, the concern of applicability of the index test is ‘low’.
Add Domain 3: comparator
The main sources of bias introduced by conducting and interpreting the comparator are blinding, defining the threshold and performing culture on frozen samples.
Blinding would be of concern if the comparator (conventional test) was undertaken following the GPP test. In that case, risk of bias should be judged as ‘high’. Blinding to the results of the verification method is of no concern, as only discordant (if any) results were verified, which means that verification took place after the comparator test.
Laboratories follow criteria for how to interpret conventional methods, which vary between tests and can be subjective (e.g. culture for bacteria and serotyping). Therefore, thresholds will vary between laboratories and individuals even with criteria specified in clinical practice. The levels of concern for the various types of tests vary as, for instance, a subjective threshold for the quantitative measure of PCRs can be pre-specified or a testing kit will report a yes/no outcome. It is of importance that studies clearly describe or reference criteria that have been used.
If not all samples were tested for all pathogens (physician chooses tests) using the comparator methods specified, the results will be highly biased, as this decision is unlikely to be independent of previous assessments and patient selection, and this question should therefore be answered with ‘no’.
Undertaking bacterial culture on previously frozen samples reduces accuracy and results in increased discrepancies. Therefore, studies that used culture of frozen samples should be classed as having a ‘high’ risk of bias.
A. Risk of bias
Were the comparator results interpreted without knowledge of the results of the index test and verification method?
If the comparator (mostly conventional tests) is undertaken following the GPP test, the risk of bias should be judged as ‘high’ unless the tests are undertaken in different laboratories.
Was a threshold explicitly pre-specified?
Conventional microbiological methods in standard clinical laboratories follow criteria in the interpretation of tests (in the UK these need to meet UK standards), which vary between tests. For PCR it can be quantitative (the number of cycles after which a positive result must be detectable at a given level), for others it is qualitative and defined by meeting a set of conditions (e.g. that something is growing under particular growth conditions AND that it has a defined appearance on staining and viewing down a microscope AND causes a particular biochemical reaction); these can be subjective.
For the assessment of threshold used in the studies, we would expect that studies report or reference the criteria used for interpretation of results in order to answer this question with ‘yes’. Otherwise, this question needs to be answered with ‘no’.
Add: was culture performed on fresh (not previously frozen) samples?
This question should be answered with ‘no’ only if the study clearly states that culture was undertaken from frozen samples.
B. Concerns about applicability
Concerns about the applicability of the study to the research question are high if the comparator methods are not consistent with the methods specified in the PHE algorithm. ‘High’ concern about applicability should, therefore, be chosen if the conventional methods include multiplexing assays, PCR and sequencing, culture confirmed by serotyping or sequencing. Concerns about applicability are also ‘high’ on study level if the study included fewer pathogens than included in the PHE algorithm and if the clinician requested the pathogens that should be tested for with the conventional test.
The definition for this study is the following:
The PHE algorithm8 consists of primary testing for Salmonella, Campylobacter, Shigella, E. coli O157, Cryptosporidium and C. difficile with additional tests for children aged < 5 years for rotavirus, adenovirus 40/41 and norovirus. Secondary testing in hospitalised adults is recommended for norovirus and parasites including Giardia, whereas people in the community should not be tested for norovirus and children aged < 5 years should also be tested for astro- and sapovirus. Furthermore, our assessment included travellers as a subgroup of interest for whom additional tests for Vibrio and Plesiomonas species are considered if returning from areas other than Western Europe, North America, Australia or New Zealand. Physicians following this algorithm use local tests that vary across the UK and may or may not request all tests detailed in the PHE algorithm, depending on symptoms and suspicion following patient assessment. In order to compare GPP testing to microbiology techniques, as outlined in the PHE syndromic algorithm for routine testing, a clear definition of the comparator is needed.
For the purpose of this review, no differentiation is being made between primary and secondary testing or differences in terms of population testing (children, adults, travellers) and seasonality (including norovirus testing only in winter months). For a sufficiently equivalent comparator, all pathogens mentioned in the syndromic PHE pathway8 for any of the populations of interest are considered. A pragmatic judgement was required regarding whether or not the number of tested pathogens in the studies is sufficiently similar to the PHE algorithm8 [i.e. at least 75% (≥ 11 of previously specified pathogens) included in study].
In terms of tests, the following was assumed – testing is classed as sufficiently equivalent to the PHE algorithm8 if the comparator in the studies includes:
-
culture or PCR for bacteria
-
PCR or enzyme immunoassay (EIA) for viruses
-
microscopy or EIA for parasites
-
PCR and/or EIA for C. difficile.
Domain 4: reference standard
As there is no independent and reliable reference standard, we would at least expect an independent and unbiased umpire test to be used in the studies to assess discordant results between index test and conventional methods [e.g. exposure, treatment effect (pathogen-specific treatment) or self-reported symptoms, previous antibiotic treatment, over-the-counter medicine].
Verification of discordant results by retesting, by optimising index test and conventional methods or by undertaking individual PCRs with primers different from primers used in the GPP assay is still biased, as PCR assays are not sufficiently different from GPP tests (i.e. index test), which are also based on PCR methodology that identifies all nucleic acids and does not differentiate between viable organisms, colonisation and dead organisms.
Blinding is of concern for the verification method, as it is undertaken following GPP and conventional testing.
A. Risk of bias
Is the reference standard likely to correctly classify the target condition?
Answer this question with ‘no’ if conventional methods were used as reference standard or if GPP was used as reference standard. Answer the question with ‘no’ if the verification method or the fair umpire was biased and insufficiently independent of either the conventional methods or the index test. This question can only be answered with ‘yes’ if a fair umpire (unbiased and independent) was used to solve discordant test results.
Were the reference standard results interpreted without knowledge of the results of the index test?
This question should be answered with ‘no’ if no blinding was clearly stated for the interpretation of the verification methods if they were undertaken after the index test.
Add: was the reference standard independent and unbiased?
This question can only be answered with ‘yes’ if a fair umpire test based on exposure or treatment was used to assess discordant results. Otherwise the question should be answered with ‘no’ and risk of bias classed as ‘high’.
B. Concerns about applicability
This question assesses whether or not the target condition identified by the reference standard matches the target condition of interest in the review. The target condition for this assessment is the pathogen causing symptoms of diarrhoea. If the reference standard detects pathogen DNA/ribonucleic acid at levels that are unlikely to be cause for symptoms or detect dead organisms, then the applicability concern is high. Therefore, the concerns about the applicability of the reference standard are ‘high’ if discrepant results were investigated with PCR-based methods and the comparator methods for at least some pathogens were conventional non-PCR microbiology tests.
Domain 5: flow and timing
A. Risk of bias
Was there an appropriate interval between index test(s) and reference standard?
As there is no disease progression or treatment effect if both tests are undertaken on the same sample, this question can be answered with ‘yes’ if it is clear that both tests were performed on the same sample. If the tests were undertaken on different samples the risk of bias should be regarded as ‘high’, as gastroenteritis is an acute condition that often resolves in a few days.
Did all discordant samples receive a reference standard?
If the study did not undertake verification of discordant results or only a proportion of discordant samples were verified, this question should be answered with ‘no’.
Did all patients receive the same reference standard?
Methods are different for different pathogens. This is not classed as differential verification as the GPP test is assessed on the pathogen level. If any of the pathogens in the study was verified with different methods, this question should be answered with ‘no’. This question should be answered with ‘yes’ if the verification method was the same for all patients for individual pathogens. For any pathogen samples that were still discrepant following initial verification, and some had to be followed up by additional, different testing, this question should be answered with ‘no’.
Add: did all samples receive the comparator methods for all pathogens considered in the study?
This question should only be answered with ‘yes’ if the study clearly states that all samples received the comparator methods for all pathogens considered in the study. This question needs to be answered with ‘no’ if the study reports that physicians requested the number of conventional tests for each sample (resulting in different totals in the 2 × 2 table across pathogens) and risk of bias should be classed as ‘high’.
Were all patients included in the analysis?
If inhibited or undetermined results were excluded from the analysis, this question should be answered with ‘no’.
Appendix 5 List of excluded full texts and abstracts with reasons
| Study reference | Reason for exclusion |
|---|---|
| Abbasi P, Kargar M, Doosti A, Mardaneh J, Ghorbani-Dalini S, Dehyadegari MA. Characterization of shiga-toxin producing E. coli (STEC) and enteropathogenic E. coli (EPEC) using multiplex real-time PCR assays for stx1, stx2, eaeA. Iranian J Microbiol 2014;6:169–74 | Not index test |
| Al-Talib H, Latif B, Mohd-Zain Z. Pentaplex PCR assay for detection of hemorrhagic bacteria from stool samples. J Clin Microbiol 2014;52:3244–9 | Not index test |
| Antikainen J, Kantele A, Pakkanen SH, Laaveri T, Riutta J, Vaara M, et al. A quantitative polymerase chain reaction assay for rapid detection of 9 pathogens directly from stools of travellers with diarrhea. Clin Gastroenterol Hepatol 2013;11:1300–7.e3 | Not index test |
| Antonishyn NA, Crozier NA, McDonald RR, Levett PN, Horsman GB, et al. Rapid detection of norovirus based on an automated extraction protocol and a real-time multiplexed single-step RT-PCR. J Clin Virol 2006;37:156–61 | Not index test |
| Assis FEA, Wolf S, Surek M, De Toni F, Souza EM, Pedrosa FO, et al. Impact of aeromonas and diarrheagenic Escherichia coli screening in patients with diarrhea in Parana, Southern Brazil. J Infect Dev Ctries 2014;8:1609–14 | Not index test |
| Barra-Carrasco J, Hernandez-Rocha C, Miranda-Cardenas C, Alvarez-Lobos M, Guzman Duran AM, Paredes-Sabja D. Diagnostic accuracy of a multiplex real-time PCR to predict Clostridium difficile ribotype 027. Anaerobe 2013;22:115–17 | Letter to editor |
| Bonkoungou IJ, Lienemann T, Martikainen O, Dembele R, Sanou I, Traore AS, et al. Diarrhoeagenic Escherichia coli detected by 16-plex PCR in children with and without diarrhoea in Burkina Faso. Clin Microbiol Infect 2012;18:901–6 | Not index test |
| Bonkoungou IJ, Lienemann T, Martikainen O, Dembele R, Sanou I, Traore AS, et al. Comparison of multiplex PCR with serogrouping and PCR-RFLP of fliC gene for the detection of enteropathogenic Escherichia coli (EPEC). Braz J Infect Dis 2011;15:365–9 | Not index test |
| Bouzari S, Jafari A, Zarepour M. Distribution of virulence related genes among enteroaggregative Escherichia coli isolates: using multiplex PCR and hybridization. Infect Genet Evolut 2005;5:79–83 | Not index test |
| Bruijnesteijn van Coppenraet LES, Dullaert-de Boer M, Ruijs GJHM, van der Reijden WA, van der Zanden AGM, Weel JFL, et al. Case–control comparison of bacterial and protozoan microorganisms associated with gastroenteritis: application of molecular detection. Clin Microbiol Infect 2015;21:592.e9–92.e19 | Not index test |
| Bubba L, Pellegrinelli L, Pariani E, Primache V, Amendola A, Binda S. A novel multiplex one-step real-time RT-PCR assay for the simultaneous identification of enterovirus and parechovirus in clinical fecal samples. J Prev Med Hyg 2015;56:E57–60 | Not index test |
| Bueris V, Sircili MP, Taddei CR, dos Santos MF, Franzolin MR, Martinez MB, et al. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Memorias do Instituto Oswaldo Cruz 2007;102:839–44 | Not index test |
| Catanzaro M, Cirone J. Real-time polymerase chain reaction testing for Clostridium difficile reduces isolation time and improves patient management in a small community hospital. Am J Infect Control 2012;40:663–6 | Not index test |
| Chavada R, Maley M. Evaluation of a commercial multiplex PCR for rapid detection of multi drug resistant Gram negative infections. Open Microbiol J 2015;9:125–35 | Not stool samples |
| Chen Y, Li Z, Han D, Cui D, Chen X, Zheng S, et al. Viral agents associated with acute diarrhea among outpatient children in southeastern China. Pediatr Infect Dis J 2013;32:e285–90 | Not index test |
| Chiu CH, Ou JT. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol 1996;34:2619–22 | Not index test |
| de Boer RF, Ott A, Kesztyus B, Kooistra-Smid AM. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol 2010;48:4140–6 | Not index test |
| de Boer RF, Wijma JJ, Schuurman T, Moedt J, Dijk-Alberts BG, Ott A, et al. Evaluation of a rapid molecular screening approach for the detection of toxigenic Clostridium difficile in general and subsequent identification of the tcdC DELTA117 mutation in human stools. J Microbiol Methods 2010;83:59–65 | Not index test |
| Di Cristanziano V, Timmen-Wego M, Lubke N, Kaiser R, Pfister H, Di Cave D, et al. Application of Luminex Gastrointestinal Pathogen Panel to human stool samples from Côte d’Ivoire. J Infect Dev Ctries 2015;9:884–9 | Diarrhoeal patients < 80% of study population [3/34 (9%)] |
| Dunbar SA. Molecular revolution entering GI diagnostic testing. MLO Med Lab Obs 2013;45:28 | Review |
| Dung TT, Phat VV, Nga TV, My PV, Duy PT, Campbell JI, et al. The validation and utility of a quantitative one-step multiplex RT real-time PCR targeting rotavirus A and norovirus. J Virol Methods 2013;187:138–43 | Not index test |
| El Metwally HAR, Ibrahim HAH, El-Athamna MN, Amer MA. Multiplex PCR for detection of diarrheagenic Escherichia coli in Egyptian children. J Med Sci 2007;7:255–62 | Not index test |
| Freedman SB, Lee BE, Louie M, Pang XL, Ali S, Chuck A, et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 2015;15:89 | No outcomes |
| Frickmann H, Warnke P, Frey C, Schmidt S, Janke C, Erkens K, et al. Surveillance of food- and smear-transmitted pathogens in European soldiers with diarrhea on deployment in the tropics: experience from the European Union Training Mission (EUTM) Mali. BioMed Res Int 2015;2015:573904 | Not index test |
| Garcia C, Chincha O, Leon M, Iglesias D, Barletta F, Mercado E, et al. High frequency of diarrheagenic Escherichia coli in human immunodeficiency virus (HIV) patients with and without diarrhea in Lima, Peru. Am J Trop Med Hyg 2010;82:1118–20 | Not index test |
| Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD. A cost benefit analysis of the Luminex xTAG Gastrointestinal Pathogen Panel for detection of infectious gastroenteritis in hospitalised patients. J Infect 2015;70:504–11 | Not test accuracy |
| Hagen RM, Adlkofer J, Acqua S, Sarpong N, Priesnitz S, Adu-Sarkodie Y, et al. Multiplex/realtime PCR in the laboratory diagnosis and for the epidemiological screening of pathogens causing diarrhea in early childhood in Ghana. Trop Med Int Health 2009;14:58 | Not index test |
| Hegde A, Ballal M, Shenoy S. Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J Med Microbiol 2012;30:279–84 | Not index test |
| Heyworth MF. Diagnostic testing for Giardia infections. Trans R Soc Trop Med Hyg 2014;108:123–5 | Review |
| Hopkins MJ, Booth JA, Cunliffe NA, Hart IJ. Validation of a real-time multiplex PCR protocol for diagnosis of viral gastroenteritis. Clin Microbiol Infect 2012;18:198–9 | Not index test |
| Hussein EM. Molecular identification of Cyclospora spp. using multiplex PCR from diarrheic children compared to others conventional methods. J Egyptian Soc Parasitol 2007;37:585–98 | Not index test |
| Jasuja J, Veit J, Fofana HKM, Nimmesgern A, Saye R, Doumbia MN, et al. Stool-based polymerase chain reaction for the diagnosis of multiple pathogens in Mali: a case–control study. Trop Med Int Health 2015;20:142 | Not index test |
| Jex AR, Stanley KK, Lo W, Littman R, Verweij JJ, Campbell BE, et al. Detection of diarrhoeal pathogens in human faeces using an automated, robotic platform. Mol Cell Probes 2012;26:11–15 | Not index test |
| Kim S, Frye JG, Hu J, Fedorka-Cray PJ, Gautom R, Boyle DS. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J Clin Microbiol 2006;44:3608–15 | Not index test |
| Li W, Zhang N, Gong P, Cao L, Li J, Su L, et al. A novel multiplex PCR coupled with Luminex assay for the simultaneous detection of Cryptosporidium spp., Cryptosporidium parvum and Giardia duodenalis. Vet Parasitol 2010;173:11–18 | Not index test |
| Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, et al. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-luminex assay. J Clin Microbiol 2012;50:98–103 | Not index test |
| Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 2014;14:716–24 | Not index test |
| Liu J, Kibiki G, Maro V, Maro A, Kumburu H, Swai N, et al. Multiplex reverse transcription PCR Luminex assay for detection and quantitation of viral agents of gastroenteritis. J Clin Virol 2011;50:308–13 | Not index test |
| Maas L, Dorigo-Zetsma JW, de Groot CJ, Bouter S, Plotz FB, van Ewijk BE. Detection of intestinal protozoa in paediatric patients with gastrointestinal symptoms by multiplex real-time PCR. Clin Microbiol Infect 2014;20:545–50 | Not index test |
| Malecki M, Schildgen V, Kamm M, Mattner F, Schildgen O. Rapid screening method for multiple gastro-enteric pathogens also detects novel EHEC O104:H4. Int J Med Microbiol 2011;301:38 | Letter to Editor |
| Navidad JF, Griswold DJ, Gradus MS, Bhattacharyya S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J Clin Microbiol 2013;51:3018–24 | Not index test (not xTAG commercialised kit) |
| Nazeer JT, El Sayed Khalifa K, von Thien H, El-Sibaei MM, Abdel-Hamid MY, Tawfik RA, et al. Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol Res 2013;112:595–601 | Not index test |
| Pavlinac PB, Onchiri FM, John-Stewart GC, Naulikha JM, Donna DM, Odundo EA, et al. Accuracy of the integrated management of childhood illness (IMCI) algorithm in identifying culture-confirmed diarrheal pathogens requiring antibiotic therapy. Am J Trop Med Hyg 2014;1:574–5 | Not index test |
| Qin X, Klein EJ, Galanakis E, Thomas AA, Stapp JR, Rich S, et al. Real-time PCR assay for detection and differentiation of shiga toxin-producing Escherichia coli from clinical samples. J Clin Microbiol 2015;53:2148–53 | Not index test |
| Siah SP, Merif J, Kaur K, Nair J, Huntington PG, Karagiannis T, et al. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology 2014;46:53–9 | Not index test |
| Sjoling A, Sadeghipoorjahromi L, Novak D, Tobias J. Detection of major diarrheagenic bacterial pathogens by multiplex PCR panels. Microbiol Res 2015;172:34–40 | Not index test |
| So CW, Kim DS, Yu ST, Cho JH, Kim JD. Acute viral gastroenteritis in children hospitalized in Iksan, Korea during December 2010–June 2011. Korean J Pediatr 2013;56:383–8 | Not index test |
| Stark D, Al-Qassab SE, Barratt JL, Stanley K, Roberts T, Marriott D, et al. Evaluation of multiplex tandem real-time PCR for detection of Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis in clinical stool samples. J Clin Microbiol 2011;49:257–62 | Not index test |
| Taniuchi M, Verweij JJ, Noor Z, Sobuz SU, Lieshout L, Petri WA Jr, et al. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg 2011;84:332–7 | Not index test |
| ten Hove R, Schuurman T, Kooistra M, Moller L, van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect 2007;13:1001–7 | Not index test |
| Tobias J, Kassem E, Rubinstein U, Bialik A, Vutukuru SR, Navaro A, et al. Involvement of main diarrheagenic Escherichia coli, with emphasis on enteroaggregative E. coli, in severe non-epidemic pediatric diarrhea in a high-income country. BMC Infect Dis 2015;15:79 | Not index test |
| Tobias J, Vutukuru SR. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol Res 2012;167:564–70 | Not index test |
| Van Lint P, De Witte E, De Henau H, De Muynck A, Verstraeten L, Van Herendael B, et al. Evaluation of a real-time multiplex PCR for the simultaneous detection of Campylobacter jejuni, Salmonella spp., Shigella spp./EIEC, and Yersinia enterocolitica in fecal samples. Eur J Clin Microbiol Infect Dis 2015;34:535–42 | Not index test |
| Van Lint P, Rossen JW, Vermeiren S, Ver Elst K, Weekx S, Van Schaeren J, et al. Detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in clinical stool samples by using multiplex real-time PCR after automated DNA isolation. Acta Clin Belg 2013;68:188–92 | Not index test |
| Verweij JJ, van Lieshout L. Intestinal parasitic infections in an industrialized country; a new focus on children with better DNA-based diagnostics. Parasitology 2011;138:1492–8 | Review |
| Wang J, Xu Z, Niu P, Zhang C, Zhang J, Guan L, et al. A two-tube multiplex reverse transcription PCR assay for simultaneous detection of viral and bacterial pathogens of infectious diarrhea. BioMed Res Int 2014;2014:648520 | Not index test |
| Wiemer D, Loderstaedt U, von Wulffen H, Priesnitz S, Fischer M, Tannich E, et al. Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in fecal samples. Int J Med Microbiol 2011;301:577–84 | Not index test |
| Study reference | Reason for exclusion |
|---|---|
| Ciardo D, Dubuis O, Burki D, Noppen C, Viollier EH. Infectious gastroenteritis: comparison of conventional and molecular methods for detection of pathogens. Clin Microbiol Infect 2012;18:501–2 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Halligan E, Bible J, Cliff P, Wilson A, Carlton-Carew L, Goldenberg S, et al. An evaluation of the Luminex xTAG gastrointestinal pathogen panel at a London teaching hospital 2011: the comparative performance of a rapid molecular multiplex assay and current standard laboratory investigations of infectious gastroenteritis. Clin Microbiol Infect 2012;18:403 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Mansuy JM, Plachot C, Mengelle C, Grouteau E, Claudet I, Kamar N, et al. Use of a multiplex molecular assay for the detection of pathogens in stools from diarrhoeic patients. Clin Microbiol Infect 2012;18:502 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Marimon JM, Montes M, Gomariz M, Cilla G, Perez-Trallero E. Diagnosis of gastroenteric infections: comparison of traditional methods with the new molecular technologies. Clin Microbiol Infect 2012;18:503 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| McMillen T, Chen J, Sun J, Nie S, Fan F, Babady N, et al. Evaluation of cepheid norovirus assay for detection of noroviruses genogroup I and II in stool. J Mol Diagnos 2014;16:730 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Vocale C, Landini MP, Sambri V. First Italian experience in clinical practice of the gastrointestinal pathogen panel using a unique multiplexing technology at a Bologna hospital. Clin Microbiol Infect 2012;18:501 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Becker S, Chatigre JK, Coulibaly JT, Herrmann M, N’Goran E, Utzinger J, et al. Luminex xTAG GPP multiplex PCR assay for the diagnosis of gastrointestinal pathogens: preliminary findings from a case–control study in Côte d’Ivoire. Int J Med Microbiol 2013;303:25–6 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Chapin K, McVeigh L, Ponraj V, Flores-Cortez E, Dickenson R. Comparison of the BioFire FilmArray gastrointestinal panel, the BD MAX enteric bacterial panel and the luminex xTAG gastrointestinal pathogen panel to traditional laboratory methods. J Mol Diagn 2014;16:738 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Chapin KC, Dickenson RA, Andrea SB, Huang R. Multiplex assay evaluation of Luminex xTAG analyte-specific reagents for diagnosis of gastrointestinal pathogens. J Mol Diagn 2012;14:681 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Eibach D, Krumkamp R, Hahn A, Sarpong N, Adu-Sarkodie Y, Leva A, et al. Application of the Luminex xTAG gastrointestinal pathogen panel for the detection of gastrointestinal pathogens in a rural African setting. Trop Med Int Health 2015;20:117 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Herding E, Hansen G. Clinical utility of the luminex xTAG GPP assay in a characterized culture negative high-risk population cohort suggests enhanced detection of infectious GI associated illness compared to conventional culture testing and hospital ordering. J Mol Diagn 2014;16:743 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Kahlau P, Malecki M, Schildgen V, Mattner F, Schildgen O. ProGastro SSCS vs Luminex GPP: a first experience in comparing two multiplex assays for gastrointestinal pathogens. J Mol Diagn 2012;14:672 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Lee SD, Elshimali Y, Lobos J, Scarsella A. Validation of luminex xTAG gastrointestinal pathogen panel with stool specimens in S.T.A.R. buffer. J Mol Diagn 2014;16:737–8 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Macpherson M, Rutherford C, Jayaratne P, Dale SE. Development of a real-time, reverse transcriptase (RT) multiplex PCR assay for the detection of rotavirus and adenovirus in faecal specimens. Can J Infect Dis Med Microbiol 2013;24:33B–4B | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Manji R, Ginocchio CC. Clinical performance assessment of the luminex xTAG investigational assay for the detection of enteric pathogens directly from clinical specimens. J Mol Diagn 2013;15:894 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Morrison S, Shennan M, Zhang H. Evaluation of the xTAG stool sample pretreatment pack using the xTAG gastrointestinal pathogen panel. Clin Microbiol Infect 2012;18:782–3 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Ohrmalm C, Yaghoubian S, Leyva L, Zhang H, Blomberg J. Evaluation of a multiplex nucleic acid test for the detection of gastrointestinal pathogens in faecal samples. Clin Microbiol Infect 2011;17:S553 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Pichon M, Bal A, Morfin F, Casalegno JS, Billaud G, Lina B, et al. Evaluation of a multiplex gastrointestinal panel. Which test for a pediatric population? J Clin Virol 2015;70:S32–3 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Rogatcheva M, Vaughn M, Wallace R, Crisp R, Alger G, Gardner J, et al. Detection of bacterial, viral, and protozoan pathogens using the FilmArray gastrointestinal (GI) system. J Mol Diagn 2012;14:669 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Sefers S, Chappell J. Evaluation of the luminex xTAG gastrointestinal pathogen panel and analyte-specific reagents for the detection of enteropathogens in stool. J Mol Diagn 2013;15:886 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Sinclair WB, Lopansri BK, Merica CB, Panahi R, Wood JS, Owen M, et al. BioFire FilmArray 2.0 throughput performance. J Mol Diagn 2015;17:846 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Twining N, Spirtovic S, Costello M, Michalov M, Mazur L. Performance of luminex xTAG gastrointestinal pathogen panel LDT assay. J Mol Diagn 2013;15:881 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Wessels E, Rusman L, Van Bussel M, Claas ECJ. Prospective application of the Luminex xTAG-GPP multiplex PCR in diagnosing infectious gastroenteritis. Clin Microbiol Infect 2012;18:503–4 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| Yaghoubian S, Himsworth D, Dey C, Lan L, Bussel MV, Claas EC. Clinical performance of xTAG GPP multiplexed gastrointestinal panel for the simultaneous detection and identification of viral, bacterial and parasitic pathogens in stool specimens. J Mol Diagn 2010;12:881 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
| York V, Petterson J, Ward PM, Green N, She RC. The value of a multiplexed gastrointestinal pathogen panel in two distinct patient populations. J Mol Diagn 2014;16:731 | Abstract, insufficient detail on validation method, comparator and/or test accuracy outcomes per pathogen tested |
Appendix 6 Ongoing trial
| Criterion | Details |
|---|---|
| Title | Implementation of a Molecular Diagnostic for Paediatric Acute Gastroenteritis: The FilmArray GI Panel IMPACT Study (IMPACT) |
| Population | Children (aged < 18 years) presenting to the emergency department or on-site urgent care centre with symptoms of gastroenteritis (e.g. diarrhoea, vomiting, nausea, etc.) with symptoms for at least 24 hours but < 14 days |
| Primary outcomes | Number of additional health-care encounters experienced related to the initial gastrointestinal episode |
| Secondary outcomes | Health-care cost analysis following implementation of the FilmArray Gastrointestinal Panel |
| Intervention | FilmArray Gastrointestinal Panel |
| Study design | Non-randomised efficacy study |
| Source | https://clinicaltrials.gov/ct2/show/NCT02248285 |
Appendix 7 Tables of 2 × 2 data by pathogen and gastrointestinal pathogen panel test
The 2 × 2 data in the tables in this appendix are reported in the format shown in Table 59.
| Conventional test + | Conventional test – | |
|---|---|---|
| GPP+ | a (+/+) | b (–/+) |
| GPP– | c (+/–) | d (–/–) |
xTAG
Viruses
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 4 | 4 | 16 | 628 | 652 |
| Deng et al. 201523 | 3 | 0 | 2 | 285 | 290 |
| Duong et al. 201624 | 23 | 5 | 2 | 449 | 479 |
| FDA 201225 | 4 | 13 | 1 | 1154 | 1172 |
| Gu et al. 201526 | 3 | 2 | 20 | 110 | 135 |
| Halligan et al. 201427 | 6 | 2 | 0 | 181 | 189 |
| Mengelle et al. 201329 | 0 | 2 | 5 | 396 | 403 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 (G1) | 9 | 0 | 0 | 642 | 651 |
| Claas et al. 201321 (G2) | 62 | 14 | 5 | 570 | 651 |
| Deng et al. 201523 (G1) | 8 | 0 | 0 | 282 | 290 |
| Deng et al. 201523 (G2) | 37 | 3 | 2 | 248 | 290 |
| Duong et al. 201624 (G1) | 15 | 5 | 2 | 457 | 479 |
| Duong et al. 201624 (G2) | 85 | 4 | 3 | 387 | 479 |
| Gu et al. 201526 | 29 | 9 | 2 | 159 | 199 |
| Halligan et al. 201427 | 66 | 79 | 14 | 1284 | 1443 |
| Mengelle et al. 201329 | 0 | 17 | 1 | 250 | 268 |
| Pankhurst et al. 201430 | 183 | 16 | 16 | 624 | 839 |
| FDA 201225 | 74 | 98 | 4 | 1023 | 1199 |
| Coste et al. 201322 | 0 | 14 | 0 | 40 | 54 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 18 | 1 | 0 | 633 | 652 |
| Deng et al. 201523 | 61 | 6 | 1 | 222 | 290 |
| Duong et al. 201624 | 117 | 4 | 9 | 349 | 479 |
| Gu et al. 201526 | 1 | 0 | 0 | 109 | 110 |
| Halligan et al. 201427 | 13 | 6 | 0 | 158 | 177 |
| Mengelle et al. 201329 | 61 | 9 | 2 | 332 | 404 |
| FDA 201225 | 2 | 2 | 0 | 1162 | 1166 |
| Coste et al. 201322 | 2 | 0 | 0 | 52 | 54 |
Bacteria
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 30 | 11 | 1 | 343 | 385 |
| Gu et al. 201526 | 44 | 8 | 4 | 107 | 163 |
| Halligan et al. 201427 | 121 | 10 | 6 | 1175 | 1312 |
| Mengelle et al. 201329 | 5 | 8 | 0 | 331 | 344 |
| Pankhurst et al. 201430 | 195 | 19 | 4 | 621 | 839 |
| FDA 201225 | 107 | 105 | 7 | 922 | 1141 |
| Coste et al. 201322 | 1 | 0 | 0 | 53 | 54 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 111 | 15 | 3 | 382 | 511 |
| Deng et al. 201523 | 20 | 16 | 0 | 254 | 290 |
| Duong et al. 201624 (PCR) | 59 | 3 | 6 | 411 | 479 |
| Duong et al. 201624 (culture) | 27 | 35 | 3 | 414 | 479 |
| Gu et al. 201526 | 0 | 0 | 0 | 112 | 112 |
| Halligan et al. 201427 | 23 | 37 | 0 | 1336 | 1396 |
| Mengelle et al. 201329 | 1 | 12 | 0 | 368 | 381 |
| Pankhurst et al. 201430 | 110 | 11 | 7 | 711 | 839 |
| FDA 201225 | 3 | 21 | 0 | 1155 | 1179 |
| Coste et al. 201322 | 2 | 12 | 0 | 40 | 54 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 15 | 5 | 1 | 407 | 428 |
| Deng et al. 201523 | 1 | 2 | 0 | 287 | 290 |
| aGu et al. 201526 | 0 | 1 | 0 | 111 | 112 |
| Halligan et al. 201427 | 3 | 2 | 0 | 1391 | 1396 |
| Mengelle et al. 201329 | 0 | 1 | 0 | 265 | 266 |
| FDA 201225 | 2 | 9 | 0 | 1158 | 1169 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 0 | 7 | 0 | 68 | 75 |
| Deng et al. 201523 | 1 | 4 | 0 | 285 | 290 |
| Mengelle et al. 201329 | 0 | 0 | 0 | 266 | 266 |
| FDA 201225 | 2 | 4 | 6 | 1156 | 1168 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 62 | 6 | 13 | 408 | 489 |
| Deng et al. 201523 | 25 | 6 | 5 | 254 | 290 |
| Duong et al. 201624 (PCR) | 84 | 128 | 9 | 258 | 479 |
| Duong et al. 201624 (culture) | 38 | 172 | 2 | 267 | 479 |
| Gu et al. 201526 | 1 | 3 | 0 | 108 | 112 |
| Halligan et al. 201427 | 11 | 36 | 0 | 1349 | 1396 |
| Mengelle et al. 201329 | 7 | 14 | 2 | 356 | 379 |
| Pankhurst et al. 201430 | 15 | 9 | 18 | 797 | 839 |
| FDA 201225 | 10 | 18 | 0 | 1143 | 1171 |
| Coste et al. 201322 | 0 | 1 | 0 | 53 | 54 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 40 | 13 | 0 | 452 | 505 |
| Deng et al. 201523 | 3 | 1 | 0 | 286 | 290 |
| Duong et al. 201624 (PCR) | 86 | 6 | 4 | 383 | 479 |
| Duong et al. 201624 (culture) | 40 | 51 | 0 | 388 | 479 |
| Gu et al. 201526 | 1 | 0 | 0 | 111 | 112 |
| Halligan et al. 201427 | 3 | 11 | 0 | 1382 | 1396 |
| Mengelle et al. 201329 | 1 | 0 | 0 | 378 | 379 |
| FDA 201225 | 2 | 17 | 0 | 1154 | 1173 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 8 | 1 | 0 | 162 | 171 |
| Mengelle et al. 201329 | 5 | 2 | 0 | 258 | 265 |
| FDA 201225 | 1 | 16 | 0 | 1153 | 1170 |
| Coste et al. 201322 | 0 | 2 | 1 | 51 | 54 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 0 | 0 | 1 | 194 | 195 |
| Deng et al. 201523 | 0 | 0 | 0 | 290 | 290 |
| FDA 201225 | 0 | 1 | 0 | 1166 | 1167 |
| Mengelle et al. 201329 | 0 | 0 | 0 | 379 | 379 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 0 | 0 | 0 | 366 | 366 |
| Deng et al. 201523 | 1 | 0 | 0 | 289 | 290 |
| Gu et al. 201526 | 0 | 0 | 0 | 38 | 38 |
| Mengelle et al. 201329 | 0 | 0 | 0 | 379 | 379 |
Parasites
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 32 | 7 | 3 | 833 | 868 |
| Gu et al. 201526 | 0 | 0 | 0 | 35 | 35 |
| Halligan et al. 201427 | 1 | 6 | 0 | 229 | 236 |
| Mengelle et al. 201329 | 0 | 1 | 0 | 117 | 118 |
| FDA 201225 | 12 | 53 | 1 | 1131 | 1197 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 6 | 6 | 0 | 845 | 857 |
| Deng et al. 201523 | 0 | 1 | 0 | 289 | 290 |
| Halligan et al. 201427 | 0 | 9 | 1 | 226 | 236 |
| FDA 201225 | 0 | 19 | 0 | 1149 | 1168 |
| Mengelle et al. 201329 | 0 | 0 | 0 | 285 | 285 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Claas et al. 201321 | 26 | 13 | 0 | 829 | 868 |
| Deng et al. 201523 | 0 | 0 | 0 | 290 | 290 |
| Gu et al. 201526 | 0 | 0 | 0 | 35 | 35 |
| Halligan et al. 201427 | 1 | 17 | 0 | 218 | 236 |
| Mengelle et al. 201329 | 0 | 0 | 0 | 118 | 118 |
| FDA 201225 | 4 | 39 | 0 | 1132 | 1175 |
| Coste et al. 201322 | 1 | 0 | 0 | 53 | 54 |
FilmArray
Viruses
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 42 | 13 | 2 | 1499 | 1556 |
| Gu et al. 201526 | 3 | 2 | 20 | 110 | 135 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 7 | 1 | 0 | 1548 | 1556 |
| Gu et al. 201526 | 4 | 0 | 6 | 189 | 199 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 52 | 18 | 3 | 1483 | 1556 |
| Gu et al. 201526 | 28 | 0 | 3 | 168 | 199 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 6 | 12 | 0 | 1538 | 1556 |
| Gu et al. 201526 | 1 | 0 | 0 | 109 | 110 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 46 | 13 | 0 | 1497 | 1556 |
| Gu et al. 201526 | 5 | 0 | 2 | 192 | 199 |
Bacteria
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 163 | 41 | 2 | 1350 | 1556 |
| Gu et al. 201526 | 39 | 5 | 9 | 110 | 163 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 34 | 24 | 1 | 1497 | 1556 |
| Gu et al. 201526 | 0 | 0 | 0 | 112 | 112 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 47 | 2 | 2 | 1505 | 1556 |
| Gu et al. 201526 | 1 | 0 | 0 | 111 | 112 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 33 | 5 | 0 | 1518 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 0 | 2 | 0 | 1554 | 1556 |
| Gu et al. 201526 | 0 | 1 | 0 | 111 | 112 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 0 | 1 | 0 | 1555 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 1 | 0 | 0 | 1555 | 1556 |
| Gu et al. 201526 | 0 | 0 | 0 | 38 | 38 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 3 | 1 | 0 | 34 | 38 |
| aGu et al. 201526 | 0 | 4 | 0 | 108 | 112 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 82 | 27 | 1 | 1446 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 314 | 34 | 3 | 1167 | 1518 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 22 | 9 | 0 | 1525 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 3 | 15 | 0 | 1538 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 31 | 6 | 0 | 1519 | 1556 |
| Gu et al. 201526 | 1 | 0 | 0 | 111 | 112 |
Parasites
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 0 | 0 | 0 | 1556 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 18 | 6 | 0 | 1532 | 1556 |
| Gu et al. 201526 | 0 | 0 | 0 | 35 | 35 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 19 | 0 | 0 | 1537 | 1556 |
| Study | a | b | c | d | Total |
|---|---|---|---|---|---|
| Buss et al. 201520 | 20 | 7 | 0 | 1529 | 1556 |
| Gu et al. 201526 | 0 | 0 | 0 | 35 | 35 |
Appendix 8 Meta-analytic outcomes of test agreement
FIGURE 43.
Negative agreement: xTAG vs. conventional testing (benchmark). ETEC, enterotoxigenic E. coli; STEC, shiga toxin-producing E. coli; RE, random-effects estimate. Reproduced from Freeman et al. 2017. 49 © 2017 Freeman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
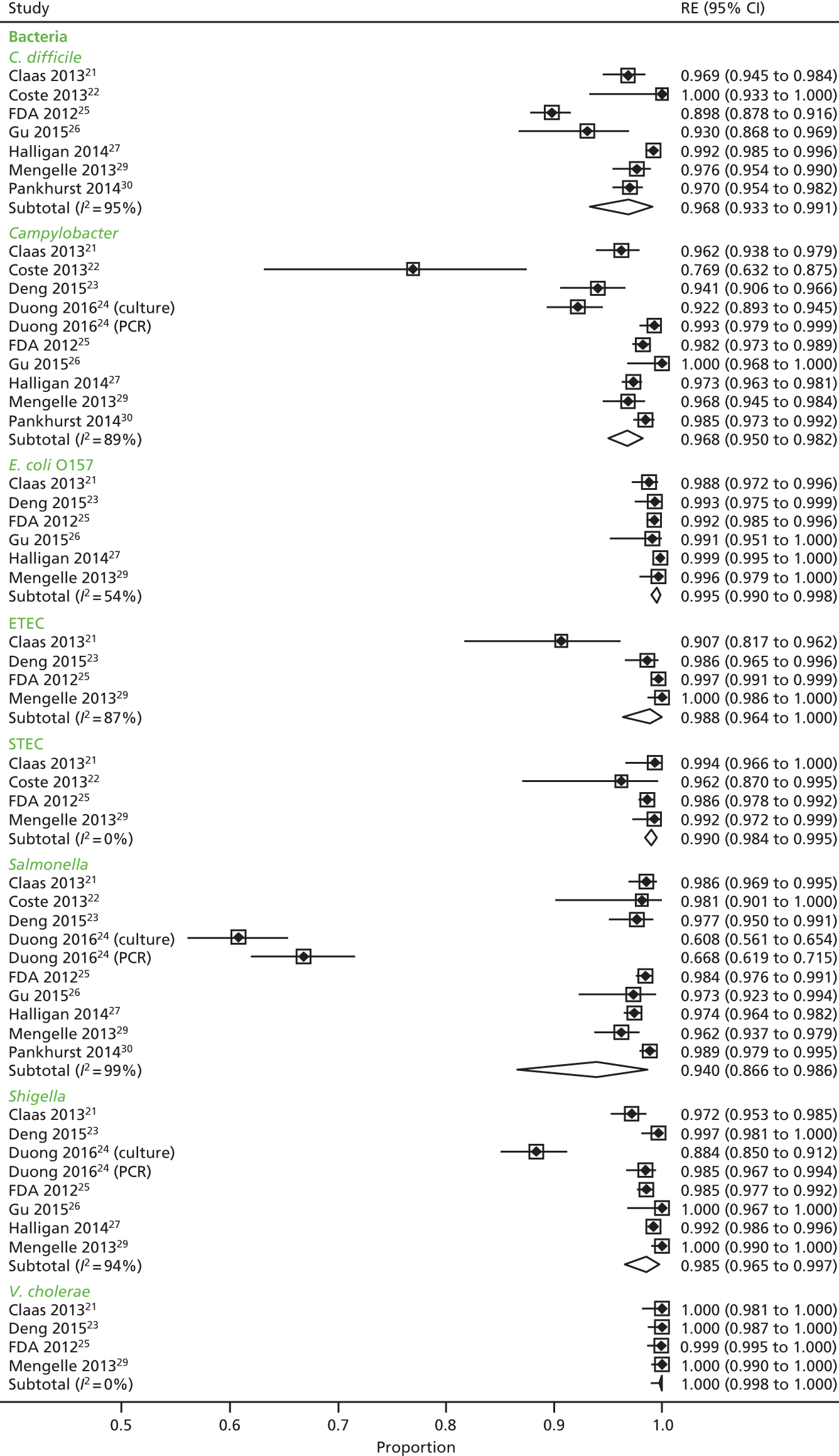

FIGURE 44.
Negative agreement: conventional testing vs. xTAG (benchmark). ETEC, enterotoxigenic E. coli; STEC, shiga toxin-producing E. coli; RE, random-effects estimate. Reproduced from Freeman et al. 2017. 49 © 2017 Freeman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
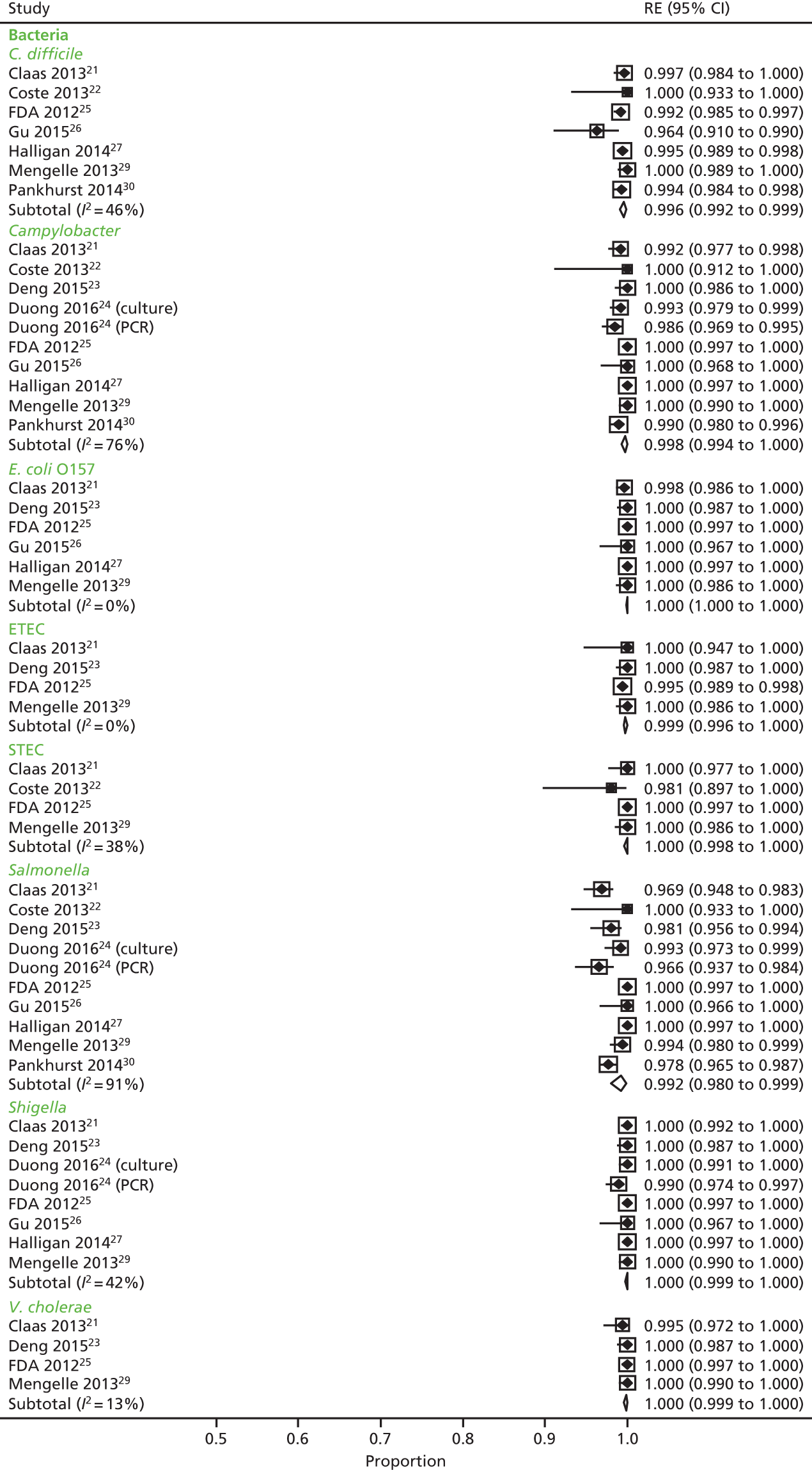

FIGURE 45.
Negative agreement: FilmArray vs. conventional testing (benchmark). EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; STEC, shiga toxin-producing E. coli; RE, random-effects estimate; p.v.c., parahaemolyticus, vulnificus and cholerae.

FIGURE 46.
Negative agreement: conventional testing vs. FilmArray (benchmark). EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; STEC, shiga toxin-producing E. coli; RE, random-effects estimate; p.v.c., parahaemolyticus, vulnificus and cholerae.
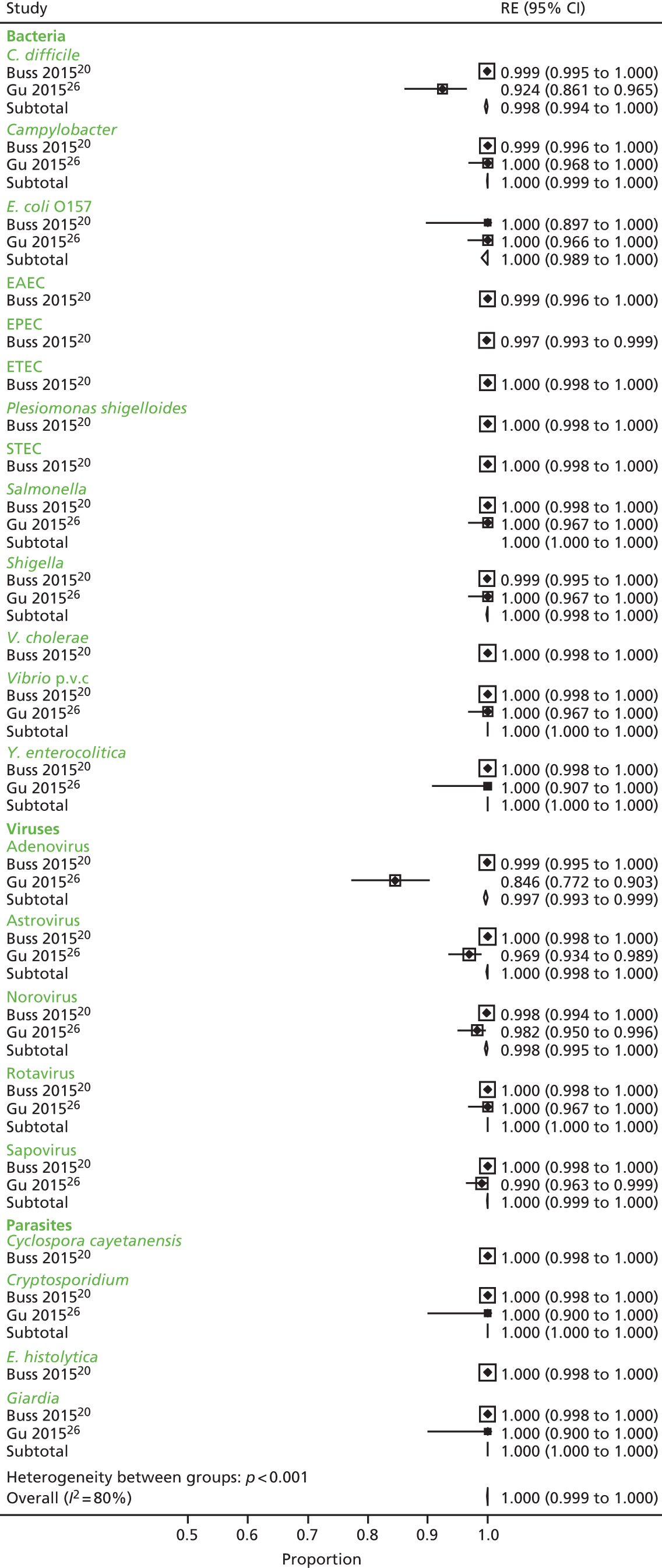
Appendix 9 Excluded gastrointestinal pathogen panel economic studies with reason
| Author (year) | Title | Journal details | Comments | Reason for exclusion |
|---|---|---|---|---|
| Bignardi and Settle (2015)53 | Can the Luminex xTAG gastrointestinal pathogen panel really save money? | Journal of Infection 71:498–9 | The author provides some comments on why they think xTAG GPP will not actually save the NHS any money | Letter to editor commenting on Goldenberg et al.56 paper. Not an economic evaluationa |
| Binnicker (2015)54 | Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness | Journal of Clinical Microbiology 53:3723–8 | The paper looks at how xTAG GPP and FilmArray work and some of the trials the two interventions are in | Not an economic evaluationa |
| Freedman et al. (2015)55 | Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics | BMC Pediatrics 15:89 | The paper proposes using xTAG GPP instead of current methods | A protocol. Not an economic evaluationa |
| Halligan et al. (2014)27 | Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study | Clinical Microbiology and Infection 20:460–7 | The paper provides some useful cost information as provided in the Goldenberg et al. study56 | Clinical paper linking to Goldenberg et al.56 paper. Not an economic evaluationa |
Appendix 10 Quality assessment of economic evaluation studies
| Assessment | Goldenberg et al.56 |
|---|---|
| Title | Y |
| Abstract | Y |
| Introduction | |
| Background and objectives | Y |
| Methods | |
| Target population and subgroups | Y |
| Setting and location | Y |
| Study perspective | Y |
| Comparators | Y |
| Time horizon | Y |
| Discount rate | N/A |
| Choice of health outcomes | Y |
| Measurement of effectiveness | N |
| Measurement and valuation of preference-based outcomes | N |
| Estimating resources and costs | Y |
| Currency, price date and conversion | P |
| Choice of model | N/A |
| Assumptions | P |
| Analytical methods | P |
| Results | |
| Study parameters | Y |
| Incremental costs and outcomes | Y |
| Characterising uncertainty | Y |
| Discussion | |
| Study findings | Y |
| Limitations | Y |
| Generalisability | N |
| Other | |
| Source of funding | Y |
| Conflicts of interest | Y |
| Philips’ criteria | Goldenberg et al.56 |
|---|---|
| Structure | |
| 1. Is there a clear statement of the decision problem? | Y |
| 2. Is the objective of the model specified and consistent with the stated decision problem? | Y |
| 3. Is the primary decision-maker specified? | Y |
| 4. Is the perspective of the model stated clearly? | Y |
| 5. Are the model inputs consistent with the stated perspective? | Y |
| 6. Has the scope of the model been stated and justified? | N |
| 7. Are the outcomes of the model consistent with the perspective, scope and overall objective of the model? | P |
| 8. Is the structure of the model consistent with a coherent theory of the health condition under evaluation? | P |
| 9. Are the sources of the data used to develop the structure of the model specified? | Y |
| 10. Are the causal relationships described by the model structure justified appropriately? | N |
| 11. Are the structural assumptions transparent and justified? | N |
| 12. Are the structural assumptions reasonable given the overall objective, perspective and scope of the model? | UNC |
| 13. Is there a clear definition of the options under evaluation? | Y |
| 14. Have all feasible and practical options been evaluated? | UNC |
| 15. Is there justification for the exclusion of feasible options? | UNC |
| 16. Is the chosen model type appropriate given the decision problem and specified causal relationships within the model? | P |
| 17. Is the time horizon of the model sufficient to reflect all important differences between the options? | Y |
| 18. Are the time horizon of the model and the duration of treatment described and justified? | UNC |
| 19. Do the disease states (state transition model) or the pathways (decision tree model) reflect the underlying biological process of the disease in question and the impact of interventions? | Y |
| 20. Is the cycle length defined and justified in terms of the natural history of disease? | N/A |
| Data | |
| 21. Are the data identification methods transparent and appropriate given the objectives of the model? | P |
| 22. When choices have been made between data sources, are these justified appropriately? | UNC |
| 23. Has particular attention been paid to identifying data for the important parameters of the model? | UNC |
| 24. Has the quality of the data been assessed appropriately? | N |
| 25. When expert opinion has been used, are the methods described and justified? | UNC |
| 26. Is the data modelling methodology based on justifiable statistical and epidemiological techniques? | UNC |
| 27. Is the choice of baseline data described and justified? | P |
| 28. Are transition probabilities calculated appropriately? | N/A |
| 29. Has a half-cycle correction been applied to both costs and outcomes? | N/A |
| 30. If not, has the omission been justified? | N/A |
| 31. If relative treatment effects have been derived from trial data, have they been synthesised using appropriate techniques? | N/A |
| 32. Have the methods and assumptions used to extrapolate short-term results to final outcomes been documented and justified? | N/A |
| 33. Have alternative extrapolation assumptions been explored through sensitivity analysis? | N/A |
| 34. Have assumptions regarding the continuing effect of treatment once treatment is complete been documented and justified? | N/A |
| 35. Have alternative assumptions regarding the continuing effect of treatment been explored through sensitivity analysis? | N/A |
| 36. Are the costs incorporated into the model justified? | Y |
| 37. Has the source for all costs been described? | Y |
| 38. Have discount rates been described and justified given the target decision-maker? | N/A |
| 39. Are the utilities incorporated into the model appropriate? | N/A |
| 40. Is the source of utility weights referenced? | N/A |
| 41. Are the methods of derivation for the utility weights justified? | N/A |
| 42. Have all data incorporated into the model been described and referenced in sufficient detail? | P |
| 43. Has the use of mutually inconsistent data been justified (i.e. are assumptions and choices appropriate)? | UNC |
| 44. Is the process of data incorporation transparent? | UNC |
| 45. If data have been incorporated as distributions, has the choice of distributions for each parameter been described and justified? | N/A |
| 46. If data have been incorporated as distributions, is it clear that second order uncertainty is reflected? | N/A |
| 47. Have the four principal types of uncertainty been addressed? | N/A |
| 48. If not, has the omission of particular forms of uncertainty been justified? | N/A |
| 49. Have methodological uncertainties been addressed by running alternative versions of the model with different methodological assumptions? | N |
| 50. Is there evidence that structural uncertainties have been addressed via sensitivity analysis? | N |
| 51. Has heterogeneity been dealt with by running the model separately for different subgroups? | N |
| 52. Are the methods of assessment of parameter uncertainty appropriate? | N/A |
| 53. If data are incorporated as point estimates, are the ranges used for sensitivity analysis stated clearly and justified? | P |
| 54. Is there evidence that the mathematical logic of the model has been tested thoroughly before use? | N |
| 55. Are any counterintuitive results from the model explained and justified? | N |
| 56. If the model has been calibrated against independent data, have any differences been explained and justified? | N |
| 57. Have the results been compared with those of previous models and any differences in results explained? | N |
Appendix 11 Additional economic analyses as requested by the National Institute for Health and Care Excellence Committee
At the request of the committee, we have changed the assumptions outlined in this appendix.
Comparator cost fixed at £20 for each of the models
For each of the models, we have changed the comparator cost from £66.18 (base-case analysis) to £20. Most results have stayed in the same direction as the base-case results presented in the report. However, for models 3 and 5, when comparing xTAG with conventional care, xTAG no longer dominates conventional care – xTAG is now slightly more expensive, although it is still slightly more effective (Table 100).
| Test | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Probability cost-effectivea | Net monetary benefit (£)a (95% CI) |
|---|---|---|---|---|---|
| Model 1: hospitalised adult patients | |||||
| xTAG | –26 (–872 to 837) | 0.00018 (–0.00072 to 0.00111) | Dominant | 0.548 | 30 (–852 to 891) |
| FilmArray | –24 (–878 to 838) | 0.00013 (–0.00076 to 0.00104) | Dominant | 0.543 | 27 (–852 to 894) |
| Model 2: hospitalised children | |||||
| xTAG | –16 (–1009 to 959) | 0.00024 (–0.00061 to 0.00112) | Dominant | 0.545 | 21 (–965 to 1021) |
| FilmArray | –43 (–942 to 885) | 0.00016 (–0.00068 to 0.00106) | Dominant | 0.571 | 47 (–890 to 957) |
| Model 3: people in the community | |||||
| xTAG | 18 (8 to 26) | 0.00003 (–0.00032 to 0.00045) | 518,112 | 0.379 | –17 (–32 to –1) |
| FilmArray | 74 (52 to 91) | 0.00002 (–0.00037 to 0.00040) | 3,264,373 | 0.069 | –74 (–97 to –46) |
| Model 4: immunocompromised patients | |||||
| xTAG | –25 (–916 to 912) | 0.00023 (–0.00086 to 0.00139) | Dominant | 0.544 | 29 (–932 to 935) |
| FilmArray | –30 (–981 to 899) | 0.00019 (–0.00088 to 0.00136) | Dominant | 0.539 | 34 (–916 to 1009) |
| Model 5: recent travellers | |||||
| xTAG | 18 (11 to 26) | 0.00005 (–0.00048 to 0.00055) | 356,931 | 0.432 | –17 (–35 to –1) |
| FilmArray | 75 (58 to 90) | 0.00002 (–0.00049 to 0.00056) | 4,203,556 | 0.112 | –75 (–99 to –51) |
Threshold analysis
Taking into account the change in direction and magnitude for models 3 and 5 when comparing xTAG with conventional care, we have computed a threshold analysis to see at which comparator cost the ICER result changes from being dominant to not dominant. For model 3 using the probabilistic results, the switching point is when the conventional cost is £36.60, and for model 5 using the probabilistic results, the switching point is when the conventional cost is £36.80 (Table 101).
| Test | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Probability cost-effectivea | Net monetary benefit (£)a (95% CI) |
|---|---|---|---|---|---|
| Model 3: people in the community | |||||
| £20 | 18 (8 to 26) | 0.00003 (–0.00032 to 0.00045) | 518,112 | 0.379 | –17 (–32 to –1) |
| £36.60 | 0 (–11 to 11) | 0.00003 (–0.00036 to 0.00042) | 1857 | 0.530 | 0 (–17 to 19) |
| Base case | –32 (–49 to –15) | 0.00003 (–0.00036 to 0.00045) | Dominant | 0.997 | 32 (9 to 56) |
| Model 5: recent travellers | |||||
| £20 | 18 (11 to 26) | 0.00005 (–0.00048 to 0.00055) | 356,931 | 0.432 | –17 (–35 to –1) |
| £36.80 | 0 (–10 to 9) | 0.00006 (–0.00043 to 0.00054) | 90 | 0.588 | 1 (–16 to 19) |
| Base case | –31 (–48 to –16) | 0.00006 (–0.00044 to 0.00055) | Dominant | 0.994 | 32 (9 to 57) |
This threshold analysis is better reflected in Figures 47 and 48, which show that the 95% CI fluctuates around zero at a threshold of £20,000 per QALY when the price of conventional testing is changed.
FIGURE 47.
Net monetary benefit (willingness-to-pay threshold £20,000/QALY) (probabilistic results): xTAG vs. conventional care (model 3).

FIGURE 48.
Net monetary benefit (willingness-to-pay threshold £20,000/QALY) (probabilistic results): for xTAG vs. conventional care (model 5).
Tornado diagram
We have not presented ICERs for the most influential parameters, as seen in Figures 21, 22, 33 and 34, as most results are dominant and it does not make sense to present ICERs. Instead, we have tabulated the net monetary benefit ratios for the most influential parameters at a threshold of £20,000 per QALY (Table 102).
| Parameters | xTAG vs. conventional care (£) | FilmArray vs. conventional care (£) | ||
|---|---|---|---|---|
| Increase by 50% | Decrease by 50% | Increase by 50% | Decrease by 50% | |
| Hospital model (model 1) | ||||
| Base-case net monetary benefit ratio | 72.10 | 63.77 | ||
| Cost of adult bed-day: general | 88.53 | 55.67 | 67.99 | 59.54 |
| Cost of adult bed-day: isolation | 74.23 | 69.98 | 100.86 | 26.67 |
| Cost of conventional test | 106.86 | 37.35 | 98.52 | 29.01 |
| Cost of xTAG or FilmArray test | 52.50 | 91.71 | 15.82 | 111.71 |
| Bed-days for GPP testing (not detect and symptoms resolve) | –575.08 | 719.28 | –605.26 | 732.80 |
| Probability of detect (first test) | 101.25 | 42.95 | 77.89 | 49.64 |
| Probability of isolates | 50.35 | 93.85 | 49.89 | 77.64 |
| Proportion of n = false positive | 20.80 | 123.41 | 43.07 | 84.47 |
| Community model (model 3) | ||||
| Base-case net monetary benefit ratio | 32.30 | –24.90 | ||
| Cost of conventional test | 67.82 | –3.23 | 10.63 | –60.43 |
| Cost of xTAG or FilmArray test | 12.67 | 51.92 | –72.94 | 23.14 |
Weighted analyses
For this analysis we have looked at what happens to the incremental costs and benefits when we have a 50% weighting for models 1 (adult hospital) and 3 (adult community). For the weighted analysis, GPP testing no longer dominates conventional care, as although it is slightly cheaper, it is slightly less effective (Table 103).
| Test | Incremental costs (95% CI) (£) | Incremental QALYs gained (95% CI) | ICER (cost per QALY gained) | Probability cost-effectivea |
|---|---|---|---|---|
| xTAG | –48 (–785 to 658) | –0.00010 (–0.00088 to 0.00059) | 490,232 | 0.689 |
| FilmArray | –13 (–746 to 643) | –0.00007 (–0.00088 to 0.00064) | 195,303 | 0.444 |
List of abbreviations
- BNF
- British National Formulary
- CE
- Conformité Européenne
- CEAC
- cost-effectiveness acceptability curve
- CHEERS
- Consolidated Health Economic Evaluation Reporting Standards
- CI
- confidence interval
- DNA
- deoxyribonucleic acid
- EIA
- enzyme immunoassay
- EQ-5D
- EuroQol-5 Dimensions
- FDA
- Food and Drug Administration
- GBP
- Great British pounds
- GDH
- glutamate dehydrogenase
- GP
- general practitioner
- GPP
- gastrointestinal pathogen panel
- ICER
- incremental cost-effectiveness ratio
- NICE
- National Institute for Health and Care Excellence
- PCR
- polymerase chain reaction
- PHE
- Public Health England
- PRISMA
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- QALD
- quality-adjusted life-day
- QALY
- quality-adjusted life-year
- QUADAS-2
- Quality Assessment of Diagnostic Accuracy Studies-2