Notes
Article history
The research reported in this issue of the journal was commissioned and funded by the HTA programme on behalf of NICE as project number 15/17/04. The protocol was agreed in January 2016. The assessment report began editorial review in September 2016 and was accepted for publication in March 2017. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Ian M Frayling reports personal fees from the UK National External Quality Assessment Scheme for Immunocytochemistry and In Situ Hybridisation (UK NEQAS ICC and ISH) outside the submitted work and is Honorary Medical Adviser to the charity Lynch Syndrome UK.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2017. This work was produced by Snowsill et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Background and definition of the decision problem
Colorectal cancer (CRC) is the second most common cause of death from cancer in the UK, with 16,187 deaths in 2012, which accounted for 10% of all cancers. 1 It is considered to be a multifactorial disease, with environment and inheritance playing varying roles in different patients. 2 The majority of individuals with CRC have sporadic disease (approximately 70–80%). However, the remaining 20–30% have a family history of the disease, with 5–6% of these having mutations involved in a known hereditary cancer syndrome. 3 The most common form of genetically defined hereditary CRC is Lynch syndrome (LS), which accounts for approximately 3.3% of these tumours. Previously referred to as hereditary non-polyposis colorectal cancer (HNPCC), LS is inherited as an autosomal dominant disorder. 4,5 Therefore, if one parent has the disease, there is a 50% chance that each of their children will inherit it. Although characterised by an increased risk of CRC, individuals with LS are also at increased risk of other cancers such as endometrial, ovarian, stomach, small intestine, hepatobiliary tract, urinary tract, brain and skin cancers. 4
It is important to identify families with LS so that risk-reducing measures can be taken to address the elevated risk and earlier onset of CRC and other cancers.
It should be noted that, although LS is characterised by a particular collection of cancers because of a mutation in one of a number of mismatch repair (MMR) genes, for simplicity, in this report ‘individuals with LS’ should be interpreted as ‘individuals born with LS-causing mutations’.
Condition and aetiology
Aetiology, pathology and prognosis
Lynch syndrome is caused by constitutional pathogenic mutations in deoxyribonucleic acid (DNA) MMR genes. MMR proteins are involved in recognising and repairing errors during DNA replication.
Although a person with LS still has a functioning MMR system (as they inherit a normal ‘wild-type’ allele from one parent in addition to the mutant allele from the parent with LS), there is a high risk that MMR function will be lost because of somatic mutation of the normal copy of the gene. This loss of MMR function during cell division leads to an inability to repair mutations such as DNA base–base mismatches and small insertions and deletions, eventually resulting in tumorigenesis. 4
In cells that have lost MMR function, mutations occur all over the genome, but especially in repetitive DNA sequences such as microsatellites. 4 Consequently, in tumours that have lost MMR function, a large number of mutations are present at microsatellite sequences (compared with surrounding normal tissue) and this is known as microsatellite instability (MSI).
The five currently known genes mutated in LS are:4
-
MutL homologue 1 (MLH1)
-
MutS homologue 2 (MSH2)
-
MutS homologue 6 (MSH6)
-
postmeiotic segregation increased 2 (PMS2)
-
epithelial cell adhesion molecule (EPCAM).
It should be noted that EPCAM, which is not a MMR gene, plays an indirect role in LS. 6 Located upstream of MSH2, deletions in the EPCAM gene have been shown to result in hypermethylation of the MSH2 promoter region, leading to a loss of MSH2 expression. 6,7 In some people it ‘switches off’ the MSH2 gene, which causes an increase in cases of CRC but not the other MSH2-associated cancers. In other individuals, the EPCAM deletion stops the MSH2 gene from working, just like a mutation in the MSH2 gene itself, in which case all of the MSH2 cancer risks are present. 7 Because of the tissue-dependent levels of epithelial cellular adhesion molecule (EpCAM) expression, carriers have a high risk of CRC, whereas extra-colonic cancers are rarely found and the risk for endometrial cancer is reduced. 8,9
Colorectal cancers associated with LS develop at a young age and are believed to arise from pre-existing discrete proximal colonic adenomas. Although people with LS develop adenomas at the same rate as individuals in the general population, the adenomas are more likely to progress to cancer. Furthermore, carcinogenesis may progress more rapidly in these patients (in 2–3 years) than in patients with sporadic adenomas (8–10 years). 10 Recent evidence also shows that, in addition to the adenoma–carcinoma pathway, CRCs in LS can arise directly from abnormal colonic crypts that have lost MMR proficiency. 11 This gives rise to cancers that do not go through an adenomatous polyp phase and therefore has implications for detection and surveillance. 11 The age at onset of CRC in LS varies by MMR gene, but is typically 42 years, that is, younger than the age at which the new bowel scope screening is gradually being introduced by the NHS (55 years) and much younger than the usual 60–74 years. 3,5
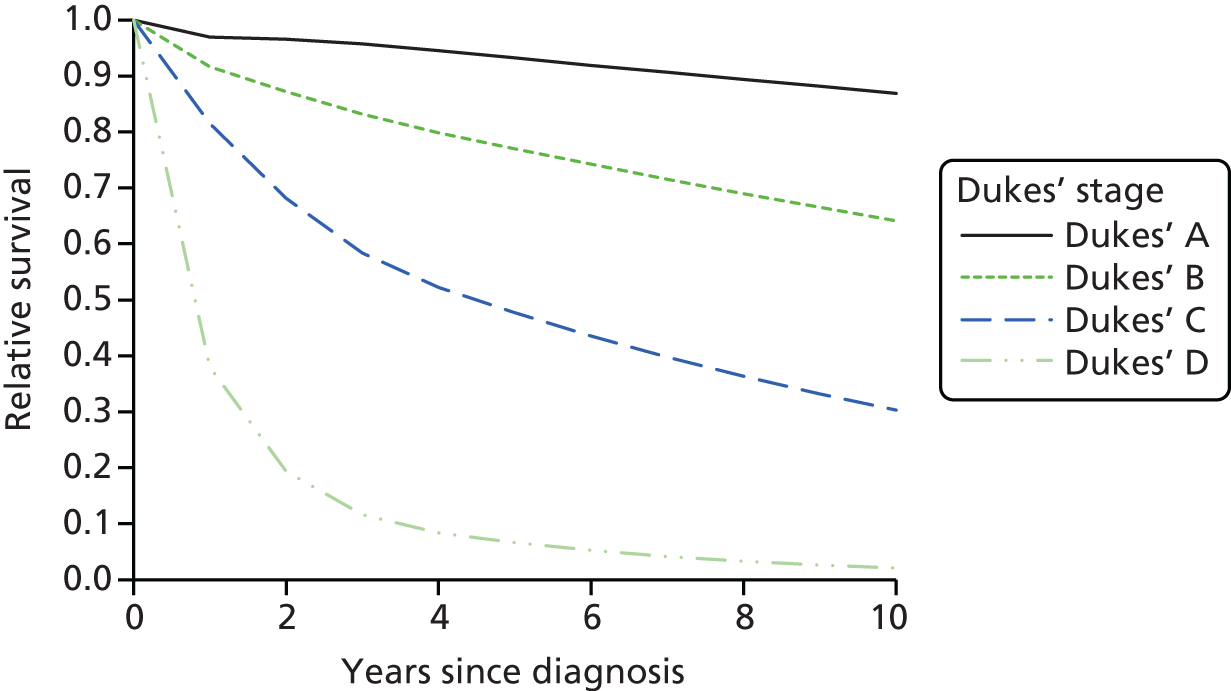
Synchronous colorectal tumours (primary tumours diagnosed within 6 months of each other) and metachronous colorectal tumours (primary tumours occurring > 6 months apart) are also more common in people with LS. 10 However, there is evidence to suggest that patients with CRC from LS families survive longer than sporadic CRC patients with same-stage tumours, which may be because of a reduced propensity to metastasise. Explanations for this include that immunological host defence mechanisms may be more active in tumours of the MSI phenotype and that the relatively high mutational load that occurs in tumours with defective DNA repair systems is detrimental to their survival. 12
Epidemiology
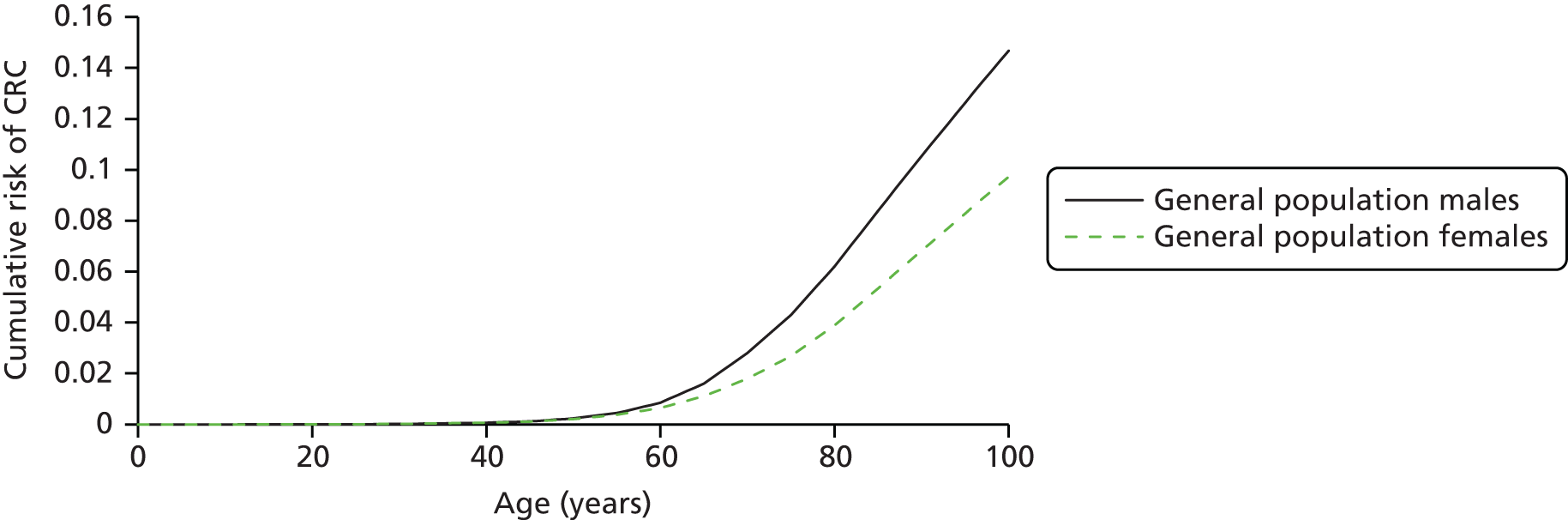
Lynch syndrome affects both men and women, although certain cancers will be specific to each sex, for example ovarian and prostate cancer. 13 However, the highest cancer risk is from CRC, with individuals with LS having a risk to age 70 years of 33–46%, as opposed to 5.5% and 7.3% for women and men, respectively, in the general population. 1,5
The average age at CRC onset is 44 years in members of families that meet the criteria for LS, whereas for the general population it is 60–65 years. 10 Two-thirds of the CRCs occur in the proximal colon (proximal to the splenic flexure) and the risk of a second primary CRC in patients is high (estimated at 16% within 10 years and 60% if the first CRC was diagnosed before the age of 25 years). 14,15
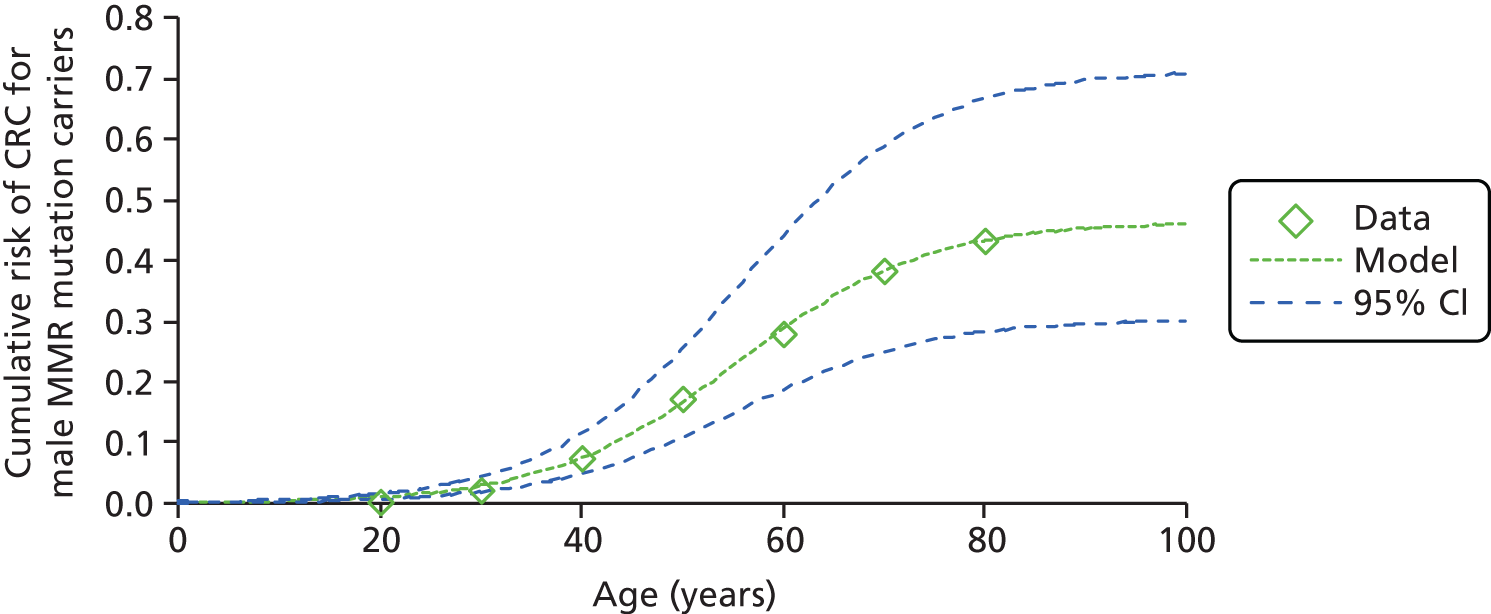
The risk of cancer in people with LS varies according to the affected MMR gene, with mutations in MLH1 and MSH2 conferring the highest cancer risk (Table 1). In individuals ascertained through family history clinics, nearly 90% of mutations are located in MLH1 and MSH2, with approximately 10% in MSH6 and PMS2. 18 Previous estimates of the cumulative risk of CRC at 70 years in MLH1 or MSH2 mutation carriers range from 22% to 74%. 18 Mutations in MSH6 and PMS2 genes have lower penetrance and different patterns of expression: MSH6 mutation carriers may have a high risk of endometrial cancer, similar to that in MLH1 and MSH2 mutation carriers, but have a later age at onset and a lower risk of CRC. 5
| Cancer | General population lifetime risk (%) | Gene | ||||||
|---|---|---|---|---|---|---|---|---|
| MLH1 or MSH2 | MSH6 | PMS2 | ||||||
| Male | Female | Risk (%) | Mean age at onset (years) | Risk (%) | Mean age at onset (years) | Risk (%) | Mean age at onset (years) | |
| Colon | 7.27 | 5.47 | 40–80 | 44–61 | 10–22 | 54 | 15–20 | 61–66 |
| Endometrial | NA | 2.44 | 25–60 | 48–62 | 16–26 | 55 | 15 | 49 |
| Stomach | 1.51 | 0.76 | 1–13 | 56 | ≤ 3 | 63 | 70–78 | |
| Ovary | NA | 1.95 | 4–24 | 42.5 | 1–11 | 46 | 42 | |
| Hepatobiliary tract | 0.5 | 1.4–4 | 50–57 | |||||
| Urinary tract | 1–4 | 54–60 | < 1 | 65 | ||||
| Small bowel | 0.01 | 3–6 | 47–49 | 54 | 59 | |||
| Brain/CNS | 1.35 | 1.37 | 1–3 | ≈50 | 45 | |||
| Sebaceous neoplasm | ||||||||
| Pancreas | 1.44 | 1.38 | ||||||
It is interesting to note that the pattern of the cancer sites associated with LS has changed over time. In the first family with the syndrome, gastric and endometrial cancer were the most common cancers whereas in the generations of the same family described by Lynch in 1971 CRC was the most frequent tumour. 19 In addition, current differences in expression of the LS between families from Western countries and families from the Far East reflect the variation in incidence of cancers in the respective populations. 19 This, and more recent work, confirms the importance of lifestyle and environmental effects in LS and strengthens the opportunity to give directed advice to those known to have LS. 20
Incidence and/or prevalence
There were 34,024 new cases of CRC diagnosed in England in 2014, 18,789 in men and 15,236 in women. 21 LS accounts for approximately 2–3% of cases of CRC, with the population prevalence estimated as 1 in 440. 3,22 However, according to Hampel and de la Chapelle,23 individuals with LS are grossly underdiagnosed. They estimate that the population incidence of LS in the USA is approximately 1 in 370, which is based on the 2.8% incidence of LS among newly diagnosed CRC patients and the 5% lifetime risk for CRC. 23 As the penetrance of a mutation in the MMR genes for CRC is approximately 50%, the incidence of LS in the general population is about double the incidence among CRC patients, or 0.028 × 0.05 × 2 = 0.0028, which is 1 in 370 individuals. 23 On this basis, an estimated 175,000 people in the UK have LS and this leads to > 1100 CRCs per year across the UK.
Although the prevalence of LS is not believed to differ significantly across countries and cultures, population-specific ‘founder’ mutations are well described, for example in Finnish and Swedish populations. A specific founder mutation in the MSH2 gene is found in 2–3% of all CRCs in Ashkenazi Jews aged < 60 years. This mutation is rarely found in the non-Ashkenazi population. In Ashkenazi individuals in whom CRC is diagnosed at age ≤ 40 years, 7% have been found to carry this mutation. Conversely, the mutation is found in < 1% of Ashkenazi individuals in whom CRC is diagnosed after age 60 years. 10
Impact of the health problem
In terms of the impact of LS on an individual, he or she may develop several characteristic clinical and pathological features: colorectal, endometrial, gastric, ovarian and other cancers, early onset of cancer, multiple independent cancers discovered simultaneously (synchronous) or apart (metachronous), more right-sided/proximal CRCs, improved survival and morphological features such as poor differentiation and Crohn’s-like infiltration of lymphocytes. 24
Frequent colonoscopy with polypectomy decreases the mortality of CRC in patients with LS. 3,5 The current screening protocol recommended by the European Hereditary Tumour Group (formerly the Mallorca Group) is for colonoscopy every 1–2 years starting from age 25 years. 19 Unfortunately, there is currently no proven surveillance regime for women at risk of endometrial or ovarian cancer. 19
The risk of gastric cancer in LS in Western countries is low. Screening by endoscopy has been suggested but is unproven and therefore gastric surveillance should be discussed only in those families that have a high incidence of this tumour. 19 The usefulness of screening of the urothelial tract is also debatable. Although urinary tract cancer patients most likely have MSH2 and MSH6 mutations, patients with MLH1 and possibly PMS2 mutations are also at risk and so a pragmatic approach has been to offer yearly urinalysis, urine cytology and renal ultrasound from age 30–35 years, but only to families in whom these cancers have been recorded. There is now evidence that the presence or absence of urinary tumours in a family does not predict their occurrence in other family members. 19,25
Genetic testing for relatives
Understandably, there may be considerable anxiety and distress associated with genetic testing for hereditary cancer syndromes. For example, the knowledge of being at increased risk of cancer but not knowing if cancer will actually develop can be particularly concerning. Furthermore, parents of children with LS may express feelings of guilt. As people may be anxious about many aspects of genetic testing and screening or whether to have risk-reducing surgery, detailed information provided by an experienced clinical geneticist with psychosocial support is essential. Therefore, it is recommended that relatives opting for genetic testing should receive one or more individual pretest counselling sessions with psychological support throughout the whole testing procedure. 19
Genetic counselling helps explain what a positive or negative result means and what the implications are for the person and his or her extended family. It can also help people understand the importance of informing their extended family about the risk of having LS and the benefits of being tested [see www.macmillan.org, www.lynch-syndrome-uk.org, www.ihavelynchsyndrome.com (accessed 13 July 2017)]. Once people fully understand the implications of being diagnosed with LS for them and their families, the associated anxiety may substantially reduce. Studies evaluating the psychological distress associated with genetic testing for LS showed that, immediately after disclosure of the test result, distress significantly increases, but this decreases again after 6 months. 19 Long-term studies have demonstrated that post-result increases in distress return to baseline by 1–3 years. 19
Surgical management
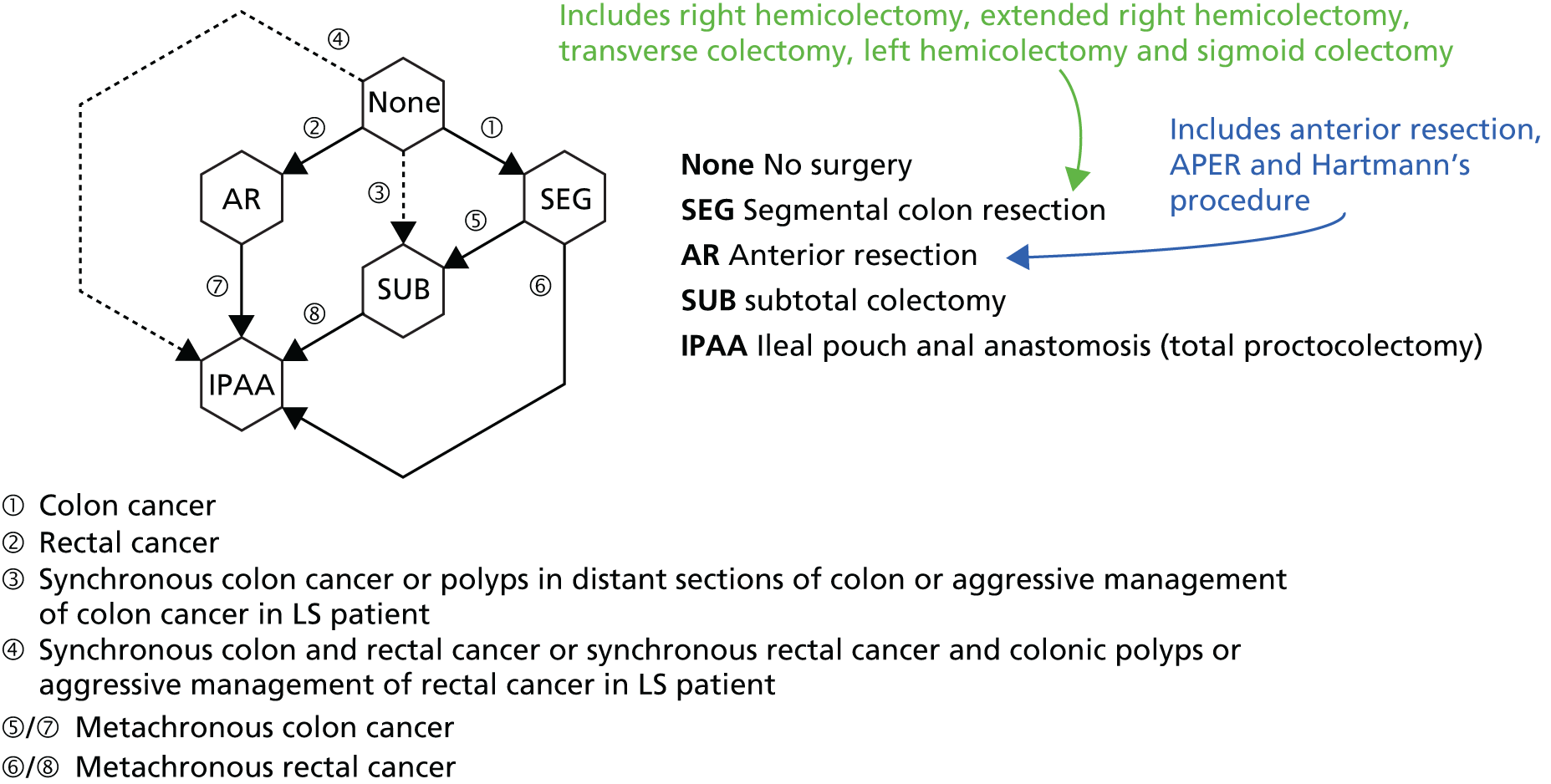
An individual with LS who does not undergo a partial or total colectomy after the first mass is diagnosed as malignant has an estimated 30–40% risk of developing a metachronous tumour within 10 years and a 50% risk of developing a metachronous tumour within 15 years. It is also now known that the risks from age 40 to 70 years of a metachronous CRC (mCRC) in those who have had surgery for an initial primary CRC are 46% for MLH1 mutation carriers, 48% for MSH2 mutation carriers and 23% for MSH6 mutation carriers. 15 This compares with risks in the general population of 3% within 10 years and 5% within 15 years. 10 Furthermore, because of the risk of a synchronous tumour in individuals with LS, the complete colon should be visualised before resection. In view of this substantial risk, it is possible that a subtotal colectomy instead of a segmental resection might be the preferred treatment in patients from LS families with a primary tumour. Vasen et al. 19 also suggest that a colectomy with ileorectal anastomosis may be considered if CRC is detected in a young patient participating in a surveillance programme.
Prophylactic surgery may also be an option for certain extracolonic cancers, for example hysterectomy and oophorectomy after childbearing is complete nearly eliminate the risk of endometrial and ovarian cancer in women with LS,26 but the potential effectiveness and place of such prophylactic surgery is now open to reconsideration given that mortality from endometrial and ovarian cancer in LS is not as high as previously thought. 3,15
Chemotherapy
Fluorouracil (5-FU)-based regimes represent the current gold standard in adjuvant chemotherapy for bowel cancer. In contrast, in vitro studies indicate that MMR-proficient cells treated with 5-FU grow more slowly than MMR-deficient cells, suggesting that a competent MMR system is a critical condition for 5-FU cytotoxicity. 19 Clinical studies on the efficacy of 5-FU in MSI-high (MSI-H) colon cancer are, however, contradictory. 19 A systematic review and meta-analysis did not find evidence that the chemotherapy response was determined by MSI status,27 but it is understood that individuals with stage II CRC may receive MSI testing or MMR immunohistochemistry (IHC) testing to aid clinical decision-making. Stage II tumours showing MSI (i.e. that are MMR deficient) are usually not treated with (5-FU-based) chemotherapy as it is associated with toxicity in some patients and is believed to be of marginal clinical benefit.
There is also increasing evidence to show that MMR-deficient cancers respond especially well to programmed cell death protein 1 inhibitors. 28
Chemoprevention
There is evidence to suggest that LS may be susceptible to environmental manipulation, as demonstrated by the decrease in the incidence of gastric cancer and also perhaps by the apparent differences in penetrance between men and women. 19 This is supported by investigations into the role of aspirin in bowel cancer prevention. Several large studies have demonstrated that aspirin reduces the risk of bowel cancer in the general population, although the mechanisms by which this occurs are unknown. 4 One study has shown that a daily dose of aspirin reduced the incidence of CRC in carriers of LS after 56 months’ follow-up. 29 Furthermore, the Colorectal Adenoma/Carcinoma Prevention Programme 2 (CAPP2) trial of aspirin prophylaxis in LS has demonstrated that aspirin treatment for up to 3 years reduces, a decade later, the overall incidence of LS-associated cancers, including CRC, by 63%. 4,29
The development of vaccines directed at tumours with MSI are also showing promise that this may be an especially good way of preventing otherwise un-addressable LS-associated cancers with a poor prognosis, as well as potentially any cancer in the general population with MSI. 15
Measurement of disease
In current practice, testing for LS in people with CRC is targeted using the Amsterdam II criteria30 and revised Bethesda guidelines31 (Table 2). The Amsterdam criteria were originally developed to identify LS for research studies and the Bethesda guidelines were developed to identify patients with CRC for evaluation for MMR deficiency through tumour testing. Both guidelines use criteria mainly based on family cancer history and age at onset.
| Amsterdam II criteria30 | Revised Bethesda guidelines31 |
|---|---|
All criteria must be met:
|
Only one criterion must be met:
|
These methods are unlikely to be sensitive enough to detect all patients with LS because family history is not always reliable or available and some people with LS may not meet all of the criteria. Furthermore, there is currently no National Institute for Health and Care Excellence (NICE) guidance on the population to be tested or the testing strategy for LS and as a result there is considerable variation in clinical practice.
Description of the technologies under assessment
Summary of the technologies
Overall, the majority of CRCs from individuals with LS genes have two distinguishing characteristics and therefore the diagnostic technologies focus on these aspects:
-
MSI – polymerase chain reaction (PCR)-based MSI testing as carried out by United Kingdom Accreditation Service (UKAS)-accredited regional genetics laboratories using validated in-house tests (including the Promega MSI Analysis System, which is licensed for research use only; any version, Promega Corporation, Madison, WI, USA).
-
Loss of expression or reduced levels of expression of the MMR proteins in the tumour compared with normal tissue – IHC testing for MMR proficiency using antibodies against MMR proteins.
Tumour-based tests
Microsatellite instability testing
As previously mentioned, MSI refers to the variety of patterns of microsatellite repeats observed when DNA is amplified from a MMR-deficient tumour compared with DNA amplified from surrounding normal colonic tissue. 4 Repetitive mono- or dinucleotide DNA sequences (microsatellites) are particularly vulnerable to defective MMR. 17
Microsatellite instability testing involves PCR amplification of standardised DNA markers from a tumour tissue sample and a matched (i.e. from the same patient) healthy tissue sample. The tissue samples must be microdissected, in which a microscope is used to aid dissection, before DNA is extracted, amplified and observed on a DNA fragment length analyser. MSI is indicated when DNA extracted from tumour tissue displays additional peaks on the analyser output (i.e. microsatellite markers of a different size) compared with normal tissue DNA. Tumour samples with microsatellite marker sizes identical to those seen in non-tumour tissue DNA are considered microsatellite stable (MSS).
Laboratories may use a panel of ≥ 10 markers and, more recently, a commercially available kit based on five mononucleotide markers has become popular as mononucleotide microsatellites may be the most sensitive markers for use in detecting MSI. 33 Table 3 provides examples of the panels used.
| Panel | Mononucleotide | Dinucleotide | Other | Notes |
|---|---|---|---|---|
| Bethesda/NCI31 | BAT-25 and BAT-26 | D2S123, D5S346 and D17S250 | If only dinucleotide repeats are mutated, test a secondary panel of microsatellite markers with mononucleotide markers to exclude MSI-low | |
| 10-marker panel34 | BAT-25, BAT-26, BAT-40, MYCL, ACTC and BAT-34C4 | D5S346, D17S250, D18S55 and D10S197 | ||
| NCI-suggested markers for secondary panel31 | BAT-40 and MYCL | |||
| Promega MSI Analysis System version 1.235 | BAT-25, BAT-26, MONO-27, NR-21 and NR-24 | Penta C and Penta D |
Instability in ≥ 30% of the markers is considered MSI-H, instability in < 30% of markers is considered MSI-low (MSI-L) and no shifts or additional peaks is considered MSS. However, if instability is observed in any mononucleotide markers, MSI may be diagnosed. For this reason, MSI testing is moving to a smaller panel of mononucleotide markers, making the process more efficient and cheaper. MSI-H is associated with LS but is also present in around 10–15% of sporadic cancers. 19
There is ongoing debate whether or not MSI-L, which appears to be more common for MSH6 mutations, should be considered as an indication of MSI. As a result, some studies do not report MSI-L separately and include it with either MSS or MSI-H. This obviously provides a challenge when comparing studies. There is also some heterogeneity in the composition of microsatellite markers in MSI panels (both in the nature and in the number of markers), which may lead to differences in test performance and/or threshold effects.
Personal communication from Ian M Frayling (Institute of Cancer and Genetics, University Hospital of Wales, Cardiff, UK, 18 August 2017) indicates that MSI test failures occur in a small proportion of tests (around 5%), largely because of technical challenges surrounding the collection of sufficient DNA from poorly fixed tumour tissue.
Mismatch repair immunohistochemistry
Mismatch repair IHC tests for the presence or absence of MMR proteins in colorectal tumour cells. Antibodies against the MMR proteins are used to stain both tumour cells and non-tumour cells (as an internal control). If nuclear staining is present for all MMR proteins, this suggests that the MMR system is intact. In contrast to MSI, a number of patterns of abnormal MMR expression are seen in Lynch/HNPCC tumours (Table 4).
| MMR mutation | Loss of (or otherwise abnormal) expression by IHC | Overall (%) | |||
|---|---|---|---|---|---|
| MLH1 (alone or in combination with PMS2) (%) | MSH2 (alone or in combination with MSH6) (%) | MSH6 (alone) (%) | PMS2 (alone) (%) | ||
| LS | |||||
| Constitutional MLH1 mutation | 11.8 | 2.0 | 13.8 | ||
| Constitutional MLH1 promoter methylationa | 0.4 | 0.4 | |||
| Constitutional MSH2 mutation | 14.2 | 0.4 | 14.6 | ||
| Constitutional EPCAM mutation | 2.0 | 2.0 | |||
| Constitutional MSH6 mutation | 0.8 | 10.2 | 11.0 | ||
| Constitutional PMS2 mutation | 5.9 | 5.9 | |||
| Not LS | |||||
| Acquired MLH1 promoter methylation | 24.0 | 24.0 | |||
| Acquired MLH1 mutation | 6.7 | 6.7 | |||
| Acquired MSH2 mutation | 2.4 | 2.4 | |||
| Unexplained | 10.2 | 5.9 | 1.6 | 1.6 | 19.3 |
| Total | 53.1 | 25.3 | 12.2 | 9.5 | 100 |
Table 4 highlights that, for some individuals with abnormal MMR IHC results in the tumour, no germ-line mutation can be found. Somatic mutations in MMR genes can occur (with attendant loss of MMR protein expression) as secondary events in other CRC predisposition syndromes, which can mimic LS, such as inherited mutations in MUTYH or POLD1. 38,39
It is also important to realise that a small proportion of possibly Lynch-related tumours do not exhibit any abnormality on analysis by IHC, even though they have lost MMR function as demonstrated by MSI. This may be because they have mutations that allow expression of a stable protein with nuclear localisation and an intact epitope but which is functionally inactive. 40
Immunohistochemistry panels may use two MMR antibodies (either MLH1 and MSH2 or MSH6 and PMS2) or four antibodies (MLH1, MSH2, MSH6 and PMS2). However, a panel with only MLH1 and MSH2 antibodies is unlikely to detect MMR deficiency in MSH6 and PMS2, will be expected to have a lower sensitivity than a four-antibody panel and will not be able to fully diagnose the pattern of abnormal expression seen in all tumours. 37 Furthermore, the heterodimeric association of proteins such that loss of MLH1 expression is almost always accompanied by loss of PMS2 expression and loss of MSH2 expression is almost always associated with loss of MSH6 acts as a very useful confirmatory finding, especially in CRCs that are prone to suboptimal fixation. 37,40 A four-antibody panel is recommended in the UK. 41
Test failure is a recognised issue with IHC and usually results from incomplete tissue fixation (a common problem with CRCs). Palomaki et al. 42 counted six test failures across a number of studies (n = 136 patients), corresponding to a failure rate of 4.4%.
BRAF V600E mutation testing and MLH1 promoter methylation testing
Around 10–15% of sporadic CRCs show MSI-H and in the vast majority of these tumours this is the result of acquired promoter methylation of the MLH1 gene, leading to loss of MLH1 protein expression. However, a small proportion of sporadic MSI-H CRCs will occur because of the loss of the MSH2, MSH6 or PMS2 gene and germ-line MLH1 hypermethylation will be observed in a very small number of CRCs as a result of LS. 40,43 Approximately half of the sporadic CRCs with MSI-H will also have a BRAF V600E mutation. This is a specific mutation in the BRAF gene that almost never occurs in tumours arising in LS.
Therefore, testing for MLH1 promoter methylation and the BRAF V600E mutation represents a way of distinguishing sporadic CRCs from LS in a proportion of MLH1-negative tumours. 40
Identification of important subgroups
First-degree relatives of a LS mutation carrier have a 50% risk of inheriting the mutation. Therefore, when the familial mutation has been identified, cascade testing should be offered to the first- and second- and, when possible, third-degree biological relatives. Studies have shown that the uptake of genetic testing in families with LS varies from 43% in the USA to 57% in the Netherlands and 75% in Finland. 19 Suggested reasons for this variation include differences in the study setting or fundamental differences between the health-care and social security systems.
As CRC in an individual with LS is likely to be diagnosed at a younger age, the prevalence of LS in people with CRC will vary across age groups. For example, the prevalence of LS falls from 8.4% at 50 years to 5.7% at 60 years and 3.8% at 70 years. 4 This is because the incidence of CRC in the general population rises more rapidly than the incidence of CRC in people with LS. The total annual incidence of cases of CRC in England in 2014 increased from 2107 at 50 years, to 5880 at 60 years and 13,823 at 70 years. 21
Current usage in the NHS
Microsatellite instability testing for the purpose of identifying LS is currently only carried out in people considered to be at high risk of having LS, that is, people with a family history of cancer and/or who are aged < 50 years at the onset of cancer. For cancer for which there is no suspicion of LS, MSI or IHC testing may yet be conducted to inform prognosis or guide therapy. Expanding diagnostic testing to all people with CRC and identification of families who could benefit from cascade genetic testing may lead to increased surveillance and consequently improved patient outcomes through earlier diagnosis and treatment. Currently, testing for LS may occur using a number of different strategies (Figure 1).
FIGURE 1.
Diagnostic test strategies for LS. MLPA, multiplex ligation-dependant probe amplification.

Anticipated costs associated with the intervention
Microsatellite instability and IHC testing are both tumour-based tests, the costs of which need to include the costs of preparing the sample, analysing the sample and reporting the test results as well as the costs of administration, transportation, additional wear and tear on machinery, training time and repeat tests. As a LS diagnosis cannot be confirmed with just MSI or IHC testing, the cost of downstream testing also impacts on the overall costs of the interventions, as well as the number of downstream tests that will be run as a result of MSI or IHC testing being indicative of LS.
The costs for constitutional DNA tests in the study by Snowsill et al. 4 were assumed to be all-inclusive; all laboratory, processing and transportation costs were assumed to be accounted for through core funding. It is not clear if this will continue with the increase in the number of tests requested. There is also the additional factor that gene sequencing costs in particular have reduced in the last 2 years in UK regional genetics laboratories with the increasing introduction of next-generation sequencing (NGS).
The costs of the diagnostic tests are detailed further in Chapter 5 (see Diagnostic tests). Unit costs for MSI and IHC testing are estimated to be £178 and £210 respectively. This is equivalent to ≈£7M (without additional testing) for a cohort of ≈34,000 individuals with newly diagnosed CRC,21 increased from an estimated ≈£2M for the same cohort under a no-reflex diagnostic testing strategy (assuming that some people with CRC will receive MSI and IHC testing for therapeutic/prognostic purposes). However, this total cost is unlikely to remain constant, as the number of individuals with newly diagnosed CRC, and the number of families with LS identified (for whom MSI and IHC diagnostic testing will be unnecessary), are likely to differ in the future.
Comparators
Constitutional deoxyribonucleic acid tests
The gold standard for the diagnosis of LS is comprehensive screening for constitutional mutations in the MMR genes and the EPCAM gene. This screening is conducted using a DNA sequencing method to detect point mutations, small insertions and small deletions and multiplex ligation-dependant probe amplification (MLPA) to detect large structural DNA abnormalities such as genomic deletions, duplications and rearrangements. 4 Sequencing is usually performed on lymphocytic DNA from a blood sample. The various forms of mutations are described in Box 1.
-
Missense: a change in one DNA base pair that results in the substitution of one amino acid for another in the protein made by a gene.
-
Nonsense: a change in one DNA base pair that results in a premature signal to stop building a protein. This type of mutation results in a shortened protein that may function improperly or not at all.
-
Insertion: changes the number of DNA bases in a gene by adding a piece of DNA. As a result, the protein made by the gene may not function properly.
-
Deletion: changes the number of DNA bases by removing a piece of DNA. Small deletions may remove one or a few base pairs within a gene whereas larger deletions can remove an entire gene or several neighbouring genes. Deletion of the DNA may alter the function of the resulting protein(s).
-
Duplication: consists of a piece of DNA that is abnormally copied one or more times. This type of mutation may alter the function of the resulting protein.
-
Frameshift mutation: occurs when the addition or loss of DNA bases changes a gene’s reading frame. A reading frame consists of groups of three bases that each code for one amino acid. A frameshift mutation shifts the grouping of these bases and changes the code for amino acids. The resulting protein is usually non-functional. Insertions, deletions and duplications can all be frameshift mutations.
-
Splice site: causes abnormal messenger ribonucleic acid (mRNA) processing, generally leading to in-frame deletions of whole exons or out-of-frame mRNA mutations leading to nonsense-mediated decay of mRNA. Mutations may be located deep in intronic sequences.
-
Promoter: mutations (occurring) in the controlling region of a gene (the promoter) leading to its non-expression. Epigenetic mutations in the promoter (i.e. abnormal methylation of CpG sites) may give rise to the same effect.
CpG, cytosine–phosphate–guanine (sequence of DNA bases).
Adapted from the US National Library of Medicine44 (public domain).
Other techniques for genetic testing in LS, as listed in Table 5, may be found in older studies.
| Test | Description | Comments |
|---|---|---|
| High-output screening techniques | Single-strand conformational polymorphism (SSCP), conformation-sensitive gel electrophoresis (CSGE), denaturing gradient gel electrophoresis (DGGE), denaturing high-performance liquid chromatography (DHPLC) | These methods use the change in chemical properties of altered DNA to differentiate it from normal DNA (now considered obsolescent/obsolete in the UK) |
| DNA sequencing | Can be used following high-output screening or as a primary approach when directed by IHC patterns | The main method used in the UK for detecting most MMR gene mutations. However, it does not reliably detect deletions or rearrangements |
| Methods to detect large structural DNA abnormalities | MLPA | MLPA is the preferred technique in the UK. Large structural DNA abnormalities are an important cause of LS (5–25% of cases, depending on the gene) but are not generally detected by high-output screening techniques or DNA sequencing. MLPA, which involves measurement of the relative copy number of DNA sequences, has evolved to become a standard approach for analysing MMR genes for deletions |
| Conversion analysis | Only a single allele is analysed at a time. This can increase the yield of genetic testing but is technically complicated, expensive and not widely available |
Although comprehensive screening for constitutional mutations should accurately detect the majority of known LS-causing mutations, there are some occasions when a novel mutation may be identified that is of uncertain significance. As this variant or mutation cannot be demonstrated to be pathological or non-pathological, it is not possible to make a diagnosis or recommendation for management, such as colorectal surveillance. However, a major advance has been the establishment by the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) of an internationally recognised reference database together with a multidisciplinary team of experts to maximise the number of mutations interpreted as being either of clinical consequence or innocuous, thus minimising the number of mutations in the ‘uncertain’ bracket. Before this work, 58% of the 12,006 mutations listed were unclassified variants; this has now been reduced to 32%, that is, those mutations that now fall into the category of ‘variants of uncertain significance’, otherwise known as class 3. 45 Hence, this now enables a variant of uncertain significance to be pursued by testing for the variant in other family members with LS-related cancers or by testing stored tumour tissue for MMR deficiency.
Care pathways
There is currently no NICE guidance on the diagnosis and management of LS. However the diagnosis and management of LS is described in several national and international guidelines:
-
British Society of Gastroenterology (BSG) (2010): Guidelines for Colorectal Cancer Screening and Surveillance in Moderate and High Risk Groups46
-
European guidelines (2013): Revised Guidelines for the Clinical Management of Lynch Syndrome (HNPCC)19
-
Bethesda guidelines (2004): Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability31
-
Amsterdam II criteria (1999): New Clinical Criteria for Hereditary Nonpolyposis Colorectal Cancer (HNPCC, Lynch syndrome) Proposed by the International Collaborative Group on HNPCC. 24
In the NHS, CRC in LS patients is generally treated as per NICE clinical guideline 13147 (Figure 2).
FIGURE 2.
Overview of pathway for CRC. Source: National Institute for Health and Care Excellence. Colorectal Cancer: the Diagnosis and Management of Colorectal Cancer. Clinical guideline CG131. 47 © National Institute for Health and Care Excellence 2017. All rights reserved and subject to NICE ‘Notice of Rights’. Available from https://pathways.nice.org.uk/pathways/colorectal-cancer. NICE guidance is prepared for the National Health Service in England. It is subject to regular review and updating and may be withdrawn.

The European Society for Medical Oncology guidelines48 are also used by clinicians in the NHS to guide treatment decisions. The guidelines state that ‘MSI/MMR may be useful to identify a small (10–15%) subset (those with microsatellite instability) of stage II colorectal cancer patients who are at a very low risk of recurrence and in whom the benefits of chemotherapy are very unlikely’ (p. vi67). 48
The Royal College of Pathologists’ minimum data set (July 2014) for CRC mandates the use of IHC or other testing for molecular features of LS in CRC or Lynch-associated cancer patients aged < 50 years at the time of diagnosis. 49 However, the results of a 2015 Bowel Cancer UK survey on reflex testing for LS in people diagnosed with bowel cancer aged < 50 years highlighted that there is variability in who receives testing among the different trusts and health boards. 50 Ideally, MSI and IHC testing will usually be conducted on tumour tissue obtained during surgical treatment or via biopsy. A histopathologist selects tissue for testing and performs microdissection for MSI testing or sectioning and staining for IHC. Microdissected samples for MSI testing are processed by a laboratory genetics centre where PCR-based MSI testing is performed and reported to the histopathologist, who in turn informs the cancer team (usually a consultant colorectal surgeon) along with recommendations for further testing.
If the results of MSI and/or IHC testing are suggestive of LS further tumour tissue-based tests may be ordered [e.g. IHC (if not already conducted), BRAF V600E mutation testing, MLH1 promoter methylation testing] or the patient may be referred directly to the clinical genetics department. At this point, the clinical genetics department will discuss the findings with the patient, describe LS and take a detailed family history (pretest genetic counselling). If the clinical genetics team and the patient agree that constitutional MMR mutation testing is appropriate then a blood sample will be sent to the genetics laboratory for testing.
If a pathogenic constitutional MMR mutation is not found, a variant of uncertain significance is found or the mutation identified is inconsistent with existing findings, the genetics team will provide appropriate counselling and further testing and propose an appropriate management strategy for the patient.
Outcomes
The accuracy of MSI and IHC testing of tumour tissue for LS has been evaluated against the reference (gold) standard of constitutional genetic testing. Clinically important outcomes relevant to test accuracy include:
-
Sensitivity – the probability of detecting LS in someone with LS:(1)Sensitivity=True positiveTrue positive+False negative=TPTP+FN.
-
Specificity – the probability of not detecting LS in someone without LS:(2)Specificity=True negativeFalse positive+True negative=TNFP+TN.
-
Likelihood ratio (LR) – the likelihood that a given test result would be expected in a patient with the target disorder compared with the likelihood that that same result would be expected in a patient without the target disorder.
-
LR for positive test result (LR+) – the relative likelihood that a positive test result occurs in people with the disease compared with those without the disease:(3)LR+=Pr(T+|D+)Pr(T+|D−)=Sensitivity1−Specificity.
-
LR for negative test result (LR–) – the relative likelihood that a negative test result occurs in people with the disease compared with those without the disease:(4)LR−=Pr(T−|D+)Pr(T−|D−)=1−SensitivitySpecificity.
-
Positive predictive value (PPV) – the probability of someone with a positive test result actually having LS:(5)PPV=TPTP+FP.
-
Negative predictive value (NPV) – the probability of someone with a negative test result actually not having LS. (6)NPV=TNTN+FN.
-
Diagnostic yield (also known as test positivity rate or apparent prevalence) – the number of positive test results divided by the number of samples.
-
Test failure (non-informative test result) rate.
Outcomes relevant to cost-effectiveness are:
-
the number of individuals with LS receiving LS surveillance
-
the number of individuals with LS not receiving LS surveillance
-
the number of individuals without LS receiving LS surveillance
-
the number of individuals without LS not receiving LS surveillance
-
the sensitivity and specificity of each diagnostic strategy (as opposed to individual tests)
-
the costs of each strategy (discounted and undiscounted), including disaggregated costs of diagnosis and outcomes
-
quality-adjusted life-years (QALYs) (discounted and undiscounted)
-
overall survival (and whether censored on reaching age 100 years)
-
CRC-, endometrial cancer- and overall cancer-free survival (and whether censored because of death or on reaching age 100 years)
-
event-free survival (and whether censored on reaching age 100 years)
-
the number of incident CRCs
-
the number of incident endometrial cancers
-
the number of colonoscopies performed.
Chapter 2 Assessment of test accuracy
Methods for reviewing effectiveness
The diagnostic accuracy of molecular MSI testing and MMR IHC (each with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing) was assessed in a systematic review of the research evidence. The review was undertaken following the principles published by the University of York Centre for Reviews and Dissemination. 51
Identification of studies
The following bibliographic databases were searched to identify relevant studies: MEDLINE, MEDLINE In-Process & Other Non-Indexed Citations, EMBASE and the Health Management Information Consortium (HMIC) (all via Ovid); Web of Science (including conference proceedings, via Thomson Reuters); and the Cochrane Database of Systematic Reviews (CDSR), Cochrane Central Register of Controlled Trials (CENTRAL) and Health Technology Assessment (HTA) database (all via The Cochrane Library). The search strategies were developed by an information specialist (SB) and consisted of population terms for LS or HNPCC and intervention terms for MSI or IHC. Methodological filters for test accuracy studies were not used to limit the study designs retrieved as these have been shown to reduce sensitivity. 52 Search results were limited by date from 2006 (searches were run in February 2016) and to English-language studies. The full search strategies for each database are reproduced in Appendix 1.
The search results were exported to EndNote X7 (Thomson Reuters, CA, USA) and deduplicated using automatic and manual checking.
To identify relevant studies published before 2006, the study by Bonis et al. 17 was screened. In addition, the studies by Palomaki et al. ,42 Snowsill et al. 4 and Vasen et al. ,19 as well as any other systematic reviews identified by the bibliographic database searches, were used to source relevant studies published before 2006 and additional studies published after 2006. For the purpose of this review, a systematic review was defined as one that includes a focused research question, explicit search criteria that are available to view, explicit inclusion/exclusion criteria, a critical appraisal of included studies, including consideration of internal and external validity of the research, and a synthesis of the included evidence (narrative or quantitative).
Items included after full-text screening were forward citation chased using Scopus (Elsevier). The reference lists of included studies were also screened for any other relevant studies.
Relevant studies were then identified in two stages. First, titles and abstracts returned by the search strategy were examined independently by two researchers (HC and TJH) and screened for possible inclusion, using prespecified inclusion and exclusion criteria (see following section). Disagreements were resolved by discussion. Full texts of studies included at the title and abstract screening stage were obtained, as were full texts of studies identified from systematic reviews and from forward and backward citation chasing. Two researchers (HC and TJH) independently examined full texts for inclusion or exclusion. Disagreements were again resolved by discussion.
Inclusion and exclusion criteria
Population
Studies of individuals with CRC were included. This included fresh samples taken from people who were newly diagnosed with CRC or samples that had been retained in storage.
The unit of assessment was individual patients. If results were presented according to individual cancers (e.g. when patients had multiple primary colorectal malignancies), when possible the earliest CRC tested with an index test was used as the unit of assessment.
Studies in which clinical or family history criteria were used to select CRC patients were eligible for inclusion under certain circumstances (see Study design).
Index tests
The index tests to be considered were:
-
molecular MSI testing with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing
-
MMR IHC with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing.
Studies in which BRAF V600E and/or MLH1 promoter methylation tests were performed only on certain patients according to their MSI or IHC test results were eligible for inclusion.
Studies were eligible for inclusion if one or more index test was assessed against a reference standard.
Reference standard
The reference standard was constitutional MMR mutation testing (for abnormalities that provide a genetic diagnosis of LS), which, as a minimum, included DNA sequencing and MLPA (or another appropriate technique for detecting large genomic abnormalities). Other appropriate techniques were Southern blot analysis, gene-targeted array-based comparative genomic hybridisation (aGCH) and NGS. However, NGS alone (i.e. without MLPA) was accepted as an includable technique for detecting large genomic abnormalities only if the study described or cited peer-reviewed methodology for identifying structural variants in LS based on output data. If no such methodology was described it was assumed that NGS would not detect structural variants and would not be an includable technique. Studies were eligible for inclusion if MLPA, Southern blot, aGCH or NGS (as described above) was conducted only when sequencing found no clearly pathogenic mutations. Studies in which IHC results directed the MMR genes to be tested were eligible for inclusion (e.g. if MLH1 was not tested when only MSH2 and MSH6 proteins were absent on IHC).
Unless the aim of a study was to investigate the test accuracy of an index test in individuals with mutations in a particular MMR gene, studies must have tested MLH1, MSH2 and MSH6 as a minimum (unless IHC results directed otherwise).
Outcomes
The outcomes assessed for index tests were:
-
sensitivity
-
specificity
-
LR+
-
LR–
-
PPV
-
NPV
-
accuracy or concordance with the reference standard: the proportion of test results correctly identified by the test, that is, the rate of agreement with the reference standard:(7)Accuracy=True positive+True negativeTotal number of subjects=TP+TNTP+FP+FN+TN,
-
diagnostic yield (also known as test positivity rate or apparent prevalence)
-
test failure (non-informative test result) rate.
Study design
Single-gate diagnostic studies with random or consecutively recruited participants were considered the optimal design for evaluating the test accuracy of MSI and IHC testing and were therefore eligible for inclusion. Two-gate diagnostic studies were also included.
Studies were included if all participants received the index test(s) and the reference standard. Studies that recruited a representative sample of all CRC patients but did not apply the reference standard to all patients were included if the reference standard was applied to all patients testing positive for one or more index test(s) and to a representative (e.g. random) sample of patients testing negative for all index tests.
Studies that limited recruitment to high-risk populations (except by applying an age limit to an otherwise population-based sample) were included only to estimate sensitivity and only if the index test(s) and reference standard had been applied to all participants.
Data abstraction strategy
Data were extracted by one reviewer (TJH) using a standardised data extraction form (see Appendix 3) and were checked by a second reviewer (HC). Disagreements were resolved by discussion, with involvement of a third reviewer if necessary. Data were then transferred to standardised tables.
Critical appraisal strategy
The methodological quality of the studies was assessed according to criteria specified by phase 3 of the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies) tool53 (see Appendix 2). This was carried out alongside data extraction using the same form.
Assessments were conducted by one reviewer (TJH) and judgements were checked by a second (HC). Any disagreement was resolved by discussion, with involvement of a third reviewer as necessary.
Methods of data synthesis
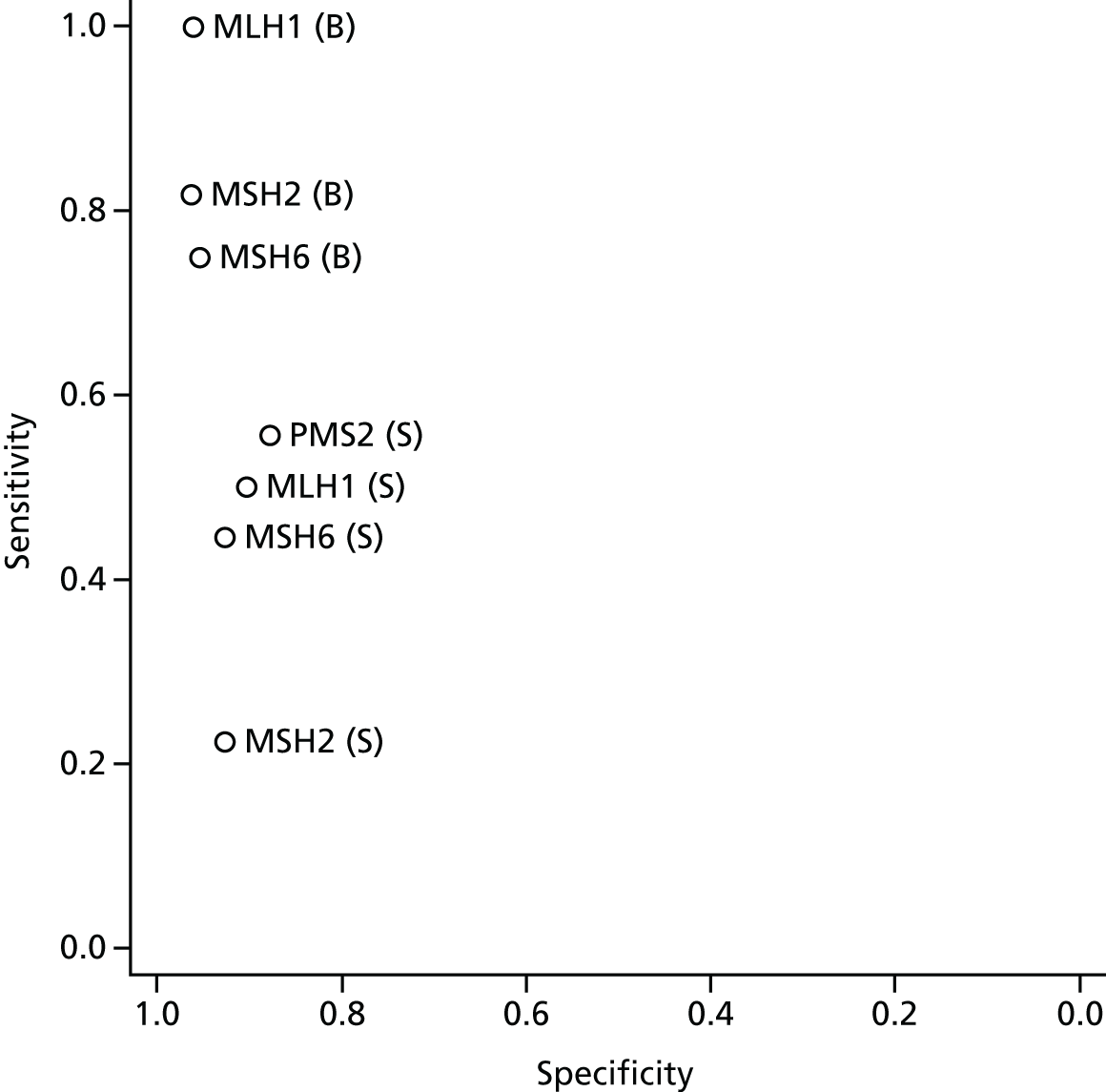
The data extracted from each study were analysed in Stata® 13 (StataCorp LP, College Station, TX, USA). For studies based on high-risk samples the sensitivity of the index test(s), with 95% confidence intervals (CIs), were calculated. For studies that were not based on high-risk samples (including studies in which the population was age limited), sensitivity, specificity, LR+, LR–, PPV and NPV, with 95% CIs, were calculated when data permitted. Of these latter studies, those that provided estimates of both sensitivity and specificity had their point estimates plotted in receiver operating characteristic (ROC) space.
The data extracted from each study, the results obtained from analysing individual studies and the quality assessment for each study are presented in structured tables and in a narrative synthesis. Any possible effects of study quality on the data are discussed.
Data from individual studies were not pooled in meta-analysis; once data from individual studies were sorted into categories (e.g. high-risk population or age-limited population; MSI-L defined as positive or defined as negative) there were insufficient methodologically homogeneous data sets to enable meaningful data pooling.
Results
Quantity and quality of research available
The electronic searches retrieved a total of 3921 unique titles and abstracts, of which 3844 articles were excluded based on screening titles and abstracts. The remaining 77 articles were requested as full texts for more in-depth screening.
After screening systematic reviews, including those that were prespecified,4,17,19,42 a further 41 articles were retrieved as full texts for in-depth screening.
Of the 118 articles retrieved as full texts (identified from electronic searches and systematic reviews), 109 were excluded. The reasons for exclusion were that the study design (n = 47), reference standard (n = 33) or outcomes (n = 3) did not match the review inclusion criteria, the article was a duplicate publication (n = 3) or the article was an abstract that both reported insufficient information to be included in the review and was unconnected to any of the included studies (n = 23). The bibliographic details of the studies retrieved as full papers and subsequently excluded, along with the reasons for their exclusion, are detailed in Appendix 3. The remaining nine studies were included.
After backward and forward citation chasing, a further 16 full-text papers were obtained, of which one study was included and 15 were excluded because the reference standard (n = 9), study design (n = 3), population (n = 2) or index test (n = 1) did not match the review inclusion criteria. The bibliographic details of these excluded studies, along with the reasons for their exclusion, are also provided in Appendix 3.
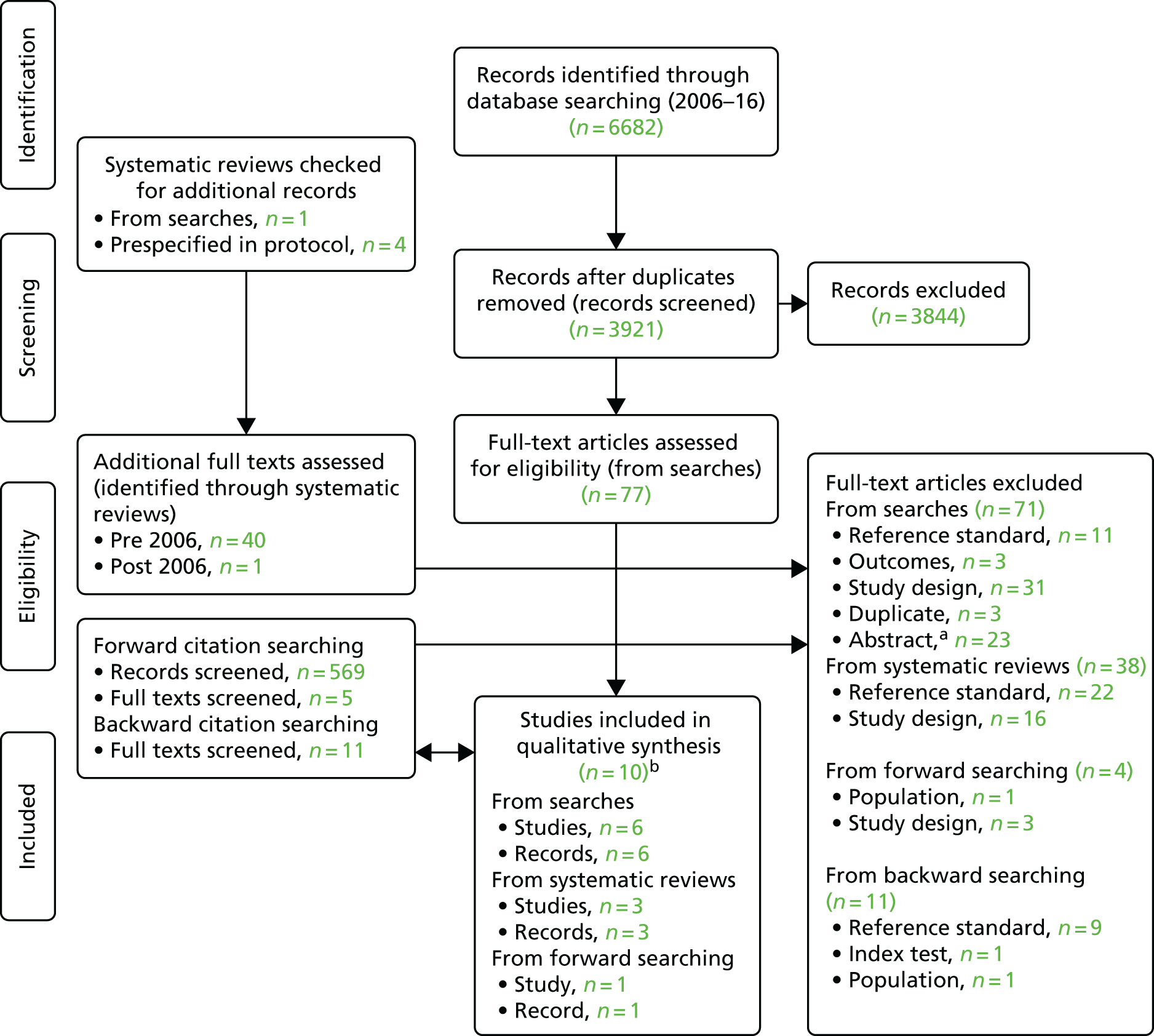
In total, therefore, 133 full-text articles were assessed, of which 10 studies34,54–62 met the review inclusion criteria. The process of study selection is shown in Figure 3. It should be noted that one of the included studies had two distinct samples (a population-based sample and a high-risk sample) and therefore had two distinct sets of data. 34 These two samples are treated separately in this review; therefore, although there are 10 included studies, there are, in fact, 11 included populations/data sets. The results from all 11 populations are included in the narrative synthesis.
FIGURE 3.
Summary of the selection process. a, Abstracts were excluded when they were not linked to an included study and did not provide sufficient methodological information to meet the review inclusion criteria or allow data to be extracted. b, One of these studies included two distinct populations,34 both of which are included in this review. Although there are 10 included studies, there are, therefore, 11 included data sets. Source: adapted from Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med 6(7):e1000097. 63 https://doi.org/10.1371/journal.pmed1000097
Description of the included studies
Four of the included studies reported data from a population-based sample. The study by Poynter et al. 34 reported data from two populations, one of which appeared to be an unselected CRC population, although this is not entirely clear because participant inclusion criteria were not reported. The other three population-based studies54–56 included CRC populations to which an age limit had been applied. All of these, and the study by Poynter et al. ,34 were single-gate studies.
The remaining studies (including the other sample reported in the study by Poynter et al. 34) were based on participants with CRC who were also selected for being at high risk for LS. Five of these studies used a single-gate design. 34,57–60 The remaining two studies used variations on a two-gate study design;61,62 in these two studies participants with positive reference standard results were recruited but no reference standard negatives were recruited, thus resembling half of a two-gate design from which sensitivity estimates could be obtained. These studies do not, therefore, have two gates and from this point forward, for clarity, they will be referred to as reference standard-positive studies.
Table 6 provides a summary of all studies included in the test accuracy review. A narrative summary of the included studies and their population characteristics is provided in the following sections.
| Study | Participants and selection | n | Reference standard | MSI testing | MSI panel | IHC | IHC proteins |
|---|---|---|---|---|---|---|---|
| Single-gate studies recruiting population-based samples | |||||||
| Barnetson et al., 200654 | Diagnosed at < 55 years of age; consecutive recruitment | Recruited, n = 1259; RS, n = 870; MSI, n = 352; IHC, n = 312–328 | DHPLC for MSH2 and MLH1. Noted variants were sequenced (as were five MSH2 exons and three MLH1 exons and all 10 MSH6 exons). MLH1 and MSH2 were assessed for deletions by MLPA | Yes | BAT25, BAT26, D2S123, D5S346 and D17S250 | Yes | MLH1, MSH2 and MSH6 |
| Limburg et al., 201155 | Diagnosed at < 50 years of age; random recruitment | Recruited, n = 195; RS, n = 189–195; IHC, n = 155 | Direct sequencing following PCR. Potential variants were confirmed by repeated PCR amplification of the indicated gene region(s) and sequence determination. MLH1 and MSH2 were assessed for deletions by Southern blotting and MLPA | No | NA | Yes | MLH1, MSH2 and MSH6 |
| aPoynter et al., 200834 | Recruitment through population-based cancer registries (population-based sample); selection process unclearb | Recruited, n = 1061; RS, n = 726; MSI, n = 1061; IHC, n = 719 | For MSH2 and MLH1 a combined approach of DHPLC/direct sequencing and MLPA was used. For MSH6, direct sequencing was used in cases with absent IHC staining of MSH6 | Yes | BAT25, BAT26, D5S346, D17S250, BAT40, MYCL, ACTC, Dl 8S55, D1OS197 and BAT34C4 | Yes | MLH1, MSH2, MSH6 and PMS2 |
| Southey et al., 200556 | Diagnosed at < 45 years of age; random recruitment | Recruited, n = 131; RS, n = 59; MSI, n = 105; IHC, n = 118 | DHPLC, PCR for direct automated sequencing, MLPA on samples from 10 patients who had tumours lacking expression of at least one MMR protein and for which no previous mutation had been identified by sequencing | Yes | BAT25, BAT26, D2S123, D5S346, D17S250, BAT40, MYB, TGFRII, IGFIIR and BAX | Yes | MLH1, MSH2, MSH6 and PMS2 |
| Single-gate studies recruiting populations at high risk for LS | |||||||
| Caldes et al., 200457 | HNPCC families selected through a clinic for familial cancer; selection process unclear | Recruited, n = 58; RS, n = 58; MSI, n = 58; IHC, n = 58c | PCR, DGGE and sequencing. MSI-H cases that were negative for mutations in MLH1, MSH2 and MSH6 by DGGE and direct sequencing were analysed for genomic deletions in MSH2 and MLH1 by Southern blotting | Yes | BAT25, BAT26, D2S123, D5S346 and D17S250 | Yes | MLH1, MSH2 and MSH6 |
| Mueller et al., 200958 | ‘Suspected LS’ participants who met Amsterdam criteria, met modified Amsterdam criteria, were ‘HNPCC-like’ or met Bethesda criteria; selection process unclear | Recruited, n = 48;d RS, n = 48; MSI, n = 48; IHC, n = 48 | Sequencing and MLPA | Yes | Five- and 10-panel markers, no further details provided | Yes | MLH1, MSH2, MSH6 and PMS2 |
| Overbeek et al., 200759 | Family history that fulfilled one of the following criteria:
|
Recruited, n = 83; RS, n = 83; MSI, n = 43; IHC, unclear | SSCP or DGGE and direct sequencing; MLPA for the detection of large deletions and duplications (confirmed by Southern blot analysis or with specific PCR using primers flanking the deletion or one of the breakpoints of a duplicated region) | Yes | BAT25, BAT26, D2S123, D5S346 and D17S250 (BAT40 was also added to the standard set of markers but it is unclear for which participants) | Yes | MLH1, MSH2, MSH6 and PMS2 |
| aPoynter et al., 200834 | Recruitment through high-risk clinics (clinic-based sample); selection process unclearb | Recruited, n = 172; RS, n = 152; MSI, n = 172; IHC, n = 157 | For MSH2 and MLH1, a combined approach of DHPLC/direct sequencing and MLPA was used. For MSH6, direct sequencing was carried out in cases with absent IHC staining of MSH6 | Yes | BAT25, BAT26, D5S346, D17S250, BAT40, MYCL, ACTC, Dl 8S55, D1OS197 and BAT34C4 | Yes | MLH1, MSH2, MSH6 and PMS2 |
| Shia et al., 200560 | Family history that fulfilled one of the following criteria:
|
Recruited, n = 83; RS, n = 83; MSI, unclear;e IHC, uncleare | DHPLC followed by direct sequencing of DNA fragments that displayed an abnormal chromatogram. Analysis for large deletions (multiplex PCR of short florescent fragments) was performed only in MSI-H tumours in which a point mutation was not detected | Yes | BAT25, BAT26, D2S123, D17S250, BAT40, PAX6 and MYCL1 | Yes | MLH1, MSH2 and MSH6 |
| Reference standard-positive studies (recruiting populations with known mutation status) | |||||||
| Hendriks et al., 200361 | Germ-line mutation in MLH1, MSH2 or MSH6; selection process unclear | Recruited, n = 45; RS, n = 45; MSI, n = 33; IHC, n = 45 | DGGE or Southern blotting | Yes | BAT25, BAT26, D2S123, D5S346, D17S250, BAT40, MSH3 and MSH6 | Yes | MLH1, MSH2 and MSH6 |
| Okkels et al., 201262 | Germ-line mutation in MSH6; consecutive recruitment | Recruited, n = 56; RS, n = 56; IHC, n = 56 | PCR and sequencing in sense and antisense directions, MLPA | No | NA | Yes | MLH1, MSH2, MSH6 and PMS2f |
Single-gate studies recruiting population-based samples
As mentioned earlier, the population-based sample included in the study by Poynter et al. 34 appeared to be completely unselected. The other three population-based studies included in this review54–56 were based on CRC populations to which an age limit had been applied. For all three of these studies this limit was for age at diagnosis (rather than age at recruitment) and was < 55 years for Barnetson et al. ,54 < 50 years for Limburg et al. 55 and < 45 years for Southey et al. 56 These four studies varied in size; the studies by Barnetson et al. 54 and Poynter et al. 34 were the largest, recruiting 1259 participants and 1061 participants respectively. The studies by Limburg et al. 55 and Southey et al. 56 recruited similar numbers of participants (n = 195 and n = 131 respectively).
With regard to the population characteristics of these four studies, all provided details on the participants’ sex and for all studies the ratio of men to women was similar (Table 7). Although the study by Poynter et al. 34 included the only unselected CRC population identified by this review, the age of the participants was not reported. The age of the participants in the other three studies ranged from a median of 37.1 years (range 24–42 years) for those receiving the reference standard in the study by Southey et al. 56 to a mean of 49.0 years (± 3.9 years) for one of the subgroups in the study by Barnetson et al. 54 These low mean and median ages, which were similar across the three studies, are unsurprising given that all three applied an age limit to participants for inclusion in the study. The four studies recruiting population-based samples all reported a low proportion of participants meeting Amsterdam II criteria, ranging from 0.1% in one of the subgroups in the study by Poynter et al. 34 to 12% in the study by Southey et al. 56 Two studies34,54 also reported the proportion of participants meeting the revised Bethesda guidelines. The specific location of the CRC was reported by Barnetson et al. 54 and Poynter et al. 34 but not by Limburg et al. 55 or Southey et al. 56 None of the studies provided details on the ethnicity of the participants.
| Study | Age (years), mean (± SD) or median (range) | Participants meeting AMS II/RBG criteria, n (%) | Sex, n (%) | Cancer location, n (%) |
|---|---|---|---|---|
| Single-gate studies recruiting population-based samples | ||||
| Barnetson et al., 200654 | Non-carrier, 48.2 (± 6.0); carrier, 42.7 (± 7.7); MLH1, 38.5 (± 8.4); MSH2, 43.8 (± 6.1); MSH6, 49.0 (± 3.9) | AMS II, 34 (4); RBG, 555 (64) | Male, 462 (53.1); female, 408 (46.9) | Non-carrier: rectum, 285 (35.2); sigmoid colon and rectosigmoid, 249 (30.7); descending colon, 37 (4.6); splenic flexure, 21 (2.6); ascending colon/hepatic flexure, 68 (8.4); caecum, 110 (13.6); appendix 10, (1.2); transverse colon, 30 (3.7); site not assessable, 22 (2.6) Carrier: rectum, 7 (18.4); sigmoid colon and rectosigmoid, 7 (15.5); descending colon, 2 (5.3); ascending colon/hepatic flexure, 10 (26.3); caecum, 9 (23.7); transverse colon, 3 (7.9) |
| Limburg et al., 201155 | 42.9 (± 6.1) | AMS II, 10 (5.1) | Male, 91 (47); female, 104 (53) | NR |
| aPoynter et al., 200834 | NR | Methylated: AMS II, 6 (0.6); RBG, 81 (7.6) Unmethylated (loss of MLH1): AMS II, 10 (0.9); RBG, 39 (3.7) Unmethylated (loss of other MMR gene): AMS II, 17 (1.6); RBG, 50 (4.7) Unmethylated (no MMR loss): AMS II, 1 (0.1); RBG, 20 (1.9) |
Methylated: male, 44 (4.1); female, 125 (11.8) Unmethylated (loss of MLH1): male, 25 (2.4); female, 26 (2.4) Unmethylated (loss of other MMR gene): male, 35 (3.3); female, 32 (3.0) Unmethylated (no MMR loss): male, 16 (1.5); female, 10 (0.9) |
Methylated: rectum, 4 (0.4); left colon, 9 (0.8); right colon, 155 (14.6) Unmethylated (loss of MLH1): rectum, 3 (0.3); left colon, 6 (0.6); right colon, 41 (3.9) Unmethylated (loss of other MMR gene): rectum, 7 (0.7); left colon, 12 (1.1); right colon, 45 (4.2) Unmethylated (no MMR loss): rectum, 15 (1.4); left colon, 8 (0.7); right colon, 40 (3.8) |
| Southey et al., 200556 | IHC, 37.2 (24–42); MSI, 37.2 (24–42); RS, 37.1 (24–42) | AMS II, 12 (9.2) | IHC: male, 59 (45.0); female, 46 (35.1) MSI: male, 59 (45.0); female, 46 (35.1) Reference standard: male, 37 (28.2); female, 22 (16.8) |
NR |
| Single-gate studies recruiting populations at high risk for LS | ||||
| Caldes et al., 200457 | NR | NR | NR | NR |
| Mueller et al., 200958 | NR | NR | NR | NR |
| Overbeek et al., 200759 | 40.7 (29–51) | NR | NR | Rectum 4, (4.8); colon, 2 (2.4); splenic flexure, 1 (1.2); ascending colon, 2 (2.4); ileocaecum, 1 (1.2) |
| bPoynter et al., 200834 | NR | NR | NR | NR |
| Shia et al., 200560 | Mean, 50.5; median, 50 (23–78) | AMS II, 42 (38.2); RBG, 9 (8.2) | Male, 48 (43.6); female, 62 (56.3) | NR |
| Reference standard-positive studies (recruiting populations with known mutation status) | ||||
| Hendriks et al., 200361 | Reference standard/IHC: MLH1, 46 (28–90); MSH2, 40 (23–61);c MSH6, 62 (26–84) MSI: MLH1, 48 (29–90); MSH2, 40 (23–61);c MSH6, 62 (26–84) |
NR | Reference standard/IHC: male, 25 (56.0); female, 20 (44.0) MSI: male, 16 (35.6); female, 18 (40.0) |
Reference standard/IHC: rectum, 1 (2.2); colon, 12 (2.7); descending colon, 1 (2.2); sigmoid colon, 4 (8.9); splenic flexure, 1 (2.2) [duodenum, 1 (2.2)];c ascending colon, 3 (6.7); caecum, 12 (26.7); hepatic flexure, 2 (4.4); transverse colon, 8 (17.8) MSI: rectum, 1 (2.2); colon, 4 (8.9); descending colon, 1 (2.2); sigmoid colon, 3 (6.7); splenic flexure, 1 (2.2); [duodenum, 1 (2.2)];c ascending colon, 2 (4.4); caecum, 10 (22.2); hepatic flexure, 2 (4.4); transverse colon, 8 (17.8) |
| Okkels et al., 201262 | NR | NR | NR | NR |
Three54–56 of the studies recruiting population-based samples assessed MSI and IHC, with the study by Limburg et al. 55 assessing only IHC (see Table 6).
Single-gate studies recruiting high-risk populations
The five single-gate studies34,57–60 that were based on high-risk populations all applied different criteria to select participants for inclusion. For two34,57 of these studies the participant inclusion criteria were unclear, although for the study by Caldes et al. 57 it was reported that participants were recruited from a familial cancer clinic and for the study by Poynter et al. 34 it was reported that participants were recruited through high-risk clinics and that some clinics selected participants with a ‘family history or early age at onset’. The remaining three single-gate studies appeared to include participants who were at high risk because of their family history (see Table 6). These five studies varied in size, with the largest study being that by Poynter et al. ,34 which recruited 172 participants, and the smallest being that by Mueller et al. ,58 which recruited 48 participants.
With regard to the population characteristics of these five studies (see Table 7), only one60 provided details on the participants’ sex (56.3% female). The age of participants was reported in two of the five studies,59,60 ranging from 29 to 51 years in the study by Overbeek et al. 59 and from 23 to 78 years in the study by Shia et al. 60 Only one60 of the five studies reported the proportion of participants meeting Amsterdam II criteria (38.2%) or the revised Bethesda guidelines (8.2%) and the specific location of the CRC was reported only by Overbeek et al. 59 None of the studies provided details on the ethnicity of the participants.
All five of the single-gate studies recruiting high-risk populations assessed both MSI and IHC (see Table 6).
Reference standard-positive studies
There were two reference standard-positive studies (i.e. studies that used a variation on a two-gate study design in which only participants who were reference standard positives were recruited). 61,62 The study by Hendriks et al. 61 recruited participants with a germ-line mutation in MLH1, MSH2 or MSH6 and assessed both MSI and IHC whereas the study by Okkels et al. 62 was focused on the identification of a germ-line mutation in MSH6 and assessed only IHC (see Table 6). It should be noted that, although the study by Okkels et al. 62 provides an assessment of the test accuracy of four proteins (MLH1, MSH2, MSH6, PMS2) and data are provided for all proteins combined, these data are not included in this review. Instead, the IHC results for MSH6 only are included.
Both of the reference standard-positive studies included in this review were quite small, with Hendriks et al. 61 recruiting 45 participants and Okkels et al. 62 recruiting 56 participants.
The study by Okkels et al. 62 did not provide details on participants’ age, sex or cancer location or the number of participants meeting Amsterdam II criteria or the revised Bethesda guidelines. The study by Hendriks et al. 61 included a similar number of men and women (44% female), participants ranged in age from 23 to 90 years and the specific locations of the CRCs were reported (see Table 7). The proportions meeting Amsterdam II criteria and the revised Bethesda guidelines were not reported. Neither of the reference standard-positive studies reported the ethnicity of the participants.
Summary of the reference standard in the included studies
Three of the four studies that recruited a population-based sample used a combination of direct sequencing and denaturing high-performance liquid chromatography (DHPLC) as the reference standard followed by MLPA to detect large genomic alterations or deletions. 34,54,56 The study by Limburg et al. 55 used direct sequencing, but not DHPLC, followed by MLPA and Southern blot analysis. All four studies investigated mutations in MLH1, MSH2 and MSH6. However, in the study by Poynter et al. ,34 direct sequencing was used only to detect MSH6 mutations in cases with absent IHC staining of MSH6 and in the study by Southey et al. 56 mutations in PMS2 were also investigated. In all four of these studies, large alterations or deletions were assessed in MLH1 and MSH2 but not MSH6, although in the study by Southey et al. 56 this was conducted only for participants who had tumours lacking expression of at least one MMR protein and for whom no previous mutation had been identified by sequencing. In addition, it should be noted that in the population-based sample reported in the study by Poynter et al. ,34 the reference standard was applied to all MSI-H and MSI-L participants and a random sample of MSS participants. Further details are provided in Table 8. It should also be noted that three34,54,55 of the four population-based studies reported on unclassified variants (i.e. mutations for which the association with LS is unclear). This can complicate the assessment of MSI in particular; mutations may be considered to be unclassified variants, with uncertain pathogenicity, because the variant may occur in cases with either MSI-H or MSS tumours. In this review, in primary analyses, unclassified variants have been counted as reference standard negatives. Secondary analyses have been conducted, as appropriate, in which unclassified variants are considered to be reference standard positives. None of the population-based studies provided sufficient data on unclassified variants to be included in secondary analyses.
| Study | Description | Participants receiving the reference standard |
|---|---|---|
| Single-gate studies recruiting population-based samples | ||
| Barnetson et al., 200654 | Germ-line DNA obtained from blood leucocytes was analysed for MLH1, MSH2 and MSH6 mutations. DHPLC was used for MSH2 and MLH1. Variants noted on chromatography were then sequenced. Mutations were confirmed by reamplification of an independent sample of DNA and resequencing in both directions. MLH1 and MSH2 were assessed for deletions by MLPA, with products separated on a genetic analyser | Mutational analysis and follow-up were complete in the total study population of 870 participants |
| Limburg et al., 201155 | DNA samples extracted from peripheral blood leucocytes received full mutation analyses of MLH1, MSH2 and MSH6. DNA from each subject was amplified by PCR and directly sequenced in the forward and reverse directions using fluorescent dye-labelled sequencing primers: MLH1, MSH2 and MSH6. All potential genetic variants were independently confirmed by repeat PCR amplification of the indicated gene region(s) and sequence determination. Large rearrangement testing for MLH1 and MSH2 was performed by Southern blot analysis in conjunction with MLPA | Germ-line MLH1, MSH2 and MSH6 sequencing data were obtained for 195 (100%), 195 (100%) and 189 (97%) subjects respectively |
| aPoynter et al., 200834 | Mutations in MSH2 and MLH1 were detected using a combined approach of DHPLC/direct sequencing and MLPA. Direct sequencing was used to detect MSH6 mutations in cases with absent IHC staining of MSH6 | Population-based and clinic-based probands with CRC were tested for mutations in the MMR genes MSH2, MLH1, MSH6 and PMS2. MMR gene mutation testing for MSH2 and MLH1 was conducted for all clinic-based probands and all MSI-H or MSI-L population-based probands and in a random sample of 300 MSS population-based probands. MMR germ-line mutation status was available for 324/374 population-based MSI-H cases, 197/223 MSI-L cases and 205/464 MSS cases |
| Southey et al., 200556 | MLH1, MSH2, MSH6 and PMS2 genes were screened for germ-line mutations using sequencing approaches or DHPLC. Confirmation of putative mutations was sought using an independent PCR for direct automated sequencing. MLPA was used to detect large genomic alterations in MLH1 and MSH2 in samples from 10 patients who had tumours lacking expression of at least one MMR protein and for whom no previous mutation had been identified by sequencing | Ninety-two of 110 participants received germ-line mutation analysis. This included participants with one or more of the following characteristics: a family history that fulfilled the Amsterdam criteria for HNPCC; having a tumour that was MSI-H or MSI-L or that lacked expression of at least one MMR protein; and presence in a random sample of 23 patients selected from those who had tumours that were MSS and did not lack expression of any MMR protein |
| Single-gate studies recruiting populations at high-risk for LS | ||
| Caldes et al., 200457 | Genomic DNA isolated from peripheral blood lymphocytes was analysed for MLH1, MSH2 and MSH6 mutations. DNA was amplified using PCR and all amplicons were subjected to DGGE or cycle sequencing. The MSI-H cases that were negative for mutations were analysed for genomic deletions in MLH1 and MSH2 by Southern blotting | Total population of 58 participants received germ-line mutation analysis |
| Mueller et al., 200958 | Limited details. Deletion analysis was performed using MLPA | Seventy-one CRC cases suspected to be LS cases were analysed for MSH2, MLH1, MSH6 and PMS2 gene defects. Mutation-negative cases were screened for MLH1 promoter methylation and mutations in PMS2 |
| Overbeek et al., 200759 | Mutation analysis of MLH1, PMS2, MSH2 and MSH6 was performed in DNA from peripheral blood lymphocytes by a combination of either single-strand conformation polymorphism analysis or DGGE and direct sequence analysis. For the detection of large deletions and duplications in MLH1, MSH2, MSH6 and PMS2, MLPA was used. All deletions and duplications were confirmed by Southern blot analysis or with a specific PCR | Mutation analysis of germ-line DNA was performed as the first test in 83 families who fulfilled clinical criteria for LS |
| aPoynter et al., 200834 | Mutations in MSH2 and MLH1 were detected using a combined approach of DHPLC/direct sequencing and MLPA. Direct sequencing was used to detect MSH6 mutations in cases with absent IHC staining of MSH6 | Population-based and clinic-based probands with CRC were tested for mutations in the MMR genes MSH2, MLH1, MSH6 and PMS2. MMR gene mutation testing for MSH2 and MLH1 was conducted for all clinic-based probands and all MSI-H or MSI-L population-based probands and in a random sample of 300 MSS population-based probands. MMR germ-line mutation status was available for 324/374 population-based MSI-H cases, 197/223 MSI-L cases and 205/464 MSS cases |
| Shia et al., 200560 | Each of the exons of MLH1, MSH2 and MSH6 was amplified by PCR and heteroduplex analyses were performed using DHPLC. DNA fragments that displayed an abnormal chromatogram were sequenced directly. Cases with tumours that exhibited MSI but in whom a point mutation was not detected were analysed for large deletions in MLH1 and MSH2 using a procedure based on multiplex PCR amplification of short fluorescent fragments | Mutation analysis of germ-line DNA was performed in 83 participants with a carcinoma |
| Reference standard-positive studies (recruiting populations with known mutation status) | ||
| Hendriks et al., 200361 | Limited details. Among the 35 HNPCC families with a known MMR defect, 27 different germ-line mutations were identified by DGGE or Southern blotting | All 45 patients (25 men and 20 women) had a known germ-line mutation in MLH1, MSH2 or MSH6 |
| Okkels et al., 201262 | Limited details. Standard sequencing of genomic DNA and MLPA | A total of 815 families were screened for MSH6 mutations |
Of the five studies that reported data for high-risk populations,34,57–60 two57,59 used a combination of sequencing and denaturing gradient gel electrophoresis (DGGE), although in the study by Overbeek et al. 59 single-strand conformation polymorphism analysis was sometimes used instead of DGGE. The study by Caldes et al. 57 followed this with Southern blot analysis to detect large deletions whereas Overbeek et al. 59 used a combination of MLPA and Southern blot analysis to detect large deletions. As reported above, Poynter et al. 34 used a combination of direct sequencing and DHPLC as the reference standard followed by MLPA to detect large genomic alterations or deletions. Similarly, Shia et al. 60 used DHPLC analysis and direct sequencing but this was followed by multiplex PCR of short fluorescent fragments for the detection of large deletions. The study by Mueller et al. 58 provided limited details on the reference standard, although it was clearly specified that MLPA was used.
All five of the single-gate studies based on high-risk populations investigated mutations in MLH1, MSH2 and MSH6,34,57–60 although, as mentioned above, Poynter et al. 34 used only direct sequencing to detect MSH6 mutations in cases with absent IHC staining of MSH6. In addition, Overbeek et al. 59 investigated mutations in PMS2 in all cases and Mueller et al. 58 investigated mutations in PMS2 only in cases that were negative for mutations in MLH1, MSH2 and MSH6. For three34,57,60 of the five studies large alterations or deletions were assessed in MLH1 and MSH2 but not MSH6. It should be noted that in the studies by Caldes et al. 57 and Shia et al. 60 it was reported that large alterations or deletions were assessed only for MSI-positive cases that were mutation negative. Mueller et al. 58 did not clearly report which genes were assessed for large alterations or deletions. In the study by Overbeek et al. 59 large deletions and duplications were assessed in MLH1, MSH2, MSH6 and PMS2. Two57,60 of these studies providing data from high-risk populations reported on unclassified variants, but only one57 provided sufficient data to be included in secondary analyses (in which unclassified variants were considered to be reference standard positives).
Both of the reference standard-positive studies provided limited details on the reference standard. 61,62 The study by Okkels et al. 62 used sequencing followed by MLPA and was focused only on MSH6. The study by Hendriks et al. 61 used DGGE followed by Southern blot analysis and assessed MLH1, MSH2 and MSH6. Both studies recruited only reference standard-positive participants and both studies recruited participants with, and reported on, unclassified variants. However, only the study by Hendriks et al. 61 provided sufficient data to be included in secondary analyses (in which unclassified variants are considered to be reference standard positives).
Quality appraisal of included studies
Quality appraisal was conducted using phase 3 of the QUADAS-2 tool53 for all 11 data sets (all 10 studies, including both the population-based and high-risk samples reported by Poynter34). Phase 3 of the QUADAS-2 tool contains four domains: patient selection, index tests, reference standard and flow and timing. The quality of the included studies is discussed in the following sections according to these domains and is summarised in Table 9.
| Domain | Item | Study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population based | High risk single gate | Other | ||||||||||
| Barnetson et al., 200654 | Limburg et al., 201155 | Southey et al., 200556 | aPoynter et al., 200834 | Caldes et al., 200457 | Mueller et al., 200958 | Overbeek et al., 200759 | aPoynter et al., 200834 | Shia et al., 200560 | Hendriks et al., 200361 | Okkels et al., 201262 | ||
| Patient selection | Was a consecutive or random sample of patients enrolled? | Y | Y | Y | U | U | U | U | U | U | U | Y |
| Was a case–control design avoided? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Nb | Nb | |
| Did the study avoid inappropriate exclusions? | Y | Y | Y | U | Y | U | U | U | U | Y | Y | |
| Could the selection of patients have introduced bias? | L | L | L | U | U | U | U | U | U | Uc | Ld | |
| Is there concern that the included patients do not match the review question? | L | L | L | L | L | L | L | L | L | L | L | |
| Index test (MSI) | Were the index test results interpreted without knowledge of the results of the reference standard? | U | U | U | U | U | U | U | U | U | U | U |
| If a threshold was used, was it prespecified? | U | U | U | U | U | U | U | U | U | U | U | |
| Could the conduct or interpretation of the index test have introduced bias? | U | U | U | U | U | U | U | U | U | U | U | |
| Is there concern that the index test, its conduct or its interpretation differs from the review question? | L | L | L | L | L | L | L | L | L | L | L | |
| Index test (IHC) | Were the index test results interpreted without knowledge of the results of the reference standard? | U | U | U | U | U | U | U | U | Y | U | U |
| If a threshold was used, was it prespecified? | U | U | U | U | U | U | U | U | U | U | U | |
| Could the conduct or interpretation of the index test have introduced bias? | U | U | U | U | U | U | U | U | U | U | U | |
| Is there concern that the index test, its conduct or its interpretation differs from the review question? | L | L | L | L | L | L | L | L | L | L | L | |
| Reference standard | Is the reference standard likely to correctly classify the target condition? | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Were the reference standard results interpreted without knowledge of the results of the index test? | U | U | U | U | U | U | U | U | U | Y | U | |
| Could the reference standard, its conduct or its interpretation have introduced bias? | U | U | U | U | U | U | U | U | U | L | U | |
| Is there concern that the target condition as defined by the reference standard does not match the review question? | L | L | L | L | L | L | L | L | L | L | L | |
| Flow and timing | Was there an appropriate interval between index test(s) and reference standard? | U | U | U | U | U | U | U | U | U | U | U |
| Did all patients receive a reference standard? | Y | Y | N | N | Y | Y | Y | Y | Y | Y | Y | |
| Did patients receive the same reference standard? | Y | U | N | N | N | U | U | N | N | U | U | |
| Were all patients included in the analysis? | N | N | N | N | N | Y | U | N | U | N | N | |
| Could the patient flow have introduced bias? | U | U | U | U | U | U | U | U | U | U | U | |
Patient selection
Four of the studies were rated as having a low risk of bias for patient selection. Three of these were population-based single-gate studies. 54–56 The other was a reference standard-positive study62 from which only sensitivity estimates could be ascertained. All four of these studies enrolled either a consecutive or a random sample of participants and avoided inappropriate exclusions.
For the remaining seven studies34,57–62 it was unclear whether patient selection could have introduced bias. In all of these studies it was unclear whether inappropriate exclusions were avoided by enrolling a consecutive or a random selection of participants.
For all studies included in the review there were no concerns about whether or not the included participants matched the review question.
Index tests
All of the studies included in the review of test accuracy evaluated IHC. With the exception of the studies by Limburg et al. 55 and Okkels et al. ,62 all studies also assessed MSI. For both of these index tests, all studies were rated as unclear with regard to whether the conduct and interpretation of the test could have introduced bias. None of the studies clearly reported whether the thresholds used were prespecified. In addition, none of the studies reported whether MSI results were interpreted without knowledge of the results of the reference standard. The study by Shia et al. 60 reported that IHC results were interpreted without knowledge of the results of the reference standard, but for the remaining studies this was not reported.
There were no concerns (in any of the studies) that the conduct or interpretation of either of the index tests was different from the review question.
Reference standard
In all of the included studies the reference standard was assessed as being likely to correctly classify the target condition. However, it should be noted that it has not been established in any study that the reference standard is 100% accurate and that there is between-study variation in the reference standard (see Summary of the reference standard in the included studies). Nevertheless, because a genetic definition of LS was used in this review (i.e. the reference standard is LS as indicated by a genetic mutation rather than, for example, LS defined by clinical criteria) and because the inclusion criteria for the reference standard were set so that only studies using the best current methods, or other similarly appropriate methods, for detecting LS-based gene defects were included (see Reference standard), the assumption remains that any specific disagreements between the reference standard and the index test are assumed to result from incorrect classification by the index test. Indeed, there were no concerns (in any of the studies) that the target condition, as defined by the reference standard, did not match the review question.
Despite this, all of the included studies apart from that by Hendriks et al. 61 were rated as unclear with regard to whether or not the conduct or interpretation of the reference standard could have introduced bias. This was because only Hendriks et al. 61 specified that the reference standard results were interpreted without knowledge of the results of the index test, with the rest of the studies not reporting this information. It is therefore unclear for the other studies34,54–60,62 whether there was any prior knowledge that could have influenced the interpretation of the reference standard.
Flow and timing
For all of the included studies it was unclear whether the flow of participants through the study could have introduced bias. In most of the included studies (including the high-risk sample reported by Poynter et al. 34), all of the participants received a reference standard,34,54,55,57–62, although only one54 of these studies provided information to indicate that all of the participants received the same reference standard. In five of these studies55,58,59,61,62 it was not clear whether all participants received the same reference standard and in three of these studies34,57,60 it was clear that not all participants received the same reference standard. Indeed, in the study by Caldes et al. ,57 only the MSI-H cases that were negative for mutations were analysed for genomic deletions, by Southern blot analysis, in MLH1 and MSH2. Similarly, in the study by Shia et al. ,60 only cases with tumours that exhibited MSI but in whom a point mutation was not detected were analysed for large deletions in MLH1 and MSH2. In the high-risk sample in Poynter et al. ,34 direct sequencing was used only to detect MSH6 mutations in cases with absent IHC staining of MSH6. However, in these three studies it was not believed that this would constitute a high risk of bias; it is acceptable for large deletions to be investigated only when a mutation is not found57,60 and for the reference standard to be directed by IHC results. 34
In the study by Southey et al. 56 and the population-based sample reported by Poynter et al. ,34 not all patients received the reference standard. In both of these samples this was because the reference standard was applied to a random sample of participants who were index test negative. In addition, in both of these samples it was clear that not all of the patients who received the reference standard received the same reference standard. In the study by Southey et al. 56 MLPA was used to detect large genomic alterations in MLH1 and MSH2 for cases with tumours lacking expression of at least one MMR protein and for whom no previous mutation had been identified by sequencing and, as with the high-risk sample in the study by Poynter et al. ,34 in the population-based sample reported by Poynter et al. ,34 direct sequencing was used to detect MSH6 mutations only in cases with absent IHC staining of MSH6. As discussed earlier, these variations in study design would not be thought to constitute a high risk of bias.
None of the studies included in the test accuracy review clearly specified the interval between the index tests and the reference standard. However, as the results for the index tests and the reference standard would be expected to be stable over time, this information is of little importance in itself; variations in timing between the index tests and the reference standard would not lead to a high risk of bias.
With regard to missing data, it was clear that in most of the studies some of the participants were excluded from the analysis,34,54–57,61,62 although details regarding the characteristics of these participants were not provided. However, the study by Mueller et al. 58 clearly specified that data were analysed from all participants who received tests. In the studies by Overbeek et al. 59 and Shia et al. 60 it was unclear whether or not all tested participants were analysed.
Quality appraisal summary
Overall, no evidence was found to indicate that any of the included studies was at a high risk of bias. Of course, the single-gate studies based on a high-risk population34,57–60 would necessarily confer a risk of bias (i.e. if the results were used to make assumptions about the general population with CRC). Indeed, when studies recruit only from high-risk populations this would obviously lead to biased estimates of PPV, NPV and yield. It would also possibly lead to biased estimates of sensitivity and specificity because of spectrum bias,64 although in a previous review this did not appear to lead to significant bias in estimates of sensitivity. 42 As a result, this was dealt with in this review by prespecifying that single-gate studies based on high-risk populations would be used only to estimate sensitivity. Ordinarily, two-gate studies would also be at risk of inflating diagnostic accuracy. 65,66 However, the two studies included in this review that were not single-gate studies were not in fact two-gate studies but reference standard-positive studies;61,62 no reference standard negatives were included in these two studies and so an unbiased estimate of sensitivity could be made.
It is important to note that an absence of evidence to suggest that the included studies were at high risk of bias does not suggest that the studies were at low risk of bias. In fact, for all studies it was unclear whether the index tests or the flow and timing of the study would have introduced bias. Similarly, in all but one study61 it was unclear whether the conduct of the reference standard would have introduced bias and only four studies54–56,62 provided sufficient information to establish that the selection of participants was unlikely to have introduced bias.
Assessment of test accuracy
The index tests included in this review (MSI and IHC) are highly susceptible to spectrum effects in populations that have been selected on the basis of clinical characteristics. Indeed, preselecting participants in this way will result in a population that differs from an unselected population in terms of the clinical predictor in a non-random way. Thus, the sensitivity and specificity estimates would likely be different among unselected CRC populations compared with CRC populations selected on the basis of age or because of characteristics that make them high risk for LS (such as a family history of LS or meeting clinical criteria for defining LS). In particular, the increased presence of MMR mutation carriers in a population would change the apparent sensitivity and specificity of the index tests. 54 However, a previous review did not find that this issue led to significant bias in estimates of sensitivity. 42
Because of this, the studies included in this review were grouped by population and the results are presented accordingly. In addition, studies recruiting high-risk populations were used only to estimate sensitivity. As previously described, four of the samples included in this review can be described as population-based samples, although only one recruited an unselected CRC population,34 with the other three recruiting age-limited populations,54–56 for which some spectrum bias may be expected. The remaining seven samples included in this review were all high risk; five of the remaining studies had a single-gate design and recruited high-risk participants,34,57–60 with the other two studies recruiting only participants who were reference standard positives. 61,62 For the five single-gate studies based on high-risk populations, only sensitivity is reported, even if data were available for other outcomes. For the two studies based on participants who were reference standard positives, only sensitivity is estimable from the data reported.
It is important to mention that none of the studies included in this review made a direct comparison between MSI and IHC. As such, results are reported separately for these tests.
Assessment of test accuracy: microsatellite instability testing
Microsatellite instability was assessed in eight of the 10 studies (nine of the 11 samples) included in the review of test accuracy: three of the four population-based samples,34,54,56 all five high-risk samples34,57–60 and one of the reference standard-positive studies. 61
Microsatellite instability testing methods
A summary of the MSI testing methods used in these studies is provided in Table 10. It is evident that a variety of between-study differences exist in the MSI testing procedures used. In addition, differences between studies in MSI testing methods were not always clear because methods were not always reported in sufficient detail. For example, three34,58,59 of the eight studies assessing MSI did not report microdissection techniques (microdissection assists in ensuring that malignant tissue that does not contain DNA from surrounding, healthy colonic tissue is analysed). A further two studies60,61 reported only very limited details about microdissection. Among the three studies54,56,57 that did report details regarding microdissection, there was some variation in the technique used (see Table 10 for details). The panel of markers used also differed between studies (see Table 10). None of the population-based studies assessed the same panel of markers. Two54,56 of the three population-based studies included an assessment of the Bethesda panel of markers (BAT25, BAT26, D2S123, D5S346, D17S250), with the study by Southey et al. 56 also assessing five additional markers (BAT40, MYB, TGFRII, IGFIIR and BAX). The other population-based study (the population-based sample included in the study by Poynter et al. 34) did not assess D2S123, even though the other Bethesda panel markers were included. This study also assessed six additional markers (BAT40, MYCL, ACTC, Dl 8S55, D1OS197 and BAT34C4). The same panel of markers was used for the single-gate, high-risk sample included in the study by Poynter et al. 34 Of the other four single-gate studies based on high-risk populations, three57–59 included the Bethesda panel of markers. However, Overbeek et al. 59 also assessed BAT40 for some participants (it is unclear which ones) and Mueller et al. 58 used a 10-marker panel (BAT26, BAT4O, Mfdl5, D2S123, APC, BAT25, D10S197, D18S58, D18S69 and MYCLJ) as well as a five-marker panel (the Bethesda panel), although it is unclear which participants received which panel of markers and which panel of markers the reported data are based on. The remaining single-gate study by Shia et al. 60 did not assess D5S346 but did assess the other markers in the Bethesda panel, as well as BAT40, PAX6 and MYCL1. It appears, therefore, that none of the single-gate studies based on high-risk samples used the same panel of markers (although this is unclear in the case of Mueller et al. 58). The reference standard-positive study that assessed MSI61 also included a different panel of markers: the Bethesda panel plus BAT40, MSH3 and MSH6.
| Study | Microdissection | MSI markers | Threshold | ||
|---|---|---|---|---|---|
| MSI-H | MSI-L | MSS | |||
| Population-based single-gate studies | |||||
| Barnetson et al., 200654 | 10-µm tumour sections; microdissection performed on purified tumour DNA and control DNA from blood or normal tissue in the section | BAT25, BAT26, D2S123, D5S346 and D17S250 | > 1 marker | 1 marker | 0 markers |
| Southey et al., 200556 | 5-µm tumour sections; microdissection performed on invasive tumour cells from paraffin-embedded archival tumour tissue stained with 1% methyl green and normal cells from colonic or lymph node tissue/DNA extracted from peripheral blood lymphocytes | BAT25, BAT26, D2S123, D5S346, D17S250, BAT40, MYB, TGFRII, IGFIIR and BAX | > 5 markers | 2–5 markers | < 2 markers |
| a,bPoynter et al., 200834 | Not reported | BAT25, BAT26, D5S346, D17S250, BAT40, MYCL, ACTC, Dl 8S55, D1OS197 and BAT34C4 | ≥ 30% | > 0% and < 30% | 0% |
| Single-gate studies recruiting high-risk samples | |||||
| Caldes et al., 200457 | 10-µm tumour sections; microdissection performed on H&E stained slides with demarked areas containing cancer cells, and corresponding areas on unmarked slides | BAT25, BAT26, D2S123, D5S346 and D17S250 | > 1 marker, or 1 marker if BAT26 | Not used | 0 markers |
| Mueller et al., 200958 | Not reported | 5- and 10-panel markersc | Not reportedd | ||
| Overbeek et al., 200759 | Not reported | BAT25, BAT26, D2S123, D5S346, D17S250 and BAT40e | Tumours categorised as positive (> 2 Bethesda panel markers) or negative | ||
| a,bPoynter et al., 200834 | Not reported | BAT25, BAT26, D5S346, D17S250, BAT40, MYCL, ACTC, Dl 8S55, D1OS197 and BAT34C4 | ≥ 30% | > 0% and < 30% | 0% |
| bShia et al., 200560 | Microdissection performed on DNA from paraffin-embedded tissue blocks. No further details reported | BAT25, BAT26, D2S123, D17S250, BAT40 and PAX6, MYCL1 | Tumours categorised as positive (≥ 30%) or negative | ||
| Reference standard-positive study | |||||
| Hendriks et al., 200361 | Microdissection not specifically reported, paired tumour and normal tissue DNA samples were used | BAT25, BAT26, D2S123, D5S346, D17S250, BAT40, MSH3 and MSH6 | > 1 Bethesda panel marker | 1 Bethesda panel marker | 0 Bethesda panel markers |
The eight studies that assessed MSI also varied in the way in which MSI was categorised (as a bimodal or a trimodal distribution) and in the thresholds used to define these categories. Indeed, of the studies included in this review, five34,54,56,58,61 defined tumours as MSI-H, MSI-L or MSS, also known as a trimodal distribution, two59,60 defined tumours as MSI positive or MSI negative, also known as a bimodal distribution, and one57 used a bimodal distribution but defined tumours as either MSI-H or MSS.
It is also unsurprising that the thresholds used to categorise tumours (as MSI-H, MSI-L or MSS or as MSI positive or MSI negative) differed between studies; the distinction between these categories is dependent on both the type and the number of microsatellites analysed and, as discussed above, the studies included in this review used various different panels of markers. 69 Indeed, with regard to the trimodal distribution of MSI, many research groups have defined MSI-H tumours as those with > 30–40% of unstable markers, MSI-L as instability lower than this threshold and MSS as no instability. 69 However, in the five studies included in the review that used the MSI-H, MSI-L and MSS categories (trimodal distribution), the thresholds used to categorise the tumours varied greatly, with one34 of these studies using the commonly used threshold of ≥ 30% of unstable markers to define MSI-H tumours, three54,56,61 using differing numbers of unstable markers to define MSI-H tumours and one58 not providing details on the thresholds used to categorise the tumours (see Table 10 for details). Of the two studies that defined tumours as positive or negative (bimodal distribution), one60 reported using a threshold of ≥ 30% of unstable markers to define MSI-positive tumours and the other59 defined MSI-positive tumours as those with more than two unstable Bethesda panel markers. The study by Caldes et al. 57 defined MSI-H tumours as those with two or more unstable Bethesda panel markers (or one marker in the case of BAT26) and MSS tumours as those showing no instability. It is not clear how cases with only one unstable marker (other than BAT26) were categorised, but in any case data are presented only for tumours that were categorised as MSS and MSI-H.
It has been asserted by Pawlik et al. 69 that a bimodal distribution of MSI (as used in the studies by Shia et al. 60 and Overbeek et al. 59) may be more useful than a trimodal distribution of MSI (as used in the studies by Barnetson et al. ,54 Southey et al. ,56 Poynter et al. ,34 Caldes et al. ,57 Mueller et al. 58 and Hendriks et al. 61). Indeed, in studies using a trimodal distribution, MSI-L can be considered as either positive or negative and, although MSI-L tumours may behave more similarly to MSS tumours in clinical and prognostic terms, the significance of MSI-L is still uncertain and would vary according to the particular markers used. 69 Clearly, a range of markers were used in the studies included in this review. Thus, for studies using a trimodal distribution of MSI, and for which data were available, the sensitivity and specificity of the MSI test were estimated with MSI-L values considered as index test positives and with MSI-L values considered as index test negatives.
Microsatellite instability-high compared with microsatellite stable and microsatellite instability-low
Three population-based samples,34,54,56 two high-risk single-gate study samples34,58 and one reference standard-positive study sample61 provided test accuracy data for MSI in which MSI-L cases were considered to be index test negative results. In the other three studies assessing MSI,57,59,60 MSI was already categorised using a bimodal distribution (MSI positive or negative in the studies by Overbeek et al. 59 and Shia et al. ;60 MSI-H or MSS in the study by Caldes et al. 57). For ease of reference, the results from these three studies are included here and again in the section reporting data in which MSI-L tumours are considered to be positive index text results.
Sensitivity was calculated for all studies that provided data in which MSI-L was considered to be a negative index test result (as well as the studies categorising MSI using a bimodal distribution), whereas specificity was calculated only for the three population-based studies. 34,54,56 These sensitivity and specificity estimates are reported in Table 11.
| Study | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) |
|---|---|---|
| Population-based single-gate studies | ||
| aPoynter et al., 200834 | 100.0 (93.9 to 100.0) | 61.1 (57.0 to 65.1) |
| Barnetson et al., 200654 | 66.7 (47.2 to 82.7) | 92.5 (89.1 to 95.2) |
| Southey et al., 200556 | 72.2 (46.5 to 90.3) | 87.8 (73.8 to 95.9) |
| Single-gate studies recruiting high-risk samples | ||
| bCaldes et al., 200457 | 79.4 (62.1 to 91.3) | – |
| Mueller et al., 200958 | 91.3 (72.0 to 98.9) | – |
| bOverbeek et al., 200759 | 90.0 (59.6 to 98.2) | – |
| cPoynter et al., 200834 | 86.8 (71.9 to 95.6) | – |
| bShia et al., 200560 | 100.0 (85.8 to 100.0) | – |
| Reference standard-positive study | ||
| Hendriks et al., 200361 | 88.0 (68.8 to 97.5) | – |
Only one study34 included a population that was unselected (not limited by age or risk) and the data from this study produced a sensitivity estimate of 100% (95% CI 93.9% to 100.0%) and a specificity estimate of 61.1% (95% CI 57.0% to 65.1%). It should be noted that in this sample and in the data reported for this study in Table 11, unclassified variants were considered to be reference standard negatives. Two of the studies that reported data in which MSI-L was considered to denote an index test negative were based on age-limited populations. 54,56 The study by Barnetson et al. 54 included unclassified variants as reference standard negative results and is reported in this section as such. In these two studies, sensitivity and specificity estimates were fairly similar, despite the fact that different panels of markers and different thresholds were used to categorise the tumours (see Table 10), with a sensitivity of 66.7% (95% CI 47.2% to 82.7%) in the study by Barnetson et al. 54 and 72.2% (95% CI 46.5% to 90.3%) in the study by Southey et al. 56 and a specificity of 92.5% (95% CI 89.1% to 95.2%) in the study by Barnetson et al. 54 and 87.8% (95% CI 73.8% to 95.9%) in the study by Southey et al. 56 (see Table 11).
For the five single-gate, high-risk samples presented in Table 11,34,57–60 sensitivity estimates ranged from 79.4% (95% CI 62.1% to 91.3%) in the study by Caldes et al. 57 to 100.0% (95% CI 85.8% to 100.0%) in the study by Shia et al. 60 (see Table 11). Two of these high-risk, single-gate studies mentioned unclassified variants57,60 and these are counted as reference standard negative results in these analyses. Between-study variation in sensitivity estimates may be the result of a variety of factors, including differences in the panel of markers used and in the MSI thresholds used to denote cases, as well as differences in the reference standard. Nevertheless, all of these sensitivity estimates were > 79%. It should be noted that Caldes et al. 57 reported that the sensitivity of MSI-H in predicting a pathogenic mutation was 96.0% (95% CI 90.0% to 100.0%) in contrast to our calculation of 79.4% (95% CI 62.1% to 91.3%). This is because Caldes et al. 57 excluded five cases from their MSI analyses that did not also have IHC data, whereas these cases were included in our calculations. The data from the reference standard-positive study that assessed MSI61 was also used to generate a sensitivity estimate; when MSI-L cases were considered to be index test negatives, sensitivity was 88.0% (95% CI 68.8% to 97.5%).
Because of a potential for spectrum bias, it would be expected that studies recruiting high-risk populations would have higher sensitivity estimates than those estimated from population-based studies. However, as discussed in a systematic review by Palomaki et al. ,42 we did not find great differences between the sensitivity estimates in the population-based studies and the sensitivity estimates in the high-risk studies. Indeed, although two of the three population-based studies produced the lowest sensitivity estimates,54,56 the other population-based study produced a sensitivity estimate of 100.0% (95% CI 93.9% to 100.0%). In fact, the two population-based studies with the lower sensitivity estimates would, in theory, be more likely to be subject to some spectrum bias than the study by Poynter et al. 34 because they are based on age-limited populations. This highlights how comparison between the studies included in this review may not be meaningful. Other factors such as the particular MSI testing methods, panel of markers and thresholds used, as well as methods used to conduct the reference standard, varied between studies.
For three34,58,59 of the five single-gate high-risk samples, data regarding methylation were reported. In the study by Poynter et al. 34 it was reported that the prevalence of MLH1 promoter methylation in MSI-H tumours was much lower in the high-risk sample than in the population-based sample (13% vs. 60%; p < 0.0001), with no MLH1 promoter methylation at all in the MSI-L or MSS tumours from the high-risk sample. The authors suggest that this may be the result of the higher frequency of clinic-based MSI-H cases with a MMR germ-line mutation and because the MSI-H cases in the population-based series were diagnosed at an older age than those in the clinic-based series (median 63 years, range 22–75 years vs. median 44 years, range 19–77 years). Despite recruiting a high-risk population, Mueller et al. 58 reported that, of the seven MSI-H cases for which no mutation was identified by the reference standard, four had somatic silencing of MLH1 and were likely to be sporadic cases. Overbeek et al. 59 did not test all participants relevant to this review for MLH1 promoter methylation; however, of those tested, none had a positive result.
The other test accuracy outcomes included in this review were LRs (LR+ and LR–), PPV and NPV, accuracy or concordance with the reference standard, diagnostic yield and test failure rates. The last three outcomes (accuracy or concordance with the reference standard, diagnostic yield and test failure rates) were not reported in any of the included studies. The other four outcomes (LR+, LR–, PPV and NPV) were calculated for the three population-based studies reporting MSI data. 34,54,56 These outcomes are reported in Table 12 and are based on data in which MSI-L was assumed to be a negative index test result and the unclassified variants reported in the studies by Barnetson et al. 54 and Poynter et al. 34 were assumed to be negative reference standard results.
| Study | LR+ (95% CI) | LR– (95% CI) | PPV (95% CI) (%) | NPV (95% CI) (%) |
|---|---|---|---|---|
| Population-based single-gate studies | ||||
| aPoynter et al., 200834 | 2.57 (2.32 to 2.85) | 0.00 (NE) | 20.8 (16.2 to 26.0) | 100.0 (99.0 to 100.0) |
| Barnetson et al., 200654 | 8.94 (5.54 to 14.20) | 0.36 (0.21 to 0.60) | 45.5 (30.4 to 61.2) | 96.8 (94.1 to 98.4) |
| Southey et al., 200556 | 5.92 (2.48 to 14.10) | 0.32 (0.15 to 0.67) | 72.2 (46.5 to 90.3) | 87.8 (73.8 to 95.9) |
As can be seen in Table 12, the results were fairly consistent among the three studies. 34,54,56 The LR+ ranged from 2.57 (95% CI 2.32 to 2.85)34 to 8.94 (95% CI 5.54 to 14.20). 54 The LR– could be estimated for only two of the studies54,56 because there were no false-negative MSI results in the study by Poynter et al. 34 LR– was similar in both studies. The PPV varied a lot more between the studies, with the lowest estimate coming from the study by Poynter et al. 34 (20.8%, 95% CI 16.2% to 26.0%) and the highest estimate coming from the study by Southey et al. 56 (72.2%, 95% CI 46.5% to 90.3%). It should be noted that the CIs generated from the study by Southey et al. 56 are relatively wide, which reflects the smaller sample size of 59, as opposed to the sample size of 638 for Poynter et al. 34 and 352 for Barnetson et al. 54 The NPV was consistent across the studies, with all estimates being > 87%.
Secondary analyses were conducted in which unclassified variants were considered to indicate a positive reference standard test result. It was not possible to include all studies reporting MSI data in these analyses (because sufficient data were not reported). Five studies34,54,57,60,61 reported unclassified variants in their assessment of MSI. Of these five studies, only two57,61 provided sufficient data to conduct secondary analyses. However, because the study by Caldes et al. 57 was based on a high-risk population and that by Hendriks et al. 61 was a reference standard-positive study, only sensitivity estimates were made.
Therefore, when unclassified variants were considered to be reference standard positives and MSI-L was considered to be an index test negative, Caldes et al. 57 reported a sensitivity of 81.6% (95% CI 65.7% to 92.3%) and Hendriks et al. 61 reported a sensitivity of 84.8% (95% CI 69.0% to 93.3%). These results were similar to those obtained when unclassified variants were considered to be reference standard negatives (79.4%, 95% CI 62.1% to 91.3% in the study by Caldes et al. 57 and 88.8%, 95% CI 68.8% to 97.5% in the study by Hendriks et al. ;61 see Table 11).
Microsatellite instability-high and microsatellite instability-low compared with microsatellite stable
When MSI-L tumours were considered to be index test positives, data were available from all three population-based samples that assessed MSI,34,54,56 two high-risk single-gate study samples34,58 and one reference standard-positive study sample. 61 As previously discussed, the other three studies assessing MSI57,59,60 already categorised MSI using a bimodal distribution (MSI positive or MSI negative in the study by Overbeek et al. 59 and Shia et al. 60; MSI-H or MSS in the study by Caldes et al. 57). The results from these three studies are included here as well as previously in Microsatellite instability-high compared with microsatellite stable and microsatellite instability-low.
Sensitivity was calculated for all studies that provided data in which MSI-L was considered to be index test positive (as well as the studies categorising MSI using a bimodal distribution). Specificity was calculated only for the three population-based study samples. 34,54,56 These sensitivity and specificity estimates are reported in Table 13. For the five studies that mentioned assessing unclassified variants in addition to pathogenic mutations,34,54,57,60,61 the unclassified variants were considered to be reference standard negatives.
| Study | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) |
|---|---|---|
| Population-based single-gate studies | ||
| aPoynter et al., 200834 | 100.0 (93.9 to 100.0) | 29.5 (25.8 to 33.4) |
| Barnetson et al., 200654 | 93.3 (77.9 to 99.2) | 84.5 (80.0 to 88.2) |
| Southey et al., 200556 | 94.4 (72.7 to 99.9) | 58.5 (42.1 to 73.7) |
| Single-gate studies recruiting high-risk samples | ||
| bCaldes et al., 200457 | 79.4 (62.1 to 91.3) | – |
| Mueller et al., 200958 | 93.1 (77.2 to 99.2) | – |
| bOverbeek et al., 200759 | 90.0 (59.6 to 98.2) | – |
| cPoynter et al., 200834 | 94.7 (82.3 to 99.4) | – |
| bShia et al., 200560 | 100.0 (85.8 to 100.0) | – |
| Reference standard-positive study | ||
| Hendriks et al., 200361 | 92.0 (74.0 to 99.0) | – |
The data from the study that included an unselected CRC population34 produced a sensitivity estimate of 100.0% (95% CI 93.9% to 100.0%) and therefore sensitivity was unchanged from when MSI-L cases were considered to be index test negatives (see Microsatellite instability-high compared with microsatellite stable and microsatellite instability-low). However, specificity was much lower at 29.5% (95% CI 25.8% to 33.4%), compared with 61.1% (95% CI 57.0% to 65.1%) when MSI-L cases were considered to be index test negatives, reflecting a large increase in false-positive results. The two studies based on age-limited populations that reported data in which MSI-L cases were considered to be index test positives reported sensitivities of 93.3% (95% CI 77.9% to 99.2%)54 and 94.4% (95% CI 72.7% to 99.9%)56 respectively. These sensitivities were higher than those estimated when MSI-L cases were considered to be index test negatives, with sensitivities of 66.7% (95% CI 47.2% to 82.7%) for the study by Barnetson et al. 54 and 72.2% (95% CI 46.5% to 90.3%) for the study by Southey et al. 56 (see Table 11). As would be expected in a trade-off between sensitivity and specificity, specificities were reduced for these two studies when MSI-L cases were considered to be index test positives. However, in the study by Barnetson et al. 54 this reduction was small, with specificity estimated as 84.5% (95% CI 80.0% to 88.2%) when MSI-L cases were considered to be index test positives (see Table 13) and 92.5% (95% CI 89.1% to 95.2%) when MSI-L cases were considered to be index test negatives (see Table 11). For Southey et al. ,56 specificity was estimated as 58.5% (95% CI 42.1% to 73.7%) when MSI-L was considered to be a positive index test result (see Table 13) and 87.8% (95% CI 73.8% to 95.9%) when MSI-L was considered to be a negative index test result (see Table 11). To further demonstrate the differences in test performance between MSI-L as a positive test result and MSI-L as a negative test result, sensitivities and specificities were graphically summarised using a summary receiver operating characteristic (SROC) curve (Figure 4). This visually elucidates the trade-off between sensitivity and specificity across the three population-based study samples used to assess MSI. 34,54,56 In this SROC curve, the unclassified variants mentioned in the studies by Poynter et al. 34 and Barnetson et al. 54 were assumed to be negative reference standard results.
FIGURE 4.
Summary receiver operating characteristic curve graph for MSI testing in which unclassified variants are assumed to be negative reference standard results. Solid black circles, Barnetson et al. ;54 solid green circles, Southey et al. ;56 open circles, Poynter et al. ;34 +, MSI-L is positive; –, MSI-L is negative.

The ideal diagnostic test would generate a point in the upper left corner of the ROC space, representing 100% sensitivity [no false negatives (FNs)] and 100% specificity [no false positives (FPs)]. The closest point to this is found for the study by Barnetson et al. 54 when MSI-L cases are considered to be index test positive results. When MSI-L cases are considered to be index test negative results, specificity improves slightly but sensitivity is reduced. 54 This was also the case for the study by Southey et al. ;56 however, because of the wider discrepancy between sensitivity and specificity, the points are further away from a perfect classification. With regard to the study by Poynter et al. ,34 the lack of FNs ensured that sensitivity remained constant, with only specificity altering according to the allocation of MSI-L results.
It is unsurprising that, on the whole, sensitivity was higher and specificity was lower when MSI-L cases were considered to be index test positive results rather than index test negative results; including MSI-L cases as index test positives essentially lowers the threshold for a positive index test result. Further to the results presented above, Barnetson et al. 54 also reported that MSI-H has a sensitivity of 83% for the detection of MLH1 mutations, 75% for the detection of MSH2 mutations and 17% for the detection of MSH6 mutations, whereas MSI-L has sensitivities of 17%, 25% and 50% respectively. Therefore, the usefulness of including MSI-L as a positive index test result will likely vary according to which gene is mutated. Indeed, according to Mueller et al. ,58 the vast majority of mutations detected are usually in the MSH2 or MLH1 gene, followed by the MSH6 gene and finally the PMS2 gene; however, it is not clear what thresholds were used in this study to define MSI-H, MSI-L and MSS. Similarly, Caldes et al. 57 suggest that mutations in MSH6 do not always produce instability in tumours but in this study an MSI-L categorisation is not used (see Table 10).
For the five single-gate, high-risk samples presented in Table 13,34,57–60 sensitivity estimates ranged from 79.4% (95% CI 62.1% to 91.3%) in the study by Caldes et al. 57 to 100.0% (85.8% to 100.0%) in the study by Shia et al. 60 when MSI-L cases were considered to be index test positive results. Therefore, all of these sensitivity estimates were > 79%. It is important to remember that, for three of these studies,57,59,60 MSI was already categorised bimodally and therefore the estimates presented in Table 13 are identical to those presented in Table 11 (and are based on the same data). For the other two single-gate, high-risk studies,34,58 sensitivities were very similar, albeit slightly higher when MSI-L cases were considered to be index test positive results (93.1%, 95% CI 77.2% to 99.2% in the study by Mueller et al. 58 and 94.7%, 95% CI 82.3% to 99.4% in the study by Poynter et al. 34) than when they were considered to be index test negative results (91.3%, 95% CI 72.0% to 98.9% in the study by Mueller et al. 58 and 86.8%, 95% CI 71.9% to 95.6% in the study by Poynter et al. ;34 see Table 11). The MSI data from the reference standard-positive study61 was also used to generate sensitivity estimates (see Table 13) and, similarly, sensitivity was slightly higher when MSI-L cases were considered to be index test positives (92.0%, 95% CI 74.0% to 99.0%) than when they were considered to be index test negatives (88.0%, 95% CI 68.8% to 97.5%; see Table 11).
Likelihood ratios (LR+ and LR–), PPV and NPV were calculated for the three population-based studies reporting MSI data34,54,56 when MSI-L cases were assumed to be positive index test results (and the unclassified variants reported in the studies by Barnetson et al. 54 and Poynter et al. 34 were assumed to be negative reference standard results). These estimates are summarised in Table 14.
| Study | LR+ (95% CI) | LR– (95% CI) | PPV (95% CI) (%) | NPV (95% CI) (%) |
|---|---|---|---|---|
| Population-based single-gate studies | ||||
| aPoynter et al., 200834 | 1.42 (1.35 to 1.50) | 0.00 (NE) | 12.6 (9.8 to 16.0) | 100.0 (97.9 to 100.0) |
| Barnetson et al., 200654 | 6.01 (4.58 to 7.89) | 0.08 (0.02 to 0.30) | 35.9 (25.3 to 47.6) | 99.3 (97.4 to 99.9) |
| Southey et al., 200556 | 2.28 (1.56 to 3.33) | 0.09 (0.01 to 0.65) | 50.0 (32.4 to 67.6) | 96.0 (79.6 to 99.9) |
For all three population-based studies, the LR+ estimate was reduced when MSI-L cases were considered to be positive index test results (1.42, 95% CI 1.35 to 1.50 in the study by Poynter et al. ;34 6.01, 95% CI 4.58 to 7.89 in the study by Barnetson et al. ;54 2.28, 95% CI 1.56 to 3.33 in the study by Southey et al. 56) compared with when they were considered to be negative index test results (2.57, 95% CI 2.32 to 2.85 in the study by Poynter et al. ;34 8.94, 95% CI 5.54 to 14.20 in the study by Barnetson et al. ;54 5.92, 95% CI 2.48 to 14.10 in the study by Southey et al. 56; see Table 12). As before, the LR– could be estimated for only two of the studies54,56 because there were no false-negative MSI results in the study by Poynter et al. 34 Again, the LR– estimate was similar in both studies54,56 and was lower when MSI-L cases were considered to be positive index test results (0.08, 95% CI 0.02 to 0.30 in the study by Barnetson et al. ;54 0.09, 95% CI 0.01 to 0.65 in the study by Southey et al. 56) than when MSI-L cases were considered to be negative index test results (0.36, 95% CI 0.21 to 0.60 in the study by Barnetson et al. ;54 0.32, 95% CI 0.15 to 0.67 in the study by Southey et al. ;56 see Table 12).
As before, PPV estimates varied a lot more between the studies, with the lowest estimate still coming from the study by Poynter et al. 34 (12.6%, 95% CI 9.8% to 16.0%) and the highest estimate still coming from the study by Southey et al. 56 (50.0%, 95% CI 32.4% to 67.6%). For all three studies, PPV estimates were lower when MSI-L cases were considered to be index test positives than when they were considered to be index test negatives (see Table 12). Conversely, NPV estimates, which were consistent across the three studies (with all estimates being > 96%), were higher when MSI-L cases were considered to be index test positives than when they were considered to be index test negatives (see Table 12). However, it should be noted that, for the unselected CRC population in the study by Poynter et al. ,34 the NPV estimate was 100% regardless of whether MSI-L cases were considered to be positive or negative index test results, although the CIs were slightly wider when MSI-L cases were considered to be positive index test results (95% CI 97.9% to 100.0%) than when they were considered to be negative index test results (95% CI 99.0% to 100.0%).
As before, secondary analyses were conducted in which unclassified variants were considered to indicate a positive reference standard test result. Although five of the included studies34,54,57,60,61 reported unclassified variants in their assessment of MSI, only one61 provided sufficient data to conduct secondary analyses. However, because this study is a reference standard-positive study, only sensitivity estimates were made.
When unclassified variants were considered to be reference standard positives and MSI-L cases were considered to be index test positives, Hendriks et al. 61 reported a sensitivity of 93.9% (95% CI 80.3% to 98.3%). As expected, because the threshold for a MSI case is essentially lowered when MSI-L is considered to be an index test positive, this sensitivity was higher than that reported when MSI-L cases were considered to be index test negatives and unclassified variants were considered to be reference standard positives (84.8%, 95% CI 69.0% to 93.3%). When comparing the results from the study by Hendriks et al. 61 when unclassified variants were considered to be reference standard positives (MSI-L as an index test positive) with those generated when unclassified variants were considered to be reference standard negatives (MSI-L as an index test positive), sensitivity was very similar (93.9%, 95% CI 80.3% to 98.3% vs. 92.0%, 95% CI 74.0% to 99.0% respectively).
Assessment of test accuracy: immunohistochemistry
Immunohistochemistry was conducted in all 10 studies (11 samples) included in the review of test accuracy. However, not all studies provided sufficient data to be included in the analyses. Indeed, despite IHC being conducted for the high-risk sample in the study by Poynter et al. 34 and the sample in the study by Mueller et al. ,58 insufficient data were provided for these samples to be included in any of the IHC analyses. For the population-based sample in the study by Poynter et al. 34 and the samples in the studies by Barnetson et al. ,54 Southey et al. ,56 Hendriks et al. 61 and Okkels et al. ,62 the IHC results were reported according to the particular protein assessed (MLH1, MSH2, MSH6 or PMS2), enabling an assessment of IHC for at least one of these individual proteins (i.e. whether an absence of a particular protein accurately identifies a mutation in a particular gene). In seven cases54–57,59–61 an overall result was provided (i.e. whether a positive IHC result, regardless of which protein this applies to, predicts a positive reference standard result).
As with the results for MSI, in primary analyses unclassified variants were considered to be reference standard negatives. When sufficient data were available, secondary analyses were conducted in which unclassified variants were considered to be reference standard positives.
Overall immunohistochemistry results
As noted above, seven studies54–57,59–61 provided sufficient data to enable an assessment of the overall test performance of IHC (i.e. whether a positive IHC result, regardless of which protein this applies to, predicts a positive reference standard result). All of these studies assessed MLH1, MSH2 and MSH6 proteins. Therefore, abnormal staining for any of these three proteins was considered to be a positive index test result. Southey et al. 56 and Overbeek et al. 59 also assessed PMS2. Therefore, for these two studies an abnormal PMS2 result was included as a positive index test result.
Three population-based studies, all based on age-limited samples,54–56 were included in the primary analyses, in which unclassified variants were considered to be reference standard negatives. For all three of these studies sensitivity estimates were made, with Limburg et al. 55 providing the lowest estimate (85.7%, 95% CI 41.2% to 99.6%) and Southey et al. 56 providing the highest estimate (100.0%, 81.5% to 100.0%). The study by Southey et al. 56 included an assessment of PMS2 as well as assessments of MLH1, MSH2 and MSH6 and it is possible that this accounted for the higher sensitivity estimate. Nevertheless, all sensitivity estimates from the population-based studies were > 85% (Table 15). For two of the population-based studies55,56 specificity estimates were also made for the overall IHC results; these were > 80% for both studies (91.9%, 95% CI 86.3% to 95.7% for Limburg et al. 55 and 80.5%, 95% CI 65.1% to 91.2% for Southey et al. ;56 see Table 15). Specificity could not be estimated for the study by Barnetson et al. 54 because overall IHC results were available only for reference standard-positive participants.
| Study | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) |
|---|---|---|
| Population-based single-gate studies | ||
| Barnetson et al., 200654 | 92.6 (76.6 to 97.9) | NE |
| Limburg et al., 201155 | 85.7 (42.1 to 99.6) | 91.9 (86.3 to 95.7) |
| Southey et al., 200556 | 100.0 (81.5 to 100.0) | 80.5 (65.1 to 91.2) |
| Single-gate studies recruiting high-risk samples | ||
| Caldes et al., 200457 | 96.4 (81.7 to 99.9) | – |
| Overbeek et al., 200759 | 87.5 (52.9 to 97.7) | – |
| Shia et al., 200560 | 80.8 (60.6 to 93.4) | – |
| Reference standard-positive study sample | ||
| Hendriks et al., 200361 | 91.7 (77.5 to 98.2) | – |
For the three high-risk, single-gate samples presented in Table 15,57,59,60 sensitivity estimates ranged from 80.8% (95% CI 60.6% to 93.4%) in the study by Shia et al. 60 to 96.4% (95% CI 81.7% to 99.9%) in the study by Caldes et al. 57 Two of these high-risk, single-gate studies mentioned unclassified variants57,60 and these were counted as reference standard negatives in these analyses. The data from the reference standard-positive study that assessed overall IHC results61 was also used to generate a sensitivity estimate (91.7%, 95% CI 77.5% to 98.2%).
Because of a potential for spectrum bias, it would be expected that studies recruiting high-risk populations would have higher sensitivity estimates than population-based studies. However, as discussed in a systematic review by Palomaki et al. ,42 and as with the MSI results reported earlier (see Assessment of test accuracy: microsatellite instability testing), we did not find great differences between the sensitivity estimates in the population-based studies and the sensitivity estimates in the high-risk studies. However, this could be because the three population-based studies with overall IHC data54–56 were based on age-limited populations and may also be subject to spectrum bias.
The other test accuracy outcomes included in this review were LRs (LR+ and LR–), PPV and NPV, accuracy or concordance with the reference standard, diagnostic yield and test failure rates. As previously mentioned, the last three outcomes (accuracy or concordance with the reference standard, diagnostic yield and test failure rates) were not reported in any of the included studies. The other four outcomes (LR+, LR–, PPV and NPV) were calculated for the two population-based studies with sufficient overall IHC data available. 55,56 These outcomes are reported in Table 16. Unclassified variants were considered to be reference standard negatives. Again, the study by Barnetson et al. 54 is not included in this table because overall IHC results were available only for reference standard-positive participants.
| Study | LR+ (95% CI) | LR– (95% CI) | PPV (95% CI) (%) | NPV (95% CI) (%) |
|---|---|---|---|---|
| Population-based single-gate studies | ||||
| Limburg et al., 201155 | 10.6 (5.7 to 19.7) | 0.16 (0.02 to 0.95) | 33.3 (13.3 to 59.0) | 99.3 (96.0 to 100.0) |
| Southey et al., 200556 | 5.1 (2.8 to 9.5) | 0.00 (NE) | 69.2 (48.2 to 85.7) | 100.0 (89.4 to 100.0) |
The LR+ was 10.6 (95% CI 5.7 to 19.7) for Limburg et al. 55 and 5.1 (95% CI 2.8 to 9.5) for Southey et al. 56 The LR– could be estimated only for Limburg et al. 55 (0.16, 95% CI 0.02 to 0.95) because there were no false-negative overall IHC results in the study by Southey et al. 56 The PPV was lower in the study by Limburg et al. 55 (33.3%, 95% CI 13.3 to 59.0) than in the study by Southey et al. 56 (69.2%, 95% CI 48.2 to 85.7). The NPV was high in both studies, with both estimates being > 99%. Again, when apparent differences in IHC performance exist between these two studies (e.g. in the PPV results) it should be considered that Southey et al. 56 included an assessment of PMS2 in their results whereas Limburg et al. 55 did not. Additionally, the specific techniques and methods used to perform the reference standard differed between the studies (see Table 8) and this may also have had an impact on the apparent test performance.
Secondary analyses were conducted in which unclassified variants were considered to be reference test positives. It was not possible to include all studies reporting IHC data in these analyses (because sufficient data were not reported); indeed, only two studies57,61 provided sufficient data to conduct secondary analyses. However, because the study by Caldes et al. 57 was based on a high-risk population and the study by Hendriks et al. 61 was a reference standard-positive study, only sensitivity was estimated.
When unclassified variants were considered to be reference standard positive results, overall IHC sensitivity was estimated to be 75.0% (95% CI 57.8% to 87.9%) in the study by Caldes et al. 57 and 88.6% (95% CI 76.0% to 95.0%) in the study by Hendriks et al. 61 For Caldes et al. 57 this represents quite a reduction in sensitivity compared with when unclassified variants were considered to be index test negative results (96.4%, 95% CI 81.7% to 99.9%), whereas for Hendriks et al. 61 sensitivity was only slightly reduced by categorising unclassified variants as reference standard positives rather than reference standard negatives (see Table 15).
Immunohistochemical analysis of individual proteins
The analyses described in the previous section (in which overall IHC results are considered) are limited in what they can demonstrate about the loss of expression of individual proteins and how this relates to pathogenic mutations. Indeed, Overbeek et al. 59 note that tumour cells of MLH1 mutation carriers generally lacked MLH1 and PMS2 according to IHC staining, those of MSH2 mutation carriers lacked MSH2 and MSH6, those of MSH6 mutation carriers lacked MSH6 and those of PMS2 mutation carriers lacked PMS2. Furthermore, Barnetson et al. 54 suggest that ‘the absence of MSH6 protein predicted mutations in MSH2 or MSH6 . . . , as did the absence of MSH2 for mutations in MSH2 or MSH6 . . . , reflecting the biologic interaction between these proteins’ (p. 2756).
Indeed, the significance of the patterns of IHC abnormality in predicting underlying genetic causes of CRC predisposition appears to be becoming clearer, if more complex (see Table 4); it is becoming apparent that not all mutations are associated with loss or abnormality of the corresponding protein and that a specific IHC abnormality cannot be assumed to be an absolute indicator of the underlying genetic defect. 37 However, it was beyond the scope of this review to use data from the included studies to attempt an assessment of which IHC results were more or less likely to predict the presence of particular pathogenic mutations (or whether particular patterns or combinations of IHC abnormality correspond to particular defects). In any case, there were insufficient individual patient data available from the population-based samples to attempt such an analysis.
However, for the population-based sample in the study by Poynter et al. 34 and the samples in the studies by Barnetson et al. ,54 Southey et al. ,56 Hendriks et al. 61 and Okkels et al. ,62 sufficient data were available to enable an assessment of IHC for at least one individual protein, in terms of whether loss of expression of that protein was an accurate test for a pathogenic mutation in the corresponding gene (regardless of whether there was also loss of expression of additional proteins). Three of these studies54,56,61 provided sufficient data to assess whether loss of protein expression of MLH1, MSH2 or MSH6 was an accurate test of a pathogenic mutation in the same gene. Southey et al. 56 additionally provided data to enable an assessment of whether loss of protein expression of PMS2 was an accurate test of a pathogenic mutation in PMS2. The study by Okkels et al. 62 was designed to assess whether lack of protein expression of MSH6 would predict a pathogenic mutation in MSH6. For the population-based sample in Poynter et al. ,34 limited IHC data were available for individual proteins. However, sufficient data were available to assess the specificity of the loss of protein expression of MLH1. Sensitivity was not calculated for this study because these data were available only for reference standard negatives. The estimates of sensitivity and specificity generated from these studies (for IHC analysis of individual proteins) are provided in Table 17.
| Study | MLH1 | MSH2 | MSH6 | PMS2 | ||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | |
| Population-based single-gate studies | ||||||||
| Poynter et al., 200834 | – | 70.6 (66.8 to 74.2) | – | – | – | – | – | – |
| Barnetson et al., 200654 | 100.0 (73.5 to 100.0) | 96.0 (93.1 to 97.9) | 81.8 (48.2 to 97.7) | 96.2 (93.5 to 98.0) | 75.0 (19.4 to 99.4) | 95.4 (92.5 to 97.4) | – | – |
| Southey et al., 200556 | 50.0 (26.0 to 74.0) | 90.2 (76.9 to 97.3) | 22.2 (6.4 to 47.6) | 92.7 (80.1 to 98.5) | 44.4 (21.5 to 69.2) | 92.7 (80.1 to 98.5) | 55.6 (30.8 to 78.5) | 87.8 (73.8 to 95.9) |
| Reference standard-positive studies | ||||||||
| Hendriks et al., 200361 | 85.7 (63.7 to 97.0) | – | 81.8 (48.2 to 97.7) | – | 75.0 (19.4 to 99.4) | – | – | – |
| Okkels et al., 201262 | – | – | – | – | 72.7 (51.8 to 86.8) | – | – | – |
Of the four studies that provided data relevant to whether loss of expression of MLH1 was an accurate test result for assessing a pathogenic mutation in MLH1, three were population-based single-gate studies34,54,56 and one was a reference standard-positive study. 61 The studies by Barnetson et al. ,54 Southey et al. 56 and Hendriks et al. 61 all provided data from which sensitivities were generated. These ranged from 50.0% (95% CI 26.0% to 74.0%) for Southey et al. 56 to 100.0% (95% CI 73.5% to 100.0%) for Barnetson et al. 54 The three population-based studies34,54,56 provided data from which specificities were generated. These ranged from 70.6% (95% CI 66.8% to 74.2%) for Poynter et al. 34 to 96.0% (95% CI 93.1% to 97.9%) for Barnetson et al. 54 The results for MSH2 were even more variable. Three studies provided data relevant to whether or not loss of expression of MSH2 was an accurate test result for assessing a pathogenic mutation in MSH2. Two of these were single-gate population-based studies54,56 and one was a reference standard-positive study. 61 Sensitivities ranged from 22.2% (95% CI 6.4% to 47.6%) for Southey et al. 56 to 81.8% (95% CI 48.2% to 97.7%) for Barnetson et al. 54 and Hendriks et al. 61 The two population-based studies54,56 provided data from which specificities were generated. These were fairly consistent across the studies, with both specificity estimates being > 92% (see Table 17).
Four studies provided data relevant to whether loss of expression of MSH6 was an accurate test result for assessing a pathogenic mutation in MSH6. Two of these were single-gate population-based studies54,56 and two were reference standard-positive studies. 61,62 Again, there was more variation in the sensitivities generated than in the specificities generated: sensitivities ranged from 44.4% (95% CI 21.5% to 69.2%) for Southey et al. 56 to 75.0% (95% CI 19.4% to 99.4%) for both Barnetson et al. 54 and Hendriks et al. ,61 whereas specificities, which were generated only for the population-based studies,54,56 were similar across studies, with both studies producing an estimate of > 92% (see Table 17). Only one study56 provided data to enable an assessment of whether loss of expression of PMS2 was an accurate test result for assessing a pathogenic mutation in PMS2, producing a sensitivity estimate of 55.6% (95% CI 30.8% to 78.5%) and a specificity estimate of 87.8 (95% CI 73.8% to 95.9%) (see Table 17).
With the exception of the sensitivity estimates generated from the study by Southey et al. ,56 it appears that the data presented in Table 17 are fairly consistent across studies. The different estimates generated from the study by Southey et al. 56 appear to be the result of higher rates of false-negative results than in the other studies. It is possible that this is because of specific between-study differences in the assessment of IHC results (a positive IHC result is a somewhat subjective judgement, made by human assessors, and so inter-rater variability may have an impact on the results). Again, it is also possible that between-study differences in the reference standard could, to some extent, account for these differences in sensitivity estimates.
This is further demonstrated in Figure 5, in which sensitivities and specificities from the studies by Barnetson et al. 54 and Southey et al. 56 are graphically summarised using a SROC curve. This visually elucidates the trade-off between sensitivity and specificity across the two studies for each of the individual proteins assessed (MLH1, MSH2 and MSH6, as well as PMS2 for Southey et al. 56). In this figure, unclassified variants were assumed to be negative reference standard results.
The other test accuracy outcomes included in this review were LRs (LR+ and LR–), PPV and NPV, accuracy or concordance with the reference standard, diagnostic yield and test failure rates. As previously mentioned, the last three outcomes (accuracy or concordance with the reference standard, diagnostic yield and test failure rates) were not reported in any of the included studies. The other four outcomes (LR+, LR–, PPV and NPV) were calculated for the two population-based studies with sufficient IHC data by protein available. 54,56
The LR+ and LR– estimates are reported in Table 18. Unclassified variants were considered to be reference standard negative results. For loss of protein expression of MLH1, the LR+ was 25.0 (95% CI 14.4 to 43.5) for Barnetson et al. 54 and 5.1 (95% CI 1.8 to 14.5) for Southey et al. 56 For loss of protein expression of MSH2, the LR+ was 21.6 (95% CI 11.6 to 40.2) for Barnetson et al. 54 and 3.0 (95% CI 0.8 to 12.2) for Southey et al. 56 For loss of protein expression in MSH6, the LR+ was 16.2 (95% CI 7.6 to 34.3) for Barnetson et al. 54 and 6.1 (95% CI 1.8 to 20.3) for Southey et al. 56 For loss of protein expression in PMS2, data were available only for Southey et al. ,56 with a LR+ of 4.6 (95% CI 1.8 to 11.4). There are several possible reasons why the LR+ estimates were higher for Barnetson et al. 54 than for Southey et al. ,56 including the fact that the study by Barnetson et al. 54 was a larger study than that by Southey et al. ,56 that the reference standard was not identical in these studies and that there is a possibility that IHC ratings may have differed across studies (inter-rater reliability).
| Study | Protein | |||||||
|---|---|---|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | |||||
| LR+ (95% CI) | LR– (95% CI) | LR+ (95% CI) | LR– (95% CI) | LR+ (95% CI) | LR– (95% CI) | LR+ (95% CI) | LR– (95% CI) | |
| Barnetson et al., 200654 | 25.0 (14.4 to 43.5) | 0.0 (NE) | 21.6 (11.6 to 40.2) | 0.2 (0.1 to 0.7) | 16.2 (7.6 to 34.3) | 0.3 (0.1 to 1.4) | – | – |
| Southey et al., 200556 | 5.1 (1.8 to 14.5) | 0.6 (0.4 to 0.9) | 3.0 (0.8 to 12.2) | 0.8 (0.7 to 1.1) | 6.1 (1.8 to 20.3) | 0.6 (0.4 to 0.9) | 4.6 (1.8 to 11.4) | 0.5 (0.3 to 0.9) |
For loss of protein expression of MLH1, the LR– was estimated for only one study56 (0.6, 95% CI 0.4 to 0.9) because there were no false-negative results in the study by Barnetson et al. 54 For loss of protein expression of MSH2, the LR– was 0.2 (95% CI 0.1 to 0.7) for Barnetson et al. 54 and 0.8 (95% CI 0.7 to 1.1) for Southey et al. 56 For loss of protein expression of MSH6, the LR– was 0.3 (95% CI 0.1 to 1.4) for Barnetson et al. 54 and 0.6 (95% CI 0.4 to 0.9) for Southey et al. 56 For loss of protein expression of PMS2, data were available only from Southey et al. ,56 with the LR– estimated as 0.5 (95% CI 0.3 to 0.9).
The PPV and NPV estimates are reported in Table 19. Again, unclassified variants were considered to be reference standard negative results. For loss of protein expression of MLH1 and MSH2, the PPV and NPV results were largely consistent across the two studies providing data,54,56 although the PPV estimates were lower in the study by Barnetson et al. 54 and the NPV estimates were lower in the study by Southey et al. 56 For loss of expression of MSH6, the NPV estimates were consistent across the two studies but the PPV estimates were vastly different, with the data from Barnetson et al. 54 resulting in a PPV of 16.7% (95% CI 3.6% to 41.4%) and the data from Southey et al. 56 resulting in a PPV of 72.7% (95% CI 39.0% to 94.0%). Although the reason for this difference is not completely clear, it is likely, in part, to be because of the very low number of true positive (TP) results (n = 3) for loss of expression of MSH6 in the study by Barnetson et al. 54 Again, only Southey et al. 56 provided data for loss of protein expression of PMS2, with the PPV estimated as 66.7% (95% CI 38.4% to 88.2%) and the NPV estimated as 81.8% (95% CI 67.3% to 91.8%).
| Study | Protein | |||||||
|---|---|---|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | |||||
| PPV (95% CI) (%) | NPV (95% CI) (%) | PPV (95% CI) (%) | NPV (95% CI) (%) | PPV (95% CI) (%) | NPV (95% CI) (%) | PPV (95% CI) (%) | NPV (95% CI) (%) | |
| Barnetson et al., 200654 | 50.0 (29.1 to 70.9) | 100.0 (98.7 to 100.0) | 42.9 (21.8 to 66.0) | 99.3 (97.7 to 99.9) | 16.7 (3.6 to 41.4) | 99.7 (96.2 to 100.0) | – | – |
| Southey et al., 200556 | 69.2 (38.6 to 90.9) | 80.4 (66.1 to 90.6) | 57.1 (18.4 to 90.1) | 73.1 (59.0 to 84.4) | 72.7 (39.0 to 94.0) | 79.2 (65.0 to 89.5) | 66.7 (38.4 to 88.2) | 81.8 (67.3 to 91.8) |
Secondary analyses were conducted in which unclassified variants were considered to be positive reference standard test results. It was not possible to include all five studies34,54,56,61,62 reporting IHC data for individual proteins in these analyses because sufficient data were not always reported. Indeed, only one study61 provided sufficient data to conduct secondary analyses and, because this study was a reference standard-positive study, only sensitivity estimates were made. These sensitivity estimates were very similar to those estimated from data in which unclassified variants were considered to be reference standard negatives.
Indeed, when unclassified variants were considered to be reference standard positives, loss of protein expression of MLH1 was estimated to have a sensitivity of 80.0% (95% CI 60.8% to 91.1%) compared with the previously reported sensitivity of 81.0% (95% CI 58.1% to 94.6%) when unclassified variants were considered to be reference test negatives. Loss of protein expression of MSH2 was estimated to have a sensitivity of 83.3% (95% CI 55.1% to 95.3%) compared with the previously reported sensitivity of 81.8% (95% CI 48.2% to 97.7%) when unclassified variants were considered to be reference test negatives. Loss of protein expression in MSH6 was estimated to have a sensitivity of 80.0% (95% CI 37.6% to 96.4%) compared with the previously reported sensitivity of 75.0% (95% CI 19.4% to 99.4%) when unclassified variants were considered to be reference test negatives (see Table 17).
Summary of results from the test accuracy review
Summary of included studies
Ten studies met the test accuracy review inclusion criteria. One of the included studies included two distinct samples (a population-based sample and a high-risk sample) and these two samples were treated separately. Thus, although 10 studies were included, 11 populations/data sets were included. The results from all 11 populations were considered.
The 11 study samples were divided as follows: four single-gate studies with population-based samples, including one apparently unselected CRC population34 and three age-limited populations;54–56 five single-gate studies based on high-risk populations;34,57–60 and two studies that used a variation on a two-gate study design61,62 in which participants with positive reference standard results were recruited but no reference standard negatives were recruited. For this report, and for clarity, these studies are termed ‘reference standard-positive studies’.
With the exception of the studies by Limburg et al. 55 and Okkels et al. ,62 all studies assessed MSI. Although IHC was conducted in all 10 studies (11 samples) included in the review, not all studies provided sufficient data to be included in the analyses. Indeed, for the high-risk sample in the study by Poynter et al. 34 and the sample in the study by Mueller et al. ,58 despite IHC being conducted, insufficient data were provided for these samples to be included in any of the IHC analyses. None of the studies made a direct comparison between MSI and IHC. As such, the results are reported separately for these tests.
There was significant methodological and clinical heterogeneity across studies. In particular, the reference standard differed between studies, as did the index tests. With regard to the reference standard there were differences in the testing methods used (including the sequencing methods used and genes tested, the techniques used to test for large genomic alterations and deletions, the genes tested for large genomic alterations and deletions and whether unclassified variants were investigated). As a result of this, pooling of data in statistical analyses was not appropriate. In addition, there were insufficient data to conduct subgroup analyses based on most of these variables. However, test performance statistics were primarily generated with unclassified variants categorised as negative reference standard results and two studies57,61 provided sufficient data to conduct secondary analyses in which unclassified variants were categorised as positive reference standard results.
Quality appraisal of all 11 data sets was conducted using phase 3 of the QUADAS-2 tool. Four of the studies were rated as having a low risk of bias for patient selection, with three of these being population-based single-gate studies54–56 and one being a reference standard-positive study62 for which only sensitivity estimates could be made. For all studies included in the review there were no concerns about whether or not the included participants matched the review question. For both index tests, all studies were rated as having an unclear risk of bias with regard to whether the conduct and interpretation of the test could have introduced bias, but there were no concerns (in any of the studies) that the conduct or interpretation of either of the index tests was different from the review question. All of the included studies apart from that by Hendriks et al. 61 were rated as unclear with regard to whether or not the conduct or interpretation of the reference standard could have introduced bias. This was because only the study by Hendriks et al. 61 specified that the reference standard results were interpreted without knowledge of the results of the index test, with the rest of the studies not reporting this information. However, in all of the included studies the reference standard was assessed as being likely to correctly classify the target condition (because a genetic definition of LS was used in this review) and there were no concerns in any of the studies that the target condition, as defined by the reference standard, did not match the review question. For all included studies it was unclear whether the flow of participants through the study could have introduced bias.
The index tests included in this review (MSI and IHC) are highly susceptible to spectrum effects in populations that have been selected on the basis of clinical characteristics. In particular, an increased presence of MMR mutation carriers in a population would change the apparent sensitivity and specificity of the index tests. 54 However, a previous review did not find that this issue led to significant bias in estimates of sensitivity. 42 Because of this, studies recruiting high-risk populations were used only to estimate sensitivity. For the four samples included in this review that can be described as population-based samples,34,54–56 sensitivity, specificity, LR+, LR–, PPV and NPV were all estimated. However, it should be noted that three of these studies recruited age-limited populations,54–56 for which some spectrum bias may be expected.
Summary of the results for microsatellite instability
There were a variety of between-study differences in the MSI testing procedures used. In addition, potential differences between studies in MSI testing methods were not always clear because the methods used were not always reported in sufficient detail. For example, three34,58,59 of the eight studies assessing MSI did not report microdissection techniques (microdissection assists in ensuring that malignant tissue that does not contain DNA from surrounding healthy colonic tissue is analysed). The other differences between studies with regard to MSI testing methods can be categorised as differences in the panel of markers used, differences in the way in which MSI was categorised (e.g. as a bimodal or a trimodal distribution) and differences in the thresholds used to categorise MSI. Indeed, none of the population-based studies assessed the same panel of markers (differences exist in both the type and the number of markers assessed). Five studies34,54,56,58,61 defined tumours as MSI-H, MSI-L or MSS, also known as a trimodal distribution, two59,60 defined tumours as MSI positive or MSI negative, also known as a bimodal distribution, and one57 used a bimodal distribution but defined tumours as either MSI-H or MSS. Of the five studies that used the MSI-H, MSI-L and MSS categories (trimodal distribution), the thresholds used to categorise the tumours varied greatly, with one study34 using the commonly used threshold of ≥ 30% of unstable markers to define MSI-H tumours, three studies54,56,61 using differing numbers of unstable markers to define MSI-H tumours and one study58 not providing details on the thresholds used to categorise the tumours as MSI-H, MSI-L or MSS. Of the two studies that defined tumours as MSI positive or MSI negative (bimodal distribution), one reported using a threshold of ≥ 30% of unstable markers to define MSI-positive tumours60 and the other defined MSI-positive tumours as those with more than two unstable Bethesda panel markers. 59 The study by Caldes et al. 57 defined MSI-H tumours as those with two or more unstable Bethesda panel markers (or one marker in the case of BAT26) and MSS tumours as those showing no instability. It is not clear how cases with only one unstable marker (other than BAT26) were categorised, but in any case data were presented only for tumours that were categorised as MSS and MSI-H.
In primary analyses unclassified variants were categorised as negative reference standard results. Six study samples (including both samples in the study by Poynter et al. 34) provided data in which MSI-L was considered to be a negative index test result. 34,54,56,58,61 The other three samples utilised a bimodal distribution of MSI. 57,59,60 Across all nine samples, when MSI-L was considered to be a negative index test result, sensitivity ranged from 66.7% (95% CI 47.2% to 82.7%) in the population-based sample reported by Barnetson et al. 54 to 100.0% (95% CI 93.9% to 100.0%) in the population-based sample reported in Poynter et al. 34 and 100% (95% CI 85.8% to 100.0%) in the high-risk sample reported in Shia et al. 60 Sensitivity increased when MSI-L was considered to be a positive index test result (for the six study samples in which a trimodal distribution of MSI was used, with data remaining unchanged for the three samples in which a bimodal distribution of MSI was used). Indeed, across the nine study samples, the lower end of the range for sensitivity increased to 79.4% (95% CI 62.1% to 91.3%) in the high-risk sample recruited by Caldes et al. ,57 with the upper end of the range still being 100%.
In primary analyses (in which unclassified variants were categorised as negative reference standard results) three population-based study samples provided data in which MSI-L was considered to be a negative index test result. 34,54,56 Across these three samples, when MSI-L was considered to be a negative index test result, specificity ranged from 61.1% (95% CI 57.0% to 65.1%) in the study by Poynter et al. 34 to 92.5% (95% CI 89.1% to 95.2%) in the study by Barnetson et al. 54 It should be noted that the study by Barnetson et al. 54 included an age-limited sample whereas the study by Poynter et al. 34 included an unselected CRC population. Specificity decreased when MSI-L was considered to be a positive index test result. Indeed, the lower end of the range decreased to 29.5% (95% CI 25.8% to 33.4%) in the study by Poynter et al. 34 and the upper end of the range decreased to 84.5% (95% CI 80.0% to 88.2%) in the study by Barnetson et al. 54
It is unsurprising that, on the whole, sensitivity was higher and specificity was lower when MSI-L was considered to be a positive result than when MSI-L was considered to be a negative result; including MSI-L as a positive result essentially lowers the threshold for a positive index test result.
For the three studies that recruited population-based samples,34,54,56 LR+, LR–, PPV and NPV were also calculated. The LR+ was reduced when MSI-L was considered to be a positive index test result (1.42, 95% CI 1.35 to 1.50 for Poynter et al. ;3 6.01, 95% CI 4.58 to 7.89 for Barnetson et al. ;54 2.28, 95% CI 1.56 to 3.33 for Southey et al. 56) compared with when MSI-L was considered to be a negative index test result (2.57, 95% CI 2.32 to 2.85 for Poynter et al. ;34 8.94, 95% CI 5.54 to 14.20 for Barnetson et al. ;54 5.92, 95% CI 2.48 to 14.10 for Southey et al. 56) The LR– could be estimated for only two of the studies54,56 because there were no false-negative MSI results in the study by Poynter et al. 34 The LR– was similar in both studies and was lower when MSI-L was considered to be a positive index test result (0.08, 95% CI 0.02 to 0.30 for Barnetson et al. ;54 0.09, 95% CI 0.01 to 0.65 for Southey et al. 56) than when it was considered to be a negative index test result (0.36, 95% CI 0.21 to 0.60 for Barnetson et al. ;54 0.32, 95% CI 0.15 to 0.67 for Southey et al. 56). PPV estimates varied a lot more between the studies but were lower when MSI-L was considered to be a positive index test result than when it was considered to be a negative index test result. Conversely, NPV estimates were consistent across the three studies and were higher when MSI-L was considered to be a positive index test result than when it was considered to be a negative index test result. However, it should be noted that, for the unselected CRC population in the study by Poynter et al. ,34 the NPV estimate was 100% regardless of whether MSI-L was considered to be a positive or a negative index test result, although the CIs were slightly wider when MSI-L was considered to a positive result (95% CI 97.9% to 100.0%) than when it was considered to be a negative result (95% CI 99.0% to 100.0%).
Secondary analyses were conducted when data permitted in which unclassified variants were considered to be positive reference standard results. When MSI-L was considered to be a negative index test result, two studies57,61 provided sufficient data to conduct secondary analyses of the sensitivity estimates. Caldes et al. 57 reported a sensitivity of 81.6% (95% CI 65.7% to 92.3%) and Hendriks et al. 61 reported a sensitivity of 84.8% (95% CI 69.0% to 93.3%). These results were similar to those obtained when unclassified variants were considered to be negative reference standard results (79.4%, 95% CI 62.1% to 91.3% for Caldes et al. ;57 88.8%, 95% CI 68.8% to 97.5% for Hendriks et al. 61). When MSI-L was considered to be a positive index test result, only one study61 provided sufficient data to conduct a secondary analysis of the sensitivity estimate. In this study, sensitivity was 93.9% (95% CI 80.3% to 98.3%), which was similar to the estimate when unclassified variants were considered to be negative reference standard results (92.0%, 95% CI 74.0% to 99.0%).
Summary of the results for immunohistochemistry
In primary analyses unclassified variants were categorised as negative reference standard results. Seven studies54–57,59–61 provided data to assess the accuracy of an overall IHC result at identifying a positive reference standard result (i.e. whether a positive IHC result, regardless of which protein this applies to, identifies a positive reference standard result). For the population-based sample in the study by Poynter et al. 34 and the samples in the studies by Barnetson et al. 54; Southey et al. ,56 Hendriks et al. 61 and Okkels et al. ,62 IHC results were reported according to the particular protein assessed (MLH1, MSH2, MSH6 or PMS2), enabling an assessment of IHC for at least one of these individual proteins (i.e. whether an absence of a particular protein accurately identifies a mutation in that particular gene).
All of the seven studies54–57,59–61 assessing the overall test performance of IHC assessed MLH1, MSH2 and MSH6. Therefore, abnormal staining for any of these three proteins was considered to be a positive index test result. However, Southey et al. 56 and Overbeek et al. 59 also assessed PMS2. Therefore, for these two studies an abnormal PMS2 result was also included as a positive index test result. Three of these studies were population-based studies,54–56 three were single-gate high-risk studies57,59,60 and one was a reference standard-positive study (in which only reference standard positives were recruited). Sensitivity estimates ranged from 80.8% (95% CI 60.6% to 93.4%) in the study by Shia et al. 60 to 100.0% (95% CI 81.5% to 100.0%) in the study by Southey et al. 56 The study by Southey et al. 56 included an assessment of PMS2 as well as assessments of MLH1, MSH2 and MSH6 and it is possible that this accounted for the higher sensitivity estimate in this study. Nevertheless, all sensitivity estimates were > 80%. Because of the potential for spectrum bias, it would be expected that studies recruiting high-risk populations would have higher sensitivity estimates than those recruiting population-based samples. However, as discussed in a systematic review by Palomaki et al. ,42 and as with the MSI results reported earlier, we did not find great differences between the sensitivity estimates in the population-based studies and those in the high-risk studies. This could be because the three population-based studies with overall IHC data54–56 were based on age-limited populations and may also be subject to spectrum bias.
For two of the population-based studies55,56 specificity, LR+, LR–, PPV and NPV were also estimated for the overall IHC results. These analyses were not conducted for the third population-based study54 because overall IHC results were available only for reference standard-positive participants. Specificity was estimated as 91.9% (95% CI 86.3% to 95.7%) for Limburg et al. 55 and 80.5% (95% CI 65.1% to 91.2%) for Southey et al. 56 LR+ was estimated as 10.6 (95% CI 5.7 to 19.7) for Limburg et al. 55 and 5.1 (95% CI 2.8 to 9.5) for Southey et al. 56 LR– could be estimated for only one study55 (0.16, 95% CI 0.02 to 0.95) because there were no false-negative overall IHC results in the study by Southey et al. 56 PPV was lower in the study by Limburg et al. 55 (33.3%, 95% CI 13.3% to 59.0%) than in the study by Southey et al. 56 (69.2%, 95% CI 48.2% to 85.7%). NPV was high in both studies, with both estimates being > 99%. Again, when apparent differences in IHC performance exist between these two studies (e.g. in the PPV results), it should be considered that Southey et al. 56 included an assessment of PMS2 in their results whereas Limburg et al. 55 did not. Additionally, the specific techniques and methods used to perform the reference standard differed between studies and this may also impact on the apparent test performance.
Secondary overall IHC analyses were conducted in which unclassified variants were considered to be positive reference standard results. Only two studies57,61 provided sufficient data to conduct these secondary analyses and, because the study by Caldes et al. 57 included a high-risk population and that by Hendriks et al. 61 was a reference standard-positive study, only sensitivity was estimated (75.0%, 95% CI 57.8% to 87.9% for Caldes et al. ;57 88.6%, 95% CI 76.0% to 95.0% for Hendriks et al. 61). For Caldes et al. 57 this represented quite a reduction in sensitivity compared with when unclassified variants were considered to be reference test negatives (96.4%, 95% CI 81.7% to 99.9%).
For the population-based sample in the study by Poynter et al. 34 and the samples in the studies by Barnetson et al. ,54 Southey et al. ,56 Hendriks et al. 61 and Okkels et al. ,62 an assessment was made of IHC for at least one individual protein. Four studies provided data relevant to whether loss of expression of MLH1 was an accurate test result for assessing a pathogenic mutation in MLH1, of which three were population-based single-gate studies34,54,56 and one was a reference standard-positive study. 61 The studies by Barnetson et al. ,54 Southey et al. 56 and Hendriks et al. 61 all provided data from which sensitivities were generated; these ranged from 50.0% (95% CI 26.0% to 74.0%) for Southey et al. 56 to 100.0% (95% CI 73.5% to 100.0%) for Barnetson et al. 54 The three population-based studies34,54,56 provided data from which specificities were generated; these ranged from 70.6% (95% CI 66.8% to 74.2%) for Poynter et al. 34 to 96.0% (95% CI 93.1% to 97.9%) for Barnetson et al. 54 The results for MSH2 were even more variable. Three studies provided data for MSH2,54,56,61 with sensitivities ranging from 22.2% (95% CI 6.4% to 47.6%) for Southey et al. 56 to 81.8% (95% CI 48.2% to 97.7%) for both Barnetson et al. 54 and Hendriks et al. 61 The two population-based studies54,56 provided data from which specificities were generated, with both being > 92%. Four studies provided data for MSH6 and, again, there was more variation in the sensitivities generated than in the specificities generated: sensitivities ranged from 44.4% (95% CI 21.5% to 69.2%) for Southey et al. 56 to 75.0% (95% CI 19.4% to 99.4%) for both Barnetson et al. 54 and Hendriks et al. ,61 whereas specificities, which were generated only for the population-based studies,54,56 were > 92% in both studies. It was clear that, for loss of expression of MLH1, MSH2 and MSH6, the sensitivity estimates generated from the study by Southey et al. 56 were lower than those in the other studies. It is possible that this is because of specific between-study differences in the assessment of IHC (a positive IHC result is a somewhat subjective judgement, made by human assessors, and so inter-rater variability may impact on the results). Again, it is also possible that between-study differences in the reference standard could, to some extent, account for these differences in sensitivity. Only the study by Southey et al. 56 provided IHC data for PMS2, with a sensitivity estimate of 55.6% (95% CI 30.8% to 78.5%) and a specificity estimate of 87.8% (95% CI 73.8% to 95.9%).
The LR+, LR–, PPV and NPV were calculated for the two population-based studies with sufficient IHC data available by protein. 54,56 For MLH1, MSH2 and MSH6, LR+ was greater in the study by Barnetson et al. 54 than in the study by Southey et al. 56 (MLH1: 25.0, 95% CI 14.4 to 43.5 for Barnetson et al. 54 and 5.1, 95% CI 1.8 to 14.5 for Southey et al. ;56 MSH2: 21.6, 95% CI 11.6 to 40.2 for Barnetson et al. 54 and 3.0, 95% CI 0.8 to 12.2 for Southey et al. ;56 MSH6: 16.2, 95% CI 7.6 to 34.3 for Barnetson et al. 54 and 6.1, 95% CI 1.8 to 20.3 for Southey et al. 56) For PMS2, data were available only from Southey et al. ,56 with the LR+ estimated as 4.6 (95% CI 1.8 to 11.4). There are several possible reasons why the LR+ estimates were higher in the study by Barnetson et al. 54 than in the study by Southey et al. ,56 including that the study by Barnetson et al. 54 was a larger study than that by Southey et al. ,56 that the reference standard was not identical in these studies and that there is a possibility that IHC ratings may have differed across studies (inter-rater reliability). For MLH1, the LR– was estimated for only one study56 (0.6, 95% CI 0.4 to 0.9) because there were no false-negative results in the study by Barnetson et al. 54 For MSH2, the LR– was 0.2 (95% CI 0.1 to 0.7) for Barnetson et al. 54 and 0.8 (95% CI 0.7 to 1.1) for Southey et al. 56 and, for MSH6, the LR– was 0.3 (95% CI 0.1 to 1.4) for Barnetson et al. 54 and 0.6 (95% CI 0.4 to 0.9) for Southey et al. 56 For loss of protein expression of PMS2, data were available only from Southey et al. ,56 with the LR– estimated as 0.5 (95% CI 0.3 to 0.9). PPV and NPV estimates for MLH1 and MSH2 were largely consistent across the two studies. 54,56 For MSH6, NPV estimates were consistent across the two studies but PPV estimates were vastly different, with data from Barnetson et al. 54 resulting in a PPV of 16.7% (95% CI 3.6% to 41.4%) and data from Southey et al. 56 resulting in a PPV of 72.7% (95% CI 39.0% to 94.0%). Although the reason for this difference is not completely clear, in part it is likely to be because of the very low number of true-positive results (n = 3) for loss of expression of MSH6 in the study by Barnetson et al. 54 Again, only Southey et al. 56 provided data for PMS2, with the PPV estimated as 66.7% (95% CI 38.4% to 88.2%) and the NPV estimated as 81.8% (95% CI 67.3% to 91.8%).
Secondary IHC analyses for individual proteins were conducted in which unclassified variants were considered to be reference standard negatives. However, only one study61 provided sufficient data to conduct secondary analyses and, because this study was a reference standard-positive study, only sensitivity was estimated. These sensitivity estimates were very similar to those estimated from data in which unclassified variants were considered to be reference standard negatives.
Chapter 3 Assessment of end-to-end studies
End-to-end studies have an important place in the health technology assessments of tests. They are broadly defined in the methods guidance as ‘studies that follow patients from testing, through treatment, to final outcomes’ (p. 70). 70 Such studies can have a wide variety of designs but, when available, randomised controlled trials (RCTs) are noted to be of great importance because they provide comparative evidence with high internal validity. If end-to-end studies are found it may avoid the need for modelling as the end-to-end studies provide a direct linkage between a testing strategy and patient outcomes, which otherwise could be achieved only by linkage in an economic model. For these reasons we specifically performed a systematic review of end-to-end studies, with a particular focus on RCTs and controlled clinical trials (CCTs), recognising a priori that such studies were unlikely to exist for an intervention as complex as screening for LS.
Methods for reviewing effectiveness
Identification of studies
The same search was performed as for the review of diagnostic accuracy studies (see Chapter 2). This was appropriate as there were no restrictions by study design. In the protocol71 we did not rule out the use of a methods filter to focus the search on intervention studies; however, this was not used to maximise the sensitivity of the searches.
Inclusion and exclusion criteria
The full inclusion criteria are provided in Table 20. Concerning study design we agreed to consider other study designs if no RCTs or CCTs were identified.
| Criteria | Include | Exclude |
|---|---|---|
| Participants | Studies of unselected or randomly selected CRC patients or CRC patients selected according to an age limit | Studies using retained samples as storage methods may adversely affect test accuracy |
| Index tests | Molecular MSI testing (with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing), including studies in which BRAF V600E and/or MLH1 promoter methylation tests were performed according to MSI test results, followed by constitutional MMR mutation testing; MMR IHC (with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing), followed by the reference standard, followed by constitutional MMR mutation testing | Studies that do not include either of the index tests, studies in which mutation testing is limited to seeking only founder mutations |
| Comparators | Index tests may be compared with each other, no testing for LS or direct constitutional MMR mutation testing | |
| Outcomes | Number of individuals receiving MSI and/or IHC testing, number of individuals receiving subsequent tumour-based tests, number of individuals receiving constitutional MMR mutation testing, number of cascade tests on relatives, number of LS diagnoses, number of colonoscopies, morbidity, mortality and/or life expectancy, costs associated with interventions and comparators, health-related quality of life | |
| Study design | RCTs, CCTs | Non-experimental, preclinical and animal studies, studies published only in abstract form, systematic reviews of RCTs or CCTs |
Screening was performed by one primary screener (CH), with a 10% random sample checked by a second reviewer (TS). In the screen, any study that appeared to investigate the impact of introducing the tests of interest on the outcomes of interest was retrieved in full, irrespective of the apparent study design or whether it appeared to be reported in abstract form only. In total, 3920 records were screened for eligibility by title and abstract.
Twenty-two articles were retrieved in full text (details can be obtained from the authors on request). These reported 20 studies as two articles were duplicate publications (both abstracts that were subsequently published in full). The final inclusion/exclusion decisions and abstraction of brief details about all articles retrieved in hard text was performed by a single reviewer (CH).
Further aspects of the methods including the data abstraction strategy, critical appraisal strategy and methods of data synthesis are not reported as none of the studies reviewed in full text were included in the review.
Results
Quantity and quality of research available
Eleven studies were excluded through failure to meet the first inclusion criterion applied, introduction of the index tests of interest. The remaining nine studies were all reported in abstract form and so were excluded because they provided insufficient information to confirm the detailed nature of the testing introduced or to perform a proper quality assessment.
Even if the criterion about publication in full had been relaxed, only three studies could have been considered for inclusion because six of the abstracts appeared to report no comparator. The three abstracts that did report a comparator were all pre–post studies measuring changes surrounding the introduction of testing for LS. None provided any contact details and so it was not possible to easily enquire whether further information or a full publication of the results was available.
Two RCTs were identified among the full-text articles reviewed. The first was a protocol from a RCT comparing whole exome sequencing with ‘current practice’ in screening for LS. 72 The second was a cluster RCT investigating the effect of quality improvement initiatives. 73 The effect of quality improvement was further examined in a time-series analysis. 74 Only one study assessed patient survival. 75 This examined whether conclusive follow-up testing, or not, following initial screen-positive IHC testing in CRC cases in a specific institution was associated with better outcome. Thus, it did not directly examine the effect of the introduction of screening for LS. In addition, this study was presented only in abstract form and limited contact details were provided.
Assessment of effectiveness
Critical review and synthesis of information
We were not able to draw any conclusions on the effectiveness of screening for LS from the systematic review of end-to-end studies.
Discussion
Although the review identified some interesting near-miss excluded studies providing evidence of indirect relevance to other aspects of the appraisal, no sufficiently relevant end-to-end studies were reported in enough detail to obviate the need for a linked evidence approach using modelling. The three studies that were closest to meeting the inclusion criteria were reported only in abstract form and it may be that if full details had been available they would still have been excluded. All three employed pre–post designs and were hence highly susceptible to bias.
The review reinforces the lack of usefulness of studies that are reported only in abstract form, which is further compounded by the fact that few abstracts provide any means to contact authors for further information. The review also invites consideration of whether or not some study designs that might currently be considered as end-to-end studies, such as pre–post studies, may be too open to bias to be worth including in a systematic review of end-to-end studies, even if they are the only evidence available. This does not preclude them being used to parameterise a model, providing the openness to bias is fully acknowledged.
We believe that the methods used in the review, particularly the extremely comprehensive search, which was not restricted by a study design filter as originally planned, make it unlikely that we have missed major items of published literature. It is possible that we may have overlooked unpublished literature, as this is extremely difficult to find in a short timescale. We did, however, fully consider conference abstracts appearing in the main bibliographic databases that we included in the search, although they did not yield useful information for the reasons already indicated. Ideally, we could have used double screening and inclusion/exclusion in duplicate; however, this did not seem to be justified after good agreement was achieved in the 10% random check of screening decisions. Similarly, although not formally checked we are confident that the limited number of decisions made on inclusion/exclusion, performed by an experienced reviewer using a well-developed set of inclusion/exclusion criteria, were accurately performed. We did not feel that checking this review step was a priority among the other tasks required to complete the health technology assessment.
Chapter 4 Assessment of existing cost-effectiveness evidence
The cost-effectiveness of using MSI and IHC testing in strategies to identify LS was assessed by conducting a systematic review of published research evidence.
Objectives
The objectives of this systematic review were to:
-
gain insights into the key drivers of cost-effectiveness in this disease area
-
obtain an overview of the alternative modelling approaches that have been adopted in this disease and treatment area
-
provide a summary of the findings of previous relevant cost–utility, cost-effectiveness and cost–benefit studies generalisable to the UK.
Methods
Study identification
This systematic review was an update of the systematic review of cost-effectiveness reported in Snowsill et al. 4
The search strategy included the following sources:
-
electronic databases – MEDLINE (via Ovid), MEDLINE In-Process & Other Non-Indexed Citations (via Ovid), EMBASE (via Ovid), Web of Science (Thomson Reuters), NHS Economic Evaluation Database (NHS EED) (The Cochrane Library) and EconLit (EBSCOhost)
-
backward citation chasing on included studies.
The searches were developed and run by an information specialist (SB) in February 2016. Search filters were used to limit the searches to economic or health utilities studies as appropriate and searches were limited to English-language studies when possible. A date limit of 2013 was used. The search strategies for each database are detailed in Appendix 1.
The database search results were exported to, and deduplicated using, EndNote X7. Deduplication was also performed using manual checking. After the reviewers had completed the screening process, the bibliographies of included papers were scrutinised for further potentially includable studies.
Titles and abstracts returned by the search strategy were examined independently by two researchers (NH and TS) and screened for possible inclusion. Disagreements were resolved by discussion. Full texts of potentially relevant studies were ordered. Full publications were assessed by the same reviewers (NH and TS) for inclusion or exclusion against prespecified criteria. Again, disagreements were resolved by discussion.
Reviewers also examined the studies included in the systematic reviews by Snowsill et al. 4 and Grosse76 for other relevant studies.
Eligibility criteria
Table 21 shows the inclusion criteria used in the current systematic review of published cost-effectiveness studies compared with the inclusion criteria used in the previous Peninsula Technology Assessment Group (PenTAG) review. 4 The inclusion criteria for the current review were narrowed to address the decision problem, meaning that searches needed to be run only for the time period after the previous PenTAG review.
| PICOS criteria | Previous PenTAG review4 | Current review |
|---|---|---|
| Population | Those who may or may not have LS | All people newly diagnosed with CRC |
| Intervention | Any of the following (including combinations): strategies to identify LS in the population, strategies to manage LS in the population, strategies to manage patients in whom LS is identified | One of MSI testing (with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing) or IHC (with or without BRAF V600E mutation testing and with or without MLH1 promoter methylation testing) |
| Comparator | Current clinical practice (may or may not include efforts to identify LS) | At least one of the included interventions, no testing or direct constitutional MMR mutation testing |
| Outcomes | Any of the following: costs, clinically relevant outcomes (e.g. life-years gained, QALYs, CRCs prevented), mutations detected | Costs and health effects measured in life-years or QALYs |
| Study type | Any of the following: decision-analytic models (with or without a cost-effectiveness component), evaluations of cost-effectiveness within trials (including cost-effectiveness, cost–utility and cost–benefit studies; no requirement for randomisation), cost or resource use studies, guidelines from national institutions, professional bodies and international bodies (including working groups) | Any of the following: decision-analytic models, economic evaluations within trials, cost or resource use studies from the UK |
In the protocol for this project71 it is stated that IHC should be treated as a comparator only and not an intervention. We updated the inclusion criteria for this review so that IHC was treated as an intervention. This means that included studies that contained IHC-based strategies did not need to also contain MSI-based strategies, as specified when IHC is treated as a comparator. This also ensured consistency between the clinical effectiveness review and the cost-effectiveness review.
Systematic reviews, if identified, were not directly included but their bibliographies were searched for potentially includable studies.
Data extraction
Data were extracted by one reviewer (NH) using the blank data extraction forms used in the previous PenTAG review. 4
The completed forms for studies included in the previous PenTAG review were reused.
The evidence base was assessed using narrative synthesis supported by summary data extraction tables, constructed for the previous PenTAG review. 4 When studies did not conduct a fully incremental cost-effectiveness analysis (e.g. if they performed a cost–consequences analysis), but it was possible to conduct such an analysis based on the reported results, this was carried out.
Currency conversion was not performed but an indication of purchasing power parity exchange rates was given and, if currency- or country-specific cost-effectiveness thresholds were supplied by the authors, these were also reported (in the original currency).
Critical appraisal
For consistency with the previous PenTAG cost-effectiveness review,4 the quality appraisal was conducted using selected criteria from the Drummond and Jefferson77 checklist. Quality appraisal was conducted by one reviewer (NH).
Additionally, a set of review-specific criteria was developed for the previous PenTAG review4 and this was adapted to reflect the current decision problem.
Results
Figure 6 shows the study flow diagram for this update review. The electronic database search for cost-effectiveness evidence identified 352 records after deduplication. All were screened by title and abstract, with 11 identified for full-text screening. All 11 full texts were retrieved and assessed for eligibility.
FIGURE 6.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram for cost-effectiveness papers. a, Snowsill et al. 4 and Grosse. 76 Source: adapted from Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med 6(7):e1000097. 63 https://doi.org/10.1371/journal.pmed1000097
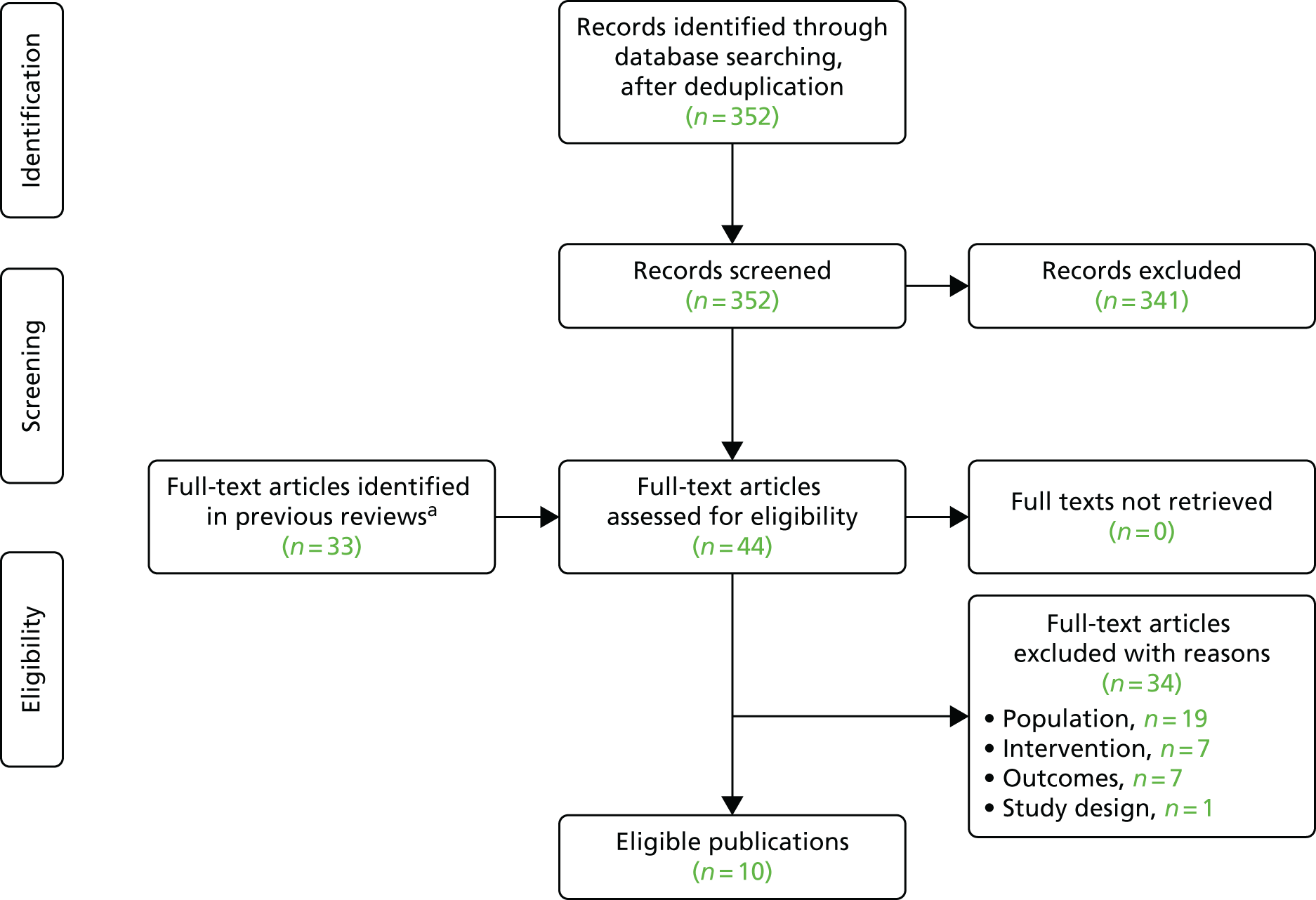
Of the 11 full texts assessed for eligibility, six4,78–82 were deemed to meet the eligibility criteria for the review.
Of the 33 additional publications identified in the previous reviews,4,76 four additional publications were deemed to meet the eligibility criteria for the review and were assessed in full. 83–86 The remaining 29 were excluded on the basis of population, intervention or outcome.
One UK-based cost study was identified87 but in this study the population was identified using clinical criteria and it was therefore ineligible for inclusion in our review.
Characteristics of the identified cost-effectiveness studies
The characteristics of the 10 included papers3,78–86 and their results are provided in Tables 22 and 23 respectively. Some studies reported results of strategies that used clinical criteria to identify LS; when possible, these results are excluded. One study is reported in abstract form. 78 One study is reported in two separate papers;4,79 these two papers have been grouped together for reporting purposes, giving a total of nine separate studies.
| Study | Setting, perspective | Population | Study purpose | Study approach | Diagnostic comparators | Treatment strategies | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ramsey et al., 200383 | USA, US health-care system | Newly diagnosed CRC patients and their siblings and children | Cost-effectiveness of strategies to identify LS | Decision model, ad hoc | Probands:Test 1Test 2Test 3NoneBethesdaMSIGTMSIGTBethesdaGTGTRelatives (proband confirmed mutation/indeterminate result): germ-line testing for siblings and children | Test 1 | Test 2 | Test 3 | None | Bethesda | MSI | GT | MSI | GT | Bethesda | GT | GT | Confirmed mutation: LS colorectal surveillance and prophylactic surgery on CRC diagnosis; indeterminate result: LS colorectal surveillance; LS negative: probands – standard care, relatives – no further action | ||||||||||||||||||||||||||||
| Test 1 | Test 2 | Test 3 | ||||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| Bethesda | MSI | GT | ||||||||||||||||||||||||||||||||||||||||||||
| MSI | GT | |||||||||||||||||||||||||||||||||||||||||||||
| Bethesda | GT | |||||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Mvundura et al., 201084 | USA, US health-care system | Newly diagnosed CRC patients and their relatives | Cost-effectiveness and cost–utility of strategies to identify LS | Decision model, ad hoc | Probands: Test 1Test 2Test 3NoneIHCBRAF (abnormal IHC for MLH1)GT (MLH1)GT (all other abnormal IHC results)IHCGTMSIGTGT Relatives (proband confirmed mutation): GT |
Test 1 | Test 2 | Test 3 | None | IHC | BRAF (abnormal IHC for MLH1) | GT (MLH1) | GT (all other abnormal IHC results) | IHC | GT | MSI | GT | GT | Relatives with confirmed mutation: colonoscopy every 1–2 years from age 20–25 years until age 79 years; LS-negative relatives (and some LS-positive relatives who declined LS surveillance): colonoscopy every 10 years from age 50 years | |||||||||||||||||||||||||||
| Test 1 | Test 2 | Test 3 | ||||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| IHC | BRAF (abnormal IHC for MLH1) | GT (MLH1) | ||||||||||||||||||||||||||||||||||||||||||||
| GT (all other abnormal IHC results) | ||||||||||||||||||||||||||||||||||||||||||||||
| IHC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| MSI | GT | |||||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Ladabaum et al., 201185 | USA, third-party payer (NIH) | Newly diagnosed CRC patients and their relatives | Cost-effectiveness of strategies to identify LS | Decision model, decision tree model, Markov subtrees | Probands: Test 1Test 2Test 3NoneCCGTCCIHCGTTBTGTGT Relatives (proband confirmed mutation): germ-line testing |
Test 1 | Test 2 | Test 3 | None | CC | GT | CC | IHC | GT | TBT | GT | GT | Persons with confirmed LS mutations or assumed LS and their FDRs: annual colonoscopy from age 25 years, women –gynaecological screening from age 35 years, prophylactic TAH-BSO at age 40 years; others: colonoscopy every 10 years from age 50 years | ||||||||||||||||||||||||||||
| Test 1 | Test 2 | Test 3 | ||||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| CC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| CC | IHC | GT | ||||||||||||||||||||||||||||||||||||||||||||
| TBT | GT | |||||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Wang et al., 201286 | USA, third-party payer (NIH) | Newly diagnosed CRC patients and their relatives | Cost–utility of strategies to identify LS (update of study by Ladabaum et al.85 to include quality of life) | Decision model, decision tree model, Markov subtrees | Probands: Test 1Test 2Test 3NoneCCGTCCIHCGTTBTGTGT Relatives (proband confirmed mutation): germ-line testing |
Test 1 | Test 2 | Test 3 | None | CC | GT | CC | IHC | GT | TBT | GT | GT | Persons with confirmed LS mutations or assumed LS and their FDRs: annual colonoscopy from age 25 years, women – gynaecological screening from age 35 years, prophylactic TAH-BSO at age 40 years; others: colonoscopy every 10 years from age 50 years | ||||||||||||||||||||||||||||
| Test 1 | Test 2 | Test 3 | ||||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| CC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| CC | IHC | GT | ||||||||||||||||||||||||||||||||||||||||||||
| TBT | GT | |||||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Gallego et al., 201478 (abstract) | USA, NR | CRC patients and their relatives | Cost-effectiveness and cost–utility of strategies to identify LS | Decision model | Probands: Test 1Test 2IHC/MSITargeted sequencingIHC/MSINGSNGS Relatives: NR |
Test 1 | Test 2 | IHC/MSI | Targeted sequencing | IHC/MSI | NGS | NGS | Relatives with LS detected: CRC surveillance | |||||||||||||||||||||||||||||||||
| Test 1 | Test 2 | |||||||||||||||||||||||||||||||||||||||||||||
| IHC/MSI | Targeted sequencing | |||||||||||||||||||||||||||||||||||||||||||||
| IHC/MSI | NGS | |||||||||||||||||||||||||||||||||||||||||||||
| NGS | ||||||||||||||||||||||||||||||||||||||||||||||
| Snowsill et al., 2014,4 Snowsill et al., 201579 | UK, NHS and PSS | Newly diagnosed CRC patients aged < 50 years and their relatives | Cost–utility of strategies to identify LS | Decision model, decision tree model and individual patient simulation model | Probands: Test 1Test 2Test 3Test 4NoneACIIIHCGTIHCBRAFGTMSIGTMSIBRAFGTMSIBRAFIHCGTIHCMSIBRAFGTGT Relatives: proband-confirmed LS – genetic testing, proband-suspected LS – FDRs assumed to have LS |
Test 1 | Test 2 | Test 3 | Test 4 | None | ACII | IHC | GT | IHC | BRAF | GT | MSI | GT | MSI | BRAF | GT | MSI | BRAF | IHC | GT | IHC | MSI | BRAF | GT | GT | Biannual colonoscopic surveillance for LS positive/suspected probands and relatives (from age 25 years), prophylactic TAH-BSO offered to women with a LS diagnosis at age 45 years | |||||||||||||||
| Test 1 | Test 2 | Test 3 | Test 4 | |||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| ACII | ||||||||||||||||||||||||||||||||||||||||||||||
| IHC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| IHC | BRAF | GT | ||||||||||||||||||||||||||||||||||||||||||||
| MSI | GT | |||||||||||||||||||||||||||||||||||||||||||||
| MSI | BRAF | GT | ||||||||||||||||||||||||||||||||||||||||||||
| MSI | BRAF | IHC | GT | |||||||||||||||||||||||||||||||||||||||||||
| IHC | MSI | BRAF | GT | |||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Barzi et al., 201580 | USA, societal | CRC patients and their relatives and the general population | Cost-effectiveness of strategies to identify LS | Decision model, decision tree and individual-level microsimulation Markov model | Probands: Test 1Test 2Test 3Test 4NoneCCGTCCIHCGTIHCGTIHC + BRAFGTMSIGTMSI + IHCGTMSI + IHC + BRAFGTGT Relatives (FDRs): GT; general population screening: PREMM > 20 years, > 25 years, > 30 years, > 35 years |
Test 1 | Test 2 | Test 3 | Test 4 | None | CC | GT | CC | IHC | GT | IHC | GT | IHC + BRAF | GT | MSI | GT | MSI + IHC | GT | MSI + IHC + BRAF | GT | GT | Annual colonoscopy from age 20 to 80 years, TAH-BSO offered at age 40 years | |||||||||||||||||||
| Test 1 | Test 2 | Test 3 | Test 4 | |||||||||||||||||||||||||||||||||||||||||||
| None | ||||||||||||||||||||||||||||||||||||||||||||||
| CC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| CC | IHC | GT | ||||||||||||||||||||||||||||||||||||||||||||
| IHC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| IHC + BRAF | GT | |||||||||||||||||||||||||||||||||||||||||||||
| MSI | GT | |||||||||||||||||||||||||||||||||||||||||||||
| MSI + IHC | GT | |||||||||||||||||||||||||||||||||||||||||||||
| MSI + IHC + BRAF | GT | |||||||||||||||||||||||||||||||||||||||||||||
| GT | ||||||||||||||||||||||||||||||||||||||||||||||
| Gallego et al., 201581 | USA, perspective not reported but appears to be US health-care system | CRC patients and their relatives | Cost-effectiveness and cost–utility of NGS in strategies to identify CRCP (including LS) | Decision model, decision tree, long-term estimates based on results from Mvundura et al.84 | Probands: standard care – IHC, BRAF, GT; intervention: NGS panels (for our purposes panel 1 = LS only); relatives: GT | Surveillance for patients and relatives | ||||||||||||||||||||||||||||||||||||||||
| Severin et al., 201582 | Germany, German statutory health insurance system | Newly diagnosed CRC patients and their FDRs | Cost-effectiveness of strategies to identify LS | Decision model, decision tree plus Markov model | (FH) IHC, MSI, GT (MSI if IHC abnormal, otherwise discharged); (FH) IHC, MSI, GT (MSI if IHC normal, otherwise progress directly to GT) | LS relatives: annual colonoscopy, aspirin chemoprevention; LS negative/unknown relatives: colonoscopy every 10 years between ages of 55 and 75 years | ||||||||||||||||||||||||||||||||||||||||
| Study | Outcomes measured | Discount rate (%) | Base-case results | Sensitivity analysis approach | Main sensitivity analysis results | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ramsey et al., 200383 | Life-years, costs, ICERs | 3 | Compared with no screening, all strategies were cost-effective ($50,000 threshold) apart from universal testing. ICERs: StrategyICER (US$/LYG)Bethesda/MSI11,865MSI394,067Bethesda441,172Universal2,553,345 |
Strategy | ICER (US$/LYG) | Bethesda/MSI | 11,865 | MSI | 394,067 | Bethesda | 441,172 | Universal | 2,553,345 | Univariate sensitivity analysis, PSA | Univariate analysis: Sensitive to survival benefit for increased surveillance in LS positives, specificity of family history/MSI, prevalence of LS in probands. PSA: Unclear which strategy was most cost-effective, but universal testing was the least cost-effective | ||||||||||||||||||||
| Strategy | ICER (US$/LYG) | ||||||||||||||||||||||||||||||||||
| Bethesda/MSI | 11,865 | ||||||||||||||||||||||||||||||||||
| MSI | 394,067 | ||||||||||||||||||||||||||||||||||
| Bethesda | 441,172 | ||||||||||||||||||||||||||||||||||
| Universal | 2,553,345 | ||||||||||||||||||||||||||||||||||
| Mvundura et al., 201084 | Life-years, costs, ICERs, QALYs | 3 | All strategies with preliminary tests were cost-effective relative to no testing. IHC followed by BRAF as preliminary tests was optimal (ICER US$22,552 per LYG) | Univariate sensitivity analysis, scenario analyses | Univariate analysis: Most sensitive to CRC risk among relatives, number of relatives per proband and compliance to surveillance. Scenario analyses: Using median laboratory prices increased costs and meant that age-targeted IHC followed by BRAF was the most cost-effective. Cascade testing reduced all ICERs relative to no testing. On average, using QALYs scaled ICERs by 1.18 LYGs per QALY | ||||||||||||||||||||||||||||||
| Ladabaum et al., 201185 | Life-years, costs, ICERs, cancer cases, cancer deaths | 3 | StrategyICER (US$/LYG)ICER (US$/LYG) excluding CC strategiesMMRpro/IHC30,600–Bethesda/IHC39,600–MMRpro41,400–Bethesda50,200–IHC (+BRAF)–36,200MSI + IHC (+BRAF)117,000108,000Universal GT293,000293,000 All other strategies dominated or extendedly dominated |
Strategy | ICER (US$/LYG) | ICER (US$/LYG) excluding CC strategies | MMRpro/IHC | 30,600 | – | Bethesda/IHC | 39,600 | – | MMRpro | 41,400 | – | Bethesda | 50,200 | – | IHC (+BRAF) | – | 36,200 | MSI + IHC (+BRAF) | 117,000 | 108,000 | Universal GT | 293,000 | 293,000 | Univariate sensitivity analysis, PSA, scenario analyses | Univariate analysis: Most sensitive to age of relative, effectiveness of LS surveillance for CRC and prevalence of LS. PSA: IHC (+BRAF) was the optimal strategy in 53% of iterations. Scenario analyses: Age limit for probands improves cost-effectiveness. Three to four relatives are needed for most strategies to be cost-effective compared with doing nothing | ||||||
| Strategy | ICER (US$/LYG) | ICER (US$/LYG) excluding CC strategies | |||||||||||||||||||||||||||||||||
| MMRpro/IHC | 30,600 | – | |||||||||||||||||||||||||||||||||
| Bethesda/IHC | 39,600 | – | |||||||||||||||||||||||||||||||||
| MMRpro | 41,400 | – | |||||||||||||||||||||||||||||||||
| Bethesda | 50,200 | – | |||||||||||||||||||||||||||||||||
| IHC (+BRAF) | – | 36,200 | |||||||||||||||||||||||||||||||||
| MSI + IHC (+BRAF) | 117,000 | 108,000 | |||||||||||||||||||||||||||||||||
| Universal GT | 293,000 | 293,000 | |||||||||||||||||||||||||||||||||
| Wang et al., 201286 | QALYs, costs, ICERs | 3 | StrategyICER (US$/QALY)ICER (US$/QALY) excluding CC strategiesMMRpro/IHC50,562–Bethesda/IHC65,347–MMRpro68,384–Bethesda82,864–IHC (+BRAF)–59,719MSI + IHC (+BRAF)193,343179,576Universal GT393,303271,219 All other strategies dominated or extendedly dominated |
Strategy | ICER (US$/QALY) | ICER (US$/QALY) excluding CC strategies | MMRpro/IHC | 50,562 | – | Bethesda/IHC | 65,347 | – | MMRpro | 68,384 | – | Bethesda | 82,864 | – | IHC (+BRAF) | – | 59,719 | MSI + IHC (+BRAF) | 193,343 | 179,576 | Universal GT | 393,303 | 271,219 | Univariate sensitivity analysis, PSA, scenario analyses | Univariate analysis: Results consistent with those of Ladabaum et al.85 PSA: IQRs narrow and within cost-effective ranges; ICERs had wide 95% CIs, reflecting wide distributions of utility estimates. Scenario analyses: Length of effect from disutility associated with GT or surveillance affected ICERs: if it was > 12 months the ICERs exponentially increased | ||||||
| Strategy | ICER (US$/QALY) | ICER (US$/QALY) excluding CC strategies | |||||||||||||||||||||||||||||||||
| MMRpro/IHC | 50,562 | – | |||||||||||||||||||||||||||||||||
| Bethesda/IHC | 65,347 | – | |||||||||||||||||||||||||||||||||
| MMRpro | 68,384 | – | |||||||||||||||||||||||||||||||||
| Bethesda | 82,864 | – | |||||||||||||||||||||||||||||||||
| IHC (+BRAF) | – | 59,719 | |||||||||||||||||||||||||||||||||
| MSI + IHC (+BRAF) | 193,343 | 179,576 | |||||||||||||||||||||||||||||||||
| Universal GT | 393,303 | 271,219 | |||||||||||||||||||||||||||||||||
| Gallego et al., 201478 | Life-years, QALYs, costs, ICERs | NR | Universal NGS testing vs. reference strategy US$196,000 per QALY gained; IHC/MSI followed by NGS US$71,000 per QALY gained | Univariate sensitivity analysis | Most influential parameters were the number of relatives tested, the prevalence of LS in CRC and the cost of CRC surveillance in relatives | ||||||||||||||||||||||||||||||
| Snowsill et al., 2014,4 Snowsill et al., 201579 | Life-years, QALYs, costs, ICERs, INHB | 3.5 | StrategyICER vs. no testing (£/QALY)ICER (£/QALY)No testing––ACII6021Extendedly dominatedIHC, GT6444DominatedIHC, BRAF, GT5831Extendedly dominatedMSI, GT5610DominatedMSI, BRAF, GT54915491MSI, BRAF, IHC, GT5774DominatedIHC, MSI, BRAF, GT760125,106Universal GT957182,962 | Strategy | ICER vs. no testing (£/QALY) | ICER (£/QALY) | No testing | – | – | ACII | 6021 | Extendedly dominated | IHC, GT | 6444 | Dominated | IHC, BRAF, GT | 5831 | Extendedly dominated | MSI, GT | 5610 | Dominated | MSI, BRAF, GT | 5491 | 5491 | MSI, BRAF, IHC, GT | 5774 | Dominated | IHC, MSI, BRAF, GT | 7601 | 25,106 | Universal GT | 9571 | 82,962 | Scenario analyses, univariate sensitivity analyses | Scenario analyses: (1) endometrial cancer excluded: cost-effectiveness of strategies improved; MSI BRAF GT strategy still most cost-effective at £4439 per QALY gained; (2) BRAF testing replaced by methylation testing: MSI GT strategy most cost-effective at £7965 per QALY gained (vs. MSI, BRAF, GT); (3) age limit increased: age 60 years, MSI BRAF GT strategy most cost-effective at £7681 per QALY gained; age 70 years, MSI BRAF GT strategy most cost-effective at £10,247 per QALY gained. Univariate analyses: Most sensitive parameters were disutilities for endometrial cancer and prophylactic TAH-BSO. Other sensitive parameters included the number of relatives, the prevalence of LS, the cost of colonoscopy complications, CRC incidence and the effectiveness of colonoscopy to prevent CRC |
| Strategy | ICER vs. no testing (£/QALY) | ICER (£/QALY) | |||||||||||||||||||||||||||||||||
| No testing | – | – | |||||||||||||||||||||||||||||||||
| ACII | 6021 | Extendedly dominated | |||||||||||||||||||||||||||||||||
| IHC, GT | 6444 | Dominated | |||||||||||||||||||||||||||||||||
| IHC, BRAF, GT | 5831 | Extendedly dominated | |||||||||||||||||||||||||||||||||
| MSI, GT | 5610 | Dominated | |||||||||||||||||||||||||||||||||
| MSI, BRAF, GT | 5491 | 5491 | |||||||||||||||||||||||||||||||||
| MSI, BRAF, IHC, GT | 5774 | Dominated | |||||||||||||||||||||||||||||||||
| IHC, MSI, BRAF, GT | 7601 | 25,106 | |||||||||||||||||||||||||||||||||
| Universal GT | 9571 | 82,962 | |||||||||||||||||||||||||||||||||
| Barzi et al., 201580 | Life-years, costs, ICERs | 3 | For scenario 3 (which included costs and benefits for probands), ignoring CC strategies, IHC followed by GT was the most cost-effective strategy: ICER ≈US$50,000 per LYG vs. no testing. Universal GT: ICER ≈US$131,000 per LYG vs. no testing, ≈US$943,000 per LYG vs. IHC followed by GT. All other tumour testing strategies dominated | Scenario analysis, univariate sensitivity analysis | Results were most sensitive to the cost of germ-line testing | ||||||||||||||||||||||||||||||
| Gallego et al., 201581 | Life-years, QALYs, costs, ICERs | 3 (benefits NR) | NGS vs. standard care US$144,235 per QALY gained | Scenario analyses, univariate sensitivity analyses | NR for panel 1 (the intervention of interest) | ||||||||||||||||||||||||||||||
| Severin et al., 201582 | Life-years, costs, ICERs | NR | StrategyICERs (€/LYG)No screeningCounselling including Bethesda, IHC, BRAF, sequencing77,268Counselling including IHC, BRAF, sequencing253,258Counselling including direct sequencing4,188,036 | Strategy | ICERs (€/LYG) | No screening | Counselling including Bethesda, IHC, BRAF, sequencing | 77,268 | Counselling including IHC, BRAF, sequencing | 253,258 | Counselling including direct sequencing | 4,188,036 | Scenario analyses, univariate sensitivity analyses, PSA | Scenario analyses: Uptake of testing by FDRs was influential on the ICER, aspirin chemoprevention had a small impact on cost-effectiveness. Univariate analysis: Sensitive to number of relatives and prevalence of LS. PSA: At a threshold of €50,000 per LYG, the no-screening strategy had an 87% chance of being considered cost-effective | |||||||||||||||||||||
| Strategy | ICERs (€/LYG) | ||||||||||||||||||||||||||||||||||
| No screening | |||||||||||||||||||||||||||||||||||
| Counselling including Bethesda, IHC, BRAF, sequencing | 77,268 | ||||||||||||||||||||||||||||||||||
| Counselling including IHC, BRAF, sequencing | 253,258 | ||||||||||||||||||||||||||||||||||
| Counselling including direct sequencing | 4,188,036 | ||||||||||||||||||||||||||||||||||
The majority of the studies (seven78,80,81,83–86 out of nine) reported results for a US population. Of the other two studies, one was carried out from the perspective of a German population82 and one was UK based. 4,79
All nine studies included strategies to identify LS in index CRC patients (probands) and relatives of CRC patients. Five studies4,79,81,85,86 identified relatives through cascade testing in their base-case analyses, whereas four studies80,82–84 appeared to include only first-degree relatives of the index CRC patient in their base-cases analyses. The method of identifying relatives was unclear in the remaining study. 78
When reported, the modelling approach appears to be similar across studies. Models were split into two sections: a diagnostic model to identify LS (often a decision tree) and a long-term model to estimate the costs and benefits associated with the outcomes of the diagnostic model.
All studies included either IHC- or MSI-based diagnostic strategies and a universal testing strategy as a comparator. Six studies4,79,80,82,84–86 included both IHC- and MSI-based strategies, allowing for a comparison between the two interventions. The optimal strategy varied across these studies and depended greatly on the willingness-to-pay threshold of the relevant country, or the main relevant comparator, and so no strategy was consistently seen to be the most cost-effective.
Lynch syndrome status was confirmed with germ-line testing of MLH1, MSH2, MSH6 and PMS2 in all studies. Gallego et al. 81 considered the use of NGS. However, instead of comparing the cost-effectiveness of strategies to diagnose LS, Gallego et al. 81 investigated the cost-effectiveness of using NGS to identify a wider range of hereditary CRC predispositions, which included LS. Therefore, this analysis did not include a strategy without testing.
All studies modelled the benefit of colonoscopic surveillance for LS-positive relatives, with most (six studies4,79–81,83,85,86) also explicitly offering the same surveillance to probands. The frequency of this surveillance was different across studies, but was usually modelled on an annual or biannual interval, based on recommended guidelines.
Four studies4,79,80,85,86 also included prevention for gynaecological cancers [prophylactic hysterectomy and bilateral salpingo-oophorectomy (H-BSO) or gynaecological screening]. Barzi et al. ,80 Ladabaum et al. 85 and Wang et al. 86 all included both endometrial cancer and ovarian cancer, whereas Snowsill et al. 4,79 included only endometrial cancer. The studies by Ladabaum et al. 85 and Wang et al. 86 were the only studies to include gynaecological screening and, in their base cases, only a cost was applied for this screening; no benefit was assumed.
One study considered the use of aspirin prevention. 82 Severin et al. 82 used data from the CAPP2 study to apply a hazard ratio (HR) of 63% for a maximum of 11 years (observation time of the CAPP2 study) to reduce the incidence of CRC in patients taking daily aspirin. We discuss this trial in more detail in Chapter 5 (see Chemoprevention). Both the cost of treatment with aspirin and the cost of complications from aspirin were included, with a total annual cost of €596 (2012 costs).
Of the nine cost-effectiveness studies, five were cost–utility studies. 4,78,79,81,84,86 These are the five studies that we focus on for the rest of the review.
Mvundura et al.84
Mvundura et al. 84 produced a decision model of newly diagnosed CRC patients and their relatives in the USA. It compared IHC (with or without BRAF V600E testing), MSI testing, no testing and universal genetic testing. Relatives were identified through genetic testing. Although the study did include both MSI and IHC testing strategies, it did not include all of the interventions identified in the NICE scope. 88
Relatives with a confirmed mutation were offered colonoscopy every 1–2 years from the age of 20–25 years until the age of 79 years. LS-negative relatives and some of those who were LS positive and who declined the intensive LS surveillance were offered colonoscopy every 10 years from the age of 50 years.
The analysis did not include LS-related cancers other than CRC or the differences in CRC incidence between males and females. It also did not include all of the comparators identified in the NICE scope. 88
In their base case Mvundura et al. 84 reported only life-years gained (LYGs) as opposed to QALYs. All strategies with preliminary tests were found to be cost-effective compared with no testing. IHC followed by BRAF V600E testing was the optimal strategy, with an incremental cost-effectiveness ratio (ICER) of US$22,552 per LYG. To turn their analysis into a cost–utility analysis, the ICERs were scaled by a factor of 1.18 LYGs per QALY. This did not impact on the order of the strategies and increased the ICERs for all strategies. This approach required a number of assumptions to be made and therefore it is unlikely to reflect the true cost per QALY gained for each testing strategy.
The model results were sensitive to CRC risk in relatives, the number of relatives and adherence to surveillance.
Wang et al.86
Wang et al. 86 provided an update of the study by Ladabaum et al. ,85 looking at the cost–utility of strategies to identify LS in newly diagnosed CRC patients and their relatives in the USA. The difference between the two models was the inclusion of utilities in the update model.
Wang et al. 86 included strategies that started with clinical criteria (ACII and revised Bethesda or prediction models) as well as those that began with tumour-based testing (IHC and/or MSI). We do not focus on the strategies that began with clinical criteria as these are not considered as part of our analysis. As with the study by Mvundura et al. ,84 not all interventions identified by the NICE scope88 were included.
The model used a decision tree with Markov subtrees to model newly diagnosed CRC patients and their relatives. For people identified with a LS mutation or with an unconfirmed diagnosis of LS (plus their first-degree relatives), annual colonoscopy (from the age of 25 years) was offered. Women were offered gynaecological screening from the age of 35 years and prophylactic total abdominal hysterectomy and bilateral salpingo-oophorectomy (TAH-BSO) from the age of 40 years. When LS was not diagnosed, people were offered 10-yearly colonoscopy from the age of 50 years.
The strategy that used IHC (with or without BRAF V600E mutation testing) was found to be the most cost-effective strategy, with an ICER of US$59,719 per QALY gained. With the exception of MSI in combination with IHC (with or without BRAF V600E testing) and universal germ-line testing (both of which had ICERs of > US$100,000 compared with the next most cost-effective strategy), all other strategies were dominated or extendedly dominated.
The results were most sensitive to the age of relatives, the effectiveness of LS surveillance for CRC and the prevalence of LS.
Gallego et al.78
Gallego et al. 78 examined the cost-effectiveness and cost–utility of strategies to identify LS in CRC patients and their relatives. Details of the modelling are scarce as this study was reported only in abstract form. The main comparison appears to look at targeted sequencing compared with NGS. It was unclear whether this study used the same underlying model as that reported in the study by Gallego et al. ,81 but the author list suggests that the two studies are likely to be related. For relatives in whom LS was detected, CRC surveillance was offered, although, again, details are not provided. The reference strategy was a combination of IHC and MSI followed by targeted sequencing. Replacing targeted sequencing with NGS gave an ICER of US$71,000 per QALY gained and a strategy of NGS for all patients compared with the reference strategy gave an ICER of US$196,000 per QALY gained.
As with most other analyses, the most influential parameters included the number of relatives tested, the prevalence of LS in CRC and the cost of CRC surveillance in relatives.
Snowsill et al.4,79
Snowsill et al. 4,79 report the previous work by PenTAG, which assessed the cost-effectiveness of strategies to diagnose LS in newly diagnosed CRC patients aged < 50 years and their relatives. Snowsill et al. 4,79 presented a cost–utility analysis from the perspective of the NHS and Personal Social Services (PSS) and included both CRC and endometrial cancer as outcomes. They stated that, when possible, they adhered to the NICE reference case. 89 The model included a decision tree diagnostic section and an individual sampling model to calculate long-term outcomes and survival for each of the diagnoses from the diagnostic section. Probands and relatives who received a confirmed (mutation-positive) diagnosis or assumed LS diagnosis (probands and relatives who were offered but who declined germ-line testing and first-degree relatives of probands who were assumed to have LS) were offered colonoscopic surveillance, with women also offered prophylactic TAH-BSO at the age of 40 years.
In this analysis, all testing strategies had ICERs of < £10,000 per QALY gained. When compared incrementally, MSI followed by BRAF V600E testing was the most cost-effective strategy at a willingness-to-pay threshold of £20,000. Scenario analyses demonstrated that increasing the age of the population increased the ICERs for each strategy compared with no testing, although MSI plus BRAF V600E testing remained the most cost-effective strategy. The model appeared most sensitive to the disutilities for endometrial cancer and prophylactic TAH-BSO. Other sensitive parameters included the number of relatives, the prevalence of LS, the cost of colonoscopy complications, CRC incidence and the effectiveness of colonoscopy to prevent CRC.
Although we believe that this previous work was the first UK-based model to assess the cost–utility of strategies to diagnose LS and has been described by Ladabaum et al. 3 as the ‘most comprehensive decision model’, this model still does not fully answer the decision problem set out in this assessment. The strategies included in the study by Snowsill et al. 4,79 are not in line with the current scope,88 with several strategies including a combination of MSI and IHC testing, and methylation testing included only as a scenario analysis, in place of BRAF V600E testing. The parameters are also now several years old and were chosen for CRC patients aged < 50 years and so they are unlikely to represent the overall population and the current clinical experience.
Gallego et al.81
Gallego et al. 81 presented a cost–utility analysis of CRC patients and their relatives from a US perspective. They used a decision tree model with long-term outcome estimates based on the results presented by Mvundura et al. 84 As such, the concerns that we have over the cost–utility analysis by Mvundura et al. 84 are equally relevant here.
In this study the strategies were IHC followed by BRAF V600E testing prior to germ-line testing. The intervention strategies were NGS panels. The relevant intervention strategy for our analysis was panel 1, which looked at LS only. CRC surveillance was offered to both probands and relatives, although details of this surveillance were not provided. As the study by Mvundura et al. 84 was stated to be the source of the long-term outcomes, we assumed that surveillance was modelled as in this study.
Next-generation sequencing for LS compared with standard care gave an ICER of US$144,235 per QALY gained. Sensitivity analyses were not reported for this scenario, as it was not the base case.
Quality of the identified cost-effectiveness studies
The results of the quality appraisal of the included studies are provided in Tables 24 and 25.
In general, reporting ranged from mixed to quite good. The study by Snowsill et al. 4,79 appeared to be the most comprehensively reported study. Very few studies considered endometrial cancer and only the studies by Barzi et al. ,80 Wang et al. 86 and Ladabaum et al. 85 considered ovarian cancer.
| Study | Criteria | ||||||
|---|---|---|---|---|---|---|---|
| The viewpoint(s) of the analysis are clearly stated and justified | The source(s) of the effectiveness estimates are stated | Methods for the estimation of quantities and unit costs are described | Currency and price date are recorded | Details of any models used are given | Time horizon of costs and benefits is stated | The ranges over which the variables are varied are justified | |
| Ramsey et al., 200383 | ✗ | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ |
| Mvundura et al., 201084 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Ladabaum et al., 201185 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Wang et al., 201286 | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Snowsill et al., 2014, 20154,79 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Barzi et al., 201580 | ✓ | ✓ | ✗ | ✗ | ✓ | ✓ | ✗ |
| Gallego et al., 201581 | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ |
| Severin et al., 201582 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Criteria | Ramsey 200383 | Mvundura 201084 | Ladabaum 201185 | Wang 201286 | Snowsill 2014, 20154,79 | Barzi 201580 | Gallego 201581 | Severin 201582 |
|---|---|---|---|---|---|---|---|---|
| Diagnosis | ||||||||
| Patients are tested for MLH1 mutations | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Patients are tested for MSH2 mutations | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Patients are tested for MSH6 mutations | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Patients are tested for PMS2 mutations | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Appropriate informed consent and counselling is included | ✓ | ✓ | ✓ | ✓ | ✓ | ? | ✓ | ✓ |
| The study considers patients declining counselling | ? | ✓ | ? | ? | ✓ | ? | ✓ | ? |
| The study considers patients declining genetic testing | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| The effect of diagnostic errors is considered | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| The study considers the impact of a national strategy on the proportion of patients who do not already know their LS status | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ | ✗ |
| Management | ||||||||
| The study considers CRC | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| The study considers endometrial cancer | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ |
| The study considers ovarian cancer | ✗ | ✗ | ✓ | ✓ | ✗ | ✓ | ✗ | ✗ |
| The study considers other LS-associated cancers | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ |
| The study considers interactions of cancers appropriately | NA | NA | ? | ? | ✗ | ? | NA | NA |
| Colonoscopic surveillance in the study is explicitly justified (e.g. by reference to guidelines or clinical practice) | ✓ | ✓ | ✗ | ✗ | ✓ | ✓ | ✗ | ✓ |
| The study considers patients declining recommended surveillance | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ? | ✓ |
| The study considers the difference in incidence of CRC between males and females | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ |
| The study considers varying incidence of LS-associated cancers between mutations of different MMR genes | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ |
| The study accounts for the improved survival of LS CRCs relative to sporadic CRCs | ? | ✓ | ✓ | ✓ | ✓ | ✓ | ? | ✓ |
| The study considers the potential psychological impact of genetic testing | ✗ | ✓ | ✗ | ✓ | ✓ | ✗ | ✗ | ✗ |
Discussion
The studies identified in this review reported a wide variety of analyses, with varying quality in reporting. No single study answered our decision problem in full and the most common reason for this was that studies did not include all of the interventions identified by the NICE scope88 or they were not carried out from a UK perspective and therefore were hard to generalise.
It is difficult to draw specific conclusions about which was the most cost-effective strategy, as this varied across studies and depended greatly on the willingness-to-pay threshold applied. Most studies stated that at least one strategy to identify LS could be cost-effective according to the perspective used and, when a universal genetic testing strategy was included, strategies that used tumour-based tests to enrich the population appeared to improve cost-effectiveness (reducing the ICERs). Most studies agreed that the effectiveness of colonoscopy screening, the number of relatives and the prevalence of LS had most impact on the cost-effectiveness of the models.
Conclusions
The economic analysis that came closest to answering the current decision problem was that by Snowsill et al. 4 However, this analysis requires updating to fully answer the current problem posed in the NICE scope. 88 Therefore, our approach was to further adapt and develop the model created by Snowsill et al. 4 to suit the current decision problem.
Chapter 5 Independent economic assessment
Methods
The economic assessment reported in this chapter builds on the model described in Snowsill et al. 4,79 Where the current model has not changed, reference is made to these articles, with direct quotations provided, and therefore these aspects are described here only briefly for clarity. Focus instead is given to alterations to the model.
Population
The base-case population specified by the NICE scope includes all newly diagnosed CRC patients (probands) and their biological relatives (henceforth referred to as ‘relatives’). This is a broader population than that previously specified in the study by Snowsill et al. ,4 which considered only CRC patients aged < 50 years and their relatives.
Four subgroups based on age (< 50, < 60, < 70 and ≥ 70 years) were also specified in the scope and are presented in this report as subgroup analyses, separate from the main base-case analysis. Parameters that have been identified as age dependent are reported as such and include the number of probands, the proportions of probands and relatives who are men and the prevalence of LS in the population. When data are unavailable for age subgroups, the base-case values are used.
Number of probands
To estimate the number of probands we used the same approach as in the study by Snowsill et al. ,4 that is, taking the number of probands from the most recent Office for National Statistics (ONS) cancer registration statistics for England. 21 The figures for the overall population and the age subgroups are provided in Table 26.
| Age subgroup (years) | Number of CRC registrations | Proportion of mena (%) |
|---|---|---|
| Base case (no age limit) | 34,025 | 55 |
| < 50 | 2107 | 52 |
| < 60 | 5880 | 55 |
| < 70 | 13,823 | 58 |
| ≥ 70 | 20,202 | 53 |
Number of relatives
The method for estimating the number of relatives was discussed in detail in Snowsill et al. 4 In summary, the number of relatives included in the model represents the possible group of identifiable relatives. For example, for probands in whom a LS mutation is identified, this is the possible number of relatives who both are contactable and may be identified through cascade testing.
The number of relatives in the study by Snowsill et al. 4 was set to five, based on previous data from Barrow et al. 90 (2.35 relatives per proband) and unpublished data provided by Ian M Frayling (Institute of Cancer and Genetics, University Hospital of Wales, Cardiff, UK, 18 December 2012, personal communication). Data recently collected from the Manchester Regional Lynch Syndrome Registry, as part of a PhD thesis investigating the diagnosis of LS,91 suggested an average of 9.95 relatives per index patient. As such, the base case number of relatives in the current model was increased slightly to six to better account for this new information. As in the study by Snowsill et al. ,4 this number was varied from 0 to 12 in univariate sensitivity analysis, to acknowledge the wide variation among the data sources. The number of relatives per proband was assumed not to alter with the age of the proband, given a paucity of evidence to demonstrate otherwise.
The Manchester Regional Lynch Syndrome Registry estimated an average of eight first-degree relatives per index patient,91 but this seems exceptionally high and so we have assumed that this figure actually represents the results of cascade testing to identify index patients. As such, our estimated number of first-degree relatives remained as 42% of all relatives (2.5 relatives per proband), which is based on a combination of published92,93 and unpublished (Ian M Frayling, personal communication) data, as reported in Snowsill et al. 4
The proportion of relatives who are male was taken from pooled analysis of unpublished data (Ian M Frayling, personal communication; Munaza Ahmed, Wessex Clinical Genetics Service, University Hospital Southampton NHS Foundation Trust, 23 January 2013, personal communication), giving a value of 38%, as used previously in the analysis reported in Snowsill et al. 4
Prevalence of Lynch syndrome
The prevalence of LS among CRC probands was estimated from the study by Hampel et al. ,92 as in the study by Snowsill et al. 4 The base-case value (2.8%) and the age subgroup values are presented in Table 27.
| Age subgroup (years) | Prevalence of LS in CRC population (%) |
|---|---|
| Base case (no age limit) | 2.8 |
| < 50 | 8.4 |
| < 60 | 5.7 |
| < 70 | 3.8 |
| ≥70 | 1.1 |
In the study by Snowsill et al. 4 it was important to subdivide the proportion of the population with LS by mutation, as several parameters relied on this information, including the sensitivity and specificity of MSI and IHC. In the current analysis, the test accuracy was assumed to apply for all mutations, as new test accuracy evidence is not available to predict the effect that these mutations may have. This is a result of restricting the test accuracy studies to those that answer the decision problem and not including studies with high-risk input populations. Mutation type does still affect some costs and therefore is still included in the model.
As in the study by Snowsill et al. ,4 the proportions for each LS mutation were taken from the supplementary evidence in the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) review,42 which stated that, for all true LS-positive patients, 32% have a MLH1 mutation, 39% have a MSH2 mutation, 14% have a MSH6 mutation and 15% have a PMS2 mutation (Table 28). Reported family registry data can differ significantly from these values, particularly with respect to the proportion of PMS2 mutations identified91,94,95 (see Table 28). This may potentially be because of PMS2 testing having historically occurred less in current practice than in the trials on which the EGAPP review based its values. Therefore, if systematic testing were more common in UK practice, these values might change. We explore the possibility of the proportion of each mutation being closer in value to the reported registry data in a sensitivity analysis, using the Manchester Regional Lynch Syndrome Registry data as these are UK-based data. 91
| Source | Gene | ||||
|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | PMS2 | Notes | |
| EGAPP review supplementary evidence42 | 32% | 39% | 14% | 15% | Based on trial data |
| Sjursen et al.94 | 18% | 50% | 26% | 6% | Registry data from Norway (up to 2009) |
| Sjursen et al.95 | 36% | 44% | 17% | 2% | Registry data from New South Wales, Australia (up to 2010) |
| Barrow91 | 40% | 46% | 11% | 2% | Registry data from Manchester, UK (up to 2013) |
For relatives of probands with LS, the proportion of relatives expected to test positive was estimated using a meta-analysis of studies (Table 29), as reported in Snowsill et al. ;4 this gave a value of 44%. This value falls below 50% for a number of reasons (Box 2).
| Study | Proportion (%) | 95% CI (%) |
|---|---|---|
| Jenkins 200693 | 44.1 | 37.0 to 51.5 |
| Hampel 200892 | 43.8 | 37.7 to 50.0 |
| Ian M Frayling (personal communication) | 40.4 | 31.5 to 49.7 |
| Munaza Ahmed (personal communication) | 45.5 | 40.1 to 50.9 |
| Random-effects meta-analysis | 44.0 | 40.7 to 47.4 |
-
De novo mutations can occur, which mean that no relatives of the index case will have the mutation [in the study by Jenkins and colleagues (2006),93 1 of 18 probands had a de novo mutation].
-
Non-paternity can occur.
-
Mortality bias can occur, meaning that mutation carriers are more likely to have died before being able to receive predictive testing.
Reproduced from Snowsill et al. (p. 143)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
Age on entry
In the base case there was no age threshold for CRC patients for screening for LS.
The age on entry for probands was based on the age distribution of CRC diagnoses. For probands without LS, the age distribution was estimated from cancer registration statistics in England from 2006 to 2014. 21,96–103 These statistics are grouped into 5-year age groups. It was assumed that registrations were uniform within these age groups.
For probands with LS, the age distribution was estimated from the parametric CRC incidence function (see Incidence rates for individuals with Lynch syndrome).
The cumulative registrations were used to estimate an empirical cumulative incidence function. The cumulative incidence function was truncated as appropriate for the different age subgroups. Figure 7 shows the age distribution of probands in the base case.
FIGURE 7.
Age distribution of simulated probands.

For relatives without LS, the age distribution was assumed to be equal to the age distribution of the general population, which was taken from the mid-2014 population estimates for England104 and mid-2014 population estimates of the very old (including centenarians) for the UK. 105
For relatives with LS, the age distribution was estimated by multiplying the population estimate for the general population by an estimate of the CRC mortality-free survival for individuals with LS (i.e. incorporating the raised incidence of CRC in relatives with LS).
As previously,4 the age distribution of relatives was right-truncated at 75 years as it is unlikely that any intervention would be offered to individuals aged > 75 years and was left-truncated at 18 years as few relatives are offered predictive testing before the age of 18 years. Figure 8 shows the age distribution of relatives in the base case.
FIGURE 8.
Age distribution of simulated relatives.
Model structure
The model structure remains broadly similar to that reported in the study by Snowsill et al. 4,79 The model consists of two distinct sections: a decision tree model to investigate the short-term outcomes of strategies to identify LS patients and an individual patient simulation model to assess the long-term implications of strategies to identify and manage LS. The model was built in Microsoft Excel® (Microsoft Corporation, Redmond, WA, USA). A brief summary of the model structure and parameters is provided in the following sections, with detailed discussion of any changes made to the model.
Diagnostic testing model
The section of the model that calculates diagnoses was built as a decision tree with no time component included. As in the study by Snowsill et al. ,4 this assumes that the diagnosis of LS in probands and relatives occurs instantaneously, although in reality this may take up to several months or years. As before, it was also assumed that probands’ treatment will not be influenced by a LS diagnosis, following the assumption that the results of LS testing will be unavailable prior to treatment. In most cases this treatment will be surgical resection with the possibility of chemotherapy or radiotherapy, depending on the stage of the cancer.
An overview of the diagnostic model is given in Figure 9. This is unchanged from that in Snowsill et al. 4
Diagnostic strategies for probands
The NICE scope88 for this project specifies diagnostic strategies for probands that are significantly altered from those reported in Snowsill et al. 4
The 10 new strategies considered are:
-
No systematic testing to identify LS (all probands assumed not to have LS).
-
IHC four-panel test for MLH1, MSH2, MSH6 and PMS2 followed by genetic testing if IHC results are abnormal.
-
IHC four-panel test followed by BRAF V600E testing for abnormal MLH1 results. Genetic testing is carried out for any other (not MLH1) abnormal IHC results or after a negative BRAF V600E test.
-
IHC four-panel test followed by MLH1 promoter methylation testing for abnormal MLH1 results. Genetic testing is carried out for any other (not MLH1) abnormal IHC results or after a negative MLH1 promoter methylation test.
-
IHC four-panel test followed by BRAF V600E testing for abnormal MLH1 results. A negative BRAF V600E test is followed by MLH1 promoter methylation testing. Genetic testing is carried out for any other (not MLH1) abnormal IHC results or after a negative MLH1 promoter methylation test.
-
MSI test followed by genetic testing for MSI-H result.
-
MSI test followed by BRAF V600E testing for MSI-H result. Genetic testing is carried out after a negative BRAF V600E test.
-
MSI test followed by MLH1 promoter methylation testing for MSI-H result. Genetic testing is carried after a negative MLH1 promoter methylation test.
-
MSI test followed by BRAF V600E testing for MSI-H result. A negative BRAF V600E test is followed by MLH1 promoter methylation testing. Genetic testing is carried after a negative MLH1 promoter methylation test.
-
Universal genetic testing (i.e. as the first and only test for all probands).
These strategies are presented in Figures 10–13.
FIGURE 10.
Proband diagnostic strategy: no testing.

FIGURE 11.
Immunohistochemistry-based diagnostic strategies for probands. GC, genetic counselling; GT, genetic testing.
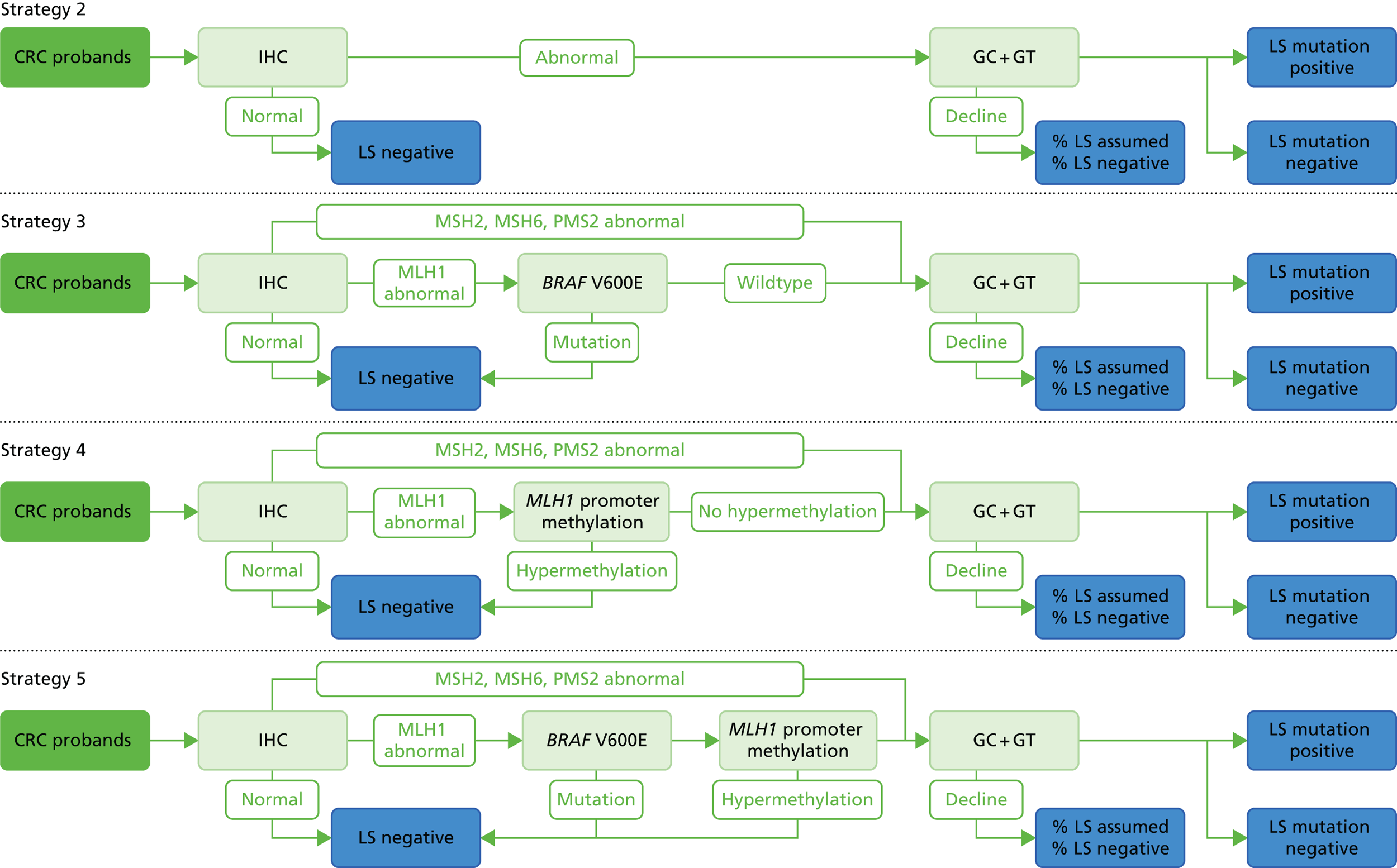
FIGURE 12.
Microsatellite instability-based diagnostic strategies for probands. GC, genetic counselling; GT, genetic testing.
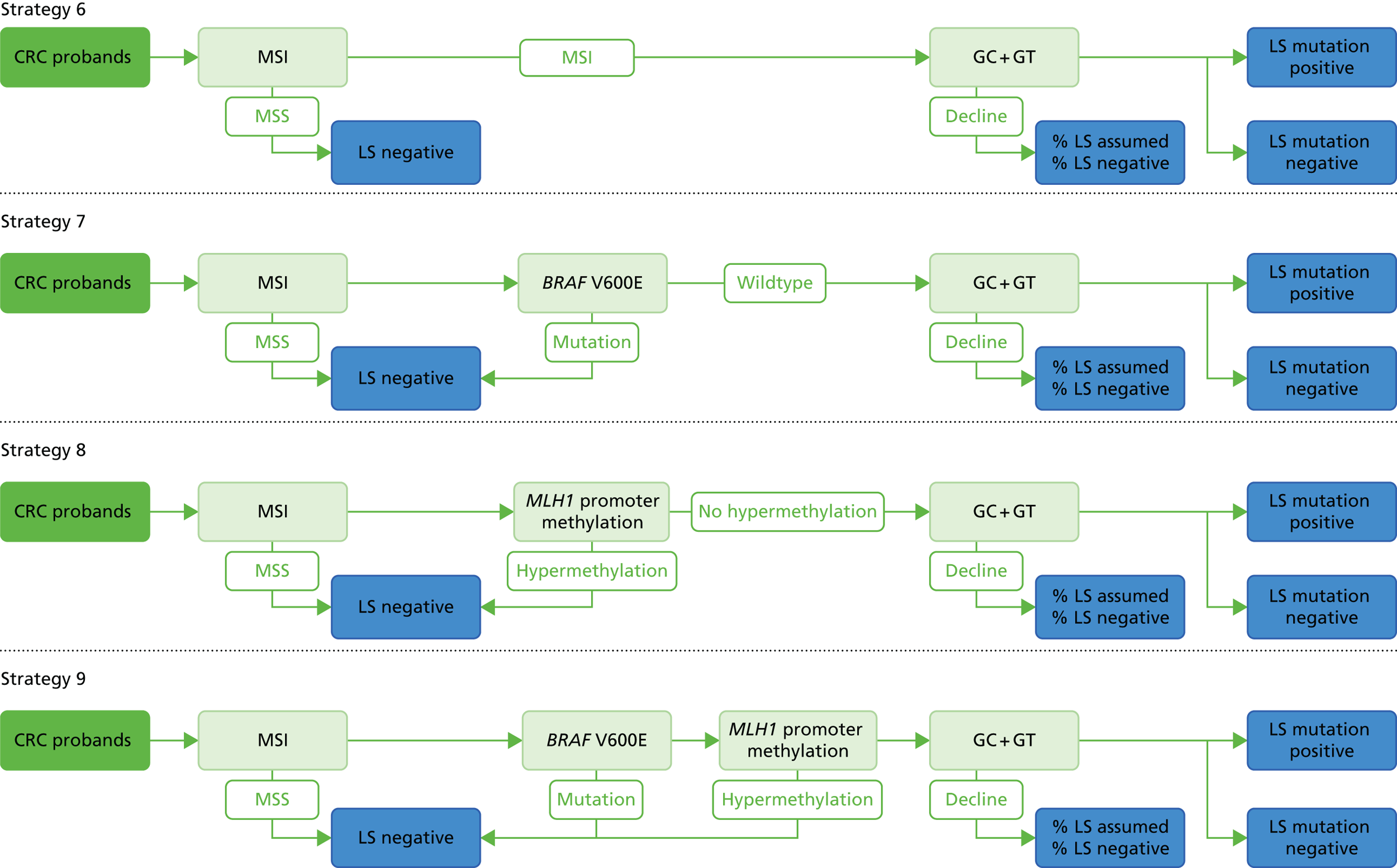
FIGURE 13.
Universal genetic testing strategy for probands. GC, genetic counselling; GT, genetic testing.
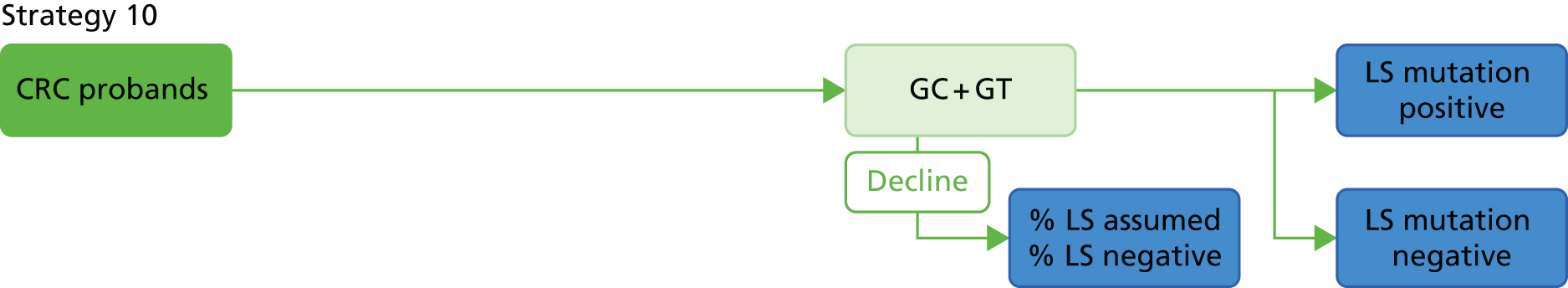
Strategies 5 and 9 include both BRAF V600E and MLH1 promoter methylation testing. These are performed in sequence with either test able to rule out LS, for example a patient with BRAF V600E would not receive further testing. The ordering of these tests was informed by discussion at the NICE scoping workshop (11 January 2016).
In clinical practice it is expected that a number of patients with stage II CRC will undergo MSI or IHC analysis to inform treatment options. To highlight this, the model includes costs for MSI or IHC testing for all stage II CRC patients (≈27% of probands) in strategy 1 (no testing), but assumes that this does not lead to any further testing for LS.
For the purposes of the base case it was assumed that a MSI result corresponds to MSI-H. This assumption was explored in a scenario analysis, in which it was assumed that a MSI result corresponds to MSI-L; this is detailed further in Model parameters.
A certain proportion of probands in the MSI strategies were assumed to receive IHC to help interpret their genetic test results. For example, a variant of uncertain significance may be identified in MSH2 and IHC was conducted to identify whether the tumour cells were MSH2 deficient (indicating that the variant is likely to be pathogenic). Clinical opinion appears to be that this proportion can be quite low [Ottie O’Brien, Northern Molecular Genetics Service, personal communication, 7 April 2016 (value of < 5%); Samantha Butler, West Midlands Regional Genetics Laboratory, personal communication, 13 July 2016 (value of 10%)], so the value was set to 5% in the base case. The impact of this parameter was explored in univariate sensitivity analysis, with the proportion varied from 0% to 10%.
The potential results of the testing strategies for patients are as reported previously:4 LS mutation positive, LS assumed and LS negative. LS mutation positive occurs when a proband receives a positive genetic test result and LS negative occurs when a proband is ruled out by one of the tests in the strategy.
In the study by Snowsill et al. ,4 the outcome of LS assumed could occur when genetic testing either was uninformative or was simply not carried out (the proband declined testing). A negative genetic test result was assumed to be uninformative for probands, meaning that, although the test did not detect LS, it did not rule it out. The Amsterdam II criteria were used as an additional test for those probands who were LS mutation negative and those who declined genetic testing to decide whether they were LS assumed. In this update to the model, any use of the Amsterdam II criteria was removed, in line with the focus of the decision problem, which does not include clinical criteria. It was also assumed in the current model that only probands who decline testing can become LS assumed, and this was set to 10% for all patients [lower than the average proportion of probands who were LS assumed after declining testing in the 2014 model (21%)]. We adjusted this number for each age subgroup using the three age limit scenarios from Snowsill et al. 4 (21% for probands aged < 50 years, 17% for probands aged < 60 years, 13% for probands aged < 70 years) and set the value for the ≥ 70 years subgroup to a significantly lower value (5%) to reflect that, in higher age groups, few people are likely to be diagnosed with LS without genetic testing.
Unlike in the study by Snowsill et al. ,4 in which testing for PMS2 was modelled only for probands who were negative for MLH1, MSH2 and MSH6 but who had a family history that was indicative of LS, in the current model it was assumed that all patients who accept genetic testing will receive testing for all four known genes (MLH1, MSH2, MSH6 and PMS2). The exception to this was that probands who follow strategies that use IHC followed by either BRAF V600E or MLH1 promoter methylation testing receive only MLH1 and PMS2 germ-line testing. Mutations related to EPCAM were assumed to be identified through testing for MSH2. This is believed to be in line with current clinical practice; when clinical practice differs from this, the model will overestimate costs associated with tumour-based testing strategies.
As in the study by Snowsill et al. ,4 only a proportion of patients diagnosed as LS positive or LS assumed accept an offer of LS surveillance.
Testing outcomes for probands
The primary outputs from the short-term model for each testing strategy that lead into the survival (i.e. long-term) section of the model are the same as those reported in Snowsill et al. 4 (Box 3).
-
Number of probands with LS receiving LS surveillance.
-
Number of probands with LS not receiving LS surveillance (probands will receive some surveillance in line with BSG guidelines;46 these are split into those identified as LS positive, but declined surveillance and those who were diagnosed LS negative.
-
Number of probands without LS receiving LS surveillance.
-
Number of probands without LS who do not receive LS surveillance (probands will receive some surveillance in line with BSG guidelines46); these are split into those identified as LS positive, but declined surveillance and those who were diagnosed LS negative.
Other outcomes include overall sensitivity and specificity of each diagnostic strategy.
Reproduced from Snowsill et al. (p. 98)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
Probands who are diagnosed with LS (either mutation positive or assumed) but who refuse surveillance may still be offered prophylactic surgery for mCRC or endometrial cancer.
Diagnostic strategies for relatives
The diagnostic strategies for relatives are unchanged from those in the study by Snowsill et al. ;4 these strategies are summarised in Figures 14–16. As with probands, relatives could decline any testing or LS surveillance offered to them.
Testing outcomes for relatives
The testing outcomes for relatives are as stated in the study by Snowsill et al. 4 (Box 4).
The primary short term model outputs are:
-
number of relatives with LS receiving LS surveillance
-
number of relatives with LS not receiving LS surveillance (split into those identified as LS positive, but declined surveillance and those who were diagnosed LS negative)
-
number of relatives without LS receiving LS surveillance
-
number of relatives without LS who do not receive surveillance (split into those identified as LS positive, but declined surveillance and those who were diagnosed LS negative)
The sensitivity and specificity for each testing strategy for relatives is recorded.
Reproduced from Snowsill et al. (p. 100)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
Long-term outcomes model
As described previously,4 long-term outcomes were modelled for all probands and relatives regardless of the diagnostic path that they follow. An individual patient sampling model was used to simulate 240,000 patients, distributed across 24 groups, representing all combinations of the variables shown in Table 30. An individual patient sampling model was justified because no patient interactions were modelled but there were too many patient states for a cohort model.
| Variable | Values |
|---|---|
| Patient type | Proband, relative |
| Actually has LS | Yes, no |
| LS diagnosis and management | Diagnosed and surveillance colonoscopy accepted, diagnosed and surveillance colonoscopy not accepted, not diagnosed |
| Sex | Male, female |
Mean long-term outcomes were then estimated for each of the 24 groups. These were then used to estimate the long-term outcomes for each of the diagnostic strategies in the decision tree.
Patients were simulated for 1 year at a time. Each patient starts each year in a particular state, which determines the events that can occur during that year. The events in turn determine the costs incurred and the state of the patient in the next year if the patient is still alive. The hazard rate for events (except elective events such as surveillance colonoscopy) was assumed to be constant during each year but could change between years.
Life-years and QALYs were calculated based on a patient’s state at the beginning of the year and any events occurring during the year, for example if a patient starts the year without cancer and develops cancer after 3 months, then the patient accrues 3 months of life with a non-cancer utility and 9 months of life with the utility decrement from cancer (assuming that the patient does die within the year).
Table 31 shows the different events included in the model. Mortality events are competing – no other events can occur after the patient dies.
| Patient group | Competing events | Non-competing events |
|---|---|---|
| All patients | General mortality | |
| Patients undergoing LS surveillance (aged 25–75 years) | Mortality following colonoscopy | Colonoscopy, adverse events (including bleeding and perforation) following colonoscopy |
| Patients with CRC (aged < 75 years) | Mortality following colonoscopy | Colonoscopy, adverse events (including bleeding and perforation) following colonoscopy |
| Patients with CRC | CRC mortality | |
| Patients with an index CRC (without mCRC) | mCRC incidence | |
| Patients without CRC | CRC incidence | |
| Women with LS without endometrial cancer | Endometrial cancer incidence | |
| Women with LS with endometrial cancer | Endometrial cancer mortality | |
| Women diagnosed with LS without endometrial cancer and without H-BSO | Mortality following prophylactic H-BSO | Commence gynaecological surveillance, stop gynaecological surveillance, decline risk reduction for gynaecological cancer, prophylactic H-BSO |
Figure 17 presents a model diagram for the long-term outcomes model, indicating the mortality events and cancer incidence events, as well as prophylactic H-BSO. Gynaecological surveillance is indicated to reduce the risk of endometrial cancer mortality, but this is only if the individual is receiving surveillance prior to cancer incidence.
FIGURE 17.
Simplified model diagram for the long-term outcomes model. EC, endometrial cancer.

Patient state
The state of each simulated individual at any time is defined by a number of properties, which collectively provide all of the information necessary to select appropriate treatment pathways and calculate risks of events. The patient state is composed of:
-
whether or not the patient is alive
-
the patient’s age (at the start of the year)
-
the patient’s sex
-
the patient’s bowel state (defined in the following section)
-
patient’s gynaecological state [see Patient’s gynaecological state (women only)]
-
the patient’s LS status and diagnosis status
-
the patient’s acceptance of LS surveillance colonoscopies if offered.
Patient’s bowel state
The patient’s bowel state encapsulates whether the patient has a clinically diagnosed CRC and the extent of any bowel surgery. Although it is possible within an individual sampling model to track a number of primary CRCs and their properties, we make the simplifying assumption that each patient will have no more than two primary CRCs throughout their life (as in other decision models, e.g. Mvundura et al. 84).
Each CRC is staged using the modified Dukes’ stage (A–D) or American Joint Committee on Cancer stage (I–IV), which are effectively equivalent. The tumour–node–metastasis (TNM) system is used clinically to give finer staging detail, but incidence and survival statistics are currently not widely available for TNM stages.
The model tracks the stage of each CRC and the time since diagnosis (to determine the hazard of CRC mortality).
We modelled two portions of the bowel: the colon and the rectum. CRCs can develop in any portion of the bowel still intact. We modelled four surgery types, based on the extent of bowel removed (Table 32). This is a small extension to the three surgery types used in the study by Maeda et al. 106 to account for the fact that rectal cancer can be the first primary cancer in our cohort. Colorectal surgery pathways gives details on surgical management.
| Surgery | Bowel removed |
|---|---|
| Segmental colon resection | Part (but not all) of the colon |
| Subtotal colectomy | All of the colon |
| Anterior resection | All of the rectum |
| Proctocolectomy | All of the colon and rectum |
All probands enter the model with an index CRC (i.e. without a mCRC). The Dukes’ stage for probands is sampled randomly using the distribution described in Model parameters, Stage on diagnosis.
Probands entering the simulation are randomly assigned a surgical state in accordance with the estimated probability that they had colon cancer compared with rectal cancer (Table 33) and the probabilities of different types of surgery for those cancers (Table 34). Table 35 gives the resulting distribution of initial surgical states for probands entering the model, according to their sex and LS status.
| Proband type | Male | Female | Source |
|---|---|---|---|
| With LS | 0.94 | 0.94 | Dinh online appendix107 |
| Without LS | ONS Cancer registration statistics, England 2013103 | ||
| Base case | 0.63 | 0.72 | |
| < 50 years | 0.61 | 0.70 | |
| < 60 years | 0.56 | 0.66 | |
| < 70 years | 0.57 | 0.68 | |
| ≥ 70 years | 0.67 | 0.75 |
| Location of CRC | Surgery (% of cases) | Source |
|---|---|---|
| Colon | Segmental resection (96%), subtotal colectomy (4%) | National Bowel Cancer Audit 2011108 |
| Rectum | Anterior resection (98%), proctocolectomy (2%) | National Bowel Cancer Audit 2011108 |
| Surgery | With LS | Without LS | ||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| Segmental resection | 0.907 | 0.907 | 0.603 | 0.696 |
| Subtotal colectomy | 0.033 | 0.033 | 0.022 | 0.026 |
| Anterior resection | 0.059 | 0.059 | 0.368 | 0.274 |
| Proctocolectomy | 0.001 | 0.001 | 0.007 | 0.005 |
Relatives enter the model without CRC, that is, they are at risk of up to two CRCs (index and metachronous). In reality, some relatives would be survivors of a previous CRC.
Table 36 provides estimates of what proportions of relatives would be survivors of previous CRCs. Estimates for relatives without LS are based on 10-year CRC prevalence data published by the (UK) National Cancer Intelligence Network,109 assuming that the proportion of CRC survivors with colon cancer is the same as the proportion of incident CRCs that are colon cancer. The prevalence of previous CRC for relatives with LS was estimated by multiplying by a scale factor of 38%/2.6% = 14.8 for males and 31%/1.7% = 18.1 for females, with 38% and 31% being estimates of the cumulative risk of CRC up to age 70 years for males and females with LS, respectively,18 and 2.6% and 1.7% being estimates of the cumulative risk of CRC up to age 70 years for males and females without LS, respectively, calculated using population, CRC incidence and CRC mortality statistics for England and Wales in 2010. 100,110–112 Again, it was assumed that the proportion of survivors with colon cancer would match the proportion of incident cases, this time estimated by Dinh et al. 107
| CRC prevalence | With LS | Without LS | ||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| None | 0.9683 | 0.9748 | 0.9979 | 0.9986 |
| Colon cancer | 0.0298 | 0.0237 | 0.0013 | 0.0010 |
| Rectal cancer | 0.0019 | 0.0015 | 0.0009 | 0.0004 |
Relatives with previous CRC would experience a higher mortality rate than relatives without previous CRC and therefore preventing a further CRC would be expected to give a smaller LYG. These CRC survivors would be likely to have early-stage CRC, to have undergone segmental resection and to be followed up for recurrence or metachronous cancer.
The model incorporates initial surgical states for relatives entering the model (Table 37; proportions based on surgical choice for people not known to have LS and the prevalence of colon and rectal cancer, as in Table 36). Most relatives have not had previous surgery (as most relatives have no previous CRC) and, of those who have undergone previous surgery, the majority have had a previous segmental resection, which imparts no risk reduction in the model. A very small number (<< 1%) of relatives enter the model having had previous surgery that does impart a risk reduction. As all of these relatives enter with a risk reduction irrespective of the diagnostic strategy, this would decrease the potential LYG of correctly identifying relatives as having LS; we therefore expect that including initial surgical states has a very small (probably negligible) negative impact on the cost-effectiveness of strategies identifying LS (i.e. ICERs for testing strategies slightly increased compared with no testing).
| Surgery | With LS | Without LS | ||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| None | 0.9683 | 0.9748 | 0.9979 | 0.9986 |
| Segmental resection | 0.0288 | 0.0229 | 0.0012 | 0.0009 |
| Subtotal colectomy | 0.0011 | 0.0008 | 0.0000 | 0.0000 |
| Anterior resection | 0.0019 | 0.0015 | 0.0009 | 0.0004 |
| Proctocolectomy | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
Patient’s gynaecological state (women only)
A patient’s gynaecological state encapsulates what risk-reducing measures she has employed and whether or not she has had endometrial cancer.
Although endometrial cancer can be staged and there are some survival estimates according to stage,113 the staging system has been changed recently and most patients are diagnosed in the early stage of cancer. Therefore, the stage of endometrial cancer is not tracked in the model; rather, the time since diagnosis is tracked.
It was assumed that patients will not get more than one primary endometrial cancer as they will undergo a hysterectomy, which is very effective at preventing endometrial cancer when used prophylactically. 26
Risk-reducing measures available to patients are gynaecological surveillance and prophylactic hysterectomy (and bilateral salpingo-oophorectomy). Patients can start and stop surveillance but prophylactic hysterectomy is irreversible and surveillance cannot be performed in patients after hysterectomy (Figure 18).
FIGURE 18.
Gynaecological state model diagram.

All simulated individuals were assumed to start without endometrial cancer.
Probands and relatives who have been diagnosed with LS may be offered surveillance or prophylactic H-BSO depending on their age.
Table 38 shows the assumed distribution of risk-reducing strategies according to age at diagnosis, based on audit data from the Northern Genetics Service (Lorraine Cowley, Principal Genetic Counsellor, Newcastle upon Tyne Hospitals NHS Foundation Trust, 20 November 2012, personal communication).
| Age (years) at LS diagnosis | No risk reduction | Surveillance | Prophylactic H-BSO |
|---|---|---|---|
| 0–34 | 1.000 | 0.000 | 0.000 |
| 35–44 | 0.200 | 0.600 | 0.200 |
| 45–59 | 0.167 | 0.458 | 0.375 |
| 60–69 | 0.000 | 0.143 | 0.857 |
| ≥ 70 | 0.143 | 0.000 | 0.857 |
Outcomes
For each of the 24 patient groups (see Long-term outcomes model), the following outcomes were recorded from the simulation:
-
costs (discounted and undiscounted)
-
QALYs (discounted and undiscounted)
-
overall survival (and whether or not censored on reaching age 100 years)
-
CRC-, endometrial cancer- and overall cancer-free survival (and whether censored because of death or on reaching age 100 years)
-
event-free survival (and whether or not censored on reaching age 100 years)
-
number of incident CRCs
-
number of incident endometrial cancers
-
number of colonoscopies performed
-
disaggregated costs (discounted and undiscounted).
Colorectal cancer
The simulation model includes four events relating to CRC:
-
index CRC incidence
-
index CRC mortality
-
mCRC incidence
-
mCRC mortality.
The incidence events transform the patient’s bowel state (e.g. index CRC incidence transforms the bowel state from ‘no CRC’ to ‘index CRC’ with a particular stage). The mortality events result in the patient dying with no further events occurring.
Figure 19 shows the 21 different bowel cancer histories included in the model.
FIGURE 19.
Colorectal cancer and mCRC incidence model diagram.

The probability that a CRC incidence event occurs within a year is dependent on the incidence rate, whereas the probability that a CRC mortality event occurs is dependent on the survival function.
Colorectal cancer incidence rates in the model are dependent on the following patient characteristics:
-
age
-
sex
-
whether or not the patient has had a previous CRC
-
time since first CRC
-
LS status
-
risk-reducing measures, that is, regular colonoscopies and aspirin, as described in Colorectal surgery pathways and Aspirin chemoprevention respectively.
Different annual incidence rates are provided for the eight combinations of sex, previous cancer (yes/no) and LS status and then risk-reducing measures are incorporated as HRs, which have a simple multiplicative effect on the incidence rate.
The incidence rates for males and females without previous cancer without LS were estimated from pooled registration statistics for CRC in England between 2006 and 2014 inclusive21,96–103 and the estimated population in the midpoints of those years. 114 Following the methodology adopted by the ONS,100 we calculated the age-specific rate of CRC incidence by dividing the number of CRC registrations within a time period by an estimate of the person-years lived during that period. Incidence figures were pooled across 5 years to achieve a large sample size but not further back than 2006 as such data may not reflect more recent developments in cancer detection and registration.
Cancer registration statistics are not provided for each year of age but for age groups, generally of 5 years. We assumed that within each of these age groups the incidence rate would remain constant. The resulting cumulative risk of CRC for individuals without LS is shown in Figure 20.
We estimated the incidence of mCRC (i.e. the incidence in individuals who had had a previous CRC) for individuals without LS by adjusting the incidence of the first CRC by a HR of 1.4 for the first 3 years after the first CRC and 1.3 for the following 7 years, as in the study by Mulder et al. 115 Mulder et al. 115 studied 10,283 Dutch patients with CRC undergoing standard follow-up. After 10 years no additional hazard was applied.
Previously,4 we conducted a literature review to identify studies from which the age-dependent incidence rates for individuals with LS could be estimated.
Subsequent reviews2,16,116 have not identified any additional studies.
Møller et al. 5 have subsequently published estimates of the cancer risk for individuals with LS while undergoing colonoscopic surveillance; this does not address the need here (i.e. for the cancer risk in the absence of interventions).
In the absence of new evidence to consider, the model includes an incidence rate for CRC based on the study by Bonadona et al. 18
A logistic model for cumulative risk was fitted to data from Bonadona et al. 18 using the following parameterisation and ordinary least-squares regression:
The 95% CIs for the cumulative risk to age 70 years from the study by Bonadona et al. 18 were used in sensitivity analyses (by varying β0 appropriately). Table 39 shows the parameters used in the base-case and sensitivity analyses and Figures 21 and 22 show graphically the fit to the data from Bonadona et al. 18
| Parameter | Base case | Sensitivity analyses (95% CI) |
|---|---|---|
| β0 | M 0.464, F 0.435 | M 0.303 to 0.715; F 0.265 to 0.697 |
| β1 | M 0.107, F 0.108 | |
| β2 | M 55.5, F 61.3 |
The CRC stage on diagnosis is an important predictor of survival. Historically, the modified Dukes’ stage (A–D) has been used for cancer staging, but it is more common now to refer to stages by Roman numerals (I–IV).
We assumed that the stage on diagnosis would be independent of age, sex, LS status and whether it was the first (index) CRC or a mCRC (Fajobi et al. 117 conclude that stages for mCRC are no worse than those for index CRC and there was no consensus on whether they might be better). We also assumed that stage on diagnosis of mCRC was independent of the stage of the index CRC and the CRC site. CRC stage was assumed to depend only on whether the person was undergoing LS surveillance colonoscopies.
The stage distributions are provided in Model parameters, Stage on diagnosis.
As stated in Patient’s bowel state we model two sections of the bowel, the colon and the rectum. We grouped rectosigmoid cancer (International Classification of Diseases, 10th Revision code C19) into rectal cancer. The site of incident CRCs was dependent on sex, whether or not the person had LS and any previous surgery (Table 40).
| Previous surgery | With LS | Without LS | |
|---|---|---|---|
| Men | Women | ||
| None | 0.94 | 0.63 | 0.72 |
| Segmental resection | 0.94 | 0.63 | 0.72 |
| Subtotal colectomy | 0.00 | 0.00 | 0.00 |
| Anterior resection | 1.00 | 1.00 | 1.00 |
| Proctocolectomy | NAa | NAa | NAa |
It was assumed that all CRCs are colon cancers following anterior resection (AR) and that all CRCs are rectal cancers following subtotal colectomy. If there is no previous surgery or a previous segmental resection the probability of the CRC being situated in the colon for a person with LS was estimated as 0.94 based on the study by Dinh et al. 107 For males and females without LS the probability of colon cancer was estimated from ONS cancer registration statistics. 100
We assumed that mortality from CRC depends on the following:
-
CRC stage at diagnosis
-
years since diagnosis
-
age at diagnosis
-
LS status (see Lynch syndrome colorectal cancer survival).
We did not consider the effect of the following on mortality from CRC:
-
patient’s sex
-
site of CRC
-
surgery for CRC.
The baseline annual rate of mortality from CRC was derived from data provided by the (UK) National Cancer Intelligence Network109 by extracting 1-, 2-, 3-, 4- and 5-year relative survival from survival curves and assuming constant rates of mortality within each year (Table 41 and Figure 23). It was assumed that the mortality rate for 4–5 years since diagnosis also applied after 5 years (Table 42).
| Years since diagnosis | Dukes’ stage | |||
|---|---|---|---|---|
| A | B | C | D | |
| 1 | 0.969 | 0.917 | 0.815 | 0.380 |
| 2 | 0.965 | 0.872 | 0.681 | 0.193 |
| 3 | 0.957 | 0.831 | 0.583 | 0.116 |
| 4 | 0.945 | 0.799 | 0.522 | 0.083 |
| 5 | 0.932 | 0.770 | 0.477 | 0.066 |
| Years since diagnosis | Dukes’ stage, n | |||
|---|---|---|---|---|
| A | B | C | D | |
| 0–1 | 3102 | 8709 | 20,460 | 96,729 |
| 1–2 | 419 | 5000 | 17,971 | 67,733 |
| 2–3 | 843 | 4761 | 15,465 | 51,116 |
| 3–4 | 1279 | 4000 | 11,060 | 32,857 |
| > 4 | 1400 | 3667 | 9068 | 23,375 |
The assumption that the mortality rate after 5 years is equal to the mortality rate for 4–5 years is likely to be a slight overestimate of CRC mortality (Table 43). The result of this would be a slight improvement in the cost-effectiveness of strategies with a high yield of LS mutations.
| Years since diagnosis | Male colon cancer | Female colon cancer | Male rectal cancer | Female rectal cancer | Model CRC |
|---|---|---|---|---|---|
| 1 | 0.730 | 0.722 | 0.788 | 0.788 | 0.757 |
| 5 | 0.544 | 0.551 | 0.546 | 0.575 | 0.530 |
| 10 | 0.501 | 0.508 | 0.473 | 0.521 | 0.421 |
The HRs for CRC mortality by age compared with CRC mortality across all ages were estimated using net survival statistics from the ONS118 and are shown in Table 44. Details of the calculations are given in appendix 6 of Snowsill et al. 4
As previously,4 mortality from mCRC was modelled by adding the mortality rates for both the index CRC and the mCRC as calculated above, assuming that mortality from the metachronous cancer would be no different from mortality from the index cancer for the same Dukes’ stage (as assumed by, for example, Mvundura et al. 84 and Dinh et al. 107). The same approach was used independent of the LS status of the patient. As the mortality rates are dependent on the time since diagnosis in the model, we kept track of time since diagnosis of the index cancer and the metachronous cancer.
As previously,4 the model assumed improved survival for individuals with LS and local CRC compared with individuals with sporadic CRC. A HR of 0.57 was applied for Dukes’ A and Dukes’ B CRC if the simulated individual had LS, based on the study by Lin et al. 119 Survival for individuals with Dukes’ C and Dukes’ D CRC was equal for patients with and without LS, based on the study by Barnetson et al. 54
Endometrial cancer
The lifetime risk of endometrial cancer in the general population is one in 41. 120 The lifetime risk in women with LS (in the absence of risk-reducing measures) is around 35%. 18 As previously,4 endometrial cancer was modelled only for women with LS.
As previously,4 and as for CRC, the incidence rates for endometrial cancer in women with LS were estimated from the study by Bonadona et al. 18
A piecewise constant hazard of endometrial cancer was used per decade of life to achieve the cumulative risk profile observed in the study by Bonadona et al. 18 and it was assumed that the incidence of endometrial cancer would be zero after age 80 years. Figure 24 shows the cumulative risk used in the model as well as the data from Bonadona et al. 18 for reference.
FIGURE 24.
Endometrial cancer incidence. Gynaecological surveillance was not assumed to affect the incidence of endometrial cancer (see Gynaecological surveillance).

Survival from uterine cancer in England and Wales has recently been estimated by the London School of Hygiene and Tropical Medicine and published by Cancer Research UK (Figure 25). 121
FIGURE 25.
Net survival for uterine cancer in England and Wales. Source: based on a graphic created by Cancer Research UK. 121

Endometrial cancer cases make up the vast majority of uterine cancer cases, so uterine cancer cases are expected to closely approximate survival of endometrial cancer.
The survival curve was used to estimate a piecewise constant rate of mortality for each year since diagnosis. After 10 years the rate of mortality is zero.
General mortality
Death from other causes was modelled by using mortality rates separately for men and women from life tables for England and Wales, 2008–10,122 adjusted to remove the proportion of mortality resulting from CRC, which was estimated by dividing the number of deaths from CRC in each age group by the total number of deaths in that age group from mortality data for England in 2010. 110 We did not adjust for mortality from endometrial cancer as this accounted for < 1% of deaths in the general population (whereas CRC accounted for 2.8% of deaths) and we did not adjust for mortality from other LS-associated cancers as these were not included in our model.
Surveillance pathways
Previously,4 a colorectal surveillance pathway was modelled based on the synthesis of a number of relevant recommendations,46,47,123 which included biennial (every 2 years) colonoscopies for individuals diagnosed with LS.
The recently published guidelines from the European Mallorca group advocate an interval of 1–2 years between colonoscopies for individuals with LS. 19
The evidence underpinning the effectiveness estimates for colonoscopic surveillance in the model is based on 3-yearly surveillance124 (see Colonoscopy).
To avoid excessive inconsistency between the effectiveness evidence and the associated costs of colonoscopic surveillance, an interval of 2 years was modelled for individuals with LS.
Colorectal cancer patients are assumed to also receive a carcinoembryonic antigen (CEA) test every 3 months for 2 years and then every 6 months for a further 3 years; computerised tomography (CT) scans of the chest, abdomen and pelvis at 12 months and 24 months; and colonoscopy at 12 months and then 5-yearly (unless diagnosed with LS). 4,47,123
The European Society for Medical Oncology guidelines recommend annual gynaecological examination, pelvic ultrasound, CA-125 (cancer antigen 125) analysis and aspiration biopsy for gynaecological surveillance, starting at age 30–35 years. 125 This was used as the basis for modelling gynaecological surveillance, with surveillance initially offered at age 35 years (or immediately if the diagnosis of LS is after the age of 35 years) and continuing until 70 years of age.
Colorectal surgery pathways
As previously,4 colorectal surgical pathways were based on published guidelines46 with input from clinical experts.
Patients undergo surgical management if they are diagnosed with CRC and the cancer is deemed to be operable (this includes surgery when intent is palliative rather than curative). We made the simplifying assumption that all patients diagnosed with CRC undergo surgical management (> 75% of patients in the National Bowel Cancer Audit108 were treated surgically). In each case, surgery would remove the bowel portion affected by the cancer and, in some cases, additional portions, depending on previous surgery and whether LS had been diagnosed (Figure 26). Our clinical expert advice was that, in general, surgery for patients without LS tends to be conservative, without a risk-reducing element (Mr Ian Daniels, Consultant Colorectal Surgeon, Royal Devon and Exeter Hospital, 24 October 2012, personal communication.)
Clinical guidelines indicate that there is a place for more aggressive surgery, with a risk-reducing element, for patients known to have LS on CRC diagnosis; in particular, Cairns et al. 46 stated that, ‘For patients with proximal tumours, colectomy and ileorectal anastomosis is most relevant’ (p. 678). Input from our clinical expert suggested that this particular guidance would rarely be followed as it was based on a low category of evidence (evidence obtained from expert committee reports or opinions or clinical experiences of respected authorities) and colonoscopic surveillance is deemed effective enough to negate the need for aggressive surgery (Mr Ian Daniels, personal communication). To resolve this disagreement we included a parameter in the model that defines the probability that more aggressive surgery would be used for LS patients, which could be varied from 0 (ignore guidelines; surgical treatment not affected by Lynch diagnosis) to 1 (full adherence to guidelines; aggressive surgery always used). Previous analyses by Snowsill et al. 4 demonstrated that altering this parameter had a minimal impact on cost-effectiveness and therefore sensitivity analyses on this parameter have not been repeated.
When surgery removes the rectum because of cancer in the rectum there are two common operations: AR, which preserves the anus, and abdominoperineal excision of the rectum (APER), which results in permanent stoma. We grouped these operations together and assumed that they are both as effective at preventing metachronous rectal cancer. Some patients would require a permanent stoma, which would affect health-related quality of life (HRQoL) and costs. Rather than modelling this on an individual patient basis we assumed an average effect across all patients.
Any subsequent surgery depends on the location of the CRC, the nature of the previous surgery and whether the patient has been diagnosed with LS (unless the parameter described above is 0) (Tables 45 and 46).
| Previous surgery | Segmental resection (%) | Subtotal colectomy (%) | AR (%) | Proctocolectomy (%) | Source |
|---|---|---|---|---|---|
| None | 96a | 4b | 0 | 0 | National Bowel Cancer Audit108 |
| Segmental resection | 0 | 100 | 0 | 0 | Assumption |
| Subtotal colectomy | NAc | NAc | NAc | NAc | Assumption |
| AR | 0 | 0 | 0 | 100 | Assumption |
| Proctocolectomy | NAc | NAc | NAc | NAc | Assumption |
| Previous surgery | Segmental resection (%) | Subtotal colectomy (%) | AR (%) | Proctocolectomy (%) | Source |
|---|---|---|---|---|---|
| None | 0 | 0 | 98a | 2b | National Bowel Cancer Audit108 |
| Segmental resection | 0 | 0 | 0 | 100 | Assumption |
| Subtotal colectomy | 0 | 0 | 0 | 100 | Assumption |
| AR | NAc | NAc | NAc | NAc | Assumption |
| Proctocolectomy | NAc | NAc | NAc | NAc | Assumption |
Surgery distributions for CRC patients diagnosed with LS were adjusted by the parameter representing the probability of aggressive surgery. If this probability is denoted as p, then for colon cancer patients:
where SEG is segmental resection and SUB is subtotal colectomy. For rectal cancer patients:
where IPAA is ileal pouch anal anastomosis.
In the base case, this parameter was set to 0 for the following reasons:
-
the clinical guidelines [e.g. the BSG/Association of Coloproctology of Great Britain and Ireland (ACPGBI) guidelines46] are not based on high-quality evidence
-
the model does not incorporate any utility decrement for more aggressive surgery, even though function can be affected (see Colorectal cancer treatment)
-
the model does not incorporate excess surgical mortality from more aggressive surgery.
Aspirin chemoprevention
Aspirin has been shown in observational studies to reduce the CRC burden126 and the CAPP2 RCT in individuals with LS mutations demonstrated a protective effect from aspirin against CRC and other LS-associated cancers in individuals completing the protocol.
As the balance of risks and benefits is favourable for prescribing regular aspirin to individuals with LS (unless contraindicated), and aspirin use in this context is highly likely to be cost-effective (and patients may also purchase aspirin over the counter if not prescribed), it was judged that aspirin chemoprevention should be included as part of the base-case analysis.
It should be noted that, at present, individuals with LS are most likely to receive aspirin as part of the ongoing Cancer Prevention Programme 3 (CaPP3) dosing study rather than by being directly prescribed aspirin.
A scenario analysis was also conducted in which aspirin was not prescribed.
Perspective, time horizon and discounting
Costs were included from a NHS and PSS perspective.
The perspective on health outcomes was all direct health effects on patients, which in this case included individuals newly diagnosed with CRC (probands) and their blood relatives who may be at risk of LS.
A lifetime time horizon was used, with a maximum age of 100 years modelled.
Costs and QALYs were discounted at 3.5% per annum (unless otherwise stated). Life-years and natural outcomes (e.g. number of CRCs) were not discounted (unless otherwise stated).
Model parameters
A summary table of all of the model parameters is provided in Appendix 4.
Treatment effectiveness and extrapolation
Diagnostic performance
Estimates of test accuracy (sensitivity and specificity) were taken from the available literature, identified through the diagnostic accuracy and cost-effectiveness literature reviews reported in Chapters 2 and 4 respectively.
In Snowsill et al. ,4 test accuracy for IHC and MSI was taken from the EGAPP review reported by Palomaki et al. 42 In the EGAPP review, studies with very different input populations were synthesised, not all of which were relevant to this review. In the current model, population-based studies identified in Chapter 234,54–56 were used to produce estimates of test accuracy. In Chapter 2 it was explained that a meta-analysis of the included studies was not conducted because of the heterogeneity of the studies. In particular, although they were population-based studies, the studies by Barnetson et al. ,54 Southey et al. 56 and Limburg et al. 55 all used age limits in their input population, which will influence the prevalence of these populations and potentially the sensitivity and specificity of the tests.
The results of these studies were synthesised using the multilevel mixed-effects logistic regression command ‘melogit’ in Stata®/SE 14.1 (StataCorp LP, Texas, USA). The code for this analysis is presented in Appendix 5. The results of this analysis (with 95% CIs), plus the EGAPP results, are presented in Table 47. The values from Snowsill et al. 4 lie well within the CIs of the new estimates, although the new point estimates for sensitivity appear higher and for specificity appear slightly lower than those reported by the EGAPP review.
| Test | Parameter | Base case, MSI = MSI-H (95% CI) | Scenario analysis, MSI = MSI-L and MSI-H (95% CI) | Source | Snowsill et al.4 base case | Source |
|---|---|---|---|---|---|---|
| MSI | Sensitivity | 0.913 (0.426 to 0.993) | 0.973 (0.893 to 0.994) | Barnetson et al.,54 Poynter et al.34 and Southey et al.56 | MLH1, MSH2 0.89; MSH6, PMS2 0.77 | Palomaki et al.42 |
| Specificity | 0.837 (0.638 to 0.937) | 0.596 (0.304 to 0.833) | 0.902 | |||
| IHC | Sensitivity | 0.962 (0.694 to 0.996) | a | Limburg et al.55 and Southey et al.56 | 0.77 | |
| Specificity | 0.884 (0.790 to 0.940) | a | 0.888 | |||
| BRAF V600E | Sensitivity | 0.96 (0.60 to 0.99) | a | Ladabaum et al.3 | 1.00 | Domingo et al.127 |
| Specificity | 0.76 (0.60 to 0.87) | a | Following MSI: 0.40; following IHC: 0.69 | Domingo et al.127 and Palomaki et al.42 | ||
| MLH1 promoter methylation | Sensitivity | 0.94 (0.79 to 0.98) | a | Ladabaum et al.3 | 0.93 | Bouzourene et al.128 and Chang et al.129 |
| Specificity | 0.75 (0.59 to 0.86) | a | 0.83 | |||
| Diagnostic genetic testing for probands | Sensitivity | MLH1, MSH2 and MSH6 0.90; PMS2 0.67 | Dinh et al.107 and Senter et al.130 | |||
| Specificity | 0.997 | |||||
| Predictive genetic testing for relatives | Sensitivity | 1.00 | Assumed4 | |||
| Specificity | 1.00 |
Because of the low number of studies (three for MSI, two for IHC) it was not possible to estimate the correlation between true sensitivity and true specificity across studies, so this parameter was removed from the estimation (i.e. independence of random effects was assumed).
As indicated in Chapter 2, this synthesis may not be entirely appropriate to determine test accuracy for the purposes of review. This is because of the small number of studies, the variation in methodology (e.g. heterogeneity in the reference standard) and the heterogeneity in the accuracy estimates (e.g. Poynter et al. 34 predicted much higher sensitivity and lower specificity for MSI than the other two studies). The small number of studies means that it is not possible to adequately explore sources of heterogeneity and it is also not possible to assess the likelihood of publication bias (e.g. using a funnel plot). However, the CIs are sufficiently wide that for modelling purposes a range likely to include the true values can be implemented in univariate sensitivity analyses.
Accuracy results for BRAF V600E and MLH1 promoter methylation testing were taken from the recent technical review by Ladabaum et al. ,3 which was identified from the searches run in the cost-effectiveness review (see Chapter 4, Study identification). In this technical review 14 studies reporting MLH1 promoter methylation testing and 11 studies reporting BRAF V600E testing were synthesised. These studies included a variety of previous tests, including MSI and IHC. These values update the values used in the study by Snowsill et al. ,4 which were based on very few studies.
The accuracy of genetic testing in probands and relatives is unchanged in the current model from the values used in Snowsill et al. 4 and is taken from published literature. Diagnostic genetic testing for PMS2 is assumed to have a lower sensitivity than testing for the other genes, given the greater complexity of the molecular analysis for this gene (which results in a lower pick-up rate) and the greater difficulties in interpreting mutations (when found) as pathogenic (given the lower penetrance of mutations in PMS2). 5,45
One other important component of the effectiveness of a strategy is the acceptance rate of the tests. Acceptance rates of diagnostic tests and their sources are reported in Table 48. With the exception of the acceptance of counselling and genetic testing for relatives, which was updated to use UK data from the Manchester Regional Lynch Syndrome Registry, as reported in Barrow,91 the acceptance rates remain unchanged from those used in the study by Snowsill et al. 4 Previously these values were taken from the study by Palomaki et al. 42 and were not UK specific.
| Test | Proband/relative | Acceptance rate (%) | Original source |
|---|---|---|---|
| MSI | Proband | 100 | Ramsey et al.,83 confirmed by expert (IF) in Snowsill et al.4 |
| IHC | Proband | 100 | Assumed |
| BRAF V600E | Proband | 100 | Assumed |
| MLH1 promoter methylation | Proband | 100 | Assumed |
| Genetic test following counselling (proband) | Proband | 90 | Ladabaum et al.85 |
| Genetic counselling (proband) | Proband | 92.5 | Clinical expert (IMF) gave range 90–95% in Snowsill et al.4 |
| Genetic test following counselling (relative) | Relative | 77 | Calculated from Manchester Familial Colorectal Cancer Registry data, reported in Barrow91 |
| Genetic counselling (relative) | Relative | 78 | Calculated from Manchester Familial Colorectal Cancer Registry data, reported in Barrow91 |
Surveillance
Colonoscopy is assumed to lead to improved health outcomes by reducing the incidence of CRC (as adenomas that could have become adenocarcinomas are identified and removed) and by improving the cancer stage distribution on diagnosis (i.e. catching the cancer earlier), which leads to improved survival.
The model assumed that the majority of patients diagnosed with LS will be offered and will accept surveillance colonoscopies.
Data from 591 individuals from the Manchester Familial Colorectal Cancer Registry, as reported in Barrow,91 were used to estimate the acceptance rates for relatives diagnosed with LS. We assumed that these rates would be the same for probands as for relatives, as in the study by Snowsill et al. 4 the acceptance of surveillance in probands was the same as or higher than that in relatives. Previously, these estimates were taken from the study by Ladabaum et al. ,85 which was a study of a US population and which therefore does not reflect the acceptance rate in the UK population. The current values appear to suggest higher acceptance rates than previously modelled. However, as these values are taken from only one UK institution and acceptance of surveillance has been previously shown to be an influential parameter with regard to cost-effectiveness, we examined the impact of these rates in sensitivity analyses.
Table 49 shows the assumed acceptance rates used in the model.
Previously,4 event times were extracted from figure 1 of Järvinen et al. 124 and used in a Cox proportional hazards regression to estimate the effectiveness of colonoscopy in reducing CRC incidence. The resulting HR estimate was 0.387 (95% CI 0.169 to 0.885).
Subsequently, other evidence has also been collected relating to this question.
The American Gastroenterological Association published a technical review on the diagnosis and management of LS in 2015. 3 This review identified five observational studies (including that by Järvinen et al. 124), which allowed estimation of the effectiveness of colonoscopy in reducing CRC incidence. The pooled effect estimate (odds ratio) was 0.23 (95% CI 0.13 to 0.41).
Møller et al. 5 reported on the cancer risks in a prospectively identified population of LS carriers, in whom surveillance is widespread. This study included 1942 LS carriers from across Europe (and Australia), with 195 of these from the UK. The authors found high rates of CRC in spite of regular surveillance.
Figure 27 illustrates the contradictory nature of the evidence. ‘Bonadona’ and ‘Møller’ are the estimated incidence rates in the studies, whereas ‘Bonadona + Järvinen’ is a counterfactual incidence rate profile obtained by applying the HR of 0.387 to the ‘Bonadona’ data. If all of these studies were conducted without bias and in similar populations and with enough statistical power, one would expect the ‘Møller’ data to closely align with the ‘Bonadona + Järvinen’ data rather than the ‘Bonadona’ data.
FIGURE 27.
Comparison of CRC incidence rates in the absence of surveillance (Bonadona) and with surveillance (Møller). Notes: assumes that 41% have MLH1 mutation, 44% have MSH2 mutations and 16% have MSH6 mutations (figures do not sum to 100% because of rounding). Sources: Bonadona et al. 18 and Møller et al. 5
There are a number of potential reasons why the results of Møller et al. 5 might not confirm the effectiveness of colonoscopy observed in the study by Järvinen et al. :124
-
the limited statistical power in both the Bonadona et al. 18 study and the Møller et al. 5 study (i.e. random chance)
-
the naive comparison of the results from the Bonadona et al. 18 study and the Møller et al. 5 study, with no adjustment for differences in populations (other than for gene distribution and age)
-
the biased effect estimate in the study by Järvinen et al. 124
It is difficult to assess the similarity of the Bonadona et al. 18 and Møller et al. 5 patient populations. The population in Bonadona et al. 18 seems to have a more even balance between men and women whereas the population in Møller et al. 5 included somewhat more women. It is possible that there are differences in the proportions of patients with clearly pathogenic mutations – Bonadona et al. 18 included 7% (35/537) of families with variants of uncertain significance, whereas Møller et al. 5 included only patients with mutations judged pathogenic by their reporting centre (although 31% of patients had mutations not reported on the Leiden Open Variant Database by October 2015).
The two main sources of potential bias in the Järvinen et al. 124 study are confounding because of:
-
self-selection of the intervention group (i.e. participants chose whether or not to receive the intervention) – this would be expected to exaggerate the effectiveness estimate as participants choosing to receive surveillance are more likely to engage in other healthy behaviours and to have other characteristics predictive of good outcomes
-
treatment switching (i.e. participants who initially declined surveillance who later opted into surveillance) – this would be expected to attenuate the effectiveness estimate.
Furthermore, the results from Järvinen et al. 124 may not generalise to current surveillance in the NHS because of developments in technology or differences in service delivery and behaviour, which could result in a different distribution of screening intervals.
However, in the absence of compelling alternative effectiveness estimates, we continued to use the HR of 0.387 from the study by Järvinen et al. 124
In a worst-case scenario analysis it was assumed that surveillance does not reduce the incidence of CRC, that is, a HR of 1 was used.
As previously,4 the stage on diagnosis was assumed to be dependent only on whether an individual has been offered and accepted LS surveillance colonoscopies. Whether the individual has a LS mutation, their previous history of cancer and their sex and age were not modelled as affecting the stage distribution.
The stage on diagnosis for individuals not accepting surveillance was estimated from national data from England. In England in 2012, 11% of CRCs were not staged (or the stage was not recorded). 131 The National Cancer Intelligence Network used multiple imputation methods, based on patient and cancer characteristics that were recorded, to estimate the stage distribution for the unstaged patients (Table 50). When this stage distribution is combined with the data for patients whose cancers were staged, the resulting distribution for stages I–IV is 17.6 : 27.0 : 29.5 : 25.9 respectively. Previously,4 a distribution of 16.4 : 31.7 : 27.1 : 24.8 for stages I–IV, respectively, was estimated from 2009/10 data by excluding patients whose cancer stage was not recorded. The effect of the change is to reduce the number of patients with stage II CRC and increase the number of patients with other stages.
| Stage | Observed data for all patients | Imputed stage distribution of ‘unknown’ | Combined data | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| I | 5255 | 15.5 | 734 | 2.16 | 5989 | 17.6 |
| II | 8402 | 24.7 | 768 | 2.26 | 9170 | 27.0 |
| III | 9258 | 27.2 | 778 | 2.29 | 10,036 | 29.5 |
| IV | 7351 | 21.6 | 1465 | 4.31 | 8816 | 25.9 |
| Unknown | 3745 | 11.0 | ||||
| Total | 34,011 | 100.0 | 3745 | 11.02 | 34,011 | 100.0 |
For individuals accepting LS surveillance colonoscopies, a stage distribution of 68.6 : 10.5 : 12.8 : 8.1 for stages I–IV, respectively, was used, as previously,4 based on data from Mecklin et al. 132
A literature review was conducted previously of the effectiveness of surveillance for endometrial and ovarian cancer. 4 Conceptually, surveillance was expected to reduce the incidence of gynaecological cancers by the detection of premalignancies, which are either individually removed or which prompt hysterectomy. Malignancies identified during surveillance were also expected to have a more favourable stage profile.
No experimental studies (e.g. RCTs) were identified, but three non-experimental studies were identified. 133–135 Two of these studies134,135 considered cohorts of women with LS eligible for gynaecological surveillance and compared patients receiving surveillance with patients refusing or not receiving surveillance during the study (Figures 28 and 29). These study designs could provide estimates of the effectiveness of surveillance in reducing the incidence of gynaecological cancer; however, they were both at high risk of bias because of potential confounding factors between the groups and neither could provide satisfactory effect size estimates because of the limited control group sizes and the limited number of events. The limited number of events also means that these studies are not informative for the stage distribution of gynaecological cancers.
FIGURE 29.
Patient flow diagram for the study by Järvinen et al. 135 EC, endometrial cancer; OC, ovarian cancer.
The study by Renkonen-Sinisalo et al. ,133 in contrast, considered patients before and after institution of a surveillance programme. The outcomes of 385 women with LS mutations were compared with the outcomes of 83 women with LS mutations who were affected by endometrial cancer before surveillance was instituted (Figure 30).
FIGURE 30.
Patient flow diagram for the study by Renkonen-Sinisalo et al. 133 EC, endometrial cancer.
This study design cannot be used to estimate the effect of surveillance on cancer incidence, but it can be used to estimate the impact of surveillance on the stage of cancer at diagnosis (Figure 31).
FIGURE 31.
International Federation of Gynecology and Obstetrics (FIGO) stage distribution of endometrial cancers in the study by Renkonen-Sinisalo et al. 133 (a) Surveillance; and (b) control.

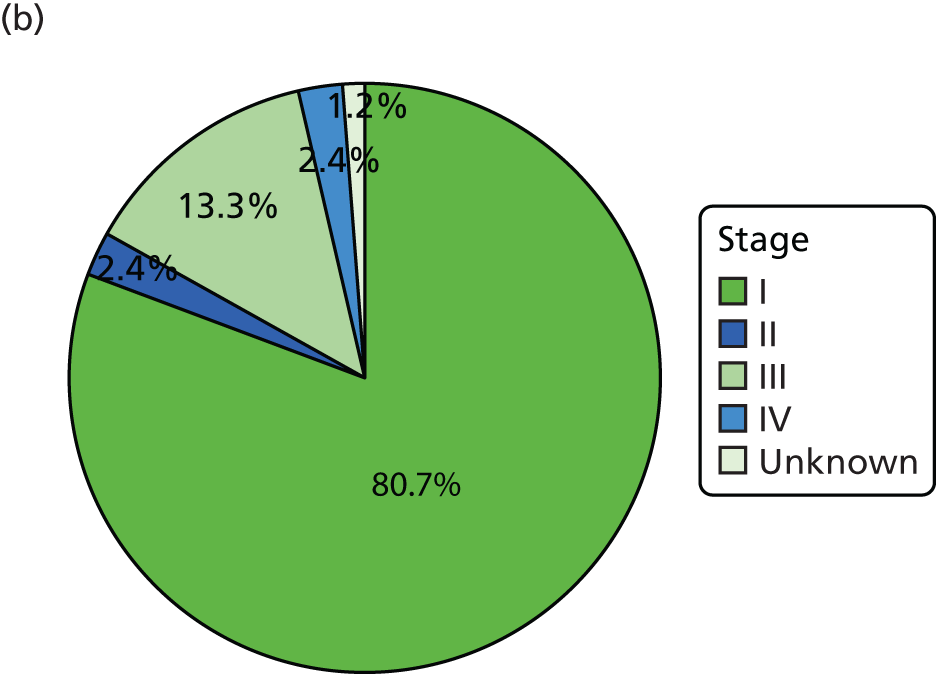
An ordered logistic regression was used to estimate the impact of surveillance. The regression coefficient for surveillance was not statistically significant (z = –0.45, p = 0.651) but was suggestive that surveillance could improve the stage distribution. Predicted stage distributions from the regression are shown in Figure 32.
FIGURE 32.
Predicted stage distribution based on the results of Renkonen-Sinisalo et al. 133 (a) Surveillance; and (b) control.


These stage distributions were not used directly but were instead used to estimate a HR for survival from endometrial cancer.
Lewin et al. 113 estimated the 5-year survival from endometrial cancer according to stage at diagnosis (Table 51). The weighted average 5-year survival for patients in the surveillance and control groups was estimated as 83.4% and 81.8%, respectively, from which a HR of 0.898 was derived.
| FIGO (1998) stage | 5-year survival (%) |
|---|---|
| I | 87.8 |
| II | 76.2 |
| III | 55.3 |
| IV | 21.1 |
Because of the significant uncertainty in this HR, a scenario analysis is included in which there is no survival benefit (i.e. no benefit at all) from gynaecological surveillance.
It should be noted that the women in the study by Renkonen-Sinisalo et al. 133 were offered surveillance with 2- or 3-year intervals, whereas current European guidelines propose annual surveillance,125 so it is possible that a study of annual surveillance could find greater effectiveness than what is modelled here.
Chemoprevention
There is evidence from the CAPP2 RCT that aspirin reduces the incidence of cancer in individuals with LS mutations. 29 In this study participants were randomised to receive aspirin (600 mg of enteric coated aspirin daily) or aspirin placebo (alongside resistant starch or starch placebo in a two-by-two factorial design) for up to 4 years.
Intention-to-treat and per-protocol analyses were conducted. Per-protocol analyses considered patients who continued to take aspirin (or aspirin placebo) for at least 2 years.
Analyses were conducted using Cox proportional hazards regression (to obtain a HR for the incidence of first cancer) and Poisson regression (to obtain an incidence rate ratio accounting for multiple cancers within each individual).
In total, 861 participants were randomised to aspirin (n = 427) or aspirin placebo (n = 434). At the time of publication the mean follow-up was 55.7 months (4.6 years), with a maximum follow-up of 128.0 months (10.7 years).
The intention-to-treat proportional hazards analysis showed a reduction in CRC incidence in those taking aspirin compared with those taking aspirin placebo (HR 0.63, 95% CI 0.35 to 1.13), but this did not reach statistical significance (p = 0.12). A number of other analyses did reach statistical significance and the authors concluded that aspirin is effective at reducing the risk of LS cancers (Table 52).
| Analysis | HR (95% CI) | Incidence rate ratio (95% CI) |
|---|---|---|
| Intention to treat | ||
| CRC | 0.63 (0.35 to 1.13) | 0.56 (0.32 to 0.99) |
| LS cancers except CRC | 0.63 (0.34 to 1.19) | 0.63 (0.34 to 1.16) |
| All LS cancers | 0.65 (0.42 to 1.00) | 0.59 (0.39 to 0.90) |
| Per protocola | ||
| CRC | 0.41 (0.19 to 0.86) | 0.37 (0.18 to 0.78) |
| LS cancers except CRC | 0.47 (0.21 to 1.06) | 0.49 (0.23 to 1.05) |
| All LS cancers | 0.45 (0.26 to 0.79) | 0.42 (0.25 to 0.72) |
Not all patients recruited to the trial (n = 1071) were randomised to aspirin or aspirin placebo; 134 were ineligible to receive aspirin or withdrew before treatment and 76 participants requested not to receive aspirin. On this basis it was estimated that 80.4% of patients would be offered and would accept aspirin chemoprevention on diagnosis of LS.
The majority (59%) of participants randomised to aspirin or aspirin placebo were treated for ≥ 2 years. It was assumed that 59% of individuals accepting aspirin chemoprevention would receive aspirin for 4 years and would have a reduced incidence of CRC and endometrial cancer (incidence rate ratios of 0.37 and 0.49 respectively), whereas the remaining individuals would receive no aspirin and would see no change in their incidence rates.
As the estimates are taken from a study with a maximum follow-up of just over 10 years, it was assumed conservatively that the duration of effect would be 10 years.
Health-related quality of life
Systematic searches were conducted for utilities associated with CRC, endometrial cancer and prophylactic hysterectomy. The literature searches for utility studies were conducted in MEDLINE (via Ovid) and EMBASE (via Ovid). Searches consisted of population terms for hysterectomy and salpingo-oophorectomy, CRC and endometrial cancer, combined with relevant utility terminology. The searches for CRC utility studies were date limited from 2005 to February 2016, with literature published prior to 2005 identified using the study by Snowsill et al. 4 The hysterectomy and salpingo-oophorectomy and the endometrial cancer searches were not date limited. Searches for each population group were conducted separately and were then combined and deduplicated using EndNote X7. The full search strategies and the number of hits per database and in total are detailed in Appendix 1.
The searches were screened (first by title and abstract and then by full text) by one reviewer (NH), who also carried out data extraction and assessed the studies for suitability for parameterising the model.
Colorectal cancer
Twelve full texts reporting CRC and related utilities were identified, six of which reported the effect of CRC on utility estimates and six of which reported the effect of CRC treatment on utilities. An overview of the first group of studies is provided in Table 53. Several studies reported multiple utility measures, but only those that are most relevant to the review are presented here.
| Study | Population | Method | Utility measure | Results | Advantages | Limitations |
|---|---|---|---|---|---|---|
| Färkkilä 2013136 | 508 Finnish CRC patients | Cross-sectional observational survey | EQ-5D | EQ-5D change from standardised GP: primary treatment (local disease 0–6 months after diagnosis) –0.033; rehabilitation (local disease 6–18 months after diagnosis) 0.064; remission (local disease > 18 months after diagnosis) 0.046; metastatic disease (receive treatment) –0.005; palliative care –0.119 | EQ-5D; study did not influence treatment decisions; uses UK TTO tariff for EQ-5D | States based on time since diagnosis, not stage at diagnosis; non-UK based; disease severity of non-responders unknown |
| Hall 2015137 | 128 (reduced to 97 at last follow-up) CRC patients (within 6 months of diagnosis) at two NHS trusts (Leeds Teaching Hospitals NHS Trust and Calderdale and Huddersfield NHS Foundation Trust) | Hospital-based survey. Routine data collection through use of online systems at three time points | EQ-5D | < 6 months post diagnosis: 0.765 (GP 0.793); 9 months: 0.802 (GP 0.793); 15 months 0.812 (GP 0.794). At no time point was the difference in quality of life from the GP statistically significant | EQ-5D; UK NHS | Only two trusts; aim of analysis was to investigate costs and quality of life; all patients had to have internet access; only one Dukes’ D patient; Dukes’ stage unknown for 75 responders; utility by Dukes’ stage not reported |
| Hung 2013138 | 134 CRC patients in Taiwan | Descriptive and longitudinal cohort study | FACT-G | FACT-G stage IV vs. I –15.16 (7.34); III vs. I 1.10 (4.31); II vs. I –0.44 (4.02) | CRC patients; difference by stage reported – all stages represented | FACT-G not EQ-5D; no comparison with population norms; Taiwan population; small sample size for some stages |
| Mhaidat 2014139 | 74 CRC patients in Jordan | Cross-sectional study | EORTC QLQ-C30, EORTC QLC-CR29 | EORTC QLQ-C30: GHS not statistically influenced by age – median 66.67 for all; GHS by stage, median (range): I 70.83 (41.67–100), II 75 (41.67–100), III 66.67 (0–100), IV 50 (16.67–91.67) | CRC patients; all stages represented and utilities reported by stage | EORTC not EQ-5D; small sample size (14–27 patients in each stage); population from Jordan |
| Stein 2014140 | 74 mCRC patients in the UK and the Netherlands | Observational, non-interventional, cross-sectional single-visit study | EQ-5D-3L | Pre-progression 0.741 ± 0.230; post-progression 0.731 ± 0.292 | Includes UK patients; EQ-5D | mCRC patients; small sample size; utilities by CRC stage not reported |
| Wong 2013141 | 381 CRC patients at Queen Mary Hospital, Hong Kong (subgroup of colorectal neoplasms patients) | Cross-sectional study | Chinese version of SF-12v2 and SF-6D | SF-6D, mean (by stage): I 0.831 ± 0.14, II 0.858 ± 0.12, III 0.817 ± 0.13, IV 0.732 ± 0.15 | Large sample; results reported by stage | Chinese population and utility measure; one institution; no comparison with GP for SF-6D |
The findings of these results were consistent with what has been previously modelled (Table 54). As such, the base case in the current study remains unchanged from the model in Snowsill et al. ,4 with no disutility assumed for individuals diagnosed with Dukes’ A, B or C and a disutility of 0.13 for individuals diagnosed with Dukes’ D. 142,143 A scenario analysis was used to investigate the possibility of an increased disutility associated with CRC, using the figures from Ness et al. ,144 as reported previously in Snowsill et al. 4 This is consistent with other cost-effectiveness analyses that have used the same sources. 145–148
| CRC stage | Base case | Sensitivity analysis | ||
|---|---|---|---|---|
| Disutility | Source | Disutility | Source | |
| Dukes’ A | 0.00 | Ramsey et al.142 | 0.11 | Ness et al.144 |
| Dukes’ B | 0.00 | Ramsey et al.142 | 0.23 | Ness et al.144 |
| Dukes’ C | 0.00 | Ramsey et al.142 | 0.26 | Ness et al.144 |
| Dukes’ D | 0.13 | Mittmann et al.143 | 0.60 | Ness et al.144 |
Four studies149–152 all reported that more extensive colorectal surgery does not appear to impact on quality of life in CRC patients with familial cancer syndromes, including patients with LS. These studies reported findings from a variety of countries (the Netherlands, USA, Australia and New Zealand), although none were UK based. The quality-of-life questionnaires used in these studies were the European Organisation for Research and Treatment of Cancer (EORTC) QLQ C-30 or Short Form questionnaire-36 items (SF-36) and so the presented utility estimates cannot be directly compared with each other or with the EuroQol-5 Dimensions (EQ-5D); however, the message appears consistent that the type of surgery does not adversely impact quality of life in CRC patients. These findings agree with those reported in Snowsill et al. 4 and, as such, in our base case we assume no disutility for more extensive surgeries.
One study153 presented Short Form questionnaire-6 Dimensions results for US CRC survivors with or without an ostomy. The authors indicated that disutility from an ostomy could be explained by other patient characteristics. No other studies reported HRQoL results for CRC survivors according to whether or not they had received an ostomy. We therefore assumed no disutility for patients with an ostomy compared with those without in our base case.
Another study154 presented SF-36 scores for CRC patients in the Netherlands who were receiving either radiotherapy or radiotherapy and chemotherapy. The results from this study suggested that the addition of chemotherapy did not have a significant impact on quality of life. No other studies reported HRQoL results for CRC survivors according to whether or not they were receiving chemotherapy. We therefore assume no disutility according to treatment in our base case.
One study155 presented SF-36 results for 100 individuals undergoing colonoscopy for various reasons and indications, including surveillance for CRC (surveillance following CRC, n = 21; family history of CRC, n = 13), in Israel. General health was found to be comparable before and after (both immediately and 1 month after) colonoscopy for non-inflammatory bowel disease patients (n = 88), including those receiving surveillance for CRC, and no single component of the SF-36 was found to be statistically significantly different after colonoscopy. No other studies were identified that reported HRQoL for individuals receiving colonoscopy. Therefore, no disutility for asymptomatic individuals resulting from colonoscopy was assumed in the base case.
Endometrial cancer
Five studies156–160 were identified that reported endometrial cancer quality-of-life estimates; two reported the PORTEC-2 (Postoperative Radiation Therapy for Endometrial Carcinoma) Phase III RCT at multiple time points,156,160 which had the largest population (n = 427 at baseline, n = 80 at 10 years’ follow-up) and longest follow-up (maximum 10 years) and compared the quality-of-life estimates for endometrial cancer with general population estimates. This trial included stage 1, high-risk endometrial cancer and reported HRQoL from the EORTC QLQ-C30. Nout et al. 156 demonstrated an equal or improved HRQoL as time progressed after diagnosis and initial treatment (TAH-BSO) compared with the general population. Although the PORTEC-2 RCT was based in the Netherlands, this finding was supported by the results of the other three studies, which were based in Italy,159 Germany157 and Turkey. 158
We therefore used the PORTEC-2 trial as our source of disutility for endometrial cancer compared with the general population. We mapped the EORTC QLQ-C30 results to the EQ-5D using the algorithm provided by Longworth et al. ,161 which has been validated by Doble and Lorgelly. 162 Longworth et al. 161 created the algorithm to map from the EORTC QLQ-C30 to the EQ-5D based on 771 patients with multiple myeloma [Velcade as Initial Standard Therapy (VISTA trial)], breast cancer and lung cancer (Vancouver Cancer clinic). Their work was funded by the UK Medical Research Council and, as such, was conducted from a UK perspective, although the EORTC QLQ-C30 data came from international sources. The algorithm provides estimates for each of the dimensions of the EQ-5D, which can then be transformed into utilities using the results from Dolan,163 which uses a UK-validated set to estimate utilities from EQ-5D data. Doble and Lorgelly162 assessed the external validity of 10 mapping algorithms that mapped from the EORTC QLQ-C30 onto the EQ-5D using data from the Cancer 2015 prospective longitudinal study. 164 This study included 1834 patients with a range of cancer tumour sites, excluding leukaemia, and a range of disease stages, including both local and metastatic disease. Doble and Lorgelly162 reported that the algorithm created by Longworth et al. 161 was one of the most computationally heavy but also performed well on a number of criteria, including extreme health states and having no statistically significant difference between observed and predicted QALYs over time.
By estimating the baseline EQ-5D utilities in the PORTEC-2 trial (0.837 general population, 0.819 external beam radiation therapy receiving patients, 0.783 vaginal brachytherapy receiving patients) and averaging the utility across the two treatments to give a utility estimate for the endometrial cancer population, the disutility for endometrial cancer was estimated to be –0.036. As the PORTEC-2 trial indicates that quality of life improves over time for endometrial cancer patients, reverting to (or exceeding) the quality of life of the general population, this disutility is applied only for the first year following an endometrial cancer diagnosis.
Prophylactic hysterectomy
No studies were identified that could inform the disutility of prophylactic hysterectomy. In the base case we assumed that there was no disutility from prophylactic hysterectomy, to reflect our belief that quality of life would be similar to or better than the long-term quality of life for endometrial cancer patients who have received hysterectomy and recovered from cancer. In the PORTEC-2 trial this was at least as good as the general population utility and therefore results in a utility decrement of 0. In sensitivity analysis we set the disutility equal to the utility decrement from endometrial cancer, under the assumption that prophylactic hysterectomy should not have a higher disutility than endometrial cancer.
Psychological impacts of Lynch syndrome testing and management on quality of life
No additional literature was identified to that in Snowsill et al. 4 to estimate the psychological impact of LS testing on individuals offered testing for LS. Box 5 describes the existing evidence, as reported in Snowsill et al. 4
Although diagnosis of LS can lead to interventions to reduce the chance of developing colorectal, gynaecological and other cancers, it can also lead to anxiety about developing these cancers and the need to make difficult decisions about whether or not to undergo risk-reducing surgeries. Furthermore, those diagnosed with LS must decide whether or not and how to inform relatives about their test results so that these relatives can consider whether or not they wish to be tested themselves. Given that anxiety is one aspect of HRQoL, such effects should be considered in the estimation of health-state utilities of probands and relatives.
We identified just a single study of the cost-effectiveness of strategies for testing for LS [Wang and colleagues (2012)86] that incorporates disutilities associated with the psychological impact of testing. In this study it was assumed that such disutilities are transient, lasting 1 year in the base-case analysis. . . . Disutilities due to testing itself and the test results were taken from the empirical study by Kuppermann and colleagues (2013).
Reproduced from Snowsill et al. (p. 162)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
The disutility associated with testing for relatives was calculated using the utilities reported by Kuppermann et al. ,165 as previously4 (Box 6), as was the disutility associated with testing for probands4 (Box 7).
[We] assume that relatives who decline testing incur a disutility over 4 months of 0.04, equal to the utility of 0.76 (siblings who undergo testing, and test negative) minus 0.72 (siblings who decline testing). This disutility reflects anxiety the relative may feel in not knowing whether or not he or she has LS, with the corresponding substantial risk of developing cancer. Next, we assume a disutility of 0.02 for male relatives who are diagnosed with LS, equal to 0.76 for siblings who undergo testing and test negative, minus 0.74 for males who are tested positive for LS. Similarly, we assume a disutility of 0.06 for women who test positive and undergo TAHBSO, equal to 0.76 minus 0.70, and a disutility of 0.09 for women who test positive and decline TAHBSO, equal to 0.76 minus 0.67. The disutility is greater for women who decline TAHBSO presumably because they know that there remains a chance that they will develop gynaecological cancers. For women who test positive but are not offered TAHBSO as they are not at the appropriate age, we assume that the disutility of testing positive will be the same as for men who test positive, i.e. 0.02.
Reproduced from Snowsill et al. (p. 163)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
Kuppermann and colleagues did not measure the utility for probands who accepted testing and were diagnosed as LS negative. In the absence of this information, we assume that these individuals have no associated disutility due to genetic testing . . . . Next, we estimate the disutility for probands of declining testing as 0.04, equal to the corresponding value for relatives. . . . Kuppermann and colleagues do not measure the utility for probands who accepted testing and were diagnosed with LS but were not offered any risk-reducing surgery, so we assume that the disutility for testing positive for male probands is the same as the disutility for male relatives, i.e. 0.02. For female probands who test positive and are not offered any risk-reducing surgery, we again assume the same disutility as for males, i.e. 0.02. For female probands who test positive and are offered prophylactic TAHBSO, we assume disutilities of 0.03 for those accepting surgery and 0.09 for those declining it. These disutilities are estimated by subtracting the utilities of 0.67 and 0.61 reported in Kuppermann and colleagues from the imagined utility of probands testing negative, which we estimate as the utility of probands declining testing (0.66) plus a utility of 0.04 for not declining testing taken from the relatives, to give a utility for probands testing negative for LS of 0.70. If a proband or relative declines testing but is still diagnosed with LS (by FH [family history] for probands or on account of being a FDR [first-degree relative] of a known carrier for relatives) and offered TAHBSO, we assume a disutility of 0.01 for probands and 0.04 for relatives (i.e. the same disutility as for probands or relatives testing positive), with an additional disutility of 0.06 for probands declining TAHBSO and 0.03 for relatives declining TAHBSO. For example, the total disutility for a proband declining testing and accepting TAHBSO would be 0.04 (declined testing) + 0.01 (offered TAHBSO) = 0.05, while the total disutility for a relative declining testing and declining TAHBSO would be 0.04 (declined testing) + 0.04 (offered TAHBSO) + 0.03 (declined TAHBSO) = 0.11.
Reproduced from Snowsill et al. (p. 164)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
Table 55 summarises the disutilities included in the model relating to the psychological impact of genetic testing.
| Result of genetic testing | Disutility | |
|---|---|---|
| Males | Females | |
| Proband | ||
| Test declined, risk reduction not offered | 0.04 | 0.04 |
| Test declined, accept risk reduction | NA | 0.05 |
| Test declined, decline risk reduction | NA | 0.11 |
| Test accepted, LS negative | 0 | 0 |
| Test accepted, LS positive, risk reduction not offered | 0.02 | 0.02 |
| Test accepted, LS positive, accept risk reduction | NA | 0.03 |
| Test accepted, LS positive, decline risk reduction | NA | 0.09 |
| Relative | ||
| Test declined, risk reduction not offered | 0.04 | 0.04 |
| Test declined, accept risk reduction | NA | 0.08 |
| Test declined, decline risk reduction | NA | 0.11 |
| Test accepted, LS negative | 0 | 0 |
| Test accepted, LS positive, risk reduction not offered | 0.02 | 0.02 |
| Test accepted, LS positive, accept risk reduction | NA | 0.06 |
| Test accepted, LS positive, decline risk reduction | NA | 0.09 |
Resources and costs
Adjustments to 2016/17 prices
Costs were inflated to 2014/15 prices using the Hospital and Community Health Services (HCHS) pay and prices index and then to 2016/17 prices according to the average inflation in the 3 most recent recorded years (Figure 33). 166
FIGURE 33.
Historical and projected inflation of the HCHS pay and prices index. a, Projection.

Resource use
As described earlier (see Surveillance pathways, Colorectal surveillance), the model assumed that individuals diagnosed with LS would undergo biennial colonoscopy. The majority of those diagnosed with LS take up surveillance: 97% of those with a confirmed pathogenic mutation and 70% of those in whom LS is suspected without a causative mutation being identified (see Surveillance, Colonoscopy).
Although acceptance is high, it is also recognised that implementation of or concordance with the surveillance regimen is imperfect, for a number of reasons (e.g. service delivery, patient circumstances and factors). It was estimated that 12.7% of planned colonoscopies would be missed, based on a Regional Familial Colorectal Cancer Registry study by Newton et al. 167 This parameter is not assumed to affect the effectiveness of colonoscopy and therefore its impact on cost-effectiveness is not as expected – increasing this parameter in a sensitivity analysis would reduce the costs of surveillance without reducing effectiveness and would therefore improve the cost-effectiveness of diagnostic strategies. For this reason, this parameter was not subjected to sensitivity analyses.
It is important (especially for individuals with LS who are more susceptible to proximal colon cancer than the general population) that colonoscopy is complete (including intubation of the caecum). For this reason it is recommended that repeat colonoscopies are conducted when colonoscopy is incomplete. The probability of any colonoscopy being incomplete and needing repeating was estimated as 7.7%, based on a national colonoscopy audit. 168 It was assumed that, at most, one repeat colonoscopy would be performed.
It was assumed that surveillance colonoscopies for LS would start at age 25 years and end at age 75 years, based on BSG/ACPGBI guidelines. 46
Based on a national colonoscopy audit168 it was assumed that, for each 100,000 colonoscopies, there would be 260 bleeding events and 40 perforation events. It was also assumed that there would be 8.3 deaths per 100,000 colonoscopies. 46
Bleeding events were modelled only if they resulted in admission and it was estimated that 21% of bleeds would result in admission. 168 Of these, approximately 18% would be ‘moderate’ bleeds and 9% would be ‘severe’ bleeds. 168
As described in Chemoprevention, 80.4% of individuals with LS are offered aspirin chemoprevention and accept it. Of these, 59.0% are concordant with the protocol and receive aspirin for 4 years, with the remaining individuals assumed to discontinue immediately. The daily dose of aspirin modelled was 600 mg.
All CRC patients were assumed to incur the cost of diagnosis once, in the year of diagnosis.
For rectal cancer patients, 79% are estimated to be operable and, of these, 42.5% are estimated to present as emergency cases, with the remaining 57.5% undergoing elective surgery. 123 Of the emergency cases, 11% are estimated to receive postoperative chemoradiotherapy (for 2 weeks). Of the elective cases, 82% are predicted by magnetic resonance imaging to have clear margins after resection and 60% of these have preoperative chemoradiotherapy. A further 4.4% have postoperative chemoradiotherapy (11% of the 40% not receiving preoperative chemoradiotherapy). All patients not predicted to have clear margins after resection receive a course of chemoradiotherapy and 88% of these go on to have surgery. 123
Post-surgery chemotherapy courses are given to 28% of patients with Dukes’ B rectal cancer and 75% of patients with Dukes’ C rectal cancer. Chemotherapy is not given to Dukes’ A patients and Dukes’ D patients receive palliative therapy, which is detailed in Palliative care.
Adjuvant chemotherapy is given to 0% of Dukes’ A, 39% of Dukes’ B and 89% of Dukes’ C colon cancer patients. 123
Dukes’ D colon cancer patients may receive downstaging chemotherapy for liver metastases, but this is costed under recurrence chemotherapy and surgery.
All patients diagnosed with CRC are assumed to receive primary surgery, as described in Colorectal surgery pathways.
In the first 5 years after diagnosis, CRC patients follow a surveillance pathway that includes CEA tests, CT scans, colonoscopies and clinical consultations. Patients are assumed to receive 3-monthly CEA tests for the first 2 years, then 6-monthly tests for the following 3 years, plus CT scans at 12 and 24 months and an annual clinical consultation. Patients are also assumed to receive a colonoscopy at 12 months and then every 5 years thereafter (unless they already receive LS surveillance colonoscopies). Table 56 shows this pathway. As in the study by Trueman et al. ,123 a weighted average cost per year was calculated.
| Year | CEA test | CT scan | Clinical consultation |
|---|---|---|---|
| 1 | 4 | 0 | 1 |
| 2 | 4 | 1 | 1 |
| 3 | 2 | 1 | 1 |
| 4 | 2 | 0 | 1 |
| 5 | 2 | 0 | 1 |
All patients dying from CRC within 5 years of diagnosis incur the cost of surgery and chemotherapy for recurrence in the year of their death.
All patients with CRC incur average stoma care costs (i.e. the model does not actually track whether a patient has a stoma or not). It was estimated from the study by Trueman et al. 123 that 67% of rectal cancer patients would require a stoma after surgery compared with 14.5% of colon cancer patients. Of these, 26.6% would be reversed, with patients not requiring long-term stoma care. On this basis it was estimated that 11% of colon cancer patients and 49% of rectal cancer patients would require a permanent stoma.
All patients dying from CRC (at any time after diagnosis) incur the cost of palliative care in the year of their death.
Women diagnosed with LS may be offered interventions to reduce their risk of developing gynaecological cancers (in the model only endometrial cancer is included).
Women with LS were assumed to be offered gynaecological surveillance at age 35 years (in line with European Society for Medical Oncology guidelines125) or at the time of their diagnosis (whichever is later).
As described earlier (see Gynaecological state on entry), the probability that a woman diagnosed with LS opts for surveillance, prophylactic surgery or no intervention, at the time of diagnosis, was estimated based on the results of the Northern Genetics Service audit (Table 57) by dividing the number of women opting for each intervention in each age range (except those aged ≤ 35 years) by the total number of women, excluding those with previous cancer or whose results were not recorded. The resulting probabilities of opting for surveillance or prophylactic surgery at the time of diagnosis are provided in Table 58.
| Age group (years) | Discussed only | Surveillance | Prophylactic surgery | Previous cancer | Not recorded |
|---|---|---|---|---|---|
| ≤ 35 | 10 | 6 | 0 | 0 | 1 |
| 36–45 | 3 | 9 | 3 | 0 | 1 |
| 46–60 | 4 | 11 | 9 | 6 | 2 |
| > 60 | 0 | 2 | 12 | 4 | 4 |
| Age at diagnosis (years) | Surveillance (%) | Prophylactic H-BSO (%) |
|---|---|---|
| < 35 | 0.0 | 0.0 |
| 35–44 | 60.0 | 20.0 |
| 45–59 | 45.8 | 37.5 |
| 60–69 | 14.3 | 85.7 |
| ≥ 70 | 0.0 | 85.7 |
Women diagnosed with LS can change their risk reduction subsequently. It was assumed that all women still receiving surveillance would stop receiving surveillance at age 70 years. It was assumed that at age 35 years women would be offered surveillance or prophylactic H-BSO and at age 45 years and 60 years women would be offered prophylactic H-BSO. The probability of receiving prophylactic H-BSO at age 45 years was estimated as 21.9% to achieve the desired increase in H-BSO between the 35–44 and 45–59 years age groups (17.5 percentage points). Similarly, it was estimated that 77.1% of women would receive prophylactic H-BSO at age 60 years (Table 59).
| Reaching age (years) | Surveillance (%) | Prophylactic H-BSO (%) |
|---|---|---|
| 35 | 60.0 | 20.0 |
| 45 | 0.0 | 21.9 |
| 60 | 0.0 | 77.1 |
As previously,4 endometrial cancer treatment was assumed to consist of surgery for all patients and adjuvant radiotherapy and/or chemotherapy for some patients.
All women diagnosed with endometrial cancer were assumed to receive surgery.
It was assumed that radiotherapy was given to 33% of stage I patients, 100% of stage II/III patients and 0% of stage IV patients. 169 It was estimated that this results in radiotherapy being given to 47% of patients overall.
It was assumed that chemotherapy was given to 0% of stage I patients, 50% of stage II/III patients and 100% of stage IV patients. 169 It was estimated that this results in chemotherapy being given to 18% of patients overall.
We modelled a combination regimen of carboplatin and paclitaxel (Table 60). This has been noted to be a popular and reasonable regimen in the British Gynaecological Cancer Society draft guidelines,170 which also state that there is currently no evidence to conclude that any adjuvant chemotherapy regimen is superior to any other.
| Day | Drug | Dose | Administration |
|---|---|---|---|
| 1 | Paclitaxel | 175 mg/m2 | IV over 3 hours |
| 1 | Carboplatin | AUC 5–6 mg · minute/ml | IV over 60 minutes |
The dose required to obtain a specific carboplatin area under the curve (AUC) is given by the Calvert formula:171
We assumed an average glomerular filtration rate of 82.7 ml/minute, based on a Belgian study of 1218 patients. 172 We therefore obtained a dose of 592.35 mg of carboplatin per cycle (assuming a target AUC of 5.5 mg · minute/ml).
We assumed an average body surface area of 1.71 m2, based on the average body surface area for women with cancer, as reported by Sacco et al. ,173 which leads to a dose of 299 mg of paclitaxel.
Unit costs
The costs of the preliminary tumour tests were obtained directly from laboratories in the UK (Table 61). When possible, these were sourced from multiple genetics services through the UK Genetic Testing Network (UKGTN) to produce costs that were representative across the UK. Unit costs from personal communications received during the completion of this report and from Snowsill et al. 4 were inflated to 2016/17 prices.
| Test | Patient | Base-case cost (£)a | Base-case source |
|---|---|---|---|
| MSI | Proband | 202 | Average from UKGTN175 (reported for Newcastle, Oxford, Birmingham and Sheffield) |
| IHC | Proband | 210 | Average of UKGTN175 (reported for University College London only), Dr Mark Arends (Department of Pathology, University of Cambridge, 31 October 2012, personal communication) and Dr Ian M Frayling (personal communication) (£220–240) |
| BRAF V600E | Proband | 119 | Average of £140 (Mr Michael Gandy, UCL Advanced Diagnostics, 14 September 2012, personal communication), £117 (East of Scotland Regional Genetic Service174) and £85 (All Wales Molecular Genetics Laboratory176) from Snowsill et al.4 and £65 (North West Regional Genetics Service175) |
| MLH1 promoter methylation testing | Proband | 136 | UKGTN175 (reported for Newcastle, Cardiff and London) |
| Proband genetic test, all four genes | Proband | 1276 | UKGTN175 (weighted average of available tests) |
| Proband genetic counselling | Proband | 64 | PSSRU166,177 and Professor Mary Porteous (South East Scotland Genetic Service, 2013, personal communication) |
| Targeted genetic test for relatives (MLH1) | Relative | 166 | UKGTN175 |
| Targeted genetic test for relatives (MSH2) | Relative | 161 | UKGTN175 |
| Targeted genetic test for relatives (MSH6) | Relative | 161 | UKGTN175 |
| Targeted genetic test for relatives (PMS2) | Relative | 165 | UKGTN175 |
| Genetic counselling for relatives | Relative | 64 | PSSRU166,177 and Professor Mary Porteous (personal communication) |
Costs for the genetic tests (for probands and relatives) were taken directly from genetic testing laboratories, as reported through the UKGTN. 175 As in the study by Snowsill et al. ,4 only costs applicable to the NHS were collected (private fees were excluded when possible) and the overall cost for each test was calculated across the laboratories that supply that test. The available genetic tests were individual sequencing tests for probands for MLH1, MSH2, MSH6 and PMS2; individual targeted tests for relatives for MLH1, MSH2, MSH6 and PMS2; a combined MLH1 and MSH2 sequencing test for probands; a combined MSH2 and MSH6 sequencing test for probands; a combined MLH1, MSH2 and MSH6 sequencing test for probands; and, at one centre, a combined sequencing test for all four genes. As the PenTAG model assumed that probands will receive testing for four genes (MLH1, MSH2, MSH6 and PMS2), the total cost for testing all four genes was based on the average of the plausible combinations of tests that can be found in current practice. This cost does not include the added costs of having to use multiple centres to produce the results. There is some indication that laboratories are increasingly testing MMR genes as part of NGS multigene panels (based on clinical opinion from Dr Ian M Frayling, 2016). The result of this is both to reduce costs and to increase the yield of conditions causative of CRC. Costs were therefore explored in univariate sensitivity analyses.
As in the study by Snowsill et al. ,4 genetic counselling was assumed to occur after initial tumour testing and before genetic testing. Genetic counselling remains a parameter without a standard unit cost. As such, the cost was calculated using the same approach as detailed in Snowsill et al. ,4 with updating to 2016/17 prices. To calculate the time and staff involved in genetic counselling, Snowsill et al. 4 corresponded with Professor Mary Porteous of the South East Scotland Genetic Service, based at the Western General Hospital in Edinburgh (2013, personal communication). Box 8 details the resulting resource use involved in genetic counselling.
In this centre genetic counselling occurred, where applicable, after the tumour tests (IHC and/or MSI) for the proband. Generally, probands received a maximum of a single 45-minute session with a band 7 counsellor before gene testing, and a 30-minute session to discuss the results. The same was also true of the relatives, though in practice the total 75 minutes could be split in various ways (for example, sometimes relatives would have a group session then return for a shorter individual session before they were tested). In this centre the cost of genetic counselling incurred for a relative of a proband was therefore the same as that for the proband.
Reproduced from Snowsill et al. (p. 168)4 under the terms of the UK government’s non-commercial licence for public sector information. 32
The cost per hour of the band 7 counsellor (£50) was taken from the 2014 edition of Unit Costs of Health and Social Care177 and uprated to 2016 costs, as no equivalent cost was available from the most recent edition. The cost per hour of a band 5 hospital nurse (£43) was taken from Unit Costs of Health and Social Care 2015166 and uprated to 2016 costs.
As different genetic centres have different approaches to genetic counselling, sensitivity analyses were performed on the cost of counselling, with the time spent on each individual halved and doubled.
The unit costs of surveillance colonoscopies for individuals with LS were estimated from NHS Reference Costs 2014–15178 and uprated to 2016/17 prices, as described in Adjustments to 2016/17 prices. The Healthcare Resource Group (HRG) codes FZ51Z, FZ52Z and FZ53Z were used as previously,4 giving a unit cost of £585.80 per colonoscopy (Table 62).
| HRG code | Description | Number of colonoscopies | Unit cost (£) | Total cost (£) |
|---|---|---|---|---|
| FZ51Z | Diagnostic Colonoscopy, ≥ 19 years | 162,933 | 519.42 | 84,630,117 |
| FZ52Z | Diagnostic Colonoscopy with Biopsy, ≥ 19 years | 153,795 | 604.02 | 92,894,549 |
| FZ53Z | Therapeutic Colonoscopy, ≥ 19 years | 116,071 | 601.86 | 69,858,823 |
| Weighted average (2014–15 prices) | 432,799 | 571.59 | 247,383,489 | |
| Weighted average (2016–17 prices) | 585.80 | |||
The unit costs of complications from colonoscopies were estimated from NHS Reference Costs 2014–15178 (Table 63).
| Complication | HRG code | Unit cost (£) | |
|---|---|---|---|
| 2014/15 | 2016/17 | ||
| Bleed requiring admission | |||
| Mild | FZ38P Gastrointestinal Bleed without Interventions, with CC Score 0–4a | 462 | 473 |
| Moderate | FZ38J–FZ38L Gastrointestinal Bleed with Single Interventiona | 1110 | 1138 |
| Severe | FZ38G–FZ38H Gastrointestinal Bleed with Multiple Interventionsb | 4287 | 4394 |
| Perforation | FZ77C–FZ77E Major Large Intestine Procedures, 19 years and overb | 4790 | 4909 |
| Death | Assumed same as perforation | 4790 | 4909 |
As previously,4 many of the costs associated with CRC in the model are based on the work of Trueman et al. 123 The main exception to this is the cost of primary surgery (see Primary surgery).
The cost of diagnosis is incurred at the time of CRC diagnosis and is estimated to be £1022 in 2016/17 prices (inflated from £790 in 2004/5 prices). 123
In addition, it was assumed that patients with stage II (Dukes’ B) colon cancer will receive MSI testing to predict their response to 5-FU-based chemotherapy, at a cost of £202 (see Table 61).
Trueman et al. 123 modelled pre/postoperative chemoradiotherapy and adjuvant chemotherapy.
In the study by Trueman et al. 123 the cost of pre/postoperative chemoradiotherapy was £2263 (2004/5 prices) and the cost for a full chemotherapy course after surgery was £11,209 (2004/5 prices).
When weighted by resource use, the relevant costs according to rectal cancer stage are shown in Table 64.
| Rectal cancer stage | Pre/postoperative chemoradiotherapy | Chemotherapy | Total (£) | ||
|---|---|---|---|---|---|
| % | Subtotal (£) | % | Subtotal (£) | ||
| Dukes’ A | 35.9 | 1049.30 | 0 | 0.00 | 1049.30 |
| Dukes’ B | 35.9 | 1049.30 | 21.8 | 3156.37 | 4205.67 |
| Dukes’ C | 35.9 | 1049.30 | 58.3 | 8454.57 | 9503.87 |
The cost of chemotherapy for colon cancer was £11,209 in 2004/5 prices in the study by Trueman et al. ,123 corresponding to a cost of £14,494 in 2016/17 prices.
As previously,4 the costs of CRC surgery were estimated according to the type of surgery performed and whether the patient had LS or not (as proximal CRCs are more common in individuals with LS). Stoma reversal costs were included as previously. 4 Unit costs were updated using NHS Reference Costs 2014–15178 and uprated to 2016/17 prices (Tables 65 and 66).
| Surgery | HRG code | Unit cost (£2014/15) (£) | Unit cost (£2016/17) (£) |
|---|---|---|---|
| Segmental resection (proximal without exteriorisation) | FZ75 Proximal Colon Procedures, 19 years and over | 6286.10 | 6442.38 |
| Segmental resection (distal without exteriorisation) | FZ76 Distal Colon Procedures, 19 years and over | 5920.68 | 6067.87 |
| Segmental resection (with exteriorisation) | FZ74 Complex Large Intestine Procedures, 19 years and over | 7671.24 | 7861.95 |
| Subtotal colectomy with ileorectal anastomosis | |||
| AR | |||
| Proctocolectomy with IPAA | |||
| Stoma reversal | FZ50 Intermediate Large Intestine Procedures, 19 years and over | 420.77 | 431.23 |
| Surgical extent | Unit cost (£)a |
|---|---|
| Segmental resection | |
| General population | 6500.58 |
| LS | 6604.77 |
| Subtotal colectomy with ileorectal anastomosis | 7878.59 |
| Rectal excision | 7938.81 |
| Proctocolectomy | 7976.66 |
We estimated that CEA tests cost £13.64 each (by inflating the cost of £10.55 from Trueman et al. 123). CT scans were assumed to include three areas with contrast, at a cost of £127.63 (inflated from £124.53 in NHS Reference Costs 2014–15178). Clinical consultations were estimated to cost £128.17 each, inflated from £125.06 in NHS Reference Costs 2014–15 (consultant-led, service code 104 colorectal surgery).
Trueman et al. 123 estimated 5 years of surveillance costs for CRC, from which we derived an average cost per person-year of £232 for rectal cancer and £229 for colon cancer, as shown in Table 67.
| Year | Rectal cancer patients | Colon cancer patients | Unit cost (£)a |
|---|---|---|---|
| 1 | 7029 | 17,209 | 182.74 |
| 2 | 4570 | 11,142 | 310.36 |
| 3 | 2533 | 7008 | 283.08 |
| 4 | 1316 | 4356 | 155.45 |
| 5 | 571 | 2337 | 155.45 |
| Total person-years | 16,019 | 42,052 | |
| Total cost (£) | 3,713,208 | 9,627,079 | |
| Average cost per person-year (£) | 231.80 | 228.93 |
The model does not distinguish between colon and rectal cancer after the year of diagnosis, so these costs were applied as an average, weighted according to whether the patient has LS or not. Corresponding costs of £229 and £230 were used for patients with and without LS respectively.
In addition to this the model includes colonoscopy every 5 years starting at 12 months, at a cost of £586 (see Colorectal surveillance).
A single cost of £11,999 was applied if a patient died within 5 years from diagnosis of rectal cancer and a single cost of £12,354 was applied if a patient died within 5 years from diagnosis of colon cancer. These costs were inflated from estimates of £9279 and £9554 (2004/5 prices) in the study by Trueman et al. 123
As before, as the model does not track the site of cancer after the year of diagnosis, these were applied as weighted averages of £12,333 and £12,236 for patients with and without LS respectively.
The annual cost of stoma care was estimated as £1279 (2004/5 prices) by Trueman et al. 123 and this was inflated to £1654 in 2016/17 prices.
Based on 11% of colon cancer and 49% of rectal cancer patients requiring a permanent stoma, annual average costs of stoma care of £214 and £388 were applied to patients with or without LS respectively.
Trueman et al. 123 estimated costs of £7703 and £7016 for palliative care for colon and rectal cancer patients respectively. These were inflated to £9961 and £9072 in 2016/17 prices and then used to estimate costs of £9907 and £9665 for patients with and without LS respectively.
Gynaecological surveillance was assumed to include annual CA-125 testing, gynaecological examination, transvaginal ultrasound and endometrial aspiration biopsy. The total annual cost was estimated to be £473.41.
The cost of CA-125 testing was estimated to be £21.71, based on an approximate cost of £20 given in a NHS news story in 2011. 179
The cost of a gynaecological examination was estimated to be £122.93. This was based on the NHS reference cost for HRG code WF01A for consultant-led, non-admitted face-to-face attendance (follow-up) in the gynaecology service (service code 502) (£119.95 in 2014/15 prices). 178
The cost of transvaginal ultrasound was estimated to be £160.65. This was based on the NHS reference cost for HRG code MA36Z (Transvaginal Ultrasound) (£156.75 in 2014/15 prices).
The cost of endometrial aspiration biopsy was estimated to be £168.12. This was based on the NHS reference cost for HRG code MA25Z (Minimal Upper Genital Tract Procedures) (£164.04 in 2014/15 prices).
The cost of prophylactic gynaecological surgery (H-BSO) was estimated to be £3428. This was based on an average cost of £3345 for HRG codes MA07E–MA07G (Major Open Upper Genital Tract Procedures) and MA08A–MA08B (Major, Laparoscopic or Endoscopic, Upper Genital Tract Procedures). 178
The cost of surgical management of endometrial cancer was estimated to be £4005. This was based on an average cost of £3907 for HRG codes MA06A–MA06C (Major, Open or Laparoscopic, Upper or Lower Genital Tract Procedures for Malignancy). 178
Havrilesky et al. 169 estimated a cost of US$7895 for a course of radiotherapy for endometrial cancer. It was estimated that this corresponded to a cost of £5870 in 2016/17 prices (based on the same methodology as previously employed4).
The Electronic Market Information Tool (eMit) database180 was used to estimate the average cost of carboplatin (4.41 pence/mg) and paclitaxel (8.09 pence/mg), reflecting average acquisition costs (weighted across pack sizes by the total number of milligrams reported).
The cost of administering the chemotherapy regimen was estimated to be £399.09 per cycle. This was based on a cost of £389.41 for HRG code SB14Z (Deliver Complex Chemotherapy, Including Prolonged Infusional Treatment, at First Attendance). 178
The total cost for a course of chemotherapy was estimated to be £1797.63 per patient receiving chemotherapy.
It was assumed that aspirin would be prescribed by general practitioners and dispensed in the community; therefore, the reference case unit cost is the list price or the price on the NHS drug tariff.
A pack of 100 enteric coated tablets of 300 mg aspirin cost £20.33 in the BNF181 and £20.34 on the NHS drug tariff. 182 The cost of £20.33 was used, which corresponds to a daily cost (600 mg) of £0.41 and an annual cost of £148.51.
Quality assurance
The independent economic assessment was conducted by extending the model from Snowsill et al. ,4 which had already been quality assured. All parts of this model had been checked by at least one developer not responsible for developing that component, using code review and black box testing.
Extensions to the previous model were highlighted as requiring checking and were then checked by the developer not responsible for developing that extension. The checking involved code review and checking that input parameters matched the described sources. A parallel build was also conducted for one component. This revealed a small discrepancy in the calculated results, which was then resolved through discussion.
Cost-effectiveness results
The population simulated in the model consists of probands (individuals diagnosed with CRC for whom different diagnostic strategies for LS may be employed) and relatives who would be identified if a LS-causing mutation were diagnosed in the proband. As the majority of probands do not have LS, the majority of relatives also do not have LS.
All costs and QALYs in the results section are discounted at 3.5% per annum (unless otherwise stated) and life-years are not discounted.
Base-case results
Characteristics of the simulated population
In the base case there are 238,175 simulated individuals (reflecting an annual cohort), of whom 34,025 (14.3%) are probands (i.e. individuals with a CRC diagnosis) and 204,150 (85.7%) are relatives of probands. Of the probands, 956 (2.8%) are expected to have LS, with 2524 (1.2%) relatives having LS (Table 68). Of the probands, 55.0% are men, whereas only 37.6% of relatives are men.
| Simulated individuals | With LS, n (%) | Without LS, n (%) | Total, n (%) |
|---|---|---|---|
| Probands | 956 (2.8) | 33,069 (97.2) | 34,025 (100) |
| Relatives | 2524 (1.2) | 201,626 (98.8) | 204,150 (100) |
| Total | 3480 | 234,695 | 238,175 |
The average age of probands at the time of diagnosis was 72.7 years for those without LS and 58.0 years for those with LS, reflecting the widely observed earlier age of CRC incidence in individuals with LS. The mean age at entry for relatives was 44.4 years without LS and 43.2 years with LS.
Cost-effectiveness results
Table 69 reports the summary cost-effectiveness results for the 10 different testing strategies. We present both ICERs compared with no testing (strategy 1) and the comparative ICERs for all strategies. The optimal strategy [highest incremental net health benefit (INHB) at a willingness-to-pay threshold of £20,000 per QALY gained] is IHC testing followed by both BRAF V600E and MLH1 promoter methylation testing (strategy 5). Universal genetic testing has the highest ICER compared with no testing, at £25,884 per QALY gained.
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing (QALY) | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,508,052 | 743,298,306 | – | – | – |
| 2. IHC | 3,510,017 | 767,955,447 | 12,553 | 731.5 | 60,967 |
| 3. IHC plus BRAF V600E | 3,509,977 | 765,532,726 | 11,553 | 812.9 | 37,495 |
| 4. IHC plus MLH1 promoter methylation | 3,509,965 | 765,535,788 | 11,672 | 793.3 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,509,937 | 764,048,240 | 11,005 | 848.0 | 11,008 |
| 6. MSI | 3,509,926 | 769,249,096 | 13,849 | 576.3 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,509,832 | 763,660,095 | 11,438 | 762.0 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,509,796 | 763,503,459 | 11,589 | 733.2 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,509,721 | 761,784,044 | 11,076 | 744.7 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,509,987 | 793,380,127 | 25,884 | –569.2 | Dominated by strategy 2 |
Figure 34 shows that, with the exception of universal genetic testing, all testing strategies are cost-effective at a willingness-to-pay threshold of £20,000 per QALY compared with the no-testing strategy (strategy 1).
FIGURE 34.
Incremental discounted costs and QALYs for all probands and relatives.

In a fully incremental analysis, four strategies are on the cost-effectiveness frontier: 1 (no testing), 2 (IHC), 3 (IHC plus BRAF V600E testing) and 5 (IHC plus BRAF V600E and MLH1 promoter methylation testing). The remaining strategies are dominated (i.e. are more costly and less effective than one or more comparator) or extendedly dominated (i.e. are more costly and less effective than a combination of other comparators).
We now examine the drivers of these cost-effectiveness results by looking at the test accuracy, life expectancy and costs of each strategy.
Test accuracy results
Figure 35 shows the number of diagnoses made by each strategy. Of most interest is the number of incorrect diagnoses. Aside from strategy 1, strategy 9 (MSI followed by BRAF V600E and MLH1 promoter methylation testing) has the highest number of FNs (people with undiagnosed LS) and the lowest number of FPs (people diagnosed with LS who do not have it). This means that strategy 9 will likely have few unnecessary LS prevention costs, but more cancer treatment costs. Universal genetic testing has the lowest number of FNs, which is likely to reduce cancer treatment costs. The highest number of FPs is found in strategy 2 (IHC) and strategy 6 (MSI), which is likely to lead to additional unnecessary surveillance costs. Only strategies in which only one test is used in a sequence have a higher false-positive rate than false-negative rate. This makes sense as multiple tests in a sequence are used to enrich a population and reduce the number of FPs.
FIGURE 35.
Number of probands and relatives identified by each strategy. For a description of each strategy see Table 69. Note: true negatives have not been shown in the interest of clarity. The number of true negatives is substantially larger than the numbers for the other three diagnoses.

The sensitivities and specificities of the different strategies are shown in Table 70. These outputs can be calculated for probands only or for the overall population (including probands and relatives). As can be seen, diagnostic performance follows a similar pattern in the two populations, but sensitivity and specificity are generally lower in the overall population. Performance is reduced through two mechanisms. First, in the event of a positive diagnosis but without identification of a causative mutation, all first-degree relatives are treated as though they have LS and more distant relatives are treated as though they do not have LS. Second, some relatives opt not to receive predictive testing for LS.
| Strategy | Probands only | Overall population | ||
|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |
| 1. No test | 0.00 | 100.00 | 0.00 | 100.00 |
| 2. IHC | 70.35 | 99.80 | 69.67 | 99.47 |
| 3. IHC plus BRAF V600E | 69.40 | 99.94 | 68.73 | 99.55 |
| 4. IHC plus MLH1 promoter methylation | 68.93 | 99.93 | 68.26 | 99.55 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 68.04 | 99.97 | 67.38 | 99.57 |
| 6. MSI | 66.80 | 99.72 | 66.17 | 99.46 |
| 7. MSI plus BRAF V600E | 64.13 | 99.93 | 63.51 | 99.58 |
| 8. MSI plus MLH1 promoter methylation | 62.79 | 99.93 | 62.18 | 99.58 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 60.28 | 99.98 | 59.69 | 99.63 |
| 10. Universal genetic testing | 71.43 | 99.98 | 71.53 | 99.55 |
Long-term clinical outcomes
Figures 36 and 37 give the estimated life expectancy for probands and relatives according to LS status. As expected, life expectancy is much higher in relatives (they are more likely to enter the model at a younger age and they are healthy at the time of entry to the model). We found that testing for LS improves the life expectancy of both relatives and probands, a probable consequence of the reduction in CRC (index and metachronous) and endometrial cancer from the preventative measures offered to patients in whom LS is diagnosed. With regard to probands, LS probands consistently have longer life expectancy than probands without LS because LS patients generally have a better prognosis for CRC than the general population.
FIGURE 36.
Life expectancy of probands.
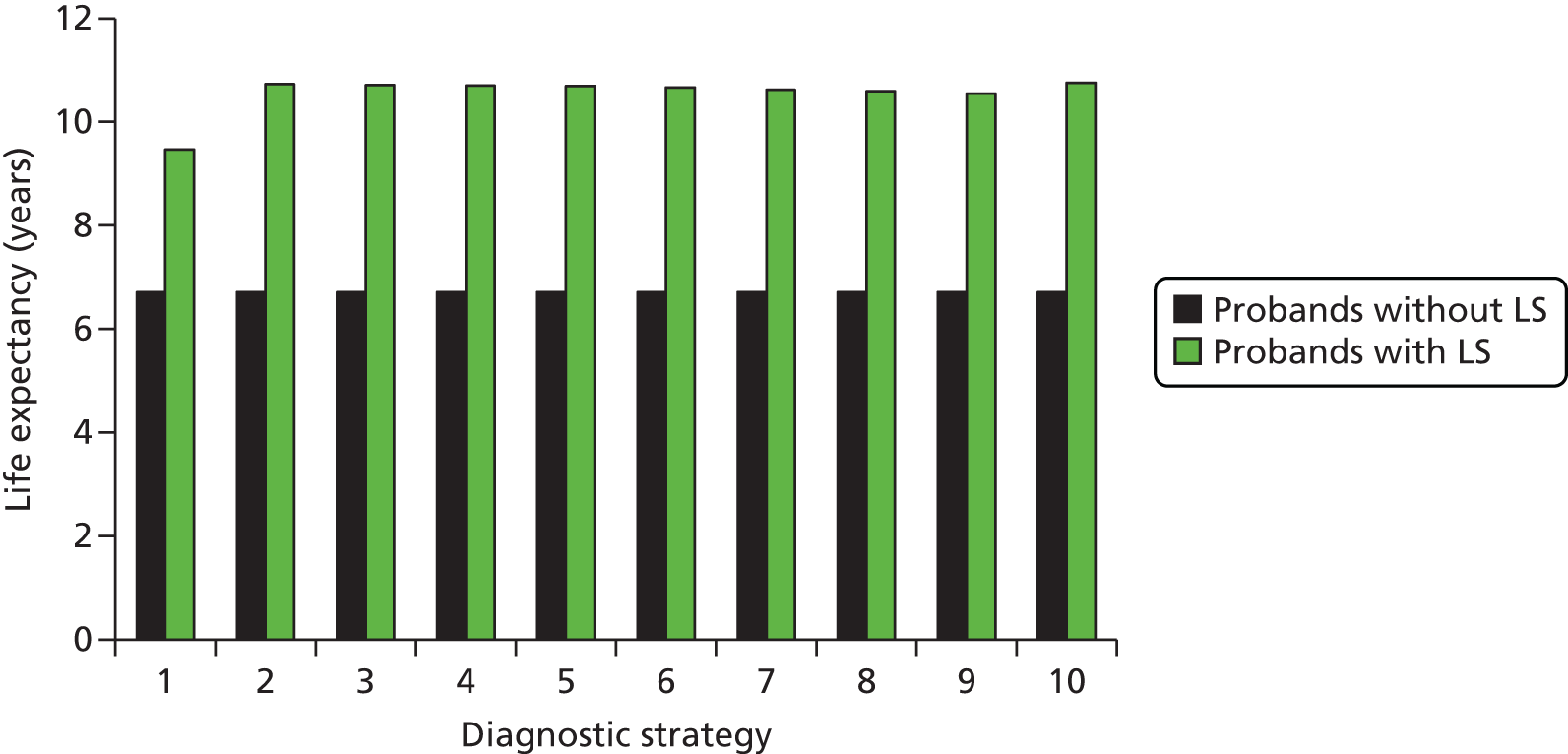
FIGURE 37.
Life expectancy of relatives.

The life expectancy of relatives is generally lower for those with LS as they are at increased risk of CRC and endometrial cancer (women only).
Kaplan–Meier graphs for alternative strategies were produced, applying weights to simulated individuals according to each strategy.
Figures 38 and 39 show the overall survival for probands and relatives with LS respectively. Kaplan–Meier graphs are not shown for individuals without LS as there is no separation of the survival functions.
FIGURE 38.
Overall survival for probands with LS.

FIGURE 39.
Overall survival for relatives with LS.

Figure 38 shows that testing for LS has no immediate impact on the overall survival of probands, as it is not assumed to improve survival for the index CRC but rather to reduce the risk of mCRC. This is why the overall survival curves do separate after a time, when the hazard of mortality from CRC has lowered.
Figure 39 shows that implementing testing results in sustained improvements in overall survival for relatives with LS. In reality, there would be expected to be a slightly greater delay before improvement (relatives are often not diagnosed with LS at the same time as the proband).
Event-free survival (i.e. the time to death, CRC incidence or endometrial cancer incidence) is also improved for probands and relatives with LS when a testing strategy is employed (Figures 40 and 41 respectively).
FIGURE 40.
Event-free survival for probands with LS.

FIGURE 41.
Event-free survival for relatives with LS.
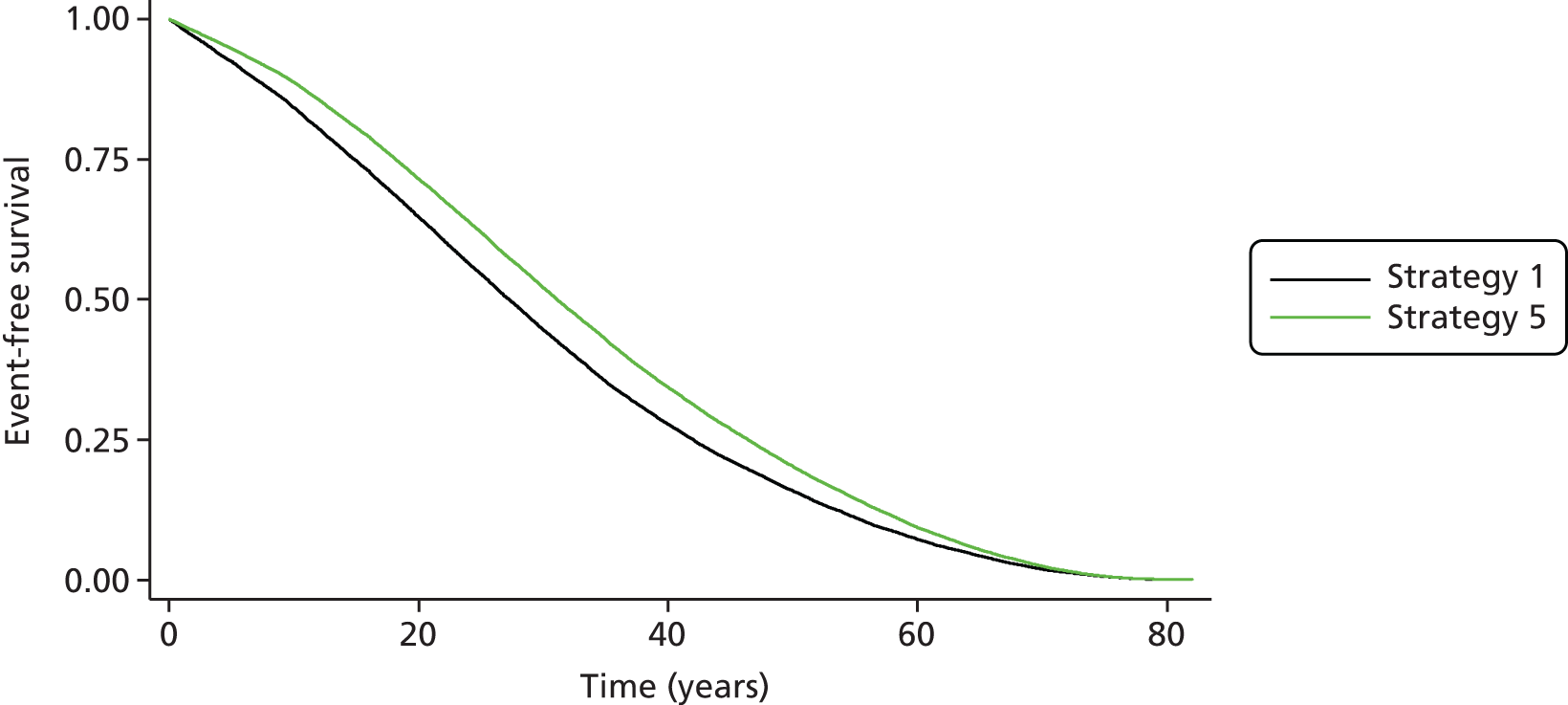
Additional outcomes
The average number of colonoscopies for individuals who actually have LS is affected significantly by the introduction of testing (Figure 42). In the absence of testing the expected number of colonoscopies is 0.42 for relatives and 1.58 for probands. Probands will receive 5-yearly colonoscopy as follow-up, as will relatives who develop CRC. With testing the number of colonoscopies increases significantly. Probands receive around 3 colonoscopies on average, whereas relatives receive 7.18–8.55 colonoscopies, depending on the strategy. The variation in the number of colonoscopies here is entirely driven by the number of false-negative diagnoses.
FIGURE 42.
Average number of colonoscopies for individuals with LS.

The average number of colonoscopies for individuals without LS is not significantly affected by the introduction of testing. There is a small increase in the average number of colonoscopies per relative. This small increase in the average number of colonoscopies does, however, correspond to a large increase in the total number of colonoscopies (Figure 43), as relatives without LS make up the largest group of modelled individuals. For each annual cohort the model predicts around 10,000 additional colonoscopies in relatives without LS (these may reasonably be considered unnecessary). This increase is attributable to the number of false-positive diagnoses.
FIGURE 43.
Total number of colonoscopies for individuals without LS.
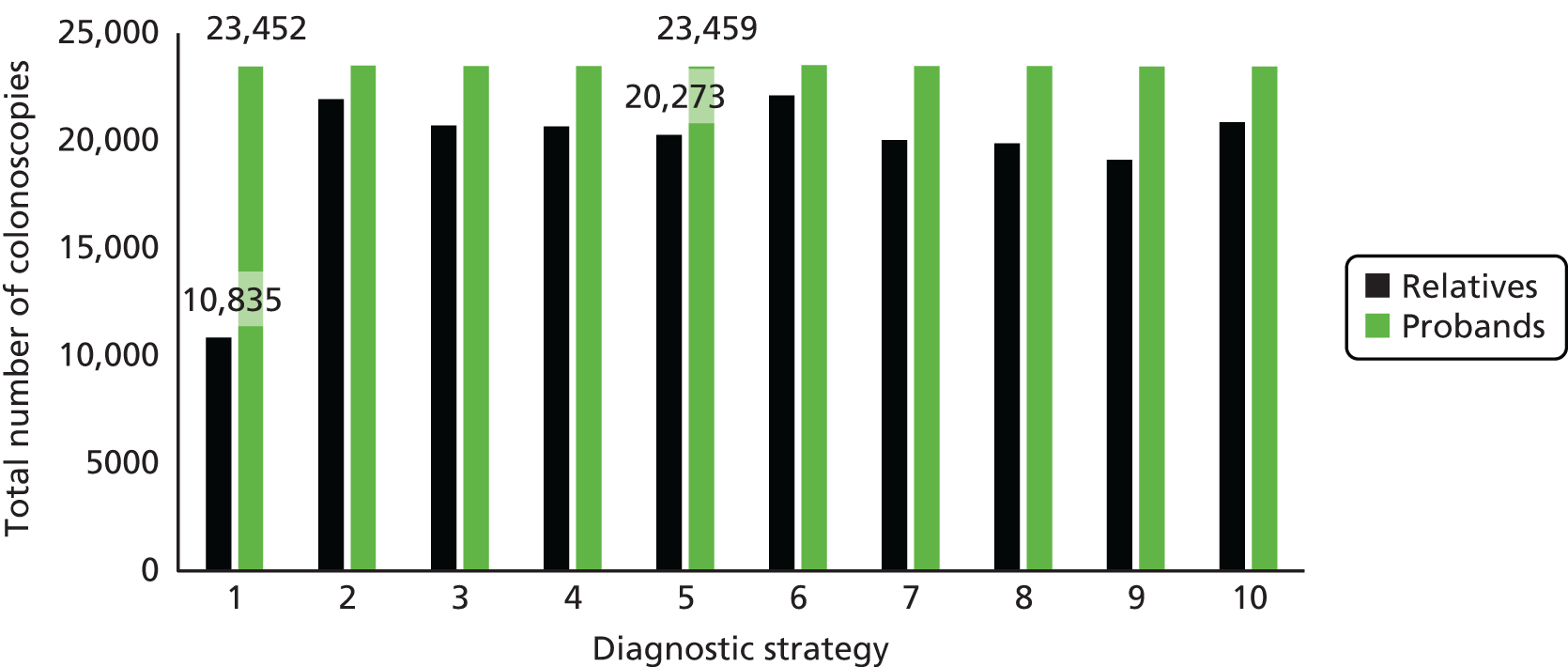
The introduction of testing for LS is expected to reduce the probability of subsequent CRC incidence for individuals with LS (Figure 44). There is very little impact on the probability of CRC incidence for individuals without LS, as testing leads to increased surveillance in these individuals only in the event of a false-positive diagnosis.
FIGURE 44.
Probability of CRC incidence for individuals with LS.

Testing is also predicted to reduce the risk of endometrial cancer for probands and relatives with LS (Figure 45). The majority of such cancers occur in relatives because of their greater number and life expectancy.
FIGURE 45.
Number of endometrial cancers in individuals with LS.
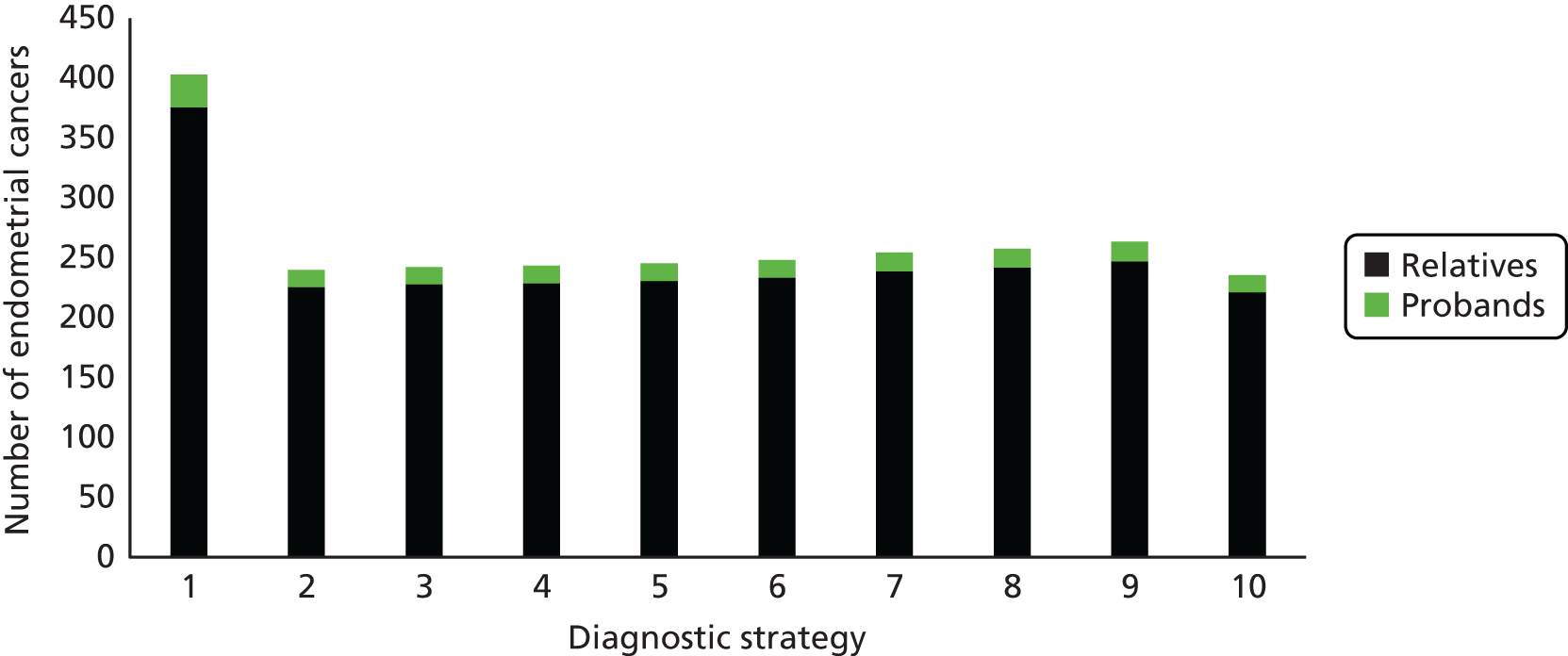
Disaggregated costs
Figure 46 shows that the largest cost component for each strategy is the long-term costs. However, given the small incremental difference between each arm, we see that the diagnostic costs are influential in terms of the overall incremental costs. We explore these in more detail in Figures 47–49.
FIGURE 46.
Summary total undiscounted costs.

FIGURE 47.
Undiscounted diagnostic costs: base case. FH, family history.
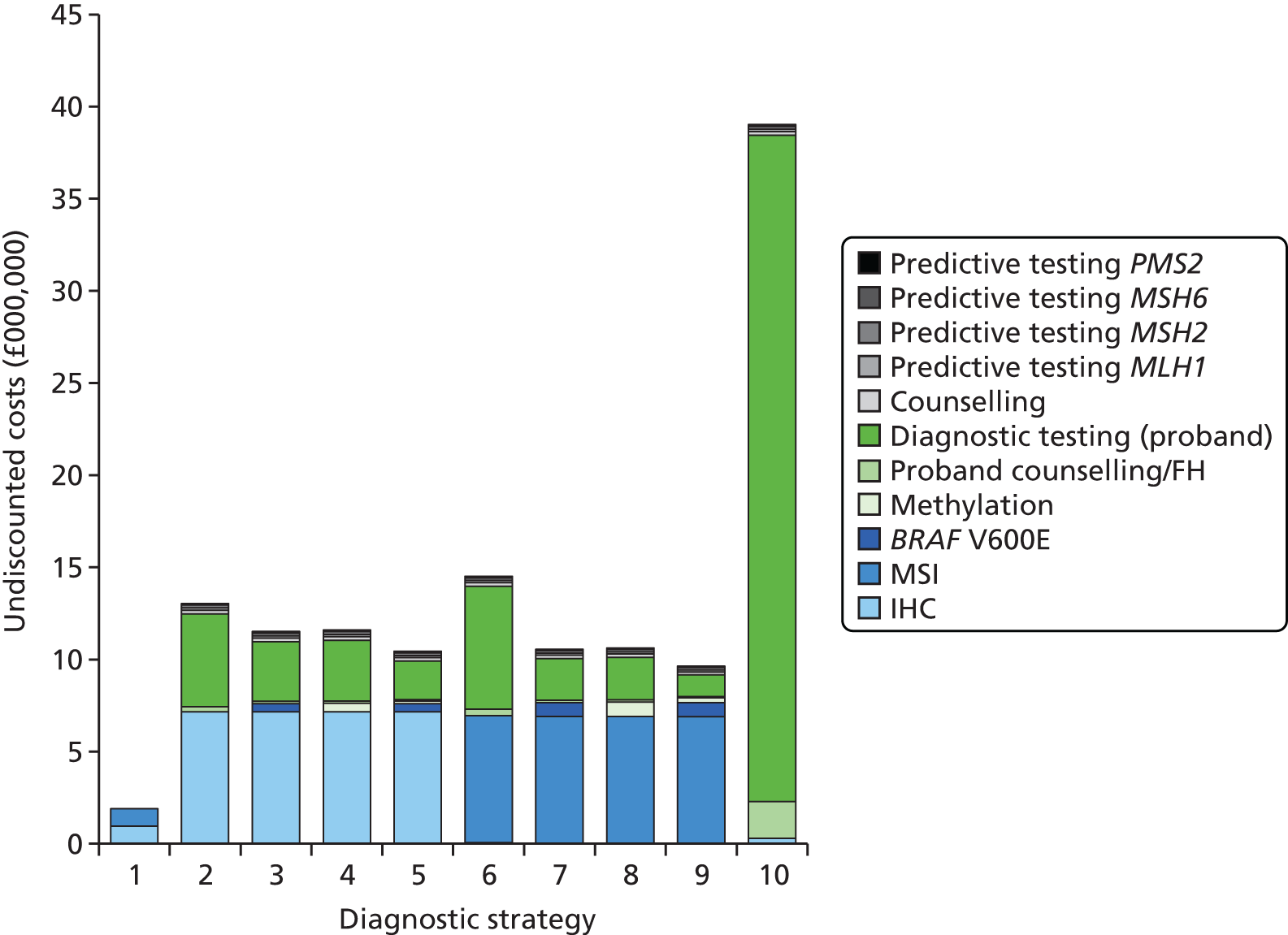
FIGURE 48.
Long-term costs: base case. EC, endometrial cancer.
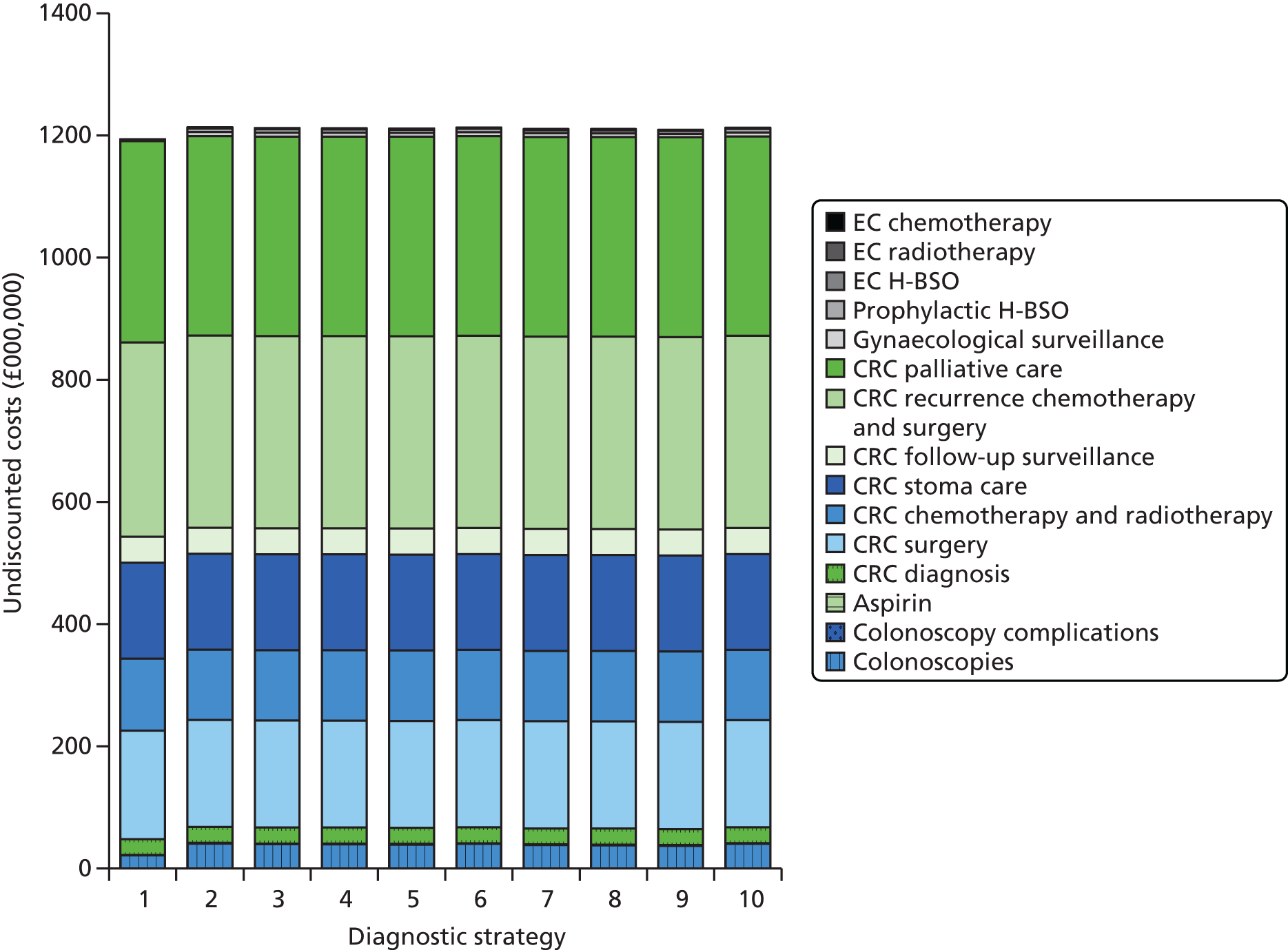
FIGURE 49.
Incremental discounted costs compared with no testing. EC, endometrial cancer. Note: the cost of aspirin is included in the CRC prevention costs although it also reduces the risk of EC.
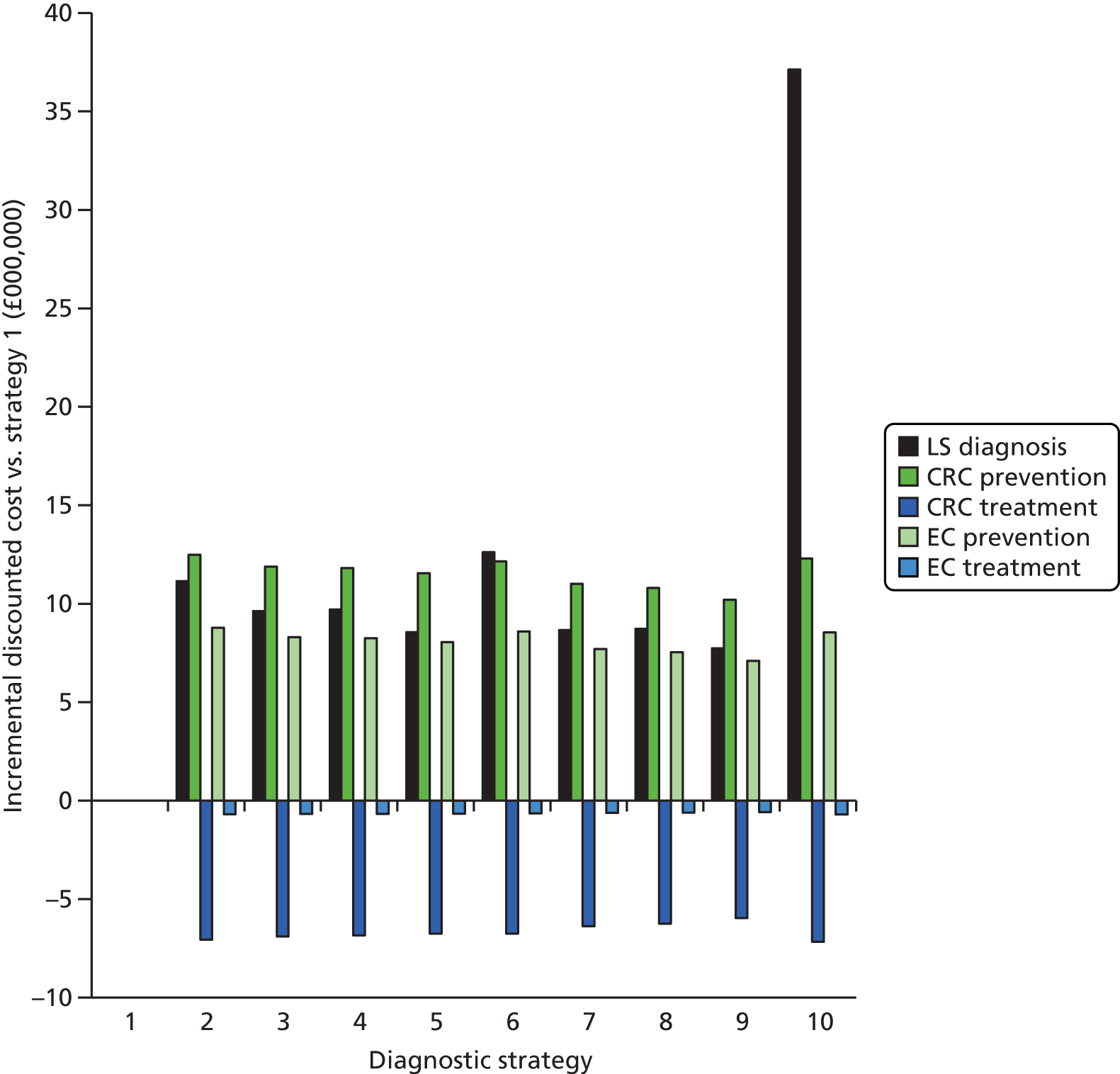
The strategy with the highest diagnostic costs is strategy 10 (universal testing) and the strategy with the lowest diagnostic costs is strategy 1 (the no-testing strategy). Strategy 10 is particularly expensive because genetic testing is the only test in the strategy and genetic testing is far more expensive than IHC and MSI testing, both of which are initial tests in the other strategies. In general, the MSI strategies have lower diagnostic costs, with the exception of strategy 6, in which MSI testing is used on its own. The larger costs here are driven by a larger proportion of probands going forward for diagnostic genetic testing. The largest cost component of every strategy is the initial test or the cost of genetic testing for the proband (see Figure 47).
Long-term costs appear to be broadly similar across the strategies (see Figure 48). However, as Figure 49 shows, this may be because the costs of cancer prevention and the costs of cancer treatment largely negate each other in each strategy, making LS diagnosis one of the main drivers of the incremental cost difference between strategies.
Subgroup analyses
In subgroup analyses, a variety of age limits applied to newly diagnosed CRC patients (probands) were considered, as requested by the NICE scope. When the age limit of probands is lowered, the prevalence of LS in probands increases (Table 71) because the incidence of CRC in the general population falls more rapidly than the incidence of CRC in people with LS. For the same reason, when the age limit is set at a minimum of 70 years, the prevalence of LS falls significantly (from 2.8% in the base case to 1.1%). The total annual incidence of CRC also changes with each age limit, with a higher incidence of CRC in people aged ≥ 70 years than in any other age group (20,202 vs. 13,823 in the those aged < 70 years). As the number of relatives per proband does not alter, this means that the overall cohort size in our analysis changes depending on the age limit used.
| Input parameter | Base case | Age-limited subgroup (years) | Source | |||
|---|---|---|---|---|---|---|
| < 50 | < 60 | < 70 | ≥ 70 | |||
| Prevalence of LS in probands (%) | 2.8 | 8.4 | 5.7 | 3.8 | 1.1 | Hampel et al.92 |
| Number of probands per annum in England | 34,025 | 2,107 | 5,880 | 13,823 | 20,202 | ONS21 |
| Proportion of probands who are male (%) | 55.6 | 51.8 | 55.5 | 59.2 | 53.0 | ONS21,96–103 |
| Proportion of probands assumed to have LS (tumour test results available only) (%) | 10 | 21 | 17 | 13 | 5 | Snowsill et al.4 model and assumptions |
| CRC incidence in male probands without LS | 0.63 | 0.61 | 0.56 | 0.57 | 0.67 | ONS103 |
| CRC incidence in female probands without LS | 0.72 | 0.70 | 0.66 | 0.68 | 0.75 | ONS103 |
All parameters that are affected by the subgroup analyses are provided in Table 71.
As shown in Table 72, the mean age of the probands in the cohort alters as expected, according to the age limit of the probands, although the age of probands without LS is consistently estimated to be higher than the age of those with LS. For the groups with lower maximum age limits the age of probands with and without LS do become more similar, reflecting the change in prevalence compared with the change in CRC incidence rates. The age of the relatives is not linked to the age of the probands, similar to the base case, and therefore the age distribution of the relatives does not alter for these analyses.
| Age-limited subgroup (years) | Mean age of probands at diagnosis (years) | |
|---|---|---|
| Without LS | With LS | |
| Base case | 72.7 | 58.0 |
| < 50 | 41.6 | 39.8 |
| < 60 | 51.1 | 47.1 |
| < 70 | 60.0 | 52.3 |
| ≥ 70 | 77.1 | 72.9 |
Summary results for the age-limited subgroups are presented in Tables 73–76. Results on the cost-effectiveness frontier were generally similar for all subgroups (and were similar to the base case), with strategy 5 remaining the optimal strategy. Larger total discounted costs and QALYs are reported for subgroups with a higher maximum age limit. This is primarily driven by the size of the cohort.
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 225,106 | 52,979,766 | – | – | – |
| 2. IHC | 225,489 | 56,319,403 | 8731 | 215.5 | 34,526 |
| 3. IHC plus BRAF V600E | 225,482 | 56,098,834 | 8293 | 220.2 | 19,903 |
| 4. IHC plus MLH1 promoter methylation | 225,481 | 56,087,979 | 8298 | 219.2 | Extendedly dominated by strategies 5 and 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 225,476 | 55,970,603 | 8090 | 220.1 | 8090 |
| 6. MSI | 225,469 | 56,333,761 | 9229 | 195.7 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 225,454 | 55,873,734 | 8304 | 203.8 | Extendedly dominated by strategies 8 and 5 |
| 8: MSI plus MLH1 promoter methylation | 225,447 | 55,831,834 | 8358 | 198.6 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 225,433 | 55,659,876 | 8184 | 193.5 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 225,490 | 57,714,180 | 12,336 | 147.1 | 1,096,665 |
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 625,933 | 142,438,302 | – | – | – |
| 2. IHC | 626,623 | 149,410,482 | 10,106 | 341.3 | 54,320 |
| 3. IHC plus BRAF V600E | 626,613 | 148,866,574 | 9454 | 358.5 | 25,681 |
| 4. IHC plus MLH1 promoter methylation | 626,609 | 148,851,044 | 9482 | 355.6 | Extendedly dominated by strategies 5 and 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 626,601 | 148,551,307 | 9156 | 362.0 | 9156 |
| 6. MSI | 626,588 | 149,549,494 | 10,857 | 299.4 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 626,562 | 148,382,032 | 9447 | 332.0 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 626,549 | 148,307,783 | 9528 | 322.6 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 626,524 | 147,905,444 | 9243 | 318.1 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 626,625 | 153,518,956 | 16,018 | 137.7 | 2,233,950 |
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 1,454,988 | 322,566,730 | – | – | – |
| 2. IHC | 1,456,078 | 334,746,310 | 11,175 | 480.9 | 59,733 |
| 3. IHC plus BRAF V600E | 1,456,059 | 333,636,101 | 10,333 | 517.8 | 31,707 |
| 4. IHC plus MLH1 promoter methylation | 1,456,053 | 333,625,144 | 10,383 | 512.1 | Extendedly dominated by strategies 5 and 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 1,456,039 | 332,980,152 | 9912 | 529.9 | 9912 |
| 6. MSI | 1,456,025 | 335,222,782 | 12,205 | 404.2 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 1,455,979 | 332,749,603 | 10,275 | 481.9 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 1,455,958 | 332,649,631 | 10,390 | 466.3 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 1,455,918 | 331,857,287 | 9984 | 466.0 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 1,456,072 | 344,821,179 | 20,528 | –28.6 | Dominated by strategy 2 |
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 2,075,242 | 430,082,601 | – | – | – |
| 2. IHC | 2,075,648 | 439,327,880 | 22,794 | –56.7 | 146,300 |
| 3. IHC plus BRAF V600E | 2,075,640 | 438,172,824 | 20,342 | –6.8 | 105,987 |
| 4. IHC plus MLH1 promoter methylation | 2,075,638 | 438,205,815 | 20,512 | –10.1 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 2,075,633 | 437,415,128 | 18,774 | 23.9 | 18,839 |
| 6. MSI | 2,075,629 | 440,240,971 | 26,305 | –121.7 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 2,075,611 | 437,375,080 | 19,789 | 3.9 | Extendedly dominated by strategies 9 and 5 |
| 8. MSI plus MLH1 promoter methylation | 2,075,603 | 437,374,272 | 20,205 | –3.7 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 2,075,588 | 436,573,720 | 18,766 | 21.3 | 18,766 |
| 10. Universal genetic testing | 2,075,621 | 454,981,317 | 65,701 | –866.0 | Dominated by strategy 2 |
Primarily because of the higher prevalence of LS in subgroups with a lower age limit, the ICERs are reduced compared with the base case. For the subgroup in which all probands are aged < 50 years, all strategies have an ICER of < £13,000 per QALY gained compared with the no testing (strategy 1). The subgroup with a minimum age limit of 70 years has the largest ICERs compared with no testing, with only strategies 5 and 9 (IHC or MSI plus BRAF V600E and MLH1 promoter methylation testing) having ICERs below a cost-effectiveness threshold of £20,000 per QALY gained. These strategies have lower ICERs because the multiple tests in sequence have enriched the population prior to diagnostic genetic testing, reducing the diagnostic costs.
The difference in ICERs is also driven by the benefit accrued by probands within the model. Probands aged < 50 years have a much higher life expectancy than probands aged > 70 years (Figures 50 and 51) and therefore the potential LYGs from diagnosing LS are higher the lower the age of the probands. However, there is still benefit in terms of life expectancy for probands in the > 70 years subgroup.
FIGURE 50.
Life expectancy of probands: maximum age 50 years.

FIGURE 51.
Life expectancy of probands: minimum age 70 years.
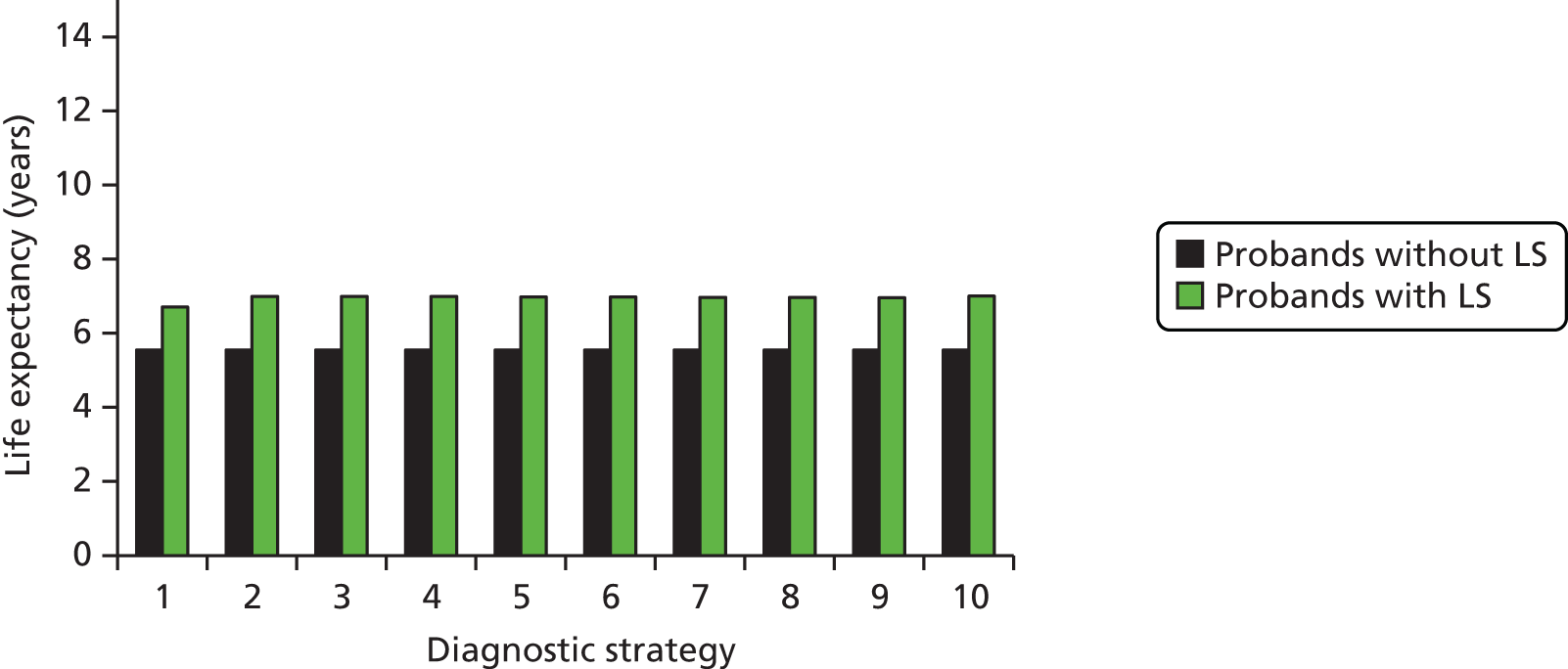
Of note, the higher total costs in the subgroups with the higher age limits are driven entirely by the larger size of the cohorts. The per-person cost is reduced in the groups with a higher age limit for probands (e.g. < £5000 for all strategies in the over 70s, but > £5900 for all strategies in the under 50s). This is primarily driven by the life expectancy of the probands in these strategies (diagnosis at an earlier age will result in higher prevention and treatment costs).
Scenario analyses
Scenario 1: microsatellite instability-low corresponds to a Lynch syndrome-positive MSI result
As previously described, different thresholds are used to decide whether a tumour has MSI. Broadly, these fall into two categories, MSI-L and MSI-H, with MSI-H having a higher level of MSI than MSI-L (but both have some level of instability). Exact measures and cut-offs can differ according to the specific test used. Further discussion of these is provided in Chapter 1 (see Microsatellite instability testing).
In the base case, only MSI-H tumours are assumed to be indicative of LS. However, clinical opinion suggests that MSI-L tumours may also be indicative of LS and we explore the impact of this in this scenario analysis. As MSI-L requires a lower threshold for instability, the number of people diagnosed with MSI increases. This has the impact of altering the accuracy of the tests as the numbers of TPs and FPs increase, whereas the numbers of true negatives (TNs) and FNs decrease. Table 77 shows that this increases sensitivity [TP/(TP + FN)] and decreases specificity [TN/(TN + FP)] compared with the base case, in which MSI-H is the chosen threshold.
| Scenario | Sensitivity (%) | Specificity (%) |
|---|---|---|
| MSI = MSI-H (base case) | 91.3 | 83.7 |
| MSI = MSI-H or MSI-L | 97.3 | 59.6 |
The cost-effectiveness results for this scenario are provided in Table 78. As expected, the cost and QALY results for strategies 1–5 and 10 are unaffected as MSI testing is not part of these diagnostic pathways. For strategies 6–9 both the discounted costs and the QALYs increase from the base case; however, the change in costs is substantially larger than the change in QALYs (an increase of £2.5–12M compared with an increase of 110–160 QALYs, depending on the strategy). Therefore, the ICERs compared with no testing for all MSI strategies are increased compared with the base case.
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,508,052 | 743,298,306 | – | – | – |
| 2. IHC | 3,510,017 | 767,955,447 | 12,553 | 731.5 | 60,967 |
| 3. IHC plus BRAF V600E | 3,509,977 | 765,532,726 | 11,553 | 812.9 | 37,495 |
| 4. IHC plus MLH1 promoter methylation | 3,509,965 | 765,535,788 | 11,626 | 800.8 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,509,937 | £764,048,240 | £11,008 | 847.5 | 11,008 |
| 6. MSI | 3,510,086 | 781,391,603 | 18,729 | 129.2 | 193,128 |
| 7. MSI plus BRAF V600E | 3,509,958 | 768,160,279 | 13,044 | 662.9 | Dominated by strategy 2 |
| 8. MSI plus MLH1 promoter methylation | 3,509,920 | 768,141,508 | 13,305 | 625.1 | Dominated by strategy 2 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,509,833 | 764,450,482 | 11,877 | 723.3 | Dominated by strategy 5 |
| 10. Universal genetic testing | 3,509,987 | 793,380,127 | 25,884 | –569.2 | Dominated by strategy 6 |
The additional costs and QALYs for the MSI strategies are driven by the increased number of probands and relatives identified as LS positive (Figure 52), leading to additional downstream costs and benefits associated with CRC and endometrial cancer risk reductions (Figure 53). Table 79 demonstrates that the sensitivity of strategies 6–9 are now increased and specificity is slightly decreased. Interestingly, in both the probands-only subpopulation and the overall population the specificity is not greatly affected, despite the specificity of MSI testing reducing to 59%. This is a result of the downstream tests in each strategy correcting for the poorer specificity of using MSI-L as being indicative of LS. Indeed, the difference in specificity from the base case falls from a difference of 0.42% (probands only) in the MSI-only strategy (strategy 6) to 0.02% when MSI is followed by both BRAF V600E and promoter methylation testing (strategy 9). Similarly, the difference in sensitivity also reduces as additional tests, with imperfect sensitivity, are added to the strategy, although the difference from the base case remains about 4% for all strategies.
FIGURE 52.
Number of probands and relatives identified by each strategy: MSI-L is indicative of LS. Note: TNs have not been shown in the interest of clarity. The number of TNs is substantially larger than the numbers with the other three diagnoses.
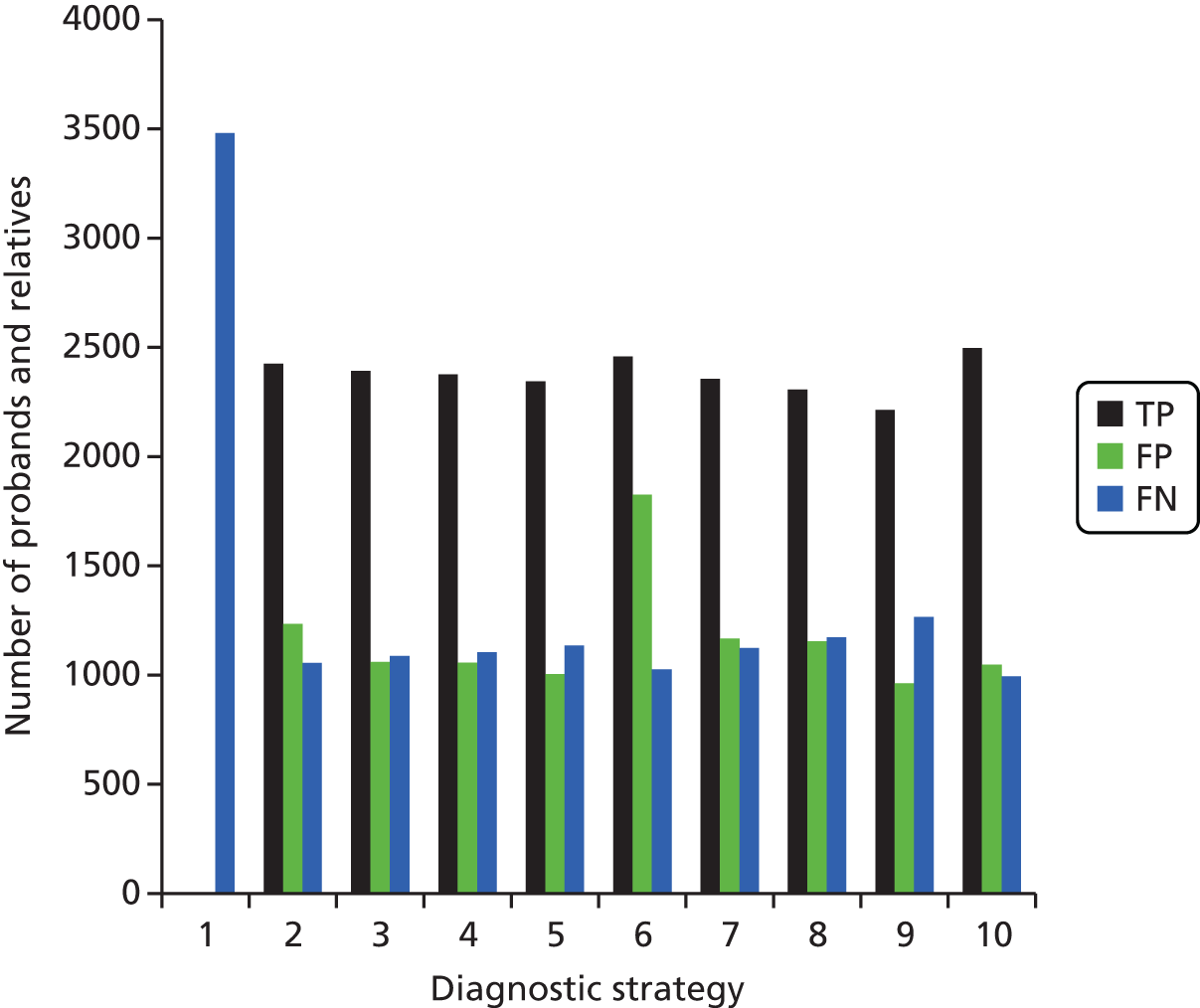
FIGURE 53.
Incremental discounted costs compared with no testing: MSI-L is indicative of LS. EC, endometrial cancer.
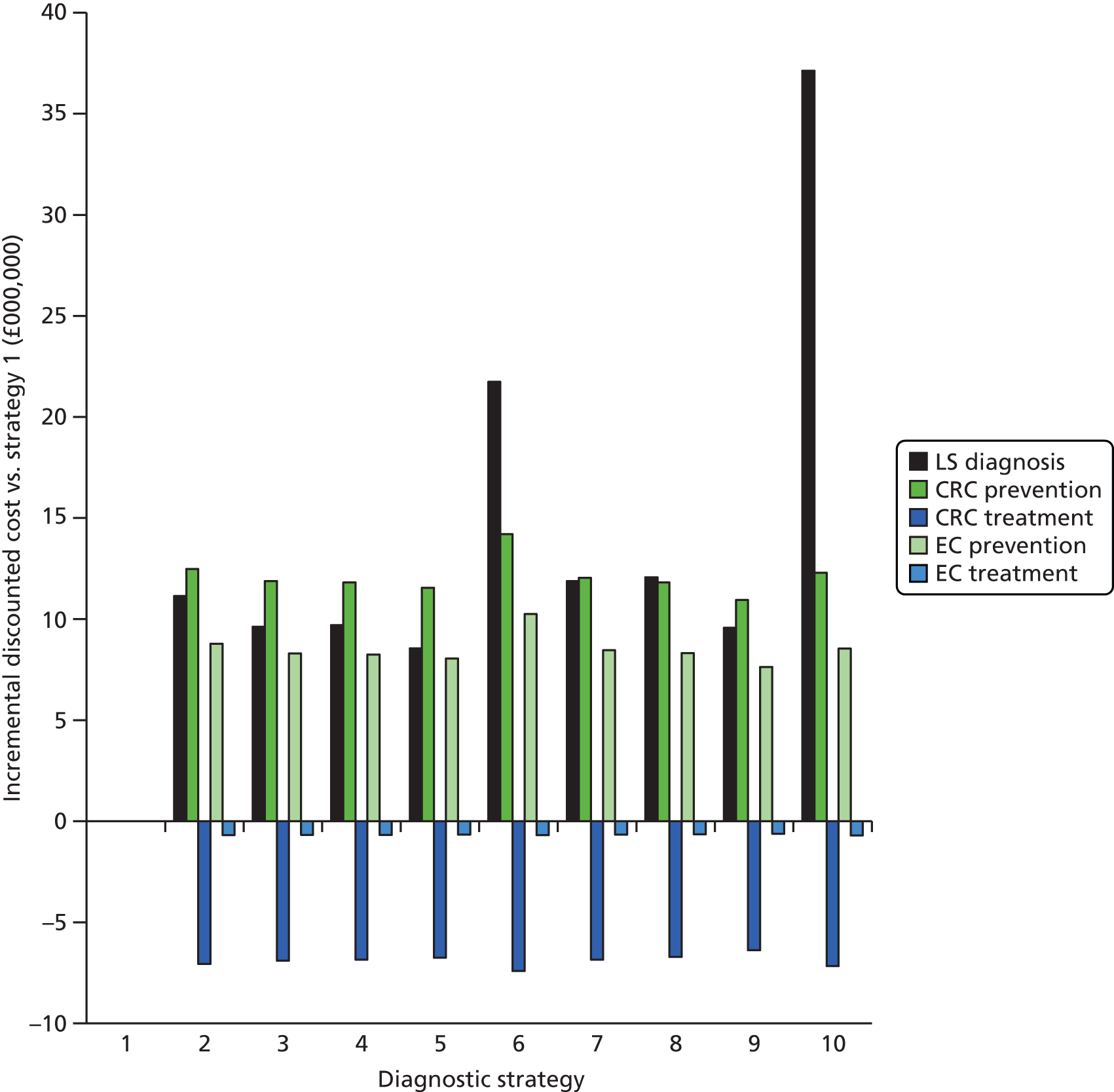
| Strategy | Probands only | Overall population | ||
|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |
| 1 | 0.00 | 100.00 | 0.00 | 100.00 |
| 2 | 70.35 | 99.80 | 69.67 | 99.47 |
| 3 | 69.40 | 99.94 | 68.73 | 99.55 |
| 4 | 68.93 | 99.93 | 68.26 | 99.55 |
| 5 | 68.04 | 99.97 | 67.38 | 99.57 |
| 6 | 71.19 | 99.30 | 70.54 | 99.22 |
| 7 | 68.35 | 99.83 | 67.69 | 99.50 |
| 8 | 66.92 | 99.83 | 66.28 | 99.51 |
| 9 | 64.24 | 99.96 | 63.62 | 99.59 |
| 10 | 71.43 | 99.98 | 71.53 | 99.55 |
Figure 54 demonstrates that using MSI-L as indicative of LS results in much higher costs of tests subsequent to MSI testing, a result of more probands receiving results indicative of LS. This leads to the higher overall costs of diagnosis, as demonstrated in Figure 53.
FIGURE 54.
Disaggregated diagnostic costs: MSI-L is indicative of LS. EC, endometrial cancer; FH, family history.
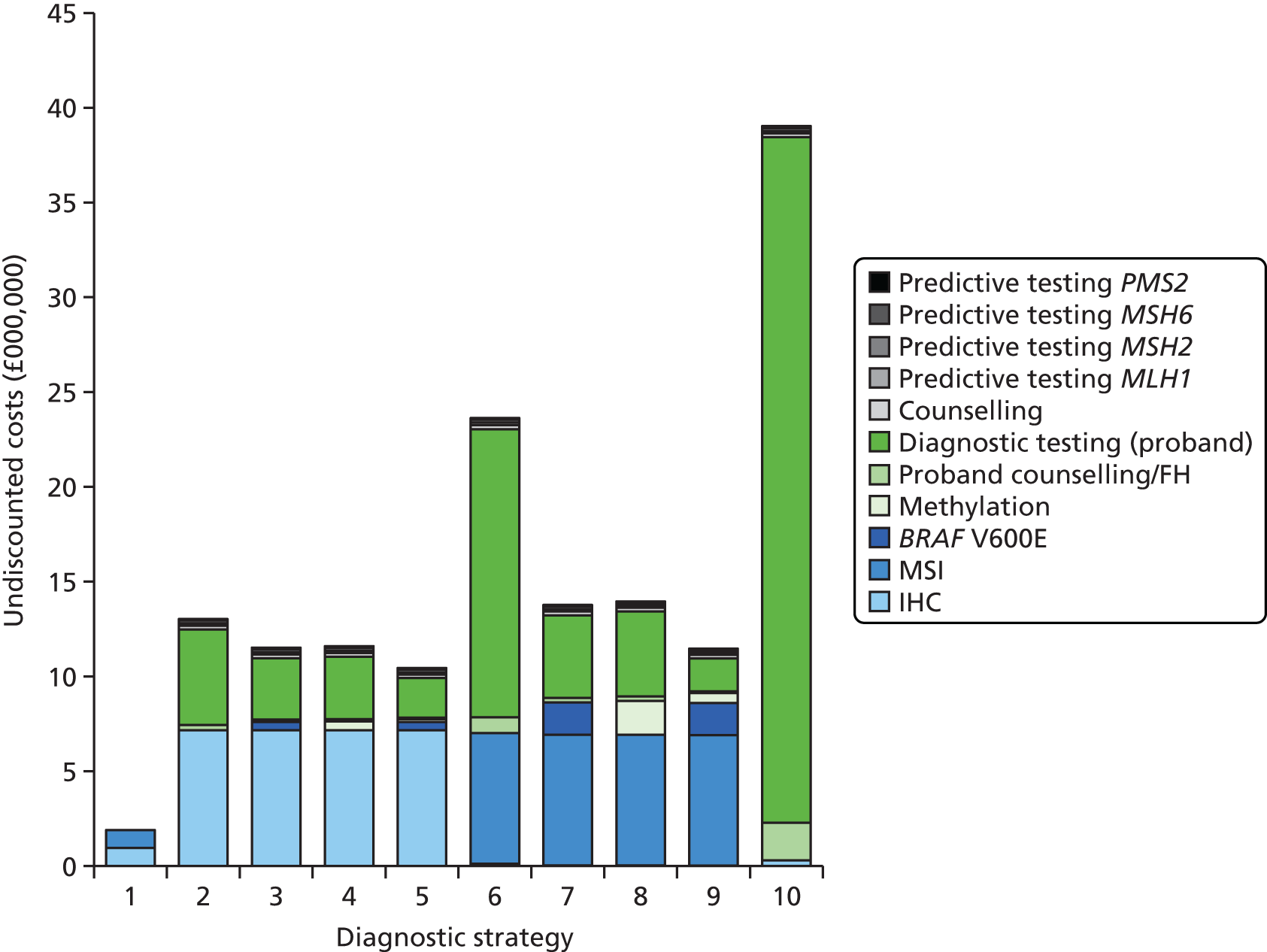
Scenario 2: aspirin removed from the model
In the base case, aspirin use for CRC prevention in LS-positive people is estimated as in the CAPP2 trial. 29 In this scenario analysis aspirin was not included as a risk-reducing component in the model (and therefore the costs and benefits associated with its use are removed).
In this scenario there is a slight reduction in QALYs for all strategies and in life expectancy for relatives compared with the base case (see Figure 37 compared with Figure 55). However, the difference in life expectancy for relatives without LS is at least partially driven by the new set of simulations, as this holds even in strategy 1, and is therefore likely to be related to the expected differences when running a new set of simulations. However, the life expectancy for probands and relatives with LS is also likely to be reduced by the increase in CRC and endometrial cancer incidence (although again this too is increased for strategy 1 compared with the base case).
FIGURE 55.
Life expectancy of relatives: aspirin removed as a risk-reducing measure.
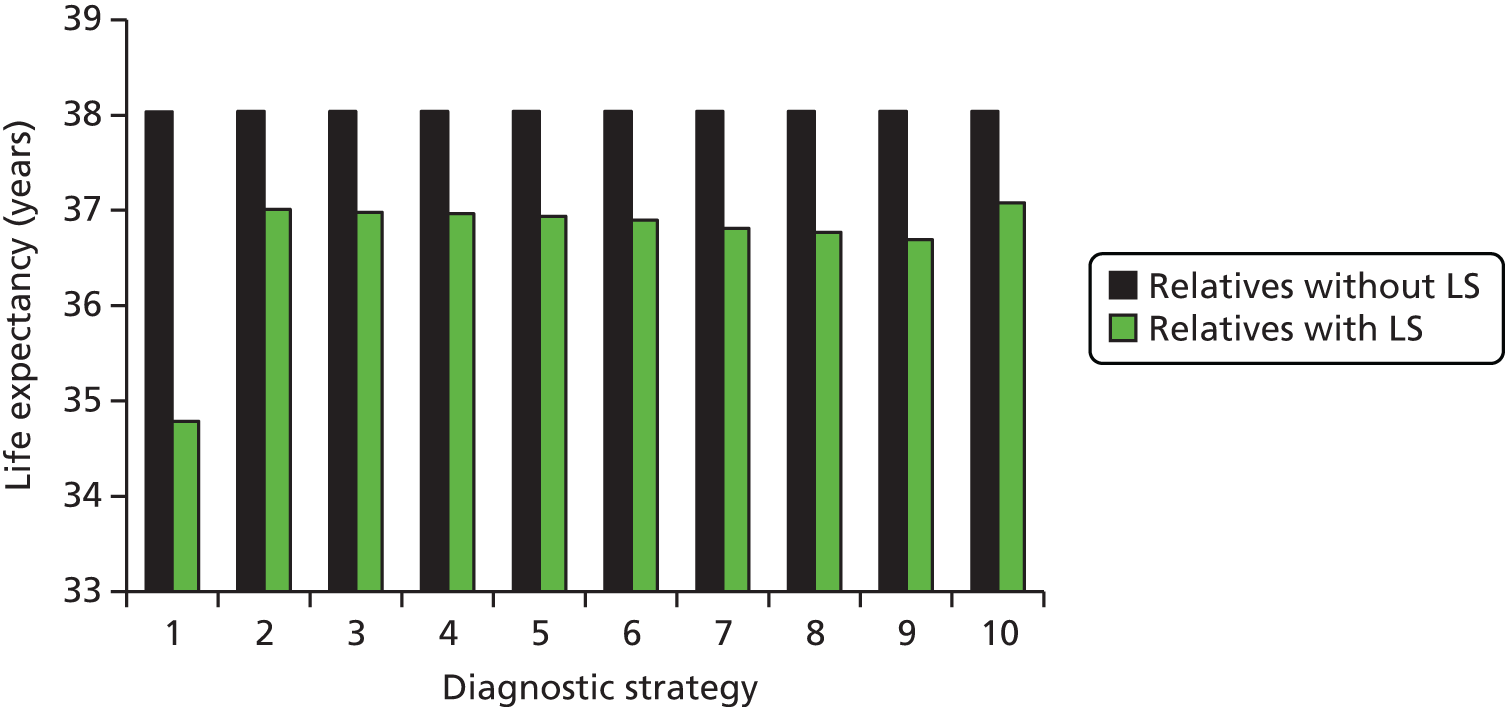
Similarly, although the cost of risk reduction is reduced, the cost of treatment for CRC and EC is increased, resulting in higher overall costs compared with the base case. Again, some of this is related to the simulations, but by comparing the incremental results of each strategy compared with no testing (Table 80), we can see that the overall outcome of removing aspirin from the model is to marginally increase the ICERs.
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,506,867 | 731,729,637 | – | – | – |
| 2. IHC | 3,508,703 | 756,230,300 | 13,350 | 610.2 | 76,621 |
| 3. IHC plus BRAF V600E | 3,508,671 | 753,822,657 | 12,248 | 699.2 | 41,422 |
| 4. IHC plus MLH1 promoter methylation | 3,508,660 | 753,826,465 | 12,326 | 687.8 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,508,636 | 752,343,957 | 11,659 | 737.4 | 11,659 |
| 6. MSI | 3,508,614 | 757,522,873 | 14,766 | 457.1 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,508,536 | 751,959,893 | 12,128 | 656.6 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,508,501 | 751,805,586 | 12,290 | 629.7 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,508,433 | 750,096,269 | £11,729 | 647.7 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,508,680 | 781,664,560 | £27,541 | –683.7 | Dominated by strategy 2 |
In this scenario, strategy 5 remains the optimal strategy, with an ICER of £11,659 per QALY gained (compared with £11,008 per QALY gained in the base case).
Scenario 3: gynaecological surveillance assumed to have no benefit
In the base case, gynaecological surveillance is offered to women diagnosed with LS and, if accepted, incurs both a cost and reduces the risk of mortality from endometrial cancer. However, the true benefit of this surveillance is disputed. As such, we investigated the impact of gynaecological surveillance, first, by assuming that the surveillance has no benefit (but still incurs a cost) (scenario 3) and, second, by removing gynaecological surveillance from the model entirely (so that no associated costs or benefits are modelled) (scenario 4).
In scenario 3, the HR for endometrial cancer survival was increased from 0.898 to 1 for women receiving surveillance.
Summary results for this scenario are presented in Table 81. Both the incremental costs and QALYs are slightly reduced compared with the base case. Although a decrease in the incremental costs may seem counterintuitive (as the cost of surveillance is still incorporated in the model), this is likely offset by a reduction in life expectancy for women in strategies 2–10 of the model (Figures 56 and 57), resulting in fewer total costs and QALYs accrued over their lifetime compared with the base case. Overall, this slight reduction in incremental costs is not as pronounced as the incremental QALY loss compared with no testing and therefore the ICERs are very slightly increased compared with the base case (as expected). The optimal strategy remains strategy 5 (IHC followed by BRAF V600E and MLH1 promoter methylation testing), with an ICER of £11,375 compared with no testing.
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,518,332 | 729,775,566 | – | – | – |
| 2. IHC | 3,520,216 | 754,377,439 | 13,053 | 654.6 | 80,413 |
| 3. IHC plus BRAF V600E | 3,520,186 | 751,943,691 | 11,954 | 746.0 | 40,972 |
| 4. IHC plus MLH1 promoter methylation | 3,520,175 | 751,947,401 | 12,031 | 734.3 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,520,150 | 750,457,773 | 11,375 | 784.1 | 11,375 |
| 6. MSI | 3,520,124 | 755,682,001 | 14,453 | 497.2 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,520,046 | 750,076,890 | 11,839 | 699.7 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,520,011 | 749,921,886 | 11,998 | 671.8 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,519,942 | 748,200,353 | 11,441 | 689.2 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,520,197 | 779,782,852 | 26,815 | –635.5 | Dominated by strategy 2 |
FIGURE 56.
Life expectancy of probands: no benefit assumed from gynaecological surveillance. Note: the increase in life expectancy for male probands is a result of the simulation run and appears more pronounced because of the scale of the figure.
FIGURE 57.
Life expectancy of relatives: no benefit assumed from gynaecological surveillance.
Scenario 4: gynaecological surveillance not included
As described in the previous section, in this scenario all gynaecological surveillance was removed from the model (i.e. the probability of being offered gynaecological surveillance becomes 0).
This scenario results in similar incremental QALY gains compared with no testing as in scenario 3 (absolute QALYs differ because this is a different simulation run), but additional reductions in incremental costs because of the removal of the cost of gynaecological surveillance (Table 82). The resulting ICERs compared with no testing are slightly reduced compared with both scenario 3 and the base case, with the exception of universal genetic testing (strategy 10), which has a larger ICER than in the base case. This strategy differs as the change in surveillance costs is less influential on the overall incremental costs, given that the cost of diagnosing LS is the main driver of the overall incremental costs compared with no testing (Figure 58).
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,524,850 | 729,116,320 | – | – | – |
| 2. IHC | 3,526,553 | 749,618,872 | 12,033 | 678.8 | 109,979 |
| 3. IHC plus BRAF V600E | 3,526,533 | 747,412,582 | 10,866 | 769.1 | 44,025 |
| 4. IHC plus MLH1 promoter methylation | 3,526,523 | 747,439,747 | 10,950 | 757.2 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,526,502 | 746,042,546 | 10,241 | 806.4 | 10,241 |
| 6. MSI | 3,526,465 | 751,010,931 | 13,553 | 520.7 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,526,406 | 745,828,322 | 10,736 | 721.0 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,526,374 | 745,742,636 | £10,909 | 692.7 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,526,314 | 744,227,411 | 10,318 | 709.0 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,526,539 | 775,139,777 | 27,239 | –611.6 | Dominated by strategy 2 |
FIGURE 58.
Incremental discounted costs compared with no testing: no gynaecological surveillance. EC, endometrial cancer.
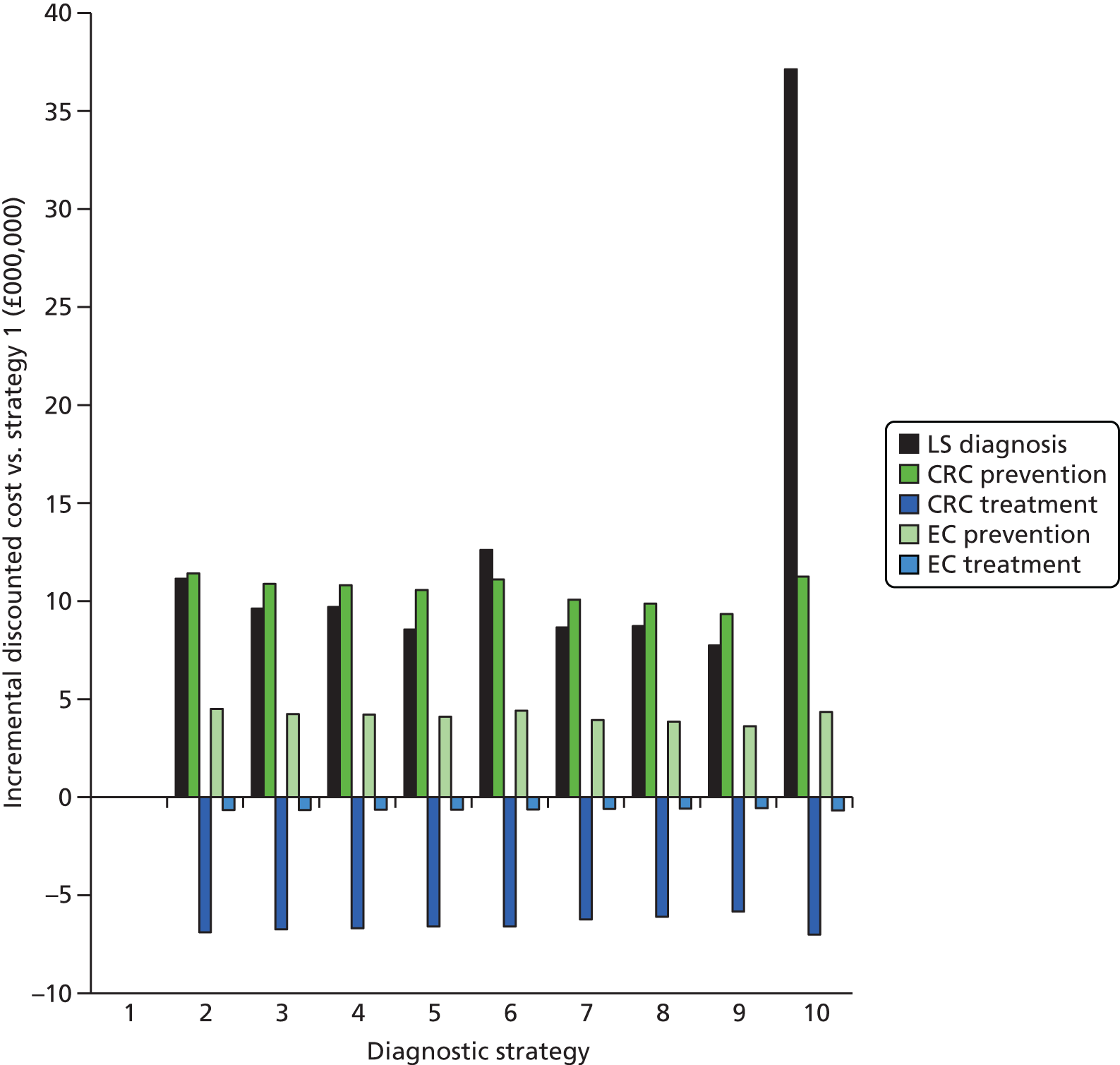
As with scenario 3, strategy 5 remains the optimal strategy, with an ICER of £10,241 per QALY gained.
Scenario 5: colorectal cancer utilities taken from Ness et al.144
In the base case, quality of life (as measured by the EQ-5D) for all CRC stages except Dukes’ D was expected to be similar to that in the general population. Dukes’ stage D CRC was expected to have a non-zero disutility of 0.13. Our review of CRC utilities provided some evidence to support this approach. However, it is important to investigate a scenario in which all stages of CRC incur a quality-of-life decrement, to assess the impact that this has on the cost-effectiveness results. The study by Ness et al. ,144 which reports CRC utilities, has been widely cited in previous cost-effectiveness analyses of CRC (including by Snowsill et al. 4) and was therefore chosen for this scenario analysis. A comparison of the disutilities between the base case and the scenario analysis is provided in Table 83.
| Stage | Base-case disutilities142,143 | Scenario analysis disutilities144 |
|---|---|---|
| Dukes A | 0 | 0.11 |
| Dukes B | 0 | 0.23 |
| Dukes C | 0 | 0.26 |
| Dukes D | 0.13 | 0.60 |
A summary of the results of this scenario analysis are provided in Table 84. As expected, the ICERs for strategies 2–10 compared with no testing are reduced compared with the base case. Incremental cost differences compared with strategy 1 remain broadly similar (again, absolute costs differ from the base case because this is a different simulation run). However, the incremental QALYs gained compared with strategy 1 are increased from the base case; for example, incremental QALYs gained in strategy 5 increase from 1885 in the base case to 2116 in this scenario compared with no testing. This reflects the additional benefit of reducing CRC incidence in probands and relatives diagnosed with LS (i.e. the avoidance of quality-of-life loss associated with CRC).
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,459,708 | 729,043,142 | – | – | – |
| 2. IHC | 3,461,906 | 753,654,444 | 11,195 | 967.9 | 62,734 |
| 3. IHC plus BRAF V600E | 3,461,867 | 751,217,817 | 10,268 | 1050.9 | 34,368 |
| 4. IHC plus MLH1 promoter methylation | 3,461,854 | 751,221,527 | 10,335 | 1037.1 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,461,824 | 749,731,129 | 9775 | 1081.9 | 9775 |
| 6. MSI | 3,461,801 | 754,960,430 | 12,380 | 797.5 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,461,705 | 749,350,705 | 10,169 | 981.6 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,461,663 | 749,195,658 | 10,305 | 948.0 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,461,582 | 747,472,833 | 9833 | 952.8 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,461,891 | 779,058,290 | 22,904 | –317.1 | Dominated by strategy 2 |
Strategy 5 remains the optimal strategy, with an ICER of £9775 per QALY gained compared with no testing.
Scenario 6: colonoscopic surveillance assumed to have no impact on colorectal cancer incidence
As discussed in Surveillance, Colorectal cancer incidence, there is some evidence suggesting that our base-case assumption for the effectiveness of colonoscopy for CRC incidence may be optimistic. We therefore investigated a ‘worst-case’ scenario in which the HR for CRC incidence while receiving colonoscopic surveillance was set to 1 for both the index CRC and mCRCs (i.e. surveillance has no impact on CRC incidence). Previously, the HRs were set as 0.387 for index cancers (applicable to only relatives) and 0.533 for metachronous cancers (applicable to probands and relatives). The costs of colonoscopic surveillance remained as in the base case.
Summary results are presented in Table 85. As expected, the ICERs are increased for all strategies, resulting from lower QALYs in strategies 2–10 than in the base case, but similar costs. This lower QALY gain is driven by the increase in CRC incidence in these arms and the resulting reduction in life expectancy. This is seen particularly for relatives (Figure 59 compared with Figure 37), who in the base case receive the most benefit from colonoscopic surveillance (particularly as the base-case HR for index CRC incidence with colonoscopic surveillance is lower than that for mCRC).
| Strategy | QALYs | Cost (£) | ICER vs. no testing (cost per QALY) (£) | INHB at WTP of £20,000 per QALY vs. no testing | Incremental ICER (cost per QALY) (£) |
|---|---|---|---|---|---|
| 1. No test | 3,516,035 | 734,308,670 | – | – | – |
| 2. IHC | 3,517,362 | 763,001,311 | 21,613 | –107.1 | 125,265 |
| 3. IHC plus BRAF V600E | 3,517,342 | 760,495,764 | 20,028 | –1.8 | 62,975 |
| 4. IHC plus MLH1 promoter methylation | 3,517,334 | 760,472,422 | 20,131 | –8.5 | Dominated by strategy 3 |
| 5. IHC plus BRAF V600E and MLH1 promoter methylation | 3,517,317 | 758,927,199 | 19,194 | 51.7 | 19,194 |
| 6. MSI | 3,517,296 | 764,111,396 | 23,625 | –228.7 | Dominated by strategy 2 |
| 7. MSI plus BRAF V600E | 3,517,244 | 758,324,753 | 19,861 | 8.4 | Extendedly dominated by strategies 8 and 5 |
| 8. MSI plus MLH1 promoter methylation | 3,517,219 | 758,092,893 | 20,087 | –5.2 | Extendedly dominated by strategies 9 and 7 |
| 9. MSI plus BRAF V600E and MLH1 promoter methylation | 3,517,171 | 756,219,424 | 19,284 | 40.7 | Extendedly dominated by strategies 1 and 5 |
| 10. Universal genetic testing | 3,517,331 | 788,502,246 | 41,796 | –1413.0 | Dominated by strategy 2 |
FIGURE 59.
Life expectancy of relatives: no impact of colonoscopic surveillance on CRC incidence.

Deterministic sensitivity analyses
In this section we present the results of the deterministic sensitivity analyses. Summary results for all analyses are provided in Table 86 and the subset of analyses that most influences the results is discussed further in the text. In Table 86 we identify the optimal strategy and report the INHB for this strategy compared with no testing. This provides a more meaningful comparison than an isolated ICER. The net health benefit (NHB) is defined as the total QALYs minus (the total costs divided by the willingness-to-pay threshold) of each strategy. Strategies with the highest NHB are found to be the optimal strategy for a chosen willingness-to-pay threshold. In real terms, as reported in Snowsill et al. ,4 ‘the total discounted QALYs for a man who lives to age 80 years, allowing for age-related quality of life, is approximately 25 (p. 215)’.
| Parameter | Base-case value | Sensitivity analyses | INHB for strategy 5 (base-case optimal strategy) vs. no testing | Optimal strategy (if different from the base case) | INHB vs. no testing at WTP of £20,000 per QALY for optimal strategy in SA (QALYs) |
|---|---|---|---|---|---|
| Base case | 847.5 | ||||
| Diagnostic parameters | |||||
| LS-positive population by gene | MLH1 32%, MSH2 39%, MSH6 14%, PMS2 15% | MLH1 40%, MSH2 46%, MSH6 11%, PMS2 2% | 889.2 | ||
| Proportion of probands requiring IHC at time of genetic test (MSI strategies only) | 5% | 0% | 847.5 | ||
| 10% | 847.5 | ||||
| Diagnostic test performance | |||||
| Sensitivity | MSI 0.913, IHC 0.962, BRAF V600E 0.96, MLH1 promoter methylation 0.94 | Lower 95% CI: MSI 0.426, IHC 0.694, BRAF V600E 0.60, MLH1 promoter methylation 0.79 | 381.8 | Strategy 4 (IHC and methylation) | 414.6 |
| Upper 95% CI: MSI 0.993, IHC 0.996, BRAF V600E 0.99, MLH1 promoter methylation 0.98 | 920.3 | Strategy 9 (MSI followed by BRAF V600E and methylation) | 924.1 | ||
| Specificity | MSI 0.837, IHC 0.884, BRAF V600E 0.76, MLH1 promoter methylation 0.75 | Lower 95% CI: MSI 0.638, IHC 0.790, BRAF V600E 0.60, MLH1 promoter methylation 0.59 | 696.4 | ||
| Upper 95% CI: MSI 0.937, IHC 0.940, BRAF V600E 0.87, MLH1 promoter methylation 0.86 | 901.3 | Strategy 3 (IHC followed by BRAF V600E) | 906.9 | ||
| Sensitivity and specificity | As above | Lower 95% CI: both sensitivity and specificity | 230.7 | ||
| Upper 95% CI: both sensitivity and specificity | 974.2 | Strategy 7 (MSI followed by BRAF V600E) | 986.8 | ||
| Acceptance of genetic testing by probands | Genetic counselling 92.5%; probability of accepting genetic testing given that have already accepted genetic counselling 90% | Genetic counselling 100%; probability of accepting genetic testing given that have already accepted genetic counselling 100% | 1105.9 | ||
| Genetic counselling 50%; probability of accepting genetic testing given that have already accepted genetic counselling 50% | 74.3 | ||||
| Acceptance of genetic testing by relatives | Genetic counselling 77.7%; probability of accepting genetic testing given that have already accepted genetic counselling 71.6% | Genetic counselling 100%; probability of accepting genetic testing given that have already accepted genetic counselling 100% | 1070.0 | ||
| Genetic counselling 50%; probability of accepting genetic testing given that have already accepted genetic counselling 50% | 670.2 | ||||
| Proportion of probands with LS assumed (following declined genetic testing) | 10% | 0% | 838.3 | ||
| 20% | 856.4 | ||||
| No LS assumed (no confirmed mutation = LS negative) | Probands who decline testing (and their relatives) and relatives who decline testing | Only confirmed mutation status is treated as LS positive | 1291.1 | ||
| Number of relatives | 6 | 0 | 46.9 | ||
| 12 | 1648.1 | ||||
| Psychological disutility of testing | Declining testing 0.04, testing LS positive 0.02 | 0 for all | 883.4 | ||
| Declining testing 0.12, testing LS positive 0.06 | 775.6 | ||||
| Diagnostic costs | |||||
| IHC | £210 | £105 | 1002.3 | ||
| £420 | 537.9 | Strategy 7 (MSI followed by BRAF V600E) | 809.3 | ||
| MSI | £202 | £101 | 824.3 | Strategy 7 (MSI followed by BRAF V600E) | 911.0 |
| £405 | 893.9 | ||||
| BRAF V600E | £119 | £60 | 858.6 | ||
| £238 | 825.3 | ||||
| MLH1 promoter methylation | £125 | £62 | 850.9 | ||
| £249 | 840.6 | ||||
| Genetic counselling probands | £63 | £32 | 849.7 | ||
| £127 | 843.1 | ||||
| Genetic testing probands | All four genes £1276, MLH1 £481, PMS2 £468 (only applied to strategies 3, 4 and 5) | All four genes £610 (cheapest testing option) | 898.9 | ||
| Genetic counselling relatives | £63 | £32 | 852.3 | ||
| £127 | 830.2 | ||||
| Genetic testing relatives | MLH1 £166, MSH2 £161, MSH6 £161, PMS2 £165 | MLH1 £83, MSH2 £80, MSH6 £81, PMS2 £83 | 856.1 | ||
| MLH1 £331, MSH2 £321, MSH6 £322, PMS2 £330 | 820.6 | ||||
| CRC parameters | |||||
| Acceptance of CRC surveillance | LS mutation 97.2%, LS assumed 70.1% (probands and relatives) | LS mutation 70.1%, LS assumed 70.1% | 593.6 | ||
| LS mutation 97.2%, LS assumed 97.2% | 948.1 | ||||
| Logistic model parameters for CRC incidence in individuals with LS | b0: male 0.464, female 0.435 | b0: male 0.303, female 0.265 | 41.5 | ||
| b0: male 0.715, female 0.697 | 1770.2 | ||||
| CRC incidence – HR for LS survival | Dukes’ A and B: 0.57, Dukes’ C and D: 1 | Dukes’ A, B, C and D: 1 | 843.9 | ||
| CRC surgery disutility | Segmental resection 0, subtotal colectomy IRA 0, rectal excision 0, proctocolectomy 0 | Segmental resection 0, subtotal colectomy IRA 0.1, rectal excision 0.1, proctocolectomy 0.1 | 875.3 | ||
| CRC-related costs | |||||
| Colonoscopy | £585.80 | £292.9 | 1112.4 | ||
| £1171.6 | 317.6 | ||||
| Colonoscopy complication | Bleeding £473–4394 (severity dependent); perforation or mortality £4909 | Bleeding £237–2197 (severity dependent); perforation or mortality £2455 | 848.9 | ||
| Bleeding £947–8788 (severity dependent); perforation or mortality £9818 | 844.8 | ||||
| Aspirin | £149 | £74 | 870.0 | ||
| £297 | 802.5 | ||||
| CRC diagnosis | £1022 | £511 | 841.5 | ||
| £2043 | 859.5 | ||||
| CRC surgery | Segmental resection GP £6514, LS £6605; subtotal colectomy IRA £7879; rectal excision £7939; proctocolectomy £7977 | Segmental resection GP £3257, LS £3302; subtotal colectomy IRA 3939; rectal excision £3969; proctocolectomy £3988 | 808.9 | ||
| Segmental resection GP £13,028, LS £13,210; subtotal colectomy IRA £15,757; rectal excision £15,878; proctocolectomy £15,953 | 924.7 | ||||
| CRC chemotherapy and radiotherapy | Colon: Dukes’ A and D £0, Dukes’ B £5653, Dukes’ C £12,900; rectal: Dukes’ A £1049, Dukes’ B £4206, Dukes’ C £9504, Dukes’ D £0 | Colon: Dukes’ A and D £0, Dukes’ B £2826, Dukes’ C £6450; rectal: Dukes’ A £525, Dukes’ B £2103, Dukes’ C £4752, Dukes’ D £0 | 810.7 | ||
| Colon: Dukes’ A and D £0, Dukes’ B £11,306, Dukes’ C £25,800; rectal: Dukes’ A £2099, Dukes’ B £8411, Dukes’ C £19,008, Dukes’ D £0 | 921.0 | ||||
| CRC stoma care | GP £388, LS £214 | GP £194, LS £107 | 845.9 | ||
| GP £776, LS £429 | 850.6 | ||||
| CRC follow-up and surveillance | GP £230, LS £229 | GP £115, LS £115 | 844.9 | ||
| GP £460, LS £458 | 852.6 | ||||
| CRC recurrence | GP £12,236, LS £12,333 | GP £6118, LS £6166 | 803.6 | ||
| GP £24,472, LS £24,666 | 935.2 | ||||
| CRC palliative care | GP £9665, LS £9907 | GP £4833, LS £4954 | 808.1 | ||
| GP £19,331, LS £19,815 | 926.2 | ||||
| EC-related parameters | |||||
| Prophylactic H-BSO disutility | 0 | 0.04 applied for 1 year (equal to base-case EC disutility) | 400.8 | ||
| EC disutility | 0.04 applied for 1 year | 0.07 applied for 1 year | 782.2 | ||
| EC-related costs | |||||
| Gynaecological screening | £473 | £237 | 946.1 | ||
| £947 | 650.1 | ||||
| Prophylactic H-BSO | £3428 | £1714 | 950.2 | ||
| £6856 | 642.1 | ||||
| EC-related H-BSO | £4005 | £2002 | 838.1 | ||
| £8009 | 866.2 | ||||
| EC radiotherapy | £2735 | £1367 | 831.2 | ||
| £5469 | 860.3 | ||||
| EC chemotherapy | £324 | £162 | 846.7 | ||
| £647 | 849.0 | ||||
| Disutility – EC risk reduction (surveillance or H-BSO) declined | Declined genetic testing and risk reduction 0.11; LS+ve and declined risk reduction 0.09 | Declined genetic testing and risk reduction 0; LS+ve and declined risk reduction 0 | 748.1 | ||
| Declined genetic testing and risk reduction 0.22; LS+ve and declined risk reduction 0.18 | 856.6 | ||||
The deterministic sensitivity analyses demonstrate that the model is sensitive to several parameters.
First, the diagnostic accuracy of the strategies affects the NHB ranking of the strategies. To investigate the impact of diagnostic test performance we considered several scenarios: altering the sensitivity of all tumour tests, altering the specificity of all tumour tests and altering the sensitivity and specificity of all tumour tests. The altered sensitivities and specificities were based on the reported or estimated 95% CIs for the tests. The aim of these analyses was to demonstrate the impact of diagnostic accuracy on the results and not necessarily to show what we believe to be a true reflection of clinical validity. Table 86 clearly shows that reducing sensitivity and specificity (either individually or jointly) not only reduces the INHB of strategies 2–9 (strategy 10 remains unchanged as the diagnostic accuracy of gene testing is unchanged), but, in the case of reduced sensitivity, also affects which strategy is optimal in terms of cost-effectiveness [strategy 4 (IHC followed by MLH1 promoter methylation) becomes the optimal strategy]. This is likely to be because of the combination of lower diagnostic costs (fewer people diagnosed as LS positive at each stage) and higher overall sensitivity for strategy 4 than strategy 5. When both the sensitivity and the specificity of each tumour test are reduced to the lowest values, the ICER for strategy 5 compared with no testing increases to £16,036 per QALY.
When sensitivity is increased for all tests, strategies with MSI testing become optimal (strategy 9 is optimal when only sensitivity is altered, strategy 7 is optimal when both sensitivity and specificity are increased). This occurs despite MSI strategies still having worse diagnostic outcomes than IHC strategies in terms of overall sensitivity and specificity and is the result of a combination of factors, including the additional costs in the IHC arms from identifying a higher rate of TPs not being entirely offset by the additional benefits.
Acceptance of genetic tests by probands also influences the cost-effectiveness results. In the base case, acceptance of genetic testing is relatively high (90% acceptance of tests following genetic counselling, with 92.5% acceptance of counselling). By setting both of these parameters to 50%, the ICER for strategy 5 compared with no testing increases to £17,767 per QALY gained. One of the reasons why this analysis is not likely to have produced an ICER of > £20,000 per QALY gained is that a proportion of probands are assumed to have LS and therefore still receive CRC and endometrial cancer risk-reducing measures.
However, the diagnosis of LS assumed is also shown to have a big impact on the results. If all LS assumed probands and relatives are instead diagnosed as LS negative, the ICER for strategy 5 decreases significantly to £5225 per QALY gained. This is likely to be because the number of FPs in the LS assumed population is similar to the number of TPs in the base case (48–65% depending on the strategy), which increases risk reduction costs without providing a significant benefit.
Of the diagnostic costs, those for IHC and MSI testing have the biggest impact on the results, as they are the most significant drivers of the overall diagnostic cost (as they are applied to all probands in the relevant arms). They also affect the costs of strategy 1, in which the costs of MSI and IHC are included for stage II colon cancers. For all analyses, adjusting the cost of IHC or MSI testing results in the ICERs remaining below the £20,000 per QALY cost-effectiveness threshold.
As in Snowsill et al. ,4 including only probands in the model increases the ICERs for all strategies (the ICER for strategy 5 vs. no testing increases to £17,921 per QALY gained) and reduces the INHB compared with no testing. Increasing the number of relatives to 12 decreases the ICERs as expected (the size of the cohort that can benefit from CRC and endometrial cancer risk reduction increases), but the impact does not appear to be significant (the ICER for strategy 5 vs. no testing decreases to £10,068 per QALY gained).
The model is also sensitive to the incidence of CRC for people with LS. With a high incidence of CRC the ICER for strategy 5 compared with no testing decreases to £6689 per QALY gained and with a low incidence of CRC the ICER for strategy 5 compared with no testing increases to £19,300 per QALY gained. This is expected as a lower CRC incidence results in fewer benefits from the risk-reducing measures.
The model is also sensitive to the cost of colonoscopy, particularly to increases in this cost. When the cost of colonoscopy is doubled, all ICERs increase compared with no testing, with the ICER for strategy 5 increasing to £16,630 per QALY gained.
One other parameter that has an impact on the results is the disutility associated with prophylactic H-BSO. In the base case the disutility was assumed to be 0. In sensitivity analysis we increased this to 0.04 for 1 year, matching the disutility associated with endometrial cancer. This decreases the INHB and increases the ICER for all strategies compared with no testing, with the ICER for strategy 5 compared with no testing increasing to £14,441 per QALY gained. As the disutility associated with prophylactic H-BSO is relatively uncertain (no literature was identified to provide estimates), it is important to recognise the impact that this parameter can have on the model.
The results are also moderately sensitive to the acceptance rate for colonoscopic surveillance. Reducing the acceptance rate for surveillance from > 90% for people with a confirmed LS mutation status to 70% (the acceptance rate for people assumed to have LS) results in a reduction in the INHB and an increase in the ICERs compared with no testing. Although we were unable to assess the acceptance rate for gynaecological surveillance in the same way, it is likely to affect the results in a similar manner as fewer LS-positive people will be receiving risk-reducing measures.
All other deterministic sensitivity analyses did not appear to significantly alter the cost-effectiveness results when applied in isolation.
Discussion
The analyses conducted suggest that screening for LS in CRC patients using tumour-based tests (in particular IHC, BRAF V600E and MLH1 promoter methylation testing) would be cost-effective at a threshold of £20,000 per QALY. Direct MMR mutation testing was predicted not to be cost-effective at any cost-effectiveness threshold as it was not on the cost-effectiveness frontier.
Subgroup analyses suggest that the use of a maximum age limit for testing does not significantly affect the cost-effectiveness of testing, although it does significantly affect the number of individuals affected. For this reason, budget impact and total health benefits are smaller when a lower age limit is used. Using a minimum age limit for testing significantly worsens cost-effectiveness but, when a minimum age of 70 years is employed, testing is still predicted to be cost-effective at a threshold of £20,000 per QALY.
Scenario and sensitivity analyses indicate that there are some assumptions or parameters to which the cost-effectiveness results are sensitive. The most important costs in this respect are the cost of colonoscopy and the costs of MSI and IHC testing. Cost-effectiveness is also sensitive to the accuracy of the tumour tests, the acceptance rate for genetic counselling and testing and the number of relatives identified through cascade testing per proband. The effectiveness of surveillance colonoscopy and the lifetime risk of CRC for people with LS are also key determinants of cost-effectiveness.
The results are expected to be valid for the NHS (i.e. the results are expected to generalise to the NHS), partially as key costs were estimated from NHS sources, including NHS reference costs. Also, survival rates for CRC and endometrial cancer were estimated from English or UK patients. Survival rates for cancers can vary even among high-income countries in Europe. The effectiveness of colonoscopic surveillance was estimated from a Finnish study and so it is possible that surveillance in the NHS may not have the same effectiveness. It is considered likely that the cancer risks estimated for individuals with LS are appropriate as they were estimated from a French population. The prevalence of LS in CRC patients was estimated from a US study, but there is no obvious reason to think that prevalence would differ markedly in the UK as neither the UK nor the USA has significant founder mutations.
Chapter 6 Assessment of factors relevant to the NHS and others
Current variability in implementation
The Royal College of Pathologists’ data set for CRC indicates the value of assessing MMR status (Box 9).
MMR status has prognostic significance, possible predictive significance and can help detect Lynch syndrome families. As such, a strong case can now be made for performing MMR immunohistochemistry in all cases of CRC. However, given the resource implications of implementing this, it is not considered a core data item for all colorectal cancers currently. We now consider MMR immunohistochemistry a core dataset item for patients under 50 years at time of diagnosis
Loughrey et al. (p. 5)49; reproduced with permission from the Royal College of Pathologists
This has been in place for 2 years now and there have been efforts from the charity Bowel Cancer UK, through freedom of information requests, to document compliance with this core item (MMR IHC for patients aged < 50 years). 183
Of the 130 hospitals in England that responded (82% of the 159 contacted), 90 (69%) indicated that all patients diagnosed with bowel cancer aged < 50 years are tested with MSI or IHC, but only 49 hospitals (54%) conducted these tests as reflex tests (i.e. automatically, without referral).
Hospitals that have not implemented routine testing cited finances and practicalities as the principal barriers to implementation. 183
This suggests that there is current variability in the level of implementation (some hospitals implement testing for CRC patients aged < 50 years whereas others do not) and the pathway (reflex testing vs. referral).
Use and impact of age limits
Age limits for testing and surveillance in LS have been suggested by numerous expert groups,19,31,49,125 usually with the intention of balancing clinical benefits against resource implications and clinical risks.
The Equality Act 2010184 applies to NICE, which is required to have due regard to the need to eliminate discrimination. 185 Age is a protected characteristic in this context.
In the context of screening for LS in CRC patients, age is a proxy for the pretest probability of having LS92 and is therefore directly related to the potential for an individual to benefit from screening. Although there are other observable characteristics that could also be used to identify a higher-risk group (such as tumour morphology associated with MMR deficiency49), assessing these would increase the complexity of the service pathway and could result in reduced implementation.
In accordance with the NICE scope,88 subgroup analyses were conducted on a number of age groups. As in the base case, strategy 5 was the cost-effective strategy in all subgroup analyses (except when an age limit of 50 years was used, in which it was a close second, assuming a cost-effectiveness threshold of £20,000 per QALY). Figure 60 shows the incremental costs and QALYs for strategy 5 compared with strategy 1 in the different analyses. The greatest INHB (at a willingness to pay of £20,000 per QALY) is obtained in the base case (848 QALYs). This is because the base case includes the largest population and the ratio of incremental costs to QALYs is relatively stable as the maximum age limit is progressively lifted.
FIGURE 60.
Incremental costs and QALYs for strategy 5 compared with strategy 1 in the base case and across different subgroup analyses.
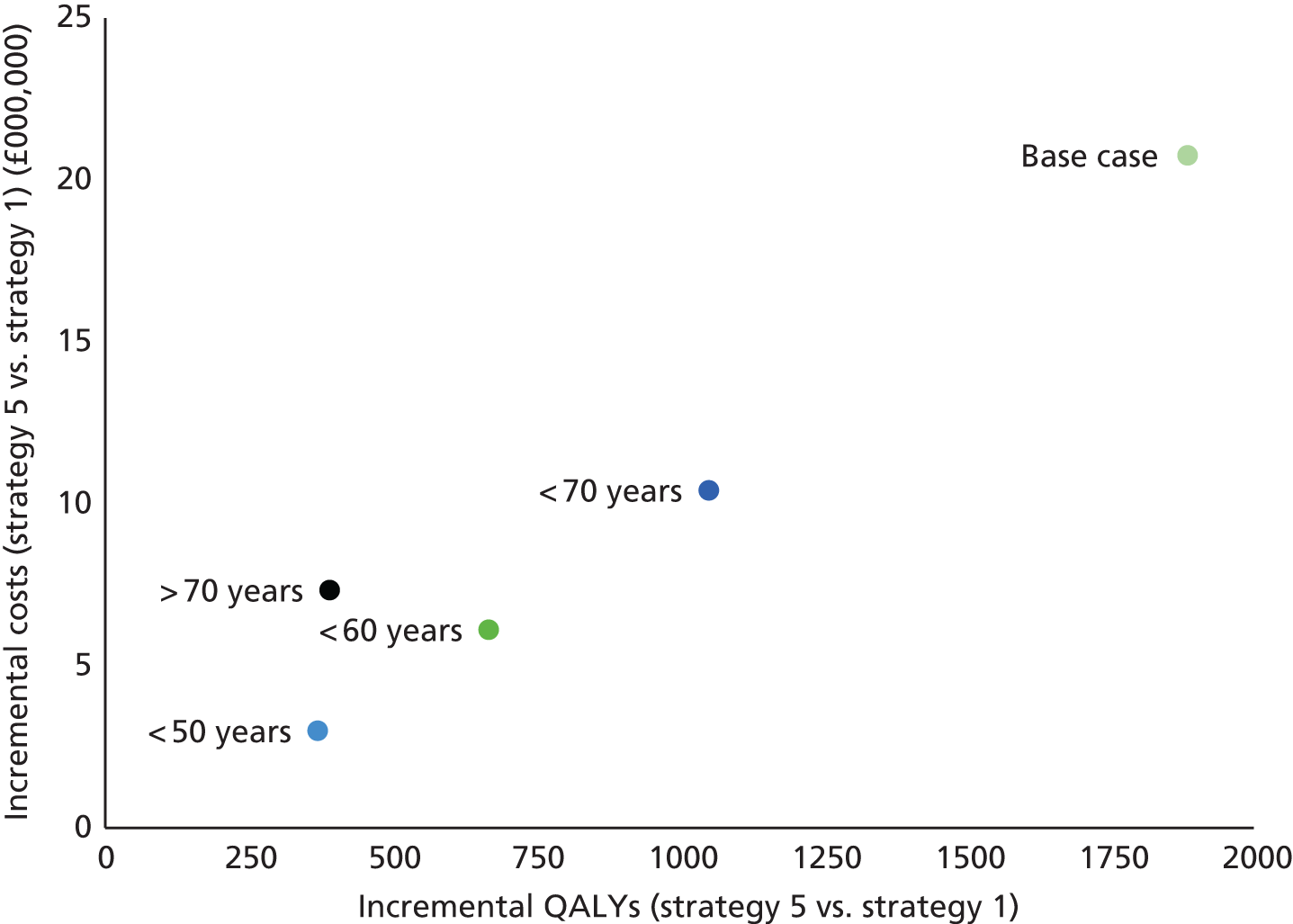
Imposing a minimum age limit of 70 years has a detrimental impact on cost-effectiveness and has not been recommended by any organisations. However, using the base-case assumptions and parameter values, testing using strategy 5 in this scenario is still cost-effective at a cost-effectiveness threshold of £20,000 per QALY (ICER £18,774 per QALY).
The decision modelling has assumed that there is no direct cost to imposing an age threshold (which is probably a reasonable assumption) but also that the imposition of an age threshold does not result in reduced compliance with reflex testing.
Chapter 7 Discussion
Review of test accuracy evidence
Findings in relation to previous studies
Ten studies met the test accuracy review inclusion criteria. One of the included studies34 had two distinct samples (a population-based sample and a high-risk sample); therefore, the 10 studies included 11 populations/samples. IHC was conducted in all 10 studies (11 samples); however, not all studies provided sufficient data to be included in the IHC analyses. Indeed, the high-risk sample in the study by Poynter et al. 34 and the sample in the study by Mueller et al. 58 provided too few data to be included in any of the IHC analyses.
A variety of studies were included:
-
four single-gate studies with population-based samples, including an unselected CRC sample34 and three age-limited populations54–56
-
five single-gate studies based on high-risk populations34,57–60
-
two studies that used a variation on a two-gate study design61,62 in which participants with positive reference standard results were recruited but no reference standard-negative participants were recruited (termed reference standard-positive studies for the purposes of this report).
Overall, there were no concerns about whether or not the included participants matched the review question. The reference standard was assessed as being likely to correctly classify the target condition and there were no concerns that the target condition, as defined by the reference standard, did not match the review question. However, all studies were rated as unclear with regard to whether the conduct and interpretation of the test could have introduced bias. It was also unclear for all studies whether the flow of participants through the study could have introduced bias.
Only four studies were rated as having a low risk of bias for patient selection. 54–56,62 However, none of the studies was rated as having a high risk of bias for patient selection, even though the index tests are highly susceptible to spectrum effects in populations that have been selected on the basis of clinical characteristics. This is because it was decided prior to conducting the review that only sensitivity estimates would be made for studies recruiting high-risk samples. This decision was based on the fact that a previous review42 did not find significant bias in estimates of sensitivity after recruitment of high-risk samples.
Evidence for microsatellite instability testing
With the exception of the studies by Limburg et al. 55 and Okkels et al. ,62 all studies assessed MSI testing.
Sensitivity results for microsatellite instability testing
Across all nine study samples (population-based and high-risk samples), sensitivity ranged from 66.7% (95% CI 47.2% to 82.7%)54 to 100.0% (95% CI 93.9% to 100.0%)34 when MSI-L was considered to be a negative result. In a previous review by Palomaki et al. 42 data from 11 studies (one age-limited sample and 10 high-risk samples) were pooled in a random-effects model and the sensitivity of MSI was reported to be 85% (95% CI 75% to 92%) for identifying MLH1 mutations and 85% (95% CI 73% to 93%) for identifying MSH2 mutations. Data were pooled from five studies to produce a sensitivity of MSI of 69% (95% CI 46% to 85%) for identifying MSH6 mutations. In all of these analyses, MSI-L was considered to be a negative index test result. It is difficult to compare the results of Palomaki et al. 42 with the results of this review, primarily because different inclusion criteria were used, in particular for the reference standard. In addition, data were not pooled in this review and were not split according to gene. Nevertheless, the sensitivities and CIs reported by Palomaki et al. 42 were similar to those in this review. Similarly, a review by Bonis et al. 17 provided an overall sensitivity of 56–100% for MSI-H compared with MSS, based on 16 studies. Again, although these results appear to be similar to those from this review, because of differences in the inclusion criteria used in the study by Bonis et al. 17 and this review any comparison made may not be particularly meaningful. For example, in the study by Bonis et al. ,17 any genetic testing was included as the reference standard and so a wider range of studies was included.
In this review of test accuracy, it was noted that sensitivity increased if MSI-L was considered to be a positive index test result. Indeed, across the nine study samples, the lower end of the range for sensitivity increased to 79.4% (95% CI 62.1% to 91.3%),57 with the higher end of the range remaining at 100%. The review by Bonis et al. 17 included seven studies in which MSI-L was considered to be a positive index test result and also found increased sensitivity estimates, with pooled data producing a summary estimate for sensitivity of 94% (95% CI 86% to 97%). Indeed, when MSI-L tumour results were excluded from the analyses, summary sensitivity was reported as 80% (95% CI 63% to 90%) because the number of positive results was reduced. As previously mentioned, it is unsurprising that including MSI-L as a positive index test result increases sensitivity because this essentially decreases the threshold for a positive test result.
Another review, by Snowsill et al,4 also reported sensitivity data for MSI testing, with sensitivity ranging from 88% to 100% when MSI-L was considered to be a positive index test result. Again, although these estimates are similar to those reported in this review, comparison may not be particularly useful because of the different inclusion criteria used. The review by Snowsill et al. 4 included nine high-risk or age-limited studies (studies were included if the participants were at risk of LS as a result of being aged < 50 years at diagnosis or because of clinical criteria or family history indicators); studies were also included in which not all patients received the reference standard; and looser criteria were applied to the reference standard than those applied in this review. It is notable, however, that despite these differences, the results are similar. This may, in part, be because of a lack of data identified in this review from unselected CRC populations.
Specificity results for microsatellite instability testing
Three population-based study samples provided data in which MSI-L was considered to be a negative index test result. 34,54,56 Across these three samples, specificity ranged from 61.1% (95% CI 57.0% to 65.1%)34 to 92.5% (95% CI 89.1% to 95.2%). 54 It should be noted that the study by Barnetson et al. 54 included an age-limited sample whereas the sample in the study by Poynter et al. 34 was based on an unselected CRC population. Six studies that provided data in which MSI-L was considered to be a negative index test result were pooled in the review by Palomaki et al. ,42 with summary specificity given as 90.2% (95% CI 87.7% to 92.7%). This result is closer to the specificity reported by Barnetson et al. 54 in this review and this may be, in part, because the study by Palomaki et al. 42 included only high-risk or age-limited populations. The review by Bonis et al. 17 reported an overall specificity of between 17% and 93% for MSI-H compared with MSS, based on 16 studies, indicating substantial heterogeneity. Again, it is difficult to compare the results from these previous reviews with the results of this review because of key differences in the inclusion criteria applied.
In this review, and as expected, specificity decreased when MSI-L was considered to be a positive index test result. The lower end of the range decreased to 29.5% (95% CI 25.8% to 33.4%)34 and the upper end of the range decreased to 84.5% (95% CI 80.0% to 88.2%). 54 Pooling data from seven studies included in the review by Bonis et al. 17 resulted in a specificity of 83% (95% CI 77% to 88%) when MSI-L was considered to be a positive index test result, whereas specificity was 88% (95% CI 83% to 91%) when MSI-L tumours were excluded. However, the authors noted that there was substantial statistical heterogeneity in both analyses. Snowsill et al. 4 reported a specificity range of 68–84% when MSI-L was considered to be a positive index test result, the upper end of which is comparable to the upper end of the range in this review. However, the review by Snowsill et al. 4 was based on high-risk or age-limited studies only and so does not include the population-based sample from the study by Poynter et al. 34
Evidence for immunohistochemistry
Seven studies54–57,59–61 provided data to assess the accuracy of an overall IHC result at identifying a positive reference standard result (i.e. whether a positive IHC result, regardless of which protein this applies to, identifies a positive reference standard result). All seven of these studies assessed MLH1, MSH2 and MSH6 proteins. The studies by Southey et al. 56 and Overbeek et al. 59 also assessed PMS2. Five studies34,54,56,61,62 split IHC data according to the particular protein assessed (MLH1, MSH2, MSH6 or PMS2).
Overall sensitivity and specificity results for immunohistochemistry
Sensitivity estimates ranged from 80.8% (95% CI 60.6% to 93.4%)60 to 100.0% (95% CI 81.5% to 100.0%)56 for IHC overall. The study by Southey et al. 56 included an assessment of PMS2 as well as MLH1, MSH2 and MSH6, which may account for the higher sensitivity estimate in this study. Nevertheless, all sensitivity estimates were > 80%. Specificity was estimated as 91.9% (95% CI 86.3% to 95.7%)55 and 80.5% (95% CI 65.1% to 91.2%). 56
Other reviews have produced similar results, but, again, it must be noted that these reviews used different inclusion criteria from the inclusion criteria used in this review. The review by Bonis et al. 17 produced a summary sensitivity of 74% (95% CI 54% to 87%) and specificity of 77% (95% CI 61% to 88%) for IHC overall, based on six studies considered to be of good or fair quality. Similar results were reported by Palomaki et al. 42 based on pooled data from three studies; overall sensitivity was estimated as 77% (95% CI 69% to 84%) and overall specificity as 88.8% (95% CI 67.6% to 94.8%), although statistical heterogeneity was noted. Similarly, without pooling data, Snowsill et al. 4 reported a sensitivity range of 73.3–100.0% and a very wide ranging specificity of 12.5–100.0%. Indeed, Snowsill et al. 4 discuss how specificity appears to be the greatest concern with regard to IHC, as the high number of false-positive results means that individuals may be told that they have LS when they do not. However, in the two studies included in this review, the specificity of IHC was > 80%. This difference is likely to occur because the specificity data from the review by Snowsill et al. 4 were based on high-risk as well as age-limited studies, whereas, to mitigate spectrum effects, the specificity data in this review were based on only two studies that recruited age-limited populations.
Sensitivity and specificity of immunohistochemistry by individual protein
For the population-based sample from Poynter et al. 34 and the study samples from Barnetson et al. ,54 Southey et al. ,56 Hendriks et al. 61 and Okkels et al. ,62 an assessment of IHC was made for at least one individual protein. With regard to estimating sensitivity, three of these studies54,56,61 provided data relevant to whether loss of expression of MLH1 was an accurate test result for assessing a pathogenic mutation in MLH1. Sensitivities ranged from 50.0% (95% CI 26.0% to 74.0%)56 to 100.0% (95% CI 73.5% to 100.0%). 54 The results for MSH2 were even more variable; the same three studies reported sensitivities ranging from 22.2% (95% CI 6.4% to 47.6%)56 to 81.8% (95% CI 48.2% to 97.7%). 54,61 Four studies provided data for MSH654,56,61,62 and, again, there was substantial variability in the results, with sensitivities ranging from 44.4% (95% CI 21.5% to 69.2%)56 to 75.0% (95% CI 19.4% to 99.4%). 54,61 Only the study by Southey et al. 56 provided IHC data for PMS2, with a sensitivity estimate of 55.6% (95% CI 30.8% to 78.5%).
The results for specificity displayed less variability. Three population-based studies34,54,56 provided data enabling the estimation of the specificity of loss of expression of MLH1 for identifying pathogenic mutations in MLH1. These specificities ranged from 70.6% (95% CI 66.8% to 74.2%)34 to 96.0% (95% CI 93.1% to 97.9%). 54 Two population-based studies54,56 provided data for MSH2 and MSH6, with all specificities being > 92%. Only the study by Southey et al. 56 provided data for PMS2, with a specificity estimate of 87.8% (95% CI 73.8% to 95.9%).
The review by Palomaki et al. 42 investigated whether overall IHC results indicated gene-specific pathogenic mutations. However, none of the previous systematic reviews identified through database searching provided an evaluation of whether loss of expression of a particular protein indicates a pathogenic mutation in that protein.
Strengths
The strengths of this systematic review are that it was conducted by an independent, experienced research team using the latest evidence and working to a prespecified protocol (PROSPERO CRD42016033879) that followed a robust methodology.
The search strategy was devised by an information specialist and did not restrict the results by study design and also included both forward and backward citation chasing. The studies were independently screened by two reviewers, with data extraction and quality appraisal performed by one reviewer and checked by a second.
Weaknesses
The relatively low prevalence of LS means that comprehensive MMR gene testing among all CRC patients in the general population would be expensive. Therefore, as mentioned previously, five34,57–60 of the 10 studies included in this review were single-gate studies based on high-risk populations. There is some concern that the increased presence of MMR mutation carriers in a population may change the apparent sensitivity and specificity of the index tests. However, we did not use these studies to estimate specificity and we did not find great differences between the studies with regard to sensitivity, although this may be because the three population-based studies with overall IHC data54–56 were based on age-limited populations and therefore may also be subject to spectrum bias. Palomaki et al. 42 also suggest that spectrum bias does not seem to have been an issue for estimating sensitivity. For example, when the population-based results for those aged < 55 years (80% for MLH1 and 82% for MSH2) are compared with the results of the other four studies that were based on strong family histories regardless of age (82% for MLH1 and 88% for MSH2), the results are remarkably similar. This may be because similar spectrum bias is obtained in the high-risk (age-unrestricted) population and the general age-limited population. The inclusion of high-risk studies in this review could therefore have introduced spectrum bias in the sensitivity estimates, although this does not appear to be the case. The inclusion of high-risk studies in this review does mean that there are several studies from which sensitivity estimates could be made, despite the low number of population-based studies identified (only one unselected CRC sample and three age-limited samples). However, the low number of population-based studies resulted in limited specificity data, as well as limited data for other outcomes (predictive values and LRs).
Although not a weakness of the review methods per se, significant methodological and clinical heterogeneity were noted across the included studies and this impacts on both the type of synthesis that could be conducted and the way in which the findings should be considered. In particular, when considering the results, it is important to note that the reference standard differed between studies, as did the index tests. With regard to the reference standard there were differences in the testing methods used, including the sequencing methods and the genes tested, the techniques used to test for large genomic alterations and deletions, the genes tested for large genomic alterations and deletions and whether unclassified variants were investigated.
For MSI testing, microdissection techniques were not always reported and the panel of markers varied, as did the thresholds and the categorisation of MSI. Indeed, none of the population-based studies in this review assessed the same panel of MSI markers. Issues with heterogeneity have also been reported in other similar reviews. For example, Bonis et al. 17 reported that they could not explain heterogeneity among the estimates of sensitivity and specificity based on ‘the overall study quality, the comprehensiveness of the genetic testing, the presence of microdissection, the use of National Cancer Institute (NCI)-recommended marker sets or whether the study used a transparent sample selection process’. As with this review, Palomaki et al. 42 found weaknesses in the studies included in their review, as well as clinical and methodological heterogeneity between studies. For example, none of the studies included in their review explicitly reported laser microdissection, which has been reported to be the optimal method for sample preparation. In addition, none of the studies reported a minimum proportion of tumour cells. Roughly half of the studies relied solely on the 1998 NCI-recommended panel, which includes only two mononucleotide markers, whereas the remaining studies utilised three or more mononucleotide markers. When the results were stratified by this quality measure, the clinical sensitivities in studies using three or more mononucleotide markers were higher than those in studies using two mononucleotide markers. 42 This provides some evidence to suggest that additional mononucleotide markers (up to five) should be included in a MSI panel for clinical testing. It was not possible to confirm this by running similar analyses in this review; to focus on the best reference standard testing methods, fewer studies were included and so insufficient homogeneous data were available to perform similar analyses. Indeed, the clinical and methodological heterogeneity in the studies included in this review precluded any statistical pooling of data. Instead, the results were discussed in a narrative synthesis.
It should also be noted that the review of test accuracy included only published studies in the English language. To enable a full evaluation of the methodological quality of included studies to be carried out, those studies published only as abstracts were also excluded.
It is possible that the estimates derived from the included studies are subject to some amount of publication bias. However, insufficient data were available from the included studies to perform a statistical assessment of publication bias.
The bibliographic searches were conducted in February 2016 and therefore it is possible that further relevant studies have been published or indexed since this date that have not been identified. There were also no attempts to review grey literature, such as technical reports, that would not have been peer reviewed.
Areas of uncertainty
The greatest area of uncertainty is in the generalisability of the results from high-risk and age-limited studies to the general CRC population. As discussed earlier, only one study sample was identified that was based on an unselected CRC population. 34 This is unsurprising given that large population-based studies in which all participants receive both the index test and the reference standard would be costly. Performing the reference standard on a random sample of index test negative participants would be methodologically acceptable and would somewhat decrease the costs of performing a population-based study in which all participants receive all tests.
Another area of uncertainty relates to the categorisation of unclassified variants [three34,54,55 of the four population-based studies reported on unclassified variants (i.e. mutations for which the association with LS is unclear)]. This can complicate the assessment of MSI, in particular, as the variants have uncertain pathogenicity and may occur in cases with either MSI-H or MSS tumours. In this review, in primary analyses, unclassified variants were counted as negative reference standard results. However, secondary analyses were also conducted, as appropriate, in which unclassified variants were considered to be positive reference standard results.
Unclassified variants: microsatellite instability
Two studies57,61 provided sufficient data to conduct secondary analyses in which unclassified variants were categorised as positive reference standard results. The study by Caldes et al. 57 included a high-risk population and the study by Hendriks et al. 61 was a reference standard-positive study and so only sensitivity estimates were made. When MSI-L was considered to be a negative index test result, Caldes et al. 57 reported a sensitivity of 81.6% (95% 65.7% to 92.3%) and Hendriks et al. 61 reported a sensitivity of 84.8% (95% CI 69.0% to 93.3%). These results were similar to those obtained when unclassified variants were considered to be negative reference standard results (79.4%, 95% CI 62.1% to 91.3%;57 88.8%, 95% CI 68.8 to 97.561).
When MSI-L was considered to be a positive index test result, only one study61 provided sufficient data to conduct a secondary analysis of the sensitivity estimate. In this study, sensitivity was 93.9% (95% CI 80.3% to 98.3%), which was similar to when unclassified variants were considered to be negative reference standard results (92.0%, 95% CI 74.0% to 99.0%).
Unclassified variants: immunohistochemistry
As for MSI, only two studies57,61 provided sufficient data to conduct secondary analyses. Again, because the study by Caldes et al. 57 included a high-risk population and the study by Hendriks et al. 61 was a reference standard-positive study, only sensitivity estimates were made. Sensitivity was 75.0% (95% CI 57.8% to 87.9%) for Caldes et al. 57 and 88.6% (95% CI 76.0% to 95.0%) for Hendriks et al. 61 For Caldes et al. 57 this represents quite a reduction in sensitivity compared with when unclassified variants were considered to be negative reference standard results (96.4%, 95% CI 81.7% to 99.9%).
Review of end-to-end studies
Findings in relation to previous studies
No studies were identified that met the inclusion criteria for the review of end-to-end studies.
The review by Bonis et al. 17 also attempted to identify end-to-end studies (their key question 1) but no studies were found.
Strengths
The review was conducted by an experienced researcher and the searches employed were very sensitive as no study design filters were employed. A second reviewer parallel screened a random sample of bibliographic records and excellent agreement was achieved. Although no studies were eventually included in the review, the prospective protocol prespecified the quality appraisal strategy to be used.
Weaknesses
The review focused on published literature indexed by bibliographic databases, meaning that grey literature was not identified. The searches were conducted in February 2016 and therefore it is possible that further relevant studies have been published or indexed since this date that have not been identified.
Areas of uncertainty
As no studies were identified for inclusion, there is no high-quality published evidence (up to February 2016) that screening for LS in CRC patients using MSI or IHC testing improves health outcomes.
Given that it is widely believed that such screening would improve health outcomes, it is unlikely that high-quality evidence will be generated in the future, for example in the form of a RCT. What evidence may be produced in the future will likely be observational and, although statistical methods may be used to remove certain biases, it is likely that there will be certain biases that cannot be adjusted for and that statistical power will be low compared with what could be produced by a RCT.
Review of existing cost-effectiveness evidence
Findings in relation to previous studies
The cost-effectiveness review found that, although no study fully answered the decision problem specified by the NICE scope,88 studies generally found that some testing strategy was cost-effective (according to the relevant perspective) and that tumour-based testing usually improved cost-effectiveness compared with direct genetic testing. The effectiveness of colonoscopy, the number of relatives receiving cascade genetic testing and the prevalence of LS in the population were generally identified as key parameters to which the results were sensitive.
These results are similar to the results of the previous review by the authors. 4
Other reviews of cost-effectiveness have been published. Grosse76 similarly found that studies usually produced one or more cost-effective strategy, but that these sometimes used age limits or clinical criteria. The number of first-degree relatives identified for cascade genetic testing and the cost of gene sequencing were found to have a significant effect on cost-effectiveness, as did the frequency and cost of surveillance colonoscopy and the inclusion of extracolonic surveillance.
Ladabaum et al. 3 also reviewed the health economic literature on tumour testing and concluded that it is estimated to be cost-effective, especially if cascade genetic testing is employed.
Strengths
This review was conducted by an independent, experienced research team using the latest evidence and working to a prespecified protocol (PROSPERO CRD42016033879). Screening was conducted independently by two reviewers.
Weaknesses
Data extraction and quality appraisal were conducted by one reviewer. This has the potential to lead to inconsistencies and misinterpretation between the published studies and the review reporting.
As with all reviews of published literature, this review is open to publication bias and so some information may have been missed. The searches were conducted in February 2016 and therefore it is possible that further relevant studies have been published or indexed since this date that have not been identified.
More recent quality appraisal tools are available than that of Drummond and Jefferson,77 which was used in this review for consistency with the previous review. 4 The Philips et al. 186 or Evers et al. 187 checklists may now be more appropriate.
Areas of uncertainty
The reporting was of mixed quality and some studies were reported only in abstract form and therefore the data extracted from the studies and the conclusions drawn are likely to reflect this.
The results of the review highlighted the need for a cost-effectiveness analysis specific to the decision problem. No individual study was able to address this and the study results could not be adequately synthesised, given the combination of different strategies and modelling techniques used and the different perspectives of the studies.
Although the included studies indicated that a strategy of diagnostic testing for LS is likely to be cost-effective for each of the described contexts, conclusions could not be drawn with regard to which strategies would be most cost-effective, particularly from the perspective of the NHS.
Independent economic assessment
Findings in relation to previous studies
The base-case estimates from the independent economic assessment suggest that testing for LS in CRC patients would be cost-effective (compared with not testing) at a willingness-to-pay threshold of £20,000 per QALY, except in the case in which patients are offered comprehensive genetic testing for LS without any tests being conducted on the tumour. In this case, testing is estimated to be cost-effective compared with no testing at a willingness-to-pay threshold of £30,000 per QALY. This universal testing strategy is, however, not predicted to be cost-effective in a fully incremental analysis (in which testing with MMR IHC, BRAF V600E testing and MLH1 hypermethylation testing is cost-effective). Indeed, it is estimated that the universal genetic testing strategy would be more expensive and less effective than another strategy.
These findings are similar to those in the previous PenTAG economic evaluation4 (in particular the scenario analysis, in which the age limit for testing was raised to 70 years), even though in the base case of this assessment there was no age limit and a number of changes have been made to the analysis (e.g. new diagnostic strategies, new parameter estimates for diagnostic accuracy, inclusion of aspirin chemoprevention and gynaecological surveillance).
The current analysis suggests that IHC-based testing would be more cost-effective than MSI-based testing, whereas the previous PenTAG analysis suggested the opposite. This is partially because the sensitivity of IHC modelled in this study is higher than that modelled previously and the specificity of MSI testing is lower. There is no compelling evidence that either test has superior diagnostic performance and the costs for both tests can vary between settings (e.g. in some hospitals a pathologist will be able to perform IHC but there will be no facility for MSI testing and samples will need to be sent elsewhere at a cost). BRAF V600E and/or MLH1 hypermethylation testing are conducted only on patients with abnormal MLH1 protein staining on IHC testing, whereas these tests are conducted for all patients with MSI (so all else being equal there would be a reduction in downstream costs if IHC were used). IHC-based testing is also required in a small minority of cases to assist in the interpretation of MMR mutation testing and therefore some individuals receiving MSI testing initially may later undergo IHC-based testing as well.
There are no other UK-based cost–utility analyses for further comparison, but the results of this study are also comparable to those of a number of studies from elsewhere. For example, Mvundura et al. 84 (a US study) found that tumour-based testing (using IHC and BRAF V600E testing) was cost-effective (ICER US$22,552 per life-year), whereas direct genetic testing was not cost-effective (ICER > US$142,289 per life-year compared with no testing and ICER US$737,025 per life-year in a fully incremental analysis). Wang et al. 86 (another US study) found that tumour-based testing may be cost-effective (ICER US$59,719 per QALY) but that direct genetic testing would not be cost-effective (ICER US$271,219 per QALY).
Strengths
The economic evaluation was conducted by an independent academic group with experience in reviewing and modelling in this disease area. The population, interventions, comparators and outcomes were based on the NICE scope. 88
The model was built by extending an existing model that had previously been quality assured and peer reviewed. The extensions mean that the model now more closely matches likely clinical practice.
The model methodology (individual patient simulation) allowed for detailed modelling, including of multiple cancers, different CRC stages, evolving strategies for gynaecological risk reduction and event probabilities that can be dependent on the time since previous events. This means that there was no need to make assumptions about constant hazards of events (i.e. exponential survival distributions).
The evidence for diagnostic test accuracy parameters was identified through systematic review. Evidence for key model parameters was identified by reviewing the published literature and critically appraising alternative sources.
Scenario and sensitivity analyses were conducted to identify key sources of decision uncertainty.
The model was quality assured by developers testing each other’s components.
Weaknesses
The weaknesses of the systematic review of test accuracy are carried through to the economic assessment, as the cost-effectiveness of the testing strategies is sensitive to the test accuracy parameter values.
The model base case used sensitivity and specificity estimates for MSI and IHC from a meta-analysis of studies identified in the systematic review of test accuracy, even though for a number of reasons meta-analysis was not felt to be appropriate for the review.
The model assumed that the diagnosis of LS in the probands and relatives occurs at the model start, that is, that there are no delays to testing and that surveillance does not commence before testing is completed.
To achieve convergence in model estimates, 240,000 individual patients were simulated. A probabilistic sensitivity analysis was not conducted because of the computational requirements of such an analysis and therefore decision uncertainty was explored only using scenario analyses and sensitivity analyses, with limited numbers of parameters varied simultaneously.
The model does not include ovarian cancer, small bowel cancer, gastric cancer or other cancers associated with LS. Of these, ovarian cancer would be most likely to have an affect on cost-effectiveness, as the model already includes the costs of risk-reducing interventions for ovarian cancer (these are the same as those for endometrial cancer) but none of the benefits.
Areas of uncertainty
There is significant uncertainty as to the true risk of CRC for individuals with LS not receiving colorectal surveillance and the true effectiveness of colorectal surveillance in reducing the risk of CRC. The cost-effectiveness of the testing strategies was sensitive to both of these parameters. Neither parameter is likely to be directly investigated in an experimental context (e.g. a RCT) for ethical reasons. The parameter values for each were chosen with care following a review of the literature and are more conservative than estimates made in other economic evaluations and systematic reviews. For example, a recent technical review3 estimated that CRC incidence would be reduced more heavily by colorectal surveillance than we have estimated and most reports quote a higher range of lifetime CRC risks than we have modelled. Univariate sensitivity analyses were conducted on these parameters but two-way sensitivity analysis was not conducted.
There remains substantial uncertainty in the estimates of test accuracy measures for MSI- and IHC-based testing, as there are few studies investigating test accuracy that are not at high risk of bias (e.g. because of applying the reference standard only when the index test is positive or some family history criteria are met). If the true sensitivity and specificity of MSI are as low as explored in sensitivity analysis, it would not be cost-effective (compared with not testing) to screen using MSI-based strategies (at a willingness-to-pay threshold of £20,000 per QALY).
It is possible that the costs of genetic counselling included in the economic evaluation are too low. The economic evaluation assumes a base-case total cost of £63 for each person offered genetic testing for LS. A specialist committee member in the NICE Diagnostics Assessment Programme process noted that the NHS reference costs for outpatient clinics in clinical genetics are significantly higher than this, with an average cost of £432 per clinic (2014/15 prices). 178 In addition, the 2013 NICE clinical guidelines on familial breast cancer188 made recommendations informed by an economic evaluation that (breast cancer-) affected individuals would require two counselling sessions in relation to BRCA mutation testing and unaffected individuals would require three counselling sessions. The costing report estimated that these would cost £125 each, whereas the economic evaluation estimated that they would cost approximately £400 each for the index individual and £300 each for a family member. A separate microcosting study of ‘standard’ cancer genetics pathways for BRCA mutation testing estimated that referral and counselling costs for an index individual would be £460 and for a relative would be £335 (based on supplementary Tables 1 and 2). 189 If these costs (uprated from 2013/14 prices) were used in our economic evaluation, testing would become less cost-effective compared with no testing, with the ICER for strategy 5 increasing from £11,008 per QALY to £11,764 per QALY. However, the Mainstreaming Cancer Genetics programme has operated successfully including only one post-test counselling session for index individuals found to have a pathogenic BRCA mutation, one pretest counselling session for relatives and one post-test counselling session for relatives with the mutation. 190,191 Such a regime would reduce costs further, as generally half or fewer of those tested for LS mutations are found to have a pathogenic MMR mutation.
It was also suggested by a specialist committee member in the NICE Diagnostics Assessment Programme process that the costs of genetic testing included in the economic evaluation are too high. Base-case costs of £1276 to test all four MMR genes and £949 to test only MLH1 and PMS2 were included in the model. The UKGTN database was searched in December 2016 and two laboratories were found to offer sequencing and copy number analysis of all four MMR genes for £650 and £860. 175 Replacing the base-case costs with £755 improves the cost-effectiveness of testing compared with no testing, with the ICER for strategy 5 decreasing from £11,008 per QALY to £10,596 per QALY.
The economic evaluation assumed that the diagnostic performance of MLH1 promoter methylation testing would be independent of the BRAF V600E status of tested individuals. This may be an unrealistic and optimistic assumption if these tumour markers are correlated, even after controlling for LS status. A literature review by Parsons et al. 192 identified three studies that evaluated MMR mutation, BRAF V600E and MLH1 promoter methylation status. A total of 107 tumours were included in these studies, of which 35 had a MMR mutation. The sensitivity and specificity of MLH1 promoter methylation to detect a MMR mutation (not conditional on BRAF V600E status) were 91.4% and 75.0% respectively. When this was limited to tumours without BRAF V600E, the specificity dropped to 60.9% (the sensitivity estimate was unchanged). Changing the economic evaluation base-case specificity of 75% for MLH1 promoter methylation testing following BRAF V600E testing to 60.9% leads to a lower cost-effectiveness for strategies 5 and 9 (which include BRAF V600E and MLH1 promoter methylation testing), with the ICER of strategy 5 increasing from £11,008 per QALY to £11,140 per QALY. Strategy 5 remains the optimal strategy assuming a willingness-to-pay threshold of £20,000 per QALY.
The economic assessment has also, by necessity, incorporated evidence from countries other than the UK. For example, the effectiveness of colonoscopic surveillance was estimated from a Finnish study,124 the prevalence of LS among CRC patients was estimated from a US study92 and the lifetime risks of CRC and endometrial cancer were estimated from a French study. 18 Although there is no evidence to suggest that these findings will not generalise to the UK, this remains an area of uncertainty.
It appears that gynaecological surveillance and aspirin chemoprevention do not have a significant impact on cost-effectiveness and therefore the uncertainty in the extent to which they are offered to patients is unlikely to translate into decision uncertainty.
Chapter 8 Conclusions
There is evidence that MSI testing and MMR IHC are effective tests to identify CRC patients who may have LS. This evidence comes from 10 published studies that were judged not to be at high risk of bias. Some studies did not include patients known not to have LS and could not therefore provide estimates of specificity for the tests. The reference standard (comprehensive genetic testing) was variable across the studies and cannot be considered a gold standard, particularly because of the number of variants of uncertain significance identified. A number of studies used clinical criteria (or other means) to select from a high-risk population for testing, which puts their results at risk of spectrum bias.
Because of the limited number of population-based studies and the heterogeneity in test accuracy estimates from these studies, it was judged that quantitative synthesis (meta-analysis) should not be conducted as part of the systematic review.
It is primarily cost considerations that prohibit a large-scale high-quality assessment of the diagnostic test accuracy of MSI- and IHC-based testing in CRC patients. Conducting both index tests and the reference standard would cost > £1000 per patient.
No end-to-end studies were identified that directly investigated the impact of screening for LS in CRC patients on long-term outcomes such as survival or cancer incidence.
It is unlikely that any high quality end-to-end studies will be conducted (e.g. RCTs or cluster RCTs) because of ethical considerations; it is believed, and supported by modelling, that identifying LS-causing mutations and offering risk-reducing interventions leads to clinical benefits for patients and so there would not be clinical equipoise.
The majority of health economic evaluations of screening for LS in CRC patients have suggested that it may be cost-effective to screen using MSI- or IHC-based testing. However, these studies may not reliably generalise and therefore a new economic evaluation was conducted as part of this study.
The results of the economic evaluation suggest that it is likely to be cost-effective (at a willingness-to-pay threshold of £20,000 per QALY) to screen CRC patients using IHC, BRAF V600E testing and MLH1 promoter methylation testing. This is estimated to extend the life expectancy of patients with LS by around 1 year and the life expectancy of their relatives with LS-causing mutations by 2 years. There are estimated to be 956 such patients (CRC patients with LS) and 2524 corresponding relatives per year. The diagnosis costs are estimated to be around £10.5M per year (compared with an estimated £1.9M cost of MSI/IHC conducted to predict 5-FU chemotherapy response). It is estimated that additional QALYs would be gained at an incremental cost of £11,000 per QALY.
The cost-effectiveness of screening for LS is estimated to be most sensitive to the:
-
effectiveness of surveillance colonoscopy in reducing CRC incidence
-
diagnostic test accuracy of MSI testing and IHC
-
proportion of patients declining genetic counselling and/or genetic testing after tumour testing
-
lifetime risk of CRC for individuals with LS-causing mutations
-
number of relatives identified through cascade testing
-
cost of colonoscopy.
Of these, the cost of colonoscopy is most precisely known (on average, the cost does vary between trusts), although the frequency of colonoscopy should be considered; the model assumes biennial colonoscopy but costs would be significantly increased if annual colonoscopy was implemented.
The number of relatives identified through cascade testing and the proportion of patients declining genetic counselling and/or genetic testing after tumour testing can be estimated retrospectively from clinical records or can be monitored prospectively, although the values employed in sensitivity analyses are likely to be worst-case scenarios.
The sensitivity and specificity of MSI and IHC testing cannot be directly monitored by introducing the tests into clinical practice (although the PPV could be estimated). As explained earlier, a comparative diagnostic test accuracy study would be relatively expensive to conduct. When the sensitivity and specificity of IHC, BRAF V600E and MLH1 promoter methylation testing are all estimated from their respective lower 95% confidence limits it is still predicted that screening is cost-effective at a willingness-to-pay threshold of £20,000 per QALY.
The effectiveness of surveillance colonoscopy and the lifetime risk of CRC for individuals with LS-causing mutations (in the absence of risk-reducing interventions) are subject to significant uncertainty, for the reason that they cannot be simply observed without bias.
The effectiveness of surveillance colonoscopy cannot be examined in a RCT for ethical reasons and is therefore generally estimated by comparing the risks observed in individuals undergoing screening with the risks in those diagnosed with LS and declining surveillance or those not diagnosed with LS but later established to have LS. Neither of these comparisons is between like groups.
The lifetime risk of CRC for individuals with LS-causing mutations but without risk-reducing interventions can generally be estimated only by studying the cancer incidence prior to diagnosis and the commencement of risk-reducing interventions and also correcting for ascertainment bias. Estimates are therefore of limited precision, even from large studies.
When either of these parameters is modelled at its extreme value, the cost-effectiveness of screening is marginal at a willingness-to-pay threshold of £20,000 per QALY (ICERs ≈£19,000 per QALY). The extreme value for the effectiveness of colonoscopy (zero effectiveness) may not be considered clinically likely, but the extreme value for the lifetime CRC risk is the lower 95% confidence limit. The base-case estimates for these parameters are from different populations from different studies, but if a study were designed to estimate both parameters there would be significant correlation in the parameter estimates, as a low observed cancer risk in those undergoing surveillance could result from surveillance being highly effective or the underlying risk already being low. This would mean that the precision in estimated cost-effectiveness would not be increased as much as if the parameters were not correlated.
One way to partially ameliorate this issue might be to delay colorectal surveillance for individuals with MSH6 or PMS2 mutations to 10–20 years later than for individuals with MLH1 or MSH2 mutations. 2,5 It is considered that this could save 5–10 colonoscopies per individual, with a minimal increase in the number of CRCs.
It is also possible that other developments in the management of individuals with LS-causing mutations or the management of CRC or endometrial cancer will lead to shifts in the cost-effectiveness of screening. For example, a vaccine targeting frameshift peptides, which are produced by MSI-H cancer cells, is considered to be a promising avenue for treating MSI-H cancer and for cancer prevention in individuals with LS. 193
We have also assumed that treatment for CRC does not include monoclonal antibody chemotherapy, which means that the costs associated with CRC may be underestimated. If CRC treatment is (or becomes) more expensive than is modelled in this report, the cost-effectiveness of screening for LS will be improved.
Recommendations for research
Screening for Lynch syndrome in endometrial cancer patients
Given the excellent survival of endometrial cancer patients (compared with colorectal and ovarian cancer patients), there is an argument that endometrial cancer patients (either early-onset or all patients) should be screened for LS, as they will stand a good chance of benefiting from colorectal surveillance and other risk-reducing interventions. It is recommended that the diagnostic test accuracy of tumour-based tests should be established in endometrial tumours and the cost-effectiveness of screening should be investigated through modelling of a similar nature to that employed in this report.
Screening for other colorectal cancer predisposition genes in colorectal cancer patients
There are other CRC predisposition genes beyond those responsible for LS. It may be economical to test CRC patients for all such genes using NGS technology, but not if tumour-based tests are used that enrich the population specifically for LS testing. An assessment of the yield of clinically actionable diagnoses based on large cancer risk gene panels compared with targeted testing for LS-causing mutations could be conducted, along with an economic evaluation of whether such screening would be cost-effective compared with screening for LS (if this is recommended).
Costs of diagnostic tests
The cost estimates for diagnostic tests have generally been reported by genetics laboratories, rather than being formally costed. It may be worthwhile to perform a full costing exercise for these tests to determine accurate costs from a NHS and PSS perspective. The potential for cost savings through the use of NGS technology should also be assessed.
Acknowledgements
We are grateful for the excellent administrative assistance of Sue Whiffin (including proofreading) and Jenny Lowe (including document retrieval for the systematic reviews). We thank Martin Hoyle for reviewing the report. We thank Ottie O’Brien (Northern Molecular Genetics Service, Newcastle upon Tyne) for providing information regarding current clinical practice and the costs of diagnostic tests; Sandi Deans [UK National External Quality Assessment Scheme (UK NEQAS), Edinburgh] for cascading questions to genetics laboratories; and Samantha Butler (West Midlands Regional Genetics Laboratory, Birmingham Women’s NHS Foundation Trust) for providing information on current clinical practice.
Contributions of authors
Tristan Snowsill (Research Fellow, Health Economic Modelling) contributed to the systematic reviews of end-to-end studies and economic evaluations, contributed to the development, implementation and checking of the independent economic assessment, provided overall project management and contributed to the writing and editing of the report.
Helen Coelho (Research Fellow, Systematic Review) led the systematic review of diagnostic test accuracy and contributed to all aspects of this review and contributed to the writing and editing of the report.
Nicola Huxley (Research Fellow, Health Economic Modelling) led the review of published cost-effectiveness studies, contributed to the development, implementation and checking of the independent economic assessment and contributed to the writing and editing of the report.
Tracey Jones-Hughes (Research Fellow, Systematic Review) contributed to all aspects of the diagnostic test accuracy systematic review, wrote the background section and contributed to the writing and editing of the rest of the report.
Simon Briscoe (Information Specialist) developed and conducted the literature searches for the systematic reviews of diagnostic test accuracy, end-to-end studies and economic evaluations and contributed to the writing and editing of the report.
Ian M Frayling (Consultant, Genetic Pathology) advised on current clinical practice and scientific understanding of Lynch syndrome and contributed to the writing and editing of the report.
Chris Hyde (Consultant, Public Health and Clinical Epidemiology) led the systematic review of end-to-end studies and contributed to the writing and editing of the report.
Data sharing statement
The authors are prepared to share a copy of the executable economic model (including the simulation results analysed in this report) on request and signing a data sharing agreement. Please contact the corresponding author. Requests for access to other data should also be addressed to the corresponding author.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Cancer Research UK. Bowel Cancer Survival Statistics. London: Cancer Research UK; 2013.
- Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on Colorectal Cancer. Gastrointest Endosc 2014;80:197-220. https://doi.org/10.1016/j.gie.2014.06.006.
- Ladabaum U, Ford JM, Martel M, Barkun AN. American Gastroenterological Association technical review on the diagnosis and management of Lynch syndrome. Gastroenterology 2015;149:783-813.e20. https://doi.org/10.1053/j.gastro.2015.07.037.
- Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, et al. A systematic review and economic evaluation of diagnostic strategies for Lynch syndrome. Health Technol Assess 2014;18. https://doi.org/10.3310/hta18580.
- Møller P, Seppälä T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut 2017;66:464-72. https://doi.org/10.1136/gutjnl-2015-309675.
- Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta 2013;1828:1989-2001. https://doi.org/10.1016/j.bbamem.2013.04.018.
- Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet 2009;41:112-17. https://doi.org/10.1038/ng.283.
- Kempers MJ, Kuiper RP, Ockeloen CW, Chappuis PO, Hutter P, Rahner N, et al. Risk of colorectal and endometrial cancers in EPCAM deletion-positive Lynch syndrome: a cohort study. Lancet Oncol 2011;12:49-55. https://doi.org/10.1016/S1470-2045(10)70265-5.
- Lynch HT, Lynch JF, Snyder CL, Riegert-Johnson D. EPCAM deletions, Lynch syndrome, and cancer risk. Lancet Oncol 2011;12:5-6. https://doi.org/10.1016/S1470-2045(10)70291-6.
- Munoz JC, Lambiase LR. Hereditary Nonpolyposis Colorectal Cancer 2015. http://emedicine.medscape.com/article/188613-overview#a5 (accessed 7 July 2016).
- Ahadova A, von Knebel Doeberitz M, Bläker H, Kloor M. CTNNB1-mutant colorectal carcinomas with immediate invasive growth: a model of interval cancers in Lynch syndrome. Fam Cancer 2016;15:579-86. https://doi.org/10.1007/s10689-016-9899-z.
- Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 1999;81:214-18. https://doi.org/10.1002/(SICI)1097-0215(19990412)81:2<214::AID-IJC8>3.0.CO;2-L.
- Haraldsdottir S, Hampel H, Wei L, Wu C, Frankel W, Bekaii-Saab T, et al. Prostate cancer incidence in males with Lynch syndrome. Genet Med 2014;16:553-7. https://doi.org/10.1038/gim.2013.193.
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group . Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 2009;11:35-41. https://doi.org/10.1097/GIM.0b013e31818fa2ff.
- Møller P, Seppälä T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, et al. Incidence of and survival after subsequent cancers in carriers of pathogenic MMR variants with previous cancer: a report from the prospective Lynch syndrome database [published online ahead of print 3 June 2016]. Gut 2016. https://doi.org/10.1136/gutjnl-2016-311403.
- Clinical Practice Guidelines in Oncology. Genetic/Familial High-Risk Assessment: Colorectal. Fort Washington, PA: National Comprehensive Cancer Network; 2015.
- Bonis PA, Trikalinos TA, Chung M, Chew P, Ip S, DeVine DA, et al. Hereditary nonpolyposis colorectal cancer: diagnostic strategies and their implications. Evid Rep Technol Assess 2007;150:1-180.
- Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011;305:2304-10. https://doi.org/10.1001/jama.2011.743.
- Vasen HF, Blanco I, Aktan-Collan K, Gopie JP, Alonso A, Aretz S, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 2013;62:812-23. https://doi.org/10.1136/gutjnl-2012-304356.
- Pande M, Lynch PM, Hopper JL, Jenkins MA, Gallinger S, Haile RW, et al. Smoking and colorectal cancer in Lynch syndrome: results from the Colon Cancer Family Registry and the University of Texas MD Anderson Cancer Center. Clin Cancer Res 2010;16:1331-9. https://doi.org/10.1158/1078-0432.CCR-09-1877.
- Cancer Registration Statistics, England: 2014. London: ONS; 2016.
- Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 2006;296:1479-87. https://doi.org/10.1001/jama.296.12.1479.
- Hampel H, de la Chapelle A. The search for unaffected individuals with Lynch syndrome: do the ends justify the means?. Cancer Prev Res 2011;4:1-5. https://doi.org/10.1158/1940-6207.CAPR-10-0345.
- Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterol 1999;116:1453-6. https://doi.org/10.1016/S0016-5085(99)70510-X.
- Desouza B. Development, Optimisation and Implementation of Clinical Management Protocols for Familial Colorectal Cancer. London: University of London; 2016.
- Schmeler KM, Lynch HT, Chen LM, Munsell MF, Soliman PT, Clark MB, et al. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med 2006;354:261-9. https://doi.org/10.1056/NEJMoa052627.
- Webber EM, Kauffman TL, O’Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer 2015;15. https://doi.org/10.1186/s12885-015-1093-4.
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509-20. https://doi.org/10.1056/NEJMoa1500596.
- Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 2011;378:2081-7. https://doi.org/10.1016/S0140-6736(11)61049-0.
- Vasen HFA, Moslein G, Alonso A, Bernstein I, Bertario L, Blanco I, et al. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer). J Med Genet 2007;44:353-62. https://doi.org/10.1136/jmg.2007.048991.
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004;96:261-8. https://doi.org/10.1093/jnci/djh034.
- Non-commercial Government Licence for Public Sector Information. Richmond: The National Archives; 2016.
- Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 1991;34:424-5. https://doi.org/10.1007/BF02053699.
- Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev 2008;17:3208-15. https://doi.org/10.1158/1055-9965.EPI-08-0512.
- Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn 2008;10:301-7. https://doi.org/10.2353/jmoldx.2008.080062.
- Ward RL, Dobbins T, Lindor NM, Rapkins RW, Hitchins MP. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med 2013;15:25-3. https://doi.org/10.1038/gim.2012.91.
- Frayling IM, Arends MJ. How can histopathologists help clinical genetics in the investigation of suspected hereditary gastrointestinal cancer?. Diagn Histopathol 2015;21:137-46. https://doi.org/10.1016/j.mpdhp.2015.04.004.
- Morak M, Heidenreich B, Keller G, Hampel H, Laner A, de la Chapelle A, et al. Biallelic MUTYH mutations can mimic Lynch syndrome. Eur J Hum Genet 2014;22:1334-7. https://doi.org/10.1038/ejhg.2014.15.
- Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet 2015;47:257-62. https://doi.org/10.1038/ng.3202.
- Ibrahim M. Immunohistochemistry of MMR protein expression. Biomed Sci 2009;53.
- Arends M, Ibrahim M, Happerfield L, Frayling I, Miller K. Interpretation of immunohistochemical analysis of mismatch repair (MMR) protein expression in tissue sections for investigation of suspected Lynch/hereditary non-polyposis colorectal cancer (HNPCC) syndrome. UK NEQAS ICC &Amp; ISH Recommendations 2008;1:1-2.
- Palomaki GE, McClain MR, Melillo S, Hampel HL, Thibodeau SN. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med 2009;11:42-65. https://doi.org/10.1097/GIM.0b013e31818fa2db.
- Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010;56:167-79. https://doi.org/10.1111/j.1365-2559.2009.03392.x.
- US National Library of Medicine . What Kinds of Gene Mutations Are Possible? n.d. https://ghr.nlm.nih.gov/primer/mutationsanddisorders/possiblemutations (accessed 13 July 2016).
- Thompson BA, Spurdle AB, Plazzer J-P, Greenblatt MS, Akagi K, Al-Mulla F, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 2014;46:107-15. https://doi.org/10.1038/ng.2854.
- Cairns SR, Scholefield JH, Steele RJ, Dunlop MG, Thomas HJ, Evans GD, et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups (update from 2002). Gut 2010;59:666-89. https://doi.org/10.1136/gut.2009.179804.
- Colorectal Cancer: The Diagnosis and Management of Colorectal Cancer. London: NICE; 2011.
- Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandalà M, Cervantes A, et al. Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24:64-72. https://doi.org/10.1093/annonc/mdt354.
- Loughrey M, Quirke P, Shepherd N. Dataset for Colorectal Cancer Histopathology Reports. London: Royal College of Pathologists; 2014.
- Kaur A, Monahan K, Schofield J, Lishman S, Alsina D, Bason N. PWE-346 Lynch syndrome and application of the RCPath colorectal cancer dataset in the United Kingdom. Gut 2015;64. https://doi.org/10.1136/gutjnl-2015-309861.792.
- Systematic Reviews: CRD’s Guidance for Undertaking Reviews in Health Care. York: Centre for Reviews and Dissemination, University of York; 2009.
- Whiting P, Westwood M, Beynon R, Burke M, Sterne JA, Glanville J. Inclusion of methodological filters in searches for diagnostic test accuracy studies misses relevant studies. J Clin Epidemiol 2011;64:602-7. https://doi.org/10.1016/j.jclinepi.2010.07.006.
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36. https://doi.org/10.7326/0003-4819-155-8-201110180-00009.
- Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 2006;354:2751-63. https://doi.org/10.1056/NEJMoa053493.
- Limburg PJ, Harmsen WS, Chen HH, Gallinger S, Haile RW, Baron JA, et al. DNA mismatch repair gene alterations in a population-based sample of young-onset colorectal cancer patients. Clin Gastroenterol Hepatol 2011;9. https://doi.org/10.1016/j.cgh.2010.10.021.
- Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, Tesoriero AA, et al. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J Clin Oncol 2005;23:6524-32. https://doi.org/10.1200/JCO.2005.04.671.
- Caldes T, Godino J, Sanchez A, Corbacho C, De La Hoya M, Asenjo JL, et al. Immunohistochemistry and microsatellite instability testing for selecting MLH1, MSH2 and MSH6 mutation carriers in hereditary non-polyposis colorectal cancer. Oncol Rep 2004;12:621-9. https://doi.org/10.3892/or.12.3.621.
- Mueller J, Gazzoli I, Bandipalliam P, Garber JE, Syngal S, Kolodner RD. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res 2009;69:7053-61. https://doi.org/10.1158/0008-5472.CAN-09-0358.
- Overbeek L, Kets C, Hebeda K, Bodmer D, Van Der Looij E, Willems R, et al. Patients with an unexplained microsatellite instable tumour have a low risk of familial cancer. Br J Cancer 2007;96:1605-12. https://doi.org/10.1038/sj.bjc.6603754.
- Shia J, Klimstra DS, Nafa K, Offit K, Guillem JG, Markowitz AJ, et al. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol 2005;29:96-104. https://doi.org/10.1097/01.pas.0000146009.85309.3b.
- Hendriks Y, Franken P, Dierssen JW, de Leeuw W, Wijnen J, Dreef E, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 2003;162:469-77. https://doi.org/10.1016/S0002-9440(10)63841-2.
- Okkels H, Lindorff-Larsen K, Thorlasius-Ussing O, Vyberg M, Lindebjerg J, Sunde L, et al. MSH6 mutations are frequent in hereditary nonpolyposis colorectal cancer families with normal pMSH6 expression as detected by immunohistochemistry. Appl Immunohistochem Mol Morphol 2012;20:470-7. https://doi.org/10.1097/PAI.0b013e318249739b.
- Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLOS Med 2009;6. https://doi.org/10.1371/journal.pmed.1000097.
- Ransohoff DF, Feinstein AR. Problems of spectrum and bias in evaluating the efficacy of diagnostic tests. N Engl J Med 1978;299:926-30. https://doi.org/10.1056/NEJM197810262991705.
- Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, van der Meulen JH, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA 1999;282:1061-6. https://doi.org/10.1001/jama.282.11.1061.
- Whiting P, Rutjes AW, Reitsma JB, Glas AS, Bossuyt PM, Kleijnen J. Sources of variation and bias in studies of diagnostic accuracy: a systematic review. Ann Intern Med 2004;140:189-202. https://doi.org/10.7326/0003-4819-140-3-200402030-00010.
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57.
- Dietmaier W, Hofstädter F. Detection of microsatellite instability by real time PCR and hybridization probe melting point analysis. Lab Invest 2001;81:1453-6. https://doi.org/10.1038/labinvest.3780358.
- Pawlik TM, Raut CP, Rodriguez-Bigas MA. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers 2004;20:199-206. https://doi.org/10.1155/2004/368680.
- Diagnostics Assessment Programme Manual. Manchester: NICE; 2011.
- Peninsula Technology Assessment Group (PenTAG) . Molecular Testing for Lynch Syndrome in People With Colorectal Cancer 2016. www.nice.org.uk/guidance/GID-DG10001/documents/final-protocol-2 (accessed 17 July 2017).
- Gallego CJ, Bennette CS, Heagerty P, Comstock B, Horike-Pyne M, Hisama F, et al. Comparative effectiveness of next generation genomic sequencing for disease diagnosis: design of a randomized controlled trial in patients with colorectal cancer/polyposis syndromes. Contemp Clin Trials 2014;39:1-8. https://doi.org/10.1016/j.cct.2014.06.016.
- Overbeek LI, Hermens RP, van Krieken JH, Adang EM, Casparie M, Nagengast FM, et al. Electronic reminders for pathologists promote recognition of patients at risk for Lynch syndrome: cluster-randomised controlled trial. Virchows Arch 2010;456:653-9. https://doi.org/10.1007/s00428-010-0907-7.
- Dineen S, Lynch PM, Rodriguez-Bigas MA, Bannon S, Taggart M, Reeves C, et al. A Prospective six sigma quality improvement trial to optimize universal screening for genetic syndrome among patients with young-onset colorectal cancer. J Natl Compr Canc Netw 2015;13:865-72. https://doi.org/10.6004/jnccn.2015.0103.
- Haraldsdottir S, Wu CSY, Hampel H, Frankel W, Pan XJ, Bekaii-Saab TS, et al. Universal screening for Lynch syndrome (LS) in colorectal cancer (CRC) and survival. J Clin Oncol 2014;32. https://doi.org/10.1200/jco.2014.32.3_suppl.415.
- Grosse SD. When is genomic testing cost-effective? Testing for Lynch syndrome in patients with newly-diagnosed colorectal cancer and their relatives. Healthcare 2015;3:860-78. https://doi.org/10.3390/healthcare3040860.
- Drummond MF, Jefferson TO. Guidelines for authors and peer reviewers of economic submissions to the BMJ. The BMJ Economic Evaluation Working Party. BMJ 1996;313:275-83. https://doi.org/10.1136/bmj.313.7052.275.
- Gallego CJ, Shirts B, Garrison L, Jarvik G, Veenstra DL. Can next generation sequencing save lives and provide a good economic value in colon cancer prevention?. Value Health 2014;17:A86-7. https://doi.org/10.1016/j.jval.2014.03.504.
- Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, et al. A model-based assessment of the cost–utility of strategies to identify Lynch syndrome in early-onset colorectal cancer patients. BMC Cancer 2015;15. https://doi.org/10.1186/s12885-015-1254-5.
- Barzi A, Sadeghi S, Kattan MW, Meropol NJ. Comparative effectiveness of screening strategies for Lynch syndrome. J Natl Cancer Inst 2015;107. https://doi.org/10.1093/jnci/djv005.
- Gallego CJ, Shirts BH, Bennette CS, Guzauskas G, Amendola LM, Horike-Pyne M, et al. Next-generation sequencing panels for the diagnosis of colorectal cancer and polyposis syndromes: a cost-effectiveness analysis. J Clin Oncol 2015;33:2084-91. https://doi.org/10.1200/JCO.2014.59.3665.
- Severin F, Stollenwerk B, Holinski-Feder E, Meyer E, Heinemann V, Giessen-Jung C, et al. Economic evaluation of genetic screening for Lynch syndrome in Germany. Genet Med 2015;17:765-73. https://doi.org/10.1038/gim.2014.190.
- Ramsey SD, Burke W, Clarke L. An economic viewpoint on alternative strategies for identifying persons with hereditary nonpolyposis colorectal cancer. Genet Med 2003;5:353-63. https://doi.org/10.1097/01.GIM.0000086626.03082.B5.
- Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med 2010;12:93-104. https://doi.org/10.1097/GIM.0b013e3181cd666c.
- Ladabaum U, Wang G, Terdiman J, Blanco A, Kuppermann M, Boland CR, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med 2011;155:69-7. https://doi.org/10.7326/0003-4819-155-2-201107190-00002.
- Wang G, Kuppermann M, Kim B, Phillips KA, Ladabaum U. Influence of patient preferences on the cost-effectiveness of screening for Lynch syndrome. J Oncol Pract 2012;8:e24s-30s. https://doi.org/10.1200/JOP.2011.000535.
- Colling R, Church DN, Carmichael J, Murphy L, East J, Risby P, et al. Screening for Lynch syndrome and referral to clinical genetics by selective mismatch repair protein immunohistochemistry testing: an audit and cost analysis. J Clin Pathol 2015;68:1036-9. https://doi.org/10.1136/jclinpath-2015-203083.
- National Institute for Health and Care Excellence . Molecular Testing for Lynch Syndrome in People With Colorectal Cancer: Final Scope n.d. www.nice.org.uk/guidance/dg27/documents/final-scope (accessed 14 July 2017).
- Guide to the Methods of Technology Appraisal 2013. London: NICE; 2013.
- Barrow E, Robinson L, Alduaij W, Shenton A, Clancy T, Lalloo F, et al. Cumulative lifetime incidence of extracolonic cancers in Lynch syndrome: a report of 121 families with proven mutations. Clin Genet 2009;75:141-9. https://doi.org/10.1111/j.1399-0004.2008.01125.x.
- Barrow P. Hereditary Colorectal Cancer: Registration, Screening and Prognostic Biomarker Analysis. Manchester: University of Manchester; 2015.
- Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 2008;26:5783-8. https://doi.org/10.1200/JCO.2008.17.5950.
- Jenkins MA, Baglietto L, Dowty JG, Van Vliet CM, Smith L, Mead LJ, et al. Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol 2006;4:489-98. https://doi.org/10.1016/j.cgh.2006.01.002.
- Sjursen W, Haukanes BI, Grindedal EM, Aarset H, Stormorken A, Engebretsen LF, et al. Current clinical criteria for Lynch syndrome are not sensitive enough to identify MSH6 mutation carriers. J Med Genet 2010;47:579-85. https://doi.org/10.1136/jmg.2010.077677.
- Sjursen W, McPhillips M, Scott RJ, Talseth-Palmer BA. Lynch syndrome mutation spectrum in New South Wales, Australia, including 55 novel mutations. Mol Genet Genomic Med 2016;4:223-31. https://doi.org/10.1002/mgg3.198.
- Cancer Statistics Registrations, England (Series MB1) – No. 37, 2006. London: ONS; 2008.
- Cancer Statistics Registrations, England (Series MB1) – No. 38, 2007. London: ONS; 2010.
- Cancer Statistics Registrations, England (Series MB1) – No. 39, 2008. London: ONS; 2010.
- Cancer Statistics Registrations, England (Series MB1) – No. 40, 2009. London: ONS; 2011.
- Cancer Statistics Registrations, England (Series MB1) - No. 41, 2010. London: ONS; 2012.
- Cancer Registration Statistics, England: 2011. London: ONS; 2013.
- Cancer Registration Statistics, England: 2012. London: ONS; 2014.
- Cancer Registration Statistics, England: 2013. London: ONS; 2015.
- Annual Mid-year Population Estimates: 2014. London: ONS; 2015.
- Mid-year Population Estimates of the Very Old (Including Centenarians): England and Wales, and United Kingdom, 2002 to 2014. London: ONS; 2015.
- Maeda T, Cannom RR, Beart RW, Etzioni DA. Decision model of segmental compared with total abdominal colectomy for colon cancer in hereditary nonpolyposis colorectal cancer. J Clin Oncol 2010;28:1175-80. https://doi.org/10.1200/JCO.2009.25.9812.
- Dinh TA, Rosner BI, Atwood JC, Boland CR, Syngal S, Vasen HFA, et al. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res 2011;4:9-22. https://doi.org/10.1158/1940-6207.CAPR-10-0262.
- Finan P, Smith J, Walker K, van der Meulen J, Greenaway K, Yelland A, et al. National Bowel Cancer Audit Annual Report. Leeds: NHS Health and Social Care Information Centre; 2011.
- Colorectal Cancer Survival by Stage. London: National Cancer Intelligence Network; 2009.
- Mortality Statistics: Deaths Registered in 2010 (Series DR). London: ONS; 2011.
- Mid-1971 to Mid-2010 Population Estimates: Quinary Age Groups for Constituent Countries in the United Kingdom; Estimated Resident Population. London: ONS; 2011.
- Cancer Incidence in Wales 2006–2010. Cardiff: Welsh Cancer Intelligence and Surveillance Unit; 2012.
- Lewin SN, Herzog TJ, Barrena Medel NI, Deutsch I, Burke WM, Sun X, et al. Comparative performance of the 2009 International Federation of Gynecology and Obstetrics’ staging system for uterine corpus cancer. Obstet Gynecol 2010;116:1141-9. https://doi.org/10.1097/AOG.0b013e3181f39849.
- Population Estimates for UK, England and Wales, Scotland and Northern Ireland. London: ONS; 2016.
- Mulder SA, Kranse R, Damhuis RA, Ouwendijk RJ, Kuipers EJ, van Leerdam ME. The incidence and risk factors of metachronous colorectal cancer: an indication for follow-up. Dis Colon Rectum 2012;55:522-31. https://doi.org/10.1097/DCR.0b013e318249db00.
- Barrow E, Hill J, Evans DG. Cancer risk in Lynch syndrome. Fam Cancer 2013;12:229-40. https://doi.org/10.1007/s10689-013-9615-1.
- Fajobi O, Yiu CY, Sen-Gupta SB, Boulos PB. Metachronous colorectal cancers. Br J Surg 1998;85:897-901. https://doi.org/10.1046/j.1365-2168.1998.00800.x.
- Cancer Survival in England – Patients Diagnosed 2006–2010 and Followed Up to 2011. London: ONS; 2012.
- Lin KM, Shashidharan M, Ternent CA, Thorson AG, Blatchford GJ, Christensen MA, et al. Colorectal and extracolonic cancer variations in MLH1/MSH2 hereditary nonpolyposis colorectal cancer kindreds and the general population. Dis Colon Rectum 1998;41:428-33. https://doi.org/10.1007/BF02235755.
- Cancer Research UK . Uterine Cancer Incidence Statistics: Lifetime Risk of Uterine Cancer 2016. www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/uterine-cancer/incidence#heading-Five (accessed 8 July 2016).
- Cancer Research UK . Uterine Cancer Survival Statistics 2014. www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/uterine-cancer/survival (accessed 8 July 2016).
- England and Wales, Interim Life Tables, 1980–82 to 2008–10. London: ONS; 2011.
- Trueman P, Lowson K, Bending M, Ganderton M, Chaplin S, Wright D, et al. Bowel Cancer Services: Costs and Benefits. York: York Health Economics Consortium; 2007.
- Järvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomäki P, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterol 2000;118:829-34. https://doi.org/10.1016/S0016-5085(00)70168-5.
- Balmana J, Balaguer F, Cervantes A, Arnold D. ESMO Guidelines Working Group . Familial risk-colorectal cancer: ESMO Clinical Practice Guidelines. Ann Oncol 2013;24:vi73-80. https://doi.org/10.1093/annonc/mdt209.
- Cuzick J, Otto F, Baron JA, Brown PH, Burn J, Greenwald P, et al. Aspirin and non-steroidal anti-inflammatory drugs for cancer prevention: an international consensus statement. Lancet Oncol 2009;10:501-7. https://doi.org/10.1016/S1470-2045(09)70035-X.
- Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet 2004;41:664-8. https://doi.org/10.1136/jmg.2004.020651.
- Bouzourene H, Hutter P, Losi L, Martin P, Benhattar J. Selection of patients with germline MLH1 mutated Lynch syndrome by determination of MLH1 methylation and BRAF mutation. Fam Cancer 2010;9:167-72. https://doi.org/10.1007/s10689-009-9302-4.
- Chang SC, Lin PC, Yang SH, Wang HS, Liang WY, Lin JK. Taiwan hospital-based detection of Lynch syndrome distinguishes 2 types of microsatellite instabilities in colorectal cancers. Surgery 2010;147:720-8. https://doi.org/10.1016/j.surg.2009.10.069.
- Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 2008;135:419-28. https://doi.org/10.1053/j.gastro.2008.04.026.
- Cancer Survival in England by Stage. London: Public Health England; 2014.
- Mecklin JP, Aarnio M, Läärä E, Kairaluoma MV, Pylvänäinen K, Peltomäki P, et al. Development of colorectal tumors in colonoscopic surveillance in Lynch syndrome. Gastroenterology 2007;133:1093-8. https://doi.org/10.1053/j.gastro.2007.08.019.
- Renkonen-Sinisalo L, Butzow R, Leminen A, Lehtovirta P, Mecklin JP, Järvinen HJ. Surveillance for endometrial cancer in hereditary nonpolyposis colorectal cancer syndrome. Int J Cancer 2007;120:821-4. https://doi.org/10.1002/ijc.22446.
- Dove-Edwin I, Boks D, Goff S, Kenter GG, Carpenter R, Vasen HF, et al. The outcome of endometrial carcinoma surveillance by ultrasound scan in women at risk of hereditary nonpolyposis colorectal carcinoma and familial colorectal carcinoma. Cancer 2002;94:1708-12. https://doi.org/10.1002/cncr.10380.
- Järvinen HJ, Renkonen-Sinisalo L, Aktan-Collan K, Peltomaki P, Aaltonen LA, Mecklin JP. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol 2009;27:4793-7. https://doi.org/10.1200/JCO.2009.23.7784.
- Färkkilä N, Sintonen H, Saarto T, Järvinen H, Hänninen J, Taari K, et al. Health-related quality of life in colorectal cancer. Colorectal Dis 2013;15:e215-22. https://doi.org/10.1111/codi.12143.
- Hall PS, Hamilton P, Hulme CT, Meads DM, Jones H, Newsham A, et al. Costs of cancer care for use in economic evaluation: a UK analysis of patient-level routine health system data. Br J Cancer 2015;112:948-56. https://doi.org/10.1038/bjc.2014.644.
- Hung HC, Chien TW, Tsay SL, Hang HM, Liang SY. Patient and clinical variables account for changes in health-related quality of life and symptom burden as treatment outcomes in colorectal cancer: a longitudinal study. Asian Pac J Cancer Prev 2013;14:1905-9. https://doi.org/10.7314/APJCP.2013.14.3.1905.
- Mhaidat NM, Al-Wedyan TJ, Alzoubi KH, Al-Efan QM, Al-Azzam SI, Balas QA, et al. Measuring quality of life among colorectal cancer patients in Jordan. J Palliat Care 2014;30:133-40.
- Stein D, Joulain F, Naoshy S, Iqbal U, Muszbek N, Payne KA, et al. Assessing health-state utility values in patients with metastatic colorectal cancer: a utility study in the United Kingdom and the Netherlands. Int J Colorectal Dis 2014;29:1203-10. https://doi.org/10.1007/s00384-014-1980-1.
- Wong CK, Lam CL, Poon JT, Kwong DL. Clinical correlates of health preference and generic health-related quality of life in patients with colorectal neoplasms. PLOS ONE 2013;8. https://doi.org/10.1371/journal.pone.0058341.
- Ramsey SD, Andersen MR, Etzioni R, Moinpour C, Peacock S, Potosky A, et al. Quality of life in survivors of colorectal carcinoma. Cancer 2000;88:1294-303. https://doi.org/10.1002/(SICI)1097-0142(20000315)88:6<1294::AID-CNCR4>3.0.CO;2-M.
- Mittmann N, Au HJ, Tu D, O’Callaghan CJ, Isogai PK, Karapetis CS, et al. Prospective cost-effectiveness analysis of cetuximab in metastatic colorectal cancer: evaluation of National Cancer Institute of Canada Clinical Trials Group CO.17 trial. J Natl Cancer Inst 2009;101:1182-92. https://doi.org/10.1093/jnci/djp232.
- Ness RM, Holmes AM, Klein R, Dittus R. Utility valuations for outcome states of colorectal cancer. Am J Gastroenterol 1999;94:1650-7. https://doi.org/10.1111/j.1572-0241.1999.01157.x.
- Wong CK, Lam CL, Wan YF, Fong DY. Cost-effectiveness simulation and analysis of colorectal cancer screening in Hong Kong Chinese population: comparison amongst colonoscopy, guaiac and immunologic fecal occult blood testing. BMC Cancer 2015;15. https://doi.org/10.1186/s12885-015-1730-y.
- Sharp L, Tilson L, Whyte S, O’Ceilleachair A, Walsh C, Usher C, et al. Cost-effectiveness of population-based screening for colorectal cancer: a comparison of guaiac-based faecal occult blood testing, faecal immunochemical testing and flexible sigmoidoscopy. Br J Cancer 2012;106:805-16. https://doi.org/10.1038/bjc.2011.580.
- Goldstein DA, Chen Q, Ayer T, Howard DH, Lipscomb J, El-Rayes BF, et al. First- and second-line bevacizumab in addition to chemotherapy for metastatic colorectal cancer: a United States-based cost-effectiveness analysis. J Clin Oncol 2015;33:1112-18. https://doi.org/10.1200/JCO.2014.58.4904.
- Hanly P, Skally M, Fenlon H, Sharp L. Cost-effectiveness of computed tomography colonography in colorectal cancer screening: a systematic review. Int J Technol Assess Health Care 2012;28:415-23. https://doi.org/10.1017/S0266462312000542.
- Haanstra JF, De Vos tot Nederveen Cappel WH, Vanhoutvin S, Cats A, Vecht J, Kleibeuker JH, et al. Quality of life after surgery for colon cancer in patients with Lynch syndrome; partial versus (sub)total colectomy. Gastroenterology 2011;1:S259-60.
- Kalady MF, Dziedzic M, Manilich E, Lynch C, McGannon E, Fay S, et al. Quality of life after surgery for colorectal cancer in HNPCC patients. Fam Cancer 2011;10.
- Pollett WG, Marion K, Moeslein G, Schneider C, Parry S, Veysey K, et al. Quality of life after surgery in individuals with familial colorectal cancer: does extended surgery have an adverse impact?. ANZ J Surg 2014;84:359-64. https://doi.org/10.1111/ans.12336.
- Veysey K, Steinke V, Engel C, Rahner N, Buttner R, Loeffler M, et al. Extended colonic resection does not impair quality of life in individuals with familial bowel cancer or polyps. Fam Cancer 2011;10.
- Hornbrook MC, Wendel CS, Coons SJ, Grant M, Herrinton LJ, Mohler MJ, et al. Complications among colorectal cancer survivors: SF-6D preference-weighted quality of life scores. Med Care 2011;49:321-6. https://doi.org/10.1097/MLR.0b013e31820194c8.
- Thong MS, Mols F, Lemmens VE, Creemers GJ, Slooter GD, van de Poll-Franse LV. Impact of chemotherapy on health status and symptom burden of colon cancer survivors: a population-based study. Eur J Cancer 2011;47:1798-807. https://doi.org/10.1016/j.ejca.2011.02.006.
- Niv Y, Bogolavski I, Ilani S, Avni I, Gal E, Vilkin A, et al. Impact of colonoscopy on quality of life. Eur J Gastroenterol Hepatol 2012;24:781-6. https://doi.org/10.1097/MEG.0b013e328352deff.
- Nout RA, Putter H, Jurgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, van der Steen-Banasik EM, et al. Five-year quality of life of endometrial cancer patients treated in the randomised Post Operative Radiation Therapy in Endometrial Cancer (PORTEC-2) trial and comparison with norm data. Eur J Cancer 2012;48:1638-48. https://doi.org/10.1016/j.ejca.2011.11.014.
- Hildebrandt T, Thiel FC, Fasching PA, Graf C, Bani MR, Loehberg CR, et al. Health utilities in gynecological oncology and mastology in Germany. Anticancer Res 2014;34:829-35.
- Goker A, Guvenal T, Yanikkerem E, Turhan A, Koyuncu FM. Quality of life in women with gynecologic cancer in Turkey. Asian Pac J Cancer Prev 2011;12:3121-8.
- Ferrandina G, Petrillo M, Mantegna G, Fuoco G, Terzano S, Venditti L, et al. Evaluation of quality of life and emotional distress in endometrial cancer patients: a 2-year prospective, longitudinal study. Gynecol Oncol 2014;133:518-25. https://doi.org/10.1016/j.ygyno.2014.03.015.
- de Boer SM, Nout RA, Jurgenliemk-Schulz IM, Jobsen JJ, Lutgens LC, van der Steen-Banasik EM, et al. Long-term impact of endometrial cancer diagnosis and treatment on health-related quality of life and cancer survivorship: results from the randomized PORTEC-2 trial. Int J Radiat Oncol Biol Phys 2015;93:797-809. https://doi.org/10.1016/j.ijrobp.2015.08.023.
- Longworth L, Yang Y, Young T, Mulhern B, Hernandez Alava M, Mukuria C, et al. Use of generic and condition-specific measures of health-related quality of life in NICE decision-making: a systematic review, statistical modelling and survey. Health Technol Assess 2014;18. https://doi.org/10.3310/hta18090.
- Doble B, Lorgelly P. Mapping the EORTC QLQ-C30 onto the EQ-5D-3L: assessing the external validity of existing mapping algorithms. Qual Life Res 2016;25:891-91. https://doi.org/10.1007/s11136-015-1116-2.
- Dolan P. Modeling valuations for EuroQol health states. Med Care 1997;35:1095-108. https://doi.org/10.1097/00005650-199711000-00002.
- Wong SQ, Fellowes A, Doig K, Ellul J, Bosma TJ, Irwin D, et al. Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients. Br J Cancer 2015;112:1411-20. https://doi.org/10.1038/bjc.2015.80.
- Kuppermann M, Wang G, Wong S, Blanco A, Conrad P, Nakagawa S, et al. Preferences for outcomes associated with decisions to undergo or forgo genetic testing for Lynch syndrome. Cancer 2013;119:215-25. https://doi.org/10.1002/cncr.27634.
- Curtis L, Burns A. Unit Costs of Health and Social Care 2015. Canterbury: Personal Social Services Research Unit, University of Kent; 2015.
- Newton K, Green K, Lalloo F, Evans DG, Hill J. Colonoscopy screening compliance and outcomes in patients with Lynch syndrome. Colorectal Dis 2015;17:38-46. https://doi.org/10.1111/codi.12778.
- Gavin DR, Valori RM, Anderson JT, Donnelly MT, Williams JG, Swarbrick ET. The national colonoscopy audit: a nationwide assessment of the quality and safety of colonoscopy in the UK. Gut 2013;62:242-9. https://doi.org/10.1136/gutjnl-2011-301848.
- Havrilesky L, Maxwell G, Myers E. Cost-effectiveness analysis of annual screening strategies for endometrial cancer. Am J Obstet Gynecol 2009;200:e1-7. https://doi.org/10.1016/j.ajog.2009.02.022.
- Sundar S, Gallos I, Gupta J, Drake A, Johnson N, Ganesan R, et al. BGCS Uterine Cancer Guidelines: Recommendations for Practice n.d. https://bgcs.org.uk/professionals/guidelines.html (accessed 26 June 2016).
- Calvert AH, Newell DR, Gumbrell LA, O’Reilly S, Burnell M, Boxall FE, et al. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol 1989;7:1748-56. https://doi.org/10.1200/JCO.1989.7.11.1748.
- Janus N, Launay-Vacher V, Byloos E, Machiels JP, Duck L, Kerger J, et al. Cancer and renal insufficiency results of the BIRMA study. Br J Cancer 2010;103:1815-21. https://doi.org/10.1038/sj.bjc.6605979.
- Sacco JJ, Botten J, Macbeth F, Bagust A, Clark P. The average body surface area of adult cancer patients in the UK: a multicentre retrospective study. PLOS ONE 2010;5. https://doi.org/10.1371/journal.pone.0008933.
- East of Scotland Regional Genetic Service . Molecular Genetics List of Disorders n.d. http://134.36.196.124/Molecular_Genetics_List_of_Disorders.htm (accessed August 2012).
- UK Genetic Testing Network . UKGTN – Promoting Gene Testing n.d. http://ukgtn.nhs.uk/ (accessed 8 July 2016).
- John M. AWMGL Service Price List: All Wales Molecular Genetics Laboratory 2012. www.wales.nhs.uk/sites3/Documents/525/MI-MGN-TestPrice_5.1.pdf (accessed 9 August 2012).
- Curtis L. Unit Costs of Health and Social Care 2014. Canterbury: Personal Social Services Research Unit, University of Kent; 2014.
- Department of Health . NHS Reference Costs 2014–15 n.d. www.gov.uk/government/publications/nhs-reference-costs-2014-to-2015 (accessed 17 July 2017).
- NHS . New Guidance for Ovarian Cancer Tests 2011. www.nhs.uk/news/2010/04April/Pages/new-advice-for-testing-for-ovarian-cancer.aspx (accessed 21 July 2016).
- Drugs and Pharmaceutical Electronic Market Information (eMit). London: Department of Health; 2016.
- British National Formulary. London: BMJ Group and Pharmaceutical Press; 2016.
- NHS Drug Tariff (June 2016). Newcastle upon Tyne: NHS Business Services Authority; 2016.
- Data Briefing: Reflex Testing for Lynch Syndrome in People Diagnosed with Bowel Cancer under the Age of 50. London: Bowel Cancer UK; 2016.
- Equality Act 2010. London: The Stationery Office; 2010.
- NICE’s Equality Objectives and Equality Programme 2016–2020. London: NICE; 2016.
- Philips Z, Ginnelly L, Sculpher M, Claxton K, Golder S. Review of guidelines for good practice in decision-analytic modelling in health technology assessment. Health Technol Assess 2004;8. https://doi.org/10.3310/hta8360.
- Evers S, Goossens M, de Vet H, van Tulder M, Ament A. Criteria list for assessment of methodological quality of economic evaluations: Consensus on Health Economic Criteria. Int J Technol Assess Health Care 2005;21:240-5.
- Familial Breast Cancer: Classification, Care and Managing Breast Cancer and Related Risks in People with a Family History of Breast Cancer. London: NICE; 2013.
- Slade I, Hanson H, George A, Kohut K, Strydom A, Wordsworth S, et al. MCG programme . A cost analysis of a cancer genetic service model in the UK. J Community Genet 2016;7:185-94. https://doi.org/10.1007/s12687-016-0266-4.
- George A, Riddell D, Seal S, Talukdar S, Mahamdallie S, Ruark E, et al. Implementing rapid, robust, cost-effective, patient-centred, routine genetic testing in ovarian cancer patients. Sci Rep 2016;6. http://dx.doi.org/10.1038/srep29506.
- Eccleston A, Bentley A, Dyer M, Strydom A, Vereecken W, George A, et al. A discrete event simulation to evaluate the cost effectiveness of germline BRCA1 and BRCA2 testing in UK women with ovarian cancer. BioRxiv 2016. https://doi.org/10.1101/060418.
- Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet 2012;49:151-7. http://dx.doi.org/10.1136/jmedgenet-2011-100714.
- Kloor M, Reuschenbach M, Karbach J, Rafiyan M, Al-Batran SE, Pauligk C, et al. Vaccination of MSI-H colorectal cancer patients with frameshift peptide antigens: a Phase I/IIa clinical trial. J Clin Oncol 2015;33.
- Niessen RC, Berends MJ, Wu Y, Sijmons RH, Hollema H, Ligtenberg MJ, et al. Identification of mismatch repair gene mutations in young parents with colorectal cancer and inpatients with multiple tumours associated with hereditary non-polyposis colorectal cancer. Gut 2006;55:1781-8. https://doi.org/10.1136/gut.2005.090159.
Appendix 1 Literature search strategies
Test accuracy searches
MEDLINE (via Ovid)
Data parameters: 1946 to January Week 3 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 1274.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | Colorectal Neoplasms, Hereditary Nonpolyposis/ |
| 14. | or/4-13 |
| 15. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 16. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 17. | 15 and 16 |
| 18. | Amsterdam criteria.tw. |
| 19. | 3 or 14 or 17 or 18 |
| 20. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 21. | (Bethesda adj3 (marker* or panel*)).tw. |
| 22. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 23. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 24. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 25. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 26. | exp Immunohistochemistry/ |
| 27. | or/20-26 |
| 28. | 19 and 27 |
| 29. | exp animals/ not humans.sh. |
| 30. | 28 not 29 |
| 31. | limit 30 to (english language and yr=‘2006 -Current’) |
MEDLINE In-Process & Other Non-indexed Citations (via Ovid)
Data parameters: 29 January 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 134.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | or/4-12 |
| 14. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 15. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 16. | 14 and 15 |
| 17. | Amsterdam criteria.tw. |
| 18. | 3 or 13 or 16 or 17 |
| 19. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 20. | (Bethesda adj3 (marker* or panel*)).tw. |
| 21. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 22. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 23. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 24. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 25. | or/19-24 |
| 26. | 18 and 25 |
| 27. | limit 26 to (english language and yr=‘2006 -Current’) |
EMBASE (via Ovid)
Data parameters: 1974 to 2016 January 29.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 2928.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | hereditary nonpolyposis colorectal cancer/ |
| 14. | or/4-13 |
| 15. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 16. | protein MLH1/ |
| 17. | protein MSH2/ |
| 18. | protein MSH6/ |
| 19. | mismatch repair protein PMS2/ |
| 20. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 21. | or/15-19 |
| 22. | 20 and 21 |
| 23. | Amsterdam criteria.tw. |
| 24. | 3 or 14 or 22 or 23 |
| 25. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 26. | (Bethesda adj3 (marker* or panel*)).tw. |
| 27. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 28. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 29. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 30. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 31. | microsatellite instability/ |
| 32. | exp Immunohistochemistry/ |
| 33. | or/25-32 |
| 34. | 24 and 33 |
| 35. | limit 34 to (english language and yr=‘2006 -Current’) |
Web of Science [Science Citation Index (SCI) and Conference Proceedings Citation Index – Science (CPCI-S)] (Thomson Reuters)
Data parameters: not applicable.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 2335.
Strategy
| 1. | TS=(lynch* near/2 syndrome) |
| 2. | TS=((lynch* near/2 famil*) and (cancer* or neoplasm*)) |
| 3. | #2 OR #1 |
| 4. | TS=(‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’) |
| 5. | TS=(HNPCC) |
| 6. | TS=(((hereditary or inherited) near/2 (colon* or colorectal*)) and (cancer or neoplasm*)) |
| 7. | TS=((hereditary near/2 nonpolyposis) and (colon* or colorectal*)) |
| 8. | TS=((hereditary near/2 non-polyposis) and (colon* or colorectal*)) |
| 9. | TS=((hereditary near/2 (cancer or neoplasm*)) and (colon* or colorectal*)) |
| 10. | TS=((Familial near/2 Nonpolyposis) and (colon* or colorectal*)) |
| 11. | TS=((Familial near/2 Non-polyposis) and (colon* or colorectal*)) |
| 12. | TS=(familial near/2 (colon* or colorectal*)) |
| 13. | #12 OR #11 OR #10 OR #9 OR #8 OR #7 OR #6 OR #5 OR #4 |
| 14. | TS=(EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) |
| 15. | TS=(colon* or colorectal* or lynch* or HNPCC or hereditary) |
| 16. | #15 AND #14 |
| 17. | TS=(Amsterdam criteria) |
| 18. | #17 OR #16 OR #13 OR #3 |
| 19. | TS=((microsatellite near/2 instabilit*) or (msi near/2 test*)) |
| 20. | TS=(Bethesda near/2 (marker* or panel*)) |
| 21. | TS=(immunohistochemistry or (IHC near/2 test*)) |
| 22. | TS=((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/2 antibod*) |
| 23. | TS=((BRAFV600E or ‘BRAF V600E’) near/2 mutation*) |
| 24. | TS=(MLH1 near/2 (methylation or hypermethylation or ‘hyper methylation’)) |
| 25. | #24 OR #23 OR #22 OR #21 OR #20 OR #19 |
| 26. | (#25 AND #18) AND LANGUAGE: (English) |
| Timespan=2006-2016 |
The Cochrane Library [Cochrane Database of Systematic Reviews (CDSR), Cochrane Central Register of Controlled Trials (CENTRAL), Health Technology Assessment (HTA) database] (Cochrane Collaboration)
Data parameters: CDSR and CENTRAL: Issue 1 of 12, January 2016; HTA database: Issue 1 of 4, January 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: CDSR = 0; CENTRAL = 9; HTA = 0.
Strategy
| 1. | (lynch* near/3 syndrome):ti or (lynch* near/3 syndrome):ab |
| 2. | ((lynch* near/3 famil*) and (cancer* or neoplasm*)):ti or ((lynch* near/3 famil*) and (cancer* or neoplasm*)):ab |
| 3. | #1 or #2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’):ti or (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’):ab |
| 5. | HNPCC:ti or HNPCC:ab |
| 6. | (((hereditary or inherited) near/3 (colon* or colorectal*)) and (cancer or neoplasm*)):ti or (((hereditary or inherited) near/3 (colon* or colorectal*)) and (cancer or neoplasm*)):ab |
| 7. | ((hereditary near/3 nonpolyposis) and (colon* or colorectal*)):ti or ((hereditary near/3 nonpolyposis) and (colon* or colorectal*)):ab |
| 8. | ((hereditary near/3 non-polyposis) and (colon* or colorectal*)):ti or ((hereditary near/3 non-polyposis) and (colon* or colorectal*)):ab |
| 9. | ((hereditary near/3 (cancer or neoplasm*)) and (colon* or colorectal*)):ti or ((hereditary near/3 (cancer or neoplasm*)) and (colon* or colorectal*)):ab |
| 10. | ((Familial near/3 Nonpolyposis) and (colon* or colorectal*)):ti or ((Familial near/3 Nonpolyposis) and (colon* or colorectal*)):ab |
| 11. | ((Familial near/3 Non-polyposis) and (colon* or colorectal*)):ti or ((Familial near/3 Non-polyposis) and (colon* or colorectal*)):ab |
| 12. | (familial near/3 (colon* or colorectal*)):ti or (familial near/3 (colon* or colorectal*)):ab |
| 13. | MeSH descriptor: [Colorectal Neoplasms, Hereditary Nonpolyposis] this term only |
| 14. | {or #4-#13} |
| 15. | ((EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) and (colon* or colorectal* or lynch* or HNPCC or hereditary)):ti or ((EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) and (colon* or colorectal* or lynch* or HNPCC or hereditary)):ab |
| 16. | Amsterdam criteria:ti or Amsterdam criteria:ab |
| 17. | #15 or #16 |
| 18. | #3 or #14 or #17 |
| 19. | ((microsatellite near/3 instabilit*) or (msi near/3 test*)):ti or ((microsatellite near/3 instabilit*) or (msi near/3 test*)):ab |
| 20. | (Bethesda near/3 (marker* or panel*)):ti or (Bethesda near/3 (marker* or panel*)):ab |
| 21. | (immunohistochemistry or (IHC near/3 test*)):ti or (immunohistochemistry or (IHC near/3 test*)):ab |
| 22. | (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/3 antibod*:ti or (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/3 antibod*:ab |
| 23. | ((BRAFV600E or ‘BRAF V600E’) near/3 mutation*):ti or ((BRAFV600E or ‘BRAF V600E’) near/3 mutation*):ab |
| 24. | (MLH1 near/3 (methylation or hypermethylation or ‘hyper methylation’)):ti or (MLH1 near/3 (methylation or hypermethylation or ‘hyper methylation’)):ab |
| 25. | MeSH descriptor: [Immunohistochemistry] explode all trees |
| 26. | {or #19-#25} |
| 27. | #18 and #26 Publication Year from 2006, in Cochrane Reviews (Reviews and Protocols), Trials and Technology Assessments |
Health Management Information Consortium (HMIC) (via Ovid)
Data parameters: 1979 to November 2015.
Date searched: 1 February 2016.
Searcher: SB.
Hits: two.
Strategy
See Strategy for MEDLINE In-Process & Other Non-Indexed Citations.
Number of hits per database and in total
| Database | Hits |
|---|---|
| MEDLINE | 1274 |
| MEDLINE In-Process & Other Non-Indexed Citations | 134 |
| EMBASE | 2928 |
| Web of Science | 2335 |
| CDSR | 0 |
| CENTRAL | 9 |
| HTA | 0 |
| HMIC | 2 |
| Total records | 6682 |
| Duplicates | 2762 |
| Total unique records | 3920 |
Cost-effectiveness searches
MEDLINE (via Ovid)
Data parameters: 1946 to January Week 3 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 85.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | Colorectal Neoplasms, Hereditary Nonpolyposis/ |
| 14. | or/4-13 |
| 15. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 16. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 17. | 15 and 16 |
| 18. | Amsterdam criteria.tw. |
| 19. | 3 or 14 or 17 or 18 |
| 20. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 21. | (Bethesda adj3 (marker* or panel*)).tw. |
| 22. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 23. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 24. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 25. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 26. | exp Immunohistochemistry/ |
| 27. | or/20-26 |
| 28. | exp Economics/ |
| 29. | ec.fs. |
| 30. | economics, medical/ |
| 31. | economics, nursing/ |
| 32. | economics, pharmaceutical/ |
| 33. | exp ‘economics, hospital’/ |
| 34. | (economic* or price or prices or pricing or priced or discount or discounts or discounted or discounting or ration* or expenditure or expenditures or budget* or afford* or pharmacoeconomic or pharmaco-economic*).tw. |
| 35. | (cba or cea or cua).ti,ab. |
| 36. | exp ‘fees and charges’/ |
| 37. | (fee or fees or charge* or preference*).tw. |
| 38. | (fiscal or funding or financial or finance).tw. |
| 39. | exp ‘costs and cost analysis’/ |
| 40. | exp Health Care Costs/ |
| 41. | cost*.tw. |
| 42. | exp decision support techniques/ |
| 43. | exp models, economic/ |
| 44. | exp Statistical Model/ |
| 45. | markov*.tw. |
| 46. | markov chains/ |
| 47. | monte carlo.tw. |
| 48. | monte carlo method/ |
| 49. | (decision adj2 (tree* or analy* or model*)).tw. |
| 50. | (survival adj3 analys*).tw. |
| 51. | ‘deductibles and coinsurance’/ |
| 52. | exp Health expenditures/ |
| 53. | uncertain*.tw. |
| 54. | uncertainty/ |
| 55. | (quality adj3 life).tw. |
| 56. | quality of life/ |
| 57. | value of life/ |
| 58. | Quality-adjusted life years/ |
| 59. | (qol* or qoly or qolys or hrqol* or qaly or qalys or qale or qales).tw. |
| 60. | (sensitivity analys* or ‘willingness to pay’ or quality-adjusted life year* or quality adjusted life year* or quality-adjusted life expectanc* or quality adjusted life expectanc*).tw. |
| 61. | utilit*.tw. |
| 62. | valu*.tw. |
| 63. | exp hospitalization/ |
| 64. | or/28-63 |
| 65. | 19 and 27 and 64 |
| 66. | Animals/ not humans.sh. |
| 67. | 65 not 66 |
| 68. | limit 67 to (english language and yr=‘2013 -Current’) |
MEDLINE In-Process & Other Non-Indexed Citations (via Ovid)
Data parameters: 29 January 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 30.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | or/4-12 |
| 14. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 15. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 16. | 14 and 15 |
| 17. | Amsterdam criteria.tw. |
| 18. | 3 or 13 or 16 or 17 |
| 19. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 20. | (Bethesda adj3 (marker* or panel*)).tw. |
| 21. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 22. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 23. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 24. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 25. | or/19-24 |
| 26. | (economic* or price or prices or pricing or priced or discount or discounts or discounted or discounting or ration* or expenditure or expenditures or budget* or afford* or pharmacoeconomic or pharmaco-economic*).tw. |
| 27. | (cba or cea or cua).ti,ab. |
| 28. | (fee or fees or charge* or preference*).tw. |
| 29. | (fiscal or funding or financial or finance).tw. |
| 30. | cost*.tw. |
| 31. | markov*.tw. |
| 32. | monte carlo.tw. |
| 33. | (decision adj2 (tree* or analy* or model*)).tw. |
| 34. | (survival adj3 analys*).tw. |
| 35. | uncertain*.tw. |
| 36. | (quality adj3 life).tw. |
| 37. | (qol* or qoly or qolys or hrqol* or qaly or qalys or qale or qales).tw. |
| 38. | (sensitivity analys* or ‘willingness to pay’ or quality-adjusted life year* or quality adjusted life year* or quality-adjusted life expectanc* or quality adjusted life expectanc*).tw. |
| 39. | utilit*.tw. |
| 40. | valu*.tw. |
| 41. | or/25-40 |
| 42. | 18 and 25 and 41 |
| 43. | limit 42 to yr=‘2013 -Current’ |
EMBASE (via Ovid)
Data parameters: 1974 to 2016 January 29.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 256.
Strategy
| 1. | (lynch* adj3 syndrome).tw. |
| 2. | ((lynch* adj3 famil*) and (cancer* or neoplasm*)).tw. |
| 3. | or/1-2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’).tw. |
| 5. | HNPCC.tw. |
| 6. | (((hereditary or inherited) adj3 (colon* or colorectal*)) and (cancer or neoplasm*)).tw. |
| 7. | ((hereditary adj3 nonpolyposis) and (colon* or colorectal*)).tw. |
| 8. | ((hereditary adj3 non-polyposis) and (colon* or colorectal*)).tw. |
| 9. | ((hereditary adj3 (cancer or neoplasm*)) and (colon* or colorectal*)).tw. |
| 10. | ((Familial adj3 Nonpolyposis) and (colon* or colorectal*)).tw. |
| 11. | ((Familial adj3 Non-polyposis) and (colon* or colorectal*)).tw. |
| 12. | (familial adj3 (colon* or colorectal*)).tw. |
| 13. | hereditary nonpolyposis colorectal cancer/ |
| 14. | or/4-13 |
| 15. | (EPCAM? or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2).tw. |
| 16. | protein MLH1/ |
| 17. | protein MSH2/ |
| 18. | protein MSH6/ |
| 19. | mismatch repair protein PMS2/ |
| 20. | (colon* or colorectal* or lynch* or HNPCC or hereditary).tw. |
| 21. | or/15-19 |
| 22. | 20 and 21 |
| 23. | Amsterdam criteria.tw. |
| 24. | 3 or 14 or 22 or 23 |
| 25. | ((microsatellite adj3 instabilit*) or (msi adj3 test*)).tw. |
| 26. | (Bethesda adj3 (marker* or panel*)).tw. |
| 27. | (immunohistochemistry or (IHC adj3 test*)).tw. |
| 28. | ((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) adj3 antibod*).tw. |
| 29. | ((BRAFV600E or ‘BRAF V600E’) adj3 mutation*).tw. |
| 30. | (MLH1 adj3 (methylation or hypermethylation or ‘hyper methylation’)).tw. |
| 31. | microsatellite instability/ |
| 32. | exp Immunohistochemistry/ |
| 33. | or/25-32 |
| 34. | exp Economics/ |
| 35. | pe.fs. |
| 36. | economics, medical/ |
| 37. | economics, nursing/ |
| 38. | economics, pharmaceutical/ |
| 39. | exp ‘economics, hospital’/ |
| 40. | (economic* or price or prices or pricing or priced or discount or discounts or discounted or discounting or ration* or expenditure or expenditures or budget* or afford* or pharmacoeconomic or pharmaco-economic*).tw. |
| 41. | (cba or cea or cua).ti,ab. |
| 42. | exp ‘fees and charges’/ |
| 43. | (fee or fees or charge* or preference*).tw. |
| 44. | (fiscal or funding or financial or finance).tw. |
| 45. | exp ‘costs and cost analysis’/ |
| 46. | exp Health Care Costs/ |
| 47. | cost*.tw. |
| 48. | exp decision support techniques/ |
| 49. | exp models, economic/ |
| 50. | exp Statistical Model/ |
| 51. | markov*.tw. |
| 52. | markov chains/ |
| 53. | monte carlo.tw. |
| 54. | monte carlo method/ |
| 55. | (decision adj2 (tree* or analy* or model*)).tw. |
| 56. | (survival adj3 analys*).tw. |
| 57. | ‘deductibles and coinsurance’/ |
| 58. | exp Health expenditures/ |
| 59. | uncertain*.tw. |
| 60. | uncertainty/ |
| 61. | (quality adj3 life).tw. |
| 62. | quality of life/ |
| 63. | value of life/ |
| 64. | Quality-adjusted life years/ |
| 65. | (qol* or qoly or qolys or hrqol* or qaly or qalys or qale or qales).tw. |
| 66. | (sensitivity analys* or ‘willingness to pay’ or quality-adjusted life year* or quality adjusted life year* or quality-adjusted life expectanc* or quality adjusted life expectanc*).tw. |
| 67. | utilit*.tw. |
| 68. | valu*.tw. |
| 69. | exp hospitalization/ |
| 70. | or/34-69 |
| 71. | 24 and 33 and 70 |
| 72. | limit 71 to (english language and yr=‘2013 -Current’) |
Web of Science [Science Citation Index (SCI) and Conference Proceedings Citation Index – Science (CPCI-S)] (Thomson Reuters)
Data parameters: not applicable.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 183.
Strategy
| 1. | TS=(lynch* near/2 syndrome) |
| 2. | TS=((lynch* near/2 famil*) and (cancer* or neoplasm*)) |
| 3. | #2 OR #1 |
| 4. | TS=(‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’) |
| 5. | TS=(HNPCC) |
| 6. | TS=(((hereditary or inherited) near/2 (colon* or colorectal*)) and (cancer or neoplasm*)) |
| 7. | TS=((hereditary near/2 nonpolyposis) and (colon* or colorectal*)) |
| 8. | TS=((hereditary near/2 non-polyposis) and (colon* or colorectal*)) |
| 9. | TS=((hereditary near/2 (cancer or neoplasm*)) and (colon* or colorectal*)) |
| 10. | TS=((Familial near/2 Nonpolyposis) and (colon* or colorectal*)) |
| 11. | TS=(familial near/2 (colon* or colorectal*)) |
| 12. | TS=((Familial near/2 Non-polyposis) and (colon* or colorectal*)) |
| 13. | #12 OR #11 OR #10 OR #9 OR #8 OR #7 OR #6 OR #5 OR #4 |
| 14. | TS=(EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) |
| 15. | TS=(colon* or colorectal* or lynch* or HNPCC or hereditary) |
| 16. | #15 AND #14 |
| 17. | TS=(Amsterdam criteria) |
| 18. | #17 OR #16 OR #13 OR #3 |
| 19. | TS=((microsatellite near/2 instabilit*) or (msi near/2 test*)) |
| 20. | TS=(Bethesda near/2 (marker* or panel*)) |
| 21. | TS=(immunohistochemistry or (IHC near/2 test*)) |
| 22. | TS=((MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/2 antibod*) |
| 23. | TS=((BRAFV600E or ‘BRAF V600E’) near/2 mutation*) |
| 24. | TS=(MLH1 near/2 (methylation or hypermethylation or ‘hyper methylation’)) |
| 25. | #24 OR #23 OR #22 OR #21 OR #20 OR #19 |
| 26. | TS=(economic* or price or prices or pricing or priced or discount or discounts or discounted or discounting or ration* or expenditure or expenditures or budget* or afford* or pharmacoeconomic or pharmaco-economic*) |
| 27. | TS=(cba or cea or cua) |
| 28. | TS=(fee or fees or charge* or preference*) |
| 29. | TS=(fiscal or funding or financial or finance) |
| 30. | TS=(cost*) |
| 31. | TS=(markov*) |
| 32. | TS=(monte carlo) |
| 33. | TS=(decision near/1 (tree* or analy* or model*)) |
| 34. | TS=(survival near/2 analys*) |
| 35. | TS=(uncertain*) |
| 36. | TS=(quality near/2 life) |
| 37. | TS=(qol* or qoly or qolys or hrqol* or qaly or qalys or qale or qales) |
| 38. | TS=(sensitivity analys* or ‘willingness to pay’ or quality-adjusted life year* or quality adjusted life year* or quality-adjusted life expectanc* or quality adjusted life expectanc*) |
| 39. | TS=(utilit*) |
| 40. | TS=(valu*) |
| 41. | #40 OR #39 OR #38 OR #37 OR #36 OR #35 OR #34 OR #33 OR #32 OR #31 OR #30 OR #29 OR #28 OR #27 OR #26 |
| 42. | #18 AND #25 AND #41 |
| Timespan=2013-2016 |
NHS Economic Evaluation Database (NHS EED) (The Cochrane Library)
Data parameters: Issue 2 of 12, February 2016.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 0.
Strategy
| 1. | (lynch* near/3 syndrome):ti or (lynch* near/3 syndrome):ab in Economic Evaluations |
| 2. | ((lynch* near/3 famil*) and (cancer* or neoplasm*)):ti or ((lynch* near/3 famil*) and (cancer* or neoplasm*)):ab in Economic Evaluations |
| 3. | #1 or #2 |
| 4. | (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’):ti or (‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’):ab in Economic Evaluations |
| 5. | HNPCC:ti or HNPCC:ab in Economic Evaluations |
| 6. | (((hereditary or inherited) near/3 (colon* or colorectal*)) and (cancer or neoplasm*)):ti or (((hereditary or inherited) near/3 (colon* or colorectal*)) and (cancer or neoplasm*)):ab in Economic Evaluations |
| 7. | ((hereditary near/3 nonpolyposis) and (colon* or colorectal*)):ti or ((hereditary near/3 nonpolyposis) and (colon* or colorectal*)):ab in Economic Evaluations |
| 8. | ((hereditary near/3 non-polyposis) and (colon* or colorectal*)):ti or ((hereditary near/3 non-polyposis) and (colon* or colorectal*)):ab in Economic Evaluations |
| 9. | ((hereditary near/3 (cancer or neoplasm*)) and (colon* or colorectal*)):ti or ((hereditary near/3 (cancer or neoplasm*)) and (colon* or colorectal*)):ab in Economic Evaluations |
| 10. | ((Familial near/3 Nonpolyposis) and (colon* or colorectal*)):ti or ((Familial near/3 Nonpolyposis) and (colon* or colorectal*)):ab in Economic Evaluations |
| 11. | ((Familial near/3 Non-polyposis) and (colon* or colorectal*)):ti or ((Familial near/3 Non-polyposis) and (colon* or colorectal*)):ab in Economic Evaluations |
| 12. | (familial near/3 (colon* or colorectal*)):ti or (familial near/3 (colon* or colorectal*)):ab in Economic Evaluations |
| 13. | MeSH descriptor: [Colorectal Neoplasms, Hereditary Nonpolyposis] this term only |
| 14. | {or #4-#13} in Economic Evaluations |
| 15. | ((EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) and (colon* or colorectal* or lynch* or HNPCC or hereditary)):ti or ((EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) and (colon* or colorectal* or lynch* or HNPCC or hereditary)):ab in Economic Evaluations |
| 16. | Amsterdam criteria:ti or Amsterdam criteria:ab in Economic Evaluations |
| 17. | #15 or #16 |
| 18. | #3 or #14 or #17 |
| 19. | ((microsatellite near/3 instabilit*) or (msi near/3 test*)):ti or ((microsatellite near/3 instabilit*) or (msi near/3 test*)):ab in Economic Evaluations |
| 20. | (Bethesda near/3 (marker* or panel*)):ti or (Bethesda near/3 (marker* or panel*)):ab in Economic Evaluations |
| 21. | (immunohistochemistry or (IHC near/3 test*)):ti or (immunohistochemistry or (IHC near/3 test*)):ab in Economic Evaluations |
| 22. | (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/3 antibod*:ti or (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) near/3 antibod*:ab in Economic Evaluations |
| 23. | ((BRAFV600E or ‘BRAF V600E’) near/3 mutation*):ti or ((BRAFV600E or ‘BRAF V600E’) near/3 mutation*):ab in Economic Evaluations |
| 24. | (MLH1 near/3 (methylation or hypermethylation or ‘hyper methylation’)):ti or (MLH1 near/3 (methylation or hypermethylation or ‘hyper methylation’)):ab in Economic Evaluations |
| 25. | MeSH descriptor: [Immunohistochemistry] explode all trees |
| 26. | {or #19-#25} |
| 27. | #18 and #26 Publication Year from 2013 to 2016, in Economic Evaluations |
EconLit (EBSCOhost)
Data parameters: not applicable.
Date searched: 1 February 2016.
Searcher: SB.
Hits: 2.
Strategy
| 1. | TI lynch* N2 syndrome OR AB lynch* N2 syndrome |
| 2. | TI ( (lynch* N2 famil*) AND (cancer* or neoplasm*) ) OR AB ( (lynch* N2 famil*) AND (cancer* or neoplasm*) ) |
| 3. | S1 OR S2 |
| 4. | TI ( ‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’ ) OR AB ( ‘Hereditary Nonpolyposis Colorectal Cancer’ or ‘Hereditary Non-polyposis Colorectal Cancer’ ) |
| 5. | TI HNPCC OR AB HNPCC |
| 6. | TI ( ((hereditary or inherited) N2 (colon* or colorectal*)) and (cancer or neoplasm*) ) OR AB ( ((hereditary or inherited) N2 (colon* or colorectal*)) and (cancer or neoplasm*) ) |
| 7. | TI ( (hereditary N2 nonpolyposis) and (colon* or colorectal*) ) OR AB ( (hereditary N2 nonpolyposis) and (colon* or colorectal*) ) |
| 8. | TI ( (hereditary N2 non-polyposis) and (colon* or colorectal*) ) OR AB ( (hereditary N2 non-polyposis) and (colon* or colorectal*) ) |
| 9. | TI ( (hereditary N2 (cancer or neoplasm*)) and (colon* or colorectal*) ) OR AB ( (hereditary N2 (cancer or neoplasm*)) and (colon* or colorectal*) ) |
| 10. | TI ( (Familial N2 Nonpolyposis) and (colon* or colorectal*) ) OR AB ( (Familial N2 Nonpolyposis) and (colon* or colorectal*) ) |
| 11. | TI ( ((Familial N2 Non-polyposis) and (colon* or colorectal*)) ) OR AB ( (Familial N2 Non-polyposis) and (colon* or colorectal*) ) |
| 12. | TI ( (familial N2 (colon* or colorectal*)) ) OR AB ( (familial N2 (colon* or colorectal*)) ) |
| 13. | S4 OR S5 OR S6 OR S7 OR S8 OR S9 OR S10 OR S11 OR S12 |
| 14. | TI ( EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2 ) OR AB ( EPCAM* or MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2 ) |
| 15. | TI ( colon* or colorectal* or lynch* or HNPCC or hereditary ) AND AB ( colon* or colorectal* or lynch* or HNPCC or hereditary ) |
| 16. | S14 AND S15 |
| 17. | TI Amsterdam criteria OR AB Amsterdam criteria |
| 18. | S3 OR S13 OR S16 OR S17 |
| 19. | TI ( (microsatellite N2 instabilit*) or (msi N2 test*) ) OR AB ( (microsatellite N2 instabilit*) or (msi N2 test*) ) |
| 20. | TI ( (Bethesda N2 (marker* or panel*)) ) OR AB ( (Bethesda N2 (marker* or panel*)) ) |
| 21. | TI ( immunohistochemistry or (IHC N2 test*) ) OR AB ( immunohistochemistry or (IHC N2 test*) ) |
| 22. | TI ( (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) N2 antibod* ) OR AB ( (MLH1 or MSH2 or MSH6 or hMSH2 or hMLH1 or hPMS2 or hMSH6 or PMS2) N2 antibod* ) |
| 23. | TI ( (BRAFV600E or ‘BRAF V600E’) N2 mutation* ) OR AB ( (BRAFV600E or ‘BRAF V600E’) N2 mutation* ) |
| 24. | TI ( (MLH1 N2 (methylation or hypermethylation or ‘hyper methylation’)) ) OR AB ( (MLH1 N2 (methylation or hypermethylation or ‘hyper methylation’)) ) |
| 25. | S19 OR S20 OR S21 OR S22 OR S23 OR S24 |
| 26. | S18 AND S25 |
Number of hits per database and in total
| Database | Hits |
|---|---|
| MEDLINE | 85 |
| MEDLINE In-Process & Non-Indexed Citations | 30 |
| EMBASE | 256 |
| Web of Science | 183 |
| NHS EED | 0 |
| EconLit | 2 |
| Total records | 556 |
| Duplicates | 204 |
| Total unique records | 352 |
Utilities searches
Hysterectomy and salpingo-oophorectomy
MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (via Ovid)
Data parameters: 1946 to present.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 13.
Strategy
| 1. | hysterectom*.tw. |
| 2. | (‘salpingo oophorectom*’ or ‘salpingo ovariectom*’ or salpingooophorectom*).tw. |
| 3. | (salpingectom* adj7 (oophorectom* or ovariectom*)).tw. |
| 4. | 1 and (2 or 3) |
| 5. | exp Hysterectomy/ |
| 6. | Salpingectomy/ |
| 7. | Ovariectomy/ |
| 8. | and/5-7 |
| 9. | 4 or 8 |
| 10. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 11. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 12. | 10 or 11 |
| 13. | 9 and 12 |
| 14. | exp animals/ not humans.sh. |
| 15. | 13 not 14 |
| 16. | limit 15 to english language |
EMBASE (via Ovid)
Data parameters: 1974 to 2016 February 24.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 27.
Strategy
| 1. | hysterectom*.tw. |
| 2. | (‘salpingo oophorectom*’ or ‘salpingo ovariectom*’ or salpingooophorectom*).tw. |
| 3. | (salpingectom* adj7 (oophorectom* or ovariectom*)).tw. |
| 4. | 1 and (2 or 3) |
| 5. | exp hysterectomy/ |
| 6. | salpingooophorectomy/ |
| 7. | and/5-6 |
| 8. | 4 or 7 |
| 9. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 10. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 11. | 9 or 10 |
| 12. | 8 and 11 |
| 13. | Limit 12 to english language |
Colorectal cancer
MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (via Ovid)
Data parameters: 1946 to present.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 757.
Strategy
| 1. | ((colorectal or colon or colorectum or colonic or rectal or rectum or bowel or intenstin*) adj3 (cancer* or carcinom* or neoplasm* or malignan* or tumo?r*)).tw. |
| 2. | (CRC or mCRC).tw. |
| 3. | exp Colorectal Neoplasms/ |
| 4. | or/1-3 |
| 5. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 6. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 7. | (‘QLQ-C30’ or ‘FACT-G’).tw. |
| 8. | or/5-7 |
| 9. | 4 and 8 |
| 10. | exp animals/ not humans.sh. |
| 11. | 9 not 10 |
| 12. | limit 11 to (english language and yr=‘2005 -Current’) |
EMBASE (via Ovid)
Data parameters: 1974 to 2016 February 24.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 1833.
Strategy
| 1. | ((colorectal or colon or colorectum or colonic or rectal or rectum or bowel or intenstin*) adj3 (cancer* or carcinom* or neoplasm* or malignan* or tumo?r*)).tw. |
| 2. | (CRC or mCRC).tw. |
| 3. | exp colon tumor/ |
| 4. | exp rectum tumor/ |
| 5. | or/1-4 |
| 6. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 7. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 8. | (‘QLQ-C30’ or ‘FACT-G’).tw. |
| 9. | or/6-8 |
| 10. | 5 and 9 |
| 11. | limit 10 to (english language and yr=‘2005 -Current’) |
Endometrial cancer
MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (via Ovid)
Data parameters: 1946 to present.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 137.
Strategy
| 1. | ((endometrial or endometrium or uterine or uterus) adj3 (cancer* or carcinom* or neoplasm* or malignan* or tumo?r*)).tw. |
| 2. | exp Endometrial Neoplasms/ |
| 3. | 1 or 2 |
| 4. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 5. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 6. | (‘QLQ-C30’ or ‘FACT-G’).tw. |
| 7. | or/4-6 |
| 8. | 3 and 7 |
| 9. | exp animals/ not humans.sh. |
| 10. | 8 not 9 |
| 11. | limit 10 to english language |
EMBASE (via Ovid)
Data parameters: 1974 to 2016 February 24.
Date searched: 25 February 2016.
Searcher: SB.
Hits: 330.
Strategy
| 1. | ((endometrial or endometrium or uterine or uterus) adj3 (cancer* or carcinom* or neoplasm* or malignan* or tumo?r*)).tw. |
| 2. | exp endometrium tumor/ |
| 3. | 1 or 2 |
| 4. | (HRQOL or HRQL or QOL or QALY*).tw. |
| 5. | (EQ-5D or EQ-5D-3L or EQ-5D-5L or EQ-VAS or SF-6D or SF-12 or SF-36 or HUI2 or HUI3 or 15D or PROMIS).tw. |
| 6. | (‘QLQ-C30’ or ‘FACT-G’).tw. |
| 7. | or/4-6 |
| 8. | 3 and 7 |
| 9. | limit 8 to english language |
Number of hits per database and in total
| Database | Hits |
|---|---|
| MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (hysterectomy) | 13 |
| EMBASE (hysterectomy) | 27 |
| MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (colorectal cancer) | 757 |
| EMBASE (colorectal cancer) | 1833 |
| MEDLINE and MEDLINE In-Process & Other Non-Indexed Citations (endometrial cancer) | 137 |
| EMBASE (endometrial cancer) | 330 |
| Total records | 3097 |
| Duplicates | 906 |
| Total unique records | 2191 |
Forward citation chasing on included test accuracy studies
Scopus (Elsevier)
Data parameters: not applicable.
Date searched: 14 April 2016.
Searcher: SB.
Number of hits per study and in total
| Study | Hits |
|---|---|
| Barnetson et al., 200654 | 258 |
| Hendriks et al., 200361 | 132 |
| Limburg et al., 201155 | 24 |
| aNiessen 2006194 | 64 |
| Okkels et al., 201262 | 5 |
| Overbeek et al., 200759 | 23 |
| Poynter et al., 200834 | 97 |
| Shia et al., 200560 | 85 |
| Southey et al., 200556 | 137 |
| Total records | 825 |
| Duplicates | 138 |
| Duplicates in search results for test accuracy studies | 118 |
| Total unique records | 569 |
Appendix 2 Quality assessment
Quality appraisal was performed using phase 3 of the QUADAS-2 tool53 [reproduced with permission; see www.quadas.org (accessed 27 June 2017)].
Phase 3: risk of bias and applicability judgments
QUADAS-2 is structured so that four key domains are each rated in terms of the risk of bias and the concern regarding applicability to the research question (as defined above). Each key domain has a set of signalling questions to help reach the judgments regarding bias and applicability.
Domain 1: patient selection
A. Risk of bias
| Describe methods of patient selection: |
-
Was a consecutive or random sample of patients enrolled? Yes/no/unclear
-
Was a case–control design avoided? Yes/no/unclear
-
Did the study avoid inappropriate exclusions? Yes/no/unclear
Could the selection of patients have introduced bias? Risk: low/high/unclear
B. Concerns regarding applicability
| Describe included patients (prior testing, presentation, intended use of index test and setting): |
Is there concern that the included patients do not match the review question? Concern: low/high/unclear
Domain 2: index test(s)
If more than one index test was used, please complete for each test.
A. Risk of bias
| Describe the index test and how it was conducted and interpreted: |
-
Were the index test results interpreted without knowledge of the results of the reference standard? Yes/no/unclear
-
If a threshold was used, was it prespecified? Yes/no/unclear
Could the conduct or interpretation of the index test have introduced bias? Risk: low/high/unclear
B. Concerns regarding applicability
Is there concern that the index test, its conduct, or interpretation differs from the review question? Concern: low/high/unclear
Domain 3: reference standard
A. Risk of bias
| Describe the reference standard and how it was conducted and interpreted: |
-
Is the reference standard likely to correctly classify the target condition? Yes/no/unclear
-
Were the reference standard results interpreted without knowledge of the results of the index test? Yes/no/unclear
Could the reference standard, its conduct or its interpretation have introduced bias? Risk: low/high/unclear
B. Concerns regarding applicability
Is there concern that the target condition as defined by the reference standard does not match the review question? Concern: low/high/unclear
Domain 4: flow and timing
A. Risk of bias
| Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 × 2 table (refer to flow diagram): Describe the time interval and any interventions between the index test(s) and the reference standard: |
-
Was there an appropriate interval between the index test(s) and the reference standard? Yes/no/unclear
-
Did all patients receive a reference standard? Yes/no/unclear
-
Did patients receive the same reference standard? Yes/no/unclear
-
Were all patients included in the analysis? Yes/no/unclear
Could the patient flow have introduced bias? Risk: low/high/unclear
Appendix 3 Tables of excluded studies with rationale
Studies excluded at the full-text stage came from four sources: electronic searches (n = 71), searching systematic reviews (n = 38), forward citation chasing (n = 4) and backward citation chasing (n = 11). The reasons for exclusion were:
-
population – the study did not recruit the population specified in the protocol
-
index test – there was no index test or the study did not use an index test as specified in the protocol
-
reference standard – there was no reference standard or the study did not include a reference standard as specified in the protocol
-
outcomes – the study did not report outcomes as specified in the protocol
-
study design – the study design was not as specified in the protocol, including when the reference standard was not given to all participants (for high-risk studies) or was not given to a representative sample of index test-negative participants (for population-based studies)
-
abstract only – the study was published only as an abstract that was not linked to an included study that was published in full
-
duplicate – the reference was a duplicate of a study that had already been assessed for inclusion but this was either missed or not evident at an earlier stage.
Full texts identified from electronic searches and excluded
| Reference | Primary reason for exclusion |
|---|---|
| Abbott DE, Cantor SB, Rodriguez-Bigas MA, Chang GJ, Lynch PM, Feig BW, et al. Detecting hereditary nonpolyposis colorectal cancer syndrome (HNPCC) in patients with colorectal cancer (CRC): optimal strategies at lower costs. J Clin Oncol 2012;30(4 Suppl.):396 | Abstract only |
| Akagi K, Kakuta M, Takahashi A, Arai Y, Nishimura Y, Yatsuoka T, et al. Molecular screening with colorectal tumor tissue for Lynch syndrome. Fam Cancer 2011;10:S43 | Study design |
| Alemayehu A, Tomkova K, Zavodna K, Ventusova K, Krivulcik T, Bujalkova M, et al. The role of clinical criteria, genetic and epigenetic alterations in Lynch-syndrome diagnosis. Neoplasma 2007;54:391–401 | Study design |
| Alenda C, Paya A, Perez L, Alcaraz E, Soto JL, Guillen C, et al. Usefulness of p16 immunohistochemistry in the diagnosis of Lynch’s syndrome. Lab Invest 2009;9:122–3A | Abstract only |
| Alvarez K, Hurtado C, Hevia MA, Wielandt AM, de la Fuente M, Church J, et al. Spectrum of MLH1 and MSH2 mutations in Chilean families with suspected Lynch syndrome. Dis Colon Rectum 2010;53:450–9. https://doi.org/10.1007/DCR.0b013e3181d0c114 | Study design |
| Barrow E, Alduaij W, Robinson L, Shenton A, Clancy T, Lalloo F, et al. Colorectal cancer in HNPCC: cumulative lifetime incidence, survival and tumour distribution. A report of 121 families with proven mutations. Clin Genet 2008;74:233–42 | Study design |
| Barrow E, Evans DG, McMahon R, Byers R, Hill J. A comparative study of quantitative immunohistochemistry and quantum dot immunohistochemistry for mutation carrier identification in Lynch Syndrome. Colorectal Dis 2009;11:15 | Abstract only |
| Barrow E, Jagger E, Brierley J, Wallace A, Evans G, Hill J, McMahon R. Semiquantitative assessment of immunohistochemistry for mismatch repair proteins in Lynch syndrome. Histopathology 2010;56:331–44. https://doi.org/10.1111/j.1365-2559.2010.03485.x | Reference standard |
| Barrow E, Evans DG, McMahon R, Hill J, Byers R. A comparative study of quantitative immunohistochemistry and quantum dot immunohistochemistry for mutation carrier identification in Lynch syndrome. J Clin Pathol 2011;64:208–14 | Reference standard |
| Bessa X, Balleste B, Andreu M, Castells A, Bellosillo B, Balaguer F, et al. A prospective, multicenter, population-based study of BRAF mutational analysis for Lynch syndrome screening. Clin Gastroenterol Hepatol 2008;6:206–14 | Study design |
| Canard G, Lefevre J, Colas C, Coulet F, Soubrier F, Svreck M, et al. Screening HNPCC syndrome in colorectal cancer: are we doing enough? Colorectal Dis 2010;12:13–14 | Study design |
| Canard G, Lefevre JH, Colas C, Coulet F, Svrcek M, Lascols O, et al. Screening for Lynch syndrome in colorectal cancer: are we doing enough? Ann Surg Oncol 2012;19:809–16. https://doi.org/10.1245/s10434-011-2014-7 | Study design |
| Castells A, Payá A, Alenda C, Rodríguez-Moranta F, Agrelo R, Andreu M, et al. Cyclooxygenase 2 expression in colorectal cancer with DNA mismatch repair deficiency. Clin Cancer Res 2006;12:1686–92 | Study design |
| Chang SC, Lin PC, Yang SH, Wang HS, Liang WY, Lin JK. Taiwan hospital-based detection of Lynch syndrome distinguishes 2 types of microsatellite instabilities in colorectal cancers. Surgery 2010;147:720–8 | Study design |
| Chew MH, Liu YQ, Tang CL. Systematic study on genetic and epimutational profile of a cohort of Amsterdam criteria-defined Lynch syndrome in Singapore. Colorectal Dis 2013;15:41 | Abstract only |
| De Leon ED, Robinson L, Euhus D, Burstein E, Sarode VR. Evaluation of Lynch syndrome by immunohistochemistry and quantitative scoring by digital image analysis as a screening tool for the diagnosis of hereditary colon cancer and correlation with genetic analysis. Gastroenterology 2014;146(5 Suppl. 1):S-346 | Abstract only |
| Dong HS, Dong KC, Kim YH, Rhee PL, Kim JJ, Dae SK, et al. Effectiveness of each Bethesda marker in defining microsatellite instability when screening for Lynch syndrome. Hepatogastroenterology 2009;56:672–6 | Study design |
| Dudley B, Brand RE, Thull D, Bahary N, Nikiforova MN, Pai RK. Germline MLH1 mutations are frequently identified in Lynch syndrome patients with colorectal and endometrial carcinoma demonstrating isolated loss of PMS2 immunohistochemical expression. Am J Surg Pathol 2015;39:1114–20 | Study design |
| Farrell MP, Clabby C, Shea RO, Green AJ, Gallagher DJ. MLH1 hypermethylation assay should be considered when evaluating mismatch repair deficient tumours. J Clin Oncol 2015;33(15 Suppl.):e12524 | Abstract only |
| Gausachs M, Mur P, Corral J, Pineda M, Gonzalez S, Benito L, et al. MLH1 promoter hypermethylation in the analytical algorithm of Lynch syndrome: a cost-effectiveness study. Eur J Hum Genet 2012;20:762–8 | Reference standard |
| Gilmour K, Azam T, Onyeador N, Lees L, Walsh K, Ingman T, et al. Multidisciplinary stratification of colorectal cancer patients for clinical and therapeutic decision making. J Pathol 2012;226:S24 | Reference standard |
| Giraldez MD, Balaguer F, Petit A, Bujanda L, Moyano S, Gonzalo V, et al. High frequency of MSH6 loss in early-onset colorectal cancer. Gastroenterology 2009;136(5 Suppl. 1):A-452 | Reference standard |
| Giraldez MD, Balaguer F, Cuatrecasas M, Munoz J, Alonso-Espinaco V, Bujanda L, et al. High frequency of MSH6 germline mutations in early-onset colorectal cancer. Gastroenterology 2010;138(5 Suppl. 1):S150 | Abstract only |
| Goel A, Rhees J, Nagasaka T, Boland CR. Somatic hypermethylation of MSH2 is a frequent event in lynch syndrome colorectal cancers. Gastroenterology 2010;138(5 Suppl. 1):S-150 | Outcomes |
| Gould-Suarez M, El-Serag HB, Musher B, Franco LM, Chen GJ. Cost-effectiveness and diagnostic effectiveness analyses of multiple algorithms for the diagnosis of Lynch syndrome. Dig Dis Sci 2014;59:2913–26 | Study design |
| Guarinos C, Castillejo A, Barbera VM, Carbonell L, Sanchez-Heras AB, Castillejo MI, et al. Impact of TACSTD1 germline deletions as Lynch syndrome causing mutations in Spanish hereditary non-polyposis colorectal cancer-suspected patients. Eur J Cancer Suppl 2010;8:45 | Abstract only |
| Guarinos C, Castillejo A, Barberá VM, Pérez-Carbonell L, Sánchez-Heras AB, Segura A, et al. EPCAM germ line deletions as causes of Lynch syndrome in Spanish patients. J Mol Diagn 2010;12:765–70. https://doi.org/10.2353/jmoldx.2010.100039 | Study design |
| Haghighi MM, Mohebbi SR, Molaei M, Ghiasi S, Fatemi R, Zali MR. Microsatellite instability markers profile in patients with colorectal cancers caused by germ-line mutations in DNA mismatch repair genes. IUBMB Life 2009;61:309 | Reference standard |
| Haraldsdottir S, Hampel H, Frankel W, Wu CSY, Pan XJ, Bekaii-Saab TS, et al. Effect of genetic counseling on detection of Lynch syndrome (LS) in colorectal cancer (CRC) patients (pts). J Clin Oncol 2014;32(3 Suppl.):419 | Abstract only |
| Idris AF, Hooper SD, Farrell MP, Nolan C, Clarke R, Berkley E, et al. Are immunohistochemical (IHC)/microsatellite instability (MSI) testing necessary as part of Lynch syndrome work-up in the era of multiplex genetic testing? J Clin Oncol 2014;32(3 Suppl.):425 | Outcomes |
| Kets CM, van Krieken J, Hebeda KM, Wezenberg SJ, Goossens M, Brunner HG, et al. Very low prevalence of germline MSH6 mutations in hereditary non-polyposis colorectal cancer suspected patients with colorectal cancer without microsatellite instability. Br J Cancer 2006;95:1678–82. https://doi.org/10.1038/sj.bjc.6603478 | Study design |
| Kim S, Kim J, Yu C, Chung H, Chun S, Lee W, et al. Germline mutations in MLH1 and MSH2 in Korean hereditary non-polyposis colorectal cancer (HNPCC) patients. J Mol Diagn 2011;13:717 | Abstract only |
| Kostina O, Rebane E, Anderson W, Kask M, Valkna A. Molecular diagnostics strategy to identify hereditary non-polyposis colorectal cancer patients. Clin Cancer Res 2010;16(7 Suppl.):B39 | Abstract only |
| Kostina O, Rebane E, Kask M, Anderson W. Molecular diagnostic strategy to identify families with hereditary non-polyposis colorectal cancer syndrome. Eur J Cancer 2011;47:S186 | Abstract only |
| Kraus C, Rau T, Lux P, Erlenbach-Wunsch K, Stohr R, Agaimy A, et al. Detection of presumably low penetrance germ line mutations associated with colorectal cancer – results of a prospective comparative study of germ line mutation analysis by next generation sequencing and classical molecular pathology testing. Med Genet 2014;26:121–2 | Abstract only |
| Lastella P, Patruno M, Forte G, Montanaro A, Di Gregorio C, Sabba C, et al. Identification and surveillance of 19 Lynch syndrome families in southern Italy: report of six novel germline mutations and a common founder mutation. Fam Cancer 2011;10:285–95 | Study design |
| Leenen CHM, Dubbink EJ, Van Lier MGF, Hulspas SM, Kuipers EJ, Van Leerdam ME, et al. Challenges and pitfalls in screening for Lynch syndrome by molecular tumor tissue analysis. Fam Cancer 2011;10:S29 | Duplicate |
| Leenen CH, Dubbink EJ, Van Lier MG, Hulspas S, Kuipers EJ, Van Leerdam M, et al. Challenges and pitfalls in screening for Lynch syndrome by molecular tumor tissue analysis. Gastroenterology 2011;140(5 Suppl. 1):S352–3 | Abstract only |
| Liu YQ, Goh XW, Tan SY, Chew MH, Tien CTL, Koh PK, et al. Molecular detection of 5 mismatch repair gene defects in Singapore cohort of Lynch syndrome families. Proc Singapore Healthcare 2012;21:S347 | Abstract only |
| Mafnas C, Martin B, Ford J, Longacre T. Lynch syndrome screening: Discordance in MMR and germline test results. Lab Invest 2015;95:177A | Abstract only |
| Mahooti S, Hampel H, LaJeunesse J, Sotamaa K, de la Chapelle A, Frankel WL. MLH1 and PMS2 protein expression in 103 colorectal carcinomas with MLH1 promoter methylation and without MLH1 or PMS2 germline mutation. Mod Pathol 2006;19:113A–113A | Abstract only |
| Marginean F, Landolfi S, Hernandez J, de torres I, Garrido M, Badia D, et al. High feasibility of hmlh1, hmsh2 and hmsh6 protein expression and microsatellite instability analysis (pentaplex system) to screen patients with clinical criteria of Lynch syndrome. Virchows Archiv 2008;452:S193–4 | Reference standard |
| Mokarram P, Rismanchi M, Alizadeh Naeeni M, Mirab Samiee S, Paryan M, Alipour A, et al. Microsatellite instability typing in serum and tissue of patients with colorectal cancer: comparing real time PCR with hybridization probe and high-performance liquid chromatography. Mol Biol Rep 2014;41:2835–44 | Reference standard |
| Molinari F, Signoroni S, Lampis A, Bertan C, Perrone F, Sala P, et al. BRAF mutation analysis is a valid tool to implement in Lynch syndrome diagnosis in patients classified according to the Bethesda guidelines. Tumori 2014;100:315–20 | Study design |
| Monzon JG, Cremin C, Armstrong L, Nuk J, Young S, Horsman DE, et al. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer 2010;126:930–9 | Study design |
| Niessen RC, Berends MJ, Wu Y, Sijmons RH, Hollema H, Ligtenberg MJ, et al. Identification of mismatch repair gene mutations in young patients with colorectal cancer and in patients with multiple tumours associated with hereditary non-polyposis colorectal cancer. Gut 2006;55:1781–8 | Study design |
| Zahary MN, Gurjeet K, Abu Hassan MR, Singh H, Venkatesh RN, Ankathil R. Mutation and protein expression analysis of mismatch repair gene, MLH1 in Malaysian Lynch syndrome patients. J Gastroenterol Hepatol 2010;25:A37 | Reference standard |
| Zahary MN, Gurjeet K, Abu Hassan MR, Singh H, Venkatesh RN, Ankathil R. Germline mutation and epimutation analysis of human mismatch repair genes, MLH1 and MSH2 in Malaysian Lynch syndrome patients. HUGO J 2011;5:243 | Reference standard |
| Orellana P, Wielandt AM, Alvarez K, Pinto E, Hurtado C, Zarate AJ, et al. Usefulness of immunohistochemistry and microsatellite instability in families with suspected Lynch syndrome. Colorectal Dis 2010;12:2 | Abstract only |
| Orellana P, Wielandt AM, Alvarez K, Hurtado C, Pinto E, Carvallo P, et al. Analysis of point mutation and genomic rearrangements in MLH1, MSH2, MSH6 and PMS2 genes in Lynch syndrome: a Chilean experience. Fam Cancer 2013;12:803–4 | Abstract only |
| Pan M, Hoodfar E, Bergoffen JA, Fulton R, Hofmeister L, Chavez A, et al. Improving detection of Lynch syndrome using a reflex immunohistochemistry algorithm for all patients with newly diagnosed colorectal cancer. J Clin Oncol 2012;30(34 Suppl.):98 | Abstract only |
| Payá A, Alenda C, Pérez-Carbonell L, Rojas E, Soto JL, Guillén C, et al. Utility of p16 immunohistochemistry for the identification of Lynch syndrome. Clin Cancer Res 2009;15:3156–62. https://doi.org/10.1158/1078-0432.CCR-08-3116 | Study design |
| Perea J, Rodriguez Y, Rueda D, Marin JC, Diaz-Tasende J, Alvaro E, et al. Early-onset colorectal cancer is an easy and effective tool to identify retrospectively Lynch syndrome. Ann Surg Oncol 2011;18:3285–91 | Study design |
| Perez-Cabornero L, Infante Sanz M, Velasco Sampedro E, Lastra Aras E, Acedo Becares A, Miner Pino C, et al. Frequency of rearrangements in Lynch syndrome cases associated with MSH2: characterization of a new deletion involving both EPCAM and the 5’ part of MSH2. Cancer Prev Res 2011;4:1556–62 | Study design |
| Pérez-Carbonell L, Alenda C, Payá A, Castillejo A, Barbera VM, Guillen C, et al. Methylation analysis of MLH1 improves the selection of patients for genetic testing in Lynch syndrome. J Mol Diagn 2010;12:498–504 | Study design |
| Pérez-Carbonell L, Ruiz-Ponte C, Bessa X, Soto JL, Castillejo A, Barbera V, et al. Comparison between routine immunohistochemistry for mismatch repair proteins versus revised Bethesda guidelines in the diagnosis of Lynch syndrome in a non-selected population of colorectal cancer patients. Gastroenterology 2010;138(5 Supp. 1):S-297 | Study design |
| Pérez-Carbonell L, Guarinos C, Soler MR, Sanchez-Fortun C, Sempere-Robles L, Ruiz-Ponte C, et al. Comparison between universal immunohistochemistry for mismatch repair proteins versus revised Bethesda guidelines in the detection of patients with Lynch syndrome. Gastroenterology 2011;140(5 Suppl. 1):S-97 | Study design |
| Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Paya A, Brea A, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut 2012;61:865–72 | Study design |
| Rea G, Magee A, Loughrey MB. Streamlining the use of IHC in identifying germline mismatch repair mutations in Lynch syndrome. Ulster Med J 2012;81:98–9 | Study design |
| Ristimaki A, Thiel A, Heinonen M, Kantonen J, Lahtinen L, Mecklin JP, et al. Immunohistochemical detection of BRAF V600E mutation in colorectal cancer and its use to identify Lynch syndrome patients. Virchows Archiv 2013;463:207–8 | Abstract only |
| Rodriguez-Soler M, Perez-Carbonell L, Guarinos C, Zapater P, Castillejo A, Barbera VM, et al. Risk of cancer in cases of suspected Lynch syndrome without germline mutation. Gastroenterology 2013;144:926–932.e1; quiz e13-4 | Study design |
| Roncari B, Pedroni M, Maffei S, Di Gregorio C, Ponti G, Scarselli A, et al. Frequency of constitutional MSH6 mutations in a consecutive series of families with clinical suspicion of HNPCC. Clin Genet 2007;72:230–7 | Study design |
| Salazar R, Tian S, Santos C, Rosenberg R, Nitsche U, Mesker WE, et al. Development and validation of a genomic signature to identify colorectal cancer patients with microsatellite instability. J Clin Oncol 2012;30(4 Suppl.):466 | Abstract only |
| Serrano M, Lage P, Belga S, Filipe B, Francisco I, Rodrigues P, et al. Bethesda criteria for microsatellite instability testing: impact on the detection of new cases of Lynch syndrome. Fam Cancer 2012;11:571–8 | Study design |
| Trano G, Sjursen W. Molecular analyses of colorectal tumours may improve the identification of Lynch syndrome related colorectal cancer. Colorectal Dis 2014;16:92 | Study design |
| Wang J, Luo MH, Zhang ZX, Zhang PD, Jiang XL, Ma DW, et al. Clinical and molecular analysis of hereditary non-polyposis colorectal cancer in Chinese colorectal cancer patients. World J Gastroenterol 2007;13:1612–17 | Reference standard |
| Warrier SK, Trainer AH, Lynch AC, Mitchell C, Hiscock R, Sawyer S, et al. Preoperative diagnosis of Lynch syndrome with DNA mismatch repair immunohistochemistry on a diagnostic biopsy. Dis Colon Rectum 2011;54:e23–24. https://doi.org/10.1097/DCR.0b013e318231db1f | Abstract only |
| You JF, Buhard O, Ligtenberg MJ, Kets CM, Niessen RC, Hofstra RM, et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer 2010;103:1840–5. https://doi.org/10.1038/sj.bjc.6605988 | Outcomes |
| Zahary MN, Kaur G, Abu Hassan MR, Singh H, Naik VR, Ankathil R. Germline mutation analysis of MLH1 and MSH2 in Malaysian Lynch syndrome patients. World J Gastroenterol 2012;18:814–20. https://doi.org/10.3748/wjg.v18.i8.814 | Study design |
Full texts identified from systematic reviews and excluded
| Reference | Primary reason for exclusion |
|---|---|
| Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 1998;338:1481–7. https://doi.org/10.1056/NEJM199805213382101 | Study design |
| Bacher JW, Flanagan LA, Smalley RL, Nassif NA, Burgart LJ, Halberg RB, et al. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers 2004;20:237–50 | Reference standard |
| Becouarn Y, Rullier A, Gorry P, Smith D, Richard-Molard B, Echinard E, et al. Value of microsatellite instability typing in detecting hereditary non-polyposis colorectal cancer. A prospective multicentric study by the Association Aquitaine Gastro. Gastroenterol Clin Biol 2005;29:667–75 | Reference standard |
| Calistri D, Presciuttini S, Buonsanti G, Radice P, Gazzoli I, Pensotti V, et al. Microsatellite instability in colorectal-cancer patients with suspected genetic predisposition. Int J Cancer 2000;89:87–91 | Study design |
| Casey G, Lindor NM, Papadopoulos N, Thibodeau SN, Moskow J, Steelman S, et al. Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA 2005;293:799–809 | Study design |
| Christensen M, Katballe N, Wikman F, Primdahl H, Sorensen FB, Laurberg S, et al. Antibody-based screening for hereditary nonpolyposis colorectal carcinoma compared with microsatellite analysis and sequencing. Cancer 2002;95:2422–30. https://doi.org/10.1002/cncr.10979 | Study design |
| Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet 2001;69:780–90. https://doi.org/10.1086/323658 | Study design |
| Curia MC, Palmirotta R, Aceto G, Messerini L, Verì MC, Crognale S, et al. Unbalanced germ-line expression of hMLH1 and hMSH2 alleles in hereditary nonpolyposis colorectal cancer. Cancer Res 1999;59:3570–5 | Reference standard |
| Debniak T, Kurzawski G, Gorski B, Kladny J, Domagala W, Lubinski J. Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer 2000;36:49–54 | Reference standard |
| Dieumegard B, Grandjouan S, Sabourin JC, Le Bihan ML, Lefrere I, Bellefqih, et al. Extensive molecular screening for hereditary non-polyposis colorectal cancer. Br J Cancer 2000;82:871–80. https://doi.org/10.1054/bjoc.1999.1014 | Reference standard |
| Drobinskaya I, Gabbert HE, Moeslein G, Mueller W. A new method for optimizing multiplex DNA microsatellite analysis in low quality archival specimens. Anticancer Res 2005;25:3251–8 | Reference standard |
| Durno C, Aronson M, Bapat B, Cohen Z, Gallinger S. Family history and molecular features of children, adolescents, and young adults with colorectal carcinoma. Gut 2005;54:1146–50 | Reference standard |
| Farrington SM, Lin-Goerke J, Ling J, Wang Y, Burczak JD, Robbins DJ, Dunlop MG. Systematic analysis of hMSH2 and hMLH1 in young colon cancer patients and controls. Am J Hum Genet 1998;63:749–59 | Reference standard |
| Giuffre G, Muller A, Brodegger T, Bocker-Edmonston T, Gebert J, Kloor M, et al. Microsatellite analysis of hereditary nonpolyposis colorectal cancer-associated colorectal adenomas by laser-assisted microdissection: correlation with mismatch repair protein expression provides new insights in early steps of tumorigenesis. J Mol Diagn 2005;7:160–70. https://doi.org/10.1016/S1525-1578(10)60542-9 | Reference standard |
| Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851–60. https://doi.org/10.1056/NEJMoa043146 | Study design |
| Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 2008;26:5783–8. https://doi.org/10.1200/JCO.2008.17.5950 | Study design |
| Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology 2004;127:17–25 | Reference standard |
| Hoedema R, Monroe T, Bos C, Palmer S, Kim D, Marvin M, Luchtefeld M. Genetic testing for hereditary nonpolyposis colorectal cancer. Am Surg 2003;69:387–91 | Reference standard |
| Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004;53:1137–44. https://doi.org/10.1136/gut.2003.037671 | Reference standard |
| Katballe N, Christensen M, Wikman FP, Ørntoft TF, Laurberg S. Frequency of hereditary non-polyposis colorectal cancer in Danish colorectal cancer patients. Gut 2002;50:43–51 | Reference standard |
| Lee SC, Guo JY, Lim R, Soo R, Koay E, Salto-Tellez M, et al. Clinical and molecular characteristics of hereditary non-polyposis colorectal cancer families in Southeast Asia. Clin Genet 2005;68:137–45 | Reference standard |
| Luo DC, Cai Q, Sun MH, Ni YZ, Ni SC, Chen ZJ, et al. Clinicopathological and molecular genetic analysis of HNPCC in China. World J Gastroenterol 2005;11:1673–9 | Reference standard |
| Menigatti M, Di Gregorio C, Borghi F, Sala E, Scarselli A, Pedroni M, et al. Methylation pattern of different regions of the MLH1 promoter and silencing of gene expression in hereditary and sporadic colorectal cancer. Genes Chromosomes Cancer 2001;31:357–61. https://doi.org/10.1002/gcc.1154 | Study design |
| Moslein G, Tester DJ, Lindor NM, Honchel R, Cunningham JM, French AJ, et al. Microsatellite instability and mutation analysis of hMSH2 and hMLH1 in patients with sporadic, familial and hereditary colorectal cancer. Hum Mol Genet 1996;5:1245–52 | Study design |
| Müller A, Giuffre G, Edmonston TB, Mathiak M, Roggendorf B, Heinmöller E, et al. Challenges and pitfalls in HNPCC screening by microsatellite analysis and immunohistochemistry. J Mol Diagn 2004;6:308–15 | Reference standard |
| Park JG, Park YJ, Wijnen JT, Vasen HF. Gene-environment interaction in hereditary nonpolyposis colorectal cancer with implications for diagnosis and genetic testing. Int J Cancer 1999;82:516–19 | Study design |
| Peel DJ, Ziogas A, Fox EA, Gildea M, Laham B, Clements E, et al. Characterization of hereditary nonpolyposis colorectal cancer families from a population-based series of cases. J Natl Cancer Inst 2000;92:1517–22 | Reference standard |
| Pinol V, Castells A, Andreu M, Castellvi-Bel S, Alenda C, Llor X, et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA 2005;293:1986–94. https://doi.org/10.1001/jama.293.16.1986 | Study design |
| Plaschke J, Engel C, Kruger S, Holinski-Feder E, Pagenstecher C, Mangold E, et al. Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J Clin Oncol 2004;22:4486–94. https://doi.org/10.1200/JCO.2004.02.033 | Reference standard |
| Plevová P, Krepelová A, Papezová M, Sedláková E, Curík R, Foretová L, et al. Immunohistochemical detection of the hMLH1 and hMSH2 proteins in hereditary non-polyposis colon cancer and sporadic colon cancer. Neoplasma 2004;51:275–84 | Reference standard |
| Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 2000;18:2193–200 | Study design |
| Stormorken AT, Bowitz-Lothe IM, Noren T, Kure E, Aase S, Wijnen J, et al. Immunohistochemistry identifies carriers of mismatch repair gene defects causing hereditary nonpolyposis colorectal cancer. J Clin Oncol 2005;23:4705–12. https://doi.org/10.1200/JCO.2005.05.180 | Study design |
| Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet 2000;37:641–5 | Study design |
| Terdiman JP, Gum JR Jr, Conrad PG, Miller GA, Weinberg V, Crawley SC, et al. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology 2001;120:21–30 | Study design |
| Trusky CL, Sepulveda AR, Hunt JL. Assessment of microsatellite instability in very small microdissected samples and in tumor samples that are contaminated with normal DNA. Diagn Mol Pathol 2006;15:63–9 | Study design |
| Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, et al. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res 2002;62:3485–92 | Reference standard |
| Wang L, Cunningham JM, Winters JL, Guenther JC, French AJ, Boardman LA, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 2003;63:5209–12 | Reference standard |
| Wolf B, Henglmueller S, Janschek E, Ilencikova D, Ludwig-Papst C, Bergmann M, et al. Spectrum of germ-line MLH1 and MSH2 mutations in Austrian patients with hereditary nonpolyposis colorectal cancer. Wien Klin Wochenschr 2005;117:269–77 | Reference standard |
Full texts identified from forward citation chasing and excluded
| Reference | Primary reason for exclusion |
|---|---|
| Jin HY, Liu X, Li VK, Ding Y, Yang B, Geng J, et al. Detection of mismatch repair gene germline mutation carrier among Chinese population with colorectal cancer. BMC Cancer 2008;8:44. https://doi.org/10.1186/1471-2407-8-44 | Study design |
| Vasen HF, Hendriks Y, de Jong AE, van Puijenbroek M, Tops C, Bröcker-Vriends AH, et al. Identification of HNPCC by molecular analysis of colorectal and endometrial tumors. Dis Markers 2004;20:207–13 | Study design |
| Wagner A, Barrows A, Wijnen JT, Van Der Klift H, Franken PF, Verkuijlen P, et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 2003;72:1088–100. https://doi.org/10.1086/373963 | Population |
| Yan HL, Hao LQ, Jin HY, Xing QH, Xue G, Mei Q, et al. Clinical features and mismatch repair genes analyses of Chinese suspected hereditary non-polypsis colorectal cancer: a cost-effective screening strategy proposal. Cancer Sci 2008;99:770–80. https://doi.org/10.1111/j.1349-7006.2008.00737.x | Study design |
Full texts identified from backwards citation chasing and excluded
| Reference | Primary reason for exclusion |
|---|---|
| Chapusot C, Martin L, Puig PL, Ponnelle T, Cheynel N, Bouvier AM, et al. What is the best way to assess microsatellite instability status in colorectal cancer? Study on a population base of 462 colorectal cancers. Am J Surg Pathol 2004;28:1553–9 | Reference standard |
| de Jong AE, van Puijenbroek M, Hendriks Y, Tops C, Wijnen J, Ausems MG, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 2004;10:972–80 | Reference standard |
| Gille JJP, Hogervorst FBL, Pals G, Wijnen JT, van Schooten RJ, Dommering CJ, et al. Genomic deletions of MSH2 and MLH1 in colorectal cancer families detected by a novel mutation detection approach. Br J Cancer 2002;87:892–7 | Index Test |
| Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043–8 | Reference standard |
| Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med 1996;2:169–74 | Reference standard |
| Mangold E, Pagenstecher C, Friedl W, Fischer HP, Merkelbach-Bruse S, Ohlendorf M, et al. Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining. J Pathol 2005;207:385–95. https://doi.org/10.1002/path.1858 | Reference standard |
| Marcus VA, Madlensky L, Gryfe R, Kim H, So K, Millar A, et al. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol 1999;23:1248–55 | Reference standard |
| Müller W, Burgart LJ, Krause-Paulus R, Thibodeau SN, Almeida M, Edmonston TB, et al. The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC) – results of an international collaborative study. Fam Cancer 2001;1:87–92 | Reference standard |
| Salahshor S, Koelble K, Rubio C, Lindblom A. Microsatellite Instability and hMLH1 and hMSH2 expression analysis in familial and sporadic colorectal cancer. Lab Invest 2001;81:535–41 | Reference standard |
| Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology 2005;128:1160–71 | Reference standard |
| Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, et al. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 2003;72:1088–100 | Population |
Appendix 4 Summary of parameters used in the health economic model
| Parameter name | Base-case value | Source |
|---|---|---|
| Population characteristics | ||
| Maximum age | 100 years | |
| Number of probands | 34,025 | ONS21 |
| Proportion of probands who are men | 55% | ONS4,21,101–103 |
| Number of relatives per proband | 6 | Assumption, based on Snowsill et al.4 and Barrow91 |
| Proportion of relatives who are first-degree relatives | 42% | Snowsill et al.4 (published and unpublished data) |
| Proportion of relatives who are men | 38% | Unpublished data reported in Snowsill et al.4 |
| Prevalence of LS | 2.8% | Hampel et al.92 |
| Proportion of LS gene mutations | ||
| MLH1 | 32% | Palomaki et al.42 |
| MSH2 | 39% | |
| MSH6 | 14% | |
| PMS2 | 15% | |
| Probability that relative has LS if proband has LS | 44% | Meta-analysis of published and unpublished data reported in Snowsill et al.4 |
| Age on entry (probands) | ||
| General population | Distribution (mean 73.0 years – model outcome) | ONS21,96–103 |
| With LS | Distribution (mean 59.2 years – model outcome) | Estimated from the parametric CRC incidence function |
| Age on entry (relatives) | ||
| General population | Distribution (mean 44.4 years – model outcome) | ONS104,105 |
| With LS | Distribution (mean 43.2 years – model outcome) | Without LS data multiplied by CRC mortality-free survival for LS. Truncated at 18 and 75 years4 |
| Diagnostic parameters | ||
| Test accuracy | ||
| MSI | ||
| Sensitivity (95% CI) | 0.913 (0.426 to 0.993) | Barnetson et al.,54 Poynter et al.,34 Southey et al.56 |
| Specificity (95% CI) | 0.837 (0.638 to 0.937) | |
| IHC | ||
| Sensitivity (95% CI) | 0.962 (0.694 to 0.996) | Limburg et al.,55 Southey et al.56 |
| Specificity (95% CI) | 0.884 (0.790 to 0.940) | |
| BRAF V600E | ||
| Sensitivity (95% CI) | 0.96 (0.60 to 0.99) | Ladabaum et al.3 |
| Specificity (95% CI) | 0.76 (0.60 to 0.87) | |
| MLH1 promoter methylation | ||
| Sensitivity (95% CI) | 0.94 (0.79 to 0.98) | Ladabaum et al.3 |
| Specificity (95% CI) | 0.75 (0.59 to 0.86) | |
| Diagnostic testing probands | ||
| Sensitivity | MLH1, MSH2, MSH6 0.90; PMS2 0.67 | Dinh et al.107 |
| Specificity | 0.997 | Senter et al.130 |
| Predictive testing relatives | ||
| Sensitivity | 1.00 | Assumed (Snowsill et al.4) |
| Specificity | 1.00 | |
| Acceptance of diagnostic tests and genetic counselling | ||
| MSI – proband | 100% | Ramsey et al.,83 confirmed by expert Dr Ian M Frayling in Snowsill et al.4 |
| IHC – proband | 100% | Assumed |
| BRAF V600E – proband | 100% | Assumed |
| MLH1 promoter methylation – proband | 100% | Assumed |
| Genetic test following counselling – proband | 90% | Ladabaum et al.85 |
| Genetic counselling – proband | 92.5% | Clinical expert (Dr Ian M Frayling) estimated range 90–95% as reported in Snowsill et al.4 |
| Genetic test following counselling – relative | 77% | Manchester Familial Colorectal Cancer Registry data reported in Barrow91 |
| Genetic counselling – relative | 78% | |
| Proportion of genetic tests in MSI strategies requiring IHC analysis | 5% | Personal communication (Ottie O’Brien and Samantha Butler) |
| Proportion of probands who decline testing assumed to have LS | 10% | Assumption based on results in Snowsill et al.4 |
| Psychological disutility associated with testing for LS | ||
| Proband | ||
| Test declined, surgery not offered | 0.04 | Kuppermann et al.,165 Snowsill et al.4 |
| Test declined, accept TAH-BSO | 0.05 (women only) | |
| Test declined, decline TAH-BSO | 0.11 (women only) | |
| Test accepted, LS negative | 0 | |
| Test accepted, LS positive, surgery not offered | 0.02 | |
| Test accepted, LS positive, accept TAH-BSO | 0.03 (women only) | |
| Test accepted, LS positive, decline TAH-BSO | 0.09 (women only) | |
| Relative | ||
| Test declined, surgery not offered | 0.04 | Kuppermann et al.,165 Snowsill et al.4 |
| Test declined, accept TAH-BSO | 0.08 (women only) | |
| Test declined, decline TAH-BSO | 0.11 (women only) | |
| Test accepted, LS negative | 0 | |
| Test accepted, LS positive, surgery not offered | 0.02 | |
| Test accepted, LS positive, accept TAH-BSO | 0.06 (women only) | |
| Test accepted, LS positive, decline TAH-BSO | 0.09 (women only) | |
| Diagnostic costs | ||
| MSI | £202 | UKGTN175 |
| IHC | £210 | UKGTN175 and personal communication (Dr Mark Arends and Dr Ian M Frayling) from Snowsill et al.4 |
| BRAF V600E | £119 | UKGTN175 and personal communication (Mr Michael Gandy), East of Scotland Regional Genetic Service174 and All Wales Molecular Genetics Laboratory176 from Snowsill et al.4 |
| MLH1 promoter methylation testing | £125 | UKGTN175 |
| Proband genetic test, all four genes | £1276 | UKGTN175 |
| Proband genetic counselling | £63 | PSSRU166,177 and personal communication with Professor Mary Porteous from Snowsill et al.4 |
| Targeted genetic test for relatives (MLH1) | £166 | UKGTN175 |
| Targeted genetic test for relatives (MSH2) | £161 | UKGTN175 |
| Targeted genetic test for relatives (MSH6) | £161 | UKGTN175 |
| Targeted genetic test for relatives (PMS2) | £165 | UKGTN175 |
| Relative genetic counselling | £63 | PSSRU166,177 and personal communication with Professor Mary Porteous from Snowsill et al.4 |
| CRC parameters | ||
| Acceptance of LS surveillance for CRC | ||
| Proband tested LS mutation positive | 97% | Manchester Familial Colorectal Cancer Registry91 |
| Proband LS assumed | 70% | |
| Relative tested LS mutation positive | 97% | |
| Relative LS assumed | 70% | |
| Start age LS surveillance colonoscopy | 25 years | Snowsill et al.4 |
| End age (years) LS surveillance colonoscopy | 75 years | |
| HR associated with surveillance colonoscopy | 0.387 | Järvinen et al.124 |
| Colonoscopy complication probability | ||
| Bleeding resulting in admission | 0.0546% | Gavin et al.168 |
| Perforation | 0.04% | |
| Death | 0.0083% | Cairns et al.46 |
| CRC stage on diagnosis | ||
| I | 17.6% | National Cancer Intelligence Network131 |
| II | 27.0% | |
| III | 29.5% | |
| IV | 25.9% | |
| CRC stage on diagnosis with LS surveillance colonoscopies | ||
| I | 68.6% | Mecklin et al.132 |
| II | 10.5% | |
| III | 12.8% | |
| IV | 8.1% | |
| Probability proband has colon cancer | ||
| With LS | ||
| Male | 0.94 | Dinh et al.107 (online appendix) |
| Female | 0.94 | |
| Without LS | ||
| Male | 0.63 | ONS103 |
| Female | 0.72 | |
| Surgery for CRC | ||
| Colon | ||
| Segmental resection | 96% | Finan et al.108 |
| Subtotal colectomy | 4% | |
| Rectum | ||
| AR | 98% | Finan et al.108 |
| Proctocolectomy | 2% | |
| Initial surgical state for probands | ||
| With LS | Segmental resection 90.7%, subtotal colectomy 3.3%, AR 5.9%, proctocolectomy 0.1% | Calculated from above |
| Without LS | ||
| Men | Segmental resection 60.3%, subtotal colectomy 2.2%, AR 36.8%, proctocolectomy 0.7% | Calculated from above |
| Women | Segmental resection 69.6%, subtotal colectomy 2.6%, AR 27.4%, proctocolectomy 0.5% | |
| Proportion of relatives who have previously had CRC | ||
| With LS | ||
| Men | None 96.8%, colon 2.98%, rectal 0.19% | Bonadona et al.,18 ONS,100,110,111 Welsh Cancer Intelligence and Surveillance Unit,112 Dinh et al.107 |
| Women | None 97.5%, colon 2.37%, rectal 0.15% | |
| Without LS | ||
| Men | None 99.8%, colon 0.13%, rectal 0.09% | National Cancer Intelligence Network109 |
| Women | None 99.9%, colon 0.10%, rectal 0.04% | |
| Initial surgical state for relatives | ||
| With LS | ||
| Men | None 96.8%, segmental resection 2.88%, subtotal colectomy 0.11%, AR 0.19%, proctocolectomy 0.00% | Calculated from above |
| Women | None 97.5%, segmental resection 2.29%, subtotal colectomy 0.08%, AR 0.15%, proctocolectomy 0.00% | |
| Without LS | ||
| Men | None 99.8%, segmental resection 0.12%, subtotal colectomy 0.00%, AR 0.09%, proctocolectomy 0.00% | Calculated from above |
| Women | None 99.9%, segmental resection 0.09%, subtotal colectomy 0.00%, AR 0.04%, proctocolectomy 0.00% | |
| Logistic model parameters for CRC incidence in individuals with LS | ||
| β0 | ||
| Men | 0.464 | Fit based on Bonadona et al.18 |
| Women | 0.435 | |
| β1 | ||
| Men | 0.107 | Fit based on Bonadona et al.18 |
| Women | 0.108 | |
| β2 | ||
| Men | 55.5 | Fit based on Bonadona et al.18 |
| Women | 61.3 | |
| Probability incident CRC is situated in the colon | ||
| With LS | ||
| No previous surgery | 0.94 | Dinh et al.107 |
| Segmental resection | 0.94 | |
| Subtotal colectomy | 0.00 | Assumption |
| AR | 1.00 | Assumption |
| Proctocolectomy | Not applicable | Assumption |
| Without LS | ||
| No previous surgery | Men 0.63, women 0.72 | ONS100 |
| Segmental resection | Men 0.63, women 0.72 | |
| Subtotal colectomy | 0.00 | Assumption |
| AR | 1.00 | Assumption |
| Proctocolectomy | Not applicable | Assumption |
| Mortality rate from CRC (per 100,000 person years) | ||
| 0–1 years since diagnosis | ||
| Dukes’ A | 3102 | Calculated from National Cancer Intelligence Network109 |
| Dukes’ B | 8709 | |
| Dukes’ C | 20,460 | |
| Dukes’ D | 96,729 | |
| 1–2 years since diagnosis | ||
| Dukes’ A | 419 | Calculated from National Cancer Intelligence Network109 |
| Dukes’ B | 5000 | |
| Dukes’ C | 17,971 | |
| Dukes’ D | 67,733 | |
| 2–3 years since diagnosis | ||
| Dukes’ A | 843 | Calculated from National Cancer Intelligence Network109 |
| Dukes’ B | 4761 | |
| Dukes’ C | 15,465 | |
| Dukes’ D | 51,116 | |
| 3–4 years since diagnosis | ||
| Dukes’ A | 1279 | Calculated from National Cancer Intelligence Network109 |
| Dukes’ B | 4000 | |
| Dukes’ C | 11,060 | |
| Dukes’ D | 32,857 | |
| > 4 years since diagnosis | ||
| Dukes’ A | 1400 | Calculated from National Cancer Intelligence Network109 |
| Dukes’ B | 3667 | |
| Dukes’ C | 9068 | |
| Dukes’ D | 23,375 | |
| HRs for CRC mortality, age at diagnosis vs. all ages | ||
| < 70 years | ||
| First year | 0.599 | Estimated using net survival statistics from the ONS;118 details in appendix 5 of Snowsill et al.4 |
| 1–4 years | 0.972 | |
| After 4 years | 1 | |
| 70–79 years | ||
| First year | 0.956 | Estimated using net survival statistics from the ONS;118 details in appendix 5 of Snowsill et al.4 |
| 1–4 years | 0.966 | |
| After 4 years | 1 | |
| ≥ 80 years | ||
| First year | 1.797 | Estimated using net survival statistics from the ONS;118 details in appendix 5 of Snowsill et al.4 |
| 1–4 years | 1.116 | |
| After 4 years | 1 | |
| HRs for CRC survival with LS | ||
| Dukes’ A and B | 0.57 | Lin et al.119 |
| Dukes’ C and D | 1 | Barnetson et al.54 |
| Disutilities associated with CRC | ||
| Dukes’ A | 0.00 | Ramsey et al.142 |
| Dukes’ B | 0.00 | |
| Dukes’ C | 0.00 | |
| Dukes’ D | 0.13 | Mittmann et al.143 |
| Disutility associated with CRC treatment (surgery, ostomies, chemotherapy) | 0 | Assumption based on literature review149–154 |
| Disutility associated with colonoscopy | 0 | Assumption based on literature review155 |
| Resource use colonoscopy | ||
| LS surveillance | Every 2 years from LS diagnosis (age 25–75 years) | Snowsill et al.4 |
| Post-CRC surveillance | Every 5 years post CRC diagnosis | Snowsill et al.4 |
| Unit cost colonoscopy | £585.80 | NHS Reference Costs 2014–15178 (HRG codes FZ51Z, FZ52Z, FZ53Z) |
| Cost of colonoscopy complications | ||
| Bleeding | ||
| Mild | £473 | NHS Reference Costs 2014–15178 (HRG code FZ38P) |
| Moderate | £1138 | NHS Reference Costs 2014–15178 (HRG codes FZ38J–FZ38L) |
| Severe | £4394 | NHS Reference Costs 2014–15178 (HRG codes FZ38G–FZ38H) |
| Perforation | £4909 | NHS Reference Costs 2014–15178 (HRG codes FZ77C–FZ77E) |
| Death | £4909 | Assumed same as perforation |
| CRC costs | ||
| Diagnosis | £1022 (+£202 if stage II for MSI testing) | Trueman et al.123 |
| Primary chemotherapy and radiotherapy | ||
| Colon | £14,494 | Trueman et al.123 |
| Rectal Dukes’ A | £1049 | |
| Rectal Dukes’ B | £4206 | |
| Rectal Dukes’ C | £9504 | |
| Primary surgery | ||
| Segmental resection | ||
| General population | £6501 | NHS Reference Costs 2014–15178 (HRG codes FZ74–76, FZ50) |
| LS | £6605 | |
| Subtotal colectomy with ileorectal anastomosis | £7879 | |
| Rectal excision | £7939 | |
| Proctocolectomy | £7977 | |
| Follow-up surveillance costs (excluding colonoscopy) per year | ||
| With LS | £229 | Trueman et al.,123 NHS Reference Costs 2014–15178 |
| Without LS | £230 | |
| Surgery and chemotherapy for recurrence | ||
| With LS | £12,333 | Trueman et al.123 |
| Without LS | £12,236 | |
| Stoma care | ||
| With LS | £214 | Trueman et al.123 |
| Without LS | £388 | |
| Palliative care | ||
| With LS | £9907 | Trueman et al.123 |
| Without LS | £9665 | |
| Endometrial cancer-related parameters | ||
| Gynaecological risk reduction for women with LS on entry | ||
| 0–34 years at diagnosis | No risk reduction 100.0%, surveillance 0.00%, prophylactic H-BSO 0.00% | Balmana et al.,125 Lorraine Cowley (personal communication) |
| 35–44 years at diagnosis | No risk reduction 20.0%, surveillance 60.0%, prophylactic H-BSO 20.0% | |
| 45–59 years at diagnosis | No risk reduction 16.7%, surveillance 45.8%, prophylactic H-BSO 37.5% | |
| 60–69 years at diagnosis | No risk reduction 0.0%, surveillance 14.3%, prophylactic H-BSO 85.7% | |
| ≥ 70 years at diagnosis | No risk reduction 14.3%, surveillance 0.0%, prophylactic H-BSO 85.7% | |
| Lifetime endometrial cancer risk in model | ||
| With LS | 35% | Bonadona et al.18 |
| Without LS | 0% | Assumption based on expected 1 in 41120 |
| Endometrial cancer survival | ||
| < 10 years since diagnosis | Piecewise constant rate of mortality for each year since diagnosis (77.5% alive at 10 years) | Cancer Research UK121 |
| ≥ 10 years since diagnosis | No further mortality | Assumed |
| Impact of gynaecological surveillance on endometrial cancer survival (hazard ratio) | 0.898 | Lewin et al.113 |
| Disutility associated with endometrial cancer | 0.036 | Nout et al.,156 Longworth et al.161 |
| Length of time endometrial cancer disutility applied | 1 year | Nout et al.156 |
| Disutility associated with prophylactic H-BSO | 0 | Assumption based on disutility of endometrial cancer treatment and long-term disutility |
| Costs related to endometrial cancer | ||
| Gynaecological surveillance | £473 | NHS,179 NHS Reference Costs 2014–15178 (HRG codes WF01A, MA36Z, MA25Z) |
| Prophylactic H-BSO | £3428 | NHS Reference Costs 2014–15178 (HRG codes MA07E–MA07G, MA08A–MA08B) |
| Surgery | £4005 | NHS Reference Costs 2014–15178 (HRG codes MA06A–MA06C) |
| Radiotherapy | £5870 | Havrilesky et al.169 |
| Adjuvant chemotherapy | £1798 | eMit database,180 NHS Reference Costs 2014–15178 (HRG code SB14Z) |
| Aspirin | ||
| Offered and accept chemoprevention | 80.4% | Burn et al.29 |
| Proportion receiving aspirin for 4 years | 59.0% | |
| Incidence rate ratio CRC | 0.37 | |
| Incidence rate ratio endometrial cancer | 0.49 | |
| Time that aspirin effect lasts for | 10 years | |
| Daily dose | 600 mg | |
| Annual cost aspirin | £149 | BNF181 |
| Other parameters | ||
| General mortality | Age dependent | England and Wales, 2008–10,122 adjusted by CRC mortality for England in 2010110 |
| Discounting | ||
| Costs | 3.5% | NICE reference case89 |
| QALYs | 3.5% | |
Appendix 5 Diagnostic meta-analysis code (Stata)
Glossary
- Adenoma
- A benign tumour formed from glandular structures in epithelial tissue.
- Adjuvant chemotherapy
- Additional cancer treatment given after the primary treatment to lower the risk that the cancer will come back.
- Amsterdam criteria
- A set of diagnostic criteria used by doctors to help identify families that are likely to have Lynch syndrome.
- Anastomosis
- A connection made surgically between adjacent blood vessels, parts of the intestine or other channels of the body.
- Anterior resection
- Removal of an area of the rectum and/or left side of the bowel.
- Array-based comparative genomic hybridisation
- A cytogenetic technology that evaluates areas of the human genome for gains or losses of chromosome segments.
- Base case
- The expected case using the assumptions deemed most likely to occur.
- Bethesda panel of microsatellite instability markers
- A panel of five microsatellite markers (BAT25, BAT26, D2S123, D5S346, D17S250) proposed in the original Bethesda guidelines to describe microsatellite instability in Lynch syndrome.
- Bimodal distribution of microsatellite instability markers
- Tumours defined as positive or negative for microsatellite instability.
- BRAF
- A human gene that makes a protein called B-raf (a member of the Raf kinase family).
- BRAF V600E
- A mutation of the BRAF gene detected in a range of carcinomas, including colorectal cancer.
- BRCA mutation
- Mutation in the BRCA1 or BRCA2 tumour-suppressor gene that causes hereditary breast–ovarian cancer syndrome.
- Cancer antigen 125 analysis
- Blood test for ovarian cancer.
- Carcinoembryonic antigen test
- Carcinoembryonic antigens are harmful substances (usually proteins) that are produced by some types of cancer. In response to the antigens, the body produces antibodies to help fight them. A carcinoembryonic antigen test is often carried out after surgery to check for carcinoembryonic antigen levels.
- Carcinogenesis
- The formation of a cancer whereby normal cells are transformed into cancer cells.
- Cascade testing
- The identification of close relatives of an individual with a disorder to determine whether the relatives are also affected or are carriers of the same disorder.
- Chemoprevention
- The use of pharmacological or natural agents that inhibit the development of invasive cancer either by blocking the deoxyribonucleic acid damage that initiates carcinogenesis or by arresting or reversing the progression of premalignant cells in which such damage has already occurred.
- Colonoscopic surveillance
- A screening programme in people at high risk of developing colorectal cancer with the aim of detecting precancerous changes early on and potentially preventing progression.
- Concordance
- The degree of agreement between two diagnostic tests.
- Conformation-sensitive gel electrophoresis
- A method for the rapid detection of single-base differences in double-stranded polymerase chain reaction products and deoxyribonucleic acid fragments.
- Confounder
- A third variable that can make it appear (sometimes incorrectly) that an observed exposure is associated with an outcome.
- Constitutional genetic testing
- Tests for mutations that affect all cells in the body and have been there since conception (also known as germ-line testing).
- Constitutional mutation
- A genetic mutation present in all cells (also known as germ-line mutation).
- Cost-effectiveness analysis
- An economic analysis that converts effects into health terms and describes the costs for additional health gains.
- Cytotoxicity
- The quality of being toxic to cells.
- Decision modelling
- A theoretical construct that allows analysis of the relationship between costs and outcomes of alternative health-care interventions.
- Decision tree
- A decision support tool that uses a tree-like graph or model of decisions and their possible consequences, including chance event outcomes, resource costs and utility.
- Deletion
- Change in the number of deoxyribonucleic acid bases by removing a piece of deoxyribonucleic acid.
- Denaturing gradient gel electrophoresis
- A molecular fingerprinting method that separates polymerase chain reaction-generated deoxyribonucleic acid products.
- Denaturing high-performance liquid chromatography
- A method of chromatography for the detection of base substitutions, small deletions or small insertions in the deoxyribonucleic acid.
- Deoxyribonucleic acid mismatch repair
- A process that corrects mismatches generated during deoxyribonucleic acid replication.
- Diagnostic yield
- The number of positive test results divided by the number of samples.
- Dinucleotide
- A molecule consisting primarily of two nucleotides.
- Discounting
- Weighting future effects on expenditure and health outcomes less heavily than effects in the present. Health economic evaluations for the National Institute for Health and Care Excellence discount future costs and benefits at a constant rate of 3.5% per year.
- Disutility
- The adverse or harmful effects associated with a particular activity or process, especially when carried out over a long period of time.
- Dukes’ stage
- The Dukes’ cancer staging system is divided into four groups: A, B, C and D. Dukes’ A is early bowel cancer and Dukes’ D is advanced bowel cancer.
- Duplication
- A piece of deoxyribonucleic acid that is abnormally copied one or more times. This type of mutation may alter the function of the resulting protein.
- Dysplasia
- The enlargement of an organ or tissue by the proliferation of cells of an abnormal type, as a developmental disorder or an early stage in the development of cancer.
- End-to-end study
- Studies that follow patients from testing through treatment to final outcomes.
- EuroQol-5 Dimensions
- A generic health-related quality-of-life questionnaire.
- Exteriorisation
- The transposition of an internal organ to the exterior of the body.
- False negative
- Incorrect negative test result – number of diseased people with a negative test result.
- False positive
- Incorrect positive test result – number of non-diseased people with a positive test result.
- First-degree relative
- A person’s parent, sibling or child.
- Frameshift mutation
- Occurs when the addition or loss of deoxyribonucleic acid bases changes a gene’s reading frame. The resulting protein is usually non-functional.
- Functional Assessment of Cancer Therapy – General
- A health-related quality-of-life questionnaire for cancer therapy.
- Germ line
- Inherited material that comes from the eggs or sperm and is passed on to offspring.
- Germ-line mutation
- A detectable and heritable variation in the lineage of germ cells, which is subsequently transferred to offspring and gives rise to a constitutional mutation.
- Hazard ratio
- A measure of how often a particular event happens in one group compared with how often it happens in another group over time.
- Hereditary non-polyposis colorectal cancer
- Previous name for Lynch syndrome, a hereditary disorder that causes an increase in the risk of several types of cancer.
- Hypermethylation
- An increase in the epigenetic methylation of cytosine and adenosine residues in deoxyribonucleic acid.
- Hysterectomy
- A surgical procedure to remove the womb (uterus).
- Immunoreactivity
- A measure of the immune reaction caused by an antigen.
- Incidence
- The rate of occurrence of new (or newly diagnosed) cases of a disease.
- Incremental cost-effectiveness ratio
- The difference in the mean costs of two interventions in the population of interest divided by the difference in the mean outcomes in the population of interest.
- Index test
- The test whose performance is being evaluated.
- Insertion
- Changes in the number of deoxyribonucleic acid bases in a gene by adding a piece of deoxyribonucleic acid. As a result, the protein made by the gene may not function properly.
- Intention-to-treat analysis
- Includes every subject who is randomised according to their randomised treatment assignment. It ignores non-compliance, protocol deviations, withdrawals and anything that happens after randomisation.
- Inter-rater variability
- The degree of agreement among raters (histopathologists for immunohistochemistry).
- Likelihood ratio
- The likelihood that a given test result would be expected in a patient with the target disorder compared with the likelihood that that same result would be expected in a patient without the target disorder.
- Locoregional metastases
- Metastasis (spread) of a cancer only within the region in which it arose.
- Markov model
- A stochastic model describing a sequence of possible events in which the probability of each event depends only on the state attained in the previous event.
- Meta-analysis
- A statistical technique used to combine the results of two or more studies and obtain a combined estimate of effect.
- Metachronous tumours
- Primary tumours not occurring at the same time (usually occurring > 6 months apart).
- Metastatic disease
- The spread of cancer from one organ or body part to another organ or body part.
- Methylation
- A process by which methyl groups are added to deoxyribonucleic acid.
- Microdissection
- Refers to a variety of techniques in which a microscope is used to assist in dissection.
- Microsatellite instability
- Abnormal patterns of microsatellite repeats observed when deoxyribonucleic acid is amplified from a tumour with defective mismatch repair compared with deoxyribonucleic acid amplified from surrounding normal tissue.
- Microsatellite stable
- No evidence of abnormal patterns of microsatellite repeats or defective mismatch repair.
- Missense
- A change in one deoxyribonucleic acid base pair that results in the substitution of one amino acid for another in the protein made by a gene.
- Mixed-effects logistic regression
- Used to model binary outcome variables, with the log odds of the outcomes modelled as a linear combination of the predictor variables when data are clustered or there are both fixed and random effects.
- Mononucleotide
- A single nucleotide molecule (with one nitrogenous base, one sugar and one phosphate group).
- Mortality bias
- When mutation carriers are more likely to have died before being able to receive predictive testing.
- Multiplex ligation-dependent probe amplification
- A multiplex polymerase chain reaction method that is capable of detecting abnormal copy numbers of up to 50 different genomic deoxyribonucleic acid or ribonucleic acid sequences and which is able to distinguish sequences differing by only one nucleotide.
- Negative predictive value
- The probability of someone with a negative test result actually not having the disease.
- Net survival
- The survival calculated from the estimated excess hazard of mortality caused by a condition.
- Next-generation sequencing
- Non-Sanger-based high-throughput deoxyribonucleic acid sequencing technologies.
- Nonsense
- A change in one deoxyribonucleic acid base pair that results in a premature signal to stop building a protein. This type of mutation results in a shortened protein that may function improperly or not at all.
- Nucleotide
- Small molecules that make up deoxyribonucleic acid strands. Nucleotides consist of a nitrogenous base, a sugar and a phosphate group.
- Optimum cut-off
- The cut-off score that demonstrates the best trade-off between sensitivity and specificity.
- Pathogenic mutation
- A mutation capable of causing disease.
- Penetrance
- The proportion of individuals carrying a particular variant of a gene (allele or genotype) that also expresses an associated trait.
- Per-protocol analysis
- A comparison of treatment groups that includes only those patients who completed the treatment that they were originally allocated to.
- Polymerase chain reaction
- A technology used for amplifying deoxyribonucleic acid sequences.
- Positive predictive value
- The probability of someone with a positive result actually having the disease.
- Predictive testing
- Testing for known mutations.
- Prevalence
- The proportion of cases in the population at a given time.
- Primary tumour
- A tumour growing at the anatomical site where tumour progression began.
- Probabilistic sensitivity analysis
- A technique to quantify the level of confidence that a decision-maker has in the conclusions of an economic evaluation.
- Proband
- The first affected family member.
- Proctocolectomy
- Surgical removal of the rectum and all or part of the colon.
- Promoter
- A region of deoxyribonucleic acid that initiates transcription of a particular gene.
- Proximal colon
- The first and middle parts of the colon.
- QLQ-C30
- A health-related quality-of-life questionnaire for cancer patients.
- QLQ-CR29
- An add-on module to the QLQ-C30 for colorectal cancer patients.
- Quality-adjusted life-year
- A measure of health gain used in economic evaluations in which survival duration is weighted or adjusted by the patient’s quality of life during the survival period.
- Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2)
- A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews.
- Receiver operating characteristic curve
- A graph that illustrates the trade-offs between sensitivity and specificity that result from varying the diagnostic threshold.
- Reference costs
- The average unit costs to the NHS of providing secondary health care to NHS patients.
- Reference standard
- The best currently available diagnostic test, against which the index test is compared.
- Reference standard-positive study
- A study that uses a variation on a two-gate study design in which only participants who are reference standard positive are recruited (authors’ own term).
- Reflex testing
- Testing performed automatically in response to patient characteristics or the results of other tests.
- Regional metastases
- The spread of cancer beyond the initial site to regional lymph nodes.
- Relative survival
- The observed survival within a group (e.g. people with colorectal cancer) as a proportion of the expected survival for a group with the same age and sex distribution.
- Revised Bethesda guidelines
- Recommendations for identifying individuals with Lynch syndrome.
- Salpingo-oophorectomy
- Surgical removal of a fallopian tube and an ovary.
- Scenario analysis
- A process of analysing possible future costs by considering alternative possible outcomes.
- Second-degree relative
- Someone who shares 25% of a person’s genes. This includes uncles, aunts, nephews, nieces, grandparents, grandchildren and half-siblings.
- Segmental resection
- Removal of the cancer, a section of normal colon on either side of the cancer and nearby lymph nodes, with the sections of the remaining colon then reattached.
- Sensitivity
- Proportion of individuals with the target disorder who have a positive test result.
- Sensitivity analysis
- A technique used to determine how different values of an independent variable impact on a particular dependent variable under a given set of assumptions.
- Short Form questionnaire-6 Dimensions (SF-6D)
- A general health-related quality-of-life tool.
- Short Form questionnaire-12 items (SF-12)
- A general health-related quality-of-life tool.
- Short Form questionnaire-36 items (SF-36)
- A general health-related quality-of-life tool.
- Single-gate study
- A test accuracy study design in which a single cohort of individuals is assessed by both the index test and the reference standard.
- Single-strand conformational polymorphism
- A conformational difference of single-stranded nucleotide sequences of identical length as induced by differences in the sequences under certain experimental conditions. This allows sequences to be distinguished by means of gel electrophoresis.
- Somatic
- Referring to the cells of the body in contrast to the germ-line cells.
- Southern blot analysis
- A procedure for identifying specific sequences of deoxyribonucleic acid in which fragments separated by gel electrophoresis are transferred directly to a second medium on which a hybridisation assay is carried out.
- Specificity
- Proportion of individuals without the target disorder who have a negative test result.
- Spectrum bias
- The phenomenon that the performance of a diagnostic test may vary in different clinical settings because each setting has a different mix of patients.
- Splenic flexure
- A curvature on the left of the transverse colon.
- Splice site
- Causes abnormal messenger ribonucleic acid processing.
- Sporadic colorectal cancer
- Colorectal cancer with no apparent hereditary component.
- Subtotal colectomy
- An operation to remove the colon, leaving the rectum behind.
- Synchronous tumours
- Primary tumours diagnosed within 6 months of each other.
- Trimodal distribution of microsatellite instability
- Threshold distribution that includes microsatellite instability-high, microsatellite instability-low and microsatellite stable.
- Tumorigenesis
- The production or formation of a tumour or tumours.
- Two-gate study
- A study that employs separate sampling schemes for diseased and non-diseased participants, with both groups being assessed by the index test.
- Unclassified variant
- A variation in a genetic sequence whose association with disease risk has not yet been established.
- Univariate analysis
- The examination of one variable at a time.
- Variant of uncertain significance
- A variation in a genetic sequence that after interpretation efforts still cannot be classified as likely pathogenic or non-pathogenic. Also called variant of unknown significance.
List of abbreviations
- 5-FU
- fluorouracil
- ACPGBI
- Association of Coloproctology of Great Britain and Ireland
- aGCH
- array-based comparative genomic hybridisation
- APER
- abdominoperineal excision of the rectum
- AR
- anterior resection
- AUC
- area under the curve
- BSG
- British Society of Gastroenterology
- CA-125
- cancer antigen 125
- CAPP2
- Colorectal Adenoma/Carcinoma Prevention Programme 2
- CCT
- controlled clinical trial
- CDSR
- Cochrane Database of Systematic Reviews
- CEA
- carcinoembryonic antigen
- CENTRAL
- Cochrane Central Register of Controlled Trials
- CI
- confidence interval
- CRC
- colorectal cancer
- CT
- computerised tomography
- DGGE
- denaturing gradient gel electrophoresis
- DHPLC
- denaturing high-performance liquid chromatography
- DNA
- deoxyribonucleic acid
- EGAPP
- Evaluation of Genomic Applications in Practice and Prevention
- eMit
- Electronic Market Information Tool
- EORTC
- European Organisation for Research and Treatment of Cancer
- EPCAM
- epithelial cell adhesion molecule
- EQ-5D
- EuroQol-5 Dimensions
- FN
- false negative
- FP
- false positive
- H-BSO
- hysterectomy and bilateral salpingo-oophorectomy
- HCHS
- Hospital and Community Health Services
- HMIC
- Health Management Information Consortium
- HNPCC
- hereditary non-polyposis colorectal cancer
- HR
- hazard ratio
- HRG
- Healthcare Resource Group
- HRQoL
- health-related quality of life
- HTA
- Health Technology Assessment
- ICER
- incremental cost-effectiveness ratio
- IHC
- immunohistochemistry
- INHB
- incremental net health benefit
- IPAA
- ileal pouch anal anastomosis
- LR
- likelihood ratio
- LR–
- likelihood ratio for negative test result
- LR+
- likelihood ratio for positive test result
- LS
- Lynch syndrome
- LYG
- life-year gained
- mCRC
- metachronous colorectal cancer
- MLH1
- MutL homologue 1
- MLPA
- multiplex ligation-dependant probe amplification
- MMR
- mismatch repair
- MSH2
- MutS homologue 2
- MSH6
- MutS homologue 6
- MSI
- microsatellite instability
- MSI-H
- microsatellite instability-high
- MSI-L
- microsatellite instability-low
- MSS
- microsatellite stable
- NCI
- National Cancer Institute
- NGS
- next-generation sequencing
- NHB
- net health benefit
- NHS EED
- NHS Economic Evaluation Database
- NICE
- National Institute for Health and Care Excellence
- NPV
- negative predictive value
- ONS
- Office for National Statistics
- PCR
- polymerase chain reaction
- PenTAG
- Peninsula Technology Assessment Group
- PMS2
- postmeiotic segregation increased 2
- PORTEC
- Postoperative Radiation Therapy for Endometrial Carcinoma
- PPV
- positive predictive value
- PSS
- Personal Social Services
- QALY
- quality-adjusted life-year
- QUADAS-2
- Quality Assessment of Diagnostic Accuracy Studies
- RCT
- randomised controlled trial
- ROC
- receiver operating characteristic
- SF-36
- Short Form questionnaire-36 items
- SROC
- summary receiver operating characteristic
- TAH-BSO
- total abdominal hysterectomy with bilateral salpingo-oophorectomy
- TN
- true negative
- TNM
- tumour–node–metastasis
- TP
- true positive
- UKGTN
- UK Genetic Testing Network