Notes
Article history
The research reported in this issue of the journal was funded by the HTA programme as award number NIHR135620. The contractual start date was in August 2022. The draft manuscript began editorial review in March 2023 and was accepted for publication in July 2023. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ manuscript and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this article.
Permissions
Copyright statement
Copyright © 2024 Carroll et al. This work was produced by Carroll et al. under the terms of a commissioning contract issued by the Secretary of State for Health and Social Care. This is an Open Access publication distributed under the terms of the Creative Commons Attribution CC BY 4.0 licence, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. See: https://creativecommons.org/licenses/by/4.0/. For attribution the title, original author(s), the publication source – NIHR Journals Library, and the DOI of the publication must be cited.
2024
Chapter 1 Background and definition of decision problem
Sections of this chapter have been reproduced from the study’s Protocol document, available at the NICE website. 1
Population
The population of interest for this appraisal is people who have had non-cardioembolic ischaemic stroke, minor stroke or transient ischaemic attack (TIA), and for whom clopidogrel treatment is being considered. Approximately 100,000 strokes occur every year in the UK and between 46,000 and 65,000 people experience a TIA. 2 Around 85% of strokes are ischaemic, occurring when the supply of blood to a part of the brain is interrupted, usually by a blocked artery. 2 It has been suggested that a TIA is not a separate pathological entity, but exists on an ischaemic stroke spectrum, constituting the mildest form. 3 Symptoms of stroke often occur suddenly and vary depending on the part of the brain being compromised. Symptoms tend to include issues with movement, speech, facial drooping and vision.
The median age for stroke in the UK is 77 years and a quarter of strokes in the UK happen in people of working age. 4 Lifestyle factors associated with stroke and TIA include smoking, alcohol and drug abuse, physical inactivity and poor diet. The presence of cardiovascular diseases, and medical conditions including diabetes mellitus, atrial fibrillation, chronic kidney disease and migraine, are also risk factors for stroke. 2 Other risk factors include previous stroke/TIA, family history of stroke, lower education and genetic or hereditary factors. Strokes are more common in people with African Caribbean or South Asian background (Stroke Association; Kings Fund). 5,6
People who have experienced a stroke or TIA are at an increased risk of further occlusive vascular events [e.g. ischaemic stroke, TIA and myocardial infarction (MI)]. 7 TIA precedes stroke in 15% of cases, providing a crucial opportunity to prevent more severe stroke. 8 Risk of stroke after TIA has been found to be approximately 8% at 7 days, 11.5% at 1 month and 17.3% at 3 months. Risk of recurrent stroke after a minor stroke has been suggested to be 11.5, 15 and 18.5%, respectively. 9 National Institute for Health and Care Excellence (NICE) TA210 recommends the use of antiplatelet medications as a preventative treatment for people who have had an ischaemic stroke or TIA. 10 This includes clopidogrel treatment and is discussed further in Place of the technology in the treatment pathway.
Target condition: clopidogrel resistance
Clopidogrel is an irreversible adenosine diphosphate (ADP)-receptor antagonist with antiplatelet properties. It is available as branded and generic preparations and has marketing authorisation for patients who have recently had an ischaemic stroke or TIA. 11
Clopidogrel is a prodrug, which needs to be converted (metabolised) into an active form by P450 CYP enzymes. 12 Inactive clopidogrel has no effect on platelet aggregation, and therefore does not prevent occlusive vascular events. A substantial proportion of the population are less able to metabolise clopidogrel to its active form, and clopidogrel does not achieve its full pharmacological effect in these patients. One of the main causes of impairment in the metabolisation process are genetic variants, mainly in the CYP2C19 gene, which encodes P450 CYP enzymes. This is known as ‘clopidogrel resistance’. In addition to genetic variations in the CYP2C19 gene, other factors that may cause or exacerbate clopidogrel resistance include taking drugs such as omeprazole, which compete for metabolism by the CYP450 system,13 and factors such as obesity, diabetes and hypertension. 14 There is also a potential role of other rare genetic changes. Thus, both genetic and clinical factors need to be considered when determining whether an individual will respond to clopidogrel treatment. Inhibition of platelet function can be measured in laboratories to assess the impact of these factors in each individual and their potential response to clopidogrel or other antiplatelet drugs, but the applicability of these tests is limited because of technical limitations and a lack of standardisation in the definitions of non-responders. 11
Genetic basis of clopidogrel resistance
Cytochrome P450 2C19 is one of the main enzymes that metabolises clopidogrel to its active form. This enzyme is encoded by the CYP2C19 gene. CYP2C19 is one of many genes associated with clopidogrel response, but it is widely recognised as being the most validated genetic determinant. 15 The CYP2C19 gene has multiple variant forms (alleles) which produce CYP2C19 enzymes. These alleles are given a star (*) number for identification. The Pharmacogene Variation Consortium (PharmVar) has outlined more than 35 star (*) allele haplotypes. 16 The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 genotype and clopidogrel therapy notes that CYP2C19 allele *1 pertains to normal function, and that *2 and *3 are the most common alleles associated with loss of function (LOF). We use the abbreviation ‘LOF allele’ from here to refer to alleles associated with LOF. A systematic review found that people who carried one or two of these alleles had an increased risk of stroke and composite vascular events in contrast to non-carriers among patients with ischaemic stroke or TIA treated with clopidogrel. 17 Some alleles, in particular allele *17, are associated with increased function. 15
A person’s genotype is their unique sequence of deoxyribonucleic acid (DNA), while their phenotype is the observable expression of this genotype. A person’s phenotype [in this case, how they will respond to (metabolise) clopidogrel] can be predicted based on their allele function combinations. Generally, people with the genotype of two normal function alleles (e.g. CYP2C19*1/*1) have the phenotype of normal metabolisers. Intermediate metabolisers (IMs) have one normal function allele and one LOF allele (e.g. CYP2C19*1/*2). Poor metabolisers (PMs) have two LOF alleles (e.g. CYP2C19*2/*3). Rapid metabolisers have one normal and one increased function allele (e.g. CYP2C19*1/*17) and those with two increased function alleles (e.g. CYP2C19*17/*17) are ultra-rapid metabolisers. 15
There are significant ethnic variations in the incidence of the different CYP2C19 alleles. Table 1 provides an overview of some of the main CYP2C19 alleles, their impact on clopidogrel metabolism and their prevalence in different populations.
| Allele | Impact on clopidogrel metabolism | Prevalence | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Global | European | African | Asian | South Asian | East Asian | Latin American | UK | ||
| *2 | LOF | 16.02 | 14.72 | 17.50 | 29.19 | 36.70 | 28.01 | 16.16 | 15.08 |
| *3 | LOF | 0.26 | 0.58 | 0.05 | 0.80 | 0.33 | 0.78 | 0.07 | 0.05 |
| *4 | LOF | 0.32 | 0.33 | 0.07 | 0.10 | 0.04 | 0.06 | 0.35 | 0.16 |
| *17 | Increased function | 19.60 | 23.13 | 22.64 | 1.80 | 7.00 | 1.00 | 16.4 | 20.89 |
Diagnostic test
This review focuses on two categories of CYP2C19 genetic testing: point-of-care tests (POCTs) and laboratory-based tests. POCTs include any analytical test carried out by a healthcare professional outside of the laboratory, although it is also possible to install near patient testing equipment in local laboratories, which may overcome challenges associated with storage of reagents. 20 These tests have the potential to deliver results more quickly than standard laboratory-based tests. The two POCTs in scope are the Genomadix Cube CYP2C19 System and the Genedrive CYP2C19 ID Kit. The Genomadix Cube test was previously known as the ‘Spartan Cube’, which is a successor to the ‘Spartan RX CYP2C 19 System’. The two Spartan tests are very similar but there are some differences: the three reaction tubes have been integrated into a single test cartridge, the swabs and test cartridges are packaged separately and the DNA analyser device is smaller. 21 There are also differences in the mechanisms used to heat and cool the samples; the storage, use and stability of the specimens on the swab; the optical system; and the test workflow. 22
Laboratory-based tests are conducted by technicians in the laboratory. In the NHS, genomic testing is generally delivered by a network of seven Genomic Laboratory Hubs. Testing for CYP2C19 is not currently included in the National Genomic Test Directory of tests commissioned by the NHS in England. Table 2 provides an overview of some of the available CYP2C19 genetic tests. The POCTs only target specific LOF alleles. Laboratory-based tests have the potential to target all LOF alleles; however, commercial kits are likely to only test for the most common variants or those with established clinical utility. However, lab-based testing would have greater flexibility to alter variants screened for as new evidence emerges.
| Name of test | Type of test | General information | CYP2C19 alleles targeted | Time to run test |
|---|---|---|---|---|
| Genomadix Cube CYP2C19 system | Point of care | Intended to be used in conjunction with clinical judgement and routine monitoring to determine therapeutic strategy for drugs metabolised by the CYP2C19 enzyme. Test kit cartridges must be stored between −15 °C and −80 °C and used within 15 minutes of removal from the freezer. Results are stored locally on a laptop connected to the device and can be exported as a portable document format (PDF). |
*2, *3, *17 | The test takes 1 hour to run for each cartridge. |
| Genedrive CYP2C19 ID Kit | Point of care | Used for qualitative in vitro molecular diagnostic tests. Test for CYP2C19 under development and likely to be available to NHS in early 2023. Results will be able to be transferred electronically to patient records by internet or through third-party middleware or printed with an optional label printer. |
*2, *3, *4, *8, *17, *35 | Less than 1 hour to run for each cartridge. |
| Sanger CYP2C19 sequencing | Laboratory | Routine genomic testing approach used in all NHS genomic laboratory hubs. This test sequences a single DNA fragment at a time. | All alleles | Depends on sample numbers and number of alleles being tested for – more will mean longer turnaround times |
| Next-generation CYP2C19 gene sequencing | Laboratory | Sequences millions of short DNA sequences in parallel. | All alleles | Quicker turnaround for large sample numbers compared to Sanger sequencing. |
| Targeted CYP2C19 gene variant | Laboratory | Targeted genotyping assay amplifies and detects specific variants in target genomic DNA. Examples include:
|
Potential to target all alleles but usually target specific alleles. | The methods of detection, equipment requirements and throughput capability vary between systems. |
Place of the technology in the treatment pathway
Guidelines on appropriate antiplatelet therapy for the secondary prevention of stroke vary. The two main guidance documents of relevance are NICE guidance NG128 on stroke and TIA4 and guidance from the Royal College of Physicians (RCP) on therapy for secondary prevention for people with stroke. 23 The treatment pathway is shown in Figure 1 for (1) adults with non-minor ischaemic stroke and (2) adults with minor stroke or TIA. Pathways are different in children and for patients with atrial fibrillation. In children, aspirin rather than clopidogrel is currently recommended to prevent recurrence. Other antiplatelets, including clopidogrel, should only be considered when there are other risk factors for cerebrovascular disease. 24 People who have disabling ischaemic stroke, and who are in atrial fibrillation, should be treated with aspirin for 2 weeks after which anticoagulation treatment should be considered. 4
FIGURE 1.
Treatment pathway for current NHS practice: (i) non-minor ischaemic stroke, (ii) minor ischaemic stroke (with NIHSS < 3) or TIA. LAMP, loop-mediated isothermal amplification; NIHSS, National Institutes of Health Stroke Scale; PCR, polymerase chain reaction; SNP, single-nucleotide polymorphisms. Doses: a, Clopidogrel 75 mg daily (after loading dose of 300 mg). b, Aspirin 300 mg daily. c, Aspirin 75 mg daily plus clopidogrel 75 mg daily (after loading dose of 300 mg).

Everyone with a suspected stroke should be admitted to a specialist acute stroke unit following assessment by first responders. NICE guidance NG128 states that within 24 hours of ischaemic stroke onset, daily aspirin 300 mg should be offered unless the individual is intolerant to aspirin. 4 Aspirin should be continued until 2 weeks after stroke symptoms begin or until discharged.
For people with high-risk TIA [often defined as patients with a risk of stroke scoring system after a suspected TIA (ABCD2) score of ≥ 4]25 or minor stroke, dual antiplatelet therapy (DAPT) of aspirin and clopidogrel is often used in line with guidance from the European Stroke Organisation, beginning with 2 weeks acute dual therapy. 26 After 2 weeks of acute treatment, NICE guidance recommends long-term antiplatelet treatment with clopidogrel monotherapy. 4 However, in practice, patients are often given dual treatment with aspirin and clopidogrel before moving to longer-term clopidogrel monotherapy. The recommended duration of dual therapy varies according to guidance from up to 21 days,9 21–90 days27 or up to 90 days. 28 This is consistent with the NICE clinical knowledge summary on secondary prevention following stroke and TIA, updated in 2022, which states that
Dual therapy with aspirin plus clopidogrel (for up to 90 days) or aspirin plus ticagrelor (for 30 days) may be initiated in secondary care for some people (for example, people at high risk of TIA, or those with intracranial stenosis) followed by antiplatelet monotherapy. 28
In those who are intolerant of aspirin, the RCP guidelines suggest that clopidogrel could be considered as initial treatment. 23
For patients with TIA that is not high risk (ABCD2 score of < 4), NICE guidance (TA210) recommends urgent treatment with modified-release dipyridamole in combination with aspirin in the first instance. 10 However, the NICE clinical knowledge summary advises clopidogrel monotherapy following acute 2-week treatment with aspirin,28 and the RCP Guidelines recommend clopidogrel regardless of stroke risk score for TIA patients. 23
Currently, genetic testing for clopidogrel resistance is not routinely performed in the NHS before using clopidogrel in ischaemic stroke or TIA patients. If genetic testing to inform preventative treatment is introduced in the NHS in people with stroke, it could take place in hospital before long-term antiplatelet treatment is started 2 weeks post ischaemic stroke, or sooner in the case of TIA. People with an allele suggesting poor or intermediate metabolism of clopidogrel could be treated with an alternative to clopidogrel, while those without these alleles would receive standard clopidogrel treatment. Alternative treatments could include the following:
-
aspirin
-
aspirin combined with dipyridamole
-
clopidogrel dose escalation (Unlicensed)
-
ticagrelor (Unlicensed).
We heard from our clinical advisors that, of these, the most likely to be used in NHS practice would be aspirin combined with dipyridamole, with a potential treatment pathway shown in Figure 2 for people with (1) non-minor ischaemic stroke and (2) minor stroke or TIA. Ticagrelor does not have marketing authorisation in the UK for secondary prevention after ischaemic stroke or TIA. However, we have heard from clinicians that it is sometimes used in high-risk patients, although it is not considered in those at high risk of bleeding due to an elevated bleeding risk. There is a suspended NICE technology appraisal on ticagrelor for preventing stroke after previous ischaemic stroke or high-risk TIA. 29 This was suspended by the company on 11 May 2021, who also withdrew their application for marketing authorisation for stroke to the European Medicines Agency (EMA) in December 2021. 29 Ticagrelor in combination with aspirin for up to 30 days is however included as a potential treatment for secondary prevention for some people (e.g. people at high risk of TIA or those with intracranial stenosis) in the 2022 NICE clinical knowledge summary on secondary prevention following stroke and TIA. 28
FIGURE 2.
Potential treatment pathway for people with CYP2C19 LOF alleles: (i) non-minor ischaemic stroke, (ii) minor stroke (with NIHSS < 3) or TIA. LAMP, Loop-mediated isothermal amplification; NIHSS, National Institutes of Health Stroke Scale. Doses: a, Modified-release dipyridamole 200 mg twice daily. b, Aspirin 300 mg daily. c, Aspirin 75 mg daily plus modified-release dipyridamole 200 mg twice daily.

Chapter 2 Objectives
Sections of this chapter have been reproduced from the study’s Protocol document, available at the NICE website. 1
The overall aim of this project is to summarise the evidence on the clinical and cost-effectiveness of genetic testing to identify clopidogrel resistance in people with non-cardioembolic ischaemic stroke or TIA. We defined the following objectives to address the overall aim:
Objective 1: Do people who have genetic testing for clopidogrel resistance, and who are treated based on these results, have a reduced risk of secondary vascular occlusive events compared to those who are not tested and are treated with clopidogrel following standard guidelines?
Objective 2: Do people who have LOF alleles associated with clopidogrel resistance have a reduced risk of secondary vascular occlusive events if treated with alternative interventions compared to treatment with clopidogrel?
Objective 3: Do people who have LOF alleles associated with clopidogrel resistance have an increased risk of secondary vascular occlusive events when treated with clopidogrel compared to patients without LOF alleles who are treated with clopidogrel?
Objective 4: What is the accuracy of point-of-care genotype tests for detecting variants associated with clopidogrel resistance?
Objective 5: What is the technical performance (other than accuracy) and cost of the different CYP2C19 genetic tests?
Objective 6: What is the cost-effectiveness of different POCT and laboratory-based genetic tests for clopidogrel resistance compared with not testing for clopidogrel resistance?
Objectives 1–3 focus on assessing whether people with LOF alleles have better outcomes if treated with alternative antiplatelet drugs. Objectives 4 and 5 evaluate the accuracy and technical performance of CYP2C19 genetic tests.
Chapter 3 Assessment of clinical effectiveness
Sections of this chapter have been reproduced from the study’s Protocol document, available at the NICE website. 1
A systematic review was conducted to summarise the evidence on the clinical effectiveness of clopidogrel genotype testing after ischaemic stroke, including minor stroke and TIA. The systematic review followed the principles outlined in the Centre for Reviews and Dissemination (CRD) guidance for undertaking reviews in health care,30 the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy31 and the NICE Health Technology Evaluations Manual. 32 The protocol was registered on the PROSPERO database (CRD42022357661) and the systematic review is reported according to PRISMA-2020 and PRISMA-DTA guidelines. 33,34 The systematic review was supplemented by a survey of manufacturers of POCTs and genomic laboratory hubs to collect information on the technical performance of the different CYPC19 genetic tests (Objective 5; Survey of laboratories).
Inclusion and exclusion criteria
Objectives 1, 2 and 3
Inclusion criteria for Objectives 1, 2 and 3 are summarised in Table 3. Studies that met these criteria were eligible for inclusion:
| Objective 1 | Objective 2 | Objective 3 | |
|---|---|---|---|
| Participants | Adults or children who have experienced an ischaemic stroke or TIA | Adults or children who have experienced an ischaemic stroke or TIA and who have one or two CYP2C19 LOF alleles associated with under metabolism of clopidogrel (e.g. *2 or *3). | Adults or children who have had an ischaemic stroke or TIA who are treated with clopidogrel alone or in combination with a second antiplatelet drug. |
| Intervention/exposure | Any CYP2C19 genotype test followed by any alternative antiplatelet drug(s) | Any alternative antiplatelet drug(s). | Presence of one or two CYP2C19 LOF alleles for metabolism of clopidogrel (e.g. *2 or *3). |
| Comparators | No testing; all patients treated with clopidogrel alone or in combination with a second antiplatelet drug | Clopidogrel alone or in combination with a second antiplatelet drug | No LOF alleles |
| Outcomes | Incidence of secondary vascular occlusive events Adverse events (e.g. bleeding or headache) Mortality Time to starting antiplatelet treatment, or to change of antiplatelet treatment Impact of test result on decisions about care Healthcare resource use (e.g. length of hospital stay) Quality of life Healthcare costs |
||
| Study design | RCTs or cohort studies | RCT or cohort studies | Cohort studies |
Objectives 4 and 5
Inclusion criteria for Objectives 4 and 5 are summarised in Table 4. Additional data for Objective 5, in particular for standard laboratory-based tests, were identified through the section Survey of laboratories. Studies that fulfilled the following criteria were eligible for inclusion:
| Participants | Adults or children who have experienced an ischaemic stroke or TIA. If insufficient studies are found in these populations, then we will include studies in other populations; we do not anticipate that test accuracy is likely to differ substantially based on population. |
| Index test | Either of the following POCT: Genomadix or Spartan Cube CYP2C19 system (referred to as ‘Genomadix Cube’). Studies of the previous version of this test, the Spartan RX CYP2C19 System and Spartan FRX CYP2C19 were also eligible. Genedrive system CYP2C19 test (referred to as ‘Genedrive test’ from here) |
| Target condition | Presence of at least one CYP2C19 LOF allele |
| Reference standard | Any reported laboratory-based reference standard for CYP2C19 |
| Outcomes | Data on sensitivity and specificity or sufficient data to construct a 2 × 2 table of test accuracy. Test failure rate; number of people with variant forms of CYP2C19 (and incidence of particular alleles); time to results; ease of use of test; cost of testing. |
| Setting | Any setting |
| Study design | Any primary study |
Study identification
Studies were identified using bibliographic and non-bibliographic search methods following the guidance in the NICE handbook. 32 We carried out two searches:
-
Search 1, undertaken on 10 August 2022, aimed to address Objectives 1, 2 and 3, taking the following form: [(search terms for Clopidogrel) AND (search terms for CYP2C19)]
-
Search 2, undertaken on 11 August 2022, aimed to address Objectives 4 and 5, taking the following form: [(terms for POCTs OR Genomadix OR Genedrive) AND (terms for CYP2C19 OR terms for Clopidogrel)]
The search strategies are reported in Appendix 1: Literature search strategies, using a search narrative. 35 They were developed by one researcher (CC) and checked by another (ET) using the peer review of electronic search strategies (PRESS) checklist. 36
Bibliographic searching
We searched the following databases from inception:
-
MEDLINE (MEDALL) via Ovid
-
EMBASE via Ovid
-
The Cochrane Central Register of Controlled Trials (CENTRAL) via Wiley
-
The Cumulative Index to Nursing and Allied Health Literature (CINAHL) via EBSCO Host
-
ECONLit via EBSCO Host
-
Health Technology Assessment (HTA) Library via the York CRD interface
-
NHS Economic Evaluation Databases (EEDs) Via the York CRD interface
-
Tufts cost-effectiveness analysis (CEA) Register via the Tufts Medical Centre website.
Non-bibliographic search methods
We also searched the following trials registry resources:
-
ClinicalTrials.gov via www.clinicaltrials.gov/
-
World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP) via www.who.int/clinical-trials-registry-platform
We screened the manufacturer submissions and their respective websites to identify additional relevant studies.
For all objectives, the reference lists of studies included at full-text screening were checked through manual review. Reference lists of any reviews (systematic or non-systematic) identified by our searches were also screened. For Objectives 4 and 5 (the accuracy review), studies fulfilling eligibility criteria at full text were forward-citation searched using the Science Citations Index (Clarivate).
Managing the searches
Data were exported to EndNote X9 for deduplication using the default deduplication settings.
Review strategy
Two reviewers independently screened titles and abstracts identified by the searches. Full copies of all reports considered potentially relevant were obtained and two reviewers independently assessed these for inclusion. Any disagreements were resolved by consensus or discussion with a third reviewer.
Data were extracted using standardised data extraction forms developed in Microsoft Access. Data extraction forms were piloted on a small sample of papers and adapted as necessary. Data were extracted by one reviewer and checked in detail by a second reviewer. Any disagreements were resolved by consensus or discussion with a third reviewer.
Objectives 1, 2 and 3
Data were extracted on the following: study name, study design [randomised controlled trial (RCT) or cohort study], objective that study addresses, funding sources (public, industry, mixed), study location, participants (type of stroke, age, sex, ethnicity), inclusion criteria, omeprazole use, number of eligible patients, number of patients recruited, CYP2C19 test details (test used, alleles tested for and definition of PM), interventions (e.g. clopidogrel, alternative antiplatelet drug) and incidence of secondary vascular occlusive events (number in intervention/exposed group and number in control group). Data were also extracted on the following secondary outcomes, where reported: adverse events (e.g. bleeding or headache), mortality, time to starting antiplatelet treatment or to change of antiplatelet treatment, impact of test result on decisions about care, healthcare resource use (e.g. length of hospital stay), quality of life and healthcare costs.
Dichotomous data were extracted as number of patients with events and/or number of events and total number of patients in each treatment arm. Data on follow-up time were also extracted. Where available, summary effect estimates together with 95% confidence intervals (CIs) and p-values for comparisons between groups together with details on the methods of analysis, any variables controlled for in the analysis and the test statistic were also extracted. None of the studies reported continuous or categorical outcome data. Where studies reported results stratified by ethnicity, these were extracted separately.
Objectives 4 and 5
Data were extracted on the following: funding (industry, non-industry, mixed), study location, start date, study design, inclusion criteria, exclusion criteria, participants (condition, age, sex, ethnicity), point-of-care genetic test (Genomadix Cube or Genedrive test) and reference standard test details (name, number tested, alleles tested for, who administered test, threshold for positive result) and accuracy data. Where reported, we also extracted data on the following secondary outcomes: test failure rate; number of people with variant forms of CYP2C19 (and incidence of particular alleles); time to results; ease of use of test; cost of testing.
Accuracy data were extracted as 2 × 2 tables comparing the POCT with a laboratory reference standard. Where 2 × 2 data were not available, data were extracted on any reported estimates of accuracy [e.g. sensitivity, specificity, area under the receiver operating characteristic curve (AUC ROC)]. Authors of studies were also contacted to request data to allow construction of 2 × 2 tables.
Each individual will have two alleles – one or both of these may be associated with LOF. As described in section Genetic basis of clopidogrel resistance, some alleles are associated with over metabolism rather than poor metabolism (e.g. *17). As no difference in treatment is recommended in people who are over metabolisers, these alleles were grouped with those that are associated with normal function. This gave three potential categories for each individual:
-
Two LOF alleles (e.g. *2/*2 or *3/*3 or *3/*2)
-
One LOF allele (e.g. *2/*1, *3/*1, *3/*17 or *2/*17)
-
Normal function (e.g. *1/*1 or *1/*17)
These categories were dichotomised into alleles that encode for normal function and those that are non-functional. A ‘positive’ test result (non-functional) was defined as the presence of at least one LOF allele. A positive reference standard was as reported in the study – either detection of any loss of allele function or detection of those alleles that are detectable by the POCT evaluated. If data were reported for both possible reference standards, then data were extracted for both of these. The reference standard was also dichotomised so that a ‘poor metaboliser’ was defined as having at least one LOF allele.
Where multiple sets of 2 × 2 data were reported in a single study, for example, for different tests, thresholds or alleles, all data were extracted.
Risk-of-bias assessment
The risk of bias (RoB) in included RCTs and controlled clinical trials (CCTs) was assessed using the RoB 2 tool. 37 Observational studies of exposure were assessed using the ROBINS-E tool. 38 Diagnostic accuracy studies were assessed using a modified version of QUADAS-2. 39 We omitted two signalling questions – ‘If a threshold was used, was it pre-specified’ in the Index Test domain, and ‘Was there an appropriate interval between index test and reference standard’ in the Flow and Timing domain. Genetic tests do not have a threshold in the standard test accuracy sense – they identify the presence or absence of certain alleles and so we considered that this question did not apply to this review. Similarly, the question on timing is not relevant for genetic tests as the allele would either be present or not and this would not change over time: therefore, the time interval between tests does not matter. We did not formally assess applicability as our research question was broad and all studies were applicable; instead, we extracted data on potential sources of variation such as population and considered these in our synthesis. Details of the tools are provided in Report Supplementary Material 1. Quality assessment was undertaken by one reviewer and checked by a second reviewer. Disagreements were resolved by consensus or discussion with a third reviewer.
Survey of laboratories
We conducted a web-based survey to gather data on the technical performance characteristics of CYP2C19 genetic tests (Objective 5). The survey was sent to seven genomic laboratory hubs who are responsible for delivering genomic testing in the NHS in England and to genomic laboratory hubs in Wales, Northern Ireland and Scotland. The survey collected information on:
-
Platforms capable of performing CYP2C19 testing available in the lab.
-
Preferred test platform for running CYP2C19.
-
Reason for preference.
-
For each platform or genetic test, we ask for information on:
-
alleles that would be tested for
-
impact of having to test for additional alleles
-
time to results
-
resources for running tests: staff time, staff grade, cost per test to run, maintenance of machines/quality assurance, additional administrative resources
-
ease of use
-
test failure rate
-
current testing capacity
-
whether faster turnaround would be possible with additional resources and what these would be
-
whether additional testing capacity would be possible with additional resources and what these would be
-
whether the test could be performed in local testing laboratories.
-
-
Facilitators and barriers to implementing testing, and what platform would be most likely to be implemented.
-
How feasible would it be to install POCTs in local laboratories and extra resources required.
Synthesis methods
For each objective, a narrative summary of all the included studies is presented. This includes a summary of the study characteristics and study quality.
Objectives 1, 2 and 3
We extracted and used hazard ratios (HRs) presented by the studies where available. For observational (cohort) studies, estimates that had been adjusted for potential confounders were used if reported, otherwise unadjusted estimates were used. When HRs were not available, they were estimated with a hazard rate analysis of event frequencies in relation to time at risk (when follow-up time was available), or from 2 × 2 tables of event numbers using complementary log–log (cloglog) transformations, assuming proportional hazards. 16 For studies with a zero cell, we applied a ‘continuity correction’, adding 0.5 to every cell. Details on how each HR estimate was obtained are presented in the Results tables for Objectives 1, 2 and 3 in Report Supplementary Material 1.
Objective 1
We did not identify sufficient data on similar intervention comparisons to carry out a meta-analysis for any outcomes for Objective 1. We provide a narrative summary of results from these studies, presented together with a forest plot showing HRs estimates comparing secondary occlusive vascular events between patients who received a genetic test and were treated accordingly, against patients with standard treatment with clopidogrel.
Objective 2
Where at least two studies evaluated the same outcome, meta-analysis was used to generate summary effect estimates for each objective. We had intended to perform random-effects meta-analyses, but insufficient data were available for this. Therefore, fixed-effect meta-analyses were performed. Forest plots were produced for each outcome showing individual and summary HRs with 95% CIs, stratified by interventions evaluated. To inform decisions on whether to conduct network meta-analyses, we drew network plots of treatment comparisons for each outcome, to assess whether networks were connected and whether loops of evidence existed. 40 Network meta-analysis was not subsequently performed for any outcome.
Objective 3
We used random-effects meta-analysis to estimate summary HRs, 95% CIs and 95% prediction intervals, for each outcome evaluated by the included studies, when at least three studies were available. Heterogeneity and inconsistency across studies were quantified using the tau and I2 statistics. A restricted maximum likelihood (REML) approach was used to estimate tau. 41 Fixed-effect meta-analyses were performed as sensitivity analyses, or as the sole analyses if only two studies were available. Funnel plots were produced for each outcome, to assess the presence of small-study effects. 42,43
We used subgroup analysis and metaregression to investigate potential heterogeneity in the HR for risk of secondary vascular occlusive events. In investigating heterogeneity, we included different vascular event outcomes (composite outcome, stroke, ischaemic stroke) in the same analyses. This allowed us to include more studies in these analyses, increasing power to detect differences in HR across variables. This was a post hoc decision based on observing that estimates of HR were very similar for these outcomes within studies that reported on two or more. For these analyses, we selected one outcome per study related to a secondary vascular event based on the following hierarchy: composite outcome, any stroke, ischaemic stroke.
We conducted subgroup analysis and univariable metaregression to explore whether the HR for risk of secondary vascular occlusive events in those with LOF compared to those with LOF alleles varied with any of the following covariates:
-
ethnicity: Asian, white, mixed, Hispanic, black or not reported (N/R) (pre-specified)
-
primary event: stroke, stroke or TIA, TIA (pre-specified)
-
RoB: high versus low (pre-specified)
-
clopidogrel regimen: clopidogrel alone (which includes clopidogrel plus initial aspirin), clopidogrel plus long-term aspirin, clopidogrel plus optional aspirin (which also includes other antiplatelets or anticoagulants) (post hoc exploratory) proton-pump inhibitor use: < 10%, 10–20%, 20–30%, 40–50%, > 50% or N/R (post hoc exploratory)
-
duration of follow-up: 3 months, 6 months, 1 year, 1–3 years, 3–5 years or N/R (post hoc exploratory)
-
loading dose (whether a higher initial dose of clopidogrel was administered): yes, no N/R
Where a study reported multiple categories (e.g. estimates stratified by ethnicity), these separate estimates were used in the relevant subgroup analyses.
Objective 4
Estimates of sensitivity and specificity of the POCTs were calculated from each set of 2 × 2 data, under the assumption that the laboratory reference standards have correctly categorised all study participants. Analyses were stratified according to POCT. Summary estimates of sensitivity and specificity together with 95% CIs were calculated using bivariate random-effects meta-analysis of sensitivity and specificity, using binomial likelihoods. 44,45 Coupled forest plots of sensitivity and specificity were used to display results from individual studies and summary estimates, to allow visual assessment of heterogeneity. Due to homogeneity of estimates across studies, heterogeneity was not formally investigated.
Objective 5
We did not identify sufficient data to carry out a meta-analysis for the secondary outcomes that address Objective 5. We provide a narrative summary of results from these studies, presented together with a summary of the results of the web-based survey (see Survey).
All analyses were conducted in Stata version 17 (StataCorp., College Station, LLC, Los Angeles, CA, United States). Coupled forest plots were produced in R, using the DTAPlots package.
Chapter 4 Results of clinical effectiveness review
Results of the searches
The process of study identification and selection is summarised in Figure 3 (Objectives 1–3) and Figure 4 (Objectives 4–5). Studies included, stratified by objective, and studies excluded at full text are reported in Appendix 2: Tables of included, ongoing or excluded studies.
FIGURE 3.
PRISMA flow chart: Objectives 1–3.
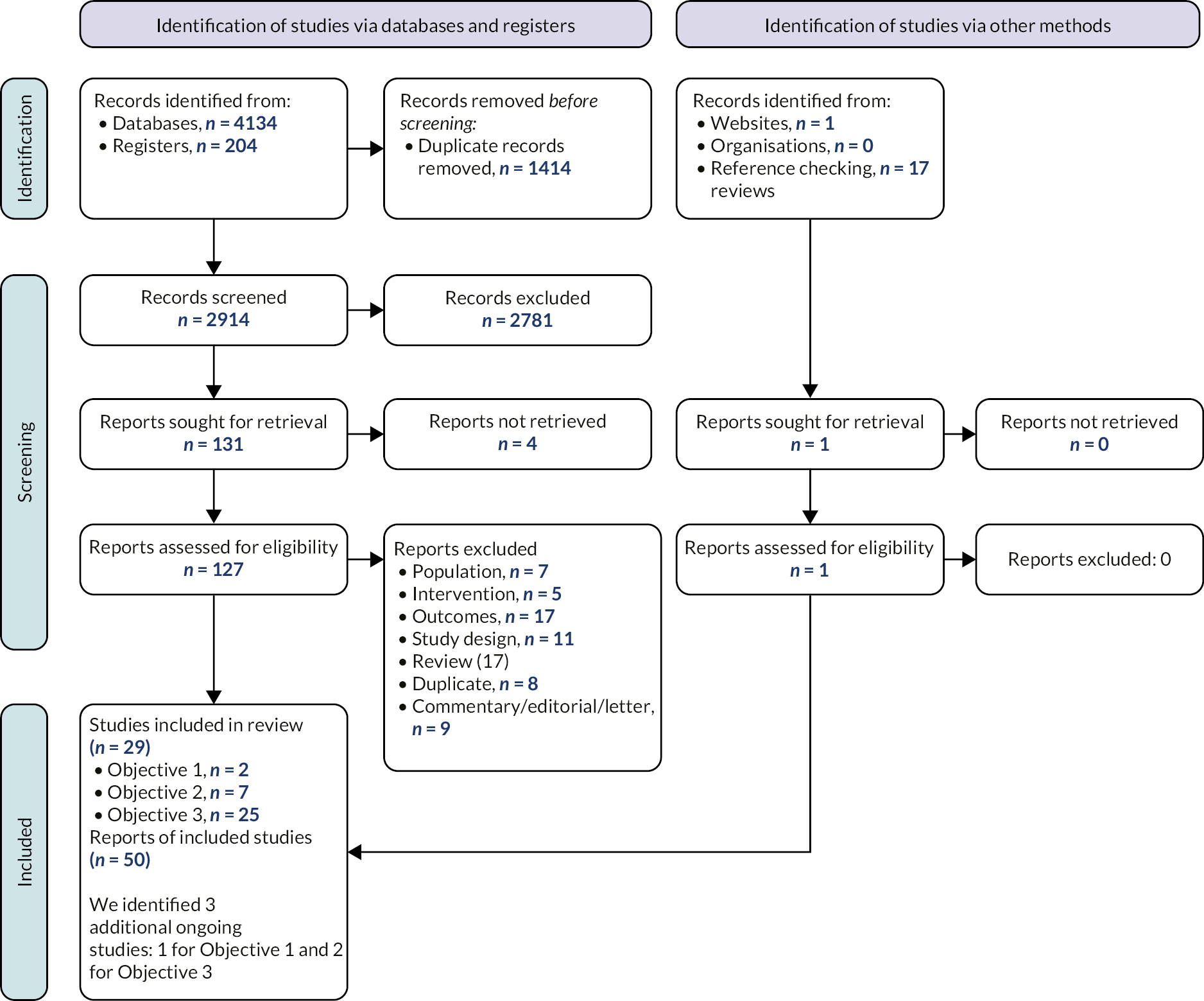
FIGURE 4.
PRISMA flow chart: Objectives 4–5. a, Three included studies include a pre-trial and a main trial, we are therefore treating these as separate studies.

Search 1: Objectives 1–3
The searches of bibliographic databases and trials registries identified 4338 references. After initial screening of titles and abstracts, 131 references were considered to be potentially relevant and ordered for full-paper screening; of these, 29 studies reported in 50 reports were included in the review: two studies for Objective 1; seven studies for Objective 2; and 25 studies for Objective 3. Five studies were included for Objectives 2 and 3. We identified three ongoing studies, one for Objective 1 and two for Objective 3.
Search 2: Objectives 4–5
The searches of bibliographic databases and trials registries identified 555 references. After initial screening of titles and abstracts, 35 references were considered to be potentially relevant and ordered for full paper screening; of these, 21 studies reported in 25 publications were included in the review. Nine studies for Objective 4 (three of these reported a pre-trial and a main trial) and 17 studies for Objective 5. Some studies were eligible for both objectives. All 21 references included in the manufacturers' submissions were identified by our searches; 4 were included in the review and 17 references did not meet inclusion criteria.
Objective 1
Two controlled trials from China were included for Objective 1. 46,47 Full details on these studies are reported in Report Supplementary Material 1. Both studies were small (80 and 190 patients) and did not provide sample size or power calculations. Duration of follow-up was 90 days in one study and 1 year in the other. One of these studies was reported in Chinese and was extracted with help from a native Chinese speaker and using Google Translate. 46 Both studies used laboratory-based testing to determine the presence of LOF alleles.
Xia et al. 46 allocated 80 patients to two groups:
-
Group A: all received clopidogrel 75 mg/day.
-
Group B: genotyped for the *1, *2, *3 and *17 alleles.
-
No LOF alleles: clopidogrel 75 mg/day (same as control).
-
One LOF allele: clopidogrel 150 mg.
-
Two LOF alleles: ticagrelor (this was recorded as ‘tigrillo’ in the English abstract but translation of the Chinese term suggested that this was ticagrelor).
-
Lan et al. 47 genotyped all participants for the *1, *2, *3 and *17 alleles. Participants were then divided into two groups (groups A and B with 90 patients in each) so that equal numbers with each potential genotype were included in each group. All patients were initially treated with clopidogrel (300 mg loading dose followed by 75 mg/day) and aspirin 100 mg day for 21 days. Treatment after this varied by intervention group and presence of LOF alleles:
-
Group A: clopidogrel 75 mg/day.
-
Group B:
-
normal metaboliser (no LOF alleles) and extensive metaboliser (EM) (1 or 2 *17 alleles): clopidogrel 75 mg/day
-
PM (1 or 2 LOF alleles): aspirin 100 mg/day.
-
This study did not technically meet inclusion criteria for Objective 1, as all patients were tested, however, as half of the tested patients were treated as if they had not been tested (i.e. standard treatment), we considered it appropriate to include this study for this objective.
Both studies enrolled patients with a stroke as a primary event. Mean age was 69 years and percentage of female participants was 38% in both studies. One study was funded by non-industry47 and the other did not report funding sources. 46
Risk of bias
Both studies46,47 were judged at high RoB for all outcomes extracted (Table 5). There was no clear information on the allocation process, and they were not randomised – the Lan study47 allocated patients so that equal numbers of each genotype were included in each group, but it was unclear how this was done. There was no evidence of a pre-registered protocol for either study. Full details on RoB assessment are presented in Report Supplementary Material 1.
| Study details | Domain | Rationale | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Overall | ||
| Lan et al. (2019)47 | ☹ | ☹ | ☹ | ☺ | 😐 | ☹ | Not randomised. Patients and carers were likely aware of the allocation, and there is no information on potential deviations, which could have affected the outcome. High proportion of loss to follow-up. No evidence of a pre-registered protocol. |
| Xia et al. (2021)46 | ☹ | ☹ | ☺ | ☺ | 😐 | ☹ | Not randomised. Patients and carers were likely aware of the allocation, and there is no information on potential deviations from the intervention, which could have affected the outcome. No evidence of a pre-registered protocol. |
Results
Incidence of secondary vascular events
Both studies46,47 presented data on incidence of secondary ischaemic stroke and MI. Xia et al. 46 reported the incidence of TIA, vascular death and a composite outcome (including stroke, TIA, MI and death). Lan et al. 47 reported data on haemorrhagic stroke. We additionally calculated a composite outcome for Lan et al., adding events for all outcomes reported. Report Supplementary Material 1 (see Table 3) provides an overview of results for incidence of secondary vascular events in these studies. Hazard ratios were N/R in these studies, so estimates were calculated from event frequencies and follow-up time. We did not meta-analyse results from these two studies due to the differences in interventions. In general, HRs suggested a reduction in composite outcomes, secondary ischaemic stroke, haemorrhagic stroke and TIA in patients tested for LOF alleles and treated accordingly, but CIs were wide and included the null (HR = 1) in all cases (Figure 5). There was no evidence of benefit in either group for vascular death or MI, although incidence of these outcomes was low (< 5%). Full details on results are presented in Report Supplementary Material 1.
FIGURE 5.
Forest plot showing HRs (95% CI) for secondary vascular events in patients treated with clopidogrel compared with patients tested for LOF alleles and offered personalised treatment.

Objective 2
Seven trials, reported in 23 full report publications, were included for Objective 2. 48–54 All studies were published in English. Two trials were restricted to patients with LOF alleles who were then randomised to different antiplatelet therapies. The other five studies were not restricted based on LOF alleles – patients were randomised to different antiplatelet strategies, and a subgroup analysis was then performed restricted to those with LOF alleles. Table 6 shows an overview of the studies included for Objective 2. Full details on the studies are reported in Report Supplementary Material 1.
| Feature | Category | Number of studies |
|---|---|---|
| Population | Stroke | 3 |
| Stroke and TIA | 4 | |
| Comparisons | (Clopidogrel 75 mg/day + aspirin 50–325 mg/day) vs. aspirin 50–325 mg/day | 1 |
| (Clopidogrel 75 mg/day + aspirin 75–200 mg/day for first 21/30 days) vs. aspirin | 2 | |
| Clopidogrel 75 mg/day vs. triflusal 300 mg twice daily | 1 | |
| Clopidogrel 75 mg/day (+aspirin 75–300 mg/day for 21 days) vs. ticagrelor 90 mg (+aspirin 75–300 mg/day for 21 days) | 2 | |
| Clopidogrel 75 mg/day (+aspirin) vs. high-dose (HD) clopidogrel 150 mg/day (+aspirin) for 21 days followed by aspirin alone | 1 | |
| Clopidogrel Loading dose | 600 mg | 1 |
| 300 mg | 4 | |
| No loading dose | 2 | |
| Design | RCT | 7 |
| Country | South Korea | 1 |
| USA | 1 | |
| China | 5 | |
| Funding | Non-industry | 5 |
| Drugs and tests provided by industry | 1 | |
| Industry – other | 1 | |
| CYP2C19 test | Seeplex CYP2C19 ACE genotyping system and Real-Q CYP2C19 genotyping kit | 1 |
| Drug Metabolism Enzyme TaqMan Allelic Discrimination assay | 1 | |
| GMEX point-of-care genotyping system | 1 | |
| Sequenom MassARRAY iPLEX platform | 2 | |
| N/R | 2 | |
| LOF alleles | CYP2C19 *2 and *3 | 6 |
| CYP2C19 *2 only | 1 | |
| Follow-up time | 90 days | 5 |
| 2–3 years (731–1095 days) | 1 | |
| 4–5 years (1461–1825 days) | 1 |
Three studies included patients who presented with stroke as their primary event, and four included patients with either stroke or TIA. Five studies took place in China and recruited patients predominantly of Chinese origin, one was done in South Korea including mostly patients of South Korean heritage and one took place in an international setting, with a majority white (67%) ethnicity. Mean age ranged from 60.8 years [standard deviation (SD) 8.7] to 64.8 years (SD N/R). The percentage of females ranged from 24% to 45%. Sample size ranged between 154 and 6412.
Three studies compared clopidogrel plus aspirin with aspirin alone. In the clopidogrel arm, one of these studies gave a one-off 300 mg loading dose of clopidogrel and aspirin only for an initial 21-day period, the second did not offer a loading dose of clopidogrel and stopped aspirin after 30 days, and the third gave a 600 mg loading dose of clopidogrel and continued the aspirin in combination with clopidogrel longer term. Two studies compared clopidogrel with ticagrelor – both studies included a 300 mg clopidogrel loading dose and an initial 21-day period when aspirin was given in addition to the clopidogrel or ticagrelor. One study compared clopidogrel with triflusal, without a loading dose in either arm. The final study compared a standard dose of clopidogrel (75 mg) with a higher dose of clopidogrel (150 mg). In this study, all patients received a 300 mg loading dose of clopidogrel and 150 mg aspirin for the first 21 days; after this, clopidogrel was stopped and patients continued treatment with 150 mg aspirin alone. One study was funded by industry organisations (drug manufacturer), one was funded by non-industry but drugs and genetic tests were supplied by industry and five were funded by non-industry organisations.
Four studies used laboratory-based genotyping tests [Seeplex CYP2C19 angiotensin-converting enzyme (ACE) genotyping system and Real-Q CYP2C19 genotyping kit, Drug Metabolism Enzyme TaqMan Allelic Discrimination Assay, and Sequenom MassARRAY increased plexing efficiency and flexibility (iPLEX) platform (Sequenom)], one used a POCT (GMEX point-of-care genotyping system) and two did not report the type of test that was used. Six studies investigated the two main LOF alleles (*2 and *3) and one study only genotyped one LOF allele (*2).
Duration of follow-up ranged across studies: five studies had a follow-up time of 90 days, one followed patients up between 2 and 3 years and one between 4 and 5 years.
Risk of bias
All outcomes assessed for every study were judged at the same level of RoB. Four of the seven studies were judged at low RoB. 48,50,52,53 One study was judged at some concerns due to lack of information on allocation concealment. Two studies were judged at high concerns, one due to lack of information on loss to follow-up, and the other due to lack of information on the randomisation process and potential deviations from the intended intervention. Table 7 provides a summary of the RoB assessment for each study; full details are provided in Report Supplementary Material 1.
| Study details | Domain | Rationale | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Overall | ||
| Chen et al. (2019)53 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Han et al. (2017)48 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Meschia et al. (2020)49 | ☺ | ☺ | ☹ | ☺ | ☺ | ☹ | No clear data on loss to follow-up, and it could potentially be related to the outcomes |
| Wang et al. (2016a)52 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Wang et al. (2021)50 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Wu et al. (2020)51 | 😐 | ☺ | ☺ | ☺ | ☺ | 😐 | No information on allocation concealment, baseline differences do not suggest a problem with the randomisation process. |
| Yi et al. (2018)54 | 😐 | ☹ | ☺ | ☺ | ☺ | ☹ | No information on allocation concealment, no data on blinding and potential deviations from the intended interventions. No information on statistical analysis |
Results
Included studies presented data on incidence of secondary vascular occlusive events, adverse events and mortality, in people who had LOF alleles associated with clopidogrel resistance and were treated with alternative interventions compared to standard treatment with clopidogrel. There were no studies reporting data on other outcomes of interest for Objective 2.
Secondary vascular occlusive events
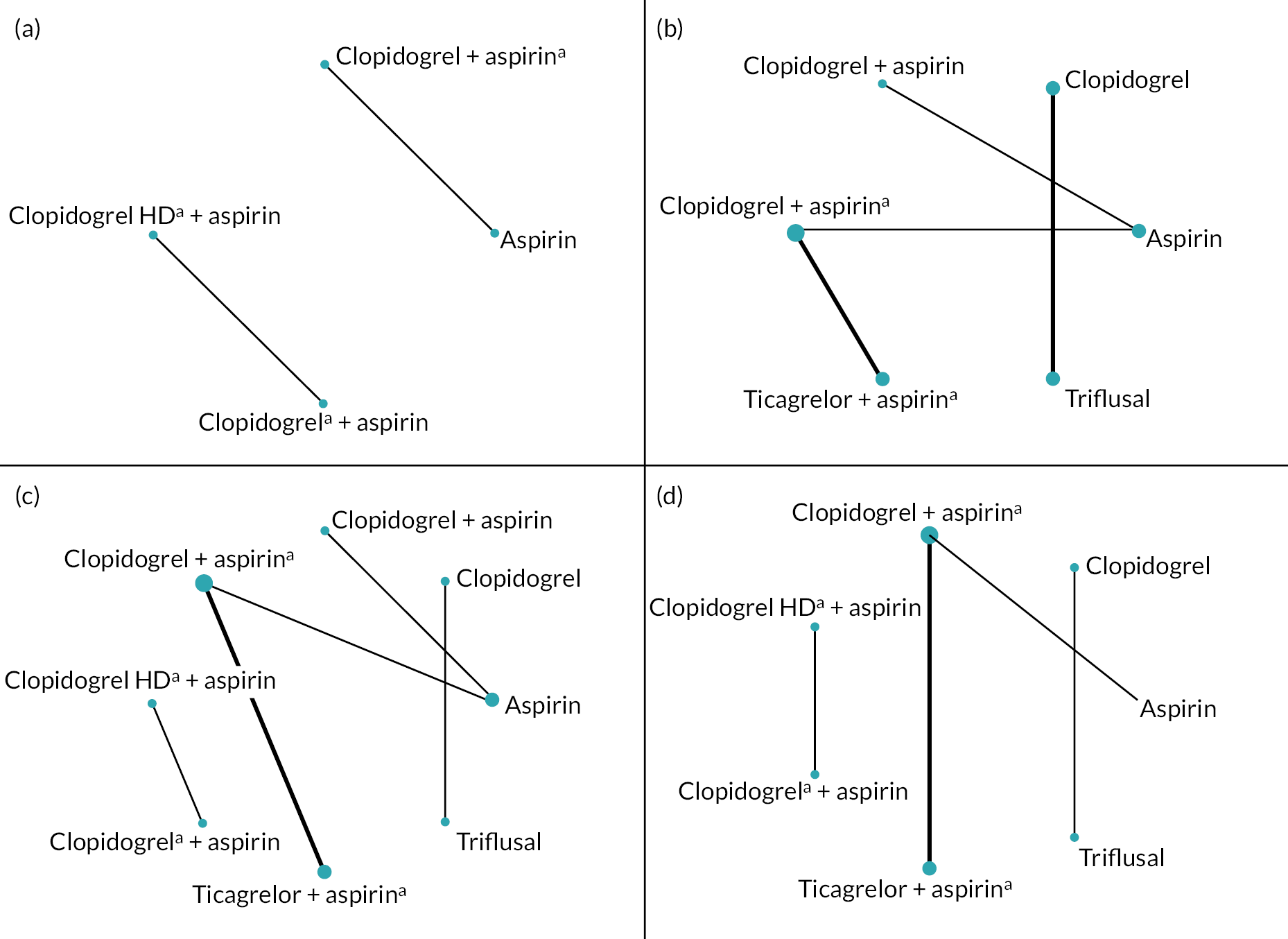
Six studies reported data on the incidence of a composite outcome of secondary vascular occlusive events (including stroke, TIA, MI and vascular death), five studies on incidence of secondary stroke, six studies on incidence of secondary ischaemic stroke, one on incidence of secondary TIA, two on secondary MI, two on secondary vascular death and two studies presented data on mortality of any cause. Figure 6 shows the network of intervention comparisons for each outcome. These are all seen to be disconnected, with no loops of evidence, so network meta-analysis was performed. As there were a maximum of two studies making any one comparison between treatments, only fixed-effect meta-analyses were performed.
FIGURE 6.
Network plots showing drug comparisons for main outcomes in Objective 2. (a) Composite outcome, (c) any stroke, (c) ischaemic stroke, (d) any bleeding. a, Drug given on a temporary basis (21–31 days).
Composite outcome of secondary vascular occlusive events
There was some evidence that treatment with alternatives to clopidogrel reduced the risk of secondary vascular events in those with LOF alleles (see Appendix 4, Figure 15). Ticagrelor was associated with a reduced incidence of secondary vascular events compared to clopidogrel [summary HR 0.76, 95% CI 0.65 to 0.90; two studies]. There was a suggestion that high-dose (HD) clopidogrel plus aspirin was associated with a reduced incidence of secondary vascular occlusive events compared to standard-dose clopidogrel plus aspirin, but CIs were wide (HR 0.18, 95% CI 0.02 to 1.52; one study). There was no difference in the incidence of vascular events amongst those taking clopidogrel alone compared to aspirin, although one other study suggested that the risk of secondary vascular events was higher for those taking aspirin alone compared to clopidogrel plus aspirin. However, this was a small study with very few events (all corresponding to ischaemic strokes), and CIs were wide (HR 3.03, 95% CI 0.83 to 11.11). There was no evidence of heterogeneity for any comparison. All summary estimates are from fixed-effects meta-analysis.
Stroke
The risk of stroke and ischaemic stroke was also reduced in those with LOF alleles taking ticagrelor compared to those taking clopidogrel (HR 0.76, 95% CI 0.63 to 0.92 for any stroke; HR 0.77, 95% CI 0.65 to 0.93 for ischaemic stroke; two studies) (see Figure 7 and Appendix 4, Figure 16). There was no evidence of a difference in stroke risk between clopidogrel and triflusal, or between clopidogrel alone and aspirin. As with the composite clinical outcome, the study that compared clopidogrel plus aspirin (vs. aspirin alone) for the duration of the study suggested that the risk of stroke was higher for aspirin alone compared to clopidogrel plus aspirin (HR 3.03, 95% CI 0.83 to 11.11). There was no evidence of heterogeneity for any comparison.
FIGURE 7.
Forest plot showing HRs (95% CI) for incidence of ischaemic stroke in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.

Other secondary efficacy outcomes
Other secondary outcomes evaluated included TIA, MI, vascular death and mortality. There were very few events for these outcomes and no statistical evidence of a difference between any of the antiplatelet strategies evaluated (see Appendix 4, Figure 17).
Adverse events
Seven studies reported data on incidence of bleeding events in those with LOF alleles treated with different antiplatelet therapies. Details on how bleeding outcomes were defined and categorised are summarised in (see Appendix 3, Table 47).
One study reported an increased risk of bleeding with ticagrelor compared to clopidogrel, while the other study that compared ticagrelor with clopidogrel found no difference in the risk of bleeding. There was no statistical evidence for differences between antiplatelet treatment strategies for any of the other comparisons or bleeding outcomes (see Appendix 4, Figures 18 and 19).
Objective 3
Twenty-five studies reported in 45 publications were included for Objective 3. 48,49,52–74 All studies were published in English. Five of the studies included for Objective 2 also provided data for Objective 3. 48,49,52–54
Table 8 provides an overview of the studies included for Objective 3. Full details on the studies are reported in Report Supplementary Material 1.
| Feature | Category | Number of studies |
|---|---|---|
| Population | Stroke | 14 |
| TIA | 1 | |
| Both | 10 | |
| Drug(s) | Clopidogrel | 15 |
| Clopidogrel + aspirin | 3 | |
| Clopidogrel + aspirin (for 14–30 days) | 5 | |
| Clopidogrel (any additional antiplatelets allowed) | 2 | |
| Clopidogrel loading dose | Yes | 4 |
| Optional | 2 | |
| No | 19 | |
| Clopidogrel dose | 75 mg | 18 |
| N/R | 7 | |
| Aspirin dose | 50–325 mg | 8 |
| Design | RCT subanalysis | 5 |
| Prospective cohort | 13 | |
| Retrospective cohort | 7 | |
| Country | South Korea | 1 |
| USA | 4 | |
| China | 13 | |
| International | 1 | |
| Czech Republic | 1 | |
| Scotland | 1 | |
| Japan | 2 | |
| Spain | 1 | |
| Turkey | 1 | |
| Funding | Non-industry | 16 |
| Drugs and tests provided by industry | 1 | |
| Industry – other | 1 | |
| Not stated | 7 | |
| CYP2C19 test | Seeplex CYP2C19 ACE genotyping system and Real-Q CYP2C19 genotyping kit | 1 |
| Drug Metabolism Enzyme TaqMan Allelic Discrimination assay | 6 | |
| Sequenom MassARRAY iPLEX platform | 4 | |
| Sequenom MassARRAY iPLEX platform and Drug Metabolism Enzyme TaqMan Allelic Discrimination assay | 2 | |
| PCR–restriction fragment length polymorphism (RFLP) | 1 | |
| Improved Multiple Ligase Detection Reaction (iMLDR) | 3 | |
| Cwbiotech | 1 | |
| Lightmix | 1 | |
| N/R | 4 | |
| Perkin Elmer Gene Amp PCR Systems 9600 | 1 | |
| LightScanner system | 1 | |
| LOF alleles | CYP2C19 *2 and *3 | 16 |
| CYP2C19 *2 | 5 | |
| N/R | 2 | |
| CYP2C19 *2, *3 and *8 | 1 | |
| CYP2C19 *2, *3, *4, *5, *6, *7 and *8 | 1 | |
| Follow-up time | 90 days | 4 |
| 180 days | 5 | |
| 365 days | 2 | |
| 1–2 years (366–730 days) | 4 | |
| 2–3 years (731–1095 days) | 1 | |
| 3–4 years (1096–1460 days) | 1 | |
| 4–5 years (1461–1825 days) | 2 | |
| N/R | 8 |
Twenty studies used a cohort design – 13 enrolled participants prospectively and 7 used a retrospective design. The five RCTs also included for Objective 2 compared standard clopidogrel therapy against an alternative and provided data for participants with and without LOF alleles. Data were extracted from these studies for the clopidogrel treatment arm only, effectively giving a cohort of patients treated with clopidogrel in whom results could be compared between those with and without LOF alleles. All studies administered clopidogrel to all patients, and outcomes were compared between those with and without LOF alleles. In 15 studies, patients received clopidogrel alone, 7 studies gave clopidogrel plus transitory aspirin (14–30 days), 3 studies administered both clopidogrel and aspirin for the duration of the study and the other two included patients taking clopidogrel, with or without other antiplatelets. In four studies, an initial loading dose of clopidogrel was given to all participants, in two studies some patients had been given an initial loading dose, and 21 studies did not give a loading dose.
Four studies had a follow-up time of 90 days, 5 followed up patients for 180 days, 2 for 365 days, four studies from 1 to 2 years, one study from 2 to 3 years, one study from 3 to 4 years and two from 4 to 5 years. Eight studies did not report follow-up time.
Most studies enrolled patients who had experienced a stroke as their primary event (14 studies), 1 study only enrolled patients who had experienced a TIA and 10 studies enrolled patients who had experienced a stroke or TIA. Most studies were conducted in Asia (13 in China, 2 in Japan and 1 in Korea), 4 studies were conducted in the USA with single studies from other countries. One study had drugs and tests provided by industry, one was sponsored by a commercial company, other studies either did not report on funding source or were funded by non-commercial organisations. A variety of different laboratory tests were used to determine CYP2C19 status – none of the studies used POCTs. The majority of studies tested for both *2 and *3 LOF alleles, five studies only tested for *2 and two did not report on which LOF alleles were tested for. Two studies tested for additional alleles as well as *2 and *3 – *8 in one study and *5, *6, *7 and *8 in the other.
Risk of bias
Nineteen studies were judged to be at low concern regarding RoB; seven studies had high concerns. Table 9 provides a summary of the RoB assessment for each study; full details are provided in Report Supplementary Material 1. Studies judged at high RoB were due to potential loss to follow-up and the potential for this to be related to the outcome (three studies), likelihood of ethnically diverse population that was not described in detail or considered in the synthesis (two studies) and selection of participants dependent on clopidogrel prescription redemption (retrospective study) which might be associated with the outcome (one study). All outcomes evaluated for each study were judged to have the same RoB.
| Study details | Domain | Rationale | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | Overall | ||
| Chen et al. (2019)53 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Diaz-Villamarin et al. (2018)55 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Fu et al. (2020)56 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Fukuma et al. (2022)57 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Han et al. (2017)48 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Hoh et al. (2016)58 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Lin et al. (2021)59 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Liu et al. (2020)60 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Lv et al. (2022)61 | ☺ | ☺ | ☺ | ☺ | ☹ | ☺ | ☺ | ☹ | High percentage of loss to follow-up, likely related to the outcome. |
| McDonough et al. (2015)62 | ☺ | ☺ | ☺ | ☺ | ☹ | ☺ | ☺ | ☹ | No data on loss to follow-up, potential missing data likely related to outcome. |
| Meschia et al. (2020)49 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Ni et al. (2017)63 | ☺ | ☺ | ☺ | ☺ | ☹ | ☺ | ☺ | ☹ | No data on loss to follow-up. Potential missing data likely to be related with the outcome. |
| Patel et al. (2021)64 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Qiu et al. (2015)65 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Sen et al. (2014)66 | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☹ | Population likely not ethnically homogeneous, no info on ethnicity, not adjusted. |
| Spokoyny et al. (2014)67 | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☹ | Ethnicity is a common cause of CYP219 variations and recurrent events – mixed population, results probably not adjusted by ethnicity. |
| Sun et al. (2015)68 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Tanaka et al. (2019)69 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Tomek et al. (2018)70 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Tornio et al. (2018)75 | ☺ | ☺ | ☹ | ☺ | ☺ | ☺ | ☺ | ☹ | Retrospective study – inclusion of participants dependent on redemption of clopidogrel prescription which is associated with the outcome. |
| Wang et al. (2016a)52 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Wang et al. (2016b)72 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Yi et al. (2018)54 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Yi et al. (2017)73 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Zhang et al. (2017)74 | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
Results
Secondary occlusive events
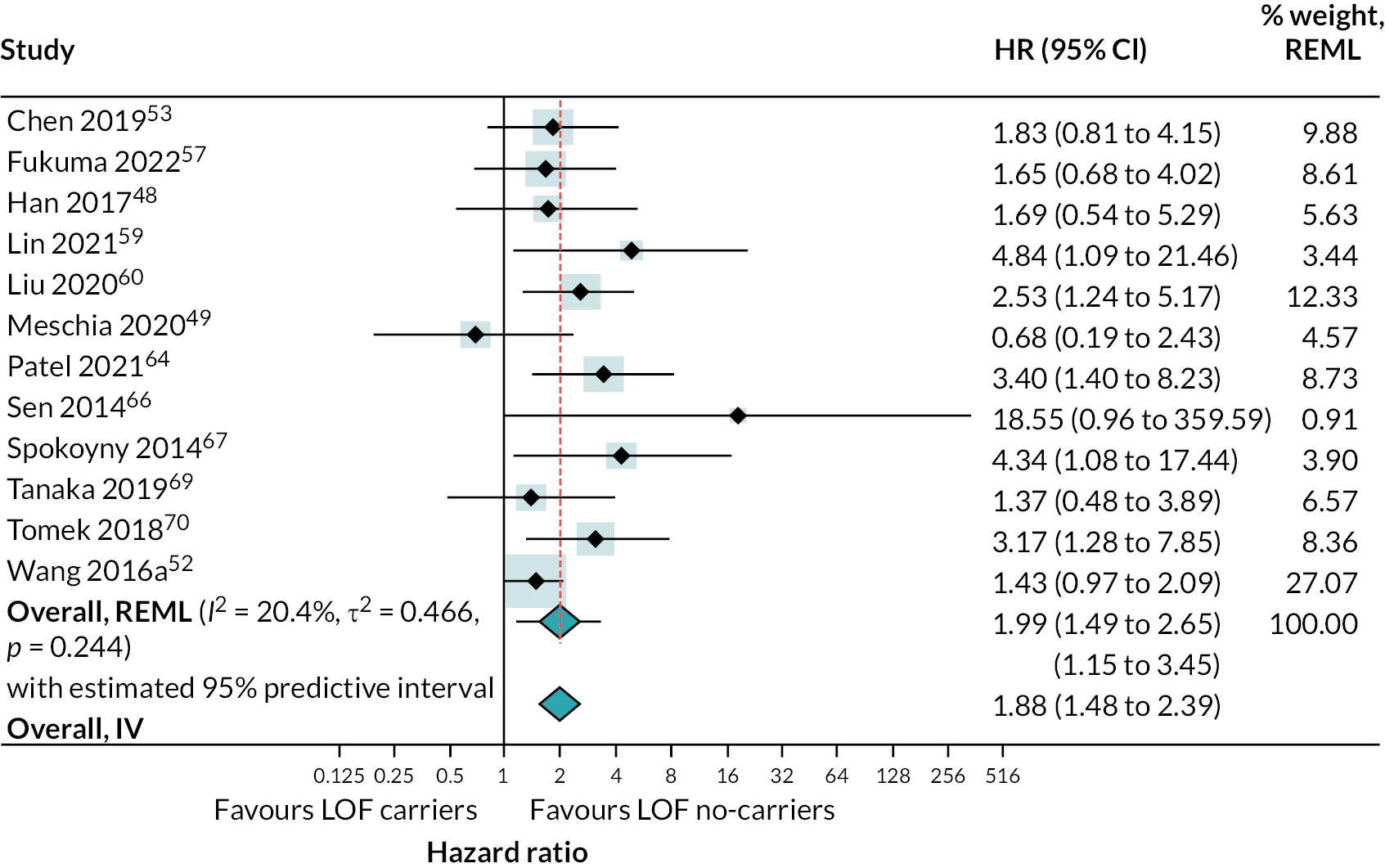
There was strong evidence that people with LOF alleles treated with clopidogrel (or clopidogrel plus aspirin) have a greater incidence of secondary vascular events (HR 1.72, 95% CI 1.43 to 2.08; 18 studies) (see Appendix 4, Figure 20), stroke (HR 1.46, 95% CI 1.09 to 1.95; 5 studies) (see Appendix 4, Figure 21) and ischaemic stroke (HR 1.99, 95% CI 1.49 to 2.65; 12 studies) (Figure 8), than those without LOF alleles (estimates from random-effects meta-analysis). There was some evidence of heterogeneity for the composite outcome of secondary vascular events (I2 = 33%; τ2 = 0.027); there was little or no evidence of heterogeneity for other outcomes. Fixed-effect meta-analysis estimates were very similar to pooled results from random-effects analyses.
FIGURE 8.
Forest plot showing HRs (95% CI) for incidence of ischaemic stroke in carriers of LOF alleles compared with non-carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin).
Secondary efficacy outcomes
There was little evidence to suggest any association between LOF alleles and secondary outcomes of mortality and TIA (see Appendix 4, Figure 22). However, these were evaluated in very few studies and there were very few events. There was evidence that the risk of vascular death is increased in patients with LOF alleles treated with clopidogrel compared to those without LOF alleles (HR 5.07, 95% CI 1.26 to 20.39).
Investigation of heterogeneity
Within studies that evaluated multiple vascular occlusive event outcomes, estimates of HR were very similar for composite outcome, stroke and ischaemic stroke (ischaemic stroke accounted for most of the secondary vascular outcomes reported in all studies). This is shown in (see Appendix 4, Figure 23). As described in the Methods, a post hoc decision was therefore made to combine data across different types of vascular event when exploring heterogeneity.
Results of univariable metaregressions are show in (see Appendix 4, Table 49). Forest plots stratified for each of these variables are provided (see Appendix 4, Figure 31). There was evidence of a reduced effect of LOF alleles in patients given a loading dose of clopidogrel relative to those who were not [ratio of hazard ratios (RHR) 0.64, 95% CI 0.43 to 0.96], in patients taking clopidogrel plus long-term aspirin relative to those taking only clopidogrel or clopidogrel plus short-term aspirin (RHR 0.47, 95% CI 0.22 to 0.96), and in studies that included patients with stroke and TIA as primary events compared with only patients with stroke (RHR 0.62, 95% CI 0.44 to 0.86). The stratified analysis based on clopidogrel regimen suggested that there was no evidence of a difference in the risk of secondary vascular events between those with and without LOF alleles when taking clopidogrel plus aspirin (HR 0.74; 95% CI 0.23 to 2.38) stratified analyses results shown in (see Appendix 4, Figures 24–31). There was no evidence of a difference between studies which included patients with TIA as primary event and those including patients with stroke, but only one study investigated TIA patients exclusively.
There was some suggestion from subgroup analyses that effects of LOF alleles may vary by ethnicity, with a possibly reduced effect in studies in mixed and Hispanic populations compared with white. However, there was considerable uncertainty in these stratified estimates, resulting in no statistical evidence for differences between LOF effect by ethnicity in the metaregression.
There was no evidence for a difference in LOF alleles effect on secondary vascular occlusive outcomes based on RoB, proton pump inhibitor (PPI) use, study location or duration of follow-up.
Investigation of small-study effects
The funnel plot showing HRs for incidence of secondary vascular occlusive outcomes in carriers of LOF alleles compared with non-carriers of LOF alleles appears symmetrical (see Appendix 4, Figure 33). This suggests that there is no evidence of small-study effects.
Adverse events
Eight studies reported data on incidence of bleeding events in those with LOF alleles treated with different antiplatelet therapies. Details on outcome definitions for each study and categories assigned for this analysis are presented in (see Appendix 3, Table 48). There was no evidence of a difference in the risk of bleeding among those with and without LOF alleles for each category of bleeding assessed: any bleeding (HR 0.98, 95% CI 0.68 to 1.40; five studies), severe bleeding (HR 0.97, 95% CI 0.42 to 2.20; five studies), haemorrhagic stroke (HR 1.34, 95% CI 0.29 to 6.20; two studies) or mild bleeding (HR 0.68, 95% CI 0.30 to 1.54; two studies). This is shown in (see Appendix 4, Figure 32). There was no evidence of heterogeneity for any of these outcomes (I2 = 0). For this reason, subgroup analyses and metaregression was not performed. Fixed-effect meta-analysis estimates were identical to pooled results from random-effects analyses.
Objective 4
Nine studies, reported in 12 publications, reported data on test accuracy of the POCTs in scope. Two studies reported separate accuracy data for a pre-trial and the main trial – these are treated as separate studies giving a total of 11 studies. 76,77 Three studies were only available as clinical trial registrations, all others were published as full reports. All studies available only as clinical trial registrations were conducted by Spartan (Genomadix), who provided additional information when requested for two of the studies. 78,79 All studies were reported in English.
All studies evaluated Spartan versions of the test. Two evaluated Spartan Cube,78,79 eight evaluated Spartan RX76,77,80–83 and one evaluated Spartan FRX. 84 These tests are considered broadly equivalent to the Genomadix Cube and so were evaluated as a single group referred to from here as ‘Genomadix (Spartan) CYP2C19 tests’, unless referring to specific tests. There were no studies on the accuracy of Genedrive.
Table 10 provides an overview of the studies included for Objective 4. Full details of the studies are reported in Report Supplementary Material 1. Five studies were funded by the test manufacturer. One study was funded by other industry organisations and one by both industry and non-industry.
| Feature | Category | Number of studies |
|---|---|---|
| POCT | Spartan (Genomadix) Cube | 2 |
| Spartan (Genomadix) RX | 8 | |
| Spartan (Genomadix) FRX | 1 | |
| Population | N/R | 3 |
| Healthy volunteers | 2 | |
| PCI | 6 | |
| Country | Canada | 6 |
| South Korea | 1 | |
| Malta | 1 | |
| Czech Republic | 1 | |
| Multicountry International (US/Canada/South Korea/Mexico) | 2 | |
| Funding | Industry – test manufacturer | 5 |
| Industry – other | 1 | |
| Non-industry | 4 | |
| Mixed (industry and non-industry) | 1 | |
| Alleles targeted | *2, *3 and *17 | 7 |
| *2 and *17 | 1 | |
| *2 only | 3 | |
| Reference standard (laboratory test) | Bidirectional sequencing | 3 |
| Direct DNA sequencing | 2 | |
| Sanger sequencing | 1 | |
| Taqman | 3 | |
| Taqman plus Sanger sequencing where POCT and Taqman discordant | 1 |
Six studies recruited patients undergoing percutaneous coronary intervention (PCI). The two pre-trials included healthy volunteers as they were pre-trial validations of the test. Three studies did not report details on the population studied – all were only available as clinical trial registrations. None of the studies were conducted in our population of interest – stroke patients.
The number of participants ranged from 879 to 2587. 76
Two studies took place in Europe, six studies in Canada, one in South Korea and two studies (reported in the same publication) were multinational conducted in USA/Canada/South Korea/Mexico.
Studies targeted different combinations of the three alleles that can be detected using Genomadix (Spartan) CYP2C19 tests (*2, *3, *17). Seven studies targeted all three LOF alleles, one targeted *2 and *17 and the remainder targeted only *2. We dichotomised results into presence of LOF alleles or no LOF alleles so that those with at least one *2 or *3 LOF allele were considered to have LOF alleles; we categorised *17 as normal function, as described in the methods. The reference standard (standard laboratory test) was bidirectional sequencing in three studies, direct DNA sequencing in two studies and Sanger sequencing in one study (all these methods can detect the presence of any LOF allele). The remaining four studies used Taqman, which can be set up with different probes to detect different LOF alleles. One of the studies used Sanger sequencing as an additional reference standard where there were discrepancies between the Genomadix Cube and Taqman results. In all studies, even those that used a reference standard that could detect any LOF alleles, the laboratory tests only targeted the same alleles as were targeted by the Genomadix Cube. Estimates of accuracy from these studies therefore show the accuracy in detecting only those variants that Genomadix (Spartan) CYP2C19 tests can detect (and in four studies only the *2 LOF allele), rather than the accuracy for the detection of any variant associated with LOF.
Risk of bias
All studies were considered at low RoB. An overview of RoB in the studies is provided in Table 11, and full details are provided in Report Supplementary Material 1. Although a variety of different populations were enrolled, and enrolment was not always consecutive, we considered that how patients were enrolled was unlikely to affect estimates of test performance. Information on whether the person interpreting the Genomadix (Spartan) CYP2C19 test was blinded to the laboratory test was N/R, although some studies did suggest that this was conducted and interpreted before the laboratory test. However, as the Genomadix (Spartan) CYP2C19 tests are objective in interpretation, blinding was considered unlikely to have influenced test interpretation. All studies used a laboratory-based reference standard – this was considered appropriate. Most of these are also objective in their interpretation and so we considered it unlikely that knowledge of the Genomadix (Spartan) CYP2C19 test results could have biased interpretation of the reference standard. There were very few patients who did not receive both index test and reference standard and so there were no concerns regarding patient flow.
| Study details | Patient selection | Index test | Reference standard | Flow and timing | Overall | Rationale for judgement |
|---|---|---|---|---|---|---|
| Baudhuin et al. (2022)76 – pre-trial | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Baudhuin et al. (2022)76 – main trial | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Choi et al. (2016)80 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| NCT0171853584 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| NCT0447357379 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| NCT0447358678 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Petrek et al. 201681,85 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Roberts et al. (2012)77 – pre-trial | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Roberts et al. (2012)77 – main trial | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| So et al. (2016)82 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
| Wirth et al. (2016)83 | ☺ | ☺ | ☺ | ☺ | ☺ | No concerns |
Results
Estimates of the accuracy of Genomadix (Spartan) CYP2C19 tests were very high – 8 of the 11 studies reported 100% sensitivity and specificity. Summary sensitivity was 100% (95% CI 94% to 100%) and summary specificity was also 100% (95% CI 99% to 100%). It was possible to extract 2 × 2 data for 9 of the 11 studies. We contacted the authors of the other two studies but did not receive a response. 76,85 For one of these studies, we were able to estimate 2 × 2 data based on data reported in the paper. 76 Data were reported on sensitivity, specificity and the total number of people tested using Spartan (Genomadix) RX (255/2641 did not have a Genomadix Cube result). Data were N/R on the number tested regarding who did and did not have LOF alleles based on the reference standard. However, information was available on the numbers with and without LOF in the total sample, we assumed that the proportion with LOF alleles would be similar in the tested subset and overall cohort and used this to estimate numbers with and without LOF in the tested sample and then applied sensitivity and specificity to the numbers to estimate 2 × 2 data. An overview of discordant results is provided in Table 12.
| Study | Genomadix test | Proportion discordant | Overview of discordant results | Impact on accuracy |
|---|---|---|---|---|
| Badhuin et al. (2022)76 – pre-trial | Spartan (Genomadix) RX | 2/373 (0.5%) | 2 discordant initially due to pre-analytical sample mix-up at testing centre. Samples recollected and retested, then concordant. | None |
| Badhuin et al. (2022)76 – main trial | Spartan (Genomadix) RX | 21/2384 (0.9%) | 21 discordant:
|
9 FN and 11 FP |
| Choi et al. (2016)80 | Spartan (Genomadix) RX | 2/119 (1.7%) | 2 discordant: *3/*17 on Spartan and *1/*3 on SNP *1/*17 on Spartan and *1/*1 on SNP |
None |
| NCT0171853584 | Spartan (Genomadix) FRX | 0/325 (0%) | None | None |
| NCT0447358678 | Spartan (Genomadix) Cube | 2/411 (0.5%) | 2 test runs discordant on 1-hour samples – 2 samples mixed up due to a sample swap of two adjacent samples: *1/*2 was called *2/*2 *2/*2 was called *1/*2 |
None |
| NCT0447357379 | Spartan (Genomadix) Cube | 0/960 | None | None |
| Petrek et al. (2016)81,85 | Spartan (Genomadix) RX | 0/53 (0%) | None | None |
| Roberts et al. (2012)77 pre-trial | Spartan (Genomadix) RX | 0/37 (0%) | N/A | None |
| Roberts et al. (2012)77 main trial | Spartan (Genomadix) RX | 1/187 (0.5%) | One incorrectly classified as *2 carrier on Spartan | 1 FP |
| So et al. (2016)82 | Spartan (Genomadix) RX | 2/102 (2%) | No details | 2 FP |
| Wirth et al. (2016)83 | Spartan (Genomadix) RX | 1/35 (2.9%) | One incorrectly classified as *2/*2 on Spartan vs. one *2 on Taqman and on GenID | None |
Seven studies reported discordant results between Genomadix (Spartan) CYP2C19 tests and the laboratory reference standard, but these only impacted estimates of accuracy in two studies as in other studies they did not affect the classification of the individual as a poor or normal metaboliser. An overview of discordant results is provided in Table 12.
Objective 5
Technical performance of point-of-care tests
Seventeen studies, reported in 24 publications, reported on the technical performance of POCTs. 76–83,86–94 Three studies reported data for both a pre-study and main study, these are included as separate studies giving a total of 20 included studies. All but one84 of the studies included for Objective 4 also provided data on test performance and so were also included for Objective 5. Two studies were available as trial registry entries only (with additional information provided by Genomadix),78,79 two were conference abstracts (with a full conference poster shared for one of these)91,94 and all others were reported as full journal articles. All studies were reported in English. Table 13 provides an overview of the studies included for Objective 5. Full details of the studies are reported in the baseline data tables and results tables presented in Report Supplementary Material 1.
| Feature | Category | Number of studies |
|---|---|---|
| Tests | Spartan (Genomadix) Cube | 2 |
| Spartan (Genomadix) RX | 17 | |
| Genedrive | 1 | |
| Population | PCI | 9 |
| N/R | 5 | |
| Healthy people | 2 | |
| Stroke | 1 | |
| STEMI | 1 | |
| Stable CAD | 1 | |
| Diagnostic coronary angiography | 1 | |
| Outcomes | Test failure rate | 10 |
| Number of people with variant forms of CYP2C19 (%) | 16 | |
| Time to results | 13 | |
| Ease of use of test | 8 | |
| Cost of testing | 2 | |
| Country | USA | 6 |
| Saudi Arabia | 1 | |
| UK | 1 | |
| Poland | 1 | |
| Canada | 5 | |
| South Korea | 1 | |
| Malta | 1 | |
| Czech Republic | 1 | |
| Multicountry in Europe (Netherlands/Italy/Belgium) | 1 | |
| Multicountry International (USA/Canada/South Korea/Mexico) | 2 | |
| Funding | Industry – test manufacturer | 5 |
| Non-industry but kits provided by manufacturer | 3 | |
| Industry – other | 1 | |
| Non-industry | 10 | |
| Mixed (industry and non-industry) | 1 | |
| N/R/unclear | 2 |
One study evaluated Genedrive,94 detailing the development of an earlier version of the test. All other studies evaluated Spartan versions of the Genomadix Cube test. Two evaluated Spartan Cube,78,79 others evaluated Spartan RX.
Five of the studies reporting on technical performance were funded by the test manufacturer. 77–79,84 One study was funded by other industry organisations and one by both industry and non-industry. Study populations and locations varied between studies. Conditions studied included stroke, coronary artery disease (CAD), healthy volunteers (in test pre-validation studies) and patients undergoing PCI. Five studies took place in Europe, 11 studies in North America,77–79,82,88–91,93 1 in South Korea, 1 in Saudi Arabia and 2 studies (reported in the same publication) were international in USA/Canada/South Korea/Mexico.
Results
Test failure rate
Ten studies, all of which evaluated Genomadix (Spartan) CYP2C19 tests, reported test failure rate (Table 14). 76–79,81,83,87,88,92,93 There was substantial variation in test failure rate across studies, from a minimum of 0.4% of tests (1/267) to a maximum of 18.9% (10/53 patients) for the initial run. In some studies, samples that failed initially were retested and a subset produced results on retesting. Terminology to describe test failures also varied across studies. Though often described as ‘inconclusive results’, studies also highlight device errors, failure during the amplification process and not identifying a genotype. Of studies that reported what they did post-test failure, most said they repeated the genotype test and highlight the need to consider this when assessing the cost of genotyping.
| Study details | Number of patients with unavailable test result | Details of missing results | Action taken post-test failure |
|---|---|---|---|
| Badhuin et al. (2022)76,95 | 172/2642 (7%) | Main trial: In 54/2642 (2%) had no Spartan result available (no definition of what this means); 118 (4%) had inconclusive results | N/R |
| Bergmeijer et al. (2014)87,96 | 39 (8%) | Inconclusive results | Sample shipped to central lab for Taqman genotyping (30 patients), repeated Spartan testing (2 patients), no further genotyping (7 patients). |
| Cavallari et al. (2018)88 | 129/931 (14%) | 56 inconclusive results, 73 device errors | One additional sample collected (113 patients), two additional samples collected (10 patients), refused sample recollection (6 patients). Nine out of 123 patients with additional sample collection had multiple inconclusive results. |
| NCT0 447358678 | 16/621 tests (2.6%) | First pass call rate: 605/621 (97.4%) | Tested samples again. |
| 2/621 tests (0.3%) | Final call pass rate: 619/621 (99.7%) | Final call rate including re-reruns of first pass failures; no information on further action. | |
| NCT0 447357379 | 9/960 tests (0.9%) | First pass call rate was 951/960 (99.1%) | Tested samples again. |
| 3/960 tests (0.3%) | Final call pass rate was 957/960 (99.7%) | Final call rate including re-reruns of first pass failures; no information on further action. | |
| Petrek et al. 201681,85 | 10/53 (18.9%) | Failure during amplification process (n = 4), inconclusive result (n = 3), only two of three alleles tested for gave results (n = 3) | N/R |
| 7/53 (13.2%) | Failure during amplification process (n = 4), inconclusive result (n = 3) – results not included where only 2/3 alleles gave a result | N/R | |
| Roberts et al. (2012)77 | 1/267 tests (0.4%) | Pre-trial: test did not identify a genotype. This is 1 test, not necessarily one patient (multiple tests done on each patient) | N/R |
| Tomaniak et al. (2017)92,97,98 | 4/34 (11.8%) | Inconclusive results | Genotyping repeated. No further information given. |
| Wirth et al. (2016)83,99 | 5/35 (14.3%) | 4 tests resulted in error (11.4% – no further details); 1 test inconclusive | The 4 tests resulting in error were repeated with a new test as per manufacturer’s instructions. The inconclusive test was not repeated as the patient had been discharged home. No further information given. |
| Zhou et al. (2017)93,100 | 25/342 (7.3%) | Main trial: 14 inconclusive results (4%), 10 failed controls (3%), 1 instrument failure (0.3%) (no further information given) | 12 patients resulted after retesting; one patient refused to recollect sample and 1 had 2 consecutive inconclusive results. No further information given. |
Number of people with variant forms of CYP2C19 (%)
Thirteen studies reported the number of people with variant forms of CYP2C19. 76,80,82,83,86,88,90–93 We defined variant forms of CYP2C19 as people with one LOF allele (IM, e.g. *2/*1) or two LOF alleles (PM, e.g. *2/*2) (see Appendix 5, Table 51) provides an overview of the number of participants with each allele combination in the studies that reported this information.
Overall, IMs were more commonly found than PMs. The allele combination *2/*1 was most frequently reported, and the *3 allele was reported less frequently than the *2 allele. The proportion of participants with variant forms varied from 15% to 64%. We would expect to see an association with ethnicity and CYP2C19 variants; however, most studies did not provide information on ethnicity and so it was not possible to investigate this association. The UK population in the 2021 census was 81% white, 9.6% Asian, 4.2 % black, 3% mixed ethnic groups and other (2.2%). 101 The five studies that reported on ethnicity had majority white ethnicity (68–100%); these reported that the proportion of people that were poor or IMs ranged from 29% to 38%. The study with the highest proportion of people with variant forms (64%) did not report on ethnicity but was conducted in South Korea and so is likely to have included a mainly Asian population.
Time to results
Thirteen studies provided information on time to results (see Appendix 5, Table 50). Ten of these studies reported data on time to results based on experience from their study: all evaluated Spartan (Genomadix) RX. 77,81–83,86–88,90–92 Seven studies reported that the turnaround time (TAT) from buccal swab to result took approximately 1 hour. Two studies reported that this took 90 minutes and one reported that it took 90–120 minutes.
Three studies reported information about time to results, but this was reported as a description of a feature of the test, rather than being a clear finding from the study itself. 80,93,94 Two of the studies evaluated the Genomadix Cube and stated that this takes one hour from sample to result. In addition, information reported by Genomadix, the test manufacturer, to the External Assessment Group (EAG), stated that the time to result for the test was 64 minutes. The study that evaluated Genedrive reported that it is ‘rapid’, taking around 40 minutes (no further information was reported). 94
Ease of use of test
Eight studies reported data on the ease of use of the test (Table 15). Five studies, all of which evaluated Spartan (Genomadix) RX, reported data on the ease of use of the POCT based on experience from their study. 76,77,81,83,88 Overall, these studies suggested that the process of using the Spartan POCT was simple, user-friendly and that it can be conducted by staff who have received minimal training. Limitations highlighted include storage conditions of the POCT,83 and that only one sample can be genotyped at a time. 88
| Study details | Ease of use of testa |
|---|---|
| Badhuin et al. (2022)76,95 | Non-laboratory-trained personnel can successfully perform rapid genotyping in a POC setting |
| Bergmeijer et al. (2014)87,96 | Description of feature of the test: Buccal swab more patient friendly than venipuncture for blood sample, but test is limited to testing *2, *3, *17 for one patient at a time per genotyping device |
| Cavallari et al. (2018)88 | Could not be used as POCT due to absence of licensed molecular medical technologist, so must be sent to central laboratory (the case for all of USA), and only a single sample genotyped at a time limiting number of patients that can be offered genotyping |
| Davis et al. (2020)89 | Description of features of the test: Barriers to implementation: time constraints, personnel requirements and coordination, storage and sample stability, samples unable to be collected by bedside nurses, patients unable to provide samples, sample recollection due to interference or improper techniques |
| Petrek et al. 201681,85 | Simple and non-invasive |
| Roberts et al. (2012)77 | Main trial: Nurses with no previous laboratory training implemented test after a 30-minute training session |
| Wirth et al. (2016)83,99 | Simple procedure, portable, convenient, no laborious preparation, minimal training required to conduct test. User-friendly interpretation with no training required. Storage conditions limit ease of use |
| McDermott et al. (2020)94 – Genedrive | Description of features of the test: Portable, rapid (~40 minutes), no cold chain, simple read out for non-specialist users |
Three studies reported further information on ease of use of POCT; however, these were reported as descriptions of features of the test rather than direct findings from the study. 87,89,94 Regarding Genomadix Cube, these corroborate the findings outlined previously but add further limitations that the test is restricted to *2/*3/*1787 and that there can be issues with sample collection, including sample recollection due to interference. 89 The study that evaluated Genedrive, noted that the test is simple, portable, rapid and does not require analytes to be frozen. 94
Cost of testing
Two studies provide information about POCT costs – one evaluated Spartan (Genomadix) RX,83 and the other evaluated Genedrive. 94 Additional information on the cost of Genomadix Cube was provided by the manufacturer (Table 16). Wirth et al. 83 estimate the cost per patient of Genomadix Cube POCT at 225 euros, compared to 13 euros for the Taqman laboratory assay and 23 euros for the GenID laboratory assay. The authors do not state how they calculated this costing. The manufacturers of the two tests shared information on costs.
| Study details | Test name | Cost of testing |
|---|---|---|
| Genomadix (test manufacturer) response to request for information | Genomadix Cube (Spartan) | Description of feature of the test: (a) Platform cost: £3500 per testing platform, (b) testing assay cost: £175 per test kit, (c) external control kits: £50 GBP per external control kit |
| Genedrive (test manufacturer) response to request for information | Genedrive System | Description of feature of the test: (a) Platform cost: 4995 GBP per testing platform, (b) testing assay cost: £100 per test kit, (c) external control kits: £100 per external control kit |
| Wirth et al. (2016)83,99 | Genomadix Cube (Spartan) | Estimated cost per patient test: 225 euros (Taqman estimated at 13 euros and GenID at 23 euros). No indication of how this was calculated. |
Survey
The survey was sent to 10 laboratories (labs) for completion – the 7 genomic laboratory hubs in England, All Wales Medical Genomics Services, Northern Ireland Regional Genetics Service and the Scottish Strategic Network for Genomic Medicine. Responses were received from 8 labs – 5 regional genomic laboratory hubs (Central and South; East; North West; South East; and North East and Yorkshire) and from the Scottish (NHS North Tayside), Welsh and Irish services. Full survey results are reported in Appendix 6.
Testing platform
Table 17 provides an overview of the test platforms that each lab reported currently having in place that would be capable of performing CYP2C19 genotyping, and the platforms identified as preferred platforms by each lab. Seven of the eight labs reported having Sanger sequencing, six also had next-generation gene sequencing; one did not report having any sequencing technology. All had at least one form of targeted CYP2C19 gene variant detection, most commonly polymerase chain rection (PCR)-based single nucleotide polymorphism (SNP) genotyping assays using fluorescent reporter systems, such as TaqMan (ThermoFisher) – this was also one of the most commonly reported reference standard in the DTA studies included for Objective 4. Preferred technologies included next generation sequencing (NGS) (two labs), MassARRAY (three labs), LAMP (three labs), PCR-based SNP genotyping assays using fluorescent reporter systems, such as TaqMan (ThermoFisher) and QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System (one lab). Note that two labs highlighted two different technologies as their preferred technology – one selected both MassARRAY and LAMP, the other selected NGS and LAMP. When asked about whether there are other platforms available that they may consider for CYP2C19 testing, one lab reported that they were currently looking at NGS Genexus due to speed and capacity. Another stated that they would use Sanger sequencing as a back-up test for when LAMP produced indiscriminate results.
| Technology | Technology available | Preferred technology | Reasons for preference |
|---|---|---|---|
| Sequencing technology | |||
| Sanger CYP2C19 sequencing | 7 | 0 | N/A |
| Next-generation CYP2C19 gene sequencing | 6 | 2 |
|
| Targeted CYP2C19 gene variant detection | |||
| PCR-based SNP genotyping assays using fluorescent reporter systems, such as TaqMan (ThermoFisher) | 6 | 1 |
|
| Other PCR-based genotyping panels that use proprietary detection methods, such as the xTAG CYP2C19 Kit v3 (Luminex) | 1 | 0 | N/A |
| Variant detection using mass spectrometry, such as MassARRAY (Agena Bioscience) | 4 | 3 |
|
| LAMP, such as the LAMP human CYP2C19 mutation KIT (LaCAR MDx Technologies) | 4 | 3 |
|
| Other: QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System | 1 | 1 |
|
Alleles targeted
Laboratories reported that the following alleles targeted by their preferred CYP2C19 test:
-
*1–3 laboratories
-
*2–5 laboratories
-
*3–5 laboratories
-
*7–4 laboratories
Three laboratories stated that they were unsure which alleles would be targeted and one stated that a NGS assay would be able to detect all sequence variants. Note that there was an error in this question on the survey so that we asked about *7 rather than *17.
Four labs stated that the test would be affected by testing for all LOF alleles compared to only testing for *2 or *3 alleles, although two highlighted that this would depend on the technology. Potential impacts included increased cost and increased TAT.
Resources required
Two labs were not able to provide any information on resources required and one lab was only able to provide an estimate of the cost of the test.
Staff time
Three labs provided an estimate of staff time to run the selected test. One, that had selected LAMP as the preferred test, estimated 1–2 days in total: 1–2 hours set-up, 2-hour analysis and 2 hours checking and reporting. The second, that selected QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System, estimated 0.5 working time equivalent (WTE) for performing test 0.5 WTE for DNA extraction 0.2 WTE for admin. The third lab selected PCR-based SNP genotyping assays using fluorescent reporter systems as their performed test and are currently performing this test estimate staff time at 22 minutes/sample.
Staff grade
Estimates of staffing grade varied in the five labs that reported on this:
-
Band 5 set-up, band 6 analysis and reporting, band 7 checking and authorisation of reports
-
Band 3 up to band 8a
-
Band 3, band 5, band 7
-
Band 3, band 4, band 5
-
Band 2, 3, 4 for laboratory work; band 7 for authorising reports
Cost
There was also variation in estimates of cost for test. Estimated costs are summarised in Table 18, which also shows the preferred technology that the estimate relates to.
| Preferred platform | Cost per test | Maintenance costs |
|---|---|---|
| MassARRAY (Agena Bioscience) | ~ £15 per test | £15k maintenance plus EQA |
| LAMP | £40 per test (reagent cost only) | N/R |
| MassARRAY or LAMP | ~£100 | N/R |
| Next-generation gene sequencing or LAMP | £100–£250 | N/R |
| QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System | ~£200 per sample. Additional costs in data analysis either by scientists or using automated calling and reporting system = £5–10 per sample | £5000 pa for qPCR machine BUT for 10,000 samples pa we would need to increase our existing DNA extraction capacity, which may mean another automated DNA extraction system = £150k capital investment |
| PCR-based SNP genotyping assays using fluorescent reporter systems, for example, TaqMan | £25.09 inc VAT – include reagents/consumables, staff time and overheads | N/R |
Additional administrative resources
Three labs highlighted additional administrative resources that would be required and one stated that they would be required but did not provide further details. One lab stated that these would not be required. Additional administrative resources required were:
-
One band 4 admin
-
Laboratory information management system (LIMS) upload to electronic care record (ECR) where link does not exist – admin support to send results and upload to ECR.
-
Preferable electronic test ordering but may require admin support for dealing with enquiries.
Ease of use
Six labs reported that their preferred test could be performed by existing staff members who have received standard training, one lab reported that the test was fully automated (LAMP) and the other that additional training would be required – this lab had selected QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System has their preferred test, which would be a new test for their lab and was the reason training would be required.
Test validity
Most labs reported that it was difficult to estimate the proportion of samples that would not return a valid result, most of those that responded stated that they expected this to be < 1%. One lab did report ~90% for this question; it appears likely that they have misinterpreted the question. One lab, which is currently performing PCR-based SNP genotyping assays using fluorescent reporter systems, reported that 5% of samples would not return a valid result.
Testing capacity and turnaround time
In the introduction to the survey, we estimated that the NHS would need to perform approximately:
-
150,000 CYP2C19 tests in the first year (assuming annual stroke incidence of 100,000 and TIA incidence of 50,000)
-
100,000 CYP2C19 tests annually after this (57,000 first strokes, 46,000 first TIAs assuming ~10–15% of those with first stroke would previously have had a TIA and already been tested).
Two laboratories reported that their current testing capacity was zero, two were unable to answer this question and another said that they would not be able to process any samples without additional staff and equipment. One laboratory reported that there were currently delivering 110 tests per week, one that they were delivering 200 tests per week and another that they could do 92 tests per run with up 2 runs per week (total 184 tests per week).
Estimated TAT from receiving a sample to returning a test result varied considerably across laboratories ranging from 24 to 72 hours (one laboratory) to > 4 weeks (one laboratory). The most common estimate (five laboratories) was 72 hours to 1 week; one laboratory estimates that results would be returned in 1–2 weeks.
Most laboratories reported that additional testing capacity and faster TAT would be possible with additional resources – one lab reported that faster TAT would not be possible (this lab had estimated TAT at 72 hours–1 week). Additional requirements included: additional staffing (six labs); increased laboratory space (two labs), increased automation (two labs) and additional equipment (four labs). One laboratory specified that staffing would need to be at all grades, another that more technical and IT staff would be needed, the others did not specify further.
Seven labs confirmed that the test could be performed in local laboratories, but most said that this would require additional staff training and/or equipment – one stated this could be done using existing staff and equipment. The laboratory that stated that the test could not be performed in local laboratories had selected a Real-Time PCR System as its preferred test.
Barriers to implementing CYP2C19 testing
The major barriers to implementing CYP2C19 testing were the scale of the predicted activity and current capacity (four labs), with one highlighting that they do not currently perform any tests of this scale in the NHS and so do not have the infrastructure for this. Staffing was also seen as a major barrier – this was highlighted by five labs. Two labs highlighted the importance of having automated/electronic laboratory systems in place. One lab, despite highlighting several barriers to implementing CYP2C19 testing, did state that it is ‘entirely possible’ to overcome these barriers. Another lab also highlighted facilitators to implementing testing including previous knowledge of pharmacogenomics testing in lab and the availability of appropriate equipment available within the department. The Scottish Tayside lab, which is currently piloting CYP2C19 testing, highlighted the following as barriers to implementing testing:
-
fixed budget for pilot and so had to confine requests to Stroke Unit and Cardiology
-
unable to accept requests from GPs
-
difficulty for some medical disciplines to understand output of genetic results
-
separate requesting and reporting systems for acute and primary care.
They also stated that strong support from stroke clinicians, specialist pharmacists and senior managers were facilitators for testing.
Implementation of rapid point-of-care tests in laboratory workflow
Six labs stated that it should be possible to implement a POCT within the laboratory workflow. One highlighted that this would not be the most efficient process for the number of samples that would need to be tested, and another that there is no precedent for this in their lab. Additional resources needed included more additional staffing (three labs) and additional freezers (one lab). Two labs stated that they would not be able to implement POCT. One explained that this would require staff to be able to drop all other duties to perform this test which would not be feasible. One of the labs that stated that it would be possible highlighted that delivering POCT would require different testing technology and cost would increase, another lab highlighted that the time for sample to be receiving in the laboratory might be an issue.
Chapter 5 Assessment of cost-effectiveness
Sections of this chapter have been adapted from the study’s Protocol document, available at the NICE website. 1
Review of economic evaluations of CYP2C19 genetic tests for clopidogrel resistance in non-cardioembolic ischaemic stroke and transient ischaemic attack patients
Review methods
We conducted a systematic review to identify previous studies on the cost-effectiveness of CYP2C19 genetic tests for guiding treatment in non-cardioembolic ischaemic stroke and TIA patients. We searched the following databases:
-
MEDLINE (MEDALL) via Ovid: 1946 to present
-
EMBASE via Ovid: 1974 to 9 August 2022 (Search 1) and 1974 to 10 August 2022 (Search 2)
-
the Cochrane Central Register of Controlled Trials (CENTRAL) via Wiley: Issue 7 of 12, July 2022
-
the Cumulative Index to Nursing and Allied Health Literature (CINAHL) via EBSCO Host: 1981 to present
-
ECONLit via EBSCO Host: 1986 to present
-
HTA Library via the York CRD interface
-
NHS EED via the York CRD interface
-
Tufts CEA Register via the Tufts Medical Centre website.
We also included any relevant papers on cost-effectiveness identified in the clinical effectiveness reviews, searched citations in relevant publications that we identified and asked experts in the field. We supplemented the searches with a targeted search for economic models of treatment for secondary prevention following non-cardioembolic ischaemic stroke or TIA. This search was undertaken in MEDLINE, EMBASE and EconLit. The search strategy for this search is reported in Appendix 1.
The quality of included cost-effectiveness studies was assessed using the Drummond checklist. 102
Sources for parameter inputs for the model were identified from previous models, the studies identified in the clinical effectiveness reviews (Objectives 1–5), and by running additional targeted searches to identify inputs to the economic model (as required). This included searching for previous network meta-analyses of antiplatelet treatments, in general non-cardioembolic ischaemic stroke and TIA populations.
Results of the review of cost-effectiveness studies for CYP2C19 testing strategies
The PRISMA flow chart showing the studies identified from the systematic review of cost-effectiveness studies for CYP2C19 testing for patients who have had a non-cardioembolic ischaemic stroke and TIA can be found in (see Appendix 7, Figure 34).
Five cost-effectiveness studies were identified for genetic testing for CYP2C19 LOF followed by antiplatelet therapy in patients suffering from TIA/minor stroke. One study was from a UK NHS perspective103 where a POCT was modelled and LOF carriers were assumed to be treated with dipyridamole–aspirin instead of clopidogrel. Three studies modelled alternative treatment with ticagrelor,104–106 which is not licensed for this indication in the UK. The studies are summarised in Appendix 7, Tables 52A and 52B.
Micieli et al.104
This economic evaluation was undertaken to assess the cost-effectiveness of the GMEX POCT for CYP2C19 LOF alleles followed by targeted DAPT compared with no testing in patients living in Canada following TIA/minor stroke. It is assumed that the modelled population meet inclusion in the CHANCE protocol with either had an acute non-disabling ischaemic stroke [National Institutes of Health Stroke Scale (NIHSS) ≤ 3] or a high-risk TIA (ABCD2 ≥ 4). 52 Under the testing strategy, patients with LOF alleles receive ticagrelor with aspirin, whereas those without LOF alleles receive clopidogrel with aspirin. In the no-testing strategy, all patients receive clopidogrel with aspirin. In all cases, it is assumed that DAPT is given for 3 weeks followed by long-term aspirin monotherapy. The analysis was performed from a Canadian health policy decision-maker’s perspective. Many of the model inputs were based on the multicentre, placebo-controlled CHANCE-2 trial comparing the modelled strategies in China50 and also from the CHANCE substudy looking at the association between CYP2C19 LOF and outcomes on clopidogrel with aspirin. 52 Both CHANCE and CHANCE-2 studies were identified in our clinical review (Objectives 2 and 3).
The decision model is a Markov state transition model for a cohort of patients average age of 65 years over a 20-year time horizon. The first stage model simulates patients’ outcomes at 90 days, where a proportion would transition into one of the following four health states:
-
Survive without clinical event.
-
Ischaemic stroke: mild [modified Rankin Scale (mRS) 0–1], moderate (mRS 0–2), severe (mRS 3–5) and fatal (mRS 6).
-
Haemorrhage: minor, major, intracerebral haemorrhage (ICH) and fatal.
-
Death.
Patients were then modelled for the reminder of the 20-year time horizon using a second stage of the Markov model which allows patients to have recurrent strokes by employing tunnel states; however, the exact form of the model was not clear from the paper. Baseline age-specific probability of death was sourced from Canadian lifetables and modified to account for severity of health states with data obtained from the ACTIVE-W trial107 of clopidogrel with irbesartan in atrial fibrillation patients. 104
The costs of medicines and the costs of clinical events were from local sources specific to Alberta. Utility values were taken from the ‘One thousand health-related quality-of-life estimates’ systematic review. 108
Testing for CYP2C19 LOF was found to be cost-effective with an incremental cost-effectiveness ratio (ICER) of 4310 Canadian dollars per quality-adjusted life-year (QALY), with a probability of being cost-effective more than 0.99 at a willingness-to-pay threshold of 50,000 Canadian dollars per QALY.
Cai et al.109
This economic evaluation was undertaken to assess the cost-effectiveness of testing for CYP2C19 LOF alleles using the Sequenom MassARRAY iPLEX laboratory test followed by targeted DAPT compared with no testing in Chinese patients following TIA/minor stroke. It is assumed that the modelled population matches that of the CHANCE study. 52 Under the testing strategy, patients with LOF alleles receive dipyridamole with aspirin, and patents without the LOF alleles receive clopidogrel with aspirin. In the no-testing strategy, all patients receive clopidogrel with aspirin. In all cases, DAPT is given for 90 days followed by long-term aspirin monotherapy. The analysis was performed from a Chinese health payer perspective.
This economic evaluation relies on two sources of clinical evidence. The efficacy of clopidogrel with aspirin according to CYP2C19 LOF allele status is taken from a subgroup analysis from the CHANCE trial,52 which was identified in our clinical review (Objectives 2 and 3). The efficacy of dipyridamole with aspirin compared with clopidogrel with aspirin is estimated using an indirect comparison via common comparator aspirin monotherapy based on an individual patient data meta-analysis of five RCTs comparing dipyridamole with aspirin versus aspirin110 and the CHANCE study comparing clopidogrel with aspirin versus aspirin. 52 The CHANCE study was included in our clinical review (Objectives 1 and 2), and also used in our economic model.
The decision model is a combination of a decision tree and a Markov state transition model for a cohort of patients for a 30-year time horizon. The decision-tree model simulates patients’ outcomes after the first 90 days, where a proportion would transition into one of the following four health states:
-
minor or no disability (mRS 0–2)
-
moderate disability (mRS 3–4)
-
severe disability (mRS 5)
-
death (mRS 6).
The Markov model covers the remainder of the 30-year time horizon patients experiencing recurrent strokes, ICH, major extracranial haemorrhage (ECH) and MI. Age-specific mortality rates for non-stroke death were derived from a published census of China and adjusted by the causes of death.
The costs of medicines were based on the retail prices according to the Beijing Municipal Commission of Development and Reform. The one-time hospitalisation costs associated with clinical events were based on the China health statistics yearbook. 111 Utility values were based on a previous CEA of clopidogrel with aspirin versus aspirin alone for patients who have had a minor stroke or TIA. 112
The ICER for the CYP2C19 LOF testing strategy was 13,552.74 Chinese Yuan per QALY gained compared with no testing, and the probability of being cost-effective was 0.96 at a willingness-to-pay threshold 72,100 Chinese Yuan per QALY.
Narasimhalu et al.105
This economic evaluation was undertaken to assess the cost-effectiveness of testing for CYP2C19 LOF alleles using the Spartan RX POCT followed by targeted antiplatelet therapy compared with no testing in Singaporean patients who have had their first ischaemic stroke. Under the testing strategy, patients with LOF alleles receive ticagrelor, whereas those without LOF alleles receive clopidogrel. In the no-testing strategy, all patients receive clopidogrel. In all cases, long-term antiplatelet monotherapy is given. The analysis was performed from a local healthcare provider’s perspective.
This economic evaluation relies on two sources of clinical evidence. Outcomes for patients on clopidogrel according to CYP2C19 LOF status (where LOF allele carriers were determined as CYP2C19*2 and CYP2C19*3) were taken from a prospective cohort study that evaluated the impact of CYP2C19 polymorphisms on stroke recurrence and other vascular events in a cohort of Chinese patients receiving clopidogrel,68 identified in our clinical review (Objective 3). Outcomes on ticagrelor were taken from the ticagrelor arm of the multinational multicentre SOCRATES trial of ticagrelor versus aspirin in a subgroup of patients with a non-cardioembolic, non-severe acute ischaemic stroke or high-risk TIA. 113
The decision model is a Markov state transition for a cohort of patients average age of 65 years over a 20-year time horizon. Patients transition into one of the following three health states:
-
non-recurrent ischaemic stroke (the starting state)
-
post-ischaemic stroke (after a recurrent stroke)
-
death.
Local rates of ischaemic stroke were sourced from the Singapore Stroke Registry data from public hospitals between 2007 and 2016. 114 The standard mortality rates at every age were obtained from life tables for the Singapore Resident Population 2017–8. The prevalence of LOF allele carriers was taken from previously reported values of 506 genomic samples of healthy Singaporean individuals. 115
The cost of the genetic test and the costs of medicines were sourced from a local hospital. The total cost of ischaemic stroke was sourced from administrative data. Utility values were sourced from an economic evaluation of primary stroke centres,38,96,116 which based the values on a survey of preferences among persons at increased risk for stroke in the USA. 117
CYP2C19 testing for LOF was found to be cost-effective with an ICER of $33,839/QALY compared with no testing, with a probability of being cost-effective of 0.78 at a willingness-to-pay threshold of $60,000/QALY.
Kremers et al.106
This economic evaluation was undertaken to assess the cost-effectiveness of POCT for CYP2C19 LOF alleles followed by targeted therapy compared with no testing in patients living in the Netherlands following minor acute stroke/TIA. Under the testing strategy, patients with LOF alleles receive aspirin monotherapy, prasugrel, ticagrelor or aspirin–dipyridamole instead of clopidogrel with aspirin, whereas in the no-testing strategy, all patients receive DAPT clopidogrel with aspirin. Clinical inputs for the model are based on published studies, but this abstract does not give further details. 118
The decision model is a Markov state transition 1-year cycle length for a cohort of patients over a lifetime horizon. Testing for CYP2C19 LOF followed by prasugrel or ticagrelor were found to be cost saving with incremental cost savings of €461 or €438, and gains of 0.01 QALYs per patient compared to no testing and treatment with clopidogrel.
Wright et al.103
This early economic evaluation was undertaken to assess the high-level cost-effectiveness of the Genedrive® CYP2C19 ID Kit POCT for CYP2C19 LOF alleles followed by targeted DAPT compared with no testing in patients living in the UK following first stroke. Under the testing strategy, patients with LOF alleles receive dipyridamole with aspirin instead of clopidogrel monotherapy, whereas in the no-testing strategy, all patients receive clopidogrel. Patients who do not tolerate clopidogrel are switched to modified release dipyridamole, or to aspirin if the modified release dipyridamole is not tolerated. The analysis was performed from the UK NHS perspective. 103
The treatment effects of clopidogrel on LOF carriers were based on a systematic review and meta-analysis of studies to assess the association between CYP2C19 genotype and clopidogrel efficacy for ischaemic stroke or TIA. 17 The treatment effects for LOF non-carriers were based on a network meta-analysis of treatments for first strokes and recurrent strokes in a general stroke/TIA population taken from the HTA of clopidogrel and modified-release dipyridamole for the prevention of occlusive vascular events for NICE TA210. 119
A decision tree and Markov model is used for a cohort of patients with average age of 67 years over a lifetime time horizon. In the first stage, the decision tree is used to capture the testing process and allocation to treatment. The proportion of patients who are LOF carries was based on data reported in the meta-analysis. 17 Patients then enter the Markov model and transition into one of the following five health states:
-
no further stroke
-
one further stroke
-
> 1 further stoke
-
vascular death
-
other cause of death.
All-cause mortality was estimated from the Office of National Statistics mortality data. 120 The costs and utility values were taken from the economic evaluation of clopidogrel and modified-release dipyridamole for the prevention of occlusive vascular events. 119
Testing for CYP2C19 LOF was found to be dominate the no-testing strategy, with lower mean costs (incremental savings of £170) and higher mean QALYs (incremental gain of 0.096 QALYs per patient), and a probability of being cost-effective more than 0.77 at a willingness-to-pay threshold of £20,000 per QALY. 103
Quality assessment of cost-effectiveness studies
The assessment of study quality of the cost-effectiveness studies using the Drummond checklist can be found in Appendix 7, Table 53. 102 In general, the studies were of high quality, although the estimation of unit costs was not clear. Kremers et al. 106 was a conference abstract; although limited detail is available, we confirmed treatment strategies via correspondence with authors.
Summary of relevance of existing evidence to this economic evaluation
Of the previous models, only Wright et al. 103 was in a UK setting using an alternative treatment (dipyridamole) that would be used in UK clinical practice for LOF carriers. However, the model inputs used by Wright et al. 103 were based on an old HTA119 and a meta-analysis17 that does not include some of the recent relevant evidence. None of the previous studies compared the different types of CYP2C19 tests. Most of the models used a decision-tree structure for the short-term impacts of CYP2C19 testing followed by a long-term Markov model, and all of the previous models found that CYP2C19 testing is likely to be cost-effective.
Results of the review of cost-effectiveness studies of secondary prevention of ischaemic stroke
To supplement the review of cost-effectiveness studies for CYP2C19 testing strategies, we also reviewed cost-effectiveness studies of secondary prevention of ischaemic stroke in a general population to help inform the structure of the long-term model for antiplatelet therapies in patients who had a previous ischaemic stroke or TIA, and also to help identify relevant evidence sources. The PRISMA flow chart showing the studies identified from this review is shown in Appendix 7, Figure 35.
We identified four relevant cost-effectiveness studies which are summarised in Appendix 7, Tables 54 and 55.
Zhou et al.121
This economic evaluation was undertaken to assess the cost-effectiveness of adding Cilostazol to aspirin or clopidogrel compared with aspirin or clopidogrel monotherapy in patients with non-cardioembolic stroke. A Markov state transition model was used to simulate a cohort of patients, average age of 70 years, over a 40-year time horizon between the following four health states:
-
neurologically intact, (score of 0 mRS)
-
mild disability (score of 1–2 on the mRS)
-
moderate to severe disability (score of 3–5 on the mRS)
-
deceased (mRS score of 6).
The analysis was performed from a US payer/Medicare perspective.
Base rates of recurrent ischaemic stroke for patients on aspirin and clopidogrel were derived from a subgroup analysis of the clopidogrel versus aspirin in patients at risk of ischaemic events (CAPRIE) trial. 122 Neurological outcomes after ICH while on antiplatelet therapy were derived from the results of the PATCH trial. 123
Treatment effects for adding cilostazol to aspirin or clopidogrel were based on the multicentre, placebo-controlled CSPS.com trial in Japan. 121 For those experiencing a recurrent ischaemic stroke or ICH, the resulting mRS state was based on the results of the POINT trial. 124
The costs of medicines and those associated with clinical events were from local sources specific to the USA. The annual costs of mild and severe health states were taken from a cost-effectiveness study of Diagnostic Strategies. 125 The health utility scores associated with disability states was calculated using US-specific preference weights multiplied by utilities derived from the Virtual International Stroke Trials Archive (VISTA). 126
Greenhalgh et al.119
This economic evaluation was undertaken to compare the cost-effectiveness of different treatment sequences with aspirin, clopidogrel and modified-release dipyridamole plus aspirin, compared with no treatment in patients who have had a recent stroke or TIA. The decision model is an individual patient simulation model over a lifetime horizon, where patient characteristics were based on data from the Health Survey of England 1996. Patients transition according to risks associated with disability status (disabled classified as mRS ≥ 3) through the following health states:
-
new fatal or non-fatal ischaemic stroke event
-
new fatal or non-fatal non-ischaemic stroke event (haemorrhagic stroke or ICH)
-
new fatal or non-fatal MI
-
death from other vascular causes
-
death from non-vascular causes.
The analysis was performed from a UK health policy decision-makers' perspective. Risk models for events and fatality due to events were based on confidential data from the CAPRIE122 and PRoFESS127 clinical trials. Treatment discontinuation is modelled by an exponential survival function which calculates the time of discontinuation for each patient. 119
The treatment effects for the first recurrent ischaemic stroke were estimated using a network meta-analysis of three clinical trials CAPRIE,122 ESPIRIT128 and PRoFESS. 127 The treatment effects for recurrent ischaemic stroke were estimated using a network meta-analysis of two clinical trials ESPS-2129 and PRoFESS. 119 Furthermore, the network meta-analyses were used to estimate death from all causes, vascular death, major bleeds and all bleeds. 119
Treatment costs were drawn from the manufacturers' submissions,130,131 stroke event costs from a UK economic burden study132 and MI event costs from the UK Prospective Diabetes Study. 133
Utility values for ischaemic stroke and related disability were taken from the PRoFESS trial,127 and the utility for MI taken from a cost-effectiveness study of the secondary prevention of stroke. 134 The utility decrement for minor bleeds was taken from a cost-effectiveness study for stroke prophylaxis in atrial fibrillation. 135 The utility decrement of dyspepsia was taken from an economic evaluation for a treatment of ankylosing spondylitis. 136 The utility decrement for ICH was taken from a cost-effectiveness study of anticoagulation for haemodialysis patients with atrial fibrillation. 137
Malinina et al.138
This economic evaluation was undertaken to assess the cost-effectiveness of DAPT clopidogrel + aspirin or extended-release dipyridamole + aspirin compared with aspirin in patients who have had a non-cardioembolic stroke or TIA. The decision model is a decision-tree model for a cohort of patients average age of 65 years over a 1-year time horizon. The analysis was performed from a US payer perspective. Treatment effects for model were estimated by calculating the relative risk reductions derived from the randomised control trials ESPS-2, MATCH, CAPRIE and a metaregression of the effects of aspirin on stroke. 138,139
The proportion of stroke survivors was calculated from the Atherosclerosis Risk in Communities Study,140 and the risk of recurrent stroke taken from the Oxfordshire Community Stroke Project. 141 The annual costs of stroke were based on a study of Medicare claims,142 and the costs of minor bleeds were taken from a cost-effectiveness study of the secondary prevention of stroke. 134 Utility values were not included in this model as the primary aim of the model was to estimate the number of stokes averted over the time period.
Jones et al.143
This economic evaluation was undertaken to compare the cost-effectiveness of different antiplatelets in patients who have had either a stroke or a TIA. Aspirin, clopidogrel, modified-release dipyridamole, modified-release dipyridamole + aspirin and aspirin are compared for patients who have had a stroke. Aspirin, modified-release dipyridamole, modified-release dipyridamole + aspirin and aspirin are compared for in patients who have had a TIA. The decision model is a Markov state transition for a cohort of patients average age of 60 years over a 40-year time horizon. The patients enter model after their initial stroke/TIA into one of the following five health states:
-
TIA (starting state for the TIA population).
-
year 1 post stroke (no further stroke/event free) (starting state for the ischaemic stroke population)
-
new stroke (recurrent stroke), either disabling or non-disabling
-
vascular death
-
non-vascular death (excluded by scenario analysis).
The analysis was performed from a UK NHS perspective. Baseline age-adjusted event rates (recurrent stroke, vascular death) for stroke patients assumed to be on treatment with aspirin were calculated from patient-level data the South London Stroke Register (SLSR). 144 Proportions of patients having disabling first stokes and disabling recurrent strokes were obtained from the ESPS-2129 trial. Baseline risks of non-vascular death were estimated from the national statistics and excluding deaths due to diseases of the circulatory system. 143
The treatment effects for the model are estimated from an indirect comparison143 which connects the evidence on the treatment effects attributable to aspirin and risks of fatal/non-fatal bleeds reported in a meta-analysis by the Antithrombotic Trialists’ Collaboration. 145 The treatment effects of clopidogrel versus aspirin were estimated from the ESPS-2129 trial, and treatment effects for dipyridamole, modified-release dipyridamole + aspirin versus aspirin were estimated from the CAPRIE trial.
The annual cost associated with stroke was derived from study describing the economic burden of stroke to the UK. 132 The authors assumed that costs associated with mild and moderate strokes were attributable to non-disabled stroke patients and that costs associated with severe stroke were attributable to disabled stroke patients. Utility values were taken from a systematic review of the effectiveness, cost-effectiveness and barriers to implementation of thrombolytic and neuroprotective therapy for acute ischaemic stroke in the NHS. 146
Model structure and methods of economic evaluation
We developed a decision-analytic model to estimate the incremental costs and QALYs for CYP2C19 genetic testing for clopidogrel resistance in patients in England and Wales who have had a non-cardioembolic ischaemic stroke or TIA where treatment with clopidogrel is being considered, compared with no genetic testing. We refer to LOF carriers as LOF patients, and LOF non-carriers as loss-of-function non-carrier (NoLOF) patients.
Populations
The model was developed for two distinct populations: patients who have had a non-minor ischaemic stroke; and patients who have had a TIA or minor stroke. This is to reflect the different treatment pathways (see Place of the technology in the treatment pathway) and different event rates in these two subpopulations. The key differences between the models are the event rates and transition probabilities, the costs and utilities for patients who have not had a recurrent stroke event (worse health state for non-minor stroke patients compared with TIA/minor stroke patients), and the time at which clopidogrel is initiated and LOF patients can benefit from targeted treatment (immediately for TIA/minor stroke patients, and after 2 weeks aspirin otherwise). Results are reported for the two subpopulations separately, and for a combined population using a weighted average over the subpopulations, according to prevalence. We conducted a scenario analysis for populations with high prevalence of clopidogrel resistance. We had planned to conduct a scenario analysis for children; however, insufficient evidence was identified to do this. We instead conduct a scenario analysis for younger adults at time of index stroke or TIA.
Genetic testing and treatment strategies and comparators
We compared different CYP2C19 testing strategies with a no-testing strategy. We included the Genomadix Cube and the Genedrive CYP2C19 point-of-care genetic tests included in the clinical effectiveness review (see Chapter 3, Objectives 1, 2 and 3), and a single laboratory-based CYP2C19 genetic test chosen to be representative of how laboratory-based tests are used in practice (based on our survey of genomic laboratory hubs; Chapter 3, Survey of laboratories). We note that there is very little information on the Genedrive POCT, so the results for this test are based on assumptions and should be interpreted with this in mind. We varied the time taken to receive results and the cost of the lab-based test in scenario analyses (see Scenario analyses). We assumed that the tests fail to provide a result in a proportion of cases (which depends on test type), and that for those cases a second test would be required, incurring additional costs.
Under the no-testing strategy, it is assumed that all patients will be treated according to the treatment pathways in Figure 1. Non-minor stroke patients receive aspirin 300 mg daily for 2 weeks (starting within 24 hours), followed by long-term clopidogrel 75 mg daily (after a loading dose of 300 mg). TIA and minor stroke patients receive either DAPT aspirin 75 mg daily plus clopidogrel 75 mg or monotherapy clopidogrel 75 mg daily (after a loading dose of 300 mg) for up to 90 days (started within 24 hours), followed by long-term monotherapy clopidogrel 75 mg daily.
For patients whose test indicates they are a CYP2C19 clopidogrel LOF carrier, we assumed that clopidogrel is replaced with DAPT aspirin 75 mg daily plus dipyridamole 200 mg twice daily as recommended by NICE guidance28 (see Figure 2). We ran scenario analyses assuming that CYP2C19 LOF carriers switch to different alternative antiplatelet therapy (Alt Tx). Based on clinical advice, other alternative antiplatelets included were low-dose aspirin 75 mg daily or ticagrelor 90 mg twice daily.
Model structure
The model structure was developed to capture the short- and long-term costs and benefits of CYP2C19 genetic testing, based on our review of previous cost-effectiveness models (see Results of the review of cost-effectiveness studies for CYP2C19 testing strategies and Results of the review of cost-effectiveness studies of secondary prevention of ischaemic stroke), results from the Chapter 3, Survey of laboratories, information provided by Genedrive and Genomadix, and in discussion with specialist members of the Diagnostic Appraisal Committee (DAC).
The model utilises a hybrid decision-tree and Markov structure. Diagnostic decisions and short-term 90-day outcomes were modelled using a decision-tree structure, and long-term outcomes were modelled using a five state Markov model. The model is split into short-term (90-day) and long-term outcomes to reflect the elevated risk of a subsequent stroke in the short term following an event, which is particularly relevant for patients who have had a TIA.
Decision tree
The decision tree (Figures 9–11) differs by test type according to the diagnostic outcomes for the test. For POCTs, there are four branches of the tree for patients who receive a true-positive (TP), false-negative (FN), true-negative (TN) and false-positive (FP) result. We assume that the lab test is a gold standard test with perfect sensitivity and specificity, so there are just two branches of the tree for LOF patients and NoLOF patients who receive appropriate test results and corresponding treatment. For the no-testing strategy, there are also two branches of the tree for LOF patients and NoLOF patients, but the event rates differ because all the LOF patients will receive clopidogrel rather than dipyridamole + aspirin. The proportion of the modelled population that are of LOF carriers is the same regardless of test.
FIGURE 9.
Point-of-care testing decision-tree branch. The timing of clopidogrel treatment will depend on the indication. Those patients who have had a TIA/minor stroke may begin dual clopidogrel–aspirin treatment immediately. Those patients who have had a major stroke may initiate with 2 weeks of aspirin before clopidogrel treatment. Alternative treatment (Alt Tx) is aspirin combined with dipyridamole in the base case, with scenarios for low-dose aspirin and ticagrelor.
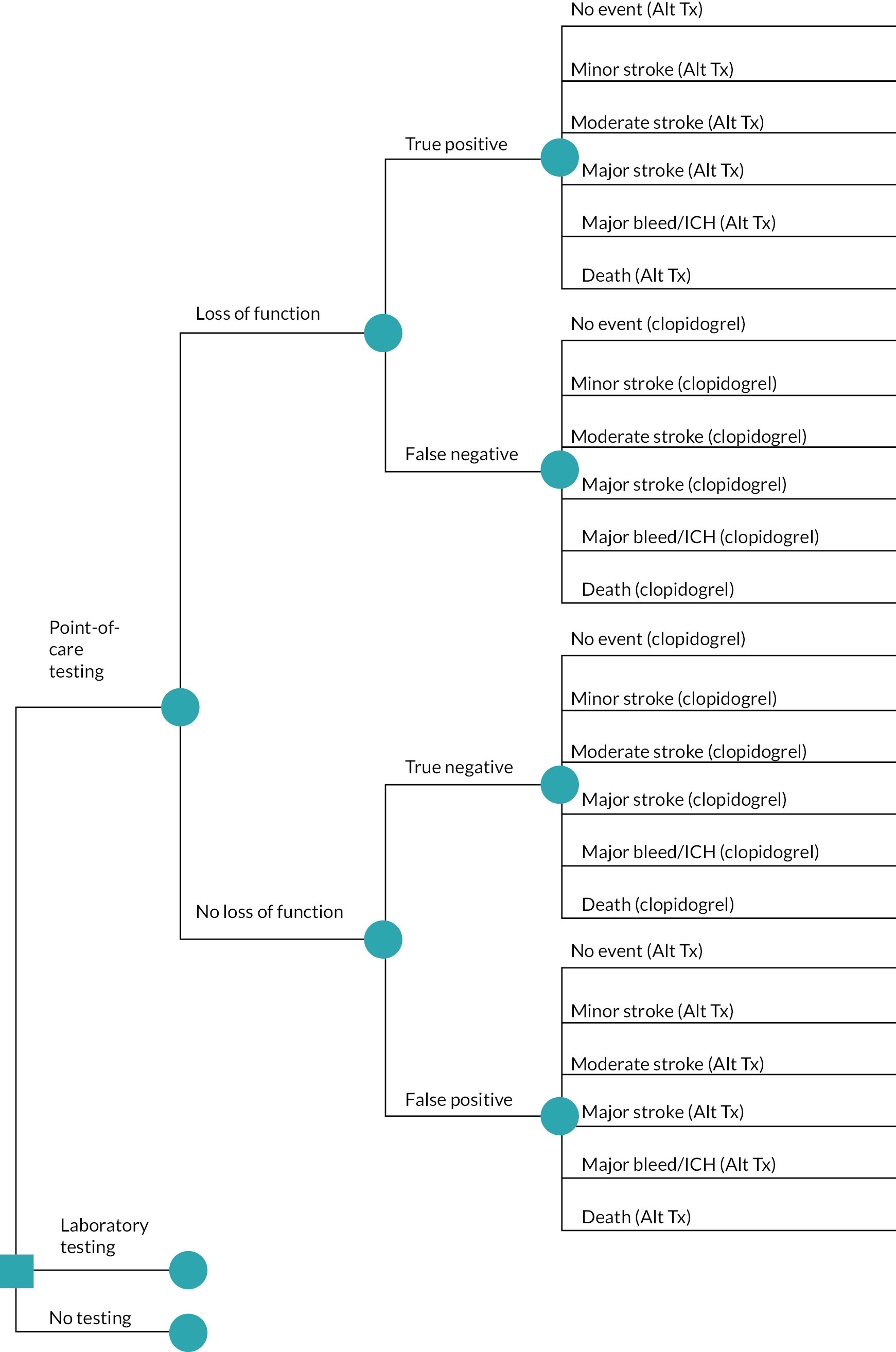
FIGURE 10.
Laboratory testing decision-tree branch. The timing of clopidogrel treatment will depend on the indication. Those patients who have had a TIA/minor stroke may begin dual clopidogrel–aspirin treatment immediately. Those patients who have had a major stroke may initiate with 2 weeks of aspirin before clopidogrel treatment. Alternative treatment (Alt Tx) is aspirin combined with dipyridamole in the base case, with scenarios for low-dose aspirin and ticagrelor.
FIGURE 11.
No test decision-tree branch. The timing of clopidogrel treatment will depend on the indication. Those patients who have had a TIA/minor stroke may begin dual clopidogrel–aspirin treatment immediately. Those patients who have had a major stroke may initiate with 2 weeks of aspirin before clopidogrel treatment. Alternative treatment (Alt Tx) is aspirin combined with dipyridamole in the base case, with scenarios for low-dose aspirin and ticagrelor

The second part of the decision tree captures the 90-day outcomes, which are the same for each diagnostic outcome branch of the tree but with different event rates depending on treatment and LOF status. The 90-day health outcomes are:
-
no further event
-
further minor stroke
-
major bleed/ICH
-
further moderate stroke
-
further major stroke
-
death.
The health outcomes are defined according to stroke severity, which correspond to disability states. Advice from clinical members of the DAC suggested the following mRS scores for the different stroke severity states:
-
TIA: mRS range 0
-
minor stroke: mRS range 0–1
-
moderate stroke: mRS range 2–3
-
major stroke: mRS range 4–5.
These are largely in line with the categories used in previous economic evaluations of CYP2C19 testing. 104,109,121 We place major bleed/ICH between further minor and further moderate stroke in terms of severity, based on clinical advice and utility estimates (see Model parameters and inputs).
Under lab-based testing, there may be a delay in receiving test results after which those identified as LOF carriers switch from clopidogrel to appropriate alternative treatment (Alt Tx). We assume that POCT results are available within a day, so that there is no delay in starting appropriate treatment for LOF carriers. Treatment switches were modelled by averaging the event rates during the short-term (90-day) part of the model according to the time spent on different treatments.
For all strategies, patients may discontinue treatment as a result of treatment-related side effects, and it is assumed that patients discontinuing would switch to low-dose aspirin monotherapy.
The model structure is the same for the non-minor ischaemic stroke and TIA/minor stroke subpopulations, but the model inputs and transition probabilities differ between populations.
As the decision tree models the first 90 days after the patient’s initial ischaemic stroke or TIA, no discount rate is applied to costs and QALYs accrued in this period.
Markov model
Health states
Following the initial decision tree, a Markov model (Figure 12) was used to model long-term patient outcomes for a cohort of patients. In the model, patients move between five possible health states: no recurrent stroke, post minor stroke, post major bleed/ICH, post moderate stroke or post major stroke. Health states were chosen based on discussion with clinical experts and inspection of the clinical and health economic literature on the most impactful and frequent events experienced by ischaemic stroke survivors. Health states differ in costs, health-related quality of life (HRQoL), mortality rate and recurrent event rates. The ordering of the health states by severity was motivated by consultation with patient and clinical experts and inspection of long-term HRQoL measurements from studies which had measured this by health state (see Model parameters and inputs).
FIGURE 12.
Long-term Markov model structure.
We assume that patients can progress to a more severe disease health state, but they cannot move from a more severe health state to a less severe state. This was based on the nature of the long-term outcomes associated with the events included in the model (strokes, major bleeds) and the chronic nature of the disease. Patients are categorised into the most severe category that they have experienced.
The proportion of patients in each health state in the first Markov time cycle is determined by the proportion in each health state at the end of the 90-day decision tree, according to true LOF status and treatment allocation.
Cohorts evaluated
The transitions in the Markov model depend on treatment and LOF status, and so four cohorts are evaluated corresponding to the paths in the decision tree:
-
patients with LOF alleles undergoing clopidogrel treatment
-
patients with LOF alleles undergoing a non-clopidogrel alternative treatment
-
patients without LOF alleles undergoing clopidogrel treatment
-
patients without LOF alleles undergoing a non-clopidogrel alternative treatment.
We assume that patients stay on the same treatment they were on at the end of the decision-tree period, and only switch treatment as a consequence of a bleeding event, which is modelled with a discontinuation rate set equal to the rate of new major bleeds/ICH each year for each treatment.
Time cycles, time horizon and discounting
The initial time cycle in the Markov model takes the length of 275¼ days, calculated as a 1 year minus the 90-day period of the decision-tree period. The second and subsequent time cycles take the length of a year (365¼ days). The Markov model utilises a lifetime time horizon and so patient outcomes in the Markov model are followed until the general population life tables end (age 100 years).
Costs and QALYs in the long-term Markov model are discounted at a rate of 3.5% per annum. Discounting begins in the second Markov time cycle which models the second year after the patient’s initial stroke or TIA.
Model outcomes
Costs are accrued in the model through health state-specific costs and treatment costs, which when summed over time spent in health states and time on treatment give total (discounted) expected costs over the model time horizon for each path in the decision tree. QALYs accrue in the model through the health state utilities summed over time spent in each health state to give (discounted) total expected QALYs over the model time horizon for each path in the decision tree. The total expected costs and QALYs of the decision-tree pathways are then averaged to calculate the total expected costs and QALYs associated with the POCTs, laboratory testing and no testing, respectively. The cost-effectiveness results are summarised with ICERs and expected net benefit.
Perspective
An NHS and personal social services (PSS) perspective was taken with a lifetime horizon, where costs and QALYs are discounted at an annual rate of 3.5%. The model includes health effects for both patients and carers. A 2022 price year was used in the base case. Costs from previous years were inflated to the 2022 price year using the Office for National Statistics’ (ONS) CPIH index.
Model parameters and inputs
Model inputs were derived from the clinical effectiveness review, our review of previous cost-effectiveness models (see Results of the review of cost-effectiveness studies for CYP2C19 testing strategies and Results of the review of cost-effectiveness studies of secondary prevention of ischaemic stroke), results from the Chapter 3, Survey of laboratories, information provided by Genedrive and Genomadix and additional targeted searches where required. Where it was necessary to make assumptions, this was based on expert opinion and scenario analyses conducted to explore the impact of these assumptions on the results.
Test performance
Time to receive test results
The clinical review found that results were available within a matter of hours for the POCTs (see Appendix 5, Table 50) and the companies confirmed that results will be available in 40 minutes to approximately 1 hour. We therefore assume that there would be no delay in patients receiving targeted therapy for the POCT strategy.
The Survey of laboratories (see Chapter 4, section Survey) indicated that there is likely to be some variability in the time until lab-based test results are available, and this would depend on capacity/resources. The most common estimate was approximately 1 week, but one lab reported it may be longer than 4 weeks. Our clinical advisors estimated it would take between 5 days and 6 weeks, which is in line with the survey responses. We assume a 1-week turnaround in the base case, and 4 weeks in a sensitivity analysis.
Test failure rate
The clinical review found variation in the test failure rate (test result unavailable) ranging from 0.4% to 18.9% for studies on the Genomadix (Spartan) CYP2C19 tests (see Table 14). Pooling results from these studies in a random-effects meta-analysis gives an estimated average failure probability of 0.08 (95% CI 0.05 to 0.15). We assume a test failure rate of 8% and describe the uncertainty around this with a beta distribution with parameters matched to the meta-analysis estimates. No studies were available for Genedrive, and so we had to make an assumption. We assumed the test failure rate for Genedrive is equal to that seen for the Genomadix Cube but note the uncertainty around this assumption. All results for Genedrive are therefore illustrative and need updating when data become available.
The Survey of laboratories (see Chapter 4, section Survey) found that most laboratories expected < 1% of samples not to give valid results, although one laboratory estimated this to be 5%. We assume that all samples give a valid result in our model. Higher teat failure rates would increase the test cost slightly. We conduct a scenario analysis to the cost of the laboratory test.
Test accuracy of point-of-care tests
The bivariate meta-analysis for the Genomadix (Spartan) CYP2C19 tests based on the studies identified in the clinical review estimates very high sensitivity of 100% (95% CI 94% to 100%) and specificity of 100% (95% CI 99% to 100%) (see Chapter 4, Results). Note, however, that these figures are based on detecting the *2 and *3 alleles that the Genomadix (Spartan) CYP2C19 tests for. There are other LOF alleles (*4, *5, *6, *7 and *8), which would not be detected by the Genomadix Cube. The prevalence of these additional alleles is very small across ethnicities,147 but may reduce the sensitivity of the test very slightly. We therefore assume a sensitivity of 99% rather than 100% in the model. Genedrive tests for more alleles (see Table 2), and so would have the potential to detect *4 and *8, but not *5, *6 and *7. There was no diagnostic test accuracy data for Genedrive, so in the absence of data and due to the very small prevalence for *4 and *8, we assumed that the diagnostic test accuracy for Genedrive was the same as for the Genomadix Cube. All results for Genedrive are therefore illustrative and need updating when data become available.
Patient characteristics, prevalence of stroke, transient ischaemic attack and CYP2C19 loss of function
Incidence of first stroke and patient characteristics
The prevalence and population characteristics of first ischaemic stroke are taken from the NICE Clinical Knowledge Summary (https://cks.nice.org.uk/topics/stroke-tia/background-information/prevalence/) and the data sources it is based upon. The Public Health England (PHE) briefing document on incidence of first strokes in 2016 found a crude incidence rate of first strokes to be 107 per 100,000 population, with 49% females. 148 The mean age for first strokes was 68.2 years for males and 73 years for females. Ethnicity was only reported in approximately half of cases, but of these, the proportions were 92%, 4%, 2.5% and 1.5% for white, Asian, black and other, respectively. Note that this distribution is different to the population split from the census due to a higher incidence of stroke in white people (due largely to differences in demographics). The PHE briefing document reports an incidence of first-ever TIA of approximately 50 per 100,000 people per year. This gives the proportion of stroke/TIA cases that are TIA to be 50/157 = 31.8%.
Prevalence of CYP2C19 loss of function
The clinical review identified a wide range of estimates of prevalence of LOF from studies of clopidogrel (see Appendix 5, Table 51), but these were not on UK populations and did not provide estimates of prevalence according to ethnicity from these studies. We ran additional searches to identify prevalence studies of LOF alleles. We found a UK-based study Pilling et al. 149 which estimated a prevalence of having at least one CYP2C19 LOF variant to be 28.7% in 7483 European-ancestry adults prescribed clopidogrel based on the UK Biobank study with genetic and linked primary care data. This is likely to be an underestimate of the prevalence of CYP2C19 LOF in the UK due to a higher prevalence in those with non-European ancestry. The CHANCE study52 found that 58.8% of Chinese patients randomised to the trial had a LOF variant. A recent large-scale analysis of CYP2C19 LOF status150 by ethnicity in the USA estimated the prevalence of intermediate or PMs of 27.2% in Europeans, 56.8% for East Asians and 31.9% for African Americans. These figures agree well with those from Pilling et al. 149 and Wang et al. 52 for European and Chinese populations, respectively. Applying these estimates to the ethnicity mix in the UK based on the PHE briefing report,148 and assuming that the prevalence for those of non-European or Asian ancestry can be assumed to be 31.9%, we obtain a prevalence estimate of 0.92 × 27.2 + 0.04 × 56.8 + 0.04 × 31.9 = 32.1% in the UK population. We use this proportion of patients who are LOF carriers in the base case and conduct a scenario analysis to a higher proportion of 56.8% as estimated in East Asians.
Transition probabilities
In the decision tree and Markov model, the event transition probabilities according to treatment and LOF status are:
for time at risk t, where λtrt,status is the event rate. We estimate the baseline event rates for patients without LOF alleles who are taking clopidogrel, λclop,NoLOP, and then estimate HRs for each treatment and LOF status relative to NoLOF on clopidogrel
The HRs (Equation 2) are applied to the baseline event rate λclop,NoLOP to obtain the hazard rate λtrt,status for use in Equation 1.
Because patients may switch treatments, the hazard will be a weighted average of the hazards for the different treatments they have taken according to time on each treatment. For example, if a patient with LOF alleles starts clopidogrel and then switches to DAPT dipyridamole + aspirin after 6 weeks, then their hazard rate will be
The transition probabilities in the Markov model were derived in exactly the same way as for the decision-tree probabilities [Equations (1)–(3)]; however, the baseline event rates are assumed to differ in the longer term, and it is assumed that after 90 days patients only switch treatment as a result of a major bleed or ICH, modelled with a discontinuation rate set to the rate of major bleed/ICH.
Baseline recurrence rates (for patients with no loss of function on clopidogrel)
We searched for large recent UK-based cohort studies to estimate the baseline event rates for the outcomes in the decision-tree model (see Figures 9–11) in patients who have experienced a stroke or TIA. We assume that event rates depend on the severity of primary stroke experienced, but that relative treatment effects (HRs) do not vary by stroke severity.
Mohan et al. 151 report stroke recurrence rates based on 2874 patients following their first stroke with 8311 person-years follow-up from the SLSR, for cases registered between 1 January 1995 and 31 December 2004. They estimate a cumulative risk of recurrence of 7.1% (95% CI 6.0% to 8.3%) in the first year, 16.2% (95% CI 14.4% to 18.1%) by 5 years and 24.5% (95% CI 21.3% to 27.9%) by 10 years. These correspond to hazard rates per person-year of 0.074, 0.044 and 0.056 in years 1, 2–5 and 6–10, respectively, computed using λ=−ln(1−pint)/tint where λint is the hazard rate, pint the probability of recurrence on the time interval and tint the length of the time interval in years.
More recent data are available from the Sentinel Stroke National Audit Programme (SSNAP). 152 SSNAP provides national audit data on stroke patients from every acute hospital in England, Wales and Northern Ireland with longitudinal data collection on outcomes for up to 6 months post stroke, plus longer-term information on stroke recurrence. We prefer to use the SSNAP data to estimate short-term recurrence rates in our model, as it is most representative of a contemporary stroke population in England and Wales and provides detailed results specifically for patients who have had an ischaemic stroke. In the SSNAP health economics report,153 recurrence probability estimates are provided for up to 5 years based on the SSNAP data for the short term and SLSR in the longer term. However, it is not clear how the SLSR data have been used to form these estimates, and they do not align with the estimates reported in Mohan et al. 151 Table 19 shows the estimated cumulative probability of recurrence, and the hazard rate per person-year on each time interval from the two sources of evidence. We use the hazard rate of 0.092 from SSNAP for the first 90 days in our model, and beyond 90 days we use a hazard rate of 0.056 which is in line with the first year from SSNAP and the longer-term data from SLSR.
| Time from index event (years) | Cumulative percentage recurrence | Time period (interval) | Recurrence rate per person-year |
|---|---|---|---|
| SLSR (Mohan et al. 2009) 151 | |||
| 1 | 7.1% (95% CI 6.0% to 8.3%) | 0–1 (1 year) | 0.074 |
| 5 | 16.2% (95% CI 14.4% to 18.1%) | 1–5 (4 years) | 0.044 |
| 10 | 24.5% (95% CI 21.3% to 27.9%) | 5–10 (5 years) | 0.056 |
| SSNAP (ischaemic stroke patients)a153 | |||
| 0.25 | 2.28% | 0–0.25 (0.25 year) | 0.092 |
| 1 | 6.37% | 0.25–1 (0.75 year) | 0.056 |
| 2 | 11.41% | 1–2 (1 year) | 0.052 |
| 3 | 19.29% | 2–3 (1 year) | 0.082 |
| 4 | 28.50% | 3–4 (1 year) | 0.097 |
| 5 | 41.05% | 4–5 (1 year) | 0.134 |
For TIA patients, we identified a recent retrospective cohort study using the Framingham Heart Study data which report recurrence rates for 435 patients who had an index first TIA. Lioutas et al. report a crude hazard rate of recurrent stroke of 1.29 per 1000 person-years and give the proportion of recurrent events occurring over time from the index TIA. 154 Based on this, we estimated an annualised hazard rate in the time periods following the index TIA (Table 20). There is an elevated rate of stroke in the first week following TIA and a high rate for the first 90 days, falling to a lower rate beyond 90 days. In the model, we use a weighted average rate per person-year of 0.0838 for the first 90 days, and 0.0064 for day 90 onwards.
| Time period (interval) | Percentage of strokes in time period (%) | Stroke rate per person-year |
|---|---|---|
| 0–7 days (7 days) | 21.5 | 0.586 |
| 8–30 days (23 days) | 9.2 | 0.076 |
| 31–90 days (60 days) | 8.5 | 0.027 |
| 91–365 days (274 days) | 12.3 | 0.009 |
| 1–5 years (4 years) | 48.5 | 0.0064 |
| Average over 0–90 days | 0.0838 |
Stroke severity
Sentinel Stroke National Audit Programme provides the breakdown of recurrent strokes into NIHSS categories. 153 We classified NIHSS 0–4 as mild, NIHSS 5–15 as moderate and NIHSS > 15 as severe to estimate the proportion of recurrent strokes that fall into each category (Table 21). We assume that the proportion of recurrent strokes in each category does not depend on the initial stroke category. However, the movement between states in the model depends on the current state, with patients attributed to the worst severity state that they have experienced.
| NIHSS range | Recurrent strokes by severity | Total recurrent strokes | Proportion |
|---|---|---|---|
| 0 | 0 | 101 | 0 |
| 1–4 (mild) | 43 | 101 | 0.426 |
| 5–15 (moderate) | 48 | 101 | 0.475 |
| 16–42 (severe) | 10 | 101 | 0.099 |
Baseline mortality rates (for patients with no loss of function on clopidogrel)
Mortality rates were assumed to depend on model state via the mRS score. The health economics report for SSNAP fits a Cox survival analysis to data from SSNAP and the SLSR to estimate survival over a 5-year time period. 153 The survival probabilities are provided for a reference category of a 65-year-old male patient with mRS = 0 following an ischaemic stroke (Table 22), from which we form the hazard rate per person-year. SSNAP also provide the HRs to adjust for age, sex and mRS status (see Table 22). We applied the HRs to the reference hazard rates, to obtain the estimated hazard for an average cohort matching our population (the population was assumed to be 49% female patients with average age 68.2 years for males and females 73 years). The HRs by mRS category only show an elevated mortality rate for those with mRS = 4 or 5, which corresponds to our severe stroke state. We therefore apply a HR (averaged over mRS = 4 and mRS = 5) to reflect the increased mortality rate for those in the severe stroke state (Table 23). For TIA, it is assumed that mortality is equal to that for mRS = 0. Mortality increases with age as patients progress through the model which we capture using the rates by age and sex based on ONS. 155
| Time (years) | Survival probability | Mortality rate (hazard) per person-year | Covariate | Hazard ratio | Confidence interval |
|---|---|---|---|---|---|
| 0 | 1 | Female | 1.001152 | (0.924 to 1.084) | |
| 0.0847 | 0.999 | 0.011812 | age (years) | 1.026459 | (1.023 to 1.030) |
| 0.506 | 0.981 | 0.043114 | mRS1 | 0.9557 | (0.822 to 1.112) |
| 0.669 | 0.977 | 0.024589 | mRS2 | 0.832645 | (0.692 to 1.003) |
| 0.93 | 0.969 | 0.030775 | mRS3 | 0.941297 | (0.834 to 1.063) |
| 1.24 | 0.962 | 0.02266 | mRS4 | 1.037715 | (0.934 to 1.153) |
| 1.55 | 0.954 | 0.02591 | mRS5 | 1.277252 | (1.113 to 1.465) |
| 1.64 | 0.95 | 0.044534 | |||
| 1.92 | 0.943 | 0.025088 | |||
| 2.1 | 0.938 | 0.027847 | |||
| 2.31 | 0.932 | 0.028657 | |||
| 2.63 | 0.921 | 0.034565 | |||
| 2.79 | 0.917 | 0.02505 | |||
| 3.03 | 0.909 | 0.033467 | |||
| 3.26 | 0.903 | 0.026166 | |||
| 3.56 | 0.896 | 0.023415 | |||
| 3.83 | 0.884 | 0.044713 | |||
| 4.24 | 0.872 | 0.029445 | |||
| 4.73 | 0.858 | 0.028773 | |||
| 4.98 | 0.851 | 0.028098 | |||
| 5 | 0.847 | 0.200401 |
| Time period | mRS 0–3 (mild/moderate stroke) | mRS 4–5 (severe stroke) |
|---|---|---|
| 0–30 days | 0.0128 | 0.0157 |
| 31–90 days | 0.0467 | 0.0574 |
| 91 days–5 years | 0.0329 | 0.0407 |
Baseline rate of major bleeds/intracerebral haemorrhage (on clopidogrel)
We assumed that bleeding and ICH adverse events do not depend on LOF status, in line with findings from the clinical review (Chapter 4, Results). We did not find any data on bleeding rates in cohort or registry data, and so we relied on evidence from large RCTs which had sufficient major bleed/ICH events for robust estimation. Based on the studies identified in recent network meta-analyses,156,157 by far the largest study reporting bleeding rates on clopidogrel monotherapy is the multicentre global PRoFESS RCT. 158 In the clopidogrel arm of PRoFESS, there were 365 major haemorrhagic events with 25,377.5 person-years follow-up (10,151 patients × 2.5 y mean follow-up), giving a hazard rate of 0.0144 per person-year. The proportion of major haemorrhagic events that were ICH was 103/365 = 0.282, and the proportion of ICH that were fatal was 29/55 = 0.527. We use these estimates for clopidogrel in the model.
Hazard ratios
The baseline event rates described above are assumed to represent patients with NoLOF taking clopidogrel monotherapy. For the model, we need to know the event rates for each treatment option for LOF and NoLOF patients for all treatments in the pathways (i.e. clopidogrel, dipyridamole + aspirin, aspirin and in the scenario analysis ticagrelor).
Ideally, we would have studies comparing the different testing and targeted treatment strategies. Objective 1 of the clinical review (see Chapter 4, Objective 1) searched for comparative studies of targeted testing and treatment strategies; however, we only found two small studies46,47 that had very limited power to estimate relative effects. Furthermore, the targeted treatment strategy varied by number of LOF alleles in Xia et al. 46 which does not align to the testing strategies in our model, and in Lan et al. 47 the targeted treatment strategy is to use aspirin 100 mg/day in LoF patients, which is not used in our base-case model.
An alternative approach is to use results from studies that compare treatment effects for LOF and No LOF patients. Objective 2 of the clinical effectiveness review (see Chapter 4, Objective 2) identified studies that compare the relative efficacy of different treatments for LOF patients. The studies relevant to the treatments in our model are the CHANCE study52 which compares clopidogrel versus aspirin for LOF and No LOF patients, and the CHANCE-2 study50 which compares ticagrelor versus clopidogrel in LOF patients. These two studies have been the main source of relative effects used in previous cost-effectiveness analyses of CYP2C19 testing. 104,109 Objective 2 also identified a phase-II study53 comparing ticagrelor versus clopidogrel by LOF status; however, this study was underpowered for the outcomes of interest for our model, and so we prefer to use results from the much larger phase-III CHANCE-2 study,50 in line with previous models of CYP2C19 testing.
For mortality, there was very limited evidence available, and the estimates that were available were very uncertain. We therefore made the assumption that differences in mortality between the treatments are a result of differences in the proportion of patients having a major stroke (which has a higher mortality rate), and the proportions of patients with a major bleed/ICH of which a proportion are fatal.
To obtain HRs for LOF carriers on clopidogrel relative to NoLOF on clopidogrel, we use the results from Objective 3 using the meta-analysis for any recurrent stroke. We assume that the rate of major bleed/ICH on clopidogrel does not vary with LOF status.
For dipyridamole plus aspirin, no comparative evidence was identified by LOF status (see Figure 6, Objective 2). However, because we do not expect outcomes on dipyridamole plus aspirin to vary by LOF status, we conducted a pragmatic literature search to identify network meta-analyses comparing treatments for secondary prevention of stroke in a general ischaemic stroke/TIA population. We identified two network meta-analyses addressing this question: Greving et al. 156 and Del Giovane et al. 157 These two recent reviews of RCTs identified a single study comparing dipyridamole plus aspirin versus clopidogrel monotherapy, the PRoFESS trial,158 which is a large global trial of 20,095 patients. We use the results from this trial to inform the relative effect of dipyridamole plus aspirin (LOF or NoLOF) relative to clopidogrel NoLOF for recurrent stroke, and major bleed/ICH.
For low-dose aspirin, the CHANCE study,52 identified in Objective 2, gives HRs for aspirin versus clopidogrel monotherapy by LOF status. Our baseline hazards are for NoLOF on clopidogrel, which we wish to estimate HRs against. For aspirin NoLOF, the CHANCE study provides this directly. For aspirin LOF, CHANCE provides a HR for aspirin versus clopidogrel in patients with LOF, HRAsp,LOFvClop,LOF. To estimate a HR for aspirin LOF versus clopidogrel NoLOF patients, we use the relation:
using the HR for LOF versus NoLOF on clopidogrel obtained from Objective 3 (see Figure 8). We assume that the rate of major bleed/ICH does not vary with the LOF status.
For ticagrelor, the CHANCE-2 study,50 identified in Objective 2, gives HRs for ticagrelor versus clopidogrel monotherapy for LOF carriers. We use the same approach as described above using Equation (4) to obtain a HR for ticagrelor LOF versus clopidogrel NoLOF [replacing Asp with Tic in (4)]. We assume that the rate of major bleed/ICH does not vary with LOF status.
The HRs used in the model for each treatment, LOF status and outcome are summarised in Table 24.
| Treatment, LOF status | HR recurrent stroke relative to clopidogrel NoLOF | Source |
|---|---|---|
| Recurrent stroke | ||
| Clopidogrel monotherapy, NoLOF | 1 | – |
| Clopidogrel monotherapy, LOF | 1.46, 95% CI (1.09 to 1.95) | Objective 3 (see Figure 8). |
| Dipyridamole + aspirin, NoLOF | 1.01, 95% CI (0.92 to 1.11) | PRoFESS158 |
| Dipyridamole + aspirin, LOF | 1.01, 95% CI (0.92 to 1.11) | PRoFESS158 |
| Aspirin, no LOF | 1.96, 95% CI (1.33 to 2.857) | CHANCE52 |
| Aspirin, LOF | 1.387, 95% CI (0.8947 to 2.054) | CHANCE52 with HR from Objective 3 (see Figure 8) |
| Ticagrelor, LOF | 1.142, 95% CI (0.7967 to 1.587) | CHANCE-250 with HR from Objective 3 (see Figure 8) |
| Ticagrelor, No LOF | 1.142, 95% CI (0.7967 to 1.587) | Assume equal to LOF |
| Major bleed/ICH | ||
| Clopidogrel monotherapy, LOF or NoLOF | 1 | Assumption that independent of LOF status |
| Aspirin + dipyridamole, LOF or NoLOF | 1.15, 95% CI (1 to 1.32) | PRoFESS158 |
| Aspirin, LOF or NoLOF | 0.637, 95% CI (1.087 to 0.373) | CHANCE52 |
| Ticagrelor, LOF or NoLOF | 0.82, 95% CI (0.34 to 1.98) | CHANCE-250 |
Uptake of targeted treatment and discontinuation rates
We heard from our clinical advisors that only a proportion of patients diagnosed as CYP2C19 LOF may receive targeted treatment for a variety of reasons, such as physician or patient preference, issues with the results not being made available to prescribers, or failure for the test to produce a result. Swen et al. 159 found that physician adoption of pharmacogenetic recommendations was for a range of genes including CYP2C19 was only 69.9%. In our base case, we assume that there is 100% uptake of alternative treatment for patients diagnosed as LOF carriers and vary this in a scenario analysis to 69.9%.
We assumed that patients may switch treatment in the short-term decision-tree model due to side effects. We used data from large RCTs to estimate the discontinuation rates (Table 25) and assume that patients discontinuing switch to aspirin monotherapy.
Health state utilities
Stroke state utilities
Three of the reviewed cost-effectiveness studies accounted for health state utility values according to whether stroke status was disabling or non-disabling;119,143,160 and the other three accounted for health state utility values according to the severity of disability. 104,109,121 Disability categories were mild (mRS 0–1), moderate (mRS 2–3) and severe (mRS 4–5) in the study by Micieli et al.;104 neurologically intact (mRS 0), mild (mRS 1–2) and moderate to severe (mRS 3–5) in the study by Zhou et al.;121 and minor or no disability (mRS 0–2), moderate disability (mRS 3–4) and severe disability (mRS 5) in the study by Cai et al. 109
Because utility studies in this disease area commonly report results by mRS, our preferred approach was to model the utility based on the mRS which could be mapped onto severity of stroke to assign appropriate utilities for the different health states in the model using the categorisation according to categorisation described in decision-tree methods.
A pragmatic literature review identified six studies which reported utility values based on the mRS,126,161–165 of which two were studies on UK patients. The study by Whynes et al. 163 reported EuroQol 5 dimensions (EQ-5D) scores (UK tariff) for 1462 acute stroke patients enrolled on the Efficacy of nitric Oxide in Stroke (ENOS) trial. The study by Rivero-Arias et al. 162 reported EQ-5D scores for 2425 stroke/TIA patients from the Oxford Vascular (OXVASC) observational study. The utility values from the two studies reporting EQ-5D utilities relevant to the UK are presented in Table 26. These values are very similar, but we use the more recent figures from Whynes et al.,163 which is line with the economic evaluation for the SSNAP. 153 For each state in the model, we assume a mRS range according to the categorisation described in decision-tree methods and attribute an average utility over the mRS range from Whynes et al. 163 (see Table 26).
| mRs | Whynes et al.163 utility (se) | Rivero-Arias et al.162 utility (se) |
|---|---|---|
| 0 | 0.93 (0.04) | 0.936 (0.003) |
| 1 | 0.85 (0.03) | 0.817 (0.004) |
| 2 | 0.71 (0.03) | 0.681 (0.004) |
| 3 | 0.55 (0.03) | 0.558 (0.006) |
| 4 | 0.28 (0.03) | 0.265 (0.006) |
| 5 | −0.15 (0.03) | −0.054 (0.005) |
The utilities for the no recurrent stroke state depend on the population, assumed the average of the utilities for mRS 0–1 in the TIA/minor stroke population, and the average of the utilities for mRS 2–3 in the non-minor stroke population.
Major bleed/intracerebral haemorrhage utilities
Two of the reviewed cost-effectiveness studies accounted for bleeds by applying a temporary utility decrement;103,119 and the other three studies accounted for ICH by assigning a health state-specific utility value;104 or allowing for ICH severity by mapping to the mRS, and then using the utility values assigned to stroke severity. 109,121 Cai et al. 109 assume a mRS range of 0–2 for ICH. Micieli et al. 104 estimate a utility of 0.62 for ICH which is a little lower than the utility for TIA/minor stroke in their model, suggesting that ICH corresponds to mRS values of 1–2. Zhou et al. 121 assume a distribution of mRS states (0–5) with an average of 3.4. Because we combine major bleed and ICH, we assume a mRS range of 1–2 in line with Cai et al. 109 and Micieli et al. 104 Major-bleed/ICH therefore has a utility similar to moderate stroke.
Carer disutilities
There can be substantial impact on the quality of life of those caring for patients who have had a stroke, which we included in our model as a utility decrement. None of the cost-effectiveness studies identified in our review included carer quality of life, and so we undertook a pragmatic literature review. Two studies were identified that reported very similar carer utility values. 166,167 The utility reported for 928 caregivers enrolled on structured training programme for caregivers of inpatients after stroke in the TRACS trial was 0.791, 95% CI (0.790 to 0.792). 167 The utility reported for 414 carers enrolled on the Organising Support for Carers of Stroke Survivors (OSCARSS) trial was 0.78, 95% CI (0.75 to 0.81). 166 Assuming that the utility for mRS = 0 is equivalent to that of the general population, the utility decrement for carers is estimated as (0.936 – 0.791) = 0.145 which is applied for one carer per patient who has experienced stroke. This included all patients in the ischaemic stroke population and all patients who experienced a minor, moderate or severe stroke in the TIA population. This meant that patients could be assigned negative QALYs if the carer’s utility decrement was greater than the patients’ health state utility.
Resource use and costs
Medicine costs
Costs of medicines used in the model are sourced from the British National Formulary (BNF) using the cheapest available option, detailed in Appendix 7, Table 56.
Test costs
The assumed costs and resources for the Genedrive and Genomadix Cube POCTs are detailed in Table 27. The per test device cost was obtained by dividing the cost of acquiring and maintaining the device by the estimated number of tests that it will conduct over its lifetime using estimates provided by the companies. We assume that an extended warranty will be taken out to cover device failure and maintenance costs within the extended warranty period. Administration costs per test were estimated by multiplying the staff time required to run a test and record the results by the average hourly rates of the staff involved. The main consumable cost required for each test is the single use test kit which is listed per unit. Periodically control tests are required which incur the cost of a single-use control kit, which we turn into a per test cost by dividing by the number of tests that would be conducted within the period between control tests.
| Genedrive | Genomadix Cube | Source | |
|---|---|---|---|
| POCTs | |||
| Point of Care Device per unit cost (ex. VAT) | £4995 | £3500 | Company |
| Test kit per unit cost (ex. VAT) | £100 | £175 | Company |
| Control kit per unit cost (ex. VAT) | £100 | £50 | Company |
| Warranty annual cost per year (after first 12 months) | £750 (5 years) | £700 (1 year) | Company |
| Device life in number of tests | 6250 (range 5000–7500) |
2000 (at least 1500) |
Company |
| Device lifetime in years | 6 years | 2 years | Company |
| Time to administer test | 10 minutes | Company | |
| Hourly rate of band 5 nurse | £13.67 | NHS Employers costs168 | |
| TOTAL COST | |||
| Costs per test | £104 | £197 | |
| Laboratory tests | Parameter | Source | |
| Device per unit cost (Agena MassARRAY iPlex) | £414,800 (range: £248,880–663,680) |
Xu et al. (2019)169 | |
| Reagent per unit cost | £40 per test | Survey of laboratories (Chapter 4, section survey) | |
| No. of tests per day | 40,000 samples | Svidnicki et al. (2015)170 Le Hellard et al. (2002)171 |
|
| Device lifetime in years | 1 year | Assumption | |
| Time to set up test (band 5 nurse) | 90 minutes | Survey of laboratories (see Chapter 4, section Survey) | |
| Time for analysis of test (band 6 nurse) | 120 minutes | ||
| Time to check and report results (band 7 nurse) | 120 minutes | ||
| Time to process test (band 5 nurse) | 10 minutes | Assumption | |
| Hourly rate of nurse (band 5) | £13.67 | NHS employers costs168 | |
| Hourly rate of nurse (band 6) | £17.00 | ||
| Hourly rate of nurse (band 7) | £21.00 | ||
| TOTAL | |||
| Cost per test | £139 | ||
The clinical review found that staff would require minimal training to conduct POCTs, and given that training costs would be incurred once for each member of staff who will then conduct many tests, the per test training cost would be negligible, and is omitted from our model. The Genomadix Cube requires freezer space, and this is likely to require purchase and maintenance of an appropriate freezer. However, the cost of this is again negligible per test and is omitted from our model.
The total cost per test using the inputs from Table 27 was £104 per Genedrive test, and £197 per Genomadix Cube test. In the absence of estimates of uncertainty around these costs, we assume a Gamma distribution with a standard deviation of 10% of the estimated total cost.
The assumed costs and resource for the laboratory test are detailed in Table 27. In the survey, the preferred platforms for conducting CYP2C19 testing were variant detection using mass spectrometry, for example, MassARRAY (Agena Bioscience) or loop-mediated isothermal amplification (LAMP), for example, LAMP (LaCAR MDx Technologies), with the former having more flexibility to test for multiple variants, and the latter being simpler and quicker to perform. We base our costs on the Agena MassARRAY iPlex and estimate a per test device cost was obtained by dividing the device cost by the estimated number of tests it can conduct over its lifetime. In the absence of information on the device lifetime, we assume a 1-year lifetime, but explore the sensitivity of results of the laboratory test cost in a threshold analysis.
Each test also incurs a reagent cost and staff costs. Most responses to the survey were unable to provide detailed staff time per test, but there was agreement that three staff (band 5, band 6 and band 7) would be involved, and the most detailed response estimated 1.5 hours of band 5 for set-up, 2 hours of band 6 for analysis and 2 hours of band 7 for checking and reporting. We assume these times for laboratory staff, plus an additional cost of a member of hospital staff (band 5) to send the test and process results. In the absence of more detailed information, staff costs for specimen collection were assumed to be equivalent between the three tests. We note that this is likely to represent only a small contribution to the overall test costs.
The total cost for the laboratory test was estimated to be £139 per lab test. In the absence of estimates of uncertainty around these costs, we assume a Gamma distribution with a standard deviation of 10% of the estimated total cost but run a sensitivity (threshold) analysis to the laboratory test cost.
Health state costs
Three of the cost-effectiveness studies reviewed modelled costs specific to the UK,103,119,143 all of which are based the health state costs from the economic burden of stroke in the UK study by Youman et al. 132 This cost-of-illness model estimates the 5-year stroke related formal and informal costs by severity; however, it is now over 20 years old. We therefore searched for and identified the more recent SSNAP study representing all stroke hospitalisations in the UK for 2016. 152 We selected this to be the base case in our model because it is more recent, captures health state costs for both in-hospital stay and out-of-hospital rehabilitation, and provides costs according to severity of stroke. 153 Mean costs are reported over a 1-year period and over a 5-year period post stroke, which allows us to capture differences in short-term and long-term costs following a recurrent stroke in Appendix 7, Table 57.
We use the 1-year costs from SSNAP (see Appendix 7, Table 57) in the first year following stroke, and annualised costs calculated from the 5-year costs from SSNAP (see Appendix 7, Table 57) for subsequent years. Health state costs are calculated according to severity as follows:
-
In the no recurrent stroke state, we assume only rehabilitation costs are incurred which are equal to the social care costs from SSNAP, using NIHSS = 0 for the TIA population and NIHSS = 5–15 for the ischaemic stroke population.
-
In the post-secondary minor stroke state, we include both NHS costs and social care costs, where the NHS costs are for NIHSS = 1–4 for both TIA and ischaemic stroke populations. Social care costs for the TIA/minor stroke population who have had a minor stroke are those for NIHSS = 1–4, whereas for the non-minor stroke who have a minor stroke, the social care costs are those for NIHSS = 15.
-
In the post-secondary moderate stroke (NIHSS = 5–15) and the post-secondary severe stroke (NIHSS = 21–42) states, both NIHS costs and social care costs are applied for the corresponding NIHSS range, and this is the same for both the TIA/minor stroke and non-minor stroke populations.
-
Major bleed/ICH costs were modelled as a single cost on the cycle when the event occurs that applied in addition to the cost of the health state the patient is in. In the absence of more recent data, the cost of a major bleed/ICH was taken from the economic evaluation conducted for NICE TA90. 143
The resulting assumed costs are shown in Table 28 for the first and subsequent years for both modelled populations in 2014 prices.
| Health states | TIA/minor stroke | Non-minor ischaemic stroke |
|---|---|---|
| Annual costs in year 1 | ||
| No secondary event | £4085 | £9741 |
| Post-secondary minor stroke | £15,864 | £19,776 |
| Post-secondary moderate stroke | £26,160 | £26,160 |
| Post-secondary major stroke | £33,445 | £33,445 |
| Post major bleed/ICH (additional single cost when event occurs) | £2010 | £2010 |
| Annual costs in subsequent years | ||
| No secondary event | £2841 | £5994 |
| Post-secondary minor stroke | £6869 | £9015 |
| Post-secondary moderate stroke | £10,154 | £10,154 |
| Post-secondary major stroke | £13,035 | £13,035 |
| Post major bleed/ICH (additional single cost when event occurs) | £2010 | £2010 |
Uncertainty
To reflect uncertainty in model inputs, we conducted probabilistic analysis, where parameter uncertainty is captured with probability distributions and simulation used to estimate expected (mean) costs, expected QALYs and ICERs. The impact of uncertainty is presented using cost-effectiveness planes and cost-effectiveness acceptability curves. One-way sensitivity analyses were performed for all key parameters. Variance in input parameters was taken from the input source where available. Where unavailable, model inputs were varied by a user-defined variation parameter to conduct deterministic and probabilistic sensitivity analysis (PSA) (set to 10% variation in results reported here).
Model validation
The model underwent internal validation by two members of the team not involved in the building of the model, following Büyükkaramikli et al. 172 The validation included face validity tests, checks of model calculations, examination of the model outputs and comparison of results with previous models.
Scenario and sensitivity analyses
We ran a wide range of scenario and sensitivity analyses to test the robustness of the findings to assumptions made in the model. A summary of the scenario analyses is given in Table 29 together with a rationale for each scenario.
| Scenario | Description | Model parameters changed | Rationale for analysis |
|---|---|---|---|
| 1 | Prevalence of clopidogrel resistance | Increased the proportion of patients with LOF variants from 32.1% to 56.8% | Prevalence of LOF variants varies across populations due to differences in ethnicity |
| 2 | Aspirin as Alt Tx for LOF patients | Patients whose test indicates LOF receive aspirin instead of dipyridamole plus aspirin. Costs and HRs for aspirin are used for the alternative treatment | Dipyridamole may not be used due to tolerability issues |
| 3 | Mean age of cohort | Mean age of cohort reduced to 40 and corresponding life-table values used | This is a long-term treatment, and so costs and benefits of targeted treatment may depend on age at index event |
| 4 | Low uptake of alternative therapy after POCT results | A probability 0.699 of receiving alternative treatment for those with LOF test result is applied. Applied to Genomadix Cube only for illustration (but effects would be similar for Genedrive and laboratory tests) | Swen et al.159 found that physician adoption of pharmacogenetic recommendations was for a range of genes including CYP2C19 was only 69.9% |
| 5 | Extended time to lab test results | For the lab test, the time spent on clopidogrel before switching to alternative treatment for LOF patients is varied to 4 weeks | Our survey found that there is a variability between labs in how quickly results are produced, and this can change with capacity |
| 6 | Ticagrelor (following DAPT ticagrelor + aspirin) as Alt Tx for LOF patients | Patients whose test indicates LOF receive ticagrelor (following DAPT ticagrelor + aspirin) instead of dipyridamole plus aspirin. Costs and HRs for ticagrelor are used for the alternative treatment | Ticagrelor has not been approved for use in England and Wales, but it may be used off-label |
| 7 | Early clopidogrel introduction | In the non-minor ischaemic stroke population, clopidogrel treatment begins immediately. LOF carriers can benefit from alternative treatment sooner | Some non-minor ischaemic stroke patients may begin clopidogrel immediately (e.g. if they are already taking aspirin) |
| 8 | Price year 2021 | Prices are inflated to 2021 prices instead of 2022 | High levels of inflation in 2022 may be impactful |
| 9 | Lab-based test costs | The cost of laboratory tests is varied in a threshold analysis | Uncertainty and heterogeneity in lab costs, which may change with changes in infrastructure |
| 10 | Genedrive efficacy analysis | The sensitivity and specificity of the Genedrive test was varied in a threshold analysis. A one-way analysis where sensitivity and specificity were set to the same rate was performed | Limited data were found reporting the efficacy of the Genedrive system in our clinical review |
A summary of inputs to the model, assumed values in the base-case analysis and in sensitivity analyses, distribution used for the probabilistic analysis and source of evidence is given in Table 30.
| Model parameter | Value in base case (sensitivity analysis) | Distribution for probabilistic analysis | Evidence source |
|---|---|---|---|
| Test performance | |||
| POCT: time to receive results | 1 day | N/A | Clinical review (see Appendix 5, Table 50) |
| Lab test: time to receive results | 1 week (4 weeks) |
N/A | Survey (see Chapter 4, Survey) |
| POCT test failure probability | 0.08 | N/A | Meta-analysis of studies identified in clinical review for Genomadix Cube (see Table 14). No data for Genedrive. |
| Lab test failure probability | 0.00 | N/A | Survey (see Chapter 4, Survey) |
| POCT: sensitivity | 99% | 95% CI (94% to 100%) Beta (33 to 0.333) |
Meta-analysis for Genomadix Cube, reduced from 100% to 99% to account for the small proportion of patients with LOF alleles not tested for. No data for Genedrive. |
| POCT: specificity | 100% | 95% CI (99 to 100) Beta (100 to 1.01) |
Meta-analysis for Genomadix Cube. No data for Genedrive. |
| Patient characteristics, prevalence of stroke, TIA and CYP2C19 LOF | |||
| Incidence of first stroke and TIA, proportion TIA | Stroke: 107 per 100,000 TIA: 50 per 100,000 P(TIA) = 31.8% |
N/A | PHE briefing report.148 |
| Patient characteristics: proportion female, mean age | P(female) = 49% Mean age females: 73 years Mean age males: 68.2 years |
N/A | PHE briefing report.173 |
| Prevalence of CYP2C19 LOF | 32.1% (56.8%) | Normal (SE = 10% of rate) |
Ionova et al. (2020)150 and PHE briefing report.148 |
| Baseline event rates (representing NoLOF patients on clopidogrel) | |||
| Stroke recurrence rates (per person-year, ppy) for stroke patients | 0–90 days 0.092 90 + days 0.056 |
Normal distribution 0–90 days (0.075–0.113) 90 + days (0.044–0.072) |
SSNAP152,153 and SLSR Mohan et al. (2009).151 |
| Stroke rates (per person-year, ppy) for TIA patients | 0–90 days 0.0838 90 + days 0.0064 |
Normal (SE = 10% of rate) |
Lioutas et al. (2021)154 |
| Proportion of recurrent stroke by severity | Minor 0.426 Moderate 0.475 Severe 0.0495 |
Dirichlet (43, 48, 10) | SSNAP152,153 |
| Mortality rate by time for mRS 0–3 | 0–30 days 0.0128 31–90 days 0.0467 90 + days 0.0331 |
Normal (SE = 10% of rate) |
SSNAP152,153 and SLSR Mohan et al. (2009).151 |
| Mortality rate by time for mRS 4–5 | 0–30 days 0.0157 31–90 days 0.0574 90 + days 0.0407 |
Normal (SE = 10% of rate) |
SSNAP152,153 and SLSR Mohan et al. (2009).151 |
| Major bleed or ICH (per person-year) | 0.0144 | Normal (SE = 10% of rate) |
PRoFESS trial158 |
| Proportion of major bleed or ICH that is ICH | 0.282 | Normal (SE = 10% of rate) |
PRoFESS trial158 |
| Proportion of ICH which is fatal | 0.527 | Normal (SE = 10% of rate) |
PRoFESS trial158 |
| Relative treatment effects (hazard ratios) all relative to clopidogrel monotherapy, NoLOF | |||
| Recurrent stroke | |||
| Clopidogrel monotherapy, LOF | 1.46 | 95% CI (1.09 to 1.95) | Objective 3 (see Figure 8) |
| Dipyridamole + aspirin, NoLOF | 1.01 | 95% CI (0.92 to 1.11) | PRoFESS158 |
| Dipyridamole + aspirin, LOF | 1.01 | 95% CI (0.92 to 1.11) | PRoFESS158 |
| Aspirin, no LOF | 1.96 | 95% CI (1.33 to 2.857) | CHANCE52 |
| Aspirin, LOF | 1.387 | 95% CI (0.8947 to 2.054) | CHANCE52 with HR from Objective 3 (see Figure 8) |
| Ticagrelor, LOF | 1.142 | 95% CI (0.7967 to 1.587) | CHANCE-250 with HR from Objective 3 (see Figure 8) |
| Ticagrelor, no LOF | 1.142 | 95% CI (0.7967 to 1.587) | Assumed equal to Ticagrelor no LOF |
| Major bleed/ICH | |||
| Clopidogrel monotherapy (LOF or NoLOF) | 1 | 1 | Assumption that independent of LOF status, in line with clinical review (see Chapter 4, Results) |
| Aspirin + dipyridamole (LOF or no LOF) | 1.15 | 95% CI (1 to 1.32) | PRoFESS158 |
| Aspirin (LOF or no LOF) | 0.637 | 95% CI (1.087 to 0.373) | CHANCE52 |
| Ticagrelor, (LOF or no LOF) | 0.82 | 95% CI (0.34 to 1.98) | CHANCE-250 |
| Treatment discontinuation | |||
| Discontinuation probability for clopidogrel | 0.106 | Normal (SE = 10% of rate) | PRoFESS trial158 |
| Discontinuation probability for DAPT dipyridamole + aspirin | 0.164 | Normal (SE = 10% of rate) | PRoFESS trial158 |
| Discontinuation probability for aspirin | 0.147 | Normal (SE = 10% of rate) | SOCRATES113 |
| Discontinuation probability for ticagrelor | 0.175 | Normal (SE = 10% of rate) | SOCRATES113 |
| Utilities | |||
| No secondary events | 0.89 | Normal distribution mean = 0.89, SE = 0.03 | Whynes et al. (2012)163 |
| Post minor stroke – mRS 0–1 | 0.89 | Normal distribution mean = 0.89, SE = 0.03 | Whynes et al. (2012)163 |
| Post major bleed/ICH | 0.62 | Normal distribution mean = 0.62, SE = 0.107 | Micieli et al. (2022)104 |
| Post moderate stroke – mRS 2–3 | 0.63 | Normal distribution mean = 0.63, SE = 0.03 | Whynes et al. (2012)163 |
| Post major stroke – mRS 4–5 | 0.065 | Normal distribution mean = 0.065, SE = 0.03 | Whynes et al. (2012)163 |
| Carer disutility for patients with moderate or major stroke | −0.145 | Normal distribution mean = −0.145, SE = 0.03 | TRACS167 |
| Test costs | |||
| Genedrive per test cost | £104 | Gamma with mean = 104 and SD = 10.4 | Company |
| Genomadix Cube per test cost | £197 | Gamma with mean 197 and SD = 19.7 | Company |
| Laboratory per test cost | £139 | Gamma with mean 139 and SD = 13.9 | Survey (see Chapter 4, Survey) |
| Health state costs (2014 prices) | |||
| Annual health state costs in year 1 (non-minor ischaemic stroke population) | |||
| No secondary event | £9741 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary minor stroke | £19,776 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary moderate stroke | £26,160 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary major stroke | £33,445 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post major bleed/ICH (additional Single cost when event occurs) | £2010 | Gamma with SD = 10% of cost | NICE TA90143 |
| Annual health state costs in year 1 (TIA/minor ischaemic stroke population) | |||
| No secondary event | £4085 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary minor stroke | £15,864 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary moderate stroke | £26,160 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary major stroke | £33,445 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post major bleed/ICH (additional single cost when event occurs) | £2010 | Gamma with SD = 10% of cost | NICE TA90143 |
| Annual health state costs subsequent years (non-minor ischaemic stroke population) | |||
| No secondary event | £5994 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary minor stroke | £9015 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary moderate stroke | £10,154 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary major stroke | £13,035 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post major bleed/ICH (additional single cost when event occurs) | £2010 | Gamma with SD = 10% of cost | NICE TA90143 |
| Annual health state costs subsequent years (TIA/minor ischaemic stroke population) | |||
| No secondary event | £2841 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary minor stroke | £6869 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary moderate stroke | £10,154 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post-secondary major stroke | £13,035 | Gamma with SD = 10% of cost | SSNAP152,153 |
| Post major bleed/ICH (additional single cost when event occurs) | £2010 | Gamma with SD = 10% of cost | NICE TA90143 |
Model results
All results are reported separately for (1) the TIA/minor stroke population and (2) the non-minor ischaemic stroke population. Key summary results are also reported for a mixed TIA/ischaemic stroke population using a weighted average using the proportions of the population in each group in Appendix 9, Figures 36–42, Tables 60–63. Due to the paucity of clinical efficacy data for the Genedrive system, we assumed that sensitivity, specificity and test failure rates are set equivalent to those for the Genomadix Cube. For this reason, the results for Genedrive should be considered illustrative only, and only key summary results are reported for Genedrive. Deterministic base-case results are outlined in the section Deterministic base-case analyses, with deterministic sensitivity analyses reported in the section Deterministic sensitivity analyses. PSA, scenario analyses and diagnostic test cost and accuracy threshold analyses are reported in sections Probabilistic analysis, Scenario analyses and Threshold analyses.
Deterministic base-case analyses
Table 31 shows the fully incremental results for the two populations. Incremental analyses sort comparators by ascending cost and assess the incremental costs and QALYs gained. Fully incremental analysis can be used to calculate the cost-effective strategy between multiple comparators. Overall total costs are lower and total QALYs are higher in the TIA/minor stroke population compared with the non-minor ischaemic stroke population. All laboratory and point-of-care CYP2C19 testing strategies dominated no testing, that is, CYP2C19 testing generated more QALYs and lower costs compared with no testing. Based on these results, Genedrive dominates both laboratory testing and the Genomadix Cube CYP2C19 test; however, the results for Genedrive are based on strong assumptions on accuracy and test performance. Omitting Genedrive, due to insufficient data, the ICER for the Genomadix Cube CYP2C19 test relative to laboratory testing was £42,123, £5023 and £24,387 in the non-minor stroke, TIA/minor stroke and mixed populations, respectively. This suggests that the Genomadix Cube CYP2C19 test is the most cost-effective option in the TIA/minor stroke population where clopidogrel is started sooner, whereas laboratory testing is more cost-effective in the non-minor stroke population.
| Treatments | Total costs £ (discounted) | Total QALYs (discounted) | Strictly dominated | Extendedly dominated | ICER (£) | ||
|---|---|---|---|---|---|---|---|
| vs. Genedrive | vs. Lab test | vs. Genomadix Cube | |||||
| Non-minor ischaemic stroke | |||||||
| POCT: Genedrive | £98,557 | 6.55 | |||||
| Laboratory genetic test | £98,563 | 6.55 | Yes | N/A | Dominated | ||
| POCT: Genomadix Cube | £98,650 | 6.55 | Yes | N/A | Dominated | £42,123 | |
| No test | £100,472 | 6.34 | Yes | N/A | Dominated | Dominated | Dominated |
| Transient ischaemic attack/minor stroke | |||||||
| POCT: Genedrive | £44,864 | 8.66 | |||||
| Laboratory genetic test | £44,936 | 8.65 | Yes | N/A | Dominated | ||
| POCT: Genomadix Cube | £44,957 | 8.66 | Yes | N/A | Dominated | £5023 | |
| No test | £46,005 | 8.58 | Yes | N/A | Dominated | Dominated | Dominated |
Table 32 reports the pairwise comparisons where the cost-effectiveness outcomes of each diagnostic strategy are compared to no testing for both populations. Total QALYs were very similar between the different testing strategies make interpretation of ICERs challenging. For this reason, we compare the CYP2C19 testing strategies in terms of net monetary benefit presented in the pairwise results in Table 32 for a willingness to pay of £20,000 per QALY, preferring tests with the highest net monetary benefit. Positive net monetary benefit indicates the intervention is cost-effective compared with the comparator diagnostic at the chosen willingness-to-pay threshold. In the non-minor ischaemic stroke population, the net monetary benefits were £6159, £6112 and £6066 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. In the TIA/minor stroke population, the expected net monetary benefits were £2737, £2584 and £2644 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. Net monetary benefit is greatest in the non-minor ischaemic stroke population due to the higher event rate in the long term, and hence greater benefit of appropriate treatment in this population. In the combined TIA/ischaemic stroke population, the net monetary benefits were £5069, £4988 and £4976 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. In all populations, net monetary benefit is similar, suggesting little difference between the tests. Omitting Genedrive, where there are insufficient data, the highest expected net benefit is for the laboratory test in the non-minor ischaemic stroke population, and the Genomadix Cube CYP2C19 test in the TIA/minor stroke population.
| Comparator vs. no test | Incremental costs (discounted) | Incremental QALYs (discounted) | ICER | Net monetary benefit |
|---|---|---|---|---|
| Non-minor ischaemic stroke | ||||
| POCT: Genedrive vs. no test | −£1915 | 0.21 | −£9027 | £6159 |
| POCT: Genomadix Cube vs. no test | −£1823 | 0.21 | −£8590 | £6066 |
| Laboratory test vs. no test | −£1909 | 0.21 | −£9084 | £6112 |
| Transient ischaemic attack/minor stroke | ||||
| POCT: Genedrive vs. no test | −£1141 | 0.08 | −£14,306 | £2737 |
| POCT: Genomadix Cube vs. no test | −£1048 | 0.08 | −£13,143 | £2644 |
| Laboratory test vs. no test | −£1069 | 0.08 | −£14,105 | £2584 |
Appendix 8, Tables 58 and 59 show for both populations, the contributions to total costs and total QALYs arising from each branch of the decision tree in the short-term (decision tree), long-term (Markov) components of the model, and in total. Sensitivity and specificity are very high, so the majority of costs and QALYs for POCTs are from TNs (the majority are NoLOF patients) followed by TPs (LOF patients).
Deterministic sensitivity analyses
We ran a deterministic sensitivity analysis by setting each parameter in turn at their lower and upper bounds (Table 30, see Scenario and sensitivity analyses) and reporting the resulting change to the ICER in a tornado plots. Results are reported for the laboratory test and Genomadix Cube versus no test and for both populations in Appendix 10, Figures 43–46. Parameters with an ICER range within £500 of the base-case ICER were excluded for brevity.
In all cases, the base-case ICER changes by < £3000 per QALY. For the non-minor ischaemic stroke population, the parameters with the biggest impact were the health state costs, and the HRs for stroke for aspirin and clopidogrel in LOF patients relative to clopidogrel NoLOF, and mortality rates. For the TIA/minor stroke population, the parameters with the biggest impact were the health state costs, HRs for stroke for aspirin and clopidogrel in LOF patients relative to clopidogrel NoLOF, HR for bleed on aspirin, health state utilities, mortality rates and prevalence of LOF carriers.
Probabilistic analysis
The sensitivity of results to uncertainty in model inputs was explored using a probabilistic analysis where uncertainty around model parameters is described by the distributions outlined in Table 30. The PSAs were run for 5000 iterations in each population (non-minor ischaemic stroke, TIA/minor stroke and mixed). In the mixed population at each iteration, the population was sampled with a probability 0.68 for non-minor ischaemic stroke 0.32 and the TIA/minor stroke population otherwise.
Results are reported in incremental cost-effectiveness planes where the incremental costs are plotted against incremental QALYs for each probabilistic iteration, with a willingness-to-pay threshold of £20,000 per QALY indicated by a dotted line. Points that are south-east of the cost-effectiveness threshold indicate that the intervention is cost-effective against the comparator at that willingness-to-pay threshold. Cost-effectiveness acceptability curves are also presented which show the probability that each testing strategy is cost-effective at each willingness-to-pay threshold assessed.
Incremental cost-effectiveness planes for the laboratory test and Genomadix Cube are reported for the non-minor ischaemic stroke population in Appendix 9, Figures 36 and 37, TIA/minor stroke population (see Figures 38 and 39) and mixed population in (see Appendix 9, Figures 40 and 41). In all cases, the spread of points lies below the willingness-to-pay threshold indicating a high certainty that both tests are cost-effective compared with no testing. Cost-effectiveness acceptability curves are reported for the non-minor ischaemic stroke population (Figure 13), TIA (Figure 14) and mixed populations (see Appendix 9, Figure 42), where Genedrive is excluded due to insufficient data. In all cases, there is a very high probability that one of the testing strategies is cost-effective, with the probability of no testing being cost-effective close to zero across all willingness-to-pay thresholds. There is uncertainty as to whether the laboratory test or Genomadix Cube CYP2C19 test is most cost-effective, but in the willingness-to-pay threshold range of £20,000–30,000, the probably of being most cost-effective is higher for laboratory tests for the non-minor stroke population, and for the Genomadix Cube CYP2C19 test for the TIA/minor stroke population. There is more certainty that the Genomadix Cube CYP2C19 test is most cost-effective in the TIA/minor stroke population, due to impact of the delay in receiving results with laboratory testing.
FIGURE 13.
Cost-effectiveness acceptability curve for the non-minor ischaemic stroke population (excluding Genedrive due to insufficient data).

FIGURE 14.
Cost-effectiveness acceptability curve for the TIA/minor stroke population (excluding Genedrive due to insufficient data).

Cost-effectiveness results from the probabilistic analysis are reported as costs and QALYs averaged over the iterations. Table 33 reports the probabilistic fully incremental analysis for the three populations. As for the deterministic analysis, all laboratory and point-of-care CYP2C19 testing strategies dominated no testing in all populations (i.e. cost less and have higher QALYs). Omitting Genedrive, due to insufficient data, the ICER for the Genomadix Cube CYP2C19 test relative to laboratory testing was £86,272, £10,797 and £46,446 in the non-minor ischaemic stroke population, TIA/minor stroke population and mixed population, respectively. This suggests that the Genomadix Cube CYP2C19 test is the most cost-effective option in the TIA/minor stroke population where clopidogrel is started sooner, whereas laboratory testing is more cost-effective in the non-minor stroke population. Note however that the differences between the tests in QALYs were very small, making the interpretation of the ICERs challenging, so we prefer to compare the tests using expected net monetary benefit.
| Treatments | Total costs £ (discounted) | Total QALYs (discounted) | Strictly dominated | Extendedly dominated | ICER (£) | ||
|---|---|---|---|---|---|---|---|
| vs. Lab test | vs. Genedrive | vs. Genomadix Cube | |||||
| Non-minor ischaemic stroke | |||||||
| Laboratory genetic test | 98,517 | 6.538 | |||||
| POCT: Genedrive | 98,523 | 6.539 | No | No | 5530 | ||
| POCT: Genomadix Cube | 98,616 | 6.539 | No | No | 86,272 | 2,985,232 | |
| No test | 100,450 | 6.324 | Yes | N/A | Dominated | Dominated | Dominated |
| Transient ischaemic attack/minor stroke | |||||||
| POCT: Genedrive | 45,016 | 8.688 | |||||
| Laboratory genetic test | 45,086 | 8.685 | Yes | N/A | Dominated | ||
| POCT: Genomadix Cube | 45,118 | 8.688 | No | No | 3,1432,806 | 10,797 | |
| No test | 46,155 | 8.598 | Yes | N/A | Dominated | Dominated | Dominated |
Table 34 reports the pairwise comparisons from the probabilistic analysis of each CYP2C19 testing strategy compared with no testing. In the non-minor ischaemic stroke population, the expected net monetary benefits were £6230 for Genedrive, £6138 for the Genomadix Cube and £6214 for the laboratory test. In the TIA/minor stroke population, the expected net monetary benefits were £2932 for Genedrive, £2829 for the Genomadix Cube and £2802 for the laboratory test. In the combined TIA/ischaemic stroke population, the expected net monetary benefits were £5211 for Genedrive, £5119 for the Genomadix Cube and £5163 for the laboratory test. In all populations, net monetary benefit is similar, suggesting little difference between the tests. Omitting Genedrive, where there are insufficient data, the highest expected net benefit is for the laboratory test in the non-minor ischaemic stroke population, and the Genomadix Cube CYP2C19 test in the TIA/minor stroke population.
| Comparator vs. no test | Incremental costs (discounted) | Incremental QALYs (discounted) | ICER | Net monetary benefit |
|---|---|---|---|---|
| Non-minor ischaemic stroke | ||||
| POCT: Genedrive vs. no test | −£1927 | 0.215 | −£9118 | £6230 |
| POCT: Genomadix Cube vs. no test | −£1835 | 0.215 | −£8650 | £6138 |
| Laboratory genetic test vs. no test | −£1933 | 0.214 | −£9214 | £6214 |
| Transient ischaemic attack/minor stroke | ||||
| POCT: Genedrive vs. no test | −£1139 | 0.090 | −£12,843 | £2932 |
| POCT: Genomadix Cube vs. no test | −£1036 | 0.090 | −£11,592 | £2829 |
| Laboratory genetic test vs. no test | −£1069 | 0.087 | −£12,472 | £2802 |
Scenario analyses
We ran a number of scenario analyses to assess the robustness of our findings to assumptions made in the model, where the rational for each is outlined in Table 29 (see Scenario and sensitivity analyses). Results from the scenario analyses for the non-minor ischaemic stroke and TIA/minor stroke populations can be found in Tables 35 and 36, respectively. For each scenario, we report the incremental costs, the incremental QALYs, ICERs and net monetary benefit. When compared against no test, the Genomadix Cube CYP2C19 test and laboratory test had a positive net monetary benefit in all scenarios and populations modelled. The overall finding that CYP2C19 testing is cost saving and generates more QALYs compared with no testing was robust in all the scenarios that we explored. The scenarios where CYP2C19 testing was most cost-effective were when prevalence of CYP2C19 LOF was high and for younger cohorts of patients. The scenarios where CYP2C19 testing was least cost-effective were when we assumed that only 69.9% of LOF patients actually receive alternative treatment, and when the alternative treatment was ticagrelor. However, CYP2C19 testing was still cost saving but with a smaller increase in QALYs.
| Genomadix cube vs. no testing | Laboratory test vs. no testing | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Incremental costs (£) (discounted) | Incremental QALYs (discounted) | ICER (£) | Net monetary benefit (£) | Incremental costs (£) (discounted) | Incremental QALYs (discounted) | ICER (£) | Net monetary benefit (£) | ||
| Deterministic base case | −1823 | 0.21 | −8590 | 6066 | −1909 | 0.21 | −9084 | 6112 | |
| 1 | Prevalence of clopidogrel resistance of 56.8% | −2941 | 0.36 | −8204 | 10,111 | −3015 | 0.36 | −8397 | 10,196 |
| 2 | Aspirin as Alt Tx for LOF patients | −1260 | 0.15 | −8227 | 4,322 | −1352 | 0.15 | −8995 | 4358 |
| 3 | Mean age of cohort (including a scenario for young people) – 40 years old | −2553 | 0.34 | −7547 | 9318 | −2645 | 0.34 | −7834 | 9398 |
| 4 | Low uptake of alternative therapy after POCT results | −822 | 0.12 | −7022 | 3162 | ||||
| 5 | Extended time to lab test results | −1879 | 0.21 | −8990 | 6058 | ||||
| 6 | Ticagrelor + aspirin as Alt Tx for LOF patients | −713 | 0.19 | −3771 | 4496 | −760 | 0.19 | −4064 | 4498 |
| 7 | Early clopidogrel introduction | −1818 | 0.21 | −8577 | 6058 | −1901 | 0.21 | −9054 | 6099 |
| 8 | Price year 2021 | −1688 | 0.21 | −7956 | 5932 | −1768 | 0.21 | −8413 | 5971 |
| Genomadix vs. no testing | Laboratory test vs. no testing | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Incremental costs (£) (discounted) | Incremental QALYs (discounted) | ICER (£) | Net monetary benefit | Incremental costs (discounted) | Incremental QALYs (discounted) | ICER | Net monetary benefit | ||
| Deterministic base case | −£1048 | 0.08 | −£13,143 | £2644 | −£1069 | 0.08 | −£14,105 | £2584 | |
| 1 | Prevalence of clopidogrel resistance of 56.8% | −£1296 | 0.12 | −£11,259 | £3598 | −£1305 | 0.11 | −£11,613 | £3551 |
| 2 | Aspirin as Alt Tx for LOF patients | −£914 | 0.07 | −£13,967 | £2223 | −£947 | 0.06 | −£15,500 | £2168 |
| 3 | Mean age of cohort (including a scenario for young people) – 40years old | −£1614 | 0.13 | −£12,851 | £4125 | −£1634 | 0.12 | −£13,395 | £4074 |
| 4 | Low uptake of alternative therapy after POCT results | −£283 | 0.03 | −£9,088 | £907 | – | – | -– | – |
| 5 | Extended time to lab test results | – | – | – | – | −£1014 | 0.07 | −£13,779 | £2485 |
| 6 | Ticagrelor + aspirin as Alt Tx for LOF patients | −£149 | 0.07 | −£2077 | £1584 | −£137 | 0.07 | −£2,026 | £1493 |
| 7 | Early clopidogrel introduction | −£1048 | 0.08 | −£13,143 | £2644 | −£1069 | 0.08 | −£14,105 | £2584 |
| 8 | Price year 2021 | −£971 | 0.08 | −£12,172 | £2567 | −£990 | 0.08 | −£13,063 | £2505 |
Threshold analyses
We conducted two threshold analyses: scenario analyses 9 and 10 as described in methods Table 29. The first threshold analysis varied the laboratory test costs (cost per test) and the other varied the diagnostic accuracy for the Genedrive. For each threshold analysis and population, we plot incremental net monetary benefit relative to the other CYP2C19 testing strategies against different values of the parameter that is varied. All net monetary benefit figures presented here calculated using a £20,000 per QALY willingness-to-pay threshold.
Diagrams presenting the incremental net benefit for the laboratory test compared against the other CYP2C19 testing strategies non-minor ischaemic stroke and TIA/minor stroke populations can be found in Appendix 10, Figures 47 and 48. In the non-minor ischaemic stroke population, the laboratory test was cost-effective versus Genedrive below £29, versus the Genomadix Cube below £184 and versus no test below £6251. In the TIA/minor stroke population, the laboratory test was found to be as cost-effective at cost-per-tests below £79 versus the Genomadix Cube, below £2723 versus no test and was strictly dominated at all costs against the Genedrive test.
Our reviews did not identify any evidence on the sensitivity and specificity of the Genedrive system. We explored the sensitivity of our results to assumptions on this by using a threshold analysis for the test accuracy of the Genedrive system. We varied sensitivity and specificity together in a one-way threshold analysis, with sensitivity and specificity set to the same value, presented in Appendix 10, Figures 49 and 50. In both populations, the Genedrive system was found to be cost-effective at sensitivity and specificity equal and over 99% versus the Genomadix Cube, equal and over 98% versus the lab test.
Summary of findings of economic evaluation
In summary, in our base case for all populations, we found that laboratory and point-of-care CYP2C19 testing strategies dominated no testing, that is, CYP2C19 testing generated more QALYs and lower costs compared with no testing. This finding was robust to the sensitivity and scenario analyses that we conducted. The scenarios where CYP2C19 testing was most cost-effective were when prevalence of CYP2C19 LOF was high and for younger cohorts of patients. The scenarios where CYP2C19 testing was least cost-effective were (1) when we assumed that only 69.9% of LOF patients actually receive alternative treatment due to reluctance to use alternative treatment or due to the test results not being visible to physicians; and (2) when the alternative treatment was ticagrelor. However, CYP2C19 testing was still cost saving in these scenarios, but with a smaller increase in QALYs.
It was challenging to compare the different types of tests due to very small differences in QALYs between the different CYP2C19 testing strategies. Because sensitivity and specificity were so high in our model, the main difference between the CYP2C19 testing strategies was their per-test cost, which suggest that a cost-minimisation approach may be appropriate. The laboratory test has the disadvantage that there could be a delay in receiving test results, and so is less cost-effective than the POCTs in the minor stroke/TIA population where clopidogrel is started immediately. However, we found our results were not sensitive to the time until test results were received. This is because the period of time waiting for test results represents only a small part of the time horizon over which costs and QALYs accrue. We conducted a threshold analysis to the cost of the laboratory test, which provides costs below which the laboratory test is cost-effective compared with the two POCTs for each population.
Due to limited information on Genedrive, we assumed all model inputs were the same as for the Genomadix Cube CYP2C19 test with the exception of the costs, which is why Genedrive appears the more cost-effective of the POCTs. The results for Genedrive should therefore be considered exploratory only and this finding may change as further evidence on the accuracy and performance of Genedrive becomes available. We conducted a threshold analysis for the diagnostic test accuracy of Genedrive, reducing sensitivity and specificity (but keeping them equal). We found that the Genedrive system was found to be cost-effective at sensitivity and specificity equal and over 99% versus the Genomadix Cube, equal and over 98% versus the lab test.
Chapter 6 Assessment of factors relevant to the NHS and other parties
We heard from our clinical advisors that only a proportion of patients diagnosed as CYP2C19 LOF may receive targeted treatment for a variety of reasons, such as physician or patient preference, issues with results not being made available to prescribers or failure for the test to produce a result. Swen et al. 159 found that physician adoption of pharmacogenetic recommendations was only 69.9% for a range of genes including CYP2C19. If CYP2C19 testing is introduced in the NHS, it may need to be accompanied with training and education of staff on the benefits of targeted treatment, and systems in place to ensure that results are visible to those prescribing antiplatelet therapy, to realise the full value for money of testing.
Our survey of laboratories indicated that while facilities are available for CYP2C19 testing, there would be capacity issues to incorporate routine testing into existing workflow. If laboratory testing is adopted, then investment would be required to ensure sufficient capacity. An alternative option that we have not modelled is to consider performing the tests in local labs rather than sending to national laboratories.
The POCTs that we evaluated only detect some CYP2C19 variants, while laboratory testing has the potential to test for more variants as research evolves on the impact of CYP2C19 genetic variants on clopidogrel metabolism. It is also worth noting that if pharmacogenetic panel testing is introduced in the future to test for a range of genetic variants associated with adverse treatment reactions, then the benefits of specific CYP2C19 testing in a TIA/ischaemic stroke population may be diminished.
The Genomadix Cube would require appropriate storage in a freezer which would require an investment in both freezers and space, if adopted.
CYP2C19 LOF is more prevalent in some populations than others, with particularly high prevalence in Asian populations. We found that testing was cost-effective (cost-saving and generating more QALYs than no testing) across the range of prevalence observed for different ethnicities, although the benefits are greater in populations with higher prevalence of CYP2C19 LOF. This should be kept in mind when considering adoption of routine testing for CYP2C19 LOF.
Chapter 7 Discussion
Statement of principal findings
Our results suggest that CYP2C19 testing followed by tailored treatment is likely to be effective and cost-effective in both adult populations modelled (non-minor ischaemic stroke and TIA/minor ischaemic stroke) providing cost savings and increased QALYs compared with not testing. We found this finding was robust to assumptions made in the model. There is uncertainty around the best test to use (laboratory or POCT), and there is no direct evidence for alternative treatment dipyridamole for patients with LOF allele status. There was no evidence in children for any of the objectives.
Evidence identified for Objectives 1–3 suggests that people with LOF alleles are more likely to experience a secondary vascular event and treatments with alternatives to clopidogrel may reduce this risk. Alternative treatments investigated included in the studies were short-term HD clopidogrel (150 mg) followed by long-term aspirin alone, ticagrelor, aspirin and triflusal. Some studies combined these either with an initial (up to 30 day) course of aspirin or with longer-term aspirin. There were no studies of dipyridamole, one of the antiplatelets that is likely to be offered as an alternative to clopidogrel in the UK, if CYP2C19 testing were to be introduced.
We only identified two small studies, both at high RoB, that evaluated a ‘test-and-treat’ strategy (Objective 1). This is the ideal study design to investigate the benefits of introducing genetic testing to identify CYP2C19 LOF alleles. There was a suggestion that testing plus treating based on LOF allele status was associated with a reduced incidence of ischaemic stroke, TIA and a compositive outcome of secondary vascular events, but CIs were wide and overlapped the null.
Objective 2 identified seven studies investigating whether people who have LOF alleles have a reduced risk of secondary vascular occlusive events if treated with alternative antiplatelet therapies compared to treatment with clopidogrel. Four were judged at low RoB, three had concerns regarding the potential for bias due to missing data and lack of information on allocation concealment. There was evidence that ticagrelor was associated with a lower risk of secondary vascular events, including ischaemic stroke, than clopidogrel. One study suggested that ticagrelor was associated with an increased risk of any bleeding event; the other found no difference in the risk of bleeding with ticagrelor compared to clopidogrel. There was no statistical evidence for differences between antiplatelet treatment strategies for any of the other comparisons or bleeding outcomes.
Objective 3 identified 25 studies (20 cohort studies and 5 trials) that compared people with and without LOF alleles, all of whom were treated with clopidogrel (alone or combined with aspirin or other antiplatelet drugs) to see whether the risk of secondary vascular occlusive events differed between groups. Six studies were judged at high RoB due to loss to follow-up that could potentially be related to incidence of vascular events. There was strong evidence that people with LOF alleles treated with clopidogrel (or clopidogrel plus short-term aspirin) have a greater incidence of secondary vascular events (HR 1.72, 95% CI 1.43 to 2.08), stroke (HR 1.46, 95% CI 1.09 to 1.95) and ischaemic stroke (HR 1.99, 95% CI 1.49 to 2.64) than those without LOF alleles. Metaregression suggested that there was evidence of a reduced effect of LOF alleles in patients given a loading dose of clopidogrel compared to those who were not; in patients taking clopidogrel plus long-term aspirin relative to those taking only clopidogrel or clopidogrel plus short-term aspirin; and in studies that included patients with stroke and TIA as primary events compared with only patients with stroke. The stratified analysis based on clopidogrel regimen suggested that there was no evidence of a difference in the risk of secondary vascular events between those taking clopidogrel plus aspirin (HR 0.92; 95% CI 0.50 to 1.74). However, the analyses based on loading dose of clopidogrel and clopidogrel regimen should be considered exploratory as analyses were not pre-specified. There was no difference in the risk of bleeding between those with and without LOF alleles (HR 0.98, 95% CI 0.68 to 1.40).
Objective 4 identified 11 relevant studies evaluating the accuracy of the POCT in scope. All evaluated Spartan versions of the Genomadix Cube – either Spartan Cube, Spartan RX or Spartan FRX, against a laboratory reference standard – there were no studies on the accuracy of Genedrive. All studies were judged at low RoB. None of the studies were conducted in a stroke population. The Genomadix (Spartan) CYP2C19 tests were found to have very high accuracy with summary estimates of sensitivity and specificity both 100% (95% CI 94% to 100% for sensitivity and 99% to 100% for specificity) for the detection of *2 and/or *3 LOF alleles. There were very few disagreements between the Genomadix Cube and laboratory-based reference standards – 8 of the 11 studies reported perfect agreement between the tests. There was no suggestion of a difference across the three different versions of the test evaluated.
Objective 5 identified 17 relevant studies evaluating the technical performance of the two POCTs in scope and conducted a survey of genomic laboratories to gather information on the technical performance of laboratory-based tests. One study evaluated Genedrive and the others all evaluated Genomadix (Spartan) CYP2C19 tests. Only one of the studies was conducted in a stroke population. There was substantial variation in estimates of test failure rate for Genomadix (Spartan) CYP2C19 tests, which ranged from 0.4% to 19% across studies – the true test failure rate is therefore unclear but has the potential to be a large proportion of samples. Some studies provided data on the prevalence of the different variant forms of CYP2C19; however, these were relatively small samples with little information on ethnicity which is a major factor determining the prevalence of LOF alleles. Studies reporting time to results for Genomadix (Spartan) CYP2C19 tests were consistent with the estimate provided by the manufacturer (64 minutes) – most studies reported that time from buccal swab to results was around 1 hour, although two studies reported higher estimates of 90 minutes and 90–120 minutes. One study of Genedrive reported that it gives results in around 40 minutes. Studies generally suggested that Genomadix (Spartan) CYP2C19 tests were simple, user friendly and can be conducted by staff who have received minimal training. Limitations highlighted include storage conditions (samples need to be frozen and stored between −15 and −80 degrees), only one sample can be genotyped at a time, and it only tests for *2, *3 and *17 alleles. The study that evaluated Genedrive, noted that the test is simple, portable, rapid and does not require analytes to be frozen, and tests for *2, *3, *4, *8 and *17 alleles. One study estimated the cost per patient test at 225 Euros for Spartan (Genomadix) RX but did not provide any information on how this estimate was reached. Genedrive and Genomadix provided information on the platform cost, assay cost and cost of external control kits, which were used in our economic model.
The survey of genomic laboratory hubs had an excellent response rate. Of the 10 labs to home the survey was sent, 8 labs, including those in Northern Ireland, Scotland and Wales, completed the survey. All but one of the labs reported that they had at least one form of sequencing technology and all had at least one form of targeted CYP2C19 gene variant detection, most commonly PCR-based SNP genotyping assays using fluorescent reporter systems, such as TaqMan (ThermoFisher). Preferred technologies for performing CYP2C19 testing varied across labs and included: NGS (two labs), MassARRAY (three labs), LAMP (three labs), PCR-based SNP genotyping assays using fluorescent reporter systems, such as TaqMan (ThermoFisher) and QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System (one lab). Resource requirements varied across labs, making it difficult to estimate the exact resources that would be required for CYP2C19 testing, and this may vary according to the specific lab test that would be used. Costs per test varied from around £15 (MassARRAY, although another lab estimated this as £100) to £250 for next-generation gene sequencing. Most labs reported that their preferred test could be performed by existing staff members with standard training or that the test was fully automated, although one lab stated that their preferred test (QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System) would be new to their lab and so training would be required. Most labs reported that it was difficult to estimate the proportion of samples that would not return a valid result, but most expected this to be < 1%. Testing capacity and TATs also varied across labs. Current testing capacity ranged from 0 to 200 tests per week, and TAT ranged from 24 to 72 hours up to 1–2 weeks, although five laboratories estimate that this would be between 72 hours and 1 week. Most laboratories reported that additional testing capacity and faster TAT would be possible with additional resources – including staff, laboratory space, increased automation and equipment. Most labs also confirmed that the test could be performed in local laboratories although additional staff training and/or equipment would be needed. The major barriers to implementing CYP2C19 testing were the scale of the predicted activity and current capacity (four labs), with one highlighting that they do not currently perform any tests of this scale in the NHS and so do not have the infrastructure for this. Most labs stated that while it should be possible to implement POCTs within the laboratory workflow, this may not be the most efficient process for the number of samples that would need to be tested.
We developed a decision-analytic model to evaluate the cost-effectiveness of POCT and laboratory tests compared with no testing (Objective 6), in two populations: (1) TIA/minor ischaemic stroke and (2) non-minor ischaemic stroke; to reflect the different treatment pathways and event rates in these populations. Results were also obtained for a mixed ischaemic stroke and TIA population. We modelled patients moving between five health states: no recurrent stroke, minor stroke, major bleed or ICH, moderate stroke and severe stroke, with a mortality rate depending on health state. A decision tree was used to capture short-term (90-day) outcomes, and a Markov model with a 1-year cycle captured outcomes beyond 90 days over a patient’s lifetime.
In our base case for all populations, we found that testing was cost-effective, with laboratory and point-of-care CYP2C19 testing strategies dominated no testing, that is, CYP2C19 testing generated more QALYs and lower costs compared with no testing. This finding was robust to assumptions made in the model. There was uncertainty, however, on the most cost-effective test (laboratory or POCTs), and it should be noted that results for Genedrive were illustrative only, and conclusions should be focused on comparisons between the laboratory test and the Genomadix Cube CYP2C19 test.
The laboratory test and POCTs gave very similar mean QALYs, and so we compare the tests in terms expected net monetary benefit at a willingness to pay of £20,000 per QALY from our probabilistic analysis, preferring tests with the highest expected net monetary benefit. In the non-minor ischaemic stroke population, the expected net monetary benefits were £6230, £6214 and £6138 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. In the TIA/minor stroke population, the expected net monetary benefits were £2932, £2802 and £2829 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. Net monetary benefit is greatest in the non-minor ischaemic stroke population due to the higher event rate in the long term, and hence greater benefit of appropriate treatment in this population. In the combined TIA/ischaemic stroke population, the expected net monetary benefits were £5211, £5163 and £5119 for Genedrive, the laboratory test and the Genomadix Cube CYP2C19 test, respectively. In all populations, expected net monetary benefit is similar, suggesting little difference between the tests. Omitting Genedrive, where there are insufficient data, the highest expected net benefit is for the laboratory test in the non-minor ischaemic stroke population, and the Genomadix Cube CYP2C19 test in the TIA/minor stroke population.
It should be noted that due to limited information on Genedrive, we assumed all model inputs were the same as for the Genomadix Cube CYP2C19 tests with the exception of the costs, which is why Genedrive appears the more cost-effective of the POCTs. The results for Genedrive should therefore be considered exploratory only and this finding may change as further evidence on the accuracy and performance of Genedrive becomes available. We conducted a threshold analysis for the diagnostic test accuracy of Genedrive, reducing sensitivity and specificity (but keeping them equal). We found that the Genedrive system was found to be cost-effective at sensitivity and specificity equal and over 99% versus the Genomadix Cube, equal and over 98% versus the lab test.
The model inputs that have the biggest impact on the cost-effectiveness results were the costs of the different stroke states, and the treatment effects for stroke in patients with CYP2C19 LOF, and the HR for major bleed/ICH on aspirin relative to clopidogrel. However, varying these parameters did not change the overall finding that CYP2C19 testing is cost saving and generates more QALYs compared with no testing. Accounting for uncertainty in a probabilistic analysis gave very similar results. Cost-effectiveness acceptability curves show that there is a high probability that one of the testing strategies is the most cost-effective, with Genedrive having the highest probability of being cost-effective. The laboratory test has a low probability of being cost-effective in the TIA/minor stroke population due to the delay in receiving results.
The overall finding that CYP2C19 testing is cost saving and generates more QALYs compared with no testing was robust in all the scenarios that we explored. The scenarios where CYP2C19 testing was most cost-effective were when prevalence of CYP2C19 LOF was high and for younger cohorts of patients. The scenarios where CYP2C19 testing was least cost-effective were when we assumed that only 69.9% of LOF patients actually receive alternative treatment, and when the alternative treatment was ticagrelor. However, CYP2C19 testing was still cost saving but with a smaller increase in QALYs.
Strengths and limitations of the assessment
Our systematic review followed published guidance on the conduct of systematic reviews and is reported according to PRISMA-2020 guidance33 and PRISMA-DTA guidance,34 making our review processes transparent and robust. The protocol was pre-registered on the PROSPERO database (CRD42022357661) and published on the NICE website for this appraisal. 1 Following protocol registration, we increased the scope of studies eligible for inclusion for Objective 4 or 5, from diagnostic cohort studies (only) to include primary studies of any design. This increased the scope of potentially eligible studies, and we consider it a strength of the work.
We conducted extensive literature searches designed to maximise retrieval of relevant studies and did not apply any language or date restrictions to these searches. We included attempts to locate ongoing and unpublished studies through searches of trial registries and screening of the manufacturers’ submissions. We defined clear, unambiguous inclusion criteria and documented reasons for exclusion of studies at full-text review and for all studies included in the manufacturers' submissions to ensure transparency in how we applied our inclusion criteria. We conducted a formal assessment of the RoB of included studies using the RoB 2 tool for RCTs,37 the ROBINS-E tool for observational studies38 and the QUADAS-2 tool for diagnostic test accuracy studies. 39 These tools are the most robust tools available for these study designs with a clear focus on the RoB within each study. We modified QUADAS-2 to exclude the assessment of applicability; the other tools do not include an applicability assessment. Instead of a formal assessment of applicability, we extracted details on information that could result in variation across studies and considered this in our synthesis of results. All stages of the review process involved at least two reviewers to minimise the RoB or error in the review. Our synthesis included a meta-analysis for Objectives 2, 3 and 4. For Objective 2, different studies evaluated different intervention comparisons. We investigated the potential to carry out a NMA, but the networks either did not connect or were a simple chain of evidence, and so a NMA would give the same results as an analysis of each comparison pair separately. Most comparisons were made by a single study except for ticagrelor versus clopidogrel and aspirin versus clopidogrel, where we pooled the results from two studies.
For Objective 3, where all studies compared outcomes in those with and without LOF alleles taking clopidogrel, there was some evidence of heterogeneity which we explored using metaregression. However, we did not pre-specify all variables that we later considered potential sources of heterogeneity; analyses based on these variables should be considered exploratory and interpreted with caution. There were too few studies to investigate differences across studies for Objectives 1 and 2. For Objective 4, all studies reported consistently high estimates of the accuracy of Genomadix (Spartan) CYP2C19 tests, and so there was no evidence of heterogeneity in outcomes across studies. We formally assessed the potential for publication bias/small-study effects for Objective 3 and found no evidence to suggest the presence of publication bias. We did not include a formal assessment of publication bias due to the small number of included studies for Objectives 1 and 2, and for Objective 4 due to the difficulties in assessing publication bias for diagnostic test accuracy studies where there is no clear threshold for ‘significance’.
We assessed the accuracy of the two POCTs in scope in relation to laboratory tests which were considered the reference standard for this appraisal, this assumes that they have 100% accuracy. Although there are a variety of laboratory tests available there is no clear ‘reference standard’ amongst these, so it would not be possible to specify a reference standard for evaluation of laboratory-based tests. Instead, we assumed that they all have 100% accuracy, following advice from our clinical experts. Given that we found the Genomadix (Spartan) CYP2C19 tests to have close to 100% accuracy (Objective 4), this assumption appears reasonable. For Objective 5, we reviewed studies of the technical performance for our two POCTs; however, for laboratory-based tests, we took a different approach conducting a survey of genomic laboratories to obtain information on the technical performance of laboratory-based tests. We considered that data obtained by a survey would be specific to our research question, provide information of direct relevance to the NHS and provide the most up-to-date data, which would be unlikely to be the case had we reviewed the existing literature on these tests. The survey of the genomic laboratories had a very high response rate with 8 of the 10 labs invited to participate completing the survey.
While there have been previous economic evaluations of the cost-effectiveness of CYP2C19 testing for targeted antiplatelet treatment for patients following a TIA or ischaemic stroke,103–106 there has only been one previous model in a UK population which compared Genedrive versus no testing. 103 Our model has a similar structure to that of Wright et al.,103 but we model stroke severity state rather than number of strokes experienced, and have used more recent evidence sources as inputs to our model. Our conclusions are in line with those found by Wright et al. 103 for Genedrive, but our model also includes the Genomadix Cube CYP2C19 test and laboratory testing options.
We made a range of assumptions in our model (Objective 6), but our scenario analyses suggest that the main conclusion that CYP2C19 testing is cost saving and generates additional QALYs was robust to these assumptions. The different testing strategies had similar QALYs under our model assumptions, and choice between them is largely one of cost minimisation and ease of implementation. To estimate per test costs, we needed to assume a lifetime of the devices which was based on estimates from Genedrive, Genomadix, and an assumption for the lab test. Some costs were excluded because they become negligible when evaluated per test, including freezers to store the Genomadix Cube CYP2C19 test, and staff training costs, and changes to processes to ensure results are recorded in patient records. However, these would represent an upfront investment if testing is adopted in the NHS. Our survey suggested different laboratories would use different platforms; we used the Agena MassARRAY iPlex as a typical platform when estimating lab test costs, but this cost is likely to vary across laboratories. We conducted a threshold analysis for the cost of the lab test which provides lab costs below which the lab test is cost-effective compared with each POCT and for each population.
In our base case, we assumed that all patients with a LOF test result would receive targeted treatment; however, Swen et al. 159 found that physician adoption of pharmacogenetic recommendations was only 69.9% for a range of genes including CYP2C19. Our scenario analysis suggests that although costs increase and QALYs decrease as uptake falls, CYP2C19 is still cost saving and generates more QALYs compared with no testing.
We assumed that the alternative treatment that CYP2C19 LOF patients would receive is dipyridamole plus aspirin, and that the efficacy of dipyridamole plus aspirin does not depend on CYP2C19 LOF status. This assumption was necessary because we did not identify any studies of dipyridamole plus aspirin in patients with CYP2C19 LOF in our reviews and instead used evidence from the PRoFESS RCT. 158 The CHANCE study52 found that the efficacy of aspirin varied with CYP2C19 LOF status, and so it may be the case that dipyridamole plus aspirin also depends on CYP2C19 LOF status. Further research would be required to assess this.
In the absence of test-and-treat studies, we used RCTs comparing treatments for LOF and NoLOF patients where these were available. For some treatments (aspirin, ticagrelor), relative to clopidogrel, we had information for LOF patients, but no information for NoLOF patients. In these cases, we used the result for LOF patients and then applied a HR for clopidogrel for LOF versus NoLOF estimated from the clinical review (Objective 3). The results were not found to be sensitive to changes in this assumption.
We included treatment discontinuation due to treatment-related adverse events, including major bleeds/ICH. It was assumed that in all cases patients would switch to low-dose aspirin following treatment discontinuation. If patients taking clopidogrel would instead switch to dipyridamole plus aspirin, this may change the cost-effectiveness results, because dipyridamole plus aspirin is more effective at preventing future strokes than aspirin. However, dipyridamole plus aspirin also has a high risk of bleeding events, so it is not clear how this would change the results.
Some patients may not take aspirin either due to drug sensitivity or patient or physician choice, and for these patients who have a CYP2C19 LOF variant the alternative treatment is likely to be dipyridamole monotherapy. We did not explicitly model these patients, but the benefit of testing is likely to be reduced for these patients due to lower efficacy of dipyridamole monotherapy relative to dipyridamole plus aspirin. 129
We incorporated the impact of stroke on carers by applying a utility decrement for carers of patients who had a moderate or major stroke (one carer per patient). We assumed that patients do not require a carer following a minor stroke or TIA. Applying the carer utility decrement to patients in the major stroke state led to negative contributions to QALYs, which we felt was reasonable given the very low quality of life for patients who have had a major stroke.
For all objectives, we took a pragmatic approach where the presence of LOF alleles was considered to be the presence of at least one *2 or *3 LOF allele – these are the two most common LOF alleles, although the prevalence of *3 (0.3%) is much lower than the prevalence of *2 (16%). Genomadix Cube only detects these LOF alleles; Genedrive also detects *4, *8 and *35, and the prevalence of these is very low (0.2% and 0.1%, respectively). Both tests are also able to identify the *17 allele, which is associated with increased function; we did not investigate how this could have impacted on outcomes (Objectives 1–3) or on the accuracy of the test (Objective 4). Laboratory tests could, in principle, detect all LOF alleles. There has been some suggestion that having one LOF allele and one increased function allele (e.g. *2/*17 or *17/*3) could, in effect, cancel each other out and lead to normal function, but there is little evidence to support this. 15 It would be possible to offer alternative treatments to rapid metabolisers, those with one normal function allele and one increased function allele (e.g. *1/*17) or ultrarapid metabolisers, those with two LOF alleles (*17/*17). There is also the potential that having two LOF alleles (e.g. *2/*2 or *2/*3) could lead to poorer metabolism of clopidogrel than having only one LOF allele (e.g. *2/*1 or *1/*2). While some studies included in the review did classify participants into normal (no LOF alleles), intermediate (one LOF allele) and poor (two LOF alleles), in this situation, we combined data across categories to dichotomise into normal and poor (intermediate and poor combined) metabolisers. For example, one of the studies included for Objective 1 applied different treatments for those carrying no LOF alleles, one LOF allele and two LOF alleles. There is potential for considerable complexity in how treatment is tailored based on CYP2C19 testing. In practice, a similar pragmatic approach to the approach that we have taken is most likely to be adopted where individuals are dichotomised into low and normal metabolisers. For example, although the CPIC guidance15 classifies people into five metaboliser categories, treatment is dichotomised so that standard treatment with clopidogrel is recommended for ultrarapid (two increased function alleles), rapid (one increased function alleles and one normal function allele) and normal metabolisers (two normal function alleles); alternative treatment is recommended for poor (two LOF alleles) or IMs (one LOF allele, one normal function allele). 15 A related limitation is that some studies only evaluated the *2 LOF allele and did not look for the presence of *3 mutations. Given the very low prevalence of *3 in most populations, this is unlikely to have had a substantial impact on results.
Our review and model focused on evaluation of the accuracy of POCTs against a laboratory reference standard. It could also be of interest to compare the accuracy of both POCT and laboratory CYP2C19 tests (genotype tests) to P2Y12-pathway specific platelet function tests (phenotype test) as a reference standard, or to consider the potential utility of these tests as an alternative to genetic testing. Platelet function testing of blood samples can assess the response to antiplatelet medication and give information on whether the phenotype (in this case, clopidogrel resistance) is encoded by the gene is expressed. These tests must be performed while a patient is taking clopidogrel. This was considered as part of the scoping process for this appraisal, but it was determined that this was not in scope. Such testing is not currently in use in the NHS, and results can be difficult to interpret and are affected by factors other than the genotype alone, for example, a person’s size. 174 While such tests would potentially have a benefit in that they can predict response to clopidogrel based on both genetic and environmental factors, the prognostic value and clinical utility of platelet reactivity testing are still unclear. 175 Further, the need for patients to have started clopidogrel testing for these tests to be used limits their potential usefulness within the setting of this appraisal.
There were a number of limitations in the evidence base. The ideal study to evaluate the potential impact of a new diagnostic test is a RCT where participants are randomised to either be tested and treated accordingly or to not test and receive standard care. We only identified two ‘test-and-treat’ studies; however, neither of these were randomised and both appeared underpowered to detect differences in secondary vascular events between groups. We therefore needed to draw on less robust types of evidence to determine whether people with LOF alleles have better outcomes if treated with alternative antiplatelet drugs. There were also very FEW data for Objective 2, which looked at whether people with LOF alleles had better outcomes when treated with alternative antiplatelet drugs compared to treatment with clopidogrel. A major limitation for our review was that none of the studies for Objective 1 or 2 evaluated dipyridamole + aspirin, the most likely alternative treatment to clopidogrel that would be offered in the NHS. However, aspirin alone may potentially be considered as an alternative treatment option in the NHS, and we did identify two studies comparing clopidogrel plus aspirin for the first 21/30 days with aspirin alone. These studies suggested that there was no difference in the risk of secondary adverse events between those taking clopidogrel plus aspirin and long-term aspirin alone, suggesting that aspirin alone may be an appropriate treatment option. Furthermore, our scenario analysis found that test and treat with aspirin as an alternative treatment was cost saving and generated more QALYs compared with no-test, although the cost savings and QALY benefits were not as great as when the alternative treatment was dipyridamole plus aspirin. While there was evidence that ticagrelor may reduce secondary vascular events compared to clopidogrel in those with LOF alleles who have had a stroke, this is not currently licensed for stroke patients in the UK. One study also suggested an increased risk of bleeding with ticagrelor compared to clopidogrel, although a second study reported no difference in bleeding risk. We ran a scenario analysis using ticagrelor as an alternative treatment for LOF patients and found that CYP2C19 testing was still cost saving and gained QALYs, but that these were less than for dipyridamole plus aspirin as the alternative treatment. Triflusal was evaluated in two studies – this is not currently a realistic treatment option as it is unavailable in the UK.
There was the lack of consistency in which outcome results were reported by the included studies. We performed meta-analyses of HRs, and estimated HRs and their standard errors (SEs) from event frequencies and follow-up time, or 2 × 2 tables of event numbers,52 where these were N/R. This relies on an assumption of proportional hazards. Although hazards of recurrent ischaemic events are variable and higher early after the primary event, it is plausible that that the ratio of hazards remains proportional. Some studies reporting HRs reported testing this assumption and found no evidence against it. 52,53 We were unable to explore relaxing this assumption based on the data available in the primary study reports. A related, and potentially more significant, limitation is that some studies did not report a mean follow-up time per arm, and so projected follow-up times had to be used instead. For these studies, real follow-up time and time at risk are likely to be lower, and could potentially differ between groups.
For Objective 3, there was some overlap between studies that treated patients with clopidogrel plus long-term aspirin and that included a clopidogrel loading dose, both characteristics found to be associated with improved outcomes in LOF carriers. There were not enough studies to assess if the effects from these interventions were independent from each other.
Uncertainties
Our systematic reviews and economic modelling have identified several areas where uncertainties remain. As highlighted above, for this appraisal, we dichotomised into poor and normal metabolisers based on the presence of at least one *2 or *3 LOF allele. All studies that evaluated the accuracy of POCT evaluated their concordance with a laboratory-based reference standard that was testing for the presence or absence of the same LOF alleles – *2 and/or *3. It is unclear how accuracy would have differed had the laboratory-based reference standard tested for the presence of any LOF allele. As *2 and *3 are the most common LOF alleles, this may have had limited impact on findings. There were insufficient data to consider the impact of testing for other LOF alleles, or to evaluate whether treatment should take a more complex approach with different categories based on different combinations of LOF, normal function and increased function alleles – there are up to five categories that could be considered for this, but the benefit of treating each of these metaboliser categories separately is unclear, and in particular how those with increased function alleles (e.g. *17) should be treated. It is also unclear exactly which alleles should be tested for, while some alleles such as *2, *3, *4 and *8 have been clearly linked to LOF, for others such as *9 and *10 the evidence based is still evolving and these are currently categories as ‘indeterminate’ or ‘likely LOF’. 15 These alleles are also much rarer than some of the other alleles and so the benefit of testing for alleles beyond those most frequently associated with LOF is unclear. This is of particular importance when considering which test would be most appropriate to implement within the NHS. Genomadix Cube only detects *2, *3 and *17, while Genedrive detects *4, *8 and *35 in addition to these alleles. There is more flexibility in which tests can be targeted by laboratory-based tests. Some, such as Sanger sequencing, will detect all variants, others will target specific LOF alleles, although probes can potentially be developed for all alleles of interest.
There have been a number of other reviews looking at the role of CYP2C19 testing to guide antiplatelet treatment. The most recent of these was the review by McDermott et al. published in 2022. 176 This non-systematic review included studies describing the interaction between CYP2C19 genotype and clinical outcomes following ischaemic stroke or TIA. The authors concluded that there was good evidence that CYP2C19 LOF allele carriers of Han Chinese ancestry have increased risk of further vascular events when treated with clopidogrel. This is in line with our findings, and although the majority of our studies (13/25 for Objective 3) were from China, we found that results from these studies were consistent with results from studies conducted in other settings. Most of the observed heterogeneity across studies for Objective 3 was explained by whether a loading dose of clopidogrel was applied and by whether clopidogrel was given alone or in combination with aspirin with a smaller difference seen between LOF and NoLOF carriers when a loading dose was applied and clopidogrel given in combination with aspirin. The McDermott review did not carry out a formal statistical synthesis of results across studies and so was not able to carry out the formal investigation of differences across studies that we conducted as part of our review. It also did not differentiate between the different objectives of the studies in the same way that we have done, which allowed us to draw conclusions both on whether people with LOF alleles have poorer outcomes when treated with clopidogrel compared to those without LOF alleles, and on whether those with LOF alleles have a reduced risk of secondary ischaemic events when treated with an alternative to clopidogrel.
Although our review showed consistently high estimates of accuracy across the included studies, a number of questions remain regarding the accuracy and technical performance of CYP2C19 POCT, including accuracy of Genomadix Cube compared to Spartan versions of the test; accuracy of Genedrive; accuracy in stroke patients; and test failure rate.
All the evaluations that contributed data on Genomadix Cube were actually Spartan versions of the test. Two of these evaluated Spartan Cube; this appears equivalent to Genomadix Cube and was renamed when Spartan Bioscience’s assets were acquired by Genomadix. Other studies evaluated Spartan RX and one study evaluated FRX. The difference between the RX and FRX versions is unclear, but it appears that FRX is either an earlier version of the RX or the test was renamed – the study of the FRX version was earlier than other evaluations. Genomadix as part of their submission to NICE explained that
the original Spartan RX CYP2C19 System and the subsequent Genomadix Cube CYP2C19 system both test for the same CYP2C19 alleles; however, the Genomadix Cube test is not a newer version of the Spartan RX test. Significant differences include the mechanisms used to heat and cool the samples, the storage, use and stability of the specimens on the swab, the optical system, and the test workflow. A direct comparison of the performance of the two systems is not available.
A related NICE methods innovation briefing on Spartan Cube explained that this was released as a successor to Spartan RX and they differ in that ‘the 3 reaction tubes are integrated into a single test cartridge, the swabs and test cartridges are packaged separately, and the DNA analyser device is smaller’. 177 It is therefore unclear whether the performance of the Genomadix Cube can be considered equivalent to that of the Spartan RX; however, there was no suggestion of a difference in performance between the two studies that evaluated Spartan Cube and those that evaluated Spartan RX. Further studies that directly compare the two tests are required to confirm this.
The only data available on Genedrive were one small unpublished early evaluation of the test that did not report data on accuracy. The manufacturer confirmed that there were no studies currently ongoing, but these were planned to start from the first quarter of 2023; no details were provided on what these studies would evaluate. They also highlighted that the Genedrive system has been reviewed by NICE for an alternative assay – the Genedrive MT-RNR1 ID Kit, under the MIB29021 and this is also currently under review as an Early Value Assessment, currently in progress. 21 Genedrive offers some potential benefits compared with Genomadix Cube: it is slightly quicker (40 vs. 60 minutes), cheaper (we estimated the cost at £104 per test for Genedrive and £197 for Genomadix), tests for a greater number of alleles – *8 and *35 in addition to the *2, *3 and *17 that are detected by Genomadix Cube. This may be of particular value in ethnicities such as those with Asian and Jewish origins where these alleles are found at higher frequencies. A further benefit of this test is that it does not require the frozen storage of test kits that is required for Genomadix Cube. It also allows patient data to be uploaded directly into patient records, where data are only stored locally with Genomadix Cube. In the absence of data on accuracy and performance of Genedrive, we assumed these would be the same as for the Genomadix Cube in our economic model, which meant that Genedrive was slightly cheaper with the same benefits as Genomadix. However, these findings are uncertain and may change when data on the accuracy and performance of Genedrive become available.
None of the studies that evaluated the accuracy of the POCT evaluated these tests in our population of interest – people who have had an ischaemic stroke or TIA. While for many tests, the population in which the test is conducted can lead to substantial variation in estimates of accuracy,178 we consider this less likely to be the case for genetic tests. With genetic markers, the LOF alleles are either present or absent and will not be affected by other factors that could influence test performance such as comorbidities, age, sex, setting and disease prevalence.
There was substantial variation in test failure rate across studies – this ranged from 0.4% to 18.9% for studies of Genomadix (Spartan) CYP2C19 tests. Reasons for this substantial variation were unclear and so it is difficult to determine the true failure rate. There was no information on the failure rate of Genedrive. Most laboratories expected their favoured laboratory test for CYP2C19 testing to have a failure rate of < 1%. Failure rate could be an important factor in deciding which CYP2C19 test to implement and so further, accurate data are required on failure rate for all tests, both POCT and laboratory based. Another factor that could influence which test should be recommended for CYP2C19 testing is how test results are recorded in patient records and communicated across settings. Genedrive allows results to be added directly to patient records; this is not possible with the Genomadix test. It is important that the results of CYP2C19 testing are not just available at the point of testing but are recorded in the patient’s record for future reference in case they may need to be prescribed clopidogrel again in the future. It is also important that test results are not lost as patients are discharged from hospital after the test is requested but before results are available. This would reduce the uptake of appropriate targeted treatment, which we found led to a large reduction in the cost savings and QALY benefits of testing in our scenario analyses.
There are proposals to implement pre-emptive panel testing across the NHS. A recent multinational cluster randomised trial of a 12-gene pharmacogenetic panel found that guided treatment based on this panel significantly reduced the incidence of clinically relevant adverse drug reactions and was feasible across different European healthcare systems organisation and settings, including the UK. 159 Panel testing may initially only be introduced in some areas as a pilot scheme but longer term could become standard – this may mean CYP2C19 status is already available for certain patients. A recent report on ‘Personalised Prescribing’ by the RCP and British Pharmacological Society produced a set of recommendations for embedding pharmacogenomics, including CYPC19 testing, in the NHS. 179 It is currently unclear whether and when this will be commissioned in NHS care, but a pilot project, the PROGRESS project, is currently underway. 180 If such a programme were implemented, then it may reduce the appropriateness of specific CYP2C19 testing. However, a potential limitation in implementing any form of pharmacogenetic testing is the potential unwillingness of clinicians to act upon pharmacogenetic test results. The multinational trial of the 12-gene panel found that up to 30% of clinicians did not accept the recommendation of a treatment change based on the genetic test. 159
We did not identify any studies comparing test-and-treat strategies with dipyridamole + aspirin as the alternative treatment, which is the most likely alternative treatment to clopidogrel that would be offered in the NHS, nor did we identify any studies comparing the efficacy of dipyridamole + aspirin compared with clopidogrel according to CYP2C19 LOF status. In order to include dipyridamole + aspirin as the alternative treatment in our model, we assumed that the efficacy of dipyridamole + aspirin does not depend on CYP2C19 LOF status. There is therefore uncertainty as to the costs and benefits of using dipyridamole + aspirin as the alternative treatment. Note, however, that CYP2C19 testing was still found to be cost saving and have increased QALYs compared to no testing in our scenarios where aspirin or ticagrelor were the alternative treatment. This suggests that a test-and-treat strategy with dipyridamole + aspirin as the alternative treatment is very likely to be cost-effective, regardless of the exact relative treatment effects by CYP2C19 LOF status.
Genetic variability is not the only factor to affect the efficacy of clopidogrel. Other factors that may be associated with clopidogrel efficacy include other drugs, for example, PPIs may inhibit clopidogrel,3 smoking status, weight, diabetes, hypertension14 and the influence of other rare genetic variants.
We did not find any evidence in a paediatric population in our searches and did not have sufficient evidence to include children in our economic model. However, in children, aspirin, rather than clopidogrel, is currently recommended to prevent recurrence of stroke, and so in that case, there would not be any benefit of testing for CYP2C19 LOF status. We ran a scenario analysis for a cohort of younger adults at index TIA/ischaemic stroke and found that the cost-saving and quality of life benefits improved. This suggests that if clopidogrel were to be considered for a child, then CYP2C19 testing is likely to be cost-effective.
Equality, diversity and inclusion
Our research was based on existing literature and so we had no control over the participants enrolled. We were broad in our inclusion criteria such that studies from any country and in any language of publication were eligible. CYP2C19 LOF is more prevalent in some populations than others and is particularly high in Asian populations. We explored ethnicity as a potential source of heterogeneity in our analyses for Objective 3, where sufficient data were available to do so, but found that overall there was no association between ethnicity and how those with and without LOF alleles respond to clopidogrel. Our economic analysis found that testing was cost-effective for the range of prevalence observed across ethnicities, suggesting that this would not introduce inequity.
Our survey received a broad response from laboratories across England, Wales, Scotland and Northern Ireland providing the full UK perspective on the potential for providing CYP2C19 testing.
Our team included researchers with a broad range of experience and expertise. The lead authors are junior researchers within Bristol TAG, who were given the opportunity to lead on the writing of this report to help develop their research skills and portfolio. They were supported by the two senior authors, professors and co-directors of Bristol TAG, who provided advice and mentorship to the junior researchers leading on the reviews and health economic modelling. The team included those with expertise in systematic reviews, health economics and medical statistics. We also included two clinical experts from the South West Genomic Medicine Service Alliance who provided clinical expertise, particularly around genetic testing. Where needed, we were able to draw on the broader expertise of the seven specialist DAC members.
Patient and public involvement
Our team included two patient representatives who have lived experience of stroke. The patients attended a meeting near the beginning of the project with the full team to share their experience of stroke and gave suggestions of important outcomes to be considered in the economic model. They also helped to ensure the findings of the review are comprehensible by giving feedback on the plain language summary.
Chapter 8 Conclusions
Implications for service provision
Our results suggest that CYP2C19 testing followed by tailored treatment is likely to be effective and cost-effective in both populations modelled (non-minor ischaemic stroke and TIA/minor ischaemic stroke) providing cost savings and increased QALYs compared with not testing. We found this finding was robust to assumptions made in the model. There is uncertainty around the best test to use (laboratory or POCT), and there is no direct evidence for alternative treatment dipyridamole for patients with LOF allele status. There was very little difference in cost-effectiveness estimates between the three tests. Omitting Genedrive, where there are insufficient data, the highest expected net benefit is for the laboratory test in the non-minor ischaemic stroke population, and the Genomadix Cube CYP2C19 test in the TIA/minor stroke population. The choice of test to be adopted is likely to be based on practical considerations as to which test would be most appropriate and practical within an NHS setting. Our survey of laboratories suggested that, given the high volume of testing required, it may be more appropriate to implement testing through the genomic labs, or in local labs to allow batching of tests; POCTs are only able to run a single test at a time. However, POCTs have the potential advantage of providing results more quickly (< 1 hour compared to around 1 week), which ensures appropriate treatment can be started as soon as possible and reduces the risk of results not being actioned if the patient has been discharged from secondary care. There are logistical difficulties to implementing POCTs in stroke care. The Genomadix Cube would require appropriate storage in a freezer which would require an investment in both freezers and space. Considerations on how results would be integrated into patient’s medical records both for future prescribing where clopidogrel may be considered, but also on the impact of CYP2C19 variants on a wide range of other medicines. Genedrive is able to add results directly to patient records, which is not currently possible for Genomadix Cube. Our survey of laboratories indicated that while facilities are available for CYP2C19 testing, there would be capacity issues to incorporate routine testing into existing workflow. There was also substantial variation in preferred laboratory test for CYP2C19 testing. If laboratory testing is adopted, this would require investment in equipment, staff, laboratory space and automation of processes.
There is some suggestion of reluctance by clinicians to act on pharmacogenetic variations. Implementation of CYP2C19 testing would therefore need to be accompanied by training and education of staff on the benefits of targeted treatment, to ensure that results are acted upon. There is also a question of what should happen to those already on clopidogrel treatment. Consideration needs to be given as to how such patients should be treated – it may be appropriate to test all those currently on clopidogrel treatment and adapt treatment as necessary.
As discussed above, there are proposals for introducing pharmacogenetic panel testing to test for a range of genetic variants associated with adverse treatment reactions. It is currently unclear when and if such programmes would be introduced in the NHS, but a pilot programme is currently in process. If such panel testing is likely to be introduced into the NHS, then the benefits of specific CYP2C19 testing in a TIA/ischaemic stroke population at this point need to be carefully considered.
Suggested research priorities
The section on uncertainties (see Uncertainties) highlights a number of areas where further research is needed. A clear gap in the evidence is on the clinical and cost-effectiveness of alternative antiplatelet strategies, in particular on dipyridamole plus aspirin, in a stroke population with LOF alleles. Further studies are required to determine the true effectiveness of dipyridamole plus aspirin in this population. The ideal study would randomise patients to be tested and treated based on LOF status – those with LOF alleles would receive dipyridamole plus aspirin, those without would receive clopidogrel. Outcomes would then be compared across groups. Such a study could incorporate a full economic analysis and be cluster randomised by hospital.
There is a lack of data on Genedrive. For the health economic model, we assumed that accuracy and test failure rate would be equivalent to that of Genomadix Cube; however, it is unclear how likely this is to be the case. Further test accuracy studies that also provide information on the technical performance of the test (e.g. test failure rate, cost, time to perform the test) are needed; Genedrive highlights that testing will be starting this quarter but details on what studies are proposed are not available. The true test failure rate of Genomadix Cube remains unclear; there was substantial variation in estimates of test failure rate. Further studies are required to determine what the test failure rate would be if the test were to be implemented into routine practice in the NHS.
The value of testing additional alleles beyond *2 and *3 is unclear, as is the clinical significance of the *17 LOF allele. Our review and economic model took a pragmatic approach where we dichotomised action based on testing such that those with at least one LOF allele were considered; however, it remains unclear whether this is the appropriate approach. Future studies should consider how those with *17 LOF allele respond to clopidogrel and whether those with two LOF alleles have worse outcomes than those with one LOF allele. This would inform whether the dichotomy used in our appraisal is the appropriate strategy to implement in practice.
Additional information
Contributions of authors
Joe Carroll (https://orcid.org/0009-0004-7006-0938) developed and coded the health economic model, produced all model results and drafted the sections of the report describing the model, sensitivity and scenario analyses, and cost-effectiveness results. He also drafted the cost-effectiveness section of the report.
Catalina Lopez Manzano (https://orcid.org/0009-0009-8883-0167) led the review of Objectives 1–3. She also drafted sections of the clinical effectiveness report.
Eve Tomlinson (https://orcid.org/0000-0002-0969-602X) led the review of Objectives 4–5, contributed as second reviewer to Objectives 1–3, and checked the literature searches. She also drafted sections of the clinical effectiveness report.
Ayman Sadek (https://orcid.org/0009-0000-6565-0790) conducted the reviews of previous economic evaluations of CYP2C19 testing for clopidogrel resistance and previous models of secondary prevention of recurrent stroke, reviewed evidence for model inputs, contributed to model validation and drafted the cost-effectiveness review and model inputs sections of the report. He also drafted the cost-effectiveness section of the report.
Chris Cooper (https://orcid.org/0000-0003-0864-5607) designed and undertook the literature searches, acted as second reviewer for Objectives 4 and 5 (including review of company submissions), and worked on the review of cost-effectiveness. He drafted the sections of the report related to searching.
Hayley E Jones (https://orcid.org/0000-0002-4265-2854) provided statistical supervision for Objectives 1–4.
Lorraine Rowsell provided a patient perspective on the project and edited the plain language summary.
John Knight provided a patient perspective on the project and edited the plain language summary.
Andrew Mumford (https://orcid.org/0000-0002-5523-511X) provided clinical advice for the project, particularly on genetic testing.
Rachel Palmer (https://orcid.org/0009-0006-6253-3330) provided clinical advice for the project, particularly on genetic testing.
William Hollingworth (https://orcid.org/0000-0002-0840-6254) contributed to model conceptualisation and protocol development.
Nicky J Welton (https://orcid.org/0000-0003-2198-3205) provided oversight of the cost-effectiveness analysis, contributing to model conceptualisation, protocol development, review of previous models, identification of inputs to the model, model validation, interpretation and discussion of results of the cost-effectiveness analysis. She also drafted the cost-effectiveness section of the report.
Penny Whiting (https://orcid.org/0000-0003-1138-5682) provided oversight of the clinical effectiveness sections of report. She drafted the clinical effectiveness sections of the protocol, led the survey of laboratories and contributed to the reviews of effectiveness (Objectives 1–3) and accuracy (Objectives 4–5). She also drafted sections of the clinical effectiveness report.
All authors were involved in commenting on the final report. Penny Whiting is the senior author and guarantor.
Acknowledgements
We would like to thank the following specialist Diagnostic Appraisal Committee (DAC) members for clinical advice relating to this project: Dr Dagan Lonsdale (St George’s, University Of London), Prof Albert Ferro (Kings College London), Dr Daniel Burrage (Whittington Health NHS Trust), Dr Alexander Doney (University Of Dundee), Dr Dheeraj Kalladka (Imperial College Healthcare NHS Trust), Mr Paresh Parmar (London North West University Healthcare NHS Trust) and Dr Salim Elyas (Royal Devon University Healthcare NHS Foundation Trust).
We would also like to thank:
Professor Katherine Payne and Dr Stuart Wright (University of Manchester) for discussions on their health economic model of CY219Y testing.
Dr Cinzia Del Giovane (UniMore, Università degli Studi di Modena e Reggio Emilia) and Dr Irene Tramacere (Fondazione I.R.C.C.S. Istituto Neurologico Carlo Besta) for providing data from their Network Meta-Analysis of antiplatelets for secondary prevention in stroke/TIA patients.
Those that responded to our survey on behalf of each of the five regional genomic laboratory hubs (Central and South; East; North West; South East; and North East and Yorkshire) and the Scottish (NHS North Tayside), Welsh and Irish services.
Charlene Wisdom-Trew, Bristol TAG, for providing administrative support.
Data-sharing statement
All data extracted for the systematic review and the results of the risk of bias assessments are provided in full in the appendices to this report. The economic model can be obtained from the corresponding author and will be shared upon reasonable request for academic collaboration.
Ethics statement
The majority of the research included in this report is secondary research and as such did not require ethical approval. The survey of genomic laboratory hubs was shared with the Health Sciences Faculty Research Ethics Officers from the University of Bristol Research Ethics Committee to determine whether ethical approval was required. They determined that the survey constituted an audit of practice/service evaluation and so ethical review was not required.
Information governance statement
There were no personal data involved in the production of this report.
Disclosure of interests
Full disclosure of interests: Completed ICMJE forms for all authors, including all related interests, are available in the toolkit on the NIHR Journals Library report publication page at https://doi.org/10.3310/PWCB4016.
Primary conflicts of interest: Joe Carroll reports consulting fees from Wickenstones Ltd. Rachel Palmer reports UKCPA Genomics Committee Membership Lead. Nicky J. Welton reports honoraria from ABPI and payment from Takeda. All other authors have no interests to disclose.
Disclaimers
This article presents independent research funded by the National Institute for Health and Care Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, the HTA programme or the Department of Health and Social Care. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, the HTA programme or the Department of Health and Social Care.
References
- Bristol TAG . Clopidogrel Genotype Testing After Ischaemic Stroke or Transient Ischaemic Attack (DAP 65): Final Protocol 2022. www.nice.org.uk/guidance/gid-dg10054/documents/final-protocol (accessed August 2022).
- The National Institute for Health and Care Excellence (NICE) . Stroke and TIA: what causes stroke and TIA? 2022. https://cks.nice.org.uk/topics/stroke-tia/background-information/causes/ (accessed July 2022).
- Zerna C, Thomalla G, Campbell BCV, Rha JH, Hill MD. Current practice and future directions in the diagnosis and acute treatment of ischaemic stroke. Lancet 2018;392:1247-56.
- The National Institute for Health and Care Excellence (NICE) . Stroke and Transient Ischaemic Attack in Over 16s: Diagnosis and Initial Management. NG128 2022. www.nice.org.uk/guidance/ng128/chapter/Context (accessed July 2022).
- Stroke Association . Are You at Risk of Stroke? n.d. www.stroke.org.uk/what-is-stroke/are-you-at-risk-of-stroke (accessed July 2022).
- The King’s Fund . The Health of People from Ethnic Minority Groups in England 2021. www.kingsfund.org.uk/publications/health-people-ethnic-minority-groups-england (accessed 13 July 2022).
- Boysen G, Truelsen T. Prevention of recurrent stroke. Neurol Sci 2000;21:67-72.
- Hankey GJ. Impact of treatment of people with transient ischaemic attacks on stroke incidence and public health. Cerebrovasc Dis 1996;6:26-33.
- Coull AJ, Lovett JK, Rothwell PM. Population based study of early risk of stroke after transient ischaemic attack or minor stroke: implications for public education and organisation of services. BMJ 2004;328.
- The National Institute for Health and Care Excellence (NICE) . Clopidogrel and Modified-Release Dipyridamole for the Prevention of Occlusive Vascular Events 2010. www.nice.org.uk/guidance/ta210/chapter/1-guidance (accessed July 2022).
- European Medicines Agency (EMA) . Plavix (Clopidogrel) n.d. www.ema.europa.eu/en/medicines/human/EPAR/plavix (accessed July 2022).
- Nguyen TA, Diodati JG, Pharand C. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol 2005;45:1157-64.
- Gilard M, Arnaud B, Cornily JC, Le Gal G, Lacut K, Le Calvez G, et al. Influence of omeprazole on the antiplatelet action of clopidogrel associated with aspirin: the randomized, double-blind OCLA (Omeprazole CLopidogrel Aspirin) study. J Am Coll Cardiol 2008;51:256-60.
- Feher G, Koltai K, Alkonyi B, Papp E, Keszthelyi Z, Kesmarky G, et al. Clopidogrel resistance: role of body mass and concomitant medications. Int J Cardiol 2007;120:188-92.
- Lee CR, Luzum JA, Sangkuhl K, Gammal RS, Sabatine MS, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin Pharmacol Ther 2022;112:959-67. https://doi.org/10.1002/cpt.2526.
- Botton MR, Whirl-Carrillo M, Del Tredici AL, Sangkuhl K, Cavallari LH, Agúndez JAG, et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther 2021;109:352-66.
- Pan Y, Chen W, Xu Y, Yi X, Han Y, Yang Q, et al. Genetic polymorphisms and clopidogrel efficacy for acute ischemic stroke or transient ischemic attack: a systematic review and meta-analysis. Circulation 2017;135:21-33.
- National Institute of Health . DbSNP Reference SNP n.d. www.ncbi.nlm.nih.gov/snp/ (accessed February 2023).
- PharmVar. Pharmacogene Variation Consortium . CYP2C19 n.d. www.pharmvar.org/gene/CYP2C19 (accessed February 2023).
- Medicines and Healthcare products Regulatory Agency . Management and Use of IVD Point of Care Test Devices 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/957967/Management_and_use_of_IVD_point_of_care_test_devices.pdf (accessed July 2022).
- National Institute of Health and Care Excellence (NICE) . Genedrive MT RNR1 ID Kit for Detecting a Genetic Variant to Guide Antibiotic Use and Prevent Hearing Loss in Babies: Early Value Assessment 2023. www.nice.org.uk/guidance/indevelopment/gid-dg10057 (accessed February 2023).
- National Institute of Health and Care Excellence (NICE) . Clopidogrel Genotype Testing After Ischaemic Stroke (Provisional Title): Request for Information (Genomadix) 2022.
- Royal College of Physician . National Clinical Guideline for Stroke 2016. www.strokeaudit.org/SupportFiles/Documents/Guidelines/2016-National-Clinical-Guideline-for-Stroke-5t-(1).aspx (accessed July 2022).
- Royal College of Paediatrics and Child Health (RCPCH) . Stroke in Childhood: Clinical Guideline for Diagnosis, Management and Rehabilitation 2021. www.rcpch.ac.uk/sites/default/files/2021-02/Stroke%20guideline%2008.04.19%20updated%202021.pdf (accessed January 2023).
- Johnston SC, Rothwell PM, Nguyen-Huynh MN, Giles MF, Elkins JS, Bernstein AL, et al. Validation and refinement of scores to predict very early stroke risk after transient ischaemic attack. Lancet 2007;369:283-92.
- Dawson J, Merwick A, Webb A, Dennis M, Ferrari J, Fonseca AC, et al. European Stroke Organisation expedited recommendation for the use of short-term dual antiplatelet therapy early after minor stroke and high-risk TIA. Eur Stroke J 2021;6:CLXXXVII-CXCI.
- Kleindorfer DO, Towfighi A, Chaturvedi S, Cockroft KM, Gutierrez J, Lombardi-Hill D, et al. 2021 Guideline for the prevention of stroke in patients with stroke and transient ischemic attack: a guideline from the American Heart Association/American Stroke Association. Stroke 2021;52:e364-467.
- The National Institute for Health and Care Excellence (NICE) . Stroke and TIA: Scenario: Secondary Prevention Following Stroke and TIA 2022. https://cks.nice.org.uk/topics/stroke-tia/management/secondary-prevention-following-stroke-tia/ (accessed July 2022).
- The National Institute for Health and Care Excellence (NICE) . [ID2689] Ticagrelor for Preventing Stroke After Previous Ischaemic Stroke or High-Risk Transient Ischaemic Attack n.d. www.nice.org.uk/guidance/indevelopment/gid-ta10663 (accessed July 2022).
- CRD’s Guidance for Undertaking Reviews in Health Care. Centre for Reviews and Dissemination. York: University of York; 2009.
- Bossuyt PM, Deeks JJ, Leeflang MM, Takwoingi Y, Flemyng E. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy. Oxford: Cochrane; 2022.
- The National Institute for Health and Care Excellence (NICE) . Developing NICE Guidelines: The Manual 2022. www.nice.org.uk/process/pmg36/chapter/how-to-use-this-manual (accessed July 2022).
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021;372.
- Sharifabadi AD, McInnes MDF, Bossuyt PMM. PRISMA-DTA: an extension of PRISMA for reporting of diagnostic test accuracy systematic reviews. Clin Chem 2018;64:985-6.
- Cooper C, Dawson S, Peters J, Varley-Campbell J, Cockcroft E, Hendon J, et al. Revisiting the need for a literature search narrative: a brief methodological note. Res Synth Methods 2018;9:361-5.
- McGowan J, Sampson M, Salzwedel DM, Cogo E, Foerster V, Lefebvre C. PRESS peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol 2016;75:40-6.
- Sterne JAC, Savović J, Page MJ, Elbers RG, Blencowe NS, Boutron I, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. British Medical Journal 2019;366.
- ROBINS-E Development Group . Risk Of Bias In Non-Randomized Studies – of Exposure (ROBINS-E) 2022. www.riskofbias.info/welcome/robins-e-tool (accessed September 2022).
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529-36.
- Dias S, Ades AE, Welton NJ, Jansen JP, Sutton AJ. Network Meta-analysis for Comparative Effectiveness Research. New Jersey: Wiley; 2018.
- Langan D, Higgins JPT, Jackson D, Bowden J, Veroniki AA, Kontopantelis E, et al. A comparison of heterogeneity variance estimators in simulated random-effects meta-analyses. Res Synth Methods 2019;10:83-98.
- Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34.
- Sterne JAC, Sutton AJ, Ioannidis JPA, Terrin N, Jones DR, Lau J, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011;343.
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005;58:982-90.
- Chu H, Cole SR. Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach (author reply). J Clin Epidemiol 2006;59:1331-2.
- Xia C, Zhang Z, He X, Liu J, Li X, Chang Q, et al. Correlation between CYP2C19 gene polymorphism and individualized medication in patients with ischemic stroke. [Chinese]. Chin J Clin Pharmacol Therapeut 2021;26:966-73.
- Lan H, Ying T, Xi-Hua S, Yi L. Anti-platelet therapy in mild cerebral infarction patients on the basis of CYP2C19 metabolizer status. Cell Transplant 2019;28:1039-44.
- Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Effects of triflusal and clopidogrel on the secondary prevention of stroke based on cytochrome P450 2C19 genotyping. J Stroke 2017;19:356-64.
- Meschia JF, Walton RL, Farrugia LP, Ross OA, Ross OA, Elm JJ, et al. Efficacy of clopidogrel for prevention of stroke based on CYP2C19 allele status in the POINT trial. Stroke 2020;51:2058-65.
- Wang Y, Meng X, Wang A, Xie X, Pan Y, Johnston SC, et al. Ticagrelor versus clopidogrel in CYP2C19 loss-of-function carriers with stroke or TIA. N Engl J Med 2021;385:2520-30.
- Wu H, Song H, Dou L, Gao B, Pan Y, Dong M, et al. Effectiveness and safety of high dose clopidogrel plus aspirin in ischemic stroke patients with the single CYP2C19 loss-of-function allele: a randomized trial. BMC Neurol 2020;20.
- Wang YL, Zhao XQ, Lin JX, Li H, Johnston SC, Lin Y, et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. JAMA 2016;316:70-8.
- Chen W, Lin Y, Meng X, Chen G, Wang Z, Wu J, et al. Ticagrelor plus aspirin versus clopidogrel plus aspirin for platelet reactivity in patients with minor stroke or transient ischaemic attack: open label, blinded endpoint, randomised controlled phase II trial. BMJ 2019;365.
- Yi X, Lin J, Zhou J, Wang Y, Huang R, Wang C. The secondary prevention of stroke according to cytochrome P450 2C19 genotype in patients with acute large-artery atherosclerosis stroke. Oncotarget 2018;9:17725-34.
- Díaz Villamarín X . Polimorfismos CYP2C19 Y Abcb1 En El Tratamiento Antiagregante Con Clopidogrel En Pacientes Con Enfermedad Arterial periférica Sometidos a Angioplastia Transluminal percutánea, Ictus O Accidente isquémico Transitorio 2018.
- Fu H, Hu P, Ma C, Peng F, He Z. Association of clopidogrel high on-treatment reactivity with clinical outcomes and gene polymorphism in acute ischemic stroke patients: an observational study. Medicine (Baltimore) 2020;99.
- Fukuma K, Yamagami H, Ihara M, Tanaka T, Miyata T, Miyata S, et al. P2Y12 reaction units and clinical outcomes in acute large artery atherosclerotic stroke: a multicenter prospective study. J Atheroscler Thromb 2022;5:39-55.
- Hoh BL, Gong Y, McDonough CW, Waters MF, Royster AJ, Sheehan TO, et al. CYP2C19 and CES1 polymorphisms and efficacy of clopidogrel and aspirin dual antiplatelet therapy in patients with symptomatic intracranial atherosclerotic disease. J Neurosurg 2016;124:1746-51.
- Lin J, Mo Y, Cai D, Mao D, Fu H, Wei D. CYP2C19 polymorphisms and clopidogrel efficacy in the secondary prevention of ischemic stroke: a retrospective observational study. Ann Palliat Med 2021;10:12171-80.
- Liu G, Yang S, Chen S. The correlation between recurrent risk and CYP2C19 gene polymorphisms in patients with ischemic stroke treated with clopidogrel for prevention. Medicine (Baltimore) 2020;99.
- Lv H, Yang Z, Wu H, Liu M, Mao X, Liu X, et al. High on-treatment platelet reactivity as predictor of long-term clinical outcomes in stroke patients with antiplatelet agents. Transl Stroke Res 2022;13:391-8.
- McDonough CW, McClure LA, Mitchell BD, Gong Y, Horenstein RB, Lewis JP, et al. CYP2C19 metabolizer status and clopidogrel efficacy in the Secondary Prevention of Small Subcortical Strokes (SPS3) study. J Am Heart Assoc 2015;4.
- Ni G, Liang C, Liu K, Cao Y, Zhang H, Tian X, et al. The effects of CES1A2 and CYP2C19 polymorphisms on responsiveness to clopidogrel and clinical outcomes among Chinese patients with acute ischemic stroke. Int J Clin Exp Med 2017;10:3190-6.
- Patel PD, Vimalathas P, Niu X, Shannon CN, Denny JC, Peterson JF, et al. CYP2C19 loss-of-function is associated with increased risk of ischemic stroke after transient ischemic attack in intracranial atherosclerotic disease. J Stroke Cerebrovasc Dis 2021;30.
- Qiu LN, Sun Y, Wang L, Han RF, Xia XS, Liu J, et al. Influence of CYP2C19 polymorphisms on platelet reactivity and clinical outcomes in ischemic stroke patients treated with clopidogrel. Eur J Pharmacol 2015;747:29-35.
- Sen HM, Silan F, Silan C, Degirmenci Y, Kamaran OHI. Effects of CYP2C19 and P2y12 gene polymorphisms on clinical results of patients using clopidogrel after acute ischemic cerebrovascular disease. Balkan J Med Genet 2014;17:37-41.
- Spokoyny I, Barazangi N, Jaramillo V, Rose J, Chen C, Wong C, et al. Reduced clopidogrel metabolism in a multiethnic population: prevalence and rates of recurrent cerebrovascular events. J Stroke Cerebrovasc Dis 2014;23:694-8.
- Sun W, Li Y, Li J, Zhang Z, Zhu W, Liu W, et al. Variant recurrent risk among stroke patients with different CYP2C19 phenotypes and treated with clopidogrel. Platelets 2015;26:558-62.
- Tanaka T, Yamagami H, Ihara M, Miyata T, Miyata S, Hamasaki T, et al. Association of CYP2C19 polymorphisms with clopidogrel reactivity and clinical outcomes in chronic ischemic stroke. Circ J 2019;83:1385-93.
- Tomek A, Mat’oska V, Frydmanova A, Magerova H, Sramek M, Paulasova-Schwabova J, et al. Impact of CYP2C19 polymorphisms on clinical outcomes and antiplatelet potency of clopidogrel in caucasian poststroke survivors. Am J Ther 2018;25:e202-12.
- Tornio A, Flynn R, Morant S, Velten E, Palmer CNA, MacDonald TM, et al. Investigating real-world clopidogrel pharmacogenetics in stroke using a bioresource linked to electronic medical records. Clin Ther 2017;39:281-86.
- Wang Y, Cai H, Zhou G, Zhang Z, Liu X. Effect of CYP2C19*2 and *3 on clinical outcome in ischemic stroke patients treated with clopidogrel. J Neurol Sci 2016;369:216-9.
- Yi X, Wang Y, Lin J, Cheng W, Zhou Q, Wang C. Interaction of CYP2C19, P2Y12, and GPIIIa variants associates with efficacy of clopidogrel and adverse events on patients with ischemic stroke. Clin Appl Thromb Hemost 2017;23:761-8.
- Zhang J, Sun H, Ming T, Liu X, Cong Y, Li F, et al. Association between platelet function and recurrent ischemic vascular events after TIA and minor stroke. Int J Clin Pharmacol Ther 2017;55:789-97.
- Tornio A, Flynn R, Morant S, Velten E, Palmer CNA, MacDonald TM, et al. Investigating real-world clopidogrel pharmacogenetics in stroke using a bioresource linked to electronic medical records. Clin Pharmacol Ther 2018;103:281-6.
- Baudhuin LM, Train LJ, Goodman SG, Lane GE, Lennon RJ, Mathew V, et al. Point of care CYP2C19 genotyping after percutaneous coronary intervention. Pharmacogenomics J 2022;22:303-7.
- Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 2012;379:1705-11.
- Spartan Bioscience Inc . Spartan Cube CYP2C19 Method Comparison Study. NCT04473586 2020. https://ClinicalTrials.gov/show/NCT04473586 (accessed November 2022).
- Spartan Bioscience Inc . Spartan Cube CYP2C19 Inter Laboratory Reproducibilty Study. NCT04473573 2020. https://ClinicalTrials.gov/show/NCT04473573 (accessed November 2022).
- Choi JL, Kim BR, Woo KS, Kim KH, Kim JM, Kim MH, et al. The diagnostic utility of the point-of-care CYP2C19 genotyping assay in patients with acute coronary syndrome dosing clopidogrel: comparison with platelet function test and SNP genotyping. Ann Clin Lab Sci 2016;46:489-94.
- Petrek M, Kocourkova L, Zizkova V, Nosek Z, Taborsky M, Petrkova J. Characterization of three CYP2C19 gene variants by MassARRAY and point of care techniques: experience from a Czech Centre. Arch Immunol Ther Exp (Warsz) 2016;64:99-107.
- So DYF, Wells GA, McPherson R, Labinaz M, Le May MR, Glover C, et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics J 2016;16:71-8.
- Wirth F, Zahra G, Xuereb RG, Barbara C, Fenech A, Azzopardi LM. Comparison of a rapid point-of-care and two laboratory-based CYP2C19*2 genotyping assays for personalisation of antiplatelet therapy. Int J Clin Pharm 2016;38:414-20.
- Spartan Bioscience Inc . Method Comparison Study of the Spartan FRX CYP2C19 Genotyping System Against Bi-Directional Sequencing. NCT01718535 2022. https://ClinicalTrials.gov/show/NCT01718535 (accessed November 2022).
- Petrkova J, Paskova L, Zizkova V, Nosek Z, Taborsky M, Petrek M. POCT for determination of basic pharmacogenetic profile for individualization of antiplatelet therapy: pilot study. Eur Heart J 2014;1.
- Al-Rubaish AM, Al-Muhanna FA, Alshehri AM, Alsulaiman AA, Alabdulali MM, Alkhamis F, et al. Prevalence of CYP2C19*2 carriers in Saudi ischemic stroke patients and the suitability of using genotyping to guide antiplatelet therapy in a university hospital setup. Drug Metab Pers Ther 2021;37:35-40.
- Bergmeijer TO, Vos GJ, Claassens DM, Janssen PW, Harms R, der Heide RV, et al. Feasibility and implementation of CYP2C19 genotyping in patients using antiplatelet therapy. Pharmacogenomics 2018;19:621-8.
- Cavallari LH, Franchi F, Rollini F, Been L, Rivas A, Agarwal M, et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med 2018;16.
- Davis BH, DeFrank G, Limdi NA, Harada S. Validation of the Spartan RXCYP2C19 genotyping assay utilizing blood samples. Clin Transl Sci 2020;13:260-4.
- Franchi F, Rollini F, Rivas J, Rivas A, Agarwal M, Briceno M, et al. Prasugrel versus ticagrelor in patients with CYP2C19 loss-of-function genotypes results of a randomized pharmacodynamic study in a feasibility investigation of rapid genetic testing. JACC Basic Transl Sci 2020;5:419-28.
- Gurbel PA, Bell R, Bliden K, Yazdani S, Taheri H, Akbari M, et al. Bedside testing of CYP2C19 genotype to guide antiplatelet therapy: implementation in the catheterization laboratory. Conference: 67th Annual Scientific Session of the American College of Cardiology and I2 Summit: Innovation in Intervention, ACC 2018;71.
- Tomaniak M, Koltowski L, Kochman J, Huczek Z, Rdzanek A, Pietrasik A, et al. Can prasugrel decrease the extent of periprocedural myocardial injury during elective percutaneous coronary intervention?. Pol Arch Intern Med 2017;127:730-40.
- Zhou Y, Armstead AR, Coshatt GM, Limdi NA, Harada S. Comparison of two point-of-care CYP2C19 genotyping assays for genotype-guided antiplatelet therapy. Ann Clin Lab Sci 2017;47:738-43.
- McDermott JH, Ainsworth S, Wright S, Sen D, Miele G, Smith CJ, et al. Development of a point of care test for CYP2C19 allowing genotype guided antiplatelet prescribing to prevent recurrent ischaemic strokes. Eur J Hum Genet 2020;28.
- Baudhuin L, Train L, Goodman S, Lane G, Lennon R, Mathew V, et al. Validation and performance of point-of-care rapid CYP2C19 genotyping in the Tailor-PCI multicenter international randomized clinical trial. J Am Coll Cardiol 2021;77.
- Bergmeijer TO, Janssen PWA, Schipper JC, Qaderdan K, Ishak M, Ruitenbeek RS, et al. CYP2C19 genotype-guided antiplatelet therapy in ST-segment elevation myocardial infarction patients – rationale and design of the Patient Outcome after primary PCI (POPular) Genetics study. Am Heart J 2014;168:16-22.e1.
- Koltowski L, Tomaniak M, Aradi D, Huczek Z, Filipiak KJ, Kochman J, et al. Optimal antiplatelet pharmacotherapy guided by bedSIDE genetic or functional testing in elective PCI patients: a pilot study: ONSIDE TEST pilot. Cardiol J 2017;24:284-92.
- Koltowski L, Aradi D, Huczek Z, Tomaniak M, Sibbing D, Filipiak KJ, et al. Study design and rationale for optimal antiplatelet pharmacotherapy guided by bedside genetic or functional TESTing in elective percutaneous coronary intervention patients (ONSIDE TEST): a prospective, open-label, randomised parallel-group multicentre trial (NCT01930773). Kardiol Pol 2016;74:372-9.
- Wirth F, Zahra G, Xuereb RG, Barbara C, Fenech A, Azzopardi LM. Comparison between a point-of-care and a laboratory-based CYP2C19 genotyping assay for pharmacist-led personalisation of antiplatelet therapy. Pharmacotherapy 2015;35:282-3.
- Zhou Y, Armstead AR, Coshatt GM, Brott BC, Sankaranarayanan A, Limdi NA, et al. Rapid CYP2C19 genotype testing: comparison between spartan RX CYP2C19 and verigene CYP2C19. J Mol Diagn 2015;17:738-43.
- Office for National Statistics . Ethnic Group, England and Wales: Census 2021 2021. www.ons.gov.uk/peoplepopulationandcommunity/culturalidentity/ethnicity/bulletins/ethnicgroupenglandandwales/census2021#:~:text=the%20%22Asian%2C%20or%20Asian%20British,was%2081.0%25%20(45.8%20million) (accessed January 2023).
- Drummond MF, Sculpher MJ, Torrance GW, O’Brien BJ, Stoddart GL. Methods for the Economic Evaluation of Health Care Programmes. Oxford: Oxford University Press; 2005.
- Wright SJ, McDermott J, Sen D, Smith C, Newman W, Payne K. Development of a Point-of-Care Genetic Test for Effective Treatment of Patients With Ischaemic Stroke and Transient Ischaemic Attacks: An Early Model-Based Cost-Effectiveness Analysis 2022. https://wellcomeopenresearch.org/articles/8-183/v1 (accessed August 2022).
- Micieli A, Singh N, Jahn B, Siebert U, Menon BK, Demchuk AM. Cost effectiveness of testing for CYP2C19 loss-of-function carriers following transient ischemic attack/minor stroke: a Canadian perspective. Int J Stroke 2022;18:416-25.
- Narasimhalu K, Ang YK, Tan DSY, De Silva DA, Tan KB. Cost effectiveness of genotype-guided antiplatelet therapy in Asian ischemic stroke patients: ticagrelor as an alternative to clopidogrel in patients with CYP2C19 loss of function mutations. Clin Drug Investig 2020;40:1063-70.
- Kremers F, Venema E, Cras T, Burke J, Roozenbeek B, Dippel D. Cost-effectiveness of CYP2C19 genotype-guided dual antiplatelet therapy for acute minor ischemic stroke or TIA. Int J Stroke 2021;16.
- The ACTIVE Writing Group on behalf of the ACTIVE Investigators . Clopidogrel plus aspirin versus oral anticoagulation for atrial fibrillation in the Atrial fibrillation Clopidogrel Trial with Irbesartan for prevention of Vascular Events (ACTIVE W): a randomised controlled trial. Lancet 2006;367:1903-12.
- Tengs TO, Wallace A. One thousand health-related quality-of-life estimates. Med Care 2000;38:583-637.
- Cai Z, Cai D, Wang R, Wang H, Yu Z, Gao F, et al. Cost-effectiveness of CYP2C19 genotyping to guide antiplatelet therapy for acute minor stroke and high-risk transient ischemic attack. Sci Rep 2021;11.
- Halkes PH, Gray LJ, Bath PM, Diener HC, Guiraud-Chaumeil B, Yatsu FM, et al. Dipyridamole plus aspirin versus aspirin alone in secondary prevention after TIA or stroke: a meta-analysis by risk. J Neurol Neurosurg Psychiatry 2008;79:1218-23.
- Ministry of Health of the People’s Republic of China . China Statistics Yearbook 2012. https://data.cnki.net/ValueSearch/ (accessed November 2022).
- Pan Y, Wang A, Liu G, Zhao X, Meng X, Zhao K, et al. Cost-effectiveness of clopidogrel-aspirin versus aspirin alone for acute transient ischemic attack and minor stroke. J Am Heart Assoc 2014;3.
- Amarenco P, Albers GW, Denison H, Easton JD, Evans SR, Held P, et al. Efficacy and safety of ticagrelor versus aspirin in acute stroke or transient ischaemic attack of atherosclerotic origin: a subgroup analysis of SOCRATES, a randomised, double-blind, controlled trial. Lancet Neurol 2017;16:301-10.
- National Registry of Diseases Office . Singapore Stroke Registry Annual Report 2016. www.nrdo.gov.sg/docs/librariesprovider3/Publications-Stroke/singapore-stroke-registry-annual-reprot-2016_upload_nrdo_website.pdf?sfvrsn=0 (accessed Novemeber 2022).
- Goh LL, Lim CW, Sim WC, Toh LX, Leong KP. Analysis of genetic variation in CYP450 genes for clinical implementation. PLOS ONE 2017;12.
- Guzauskas GF, Boudreau DM, Villa KF, Levine SR, Veenstra DL. The cost-effectiveness of primary stroke centers for acute stroke care. Stroke 2012;43:1617-23.
- Samsa GP, Reutter RA, Parmigiani G, Ancukiewicz M, Abrahamse P, Lipscomb J, et al. Performing cost-effectiveness analysis by integrating randomized trial data with a comprehensive decision model: application to treatment of acute ischemic stroke. J Clin Epidemiol 1999;52:259-71.
- Kremers F. Cost-Effectiveness of CYP2C19 Genotype-Guided Dual Antiplatelet Therapy for Acute Minor Ischemic Stroke or TIA – Updated Abstract via Correspondence With Author 2022.
- Greenhalgh J, Bagust A, Boland A, Martin Saborido C, Oyee J, Blundell M, et al. Clopidogrel and modified-release dipyridamole for the prevention of occlusive vascular events (review of Technology Appraisal No. 90): a systematic review and economic analysis. Health Technol Assess 2011;15:1-178.
- Office for National Statistics . Office for National Statistics. Population Estimates for the UK, England and Wales, Scotland and Northern Ireland 2019. www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/bulletins/annualmidyearpopulationestimates/mid2018 (accessed March 2022).
- Zhou LW, Kraler L, de Havenon A, Lansberg MG. Cost-effectiveness of cilostazol added to aspirin or clopidogrel for secondary prevention after noncardioembolic stroke. J Am Heart Assoc 2022;11.
- CAPRIE Steering Committee . A randomised, blinded, trial of clopidogrel versus aspirin in patients at risk of ischaemic events (CAPRIE). CAPRIE Steering Committee. Lancet 1996;348:1329-39.
- Baharoglu MI, Cordonnier C, Salman RA-S, de Gans K, Koopman MM, Brand A, et al. Platelet transfusion versus standard care after acute stroke due to spontaneous cerebral haemorrhage associated with antiplatelet therapy (PATCH): a randomised, open-label, phase 3 trial. Lancet 2016;387:2605-13.
- Cucchiara B, Elm J, Easton JD, Coutts SB, Willey JZ, Biros MH, et al. Disability after minor stroke and transient ischemic attack in the POINT trial. Stroke 2020;51:792-9.
- Buskens E, Nederkoorn PJ, Buijs-Van Der Woude T, Mali WPTM, Kappelle LJ, Eikelboom BC, et al. Imaging of carotid arteries in symptomatic patients: cost-effectiveness of diagnostic strategies. Radiology 2004;233:101-12.
- Ali M, MacIsaac R, Quinn TJ, Bath PM, Veenstra DL, Xu Y, et al. Dependency and health utilities in stroke: data to inform cost-effectiveness analyses. Eur Stroke J 2016;2:70-6.
- Diener H-C, Sacco RL, Yusuf S, Cotton D, Ôunpuu S, Lawton WA, et al. Effects of aspirin plus extended-release dipyridamole versus clopidogrel and telmisartan on disability and cognitive function after recurrent stroke in patients with ischaemic stroke in the Prevention Regimen for Effectively Avoiding Second Strokes (PRoFESS) trial: a double-blind, active and placebo-controlled study. Lancet Neurol 2008;7:875-84.
- Halkes PH, van Gijn J, Kappelle LJ, Koudstaal PJ, Algra A. Aspirin plus dipyridamole versus aspirin alone after cerebral ischaemia of arterial origin (ESPRIT): randomised controlled trial. Lancet 2006;367:1665-73.
- Diener HC, Cunha L, Forbes C, Sivenius J, Smets P, Lowenthal A. European Stroke Prevention Study. 2. Dipyridamole and acetylsalicylic acid in the secondary prevention of stroke. J Neurol Sci 1996;143:1-13.
- Boehringer Ingelheim . Submission to NICE to Support the Review of Health Technology Appraisal 90: Clopidogrel and Modified-Release Dipyridamole and Aspirin for the Prevention of Occlusive Vascular Events 2009.
- Sanofi-aventis and Bristol-Myers Squibb . The Use of Clopidogrel Hydrogen Sulphate (Plavix) in Patients With Atherosclerotic Disease: Manufacturer Submission 2009.
- Youman P, Wilson K, Harraf F, Kalra L. The economic burden of stroke in the United Kingdom. PharmacoEcon 2003;21:43-50.
- Clarke P, Gray A, Legood R, Briggs A, Holman R. The impact of diabetes-related complications on healthcare costs: results from the United Kingdom Prospective Diabetes Study (UKPDS Study No. 65). Diabet Med 2003;20:442-50.
- Schleinitz MD, Weiss JP, Owens DK. Clopidogrel versus aspirin for secondary prophylaxis of vascular events: a cost-effectiveness analysis. Am J Med 2004;116:797-806.
- O’Brien CL, Gage BF. Costs and effectiveness of ximelagatran for stroke prophylaxis in chronic atrial fibrillation. JAMA 2005;293:699-706.
- Jansen JP, Pellissier J, Choy EH, Ostor A, Nash JT, Bacon P, et al. Economic evaluation of etoricoxib versus non-selective NSAIDs in the treatment of ankylosing spondylitis in the UK. Curr Med Res Opin 2007;23:3069-78.
- Quinn RR, Naimark DM, Oliver MJ, Bayoumi AM. Should hemodialysis patients with atrial fibrillation undergo systemic anticoagulation? A cost–utility analysis. Am J Kidney Dis 2007;50:421-32.
- Malinina D, Zema C, Sander S, Serebruany V. Cost–effectiveness of antiplatelet therapy for secondary stroke prevention. Expert Rev Pharmacoecon Outcomes Res 2007;7:357-63.
- Johnson ES, Lanes SF, Wentworth CE, Satterfield MH, Abebe BL, Dicker LW. A metaregression analysis of the dose – response effect of aspirin on stroke. Arch Intern Med 1999;159:1248-53.
- Rosamond WD, Folsom AR, Chambless LE, Wang CH, McGovern PG, Howard G, et al. Stroke incidence and survival among middle-aged adults: 9-year follow-up of the Atherosclerosis Risk in Communities (ARIC) cohort. Stroke 1999;30:736-43.
- Burn J, Dennis M, Bamford J, Sandercock P, Wade D, Warlow C. Long-term risk of recurrent stroke after a first-ever stroke. the Oxfordshire Community Stroke Project. Stroke 1994;25:333-7.
- Samsa GP, Bian J, Lipscomb J, Matchar DB. Epidemiology of recurrent cerebral infarction: a medicare claims-based comparison of first and recurrent strokes on 2-year survival and cost. Stroke 1999;30:338-49.
- Jones L, Griffin S, Palmer S, Main C, Orton V, Sculpher M, et al. Clinical effectiveness and cost-effectiveness of clopidogrel and modified-release dipyridamole in the secondary prevention of occlusive vascular events: a systematic review and economic evaluation. Health Technol Assess 2004;8:iii-iv.
- Wolfe CD, Rudd AG, Howard R, Coshall C, Stewart J, Lawrence E, et al. Incidence and case fatality rates of stroke subtypes in a multiethnic population: the South London Stroke Register. J Neurol Neurosurg Psychiatry 2002;72:211-6.
- Antithrombotic Trialists’ Collaboration . Collaborative meta-analysis of randomised trials of antiplatelet therapy for prevention of death, myocardial infarction, and stroke in high risk patients. BMJ 2002;324:71-86.
- Sandercock P, Berge E, Dennis M, Forbes J, Hand P, Kwan J, et al. A systematic review of the effectiveness, cost-effectiveness and barriers to implementation of thrombolytic and neuroprotective therapy for acute ischaemic stroke in the NHS. Health Technol Assess 2002;6:1-112.
- PharmGKB . Gene-Specific Information Tables for CYP2C19 n.d. www.pharmgkb.org/page/cyp2c19RefMaterials (accessed July 2022).
- Amin AM, Sheau Chin L, Teh CH, Mostafa H, Mohamed Noor DA, Abdul Kader M, et al. Pharmacometabolomics analysis of plasma to phenotype clopidogrel high on treatment platelets reactivity in coronary artery disease patients. Eur J Pharm Sci 2018;117:351-6.
- Pilling LC, Turkmen D, Fullalove H, Atkins JL, Delgado J, Kuo CL, et al. Analysis of CYP2C19 genetic variants with ischaemic events in UK patients prescribed clopidogrel in primary care: a retrospective cohort study. BMJ Open 2021;11.
- Ionova Y, Ashenhurst J, Zhan J, Nhan H, Kosinski C, Tamraz B, et al. CYP2C19 allele frequencies in over 2.2 million direct-to-consumer genetics research participants and the potential implication for prescriptions in a large health system. Clin Transl Sci 2020;13:1298-306.
- Mohan KM, Crichton SL, Grieve AP, Rudd AG, Wolfe CDA, Heuschmann PU. Frequency and predictors for the risk of stroke recurrence up to 10 years after stroke: the South London Stroke Register. J Neurol Neurosurg Psychiatry 2009;80:1012-8.
- Sentinel Stroke National Audit Programme (SSNAP) . SSNAP Annual Portfolio for April 2021–March 2022 Admissions and Discharges 2021. www.strokeaudit.org/results/Clinical-audit/National-Results.aspx (accessed November 2022).
- National Guideline Centre & SSNAP . Sentinel Stroke National Audit Program. Cost and Cost-Effectiveness Analysis 2016. www.strokeaudit.org/SupportFiles/Documents/Health-Economics/Health-economic-report-2016.aspx (accessed December 2022).
- Lioutas V-A, Ivan CS, Himali JJ, Aparicio HJ, Leveille T, Romero JR, et al. Incidence of transient ischemic attack and association with long-term risk of stroke. J AMA 2021;325:373-81.
- Office for National Statistics . National Life Tables – Life Expectancy in the UK: 2018 to 2020 2021. www.ons.gov.uk/releases/nationallifetableslifeexpectancyintheuk2018to2020 (accessed January 2022).
- Greving JP, Diener HC, Reitsma JB, Bath PM, Csiba L, Hacke W, et al. Antiplatelet therapy after noncardioembolic stroke. Stroke 2019;50:1812-8.
- Del Giovane C, Boncoraglio GB, Bertu L, Banzi R, Tramacere I. Antiplatelet drugs for secondary prevention in patients with ischemic stroke or transient ischemic attack: a systematic review and network meta-analysis. BMC Neurol 2021;21.
- Sacco RL, Diener HC, Yusuf S, Cotton D, Ounpuu S, Lawton WA, et al. Aspirin and extended-release dipyridamole versus clopidogrel for recurrent stroke. N Engl J Med 2008;359:1238-51.
- Swen JJ, van der Wouden CH, Manson LEN, Abdullah-Koolmees H, Blagec K, Blagus T, et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet 2023;401:347-56.
- Narasimhalu K, Ang YK, Tan D, De Silva D, Tan K. Cost effectiveness of CYP2C19 genetic testing in guiding antiplatelet choice in patients with ischemic stroke – an Asian perspective. Int J Stroke 2020;15.
- Haacke C, Althaus A, Spottke A, Siebert U, Back T, Dodel R. Long-term outcome after stroke: evaluating health-related quality of life using utility measurement. Stroke 2006;37:2218-9.
- Rivero-Arias O, Ouellet M, Gray A, Wolstenholme J, Rothwell PM, Luengo-Fernandez R. Mapping the modified Rankin Scale (mRS) measurement into the generic EuroQol (EQ-5D) health outcome. Med Decis Making 2010;30:341-54.
- Whynes DK, Sprigg N, Selby J, Berge E, Bath PM. Testing for differential item functioning within the EQ-5D. Med Decis Making 2012;33:252-60.
- Wang YL, Pan YS, Zhao XQ, Wang D, Johnston SC, Liu LP, et al. Recurrent stroke was associated with poor quality of life in patients with transient ischemic attack or minor stroke: finding from the CHANCE trial. CNS Neurosci Ther 2014;20:1029-35.
- Wang X, Moullaali TJ, Li Q, Berge E, Robinson TG, Lindley R, et al. Utility-weighted modified Rankin Scale scores for the assessment of stroke outcome: pooled analysis of 20,000+ patients. Stroke 2020;51:2411-7.
- Patchwood E, Woodward-Nutt K, Rhodes SA, Batistatou E, Camacho E, Knowles S, et al. Organising Support for Carers of Stroke Survivors (OSCARSS): a cluster randomised controlled trial with economic evaluation. BMJ Open 2021;11.
- Forster A, Dickerson J, Young J, Patel A, Kalra L, Nixon J. A Cluster Randomised Controlled Trial and Economic Evaluation of a Structured Training Programme for Caregivers of Inpatients After Stroke: The TRACS Trial. Southampton: NIHR Journals Library; 2013.
- NHS Employers . Hourly Pay Scales 2021 22 2021. www.nhsemployers.org/articles/hourly-pay-scales-202122 (accessed January 2023).
- Xu Z, Li Y, Dai S, Liu J, Pan J, Jiang Y. Comparison between MassARRAY and pyrosequencing for CYP2C19 and ABCB1 gene variants of clopidogrel efficiency genotyping. Mol Membr Biol 2019;35:1-8.
- Svidnicki MCCC, Silva-Costa SM, Ramos PZ, dos Santos NZP, Martins FTA, Castilho AM, et al. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX® technology. BMC Med Genet 2015;16.
- Le Hellard S, Ballereau SJ, Visscher PM, Torrance HS, Pinson J, Morris SW, et al. SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi automated method for data storage and analysis. Nucleic Acids Res 2002;30.
- Büyükkaramikli NC, Rutten-van Mölken MPMH, Severens JL, Al M. TECH-VER: a verification checklist to reduce errors in models and improve their credibility. Pharmacoeconomics 2019;37:1391-408.
- Public Health England . Briefing Document: First Incidence of Stroke. Estimates for England 2007 to 2016 2018. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/678444/Stroke_incidence_briefing_document_2018.pdf (accessed January 2023).
- National Institute of Health and Care Excellence (NICE) . Clopidogrel Genotype Testing After Ischaemic Stroke (Provisional Title) Draft Scope 2022.
- Dahabreh IJ, Moorthy D, Lamont LJ, Chen ML, Kent DM, Joseph L. Testing of CYP2C19 Variants and Platelet Reactivity for Guiding Antiplatelet Treatment. Agency for Healthcare Research and Quality (AHRQ); 2013.
- McDermott JH, Leach M, Sen D, Smith CJ, Newman WG, Bath PM. The role of CYP2C19 genotyping to guide antiplatelet therapy following ischemic stroke or transient ischemic attack. Expert Rev Clin Pharmacol 2022;15:811-25.
- National Institute for Health and Care Excellence (NICE) . Spartan RX Point-of-Care CYP2C19 Test to Guide Treatment in Acute Coronary Syndrome 2020. www.nice.org.uk/advice/mib223 (accessed January 2023).
- Whiting PF, Rutjes AWS, Westwood ME, Mallett S. A systematic review classifies sources of bias and variation in diagnostic test accuracy studies. J Clin Epidemiol 2013;66:1093-104.
- Royal College of Physicians, British Pharmacological Society joint working party . Personalised Prescribing: Using Pharmacogenomics to Improve Paqtient Outcomes 2022. www.bps.ac.uk/getmedia/b43a3dca-1bbf-4bff-9379-20bef9349a8c/Personalised-prescribing-full-report.pdf.aspx (accessed February 2023).
- NHS East Genomics . PROGRESS Programme – Pharmacogenomics Roll Out and Gauging Response to Service n.d. www.eastgenomics.nhs.uk/about-us/genomic-medicine-service-alliance/Transformation_Projects/progress-programme-pharmacogenomics-roll-out-and-gauging-response-to-service/ (accessed February 2023).
- Wang Y, Lin Y, Meng X, Chen W, Chen G, Wang Z, et al. Effect of ticagrelor with clopidogrel on high on-treatment platelet reactivity in acute stroke or transient ischemic attack (PRINCE) trial: rationale and design. Int J Stroke 2017;12:321-5.
- Zhou M, Chen W, Pan Y, Lin Y, Meng X, Zhao X, et al. Antiplatelet effect of ticagrelor with aspirin in acute minor stroke and transient ischemic attack stratified by CYP2C19 metabolizer status: subgroup analysis of the PRINCE trial. Aging (Albany, NY) 2020;13:3994-4006.
- Gangnam Severance Hospital . Comparison of Triflusal and Clopidogrel in Secondary Prevention of Stroke Based on the Genotyping. NCT01174693 2015. https://ClinicalTrials.gov/show/NCT01174693 (accessed September 2022).
- Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Protocol for the comparison of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping (MASETRO study): a multicenter, randomized, open-label, parallel-group trial. Int J Stroke 2016;11:485-91.
- Han SW, Park JH. Prevalence of CYP2C19 alleles in the maestro study participants. Eur Stroke J 2017;2.
- Han SW, Park JH, Cheon KY, Lee KY. Verifynow P2Y12 assay with regard to cytochrome P450 2C19 polymorphisms and stroke recurrence. Stroke Conference: American Heart Association American Stroke Association 2018 International Stroke Conference and State-of-the-Science Stroke Nursing Symposium, United States 2018;49.
- Lee KY, Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, et al. Effects of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping. Stroke 2017;48.
- Wang Y, Johnston C, Bath PM, Meng X, Jing J, Xie X, et al. Clopidogrel with aspirin in high-risk patients with acute non-disabling cerebrovascular events II (CHANCE-2): rationale and design of a multicentre randomised trial. Stroke Vasc Neurol 2021;6:280-5.
- Pan Y, Meng X, Jin A, Johnston SC, Li H, Bath PM, et al. Time course for benefit and risk with ticagrelor and aspirin in individuals with acute ischemic stroke or transient ischemic attack who carry CYP2C19 loss-of-function alleles: a secondary analysis of the CHANCE-2 randomized clinical trial. JAMA Neurol 2022;79:739-45.
- Joundi RA. In patients with stroke or TIA and CYP2C19 loss-of-function alleles, ticagrelor vs. clopidogrel reduced 90-d stroke. Ann Intern Med 2022;175.
- Wang A, Meng X, Tian X, Johnston SC, Li H, Bath PM, et al. Bleeding risk of dual antiplatelet therapy after minor stroke or transient ischemic attack. Ann Neurol 2022;91:380-8.
- Beijing Tiantan Hospital . Clopidogrel With Aspirin in High-Risk Patients With Acute Non-Disabling Cerebrovascular Events II. NCT04078737 2021. https://ClinicalTrials.gov/show/NCT04078737 (accessed September 2022).
- Xu J, Wang A, Wangqin R, Mo J, Chen Z, Dai L, et al. Efficacy of clopidogrel for stroke depends on CYP2C19 genotype and risk profile. Ann Neurol 2019;86:419-26.
- Pan Y, Wangqin R, Li H, Meng X, Johnston SC, Simon T, et al. F2R polymorphisms and clopidogrel efficacy and safety in patients with minor stroke or TIA. Neurology 2021;96:e1-9.
- Wu Y, Zhou Y, Pan Y, Zhao X, Liu L, Wang D, et al. Impact of CYP2C19 polymorphism in prognosis of minor stroke or TIA patients with declined eGFR on dual antiplatelet therapy: CHANCE substudy. Pharmacogenomics J 2018;18:713-20.
- Davila-Fajardo CL, Diaz-Villamarin X, Blanquez-Martinez D, Caballero-Romero A, Antunez-Rodriguez A, Perez-Campos M, et al. Implementing CYP2C19 genotyping in our daily clinical practice. Int J Clin Pharm 2018;40:256-7.
- Diaz-Villamarin X, Davila-Fajardo CL, Blanquez-Martinez D, Fernandez-Gomez E, Antunez-Rodriguez A, Raquel AS. CYP2C19 SNP’s influence on clopidogrel response in cerebrovascular disease patients: final results. Eur J Hosp Pharm 2019;26.
- Fukuma K, Yamagami H, Kamiyama K, Enomoto Y, Furui E, Manabe Y, et al. Association between CYP2C19 genetic polymorphisms and clinical outcome in acute atherothrombotic stroke: a sub-analysis of the praise study. Eur Stroke J 2017;2:98-476.
- Han Y, Lv H, Wu H, Liu M, Liu Q, Huo Y, et al. The value of platelet reactivity and genetic polymorphism in predicting long-term clinical outcomes in stroke patients. Circ Res 2019;125:e99-111.
- Simpkins AN, McDonough CW, McClure LA, Mitchell BD, Shuldiner AR, Benavente OR, et al. Secondary stroke prevention with aspirin and clopidogrel in CYP2C19*17 carriers increases risk of major non-CNS bleeding. Stroke Conference: American Heart Association American StrokeAssociation 2019;50.
- Spokoyny I, Barazangi N, Jaramillo V, Rose J, Chen C, Wong C, et al. Reduced CYP219 clopidogrel metabolism in a multiethnic population: prevalance and associated rates of recurrent cerebrovascular events. Neurology Conference: 65th American Academy of Neurology Annual Meeting, San Diego, CA, United States Conference Publication 2013;80.
- National Cerebral and Cardiovascular Center . The Influence of CYP2C19 Polymorphism and Clinical Outcomes in Stroke Patients. NCT02711410 2016. https://ClinicalTrials.gov/show/NCT02711410 (accessed September 2022).
- Doney A, Palmer C, Morant S, Flynn R, MacDonald T. Impact of CYP2C19 genotype in ischaemic stroke patients treated with clopidogrel. Int J Stroke 2015;5:1608-20.
- Wang J, Han M, Kuang J, Tu J, Starcevich K, Gao P, et al. Personalized antiplatelet therapy based on clopidogrel/aspirin resistance tests in acute ischemic stroke and transient ischemic attack: study protocol of a multi-center, single-blinded and randomized controlled trial. Contemp Clin Trials 2021;108.
- Gangnam Severance Hospital . Clopidogrel Preventive Effect Based on CYP2C19 Genotype in Ischemic Stroke. NCT04072705 2022. https://ClinicalTrials.gov/show/NCT04072705 (accessed September 2022).
- Zhang XG, Zhu XQ, Xue J, Li ZZ, Jiang HY, Hu L, et al. Personalised antiplatelet therapy based on pharmacogenomics in acute ischaemic minor stroke and transient ischaemic attack: study protocol for a randomised controlled trial. BMJ Open 2019;9.
Appendix 1 Literature search strategies
Effectiveness and accuracy searches
We used two searches: one for Objectives 1–3 and a separate search for Objectives 4 and 5. The searches were not limited by study design, date of publication, or language. This allowed us to use these searches to identify studies for our review of cost-effectiveness.
Objectives 1, 2 and 3
| Resource | N |
|---|---|
| MEDLINE | 1330 |
| EMBASE | 2334 |
| CENTRAL | 379 |
| CINAHL | 82 |
| CTG | 115 |
| ICTRP | 45 |
| ECONLit | 2 |
| HTA Library | 3 |
| NHS EED | 4 |
| Tufts CEA Register | 44 |
| Total | 4328 |
| - Duplicates | - 1414 |
| To screen | 2914 |
Database: MEDLINE (MEDALL)
Host: Ovid
Data parameters: 1946 to present
Date of search: 10 August 2022
| # | Searches | Results |
|---|---|---|
| 1 | Clopidogrel/ | 9894 |
| 2 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”).ti,ab,kw,rn,tn,dq,dy,cn. | 13,992 |
| 3 | 1 or 2 | 15,893 |
| 4 | Cytochrome P-450 CYP2C19/ | 3314 |
| 5 | (CYP2C19* or cypiic19* or “Cytochrome P-450”).ti,ab,kw,kf. | 20,356 |
| 6 | 4 or 5 | 20,904 |
| 7 | 3 and 6 | 1330 |
Search narrative
Lines 1–2: search for Clopidogrel. Line 1 focuses on the controlled indexing term for Clopidogrel. The/indicates that this is a controlled indexing term. The free-text terms search in the following fields: ti = title; ab = abstract; kw = author keyword; kf = key field; and; ot = original title. The structure in line 2 is the prevailing name of the intervention (clopidogrel), followed by alternate brand names or synonyms (e.g. duoplavin, plavix, zyllt) anatomical therapeutic chemical (ATC) codes or UNII code (A74586SNO7), and the Chemical Abstracts Service (CAS) registry number (“113665-84-2”).
Lines 4–5: search for CYP2C19 Genotype. We have truncated CYP2C19 (using the * marker) to identify CYP2C19*2, CYP2C19*3, and CYP2C9*17, and other alleles.
Line 7 combines the search: Line 3 – terms for Clopidogrel AND Line 6 – terms for CYP2C19
Database: EMBASE
Host: Ovid
Data parameters: 1974 to 9 August 2022
Date of search: 10 August 2022
| # | Searches | Results |
|---|---|---|
| 1 | *clopidogrel/ | 12,222 |
| 2 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”).ti,ab,kw,rn,tn,dq,dy,cn. | 71,596 |
| 3 | 1 or 2 | 71,596 |
| 4 | *cytochrome P450 2C19/ | 2197 |
| 5 | (CYP2C19* or cypiic19* or “Cytochrome P-450”).ti,ab,kw,kf. | 25,516 |
| 6 | 4 or 5 | 25,632 |
| 7 | 3 and 6 | 2324 |
Database: CENTRAL
Host: Wiley interface
Data parameters: Issue 7 of 12, July 2022
Date of search: 10 August 2022
| ID | Query | Results |
|---|---|---|
| #1 | [mh ^Clopidogrel] | 2166 |
| #2 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”):ti,ab,kw | 5907 |
| #3 | #1 or #2 | 5907 |
| #4 | MeSH descriptor: [Cytochrome P-450 CYP2C19] this term only | 362 |
| #5 | (CYP2C19* or cypiic19* or “Cytochrome P-450”):ti,ab,kw | 362 |
| #6 | #4 or #5 | 2496 |
| #7 | #3 AND #6 | 379 |
Database: CINAHL
Host: Ovid
Data parameters: 1981 to present
Date of search: 10 August 2022
| # | Query | Results |
|---|---|---|
| S1 | TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) | 4401 |
| S2 | TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) | 1092 |
| S3 | S1 AND S2 | 273 |
| S4 | S1 AND S2 | 82 |
Database: ClinicalTrials.gov
Host: www.clinicaltrials.gov/ct2/results/refine?show_xprt=Y
Date of search: 10 August 2022
115 Studies found for: (clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR EXPAND[Concept] “R 130964” OR EXPAND[Concept] “R-130964” OR R130964 OR EXPAND[Concept] “SR 25990” OR EXPAND[Concept] “SR-25990” OR SR25990 OR A74586SNO7 OR EXPAND[Concept] “113665-84-2”) AND (CYP2C19 OR cypiic19 OR EXPAND[Concept] “Cytochrome P-450”)
Database: WHO ICTRP
Host: https://trialsearch.who.int/Default.aspx
Date of search: 10 August 2022
(clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (CYP2C19 OR cypiic19 OR “Cytochrome P-450”)
Database: ECONLit
Host: EBSCOhost
Data parameters: 1886 to present
Date of search: 10 August 2022
| # | Query | Results |
|---|---|---|
| S1 | TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) | 8 |
| S2 | TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) | 2 |
| S3 | S1 AND S2 | 2 |
Database: HTA Library
Host: www.crd.york.ac.uk/CRDWeb/HomePage.asp
Date of search: 10 August 2022
(((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or R 130964 or R-130964 or R130964 or SR 25990 or SR-25990 or SR25990 or A74586SNO7 or 113665-84-2)) AND ((CYP2C19* or cypiic19* or Cytochrome P-450))) and (Project record:ZDT OR Full publication record:ZDT) IN HTA
Database: NHS EED
Host: www.crd.york.ac.uk/CRDWeb/HomePage.asp
Date of search: 10 August 2022
(((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or R 130964 or R-130964 or R130964 or SR 25990 or SR-25990 or SR25990 or A74586SNO7 or 113665-84-2)) AND ((CYP2C19* or cypiic19* or Cytochrome P-450))) and ((Economic evaluation:ZDT and Bibliographic:ZPS) OR (Economic evaluation:ZDT and Abstract:ZPS)) IN NHS EED
Database: Tufts CEA Register
Host: https://cear.tuftsmedicalcenter.org/about
Date of search: 10 August 2022
Methods
Clopidogrel AND CYP2C19 n = 20
Ratios
Clopidogrel AND CYP2C19 n = 9
Utilities
Clopidogrel AND CYP2C19 n = 15
Objectives 4 and 5
| Resource | N |
|---|---|
| MEDLINE | 92 |
| EMBASE | 296 |
| CENTRAL | 51 |
| CINAHL | 8 |
| CTG | 93 |
| ICTRP | 13 |
| ECONLit | 0 |
| HTA Library | 2 |
| NHS EED | 0 |
| Tufts CEA Register | 0 |
| Total | 555 |
| - Duplicates | -107 |
| To screen | 448 |
Database: MEDLINE (MEDALL)
Host: Ovid
Data parameters: 1946 to present
Date of search: 11 August 2022
| # | Searches | Results |
|---|---|---|
| 1 | Point-of-Care Testing/ | 3652 |
| 2 | ((Point of Care adj2 test*) or POCT).ti,ab,kf,kw. | 9296 |
| 3 | (Genomadix* or Genedrive or Spartan).ti,ab,hw,kf,kw. | 340 |
| 4 | 1 or 2 or 3 | 11,544 |
| 5 | Cytochrome P-450 CYP2C19/ | 3312 |
| 6 | (CYP2C19* or cypiic19* or “Cytochrome P-450”).ti,ab,kw,kf. | 20,353 |
| 7 | 5 or 6 | 20,901 |
| 8 | Clopidogrel/ | 9890 |
| 9 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”).ti,ab,kw,kf. |
13,984 |
| 10 | 8 or 9 | 15,885 |
| 11 | 4 and (7 or 10) | 92 |
Search narrative
Line 1: focuses on the controlled indexing term for Point of Care Testing. The/indicates that this is a controlled indexing term. The free-text terms (Line 2 or Line 3) search in the following fields: ti = title; ab = abstract; kw = author keyword; kf = key field; hw = heading word.
Lines 5–7: Terms for CYP2C19. The free-text terms are truncated using the * marker. Truncation ensures that the root word and other possible variations are identified and returned by the search. We have truncated CYP2C19 (using the * marker again) to identify CYP2C19*2, CYP2C19*3, and CYP2C9*17, and other alleles.
Lines 8 or 9: Terms for clopidogrel
Line 11: combines terms for Point of Care testing AND terms for CYP2C19 OR terms for clopidogrel.
Database: EMBASE
Host: Ovid
Data parameters: 1974 to 10 August 2022
Date of search: 11 August 2022
| # | Searches | Results |
|---|---|---|
| 1 | *“point of care testing”/ | 6784 |
| 2 | ((Point of Care adj2 test*) or POCT).ti,ab,kf,kw. | 12,848 |
| 3 | (Genomadix* or Genedrive or Spartan).ti,ab,kf,kw. | 558 |
| 4 | 1 or 2 or 3 | 16,726 |
| 5 | *cytochrome P450 2C19/ | 2200 |
| 6 | (CYP2C19* or cypiic19* or “Cytochrome P-450”).ti,ab,kw,kf. | 25,520 |
| 7 | 5 or 6 | 25,636 |
| 8 | *clopidogrel/ | 12,224 |
| 9 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”).ti,ab,kw,rn,tn,dq,dy,cn. | 71,601 |
| 10 | 8 or 9 | 71,601 |
| 11 | 4 and (7 or 10) | 296 |
Database: CENTRAL
Host: Wiley interface
Data parameters: Issue 7 of 12, July 2022
Date of search: 11 August 2022
| ID | Query | Results |
|---|---|---|
| #1 | [mh ^Clopidogrel] | 2166 |
| #2 | (clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”):ti,ab,kw | 5907 |
| #3 | #1 or #2 | 5907 |
| #4 | MeSH descriptor: [Cytochrome P-450 CYP2C19] this term only | 362 |
| #5 | (CYP2C19* or cypiic19* or “Cytochrome P-450”):ti,ab,kw | 2496 |
| #6 | #4 or #5 | 24,96 |
| #7 | #3 OR #6 | 8024 |
| #8 | MeSH descriptor: [Point-of-Care Testing] this term only | 101 |
| #9 | ((Point of Care NEAR/2 test*) or POCT):ti,ab,kw | 949 |
| #10 | (Genomadix* or Genedrive or Spartan) | 161 |
| #11 | #8 OR #9 OR #10 | 1103 |
| #12 | #7 AND #11 | 51 |
Database: CINAHL
Host: Ovid
Data parameters: 1981 to present
Date of search: 11 August 2022
| # | Query | Results |
|---|---|---|
| S1 | (MH “Point-of-Care Testing”) | 4414 |
| S2 | TI (((Point of Care N2 test*) or POCT)) OR AB (((Point of Care N2 test*) or POCT)) | 3304 |
| S3 | TI ((Genomadix* or Genedrive or Spartan)) OR AB ((Genomadix* or Genedrive or Spartan).) | 98 |
| S4 | S1 OR S2 OR S3 | 6432 |
| S5 | TI (TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”))) OR AB (TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”))) | 1092 |
| S6 | TI (TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”))) OR AB (TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”))) | 4402 |
| S7 | S5 OR S6 | 5221 |
| S8 | S4 AND S7 | 28 |
| S9 | S4 AND S7 | 8 |
Database: CTG
Host: www.clinicaltrials.gov/ct2/results/refine?show_xprt=Y
Date of search: 11 August 2022
Search a
((clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Search b
((CYP2C19 OR cypiic19 OR “Cytochrome P-450”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Database: ICTRP
Host: https://trialsearch.who.int/Default.aspx
Data parameters: 1946 to present
Date of search: 11 August 2022
Search a
((clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Search b
((CYP2C19 OR cypiic19 OR “Cytochrome P-450”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Database: ECONLit
Host: EBSCOhost
Data parameters: 1886 to present
Date of search: 11 August 2022
| # | Query | Results |
|---|---|---|
| S1 | (MH “Point-of-Care Testing”) | 0 |
| S2 | TI (((Point of Care N2 test*) or POCT)) OR AB (((Point of Care N2 test*) or POCT)) | 5 |
| S3 | TI ((Genomadix* or Genedrive or Spartan)) OR AB ((Genomadix* or Genedrive or Spartan).) | 10 |
| S4 | S1 OR S2 OR S3 | 15 |
| S5 | TI (TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”))) OR AB (TI ((CYP2C19* or cypiic19* or “Cytochrome P-450”)) OR AB ((CYP2C19* or cypiic19* or “Cytochrome P-450”))) | 2 |
| S6 | TI (TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”))) OR AB (TI ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”)) OR AB ((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or “R 130964” or “R-130964” or R130964 or “SR 25990” or “SR-25990” or SR25990 or A74586SNO7 or “113665-84-2”))) | 8 |
| S7 | S5 OR S6 | 8 |
| S8 | S4 AND S7 | 0 |
| S9 | S4 AND S7 | 0 |
Database: HTA Library
Host: www.crd.york.ac.uk/CRDWeb/HomePage.asp
Date of search: 11 August 2022
Results for: ((((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or R 130964 or R-130964 or R130964 or SR 25990 or SR-25990 or SR25990 or A74586SNO7 or 113665-84-2) OR (CYP2C19* or cypiic19* or Cytochrome P-450))) AND ((Genomadix* OR Genedrive OR Spartan OR Point of Care))) and (Project record:ZDT OR Full publication record:ZDT) IN HTA
Database: NHS EED
Host: www.crd.york.ac.uk/CRDWeb/HomePage.asp
Date of search: 11 August 2022
Results for: ((((clopidogrel* or clopidogrelum or M-clopidogrel or Nra-clopidogrel or grepid or iscover or duoplavin* or plavi* or Plavix* or zyllt* or R 130964 or R-130964 or R130964 or SR 25990 or SR-25990 or SR25990 or A74586SNO7 or 113665-84-2) OR (CYP2C19* or cypiic19* or Cytochrome P-450))) AND ((Genomadix* OR Genedrive OR Spartan OR Point of Care))) and ((Economic evaluation:ZDT and Bibliographic:ZPS) OR (Economic evaluation:ZDT and Abstract:ZPS)) IN NHS EED
Database: Tufts CEA Register
Host: https://cear.tuftsmedicalcenter.org/about
Date of search: 11 August 2022
Methods
-
(Genomadix* OR Genedrive OR Spartan) n = 0
-
((clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
-
((CYP2C19 OR cypiic19 OR “Cytochrome P-450”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Ratios
-
(Genomadix* OR Genedrive OR Spartan) n = 0
-
((clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
-
((CYP2C19 OR cypiic19 OR “Cytochrome P-450”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Utilities
-
(Genomadix* OR Genedrive OR Spartan) n = 0
-
((clopidogrel OR clopidogrelum OR M-clopidogrel OR Nra-clopidogrel OR grepid OR iscover OR duoplavin OR plavi OR Plavix OR zyllt OR “R 130964” OR “R-130964” OR R130964 OR “SR 25990” OR “SR-25990” OR SR25990 OR A74586SNO7 OR “113665-84-2”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
-
((CYP2C19 OR cypiic19 OR “Cytochrome P-450”) AND (Genomadix* OR Genedrive OR Spartan OR “point of care”))
Supplemental cost-effectiveness searches
| Resource | N |
|---|---|
| MEDLINE | 48 |
| EMBASE | 43 |
| Econlit | 2 |
| Total | 93 |
| - duplicates | −34 |
| Total to screen | 59 to screen |
Database: MEDLINE (MEDALL)
Host: Ovid
Data parameters: 1946 to present
Date of search: 17 October 2022
| # | Searches | Results |
|---|---|---|
| 1 | *Ischemic Stroke/and second*.ti,ab,kw,kf. | 939 |
| 2 | (second* adj2 (stroke or ischemic)).ti,ab,kw,kf. | 4404 |
| 3 | 1 or 2 | 5198 |
| 4 | *economics/or exp *“costs and cost analysis”/ | 89,042 |
| 5 | ((cost adj2 effectiveness) or (economic adj2 evaluation*)).ti,ab,kw,kf. | 78,596 |
| 6 | 4 or 5 | 155,602 |
| 7 | (2012* or 2013* or 2014* or 2015* or 2016* or 2017* or 2018* or 2019* or 2020* or 2021* or 2022*).dt,dp,ed,ep,yr. | 1,365,3990 |
| 8 | 3 and 6 and 7 | 48 |
Database: EMBASE
Host: Ovid
Data parameters: 1980–2022 Week 41
Date of search: 17 October 2022
| # | Searches | Results |
|---|---|---|
| 1 | *Ischemic Stroke/and second*.ti,ab,kw,kf. | 713 |
| 2 | (second* adj2 (stroke or ischemic)).ti,ab,kw,kf. | 7718 |
| 3 | 1 or 2 | 8275 |
| 4 | *economics/or *“cost-effectiveness analysis”/ | 62,335 |
| 5 | ((cost adj2 effectiveness) or (economic adj2 evaluation*)).ti,ab,kw,kf. | 116,387 |
| 6 | 4 or 5 | 147,059 |
| 7 | (2012* or 2013* or 2014* or 2015* or 2016* or 2017* or 2018* or 2019* or 2020* or 2021* or 2022*).yr. | 1,713,4825 |
| 8 | 3 and 6 and 7 | 76 |
| 9 | limit 8 to embase | 43 |
Database: EconLit
Host: EBSCOhost
Data parameters: 1981 to current
Date of search: 17 October 2022
| # | Query | Results |
|---|---|---|
| 1 | AB ((second* N2 (stroke or ischemic))) OR TI ((second* N2 (stroke or ischemic))) | 2 |
Appendix 2 Tables of included, ongoing or excluded studies
Studies included in the review showing primary and secondary reports
Primary reports are the primary publication for the study and are used to refer to that study throughout text and tables.
| Study name | Primary report | Secondary reports |
|---|---|---|
| N/A | Lan H, Ying T, Xi-Hua S, Yi L. Anti-platelet therapy in mild cerebral infarction patients on the basis of CYP2C19 metabolizer status. Cell Transplantation 2019;28(8).47 | None |
| N/A | Xia C, Zhang Z, He X, Liu J, Li X, Chang Q, et al. Correlation between CYP2C19 gene polymorphism and individualized medication in patients with ischemic stroke. [Chinese]. Chinese Journal of Clinical Pharmacology and Therapeutics 2021;26(3).46 | None |
| Study name | Primary report | Secondary reports |
|---|---|---|
| PRINCE | Chen W, Lin Y, Meng X, Chen G, Wang Z, Wu J, et al. Ticagrelor plus aspirin versus clopidogrel plus aspirin for platelet reactivity in patients with minor stroke or transient ischaemic attack: open label, blinded endpoint, randomised controlled phase II trial. British Medical Journal 2019;365.53 | Wang Y, Lin Y, Meng X, Chen W, Chen G, Wang Z, et al. Effect of ticagrelor with clopidogrel on high on-treatment platelet reactivity in acute stroke or transient ischemic attack (PRINCE) trial: rationale and design. International Journal of Stroke 2017;12(3).181 Zhou M, Chen W, Pan Y, Lin Y, Meng X, Zhao X, et al. Antiplatelet effect of ticagrelor with aspirin in acute minor stroke and transient ischemic attack stratified by CYP2C19 metabolizer status: subgroup analysis of the PRINCE trial. Aging 2020;13(3).182 |
| MASETRO | Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Effects of triflusal and clopidogrel on the secondary prevention of stroke based on cytochrome P450 2C19 genotyping. Journal of Stroke 2017;19(3).48 | Gangnam Severance Hospital. Comparison of Triflusal and Clopidogrel in Secondary Prevention of Stroke Based on the Genotyping. NCT01174693. 2015. URL: https://ClinicalTrials.gov/show/NCT01174693 (accessed September 2022).183 Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Protocol for the comparison of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping (MASETRO study): a multicenter, randomized, open-label, parallel-group trial. International Journal of Stroke 2016;11(4).184 Han SW, Park JH. Prevalence of CYP2C19 alleles in the maestro study participants. European Stroke Journal 2017;2(1 Suppl. 1).185 Han SW, Park JH, Cheon KY, Lee KY. Verifynow P2Y12 assay with regard to cytochrome P450 2C19 polymorphisms and stroke recurrence. Stroke Conference: American Heart Association/American Stroke Association 2018 International Stroke2018;49(Suppl. 1).186 Lee KY, Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, et al. Effects of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping. Stroke 2017;48.187 |
| POINT | Meschia JF, Walton RL, Farrugia LP, Ross OA, Ross OA, Elm JJ, et al. Efficacy of clopidogrel for prevention of stroke based on CYP2C19 allele status in the POINT trial. Stroke 2020;51(7).49 | None |
| N/A | Wang Y, Meng X, Wang A, Xie X, Pan Y, Johnston SC, et al. Ticagrelor versus clopidogrel in CYP2C19 loss-of-function carriers with stroke or TIA. New England Journal of Medicine 2021;385(27).50 | Wang Y, Johnston C, Bath PM, Meng X, Jing J, Xie X, et al. Clopidogrel with aspirin in High-risk patients with Acute Non-disabling Cerebrovascular Events II (CHANCE-2): rationale and design of a multicentre randomised trial. Stroke and Vascular Neurology 2021;6(2).188 Pan Y, Meng X, Jin A, Johnston SC, Li H, Bath PM, et al. Time course for benefit and risk with ticagrelor and aspirin in individuals with acute ischemic stroke or transient ischemic attack who carry CYP2C19 loss-of-function alleles: a secondary analysis of the CHANCE-2 randomized clinical trial. JAMA Neurology 2022.189 Joundi RA. In patients with stroke or TIA and CYP2C19 loss-of-function alleles, ticagrelor vs. clopidogrel reduced 90-d stroke. Annals of Internal Medicine 2022;175(3).190 Wang A, Meng X, Tian X, Johnston SC, Li H, Bath PM, et al. Bleeding risk of dual antiplatelet therapy after minor stroke or transient ischemic attack. Annals of Neurology 2022;91(3).191 |
| N/A | Wu H, Song H, Dou L, Gao B, Pan Y, Dong M, et al. Effectiveness and safety of high dose clopidogrel plus aspirin in ischemic stroke patients with the single CYP2C19 loss-of-function allele: a randomized trial. BMC Neurology 2020;20(1).51 | None |
| N/A | Yi X, Lin J, Zhou J, Wang Y, Huang R, Wang C. The secondary prevention of stroke according to cytochrome P450 2C19 genotype in patients with acute large-artery atherosclerosis stroke. Oncotarget 2018;9(25).54 | None |
| CHANCE | Zhao X, Lin J, Li H, Johnston SC, Lin Y, Pan Y, et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. Journal of the American Medical Association 2016;316(1).52 | Beijing Tiantan Hospital. Clopidogrel With Aspirin in High-risk Patients With Acute Non-disabling Cerebrovascular Events II. NCT04078737. 2021. URL: https://ClinicalTrials.gov/show/NCT04078737 (accessed September 2022).192 Xu J, Wang A, Wangqin R, Mo J, Chen Z, Dai L, et al. Efficacy of clopidogrel for stroke depends on CYP2C19 genotype and risk profile. Annals of Neurology 2019;86(3).193 Zhang J, Sun H, Ming T, Liu X, Cong Y, Li F, et al. Association between platelet function and recurrent ischemic vascular events after TIA and minor stroke. International Journal of Clinical Pharmacology and Therapeutics 2017;55(10).74 Pan Y, Wangqin R, Li H, Meng X, Johnston SC, Simon T, et al. F2R polymorphisms and clopidogrel efficacy and safety in patients with minor stroke or TIA. Neurology 2021;96(1).194 Wu Y, Zhou Y, Pan Y, Zhao X, Liu L, Wang D, et al. Impact of CYP2C19 polymorphism in prognosis of minor stroke or TIA patients with declined eGFR on dual antiplatelet therapy: CHANCE substudy. Pharmacogenomics Journal 2018;18(6).195 |
| Study name | Primary report | Secondary reports |
|---|---|---|
| PRINCE | Chen W, Lin Y, Meng X, Chen G, Wang Z, Wu J, et al. Ticagrelor plus aspirin versus clopidogrel plus aspirin for platelet reactivity in patients with minor stroke or transient ischaemic attack: open label, blinded endpoint, randomised controlled phase II trial. British Medical Journal 2019;365.53 | Wang Y, Lin Y, Meng X, Chen W, Chen G, Wang Z, et al. Effect of ticagrelor with clopidogrel on high on-treatment platelet reactivity in acute stroke or transient ischemic attack (PRINCE) trial: rationale and design. International Journal of Stroke 2017;12(3).181 Zhou M, Chen W, Pan Y, Lin Y, Meng X, Zhao X, et al. Antiplatelet effect of ticagrelor with aspirin in acute minor stroke and transient ischemic attack stratified by CYP2C19 metabolizer status: subgroup analysis of the PRINCE trial. Aging 2020;13(3).182 |
| N/A | Diaz-Villamarin X, Davila-Fajardo CL, Martinez-Gonzalez LJ, Rodriguez-Delgado A, Villegas-Rodriguez I, Cabeza-Barrera J. CYP2C19*2, *3 polymorphisms in the response to clopidogrel after percutaneous transluminal angioplasty or stroke. International Journal of Clinical Pharmacy 2017;39(1).196 | Diaz-Villamarin X, Davila-Fajardo CL, Blanquez-Martinez D, Fernandez-Gomez E, Antunez-Rodriguez A, Raquel AS. CYP2C19 SNP’s influence on clopidogrel response in cerebrovascular disease patients: final results. European Journal of Hospital Pharmacy 2019;26(Suppl. 1).197 |
| N/A | Fu H, Hu P, Ma C, Peng F, He Z. Association of clopidogrel high on-treatment reactivity with clinical outcomes and gene polymorphism in acute ischemic stroke patients: an observational study. Medicine 2020;99(15).56 | None |
| N/A | Fukuma K, Yamagami H, Ihara M, Tanaka T, Miyata T, Miyata S, et al. P2Y12 reaction units and clinical outcomes in acute large artery atherosclerotic stroke: a multicenter prospective study. Journal of Atherosclerosis and Thrombosis 2022;5.198 | Fukuma K, Yamagami H, Kamiyama K, Enomoto Y, Furui E, Manabe Y, et al. Association between CYP2C19 genetic polymorphisms and clinical outcome in acute atherothrombotic stroke: a sub-analysis of the praise study. European Stroke Journal 2017;2(1 Suppl. 1).198 |
| MAESTRO | Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Effects of triflusal and clopidogrel on the secondary prevention of stroke based on cytochrome P450 2C19 genotyping. Journal of Stroke 2017;19(3).48 | Gangnam Severance Hospital. Comparison of Triflusal and Clopidogrel in Secondary Prevention of Stroke Based on the Genotyping. NCT01174693. 2015. URL: https://ClinicalTrials.gov/show/NCT01174693 (accessed September 2022).183 Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, Oh SH, et al. Protocol for the comparison of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping (MASETRO study): a multicenter, randomized, open-label, parallel-group trial. International Journal of Stroke 2016;11(4).184 Han SW, Park JH. Prevalence of CYP2C19 alleles in the maestro study participants. European Stroke Journal 2017;2(1 Suppl. 1).185 Han SW, Park JH, Cheon KY, Lee KY. Verifynow P2Y12 assay with regard to cytochrome P450 2C19 polymorphisms and stroke recurrence. Stroke 2018;49(Suppl. 1).186 Lee KY, Han SW, Kim YJ, Ahn SH, Seo WK, Yu S, et al. Effects of triflusal and clopidogrel in secondary prevention of stroke based on cytochrome P450 2C19 genotyping. Stroke 2017;48.187 |
| N/A | Hoh BL, Gong Y, McDonough CW, Waters MF, Royster AJ, Sheehan TO, et al. CYP2C19 and CES1 polymorphisms and efficacy of clopidogrel and aspirin dual antiplatelet therapy in patients with symptomatic intracranial atherosclerotic disease. Journal of Neurosurgery 2016;124(6).58 | None |
| N/A | Lin J, Mo Y, Cai, Mao D, Fu H, Wei D. CYP2C19 polymorphisms and clopidogrel efficacy in the secondary prevention of ischemic stroke: a retrospective observational study. Annals of Palliative Medicine 2021;10(12).59 | None |
| N/A | Liu G, Yang S, Chen S. The correlation between recurrent risk and CYP2C19 gene polymorphisms in patients with ischemic stroke treated with clopidogrel for prevention. Medicine 2020;99(11).60 | None |
| N/A | Lv H, Yang Z, Wu H, Liu M, Mao X, Liu X, et al. High on-treatment platelet reactivity as predictor of long-term clinical outcomes in stroke patients with antiplatelet agents. Translational Stroke Research 2022;13(3).61 | Han Y, Lv H, Wu H, Liu M, Liu Q, Huo Y, et al. The value of platelet reactivity and genetic polymorphism in predicting long-term clinical outcomes in stroke patients. Circulation Research 2019;125(12).199 |
| N/A | McDonough CW, McClure LA, Mitchell BD, Gong Y, Horenstein RB, Lewis JP, et al. CYP2C19 metabolizer status and clopidogrel efficacy in the Secondary Prevention of Small Subcortical Strokes (SPS3) study. Journal of the American Heart Association 2015;4(6).62 | Simpkins AN, McDonough CW, McClure LA, Mitchell BD, Shuldiner AR, Benavente OR, et al. Secondary stroke prevention with aspirin and clopidogrel in CYP2C19*17 carriers increases risk of major non-CNS bleeding. Stroke Conference: American Heart Association/American Stroke Association 2019;50(Suppl. 1).200 |
| POINT | Meschia JF, Walton RL, Farrugia LP, Ross OA, Ross OA, Elm JJ, et al. Efficacy of clopidogrel for prevention of stroke based on CYP2C19 allele status in the POINT trial. Stroke 2020;51(7).49 | None |
| N/A | Ni G, Liang C, Liu K, Cao Y, Zhang H, Tian X, et al. The effects of CES1A2 and CYP2C19 polymorphisms on responsiveness to clopidogrel and clinical outcomes among Chinese patients with acute ischemic stroke. International Journal of Clinical and Experimental Medicine 2017;10(2).63 | None |
| N/A | Patel PD, Vimalathas P, Niu X, Shannon CN, Denny JC, Peterson JF, et al. CYP2C19 Loss-of-function is associated with increased risk of ischemic stroke after transient ischemic attack in intracranial atherosclerotic Disease. Journal of Stroke and Cerebrovascular Diseases 2021;30(2).64 | None |
| N/A | Qiu LN, Sun Y, Wang L, Han RF, Xia XS, Liu J, et al. Influence of CYP2C19 polymorphisms on platelet reactivity and clinical outcomes in ischemic stroke patients treated with clopidogrel. European Journal of Pharmacology 2015;747.65 | None |
| N/A | Sen HM, Silan F, Silan C, Degirmenci Y, Ozisik Kamaran HI. Effects of CYP2C19 and P2Y12 gene polymorphisms on clinical results of patients using clopidogrel after acute ischemic cerebrovascular disease. Balkan Journal of Medical Genetics 2014;17(2).66 | None |
| N/A | Spokoyny I, Barazangi N, Jaramillo V, Rose J, Chen C, Wong C, et al. Reduced clopidogrel metabolism in a multiethnic population: prevalence and rates of recurrent cerebrovascular events. Journal of Stroke and Cerebrovascular Diseases 2014;23(4).67 | Spokoyny I, Barazangi N, Jaramillo V, Rose J, Chen C, Wong C, et al. Reduced CYP219 clopidogrel metabolism in a multiethnic population: prevalence and associated rates of recurrent cerebrovascular events. Neurology 2013;80(1).201 |
| Sun W, Li Y, Li J, Zhang Z, Zhu W, Liu W, et al. Variant recurrent risk among stroke patients with different CYP2C19 phenotypes and treated with clopidogrel. Platelets 2015;26(6).68 | ||
| N/A | Tanaka T, Yamagami H, Ihara M, Miyata T, Miyata S, Hamasaki T, et al. Association of CYP2C19 polymorphisms with clopidogrel reactivity and clinical outcomes in chronic ischemic stroke. Circulation Journal 2019;83(6).69 | National Cerebral and Cardiovascular Center. The Influence of CYP2C19 Polymorphism and Clinical Outcomes in Stroke Patients. NCT02711410; 2016. URL: https://ClinicalTrials.gov/show/NCT02711410 (accessed September 2022).202 |
| N/A | Tomek A, Mat’oska V, Frydmanova A, Magerova H, Sramek M, Paulasova-Schwabova J, et al. Impact of CYP2C19 polymorphisms on clinical outcomes and antiplatelet potency of clopidogrel in Caucasian poststroke survivors. American Journal of Therapeutics 2018;25(2).70 | None |
| N/A | Tornio A, Flynn R, Morant S, Velten E, Palmer CNA, MacDonald TM, et al. Investigating real-world clopidogrel pharmacogenetics in stroke using a bioresource linked to electronic medical records. Clinical Therapeutics 2017;39(8 Suppl. 1).71 | Doney A, Palmer C, Morant S, Flynn R, MacDonald T. Impact of CYP2C19 genotype in ischaemic stroke patients treated with clopidogrel. International Journal of Stroke 2015;5.203 Tornio A, Flynn R, Morant S, Velten E, Palmer CNA, MacDonald TM, et al. Investigating real-world clopidogrel pharmacogenetics in stroke using a bioresource linked to electronic medical records. Clinical Therapeutics 2017;39(1).71 |
| N/A | Wang Y, Cai H, Zhou G, Zhang Z, Liu X. Effect of CYP2C19*2 and *3 on clinical outcome in ischemic stroke patients treated with clopidogrel. Journal of the Neurological Sciences 2016;369.72 | None |
| N/A | Yi X, Wang Y, Lin J, Cheng W, Zhou Q, Wang C. Interaction of CYP2C19, P2Y12, and GPIIIa Variants associates with efficacy of clopidogrel and adverse events on patients with ischemic stroke. Clinical and Applied Thrombosis/Hemostasis 2017;23(7).73 | None |
| N/A | Yi X, Lin J, Zhou J, Wang Y, Huang R, Wang C. The secondary prevention of stroke according to cytochrome P450 2C19 genotype in patients with acute large-artery atherosclerosis stroke. Oncotarget 2018;9(25).54 | None |
| N/A | Zhang J, Sun H, Ming T, Liu X, Cong Y, Li F, et al. Association between platelet function and recurrent ischemic vascular events after TIA and minor stroke. International Journal of Clinical Pharmacology and Therapeutics 2017;55(10).74 | None |
| CHANCE | Zhao X, Lin J, Li H, Johnston SC, Lin Y, Pan Y, et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. Journal of the American Medical Association 2016;316(1).52 | Beijing Tiantan Hospital. Clopidogrel With Aspirin in High-risk Patients With Acute Non-disabling Cerebrovascular Events II. NCT04078737. 2021. URL: https://ClinicalTrials.gov/show/NCT04078737 (accessed September 2022).192 Xu J, Wang A, Wangqin R, Mo J, Chen Z, Dai L, et al. Efficacy of clopidogrel for stroke depends on CYP2C19 genotype and risk profile. Annals of Neurology 2019;86(3).193 Zhang J, Sun H, Ming T, Liu X, Cong Y, Li F, et al. Association between platelet function and recurrent ischemic vascular events after TIA and minor stroke. International Journal of Clinical Pharmacology and Therapeutics 2017;55(10).74 Pan Y, Wangqin R, Li H, Meng X, Johnston SC, Simon T, et al. F2R polymorphisms and clopidogrel efficacy and safety in patients with minor stroke or TIA. Neurology 2021;96(1).194 Wu Y, Zhou Y, Pan Y, Zhao X, Liu L, Wang D, et al. Impact of CYP2C19 polymorphism in prognosis of minor stroke or TIA patients with declined eGFR on dual antiplatelet therapy: CHANCE substudy. Pharmacogenomics Journal 2018;18(6).195 |
| Study name | Primary report | Secondary reports |
|---|---|---|
| TAILOR-PCI (pre-trial and main trial) | Baudhuin LM, Train LJ, Goodman SG, Lane GE, Lennon RJ, Mathew V, et al. Point of care CYP2C19 genotyping after percutaneous coronary intervention. Pharmacogenomics Journal 2022;22.76 | Baudhuin L, Train L, Goodman S, Lane G, Lennon R, Mathew V, et al. Validation and performance of point-of-care rapid CYP2C19 genotyping in the tailor-pci multicenter international randomized clinical trial. Journal of the American College of Cardiology 2021;77(18).95 |
| N/A | Choi JL, Kim BR, Woo KS, Kim KH, Kim JM, Kim MH, et al. The diagnostic utility of the point-of-care CYP2C19 genotyping assay in patients with acute coronary syndrome dosing clopidogrel: comparison with platelet function test and SNP genotyping. Annals of Clinical and Laboratory Science 2016;46(5).80 | None |
| N/A | Petrek M, Kocourkova L, Zizkova V, Nosek Z, Taborsky M, Petrkova J. Characterization of three CYP2C19 gene variants by MassARRAY and point of care techniques: experience from a Czech Centre. Archivum Immunologiae et Therapiae Experimentalis 2016;64(Suppl 1).81 | Petrkova J, Paskova L, Zizkova V, Nosek Z, Taborsky M, Petrek M. POCT for determination of basic pharmacogenetic profile for individualization of antiplatelet therapy: pilot study. European Heart Journal 2014;1.85 |
| N/A | So DYF, Wells GA, McPherson R, Labinaz M, Le May MR, Glover C, et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics Journal 2016;16(1).82 | None |
| N/A | Spartan Bioscience Inc. Method Comparison Study of the Spartan FRX CYP2C19 Genotyping System Against Bi-directional Sequencing. NCT01718535. 2022. URL: https://ClinicalTrials.gov/show/NCT01718535 (accessed November 2022).84 | None |
| N/A | Spartan Bioscience Inc. Spartan Cube CYP2C19 Inter Laboratory Reproducibilty Study. NCT04473573. 2020. URL: https://ClinicalTrials.gov/show/NCT04473573 (accessed November 2022).79 | None |
| N/A | Spartan Bioscience Inc. Spartan Cube CYP2C19 Method Comparison Study. NCT04473586. 2020. URL: https://ClinicalTrials.gov/show/NCT04473586 (accessed November 2022).78 | None |
| N/A | Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 2012;379(9827).77 | None |
| N/A | Wirth F, Zahra G, Xuereb RG, Barbara C, Fenech A, Azzopardi LM. Comparison of a rapid point-of-care and two laboratory-based CYP2C19*2 genotyping assays for personalisation of antiplatelet therapy. International Journal of Clinical Pharmacy 2016;38(2).83 | Wirth F, Zahra G, Xuereb RG, Barbara C, Fenech A, Azzopardi LM. Comparison between a point-of-care and a laboratory-based CYP2C19 genotyping assay for pharmacist-led personalisation of antiplatelet therapy. Pharmacotherapy 2015;35(11).99 |
| Study name | Primary report | Secondary reports |
|---|---|---|
| N/A | Al-Rubaish AM, Al-Muhanna FA, Alshehri AM, Alsulaiman AA, Alabdulali MM, Alkhamis F, et al. Prevalence of CYP2C19*2 carriers in Saudi ischemic stroke patients and the suitability of using genotyping to guide antiplatelet therapy in a university hospital setup. Drug Metabolism and Personalized Therapy 2021;37(1).86 | None |
| TAILOR-PCI | Baudhuin LM, Train LJ, Goodman SG, Lane GE, Lennon RJ, Mathew V, et al. Point of care CYP2C19 genotyping after percutaneous coronary intervention. Pharmacogenomics Journal 2022;22.76 | Baudhuin L, Train L, Goodman S, Lane G, Lennon R, Mathew V, et al. Validation and performance of point-of-care rapid CYP2C19 genotyping in the tailor-pci multicenter international randomized clinical trial. Journal of the American College of Cardiology 2021;77(18).95 |
| N/A | Bergmeijer TO, Vos GJ, Claassens DM, Janssen PW, Harms R, der Heide RV, et al. Feasibility and implementation of CYP2C19 genotyping in patients using antiplatelet therapy. Pharmacogenomics 2018;19(7).87 | Bergmeijer TO, Janssen PWA, Schipper JC, Qaderdan K, Ishak M, Ruitenbeek RS, et al. CYP2C19 genotype-guided antiplatelet therapy in ST-segment elevation myocardial infarction patients-rationale and design of the patient outcome after primary PCI (POPular) genetics study. American Heart Journal 2014;168(1).96 |
| N/A | Cavallari LH, Franchi F, Rollini F, Been L, Rivas A, Agarwal M, et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. Journal of Translational Medicine 2018;16(1).88 | None |
| N/A | Choi JL, Kim BR, Woo KS, Kim KH, Kim JM, Kim MH, et al. The diagnostic utility of the point-of-care CYP2C19 genotyping assay in patients with acute coronary syndrome dosing clopidogrel: comparison with platelet function test and SNP genotyping. Annals of Clinical and Laboratory Science 2016;46(5).80 | None |
| N/A | Davis BH, DeFrank G, Limdi NA, Harada S. Validation of the Spartan RXCYP2C19 genotyping assay utilizing blood samples. Clinical and Translational Science 2020;13(2).89 | None |
| N/A | Franchi F, Rollini F, Rivas J, Rivas A, Agarwal M, Briceno M, et al. Prasugrel versus ticagrelor in patients with CYP2C19 loss-of-function genotypes results of a randomized pharmacodynamic study in a feasibility investigation of rapid genetic testing. JACC – Basic to Translational Science 2020;5(5).90 | None |
| N/A | Gurbel PA, Bell R, Bliden K, Yazdani S, Taheri H, Akbari M, et al. Bedside testing of CYP2C19 genotype to guide antiplatelet therapy: implementation in the catheterization laboratory. Journal of the American College of Cardiology Conference: 67th Annual Scientific Session of the American College of Cardiology and i2 Summit: Innovation in Intervention, ACC 2018;71(Suppl. 1).91 | None |
| N/A | McDermott JH, Ainsworth S, Wright S, Sen D, Miele G, Smith CJ, et al. Development of a point of care test for CYP2C19 allowing genotype guided antiplatelet prescribing to prevent recurrent ischaemic strokes. European Journal of Human Genetics 2020;28(Suppl. 1).94 | None |
| N/A | Petrek M, Kocourkova L, Zizkova V, Nosek Z, Taborsky M, Petrkova J. Characterization of three CYP2C19 gene variants by MassARRAY and point of care techniques: experience from a Czech Centre. Archivum Immunologiae et Therapiae Experimentalis 2016;64(Suppl. 1).81 | Petrkova J, Paskova L, Zizkova V, Nosek Z, Taborsky M, Petrek M. POCT for determination of basic pharmacogenetic profile for individualization of antiplatelet therapy: pilot study. European Heart Journal 2014;1.85 |
| N/A | Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 2012;379(9827).77 | None |
| N/A | So DYF, Wells GA, McPherson R, Labinaz M, Le May MR, Glover C, et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics Journal 2016;16(1).82 | None |
| N/A | Spartan Bioscience Inc. Spartan Cube CYP2C19 Method Comparison Study. NCT04473586. 2020. URL: https://ClinicalTrials.gov/show/NCT04473586 (accessed November 2022).78 | None |
| N/A | Spartan Bioscience Inc. Spartan Cube CYP2C19 Inter Laboratory Reproducibility Study. NCT04473573. 2020. URL: https://ClinicalTrials.gov/show/NCT04473573 (accessed November 2022).79 | None |
| N/A | Tomaniak M, Koltowski L, Kochman J, Huczek Z, Rdzanek A, Pietrasik A, et al. Can prasugrel decrease the extent of periprocedural myocardial injury during elective percutaneous coronary intervention? Polish Archives of Internal Medicine-Polskie Archiwum Medycyny Wewnetrznej 2017;127(11)92 | None |
| N/A | Zhou Y, Armstead AR, Coshatt GM, Limdi NA, Harada S. Comparison of two point-of-care CYP2C19 genotyping assays for genotype-guided antiplatelet therapy. Annals of Clinical and Laboratory Science 2017;47(6).93 | Zhou Y, Armstead AR, Coshatt GM, Brott BC, Sankaranarayanan A, Limdi NA, et al. Rapid CYP2C19 genotype testing: comparison between spartan RX CYP2C19 and verigene CYP2C19. Journal of Molecular Diagnostics 2015;17(6).100 |
Ongoing studies
| Objective | Report |
|---|---|
| 1 | Wang J, Han M, Kuang J, Tu J, Starcevich K, Gao P, et al. Personalized antiplatelet therapy based on clopidogrel/aspirin resistance tests in acute ischemic stroke and transient ischemic attack: study protocol of a multi-center, single-blinded and randomized controlled trial. Contemporary Clinical Trials 2021;108204 |
| 3 | Gangnam Severance Hospital. Clopidogrel Preventive Effect Based on CYP2C19 Genotype in Ischemic Stroke. NCT04072705. 2022. URL: https://ClinicalTrials.gov/show/NCT04072705 (accessed September 2022)205 |
| 3 | Zhang XG, Zhu XQ, Xue J, Li ZZ, Jiang HY, Hu L, et al. Personalised antiplatelet therapy based on pharmacogenomics in acute ischaemic minor stroke and TIA: study protocol for a randomised controlled trial. BMJ Open 2019;9(5)206 |
Studies excluded at full-text screening (Objectives 1–3)
Reasons for exclusion align with the study selection protocol above.
| Study details | Reason for exclusion |
|---|---|
| 1. Al-Rubaish AM, Al-Muhanna FA, Alshehri AM, Alsulaiman AA, Alabdulali MM, Alkhamis F, et al. Prevalence of CYP2C19*2 carriers in Saudi ischemic stroke patients and the suitability of using genotyping to guide antiplatelet therapy in a university hospital setup. Drug Metabolism and Personalized Therapy 2022;37(1). | Intervention: no intervention listed in the protocol (obj1/2) or presence of *2 or *3 LOF alleles |
| 2. Alakbarzade V, Huang X, Drury S, Chis Ster I, McEntagert M, Pereira AC. High on-clopidogrel platelet reactivity in stroke: a systematic review and meta-analysis. European Stroke Journal 2019;4(Suppl. 1). | Publication type: systematic review |
| 3. Alakbarzade V, Huang X, Ster IC, McEntagart M, Pereira AC. High on-clopidogrel platelet reactivity in ischaemic stroke or transient ischaemic attack: systematic review and meta-analysis. Journal of Stroke and Cerebrovascular Diseases 2020;29(7) | Publication type: systematic review |
| 4. Ali Z, Elewa H. The effect of CYP2C19 and nongenetic factors on clopidogrel responsiveness in the MENA region: a systematic review. Clinical and Applied Thrombosis/Hemostasis 2019;25 | Publication type: systematic review |
| 5. Alkattan A, Almutairi Y, Alsalameen E, Alkhalifah A, Alghanim F. The CYP2C19 genotypes and its effect on clopidogrel as an anti-platelet drug among the Arab population. Indian Journal of Pharmacology 2021;53(1). | Publication type: commentary |
| 6. Alkattan A, Alsalameen E. Polymorphisms of genes related to phase-I metabolic enzymes affecting the clinical efficacy and safety of clopidogrel treatment. Expert Opinion on Drug Metabolism and Toxicology 2021;17(6). | Publication type: review |
| 7. Alrajeh KY, Roman YM. The frequency of major CYP2C19 genetic polymorphisms in women of Asian, Native Hawaiian and Pacific Islander subgroups. Personalized Medicine 2022;19(4). | Outcomes: did not report on any outcomes specified in the inclusion criteria. |
| 8. Bauer T, Bouman HJ, Van Werkum JW, Ford NF, Ten Berg JM, Taubert D. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. British Medical Journal 2011;343(7819) (no pagination). | Publication type: systematic review |
| 9. Bhopalwala AM, Hong RA, Khan ZR, Valentin MR, Badawi RA. Routine screening for CYP2C19 polymorphisms for patients being treated with clopidogrel is not recommended. Hawai’i Journal of Medicine and Public Health: A Journal of Asia Pacific Medicine & Public Health 2015;74(1). | Publication type: review |
| 10. Bo H, De-Jun C, Ying R, Bin H, Da-Ping Y, Xun Z. Effect of cytochrome P450 2C19*17 allelic variant on cardiovascular and cerebrovascular outcomes in clopidogrel-treated patients: a systematic review and meta-analysis. Journal of Research in Medical Sciences 2017;22. | Publication type: systematic review |
| 11. Borchard-Tuch C. Transient ischemic attacks/stroke: CYP2C19 variations induce reduced effectiveness of clopidogrel. Medizinische Monatsschrift fur Pharmazeuten 2017;40(6). | Publication type: commentary |
| 12 Calcagno S, Di Pietro R, Biondi-Zoccai G, Versaci F. Do We Really Need Routine CYP2C19 Genotyping? JACC: Cardiovascular Interventions 2020;13(9). | Publication type: letter |
| 13. Castrichini M, Luzum JA, Pereira N. Pharmacogenetics of antiplatelet therapy. Annual Review of Pharmacology and Toxicology 2022;1. | Publication type: review |
| 14. Chen YB, Zhou ZY, Li GM, Xiao CX, Yu WB, Zhong SL, et al. Influences of an NR1I2 polymorphism on heterogeneous antiplatelet reactivity responses to clopidogrel and clinical outcomes in acute ischemic stroke patients. Acta Pharmacologica Sinica 2019;40(6). | Intervention: no intervention listed in the protocol (obj1/2) |
| 15. Chi NF, Wang SJ. CYP2C19 loss-of-function alleles: a common but overlooked problem associated with clopidogrel resistance. Journal of the Chinese Medical Association 2019;82(10). | Publication type: editorial |
| 16. Department of Neurology, Zhujiang Hospital of Southern Medical University. The Efficacy of Clopidogrel on Patients of Different CYP2C19 Genotypes Under the Guidance of Thromboelastogram. ChiCTR1800015314. 2018. URL: www.cochranelibrary.com/central/doi/10.1002/central/CN-01898880/full (accessed September 2022). | Outcomes: did not report on any outcomes specified in the inclusion criteria. |
| 17. Dong Y, Cheng X, Dong Q. Letter by Dong et al. regarding article, ‘CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China’. Stroke 2013;44(9). | Publication type: letter |
| 18. Ellithi M, Baye J, Wilke RA. CYP2C19 genotype-guided antiplatelet therapy: promises and pitfalls. Pharmacogenomics 2020;21(12). | Publication type: review |
| 19. Fang L, Zhao Y, Wang N, Yang Z, Huang H, Lin M. Association of CYP2C19 gene polymorphisms with long-term recurrent risk of ischemic stroke among ethnic Han Chinese from Fujian. [Chinese]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chinese journal of medical genetics 2015;32(6). | Unretrievable |
| 20. Geisler T, Bigalke B, Schwab M. CYP2C19 genotype and outcomes of clopidogrel treatment [1]. New England Journal of Medicine 2011;364(5). | Publication type: letter |
| 21. Han SW, Park JH, Kim K, Lee KY. Influence of smoking on the effect of clopidogrel may be dependent on CYP2C19 polymorphisms. International Journal of Stroke 2018;13(2 Suppl. 1). | Intervention: not the right test and there is no report of LOF alleles |
| 22. Han Y, Lv HH, Liu X, Dong Q, Yang XL, Li SX, et al. Influence of Genetic polymorphisms on clopidogrel response and clinical outcomes in patients with acute ischemic stroke CYP2C19 genotype on clopidogrel response. CNS Neuroscience and Therapeutics 2015;21(9). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 23. Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. Journal of the American Medical Association 2011;306(24). | Publication type: systematic review |
| 24. Hulot JS, Collet JP, Silvain J, Pena A, Bellemain-Appaix A, Barthelemy O, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration. A systematic meta-analysis. Journal of the American College of Cardiology 2010;56(2). | Publication type: systematic review |
| 25. Jeong TD, Kim SM, Kim HJ, Lee W, Kwon SU, Min WK, et al. CYP2C19 genotype and early ischemic lesion recurrence in stroke patients treated with clopidogrel. Journal of Stroke and Cerebrovascular Diseases 2015;24(2). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 26. Jia DM, Chen ZB, Zhang MJ, Yang WJ, Jin JL, Xia YQ, et al. CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China. Stroke 2013;44(6). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 27. Joob B, Wiwanitkit V. CYP2C19*2 polymorphism and clopidogrel resistance. Archivos de Cardiologia de Mexico 2020;90(4). | Publication type: letter |
| 28. Kitazono T, Ikeda Y, Nishikawa M, Yoshiba S, Abe K, Ogawa A. Influence of cytochrome P450 polymorphisms on the antiplatelet effects of prasugrel in patients with non-cardioembolic stroke previously treated with clopidogrel. Journal of Thrombosis and Thrombolysis 2018;46(4). | Outcomes: did no report on any relevant outcomes for comparison of interest specified in the inclusion criteria by genotype |
| 29. Kremers F, Van Den Biggelaar J, Lingsma H, Roozenbeek B, Dippel D. Effect of clopidogrel in CYP2C19 polymorphisms for prevention of vascular events in patients with cardiovascular disease or recent TIA or minor ischemic stroke. European Stroke Journal 2021;6(1 SUPPL). | Publication type: systematic review |
| 30. Kreutzkamp B. Secondary prevention with clopidogrel: CYP2C19*2 gene variant as a common cause of treatment failure. [German]. Arzneimitteltherapie 2009;27(9). | Publication type: commentary |
| 31. Lan H, Ying T, Xi-Hua S, Yi L. Anti-platelet therapy in mild cerebral infarction patients on the basis of CYP2C19 metabolizer status. Cell Transplantation 2019;28(8). | Intervention: no intervention listed in the protocol (obj1/2) and presence of *2 or *3 LOF alleles |
| 32. Lee BC, Oh MS, Yu KH, Kwon KH, Kim BS. CYP2C19 variants do not associated with clinical efficacy of clopidogrel in Korean stroke survivors. Cerebrovascular Diseases 2012;2. | Unretrievable |
| 33. Li C, Jia W, Li J, Li F, Ma J, Zhou L. Association with CYP2C19 polymorphisms and clopidogrel in treatment of elderly stroke patients. BMC Neurology 2021;21(1). | Intervention: no relevant comparisons of interest |
| 34. Lin J, Han Z, Wang C, Yi X, Chai Z, Zhou Q, et al. Dual therapy with clopidogrel and aspirin prevents early neurological deterioration in ischemic stroke patients carrying CYP2C19*2 reduced-function alleles. European Journal of Clinical Pharmacology 2018;74(9). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 35. Lin YJ, Li JW, Zhang MJ, Qian L, Yang WJ, Zhang CL, et al. The association between CYP2C19 genotype and of in-stent restenosis among patients with vertebral artery stent treatment. CNS Neuroscience and Therapeutics 2014;20(2). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 36. Liu YP, Hao PP, Zhang MX, Zhang C, Gao F, Zhang Y, et al. Association of genetic variants in CYP2C19 and adverse clinical outcomes after treatment with clopidogrel: an updated meta-analysis. Thrombosis Research 2011;128(6). | Publication type: letter |
| 37. Lun R, Zitikyte G, Roy DC, Dhaliwal S, Hutton B, Dowlatshahi D. Ticagrelor and aspirin vs. clopidogrel and aspirin in patients with minor ischemic stroke or transient ischemic attack (TIA) – an updated network meta-analysis. European Stroke Journal 2022;7(1 Suppl.). | Publication type: systematic review |
| 38. Lyu SQ, Yang YM, Zhu J, Wang J, Wu S, Zhang H, et al. The efficacy and safety of CYP2C19 genotype-guided antiplatelet therapy compared with conventional antiplatelet therapy in patients with acute coronary syndrome or undergoing percutaneous coronary intervention: a meta-analysis of randomized controlled trials. Platelets 2020;31(8). | Publication type: systematic review |
| 39. Maeda A. Different influences of CYP2C19 gene polymorphisms on the antiplatelet effect of clopidogrel and ticlopidine. [Japanese]. Japanese Journal of Clinical Pharmacology and Therapeutics 2011;42(3). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 40. Malik AH, Gupta R, Chakraborty S, Mahajan P, Bandyopadhyay D, Yandrapalli S, et al. Effect of genotype-guided oral P2Y12 inhibitor selection after percutaneous coronary intervention: a systematic review and meta-analysis of randomized clinical trials. Cardiovascular Revascularization Medicine 2022;41. | Publication type: systematic review |
| 41. Mao L, Jian C, Changzhi L, Dan H, Suihua H, Wenyi T, et al. Cytochrome CYP2C19 polymorphism and risk of adverse clinical events in clopidogrel-treated patients: a meta-analysis based on 23,035 subjects. Archives of cardiovascular diseases 2013;106(10). | Publication type: systematic review |
| 42. McDermott JH, Leach M, Sen D, Smith CJ, Newman WG, Bath PM. The role of CYP2C19 genotyping to guide antiplatelet therapy following ischemic stroke or transient ischemic attack. Expert Review of Clinical Pharmacology 2022;1. | Publication type: literature review |
| 43. Medco Health Solutions Inc. Genotype Guided Comparison of Clopidogrel and Prasugrel Outcomes Study (GeCCO). NCT00995514. 2012. URL: https://ClinicalTrials.gov/show/NCT00995514 (accessed September 2022). | Population: no report of stroke or TIA |
| 44. Minderhoud C, Otten LS, Hilkens PHE, van den Broek MPH, Harmsze AM. Increased frequency of CYP2C19 loss-of-function alleles in clopidogrel-treated patients with recurrent cerebral ischemia. British Journal of Clinical Pharmacology 2022;88(7). | Study design: not a RCT or cohort study |
| 47. Niu X, Mao L, Huang Y, Baral S, Li JY, Gao Y, et al. CYP2C19 polymorphism and clinical outcomes among patients of different races treated with clopidogrel: a systematic review and meta-analysis. Journal of Huazhong University of Science and Technology Medical Sciences 2015;35(2). | Publication type: systematic review |
| 48. Pan Y, Chen W, Xu Y, Yi X, Han Y, Yang Q, et al. Genetic polymorphisms and clopidogrel efficacy for acute ischemic stroke or transient ischemic attack. Circulation 2017;135(1). | Publication type: systematic review |
| 49. Pilling LC, Turkmen D, Fullalove H, Atkins JL, Delgado J, Kuo CL, et al. Analysis of CYP2C19 genetic variants with ischaemic events in UK patients prescribed clopidogrel in primary care: a retrospective cohort study. BMJ Open 2021;11(12). | Population – not exclusively stroke/TIA population and data N/R separately for subset that were. |
| 50. Siasos G, Tousoulis D, Stefanadis C. CYP2C19 genotype and outcomes of clopidogrel treatment [2]. New England Journal of Medicine 2011;364(5). | Publication type: letter |
| 51. Sienkiewicz-Oleszkiewicz B, Wiela-Hojenska A. CYP2C19 polymorphism in relation to the pharmacotherapy optimization of commonly used drugs. Pharmazie 2018;73(11). | Publication type: literature review |
| 52. Singh A, Coy K, Schmidtman D, Stys A, Stys T. Impact of clopidogrel metabolizer status on incidence of gastrointestinal bleed. Circulation Conference: American Hearts Association’s 2021;144(Suppl. 1). | Population: no report of stroke or TIA |
| 53. Sofi F, Giusti B, Gori A, Marcucci R, Abbate R, Gensini G. Cytochrome P450 2C19 polymorphism and cardiovascular recurrences in patients under clopidogrel treatment: a meta-analysis. Journal of Thrombosis and Haemostasis 2009;7(S2). (The review was reported in publication: Sofi, F., Giusti, B., Marcucci, R. et al. Cytochrome P450 2C19*2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J 11) |
Publication type: systematic review |
| 54. Sofi F, Giusti B, Gori AM, Cesari F, Marcucci R, Abbate R, et al. Cytochrome P450 2c19 polymorphism and cardiovascular recurrences in patients under clopidogrel treatment: a meta-analysis. European Journal of Cardiovascular Prevention and Rehabilitation 2010;2. | Publication type: systematic review |
| 55. Song TJ, Kim J, Han SW, Kim YD, Lee JY, Ahn SH, et al. Clopidogrel preventive effect based on cytochrome P450 2C19 genotype in ischaemic stroke: protocol for multicentre observational study. BMJ Open 2020;10(8). | Publication type: Protocol |
| 56. Sorich MJ, Polasek TM, Wiese MD. Systematic review and meta-analysis of the association between cytochrome P450 2C19 genotype and bleeding. Thrombosis and Haemostasis 2012;108(1). | Publication type: letter |
| 57. Stanley A, Beoris M, Austria A, Baca AA, Amos Wilson J, Garces JA, et al. Clopidogrel utilization in a U.S. population: a pharmacogenetic and metabolic overview of patient eligibility. Journal of Molecular Diagnostics 2015;17(6). | Outcomes: did not report on any outcomes specified in the inclusion criteria. |
| 58. University of Puerto Rico. A Genomic Approach for Clopidogrel in Caribbean Hispanics. NCT03419325. 2022. URL: https://ClinicalTrials.gov/show/NCT03419325 (accessed September 2022). | Population: no report of stroke or TIA. |
| 59. Wang D, Li L, Jiang J, Zhang Q, Liu M, Liu Y, et al. Age-dependent association of CYP2C19 polymorphisms with clinical outcome of clopidogrel therapy in minor stroke patients with large-artery atherosclerosis. European Journal of Clinical Pharmacology 2020;76(9). | Outcomes: did not report on any outcomes specified in the inclusion criteria. |
| 60. Xie Q, Xiang Q, Liu Z, Mu G, Zhou S, Zhang Z, et al. Effect of CYP2C19 genetic polymorphism on the pharmacodynamics and clinical outcomes for patients treated with ticagrelor: a systematic review with qualitative and quantitative meta-analysis. BMC Cardiovascular Disorders 2022;22(1). | Publication type: systematic review |
| 61. Xu H, Xu B, Zu Q, Zhao Y, Gao P, Yu Y. Effects of CYP2C19 gene polymorphism on the clinical prognosis of clopidogrel in elderly patients with acute cerebral infarction. [Chinese]. Chinese Journal of Clinical Pharmacology and Therapeutics 2020;25(9). | Unretrievable |
| 62. Yamaguchi Y, Abe T, Sato Y, Matsubara Y, Moriki T, Murata M. Effects of VerifyNow P2Y12 test and CYP2C19*2 testing on clinical outcomes of patients with cardiovascular disease: A systematic review and meta-analysis. Platelets 2013;24(5). | Publication type: systematic review |
| 63. Yang L, Xie J, Liu Y, Hu X. Correlation between the genetic polymorphism of CYP2C19*2, *3 and the clinical efficacy of clopidogrel: a systematic review. [Chinese]. Chinese Journal of Evidence-Based Medicine 2012;12(9). | Unretrievable |
| 64. Yang Y, Chen W, Pan Y, Yan H, Meng X, Liu L, et al. Ticagrelor is superior to clopidogrel in inhibiting platelet reactivity in patients with minor stroke or TIA. Frontiers in Neurology 2020;11 (no pagination). | Outcomes: did not report on any outcomes specified in the inclusion criteria by genotype |
| 65. Yi X, Zhou Q, Wang C, Lin J, Chai Z. Aspirin plus clopidogrel may reduce the risk of early neurologic deterioration in ischemic stroke patients carrying CYP2C19*2 reduced-function alleles. Journal of Neurology 2018;265(10). | Outcomes: did not report on any outcomes specified in the inclusion criteria by genotype |
| 66. New Zealand University Hospital. Clopidogrel Response and CYP2C19 Genotype in Ischemic Stroke Patients (CLOGIS). NCT03385538. 2017. URL: https://ClinicalTrials.gov/show/NCT03385538 (accessed September 2022). | Outcomes: did not report on any outcomes specified in the inclusion criteria |
| 67. Zhang H, Xiang Q, Liu Z, Mu G, Xie Q, Zhou S, et al. Genotype-guided antiplatelet treatment versus conventional therapy: a systematic review and meta-analysis. British Journal of Clinical Pharmacology 2021;87(5). | Publication type: systematic review |
| 68. Zhang S, Lai X, Li W, Xiong Z, Xu A, Xu A, et al. VASP phosphorylation and genetic polymorphism for clopidogrel resistance in Chinese patients with non-cardioembolic ischemic stroke. Thrombosis Research 2014;134(6). | Publication type: systematic review |
| 69. Zhang X, Jing J, Zhao X, Liu L, Wang A, Pan Y, et al. No rebound effect after a course of clopidogrel in patients with acute TIA or minor stroke. Neurological Research 2022. | Exposure: compares patients who continued treatment vs. patients who stopped treatment |
| 70. Zheng L, Yang C, Xiang L, Hao Z. Genotype-guided antiplatelet therapy compared with conventional therapy for patients with acute coronary syndromes: a systematic review and meta-analysis. Biomarkers 2019;24(6). | Publication type: systematic review |
| 71. Zhu Y, Moriarty JP, Swanson KM, Takahashi PY, Bielinski SJ, Weinshilboum R, et al. PCV10 a model-based cost-effectiveness analysis of pharmacogenomic panel testing in cardiovascular disease management: preemptive, reactive or none? Value in Health Regional Issues 2020;22(Suppl.). | Study design: cost-effectiveness study |
Studies excluded at full-text screening (Objectives 4–5)
| Study details | Reason for exclusion |
|---|---|
| Biswas M. Global distribution of CYP2C19 risk phenotypes affecting safety and effectiveness of medications. Pharmacogenomics 2021;21(2):190–9. | Not a primary study: secondary reanalysis of a data set |
| Capodanno D, Angiolillo DJ, Lennon RJ, Goodman SG, Kim SW, O’Cochlain F, et al. ABCD-GENE score and clinical outcomes following percutaneous coronary intervention: insights from the TAILOR-PCI trial. Journal of the American Heart Association 2022;11(4). | Used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Chen X, Xu J, Chen S, Dong Q, Dong Y. Dual antiplatelet therapy with ticagrelor may increase the risk of all bleeding events in patients with minor strokes or high risk TIAs: a meta- analysis [published online ahead of print, 2022 Mar 3]. Stroke and Vascular Neurology 2022; | Publication type: systematic review |
| Claassens DMF, Bergmeijer TO, Vos GJA, Hermanides RS, t Hof A, van der Harst P, et al. Clopidogrel versus ticagrelor or prasugrel after primary percutaneous coronary intervention according to CYP2C19 genotype a poular genetics subanalysis. Circulation-Cardiovascular Interventions 2021;14(4). | Used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Claassens DMF, Gimbel ME, Bergmeijer TO, Vos GJA, Hermanides RS, van der Harst P, et al. Clopidogrel in noncarriers of CYP2C19 loss-of-function alleles versus ticagrelor in elderly patients with acute coronary syndrome: a pre-specified sub analysis from the POPular Genetics and POPular Age trials CYP2C19 alleles in elderly patients. International Journal of Cardiology 2021;334. | Used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Claassens DMF, Vos GJA, Bergmeijer TO, Hermanides RS, van ‘t Hof AWJ, van der Harst P, et al. A genotype-guided strategy for oral P2Y(12) inhibitors in primary PCI. New England Journal of Medicine 2019;381(17). | Used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Collet JP, Kerneis M, Hulot JS, O’Connor SA, Silvain J, Mansencal N, et al. Point-of-care genetic profiling and/or platelet function testing in acute coronary syndrome. Thrombosis and Haemostasis 2016;115(2). | Test: POCT out of scope (Verigene test) |
| Dawson J, Merwick A, Webb A, Dennis M, Ferrari J, Fonseca AC, et al. European Stroke Organisation expedited recommendation for the use of short-term dual antiplatelet therapy early after minor stroke and high-risk TIA. European Stroke Journal 2021;6(2):VI. | Not a primary study: guideline |
| Erlinge D, James S, Duvvuru S, Jakubowski J, Wanger H, Varenhorst C, et al. Point-of-care genetic testing of eleven CYP2C19 single nucleotide polymorphisms identifies extensive and reduced metabolizers of clopidogrel with high accuracy in patients with coronary artery disease. Journal of the American College of Cardiology 2012;17. | Test: POCT out of scope (Verigene test) |
| Franchi F, Rollini F. Genotype-guided antiplatelet therapy in patients with coronary artery disease. JACC-Cardiovascular Interventions 2021;14(7). | Publication type: editorial |
| Gurbel PA, Bliden KP, Antonino M, Verma A, Jeong YH, Tantry US. First validation of point-of-care CYP2C19 genetic testing in patients undergoing coronary angiography with the Verigene nucleic acid assay. Journal of the American College of Cardiology 2011;1. | Test: POCT out of scope (Verigene test) |
| Hao Q, Tampi M, O’Donnell M, Foroutan F, Siemieniuk RA, Guyatt G. Clopidogrel plus aspirin versus aspirin alone for acute minor ischaemic stroke or high risk transient ischaemic attack: systematic review and meta-analysis. British Medical Journal 2018;363:k5108. | Publication type: systematic review |
| Hulot JS, Chevalier B, Belle L, Cayla G, Khalife K, Funck F, et al. Routine CYP2C19 genotyping to adjust thienopyridine treatment after primary PCI for STEMI results of the GIANT study. JACC-Cardiovascular Interventions 2020;13(5). | Test: no POCT used |
| Hulot JS, Collet JP, Cayla G, Silvain J, Allanic F, Bellemain-Appaix A, et al. CYP2C19 but not PON1 genetic variants influence clopidogrel pharmacokinetics, pharmacodynamics, and clinical efficacy in post-myocardial infarction patients. Circulation-Cardiovascular Interventions 2011;4(5). | Test: no POCT used |
| Imam Abdulrahman Bin Faisal University. Bedside Testing of CYP2C19 Gene for Treatment of Patients With PCI With Antiplatelet Therapy. NCT01823185.2016. URL: https://ClinicalTrials.gov/show/NCT01823185 (accessed November 2022). | Test: used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Johnston SC, Amarenco P, Albers GW, Denison H, Easton JD, Evans SR, et al. Ticagrelor versus aspirin in acute stroke or transient ischemic attack. New England Journal of Medicine 2016;375(1):35–43. | Test: no POCT used |
| Kim HK, Sibbing D, Jeong YH. Effect of genotype-guided strategy in East Asian vs. Caucasian patients after percutaneous coronary intervention: insight from the TAILOR-PCI trial. Journal of Thoracic Disease 2020;12(12). | Not a primary study |
| Koltowski L, Aradi D, Huczek Z, Tomaniak M, Sibbing D, Filipiak KJ, et al. Study design and rationale for Optimal aNtiplatelet pharmacotherapy guided by bedSIDE genetic or functional TESTing in elective percutaneous coronary intervention patients (ONSIDE TEST): a prospective, open-label, randomised parallel-group multicentre trial. Kardiologia Polska 2016;74(4). | Not a primary study |
| Lee CR, Luzum JA, Sangkuhl K, et al. Clinical pharmacogenetics implementation consortium guideline for CYP2C19 genotype and clopidogrel therapy: 2022 update [published online ahead of print, 2022 Jan 16]. Clinical Pharmacology and Therapeutics. 2022; https://doi.org/10.1002/cpt.2526. | Not a primary study |
| Li YJ, Chen X, Tao LN, Hu XY, Wang XL, Song YQ. Association between CYP2C19 polymorphisms and clinical outcomes in patients undergoing stent procedure for cerebral artery stenosis. Sci Rep. 2021;11(1):5974. Published 2021 Mar 16. | Test: no POCT used |
| Li Y-J, Chen X, Tao L-N, Hu X-Y, Wang X-L, Song Y-Q. Association between CYP2C19 polymorphisms and clinical outcomes in patients undergoing stent procedure for cerebral artery stenosis. Scientific Reports 2021;11(1):5974. | Test: no POCT used |
| Luengo-Fernandez R, Violato M, Candio P, Leal J. Economic burden of stroke across Europe: a population-based cost analysis. European Stroke Journal 2020;5(1):17–25. | Test: not an evaluation of a POCT (cost-analysis) |
| Madan M, Abbott JD, Lennon R, So DYF, MacDougall AM, McLaughlin MA, et al. Sex-specific differences in clinical outcomes after percutaneous coronary intervention: insights from the TAILOR-PCI trial. Journal of the American Heart Association 2022;11(12). | Test: used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Marziliano N, Notarangelo MF, Cereda M, Caporale V, Coppini L, Demola MA, et al. Rapid and portable, lab-on-chip, point-of-care genotyping for evaluating clopidogrel metabolism. International Journal of Clinical Chemistry 2015;Part B. 451. | Population: not our cohort of interest |
| Meng X, Wang A, Zhang G, Niu S, Li W, Han S, et al. Analytical validation of GMEX rapid point-of-care CYP2C19 genotyping system for the CHANCE-2 trial. Stroke and Vascular Neurology 2021;6(2). | Test: POCT out of scope (GMEX system) |
| Anita Patel, Vladislav Berdunov, Derek King, Zahidul Quayyum, Raphael Wittenberg, Martin Knapp. Current, future and avoidable costs of stroke in the UK. London: Centre for Primary Care & Public Health, Queen Mary University of London, and the Personal Social Services Research Unit; 2017. www.stroke.org.uk/sites/default/files/costs_of_stroke_in_the_uk_summary_report_0.pdf (accessed November 2022) | Not a primary study: published report |
| Pereira NL, Avram R, So DY, Iturriaga E, Byrne J, Lennon RJ, et al. Rationale and design of the TAILOR-PCI digital study: transitioning a randomized controlled trial to a digital registry. American Heart Journal 2021;232 | Not a primary study: paper describing the TAILOR-PCI study |
| Pereira NL, Rihal CS, So DYF, Rosenberg Y, Lennon RJ, Mathew V, et al. Clopidogrel Pharmacogenetics state-of-the-art review and the TAILOR-PCI study. Circulation-Cardiovascular Interventions 2019;12(4). | Not a primary study: literature review |
| Pereira NL, Farkouh ME, So D, Lennon R, Geller N, Mathew V, et al. Effect of genotype-guided oral P2Y12 inhibitor selection vs. conventional clopidogrel therapy on ischemic outcomes after percutaneous coronary intervention: the TAILOR-PCI randomized clinical trial. Journal of the American Medical Association 2020;324(8). | Population: no extra data on cohort of interest |
| Pilling LC, Türkmen D, Fullalove H, et al. Analysis of CYP2C19 genetic variants with ischaemic events in UK patients prescribed clopidogrel in primary care: a retrospective cohort study. BMJ Open 2021;11(12):e053905. Published 2021 Dec 13. | Test: no POCT used |
| Stimpfle F, Karathanos A, Droppa M, et al. Impact of point-of-care testing for CYP2C19 on platelet inhibition in patients with acute coronary syndrome and early dual antiplatelet therapy in the emergency setting. Thrombosis Research 2014;134(1):105–10. | Used Genomadix Cube but did not report an evaluation of the test or outcomes of interest |
| Spartan Bioscience Inc. Spartan FRX Project Reproducibility Study. NCT01676298. URL: https://ClinicalTrials.gov/show/NCT01676298 (accessed November 2022). | Study design: analytical validity study |
| Uchino K. Guideline: starting dual antiplatelet therapy ≤ 24 h after high-risk TIA or minor ischemic stroke is recommended. Annals of Internal Medicine. 2019 16;170(8):JC38. | Not a primary study: guideline |
| Wafa HA, Wolfe CDA, Emmett E, Roth GA, Johnson CO, Wang Y. Burden of stroke in Europe: thirty-year projections of incidence, prevalence, deaths, and disability-adjusted life years. Stroke 2020;51(8):2418–27. https://doi.org/10.1161/STROKEAHA.120.029606. Epub 2020 Jul 10. | Not a primary study: review of prevenance |
| Wang YJ, Meng X, Wang AX, Xie XW, Pan YS, Johnston SC, et al. Ticagrelor versus clopidogrel in CYP2C19 loss-of-function carriers with stroke or TIA. New England Journal of Medicine 2021;385(27). | Test: POCT out of scope (GMEX system) |
| Wang YL, Zhao XQ, Lin JX, Li H, Johnston SC, Lin Y, et al. Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. Journal of the American Medical Association 2016;316(1). | Test: no POCT used |
| Zhang LC, Ma XW, You GL, Zhang XQ, Fu QH. A novel multiplex HRM assay to detect clopidogrel resistance. Scientific Reports 2017;7. | Test: test out of scope (Multiplex HRM Assay) |
Studies included in manufacturers’ submissions
Below we tabulate how studies reported in submissions were handled. These tables/studies apply to the review of test accuracy (Objective 4 or 5).
No studies were included in the manufacturers’ submissions.
| Study details | Decision | Reason for exclusion |
|---|---|---|
| Al-Rubaish AM, Al-Muhanna FA, et al. Prevalence of CYP2C19*2 carriers in Saudi ischemic stroke patients and the suitability of using genotyping to guide antiplatelet therapy in a university hospital setup. Drug Metabolism and Personalized Therapy. 2021 Jul 8;37(1). | Included | N/A |
| Baudhuin LM, Train LJ, et al. Point of care CYP2C19 genotyping after percutaneous coronary intervention. Pharmacogenomics 2022 Apr 21. | Included | N/A |
| Roberts JD, Wells GA, Le May MR, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379(9827). | Included | N/A |
| Zhou Y, Armstead AR, Coshatt GM, Limdi NA, Harada S. Comparison of two point-of-care CYP2C19 genotyping assays for genotype-guided antiplatelet therapy. Ann Clin Lab Sci. 2017 Nov;47(6):738–43. | Included | N/A |
| Biswas M. Global distribution of CYP2C19 risk phenotypes affecting safety and effectiveness of medications. Pharmacogenomics 2021;21(2). | Excluded | Not a primary study: prevalence study (no data) |
| Chen X, Xu J, Chen S, Dong Q, Dong Y. Dual antiplatelet therapy with ticagrelor may increase the risk of all bleeding events in patients with minor strokes or high risk TIAs: a meta-analysis [published online ahead of print, 2022 Mar 3]. Stroke and Vascular Neurology 2022; | Excluded | Publication type: meta-analysis |
| Claassens DMF, Vos GJA, Ten Berg JM et al. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. New England Journal of Medicine 2019 Oct 24;381(17). | Excluded | Outcomes: no outcome data for Genomadix |
| Dawson J, Merwick Á, Webb A, et al. European Stroke Organisation expedited recommendation for the use of short-term dual antiplatelet therapy early after minor stroke and high-risk TIA. European Stroke Journal 2021;6(2). | Excluded | Not a primary study: guideline |
| Hao Q, Tampi M, O’Donnell M, Foroutan F, Siemieniuk RA, Guyatt G. Clopidogrel plus aspirin versus aspirin alone for acute minor ischaemic stroke or high risk transient ischaemic attack: systematic review and meta-analysis. British Medical Journal 2018;363. | Excluded | Publication type: systematic review |
| Johnston SC, Amarenco P, Albers GW, Denison H, Easton JD, Evans SR, et al.; SOCRATES Steering Committee and Investigators. Ticagrelor versus aspirin in acute stroke or transient ischemic attack. New England Journal of Medicine 2016 Jul 7;375(1). | Excluded | Test: no reported CYP2C19 genotyping |
| Lee CR, Luzum JA, Sangkuhl K, et al. Clinical pharmacogenetics implementation consortium guideline for CYP2C19 genotype and clopidogrel therapy: 2022 update [published online ahead of print, 2022 Jan 16]. Clinical Pharmacology and Therapeutic. | Excluded | Not a primary study: guideline |
| Li C, Jia W, Li J, Li F, Ma J, Zhou L. Association with CYP2C19 polymorphisms and clopidogrel in treatment of elderly stroke patients. BMC Neurology 2021;21(1):104. | Excluded | Test: no reported CYP2C19 genotyping |
| Li YJ, Chen X, Tao LN, Hu XY, Wang XL, Song YQ. Association between CYP2C19 polymorphisms and clinical outcomes in patients undergoing stent procedure for cerebral artery stenosis. Scientific Reports 2021;11(1). | Excluded | Test: study did not report an evaluation of POCT in scope |
| Luengo-Fernandez R, Violato M, Candio P, Leal J. Economic burden of stroke across Europe: a population-based cost analysis. European Stroke Journal 2020;5(1). | Excluded | Test/ Outcomes: no reported CYP2C19 genotyping and study reports cost outcomes |
| Patel A, Berdunov V, King D, Quayyum Z, Wittenberg R, Knapp M. Current, Future and Avoidable Costs of Stroke in the UK. London: Centre for Primary Care & Public Health, Queen Mary University of London, and the Personal Social Services Research Unit; 2017. www.stroke.org.uk/sites/default/files/costs_of_stroke_in_the_uk_summary_report_0.pdf (accessed November 2022) | Excluded | Not a primary study: report |
| Pereira NL, Farkouh ME, So D, et al. Effect of genotype-guided oral P2Y12 inhibitor selection vs. conventional clopidogrel therapy on ischemic outcomes after percutaneous coronary intervention: the TAILOR-PCI randomized clinical trial. Journal of the American Medical Association 2020;324(8). | Excluded | Population: study cohort not of interest |
| Pilling LC, Türkmen D, Fullalove H, et al. Analysis of CYP2C19 genetic variants with ischaemic events in UK patients prescribed clopidogrel in primary care: a retrospective cohort study. BMJ Open 2021;11(12). | Excluded | Test: no reported CYP2C19 genotyping |
| Stimpfle F, Karathanos A, Droppa M, et al. Impact of point-of-care testing for CYP2C19 on platelet inhibition in patients with acute coronary syndrome and early dual antiplatelet therapy in the emergency setting. Thrombosis Research. 2014;134(1). | Excluded | Outcomes: no outcome data for Genomadix |
| Uchino K. Guideline: starting dual antiplatelet therapy ≤ 24 h after high-risk TIA or minor ischemic stroke is recommended. Annanls of Internal Medicine 2019 Apr 16;170(8). | Excluded | Not a primary study: guideline |
| Wafa HA, Wolfe CDA, Emmett E, Roth GA, Johnson CO, Wang Y. Burden of stroke in Europe: thirty-year projections of incidence, prevalence, deaths, and disability-adjusted life years. Stroke 2020;51(8). | Excluded | Not a primary study: modelling study. |
| Wang Y, Meng X, Wang A, et al. Ticagrelor vs. clopidogrel in CYP2C19 loss-of-function carriers with stroke or TIA. NEJM 385:2520–30. Published October 28, 2021. | Excluded | Test: study did not report an evaluation of POCT in scope |
Studies excluded at full-text screening (review of cost-effectiveness)
| Study details | Reason for exclusion |
|---|---|
| Bereza BG, Coyle D, So DY, Kadziola Z, Wells G, Grootendorst P, et al. Stated preferences for attributes of a CYP2C19 pharmacogenetic test among the general population presented with a hypothetical acute coronary syndrome scenario. Clinico Economics and Outcomes Research 2020;12. | Population/study type: population is ACS and study is not a CEA |
| Dong OM, Friede KA, Chanfreau-Coffinier C, Voora D. Cost-effectiveness of CYP2C19-guided P2Y(12) inhibitors in veterans undergoing percutaneous coronary intervention for acute coronary syndromes. European Heart Journal-Quality of Care and Clinical Outcomes; 10. | Population: PCI |
| Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai WR, et al. Diagnostic point-of-care tests in resource-limited settings. Lancet Infectious Diseases 2014;14(3). | Study type: not a CEA |
| Jiang M, You JHS. Cost-effectiveness analysis of personalized antiplatelet therapy in patients with acute coronary syndrome. Pharmacogenomics 2016;17(7). | Population: ACS |
| Kim K, Touchette DR, Cavallari LH, Ardati AK, DiDomenico RJ. Cost-effectiveness of strategies to personalize the selection of P2Y(12) inhibitors in patients with acute coronary syndrome. Cardiovascular Drugs and Therapy 2019;33(5). | Population: ACS |
| Lala A, Berger JS, Sharma G, Hochman JS, Scott Braithwaite R, Ladapo JA. Genetic testing in patients with acute coronary syndrome undergoing percutaneous coronary intervention: a cost-effectiveness analysis. Journal of Thrombosis and Haemostasis 2013;11(1). | Population: ACS |
| Pourdjabbar A, Hibbert B, Chong AY, Le May MR, Labinaz M, Simard T, et al. A randomised study for optimising crossover from ticagrelor to clopidogrel in patients with acute coronary syndrome The CAPITAL OPTI-CROSS Study. Thrombosis and Haemostasis 2017;117(2). | Population/study type: population is ACS and study is not a CEA |
Appendix 3 Adverse events definitions
| Study | Assigned category | |||
|---|---|---|---|---|
| Mild bleeding | Moderate bleeding | Severe bleeding | Any bleeding | |
| Chen et al. (2019)53 | N/R | N/R | N/R | Any bleeding or haemorrhage episodes reported |
| Han et al. (2017)48 | N/R | N/R | N/R | |
| Meschia et al. (2020)49 | All haemorrhagic events leading to interruption of therapy but not classifiable as major haemorrhagic events | N/R | A haemorrhagic event that results in clinically significant disability, symptomatic ICH, intraocular bleeding causing loss of vision, the need for a transfusion of two or more units of red cells or the equivalent amount of whole blood, or the need for hospitalisation | |
| Wang et al. (2016)52 | Bleeding that does not meet criteria for other categories | Requiring blood transfusion but not resulting in haemodynamic compromise | ICH or haemorrhage resulting in substantial haemodynamic compromise requiring treatment | |
| Wang et al. (2021)50 | ||||
| Yi et al. (2018)54 | ||||
| Wu et al. (2020)51 | N/R | N/R | N/R | |
| Study | Assigned category | |||
|---|---|---|---|---|
| Mild bleeding | Moderate bleeding | Severe bleeding | Any bleeding | |
| Chen et al. (2019)53 | N/R | N/R | N/R | Any bleeding or haemorrhage episodes reported |
| Han et al. (2017)48 | N/R | N/R | N/R | |
| Lin et al. (2014)59 | N/R | N/R | N/R | |
| McDonough et al. (2015)62 | N/R | N/R | Major bleeding: serious or life-threatening bleeding requiring transfusion of red cells or surgery or resulting in permanent functional sequelae or death. | |
| Meschia et al. (2020)49 | Minor haemorrhage: a haemorrhagic events leading to interruption of therapy but not classifiable as major haemorrhagic events | N/R | Major haemorrhage: a haemorrhagic event that results in clinically significant disability, symptomatic ICH, intraocular bleeding causing loss of vision, the need for a transfusion of two or more units of red cells or the equivalent amount of whole blood, or the need for hospitalisation | |
| Sun et al. (2015)68 | Mild bleeding: bleeding that does not meet criteria for other categories | Moderate bleeding: requiring blood transfusion but not resulting in haemodynamic compromise | Severe bleeding: ICH or haemorrhage resulting in substantial haemodynamic compromise requiring treatment | |
| Yi et al. (2018)54 | ||||
| Wang et al. (2016a)52 | ||||
| Wang et al. (2021)50 | ||||
| Tanaka et al. (2019)69 | N/R | N/R | Major bleeding: ICH, documented retroperitoneal bleed, or bleeding episodes that caused ≥ 2 g/dL decline in the haemoglobin level or required at least 1 unit of red cell transfusion. | |
Appendix 4 Additional analysis for Objectives 2 and 3
Forest plots for Objective 2
FIGURE 15.
Forest plot showing HRs (95% CI) for incidence of a composite of secondary vascular events in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.

FIGURE 16.
Forest plot showing HRs (95% CI) for incidence of any stroke in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.

FIGURE 17.
Forest plot showing HRs (95% CI) for incidence of other secondary vascular event outcomes in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.

FIGURE 18.
Forest plot showing HRs (95% CI) for incidence of any bleeding events in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.

FIGURE 19.
Forest plot showing HRs (95% CI) for incidence of other secondary adverse events in carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin) compared with an alternative antiplatelet regimen.
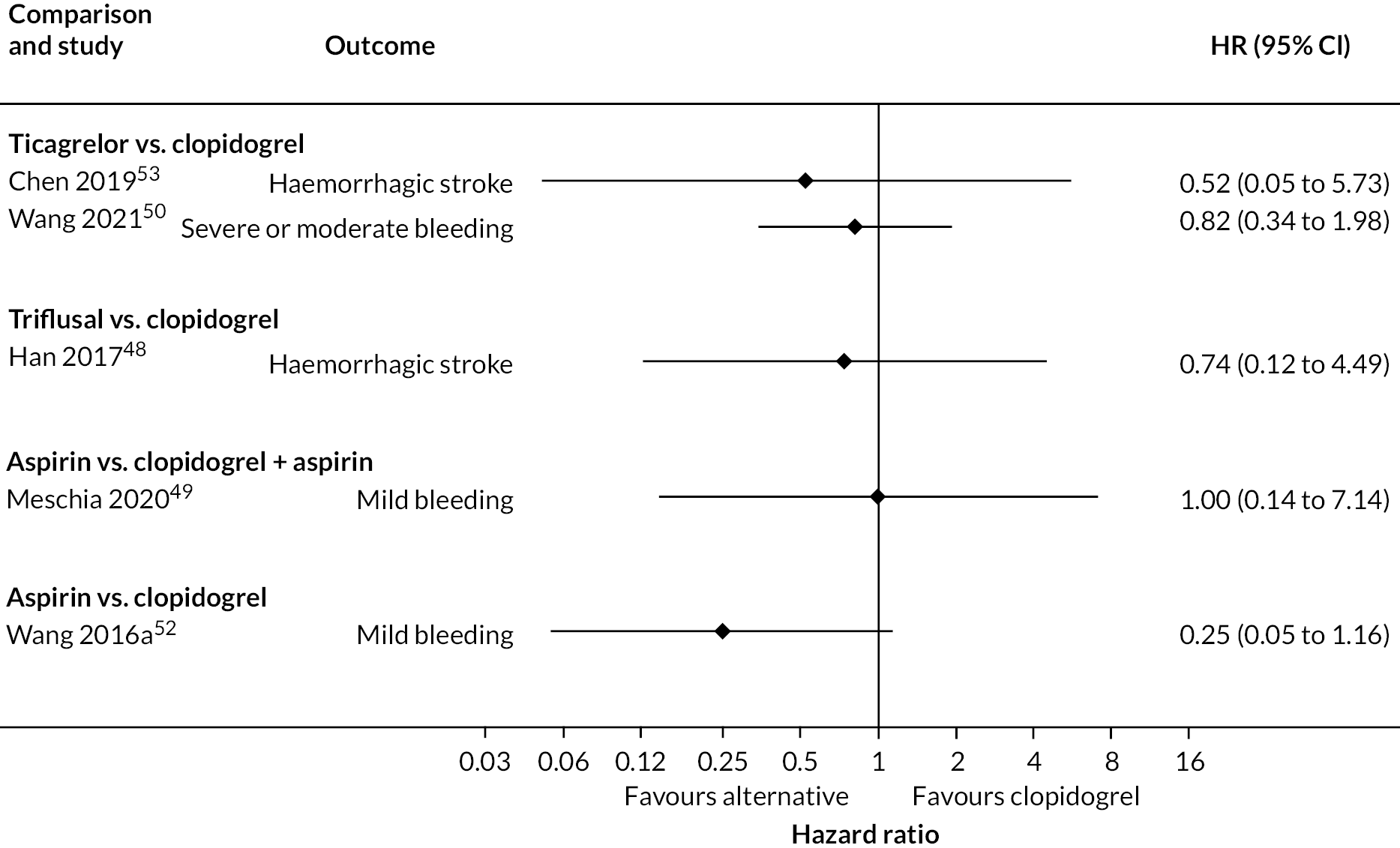
Additional Forest Plots for Objective 3
FIGURE 20.
Forest plot showing HRs (95% CI) for incidence of a composite outcome of secondary vascular events in carriers of LOF alleles compared with non-carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin).
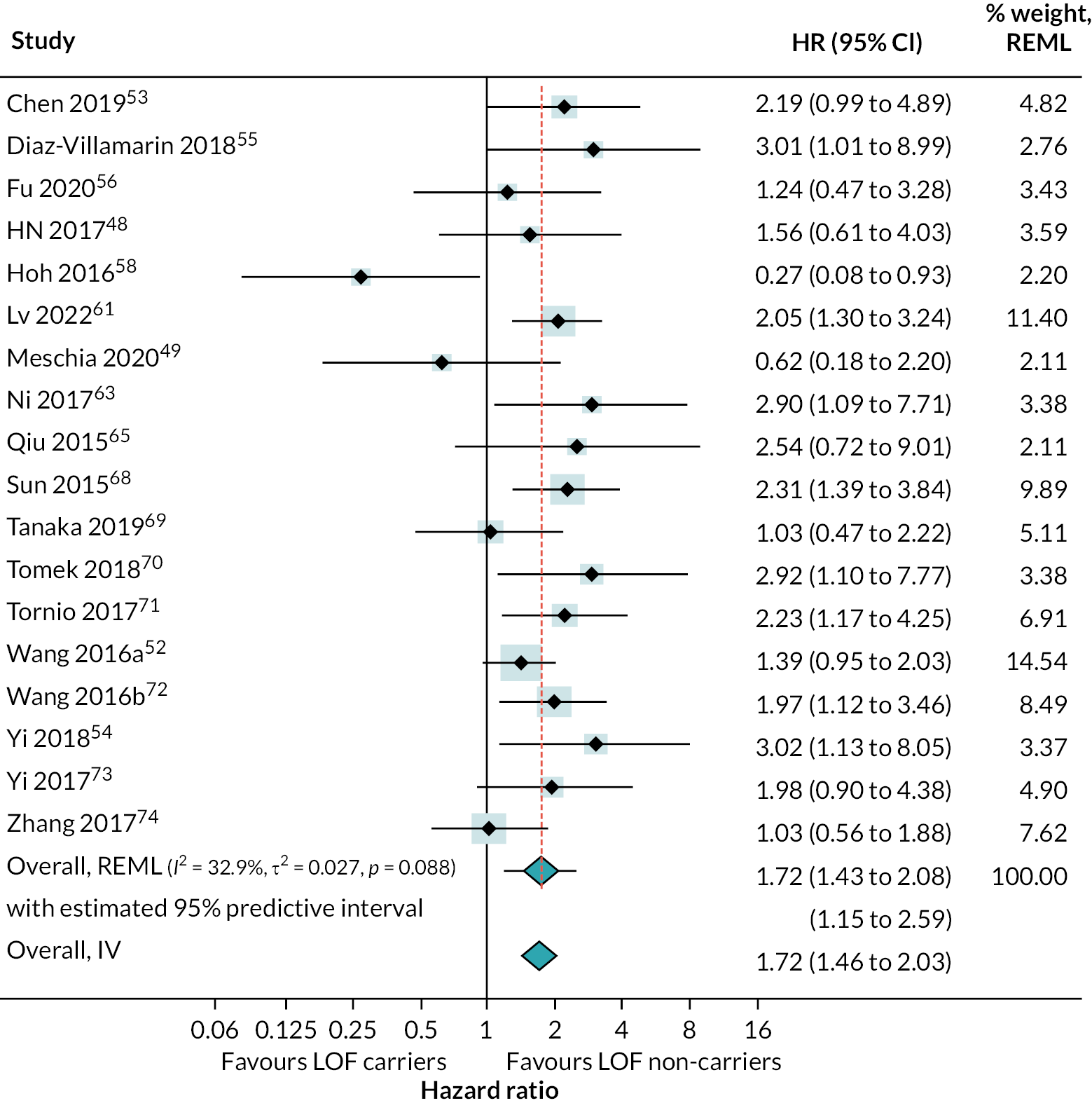
FIGURE 21.
Forest plot showing HRs (95% CI) for incidence of any stroke in carriers of LOF alleles compared with non-carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin).
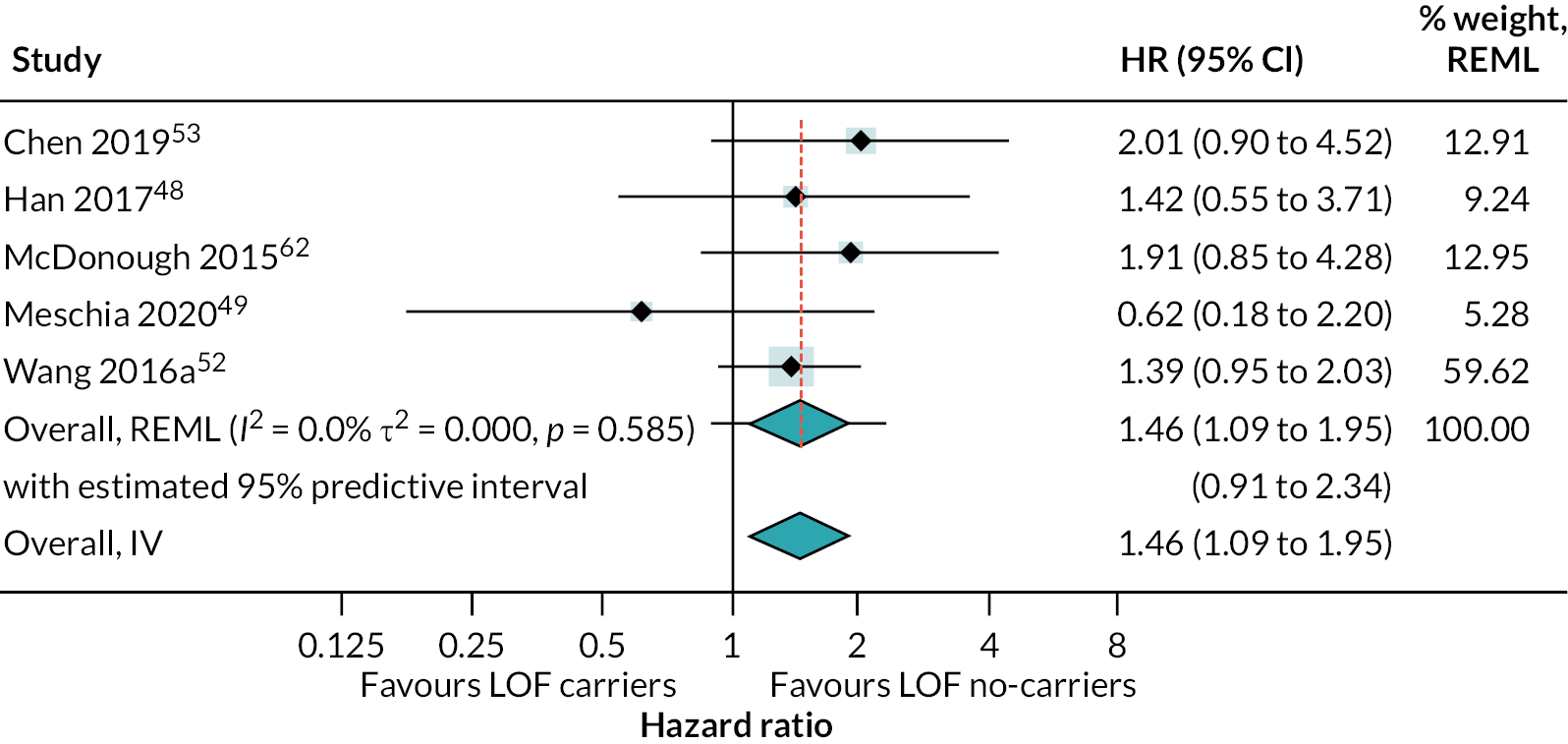
FIGURE 22.
Forest plot showing HRs (95% CI) for incidence of secondary vascular occlusive outcomes in carriers of LOF alleles compared with non-carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin).
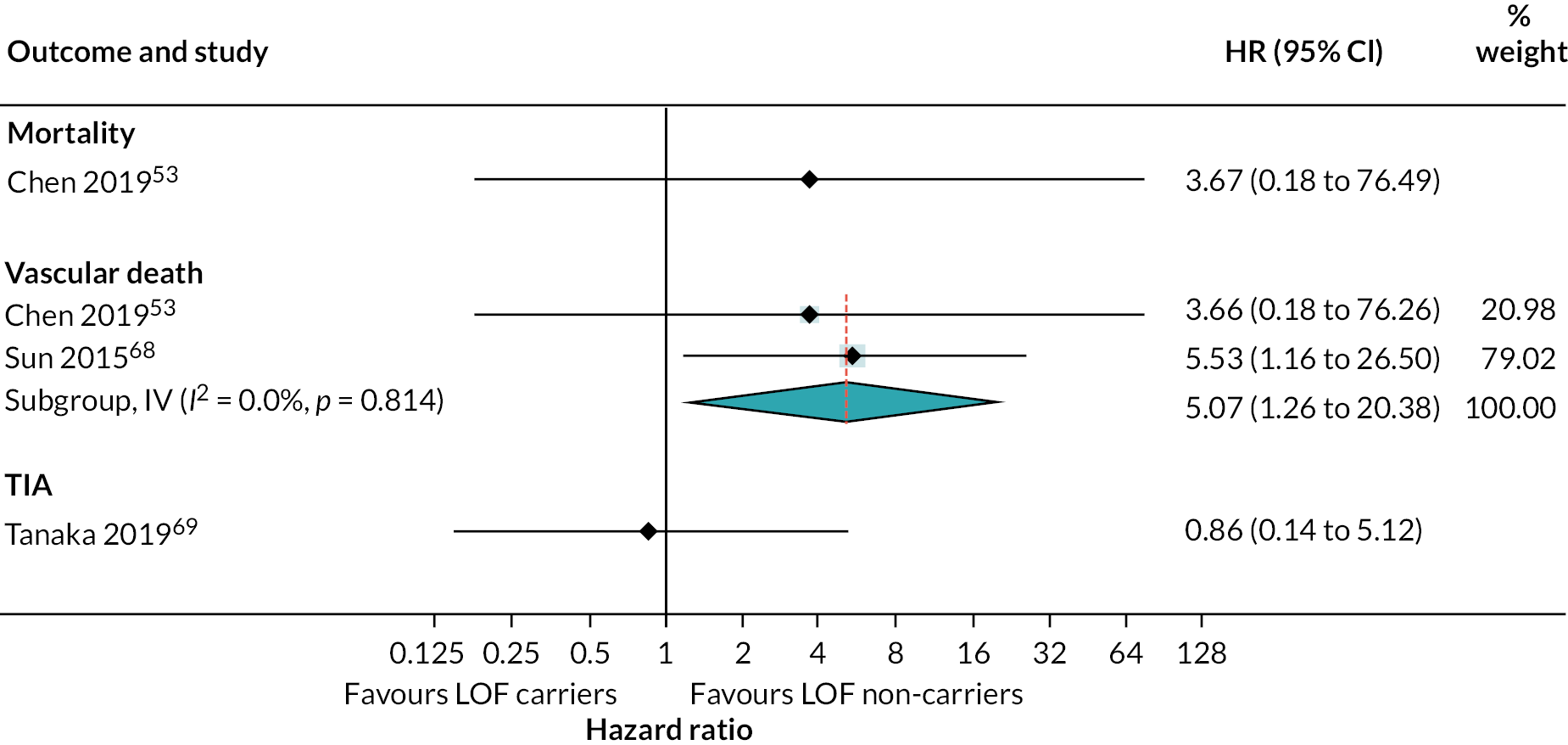
Consistency in estimates of secondary vascular events across studies
FIGURE 23.
Forest plots showing consistency in estimates of secondary vascular events across studies that evaluated multiple vascular occlusive event outcomes.

Metaregression analyses
| Covariate | Group | RHR | 95% CI | p-value | τ2 | I2 (%) | R2 (%) |
|---|---|---|---|---|---|---|---|
| Ethnicity | White | 1 | Reference | 0.02 | 30 | 33 | |
| Asian | 0.67 | 0.36 to 1.26 | 0.20 | ||||
| Mixed | 0.54 | 0.22 to 1.34 | 0.17 | ||||
| Black | 0.72 | 0.15 to 3.47 | 0.67 | ||||
| Hispanic | 0.10 | 0.00 to 3.23 | 0.182 | ||||
| N/R | 6.90 | 0.19 to 258.00 | 0.25 | ||||
| Regimen | Clopidogrel | 1 | Reference | 0.03 | 23 | 0 | |
| Clopidogrel + optional aspirin | 1.22 | 0.56 to 2.63 | 0.604 | ||||
| Clopidogrel + aspirin | 0.47 | 0.22 to 0.96 | 0.04 | ||||
| Loading dose | No loading dose | 1 | Reference | 0.00 | 19 | 100 | |
| Loading dose | 0.64 | 0.43 to 0.96 | 0.03 | ||||
| Loading dose optional | 1.15 | 0.54 to 2.49 | 0.70 | ||||
| RoB | Low risk | 1 | Reference | 0.02 | 27 | 14 | |
| High risk | 1.33 | 0.84 to 2.12 | 0.21 | ||||
| Primary event | Stroke | 1 | Reference | 0.00 | 3 | 100 | |
| Stroke or TIA | 0.62 | 0.44 to 0.86 | < 0.01 | ||||
| TIA | 1.53 | 0.58 to 4.06 | 0.38 | ||||
| PPI use | 0–10% | 1 | Reference | 0.03 | 17 | 0 | |
| 10–20% | 0.99 | 0.58 to 1.69 | 0.98 | ||||
| 20–30% | 1.33 | 0.64 to 2.80 | 0.43 | ||||
| 40–50% | 1.51 | 0.57 to 4.00 | 0.39 | ||||
| 50–60% | 0.15 | 0.04 to 0.60 | 0.01 | ||||
| N/R | 1.02 | 0.43 to 1.62 | 0.93 | ||||
| Follow-up time | 3 months | 1 | Reference | 0.01 | 22 | 57.61 | |
| 6 months | 1.11 | 0.62 to 2.01 | 0.71 | ||||
| 1 year | 0.61 | 0.18 to 2.03 | 0.40 | ||||
| 1–3 years | 1.34 | 0.77 to 2.35 | 0.28 | ||||
| 3–5 years | 1.47 | 0.79 to 2.72 | 0.20 | ||||
| N/R | 1.86 | 1.01, 3.44 | 0.05 | ||||
| Study location | Europe | 1 | Reference | 0.04 | 32 | 0 | |
| China | 0.75 | 0.38 to 1.48 | 0.39 | ||||
| Asia | 0.53 | 0.21 to 1.29 | 0.15 | ||||
| USA | 0.56 | 0.22 to 1.45 | 0.22 | ||||
| International | 0.75 | 0.22 to 2.55 | 0.63 | ||||
| Turkey | 7.26 | 0.21 to 255,00 | 0.26 |
Stratified analyses for risk of secondary vascular occlusive events in carriers of loss-of-function compared with non-carriers
FIGURE 24.
Forest plot showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by ethnicity. Note: Results split by ethnicity from McDonough et al. (2015)62 were considered more appropriate for this analysis and used here instead of results for the whole population.

FIGURE 25.
Forest plot showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by drug regimen.
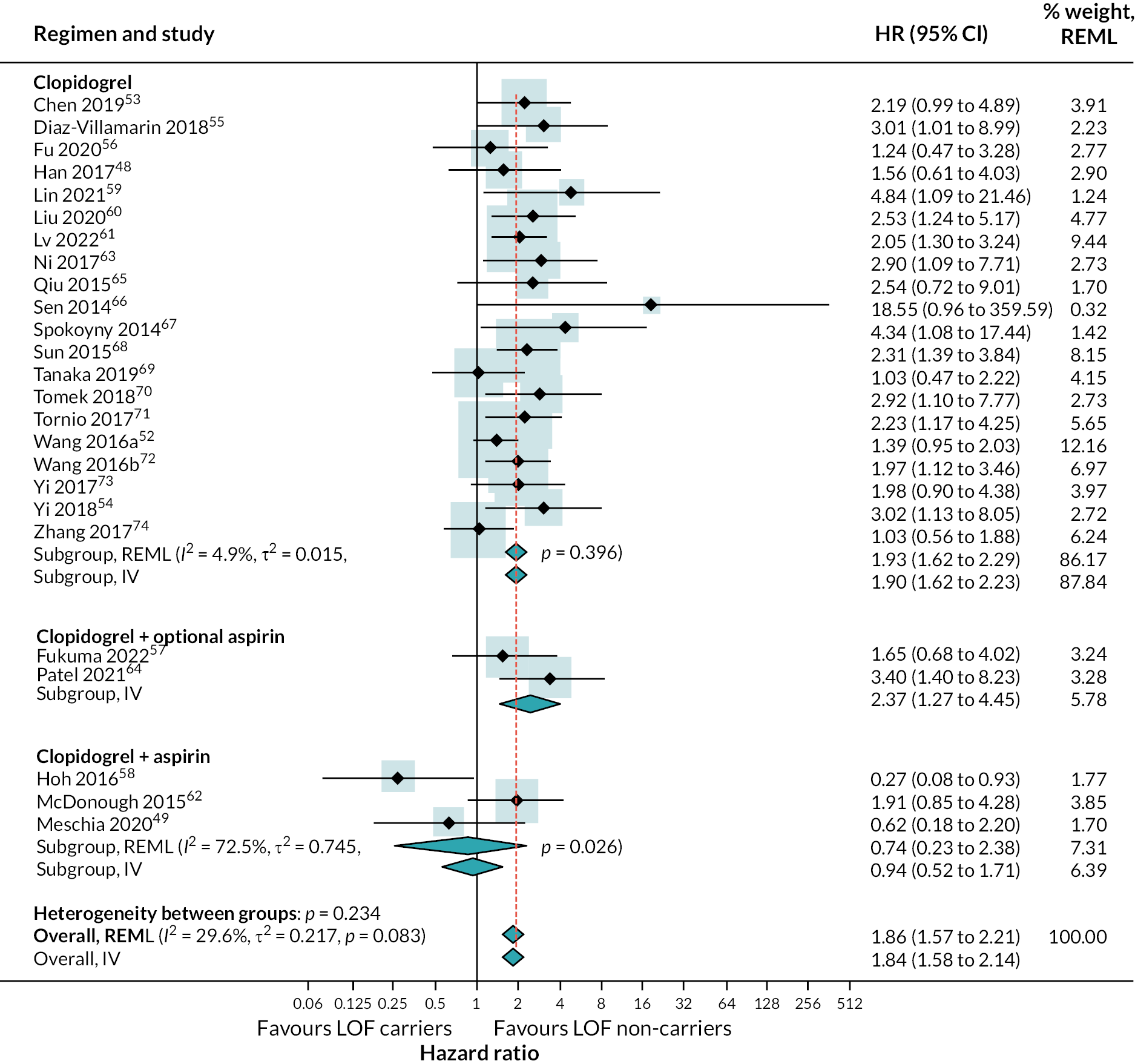
FIGURE 26.
Forest plot showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by inclusion of a loading dose of clopidogrel in the study’s regimen.
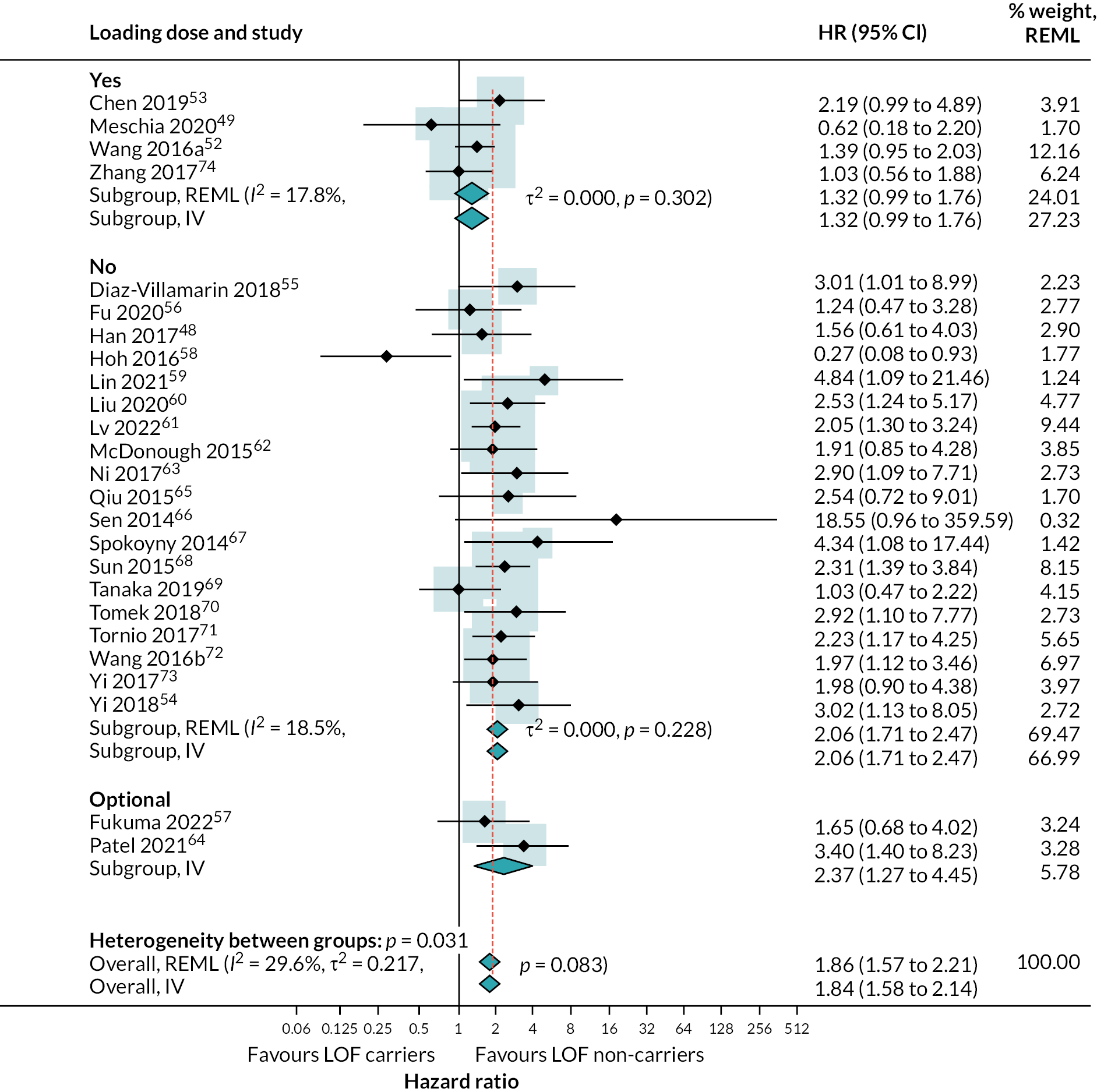
FIGURE 27.
Forest plot showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by type of qualifying primary occlusive vascular event.

FIGURE 28.
Forest plot showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by percentage of patients taking concomitant PPI.
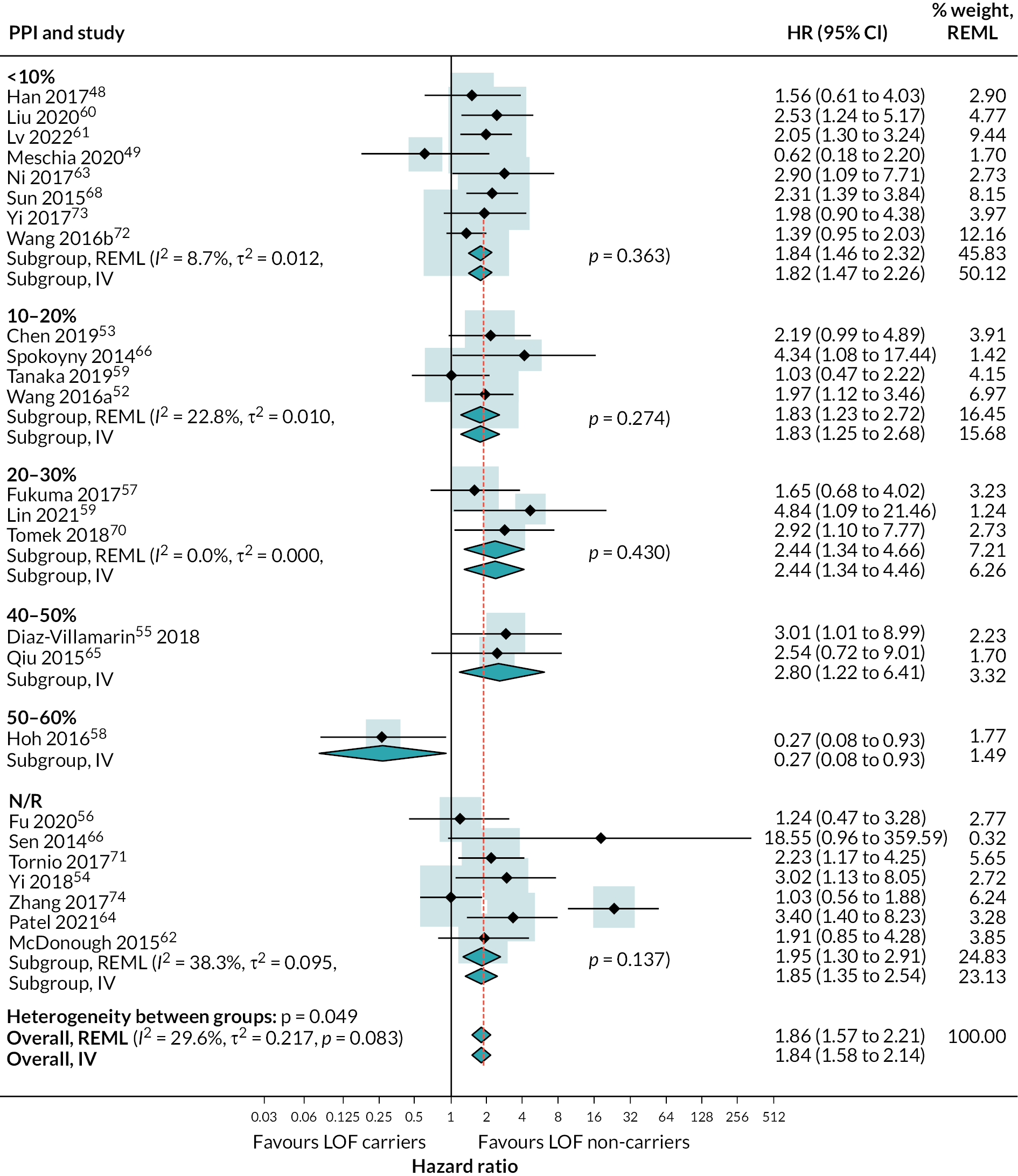
FIGURE 29.
Forest plots showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by follow-up time.

FIGURE 30.
Forest plots showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by assessed RoB.
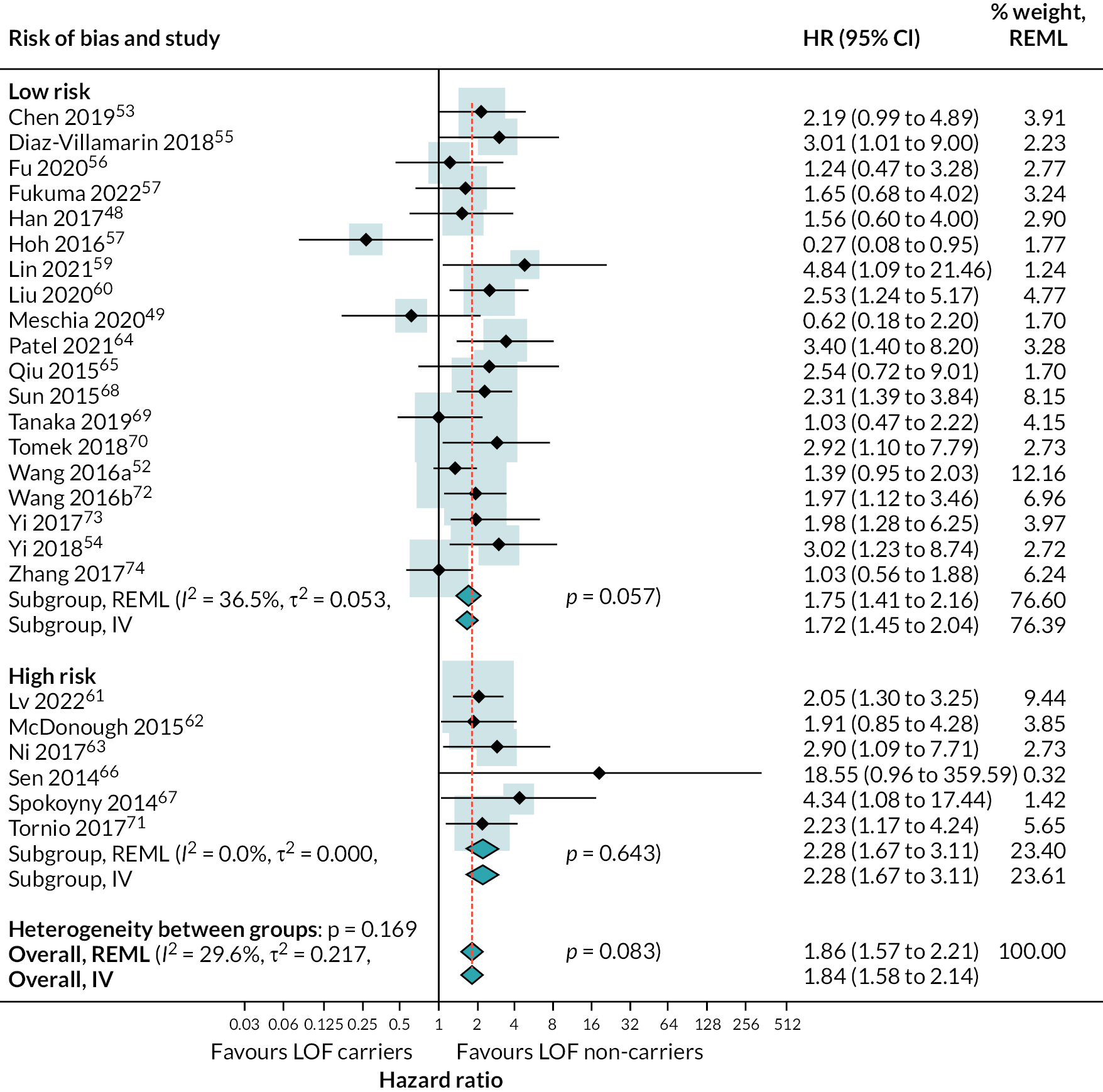
FIGURE 31.
Forest plots showing HRs (95% CI) for risk of secondary vascular occlusive events in carriers of LOF compared with non-carriers, stratified by country or region.

Risk of adverse events
FIGURE 32.
Forest plot showing HRs (95% CI) for incidence of secondary adverse effect outcomes in carriers of LOF alleles compared with non-carriers of LOF alleles receiving standard therapy with clopidogrel (or clopidogrel + aspirin).
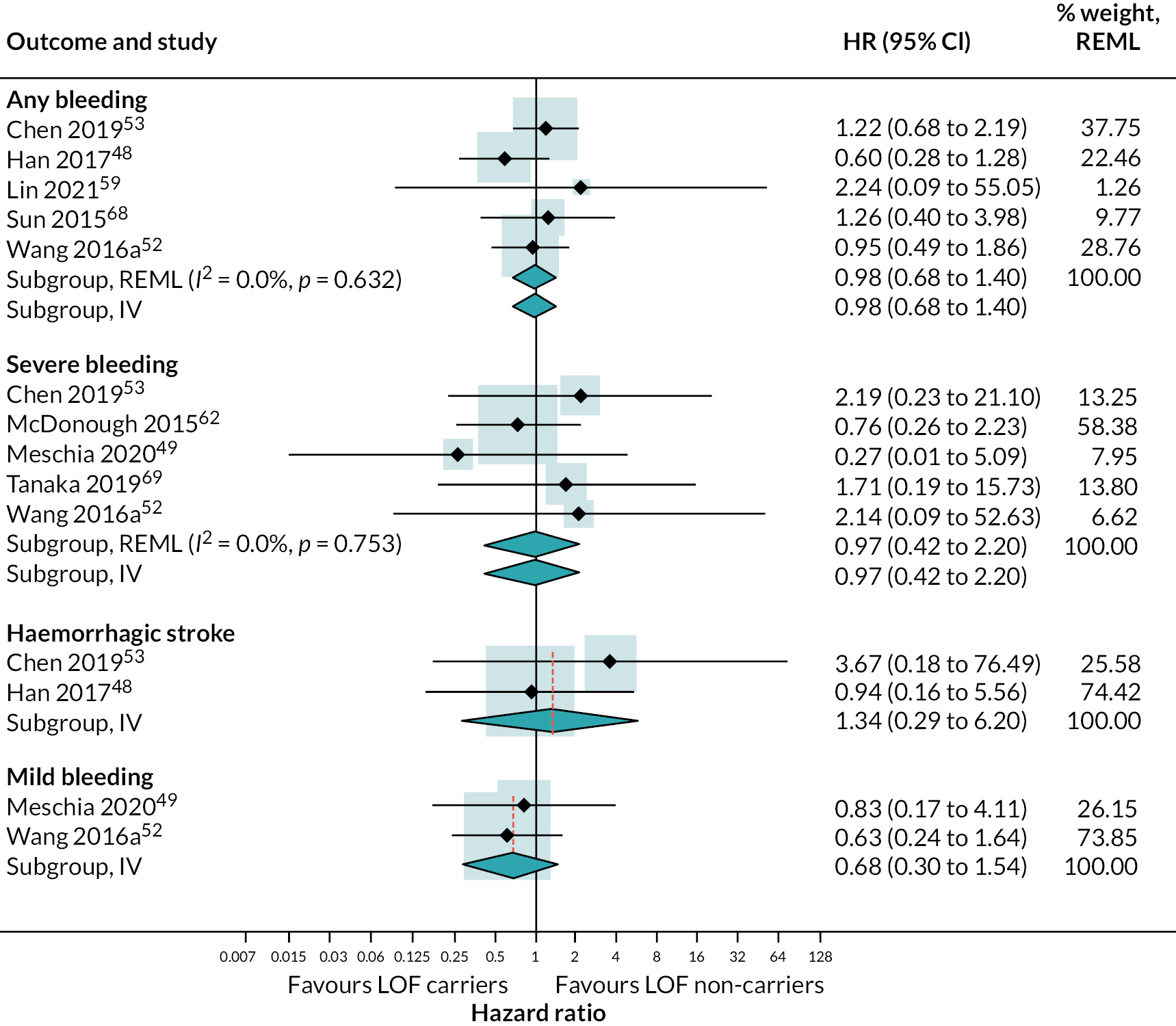
FIGURE 33.
Funnel plot of HRs for incidence of secondary vascular occlusive outcomes in carriers of LOF alleles compared with non-carriers of LOF alleles.

Appendix 5 Additional tables for Objective 5
| Study details | Time to results |
|---|---|
| Al-Rubaish et al. (2021)86 | First 50 patients: 90–120 minutes to complete the results |
| Bergmeijer et al. (2014)87,96 | Result available within 1 hour after collection of buccal swab |
| Cavallari et al. (2018)88 | For all patients genotyped: median genotype test TAT was 96 minutes [interquartile range (IQR) of 78–144] |
| Choi et al. (2016)80 | Description of feature of the test: time from sample to result ~60 minutes |
| Franchi et al. (2020)90 | Allele status within 1 hour – readily available when the decision on choice of oral P2Y12-inhibiting therapy most commonly occurs |
| Gurbel et al. (2018)91 | Results available in all patients within 90 minutes |
| Petrek et al. 201681,85 | TAT (from buccal swab sampling to result print-out) was 60 minutes |
| Roberts et al. (2012)77 | Main trial: within 60 minutes from test activation |
| So et al. (2016)82 | Within 55 minutes of test carrier status for all alleles was available |
| Genomadix (test manufacturer) response to request for information | Description of feature of the test: time to result is 64 minutes. |
| Tomaniak et al. (2017)92,97,98 | Mean (SD): 56 minutes (11), from material collection to the testing results |
| Wirth et al. (2016)83,99 | Collection of sample to genotyping result within 1 hour |
| Zhou et al. (2017)93,100 | Description of feature of the test (pre-trial and main trial): results are returned in 1 hour TAT |
| McDermott et al. (2020)94 – Genedrive | Description of feature of the test: ~40 minutes |
| Study details | Country | Ethnicity | Number of people with variant forms of CYP2C19 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *2/*2 | *3/*3 | *3/*2 | *2/*1 | *3/*1 | *3/*17 | *2/*17 | PM* | IM* | Total number with variant forms (%) | Comments | |||
| Al-Rubaish et al. (2021)86 | Saudi Arabia | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | 54 (21.1%) | Either *1/*2 or *2/*2 |
| Badhuin et al. (2022)76,95 | USA, Canada, South Korea, Mexico | N/R | 19 | 1 | 5 | 96 | 7 | 0 | 23 | 25 | 126 | 151/373 (40%) | Pre-trial |
| USA, Canada, South Korea, Mexico | 68% white, 23% East Asian | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | 837/2587 (32%) | Main trial | |
| Cavallari et al. (2018)88 | USA | White 74.5%, black 23.7%, Asian 0.8%, other or N/R 1%. | 7 | N/R | N/R | N/R | N/R | N/R | N/R | 7 | 106 | 113/392 (29%) | |
| Choi et al. (2016)80 | South Korea | N/R | 11 | 1 | 10 | 40 | 13 | 1 | 0 | 22 | 54 | 76 (63.9%) | |
| Franchi et al. (2020)90 | USA | N/R | 20 | 0 | 0 | 189 | 1 | 0 | 32 | 20 | 222 | 242/781 (28.5%) | |
| Gurbel et al. (2018)91 | USA | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | 11 | 157 | 168/578 (29%) | |
| Roberts et al. (2012)77 | Canada | 95% white ethnic origin | 7 | N/R | N/R | N/R | N/R | N/R | N/R | 7 | 39 | 46/187 (25%) | Main trial |
| So et al. (2016)82 | Canada | 91% Caucasian | 4 | 0 | 0 | 33 | 0 | 0 | 0 | 4 | 33 | 37 (36%) | |
| Tomaniak et al. (2017)92,97,98 | Poland | N/R | N/R | N/R | N/R | N/R | N/R | N/R | N/R | 2 | 12 | 14 (14.83%) | |
| Wirth et al. (2016)83,99 | Malta | 100% Caucasian | 1 | N/R | N/R | N/R | N/R | N/R | N/R | 1 | 12 | 13/34 (38%) | The 12 IMs had one copy of the *2 allele |
| Zhou et al. (2017)93,100 | USA | N/R | 0 | 0 | 2 | 4 | 0 | 0 | 1 | 2 | 5 | 7/12 (58%) | Pre-trial |
| USA | N/R | 10 | 0 | 1 | 61 | 0 | 0 | 27 | 11 | 88 | 99 (37%) | Main trial | |
Appendix 6 Survey results
a. Which of the following test platforms that would be capable of performing CYP2C19 genotypying does your laboratory have (even if not currently in use for this purpose)?
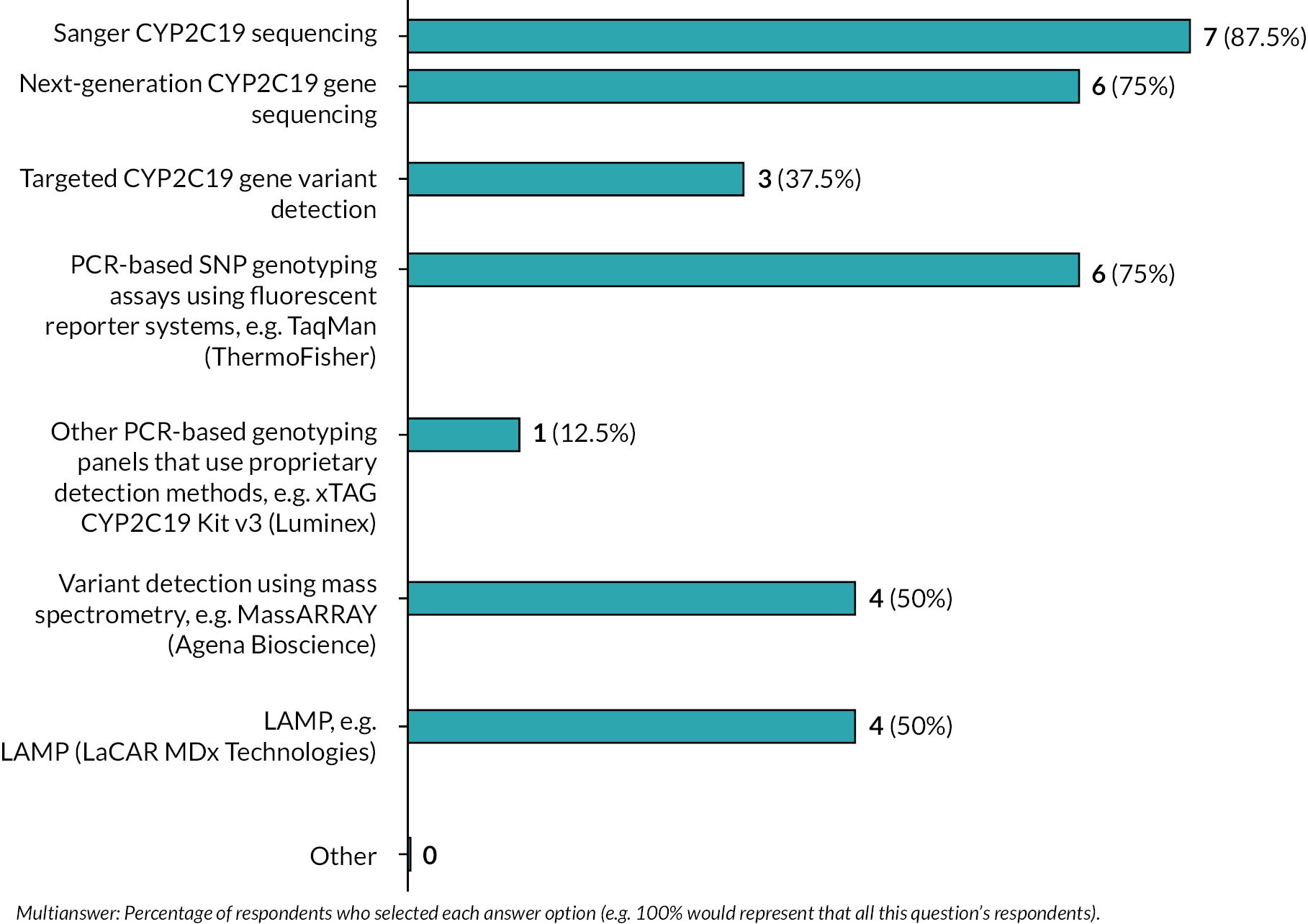
b. Of the tests that you have in your laboratory, which would be your preferred platform for running CYP2C19 testing in your laboratory if you were needing to run an estimated 10,000 tests per year. Note: This is a very rough approximation of the number of tests that each laboratory hub would need to run based on a total estimate of 100–150,000 tests per year across the UK – the exact number would be dependent on your catchment population.

If you selected other, please specify:
QuantStudio 12K Flex Real-Time PCR System or X9 Real-Time PCR System
Please briefly summarise why you would prefer this platform:
An ideal platform for targeted variant detection: ability to target multiple variants in a single assay applying automated PCR prep and automated genotype calling [validated within our lab for hemochromatosis gene (HFE) and dihydropyrimidine dehydrogenase (DPYD) testing on this platform], reduced TAT and reduces the necessary staff resources.
The ability to PCR direct from blood is also feasible for this technology (in validation for HFE and DPYD within this lab).
This Hub is also implementing a new LIMS system which will enable automated reporting from the genotype report generated by the Agena software.
Can be done directly from blood and does not require extraction; easy method to set up and automate.
Efficiency and cost and TAT
The technique used would depend on the number of variants requiring testing and which variants they are. For both Agena MassARRAY and LAMP commercial kits are available for CYP2C19 testing but offering different variants. MassARRAY offers *2,*8 and *17. The current LaCAR test covers *2,*3 and *17. It is possible to design bespoke assays, this is likely easier with the MassARRAY.
If the CYP2C19 assay was combined with other testing, the MassARRAY is probably better suited for covering increased numbers of variants.
The benefit of the LAMP assay is the speed and lack of need for a DNA extraction.
For a wider panel, a NGS solution might be worth considering.
Higher throughput
High throughput and massively parallel. Automated bioinformatics analysis. Pre-existing workflows established.
These instruments have higher throughput and can have automated loading. For example, the X9 can test 96 samples for 96 different SNPs in a 2-hour run.
-
Cost-effective
-
Time-efficient
-
Minimal staff time
-
Two-step process
-
High throughput
-
Robust technology
-
Simple analysis and reporting
Are there other platforms available that you would ideally use for CYP2C19 testing, if so please name and briefly explain why you think this would be better than the test you selected as your preferred test above:
Sanger would be used for those indiscriminate calls by LAMP – back-up test
No
NGS Genexus – looking at this option due to speed and capacity
c. Which alleles would you test for in a request for a CYP2C19 test?

If you selected other, please specify:
A NGS assay would be able to detect all sequence variants associated with the disorder provided there is no pseudogene interference with CYP2C19.
d. Would the test be affected by testing for all LOF alleles compared to only testing for *2 or *3 alleles?

If yes, how would this affect the test (e.g. longer TAT, greater cost)?
Increased cost
Longer TAT and cost as well as staff resource to deliver testing.
It could potentially impact the choice of technology chosen. This would impact the cost and TAT.
This depends on the chosen method as does the question below
Possibly greater TAT due to more variants being assessed. Cost is highly dependent on number of samples tested.
e. What would be the estimated time from receiving a sample to result data being returned to the person that requested the test?

f. To help estimate the cost of introducing CYP2C19 testing, please could you give an estimate for each of the following:
Staff time
1–2 days, 1–2 hours set-up, 2-hour analysis, 2 hours checking and reporting.
Reception, extraction, workflow, reporting
Unable to comment
1 × band 3, 1 × band 5 and 1 × band 7 WTEs
Unable to provide at this time.
-
0.5 WTE for performing test
-
0.5 WTE for DNA extraction
-
0.2 WTE for admin
22 minutes/sample
Staff grade
Band 5 set-up, band 6 analysis and reporting band 7 checking and authorisation of reports.
Band 3 up to band 8a
Unable to comment
1 × band 3, 1 × band 5 and 1 × band 7 WTEs
Unable to provide at this time.
-
Band 5
-
Band 4
-
Band 3
AfC bands 2, 3, 4 for laboratory work
AfC band 7 authorising reports
Cost per test to run
£40 per test (reagent cost only)
~£15 per test
~£100
Again method dependent – £100–250
Unable to provide at this time.
~£200 per sample
There would be additional costs in data analysis either by scientists or using automated calling and reporting system = £5–10 per sample
£25.09 inc VAT (reagents/consumables, staff time, and overheads)
Maintenance of machines/quality assurance
Monitoring of PCR instrumentation, inclusion and monitoring of internal quality controls, participation in external quality assessment (EQA) or interlaboratory sample exchange, United Kingdom Accreditation Service (UKAS) accreditation.
£15k maintenance and yes EQA (??)
Unable to comment
Unable to provide at this time.
£5000 pa for qPCR machine BUT for 10,000 sample pa we would need to increase our existing DNA extraction capacity, which may mean another automated DNA extraction system = £150k capital investment
Additional administrative resources to record test result
LIMs upload to ECR where link does not exist – admin support to send results and upload to ECR.
Yes??
Unable to comment
1 × band 4 admin
Unable to provide at this time.
Preferable electronic test ordering but may require admin support for dealing with enquiries
None
g. How easy is the test to perform?
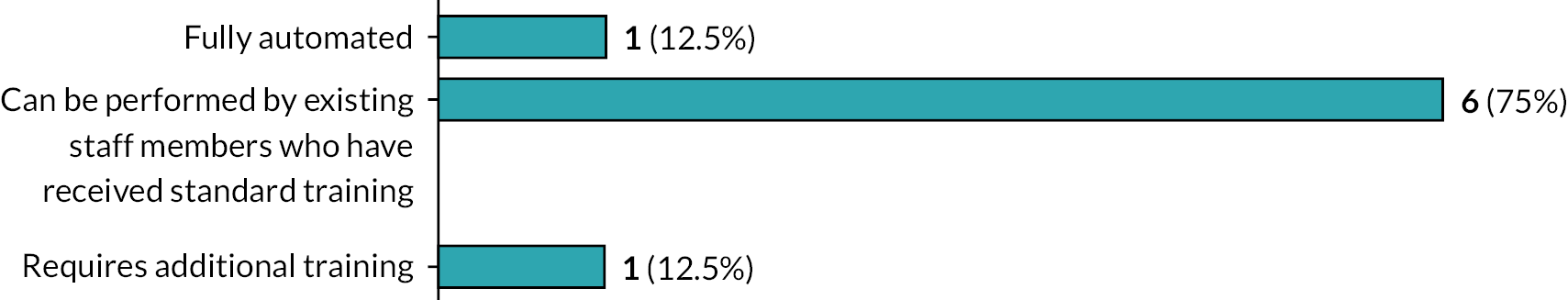
If additional training would be required, please provide a brief summary of what this would entail
No. Technology in use within the laboratory.
Either a MassARRAY or LAMP could be carried out by existing trained staff. Further staff may need training due to increased use of particular technology and all staff would need small amount of training for any differences between the current assays used and the
Any new tests require training for staff to perform the test, operate the instrument and interpret data if required
Could you give an estimate of the proportion of samples that would not return a valid result?
< 1% based on current targeted testing for germline variants using the Agena/matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) platform.
~90%
Less than 1%
This will depend on the particular assay used but should be < 1%
Difficult to say – under 1%
An experimental validation would be required to set the testing up. This would identify and resolve any assay-related issues. Once validated the test would then expect to have a low fail rate – that is < 1% samples assuming the test is performed on DNA extracted from blood.
< 1–3% but this would need to be validated if a new test and equipment is required
5%
h. Please estimate your current testing capacity – estimated number of tests of this type that could be performed in your laboratory at present in a 1-week period
0
92 per run – up to × 2 weeks
Unable to comment
Currently deliver 110 per week
Zero
We would not be able to process any samples without additional staff and equipment
Up to 200 tests/week
i. Would a faster turnaround be possible with additional resources?

If yes, what resources would be required?
Additional staff
Additional staffing at all grades
Could run every day
More staff and equipment. This may require additional lab space.
This would depend on the test used
More technical staff and potentially additional instruments to increase capacity and allow more automation
Additional staff
More automation and lablims support – as above
j. Would additional testing capacity (i.e. greater number of tests) be possible with additional resources?

If yes, what resources would be required?
Based on predicted numbers:
Additional staff
Automation platform
Increased laboratory space
Chip prep module for the MALDI-TOF machine
Additional staffing at all grades
Extraction etc etc
More staff and equipment. This may require additional lab space.
This would depend on the test used.
More technical/IT staff and potentially additional instruments to increase capacity and allow more automation
Additional staff
More automation and lablims support – see above
More staff
Liquid handling platform to automate DNA dilutions
Additional QuantStudio5
k. Could the test be performed in local testing laboratories?

l. What do you see as the major facilitators and barriers to implementing CYP2C19 testing within your region?
Given scale of predicted activity:
Test ordering should ideally be electronic (currently not possible within LIMS)
Sample receipt (additional space required to manage sample numbers)
Additional staff dedicated to sample processing.
Validation time required for automated processing, genotyping and reporting. All of which is entirely possible and in progress for smaller scale tests) but requires additional staff resources.
Staff resource is the major barrier to implementing this test. Loop-mediated isothermal amplification testing is currently being successfully used to deliver HFE testing so would be easy to implement where there sufficient staffing in place
Reporting guidelines
Facilitators: previous knowledge of pharmacogenomics testing in lab (technical staff and Clinical scientists) and within genomic laboratory hubs/genomic medicine service alliance (GLH/GMSA).
Appropriate equipment available within the department, although capacity would need to be reviewed.
Barriers: ensuring awareness of testing with all clinicians across the geography.
Capacity of laboratory to perform test alongside other clinical requirements
Throughput on existing instruments and staffing
We do not currently perform any tests of this scale in the NHS, so do not have the infrastructure. We need automation, lablims support and skilled staff
Laboratory staffing resources
Facilitators
Strong support from Stroke Clinicians, Specialist Pharmacist and Senior Managers within Trust
Barriers
Fixed budget for pilot, so had to confine requests to Stoke Unit and Cardiology
Unable to accept requests from GPs
Difficulty for some medical disciplines to understand output of genetic results
Separate requesting and reporting systems for acute and primary care
m. Would it be possible to implement a rapid POCT (input = buccal swab for a single patient) in your lab workflow?
Yes
Not at this point
No
It should be possible to implement this test within the lab workflow
Time for sample to be received in the laboratory might be an issue
May require extra freezers to hold kits under appropriate conditions
Yes with staff
In principle, yes, although there is no precedent for this in our lab
POC is not the most efficient process for the number of samples that would need to be tested per week (192 per week based on processing 10,000 samples in 52 weeks). Samples would be batched and not tested one by one as using POC
Yes
If not, why/what extra resources might you need?
Staff
This would require staff to be able to drop all other duties to perform this test. This is not currently feasible with the staffing levels in the department. Other duties considered necessary would not be completed
POC platform
Staff to support
See above
Not enough staff to deliver rapid POCT
Extraction and genotyping processes not suited to service based out with the lab
Delivering POCT would require different testing technology and cost would increase
Appendix 7 Economic review: additional tables/figures and model input sources
FIGURE 34.
PRISMA diagram showing the studies identified in the systematic review of cost-effectiveness studies for CYP2C19 testing for patients who have had a non-cardioembolic ischaemic stroke or TIA.
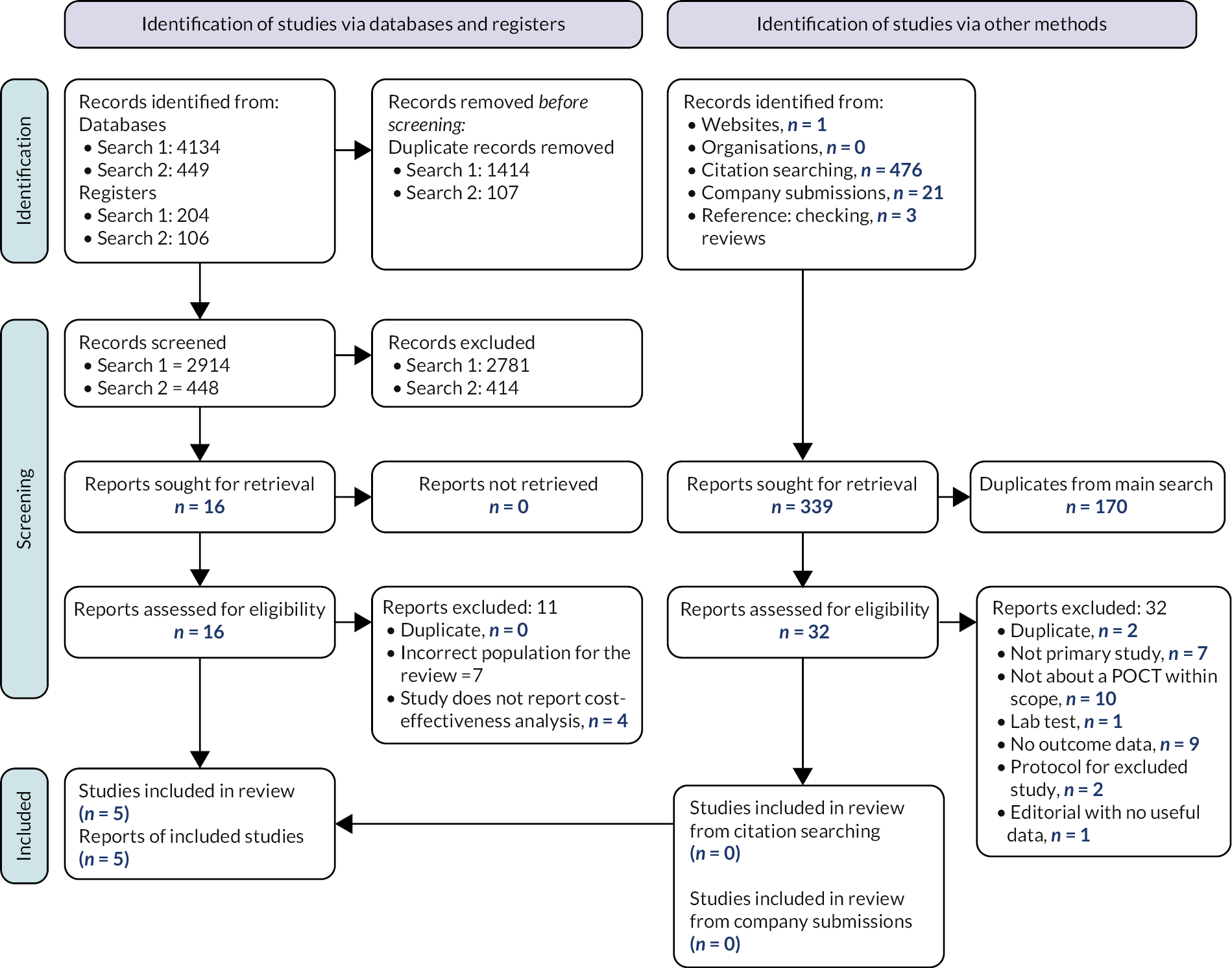
| Author, year | Setting | Population | Model type | Time horizon | Interventions | Comparators | Perspective |
|---|---|---|---|---|---|---|---|
| Micieli (2022)104 | Canada | TIA/minor stroke patients | Markov cohort model | Lifetime 20 years (30-day cycles) |
Genetic test and treat GMEX POCT LOF non-carriers: aspirin–clopidogrel. LOF carriers: DAPT aspirin (75–300 mg on day 1 followed by 75 mg daily) w/ticagrelor (180 mg on day 1 followed by 90 mg twice daily) for 3 weeks, followed by single antiplatelet aspirin. |
Treat (no test) DAPT aspirin (75–300 mg on day 1 followed by 75 mg daily) w/clopidogrel (300 mg on day 1 followed by 75 mg daily) for 3 weeks, followed by single antiplatelet aspirin. |
Federal, Provincial, and Territorial Ministries of Health |
| Cai (2021)109 | China | Acute minor stroke or high-risk TIA patients | Decision tree and Markov model | Lifetime 30 years | Genetic test and treat Sequenom MassARRAY iPLEX Lab Test LOF non-carriers: clopidogrel (300 mg on day 1 followed by 75 mg daily) for 3 months plus aspirin (75–300 mg on day 1 followed by 75 mg daily) for 21 days. LOF carriers: dipyridamole – aspirin sustained-release capsule (200 mg, 25 mg, twice daily) for 3 months. |
Treat (no test) Clopidogrel (300 mg on day 1 followed by 75 mg daily) for 3 months plus aspirin (75–300 mg on day 1 followed by 75 mg daily) for 21 days. |
Healthcare payer |
| Narasimhalu (2020)105 | Singapore | Ischaemic stroke patients | Markov model | Lifetime 20 years | Genetic test and treat Spartan RX POCT LOF non-carriers: clopidogrel (300 mg on day 1 followed by 75 mg daily). LOF carriers: ticagrelor (180 mg on day 1 followed by 90 mg twice daily). |
Treat (no test) Clopidogrel (300 mg on day 1 followed by 75 mg daily). |
Local Healthcare |
| Kremers (2021)106 | Netherlands | Minor acute ischaemic stroke/TIA patients | Markov model | Lifetime (1-year cycles) | Genetic test and treat LOF non-carriers: DAPT clopidogrel–aspirin. LOF carriers: receive either aspirin monotherapy, prasugrel, ticagrelor or DAPT aspirin–dipyridamole. |
Treat (no test) DAPT clopidogrel–aspirin for 3 weeks followed by lifelong clopidogrel monotherapy. |
N/A |
| Wright (2022)103 | UK | Patients suffered first stroke | Decision tree and Markov model | Lifetime | Genetic test and treat Genedrive® CYP2C19 ID Kit LOF non-carriers: clopidogrel, if intolerant, switch to modified release. dipyridamole plus aspirin. LOF carriers: modified release dipyridamole plus aspirin. |
Treat only (no genetic test) Clopidogrel, if intolerant, switch to modified-release dipyridamole plus aspirin. |
NHS |
| Author, year | Discount rate | Health states | Source of effectiveness information | Results |
|---|---|---|---|---|
| Micieli (2022)104 | 1.50% |
|
CHANCE trial subgroup data for non-carriers of LOF, and CHANCE-2 trial for LOF carriers. | The ICER for the CYP2C19 testing strategy was CA$4310 per QALY compared with no testing. CYP2C19 testing was cost-effective in more than 99.99% of simulations using a willingness-to-pay threshold of CA$50,000 per QALY. |
| Cai (2021)109 | 3% |
|
CYP2C19 subgroup data from the CHANCE trial. A meta-analysis of five RCTs of DAPT dipyridamole–aspirin vs. aspirin for secondary prevention after TIA or stroke. |
The ICER for the CYP2C19 testing strategy was CNY 13,552.74 (US$1931) QALY compared with no testing. CYP2C19 testing was cost-effective in more than 95.7% of simulations at a willingness-to-pay threshold CNY 72,100 (US$10,300) per QALY. |
| Narasimhalu (2020)105 | 3% | No recurrent ischaemic stroke Post recurrent ischaemic stroke Death |
The SOCRATES trial for ticagrelor. For clopidogrel a prospective cohort study on the recurrent risk among stroke patients with CYP2C19 phenotypes treated with clopidogrel from the Nanjing Stroke Registry Program. | The ICER for the CYP2C19 testing strategy was S$33,839/QALY compared with no testing. CYP2C19 testing was cost-effective in more than 77.68% of simulations at a willingness-to-pay threshold S$60,000/QALY threshold. |
| Kremers (2022)106 | N/A | N/A | N/A | Testing for CYP2C19 LOF followed by prasugrel or ticagrelor was found to be cost saving with incremental cost savings of €461 or €438, and gains of 0.01 QALYs per patient compared to no testing and treatment with clopidogrel. Probabilistic analysis results were N/R. |
| Wright (2022)103 | 3.50% | No further stroke One further stroke > 1 further stroke Vascular death Other cause of death |
A meta-analysis investigating the effect of treatment with clopidogrel on stroke or TIA patients who are LOF carriers compared to LOF non- carriers. A systematic review and economic evaluation of the clinical and cost-effectiveness of clopidogrel and modified-release dipyridamole in the secondary prevention of occlusive vascular events |
The CYP2C19 testing strategy was cost saving when compared with no testing. Incremental savings of £170 and gain of 0.096 QALYs per patient. The PSA demonstrated that testing was cost-effective in 77% of simulations at a willingness-to-pay threshold £20,000/QALY threshold. |
| Micieli (2022)104 | Cai (2021)109 | Narasimhalu (2020)105 | Kremers (2021)106 | Wright (2022)103 | |
|---|---|---|---|---|---|
| Study design | |||||
| The research question is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The economic importance of the research question is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The viewpoint(s) of the analysis are clearly stated and justified. | ✓ | ✓ | ✓ | ✗ | ✓ |
| The rationale for choosing alternative programmes or interventions compared is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The alternatives being compared are clearly described | ✓ | ✓ | ✓ | ✓ | ✓ |
| The form of economic evaluation used is stated | ✓ | ✓ | ✓ | ✓ | ✓ |
| The choice of form of economic evaluation is justified in relation to the questions addressed | ✓ | ✓ | ✓ | ✗ | ✓ |
| Data collection | |||||
| The source(s) of effectiveness estimates used are stated | ✓ | ✓ | ✓ | ✗ | ✓ |
| Details of the design and results of effectiveness study are given (if based on a single study) | ✓ | ✓ | ✓ | ✗ | ✓ |
| Details of the methods of synthesis or meta-analysis of estimates are given (if based on a synthesis of a number of effectiveness studies) | N/A | N/A | N/A | N/A | N/A |
| The primary outcome measure(s) for the economic evaluation are clearly stated | ✓ | ✓ | ✓ | ✗ | ✓ |
| Methods to value benefits are stated | ✓ | ✓ | ✓ | ✗ | ✓ |
| Details of the subjects from whom valuations were obtained were given | ✓ | ✓ | ✓ | ✗ | ✓ |
| Productivity changes (if included) are reported separately | N/A | N/A | N/A | N/A | N/A |
| The relevance of productivity changes to the study question is discussed | N/A | N/A | N/A | N/A | N/A |
| Quantities of resource use are reported separately from their unit costs | ✗ | ✗ | ✗ | ✗ | ✗ |
| Methods for the estimation of quantities and unit costs are described | ✗ | ✗ | ✗ | ✗ | ✗ |
| Currency and price data are recorded | ✓ | ✓ | ✓ | ✓ | ✓ |
| Details of currency of price adjustments for inflation or currency conversion are given | ✓ | ✓ | ✓ | ✗ | ✓ |
| Details of any model used are given | ✓ | ✓ | ✓ | ✗ | ✓ |
| The choice of model used and the key parameters on which it is based are justified | ✓ | ✓ | ✓ | ✗ | ✓ |
| Analysis and interpretation of results | |||||
| Time horizon of costs and benefits is stated | ✓ | ✓ | ✓ | ✗ | ✓ |
| The discount rate(s) is stated | ✓ | ✓ | ✓ | ✗ | ✓ |
| The choice of discount rate(s) is justified | ✓ | ✓ | ✓ | ✗ | ✓ |
| An explanation is given if costs and benefits are not discounted | N/A | N/A | N/A | ✗ | N/A |
| Details of statistical tests and Cis are given for stochastic data | ✓ | ✓ | ✓ | ✗ | ✓ |
| The approach to sensitivity analysis is given | ✓ | ✓ | ✓ | ✗ | ✓ |
| The choice of variables for sensitivity analysis is justified | ✓ | ✓ | ✓ | ✗ | ✓ |
| The ranges over which the variables are varied are justified | ✓ | ✓ | ✓ | ✗ | ✓ |
| Relevant alternatives are compared | ✓ | ✓ | ✓ | ✓ | ✓ |
| Incremental analysis is reported | ✓ | ✓ | ✓ | ✓ | ✓ |
| Major outcomes are presented in a disaggregated as well as aggregated form | N/A | N/A | N/A | N/A | N/A |
| The answer to the study question is given | ✓ | ✓ | ✓ | ✓ | ✓ |
| Conclusions follow from the data reported | ✓ | ✓ | ✓ | ✓ | ✓ |
| Conclusions are accompanied by the appropriate caveats | ✓ | ✓ | ✓ | ✗ | ✓ |
FIGURE 35.
PRISMA diagram showing the studies identified in the supplementary review of cost-effectiveness studies of secondary prevention ischaemic stroke in a general population.
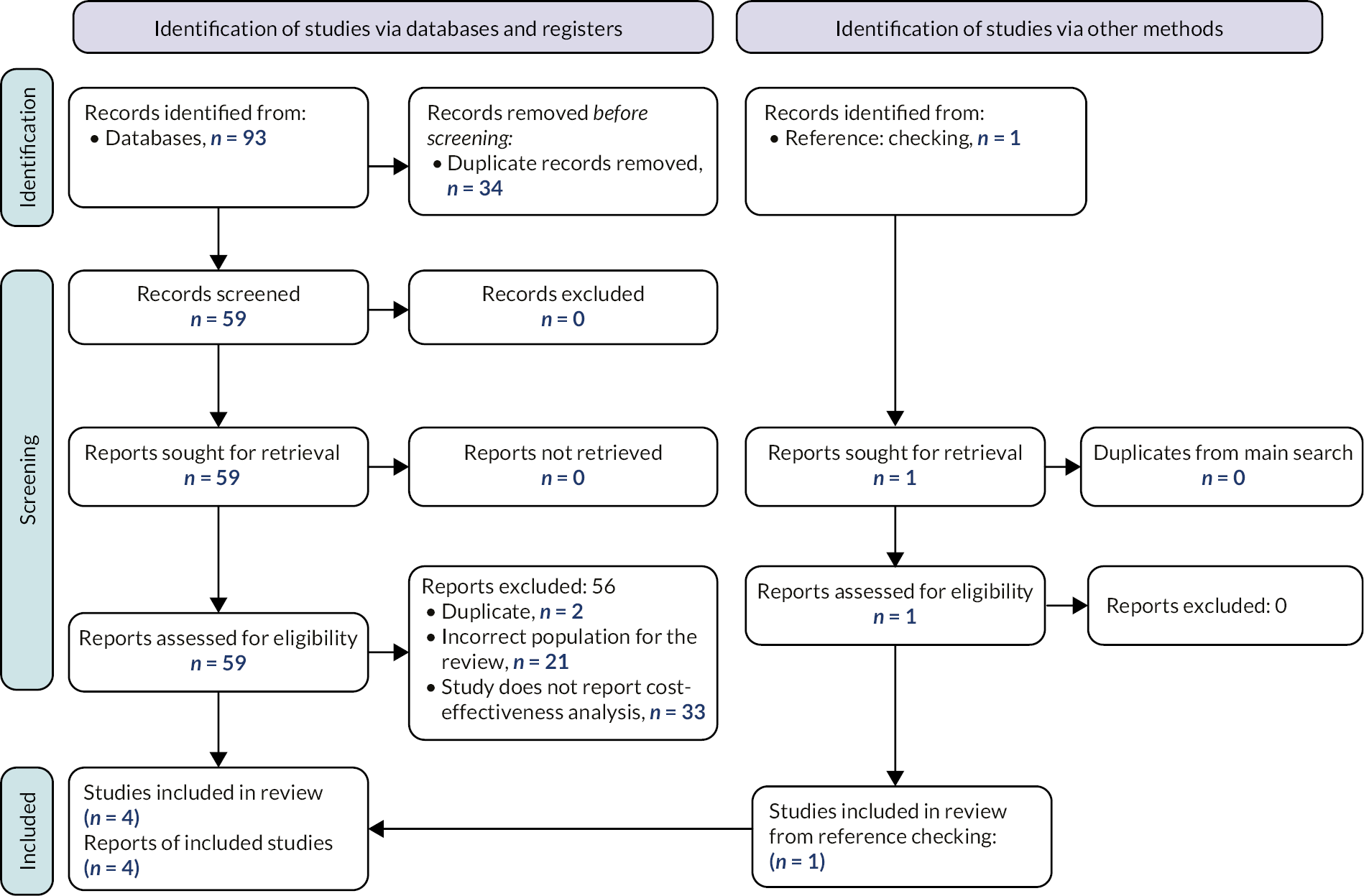
| Author, year | Setting | Population | Model type | Time horizon | Interventions | Comparators/reference treatment strategy | Perspective |
|---|---|---|---|---|---|---|---|
| Zhou (2022) 121 | USA | Patients with non-cardioembolic stroke | Markov model decision tree | Lifetime (1-year cycles) |
DAPT Cilostazol + aspirin or cilostazol + clopidogrel |
Aspirin or clopidogrel | US payer/Medicare |
| Greenhalgh (2011)119 TA 210 stroke and TIA subgroup |
UK | Patients who experienced a recent stroke or TIA | Patient-level simulation model | Lifetime | Sequences of the following treatments: clopidogrel, aspirin, modified-release dipyridamole + aspirin |
No treatment | NHS, PSS |
| Malinina (2007) 138 | USA | Patients who experienced a non-cardioembolic stroke or TIA | Unclear | 1 year | DAPT Clopidogrel + aspirin or extended-release dipyridamole + aspirin |
Aspirin | Third-party payer perspective |
| Jones (2004)143 TA 90 stroke and TIA subgroup |
UK | Patients who experienced stroke or TIA | Markov model | Lifetime (40 years) | Stroke Clopidogrel modified-release dipyridamole modified-release dipyridamole + aspirin TIA modified-release dipyridamole modified-release dipyridamole + aspirin |
Aspirin | NHS |
| Author, year | Setting | Population | Model type | Time horizon | Interventions | Comparators/reference treatment strategy | Perspective |
|---|---|---|---|---|---|---|---|
| Zhou (2022)121 | USA | Patients with non-cardioembolic stroke | Markov model decision tree | Lifetime (1-year cycles) |
DAPT cilostazol + aspirin or cilostazol + clopidogrel |
Aspirin or clopidogrel | US payer/Medicare |
| Greenhalgh (2011)119 TA 210 stroke and TIA subgroup |
UK | Patients who experienced a recent stroke or TIA | Patient-level simulation model | Lifetime | Sequences of the following treatments: clopidogrel, aspirin, modified-release dipyridamole + aspirin |
No treatment | NHS, PSS |
| Malinina (2007)138 | USA | Patients who experienced a non-cardioembolic stroke or TIA | Unclear | 1 year | DAPT clopidogrel + aspirin or extended-release dipyridamole + aspirin |
Aspirin | Third-party payer perspective |
| Jones (2004)143 TA 90 stroke and TIA subgroup |
UK | Patients who experienced stroke or TIA | Markov model | Lifetime (40 years) | Stroke clopidogrel modified-release dipyridamole modified-release dipyridamole + aspirin TIA modified-release dipyridamole modified-release dipyridamole + aspirin |
Aspirin | NHS |
| Treatment | Dose per day (mg) | Cost (£) per day |
|---|---|---|
| Aspirin | 300 | 0.1071 |
| 75 | 0.0268 | |
| Clopidogrel | 300 | 0.1757 |
| 75 | 0.0439 | |
| Modified-release dipyridamole | 400 | 0.4383 |
| 200 | 0.2192 | |
| Ticagrelor | 180 | 1.950 |
| Stroke severity (NIHSS) | 1-year costs | 5-year costs | ||
|---|---|---|---|---|
| Mean NHS costs | Mean social costs | Mean NHS costs | Mean social care costs | |
| No stroke (0) | £8632 | £4085 | £13,702 | £14,204 |
| Minor stroke (1–4) | £10,035 | £5829 | £15,103 | £19,244 |
| Moderate stroke (5–15) | £16,419 | £9741 | £20,799 | £29,972 |
| Moderate/severe stroke (16–20) | £20,061 | £16,179 | £23,180 | £47,898 |
| Severe stroke (21–42) | £17,382 | £16,063 | £19,368 | £45,809 |
| Branches | Short-term outcomes | Long-term outcomes (Discounted) | Total (discounted) | |||
|---|---|---|---|---|---|---|
| Costs (£) | QALYs | Costs (£) | QALYs | Costs (£) | QALYs | |
| POCT; LOF; TP | 1058 | 0.06 | 30,231 | 2.00 | 31,289 | 2.06 |
| POCT; LOF; FN | 11 | 0.00 | 322 | 0.02 | 332 | 0.02 |
| POCT; no LOF; TN | 2223 | 0.13 | 64,713 | 4.34 | 66,936 | 4.48 |
| POCT; no LOF; FP | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Lab test; LOF | 1090 | 0.06 | 30,544 | 2.02 | 31,634 | 2.08 |
| Lab test; no LOF | 2291 | 0.13 | 64,639 | 4.34 | 66,929 | 4.47 |
| No test; true status LOF | 1048 | 0.06 | 32,167 | 1.82 | 33,215 | 1.88 |
| No test; true status no LOF | 2194 | 0.13 | 65,063 | 4.33 | 67,257 | 4.46 |
| Branches | Short-term outcomes | Long-term outcomes (discounted) | Total (discounted) | |||
|---|---|---|---|---|---|---|
| Costs (£) | QALYs | Costs (£) | QALYs | Costs (£) | QALYs | |
| PoC test; LOF; TP | 422 | 0.06 | 13,770 | 2.71 | 14,192 | 2.78 |
| PoC test; LOF; FN | 4 | 0.00 | 145 | 0.03 | 150 | 0.03 |
| PoC test; no LOF; TN | 862 | 0.14 | 29,660 | 5.72 | 30,522 | 5.85 |
| PoC test; no LOF; FP | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Lab test; LOF | 447 | 0.06 | 13,928 | 2.74 | 14,375 | 2.80 |
| Lab test; no LOF | 926 | 0.14 | 29,636 | 5.71 | 30,562 | 5.85 |
| No test; true status LOF | 405 | 0.06 | 14,519 | 2.68 | 14,924 | 2.75 |
| No test; true status no LOF | 829 | 0.14 | 30,252 | 5.69 | 31,081 | 5.83 |
Appendix 8 Additional cost-effectiveness results
Tables 58 and 59 show, for both populations, the contributions to total costs and total QALYs arising from each branch of the decision tree in the short-term (decision tree), long-term (Markov) components of the model, and in total. These figures incorporate the probability of taking each branch for each testing strategy. Total costs and QALYs are used to calculate cost-effectiveness outcomes reported in Tables 47–52B. Sensitivity and specificity are very high, so the majority of costs and QALYs for POCTs are from TNs (the majority are NoLOF patients) followed by TPs (LOF patients).
Appendix 9 Incremental cost-effectiveness planes and cost-effectiveness results for the mixed transient ischaemic attack/ischaemic stroke population
| Treatments | Total costs (£) (discounted) | Total QALYs (discounted) | Strictly dominated | Extendedly dominated | ICER (£) | ||
|---|---|---|---|---|---|---|---|
| vs. Genedrive | vs. Lab test | vs. Genomadix Cube | |||||
| POCT: Genedrive | 81,457 | 7.99 | |||||
| Laboratory genetic test | 81,485 | 7.98 | Yes | N/A | Dominated | ||
| POCT: Genomadix Cube | 81,550 | 7.99 | Yes | N/A | Dominated | 24,387 | |
| No test | 83,126 | 7.87 | Yes | N/A | Dominated | Dominated | Dominated |
| Comparator vs. no test | Incremental costs (discounted) | Incremental QALYs (discounted) | ICER | Net monetary benefit |
|---|---|---|---|---|
| POCT: Genedrive vs. no test | −£1669 | 0.17 | −£9816 | £5069 |
| POCT: Genomadix Cube vs. no test | −£1576 | 0.17 | −£9270 | £4976 |
| Laboratory test vs. no test | −£1641 | 0.17 | −£9808 | £4988 |
FIGURE 36.
Incremental cost-effectiveness plane for laboratory test vs. no test for the non-minor ischaemic stroke population. WTP, willingness to pay.
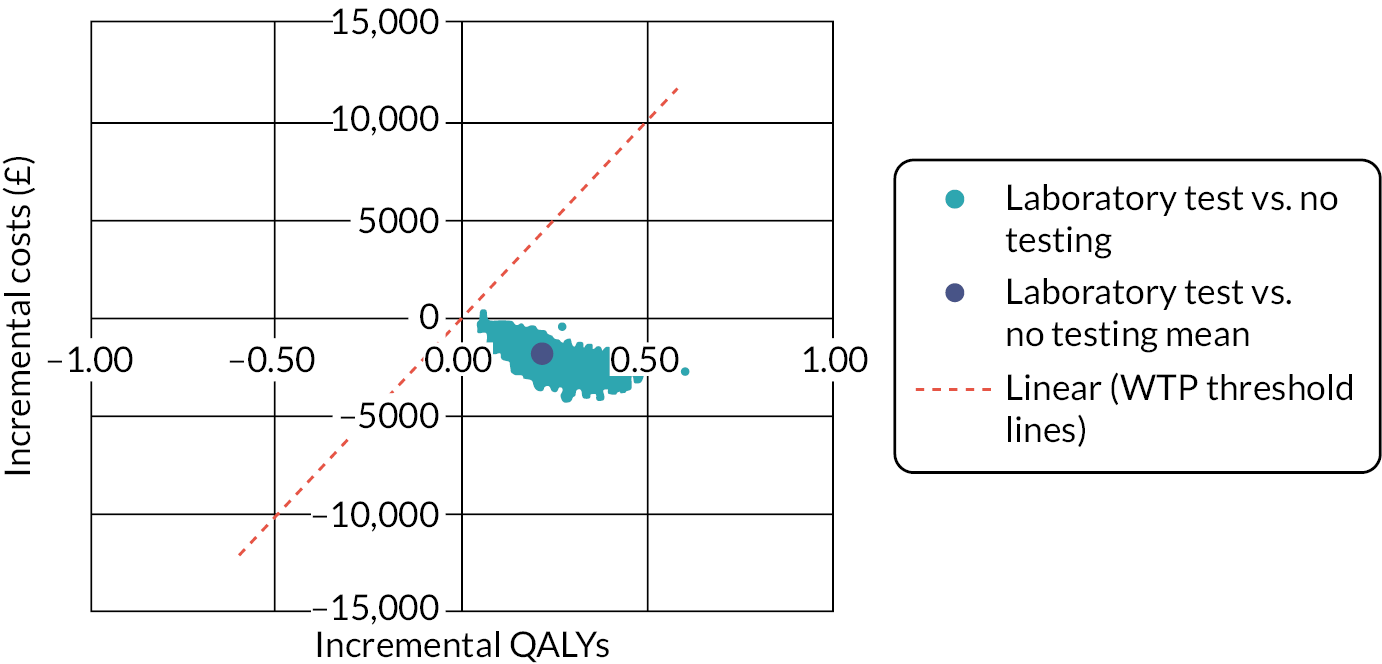
FIGURE 37.
Incremental cost-effectiveness plane for Genomadix Cube vs. no test for the non-minor ischaemic stroke population. WTP, willingness to pay.
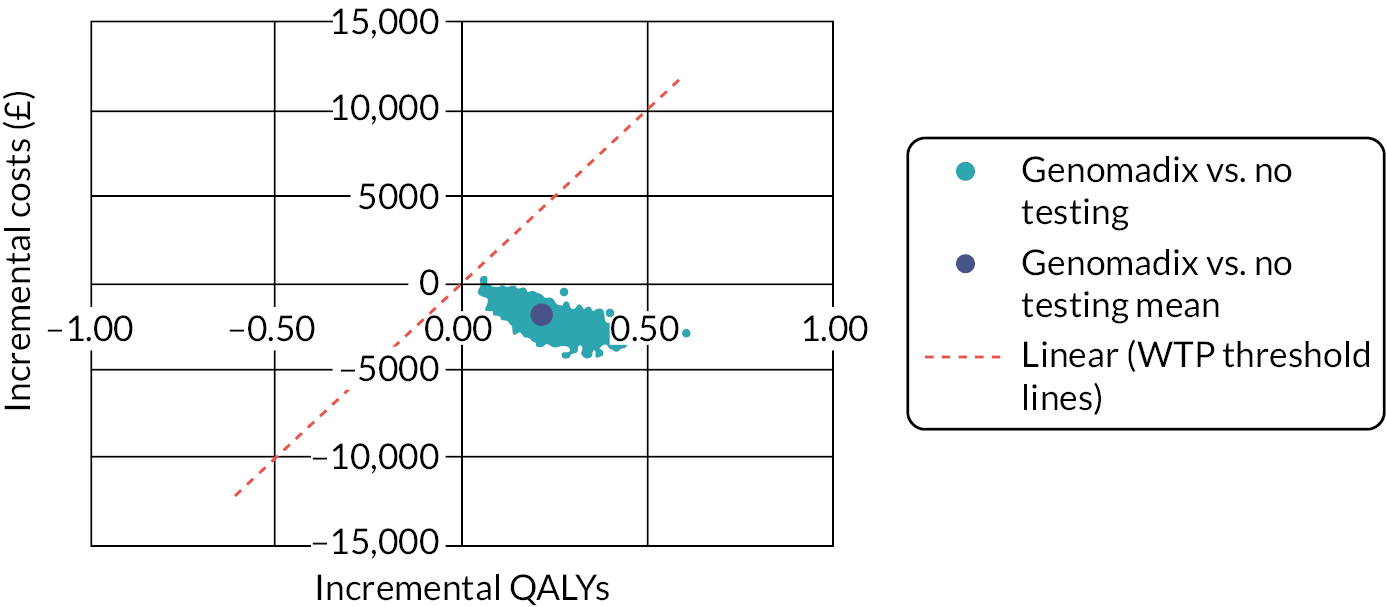
FIGURE 38.
Incremental cost-effectiveness plane for laboratory test vs. no test for the TIA/minor stroke population. WTP, willingness to pay.
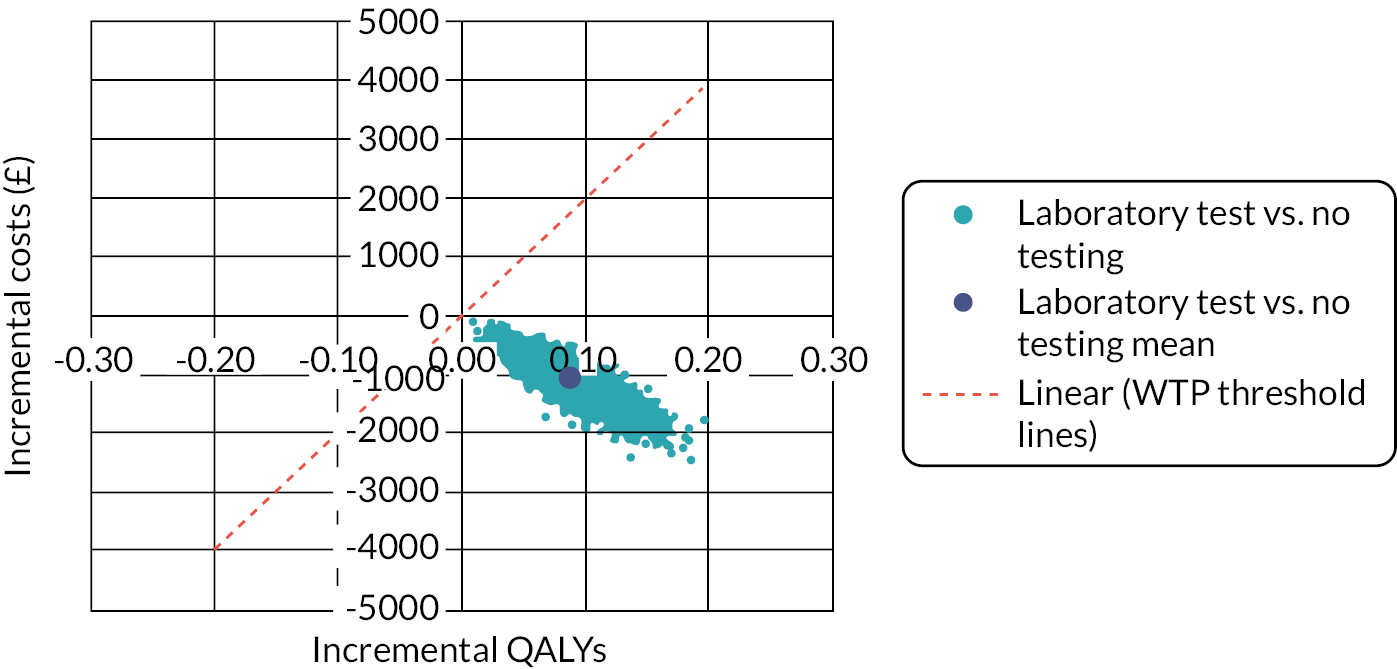
FIGURE 39.
Incremental cost-effectiveness plane for the Genomadix Cube vs. no test for the TIA/minor stroke population. WTP, willingness to pay.

FIGURE 40.
Incremental cost-effectiveness plane for laboratory test vs. no test for the mixed population. WTP, willingness to pay.

FIGURE 41.
Incremental cost-effectiveness plane for the Genomadix Cube vs. no test for the mixed population. WTP, willingness to pay.

FIGURE 42.
Cost-effectiveness acceptability curve for the mixed population (excluding Genedrive due to insufficient data).
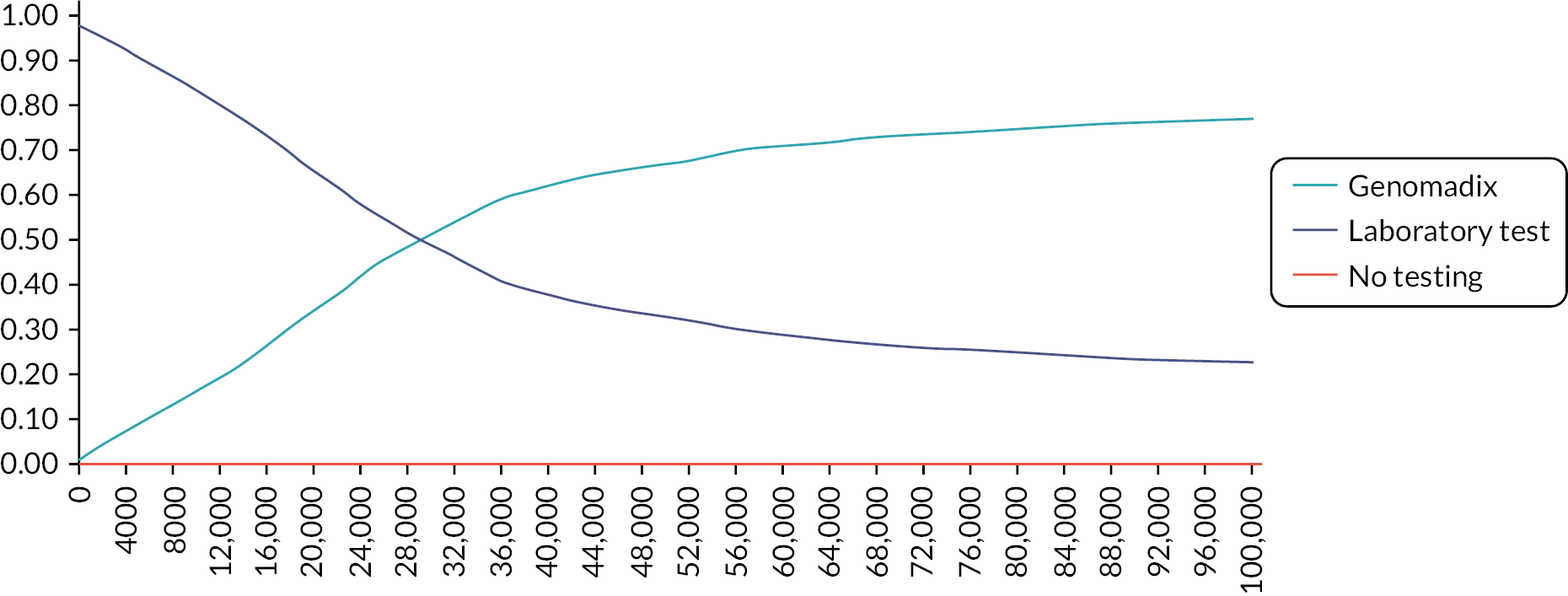
| Treatments | Total costs (£) (discounted) | Total QALYs (discounted) | Strictly dominated | Extendedly dominated | ICER (£) | ||
|---|---|---|---|---|---|---|---|
| vs. Genedrive | vs. Lab test | vs. Genomadix Cube | |||||
| POCT: Genedrive | 81,341 | 7.106 | |||||
| Laboratory genetic test | 81,356 | 7.104 | Yes | N/A | Dominated | ||
| POCT: Genomadix Cube | 81,433 | 7.106 | No | No | 2,172,044 | 46,446 | |
| No test | 83,031 | 6.930 | Yes | N/A | Dominated | Dominated | Dominated |
| Comparator vs. no test | Incremental costs (discounted) | Incremental QALYs (discounted) | ICER | Net monetary benefit |
|---|---|---|---|---|
| POCT: Genedrive vs. no test | −£1691 | 0.176 | −£12,255 | £5211 |
| POCT: Genomadix Cube vs. no test | −£1598 | 0.176 | −£11,450 | £5119 |
| Laboratory genetic test vs. no test | −£1675 | 0.174 | −£12,348 | £5163 |
Appendix 10 Additional sensitivity analyses
FIGURE 43.
Deterministic sensitivity analysis results for parameters with largest impact on the ICER for laboratory test vs. no test for the non-minor ischaemic stroke population.

FIGURE 44.
Deterministic sensitivity analysis results for parameters with largest impact on the ICER for laboratory test vs. no test for the TIA/minor stroke population.
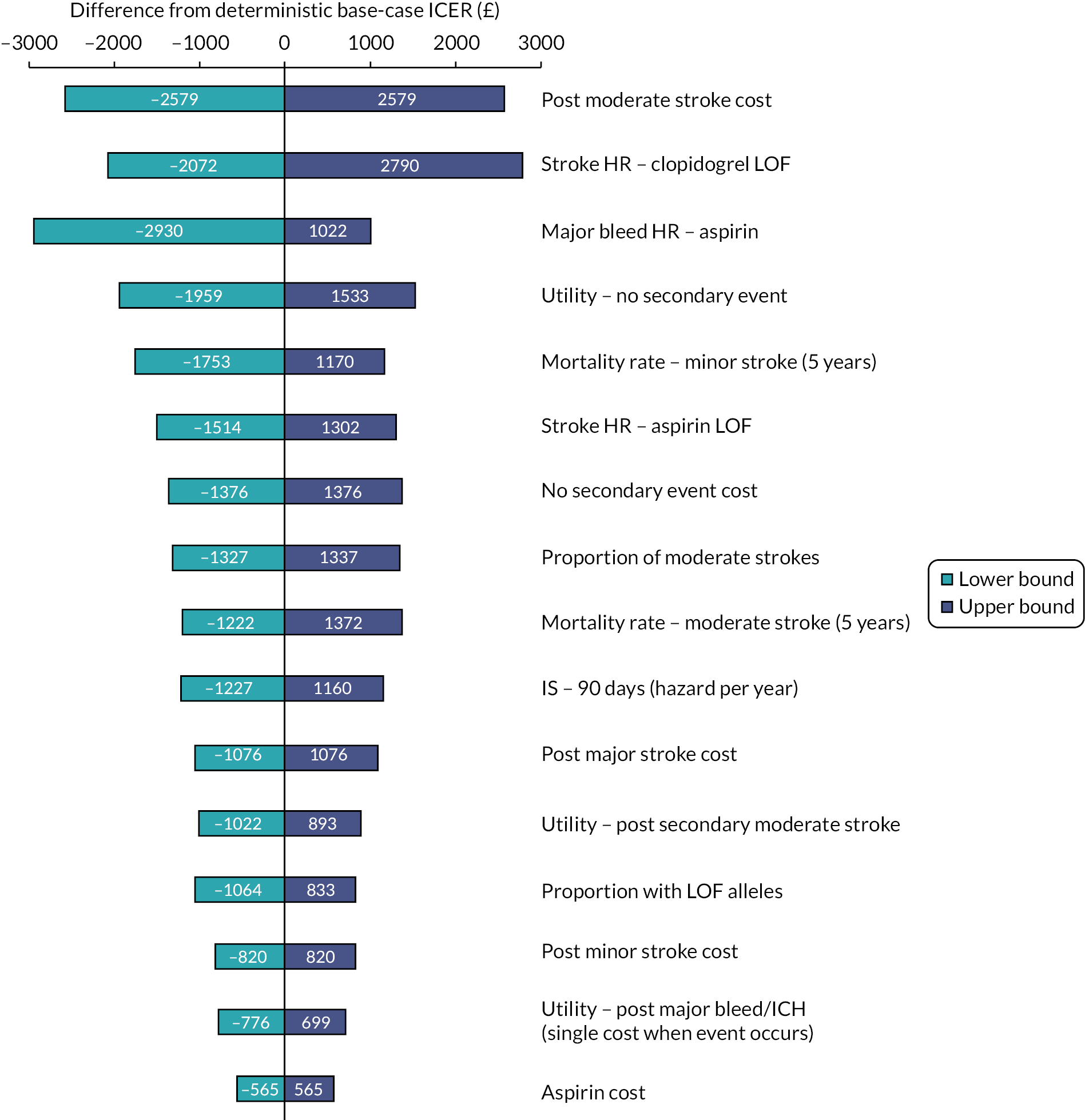
FIGURE 45.
Deterministic sensitivity analysis results for parameters with largest impact on the ICER for Genomadix Cube vs. no test for the non-minor ischaemic stroke population.
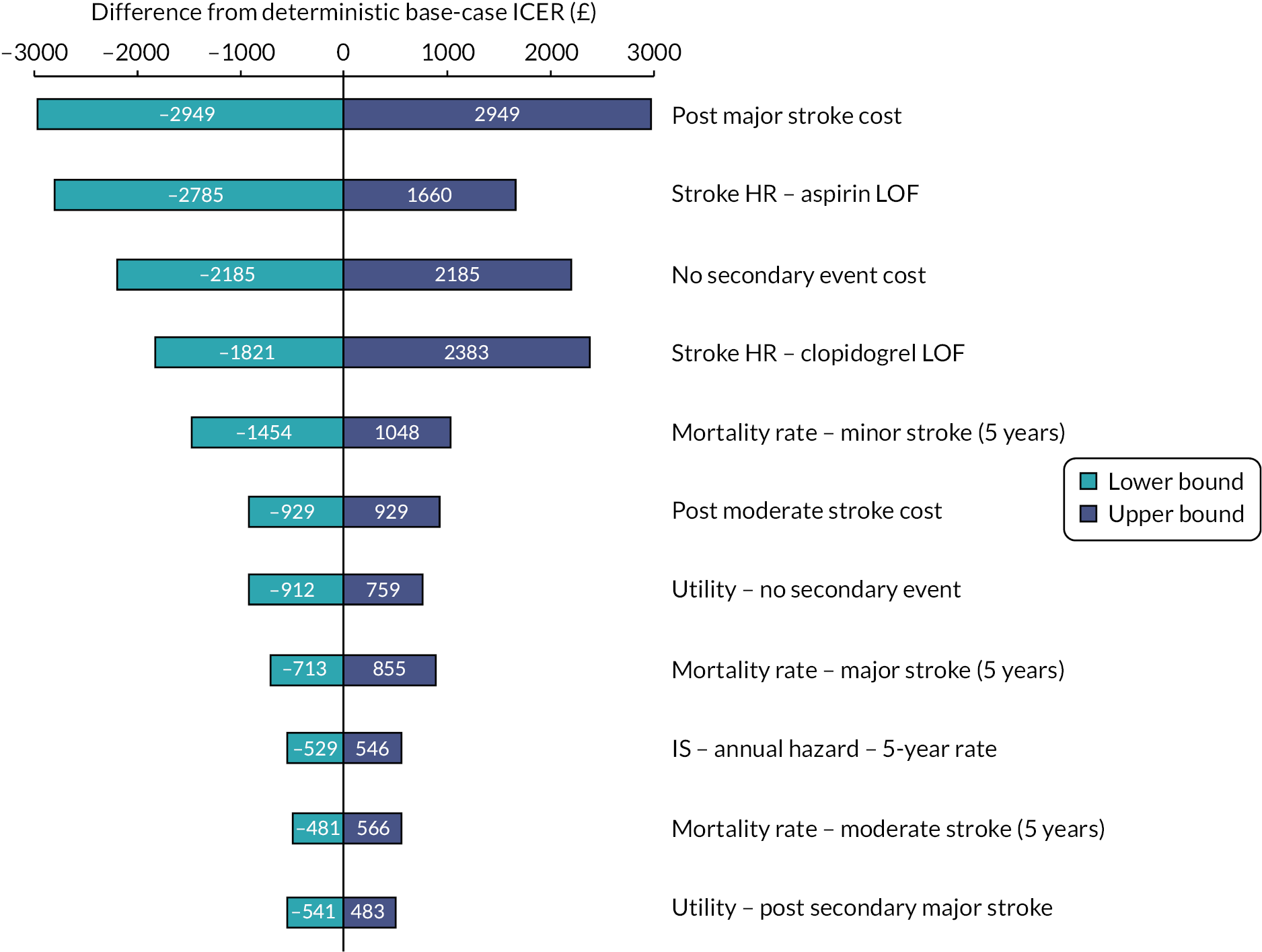
FIGURE 46.
Deterministic sensitivity analysis results for parameters with largest impact on the ICER for Genomadix Cube vs. no test for the TIA/minor ischaemic stroke population.

Threshold diagrams
FIGURE 47.
Incremental net monetary benefit of the laboratory test vs. other testing strategies by laboratory test cost for the non-minor ischaemic stroke population.
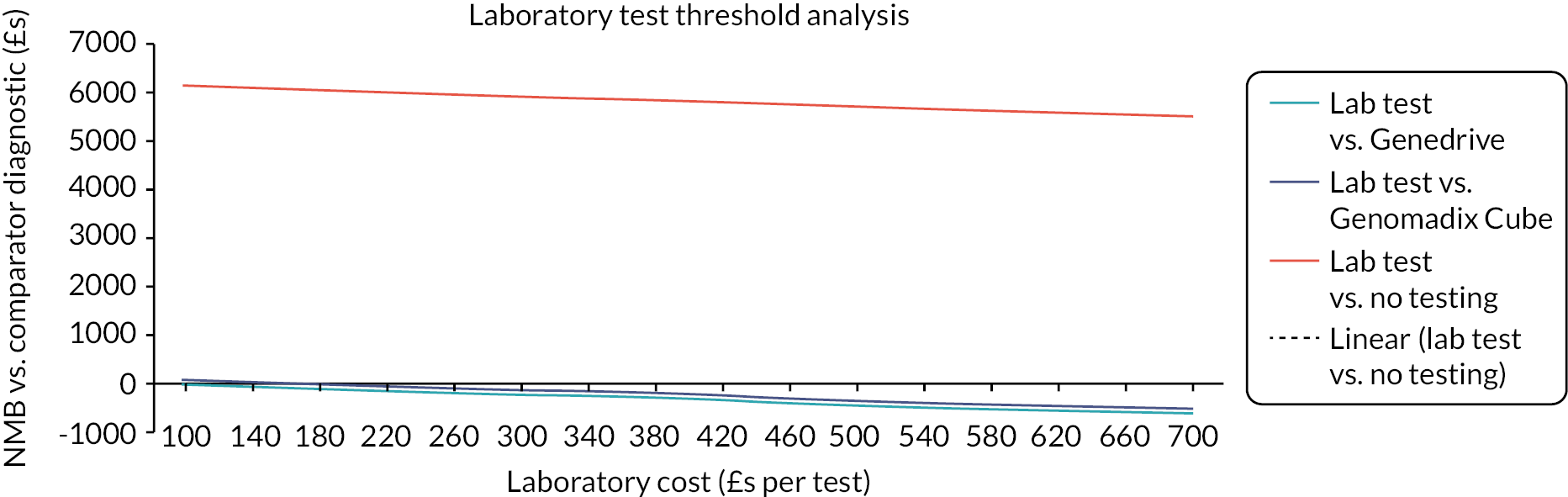
FIGURE 48.
Incremental net monetary benefit of the laboratory test vs. other testing strategies by laboratory test cost for the TIA/minor stroke population.

FIGURE 49.
Incremental net monetary benefit of the Genedrive vs. other testing strategies by Genedrive test accuracy for the net monetary benefit of the non-minor ischaemic stroke population.
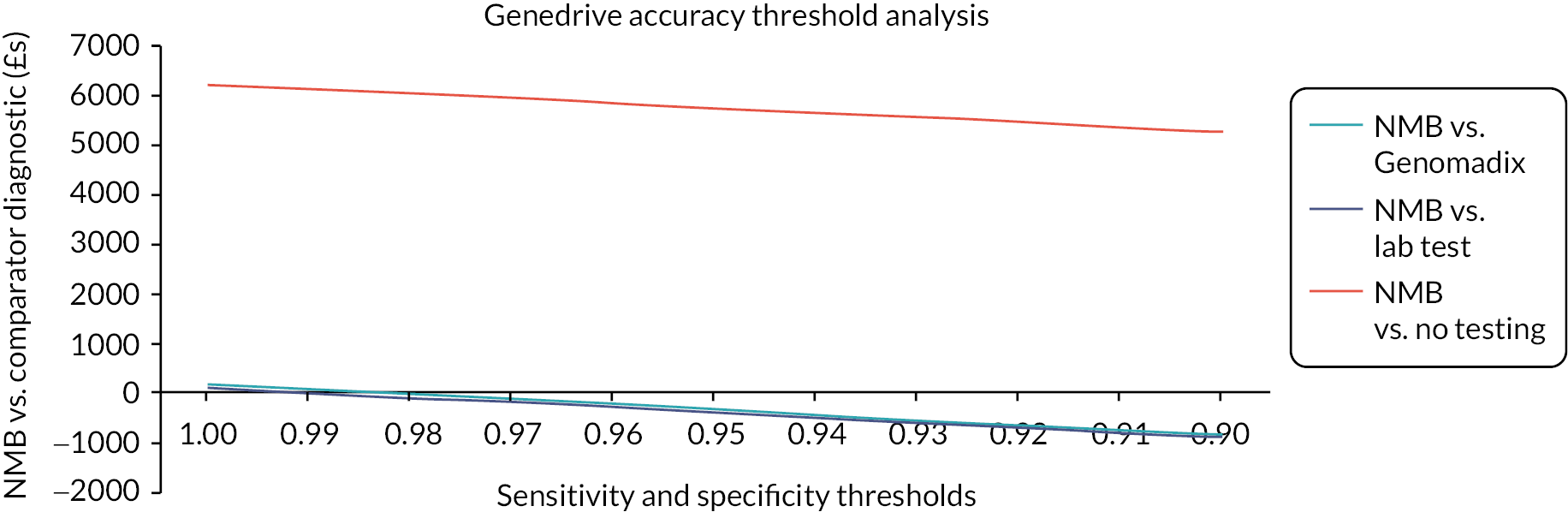
FIGURE 50.
Incremental net monetary benefit of the Genedrive vs. other testing strategies by Genedrive test accuracy for the TIA/minor stroke population.
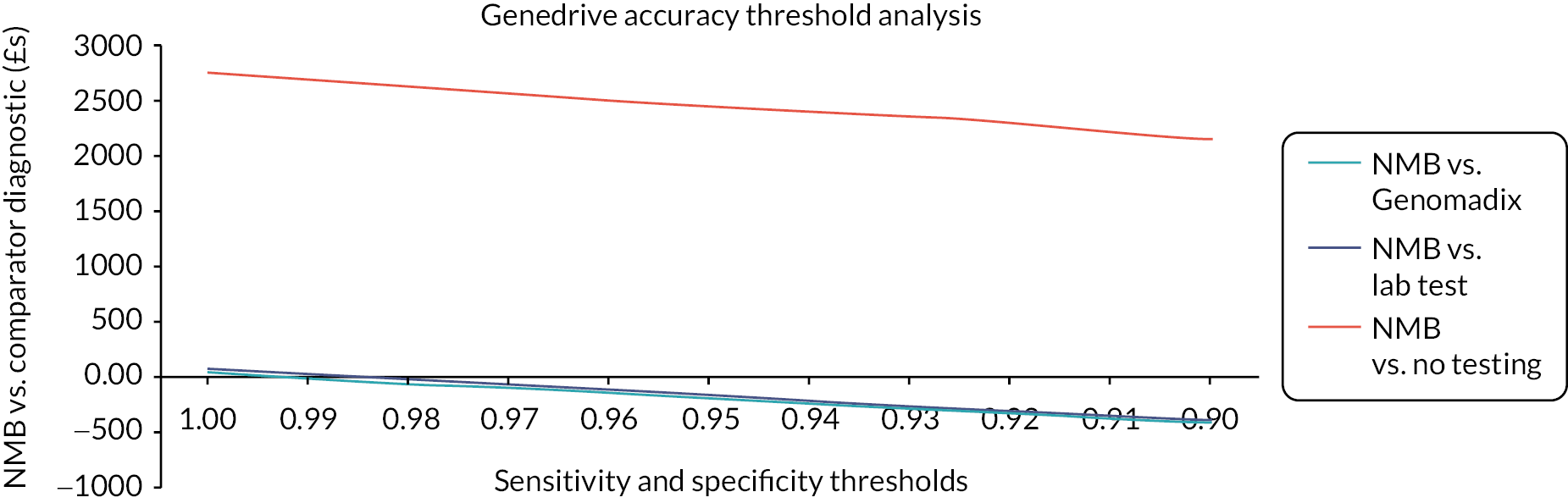
List of abbreviations
- ABCD2
- risk of stroke scoring system after a suspected transient ischaemic attack
- ACE
- angiotensin-converting enzyme
- Alt Tx
- alternative treatment
- CAD
- coronary artery disease
- CAPRIE
- clopidogrel versus aspirin in patients at risk of ischaemic events
- CEA
- cost-effectiveness analysis
- CPIC
- Clinical Pharmacogenetics Implementation Consortium
- CRD
- Centre for Reviews and Dissemination
- DAC
- Diagnostic Appraisal Committee
- DAPT
- dual antiplatelet therapy
- DNA
- deoxyribonucleic acid
- DPYD
- dihydropyrimidine dehydrogenase
- ECR
- electronic care record
- EED
- Economic Evaluations Database
- EQ-5D
- EuroQol 5 dimensions
- EQA
- external quality assessment
- FN
- false negative
- FP
- false positive
- HD
- high dose
- HFE
- hemochromatosis gene
- HR
- hazard ratio
- HRQoL
- health-related quality of life
- HTA
- Health Technology Assessment
- ICER
- incremental cost-effectiveness ratio
- ICH
- intracerebral haemorrhage
- ICTRP
- International Clinical Trials Registry Platform
- IM
- intermediate metaboliser
- iPLEX
- increased plexing efficiency and flexibility
- LAMP
- loop-mediated isothermal amplification
- LIMS
- laboratory information management system
- LOF
- loss of function
- MALDI-TOF
- matrix-assisted laser desorption/ionisation-time of flight
- MI
- myocardial infarction
- mRS
- modified Rankin Scale
- NGS
- next-generation sequencing
- NICE
- National Institute for Health and Care Excellence
- NIHSS
- National Institutes of Health Stroke Scale
- NoLOF
- loss-of-function non-carriers
- N/R
- not reported
- ONS
- Office for National Statistics
- PCI
- percutaneous coronary intervention
- PCR
- polymerase chain reaction
- PHE
- Public Health England
- PM
- poor metaboliser
- POCT
- point-of-care test
- PPI
- proton pump inhibitor
- PSA
- probabilistic sensitivity analysis
- PSS
- personal social services
- QALY
- quality-adjusted life-year
- RCP
- Royal College of Physicians
- RCT
- randomised controlled trial
- REML
- restricted maximum likelihood
- RoB
- risk of bias
- SLSR
- South London Stroke Registry
- SNP
- single-nucleotide polymorphisms
- SSNAP
- Sentinel Stroke National Audit Programme
- STEMI
- ST-segment elevation myocardial infarction
- TAT
- turnaround time
- TIA
- transient ischaemic attack
- TN
- true negative
- TP
- true positive
- WHO
- World Health Organization
- WTE
- working time equivalent
Notes
Supplementary material can be found on the NIHR Journals Library report page (https://doi.org/10.3310/PWCB4016).
Supplementary material has been provided by the authors to support the report and any files provided at submission will have been seen by peer reviewers, but not extensively reviewed. Any supplementary material provided at a later stage in the process may not have been peer reviewed.
