Notes
Article history
The research reported in this issue of the journal was funded by the HTA programme as project number 04/33/01. The contractual start date was in September 2006. The draft report began editorial review in March 2015 and was accepted for publication in December 2015. The authors have been wholly responsible for all data collection, analysis and interpretation, and for writing up their work. The HTA editors and publisher have tried to ensure the accuracy of the authors’ report and would like to thank the reviewers for their constructive comments on the draft document. However, they do not accept liability for damages or losses arising from material published in this report.
Declared competing interests of authors
Wendy Atkin receives funds from Cancer Research UK (Population Research Committee – Programme Award C8171/A16894). Jonathan Myles also receives funds from Cancer Research UK and the National Institute for Health Research (NIHR) Health Technology Assessment (HTA). Jonathan Myles was part funded by the following NIHR HTA awards: 11/136/120 and 09/22/192. Andrew Veitch has received expenses-only sponsorship from Boston Scientific and Norgine to attend Digestive Diseases Week 2015, Washington, DC, USA, and Digestive Diseases Federation 2015, London, UK.
Permissions
Copyright statement
© Queen’s Printer and Controller of HMSO 2017. This work was produced by Atkin et al. under the terms of a commissioning contract issued by the Secretary of State for Health. This issue may be freely reproduced for the purposes of private research and study and extracts (or indeed, the full report) may be included in professional journals provided that suitable acknowledgement is made and the reproduction is not associated with any form of advertising. Applications for commercial reproduction should be addressed to: NIHR Journals Library, National Institute for Health Research, Evaluation, Trials and Studies Coordinating Centre, Alpha House, University of Southampton Science Park, Southampton SO16 7NS, UK.
Chapter 1 Introduction
The call for proposal
This project was undertaken in response to a call for proposals by the National Institute for Health Research (NIHR) Health Technology Assessment (HTA) programme in anticipation of an unsustainable increase in requirements for surveillance colonoscopy with the impending introduction of the national Bowel Cancer Screening Programme (BCSP) in 2006. There was real concern that an increase in adenoma detection from the BCSP would diagnose many people as intermediate risk (IR), with a consequent impact on endoscopy resources. Therefore, a call was issued to determine the optimum frequency of colonoscopic follow-up in patients who were identified with intermediate-grade adenomas.
The current UK surveillance guideline was developed in 2002 and defines three risk groups (low, intermediate and high risk) with different surveillance recommendations. 1 From existing evidence it was suggested that, for the low-risk group, colonoscopy surveillance might not be necessary, whereas for the high-risk group surveillance was definitely indicated with an additional clearing examination 12 months after initial diagnosis (but this group constitutes only around 10% of people with adenomas2). The IR group, representing around 40% of patients with adenomas, was recommended to have a 3-yearly surveillance colonoscopy. However, this recommendation was based on limited evidence to indicate the optimum surveillance interval and the need for repeated surveillance. 1
Available evidence suggested that it might be safe to stop surveillance in the IR group after one or two negative examinations, depending on the age of the patient and the quality of the examination. Importantly, it was also proposed that patients with intermediate adenomas (IAs) may vary in their risk of developing colorectal cancer (CRC) and that there might be subgroups with different surveillance requirements. 3 The need to determine the optimum frequency of colonoscopic follow-up in IR patients was identified as a priority by the Department of Health (DH).
Rationale
Colonoscopy is the most widely used procedure for investigating colonic symptoms, and for surveillance of people at increased risk of CRC because of a personal or family history of CRC or adenomas. It is widely accepted that most CRCs develop from adenomatous polyps,4–7 and that the detection and removal (polypectomy) of these precursors through screening or surveillance reduces the risk of CRC. 8–13 Adenomas are very common and tend to recur. As such, the future risk of CRC after polypectomy is thought to depend on findings during baseline colonoscopy, particularly the number, size and histological grade of removed adenomas,3,14,15 as well as the completeness of examination and clearance of prevalent adenomas. This evidence was used to stratify patients into risk groups, each with different colonoscopic surveillance recommendations. 1,16–18
Since Atkin et al. 3 first suggested a variability in risk of CRC after adenoma removal in 1992, many countries have developed adenoma surveillance guidelines, most of which are based on either the UK or US guidelines. The indication for surveillance depends primarily on the presumed risk of recurrence of advanced adenomas (AAs),15,19–23 and development of CRC, and also by age, comorbidity and patient compliance. The current UK surveillance guideline was first commissioned and developed by the British Society of Gastroenterology (BSG) in 2002 and has since been adopted by the National Institute for Health and Care Excellence (NICE) and the European Union (EU) (Figure 1). 24 Both UK and US guidelines identify three risk groups, but the definitions and surveillance recommendations differ slightly. 1,2 Both guidelines identify a low-risk group, for which no surveillance or 5-yearly surveillance is recommended; an intermediate-(UK)/higher-risk (US) group, for which 3-yearly surveillance is recommended; and a high-risk group, for which additional colonoscopy is recommended. In the UK, the guideline specifies a single clearing colonoscopy at 12 months before continuing on 3-yearly surveillance. 1,2,16,17,25
FIGURE 1.
UK adenoma surveillance guidelines 2002. Reproduced from Surveillance guidelines after removal of colorectal adenomatous polyps. Atkin WS, Saunders BP, Gut, vol. 51, pp. V6–9, 2002,1 with permission from BMJ Publishing Group Ltd.
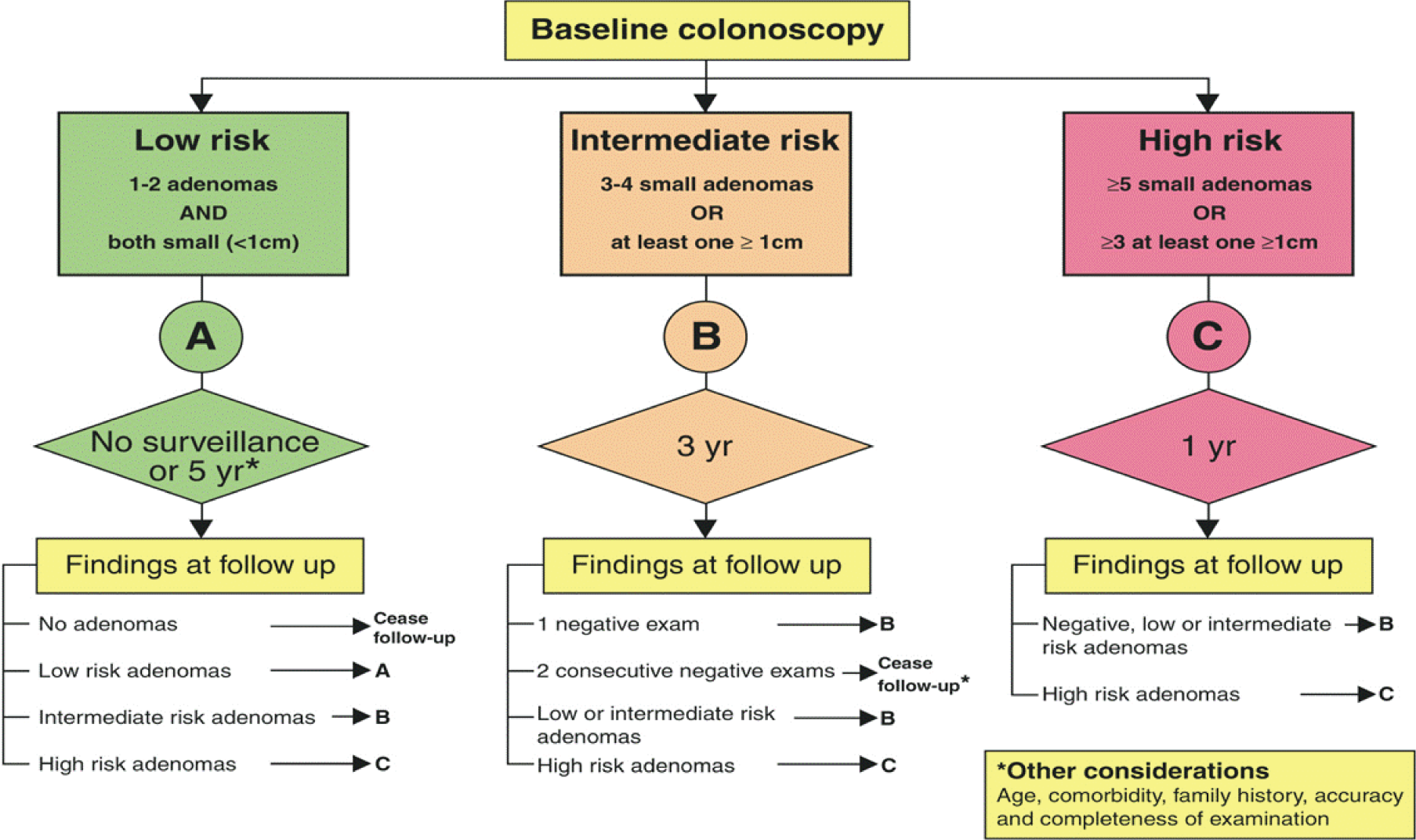
The fact that guidelines vary – particularly in defining the IR group and their surveillance recommendation18,26 – is indicative of the uncertainty about the optimum adenoma surveillance regime. After adenoma removal, some patients have a risk of CRC similar to, or lower than, that of the general population,3,27,28 implying that not all patients are at sufficient risk to warrant surveillance. 3,14,27–31 The IR 3-yearly surveillance regime is based on results of the National Polyp Study,20 which compared two follow-up colonoscopies with one follow-up colonoscopy within 3 years and found no difference in the detection of adenomas with advanced pathology. Two other studies32,33 also found the incidence of adenomas with advanced pathology to be similar regardless of interval length. However, another trial found a non-significantly higher risk of CRC in patients who were examined at 4 years than in those examined at 2 years. 29,34
As colonoscopy is both costly and invasive, surveillance should be undertaken only in those who are at increased risk and at the minimum frequency required to provide adequate protection against the development of cancer. 25 There is evidence of both over- and underutilisation of colonoscopy, and a potential for more efficient allocation of endoscopy resources. 35 The IR group comprises nearly 20% of those subjects participating in the BCSP who undergo colonoscopy for a positive test,36 and nearly half of adenoma patients,2 yet no study has yet systematically examined whether or not there is heterogeneity in risk among patients who are currently offered 3-yearly surveillance. We sought to address the unanswered questions surrounding the current IR group surveillance strategy, that is:
-
What is the effect of interval length on detection rates of AA and CRC at follow-up examinations in IR patients?
-
Are there subgroups of IR patients who do not require surveillance, or who require only one follow-up? Similarly, are there are subgroups that might benefit from shorter or longer surveillance intervals?
-
Does the risk of AA or CRC at first and second follow-ups vary by patient, procedural and polyp characteristics, and surveillance interval length?
-
Can we define factors that affect the risk of CRC after baseline in IR patients, for example number of surveillance visits, patient/procedural/polyp characteristics?
Background to the design
As a randomised controlled trial (RCT) or prospective observational study would take many years to complete, the use of pre-existing hospital patient data in a retrospective cohort study was the recommended design. It was thought that this method would be quicker, cheaper and more convenient. In addition, the use of such hospital patient data ensured that there would be sufficient variation in adenoma surveillance intervals to enable comparison between them. This may not have been possible with data collected prospectively because of the widespread adoption of UK surveillance guidelines. Furthermore, longer patient follow-up times could also be obtained in this retrospective study design.
We also requested access to data from researchers of a number of screening studies on findings at surveillance colonoscopy. Eight screening data sets were identified; however, only three provided adequate data for our analyses (see Chapter 4, Screening data set, Background).
At the time there was no systematic call or recall of patients in adenoma surveillance, so the principal investigator (PI) also wrote to the manufacturers of the patient management systems that were used to manage patient data in hospitals in the UK NHS. The manufacturers were able to identify hospitals that had used their software for a sufficiently long period of time. These hospitals were contacted and were provided with a questionnaire to complete in order to determine their suitability for the study.
Aim and objectives
The overall aim was to examine the optimum frequency of surveillance in patients who were found to have IR adenomas and assess the risks and benefits with respect to prevention of cancer/AA; anxiety, morbidity and mortality; costs and cost-effectiveness; and implications for the NHS.
The primary objective was to assess whether or not there was substantial heterogeneity in the detection of AA or CRC according to baseline characteristics and interval to first follow-up colonoscopy. The study planned to determine if there was a subgroup of IR patients who do not require surveillance and whether or not the size of this group is clinically significant. Finally, the study examined whether subgroups could be identified for which the currently recommended 3-year interval is too long, or for which the interval can be safely extended, or if there is a group that requires a second examination but no further follow-up.
An economic analysis aimed to estimate the incremental cost-effectiveness of alternative adenoma follow-up strategies, including a policy of no follow-up for individuals who have intermediate-grade adenomas. It also planned to estimate the impact of alternative adenoma follow-up strategies on colonoscopy services, and the total cost impact of alternative adenoma follow-up strategies in England and Wales.
A psychological impact analysis aimed to examine the anxiety-inducing effects of colonoscopic surveillance or being informed that colonoscopy surveillance is required.
Study design and setting
This was a retrospective cohort study using data from two sources. A cohort of patients attending UK NHS hospitals for diagnostic or surveillance endoscopy formed the largest data set – termed the ‘hospital data set’. Three smaller data sets were obtained from a research or screening setting, and involved average-risk individuals undergoing screening: one from a UK screening trial, a second from a UK pilot screening programme and a third from a US health surveillance programme – termed the ‘screening data sets’. The core results were derived from the hospital data set, as there were difficulties in obtaining additional screening data sets, and limited data completeness in the screening data sets that we were able to obtain. A health-economics evaluation and psychological study were also conducted.
Structure of this report
The findings of the hospital and screening data sets are reported and discussed in Chapters 3 and 4, respectively. The methods, which were largely the same for the two data sets, are described in Chapter 2, with any additional methods unique to the screening data set described in Chapter 4. The health-economic evaluation is reported in Chapter 5 and the psychological study is reported in Chapter 6. Finally, Chapter 7 presents a synthesis of results from the preceding chapters, as well as strengths/limitations and future work.
Chapter 2 Methods
Hospital selection
The hospital data set comprised routine gastrointestinal endoscopy and pathology data for patients having diagnostic and surveillance procedures. Participating hospitals were required to have recorded endoscopy and pathology data electronically for at least 6 years prior to the study start in 2006. After contacting endoscopy and pathology database manufacturers, 28 NHS hospitals were identified as meeting these criteria, and their participation in the study was requested. A number of hospitals were excluded because of difficulties with data extraction and data quality issues (see Data collection from hospitals, below). In total, 18 hospitals were included in the study. Two of these merged into the Imperial College Healthcare Trust (Charing Cross and Hammersmith hospitals) and thus there were 17 hospital sites included; these are listed in Table 1.
| Trust | Hospital | Study name | Study code | Collection dates |
|---|---|---|---|---|
| Brighton & Sussex University Hospitals NHS Trust | Royal Sussex County Hospital | Brighton | BRI | May 2001 to April 2008 |
| North Cumbria Acute Hospitals Trust | Cumberland Infirmary | Cumberland | CI | August 1998 to September 2009 |
| Imperial College Healthcare NHS Trust | Charing Cross Hospital/Hammersmith Hospital | Charing Cross/Hammersmith | CX/HH | October 1997 to November 2007 |
| Greater Glasgow and Clyde NHS Trust | Glasgow Royal Infirmary | Glasgow | GRI | May 1996 to August 2009 |
| University Hospitals of Leicester NHS Trust | Leicester General Hospital | Leicester | LGH | April 1998 to March 2008 |
| Royal Liverpool and Broadgreen University Hospitals Trust | Royal Liverpool University Hospital | Liverpool | RLUH | January 2000 to October 2009 |
| Royal Wolverhampton Hospitals NHS Trust | New Cross Hospital | New Cross | NC | January 1993 to November 2007 |
| University Hospital of North Tees Trust | University Hospital of North Tees | North Tees | NT | June 1986 to December 2006 |
| Queen Elizabeth Hospital NHS Trust | Queen Elizabeth Hospital | Queen Elizabeth | QEW | March 1999 to May 2006 |
| Queen Mary’s Sidcup NHS Trust | Queen Mary’s Hospital | Queen Mary’s | QMH | October 1998 to July 2009 |
| Shrewsbury and Telford Hospitals NHS Trust | Royal Shrewsbury Hospital | Shrewsbury | SH | January 2002 to September 2009 |
| St George’s Healthcare NHS Trust | St George’s Hospital | St George’s | SGH | February 1992 to July 2009 |
| London North West Healthcare NHS Trust | St Mark’s Hospital | St Mark’s | SMH | January 1985 to July 2007 |
| Imperial College Healthcare NHS Trust | St Mary’s Hospital | St Mary’s | ICMS | December 1984 to July 2010 |
| Royal Surrey County Hospital NHS Trust | Royal Surrey County Hospital | Surrey | SCH | September 1997 to May 2010 |
| South Devon Healthcare NHS Foundation Trust | Torbay District General Hospital | Torbay | TDG | November 2000 to August 2007 |
| Yeovil District Hospital Foundation Trust | Yeovil District Hospital | Yeovil | YDH | February 1997 to May 2008 |
Patient eligibility
Inclusion criteria
Patients with IR adenoma(s) and a baseline colonoscopy were eligible for inclusion in the study. Following the UK guideline, IR patients were defined as those with three or four small adenomas (of < 10 mm) or one or two adenomas, at least one of which was large (≥ 10 mm).
Exclusion criteria
Patients were excluded for having certain conditions if the condition increased their risk of CRC or could have led to an abnormal pattern of surveillance. Some diagnoses resulted in exclusion regardless of when they occurred, for example hereditary non-polyposis colorectal cancer (HNPCC), a genetic condition that confers an increased risk of cancer throughout an individual’s lifetime. Other conditions resulted in exclusion only if they were diagnosed at, or prior to, baseline, or, in other cases, patients were censored after diagnosis of a particular condition rather than excluded altogether.
Patients were excluded if they had any of the following diagnoses at, or prior to, baseline:
-
CRC or inflammatory bowel disease (IBD)
-
resection/anastomosis
-
volvulus.
Patients were excluded if they had the any of the following diagnoses at any time:
-
family history of familial adenomatous polyposis (FAP)
-
HNPCC
-
Cowden syndrome
-
juvenile or hamartomatous polyps.
Patients with polyposis could be excluded depending on polyposis type and time of diagnosis. Details of time-dependent exclusions for polyposis and colitis can be found later in the report (see Appendix 5).
Patients were also excluded if they had no baseline colonoscopy, or had one or more procedures without a date, or had more than 40 endoscopic procedures recorded.
Research governance
Data were collected from hospitals in England and Scotland. The following research governance approvals were obtained to permit data collection and follow-up via external agencies:
-
Approval was granted from the Royal Free Research Ethics Committee (REC) for the study throughout the UK (REC reference 06/Q0501/45). The REC agreed that all sites should be exempt from site-specific assessment. Further approval was granted for substantial amendments to allow changes to database hosting arrangements and logistical arrangements for data collection and follow-up.
-
Approval to access patient identifiable information without consent in England was granted from the Patient Information Advisory Group (PIAG) [later the National Information Governance Board (NIGB) and currently the Ethics and Confidentiality Committee at the Health Research Authority] in accordance with Section 60 of the Health and Social Care Act 200137 (re-enacted by Section 251 of the NHS Act 200638). Further approval was granted for substantial amendments to allow data to be extracted and anonymised, and to link identifiable information obtained from multiple sources including hospital endoscopy and pathology databases, the Hospital Episode Statistics (HES) database, and databases held by the Office for National Statistics (ONS), National Health Service Information Centre (NHSIC) [subsequently the Health and Social Care Information Centre (HSCIC)] and National Health Service Cancer Registries (NHSCR) [reference PIAG 1–05(e)/2006]. This was necessary because of the retrospective nature of the study and the large number of patients involved. Support was favourable based on the study’s System Level Security Policy and compliance with Imperial College’s policy on data handling and storage, and a recommendation from the Caldicott Guardian for the North West London Hospitals NHS Trust, who approved the arrangements to ensure patient confidentiality and anonymity. In Scotland, similar approvals were obtained in 2013 from the Community Health Index Advisory Group. Permission was granted to use the Community Health Index (CHI) to enable the Information Services Division to clean the patient information within the study data set, and to match identifiable information to data from the cancer and death registries.
-
Research approval for the study was obtained from all relevant NHS care organisations for the study sites, which were provided with the ethical approval documentation and the study protocol. As none of the members of the study team had a contractual relationship with the NHS, honorary contracts/letters of access were applied for and obtained for staff who were required to carry out work at the various study sites, in agreement with the Research Governance Framework.
-
Where necessary, applications were made to the custodians of external data sets to enable specific researchers to access information controlled by external sources and to allow the study data set to be linked to external data sets. In England, researcher status was approved and obtained from the ONS and NHSIC for individual researchers, and applications to use individual records for medical research were made to the NHSIC and the UK Association of Cancer Registries (UKACR). In Scotland, the Privacy Advisory Committee (PAC) granted approval for patient record linkage with NHS National Services Scotland (NSS) using CHI numbers, so that patients in Glasgow could be followed up to obtain details of cancers and deaths (reference PAC Application 66/11).
To ensure that patient confidentiality was maintained throughout the study, no patient-identifiable information – except date of birth – was stored on the study database. All patient identifiers were left at the individual study sites in secure locations, and all information kept at the trial office was in a pseudo-anonymised format. In addition, access to the Oracle database was controlled by username and password security, as well as a firewall that restricted access to the database server to a limited number of IP addresses. The majority of computers in the trial office were given access to the database, whereas specific access to the data via the Oracle Application Express (APEX) version 3.2.1.00.10 (Oracle Corporation, Redwood City, CA, USA) coding application was controlled via APEX’s built-in user management facility.
Data collection from hospitals
The data were extracted from hospital endoscopy and pathology databases by the study programmer. A minority of databases had an interface that permitted bulk extraction of the data according to specific criteria and in these cases data were extracted with assistance from hospital staff who were familiar with the systems. However, for most endoscopy and pathology databases, the application interface was not designed for bulk data extraction, so data extraction and processing was complex, with a number of problems encountered, for example:
-
When the maintenance and support of the endoscopy and pathology databases had been outsourced to the database manufacturers, often only they could help with extracting the data or by writing software enabling the study programmer to do so.
-
Specialist support was required when data were held on legacy systems.
-
Information technology (IT) staff at the hospitals sometimes had to restore archived data temporarily so that they could be extracted.
-
Most hospitals had replaced databases over the years, and therefore some data overlapped or were duplicated (e.g. the same patient had records on more than one system).
-
Sometimes several hospital visits were necessary to extract data from multiple databases, at the convenience of the local IT experts.
-
The data outputs from these databases were in a combination of structured and unstructured formats. Structured data could be easily cleaned and converted into a standardised format for uploading. Unstructured data (usually large text fields) needed bespoke programs written to extract, clean and convert the data into a suitable format.
Owing to various technical difficulties with data extraction, inability to access databases, partial availability of electronic data, unreliable Systematized Nomenclature of Medicine (SNOMED) coding and logistical difficulties due to local staff availability, nine hospitals were excluded from the study (Table 2). From the 17 hospitals that were included in the study, data were extracted from 27 endoscopy and 29 pathology systems. (A summary of data collection at each hospital can be seen in Appendix 1.)
| Hospital | Reason for exclusion | Summary | Details |
|---|---|---|---|
| Blackpool Victoria | Software/data collection issue | Incompatibility of old vs. new software systems for importing endoscopy data Difficulties in bulk data transfer from pathology reports |
Following the creation of extraction programs and test runs, the statistics program was unable to extract all of the necessary endoscopy data. The main endoscopy reports could not be extracted from the older EndoScribe data imported into the newer ADAM system The pathology system did not allow the uploading of pathology reports in bulk |
| Bradford Royal Infirmary | Software/data collection issue | Old software systems used to record data Difficulties in bulk data transfer |
Pre-2005 SNOMED coding for pathology data was unreliable The Co-Path system proved to be problematic and complex to extract multiple records – it would have taken too long and would have slowed down the system for the hospital |
| City Hospital, Birmingham | Difficulties in obtaining R&D approval | Delays and limited resources | The study encountered long delays in R&D approval, owing to staff shortages in the R&D department and time-consuming internal procedures |
| George Eliot, Nuneaton | Software/data collection issue | Old software systems used to record data Difficulties in bulk data transfer |
The same difficulties with the Co-Path system as encountered with Bradford Royal Infirmary |
| King George, Ilford | Data collection issue | Impractical to extract the data Difficulties in bulk data transfer |
The majority of older pathology data were initially inaccessible because of software licensing issues. When access was achieved, the reports could be accessed only one at a time, making it impractical to extract the data |
| Norfolk and Norwich University Hospital | Technical issues Missing data |
Raw endoscopy and pathology data were extracted between November 2009 and August 2010 from the Micromed, EndoScribe and Scribe databases and the pathology database over several visits to the hospital, and partly cleaned and anonymised. However, on subsequent visits to complete the task the data had been misplaced Re-extraction would have been costly and time-consuming |
|
| Pinderfields, Yorkshire | Software/data collection issue | Incomplete data | The data provided by a new HICSS system (Ascribe Ltd, now EMIS Group plc, Leeds, UK) were lacking any endoscopy procedures other than colonoscopy (e.g. sigmoidoscopy and rigid sigmoidoscopy) |
| Queen Alexandra, Portsmouth | Missing data | Missing pathology data 2006–8 | |
| University Hospitals of North Staffordshire NHS Trust | Software/data collection issue | Difficulties in bulk data transfer | Not possible to do a bulk data extraction at this hospital |
Data extraction
Endoscopy data
Endoscopy databases were searched first in order to identify patients undergoing colonic examinations, as the pathology databases contained a wide range of extracolonic samples. Before removal from the hospital, the extracted data were split into patient identifiers and endoscopic data. Patient identifiers included surname and forename(s), hospital number(s), NHS number, gender, postcode and date of birth. Endoscopic data included date of procedure, type of procedure, indications, endoscopist name, endoscopist comments, polyp information (such as size, shape, location, information on any biopsies taken), segment reached, quality of bowel preparation, complications encountered, diagnosis and any other information. The list of patient identifiers was cleaned to remove errors, inconsistencies and duplicates, and a unique study number was assigned to every patient. Study numbers were made up of a three-letter code, representing the hospital, followed by a six-digit number.
Pathology data
Pathology databases were searched for reports on colorectal lesions. The preferred search method used in most hospitals was SNOMED (College of American Pathologists), which defined the site and type of colonic lesions present. When this was not possible, Systematized Nomenclature of Pathology (SNOP) (College of American Pathologists) codes (four-digit versions of SNOMED codes), keywords or SNOMED International 3.0 codes (College of American Pathologists) were used (see Appendix 1 for details of the methods used to collect pathology data at each centre). Initial validation checks were performed to ensure that the pathology extract included the date of report, unique report number, type of procedure where specimens were taken, number of specimens and histological details.
Linking endoscopy and pathology data
Patients identified from endoscopy records were matched to their pathology records using a combination of hospital number, name and date of birth. Patient study numbers were then assigned to the matched pathology records. Manual inspection of the data and preliminary analyses were performed at the hospital to check that a sufficient number of pathology records were linked to a patient and that they occurred on, or near, the date of an endoscopy. When there was cause for concern (e.g. very few endoscopies linked to pathology reports; or endoscopies at which a biopsy was taken did not have an associated pathology report; or a large number of pathology reports could not be linked to an endoscopy, suggesting that the endoscopy extract did not retrieve all records), further investigations were undertaken and the data were re-extracted from the endoscopy and pathology systems where necessary.
Pseudo-anonymising data
In order to maintain patient confidentiality in accordance with the EU Directive on Good Clinical Practice (Directive 2005/28/EC), the Data Protection Act 199839 and the NHS Caldicott Principles, all patient identifiers except date of birth were removed from the pathology and endoscopy data, and the anonymised data were encrypted before being removed from the hospital.
A ‘patient-linking-file’ in Microsoft’s .xls or .xlsx format (Microsoft Corporation, Redmond, WA, USA), storing each patient’s identifiable information and study number was created, encrypted, and left at each hospital site. The raw endoscopy and pathology data and patient-linking-file were copied on to CDs and stored in secure locations at the hospital under supervision of the local PI.
Development of the master database
A master database was created to store the data in a standardised, structured format. To facilitate the statistical analysis, the data had to be classified into quantitative and qualitative variables, ensuring that data from different hospitals were classified in the same way, as there was wide variation in the raw data (e.g. field names were different; some data were coded or semi-coded, whereas other data were in free-text fields; and data types varied). The PI, study researchers, statistician and study programmer defined the data requirements for the study and designed the structure of the master database (see Appendix 2). The master database was designed to store the following:
-
the original source data (to safeguard against data loss during coding)
-
fields to store structured data that had been automatically extracted, cleaned and standardised using bespoke programs
-
fields to store the structured data which were manually coded.
Reference data (sometimes referred to as look-up tables) were used to categorise and define permissible values for data fields on the database. This method restricted the values to be recorded in a data field, thereby preventing coding errors and also ensuring uniformity of data from different hospitals.
It was necessary to transform the variety of data received from different hospitals into a standardised data set. As the volume of records was very large, it was necessary to code and categorise the data as much as possible using automatic coding without compromising the integrity of the data. Programs were developed to transform, clean and automatically code the data where possible. This involved several steps:
-
identifying the fields containing information required for the study, taking into account varying field names, data types and value representations
-
extracting information from free-text fields using programming techniques such as ‘regular expressions’ and ‘fuzzy matching’, and translating them into the codes used on the master database
-
translating values in the raw data into the codes used on the master database if the information was already in a coded structured format
-
identifying and consolidating overlapping data, and removing any redundancies (e.g. the same endoscopy or pathology reports extracted from two different systems)
-
identifying errors in the data and validating and correcting them (e.g. misspellings, different date formats, accounting for false-positive matches)
-
transforming polyp data to fit the structure of the polyp table in the master database. Some raw data sets had structured data on polyps (i.e. each polyp was represented as a separate table record); for other data sets, the study programmer had to separate the data into individual records.
After data transformation and cleaning, the raw data and structured data were uploaded to the master database, ensuring that the data were linked correctly across tables. Exclusions of ineligible patients were made automatically where possible, using programming techniques such as ‘regular expressions’ and ‘fuzzy matching’ to identify relevant keywords or phrases in the reports. Approximately 17% of patients were excluded using automatic exclusions.
Manual data coding
The records of the remaining 83% patients who had not been auto-excluded were manually interpreted and coded; this also involved checking the automatic coding on these records.
A web-based coding application with a graphical user interface was developed using APEX, allowing the study researchers to read, interpret and code the information in the database efficiently. This was called the Endoscopy and Pathology Reports Application (EPRA). The development of the EPRA evolved over time as new data items were encountered at different hospitals, and as processes for coding and analysing the data were developed. A change log of new features was maintained on the study database and updated when a new version of the EPRA was released. (Details and screenshots of the EPRA are provided in Appendix 3.)
Documents detailing standard operating procedures (SOPs) were produced to ensure standardised coding methods between study researchers. SOPs covered all basic coding methods, rules for coding individual fields within the database, and more complex processes used for tasks such as polyp numbering (see below). All SOPs can be found in Appendix 4.
A specific study researcher was allocated a patient’s complete set of records to ensure that the study researcher had access to all available information. The study researchers were responsible for:
-
checking and correcting data that had been automatically coded
-
checking that endoscopy and pathology records were properly linked
-
coding the raw endoscopy and pathology data into structured data
-
creating individual polyp records from the data provided in endoscopy reports. In some cases, the study researchers found that polyps were described as groups rather than as individual polyps. This is discussed further below
-
raising queries on records that could not be fully coded due to incomplete or insufficient information
-
creating a blank ‘pathology-based procedure report’ in cases for which the pathology record had no linked procedure report. Clinical information available in the pathology report was used to deduce details about the procedure from which the histological sample was obtained.
Coding accuracy and data interpretation were monitored to maintain consistency, using the following methods:
-
Study researchers systematically reviewed a blinded random sample of records that had been coded by other study researchers.
-
Regular meetings and continuous discussion/feedback were used to ensure uniformity of coding.
Records that had not been coded because of incomplete or insufficient information were reviewed by the study researchers, and further data were obtained from hospitals, where possible, in order to complete the coding.
Polyp numbering
A polyp found at one endoscopy examination that was not removed or only partially removed could be seen again at a later examination. To ensure that there was no double-counting of polyps, each polyp was assigned a unique polyp number that could be used to link sightings of the same polyp at different examinations. This process was called ‘polyp numbering’.
In approximately 17,000 patients, polyps were found on at least two occasions and were reviewed for manual polyp numbering. All sightings of an individual polyp were assigned the same unique polyp number. Each polyp was also assigned a match probability (to the nearest 10%), to indicate the degree of certainty that two polyps were the same lesion. The polyp that appeared to have the greatest number of matches with a high degree of certainty was chosen as a reference polyp, and all possible matches were considered in relation to this polyp. Polyp numbering guidelines were used to match polyps accurately and methodically, using all of the available information from the endoscopy and pathology reports. Particular attention was given to the following factors, listed in order of importance. Sightings at different examinations were considered more likely to be the same polyp if:
-
they occurred in the same segment of the colon, or in adjacent segments
-
there was an indication that the polyp at the earlier examination was not removed or was only partially removed
-
the quality of bowel preparation at the first examination was poor, making it less likely that a lesion would be removed
-
the lesions had similar grades of dysplasia
-
the lesions were the same histological type
-
the lesions had similar degrees of villousness.
Quality checks were carried out by the study researchers, who manually reviewed and checked a random sample of records for which polyps had been numbered by other study researchers.
Polyps matched with an arbitrary probability of ≥ 70%, using the above criteria, were considered the same lesion. More details on polyp numbering can be found in Appendix 6.
Polyp groups
Sometimes endoscopy reports described groups of polyps using terms such as ‘several’, ‘many’ and ‘multiple’, rather than individual polyps. During manual coding, specific fields were used to record this information. Each group of polyps was recorded as a single record and populated with information such as site, shape and histology, where this was common to all polyps within the group. Descriptions of the size and number of polyps in the group (e.g. ‘tiny’, ‘multiple’) were recorded. Where information was given for an individual polyp within the group, a polyp record was created and linked to the group record. The whole group (and the individual polyps linked to it) was allocated a unique group number.
Patients with multiple polyps could have groups of polyps seen at more than one examination, and a group of polyps seen at a later examination could include some or all of the polyps seen at a previous examination. In order to link groups of polyps seen at more than one examination, a separate group linking number was assigned to each group of polyps. This task was completed after all polyp groups had been recorded for a patient. Groups of polyps seen at more than one examination were matched and a probability was assigned, indicating the study researcher’s certainty that groups of polyps seen at separate examinations were of the same group.
The records for groups of polyps were then expanded into individual polyp records so that they could be analysed. An estimate of the number of polyps in each group was deduced from a value coded for the approximate number where available; otherwise the average of the minimum and maximum number of polyps recorded by the endoscopist was used. Alternatively, a numeric value was estimated for each vague number description (e.g. ‘some’, ‘several’, ‘few’), taking the average value for all groups in which both the specific descriptor and a numeric value was reported; these values used to define the number of polyps in the such groups are shown in Table 3.
| Description | Estimated no. of polyps |
|---|---|
| A few | 3 |
| Some | 3 |
| A number of | 3 |
| Several | 3 |
| Many | 5 |
| Multiple | 5 |
Additional information on the number of individual polyps seen at previous or subsequent examinations which were considered to be part of the same group was used to refine the estimate of the number of polyps in that group (see Appendix 6).
Once a final estimate had been derived for the total number of polyps in each group at each examination, a program was written to create individual polyp records. Where a polyp record was created based on the presence of a polyp at a previous or subsequent examination, the program assigned the same polyp number to the new polyp record, to show they were the same lesion.
Creating summary values for polyp characteristics
Most polyps were seen and removed at a single examination, and information about a polyp’s features was available from a single endoscopy and pathology report. Alternatively, a polyp might be seen at more than one examination with descriptive information contained in numerous endoscopy and pathology reports. In both of these scenarios, a single polyp characteristic might be coded for in multiple data fields. It was therefore necessary to create summary values for each lesion, taking into account information provided in reports on polyp characteristics at individual examinations and across examinations. However, the following issues had to be resolved first.
Missing polyp information
When information on a polyp characteristic was missing from the endoscopy report, it was sometimes possible to obtain supplementary information from the pathology report, or from other examinations at which the same polyp was detected.
Inconsistent polyp information
Polyp information reported in an endoscopy and pathology report for a specific examination could be inconsistent. Similarly, information reported across multiple sightings of a single polyp could be inconsistent. Sometimes it was clear from available information that an inconsistency was due to a coding or transcriptional error in one or more hospital reports. Rules were identified to determine which data items were likely to be errors; these records were manually reviewed and errors corrected where possible. Inconsistencies that could not be explained by error were resolved using hierarchies of rules (see Appendices 7 and 8).
Vague polyp information
Wherever possible, information on polyp characteristics was recorded on the database exactly as reported in endoscopy or pathology reports, and usually precise values were provided. However, in rare cases the observations recorded in the endoscopy report about size and location could be vague; for example size could be merely described as tiny or < 10 mm, and location could be described using a range of values; this was particularly problematic when there were multiple lesions seen at an examination. These vague descriptions of size, and ranges of values for size and location, were recorded in specific fields on the database. Rules were defined to derive a summary value for size and location of each individual polyp at an examination by combining all the available information. These rules are described in greater detail in this section and in Appendices 7 and 8.
Summary values were determined for polyp size, histological features, location and shape, and were derived separately for each visit. Summary values for polyp characteristics were derived using hierarchies of rules. In general, three stages were involved:
-
data cleaning to identify, review and resolve any errors in the polyp data
-
assessment of polyp characteristics at a single examination
-
assessment of polyp characteristics across examinations within a visit, if a polyp had multiple sightings.
Size
Polyp size information was recorded in several fields on the database, as shown in Table 4.
| Size field | Variable namea | Description | Derived valuesa |
|---|---|---|---|
| Endoscopy size | ENDO_SIZE | Field used to record the exact size of a polyp (in millimetres), when described precisely in the endoscopy report | Derived endoscopy sizeENDO_SIZE, ENDO_SIZE_MIN and ENDO_SIZE_MAX were combined using a hierarchy of rules to give a single derived endoscopy size for each sighting of a polyp |
| Minimum endoscopy size | ENDO_SIZE_MIN | Field used to record the minimum size of a polyp when a size range was described in the endoscopy report (e.g. 8–10 mm) | |
| Maximum endoscopy size | ENDO_SIZE_MAX | Field used to record the maximum size of a polyp when a size range was described in the endoscopy report | |
| Endoscopy size descriptor | ENDO_SIZE_OTHER | Field used to record the size of a polyp when it was described in vague terms in the endoscopy report (e.g. tiny, > 10 mm, < 5 mm, etc.) | Derived endoscopy size descriptorA numeric value was derived from a description (see Table 5) |
| Pathology size | PATH_SIZE | Field used to record the exact size of a polyp or biopsy specimen as described by the pathologist in the pathology report | Derived pathology sizeThe precise size given in most pathology reports is used |
Exact sizes (endoscopy or pathology size) were available for 65% of polyps (of all types, including adenomas) but 8% of polyps had a numeric size with a minor discrepancy, or a size range (minimum and maximum endoscopy size); both of these issues were resolved to give an accurate size. In only 6% of polyps was the size estimated based on a qualitative size description (endoscopy size descriptor). This does not account for other sightings of the same polyp, so the proportion without a precise, numerical size is likely to have been even smaller than this. These proportions relate to all patients for whom we had data, rather than just IR patients, as adenoma risk groups could not be discerned until summary values for size had been defined.
The ‘endoscopy size descriptor field’ was used in cases for which a ‘vague’, qualitative or approximate size description was given in the endoscopy report. A numerical value was derived for each size description by analysing reports in which both qualitative size descriptions and a precise numerical size were given. The median and interquartile range (IQR) were calculated for each numeric size field and cross-tabulated against associated categories of the endoscopy size descriptor field, as shown in Table 5.
| Endoscopy size descriptor category | Endoscopy size (mm) | Derived value size | Rationale for derived value size | |
|---|---|---|---|---|
| Median (IQR) | n | |||
| Tiny | 3 (2–3) | 660 | 3 mm | Used the median |
| Small | 3 (3–5) | 1574 | 5 mm | Used the larger value of 5 mm to draw a distinction between ‘Small’ and ‘Tiny’ |
| < 5 mm | 3 (2–3) | 35 | 3 mm | Used the median |
| 5–9 mm | n/a | 0 | 7 mm | No examples so took the halfway point |
| < 10 | 8 (8–8) | 3 | 8 mm | Used the median of available examples |
| ≥ 10 mm | 15 (13–15) | 79 | 15 mm | Used the median |
| Large | 20 (12–30) | 2701 | 20 | Used the median |
The endoscopy size and minimum and maximum endoscopy sizes were combined using a hierarchy of rules to give a derived endoscopy size for each sighting of a polyp (see Appendices 7 and 8 for details). The numerical values assigned to the endoscopy size descriptor field were used as the derived endoscopy size descriptor. Most pathology reports provided a precise size, which was coded for each individual polyp biopsied or resected at an examination – this was taken as the derived pathology size. Derived endoscopy and pathology sizes were automatically assigned when possible. Study researchers manually reviewed polyps for which derived endoscopy and pathology sizes could not be assigned automatically.
Finally, the three derived polyp sizes – derived endoscopy size, derived endoscopy size descriptor and derived pathology size – were compared across examinations within a visit, and the largest of each derived size was identified. The largest derived sizes were compared and the largest of these was used as the summary polyp size. The only time the largest size was not used was if it was the derived endoscopy size descriptor, and the derived endoscopy and derived pathology size was also available, which were considered more accurate. Full details of these methods are provided in Appendices 7 and 8.
Histopathology
In all patients for whom data were collected, including low-, intermediate- and high-risk patients, histological data were available for 66% of polyps (all types of polyps, including adenomas). In some cases (34%), data on histological features of a polyp were missing because no biopsies were taken, a pathology report could not be identified at the hospital, or the polyp in question was not retrieved at endoscopy or not mentioned in the pathology report. This value does not account for other sightings of polyps – they may have been removed at another examination – and we would expect some of these polyps to have been insignificant and therefore not excised. Rules were applied to the data to resolve such issues and derive polyp histology, where possible. First, if a polyp had any degree of villousness or dysplasia coded then the polyp was assumed to be an adenoma. If the polyp was ≥ 10 mm in size and no histology was recorded at any sighting of the polyp, the histology was set to ‘specimen not seen’ or ‘not able to diagnose’. If the polyp without histology was ≥ 10 mm in size and the patient had at least one adenoma recorded then the polyp was then assumed to be an adenoma.
A polyp seen at more than one examination may not have been diagnosed as an adenoma until a later sighting. As baseline started from first sighting of an adenoma, it was necessary to apply adenomatous histology back to earlier sightings, provided that the sighting without histology occurred no more than 3 years prior to the adenoma diagnosis. Adenomatous histology was applied to earlier sightings of a polyp only if the histology for the earlier sighting was unknown or recorded as hyperplastic, granulation tissue, previous polypectomy site, normal mucosa, not possible to diagnose, or specimen not seen; this ensured that histology of greater severity than an adenoma was not overwritten.
A single polyp seen at more than one examination could have different histological features recorded at each sighting. To resolve these inconsistencies, histological types encountered in the study were split into two groups: group 1 consisted of the outcomes of interest (CRCs and adenomas), along with all histological types that could potentially occur in such lesions over time (Table 6); group 2 consisted of all other histological types (data not presented). Group 1 histological types were listed from most to least severe within the following groups – CRC, possible CRC, benign lesion, no polyp features/not possible to diagnose – as shown in Table 6. When there was no clear-cut order in terms of malignant potential, histological types were arbitrarily ordered by the specificity of the description. Initially, polyps with histology from groups 1 and 2 recorded at different sightings were reviewed to check whether or not there was a reporting or coding error. Then, for remaining polyps with histology from both groups, group 1 histology took precedence for the purpose of this study, except when the group 1 histology was uncertain or unimportant (e.g. ‘normal mucosa’, ‘granulation tissue’, ‘previous polypectomy site’, ‘not possible to diagnose’ or ‘specimen not seen’), in which case the group 2 histology took precedence.
| Category | POLYP_TYPE |
|---|---|
| CRC | Cancer with remnant of sessile serrated lesion |
| Cancer with remnant of mixed/serrated adenoma | |
| Cancer with remnant of mixed adenoma | |
| Cancer with remnant of serrated adenoma | |
| Cancer with remnant of adenoma | |
| Cancer | |
| Possible CRC | Cancer or adenoma with HGD? (cancer in dispute) |
| Cancer with unknown primary | |
| Possible cancer (suspicious features but may be non-adenomatous) | |
| Benign lesion | Sessile serrated lesion |
| Mixed polyp (adenomatous and metaplastic features) | |
| Serrated adenoma | |
| Adenoma/assumed adenoma | |
| Unicryptal adenoma | |
| Metaplastic/hyperplastic polyp | |
| No polyp features/not possible to diagnose | Previous polypectomy site |
| Granulation tissue | |
| Normal mucosa | |
| Not possible to diagnose | |
| Specimen not seen |
The histology of adenomas was further defined using their greatest degree of villousness and worst dysplasia recorded within a visit.
Polyp location
The following rules were used to define a value for polyp location across all visits:
-
Where the segment was recorded at a surgical procedure, this took precedence over any segment recorded at other types of procedure.
-
If there was no surgical procedure, the most frequently described segment was taken.
-
If no segment was mentioned more frequently than another, the most distal segment was taken.
-
In cases where a segment range was given, the following rules were applied:
-
If only one range was described, the most proximal and distal segments were recorded on the database as the true range (proximal defined as descending colon to terminal ileum; distal defined as anus to sigmoid colon).
-
If several site ranges were described, the smallest segment range was used as the true range, provided that the difference in the position of the most proximal and distal segments in the range was ≤ 2. Table 7 was used to allocate a position to each segment in order to calculate this difference. If the segment range differed by more than two, the records were manually reviewed to reach a decision.
-
| Position | Segment |
|---|---|
| 1 | Ileum |
| 2 | Caecum |
| 3 | Ascending colon |
| 4 | Hepatic flexure |
| 5 | Transverse colon |
| 6 | Splenic flexure |
| 7 | Descending colon |
| 8 | Sigmoid colon |
| 9 | Rectosigmoid |
| 10 | Rectum |
| 11 | Anus |
Polyp shape
It was unclear if the most appropriate method for assigning the true shape of a polyp would be to use an order of precedence, as with other polyp characteristics, or if the first description might be the most accurate, as the shape of a polyp may have been altered once it was biopsied/resected. Shape values included flat, sessile, pedunculated or subpedunculated. It was decided that the first recorded shape of a lesion would be used.
Procedure information
Procedure date and order
In most cases, the procedure date was simply the date of the endoscopy. However, if the endoscopy report was not available, the pathology report was used to derive the examination date. Up to three dates might be specified on the pathology report. The examination date was derived using the following order of precedence:
-
date that the biopsy specimen was taken
-
date that the biopsy specimen was received at the laboratory
-
date of the pathologist’s report.
In the rare cases when it was not possible to derive a procedure date, the patient was excluded from the study. Where procedures occurred on the same day, the reasons were specified and the examinations were numbered to assign an order, otherwise it was specified why it was not possible to do so.
Procedure type
The master database contained two types of procedural report: endoscopy reports extracted directly from endoscopy databases and pathology-based procedure reports generated using clinical and procedural information from the pathology report. The latter were created by study researchers in cases when no endoscopy report was available.
In cases where the procedure type was not reported or not specified (e.g. ‘endoscopy’), procedure type was derived by applying a hierarchy of rules based on available information. For example, when a procedure type was unknown, yet there was evidence that the transverse colon or beyond was reached, the procedure type was probably a colonoscopy. When information such as bowel preparation and depth of insertion was given, the procedure was probably a colonoscopy or flexible sigmoidoscopy (FS). Likewise, if a lesion with a size of ≥ 10 mm was removed, or multiple adenomas were removed, the procedure was also probably a colonoscopy or FS. Full details of rules for deriving procedure type are given in Appendix 7.
For some patients, it remained uncertain whether or not they had a baseline colonoscopy even after procedure type was derived (i.e. derived procedure type was ‘colonoscopy or FS’ at baseline). As patients had to have a baseline colonoscopy for inclusion in the study (a baseline colonoscopy was necessary to accurately stratify patients into risk groups), procedures that were derived as ‘colonoscopy or FS’ were reclassified as colonoscopies, based on adenoma risk group and type/timing of follow-up examinations. For example, patients with a derived procedure type of ‘colonoscopy or FS’ at baseline who were classified as IR or high risk (see Defining adenoma surveillance risk groups, below) were assumed to have had a colonoscopy at some point during baseline, and so the derived procedure was relabelled as such (see Appendix 7). Unlike baseline examinations, no derived procedure types were relabelled at follow-up examinations. Instead, for each follow-up visit, the most complete whole-colon examination available was defined using a hierarchy from ‘complete colonoscopy’ to ‘unknown procedure type’.
For patients without a baseline colonoscopy who had a colonoscopy at follow-up visit 1 (FUV1), the baseline visit was shifted so that FUV1 became the baseline visit and the original baseline visit became a ‘prior’ visit. To ensure that risk was not underestimated as a result of shifting baseline in this way, any adenomas found at prior examinations (original baseline) were used to determine risk as well as those found during the baseline visit.
Colonoscopy quality
Where there were several colonoscopies within a visit, the most complete examination and the best bowel preparation achieved at any colonoscopy was taken as the highest quality examination achieved at that visit. The quality and completeness of a colonoscopy was assessed, based on the segment of the colon reached, the most proximal polyp site, the quality of bowel cleansing prior to the examination and whether or not the examination was marked as incomplete.
The quality of the colonoscopy was important for defining visits (see Defining baseline and surveillance visits, below) as well as being a potential risk factor in the final data analysis.
Defining baseline and surveillance visits
For the purposes of this study, a ‘visit’ (baseline or follow-up) was defined as one or more examinations, performed in close succession, with the aim of completing a full examination of the colon and removing all detected lesions. This is based on the assumption that a single endoscopy is not always sufficient to visualise the entire colon (e.g. owing to poor bowel preparation) or to remove large, numerous or residual lesions.
Lesions found during the baseline visit were used to classify baseline risk of CRC and to stratify patients into adenoma surveillance risk groups (see Defining adenoma surveillance risk groups, below). In addition, certain diagnoses during baseline rendered the patient ineligible for the study, including CRC (see Patient eligibility, above). Follow-up visits were then defined around the baseline visit, with the length of time between visits being used to determine surveillance intervals.
Baseline visit
The baseline visit included the examination with the first adenoma sighting and any completion examinations that occurred within the subsequent 11 months. For high-risk adenomas, surveillance examinations are scheduled 1 year after the initial examination, in accordance with UK surveillance guidelines, so 11 months was chosen as the most appropriate time frame to capture any completion examinations into the baseline visit, without including high-risk follow-up examinations. After including all examinations within 11 months, a small proportion of patients had additional procedures that occurred shortly after the ‘latest’ baseline examination and thus needed to be included into the baseline visit. Baseline was therefore extended a second time, to include any examinations within 6 months of the latest baseline examination. Finally, in a handful of special scenarios, a third repeated extension was performed to capture examinations within 6 or 9 months of the latest baseline examination (6 months if the latest baseline examination was a colonoscopy and 9 months if it was a sigmoidoscopy). These rare cases included scenarios for which:
-
the latest baseline examination was incomplete
-
quality of bowel preparation at the latest baseline examination was poor
-
a large polyp (≥ 15 mm) was seen at the latest baseline examination
-
the same polyp was seen at the latest baseline examination and the next examination, which occurred within 6/9 months
-
the latest baseline examination was followed directly by a surgical examination.
After the extension of baseline, the length of baseline was assessed; only 2% of patients with IR adenomas had a baseline that exceeded 11 months in length.
Surveillance visits
A surveillance or follow-up visit was defined using similar rules for baseline. A follow-up visit comprised the first examination after baseline (or after a follow-up visit) and any further examinations within the subsequent 11 months. As with the baseline visit, the final examination in a follow-up visit was identified, and the follow-up visit was extended as necessary, using the same criteria as for the extension of baseline. This procedure was repeated until all examinations had been grouped into a follow-up visit. Visits following a diagnosis of CRC, volvulus or resection/anastomosis were censored, as patient follow-up would be affected by such diagnoses.
Surveillance interval
Surveillance intervals were timed from the last most complete examination of one visit to the first examination of the next visit, as defined in the NHS BCSP. 40
Defining adenoma surveillance risk groups
Once the baseline visit and true polyp values were defined, patients could then be stratified into adenoma surveillance risk groups. The risk groups were defined using the criteria for stratification of patients as low risk, intermediate risk or high risk, as described in the current UK Guideline (adopted by NICE). These definitions were applied based on all adenomas found within the baseline visit, and are given below. In addition, patients who could not be classified into a specific adenoma risk group were grouped into broader categories.
-
Low risk One or two small (< 10 mm) adenomas [no large (≥ 10 mm) adenomas or adenomas of unspecified size].
-
Intermediate risk Three or four small adenomas (no large adenomas or adenomas of unspecified size) or one or two adenomas, of which at least one is large.
-
High risk Five or more adenomas (any or unknown size) or three or more adenomas, of which at least one is large.
-
Low/intermediate risk One adenoma of unknown size or two adenomas, of which none is large but one or more has an unknown size.
-
Intermediate/high risk Three or four adenomas, of which none is large but one or more has an unknown size.
Patient follow-up
We matched our study patient data with records from external repositories of national patient data: HSCIC, NHSCR Scotland and NSS in order to achieve the following:
-
List clean patient records obtained from hospitals To correct patient information that had been entered incorrectly into hospital databases.
-
Identify duplicate records across hospitals Patients who had procedures at more than one hospital, and would have been allocated a different study number for each hospital where their data were collected.
-
Identify duplicate records in the same hospital Some patients were seen at the same hospital but, as a result of variations in patient identifiers, they had not been identified as the same patient.
-
Obtain cancers and deaths data Necessary to determine the incidence of CRC and the mortality status of patients in the cohort. The HSCIC provided the cancers and mortality data for patients residing in England or patients who resided in Scotland and had moved to England; the NHSCR provided the cancers and mortality data on patients who resided in England but had now moved to Scotland (the NHSCR and HSCIC work in partnership); and the NSS provided us cancers and mortality data on patients who resided in Scotland.
List cleaning
The patient-linking-file that was left at each hospital by the study programmer inevitably contained some patient identifiers that had been entered incorrectly into the hospital databases. It was therefore necessary to use the HSCIC/NHSCR’s list cleaning and tracing service and NSS’s linking service to validate and link the patient records to their database.
When the data sets were sent to HSCIC/NHSCR, no match was found for 5% of the patients. This revealed a limitation of having missing information, in that there was a higher chance of the supplied information matching more than one patient on the HSCIC database, resulting in rejection of a match. The study programmer worked closely with HSCIC to create bespoke matching algorithms that accounted for minor differences in dates of birth, names and NHS numbers in order to get the correct match, and, in some cases, additional data were collected from hospital to resolve the differences. Ultimately, matches were found for 99.65% of all 253,798 patients by the HSCIC/NHSCR and NSS.
Duplicate patient records
The national data repositories provided a list of duplicate patients found across the cohort (including patients from England/Wales and Scotland). When all of the duplicates had been identified, each set of duplicate records on our master database were merged into one record and an audit log was kept to show which records had been merged.
Cancer matching
Cancer and mortality (deaths) data for patients in our cohort were obtained from national patient data repositories (HSCIC, NHSCR Scotland and NSS). These data had to be added to the master database, taking into account the patient and cancer data already present, to ensure that there was no duplicated or missing data. This process was termed ‘cancer matching’.
To identify duplicate records of cancers, a program was written to identify CRCs in the national repositories’ data set and link them with the procedures and individual polyp records (including cancers) on the master database. Data quality checks were carried out, and samples of records that had been linked automatically were manually reviewed.
Cancers were linked to individual polyp records based on a hierarchy involving the cancer diagnosis date from the external source, the date the polyp or cancer was identified in the hospital data, the location of lesions, the polyp number, the time between the date of cancer diagnosis, and the date of the procedure during which the lesion was identified. For cancers reported in hospital pathology reports but not in national repositories, the hospital data were accepted as conclusive evidence of cancer, except when the histology was recorded by the study researcher as ‘cancer in dispute’ or ‘cancer query’.
The histology for cancers recorded on the study database as ‘in situ’ cancers and ‘cancers in dispute’ was compared with data from the national registries and automatically reclassified if necessary, using a hierarchy of rules. For example, polyps mapped to an in situ cancer from external sources were reclassified as ‘assume adenoma’ with high-grade dysplasia (HGD) if they were not already coded as such. Similarly, polyps in the database not mapped to a cancer from external sources, but with a histology recorded as ‘cancer in dispute’, were reclassified as ‘assume adenoma’ with HGD. A full list of the rules is given in Appendix 7 (see rule 13).
Values for the cancer diagnosis date, and site of the cancer, were assigned by comparing the data recorded on the study database with data from the national registries and applying a hierarchy of rules to arrive at the true value. For example, if the external cancer date preceded the mapped endoscopy date then the external date was used. Likewise for site, if no site was given in the mapped endoscopy data (or the site was non-specific), the site used in the external cancer data was used. A full list of the rules is given in Appendix 7 (see rule 11).
Variables
Outcomes
The primary outcome measures were adenoma, AA and CRC detected at the first and second follow-up visits, and CRC incidence after baseline, and after first follow-up. Previously seen lesions were excluded from some analyses, as they were thought to be a proxy measure for patients undergoing polypectomy site surveillance, and confounded the analysis. Outcomes that had not been seen at a previous visit were termed ‘new’ outcomes (see Chapter 3, New and previously seen lesions at first follow-up).
Advanced adenoma was defined as an adenoma of ≥ 10 mm, or with villous or tubulovillous histology, or HGD. CRCs were ascertained using pathological data recorded on the study database and International Statistical Classification of Diseases and Related Health Problems (ICD) codes in data from national repositories. To determine which cancers from the national repositories were outcomes of interest, they were grouped according to site and morphology (details are given in Appendix 7, rules 11 and 13). Only cancers from national repositories that fell into the site groups ‘malignant lesions of the colon/rectum’ and certain ‘in situ neoplasm – colon’ were selected for the study. Specifically, outcomes included adenocarcinomas of the colorectum and carcinomas with unspecified morphology located between the rectum and caecum that were assumed to be adenocarcinomas. Cancers with unspecified morphology located at sites related to the anus were likely to be squamous cell carcinomas and were therefore not classed as outcomes unless they were linked to a rectal lesion, in which case they were assumed to be adenocarcinomas. CRCs reported as a cause of death in national repositories were classed as outcomes if the patient did not have cancer recorded in the cancer registry or hospital data.
Colorectal cancer sites were defined by the World Health Organization (WHO) ICD versions ICD-8, ICD-9 and ICD-10, and included site codes C18–C20 (www.who.int/classifications/icd/en/). Morphology of colorectal neoplasia was coded with the Manual of Tumor Nomenclature and Coding codes,41 the WHO International Classification of Diseases for Oncology (ICD-O) ICD-O-1 codes42 and ICD-O-2 codes. 43
Exposures
The main exposures of interest were the length of the surveillance interval between baseline and first follow-up, and between the first and second follow-ups. The surveillance interval was defined as the period of time from the last most complete colonic examination at one visit to the first examination of the next visit (as in the NHS BCSP). 40 In order to define interval, a patient’s examinations were split into baseline and follow-up visits (see Defining baseline and surveillance visits, above). Interval length was then calculated and converted into a categorical variable with seven groups: > 18 months, 2, 3, 4, 5 and 6 years (all ± 6 months) and ≥ 6.5 years. Patients with the shortest interval were used as a reference group to compare with those who were exposed to a longer interval.
The other exposure of interest was the effect of adenoma surveillance on risk of CRC after baseline. Patients who attended at least one follow-up visit at which cancer was not diagnosed were considered to be exposed to surveillance.
Risk factors and potential confounders
Patient, procedural and polyp characteristics at baseline and follow-up were assessed as a priori risk factors and confounders; these included age and gender, examination quality (based on completeness of examination, quality of bowel preparation and difficulties encountered), calendar year of examination and hospital attended, and the number, size, location and histology of polyps and adenomas, villousness and dysplasia. All potential risk factors and confounders examined are listed and defined in Table 8.
| Factor | Definition |
|---|---|
| Number of adenomas | Total number of adenomas seen during a visit |
| Size of adenoma | Size of the largest adenoma seen during visit |
| Villousness of adenoma | Worst degree of villousness of an adenoma seen during visit |
| Dysplasia of adenoma | Worst degree of dysplasia of an adenoma seen during visit |
| Distal or proximal adenomas | Detection of distal or proximal adenoma(s) at a visit. Proximal defined as descending colon to terminal ileum; distal defined as anus to sigmoid colon |
| Distal or proximal polyps | Detection of distal or proximal polyp(s) of any type, including adenomas, at a visit. Proximal defined as descending colon to terminal ileum; distal defined as anus to sigmoid colon |
| Age, years | Age of patient at time of visit |
| Gender | Gender of patient |
| Length of visit | Total length of a visit (in days, months or years) |
| Number of examinations | Total number of examinations that make up a visit |
| Most complete examination | Most complete procedure during visit (at baseline this was based on colonoscopy). Completeness was determined from segment reached by scope or location of polyp(s). A complete colonoscopy was one during which the scope reached, or polyps were found in, the caecum or beyond. If no colonoscopy was performed during the visit then the next most complete procedure type was used |
| Best bowel preparation at colonoscopy | Best bowel preparation at a colonoscopy during a visit. If there was no colonoscopy then this was classified as ‘no known colonoscopy’ |
| Difficult examination | Composite variable of examination quality. Ascertained from endoscopy report information. Coded ‘yes’ if there was poor bowel preparation, the maximum segment was not reached (i.e. caecum for colonoscopy, sigmoid colon for sigmoidoscopy) and another indicator of poor examination quality was provided, such as patient discomfort, looping, technical difficulty, equipment failure, etc. |
| Number of sightings of a unique adenoma | The greatest number of times an adenoma was seen during a visit |
| Number of hyperplastic polyps | Total number of hyperplastic polyps in a visit |
| Number of large hyperplastic polyps | Total number of large (≥ 10 mm) hyperplastic polyps in a visit |
| Number of polyps with unknown histology | Total number of polyps for which there is no histology available |
| Calendar year | Year during which the visit took place |
| Hospital | Hospital at which the visit took place |
| Family history of cancer/CRC reported | Patient has a family history of cancer or CRC indicated at an examination during or prior to a visit |
Grouping of variables
All of the aforementioned risk factors were considered separately for the baseline visit and FUV1. In some instances it was necessary to add an additional level to a variable; for example, at FUV1 some patients did not have any adenomas or a colonoscopy, whereas at baseline every individual had an adenoma and a colonoscopy. The quantitative variables interval length and calendar year were grouped into categorical variables for some analyses and used in their continuous form in other circumstances. The remaining quantitative variables (visit length, age, adenoma size, number of examinations, number of sightings of a unique adenoma and numbers of specific polyp types) were grouped into categorical variables. Standard categorisations were created for all categorical variables and these were used in the presentation of univariable results. When appropriate, the process of selecting risk factors for inclusion in multivariable models involved the investigation of the categorisation of some variables, and the final categorisation was selected by evaluating the difference in effect between levels of the variable. When data were missing for a particular variable, an ‘unknown’ category was created in order to avoid losing patients from the models, particularly those models adjusting for several confounders. Models were tested with, and without including, the ‘unknown’ category to assess the difference it made.
Study size
Sample size requirements were based on the comparison of the rates of detection of AA or CRC at first follow-up at two different intervals, using heterogeneity in practice with respect to follow-up intervals. It was deemed plausible that 5% of subjects would have an intermediate- or high-risk lesion at first follow-up at 4–6 years and 3% at 2–4 years. 44,45 For 90% power to detect this rate in the sample at the 5% significance level in a two-sided test, it was estimated that 4400 subjects with at least one follow-up examination were required. For second or subsequent follow-up, the more relaxed criterion to estimate the detection rate within 1% in either direction was applied. It was anticipated that 3% of subjects would have intermediate- or high-risk lesions at second or subsequent follow-up. This required 1200 subjects with at least two follow-up endoscopies.
Consideration was given to the fact that the sample size was required to provide relatively low coefficients of variation of the test sensitivity (S) and λ2, the rate of progression to clinical CRC, so as to enable the comparison of different intervals between follow-up with respect to rates of cancers that accrued. In order to use these with confidence to predict effects of different follow-up policies, a high degree of precision in estimation of S and λ2 was required. It was therefore stipulated that both have coefficients of variation of no more than 30% [i.e. the standard error (SE) of each estimate has magnitude no larger than 30% of the value of the estimate]. Closed-form estimation was not possible for these quantities and it was difficult to predict the variability of the estimates. Work by Chen et al. 46 and Wong et al. 47 suggests that, with around 30 events, coefficients of variation of ≤ 30% may be achieved if the rate of progression is small (≤ 0.2 per annum). Stratification or the introduction of covariates would reduce the precision and therefore the aim was to recruit a cohort with a total of 60 CRCs.
Stryker et al. 48 found rates of progression in untreated adenomas suggestive of a λ2 of around 0.01 for progression to CRC. Atkin et al. 3 studied a wide case mix of treated polyps at entry (corresponding to the situation in this project), and suggested a rate of around 2 per 1000 per year after colonoscopy overall and around 4.5 per 1000 per year for the high-risk subgroup. Thus, in the literature at the time of the call for proposal, the rate ranged from 2 to 10 per 1000 per year.
It was assumed that the underlying risk of CRC in the cohort would be considerably higher than the population risk, but that the relative risk might be brought down by the protection of endoscopic examination to between one and two times the population risk in males aged ≥ 50 years. This meant that there would be between 2.5 and 5 end points per 1000 per year. In total, therefore, between 12,000 and 24,000 person-years (pys) of follow-up after endoscopy episodes would be required. Assuming an average of 4 years’ observation, this required recruiting cohorts to a total of 6000 subjects. A failsafe strategy to recruit 10,000 was proposed.
Statistical methods
The statistical analysis strategy was split into three main stages: analysis of (1) first follow-up findings in relation to baseline findings; (2) second follow-up findings in relation to baseline and first follow-up findings; and (3) incidence of CRC after baseline in relation to risk factors and exposure to surveillance. The analysis of findings at follow-up aimed to ascertain whether or not there was substantial heterogeneity of results at subsequent examination, in terms of detection rates of AA or CRC, according to risk factors and confounders, and interval to follow-up colonoscopy. The analysis of the incidence of CRCs after baseline aimed to determine the effect of surveillance on long-term CRC risk and to identify independent risk factors for incident cancer. All tests were two-tailed with significance assigned at 5%. In all instances, adjusted effect estimates from multivariable analyses should be considered superior to unadjusted effect estimates reported in univariable analyses. Analyses were performed with Stata/IC version 13.1 (StataCorp LP, College Station, TX, USA).
The distribution of baseline characteristics among patients with and without follow-up visits was compared using chi-squared tests.
Follow-up visit 1 findings in relation to baseline findings
Initially, findings at FUV1 were investigated, considering any adenomas, AAs or CRCs, with a focus on AA and CRC outcomes. The relationship between baseline risk factors and findings at FUV1 was modelled using univariable logistic regression to estimate unadjusted odds ratios (OR). The association between interval length and baseline risk factors was evaluated using chi-squared tests. The relationship between interval from baseline to FUV1 and outcomes at FUV1 was explored both with and without adjustment for baseline risk factors using logistic regression models. Many risk factors for AA and CRC that were potential confounders were known already, based on the substantial body of evidence in the literature. 15,22,23 Owing to the large number of potential confounders, backwards stepwise logistic regression models and likelihood ratio tests (LRTs) were used to identify important confounders to be included in the models, with the significance level for inclusion set at 5%. Interval, our main variable of interest, was constrained to be included in all models. Models were also constructed to consider only ‘new’ outcomes, meaning that those lesions had not been previously seen before FUV1. Separate models for all outcomes were constructed for interval considered as a continuous variable and as a categorical variable. Effect modification of the association between interval and new findings at FUV1 by age and gender were investigated by fitting models with interaction parameters and performing a test for interaction; effect modification was investigated for only interval as a continuous variable, as this enabled examination of potential trends, and it was unlikely to be of any practical use to know whether or not the effect of interval was significantly different in a particular age group if there was no trend in the effect.
Risk factors associated with an interval of < 2 years were identified using logistic regression, and a backwards stepwise logistic regression model was used to identify independent predictors of an interval of < 2 years. All p-values from models were calculated using LRTs.
Follow-up visit 2 findings in relation to baseline and follow-up visit 1 findings
A similar approach to the analysis of outcomes at FUV1 was adopted for outcomes at follow-up visit 2 (FUV2). The relationships between FUV1 risk factors and AA and CRC at FUV2, and between baseline risk factors and AA and CRC at FUV2, were modelled using univariable logistic regression for each confounder separately. Owing to the small number of CRCs detected at FUV2, AA and CRC were grouped together and the outcome of interest was advanced neoplasia (AN). The relationship between interval from FUV1 to FUV2 and detection of AN at FUV2 was explored both with and without adjustment for FUV1 risk factors, baseline risk factors (including interval from baseline to FUV1) and cumulative baseline and FUV1 risk factors using logistic regression models. Backwards stepwise logistic regression models and LRTs were used to identify important confounders to be included in the models, with the significance level for inclusion set at 5%. The chosen confounders from each of these models were then added to a stepwise model to identify the most important factors. To compare the model fit of each of the constructed logistic regression models, pseudo R-squared values and the Akaike information criterion (AIC) were calculated. As before, our main variable of interest (interval to FUV2) was constrained to be included in all models. The complete model selection process was performed separately for interval considered as a continuous variable and as a categorical variable. All models considered only ‘new’ outcomes, that is lesions that had not been previously seen before FUV2. Effect modification of the association between continuous interval and new AN at FUV2 by age and gender was investigated by fitting models with interaction parameters and performing a test for interaction.
Colorectal cancer incidence after baseline
In the analysis of CRC incidence after baseline, for patients matched to national sources the cut-off for follow-up was either 31 December 2011 or 30 June 2012 (depending on the data source), and for unmatched patients it was the date of the patient’s last recorded procedure. All time-to-event data were censored at first CRC diagnosis, death, emigration or end of follow-up.
For the analysis of incidence following baseline, time at risk started from the latest most complete colonoscopy in baseline, and for the analysis of incidence following FUV1, time at risk started on the date of the first procedure in FUV1. If CRC was diagnosed at a follow-up visit, the follow-up visit was not included as a visit, as it did not offer any protection against CRC. Incident CRC outcomes included ‘new’ CRCs only, that is cancers arising in lesions that had not been seen at baseline.
‘One minus the Kaplan–Meier estimator of the survival function’ was used to illustrate the time to cancer diagnosis and to estimate the cumulative risk of cancer with 95% confidence intervals (CIs) at 3, 5 and 10 years. The effects of surveillance and patient, procedural and polyp characteristics at baseline and follow-up on long-term CRC incidence were examined using Cox proportional hazards models. Univariable models were used to estimate unadjusted hazard ratios (HRs). Independent predictors of cancer incidence were identified in a multivariable Cox proportional hazards model, using backward stepwise selection with a p-value of < 0.05 in the LRT to determine the retention of variables in the final model. The number of follow-up visits was included as a time-varying covariate and, as our main variable of interest, was constrained to be included in all adjusted models. Effect modification of the association between surveillance and long-term CRC risk by age and gender was investigated by fitting models with interaction parameters and performing a test for interaction.
For the analysis of risk after baseline, only baseline risk factors were considered. For the analysis of risk following FUV1, separate models were built, considering baseline factors only, FUV1 factors only and cumulative factors only. The risk factors identified from these models were then considered together and a final model selected. All p-values from models were calculated using LRTs.
The incidence of CRC was compared with that expected in the general population. Observed pys at risk were calculated by gender and 5-year age group. Expected numbers of CRC cases were calculated by multiplying the observed gender- and age-specific number of pys by the gender- and age-specific incidence in the general population of England in 2007. The ratio of observed to expected cases was reported as a standardised incidence ratio (SIR), and 95% CIs were computed assuming an exact Poisson distribution.
Sensitivity analyses and internal validation
We conducted several sensitivity analyses to investigate whether or not our methods were robust and did not introduce bias into the results. To assess the methods we used to define baseline, follow-up visits and interval, we restricted analyses of the effect of interval on the finding of new AA and CRC at FUV1 and the effect of surveillance on long-term CRC incidence after baseline: first, to patients with only one colonoscopy in baseline, and, second, to patients who had at least one complete colonoscopy at FUV1. To examine whether or not the definition of AA that was used had an impact on results, we performed a sensitivity analysis for the outcome of new AA detection at FUV1 with a definition of AA that excluded villous or tubulovillous histology, that is with AA defined as an adenoma with HGD or with a size of ≥ 10 mm. Finally, we conducted sensitivity analyses of the effect of surveillance on long-term CRC incidence when the cohort was restricted to patients who had at least 5 years and at least 7 years of time in which hospital data had been collected; this was to examine the possible effect of misclassification of attendance at follow-up visits on the estimated effect of surveillance.
To assess the predictive ability of the multivariable logistic models for the outcomes of new findings at follow-up and the multivariable Cox regression models for the analysis of long-term cancer incidence, we performed internal validation using k-fold cross-validation with k = 10. 49 For each model, the linear predictors were used to construct receiver operating characteristic (ROC) curves for each of the 10 validation sets, and the area under the ROC curve and its SE were calculated for each; the inverse variance weighted mean ROC curve and area below the curve were then calculated from these.
Chapter 3 Hospital data set: results and discussion
Routine endoscopy and pathology records for 253,798 patients were assessed; 174,978 were excluded as no adenomas were reported: 45,716 were found to be ineligible as a result of colonic conditions, 2752 had no colonoscopy at baseline 92 had missing procedure dates and one had > 40 examinations, leaving 30,259 eligible patients with a histologically confirmed adenoma at baseline. A total of 11,995 (40%) eligible patients were classified as having IR adenomas, of whom 51 IR patients were lost to follow-up (could not be matched with national cancer registry data or embarked before the end of baseline), leaving 11,944 patients for the analysis (Figure 2).
FIGURE 2.
Participant flow diagram. Adapted from Atkin et al. 50 © The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Reprinted with permission from Elsevier (The Lancet,2017, http://dx.doi.org/10.1016/S1470-2045(17)30187-0).

Baseline characteristics of all intermediate-risk patients and those with follow-up
We examined demographic, procedural, adenoma and polyp characteristics at baseline, and date and place of the baseline visit, for all 11,944 eligible IR patients. A total of 4608 (39%) patients had at least one follow-up visit and all patients were followed using NHS data and national cancer registries and deaths data (see Long-term cancer risk, below). We first assessed whether or not patients with and without follow-up visits after baseline differed in order to determine the risk of selection bias in analysis of findings at, and subsequent to, follow-up visits.
Table 9 describes demographic and procedural characteristics at baseline, and date of the baseline visit. The median age of the whole cohort of IR patients was 66.7 years (IQR 58.4–74.0 years) and 55% were male. Those who attended a follow-up were younger, on average, than those who did not (mean = 63.3 vs. 67.3 years; p < 0.001), but there was no difference by gender (p = 0.852). Most baseline examinations occurred between 2000 and 2010 (84%), but patients attending follow-up had their baseline visits and, consequently, their adenomas diagnosed significantly earlier than those without follow-up. The absolute differences between hospitals in the proportion having follow-up were small, but because of large numbers the results were significant (results not presented: p < 0.001).
| Baseline factor | All IR patients (N = 11,944) | Patients with one or more follow-up visits (N = 4608) | Patients with no follow-up visits (N = 7336) | p-value (chi-squared test) | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||
| Age (years) | < 55 | 2122 | 17.77 | 1025 | 22.24 | 1097 | 14.95 | < 0.001 |
| ≥ 55 and < 60 | 1321 | 11.06 | 622 | 13.50 | 699 | 9.53 | ||
| ≥ 60 and < 65 | 1858 | 15.56 | 788 | 17.10 | 1070 | 14.59 | ||
| ≥ 65 and < 70 | 2171 | 18.18 | 813 | 17.64 | 1358 | 18.51 | ||
| ≥ 70 and < 75 | 1786 | 14.95 | 714 | 15.49 | 1072 | 14.61 | ||
| ≥ 75 and < 80 | 1416 | 11.86 | 413 | 8.96 | 1003 | 13.67 | ||
| ≥ 80 | 1270 | 10.63 | 233 | 5.06 | 1037 | 14.14 | ||
| Gender | Male | 6625 | 55.47 | 2551 | 55.36 | 4074 | 55.53 | 0.852 |
| Female | 5319 | 44.53 | 2057 | 44.64 | 3262 | 44.47 | ||
| Family history of cancer | No | 11,445 | 95.82 | 4368 | 94.79 | 7077 | 96.47 | < 0.001 |
| Yes | 499 | 4.18 | 240 | 5.21 | 259 | 3.53 | ||
| Year of baseline | 1985–9 | 112 | 0.94 | 98 | 2.13 | 14 | 0.19 | < 0.001 |
| 1990–4 | 327 | 2.74 | 241 | 5.23 | 86 | 1.17 | ||
| 1995–9 | 1430 | 11.97 | 1030 | 22.35 | 400 | 5.45 | ||
| 2000–4 | 4251 | 35.59 | 2317 | 50.28 | 1934 | 26.36 | ||
| 2005–10 | 5824 | 48.76 | 922 | 20.01 | 4902 | 66.82 | ||
| Length of baseline visit | 1 day | 6836 | 57.23 | 2496 | 54.17 | 4340 | 59.16 | < 0.001 |
| 2–30 days | 734 | 6.15 | 246 | 5.34 | 488 | 6.65 | ||
| 1–3 months | 1643 | 13.76 | 664 | 14.41 | 979 | 13.35 | ||
| 3–6 months | 1382 | 11.57 | 595 | 12.91 | 787 | 10.73 | ||
| 6–12 months | 1177 | 9.85 | 508 | 11.02 | 669 | 9.12 | ||
| 1–2 years | 160 | 1.34 | 91 | 1.97 | 69 | 0.94 | ||
| 2–3 years | 8 | 0.07 | 5 | 0.11 | 3 | 0.04 | ||
| 3–4 years | 4 | 0.03 | 3 | 0.07 | 1 | 0.01 | ||
| Number of examinations in baseline visit | 1 | 6826 | 57.15 | 2489 | 54.01 | 4337 | 59.12 | < 0.001 |
| 2 | 3788 | 31.71 | 1518 | 32.94 | 2270 | 30.94 | ||
| 3 | 908 | 7.60 | 392 | 8.51 | 516 | 7.03 | ||
| 4+ | 422 | 3.53 | 209 | 4.54 | 213 | 2.90 | ||
| Most complete colonoscopy | Complete | 9016 | 75.49 | 2973 | 64.52 | 6043 | 82.37 | < 0.001 |
| Incomplete | 1601 | 13.40 | 1157 | 25.11 | 444 | 6.05 | ||
| Unknown | 1327 | 11.11 | 478 | 10.37 | 849 | 11.57 | ||
| Best bowel preparation at colonoscopy | Excellent | 246 | 2.06 | 92 | 2.00 | 154 | 2.10 | < 0.001 |
| Good | 3710 | 31.06 | 1309 | 28.41 | 2401 | 32.73 | ||
| Satisfactory | 1922 | 16.09 | 487 | 10.57 | 1435 | 19.56 | ||
| Poor | 671 | 5.62 | 194 | 4.21 | 477 | 6.50 | ||
| Unknown | 5395 | 45.17 | 2526 | 54.82 | 2869 | 39.11 | ||
| Difficult examinationa | No | 11,229 | 94.01 | 4387 | 95.20 | 6842 | 93.27 | < 0.001 |
| Yes | 715 | 5.99 | 221 | 4.80 | 494 | 6.73 | ||
More than half of patients had a 1-day baseline visit consisting of a single colonoscopy; however, 39% of patients required two or three examinations during their baseline visit, and 12 patients had a long baseline visit of ≥ 2 years, mainly to treat a large, recurring lesion (which was distally located in most cases). Patients attending follow-up tended to have more baseline examinations and a longer duration of the baseline visit, although absolute differences were small.
All patients had at least one baseline colonoscopy and 75% were reported to have had a complete colonoscopy. In around 50% of patients, the ‘best’ bowel preparation at a baseline colonoscopy (some individuals had more than one) was deemed to be satisfactory or better, and was described as poor in only 6% of cases; however, the quality of the bowel preparation was unknown for 45% of patients. In addition, 6% of patients were reported to have had a difficult examination at baseline: a composite measure of examination quality that indicated an incomplete examination with poor preparation and additional difficulties encountered. Patients who attended follow-up were more likely to have missing data on bowel preparation (p < 0.001) and less likely to have had a complete colonoscopy (p < 0.001) at baseline than those without follow-up.
Table 10 describes the characteristics of the adenomas and polyps diagnosed during the baseline visit. Patients defined as IR according to the UK Adenoma Surveillance guideline16 could not have had more than four adenomas at baseline otherwise they would have been classified as high risk. Owing to the use of adenoma size and number in the definition of IR, these characteristics were associated, and most patients had one large adenoma as opposed to three or four small ones (66% vs. 9%). In 37% of patients, the largest baseline adenoma was between 10 and 14 mm, whereas 34% had an adenoma of > 20 mm in size. In addition, 17% of patients had a baseline adenoma with HGD, whereas 10% had an adenoma with villous histology; 80% had an adenoma in the distal colon or rectum and 31% had a proximal adenoma, whereas 14% had adenomas in both regions. In most patients, adenomas were seen just once during baseline (74%); however, in some patients, a single adenoma was seen multiple times. The distribution of adenoma characteristics was significantly different for those with and without follow-up, but the absolute differences were small.
| Baseline factor | All IR patients (N = 11,944) | Patients with one or more follow-up visits (N = 4608) | Patients with no follow-up visits (N = 7336) | p-value (chi-squared test) | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||
| Adenoma characteristics | ||||||||
| Number | 1 | 7842 | 65.66 | 3107 | 67.43 | 4735 | 64.54 | < 0.001 |
| 2 | 3073 | 25.73 | 1151 | 24.98 | 1,922 | 26.20 | ||
| 3 | 748 | 6.26 | 240 | 5.21 | 508 | 6.92 | ||
| 4 | 281 | 2.35 | 110 | 2.39 | 171 | 2.33 | ||
| Largest size (mm) | < 10 | 1029 | 8.62 | 350 | 7.60 | 679 | 9.26 | < 0.001 |
| 10–14 | 4417 | 36.98 | 1577 | 34.22 | 2840 | 38.71 | ||
| 15–19 | 2440 | 20.43 | 953 | 20.68 | 1487 | 20.27 | ||
| ≥ 20 | 4058 | 33.98 | 1728 | 37.50 | 2330 | 31.76 | ||
| Worst histology | Tubular | 4742 | 39.70 | 1723 | 37.39 | 3019 | 41.15 | < 0.001 |
| Tubulovillous | 5576 | 46.68 | 2136 | 46.35 | 3440 | 46.89 | ||
| Villous | 1142 | 9.56 | 459 | 9.96 | 683 | 9.31 | ||
| Unknown | 484 | 4.05 | 290 | 6.29 | 194 | 2.64 | ||
| Worst dysplasia | Low grade | 9476 | 79.34 | 3427 | 74.37 | 6049 | 82.46 | < 0.001 |
| High grade | 1994 | 16.69 | 850 | 18.45 | 1144 | 15.59 | ||
| Unknown | 474 | 3.97 | 331 | 7.18 | 143 | 1.95 | ||
| Location | Distal only | 7831 | 65.56 | 3070 | 66.62 | 4761 | 64.90 | < 0.001 |
| Proximal only | 1985 | 16.62 | 681 | 14.78 | 1304 | 17.78 | ||
| Distal and proximal | 1665 | 13.94 | 601 | 13.04 | 1064 | 14.50 | ||
| Unknown | 463 | 3.88 | 256 | 5.56 | 207 | 2.82 | ||
| Distal | No | 2448 | 20.50 | 937 | 20.33 | 1511 | 20.60 | 0.729 |
| Yes | 9496 | 79.50 | 3671 | 79.67 | 5825 | 79.40 | ||
| Proximal | No | 8294 | 69.44 | 3326 | 72.18 | 4968 | 67.72 | < 0.001 |
| Yes | 3650 | 30.56 | 1282 | 27.82 | 2368 | 32.28 | ||
| Number of sightings of a single adenoma | 1 | 8807 | 73.74 | 3311 | 71.85 | 5496 | 74.92 | < 0.001 |
| 2 | 2548 | 21.33 | 1005 | 21.81 | 1543 | 21.03 | ||
| 3 | 390 | 3.27 | 182 | 3.95 | 208 | 2.84 | ||
| 4 | 108 | 0.90 | 63 | 1.37 | 45 | 0.61 | ||
| 5+ | 91 | 0.76 | 47 | 1.02 | 44 | 0.60 | ||
| Polyp characteristics (all types) | ||||||||
| Number of hyperplastic polyps | 0 | 9874 | 82.67 | 3743 | 81.23 | 6131 | 83.57 | 0.005 |
| 1 | 1307 | 10.94 | 541 | 11.74 | 766 | 10.44 | ||
| 2 | 405 | 3.39 | 159 | 3.45 | 246 | 3.35 | ||
| 3 | 152 | 1.27 | 64 | 1.39 | 88 | 1.20 | ||
| 4 | 76 | 0.64 | 38 | 0.82 | 38 | 0.52 | ||
| 5+ | 130 | 1.09 | 63 | 1.37 | 67 | 0.91 | ||
| Number of large hyperplastic polyps | 0 | 11,761 | 98.47 | 4525 | 98.20 | 7236 | 98.64 | 0.232 |
| 1 | 168 | 1.41 | 75 | 1.63 | 93 | 1.27 | ||
| 2 | 10 | 0.08 | 6 | 0.13 | 4 | 0.05 | ||
| 3 | 3 | 0.03 | 1 | 0.02 | 2 | 0.03 | ||
| 4 | 1 | 0.01 | 1 | 0.02 | 0 | 0.00 | ||
| 5 | 1 | 0.01 | 0 | 0.00 | 1 | 0.01 | ||
| Number of polyps with unknown histology | 0 | 9322 | 78.05 | 3593 | 77.97 | 5729 | 78.09 | 0.004 |
| 1 | 1510 | 12.64 | 556 | 12.07 | 954 | 13.00 | ||
| 2 | 517 | 4.33 | 187 | 4.06 | 330 | 4.50 | ||
| 3 | 249 | 2.08 | 108 | 2.34 | 141 | 1.92 | ||
| 4 | 129 | 1.08 | 63 | 1.37 | 66 | 0.90 | ||
| 5+ | 217 | 1.82 | 101 | 2.19 | 116 | 1.58 | ||
| Distal polyp | No | 1980 | 16.58 | 739 | 16.04 | 1241 | 16.92 | 0.208 |
| Yes | 9964 | 83.42 | 3869 | 83.96 | 6095 | 83.08 | ||
| Proximal polyp | No | 7369 | 61.70 | 2940 | 63.80 | 4429 | 60.37 | < 0.001 |
| Yes | 4575 | 38.30 | 1668 | 36.20 | 2907 | 39.63 | ||
Baseline polyp characteristics (including number, location and type) were considered as potential risk factors for findings at follow-up. In addition to their IR adenoma(s), 17% had hyperplastic polyps and 2% had large (≥ 10 mm) hyperplastic polyps found at baseline. In total, 83% of patients had a distal polyp, 38% had a proximal polyp and 25% had polyps in both regions. Polyp characteristics in those with and without follow-up were generally similar; however, a greater proportion of patients without follow-up had proximal polyps (p < 0.001).
Hospitals data set: patients attending follow-up visits
Table 11 describes the amount of follow-up in the hospital cohort. A total of 4608 patients had at least one follow-up visit and 1635 had two. Only 555 patients had three or more follow-up visits, so analyses of findings at follow-up were restricted to the first and second follow-up visits in which there were sufficient numbers of outcomes (see Table 11).
| Number of follow-up visits | Number of patients | Cumulative number of patients who had at least 1, 2, 3 . . . x examinations | |
|---|---|---|---|
| n | % | ||
| 1 | 2973 | 64.52 | 4608 |
| 2 | 1080 | 23.44 | 1635 |
| 3 | 354 | 7.68 | 555 |
| 4 | 135 | 2.93 | 201 |
| 5 | 45 | 0.98 | 66 |
| 6 | 14 | 0.30 | 21 |
| 7 | 2 | 0.04 | 7 |
| 8 | 2 | 0.04 | 5 |
| 9 | 2 | 0.04 | 3 |
| 10 | 1 | 0.02 | 1 |
| Total | 4608 | 100.00 | 4608 |
Table 12 shows the intervals to visits in patients having follow-up. Almost 60% of patients returned for their FUV1 earlier than the 3-year interval currently recommended for people with IR adenomas. The interval between baseline and first follow-up was < 3 years in 59% of patients, 3–4 years in 31% of patients and ≥ 5 years in 10% of patients. With regard to the interval between the first and second follow-up visits, once again, most patients (47%) had an interval of < 3 years but a greater proportion (41%) of patients had an interval of 3–4 years. Excluding the outliers with six or more follow-ups, the proportion of patients with a short interval of < 18 months tended to decrease with increasing number of follow-up visits.
| Follow-up visit number | Number of patients with varying interval lengthsa | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsb | 3 yearsb | 4 yearsb | 5 yearsb | 6 yearsb | ≥ 6.5 years | ||
| 1 | 1760 (38.19) | 976 (21.18) | 1057 (22.94) | 355 (7.70) | 217 (4.71) | 123 (2.67) | 120 (2.60) | 4608 (100) |
| 2 | 397 (24.28) | 376 (23.00) | 518 (31.68) | 152 (9.30) | 131 (8.01) | 31 (1.90) | 30 (1.83) | 1635 (100) |
| 3 | 131 (23.60) | 110 (19.82) | 191 (34.41) | 51 (9.19) | 42 (7.57) | 17 (3.06) | 13 (2.34) | 555 (100) |
| 4 | 48 (23.88) | 45 (22.39) | 65 (32.34) | 22 (10.95) | 20 (9.95) | 1 (0.50) | 0 (0) | 201 (100) |
| 5 | 22 (33.33) | 12 (18.18) | 23 (34.85) | 4 (6.06) | 5 (7.58) | 0 (0) | 0 (0) | 66 (100) |
| 6 | 2 (9.52) | 7 (33.33) | 7 (33.33) | 3 (14.29) | 2 (9.52) | 0 (0) | 0 (0) | 21 (100) |
| 7 | 1 (14.29) | 3 (42.86) | 2 (28.57) | 1 (14.29) | 0 (0) | 0 (0) | 0 (0) | 7 (100) |
| 8 | 2 (40.00) | 1 (20.00) | 1 (20.00) | 0 (0) | 0 (0) | 0 (0) | 1 (20.00) | 5 (100) |
| 9 | 1 (33.33) | 1 (33.33) | 1 (33.33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (100) |
| 10 | 0 (0) | 1 (100.00) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| Total | 2364 (33.29) | 1532 (21.57) | 1865 (26.26) | 588 (8.28) | 417 (5.87) | 172 (2.42) | 164 (2.31) | 7102 (100) |
First follow-up visit
Examinations and findings
Table 13 shows the proportion of patients found to have adenomas (all types), AA and CRC at FUV1 according to interval from baseline. Overall, 1605 (35%) patients had adenomas, 723 (16%) had AA and 84 (2%) had CRC detected at FUV1. The proportion of patients with adenomas was relatively constant across different intervals and ranged from 34% to 40%, whereas the proportion of patients with AA showed more variation, ranging from 14% to 26%, and the proportion with CRC ranged from 0.5% to 5%. The proportion of patients with CRC detected at FUV1 tended to increase with increasing interval to FUV1.
| Interval baseline to first follow-up | IR patients | Findings at FUV1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Adenoma | AA | CRC | ||||||
| N a | % | n | % | n | % | n | % | |
| < 18 months | 1760 | 38.19 | 595 | 33.81 | 268 | 15.23 | 29 | 1.65 |
| 2 yearsb | 976 | 21.18 | 349 | 35.76 | 165 | 16.91 | 25 | 2.56 |
| 3 yearsb | 1057 | 22.94 | 360 | 34.06 | 151 | 14.29 | 6 | 0.57 |
| 4 yearsb | 355 | 7.70 | 123 | 34.65 | 56 | 15.77 | 9 | 2.54 |
| 5 yearsb | 217 | 4.71 | 85 | 39.17 | 34 | 15.67 | 4 | 1.84 |
| 6 yearsb | 123 | 2.67 | 49 | 39.84 | 18 | 14.63 | 5 | 4.07 |
| ≥ 6.5 years | 120 | 2.60 | 44 | 36.67 | 31 | 25.83 | 6 | 5.00 |
| Total | 4608 | 100.00 | 1605 | 34.83 | 723 | 15.69 | 84 | 1.82 |
Table 14 describes examinations undertaken during FUV1. For most patients, FUV1 comprised a single examination (88%) and in 72% of patients the most complete examination was a complete colonoscopy.
| Examinations during FUV1 | IR patients (N = 4608) | ||
|---|---|---|---|
| n | % | ||
| Length of FUV1 | 1 day | 4068 | 88.00 |
| 2–30 days | 64 | 1.39 | |
| 1–3 months | 126 | 2.73 | |
| 3–6 months | 149 | 3.23 | |
| 6–12 months | 162 | 3.52 | |
| 1–2 years | 38 | 0.82 | |
| 2–3 years | 1 | 0.02 | |
| Number of examinations during FUV1 | 1 | 4060 | 88.00 |
| 2 | 394 | 8.55 | |
| 3 | 101 | 2.19 | |
| 4+ | 53 | 1.15 | |
| Most complete examination during FUV1 | Complete colonoscopy | 3299 | 72.00 |
| Colonoscopy of unknown completeness | 259 | 6.00 | |
| Incomplete colonoscopy | 404 | 8.77 | |
| Colonoscopy or FS | 192 | 4.17 | |
| FS | 326 | 7.07 | |
| Colonoscopy, FS or rigid sigmoidoscopy | 103 | 2.24 | |
| Surgery | 16 | 0.35 | |
| Unknown | 9 | 0.20 | |
Baseline risk factors for findings at first follow-up
Using univariable analyses, we investigated the crude associations of baseline demographic, procedural, adenoma and polyp characteristics with findings at FUV1 in order to identify risk factors for adenomas, AA and CRC and to assess potential confounders of the association between interval and outcomes.
Demographic and procedural characteristics
Table 15 details the crude effect of baseline demographic and procedural characteristics on the odds of having adenomas (all types), AA or CRC found at FUV1.
| Baseline factors | All IR patients | IR patients with findings at FUV1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N = 4608 | Adenoma(s) (N = 1605) | AA(s) (N = 723) | CRC(s) (N = 84) | |||||||||||
| n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Age (years) | < 55 | 1025 | 296 | 28.88 | 1 | < 0.0001 | 107 | 10.44 | 1 | < 0.0001 | 8 | 0.78 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 622 | 240 | 38.59 | 1.55 (1.25 to 1.91) | 105 | 16.88 | 1.74 (1.30 to 2.33) | 2 | 0.32 | 0.41 (0.09 to 1.94) | ||||
| ≥ 60 and < 65 | 788 | 278 | 35.28 | 1.34 (1.10 to 1.64) | 116 | 14.72 | 1.48 (1.12 to 1.96) | 9 | 1.14 | 1.47 (0.56 to 3.82) | ||||
| ≥ 65 and < 70 | 813 | 270 | 33.21 | 1.22 (1.00 to 1.49) | 118 | 14.51 | 1.46 (1.10 to 1.93) | 15 | 1.85 | 2.39 (1.01 to 5.66) | ||||
| ≥ 70 and < 75 | 714 | 288 | 40.34 | 1.67 (1.36 to 2.04) | 137 | 19.19 | 2.04 (1.55 to 2.68) | 16 | 2.24 | 2.91 (1.24 to 6.85) | ||||
| ≥ 75 and < 80 | 413 | 148 | 35.84 | 1.38 (1.08 to 1.75) | 87 | 21.07 | 2.29 (1.68 to 3.12) | 21 | 5.08 | 6.81 (2.99 to 15.5) | ||||
| ≥ 80 | 233 | 85 | 36.48 | 1.41 (1.05 to 1.91) | 53 | 22.75 | 2.53 (1.75 to 3.64) | 13 | 5.58 | 7.51 (3.08 to 18.34) | ||||
| Gender | Male | 2551 | 960 | 37.63 | 1 | < 0.0001 | 398 | 15.60 | 1.00 | 0.8543 | 53 | 2.08 | 1 | 0.147 |
| Female | 2057 | 645 | 31.36 | 0.76 (0.67 to 0.86) | 325 | 15.80 | 1.02 (0.87 to 1.19) | 31 | 1.51 | 0.72 (0.46 to 1.13) | ||||
| Family history of cancer | No | 4368 | 1535 | 35.14 | 1 | 0.0552 | 695 | 15.91 | 1 | 0.0678 | 81 | 1.85 | 1 | 0.4716 |
| Yes | 240 | 70 | 29.17 | 0.76 (0.57 to 1.01) | 28 | 11.67 | 0.70 (0.47 to 1.04) | 3 | 1.25 | 0.67 (0.21 to 2.14) | ||||
| Year of baseline | 1985–9 | 98 | 25 | 25.51 | 0.63 (0.40 to 1.00) | 0.0004 | 16 | 16.33 | 1.06 (0.61 to 1.83) | 0.9580 | 0 | 0.00 | n/a | 0.1283 |
| 1990–4 | 241 | 67 | 27.80 | 0.71 (0.53 to 0.95) | 42 | 17.43 | 1.14 (0.80 to 1.62) | 8 | 3.32 | 2.24 (1.03 to 4.88) | ||||
| 1995–9 | 1030 | 333 | 32.33 | 0.88 (0.75 to 1.02) | 162 | 15.73 | 1.01 (0.83 to 1.24) | 25 | 2.43 | 1.62 (0.97 to 2.72) | ||||
| 2000–4 | 2317 | 818 | 35.30 | 1 | 361 | 15.58 | 1 | 35 | 1.51 | 1 | ||||
| 2005–10 | 922 | 362 | 39.26 | 1.18 (1.01 to 1.39) | 142 | 15.40 | 0.99 (0.80 to 1.22) | 16 | 1.74 | 1.15 (0.63 to 2.09) | ||||
| Length of baseline visit | 1 day | 2496 | 877 | 35.14 | 1 | 0.0037 | 342 | 13.70 | 1 | < 0.0001 | 37 | 1.48 | 1 | 0.3993 |
| 2–30 days | 246 | 68 | 27.64 | 0.71 (0.53 to 0.94) | 33 | 13.41 | 0.98 (0.66 to 1.43) | 7 | 2.85 | 1.95 (0.86 to 4.41) | ||||
| 1–3 months | 664 | 245 | 36.90 | 1.08 (0.90 to 1.29) | 111 | 16.72 | 1.26 (1.00 to 1.60) | 12 | 1.81 | 1.22 (0.63 to 2.36) | ||||
| 3–6 months | 595 | 184 | 30.92 | 0.83 (0.68 to 1.00) | 99 | 16.64 | 1.26 (0.98 to 1.60) | 15 | 2.52 | 1.72 (0.94 to 3.15) | ||||
| 6 to 12 months | 508 | 185 | 36.42 | 1.06 (0.87 to 1.29) | 107 | 21.06 | 1.68 (1.32 to 2.14) | 10 | 1.97 | 1.33 (0.66 to 2.7) | ||||
| ≥ 12 months | 99 | 46 | 46.46 | 1.60 (1.07 to 2.40) | 31 | 31.31 | 2.87 (1.85 to 4.46) | 3 | 3.03 | 2.08 (0.63 to 6.85) | ||||
| Number of examinations in baseline visit | 1 | 2489 | 877 | 35.24 | 1 | 0.0228 | 342 | 13.74 | 1 | < 0.0001 | 37 | 1.49 | 1 | 0.1245 |
| 2 | 1518 | 499 | 32.87 | 0.90 (0.79 to 1.03) | 231 | 15.22 | 1.13 (0.94 to 1.35) | 29 | 1.91 | 1.29 (0.79 to 2.11) | ||||
| 3 | 392 | 138 | 35.20 | 1.00 (0.80 to 1.25) | 84 | 21.43 | 1.71 (1.31 to 2.24) | 11 | 2.81 | 1.91 (0.97 to 3.78) | ||||
| 4+ | 209 | 91 | 43.54 | 1.42 (1.07 to 1.89) | 66 | 31.58 | 2.90 (2.12 to 3.96) | 7 | 3.35 | 2.30 (1.01 to 5.22) | ||||
| Most complete colonoscopy | Complete | 2973 | 966 | 32.49 | 1 | < 0.0001 | 388 | 13.05 | 1 | < 0.0001 | 41 | 1.38 | 1 | < 0.0001 |
| Unknown | 1157 | 481 | 41.57 | 1.48 (1.29 to 1.70) | 244 | 21.09 | 1.78 (1.49 to 2.13) | 16 | 1.38 | 1.00 (0.56 to 1.79) | ||||
| Incomplete | 478 | 158 | 33.05 | 1.03 (0.84 to 1.26) | 91 | 19.04 | 1.57 (1.22 to 2.02) | 27 | 5.65 | 4.28 (2.61 to 7.03) | ||||
| Best bowel preparation at colonoscopy | Excellent/good | 1401 | 483 | 34.48 | 1 | 0.4337 | 181 | 12.92 | 1 | 0.0032 | 26 | 1.86 | 1 | 0.0229 |
| Satisfactory | 487 | 179 | 36.76 | 1.10 (0.89 to 1.37) | 73 | 14.99 | 1.19 (0.89 to 1.59) | 6 | 1.23 | 0.66 (0.27 to 1.61) | ||||
| Poor | 194 | 76 | 39.18 | 1.22 (0.90 to 1.67) | 36 | 18.56 | 1.54 (1.04 to 2.28) | 10 | 5.15 | 2.87 (1.36 to 6.06) | ||||
| Unknown | 2526 | 867 | 34.32 | 0.99 (0.87 to 1.14) | 433 | 17.14 | 1.39 (1.16 to 1.68) | 42 | 1.66 | 0.89 (0.55 to 1.46) | ||||
| Difficult examination | No | 4387 | 1551 | 35.35 | 1 | 0.0006 | 690 | 15.73 | 1 | 0.7493 | 73 | 1.66 | 1 | 0.0027 |
| Yes | 221 | 54 | 24.43 | 0.59 (0.43 to 0.81) | 33 | 14.93 | 0.94 (0.64 to 1.37) | 11 | 4.98 | 3.10 (1.62 to 5.92) | ||||
Adenomas (all types)
Patients aged ≥ 55 years were more likely to have an adenoma found at FUV1 than those aged < 55 years; however, no clear trend was seen after the age of 55 years. Women were 24% less likely to have an adenoma detected (OR 0.76, 95% CI 0.67 to 0.86). Patients with a family history of cancer had a non-significant 24% lower risk of adenoma (OR 0.76, 95% CI 0.57 to 1.01). The odds of detecting adenomas at FUV1 were greater in those with later baseline visits (p = 0.0004).
Patients with a baseline visit of longer than 12 months or with four or more baseline examinations were significantly more likely to have an adenoma detected. The association between the completeness of colonoscopy and risk of detection of one or more adenomas was difficult to interpret when no evidence was found of an association between adenoma detection and quality of bowel preparation. However, having a difficult examination at baseline – a composite measure of different aspects of examination quality including completeness and preparation – was associated with a significantly lower odds of having an adenoma detected at FUV1 (OR 0.59, 95% CI 0.43 to 0.81).
Advanced adenomas
The odds of detecting AA at FUV1 significantly increased with increasing age (p < 0.0001). There was no association with gender, or with year of the baseline visit.
There was a tendency for the AA detection rate to increase with increasing number of baseline examinations or a longer duration of the baseline visit, with patients whose baseline visit was 12 months or longer or who had four or more examinations having an almost threefold increased odds (OR 2.87, 95% CI 1.85 to 4.46, and OR 2.9, 95% CI 2.12 to 3.96, respectively). The odds of detecting AA were 57% greater among those with only an incomplete baseline colonoscopy (OR 1.57, 95% CI 1.22 to 2.02) and 78% greater in patients with a colonoscopy of unknown completeness (OR 1.78, 95% CI 1.49 to 2.13). Bowel preparation quality was also predictive of having AA at FUV1.
Colorectal cancers
Only 84 CRCs were detected at FUV1; therefore, although significant associations with baseline risk factors were seen, estimates were imprecise and CIs were wide. There was a strong relationship between increasing age and CRC at FUV1, with a more than sixfold greater odds in patients aged ≥ 75 years (OR 6.81, 95% CI 2.99 to 15.50, for those aged 75–80 years and OR 7.51, 95% CI 3.08 to 18.34, for those aged ≥ 80 years). No significant associations were found between gender, family history of cancer, year of baseline, length of baseline or number of examinations in baseline.
There was strong evidence of an association between having an incomplete colonoscopy or poor bowel preparation or a difficult examination at baseline and increased odds of detecting CRC at FUV1, with odds increased by three- to fourfold.
Adenoma and polyp characteristics
Table 16 describes the crude relationship between characteristics of adenomas and polyps detected at baseline and adenomas, AA and CRC at FUV1.
| Baseline factors | Number of IR patients (N = 4608) | IR patients with findings at first follow-up | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adenoma(s) | AA(s) | CRC(s) | ||||||||||||
| n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | ||||||||||||||
| Number | 1 | 3107 | 972 | 31.28 | 1 | < 0.0001 | 470 | 15.13 | 1 | 0.0068 | 62 | 2.00 | 1 | 0.1166 |
| 2 | 1151 | 475 | 41.27 | 1.54 (1.34 to 1.77) | 211 | 18.33 | 1.26 (1.05 to 1.51) | 21 | 1.82 | 0.91 (0.55 to 1.50) | ||||
| 3 | 240 | 104 | 43.33 | 1.68 (1.29 to 2.19) | 25 | 10.42 | 0.65 (0.43 to 1.00) | 1 | 0.42 | 0.21 (0.03 to 1.49) | ||||
| 4 | 110 | 54 | 49.09 | 2.12 (1.45 to 3.10) | 17 | 15.45 | 1.03 (0.61 to 1.74) | 0 | 0.00 | n/a | ||||
| Largest size (mm) | < 10 | 350 | 158 | 45.14 | 1 | < 0.0001 | 42 | 12.00 | 1 | < 0.0001 | 1 | 0.29 | 1 | 0.0361 |
| 10–14 | 1577 | 507 | 32.15 | 0.58 (0.45 to 0.73) | 190 | 12.05 | 1.00 (0.70 to 1.43) | 29 | 1.84 | 6.54 (0.89 to 48.16) | ||||
| 15–19 | 953 | 300 | 31.48 | 0.56 (0.43 to 0.72) | 122 | 12.80 | 1.08 (0.74 to 1.57) | 16 | 1.68 | 5.96 (0.79 to 45.10) | ||||
| ≥ 20 | 1728 | 640 | 37.04 | 0.71 (0.57 to 0.90) | 369 | 21.35 | 1.99 (1.41 to 2.80) | 38 | 2.20 | 7.85 (1.07 to 57.34) | ||||
| Worst histology | Tubular | 1723 | 548 | 31.80 | 1 | < 0.0001 | 171 | 9.92 | 1 | < 0.0001 | 18 | 1.04 | 1 | 0.0004 |
| Tubulovillous | 2136 | 738 | 34.55 | 1.13 (0.99 to 1.30) | 374 | 17.51 | 1.93 (1.59 to 2.34) | 39 | 1.83 | 1.76 (1.00 to 3.09) | ||||
| Villous | 459 | 173 | 37.69 | 1.30 (1.05 to 1.61) | 115 | 25.05 | 3.03 (2.33 to 3.95) | 19 | 4.14 | 4.09 (2.13 to 7.86) | ||||
| Unknown | 290 | 146 | 50.34 | 2.17 (1.69 to 2.80) | 63 | 21.72 | 2.52 (1.83 to 3.47) | 8 | 2.76 | 2.69 (1.16 to 6.24) | ||||
| Worst dysplasia | Low grade | 3427 | 1144 | 33.38 | 1 | < 0.0001 | 483 | 14.09 | 1 | < 0.0001 | 51 | 1.49 | 1 | 0.0144 |
| High grade | 850 | 297 | 34.94 | 1.07 (0.92 to 1.26) | 162 | 19.06 | 1.44 (1.18 to 1.75) | 26 | 3.06 | 2.09 (1.29 to 3.37) | ||||
| Unknown | 331 | 164 | 49.55 | 1.96 (1.56 to 2.46) | 78 | 23.56 | 1.88 (1.43 to 2.47) | 7 | 2.11 | 1.43 (0.64 to 3.18) | ||||
| Location | Distal only | 3070 | 1005 | 32.74 | 1 | < 0.0001 | 486 | 15.83 | 1 | 0.6382 | 60 | 1.95 | 1 | 0.9786 |
| Proximal only | 681 | 256 | 37.59 | 1.24 (1.04 to 1.47) | 108 | 15.86 | 1.00 (0.80 to 1.26) | 13 | 1.91 | 0.98 (0.53 to 1.79) | ||||
| Distal and proximal | 601 | 262 | 43.59 | 1.59 (1.33 to 1.90) | 96 | 15.97 | 1.01 (0.80 to 1.28) | 11 | 1.83 | 0.94 (0.49 to 1.79) | ||||
| Unknown | 256 | 82 | 32.03 | 0.97 (0.74 to 1.27) | 33 | 12.89 | 0.79 (0.54 to 1.15) | 0 | 0.00 | n/a | ||||
| Distal | No | 937 | 338 | 36.07 | 1 | 0.3723 | 141 | 15.05 | 1 | 0.5432 | 13 | 1.39 | 1 | 0.2488 |
| Yes | 3671 | 1267 | 34.51 | 0.93 (0.80 to 1.08) | 582 | 15.85 | 1.06 (0.87 to 1.3) | 71 | 1.93 | 1.40 (0.77 to 2.54) | ||||
| Proximal | No | 3326 | 1087 | 32.68 | 1 | < 0.0001 | 519 | 15.60 | 1 | 0.7968 | 60 | 1.80 | 1 | 0.8773 |
| Yes | 1282 | 518 | 40.41 | 1.40 (1.22 to 1.6) | 204 | 15.91 | 1.02 (0.86 to 1.22) | 24 | 1.87 | 1.04 (0.64 to 1.67) | ||||
| Number of sightings of a single adenoma | 1 | 3311 | 1102 | 33.28 | 1 | 0.0001 | 433 | 13.08 | 1 | < 0.0001 | 49 | 1.48 | 1 | 0.0412 |
| 2 | 1005 | 381 | 37.91 | 1.22 (1.06 to 1.42) | 196 | 19.50 | 1.61 (1.34 to 1.94) | 23 | 2.29 | 1.56 (0.95 to 2.57) | ||||
| 3 | 182 | 65 | 35.71 | 1.11 (0.82 to 1.52) | 50 | 27.47 | 2.52 (1.79 to 3.54) | 8 | 4.40 | 3.06 (1.43 to 6.56) | ||||
| 4 | 63 | 29 | 46.03 | 1.71 (1.04 to 2.82) | 21 | 33.33 | 3.32 (1.95 to 5.67) | 2 | 3.17 | 2.18 (0.52 to 9.18) | ||||
| 5+ | 47 | 28 | 59.57 | 2.95 (1.64 to 5.31) | 23 | 48.94 | 6.37 (3.56 to 11.39) | 2 | 4.26 | 2.96 (0.7 to 12.54) | ||||
| Polyp characteristics | ||||||||||||||
| Number of hyperplastic polyps | 0 | 3743 | 1297 | 34.65 | 1 | 0.0823 | 609 | 16.27 | 1 | 0.1341 | 68 | 1.82 | 1 | 0.9038 |
| 1 | 541 | 190 | 35.12 | 1.02 (0.85 to 1.23) | 69 | 12.75 | 0.75 (0.58 to 0.98) | 11 | 2.03 | 1.12 (0.59 to 2.13) | ||||
| 2 | 159 | 62 | 38.99 | 1.21 (0.87 to 1.67) | 25 | 15.72 | 0.96 (0.62 to 1.48) | 4 | 2.52 | 1.39 (0.50 to 3.87) | ||||
| 3 | 64 | 16 | 25.00 | 0.63 (0.36 to 1.11) | 5 | 7.81 | 0.44 (0.17 to 1.09) | 0 | 0.00 | n/a | ||||
| 4 | 38 | 10 | 26.32 | 0.67 (0.33 to 1.39) | 5 | 13.16 | 0.78 (0.30 to 2.01) | 1 | 2.63 | 1.46 (0.20 to 10.8) | ||||
| 5+ | 63 | 30 | 47.62 | 1.71 (1.04 to 2.82) | 10 | 15.87 | 0.97 (0.49 to 1.92) | 0 | 0.00 | n/a | ||||
| Any large hyperplastic polyps? | No | 4525 | 1576 | 34.83 | 1 | 0.9832 | 705 | 15.58 | 1 | 0.1471 | 83 | 1.83 | 1 | 0.6513 |
| Yes | 83 | 29 | 34.94 | 1.00 (0.64 to 1.58) | 18 | 21.69 | 1.50 (0.88 to 2.54) | 1 | 1.20 | 0.65 (0.09 to 4.75) | ||||
| Number of polyps with unknown histology | 0 | 3593 | 1189 | 33.09 | 1 | < 0.0001 | 567 | 15.78 | 1 | 0.1511 | 66 | 1.84 | 1 | 0.6976 |
| 1 | 556 | 212 | 38.13 | 1.25 (1.04 to 1.50) | 74 | 13.31 | 0.82 (0.63 to 1.06) | 11 | 1.98 | 1.08 (0.57 to 2.06) | ||||
| 2 | 187 | 87 | 46.52 | 1.76 (1.31 to 2.36) | 36 | 19.25 | 1.27 (0.87 to 1.85) | 1 | 0.53 | 0.29 (0.04 to 2.08) | ||||
| 3 | 108 | 48 | 44.44 | 1.62 (1.10 to 2.38) | 20 | 18.52 | 1.21 (0.74 to 1.99) | 2 | 1.85 | 1.01 (0.24 to 4.17) | ||||
| 4 | 63 | 20 | 31.75 | 0.94 (0.55 to 1.61) | 6 | 9.52 | 0.56 (0.24 to 1.31) | 2 | 3.17 | 1.75 (0.42 to 7.32) | ||||
| 5+ | 101 | 49 | 48.51 | 1.91 (1.28 to 2.83) | 20 | 19.80 | 1.32 (0.80 to 2.17) | 2 | 1.98 | 1.08 (0.26 to 4.47) | ||||
| Location of polyps | Distal only | 2716 | 873 | 32.14 | 1 | < 0.0001 | 431 | 15.87 | 1 | 0.5608 | 50 | 1.84 | 1 | 0.8835 |
| Proximal only | 515 | 199 | 38.64 | 1.33 (1.09 to 1.62) | 88 | 17.09 | 1.09 (0.85 to 1.41) | 10 | 1.94 | 1.06 (0.53 to 2.10) | ||||
| Distal and proximal | 1153 | 461 | 39.98 | 1.41 (1.22 to 1.62) | 174 | 15.09 | 0.94 (0.78 to 1.14) | 24 | 2.08 | 1.13 (0.69 to 1.85) | ||||
| Unknown | 224 | 72 | 32.14 | 1.00 (0.75 to 1.34) | 30 | 13.39 | 0.82 (0.55 to 1.22) | 0 | 0.00 | n/a | ||||
| Distal polyp | No | 739 | 271 | 36.67 | 1 | 0.2533 | 118 | 15.97 | 1 | 0.8213 | 10 | 1.35 | 1 | 0.2792 |
| Yes | 3869 | 1334 | 34.48 | 0.91 (0.77 to 1.07) | 605 | 15.64 | 0.98 (0.79 to 1.21) | 74 | 1.91 | 1.42 (0.73 to 2.76) | ||||
| Proximal polyp | No | 2940 | 945 | 32.14 | 1 | < 0.0001 | 461 | 15.68 | 1 | 0.9806 | 50 | 1.70 | 1 | 0.4138 |
| Yes | 1668 | 660 | 39.57 | 1.38 (1.22 to 1.57) | 262 | 15.71 | 1.00 (0.85 to 1.18) | 34 | 2.04 | 1.20 (0.77 to 1.87) | ||||
Adenomas (all types)
Associations between adenoma detection at FUV1 and the number, size, histology and dysplasia of adenomas detected at baseline were all highly significant (p < 0.0001). Increasing number of adenomas, villous histology and small size (< 10 mm), as opposed to larger size, were associated with a greater odds of having adenomas at FUV1, whereas the association with dysplasia was difficult to interpret. Patients with both a distal and proximal adenoma at baseline had a significant 59% increased odds of having an adenoma detected at FUV1 (OR 1.59, 95% CI 1.33 to 1.90); however, this relationship was probably confounded by the number of adenomas and, when considering proximal location separately, patients with any proximal adenoma at baseline had a 40% greater odds of having an adenoma at FUV1 (OR 1.40, 95% CI 1.22 to 1.60). There was also evidence that patients who had multiple sightings of an individual adenoma during baseline were more likely to have an adenoma detected at FUV1, with a large effect size and highly significant p-value (p = 0.0001). Detection of a proximal polyp at baseline conferred a significant 38% increased odds (OR 1.38, 95% CI 1.22 to 1.57).
Advanced adenomas
There was strong evidence that detection of an adenoma of ≥ 20 mm, with villous or tubulovillous histology or with HGD at baseline, was associated with an increased odds of AA at FUV1 – villous histology had a particularly strong effect (OR 3.03, 95% CI 2.33 to 3.95). The number of adenomas was significantly associated with detection of AA (p = 0.0068), but no clear trend was discernible. Multiple sightings of an adenoma at different examinations during baseline was highly predictive and five or more sightings conferred a more than sixfold increased odds (OR 6.37, 95% CI 3.56 to 11.39). Adenoma location had no effect on the likelihood of having AA at FUV1. There was no relationship between AA and any polyp-related variables.
Colorectal cancer
With only 84 CRCs detected at FUV1, CIs for associations between CRC and baseline adenoma and polyp characteristics were wide; nevertheless, several significant associations were found. Villous histology and HGD at baseline were significantly associated with increased odds of CRC at FUV1: patients with a villous adenoma were four times more likely to have CRC at FUV1 than those with a tubular adenoma (OR 4.09, 95% CI 2.13 to 7.86), whereas HGD at baseline doubled the odds of CRC (OR 2.09, 95% CI 1.29 to 3.37). Larger adenoma size appeared to confer an increased odds of CRC but, despite reaching statistical significance (p = 0.0361), the imprecision of the measures of effect prevented firm conclusions from being drawn. Multiple sightings of an adenoma during baseline was significantly associated with increased odds of CRC (p = 0.0412) but adenoma location had no effect. No polyp characteristics were associated with finding CRC at FUV1.
Baseline risk factors and interval
We explored the relationship between baseline risk factors and length of the interval between baseline and FUV1 to assess whether or not any factors could be acting as confounders of the association between findings at FUV1 and interval (Tables 17 and 18).
| Baseline risk factor | Category | Interval between baseline and first follow-up | Total | p-value (chi squared) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| Total | 1760 (38.19) | 976 (21.18) | 1057 (22.94) | 355 (7.7) | 217 (4.71) | 123 (2.67) | 120 (2.6) | 4608 (100) | n/a | |
| Age (years) | < 55 | 350 (34.15) | 202 (19.71) | 257 (25.07) | 95 (9.27) | 56 (5.46) | 32 (3.12) | 33 (3.22) | 1025 (100) | < 0.001 |
| ≥ 55 and < 60 | 211 (33.92) | 117 (18.81) | 164 (26.37) | 56 (9) | 36 (5.79) | 15 (2.41) | 23 (3.7) | 622 (100) | ||
| ≥ 60 and < 65 | 297 (37.69) | 170 (21.57) | 187 (23.73) | 56 (7.11) | 40 (5.08) | 24 (3.05) | 14 (1.78) | 788 (100) | ||
| ≥ 65 and < 70 | 308 (37.88) | 159 (19.56) | 196 (24.11) | 64 (7.87) | 39 (4.8) | 25 (3.08) | 22 (2.71) | 813 (100) | ||
| ≥ 70 and < 75 | 306 (42.86) | 165 (23.11) | 145 (20.31) | 41 (5.74) | 26 (3.64) | 13 (1.82) | 18 (2.52) | 714 (100) | ||
| ≥ 75 and < 80 | 185 (44.79) | 94 (22.76) | 74 (17.92) | 27 (6.54) | 16 (3.87) | 10 (2.42) | 7 (1.69) | 413 (100) | ||
| ≥ 80 | 103 (44.21) | 69 (29.61) | 34 (14.59) | 16 (6.87) | 4 (1.72) | 4 (1.72) | 3 (1.29) | 233 (100) | ||
| Year of baseline | 1985–9 | 44 (44.90) | 13 (13.27) | 19 (19.39) | 9 (9.18) | 6 (6.12) | 3 (3.06) | 4 (4.08) | 98 (100) | < 0.001 |
| 1990–4 | 73 (30.29) | 55 (22.82) | 30 (12.45) | 27 (11.2) | 15 (6.22) | 10 (4.15) | 31 (12.86) | 241 (100) | ||
| 1995–9 | 387 (37.57) | 218 (21.17) | 192 (18.64) | 96 (9.32) | 41 (3.98) | 40 (3.88) | 56 (5.44) | 1030 (100) | ||
| 2000–4 | 714 (30.82) | 484 (20.89) | 673 (29.05) | 194 (8.37) | 153 (6.6) | 70 (3.02) | 29 (1.25) | 2317 (100) | ||
| 2005–10 | 542 (58.79) | 206 (22.34) | 143 (15.51) | 29 (3.15) | 2 (0.22) | 0 (0) | 0 (0) | 922 (100) | ||
| Length of baseline visit | 1 day | 936 (37.5) | 468 (18.75) | 609 (24.4) | 205 (8.21) | 124 (4.97) | 73 (2.92) | 81 (3.25) | 2496 (100) | < 0.001 |
| 2–30 days | 112 (45.53) | 47 (19.11) | 51 (20.73) | 14 (5.69) | 8 (3.25) | 5 (2.03) | 9 (3.66) | 246 (100) | ||
| 1–3 months | 285 (42.92) | 113 (17.02) | 156 (23.49) | 40 (6.02) | 42 (6.33) | 18 (2.71) | 10 (1.51) | 664 (100) | ||
| 3–6 months | 251 (42.18) | 136 (22.86) | 124 (20.84) | 43 (7.23) | 20 (3.36) | 14 (2.35) | 7 (1.18) | 595 (100) | ||
| 6–12 months | 155 (30.51) | 174 (34.25) | 94 (18.5) | 44 (8.66) | 18 (3.54) | 11 (2.17) | 12 (2.36) | 508 (100) | ||
| ≥ 12 months | 21 (21.21) | 38 (38.38) | 23 (23.23) | 9 (9.09) | 5 (5.05) | 2 (2.02) | 1 (1.01) | 99 (100) | ||
| Number of baseline examinations | 1 | 932 (37.44) | 467 (18.76) | 609 (24.47) | 203 (8.16) | 124 (4.98) | 73 (2.93) | 81 (3.25) | 2489 (100) | < 0.001 |
| 2 | 592 (39) | 313 (20.62) | 347 (22.86) | 114 (7.51) | 79 (5.2) | 41 (2.7) | 32 (2.11) | 1518 (100) | ||
| 3 | 167 (42.6) | 114 (29.08) | 67 (17.09) | 21 (5.36) | 9 (2.3) | 7 (1.79) | 7 (1.79) | 392 (100) | ||
| 4+ | 69 (33.01) | 82 (39.23) | 34 (16.27) | 17 (8.13) | 5 (2.39) | 2 (0.96) | 0 (0) | 209 (100) | ||
| Most complete colonoscopy | Complete | 1254 (42.18) | 618 (20.79) | 715 (24.05) | 180 (6.05) | 118 (3.97) | 55 (1.85) | 33 (1.11) | 2973 (100) | < 0.001 |
| Unknown | 318 (27.48) | 237 (20.48) | 257 (22.21) | 148 (12.79) | 76 (6.57) | 50 (4.32) | 71 (6.14) | 1157 (100) | ||
| Incomplete | 188 (39.33) | 121 (25.31) | 85 (17.78) | 27 (5.65) | 23 (4.81) | 18 (3.77) | 16 (3.35) | 478 (100) | ||
| Best bowel preparation at baseline colonoscopy | Excellent/good | 551 (39.33) | 292 (20.84) | 317 (22.63) | 122 (8.71) | 67 (4.78) | 43 (3.07) | 9 (0.64) | 1401 (100) | < 0.001 |
| Satisfactory | 206 (42.3) | 114 (23.41) | 105 (21.56) | 26 (5.34) | 23 (4.72) | 9 (1.85) | 4 (0.82) | 487 (100) | ||
| Poor | 85 (43.81) | 52 (26.8) | 38 (19.59) | 10 (5.15) | 6 (3.09) | 1 (0.52) | 2 (1.03) | 194 (100) | ||
| Unknown | 918 (36.34) | 518 (20.51) | 597 (23.63) | 197 (7.8) | 121 (4.79) | 70 (2.77) | 105 (4.16) | 2526 (100) | ||
| Baseline risk factor | Category | Interval between baseline and first follow-up | Total | p-value (chi squared) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| Total | 1760 (38.19) | 976 (21.18) | 1057 (22.94) | 355 (7.7) | 217 (4.71) | 123 (2.67) | 120 (2.6) | 4608 (100) | n/a | |
| Adenoma characteristics | ||||||||||
| Number | 1 | 1158 (37.27) | 663 (21.34) | 698 (22.47) | 251 (8.08) | 164 (5.28) | 84 (2.7) | 89 (2.86) | 3107 (100) | 0.006 |
| 2 | 488 (42.4) | 241 (20.94) | 256 (22.24) | 70 (6.08) | 37 (3.21) | 33 (2.87) | 26 (2.26) | 1151 (100) | ||
| 3 | 77 (32.08) | 52 (21.67) | 67 (27.92) | 24 (10) | 12 (5) | 4 (1.67) | 4 (1.67) | 240 (100) | ||
| 4 | 37 (33.64) | 20 (18.18) | 36 (32.73) | 10 (9.09) | 4 (3.64) | 2 (1.82) | 1 (0.91) | 110 (100) | ||
| Largest size (mm) | < 10 | 114 (32.57) | 72 (20.57) | 103 (29.43) | 34 (9.71) | 16 (4.57) | 6 (1.71) | 5 (1.43) | 350 (100) | < 0.001 |
| 10–14 | 548 (34.75) | 305 (19.34) | 404 (25.62) | 127 (8.05) | 90 (5.71) | 50 (3.17) | 53 (3.36) | 1577 (100) | ||
| 15–19 | 348 (36.52) | 191 (20.04) | 223 (23.4) | 74 (7.76) | 57 (5.98) | 31 (3.25) | 29 (3.04) | 953 (100) | ||
| ≥ 20 | 750 (43.4) | 408 (23.61) | 327 (18.92) | 120 (6.94) | 54 (3.13) | 36 (2.08) | 33 (1.91) | 1728 (100) | ||
| Worst histology | Tubular | 621 (36.04) | 350 (20.31) | 451 (26.18) | 132 (7.66) | 91 (5.28) | 39 (2.26) | 39 (2.26) | 1723 (100) | < 0.001 |
| Tubulovillous | 828 (38.76) | 452 (21.16) | 477 (22.33) | 171 (8.01) | 95 (4.45) | 59 (2.76) | 54 (2.53) | 2136 (100) | ||
| Villous | 192 (41.83) | 124 (27.02) | 64 (13.94) | 36 (7.84) | 15 (3.27) | 15 (3.27) | 13 (2.83) | 459 (100) | ||
| Unknown | 119 (41.03) | 50 (17.24) | 65 (22.41) | 16 (5.52) | 16 (5.52) | 10 (3.45) | 14 (4.83) | 290 (100) | ||
| Worst dysplasia | Low grade | 1252 (36.53) | 717 (20.92) | 842 (24.57) | 269 (7.85) | 168 (4.9) | 100 (2.92) | 79 (2.31) | 3427 (100) | < 0.001 |
| High grade | 394 (46.35) | 192 (22.59) | 161 (18.94) | 46 (5.41) | 25 (2.94) | 14 (1.65) | 18 (2.12) | 850 (100) | ||
| Unknown | 114 (34.44) | 67 (20.24) | 54 (16.31) | 40 (12.08) | 24 (7.25) | 9 (2.72) | 23 (6.95) | 331 (100) | ||
| Proximal | No | 1213 (36.47) | 715 (21.5) | 766 (23.03) | 271 (8.15) | 168 (5.05) | 95 (2.86) | 98 (2.95) | 3326 (100) | 0.001 |
| Yes | 547 (42.67) | 261 (20.36) | 291 (22.7) | 84 (6.55) | 49 (3.82) | 28 (2.18) | 22 (1.72) | 1282 (100) | ||
| Number of sightings of a unique adenoma | 1 | 1216 (36.73) | 652 (19.69) | 807 (24.37) | 267 (8.06) | 176 (5.32) | 94 (2.84) | 99 (2.99) | 3311 (100) | < 0.001 |
| 2 | 428 (42.59) | 222 (22.09) | 211 (21) | 68 (6.77) | 33 (3.28) | 24 (2.39) | 19 (1.89) | 1005 (100) | ||
| 3 | 87 (47.8) | 50 (27.47) | 22 (12.09) | 11 (6.04) | 7 (3.85) | 3 (1.65) | 2 (1.1) | 182 (100) | ||
| 4 | 19 (30.16) | 31 (49.21) | 8 (12.7) | 4 (6.35) | 0 (0) | 1 (1.59) | 0 (0) | 63 (100) | ||
| 5+ | 10 (21.28) | 21 (44.68) | 9 (19.15) | 5 (10.64) | 1 (2.13) | 1 (2.13) | 0 (0) | 47 (100) | ||
| Polyp characteristics | ||||||||||
| Number of hyperplastic polyps | 0 | 1431 (38.23) | 782 (20.89) | 869 (23.22) | 283 (7.56) | 177 (4.73) | 93 (2.48) | 108 (2.89) | 3743 (100) | 0.010 |
| 1 | 192 (35.49) | 118 (21.81) | 139 (25.69) | 39 (7.21) | 19 (3.51) | 23 (4.25) | 11 (2.03) | 541 (100) | ||
| 2 | 64 (40.25) | 41 (25.79) | 22 (13.84) | 17 (10.69) | 9 (5.66) | 5 (3.14) | 1 (0.63) | 159 (100) | ||
| 3 | 24 (37.5) | 19 (29.69) | 9 (14.06) | 4 (6.25) | 6 (9.38) | 2 (3.13) | 0 (0) | 64 (100) | ||
| 4 | 17 (44.74) | 5 (13.16) | 7 (18.42) | 7 (18.42) | 2 (5.26) | 0 (0) | 0 (0) | 38 (100) | ||
| 5+ | 32 (50.79) | 11 (17.46) | 11 (17.46) | 5 (7.94) | 4 (6.35) | 0 (0) | 0 (0) | 63 (100) | ||
| Location of polyps | Distal only | 1006 (37.04) | 591 (21.76) | 616 (22.68) | 215 (7.92) | 132 (4.86) | 79 (2.91) | 77 (2.84) | 2716 (100) | < 0.001 |
| Proximal only | 226 (43.88) | 93 (18.06) | 119 (23.11) | 37 (7.18) | 22 (4.27) | 12 (2.33) | 6 (1.17) | 515 (100) | ||
| Distal and proximal | 483 (41.89) | 247 (21.42) | 269 (23.33) | 72 (6.24) | 40 (3.47) | 22 (1.91) | 20 (1.73) | 1153 (100) | ||
| Unknown | 45 (20.09) | 45 (20.09) | 53 (23.66) | 31 (13.84) | 23 (10.27) | 10 (4.46) | 17 (7.59) | 224 (100) | ||
| Proximal polyp | No | 1051 (35.75) | 636 (21.63) | 669 (22.76) | 246 (8.37) | 155 (5.27) | 89 (3.03) | 94 (3.2) | 2940 (100) | < 0.001 |
| Yes | 709 (42.51) | 340 (20.38) | 388 (23.26) | 109 (6.53) | 62 (3.72) | 34 (2.04) | 26 (1.56) | 1668 (100) | ||
All factors were highly significantly associated with interval at the 1% level except for gender (p = 0.462), family history of cancer (p = 0.067), a difficult examination (p = 0.150), large hyperplastic polyps (p = 0.645), number of polyps with unknown histology (p = 0.586), distal adenomas (p = 0.353) and distal polyps (p = 0.105). Results for non-significant factors are not presented here.
Patients of an older age, with an incomplete colonoscopy, poor bowel preparation, a large adenoma (≥ 20 mm), an adenoma with villous histology or HGD, a proximal adenoma or polyp, or multiple sightings of a unique adenoma at baseline tended to have a shorter interval. As all of these features were also associated with increased odds of finding an adenoma, AA or CRC at FUV1, they could potentially be confounding the association between findings at FUV1 and interval.
Effects of interval on findings at follow-up visit 1
Univariable analysis
The effect of interval on findings at the first follow-up was examined using univariable and multivariable analyses. Tables 19–21 show the crude and adjusted associations between interval and adenomas (advanced and non-advanced), AA and CRC at the first follow-up. The univariable analysis provided no evidence of an association between adenomas and interval, with large p-values, small effect estimates close to 1, and 95% CIs that included 1. Similarly, no relationship was observed between AA and interval. For CRC, there was evidence of an association with interval: with interval modelled as a categorical variable, there was evidence of a dose–response effect with a more than threefold increased odds of CRC with an interval of 6.5 years or longer (OR 3.14, 95% CI 1.28 to 7.72), and with interval modelled as a continuous variable, a 13% increased odds of CRC for every year increase in interval (OR 1.13, 95% CI 1.03 to 1.25).
| Baseline risk factor | Category | Univariable analysis: adenoma (all types) | Multivariable analyses: adenoma (all types) | ||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4608) | Model 2 – interval as continuous (n = 4608) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval | < 18 months | 1.00 | 0.5768 | 1.00 | 0.1848 | n/a | |
| 2 yearsa | 1.09 (0.93 to 1.28) | 1.05 (0.88 to 1.25) | |||||
| 3 yearsa | 1.01 (0.86 to 1.19) | 1.06 (0.89 to 1.25) | |||||
| 4 yearsa | 1.04 (0.82 to 1.32) | 1.08 (0.84 to 1.40) | |||||
| 5 yearsa | 1.26 (0.94 to 1.68) | 1.40 (1.03 to 1.90) | |||||
| 6 yearsa | 1.30 (0.89 to 1.88) | 1.42 (0.95 to 2.10) | |||||
| ≥ 6.5 years | 1.13 (0.77 to 1.66) | 1.47 (0.98 to 2.21) | |||||
| Per year increase | 1.03 (0.99 to 1.07) | 0.11 | n/a | 1.06 (1.02 to 1.11) | 0.0024 | ||
| Age (years) | < 55 | 1.00 | < 0.0001 | 1.00 | 0.0005 | 1.00 | 0.0004 |
| ≥ 55 and < 60 | 1.55 (1.25 to 1.91) | 1.46 (1.17 to 1.81) | 1.46 (1.17 to 1.81) | ||||
| ≥ 60 and < 65 | 1.34 (1.10 to 1.64) | 1.27 (1.04 to 1.57) | 1.28 (1.04 to 1.57) | ||||
| ≥ 65 and < 70 | 1.22 (1.00 to 1.49) | 1.10 (0.89 to 1.35) | 1.10 (0.89 to 1.35) | ||||
| ≥ 70 and < 75 | 1.67 (1.36 to 2.04) | 1.56 (1.27 to 1.93) | 1.57 (1.27 to 1.94) | ||||
| ≥ 75 and < 80 | 1.38 (1.08 to 1.75) | 1.33 (1.03 to 1.71) | 1.34 (1.04 to 1.72) | ||||
| ≥ 80 | 1.41 (1.05 to 1.91) | 1.25 (0.91 to 1.71) | 1.26 (0.92 to 1.72) | ||||
| Gender | Male | 1.00 | < 0.0001 | 1.00 | 0.0005 | 1.00 | 0.0004 |
| Female | 0.76 (0.67 to 0.86) | 0.79 (0.70 to 0.90) | 0.79 (0.70 to 0.90) | ||||
| Year of baseline | Per year increase | 1.03 (1.02 to 1.05) | < 0.0001 | 1.05 (1.03 to 1.07) | < 0.0001 | 1.05 (1.03 to 1.07) | < 0.0001 |
| Length of baseline visit | 1 day | 1.00 | 0.0037 | 1.00 | 0.0001 | 1.00 | 0.0001 |
| 2–30 days | 0.71 (0.53 to 0.94) | 0.47 (0.33 to 0.66) | 0.47 (0.33 to 0.66) | ||||
| 1–3 months | 1.08 (0.90 to 1.29) | 0.84 (0.67 to 1.06) | 0.84 (0.67 to 1.06) | ||||
| 3–6 months | 0.83 (0.68 to 1.00) | 0.67 (0.53 to 0.84) | 0.67 (0.53 to 0.84) | ||||
| 6–12 months | 1.06 (0.87 to 1.29) | 0.83 (0.65 to 1.06) | 0.83 (0.65 to 1.06) | ||||
| ≥ 12 months | 1.60 (1.07 to 2.40) | 0.78 (0.46 to 1.33) | 0.78 (0.46 to 1.32) | ||||
| Most complete colonoscopy | Complete | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| Incomplete/unknown | 1.33 (1.18 to 1.51) | 1.64 (1.41 to 1.92) | 1.64 (1.40 to 1.91) | ||||
| Difficult examination | No | 1.00 | 0.0006 | 1.00 | 0.0001 | 1.00 | 0.0001 |
| Yes | 0.59 (0.43 to 0.81) | 0.53 (0.38 to 0.74) | 0.54 (0.39 to 0.75) | ||||
| Number of adenomas | 1 | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| 2 | 1.54 (1.34 to 1.77) | 1.43 (1.23 to 1.67) | 1.43 (1.23 to 1.66) | ||||
| 3 | 1.68 (1.29 to 2.19) | 1.57 (1.18 to 2.11) | 1.57 (1.17 to 2.10) | ||||
| 4 | 2.12 (1.45 to 3.10) | 1.94 (1.29 to 2.91) | 1.92 (1.28 to 2.89) | ||||
| Largest adenoma (mm) | < 20 | 1.00 | 0.0151 | 1.00 | 0.0067 | 1.00 | 0.0062 |
| ≥ 20 | 1.17 (1.03 to 1.32) | 1.22 (1.06 to 1.4) | 1.22 (1.06 to 1.41) | ||||
| Number of sightings of a unique adenoma | 1 | 1.00 | 0.0001 | 1.00 | 0.0002 | 1.00 | 0.0002 |
| 2 | 1.22 (1.06 to 1.42) | 1.5 (1.22 to 1.85) | 1.50 (1.22 to 1.85) | ||||
| 3 | 1.11 (0.82 to 1.52) | 1.29 (0.90 to 1.85) | 1.30 (0.90 to 1.86) | ||||
| 4 | 1.71 (1.04 to 2.82) | 1.88 (1.07 to 3.32) | 1.89 (1.07 to 3.34) | ||||
| 5+ | 2.95 (1.64 to 5.31) | 3.13 (1.53 to 6.39) | 3.13 (1.53 to 6.41) | ||||
| Number of unknown histology polyps | 0 | 1.00 | < 0.0001 | 1.00 | 0.0065 | 1.00 | 0.0056 |
| 1 | 1.25 (1.04 to 1.50) | 1.17 (0.96 to 1.43) | 1.17 (0.96 to 1.43) | ||||
| 2 | 1.76 (1.31 to 2.36) | 1.54 (1.13 to 2.11) | 1.56 (1.14 to 2.13) | ||||
| 3 | 1.62 (1.10 to 2.38) | 1.35 (0.89 to 2.03) | 1.35 (0.89 to 2.03) | ||||
| 4 | 0.94 (0.55 to 1.61) | 0.79 (0.46 to 1.39) | 0.80 (0.46 to 1.39) | ||||
| 5+ | 1.91 (1.28 to 2.83) | 1.73 (1.14 to 2.63) | 1.74 (1.15 to 2.65) | ||||
| Proximal polyp | No | 1.00 | < 0.0001 | 1.00 | 0.0057 | 1.00 | 0.0053 |
| Yes | 1.38 (1.22 to 1.57) | 1.24 (1.06 to 1.44) | 1.24 (1.07 to 1.44) | ||||
| Baseline risk factor | Category | Univariable analysis: AA | Multivariable analyses: AA | ||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4318) | Model 2 – interval as continuous (n = 4318) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval | < 18 months | 1.00 | 0.083 | 1.00 | 0.7658 | n/a | |
| 2 yearsa | 1.13 (0.92 to 1.40) | 1.05 (0.83 to 1.33) | |||||
| 3 yearsa | 0.93 (0.75 to 1.15) | 1.06 (0.83 to 1.34) | |||||
| 4 yearsa | 1.04 (0.76 to 1.43) | 1.10 (0.78 to 1.55) | |||||
| 5 yearsa | 1.03 (0.70 to 1.53) | 1.24 (0.81 to 1.91) | |||||
| 6 yearsa | 0.95 (0.57 to 1.60) | 0.90 (0.51 to 1.61) | |||||
| ≥ 6.5 years | 1.94 (1.26 to 2.98) | 1.52 (0.90 to 2.57) | |||||
| Per year increase | 1.04 (0.99 to 1.09) | 0.1103 | n/a | 1.03 (0.98 to 1.09) | 0.2513 | ||
| Age (years) | < 55 | 1.00 | < 0.0001 | 1.00 | 0.0014 | 1.00 | 0.0013 |
| ≥ 55 and < 60 | 1.74 (1.30 to 2.33) | 1.56 (1.14 to 2.13) | 1.57 (1.15 to 2.15) | ||||
| ≥ 60 and < 65 | 1.48 (1.12 to 1.96) | 1.37 (1.01 to 1.85) | 1.37 (1.01 to 1.86) | ||||
| ≥ 65 and < 70 | 1.46 (1.10 to 1.93) | 1.24 (0.91 to 1.68) | 1.24 (0.92 to 1.68) | ||||
| ≥ 70 and < 75 | 2.04 (1.55 to 2.68) | 1.74 (1.30 to 2.35) | 1.75 (1.30 to 2.35) | ||||
| ≥ 75 and < 80 | 2.29 (1.68 to 3.12) | 1.81 (1.28 to 2.54) | 1.81 (1.28 to 2.54) | ||||
| ≥ 80 | 2.53 (1.75 to 3.64) | 1.81 (1.21 to 2.70) | 1.81 (1.21 to 2.71) | ||||
| Most complete colonoscopy | Complete | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| Incomplete/unknown | 1.72 (1.46 to 2.02) | 1.91 (1.57 to 2.32) | 1.92 (1.58 to 2.33) | ||||
| Number of adenomas | 1 | 1.00 | 0.0068 | 1.00 | 0.0358 | 1.00 | 0.0377 |
| 2 | 1.26 (1.05 to 1.51) | 1.25 (1.02 to 1.54) | 1.25 (1.02 to 1.53) | ||||
| 3 | 0.65 (0.43 to 1.00) | 1.22 (0.76 to 1.96) | 1.22 (0.76 to 1.96) | ||||
| 4 | 1.03 (0.61 to 1.74) | 2.05 (1.14 to 3.67) | 2.05 (1.14 to 3.67) | ||||
| Largest adenoma (mm) | < 20 | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| ≥ 20 | 1.94 (1.65 to 2.27) | 1.69 (1.39 to 2.06) | 1.69 (1.39 to 2.06) | ||||
| Worst adenoma histology | Tubular | 1.00 | < 0.0001 | 1.00 | 0.0016 | 1.00 | 0.0016 |
| Tubulovillous | 1.93 (1.59 to 2.34) | 1.42 (1.15 to 1.76) | 1.42 (1.15 to 1.75) | ||||
| Villous | 3.03 (2.33 to 3.95) | 1.59 (1.17 to 2.18) | 1.59 (1.17 to 2.17) | ||||
| Proximal polyp | No | 1.00 | 0.98 | 1.00 | 0.0182 | 1.00 | 0.0188 |
| Yes | 1.00 (0.85 to 1.18) | 1.28 (1.04 to 1.58) | 1.28 (1.04 to 1.58) | ||||
| Maximum number of sightings of a unique adenoma | 1 | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| 2 | 1.61 (1.34 to 1.94) | 1.37 (1.11 to 1.69) | 1.37 (1.11 to 1.69) | ||||
| 3 | 2.52 (1.79 to 3.54) | 1.76 (1.20 to 2.56) | 1.76 (1.20 to 2.56) | ||||
| 4 | 3.32 (1.95 to 5.67) | 2.49 (1.40 to 4.44) | 2.49 (1.40 to 4.43) | ||||
| 5+ | 6.37 (3.56 to 11.39) | 3.79 (1.97 to 7.27) | 3.79 (1.98 to 7.25) | ||||
| Number of hyperplastic polyps | 0 | 1.00 | 0.1341 | 1.00 | 0.0153 | 1.00 | 0.0147 |
| 1 | 0.75 (0.58 to 0.98) | 0.77 (0.57 to 1.03) | 0.76 (0.57 to 1.03) | ||||
| 2 | 0.96 (0.62 to 1.48) | 0.99 (0.62 to 1.58) | 0.99 (0.62 to 1.59) | ||||
| 3 | 0.44 (0.17 to 1.09) | 0.15 (0.04 to 0.65) | 0.15 (0.04 to 0.65) | ||||
| 4 | 0.78 (0.30 to 2.01) | 0.61 (0.20 to 1.83) | 0.61 (0.20 to 1.83) | ||||
| 5 | 0.97 (0.49 to 1.92) | 1.00 (0.48 to 2.08) | 1.00 (0.48 to 2.09) | ||||
| Large hyperplastic polyp | No | 1.00 | 0.1471 | 1.00 | 0.0037 | 1.00 | 0.004 |
| Yes | 1.50 (0.88 to 2.54) | 2.58 (1.40 to 4.73) | 2.56 (1.39 to 4.69) | ||||
| Baseline risk factor | Category | Univariable analysis: CRC | Multivariable analyses: CRC | ||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4186) | Model 2 – interval as continuous (n = 4186) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval | < 18 months | 1.00 | 0.0006 | 1.00 | 0.0049 | n/a | |
| 2 yearsa | 1.57 (0.91 to 2.69) | 1.69 (0.94 to 3.03) | |||||
| 3 yearsa | 0.34 (0.14 to 0.82) | 0.53 (0.22 to 1.31) | |||||
| 4 yearsa | 1.55 (0.73 to 3.31) | 2.46 (1.12 to 5.44) | |||||
| 5 yearsa | 1.12 (0.39 to 3.22) | 2.08 (0.70 to 6.18) | |||||
| 6 yearsa | 2.53 (0.96 to 6.65) | 3.02 (1.00 to 9.10) | |||||
| ≥ 6.5 years | 3.14 (1.28 to 7.72) | 4.12 (1.37 to 12.41) | |||||
| Per year increase | 1.13 (1.03 to 1.25) | 0.0232 | n/a | 1.21 (1.08 to 1.37) | 0.0040 | ||
| Age (years) | < 55 | 1.00 | < 0.0001 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| ≥ 55 and < 60 | 0.41 (0.09 to 1.94) | 0.20 (0.02 to 1.61) | 0.20 (0.02 to 1.57) | ||||
| ≥ 60 and < 65 | 1.47 (0.56 to 3.82) | 1.18 (0.44 to 3.18) | 1.16 (0.43 to 3.12) | ||||
| ≥ 65 and < 70 | 2.39 (1.01 to 5.66) | 1.86 (0.76 to 4.55) | 1.86 (0.76 to 4.56) | ||||
| ≥ 70 and < 75 | 2.91 (1.24 to 6.85) | 2.11 (0.86 to 5.19) | 2.18 (0.89 to 5.35) | ||||
| ≥ 75 and < 80 | 6.81 (2.99 to 15.5) | 5.92 (2.56 to 13.71) | 6.21 (2.69 to 14.35) | ||||
| ≥ 80 | 7.51 (3.08 to 18.34) | 4.68 (1.81 to 12.07) | 5.03 (1.96 to 12.94) | ||||
| Best bowel preparation | Excellent/good/satisfactory/unknown | 1.00 | 0.0033 | 1.00 | 0.0022 | 1.00 | 0.006 |
| Poor | 3.19 (1.62 to 6.27) | 3.80 (1.79 to 8.05) | 3.30 (1.54 to 7.07) | ||||
| Difficult examination | No | 1.00 | 0.0027 | n/a | 1.00 | 0.0425 | |
| Yes | 3.10 (1.62 to 5.92) | 2.32 (1.10 to 4.89) | |||||
| Worst adenoma histology | Tubular | 1.00 | 0.0002 | 1.00 | 0.0302 | 1.00 | 0.0098 |
| Tubulovillous | 1.76 (1.00 to 3.09) | 1.46 (0.82 to 2.60) | 1.52 (0.85 to 2.71) | ||||
| Villous | 4.09 (2.13 to 7.86) | 2.55 (1.28 to 5.08) | 2.94 (1.48 to 5.82) | ||||
| Worst adenoma dysplasia | Low grade | 1.00 | 0.0039 | 1.00 | 0.0258 | 1.00 | 0.0379 |
| High grade | 2.09 (1.29 to 3.37) | 1.81 (1.09 to 3.02) | 1.74 (1.04 to 2.90) | ||||
Multivariable analysis
To identify independent risk factors for having adenomas, AA or CRC at FUV1, and to adjust the effect of interval for potential confounding factors, multivariable logistic regression was used. Interval was first modelled as a categorical variable (model 1) and then as a continuous variable (model 2). Results of the models for adenomas (all types), AA and CRC are shown in Tables 19–21, respectively.
Adenomas (all types)
Comparison of crude and adjusted estimates for the effect of interval on adenoma findings at FUV1 showed evidence of weak negative confounding, with the effect masked slightly by that of covariates. After adjustment for covariates, the association between interval and detection of adenomas was strengthened, but there was considerable overlap between the 95% CIs (with interval as a categorical variable), most of which included 1, and statistical significance was reached only when interval was modelled as a continuous variable. The latter model showed 6% greater odds of adenomas at FUV1 per year increase in interval (OR 1.06, 95% CI 1.02 to 1.11; p = 0.0024).
A number of baseline characteristics were found to be independent risk factors for having an adenoma detected at FUV1. These included older age, male gender, later year of baseline, no complete colonoscopy and presence of multiple adenomas at baseline (all p < 0.001). Effect estimates for specific age categories should be interpreted with caution owing to their imprecision. Other risk factors included the presence of an adenoma of ≥ 20 mm or a proximal polyp, whereas odds were lower in patients with a difficult baseline examination (composite variable for an incomplete examination with poor bowel preparation and additional difficulties). Patients with a baseline visit of more than 1 day were significantly less likely to have an adenoma detected at FUV1 (p < 0.001); however, there was considerable overlap between 95% CIs, some of which included 1, which made interpretation difficult. Multiple sightings of an adenoma was a strong risk factor for the detection of adenomas, and having the same adenoma seen five or more times increased odds more than threefold (OR 3.13, 95% CI 1.53 to 6.39).
Models 1 and 2, which used interval as a categorical variable and continuous variable, respectively, were very similar and selected the same variables. Crude and adjusted estimates of effect were similar for all variables except length of baseline visit and most complete colonoscopy.
Advanced adenomas
There was little evidence of a relationship between interval and AA at FUV1, both before and after adjusting for other factors; the test statistics were non-significant and all but one 95% CI included 1, although there was a tendency towards increasing odds with increasing interval.
After adjusting for the effects of covariates, older age, no complete colonoscopy and the presence of an adenoma of ≥ 20 mm at baseline were highly predictive of AA detection at FUV1 (all p < 0.001). Other risk factors included the presence of a proximal polyp, an adenoma with villous or tubulovillous histology, a large (≥ 10 mm) hyperplastic polyp or multiple adenomas at baseline. Multiple sightings of a unique adenoma at baseline was a strong risk factor for AA at FUV1; a dose–response effect was demonstrated and five or more sightings was associated with an almost fourfold greater odds of AA (OR 3.79, 95% CI 2.0 to 7.3). The two models, examining interval as a categorical and continuous variable, were very similar and selected the same variables.
When comparing crude and adjusted estimates, the effects of age, adenoma size and histology, and number of sightings of an adenoma were exaggerated before adjustment, suggesting positive confounding by covariates in the models. There was also evidence of negative confounding, with no effect of proximal polyps, and a smaller effect of completeness of colonoscopy, number of adenomas and large hyperplastic polyps before adjustment for other factors. Number of hyperplastic polyps and presence of a large hyperplastic polyp or a proximal polyp at baseline were significantly associated with AA only after adjustment.
Colorectal cancer
A longer interval was significantly associated with increased odds of CRC detection at FUV1, both before and after adjustment, regardless of whether interval was modelled as a continuous or categorical variable. After adjustment for covariates there was 21% greater odds of finding CRC per year increase in interval (OR 1.21, 95% CI 1.08 to 1.37; p = 0.0040). There was evidence of weak negative confounding as the effect of interval became stronger after adjusting for other factors.
Independent baseline risk factors for CRC at FUV1 included older age, the detection of an adenoma with villous or tubulovillous histology or with HGD, poor bowel preparation and a difficult examination (all p < 0.05), the last of which was significant only in model 2, with interval as a continuous variable. There was evidence of positive and negative confounding; the effects of histology, dysplasia and a difficult examination on CRC were attenuated after adjustment, whereas the effect of bowel preparation was strengthened slightly.
Baseline risk factors for a short interval
Unexpectedly, little evidence of an association was found between interval and detection of adenomas or AA at FUV1, even after adjusting for a number of covariates. As a large proportion of patients returned sooner than expected for their first follow-up, crude and adjusted estimates of the effect of baseline characteristics on interval length were calculated to allow a more detailed examination of baseline predictors of a short interval. An arbitrary cut-off of 2 years from baseline was used to classify patients as having a short interval, as this was the median interval length to FUV1 in the hospital cohort. A logistic regression model was used and factors that were not significant in the model at the 95% level were not included in the final model, and were therefore not adjusted for. Table 22 shows baseline risk factors for a short interval.
| Baseline predictors | Interval from baseline to first follow-up of ≤ 2 years | ||||
|---|---|---|---|---|---|
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Age (years) | < 55 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| ≥ 55 and < 60 | 1.04 (0.85 to 1.27) | 1.02 (0.82 to 1.27) | |||
| ≥ 60 and < 65 | 1.31 (1.09 to 1.58) | 1.19 (0.97 to 1.46) | |||
| ≥ 65 and < 70 | 1.18 (0.98 to 1.42) | 1.11 (0.91 to 1.36) | |||
| ≥ 70 and < 75 | 1.59 (1.31 to 1.93) | 1.47 (1.20 to 1.82) | |||
| ≥ 75 and < 80 | 1.66 (1.32 to 2.09) | 1.45 (1.13 to 1.87) | |||
| ≥ 80 | 2.32 (1.72 to 3.11) | 1.94 (1.40 to 2.67) | |||
| Calendar year of baseline | 1-year increase | 1.06 (1.05 to 1.08) | < 0.0001 | 1.06 (1.04 to 1.08) | < 0.0001 |
| Length of baseline visit | 1 day | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| 2–30 days | 1.57 (1.21 to 2.05) | 1.13 (0.82 to 1.57) | |||
| 1–3 months | 1.30 (1.10 to 1.55) | 0.99 (0.79 to 1.23) | |||
| 3–6 months | 1.53 (1.28 to 1.84) | 1.05 (0.84 to 1.31) | |||
| 6–12 months | 1.30 (1.07 to 1.57) | 0.85 (0.67 to 1.08) | |||
| ≥ 12 months | 0.65 (0.43 to 0.98) | 0.26 (0.15 to 0.45) | |||
| Most complete colonoscopy | Complete | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| Incomplete/unknown | 0.63 (0.55 to 0.71) | 0.71 (0.61 to 0.82) | |||
| Largest adenoma (mm) | < 10 | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| 10–14 | 1.09 (0.86 to 1.38) | 1.29 (0.99 to 1.67) | |||
| 15–19 | 1.22 (0.95 to 1.56) | 1.44 (1.09 to 1.90) | |||
| ≥ 20 | 1.78 (1.42 to 2.25) | 1.80 (1.37 to 2.35) | |||
| Worst adenoma dysplasia | Low grade | 1.00 | < 0.0001 | 1.00 | < 0.0001 |
| High grade | 1.62 (1.39 to 1.89) | 1.42 (1.20 to 1.68) | |||
| Number of sightings of a unique adenoma | 1 | 1.00 | < 0.0001 | 1.00 | 0.0003 |
| 2 | 1.45 (1.26 to 1.67) | 1.34 (1.09 to 1.64) | |||
| 3 | 2.10 (1.53 to 2.87) | 1.93 (1.33 to 2.80) | |||
| 4 | 1.93 (1.15 to 3.24) | 2.55 (1.38 to 4.72) | |||
| 5+ | 0.82 (0.46 to 1.47) | 1.61 (0.77 to 3.34) | |||
| Proximal polyp | No | 1.00 | < 0.0001 | 1.00 | 0.0005 |
| Yes | 1.31 (1.16 to 1.48) | 1.29 (1.12 to 1.49) | |||
Age was significantly associated with a short interval (p < 0.0001), before and after adjustment for confounding, with a tendency towards an increasing odds of a short interval with increasing age. After adjustment, there was a 6% greater odds of a short interval per year increase in the calendar year of the baseline visit (OR 1.06, 95% CI 1.04 to 1.08) and odds also increased for patients with multiple sightings of a single adenoma (p < 0.0003). Conversely, patients without a complete colonoscopy at baseline were significantly less likely to return early, possibly as a result of the experience of a difficult examination (OR 0.71, 95% CI 0.61 to 0.82). Patients with a longer baseline visit were less likely to have a short surveillance interval; however, most 95% CIs included 1 so it was not possible to discern a real effect (p < 0.0001); these results may be affected by adjustment for multiple sightings of an adenoma, as before adjustment there was a positive association between length of baseline and a short interval. Having a large adenoma (≥ 10 mm), an adenoma with HGD or a proximal polyp were also risk factors for a short interval.
The independent predictors of a short interval were also identified as risk factors for finding an adenoma, AA or CRC at FUV1. However, adjustment for these factors made little difference to the effect estimates for interval, and did not reveal an association between interval and adenoma (only associated when interval was modelled as a continuous variable) or AA at FUV1. One possibility is that an unmeasured confounder closely linked to a factor(s) associated with the outcome and exposure may have increased the risk of a short interval and of having adenomas, AA or CRC at FUV1. This would cause an exaggerated effect of a short interval on risk, resulting in a diminished effect of interval length overall. Multiple sightings of a single adenoma at baseline was identified as a strong risk factor for a short interval and for finding an adenoma or AA at FUV1, so it is possible that this factor was acting as a proxy measure for an important, unmeasured confounder. This possibility is explored in detail in the next section.
New and previously seen lesions at first follow-up
As described in the previous section, we hypothesised that an unmeasured confounder was masking the association between interval and the detection of adenoma, AA or CRC at FUV1. It was possible that a proportion of individuals had a short interval because they were undergoing polypectomy site surveillance. Such patients would have a large adenoma, probably seen multiple times during baseline for repeated treatment, and possibly with advanced features such as HGD. Polypectomy site surveillance would be carried out to check the site of a large lesion that might not have been completely removed at baseline, rather than to check for the occurrence of newly developed lesions or lesions missed at baseline (possibly because of a poor-quality examination). The UK Adenoma Surveillance guideline16 assumes that all detected lesions are removed at baseline before surveillance begins, and includes recommendations for the treatment and surveillance of incompletely removed lesions. Such patients may require repeated treatment over a number of examinations in order to achieve complete removal and are then expected to return for a further examination(s) to check the polyp site.
We hypothesised that patients undergoing polypectomy site surveillance would be more likely not only to return for follow-up sooner, but also to have a finding detected at FUV1 – the lesion under polypectomy site surveillance. This could potentially confound the relationship between interval and detection of an adenoma or AA at FUV1. Although difficult to recognise such cases from a retrospective series, it was thought that lesions detected at FUV1 which were previously seen at baseline (i.e. the same lesion) were more likely to have been found as a result of polypectomy site surveillance. The distribution of new and previously seen outcomes by interval was examined to determine whether or not this was likely to be the case.
Tables 23–25 show a breakdown of IR patients by interval length and outcome status. Patients were stratified into four groups: (1) those with no findings at FUV1; (2) those who have only a previously seen finding; (3) those with both previously seen and new findings; and (4) those who have only a new finding. The number of patients within each stratum was then assessed to determine whether or not it was appropriate to exclude previously seen findings from the analyses.
| Adenoma status | Interval from baseline to first follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 1165 (66.19) | 627 (64.24) | 697 (65.94) | 232 (65.35) | 132 (60.83) | 74 (60.16) | 76 (63.33) | 3003 (65.17) |
| Previously seen only | 184 (10.45) | 85 (8.71) | 45 (4.26) | 13 (3.66) | 4 (1.84) | 0 (0) | 3 (2.50) | 334 (7.25) |
| New and previously seen | 48 (2.73) | 25 (2.56) | 15 (1.42) | 2 (0.56) | 2 (0.92) | 2 (1.63) | 0 (0) | 94 (2.04) |
| New only | 363 (20.63) | 239 (24.49) | 300 (28.38) | 108 (30.42) | 79 (36.41) | 47 (38.21) | 41 (34.17) | 1177 (25.54) |
| Total | 1760 (100) | 976 (100) | 1057 (100) | 355 (100) | 217 (100) | 123 (100) | 120 (100) | 4608 (100) |
| AA status | Interval from baseline to first follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 1492 (84.77) | 811 (83.09) | 906 (85.71) | 299 (84.23) | 183 (84.33) | 105 (85.37) | 89 (74.17) | 3885 (84.31) |
| Previously seen only | 132 (7.50) | 75 (7.68) | 46 (4.35) | 11 (3.10) | 5 (2.30) | 0 (0) | 3 (2.50) | 272 (5.90) |
| New and previously seen | 21 (1.19) | 9 (0.92) | 2 (0.19) | 1 (0.28) | 1 (0.46) | 2 (1.63) | 0 (0) | 36 (0.78) |
| New only | 115 (6.53) | 81 (8.30) | 103 (9.74) | 44 (12.39) | 28 (12.9) | 16 (13.01) | 28 (23.33) | 415 (9.01) |
| Total | 1760 (100) | 976 (100) | 1057 (100) | 355 (100) | 217 (100) | 123 (100) | 120 (100) | 4608 (100) |
| CRC status | Interval from baseline to first follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 1731 (98.35) | 951 (97.44) | 1051 (99.43) | 346 (97.46) | 213 (98.16) | 118 (95.93) | 114 (95.00) | 4524 (98.18) |
| Previously seen only | 14 (0.80) | 16 (1.64) | 1 (0.09) | 0 (0) | 0 (0) | 1 (0.81) | 0 (0) | 32 (0.69) |
| New and previously seen | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.81) | 0 (0) | 1 (0.02) |
| New only | 15 (0.85) | 9 (0.92) | 5 (0.47) | 9 (2.54) | 4 (1.84) | 3 (2.44) | 6 (5) | 51 (1.11) |
| Total | 1760 (100) | 976 (100) | 1057 (100) | 355 (100) | 217 (100) | 123 (100) | 120 (100) | 4608 (100) |
After stratifying patients by interval and outcome status, increasing interval length was associated with increased detection of new findings. Patients with a shorter interval had a greater proportion of previously seen lesions detected than those with a longer interval. No such trend was seen among the ‘new and previously seen’ findings group, although there were only a small number of patients in this group: 2% with adenomas, 1% with AA and < 1% with CRC.
Previously seen lesions detected at the first follow-up were most likely to represent lesions undergoing polypectomy site surveillance when found in patients with a short interval between baseline and follow-up. As interval length increased, it became less certain whether or not this was the case. Logistic regression was performed using any findings (see Tables 19–21) and then using only new findings at FUV1 (Tables 26–30), having removed all previously seen findings.
All previously seen lesions were removed regardless of the interval length, rather than just those detected in patients with a short interval to FUV1, in order to avoid the introduction of bias into the data set. If only previously seen lesions in patients with short interval were removed from the analysis then this could artificially increase the odds of an outcome among patients with a longer surveillance interval, which would overestimate the effect of interval.
Effect of interval on new findings at first follow-up
After removal of previously seen lesions to adjust for the confounding effect of polypectomy site surveillance, the association of interval with new findings at the first follow-up was examined using univariable and multivariable analyses.
Univariable analysis
Table 26 shows the crude association between interval and new findings (adenomas, AA and CRC) at the first follow-up – the effect of interval length was stronger than in the univariable analysis of all findings (new and previously seen lesions) (compare Table 26 and Table 27). There was strong evidence of an association between interval length and adenomas, AA and CRC at FUV1 (p < 0.0005), with an apparent dose–response effect on all outcomes (see Table 26). When all findings (both new and previously seen) at FUV1 were analysed, interval was associated with CRC only at FUV1. This suggests that, as predicted, lesions undergoing polypectomy site surveillance were masking the association between interval and findings at FUV1. Although CIs for new outcomes overlap somewhat, they rarely include 1, suggesting that there is a true association in the population.
| Interval from baseline to first follow-up | New findings at first follow-up | |||||
|---|---|---|---|---|---|---|
| Adenoma | AA | CRC | ||||
| Unadjusted OR (95% CI) | p-value (LRT) | Unadjusted OR (95% CI) | p-value (LRT) | Unadjusted OR (95% CI) | p-value (LRT) | |
| < 18 months | 1 | < 0.0001 | 1 | < 0.0001 | 1 | 0.0004 |
| 2 yearsa | 1.22 (1.02 to 1.46) | 1.21 (0.92 to 1.60) | 1.08 (0.47 to 2.48) | |||
| 3 yearsa | 1.39 (1.17 to 1.65) | 1.32 (1.01 to 1.72) | 0.55 (0.20 to 1.53) | |||
| 4 yearsa | 1.47 (1.15 to 1.89) | 1.73 (1.21 to 2.48) | 3.03 (1.31 to 6.97) | |||
| 5 yearsa | 1.95 (1.45 to 2.63) | 1.84 (1.20 to 2.83) | 2.18 (0.72 to 6.64) | |||
| 6 yearsa | 2.17 (1.49 to 3.17) | 2.05 (1.21 to 3.48) | 3.91 (1.28 to 11.97) | |||
| ≥ 6.5 years | 1.70 (1.15 to 2.52) | 3.63 (2.30 to 5.74) | 6.12 (2.33 to 16.08) | |||
| Interval (per year increase) | 1.12 (1.08 to 1.16) | < 0.0001 | 1.16 (1.11 to 1.22) | < 0.0001 | 1.27 (1.16 to 1.40) | < 0.0001 |
| Interval from baseline to first follow-up | Findings at FUV1 | |||||
|---|---|---|---|---|---|---|
| Adenoma | AA | CRC | ||||
| Unadjusted OR (95% CI) | p-value (LRT) | Unadjusted OR (95% CI) | p-value (LRT) | Unadjusted OR (95% CI) | p-value (LRT) | |
| < 18 months | 1 | 0.5768 | 1 | 0.0830 | 1 | 0.0006 |
| 2 yearsa | 1.09 (0.93 to 1.28) | 1.13 (0.92 to 1.40) | 1.57 (0.91 to 2.69) | |||
| 3 yearsa | 1.01 (0.86 to 1.19) | 0.93 (0.75 to 1.15) | 0.34 (0.14 to 0.82) | |||
| 4 yearsa | 1.04 (0.82 to 1.32) | 1.04 (0.76 to 1.43) | 1.55 (0.73 to 3.31) | |||
| 5 yearsa | 1.26 (0.94 to 1.68) | 1.03 (0.70 to 1.53) | 1.12 (0.39 to 3.22) | |||
| 6 yearsa | 1.30 (0.89 to 1.88) | 0.95 (0.57 to 1.60) | 2.53 (0.96 to 6.65) | |||
| ≥ 6.5 years | 1.13 (0.77 to 1.66) | 1.94 (1.26 to 2.98) | 3.14 (1.28 to 7.72) | |||
| Interval (per year increase) | 1.03 (0.99 to 1.07) | 0.1117 | 1.04 (0.99 to 1.09) | 0.1103 | 1.13 (1.03 to 1.25) | 0.0232 |
Multivariable analysis
Logistic regression was used to identify independent risk factors for having new adenomas, AA or CRC at FUV1, and to adjust the effect of interval for potential confounding factors. Interval was first modelled as a categorical variable (model 1) and then as a continuous variable (model 2). Results of the models for new adenomas (advanced and non-advanced), AA and CRC are shown in Tables 28–30, respectively.
| Baseline risk factor | Category | Number of IR patients (N = 4608) | Univariable analysis: new adenoma (all types) | Multivariable analyses: new adenoma (all types) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4608) | Model 2 – interval as continuous (n = 4608) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1760 | 411 (23.35) | 1 | < 0.0001 | 1 | < 0.0001 | n/a | |
| 2 yearsa | 976 | 264 (27.05) | 1.22 (1.02 to 1.46) | 1.26 (1.04 to 1.52) | |||||
| 3 yearsa | 1057 | 315 (29.80) | 1.39 (1.17 to 1.65) | 1.43 (1.19 to 1.71) | |||||
| 4 yearsa | 355 | 110 (30.99) | 1.47 (1.15 to 1.89) | 1.58 (1.21 to 2.06) | |||||
| 5 yearsa | 217 | 81 (37.33) | 1.95 (1.45 to 2.63) | 2.12 (1.54 to 2.91) | |||||
| 6 yearsa | 123 | 49 (39.84) | 2.17 (1.49 to 3.17) | 2.46 (1.65 to 3.68) | |||||
| ≥ 6.5 years | 120 | 41 (34.17) | 1.70 (1.15 to 2.52) | 2.27 (1.49 to 3.46) | |||||
| Per year increase | n/a | n/a | 1.12 (1.08 to 1.16) | < 0.0001 | n/a | 1.16 (1.11 to 1.21) | < 0.0001 | ||
| Age (years) | < 55 | 1025 | 243 (23.71) | 1 | 0.0002 | 1 | 0.0009 | 1 | 0.0009 |
| ≥ 55 and < 60 | 622 | 205 (32.96) | 1.58 (1.27 to 1.97) | 1.51 (1.20 to 1.90) | 1.51 (1.20 to 1.90) | ||||
| ≥ 60 and < 65 | 788 | 230 (29.19) | 1.33 (1.07 to 1.64) | 1.30 (1.05 to 1.63) | 1.31 (1.05 to 1.63) | ||||
| ≥ 65 and < 70 | 813 | 212 (26.08) | 1.14 (0.92 to 1.40) | 1.06 (0.85 to 1.33) | 1.06 (0.85 to 1.32) | ||||
| ≥ 70 and < 75 | 714 | 219 (30.67) | 1.42 (1.15 to 1.76) | 1.47 (1.17 to 1.84) | 1.46 (1.16 to 1.83) | ||||
| ≥ 75 and < 80 | 413 | 111 (26.88) | 1.18 (0.91 to 1.54) | 1.27 (0.97 to 1.68) | 1.27 (0.97 to 1.67) | ||||
| ≥ 80 | 233 | 51 (21.89) | 0.90 (0.64 to 1.27) | 0.97 (0.68 to 1.39) | 0.97 (0.68 to 1.39) | ||||
| Gender | Male | 2551 | 794 (31.13) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| Female | 2057 | 477 (23.19) | 0.67 (0.59 to 0.76) | 0.71 (0.61 to 0.81) | 0.70 (0.61 to 0.81) | ||||
| Year of baseline | Per year increase | n/a | n/a | 1.03 (1.02 to 1.05) | 0.0001 | 1.06 (1.04 to 1.08) | < 0.0001 | 1.06 (1.04 to 1.08) | < 0.0001 |
| Length of baseline visit | 1 day | 2496 | 745 (29.85) | 1 | 0.0002 | 1 | 0.0029 | 1 | 0.0055 |
| 2–30 days | 246 | 55 (22.36) | 0.68 (0.50 to 0.92) | 0.56 (0.39 to 0.81) | 0.55 (0.38 to 0.80) | ||||
| 1–3 months | 664 | 190 (28.61) | 0.94 (0.78 to 1.14) | 0.89 (0.70 to 1.14) | 0.90 (0.70 to 1.14) | ||||
| 3–6 months | 595 | 132 (22.18) | 0.67 (0.54 to 0.83) | 0.68 (0.53 to 0.88) | 0.69 (0.54 to 0.88) | ||||
| 6–12 months | 508 | 130 (25.59) | 0.81 (0.65 to 1.00) | 0.82 (0.63 to 1.07) | 0.83 (0.64 to 1.08) | ||||
| ≥ 12 months | 99 | 19 (19.19) | 0.56 (0.34 to 0.93) | 0.62 (0.33 to 1.19) | 0.64 (0.34 to 1.21) | ||||
| Most complete colonoscopy | Complete | 2973 | 771 (25.93) | 1 | 0.0008 | 1 | < 0.0001 | 1 | < 0.0001 |
| Incomplete/unknown | 1635 | 500 (30.58) | 1.26 (1.10 to 1.44) | 1.53 (1.30 to 1.80) | 1.52 (1.29 to 1.80) | ||||
| Difficult examination | No | 4387 | 1232 (28.08) | 1 | 0.0004 | 1 | 0.0013 | 1 | 0.0016 |
| Yes | 221 | 39 (17.65) | 0.55 (0.39 to 0.78) | 0.56 (0.39 to 0.81) | 0.57 (0.39 to 0.82) | ||||
| Number of adenomas | 1 | 3107 | 756 (24.33) | 1 | < 0.0001 | 1 | 0.0002 | 1 | 0.0002 |
| 2 | 1151 | 375 (32.58) | 1.50 (1.30 to 1.74) | 1.38 (1.18 to 1.63) | 1.38 (1.17 to 1.62) | ||||
| 3 | 240 | 95 (39.58) | 2.04 (1.55 to 2.67) | 1.50 (1.11 to 2.01) | 1.51 (1.12 to 2.02) | ||||
| 4 | 110 | 45 (40.91) | 2.15 (1.46 to 3.18) | 1.50 (0.99 to 2.27) | 1.51 (1.00 to 2.28) | ||||
| Number of sightings of a unique adenoma | 1 | 3311 | 939 (28.36) | 1 | 0.0008 | 1 | 0.0301 | 1 | 0.03027 |
| 2 | 1005 | 282 (28.06) | 0.99 (0.84 to 1.15) | 1.22 (0.98 to 1.53) | 1.22 (0.98 to 1.52) | ||||
| 3 | 182 | 30 (16.48) | 0.50 (0.33 to 0.74) | 0.66 (0.42 to 1.03) | 0.65 (0.42 to 1.02) | ||||
| 4 | 63 | 11 (17.46) | 0.53 (0.28 to 1.03) | 0.70 (0.34 to 1.44) | 0.70 (0.34 to 1.44) | ||||
| 5+ | 47 | 9 (19.15) | 0.60 (0.29 to 1.24) | 0.81 (0.34 to 1.94) | 0.82 (0.34 to 1.95) | ||||
| Number of polyps with unknown histology | 0 | 3593 | 915 (25.47) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| 1 | 556 | 179 (32.19) | 1.39 (1.15 to 1.69) | 1.28 (1.04 to 1.58) | 1.28 (1.04 to 1.58) | ||||
| 2 | 187 | 78 (41.71) | 2.09 (1.55 to 2.83) | 1.87 (1.36 to 2.59) | 1.87 (1.36 to 2.59) | ||||
| 3 | 108 | 39 (36.11) | 1.65 (1.11 to 2.47) | 1.44 (0.94 to 2.21) | 1.41 (0.92 to 2.15) | ||||
| 4 | 63 | 14 (22.22) | 0.84 (0.46 to 1.52) | 0.68 (0.36 to 1.26) | 0.69 (0.37 to 1.28) | ||||
| 5+ | 101 | 46 (45.54) | 2.45 (1.64 to 3.65) | 2.27 (1.48 to 3.47) | 2.26 (1.48 to 3.46) | ||||
| Proximal polyps | No | 2940 | 718 (24.42) | 1 | < 0.0001 | 1 | 0.0032 | 1 | 0.0035 |
| Yes | 1668 | 553 (33.15) | 1.53 (1.34 to 1.75) | 1.27 (1.08 to 1.49) | 1.27 (1.08 to 1.49) | ||||
| Baseline risk factor | Category | Number of patients (N = 4608) | Univariable analysis: new AA | Multivariable analyses: new AA | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4608) | Model 2 – interval as continuous (n = 4608) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1760 | 136 (7.73) | 1 | < 0.0001 | 1 | < 0.0001 | n/a | |
| 2 yearsa | 976 | 90 (9.22) | 1.21 (0.92 to 1.60) | 1.18 (0.89 to 1.57) | |||||
| 3 yearsa | 1057 | 105 (9.93) | 1.32 (1.01 to 1.72) | 1.42 (1.08 to 1.88) | |||||
| 4 yearsa | 355 | 45 (12.68) | 1.73 (1.21 to 2.48) | 1.80 (1.24 to 2.61) | |||||
| 5 yearsa | 217 | 29 (13.36) | 1.84 (1.20 to 2.83) | 1.88 (1.20 to 2.94) | |||||
| 6 yearsa | 123 | 18 (14.63) | 2.05 (1.21 to 3.48) | 2.09 (1.21 to 3.62) | |||||
| ≥ 6.5 years | 120 | 28 (23.33) | 3.63 (2.30 to 5.74) | 3.79 (2.33 to 6.16) | |||||
| Per year increase | n/a | n/a | 1.16 (1.11 to 1.22) | < 0.0001 | n/a | 1.18 (1.12 to 1.24) | < 0.0001 | ||
| Age (years) | < 55 | 1025 | 66 (6.44) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 622 | 79 (12.70) | 2.11 (1.50 to 2.98) | 2.11 (1.49 to 3.00) | 2.13 (1.50 to 3.02) | ||||
| ≥ 60 and < 65 | 788 | 76 (9.64) | 1.55 (1.10 to 2.19) | 1.66 (1.17 to 2.36) | 1.65 (1.16 to 2.34) | ||||
| ≥ 65 and < 70 | 813 | 72 (8.86) | 1.41 (1.00 to 2.00) | 1.40 (0.98 to 2.00) | 1.40 (0.98 to 2.00) | ||||
| ≥ 70 and < 75 | 714 | 81 (11.34) | 1.86 (1.32 to 2.61) | 1.95 (1.38 to 2.77) | 1.95 (1.38 to 2.76) | ||||
| ≥ 75 and < 80 | 413 | 57 (13.80) | 2.33 (1.60 to 3.38) | 2.44 (1.66 to 3.59) | 2.44 (1.66 to 3.59) | ||||
| ≥ 80 | 233 | 20 (8.58) | 1.36 (0.81 to 2.30) | 1.41 (0.82 to 2.41) | 1.42 (0.83 to 2.42) | ||||
| Most complete colonoscopy | Complete | 2973 | 244 (8.21) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| Incomplete/unknown | 1635 | 207 (12.66) | 1.62 (1.33 to 1.97) | 1.69 (1.34 to 2.11) | 1.69 (1.35 to 2.12) | ||||
| Largest adenoma (mm) | < 20 | 2880 | 265 (9.20) | 1 | 0.0857 | 1 | 0.0101 | 1 | 0.012 |
| ≥ 20 | 1728 | 186 (10.76) | 1.19 (0.98 to 1.45) | 1.31 (1.07 to 1.62) | 1.30 (1.06 to 1.60) | ||||
| Large hyperplastic polyp | No | 4525 | 436 (9.64) | 1 | 0.0199 | 1 | 0.0211 | 1 | 0.0198 |
| Yes | 83 | 15 (18.07) | 2.07 (1.17 to 3.65) | 2.10 (1.17 to 3.78) | 2.11 (1.17 to 3.80) | ||||
| Proximal polyp | No | 2940 | 265 (9.01) | 1 | 0.0199 | 1 | < 0.0001 | 1 | < 0.0001 |
| Yes | 1668 | 186 (11.15) | 1.27 (1.04 to 1.54) | 1.61 (1.30 to 2.00) | 1.61 (1.30 to 2.00) | ||||
| Baseline risk factor | Category | Number of patients (N = 4608) | Univariate analysis: new CRC | Multivariable analyses: new CRC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4608) | Model 2 – interval as continuous (n = 4608) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1760 | 15 (0.85) | 1 | 0.0004 | 1 | < 0.0001 | n/a | |
| 2 yearsa | 976 | 9 (0.92) | 1.08 (0.47 to 2.48) | 1.08 (0.47 to 2.5) | |||||
| 3 yearsa | 1057 | 5 (0.47) | 0.55 (0.20 to 1.53) | 0.69 (0.25 to 1.91) | |||||
| 4 yearsa | 355 | 9 (2.54) | 3.03 (1.31 to 6.97) | 4.02 (1.72 to 9.42) | |||||
| 5 yearsa | 217 | 4 (1.84) | 2.18 (0.72 to 6.64) | 3.11 (1 to 9.64) | |||||
| 6 yearsa | 123 | 4 (3.25) | 3.91 (1.28 to 11.97) | 5.56 (1.77 to 17.44) | |||||
| ≥ 6.5 years | 120 | 6 (5.00) | 6.12 (2.33 to 16.08) | 9.08 (3.36 to 24.59) | |||||
| Per year increase | n/a | n/a | 1.27 (1.16 to 1.40) | < 0.0001 | n/a | 1.32 (1.20 to 1.46) | < 0.0001 | ||
| Age (years) | < 60 | 1647 | 7 (0.43) | 1 | 0.0002 | 1 | < 0.0001 | 1 | < 0.0001 |
| ≥ 60 and < 65 | 788 | 5 (0.63) | 1.50 (0.47 to 4.73) | 1.54 (0.48 to 4.91) | 1.39 (0.43 to 4.47) | ||||
| ≥ 65 and < 70 | 813 | 10 (1.23) | 2.92 (1.11 to 7.69) | 2.86 (1.07 to 7.60) | 2.79 (1.05 to 7.44) | ||||
| ≥ 70 and < 75 | 714 | 12 (1.68) | 4.00 (1.57 to 10.21) | 4.54 (1.76 to 11.69) | 4.44 (1.72 to 11.43) | ||||
| ≥ 75 and < 80 | 413 | 12 (2.91) | 7.01 (2.74 to 17.92) | 8.37 (3.23 to 21.67) | 8.30 (3.21 to 21.49) | ||||
| ≥ 80 | 233 | 6 (2.58) | 6.19 (2.06 to 18.59) | 7.03 (2.29 to 21.56) | 7.00 (2.29 to 21.41) | ||||
| Most complete colonoscopy | Complete | 2973 | 25 (0.84) | 1 | 0.0149 | n/a | 1 | 0.0384 | |
| Incomplete/unknown | 1635 | 27 (1.65) | 1.98 (1.15 to 3.42) | 1.89 (1.04 to 3.44) | |||||
| Best bowel preparation | Excellent/good/satisfactory/unknown | 4414 | 44 (1) | 1 | 0.0016 | 1 | 0.0005 | 1 | 0.0008 |
| Poor | 194 | 8 (4.12) | 4.27 (1.98 to 9.20) | 5.31 (2.39 to 11.81) | 4.93 (2.22 to 10.93) | ||||
| Proximal polyp | No | 2940 | 29 (0.99) | 1 | 0.2316 | n/a | 1 | 0.0462 | |
| Yes | 1668 | 23 (1.38) | 1.40 (0.81 to 2.43) | 1.84 (1.02 to 3.33) | |||||
Adenomas (all types)
After adjusting for the effects of covariates, the independent association between interval and new adenomas at FUV1 remained highly significant (p < 0.0001), with 16% increased odds of having a new adenoma per year increase in interval length (OR 1.16, 95% CI 1.11 to 1.21). The effect estimates were precise, and there was an apparent dose–response effect, providing strong evidence of an association. There was also evidence of weak negative confounding as the effect was strengthened slightly after adjustment.
Independent risk factors for new adenomas at FUV1 included older age, male gender, later date of baseline, no complete colonoscopy, and the presence of hyperplastic polyps, proximal polyps or multiple adenomas at baseline (all p < 0.004). A difficult examination or a baseline visit of longer than 1 day both appeared to confer a lower chance of having a new adenoma; however, the association with length of visit was irregular. The effect of number of sightings of an adenoma at baseline was somewhat difficult to interpret, as two sightings conferred 22% greater odds, whereas three or more sightings were associated with lower odds of having a new adenoma at FUV1.
Models 1 and 2, with interval as categorical and continuous, were very similar and used the same covariates. All covariates were significant before and after adjustment, with little evidence of confounding. After removal of previously seen findings, larger size of baseline adenoma was no longer predictive of finding new adenomas at FUV1.
Advanced adenomas
There was strong evidence of a significant association between interval and new AA at FUV1: effect estimates were precise and demonstrated a dose–response effect, with an 18% increased odds of new AA at FUV1 per year increase in interval (OR 1.18, 95% CI 1.12 to 1.24; p < 0.0001). Adjustment for covariates had little impact on the effect of interval.
Baseline risk factors for new AA at FUV1 included no complete colonoscopy (OR 1.69, 95% CI 1.34 to 2.11), and the presence of an adenoma of ≥ 20 mm (OR 1.30, 95% CI 1.06 to 1.60), a proximal polyp (OR 1.61, 95% CI 1.30 to 2.00) or a large hyperplastic polyp (OR 2.11, 95% CI 1.71 to 3.80). Older age also conferred greater odds of new AA, although the association with increasing age was irregular.
Models 1 and 2 were quite similar. All risk factors were significantly associated with new AA before and after adjustment except for largest adenoma, the effect of which was strengthened after adjustment. After excluding previously seen lesions, histology, number of sightings of a single adenoma, and number of adenomas or of hyperplastic polyps were no longer predictive of new AA at FUV1.
Colorectal cancer
After adjusting for the effects of covariates, the effect of interval length on new CRC at FUV1 was strengthened, with a more than fivefold greater odds of CRC among those with an interval of ≥ 6 years, and a 32% increase in odds per year increase in interval (OR 1.32, 95% CI 1.20 to 1.46). Owing to the small number of new CRC outcomes, measures of effect for some strata of interval were imprecise; however, the large effect sizes, tendency towards a dose–response effect and highly significant p-value provide strong evidence of an association.
Older age and poor bowel preparation (p = 0.0005) were highly significant risk factors for new CRC at FUV1. Although effect estimates were imprecise for individual categories, risk tended to increase with age and the estimated increase in odds was sevenfold or greater for those aged ≥ 75 years and was fivefold greater for poor bowel preparation. In model 2 (with interval as continuous), the absence of a complete colonoscopy and the presence of proximal polyps at baseline were weakly associated with new CRC at FUV1.
There was some evidence of negative confounding as the effects of age, best bowel preparation and proximal polyp were strengthened after adjusting for covariates. Histology and dysplasia of baseline adenomas were no longer significantly associated with CRC at FUV1 after removal of previously seen lesions from the analysis.
Effect modification of the association between interval and new findings at follow-up visit 1
We proposed that there might be an interaction between interval and age or gender. We investigated interactions with interval to follow-up only as a continuous variable, as these results were more intuitive and enabled the examination of potential trends.
There was no evidence of effect modification by age group or gender on new adenomas (Figure 3). There was some evidence of effect modification for the finding of new AA at FUV1 (Figure 4). By age group, the test for interaction was highly significant (p = 0.0100), although there was no clear trend in the ORs. Increasing the interval had the greatest effect in the < 55 years age group but the decrease in effect was not monotonic (Figure 4). To test for a trend in the ORs, an interaction was fitted between interval and continuous age group; the p-value was 0.8987. Thus, the effect of interval differed between the categorical age groups, but there was no trend in the effect with increasing age group. By gender, the ORs suggested that increasing interval had a stronger effect in men than in women, but this difference was not statistically significant (p = 0.0663). There was no evidence of effect modification on new CRC (Figure 5).
FIGURE 3.
Effect of a 1-year increase in interval on the detection of new adenoma at FUV1 by age group and gender. a, ORs adjusted for the other covariates included in model 2 in Table 28.
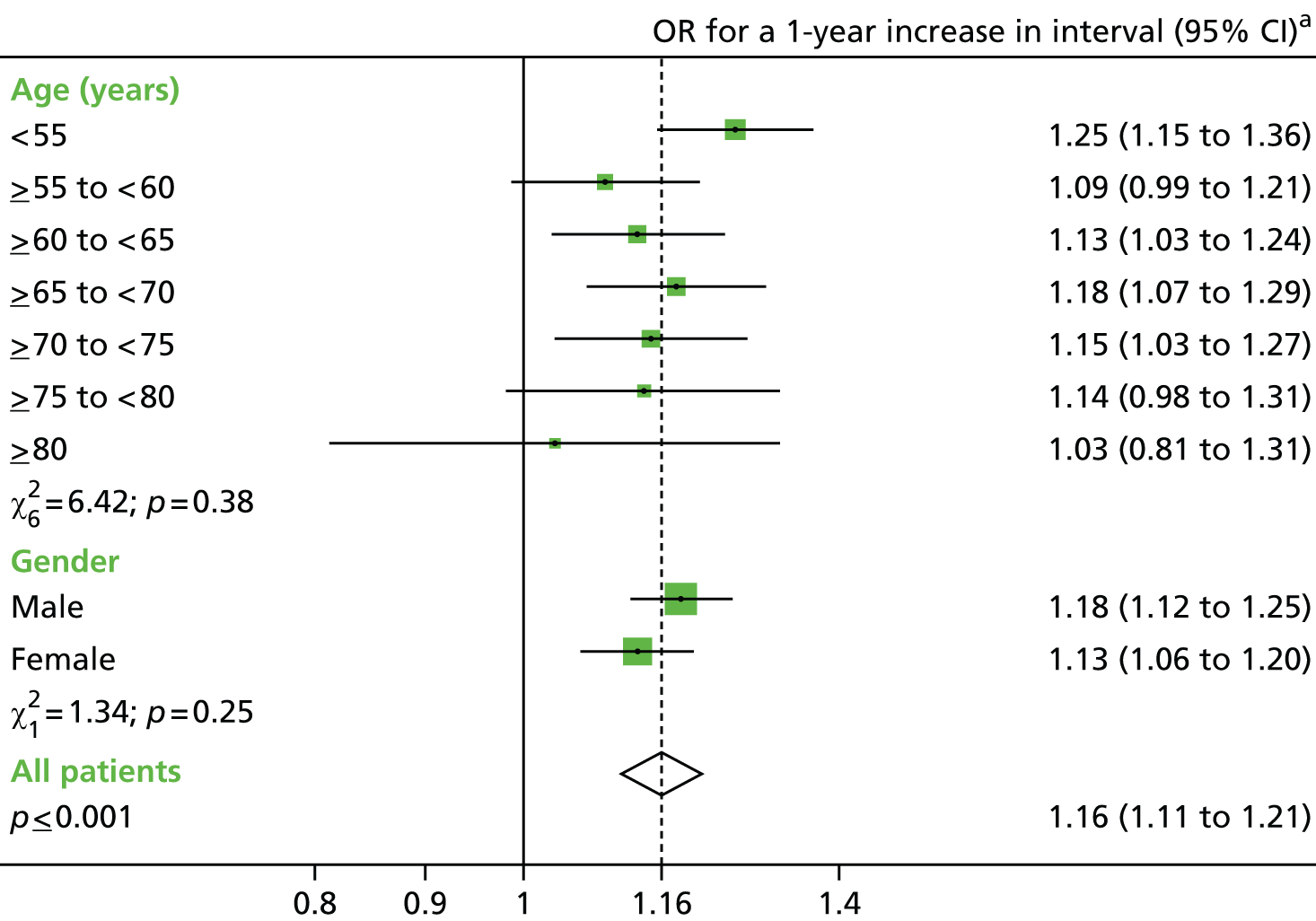
FIGURE 4.
Effect of a 1-year increase in interval on the detection of new AA at FUV1 by age group and gender. a, ORs adjusted for the other covariates included in model 2 in Table 29.
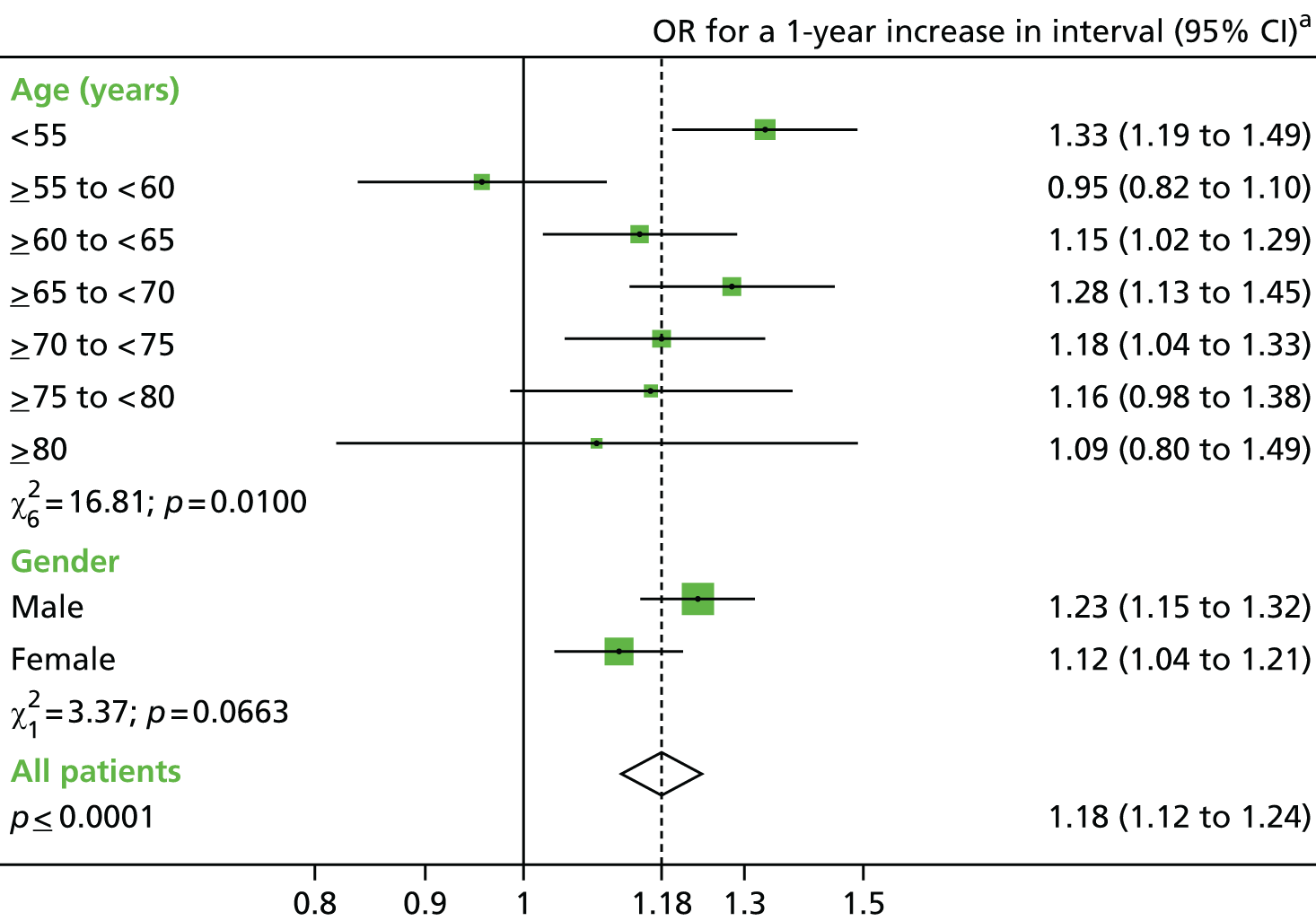
FIGURE 5.
Effect of a 1-year increase in interval on the detection of new CRC at FUV1 by age group and gender. a, ORs adjusted for the other covariates included in model 2 in Table 30.
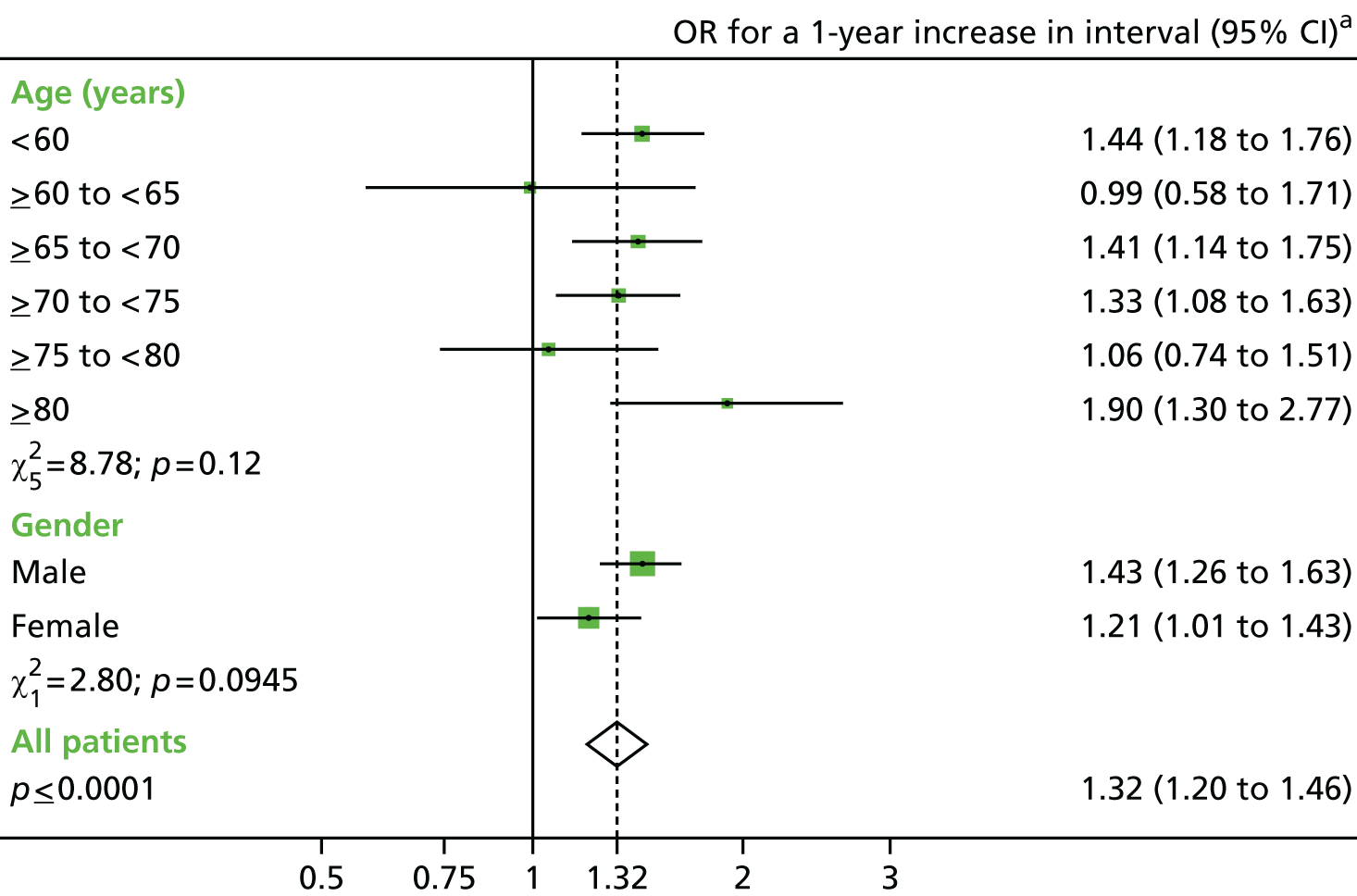
Although we detected a significant interaction between interval and age group, we did not model the interaction parameter in previously presented results, as it is likely to be impractical to offer different surveillance strategies based on age or gender in a clinical setting.
Second follow-up visit
Characteristics and findings
Of the 4608 patients who attended FUV1, 1635 (36%) patients returned for FUV2 during our data collection period and were not censored for cancer diagnosed at first follow-up.
Table 31 details the findings (new and previously seen) at FUV2 according to the interval between FUV1 and FUV2. Adenomas were detected in 527 (32%) patients, AA in 232 (14%) patients and CRC in 17 (1%) patients. The adenoma detection rate was high regardless of interval, varying from 30% to 43%, whereas the proportion of patients with AA varied from 8% to 20%, and the proportion with CRC varied from none to 3%. There was little evidence of a trend in findings with increasing interval for any of the outcomes.
| Interval FUV1 to FUV2 | Number of IR patients (N = 1635) | Patients with findingsa at FUV2 | |||||
|---|---|---|---|---|---|---|---|
| Adenoma(s) | AA(s) | CRC(s) | |||||
| n | % (n/N) | n | % (n/N) | n | % (n/N) | ||
| < 18 months | 397 | 144 | 36.27 | 79 | 19.90 | 4 | 1.01 |
| 2 yearsb | 376 | 116 | 30.85 | 55 | 14.63 | 5 | 1.33 |
| 3 yearsb | 518 | 153 | 29.54 | 63 | 12.16 | 3 | 0.58 |
| 4 yearsb | 152 | 46 | 30.26 | 14 | 9.21 | 2 | 1.32 |
| 5 yearsb | 131 | 44 | 33.59 | 11 | 8.40 | 2 | 1.53 |
| 6 yearsb | 31 | 11 | 35.48 | 4 | 12.90 | 0 | 0 |
| ≥ 6.5 years | 30 | 13 | 43.33 | 6 | 20.00 | 1 | 3.33 |
| Total | 1635 | 527 | 32.23 | 232 | 14.19 | 17 | 1.04 |
Table 32 shows the examinations undertaken at FUV2. In 89% of patients, FUV2 comprised a single procedure and was completed in 1 day. The most complete examination that we were able to glean from information provided on procedure type and polyp location was a complete colonoscopy in 74% of cases, and an incomplete colonoscopy in 15%. A further 5% are likely to have had a colonoscopy, as this is the most common procedure to offer patients undergoing surveillance in the UK, but 6% had only a FS.
| Characteristic | Category | IR patients (N = 1635) | |
|---|---|---|---|
| n | % | ||
| Number of examinations | 1 | 1460 | 89.30 |
| 2 | 125 | 7.65 | |
| 3 | 27 | 1.65 | |
| 4+ | 23 | 1.41 | |
| Length of FUV2 | 1 day | 1461 | 89.36 |
| 2–30 days | 14 | 0.86 | |
| 1–3 months | 36 | 2.20 | |
| 3–6 months | 44 | 2.69 | |
| 6–12 months | 61 | 3.73 | |
| 1–2 years | 16 | 0.98 | |
| 3–4 years | 3 | 0.18 | |
| Most complete examination at FUV2 | Complete colonoscopy | 1206 | 73.76 |
| Colonoscopy not known to be complete | 241 | 14.74 | |
| Colonoscopy or FS | 47 | 2.87 | |
| FS | 106 | 6.48 | |
| Colonoscopy, FS or rigid sigmoidoscopy | 28 | 1.71 | |
| Surgery | 6 | 0.37 | |
| Unknown procedure type | 1 | 0.06 | |
New and previously seen lesions at second follow-up
Tables 33–35 show the status of findings at FUV2 – whether or not a lesion had been seen at a previous visit – stratified by the interval from FUV1 to FUV2. Similar to findings at FUV1, there was a trend towards an increasing proportion of new findings in patients with a longer interval, and a greater proportion of previously seen lesions in those with a shorter interval.
| Adenoma status | Interval to from first to second follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 253 (63.73) | 260 (69.15) | 365 (70.46) | 106 (69.74) | 87 (66.41) | 20 (64.52) | 17 (56.67) | 1108 (67.77) |
| Previously seen only | 56 (14.11) | 23 (6.12) | 16 (3.09) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 95 (5.81) |
| New and previously seen | 13 (3.27) | 9 (2.39) | 6 (1.16) | 2 (1.32) | 0 (0) | 1 (3.23) | 0 (0) | 31(1.90) |
| New only | 75 (18.89) | 84 (22.34) | 131 (25.29) | 44 (28.95) | 44 (33.59) | 10 (32.26) | 13 (43.33) | 401 (24.53) |
| Total | 397 (100) | 376 (100) | 518 (100) | 152 (100) | 131 (100) | 31 (100) | 30 (100) | 1635 (100) |
| AA status | Interval to from first to second follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 318 (80.10) | 321 (85.37) | 455 (87.84) | 138 (90.79) | 120 (91.6) | 27 (87.10) | 24 (80) | 1403 (85.81) |
| Previously seen only | 51 (12.85) | 23 (6.12) | 12 (2.32) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 86 (5.26) |
| New and previously seen | 4 (1.01) | 5 (1.33) | 5 (0.97) | 0 (0) | 0 (0) | 1 (3.23) | 0 (0) | 15 (0.92) |
| New only | 24 (6.05) | 27 (7.18) | 46 (8.88) | 14 (9.21) | 11 (8.40) | 3 (9.68) | 6 (20) | 131 (8.01) |
| Total | 397 (100) | 376 (100) | 518 (100) | 152 (100) | 131 (100) | 31 (100) | 30 (100) | 1635 (100) |
| CRC status | Interval to from first to second follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 393 (98.99) | 371 (98.67) | 515 (99.42) | 150 (98.68) | 129 (98.47) | 31 (100) | 29 (96.67) | 1618 (98.96) |
| Previously seen only | 2 (0.50) | 1 (0.27) | 2 (0.39) | 1 (0.66) | 2 (1.53) | 0 (0) | 0 (0) | 8 (0.49) |
| New and previously seen | 2 (0.50) | 4 (1.06) | 1 (0.19) | 1 (0.66) | 0 (0) | 0 (0) | 1 (3.33) | 9 (0.55) |
| Total | 397 (100) | 376 (100) | 518 (100) | 152 (100) | 131 (100) | 31 (100) | 30 (100) | 1635 (100) |
Based on these observations, all subsequent analyses of findings at FUV2 included only new findings, so as to allow the association between interval to FUV2 and finding at FUV2 to be examined without any confounding effects of polypectomy site surveillance, as was done in the analysis of new findings at FUV1.
The proportion of patients with new adenomas was high at both the first and second follow-ups, regardless of interval length. The high detection rate of adenomas meant that this outcome was not informative in terms of identifying an optimum surveillance strategy. For this reason, adenomas were not considered as an end point in subsequent analyses for FUV2, and only AA or CRC were used as outcomes.
Follow-up visit 1 risk factors for new advanced adenomas and colorectal cancer at follow-up visit 2
Univariable analyses were performed to assess the relationship between FUV1 characteristics and detection of new AA and CRC at FUV2. Table 36 describes new AA and CRC incidence at FUV2 according to patient characteristics and examinations at FUV1. Most patient or procedural characteristics were not significantly predictive. There was weak evidence that suboptimal bowel preparation increased the odds of new AA (p = 0.0178), but again 95% CIs included 1. There was some evidence of an association between new CRC at FUV2 and a difficult examination at FUV1 (OR 5.99, 95% CI 1.22 to 29.35; p = 0.0636).
| First follow-up characteristics | Number of patients (N = 1635) | New AA at second follow-up | New CRC at second follow-up | |||||
|---|---|---|---|---|---|---|---|---|
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | n (%) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Age (years) at first follow-up | < 55 | 329 | 25 (7.6) | 1 | 0.7718 | 2 (0.61) | 1 | 0.7998 |
| ≥ 55 and < 60 | 256 | 21 (8.2) | 1.09 (0.59 to 1.99) | 0 (0) | n/a | |||
| ≥ 60 and < 65 | 279 | 30 (10.75) | 1.47 (0.84 to 2.56) | 2 (0.72) | 1.18 (0.17 to 8.44) | |||
| ≥ 65 and < 70 | 305 | 31 (10.16) | 1.38 (0.79 to 2.39) | 1 (0.33) | 0.54 (0.05 to 5.96) | |||
| ≥ 70 and < 75 | 253 | 19 (7.51) | 0.99 (0.53 to 1.84) | 1 (0.4) | 0.65 (0.06 to 7.20) | |||
| ≥ 75 and < 80 | 142 | 13 (9.15) | 1.23 (0.61 to 2.47) | 2 (1.41) | 2.34 (0.33 to 16.75) | |||
| ≥ 80 | 71 | 7 (9.86) | 1.33 (0.55 to 3.21) | 1 (1.41) | 2.34 (0.21 to 26.12) | |||
| Gender | Male | 956 | 90 (9.41) | 1 | 0.4132 | 5 (0.52) | 1 | 0.8592 |
| Female | 679 | 56 (8.25) | 0.86 (0.61 to 1.23) | 4 (0.59) | 1.13 (0.30 to 4.21) | |||
| Family history of cancer or CRC | No | 1523 | 137 (9) | 1 | 0.7273 | 8 (0.53) | 1 | 0.6393 |
| Yes | 112 | 9 (8.04) | 0.88 (0.44 to 1.79) | 1 (0.89) | 1.71 (0.21 to 13.76) | |||
| Year of first follow-up | 1985–94 | 125 | 12 (9.6) | 1 | 0.6721 | 1 (0.8) | 1 | 0.6372 |
| 1995–9 | 355 | 29 (8.17) | 0.84 (0.41 to 1.70) | 1 (0.28) | 0.35 (0.02 to 5.64) | |||
| 2000–4 | 855 | 73 (8.54) | 0.88 (0.46 to 1.67) | 4 (0.47) | 0.58 (0.06 to 5.26) | |||
| 2005–9 | 300 | 32 (10.67) | 1.12 (0.56 to 2.26) | 3 (1) | 1.25 (0.13 to 12.16) | |||
| Length of visit | 1 day | 1422 | 119 (8.37) | 1 | 0.3713 | 8 (0.56) | 1 | 0.3574 |
| 2–30 days | 18 | 2 (11.11) | 1.37 (0.31 to 6.02) | 0 (0) | n/a | |||
| 1–3 months | 37 | 5 (13.51) | 1.71 (0.65 to 4.47) | 0 (0) | n/a | |||
| 3–6 months | 58 | 5 (8.62) | 1.03 (0.41 to 2.63) | 1 (1.72) | 3.10 (0.38 to 25.21) | |||
| 6–12 months | 78 | 12 (15.38) | 1.99 (1.05 to 3.79) | 0 (0) | n/a | |||
| ≥ 12 months | 22 | 3 (13.64) | 1.73 (0.50 to 5.93) | 0 (0) | n/a | |||
| Number of examinations in visit | 1 | 1420 | 119 (8.38) | 1 | 0.147 | 8 (0.56) | 1 | 0.8762 |
| 2 | 150 | 16 (10.67) | 1.31 (0.75 to 2.27) | 1 (0.67) | 1.18 (0.15 to 9.54) | |||
| 3 | 44 | 8 (18.18) | 2.43 (1.10 to 5.35) | 0 (0) | n/a | |||
| 4+ | 21 | 3 (14.29) | 1.82 (0.53 to 6.28) | 0 (0) | n/a | |||
| Most complete examination | Complete colonoscopy | 1087 | 101 (9.29) | 1 | 0.7689 | 4 (0.37) | 1 | 0.2073 |
| Colonoscopy of unknown completeness | 130 | 8 (6.15) | 0.64 (0.30 to 1.35) | 1 (0.77) | 2.10 (0.23 to 18.92) | |||
| Incomplete colonoscopy | 145 | 12 (8.28) | 0.88 (0.47 to 1.65) | 3 (2.07) | 5.72 (1.27 to 25.82) | |||
| Colonoscopy or FS | 106 | 11 (10.38) | 1.13 (0.59 to 2.18) | 0 (0) | n/a | |||
| FS | 108 | 8 (7.41) | 0.78 (0.37 to 1.65) | 1 (0.93) | 2.53 (0.28 to 22.84) | |||
| Colonoscopy or flexible or rigid sigmoidoscopy | 53 | 6 (11.32) | 1.25 (0.52 to 2.99) | 0 (0) | n/a | |||
| Surgery | 2 | 0 (0) | n/a | 0 (0) | n/a | |||
| Unknown | 4 | 0 (0) | n/a | 0 (0) | n/a | |||
| Best bowel preparation at colonoscopy | Excellent/good | 464 | 31 (6.68) | 1 | 0.0178 | 2 (0.43) | 1 | 0.8971 |
| Satisfactory | 169 | 27 (15.98) | 2.66 (1.53 to 4.60) | 1 (0.59) | 1.38 (0.12 to 15.26) | |||
| Poor | 68 | 5 (7.35) | 1.11 (0.42 to 2.96) | 1 (1.47) | 3.45 (0.31 to 38.54) | |||
| Unknown | 661 | 58 (8.77) | 1.34 (0.85 to 2.11) | 4 (0.61) | 1.41 (0.26 to 7.71) | |||
| No known colonoscopy | 273 | 25 (9.16) | 1.41 (0.81 to 2.44) | 1 (0.37) | 0.85 (0.08 to 9.41) | |||
| Difficult examination | No | 1559 | 136 (8.72) | 1 | 0.2117 | 7 (0.45) | 1 | 0.0636 |
| Yes | 76 | 10 (13.16) | 1.59 (0.80 to 3.15) | 2 (2.63) | 5.99 (1.22 to 29.35) | |||
Table 37 describes new AA and new CRC incidence at FUV2 according to characteristics of adenomas and polyps detected at FUV1. There was a tendency towards increasing odds of new AA at FUV2 with increasing number and size of adenomas, severity of histology and proximal location of adenomas at FUV1, as well as in the presence of proximal polyps or polyps of unknown histology. Odds of new AA tended to increase with repeated sightings of an adenoma during FUV1 but the association was not significant (p = 0.0609). No significant relationship was found between new CRC at FUV2 and characteristics of adenomas or polyps seen at FUV1, a finding that was most likely due to the very small number of CRC outcomes at FUV2.
| First follow-up characteristics | Number of patients (N = 1635) | New AA at second follow-up | New CRC at second follow-up | |||||
|---|---|---|---|---|---|---|---|---|
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | n (%) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | ||||||||
| Number | 0 | 1010 | 75 (7.43) | 1 | 0.0104 | 6 (0.59) | 1 | 0.8868 |
| 1 | 429 | 39 (9.09) | 1.25 (0.83 to 1.87) | 2 (0.47) | 0.78 (0.16 to 3.90) | |||
| 2 | 117 | 20 (17.09) | 2.57 (1.50 to 4.39) | 1 (0.85) | 1.44 (0.17 to 12.09) | |||
| 3 | 37 | 4 (10.81) | 1.51 (0.52 to 4.38) | 0 | n/a | |||
| 4 | 21 | 4 (19.05) | 2.93 (0.96 to 8.94) | 0 | n/a | |||
| 5+ | 21 | 4 (19.05) | 2.93 (0.96 to 8.94) | 0 | n/a | |||
| Largest size (mm) | No adenomas | 1010 | 75 (7.43) | 1 | 0.0066 | 6 (0.59) | 1 | 0.6889 |
| < 10 | 379 | 36 (9.50) | 1.31 (0.86 to 1.98) | 3 (0.79) | 1.34 (0.33 to 5.37) | |||
| 10–14 | 95 | 8 (8.42) | 1.15 (0.54 to 2.45) | 0 (0) | n/a | |||
| 15–19 | 52 | 7 (13.46) | 1.94 (0.85 to 4.45) | 0 (0) | n/a | |||
| ≥ 20 | 75 | 15 (20.00) | 3.12 (1.69 to 5.75) | 0 (0) | n/a | |||
| Unknown | 24 | 5 (20.83) | 3.28 (1.19 to 9.03) | 0 (0) | n/a | |||
| Worst histology | No adenomas | 1010 | 75 (7.43) | 1 | 0.0476 | 6 (0.59) | 1 | 0.7327 |
| Tubular | 340 | 33 (9.71) | 1.34 (0.87 to 2.06) | 2 (0.59) | 0.99 (0.20 to 4.93) | |||
| Tubulovillous | 156 | 19 (12.18) | 1.73 (1.01 to 2.95) | 0 (0) | n/a | |||
| Villous | 66 | 10 (15.15) | 2.23 (1.09 to 4.54) | 1 (1.52) | 2.57 (0.31 to 21.70) | |||
| Unknown | 63 | 9 (14.29) | 2.08 (0.99 to 4.37) | 0 (0) | n/a | |||
| Worst dysplasia | No adenomas | 1010 | 75 (7.43) | 1 | 0.0387 | 6 (0.59) | 1 | 0.9933 |
| Low grade | 508 | 61 (12.01) | 1.70 (1.19 to 2.43) | 3 (0.59) | 0.99 (0.25 to 3.99) | |||
| High grade | 46 | 4 (8.7) | 1.19 (0.41 to 3.40) | 0 (0) | n/a | |||
| Unknown | 71 | 6 (8.45) | 1.15 (0.48 to 2.74) | 0 (0) | n/a | |||
| Distal adenomas | No adenomas | 1010 | 75 (7.43) | 1 | 0.0274 | 6 (0.59) | 1 | 0.5904 |
| No | 242 | 27 (11.16) | 1.57 (0.98 to 2.49) | 2 (0.83) | 1.39 (0.28 to 6.95) | |||
| Yes | 383 | 44 (11.49) | 1.62 (1.09 to 2.40) | 1 (0.26) | 0.44 (0.05 to 3.65) | |||
| Proximal adenomas | No adenomas | 1010 | 75 (7.43) | 1 | 0.0115 | 6 (0.59) | 1 | 0.4909 |
| No | 319 | 31 (9.72) | 1.34 (0.87 to 2.08) | 0 (0) | n/a | |||
| Yes | 306 | 40 (13.07) | 1.87 (1.25 to 2.82) | 3 (0.98) | 1.66 (0.41 to 6.66) | |||
| Number of sightings of a single adenoma | No adenomas | 1010 | 75 (7.43) | 1 | 0.0609 | 6 (0.59) | 1 | 0.9431 |
| 1 | 531 | 60 (11.3) | 1.59 (1.11 to 2.27) | 3 (0.56) | 0.95 (0.24 to 3.82) | |||
| 2 | 60 | 6 (10) | 1.39 (0.58 to 3.32) | 0 (0) | n/a | |||
| 3 | 20 | 3 (15) | 2.20 (0.63 to 7.68) | 0 (0) | n/a | |||
| 4 | 8 | 2 (25) | 4.16 (0.82 to 20.95) | 0 (0) | n/a | |||
| 5+ | 6 | 0 (0) | n/a | 0 (0) | n/a | |||
| Polyp characteristics | ||||||||
| Number of hyperplastic polyps | 0 | 1357 | 124 (9.14) | 1 | 0.6929 | 9 (0.66) | 1 | n/a |
| 1 | 173 | 13 (7.51) | 0.81 (0.45 to 1.46) | 0 (0) | n/a | |||
| 2 | 63 | 4 (6.35) | 0.67 (0.24 to 1.89) | 0 (0) | n/a | |||
| 3 | 17 | 1 (5.88) | 0.62 (0.08 to 4.73) | 0 (0) | n/a | |||
| 4 | 15 | 3 (20) | 2.49 (0.69 to 8.93) | 0 (0) | n/a | |||
| 5+ | 10 | 1 (10) | 1.10 (0.14 to 8.79) | 0 (0) | n/a | |||
| Large hyperplastic polyp | No | 1618 | 142 (8.78) | 1 | 0.0712 | 9 (0.56) | 1 | n/a |
| Yes | 17 | 4 (23.53) | 3.20 (1.03 to 9.94) | 0 (0) | n/a | |||
| Number of polyps with unknown histology | 0 | 1330 | 105 (7.89) | 1 | 0.0004 | 7 (0.53) | 1 | 0.6705 |
| 1 | 180 | 23 (12.78) | 1.71 (1.06 to 2.76) | 1 (0.56) | 1.06 (0.13 to 8.63) | |||
| 2 | 64 | 6 (9.38) | 1.21 (0.51 to 2.86) | 1 (1.56) | 3.00 (0.36 to 24.76) | |||
| 3 | 25 | 3 (12) | 1.59 (0.47 to 5.40) | 0 (0) | n/a | |||
| 4 | 13 | 0 (0) | n/a | 0 (0) | n/a | |||
| 5+ | 23 | 9 (39.13) | 7.50 (3.17 to 17.74) | 0 (0) | n/a | |||
| Distal polyps | No polyps | 667 | 44 (6.6) | 1 | 0.0046 | 5 (0.75) | 1 | 0.643 |
| No | 306 | 40 (13.07) | 2.13 (1.36 to 3.34) | 1 (0.33) | 0.43 (0.05 to 3.73) | |||
| Yes | 662 | 62 (9.37) | 1.46 (0.98 to 2.19) | 3 (0.45) | 0.60 (0.14 to 2.53) | |||
| Proximal polyps | No polyps | 667 | 44 (6.6) | 1 | 0.0006 | 5 (0.75) | 1 | 0.372 |
| No | 499 | 40 (8.02) | 1.23 (0.79 to 1.93) | 1 (0.2) | 0.27 (0.03 to 2.28) | |||
| Yes | 469 | 62 (13.22) | 2.16 (1.44 to 3.24) | 3 (0.64) | 0.85 (0.20 to 3.58) | |||
Baseline risk factors for new advanced adenomas and colorectal cancer at the second follow-up visit
The crude association of baseline characteristics with new findings at FUV2 was investigated.
Table 38 describes crude associations of patient and procedural characteristics at baseline with new AA or CRC at FUV2. Patients with an incomplete baseline colonoscopy had a twofold increased odds of AA at FUV2 (OR 2.03, 95% CI 1.24 to 3.33). There was a tendency for increased odds of AA at FUV2 with increasing interval length between baseline and FUV1, although effect estimates were imprecise and most 95% CIs included 1 (p = 0.0212). No other factors appeared to be associated with new AA at FUV2. There was little evidence of an association between any patient or procedural characteristics at baseline and detection of new CRC at FUV2, as estimates were extremely imprecise with wide CIs and non-significant test statistics.
| Baseline factors | Number of patients (N = 1635) | New AA at the second follow-up | New CRC at the second follow-up | |||||
|---|---|---|---|---|---|---|---|---|
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | n (%) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Family history of cancer | No | 1540 | 140 (9.09) | 1.00 | 0.3349 | 8 (0.52) | 1.00 | 0.5417 |
| Yes | 95 | 6 (6.32) | 0.67 (0.29 to 1.57) | 1 (1.05) | 2.04 (0.25 to 16.46) | |||
| Calendar year of baseline visit | 1985–94 | 218 | 23 (10.55) | 1.00 | 0.0644 | 1 (0.46) | 1.00 | 0.8971 |
| 1995–9 | 607 | 40 (6.59) | 0.60 (0.35 to 1.02) | 3 (0.49) | 1.08 (0.11 to 10.42) | |||
| 2000–4 | 730 | 73 (10) | 0.94 (0.57 to 1.55) | 4 (0.55) | 1.20 (0.13 to 10.75) | |||
| 2005–10 | 80 | 10 (12.5) | 1.21 (0.55 to 2.67) | 1 (1.25) | 2.75 (0.17 to 44.44) | |||
| Length of visit | 1 day | 904 | 77 (8.52) | 1.00 | 0.9364 | 4 (0.44) | 1.00 | 0.7511 |
| 2–30 days | 75 | 7 (9.33) | 1.11 (0.49 to 2.49) | 0 (0) | n/a | |||
| 1–3 months | 230 | 23 (10) | 1.19 (0.73 to 1.95) | 2 (0.87) | 1.97 (0.36 to 10.84) | |||
| 3–6 months | 195 | 16 (8.21) | 0.96 (0.55 to 1.68) | 1 (0.51) | 1.16 (0.13 to 10.43) | |||
| 6–12 months | 191 | 20 (10.47) | 1.26 (0.75 to 2.11) | 2 (1.05) | 2.38 (0.43 to 13.09) | |||
| ≥ 12 months | 40 | 3 (7.5) | 0.87 (0.26 to 2.89) | 0 (0) | n/a | |||
| Number of examinations in visit | 1 | 900 | 76 (8.44) | 1.00 | 0.2478 | 4 (0.44) | 1.00 | 0.7108 |
| 2 | 512 | 45 (8.79) | 1.04 (0.71 to 1.54) | 4 (0.78) | 1.76 (0.44 to 7.08) | |||
| 3 | 138 | 19 (13.77) | 1.73 (1.01 to 2.97) | 1 (0.72) | 1.64 (0.18 to 14.74) | |||
| 4+ | 85 | 6 (7.06) | 0.82 (0.35 to 1.95) | 0 (0) | 1 (0 to 0) | |||
| Most complete colonoscopy | Complete | 944 | 75 (7.94) | 1.00 | 0.0278 | 5 (0.53) | 1.00 | 0.0896 |
| Unknown completeness | 530 | 47 (8.87) | 1.13 (0.77 to 1.65) | 1 (0.19) | 0.36 (0.04 to 3.05) | |||
| Incomplete | 161 | 24 (14.91) | 2.03 (1.24 to 3.33) | 3 (1.86) | 3.57 (0.84 to 15.07) | |||
| Best bowel preparation at colonoscopy | Excellent/good | 465 | 36 (7.74) | 1.00 | 0.6065 | 2 (0.43) | 1.00 | 0.8677 |
| Satisfactory | 127 | 10 (7.87) | 1.02 (0.49 to 2.11) | 1 (0.79) | 1.84 (0.17 to 20.43) | |||
| Poor | 41 | 3 (7.32) | 0.94 (0.28 to 3.20) | 0 (0) | n/a | |||
| Unknown | 1002 | 97 (9.68) | 1.28 (0.86 to 1.90) | 6 (0.6) | 1.39 (0.28 to 6.94) | |||
| Difficult examination | No | 1575 | 138 (8.76) | 1.00 | 0.2516 | 9 (0.57) | n/a | n/a |
| Yes | 60 | 8 (13.33) | 1.6 (0.75 to 3.44) | 0 (0) | ||||
| Interval from baseline to first follow-up | < 18 months | 783 | 73 (9.32) | 1.00 | 0.0212 | 4 (0.51) | 1.00 | 0.4096 |
| 2 yearsa | 379 | 34 (8.97) | 0.96 (0.63 to 1.47) | 1 (0.26) | 0.52 (0.06 to 4.63) | |||
| 3 yearsa | 272 | 22 (8.09) | 0.86 (0.52 to 1.41) | 2 (0.74) | 1.44 (0.26 to 7.92) | |||
| 4 yearsa | 109 | 3 (2.75) | 0.28 (0.09 to 0.89) | 2 (1.83) | 3.64 (0.66 to 20.11) | |||
| 5 yearsa | 45 | 7 (15.56) | 1.79 (0.77 to 4.16) | 0 (0) | n/a | |||
| 6 yearsa | 22 | 1 (4.55) | 0.46 (0.06 to 3.49) | 0 (0) | n/a | |||
| ≥ 6.5 years | 25 | 6 (24) | 3.07 (1.19 to 7.93) | 0 (0) | n/a | |||
Table 39 describes the characteristics of polyps and adenomas detected at baseline, by whether patients had new AA or CRC found at FUV2. There was no association between any baseline adenoma or polyp characteristic and detection of new AA at FUV2. Similarly, no baseline polyp characteristic was a significant predictor of new CRC at FUV2, although this was affected by the small number of CRCs found at FUV2.
| Baseline factors | Number of patients (N = 1635) | New AA at the second follow-up | New CRC at the second follow-up | |||||
|---|---|---|---|---|---|---|---|---|
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | n (%) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | ||||||||
| Number | 1 | 1138 | 92 (8.08) | 1.00 | 0.3000 | 7 (0.62) | 1.00 | 0.8249 |
| 2 | 387 | 44 (11.37) | 1.46 (1.00 to 2.13) | 2 (0.52) | 0.84 (0.17 to 4.06) | |||
| 3 | 67 | 6 (8.96) | 1.12 (0.47 to 2.66) | 0 (0) | n/a | |||
| 4 | 43 | 4 (9.3) | 1.17 (0.41 to 3.34) | 0 (0) | n/a | |||
| Largest size (mm) | < 10 | 110 | 10 (9.09) | 1.00 | 0.9819 | 0 (0) | n/a | 0.3931 |
| 10–14 | 523 | 45 (8.6) | 0.94 (0.46 to 1.93) | 5 (0.96) | 1.00 | |||
| 15–19 | 343 | 30 (8.75) | 0.96 (0.45 to 2.03) | 1 (0.29) | 0.30 (0.04 to 2.60) | |||
| ≥ 20 | 659 | 61 (9.26) | 1.02 (0.51 to 2.06) | 3 (0.46) | 0.47 (0.11 to 1.99) | |||
| Worst histology | Tubular | 562 | 42 (7.47) | 1.00 | 0.1129 | 3 (0.53) | 1.00 | 0.7065 |
| Tubulovillous | 740 | 63 (8.51) | 1.15 (0.77 to 1.73) | 3 (0.41) | 0.76 (0.15 to 3.77) | |||
| Villous | 175 | 21 (12) | 1.69 (0.97 to 2.94) | 1 (0.57) | 1.07 (0.11 to 10.36) | |||
| Unknown | 158 | 20 (12.66) | 1.79 (1.02 to 3.16) | 2 (1.27) | 2.39 (0.40 to 14.42) | |||
| Worst dysplasia | Low grade | 1152 | 102 (8.85) | 1.00 | 0.0907 | 7 (0.61) | 1.00 | 0.4685 |
| High grade | 305 | 21 (6.89) | 0.76 (0.47 to 1.24) | 0 (0) | n/a | |||
| Unknown | 178 | 23 (12.92) | 1.53 (0.94 to 2.48) | 2 (1.12) | 1.86 (0.38 to 9.02) | |||
| Distal | No | 339 | 27 (7.96) | 1.00 | 0.4785 | 2 (0.59) | 1.00 | 0.9128 |
| Yes | 1296 | 119 (9.18) | 1.17 (0.76 to 1.81) | 7 (0.54) | 0.92 (0.19 to 4.42) | |||
| Proximal | No | 1227 | 108 (8.8) | 1.00 | 0.7545 | 7 (0.57) | 1.00 | 0.8475 |
| Yes | 408 | 38 (9.31) | 1.06 (0.72 to 1.57) | 2 (0.49) | 0.86 (0.18 to 4.15) | |||
| Number of sightings of a unique adenoma | 1 | 1171 | 99 (8.45) | 1.00 | 0.7397 | 5 (0.43) | 1.00 | 0.1454 |
| 2 | 341 | 35 (10.26) | 1.24 (0.83 to 1.86) | 4 (1.17) | 2.77 (0.74 to 10.37) | |||
| 3 | 75 | 7 (9.33) | 1.11 (0.50 to 2.49) | 0 (0) | n/a | |||
| 4 | 30 | 4 (13.33) | 1.67 (0.57 to 4.87) | 0 (0) | n/a | |||
| 5+ | 18 | 1 (5.56) | 0.64 (0.08 to 4.84) | 0 (0) | n/a | |||
| Polyp characteristics | ||||||||
| Number of hyperplastic polyps | 0 | 1325 | 110 (8.3) | 1.00 | 0.3793 | 8 (0.6) | 1.00 | 0.8990 |
| 1 | 189 | 22 (11.64) | 1.46 (0.90 to 2.36) | 1 (0.53) | 0.88 (0.11 to 7.04) | |||
| 2 | 55 | 4 (7.27) | 0.87 (0.31 to 2.44) | 0 (0) | n/a | |||
| 3 | 24 | 3 (12.5) | 1.58 (0.46 to 5.37) | 0 (0) | n/a | |||
| 4 | 11 | 2 (18.18) | 2.45 (0.52 to 11.5) | 0 (0) | n/a | |||
| 5+ | 31 | 5 (16.13) | 2.12 (0.80 to 5.64) | 0 (0) | n/a | |||
| Any large hyperplastic polyps | No | 1603 | 141 (8.8) | 1.00 | 0.2196 | 9 (0.56) | 1.00 | n/a |
| Yes | 32 | 5 (15.63) | 1.92 (0.73 to 5.06) | 0 (0) | n/a | |||
| Number of polyps with unknown histology | 0 | 1297 | 104 (8.02) | 1.00 | 0.1923 | 7 (0.54) | 1.00 | 0.4981 |
| 1 | 174 | 20 (11.49) | 1.49 (0.90 to 2.47) | 1 (0.57) | 1.07 (0.13 to 8.71) | |||
| 2 | 56 | 8 (14.29) | 1.91 (0.88 to 4.15) | 0 (0) | n/a | |||
| 3 | 41 | 5 (12.2) | 1.59 (0.61 to 4.15) | 1 (2.44) | 4.61 (0.55 to 38.34) | |||
| 4 | 25 | 2 (8) | 1.00 (0.23 to 4.29) | 0 (0) | n/a | |||
| 5+ | 42 | 7 (16.67) | 2.29 (0.99 to 5.29) | 0 (0) | n/a | |||
| Distal polyps | No | 268 | 20 (7.46) | 1.00 | 0.3468 | 2 (0.75) | 1.00 | 0.6495 |
| Yes | 1367 | 126 (9.22) | 1.26 (0.77 to 2.06) | 7 (0.51) | 0.68 (0.14 to 3.31) | |||
| Proximal polyps | No | 1087 | 92 (8.46) | 1.00 | 0.3555 | 5 (0.46) | 1.00 | 0.4955 |
| Yes | 548 | 54 (9.85) | 1.18 (0.83 to 1.68) | 4 (0.73) | 1.59 (0.43 to 5.95) | |||
Follow-up visit 1 risk factors and interval
The association between FUV1 risk factors and interval between FUV1 and FUV2 was examined to identify potential confounders of the association between interval and new AA or CRC at FUV2 (see Appendix 9 for tables of results). Most FUV1 characteristics were significantly associated with interval at the 1% level. A greater proportion of patients of an older age – or with a FS, poor bowel preparation, a difficult examination at FUV1 or a long visit comprising multiple examinations – had a shorter interval. Additionally, a greater proportion of patients with multiple adenomas, multiple sightings of a single adenoma, detection of an adenoma of a larger size or with villous histology or severe dysplasia had a shorter interval.
Effect of interval on new findings at second follow-up
The effect of interval to second follow-up on new findings at FUV2 was examined using univariable and multivariable analyses. As so few CRCs were found at FUV2, new AA and CRC were combined and new AN was treated as the outcome measure instead.
Table 40 shows the association between interval from FUV1 to FUV2 and new AN at the second follow-up. In the crude analysis, there was a tendency towards increasing odds of new AN with increasing interval to FUV2; however, the relationship was not statistically significant (p = 0.2313) and most 95% CIs included 1. When interval was modelled as a continuous variable, there was a borderline significant 11% increased odds for every year increase in interval (OR 1.11, 95% CI 1.00 to 1.24; p = 0.0501).
| Baseline and FUV1 risk factors | Category | Number of patients (N = 1635) | Univariable analysis: new AN | Multivariable analyses: new AN | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 1635) | Model 2 – interval as continuous (n = 1635) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval FUV1 to FUV2 | < 18 months | 397 | 29 (7.30) | 1 | 0.2313 | 1 | 0.0164 | n/a | |
| 2 yearsa | 376 | 35 (9.31) | 1.30 (0.78 to 2.18) | 1.62 (0.93 to 2.81) | |||||
| 3 yearsa | 518 | 52 (10.04) | 1.42 (0.88 to 2.28) | 2.02 (1.19 to 3.42) | |||||
| 4 yearsa | 152 | 15 (9.87) | 1.39 (0.72 to 2.67) | 2.45 (1.20 to 5.00) | |||||
| 5 yearsa | 131 | 11 (8.40) | 1.16 (0.56 to 2.40) | 2.01 (0.91 to 4.44) | |||||
| 6 yearsa | 31 | 4 (12.90) | 1.88 (0.62 to 5.74) | 2.76 (0.84 to 9.12) | |||||
| ≥ 6.5 years | 30 | 7 (23.33) | 3.86 (1.53 to 9.76) | 5.95 (2.15 to 16.46) | |||||
| Per year increase | 1.11 (1 to 1.24) | 0.0501 | n/a | 1.22 (1.09 to 1.36) | 0.0010 | ||||
| Most complete baseline colonoscopy | Complete | 944 | 78 (8.26) | 1 | 0.0065 | 1 | 0.0099 | 1 | 0.0082 |
| Unknown completeness | 530 | 48 (9.06) | 1.11 (0.76 to 1.61) | 1.06 (0.64 to 1.73) | 1.05 (0.64 to 1.71) | ||||
| Incomplete | 161 | 27 (16.77) | 2.24 (1.39 to 3.59) | 2.33 (1.37 to 3.97) | 2.36 (1.39 to 4.00) | ||||
| Largest adenoma at FUV1 (mm) | No adenomas | 1010 | 79 (7.82) | 1 | 0.0028 | 1 | 0.0112 | 1 | 0.0159 |
| < 20 | 526 | 54 (10.27) | 1.35 (0.94 to 1.94) | 0.98 (0.54 to 1.76) | 0.93 (0.52 to 1.67) | ||||
| ≥ 20 | 75 | 15 (20.00) | 2.95 (1.60 to 5.43) | 3.20 (1.42 to 7.21) | 2.93 (1.32 to 6.51) | ||||
| Unknown | 24 | 5 (20.83) | 3.10 (1.13 to 8.53) | 2.31 (0.71 to 7.50) | 2.14 (0.67 to 6.91) | ||||
| Proximal polyp at FUV1 | No polyps | 667 | 47 (7.05) | 1 | 0.0005 | 1 | 0.0286 | 1 | 0.0293 |
| No | 499 | 41 (8.22) | 1.18 (0.76 to 1.83) | 0.75 (0.41 to 1.36) | 0.76 (0.42 to 1.39) | ||||
| Yes | 469 | 65 (13.86) | 2.12 (1.43 to 3.15) | 1.39 (0.72 to 2.67) | 1.42 (0.74 to 2.71) | ||||
| Number of polyps with unknown histology at FUV1 | 0 | 1330 | 110 (8.27) | 1 | 0.0001 | 1 | 0.001 | 1 | 0.0012 |
| 1–4 | 282 | 34 (12.06) | 1.52 (1.01 to 2.29) | 1.39 (0.83 to 2.33) | 1.34 (0.80 to 2.23) | ||||
| 5+ | 23 | 9 (39.13) | 7.13 (3.02 to 16.85) | 7.16 (2.69 to 19.06) | 6.94 (2.63 to 18.26) | ||||
| Number of adenomas at baseline and FUV1 | 1 | 844 | 58 (6.87) | 1 | 0.0003 | 1 | 0.0183 | 1 | 0.0164 |
| 2+ | 791 | 95 (12.01) | 1.85 (1.31 to 2.60) | 1.71 (1.10 to 2.66) | 1.72 (1.11 to 2.67) | ||||
| Number of polyps with unknown histology at baseline | 0 | 1297 | 109 (8.40) | 1 | 0.0124 | 1 | 0.0256 | 1 | 0.0286 |
| 1+ | 338 | 44 (13.02) | 1.63 (1.12 to 2.37) | 1.65 (1.07 to 2.55) | 1.63 (1.06 to 2.51) | ||||
Logistic regression was used to identify independent risk factors for having new AN at FUV2, and to adjust the effect of interval for covariates. Interval was modelled as a categorical variable (model 1) and as a continuous variable (model 2). Appendix 9 contains details of the models fitted. Baseline and FUV1 risk factors were adjusted for in turn (models A and B) and in combination (model C), for interval as a categorical and continuous variable. The cumulative effect of factors across baseline and FUV1 were also adjusted for (model D), as well as a combination of individual and cumulative baseline and FUV1 factors (model E), with interval as a categorical and continuous variable. When the fits of models A–E were compared, with interval as categorical or continuous, model E was found to be the best in terms of its fit to the data. Measures of fit used to assess the models were the AIC and the Bayesian information criterion (BIC) (see Appendix 9 for additional results from other models and measures of fit).
After adjusting for covariates (model E, with interval as categorical and continuous), the effect of interval was strengthened and the association with new AN at FUV2 became statistically significant, with evidence of negative confounding. When interval was modelled as a categorical variable (model 1), there was an increased odds of new AN with increasing interval length (p = 0.0164) and a 22% increased odds per year increase in interval was seen when interval was modelled as a continuous variable (OR 1.22, 95% CI 1.09 to 1.36; p = 0.001); although some effect estimates were imprecise, the small p-values, large effect sizes and tendency towards a dose–response relationship provided strong evidence of an association.
Other risk factors for AN at FUV2 included the detection of a ≥ 20 mm adenoma, proximal polyp or multiple polyps with unknown histology at FUV1, or an incomplete colonoscopy or one or more polyps with unknown histology at baseline. The detection of two or more adenomas across baseline and FUV1 (cumulative) was also associated with an increased odds of AN at FUV2 (p < 0.02). Models 1 and 2 were very similar, with the same risk factors identified in each.
Effect modification of the association between interval and new findings at follow-up visit 2
A priori, we had proposed that there might be a difference in the effect of interval length on findings by age or gender at FUV2 and, to investigate this, we fitted an interaction between continuous interval and age group or gender for the outcome of new AN at FUV2. Results are presented in Figure 6; there were no significant differences between age groups or between males and females in the effect of increasing interval length.
FIGURE 6.
Effect of a 1-year increase in interval on the detection of new AN at FUV2 by age group and gender. a, ORs adjusted for the other covariates included in model 2 in Table 40.
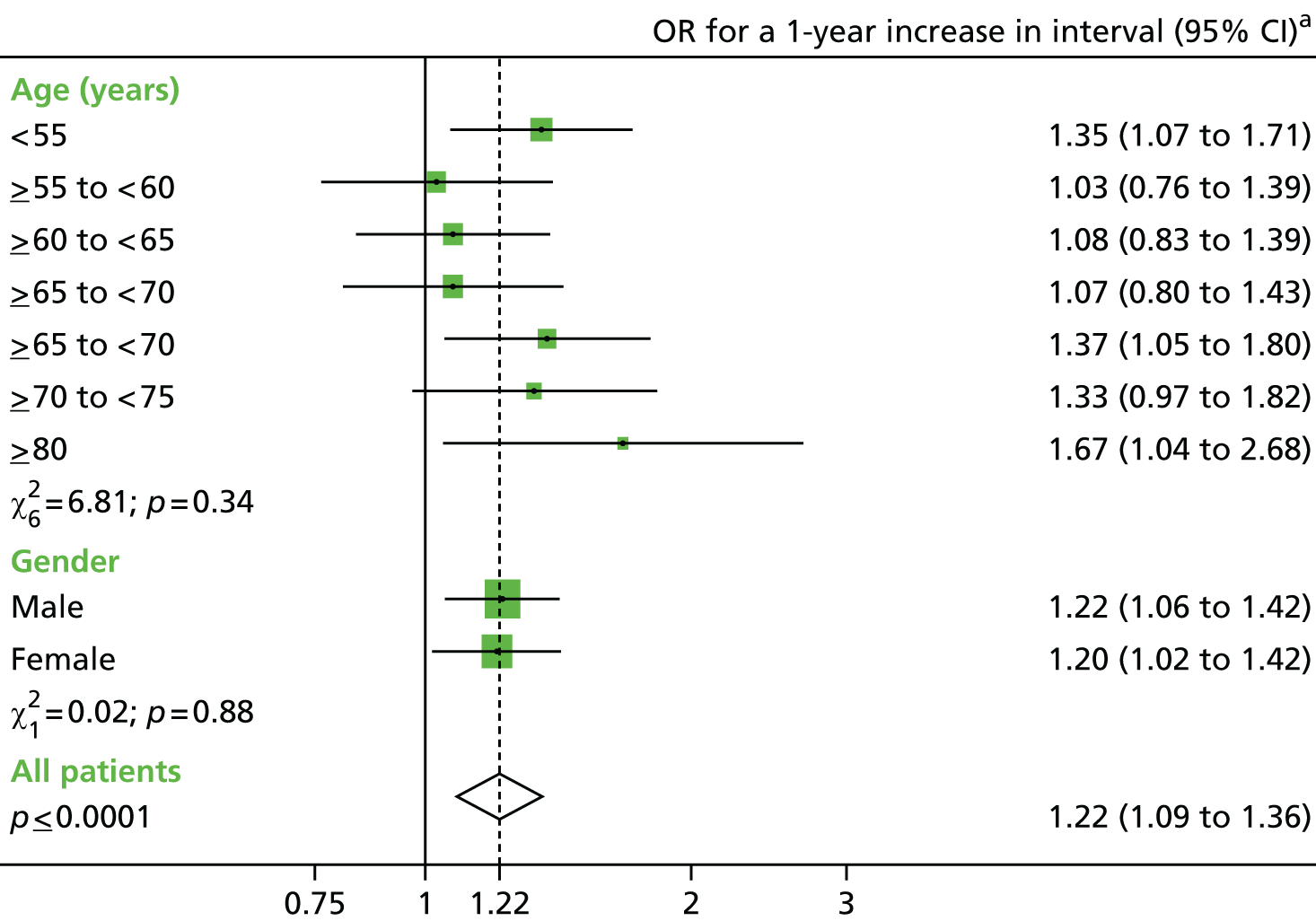
Long-term cancer risk
A survival analysis was used to assess the incidence of CRC after both baseline (see Colorectal cancer risk after baseline, below) and FUV1 (see Colorectal cancer risk after the first follow-up visit, below) to determine the combined effects on future CRC risk of surveillance visits and baseline findings for the former and surveillance visits and both baseline and first follow-up findings for the latter.
The entire IA cohort comprised 11,944 patients for the analysis of CRC incidence after baseline and 4517 patients with at least one follow-up – who remained free of CRC at FUV1 – for the analysis of CRC incidence after FUV1.
The cohort was analysed using all observation time after baseline to assess whether or not surveillance had a protective effect against CRC. If CRC was diagnosed at a follow-up visit, that follow-up visit was not counted, as it could not have offered any protection against CRC.
Colorectal cancer risk after baseline
Overall, 168 CRCs developed during 81,442 pys of observation time after baseline (median 6.0 years, IQR 3.8–9.2 years), giving an incidence rate of 206 (95% CI 177 to 240) per 100,000 pys at risk.
Univariable analysis
The relationship between patient, procedural and polyp characteristics and long-term CRC incidence was first investigated by determining incidence rates of CRC after baseline and crude HRs.
Table 41 shows CRC incidence stratified by baseline demographic and procedural characteristics. Older age was a strong predictor of CRC (p < 0.0001), with a more than fourfold increased rate among those aged 75–80 years (HR 4.79, 95% CI 2.71 to 8.84). Patients whose best baseline colonoscopy was incomplete were at an almost threefold increased risk (HR 2.78, 95% CI 1.94 to 3.98), and those with only poor preparation at baseline had a more than twofold increased risk of CRC (HR 2.40, 95% CI 1.32 to 4.39), although the overall effect of bowel preparation was not significant (p = 0.0597). Similarly, patients with a difficult examination had twice the rate of CRC (HR 2.06, 95% CI 1.25 to 3.41). No association was found between CRC and gender, family history of cancer, year of baseline, length of baseline, number of examinations in the baseline visit or hospital attended (results for hospital not presented).
| Baseline factors | Number of patients (N = 11,944) | IR patients with long-term follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) | |||||||
| pys | n = 168 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Age (years) at baseline | < 55 | 2122 | 17,900.05 | 19 | 106.14 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 1321 | 10,475.16 | 10 | 95.46 | 0.95 (0.44 to 2.04) | ||
| ≥ 60 and < 65 | 1858 | 13,308.84 | 20 | 150.28 | 1.53 (0.81 to 2.87) | ||
| ≥ 65 and < 70 | 2171 | 14,190.37 | 39 | 274.83 | 2.95 (1.70 to 5.14) | ||
| ≥ 70 and < 75 | 1786 | 11,579.17 | 27 | 233.18 | 2.54 (1.40 to 4.59) | ||
| ≥ 75 and < 80 | 1416 | 8108.39 | 34 | 419.32 | 4.79 (2.71 to 8.48) | ||
| ≥ 80 | 1270 | 5879.72 | 19 | 323.14 | 4.00 (2.09 to 7.66) | ||
| Gender | Male | 6625 | 44,061.76 | 95 | 215.61 | 1 | 0.4955 |
| Female | 5319 | 37,379.96 | 73 | 195.29 | 0.90 (0.66 to 1.22) | ||
| Family history of cancer | No | 11,445 | 77,544.37 | 160 | 206.33 | 1 | 0.9365 |
| Yes | 499 | 3897.34 | 8 | 205.27 | 0.97 (0.48 to 1.98) | ||
| Calendar year of baseline | 1985–94 | 439 | 6400.35 | 22 | 343.73 | 1 | 0.2389 |
| 1995–9 | 1430 | 15,648.51 | 43 | 274.79 | 0.85 (0.49 to 1.48) | ||
| 2000–4 | 4251 | 33,510.37 | 64 | 190.99 | 0.66 (0.38 to 1.15) | ||
| 2005–10 | 5824 | 25,882.49 | 39 | 150.68 | 0.57 (0.31 to 1.04) | ||
| Length of baseline visit | 1 day | 6836 | 46,087.39 | 83 | 180.09 | 1 | 0.5751 |
| 2–30 days | 734 | 4481.26 | 11 | 245.47 | 1.41 (0.75 to 2.64) | ||
| 1–3 months | 1643 | 11,217.87 | 26 | 231.77 | 1.32 (0.85 to 2.05) | ||
| 3–6 months | 1382 | 9815.99 | 24 | 244.50 | 1.37 (0.87 to 2.16) | ||
| 6–12 months | 1177 | 8560.31 | 21 | 245.32 | 1.35 (0.84 to 2.19) | ||
| ≥ 12 months | 172 | 1278.89 | 3 | 234.58 | 1.28 (0.41 to 4.06) | ||
| Number of examinations in baseline visit | 1 | 6826 | 45,984.04 | 83 | 180.50 | 1 | 0.1909 |
| 2 | 3788 | 26,357.29 | 64 | 242.82 | 1.36 (0.98 to 1.88) | ||
| 3 | 908 | 6200.41 | 12 | 193.54 | 1.09 (0.60 to 2.00) | ||
| 4+ | 422 | 2899.97 | 9 | 310.35 | 1.74 (0.87 to 3.46) | ||
| Completeness of colonoscopy | Complete | 9016 | 56,749.44 | 95 | 167.40 | 1 | < 0.0001 |
| Unknown | 1601 | 15,605.39 | 29 | 185.83 | 0.97 (0.63 to 1.48) | ||
| Incomplete | 1327 | 9086.89 | 44 | 484.21 | 2.78 (1.94 to 3.98) | ||
| Best bowel preparation at colonoscopy | Excellent/good | 3956 | 26,442.16 | 44 | 166.40 | 1 | 0.0597 |
| Satisfactory | 1922 | 10,317.55 | 22 | 213.23 | 1.39 (0.83 to 2.33) | ||
| Poor | 671 | 3660.88 | 14 | 382.42 | 2.40 (1.32 to 4.39) | ||
| Unknown | 5395 | 41,021.13 | 88 | 214.52 | 1.22 (0.85 to 1.75) | ||
| Difficult examination | No | 11,229 | 77,084.73 | 151 | 195.89 | 1 | 0.0101 |
| Yes | 715 | 4356.99 | 17 | 390.18 | 2.06 (1.25 to 3.41) | ||
Table 42 shows CRC incidence stratified by adenoma or polyp characteristics at baseline. Detection of an adenoma with HGD (HR 1.76, 95% CI 1.23–2.53) or a proximally located polyp (HR 1.53, 95% CI 1.13 to 2.08; p = 0.0066) or adenoma (HR 1.55, 95% CI 1.13 to 2.12; p = 0.0082) were significant predictors. Tubulovillous or villous histology and unknown histology were significantly associated with increased CRC risk. There was weak evidence that a large adenoma increased risk of CRC with a tendency towards increasing risk of CRC with increasing size.
| Baseline factors | Number of patients (N = 11,944) | IR patients with long-term follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) | |||||||
| pys | n = 168 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | |||||||
| Number | 1 | 7842 | 54,992.05 | 115 | 209.12 | 1 | 0.0816 |
| 2 | 3073 | 19,841.53 | 47 | 236.88 | 1.18 (0.84 to 1.65) | ||
| 3 | 748 | 4701.77 | 5 | 106.34 | 0.53 (0.22 to 1.30) | ||
| 4 | 281 | 1906.36 | 1 | 52.46 | 0.26 (0.04 to 1.85) | ||
| Largest size (mm) | < 10 | 1029 | 6608.13 | 6 | 90.80 | 1 | 0.0760 |
| 10–14 | 4417 | 29,913.84 | 61 | 203.92 | 2.19 (0.95 to 5.07) | ||
| 15–19 | 2440 | 16,965.60 | 33 | 194.51 | 2.07 (0.87 to 4.94) | ||
| ≥ 20 | 4058 | 27,954.14 | 68 | 243.26 | 2.60 (1.13 to 6.00) | ||
| Worst histology | Tubular | 4742 | 32,214.79 | 48 | 149.00 | 1 | 0.0098 |
| Tubulovillous | 5576 | 37,064.54 | 83 | 223.93 | 1.51 (1.06 to 2.16) | ||
| Villous | 1142 | 7611.79 | 18 | 236.48 | 1.59 (0.92 to 2.73) | ||
| Unknown | 484 | 4550.59 | 19 | 417.53 | 2.44 (1.41 to 4.21) | ||
| Worst dysplasia | Low grade | 9476 | 63,137.92 | 111 | 175.81 | 1 | 0.0077 |
| High grade | 1994 | 12,964.32 | 40 | 308.54 | 1.76 (1.23 to 2.53) | ||
| Unknown | 474 | 5339.47 | 17 | 318.38 | 1.51 (0.89 to 2.57) | ||
| Distal | No | 2448 | 16,295.31 | 37 | 227.06 | 1 | 0.4977 |
| Yes | 9496 | 65,146.41 | 131 | 201.09 | 0.88 (0.61 to 1.27) | ||
| Proximal | No | 8294 | 58,758.82 | 107 | 182.10 | 1 | 0.0082 |
| Yes | 3650 | 22,682.89 | 61 | 268.93 | 1.55 (1.13 to 2.12) | ||
| Number of sightings of a unique adenoma | 1 | 8807 | 60,393.43 | 118 | 195.39 | 1 | 0.4385 |
| 2 | 2548 | 16,965.28 | 38 | 223.99 | 1.17 (0.81 to 1.69) | ||
| 3 | 390 | 2696.76 | 6 | 222.49 | 1.15 (0.51 to 2.61) | ||
| 4 | 108 | 764.42 | 4 | 523.27 | 2.70 (1.00 to 7.31) | ||
| 5+ | 91 | 621.82 | 2 | 321.64 | 1.64 (0.41 to 6.65) | ||
| Polyp characteristics | |||||||
| Number of hyperplastic polyps | 0 | 9874 | 67,518.89 | 143 | 211.79 | 1 | 0.7086 |
| 1 | 1307 | 8862.23 | 18 | 203.11 | 0.98 (0.60 to 1.60) | ||
| 2 | 405 | 2656.32 | 3 | 112.94 | 0.55 (0.18 to 1.72) | ||
| 3 | 152 | 1002.09 | 1 | 99.79 | 0.49 (0.07 to 3.53) | ||
| 4 | 76 | 520.54 | 2 | 384.21 | 1.83 (0.45 to 7.40) | ||
| 5+ | 130 | 881.65 | 1 | 113.42 | 0.55 (0.08 to 3.92) | ||
| Any large hyperplastic polyps? | No | 11,761 | 80,155.72 | 166 | 207.10 | 1 | 0.6897 |
| Yes | 183 | 1286.00 | 2 | 155.52 | 0.76 (0.19 to 3.07) | ||
| Number of polyps with unknown histology | 0 | 9322 | 64,395.55 | 135 | 209.64 | 1 | 0.6239 |
| 1 | 1510 | 9781.43 | 15 | 153.35 | 0.75 (0.44 to 1.27) | ||
| 2 | 517 | 3266.29 | 8 | 244.93 | 1.21 (0.59 to 2.46) | ||
| 3 | 249 | 1650.11 | 5 | 303.01 | 1.45 (0.59 to 3.55) | ||
| 4 | 129 | 849.83 | 3 | 353.01 | 1.68 (0.53 to 5.26) | ||
| 5+ | 217 | 1498.49 | 2 | 133.47 | 0.62 (0.15 to 2.50) | ||
| Distal polyp | No | 1980 | 13,188.16 | 32 | 242.64 | 1 | 0.3053 |
| Yes | 9964 | 68,253.55 | 136 | 199.26 | 0.81 (0.55 to 1.20) | ||
| Proximal polyp | No | 7369 | 52,583.80 | 93 | 176.86 | 1 | 0.0066 |
| Yes | 4575 | 28,857.92 | 75 | 259.89 | 1.53 (1.13 to 2.08) | ||
The unadjusted effect of surveillance on CRC incidence after baseline is presented in Table 43. Surveillance was found to have a significant protective effect on future CRC risk, with a 46% reduction in risk with one follow-up visit (HR 0.54, 95% CI 0.37 to 0.80) and a 61% reduction with two or more visits (HR 0.39, 95% CI 0.22 to 0.66), both in comparison with no follow-up visits.
| Number of follow-up visits after baselinea | Number of patients (N = 11,944) | IR patients with long-term follow-up | ||||
|---|---|---|---|---|---|---|
| CRC(s) | ||||||
| pys | n = 168 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | ||
| 0 | 7427 | 48,891.70 | 108 | 220.90 | 1 | 0.0002 |
| 1 | 2901 | 21,030.19 | 38 | 180.69 | 0.54 (0.37 to 0.80) | |
| 2+ | 1616 | 11,519.83 | 22 | 190.97 | 0.39 (0.22 to 0.66) | |
Multivariable analysis
Cox proportional hazards regression modelling was used to examine the effect of surveillance on CRC risk, controlling for the confounding effects of baseline factors.
Table 44 presents the results of the Cox regression using the full cohort and all available follow-up time from the baseline visit. The model provided strong evidence of the beneficial effect of surveillance (p = 0.0001), with a significant 49% lower CRC incidence with one follow-up visit compared with no surveillance (HR 0.51, 95% CI 0.34 to 0.77). Having more than one surveillance examination offered additional protection against CRC, with a 68% lower incidence after attendance at two or more follow-ups (HR 0.32, 95% CI 0.17 to 0.61). As there was only a further 19% reduction in incidence associated with two or more follow-ups, much of the protective effect appeared to be contributed by the initial follow-up examination.
| Baseline risk factor | Category | Adjusted HR (95% CI) | p-value (LRT) |
|---|---|---|---|
| Number of follow-up visits after baselineb | 0 | 1 | 0.0001 |
| 1 | 0.51 (0.34 to 0.77) | ||
| 2+ | 0.32 (0.17 to 0.61) | ||
| Largest adenoma (mm) | < 10 | 1 | 0.0177 |
| 10–19 | 2.93 (1.18 to 7.31) | ||
| ≥ 20 | 3.16 (1.24 to 8.02) | ||
| Worst adenoma dysplasia | Low grade | 1 | 0.0107 |
| High grade | 1.66 (1.14 to 2.41) | ||
| Completeness of colonoscopy | Complete | 1 | 0.0002 |
| Incomplete/unknown | 1.92 (1.37 to 2.69) | ||
| Proximal polyps | No | 1 | 0.0002 |
| Yes | 1.91 (1.37 to 2.68) | ||
| Age (years) at baseline | < 55 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 0.96 (0.42 to 2.17) | ||
| ≥ 60 and < 65 | 1.42 (0.72 to 2.82) | ||
| ≥ 65 and < 70 | 2.50 (1.37 to 4.58) | ||
| ≥ 70 and < 75 | 2.47 (1.32 to 4.63) | ||
| ≥ 75 and < 80 | 3.92 (2.13 to 7.22) | ||
| ≥ 80 | 3.23 (1.64 to 6.38) |
An increased rate of CRC was independently associated with older age, as well as with having an incomplete colonoscopy or proximal polyps at baseline; both of the latter were estimated to confer an almost twofold increase in risk (see Table 44; p < 0.0001). HGD and large adenoma size were also independently predictive.
Colorectal cancer risk after the first follow-up visit
To assess the effect of additional surveillance on CRC risk after FUV1 accounting for findings at both baseline and FUV1, an analysis was performed using 4517 patients who had at least one follow-up visit and were free of CRC at their first follow-up. In these patients, 60 CRCs were diagnosed during 32,550 pys of follow-up time (184 per 100,000 pys); 38 CRCs were diagnosed after the occurrence of just one follow-up and 22 after two or more.
Univariable analysis
We first examined the effect of factors found at FUV1 on CRC risk after FUV1 and then examined whether or not any baseline factors could have affected risk.
Effect of follow-up visit 1 factors on future risk of colorectal cancer
Colorectal cancer incidence after FUV1 was stratified by demographic and procedural characteristics at FUV1 (Table 45). Older age was strongly associated with increased CRC risk (p = 0.0067), as was having a difficult examination (HR 3.98, 95% CI 2.02 to 7.88). There was some evidence of an association with number of examinations at FUV1 (p = 0.0197), although the effect estimates were imprecise. No other FUV1 risk factors were significant.
| First follow-up factors | Number of patients (N = 4517) | IR patients with long-term follow-up after first follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) after first follow-up | |||||||
| pys | n = 60 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Age (years) at first follow-up | < 55 | 722 | 6221.02 | 6 | 96.45 | 1 | 0.0067 |
| ≥ 55 and < 60 | 577 | 4714.19 | 6 | 127.28 | 1.39 (0.45 to 4.30) | ||
| ≥ 60 and < 65 | 720 | 5486.11 | 5 | 91.14 | 1.04 (0.32 to 3.43) | ||
| ≥ 65 and < 70 | 773 | 5746.65 | 14 | 243.62 | 2.93 (1.12 to 7.70) | ||
| ≥ 70 and < 75 | 805 | 5312.84 | 15 | 282.33 | 3.66 (1.40 to 9.59) | ||
| ≥ 75 and < 80 | 530 | 3193.95 | 8 | 250.47 | 3.45 (1.17 to 10.14) | ||
| ≥ 80 | 390 | 1875.26 | 6 | 319.96 | 4.97 (1.56 to 15.84) | ||
| Gender | Male | 2493 | 17,845.15 | 32 | 179.32 | 1 | 0.7762 |
| Female | 2024 | 14,704.87 | 28 | 190.41 | 1.08 (0.65 to 1.79) | ||
| Family history of cancer | No | 4222 | 30,273.11 | 56 | 184.98 | 1 | 0.9207 |
| Yes | 295 | 2276.91 | 4 | 175.68 | 0.95 (0.34 to 2.62) | ||
| Year of first follow-up | 1985–94 | 159 | 2461 | 9 | 365.7 | 1 | 0.7501 |
| 1995–9 | 544 | 6206.05 | 13 | 209.47 | 0.69 (0.26 to 1.79) | ||
| 2000–4 | 1601 | 13,311.8 | 21 | 157.75 | 0.63 (0.25 to 1.64) | ||
| 2005–9 | 2213 | 10,571.17 | 17 | 160.81 | 0.83 (0.29 to 2.32) | ||
| Length of visit | 1 day | 4041 | 29,240.39 | 51 | 174.42 | 1 | 0.0512 |
| 2–30 days | 54 | 314.53 | 0 | 0 | n/a | ||
| 1–3 months | 99 | 669.45 | 0 | 0 | n/a | ||
| 3–6 months | 137 | 943.26 | 5 | 530.08 | 3.21 (1.28 to 8.05) | ||
| 6–12 months | 152 | 1133.83 | 4 | 352.79 | 2.05 (0.74 to 5.68) | ||
| ≥ 12 months | 34 | 248.55 | 0 | 0 | n/a | ||
| Number of examinations in visit | 1 | 4033 | 29,184.02 | 51 | 174.75 | 1 | 0.0197 |
| 2 | 355 | 2477.71 | 5 | 201.8 | 1.16 (0.46 to 2.92) | ||
| 3 | 87 | 576.87 | 4 | 693.39 | 4.27 (1.54 to 11.83) | ||
| 4+ | 42 | 311.41 | 0 | 0 | n/a | ||
| Most complete examination | Complete colonoscopy | 3258 | 22,886.03 | 36 | 157.3 | 1 | 0.1061 |
| Colonoscopy of unknown completeness | 250 | 2266.09 | 3 | 132.39 | 0.75 (0.23 to 2.45) | ||
| Incomplete colonoscopy | 390 | 3077.3 | 12 | 389.95 | 2.27 (1.18 to 4.38) | ||
| Colonoscopy or FS | 181 | 1519.34 | 2 | 131.64 | 0.71 (0.17 to 2.98) | ||
| FS | 317 | 1896.99 | 3 | 158.15 | 1.04 (0.32 to 3.39) | ||
| Colonoscopy or flexible or rigid sigmoidoscopy | 100 | 774 | 2 | 258.4 | 1.49 (0.36 to 6.21) | ||
| Surgery | 12 | 44.91 | 1 | 2226.86 | 17.27 (2.35 to 126.74) | ||
| Unknown | 9 | 85.36 | 1 | 1171.46 | 6.26 (0.85 to 46.25) | ||
| Best bowel preparation at colonoscopy | Excellent/good | 1274 | 9241.46 | 13 | 140.67 | 1 | 0.5331 |
| Satisfactory | 617 | 3758.6 | 4 | 106.42 | 0.82 (0.27 to 2.53) | ||
| Poor | 240 | 1519.47 | 2 | 131.62 | 0.98 (0.22 to 4.33) | ||
| Unknown | 1767 | 13,709.89 | 32 | 233.41 | 1.57 (0.82 to 3.00) | ||
| No known colonoscopy | 619 | 4320.61 | 9 | 208.3 | 1.40 (0.60 to 3.30) | ||
| Difficult examination | No | 4285 | 30,947.19 | 50 | 161.57 | 1 | 0.0007 |
| Yes | 232 | 1602.83 | 10 | 623.9 | 3.98 (2.02 to 7.88) | ||
Colorectal cancer incidence after FUV1 was also stratified by characteristics of adenomas and polyps detected at FUV1 (Table 46). The only feature that was significantly associated with increased CRC incidence was the detection of a proximal polyp at FUV1 (HR 1.90, 95% CI 1.08 to 3.35). The detection of an adenoma with tubulovillous (but not villous) histology was associated with a borderline significant increased risk of CRC after FUV1 (HR 2.83, 95% CI 1.40 to 5.76; overall p = 0.0693). Although non-significant, there was a tendency towards an increased risk of CRC after FUV1 with the detection of multiple adenomas, a large adenoma or an adenoma with HGD at FUV1. Imprecision of effect estimates precluded meaningful interpretation for most factors.
| First follow-up factors | Number of patients (N = 4517) | IR patients with long-term follow-up after first follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) after first follow-up | |||||||
| pys | n = 60 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | |||||||
| Number | 0 | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.5336 |
| 1 | 1082 | 7450.29 | 18 | 241.6 | 1.61 (0.91 to 2.86) | ||
| 2 | 315 | 2039.62 | 4 | 196.11 | 1.38 (0.49 to 3.89) | ||
| 3 | 106 | 681.63 | 1 | 146.71 | 0.97 (0.13 to 7.08) | ||
| 4 | 34 | 218.7 | 1 | 457.24 | 3.20 (0.44 to 23.44) | ||
| 5+ | 40 | 237.76 | 1 | 420.6 | 3.11 (0.42 to 22.80) | ||
| Largest size (mm) | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.3690 |
| < 10 | 1013 | 6804.36 | 13 | 191.05 | 1.29 (0.68 to 2.45) | ||
| 10–14 | 213 | 1461.97 | 5 | 342 | 2.32 (0.91 to 5.94) | ||
| 15–19 | 115 | 749.75 | 1 | 133.38 | 0.94 (0.13 to 6.87) | ||
| ≥ 20 | 182 | 1188.45 | 4 | 336.57 | 2.26 (0.80 to 6.37) | ||
| Unknown | 54 | 423.48 | 2 | 472.28 | 2.65 (0.63 to 11.11) | ||
| Worst histology | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.0693 |
| Tubular | 946 | 6291.36 | 10 | 158.95 | 1.08 (0.53 to 2.19) | ||
| Tubulovillous | 372 | 2452.04 | 10 | 407.82 | 2.83 (1.40 to 5.76) | ||
| Villous | 122 | 839.62 | 1 | 119.1 | 0.78 (0.11 to 5.70) | ||
| Unknown | 137 | 1044.98 | 4 | 382.78 | 2.39 (0.85 to 6.73) | ||
| Worst dysplasia | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.3742 |
| Low grade | 1353 | 8802.29 | 20 | 227.21 | 1.57 (0.90 to 2.74) | ||
| High grade | 101 | 669.8 | 2 | 298.6 | 2.05 (0.49 to 8.56) | ||
| Unknown | 123 | 1155.92 | 3 | 259.53 | 1.46 (0.45 to 4.77) | ||
| Distal | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.0607 |
| No | 718 | 4714.59 | 15 | 318.16 | 2.18 (1.18 to 4.00) | ||
| Yes | 859 | 5913.42 | 10 | 169.11 | 1.13 (0.56 to 2.29) | ||
| Proximal | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.1472 |
| No | 718 | 5111.38 | 10 | 195.64 | 1.29 (0.64 to 2.61) | ||
| Yes | 859 | 5516.63 | 15 | 271.91 | 1.88 (1.02 to 3.45) | ||
| Number of sightings of a single adenoma | No adenomas | 2940 | 21,922.01 | 35 | 159.66 | 1 | 0.4693 |
| 1 | 1381 | 9312.05 | 21 | 225.51 | 1.52 (0.88 to 2.62) | ||
| 2 | 138 | 895.1 | 3 | 335.16 | 2.32 (0.71 to 7.58) | ||
| 3 | 35 | 253.76 | 1 | 394.08 | 2.55 (0.35 to 18.65) | ||
| 4 | 16 | 114.29 | 0 | 0 | n/a | ||
| 5+ | 7 | 52.82 | 0 | 0 | n/a | ||
| Polyp characteristics | |||||||
| Number of hyperplastic polyps | 0 | 3743 | 27,365.8 | 51 | 186.36 | 1 | 0.6502 |
| 1 | 496 | 3364.3 | 5 | 148.62 | 0.85 (0.34 to 2.15) | ||
| 2 | 160 | 1067.64 | 3 | 280.99 | 1.66 (0.52 to 5.35) | ||
| 3 | 58 | 363.43 | 0 | 0 | n/a | ||
| 4 | 24 | 185.01 | 0 | 0 | n/a | ||
| 5+ | 36 | 203.85 | 1 | 490.56 | 3.14 (0.43 to 22.86) | ||
| Any large hyperplastic polyps? | No | 4477 | 32,294.35 | 60 | 185.79 | n/a | n/a |
| Yes | 40 | 255.67 | 0 | 0 | |||
| Number of polyps with unknown histology | 0 | 3742 | 27,017.94 | 47 | 173.96 | 1 | 0.8852 |
| 1 | 478 | 3395.46 | 9 | 265.06 | 1.48 (0.73 to 3.03) | ||
| 2 | 142 | 1081.72 | 2 | 184.89 | 1.00 (0.24 to 4.13) | ||
| 3 | 70 | 467.01 | 1 | 214.13 | 1.23 (0.17 to 8.93) | ||
| 4 | 31 | 200.61 | 0 | 0 | n/a | ||
| 5+ | 54 | 387.28 | 1 | 258.21 | 1.5 (0.21 to 10.85) | ||
| Distal polyp | No polyps | 2000 | 15,179.5 | 24 | 158.11 | 1 | 0.1295 |
| No | 848 | 5883.19 | 17 | 288.96 | 1.90 (1.02 to 3.55) | ||
| Yes | 1669 | 11,487.33 | 19 | 165.4 | 1.11 (0.61 to 2.03) | ||
| Proximal polyp | No polyps | 2000 | 15,179.5 | 24 | 158.11 | 1 | 0.0042 |
| No | 1230 | 8816.78 | 12 | 136.1 | 0.90 (0.45 to 1.80) | ||
| Yes | 1287 | 8553.74 | 24 | 280.58 | 1.90 (1.08 to 3.35) | ||
Effect of baseline factors on future risk of colorectal cancer
Few baseline factors were associated with CRC risk after FUV1 in univariable analyses (Table 47). There was a tendency towards an increasing risk of CRC with increasing interval between baseline and FUV1; however, the effect estimates were imprecise, with most 95% CIs crossing 1. Results also indicated an increased risk of CRC in patients with unknown bowel preparation quality, no complete colonoscopy or a difficult examination at baseline, but the associations were non-significant and most 95% CIs included 1.
| Baseline factors | Number of patients (N = 4517) | IR patients with long-term follow-up after first follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) after first follow-up | |||||||
| pys | n = 60 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Family history of cancer | No | 4281 | 30,696.05 | 56 | 182.43 | 1 | 0.7605 |
| Yes | 236 | 1853.97 | 4 | 215.75 | 1.18 (0.43 to 3.24) | ||
| Calendar year of baseline visit | 1985–94 | 330 | 4473.85 | 16 | 357.63 | 1 | 0.1929 |
| 1995–9 | 1004 | 9703.32 | 20 | 206.12 | 0.64 (0.31 to 1.32) | ||
| 2000–4 | 2279 | 14,814.79 | 22 | 148.5 | 0.54 (0.26 to 1.15) | ||
| 2005–10 | 904 | 3558.05 | 2 | 56.21 | 0.23 (0.05 to 1.10) | ||
| Length of visit | 1 day | 2455 | 18,050.92 | 30 | 166.2 | 1.00 | 0.9122 |
| 2–30 days | 239 | 1667.52 | 4 | 239.88 | 1.47 (0.52 to 4.18) | ||
| 1–3 months | 651 | 4483.72 | 8 | 178.42 | 1.11 (0.51 to 2.43) | ||
| 3–6 months | 579 | 4163.45 | 9 | 216.17 | 1.33 (0.63 to 2.81) | ||
| 6–12 months | 497 | 3552.47 | 7 | 197.05 | 1.2 (0.53 to 2.73) | ||
| ≥ 12 months | 96 | 631.95 | 2 | 316.48 | 1.98 (0.47 to 8.3) | ||
| Number of examinations in visit | 1 | 2448 | 17,977.1 | 30 | 166.88 | 1 | 0.3596 |
| 2 | 1487 | 10,618.22 | 22 | 207.19 | 1.25 (0.72 to 2.17) | ||
| 3 | 381 | 2589.04 | 3 | 115.87 | 0.73 (0.22 to 2.41) | ||
| 4+ | 201 | 1365.66 | 5 | 366.12 | 2.24 (0.87 to 5.79) | ||
| Most complete colonoscopy | Complete | 2926 | 19,853.68 | 32 | 161.18 | 1 | 0.5691 |
| Unknown completeness | 1140 | 9260.95 | 19 | 205.16 | 1.16 (0.66 to 2.06) | ||
| Incomplete | 451 | 3435.39 | 9 | 261.98 | 1.50 (0.71 to 3.15) | ||
| Best bowel preparation at colonoscopy | Excellent/good | 1371 | 9689.83 | 11 | 113.52 | 1 | 0.0560 |
| Satisfactory | 480 | 2695.92 | 2 | 74.19 | 0.73 (0.16 to 3.30) | ||
| Poor | 184 | 1165.3 | 1 | 85.81 | 0.78 (0.10 to 6.07) | ||
| Unknown | 2482 | 18,998.96 | 46 | 242.12 | 2.04 (1.06 to 3.95) | ||
| Difficult examination | No | 4308 | 31,127 | 55 | 176.70 | 1 | 0.1674 |
| Yes | 209 | 1423.02 | 5 | 351.37 | 2.03 (0.81 to 5.09) | ||
| Interval from baseline to first follow-up | < 18 months | 1727 | 13,330.97 | 23 | 172.53 | 1 | 0.0089 |
| 2 yearsa | 949 | 7105.08 | 11 | 154.82 | 0.93 (0.45 to 1.90) | ||
| 3 yearsa | 1051 | 6997.94 | 15 | 214.35 | 1.38 (0.72 to 2.66) | ||
| 4 yearsa | 345 | 2449.78 | 5 | 204.1 | 1.27 (0.48 to 3.36) | ||
| 5 yearsa | 213 | 1358.88 | 1 | 73.59 | 0.48 (0.07 to 3.59) | ||
| 6 yearsa | 118 | 660.13 | 5 | 757.43 | 5.15 (1.94 to 13.65) | ||
| ≥ 6.5 years | 114 | 647.25 | 0 | 0 | n/a | ||
Table 48 shows the CRC incidence after FUV1 stratified by characteristics of adenomas and polyps detected at baseline. The only factor which reached statistical significance was the number of sightings of a unique adenoma (p = 0.0494); risk tended to increase with increased viewings but interpretation was difficult because of a lack of precision. Although no other baseline adenoma or polyp risk factors reached statistical significance and ORs were imprecise, there was a tendency towards an increased risk of CRC after FUV1 with the detection of a large (≥ 10 mm) adenoma, an adenoma with tubulovillous or villous histology, a proximal adenoma or polyp, or multiple polyps with unknown histology at baseline.
| Baseline factors | Number of patients (N = 4517) | IR patients with long-term follow-up after first follow-up | |||||
|---|---|---|---|---|---|---|---|
| CRC(s) after first follow-up | |||||||
| pys | n = 60 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |||
| Adenoma characteristics | |||||||
| Number | 1 | 3040 | 22,527.05 | 45 | 199.76 | 1 | 0.4002 |
| 2 | 1129 | 7717.5 | 14 | 181.41 | 0.95 (0.52 to 1.73) | ||
| 3 | 238 | 1494.08 | 0 | 0 | n/a | ||
| 4 | 110 | 811.39 | 1 | 123.24 | 0.63 (0.09 to 4.55) | ||
| Largest size (mm) | < 10 | 2305.47 | 1 | 43.38 | 90.80 | 1 | 0.1103 |
| 10–14 | 11,142.76 | 17 | 152.57 | 203.92 | 3.41 (0.45 to 25.64) | ||
| 15–19 | 6862.11 | 12 | 174.87 | 194.51 | 3.81 (0.49 to 29.32) | ||
| ≥ 20 | 12,239.68 | 30 | 245.1 | 243.26 | 5.36 (0.73 to 39.36) | ||
| Worst histology | Tubular | 1700 | 12,040.66 | 17 | 141.19 | 1 | 0.4971 |
| Tubulovillous | 2096 | 14,676.49 | 27 | 183.97 | 1.31 (0.72 to 2.41) | ||
| Villous | 440 | 3104.81 | 7 | 225.46 | 1.58 (0.65 to 3.80) | ||
| Unknown | 281 | 2728.06 | 9 | 329.91 | 1.85 (0.79 to 4.30) | ||
| Worst dysplasia | Low grade | 3372 | 23,754.64 | 39 | 164.18 | 1 | 0.1088 |
| High grade | 822 | 5592.51 | 8 | 143.05 | 0.88 (0.41 to 1.89) | ||
| Unknown | 323 | 3202.87 | 13 | 405.89 | 2.04 (1.05 to 3.94) | ||
| Distal | No | 921 | 6512.4 | 11 | 168.91 | 1 | 0.7652 |
| Yes | 3596 | 26,037.62 | 49 | 188.19 | 1.10 (0.57 to 2.12) | ||
| Proximal | No | 3261 | 24,154.31 | 42 | 173.88 | 1.00 | 0.3847 |
| Yes | 1256 | 8395.71 | 18 | 214.40 | 1.28 (0.74 to 2.23) | ||
| Number of sightings of a unique adenoma | 1 | 3256 | 23,786.29 | 40 | 168.16 | 1 | 0.0494 |
| 2 | 981 | 6797.85 | 16 | 235.37 | 1.43 (0.80 to 2.55) | ||
| 3 | 174 | 1255.55 | 0 | 0 | n/a | ||
| 4 | 61 | 399.74 | 2 | 500.33 | 3.19 (0.77 to 13.22) | ||
| 5+ | 45 | 310.58 | 2 | 643.95 | 3.70 (0.89 to 15.33) | ||
| Polyp characteristics | |||||||
| Number of hyperplastic polyps | 0 | 3669 | 26,896.79 | 55 | 204.49 | 1 | 0.4610 |
| 1 | 529 | 3547.39 | 3 | 84.57 | 0.44 (0.14 to 1.40) | ||
| 2 | 155 | 1039.77 | 0 | 0 | n/a | ||
| 3 | 64 | 429.76 | 1 | 232.69 | 1.22 (0.17 to 8.82) | ||
| 4 | 37 | 227.95 | 0 | 0 | n/a | ||
| 5+ | 63 | 408.36 | 1 | 244.88 | 1.30 (0.18 to 9.40) | ||
| Any large hyperplastic polyps? | No | 4435 | 31,994.36 | 59 | 184.41 | 1 | 0.9814 |
| Yes | 82 | 555.66 | 1 | 179.97 | 1.02 (0.14 to 7.40) | ||
| Number of polyps with unknown histology | 0 | 3523 | 25,765.33 | 49 | 190.18 | 1 | 0.1700 |
| 1 | 544 | 3662.15 | 3 | 81.92 | 0.44 (0.14 to 1.41) | ||
| 2 | 184 | 1230.67 | 4 | 325.03 | 1.78 (0.64 to 4.94) | ||
| 3 | 106 | 755.28 | 2 | 264.8 | 1.40 (0.34 to 5.78) | ||
| 4 | 61 | 400.04 | 2 | 499.95 | 2.81 (0.68 to 11.59) | ||
| 5+ | 99 | 736.56 | 0 | 0 | n/a | ||
| Distal polyp | No | 727 | 5149.74 | 10 | 194.18 | 1 | 0.8231 |
| Yes | 3790 | 27,400.28 | 50 | 182.48 | 0.92 (0.47 to 1.82) | ||
| Proximal polyp | No | 2886 | 21,527.35 | 37 | 171.87 | 1 | 0.3835 |
| Yes | 1631 | 11,022.66 | 23 | 208.66 | 1.26 (0.75 to 2.13) | ||
Effect of surveillance on future risk of colorectal cancer
In a univariable analysis (Table 49), additional surveillance was estimated to reduce the risk of CRC after FUV1 by 46% compared with no additional surveillance (HR 0.54, 95% CI 0.30 to 1.00); the p-value from the LRT indicated significance (p = 0.0462), although the 95% CI included 1.
| Number of follow-up visits after baseline (including first follow-up)a | Number of patients (N = 4517) | IR patients with long-term follow-up | ||||
|---|---|---|---|---|---|---|
| CRC(s) | ||||||
| pys | n = 60 | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | ||
| 1 | 2901 | 21,030.19 | 38 | 180.69 | 1 | 0.0462 |
| 2+ | 1616 | 11,519.83 | 22 | 190.97 | 0.54 (0.30 to 1.00) | |
Multivariable analysis
In order to build a final multivariable model for the analysis of risk following FUV1, separate models were first built considering baseline factors only, FUV1 factors only and cumulative factors only. The significant risk factors identified from these models were then considered together and a final model was chosen.
Table 50 presents the results of the selected final Cox regression model of CRC incidence after the first follow-up. There was a 41% reduction in CRC incidence with two or more follow-up visits compared with only one follow-up, although this was not shown to be statistically significant (HR 0.59, 95% CI 0.32 to 1.10; p = 0.0923).
| Risk factor | Category | Adjusted HR (95% CI) | p-value (LRT) |
|---|---|---|---|
| Number of follow-up visits after baseline (including first follow-up)a | 1 | 1 | 0.0923 |
| 2+ | 0.59 (0.32 to 1.10) | ||
| Age (years) at first follow-up | < 55 | 1 | 0.0151 |
| 55–59 | 1.35 (0.43 to 4.18) | ||
| 60–64 | 1.01 (0.31 to 3.32) | ||
| 65–69 | 2.77 (1.05 to 7.30) | ||
| 70–74 | 3.42 (1.29 to 9.03) | ||
| 75–79 | 2.98 (1.01 to 8.84) | ||
| ≥ 80 | 4.66 (1.45 to 15.01) | ||
| Difficult examination at baseline or first follow-upb | No | 1 | 0.0066 |
| Yes | 2.67 (1.4 to 5.08) | ||
| Proximal polyps at first follow-up | No polyps | 1 | 0.0111 |
| No | 0.97 (0.48 to 1.96) | ||
| Yes | 2.28 (1.28 to 4.07) | ||
| Largest adenoma at baseline, mm | < 10 | 1 | 0.0418 |
| 10–19 | 3.96 (0.54 to 29.28) | ||
| ≥ 20 | 5.89 (0.80 to 43.55) |
Independent risk factors for CRC after FUV1 included older age at FUV1, the presence of proximal polyps at FUV1 or a difficult examination at either baseline or FUV1. There was weak evidence that a large adenoma (≥ 10 mm) at baseline also increased CRC incidence after FUV1 [although the HRs suggested a large effect, they had extremely wide CIs that included 1 and the LRT statistic was only borderline significant (p = 0.0418)].
Effect modification of the association between surveillance and colorectal cancer incidence
As for findings at follow-up, we hypothesised that there might be differences in the effect of surveillance by age and gender. However, models with interactions terms included demonstrate no evidence of any significant difference in the effect of surveillance by age or gender (Table 51).
| Characteristic | Number of follow-up visits after baselinea | New CRC after baseline | New CRC after FUV1 | |||
|---|---|---|---|---|---|---|
| HR (95% CI)b | p-valuec | HR (95% CI)b | p-valuec | |||
| Age (years)d | < 60 | 0 | 1 | 0.2116 | n/a | 0.8669 |
| 1 | 0.22 (0.06 to 0.77) | 1 | ||||
| 2+ | 0.45 (0.17 to 1.20) | 0.75 (0.22 to 2.55) | ||||
| ≥ 60 and < 75 | 0 | 1 | n/a | |||
| 1 | 0.69 (0.41 to 1.15) | 1 | ||||
| 2+ | 0.25 (0.10 to 0.61) | 0.52 (0.24 to 1.12) | ||||
| ≥ 75 | 0 | 1 | n/a | |||
| 1 | 0.38 (0.16 to 0.89) | 1 | ||||
| 2+ | 0.36 (0.09 to 1.52) | 0.56 (0.15 to 2.04) | ||||
| Gender | Male | 0 | 1 | 0.1353 | n/a | 0.1285 |
| 1 | 0.45 (0.26 to 0.79) | 1 | ||||
| 2+ | 0.18 (0.07 to 0.46) | 0.40 (0.18 to 0.91) | ||||
| Female | 0 | 1 | n/a | |||
| 1 | 0.58 (0.32 to 1.05) | 1 | ||||
| 2+ | 0.56 (0.26 to 1.21) | 0.91 (0.41 to 2.05) | ||||
Absolute risk of colorectal cancer in intermediate-risk patients
The absolute risk of CRC was assessed using cumulative incidence rates calculated using different subsets of the cohort and varying periods of observation time, as presented in Table 52 and illustrated in Figure 7. It should be noted that the results presented in this section for observation time partitioned by the occurrence of surveillance may be contaminated, as some patients could have had surveillance that we do not know about; this would artificially reduce the estimate of pre-surveillance risk and possibly underestimate the effect of surveillance. Sensitivity analyses that assess the impact of this potential misclassification can be found below (see Sensitivity analyses and internal validation).
| Observation time | Number of patients | Cumulative incidence at | Number of observed CRCs | Number of expected CRCs | SIRa (95% CI) | ||
|---|---|---|---|---|---|---|---|
| 3 years, % (95% CI) | 5 years, % (95% CI) | 10 years, % (95% CI) | |||||
| Observation time free of surveillance in all IR patients (censored at first follow-up) | |||||||
| Total | 11,944 | 0.5 (0.4 to 0.7) | 1.1 (0.9 to 1.4) | 2.9 (2.2 to 3.9) | 108 | 102 | 1.06 (0.87 to 1.28) |
| All observation time in all IR patients | |||||||
| Total | 11,944 | 0.5 (0.3 to 0.6) | 0.9 (0.7 to 1.1) | 2.1 (1.7 to 2.5) | 168 | 172 | 0.98 (0.84 to 1.14) |
| Observation time free of further surveillance after FUV1 in IR patients with one or more follow-up visits (censored at second follow-up) | |||||||
| Total | 4517 | 0.4 (0.2 to 0.6) | 0.8 (0.5 to 1.2) | 2.3 (1.5 to 3.7) | 38 | 44 | 0.87 (0.61 to 1.19) |
| All observation time after FUV1 in IR patients with one follow-up visit | |||||||
| Total | 4517 | 0.3 (0.2 to 0.5) | 0.7 (0.5 to 1.0) | 1.9 (1.4 to 2.6) | 60 | 70 | 0.86 (0.65 to 1.10) |
FIGURE 7.
Kaplan–Meier estimates of cumulative incidence. CRC incidence after baseline censoring at first follow-up (a) and using all follow-up time (b); CRC incidence after the first follow-up censoring at the second follow-up (c) and using all subsequent follow-up time (d).
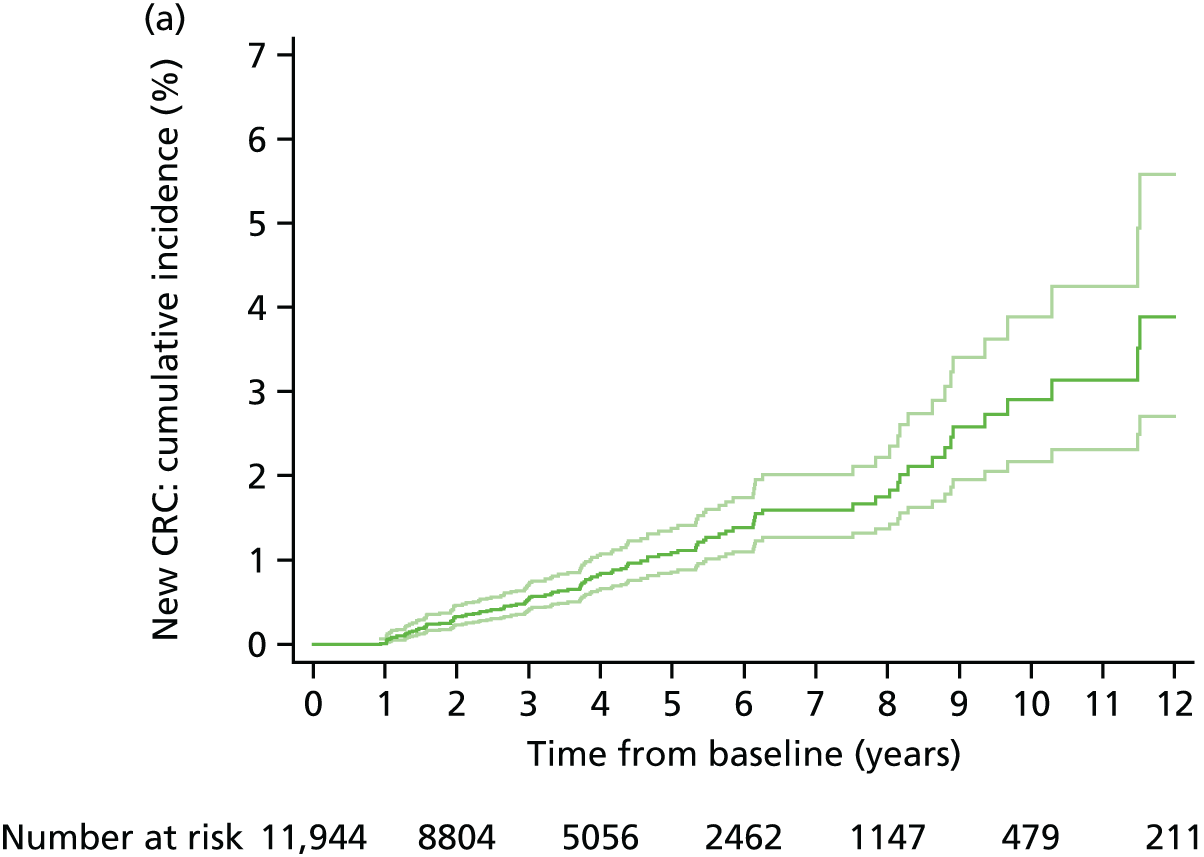
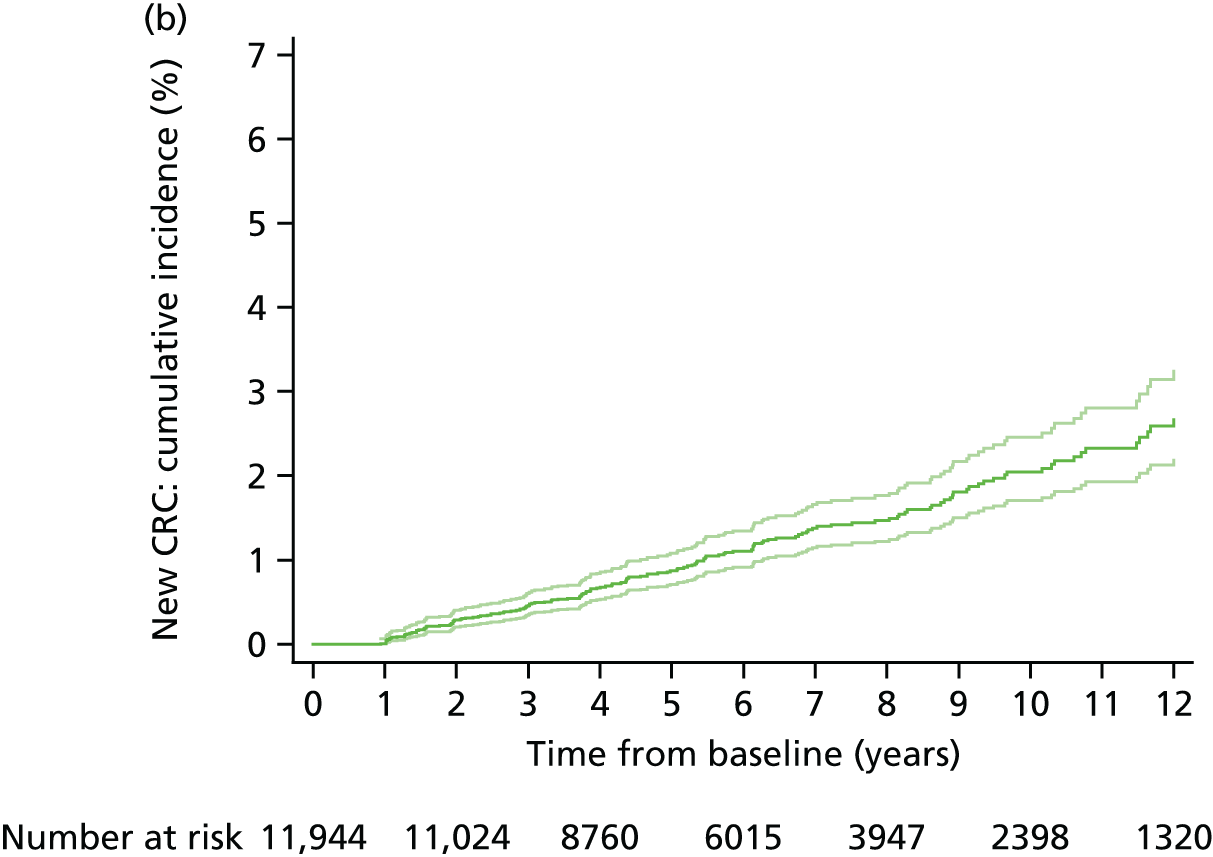


Cumulative incidence in the absence of surveillance was assessed by censoring the cohort at FUV1 (see Figure 7a). The cumulative incidence of CRC at 3, 5 and 10 years, respectively, was 0.5%, 1.1% and 2.9%, and CRC incidence was slightly, but non-significantly, higher than that of the general population.
Cumulative incidence in the whole cohort allowing for the effect of any surveillance was 0.5%, 0.9% and 2.1% at 3, 5 and 10 years, respectively (see Figure 7b). The cumulative incidence of CRC at 10 years was reduced to 2.1%, and CRC incidence was the same as the general population level; these results must be interpreted in the context of the fact that only 39% of the cohort were known to have had at least one surveillance visit.
Cumulative incidence after a single surveillance was assessed by focusing on the cohort of IR patients who attended follow-up and censoring at FUV2 to remove effects of additional surveillance (see Figure 7c). Compared with pre-surveillance risk, the cumulative incidence of CRC at 10 years was lowered to 2.3% after a single visit, and the CRC incidence was slightly lower than the general population, although not significantly so.
Finally, we assessed absolute risk of CRC after one or more surveillance visits using all observation time after the first follow-up in patients who attended one follow-up or more (see Figure 7d). Compared with the analysis that censored the cohort at the second follow-up, by including the effect of additional surveillance after the first follow-up, the cumulative incidence of CRC at 10 years was lowered to 1.9%, and CRC incidence remained slightly lower than in the general population.
Lower- and higher-intermediate-risk subgroups
Lower- and higher-intermediate-risk subgroups after baseline
To assess whether or not heterogeneity exists within the IR group, and to see if all IR patients benefit from surveillance after baseline, the cohort was divided into lower IR (LIR) and higher IR (HIR) subgroups. These subgroups were defined using risk factors for CRC identified in the Cox regression model of CRC risk after baseline (see Table 44); a HIR subgroup was defined to include patients with any of the following baseline characteristics: an adenoma of ≥ 20 mm or with HGD, proximal polyps, no complete colonoscopy or poor bowel preparation. All other patients were assigned to the LIR subgroup.
Older age was not used to define risk subgroups despite it being identified as a risk factor for CRC, as age has practical implications for surveillance, with risks of complications increasing with age, and surveillance often ceasing in patients aged ≥ 75 years. Bowel preparation quality was used despite it not being predictive in the Cox model as it was a risk factor for finding CRC at FUV1 (see Table 30) and has a crucial effect on examination quality. Although patients with an adenoma of ≥ 10 mm displayed similar risk to those with an adenoma of ≥ 20 mm, only the latter size was used to define a higher-risk subgroup, as almost all patients (91%) had a lesion of ≥ 10 mm; this led to their classification as IR and, thus, if ‘≥ 10 mm’ was used it would not be discriminant.
Based on this definition, the baseline subgroups comprised 2679 (22.4%) LIR and 9265 (77.6%) HIR patients (Table 53). In the HIR subgroup, attendance at one or more follow-up visits was associated with a significantly lower CRC risk than with no follow-up, with a 50% reduction for one follow-up (HR 0.50, 95% CI 0.34 to 0.76) and a 64% reduction for two or more (HR 0.36, 95% CI 0.20 to 0.62). Among patients in the LIR subgroup, the benefit of surveillance was less clear and, because of the small number of CRC end points, statistical significance was not reached and effect estimates were imprecise, although the attendance at one or more follow-up visits was associated with a non-significant reduction in CRC risk [38% reduction for one (HR 0.62, 95% CI 0.16 to 2.43); 71% reduction for two or more follow-ups (HR 0.29, 95% CI 0.03 to 2.82)].
| IR subgroup | Number of patients | % | pys | Patients with CRC (n) | Rate/100,000 pys (95% CI) | Effect of surveillance | ||
|---|---|---|---|---|---|---|---|---|
| Number of follow-up visits after baselinea | Unadjusted HR (95% CI) | p-value (LRT) | ||||||
| LIR | 2679 | 22.4 | 17,615 | 13 | 74 (43 to 127) | 0 | 1 | 0.47 |
| 1 | 0.62 (0.16 to 2.43) | |||||||
| 2+ | 0.29 (0.03 to 2.82) | |||||||
| HIRb | 9265 | 77.6 | 63,827 | 155 | 243 (208 to 284) | 0 | 1 | 0.0001 |
| 1 | 0.50 (0.34 to 0.76) | |||||||
| 2+ | 0.36 (0.20 to 0.62) | |||||||
Table 54 shows the differences between patients in the HIR and LIR subgroups. Patients in the HIR subgroup had significantly more follow-up visits than those in the LIR subgroup; however, the median follow-up time was similar (6.1 years in the HIR subgroup vs. 5.7 years in the LIR subgroup). The HIR subgroup was also older and had their baseline visit earlier on average; despite reaching statistical significance, these differences were small.
| Factor | Number of patients | LIR subgroup | HIR subgroupa | p-value (chi-squared test) | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Total | 11,944 | 2679 | 9265 | |||
| Number of follow-up visits | < 0.001 | |||||
| 0 | 7427 | 1909 | 71.3 | 5518 | 59.6 | |
| 1 | 2880 | 515 | 19.2 | 2365 | 25.5 | |
| 2 | 1074 | 184 | 6.9 | 890 | 9.6 | |
| 3+ | 563 | 71 | 2.7 | 492 | 5.3 | |
| Age (years) at first adenoma detection | < 0.001 | |||||
| < 55 | 2122 | 572 | 21.4 | 1550 | 16.7 | |
| ≥ 55 and < 60 | 1321 | 311 | 11.6 | 1010 | 10.9 | |
| ≥ 60 and < 65 | 1858 | 439 | 16.4 | 1419 | 15.3 | |
| ≥ 65 and < 70 | 2171 | 473 | 17.7 | 1698 | 18.3 | |
| ≥ 70 and < 75 | 1786 | 403 | 15.0 | 1383 | 14.9 | |
| ≥ 75 and < 80 | 1416 | 260 | 9.7 | 1156 | 12.5 | |
| ≥ 80 | 1270 | 221 | 8.2 | 1049 | 11.3 | |
| Year of baseline | < 0.001 | |||||
| 1985–94 | 439 | 71 | 2.7 | 368 | 4.0 | |
| 1995–9 | 1430 | 283 | 10.6 | 1147 | 12.4 | |
| 2000–4 | 4251 | 825 | 30.8 | 3426 | 37.0 | |
| 2005–10 | 5824 | 1500 | 56.0 | 4324 | 46.7 | |
Lower- and higher-intermediate-risk subgroups after the first follow-up visit
The HIR and LIR subgroups were redefined after FUV1 incorporating findings at both baseline and FUV1. Specifically, the HIR subgroup was classified as patients with any of the following: an adenoma of ≥ 10 mm or with HGD, proximal polyps, no complete colonoscopy or poor bowel preparation at FUV1, or an adenoma of ≥ 20 mm at baseline.
The risk subgroups after FUV1 comprised 1246 (27.6%) LIR patients and 3271 (72.4%) HIR patients (Table 55). In the LIR subgroup, attendance for additional surveillance after FUV1 was associated with a non-significant increased risk compared with only one follow-up visit (HR 1.20, 95% CI 0.14 to 10.31; p = 0.87), although the effect estimate was very imprecise. By comparison, in the HIR subgroup attendance for additional surveillance conferred a significant 53% reduction in CRC risk (HR 0.47, 95% CI 0.25 to 0.87; p = 0.0155).
| IR subgroup | N | % | pys | Patients with CRC (n) | Rate/100,000 pys (95% CI) | Effect of surveillance | ||
|---|---|---|---|---|---|---|---|---|
| Number of follow-up visits after baseline (including first follow-up)a | Unadjusted hazard ratio (95% CI) | p-value (LRT) | ||||||
| LIR | 1246 | 27.6 | 9268 | 6 | 65 (29 to 144) | 1 | 1 | 0.87 |
| 2+ | 1.20 (0.14 to 10.31) | |||||||
| HIRb | 3271 | 72.4 | 23,282 | 54 | 232 (118 to 303) | 1 | 1 | 0.0155 |
| 2+ | 0.47 (0.25 to 0.87) | |||||||
Absolute risk of colorectal cancer in lower- and higher-intermediate-risk subgroups
The absolute risk of CRC was assessed using cumulative incidence rates and SIRs, calculated using different subsets of the cohort and varying periods of observation time, as presented in Table 56. The pre-surveillance standardised CRC incidence in the LIR group was 60% below that of the general population, whereas the HIR group had a 26% higher incidence. This large difference in cancer risk between subgroups was also reflected in the 10-year cumulative incidence of CRC, which was 3.6% in the HIR subgroup compared with 1.0% in the LIR subgroup (Figure 8a).
| Observation time | Number of patients | Cumulative incidence at | Number of observed CRCs | Number of expected CRCs | SIRa (95% CI) | ||
|---|---|---|---|---|---|---|---|
| 3 years, % (95% CI) | 5 years, % (95% CI) | 10 years, % (95% CI) | |||||
| Observation time free of surveillance in all IR patients (censored at first follow-up) | |||||||
| Lower-risk subgroup | 2679 | 0.1 (0 to 0.4) | 0.4 (0.2 to 1.0) | 1.0 (0.4 to 2.4) | 9 | 23 | 0.39 (0.18 to 0.75) |
| Higher-risk subgroupb | 9265 | 0.7 (0.5 to 0.9) | 1.3 (1.0 to 1.7) | 3.6 (2.6 to 4.8) | 99 | 79 | 1.26 (1.02 to 1.53) |
| All observation time in all IR patients | |||||||
| Lower-risk subgroup | 2679 | 0.1 (0 to 0.4) | 0.4 (0.2 to 0.8) | 0.6 (0.3 to 1.2) | 13 | 34 | 0.38 (0.20 to 0.65) |
| Higher-risk subgroupb | 9265 | 0.6 (0.4 to 0.7) | 1.0 (0.8 to 1.3) | 2.4 (2.0 to 2.9) | 155 | 138 | 1.13 (0.96 to 1.32) |
| Observation time free of further surveillance after FUV1 in IR patients with one or more follow-up visits (censored at second follow-up) | |||||||
| Lower-risk subgroup | 1246 | 0.2 (0 to 0.7) | 0.3 (0.1 to 1.0) | 0.5 (0.2 to 1.4) | 4 | 13 | 0.32 (0.09 to 0.81) |
| Higher-risk subgroupc | 3271 | 0.4 (0.2 to 0.8) | 1 (0.6 to 1.6) | 3.3 (2.0 to 5.2) | 34 | 31 | 1.09 (0.75 to 1.52) |
| All observation time after FUV1 in IR patients with one or more follow-up visits | |||||||
| Lower-risk subgroup | 1246 | 0.2 (0 to 0.7) | 0.4 (0.1 to 0.9) | 0.5 (0.2 to 1.1) | 6 | 19 | 0.32 (0.12 to 0.70) |
| Higher-risk subgroupc | 3271 | 0.4 (0.2 to 0.7) | 0.8 (0.5 to 1.2) | 2.5 (1.8 to 3.5) | 54 | 51 | 1.05 (0.79 to 1.37) |
FIGURE 8.
Kaplan–Meier estimates of cumulative incidence in HIR and LIR subgroups. CRC incidence after baseline censoring at first follow-up (a) and using all follow-up time (b); CRC incidence after the first follow-up censoring at the second follow-up (c) and using all subsequent follow-up time (d). HR represents the HIR subgroup, and LR represents the LIR subgroup.
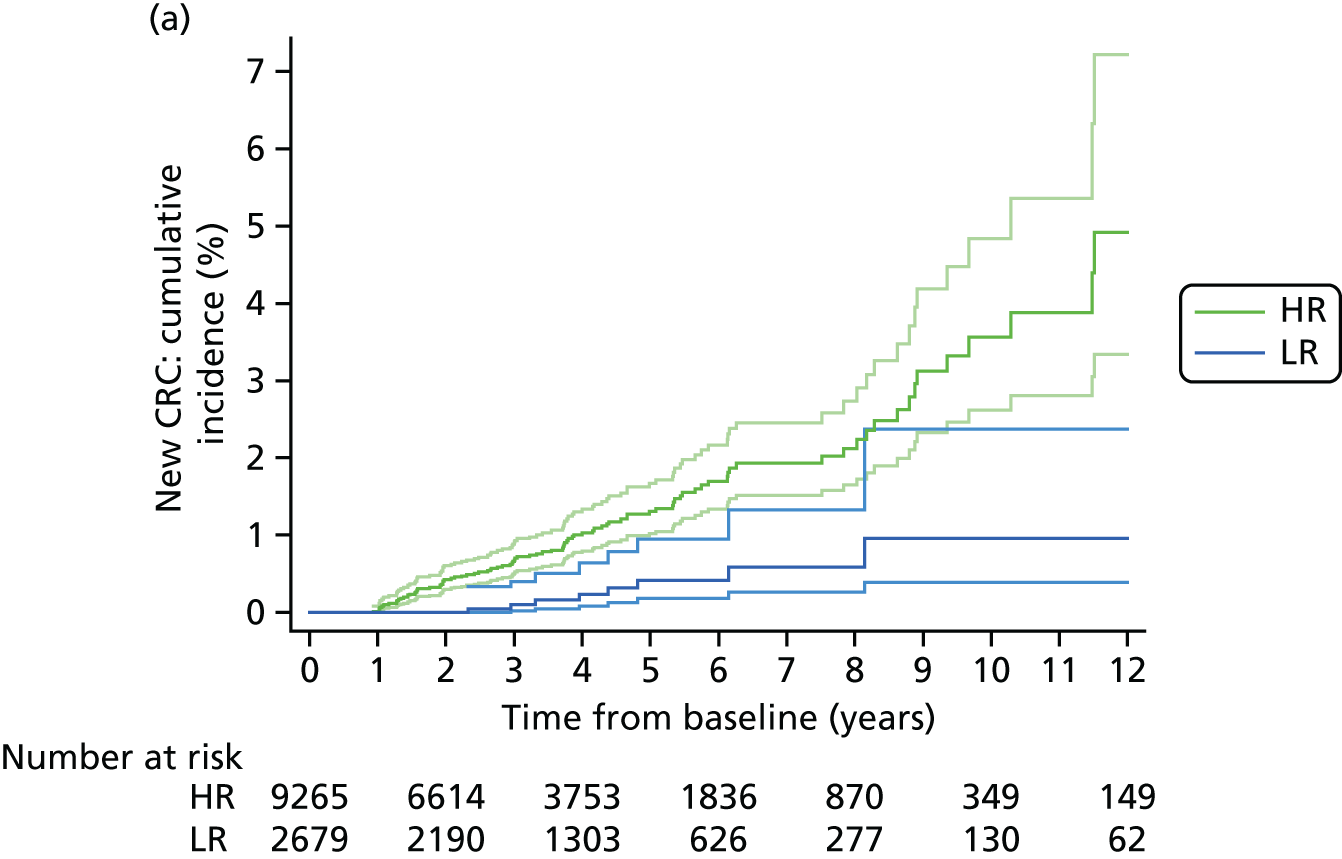
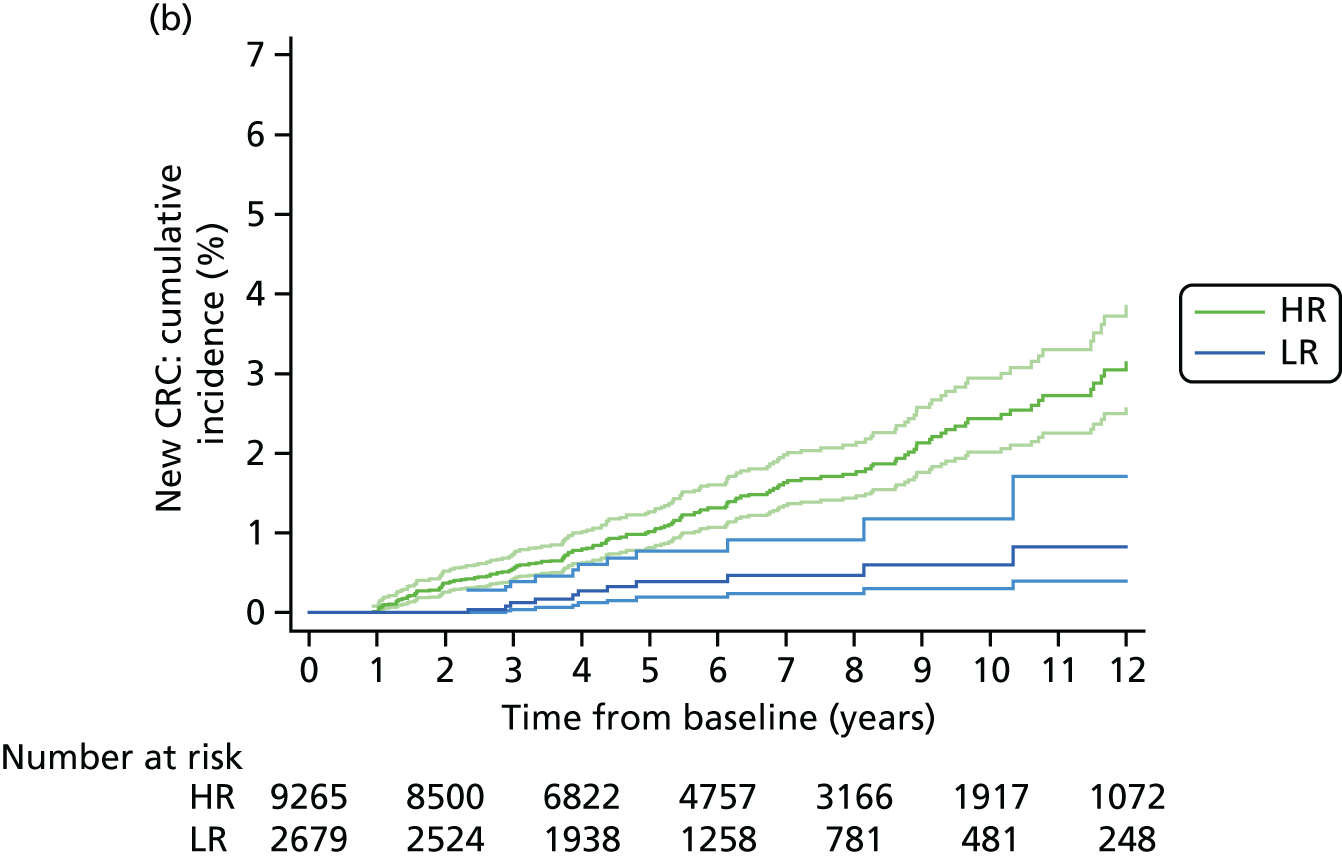


Including the effect of any surveillance, the CRC incidence in the LIR group remained around 60% lower than that of the general population, whereas the CRC incidence in the HIR subgroup was reduced to a level 13% higher than the general population. The 10-year cumulative incidence of CRC in the HIR subgroup was 2.4%, compared with 0.6% in the LIR subgroup (see Figure 8b).
We assessed the effect of just one follow-up visit on CRC risk by focusing on the cohort of IR patients who attended follow-up and censoring at FUV2 to remove effects of additional surveillance (see Figure 8c); risk groups were revised to incorporate findings at both baseline and FUV1. Compared with pre-surveillance risk, the 10-year cumulative incidence of CRC after a single surveillance visit was lower, at 0.5% and 3.3% in the LIR and HIR subgroups, respectively. In the low-risk subgroup, the standardised CRC incidence after one follow-up visit was slightly lower than the pre-surveillance level at 68% below the general population level. Similarly, for the HIR group, a single surveillance visit reduced the standardised, pre-surveillance CRC incidence closer to that of the general population.
When the effect of additional surveillance after FUV1 was included, the 10-year cumulative incidence of CRC in the HIR subgroup was 2.5% (see Figure 8d), compared with 3.3% when censoring at FUV2. The standardised CRC incidence in the LIR subgroup remained unchanged – it was significantly lower than the general population – whereas CRC incidence in the HIR group was further reduced to a level comparable with that of the general population.
Findings at follow-up examinations in lower- and higher-intermediate-risk subgroups
Lower- and higher-risk subgroups, derived from the Cox proportional hazards models for long-term CRC risk (see Tables 44 and 50), were applied to findings at FUV1 and FUV2 to determine if the criteria used to define the subgroups were discriminant in terms of risks of detecting AN at follow-up visits.
At FUV1, AN was detected in 6.2% of the LIR subgroup compared with 11.6% of the HIR subgroup (see Table 57; OR 1.99, 95% CI 1.46 to 2.71; p < 0.0001); this suggests that risk factors for CRC after baseline are also discriminant in terms of risk of AN at FUV1. At FUV2, new ANs was detected in 7.4% and 10.0% of the LIR and HIR subgroups, respectively (OR 1.38, 95% CI 0.91 to 2.10; p = 0.1245); thus, the risk groups were not discriminant for findings at FUV2, possibly owing to a lack of power because of the small number of end points detected.
The effect of interval in the LIR and HIR subgroups was examined (Table 57). At FUV1, there was a highly significant association between longer interval and new AN in the HIR subgroup (p < 0.0001); in the LIR subgroup, the trend was only borderline significant, possibly because of a paucity of end points (p = 0.0433). At FUV2, there was an association between interval and new AN in the HIR subgroup (p = 0.0191), but not the LIR subgroup (p = 0.4573).
| Interval to first follow-up | FUV1 | FUV2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower-risk subgroup | Higher-risk subgroupa | Lower-risk subgroup | Higher-risk subgroupb | |||||||||
| Number with FUV1 | Number of AN (CRC) | % | Number with FUV1 | Number of AN (CRC) | % | Number with FUV2 | Number of AN (CRC) | % | Number with FUV2 | Number of AN (CRC) | % | |
| < 18 months | 264 | 12 | 4.55 | 1496 | 134 (15) | 8.96 | 43 | 1 (1) | 2.33 | 354 | 28 (1) | 7.91 |
| 2 yearsc | 147 | 12 | 8.16 | 829 | 87 (9) | 10.49 | 81 | 6 | 7.41 | 295 | 29 (4) | 9.83 |
| 3 yearsc | 227 | 9 | 3.96 | 830 | 101 (5) | 12.17 | 164 | 15 | 9.15 | 354 | 37 (1) | 10.45 |
| 4 yearsc | 50 | 7 (1) | 14.00 | 305 | 45 (8) | 14.75 | 40 | 3 | 7.50 | 112 | 12 (1) | 10.71 |
| 5 yearsc | 53 | 3 | 5.66 | 164 | 30 (4) | 18.29 | 59 | 3 | 5.08 | 72 | 8 | 11.11 |
| 6 yearsc | 19 | 2 (1) | 10.53 | 104 | 19 (3) | 18.27 | 9 | 0 | 0 | 22 | 4 | 18.18 |
| ≥ 6.5 years | 15 | 3 | 20.00 | 105 | 29 (6) | 27.62 | 8 | 2 | 25.00 | 22 | 5 (1) | 22.73 |
| Total | 775 | 48 (2) | 6.19 | 3833 | 445 (50) | 11.61 | 404 | 30 (1) | 7.43 | 1231 | 123 (8) | 9.99 |
| pd = 0.0433 | pd < 0.0001 | pd = 0.4573 | pd = 0.0191 | |||||||||
| OR for higher-intermediate vs. LIR subgroup (95% CI), p-value | 1.99 (1.46 to 2.71), < 0.0001 | 1.38 (0.91 to 2.10), 0.1245 | ||||||||||
As interval had a strong effect on findings at both the first and second follow-ups in the HIR subgroup, the effect of interval on findings at FUV1 was assessed in patients with HIR polyp factors only, HIR procedure quality factors only, or both (Table 58). Interval had a significant effect in all subsets of the HIR group. Although the test for trend was not as significant in patients who were classified as high risk based on examination factors only, this is probably the result of the smaller size of this group.
| Interval | FUV1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HIR subgroup | |||||||||
| Polyp characteristics only | Poor examination only | Poor examination and polyp characteristics | |||||||
| Number with FUV1 | Number of AN (CRC) | % | Number with FUV1 | Number of AN (CRC) | % | Number with FUV1 | Number of AN (CRC) | % | |
| < 18 months | 926 | 71 (7) | 7.7 | 204 | 21 (3) | 10.3 | 366 | 42 (5) | 11.5 |
| 2 yearsa | 436 | 40 (3) | 9.2 | 149 | 14 (2) | 9.4 | 244 | 33 (4) | 13.5 |
| 3 yearsa | 465 | 59 (3) | 12.7 | 177 | 17 | 9.6 | 188 | 25 (2) | 13.3 |
| 4 yearsa | 125 | 18 (2) | 14.4 | 87 | 12 (3) | 13.8 | 93 | 15 (3) | 16.1 |
| 5 yearsa | 61 | 9 (2) | 14.8 | 52 | 10 (1) | 19.2 | 51 | 11 (1) | 21.6 |
| 6 yearsa | 36 | 6 | 16.7 | 34 | 4 | 11.8 | 34 | 9 (3) | 26.5 |
| ≥ 6.5 years | 18 | 5 (1) | 27.8 | 45 | 10 | 22.2 | 42 | 14 (5) | 33.3 |
| Total | 2067 | 208 (18) | 10.1 | 748 | 88 (9) | 11.8 | 1018 | 149 (23) | 14.6 |
| pb < 0.0001 | pb = 0.0137 | pb < 0.0001 | |||||||
Sensitivity analyses and internal validation
Owing to the complex nature of the hospital data set and the rules used to define baseline and follow-up visits and interval, a number of sensitivity analyses were carried out to examine whether or not our methods were robust and did not introduce bias into the study sample. Specifically, we restricted analyses of the effect of interval on AA and CRC detection rates at FUV1, and the effect of surveillance on long-term CRC risk after baseline to:
-
patients whose baseline visit comprised only a single colonoscopy – this verifies whether or not rules used to define and extend the baseline visit were adequate or if they introduced bias into the calculation of surveillance interval length
-
patients with a complete colonoscopy at FUV1 – this assesses whether or not results were biased by the inclusion of patients without a complete colonoscopy at FUV1 (arguably the lack of a complete colonoscopy at FUV1 may mean that the surveillance visit was not as effective, as the whole colon could not be examined).
To check whether or not the definition of AA used had affected our results, a sensitivity analysis of the effect of interval on AA detection rates at FUV1 was also performed, with the definition of AA changed to a large (≥ 10 mm) adenoma or an adenoma with HGD (i.e. excluding villous or tubulovillous histology).
We also undertook sensitivity analyses to investigate the potential for misclassification of surveillance attendance, and how this may have impacted on the apparent effect of surveillance on CRC risk after baseline. Owing to a gap between the end of hospital data collection and patient follow-up with cancer registries, around 50% of the hospital cohort had follow-up time after the end of data collection during which they may have attended surveillance visits for which we were not able to collect reports. We hypothesised that any such misclassification would be non-differential and would therefore result in underestimation of the effect of surveillance. In addition, we may have potentially underestimated pre-surveillance CRC risk after baseline owing to contamination of the no-surveillance group with patients who had in fact attended one or more follow-ups. To investigate this, we restricted analyses of CRC risk after baseline to patients with at least 5 years and at least 7 years of hospital data collection, among whom any misclassification of surveillance attendance is extremely unlikely.
Patients with a single baseline colonoscopy
When the cohort was restricted to patients whose baseline visit comprised a single colonoscopy only (n = 2489), the overall AA detection rate at FUV1 was 10.1%, compared with 9.8% in all patients who attended follow-up (n = 4608). In addition, the effects of interval and baseline risk factors on the odds of detecting AA at FUV1 were similar to the effects observed in the main analysis (compare Table 59 and Table 29).
| Baseline risk factor | Category | Number of patients (N = 2489) | Univariable analysis: new AA | Multivariable analyses: new AA | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 2489) | Model 2 – interval as continuous (n = 2489) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 932 | 72 (7.73) | 1 | 0.0003 | 1 | 0.0005 | n/a | |
| 2 yearsa | 467 | 46 (9.85) | 1.31 (0.89 to 1.92) | 1.24 (0.83 to 1.86) | |||||
| 3 yearsa | 609 | 62 (10.18) | 1.35 (0.95 to 1.93) | 1.42 (0.98 to 2.07) | |||||
| 4 yearsa | 203 | 20 (9.85) | 1.31 (0.78 to 2.20) | 1.18 (0.68 to 2.05) | |||||
| 5 yearsa | 124 | 21 (16.94) | 2.44 (1.44 to 4.13) | 2.42 (1.38 to 4.24) | |||||
| 6 yearsa | 73 | 11 (15.07) | 2.12 (1.07 to 4.20) | 1.97 (0.96 to 4.07) | |||||
| ≥ 6.5 years | 81 | 19 (23.46) | 3.66 (2.08 to 6.46) | 3.94 (2.14 to 7.24) | |||||
| Per year increase | n/a | n/a | 1.16 (1.09 to 1.23) | < 0.0001 | n/a | 1.17 (1.09 to 1.25) | < 0.0001 | ||
| Age (years) | < 55 | 590 | 38 (6.44) | 1 | 0.0014 | 1 | 0.0028 | 1 | 0.0030 |
| ≥ 55 and < 60 | 355 | 49 (13.8) | 2.33 (1.49 to 3.63) | 2.34 (1.48 to 3.71) | 2.31 (1.46 to 3.66) | ||||
| ≥ 60 and < 65 | 431 | 38 (8.82) | 1.40 (0.88 to 2.24) | 1.39 (0.86 to 2.26) | 1.38 (0.85 to 2.23) | ||||
| ≥ 65 and < 70 | 447 | 44 (9.84) | 1.59 (1.01 to 2.49) | 1.42 (0.89 to 2.26) | 1.41 (0.88 to 2.25) | ||||
| ≥ 70 and < 75 | 353 | 42 (11.9) | 1.96 (1.24 to 3.11) | 1.82 (1.13 to 2.94) | 1.82 (1.13 to 2.92) | ||||
| ≥ 75 and < 80 | 208 | 31 (14.9) | 2.54 (1.54 to 4.21) | 2.51 (1.49 to 4.26) | 2.5 (1.48 to 4.22) | ||||
| ≥ 80 | 105 | 9 (8.57) | 1.36 (0.64 to 2.91) | 1.45 (0.66 to 3.18) | 1.42 (0.65 to 3.12) | ||||
| Most complete colonoscopy | Complete | 1507 | 130 (8.63) | 1 | 0.0030 | 1 | 0.0018 | 1 | 0.0015 |
| Incomplete/unknown | 982 | 121 (12.32) | 1.49 (1.15 to 1.93) | 1.65 (1.20 to 2.25) | 1.65 (1.21 to 2.26) | ||||
| Largest adenoma (mm) | < 20 | 1827 | 178 (9.74) | 1 | 0.3511 | 1 | 0.1966 | 1 | 0.2275 |
| ≥ 20 | 662 | 73 (11.03) | 1.15 (0.86 to 1.53) | 1.22 (0.90 to 1.66) | 1.21 (0.89 to 1.63) | ||||
| Large hyperplastic polyp | No | 2454 | 242 (9.86) | 1 | 0.0079 | 1 | 0.0139 | 1 | 0.0151 |
| Yes | 35 | 9 (25.71) | 3.16 (1.47 to 6.83) | 3.06 (1.34 to 6.98) | 3.01 (1.32 to 6.85) | ||||
| Proximal polyp | No | 1592 | 140 (8.79) | 1 | 0.0049 | 1 | < 0.0001 | 1 | < 0.0001 |
| Yes | 897 | 111 (12.37) | 1.46 (1.13 to 1.91) | 1.92 (1.43 to 2.58) | 1.91 (1.42 to 2.56) | ||||
When the cohort was restricted to patients with a single colonoscopy at baseline, the CRC detection rate at FUV1 was the same as in the main analysis, at 1.1%. The effect of interval on CRC detection at FUV1 differed somewhat in the sensitivity analysis; although some ORs differed to those estimated in the main analysis, the trends observed were comparable (compare Table 60 and Table 30). The smaller number of end points available in the sensitivity analysis meant that there was a greater degree of imprecision and the results were less statistically significant than those of the main analysis.
| Baseline risk factor | Category | Number of patients (N = 2489) | Univariable analysis: new CRC | Multivariable analyses: new CRC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 2489) | Model 2 – interval as continuous (n = 2489) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 932 | 10 (1.07) | 1 | 0.0031 | 1 | 0.0022 | n/a | |
| 2 yearsa | 467 | 4 (0.86) | 0.80 (0.25 to 2.55) | 0.82 (0.25 to 2.65) | |||||
| 3 yearsa | 609 | 1 (0.16) | 0.15 (0.02 to 1.19) | 0.20 (0.03 to 1.59) | |||||
| 4 yearsa | 203 | 4 (1.97) | 1.85 (0.58 to 5.97) | 2.56 (0.78 to 8.45) | |||||
| 5 yearsa | 124 | 1 (0.81) | 0.75 (0.01 to 5.91) | 1.00 (0.12 to 8.00) | |||||
| 6 yearsa | 73 | 2 (2.74) | 2.6 (0.56 to 12.08) | 3.41 (0.71 to 16.38) | |||||
| ≥ 6.5 years | 81 | 5 (6.17) | 6.07 (2.02 to 18.2) | 8.27 (2.63 to 26.01) | |||||
| Per year increase | n/a | n/a | 1.27 (1.13 to 1.43) | 0.0011 | n/a | 1.31 (1.15 to 1.49) | 0.0004 | ||
| Age (years) | < 60 | 945 | 2 (0.21) | 1 | 0.0001 | 1 | 0.0001 | 1 | < 0.0001 |
| ≥ 60 and < 65 | 431 | 2 (0.46) | 2.20 (0.31 to 15.66) | 2.44 (0.34 to 17.63) | 1.89 (0.26 to 13.89) | ||||
| ≥ 65 and < 70 | 447 | 4 (0.89) | 4.26 (0.78 to 23.33) | 3.76 (0.68 to 20.90) | 3.63 (0.65 to 20.23) | ||||
| ≥ 70 and < 75 | 353 | 8 (2.27) | 10.93 (2.31 to 51.74) | 11.62 (2.43 to 55.55) | 11.01 (2.3 to 52.63) | ||||
| ≥ 75 and < 80 | 208 | 7 (3.37) | 16.42 (3.39 to 79.63) | 17.91 (3.64 to 88.22) | 17.95 (3.67 to 87.7) | ||||
| ≥ 80 | 105 | 4 (3.81) | 18.67 (3.38 to 103.22) | 19.00 (3.36 to 107.49) | 20.04 (3.57 to 112.35) | ||||
| Most complete colonoscopy | Complete | 1507 | 13 (0.86) | 1 | 0.1906 | n/a | 1 | 0.2402 | |
| Incomplete/unknown | 982 | 14 (1.43) | 1.66 (0.78 to 3.55) | 1.65 (0.72 to 3.81) | |||||
| Best bowel preparation | Excellent/good/satisfactory/unknown | 2396 | 24 (1) | 1 | 0.0968 | 1 | 0.0643 | 1 | 0.0627 |
| Poor | 93 | 3 (3.23) | 3.29 (0.97 to 11.14) | 4.03 (1.12 to 14.45) | 4.05 (1.14 to 14.41) | ||||
| Proximal polyp | No | 1592 | 14 (0.88) | 1 | 0.1957 | n/a | 1 | 0.0993 | |
| Yes | 897 | 13 (1.45) | 1.66 (0.78 to 3.54) | 2.00 (0.88 to 4.54) | |||||
In the sensitivity analysis of CRC risk after baseline restricted to patients with a single baseline colonoscopy (n = 6500), 72 CRCs were diagnosed, compared with 168 CRCs in 11,944 patients in the main analysis. The effect of a single surveillance visit remained the same, and the effect of two or more was similar; however, because of the reduction in the number of end points, the ORs were less precise and the effect of surveillance was only borderline significant (compare Table 61 and Table 44). Similar trends were seen among baseline risk factors and CRC risk after baseline.
| Baseline risk factor | Category | Adjusted HR (95% CI) | p-value (LRT) |
|---|---|---|---|
| Number of follow-up visits after baselinea | 0 | 1 | 0.0484 |
| 1 | 0.51 (0.27 to 0.96) | ||
| 2+ | 0.44 (0.18 to 1.08) | ||
| Largest adenoma (mm) | < 10 | 1 | 0.0050 |
| 10–19 | 9.56 (1.3 to 70.3) | ||
| ≥ 20 | 8.02 (1.05 to 61.41) | ||
| Worst adenoma dysplasia | Low grade | 1 | 0.0023 |
| High grade | 2.48 (1.44 to 4.26) | ||
| Completeness of colonoscopy | Complete | 1 | 0.0008 |
| Incomplete/unknown | 2.33 (1.44 to 3.79) | ||
| Proximal polyps | No | 1 | 0.0165 |
| Yes | 1.82 (1.13 to 2.95) | ||
| Age (years) at baseline | < 55 | 1 | 0.0210 |
| ≥ 55 and < 60 | 1.11 (0.36 to 3.4) | ||
| ≥ 60 and < 65 | 1.57 (0.6 to 4.09) | ||
| ≥ 65 and < 70 | 2.35 (0.98 to 5.62) | ||
| ≥ 70 and < 75 | 2.86 (1.17 to 7.01) | ||
| ≥ 75 and < 80 | 3.84 (1.56 to 9.46) | ||
| ≥ 80 | 3.37 (1.26 to 9.01) |
The similarity between these sensitivity analyses and the main analyses suggests that the methods used to define and, in some cases, extend the baseline visit did not introduce bias into the data.
Patients with a complete colonoscopy at follow-up visit 1
Analyses of AA and CRC at FUV1 and CRC risk after baseline were repeated, this time restricting the cohort to patients with a complete colonoscopy at FUV1. The detection rate of AA at FUV1 in the restricted cohort (n = 3299) was 10.5%, which was marginally higher than the 9.8% detection rate in the full cohort. The effects of interval and baseline risk factors on AA detection were similar to the effects observed in the main analysis, apart from adenoma size, which was no longer statistically significant (compare Table 62 and Table 29).
| Baseline risk factor | Category | Number of patients (N = 3299) | Univariable analysis: new AA | Multivariable analyses: new AA | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 3299) | Model 2 – interval as continuous (n = 3299) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1225 | 99 (8.08) | 1 | 0.0001 | 1 | 0.0004 | n/a | |
| 2 yearsa | 628 | 65 (10.35) | 1.31 (0.95 to 1.82) | 1.28 (0.91 to 1.80) | |||||
| 3 yearsa | 831 | 86 (10.35) | 1.31 (0.97 to 1.78) | 1.46 (1.06 to 2.00) | |||||
| 4 yearsa | 254 | 34 (13.39) | 1.76 (1.16 to 2.66) | 1.74 (1.12 to 2.72) | |||||
| 5 yearsa | 174 | 25 (14.37) | 1.91 (1.19 to 3.06) | 1.84 (1.11 to 3.03) | |||||
| 6 yearsa | 100 | 15 (15) | 2.01 (1.12 to 3.61) | 1.99 (1.07 to 3.69) | |||||
| ≥ 6.5 years | 87 | 21 (24.14) | 3.62 (2.13 to 6.16) | 3.78 (2.13 to 6.74) | |||||
| Per year increase | n/a | n/a | 1.15 (1.08 to 1.21) | < 0.0001 | n/a | 1.15 (1.08 to 1.23) | < 0.0001 | ||
| Age (years) | < 55 | 772 | 53 (6.87) | 1 | 0.0001 | 1 | 0.0001 | 1 | 0.0001 |
| ≥ 55 and < 60 | 468 | 59 (12.61) | 1.96 (1.32 to 2.89) | 1.86 (1.24 to 2.78) | 1.87 (1.25 to 2.80) | ||||
| ≥ 60 and < 65 | 580 | 55 (9.48) | 1.42 (0.96 to 2.11) | 1.46 (0.98 to 2.19) | 1.44 (0.96 to 2.16) | ||||
| ≥ 65 and < 70 | 592 | 59 (9.97) | 1.50 (1.02 to 2.21) | 1.42 (0.95 to 2.12) | 1.42 (0.95 to 2.11) | ||||
| ≥ 70 and < 75 | 509 | 62 (12.18) | 1.88 (1.28 to 2.77) | 1.99 (1.33 to 2.96) | 1.98 (1.33 to 2.94) | ||||
| ≥ 75 and < 80 | 263 | 46 (17.49) | 2.88 (1.88 to 4.39) | 3.03 (1.94 to 4.72) | 2.99 (1.92 to 4.65) | ||||
| ≥ 80 | 115 | 11 (9.57) | 1.43 (0.73 to 2.84) | 1.67 (0.83 to 3.38) | 1.65 (0.82 to 3.34) | ||||
| Most complete colonoscopy | Complete | 2333 | 206 (8.83) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| Incomplete/unknown | 966 | 139 (14.39) | 1.74 (1.38 to 2.18) | 1.78 (1.36 to 2.33) | 1.81 (1.38 to 2.37) | ||||
| Largest adenoma (mm) | < 20 | 2122 | 212 (9.99) | 1 | 0.2414 | 1 | 0.1237 | 1 | 0.1408 |
| ≥ 20 | 1177 | 133 (11.3) | 1.15 (0.91 to 1.44) | 1.21 (0.95 to 1.54) | 1.20 (0.94 to 1.52) | ||||
| Large hyperplastic polyp | No | 3231 | 330 (10.21) | 1 | 0.0050 | 1 | 0.0040 | 1 | 0.0039 |
| Yes | 68 | 15 (22.06) | 2.49 (1.39 to 4.46) | 2.65 (1.43 to 4.89) | 2.65 (1.44 to 4.88) | ||||
| Proximal polyp | No | 2039 | 192 (9.42) | 1 | 0.0136 | 1 | < 0.0001 | 1 | < 0.0001 |
| Yes | 1260 | 153 (12.14) | 1.33 (1.06 to 1.67) | 1.70 (1.33 to 2.19) | 1.70 (1.33 to 2.18) | ||||
When the analysis of CRC at FUV1 was carried out using only patients with a complete colonoscopy at FUV1, the CRC detection rate was slightly lower, at 0.7%, compared with 1.1% in the main analysis. The effect of surveillance interval was weaker and only borderline significant compared with the highly significant effect obtained when all 4608 patients were analysed (compare Table 63 and Table 30).
| Baseline risk factor | Category | Number of patients (N = 3299) | Univariable analysis: new CRC | Multivariable analyses: new CRC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 3299) | Model 2 – interval as continuous (n = 3299) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1225 | 11 (0.9) | 1 | 0.0403 | 1 | 0.0491 | n/a | |
| 2 yearsa | 628 | 2 (0.32) | 0.35 (0.08 to 1.6) | 0.36 (0.08 to 1.65) | |||||
| 3 yearsa | 831 | 2 (0.24) | 0.27 (0.06 to 1.2) | 0.33 (0.07 to 1.49) | |||||
| 4 yearsa | 254 | 5 (1.97) | 2.22 (0.76 to 6.43) | 2.99 (1.01 to 8.92) | |||||
| 5 yearsa | 174 | 1 (0.57) | 0.64 (0.08 to 4.97) | 0.91 (0.11 to 7.20) | |||||
| 6 yearsa | 100 | 2 (2) | 2.25 (0.49 to 10.3) | 3.22 (0.68 to 15.2) | |||||
| ≥ 6.5 years | 87 | 1 (1.15) | 1.28 (0.16 to 10.06) | 1.84 (0.23 to 14.73) | |||||
| Per year increase | n/a | n/a | 1.10 (0.91 to 1.34) | 0.3467 | n/a | 1.15 (0.95 to 1.4) | 0.1838 | ||
| Age (years) | < 60 | 1240 | 4 (0.32) | 1 | 0.0551 | 1 | 0.0387 | 1 | 0.0435 |
| ≥ 60 and < 65 | 580 | 3 (0.52) | 1.61 (0.36 to 7.20) | 1.46 (0.32 to 6.64) | 1.52 (0.34 to 6.84) | ||||
| ≥ 65 and < 70 | 592 | 4 (0.68) | 2.10 (0.52 to 8.43) | 1.77 (0.44 to 7.21) | 1.86 (0.46 to 7.52) | ||||
| ≥ 70 and < 75 | 509 | 6 (1.18) | 3.69 (1.04 to 13.12) | 3.79 (1.05 to 13.67) | 3.68 (1.03 to 13.20) | ||||
| ≥ 75 and < 80 | 263 | 6 (2.28) | 7.21 (2.02 to 25.75) | 7.80 (2.15 to 28.27) | 7.72 (2.14 to 27.81) | ||||
| ≥ 80 | 115 | 1 (0.87) | 2.71 (0.30 to 24.45) | 2.47 (0.27 to 22.85) | 2.40 (0.26 to 22.11) | ||||
| Most complete colonoscopy | Complete | 2333 | 15 (0.64) | 1 | 0.3857 | n/a | 1 | 0.3723 | |
| Incomplete/unknown | 966 | 9 (0.93) | 1.45 (0.63 to 3.33) | 1.52 (0.62 to 3.73) | |||||
| Best bowel preparation | Excellent/good/satisfactory/unknown | 3174 | 20 (0.63) | 1 | 0.0129 | 1 | 0.0075 | 1 | 0.0098 |
| Poor | 125 | 4 (3.2) | 5.21 (1.75 to 15.49) | 6.34 (2.03 to 19.79) | 5.79 (1.89 to 17.69) | ||||
| Proximal polyp | No | 2039 | 12 (0.59) | 1 | 0.2389 | n/a | 1 | 0.1963 | |
| Yes | 1260 | 12 (0.95) | 1.62 (0.73 to 3.63) | 1.75 (0.75 to 4.1) | |||||
In the analysis of CRC risk after baseline in patients with a complete colonoscopy at FUV1 (n = 10,685), there were 144 CRCs diagnosed, which was only slightly less than the 168 CRCs in the full cohort. Consequently, the effect of surveillance was almost identical in both analyses (compare Table 64 and Table 44). The effects of baseline risk factors on CRC risk were similar to the main analysis, except for the effect of a large adenoma, which was slightly weaker and only borderline significant in the sensitivity analysis.
| Baseline risk factor | Category | Adjusted HR (95% CI) | p-value (LRT) |
|---|---|---|---|
| Number of follow-up visits after baselinea | 0 | 1 | 0.0003 |
| 1 | 0.51 (0.32 to 0.81) | ||
| 2+ | 0.28 (0.12 to 0.64) | ||
| Largest adenoma (mm) | < 10 | 1 | 0.0455 |
| 10–19 | 2.50 (1.00 to 6.27) | ||
| ≥ 20 | 2.84 (1.11 to 7.26) | ||
| Worst adenoma dysplasia | Low grade | 1 | 0.0027 |
| High grade | 1.86 (1.26 to 2.76) | ||
| Completeness of colonoscopy | Complete | 1 | < 0.0001 |
| Incomplete/unknown | 2.16 (1.51 to 3.08) | ||
| Proximal polyps | No | 1 | 0.0003 |
| Yes | 1.95 (1.37 to 2.77) | ||
| Age (years) at baseline | < 55 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 0.92 (0.39 to 2.18) | ||
| ≥ 60 and < 65 | 1.07 (0.50 to 2.28) | ||
| ≥ 65 and < 70 | 2.41 (1.29 to 4.50) | ||
| ≥ 70 and < 75 | 2.35 (1.23 to 4.51) | ||
| ≥ 75 and < 80 | 3.37 (1.78 to 6.38) | ||
| ≥ 80 | 2.93 (1.45 to 5.89) |
The fact that these sensitivity analyses and the main analyses demonstrated comparable effects of interval and surveillance suggests that the inclusion of patients without a complete colonoscopy at FUV1 did not bias the results in any way.
Redefining advanced adenoma
When the definition of AA was altered to include only adenomas of ≥ 10 mm or with HGD, 324 patients had AA detected at FUV1 and the AA detection rate was 7.0%, compared with 415 and 9.8% when villous or tubulovillous histology was included in the definition. The effect of interval was slightly stronger when AA was redefined, with a trend of increasing odds of AA at FUV1 with increasing interval length as observed in the main analysis (compare Tables 65 and 29). The associations between baseline risk factors and AA at FUV1 were similar in both sets of analyses, except for adenoma size, which was only weakly associated with AA at FUV1 using the restricted definition. These findings suggest that the inclusion of villous or tubulovillous histology in the definition of AA is appropriate and did not bias our results for the effect of interval on AA at FUV1.
| Baseline risk factor | Category | Number of patients (N = 4608) | Univariable analysis: new AA | Multivariable analyses: new AA | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 4608) | Model 2 – interval as continuous (n = 4608) | ||||||||
| n (%) | Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | |||
| Interval | < 18 months | 1760 | 89 (5.06) | 1 | < 0.0001 | 1 | < 0.0001 | n/a | |
| 2 yearsa | 976 | 66 (6.76) | 1.36 (0.98 to 1.89) | 1.36 (0.98 to 1.91) | |||||
| 3 yearsa | 1057 | 74 (7.00) | 1.41 (1.03 to 1.94) | 1.53 (1.10 to 2.13) | |||||
| 4 yearsa | 355 | 34 (9.58) | 1.99 (1.32 to 3.00) | 2.13 (1.39 to 3.27) | |||||
| 5 yearsa | 217 | 22 (10.14) | 2.12 (1.30 to 3.46) | 2.31 (1.39 to 3.85) | |||||
| 6 yearsa | 123 | 15 (12.20) | 2.61 (1.46 to 4.66) | 2.66 (1.46 to 4.86) | |||||
| ≥ 6.5 years | 120 | 24 (20.00) | 4.69 (2.86 to 7.70) | 5.15 (3.04 to 8.73) | |||||
| Per year increase | n/a | n/a | 1.20 (1.14 to 1.27) | < 0.0001 | n/a | 1.23 (1.16 to 1.3) | < 0.0001 | ||
| Age (years) | < 55 | 1025 | 40 (3.90) | 1 | < 0.0001 | 1 | < 0.0001 | 1 | < 0.0001 |
| ≥ 55 and < 60 | 622 | 60 (9.65) | 2.63 (1.74 to 3.97) | 2.63 (1.73 to 4.01) | 2.66 (1.75 to 4.06) | ||||
| ≥ 60 and < 65 | 788 | 61 (7.74) | 2.07 (1.37 to 3.11) | 2.27 (1.50 to 3.45) | 2.26 (1.49 to 3.44) | ||||
| ≥ 65 and < 70 | 813 | 52 (6.40) | 1.68 (1.10 to 2.57) | 1.67 (1.09 to 2.57) | 1.69 (1.00.1 to 2.6) | ||||
| ≥ 70 and < 75 | 714 | 56 (7.84) | 2.10 (1.38 to 3.18) | 2.28 (1.49 to 3.49) | 2.29 (1.49 to 3.52) | ||||
| ≥ 75 and < 80 | 413 | 42 (10.17) | 2.79 (1.78 to 4.37) | 3.05 (1.92 to 4.84) | 3.08 (1.94 to 4.89) | ||||
| ≥ 80 | 233 | 13 (5.58) | 1.46 (0.77 to 2.77) | 1.63 (0.84 to 3.14) | 1.66 (0.86 to 3.21) | ||||
| Most complete colonoscopy | Complete | 2973 | 177 (5.95) | 1 | 0.0001 | 1 | 0.0002 | 1 | 0.0002 |
| Incomplete/unknown | 1635 | 147 (8.99) | 1.56 (1.24 to 1.96) | 1.64 (1.26 to 2.14) | 1.65 (1.27 to 2.15) | ||||
| Largest adenoma (mm) | < 20 | 2880 | 194 (6.74) | 1 | 0.3137 | 1 | 0.0631 | 1 | 0.0678 |
| ≥ 20 | 1728 | 130 (7.52) | 1.13 (0.89 to 1.42) | 1.26 (0.99 to 1.6) | 1.25 (0.98 to 1.59) | ||||
| Large hyperplastic polyp | No | 4525 | 311 (6.87) | 1 | 0.0067 | 1 | 0.0081 | 1 | 0.0074 |
| Yes | 83 | 13 (15.66) | 2.52 (1.38 to 4.60) | 2.52 (1.35 to 4.72) | 2.55 (1.36 to 4.76) | ||||
| Proximal polyp | No | 2940 | 187 (6.36) | 1 | 0.0192 | 1 | 0.0002 | 1 | 0.0001 |
| Yes | 1668 | 137 (8.21) | 1.32 (1.05 to 1.66) | 1.63 (1.27 to 2.09) | 1.63 (1.27 to 2.09) | ||||
Patients with specified minimum years of hospital data collected
To assess whether or not potential misclassification of surveillance attendance may have impacted on the apparent effect of surveillance on CRC risk after baseline, sensitivity analyses were restricted to patients with at least 5 years (n = 4854) and at least 7 years (n = 3055) of hospital data. We were able to look only at the effect of one or more surveillance visits compared with none because of the small numbers in the LIR subgroup when restricting the analysis to patients with at least 7 years of hospital data.
Compared with the analysis using all patients (n = 11,944), the effect of surveillance was slightly stronger when the cohort was restricted (Table 66). This underestimation of the effect of surveillance suggests non-differential misclassification of surveillance attendance between patients with and without CRC, with some contamination among patients classified as having no surveillance.
| Number of surveillance visits after baselinea | Cohort | |||||
|---|---|---|---|---|---|---|
| Full | With ≥ 5 years of hospital data | With ≥ 7 years of hospital data | ||||
| Univariable HR (95% CI) | p-value (LRT) | Univariable HR (95% CI) | p-value (LRT) | Univariable HR (95% CI) | p-value (LRT) | |
| Total | ||||||
| N = 11,944, cases = 168 | N = 4854, cases = 108 | N = 3055, cases = 83 | ||||
| 0.0001 | 0.0003 | |||||
| 0 | 1 | 1 | 1 | 0.0005 | ||
| 1+ | 0.49 (0.34 to 0.70) | 0.46 (0.30 to 0.70) | 0.43 (0.27 to 0.69) | |||
| LIR subgroup | ||||||
| N = 2679, cases = 13 | N = 936, cases = 7 | N = 578, cases = 4 | ||||
| 0.30 | 0.32 | |||||
| 0 | 1 | 1 | 1 | 0.1228 | ||
| 1+ | 0.51 (0.14 to 1.89) | 0.45 (0.09 to 2.20) | 0.18 (0.02 to 1.89) | |||
| HIR subgroup | ||||||
| N = 9265, cases = 155 | N = 3918, cases = 101 | N = 2477, cases = 79 | ||||
| < 0.0001 | 0.0002 | |||||
| 0 | 1 | 1 | 1 | 0.0002 | ||
| 1+ | 0.45 (0.31 to 0.66) | 0.43 (0.28 to 0.67) | 0.41 (0.25 to 0.67) | |||
In the LIR and HIR subgroups, an underestimation of the effect of surveillance was also observed (see Table 66). When the analysis was restricted to patients with at least 5 years of hospital data, the effect of one or more surveillance visits was slightly underestimated in both risk subgroups. When the cohort was restricted to patients with at least 7 years of hospital data, the effect of one or more surveillance visits was only slightly stronger in the HIR subgroup but considerably so in the LIR subgroup, although the small number of end points and imprecision in the latter group precluded interpretation. However, if we have truly underestimated the effect substantially in our main analyses as a result of misclassification it may be that surveillance is of considerable benefit in the LIR subgroup.
When pre-surveillance CRC risk was examined using sensitivity analyses restricted to patients with at least 5 years or at least 7 years of hospital data, there was evidence to suggest that pre-surveillance risk had been underestimated in the main analyses (Table 67). Overall, patients with at least 7 years of follow-up were at 54% higher risk of CRC than the general population, as opposed to the 6% higher risk estimated previously.
| Cohort | Full cohort, N = 11,944: observation time free of surveillance | Cohort with ≥ 5 years of hospital data, N = 4854: observation time free of surveillance | Cohort with ≥ 7 years of hospital data, N = 3055: observation time free of surveillance | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of observed CRCs | Number of expected CRCs | SIR (95% CI) | Number of observed CRCs | Number of expected CRCs | SIR (95% CI) | Number of observed CRCs | Number of expected CRCs | SIR (95% CI) | |
| Total | 108 | 102 | 1.06 (0.87 to 1.28) | 57 | 43 | 1.33 (1.01 to 1.72) | 40 | 26 | 1.54 (1.10 to 2.09) |
| LIR subgroup | 9 | 23 | 0.39 (0.18 to 0.75) | 4 | 9 | 0.46 (0.13 to 1.18) | 3 | 5 | 0.57 (0.12 to 1.68) |
| HIR subgroup | 99 | 79 | 1.26 (1.02 to 1.53) | 53 | 34 | 1.55 (1.16 to 2.02) | 37 | 21 | 1.78 (1.25 to 2.45) |
The misclassification of surveillance attendance had a great effect on the SIR in the HIR group, the pre-surveillance CRC risk of which became considerably greater than in the general population (see Table 67). In the LIR subgroup the pre-surveillance CRC risk also appeared to have been underestimated, with the SIR for LIR patients with at least 7 years of hospital data suggesting a 43% lower CRC incidence than in the general population. Owing to the small number of end points the SIRs for the LIR subgroup were imprecise, limiting interpretation.
Internal validation of models
To assess the performance of the multivariable logistic models for the outcomes of new findings at follow-up and the multivariable Cox regression models for the analysis of long-term cancer incidence, we performed internal validation using k-fold cross-validation with k = 10.
The 10-fold cross-validation results for the models for findings at FUV1 and FUV2 are presented in Figure 9 and Table 68. The presented results for new adenoma, AA and CRC at FUV1 and new AN at FUV2 correspond to models presented in Tables 28–30 and Table 40, respectively. For these models, the weighted mean area under the ROC curve ranged from 64.5% to 87.5%, with greater variation seen for the models for new CRC at FUV1 and new AN at FUV2. The predictions from the models for new CRC at FUV1 performed the best at discriminating between patients with and without new findings, but all models showed some ability to discriminate.
FIGURE 9.
Receiver operator characteristic curves for models of new findings at follow-up. ROC curves for each of the 10 cross-validation sets (black) and the weighted mean of the 10 cross-validation ROC curves (green) for the outcome of new adenoma at FUV1, with interval modelled as categorical (a) and as continuous (b); the outcome of new AA at FUV1, with interval modelled as categorical (c) and as continuous (d); the outcome of new CRC at FUV1, with interval modelled as categorical (e) and as continuous (f); and the outcome of new AN at FUV2, with interval modelled as categorical (g) and as continuous (h).
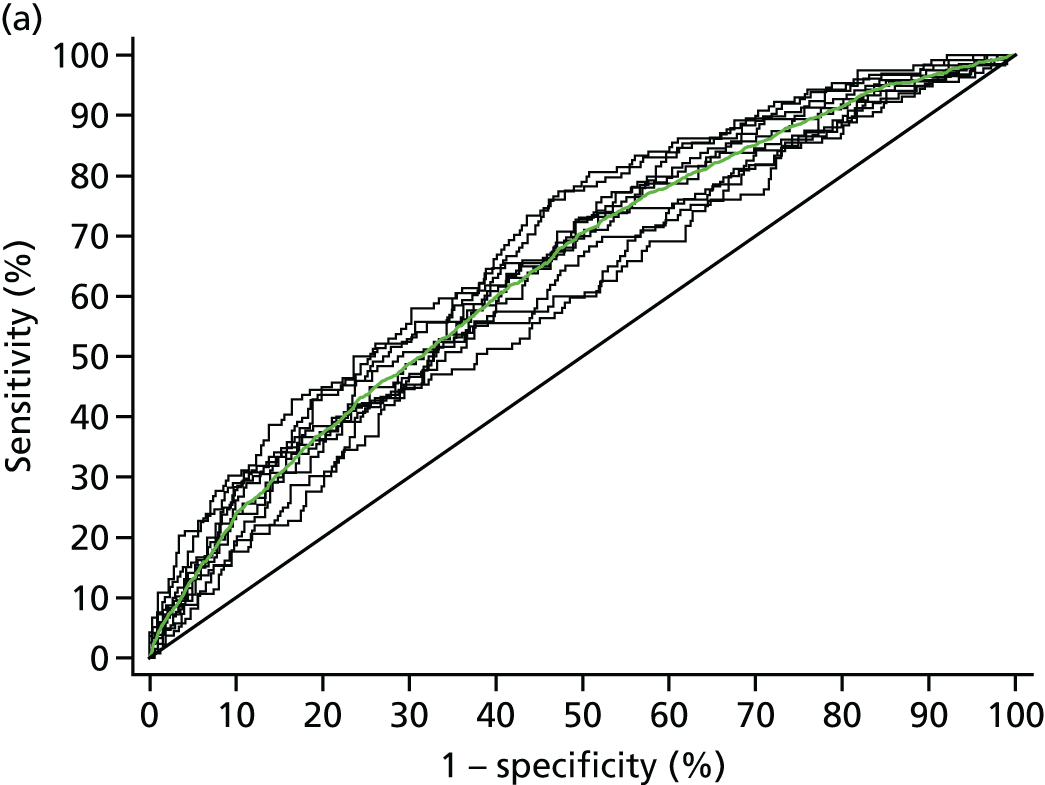





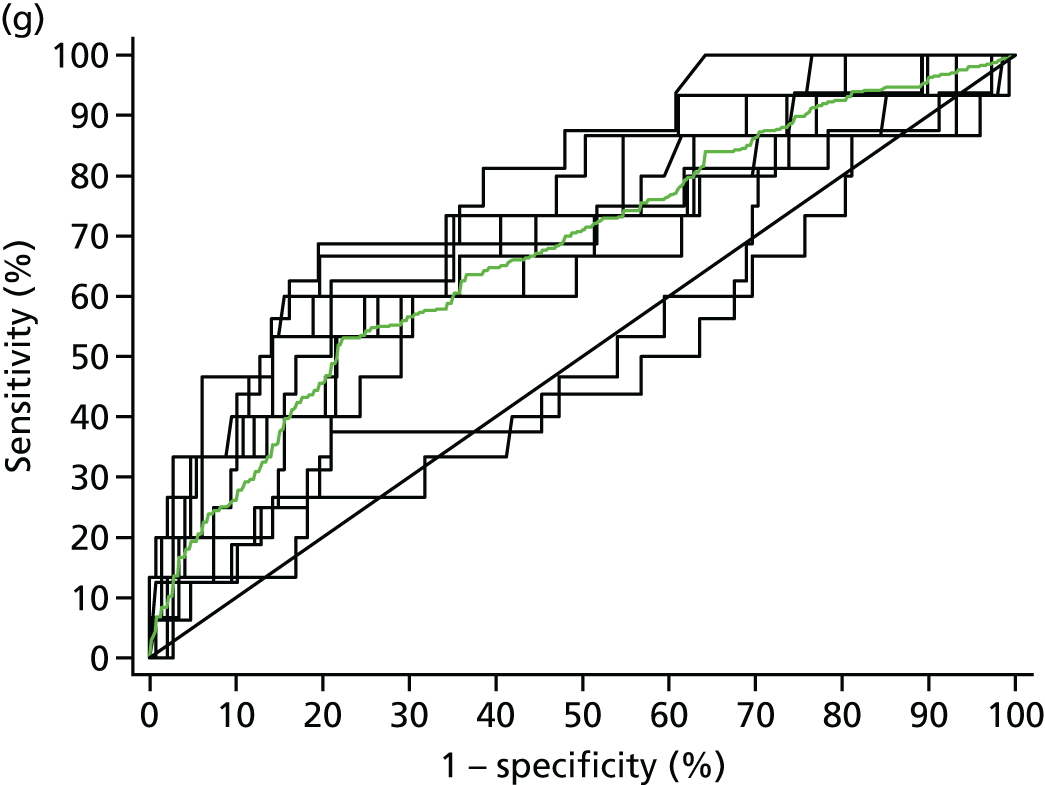

| Validation set | New adenoma at FUV1 | New AA at FUV1 | New CRC at FUV1 | New AN at FUV2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Categorical interval | Continuous interval | Categorical interval | Continuous interval | Categorical interval | Continuous interval | Categorical interval | Continuous interval | |||||||||
| Area under ROC curve | Area under ROC curve | Area under ROC curve | Area under ROC curve | Area under ROC curve | Area under ROC curve | Area under ROC curve | Area under ROC curve | |||||||||
| SE | % | SE | % | SE | % | SE | % | SE | % | SE | % | SE | % | SE | % | |
| 1 | 65.4 | 2.7 | 66.2 | 2.7 | 57.4 | 4.5 | 62.4 | 4.1 | 59.5 | 11.1 | 83.2 | 7.5 | 71.5 | 8.5 | 67.2 | 6.9 |
| 2 | 63.7 | 2.8 | 61.0 | 3.0 | 67.4 | 4.3 | 63.9 | 3.9 | 73.6 | 12.6 | 94.0 | 4.1 | 68.8 | 8.6 | 74.2 | 7.9 |
| 3 | 63.0 | 3.0 | 64.9 | 2.8 | 61.4 | 4.6 | 66.4 | 4.1 | 78.1 | 9.4 | 60.8 | 12.8 | 67.9 | 9.2 | 60.9 | 8.7 |
| 4 | 67.3 | 2.7 | 64.5 | 2.8 | 68.3 | 3.9 | 64.7 | 4.2 | 75.9 | 10.3 | 82.1 | 6.7 | 73.3 | 5.5 | 73.4 | 5.7 |
| 5 | 67.7 | 2.7 | 64.5 | 2.9 | 65.4 | 4.2 | 70.8 | 4.4 | 80.0 | 7.0 | 69.8 | 13.6 | 73.3 | 6.4 | 68.5 | 7.9 |
| 6 | 68.1 | 2.8 | 67.2 | 2.8 | 67.3 | 4.0 | 67.6 | 4.5 | 70.2 | 9.2 | 59.7 | 9.6 | 48.8 | 8.4 | 65.6 | 8.1 |
| 7 | 61.9 | 3.0 | 62.4 | 3.0 | 71.3 | 4.9 | 58.2 | 4.6 | 94.5 | 2.2 | 71.7 | 10.9 | 51.4 | 8.4 | 66.6 | 8.4 |
| 8 | 64.1 | 2.9 | 64.7 | 2.9 | 63.2 | 4.3 | 60.0 | 4.8 | 77.7 | 9.9 | 83.0 | 6.8 | 66.0 | 7.2 | 61.8 | 9.8 |
| 9 | 60.3 | 3.1 | 63.0 | 2.8 | 66.2 | 4.1 | 61.7 | 4.3 | 59.1 | 9.6 | 61.6 | 12.8 | 70.6 | 7.4 | 66.4 | 7.9 |
| 10 | 62.1 | 2.8 | 68.6 | 2.7 | 63.0 | 4.1 | 69.0 | 3.7 | 76.5 | 8.5 | 88.5 | 4.2 | 71.2 | 7.7 | 69.4 | 7.2 |
| Weighted mean | 64.5 | 0.9 | 64.8 | 0.9 | 65.1 | 1.3 | 64.7 | 1.3 | 87.5 | 1.8 | 84.2 | 2.2 | 67.5 | 2.4 | 68.3 | 2.4 |
The cross-validation results for the models for long-term CRC incidence are presented in Figure 10 and Table 69. The results for incidence after baseline correspond to the model in Table 44 and results for incidence after FUV1 correspond with the model in Table 50. The predictions from both models demonstrated some ability to discriminate between patients who were and were not diagnosed with CRC, with weighted mean areas under the ROC curve of 65–66%.
FIGURE 10.
Receiver operator characteristic curves for models of long-term CRC incidence. ROC curves for each of the 10 cross-validation sets (black) and the weighted mean of the 10 cross-validation ROC curves (green) for the outcome of long-term CRC incidence after baseline (a) and after FUV1 (b).


| Validation set | CRC incidence | |||
|---|---|---|---|---|
| After baseline | After FUV1 | |||
| Area under ROC curve | Area under ROC curve | |||
| SE | % | SE | % | |
| 1 | 61.8 | 8.3 | 53.6 | 15.2 |
| 2 | 62.2 | 6.5 | 78.3 | 6.9 |
| 3 | 62.5 | 9.0 | 47.5 | 16.4 |
| 4 | 60.5 | 6.4 | 60.9 | 10.4 |
| 5 | 69.2 | 7.0 | 52.2 | 13.0 |
| 6 | 66.2 | 7.1 | 65.8 | 18.5 |
| 7 | 62.4 | 7.4 | 60.4 | 12.1 |
| 8 | 61.1 | 8.8 | 78.0 | 9.2 |
| 9 | 66.6 | 7.3 | 66.6 | 8.3 |
| 10 | 76.3 | 7.0 | 43.3 | 12.1 |
| Weighted mean | 65.1 | 2.3 | 65.6 | 3.4 |
Discussion: hospital data set
Key findings
Does surveillance provide any benefit in terms of long-term cancer risk?
We found strong evidence that surveillance confers substantial benefit on IR patients by lowering their future risk of CRC. Overall, the first surveillance visit appeared to offer most protection, and the benefit of additional surveillance was not entirely clear. Having two or more surveillance visits was associated with an increased reduction in CRC risk, but the effect in all IR patients was not significant.
Is there a group that does not require a follow-up examination or for which a second follow-up examination might be omitted?
There appeared to be heterogeneity among IR patients in terms of long-term CRC risk and surveillance needs. When patients were subdivided into HIR and LIR subgroups based on baseline polyp and procedural predictors of CRC risk, the subgroups were discriminant in terms of future CRC risk. In the HIR subgroup, one follow-up was associated with a strong protective effect against risk of CRC after baseline, and additional surveillance provided significant further benefit compared with one follow-up alone. Before the first surveillance visit, HIR patients were at 26% increased risk of CRC compared with general population; after allowing for the effect of follow-up by including observation time after surveillance, the incidence rate of CRC was reduced but was still 13% greater than in the general population, suggesting that continued surveillance is beneficial for HIR patients.
By comparison, the effect of surveillance in the LIR subgroup was less clear, although there was a trend towards a reduction in CRC risk with a single follow-up, the results were not statistically significant. The pre-surveillance (post-baseline) incidence rate of CRC was significantly lower than that of the general population. When accounting for the effect of surveillance, there was a small reduction in the 10-year cumulative incidence of CRC, and the LIR subgroup remained at lower risk than the general population. Additional surveillance beyond one follow-up did not appear to provide any further reduction in risk in the LIR subgroup.
Is a 3-year surveillance interval appropriate for intermediate-risk patients?
Findings from the hospital data set suggest that the current surveillance interval of 3 years is appropriate for the majority of IR patients. At FUV1 the odds of detecting an AA or CRC increased significantly with increasing interval length. CRC detection rates were < 1% before the interval extended beyond 3 years, while AA detection rates were around 9% during this time. The proportion of patients with adenomas remained relatively constant across intervals (30–40%), making adenomas an uninformative outcome for specifying recommended follow-up intervals.
Based on these data we suggest that, in order to prevent delayed diagnosis of missed CRC or development of new CRC, the first surveillance examination should be performed no later than 3 years after baseline. In our data set, surveillance at 3 years would have been worthwhile, as there was an adequate yield of AA but the detection rate of CRC was low. Our results do not suggest that surveillance needs to be done earlier in most of this group of patients, as there was little increase in rates of AA in the first 3 years and a low rate of CRC.
Similarly, at FUV2 we found no evidence to suggest that the current 3-year interval between the first and second follow-ups is inappropriate. There was a significant increase in AN detection at FUV2 with increasing interval length, and more than twice the odds of AN with an interval of > 3 years.
Is there a group that needs a shorter interval to the first or second follow-up examination, or for which follow-up could be postponed?
The LIR and HIR subgroups derived from models for long-term CRC risk (see Lower- and higher-intermediate-risk subgroups, above) were discriminant when applied to findings at FUV1, but not FUV2. The detection of AN at FUV1 increased with increasing interval length in both risk subgroups, but more significantly so in the HIR subgroup. In the LIR subgroup, the AN detection rate was < 10% until the interval exceeded 3 years, providing further evidence in favour of a 3-year interval. In the HIR subgroup, the AN detection rate was 12% at 3 years, and a considerable proportion of CRCs occurred early; however, CRC incidence was only 1% before 3 years, so any gains from a shorter interval are likely to be small. An exception might be made for patients with incompletely removed lesions or a poor examination at baselines in whom a repeat examination soon after baseline might be appropriate. An association between increasing interval and AN at FUV2 was seen in the HIR subgroup but not the LIR subgroup, possibly because of a lack of power in the latter. A 3-year interval to FUV2 appeared to be the most appropriate for the HIR subgroup for the same reasons as for FUV1, but our data do not permit us to deduce the interval to FUV2 in the LIR subgroup if surveillance is offered at all.
Risk factors
Older age, poor bowel preparation quality, incomplete colonoscopy, a large adenoma (≥ 20 mm) and proximal polyps were identified as risk factors in a number of analyses of the hospital data set. Baseline risk factors for AA at FUV1 were similar to those for CRC risk after baseline, providing evidence of the validity of AA detection at follow-up as a short-term surrogate outcome for future risk of CRC. Risk factors common to both analyses included older age, large adenoma size (≥ 20 mm), proximal polyps and completeness of colonoscopy. HGD was associated with long-term CRC risk after baseline but not findings at FUV1, whereas having a large hyperplastic polyp was a risk factor for AA at FUV1 but not for CRC after baseline. Finding AA or CRC at FUV2, and future CRC risk after follow-up, were affected by both baseline and FUV1 risk factors.
Some of these risk factors have been previously reported in the literature on multiple occasions;3,23,32,51 however, a lesser known risk factor is the presence of polyps in the proximal colon, which was associated with a twofold increased risk of CRC in our cohort. An increased risk of AA at first surveillance in patients with proximal adenomas at baseline has been noted by some authors,32,52 but, to our knowledge, no study has identified this as a risk factor for CRC. The presence of proximal polyps may indicate a different biological pathway, such as the serrated pathway, whereby patients may produce lesions with rapid malignant transformation or be more likely to develop hard-to-find cancers in the proximal colon. 53
Examination quality
Colonoscopy completeness and quality of bowel preparation were important predictors of CRC in our cohort. Since the introduction of the national quality assessment tool in 2004 (UK National Endoscopy Training Programme and the Global Endoscopy Rating Scale), there has been a substantial improvement in colonoscopy quality, as assessed by ability to reach the caecum, and polyp detection rates, a measure of meticulousness in examining the colonic mucosa. 54,55 Furthermore, a study using data from the English National Cancer Repository56 found a significant 27% decline in cancers diagnosed within 3 years of colonoscopy between 2001 and 2008; such cancers are assumed to have arisen from missed or incompletely removed lesions.
An incomplete colonoscopy might be due to a number of factors, from endoscopist performance to patient characteristics such as older age or having prior abdominal or pelvic surgery. 57,58 Risk factors for poor bowel preparation quality include older age, overweight, diabetes and other comorbidities. 59,60 In those individuals for whom colonoscopy is difficult and therefore unsuccessful, it is probably inappropriate to recommend repeated colonoscopic surveillance; alternative surveillance strategies need to be explored.
The NICE colonoscopy guidelines advise that a repeat examination is performed in cases of suboptimal bowel preparation;24 however, paradoxically, this may result in reluctance on the part of an endoscopist to categorise bowel preparation quality as poor, particularly if the patient has been difficult to examine. There is only weak evidence on how to salvage a procedure when bowel preparation is found to be inadequate. 61 However, the importance of achieving good bowel preparation cannot be overstated; a systematic review62 found that adenoma detection rates were significantly higher in patients with adequate or good-quality preparation compared with poor-quality preparation.
Strengths and limitations
A major strength of the hospital data set was the wide variation in surveillance interval length. Following the adoption of national surveillance guidelines that prescribe set intervals,1,16,24 this feature is unlikely to be seen in future data sets examining adenoma follow-up.
Another achievement of the investigation was the creation of a high-quality data set despite the numerous difficulties encountered. The data used for the study were usually in a format that was not intended for research purposes, and required extensive cleaning. Thorough data collection and meticulous data coding enabled the ascertainment of detailed patient, procedural and polyp characteristics, the accuracy of which was often corroborated through the use of more than one source of information, for example endoscopic and pathological information, or multiple procedure reports. A major strength was that the raw source data have been retained for verification of our cleaning processes.
Extensive data cleaning was used to resolve transcriptional errors in, or discrepancies between, reports. Most data cleaning tasks were performed manually to ensure accuracy and avoid assumptions inherent to automation. Any inconsistencies were carefully scrutinised before being corrected. To account for changes in pathological classifications over the follow-up period, all lesions were classified using standardised, up-to-date terminology from the EU guideline for quality assurance in CRC screening and diagnosis. 63 Manual coding errors were minimised through comprehensive data consistency and validity checks, and standardised coding procedures, ensuring data accuracy.
In terms of missing data, as a result of extensive work on recorded data, the number and size of adenomas at baseline were complete, and only 4% of patients were missing data on histology or dysplasia. Completeness of baseline colonoscopy was unknown in only 14% of patients, but quality of bowel preparation was missing in 39% of patients and it is known that the reporting of the bowel preparation quality is subjective. 64 The ‘unknown’ category was retained in analyses to avoid the introduction of bias, with the assumption that it probably comprised a mixture of examinations with both suboptimal and good-quality bowel preparation.
Despite our rigorous coding and data cleaning methods, some measurement error and misclassification of exposures and confounding factors is to be expected given the nature of the routine data used; however, any misclassification should be non-differential and, thus, may have resulted in the underestimation of the effect of surveillance and interval. The potential effect of misdetermination of intervals was dealt with in the sensitivity analysis reported above (see Sensitivity analyses and internal validation). A number of studies have highlighted the potential for polyp type, size, histology, dysplasia and number to be measured inaccurately. Existing literature was used to estimate the potential effect of measurement error on our study findings. In terms of measurement error of pathological attributes of adenomas, a number of sources are available. 65–69 These indicate excellent histopathological measurement of adenoma size, with interobserver correlations among measures of the order of 0.98 and kappa values of the order of 0.85–0.90. 66,67 This would confer around a 2% bias in the estimates of logistic and Cox regression coefficients,70 and around the same proportionate increase in size of 95% CIs – see Spiegelman et al. 70 for mathematical details. Endoscopic determination of size is likely to be more subject to error. 66,68 Determination of grade of dysplasia is generally observed to be good, with kappa values of the order of 0.6 and interobserve agreement as high as 94%. 65,67 Most studies find that determination of villous status (and further classification of villous adenomas) is subject to a greater degree of measurement error, with kappa values of 0.40–0.60. 65,67,69 However, in our sensitivity analyses, we found that this was not crucial to our results. Thus, it is likely that measurement error of the pathological attributes of the adenomas, although not negligible, does not significantly alter the interpretation of our results.
Another limitation of our study is that, although some patients were censored before the end of hospital data collection, the majority had follow-up time after the end of data collection, during which they may have attended surveillance visits. Patients who were aged ≥ 73 years at the end of data collection and who had two or more follow-up visits, would not be affected in our analysis. Patients aged < 73 years with only one or no surveillance visit, recorded at the end of data collection (≈50% of our cohort) could have an underestimated number of surveillance visits resulting in the underestimation of the effect of surveillance on CRC risk. This issue was addressed in sensitivity analyses restricted to patients with at least 5 years and at least 7 years of hospital data; the sensitivity analyses suggested that, as expected, the effect of surveillance may have been underestimated.
In addition, we may have underestimated pre-surveillance risk after baseline if some patients classified as having no surveillance did, in fact, attend one or more follow-ups, which thus lowered their risk and contaminated the ‘pre-surveillance’ group. This was of particular concern in the LIR subgroup, as their pre-surveillance risk was very low compared with the general population (SIR = 0.39, 95% CI 0.18 to 0.75). When sensitivity analyses restricted to patients with at least 5 years or at least 7 years of hospital data were undertaken, the results suggested that, although the pre-surveillance CRC risk may have been underestimated as a result of misclassification of surveillance attendance, the LIR subgroup still appeared to be at substantially lower risk than the general population; however, SIRs were imprecise because of the small number of end points.
With regard to bias, we believe our methods to be relatively robust. National registries were consulted to accurately trace almost all patients’ mortality and cancer status. Together with extensive interrogation of follow-up data, this prevented loss to follow-up and limited the risk of selection bias and outcome misclassification, ensuring the quality of the data set. Similarly, selection bias due to non-response was not applicable because of the retrospective nature of the study – we extracted all available endoscopy and pathology data on every adenoma patient who underwent an endoscopy between specific dates at all study sites.
Exposed and unexposed groups (i.e. patients with and without follow-up, or with differing surveillance intervals) were both drawn from the same hospital databases of patients presenting during similar time periods, ensuring comparability. Potential selection bias could have resulted from the fact that those who attended surveillance differed from those who did not in terms of patient, procedural and polyp characteristics; however, the actual differences between patients with and without follow-up examinations were generally small. The most notable differences were in age, completeness of colonoscopy, bowel preparation and year of entry. Similarly, those with a short interval differed from those with a long interval. As factors associated with attendance at one or more follow-up visits, or with a short interval, were also risk factors for CRC after baseline and for AA and CRC at follow-up, then any selection bias should have resulted in an underestimate of the effect of interval length or surveillance on CRC risk.
A complex set of rules was generated to group examinations into visits and intervals using the NHS BCSP guideline. 71 In general, most patients had a visit that consisted of a single colonoscopy, and the baseline visit was extended beyond 11 months in only 2% of cases. With no clear surveillance recommendations or reasons for the examination provided in the reports, this was the best available method. Some error in the classification of interval length is to be expected; however, any misclassification is likely to be non-differential and so should result in only-ran underestimate of the effect of interval. We undertook sensitivity analyses using only those patients whose baseline visit comprised a single examination, or patients with a complete colonoscopy at follow-up, and we found no noteworthy differences between these and our main analyses, suggesting that the methods used to extend baseline and define visits and intervals were satisfactory and did not introduce any form of bias into the data.
All a priori confounders – for example adenoma size, dysplasia and age – were adjusted for in multivariable regression analyses in order to remove any potential confounding effects that may have obscured the associations of interest. In general, there was no confounding or evidence of only weak confounding in most multivariable analyses. However, a particularly strong confounding factor was identified when assessing the effect of interval on findings at follow-up. We found that patients at higher risk of AN at follow-up were also more likely to have had a short interval, which appeared to be due to recall for polypectomy site surveillance or continued treatment of a large lesion. This biased the effect of interval, making a shorter interval appear risky and diminishing the effect of interval overall. This confounding effect was adjusted for by removing all previously seen lesions from our analyses, as a prior sighting was the best proxy measure that could be identified in the data available. Despite this, there is some risk of residual confounding, which may have caused us to underestimate the effect of interval.
In cases for which only paper records were available (prior to the 1990s) a few early endoscopy examination(s) may have been missed; however, as most baseline data fell between 2000 and 2010 (84%) any missing data on prior examinations are likely to be negligible. We may have missed baseline or follow-up examinations for patients who were treated at a hospital that was not included in the study. This was an unavoidable problem inherent to the retrospective methods used, which could have resulted in the incorrect classification of baseline, surveillance visits or risk groups. Owing to the wide geographic coverage of the study, we believe that this is unlikely to affect many patients, if any. It should be noted that, although follow-up examinations were assumed to be for surveillance, some may have been for symptomatic purposes.
Research using pseudo-anonymised data
A number of problems were encountered as a result of the use of pseudo-anonymised data in the hospital data set. Our patient identifier lists, consisting of surname and forename(s), hospital number(s), NHS number, gender, postcode and date of birth, were created from data held on endoscopy and pathology systems. Inevitably, some of these patient identifiers were subject to data entry errors, such as spelling, incorrect recording, spaces in the NHS numbers or transpositions errors, and not all of the patient identifiers were available from the systems from which the data were extracted.
When carrying out patient follow-up, we found that a high percentage of our records could not be matched by the HSCIC, as their algorithms were designed for cleaned data sets. Not having the patient identifiers significantly limited our ability to correct the errors and complete the missing information. Moreover, the HSCIC algorithms took very few fields into account in order to find a match which, in around 7% of cases, resulted in either no match being found or multiple matches, which the HSCIC also classifies as no match.
We worked closely with the HSCIC over several months to develop new algorithms to look at multiple combinations of patient identifiers to find the match. Multiple checks were then done on the validity of the matches in order to achieve a high match rate while avoiding compromising data integrity.
In around 2.5% of cases it was necessary to ask staff at individual hospitals to use the patient-identifiable information they held (particularly the hospital number, which the HSCIC cannot use) to complete the missing information on any cases that could not be matched with certainty after all of the algorithms had been applied. The HSCIC then used the new information from the hospitals to re-match these cases. It was very difficult to get the already overstretched hospital staff to do this work for us. Finally, where it was not possible to find a match, the HSCIC performed manual ‘operator’ matches.
We also had to ensure that the individuals responsible for safeguarding the patient-linking-files at the hospital were still contactable and that in the event that they moved, the information was passed to another hospital staff member, who safeguarded the data.
The problem of following up patients to obtain information on cancers and deaths is an aspect of research that would greatly benefit from an improved ability to use non-anonymised data, or from better access to patient-identifiable information. For future follow-up, we would ideally like to hold all of the data collected, including patient identifiers, in one secure location, but current site-specific restrictions prevent the data from being held centrally; each hospital trust has its own regulations and requires the data to be held within the hospital environment. Currently, our latest cleaned data are held only by the HSCIC; if they were required to un-flag our patients, it would take many months to collate, clean and match again. The re-matching of some patients might not even be possible, as the original data collected were supplemented with information from hospital databases, some of which were quite old and may no longer be available if the databases are decommissioned. It is also possible that with tightening regulations, this process would not be permitted. Carrying out detailed research using pseudo-anonymised data is extremely challenging and has not proved to be the simple, time- and cost-saving exercise that may have been envisaged when the call for proposal using retrospective data was originally made.
Chapter 4 Screening data set: results and discussion
Background
The study team had knowledge of several large data sets collected within screening studies and programmes, which contained data on individuals who were under surveillance and were believed to have been followed up. Seven screening data sets were originally identified for inclusion; however, four were excluded for reasons given in Table 70, leaving only three which were deemed of sufficient size and quality for analysis: the UK Flexible Sigmoidoscopy Screening Trial (UKFSST), the English Bowel Cancer Screening Pilot (EP) and the Kaiser Permanente Colon Cancer Prevention Program (KP).
| Research data set | Author | Exclusion reason |
|---|---|---|
| Veteran Affairs Study | Lieberman et al. 200072 | Permission to access the data was not granted owing to concerns over data security |
| Nottingham Trial of Faecal Occult Blood Testing | Scholefield and Moss 200273 | Permission was denied for collection of data on follow-up examinations; many of the data were available only in paper records and would have been expensive and lengthy to retrieve |
| Scottish Bowel Cancer Screening Pilot | Alexander 200374 | Endoscopy and pathology data were not linked and would have had to be linked manually. There were many repeat examinations with the same information but varying dates, and over 1000 pathology reports with no corresponding endoscopy report |
| Italian FS Screening Trial (SCORE) | Segnan et al. 200275 | In this one-off FS screening trial, 17,148 men and women were invited and 9911 had FS screening in six centres. However, baseline information and cancer registry information were obtained for only one centre, which included only 194 subjects referred for colonoscopy following screening. This data set was deemed too small for inclusion |
UK Flexible Sigmoidoscopy Screening Trial
The UKFSST aimed to examine the efficacy of a single FS screening in reducing CRC incidence and mortality rates. The trial randomised 170,432 men and women aged between 55 and 64 years to either FS screening or usual care, which at the time meant no CRC screening. 10 A total of 40,674 participants was screened by FS in 14 UK centres. Individuals undergoing FS screening who were found to have a large (≥ 10 mm) lesion, three or more adenomas, villous or tubulovillous histology, severe dysplasia, malignant disease or ≥ 20 hyperplastic polyps above the rectum were offered colonoscopy surveillance. The cohort was followed up using records held by the ONS and cancer registries for incidence of CRC and deaths. Follow-up data were available until 31 December 2012.
English Bowel Cancer Screening Pilot
This study was commissioned by the DH in 1999 to determine the feasibility of CRC screening using a guaiac faecal occult blood test (gFOBT) in the UK. The pilot included two sites, one in Scotland and one in England, and ran from 2000 to 2002. 74 Men and women aged 50–69 years, registered with a NHS general practitioner (GP), were invited to complete a gFOBT, and, by 2003, 189,319 subjects in England and 297,036 in Scotland had been invited for screening, with an uptake rate of around 60%. In the EP, individuals who tested gFOBT positive were offered a meeting with a specialist screening practitioner at one of three pilot centres, who assessed their fitness for colonoscopy. In total, 82% of referred participants attended their colonoscopy. Pseudo-anonymised data on baseline and follow-up colonoscopies for those offered surveillance were available to 2012. Patient identifiers were sent directly to the ONS to obtain cancer and mortality data; data were available until 30 June 2012.
Both the UKFSST and the EP had data available for the significant risk factors for AA and CRC that were identified in the analysis of the hospital data set.
Kaiser Permanente Colon Cancer Prevention Program
The Northern California Kaiser Permanente Medical Care Program began its Colon Cancer Prevention Program in 1994, with the aim of offering sigmoidoscopy screening to all members aged ≥ 50 years once every 10 years. 76,77 The KP data set with which we were provided comprised all participants with a baseline sigmoidoscopy between January 1994 and December 1995 who then had a baseline colonoscopy within 6 months of sigmoidoscopy and at least 1 year of subsequent follow-up. Follow-up data on CRCs and deaths were available up to 31 December 2006 or until the date the participant left the program, if earlier.
Methods
Rules used to derive variables for the hospital data set (see Chapter 2, Creating summary values for polyp characteristics, Procedure information and Defining baseline and surveillance visits), including baseline and follow-up visits, and polyp and procedural characteristics, were applied to the screening data set. The KP data set did not contain information regarding quality of baseline colonoscopy; therefore, all subjects in the KP cohort were assumed to have had a complete colonoscopy with good bowel preparation at baseline. Analyses were performed with Stata/IC 13.1.
The baseline characteristics of IR subjects in the hospital and screening data sets, in the individual screening cohorts, and in screening participants with and without follow-up, were compared. The distribution of baseline characteristics among patients with and without follow-up visits in the screening data set was compared using chi-squared tests.
In the analysis of long-term CRC risk after baseline, the cut-off for follow-up was the end date of follow-up data availability in each of the individual screening cohorts. All time-to-event data were censored at first CRC diagnosis, death, emigration, end of program participation (KP data set only) or end of follow-up. Time at risk started from the latest most complete colonoscopy in baseline and, for the analysis of incidence following FUV1, time at risk started on the date of the first procedure in FUV1. If CRC was diagnosed at a follow-up visit, the follow-up visit was not included as a visit, as it did not offer any protection against CRC. ‘One minus the Kaplan–Meier estimator of the survival function’ was used to illustrate the time to cancer diagnosis and to estimate the cumulative risk of cancer with 95% CIs at 3, 5 and 10 years.
The effects of surveillance and baseline risk factors for CRC identified in the hospital data set on long-term CRC incidence in the screening data set were examined. Univariable Cox proportional hazards models were used to estimate unadjusted HRs. Independent predictors of cancer incidence identified in the hospital data set were fitted to the screening data set using a multivariable Cox proportional hazards model, with the number of follow-up visits included as a time-varying covariate.
Observed pys at risk were calculated by gender and 5-year age group. Expected numbers of CRC cases were calculated by multiplying the observed gender- and age-specific number of pys by the gender- and age-specific incidence in the general population of England in 2007. The ratio of observed to expected cases was reported as a SIR, and 95% CIs were computed assuming an exact Poisson distribution.
Findings at FUV1 were investigated. The relationship between interval from baseline to FUV1 and new AA and CRC at FUV1 was explored, both with and without adjustment for baseline risk factors identified in the hospital data set. Logistic regression models were fitted to the pooled screening data set in order to assess whether or not baseline risk factors identified in the hospital data set were predictive of advanced findings at first follow-up in screening populations. The predictive ability of these models for new AA and CRC in the screening data set was assessed using receiver operating characteristic (ROC) curves.
Screening data set: comparison of results with the hospital data set
Baseline characteristics of screening participants with intermediate-risk adenomas
In the pooled screening data set, there were 2352 subjects with IR adenomas: 796 in the UKFSST cohort, 407 in the EP cohort and 625 in the KP cohort. This compares with 11,944 in the hospital data set.
Table 71 compares the distribution of baseline demographic characteristics and risk factors for finding new AA or new cancers at FUV1 identified from the hospital data set models in the different data sets and cohorts. Participants in the pooled screening data set were younger: > 20% of patients in the hospital cohort were aged > 75 years, compared with negligible numbers in all of the screening cohorts. Individuals in the screening data set were also more likely to have had a better-quality baseline colonoscopy; this was despite almost 80% of baseline examinations being done prior to 2000, compared with only 16% in the hospital patients. Furthermore, the adenomas detected at baseline in the screening participants were less likely to be large (≥ 20 mm) or have HGD.
| Baseline risk factor | Cohort | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital (N = 11,944) | Pooled screening (N = 2352) | EP (N = 490) | UKFSST (N = 952) | KPa (N = 910) | |||||||
| n | % | n | % | n | % | n | % | n | % | ||
| Age (years) at start of baseline | < 55 | 2122 | 17.8 | 232 | 9.9 | 69 | 14.1 | 0 | 0 | 163 | 17.9 |
| ≥ 55 and < 60 | 1321 | 11.1 | 669 | 28.4 | 95 | 19.4 | 381 | 40.0 | 193 | 21.2 | |
| ≥ 60 and < 65 | 1858 | 15.6 | 855 | 36.4 | 159 | 32.4 | 500 | 52.5 | 196 | 21.5 | |
| ≥ 65 and < 70 | 2171 | 18.2 | 410 | 17.4 | 163 | 33.3 | 71 | 7.5 | 176 | 19.3 | |
| ≥ 70 and < 75 | 1786 | 15.0 | 125 | 5.3 | 3 | 0.6 | 0 | 0 | 122 | 13.4 | |
| ≥ 75 and < 80 | 1416 | 11.9 | 51 | 2.2 | 1 | 0.2 | 0 | 0 | 50 | 5.5 | |
| ≥ 80 | 1270 | 10.6 | 10 | 0.4 | 0 | 0 | 0 | 0 | 10 | 1.1 | |
| Gender | Male | 6625 | 55.5 | 1595 | 67.8 | 327 | 66.7 | 655 | 68.8 | 613 | 67.4 |
| Female | 5319 | 44.5 | 757 | 32.2 | 163 | 33.3 | 297 | 31.2 | 297 | 32.6 | |
| Year of baseline | 1980–94 | 439 | 3.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1995–9 | 1430 | 12.0 | 1861 | 79.1 | 0 | 0 | 951 | 99.9 | 910 | 100 | |
| 2000–4 | 4251 | 35.6 | 395 | 16.8 | 394 | 80.4 | 1 | 0.1 | 0 | 0 | |
| 2005–10 | 5824 | 48.8 | 96 | 4.1 | 96 | 19.6 | 0 | 0 | 0 | 0 | |
| Most complete colonoscopy | Complete | 9016 | 75.5 | 2261 | 96.1 | 475 | 96.9 | 876 | 92.0 | 910 | 100 |
| Incomplete/unknown | 2928 | 24.5 | 91 | 3.9 | 15 | 3.1 | 76 | 8.0 | 0 | 0 | |
| Best bowel preparation at colonoscopy | Excellent/good/satisfactory/unknown | 11,273 | 94.4 | 2303 | 97.9 | 482 | 98.4 | 911 | 95.7 | 910 | 100 |
| Poor | 671 | 5.6 | 49 | 2.1 | 8 | 1.6 | 41 | 4.3 | 0 | 0 | |
| Largest adenoma (mm) | < 10 | 1029 | 8.6 | 272 | 11.6 | 30 | 6.1 | 95 | 10.0 | 147 | 16.1 |
| 10–14 | 4417 | 37.0 | 1108 | 47.1 | 192 | 39.2 | 429 | 45.1 | 487 | 53.5 | |
| 15–19 | 2440 | 20.4 | 512 | 21.8 | 144 | 29.4 | 210 | 22.1 | 158 | 17.4 | |
| ≥ 20 | 4058 | 34.0 | 460 | 19.6 | 124 | 25.3 | 218 | 22.9 | 118 | 13.0 | |
| Worst adenoma histology | Tubular | 4742 | 39.7 | 1146 | 48.7 | 112 | 22.9 | 468 | 49.2 | 566 | 62.2 |
| Tubulovillous | 5576 | 46.7 | 1020 | 43.4 | 340 | 69.4 | 396 | 41.6 | 284 | 31.2 | |
| Villous | 1142 | 9.6 | 153 | 6.5 | 30 | 6.1 | 63 | 6.6 | 60 | 6.6 | |
| Unknown | 484 | 4.0 | 33 | 1.4 | 8 | 1.6 | 25 | 2.6 | 0 | 0 | |
| Worst adenoma dysplasia | Low grade | 9476 | 79.3 | 2071 | 88.1 | 389 | 79.4 | 811 | 85.2 | 871 | 95.7 |
| High grade | 1994 | 16.7 | 260 | 11.0 | 100 | 20.4 | 121 | 12.7 | 39 | 4.3 | |
| Unknown | 474 | 4.0 | 21 | 0.9 | 1 | 0.2 | 20 | 2.1 | 0 | 0 | |
| Distal polyps | No | 1980 | 16.6 | 98 | 4.2 | 31 | 6.3 | 33 | 3.5 | 34 | 3.7 |
| Yes | 9964 | 83.4 | 2254 | 95.8 | 459 | 93.7 | 919 | 96.5 | 876 | 96.3 | |
| Proximal polyps | No | 7369 | 61.7 | 1682 | 71.5 | 348 | 71.0 | 709 | 74.5 | 625 | 68.7 |
| Yes | 4575 | 38.3 | 670 | 28.5 | 142 | 29.0 | 243 | 25.5 | 285 | 31.3 | |
| Largest hyperplastic polyp (mm) | < 10 or none | 11,761 | 98.5 | 2284 | 97.1 | 484 | 98.8 | 913 | 95.9 | 887 | 97.5 |
| ≥ 10 | 183 | 1.5 | 68 | 2.9 | 6 | 1.2 | 39 | 4.1 | 23 | 2.5 | |
The three screening cohorts also differed in several respects. There were differences in the age at which screening was offered in the different cohorts: age 55–65 years in the UKFSST, age 50–69 years in the EP, but a wider age range in the KP cohort. Thus, the UKSST participants tended to be younger than in the KP and EP cohorts. The EP participants had their baseline between 2000 and 2010, whereas in the KP and UKFSST cohorts almost all were between 1995 and 1999. Examination quality was slightly worse in the UKFSST than in the EP or KP; however, data on examination quality were missing in the KP cohort, so all participants were assumed to have had a complete colonoscopy with at least satisfactory bowel preparation at baseline. Adenomas detected in the EP tended to be larger, and a much higher proportion had tubulovillous histology (69.4% EP vs. 41.6% UKFSST and 31.2% KP) or HGD (20.4% EP vs. 12.7% UKFSST and 4.3% KP).
Patients with and without surveillance
We examined the distribution of the baseline characteristics among screening participants with and without surveillance after baseline to determine the risk of selection bias in analysis of findings at and subsequent to follow-up visits (Table 72). Three-quarters of screening (1828) participants attended at least one follow-up and the remaining 524 were followed using external cancer and deaths data only.
| Baseline risk factor | Participants with one or more surveillance visits (N = 1828) | Participants with no surveillance visits (N = 524) | p-value (chi-squared test) | |||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Age (years) at start of baseline | < 55 | 56 | 10.7 | 176 | 9.6 | < 0.0001 |
| ≥ 55 and < 60 | 119 | 22.7 | 550 | 30.1 | ||
| ≥ 60 and < 65 | 163 | 31.1 | 692 | 37.9 | ||
| ≥ 65 and < 70 | 97 | 18.5 | 313 | 17.1 | ||
| ≥ 70 and < 75 | 52 | 9.9 | 73 | 4.0 | ||
| ≥ 75 and < 80 | 27 | 5.2 | 24 | 1.3 | ||
| ≥ 80 | 10 | 1.9 | 0 | 0 | ||
| Gender | Male | 1232 | 67.4 | 363 | 69.3 | 0.4171 |
| Female | 596 | 32.6 | 161 | 30.7 | ||
| Most complete colonoscopy | Complete | 1760 | 96.3 | 501 | 95.6 | 0.4836 |
| Incomplete/unknown | 68 | 3.7 | 23 | 4.4 | ||
| Best bowel preparation at colonoscopy | Excellent/good/satisfactory/unknown | 1788 | 97.8 | 515 | 98.3 | 0.5061 |
| Poor | 40 | 2.2 | 9 | 1.7 | ||
| Largest adenoma (mm) | < 10 | 201 | 11.0 | 71 | 13.5 | 0.0187 |
| 10–14 | 841 | 46.0 | 267 | 51.0 | ||
| 15–19 | 415 | 22.7 | 97 | 18.5 | ||
| ≥ 20 | 371 | 20.3 | 89 | 17.0 | ||
| Worst adenoma histology | Tubular | 843 | 46.1 | 303 | 57.8 | < 0.0001 |
| Tubulovillous | 843 | 46.1 | 177 | 33.8 | ||
| Villous | 116 | 6.4 | 37 | 7.1 | ||
| Unknown | 26 | 1.4 | 7 | 1.3 | ||
| Worst adenoma dysplasia | Low grade | 1590 | 87.0 | 481 | 91.8 | 0.0070 |
| High grade | 222 | 12.1 | 38 | 7.2 | ||
| Unknown | 16 | 0.9 | 5 | 1.0 | ||
| Distal polyps | No | 70 | 3.8 | 28 | 5.3 | 0.1262 |
| Yes | 1758 | 96.2 | 496 | 94.7 | ||
| Proximal polyps | No | 1311 | 71.7 | 371 | 70.8 | 0.6821 |
| Yes | 517 | 28.3 | 153 | 29.2 | ||
| Largest hyperplastic polyp (mm) | < 10 or none | 1778 | 97.3 | 506 | 96.6 | 0.3993 |
| ≥ 10 | 50 | 2.7 | 18 | 3.4 | ||
Those attending surveillance were younger, on average, than those who did not attend [mean 61.5 years (SD 5.2) vs. mean 63.4 years (SD 7.1); p < 0.001), but there was no difference by gender or the quality of baseline colonoscopy. Attenders were more likely to have a large adenoma (p = 0.0187), an adenoma with tubulovillous histology (p < 0.0001) or an adenoma with HGD at baseline (p = 0.0070); however, the proportions with proximal polyps or large (≥ 10 mm) hyperplastic polyps were similar.
Table 73 describes the number of follow-up visits in the screening and hospital data sets. Almost 80% of screening participants had at least one follow-up examination, compared with < 40% of the hospital patients, and 43% had at least two follow-ups, compared with only 14% of the hospital patients. The amount of follow-up was relatively similar in the EP and UKFSST cohorts, whereas the KP cohort had a greater proportion of participants without any follow-up (31% vs. 16–17%).
| Number of follow-up visits | Data set | Cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hospital (N = 11,944) | Pooled screening (N = 2352) | EP (N = 490) | UKFSST (N = 952) | KP (N = 910) | ||||||
| n | % | n | % | n | % | n | % | n | % | |
| None | 7336 | 61.4 | 524 | 22.3 | 83 | 16.9 | 156 | 16.4 | 285 | 31.3 |
| 1 | 2973 | 24.9 | 817 | 34.7 | 158 | 32.2 | 262 | 27.5 | 397 | 43.6 |
| 2 | 1080 | 9.0 | 723 | 30.7 | 189 | 38.6 | 338 | 35.5 | 196 | 21.5 |
| 3 | 354 | 3.0 | 235 | 10.0 | 52 | 10.6 | 153 | 16.1 | 30 | 3.3 |
| 4 | 135 | 1.1 | 42 | 1.8 | 6 | 1.2 | 34 | 3.6 | 2 | 0.2 |
| 5 | 45 | 0.4 | 10 | 0.4 | 2 | 0.4 | 8 | 0.8 | 0 | 0 |
| 6–10 | 21 | 0.2 | 1 | 0.04 | 0 | 0 | 1 | 0.1 | 0 | 0 |
Long-term cancer risk
A survival analysis was undertaken to assess CRC incidence after baseline and after FUV1, to account for length of follow-up time in each patient and allow investigation of the effect of surveillance on CRC risk. The median follow-up time was greater in the screening data set than in the hospital data set (11.2 years vs. 6.0 years). Among 2352 screening participants, 32 CRCs developed during 25,745 pys, with an overall incidence rate of 124.3 per 100,000 pys (95% CI 87.9 to 175.8 pys); this compares with a rate of 206.3 per 100,000 in the hospital data set (Table 74). When formally compared, there was a borderline significant, 31% lower risk of CRC in the screening data set, after adjusting for age and number of follow-up visits (p = 0.0693). Among the screening cohorts, the EP had the highest CRC incidence, which was not dissimilar to the hospital data set, followed by the UKFSST cohort; the KP cohort had a considerably lower incidence rate of CRC. The Kaplan–Meier curves in Figure 11 illustrate how the risks differ in the screening and hospital data sets (see Figure 11a) and also demonstrate how risks differ between screening data set cohorts; notably, cancers were diagnosed in the EP cohort earlier than in the UKFSST and KP cohorts (see Figure 11b).
| Cohort | Follow-up time, years: median (IQR) | pys | Number with CRC | Rate (per 100,000 pys) (95% CI) | Adjusted HR (95% CI)a | p-value (LRT) |
|---|---|---|---|---|---|---|
| Hospital | 6.0 (3.8–9.2) | 81,441.7 | 168 | 206.3 (177.3 to 240.0) | 1 | 0.0693 |
| Screening | 11.2 (9.0–14.2) | 25,745.0 | 32 | 124.3 (87.9 to 175.8) | 0.69 (0.46 to 1.04) | |
| EP | 9.6 (7.3–10.6) | 4264.4 | 8 | 187.6 (93.8 to 375.1) | n/a | n/a |
| UKFSST | 14.5 (13.8–15.1) | 12,777.5 | 17 | 133.0 (82.7 to 214.0) | ||
| KP | 10.9 (8.4–11.4) | 8703.1 | 7 | 80.4 (38.3 to 168.7) |
FIGURE 11.
Kaplan–Meier estimate of cumulative incidence by data set (a) and screening cohorts (b). Effect of surveillance on CRC risk in the screening data set.

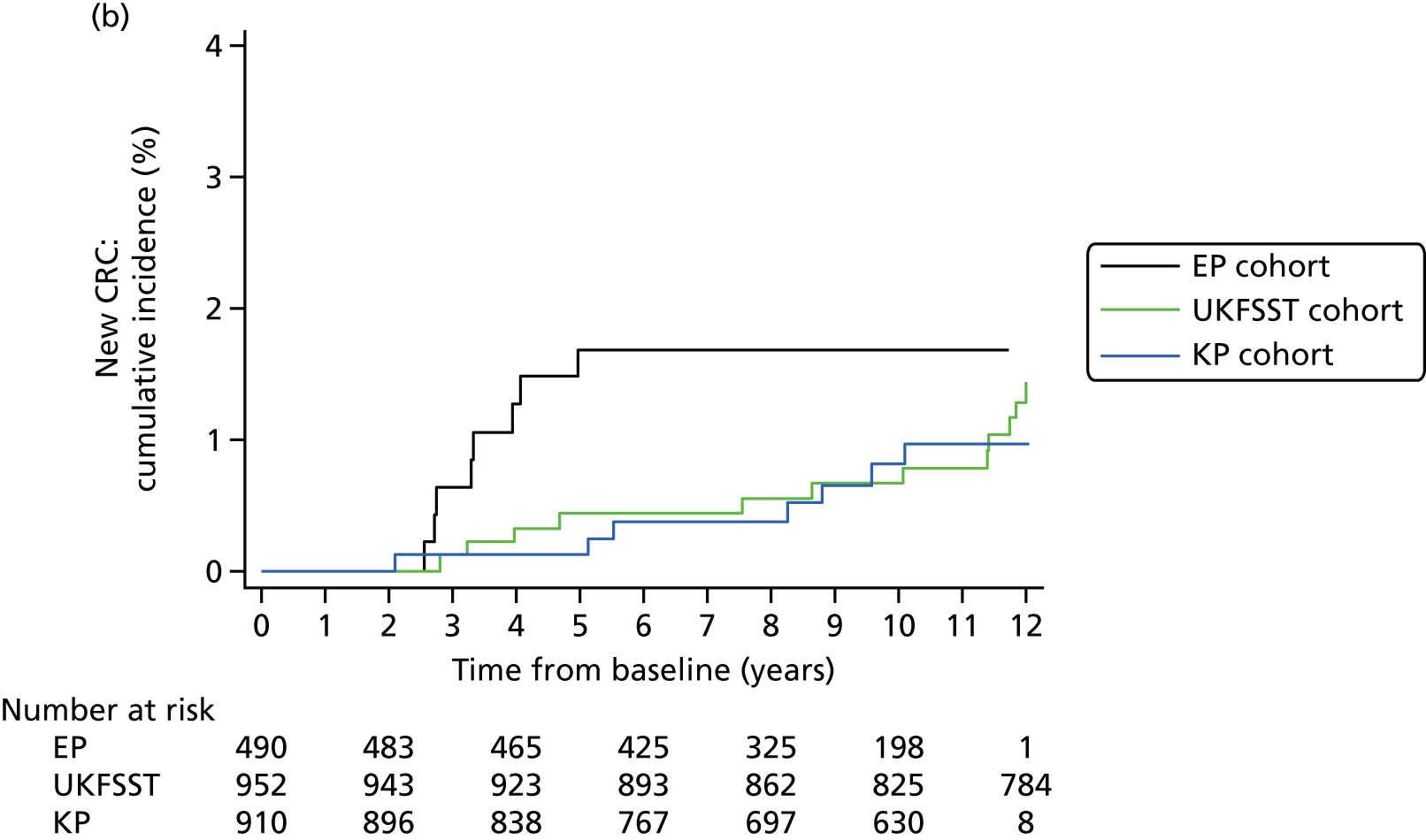
Table 75 shows the crude effects of surveillance and baseline factors on incidence of CRC after baseline in the pooled screening data set. A single surveillance examination was associated with a significant 72% lower risk of CRC (HR 0.28, 95% CI 0.10 to 0.72).
| Risk factor | pys | Number with CRC | Rate (per 100,000 pys) | Unadjusted HR (95% CI) | p-value (LRT) | |
|---|---|---|---|---|---|---|
| Number of follow-up visits after baselinea | 0 | 11,198.7 | 18 | 160.7 | 1 | 0.0154 |
| 1 | 8848.4 | 6 | 67.8 | 0.28 (0.10 to 0.72) | ||
| 2+ | 5697.9 | 8 | 140.4 | 0.36 (0.13 to 0.97) | ||
| Age (years) at baseline | < 55 | 2240.8 | 1 | 44.6 | 1 | 0.2889 |
| ≥ 55 and < 60 | 8009.6 | 7 | 87.4 | 1.44 (0.17 to 11.95) | ||
| ≥ 60 and < 65 | 9965.0 | 14 | 140.5 | 2.35 (0.30 to 18.21) | ||
| ≥ 65 and < 70 | 3907.5 | 8 | 204.7 | 4.14 (0.52 to 33.19) | ||
| ≥ 70 and < 75 | 1129.9 | 2 | 177.0 | 2.76 (0.25 to 30.41) | ||
| ≥ 75 and < 80 | 433.5 | 0 | 0 | |||
| ≥ 80 | 58.8 | 0 | 0 | |||
| Gender | Male | 17,223.9 | 20 | 116.1 | 1 | 0.6062 |
| Female | 8521.1 | 12 | 140.8 | 1.21 (0.59 to 2.47) | ||
| Most complete colonoscopy | Complete | 24,605.5 | 31 | 126.0 | 1 | 0.5845 |
| Incomplete/unknown | 1139.5 | 1 | 87.8 | 0.60 (0.08 to 4.41) | ||
| Best bowel preparation at colonoscopy | Excellent/good/satisfactory/unknown | 25,164.0 | 32 | 127.2 | n/a | n/a |
| Poor | 581.0 | 0 | 0 | |||
| Largest baseline adenoma, mm | < 10 | 2870.4 | 3 | 104.5 | 1 | 0.5979 |
| 10–19 | 17,721.7 | 20 | 112.9 | 1.05 (0.31 to 3.54) | ||
| ≥ 20 | 5152.9 | 9 | 174.7 | 1.57 (0.43 to 5.82) | ||
| Worst adenoma histology | Tubular | 12,508.8 | 13 | 103.9 | 1 | 0.0747 |
| Tubulovillous | 11,113.1 | 13 | 117.0 | 1.15 (0.53 to 2.47) | ||
| Villous | 1714.5 | 6 | 350.0 | 3.39 (1.29 to 8.93) | ||
| Unknown | 408.7 | 0 | 0 | n/a | ||
| Worst adenoma dysplasia | Low grade | 22,540.5 | 25 | 110.9 | 1 | 0.1029 |
| High grade | 2926.3 | 7 | 239.2 | 2.12 (0.92 to 4.91) | ||
| Unknown | 278.2 | 0 | 0 | n/a | ||
| Distal polyps | No | 1050.3 | 0 | 0 | n/a | n/a |
| Yes | 24,694.7 | 32 | 129.6 | |||
| Proximal polyps | No | 18,554.9 | 21 | 113.2 | 1 | 0.3958 |
| Yes | 7190.1 | 11 | 153.0 | 1.38 (0.67 to 2.86) | ||
| Largest hyperplastic polyp (mm) | < 10 or none | 25,010.4 | 31 | 123.9 | 1 | 0.9370 |
| ≥ 10 | 734.6 | 1 | 136.1 | 1.08 (0.15 to 7.95) | ||
As no procedural or polyp characteristics were predictive of CRC in the screening data set, risk factors identified in the hospital data set (including older age, incomplete colonoscopy, poor bowel preparation, large adenoma size, HGD, villous histology, proximal polyps and large hyperplastic polyp) were applied to the screening data set. These factors showed a tendency towards an increased risk of CRC, although the results were not significant and CIs for the HRs were wide and included 1 (probably because of the small number of CRC outcomes in the screening data set).
Cox proportional hazards regression was used to examine the effect of surveillance on CRC risk, controlling for potential confounding factors, with number of follow-up visits modelled as a time-varying covariate (Table 76). As no polyp or procedure factors were predictive of CRC in univariable analyses (see Table 75), the model was fitted using the set of risk factors for CRC identified from the hospital data set (see Table 44).
| Baseline risk factorsb | Category | Adjusted HR (95% CI) | p-value (LRT) |
|---|---|---|---|
| Number of follow-up visits after baselinec | 0 | 1.00 | 0.0123 |
| 1 | 0.27 (0.10 to 0.71) | ||
| 2+ | 0.33 (0.12 to 0.90) | ||
| Largest adenoma (mm) | < 10 | 1.00 | 0.6826 |
| 10–19 | 1.11 (0.32 to 3.85) | ||
| ≥ 20 | 1.57 (0.40 to 6.15) | ||
| Worst adenoma dysplasia | Low grade | 1.00 | 0.0900 |
| High grade | 2.26 (0.94 to 5.43) | ||
| Completeness of colonoscopy | Complete | 1.00 | 0.4519 |
| Incomplete/unknown | 0.50 (0.07 to 3.74) | ||
| Proximal polyp | No | 1.00 | 0.3793 |
| Yes | 1.41 (0.67 to 2.97) | ||
| Age (years) | < 55 | 1.00 | 0.3299 |
| ≥ 55 and < 60 | 1.67 (0.20 to 13.88) | ||
| ≥ 60 and < 65 | 2.64 (0.34 to 20.53) | ||
| ≥ 65 and < 70 | 4.41 (0.55 to 35.41) | ||
| ≥ 70 | 2.44 (0.22 to 27.00) |
The screening data set results provided further evidence of the benefit of surveillance in IR patients, with one follow-up visit conferring a significant 73% reduction in the rate of CRC (HR 0.27, 95% CI 0.10 to 0.71), after adjusting for covariates (adjusted effect estimates were similar to the crude estimates, suggesting little confounding).
None of the baseline risk factors for CRC identified in the hospital data set were significantly predictive of CRC in the screening cohort, before or after adjustment for covariates. The ORs suggested that older age, proximal polyps, a large adenoma, or an adenoma with HGD increased the risk of CRC; however, 95% CIs were wide, precluding interpretation.
Lower- and higher-intermediate-risk subgroups
The screening data set was divided into LIR and HIR subgroups using the definition derived from risk factors for CRC after baseline identified in the Cox regression model of CRC risk in the hospital data set (see Chapter 3, Lower- and higher-intermediate-risk subgroups, and Table 44). The HIR subgroup was defined to include participants with any of the following baseline characteristics: an adenoma of ≥ 20 mm or with HGD, proximal polyps, no complete colonoscopy or poor bowel preparation. All other participants were assigned to the LIR subgroup.
Table 77 compares the proportion of participants and CRC incidence in the LIR and HIR subgroups by data set, with the HIR subgroup stratified by polyp and procedural risk factors. In the screening data set, the HIR and LIR subgroups comprised 51% and 49% of participants, respectively. By contrast, in the hospital cohort the LIR subgroup comprised only 22%.
| Cohort | n | % | pys | Number with CRC | Rate (95% CI) (per 100,000 pys) | HR (95% CI) | p-value (LRT) | |
|---|---|---|---|---|---|---|---|---|
| Hospital (N = 11,944) | Higher-risk subgroupa | 9265 | 77.6 | 63,826.9 | 155 | 242.8 (207.5 to 284.2) | 1 | < 0.0001 |
| Lower-risk subgroup | 2679 | 22.4 | 17,614.8 | 13 | 73.8 (42.9 to 127.1) | 0.31 (0.18 to 0.55) | ||
| Higher-risk subgroup classifications | ||||||||
| Polyp factors onlyb | 5874 | 49.2 | 36,693.9 | 74 | 201.7 (160.6 to 253.3) | |||
| Poor examination onlyc | 1391 | 11.6 | 11,753.7 | 26 | 221.2 (150.6 to 324.9) | |||
| Polyp factors and poor examinationd | 2000 | 16.7 | 15,379.3 | 55 | 357.6 (274.6 to 465.8) | |||
| Screening (N = 2352) | Higher-risk subgroupa | 1200 | 51.0 | 13,190.0 | 20 | 151.6 (97.8 to 235.0) | 1 | 0.2215 |
| Lower-risk subgroup | 1152 | 49.0 | 12,555.0 | 12 | 95.6 (54.3 to 168.3) | 0.64 (0.31 to 1.32) | ||
| Higher-risk subgroup classifications | ||||||||
| Polyp factors onlyb | 1071 | 45.5 | 11,615.3 | 19 | 163.6 (104.3 to 256.4) | |||
| Poor examination onlyc | 63 | 2.7 | 769.1 | 0 | 0 | |||
| Polyp factors and poor examinationd | 66 | 2.8 | 805.5 | 1 | 124.1 (17.5 to 881.3) | |||
| Restricted to age 55–69 years | ||||||||
| Hospital (N = 5350) | Higher-risk subgroupa | 4127 | 77.1 | 29,806.6 | 61 | 204.7 (159.2 to 263.0) | 1 | 0.0486 |
| Lower-risk subgroup | 1223 | 22.9 | 8167.8 | 8 | 97.9 (49.0 to 195.9) | 0.51 (0.24 to 1.06) | ||
| Higher-risk subgroup reasons | ||||||||
| Polyp factors onlyb | 2725 | 50.9 | 17,325.7 | 26 | 150.1 (102.2 to 220.4) | |||
| Poor examination onlyc | 603 | 11.3 | 5560.6 | 12 | 215.8 (122.6 to 380.0) | |||
| Polyp factors and poor examinationd | 799 | 14.9 | 6920.3 | 23 | 332.4 (220.9 to 500.1) | |||
| Screening (N = 1934) | Higher-risk subgroupa | 1017 | 52.6 | 11,498.3 | 19 | 165.2 (105.4 to 259.1) | 1 | 0.1717 |
| Lower-risk subgroup | 917 | 47.4 | 10,383.7 | 10 | 96.3 (51.8 to 179.0) | 0.59 (0.28 to 1.28) | ||
| Higher-risk subgroup classifications | ||||||||
| Polyp factors onlyb | 893 | 46.2 | 9966.2 | 18 | 180.6 (113.8 to 286.7) | |||
| Poor examination onlyc | 62 | 3.2 | 757.7 | 0 | 0 | |||
| Polyp factors and poor examinationd | 62 | 3.2 | 774.4 | 1 | 129.1 (18.2 to 916.7) | |||
In the screening data set, incidence of CRC after baseline was 36% lower in the LIR subgroup than in the HIR subgroup but the risk subgroups were not significantly different (HR 0.64, 95% CI 0.31 to 1.32; p = 0.22); by comparison, in the hospital data set the LIR subgroup was at significantly lower risk (HR 0.31, 95% CI 0.18 to 0.55; p < 0.0001). As results may have been affected by differences in age between the hospital and screening data sets, analyses were repeated, restricting the analysis to participants aged 55–69 years. This had little effect on the screening data set, but CRC incidence was reduced in the high-risk subgroup of the hospital data set, from a rate of 242.8 to 204.7 per 100,000 pys. As a consequence, differences between the higher- and lower-risk subgroups in the hospital and screening data sets became similar (HR 0.51, 95% CI 0.24 to 1.06, and HR 0.59, 95% CI 0.28 to 1.28, respectively). These results suggest that people in the lower-risk group are at approximately 40–50% lower risk than those in the higher-risk group.
With regard to specific higher-risk polyp factors, approximately 50% of subjects in both the screening and hospital data sets were classified as HIR because of polyp characteristics alone (HGD, adenoma of ≥ 20 mm or proximal polyps at baseline). Among these subjects, the incidence rate of CRC after baseline was 201.7 and 163.6 per 100,000 pys in the hospital and screening data sets, respectively. When the HIR subgroup was restricted to participants/patients aged 55–69 years and those with only polyp risk factors, the rate of CRC became more similar in the hospital and screening data sets (150.1 vs. 180.6 per 100,000 pys).
The main difference between data sets, apart from age, was the proportion with a poor examination. In the screening data set only 2.7% of all participants were classified as HIR based solely on examination factors (an incomplete colonoscopy or poor bowel preparation), compared with 11.6% of the hospital data set. Similarly, the proportion of participants in the screening data set who were classified as HIR owing to both polyp and procedural factors was smaller than in the hospital data set (2.8% vs. 16.7%). Among these subjects, the incidence rate of CRC after baseline was 124.1 and 357.6 per 100,000 pys in the screening and hospital data sets, respectively. Restricting the analysis by age had little impact on these rates.
Effect of surveillance in the lower- and higher-intermediate-risk subgroups
The effect of surveillance in the LIR and HIR subgroups was examined, stratifying the HIR subgroup by risk factor, and restricting the analyses by age (Table 78). In the screening data set, there was a 72% lower risk of CRC with one follow-up visit than with no follow-up in the HIR subgroup, although the effect of surveillance was only borderline significant (HR 0.28, 95% CI 0.09 to 0.92; overall p = 0.0508). Attendance at additional follow-ups (two or more) did not appear to provide any further benefit. When the HIR subgroup was restricted to participants with HIR polyp characteristics only, the effect of surveillance was very similar to the effect in the HIR subgroup overall. Surveillance also had a comparable effect in the LIR subgroup, although the effect estimates were imprecise and the association was non-significant (HR 0.25, 95% CI 0.05 to 1.30, for one follow-up visit; overall p = 0.2084), probably as a result of the smaller number of CRC end points in the LIR subgroup.
| Cohort | n | % | Effect of surveillance | |||
|---|---|---|---|---|---|---|
| Number of follow-up visits after baselinea | Unadjusted HR (95% CI) | p-value (LRT) | ||||
| Hospital (N = 11,944) | Higher-risk subgroupb | 9265 | 77.6 | 0 | 1 | 0.0001 |
| 1 | 0.50 (0.34 to 0.76) | |||||
| 2+ | 0.36 (0.20 to 0.62) | |||||
| Lower-risk subgroup | 2679 | 22.4 | 0 | 1 | 0.4741 | |
| 1 | 0.62 (0.16 to 2.43) | |||||
| 2+ | 0.29 (0.03 to 2.82) | |||||
| Higher-risk subgroup classifications | ||||||
| Polyp factors onlyc | 5874 | 49.2 | 0 | 1 | 0.0078 | |
| 1 | 0.79 (0.46 to 1.36) | |||||
| 2+ | 0.26 (0.10 to 0.66) | |||||
| Poor examination onlyd | 1391 | 11.6 | 0 | 1 | 0.3943 | |
| 1 | 0.51 (0.18 to 1.41) | |||||
| 2+ | 0.82 (0.25 to 2.67) | |||||
| Polyp factors and poor examinatione | 2000 | 16.7 | 0 | 1 | 0.0001 | |
| 1 | 0.21 (0.09 to 0.49) | |||||
| 2+ | 0.28 (0.11 to 0.67) | |||||
| Screening (N = 2352) | Higher-risk subgroupb | 1200 | 51.0 | 0 | 1 | 0.0508 |
| 1 | 0.28 (0.09 to 0.92) | |||||
| 2+ | 0.30 (0.09 to 1.01) | |||||
| Lower-risk subgroup | 1152 | 49.0 | 0 | 1 | 0.2084 | |
| 1 | 0.25 (0.05 to 1.30) | |||||
| 2+ | 0.46 (0.09 to 2.46) | |||||
| Higher-risk subgroup classifications | ||||||
| Polyp factors onlyc | 1071 | 45.5 | 0 | 1 | 0.0647 | |
| 1 | 0.30 (0.09 to 0.98) | |||||
| 2+ | 0.30 (0.09 to 1.02) | |||||
| Restricted to age 55–69 years | ||||||
| Hospital (N = 5350) | Higher-risk subgroupb | 4127 | 77.1 | 0 | 1 | 0.1186 |
| 1 | 0.58 (0.30 to 1.10) | |||||
| 2+ | 0.48 (0.21 to 1.07) | |||||
| Lower-risk subgroup | 1223 | 22.9 | 0 | 1 | 0.914 | |
| 1+ | 0.92 (0.19 to 4.39) | |||||
| Higher-risk subgroup classifications | ||||||
| Polyp factors onlyc | 2725 | 50.9 | 0 | 1 | 0.1769 | |
| 1 | 1.22 (0.49 to 3.01) | |||||
| 2+ | 0.36 (0.08 to 1.57) | |||||
| Screening (N = 1934) | Higher-risk subgroupb | 1017 | 52.6 | 0 | 1 | 0.0743 |
| 1 | 0.29 (0.09 to 0.97) | |||||
| 2+ | 0.32 (0.09 to 1.12) | |||||
| Lower-risk subgroup | 917 | 47.4 | 0 | 1 | 0.1571 | |
| 1 | 0.15 (0.02 to 1.41) | |||||
| 2+ | 0.53 (0.08 to 3.28) | |||||
| Higher-risk subgroup classifications | ||||||
| Polyp factors onlyc | 893 | 46.2 | 0 | 1 | 0.0943 | |
| 1 | 0.31 (0.09 to 1.05) | |||||
| 2+ | 0.32 (0.09 to 1.12) | |||||
By contrast, in the hospital data set a single follow-up visit appeared to provide less of a reduction in risk in the HIR subgroup, although the effect was considerable and highly significant (for one follow-up visit HR 0.50, 95% CI 0.34 to 0.76) and additional surveillance was associated with a further 14% reduction in risk. Similarly, the effect of surveillance in the LIR subgroup was also smaller than in the screening data set, but interpretation was still limited by imprecision and lack of power (p = 0.4741; HR 0.63, 95% CI 0.16 to 2.43, for one follow-up visit).
When the screening data set was restricted to participants aged 55–69 years, a single follow-up appeared to have a greater effect in the LIR subgroup, but, again, the effect estimates were imprecise and results were non-significant (HR 0.15, 95% CI 0.02 to 1.41). In the hospital data set restricted by age, it was difficult to make inferences regarding the effect of surveillance in patients with HIR polyp characteristics only or in the LIR subgroup owing to imprecision.
Absolute risk of colorectal cancer in the screening data set
The cumulative incidence of CRC after baseline, and after the first follow-up, was examined in the screening data set, and risk of CRC was compared with that of the general population (Table 79). A total of 1816 participants with at least one follow-up, and who remained free of CRC at FUV1, were used for the analysis of CRC incidence after FUV1. If CRC was diagnosed at a follow-up visit, the follow-up visit was not included as a visit, as it did not offer any protection against CRC.
| Cohort | Number of patients | pys | CRC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cumulative incidence, % (95% CI) at: | Number of observed cases | Number of expected cases | SIRb (95% CI) | ||||||
| 3 years | 5 years | 10 years | |||||||
| Hospital | Total | 11,944 | 48,891.7 | 0.5 (0.4 to 0.7) | 1.1 (0.9 to 1.4) | 2.9 (2.2 to 3.9) | 108 | 102 | 1.06 (0.87 to 1.28) |
| Lower-risk subgroup | 2679 | 12,021.0 | 0.1 (0.03 to 0.4) | 0.4 (0.2 to 1.0) | 1.0 (0.4 to 2.4) | 9 | 23 | 0.39 (0.18 to 0.75) | |
| Higher-risk subgroupc | 9265 | 36,870.7 | 0.7 (0.5 to 0.9) | 1.3 (1.0 to 1.7) | 3.6 (2.6 to 4.8) | 99 | 79 | 1.26 (1.02 to 1.53) | |
| Screening | Total | 2352 | 11,198.7 | 0.3 (0.1 to 0.7) | 0.6 (0.3 to 1.2) | 1.9 (1.0 to 3.5) | 18 | 20 | 0.90 (0.53 to 1.42) |
| Lower-risk subgroup | 1152 | 5890.9 | 0.2 (0.05 to 0.8) | 0.5 (0.2 to 1.3) | 1.1 (0.4 to 2.6) | 7 | 10 | 0.67 (0.27 to 1.38) | |
| Higher-risk subgroupc | 1200 | 5307.8 | 0.4 (0.1 to 1.1) | 0.8 (0.3 to 2.0) | 2.7 (1.2 to 6.3) | 11 | 10 | 1.15 (0.57 to 2.06) | |
Absolute risk of CRC in the screening and hospital data sets, in the absence of surveillance, was assessed by censoring at FUV1 (see Table 79). In the screening data set, the cumulative incidence of CRC at 10 years was 1.9% overall, and 1.1% and 2.7%, respectively, in LIR and HIR subgroups. In the hospital data set, equivalent rates were 2.9%, 1.0% and 3.6%, respectively, so slightly higher than in the screening data set. Age- and gender-standardised incidence rates in the absence of surveillance were 0.90, 0.67 and 1.15, overall and in the HIR, and LIR groups, respectively, in the screening data set. Equivalent figures for the hospital data set were 1.06, 0.39 and 1.26, respectively. Thus, there were small differences between the LIRs in the screening and hospital data sets.
Using all observation time, risk of CRC was determined allowing for the effect of surveillance in those who attended (Table 80); comparisons between the hospital and screening data sets must be interpreted with caution because of the greater number of follow-up visits in the screening data set. Compared with pre-surveillance risk, the cumulative incidence of CRC at 10 years was reduced to 0.9%, and incidence was significantly lower in the screening data set than in the general population (SIR = 0.61, 95% CI 0.41 to 0.85). The 10-year cumulative incidence of CRC in the LIR subgroup was 0.8%, compared with 1.1% in the HIR subgroup. Incidence in the LIR subgroup was reduced to around 50% of that of the general population level, and was 26% lower in the HIR subgroup, although not significantly so (the 95% CI included 1). In comparison with the screening data set, CRC risk in the HIR subgroup of the hospital data set remained above the general population level despite allowing for the effect of surveillance (with the caveat that less surveillance occurred in the hospital data set), whereas risk in the LIR subgroup was slightly lower than that of the screening data set.
| Cohort | N | pys | CRC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cumulative incidence, % (95% CI), at | Number of observed cases | Number of expected cases | SIRa (95% CI) | ||||||
| 3 years | 5 years | 10 years | |||||||
| Hospital | Total | 11,944 | 81,441.7 | 0.5 (0.3 to 0.6) | 0.9 (0.7 to 1.1) | 2.1 (1.7 to 2.5) | 168 | 172 | 0.98 (0.84 to 1.14) |
| Lower-risk subgroup | 2679 | 17,614.8 | 0.1 (0 to 0.4) | 0.4 (0.2 to 0.8) | 0.6 (0.3 to 1.2) | 13 | 34 | 0.38 (0.20 to 0.65) | |
| Higher-risk subgroupb | 9265 | 63,826.9 | 0.6 (0.4 to 0.7) | 1.0 (0.8 to 1.3) | 2.4 (2.0 to 2.9) | 155 | 138 | 1.13 (0.96 to 1.32) | |
| Screening | Total | 2352 | 25,745.0 | 0.2 (0.09 to 0.5) | 0.6 (0.3 to 1.0) | 0.9 (0.6 to 1.5) | 32 | 53 | 0.61 (0.41 to 0.85) |
| Lower-risk subgroup | 1152 | 12,555.0 | 0.2 (0.04 to 0.7) | 0.4 (0.1 to 1.0) | 0.8 (0.4 to 1.5) | 12 | 26 | 0.47 (0.24 to 0.81) | |
| Higher-risk subgroupb | 1200 | 13,190.0 | 0.3 (0.08 to 0.8) | 0.8 (0.4 to 1.5) | 1.1 (0.6 to 2.0) | 20 | 27 | 0.74 (0.45 to 1.14) | |
Patients attending follow-up visits
In the screening data set, 1828 participants attended one or more follow-up visits. The effect of interval, and baseline factors on the detection of AA or CRC at FUV1 was examined using univariable and multivariable analyses.
The number of follow-ups and intervals to successive follow-ups were examined (Table 81). Overall, the proportion with an interval between 3 and 4 years remained relatively constant, varying between 45% and 60% to different follow-ups, and showing no trend with increasing follow-up visit number.
| Follow-up visit no. | Number (%) of patients with varying interval lengthsa | |||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsb | 3 yearsb | 4 yearsb | 5 yearsb | 6 yearsb | ≥ 6.5 years | Total | |
| 1 | 304 (16.63) | 132 (7.22) | 530 (28.99) | 367 (20.08) | 276 (15.10) | 90 (4.92) | 129 (7.06) | 1828 (100) |
| 2 | 64 (6.34) | 93 (9.22) | 375 (37.17) | 231 (22.89) | 149 (14.77) | 64 (6.34) | 33 (3.27) | 1009 (100) |
| 3 | 32 (11.15) | 25 (8.71) | 130 (45.30) | 37 (12.89) | 41 (14.29) | 17 (5.92) | 5 (1.74) | 287 (100) |
| 4 | 7 (13.46) | 10 (19.23) | 19 (36.54) | 6 (11.54) | 6 (11.54) | 2 (3.85) | 2 (3.85) | 52 (100) |
| 5 | 2 (18.18) | 1 (9.09) | 3 (27.27) | 2 (18.18) | 3 (27.27) | 0 (0) | 0 (0) | 11 (100) |
| 6 | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 21 (100) |
The interval to first follow-up varied considerably between the individual screening cohorts (Table 82). The KP cohort tended to have a longer interval, with most participants (29.9%) returning at 5 years. By contrast, most UKFSST and EP participants had an interval of 3–4 years (59% and 54%), and a greater proportion returned in < 18 months than in the KP cohort.
| Interval baseline to first follow-up | Pooled screening data set (N = 1828) | Cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| EP (N = 407) | UKFSST (N = 796) | KP (N = 625) | ||||||
| n | % | n | % | n | % | n | % | |
| < 18 months | 304 | 16.6 | 80 | 19.7 | 217 | 27.3 | 7 | 1.1 |
| 2 yearsa | 132 | 7.2 | 41 | 10.1 | 60 | 7.5 | 31 | 5.0 |
| 3 yearsa | 530 | 29.0 | 145 | 35.6 | 267 | 33.5 | 118 | 18.9 |
| 4 yearsa | 367 | 20.1 | 75 | 18.4 | 208 | 26.1 | 84 | 13.4 |
| 5 yearsa | 276 | 15.1 | 51 | 12.5 | 38 | 4.8 | 187 | 29.9 |
| 6 yearsa | 90 | 4.9 | 8 | 2.0 | 3 | 0.4 | 79 | 12.6 |
| ≥ 6.5 years | 129 | 7.1 | 7 | 1.7 | 3 | 0.4 | 119 | 19.0 |
Findings at the first follow-up
In the analysis of findings at follow-up in the hospital data set, we found that the association between interval and findings was being masked by the effect of polypectomy site surveillance. Patients undergoing polypectomy site surveillance were more likely to have the same lesion seen again at FUV1 and were also more likely to return sooner for their first follow-up. To adjust for this confounding effect, all findings at FUV1 that had been previously seen were removed from the analysis in the hospital data set. We examined findings at FUV1 in the screening data set stratified by whether or not they had been seen previously. No previously seen cancers were detected at FUV1 so we restricted analysis to AA to determine whether or not patients with a short interval were more likely to have a previously seen lesion (Table 83). Very few participants had only a previously seen AA detected at FUV1 (0.7%), but the proportion of patients with a previously seen AA was greater among those with a shorter interval. Consequently, all analyses of findings at FUV1 in the screening data set considered only new outcomes.
| AA status | Interval from baseline to first follow-up, n (%) | Total, n (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||
| None | 287 (94.4) | 123 (93.2) | 503 (94.9) | 351 (95.6) | 262 (94.9) | 87 (96.7) | 126 (97.7) | 1739 (95.1) |
| Previously seen only | 3 (1.0) | 3 (2.3) | 4 (0.7) | 2 (0.5) | 1 (0.4) | 0 (0) | 0 (0) | 13 (0.7) |
| Previously seen and new | 1 (0.3) | 0 (0) | 1 (0.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (0.1) |
| New only | 13 (4.3) | 6 (4.5) | 22 (4.2) | 14 (3.8) | 13 (4.7) | 3 (3.3) | 3 (2.3) | 74 (4.0) |
| Total | 304 (100) | 132 (100) | 530 (100) | 367 (100) | 276 (100) | 90 (100) | 129 (100) | 1828 (100) |
The detection of new AA and CRC at FUV1 in the hospital and screening data sets, and individual screening cohorts, was compared (Table 84). A new AA was detected at FUV1 in only 4.2% compared with 9.8% in the screening and hospital data sets, respectively (p < 0.0001). The proportion with new CRC was slightly lower in the screening data set than in the hospital data set (0.7% vs. 1.1%; p = 0.0852).
| New findings at first follow-up | Data set | p-value (chi-squared test) | Cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hospital (N = 4608) | Pooled screening (N = 1828) | EP (N = 407) | UKFSST (N = 796) | KP (N = 625) | |||||||
| n | % | n | % | n | % | n | % | n | % | ||
| AA | 451 | 9.8 | 76 | 4.2 | < 0.0001 | 26 | 6.4 | 37 | 4.6 | 13 | 2.1 |
| CRC | 52 | 1.1 | 12 | 0.7 | 0.0852 | 4 | 1.0 | 3 | 0.4 | 5 | 0.8 |
The incidence of new AA and CRC at FUV1 was examined in the LIR and HIR subgroups, as defined using risk factors for long-term CRC identified in the hospital data set (Table 85). In the hospital data set, the odds of both new AA and CRC were significantly lower in the LIR subgroup compared with the HIR subgroup. Results were similar when the analysis was restricted by age. Thus, in the hospital data set the risk factors identified for longer-term CRC after baseline were predictive of findings at first follow-up. In the screening data set, however, the risk factors for long-term CRC risk were not predictive, either overall or in the age-restricted group.
| Cohort | Number of patients | % | New findings at FUV1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | CRC | ||||||||||||
| n | % (n/N) | OR | 95% CI | p-value (LRT) | n | % (n/N) | OR | 95% CI | p-value (LRT) | ||||
| Hospital (N = 4608) | Higher-risk subgroupa | 3833 | 83.2 | 405 | 10.6 | 1 | < 0.0001 | 50 | 1.3 | 1 | 0.0032 | ||
| Lower-risk subgroup | 775 | 16.8 | 46 | 5.9 | 0.53 | 0.39 to 0.73 | 2 | 0.3 | 0.20 | 0.05 to 0.81 | |||
| Screening (N = 1828) | Higher-risk subgroupa | 947 | 51.8 | 43 | 4.5 | 1 | 0.3940 | 6 | 0.6 | 1 | 0.9001 | ||
| Lower-risk subgroup | 881 | 48.2 | 33 | 3.7 | 0.82 | 0.51 to 1.30 | 6 | 0.7 | 1.08 | 0.35 to 3.35 | |||
| Restricted to ages 55–69 years | |||||||||||||
| Hospital (N = 2223) | Higher-risk subgroupa | 1852 | 83.3 | 203 | 11.0 | 1 | 0.0060 | 14 | 0.8 | 1 | 0.2448 | ||
| Lower-risk subgroup | 371 | 16.7 | 24 | 6.5 | 0.56 | 0.36 to 0.87 | 1 | 0.3 | 0.35 | 0.05 to 2.71 | |||
| Screening (N = 1555) | Higher-risk subgroupa | 829 | 53.3 | 35 | 4.2 | 1 | 0.9297 | 5 | 0.6 | 1 | 0.8334 | ||
| Lower-risk subgroup | 726 | 46.7 | 30 | 4.1 | 0.98 | 0.59 to 1.61 | 5 | 0.7 | 1.14 | 0.33 to 3.96 | |||
Effect of interval on findings at follow-up
Table 86 shows the crude effect of interval to FUV1 in the screening data set on the detection of new AA or CRC at FUV1. No associations were found, and the proportion with new AA remained relatively constant with intervals of < 18 months to 6 years, ranging from 3.3% to 4.7%. Very few CRCs were detected and they were irregularly distributed, varying from 0.3% to 1.1% across intervals of between 2 and 6 years.
| Interval baseline to first follow-up | Number of patients (N = 1828) | Pooled screening cohort | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AA(s) | CRC(s) | ||||||||
| n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | ||
| < 18 months | 304 | 14 | 4.6 | 1 | 0.9193 | 0 | 0 | 1 | 0.3043 |
| 2 yearsa | 132 | 6 | 4.5 | 0.99 (0.37 to 2.63) | 1 | 0.8 | |||
| 3 yearsa | 530 | 23 | 4.3 | 0.94 (0.48 to 1.85) | 6 | 1.1 | 4.98 (0.60 to 41.53) | ||
| 4 yearsa | 367 | 14 | 3.8 | 0.82 (0.39 to 1.75) | 1 | 0.3 | 1.19 (0.07 to 19.07) | ||
| 5 yearsa | 276 | 13 | 4.7 | 1.02 (0.47 to 2.22) | 1 | 0.4 | 1.58 (0.10 to 25.39) | ||
| 6 yearsa | 90 | 3 | 3.3 | 0.71 (0.20 to 2.54) | 1 | 1.1 | 4.89 (0.30 to 78.88) | ||
| ≥ 6.5 years | 129 | 3 | 2.3 | 0.49 (0.14 to 1.75) | 2 | 1.6 | 6.85 (0.62 to 76.16) | ||
We also investigated the crude associations of baseline characteristics with findings at FUV1 in order to identify risk factors for new AA and CRC and to assess potential confounders of the association between interval and outcomes in the screening data set (Table 87). Detection of a large adenoma at baseline was significantly associated with increased odds of AA at FUV1 (p = 0.0278); although estimates were imprecise, a ≥ 20 mm adenoma was associated with an almost threefold increase (OR 2.83, 95% CI 1.07 to 7.52). No other risk factor was significant for either AA or CRC, although there was a tendency towards greater odds of AA with an incomplete colonoscopy, poor bowel preparation, an adenoma with villous histology, or a large hyperplastic polyp, and increased odds of CRC with older age, female gender, an incomplete colonoscopy or the detection of an adenoma with HGD at baseline. The number of outcomes in these analyses was too small to enable meaningful associations to be assessed.
| Baseline risk factors | New advanced finding at FUV1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of patients (N = 1828) | AA | CRC | ||||||||
| n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | n | % (n/N) | Unadjusted OR (95% CI) | p-value (LRT) | |||
| Age at start of baseline (years) | < 60 | 726 | 29 | 4.0 | 1 | 0.4514 | 2 | 0.3 | 1 | 0.1411 |
| ≥ 60 and < 65 | 692 | 34 | 4.9 | 1.24 (0.75 to 2.06) | 4 | 0.6 | 2.10 (0.38 to 11.53) | |||
| ≥ 65 and < 70 | 313 | 11 | 3.5 | 0.88 (0.43 to 1.78) | 4 | 1.3 | 4.69 (0.85 to 25.72) | |||
| ≥ 70 | 97 | 2 | 2.1 | 0.51 (0.12 to 2.15) | 2 | 2.1 | 7.62 (1.06 to 54.7) | |||
| Gender | Male | 1232 | 56 | 4.6 | 1 | 0.2239 | 7 | 0.6 | 1 | 0.5101 |
| Female | 596 | 20 | 3.4 | 0.73 (0.43 to 1.23) | 5 | 0.8 | 1.48 (0.47 to 4.68) | |||
| Completeness of colonoscopy | Complete | 1760 | 72 | 4.1 | 1 | 0.4923 | 11 | 0.6 | 1.00 | 0.4634 |
| Incomplete | 68 | 4 | 5.9 | 1.47 (0.52 to 4.13) | 1 | 1.5 | 2.37 (0.30 to 18.65) | |||
| Best bowel preparation at colonoscopy | Excellent/good/satisfactory/unknown | 1788 | 72 | 4.0 | 1 | 0.1089 | 12 | 0.7 | n/a | |
| Poor | 40 | 4 | 10.0 | 2.65 (0.92 to 7.64) | 0 | 0 | ||||
| Largest adenoma (mm) | < 10 | 201 | 5 | 2.5 | 1 | 0.0278 | 3 | 1.5 | 1 | 0.5401 |
| 10–14 | 841 | 27 | 3.2 | 1.30 (0.49 to 3.42) | 4 | 0.5 | 0.32 (0.07 to 1.42) | |||
| 15–19 | 415 | 19 | 4.6 | 1.88 (0.69 to 5.11) | 3 | 0.7 | 0.48 (0.10 to 2.40) | |||
| ≥ 20 | 371 | 25 | 6.7 | 2.83 (1.07 to 7.52) | 2 | 0.5 | 0.36 (0.06 to 2.16) | |||
| Worst adenoma histology | Tubular | 843 | 28 | 3.3 | 1 | 0.1131 | 6 | 0.7 | 1 | 0.9900 |
| Tubulovillous | 843 | 37 | 4.4 | 1.34 (0.81 to 2.20) | 6 | 0.7 | 1.00 (0.32 to 3.11) | |||
| Villous | 116 | 8 | 6.9 | 2.16 (0.96 to 4.85) | 0 | 0 | n/a | |||
| Unknown | 26 | 3 | 11.5 | 3.80 (1.08 to 13.39) | 0 | 0 | n/a | |||
| Worst adenoma dysplasia | Low grade | 1590 | 65 | 4.1 | 1 | 0.3909 | 10 | 0.6 | 1 | 0.6548 |
| High grade | 222 | 9 | 4.1 | 0.99 (0.49 to 2.02) | 2 | 0.9 | 1.44 (0.03 to 6.60) | |||
| Unknown | 16 | 2 | 12.5 | 3.35 (0.75 to 15.05) | 0 | 0 | ||||
| Distal polyps | No | 70 | 2 | 2.9 | 1 | 0.5574 | 0 | 0 | ||
| Yes | 1758 | 74 | 4.2 | 1.49 (0.36 to 6.21) | 12 | 0.7 | n/a | |||
| Proximal polyps | No | 1311 | 54 | 4.1 | 1 | 0.8956 | 9 | 0.7 | 1.00 | 0.7976 |
| Yes | 517 | 22 | 4.3 | 1.03 (0.62 to 1.72) | 3 | 0.6 | 0.84 (0.23 to 3.13) | |||
| Largest hyperplastic polyp (mm) | < 10 or none | 1778 | 71 | 4.0 | 1 | 0.0720 | 12 | 0.7 | n/a | |
| ≥ 10 | 50 | 5 | 10.0 | 2.67 (1.03 to 6.93) | 0 | 0 | ||||
Logistic regression was performed using the same set of predictors identified in the models of findings at FUV1 in the hospital data set in order to compare the estimated coefficients from the screening and hospital models (Table 88). The same variables were used, as almost nothing was predictive in the screening data set. The hospital model for AA included continuous interval, proximal polyps, age, completeness of colonoscopy, largest adenoma size (< 20 mm, ≥ 20 mm) and large hyperplastic polyps (see Table 29). The model for new CRC included the same factors except for adenoma size and large hyperplastic polyps (see Table 30), and also included quality of bowel preparation. Owing to the narrower range of ages in the screening data set, 24 subjects aged > 75 years were removed from the model, as no events occurred. Among the subjects included in the model for new CRC, no cancer occurred in those whose best colonoscopy bowel preparation was poor; thus, this predictor was not included in the model, as it would predict the outcome perfectly. There was no effect of interval on the odds of new AA or CRC, after adjusting for covariates, and none of the independent risk factors for findings at FUV1 identified in the hospital data set was predictive of findings at FUV1 in the screening data set.
| Risk factora | New findings at FUV1 | ||||
|---|---|---|---|---|---|
| AA | CRC | ||||
| Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval from baseline to first follow-up | 1-year increase | 0.98 (0.86 to 1.12) | 0.8192 | 1.12 (0.84 to 1.48) | 0.4635 |
| Proximal polyps at baseline | No | 1 | 0.9978 | 1 | 0.8482 |
| Yes | 1.00 (0.60 to 1.68) | 0.88 (0.23 to 3.30) | |||
| Age (years) at start of baseline | < 60 | 1 | 0.644 | 1 | 0.1581 |
| ≥ 60 and < 65 | 1.20 (0.72 to 2.01) | 2.14 (0.39 to 11.83) | |||
| ≥ 65 and < 70 | 0.91 (0.45 to 1.85) | 4.64 (0.84 to 25.53) | |||
| ≥ 70 | 0.58 (0.13 to 2.48) | 7.23 (0.98 to 53.52) | |||
| Completeness of baseline colonoscopy | Complete | 1 | 0.6132 | 1 | 0.3779 |
| Incomplete/unknown | 1.32 (0.46 to 3.80) | 2.95 (0.36 to 24.19) | |||
| Largest baseline adenoma (mm) | < 20 | 1 | 0.0173 | n/a | |
| ≥ 20 | 1.90 (1.14 to 3.18) | ||||
| Large hyperplastic polyp at baseline | No | 1 | 0.0819 | n/a | |
| Yes | 2.63 (0.98 to 7.01) | ||||
The ROC curves for the above models were plotted. For new AA, the area under the curve was 0.60 (95% CI 0.53 to 0.67); thus, the variables in the model were somewhat predictive of new AA at first follow-up in the screening data set (Figure 12). For new CRC, the area under the curve was 0.71 (95% CI 0.56 to 0.86), which demonstrated that interval length, examination quality and older age were fair predictors of CRC at first follow-up in the screening data set (Figure 13).
FIGURE 12.
Receiver operating characteristic curve from logistic regression model of the application to the screening data set of the hospital data set risk factors for new AA at the first follow-up. Area under ROC curve = 0.6013.

FIGURE 13.
Receiver operating characteristic curve from logistic regression model of the application to the screening data set of the hospital data set risk factors for new CRC at the first follow-up. Area under ROC curve = 0.7117.

Discussion: screening data set
Key findings
Is a 3-year interval appropriate? Is there a group that needs a shorter interval to the first or second follow-up examination, or for which follow-up could be postponed?
In the screening data set, there was no association between interval and AA or CRC detection at FUV1, before or after adjustment for covariates, possibly due to the lack of variation in interval length. Thus evidence from the screening data set was uninformative in terms of appropriateness of the 3-year interval for IR patients.
In screening programmes, interval length tends to be prescribed, so any variation in interval length in our screening data set tended to be between, rather than within, cohorts; for example, the KP cohort tended to have a longer interval, of around 5 years, and also had a considerably lower rate of CRC, whereas in the EP and UKFSST cohorts a shorter interval of around 3 years was more common and there were higher rates of CRC, thus cancelling out any potential effects of interval when the cohorts were pooled. Similarly, HIR and LIR subgroups derived from the hospital data set were not discriminant when applied to findings at FUV1 in the screening data set. We were therefore unable to validate our findings regarding the optimum interval inferred from the hospital data set using the screening data set.
When the risk subgroups were applied to findings at first follow-up in the screening data set there were slightly lower odds of new AA in the LIR subgroup, but higher odds of new CRC, further reflecting the fact that the screening data set did not validate the hospital data set in terms of findings at follow-up.
Does surveillance provide any benefit in terms of long-term cancer risk? Is there a group that does not require a follow-up examination or for which a second follow-up examination might be omitted?
Results from the screening data set validated findings of a protective effect of a single surveillance visit in the hospital data set, with a significant 73% lower risk of CRC observed after one follow-up. Additional surveillance did not appear to provide any further protection, possibly because the screening participants were already at lower risk as a consequence of their younger age and better-quality examinations. Although pre-surveillance CRC incidence in the screening data set was not significantly different from that of the general population, when observation time after surveillance was included in survival analyses the CRC incidence became significantly lower than the general population rate, providing further evidence that surveillance is effective in reducing cancer risk.
When the risk subgroups defined using CRC risk factors derived from the hospital data set were applied to the screening data set, the subgroups did not differ significantly. The absence of a significant difference between the risk subgroups in the screening data set may be a result of the small number of outcomes, and this lack of power prohibited conclusions regarding the effect of surveillance in the LIR subgroup from being drawn; although there was a trend towards a reduction in CRC risk with a single follow-up in the LIR subgroup, the results were not statistically significant. In the HIR subgroup a single surveillance had a strong protective effect, as in the whole screening cohort, and additional surveillance did not seem to offer further protection.
To account for differences in age between the hospital and screening cohorts, some analyses were restricted to patients aged 55–69 years. This did not have an impact on the risk of CRC in the HIR subgroup compared with the LIR subgroup in the screening data set, which remained non-significantly different, nor did it change the effect of surveillance in the risk subgroups. Restriction by age, however, did cause the rate of CRC in the LIR subgroup to become very similar between the hospital and screening data sets, which was surprising, as the overall rate of CRC was higher in the hospital data set. Furthermore, in patients aged 55–69 years who were defined as HIR only because of polyp risk factors, the rate of CRC was similar in both the hospital and screening data sets, suggesting that the polyp risk factors for CRC identified from the hospital data set are discriminant and relevant when applied to a screening population.
Risk factors
No baseline procedural or patient characteristics were independent predictors of AA or CRC at follow-up or of long-term CRC risk in the screening data set. Consequently, models were built using risk factors identified in the hospital data set.
Strengths and limitations
A major limitation of the screening data set was the lack of variation in interval length, which meant that no effect of interval was observed and no conclusions could be drawn from the screening data set regarding the optimum surveillance interval for patients with IR adenomas. Another limitation was the small number of CRC end points in the screening data set, which meant that the LIR and HIR subgroups were not significantly different in terms of CRC risk and were not discriminant in terms of findings at follow-up.
Although the data were mostly complete, assumptions had to be made for the KP cohort about examination quality owing to a lack of such data. This may have resulted in misclassification of bowel preparation and completeness of colonoscopy, but, as all patients were assumed to have both satisfactory preparation and a complete examination, any misclassification should be non-differential.
A strength of the screening data set was that follow-up time was substantially longer than in the hospital data set, which is preferable for survival analysis, although the number of cancers diagnosed was small. Additionally, despite the limitations and major differences in patient characteristics between the screening and hospital data sets, the screening data set validated our finding from the hospital data set of the protective effect of surveillance in IR patients. Additionally, results from the screening data set supported hospital data set results, which indicated that most protection comes from the first follow-up and that the LIR subgroup may not require surveillance.
Chapter 5 Health-economic evaluation of alternative surveillance strategies for patients in whom intermediate-grade adenomas have been detected
Introduction
This chapter details the methods and results of a model-based health-economic evaluation of alternative strategies for the surveillance of individuals in whom intermediate-grade adenomas have been detected. The chapter is set out in the following sections:
-
Economic analysis scope sets out the scope of the health-economic analysis.
-
Conceptual and implemented model structure details the conceptual logic and structure of the health-economic model.
-
Evidence used to inform the model parameters details the evidence used to inform the model’s input parameters.
-
Model evaluation methods details the model evaluation methods.
-
Model verification and validation methods details the methods used to ensure the credibility of the health-economic model.
-
Health-economic results presents the results of the analysis.
The discussion and conclusions of the analysis then follow at the end of the chapter.
Economic analysis scope
The main research question addressed by the economic evaluation is ‘what is the optimal strategy for the surveillance of individuals in whom intermediate-grade adenomas have been detected?’. The scope of the health-economic analysis is summarised in Table 89.
| Population | Patients in whom intermediate-grade adenomas have been detected |
|---|---|
| Interventions and comparators | S1. 3-yearly colonoscopy, maximum surveillance age = 75 years |
| S2. 5-yearly colonoscopy, maximum surveillance age = 75 years | |
| S3. 10-yearly colonoscopy, maximum surveillance age = 75 years | |
| S4. 3-yearly colonoscopy, no maximum surveillance age | |
| S5. 5-yearly colonoscopy, no maximum surveillance age | |
| S6. 10-yearly colonoscopy, no maximum surveillance age | |
| S7. Once-only colonoscopy 3 years post baseline, maximum surveillance age = 75 years | |
| S8. Once-only colonoscopy 5 years post baseline, maximum surveillance age = 75 years | |
| S9. Once-only colonoscopy 10 years post baseline, maximum surveillance age = 75 years | |
| S10. Once-only colonoscopy 3 years post baseline, no maximum surveillance age | |
| S11. Once-only colonoscopy 5 years post baseline, no maximum surveillance age | |
| S12. Once-only colonoscopy 10 years post baseline, no maximum surveillance age | |
| S13. No colonoscopy | |
| Outcome | Incremental cost per QALY gained |
| Time horizon | Patients’ remaining lifetime |
| Perspective | NHS and PSS |
| Discount rate | 3.5% per year |
| Price year | 2012–13 |
The population included in the health-economic analysis relates to individuals in whom intermediate-grade adenomatous polyps have been detected. Thirteen alternative surveillance strategies were evaluated using the model; these options were formulated through discussion among the research team, taking into account a range of alternative surveillance intervals and the presence/absence of a cut-off for eligibility based on patient age. The options evaluated are 3-yearly, 5-yearly and 10-yearly colonoscopic surveillance with/without a maximum age cut-off of age 75 years (options S1–6), once-only colonoscopic surveillance with/without a maximum age cut-off age of 75 years (options S7–12) and no surveillance following the baseline visit (option S13). The economic evaluation takes the form of a cost–utility analysis whereby the primary health-economic outcome is defined in terms of the incremental cost per quality-adjusted life-year (QALY) gained. Costs and health outcomes are evaluated from the perspective of the NHS and Personal Social Services (PSS) over the patients’ remaining lifetime. In line with current recommendations,78 all costs and health outcomes are discounted at a rate of 3.5% per year. All costs were valued at 2012–13 prices.
Conceptual and implemented model structure
Conceptual model
Figures 14 and 15 present problem-orientated conceptual models79 of the natural history of CRC and pathways for adenoma surveillance, respectively. These conceptual models form the basis of the implemented health-economic model described below (see Implemented health-economic model structure and assumptions).
FIGURE 14.
Colorectal cancer conceptual disease logic model.
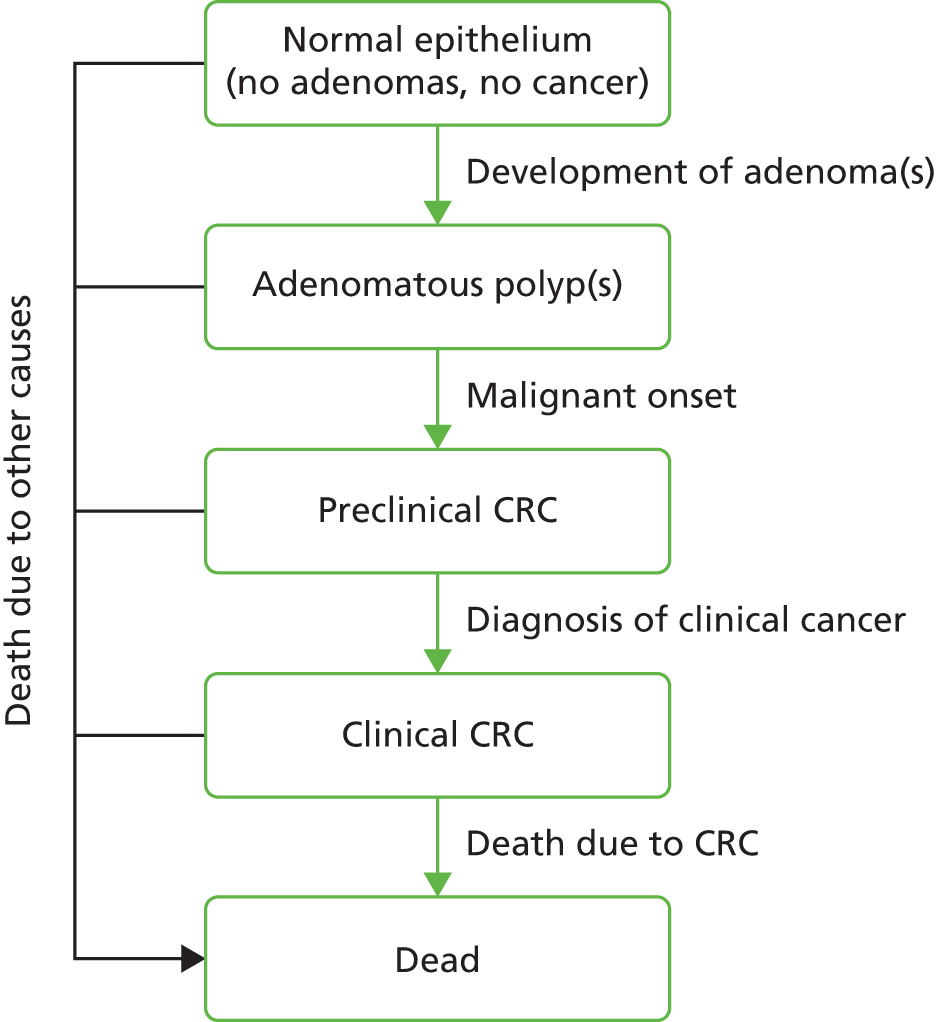
FIGURE 15.
Conceptual service pathways model. COL, colonoscopy.

It is widely accepted that the majority of CRCs follow the adenoma–carcinoma sequence whereby malignant neoplasia develops from pre-existing pre-malignant adenomatous polyps within the bowel (see Figure 14). There is indirect evidence to suggest that a smaller proportion of cancers may arise de novo, although there has historically been both controversy and uncertainty surrounding these competing theories of disease natural history. 80 The development CRC is associated with a reduction in health-related quality of life (HRQoL) and survival.
On detection of the initial adenomatous polyp(s) at the baseline visit (see Figure 15), either as a consequence of follow-up of a positive colorectal screening test or through opportunistic examination, the identified adenoma(s) would be removed, most likely via polypectomy (although surgery may be required in a small proportion of cases). Patients would subsequently be considered adenoma free, although it is possible that other lesions (adenomas and/or cancer) had been previously missed at the baseline visit. While undergoing adenoma surveillance, patients would no longer be invited to attend CRC screening. Patients may subsequently develop further adenomatous polyps and CRC. Depending on the surveillance schedule adopted, further colonoscopic examination and intervention may interrupt this natural history process. Subsequent adenomas identified at colonoscopic surveillance would be removed, thereby reducing the risk of CRC. 10 Given the imperfect test sensitivity of colonoscopy,81–83 a proportion of adenomas present in the bowel may be missed. The identification of preclinical (undiagnosed) CRC at colonoscopy surveillance would lead to the patient being referred for further investigations and treatment; again, a proportion of previously undetected cancers may be missed as a result of imperfect test sensitivity. Colonoscopy is associated with a number of relatively infrequent complications including lower gastrointestinal bleeding and perforation. Bleeding is most likely to be managed conservatively, necessitating an overnight stay in hospital. Perforation may be managed conservatively or in some cases may require more immediate surgical intervention. 84 In a small proportion of cases, bowel perforation may be fatal. The diagnosis and subsequent treatment of CRC may lead to improvements in survival and HRQoL; the management of the disease and the outlook for patients is strongly influenced by stage at diagnosis.
Implemented health-economic model structure and assumptions
The health-economic model was implemented as a next-event patient-level simulation using SIMUL8® software (SIMUL8 Corporation, Boston, MA, USA). Figures 16 and 17 present the implemented simulation model structure and logic. The model comprises five mutually exclusive health states: (1) no adenomas; (2) adenomas; (3) preclinical CRC; (4) diagnosed CRC; and (5) dead. The structure of the health-economic model was defined in line with that of the multistate model (MSM) analysis (see Baseline model of natural history progression and colonoscopy test characteristics, below), which, in turn, was defined according to the patients’ true underlying histology and colonoscopy test characteristics. Differential prognoses by adenoma type and cancer stage are not captured within the model. Health states are defined according to the index lesion (i.e. the most AA or cancer present within the patient’s bowel). Disease natural history is assumed to follow the adenoma–carcinoma sequence, as illustrated in Figure 14. The model does not allow for the development of de novo cancers without patients first developing one or more prior adenomas.
FIGURE 16.
Implemented simulation model. COL, colonoscopy.
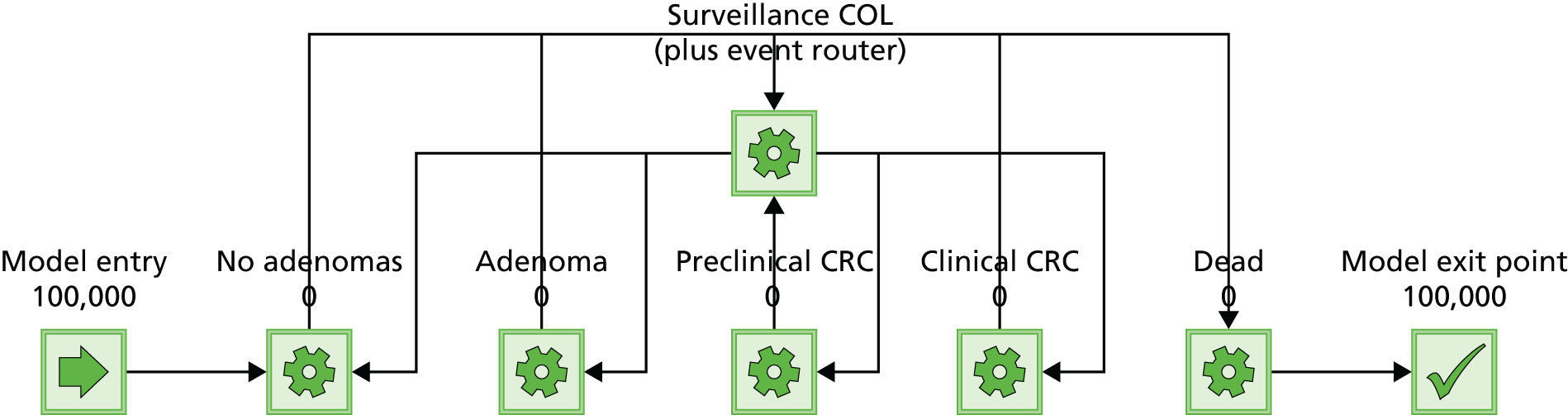
FIGURE 17.
Simulation model logic. COL, colonoscopy; ID, identification number; LYG, life-year gained; TTNE, time to next event.

The model simulates the experience of patients from the identification and removal of the index adenoma(s) at the baseline visit through to the development of further adenomas and CRC, and the impact of alternative colonoscopic surveillance options on this natural history process, as described above (see Conceptual model). Patients enter the model following the detection and removal of intermediate-grade adenomatous polyps at their baseline colonoscopy visit. Patient-level characteristics (age, life expectancy, time to disease progression, time to next surveillance colonoscopy) are then sampled and assigned to each patient. Four competing events are simulated: (1) death as a result of other causes; (2) progression to next most advanced health state; (3) attendance at next scheduled surveillance colonoscopy (provided the patient is still eligible); and (4) death as a result of undiagnosed/diagnosed CRC. The next event is determined according to the minimum of patient-specific time-to-event outcomes for these four competing events.
Patients progress through the natural history component of the model until they (1) die from other causes, (2) die as a consequence of undiagnosed/diagnosed CRC or (3) attend their next scheduled surveillance colonoscopy. At surveillance colonoscopy, patients without significant colorectal histology are assumed to receive negative colonoscopic findings and remain in the ‘no adenomas’ state. For patients with previously undetected adenomas, their adenomas may be identified by colonoscopy and subsequently removed via polypectomy; these patients subsequently transit back to the ‘no adenomas’ state. In a proportion of patients, adenomas present in the patient’s bowel may be missed at any colonoscopy visit. For patients with previously undetected CRC, the tumour may be identified by colonoscopy, after which the patient would go on to receive further investigation and treatment. In a proportion of these patients, the presence of preclinical cancer will be missed by colonoscopy. The probability of detecting lesions is determined by the test sensitivities estimated using the MSM (see Baseline model of natural history progression and colonoscopy test characteristics, below). Patients with undiagnosed CRC may present symptomatically at any point in time; these patients are assumed to progress immediately to the clinical cancer state.
Health utilities are defined according to the presence/absence of cancer without differentiation according to cancer stage. The model assumes that neither the prior detection of adenomas nor the incidence of complications following colonoscopy impacts on HRQoL; this is consistent with previous economic models of CRC screening. 85–87 In addition, the base-case analysis of the model assumes that different health utilities are applied at the point of development of CRC. It is possible, however, that a patient’s HRQoL would be affected by the diagnosis of clinical cancer rather than the development of preclinical disease. These assumptions are tested in the sensitivity analyses (see Probabilistic scenario analysis, below). The total QALY gain associated with each surveillance strategy is determined by the resulting incidence of CRC and the overall amount of time spent alive with/without cancer.
The model includes the costs associated with colonoscopy, lifetime costs associated with the diagnosis and management of CRC, and the costs of managing complications of colonoscopy. Costs are accrued as patients undergo colonoscopy and at the point of diagnosis of CRC. Cost profiles for the simulated population differ according to the surveillance option under consideration and its impact on the incidence and timing of cancer diagnosis.
The visual logic code underpinning the simulation is provided in Appendix 10.
The model makes the following key assumptions:
-
Natural history disease progression follows the adenoma–carcinoma sequence. The model does not allow for the development of de novo cancers.
-
Disease states are defined according to the presence/absence of adenomas and cancer and whether or not cancer is clinically diagnosed.
-
At any point in time, patients may have more than one lesion within their bowel (multiple adenomas or synchronous adenomas and cancer); health-state occupancy and colonoscopy test sensitivity are defined according to the index lesion (the most AA/cancer).
-
With the exception of CRC survival, progression rates through the natural history model are constant with respect to time.
-
The sensitivity of colonoscopy is imperfect and differs for adenomas and cancers.
-
The specificity of colonoscopy is perfect; false-positive test results are not considered within the model.
-
Patients are sufficiently fit to undergo colonoscopy (in reality, a small proportion of patients may undergo alternative diagnostic tests).
-
Compliance with scheduled investigations is perfect.
-
Adenoma recurrence rates are independent of the number and characteristics of previous adenomas removed.
-
Surveillance colonoscopy is associated with risks of perforations and gastrointestinal bleeds.
-
Bowel perforation may be fatal.
-
The survival of patients with undiagnosed CRC is half that of patients with diagnosed CRC. This assumption is necessary, as it is not possible to observe this rate and the MSM censored patients for the event of death.
-
Surveillance colonoscopy does not have an impact on the stage distribution of patients with diagnosed CRC.
-
Patients undergoing surveillance colonoscopy will not be eligible to participate in general population CRC screening programmes.
-
The presence of preclinical cancer impacts on survival and HRQoL.
-
The presence of diagnosed cancer impacts on survival and HRQoL.
Evidence used to inform the model parameters
Summary of evidence used to inform the model parameters
Table 90 summarises the evidence sources used to inform the parameter values assumed within the health-economic model. These are described in further detail below.
| Model parameter group | Source |
|---|---|
| Age distribution for patients with detected adenomas | Statistical analysis of hospital dataa |
| Gender distribution for patients with detected adenomas | Statistical analysis of hospital dataa |
| Baseline transition rates between no adenoma(s), adenoma(s), preclinical CRC and clinical CRC | MSM of hospital datab |
| Colonoscopy test characteristics (sensitivity and specificities for adenomas and cancer) | MSM of hospital datab |
| Health utility general population | Ara and Brazier 201088 |
| Health utility CRC | Djalalov et al. 201489 |
| Age- and gender-specific other cause mortality | ONS interim life tables 2010–1290 |
| CRC survival | National Bowel Cancer Audit Annual Report 201391 |
| Probability of bleed due to colonoscopy | EP |
| Probability of perforation due to colonoscopy | EP |
| Probability of death following perforation | Gatto et al. 200392 |
| Cost CRC (lifetime) | Whyte et al. 201286 |
| Cost surveillance colonoscopy | NHS Reference Costs 2012–13 93 |
| Cost management of bleeding | NHS Reference Costs 2012–13 93 |
| Cost management of perforation | NHS Reference Costs 2012–13 93 |
Table 91 summarises the parameter values applied directly within the model.
| Parameter | Distribution | Mean | Parameter | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| Sojourn no adenoma to adenoma (years) | MSM posterior | 9.64 | 9.64 | 7.25 | 11.97 |
| Sojourn adenoma to preclinical cancer (years) | MSM posterior | 23.48 | 23.48 | 10.95 | 43.97 |
| Sojourn preclinical to clinical cancer (years) | MSM posterior | 2.15 | 2.15 | 0.46 | 7.46 |
| Relative survival preclinical vs. clinical | Beta | 0.50 | 7.00 | 7.00 | – |
| Transition probability clinical cancer to dead | Weibull (MVN) | – | 1.08 | 4.67 | – |
| Specificity adenomas/cancer | Fixed | 1.00 | 1.00 | – | – |
| Sensitivity adenomas | MSM posterior | 0.77 | 0.77 | 0.70 | 0.83 |
| Sensitivity cancer | MSM posterior | 0.88 | 0.88 | 0.50 | 0.99 |
| Probability complication | Beta | 0.01 | 26.00 | 3664.00 | – |
| Probability complication is perforation | Beta | 0.08 | 2.00 | 24.00 | – |
| Probability death | perforation | Beta | 0.05 | 4.00 | 73.00 | – |
| HRQoL no cancer | MVN | 0.81 | 0.81 | – | – |
| HRQoL preclinical cancer | Beta | 0.73 | 358.98 | 132.77 | – |
| HRQoL clinical cancer | Beta | 0.73 | 358.98 | 132.77 | – |
| Cost surveillance COL | Normal | £514.38 | £514.38 | £16.80 | – |
| Cost cancer (lifetime) | Normal | £20,212.59 | £20,212.59 | £3031.89 | – |
| Cost perforation | Normal | £4170.95 | £4170.95 | £464.59 | – |
| Cost bleed | Normal | £192.41 | £192.41 | £29.47 | – |
| Distribution key | |||||
| Distribution | Parameter | ||||
| 1 | 2 | 3 | |||
| Posterior | Mean | 2.5th percentile | 97.5th percentile | ||
| MVN | Mean | Covariance not shown | |||
| Beta | Alpha | Beta | n/a | ||
| Normal | Mean | SE | n/a | ||
| Weibull | Alpha | Beta | n/a | ||
Patient characteristics
The model takes account of two initial patient characteristics: age and gender. These variables influence life expectancy and HRQoL for patients without cancer. Patient age and gender were based on the statistical analysis of the hospital data presented in Chapter 3 (see Baseline characteristics of all intermediate risk patients and those with follow-up). Within the model, patients aged < 50 years were assumed to be distributed equally within the age interval 30–49 years. Age and gender distributions were held fixed within the simulation; uncertainty surrounding these data was not considered.
Baseline model of natural history progression and colonoscopy test characteristics
The baseline natural history progression rates and colonoscopy test characteristics were derived through the development of a multistate model-based analysis of hospital data detailed in Chapter 3 (see First follow-up visit). Data were available on first colonoscopy after baseline for 4608 subjects. Data for each subject consisted of the time since baseline examination and the state allocated to the subject: normal, adenoma or presymptomatic cancer (the presymptomatic nature was not certain but the assumption had to be made for estimation to be possible). If a subject had a colonoscopy between (N – 1) and N years then their time to colonoscopy was approximated by (N – 1/2) years. The total number of subjects who had a colonoscopy between (N – 1) and N years after baseline, and the number of these observed to have presymptomatic cancer, or adenomas, for each N, are given in Table 92.
| Years to colonoscopy | Cancers | Adenomas | Total colonoscopies |
|---|---|---|---|
| 1 | 4 | 72 | 384 |
| 2 | 17 | 482 | 1936 |
| 3 | 5 | 230 | 813 |
| 4 | 6 | 270 | 881 |
| 5 | 8 | 84 | 233 |
| 6 | 3 | 72 | 188 |
| 7 | 3 | 29 | 84 |
| 8 | 1 | 13 | 33 |
| 9 | 2 | 9 | 28 |
| 10 | 1 | 2 | 11 |
| 11 | 1 | 2 | 5 |
| 12 | 0 | 1 | 1 |
| 13 | 0 | 0 | 1 |
| 14 | 0 | 1 | 3 |
| 15 | 1 | 3 | 4 |
| 16 | 0 | 1 | 1 |
| 18 | 0 | 0 | 1 |
| 20 | 0 | 0 | 1 |
The structure of the MSM is shown in Figure 18.
FIGURE 18.
Structure of the multistate model.

The multistate model comprises four natural history states:
-
O state (no adenomas or cancer)
-
A adenoma
-
C presymptomatic cancer
-
S symptomatic cancer.
Subjects who are truly in state O have no adenoma or cancer.
Subjects who are truly in state A have an adenoma that is correctly observed at a screen with probability Sa; they are otherwise observed to be in state O.
Subjects who are truly in state C have cancer but are asymptomatic, and at a screen are detected to have cancer with probability Sc; they are otherwise observed to be in state O.
Thus, a subject may be observed to be in state O but may truly be in one of states O, A or C. The model assumes that subjects undergo the transitions O → A, A → C and C → S as time-homogeneous Poisson processes with rates λ0, λ1 and λ2, respectively.
Let the probability that a subject is observed to be in state X at time t after their clearance be p(X^; t), let the probability that a subject is truly in state X at time t after clearance be p(X; t) and let the probability that a subject is truly in state V at time t after clearance given that they were truly in state U at clearance be given by p(U, V; t). Finally, let the probability that a subject observed to be in state O at clearance is truly in state X at that time be given by p0(X).
Subjects are given a screen at clearance and entered into the study only if they are observed to be in state O. The probability of a subject being observed in state C at a time t after clearance is given by:
which can be shown to be equal to:
where:
Similarly, the probability of a subject being observed in state A at a time t after clearance is given by:
which can be shown to be equal to:
We wish to estimate x = (Sa,Sc,λ0,λ1,λ2). Under the Bayesian paradigm, inference can be performed by obtaining the posterior distribution:
where D is the data obtained, and Pr(x) is the prior distribution on x. We use a non-informative proper prior for Pr(x).
where U(;c,d) is a uniform distribution over the real interval (c,d). The likelihood, Pr(D|x), is given by:
where subject i is observed to be in state Vl, at time ti after clearance.
This inference can be performed using the program STAN which obtains samples from an approximation to the posteriors density p(x) by the method of Hamiltonian Monte Carlo. We used four chains of 1000 samples, taking 500 samples in each chain as ‘burn-in’ and using the remaining 500, and checking convergence through examination of the Gelman and Rubin diagnostic. Sample medians of each component of were taken as the estimates of the parameters and sample 2.5% and 97.5% quantiles were used to obtain their credible intervals. Within the health-economic analysis, the Convergence Diagnostic and Output Analysis samples, which are the correlated parameter samples from the joint posterior distribution from the model, were used directly. Table 93 presents the posterior median and 95% credible intervals for the estimated MSM parameters.
| Parameter | Posterior median | 95% credible interval |
|---|---|---|
| λ0 | 0.104 | 0.084 to 0.132 |
| λ1 | 0.045 | 0.024 to 0.083 |
| λ2 | 0.687 | 0.146 to 1.854 |
| S a | 0.776 | 0.719 to 0.824 |
| S c | 0.917 | 0.65 to 0.985 |
Other-cause mortality
Other-cause mortality rates were modelled using 2012 interim life expectancy tables published by the ONS. 90 Remaining life expectancy conditional on individual patient age and gender was modelled using non-parametric distributions.
Survival of patients with colorectal cancer
The MSM (see Baseline model of natural history progression and colonoscopy test characteristics, above) did not include data on other-cause mortality or deaths due to CRC; within the statistical analysis these data were effectively treated as censored. As such, evidence from other sources was required to inform the survival duration for patients with preclinical CRC and for patients with diagnosed CRC. As noted above in Implemented health-economic model structure and assumptions, the survival of patients with CRC was not split by stage within the model. A number of options exist from which estimates of survival for patients with CRC could be estimated: these include RCTs, cancer registries, model-based syntheses and clinical audits. Registry data are not ideal, as these typically report relative survival rates relative to an age-matched population over a finite time period. Without a baseline survival estimate for patients without cancer these are difficult to interpret, and the appropriateness of extrapolating relative survival rates beyond the specified period (typically 1, 3 or 5 years) may be questionable. RCT evidence may provide an alternative source of evidence of survival estimates; however, such data tend to be upwardly biased because of a lack of representativeness of the sample, for example owing to younger age, performance status and other potential confounders. In addition, most CRC trials report outcomes for specific treatments and patient groups at a specific point in the treatment pathway rather than reporting survival outcomes from diagnosis for the CRC population as a whole.
In light of these problems, the health-economic model instead utilises reported survival data collected as part of the 2013 National Bowel Cancer Audit Programme (NBOCAP). 91 Absolute overall survival estimates were available for 2 years from the NBOCAP. The key assumption resulting from the use of these data is that the stage of patients within the audit is representative of the stage of patients progressing to CRC within the modelled population.
The available survival data were digitised using Engauge® 4.7 software (Engauge Digitizer Open Source Project, Torrance, CA, USA). The digitised data were used to reproduce the underlying patient-level time-to-event data using methods reported by Guyot et al. 94 within the software package R 0.98.977 (RStudio) (Table 94). Given the high event rates reported within the audit report, the analysis assumes zero censoring.
| Time (years) | Number censored | Number who died | Number at risk | S(t) | S(ti) |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 50,130 | – | 1 |
| 0.25 | 0 | 5013 | 45,117 | 0.90 | 0.90 |
| 0.50 | 0 | 2005 | 43,112 | 0.96 | 0.86 |
| 1.00 | 0 | 4011 | 39,101 | 0.91 | 0.78 |
| 1.50 | 0 | 3007 | 36,094 | 0.92 | 0.72 |
| 2.00 | 0 | 2507 | 33,587 | 0.93 | 0.67 |
Regression methods were used to fit a range of alternative candidate parametric survivor functions to the replicated patient-level time-to-event data. The candidate survivor functions included exponential, Weibull, Gompertz, log-normal and log-logistic models. Model discrimination was undertaken by examining log-hazard plots and through examination of the AIC and the BIC statistics for each model. The plausibility of the extrapolated portion of each curve was examined through comparison with other less recent but more complete audit data reported for the Wessex region. 95 Figure 19 presents the empirical Kaplan–Meier estimate together with the fitted exponential, Weibull, Gompertz, log-normal and log-logistic survivor functions. Table 95 presents the AIC and BIC statistics for each model.
FIGURE 19.
Empirical and modelled survivor functions for overall survival.

| Parametric model | AIC | BIC |
|---|---|---|
| Exponential | 97,202.41 | 97,211.23 |
| Weibull | 97,107.04 | 97,124.69 |
| Gompertz | 97,193.89 | 97,211.53 |
| Log-normal | 96,978.51 | 96,996.15 |
| Log-logistic | 96,105.23 | 96,122.87 |
As shown in Figure 19, within the observable period, all five of the candidate survivor functions appear to provide a reasonable fit to the observed data; this is also reflected in the similarity of the AIC and BIC statistics for each survival model. Unsurprisingly, as the log-logistic and log-normal are statistically similar models, they produce very similar estimates. These two models appear implausible, however, as both suggest that around 15% of patients will still be alive at 30 years post diagnosis. These models were ruled out on this basis. The extrapolated portions of the remaining exponential Gompertz and Weibull survivor functions appear to be broadly similar. The Weibull model was selected for inclusion in the health-economic model because of its lower AIC and BIC values. The mean survival for the modelled Weibull survivor function is estimated to be approximately 4.54 years. Uncertainty surrounding the correlated parameters of the Weibull survivor function was sampled from a multivariate normal (MVN) distribution using the estimated variance–covariance matrix for the model.
It is important to note that the survival of patients with undiagnosed CRC is not directly observable and could not be indirectly estimated using the MSM. Instead, this estimate was based on an assumption. Within the model, the survival of patients with undiagnosed CRC was assumed to be half of that for patients with diagnosed CRC (this assumption was varied within the simple sensitivity analysis – see Probabilistic scenario analysis, below).
Health-related quality of life
The definition of HRQoL states within the model was guided by the states included in the MSM (see Baseline model of natural history progression and colonoscopy test characteristics, above). The MSM defines histology according to the presence/absence of adenomas and the clinical status of any CRC present (either diagnosed or not).
A systematic review was planned to inform the HRQoL parameters within the health-economic model. Systematic searches for studies, utilising the Health Utilities Index, the Short Form questionnaire-6 Dimensions and the European Quality of Life-5 Dimensions questionnaires, were undertaken based on a previously published CRC search strategy96 across 15 electronic databases:
-
MEDLINE and MEDLINE in Process & Other Non-Indexed citations (via Ovid)
-
EMBASE (via Ovid)
-
Cumulative Index to Nursing and Allied Health Literature (via EBSCOhost)
-
BIOSIS previews (via WoK)
-
Science Citation Index (via Web of Science)
-
Cochrane Database of Systematic Reviews (Cochrane)
-
Cochrane Central Register of Controlled Trials (Cochrane)
-
Database of Abstracts of Reviews of Effects (Cochrane)
-
NHS Health Economic Evaluation Database (Cochrane)
-
HTA Database (Cochrane)
-
EconLit (via Ovid)
-
Web of Science (via WoK)
-
Conference Proceedings index (Web of Science via WoK)
-
ProQuest Dissertations and Theses (ProQuest)
-
Tufts (Cost-effectiveness Analysis Registry).
However, once the searches were complete and the sifting process was under way, the authors were alerted to the publication of another systematic review of preference-based HRQoL values in 2014, by Djalalov et al. 89 This study89 had also adopted the same broad search strategy as our review (see Appendix 11). Consequently, our systematic review was aborted and estimates from Djalalov et al. 89 were instead used to inform estimates of quality of life for patients with CRC within the health-economic model. The model assumes an overall value of 0.73 for CRC. Uncertainty surrounding this estimate was modelled using a beta distribution assuming a SE of 0.03. For patients without CRC, a health-utility value of HRQoL 0.81 was estimated based on general population utilities reported by Ara and Brazier,88 and the distribution according to the age and gender weights in the intermediate-grade adenomas data set. Uncertainty surrounding this parameter was estimated using a MVN distribution.
Probability of complications associated with colonoscopy
The probability of experiencing complications of colonoscopy was taken from the first round of the English Bowel Screening Pilot (Table 96).
| Complication | Number of events | Total number of colonoscopies performed |
|---|---|---|
| Perforations | 2 | 3690 |
| Other (considered as bleeding) | 24 | |
| Total | 26 |
Within the prevalence round of the screening pilot, two perforations were reported in 3690 colonoscopies; a probability of perforation due to colonoscopy of 0.0005 was assumed in the model. Similarly, a further 24 non-fatal events involving bleeding and/or abdominal pain were reported; a probability of bleeding of 0.006 was assumed in the model. The probability of death following perforation was taken from a study by Gatto et al. 92 (4 of 77 perforations, probability = 5.19%). Uncertainty surrounding these parameters was characterised using beta distributions.
Costs associated with surveillance and the diagnosis and management of colorectal cancer
The model includes the costs of colonoscopy, the costs of managing complications of colonoscopy and the lifetime costs associated with the diagnosis and management of CRC (Table 97). The costs of colonoscopy and associated complications were taken from NHS Reference Costs 2012–13. 93 Estimates of the lifetime costs of diagnosis and management of CRC were taken from a previous modelling study reported by Whyte et al. 86 and adjusted according to the stage distribution reported in the 2011 report of the NBOCAP. 97
| Cost parameter | Mean | SE | Source |
|---|---|---|---|
| Colonoscopy | 514.38 | 16.80 | NHS Reference Costs 2012–13,93 day case FZ15Z, diagnostic colonoscopy, ≥ 19 years |
| Management of perforation | 1896.65 | 211.26 | NHS Reference Costs 2012–13,93 elective inpatient WA12D, complications of procedures with critical care score 0 |
| Management of bleed | 192.41 | 29.47 | NHS Reference Costs 2012–13,93 elective inpatient (excess bed-day) gastrointestinal bleed, without interventions, with critical care score 0–4 |
| Lifetime cost of CRC diagnosis and management | 20,212.59 | 3031.89 | Whyte et al. 201286 |
Standard errors surrounding the reference cost estimates were estimated using the IQRs and number of data submissions. The SE for the lifetime cost of CRC diagnosis and management was assumed to be 15% of the mean. Uncertainty surrounding cost parameters was characterised using normal distributions.
Model evaluation methods
Model stability testing
Given that the model adopts a simulation approach, it is necessary to determine how many patient runs are sufficient to produce stable results. Strategy-specific life-years gained (LYGs), QALYs and total costs were evaluated using simulation cohort sizes ranging from 1000 patients to 1 million patients for three alternative surveillance options (3-yearly surveillance, no age cut-off; 5-yearly surveillance, no age cut-off; and 10-yearly surveillance, no age cut-off).
Figures 20–22 present the per cent deviation in total LYGs, total QALYs and total costs for each cohort size relative to a baseline size of 1 million patients.
FIGURE 20.
Model stability testing: 3-yearly surveillance, no age cut-off.

FIGURE 21.
Model stability testing: 5-yearly surveillance, no age cut-off.

FIGURE 22.
Model stability testing: 10-yearly surveillance, no age cut-off.
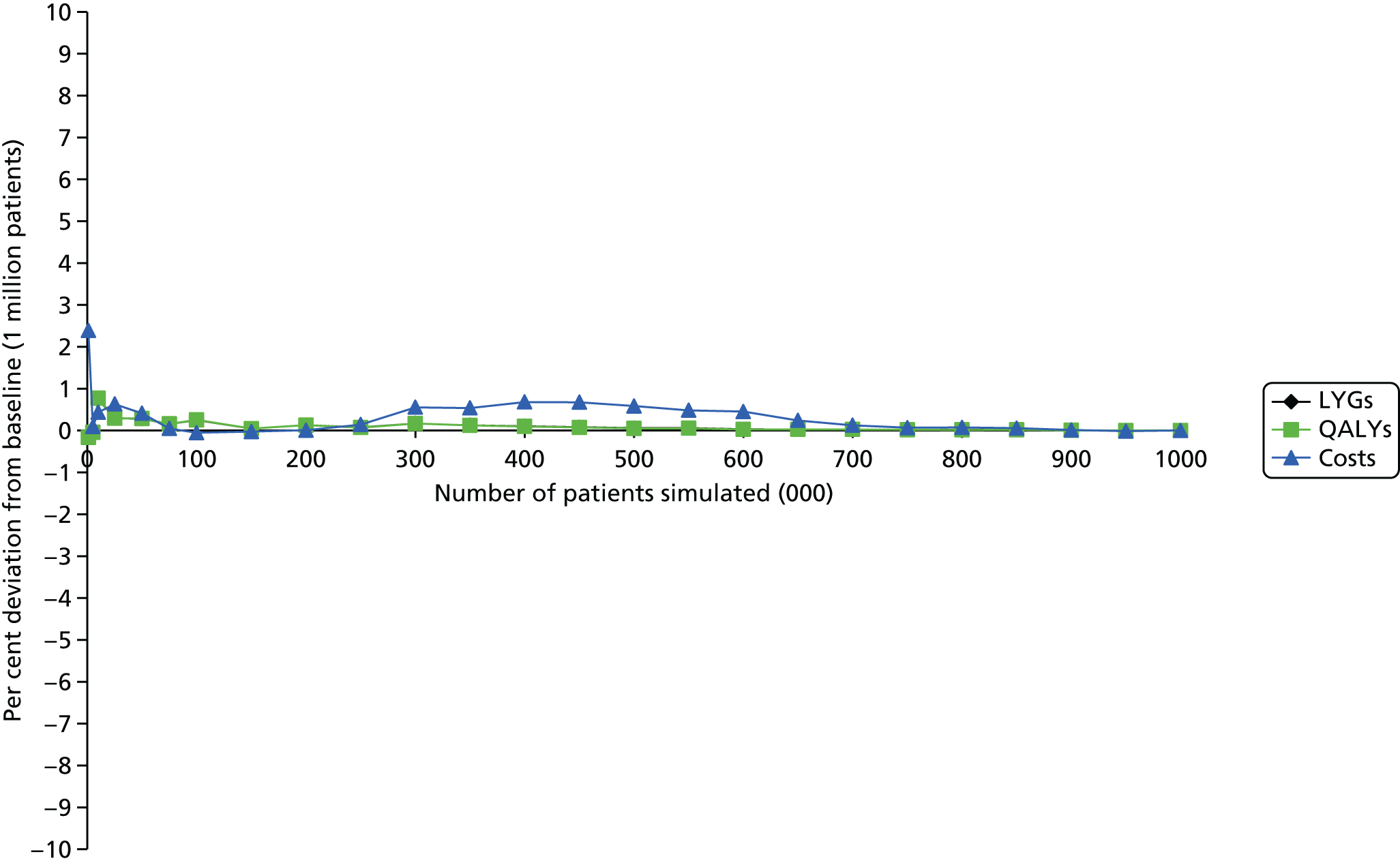
The stability test analyses presented in Figures 20–22 indicate that the total LYGs, total QALYs and total costs are very stable for sample sizes of ≥ 50,000 patients (< 1% deviation from a baseline of 1 million patients). In order to ensure stability in the model results, a total sample size of 300,000 patients was conservatively selected for all probabilistic analyses.
Probabilistic model evaluation and uncertainty analysis
The model was evaluated probabilistically over 1000 Monte Carlo samples, each of which comprised 300,000 individually sampled patients. Absolute estimates of the total cost and health gains associated with each surveillance strategy were based on the expectation of the mean. The incremental cost-effectiveness of alternative surveillance strategies was evaluated using standard cost-effectiveness decision rules,98 whereby each option was compared with its next best alternative. Options that were dominated or extendedly dominated were ruled out of the analysis. Uncertainty surrounding the incremental costs and health outcomes was represented using cost-effectiveness planes and cost-effectiveness acceptability curves (CEACs); these present the probability that each option produces the greatest net benefit at a given willingness-to-pay (WTP) threshold.
It is important to note that some of the model parameters are subject to considerable uncertainty. In particular, the expected survival duration and health utility for patients with undiagnosed preclinical cancer are unobservable; these parameters have been populated using assumptions. Within the base-case analysis, the health utility for patients with preclinical cancer is assumed to be equal to that of patients with clinically diagnosed disease, whereas their expected survival is assumed to be equal to half of that for diagnosed patients. In order to examine the importance of this uncertainty on the model results, five alternative probabilistic scenarios were evaluated:
-
Probabilistic scenario analysis 1 – preclinical cancer utility = clinical cancer utility, preclinical survival = 0.33 × clinical survival.
-
Probabilistic scenario analysis 2 – preclinical cancer utility = clinical cancer utility, preclinical survival = 0.67 × clinical survival.
-
Probabilistic scenario analysis 3 – preclinical cancer utility = general population utility, preclinical survival = 0.33 × clinical survival.
-
Probabilistic scenario analysis 4 – preclinical cancer utility = general population utility, preclinical survival = 0.50 × clinical survival.
-
Probabilistic scenario analysis 5 – preclinical cancer utility = general population utility, preclinical survival = 0.67 × clinical survival.
In addition to the probabilistic analysis, simple one-way sensitivity analyses were undertaken to examine the impact of individual parameters on the results of the economic analysis (Table 98); these analyses were undertaken using point estimates of parameters rather than using the probabilistic model.
| Scenario number | Description |
|---|---|
| 1 | Base case using point estimates of parameters |
| 2 | Test sensitivity for adenomas increased by 5 percentage points |
| 3 | Test sensitivity for adenomas decreased by 5 percentage points |
| 4 | Test sensitivity for cancer increased by 5 percentage points |
| 5 | Test sensitivity for cancer decreased by 5 percentage points |
| 6 | Test sensitivity for adenomas and cancer increased by 5 percentage points |
| 7 | Test sensitivity for adenomas and cancer decreased by 5 percentage points |
| 8 | Survival of patients with preclinical CRC +25% |
| 9 | Survival of patients with preclinical CRC –25% |
| 10 | All cost estimates +25% |
| 11 | All cost estimates –25% |
Value of information analysis
Alongside the cost-effectiveness analysis, value of information (VOI) methods were used to estimate the monetary value of eliminating existing uncertainty through undertaking further research. VOI analysis can be used to assess the value of additional information on all parameters [global expected value of perfect information (EVPI)] or on specific parameter groups or individual parameters [expected value of partial perfect information (EVPPI)], thereby enabling the prioritisation of further research through pursuing research projects, the additional information of which is expected to yield the greatest pay-off in terms of expected net benefit. Uncertainty in model parameters indicates that there is a possibility of selecting a suboptimal strategy, and hence the VOI is high in situations where the additional information gained from further research would lead to a switch away from the strategy adopted given current information. Similarly, if further research on a specific parameter would not lead to a switch in adoption decisions, there is no value in conducting such research. VOI analysis can therefore be considered as a useful tool in placing a ceiling on the monetary value of further research, whereas EVPPI can be used to provide a basis for informing the design of clinical trials and other studies. Within the analysis, the global EVPI (i.e. the monetary value of eliminating all uncertainty in the model parameters) was estimated using standard methods. 99 The EVPPI for all individual parameters was estimated using non-linear regression methods reported by Strong and Oakley100
Model verification and validation methods
A number of efforts were made to ensure the credibility of the health-economic analysis and the underlying model on which this was based; these methods were based on the taxonomy of model errors and other threats to model credibility recently published by Tappenden and Chilcott. 101
-
The characterisation of the natural history of CRC was guided by previous models of the adenoma–carcinoma sequence and the feasibility of the multistate modelling given available hospital data (see Chapter 3, First follow-up visit).
-
The model is based on formal conceptual modelling:79 this helps to draw out key assumptions, abstractions and simplifications in the final model.
-
All model inputs were double-checked against the original input material.
-
The model was developed and evaluated in line with the current NICE reference for economic evaluations. 78
-
Black-box testing was undertaken to ensure that the model was behaving as intended. This included automated and manual checking check by two researchers (PT and BK) to ensure that both the developed conceptual models are sufficient to meet the objectives of the evaluation and credibility was not lost during the translation from the conceptual to the mathematical model.
-
Grey-box validation was undertaken to examine whether or not intermediate outcomes recorded within the model (e.g. number of deaths due to various causes, number of patients with symptomatic/surveillance-detected cancers) met predetermined expectations.
-
White- and grey-box validation were undertaken by checking the integrity of the model programming, running tests with isolated parts of the code by two researchers (PT and BK), and analysing simulated patient pathways for credibility.
-
Additional validation checks were undertaken including checking that any logical relationships between model inputs (such as correlations or monotonic relationships) were preserved and sampled input values did not violate boundary constraints (such as sampling negative survival times), and assessing the performance of the model under extreme input values.
-
Any errors or omissions that were identified as a result of the validation checks were rectified, and the validations checks were repeated, leading to a process of iterative model improvement.
-
The model and associated economic analysis were peer reviewed by methodological experts.
Health-economic results
Base-case analysis
Central estimates of cost-effectiveness: base-case analysis (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.50)
Table 99 presents the central estimates of cost-effectiveness based on the probabilistic version of the model. The analysis indicates that more frequent surveillance is associated with greater health gains (irrespective of whether or not an age cut-off is applied). Of the 13 options included in the analysis, option S4 (3-yearly ongoing surveillance with no age cut-off) is expected to be the most effective. Options that include once-only surveillance or no surveillance are expected to be dominated by more effective and less expensive options. Option S6 (10-yearly ongoing surveillance with no age cut-off) is expected to be ruled out as a result of extended dominance; this means that greater health gains could be achieved by funding a combination of other non-dominated options. The incremental cost-effectiveness ratios (ICERs) for the remaining non-dominated options are all very low (< £3000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option (option S1: 3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £2748 per QALY gained.
It should be noted that when compared marginally against the no-surveillance strategy (option S13), all surveillance options either dominate or have an ICER that is < £1000 per QALY gained.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 9.66 | 3706 | 0.12 | 336 | 2748.10 |
| S1: 3-yearly ongoing, maximum age 75 years | 9.54 | 3370 | 0.07 | 152 | 2079.73 |
| S5: 5-yearly ongoing, no maximum age | 9.47 | 3218 | 0.08 | 113 | 1355.74 |
| S2: 5-yearly ongoing, maximum age 75 years | 9.38 | 3105 | 0.21 | 87 | 422.78 |
| S6: 10-yearly ongoing, no maximum age | 9.20 | 3039 | – | – | Ext dom |
| S11: Once only at 5 years, no maximum age | 9.18 | 3128 | – | – | Dominated |
| S3: 10-yearly ongoing, maximum age 75 years | 9.18 | 3018 | – | – | – |
| S8: Once only at 5 years, maximum age 75 years | 9.17 | 3106 | – | – | Dominated |
| S10: Once only at 3 years, no maximum age | 9.14 | 3235 | – | – | Dominated |
| S12: Once only at 10 years, no maximum age | 9.14 | 3040 | – | – | Dominated |
| S9: Once only at 10 years, maximum age 75 years | 9.13 | 3031 | – | – | Dominated |
| S7: Once only at 3 years, maximum age 75 years | 9.13 | 3196 | – | – | Dominated |
| S13: No surveillance | 8.95 | 3083 | – | – | Dominated |
Table 100 presents a breakdown of the model results in terms of QALYs gained pre and post cancer diagnosis, cost components and number of additional surveillance colonoscopies for individuals identified as having intermediate-grade adenomas.
| Option | Non-cancer QALYs | Cancer QALYs | Surveillance costs (£) | Complication costs (£) | Cancer costs (£) | Number of surveillance colonoscopies |
|---|---|---|---|---|---|---|
| S1 | 9.29 | 0.25 | 1542 | 7 | 1822 | 2.79 |
| S2 | 9.09 | 0.29 | 1075 | 4 | 2027 | 1.55 |
| S3 | 8.84 | 0.33 | 755 | 2 | 2262 | 0.71 |
| S4 | 9.45 | 0.21 | 2274 | 12 | 1421 | 5.24 |
| S5 | 9.20 | 0.26 | 1443 | 6 | 1769 | 2.82 |
| S6 | 8.87 | 0.33 | 847 | 2 | 2191 | 1.08 |
| S7 | 8.79 | 0.34 | 855 | 2 | 2340 | 0.73 |
| S8 | 8.83 | 0.34 | 815 | 2 | 2290 | 0.69 |
| S9 | 8.79 | 0.34 | 718 | 1 | 2312 | 0.56 |
| S10 | 8.80 | 0.34 | 935 | 3 | 2298 | 0.90 |
| S11 | 8.84 | 0.33 | 876 | 2 | 2250 | 0.83 |
| S12 | 8.80 | 0.34 | 743 | 2 | 2296 | 0.62 |
| S13 | 8.57 | 0.38 | 515 | 0 | 2569 | 0.00 |
The results presented in Table 100 indicate that the differences in the costs of additional surveillance colonoscopies between the options are small, ranging from £515 (baseline colonoscopy only) to £2274 per patient. Although option S4 is associated with the greatest number of additional surveillance colonoscopies, and hence the greatest cost of surveillance, this option is also associated with the greatest reduction in cancer costs and produces the greatest QALY gains.
Probabilistic sensitivity analysis (base case)
Figure 23 presents an absolute cost-effectiveness plane for the 13 options included in the economic analysis. The plane indicates that the costs and QALYs are broadly similar between all 13 surveillance options.
FIGURE 23.
Cost-effectiveness plane (absolute costs and QALYs).

Figure 24 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is also approximately 1.0.
FIGURE 24.
Base-case analysis, CEACs (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.50).

Expected value of perfect information analysis
The per-patient global EVPI was calculated; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero. This suggests that, given the model structure and the data used to inform it, undertaking further research to eliminate current levels of uncertainty would not change the adoption decision; hence, the value of undertaking further research is zero.
Probabilistic scenario analysis
Scenario analysis 1 (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.33)
A separate probabilistic analysis was undertaken assuming that the relative survival of patients with preclinical CRC is equal to one-third of the survival for patients with clinically diagnosed CRC, and assuming that the utility of patients with preclinical CRC is equal to that of patients with diagnosed CRC. Table 101 presents the central estimates of cost-effectiveness for this scenario.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 14.22 | 5137 | 0.30 | 627 | 2111 |
| S1: 3-yearly ongoing, maximum age 75 years | 13.92 | 4511 | 0.15 | 192 | 1302 |
| S5: 5-yearly ongoing, no maximum age | 13.78 | 4319 | 0.21 | 224 | 1057 |
| S2: 5-yearly ongoing, maximum age 75 years | 13.56 | 4094 | 0.47 | 182 | 385 |
| S6: 10-yearly ongoing, no maximum age | 13.16 | 3956 | – | – | Ext dom |
| S3: 10-yearly ongoing, maximum age 75 years | 13.09 | 3912 | – | – | – |
| S11: Once only at 5 years, no maximum age | 12.96 | 4069 | – | – | Dominated |
| S8: Once only at 5 years, maximum age 75 years | 12.95 | 4042 | – | – | Dominated |
| S12: Once only at 10 years, no maximum age | 12.94 | 3944 | – | – | Dominated |
| S9: Once only at 10 years, maximum age 75 years | 12.93 | 3930 | – | – | Dominated |
| S10: Once only at 3 years, no maximum age | 12.87 | 4180 | – | – | Dominated |
| S7: Once only at 3 years, maximum age 75 years | 12.85 | 4135 | – | – | Dominated |
| S13: No surveillance | 12.51 | 3971 | – | – | Dominated |
The results of scenario analysis 1 are similar to those presented in the base-case analysis. Options that include once-only surveillance or no surveillance are expected to be ruled out because of dominance. Option S6 (10-yearly ongoing surveillance with no age cut-off) is expected to be ruled out because of extended dominance (greater health gains could be achieved by funding a combination of other non-dominated options). The ICERs for the remaining non-dominated options are all very low (< £3000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option (option S1: 3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £2111 per QALY gained.
Figure 25 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is also approximately 1.0.
FIGURE 25.
Scenario analysis 1, CEACs (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.33).

The per-patient global EVPI was calculated for this scenario; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero.
Scenario analysis 2 (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.67)
A separate probabilistic analysis was undertaken assuming that the relative survival of patients with preclinical CRC is equal to two-thirds of the survival for patients with clinically diagnosed CRC, and assuming that the utility of patients with preclinical CRC is equal to that of patients with diagnosed CRC. Table 102 presents the central estimates of cost-effectiveness for this scenario.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 14.27 | 5575 | 0.25 | 392 | 1574 |
| S1: 3-yearly ongoing, maximum age 75 years | 14.02 | 5183 | 0.16 | 251 | 1551 |
| S5: 5-yearly ongoing, no maximum age | 13.86 | 4932 | 0.18 | 69 | 385 |
| S2: 5-yearly ongoing, maximum age 75 years | 13.68 | 4864 | 0.38 | 80 | 209 |
| S6: 10-yearly ongoing, no maximum age | 13.30 | 4783 | – | – | – |
| S3: 10-yearly ongoing, maximum age 75 years | 13.24 | 4790 | – | – | Dominated |
| S11: Once only 5 years, no maximum age | 13.12 | 4985 | – | – | Dominated |
| S8: Once only 5 years, maximum age 75 years | 13.11 | 4973 | – | – | Dominated |
| S12: Once only 10 years, no maximum age | 13.10 | 4857 | – | – | Dominated |
| S9: Once only 10 years, maximum age 75 years | 13.09 | 4848 | – | – | Dominated |
| S10: Once only 3 years, no maximum age | 13.04 | 5123 | – | – | Dominated |
| S7: Once only 3 years, maximum age 75 years | 13.02 | 5095 | – | – | Dominated |
| S13: No surveillance | 12.70 | 5018 | – | – | Dominated |
The results for scenario analysis 2 are similar to those presented in the base-case analysis. Options that include once-only surveillance or no surveillance are expected to be ruled out because of dominance. Option S3 (1-yearly ongoing surveillance with no age cut-off) is also expected to be dominated. The ICERs for the remaining non-dominated options are all very low (< £2000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option (option S1: 3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £1574 per QALY gained.
Figure 26 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is also approximately 1.0.
FIGURE 26.
Scenario analysis 2, CEACs (probabilistic, utility preclinical cancer = utility clinical cancer, survival preclinical cancer = survival clinical cancer × 0.67).

The per-patient global EVPI was calculated for this scenario; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero.
Scenario analysis 3 (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.33)
A separate probabilistic analysis was undertaken assuming that the relative survival of patients with preclinical CRC is equal to one-third of the survival for patients with clinically diagnosed CRC, and assuming that the utility of patients with preclinical CRC is equal to that of the general population. Table 103 presents the central estimates of cost-effectiveness for this scenario.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 14.23 | 5137 | 0.29 | £627 | 2152 |
| S1: 3-yearly ongoing, maximum age 75 years | 13.93 | 4511 | 0.15 | £192 | 1280 |
| S5: 5-yearly ongoing, no maximum age | 13.78 | 4319 | 0.21 | £224 | 1078 |
| S2: 5-yearly ongoing, maximum age 75 years | 13.58 | 4094 | 0.47 | £182 | 386 |
| S6: 10-yearly ongoing, no maximum age | 13.18 | 3956 | – | – | Ext dom |
| S3: 10-yearly ongoing, maximum age 75 years | 13.11 | 3912 | – | – | – |
| S11: Once only 5 years, no maximum age | 12.98 | 4069 | – | – | Dominated |
| S8: Once only 5 years, maximum age 75 years | 12.96 | 4042 | – | – | Dominated |
| S12: Once only 10 years, no maximum age | 12.96 | 3944 | – | – | Dominated |
| S9: Once only 10 years, maximum age 75 years | 12.95 | 3930 | – | – | Dominated |
| S10: Once only 3 years, no maximum age | 12.89 | 4180 | – | – | Dominated |
| S7: Once only 3 years, maximum age 75 years | 12.87 | 4135 | – | – | Dominated |
| S13: No surveillance | 12.53 | 3971 | – | – | Dominated |
Options that include once-only surveillance or no surveillance are expected to be ruled out because of dominance. Option S6 (10-yearly ongoing surveillance with no age cut-off) is expected to be ruled out because of extended dominance. The ICERs for the remaining non-dominated options are all very low (< £3000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option (option S1: 3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £2152 per QALY gained.
Figure 27 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off will produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off will produce the greatest net benefit is also approximately 1.0.
FIGURE 27.
Scenario analysis 3, CEACs (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.33).

The per-patient global EVPI was calculated for this scenario; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero.
Scenario analysis 4 (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.50)
A separate probabilistic analysis was undertaken assuming that the relative survival of patients with preclinical CRC is equal to half of the survival for patients with clinically diagnosed CRC, and assuming that the utility of patients with preclinical CRC is equal to that of the general population. Table 104 presents the central estimates of cost-effectiveness for this scenario.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 14.26 | 5409 | 0.26 | 491 | 1872 |
| S1: 3-yearly ongoing, maximum age 75 years | 14.00 | 4918 | 0.16 | 225 | 1420 |
| S5: 5-yearly ongoing, no maximum age | 13.84 | 4693 | 0.19 | 135 | 725 |
| S2: 5-yearly ongoing, maximum age 75 years | 13.65 | 4558 | 0.39 | 107 | 275 |
| S6: 10-yearly ongoing, no maximum age | 13.26 | 4451 | 0.07 | 15 | 224 |
| S3: 10-yearly ongoing, maximum age 75 years | 13.20 | 4436 | – | – | – |
| S11: Once only 5 years, no maximum age | 13.08 | 4614 | – | – | Dominated |
| S8: Once only 5 years, maximum age 75 years | 13.06 | 4595 | – | – | Dominated |
| S12: Once only 10 years, no maximum age | 13.06 | 4486 | – | – | Dominated |
| S9: Once only 10 years, maximum age 75 years | 13.05 | 4476 | – | – | Dominated |
| S10: Once only 3 years, no maximum age | 12.99 | 4739 | – | – | Dominated |
| S7: Once only 3 years, maximum age 75 years | 12.98 | 4704 | – | – | Dominated |
| S13: No surveillance | 12.65 | 4589 | – | – | Dominated |
The results for this scenario are similar to those presented in the base-case analysis. Options that include once-only surveillance or no surveillance are expected to be ruled out because of dominance. The ICERs for the remaining non-dominated options are all very low (< £2000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option (option S1: 3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £1872 per QALY gained.
Figure 28 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is also approximately 1.0.
FIGURE 28.
Scenario analysis 4, CEACs (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.50).
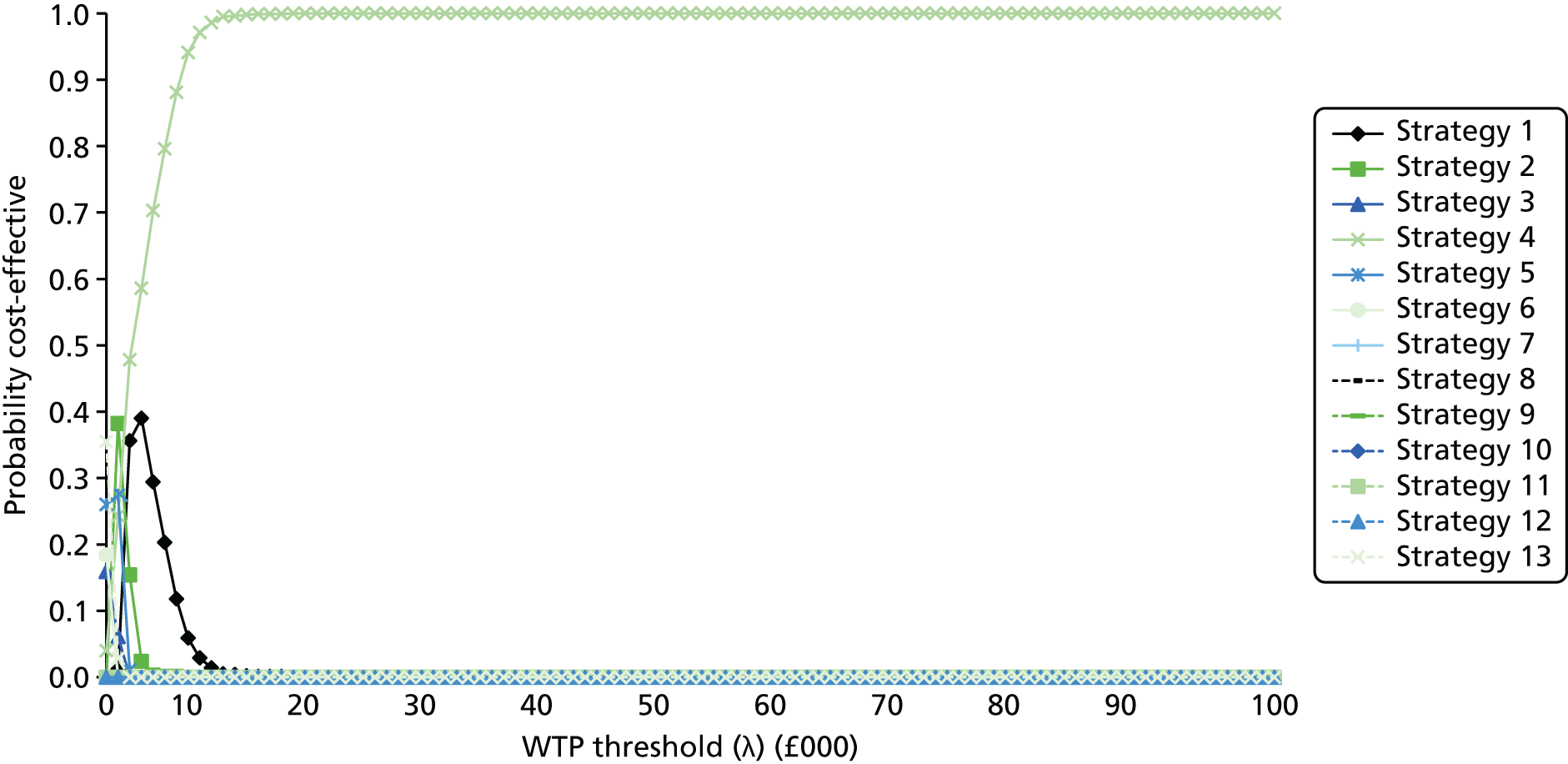
The per-patient global EVPI was calculated for this scenario; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero.
Scenario analysis 5 (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.67)
A separate probabilistic analysis was undertaken assuming that the relative survival of patients with preclinical CRC is equal to two-thirds of the survival for patients with clinically diagnosed CRC, and assuming that the utility of patients with preclinical CRC is equal to that of the general population. Table 105 presents the central estimates of cost-effectiveness for this scenario.
| Option | QALYs | Costs (£) | Incremental QALYs | Incremental cost (£) | ICER (£) |
|---|---|---|---|---|---|
| S4: 3-yearly ongoing, no maximum age | 14.28 | 5575 | 0.24 | 392 | 1627 |
| S1: 3-yearly ongoing, maximum age 75 years | 14.04 | 5183 | 0.17 | 251 | 1514 |
| S5: 5-yearly ongoing, no maximum age | 13.87 | 4932 | 0.17 | 69 | 399 |
| S2: 5-yearly ongoing, maximum age 75 years | 13.70 | 4864 | 0.38 | 80 | 209 |
| S6: 10-yearly ongoing, no maximum age | 13.32 | 4783 | – | – | – |
| S3: 10-yearly ongoing, maximum age 75 years | 13.26 | 4790 | – | – | Dominated |
| S11: Once only 5 years, no maximum age | 13.14 | 4985 | – | – | Dominated |
| S8: Once only 5 years, maximum age 75 years | 13.13 | 4973 | – | – | Dominated |
| S12: Once only 10 years, no maximum age | 13.12 | 4857 | – | – | Dominated |
| S9: Once only 10 years, maximum age 75 years | 13.11 | 4848 | – | – | Dominated |
| S10: Once only 3 years, no maximum age | 13.06 | 5123 | – | – | Dominated |
| S7: Once only 3 years, maximum age 75 years | 13.05 | 5095 | – | – | Dominated |
| S13: No surveillance | 12.72 | 5018 | – | – | Dominated |
Options that include once-only surveillance or no surveillance are expected to be ruled out because of dominance. The ICERs for the remaining non-dominated options are all very low (< £2000 per QALY gained). The ICER for the most effective option, option S4 (3-yearly ongoing surveillance with no maximum age cut-off), compared with the next most effective option, option S1 (3-yearly ongoing surveillance up to age 75 years) is expected to be approximately £1627 per QALY gained.
Figure 29 presents CEACs for each of the 13 options. Assuming a WTP threshold of £20,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is approximately 1.0. Assuming a WTP threshold of £30,000 per QALY gained, the probability that 3-yearly ongoing surveillance with no age cut-off is expected to produce the greatest net benefit is also approximately 1.0.
FIGURE 29.
Scenario analysis 5, CEACs (probabilistic, utility preclinical cancer = utility general population, survival preclinical cancer = survival clinical cancer × 0.67).

The per-patient global EVPI was calculated for this scenario; assuming thresholds of £20,000 per QALY gained and £30,000 per QALY gained, the global EVPI is expected to be zero. As the global EVPI is expected to be zero, the partial EVPPIs for individual parameters are also zero.
Simple sensitivity analysis
Table 106 presents the results of the simple sensitivity analysis of the base-case model.
| Option/analysis | Incremental cost (£) per QALY gained | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Point estimates | Sens polyps +5% | Sens polyps –5% | Sens CRC +5% | Sens CRC –5% | All sens +5% | All sens –5% | CRC survival+50% | CRC survival –50% | Costs +25% | Costs –25% | |
| S1: 3-yearly ongoing, maximum age 75 years | 1996 | 2077 | 2258 | 2195 | 2051 | 2129 | 2051 | 2109 | 2413 | 2495 | 1497 |
| S2: 5-yearly ongoing, maximum age 75 years | 678 | 707 | 749 | 673 | 699 | 743 | 699 | 617 | 837 | 862 | 517 |
| S3: 10-yearly ongoing, maximum age 75 years | Ext dom | – | 603 | 220 | 137 | 21 | 137 | – | 342 | 790 | 474 |
| S4: 3-yearly ongoing, no maximum agea | 4665 | 3816 | 4089 | 3950 | 4386 | 3848 | 4386 | 4170 | 3675 | 5832 | 3499 |
| S5: 5-yearly ongoing, no maximum age | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom |
| S6: 10-yearly ongoing, no maximum age | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom | Ext dom |
| S7: Once only at 3 years, maximum age 75 years | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom |
| S8: Once only at 5 years, maximum age 75 years | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom |
| S9: Once only at 10 years, maximum age 75 years | 65 | Dom | 205 | Ext dom | Ext dom | Dom | Ext dom | Dom | Ext dom | 81 | – |
| S10: Once only at 3 years, no maximum age | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom |
| S11: Once only at 5 years, no maximum age | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom |
| S12: Once only at 10 years, no maximum age | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom | Dom |
| S13: No surveillance | – | Dom | – | – | – | – | – | Dom | – | – | Dom |
The simple sensitivity analysis consistently suggests that option S4 (3-yearly ongoing surveillance with no maximum age cut-off) is expected to produce the greatest number of QALYs. The ICER for this option is < £6000 per QALY gained across all scenarios. It is also noteworthy that none of the deterministic ICERs for any of the surveillance options exceeds £5000 per QALY gained.
Impact on NHS expenditure
Given that the alternative surveillance options considered within the health-economic analysis include different intervals and age cut-offs, together with expected changes to the NHS BCSP that may alter the specific populations eligible for screening and the screening modality used, estimating the expected impact of the alternative surveillance options on the NHS budget is difficult. Based on data from the NHS BCSP reported by Kearns et al. ,102 the positivity rate for the initial FOBT screen during the period 2008–11 is estimated to be 2.21%. According to this same source, 909,839 usable kits were returned in 2011, which implies a total of 20,097 individuals testing positive and requiring further investigation. Assuming that 17% of these patients are classed as having IR adenomas,36 this would suggest an incident population of around 3417 patients each year. However, the lifetime incremental cost of the alternative surveillance options relative to a policy of no adenoma surveillance is small (–£59 to £1166); hence, it is likely that the overall impact of alternative adenomas surveillance options on NHS expenditures each year will also be relatively small, and may even produce small cost-savings depending on which option is pursued. Attributing the entire lifetime incremental cost for option S4 (3-yearly ongoing surveillance with no maximum age) against a policy of no adenoma surveillance to the estimated incident IR population suggests a maximum budget impact of around £4M over the lifetime of 3417 incident patients.
Discussion
Summary of cost-effectiveness findings
This chapter has presented the methods and results of a health-economic simulation model used to assess the cost-effectiveness of 13 surveillance options for patients with intermediate-grade adenomatous polyps. The model results suggest that 3-yearly ongoing colonoscopic surveillance without an age cut-off is expected to produce the greatest health gain. The ICER for this option (compared against the same strategy with an age cut-off of 75 years) is expected to be approximately £2748 per QALY gained. The probabilistic sensitivity analysis indicates that, assuming a WTP threshold of £20,000 per QALY gained, the probability that this option produces the greatest expected net benefit is approximately 1.0.
When compared marginally against the no-surveillance strategy, all surveillance options either dominate or have an ICER which is < £1000 per QALY gained.
Several alternative probabilistic scenario analyses were undertaken to explore uncertainty surrounding unobservable parameters relating to the survival and HRQoL impacts of preclinical cancer. However, the results were similar to the base-case findings; hence, the economic conclusions of the analysis are not strongly influenced by these assumptions. The simple one-way sensitivity analyses also support the base-case results. Given the favourable cost-effectiveness profile of 3-yearly ongoing surveillance and the limited decision uncertainty, the global EVPI and individual parameter EVPPIs estimated by the model are approximately zero.
Limitations and uncertainties
The interpretation of the findings of the economic analysis presented here requires some consideration of limitations of the model, as well as the uncertainties within the current evidence base. These are discussed below.
Assumptions regarding the natural history of colorectal cancer
The model assumes that all cancers arise from prior adenomas. However, there is indirect evidence to suggest that a proportion of CRCs arise de novo rather than as a consequence of the adenoma–carcinoma sequence. This theory of disease natural history was not considered within the MSM and hence could not be incorporated into the health-economic analysis, and its impact on the cost-effectiveness of the alternative surveillance options is unclear. Furthermore, given better data, it may have been possible to produce a more accurate representation of the natural history process, for example by capturing different malignant potential of different types and numbers of adenomas present in the bowel, perhaps including the use of frailty models to distinguish between those adenomas which will develop into cancer and those which will not. It is also noteworthy that the MSM includes states for preclinical and clinical cancer but does not differentiate by cancer stage; the model is ‘blunt’ in the sense that surveillance may result in a shift in stage distribution at diagnosis and thus lead to different estimates of diagnosis and treatment costs; this potential effect is not reflected within the model.
Assumptions regarding adenoma surveillance programmes
The model includes only the possibility of premalignant adenoma detection through surveillance; in reality adenomas may be detected in a small number of patients through symptomatic or opportunistic presentation. Furthermore, the model does not include the possibility of participation in national screening programmes following discharge of a planned adenoma surveillance programme. In addition, all patients are assumed to attend their scheduled surveillance visits and all are assumed to be sufficiently fit to undergo colonoscopy. These assumptions may be somewhat optimistic and, as a consequence, the model may overestimate the clinical benefits and cost-effectiveness of surveillance programmes in comparison with what may be expected to be seen in usual clinical practice.
Prognosis of patients with preclinical/clinical colorectal cancer
The model uses survival data derived from the NBOCAP to inform parameters relating to the expected survival of patients with CRC. These data were required as the MSM censored patients for death. The NBOCAP data are subject, however, to considerable censoring; hence, there is uncertainty surrounding their long-term extrapolation. Although the sensitivity analysis suggests that this does not strongly influence the model results, further data in the future may enable a more accurate evaluation of alternative adenoma surveillance strategies.
Surveillance options considered
The model evaluation includes two types of surveillance option: (1) ongoing surveillance with a fixed interval between scheduled visits; and (2) once-only surveillance. These options were agreed by the project team prior to implementation of the health-economic model. The model is capable, however, of considering more complex surveillance strategies, for example schedules with different time intervals or schedules that take into account previous histological findings or numbers of attendances. It may be possible that an alternative surveillance option, which has not been defined for inclusion in this analysis, could produce greater expected health gains than those estimated within this analysis.
It is also important to recognise that surveillance options which include more frequent ongoing visits will put greater stress on endoscopy services. Although 3-yearly ongoing surveillance appears to be the most effective strategy, and such health gains are expected to be achieved at a cost which may be acceptable to NHS policy-makers, it may be preferable to pursue less effective strategies that also offer a favourable cost-effectiveness profile.
Conclusions
Of the 13 surveillance options considered, 3-yearly ongoing colonoscopic surveillance without an age cut-off is expected to produce the greatest health gain. The ICER for this option (compared with the same strategy with an age cut-off of 75 years) is expected to be < £3000 per QALY gained.
Chapter 6 Psychological study: examination of anxiety levels
Introduction
Existing studies of the psychological effects of CRC screening have shown few adverse effects of faecal occult blood testing,103 FS104 or colonoscopy105 among the screened population as a whole. However, there has been little research looking at the psychological impact of entering a surveillance programme after screening.
Surveillance may have a positive psychological effect by giving patients a sense of ongoing care, along with reassurance that they do not have bowel cancer. However, colonoscopy is widely seen as an uncomfortable and embarrassing procedure,106 and regular screening might be a reminder of cancer risks.
In the IA study, we were asked to investigate the psychological impact of surveillance using retrospective data only. We therefore undertook additional analyses of the UKFSST data to answer this question.
Methods
Sample
Participants were men and women aged 55–64 years, at average risk of CRC, invited for screening in 12 of the 14 study centres in the UKFSST. 107 Exclusion criteria for the trial were:
-
inability to provide consent
-
personal history of CRC, adenomas, or IBD
-
strong family history of CRC (two or more close relatives affected)
-
severe or terminal disease with a life expectancy of < 5 years
-
presence of a temporary health problem that would prevent the patient from having FS
-
sigmoidoscopy or colonoscopy within the previous 3 years.
Patients eligible for inclusion in the trial were sent a letter by their GP, with an information leaflet on CRC and FS screening, and were asked whether or not they would accept the offer of screening if invited. Respondents who replied saying that they were interested in screening were randomly allocated to screening or to usual care. 107 Data from the pilot and the first study centre were excluded from the analyses because different post-screening questionnaires had been used.
In the UKFSST, individuals with one to two small tubular adenomas were considered ‘lower risk’ and were not offered colonoscopy after their polyps were removed at FS, whereas those with more numerous tubular adenomas or AA were offered colonoscopy. The majority of these individuals were offered surveillance colonoscopy in accordance with a prescribed protocol, similar to the British Society of Gastroenterology guidelines for the management of ‘higher risk’ cases. 1 Outcomes data were available for 35,891 individuals. Of these:
-
26,573 had no polyps detected
-
7401 had lower-risk polyps, removed at FS
-
183 had higher-risk polyps, were referred to colonoscopy and then discharged
-
1543 had higher-risk polyps, were referred to colonoscopy and then recommended for ongoing colonoscopic surveillance
-
62 were considered higher risk but colonoscopy was not performed for a variety of reasons (e.g. they were too ill or did not want to undergo the procedure); a further six were referred straight to surgery and 123 were diagnosed with cancer – these 191 were excluded, leaving a total sample of 35,700.
Participants were sent a detailed questionnaire 3–6 months after screening, by which time they had been told whether or not they needed surveillance. The response rate to this questionnaire was 90% (Figure 30). Completion of the post-screening questionnaire was higher among women, older people and people with lower levels of socioeconomic deprivation.
FIGURE 30.
Trial profile.
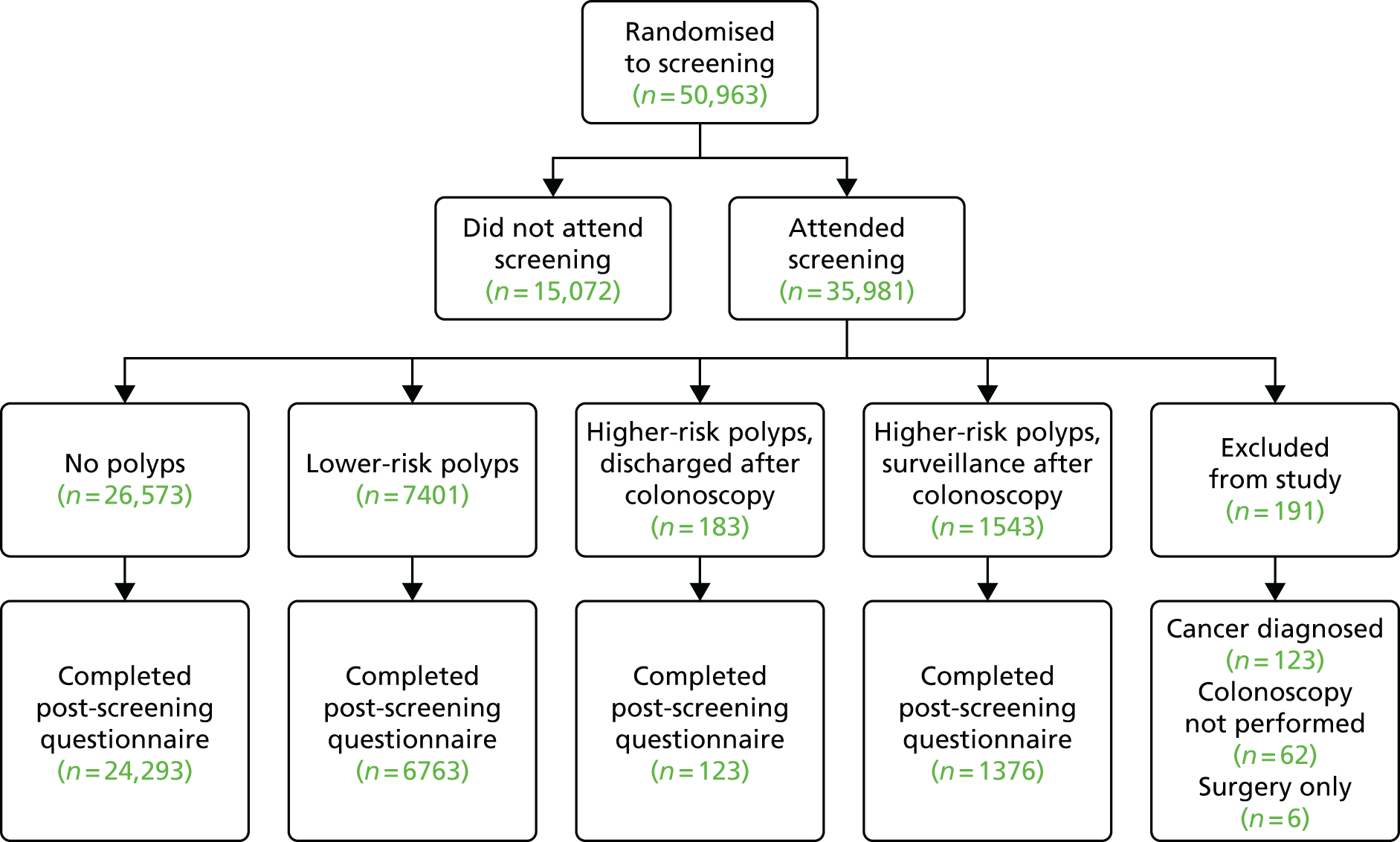
A subsample (n = 6389) had also completed a detailed questionnaire 6 months prior to screening attendance, making it possible to compare pre- and post-screening results in this group. However, the subsample contained only 20 individuals who were discharged following colonoscopy, so these 20 patients were excluded from longitudinal analyses.
Measures
Demographic variables including date of birth, gender and postcode were supplied by GP practices. Socioeconomic deprivation was indexed by converting postcode into Townsend Index scores108 (an established indicator of area-based socioeconomic deprivation in England) for all centres except Glasgow.
Primary outcome variables
-
Bowel cancer worry was assessed before and after screening using the question: ‘How worried are you about getting bowel cancer?’ Response options were on a four-point Likert scale: ‘not worried at all’, ‘a bit worried’, ‘quite worried’ and ‘very worried’.
-
Psychological distress was measured post screening using the 12-item version of the General Health Questionnaire (GHQ-12). 109 This assesses how people have felt in the preceding 3 months, with a four-item response scale: ‘better/more than usual’, ‘same as usual’, ‘less than usual’ and ‘much less than usual’. Responses were scored 0–3 and summed to produce a scale from 0 to 36, with higher scores indicating greater distress.
-
Positive emotional consequences of screening were assessed post screening using three items from the positive emotional subscale of the Psychological Consequences of screening Questionnaire (PCQ). 110 These were: ‘Do you think that your experience of having the Flexi-Scope test has . . .’ : ‘made you feel more hopeful about the future?’; ‘made you feel less anxious about bowel cancer?’; ‘given you a greater sense of wellbeing?’. Response options were on a four-point Likert scale (‘not at all’; ‘a little bit’; ‘quite a bit’; and ‘a great deal’) and were summed to produce a score from 3 to 12, with higher scores indicating more positive consequences. Cronbach’s alpha for the emotional items was 0.81, which is similar to the value of 0.89 reported for the full 10-item scale (containing positive and negative emotional items).
Secondary outcome variables
-
Reassurance was assessed post screening using a single item on reassurance from the PCQ: ‘Do you think that your experience of having the Flexi-Scope test has given you a sense of reassurance that you do not have bowel cancer?’ Response options were on a four-point Likert scale: ‘not at all’, ‘a little bit’, ‘quite a bit’ and ‘a great deal’.
-
Generalised anxiety was measured before and after screening using the six-item version of the Spielberger State–Trait Anxiety Inventory. 111 This asks people to consider how they are feeling ‘right now’ and includes items such as ‘I feel calm’ or ‘I am tense’. Responses were added giving a score of between 6 and 24, with higher scores indicating higher levels of anxiety.
-
Bowel symptoms were assessed before and after screening using the stem question: ‘Because we are studying bowel screening, we would like to know how often people get these bowel symptoms. In the last three months have you . . .’ : ‘been constipated’; ‘had haemorrhoids (piles)’; ‘had diarrhoea’; ‘been troubled with wind’; ‘had pains in the abdomen (gut)’; ‘had bowel incontinence’; ‘noticed blood in your stools’. Response options were: ‘no’, ‘occasionally’ or ‘frequently’. People were classified as having ‘one or more’ bowel symptoms (if they replied ‘occasionally’ or ‘frequently’ to any of the questions) or ‘none’.
-
GP attendance was measured before and after screening using the question: ‘About how many times have you been to see your GP in the last 3 months?’ Response options were ‘have not been’, ‘once’, ‘twice’, ‘three or more times’.
Statistical analysis
Between-group analyses of variance (ANOVAs) were used to test for differences between screening outcome groups. Contrasts were used to examine whether or not the surveillance group differed from the three other outcome groups. Changes over time (pre- to post-screening) were assessed using repeated-measures ANOVA with time as a within-subjects variable and screening outcome group as a between-subjects variable.
Results
Demographic variables
Outcome groups differed significantly by age and gender. The groups with lower-risk polyps and higher-risk polyps assigned to surveillance were slightly older than the group with no polyps (average age – no polyps, 60.3 years; lower-risk polyps, 60.5 years; higher-risk polyps, discharged, 60.5 years; higher-risk polyps, surveillance, 60.7 years). All three groups with polyps had a higher proportion of men (no polyps: 46% men; lower risk: 62%; higher risk, discharged: 62%; higher risk, surveillance: 68%). Socioeconomic deprivation was not associated with the presence or absence of polyps. Age and gender were entered into all analyses as independent variables, but results are reported only when age or gender moderated the psychological impact associated with different screening outcomes.
Post-screening scores
Primary outcome variables
Bowel cancer worry differed across the four groups (F3,31904 = 16.3; p < 0.001) but planned contrasts showed higher worry in the surveillance group than in the no-polyps group, and no significant differences between the surveillance group and the other two groups with polyps. Although general psychological distress (GHQ) differed by outcome group (F3,32055 = 2.66; p < 0.05), the surveillance group reported lower distress than the groups with no polyps or lower-risk polyps (Table 107).
| Variables | No polyps (n = 24,293) | Lower-risk polyps, no colonoscopy (n = 6763) | Higher-risk polyps, discharged after colonoscopy (n = 123) | Higher-risk polyps, surveillance after colonoscopy (n = 1376) | |
|---|---|---|---|---|---|
| Primary outcome | |||||
| Bowel cancer worry | 1–4 (higher scores indicate greater worry) | 1.91 (0.01)* | 1.99 (0.01) | 1.90 (0.08) | 1.96 (0.02) |
| Psychological distress | 0–36 (higher scores indicate greater distress: a score of 24 = cut-off for psychiatric illness) | 8.91 (0.03)** | 8.87 (0.05)* | 8.83 (0.36) | 8.58 (0.11) |
| Positive emotional consequences of screening | 3–12 (higher scores indicate greater positive consequences) | 8.16 (0.02)*** | 8.10 (0.03)*** | 8.01 (0.24)* | 8.53 (0.07) |
| Reassurance do not have bowel cancer | 1–4 (higher scores indicate greater reassurance) | 3.53 (0.01) | 3.45 (0.01)* | 3.48 (0.07) | 3.50 (0.02) |
| Secondary outcome | |||||
| Generalised anxiety | 6–24 (higher scores indicate greater anxiety) | 9.85 (0.03)*** | 9.96 (0.05)*** | 9.52 (0.37) | 9.35 (0.12) |
| Bowel symptoms | 0–7 (higher scores indicate a greater number of symptoms) | 1.54 (0.01)*** | 1.56 (0.02)** | 1.71 (0.14) | 1.70 (0.04) |
| GP visits | 1–4 (higher scores indicate more frequent GP visits) | 1.97 (0.01) | 1.94 (0.01) | 2.20 (0.10)** | 1.94 (0.03) |
There were significant differences across the groups in reported emotional consequences of screening (F3,31971 = 9.37; p < 0.001), with the surveillance group reporting higher positive consequences of screening than all the other groups (see Table 107). They also reported higher reassurance than the lower-risk group, although reassurance scores did not differ from the two remaining outcome groups. There was a significant age-by-group interaction (F3,32054 = 3.38; p < 0.05), with no age differences in reassurance among people with no polyps or lower-risk polyps, but greater reassurance among those aged > 60 years who were assigned to surveillance (F1,1349 = 6.78; p < 0.01) (see Table 107).
Secondary outcome variables
Anxiety differed across outcome groups (F3,31667 = 7.22; p < 0.001), with people assigned to surveillance reporting significantly lower post-screening anxiety than the groups with no or lower-risk polyps (see Table 107). There were also group differences in bowel symptoms (F3,288869 = 5.73; p < 0.001) and GP visits (F3,30601 = 3.60; p < 0.05), with the surveillance group reporting more bowel symptoms post screening than the groups with no or lower-risk polyps. However, they reported fewer GP visits than the group discharged following colonoscopy, and did not differ on this measure from the groups with no or lower-risk polyps.
Pre- and post-screening measures
In order to check whether or not post-screening group differences simply reflected pre-screening group differences, and to see whether or not factors such as worry increased or decreased following screening, we examined longitudinal changes in a subsample. As noted above, these analyses excluded the group discharged following colonoscopy because the sample size was too small.
There were no differences between outcome groups on pre-screening bowel cancer worry, generalised anxiety, number of bowel symptoms reported or frequency of GP visits. Bowel cancer worry declined over time (F1,6440 = 45.9; p < 0.001), with a significant interaction between change in worry over time and outcome group (F2,6440 = 4.11; p < 0.05). However, reductions in worry in the surveillance group were greater than in the lower-risk group, although the difference was not significant (F1,1743 = 3.34; p < 0.10) and no difference was observed between the surveillance and the no-polyps group (Table 108).
| Outcome | No polyps (n = 24,293) | Lower-risk polyps, no colonoscopy (n = 6763) | Higher-risk polyps, surveillance after colonoscopy (n = 1376) | |||
|---|---|---|---|---|---|---|
| Bowel cancer worry (1–4, higher scores indicate greater worry) | ||||||
| Pre | 2 | 0.00 (0.01) | 2 | 0.05 (0.02) | 2 | 0.06 (0.05) |
| Post | 1 | 0.86 (0.01) | 1 | 0.98 (0.02) | 1 | 0.88 (0.05) |
| Change | 0 | 0.14 | 0 | 0.07 | 0 | 0.18 |
| Generalised anxiety (6–24, higher scores indicate greater anxiety) | ||||||
| Pre | 10 | 0.10 (0.05) | 10 | 0.09 (0.10) | 10 | 0.49 (0.23) |
| Post | 9 | 0.63 (0.06) | 9 | 0.80 (0.10) | 9 | 0.41 (0.25) |
| Change | 0 | 0.47* | 0 | 0.29** | 1 | 0.08 |
| Bowel symptoms (0–7, higher scores indicate a greater number of symptoms) | ||||||
| Pre | 2 | 0.11 (0.02) | 2 | 0.01 (0.04) | 2 | 0.14 (0.10) |
| Post | 1 | 0.60 (0.02) | 1 | 0.53 (0.04) | 1 | 0.82 (0.09) |
| Change | 0 | 0.51* | 0 | 0.48 | 0 | 0.32 |
| GP visits (1–4, higher scores indicate more frequent GP visits) | ||||||
| Pre | 1 | 0.97 (0.02) | 1 | 0.96 (0.03) | 2 | 0.02 (0.07) |
| Post | 1 | 0.90 (0.02) | 1 | 0.83 (0.03) | 1 | 0.98 (0.07) |
| Change | 0 | 0.07 | 0 | 0.13 | 0 | 0.04 |
There was a significant reduction in anxiety over time (F1,6377 = 41.6; p < 0.001) and also a significant interaction with group (F2,6377 = 4.29; p < 0.05), with the surveillance group showing a greater reduction in anxiety than both the lower-risk group (F1,1724 = 8.03; p < 0.01) and the no-polyps group (F1,4926 = 5.53; p < 0.05) (see Table 108).
Although the number of reported bowel symptoms decreased following screening (F1,5662 = 181.2; p < 0.001), the interaction between change over time and screening outcome group was not significant. Pairwise comparisons indicated smaller reductions in symptoms in the surveillance group than in the no-polyps group (F1,4367 = 4.38; p < 0.05) but the former was similar to that observed in the lower-risk group. Self-reported frequency of GP visits decreased significantly over time (F1,5998 = 8.80; p < 0.01) but the interaction between change over time and outcome group was not significant (see Table 108).
Discussion
Following screening, we found that bowel cancer worry was higher in patients undergoing surveillance than in those who had no polyps detected, but did not differ from the level observed in the group found to have lower-risk polyps, suggesting that bowel cancer worry was related to the detection of growths in the bowel rather than to colonoscopic surveillance itself. In addition, the decline in bowel cancer worry in the surveillance group over time was equivalent to that observed in the group who had no polyps found and did not undergo surveillance, supporting the idea that surveillance did not make people worry about bowel cancer. On other measures, there was evidence of beneficial psychological outcomes in the surveillance group, who reported lower general distress and more positive consequences of screening, and had greater reductions in anxiety than both the no-polyps group and the lower-risk group. This suggests that the positive consequences of screening may extend beyond the removal of polyps and are perceived as greater among people assigned to colonoscopic surveillance. People assigned to surveillance were also more reassured that they did not have bowel cancer than the lower-risk group (who had polyps detected but no colonoscopy) and levels of reassurance were similar to those observed among people with no polyps found, indicating that a programme of surveillance following the detection of polyps offers as much reassurance as having no polyps detected in the first place.
No differences were observed between the group assigned to colonoscopic surveillance and people discharged following colonoscopy in post-screening bowel cancer worry, psychological distress, reassurance that they did not have bowel cancer, anxiety or number of bowel symptoms. However, the group discharged following colonoscopy reported fewer positive consequences of screening and a greater frequency of GP visits than the surveillance group. That said, relatively few people were discharged following colonoscopy so comparisons may have been underpowered to detect small differences.
Limitations of the study include the lower response rates among men, younger people and people from areas with higher socioeconomic deprivation, and we cannot rule out the possibility that these groups might experience a higher proportion of adverse consequences following screening. However, with the exception of the group discharged following colonoscopy, completion rates were similar across the three outcome groups, so it seems unlikely that this would have affected the pattern of results observed in the present study. The lower response rate in the group who were discharged following colonoscopy is more problematic, and might have contributed to the failure to find differences between them and the group assigned to surveillance.
Information on the demographic characteristics of the sample was limited to age, gender and socioeconomic deprivation, leaving open the possibility that the screening outcome groups may have differed in other ways that could have influenced their responses. For example, differences between the screening outcome groups in dispositional optimism or depression may have led to greater endorsement of positive or negative items, respectively. But, if this were the case, differences between the screening outcome groups on pre-screening bowel cancer worry and anxiety should have been apparent, and no such differences were observed. In addition, the pattern of findings varied across measures, suggesting that global response biases are an unlikely explanation for the effects observed.
No measures of general quality of life were taken, so it is unclear whether or not surveillance colonoscopy might have a more general impact. However, both screening- and cancer-specific concerns were assessed, and previous research has tended to show stronger effects when specific rather than general measures of psychological impact are used. 112 It therefore seems unlikely that adverse psychological effects associated with surveillance colonoscopy went undetected, although effects on social or physical aspects of life may have done.
Additional limitations of this study include its focus on people who had taken up the offer of an invitation to be screened and who may therefore have more positive views on medical testing than other members of the general population. Consequently, the results may not be generalisable to a higher-risk group who are explicitly recommended to have screening, and among whom attitudes towards medical surveillance and testing may be less positive. In addition, we assessed only the relatively short-term psychological impact of screening and therefore do not know whether or not the observed positive consequences persist over time.
A further limitation was the use of single-item measures for a number of the outcome variables, which are less reliable than multiple-item scales and may have reduced the likelihood of detecting significant differences. Although the large sample size compensates for this limitation to some extent, future research could use improved measures of reassurance and use of health care to verify the findings observed here.
The results of the current study are broadly reassuring and show that referral for colonoscopic surveillance is not associated with adverse psychological consequences. Although post-screening bowel cancer worry and bowel symptoms were higher in people assigned to surveillance, they declined over time and were equivalent to levels observed in the other two groups found to have polyps. This suggests they were a result of polyp detection rather than surveillance per se. Overall, surveillance itself appeared to be associated with improved psychological well-being.
Chapter 7 Synthesis
Current UK guidelines recommend 3-yearly surveillance for patients classified as being at IR based on characteristics of adenomas that were found at baseline colonoscopy. The IR group represents 17% of people testing positive in the NHS BCSP,36 and around 40% of patients with adenomas in the hospital data set in this study, with consequent demand on colonoscopy resources, yet the heterogeneity of the group in terms of cancer risk and surveillance requirements has not been investigated. We therefore sought to examine the effect of surveillance interval length on detection rates of AA and CRC in patients attending follow-up after removal of IR adenomas, and of surveillance colonoscopy on CRC risk after baseline. We investigated whether or not there exist subgroups of patients who might not benefit from surveillance, or in whom a single surveillance examination might suffice, and also assessed whether or not the interval to surveillance should be shortened or could be safely extended in certain patients.
Hospital and screening data sets
To investigate these questions we used two main sources of data. The first, the hospital data set, represented a symptomatic population; being the largest data set (n = 11,944), this formed the core of our study. The second, smaller, screening data set comprised an asymptomatic screening population (n = 2352) from three individual screening studies. There were several differences between the hospital and screening data sets. Patients in the hospital data set were older, and examinations were generally more recent (2005–10), and, as a result, the average follow-up time for CRC risk was shorter than for the screening data (6.0 years vs. 11.0 years). The hospital cohort had more adenomas that were large (≥ 20 mm) or had HGD, more reports of poor-quality examinations, and lower follow-up attendance (40% hospital data set vs. 80% screening data set; 14% vs. 43% with two or more follow-ups). Because the hospital data set was larger, this data set was used to identify risk factors at baseline and FUV1 for future CRC risk and for findings at FUV1 and FUV2, and thereby to identify HIR and LIR subgroups.
In the hospital data set, 20% of baseline examinations and 50% of follow-up procedures took place after 2004. Since then when examination quality has probably improved, as demonstrated by the declining proportion of patients with an incomplete colonoscopy in our data over time; however, there was no evidence to suggest that bowel preparation quality had improved. Examination quality was better in the screening data set than in the hospital data set (96% with complete colonoscopy vs. 76%). We have shown that this difference partly explains the larger size of the LIR subgroup and lower rate of CRC in the screening data set, and may also account for the lower detection rates of AA and CRC at FUV1.
Main findings
Is a 3-year interval appropriate? Is there a group that needs a shorter interval to the first or second follow-up examination, or in which follow-up could be postponed?
Our results from the hospital data set suggest that the current surveillance interval of 3 years is suitable for the majority of IR patients, as detection rates of CRC were < 1% before the interval extended beyond 3 years, and detection rates of AA were around 9% during this time, making the examination at 3 years worthwhile. The data do not indicate that surveillance needs to be done earlier in this group of patients, as in the first 3 years there was little increase in rates of AA and a low rate of CRC, so any gains from a shorter interval are likely to be very small.
In the hospital data set, at FUV1 the odds of detecting AA or CRC increased significantly with increasing with interval length. In contrast, in the screening data set there was no association between interval and findings at FUV1, before or after adjustment for covariates, possibly due to the lack of variation in interval length within cohorts, as in a screening programme, recommended surveillance intervals are usually fixed. Also, the KP cohort tended to have a longer interval and a lower rate of CRC, whereas in the EP and UKFSST cohorts a shorter interval was more common and there were higher rates of CRC, thus cancelling out any potential effects of interval when the cohorts were pooled. We were therefore unable to validate our findings regarding the optimum interval inferred from the hospital data set using the screening data set.
Does surveillance provide any benefit in terms of long-term cancer risk? Is there a group who do not require a follow-up examination or in whom a second follow-up examination might be omitted?
We found strong evidence that a single surveillance visit conferred substantial benefit to IR patients by lowering their future risk of CRC, in both the hospital and screening data sets. In the hospital data set, one follow-up conferred a significant 49% reduced risk of CRC after baseline, and results from the screening data set validated this finding, with a significant 73% lower risk of CRC observed after one follow-up.
The first surveillance visit appeared to offer most of the protection, and the benefit of additional surveillance was less clear. In the hospital data set, having two or more follow-up visits was associated with an additional reduction in CRC risk, whereas in the screening data set additional surveillance did not appear to provide any further protection, possibly because the screening participants were already at lower risk as a consequence of their younger age and better-quality examinations.
There did appear to be heterogeneity among IR patients in terms of long-term CRC risk and surveillance needs. The LIR and HIR subgroups (based on polyp and procedural predictors of CRC risk) were discriminant in terms of future CRC risk in both the hospital and screening data sets, although the difference was not statistically significant in the screening data set. In both data sets, in the HIR subgroup a single surveillance visit was associated with a strong protective effect against risk of CRC, and additional surveillance appeared to provide further benefit.
By contrast, in both data sets, in the LIR subgroup the first surveillance visit was associated with a small, non-significant reduction in risk, and additional surveillance did not appear to provide any further protection. In addition, the pre-surveillance (post-baseline) standardised incidence rate of CRC in the LIR subgroup was lower than that of the general population, significantly so in the hospital data set.
We conclude that those in the HIR subgroup are likely to benefit from additional surveillance after the first follow-up, but, for the LIR group, additional surveillance, or potentially any surveillance, may be unnecessary. The HIR subgroup is at higher risk of CRC than the general population, and continued surveillance appears to reduce this risk close to the population level. The small number of CRC end points in the LIR subgroup in both data sets prevented us from concluding whether or not this group would benefit from surveillance. Indeed, as the LIR subgroup were already at lower risk than the general population, the question remains as to what level of surveillance should they be offered in the future.
Economic evaluation
A health-economic simulation model, which assessed the cost-effectiveness of 13 surveillance options for patients with intermediate-grade adenomas, suggested that 3-yearly ongoing colonoscopic surveillance without an age cut-off would produce the greatest health gain. The ICER for this option (compared against the same strategy with an age cut-off of 75 years) was expected to be approximately £2748 per QALY gained. Although 3-yearly ongoing surveillance appears to be the most effective strategy and health gains are expected to be achieved at a cost which may be acceptable to NHS policy-makers, it may be preferable to pursue less effective strategies that also offer a favourable cost-effectiveness profile. As such, the potential to safely stop surveillance in the LIR subgroup remains an important and pertinent finding that warrants further investigation.
Psychological study
The results of the psychological study demonstrated that referral for colonoscopic surveillance is not associated with adverse psychological consequences. Although post-screening bowel cancer worry and bowel symptoms were higher in people assigned to surveillance, they declined over time and were equivalent to levels observed in the other two groups found to have polyps. This suggests that they were a result of polyp detection rather than surveillance per se. Overall, surveillance itself appeared to be associated with improved psychological well-being and, consequently, were colonoscopic surveillance to be stopped in a subset of IR patients it may be necessary to consider offering an alternative intervention, such as a faecal immunochemical test, to patients in order to provide reassurance.
Factors affecting surveillance
Examination quality
Colonoscopy completeness and quality of bowel preparation were important predictors of CRC in our cohort. There has been a substantial improvement in endoscopy quality in the past 10 years; however, there needs to be clear guidance on how to manage patients who are found to have adenomas warranting surveillance and who have suboptimal examinations. Although colonoscopy guidelines advise that a repeat examination is performed in such cases,24 for those individuals in whom colonoscopy is difficult and therefore unsuccessful it is probably inappropriate to recommend repeated colonoscopic surveillance; alternative whole-bowel investigations should be offered instead.
Missed and incompletely resected lesions
Important lesions found at surveillance colonoscopy are thought to be a mixture of missed lesions, incompletely excised baseline lesions and newly developing lesions. 17,113 A systematic review of six studies,114 including 465 patients, found an adenoma miss rate of up to 7% for adenomas of ≥ 10 mm, 18% for adenomas of 6–9 mm and 35% for small (≤ 5 mm) adenomas, with an overall miss rate of one in five polyps; flat and sessile serrated lesions were especially likely to be missed. 115,116 The potential for lesions missed at baseline to develop into post-colonoscopy CRCs,117,118 and the possibility that missed baseline lesions may cause underestimation of risk and improper surveillance recommendations – the Will Rogers phenomenon119 – should be taken into account when considering how soon to perform surveillance after baseline. For this reason, if a patient is deemed to be at sufficient risk to warrant surveillance, the first surveillance should not be delayed.
Compliance
Another factor to consider when determining appropriate intervals is the effect on patient adherence to surveillance. In light of this, we might ask whether surveillance should be performed soon after baseline or not, and what are the possible reasons for carrying out surveillance sooner. Compliance with surveillance colonoscopy is not extensively studied, although the limited studies available suggest that compliance rates in studies of post-polypectomy surveillance range from 66% to 85%. 8,33,120–122 We were not able to examine compliance in our study, which recorded only if a person attended follow-up and not if they were scheduled to do so.
Compliance with surveillance may be improved by inviting patients to return sooner. In the US National Polyp Study,8 compliance was slightly higher in patients whose first examination was at 1 year than in those first examined at 3 years (83% vs. 78%), providing some evidence that a shorter interval may be beneficial in terms of improving adherence. A randomised trial in England found that patients with a 2- or 5-year surveillance interval had compliance rates of 86% and 74%, respectively, although non-attenders were pursued using postal reminders and a letter to GPs, which may have improved attendance in both groups. 33 A larger proportion of patients in the 5-year interval group were lost to follow-up due to death or because they reached 75 years of age, making them ineligible for surveillance; thus, it is difficult to discern the effect of interval on compliance in this study. However, this study does show that if surveillance is to be stopped at age 75 years then surveillance for patients aged > 70 years at initial adenoma detection should not have their surveillance examinations delayed if they are fit and at high enough risk.
Strengths and limitations
A major strength of this study was the wide variation in surveillance interval length, which is a unique feature of the hospital data set. Another achievement, in terms of the hospital data set, was the creation of a high-quality data set despite the numerous difficulties encountered, which demonstrates that, although problematic, it is possible to use data from routine secondary care sources and obtain reliable results, as evidenced by validation of our results in the screening data set.
Apart from data on examination quality in both the hospital and screening data sets, there were very few missing data and this, together with the use of extensive data cleaning, reduced the risk of bias arising from measurement error or misclassification. Owing to the nature of the routine data that made up the hospital data set, some measurement error and misclassification is inevitable; however, this would most likely have been non-differential and thus would have resulted in the underestimation of the effect of surveillance and interval length. Furthermore, follow-up was complete for almost all patients, limiting the potential for selection bias, and the study had wide geographic coverage, which improves external validity.
Sensitivity analyses described in Chapter 3 (see Sensitivity analyses and internal validation) provided evidence of the relative robustness of the methods used to define visits and intervals, to extend baseline and to define the outcome of AA. The results of these analyses did not differ substantially from the main analyses and thus suggested that the applied methods did not introduce bias.
A limitation of our study is that in the hospital data set, although some patients were censored before the end of hospital data collection, the majority had follow-up time after the end of data collection in which they may have attended surveillance visits. Sensitivity analyses restricted to patients with at least 5 years of follow-up indicate that the effect of surveillance on CRC risk and absolute pre-surveillance risk in the cohort is likely to have been underestimated as a consequence of contamination of the no-surveillance group. This could be remedied through the collection of additional hospital data in the future.
One limitation of the economic analysis is that the model assumed that all cancers arise from prior adenomas when there is indirect evidence to suggest that a proportion of CRCs arise de novo rather than as a consequence of the adenoma–carcinoma sequence. Furthermore, given better data, frailty models – to distinguish between those adenomas that will develop into cancer and those that will not – may have produced a more accurate representation of the natural history process. Additionally, the MSM did not differentiate by cancer stage, which may result in different estimates of diagnosis and treatment costs. The model also assumed adenoma detection through surveillance, but, in reality, adenomas may also be detected through symptomatic or opportunistic presentation. Furthermore, the model may overestimate the clinical benefits and cost-effectiveness of surveillance programmes in comparison with usual clinical practice as all patients were assumed to attend their scheduled surveillance visits and be sufficiently fit to undergo colonoscopy. Finally, the model assessed only ongoing surveillance with a fixed interval or once-only surveillance; however, it may be possible that alternative surveillance options that were not defined for inclusion in this analysis could produce greater expected health gains than those estimated within this analysis.
Implications for health care
Although policy changes cannot be recommended, we believe that the findings of this study provide high-quality evidence which will allow policy-makers to review current surveillance guidelines for individuals with intermediate-grade adenomas. The results suggest that, although the current 3-year interval is appropriate for most, patients with certain baseline characteristics may not benefit from additional surveillance beyond the first follow-up, offering the potential to alleviate pressure on endoscopy services. In terms of the implications that this research may have for health care, the difficulties encountered during the study indicate that improved reporting techniques and standardisation across hospital endoscopy and pathology systems would enable the more effective use of such data for research purposes in the future. We feel that this is an important point, as hospital endoscopy and pathology data contain invaluable information that should be recorded bearing future research in mind.
Recommendations for future research
Future studies are needed to validate whether or not the long-term CRC risk factors identified in this study are predictors of findings at surveillance, as suggested by results from our hospital data set. Research is also needed to further examine whether or not surveillance is of equal benefit to all IR patients, and whether or not one surveillance visit provides adequate protection for some.
A number of outstanding research questions can be investigated using this high-quality data set; such investigations should be relatively straightforward, as all data are coded and analysable. Future research could investigate the optimum surveillance strategies for low- and high-risk groups, both of which are present in large numbers within the original study cohort. Specifically, we plan to use the data set to determine the optimum frequency of surveillance in patients with high-risk adenomas and the necessity for the 1-year post-baseline colonoscopy currently recommended, and to verify whether or not the low-risk group is of low enough risk of CRC to justify offering no surveillance. Research into the effect of hyperplastic polyps and other serrated lesions on the risk of AN will also be possible using this rich data set, and we will examine the modifying effect of hyperplastic and serrated polyps on future risk of CRC in patients with low-, intermediate- or high-risk adenomas and identify appropriate surveillance intervals.
We will also investigate the feasibility of collecting information on follow-up examinations for the last 5 years, during which time we followed the hospital cohort using national sources but did not have the resources to collect further data.
Chapter 8 Conclusions
Our results provide strong evidence that a single surveillance visit confers substantial benefit to IR patients by lowering their future risk of CRC. IR patients showed heterogeneity in terms of their surveillance needs; in the hospital data set a HIR subgroup benefited from additional surveillance after the first follow-up examination, but in the LIR subgroup additional surveillance may be unnecessary, as patients in this subgroup were at a substantial 60% lower risk than the general population, even before their first surveillance visit.
The screening data set results validated findings from the survival analysis in the hospital cohort, providing further evidence of the benefit of surveillance in IR patients. Risk subgroups derived from risk factors for longer-term CRC in the hospital data set were discriminant in terms of CRC risk in the screening data set, but did not discriminate between individuals with and without findings at the first follow-up, probably because individuals in the screening data sets were at lower risk and had fewer outcomes.
Referral for colonoscopic surveillance was not associated with adverse psychological consequences. Rather, bowel cancer worry was seemingly related to the detection of growths in the bowel rather than to colonoscopic surveillance itself. Overall, surveillance itself appeared to be associated with improved psychological well-being.
Although 3-yearly ongoing surveillance appears to be the most effective strategy, and such health gains are expected to be achieved at a cost which may be acceptable to NHS policy-makers, it may be preferable to pursue less effective strategies that also offer a favourable cost-effectiveness profile.
Future studies should be undertaken to further validate the findings of this study, and confirm whether or not surveillance is truly of benefit to all IR patients, and whether or not one surveillance visit provides adequate protection for some.
Acknowledgements
The investigators are grateful to the people listed below for their involvement in this project. In addition to those named, we would like to thank all of the administrative staff and nurses within endoscopy, pathology and ICT departments at all hospitals that were involved in this study.
The work of the Cancer Screening and Prevention Research Group (CSPRG) at Imperial College is also supported by The Bobby Moore Fund for Cancer Research UK (C8171/A16894).
Role of the funding source
The funder stipulated a retrospective cohort design but had no involvement in the collection, analysis, or interpretation of data, in writing the report, or in the decision to submit for publication.
Contributions of authors
Wendy Atkin was the Chief Investigator for the study.
Wendy Atkin, Amy Brenner, Katherine Wooldrage, Jonathan Myles and Stephen W Duffy were responsible for data analysis and interpretation.
Paul Tappenden was responsible for the economic analysis, assisted by Jessica Martin and Stephen W Duffy.
Amy Brenner, Urvi Shah, Paul Greliak, Kevin Pack, Ann Thomson, Sajith Perera and Jill Wood were responsible for collecting, coding and cleaning the data.
Amy Brenner and Fiona Lucas prepared the final report.
Wendy Atkin, Ines Kralj-Hans, Anne Miles, Jane Wardle, Paul Tappenden and Stephen W Duffy were responsible for obtaining funding and research approvals, and study design.
Anne Miles and Jane Wardle were responsible for the psychological analyses.
Benjamin Kearns checked the final model and helped to resolve problems in its implementation.
Wendy Atkin, Amy Brenner, Katherine Wooldrage, Urvi Shah and Fiona Lucas reviewed and edited the report.
Andy Veitch offered clinical advice throughout the study, and acted as chair of the Trial Steering Committee.
Trial Steering Committee
Professor Wendy Atkin (PI), Dr Andrew Veitch (Chairperson), Ms Lynn Faulds-Wood (patient representative), Ms Helen Watson (patient representative), Ms Fay Cafferty, Professor Stephen Duffy, Professor Allan Hackshaw, Professor Jeremy Jass, Professor Sue Moss, Professor Marco Novelli, Professor Matt Rutter and Professor Chris Todd.
Patient and public involvement
Throughout the study, two patient and public involvement representatives sat on the Trial Steering Committee and provided relevant insight into the views of patients with regards to the study, and wider issues relating to bowel cancer diagnosis, screening and surveillance.
Trial office staff
Professor W Atkin (Chief Investigator), Miss K Wooldrage (Senior Statistician), Miss J Martin (Statistician), Mrs U Shah (Data Analyst), Mr S Perera (Study Programmer), Dr I Kralj-Hans (Research Manager), Mrs E MacRae (Trial Manager), Miss A Thomson (Research Assistant), Miss A Brenner (Research Assistant), Mr K Pack (Senior Data Clerk), Mr P Greliak (Senior Data Clerk), Miss J Monger (Data Clerk), Mrs J Wood (Data Clerk), Ms A Verjee (Projects Manager), Dr C Monk (Projects Manager), Dr L J Turner (Projects Manager), Mrs R Howe (Projects Administrator), Mrs E Coles (Projects Administrator), Dr F R Lucas (Science Writer) and Mr E Dadswell (Data Manager).
Collaborators
Statistics Professor S Duffy (Statistician), J Myles (Statistician).
Health Psychology Professor J Wardle (Clinical Psychologist), Dr A Miles (Health Psychologist).
Health Economics Dr P Tappenden (Health Economist), Mr B Kearns (Statistician).
Research data sets Dr Theodore R Levin (Chief of Gastroenterology, Kaiser Permanente Medical Center, Walnut Creek, CA, USA) and Dr Carol Conell (Senior Data Consultant, Kaiser Permanente, Oakland, CA, USA). Ms Pat Ramsell (Lead Nurse) and Linda Stretton, Natalie Harrold and Carol Wheatley (Screening Sisters), BCSP, c/o Endoscopy Unit, University Hospitals Coventry and Warwickshire NHS Trust.
NIHR–HTA grant co-applicants
Professor W Atkin (Chief Investigator), Dr T R Levin (Chief of Gastroenterology, Kaiser Permanente Medical Center), Dr P Tappenden (Health Economist), Dr A Miles (Health Psychologist), Professor David Lieberman (Chief, Division of Gastroenterology and Hepatology, Oregon University, Portland, OR, USA), Professor David Weller (Head of General Practice, Centre for Population Health Sciences, University of Edinburgh, UK), Professor S Duffy (Statistician), Professor J Wardle (Clinical Psychologist) and Professor S Moss (Statistician).
Principal investigators
Royal Sussex County Hospital, Brighton: Dr Stuart Cairns.
Cumberland Infirmary, Carlisle: Dr Denis Burke.
Charing Cross Hospital, Hammersmith Hospitals and St Mary’s, London: Mr Paul Ziprin/Dr Geoff Smith.
Glasgow Royal Infirmary, Glasgow: Mr John Anderson.
Leicester General Hospital, Leicester: Dr John de Caestecker.
Royal Liverpool University Hospital, Liverpool: Dr Michael Burkitt and Professor Tony Morris.
New Cross Hospital, Wolverhampton: Dr Andrew Veitch.
University Hospital of North Tees, Stockton-on-Tees: Professor Matt Rutter.
Queen Elizabeth Hospital, Woolwich, London: Dr Alastair McNair.
Queen Mary’s Hospital, Sidcup, Kent: Dr Howard Curtis.
Royal Shrewsbury Hospital, Shropshire: Dr Mark Smith.
St George’s Hospital, Tooting, London: Mr Roger Leicester.
St Mark’s Hospital, Harrow, London: Professor Wendy Atkin.
Royal Surrey County Hospital, Surrey: Mr John Stebbing.
Torbay District General Hospital, Devon: Dr Mark Feeney.
Yeovil District Hospital, Somerset: Mrs Sue Bulley.
Norfolk and Norwich University Hospital: Dr Richard Tighe.
Consultant clinicians, surgeons and endoscopists
Dr Stuart Cairns,* Consultant Gastroenterologist (Royal Sussex County Hospital, Brighton).
Dr Denis Burke,* Consultant Gastroenterologist (Cumberland Infirmary, Carlisle).
Dr Geoff Smith,* Consultant Gastroenterologist (Charing Cross and Hammersmith Hospitals, London).
Mr Paul Ziprin* Consultant Surgeon (St Mary’s Hospital, London).
Dr Bola Makinwa, Endoscopist (St Mary’s Hospital, London).
Dr John de Caestecker,* Consultant Gastroenterologist (Leicester General Hospital, Leicester).
Professor Tony Morris,* Consultant Gastroenterologist (Royal Liverpool University Hospital, Liverpool).
Dr Andrew Veitch* Consultant Gastroenterologist (New Cross Hospital, Wolverhampton).
Professor Matt Rutter,* Consultant Gastroenterologist (University Hospital North Tees, Stockton-on-Tees).
Dr Matthew Foxton, Consultant Gastroenterologist (Queen Elizabeth Hospital, Woolwich, London).
Dr Alistair McNair,* Consultant Gastroenterologist (Queen Elizabeth Hospital, Woolwich, London).
Dr Howard Curtis* Consultant Gastroenterologist (Queen Mary’s Hospital, Sidcup).
Dr Mark Smith,* Consultant Gastroenterologist (Royal Shrewsbury Hospital, Shropshire).
Mr Roger Leicester,* Consultant Surgeon (St George’s Hospital, Tooting, London).
Dr Chris Groves, Consultant Gastroenterologist (St George’s Hospital, Tooting, London).
Professor Brian Saunders, Consultant Gastroenterologist (St Mark’s Hospital, Harrow, London).
Professor John Northover, Consultant Surgeon (St Mark’s Hospital, Harrow, London).
Mr John Stebbing,* Consultant Surgeon (Royal Surrey County Hospital, Surrey).
Dr Mark Feeney,* Consultant Gastroenterologist (Torbay District General Hospital, Devon).
Dr Richard Tighe,* Consultant Gastroenterologist (Norfolk and Norwich University Hospital).
(*PIs for study centres.)
Other
Wendy Harman (Senior Research Nurse), Nigel Loaring (Deputy Head BMS – Cellular Pathology), Dr Andrew Rainey (Consultant Histopathologist): Royal Sussex County Hospital, Brighton.
Dr Fergus Young (Pathologist), Judith Johnston (PA to Dr Burke): Cumberland Infirmary, Carlisle.
Catherine Bilter (Scorpio), Dr Patrizia Cohen (Consultant Pathologist), Bob Chapman (Osirus pathology system creator), Rob Goldin (Clinical Lead Pathology), Alan Hales (Scorpio), Yusuf Mangera (IT), David Peston (Pathology Laboratory Manager), Laurell Ramsay (System Administrator for Scorpio), Philip Robinson (Information Governance Manager), Craig Rothwell (Co-Path Account Manager – Sunquest): Charing Cross Hospital and Hammersmith Hospitals, London
Andrew Hay (Laboratory Manager, Cellular Pathology), George Philp (Isoft Portfolio Manager – Telepath), Arthur Sanchez (Scorpio): St Mary’s Hospital, London.
Keith Carter (Product Manager for GIScribe), Mary Docherty (Senior endoscopy nurse), Jane Hair (NHS Greater Glasgow & Clyde Bio-repository Deputy Director), Ian Holloway (Isoft/GIScribes), Sandy Kerr (Senior Support Analyst), Peter Livesey (IT contact for GIScribes), George Philp (Isoft Portfolio Manager – Telepath), Gordon Reid (Endoscopy nurse): Glasgow Royal Infirmary, Glasgow.
Sue Carvell (Pathology), Sally Moulds and Vicky Wood (Secretaries to John de Caestecker): Leicester General Hospital, Leicester
Steve Bradburn (Pathology IT Systems), Dr Michael Burkitt (Academic Clinical Lecturer in Gastroenterology, University of Liverpool), Professor Martin Lombard (Clinical Director Gastroenterology): Royal Liverpool University Hospital, Liverpool.
Roy Cooper (Histopathology Administrator), Richard Tomlin (Technidata consultant): New Cross Hospital, Wolverhampton.
Kevin Downes (Pathology IT & Information Manager), Sharron Pooley (Pathology): University Hospital of North Tees, Stockton-on-Tees.
Dr Thelma Pinto (Pathology), John Pullicino (IT): Queen Elizabeth Hospital, Woolwich, London.
Sandra Applegate (Cellular Pathology), Martin Liggins (APEX consultant), Richard Mainwaring-Burton (Consultant Biochemist), Gemma Thomson (Sales Desk Account Executive) – Queen Mary’s Hospital, Sidcup, Kent.
Marion Adams (R&D/Clinical Trials Manager), Lynn Atkin (now Lead Nurse for Women and Children’s Care), Jenny Ellis (Endoscopy Sister), Wendy Harper (PA to Mark Smith), Professor Archie Malcolm (Pathology Department), George Philp (Isoft Portfolio Manager – Telepath), Kath Williams (Deputy Head Biomedical Scientist – Pathology) – Royal Shrewsbury Hospital, Shropshire.
Angela Cooke (Quality Manager/Cellular Pathology), Dr Caroline Finlayson, Scott Johnson (Pathology Office Manager), Martin Liggins (APEX consultant), Tina Martin (Endoscopy Administrator Department), Anne Morris (Pathology General Manager): St George’s Hospital, Tooting, London.
David Smith (Pathology Laboratory Manager): St Mark’s Hospital, Harrow, London.
Vanessa Bollons (Senior Endoscopy Sister), Simon Dowd (Pathology Manager), Dianne Macleod (Endoscopy Manager), Tasmin Patel (Data Analyst (Endoscopy), George Philp (Isoft Portfolio Manager – Telepath), Richard Rivett (Micromed Consultant): Royal Surrey County Hospital, Surrey.
Dr Rachel Ali (Local Data Administrator): Torbay District General Hospital, Devon.
Mrs Sue Bulley (Research & Development & Clinical Trials Manager), Jane Johnston (Senior Clinical Performance Analyst), Garry Sweet (Pathology Laboratory Manager): Yeovil District Hospital, Somerset.
Michelle Bacon (Data Quality Manager), Ben Goss (IT support engineer), Richard Harris (Information Governance & IT security Manager), Sarah Pond (Richard Tighe’s project manager), Virginia Sams (Consultant Pathologist), Iain Sheriffs (Chief Biomedical Scientist, Pathology Department): Norfolk and Norwich University Hospital, Norwich.
Jenny Gibson (Service Support Administrator) and David Cronin (Data Applications Senior Operational Delivery Manager): Data Access and Request Service (DARS), HSCIC.
Publications
Cafferty FH, Wong JM, Yen AM, Duffy S, Atkin WS, Chen TH. Findings at follow-up endoscopies in subjects with suspected colorectal abnormalities: effects of baseline findings and time to follow-up. Cancer J 2007;13:263–70.
Atkin W, Wooldrage K, Brenner A, Martin J, Shah U, Perera S, et al. Adenoma surveillance and colorectal cancer incidence: a retrospective, multicentre, cohort study [published online ahead of print 27 April 2017]. Lancet Oncol 2017.
Data sharing statement
Data sharing requests should be directed to the corresponding author.
Disclaimers
This report presents independent research funded by the National Institute for Health Research (NIHR). The views and opinions expressed by authors in this publication are those of the authors and do not necessarily reflect those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health. If there are verbatim quotations included in this publication the views and opinions expressed by the interviewees are those of the interviewees and do not necessarily reflect those of the authors, those of the NHS, the NIHR, NETSCC, the HTA programme or the Department of Health.
References
- Atkin WS, Saunders BP. Surveillance guidelines after removal of colorectal adenomatous polyps. Gut 2002;51:V6-9. http://dx.doi.org/10.1136/gut.51.suppl_5.v6.
- Martínez ME, Thompson P, Messer K, Ashbeck EL, Lieberman DA, Baron JA, et al. One-year risk for advanced colorectal neoplasia: U.S. versus U.K. risk-stratification guidelines. Ann Intern Med 2012;157:856-64. http://dx.doi.org/10.7326/0003-4819-157-12-201212180-00005.
- Atkin WS, Morson BC, Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N Engl J Med 1992;326:658-62. http://dx.doi.org/10.1056/NEJM199203053261002.
- Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer 1975;36:2251-70. http://dx.doi.org/10.1002/cncr.2820360944.
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525-32. http://dx.doi.org/10.1056/NEJM198809013190901.
- Morson BC, Bussey HJ. Magnitude of risk for cancer in patients with colorectal adenomas. Br J Surg 1985;72:S23-5. http://dx.doi.org/10.1002/bjs.1800721315.
- Bussey HJ, Wallace MH, Morson BC. Metachronous carcinoma of the large intestine and intestinal polyps. Proc R Soc Med 1967;60:208-10.
- Winawer SJ, Zauber AG, Ho MN, O’Brien MJ, Gottlieb LS, Sternberg SS, et al. Prevention of colorectal-cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med 1993;329:1977-81. http://dx.doi.org/10.1056/NEJM199312303292701.
- Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal cancer deaths. Obstet Gynecol Surv 2012;67:355-6. http://dx.doi.org/10.1097/OGX.0b013e31825bc1f5.
- Atkin WS, Edwards R, Kralj-Hans I, Wooldrage K, Hart AR, Northover JM, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet 2010;375:1624-33. http://dx.doi.org/10.1016/S0140-6736(10)60551-X.
- Segnan N, Armaroli P, Bonelli L, Risio M, Sciallero S, Zappa M, et al. Once-only sigmoidoscopy in colorectal cancer screening: follow-up findings of the Italian Randomized Controlled Trial-SCORE. J Natl Cancer Inst 2011;103:1310-22. http://dx.doi.org/10.1093/jnci/djr284.
- Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med 2012;366:2345-57. http://dx.doi.org/10.1056/NEJMoa1114635.
- Nishihara R, Lochhead P, Wu K, Morikawa T, Fuchs CS, Giovannucci E, et al. Long-term risk of colorectal cancer risk after lower endoscopy and polypectomy. Gastroenterology 2012;142. http://dx.doi.org/10.1016/S0016-5085(12)60418-1.
- Cottet V, Jooste V, Fournel I, Bouvier AM, Faivre J, Bonithon-Kopp C. Long-term risk of colorectal cancer after adenoma removal: a population-based cohort study. Gut 2012;61:1180-6. http://dx.doi.org/10.1136/gutjnl-2011-300295.
- Van Stolk RU, Beck GJ, Baron JA, Haile R, Summers R. Adenoma characteristics at first colonoscopy as predictors of adenoma recurrence and characteristics at follow-up. The Polyp Prevention Study Group. Gastroenterology 1998;115:13-8. http://dx.doi.org/10.1016/S0016-5085(98)70359-2.
- Cairns SR, Scholefield JH, Steele RJ, Dunlop MG, Thomas HJ, Evans GD, et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups (update from 2002). Gut 2010;59:666-89. http://dx.doi.org/10.1136/gut.2009.179804.
- Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR, et al. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012;143:844-57. http://dx.doi.org/10.1053/j.gastro.2012.06.001.
- Hassan C, Quintero E, Dumonceau JM, Regula J, Brandão C, Chaussade S, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2013;45:842-51. http://dx.doi.org/10.1055/s-0033-1344548.
- Wegener M, Börsch G, Schmidt G. Colorectal adenomas. Distribution, incidence of malignant transformation, and rate of recurrence. Dis Colon Rectum 1986;29:383-7. http://dx.doi.org/10.1007/BF02555053.
- Winawer SJ, Zauber AG, O’Brien MJ, Ho MN, Gottlieb L, Sternberg SS, et al. Randomized comparison of surveillance intervals after colonoscopic removal of newly diagnosed adenomatous polyps. The National Polyp Study Workgroup. N Engl J Med 1993;328:901-6. http://dx.doi.org/10.1056/NEJM199304013281301.
- Neugut AI, Jacobson JS, Ahsan H, Santos J, Garbowski GC, Forde KA, et al. Incidence and recurrence rates of colorectal adenomas: a prospective study. Gastroenterology 1995;108:402-8. http://dx.doi.org/10.1016/0016-5085(95)90066-7.
- Noshirwani KC, van Stolk RU, Rybicki LA, Beck GJ. Adenoma size and number are predictive of adenoma recurrence: implications for surveillance colonoscopy. Gastrointest Endosc 2000;51:433-7. http://dx.doi.org/10.1016/S0016-5107(00)70444-5.
- Martinez ME, Sampliner R, Marshall JR, Bhattacharyya AK, Reid ME, Alberts DS. Adenoma characteristics at baseline colonoscopy as risk factors for recurrence of advanced adenomas. Gastroenterology 2001;120:1077-83. http://dx.doi.org/10.1053/gast.2001.23247.
- Colonoscopic Surveillance for Prevention of Colorectal Cancer in People with Ulcerative Colitis, Crohn’s disease or Adenomas. London: NICE; 2011.
- Atkin WS, Valori R, Kuipers EJ, Hoff G, Senore C, Segnan N, et al. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First edition: Colonoscopic surveillance following adenoma removal. Endoscopy 2012;44:E151-63.
- Hoff G, Sauar J, Hofstad B, Vatn MH. The Norwegian guidelines for surveillance after polypectomy: 10-year intervals. Scand J Gastroenterol 1996;31:834-6. http://dx.doi.org/10.3109/00365529609051989.
- Citarda F, Tomaselli G, Capocaccia R, Barcherini S, Crespi M. Efficacy in standard clinical practice of colonoscopic polypectomy in reducing colorectal cancer incidence. Gut 2001;48:812-15. http://dx.doi.org/10.1136/gut.48.6.812.
- Bertario L, Russo A, Sala P, Pizzetti P, Ballardini G, Andreola S, et al. Predictors of metachronous colorectal neoplasms in sporadic adenoma patients. Int J Cancer 2003;105:82-7. http://dx.doi.org/10.1002/ijc.11036.
- Jørgensen OD, Kronborg O, Fenger C. The Funen Adenoma Follow-up Study. Incidence and death from colorectal carcinoma in an adenoma surveillance program. Scand J Gastroenterol 1993;28:869-74. http://dx.doi.org/10.3109/00365529309103127.
- Loeve F, van Ballegooijen M, Snel P, Habbema JD. Colorectal cancer risk after colonoscopic polypectomy: a population-based study and literature search. Eur J Cancer 2005;41:416-22. http://dx.doi.org/10.1016/j.ejca.2004.11.007.
- Løberg M, Kalager M, Holme Ø, Hoff G, Adami HO, Bretthauer M. Long-term colorectal-cancer mortality after adenoma removal. N Engl J Med 2014;371:799-807. http://dx.doi.org/10.1056/NEJMoa1315870.
- Pinsky PF, Schoen RE, Weissfeld JL, Church T, Yokochi LA, Doria-Rose VP, et al. The yield of surveillance colonoscopy by adenoma history and time to examination. Clin Gastroenterol Hepatol 2009;7:86-92. http://dx.doi.org/10.1016/j.cgh.2008.07.014.
- Lund JN, Scholefield JH, Grainge MJ, Smith SJ, Mangham C, Armitage NC, et al. Risks, costs, and compliance limit colorectal adenoma surveillance: lessons from a randomised trial. Gut 2001;49:91-6. http://dx.doi.org/10.1136/gut.49.1.91.
- Kronborg O, Jørgensen OD, Fenger C, Rasmussen M. Three randomized long-term surveillance trials in patients with sporadic colorectal adenomas. Scand J Gastroenterol 2006;41:737-43. http://dx.doi.org/10.1080/00365520500442666.
- Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Reding DJ, Hayes RB, et al. Utilization of surveillance colonoscopy in community practice. Gastroenterology 2010;138:73-81. http://dx.doi.org/10.1053/j.gastro.2009.09.062.
- Bevan R, Lee TJ, Nickerson C, Rubin G, Rees CJ. Non-neoplastic findings at colonoscopy after positive faecal occult blood testing: data from the English Bowel Cancer Screening Programme. J Med Screen 2014;21:89-94. http://dx.doi.org/10.1177/0969141314528889.
- Health and Social Care Act 2001. London: The Stationery Office; 2001.
- National Health Service Act 2006. London: The Stationery Office; 2006.
- Data Protection Act 1998. London: The Stationery Office; 1998.
- Adenoma Surveillance. Sheffield: NHS Cancer Screening Programmes; 2009.
- Percy CL, Berg JW, Thomas LB. Manual of Tumor Nomenclature and Coding. Washington, DC: American Cancer Society; 1968.
- International Classification of Diseases for Oncology. Geneva: WHO; 1976.
- International Classification of Diseases for Oncology. Geneva: WHO; 1990.
- Huang EH, Whelan RL, Gleason NR, Maeda JS, Terry MB, Lee SW, et al. Increased incidence of colorectal adenomas in follow-up evaluation of patients with newly diagnosed hyperplastic polyps. Surg Endosc 2001;15:646-8. http://dx.doi.org/10.1007/s004640000389.
- Blumberg D, Opelka FG, Hicks TC, Timmcke AE, Beck DE. Significance of a normal surveillance colonoscopy in patients with a history of adenomatous polyps. Dis Colon Rectum 2000;43:1084-91. http://dx.doi.org/10.1007/BF02236554.
- Chen TH, Chiu YH, Luh DL, Yen MF, Wu HM, Chen LS, et al. Community-based multiple screening model: design, implementation, and analysis of 42,387 participants. Cancer 2004;100:1734-43. http://dx.doi.org/10.1002/cncr.20171.
- Wong JM, Yen MF, Lai MS, Duffy SW, Smith RA, Chen TH. Progression rates of colorectal cancer by Dukes’ stage in a high-risk group: analysis of selective colorectal cancer screening. Cancer J 2004;10:160-9. http://dx.doi.org/10.1097/00130404-200405000-00005.
- Stryker SJ, Wolff BG, Culp CE, Libbe SD, Ilstrup DM, MacCarty RL. Natural history of untreated colonic polyps. Gastroenterology 1987;93:1009-13. http://dx.doi.org/10.1016/0016-5085(87)90563-4.
- Pal A, Provenzano E, Duffy SW, Pinder SE, Purushotham AD. A model for predicting non-sentinel lymph node metastatic disease when the sentinel lymph node is positive. Br J Surg 2008;95:302-9. http://dx.doi.org/10.1002/bjs.5943.
- Atkin W, Wooldrage K, Brenner A, Martin J, Shah U, Perera S, et al. Adenoma surveillance and colorectal cancer incidence: a retrospective, multicentre, cohort study [published online ahead of print 27 April 2017]. Lancet Oncol 2017. http://dx.doi.org/10.1016/S1470-2045(17)30187-0.
- de Jonge V, Sint Nicolaas J, van Leerdam ME, Kuipers EJ, Veldhuyzen van Zanten SJ. Systematic literature review and pooled analyses of risk factors for finding adenomas at surveillance colonoscopy. Endoscopy 2011;43:560-72. http://dx.doi.org/10.1055/s-0030-1256306.
- Majumdar D, Hungin A, Wilson D, Nickerson C, Rutter M. Predictors of advanced neoplasia at surveillance in screening population: a study of all high and intermediate risk group subjects in first six years of NHS BCSP. Presentation no. OC-044. Gut 2014;63:A21-2. http://dx.doi.org/10.1136/gutjnl-2014-307263.44.
- Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010;138:2088-100. http://dx.doi.org/10.1053/j.gastro.2009.12.066.
- Valori R. Quality improvements in endoscopy in England. Tech Gastrointest Endosc 2012;14:63-72. http://dx.doi.org/10.1016/j.tgie.2011.11.001.
- Gavin DR, Valori RM, Anderson JT, Donnelly MT, Williams JG, Swarbrick ET. The national colonoscopy audit: a nationwide assessment of the quality and safety of colonoscopy in the UK. Gut 2013;62:242-9. http://dx.doi.org/10.1136/gutjnl-2011-301848.
- Valori RM, Morris EJ, Thomas JD, Rutter M. Rates of post colonoscopy colorectal cancer (PCCRC) are significantly affected by methodology, but are nevertheless declining in the English National Health Service (NHS). Abstract no. Tu1485. Gastrointest Endosc 2014;79. http://dx.doi.org/10.1016/j.gie.2014.02.931.
- Shah HA, Paszat LF, Saskin R, Stukel TA, Rabeneck L. Factors associated with incomplete colonoscopy: a population-based study. Gastroenterology 2007;132:2297-303. http://dx.doi.org/10.1053/j.gastro.2007.03.032.
- Cirocco WC, Rusin LC. Factors that predict incomplete colonoscopy. Dis Colon Rectum 1995;38:964-8. http://dx.doi.org/10.1007/BF02049733.
- Hassan C, Fuccio L, Bruno M, Pagano N, Spada C, Carrara S, et al. A predictive model identifies patients most likely to have inadequate bowel preparation for colonoscopy. Clin Gastroenterol Hepatol 2012;10:501-6. http://dx.doi.org/10.1016/j.cgh.2011.12.037.
- Serper M, Gawron AJ, Smith SG, Pandit AA, Dahlke AR, Bojarski EA, et al. Patient factors that affect quality of colonoscopy preparation. Clin Gastroenterol Hepatol 2014;12:451-7. http://dx.doi.org/10.1016/j.cgh.2013.07.036.
- Johnson DA, Barkun AN, Cohen LB, Dominitz JA, Kaltenbach T, Martel M, et al. Optimizing adequacy of bowel cleansing for colonoscopy: recommendations from the US Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol 2014;109:1528-45. http://dx.doi.org/10.1053/j.gastro.2014.07.002.
- Clark BT, Rustagi T, Laine L. What level of bowel prep quality requires early repeat colonoscopy: systematic review and meta-analysis of the impact of preparation quality on adenoma detection rate. Am J Gastroenterol 2014;109:1714-23. http://dx.doi.org/10.1038/ajg.2014.232.
- Vieth M, Quirke P, Lambert R, von Karsa L, Risio M. European guidelines for quality assurance in colorectal cancer screening and diagnosis. First edition – Annotations of colorectal lesions. Endoscopy 2012;44:E131-9.
- Thomas-Gibson S, Rogers P, Cooper S, Man R, Rutter MD, Suzuki N, et al. Judgement of the quality of bowel preparation at screening flexible sigmoidoscopy is associated with variability in adenoma detection rates. Endoscopy 2006;38:456-60. http://dx.doi.org/10.1055/s-2006-925259.
- Foss FA, Milkins S, McGregor AH. Inter-observer variability in the histological assessment of colorectal polyps detected through the NHS Bowel Cancer Screening Programme. Histopathology 2012;61:47-52. http://dx.doi.org/10.1111/j.1365-2559.2011.04154.x.
- Levene Y, Hutchinson JM, Tinkler-Hundal E, Quirke P, West NP. The correlation between endoscopic and histopathological measurements in colorectal polyps. Histopathology 2015;66:485-90. http://dx.doi.org/10.1111/his.12472.
- van Putten PG, Hol L, van Dekken H, Han van Krieken J, van Ballegooijen M, Kuipers EJ, et al. Inter-observer variation in the histological diagnosis of polyps in colorectal cancer screening. Histopathology 2011;58:974-81. http://dx.doi.org/10.1111/j.1365-2559.2011.03822.x.
- Rex DK, Rabinovitz R. Variable interpretation of polyp size by using open forceps by experienced colonoscopists. Gastrointest Endosc 2014;79:402-7. http://dx.doi.org/10.1016/j.gie.2013.08.030.
- Costantini M, Sciallero S, Giannini A, Gatteschi B, Rinaldi P, Lanzanova G, et al. Interobserver agreement in the histologic diagnosis of colorectal polyps. the experience of the multicenter adenoma colorectal study (SMAC). J Clin Epidemiol 2003;56:209-14. http://dx.doi.org/10.1016/S0895-4356(02)00587-5.
- Spiegelman D, McDermott A, Rosner B. Regression calibration method for correcting measurement-error bias in nutritional epidemiology. Am J Clin Nutr 1997;65:1179-86.
- NHS Bowel Cancer Screening Programme (NHS BCSP) . Adenoma Surveillance 2009. www.gov.uk/government/uploads/system/uploads/attachment_data/file/469909/BCSP_Guidance_Note_No_1_Adenoma_Surveillance_uploaded_211015.pdf (accessed February 2014).
- Lieberman DA, Weiss DG, Bond JH, Ahnen DJ, Garewal H, Chejfec G. Use of colonoscopy to screen asymptomatic adults for colorectal cancer. Veterans Affairs Cooperative Study Group 380. N Engl J Med 2000;343:162-8. http://dx.doi.org/10.1056/NEJM200007203430301.
- Scholefield JH, Moss SM. Faecal occult blood screening for colorectal cancer. J Med Screen 2002;9:54-5. http://dx.doi.org/10.1136/jms.9.2.54.
- Alexander F, Weller D. Evaluation of the UK Colorectal Cancer Screening Pilot: Final Report. Edinburgh: UK CRC Screening Pilot Evaluation Team; 2003.
- Segnan N, Senore C, Andreoni B, Aste H, Bonelli L, Crosta C, et al. Baseline findings of the Italian multicenter randomized controlled trial of once-only sigmoidoscopy–SCORE. J Natl Cancer Inst 2002;94:1763-72. http://dx.doi.org/10.1093/jnci/94.23.1763.
- Palitz AM, Selby JV, Grossman S, Finkler LJ, Bevc M, Kehr C, et al. The Colon Cancer Prevention Program (CoCaP): rationale, implementation, and preliminary results. HMO Pract 1997;11:5-12.
- Levin TR, Palitz A, Grossman S, Conell C, Finkler L, Ackerson L, et al. Predicting advanced proximal colonic neoplasia with screening sigmoidoscopy. JAMA 1999;281:1611-17. http://dx.doi.org/10.1001/jama.281.17.1611.
- Guide to the Methods of Technology Appraisal. London: NICE; 2013.
- Tappenden P, Culyer AJ. Encyclopedia of Health Economics. Amsterdam: Elsevier Science Publishing; 2014.
- Chen CD, Yen MF, Wang WM, Wong JM, Chen TH. A case-cohort study for the disease natural history of adenoma–carcinoma and de novo carcinoma and surveillance of colon and rectum after polypectomy: implication for efficacy of colonoscopy. Br J Cancer 2003;88:1866-73. http://dx.doi.org/10.1038/sj.bjc.6601007.
- Rex DK, Rahmani EY, Haseman JH, Lemmel GT, Kaster S, Buckley JS. Relative sensitivity of colonoscopy and barium enema for detection of colorectal cancer in clinical practice. Gastroenterology 1997;112:17-23. http://dx.doi.org/10.1016/S0016-5085(97)70213-0.
- Hixson LJ, Fennerty MB, Sampliner RE, Garewal HS. Prospective blinded trial of the colonoscopic miss-rate of large colorectal polyps. Gastrointest Endosc 1991;37:125-7. http://dx.doi.org/10.1016/S0016-5107(91)70668-8.
- Bressler B, Paszat LF, Vinden C, Li C, He J, Rabeneck L. Colonoscopic miss rates for right-sided colon cancer: a population-based analysis. Gastroenterology 2004;127:452-6. http://dx.doi.org/10.1053/j.gastro.2004.05.032.
- Phillips RKS. Colorectal Surgery: A Companion to Specialist Surgical Practice. Saunders Elsevier; 2005.
- Tappenden P, Chilcott J, Eggington S, Patnick J, Sakai H, Karnon J. Option appraisal of population-based colorectal cancer screening programmes in England. Gut 2007;56:677-84. http://dx.doi.org/10.1136/gut.2006.095109.
- Whyte S, Chilcott J, Halloran S. Reappraisal of the options for colorectal cancer screening in England. Colorectal Dis 2012;14:e547-61. http://dx.doi.org/10.1111/j.1463-1318.2012.03014.x.
- Lee D, Muston D, Sweet A, Cunningham C, Slater A, Lock K. Cost effectiveness of CT colonography for UK NHS colorectal cancer screening of asymptomatic adults aged 60-69 years. Appl Health Econ Health Policy 2010;8:141-54. http://dx.doi.org/10.2165/11535650-000000000-00000.
- Ara R, Brazier JE. Populating an economic model with health state utility values: moving toward better practice. Value Health 2010;13:509-18. http://dx.doi.org/10.1111/j.1524-4733.2010.00700.x.
- Djalalov S, Rabeneck L, Tomlinson G, Bremner KE, Hilsden R, Hoch JS. A review and meta-analysis of colorectal cancer utilities. Med Decis Making 2014;34:809-18. http://dx.doi.org/10.1177/0272989X14536779.
- National Life Tables, United Kingdom: 2010–2012. London: ONS; 2014.
- Scott N, Hill J, Smith J, Walker K, Kuryba A, van der Meulen J, et al. National Bowel Cancer Audit Annual Report. Leeds: Health and Social Care Information Centre; 2013.
- Gatto NM, Frucht H, Sundararajan V, Jacobson JS, Grann VR, Neugut AI. Risk of perforation after colonoscopy and sigmoidoscopy: a population-based study. J Natl Cancer Inst 2003;95:230-6. http://dx.doi.org/10.1093/jnci/95.3.230.
- NHS Reference Costs 2012–13. London: DH; 2014.
- Guyot P, Ades AE, Ouwens MJ, Welton NJ. Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC Med Res Methodol 2012;12. http://dx.doi.org/10.1186/1471-2288-12-9.
- Wrigley H, Roderick P, George S, Smith J, Mullee M, Goddard J. Inequalities in survival from colorectal cancer: a comparison of the impact of deprivation, treatment, and host factors on observed and cause specific survival. J Epidemiol Community Health 2003;57:301-9. http://dx.doi.org/10.1136/jech.57.4.301.
- Tappenden P, Jones R, Paisley S, Carroll C. Systematic review and economic evaluation of bevacizumab and cetuximab for the treatment of metastatic colorectal cancer. Health Technol Assess 2007;11. http://dx.doi.org/10.3310/hta11120.
- Finan P, Smith J, Walker K, vander Meulan J, Greenaway K, Yelland A, et al. National Bowel Cancer Audit Annual Report. Leeds: Health and Social Care Information Centre; 2011.
- Johannesson M, Weinstein MC. On the decision rules of cost-effectiveness analysis. J Health Econ 1993;12:459-67. http://dx.doi.org/10.1016/0167-6296(93)90005-Y.
- Briggs A, Claxton K, Sculpher M. Decision Modelling For Health Economic Evaluation. Oxford: Oxford University Press; 2006.
- Strong M, Oakley JE. An efficient method for computing single-parameter partial expected value of perfect information. Med Decis Making 2013;33:755-66. http://dx.doi.org/10.1177/0272989X12465123.
- Tappenden P, Chilcott JB. Avoiding and identifying errors and other threats to the credibility of health economic models. Pharmacoeconomics 2014;32:967-79. http://dx.doi.org/10.1007/s40273-014-0186-2.
- Kearns B, Whyte S, Chilcott J, Patnick J. Guaiac faecal occult blood test performance at initial and repeat screens in the English Bowel Cancer Screening Programme. Br J Cancer 2014;111:1734-41. http://dx.doi.org/10.1038/bjc.2014.469.
- Parker MA, Robinson MH, Scholefield JH, Hardcastle JD. Psychiatric morbidity and screening for colorectal cancer. J Med Screen 2002;9:7-10. http://dx.doi.org/10.1136/jms.9.1.7.
- Thiis-Evensen E, Wilhelmsen I, Hoff GS, Blomhoff S, Sauar J. The psychologic effect of attending a screening program for colorectal polyps. Scand J Gastroenterol 1999;34:103-9. http://dx.doi.org/10.1080/00365529950172916.
- Taupin D, Chambers SL, Corbett M, Shadbolt B. Colonoscopic screening for colorectal cancer improves quality of life measures: a population-based screening study. Health Qual Life Outcomes 2006;4. http://dx.doi.org/10.1186/1477-7525-4-82.
- Schroy PC, Heeren TC. Patient perceptions of stool-based DNA testing for colorectal cancer screening. Am J Prev Med 2005;28:208-14. http://dx.doi.org/10.1016/j.amepre.2004.10.008.
- Atkin WS, Edwards R, Wardle J, Northover JM, Sutton S, Hart AR, et al. Design of a multicentre randomised trial to evaluate flexible sigmoidoscopy in colorectal cancer screening. J Med Screen 2001;8:137-44. http://dx.doi.org/10.1136/jms.8.3.137.
- Mackenbach JP. Health and deprivation. Inequality and the North. Health Policy 1988;10. http://dx.doi.org/10.1016/0168-8510(88)90006-1.
- Goldberg DP, Gater R, Sartorius N, Ustun TB, Piccinelli M, Gureje O, et al. The validity of two versions of the GHQ in the WHO study of mental illness in general health care. Psychol Med 1997;27:191-7. http://dx.doi.org/10.1017/S0033291796004242.
- Cockburn J, De Luise T, Hurley S, Clover K. Development and validation of the PCQ: a questionnaire to measure the psychological consequences of screening mammography. Soc Sci Med 1992;34:1129-34. http://dx.doi.org/10.1016/0277-9536(92)90286-Y.
- Marteau TM, Bekker H. The development of a six-item short-form of the state scale of the Spielberger State-Trait Anxiety Inventory (STAI). Br J Clin Psychol 1992;31:301-6. http://dx.doi.org/10.1111/j.2044-8260.1992.tb00997.x.
- Miles A, Waller J, Wardle J, Miller SM, Bowen DJ, Croyle RT, et al. Handbook of Cancer Control and Behavioral Science: A Resource for Researchers, Practitioners, and Policy Makers. Washington, DC: American Psychological Association; 2009.
- Huang Y, Gong W, Su B, Zhi F, Liu S, Jiang B. Risk and cause of interval colorectal cancer after colonoscopic polypectomy. Digestion 2012;86:148-54. http://dx.doi.org/10.1159/000338680.
- Van Rijn JC, Reitsma JB, Stoker J, Bossuyt PM, van Deventer SJ, Dekker E. Polyp miss rate determined by tandem colonoscopy: a systematic review. Am J Gastroenterol 2006;101:343-50. http://dx.doi.org/10.1111/j.1572-0241.2006.00390.x.
- Heresbach D, Barrioz T, Lapalus MG, Coumaros D, Bauret P, Potier P, et al. Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy 2008;40:284-90. http://dx.doi.org/10.1055/s-2007-995618.
- Xiang L, Zhan Q, Zhao XH, Wang YD, An SL, Xu YZ, et al. Risk factors associated with missed colorectal flat adenoma: a multicenter retrospective tandem colonoscopy study. World J Gastroenterol 2014;20:10927-37. http://dx.doi.org/10.3748/wjg.v20.i31.10927.
- Robertson DJ, Lieberman DA, Winawer SJ, Ahnen D, Greenberg ER, Baron JA, et al. Interval cancer after total colonoscopy: results from a pooled analysis of eight studies. Gastroenterology 2008;134:A111-12. http://dx.doi.org/10.1016/S0016-5085(08)60520-X.
- Pabby A, Schoen RE, Weissfeld JL, Burt R, Kikendall JW, Lance P, et al. Analysis of colorectal cancer occurrence during surveillance colonoscopy in the dietary Polyp Prevention Trial. Gastrointest Endosc 2005;61:385-91. http://dx.doi.org/10.1016/S0016-5107(04)02765-8.
- Feinstein AR, Sosin DM, Wells CK. The Will Rogers phenomenon. Stage migration and new diagnostic techniques as a source of misleading statistics for survival in cancer. N Engl J Med 1985;312:1604-8. http://dx.doi.org/10.1056/NEJM198506203122504.
- Siddiqui AA, Patel A, Huerta S. Determinants of compliance with colonoscopy in patients with adenomatous colon polyps in a veteran population. Aliment Pharmacol Ther 2006;24:1623-30. http://dx.doi.org/10.1111/j.1365-2036.2006.03176.x.
- Brueckl WM, Fritsche B, Seifert B, Boxberger F, Albrecht H, Croner RS, et al. Non-compliance in surveillance for patients with previous resection of large (> or = 1 cm) colorectal adenomas. World J Gastroenterol 2006;12:7313-18. http://dx.doi.org/10.3748/wjg.v12.i45.7313.
- Colquhoun P, Chen HC, Kim JI, Efron J, Weiss EG, Nogueras JJ, et al. High compliance rates observed for follow up colonoscopy post polypectomy are achievable outside of clinical trials: efficacy of polypectomy is not reduced by low compliance for follow up. Colorectal Dis 2004;6:158-61. http://dx.doi.org/10.1111/j.1463-1318.2004.00585.x.
Appendix 1 Hospital data collection from endoscopy and pathology databases
Data were bulk extracted from hospital endoscopy and pathology databases. This document contains a summary of systems from which data were extracted and the methods used.
Endoscopy and pathology data extraction summary
The following is a summary of the data extracted from each hospital’s endoscopy and pathology databases, with an explanation of the information recorded in each field.

Royal Sussex County Hospital, Brighton and Sussex University Hospitals NHS Trust
| Endoscopy systems in use at the hospital: | Unisoft (2001–Apr 2008) |
| Extraction dates: | May 2001–Apr 2008 |
| Extraction issues on endoscopy systems: | The hospital staff had to set up Unisoft’s GI auditors’ kit in order to extract the data |
| Pathology systems in use at the hospital: | Radius (1986–2005) WinPath (2005–Apr 2008) |
| Extraction dates: | Jan 2000–Apr 2008 |
| Methods used to extract the pathology data: | SNOMED codes Version 2 |
| Missing pathology manually collected later: | May 2001–Apr 2008 |
| Extraction issues on pathology systems: | The WinPath system went live in May 2005 so previous data were extracted from legacy systems. From other investigations we think the legacy system might be Radius, based on seeing Radius in use at other hospitals |
Charing Cross Hospital and Hammersmith Hospital, Imperial College Healthcare NHS Trust
| Endoscopy systems in use at the hospital: | EndoScribe (Sept 1997–Nov 2007) migrated and extracted via Scorpio |
| Extraction dates: | Sep 1997–Nov 2007 |
| Extraction issues on endoscopy systems: | The data from EndoScribe had been migrated to Scorpio so the data were extracted via Scorpio |
| Pathology systems in use at the hospital: | Osiris (Jan 1992–Apr 2002) WinPath (1990–2004) Co-Path (2004–Apr 2011) |
| Extraction dates: | Feb 1990–Jul 2010 |
| Methods used to extract the pathology data: | Osiris – SNOP codes (four-digit versions of SNOMED codes) Co-Path – search terms used: adenocarcinoma, adenoma, anastomosis, angiodysplasia, anus, benign tumour, bowel, cancer, carcinoid, colitis, colon, Crohn’s disease, diverticular disease, diverticulosis, FAP, haemorrhoids, hemicolon, IBD, malignant, melaena, melanosis coli, piles, polyp, polyposis, polyps, proctitis, prolapse, rectum, stricture, suspected IBD, tumour, volvulus WinPath – this was an old archive database in Microsoft Access® (Microsoft Corporation, Redmond, WA, USA) format. We extracted the relevant pathology from this |
| Missing pathology manually collected later: | Jan 1993–Apr 2011 |
| Extraction issues on pathology systems: | Osiris and WinPath were used in parallel between 1992 and 2002, so any overlapping records were merged |
| Other extraction issues: | Hammersmith and Charing Cross hospitals had overlapping data so they were combined as one centre |
St Mary’s Hospital, Imperial College Healthcare NHS Trust
| Endoscopy systems in use at the hospital: | Micromed (1980–2007) Scorpio (2007–Aug 2010) |
| Extraction dates: | Dec 1984–Aug 2010 |
| Extraction issues on endoscopy systems: | Scorpio data were extracted after 6 p.m. to ensure that it did not slow the system down. Extracts for colonoscopy and FS were done separately and broken down into years There was only one computer with Micromed on it, which was located in the endoscopy reception and could be used only after hours, as there were concerns that there was no back-up of the data for the system. Our team had to work within these restricted time constraints to extract the data |
| Pathology systems in use at the hospital: | Telepath (1988–Nov 2010) |
| Extraction dates: | 1990–Jul 2010 |
| Methods used to extract the pathology data: | |
| Missing pathology manually collected later: | 1987–Nov 2010 |
| Extraction issues on pathology systems: | Used Telepath SIFT search for SNOP CODES (four-digit versions of SNOMED codes) |
Cumberland Infirmary, North Cumbria University Hospitals NHS Trust
| Endoscopy systems in use at the hospital: | EndoScribe (1998–Sep 2009) |
| Extraction dates: | Jul 1998–Sep 2009 |
| Extraction issues on endoscopy systems: | Two searches were performed on EndoScribe, one for colonoscopy and one for sigmoidoscopy. Both searches extracted the same fields. Test runs were carried out to determine which fields contained useful information and 20,000 records were extracted |
| Pathology systems in use at the hospital: | Telepath (1988–Dec 2009) |
| Extraction dates: | Jul 1998–Sep 2009 |
| Methods used to extract the pathology data: | Used Telepath SIFT search for SNOP CODES (4-digit versions of SNOMED codes) |
| Missing pathology manually collected later: | Oct 1994–Dec 2009 |
| Extraction issues on pathology systems: | The study programmer was trained by iSoft, which manufactures Telepath |
Glasgow Royal Infirmary, NHS Greater Glasgow and Clyde
| Endoscopy systems in use at the hospital: | EndoScribe (1995–2005) GIScribe (2003–2007) Unisoft (2007–Sep 2009) |
| Extraction dates: | Apr 1996–Sep 2009 |
| Extraction issues on endoscopy systems: | EndoScribe and GIScribe were used concurrently for approximately a 2-year period (2002–2005), so during this period data could appear on either system. A return visit was necessary to retrieve missing data from EndoScribe that was assumed to be on GIScribe |
| Pathology systems in use at the hospital: | Telepath (1987–Nov 2011) |
| Extraction dates: | Jan 1996–Sep 2009 |
| Methods used to extract the pathology data: | Used Telepath SIFT search for SNOP CODES (4-digit versions of SNOMED codes) |
| Missing pathology manually collected later: | Aug 1992–Nov 2011 |
| Extraction issues on pathology systems: | The study programme was trained by iSoft, which manufactures Telepath |
Leicester General Hospital, University Hospitals of Leicester NHS Trust
| Endoscopy systems in use at the hospital: | Unisoft (1998–present) |
| Extraction dates: | 1988–Apr 2008 |
| Extraction issues on endoscopy systems: | Unisoft created a bespoke query for the study programmer to extract the data from the endoscopy database. This was installed at the hospital using the Unisoft GI Auditor’s Kit |
| Pathology systems in use at the hospital: | iLaboratory (iLab) (previously called APEX), supported by iSOFT (1988–Dec 2009) |
| Extraction dates: | 1988–Apr 2008 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | May 1997–Dec 2009 |
| Extraction issues on pathology systems: | Data were extracted in the form of CSV files, one for each year from 1997 to 2007. The format of the CSV files was difficult to work with, as each line of the report was on a new row, so the study programmer wrote a program to format and extract the data |
New Cross Hospital, Royal Wolverhampton Hospitals NHS Trust
| Endoscopy systems in use at the hospital: | Micromed (1992–2007) |
| Extraction dates: | 1993–2007 |
| Extraction issues on endoscopy systems: | The system was used consistently from only 1993 onwards. Instructions for extracting data were obtained from the designer of the database, Medical Systems |
| Pathology systems in use at the hospital: | Radius (1988–1999) APEX (2000–Apr 2010) |
| Extraction dates: | 1993–Nov 2007 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | Jan 1986–Apr 2010 |
| Extraction issues on pathology systems: | APEX data were extracted in December 2007, but we were unable to extract free-text information and had to wait until the data were migrated to the Technidata system. Data were re-extracted from Technidata in May 2010 to complete the missing information. Technidata were still undergoing data cleaning so we recollected the data in April 2011, as some of the previously corrected records were corrupt |
University Hospital of North Tees, North Tees and Hartlepool NHS Foundation Trust
| Endoscopy systems in use at the hospital: | Micromed (1986–2006) Unisoft (2006–2008) |
| Extraction dates: | 1986–2006 |
| Extraction issues on endoscopy systems: | Instructions for extracting data from MicroMed were provided by the company that designed the system |
| Pathology systems in use at the hospital: | Clynisis and Pathlan LIMS (Jan 1997–Dec 2001) QuadraMed OmniLab LIMS (Mar 2004–Oct 2009) |
| Extraction dates: | 2004–2007 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | Dec 1996–Oct 2009 |
| Extraction issues on pathology systems: | The histology system moved from Hartlepool to North Tees in January 2002 and OmniLab was not used until March 2004. Between these dates, they used a William Woodard system at North Tees, but the system was no longer operational so data could not be bulk extracted from it. In August 2010 we recontacted the hospital and found that they had data on OmniLab from 2002 onwards. The data between 2002 and March 2004 were then manually collected by the study researchers. We were unable to match any of the Pathlan data to the endoscopy data. This centre had a large number of endoscopy reports with missing pathology, so the additional information was collected as part of the missing pathology data collection by the study researchers |
Queen Elizabeth Hospital, South London Healthcare NHS Trust
| Endoscopy systems in use at the hospital: | EndoScribe (1999–Jun 2006) ADAM (2006–present): not used |
| Extraction dates: | 1999–Jun 2006 |
| Extraction issues on endoscopy systems: | On the ADAM system, the body of the report with the details was not exported by default so a decision was made not to use these data. Data from EndoScribe were extracted using a global search for the word ‘polyp’ |
| Pathology systems in use at the hospital: | HBO (2000–2006) Clinysis (2006–Apr 2011) |
| Extraction dates: | 2000–2006 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | Nov 1999–Apr 2011 |
| Extraction issues on pathology systems: | Clinysis was the system in use at the time of data collection. We tried to extract the data using SNOMED codes but the extracted data were limited and not usable. It was therefore decided that only data from HBO would be used. Clinysis was used for the missing pathology data collection |
Queen Mary’s Hospital, South London Healthcare NHS Trust
| Endoscopy systems in use at the hospital: | Micromed (1985–Jan 2006) Unisoft (2003–Jul 2009) |
| Extraction dates: | Oct 1988–Jul 2009 |
| Extraction issues on endoscopy systems: | It was necessary to restore the old Micromed database from a back-up tape. The systems were used in parallel between 2003 and 2006, so duplicate records were merged |
| Pathology systems in use at the hospital: | Data prior to APEX had been migrated to APEX (1988–1994) APEX (1994–Jan 2011) |
| Extraction dates: | Jul 1987–Dec 2009 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | May 1988–Jan 2011 |
| Extraction issues on pathology systems: | An iSoft consultant was hired to extract data from the APEX system |
Royal Liverpool University Hospital, Royal Liverpool and Broadgreen University Hospitals NHS Trust
| Endoscopy systems in use at the hospital: | Bespoke system (1991–2006) Unisoft (2006–Oct 2009) |
| Extraction dates: | Jan 2000–Oct 2009 |
| Extraction issues on endoscopy systems: | Hospital staff set up the system to extract the data |
| Pathology systems in use at the hospital: | Telepath (1996–Nov 2010) |
| Extraction dates: | Jan 2000–Oct 2009 |
| Methods used to extract the pathology data: | SNOMED codes Version 2 |
| Missing pathology manually collected later: | May 1991–Nov 2010 |
| Extraction issues on pathology systems: | Hospital staff helped the study programmer to extract the data |
Royal Surrey County Hospital, Royal Surrey County Hospital NHS Foundation Trust
| Endoscopy systems in use at the hospital: | Micromed (1997–May 2010) |
| Extraction dates: | Sep 1997–May 2010 |
| Extraction issues on endoscopy systems: | Micromed was used from 1988 but the records up to 1996 were lost. Searches produced errors that caused the system to crash. Data extraction had to be performed by hospital staff in batches, each batch consisting of all the records for a 6-month period |
| Pathology systems in use at the hospital: | Bespoke System (1991–2001) that existed on Telepath Telepath (2002–Sep 2009) Clinisys WinPath (Oct 2009–Nov 2010) |
| Extraction dates: | 1997–Sep 2009 |
| Methods used to extract the pathology data: | Combination of branched searches and using wildcards to search on words such as adenoma’, ‘polyp’, ‘tumour’, colorectal sites, plus SNOMED codes version 2 |
| Missing pathology manually collected later: | Jun 1996–Nov 2010 |
| Extraction issues on pathology systems: | Data were only bulk extracted from the Telepath system at Royal Surrey, which had some years for which the data were not SNOMED coded. We therefore had to conduct separate searches on SNOMED codes, branded searches, and wild card searches |
St George’s Hospital, St George’s Healthcare NHS Trust
| Endoscopy systems in use at the hospital: | Micromed (1992–Jul 2009) |
| Extraction dates: | Feb 1992–Jul 2009 |
| Extraction issues on endoscopy systems: | Instructions for extracting data were obtained from the designer of the database, Medical Systems |
| Pathology systems in use at the hospital: | Other electronic system (1988–1993) iLaboratory (iLab) (previously called APEX), supported by iSOFT (1994–Jul 2009) |
| Extraction dates: | Aug 1997–Dec 2009 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | Feb 1992–July 2009 |
| Extraction issues on pathology systems: | In July 2009, the body of the pathology report could not be extracted. An iSoft consultant was hired to extract the data for us in January 2010 to complete the missing information |
Royal Shrewsbury Hospital, Shrewsbury and Telford Hospital NHS Trust
| Endoscopy systems in use at the hospital: | EndoScribe (2001–2009) |
| Extraction dates: | Nov 2001–Sep 2009 |
| Extraction issues on endoscopy systems: | The study programmer was familiar with the system |
| Pathology systems in use at the hospital: | Telepath (1992–2010) |
| Extraction dates: | 2000–Sept 2009 |
| Methods used to extract the pathology data: | SNOP CODES (four-digit versions of SNOMED codes) |
| Missing pathology manually collected later: | Jan 2002–Sep 2009 |
| Extraction issues on pathology systems: | The study programmer required training from iSoft to extract the data from Telepath. Pathology records were collected only as far back as the endoscopy data were documented |
St Mark’s Hospital, North West London Hospitals NHS Trust
| Endoscopy systems in use at the hospital: | Metabase (1972–2003) Endosoft (2004–present) |
| Extraction dates: | 1972–Jul 2007 |
| Extraction issues on endoscopy systems: | Following data extraction from Metabase, the computer on which Metabase was stored stopped working and is no longer accessible. Luckily the data that we required had been extracted before the computer stopped working |
| Pathology systems in use at the hospital: | Cerner Classic and City Road (1989–Oct 2002) Cerner LIMS (Oct 2002–May 2011) |
| Extraction dates: | Aug 1987–31 Oct 2006 |
| Methods used to extract the pathology data: | SNOMED codes version 2 |
| Missing pathology manually collected later: | Nov 1986–May 2011 |
| Extraction issues on pathology systems: | The records from the different systems were merged |
Torbay Hospital, South Devon Healthcare NHS Foundation Trust
| Endoscopy systems in use at the hospital: | EndoScribe (2000–2007) Scorpio (2007–present) |
| Extraction dates: | Oct 2000–Aug 2007 |
| Extraction issues on endoscopy systems: | Hospital staff helped with the extraction of data |
| Pathology systems in use at the hospital: | EDS (1998–2009) WinPath (2010–Oct 2010) |
| Extraction dates: | Jun 1999–Feb 2008 |
| Methods used to extract the pathology data: | Search on the words ‘polyp’ and ‘adenoma’ |
| Missing pathology manually collected later: | Jan 1998–Oct 2010 |
| Extraction issues on pathology systems: | Initially the search on the word ‘polyp’ did not pick up all relevant reports, so the study programmer revisited the hospital to extract reports using the search terms ‘polyp’ and ‘adenoma’ |
Yeovil District Hospital, Yeovil District Hospital NHS Foundation Trust
| Endoscopy systems in use at the hospital: | EndoScribe (1997–2008) |
| Extraction dates: | Feb 1997–May 2008 |
| Extraction issues on endoscopy systems: | Some technical issues encountered with special characters in the files making the data transformation process difficult |
| Pathology systems in use at the hospital: | PathoSys (1997–Sep 2010) |
| Extraction dates: | Jan 1998–Dec 2007 |
| Methods used to extract the pathology data: | Hospital used a combination of search by SNOMED version 3.5, which included anything coded as a colorectal polyp of any type, with any associated M codes that would include carcinoma |
| Missing pathology manually collected later: | Feb 1997–Sep 2010 |
| Extraction issues on pathology systems: | The pathology search was difficult as it was carried out over several months and involved a number of manipulations, first as a text file and then in Microsoft Excel® (Microsoft Corporation, Redmond, WA, USA). One out of the four pathology files extracted did not have hospital numbers, so the matching was done using name and date of birth |
Systematized Nomenclature of Medicine codes version 2
The following is a comprehensive list of SNOMED version 2 codes that were sent to hospital pathology departments in order to extract the data.
Yeovil District Hospital did not use SNOMED version 2, so the hospital staff used the SNOMED version 2 codes that we supplied and searched for the corresponding SNOMED version 3.5 codes.
Some hospitals searched for SNOP codes, which are the first four digits of the SNOMED version 2 codes that we supplied, without the initial T or M.
Codes for lesion site
| Code | Description |
|---|---|
| T-68010 | Rectal mucous membrane |
| T-68002 | Perineal flexure of rectum |
| T-68001 | Sacral flexure of rectum |
| T-68000 | Rectum, NOS |
| T-67995 | Descending colon and sigmoid colon, CS |
| T-67995 | Left hemicolon |
| T-67995 | Left colon |
| T-67990 | Sigmoido-sigmoidocolic, CS |
| T-67990 | Sigmoid colon and sigmoid colon, CS |
| T-67980 | Colo-sigmoidocolic, CS |
| T-67980 | Colon and sigmoid colon, CS |
| T-67970 | Caeco-sigmoidocolic, CS |
| T-67970 | Caecum and sigmoid colon, CS |
| T-67965 | Caecum and ascending colon, CS |
| T-67965 | Right hemicolon |
| T-67965 | Right colon |
| T-67960 | Colo-caecal, CS |
| T-67960 | Colon and caecum, CS |
| T-67950 | Colon and skin, CS |
| T-67940 | Caecum and abdominal wall, CS |
| T-67930 | Sigmoid colon and abdominal wall, CS |
| T-67920 | Colo-rectal, CS |
| T-67920 | Colon and rectum, CS |
| T-67910 | Colon and abdominal wall, CS |
| T-67900 | Colo-colic, CS |
| T-67900 | Colon and colon, CS |
| T-67860 | Pericolic tissue |
| T-67850 | Phrenicocolic ligament |
| T-67840 | Mesentery of sigmoid colon |
| T-67830 | Mesentery of descending colon |
| T-67820 | Transverse mesocolon |
| T-67810 | Mesentery of ascending colon |
| T-67800 | Mesocolon, NOS |
| T-67800 | Mesentery of colon, NOS |
| T-67700 | Sigmoid colon |
| T-67600 | Descending colon |
| T-67500 | Splenic flexure of colon |
| T-67500 | Left colic flexure |
| T-67400 | Transverse colon |
| T-67300 | Hepatic flexure of colon |
| T-67300 | Right colic flexure |
| T-67200 | Ascending colon |
| T-67120 | Frenulum of ileocaecal valve |
| T-67110 | Ileocaecal ostium |
| T-67100 | Caecum |
| T-67090 | Colonic serosa |
| T-67080 | Colonic subserosa |
| T-67073 | Tenia libera |
| T-67072 | Tenia omentalis |
| T-67071 | Tenia mesocolica |
| T-67070 | Tenia coli |
| T-67060 | Appendix epiploica |
| T-67050 | Colonic solitary lymphoid nodule |
| T-67045 | Haustra of colon |
| T-67045 | Colonic haustra |
| T-67042 | Colonic muscularis propria, circular layer |
| T-67041 | Colonic muscularis propria, longitudinal layer |
| T-67040 | Colonic muscularis propria |
| T-67030 | Colonic submucosa |
| T-67020 | Colonic crypt of Lieberkühn |
| T-67016 | Colonic lamina propria |
| T-67015 | Colonic epithelium |
| T-67012 | Colonic gland, NOS |
| T-67011 | Lamina muscularis of colonic mucous membrane |
| T-67010 | Colonic mucous membrane |
| T-67000 | Colon, NOS |
| T-67000 | Large bowel, NOS |
| T-67000 | Large intestine |
Codes for lesion type
| Code | Description |
|---|---|
| M-74008 | Dysplasia, severe |
| M-74007 | Dysplasia, moderate |
| M-74006 | Dysplasia, mild |
| M-74005 | Dysplasia, atypical |
| M-74003 | Severe dysplasia (morphological abnormality) |
| M-74002 | Moderate dysplasia (morphological abnormality) |
| M-74001 | Mild dysplasia (morphological abnormality) |
| M-74000 | Dysplasia (morphological abnormality) |
| M-72041 | Tubulovillous adenoma |
| M-72042 | Hyperplastic polyp (morphological abnormality) |
| M-72040 | Polypoid hyperplasia (morphological abnormality) |
| M-76800 | Polyp |
| M-76801 | Polyp, sessile |
| M-76802 | Polyp, pedunculated |
| M-76803 | Polyp, atypical |
| M-76804 | Polyp, ulcerated |
| M-76805 | Polyp, inflamed |
| M-76806 | Polyp, vascular |
| M-76807 | Polyp, hyalinised |
| M-76808 | Polyp, myxoid |
| M-76809 | Polyp, multiple |
| M-76810 | Polyp, fibroepithelial |
| M-76820 | Inflammatory polyp |
| M-7680A | Inflammatory polyp [dup] (morphological abnormality) |
| M-82633 | Adenocarcinoma in tubulovillous adenoma (morphological abnormality) |
| M-82632 | Adenocarcinoma in situ in tubulovillous adenoma (morphological abnormality) |
| M-82630 | Tubulovillous adenoma (morphological abnormality) |
| M-82623 | Villous adenocarcinoma (morphological abnormality) |
| M-82613 | Adenocarcinoma in villous adenoma (morphological abnormality) |
| M-82612 | Adenocarcinoma in situ in villous adenoma (morphological abnormality) |
| M-82611 | Villous adenoma, NOS |
| M-82610 | Villous adenoma (morphological abnormality) |
| M-82603 | Papillary adenocarcinoma (morphological abnormality) |
| M-82600 | Papillary adenoma (morphological abnormality) |
| M-82553 | Adenocarcinoma with mixed subtypes (morphological abnormality) |
| M-82453 | Adenocarcinoid tumour (morphological abnormality) |
| M-82451 | Tubular carcinoid (morphological abnormality) |
| M-82443 | Composite carcinoid (morphological abnormality) |
| M-82433 | Goblet cell carcinoid (morphological abnormality) |
| M-82423 | Enterochromaffin-like cell tumour, malignant (morphological abnormality) |
| M-82421 | Enterochromaffin-like cell carcinoid (morphological abnormality) |
| M-82413 | Enterochromaffin cell carcinoid (morphological abnormality) |
| M-82411 | Carcinoid tumour, argentaffin, NOS |
| M-82403 | Carcinoid tumour (except of appendix, M-82401) (morphological abnormality) |
| M-82401 | Carcinoid tumour of uncertain malignant potential (morphological abnormality) |
| M-82313 | Carcinoma simplex (morphological abnormality) |
| M-82303 | Solid carcinoma (morphological abnormality) |
| M-82302 | Ductal carcinoma in situ, solid type (morphological abnormality) |
| M-82213 | Adenocarcinoma in multiple adenomatous polyps (morphological abnormality) |
| M-82210 | Multiple adenomatous polyps (morphological abnormality) |
| M-82203 | Adenocarcinoma in adenomatous polyposis coli (morphological abnormality) |
| M-82200 | Adenomatous polyposis coli (morphological abnormality) |
| M-82153 | Adenocarcinoma of anal glands (morphological abnormality) |
| M-82143 | Parietal cell carcinoma (morphological abnormality) |
| M-82130 | Serrated adenoma (morphological abnormality) |
| M-82120 | Flat adenoma (morphological abnormality) |
| M-82113 | Tubular adenocarcinoma (morphological abnormality) |
| M-82110 | Tubular adenoma (morphological abnormality) |
| M-82103 | Adenocarcinoma in adenomatous polyp (morphological abnormality) |
| M-82102 | Adenocarcinoma in situ in adenomatous polyp (morphological abnormality) |
| M-82100 | Adenomatous polyp (morphological abnormality) |
| M-814FF | Adenoma AND/OR adenocarcinoma (morphological abnormality) |
| M-81490 | Canalicular adenoma (morphological abnormality) |
| M-81482 | Glandular intraepithelial neoplasia, grade III (morphological abnormality) |
| M-81473 | Basal cell adenocarcinoma (morphological abnormality) |
| M-81470 | Basal cell adenoma (morphological abnormality) |
| M-81460 | Monomorphic adenoma (morphological abnormality) |
| M-81453 | Carcinoma, diffuse type (morphological abnormality) |
| M-81443 | Adenocarcinoma, intestinal type (morphological abnormality) |
| M-81433 | Superficial spreading adenocarcinoma (morphological abnormality) |
| M-81423 | Linitis plastica (morphological abnormality) |
| M-81413 | Scirrhous adenocarcinoma (morphological abnormality) |
| M-81406 | Adenocarcinoma, metastatic (morphological abnormality) |
| M-81403 | Adenocarcinoma, no subtype (morphological abnormality) |
| M-81402 | Adenocarcinoma in situ (morphological abnormality) |
| M-81401 | Atypical adenoma (morphological abnormality) |
| M-81400 | Adenoma, no subtype (morphological abnormality) |
| M-80103 | Carcinoma, no subtype (morphological abnormality) |
| M-88500 | Lipoma, no ICD-O subtype (morphological abnormality) |
| M-88501 | Atypical lipoma (morphological abnormality) |
| M-95903 | Malignant lymphoma, no ICD-O subtype (morphological abnormality) |
| M-95906 | Malignant lymphoma, metastatic (morphological abnormality) |
| M-84800 | Mucinous adenoma (morphological abnormality) |
| M-84803 | Mucinous adenocarcinoma (morphological abnormality) |
| M-84903 | Signet ring cell carcinoma (morphological abnormality) |
| M-80203 | Carcinoma, undifferentiated (morphological abnormality) |
| M-80413 | Small cell carcinoma (morphological abnormality) |
| M-76880 | Lymphoid polyp (morphological abnormality) |
| M-75630 | Hamartomatous polyp (morphological abnormality) |
| M-75640 | Juvenile polyp (morphological abnormality) |
| M-44440 | Granulomatous polyp, a polyp showing granulomatous inflammation (morphological abnormality) |
Appendix 2 Study database, entity relationship diagram and data dictionary
Intermediate adenoma database: entity relationship diagram

Patients
Patients loaded to the IA database.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| STUDY_NUMBER | VARCHAR2(10) | PRIMARY KEY, unique identifier assigned to each patient, starts with hospital code | ||
| GENDER | VARCHAR2(1) | Y | Gender of patient | |
| DOB | DATE | Y | Date of birth of patient | |
| EXCLUDED | NUMBER(1) | Y | 0 | If set to 1 this patient has been excluded from the study |
| HOSPITAL | VARCHAR2(4) | Y | Hospital the patient attended. REFERENCE DATA:XHOSPITALS | |
| ANALYSED | NUMBER(1) | Y | 0 | Set by study researchers – if set to 1 it means the record has been fully analysed and coded |
| ASSIGNED_TO | VARCHAR2(10) | Y | The study researcher responsible for coding this record. REFERENCE DATA:CODERS (not listed in this document) | |
| COMMENTS | CLOB | Y | Study researchers’ comments | |
| EXCLUSION_REASON | NUMBER(2) | Y | Reason patient has been excluded. REFERENCE DATA:XEXCLUSION | |
| PROVISIONAL_EXCLUSION | NUMBER(1) | Y | 0 | Set to 1 if patient matches provisional exclusion criteria – no polyps, only one endoscopy examination, or no linked or unlinked pathology |
| ANALYSED_TIME | DATE | Y | Time when the record was analysed – set by trigger PATIENT_ANALYSE_TIME | |
| REVIEW | NUMBER(1) | Y | Redundant field – set to 1 if patient is being reviewed, set to 2 once review finished | |
| EXCLUDED_BY | VARCHAR2(1) | Y | A = Automatically excluded by program; C = Coder Excluded | |
| EXCLUDE_TIME | DATE | Y | Time when the record was excluded, set by trigger PATIENT_EXCLUDE_TIME | |
| ALL_VIEW | NUMBER(1) | Y | 0 | If set to 1, any study researcher can view this patient’s record in the EPR application |
| OLD_ASSIGNED_TO | VARCHAR2(100) | Y | Redundant field – stores the name of the study researcher who originally coded or queried the record, if the record is now assigned to a new study researcher | |
| REVIEW_PN | NUMBER(1) | Y | Redundant field – used to carry out quality checks on polyp numbering | |
| DEVELOPER_NOTES | VARCHAR2(500) | Y | Used by the developer to record notes specific to the record | |
| MASTER_STUDYNUMBER | VARCHAR2(10) | Y | The master study number of the patient after the patient record had been merged with other records | |
| DUPLICATE_MERGED_ON | DATE | Y | Date when a duplicate record was merged – only recorded for duplicate patients | |
| REASON_MERGED | VARCHAR2(2000) | Y | Reason why a record was merged | |
| CHECKED_DUPLICATE | VARCHAR2(1) | Y | Redundant field – used by study researchers to indicate that they had checked a merged record | |
| MERGE_CODER_COMMENTS | VARCHAR2(4000) | Y | Comments recorded by study researchers for merged patients | |
| FINAL_MASTER | VARCHAR2(20) | Y | The final study number of the patient after the patient record had been merged with duplicate records across all centres |
Primary key
| Name | Columns |
|---|---|
| PATIENT_PK | STUDY_NUMBER |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| PATIENT_EXCLUSION_REASON | EXCLUSION_REASON | XEXCLUSION | EXCLUSION_ID |
| PATIENT_FK_CODER | ASSIGNED_TO | CODERS | USERNAME |
| PATIENT_FK_HOSPITAL | HOSPITAL | XHOSPITALS | HOSPITAL_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| PATIENT_ANAYLSEDTIME | ANALYSED_TIME | Normal |
| PATIENT_ASSIGNEDTO | ASSIGNED_TO | Normal |
| PATIENT_PK | STUDY_NUMBER | Unique |
Patient conditions
Coded information to show whether a patient had a cancer or resection at first examination.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| PATIENT_CONDITIONS_ID | NUMBER(10) | Unique ID for the PATIENT_CONDITIONS table. This table stores information on conditions and other features about a patient | ||
| STUDY_NUMBER | VARCHAR2(10) | Unique study number for a patient | ||
| PATIENT_CONDITIONS_TYPE_ID | NUMBER(3) | Code for the patient condition |
Primary key
| Name | Columns |
|---|---|
| PATIENT_CONDITIONS_PK | PATIENT_CONDITIONS_ID |
Unique keys
| Name | Columns |
|---|---|
| PATIENT_CONDITIONS_UNIQUE | STUDY_NUMBER,PATIENT_CONDITIONS_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| PATIENT_CONDITIONS_FK_SN | STUDY_NUMBER | PATIENT | STUDY_NUMBER |
Indexes
| Name | Columns | Type |
|---|---|---|
| PATIENT_CONDITIONS_PK | PATIENT_CONDITIONS_ID | Unique |
| PATIENT_CONDITIONS_UNIQUE | STUDY_NUMBER,PATIENT_CONDITIONS_TYPE_ID | Unique |
Endoscopy
Endoscopy examinations undertaken by patient.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| ENDO_ID | VARCHAR2(13) | PRIMARY KEY, unique identifier assigned to each endoscopy record, Trigger – NEW_ENDO_ID, start with E(normal) or EP (phantom endoscopy), incorporates hospital code | ||
| STUDY_NUMBER | VARCHAR2(10) | Study number of the patient to whom the endoscopy examination belongs | ||
| PROCEDURE_DATE | DATE | Y | Date of procedure | |
| INDICATIONS | VARCHAR2(1000) | Y | Raw textual data – indications for examination | |
| SEGMENT_REACHED_OLD | VARCHAR2(1000) | Y | Raw textual data – segment of the colon reached | |
| COMPLICATIONS | VARCHAR2(4000) | Y | Raw textual data – complications encountered during examination | |
| BOWEL_PREP_OLD | VARCHAR2(1000) | Y | Raw textual data – bowel preparation details | |
| BIOPSY_TEXT | VARCHAR2(4000) | Y | Raw textual data – biopsies taken at examination | |
| DIAGNOSIS | VARCHAR2(1000) | Y | Raw textual data – diagnosis at examination | |
| DIAGNOSIS_REPORT | VARCHAR2(4000) | Y | Raw textual data – main report text for examination | |
| ADDITIONAL_DETAILS | VARCHAR2(4000) | Y | Raw textual data – additional endoscopy report details | |
| FURTHER_MANAGMENT | VARCHAR2(4000) | Y | Raw textual data – further management details | |
| DUPLICATE | NUMBER(1) | Y | 0 | Redundant field – set to 1 if a possible duplicate record |
| LINKED | NUMBER(1) | Y | Redundant field – set to 1 if examination is linked to a pathology report | |
| ENDOSCOPIST | VARCHAR2(100) | Y | Raw textual data – name of endoscopist who performed examination | |
| ENDOSCOPIST_COMMENTS | VARCHAR2(4000) | Y | Raw textual data – any additional comments from endoscopist | |
| PROCEDURE_TYPE_OLD | VARCHAR2(4000) | Y | Raw textual data – procedure type | |
| PHANTOM_PATH_ID | VARCHAR2(20) | Y | The path_id of the pathology record in cases where no procedure report was available. A blank endoscopy report was created and linked to the pathology record | |
| BOWEL_PREP | NUMBER(1) | Y | Coded bowel preparation value REFERENCE DATA:XBOWEL_PREP |
|
| PROCEDURE_TYPE | NUMBER(2) | Y | Coded field – procedure type REFERENCE DATA:XPROCEDURE |
|
| DISTANCE_REACHED | NUMBER(3) | Y | Coded field – distance reached by endoscope (in cm) | |
| SEGMENT_REACHED | NUMBER(2) | Y | Coded field – segment reached by endoscopy REFERENCE DATA:XBOWEL_SEGMENT |
|
| RESECTION | NUMBER(1) | Y | 0 | Coded field – set to 1 if the patient has a resection noted |
| CODED | NUMBER(1) | Y | 0 | Coded field – set to 1 by study researcher when they had completed coding and analysing the endoscopy record |
| BIOPSY_NON_POLYP | NUMBER(1) | Y | Coded field set by study researcher to indicate that a non-polyp biopsy was taken at the examination | |
| RESECTION_OLD | VARCHAR2(4000) | Y | Raw textual data – information on resection | |
| COMMENTS | VARCHAR2(4000) | Y | Study researcher’s comments | |
| QUERY_TIME | DATE | Y | Redundant field – time when a query was set, updated by the trigger ENDO_QUERY_TIME | |
| REQUERY | NUMBER(1) | Y | 0 | Redundant field – set to 1 if study researcher has come back to query and still cannot resolve it |
| REVIEW_NOTES | VARCHAR2(4000) | Y | Redundant field – old review notes field | |
| EXAM_NUMBER | VARCHAR2(3) | Y | Coded field – order of examinations on the same day. Only same-day examinations are numbered, and field used only if patient has no examinations without a date | |
| EXAM_NUMBER_UNKNOWN | VARCHAR2(3) | Y | Coded field – reason why study researcher was unable to allocate a ranking for the examination. If this is recorded, it may invalidate the ranking on EXAM_RANKING for this patient REFERENCE DATA: EXAM_NUMBER_UNKNOWN |
|
| REASON_EXAM_SAME_DAY | VARCHAR2(3) | Y | Coded field – reason why examination happened on the same day as another examination REFERENCE DATA:REASON_EXAM_SAME_DAY |
|
| EXAM_RANKING | VARCHAR2(3) | Y | Coded field – ranking of all examinations for the patient. Used where the patient has at least one examination without a date. This field overrides EXAM_NUMBER | |
| OPERATION_TYPE | VARCHAR2(2) | Y | Coded field – used for surgery to record operation type REFERENCE DATA:OPERATION_TYPE |
|
| RESECTION_SPECIMEN_LENGTH | VARCHAR2(200) | Y | Coded field – specimen length at resection (mm) | |
| PATH_MISS_10MM | NUMBER(1) | Y | Redundant coded field – pathology missing query suboption: ‘> 10 mm’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| PATH_MISS_SENTLAB | NUMBER(1) | Y | Redundant coded field – pathology missing query suboption: ‘sent to lab’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| PATH_MISS_BIOPSYTEXT | NUMBER(1) | Y | Redundant coded field – pathology missing query suboption: ‘in biopsy text’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| PATH_MISS_CANCERINDICATED | NUMBER(1) | Y | Redundant coded field – pathology missing query suboption: ‘cancer has been indicated’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_SUPREPORTMISS | NUMBER(1) | Y | Redundant coded field – general query suboption: ‘supplementary report missing’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_POSSCANCER | NUMBER(1) | Y | Redundant coded field – general query suboption: ‘possible cancer’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_WHENCANCER | NUMBER(1) | Y | Redundant coded field – general query suboption: ‘when was cancer?’ Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_WHENRESECT | NUMBER(1) | Y | Redundant coded field – general query suboption: ‘when was resection?’ Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_POLYPNUM | NUMBER(1) | Y | Redundant coded field – general query suboption: ‘polyp numbers’. Most queries were resolved and unresolved queries were recorded in the table NFEATURE | |
| GEN_TERMUNSURE | NUMBER(1) | Y | Redundant coded field – general query suboption, unsure of terminology. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| APPCODE_TRUNC | NUMBER(1) | Y | Redundant coded field – application coding error query suboption for truncated pathology report. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| APPCODE_BLANKPATH | NUMBER(1) | Y | Redundant coded field – application coding error query suboption for blank pathology report. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| DISCUSS_HNPCC | NUMBER(1) | Y | Redundant coded field – Discuss query suboption, hereditary non-polyposis CRC (HNPCC)? Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| DISCUSS_COLITIS | NUMBER(1) | Y | Redundant coded field – discuss query suboption, colitis? Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| DISCUSS_HOWCODE | NUMBER(1) | Y | Redundant coded field – discuss query suboption, how to code? Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXL_CANCER1ST | NUMBER(1) | Y | Redundant coded field – exclusion query suboption, cancer first examination. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXL_RESECTION1ST | NUMBER(1) | Y | Redundant coded field – exclusion query suboption, resection first examination. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXL_HNPCC | NUMBER(1) | Y | Redundant coded field – exclusion query suboption, HNPCC. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXL_POLYPOSIS | NUMBER(1) | Y | Redundant coded field – exclusion query suboption, polyposis. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXL_COLITIS | NUMBER(1) | Y | Redundant coded field – exclusion query suboption, colitis. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| EXAM_ORDER | NUMBER(3) | Y | Coded field – chronological ranking of examinations. Lowest number corresponds to earliest examination | |
| NOTES | VARCHAR2(4000) | Y | Study researcher or study programmer’s notes related to this record | |
| APPCODE_IRRELEVANT | NUMBER(1) | Y | Redundant coded field – application coding error query suboption used for irrelevant endoscopy. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| APPCODE_DUPLICATE | NUMBER(1) | Y | Redundant coded field – application coding error query suboption used for duplicate endoscopy. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| APPCODE_TRUNC_ENDOSCOPY | NUMBER(1) | Y | Redundant coded field – application coding error query suboption – used for truncated report. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| APPCODE_BLANK_ENDO | NUMBER(1) | Y | Redundant coded field – application coding error query suboption – used for blank endoscopy. Most queries were resolved and unresolved queries are recorded on table NFEATURE | |
| POLYP_MATCHING_QUERY | VARCHAR2(1) | Y | Coded field – used for marking cases where the endoscopy and pathology information does not link the polyps clearly | |
| OLD_STUDY_NUMBER | VARCHAR2(10) | Y | Used for merging records. This is the original study number associated with this endoscopy. It may be the same as field STUDY_NUMBER if the record has not been merged to another master record | |
| SOURCE_HOSPITAL | VARCHAR2(10) | Y | Hospital where the data were extracted from. It may be different to the hospital of the master study number if cross centre merging was done |
Primary key
| Name | Columns |
|---|---|
| ENDOSCOPY_PK | ENDO_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| ENDOSCOPY_BOWEL_PREP | BOWEL_PREP | XBOWEL_PREP | PREP_ID |
| ENDOSCOPY_FK | STUDY_NUMBER | PATIENT | STUDY_NUMBER |
| ENDOSCOPY_FK_QUERY | QUERY | XQUERY | QUERY_ID |
| ENDOSCOPY_PROCEDURE | PROCEDURE_TYPE | XPROCEDURE | PROCEDURE_ID |
| ENDOSCOPY_SEGMENT_REACHED | SEGMENT_REACHED | XBOWEL_SEGMENT | SEGMENT_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| ENDOSCOPY_PK | ENDO_ID | Unique |
| ENDOSCOPY_QUERYTIME | QUERY_TIME | Normal |
| ENDOSCOPY_STUDYNUMBER | STUDY_NUMBER | Normal |
Diagnosis
Coded diagnosis information for each endoscopy report.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| DIAGNOSIS_ID | NUMBER(10) | PRIMARY KEY, unique identifier assigned to each indication sub-type | ||
| ENDO_ID | VARCHAR2(13) | Endoscopy ID of report this diagnosis value is linked to | ||
| DIAGNOSIS_TYPE_ID | NUMBER(3) | Diagnosis type, REFERENCE DATA:XDIAGNOSIS_TYPES |
Primary key
| Name | Columns |
|---|---|
| DIAGNOSIS_PK | DIAGNOSIS_ID |
Unique keys
| Name | Columns |
|---|---|
| DIAGNOSIS_UNIQUE | ENDO_ID,DIAGNOSIS_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| DIAGNOSIS_FK_ENDO | ENDO_ID | ENDOSCOPY | ENDO_ID |
| DIAGNOSIS_FK_XDIAGTYPE | DIAGNOSIS_TYPE_ID | XDIAGNOSIS_TYPES | DIAGNOSIS_TYPE_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| DIAGNOSIS_ID_ENDOID | ENDO_ID | Normal |
| DIAGNOSIS_PK | DIAGNOSIS_ID | Unique |
| DIAGNOSIS_UNIQUE | ENDO_ID,DIAGNOSIS_TYPE_ID | Unique |
Diagnosis types
Coded diagnosis subtypes for each endoscopy report (e.g. colitis and polyposis subtypes).
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| ID | NUMBER(10) | PRIMARY KEY, unique identifier assigned to each diagnosis subtype | ||
| DIAGNOSIS_TYPE_ID | NUMBER(3) | Y | Diagnosis type for which this subtype had been defined, REFERENCE DATA:XDIAGNOSIS_TYPES | |
| ENDO_ID | VARCHAR2(13) | Y | Endoscopy ID of report to which this diagnosis value is linked | |
| DIAGNOSIS_SUB_TYPE_ID | NUMBER(3) | Y | Diagnosis sub-type, REFERENCE DATA:XDIAGNOSIS_SUB_TYPES |
Primary key
| Name | Columns |
|---|---|
| DIAGNOSIS_SUB_TYPE_PK | ID |
Unique keys
| Name | Columns |
|---|---|
| DIAGNOSIS_SUB_TYPE_UNIQUE | DIAGNOSIS_TYPE_ID,ENDO_ID,DIAGNOSIS_SUB_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| DIAGNOSIS_SUB_TYPE_FK_ENDO | ENDO_ID,DIAGNOSIS_TYPE_ID | DIAGNOSIS | ENDO_ID,DIAGNOSIS_TYPE_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| DIAGNOSIS_SUB_TYPE_PK | ID | Unique |
| DIAGNOSIS_SUB_TYPE_UNIQUE | DIAGNOSIS_TYPE_ID,ENDO_ID,DIAGNOSIS_SUB_TYPE_ID | Unique |
Indications
Coded indication information for each endoscopy report.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| INDICATION_ID | NUMBER(10) | PRIMARY KEY, unique identifier assigned to each indication | ||
| ENDO_ID | VARCHAR2(13) | Endoscopy ID of report to which this indication value is linked | ||
| INDICATION_TYPE_ID | NUMBER(3) | Indication Type. REFERENCE DATA:XINDICATION_TYPES |
Primary key
| Name | Columns |
|---|---|
| INDICATIONS_PK | INDICATION_ID |
Unique keys
| Name | Columns |
|---|---|
| INDICATIONS_UNIQUE | ENDO_ID,INDICATION_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| INDICATIONS_FK_ENDO | ENDO_ID | ENDOSCOPY | ENDO_ID |
| INDICATIONS_FK_XINDTYPE | INDICATION_TYPE_ID | XINDICATION_TYPES | INDICATION_TYPE_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| INDICATIONS_ID_ENDOID | ENDO_ID | Normal |
| INDICATIONS_PK | INDICATION_ID | Unique |
| INDICATIONS_UNIQUE | ENDO_ID,INDICATION_TYPE_ID | Unique |
Indication SUB_TYPES
Coded indication information for each endoscopy report.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| INDICATION_ID | NUMBER(10) | PRIMARY KEY, unique identifier assigned to each indication | ||
| ENDO_ID | VARCHAR2(13) | Endoscopy ID of report to which this indication value is linked | ||
| INDICATION_TYPE_ID | NUMBER(3) | Indication Type. REFERENCE DATA:XINDICATION_TYPES |
Primary key
| Name | Columns |
|---|---|
| INDICATIONS_PK | INDICATION_ID |
Unique keys
| Name | Columns |
|---|---|
| INDICATIONS_UNIQUE | ENDO_ID,INDICATION_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| INDICATIONS_FK_ENDO | ENDO_ID | ENDOSCOPY | ENDO_ID |
| INDICATIONS_FK_XINDTYPE | INDICATION_TYPE_ID | XINDICATION_TYPES | INDICATION_TYPE_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| INDICATIONS_ID_ENDOID | ENDO_ID | Normal |
| INDICATIONS_PK | INDICATION_ID | Unique |
| INDICATIONS_UNIQUE | ENDO_ID,INDICATION_TYPE_ID | Unique |
NFEATURE
Coded features for each endoscopy report – used to record queries, errors, medical conditions, and so on.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| NFEATURE_ID | NUMBER(10) | PRIMARY KEY, unique identifier assigned to each notable feature | ||
| ENDO_ID | VARCHAR2(13) | Endoscopy ID of report that this notable feature is linked to | ||
| NFEATURE_TYPE_ID | NUMBER(3) | Notable feature type, REFERENCE DATA:XNFEATURE_TYPES |
Primary key
| Name | Columns |
|---|---|
| NFEATURE_PK | NFEATURE_ID |
Unique keys
| Name | Columns |
|---|---|
| NFEATURE_UNIQUE | ENDO_ID,NFEATURE_TYPE_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| NFEATURE_FK_ENDO | ENDO_ID | ENDOSCOPY | ENDO_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| NFEATURE_ID_ENDOID | ENDO_ID | Normal |
| NFEATURE_PK | NFEATURE_ID | Unique |
| NFEATURE_UNIQUE | ENDO_ID,NFEATURE_TYPE_ID | Unique |
TABLE: PATHOLOGY
Columns
| Name | Type | Optional | Default | Comments | |
|---|---|---|---|---|---|
| PATH_ID | VARCHAR2(20) | PRIMARY KEY, unique pathology report ID | |||
| STUDY_NUMBER | VARCHAR2(10) | Y | Study number of patient this pathology report belongs to (can be blank if no endoscopy examination from that patient) | ||
| PATHOLOGIST | VARCHAR2(1000) | Y | Raw textual data – name of pathologist who carried out report | ||
| REPORT | CLOB | Y | Raw textual data – main body of pathology report | ||
| ADDITIONAL_REPORT | CLOB | Y | Raw textual data – additional pathology report information | ||
| MICROSCOPIC_DESCRIPTION | CLOB | Y | Raw textual data – microscopic description of biopsy specimen | ||
| CLINICAL_HISTORY | CLOB | Y | Raw textual data – clinical history of patient this specimen came from | ||
| SPECIMEN | CLOB | Y | Raw textual data – description of biopsy specimen (often called macroscopic report) | ||
| CONCLUSION | CLOB | Y | Raw textual data – conclusion of report | ||
| SPECIMEN_TYPE | VARCHAR2(4000) | Y | Raw textual data – type of specimen (e.g. polyp biopsy) | ||
| COMMENTS | VARCHAR2(4000) | Y | Raw textual data – pathologist comments | ||
| LOCATION | VARCHAR2(4000) | Y | Raw textual data – location the specimen came from (often name of hospital ward or department) | ||
| COLLECTION_DATE | DATE | Y | Coded field – collection date of biopsy specimen | ||
| RECEIVE_DATE | DATE | Y | Coded field – date biopsy specimen received at laboratory (may be different from collection data of specimen) | ||
| REPORT_DATE | DATE | Y | Coded field – date report written by pathologist | ||
| REQUESTED_DATE | DATE | Y | Coded field – date pathology was requested | ||
| NAME_MATCHED | NUMBER(1) | Y | 0 | Coded field – set to 1 if the pathology report was matched by name (and DOB) to an endoscopy patient (and not by hospital number which is more accurate) | |
| DUPLICATE | NUMBER(1) | Y | Coded field – set to 1 if possible duplicate pathology report | ||
| EXCLUDED | NUMBER(1) | Y | 0 | Coded field – reason pathology report has been excluded, REFERENCE DATA:XPATH_EXCLUSION | |
| HOSPITAL | VARCHAR2(4) | Coded field – hospital this report belongs to, REFERENCE DATA:XHOSPITALS | |||
| MATCHING_ERROR | NUMBER(1) | Y | Redundant field – set to 1 if possible error made when matching this pathology report to a endoscopy report | ||
| ENDO_ID | VARCHAR2(13) | Y | Linked field – endoscopy ID of endoscopy report this pathology report is linked to (report of biopsy taken from that endoscopy examination) | ||
| QUERY | NUMBER(2) | Y | Coded field – query study researcher had regarding pathology report, RFERENCE DATA:XPATH_QUERY (used with unlinked path report) | ||
| NORMAL_MUCOSA | NUMBER(1) | Y | Coded field – set to 1 if all pathology report states is normal mucosa found – used with unlinked path reports | ||
| MAN_COLLECTED_PATH | NUMBER | Y | Record collected manually by coders by visiting hospitals. REFERENCE DATA:MAN_COLLECTED_PATH | ||
| NOTES | VARCHAR2(4000) | Y | Study researcher or study programmer’s notes related to the pathology | ||
| PATHOLOGY_SOURCE | VARCHAR2(255) | Y | Recorded for some reports to show where the pathology data were extracted from | ||
| OLD_STUDY_NUMBER | VARCHAR2(10) | Y | Study number associated with this pathology report prior to merging. If it is not the same as STUDY_NUMBER then it was not merged or was the master record | ||
Primary key
| Name | Columns |
|---|---|
| PATHOLOGY_PK | PATH_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| PATHOLOGY_CON_EXCLUDE | EXCLUDED | XPATH_EXCLUSION | EXCLUSION_ID |
| PATHOLOGY_CON_HOSPITAL | HOSPITAL | XHOSPITALS | HOSPITAL_ID |
| PATHOLOGY_CON_QUERY | QUERY | XPATH_QUERY | QUERY_ID |
| PATHOLOGY_ENDOFK | ENDO_ID | ENDOSCOPY | ENDO_ID |
| PATHOLOGY_FK_PAT | STUDY_NUMBER | PATIENT | STUDY_NUMBER |
Indexes
| Name | Columns | Type |
|---|---|---|
| PATHOLOGY_ID_ENDOID | ENDO_ID | Normal |
| PATHOLOGY_PK | PATH_ID | Unique |
| PATHOLOGY_SN | STUDY_NUMBER | Normal |
POLYP
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| POLYP_ID | VARCHAR2(20) | PRIMARY KEY, unique identifier assigned to each polyp record. A trigger created value, starts with P (normal) or PP (Phantom polyp) also has code for hospital embedded Trigger – POLYP_T1 |
||
| STUDY_NUMBER | VARCHAR2(10) | Study number of patient to whom this polyp belongs | ||
| ENDO_ID | VARCHAR2(13) | Linked field – endoscopy ID of report to which this polyp is linked | ||
| ENDO_SEGMENT_OLD | VARCHAR2(1000) | Y | Raw textual data – location of the polyp within the colon as per the endoscopy report | |
| ENDO_SHAPE_OLD | VARCHAR2(50) | Y | Raw textual data – shape of the polyp as per the endoscopy report | |
| HOSPITAL | VARCHAR2(4) | Y | Hospital of patient REFERENCE DATA: XHOSPITALS |
|
| ENDO_DISTANCE | NUMBER(3) | Y | Coded field – distance polyp found into bowel (cm) | |
| ENDO_SIZE | NUMBER(4,1) | Y | Coded field – size of polyp (mm) | |
| ASSUME_ENDO_SIZE | NUMBER(4,1) | Y | Coded field – override the ENDO_SIZE by this size. The study researcher believes this is the most appropriate size | |
| ENDO_SIZE_OTHER | NUMBER(2) | Y | Coded field – other size of polyp (described in words rather than an actual size) REFERENCE DATA:XPOLYP_SIZE |
|
| ASSUME_ENDO_SIZE_OTHER | NUMBER(2) | Y | Coded field – override the ENDO_SIZE_OTHER by this size. The study researcher believes this is the most appropriate size | |
| ENDO_SIZE_MIN | NUMBER(4,1) | Y | Coded field – minimum size of polyp (mm) | |
| ENDO_SIZE_MAX | NUMBER(4,1) | Y | Coded field – maximum size of polyp (mm) | |
| ENDO_SHAPE | NUMBER(2) | Y | Coded field – shape of polyp from the endoscopy report REFERENCE DATA:XPOLYP_SHAPE |
|
| ENDO_SEGMENT | NUMBER(2) | Y | Coded field – location of the polyp within the colon as per the endoscopy report REFERENCE DATA:XBOWEL_SEGMENT |
|
| ENDO_SEGMENT_TO | NUMBER(2) | Y | Coded field – location of the polyp within the colon. Only used when a site range is given rather than a specific site and the range is recorded on fields ENDO_SEGMENT and ENDO_SEGMENT_TO REFERENCE DATA:XBOWEL_SEGMENT |
|
| CLINICAL_HISTOLOGY | VARCHAR2(3) | Y | Coded field – clinical histology for a polyp REFERENCE DATA:CLINICAL_HISTOLOGY |
|
| ENDO_QUANTITY_OTHER | NUMBER(2) | Y | Coded field – recorded when a group of polyps is recorded as an individual polyp row when the exact number of polyps is unknown. This is a description value of the quantity of polyps in the group REFERENCE DATA:XPOLYP_NUMBERS |
|
| MIN_QTY | NUMBER | Y | Coded field – minimum quantity of polyps. Only used when the polyp record represented a group of polyps | |
| MAX_QTY | NUMBER | Y | Coded field – maximum quantity of polyps. Only used when the polyp record represented a group of polyps | |
| APPROX_QTY | NUMBER | Y | Coded field – recorded where a group of polyps is recorded as an individual polyp row when the approximate number of polyps is unknown | |
| PHANTOM_PATH_ID | VARCHAR2(20) | Y | Linked field – the pathology report linked to this polyp where the polyp was created from a pathology report that has no corresponding endoscopy report | |
| REMOVAL_METHOD | NUMBER(2) | Y | Coded field – method of polyp removal REFERENCE DATA:XEXCISION_METHOD |
|
| EXCISION_COMPLETE | NUMBER(1) | Y | Coded field – whether or not the polyp was completely excise REFERENCE DATA:EXCISION_COMPLETE |
|
| PIECEMEAL_BIOPSY | NUMBER(1) | Y | Coded field – set to 1 if polyp was biopsied in a piecemeal fashion | |
| MAX_BIOPSY_SIZE | NUMBER(3) | Y | Coded field – maximum size given of biopsy by pathologist (mm) | |
| PATH_SIZE | NUMBER(4,1) | Y | Coded field – size of polyp (mm) from pathology report | |
| ASSUME_PATH_SIZE | NUMBER(4,1) | Y | Coded field – override the PATH_SIZE by this size. The study researcher believes this is the most appropriate size | |
| PATH_SHAPE | NUMBER(2) | Y | Coded field – shape of polyp from the pathology report REFERENCE DATA:XPOLYP_SHAPE |
|
| PATH_HISTOLOGY | NUMBER(3) | Y | Coded field – polyp type or classification REFERENCE DATA:XPOLYP_HISTOLOGY |
|
| ASSUME_PATH_HISTOLOGY | NUMBER(3) | Y | Coded field – used to correct histology on the individual polyp rows where the study researcher believed that the pathology information was erroneous or incorrect | |
| PATH_DYSPLASIA | NUMBER(2) | Y | Coded field – dysplasia of polyp REFERENCE DATA:XDYSPLASIA |
|
| PATH_COMMENTS | VARCHAR2(4000) | Y | Redundant field – comments about polyp from pathology report | |
| PATH_ADENOMA_TYPE | NUMBER(2) | Y | Coded field – morphology of adenomatous tissues REFERENCE DATA:XADENOMA_TYPE |
|
| FATE_OF_BIOPSY | NUMBER(2) | Y | Coded field – fate of polyp biopsy REFERENCE DATA:XBIOPSY_FATE |
|
| EXCISION_EXTENT | NUMBER(1) | Y | Coded field – extent of polyp excision REFERENCE DATA:XEXCISION_EXTENT |
|
| FRAGMENT_NO | NUMBER(3) | Y | Coded field – number of relevant fragments of adenoma or polyp found at pathology | |
| SERRATION | VARCHAR2(2) | Y | Coded field – if the polyp had serration features | |
| ENDO_PATH_MAPPING | VARCHAR2(3) | Y | Coded field – field used to record the rule used for matching pathology to polyp where it was not apparent | |
| PATH_MULTI_ENDO_LINK | VARCHAR2(16) | Y | Coded field – used to link an individual polyp to a group of polyps recorded at the same examination (i.e. the group will be recorded as an individual polyp row and will have a value in the ENDO_QUANTITY_OTHER field). Shows that the polyp is part of a group | |
| MULTIPLE_POLYP_GROUP | NUMBER(5) | Y | Coded field – used to classify group of polyps and individual polyps within the group seen at the same examination | |
| MULTIPLE_GROUP_LINKING | NUMBER(5) | Y | Coded field – used to allocate a unique group number for a set of polyps seen across many examinations REFERENCE DATA:MULTIPLE_GROUP_LINKING |
|
| MULTIPLEGROUP_MATCHPROB | NUMBER(5) | Y | Coded field – certainty that polyp has been matched correctly – recorded as a percentage REFERENCE DATA: POLYP_MATCH_PROB |
|
| POLYP_NUMBERED | VARCHAR2(1) | Y | Coded field -Y or N flag showing whether polyp has been numbered or not. REFERENCE DATA:POLYP_NUMBERED | |
| POLYP_NUMBER | NUMBER(3) | Y | Coded field – all occurrences of the individual polyps seen at different examinations were assigned the same number | |
| MATCH_PROBABILITY | NUMBER(3) | Y | Coded field – certainty that polyp has been matched correctly – recorded as a percentage REFERENCE DATA: POLYP_MATCH_PROB |
|
| DERIVED_POLYP_NUMBER | NUMBER(20) | Y | Derived field – each polyp allocated a unique number. If the polyp was sighted again that it was given the same DERIVED_POLYP_NUMBER | |
| DERIVED_ENDO_RANGE | VARCHAR2(2000) | Y | Derived field – the study programmer wrote a program to automatically derive an endoscopy size from the different size values given at endoscopy where possible | |
| DERIVED_ENDO_RANGE_GROUP | VARCHAR2(2000) | Y | Derived field – method used to derive DERIVED_ENDO_RANGE | |
| DERIVED_ENDO_SIZE | VARCHAR2(100) | Y | Derived field – the study programmer wrote a program to automatically derive an endoscopy size from the different size values given at endoscopy after the size values had been reviewed by the study researchers and corrections had been made | |
| DERIVED_ENDO_SIZE_SOURCE | VARCHAR2(400) | Y | Derived field – method used to derive DERIVED_ENDO_SIZE | |
| DERIVED_ENDO_SIZE_OTHER | NUMBER(2) | Y | Derived field – derived from the fields ASSUME_ENDO_SIZE_OTHER and ENDO_SIZE_OTHER with ASSUME_ENDO_SIZE_OTHER always taking precedence over ENDO_SIZE_OTHER | |
| DERIVED_ENDOSIZE_OTHER_SOURCE | VARCHAR2(4000) | Y | Derived field – which field the size was taken from (i.e. ASSUME_ENDO_SIZE_OTHER or ENDO_SIZE_OTHER) | |
| DERIVED_PATH_SIZE | NUMBER(4,1) | Y | Derived field – derived from the fields ASSUME_PATH_SIZE and PATH_SIZE (ASSUME_PATH_SIZE took precedence) | |
| DERIVED_PATH_SIZE_SOURCE | VARCHAR2(4000) | Y | Coded field – field from which the size DERIVED_PATH_SIZE was derived |
Primary key
| Name | Columns |
|---|---|
| POLYP_PK | POLYP_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| POLYP_BIOPSY | REMOVAL_METHOD | XEXCISION_METHOD | METHOD_ID |
| POLYP_DYSPLASIA | PATH_DYSPLASIA | XDYSPLASIA | DYSPLASIA_ID |
| POLYP_ENDO_SHAPE | ENDO_SHAPE | XPOLYP_SHAPE | POLYPSHAPE_ID |
| POLYP_ENDO_SIZE_OTHER | ENDO_SIZE_OTHER | XPOLYP_SIZE | POLYPSIZE_ID |
| POLYP_FK_ADENOMATYPE | PATH_ADENOMA_TYPE | XADENOMA_TYPE | ADENOMA_TYPE_ID |
| POLYP_FK_BIOPSYFATE | FATE_OF_BIOPSY | XBIOPSY_FATE | BIOPSY_FATE_ID |
| POLYP_FK_ENDO | ENDO_ID | ENDOSCOPY | ENDO_ID |
| POLYP_FK_EXCISIONEXTENT | EXCISION_EXTENT | XEXCISION_EXTENT | EXCISION_ID |
| POLYP_FK_HOSPITAL | HOSPITAL | XHOSPITALS | HOSPITAL_ID |
| POLYP_FK_PATEINT | STUDY_NUMBER | PATIENT | STUDY_NUMBER |
| POLYP_HISTOLOGY | PATH_HISTOLOGY | XPOLYP_HISTOLOGY | POLYPTYPE_ID |
| POLYP_PATH_SHAPE | PATH_SHAPE | XPOLYP_SHAPE | POLYPSHAPE_ID |
| POLYP_QUANTITY_OTHER | ENDO_QUANTITY_OTHER | XPOLYP_NUMBERS | POLYPNUMBERS_ID |
| POLYP_SEGMENT | ENDO_SEGMENT | XBOWEL_SEGMENT | SEGMENT_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| MP_POLYP_GROUP | MULTIPLE_POLYP_GROUP,1 | Normal |
| MP_POLYP_GROUP_LINKING | MULTIPLE_GROUP_LINKING,1 | Normal |
| POLYP_ID_ENDOID | ENDO_ID | Normal |
| POLYP_ID_STUDYNUMBER | STUDY_NUMBER | Normal |
| POLYP_PK | POLYP_ID | Unique |
DERIVED_MP_POLYPS
A copy of the POLYP table for all multiple polyp patients who had at least one endo quantity row. The endo quantity rows have been multiplied out on this table.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| POLYP_ID | VARCHAR2(20) | PRIMARY KEY, unique identifier assigned to each polyp record. A trigger created value, starts with P (normal) or PP (Phantom polyp) also has code for hospital embedded. Trigger – POLYP_T1 | ||
| STUDY_NUMBER | VARCHAR2(10) | Study number of patient to whom this polyp belongs | ||
| ENDO_ID | VARCHAR2(13) | Linked field – endoscopy ID of report to which this polyp is linked | ||
| ENDO_SEGMENT_OLD | VARCHAR2(1000) | Y | Raw textual data – location of the polyp within the colon as per the endoscopy report | |
| ENDO_SHAPE_OLD | VARCHAR2(50) | Y | Raw textual data – shape of the polyp as per the endoscopy report | |
| HOSPITAL | VARCHAR2(4) | Y | Hospital of patient REFERENCE DATA: XHOSPITALS |
|
| ENDO_DISTANCE | NUMBER(3) | Y | Coded field – distance polyp found into bowel (cm) | |
| ENDO_SIZE | NUMBER(4,1) | Y | Coded field – size of polyp (mm) | |
| ASSUME_ENDO_SIZE | NUMBER(4,1) | Y | Coded field – override the ENDO_SIZE by this size. The study researcher believes this is the most appropriate size | |
| ENDO_SIZE_OTHER | NUMBER(2) | Y | Coded field – other size of polyp (described in words rather than an actual size) REFERENCE DATA:XPOLYP_SIZE |
|
| ASSUME_ENDO_SIZE_OTHER | NUMBER(2) | Y | Coded field – override the ENDO_SIZE_OTHER by this size. The study researcher believes this is the most appropriate size | |
| ENDO_SIZE_MIN | NUMBER(4,1) | Y | Coded field – minimum size of polyp (mm) | |
| ENDO_SIZE_MAX | NUMBER(4,1) | Y | Coded field – maximum size of polyp (mm) | |
| ENDO_SHAPE | NUMBER(2) | Y | Coded field – shape of polyp from the endoscopy report REFERENCE DATA:XPOLYP_SHAPE |
|
| ENDO_SEGMENT | NUMBER(2) | Y | Coded field – location of the polyp within the colon as per the endoscopy report REFERENCE DATA:XBOWEL_SEGMENT |
|
| ENDO_SEGMENT_TO | NUMBER(2) | Y | Coded field – location of the polyp within the colon. Only used when a site range is given rather than a specific site and the range is recorded on fields ENDO_SEGMENT and ENDO_SEGMENT_TO REFERENCE DATA:XBOWEL_SEGMENT |
|
| CLINICAL_HISTOLOGY | VARCHAR2(3) | Y | Coded field – clinical histology for a polyp REFERENCE DATA:CLINICAL_HISTOLOGY |
|
| ENDO_QUANTITY_OTHER | NUMBER(2) | Y | Coded field – recorded where a group of polyps is recorded as an individual polyp row where the exact number of polyps is unknown. This is a description value of the quantity of polyps in the group REFERENCE DATA:XPOLYP_NUMBERS |
|
| MIN_QTY | NUMBER | Y | Coded field – minimum quantity of polyps. Only used when the polyp record represented a group of polyps | |
| MAX_QTY | NUMBER | Y | Coded field – maximum quantity of polyps. Only used when the polyp record represented a group of polyps | |
| APPROX_QTY | NUMBER | Y | Coded field – recorded where a group of polyps is recorded as an individual polyp row where the approximate number of polyps is unknown | |
| PHANTOM_PATH_ID | VARCHAR2(20) | Y | Linked field – the pathology report linked to this polyp where the polyp was created from a pathology report that has no corresponding endoscopy report | |
| REMOVAL_METHOD | NUMBER(2) | Y | Coded field – method of polyp removal REFERENCE DATA:XEXCISION_METHOD |
|
| EXCISION_COMPLETE | NUMBER(1) | Y | Coded field – whether or not the polyp was completely excise REFERENCE DATA:EXCISION_COMPLETE |
|
| PIECEMEAL_BIOPSY | NUMBER(1) | Y | Coded field – set to 1 if polyp was biopsied in a piecemeal fashion | |
| MAX_BIOPSY_SIZE | NUMBER(3) | Y | Coded field – maximum size given of biopsy by pathologist (mm) | |
| ENDO_COMMENTS | VARCHAR2(4000) | Y | Redundant field – study researcher comments about polyps from endoscopy examination | |
| PATH_SIZE | NUMBER(4,1) | Y | Coded field – size of polyp (mm) from pathology report | |
| ASSUME_PATH_SIZE | NUMBER(4,1) | Y | Coded field – override the PATH_SIZE by this size. The study researcher believes this is the most appropriate size | |
| PATH_SHAPE | NUMBER(2) | Y | Coded field – shape of polyp from the pathology report REFERENCE DATA:XPOLYP_SHAPE |
|
| PATH_HISTOLOGY | NUMBER(3) | Y | Coded field – polyp type or classification REFERENCE DATA:XPOLYP_HISTOLOGY |
|
| ASSUME_PATH_HISTOLOGY | NUMBER(3) | Y | Coded field – used to correct histology on the individual polyp rows where the study researcher believed that the pathology information was erroneous or incorrect | |
| PATH_DYSPLASIA | NUMBER(2) | Y | Coded field – dysplasia of polyp REFERENCE DATA:XDYSPLASIA |
|
| PATH_COMMENTS | VARCHAR2(4000) | Y | Redundant field – comments about polyp from pathology report | |
| PATH_ADENOMA_TYPE | NUMBER(2) | Y | Coded field – morphology of adenomatous tissues REFERENCE DATA:XADENOMA_TYPE |
|
| FATE_OF_BIOPSY | NUMBER(2) | Y | Coded field – fate of polyp biopsy REFERENCE DATA:XBIOPSY_FATE |
|
| EXCISION_EXTENT | NUMBER(1) | Y | Coded field – extent of polyp excision REFERENCE DATA:XEXCISION_EXTENT |
|
| FRAGMENT_NO | NUMBER(3) | Y | Coded field – number of relevant fragments of adenoma or polyp found at pathology | |
| SERRATION | VARCHAR2(2) | Y | Coded field – if the polyp had serration features | |
| ENDO_PATH_MAPPING | VARCHAR2(3) | Y | Coded field – field used to record the rule used for matching pathology to polyp when it was not apparent | |
| PATH_MULTI_ENDO_LINK | VARCHAR2(16) | Y | Coded field – used to link an individual polyp to a group of polyps recorded at the same examination (i.e. the group will be recorded as an individual polyp row and will have a value in the ENDO_QUANTITY_OTHER field). Shows that the polyp is part of a group | |
| MULTIPLE_POLYP_GROUP | NUMBER(5) | Y | Coded field – used to classify group of polyps and individual polyps within the group seen at the same examination | |
| MULTIPLE_GROUP_LINKING | NUMBER(5) | Y | Coded field – used to allocate a unique group number for a set of polyps seen across many examinations REFERENCE DATA:MULTIPLE_GROUP_LINKING |
|
| MULTIPLEGROUP_MATCHPROB | NUMBER(5) | Y | Coded field – certainty that polyp has been matched correctly – recorded as a percentage REFERENCE DATA: POLYP_MATCH_PROB |
|
| POLYP_NUMBERED | VARCHAR2(1) | Y | Coded field – Y or N flag showing whether polyp has been numbered or not REFERENCE DATA:POLYP_NUMBERED |
|
| POLYP_NUMBER | NUMBER(3) | Y | Coded field – all occurrences of the individual polyps seen at different examinations were assigned the same number | |
| NUMBERED_TIME | DATE | Y | Should not be used to find the date when the polyp was numbered. Gets updated if a polyp has a number when the polyp is updated on any screen | |
| MATCH_PROBABILITY | NUMBER(3) | Y | Coded field – certainty that polyp has been matched correctly – recorded as a percentage REFERENCE DATA: POLYP_MATCH_PROB |
|
| DERIVED_POLYP_NUMBER | NUMBER(20) | Y | Derived field – each polyp allocated a unique number. If the polyp was sighted again it was given the same DERIVED_POLYP_NUMBER | |
| DERIVED_ENDO_RANGE | VARCHAR2(2000) | Y | Derived field – the study programmer wrote a program to automatically derive an endoscopy size from the different size values given at endoscopy where possible | |
| DERIVED_ENDO_RANGE_GROUP | VARCHAR2(2000) | Y | Derived field – method used to derive DERIVED_ENDO_RANGE | |
| DERIVED_ENDO_SIZE | VARCHAR2(100) | Y | Derived field – the study programmer wrote a program to automatically derive an endoscopy size from the different size values given at endoscopy after the size values had been reviewed by the study researchers and corrections had been made | |
| DERIVED_ENDO_SIZE_SOURCE | VARCHAR2(400) | Y | Derived field – method used to derive DERIVED_ENDO_SIZE | |
| DERIVED_ENDO_SIZE_OTHER | NUMBER(2) | Y | Derived field – derived from the fields ASSUME_ENDO_SIZE_OTHER and ENDO_SIZE_OTHER with ASSUME_ENDO_SIZE_OTHER always taking precedence over ENDO_SIZE_OTHER | |
| DERIVED_ENDOSIZE_OTHER_SOURCE | VARCHAR2(4000) | Y | Derived field – which field the size was taken from (i.e. ASSUME_ENDO_SIZE_OTHER or ENDO_SIZE_OTHER) | |
| DERIVED_PATH_SIZE | NUMBER(4,1) | Y | Derived field – derived from the fields ASSUME_PATH_SIZE and PATH_SIZE (ASSUME_PATH_SIZE took precedence) | |
| DERIVED_PATH_SIZE_SOURCE | VARCHAR2(4000) | Y | Coded field – field from which the size DERIVED_PATH_SIZE was derived | |
| NEW_POLYP | NUMBER | Y | A unique number allocated to every new polyp created when an endo quantity row (multiple polyp row) was separated out into individual polyps. Shows which polyps were automatically generated | |
| NEW_POLYP_STATUS | VARCHAR2(30) | Y | Allocated automatically when an endo quantity row (multiple polyp row) was separated out into individual polyps. This value was used to indicate whether the row was created based on an existing polyp or new polyp |
Primary key
| Name | Columns |
|---|---|
| DERIVED_MP_POLYPS_PK | POLYP_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| DERIVED_MP_POLYPS_FK | ENDO_ID | ENDOSCOPY | ENDO_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| DERIVED_MP_POLYPS_ID_ENDOID | ENDO_ID | Normal |
| DERIVED_MP_POLYPS_PK | POLYP_ID | Unique |
| DERIVED_MP_POLYPS_STUDYNUMBER | STUDY_NUMBER | Normal |
DERIVED_MP_SUMMARY
A table derived using a program to summarise the values of variables used in estimating the number of polyps within an endo quantity row.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| STUDY_NUMBER | VARCHAR2(10) | Study number of the patient | ||
| POLYP_ID | VARCHAR2(20) | Polyp ID of an endo quantity row (group of polyps) | ||
| POLYP_GROUP | NUMBER(5) | Y | Coded field – used to classify group of polyps and individual polyps within the group seen at the same examination | |
| GROUP_LINKING | NUMBER(5) | Y | Coded field – used to allocate a unique group number for a set of polyps seen across many examinations REFERENCE DATA:MULTIPLE_GROUP_LINKING |
|
| TOTALPOLYPS | VARCHAR2(4000) | Y | Derived field – the total number of unique polyps seen for the endo quantity row across all examinations. It was derived by taking into account number of multilinked polyps, the number of unique polyps with the same MULTIPLE_GROUP_LINKING seen at other examinations and deducting any polyps with the same MULTIPLE_GROUP_LINKING that had been excised prior to the procedure date of the endo quantity row | |
| SAMEEXAMPOLYPS | VARCHAR2(4000) | Y | Derived field – the total number of multilinked polyps at the same examination for the endo quantity row | |
| POLYPCOUNTEXAM | VARCHAR2(4000) | Y | Derived field – this was the total number of polyps observed at the same examination as the endo quantity row | |
| POLYPCOUNTGROUP | VARCHAR2(4000) | Y | Derived field –this was the total number of polyps seen at the same examination where the MULTIPLE_POLYP_GROUP or MULTIPLE_GROUP_LINKING of the endo quantity row matched the individual and multilinked polyps | |
| POLYPCOUNTGROUP_OTHERS | VARCHAR2(4000) | Y | Derived field – this was the total number of multilinked polyps seen at the same examination where the MULTIPLE_POLYP_GROUP and MULTIPLE_GROUP_LINKING of the endo quantity row and polyps match | |
| ESTIMATED_QUANTITY | NUMBER | Y | Derived field – ENDO_QUANTITY_OTHER field translated into actual values. The following quantities were assigned to each option: few – 3, some – 3, num of – 3, several – 3, many – 5, multiple – 5 | |
| MIN_QTY | NUMBER | Y | Coded field – minimum quantity of polyps in the group recorded by the study researcher | |
| MAX_QTY | NUMBER | Y | Coded field – maximum quantity of polyps in the group recorded by the study researcher | |
| APPROX_QTY | NUMBER | Y | Coded field – approximate quantity of polyps recorded by the study researcher | |
| EXCISION_EXTENT | NUMBER(1) | Y | Coded field – extent of polyp excision REFERENCE DATA:XEXCISION_EXTENT |
|
| MAX_ROWS | VARCHAR2(4000) | Y | Derived field – this was the total number of polyps that should be multilinked to the ENDO QUANTITY ROW. The total included the existing multi linked rows and any other rows that made up the group of the endo quantity row. A number of rules were used to derive this | |
| MAX_ROWS_DATASOURCE | VARCHAR2(4000) | Y | Derived field – records the name of the field used to obtain the MAX_ROWS for this endo quantity row | |
| ROWSADD | VARCHAR2(4000) | Y | Derived field – total number of polyps that would be added when the endo quantity row was multiplied out, and included the number of new polyps that are created from existing polyps observed at other examinations, and also completely new polyps that were not observed but guesstimated based on the information available | |
| ROWSADD_OVERRIDE | VARCHAR2(4000) | Y | Derived field – only applied for cases where there were more than one endo quantity rows at the same examination with the same MULTIPLE_GROUP_LINKING. If the MAX_ROWS was taken from TOTAL_POLYPS for the more than one endo quantity rows with the same MULTIPLE_GROUP_LINKING at the same examination, then it was important to ensure that a ratio was used to divide the TOTAL_POLYPS among those endo quantity rows after deducting any multilinked polyps. The program calculated the TOTAL_POLYPS minus POLYPCOUNTGROUP and then divided that based on ESTIMATED_QUANTITY |
Primary key
| Name | Columns |
|---|---|
| POLYPID_PK | POLYP_ID |
Unique keys
| Name | Columns |
|---|---|
| DERIVED_MP_SUMMARY_UNIQUE | STUDY_NUMBER,POLYP_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| DERIVED_MP_SUMMARY_FK | POLYP_ID | POLYP | POLYP_ID |
Indexes
| Name | Columns | Type |
|---|---|---|
| DERIVED_MP_SUMMARY_UNIQUE | STUDY_NUMBER,POLYP_ID | Unique |
| POLYPID_PK | POLYP_ID | Unique |
ALLCANCERS_OTHERSOURCES
Cancers from external sources and their mapping to the endoscopy and polyp tables.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| OURCANCERID | VARCHAR2(200) | Our unique ID for each cancer | ||
| STUDY_NUMBER | VARCHAR2(10) | Y | Unique study number | |
| TABLERECORDID | NUMBER | Y | ID of the cancer from the source table | |
| DATA_SOURCE | VARCHAR2(200) | Y | Data source to say which table the data come from | |
| LOAD_DATE | DATE | Y | When data were loaded | |
| EVENT_TYPE | VARCHAR2(30) | Y | Event type as supplied by the HSCIC/NHSCR | |
| LAT_DOB_DATE | DATE | Y | Latest date of birth as supplied by the HSCIC/NHSCR | |
| LAT_GENDER | VARCHAR2(10) | Y | Latest gender as supplied by the HSCIC/NHSCR | |
| EVENT_DATE | DATE | Y | Cancer diagnosis date as supplied by the HSCIC/NHSCR | |
| DEATH_REG_NO_CASITE | VARCHAR2(100) | Y | Cancer site as supplied by the HSCIC/NHSCR | |
| SITEICD9 | VARCHAR2(100) | Y | Cancer site as supplied by NSS | |
| SITEICD10 | VARCHAR2(100) | Y | Cancer site as supplied by the NSS | |
| MORPHOLOGYTYPE | VARCHAR2(100) | Y | Cancer morphology code supplied by the HSCIC/NHSCR | |
| TYPEICDO | VARCHAR2(100) | Y | Cancer morphology code supplied by the NSS | |
| TYPEICDO2 | VARCHAR2(100) | Y | Cancer morphology code supplied by the NSS | |
| TYPEICDO3 | VARCHAR2(100) | Y | Cancer morphology code supplied by the NSS | |
| LAST_UPLOADED | DATE | Y | When we received the data | |
| BASELINE_DETAILS | VARCHAR2(4000) | Y | Baseline placement of the examination using baseline dates supplied by statistician | |
| ON_IA_BASELINE | VARCHAR2(1) | Y | Whether the cancer is part of the adenoma cohort | |
| NEAREST_ENDO_ID | VARCHAR2(500) | Y | Derived automatched ENDO_ID nearest to the cancer | |
| NEAREST_POLYP_ID | VARCHAR2(500) | Y | Derived automatched POLYP_ID nearest to the cancer within the endoscopy corresponding to the NEAREST_ENDO_ID | |
| PROXIM_NEAREST_POLYP_ID | VARCHAR2(4000) | Y | Derived proximity of the NEAREST_POLYP_ID using our bowel segment | |
| DERIVED_COLORECTAL_BYSITE | VARCHAR2(4000) | Y | Derived classification of cancer groups | |
| NEAREST_CANCER_ENDO_ID | VARCHAR2(500) | Y | Derived automatched ENDO_ID when cancer was rematched using different rules for the second time | |
| NEAREST_CANCER_POLYP_ID | VARCHAR2(500) | Y | Derived automatched POLYP_ID when cancer was rematched using different rules for the second time | |
| SOURCE_NEAREST_CANCER_ENDO_ID | VARCHAR2(2000) | Y | Derived reason on why cancer was rematched second time to an endoscopy | |
| PROXIM_NEAREST_CAN_POLYP_ID | VARCHAR2(4000) | Y | Derived proximity of the NEAREST_CANCER_POLYP_ID using our bowel segment | |
| DERIVED_PROXIMITY | NUMBER(3) | Y | Derived proximity of cancer calculated using our bowel segment | |
| CODER_ENDO_ID | VARCHAR2(4000) | Y | Study researcher manually reviewed record and mapped cancer to this ENDO_ID | |
| CODER_POLYP_ID | VARCHAR2(4000) | Y | Study researcher manually reviewed record and mapped cancer to this POLYP_ID | |
| CANCER_MATCHING | NUMBER(3) | Y | Study researcher manually reviewed the record and classified their findings into one of these categories | |
| CANCERSITE_DESC | VARCHAR2(4000) | Y | Used for the EPR screens to display the cancer site description for the study researcher | |
| MORPHOLOGY_DESC | VARCHAR2(4000) | Y | Used for the EPR screens to display the cancer type description or the study researcher | |
| DERIVED_ENDO_ID | VARCHAR2(50) | Y | Derived from NEAREST_ENDO_ID; NEAREST_CANCER_ENDO_ID AND CODER_ENDO_ID | |
| DERIVED_POLYP_ID | VARCHAR2(50) | Y | Derived from NEAREST_POLYP_ID; NEAREST_CANCER_POLYP_ID AND CODER_POLYP_ID | |
| DERIVED_METHOD | VARCHAR2(500) | Y | Methods use to obtain the DERIVED_ENDO__ID AND DERIVED_POLYP_ID | |
| DERIVED_EXCLUDE_CANCER | VARCHAR2(5) | Y | Derived field – cancers were excluded based on the study researcher’s cancer matching classification, such as duplicate or not CRC | |
| DEVELOPER_NOTES | VARCHAR2(4000) | Y | Additional information about the data added by the study programmer | |
| SITE_CODING_SYSTEM | VARCHAR2(15) | Y | Derived field – ICD coding system for the derived cancer site | |
| TYPE_CODING_SYSTEM | VARCHAR2(15) | Y | Derived field – ICD coding system for the derived morphology code | |
| DERIVED_SITEGROUPING | VARCHAR2(4000) | Y | Derived field – the cancer site grouping | |
| DERIVED_MORPHGROUPING | VARCHAR2(4000) | Y | The morphology code grouping | |
| DERIVED_SITE_MORPH_DECISION | VARCHAR2(4000) | Y | Derived field – cancer outcome derived using combinations of cancer site and morphology groupings for the HSCIC/NSS/NHSCR cancer | |
| DERIVED_FINAL_POLYP_ID | VARCHAR2(50) | Y | Derived field – this is the final mapping of our polyp to the HSCIC/NSS/NHSCR cancer for the IA study derived using various rules. It is populated only when the cancer is an outcome and not excluded | |
| DERIVED_FINAL_POLYP_SOURCE | VARCHAR2(500) | Y | Derived field – provides the rule or method used to get the DERIVED_FINAL_POLYP_ID | |
| DERIVED_TRUE_CANCERDATE_SOURCE | VARCHAR2(2000) | Y | Derived field – cancer date that should be used for analysis – HSCIC/NSS/NHSCR one or our mapped polyp data | |
| DERIVED_TRUE_SITE_SOURCE | VARCHAR2(2000) | Y | Derived field – cancer site that should be used for analysis – HSCIC/NSS/NHSCR one or our mapped polyp data | |
| DERIVED_TRUE_MORPHOLOGY_SOURCE | VARCHAR2(2000) | Y | Derived field – cancer morphology that should be used for analysis – HSCIC/NSS/NHSCR one or our mapped polyp data | |
| DERIVED_FINAL_ENDO_ID | VARCHAR2(50) | Y | Derived field – this is the final mapping of our endoscopy to the HSCIC/NSS/NHSCR cancer for the IA study derived using various rules. Its only populated where the cancer is an outcome and not excluded | |
| DERIVED_FINAL_ENDO_ID_SOURCE | VARCHAR2(500) | Y | Derived field – provides the rule or method used to get the DERIVED_FINAL_ENDO_ID | |
| DERIVED_FINAL_OUTCOME | VARCHAR2(500) | Y | Derived field – cancer outcome derived using combinations of cancer site and morphology groupings and our polyp data | |
| DERIVED_FINAL_OUTCOME_SOURCE | VARCHAR2(500) | Y | Derived field – where the final outcome was derived from – HSCIC/NSS/NHSCR one or our mapped polyp data | |
| DERIVED_DEATHS_OUTCOME | VARCHAR2(500) | Y | Derived field – populated where the cancer is ascertained from a death report and information about the cancer classification | |
| DERIVED_DEATHS_OUTCOME_REASON | VARCHAR2(500) | Y | Derived field – populated where the cancer is ascertained from a death report and the reason why this death was included as a cancer outcome | |
| DERIVED_SITE_CODE | VARCHAR2(20) | Y | Derived field – final cancer site code for this cancer derived from DEATH_REG_NO_CASITE; SITEICD10 AND SITEICD9 | |
| DERIVED_MORPHOLOGY_CODE | VARCHAR2(20) | Y | Derived field – final morphology code for this cancer derived from: MORPHOLOGYTYPE; TYPEICDO3; TYPEICDO2; and TYPEICDO |
Primary key
| Name | Columns |
|---|---|
| OURCANCERID_PK | OURCANCERID |
Unique keys
| Name | Columns |
|---|---|
| ALLCANCERS_OTHERSOURCES_UNIQUE | STUDY_NUMBER,OURCANCERID |
Indexes
| Name | Columns | Type |
|---|---|---|
| ALLCANCERS_OTHERSOURCES_UNIQUE | STUDY_NUMBER,OURCANCERID | Unique |
| OURCANCERID_PK | OURCANCERID | Unique |
DATA_CLEANING_GROUPS
The reference table describing the detailed action for all incidents (data-cleaning tasks assigned to study researchers) after the manual coding had been done. The actual records reviewed are on table REVIEW_DATA_CLEANING.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| INCIDENT | VARCHAR2(50) | The incident number of the task being reviewed | ||
| REVIEW_GROUP | VARCHAR2(50) | The review group is a group within the same incident with slightly different criteria for review | ||
| ACTION | VARCHAR2(4000) | Y | Action describing what needs to be reviewed | |
| DEVELOPER_COMMENT | CLOB | Y | Study programmer comments |
Primary key
| Name | Columns |
|---|---|
| DATA_CLEANING_GROUPS_PK | INCIDENT,REVIEW_GROUP |
Indexes
| Name | Columns | Type |
|---|---|---|
| DATA_CLEANING_GROUPS_PK | INCIDENT,REVIEW_GROUP | Unique |
REVIEW_DATA_CLEANING
Records reviewed by study researchers on the EPR application in relation to specific data cleaning tasks. The task was known as the incident. The details of the task can be obtained by linking this table to table DATA_CLEANING_GROUPS.
Columns
| Name | Type | Optional | Default | Comments |
|---|---|---|---|---|
| CLEANING_ID | VARCHAR2(100) | Unique ID for the cleaning task | ||
| TABLE_NAME | VARCHAR2(50) | The table containing the unique identifier of the record being reviewed | ||
| STUDY_NUMBER | VARCHAR2(10) | The patient unique study number | ||
| UNIQUE_COLUMN_NAME | VARCHAR2(50) | The column within the table contained in TABLE_NAME which has the unique identifier of the record being reviewed | ||
| UNIQUE_RECORD_ID | VARCHAR2(50) | The unique identifier of the record being reviewed within table contained in TABLE_NAME and column contained in UNIQUE_COLUMN_NAME | ||
| INCIDENT | VARCHAR2(50) | Y | The incident number (task number). This can be linked to the DATA_CLEANING_GROUPS table to get full description of task | |
| REVIEW_GROUP | VARCHAR2(50) | Y | Relates to the REVIEW_GROUP on the DATA_CLEANING_GROUPS | |
| CLEANING_DESC | VARCHAR2(4000) | Y | Additional cleaning instructions specific to this record | |
| CHECK_COMPLETE | VARCHAR2(1) | Y | Y means that the record has been reviewed for this task. Blank or N means not reviewed | |
| REVIEW_METHOD | VARCHAR2(30) | Some records are reviewed via spreadsheets and uploaded by the database manager and others are reviewed via the screens – APEX | ||
| REVIEW_COMMENTS | VARCHAR2(4000) | Y | Other comments from the study programmer | |
| APP_USER_LAST_UPD_USER | VARCHAR2(60) | Y | Study researcher who reviewed the record | |
| APP_USER_LAST_UPD_DATE | DATE | Y | Date when the record was the record was reviewed by the study researcher | |
| RESTR_APP_USER | VARCHAR2(20) | Y | Temporary access given to this study researcher for review. The patient record would be assigned to a different study researcher | |
| POLYP_ID | VARCHAR2(20) | Y | The polyp ID is provided where the review is specific to a polyp | |
| PATH_ID | VARCHAR2(20) | Y | Pathology ID when the record being reviewed is linked to pathology | |
| PHANTOM_PATH_ID | VARCHAR2(20) | Y | Pathology ID when the record being reviewed is linked to pathology and where no endoscopy report was available | |
| CLEANINGNOTES | VARCHAR2(4000) | Y | Study researcher’s notes | |
| CANCER_ID | VARCHAR2(400) | Y | Recorded for some cases where a cancer record was being reviewed |
Primary key
| Name | Columns |
|---|---|
| REVIEW_DATA_CLEANING_PK | CLEANING_ID |
Foreign keys
| Name | Columns | Referencing table | Columns |
|---|---|---|---|
| REVIEW_DATA_CLEANING_FK_SN | STUDY_NUMBER | PATIENT | STUDY_NUMBER |
| REVIEW_DATA_CLEANING_REF | INCIDENT,REVIEW_GROUP | DATA_CLEANING_GROUPS | INCIDENT,REVIEW_GROUP |
Indexes
| Name | Columns | Type |
|---|---|---|
| REVIEW_DATA_CLEANING_PK | CLEANING_ID | Unique |
Reference data
| Reference data domain | Code | Description | Definition (populated only if further explanation of field is required) |
|---|---|---|---|
| CANCER_MATCHING | 1 | Matched correctly | |
| CANCER_MATCHING | 2 | Not matched correctly | |
| CANCER_MATCHING | 3 | Not matched correctly – missing polyp | |
| CANCER_MATCHING | 4 | Not matched correctly – missing examination | |
| CANCER_MATCHING | 5 | New polyp created | |
| CANCER_MATCHING | 6 | Not a CRC | |
| CANCER_MATCHING | 7 | Looks like NHSIC/NHSCR have a duplicate cancer | |
| CANCER_MATCHING | 8 | Matched by coder | |
| CANCER_MATCHING | 9 | Coder confirmed cancer is prior to first examination recorded on database | |
| CANCER_MATCHING | 10 | Coder confirmed cancer is after last examination recorded on the database | |
| CANCER_MATCHING | 11 | HSCIC/NHSCR should have classified this as a CRC | |
| CANCER_MATCHING | 12 | No indication of cancer on our records | |
| CANCER_MATCHING | 13 | Exclude this case as segment appears incorrect. (Only use when data source is deaths) | |
| CANCER_MATCHING | 14 | This is an in situ that cannot be mapped to our polyp | |
| CLINICAL_HISTOLOGY | 1 | Adenoma | |
| CLINICAL_HISTOLOGY | 2 | Metaplastic/hyperplastic | |
| CLINICAL_HISTOLOGY | 3 | Serrated adenoma | |
| CLINICAL_HISTOLOGY | 4 | Leiomyoma | |
| CLINICAL_HISTOLOGY | 5 | Inflammatory | |
| CLINICAL_HISTOLOGY | 6 | Normal mucosa | |
| CLINICAL_HISTOLOGY | 8 | Carcinoid/neuroendocrine tumour | |
| CLINICAL_HISTOLOGY | 10 | Juvenile polyp | |
| CLINICAL_HISTOLOGY | 11 | Mucosal prolapse | |
| CLINICAL_HISTOLOGY | 14 | Ulcer | |
| CLINICAL_HISTOLOGY | 15 | Inflammation | |
| CLINICAL_HISTOLOGY | 16 | Melanosis coli | |
| CLINICAL_HISTOLOGY | 17 | Submucosal haematoma | |
| CLINICAL_HISTOLOGY | 19 | Angiodysplasia | |
| CLINICAL_HISTOLOGY | 20 | Ischaemia | |
| CLINICAL_HISTOLOGY | 21 | Xanthoma | |
| CLINICAL_HISTOLOGY | 22 | Oedema | |
| CLINICAL_HISTOLOGY | 23 | Regenerative polyp | |
| CLINICAL_HISTOLOGY | 24 | Hamartomatous polyp | |
| CLINICAL_HISTOLOGY | 25 | Haemangioma | |
| CLINICAL_HISTOLOGY | 26 | Non-Hodgkin’s lymphoma | |
| CLINICAL_HISTOLOGY | 27 | Fibroepithelial polyp | |
| CLINICAL_HISTOLOGY | 28 | Crohn’s disease | |
| CLINICAL_HISTOLOGY | 29 | Neurofibromatosis | |
| CLINICAL_HISTOLOGY | 30 | Colitis | |
| CLINICAL_HISTOLOGY | 31 | Lipoma | |
| CLINICAL_HISTOLOGY | 32 | Pseudolipomatus | |
| CLINICAL_HISTOLOGY | 33 | Spirochaetosis | |
| CLINICAL_HISTOLOGY | 34 | Granulation tissue | |
| CLINICAL_HISTOLOGY | 35 | Gastric heterotopia | |
| CLINICAL_HISTOLOGY | 36 | Cap polyp | |
| CLINICAL_HISTOLOGY | 37 | Lymphoid polyp | |
| CLINICAL_HISTOLOGY | 39 | Previous polypectomy site | |
| CLINICAL_HISTOLOGY | 40 | Ganglioneuromatosis | |
| CLINICAL_HISTOLOGY | 41 | Amyloid | |
| CLINICAL_HISTOLOGY | 43 | Congestion | |
| CLINICAL_HISTOLOGY | 44 | Lymphangiectasia | |
| CLINICAL_HISTOLOGY | 45 | Proctitis | |
| CLINICAL_HISTOLOGY | 50 | Cancer | |
| CLINICAL_HISTOLOGY | 51 | Cancer + adenoma | |
| CLINICAL_HISTOLOGY | 52 | Cancer in dispute | |
| CLINICAL_HISTOLOGY | 53 | Mixed adenoma/metastases | |
| CLINICAL_HISTOLOGY | 56 | Unicryptal adenoma | |
| CLINICAL_HISTOLOGY | 57 | Metastases – another site | |
| CLINICAL_HISTOLOGY | 58 | Cancer + serrated adenoma | |
| CLINICAL_HISTOLOGY | 59 | Cancer + mixed adenoma | |
| CLINICAL_HISTOLOGY | 60 | METS/tumour – infiltrating | |
| CLINICAL_HISTOLOGY | 61 | Squamous cell carcinoma | |
| CLINICAL_HISTOLOGY | 62 | Cancer query | |
| CLINICAL_HISTOLOGY | 63 | Gastrointestinal stromal tumour | |
| CLINICAL_HISTOLOGY | 64 | Sarcoma | |
| CLINICAL_HISTOLOGY | 65 | Unknown primary | |
| CLINICAL_HISTOLOGY | 66 | Anaplastic/undifferentiated carcinoma | |
| CLINICAL_HISTOLOGY | 67 | Basaloid /cloacogenic cancer | |
| CLINICAL_HISTOLOGY | 68 | Sessile serrated lesion | |
| CLINICAL_HISTOLOGY | 69 | Cancer + sessile serrated lesion | |
| CLINICAL_HISTOLOGY | 70 | Granular cell tumour | |
| CLINICAL_HISTOLOGY | 71 | Melanoma | |
| CLINICAL_HISTOLOGY | 72 | Anal wart | |
| CLINICAL_HISTOLOGY | 90 | Not possible to diagnose | |
| CLINICAL_HISTOLOGY | 91 | Specimen not seen | |
| ENDO_PATH_MAPPING | 1 | Rule 1 – histology size rule | |
| ENDO_PATH_MAPPING | 2 | Rule 2 – hyperplastic distal ≤ 5 mm rule | |
| ENDO_PATH_MAPPING | 3 | Rule 3 – excision method rule | |
| ENDO_PATH_MAPPING | 4 | Rule 4 – specimen labels rule | |
| ENDO_PATH_MAPPING | 5 | Rule 5 – other sighting | |
| EXAM_NUMBER_UNKNOWN | 1 | Could not rank | |
| EXAM_NUMBER_UNKNOWN | 2 | Examination is blank | |
| EXCISION_COMPLETE | 1 | Complete | |
| EXCISION_COMPLETE | 2 | Incomplete | |
| EXCISION_COMPLETE | 3 | Uncertain | |
| MAN_COLLECTED_PATH | 0 | No | |
| MAN_COLLECTED_PATH | 1 | Main pathology | |
| MAN_COLLECTED_PATH | 2 | Other pathology | |
| MULTIPLE_GROUP_LINKING | 1 | GROUP1 | |
| MULTIPLE_GROUP_LINKING | 2 | GROUP2 | |
| MULTIPLE_GROUP_LINKING | 3 | GROUP3 | |
| MULTIPLE_GROUP_LINKING | 4 | GROUP4 | |
| MULTIPLE_GROUP_LINKING | 5 | GROUP5 | |
| MULTIPLE_GROUP_LINKING | 6 | GROUP6 | |
| MULTIPLE_GROUP_LINKING | 7 | GROUP7 | |
| MULTIPLE_GROUP_LINKING | 8 | GROUP8 | |
| MULTIPLE_GROUP_LINKING | 9 | GROUP9 | |
| MULTIPLE_GROUP_LINKING | 10 | GROUP10 | |
| MULTIPLE_GROUP_LINKING | 11 | GROUP11 | |
| MULTIPLE_GROUP_LINKING | 12 | GROUP12 | |
| MULTIPLE_GROUP_LINKING | 13 | GROUP13 | |
| MULTIPLE_GROUP_LINKING | 14 | GROUP14 | |
| MULTIPLE_GROUP_LINKING | 15 | GROUP15 | |
| MULTIPLE_GROUP_LINKING | 16 | GROUP16 | |
| MULTIPLE_GROUP_LINKING | 17 | GROUP17 | |
| MULTIPLE_GROUP_LINKING | 18 | GROUP18 | |
| MULTIPLE_GROUP_LINKING | 19 | GROUP19 | |
| MULTIPLE_GROUP_LINKING | 20 | GROUP20 | |
| MULTIPLE_GROUP_LINKING | 21 | GROUP21 | |
| MULTIPLE_GROUP_LINKING | 22 | GROUP22 | |
| MULTIPLE_GROUP_LINKING | 23 | GROUP23 | |
| MULTIPLE_GROUP_LINKING | 24 | GROUP24 | |
| MULTIPLE_GROUP_LINKING | 25 | GROUP25 | |
| MULTIPLE_POLYP_GROUP | 1 | GROUP1 | |
| MULTIPLE_POLYP_GROUP | 2 | GROUP2 | |
| MULTIPLE_POLYP_GROUP | 3 | GROUP3 | |
| MULTIPLE_POLYP_GROUP | 4 | GROUP4 | |
| MULTIPLE_POLYP_GROUP | 5 | GROUP5 | |
| MULTIPLE_POLYP_GROUP | 6 | GROUP6 | |
| MULTIPLE_POLYP_GROUP | 7 | GROUP7 | |
| MULTIPLE_POLYP_GROUP | 8 | GROUP8 | |
| MULTIPLE_POLYP_GROUP | 9 | GROUP9 | |
| MULTIPLE_POLYP_GROUP | 10 | GROUP10 | |
| MULTIPLE_POLYP_GROUP | 11 | GROUP11 | |
| MULTIPLE_POLYP_GROUP | 12 | GROUP12 | |
| MULTIPLE_POLYP_GROUP | 13 | GROUP13 | |
| MULTIPLE_POLYP_GROUP | 14 | GROUP14 | |
| MULTIPLE_POLYP_GROUP | 15 | GROUP15 | |
| MULTIPLE_POLYP_GROUP | 16 | GROUP16 | |
| MULTIPLE_POLYP_GROUP | 17 | GROUP17 | |
| MULTIPLE_POLYP_GROUP | 18 | GROUP18 | |
| MULTIPLE_POLYP_GROUP | 19 | GROUP19 | |
| MULTIPLE_POLYP_GROUP | 20 | GROUP20 | |
| MULTIPLE_POLYP_GROUP | 21 | GROUP21 | |
| MULTIPLE_POLYP_GROUP | 22 | GROUP22 | |
| MULTIPLE_POLYP_GROUP | 23 | GROUP23 | |
| MULTIPLE_POLYP_GROUP | 24 | GROUP24 | |
| MULTIPLE_POLYP_GROUP | 25 | GROUP25 | |
| MULTIPLE_POLYP_GROUP | 26 | GROUP26 | |
| OPERATION_TYPE | 1 | Right hemicolectomy | |
| OPERATION_TYPE | 2 | Left hemicolectomy | |
| OPERATION_TYPE | 3 | Transanal resection | |
| OPERATION_TYPE | 4 | Transanal excision | |
| OPERATION_TYPE | 5 | Anterior resection | |
| OPERATION_TYPE | 6 | Sigmoid resection | |
| OPERATION_TYPE | 7 | Sigmoid colectomy | |
| OPERATION_TYPE | 8 | Extended right hemicolectomy | |
| OPERATION_TYPE | 9 | Extended left hemicolectomy | |
| OPERATION_TYPE | 10 | Subtotal colectomy | |
| OPERATION_TYPE | 11 | Abdominoperineal resection | |
| OPERATION_TYPE | 12 | Transverse colectomy | |
| OPERATION_TYPE | 13 | High anterior resection | |
| OPERATION_TYPE | 14 | Low anterior resection | |
| OPERATION_TYPE | 15 | Total colectomy | |
| OPERATION_TYPE | 16 | Laparotomy with colostomy | |
| OPERATION_TYPE | 17 | Hartmann’s procedure | |
| OPERATION_TYPE | 18 | Laparotomy | |
| OPERATION_TYPE | 19 | Laparoscopic anterior resection | |
| OPERATION_TYPE | 20 | Proctocolectomy | |
| OPERATION_TYPE | 21 | TME | |
| OPERATION_TYPE | 22 | TEMS | |
| OPERATION_TYPE | 23 | Laparoscopic subtotal colectomy | |
| OPERATION_TYPE | 24 | Laparoscopy assisted right hemicolectomy | |
| OPERATION_TYPE | 25 | Ileorectal anastomosis | |
| OPERATION_TYPE | 26 | Laparoscopic assisted resection of sigmoid colon | |
| OPERATION_TYPE | 27 | Right hemicolectomy and laparotomy | |
| OPERATION_TYPE | 28 | Ileosigmoid bypass | |
| OPERATION_TYPE | 29 | Panproctocolectomy | |
| OPERATION_TYPE | 30 | Laparoscopy-assisted left hemicolectomy | |
| OPERATION_TYPE | 31 | Subtotal proctocolorectomy | |
| OPERATION_TYPE | 32 | Subtotal proctocolectomy | |
| OPERATION_TYPE | 33 | Proctosigmoidectomy | |
| OPERATION_TYPE | 34 | Resection – other | |
| OPERATION_TYPE | 35 | Non-resection surgery | |
| OPERATION_TYPE | 36 | Completion colectomy | |
| OPERATION_TYPE | 37 | Double resection | |
| OPERATION_TYPE | 38 | Proctectomy | |
| OPERATION_TYPE | 39 | Not surgery | |
| OPERATION_TYPE | 40 | Transverse colectomy | |
| OPERATION_TYPE | 41 | Laparoscopic caecectomy | |
| OPERATION_TYPE | 42 | Local excision | |
| OPERATION_TYPE | 43 | Extended transverse colectomy | |
| OPERATION_TYPE | 44 | Colectomy | |
| OPERATION_TYPE | 45 | Transverse colon resection | |
| OPERATION_TYPE | 46 | Ileocolic anastomosis | |
| OPERATION_TYPE | 47 | Stapled excision of polyp | |
| OPERATION_TYPE | 48 | Colostomy | |
| OPERATION_TYPE | 49 | Partial colectomy | |
| OPERATION_TYPE | 50 | Posterior mesorectal resection | |
| OPERATION_TYPE | 51 | Rectosigmoid resection | |
| OPERATION_TYPE | 52 | Anterior resection and right hemicolectomy | |
| OPERATION_TYPE | 53 | Right hemicolectomy and sigmoid resection | |
| OPERATION_TYPE | 54 | Left hemicolectomy and AP resection | |
| POLYP_MATCH_PROB | 10 | 10% | |
| POLYP_MATCH_PROB | 20 | 20% | |
| POLYP_MATCH_PROB | 30 | 30% | |
| POLYP_MATCH_PROB | 40 | 40% | |
| POLYP_MATCH_PROB | 50 | 50% | |
| POLYP_MATCH_PROB | 60 | 60% | |
| POLYP_MATCH_PROB | 70 | 70% | |
| POLYP_MATCH_PROB | 80 | 80% | |
| POLYP_MATCH_PROB | 90 | 90% | |
| POLYP_MATCH_PROB | 100 | 100% | |
| POLYP_NUMBERED | N | N | |
| POLYP_NUMBERED | Y | Y | |
| REASON_EXAM_SAME_DAY | 1 | Emergency surgery | |
| REASON_EXAM_SAME_DAY | 2 | First procedure abandoned | |
| REASON_EXAM_SAME_DAY | 3 | First procedure incomplete | |
| REASON_EXAM_SAME_DAY | 4 | This examination may not belong to this patient | |
| REASON_EXAM_SAME_DAY | 5 | Follow-on examination | |
| REASON_EXAM_SAME_DAY | 6 | From same procedure | |
| REASON_EXAM_SAME_DAY | 7 | Unknown | |
| REVIEW_METHOD | APEX | Reviewed by coders via APEX | |
| REVIEW_METHOD | SPREAD | Reviewed on spreadsheet | |
| REVIEW_NUMBERED | N | N | |
| REVIEW_NUMBERED | Y | Y | |
| SERRATION | 1 | Confirmed | |
| SERRATION | 2 | Possible | |
| XADENOMA_TYPE | 1 | Tubular | The pathologist describes the morphology of the adenomatous tissue as tubular |
| XADENOMA_TYPE | 3 | Tub-vill | The pathologist describes the morphology of the adenomatous tissue as tubulovillous |
| XADENOMA_TYPE | 2 | Villous | The pathologist describes the morphology of the adenomatous tissue as villous |
| XBIOPSY_FATE | 1 | Retrieved | To be used when the polyp has been collected by the endoscopist from the colon after excision, and not left inside the colon. This may or may not have been sent to pathology |
| XBIOPSY_FATE | 2 | Burnt off | Method of removal that destroys the polyp in situ |
| XBIOPSY_FATE | 4 | Unknown | To be used when the coder is unsure what has happened to the polyp post excision/biopsy |
| XBIOPSY_FATE | 5 | Not retrieved | The specimen was lost after removal either inside the patient or outside the patient on collection. A specimen/biopsy may still have been sent to pathology |
| XBOWEL_PREP | 1 | Poor | |
| XBOWEL_PREP | 2 | Satisfactory | |
| XBOWEL_PREP | 3 | Good | |
| XBOWEL_PREP | 4 | Excellent | |
| XBOWEL_SEGMENT | 33 | Anus | An |
| XBOWEL_SEGMENT | 5 | Splenic flexure | SF |
| XBOWEL_SEGMENT | 6 | Transverse colon | TC |
| XBOWEL_SEGMENT | 25 | Transverse colon (distal) | TC(d) |
| XBOWEL_SEGMENT | 26 | Transverse colon (mid) | TC(m) |
| XBOWEL_SEGMENT | 27 | Transverse colon (proximal) | TC(p) |
| XBOWEL_SEGMENT | 7 | Hepatic flexure | HF |
| XBOWEL_SEGMENT | 8 | Ascending colon | AC |
| XBOWEL_SEGMENT | 29 | Ascending colon (distal) | AC(d) |
| XBOWEL_SEGMENT | 36 | Ascending colon (mid) | AC(m) |
| XBOWEL_SEGMENT | 28 | Ascending colon (proximal) | AC(p) |
| XBOWEL_SEGMENT | 9 | Caecum | CM |
| XBOWEL_SEGMENT | 1 | Rectum | RM |
| XBOWEL_SEGMENT | 37 | Rectum (distal) | RM(d) |
| XBOWEL_SEGMENT | 38 | Rectum (mid) | RM(m) |
| XBOWEL_SEGMENT | 39 | Rectum (proximal) | RM(p) |
| XBOWEL_SEGMENT | 34 | Ileocaecal valve | ICV |
| XBOWEL_SEGMENT | 10 | Terminal Ileum | TI |
| XBOWEL_SEGMENT | 2 | Rectosigmoid | RS |
| XBOWEL_SEGMENT | 3 | Sigmoid colon | SC |
| XBOWEL_SEGMENT | 24 | Sigmoid colon (distal) | SC(d) |
| XBOWEL_SEGMENT | 40 | Sigmoid colon (mid) | SC(m) |
| XBOWEL_SEGMENT | 23 | Sigmoid colon (proximal) | SC(p) |
| XBOWEL_SEGMENT | 4 | Descending colon | DC |
| XBOWEL_SEGMENT | 22 | Descending colon (distal) | DC(d) |
| XBOWEL_SEGMENT | 41 | Descending colon (mid) | DC(m) |
| XBOWEL_SEGMENT | 21 | Descending colon (proximal) | DC(p) |
| XBOWEL_SEGMENT | 11 | Anastomosis | Anas |
| XBOWEL_SEGMENT | 31 | Colostomy | Colos |
| XBOWEL_SEGMENT | 32 | Neoterminal ileum | TI Neo |
| XBOWEL_SEGMENT | 35 | Ileal pouch | Il Po |
| XBOWEL_SEGMENT | 42 | Pre pouch ileum | Il Pre-Po |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 1 | Confirmed colitis | Confirmed colitis |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 2 | Ulcerative colitis | Chronic form of IBD characterised by ulceration of the colon and rectum |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 3 | Microscopic colitis | Refers to both collagenous colitis and lymphocytic colitis, characterised by increase in inflammatory cells |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 4 | Lymphocytic colitis | Subtype of microscopic colitis, characterised by chronic non-bloody watery diarrhoea and an accumulation of lymphocytes in the colonic mucosa and lamina propria |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 5 | Collagenous colitis | Subtype of microscopic colitis, characterised by chronic watery diarrhoea, rectal bleeding and deposition of collagen in the lamina propria |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 6 | Healed colitis | Redundant code – not available on the screen |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 7 | History of colitis | Redundant code – not available on the screen |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 8 | Ischaemic colitis | Inflammation and injury of the colon as a result of inadequate blood supply |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 9 | Diversion colitis | Inflammation in a non-functioning colonic pouch occurring as a complication of ileostomy or colostomy, often within the year following the surgery |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 10 | Infective colitis | Inflammation of the colon caused by bacterial or viral infection, commonly due to Clostridium difficile |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 11 | Chemical colitis | Inflammation of the colon caused by the introduction of harsh chemicals by an enema or other procedure |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 12 | Pseudomembranous colitis | Subtype of infectious colitis, characterised by the formation of pseudomembranes |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 13 | Drug-induced colitis | Inflammation of the colon as a result of treatment with various types of drug, (e.g. NSAIDs, anticoagulants, SSRIs) |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 14 | Radiation colitis | Inflammation and damage of the colon as a result of exposure to X-rays or radiation; commonly occurs after radiation therapy for cancer |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 15 | Reactive colitis | Redundant code – not available on the screen |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 16 | Procedural/enema related | Inflammation of the colon as a result of the endoscopic procedure |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 17 | Possible colitis | Possible colitis [of a certain type(s)] |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 18 | Antibiotic-associated colitis | Inflammation of the colon as a result of antimicrobial therapy |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 19 | Indeterminate colitis | Colitis that has features of both Crohn’s disease and ulcerative colitis |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 20 | Atypical colitis | Colitis that does not conform to criteria for accepted types of colitis |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 21 | Other colitis 1 | Other colitis 1 spare |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 22 | Other colitis 2 | Other colitis 2 spare |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 23 | Ulcerative proctitis | Ulcerative proctitis is the least severe form of IBD |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 25 | Radiation proctitis – autocoded | |
| XDIAGNOSIS_SUB_TYPES_COLITIS | 99 | Rule out colitis | Rule out colitis |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 1 | Confirmed polyposis | Confirmed polyposis |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 2 | FAP | Presence of multiple adenomas |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 3 | Juvenile polyposis | Presence of juvenile polyps |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 4 | PJ polyposis | Presence of Peutz–Jeghers type (hamartomatous) of polyps; PJS |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 5 | Hyperplastic polyposis | Presence of multiple hyperplastic polyps |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 6 | Possible | Possible polyposis [of a certain type(s)] |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 7 | Serrated polyposis | Presence of multiple serrated polyps |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 8 | Lymphoid polyposis | Presence of multiple lymphoid polyps |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 9 | Cap polyposis | Presence of multiple cap polyps |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 10 | Other polyposis 1 | Other polyposis – spare 1 |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 11 | Other polyposis 2 | Other polyposis – spare 2 |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 12 | MAP | MYH-associated polyposis: mutations in the MUTYH gene cause an autosomal recessive form of FAP (also called MUTYH-associated polyposis). Polyps caused by mutated MYH do not appear until adulthood and are less numerous than those found in patients with APC gene mutations |
| XDIAGNOSIS_SUB_TYPES_POLYPOSIS | 99 | Rule out polyposis | Rule out polyposis |
| XDIAGNOSIS_TYPES | 1 | Polyps | Endoscopist has found polyps in the bowel during the examination |
| XDIAGNOSIS_TYPES | 2 | Diverticular disease | Endoscopist has observed uncomplicated diverticula in the colon (diverticulosis) or inflamed diverticular (diverticulitis) |
| XDIAGNOSIS_TYPES | 3 | Haemorrhoids | Endoscopist has observed haemorrhoids/piles |
| XDIAGNOSIS_TYPES | 4 | Colitis | The endoscopist has specified the presence of colitis from the observations made during the examination |
| XDIAGNOSIS_TYPES | 5 | Cancer | Malignant neoplasm |
| XDIAGNOSIS_TYPES | 6 | Crohn’s disease | The endoscopist strongly suspects the patient has Crohn’s disease or is known to already have Crohn’s disease |
| XDIAGNOSIS_TYPES | 7 | Proctitis | Endoscopist has observed Inflammation of the lining of the rectum and anus. Telangiectasia is a similar condition involving the blood vessels and can be classified under proctitis |
| XDIAGNOSIS_TYPES | 8 | Piles | See haemorrhoids |
| XDIAGNOSIS_TYPES | 9 | Anastomosis | Endoscopist has mentioned the anastomosis which is the surgical reconnection of two parts of the colon post resection |
| XDIAGNOSIS_TYPES | 10 | Benign tumour | Endoscopist has observed an abnormal growth/neoplasm that they feel lacks the malignant qualities of cancer |
| XDIAGNOSIS_TYPES | 11 | Volvulus | Endoscopist has observed a life threatening bowel obstruction where the bowel twists on itself |
| XDIAGNOSIS_TYPES | 12 | Angiodysplasia | Endoscopist has observed areas of vascular malformation in the gut and can be a common cause of unexplained bleeding in the colon |
| XDIAGNOSIS_TYPES | 13 | Melaena | Black, ‘tarry’ faeces that are associated with gastrointestinal haemorrhage. There are some inconsistencies in the coding of this field |
| XDIAGNOSIS_TYPES | 14 | Incomplete examination | Any description that suggests the examination was not fully compete due to physical or technical difficulties, (e.g.:
|
| XDIAGNOSIS_TYPES | 15 | Suspected IBD | Endoscopist has seen features that are suspicious of/indicating IBD |
| XDIAGNOSIS_TYPES | 16 | IBD | IBD – a group of conditions affecting the colon and small intestine, including Crohn’s disease and ulcerative colitis |
| XDIAGNOSIS_TYPES | 17 | Ulcers/ulceration | An inflammatory and often suppurating lesion on the skin or an internal mucous surface resulting in necrosis of tissue |
| XDIAGNOSIS_TYPES | 18 | Colonic obstruction | Obstruction of the colon, preventing the normal transit of the products of digestion There are some inconsistencies in the coding of this field |
| XDIAGNOSIS_TYPES | 19 | Fissure | A crack or tear in the tissue There are some inconsistencies in the coding of this field |
| XDIAGNOSIS_TYPES | 20 | Strictures | An abnormal narrowing of the colon. The stricture may be due for example to scar tissue or to a tumour. Stricture refers to both the process of narrowing and the narrowed part itself There are some inconsistencies in the coding of this field |
| XDIAGNOSIS_TYPES | 21 | FAP – redundant do not use | FAP. Redundant code – see incident 350 |
| XDIAGNOSIS_TYPES | 22 | Polyposis | A hereditary disease in which numerous polyps erupt in a part of the body, especially on the lining of the colon and rectum, and often become malignant |
| XDIAGNOSIS_TYPES | 23 | Prolapse | The falling down or slipping of a body part from its usual position or relations There are some inconsistencies in the coding of this field |
| XDIAGNOSIS_TYPES | 24 | Non-exclusion colitis | Colitis that does not fit within the exclusion criteria |
| XDIAGNOSIS_TYPES | 25 | Possible Crohn’s disease | The possible presence of Crohn’s disease in the colon |
| XDIAGNOSIS_TYPES | 43 | Radiation and proctitis found | Words radiation and proctitis found |
| XDIAGNOSIS_TYPES | 44 | Radiation and ulcers found | Words radiation and ulcers found |
| XDYSPLASIA | 1 | Mild | The pathologist describes mild dysplasia/atypia |
| XDYSPLASIA | 2 | Moderate | The pathologist describes moderate or focally moderate dysplasia/atypia |
| XDYSPLASIA | 3 | Low grade | The pathologist describes low-grade dysplasia/atypia. This is synonymous with either mild or moderate dysplasia. Use only when specified |
| XDYSPLASIA | 4 | High grade | The pathologist describes HGD/atypia. This is synonymous with severe dysplasia. Use only when specified |
| XDYSPLASIA | 5 | Severe | The pathologist describes severe or focally dysplasia/atypia |
| XDYSPLASIA | 6 | Intramucosal cancer | The pathologist describes presence of intramucosal cancer |
| XDYSPLASIA | 7 | Intramucosal cancer in dispute | Intramucosal cancer in dispute |
| XEXCISION_EXTENT | 1 | Excised | Endoscopist believes he has removed the polyp from the colon |
| XEXCISION_EXTENT | 2 | Partially excised | Endoscopist has specified that they have only removed part of the polyp |
| XEXCISION_EXTENT | 3 | Not excised | Endoscopist has not excised the polyp. They may still have biopsied the polyp for pathology |
| XEXCISION_METHOD | 1 | Cold biopsy | Also Cold Bx/B’x. Insert if specified by endoscopist or pathologist |
| XEXCISION_METHOD | 2 | Hot biopsy | Also Hot Bx/B’x. Insert if specified by endoscopist or pathologist |
| XEXCISION_METHOD | 3 | Snare | A wire loop device designed to slip over a polyp and, on closure, result in removal of the polyp |
| XEXCISION_METHOD | 4 | Cold snare | Insert if specified by endoscopist or pathologist |
| XEXCISION_METHOD | 5 | Hot snare | Insert if specified by endoscopist or pathologist |
| XEXCISION_METHOD | 7 | EMR | Endoscopic mucosal resection. Insert if specified by endoscopist or pathologist |
| XEXCISION_METHOD | 9 | Unknown | Used when you know removal or biopsy has taken place but the method is unspecified |
| XEXCISION_METHOD | 10 | APC | Argon plasma coagulation |
| XEXCLUSION | 1 | Crohn’s disease | Chronic inflammatory disease that can affect any part of the intestinal tract |
| XEXCLUSION | 2 | Ulcerative colitis | Chronic digestive disease characterised by inflammation of the colon that includes characteristic ulcers, or open sores |
| XEXCLUSION | 3 | Colitis | Chronic digestive disease characterised by inflammation of the colon |
| XEXCLUSION | 4 | FAP | Inherited condition in which numerous polyps form mainly in the epithelium of the large intestine |
| XEXCLUSION | 5 | Family Hx FAP | History of family members who have suffered with FAP |
| XEXCLUSION | 6 | Polyposis | Colon has a very large number of polyps lining a large proportion of the colons surface |
| XEXCLUSION | 7 | P-J polyposis | Hereditary intestinal polyposis syndrome characterised by the development of benign hamartomatous polyps in the gastrointestinal tract |
| XEXCLUSION | 8 | Cancer first examination | Malignant findings in the colon that have been confirmed by pathological assessment linked to the first endoscopic examination for that patient in our records |
| XEXCLUSION | 9 | Resection first examination | Partial or complete removal of the colon occurring before first endoscopic examination for that patient in our records |
| XEXCLUSION | 10 | IBD | IBD |
| XEXCLUSION | 21 | HNPCC | Familial predisposition indicates elevated risk for polyp and/or bowel cancer development by genetic influence |
| XEXCLUSION | 97 | No bowel examinations recorded | Excluded as none of the endoscopies are a bowel examination for this patient |
| XEXCLUSION | 98 | Other – see developer notes | Excluded as records without endoscopies – see developer’s notes |
| XEXCLUSION | 99 | Other – see comments | Other condition not listed here that may exclude patient from study |
| XHOSPITALS | BRI | Brighton Hospital | |
| XHOSPITALS | CI | Cumberland Infirmary | |
| XHOSPITALS | CX | Charing Cross Hospital | |
| XHOSPITALS | GRI | Glasgow Royal Infirmary | |
| XHOSPITALS | ICMS | St Mary’s – Imperial Trust | |
| XHOSPITALS | LGH | Leicester General Hospital | |
| XHOSPITALS | NC | New Cross | |
| XHOSPITALS | NT | North Tees | |
| XHOSPITALS | QEW | Queen Elizabeth Hospital | |
| XHOSPITALS | QMH | Queen Mary’s Hospital | |
| XHOSPITALS | RLUH | Liverpool University Hospital | |
| XHOSPITALS | SCH | Royal Surrey County Hospital | |
| XHOSPITALS | SGH | St George’s | |
| XHOSPITALS | SH | Shrewsbury Hospital | |
| XHOSPITALS | SMH | St Mark’s Hospital | |
| XHOSPITALS | TDG | Torbay District General Hospital | |
| XHOSPITALS | YDH | Yeovil District Hospital | |
| XINDICATION_TYPES | 1 | Polyps | The patient has had previous polyps |
| XINDICATION_TYPES | 2 | Diverticular disease | Presence of uncomplicated diverticula in the colon (diverticulosis) or inflamed diverticular (diverticulitis) |
| XINDICATION_TYPES | 3 | Haemorrhoids | Swelling and inflammation of the veins in the rectum and anus. This is the same as piles |
| XINDICATION_TYPES | 4 | Colitis | The endoscopist has specified the presence of colitis prior to the procedure. Colitis is a chronic digestive disease characterised by inflammation of the colon. Inflammation alone does not warrant a classification of colitis |
| XINDICATION_TYPES | 5 | Carcinoma | The patient has had a carcinoma prior to the examination in question |
| XINDICATION_TYPES | 6 | Cancer | The patient has had a cancer prior to the examination in question |
| XINDICATION_TYPES | 7 | Crohn’s disease | Chronic inflammatory disease which can affect any part of the gastrointestinal tract (It is a member of the colitis family but is defined separately here for clarity) |
| XINDICATION_TYPES | 8 | Anaemia | Deficiency of haemoglobin in the blood |
| XINDICATION_TYPES | 9 | Diarrhoea | Frequent loose or liquid bowel movements |
| XINDICATION_TYPES | 10 | Abdominal pain | Generalised pain in the abdominal region. LIF and RIF pain can also be classified as abdominal pain |
| XINDICATION_TYPES | 11 | Rectal bleeding | Bleeding appearing to come from the rectum |
| XINDICATION_TYPES | 12 | Abnormal barium enema | Abnormal findings on a barium enema prior to examination in question |
| XINDICATION_TYPES | 13 | Abnormal CT | Abnormal findings on a CT scan prior to examination in question |
| XINDICATION_TYPES | 14 | Abnormal sigmoidoscopy | Abnormal findings on a sigmoidoscopy prior to examination in question |
| XINDICATION_TYPES | 15 | Family history of CRC | Patient has relatives who have had CRC There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 16 | Hereditary non-polyposis CRC | An inherited condition with which there is a very high chance of getting CRC |
| XINDICATION_TYPES | 17 | Change in bowel habit | The patient is experiencing a change in the frequency of bowel movements compared with normal |
| XINDICATION_TYPES | 18 | Polyposis | A condition through which a person suffers with a large number of polyps coating a large surface area throughout the colon |
| XINDICATION_TYPES | 19 | Constipation | Hard faeces that are difficult to expel, often accompanied by a reduction in the frequency of bowel movements |
| XINDICATION_TYPES | 20 | Rectal_Mass | Palpable mass in the area of the rectum |
| XINDICATION_TYPES | 21 | Abdominal mass | Palpable mass in the area of the abdomen |
| XINDICATION_TYPES | 22 | Weight loss | Uncharacteristic loss of body mass |
| XINDICATION_TYPES | 23 | Positive faecal occult blood test | Also FOBT or FOB. Positive result in screening test for ‘unseen’ blood |
| XINDICATION_TYPES | 24 | Rectal pain | Pain in the rectum |
| XINDICATION_TYPES | 25 | Query polyps | To test for the presence of polyps/suspect presence of polyps |
| XINDICATION_TYPES | 26 | Query colitis | To test for the presence of colitis/suspect possible colitis |
| XINDICATION_TYPES | 27 | Query Crohn’s disease | To test for the presence of Crohn’s disease/suspect possible Crohn’s disease |
| XINDICATION_TYPES | 28 | Mucus discharge | Mucus that passes out of the rectum from a source in the bowel |
| XINDICATION_TYPES | 29 | Bowel Cancer Screening Programme | The patient is having an endoscopy examination as part of the national Bowel Cancer Screening Programme. Could also be written as BCSP Please note that not all records coded with this indication as part of the national Bowel Cancer Screening Programme |
| XINDICATION_TYPES | 30 | Volvulus | Life-threatening bowel obstruction where the bowel twists on itself |
| XINDICATION_TYPES | 31 | Incontinence | Involuntary leakage of faeces There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 32 | Melena | Black, ‘tarry’ faeces, which are associated with gastrointestinal haemorrhage There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 33 | Tenesmus | Feeling of the need to evacuate the bowels, with little or no stool passed There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 34 | Query cancer | To test for the presence of cancer |
| XINDICATION_TYPES | 35 | Family History of Cancer | A history of cancer within the family, indicating a hereditary risk There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 36 | IBD | Inflammatory bowel disease – a group of conditions affecting the colon and small intestine, including; Crohn’s disease and ulcerative colitis |
| XINDICATION_TYPES | 37 | Ulcers/ulceration | An inflammatory and often suppurating lesion on the skin or an internal mucous surface resulting in necrosis of tissue |
| XINDICATION_TYPES | 38 | Colonic obstruction | Obstruction of the colon, preventing the normal transit of the products of digestion There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 39 | Fissure | A crack or tear in the tissue There are some inconsistencies in the coding of this field |
| XINDICATION_TYPES | 40 | Query polyposis | To test for the presence of polyposis |
| XINDICATION_TYPES | 41 | Family Hx of FAP | Family history of FAP |
| XINDICATION_TYPES | 42 | Cowden syndrome | A rare inherited disorder characterised by multiple tumour-like growths called hamartomas and an increased risk of developing CRC |
| XINDICATION_TYPES | 43 | Radiation and proctitis found | Words radiation and proctitis found |
| XINDICATION_TYPES | 44 | Radiation and ulcers found | Words radiation and ulcers found |
| XNFEATURE_TYPES | 1 | Pathology – blank | |
| XNFEATURE_TYPES | 2 | Pathology – truncated | |
| XNFEATURE_TYPES | 3 | Endoscopy – irrelevant | |
| XNFEATURE_TYPES | 4 | Endoscopy – duplicate | |
| XNFEATURE_TYPES | 5 | Endoscopy – truncated | |
| XNFEATURE_TYPES | 6 | Endoscopy – blank | |
| XNFEATURE_TYPES | 7 | Condition – possible cancer first examination | |
| XNFEATURE_TYPES | 8 | Condition – possible resection first examination | |
| XNFEATURE_TYPES | 9 | Condition – possible HNPCC | |
| XNFEATURE_TYPES | 10 | Condition – possible polyposis | |
| XNFEATURE_TYPES | 11 | Condition – possible IBD | |
| XNFEATURE_TYPES | 12 | General – supplementary report missing | |
| XNFEATURE_TYPES | 13 | General – possible cancer | |
| XNFEATURE_TYPES | 14 | General – when cancer | |
| XNFEATURE_TYPES | 15 | General – when resection | |
| XNFEATURE_TYPES | 16 | General – polyp numbers | |
| XNFEATURE_TYPES | 17 | General – unsure terminology | |
| XNFEATURE_TYPES | 18 | Pathology linking | |
| XNFEATURE_TYPES | 19 | Pathology missing – sent to laboratory/await pathology stated | |
| XNFEATURE_TYPES | 20 | Pathology missing – large polyp of ≥ 10 mm | |
| XNFEATURE_TYPES | 21 | Pathology missing – biopsy indicator | |
| XNFEATURE_TYPES | 22 | Pathology missing – cancer/tumour indicated | |
| XNFEATURE_TYPES | 23 | Polyp matching | |
| XNFEATURE_TYPES | 24 | Refer back to | |
| XNFEATURE_TYPES | 25 | Discuss – IBD | |
| XNFEATURE_TYPES | 26 | Discuss – how best to code | |
| XNFEATURE_TYPES | 27 | Discuss – HNPCC | |
| XNFEATURE_TYPES | 28 | Pathology – unclear specimen origin | |
| XPATH_EXCLUSION | 0 | – | |
| XPATH_EXCLUSION | 1 | General | |
| XPATH_EXCLUSION | 2 | Not relevant pathology | |
| XPATH_EXCLUSION | 3 | Duplicate | |
| XPATH_QUERY | 1 | Blank pathology | |
| XPATH_QUERY | 2 | Truncated pathology report | |
| XPATH_QUERY | 3 | Unclear specimen origin | |
| XPATH_QUERY | 4 | Possible link | |
| XPATIENT_CONDITIONS_TYPES | 8 | Cancer first examination | |
| XPATIENT_CONDITIONS_TYPES | 9 | Resection first examination | |
| XPOLYP_HISTOLOGY | 1 | Adenoma | Pathologist specifies adenoma/adenomatous polyp. Benign dysplastic colonic tumour. Can progress to become malignant |
| XPOLYP_HISTOLOGY | 2 | Metaplastic/hyperplastic | Benign non-dysplastic polyps with lengthening and cystic dilation of mucosal glands. Hyperplastic and metaplastic are synonymous with each other |
| XPOLYP_HISTOLOGY | 3 | Serrated adenoma | Benign dysplastic colonic tumour, which has a serrated appearance under the microscope |
| XPOLYP_HISTOLOGY | 4 | Leiomyoma | Benign neoplasm of smooth muscle |
| XPOLYP_HISTOLOGY | 5 | Inflammatory | Inflammatory polyp |
| XPOLYP_HISTOLOGY | 6 | Normal mucosa | Specimen shows no signs of a polyp and levels of dysplasia/atypia are within normal limits |
| XPOLYP_HISTOLOGY | 8 | Carcinoid/neuroendocrine tumour | Tumour originating from the neuroendocrine system |
| XPOLYP_HISTOLOGY | 10 | Juvenile polyp | Rare form of large bowel polyp |
| XPOLYP_HISTOLOGY | 11 | Mucosal prolapse | Slippage of mucosa |
| XPOLYP_HISTOLOGY | 14 | Ulcer | A break in the lining of the digestive tract that fails to heal naturally |
| XPOLYP_HISTOLOGY | 15 | Inflammation | Generalised inflammation of mucosa |
| XPOLYP_HISTOLOGY | 16 | Melanosis coli | Pigmentation of the wall of the colon, not associated with any disease pathway |
| XPOLYP_HISTOLOGY | 17 | Submucosal haematoma | Result of bleeding outside of the blood vessels |
| XPOLYP_HISTOLOGY | 19 | Angiodysplasia | Vascular malformation in the gut which can often cause bleeding into the colon |
| XPOLYP_HISTOLOGY | 20 | Ischaemia | Restriction of blood supply |
| XPOLYP_HISTOLOGY | 21 | Xanthoma | Fatty deposits under the skin or mucosa causing yellow bumps |
| XPOLYP_HISTOLOGY | 22 | Oedema | Swelling due to accumulation of fluids |
| XPOLYP_HISTOLOGY | 23 | Regenerative polyp | Hyperplastic polyp of the gastric mucosa |
| XPOLYP_HISTOLOGY | 24 | Hamartomatous polyp | Benign mucosal polyps usually found in the jejunum and ileum (small bowel) |
| XPOLYP_HISTOLOGY | 25 | Haemangioma | Benign noncancerous tumour composed of rapidly proliferating blood vessels |
| XPOLYP_HISTOLOGY | 26 | Non-Hodgkin’s lymphoma | NHL: A diverse group of blood cancers that include any kind of lymphoma except Hodgkin’s lymphomas |
| XPOLYP_HISTOLOGY | 27 | Fibroepithelial polyp | Benign cutaneous lesion/skin tag |
| XPOLYP_HISTOLOGY | 28 | Crohn’s disease | An autoimmune inflammatory disease that can affect any part of the gastrointestinal tract. If you notice Crohn’s disease, this person should be excluded from the study using the query section |
| XPOLYP_HISTOLOGY | 29 | Neurofibromatosis | Genetically inherited disease in which nerve fibres grow tumours |
| XPOLYP_HISTOLOGY | 30 | Colitis | Chronic bowel disease characterised by inflammation of the colon. Only to be used if colitis is specified, there is a separate option for generalised inflammation. If you notice colitis then this person should be excluded from the study using the query section |
| XPOLYP_HISTOLOGY | 31 | Lipoma | Benign tumour composed of fatty tissue |
| XPOLYP_HISTOLOGY | 32 | Pseudolipomatus | Artifactual microscopic change in tissues that resembles fatty infiltration |
| XPOLYP_HISTOLOGY | 33 | Spirochaetosis | A type of bacterial infection of the colon |
| XPOLYP_HISTOLOGY | 34 | Granulation tissue | Tissue that replaces fibrin clots during the healing of tissue |
| XPOLYP_HISTOLOGY | 35 | Gastric heterotopia | Normal gastric mucosa seen elsewhere in the body |
| XPOLYP_HISTOLOGY | 36 | Cap polyp | Inflammatory polyp with a ‘cap’ of debris or granulation tissue |
| XPOLYP_HISTOLOGY | 37 | Lymphoid polyp | Benign polyps occurring when lymphoid follicles are present in the colon |
| XPOLYP_HISTOLOGY | 39 | Previous polypectomy site | Appears to be tissue from the site where a previous polyp was removed |
| XPOLYP_HISTOLOGY | 40 | Ganglioneuromatosis | Tumours arising from the nervous system |
| XPOLYP_HISTOLOGY | 41 | Amyloid | Insoluble fibrous protein aggregates |
| XPOLYP_HISTOLOGY | 43 | Congestion | Mucosal cells appear congested |
| XPOLYP_HISTOLOGY | 44 | Lymphangiectasia | Intestinal disease characterised by lymphatic dilatation |
| XPOLYP_HISTOLOGY | 45 | Proctitis | Inflammation of the lining of the anus and rectum |
| XPOLYP_HISTOLOGY | 50 | Cancer | Malignant neoplasm/moderately differentiated adenoncarcinoma/carcinoma. If you notice cancer and it is the patient’s first dated endoscopy record then this person should be excluded from the study using the query section |
| XPOLYP_HISTOLOGY | 51 | CA + adenoma | Moderately differentiated carcinoma/cancer/malignant cell types, seen to be arising from an adenoma |
| XPOLYP_HISTOLOGY | 52 | CA in dispute | Cancer is suspected/pathologist is suspicious of malignancy or there is a difference of option on the diagnosis |
| XPOLYP_HISTOLOGY | 53 | Mixed adenomas/metastases | Polyp displaying characteristics of an adenoma and those of a metaplastic polyp. This is rare |
| XPOLYP_HISTOLOGY | 55 | CA + mixed/serrated adenoma | Moderately differentiated carcinoma/cancer/malignant cell types, seen to be arising from a serrated adenoma |
| XPOLYP_HISTOLOGY | 56 | Unicryptal adenoma | Very early beginning of adenoma growth |
| XPOLYP_HISTOLOGY | 57 | Metastases – another site | Malignant material found in the colon that is not from a primary bowel cancer and originates elsewhere in the body |
| XPOLYP_HISTOLOGY | 58 | CA + serrated adenoma | Carcinoma/cancer/malignant/invasive cell types, seen to be arising from a serrated polyp or adenoma |
| XPOLYP_HISTOLOGY | 59 | CA + mixed adenoma | Carcinoma/cancer/malignant/invasive cell types, seen to be arising from a mixed polyp or adenoma |
| XPOLYP_HISTOLOGY | 60 | METS/tumour – infiltrating | Malignant material that is infiltrating into the colon from a tumour outside the colon (if unsure use ‘Mets from another site’) |
| XPOLYP_HISTOLOGY | 61 | Squamous cell carcinoma | Skin cancer normally found in the anus, but may be reported as rectal. Code as squamous cell carcinoma |
| XPOLYP_HISTOLOGY | 62 | Cancer query | If a pathologist mention or suspects but is not able to confirm a diagnosis of cancer/malignancy |
| XPOLYP_HISTOLOGY | 63 | GIST | Gastrointestinal stromal tumour |
| XPOLYP_HISTOLOGY | 64 | Sarcoma | Cancerous tumour of soft tissue |
| XPOLYP_HISTOLOGY | 65 | Unknown primary | Confirmed malignancy of an unclear or unknown primary |
| XPOLYP_HISTOLOGY | 66 | Anaplastic/undifferentiated carcinoma | A rare type of cancer often diagnosed at advanced stage, usually found in the small intestine |
| XPOLYP_HISTOLOGY | 67 | Basaloid /cloacogenic cancer | Sometimes listed as a subclass of squamous cell cancers. They develop in the transitional zone, also called the cloaca. These cancers look slightly different under the microscope, but they behave and are treated like other squamous cell carcinomas of the anal canal |
| XPOLYP_HISTOLOGY | 68 | Sessile serrated lesion | A serrated polyp not of the traditional serrated adenoma type. Usually, but not always, without dysplasia |
| XPOLYP_HISTOLOGY | 69 | CA + sessile serrated lesion | Cancer arising in a polyp of the sessile serrated lesion type |
| XPOLYP_HISTOLOGY | 70 | Granular cell tumour | Usually benign, circumscribed, tumour-like lesion of soft tissue, particularly of the tongue, composed of large cells with prominent granular cytoplasm |
| XPOLYP_HISTOLOGY | 71 | Melanoma | Anorectal melanoma is melanoma affecting the anus and/or rectum. Melanoma is a cancer that develops from cells called melanocytes |
| XPOLYP_HISTOLOGY | 72 | Anal wart | A growth, also known as condyloma, found at the anus/rectal opening caused by HPV. Some HPV strains have been associated with increased risk of anal cancer |
| XPOLYP_HISTOLOGY | 90 | Not possible to diagnose | The polyp sample is too small or too damaged on removal to reliably diagnose the specimen |
| XPOLYP_HISTOLOGY | 91 | Specimen not seen | No evidence of a specimen in the pot received at pathology |
| XPOLYP_NUMBERS | 1 | Few | |
| XPOLYP_NUMBERS | 2 | Some | |
| XPOLYP_NUMBERS | 3 | Num of | |
| XPOLYP_NUMBERS | 4 | Several | |
| XPOLYP_NUMBERS | 5 | Many | |
| XPOLYP_NUMBERS | 6 | Multiple | |
| XPOLYP_SHAPE | 10 | Pedunc | The polyp observed was on a stalk |
| XPOLYP_SHAPE | 20 | Sessile | Polyp with no stalk |
| XPOLYP_SHAPE | 30 | Flat | Polyp that is flat on the surface of the bowel |
| XPOLYP_SHAPE | 40 | Pseudo | A mass that has the appearance of a polyp but is not |
| XPOLYP_SHAPE | 50 | Sub ped | Avoid using this option |
| XPOLYP_SIZE | 1 | Tiny | |
| XPOLYP_SIZE | 2 | Small | |
| XPOLYP_SIZE | 3 | < 5 mm | |
| XPOLYP_SIZE | 4 | 5–9 mm | |
| XPOLYP_SIZE | 5 | > 10 mm | |
| XPOLYP_SIZE | 6 | Large | |
| XPOLYP_SIZE | 7 | < 10 mm | |
| XPROCEDURE | 1 | Colonoscopy | The endoscopic examination of the whole of the large colon and the distal part of the small bowel with a camera on a tube passed through the anus |
| XPROCEDURE | 2 | FS | The endoscopic examination of the large intestine from the rectum to the distal sigmoid using a flexible scope |
| XPROCEDURE | 3 | Proctoscopy | Short ridged metal tube is inserted into the rectum, anal cavity or sigmoid to enable direct visualisation of the area |
| XPROCEDURE | 4 | Rigid sigmoidoscopy | The endoscopic examination of the large intestine from the rectum to the distal sigmoid using a rigid scope |
| XPROCEDURE | 5 | Sigmoidoscopy | Examination of the large colon up until the sigmoid colon |
| XPROCEDURE | 6 | Surgical | |
| XPROCEDURE | 7 | Endoscopy | Endoscopy |
| XQUERY | 1 | General | To be used for any other query, particularly if you feel it warrants discussion with other members of the team. When using a general query, you should describe the nature of the query in the comments box below the query field |
| XQUERY | 2 | Pathology linking | To be used when a pathology report appears to be linked incorrectly to an endoscopy record |
| XQUERY | 3 | Application coding error | To be used when indicating that options or drop-down menus will be changed at a later date by the database administrator and you will return at a later date to finish coding |
| XQUERY | 4 | Polyp matching | To be used when coder is unable to match pathological information to the polyps in the list due to lack of detail or clarity |
| XQUERY | 5 | Exclude | To be used when a patient’s endoscopy or pathology report indicates the presence of study exclusion criteria. For details on the exclusion criteria see the ‘Exclusion SOP’ |
| XQUERY | 6 | Pathology missing | To be used when there has clearly been a biopsy/excision that is awaiting histology and does not appear to have a linked pathology report |
| XQUERY | 7 | Refer back to | To be used when coder wants to come back to a record at a later time |
| XQUERY | 8 | Discuss | To be used when a endoscopy record should be discussed in a coder’s meeting |
| XQUERY_POLYP_NUM | 1 | Other | To be used for any other query, particularly if you feel it warrants discussion with other members of the team. When using a general query, you should describe the nature of the query in the comments box below the query field |
| XQUERY_POLYP_NUM | 2 | Resolved | When the query has been resolved |
| XQUERY_POLYP_NUM | 3 | Multiple quantity – other rows | When patient has unspecified number of polyps |
| XQUERY_POLYP_NUM | 4 | Multiple polyps/polyposis/HNPCC | Multiple polyps/polyposis/HNPCC |
| XQUERY_POLYP_NUM | 5 | Lack of information | Lack of information |
| XQUERY_POLYP_NUM | 6 | Polyp matching query | Polyp matching query |
Appendix 3 Endoscopy and Pathology Report Application
Appendix 4 Standard Operating Procedures
This appendix consists of all SOPs and related documents, as listed below.
-
Intermediate Adenoma Coding Application SOP.
-
Coding Reference Document.
-
Exclusion SOP.
-
Checking Coder Excluded Records SOP.
-
Coders Reference Document – Phantom Reports.
-
Phantom Endoscopy SOP.
-
Review SOP.
-
Polyposis and Colitis Reclassification Review SOP.
-
Examination Numbering and Multiple Row Review SOP.
-
Summary of Changes to Coding Documents (Aug 2009).
-
Summary of Changes to Coding Documents (Sep 2009).
-
Summary of Changes to Coding Documents (Oct 2009).
-
Missing Pathology Collection SOP.
-
Visit Checklist.
-
Missing Pathology Coding SOP.
-
Polyp Matching Re-queries – Coding Rules.
-
Polyp Numbering SOP.
-
March 2012 – SOP Updates and Amendments.
-
Cancer Reclassification Review SOP.
-
ONS Encryption SOP.
Appendix 5 Colitis and polyposis exclusion criteria
Colitis subtypes and exclusion criteria
| Hierarchy | Status | Condition | Time dependency | Exclude? | Diagnosis codes | Indication codes |
|---|---|---|---|---|---|---|
| 1 | Confirmed diagnosis – known subtype | Subtype: 2 – ulcerative colitis; 3 – microscopic colitis; 4 – lymphocytic colitis; 5 – collagenous colitis; 8 – ischaemic colitis; 14 – radiation colitis; 19 – indeterminate colitis; 23 – ulcerative proctitis; 25 – radiation proctitis | Prior or baseline | Yes | Diagnosis = 4 + subtypes 2, 3, 4, 5, 8, 14, 19, 23, 25 | Indication = 4 + subtypes 2, 3, 4, 5, 8, 14, 19, 23, 25 |
| 2 | Confirmed diagnosis – known subtype | Subtype: 9 – diversion colitis; 10 – infective colitis; 11 – chemical colitis; 12 – pseudomembranous colitis; 13 – drug-induced colitis; 16 – procedural/enema related; 18 – antibiotic-associated colitis; 20 – atypical colitis | Prior or baseline | No | Diagnosis = 4 + subtypes 9, 10, 11, 12, 13, 16, 18, 20 | Indication = 4 + subtypes 9, 10, 11, 12, 13, 16, 18, 20 |
| 3 | Confirmed diagnosis – possible subtype(s) | Any subtype(s) | Prior or baseline | Yes | Diagnosis = 4 + subtypes 2, 3, 4, 5, 8, 9, 10, 11, 12, 13, 14, 16, 18, 19, 20, 23, 25 | n/a |
| 4 | Confirmed diagnosis – no subtypes | No subtype | Prior or baseline | Yes | Diagnosis = 4 + subtype = missing | Indication = 4 + subtype = missing |
Polyposis subtypes and exclusion criteria
| Hierarchy | Status | Condition | Time dependency | Exclude? | Diagnosis codes | Indication codes |
|---|---|---|---|---|---|---|
| 1 | Confirmed diagnosis – known subtype | 5 – Hyperplastic polyposis; 7 – serrated polyposis; 9 – CAP polyposis | Prior or baseline | Yes | Diagnosis = 22 + subtypes 5, 7, 9 | Indication = 18 + subtypes 5, 7, 9 |
| 2 | Confirmed diagnosis – known subtype | 2 – FAP; 3 – juvenile polyposis; 4 – PJ polyposis; 12 – MAP | Any time | Yes | Diagnosis = 22 + subtypes 2, 3, 4, 12 | Indication = 18 + subtypes 2, 3, 4, 12 |
| 3 | Confirmed diagnosis – known subtype | 8 – Lymphoid polyposis | Any time | No | Diagnosis = 22 + subtype 8 | Indication = 18 + subtype 8 |
| 4 | Confirmed diagnosis – possible subtype(s) | 5 – Hyperplastic polyposis; 7 – serrated polyposis; 9 – CAP polyposis; 8 – lymphoid polyposis | Prior or baseline | Yes | Diagnosis = 22 + subtypes 5, 7, 8, 9 | n/a |
| 5 | Confirmed diagnosis – possible subtype(s) | 2 – FAP; 3 – juvenile polyposis; 4 – PJ polyposis; 12 – MAP | Any time | Yes | Diagnosis = 22 + subtypes 2, 3, 4, 12 | n/a |
| 6 | Confirmed diagnosis – no subtypes | No subtype | Any time | Yes | Diagnosis = 22 + subtype = missing | Indication = 18 + subtype = missing |
| 7 | Unconfirmed diagnosis | Any subtype or no subtype | Any time | Yes | Diagnosis = 22 | n/a |
Appendix 6 Polyp numbering
Polyp numbering
Sometimes a polyp that was found at one endoscopy was seen again at a later endoscopy. This was because the polyp was not removed or was only partially removed (i.e. de-bulked or biopsied). Alternatively, some residual polyp may have been left intact accidentally or there may have been regrowth after the polyp was thought to have been completely excised.
It was necessary to link these polyps to ensure that the same polyp was not counted as separate polyps. This process was called ‘polyp numbering’.
Numbering individual polyps
The study programmer added the POLYP_NUMBER and MATCH_PROBABILITY fields to the POLYP table and added a new screen to the Endoscopy and Pathology Report (EPR) application. Approximately 17,000 patients required manual polyp numbering. These were patients who had two or more examinations with polyps found on at least two occasions.
The study researchers reviewed these patients and all occurrences of the individual polyps seen at different examinations (including individual polyps within multiple polyp groups) were assigned the same number in the POLYP_NUMBER field and a percentage was assigned in the MATCH_PROBABILITY field to indicate the certainty that polyps were the same unique lesion. A single polyp was used as a reference from which all other possible matches were then based. All information provided in the endoscopy and pathology reports was used to identify unique polyps across all examinations. Polyp numbering guidelines were used to match the polyps accurately and methodically, with particular attention given to the following factors, listed in order of importance:
-
segment (proximity)
-
indication that polyp was not removed or was partially removed (i.e. de-bulking, discomfort)
-
excision extent/excision complete
-
bowel preparation
-
method of excision
-
size
-
dysplasia
-
adenoma type
-
histology.
It was necessary to allocate a polyp number only if the polyp was seen multiple times. This was a complex process and the study researcher took into account all the polyp fields, information from the endoscopy and pathology reports and used their own judgement to decide whether it was in fact the same polyp or a different one. Manual quality checks were done by the study researchers, who reviewed and checked a random sample of records that had been polyp numbered by other study researchers. Automatic checks were also done to identify cases with the same POLYP_NUMBER which had large difference such as sizes, segments of the polyps and so on and such records were manually reviewed and corrected where necessary.
A new field called DERIVED_POLYP_NUMBER was created in the polyp table. As described previously, POLYP_NUMBER was assigned only if the polyp was seen across other examinations. In order to analyse the data and derive other fields, it was necessary to allocate a unique number to each polyp. After manual polyp numbering was completed, analysis on the MATCH_PROBABILITY was conducted by the statistician. It was decided that only polyps with a MATCH_PROBABILITY of ≥ 70% would be considered the same lesion. The study programmer wrote a program to assign a DERIVED_POLYP_NUMBER which was a unique number for every polyp across all examinations. If the polyp had a POLYP_NUMBER assigned to it then all occurrences of that polyp were assigned the same DERIVED_POLYP_NUMBER if the MATCH_PROBABILITY was ≥ 70%. The DERIVED_POLYP_NUMBER and POLYP_NUMBER did not always match. However, if polyps had the same POLYP_NUMBER and MATCH_PROBABILITY was ≥ 70% they would all have the same DERIVED_POLYP_NUMBER.
Polyp groups
Coding multiple polyps
To simplify the concept of ‘multiple polyps’ the following definitions have been used to describe the different types of polyp rows recorded by the study researchers in the POLYP table, based on how they were observed.
-
Endo quantity row An individual polyp row was created in the POLYP table and used to record a group of polyps. The ENDO_QUANTITY_OTHER was set to values such as ‘many’, ‘several’, ‘multiple’, as described in the endoscopy or pathology report.
-
Multilinked polyp An individual polyp row was created in the POLYP table but the polyp was also part of a group of polyps (i.e. it belonged to an ‘endo quantity row’). The PATH_MULTI_ENDO_LINK would match the POLYP_ID of the ‘endo quantity row’, showing that they are part of a larger group defined in the ‘endo quantity row’.
-
Individual polyps Polyps that were not part of the group. In some cases, polyps could be recorded as an individual polyp at a particular examination, but may have been seen as part of a group at a previous or subsequent examination.
During the manual coding phase of the study, the study programmer created fields ENDO_QUANTITY_OTHER, PATH_MULTI_ENDO_LINK, MULTIPLE_POLYP_GROUP and MULTIPLE_POLYP_GROUP_LINKING in the POLYP table so that the study researchers could record this information. A group of polyps was recorded as an individual polyp row called an ‘endo quantity row’ and populated with information common to all polyps within that group such as segment, shape, histology, and so on, and the ENDO_QUANTITY_OTHER field was used to record the descriptive quantity of polyps in that group such as ‘tiny’, ‘multiple’, etc. Additional individual polyp rows were created for any polyps within the group for which additional information was available, and the POLYP_ID of the ‘endo quantity row’ was selected in the PATH_MULTI_ENDO_LINK field for these polyps to link them to the ‘endo quantity row’. The whole group (i.e. the ‘endo quantity row’), and the individual polyps linked to it, were allocated the same unique MULTIPLE_POLYP_GROUP. This method was used to record groups of polyps seen at the same examination.
For example, if the endoscopy description said ‘In the distal sigmoid colon multiple sessile polyps were found. The largest was 2 mm; 2 of these were excised and 1 was retrieved’ then one ‘endo quantity row’ would be created as shown in the record highlighted in red (Figure 31), and the ENDO_QUANTITY_OTHER would be set to ‘multiple’. Multilinked polyps would be created as per the records highlighted in green (see Figure 31) below. The records in green would be linked to the record in red and their PATH_MULTI_ENDO_LINK would match the POLYP_ID of the record in red. All the records in the group would be allocated the same MULTIPLE_POLYP_GROUP, in this case ‘Group 2’.
FIGURE 31.
Coding multiple row polyps.

When all of the groups of polyps had been recorded for a patient, the study researchers reviewed all of the groups of polyps and individual polyps that may have been observed within a group at the same or other examinations, and linked them all using the MULTIPLE_POLYP_GROUP_LINKING field.
The MULTIPLE_GROUP_LINKING field was labelled as ‘Multiple Group No.’ on the EPR application. It was used to allocate a unique group number for a set of polyps seen across many examinations. The MULTIPLE_POLYP_GROUP where the polyp group or individual polyp was first seen was used to populate the MULTIPLE_GROUP_LINKING for all observations of the polyp at different examinations.
The study researcher selected a designated group that was a MULTIPLE_POLYP_GROUP starting with ‘Group1’, identified all occurrences of the polyps within that group and allocated the same MULTIPLE_GROUP_LINKING to them. The study researcher then moved on to the next ‘endo quantity row’ for which no MULTIPLE_GROUP_LINKING had been assigned, and this became the next designated group. The following rules were used to assign the designated group to the MULTIPLE_GROUP_LINKING field for the following polyp rows:
-
All polyp rows where PATH_MULTI_ENDO_LINK matched the POLYP_ID of the ‘endo quantity row’ and where the MULTIPLE_POLYP_GROUP matched the designated group.
-
All polyp rows (groups and individual polyps) where the polyps appeared to be the same as the polyps for which the MULTIPLE_GROUP_LINKING had already been assigned and matched the designated group. The match probability was also recorded on the MULTIPLEGROUP_MATCHPROB field to show how certain the study researcher was that it was the same MULTIPLE_GROUP_LINKING.
-
All ‘endo quantity rows’ that appeared to be the same as the polyps for which the MULTIPLE_GROUP_LINKING had already been assigned and all multilinked polyps rows where PATH_MULTI_ENDO_LINK matched the POLYP_ID of the associated ‘endo quantity row’.
The following example shows how the study programmer allocated the MULTIPLE_POLYP_GROUP and MULTIPLE_POLYP_GROUP_LINKING. The study researcher allocated a unique group number to each set of polyps seen at a particular examination. The following is a screenshot from the EPR application, which shows how the MULTIPLE_POLYP_GROUP was assigned.
FIGURE 32.
Screenshot showing multiple groups and group linking.
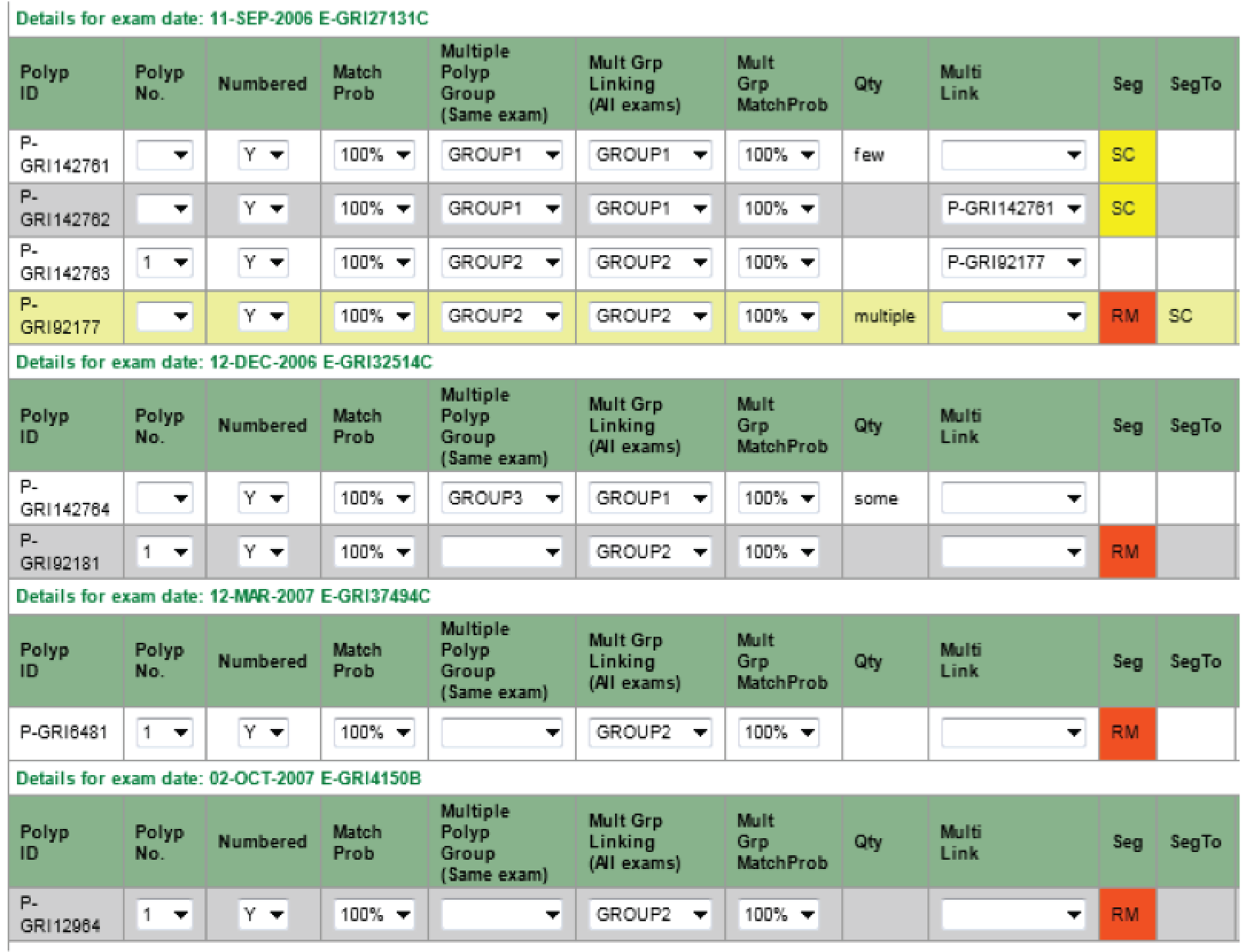
For example, two groups were identified for the examination 11-SEP-2006:
-
Polyp P-GRI142761 is the polyp with quantity ‘few’ and it is also multilinked to polyp P-GRI142762 so they were both assigned ‘GROUP1’.
-
Polyp P-GRI92177 is the polyp with quantity ‘multiple’ and it is also multilinked to polyp P-GRI142763 so they were both assigned ‘GROUP2’.
For examination 12-DEC-2006 one group was identified:
-
Polyp P-GRI142764 is the polyp with quantity ‘some’ and therefore assigned ‘GROUP3’.
Once all the polyps have been created and the MULTIPLE_POLYP_GROUP had been allocated, the MULTIPLE_GROUP_LINKING was done as follows:
-
Starting with MULTIPLE_POLYP_GROUP ‘Group1’ (designated group), the ‘endo quantity row’ and the polyps multi-linked to it were assigned ‘Group1’ on the MULTIPLE_GROUP_LINKING field. This included polyps P-GRI142761 and P-GRI142762. Any polyps that appeared to be the same as the polyps in group P-GRI142761 or polyp P-GRI142762 were also assigned ‘Group1’ in the MULTIPLE_GROUP_LINKING field (i.e. polyp P-GRI42764).
-
Moving to the next designated group ‘Group2’, polyp P-GRI92177 was the polyp with quantity ‘multiple’ and it is also multi-linked to polyp P-GRI142763, so they were both assigned ‘GROUP2’ in the MULTIPLE_GROUP_LINKING field. Polyps P-GRI92181, P-GRI6481 and P-GRI12964 all appeared to be the same polyps are the ones already in MULTIPLE_GROUP_LINKING ‘Group 2’, so they were assigned as ‘GROUP2’ in the MULTIPLE_GROUP_LINKING field.
Multiplying out the endo quantity rows
The process of estimating the number of polyps within an ‘endo quantity row’ and separating them out into individual polyps is known as ‘multiplying out the endo quantity row’. The method to calculate this took into account all of the different scenarios in which a polyp is observed across many examinations. A summary table called ‘DERIVED_MP_SUMMARY’ was generated, which contained the POLYP_ID, STUDY_NUMBER, MULTIPLE_POLYP_GROUP, MULTIPLE_GROUP_LINKING of each ‘endo_quantity_row’ and the following fields (some were derived):
-
APPROX_QTY Approximate quantity of polyps recorded by the study researcher.
-
MIN_QTY Minimum quantity of polyps in the group recorded by the study researcher.
-
MAX_QTY Maximum quantity of polyps in the group recorded by the study researcher.
-
TOTAL_POLYPS The total number of unique polyps seen for the ‘endo quantity row’ across all examinations. It was derived by taking into account number of multi-linked polyps, the number of unique polyps with the same MULTIPLE_GROUP_LINKING seen at other examinations, and deducting any polyps with the same MULTIPLE_GROUP_LINKING that had been excised prior to the procedure date of the ‘endo quantity row’.
-
SAMEEXAMPOLYPS The total number of multi-linked polyps at the same examination for the ‘endo_quantity_row’.
-
ESTIMATED_QUANTITY The ‘ENDO_QUANTITY_OTHER’ field contained categories like few, some, several and so on. It was necessary to translate these categories into values for analysis. The statistician conducted an analysis by comparing all of the records for which the ‘ENDO_QUANTITY_OTHER’, ‘MIN_QTY’ and ‘MAX_QTY’ fields had been recorded. The results of the analysis were discussed by study researchers in order to arrive at an ESTIMATED_QUANTITY for each option. This provided an indication of the number of records in the group especially when there was no other indication of quantity. The following quantities were assigned to each option.
| ENDO_QUANTITY_OTHER | ESTIMATED_QUANTITY |
|---|---|
| Few | 3 |
| Some | 3 |
| Number of | 3 |
| Several | 3 |
| Many | 5 |
| Multiple | 5 |
-
Use the largest of TOTAL_POLYPS, SAME_EXAM_POLYPS and MAX_QTY if MAX_QTY > 0.
-
Use the largest of MIN_QTY, ESTIMATED_QUANTITY, SAME_EXAM_POLYPS and TOTAL_POLYPS if MIN_QTY > 0 and if APPROX_QTY and MAX_QTY are blank.
-
Use the largest of, ESTIMATED_QUANTITY, SAME_EXAM_POLYPS and TOTAL_POLYPS if the TOTAL_POLYPS > 0 or SAME_EXAM_POLYPS > 0.
-
MAX_ROWS_DATASOURCE This was used to record the field assigned to the MAX_ROWS based on the above rules.
-
ROWSADD This was a derived field and was the total number of polyps that would be added to the ‘endo quantity row’, and included the number of new polyps that are created from polyps observed at other examinations and also completely new polyps that were not observed but estimated based on the information available.
The following rules were used to calculate the ROWSADD field:
-
Sometimes the ‘endo quantity row’ was marked as partially excised i.e. EXCISION_EXTENT = 2. In this scenario, the study researchers thought that there may have been more polyps than the ones multilinked to the ENDO QUANTITY ROW, so the study programmer took a conservative estimate and added one row in such cases:
-
If the MAXROWS = SAMEEXAMPOLYPS so ROWSADD = 0
-
If the MAXROWS = SAMEEXAMPOLYPS and EXCISION_EXTENT <> 2 then ROWSADD = 0
-
-
If the POLYPCOUNTGROUP_OTHERS > 0 and the MAX_ROWS_DATASOURCE = ’TOTAL_POLYPS’ then ROWSADD = MAXROWS – (SAMEEXAMPOLYPS + POLYPCOUNTGROUP_OTHERS). This ensured that that any polyps that already existed within that examination were excluded as the ‘TOTAL_POLYPS’ included them. Otherwise, the ROWSADD = MAXROWS – (SAMEEXAMPOLYPS), as all of the other fields included the SAMEEXAMPOLYPS in their total.
-
ROWSADD_OVERRIDE This override was applied only for cases for which there were more than one ‘endo quantity rows’ at the same examination with the same MULTIPLE_GROUP_LINKING. If the MAX_ROWS_DATASOURCE = ’TOTAL_POLYPS’ for the more than one ‘endo quantity rows’ with the same MULTIPLE_GROUP_LINKING at the same examination then it was important to ensure that a ratio was used to divide the TOTAL_POLYPS among those ‘endo quantity rows’ after deducting any multilinked polyps. The program calculated the ‘TOTAL_POLYPS’ minus POLYPCOUNTGROUP and then divided that based on ESTIMATED_QUANTITY.
Creating the DERIVED_MP_POLYPS table
The process of separating out the ‘endo quantity rows’ was based on estimates so it was decided that a new table called DERIVED_MP_POLYPS would be used for this without changing any of the data on the POLYP table. The table DERIVED_MP_POLYPS was created, which contained a copy of the POLYP table for all patients who had at least one ‘endo quantity row’. For patients with ‘endo quantity rows’, this table was used as the master data set for polyp information. The following rules were applied for this migration of data:
-
The MULTIPLE_GROUP_LINKING was set to null if the group match probability was < 70%.
-
If PROCEDURE_DATE was blank then it was populated from the endoscopy or pathology table.
-
The following fields recorded on the ‘endo quantity row’ were copied to the multilinked polyps in sets unless the fields on the multilinked polyp already had some data in at least one of the values in the set.
-
EXCISION_EXTENT = 1
-
PATH_HISTOLOGY, PATH_DYSPLASIA, PATH_ADENOMA_TYPE, SERRATION, ASSUME_PATH_HISTOLOGY
-
FATE_OF_BIOPSY and REMOVAL_METHOD
-
ENDO_SHAPE, ENDO_SHAPE_OLD and PATH_SHAPE
-
ENDO_SEGMENT, ENDO_SEGMENT_TO, ENDO_SEGMENT_OLD and ENDO_DISTANCE
-
ENDO_SIZE, ENDO_SIZE_OTHER, ASSUME_ENDO_SIZE and ASSUME_ENDO_SIZE_OTHER
-
PATH_SIZE and ASSUME_PATH_SIZE
-
ENDO_SIZE_MAX and ENDO_SIZE_MIN
-
CLINICAL_HISTOLOGY
-
EXCISION_COMPLETE
-
HOSPITAL
-
Creating new polyp rows from existing polyp rows
An ‘endo quantity row’ contained groups of polyps but some of the polyps may have been recorded as individual or multilinked polyps at prior or subsequent examinations. It was important to ensure that any new polyps created from existing polyps were also assigned the same DERIVED_POLYP_NUMBER and MULTIPLE_GROUP_LINKING to ensure that they were linked.
The new polyps were also multilinked to the ‘endo_quantity_row’. A new field called NEW_POLYP_STATUS was also created and assigned the value ‘EXISTING POLYP’ to indicate that the row was created based on an existing polyp.
Any existing polyps at previous or subsequent examinations that met the following criteria were used to create the new polyps:
-
The MULTIPLE_GROUP_LINKING of the ‘endo quantity row’ matched the MULTIPLE_GROUP_LINKING of the existing polyp at prior or subsequent examination.
-
The existing polyp did not have the same ENDO_ID as the ‘endo quantity row’.
-
If the existing polyp has EXCISION_EXTENT = 1 or EXCISION_COMPLETE = 1 and if the polyp was seen at an examination prior to the examination of the ‘endo quantity row’ then it was not used. However, the EXCISION information was not always reported properly. Therefore, if the polyp was seen again later then it was used.
-
If there was an existing polyp with the same DERIVED_POLYP_NUMBER as another polyp recorded on the same examination as the ‘endo_quantity_row’ then it was not used.
-
Sometimes the existing polyp was a new polyp created at a prior examination when the ‘endo quantity row’ of that examination was separated into rows. In such a case it was assumed then it was the same polyp seen again (unless excised previously) and it was allocated the same DERIVED_POLYP_NUMBER. This ensured that the numbers of polyps were not overestimated.
If the polyp was created from an existing polyp then only the DERIVED_POLYP_NUMBER and MULTIPLE_GROUP_LINKING were used from the existing POLYP to create the new polyp. It would have been incorrect to copy all of the information from the existing polyps, as the polyp data could change over the course of the examinations. It was deemed more appropriate to use the information from ‘endo quantity row’ as the information recorded against it was applied to all polyps in the group. It was also agreed that when the statistician would derive the true values for polyps across all examinations and the data on actual observed polyps would take precedence over the new polyps created.
The following fields are copied from the ‘endo quantity row’ as they were common to all polyps in the group: MULTIPLE_POLYP_GROUP, STUDY_NUMBER, ENDO_ID, PROCEDURE_DATE, CLINICAL_HISTOLOGY, PATH_HISTOLOGY, PATH_DYSPLASIA, PATH_ADENOMA_TYPE, SERRATION, ASSUME_PATH_HISTOLOGY, FATE_OF_BIOPSY, REMOVAL_METHOD, ENDO_SHAPE, ENDO_SHAPE_OLD, PATH_SHAPE, ENDO_SEGMENT_TO, ENDO_SEGMENT_OLD, ENDO_DISTANCE, ENDO_SEGMENT, EXCISION_COMPLETE, HOSPITAL, ENDO_SIZE, ENDO_SIZE_OTHER, ASSUME_ENDO_SIZE, ASSUME_ENDO_SIZE_OTHER, PATH_SIZE, ENDO_SIZE_MAX, ENDO_SIZE_MIN. The EXCISION_EXTENT was copied only if it was set to 1. The POLYP_ID of the ‘endo quantity row’ was copied to the PATH_MULTI_ENDO_LINK of the new polyp.
The ‘POLYP_ID’ was generated from the POLYP_ID of the ‘endo quantity row’ and by appending ‘-V’ followed by a unique number for the new polyp. This showed that it was multiplied out from the ‘endo quantity row’.
Creating new polyp rows
The number of rows created from existing polyps was deducted from ROWSADD or ROWSADD_OVERRIDE to calculate the number of completely new polyps to be added. Based on this, new polyps were created where necessary.
A new unique DERIVED_POLYP_NUMBER was generated for every new polyp created. The following fields are copied from the ENDO QUANTITY ROW: MULTIPLE_POLYP_GROUP, MULTIPLE_GROUP_LINKING, MULTIPLEGROUP_MATCHPROB, STUDY_NUMBER, ENDO_ID, PROCEDURE_DATE, CLINICAL_HISTOLOGY, PATH_HISTOLOGY, PATH_DYSPLASIA, PATH_ADENOMA_TYPE, SERRATION, ASSUME_PATH_HISTOLOGY, FATE_OF_BIOPSY, REMOVAL_METHOD, ENDO_SHAPE, ENDO_SHAPE_OLD, PATH_SHAPE, ENDO_SEGMENT_TO, ENDO_SEGMENT_OLD, ENDO_DISTANCE, ENDO_SEGMENT, EXCISION_COMPLETE, HOSPITAL, ENDO_SIZE, ENDO_SIZE_OTHER, ASSUME_ENDO_SIZE, ASSUME_ENDO_SIZE_OTHER, PATH_SIZE, ENDO_SIZE_MAX, ENDO_SIZE_MIN. The EXCISION_EXTENT was only copied if it is set to 1. The POLYP_ID of the ‘endo quantity row’ was copied to the PATH_MULTI_ENDO_LINK of the new polyp.
The polyp ID was created from the POLYP_ID of the ENDO QUANTITY ROW by appending ‘-V’ followed by a unique number for the new polyps. This showed that it was multiplied out from the ENDO QUANTITY ROW.
When the fields such as TOTAL_POLYPS were derived, the figure was based on observed polyps. However, it did not take into account any new polyps created at every examination, based on our estimates and calculations. Factoring this into the algorithm would have made it very complex and it was very likely the number of polyps would have been overestimated. It was decided that the estimates would be based on actual polyps observed and would not include any new polyp rows created as a result of multiplying out rows.
Appendix 7 Rules applied for data analysis
Rules defined before data were received by statistician
Multiple polyp group numbering and multiplication out of rows
Sometimes, an endoscopist referred to the polyps seen at an examination in a vague manner, meaning that it was unclear precisely how many polyps were seen. All details available for a group of polyps were coded within a single ‘Multiple polyp row’ to deal with this issue. Coders would set the polyp field ‘endo quantity other’ to a value such as multiple, some or few, depending on the report, so that the single polyp row could represent a group of polyps. Such polyp groups were termed ‘endo quantity rows’. Additional fields were used to indicate the number of polyps that the endo quantity row represented, where such information was available; this included minimum, maximum and approximate number of polyps. In addition to a multiple polyp row, an individual may have had other lesions. Furthermore, in some cases it was possible to glean more in-depth polyp details from the pathology, some of which related to individual polyps within the multiple polyp row. A polyp row would be added to record these specific polyp details and then that row would be ‘multilinked’ to the endo quantity row to demonstrate that it was part of the group of polyps described at endoscopy.
All polyps found at an examination with a multiple polyp row were termed ‘multiple row groups’. Within a multiple row group there may be:
-
Endo quantity rows Polyps with endo quantity other value (i.e. many, several, multiple).
-
Multilinked polyps Polyp rows containing further details on specific polyps of the endo quantity row which are linked to the endo quantity row by the polyp_id of the endo quantity row (i.e. polyps at the same examination).
-
Individual linked polyps Polyps within the endo quantity rows that are seen at other examinations. They will be within the same linking group (MULTIPLE_GROUP_LINKING) as explained later.
-
Individual polyps Polyps that are never part of the group.
When the initial polyp numbering was done, coders used the polyp numbering field to associate groups of polyps to other groups, as well as to individual polyps. It was therefore difficult to identify whether or not individual polyps within groups found at different examinations were the same. As a result, two fields were created: MULTIPLE_POLYP_GROUP (Multiple Polyp Group at the same examination) was used to allocate a unique group number to each set of polyps seen at a particular examination, and MULTIPLE_GROUP_LINKING (Multiple Group No – across all examinations) was used to allocate a unique group number for a set of polyps seen across many examinations.
Some automatic coding was performed. A unique group number was allocated to each set of polyps seen at a particular examination for the MULTIPLE_POLYP_GROUP field. For the MULTIPLE_GROUP_LINKING, a unique group number was allocated for a set of polyps seen across many examinations and in most cases it was associated with the multiple polyp group where the polyp set was seen first time. The program iterated through each MULTIPLE_POLYP_GROUP starting with GROUP1 and grouped all polyps within that set which had been seen at any examination and allocated the MULTIPLE_GROUP_LINKING number. The following rules were used to associate individual polyps to groups.
Any polyps:
-
that were ‘multilinked’ to the group
-
with the same polyp number to the first group they were seen
-
with the same polyp number as another multiple row that had linked polyps.
The polyp number was reset for all polyps that were part of a multiple polyp group and the data clerks then had to identify individual polyps that were part of the same MULTIPLE_GROUP_LINKING group across all examinations, and allocate the same polyp number to them.
Within a ‘Multiple row group’ there may be:
-
Endo quantity rows (e.g. many, several, multiple).
-
Multilinked polyps – polyp rows containing further details on specific polyps of the ‘endo quantity row’ which are linked by the polyp_id (i.e. polyps at the same examination).
-
Individual linked polyps – polyps within the ‘endo quantity rows’ that are seen at other examinations.
-
Individual polyps – polyps that are never part of the group.
It was decided that in order to analyse these records, each endo quantity row had to be ‘multiplied out’ into single polyp rows, and the data available and some rules were used to calculate the number of polyp rows to be added. If the group of polyps was linked to groups that had been seen previously and not fully excised then a method was used to ensure that the same polyp number was allocated to those polyps to show the reoccurrence. The information recorded on the endo quantity row (such as segment, histology, size, etc.) was also copied over to the new polyps as it applied to all the polyps in the group.
A variable called ‘total polyps’, which was the number of unique polyps seen for the endo quantity rows across all examinations was calculated using multilinked polyps, individual linked polyps and derived_polyp_number. The variable ‘same examination polyps’ field was calculated to provide the total number of multilinked polyps present at the same examination.
First, the following estimated quantities were assigned based on the endo quantity other field:
| ENDO_QUANTITY_OTHER | ESTIMATED_QUANTITY |
|---|---|
| Few | 3 |
| Some | 3 |
| Number of | 3 |
| Several | 3 |
| Many | 5 |
| Multiple | 5 |
The following hierarchy was then used to calculate the maximum number of polyps ‘max rows’ that made up the multiple rows.
Use:
-
the largest of total polyps, same examination polyps and approximate quantity only if approximate quantity > 0
-
the largest of total polyps, same examination polyps and ‘average of minimum quantity and maximum quantity‘ only if minimum quantity > 0 and maximum quantity > 0
-
the largest of total polyps, same examination polyps and maximum quantity if maximum quantity > 0
-
the largest of minimum quantity, estimated quantity, same examination polyps and total polyps if minimum quantity > 0 and if approximate quantity and maximum quantity are blank
-
the largest of estimated quantity, same examination polyps and total polyps if the total polyps > 0 or same examination polyps > 0
-
the maximum of estimated quantity.
The ‘max rows’ were then used to assess how many new rows should be created. The following rules were used:
-
If max rows = same examination polyps and excision extent is ‘partially excised’ then add one new polyp row.
-
If max rows = same examination polyps and excision extent is not ‘partially excised’ then no rows to add.
-
Otherwise new polyp rows = (max rows – same examination polyps).
Derived examination date created
Endoscopy and pathology reports sometimes had different types of date associated with them. In addition, where the endoscopy report was missing, a pathology based procedure report was created and the date of procedure was based on the pathology report. In order to define a true ‘examination date’ for every procedure, these dates were put into the following order of precedence, from most to least important.
Date:
-
of procedure
-
of collection of biopsy specimen
-
on which biopsy specimen was received at laboratory
-
on which report was written by pathologist.
Derived examination numbers created
When it was unclear what order a patient’s examinations occurred in, manual examination numbering was done by coders. This was necessary for patients with one or more missing examination dates or with two or more procedures on the same date. Following manual examination numbering, a ‘derived_exam_number’ was then created to order all remaining examinations, accounting for any examination numbering that was assigned by coders. It was necessary to understand the order in which examinations occurred in to be able to group them into visits and accurately assign risk groups.
Derived polyp segments created
Sometimes the segment recorded across sightings of the same unique polyp varied. For example, at one examination the polyp may be described as a sigmoid lesion, whereas at another it may be in the rectum. In order to define a true polyp segment, two derived polyp segment fields were created. The ‘derived_polyp_segment’ field contained either the segment the polyp was found in or, if a range of segments was given, the most distal segment. If a range of segments was recorded, ‘derived_polyp_segment_to’ contained the most proximal segment.
Derived polyp numbering created
Sometimes a polyp that was found at one endoscopy was seen again at a later endoscopy. This was because the polyp was not removed or was only partially removed (i.e. de-bulked or biopsied). Alternatively, some residual polyp may have been left intact or there may have been re-growth after presumed complete excision. A number of patients required manual polyp numbering in order to ensure that a unique polyp was not counted more than once. A ‘match probability’ was used to indicate the degree of certainty that matched polyps were the same unique lesion. A single polyp was used as a reference from which all other possible matches were then based. All information provided in the endoscopy and pathology reports was used to identify unique polyps that needed to be matched. Polyp numbering guidelines were used to match polyps accurately and methodically, with particular attention given to the following factors, listed in order of importance:
-
segment (proximity)
-
length of time passed between examinations
-
indication that polyp was not removed or was partially removed (i.e. de-bulking, discomfort)
-
excision extent/excision complete
-
bowel preparation
-
method of excision
-
size
-
dysplasia
-
adenoma type
-
histology.
After manual polyp numbering was completed, a cut-off of 70% was chosen following a review of the data, meaning that, for each patient, polyps numbered the same with a match probability of ≥ 70% were considered to be the same lesion. All other polyps in that patient were then assigned a unique number that was stored in the ‘derived polyp number’ field.
Three derived polyp size fields created
On the study database, size was recorded using a number of fields in order to accommodate the different types of information relating to polyp size that were supplied in endoscopy and pathology reports. The following endoscopy and pathology sizes were recorded on the database: assume_endo_size, endo_size, endo_size_min, endo_size_max, endo_size_other, assume_endo_size_other, path_size and assume_path_size. In addition to these, eight new fields were created on the polyp table to record derived polyp sizes that were based on the original size fields. There were three main derived size fields: derived_endo_size, derived_endo_size_other and derived_path_size, which were used to define true polyp size [see Rules applied after data compilation, below (rule 14: True values for polyps found during and prior to baseline, part A – true values]. All of the derived fields are described below:
-
derived_endo_size – derived from the fields assume_endo_size, endo_size, endo_size_min, endo_size_max and derived_endo_range (assume_endo_size took precedence over endo_size)
-
derived_endo_size_source – shows which field the size was taken from
-
derived_endo_size_other – derived from the fields assume_endo_size_other and endo_size_other (assume_endo_size_other took precedence)
-
derived_endosize_other_source – shows which field the size was taken from
-
derived_path_size – derived from the fields assume_path_size and path_size (assume_path_size took precedence)
-
derived_path_size_source – shows which field the size was taken from
-
derived_endo_range – complex field to deal with polyp groups, see rules below.
The rules used to define these derived sizes were numerous and complex, particularly for derived_endo_size.
Endoscopy sizes
The derived_endo_size was assigned using an algorithm that took into consideration assume_endo_size, endo_size, endo_size_min and endo_size_maximum. It was particularly difficult to define the use of endo_size_max and endo_size_min in the algorithm.
For all cases where endo_size_max was the only size recorded, it was suggested that no size should be assigned, as this field was thought to be too unreliable. It was problematic to assign a derived_endo_size to polyps that only had an endo_size_max and no other size because when an endoscopist used endo_size_max it was often to describe the sizes of a group of polyps. Thus, endo_size_max may have truly applied to only one of the polyps in a group, resulting in an inaccurate true polyp size being assigned to other lesions in the group, if it were used. This would then lead to such polyps being classified as higher risk than they really were. If endo_size_max was ≤ 5 mm then it was assumed to be the correct size, as the size was only small. In cases for which there was only one polyp, it was also assumed that the polyp’s size was endo_size_maximum.
Alternatively, in some cases, a polyp had already been assigned endo_size_max as an actual endo_size; if a patient had endo_size_max applied to a group of polyps and an individual polyp at that examination had an actual endo_size identical to the endo_size_max, and the polyps were all in the same segment or segment range or had no segment, then it was assumed that an individual polyp had already been assigned the maximum size and thus the other polyps in the group were deemed to be of an ‘unknown’ size. This was done automatically (see derived endo range). Coders reviewed the remaining cases for which a group of polyps had only endo_size_max in order to determine which polyp was the largest in the group. Others in the group were then be deemed to be of an ‘unknown’ size.
Some polyps had only endo_size_min and endo_size_max available to assign derived_endo_size. These polyps were also problematic, as it was not appropriate to assign a true polyp size by calculating an average of the two sizes, particularly in cases for which the endo_size_min and endo_size_max differed considerably. As with endo_size_max, the endo_size_min and endo_size_max fields tended to be used to when a group of polyps was seen at endoscopy. There were two particular scenarios: scenario A occurred when an endoscopy report provided specific size (and site) details for individual polyps; however, a size range was used by coders so that pathology could be assigned to a polyp, as the pathology report did not give sufficient detail to determine to which polyp the histology belonged. Scenario B arose when the endoscopy report gave only broad details of size (and site) for a group of polyps, so a size range had to be used because no individual polyp details were given, for example 10 polyps of between 5 and 25 mm. Polyps that fell into each scenario had to be reviewed manually to try and assign more specific sizes, wherever possible.
For scenario A
-
When endoscopy reports gave the actual size (and site) of individual polyps, the specific size and site details were put into to the actual_endo_size (and segment) field.
In order to assign histology to specific polyps, rules were devised and it was proposed that an additional field called ‘endo path mapping’ was added. Using this new field, the specific rule used to assign the histology could be set for any polyps that were assumed to be related to a specific histological description within the pathology report.
Pathology was assigned to a specific polyp using the following criteria:
-
Histology most likely to be associated with a large polyp was assigned to the largest lesion. Specifically, the largest polyp was villous, severely dysplastic, tubulovillous and, finally, mild/moderately dysplastic (in order of histological features most predictive of largest size).
-
In general, hyperplastic polyp pathology was assigned to the most distal lesion but only if this lesion was < 5 mm.
-
Excision extent – polyps that were snared were assumed to be larger than those that were hot biopsied.
-
When the rules above could not be used then it was assumed that specimen labels (i.e. 1–10/A-E, went from the most proximal to most distal site).
For scenario B
-
When the endoscopy report only gave a range of sizes and segments for a group of polyps, coders tried to assign one lesion the smallest size and one the largest, using the same rules for assuming histology.
-
In cases for which there were only two polyps, this resolved size for both polyps. In cases for which there were more than two polyps, coders tried to determine an assumed size for other polyps in the group.
-
If it was not possible to assign the smallest and largest size, then coders tried to assign one lesion the largest size.
-
If it was impossible to assume specific size for any polyps then size ranges were left. The sizes of polyps that were not assigned an assumed endo size were deemed ‘unknown’.
Specifically, the following records were reviewed:
-
endo_size_min is < 10 mm, endo_size_max is ≥ 10 mm AND sizes differ by ≥ 3 mm
-
endo_size_min is ≥ 10 mm, endo_size_max is ≥ 15 mm AND sizes differ by ≥ 5 mm
-
endo_size_min is ≤ 5 mm, endo_size_max is 6–9 mm AND sizes differ by ≥ 2 mm
-
endo_size_min is > 5 mm, endo_size_max is ≤ 9 mm AND sizes differ by ≥ 3 mm.
For cases that were not reviewed, an average was assigned to all polyps for which endo_size_min and endo_size_max did not differ considerably, whereas ‘unknown’ was assigned to all polyps in the review that were not assigned an assumed size. An average was also assigned to any cases with just one polyp with endo_size_min and endo_size_max.
Derived endo range
The derived_endo_range field was used to automatically derive sizes for polyp groups (polyps found at the same examination) made up of polyps with an endo_size_min or endo_size_max and another polyp(s) with the same endo_size or assume_endo_size as the endo_size_max of the other polyps in the group, all of which had the same segment. The segments and sizes had to match in order to assign the derived_endo_range, as otherwise one could not be sure that all of the polyps were part of the same group. It was not possible to calculate a derived_endo_range for polyps with both endo_size_min and endo_size_max, or in cases where the segments of the polyps in the group were different. Cleaning incident 404 was created to review such records manually, and, once reviewed, the derived_endo_range was set to blank. The rules used for derived_endo_range were as follows.
-
Group A1: Polyp with an endo_size_max of ≤ 5 mm and no endo_size_min. The derived_endo_range was set to endo_size_max.
-
Group A2: Polyp with an endo_size_min of ≤ 5 mm and no endo_size_max. The derived_endo_range was set to endo_size_min.
-
Group X1: Polyp with an endo_size_min and endo_size_max that both matched the assume_endo_size or endo_size of polyps at the same endoscopy with the same segment. Such polyps were allocated their assume_endo_size or endo_size and the other polyps with that range were allocated ‘unknown size’ as the derived_endo_range.
-
Group Y1: Polyp with an endo_size_max that matched the assume_endo_size or endo_size of another polyp at the same examination, but the segments did not match. The record was reviewed manually. The derived_endo_range was set to assume_endo_size or endo_size for the polyp that had this information and ‘unknown’ for the other polyps at the endoscopy with the same endo_size_max and no assume_endo_size or endo_size and any segment.
-
Groups B, C and D set the derived_endo_range to endo_size for the main polyp and ‘unknown’ for the rest of the group when there was only endo_size_max (not endo_size_min) and it matched the endo_size of the main polyp in the group.
-
B was applied for groups when just endo_segment for all polyps in the set.
-
C was applied for groups when endo_segment and endo_segment_to were recorded for all polyps in the set.
-
D was applied for groups when neither endo_segment and endo_segment_to were recorded for all polyps in the set.
-
-
Groups E, F and G set the derived_endo_range to endo_size for the main polyp and ‘unknown’ for the rest of the group when there was only endo_size_min (not endo_size_max) and it matched the endo_size of the main polyp in the group.
-
E was applied for groups when just endo_segment was recorded.
-
F was applied for groups when endo_segment and endo_segment_to were recorded.
-
G was applied for groups when neither endo_segment and endo_segment_to were recorded.
-
-
Groups H, I and J set the derived_endo_range to assume_endo_size for the main polyp and ‘unknown’ for the rest of the group when there was only endo_size_max (not endo_size_min) and it matched the assume_endo_size of the main polyp in the group.
-
H was applied for groups when just endo_segment was recorded.
-
I was applied for groups when endo_segment and endo_segment_to were recorded.
-
J is applied for groups when both endo_segment and endo_segment_to are blank for all polyps in the set.
-
-
Groups K, L and M set the derived_endo_range to assume_endo_size for the main polyp and ‘unknown’ for the rest of the group when there was only endo_size_min (not endo_size_max) and it matched the assume_endo_size of the main polyp in the group.
-
K was applied for groups when just endo_segment was recorded.
-
L was applied for groups when endo_segment and endo_segment_to were recorded.
-
M was applied for groups when neither endo_segment and endo_segment_to were recorded.
-
-
The derived_endo_range was set to blank if the group had been reviewed as part of incident 404.
Size discrepancies
Although work on polyp size was being undertaken, cases were identified that had large discrepancies between the sizes recorded in different fields, at the same examination and at different examinations (for a unique polyp). Although most discrepancies seemed to be due to different measuring techniques used by endoscopists and pathologists, some reports had a coding or transcription error/typo and one of the sizes could be amended. As 10 mm is an important cut-off point at which a polyp becomes classified as high risk, for discrepancies at the same examination there was a manual review of cases for:
-
endo_size 10–20 mm and path_size ≥ 40 mm
-
endo_size < 10 mm and path_size ≥ 20 mm
-
endo_size_other = tiny, small, < 5 mm, 5–9 mm, ≤ 10 mm and pathology size ≥ 20 mm.
At different examinations the following were reviewed (polyps matched with a probability of ≥ 70%):
-
endo_size < 10 mm at any examination and path_size ≥ 30 mm at any subsequent examination
-
endo_size < 10 mm at any examination and endo_size ≥ 30 mm at any subsequent examination
-
endo_size 10–20 mm at any examination and path_size ≥ 40 mm at any subsequent examination
-
endo_size 10–20 mm at any examination and endo_size ≥ 40 mm at any subsequent examination
-
endo_size_other = tiny, small, < 5 mm, 5–9 mm, ≤ 10 mm at any examination and endo_size or path_size ≥ 30 mm at any subsequent examination.
Two new fields were added to the database to accommodate the review: ‘assumed endo size’ and ‘assumed path size’. The correct ‘assumed’ size was coded in the appropriate field, which took precedence over endo_size or path_size. When doing the review, in order to identify and resolve the size errors, particular attention was paid to the excision method (i.e. was the method most viable for the removal of a large polyp or a small polyp) and the follow-up and patient’s history in general.
Additionally, for certain centres it seemed that endoscopy size was always recorded using the term ‘max endo size’, so these cases were identified and reviewed in order for the size to be correctly assigned to the endoscopy size field instead.
Derived endoscopy size
Once the derived_endo_range was generated and size discrepancies were reviewed and corrected where necessary, rules were applied to obtain the derived_endo_size. The first set of rules were applied to any polyps that were not a quantity row (i.e. endo_quantity_other has a value) or a multilinked polyp (i.e. path_multi_endo_link has been set).
-
Assume_endo_size took precedence over all endoscopy sizes, as this was field used to indicate any corrected sizes that should be used.
-
Endo_size took precedence over all endoscopy sizes except assume endo size, as this was deemed to be the next most accurate size available.
-
The automated derived_endo_range took precedence over all endoscopy sizes except assume_endo_size and endo_size, as this field took account of issues surrounding endo_size_min and endo_size_max and the way in which they were used to describe groups of polyps.
-
If there was only one polyp at an examination with endo_size_max and no other size then derived_endo_size must use this value.
-
If there was only one polyp at an examination with endo_size_min and no other size then derived_endo_size must use this value.
-
If there was only one polyp at an examination with just endo_size_min and endo_size_max then the average of endo_size_min and endo_size_max was used, as it was clear that this applied to only the one polyp and not to a group.
-
A polyp with only endo_size_min, and endo_size_max which was not reviewed, was applied an average of endo_size_min endo_size_min.
-
A polyp that was reviewed and did not have endo_size and assume_endo_size was set as Unknown.
-
For a polyp that was reviewed and did not yet have derived_endo_size, which had no endo_size_min and endo_size_max ≤ 5, the derived_endo_size was set to the endo_size_max.
-
For a polyp which was reviewed and did not yet have derived_endo_size, which had no endo_size_max and endo_size_min ≤ 5, the derived_endo_size was set to the endo_size_min.
-
A polyp that did not yet have a derived endo size and was not in IA Cohort was not allocated a size. This was because, in order to classify derived_endo_size for such cases, all patients would have to be reviewed under incident 404E and 404F, not just those in the IA cohort.
Slightly different rules were applied to patients with multiple polyps (patients with a ‘quantity other’ or multilinking, when an individual polyp row represents more than one polyp). A mp_set (multiple polyp set) was defined as a group of polyps at the same examination, which was made up of a polyp with a quantity row (i.e. endo_quantity_other has a value) and another group of polyps that were multilinked to it (i.e. path_multi_endo_link of these polyps is equal to the polyp_id of the quantity row). The following additional rules were also applied to this group.
-
Allocated the endo_size_min as derived_endo_size if the polyp was not a quantity row or a multilinked polyp. The derived_endo_size was a not yet set and the endo_size_max was null. The endo_size_min did not match any endo_size, assume_endo_size or endo_size_min of another polyp at the same endoscopy.
-
Allocated the endo_size_max as derived_endo_size if the polyp was not a quantity row or a multilinked polyp. The derived_endo_size as a not yet set and the endo_size_min was null. The endo_size_max did not match any endo_size, assume_endo_size or endo_size_max of another polyp at the same endoscopy.
-
Allocated the average of endo_size_min and endo_size_max as derived_endo_size if the polyp was not a quantity row or a multilinked polyp. The derived_endo_size was not yet set and there were no other polyps at the same endoscopy.
-
Allocated the average of endo_size_min and endo_size_max as derived_endo_size to all polyps when size was ≤ 5 except for any polyps that already had a minimum/maximum allocated. Records with unknown size were overwritten when this applied (derived_endo_range remained as unknown so it is clear when this was done).
-
Derived_endo_size was set to endo_size_min when there was only endo_size_min and it was ≤ 5. This size was allocated to all polyps in the group unless they already had a derived value. Records with unknown size were overwritten when this applied (derived_endo_range remained as unknown so it is clear when this was done).
-
Derived_endo_size was set to enod_size_max where there was only endo_size_max and it was ≤ 5. This size was allocated to all polyps in the group unless they already had a derived value. Records with unknown size were overwritten when this applied (derived_endo_range remained as unknown so it is clear when this was done).
-
An average of endo_size_min and endo_size_max was allocated to all polyps that were still without derived_endo_size, irrespective of the magnitude of these sizes, except for any polyps that already had a minimum/maximum allocated. It did not overwrite records with ‘unknown’ when this applied.
Derived endoscopy size other
Derived_endo_size_other was derived from the fields’ assume_endo_size_other and endo_size_other with assume_endo_size_other always taking precedence over endo_size_other, as this field was used to code corrected ‘assumed’ sizes. Derived_endosize_other_source shows the field from which the size was taken.
Derived pathology size
Derived_path_size was derived from the fields’ assume_path_size and path_size with assume_path_size always taking precedence over path_size, as this field was used to code corrected ‘assumed’ sizes. Derived_path_size_source shows the field from which the size was taken.
Data compilation in Stata
Dropped patients without endoscopies
Patients who did not have any endoscopies present on the study database were removed before analysis, as there was no way to ensure that the patient had a colonoscopy. As such, their baseline risk could not have been calculated with much accuracy, as there was no way to guarantee that their entire colon had been examined. Additionally, important clinical information may have been missing, for example genetic predisposition to CRC or FAP. Such patients would originally have had some form of endoscopic procedure on the database otherwise their pathology report(s) would not have been extracted from the hospital; however, this was then removed at a later date, most likely because it was for an upper gastrointestinal procedure.
Dropped auto-excluded patients
Some patients were auto-excluded prior to manual coding based on words found within their endoscopy or pathology reports, such as cancer, FAP or HNPCC. These patients were dropped from the analysis, as they had not been coded.
Rules applied after data compilation
-
Numeric endoscopy size other.
-
Derived procedure type.
-
Derived pathology histology.
-
Relabel as adenomas based on villousness/dysplasia.
-
Choose patients with one or more adenomas found.
-
Exclude any patients with missing examination dates.
-
Relabel any ≥ 10-mm polyps without histology as assume adenoma.
-
Apply baseline rules: define prior, baseline, follow-up.
-
Extend baseline.
-
Redefine baseline for patients without baseline colonoscopy.
-
Cancer matching rules.
-
Apply exclusions for conditions.
-
Adding in situ cancers and recoding histology in relation to cancer/in situ cancer.
-
True values for polyps found during, and prior to, baseline.
-
True values to be applied across all sightings of polyps.
-
Baseline risk groups.
-
Relabel procedure types of baseline endoscopies.
-
Indicate that patients do not have confirmed baseline colonoscopy.
-
Follow-up visit numbering.
-
True values for polyps found during follow-up visits.
-
Start/end date collecting records at each centre.
-
Cancer and AA end points.
-
Procedure types at follow-up visits.
-
Visit date and visit intervals.
-
Censoring of examinations after patient diagnosed with certain conditions.
-
Deaths information.
-
Tracing information.
-
Map previously seen cancers from external sources.
For each rule, a lay description is provided followed by the statistician’s technical description.
Rule 1: numeric endoscopy size other
On the study database, size was recorded using a number of fields to accommodate the different types of information relating to polyp size that were supplied in endoscopy and pathology reports. The following endoscopy and pathology sizes were recorded on the database: assume endo size, endo size, endo size min, endo size max, endo size other, assume endo size other, path size, assume path size.
The ‘endoscopy size descriptor field’ was used in cases for which a ‘vague’, qualitative or approximate size description was given in the endoscopy report. A numerical value was derived for each size description by analysing reports for which both qualitative size descriptions and a precise numerical size was given. The median and IQR was calculated for each numeric size field and cross-tabulated against associated categories of the endoscopy size descriptor field, as shown in the table below.
| Endoscopy size descriptor category (mm) | Endoscopy size (mm) | Derived value size (mm) | Rationale for derived value size | |
|---|---|---|---|---|
| Median (IQR) | n | |||
| Tiny | 3 (2–3) | 660 | 3 | Used the median |
| Small | 3 (3–5) | 1574 | 5 | Used the larger value of 5 mm to draw a distinction between Small and Tiny |
| < 5 | 3 (2–3) | 35 | 3 | Used the median |
| 5–9 | n/a | 0 | 7 | No examples so took the halfway point |
| < 10 | 8 (8–8) | 3 | 8 | Used the median of available examples |
| ≥ 10 | 15 (13–15) | 79 | 15 | Used the median |
| Large | 20 (12–30) | 2701 | 20 | Used the median |
At analysis, this field had to be assigned quantitative values to enable the classification of each patient’s baseline risk. Analyses were performed to compare the actual sizes recorded in ‘endo size’ and ‘path size’ with the values recorded in ‘endo size other’, wherever possible. After lengthy discussion, specific sizes were assigned to each ‘endo size other’ value based on this, and on rationality, as shown above.
Rule 2: derived procedure type
The study database contained two types of examination reports: those that originated from endoscopy databases at study hospitals and those that were manually generated from histology reports (taken from pathology databases) using any clinical and procedural information available. The latter, which were termed pathology-based procedure reports, did not exist on endoscopy databases, whereas a histology report was extracted, no corresponding endoscopy report was identified. Both of these types of examination report had a range of procedure types, such as proctoscopy, sigmoidoscopy, colonoscopy and surgery. In some cases, however, there was no procedure type described in the report, so the procedure was deemed an ‘unknown’ procedure. There were also examinations which were vaguely described as being an endoscopy.
The second rule applied to the data was to relabel certain procedure types and create a ‘derived procedure type’, where appropriate, with particular focus on those examinations with an unknown procedure type or examinations which appeared to have an incorrect procedure type given the segment of the colon that was visualised. (Colonoscopy is abbreviated to ‘col’, flexible sigmoidoscopy to ‘flexi-sig’ and rigid sigmoidoscopy to ‘rigid sig’ for the purpose of this rule.)
First, all pathology based procedure reports were relabelled with the group term ‘col, flexi-sig or rigid sig’. Comparatively, all ‘true’ endoscopies (i.e. ones taken from the endoscopy database) were relabelled with the term ‘col or flexi-sig’. Next, all proctoscopies were relabelled as ‘rigid sig’ because both procedures reach a similar segment of the bowel and proctoscopies are extremely rare. Then, any sigmoidoscopies that were obtained from endoscopy databases were relabelled as ‘flexi-sig’, and sigmoidoscopies that were pathology based procedure reports were called ‘flexi-sig or rigid sig’.
Unknown procedures obtained directly from an endoscopy database were relabelled as ‘col or flexi-sig’. Unknown procedures that were pathology based procedure reports were relabelled in this way only if they had bowel preparation/segment reached/distance reached fields completed, as such information clearly indicated that the procedure must have been endoscopic in nature. In addition, unknown procedures that were pathology-based procedure reports and did not have certain notable features (pathology blank/pathology truncated/pathology unclear specimen origin) were relabelled as ‘col, flexi-sig or rigid sig’, as although the possibility of a rigid sigmoidoscopy could not be ruled out, a surgical procedure could be. It was possible to identify a pathology report that was obtained from a surgical procedure, as long as the report was complete.
No procedures taken from endoscopy databases were relabelled as rigid sigmoidoscopies because this type of procedure was rare and is unlikely to have been recorded on the endoscopy database, as it would most likely have taken place in outpatient clinic rather than at an endoscopy clinic.
Any procedures that were consequently labelled as ‘col, flexi-sig or rigid sig’ as a result of the above rules were then relabelled as ‘col or flexi-sig’ if a large lesion (≥ 10 mm) or three or more adenomas were removed at that procedure, if polyps were seen in the sigmoid colon or beyond or if the procedure reached the sigmoid colon or more beyond (determined from the ‘segment reached’ field), as these features ruled out the possibility of a rigid sigmoidoscopy. Additionally, if the above criteria were satisfied, all procedures labelled as ‘flexi-sig or rigid sig’ were subsequently relabelled as ‘flexi-sig’.
Finally, all procedures labelled as ‘flexi-sig’, ‘rigid sig’, ‘flexi-sig or rigid sig’, ‘col or flexi-sig’ or ‘col, flexi-sig or rigid sig’, or endoscopies which reached the transverse colon or beyond, or had polyps found in this region of the bowel, were relabelled as colonoscopies, as it is unlikely that a sigmoidoscopy of any type would have reached so far into the bowel.
Create “derived_procedure_type” which is used from here onwards when referring to procedure types.
Note: “phantoms” are examinations that were not on the endoscopy database – a phantom endoscopy examination was created for the pathology we have.
-
Endoscopy examinations that are phantoms are relabelled as “col, flexi-sig or rigid sig”
-
Endoscopy examinations that are not phantoms are relabelled as “col or flexi-sig”
-
Procotoscopies are relabelled as “rigid sig”
-
Sigmoidoscopies that are not phantoms are relabelled as “flexi-sig”
-
Sigmoidoscopies that are phantoms are relabelled as “flexi-sig or rigid sig”
-
Procedures labelled as unknown that are not phantoms are relabelled as “col or flexi-sig”
-
Procedures labelled as unknown that are phantoms are relabelled as “col or flexi-sig” if they have bowel preparation/segment reached/distance reached fields completed
-
Procedures labelled as unknown that are phantoms are relabelled as “col, flexi-sig or rigid sig” if they do not have any of the following notable features – pathology blank/pathology truncated/pathology unclear specimen origin
-
Procedures labelled as “col, flexi-sig or rigid sig” or can be relabelled as “col or flexi-sig” if ≥ 10 mm lesion removed or 3+ adenomas removed or polyps in SC or more proximal or segment reached is SC or more proximal
-
Procedures labelled as “flexi-sig or rigid sig” can be relabelled as “flexi-sig” if ≥ 10 mm lesion removed or 3+ adenomas removed or polyps in SC or more proximal or segment reached is SC or more proximal
-
Procedures labelled as “flexi-sig”, “rigid-sig”, “flexi-sig or rigid sig”, “col or flexi-sig”, “col, flexi-sig or rigid sig” that reach/have polyps found in TC or more proximal are relabelled as “colonoscopy”
Note: phantom endoscopies were later renamed pathology-based procedure reports.
Rule 3: derived pathology/histology
In some cases, coders assigned assumed histology if it appeared that the actual histology was inaccurate or incorrect in some way. This generally occurred during cleaning incidents to prepare the data for analysis. In particular, assumed histology was assigned when reviewing groups of polyps, as these were sometimes vaguely described by the endoscopist and/or pathologist, making it difficult to assign histology to specific polyps. Rules were used to resolve such cases, which are described below (see rule 14).
In terms of analysis, the histology field was always overridden with the assumed histology, if it was present, which was used to create a field called derived_path_histology. In addition, if histology was coded as an adenoma or cancer, and assumed histology was also present, the adenoma type (villousness) and dysplasia were set to blank. As the polyp was assumed to be something other than an adenoma or cancer, it would no longer be appropriate for it to be associated with the other histological features that it was previously thought to possess.
Replace histology (“path_histology”) with assumed histology assigned by coders (“assume_path_histology”) if assumed histology is completed, to create a field called “derived_path_histology”.
Replace the adenoma type (“path_adenoma_type”) and dysplasia (“path_dysplasia”) as blank if histology (“path_histology”) is adenoma or cancer (codes 69, 55, 59, 58, 51, 50, 52, 65, 62, 53, 3, 1, 56) and assumed histology (“assume_path_histology”) is not adenoma or cancer. This is to correct cases where the adenoma type and dysplasia is no longer applicable (e.g. adenoma → inflammation).
Rule 4: relabel as adenomas based on villousness/dysplasia
Owing to missing or incomplete data in pathology reports, some polyps did not have any value coded in their histology field. In such cases, if the polyp had an adenoma type (villousness) and/or dysplasia type coded, it was assumed that the polyp was an adenoma and the histology was relabelled as ‘assume_adenoma’. This was a fair assumption, as villousness and dysplasia are both typical histological features of a classical adenoma.
Based on this, polyps with a histology value of ‘not possible to diagnose’ that had an adenoma type (villousness) and/or dysplasia type coded were also assumed to be adenomas as were relabelled as ‘assume_adenoma’.
This rule was applied regardless of whether or not the polyp had histology at another sighting or if the patient had an adenoma at any time. The histology was not relabelled for any matched polyps (only done to polyps that fitted the criteria above).
Relabel as “assume adenoma”, any polyps without histology or with histology “not possible to diagnose” (“derived_path_histology”), and which have villousness (“path_adenoma_type”) and/or dysplasia (“path_dysplasia”) completed (any value), regardless of whether that polyp has histology at another time or whether patient has an adenoma at any time. Only the polyp sightings with these criteria are relabelled and not all matched polyps.
Create variable called “path_hist_incl_assume_adenoma” where “assumed adenomas” are assigned value 97.
Rule 5: choose patients with one or more adenomas found
For inclusion in the IA cohort, patients had to have at least one adenoma present, with a pathology report confirming that the polyp was indeed adenomatous in nature. This was essential, as the study’s aim was to examine surveillance intervals in individuals with IR adenomas.
Patients must have at least one adenoma found: pathology for adenoma, serrated adenoma, mixed adenoma/hyperplastic, unicryptal adenoma or assumed adenoma (‘path_hist_incl_assume_adenoma’ = 1, 3, 53, 56, 97).
Rule 6: exclude any patients with missing examination dates
There were a number of examinations without a date, which coders reviewed when doing examination numbering. A large proportion of outstanding cases occurred at a single hospital (St Mark’s Hospital) so an attempt was made to find the examination date by visiting the hospital again to examine the Patient Administration System. Despite this, it was not possible to classify examinations without a date that had remained unnumbered following the examination numbering review. It was therefore necessary to exclude patients with an examination that was missing procedure data, as it was not possible to group such examinations into visits, which was essential to classify surveillance intervals accurately.
Remove any patients where 1 + endoscopy is missing its date.
Rule 7: relabel any ≥ 10-mm polyps without histology as assume adenoma
Another rule, similar to rule 4, was used to assign histology to polyps that were missing this information. Any polyps that were ≥ 10 mm in size with no histology or histology of ‘unable to diagnose’ were assumed to be adenomas, so the histology was relabelled as ‘assume_adenoma’. A size of ≥ 10 mm was assessed using the derived size fields (derived_endo_size, derived_endo_size_other and derived_path_size).
This rule was applied to only those patients with at least one actual adenoma. Relabelling of an individual polyp’s histology was done only if the polyp in question had no histology other than ‘not possible to diagnose’ or was missing histology at all of its sightings. The histology was relabelled for only the polyp that fitted the above criteria, not for any matched polyps.
Any ≥ 10-mm polyps without histology or histology recorded as “specimen not seen” or “not able to diagnose” will be relabelled as “assume adenoma” (use “path_hist_incl_assume_adenoma”). Relabelling occurs only for polyps which have no histology or “specimen not seen” or “not able to diagnose” at all of their sightings (use “derived_polyp_number” and “path_hist_incl_assume_adenoma”). Only the polyp sightings with these criteria are relabelled and not all matched polyps. Use derived sizes (“derived_endo_size”, “derived_path_size” or “derived_endo_size_other”) to determine size (i.e. if any of these ≥ 10 mm).
Note: this rule is only applied to patients who have at least one adenoma already.
Rule 8: apply baseline rules: define prior, baseline, follow-up
Pre-baseline checks
First, before baseline was defined, checks were carried out to ensure that all patients fulfilled the required criteria. Namely, a check was done to make sure that there were no polyps with both group1 and group 2 histology, or polyps with more than one type of group 2 histology (see rule 14). Any such histology combinations were had to be checked by coders and corrected; however, the following exceptions were allowed:
-
Polyps with group 2 histology of only one type and group 1 histology of ‘normal mucosa’, ‘granulation tissue’, ‘previous polypectomy site’, ‘not possible to diagnose’ or ‘specimen not seen’. In these cases the group 1 histology was removed.
-
Polyps with group 1 and group 2 histology (not of the combinations above) that was caused by multiplication out of multiple rows (see rule 1). In these cases the group 2 histology was removed.
-
Polyp with histology of ‘anal wart’ and ‘squamous cell carcinoma’ (both are group 2) at different sightings. Such cases were acceptable as long as these different histology types did not both occur within baseline or within follow-up (time between the occurrence of each one was estimated).
-
Polyps with histology of ‘inflammation’ and ‘oedema’ at different sightings (both group 2). In these cases inflammation took precedence over oedema.
-
Checks to do for polyps seen at more than one sighting:
Before apply baseline check that a polyp across all its sightings does not have both group 1 and group 2 histology (see lists below) or if polyp only has group 2 histology it does not have more than one type of group 2 histology (use “path_hist_incl_assume_adenoma”).
Exceptions to this rule are:
if a polyp has reported both group 2 histology (any but only one type) and group 1 histology from normal mucosa, granulation tissue, previous polypectomy site, not possible to diagnose or specimen not seen (codes 6, 34, 39, 90, 91) → remove group 1 histology from “path_hist_incl_assume_adenoma”
if a polyp has group 1 and group 2 histology (not of the combinations above) which is caused by multiplication out of multiple rows → remove group 2 histology from “path_hist_incl_assume_adenoma”
if a polyp is reported as anal wart (code 72) and SCC (code 61) at different sightings (both group 2) → ok as long as not both within baseline or both within follow-up (estimate time apart)
if a polyp is reported as inflammation (code 15) and oedema (code 22) at different sightings (both group 2) → inflammation takes precedence.
Any further combinations across sightings that are not listed as exceptions are checked by the coders and corrected.
Defining prior, baseline and follow-up visits
The baseline period had to be carefully defined, as the lesions found during this time frame were used to classify each patient’s baseline risk of CRC and stratify them into risk groups (i.e. low, intermediate or high). Prior and follow-up visits were then defined around the baseline period, with the length of time between baseline and follow-up visits being used to determine surveillance intervals. Specifically, prior examinations were defined as any examinations that occurred before the baseline visit, and follow-up examinations were defined as any examinations that occurred after the baseline visit.
The baseline visit was defined as a period of time starting from the earliest examination at which an adenoma or ‘assume adenoma’ was present. Sometimes polyps were seen but not diagnosed as adenomas until a later examination. Using polyp matching to identify such cases, baseline was shifted back to the first sighting of the adenoma if the prior matched polyp had histology of hyperplastic polyp, previous polypectomy site, granulation tissue, normal mucosa, was not possible to diagnose or specimen not seen or had no histology. Thus the first sighting was used to define the start of baseline rather than the first diagnosis of adenoma. This rule was applied as long as the matched lesions occurred within 3 years of one another. A relatively simple rule was then applied whereby all examinations within 11 months following the first adenoma were included within baseline. Using ‘within 1 year’ as a time frame may have resulted in surveillance examinations for high-risk individuals being included within baseline (as these are given 1 year after the initial examination), so 11 months was deemed to be the most appropriate time frame. Thus, baseline was defined as the first examination with an adenoma and any examinations within the subsequent 11 months.
Using this definition, the baseline period was then extended backwards from the first occurrence of an adenoma based on the time between prior examinations and the first baseline examination. In addition, the baseline period was also extended forwards using certain criteria (see rule 9).
-
Consideration for setting start of baseline in cases where adenomas seen at more than one sighting:
There are situations where a polyp is sighted at more than one examination. The polyp may not been assigned histology of an adenoma until a later sighting, for example, if at earlier sightings the polyp was not biopsied/excised and sent to pathology. We set baseline at the time of the first adenoma. Therefore, we apply adenoma histology backwards in certain situations as explained below to assure we set baseline at the time of first adenoma.
Polyps which are seen across multiple examinations and assigned adenoma histology (“path_hist_incl_assume_adenoma” = 1, 3, 53, 56, 97) at some time are identified. If at prior sightings to adenoma histology being assigned, the polyp has histology assigned from hyperplastic, previous polypectomy site, granulation tissue, normal mucosa, not possible to diagnose or specimen not seen histology (“path_hist_incl_assume_adenoma” = 2, 6, 34, 39, 90, 91) or the histology is unknown then adenoma histology is applied backwards. Adenoma histology is only applied back over 3 years at the most. If the histology at any of the prior sightings includes any other histology types than those listed above, the adenoma histology is not applied backwards (this avoids overwriting histology higher in the precedence list than an adenoma, e.g. cancer, sessile serrated lesion).
-
Assigning baseline, prior and follow-up:
Baseline starts at the earliest histology for an adenoma/assume adenoma (“path_hist_incl_assume_adenoma” = 1, 3, 53, 56, 97) for the patient. Baseline then extends 11 months (335 days) from this point.
Prior examinations are any examinations before detection of first adenoma.
Follow-up examinations are any examinations after 11 months after first adenoma.
Flags have been created to indicate prior, baseline and follow-up for each examination for patients who have one or more adenomas detected.
Rule 9: extend baseline
Extending baseline backwards
Once baseline was defined, it was then extended backwards to include prior examinations that satisfied certain criteria.
First, if a prior colonoscopy occurred within 11 months of the first baseline examination then it was included in the baseline visit. This rule was implemented because it is unusual to perform a surveillance examination at ≤ 1 year after a previous colonoscopy, so it seemed more likely that both procedures were related and thus part of the same ‘visit’. Baseline was extended backwards regardless of whether or not the patient had a colonoscopy in his/her original baseline, and any other examinations that occurred between the prior colonoscopy within 11 months and the examination with the first adenoma also became part of the baseline visit. This rule was not applied to prior colonoscopies with a clinical indication of a family history of cancer, as more frequent surveillance or screening would be feasible for such patients. The baseline visit was extended backwards only once, i.e. inclusion of prior examinations in relation to the extended baseline examinations was not considered.
Extending baseline backwards:
-
Extend baseline backwards if patient has a prior colonoscopy within 11 months of the first baseline examination.
-
Only extend backwards to colonoscopies that do not have indication of family history of cancer or CRC (‘indication’ = 15 35).
-
Baseline should be extended regardless of whether or not the patient has a colonoscopy in their current baseline.
-
Any other examinations that occur between the prior colonoscopy within 11 months and the first baseline examination will also become part of baseline clearance.
-
Extension of baseline only occurs once.
Extending baseline forwards
There were a number of scenarios for which it was necessary to extend the baseline visit forwards. This was done using a combination of timing, examination findings and characteristics, including features of the last baseline examination such as large polyps, an incomplete procedure and poor bowel preparation. Additionally, if a polyp found at the last baseline examination was matched to a polyp found shortly after baseline or if there was a surgical procedure shortly after then baseline was extended.
Some patients had examinations that occurred within 1 year of the last baseline examination. As endoscopic procedures can be delayed for logistical and medical reasons, any examinations that occurred within 9 months were likely to be part of the baseline visit because surveillance procedures should not have been carried out this soon. On the other hand, examinations that occurred within 9–11 months after the last baseline examination could potentially have been high risk surveillance examinations. Bearing this in mind, baseline was first extended to include all examinations which occurred within 6 months of the final baseline examination.
Additional criteria were then used to determine whether examinations that occurred 6–9 months after the final examination in the baseline period should be included in baseline or left as follow-up. Baseline was extended to include such examinations if:
-
the last baseline examination is incomplete
-
the last baseline examination has poor bowel preparation
-
a large polyp (≥ 15 mm) is seen at the last baseline examination
-
the same polyp is seen at the last baseline examination and examination occurring within 6–9 months
-
the first examination after the last baseline examination is surgical.
In terms of patients with a surgical examination 6–9 months after the final baseline examination, it was probable that the surgery was performed based on findings of the earlier examinations, so it was logical to extend baseline so that it included the surgery. In cases when a polyp found at the last baseline examination was large or was seen again at an examination within 6–9 months (i.e. matched polyps), the latter examination was likely to have been done to assess the polyp excision site or perform additional polyp removal. Finally, if the final baseline examination was low quality (incomplete or poor preparation) then it was feasible for another examination to have been performed to ensure that the bowel was properly examined, and this would still be part of the baseline visit.
After applying the rules for the extension of baseline backwards and forwards, the length of baseline was assessed and only a small proportion of patients had an unusually long baseline, whereas the majority were no more than 11 months in length. As with the backwards extension of baseline to include prior examinations, baseline was also extended forwards on only one occasion.
Extending baseline forwards:
-
Any examinations within 6 months of last baseline examination* will be included in baseline.
-
Include all examinations which occur 6–9 months after last baseline examination* if:
-
the patient’s last baseline examination* is incomplete (diagnosis = 14)
-
the patient’s last baseline examination* has poor preparation (bowel_prep = 1)
-
the patient’s last baseline examination* finds a polyp ≥ 15 mm (if derived_endo_size, derived_path_size or derived_endo_size_other are ≥ 15 mm)
-
the patient has the same polyp seen at their last baseline examination* as an examination occurring within 6–9 months
-
the patient’s first examination after last baseline examination* is surgical and it occurs at 6–9 months after last baseline examination*
-
-
Extension of baseline only occurs once.
*last baseline examination refers to last examination in baseline where baseline is as originally defined to include any examinations within 11 months from first adenoma.
Further extension forward of baseline to account for large polyps being removed over many examinations:
-
Define date of last examination in baseline.
-
Define whether or not lesions ≥ 15 mm (distal/proximal/any) found during baseline as it currently is assigned.
-
Include any colonoscopy procedures into baseline if they occur within 6 months of last examination in baseline and a lesion ≥ 15 mm more proximal than SC is reported during baseline. ∼
-
Redefine date of last examination in baseline. ∼
-
Include any FS/sigmoidoscopy/rigid sigmoidoscopy/unknown/endoscopy procedures into baseline if they occur within 9 months of last examination in baseline and a lesion ≥ 15 mm in SC or more distal is reported during baseline. ∼
-
Redefine date of last examination in baseline. ∼
-
Include any surgery procedures into baseline if they occur within 9 months of last examination in baseline and a lesion ≥ 15 mm is reported during baseline. ∼
-
Redefine date of last examination in baseline. ∼
-
Repeat the steps indicated with a ∼ until no further examinations are included into baseline.
Now baseline has been extended, we assume that the next examination a patient attends after baseline is follow-up surveillance (or symptomatic).
Rule 10: redefine baseline for patients without baseline colonoscopy
In order to accurately stratify patients into risk groups based on findings at baseline, it was necessary for patients to have a colonoscopy at baseline. Patients without a ‘definite’ colonoscopy at baseline were therefore reviewed. First, the procedure types of such patients were refined further. Any procedures that occurred during baseline that had been relabelled as ‘col or flexi-sig’ or ‘col, flexi-sig or rigid sig’ as a result of Rule 2 were deemed to be a colonoscopy, and relabelled as such, if the patient had significant adenomas during baseline (≥ 3, ≥ 10 mm, tubulovillous or villous histology, or HGD). Such procedures were most likely to be a colonoscopy rather than a sigmoidoscopy if any of these criteria was fulfilled.
Of the remaining patients without a ‘certain’ colonoscopy at baseline, for those who had a colonoscopy at their FUV1 it was decided that the follow-up visit should become the baseline visit and the original baseline should become a prior visit. In order to do this, the FUV1 had to be defined beforehand (see rule 21, below).
No changes were made for patients with no ‘certain’ colonoscopy during baseline or at their FUV1, and those with a colonoscopy at baseline.
To ensure that risk was not underestimated as a result of shifting the baseline to follow-up, any adenomas found at prior examinations were used to determine risk as well as those found during the baseline visit.
For patients without a certain baseline “colonoscopy” (derived_procedure_type=1):
-
If patient has “colonoscopy or flexi-sig” or “colonoscopy, flexi-sig or rigid-sig” examinations during baseline and significant adenomas during baseline (3 + adenomas, adenoma ≥ 10 mm, tubulovillous/villous histology or HGD), it is likely that this patient would have had a colonoscopy during baseline. Relabel first of either “colonoscopy or flexi-sig” or “colonoscopy, flexi-sig or rigid-sig” examination at baseline as colonoscopy.
-
Define follow-up visit 1 (see rule 21). If patient still does not have certain “colonoscopy” during baseline but has a “colonoscopy” during follow-up visit 1, follow-up visit 1 is relabelled as baseline and baseline is relabelled as prior.
Note: when baseline risk is defined, it will now need to count adenomas found at prior or baseline.
From the rules above we now have examinations divided into three time periods:
-
BASELINE
-
PRIOR (examinations prior to BASELINE)
-
FOLLOWUP (examinations after BASELINE).
Rule 11: cancer matching rules
Cancer data were obtained from a number of sources. First, information on cancers present in endoscopy and pathology reports was recorded on the study database. Then additional cancer data were obtained from external sources (HSCIC and NSS). These data had to be added into the study database, taking into account the patient and cancer data that were already present to ensure that there was no duplicated or missing data. This process was termed ‘cancer matching’.
In some cases, cancers in the hospital data were ‘missed’ by external sources (i.e. polyp rows with cancer histology on the study database could not be matched to a HSCIC cancer). These cases were queried with HSCIC and NSS; however, it was decided that for all outstanding cases the hospital pathology data should be accepted as conclusive evidence of cancer unless the histology was ‘cancer in dispute’ or ‘cancer query’. In the latter case, the evidence had to be deemed inconclusive and the lesion was regarded as an assumed adenoma with HGD.
Alternatively, sometimes there was no pathology to confirm cancer HSCIC/NSS did not report a cancer; however there was reason to believe that the patient had either a previous cancer or a cancer at an endoscopy. Such information was recorded in the notable features (unsolved queries), indications and diagnosis fields. It was decided that, without confirmation from external sources, any notable feature, diagnosis or indication of cancer should not be counted as a cancer.
Cancer outcomes
For the purposes of the study, the only cancer outcome of interest was adenocarcinoma of the colorectum. For the hospital data, it was fairly straightforward to identify such cases: cancer outcomes were defined using specific cancer histology codes (codes 50, 51, 55, 58, 59, 69). However, in order to determine which cancers from external sources were outcomes of interest, they all had to be grouped based on their morphology (Derived Morph Grouping derived from morphology codes) and site (Derived Site Grouping derived from site codes). Cancers were first selected using site codes and then further refined by morphology codes.
For cancers from external sources, Derived Morph Group related to morphology codes of lesions that were seen in the colorectum, although they may not have originated from there. It consisted of the following categories:
-
adenocarcinomas
-
non-adenocarcinoma cancers
-
cancers of unknown morphology
-
benign lesions
-
no morphology.
Derived Site Group related to site codes and consisted of the following three categories:
-
colon/rectum/anus
-
ill defined/unspecified site
-
other sites (non-colorectal).
As site codes contained morphological information, and some external cancers did not have appropriate information on morphology, the Derived Site Group was made up of 12 subgroups in total, with groups 1–3 being split into malignant, in situ, benign and unknown/unspecified.
| Derived Site Grouping (only malignant) | Derived Morphology Grouping | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3a | 4 | 5 | |
| 1 | Potential outcome (known adenocarcinoma) | Not an outcome | Potential outcome (assume adenocarcinoma) | Not an outcome | Potential outcome (assume adenocarcinoma) |
Only cancers from external sources that fell into in the first site group (malignant lesions of the colon/rectum) were selected for the study. Of these, any cancers with relevant morphology codes were considered to be outcomes and were thus added to the ‘cancer outcomes’ file (these are highlighted in the table above). More specifically, adenocarcinomas (group 1) in the colorectum were counted as outcomes. Cancers with unknown morphology (group 3) located between the rectum and caecum were assumed to be adenocarcinomas and thus counted as cancer outcomes. Cancers without morphology (group 5) were also deemed to be potential outcomes (assumed adenocarcinomas) based on site. If such cancers were located at sites relating to the anus then they were not included as cancer outcomes. This is because it was assumed that cancers in such sites were likely to be squamous cell carcinomas unless they were mapped to a rectal lesion without histology, in which cases they may have been adenocarcinomas and were included as cancer outcomes.
All cancers in the ‘cancer outcomes’ file were either automatically or manually mapped to examinations and/or polyps in the study database, wherever possible. In some cases, it was possible to map the cancer to more than one polyp row if, for example, the cancer was seen at more than one examination. The final polyp that the external cancer was mapped to was based on a hierarchy involving examination date, polyp numbering and the time between the external cancer date and the hospital procedure dates.
The true cancer date and site were also chosen using hierarchies. In terms of date, if the external cancer date preceded the mapped endoscopy date then the external date was used; if the mapped endoscopy date predated the external date then the date nearest to the external cancer event date was used. In terms of site, the site given in the external cancer data were used if no site was given in the mapped endoscopy (or site was non-specific) or if there were > 15 days between the dates of each data source. The site from the mapped endoscopy was used if it was available and the endoscopy contained the earliest polyp or nearest polyp to the cancer, or if the site given by the external cancer source was non-specific.
Cancer exclusions
With regards to cancer exclusions, all patients with a CRC were excluded from the study if the true cancer date occurred before the first examination on the study database, at prior examinations or within the baseline period.
Cancers from death data
Malignant CRCs reported in cause of death data from external sources were considered outcomes and added to the ‘cancer outcomes’ file if the patient did not have a cancer outcome reported already by externally sourced cancer registry information and did not have a cancer outcome reported from hospital data in the study database.
-
Chosen malignant colorectal cancers from external sources to add to our data
The “cancer outcomes” file contains cancers from the “allcancers” file which we consider outcomes (malignant colorectal cancers to be used for exclusions and endpoints) for the IA cohort (the patients remaining after rule 6 applied).
Cancers considered outcomes from external sources and added to the “cancer outcomes” file are highlighted in the table below:
| Derived Site Grouping (only malignant) | Derived Morphology Grouping | ||||
|---|---|---|---|---|---|
| 1 – Adenocarcinomas | 2 – Non-adenocarcinoma cancers | 3 – Cancers of unknown morphologya | 4 – Benign lesions | 5 – No morphology | |
| Group 1 – malignant neoplasm – colonb | Potential outcome (known adenocarcinoma) | Not an outcome | Potential outcome (assume adenocarcinoma) | Not an outcome | Potential outcome (assume adenocarcinoma) |
The histology codes we consider to be evidence of a cancer outcome in our data are shown in Appendix 2, Reference data table in the appendix (using field “path_hist_incl_assume_adenoma”).
Malignant colorectal cancers reported in cause of death from external sources were considered outcomes and added to the “cancer outcomes” file if the patient did not have a cancer outcome reported already external sourced cancer registry information and did not have a cancer outcome reported in our database.
The “cancer outcomes” file does not include cancers marked as “not CRC”, “no indication of cancer” or “duplicate cancer” by the coders. The “cancer outcomes” file does not include cancers which are not cancer outcomes in the external data but are mapped to something that is a cancer outcome in our data. This is because if either the external data or our data reports a cancer outcome this takes precedence even if the other source reports it is not a cancer outcome. All the cancers in our database have been double checked by coders so we can be confident of our report of cancer even if no report of cancer was given by external sources or if there is discrepancy between our report of cancer and the report of cancer by external sources (i.e. we report a cancer and they report an in situ lesion).
Also removed are cancers situated in the anus or appendix which are not adenocarcinomas (i.e. they are not morphology group 1).
All cancers in the “cancer outcomes” have been attempted to be mapped to examinations and/or polyps in our database.
-
Assigning final mapped polyp, true date and true site (and sources)
Following this, the final mapped polyp, true date, true site and true morphology were assigned as explained below. The final polyp that the external cancer is mapped to in our database (if applicable) is chosen by hierarchy and the source is assigned:
-
Find the earliest polyp with the same derived polyp number as polyp originally mapped to which has histology for cancer (codes 50, 51, 55, 58, 59, 69) and occurs earlier or on the same date as the external cancer
-
Source = “earliest polyp”
-
Find the polyp (any histology) with the same derived polyp number as the polyp originally mapped to, which occurs nearest to the external cancer (within 15 days)
-
Source = “nearest polyp to cancer”
-
Find the polyp (any histology) with the same derived polyp number as polyp originally mapped to which occurs nearest to the external cancer (more than 15 days from the external cancer)
Source = “nearest polyp to cancer but difference greater than 15 days”
The true date for the cancer is chosen by hierarchy:
-
Choose the date of the final mapped polyp from our data if the source of the final polyp ID is “earliest polyp” or “nearest polyp to cancer”. This means that the date of the final mapped polyp is chosen in these scenarios:
-
If the final mapped polyp has cancer histology(codes 50, 51, 55, 58, 59, 69) and occurs earlier than or on the same date as the external cancer.
-
If the final mapped polyp has cancer histology(codes 50, 51, 55, 58, 59, 69) and occurs up to 15 days later than the external cancer.
-
If the final mapped polyp does not have cancer histology but occurs within 15 days (either side) of the external cancer.
-
-
Choose the date of the external cancer if the source of the final polyp ID is “nearest polyp to cancer but difference greater than 15 days” or there is no final mapped polyp. This means that the date of the external cancer is chosen in the following scenarios:
-
If the final mapped polyp in our data has cancer histology (codes 50, 51, 55, 58, 59, 69) but occurs more than 15 days later than the external cancer date.
-
If the final mapped polyp does not have cancer histology and occurs more than 15 days before or after the external cancer date.
-
If there is no polyp mapped.
-
The true site for the cancer is chosen by hierarchy:
-
Choose the site of the mapped polyp from our data if the site of the final mapped polyp is not missing and the source of the final mapped polyp is “earliest polyp” or “nearest polyp to cancer”.
-
Choose the site of the external record if site of external cancer is not missing and source of the final mapped polyp is “nearest polyp to cancer but difference greater than 15 days” or if no polyp is mapped.
-
Choose site of external record if site is missing.
-
Overwrite with site from external record if site from our data is a range of segments and site from external record is a site code for a malignant cancer of colon and is specific [i.e. not C189 (ICD10), C188 (ICD10), 1538 (ICD9), 1539 (ICD9), 1538 (ICD8)].
-
Overwrite with site from our data if site from external source is not specific [i.e. one of C189 (ICD10), C188 (ICD10), 1538 (ICD9), 1539 (ICD9), 1538 (ICD8)] and our site is not missing.
-
Further considerations
Cases where external cancer is mapped to an endoscopy in our database but it is not mapped to a polyp (currently from above rules for true date and site the source of date and site for all these records is external source as no mapped polyp):
-
if mapped endoscopy is within 15 days of external date of cancer and mapped endoscopy is during BASELINE or PRIOR then mark cancer to exclude patient
-
if mapped endoscopy is within 15 days of external date of cancer and mapped endoscopy is during FOLLOWUP then mark to create a new polyp at that examination for the cancer
-
if mapped endoscopy is outside 15 days of external date of cancer then true cancer date and timings (determined by external sources) will be used and cancer will either be excluded or a new examination created for it.
If the true site of the cancer is denoted as ILEUM from our data but in the external data it is recorded as CAECUM, replace as CAECUM (and replace source of site accordingly).
If the true site of the cancer is denoted as ANUS from our data but in the external data it is recorded as RECTUM or RM/ANUS OVERLAPPING LESION, replace as RECTUM if the morphology of the cancer is adenocarcinoma (group 1 morphology) (and replace source of site accordingly).
Note: adding the cancer records to our data (section e) creates new examinations which may fall in PRIOR, BASELINE or FOLLOW-UP (i.e. within the range of examinations we have for the patient) but may also fall BEFORE FIRST EXAM ON OUR DATABASE or AFTER LAST EXAM ON OUR DATABASE (i.e. outside the range of examinations we have for the patient).
Check in “allcancers” file → The “allcancers” file contains all cancers received from external sources. Check in “allcancers” file for any cases where two or more cancers are mapped to the same polyp ID – any cases like this need to be resolved.
-
Adding external cancer outcomes to our data and identifying extra cancer outcomes in our data.
The cancer outcomes were added to our data as follows:
Group 1: identify patients to exclude-
Mark patient to be excluded (MERGE TO EXISTING POLYP) if:
-
true date of cancer occurs during “PRIOR”, “BASELINE” or “Before first endoscopy on our database”
OR
-
external cancer is mapped to an endoscopy but not to a polyp in our and the mapped endoscopy is within 15 days of external date of cancer and mapped endoscopy is during BASELINE or PRIOR.
-
-
If cancer histology codes (50, 51, 55, 58, 59, 69) on our database during “PRIOR” or “BASELINE” and patient not already marked to exclude from part (a) then mark patient to be excluded (IDENTIFY IN OUR DATA).
-
If new examination created falls within definition of baseline (see rules 8, 9 and 10) and patient not already marked to exclude from parts (a) or (b) (IDENTIFY IN OUR DATA).
If true date occurs during “FOLLOWUP” or “After last endoscopy on our database” and true date source is “OURPOLYP” then use mapped polyp_id to map exactly to data (MERGE TO EXISTING POLYP).
Group 3: cancers from external sources that can be merged directly to a polyp in our database – create a new polyp at an existing examinationIf external cancer is mapped to an endoscopy in our database but it is not mapped to a polyp and the mapped endoscopy is within 15 days of external date of cancer and mapped endoscopy is during FOLLOWUP then create a new polyp at that examination for the cancer (APPEND – CREATE NEW POLYP).
Group 4: cancers from external sources that cannot be merged directly to a polyp in our database – create a new examinationIf true date occurs during “FOLLOWUP” or “After last endoscopy on our database” and true date source is “EXTCANCER” then create new examination (APPEND – CREATE NEW EXAM).
Group 5: cancers identified in our database during follow-up but not identified from external sourcesIdentify any cancers in our database (codes 50, 51, 55, 58, 59, 69) occurring during follow-up and not reported from external source (use polyp numbering of any mapped polyps) (IDENTIFY IN OUR DATA).
Rule 12: apply exclusions for conditions
For inclusion in the analysis, patients had to have at least one adenoma found, whereas exclusion criteria were based on conditions that would either result in increased risk of CRC or abnormal surveillance. Once conditions of importance were identified, the time period during which they occurred was then considered. In some cases it was necessary to censor patients only after the occurrence of the condition or exclude them only if the condition occurred during prior or baseline examinations. For others it was more appropriate to exclude the patient regardless of when the condition was identified, for example HNPCC is a genetic condition that confers an increased risk of cancer throughout an individual’s lifetime.
The presence of an ‘exclusion condition’ was identified using different fields within the database including patient condition, diagnosis, indication, pathology and procedure type. During manual coding, patients were marked as excluded by coders; however, this information was transferred to a patient condition field and the original exclusion field was removed so that the final exclusion criteria could be applied in a more accurate and systematic way. The patient conditions, diagnosis and indications fields were also cleaned to prevent duplication of data and also to make it possible to determine the timing of exclusion conditions, thus simplifying the application of the exclusions criteria.
Patients with any of the following conditions were excluded from the analysis:
-
Colitis at prior or baseline examinations Diagnosis/indication of colitis, pathology for colitis.
-
IBD/Crohn’s disease at prior or baseline examinations Diagnosis/indication of Crohn’s disease or IBD, pathology for Crohn’s disease.
-
Polyposis (mixture of at any time and at prior or baseline examinations) Diagnosis/indication of polyposis (see Appendix 5, Polyposis subtypes and exclusion criteria), 100-plus adenomas at any time (FAP).
-
Family history of FAP at any time Indication of family history of FAP.
-
HNPCC at any time Indication of HNPCC.
-
Cowden syndrome at any time Indication of Cowden syndrome.
-
Volvulus at prior or baseline examinations Diagnosis/indication of volvulus.
-
Resection/anastomosis at prior or baseline examinations Surgical procedure type, patient condition of resection at first examination, diagnosis of anastomosis.
-
Juvenile polyps or hamartomatous polyps at any time Pathology for juvenile or hamartomatous polyps.
-
Radiation plus proctitis at prior or baseline examinations Diagnosis/indication of radiation plus proctitis
-
Derived radiation colitis at prior or baseline examinations (ulcers plus radiation) Diagnosis/indication of radiation plus ulcers.
-
More than 40 examinations recorded.
-
Diagnosis of CRC (adenocarcinoma) during or prior to baseline visit.
The exclusion criteria for cancer were more complex, as they had to take into account the cancer morphology as well as any cancers reported by HSCIC. In some cases it was not possible to be certain whether or not a patient had cancer. Problematic histology values were ‘cancer in dispute’, ‘cancer query’ and ‘unknown primary’. Cancer values coded in the study database within the diagnosis, indication and patient condition fields had to be cross-checked cancer with HSCIC cancers, and certain field over-riding others. Cancers were grouped based on their site and morphology, which were determined using endoscopic and histological data coded in the study database as well as ICD codes associated with HSCIC cancers (see section 11, above).
Other complex exclusion criteria were required for patients with polyposis and colitis. There were many different subtypes of both conditions in the database, and while some cases were confirmed by pathology or endoscopy reports, others had an uncertain diagnosis in terms of both presence and type. As a result, each combination of subtype and level of certainty was dealt with individually (see Appendix 5), to ensure that patients were not excluded unnecessarily, since some subtypes were mild or temporary and therefore did not require exclusion. In some cases, if polyposis or colitis was proposed but never confirmed, it was not deemed necessary to exclude such cases.
Patient must not have
-
colitis at prior or baseline examinations
-
diagnosis/indication of colitis (diagnosis = 4/indication = 4 + subtypes list) – see below for rules
-
pathology for colitis (path_hist_incl_assume_adenoma = 30).
-
-
IBD/Crohn’s at prior or baseline examinations
-
diagnosis of crohn’s or IBD (diagnosis = 6, 16)
-
indication of crohn’s or IBD (indication = 7, 36)
-
pathology for crohn’s (path_hist_incl_assume_adenoma = 28).
-
-
polyposis – mixture of at any time and at prior or baseline examinations
-
diagnosis/indication of polyposis (diagnosis = 22/indication = 18 + subtypes list) – see below for rules
-
presence of more than 100 adenomatous polyps of the colon and rectum at any time (FAP) (path_hist_incl_assume_adenoma = 1, 3, 53, 56 or assume adenoma).
-
-
family history of FAP at any time
-
indication of family history of FAP (indication = 41).
-
-
HNPCC at any time
-
indication of HNPCC (indication = 16).
-
-
Cowden syndrome at any time
-
indication of Cowden syndrome (indication = 42).
-
-
volvulus at prior or baseline examinations
-
diagnosis of volvulus (diagnosis = 11)
-
indication of volvulus (indication = 30).
-
-
resection/anastomosis at prior or baseline examinations
-
operation type (operation_type = 1, 2, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 36, 37, 38, 40, 41, 43, 44, 45, 46, 48, 49, 50, 51, 52, 53, 54)
-
patient condition of resection at first examination (patient_condition = 9)
-
diagnosis of anastomosis (diagnosis = 9).
-
-
juvenile polyps or hamartomatous polyps at any time
-
pathology for juvenile or hamartomatous polyps (path_hist_incl_assume_adenoma = 10, 24).
-
-
radiation + proctitis at prior or baseline examinations
-
diagnosis of radiation + proctitis (diagnosis = 43)
-
indication of radiation + proctitis (indication = 43).
-
-
ulcers + radiation = derived radiation colitis at prior or baseline examinations
-
diagnosis of radiation + ulcers (diagnosis = 44)
-
indication of radiation + ulcers (indication = 44).
-
-
> 40 examinations recorded.
-
diagnosis of cancer during or prior to baseline visit.
Rule 13: add in situ cancers and recoding histology in relation to cancer, in situ cancer and ‘cancer in dispute’
In situ cancers (lesions with HGD) were dealt with in a similar way to cancers (see rule 11). Adenocarcinoma in situ lesions obtained from external sources were relevant to the study (advanced adenomas were outcomes of interest) and thus had to be mapped to hospital data in the study database, wherever possible. The in situ cancers of interest, for the purposes of the study, are highlighted in the table below.
| Derived Site Grouping | Derived Morphology Grouping | ||||
|---|---|---|---|---|---|
| 1 – Adenocarcinomas | 2 – Non-adenocarcinoma cancers | 3 – Cancers of unknown morphology | 4 – Benign lesions | 5 – No morphology | |
| Group 1 – in situ neoplasm – colona | Include in situ | Do not include in situ | Include in situ | Include in situ | Include in situ |
In situ lesions located in the anus, which were not adenocarcinoma in situ and were not mapped to an adenoma or cancer in the hospital data, were not considered an outcome of interest, as they were likely to be squamous cell carcinoma in situ. In situ lesions in site group 1 (in situ neoplasm of the colon) with non-adenocarcinoma morphology (morphology group 2) were not counted as outcomes either, or were in situ lesions of unspecified intestinal site or those with uncertain/unspecified behaviour.
The final polyp that the external in situ lesion was mapped to and the true date and site were determined as before (see rule 11) using hierarchies.
The histology for cancers, in situ cancers and ‘cancer in disputes’ from hospital data were recoded if an in situ cancer from external sources had been mapped to it. All polyps mapped to an in situ from external sources were recoded to ‘assume adenoma’ with HGD, in line with the most recent nomenclature for such lesions, if they were not already coded as such. Dysplasia was recoded to HGD only if it was not already listed as HGD, severe dysplasia, ‘intra-mucosal cancer in dispute’ or intra-mucosal cancer. Additionally, polyps in the database with a histology of ‘cancer in dispute’ were recoded to ‘assume adenoma’ with HGD if they were not mapped to a cancer from external sources (assumed not to be cancer). Any polyps coded as ‘cancer query’ or ‘unknown primary’ were resolved, where possible, by looking for information to update their histology in the ‘allcancers’ file (i.e. non outcomes may be mapped to them).
Finally, all records of malignant CRC indicated from an external source, if not already assigned a cancer code in our data, were assigned code 50 (cancer). The dysplasia field not changed in such cases and was ignored for use with cancer histology (it can denote the dysplasia of adenoma sections of the cancer for other work).
-
Chosen in situ cancers from external sources to add to our data:
In situs to be added to our data from external sources in the “allcancers” file are highlighted in the table below (selected for patients remaining in cohort after rule 12):
| Derived Site Grouping | Derived Morphology Grouping | ||||
|---|---|---|---|---|---|
| 1 – Adenocarcinomas | 2 – Non-adenocarcinoma cancers | 3 – Cancers of unknown morphology | 4 – Benign lesions | 5 – No morphology | |
| Group 1 – in situ neoplasm – colona | Include in situ | Do not include in situ | Include in situ | Include in situ | Include in situ |
In situ lesions to be removed from those selected in the table above:
-
In situ lesions which are mapped to cancers in our database (“path_hist_incl_assume_adenoma“ = 50, 51, 55, 58, 59, 69) as the diagnosis of cancer in our database will take precedence.
-
In situ lesions of the anus which are not adenocarcinoma in situ and are not mapped to an adenoma or cancer in our data.
-
In situ lesions which are marked to exclude by the coder.
In situ lesions not included to be added to our data:
-
In situ lesions in site “group 1 – in-situ neoplasm of the colon” where morphology is non-adenocarcinoma.
-
In situ lesions (denoted by site code) of unspecified intestinal site.
-
In situ lesions (denoted by morphology code) of colon with uncertain/unspecified behaviour.
-
All in situs to be added have been attempted to be mapped to examinations and/or polyps in our database.
-
Assigning final mapped polyp, true date and true site (and sources)
Following this, the final mapped polyp, true date and true site were assigned as explained below.
The final polyp that the external in situ is mapped to in our database (if applicable) is chosen by hierarchy and the source is assigned (this is calculated by Study programmer):
-
Find the earliest polyp with the same derived polyp number as polyp originally mapped to which has histology for cancer or in situ* and occurs earlier or on the same date as the external record. Source = “in-situ earliest polyp”
-
*“path_hist_incl_assume_adenoma“== 50, 51, 55, 58, 59, 69 (cancers)
-
OR “path_hist_incl_assume_adenoma“== 52 (cancer in dispute)
-
OR “path_hist_incl_assume_adenoma“== 1, 3, 53, 56 (adenoma) and path_dysplasia=4, 5, 6, 7 (HGD, severe dysplasia, IM cancer in dispute or IM cancer)
-
OR “path_hist_incl_assume_adenoma“==missing or 90 (not possible to diagnose) and path_dysplasia =4, 5, 6, 7 (HGD, severe dysplasia, IM cancer in dispute or IM cancer) (this is to allow for those polyps with HGD which are recoded as assume adenoma once in my data).
-
-
Find the polyp (any histology) with the same derived polyp number as the polyp originally mapped to, which occurs nearest to the external record (within 15 days). Source = “in-situ nearest polyp to cancer”.
-
Find the polyp (any histology) with the same derived polyp number as polyp originally mapped to which occurs nearest to the external record (more than 15 days from the external record). Source=”in-situ nearest polyp to cancer but difference greater than 15 days”.
The true date for the in situ is chosen by hierarchy:
-
Choose the date of the final mapped polyp from our data if the source of the final polyp ID is “in-situ earliest polyp” or “in-situ nearest polyp to cancer”.
This means that the date of the final mapped polyp is chosen in these scenarios:
-
If the final mapped polyp has cancer or in situ histology and occurs earlier than or on the same date as the external record.
-
If the final mapped polyp has cancer or in situ histology and occurs up to 15 days later than the external record.
-
If the final mapped polyp does not have cancer histology but occurs within 15 days (either side) of the external record.
-
-
Choose the date of the external record if the source of the final polyp ID is “in-situ nearest polyp to cancer but difference greater than 15 days” or there is no final mapped polyp.
This means that the date of the external cancer is chosen in the following scenarios:
-
If the final mapped polyp in our data has cancer or in situ but occurs more than 15 days later than the date of the external record.
-
If the final mapped polyp does not have cancer or in situ histology and occurs more than 15 days before or after the date of the external record.
-
If there is no polyp mapped.
The true site for the in situ is chosen by hierarchy:
-
Choose the site of the mapped polyp from our data if the site of the final mapped polyp is not missing and the source of the final mapped polyp is “in-situ earliest polyp” or “in-situ nearest polyp to cancer”.
-
Choose the site of the external record if site of external record is not missing and source of the final mapped polyp is “in-situ nearest polyp to cancer but difference greater than 15 days” or if no polyp is mapped.
-
Choose site of external record if site is missing.
-
Overwrite with site from external source if site from our data is a range of segments and site from external record is specific [i.e. not D010 (ICD10)].
-
Overwrite with site from our data if site from external record is not specific [i.e. D010 (ICD10)] and our site is not missing.
-
Further considerations
Check if any of the newly added examinations for in situ lesions occurs during the definition of baseline. If so these examinations should be relabelled as baseline.
-
Adding external cancer outcomes to our data and identifying extra cancer outcomes in our data
The cancer outcomes were added to our data as follows:
Group 1: In situ lesions from external sources that can be merged directly to a polyp in our databaseIf true date source is “OURPOLYP” then use mapped polyp_id to map exactly to data (MERGE TO EXISTING POLYP)
Group 2: In situ lesions from external sources that cannot be merged directly to a polyp in our databaseIf true date source is “EXTCANCER” then create new examination (APPEND – CREATE NEW EXAM)
-
Recoding histology in relation to cancers, in situs and cancer in disputes
-
Recode all polyps which are marked as an in situ from external data, to adenoma+HGD, if not already recorded as adenoma+HGD in database. Also at this time recode polyp segment in our database as true segment of in situ.
-
Recode “cancer in dispute” to “assume adenoma” with HGD if not marked as cancer from external data (only change dysplasia if not already HGD etc.).
-
Resolve any “cancer query” and “unknown primary” polyps in our database by looking for information to update their histology in the “allcancers” file (i.e. non outcomes may be mapped to them). Mark these polyps so we know information has been updated.
-
All records of malignant colorectal cancer (either indicated from external source or codes 50, 51, 55, 58, 59, 69 in our data), if not already assigned a cancer code should be assigned code 50 (cancer). The dysplasia field will be left as it is and ignored for use with cancer histology (it can denote the dysplasia of adenoma sections of the cancer for other work). Also at this time recode polyp segment in our database as true segment of in situ.
-
Rule 14: True values for polyps found during and prior to baseline
True polyp values had to be determined for a number of polyp characteristics including size, histology, villousness, dysplasia, segment and shape, as a unique polyp was sometimes seen on a number of occasions, with different data recorded at each sighting. True polyp values for size, dysplasia, histology and villousness were applied across prior and baseline examinations, whereas true polyp values for these characteristics at follow-up visits were applied separate to prior and baseline examinations in order to see how lesions changed over time. Conversely, segment and shape were applied across all sightings, as these features were expected to remain constant over time (these are described in rule 15).
A. True size
Assigning size for polyps with multiple sightings across baseline and follow-up visits
In the simplest circumstance, a polyp was seen only once; however, sometimes there were multiple sightings of a single polyp within and/or across visits. True size was assigned separately for baseline and each follow-up visit, as polyps may have grown over time.
Once size discrepancies had been resolved and the three derived size fields had been assigned to each polyp sighting, the true polyp size could be determined. There was debate surrounding whether or not endoscopy size (derived_endo_size) was likely to be the most reliable derived size field, or if pathology size (derived_path_size) would be more accurate. Sometimes only a biopsy was sent to pathology, which would therefore not be representative of the true size of the lesion resulting in the lesion being classified as lower risk that it really was, whereas some endoscopy sizes (max, min, other) were only approximate values. It was decided that the largest of the derived sizes should be used, as there were advantages and disadvantages of both. Once the three derived size fields were defined for each polyp, there were eight possible combinations. Based on these, true polyp size was assigned as follows:
-
Group A Derived pathology size only – use.
-
Group B Derived endoscopy size other only – use.
-
Group C Derived endoscopy size only – use.
-
Group D Derived endoscopy size other and path size – use largest.
-
Group E Derived endoscopy size and path size – use largest.
-
Group F Derived endoscopy size and endoscopy size other – use derived endoscopy size.
-
Group G All three derived sizes – use largest of derived endoscopy and path size (ignore derived endoscopy size other).
-
Group H* No sizes – apply derived unknown range size where available.
Polyps with an unknown size assigned had no size at any sighting (Group H*). If there was no other size available, polyps were given a size (derived unknown range) using min/max values where appropriate. The derived unknown range was assigned once other sightings were accounted for, using the rules below:
-
no endoscopy size min, endoscopy size max of 6–9 mm, give assume endoscopy size other ‘small’.
-
min and max, both ≤ 9 mm, give average of min and max in terms of rule b: it was deemed more accurate to assign an average as (max – 1 + min)/2 as opposed to simply taking an average of the maximum and minimum values given in the range, in order to avoid making an overestimation of size
-
min and max, difference ≤ 2 mm, give average of min and max.
-
min = 40 and max = 50, give average.
-
min = 20 and max = 30, give average.
-
min = 10 and max = 12–20, give average.
Use values for true size from baseline sightings of polyp and prior to baseline sightings of polyp.
To define true size use:
-
largest endoscopy size = largest “derived_endo_size_numeric”
-
largest other endoscopy size = largest “derived_endo_size_other_numeric”
-
largest pathology size = largest “derived_path_size”.
-
These fields are derived by calculating largest of each of these derived sizes over baseline sightings of polyp and prior to baseline sightings of polyp.
Combinations of derived sizes for each unique polyp during baseline:
| Group | “largest endoscopy size” | “largest endoscopy size other” | “largest pathology size” | How to derive true polyps size |
|---|---|---|---|---|
| A | 0 | 0 | 1 | Use “largest pathology size” |
| B | 0 | 1 | 0 | Use “largest endoscopy size other” |
| C | 1 | 0 | 0 | Use “largest endoscopy size” |
| D | 0 | 1 | 1 | Use largest of “largest endoscopy size other” and “largest pathology size” |
| E | 1 | 0 | 1 | Use largest of “largest endoscopy size” and “largest pathology size” |
| F | 1 | 1 | 0 | Use “largest endoscopy size” |
| G | 1 | 1 | 1 | Use largest of “largest endoscopy size” and “largest pathology size” |
| H | 0 | 0 | 0 | “Largest endoscopy size”, “largest endoscopy other size” and “largest pathology size” are missing – use rules belowa |
Over baseline sightings of polyp and prior sightings of polyp (if within 3 years of start of baseline) – define “largest endoscopy size minimum” and “largest endoscopy size maximum”.
Note: Maximum/minimum endoscopy size(s) have already been applied to polyp(s) in the group to which the maximum/minimum endoscopy size(s) apply to:
-
“Largest endoscopy size minimum” is missing, “largest endoscopy size maximum” > 5 mm and < 10 mm, assign true size as “small” (which currently = 5 mm)
-
“Largest endoscopy size minimum” and “largest endoscopy size maximum” both < 10 mm, assign true size as average of “largest endoscopy size minimum” and (“largest endoscopy size maximum” – 1 mm)
-
Difference between “largest endoscopy size minimum” and “largest endoscopy size maximum” ≤ 2 mm, assign true size as average of “largest endoscopy size minimum” and “largest endoscopy size maximum”
-
“Largest endoscopy size minimum” and “largest endoscopy size maximum” both ≥ 20 mm and difference ≤ 10, assign true size as average of “largest endoscopy size minimum” and “largest endoscopy size maximum”
-
“Largest endoscopy size minimum” and “largest endoscopy size maximum” both ≥ 10 mm and < 20 mm, assign true size as average of “largest endoscopy size minimum” and “largest endoscopy size maximum”.
B. True histology
It was decided that an order of precedence should be used to define true histology, going from most to least severe. The order in which unique polyps with different histology types across examinations were seen did not affect how the true histology was assigned, i.e. hierarchy was applied in the same way for cases, with the worst histology taking precedence.
All histology types were split into two groups. Group 1 was composed of all ‘relevant’ histology that one would expect to be matched (i.e. adenoma and adenocarcinoma). Group 2 was made up of all ‘irrelevant’ histology that one would not expect to be matched to a group 1 histology (i.e. inflammatory polyp). Group 1 histology types were then put into an order of severity/importance. Polyps with unusual combinations of groups 1 and 2 histology types were reviewed, for example polyp is an adenoma at one sighting but a lipoma at another sighting. The order of precedence for group 1 was determined as follows, from most to least severe:
| POLYPTYPE_ID | POLYP_TYPE |
|---|---|
| 69 | CANCER + SESSILE SERRATED LESION |
| 55 | CANCER + MIXED/SERRATED ADENO |
| 59 | CANCER + MIXED ADENOMA |
| 58 | CANCER + SERRATED ADENOMA |
| 51 | CANCER + ADENOMA |
| 50 | CANCER |
| 52 | CANCER IN DISPUTE |
| 65 | UNKNOWN PRIMARY |
| 62 | CANCER QUERY |
| 68 | SESSILE SERRATED LESION |
| 53 | MIXED ADEN/META |
| 3 | SERRATED ADENOMA |
| 1 | ADENOMA |
| 97 | ASSUME ADENOMA |
| 56 | UNICRYPTAL ADENOMA |
| 2 | METAPLASTIC/HYPERPLASTIC |
| 39 | PREVIOUS POLYPECTOMY SITE |
| 34 | GRANULATION TISSUE |
| 6 | NORMAL MUCOSA |
| 90 | Not Possible To Diagnose |
| 91 | Specimen Not Seen |
There was some debate about how to treat sessile serrated lesions (SSL), since if SSLs took precedence over all types of adenoma, this may have affected the number of patients in the IA cohort. SSLs took precedence over all types of adenoma. Investigations were carried out to determine the number of patients who would be lost from the IA cohort as a result of this rule and consequently it was decided that the order of precedence should be left as such, since only a small number of patients were affected.
Group 2 histology is shown below:
| POLYPTYPE_ID | POLYP_TYPE |
|---|---|
| 8 | CARCINOID/NEUROENDOCRINE TUMOUR |
| 67 | BASALOID/CLOACOGENIC CANCER |
| 66 | ANAPLASTIC/UNDIFFERENTIATED CARCINOMA |
| 70 | GRANULAR CELL TUMOUR |
| 60 | METS/TUMOUR – INFILTRATING |
| 61 | SQUAMOUS CELL CARCINOMA |
| 63 | GASTROINTESTINAL STROMAL TUMOUR |
| 4 | LEIOMYOMA |
| 64 | SARCOMA |
| 26 | NON-HODGKINS LYMPHOMA |
| 71 | MELANOMA |
| 57 | METASTASES – ANOTHER SITE |
| 24 | HAMARTOMATOUS POLYP |
| 10 | JUVENILE POLYP |
| 5 | INFLAMMATORY |
| 36 | CAP POLYP |
| 23 | REGENERATIVE POLYP |
| 37 | LYMPHOID POLYP |
| 27 | FIBROEPITHLIAL POLYP |
| 17 | SUBMUCOSAL HAEMATOMA |
| 31 | LIPOMA |
| 25 | HAEMANGIOMA |
| 21 | XANTHOMA |
| 22 | OEDEMA |
| 41 | AMYLOID |
| 29 | NEUROFIBROMATOSIS |
| 40 | GANGLIONEUROMATOSIS |
| 32 | PSEUDOLIPOMATUS |
| 33 | SPIROCHAETOSIS |
| 19 | ANGIODYSPLASIA |
| 44 | LYMPHANGIECTASIA |
| 20 | ISCHAEMIA |
| 28 | CROHN’S DISEASE |
| 30 | COLITIS |
| 45 | PROCTITIS |
| 15 | INFLAMMATION |
| 14 | ULCER |
| 11 | MUCOSAL PROLAPSE |
| 35 | GASTRIC HETEROTOPIA |
| 16 | MELANOSIS COLI |
| 43 | CONGESTION |
Matched polyps with two or more histology types from group 2 and no histology from group 1 did not have a true histology assigned, as these were not of interest in terms of the study. Meanwhile, matched polyps with histology types from both groups 1 and 2 were reviewed in order to determine the correct histology type. An ‘assume histology’ field was used to do the review, so it was clear which type was considered correct based on the report details.
There were a number of polyps with no histology at any sighting. For polyps with no histology at baseline, if available, the worst prior histology was matched to such polyps if the prior examination was within 3 years of the baseline polyp.
Unique polyps should not have both group 1 and group 2 histology or more than one type of group 2 histology (use “path_hist_incl_assume_adenoma”).
Exceptions to this rule are:
-
if a polyp has reported both group 2 histology (any but only one type) and group 1 histology from normal mucosa, granulation tissue, previous polypectomy site, not possible to diagnose or specimen not seen (codes 6,34,39,90,91) → remove group 1 histology from “path_hist_incl_assume_adenoma”
-
if a polyp has group 1 and group 2 histology (not of the combinations above) which is caused by multiplication out of multiple rows → remove group 2 histology from “path_hist_incl_assume_adenoma”
-
if a polyp is reported as anal wart (code 72) and SCC (code 61) at different sightings (both group 2) → ok as long as not both within baseline or both within follow-up (estimate time apart)
-
if a polyp is reported as inflammation (code 15) and oedema (code 22) at different sightings (both group 2) → remove oedema from “path_hist_incl_assume_adenoma”.
If group 1 histology is reported for a unique polyp during or prior to baseline, this is assigned as the true histology according to the table of precedence below for group 1 histology.
If group 2 histology is reported for a polyp during or prior to baseline, this histology is then assigned as the true histology for baseline.
C. True villousness
Again, it was decided that an order of precedence should be used to define true villousness, going from most to least severe, as follows:
-
villous
-
tubulovillous
-
tubular.
True villousness values were applied across polyps at prior and baseline examinations (and then separately for polyps at each follow-up visit). There were a small number of cases with non-adenomatous histology that had villousness coded, which were reviewed as appropriate, for example a case with amyloid histology and villousness was reviewed as it was likely that the coder had meant to code an adenoma.
Villousness recorded as tubular, tubulovillous or villous in database.
True villousness is derived by calculating worst villousness over all sightings of the polyp during and prior to baseline.
D. True dysplasia
As was the case for villousness and histology, true dysplasia was also assigned based an order of precedence from most to least severe, as follows:
-
intramucosal cancer
-
intramucosal cancer in dispute
-
high grade/severe
-
moderate
-
low grade
-
mild.
The following values were grouped into either high or low grade:
-
high grade high grade, severe
-
low grade moderate, low grade, mild.
True dysplasia values were also applied across polyps at prior and baseline examinations, and separately for polyps at follow-up visits.
Dysplasia recorded as low grade, mild, moderate, high grade, severe, intramucosal cancer in dispute and IM cancer.
True dysplasia is derived by calculating worst dysplasia over all sightings of the polyp during and prior to baseline.
Mild and moderate are classified as low grade and severe is classified as high grade.
Rule 15: True values to be applied across all sightings of polyps
True polyp segment and shape were applied across all sightings of a polyp as these features were expected to remain constant over time.
A. True segment
The following rules were determined to define true polyp segment:
-
Surgical segment (i.e. polyp segment given at a surgical procedure) took precedence, as this was likely to be the most precise way to identify a polyp’s location.
-
If there was no surgical procedure then the most frequently described segment was assigned as the true segment.
-
If no segment was given more frequently than another, the most distal segment was assigned as the true segment.
-
In cases when a segment range was used:
-
if matched to a polyp with a single segment, assigned that segment
-
if not, assigned a range (provisional, need to investigate these cases).
When the true segment range was within a segment group, i.e. ascending colon (distal) to ascending colon (proximal) then true polyp segment was used instead, based on segment group (i.e. ascending colon). In addition, for a polyp with multiple segment ranges at different sightings but no actual segment, the smallest of all the ranges was used. Segments were then defined as follows: rectum to sigmoid colon = distal; descending colon to hepatic flexure = mid; and ascending colon to terminal ileum = proximal. True segment values were applied across all examinations, as the location of a polyp should remain constant over time.
B. True shape
It was unclear whether true polyp shape would be best assigned based on an order of precedence as with other characteristics or if the first recording of shape might be a more accurate method, as shape may have looked different once a polyp was biopsied or if complete excision was attempted. Shape values were flat, sessile, pedunculated, subpedunculated and pseudo. If an order of precedence were to be used, there was some discussion over whether or not pedunculated or sessile shape should take precedence when a lesion was described as being both shapes at different sightings. Analyses were undertaken to compare both methods and it was decided that it was more appropriate to assign the first recorded shape of a lesion as the true shape. In addition, any polyps with shape ‘subpedunculated’ were changed to ‘pedunculated’.
True polyp segment and shape to be applied across all sightings of polyp (not just in baseline).
Group segment information ignoring distal/mid/proximal information within segment.
If no ranges of segments are given for the polyp at any sighting, the order of precedence for assigning true segment is:
-
Surgical segment (if available).
-
Most frequently used segment (if any segments repeated 2 + times and repeated more than any other segment).
-
Most distal segment (maximum from ordered segment list).
If range of segments is given for the polyp at one or more sightings the order of precedence for assigning true segment is:
-
Use actual segment, if given at any time (use precedence rules above to decide which actual segment – surgical, frequent, distal).
-
No actual segments given and only one range – assign that range.
-
No actual segments given and several ranges of segments – assign the smallest of the segment ranges as long as “polyp segment from (ordered)” OR “polyp segment to (ordered)” of each of the ranges are ≤ 2 segments different.
Ordered segment list (proximal to distal):
| 1 | Ileum |
| 2 | Caecum |
| 3 | Ascending colon |
| 4 | Hepatic flexure |
| 5 | Transverse colon |
| 6 | Splenic flexure |
| 7 | Descending colon |
| 8 | Sigmoid colon |
| 9 | Rectosigmoid |
| 10 | Rectum |
| 11 | Anus |
Where true segment range is within a segment group e.g. ascending colon distal to ascending colon proximal are both ascending colon, “true polyp segment” will be changed to ascending colon and “true polyp segment to” will be set as missing.
Note: do not override true polyp segments with true segments for cancer/in situ. Use true cancer segments in rules above to define true polyp segments. After this, do not use true cancer/in situ segments for site of cancer, instead refer to true polyp segments (i.e. true cancer segment will not be same as true polyp segment in some cases).
Any records recorded as sub-pedunculated changed to pedunculated.
If pathology shape is recorded and no endoscopy shape, replace endoscopy shape with pathology shape.
Use first recording of shape as true shape.
True shape to be applied across all sightings of polyp.
Rule 16: baseline risk groups
The baseline risk groups were defined using the criteria for stratification of patients as low, intermediate and high risk as described in the current EU Guidelines for Quality Assurance in Colorectal Cancer Screening and Diagnosis. These definitions were applied using all adenomas found at prior and baseline examinations, and are as follows:
-
Low risk One or two small adenomas (no large adenomas or adenomas without size).
-
Intermediate risk Three or four small adenomas (no large adenomas or adenomas without size) or one or two adenomas of which at least one is large.
-
High risk Five or more adenomas (any or unknown size) or three or more adenomas of which at least one is large.
-
Low/intermediate risk One adenoma of unknown size or two adenomas of which none is large but one or more has unknown size.
-
Intermediate/high risk Three or four adenomas of which none is large but one or more has unknown size.
Small adenomas were those of < 10 mm in size, whereas large adenomas were those of ≥ 10 mm.
Baseline risk groups count adenomas recorded at baseline or prior to baseline or before first examination on database.
Define true histology and true size for unique polyps using values from PRIOR and BASELINE and BEFORE FIRST EXAM ON DATABASE together (to account for adenomas that occur or are seen prior to baseline).
These risk groups are defined at this point to be used for rule 18 (relabeling procedure types of baseline endoscopies) and so still include patients who currently do not have baseline colonoscopy.
Risk group definitions:
-
Low risk – 1 or 2 small adenomas (no large adenomas or adenomas without size)
-
Intermediate risk – 3 or 4 small adenomas (no large adenomas or adenomas without size) OR 1 or 2 adenomas of which at least 1 is large
-
High risk – 5 or more adenomas (any or unknown size) OR 3 or more adenomas of which at least 1 is large
-
Low/intermediate risk – 1 adenoma of unknown size OR 2 adenomas of which none is large but 1 or more has unknown size
-
Intermediate/high risk – 3 or 4 adenomas of which none is large but 1 or more has unknown size.
*small adenomas < 10mm, large adenomas ≥ 10mm.
*adenomas histology codes used “path_hist_incl_assume_adenoma”= 1, 3, 53, 56, 97.
Note: sessile serrated lesion not included in baseline risk groups.
Rule 17: relabelling unclassified endoscopies during baseline
To enable accurate classification of an individual’s baseline risk of CRC, it was preferable that a complete colonoscopy was performed during the baseline period. In some cases, there were baseline examinations with no procedure type given. In other cases, examinations were vaguely referred to as endoscopies. Such procedures were reclassified into more specific procedure types (derived procedure type) in rules 1 and 10.
Patients with no certain colonoscopy at baseline, who instead had the non-specific procedure type of ‘colonoscopy or flexi-sig’, were reviewed with the aim of identifying those in whom such a procedure could be assumed to be a colonoscopy based on certain criteria. These patients had many different combinations of specific examinations (i.e. flexi-sig) and non-specific derived procedure types, as well as having only non-specific derived procedure types during baseline.
Procedures were reclassified based on CRC risk and type of follow-up examination. If high-risk adenomas were found then one could assume that the patient was likely to have had a colonoscopy at some point during baseline. If a patient had at least one follow-up colonoscopy or ‘colonoscopy or flexi-sig’ examination, it was also deemed likely that they had had a colonoscopy at baseline, whereas for those with no follow-up colonoscopy or ‘colonoscopy or flexi-sig’, it was possible that the non-specific baseline procedure could have been a FS or a colonoscopy. If a patient had a follow-up examination within 7 years and the patient had low-risk baseline lesions, one can assume that the follow-up examination was a surveillance examination. The follow-up examination should be within 7 years for this to be feasible because 5 years is specified as the low-risk surveillance interval in the guidelines and patients may delay their appointment for a number of reasons, thus 7 years ought to include all surveillance examinations. If a patient had low-risk polyps and no follow-up colonoscopy, one cannot assume that the ‘colonoscopy or flexi-sig’ is a colonoscopy.
Specifically, patients were first split into two groups: those with one ‘colonoscopy or flexi-sig’ at baseline only (no other examinations at baseline) and those with two or more ‘colonoscopy or flexi-sig’ examinations at baseline or other examination types. In both groups, a baseline ‘colonoscopy or flexi-sig’ examination was assumed to be a colonoscopy and relabelled as such if the patient was intermediate, high or intermediate/high risk. Additionally, patients who were low or low/intermediate risk, who had at least one follow-up colonoscopy or ‘colonoscopy or flexi-sig’ examination within 7 years of their last baseline examination, also had their baseline ‘colonoscopy or flexi-sig’ examination relabelled as a colonoscopy. This is because such patients were likely to be under some sort of colonoscopic surveillance. In cases when there were two or more ‘colonoscopy or flexi-sig’ examinations during baseline and the patient met the criteria to have a ‘colonoscopy or flexi-sig’ examination relabelled (as described above), only the first ‘colonoscopy or flexi-sig’ examination during baseline was relabelled as a colonoscopy.
Analyses into examinations sequences showed that colonoscopy tended to follow sigmoidoscopy, except where a large lesion was detected and the subsequent examination was undertaken to check the completeness of polypectomy. These analyses also demonstrated that flexi-sig followed by flexi-sig was fairly uncommon compared with colonoscopy followed by colonoscopy. Consequently, in those with two examinations at baseline, one of which was a form of sigmoidoscopy, it was assumed that the ‘colonoscopy or flexi-sig’ examination was a colonoscopy if it occurred after the sigmoidoscopy.
Relabelling for patients with no baseline colonoscopies
-
One ‘colonoscopy or flexi-sig’ examination only at baseline (no other examinations)
Relabel baseline ‘colonoscopy or flexi-sig’ examination as a colonoscopy if:
-
patient is intermediate, high or intermediate/high risk, regardless of follow-up
-
patient is low or low/intermediate risk and at least one follow-up examination is a colonoscopy within 7 years of last baseline examination
-
patient is low or low/intermediate risk and at least one follow-up examination is ‘colonoscopy or flexi-sig’ examination within 7 years of last baseline examination.
-
-
Two or more ‘colonoscopy or flexi-sig’ examinations at baseline OR one or more ‘colonoscopy or flexi-sig’ examination with other examinations at baseline
Relabel baseline ‘colonoscopy or flexi-sig’ examination as a colonoscopy if:
-
patient is intermediate, high or intermediate/high risk, regardless of follow-up
-
patient is low or low/intermediate risk and at least one follow-up examination is a colonoscopy within 7 years of last baseline examination
-
patient is low or low/intermediate risk and at least one follow-up examination is ‘colonoscopy or flexi-sig’ within 7 years of last baseline examination.
-
Note: in cases where there are 2 + ‘colonoscopy or flexi-sig’ examinations during baseline and patient meets criteria to have a ‘colonoscopy or flexi-sig’ examination relabelled (as above), only the first ‘colonoscopy or flexi-sig’ examination during baseline will be relabelled as colonoscopy.
-
With any remaining patients in this group, relabel ‘colonoscopy or flexi-sig’ examination as a colonoscopy if two examinations at baseline only, where first is ‘flexi-sig’ or ‘flexi-sig or rigid-sig’ examination and second is a ‘colonoscopy or flexi-sig’ examination.
Rule 18: indicate which patients do not have baseline colonoscopy
Patients without a colonoscopy during their baseline visit were problematic because their true risk could not be accurately assessed without an initial examination of the whole colon. It was decided that any such patients should be marked so that separate analyses can be performed with the exclusion or inclusion of such patients. This also enabled stratification by procedure type at baseline in order to assess whether there is an association between risk and examination type. Thus, the most complete full colon examination available at baseline was defined using a hierarchy based on procedure type, segment reached, polyp site and whether or not the procedure was marked as incomplete or not.
Indicate which patients do not have a colonoscopy during baseline. We cannot accurately assess their baseline risk group without the entire colon being seen so they will be excluded from the some analyses.
Defined for each follow-up visit the ‘most complete full colon examination available’ at baseline using the following hierarchy:
-
complete ‘colonoscopy’
-
(define complete as reaching/recording polyps in CM or IL (segment_reached/true_segment/true_segment_to = 9, 34,10) and not marked as incomplete (diagnosis = 14) in diagnosis fields)
-
‘colonoscopy’ that is not complete or unknown whether complete
-
(does not reach/no polyps recorded in CM/IL, no info. on segment reached or marked as incomplete)
-
‘colonoscopy or flexi-sig’
-
‘flexi-sig’
-
‘colonoscopy, flexi-sig or rigid-sig’
-
‘flexi-sig or rigid-sig’
-
‘rigid-sig’
-
‘surgery’
-
‘unknown’.
Rule 19: follow-up visit numbering
The rules used to define baseline were also applied to follow-up visits, regardless of the type of follow-up procedures at each visit. The first examination after baseline or after the previous visit and any examinations within 11 month of that first examination were defined as the next follow-up visit. The final examination in a visit was identified and the visit was then extended forwards using the same criteria as those used for the extension of the baseline period. This procedure was repeated until all of the examinations had been grouped into visits.
The same rules are used for defining follow-up visits as used at baseline, regardless of types of follow-up procedures. These rules are as follows (written for defining follow-up visit 1 but they are the same for defining each follow-up visit):
-
Define the first examination after the end of baseline, as follow-up visit 1.
Include any examinations within 11 months of this examination in follow-up visit 1.
Define the last examination in follow-up visit 1 so far to use for parts (b) and (c)*.
-
Any examinations within 6 months of last examination in follow-up visit 1*will be included in follow-up visit 1.
This rule is only applied once.
-
Extension of the follow-up visit to account for various scenarios where further examinations may be performed as part of the same visit e.g. incomplete examinations, poor bowel preparation, large polyps, surgery to remove lesions.
Include all examinations which occur 6–9 months after last examination in follow-up visit 1* if:
-
the last examination in follow-up visit 1* is incomplete
-
the last examination in follow-up visit 1* has poor preparation
-
the last examination in follow-up visit 1* finds a ≥ 15-mm polyp (use derived_endo_size, derived_path_size or derived_endo_size_other)
-
the same polyp is seen at the last examination in follow-up visit 1* as an examination occurring within 6–9 months after the end of FU visit 1
-
the first examination after follow-up visit 1 is surgical and it occurs at 6-9 months after the end of follow-up visit 1*
These rules are only applied once.
-
Further extension of the follow-up visit to account for large polyps being removed over many examinations.
-
Define date of last examination in follow-up visit 1.
-
Define whether ≥ 15 mm lesions (distal/proximal/any) found anytime during follow-up visit 1 (as follow-up visit 1 is currently assigned).
-
Include any colonoscopy procedures into follow-up visit 1 if they occur within 6 months of last examination in follow-up visit 1 and a ≥ 15 mm lesion more proximal than SC is reported during follow-up visit 1.∼
-
Redefine date of last examination in follow-up visit 1. ∼
-
Include any FS/sigmoidoscopy/rigid sigmoidoscopy/unknown/endoscopy procedures into follow-up visit 1 if they occur within 9 months of last examination in follow-up visit 1 and a ≥ 15 mm lesion in SC or more distal is reported during follow-up visit 1. ∼
-
Redefine date of last examination in follow-up visit 1. ∼
-
Include any surgery procedures into follow-up visit 1 if they occur within 9 months of last examination in follow-up visit 1 and a ≥ 15 mm lesion is reported during follow-up visit 1. ∼
-
Redefine date of last examination in follow-up visit 1. ∼
-
Repeat the steps indicated with ∼ until no further examinations are included into follow-up visit 1.
-
Rule 20: true values for polyps found during follow-up visits
The same rules were applied as those used at prior and baseline examinations (see rule 14), but they were applied across polyps only within each follow-up visit, so any changes in polyp characteristics over time could be identified.
A. True size: same rules as above for baseline but only applied across polyps within each follow-up visit.
B. True histology: same rules as above for baseline but only applied across polyps within each follow-up visit.
C. True villousness: same rules as above for baseline but only applied across polyps within each follow-up visit.
D. True dysplasia: same rules as above for baseline but only applied across polyps within each follow-up visit.
Rule 21: start/end date of endoscopy records at each centre
The start and end dates of endoscopy records for each hospital were determined data, based on number of endoscopies extracted over time. The number of endoscopy examinations on the study database was plotted by month for each hospital. The plots produced were assessed, and the first and last month in which the number of endoscopies became steady were identified. The chosen start and end dates were based on the earliest/latest examination in the chosen months. It was assumed that, for each patient, all endoscopies that occurred between these dates were present in the study database.
The start and end dates of endoscopy records we will have for each hospital was decided by:
-
Plotting by month the number of examinations from the endoscopy database (i.e. not pathology-based procedure reports) and choosing the month at which the number of examinations was steady (near the start of collection not many records may have been entered onto the electronic database or near the end of collection completed examinations may have been waiting to be entered onto the database).
-
The chosen start and end dates of examinations we have for each hospital was set to the start/end of the chosen months the earliest/latest examination was midway through a month.
Therefore, we assume that between the chosen start/end dates we have all the endoscopy exams that occurred at that hospital between those dates.
| Hospital | Dates of exams | Chosen start/end dates |
|---|---|---|
| St Mark’s (SMH) | 03/01/1972 – 20/07/2010 | 01/01/1985 – 31/07/2007 |
| Leicester (LGH) | 26/05/1988 – 08/04/2008 | 01/04/1998 – 31/03/2008 |
| Brighton (BRI) | 03/05/2001 – 23/04/2008 | 03/05/2001 – 23/04/2008 |
| Torbay (TDG) | 18/05/1993 – 24/11/2009 | 01/11/2000 – 31/08/2007 |
| Yeovil (YDH) | 16/10/1989 – 06/06/2008 | 01/02/1997 – 31/05/2008 |
| North Tees (NT) | 06/06/1986 – 12/03/2010 | 06/06/1986 – 31/12/2006 |
| Queen Elizabeth (QEW) | 31/01/1990 – 27/06/2009 | 01/03/1999 – 31/05/2006 |
| Cumberland (CI) | 05/05/1994 – 28/01/2010 | 01/08/1998 – 30/09/2009 |
| Shrewsbury (SH) | 06/02/1995 – 30/09/2009 | 01/01/2002 – 30/09/2009 |
| Liverpool (RLUH) | 17/08/1999 – 21/10/2009 | 01/01/2000 – 21/10/2009 |
| Glasgow (GRI) | 01/04/1996 – 08/09/2009 | 01/05/1996 – 31/08/2009 |
| St Georges (SGH) | 15/04/1987 – 29/05/2010 | 01/02/1992 – 31/07/2009 |
| Sidcup (QMH) | 06/01/1988 – 14/07/2009 | 01/10/1998 – 14/07/2009 |
| New Cross (NC) | 04/01/1993 – 23/11/2007 | 04/01/1993 – 23/11/2007 |
| Surrey (SCH) | 25/07/1985 – 29/05/2010 | 01/09/1997 – 29/05/2010 |
| St Mary’s (ICM) | 06/04/1980 – 07/08/2010 | 01/12/1984 – 31/07/2010 |
| Charing Cross & Hammersmith (CX or HH) | 09/06/1994 – 27/11/2007 | 01/10/1997 – 27/11/2007 |
Rule 22: cancer and advanced adenoma end points
For the purposes of analysis, records with end points of interest (CRC and advanced adenomas) were flagged. Advanced adenomas were defined as adenomas of ≥ 10 mm or with HGD or villous/tubulovillous histology. The earliest record of each unique cancer or advanced adenomas was then identified and assigned timing in relation to other examinations.
-
Cancers
-
Indicate earliest record of each unique cancer now that true values have been defined.
-
Note: this may not be the same as the original polyp recorded as cancer in our data or from external data due to true values applied across each follow-up visit.
-
Define which patients report cancer and which visits report these first records of unique cancer.
-
Assign timing of cancers relative to other follow-up exams and collection dates of records at each hospital. To decide which to use as end points in analyses.
-
-
AAs
-
Define advanced adenomas as adenomas ≥ 10 mm or HGD (or more severe) or tubulovillous/villous.
-
Indicate earliest record of each unique advanced adenoma now that true values have been defined.
-
NB this may not be the same as the original polyp recorded as AA in our data or from external data due to true values applied across each follow-up visit.
-
Define which patients report AA(s) and which visits report these first records of unique AA.
-
Assign timing of AAs relative to other follow-up exams and collection dates of records at each hospital. To decide which to use as end points in analyses.
-
Rule 23: procedure types at follow-up visits
It was decided that no follow-up examinations would have their procedure types relabelled, unlike those at prior and baseline examinations. Instead, for each follow-up visit the ‘most complete full colon exam’ available was defined using a hierarchy from complete colonoscopy to unknown procedure type. The completeness of a colonoscopy was defined based on the segment reached, most proximal polyp site and whether or not the examination was marked as incomplete or not.
Decided not to relabel any follow-up procedure types.
Defined for each follow-up visit the “most complete full colon exam available” using hierarchy:
-
complete colonoscopy (define complete as reaching CM or IL (codes 9, 34,10) and not marked as incomplete (diagnosis = 14) in diagnosis fields)
-
colonoscopy that is not complete or unknown whether complete (does not reach CM/IL, no information on segment reached or marked as incomplete)
-
endoscopy
-
FS
-
sigmoidoscopy
-
rigid sig
-
surgery
-
unknown.
Rule 24: visit date and visit intervals
A visit date was defined as the earliest examination date in each visit; this was defined only for baseline and follow-up visits, not those prior to baseline. Then visit intervals were timed from the last, most complete examination of one visit to the first examination of the next visit.
Visit intervals:
Create visit date as earliest examination date of each visit.
Note: visit date is defined for baseline visits and visits after baseline (not those prior to baseline).
Visit intervals are timed from the last most complete examination of one visit to the first examination of the next visit.
Rule 25: censoring of visits after patient diagnosed with certain conditions
It was decided that visits following a diagnosis of CRC, volvulus and resection/anastomosis should be censored. Contrastingly, any visits that occur after a diagnosis of colitis, IBD/Crohn’s disease, polyposis, radiation and proctitis, and ulcers and radiation are not censored.
Censor visits after:
-
malignant colorectal cancer diagnosis
-
resection/anastomosis
-
volvulus.
Note: censor the visits after the visit at which any of these occur during.
No censoring for colitis, IBD/Crohn’s, polyposis, radiation + proctitis or ulcers + radiation if they occur after baseline.
Rule 26: deaths
Any cases for which there was more than one date of death for a patient were resolved, and the date of death for patients who were traced and had died was added to the study database.
Rule 27: tracing
Patients were traced using data from external sources – including HSCIC, NHSCR and NSS – to obtain follow-up data on cancers and deaths, verify patient information such date of birth and gender, and identify duplicate patients on the study database.
Use tracing file from Study programmer → “DERIVED_MR1201_TRACING_DATA”.
(a) One record per patient: There is more than one record for some patients. Study programmer has marked in this dataset which records to remove so that we only have one record per patient. A patient may have more than one record because they have been traced by more than one source (traced by NSS and HSCIC – choose NSS record if GRI patient and HSCIC record if not GRI patient) or because several patients have been merged together (so the duplicate patients need to be removed). These records are removed and we are left with a file that is one record per patient.
(b) Date of birth and gender: True date of birth and true gender calculated by Study programmer. These fields are derived in HSCIC and NSS fields separately before the files are merged together. Study programmer uses hierarchy to decide true values. In HSCIC file – hierarchy 1) MMP 2) HSCIC 3) hospital data 4) patient table. In NSS file – hierarchy 1) NSS 2) hospital data 3) patient table (NB NSS does not have MMP data, MMP data is from HSCIC – it is latest info for ENG/WALES cases). Check the true date of birth is not after the exams we have for that patient and check that any occurring after 1990 or before 1900 look valid.
(c) Embarkation, cancelled cipher, registered in Northern Ireland, registered in Scotland:
Patients are listed as “registered in Scotland” means that the patient has moved to Scotland and we have the date to show when they moved. In these cases we have all the England/Wales cancers and deaths up to the point they moved to Scotland and we also have cancers and death from NHSCR after they moved to Scotland. Therefore, for all “registered in Scotland” patients as long as they are subsequently “flagged in Scotland” we will know about all their cancers and deaths. In our data all patients indicated as “registered in Scotland” are also “flagged in Scotland”.
Patients are listed as “registered in Northern Ireland” meaning that the patient has moved to Northern Ireland and we have the date to show when they moved. In these cases we have all the England/Wales cancers and deaths up to the point they moved to Northern but not after that, unless they re-enter the NHS.
Patients are listed as “embarked” which is when a patient moves abroad and notifies their GP and we have the date they moved. In these cases we have all the England/Wales cancers and deaths up to the point they moved but not after that, unless they re-enter the NHS. A death may become available if it is registered with the consulate in the country, however, this would only be fact of death, no cause would be available.
Patients are listed as “cancelled cipher” which is when a patient has exited the NHS but the reason is unknown (may have moved abroad or moved into private care without informing their GP) and we have the date they moved. In these cases we have all the England/Wales cancers and deaths up to the point they moved but not after that, unless they re-enter the NHS.
Check that all patients registered in Scotland are also flagged in Scotland. Check that each patient does not have more than one of four variables above.
(d) Checking dates
Compare the following dates to check they are valid:
-
date of cancelled cipher
-
date of embarkation
-
date of registration in Northern Ireland
-
date of death
-
date of cancer
-
date of exams on our database.
Create “embarkation censor” that censors exams that occur after embarkation/cancelled cipher/registration in NI.
(e) Cancers and deaths after the cut off time for ascertainment.
For each patient we define a cut-off date for ascertainment of cancers and a cut-off date for ascertainment of deaths (i.e. a date for that source that we know ascertainment of cancers or deaths is complete until by that source). The cut off dates we use are shown in the table below.
| Dataset | Completeness | Date we received the last dataset from the source | Completeness date we have used |
|---|---|---|---|
| HSCIC Cancers | 6–12 months behind | 09/07/2013 | 30/06/2012 |
| HSCIC Deaths | 3 weeks behind | 09/07/2013 | 18/06/2013 |
| NSS Cancers | Completion date for 2011 incidence is 31st December 2012 in accordance with UKACR guidelines. 85–100% complete for end of 2011 by November 2012 | 07/11/2012 | 31/12/2011 |
| NSS Deaths | They currently get a weekly update of data which are considered provisional. In August of each year we get an update for the previous year after which the data are considered complete so 2011 deaths data will be complete by the end of August 2012 | 07/11/2012 | 31/12/2011 |
| NHSCR Cancers | ISD can take up to 2 years to compile a Scottish cancer registration. Their database is from 1958. NHSCR receive these monthly | 12/09/2013 | 31/12/2011 |
| NHSCR Deaths | Scottish deaths are usually registered within 3 weeks of the event. NHSCR obtain coded death copies each week, within a few weeks (no more than 9) of registration | 12/09/2013 | 22/08/2013 |
As a patient may have been traced by multiple sources we use a hierarchy to obtain the most recent cut-off date for cancer ascertainment and for death ascertainment for the patient.
For cancer ascertainment date:
-
if any sources are HSCIC, use 30/06/2012
-
if any sources are NHSCR, use 31/12/2011
-
if any sources are NSS, use 31/12/2011.
For death ascertainment date:
-
if any sources are NHSCR, use 22/08/2013
-
if any sources are HSCIC, use 18/06/2013
-
if any sources are NSS, use 31/12/2011.
Any cancers and deaths after the appropriate cut off point are then marked to be censored in the dataset (“trace censor cancer” and “trace censor death”).
(f) True date of birth and gender: Replace date of birth and gender in our dataset using true values.
(g) Follow-up date for cancer and death for each patient. Create date patient is followed until for cancer:
-
Earliest of cancer diagnosis, death, embarkation, cancelled cipher and registration in NI, if any of these occurred and if they occurred before the cancer ascertainment date.
-
If none of these events occurred and the patient was traced, the cancer ascertainment date was used.
-
If none of these events occurred and the patient was not traced, the last examination date from our database was used.
Create date patient is followed until for deaths:
-
Earliest of death, embarkation, cancelled cipher and registration in NI, if any of these occurred and if they occurred before the death ascertainment date.
-
If none of these events occurred and the patient was traced, the death ascertainment date was used .
-
If none of these events occurred and the patient was not traced, the last examination date from our database was used.
Check these are correct for groups corrected in section (d).
Calculate follow-up time for cancer and follow-up time for death for each patient.
Rule 28: map previously seen cancers from external sources
When external cancers were initially mapped to polyps on the study database, the criteria used for matching ensured that only polyps with cancer-related pathology were matched. Any unmapped cancers were treated as new cancers that had not been seen previously.
Some of the cancers were not mapped to previously seen lesions, as they did not have evidence of cancerous histology. In June 2014, it was decided that some of the large adenomas probably developed into the cancer that was reported by the HSCIC. The statistician, study researchers and principal investigator came up with new rules that could be applied to identify these cases and link the external cancer to a polyp on the POLYP table on the database.
The following rule (rule 1) was applied to all patients in the IA cohort (those who are IR patients with a baseline colonoscopy).
-
There must be a polyp record that matches the external cancer when the following criteria are applied:
-
external cancer and polyp segments must have close proximity (i.e. not more than two-tenths of the colon away)
-
polyp must be ≥ 15 mm in size:
-
DERIVED_ENDO_SIZE ≥ 15 OR
-
DERIVED_PATH_SIZE ≥ 15 OR
-
DERIVED_ENDO_SIZE_OTHER is > 10 mm (i.e. 15 mm) or large (i.e. 20 mm)
-
STAT_TRUE_SIZE ≥ 15 (this is the true size derived by statistician and recorded on the POLYP table)
-
-
-
the pathology of the polyp must be adenoma related, i.e. codes (1, 3, 51, 56, 58, 59).
| POLYPTYPE_ID | POLYP_TYPE |
|---|---|
| 1 | ADENOMA |
| 3 | SERRATED ADENOMA |
| 51 | CA + ADENOMA |
| 56 | UNICRYPTAL ADENOMA |
| 58 | CA + SERRATED ADENOMA |
| 59 | CA + MIXED ADENOMA |
In either of these fields on the POLYP table: PATH_HISTOLOGY, ASSUME_PATH_HISTOLOGY or STAT_TRUE_HIST (true histology derived by statistician and recorded on the polyp table):
-
The polyp must have occurred within the baseline dates.
-
The difference in days between the external cancer and the polyp must be within 5 years (i.e. 1826 days).
-
There must be at least one other occurrence of the polyp (i.e. same derived polyp number) within 5 years from the external cancer date (i.e. 1826 days at baseline or follow up).
A random sample of rule 1 records were reviewed so that we could be confident that the criteria used were successful in identifying previously seen lesions that were likely to have developed into the cancer from external sources.
Some other rules were also tested (rules 2 and 3), and records found from rule 2 and rule 3 matching were manually reviewed but there was not enough evidence to show that the cancer matched a previously seen lesion. Therefore, it was decided that only rule 1 would be applied for matching cancers.
Appendix 8 Deriving sizes for individual polyps
Polyp sizes
During the manual coding phase the following size fields were recorded on the POLYP table in order to accommodate the different types of information relating to polyp size that were supplied in endoscopy and pathology reports.
-
ENDO_SIZE An actual size in mm of the polyp or group of polyps as described on the endoscopy report. In some cases the ENDO_SIZE was either provided or an exact size was not provided, as it was described as part of a group of polyps (described later) or looked inaccurate (e.g. transcribing error). The study researchers reviewed the endoscopy report, pathology report and the occurrence of the polyp across examinations in order to provide the best guess of the endoscopy size for an individual polyp that was recorded in the field. ASSUME_ENDO_SIZE. ASSUME_ENDO_SIZE took precedence over ENDO_SIZE.
-
ENDO_SIZE_MAX The maximum size in mm of the polyp or group of polyps as described on the endoscopy report.
-
ENDO_SIZE_MIN The minimum size in mm of the polyp or group of polyps as described on the endoscopy report.
-
ENDO_SIZE_OTHER The category for the size of the polyp or group of polyps as described on the endoscopy report, for example tiny, > 10 mm, < 5 mm, and so on. In some cases the ENDO_SIZE_OTHER was not provided or was provided but was inaccurate. If no other sizes were available, the study researcher reviewed the endoscopy report, pathology report and the occurrence of the polyp across examinations in order to provide the best guess of the ENDO_SIZE_OTHER for an individual polyp and recorded this in the ASSUME_ENDO_SIZE_OTHER field, which took precedence over ENDO_SIZE_OTHER.
-
MAX_BIOPSY_SIZE The maximum size in millimetres of a given biopsy described by the pathologist on the pathology report.
-
PATH_SIZE An actual biopsy size in millimetres of a given biopsy described by the pathologist on the pathology report. On rare occasions, if the PATH_SIZE was not provided or looked inaccurate, the study researcher reviewed the endoscopy report, pathology report and the occurrence of the polyp across examinations in order to provide the best guess of the pathology size for an individual polyp which was recorded in a field called ASSUME_PATH_SIZE and took precedence over PATH_SIZE.
Polyp set
Some polyp sizes were recorded as polyp sets and made up of:
-
Collection of polyps, each with the same ENDO_SIZE_MIN and ENDO_SIZE_MAX range
-
One individual polyp seen on the same date whose ENDO_SIZE or ASSUME_ENDO_SIZE matched the ENDO_SIZE_MAX or ENDO_SIZE_MIN of the collection of polyps. The polyp may or may not have an ENDO_SIZE_MIN and ENDO_SIZE_MAX range. OR Two individual polyps seen on the same date where the ENDO_SIZE or ASSUME_ENDO_SIZE of one polyp matched the ENDO_SIZE_MAX of the collection of polyps and the ENDO_SIZE or ASSUME_ENDO_SIZE of the other polyp matched the ENDO_SIZE_MIN of the collection of polyps. The polyps may or may not have an ENDO_SIZE_MIN and ENDO_SIZE_MAX range, or
-
There were no individual polyps with an ENDO_SIZE or ASSUME_ENDO_SIZE recorded.
The screenshot below shows an example of a polyp set with two individual polyps.
FIGURE 33.
Screenshot showing polyp sets.

Derived polyp sizes
There following size fields were derived:
-
DERIVED_ENDO_SIZE Derived from the fields ASSUME_ENDO_SIZE, ENDO_SIZE, ENDO_SIZE_MIN, ENDO_SIZE_MAX and DERIVED_ENDO_RANGE (ASSUME_ENDO_SIZE took precedence over ENDO_SIZE).
-
DERIVED_ENDO_SIZE_SOURCE Shows the field from which the DERIVED_ENDO_SIZE was derived.
-
DERIVED_ENDO_SIZE_OTHER Derived from the fields ASSUME_ENDO_SIZE_OTHER and ENDO_SIZE_OTHER (ASSUME_ENDO_SIZE_OTHER took precedence).
-
DERIVED_ENDOSIZE_OTHER_SOURCE Shows the field from which the DERIVED_ENDO_SIZE_OTHER was derived.
-
DERIVED_PATH_SIZE Derived from the fields ASSUME_PATH_SIZE and PATH_SIZE (ASSUME_PATH_SIZE took precedence).
-
DERIVED_PATH_SIZE_SOURCE Shows the field from which the DERIVED_PATH_SIZE was derived.
Deriving endoscopy sizes for individual polyp rows
It was necessary to derive one endoscopy size called DERIVED_ENDO_SIZE where a size value was available from ENDO_SIZE, ENDO_SIZE_MIN and ENDO_SIZE_MAX but on initial analysis of the data the following limitations were identified and decisions were made to overcome them.
Only ENDO_SIZE_MAX was recorded
For some polyp rows only ENDO_SIZE_MAX was recorded. It was known that when an endoscopist used ENDO_SIZE_MAX, it was often to describe the maximum size within a collection of polyps. If the size was large and used for all the polyps in the collection to get the DERIVED_ENDO_SIZE, it could have led to such polyps being classified as higher risk. The following decisions were made to allocate the DERIVED_ENDO_SIZE based on different scenarios:
-
If the ENDO_SIZE_MAX was only allocated to one polyp as opposed to a collection of polyps, then the ENDO_SIZE_MAX was assumed to be its correct size.
-
If the ENDO_SIZE_MAX ≤ 5 mm for a collection of polyps, then it was assumed to be the correct size as it was small so DERIVED_ENDO_SIZE would be set to ENDO_SIZE_MAX.
-
If the ENDO_SIZE_MAX > 5 mm for a collection of polyps and it matched the ENDO_SIZE or ASSUME_ENDO_SIZE of an individual polyp in the set and all the segments in the polyp set matched (ENDO_SEGMENT and ENDO_SEGMENT_TO), then the DERIVED_ENDO_SIZE of the individual polyp was set to ENDO_SIZE_MAX and the DERIVED_ENDO_SIZE of the remaining polyps in the polyp set were set to ‘UNKNOWN’.
-
If the ENDO_SIZE_MAX > 5 mm for a collection of polyps and none of the polyps in the polyp set had an ENDO_SIZE or ASSUME_ENDO_SIZE recorded, then the polyps set was reviewed by the study researchers and they allocated an ASSUME_ENDO_SIZE where possible. The DERIVED_ENDO_SIZE was then taken from ASSUME_ENDO_SIZE where available and the DERIVED_ENDO_SIZE of the remaining polyps were set to ‘UNKNOWN’.
ENDO_SIZE_MIN and ENDO_SIZE_MAX only recorded
For some polyp rows only ENDO_SIZE_MIN and ENDO_SIZE_MAX were recorded. It would not have been appropriate to assign a DERIVED_ENDO_SIZE by calculating an average of the two sizes, particularly in cases where the ENDO_SIZE_MIN and ENDO_SIZE_MAX differed considerably.
The ENDO_SIZE_MIN and ENDO_SIZE_MAX were recorded in 2 different scenarios:
-
Scenario A – Some endoscopy reports had specific size and site details for individual polyps. However, ENDO_SIZE_MIN and ENDO_SIZE_MAX were used by study researchers so that pathology could be assigned to a polyp, as the pathology report did not give sufficient detail to determine which polyp the histology belonged to.
-
Scenario B – Some endoscopy reports only gave broad details of size (and site) for a collection of polyps, so ENDO_SIZE_MIN and ENDO_SIZE_MAX had to be used because no individual polyp details were given e.g. 10 polyps between 5–25 mm.
The following decisions were made to allocate the DERIVED_ENDO_SIZE based on different scenarios:
-
For both scenarios, if the ENDO_SIZE or ASSUMED_ENDO_SIZE of two individual polyps in the set matched the ENDO_SIZE_MIN and ENDO_SIZE_MAX of the polyp set, respectively, and all the segments in the polyp set matched (ENDO_SEGMENT and ENDO_SEGMENT_TO), then the DERIVED_ENDO_SIZE of the individual polyp(s) was set to the ENDO_SIZE or ASSUME_ENDO_SIZE and the DERIVED_ENDO_SIZE of the remaining polyps in the polyp set were set to ‘UNKNOWN’.
-
The study researcher created an ENDO_PATH_MAPPING field on the POLYP table, which was used to record the rule used for matching pathology to the polyp. For scenario A and B, if the DERIVED_ENDO_SIZE had not been allocated, the study researchers used the rules for ‘endo path mapping’ (described later) to recode the pathology of the polyps and the ENDO_SIZE when available. The ENDO_SIZE_MIN and ENDO_SIZE_MAX were set to blank if the ENDO_SIZE was recorded.
-
For scenario B, the study researchers tried to assign a value to ASSUME_ENDO_SIZE. They tried to assign one lesion the ENDO_SIZE_MIN and one lesion the ENDO_SIZE_MAX when possible. They also tried to assign a size to the other polyps if possible. If the ASSUME_ENDO_SIZE was not assigned then the DERIVED_ENDO_SIZE was deemed ‘UNKNOWN’, otherwise it was assigned to the DERIVED_ENDO_SIZE.
-
An average was assigned to DERIVED_ENDO_SIZE for any cases with just one polyp with ENDO_SIZE_MIN and ENDO_SIZE_MAX.
-
For cases that were not reviewed, an average was assigned to DERIVED_ENDO_SIZE for all polyps for which ENDO_SIZE_MIN and ENDO_SIZE_MAX did not differ considerably.
-
For cases which were reviewed, ‘UNKNOWN’ was assigned to DERIVED_ENDO_SIZE for all polyps in the review that were not assigned an ASSUME_ENDO_SIZE. Otherwise the ASSUME_ENDO_SIZE was assigned to DERIVED_ENDO_SIZE.
Deriving endoscopy size for a collection of polyps based on pathology information (‘endo path mapping’)
These rules were applied where the ENDO_SIZE_MIN and ENDO_SIZE_MAX were recorded for a collection of polyps and in some cases the actual size and pathology information was available. Majority of these records were ‘Scenario A’ type records described earlier. It was decided that such recorded needed to be recoded and specifically the following records were reviewed:
-
ENDO_SIZE_MIN is < 10 mm, ENDO_SIZE_MAX is ≥ 10 mm AND sizes differ by ≥ 3 mm
-
ENDO_SIZE_MIN is ≥ 10 mm, ENDO_SIZE_MAX is ≥ 15 mm AND sizes differ by ≥ 5 mm
-
ENDO_SIZE_MIN is ≤ 5 mm, ENDO_SIZE_MAX is 6–9 mm AND sizes differ by ≥ 2 mm
-
ENDO_SIZE_MIN is > 5 mm, ENDO_SIZE_MAX is ≤ 9 mm AND sizes differ by ≥ 3 mm
The following rules were applied when recoding these cases:
-
The study researcher put the specific size and segment details back into the ENDO_SIZE and ENDO_SEGMENT fields, removing size ranges and ‘SEGMENT_TO’ as appropriate.
-
Other pathology was recoded where specific information was available.
-
The histology rules were applied and the ENDO_PATH_MAPPING was used to record the rule applied:
-
Rule 1 – Histology size – Assign histology most likely to be associated with a large polyp to the largest lesions. Specifically, largest polyp will be a) villous, b) severely dysplastic, c) tubulovillous, d) mild/moderately dysplastic. These are in order of histological features most predictive of largest size.
-
Rule 2 – Hyperplastic Distal ≤ 5 mm – Hyperplastic polyp pathology should be assigned to the most distal lesion but ONLY if this lesion is ≤ 5 mm.
-
Rule 3 – Excision Method – Excision extent – assume that polyps which were snared are larger than those that were hot biopsied.
-
Rule 4 – Specimen Labels – Specimen labels i.e. 1–10/A-E, go from the most proximal to most distal site.
-
Rule 5 – Other Sighting – Where a polyp is seen at a prior or subsequent sighting which has pathology. Polyp numbering is considered for this.
-
-
The polyp numbering was checked to ensure it was still correct after the polyp information had been amended.
Automatic deriving of ENDO_SIZE for a polyp set
The study programmer used a number of rules to automatically assign an endoscopy size to all the polyps in the set (the collection and the individual polyps) and this size was recorded on the DERIVED_ENDO_RANGE field. The method used to derive this size was recorded on the DERIVED_ENDO_RANGE_GROUP. It was only possible to do this automatically where the segment of the collection of polyps matched the segment of the individual polyp. This was done before the study researchers manually reviewed any records for size including the records where the ‘endo path mapping’ rules were applied.
The following rules were used to assign the size to the DERIVED_ENDO_RANGE field.
-
Group A1: Polyp with an ENDO_SIZE_MAX ≤ 5 mm and no ENDO_SIZE_MIN. The DERIVED_ENDO_RANGE was set to ENDO_SIZE_MAX.
-
Group A2: Polyp with an ENDO_SIZE_MIN ≤ 5 mm and no ENDO_SIZE_MAX. The DERIVED_ENDO_RANGE was set to ENDO_SIZE_MIN.
-
Group X1: Polyp with an ENDO_SIZE_MIN and ENDO_SIZE_MAX that both matched the ASSUME_ENDO_SIZE or ENDO_SIZE of collection of polyps in the polyp set and all the segments of the polyp set were the same. Such polyps were allocated their ASSUME_ENDO_SIZE or ENDO_SIZE and the other polyps with that range were allocated ‘UNKNOWN’ size as the DERIVED_ENDO_RANGE.
-
Group Y1: Collection of polyps with an ENDO_SIZE_MAX that matched the ASSUME_ENDO_SIZE or ENDO_SIZE of an individual polyp in the polyp set, but the segments did not match. The record was reviewed manually by the study researchers. The DERIVED_ENDO_RANGE was set to ASSUME_ENDO_SIZE or ENDO_SIZE for the polyp that had this information (after study researchers had populated this information) and ‘UNKNOWN’ for the other polyps in the polyp set.
-
Groups B, C and D set the DERIVED_ENDO_RANGE to ENDO_SIZE for the individual polyp in the polyp set and ‘UNKNOWN’ for the rest of the polyp collection where there was only ENDO_SIZE_MAX and it matched the ENDO_SIZE of the individual polyp in the group.
-
B was applied for polyp sets where just ENDO_SEGMENT was recorded for all polyps in the set
-
C was applied for groups where ENDO_SEGMENT and ENDO_SEGMENT_TO were recorded for all polyps in the set
-
D was applied for groups where neither ENDO_SEGMENT and ENDO_SEGMENT_TO were recorded for all polyps in the set
-
-
Groups E, F and G set the DERIVED_ENDO_RANGE to ENDO_SIZE for the individual polyp in the set and ‘UNKNOWN’ for the rest of the polyp collection where there was only ENDO_SIZE_MIN and it matched the ENDO_SIZE of the individual polyp in the polyp set.
-
E was applied for groups where just ENDO_SEGMENT was recorded
-
F was applied for groups where ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
G was applied for groups where neither ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
-
Groups H, I and J set the DERIVED_ENDO_RANGE to ASSUME_ENDO_SIZE for the individual polyp and ‘UNKNOWN’ for the rest of the polyp collection where there was only ENDO_SIZE_MAX and it matched the ASSUME_ENDO_SIZE of the individual polyp in the polyp set.
-
H was applied for groups where just ENDO_SEGMENT was recorded
-
I was applied for groups where ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
J is applied for groups where neither ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
-
Groups K, L and M set the DERIVED_ENDO_RANGE to ASSUME_ENDO_SIZE for the individual polyp and ‘UNKNOWN’ for the rest of the polyp collection where there was only ENDO_SIZE_MIN and it matched the ASSUME_ENDO_SIZE of the of the individual polyp in the polyp set.
-
K was applied for groups where just ENDO_SEGMENT was recorded
-
L was applied for groups where ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
M was applied for groups where neither ENDO_SEGMENT and ENDO_SEGMENT _TO were recorded
-
Deriving endoscopy size for endo quantity rows and multilinked polyps
For ‘endo quantity rows’ and multilinked polyps, the following rules were applied later to derive the DERIVED_ENDO_RANGE. The DERIVED_ENDO_RANGE_GROUP and DERIVED_ENDO_RANGE was set to blank for these type of polyps and the following rules are used to re-derive them. A MP_SET (multiple polyp set) was defined as a group of polyps at the same examination which was made up of a polyp with an ‘endo quantity row’ (i.e. ENDO_QUANTITY_OTHER has a value) and another group of polyps that were multilinked to it (i.e. PATH_MULTI_ENDO_LINK of these polyps is equal to the POLYP_ID of the ‘endo quantity row’.)
-
Group A1 – sets the DERIVED_ENDO_RANGE to ENDO_SIZE_MAX where the ENDO_SIZE_MAX ≤ 5 mm and no ENDO_SIZE_MIN.
-
Group A2 – sets the DERIVED_ENDO_RANGE to ENDO_SIZE_MIN where the ENDO_SIZE_MIN ≤ 5 mm and no ENDO_SIZE_MAX.
-
Group Q1 – set the DERIVED_ENDO_RANGE to UNKNOWN for remaining polyps in the MP_SET if both the ENDO_SIZE_MIN and ENDO_SIZE_MAX of the ‘endo quantity row’ had been allocated to other multilinked polyps in the MP_SET.
-
Group Q2 – set the DERIVED_ENDO_RANGE to UNKNOWN for remaining polyps if the ENDO_SIZE_MAX of the ‘endo quantity row’ was null and ENDO_SIZE_MIN of the ‘endo quantity row’ has been allocated to a multilinked polyp in the MP_SET.
-
Group Q3 – set the DERIVED_ENDO_RANGE to UNKNOWN for remaining polyps if the ENDO_SIZE_MIN of the ‘endo quantity row’ is null and ENDO_SIZE_MAX of the ‘endo quantity row’ had been allocated to a multilinked polyp in the MP_SET.
-
Group Q4a – if the ENDO_SIZE_MIN of the ‘endo quantity row’ was null and ENDO_SIZE_MAX of the ‘endo quantity row’ > 5 mm had not been allocated to a multilinked polyp in the MP_SET, a programme was used to identify the polyp that the size could be allocated to and the DERIVED_ENDO_SIZE was allocated to that polyp.
-
Group Q4b – set the DERIVED_ENDO_RANGE to UNKNOWN for all the remaining polyps left in the MP_SET after the ENDO_SIZE_MAX has been assigned in Group4a.
-
Group 5a – where the ENDO_SIZE_MAX of the ‘endo quantity row’ was null and ENDO_SIZE_MIN of the ‘endo quantity row’ > 5 mm had not been allocated to a multilinked polyp in the MP_SET, a programme was used to identify the polyp and the DERIVED_ENDO_SIZE was allocated to that polyp.
-
Group 5b – set the DERIVED_ENDO_RANGE to UNKNOWN for all the remaining polyps left in the MP_SET after the ENDO_SIZE_MIN had been assigned in Group5a.
-
Group Q6a – where the ENDO_SIZE_MAX and ENDO_SIZE_MIN of the ‘endo quantity row’ was not null. If the ENDO_SIZE_MAX has not been allocated to a multilinked polyp in the MP_SET, a programme was used to identify the polyp and the DERIVED_ENDO_SIZE was allocated to that polyp.
-
Group Q7a – if the ENDO_SIZE_MAX and ENDO_SIZE_MIN of the ‘endo quantity row’ was not null and if the ENDO_SIZE_MIN has not been allocated to a multilinked polyp in the MP_SET, a programme was used to identify the polyp and the DERIVED_ENDO_SIZE was allocated to that polyp.
-
Group Q6b and Q7b – set the DERIVED_ENDO_RANGE to UNKNOWN For all the remaining polyps left in the MP_SET after the ENDO_SIZE_MIN and ENDO_SIZE_MAX had been assigned in Groups 6a and 7a above.
Manual review of size
After the DERIVED_ENDO_RANGE had been automatically assigned, the study researchers manually reviewed the some of the polyp rows where a size could not be automatically assigned. This manual review of records has already been discussed earlier under this section on ‘Deriving endoscopy sizes’. Where possible a size was assigned to ENDO_SIZE and ASSUME_ENDO_SIZE. In very few cases where (about 22 patients), the ASSUME_ENDO_SIZE_OTHER was sometimes recorded.
In addition to this, polyp records with other size discrepancies were also manually reviewed by the study researcher. Although most discrepancies seemed to be due to different measuring techniques used by endoscopists and pathologists, some reports had a coding, transcription or typographical errors and the sizes were corrected in such cases. In some cases the study researchers were able to decide what the correct size must be and recorded it in the ASSUME_ENDO_SIZE or ASSUME_PATH_SIZE fields as appropriate. Additionally, for certain centres it seemed that endoscopy size was always recorded using ENDO_SIZE_MAX so these cases were identified and reviewed in order for the size to be correctly assigned to the ENDO_SIZE field instead. For these reviews, particular attention was paid to the excision method i.e. was the method most viable for the removal of a large polyp or a small polyp, and the follow-up and patient’s history in general.
-
Size discrepancy at the same examination – As 10 mm was an important cut-off point where a polyp becomes classified as high risk, for discrepancies at the same examination there was a manual review of cases where:
-
ENDO_SIZE10–20 mm and PATH_SIZE ≥ 40 mm
-
ENDO_SIZE< 10 mm & PATH_SIZE ≥ 20 mm
-
Endo_size_other = tiny, small, < 5 mm, 5–9 mm, ≤ 10 mm and pathology size ≥ 20 mm
-
-
Size discrepancy across examinations – If the polyp was seen across other examinations (i.e. had the same POLYP_NUMBER and the MATCH_PROBABILITY ≥ 70%) then there was a manual review of cases where:
-
ENDO_SIZE< 10 mm at ANY examination and PATH_SIZE ≥ 30 mm at ANY SUBSEQUENT examination
-
ENDO_SIZE< 10 mm at ANY examination and endo_size ≥ 30 mm at ANY SUBSEQUENT examination
-
ENDO_SIZE was 10–20 mm at ANY examination and PATH_SIZE≥ 40 mm at ANY SUBSEQUENT examination
-
ENDO_SIZE was 10–20 mm at ANY examination and ENDO_SIZE ≥ 40 mm at ANY SUBSEQUENT examination
-
DERIVED_ENDO_SIZE for patients without multiple polyp rows
Once the DERIVED_ENDO_RANGE was generated and size discrepancies were reviewed and corrected where necessary, the study programmer used a programme and applied further rules to derive the field DERIVED_ENDO_SIZE. The first set of rules were applied to any polyps that were not an ‘endo quantity row’ (i.e. ENDO_QUANTITY_ OTHER has a value) or a multilinked polyp (i.e. PATH_MULTI_ENDO_LINK has been set). The DERIVED_ENDO_RANGE was set to blank for any polyps that had been manually reviewed after it was derived. The following rules were applied, the size was recorded on field DERIVED_ENDO_SIZE, the rule used was recorded on DERIVED_ENDO_SIZE_SOURCE and the field used for the size e.g. ENDO_SIZE, ASSUME_ENDO_SIZE was copied to DERIVED_ENDO_SIZE_SOURCE_OTHER.
-
Rule 1 – ASSUME_ENDO_SIZE took precedence over all endoscopy sizes, as this was field used to indicate any corrected sizes that should be used.
-
Rule 2 – ENDO_SIZE took precedence over all endoscopy sizes except ASSUME_ENDO_SIZE, as this was deemed to be the next most accurate size available.
-
Rule 3 – The automated DERIVED_ENDO_RANGE took precedence over all endoscopy sizes except ASSUME_ENDO_SIZE and ENDO_SIZE.
-
Rule 4 – If there was only one polyp at an examination with ENDO_SIZE_MAX and no other size, the DERIVED_ENDO_SIZE was set to ENDO_SIZE_MAX.
-
Rule 5 – If there was only one polyp at an examination with ENDO_SIZE_MIN and no other size, the DERIVED_ENDO_SIZE was set to ENDO_SIZE_MIN.
-
Rule 6 – If there was only one polyp at an examination with just ENDO_SIZE_MIN and ENDO_SIZE_MAX then the average of ENDO_SIZE_MIN and ENDO_SIZE_MAX was used.
-
Rule 7 – An average of ENDO_SIZE_MIN and ENDO_SIZE_MIN was used for DERIVED_ENDO_SIZE if the polyp was not. Polyps were not reviewed when the size discrepancies were quite small so applying an average was deemed to be acceptable in this scenario.
-
Rule 8 – If the polyp was reviewed and did not have ENDO_SIZE and ASSUME_ENDO_SIZE then the DERIVED_ENDO_SIZE was set as ‘UNKNOWN’.
-
Rule 9 – If the polyp was reviewed and did not yet have DERIVED_ENDO_SIZE or ENDO_SIZE_MIN and ENDO_SIZE_MAX ≤ 5, the DERIVED_ENDO_SIZE was set to the ENDO_SIZE_MAX.
-
Rule 10 – If the polyp was reviewed and did not yet have DERIVED_ENDO_SIZE or ENDO_SIZE_MAX and ENDO_SIZE_MIN ≤ 5, the DERIVED_ENDO_SIZE was set to the ENDO_SIZE_MIN.
DERIVED_ENDO_SIZE for patients with multiple polyp rows
Slightly different rules were applied to multiple polyp patients.
Rules applied to polyps that were not an ‘endo quantity row’ or a multilinked polyp
The normal rules 1 to 11 described above were applied to the all polyps which were not an ‘endo quantity row’ or a multilinked polyp. The following additional rules were also applied to this group.
-
Rule 12 – If the polyp was not an ‘endo quantity row’ or a multilinked polyp, the DERIVED_ENDO_SIZE was blank, the ENDO_SIZE_MAX was blank, the ENDO_SIZE_MIN did not match any ENDO_SIZE, ASSUME_ENDO_SIZE or ENDO_SIZE_MIN of another polyp at the same endoscopy, then the DERIVED_ENDO_SIZE was set to ENDO_SIZE_MIN.
-
Rule 13 – If the polyp was not an ‘endo quantity row’ or a multilinked polyp, the DERIVED_ENDO_SIZE was blank, the ENDO_SIZE_MIN was blank, the ENDO_SIZE_MAX did not match any ENDO_SIZE, ASSUME_ENDO_SIZE or ENDO_SIZE_MAX of another polyp at the same endoscopy, then the DERIVED_ENDO_SIZE was set to ENDO_SIZE_MAX.
-
Rule 14 – If the polyp was not an ‘endo quantity row’ or a multilinked polyp, the DERIVED_ENDO_SIZE was blank and there were no other polyps at the same endoscopy, the DERIVED_ENDO_SIZE was set to the average of ENDO_SIZE_MIN and ENDO_SIZE_MAX.
-
Rule 15 – If the polyp was not an ‘endo quantity row’ or a multilinked polyp, the DERIVED_ENDO_SIZE was blank and the polyp sizes were ≤ 5, the DERIVED_ENDO_SIZE was set to the average of ENDO_SIZE_MIN and ENDO_SIZE_MAX unless the ENDO_SIZE or ASSUME_ENDO_SIZE have a value. Records with unknown size were overwritten where this applied (DERIVED_ENDO_RANGE remained as UNKNOWN so it is clear when this was done).
-
Rule 16 – DERIVED_ENDO_SIZE was set to ENDO_SIZE_MIN where there was only ENDO_SIZE_MIN and it was ≤ 5. This size was allocated to all polyps in the group unless they already had a DERIVED_ENDO_SIZE. Records with unknown size were overwritten where this applied (DERIVED_ENDO_RANGE remained as UNKNOWN so it is clear when this was done).
-
Rule 17 – DERIVED_ENDO_SIZE was set to ENDO_SIZE_MAX where there was only ENDO_SIZE_MAX and it was ≤ 5. This size was allocated to all polyps in the group unless they already had a DERIVED_ENDO_SIZE. Records with unknown size were overwritten where this applied (DERIVED_ENDO_RANGE remained as UNKNOWN so it is clear when this was done).
Rules applied to ‘endo quantity rows’ or multilinked polyps
The DERIVED_ENDO_SIZE, DERIVED_ENDO_SIZE_SOURCE and DERIVED_ENDO_SIZE_SOURCE_OTHER were reset to blank for ‘endo quantity rows’ and multilinked polyps. Rules 1–6 above were re-applied and then rules 15–17 were applied. Finally rule 18 below was applied.
-
Rule 18 – An average of ENDO_SIZE_MIN and ENDO_SIZE_MAX was allocated to all remaining polyps without a DERIVED_ENDO_SIZE, irrespective of the magnitude of these sizes except for any polyps that already had a ENDO_SIZE or ASSUME_ENDO_SIZE allocated. It did not overwrite records with UNKNOWN.
Derived endoscopy size other
In some instances, the endoscopist gave a vague description of endoscopic size using terms such as ‘large’ and ‘small’. As such, the field ENDO_SIZE_OTHER was created so these data could be coded. At analysis, this field had to be assigned quantitative values to enable the classification of each patient’s baseline risk. Analyses were performed to compare the actual sizes recorded in ENDO_SIZE and PATH_SIZE with the values recorded in ENDO_SIZE_OTHER, wherever possible. After lengthy discussion, specific sizes were assigned to each ENDO_SIZE_OTHER value based on this as shown above.
| ENDO_SIZE_OTHER/ASSUME_ENDO_SIZE_OTHER/DERIVED_ENDO_SIZE_OTHER | Description | Assigned value for analysis |
|---|---|---|
| 1 | Tiny | 3 mm |
| 2 | Small | 5 mm |
| 3 | < 5 mm | 3 mm |
| 4 | 5–9 mm | 7 mm |
| 5 | > 10 mm | 15 mm |
| 6 | Large | 20 mm |
| 7 | < 10 mm | 8 mm |
DERIVED_ENDO_SIZE_OTHER was derived from the fields ASSUME_ENDO_SIZE_OTHER and ENDO_SIZE_OTHER with ASSUME_ENDO_SIZE_OTHER always taking precedence over ENDO_SIZE_OTHER, as this field was used to code corrected ‘ASSUMED’ sizes.
Derived pathology size for individual polyp rows
DERIVED_PATH_SIZE was derived from the fields ASSUME_PATH_SIZE and PATH_SIZE with ASSUME_PATH_SIZE always taking precedence over PATH_SIZE, as this field was used to code corrected ‘assumed’ sizes. DERIVED_PATH_SIZE_SOURCE showed the field from which the size was taken.
Appendix 9 Additional results tables
Follow-up visit 1 risk factors and interval between first and second follow-up
| FUV1 risk factors | Interval from first to second follow-up, n (%) | Total (%) | p-value (chi-squared) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||||
| Demographic characteristics | ||||||||||
| Age (years) | < 55 | 60 (18.24) | 83 (25.23) | 114 (34.65) | 35 (10.64) | 23 (6.99) | 7 (2.13) | 7 (2.13) | 329 (100) | < 0.001 |
| ≥ 55 and < 60 | 47 (18.36) | 51 (19.92) | 93 (36.33) | 27 (10.55) | 32 (12.5) | 3 (1.17) | 3 (1.17) | 256 (100) | ||
| ≥ 60 and < 65 | 53 (19.00) | 57 (20.43) | 101 (36.2) | 31 (11.11) | 22 (7.89) | 9 (3.23) | 6 (2.15) | 279 (100) | ||
| ≥ 65 and < 70 | 84 (27.54) | 62 (20.33) | 98 (32.13) | 33 (10.82) | 23 (7.54) | 3 (0.98) | 2 (0.66) | 305 (100) | ||
| ≥ 70 and < 75 | 74 (29.25) | 70 (27.67) | 66 (26.09) | 13 (5.14) | 18 (7.11) | 8 (3.16) | 4 (1.58) | 253 (100) | ||
| ≥ 75 and < 80 | 43 (30.28) | 39 (27.46) | 37 (26.06) | 10 (7.04) | 6 (4.23) | 0 (0) | 7 (4.93) | 142 (100) | ||
| ≥ 80 | 36 (50.7) | 14 (19.72) | 9 (12.68) | 3 (4.23) | 7 (9.86) | 1 (1.41) | 1 (1.41) | 71 (100) | ||
| Gender | Male | 239 (25) | 217 (22.7) | 299 (31.28) | 86 (9) | 81 (8.47) | 20 (2.09) | 14 (1.46) | 956 (100) | 0.715 |
| Female | 158 (23.27) | 159 (23.42) | 219 (32.25) | 66 (9.72) | 50 (7.36) | 11 (1.62) | 16 (2.36) | 679 (100) | ||
| Family history of cancer/CRC | No | 382 (25.08) | 350 (22.98) | 468 (30.73) | 142 (9.32) | 126 (8.27) | 27 (1.77) | 28 (1.84) | 1523 (100) | 0.014 |
| Yes | 15 (13.39) | 26 (23.21) | 50 (44.64) | 10 (8.93) | 5 (4.46) | 4 (3.57) | 2 (1.79) | 112 (100) | ||
| Year of visit | 1985–94 | 39 (31.2) | 26 (20.8) | 25 (20) | 14 (11.2) | 13 (10.4) | 2 (1.6) | 6 (4.8) | 125 (100) | < 0.001 |
| 1995–9 | 68 (19.15) | 69 (19.44) | 116 (32.68) | 41 (11.55) | 33 (9.3) | 10 (2.82) | 18 (5.07) | 355 (100) | ||
| 2000–4 | 165 (19.3) | 182 (21.29) | 312 (36.49) | 88 (10.29) | 83 (9.71) | 19 (2.22) | 6 (0.7) | 855 (100) | ||
| 2005–9 | 125 (41.67) | 99 (33) | 65 (21.67) | 9 (3) | 2 (0.67) | 0 (0) | 0 (0) | 300 (100) | ||
| Procedural characteristics | ||||||||||
| Most complete examination | Complete colonoscopy | 238 (21.9) | 239 (21.99) | 367 (33.76) | 102 (9.38) | 98 (9.02) | 22 (2.02) | 21 (1.93) | 1087 (100) | < 0.001 |
| Colonoscopy of unknown completeness | 20 (15.38) | 23 (17.69) | 48 (36.92) | 22 (16.92) | 10 (7.69) | 5 (3.85) | 2 (1.54) | 130 (100) | ||
| Incomplete colonoscopy | 44 (30.34) | 38 (26.21) | 42 (28.97) | 8 (5.52) | 7 (4.83) | 2 (1.38) | 4 (2.76) | 145 (100) | ||
| Colonoscopy or FS | 31 (29.25) | 29 (27.36) | 22 (20.75) | 11 (10.38) | 11 (10.38) | 1 (0.94) | 1 (0.94) | 106 (100) | ||
| FS | 49 (45.37) | 35 (32.41) | 18 (16.67) | 5 (4.63) | 1 (0.93) | 0 (0) | 0 (0) | 108 (100) | ||
| Colonoscopy, or flexible or rigid sigmoidoscopy | 12 (22.64) | 9 (16.98) | 21 (39.62) | 4 (7.55) | 4 (7.55) | 1 (1.89) | 2 (3.77) | 53 (100) | ||
| Surgery | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | ||
| Unknown | 1 (25) | 3 (75) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (100) | ||
| Best bowel preparation at colonoscopy | Excellent/good | 92 (19.83) | 93 (20.04) | 164 (35.34) | 58 (12.5) | 42 (9.05) | 11 (2.37) | 4 (0.86) | 464 (100) | < 0.001 |
| Satisfactory | 45 (26.63) | 39 (23.08) | 57 (33.73) | 18 (10.65) | 6 (3.55) | 3 (1.78) | 1 (0.59) | 169 (100) | ||
| Poor | 23 (33.82) | 14 (20.59) | 20 (29.41) | 5 (7.35) | 4 (5.88) | 1 (1.47) | 1 (1.47) | 68 (100) | ||
| Unknown | 142 (21.48) | 154 (23.3) | 216 (32.68) | 51 (7.72) | 63 (9.53) | 14 (2.12) | 21 (3.18) | 661 (100) | ||
| No known colonoscopy | 95 (34.8) | 76 (27.84) | 61 (22.34) | 20 (7.33) | 16 (5.86) | 2 (0.73) | 3 (1.1) | 273 (100) | ||
| Difficult examination | No | 373 (23.93) | 355 (22.77) | 498 (31.94) | 150 (9.62) | 127 (8.15) | 29 (1.86) | 27 (1.73) | 1559 (100) | 0.127 |
| Yes | 24 (31.58) | 21 (27.63) | 20 (26.32) | 2 (2.63) | 4 (5.26) | 2 (2.63) | 3 (3.95) | 76 (100) | ||
| Length of visit | 1 day | 303 (21.31) | 315 (22.15) | 484 (34.04) | 139 (9.77) | 125 (8.79) | 27 (1.9) | 29 (2.04) | 1422 (100) | < 0.001 |
| 2–30 days | 9 (50) | 6 (33.33) | 0 (0) | 1 (5.56) | 1 (5.56) | 0 (0) | 1 (5.56) | 18 (100) | ||
| 1–3 months | 21 (56.76) | 8 (21.62) | 4 (10.81) | 2 (5.41) | 1 (2.7) | 1 (2.7) | 0 (0) | 37 (100) | ||
| 3–6 months | 21 (36.21) | 18 (31.03) | 8 (13.79) | 6 (10.34) | 3 (5.17) | 2 (3.45) | 0 (0) | 58 (100) | ||
| 6–12 months | 26 (33.33) | 27 (34.62) | 20 (25.64) | 3 (3.85) | 1 (1.28) | 1 (1.28) | 0 (0) | 78 (100) | ||
| ≥ 1 year | 17 (77.27) | 2 (9.09) | 2 (9.09) | 1 (4.55) | 0 (0) | 0 (0) | 0 (0) | 22 (100) | ||
| Number of examinations in visit | 1 | 303 (21.34) | 314 (22.11) | 483 (34.01) | 139 (9.79) | 125 (8.8) | 27 (1.9) | 29 (2.04) | 1420 (100) | < 0.001 |
| 2 | 61 (40.67) | 46 (30.67) | 22 (14.67) | 12 (8) | 5 (3.33) | 3 (2) | 1 (0.67) | 150 (100) | ||
| 3 | 22 (50) | 10 (22.73) | 10 (22.73) | 0 (0) | 1 (2.27) | 1 (2.27) | 0 (0) | 44 (100) | ||
| 4+ | 11 (52.38) | 6 (28.57) | 3 (14.29) | 1 (4.76) | 0 (0) | 0 (0) | 0 (0) | 21 (100) | ||
| FUV1 risk factors | Interval from first to second follow-up, n (%) | Total, n (%) | p-value (chi-squared) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| < 18 months | 2 yearsa | 3 yearsa | 4 yearsa | 5 yearsa | 6 yearsa | ≥ 6.5 years | ||||
| Adenoma characteristics | ||||||||||
| Number | 0 | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| 1 | 160 (37.3) | 102 (23.78) | 97 (22.61) | 35 (8.16) | 24 (5.59) | 6 (1.4) | 5 (1.17) | 429 (100) | ||
| 2 | 33 (28.21) | 33 (28.21) | 37 (31.62) | 8 (6.84) | 3 (2.56) | 1 (0.85) | 2 (1.71) | 117 (100) | ||
| 3 | 16 (43.24) | 10 (27.03) | 6 (16.22) | 4 (10.81) | 0 (0) | 0 (0) | 1 (2.7) | 37 (100) | ||
| 4 | 12 (57.14) | 4 (19.05) | 0 (0) | 1 (4.76) | 4 (19.05) | 0 (0) | 0 (0) | 21 (100) | ||
| 5+ | 15 (71.43) | 4 (19.05) | 2 (9.52) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 21 (100) | ||
| Largest size (mm) | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| < 10 | 113 (29.82) | 81 (21.37) | 110 (29.02) | 39 (10.29) | 23 (6.07) | 6 (1.58) | 7 (1.85) | 379 (100) | ||
| 10–14 | 45 (47.37) | 25 (26.32) | 17 (17.89) | 3 (3.16) | 4 (4.21) | 0 (0) | 1 (1.05) | 95 (100) | ||
| 15–19 | 31 (59.62) | 15 (28.85) | 3 (5.77) | 2 (3.85) | 1 (1.92) | 0 (0) | 0 (0) | 52 (100) | ||
| ≥ 20 | 38 (50.67) | 25 (33.33) | 9 (12) | 2 (2.67) | 1 (1.33) | 0 (0) | 0 (0) | 75 (100) | ||
| Unknown | 9 (37.5) | 7 (29.17) | 3 (12.5) | 2 (8.33) | 2 (8.33) | 1 (4.17) | 0 (0) | 24 (100) | ||
| Worst histology | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| Tubular | 111 (32.65) | 81 (23.82) | 87 (25.59) | 34 (10) | 18 (5.29) | 5 (1.47) | 4 (1.18) | 340 (100) | ||
| Tubulovillous | 68 (43.59) | 39 (25) | 31 (19.87) | 10 (6.41) | 7 (4.49) | 0 (0) | 1 (0.64) | 156 (100) | ||
| Villous | 35 (53.03) | 20 (30.3) | 8 (12.12) | 0 (0) | 2 (3.03) | 0 (0) | 1 (1.52) | 66 (100) | ||
| Unknown | 22 (34.92) | 13 (20.63) | 16 (25.4) | 4 (6.35) | 4 (6.35) | 2 (3.17) | 2 (3.17) | 63 (100) | ||
| Worst dysplasia | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| Low grade | 187 (36.81) | 120 (23.62) | 120 (23.62) | 41 (8.07) | 27 (5.31) | 7 (1.38) | 6 (1.18) | 508 (100) | ||
| High grade | 23 (50) | 15 (32.61) | 7 (15.22) | 1 (2.17) | 0 (0) | 0 (0) | 0 (0) | 46 (100) | ||
| Unknown | 26 (36.62) | 18 (25.35) | 15 (21.13) | 6 (8.45) | 4 (5.63) | 0 (0) | 2 (2.82) | 71 (100) | ||
| Proximal adenoma | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| No | 119 (37.3) | 89 (27.9) | 69 (21.63) | 20 (6.27) | 15 (4.7) | 5 (1.57) | 2 (0.63) | 319 (100) | ||
| Yes | 117 (38.24) | 64 (20.92) | 73 (23.86) | 28 (9.15) | 16 (5.23) | 2 (0.65) | 6 (1.96) | 306 (100) | ||
| Distal adenoma | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| No | 93 (38.43) | 46 (19.01) | 58 (23.97) | 22 (9.09) | 15 (6.2) | 3 (1.24) | 5 (2.07) | 242 (100) | ||
| Yes | 143 (37.34) | 107 (27.94) | 84 (21.93) | 26 (6.79) | 16 (4.18) | 4 (1.04) | 3 (0.78) | 383 (100) | ||
| Number of sightings of a single adenoma | No adenomas | 161 (15.94) | 223 (22.08) | 376 (37.23) | 104 (10.3) | 100 (9.9) | 24 (2.38) | 22 (2.18) | 1010 (100) | < 0.001 |
| 1 | 185 (34.84) | 130 (24.48) | 127 (23.92) | 45 (8.47) | 29 (5.46) | 7 (1.32) | 8 (1.51) | 531 (100) | ||
| 2 | 30 (50) | 14 (23.33) | 11 (18.33) | 3 (5) | 2 (3.33) | 0 (0) | 0 (0) | 60 (100) | ||
| 3 | 11 (55) | 6 (30) | 3 (15) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 20 (100) | ||
| 4 | 6 (75) | 1 (12.5) | 1 (12.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 8 (100) | ||
| 5+ | 4 (66.67) | 2 (33.33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (100) | ||
| Polyp characteristics (all types) | ||||||||||
| Proximal polyps | No polyps | 83 (12.44) | 146 (21.89) | 266 (39.88) | 67 (10.04) | 75 (11.24) | 14 (2.1) | 16 (2.4) | 667 (100) | < 0.001 |
| No | 150 (30.06) | 126 (25.25) | 131 (26.25) | 46 (9.22) | 29 (5.81) | 12 (2.4) | 5 (1) | 499 (100) | ||
| Yes | 164 (34.97) | 104 (22.17) | 121 (25.8) | 39 (8.32) | 27 (5.76) | 5 (1.07) | 9 (1.92) | 469 (100) | ||
| Distal polyps | No polyps | 83 (12.44) | 146 (21.89) | 266 (39.88) | 67 (10.04) | 75 (11.24) | 14 (2.1) | 16 (2.4) | 667 (100) | < 0.001 |
| No | 92 (30.07) | 67 (21.9) | 85 (27.78) | 24 (7.84) | 23 (7.52) | 8 (2.61) | 7 (2.29) | 306 (100) | ||
| Yes | 222 (33.53) | 163 (24.62) | 167 (25.23) | 61 (9.21) | 33 (4.98) | 9 (1.36) | 7 (1.06) | 662 (100) | ||
| Number of hyperplastic polyps | 0 | 313 (23.07) | 313 (23.07) | 439 (32.35) | 121 (8.92) | 117 (8.62) | 26 (1.92) | 28 (2.06) | 1357 (100) | 0.356 |
| 1 | 52 (30.06) | 45 (26.01) | 48 (27.75) | 15 (8.67) | 8 (4.62) | 4 (2.31) | 1 (0.58) | 173 (100) | ||
| 2 | 21 (33.33) | 11 (17.46) | 14 (22.22) | 12 (19.05) | 3 (4.76) | 1 (1.59) | 1 (1.59) | 63 (100) | ||
| 3 | 3 (17.65) | 4 (23.53) | 7 (41.18) | 2 (11.76) | 1 (5.88) | 0 (0) | 0 (0) | 17 (100) | ||
| 4 | 3 (20) | 2 (13.33) | 6 (40) | 2 (13.33) | 2 (13.33) | 0 (0) | 0 (0) | 15 (100) | ||
| 5+ | 5 (50) | 1 (10) | 4 (40) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 10 (100) | ||
| Any large hyperplastic polyps | No | 391 (24.17) | 372 (22.99) | 513 (31.71) | 151 (9.33) | 131 (8.1) | 30 (1.85) | 30 (1.85) | 1618 (100) | 0.645 |
| Yes | 6 (35.29) | 4 (23.53) | 5 (29.41) | 1 (5.88) | 0 (0) | 1 (5.88) | 0 (0) | 17 (100) | ||
| Number of polyps with unknown histology | 0 | 290 (21.8) | 314 (23.61) | 435 (32.71) | 131 (9.85) | 111 (8.35) | 24 (1.8) | 25 (1.88) | 1330 (100) | 0.007 |
| 1 | 53 (29.44) | 37 (20.56) | 49 (27.22) | 18 (10) | 14 (7.78) | 4 (2.22) | 5 (2.78) | 180 (100) | ||
| 2 | 31 (48.44) | 12 (18.75) | 15 (23.44) | 1 (1.56) | 4 (6.25) | 1 (1.56) | 0 (0) | 64 (100) | ||
| 3 | 10 (40) | 9 (36) | 5 (20) | 0 (0) | 0 (0) | 1 (4) | 0 (0) | 25 (100) | ||
| 4 | 6 (46.15) | 1 (7.69) | 5 (38.46) | 1 (7.69) | 0 (0) | 0 (0) | 0 (0) | 13 (100) | ||
| 5+ | 7 (30.43) | 3 (13.04) | 9 (39.13) | 1 (4.35) | 2 (8.7) | 1 (4.35) | 0 (0) | 23 (100) | ||
Effect of interval on new advanced neoplasia at the second follow-up visit
Five models were fitted, adjusting for different covariates:
-
Model A Adjusted for FUV1 risk factors.
-
Model B Adjusted for baseline risk factors.
-
Model C Adjusted for baseline and FUV1 risk factors.
-
Model D Adjusted for cumulative baseline and FUV1 risk factors.
-
model E Adjusted for baseline, FUV1 and cumulative risk factors.
Model E had the best fit to the data and is presented in Chapter 3, Second follow-up visit, Effect of interval on new findings at second follow-up of the IA Study monograph. All other models are shown below.
| FUV1 risk factors | Univariate analysis: new AN | Multivariate analyses: new AN | |||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 1635) | Model 2 – interval as continuous (n = 1635) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval FUV1 to FUV2 | < 18 months | 1.00 | 0.2313 | 1.00 | 0.0168 | n/a | |
| 2 yearsa | 1.3 (0.78 to 2.18) | 1.7 (0.98 to 2.93) | |||||
| 3 yearsa | 1.42 (0.88 to 2.28) | 2.01 (1.19 to 3.39) | |||||
| 4 yearsa | 1.39 (0.72 to 2.67) | 2.38 (1.18 to 4.81) | |||||
| 5 yearsa | 1.16 (0.56 to 2.4) | 1.87 (0.85 to 4.09) | |||||
| 6 yearsa | 1.88 (0.62 to 5.74) | 2.7 (0.83 to 8.76) | |||||
| ≥ 6.5 years | 3.86 (1.53 to 9.76) | 5.89 (2.18 to 15.92) | |||||
| Interval (per year increase) | 1.11 (1 to 1.24) | 0.0501 | n/a | 1.2 (1.08 to 1.34) | 0.0014 | ||
| Largest adenoma at FUV1 (mm) | No adenomas | 1.00 | 0.0028 | 1.00 | 0.0038 | 1.00 | 0.0061 |
| < 20 | 1.35 (0.94 to 1.94) | 1.21 (0.71 to 2.05) | 1.16 (0.69 to 1.97) | ||||
| ≥ 20 | 2.95 (1.6 to 5.43) | 3.73 (1.72 to 8.1) | 3.44 (1.61 to 7.38) | ||||
| Unknown | 3.1 (1.13 to 8.53) | 3.41 (1.09 to 10.63) | 3.18 (1.02 to 9.86) | ||||
| Proximal polyps at FUV1 | No polyps | 1.00 | 0.0005 | 1.00 | 0.0062 | 1.00 | 0.0066 |
| No | 1.18 (0.76 to 1.83) | 0.73 (0.4 to 1.34) | 0.75 (0.41 to 1.36) | ||||
| Yes | 2.12 (1.43 to 3.15) | 1.53 (0.81 to 2.9) | 1.55 (0.82 to 2.93) | ||||
| Number of polyps with unknown histology at FUV1 | 0 | 1.00 | 0.0001 | 1.00 | 0.0006 | 1.00 | 0.0008 |
| 1–4 | 1.52 (1.01 to 2.29) | 1.47 (0.88 to 2.44) | 1.41 (0.85 to 2.33) | ||||
| 5+ | 7.13 (3.02 to 16.85) | 7.37 (2.79 to 19.43) | 7.09 (2.72 to 18.43) | ||||
| Baseline risk factors | Univariate analysis: new AN | Multivariate analyses: new AN | |||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 1635) | Model 2 – interval as continuous (n = 1635) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval FUV1 to FUV2 | < 18 months | 1 | 0.2313 | 1.00 | 0.2082 | n/a | |
| 2 yearsa | 1.3 (0.78 to 2.18) | 1.34 (0.79 to 2.27) | |||||
| 3 yearsa | 1.42 (0.88 to 2.28) | 1.46 (0.9 to 2.38) | |||||
| 4 yearsa | 1.39 (0.72 to 2.67) | 1.73 (0.88 to 3.41) | |||||
| 5 yearsa | 1.16 (0.56 to 2.4) | 1.44 (0.68 to 3.06) | |||||
| 6 yearsa | 1.88 (0.62 to 5.74) | 1.81 (0.57 to 5.76) | |||||
| ≥ 6.5 years | 3.86 (1.53 to 9.76) | 4.1 (1.52 to 11.08) | |||||
| Interval (per year increase) | 1.11 (1 to 1.24) | 0.0501 | n/a | 1.14 (1.02 to 1.27) | 0.021 | ||
| Number of polyps with unknown histology at baseline | 0 | 1 | 0.0124 | 1.00 | 0.0028 | 1.00 | 0.0032 |
| 1+ | 1.63 (1.12 to 2.37) | 1.9 (1.26 to 2.88) | 1.89 (1.25 to 2.85) | ||||
| Most complete baseline colonoscopy | Complete | 1.00 | 0.0065 | 1.00 | 0.0197 | 1.00 | 0.0155 |
| Unknown completeness | 1.11 (0.76 to 1.61) | 1.08 (0.67 to 1.72) | 1.07 (0.67 to 1.72) | ||||
| Incomplete | 2.24 (1.39 to 3.59) | 2.11 (1.27 to 3.51) | 2.15 (1.3 to 3.57) | ||||
| Baseline and FUV1 risk factors | Univariate analysis: new AN | Multivariate analyses: new AN | |||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 1635) | Model 2 – interval as continuous (n = 1635) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval FUV1 to FUV2 | < 18 months | 1 | 0.2313 | 1.00 | 0.0152 | n/a | |
| 2 yearsa | 1.3 (0.78 to 2.18) | 1.66 (0.96 to 2.88) | |||||
| 3 yearsa | 1.42 (0.88 to 2.28) | 2.04 (1.2 to 3.46) | |||||
| 4 yearsa | 1.39 (0.72 to 2.67) | 2.5 (1.23 to 5.11) | |||||
| 5 yearsa | 1.16 (0.56 to 2.4) | 2.06 (0.93 to 4.53) | |||||
| 6 yearsa | 1.88 (0.62 to 5.74) | 2.78 (0.85 to 9.1) | |||||
| ≥ 6.5 years | 3.86 (1.53 to 9.76) | 5.93 (2.14 to 16.45) | |||||
| Interval (per year increase) | 1.11 (1 to 1.24) | 0.0501 | n/a | 1.21 (1.09 to 1.36) | 0.001 | ||
| Number of polyps with unknown histology at baseline | 0 | 1 | 0.0124 | 1.00 | 0.0002 | 1.00 | 0.0189 |
| 1+ | 1.63 (1.12 to 2.37) | 1.71 (1.11 to 2.63) | 1.69 (1.1 to 2.6) | ||||
| Most complete colonoscopy at baseline | Complete | 1.00 | 0.0065 | 1.00 | 0.0139 | 1.00 | 0.0114 |
| Unknown completeness | 1.11 (0.76 to 1.61) | 1.01 (0.62 to 1.65) | 1.01 (0.62 to 1.64) | ||||
| Incomplete | 2.24 (1.39 to 3.59) | 2.23 (1.31 to 3.78) | 2.25 (1.33 to 3.81) | ||||
| Largest adenoma at FUV1 (mm) | No adenomas | 1.00 | 0.0028 | 1.00 | 0.0054 | 1.00 | 0.009 |
| < 20 | 1.35 (0.94 to 1.94) | 1.32 (0.77 to 2.26) | 1.26 (0.74 to 2.15) | ||||
| ≥ 20 | 2.95 (1.6 to 5.43) | 3.94 (1.79 to 8.69) | 3.6 (1.66 to 7.83) | ||||
| Unknown | 3.1 (1.13 to 8.53) | 3.03 (0.96 to 9.6) | 2.82 (0.9 to 8.86) | ||||
| Proximal polyps at FUV1 | No polyps | 1.00 | 0.0005 | 1.00 | 0.0134 | 1.00 | 0.0132 |
| No | 1.18 (0.76 to 1.83) | 0.74 (0.4 to 1.35) | 0.75 (0.41 to 1.38) | ||||
| Yes | 2.12 (1.43 to 3.15) | 1.47 (0.77 to 2.81) | 1.49 (0.78 to 2.84) | ||||
| Number of polyps with unknown histology at FUV1 | 0 | 1.00 | 0.0001 | 1.00 | 0.0008 | 1.00 | 0.001 |
| 1–4 | 1.52 (1.01 to 2.29) | 1.39 (0.83 to 2.32) | 1.33 (0.8 to 2.22) | ||||
| 5+ | 7.13 (3.02 to 16.85) | 7.31 (2.75 to 19.4) | 7.06 (2.69 to 18.51) | ||||
| Cumulative baseline and FUV1 risk factors | Univariate analysis: new AN | Multivariate analyses: new AN | |||||
|---|---|---|---|---|---|---|---|
| Model 1 – interval as categorical (n = 1635) | Model 2 – interval as continuous (n = 1635) | ||||||
| Unadjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | Adjusted OR (95% CI) | p-value (LRT) | ||
| Interval FUV1 to FUV2 | < 18 months | 1 | 0.2313 | 1.00 | 0.0413 | n/a | |
| 2 yearsa | 1.3 (0.78 to 2.18) | 1.51 (0.89 to 2.57) | |||||
| 3 yearsa | 1.42 (0.88 to 2.28) | 1.77 (1.07 to 2.91) | |||||
| 4 yearsa | 1.39 (0.72 to 2.67) | 2.04 (1.03 to 4.05) | |||||
| 5 yearsa | 1.16 (0.56 to 2.4) | 1.66 (0.77 to 3.56) | |||||
| 6 yearsa | 1.88 (0.62 to 5.74) | 2.26 (0.7 to 7.29) | |||||
| ≥ 6.5 years | 3.86 (1.53 to 9.76) | 5.42 (2.01 to 14.58) | |||||
| Interval (per year increase) | 1.11 (1 to 1.24) | 0.0501 | n/a | 1.19 (1.06 to 1.32) | 0.0028 | ||
| Number of adenomas at baseline and FUV1 | 1 | 1.00 | 0.0003 | 1.00 | 0.0005 | 1.00 | 0.0005 |
| 2+ | 1.85 (1.31 to 2.6) | 1.88 (1.31 to 2.69) | 1.86 (1.3 to 2.66) | ||||
| Number of polyps with unknown histology at baseline and FUV1 | 0 | 1.00 | 0.0009 | 1.00 | 0.0004 | 1.00 | 0.0005 |
| 1–4 | 1.46 (1.01 to 2.11) | 1.54 (1.04 to 2.26) | 1.51 (1.03 to 2.22) | ||||
| 5+ | 3.13 (1.73 to 5.66) | 3.65 (1.92 to 6.93) | 3.58 (1.89 to 6.76) | ||||
Measures of fit (advanced neoplasia at follow-up visit 2: logistic regression models)
Five models:
-
Model A Adjusted for FUV1 risk factors.
-
Model B Adjusted for baseline risk factors.
-
Model C Adjusted for baseline and FUV1 risk factors.
-
Model D Adjusted for cumulative baseline and FUV1 risk factors.
-
Model E Adjusted for individual and cumulative baseline and FUV1 risk factors.
| Logistic regression model for effect of interval on new AN at second follow-up (adjusted for) | Measure of goodness of fit | |||
|---|---|---|---|---|
| Interval as a categorical variable | Interval as a continuous variable | |||
| AIC | BIC | AIC | BIC | |
| A (FUV1 factors) | 980.70 | 1142.68 | 975.91 | 1110.90 |
| B (baseline factors) | 1001.49 | 1141.87 | 994.59 | 1107.98 |
| C (baseline and FUV1 factors) | 971.71 | 1149.89 | 966.60 | 1117.79 |
| D (cumulative baseline and FUV1 factors) | 988.93 | 1129.31 | 983.10 | 1096.49 |
| E (individual and cumulative baseline and FUV1 factors) | 968.15 | 1151.73 | 962.84 | 1119.42 |
Appendix 10 Visual logic code for health-economic model
VL SECTION: No adenomas Work Complete Logic
‘This workcentre determines which of the following competing events will occur first: (1) other-cause mortality (2) progression to adenomas (3) attend surveillance colonoscopy
‘Determine next event
IF TimeToOCM_lbl < TimeToProgression_lbl
IF TimeToOCM_lbl < TimeToNextCOL_lbl
IF TimeToOCM_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 1
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToProgression_lbl < TimeToNextCOL_lbl
IF TimeToProgression_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 2
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
‘Set next TTNE
IF NextEvent_lbl = 1
SET TTNE_lbl = TimeToOCM_lbl
ELSE IF NextEvent_lbl = 2
SET TTNE_lbl = TimeToProgression_lbl
ELSE IF NextEvent_lbl = 3
SET TTNE_lbl = TimeToNextCOL_lbl
ELSE IF NextEvent_lbl = 4
SET TTNE_lbl = TimeToCancerDeath_lbl
VL SECTION: Adenoma Work Complete Logic
‘Competing events: (1) other-cause mortality (2) progression to adenomas (3) attend surveillance colonoscopy
‘Determine next event
IF TimeToOCM_lbl < TimeToProgression_lbl
IF TimeToOCM_lbl < TimeToNextCOL_lbl
IF TimeToOCM_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 1
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToProgression_lbl < TimeToNextCOL_lbl
IF TimeToProgression_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 2
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
‘Set next TTNE
IF NextEvent_lbl = 1
SET TTNE_lbl = TimeToOCM_lbl
ELSE IF NextEvent_lbl = 2
SET TTNE_lbl = TimeToProgression_lbl
ELSE IF NextEvent_lbl = 3
SET TTNE_lbl = TimeToNextCOL_lbl
ELSE IF NextEvent_lbl = 4
SET TTNE_lbl = TimeToCancerDeath_lbl
VL SECTION: Preclinical CRC Work Complete Logic
‘Competing events: (1) other-cause mortality (2) progression to adenomas (3) attend surveillance colonoscopy
‘Determine next event
IF TimeToOCM_lbl < TimeToProgression_lbl
IF TimeToOCM_lbl < TimeToNextCOL_lbl
IF TimeToOCM_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 1
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToProgression_lbl < TimeToNextCOL_lbl
IF TimeToProgression_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 2
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
‘Set next TTNE
IF NextEvent_lbl = 1
SET TTNE_lbl = TimeToOCM_lbl
ELSE IF NextEvent_lbl = 2
SET TTNE_lbl = TimeToProgression_lbl
ELSE IF NextEvent_lbl = 3
SET TTNE_lbl = TimeToNextCOL_lbl
ELSE IF NextEvent_lbl = 4
SET TTNE_lbl = TimeToCancerDeath_lbl
VL SECTION: Reset Logic
‘Obeyed just after all simulation objects are initialized at time zero
VL SECTION: Clinical CRC Work Complete Logic
‘Competing events: (1) other-cause mortality (2) progression to adenomas (3) attend surveillance colonoscopy
‘Determine next event
IF TimeToOCM_lbl < TimeToProgression_lbl
IF TimeToOCM_lbl < TimeToNextCOL_lbl
IF TimeToOCM_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 1
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToProgression_lbl < TimeToNextCOL_lbl
IF TimeToProgression_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 2
ELSE
SET NextEvent_lbl = 4
ELSE
IF TimeToNextCOL_lbl < TimeToCancerDeath_lbl
SET NextEvent_lbl = 3
ELSE
SET NextEvent_lbl = 4
‘Set next TTNE
IF NextEvent_lbl = 1
SET TTNE_lbl = TimeToOCM_lbl
ELSE IF NextEvent_lbl = 2
SET TTNE_lbl = TimeToProgression_lbl
ELSE IF NextEvent_lbl = 3
SET TTNE_lbl = TimeToNextCOL_lbl
ELSE IF NextEvent_lbl = 4
SET TTNE_lbl = TimeToCancerDeath_lbl
VL SECTION: End Run Logic
‘Obeyed when the simulation reaches end of "Results Collection Period"
SET Model entry.Interarrival Time = 0.00001
IF SOUR_nbr < MaxSOUR_nbr
SET SOUR_nbr = SOUR_nbr+1
Reset before next run
RunModel 10000
VL SECTION: Dead Work Complete Logic
‘This workcentre records all health gains and stores model results
‘Record patient life years gained
SET UndiscountedLYGs_lbl = TimeStampDeath_lbl-TimeStampModelEntry_lbl
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDeath_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampModelEntry_lbl]
SET DiscountedLYGs_lbl = DiscountedLYGs_lbl+[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]
‘Record patient QALY gains
IF histology_lbl <= 2
‘Calculate undiscounted QALY gains (never develop cancer)
SET temp1_lbl = TimeStampDeath_lbl-TimeStampModelEntry_lbl
SET UndiscountedQALYs_lbl = temp1_lbl*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]
‘Calculate discounted QALY gains
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDeath_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampModelEntry_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]]
ELSE IF histology_lbl = 3
‘Calculate undiscounted QALY gains (develop preclinical cancer only)
SET temp1_lbl = TimeStampDevelopCancer_lbl-TimeStampModelEntry_lbl
SET temp2_lbl = TimeStampDeath_lbl-TimeStampDevelopCancer_lbl
SET UndiscountedQALYs_lbl = UndiscountedQALYs_lbl+[temp1_lbl*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]]
SET UndiscountedQALYs_lbl = UndiscountedQALYs_lbl+[temp2_lbl*Params_ss[16,SSrowoffset_nbr+SOUR_nbr]]
‘Calculate discounted QALY gains
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDevelopCancer_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampModelEntry_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]]
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDeath_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampDevelopCancer_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[16,SSrowoffset_nbr+SOUR_nbr]]
ELSE IF histology_lbl = 4
‘Calculate undiscounted QALY gains (develop clinical cancer)
SET temp1_lbl = TimeStampDevelopCancer_lbl-TimeStampModelEntry_lbl
SET temp2_lbl = TimeStampDiagnosedCancer_lbl-TimeStampDevelopCancer_lbl
SET temp3_lbl = TimeStampDeath_lbl-TimeStampDiagnosedCancer_lbl
SET UndiscountedQALYs_lbl = UndiscountedQALYs_lbl+[temp1_lbl*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]]
SET UndiscountedQALYs_lbl = UndiscountedQALYs_lbl+[temp2_lbl*Params_ss[16,SSrowoffset_nbr+SOUR_nbr]]
SET UndiscountedQALYs_lbl = UndiscountedQALYs_lbl+[temp3_lbl*Params_ss[17,SSrowoffset_nbr+SOUR_nbr]]
‘Calculate discounted QALY gains
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDevelopCancer_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampModelEntry_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[15,SSrowoffset_nbr+SOUR_nbr]]
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDiagnosedCancer_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampDevelopCancer_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[16,SSrowoffset_nbr+SOUR_nbr]]
SET DiscTemp1_lbl = EXP[DRi_QALYs_nbr*TimeStampDeath_lbl]
SET DiscTemp2_lbl = EXP[DRi_QALYs_nbr*TimeStampDiagnosedCancer_lbl]
SET DiscountedQALYs_lbl = DiscountedQALYs_lbl+[[[1/DRi_QALYs_nbr]*[DiscTemp1_lbl-DiscTemp2_lbl]]*Params_ss[17,SSrowoffset_nbr+SOUR_nbr]]
‘Store patient costs and QALYs in aggregate worksheet
SET ModelResults_ss[2,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[2,SSrowoffset_nbr+SOUR_nbr]+UndiscountedLYGs_lbl
SET ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]+DiscountedLYGs_lbl
SET ModelResults_ss[4,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[4,SSrowoffset_nbr+SOUR_nbr]+UndiscountedQALYs_lbl
SET ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]+DiscountedQALYs_lbl
SET ModelResults_ss[6,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[6,SSrowoffset_nbr+SOUR_nbr]+UndiscountedCost_lbl
SET ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]+DiscountedCost_lbl
‘Store patient diary (if selected)
IF StorePatientDiary_nbr = 1
SET PatientDiary_ss[13+[15+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedLYGs_lbl
SET PatientDiary_ss[13+[16+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedQALYs_lbl
SET PatientDiary_ss[13+[17+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedCost_lbl
SET PatientDiary_ss[13+[18+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedLYGs_lbl
SET PatientDiary_ss[13+[19+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedQALYs_lbl
SET PatientDiary_ss[13+[20+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedCost_lbl
‘Store intermediate outcomes (if selected)
IF StoreIntermediateOutcomes_nbr = 1
SET ModelResults_ss[9,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[9,SSrowoffset_nbr+SOUR_nbr]+COLindex_lbl
IF histology_lbl <= 2
SET ModelResults_ss[10,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[10,SSrowoffset_nbr+SOUR_nbr]+1
ELSE IF histology_lbl = 3
SET ModelResults_ss[11,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[11,SSrowoffset_nbr+SOUR_nbr]+1
ELSE
SET ModelResults_ss[12,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[12,SSrowoffset_nbr+SOUR_nbr]+1
IF CoD_lbl = 1
SET ModelResults_ss[13,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[13,SSrowoffset_nbr+SOUR_nbr]+1
ELSE IF CoD_lbl = 2
SET ModelResults_ss[14,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[14,SSrowoffset_nbr+SOUR_nbr]+1
ELSE IF CoD_lbl = 3
SET ModelResults_ss[15,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[15,SSrowoffset_nbr+SOUR_nbr]+1
IF AdenomaHistory_lbl = 1
SET ModelResults_ss[16,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[16,SSrowoffset_nbr+SOUR_nbr]+1
IF Cancerhistory_lbl = 2
SET ModelResults_ss[17,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[17,SSrowoffset_nbr+SOUR_nbr]+1
ELSE IF Cancerhistory_lbl = 1
SET ModelResults_ss[18,SSrowoffset_nbr+SOUR_nbr] = ModelResults_ss[18,SSrowoffset_nbr+SOUR_nbr]+1
‘Check stability
IF StoreStabilityTest_nbr = 1
IF StabilityTemp_nbr = StabilityTest_ss[2,3]
SET StabilityTest_ss[3,3] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,3] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,3] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,4]
SET StabilityTest_ss[3,4] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,4] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,4] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,5]
SET StabilityTest_ss[3,5] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,5] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,5] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,6]
SET StabilityTest_ss[3,6] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,6] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,6] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,7]
SET StabilityTest_ss[3,7] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,7] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,7] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,8]
SET StabilityTest_ss[3,8] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,8] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,8] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,9]
SET StabilityTest_ss[3,9] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,9] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,9] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,10]
SET StabilityTest_ss[3,10] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,10] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,10] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,11]
SET StabilityTest_ss[3,11] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,11] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,11] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,12]
SET StabilityTest_ss[3,12] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,12] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,12] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,13]
SET StabilityTest_ss[3,13] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,13] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,13] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,14]
SET StabilityTest_ss[3,14] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,14] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,14] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,15]
SET StabilityTest_ss[3,15] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,15] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,15] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,16]
SET StabilityTest_ss[3,16] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,16] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,16] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,17]
SET StabilityTest_ss[3,17] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,17] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,17] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,18]
SET StabilityTest_ss[3,18] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,18] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,18] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,19]
SET StabilityTest_ss[3,19] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,19] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,19] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,20]
SET StabilityTest_ss[3,20] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,20] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,20] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,21]
SET StabilityTest_ss[3,21] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,21] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,21] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,22]
SET StabilityTest_ss[3,22] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,22] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,22] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,23]
SET StabilityTest_ss[3,23] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,23] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,23] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,24]
SET StabilityTest_ss[3,24] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,24] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,24] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,25]
SET StabilityTest_ss[3,25] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,25] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,25] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,26]
SET StabilityTest_ss[3,26] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,26] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,26] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
ELSE IF StabilityTemp_nbr = StabilityTest_ss[2,27]
SET StabilityTest_ss[3,27] = ModelResults_ss[3,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[4,27] = ModelResults_ss[5,SSrowoffset_nbr+SOUR_nbr]
SET StabilityTest_ss[5,27] = ModelResults_ss[7,SSrowoffset_nbr+SOUR_nbr]
‘Track simulation progress
SET PercentRunComplete = [UniqId_lbl/PatientsPerRun_nbr]*100
SET PercentPSAComplete = [SOUR_nbr/MaxSOUR_nbr]*100
Setup Progress Bar SOUR_nbr , MaxSOUR_nbr
Set Progress Bar SOUR_nbr
VL SECTION: Reset model On OK Dialog
‘Determine selected surveillance option timings
SET COLInterval1_nbr = SurveillanceOptions_ss[4,2+SelectedSurveillanceOption_nbr]
SET COLInterval2_nbr = SurveillanceOptions_ss[5,2+SelectedSurveillanceOption_nbr]
SET MaxCOLage_nbr = SurveillanceOptions_ss[6,2+SelectedSurveillanceOption_nbr]
‘Reset model
SET SOUR_nbr = 1
Clear Sheet Area PatientDiary_ss[1,2] , 1000 , 100000
Clear Sheet Area ModelResults_ss[2,8] , 1000 , 100000
Clear Sheet Area StabilityTest_ss[3,3] , 100 , 100
Reset before next run
Reset Clock 1
VL SECTION: Surveillance COL (plus event router) Work Complete Logic
‘This workcentre deals with events in the simulation (death, progression, surveillance COL and related complications)
‘Set random numbers
SET rand1_nbr = RANDOM[0]
SET rand2_nbr = RANDOM[0]
SET rand3_nbr = RANDOM[0]
SET rand4_nbr = RANDOM[0]
SET rand5_nbr = RANDOM[0]
‘Update patient age (age at previous event + TTNE interval)
SET PatientAge_lbl = PatientAge_lbl+TTNE_lbl
‘Event 1 - patient dies of other causes (timestamp death, record cause of death, route to dead workcentre)
IF NextEvent_lbl = 1
SET TimeStampDeath_lbl = PatientAge_lbl-PatientStartAge_lbl
SET CoD_lbl = 1
SET Router_lbl = 1
‘Event 2 - patient progresses (increase histology by 1, route to next state and update next event times)
ELSE IF NextEvent_lbl = 2
SET histology_lbl = histology_lbl+1
IF histology_lbl = 2
SET Router_lbl = 3
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TPAdenomaToPreclinicalCRC_dst
IF TimeToNextCOL_lbl = LargeN_nbr
SET TimeToNextCOL_lbl = LargeN_nbr
ELSE
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl-TTNE_lbl
SET TimeToCancerDeath_lbl = LargeN_nbr
ELSE IF histology_lbl = 3
SET TimeStampDevelopCancer_lbl = PatientAge_lbl-PatientStartAge_lbl
SET Router_lbl = 4
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TPPreclinicalCRCToClinicalCRC_dst
IF TimeToNextCOL_lbl = LargeN_nbr
SET TimeToNextCOL_lbl = LargeN_nbr
ELSE
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl-TTNE_lbl
SET TimeToCancerDeath_lbl = CancerDeathTemp2_lbl
ELSE IF histology_lbl = 4
SET TimeStampDiagnosedCancer_lbl = PatientAge_lbl-PatientStartAge_lbl
SET Cancerhistory_lbl = 1
SET Router_lbl = 5
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = LargeN_nbr
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = CancerDeathTemp1_lbl
‘Event 3 - patient undergoes surveillance COL (update next COL interval, determine COL findings, route conditional on findings, update histology, update event times)
ELSE IF NextEvent_lbl = 3
SET COLindex_lbl = COLindex_lbl+1
IF histology_lbl = 1
IF rand1_nbr > Params_ss[8,SSrowoffset_nbr+SOUR_nbr]
SET Surveillancefindings_lbl = 1
SET histology_lbl = histology_lbl
SET Router_lbl = 2
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TimeToProgression_lbl-TTNE_lbl
SET TimeToNextCOL_lbl = COLInterval2_nbr
IF PatientAge_lbl+TimeToNextCOL_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = LargeN_nbr
ELSE
SET Surveillancefindings_lbl = 2
SET histology_lbl = histology_lbl
SET Router_lbl = 2
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TimeToProgression_lbl-TTNE_lbl
SET TimeToNextCOL_lbl = COLInterval2_nbr
IF PatientAge_lbl+TimeToNextCOL_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = LargeN_nbr
ELSE IF histology_lbl = 2
IF rand1_nbr < Params_ss[9,SSrowoffset_nbr+SOUR_nbr]
SET Surveillancefindings_lbl = 3
SET AdenomaHistory_lbl = 1
SET histology_lbl = 1
SET Router_lbl = 2
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TPNoAdenomaToAdenoma_dst
SET TimeToNextCOL_lbl = COLInterval2_nbr
IF PatientAge_lbl+TimeToNextCOL_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = LargeN_nbr
ELSE
SET Surveillancefindings_lbl = 4
SET histology_lbl = histology_lbl
SET Router_lbl = 3
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TimeToProgression_lbl-TTNE_lbl
SET TimeToNextCOL_lbl = COLInterval2_nbr
IF PatientAge_lbl+TimeToNextCOL_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = LargeN_nbr
ELSE IF histology_lbl = 3
IF rand1_nbr < Params_ss[10,SSrowoffset_nbr+SOUR_nbr]
SET Surveillancefindings_lbl = 5
SET TimeStampDiagnosedCancer_lbl = PatientAge_lbl-PatientStartAge_lbl
SET Cancerhistory_lbl = 2
SET histology_lbl = 4
SET Router_lbl = 5
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = LargeN_nbr
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = CancerDeathTemp1_lbl
ELSE
SET Surveillancefindings_lbl = 6
SET histology_lbl = histology_lbl
SET Router_lbl = 4
SET TimeToOCM_lbl = TimeToOCM_lbl-TTNE_lbl
SET TimeToProgression_lbl = TimeToProgression_lbl-TTNE_lbl
SET TimeToNextCOL_lbl = COLInterval2_nbr
IF PatientAge_lbl+TimeToNextCOL_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = TimeToNextCOL_lbl
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = TimeToCancerDeath_lbl-TTNE_lbl
‘Determine whether patient has complications (perforation or bleed), risk of death and associated costs
IF rand2_nbr < Params_ss[11,SSrowoffset_nbr+SOUR_nbr]
IF rand3_nbr < Params_ss[12,SSrowoffset_nbr+SOUR_nbr]
IF rand4_nbr < Params_ss[13,SSrowoffset_nbr+SOUR_nbr]
SET TimeStampDeath_lbl = PatientAge_lbl-PatientStartAge_lbl
SET CoD_lbl = 3
SET Router_lbl = 1
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[20,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[20,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
ELSE
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[20,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[20,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
ELSE
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[21,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[21,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
‘Patient dies as a consequence of their colorectal cancer (timestamp death)
ELSE IF NextEvent_lbl = 4
SET TimeStampDeath_lbl = PatientAge_lbl-PatientStartAge_lbl
SET CoD_lbl = 2
SET Router_lbl = 1
‘Add undiscounted and discounted costs (surveillance COL and lifetime cancer costs)
IF NextEvent_lbl = 3
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[18,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[18,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
IF histology_lbl = 4
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[19,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[19,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
IF NextEvent_lbl = 2
IF histology_lbl = 4
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[19,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[19,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
‘Store patient diary (if selected)
IF StorePatientDiary_nbr = 1
SET EventCount_lbl = EventCount_lbl+1
SET PatientDiary_ss[13+[1+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = PatientAge_lbl
SET PatientDiary_ss[13+[2+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = NextEvent_lbl
SET PatientDiary_ss[13+[3+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TTNE_lbl
SET PatientDiary_ss[13+[4+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeStampDevelopCancer_lbl
SET PatientDiary_ss[13+[5+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeStampDiagnosedCancer_lbl
SET PatientDiary_ss[13+[6+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeStampDeath_lbl
SET PatientDiary_ss[13+[7+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = CoD_lbl
SET PatientDiary_ss[13+[8+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = histology_lbl
SET PatientDiary_ss[13+[9+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = Surveillancefindings_lbl
SET PatientDiary_ss[13+[10+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = COLindex_lbl
SET PatientDiary_ss[13+[11+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeToOCM_lbl
SET PatientDiary_ss[13+[12+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeToProgression_lbl
SET PatientDiary_ss[13+[13+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeToNextCOL_lbl
SET PatientDiary_ss[13+[14+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = TimeToCancerDeath_lbl
SET PatientDiary_ss[13+[15+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedLYGs_lbl
SET PatientDiary_ss[13+[16+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedQALYs_lbl
SET PatientDiary_ss[13+[17+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = UndiscountedCost_lbl
SET PatientDiary_ss[13+[18+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedLYGs_lbl
SET PatientDiary_ss[13+[19+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedQALYs_lbl
SET PatientDiary_ss[13+[20+[[EventCount_lbl-1]*20]],1+UniqId_lbl] = DiscountedCost_lbl
‘ResetSurveillanceFindings
SET Surveillancefindings_lbl = 0
VL SECTION: Model entry Entry Logic
‘PATIENT ENTRY - this workcentre sets the intial characteristics of the patient cohort
‘Set random numbers
SET rand1_nbr = RANDOM[0]
‘Determine simulation running conditions
IF Model entry.Arrived Count = PatientsPerRun_nbr
SET Model entry.Interarrival Time = LargeN_nbr
ELSE
SET Model entry.Interarrival Time = Model entry.Interarrival Time
‘Timestamp model entry
SET TimeStampModelEntry_lbl = 0
‘Set patient characteristics (start age, current age, histology, time to next COL, life expectancy)
SET PatientStartAge_lbl = PatientAge_dst
SET PatientAge_lbl = PatientStartAge_lbl
SET Sex_lbl = Sex_dst
SET histology_lbl = 1
IF Sex_lbl = 2
IF PatientStartAge_lbl = 0
SET LifeExpectancy_lbl = GenPopDeathFemales0_dst
ELSE IF PatientStartAge_lbl = 1
SET LifeExpectancy_lbl = GenPopDeathFemales1_dst
ELSE IF PatientStartAge_lbl = 2
SET LifeExpectancy_lbl = GenPopDeathFemales2_dst
ELSE IF PatientStartAge_lbl = 3
SET LifeExpectancy_lbl = GenPopDeathFemales3_dst
ELSE IF PatientStartAge_lbl = 4
SET LifeExpectancy_lbl = GenPopDeathFemales4_dst
ELSE IF PatientStartAge_lbl = 5
SET LifeExpectancy_lbl = GenPopDeathFemales5_dst
ELSE IF PatientStartAge_lbl = 6
SET LifeExpectancy_lbl = GenPopDeathFemales6_dst
ELSE IF PatientStartAge_lbl = 7
SET LifeExpectancy_lbl = GenPopDeathFemales7_dst
ELSE IF PatientStartAge_lbl = 8
SET LifeExpectancy_lbl = GenPopDeathFemales8_dst
ELSE IF PatientStartAge_lbl = 9
SET LifeExpectancy_lbl = GenPopDeathFemales9_dst
ELSE IF PatientStartAge_lbl = 10
SET LifeExpectancy_lbl = GenPopDeathFemales10_dst
ELSE IF PatientStartAge_lbl = 11
SET LifeExpectancy_lbl = GenPopDeathFemales11_dst
ELSE IF PatientStartAge_lbl = 12
SET LifeExpectancy_lbl = GenPopDeathFemales12_dst
ELSE IF PatientStartAge_lbl = 13
SET LifeExpectancy_lbl = GenPopDeathFemales13_dst
ELSE IF PatientStartAge_lbl = 14
SET LifeExpectancy_lbl = GenPopDeathFemales14_dst
ELSE IF PatientStartAge_lbl = 15
SET LifeExpectancy_lbl = GenPopDeathFemales15_dst
ELSE IF PatientStartAge_lbl = 16
SET LifeExpectancy_lbl = GenPopDeathFemales16_dst
ELSE IF PatientStartAge_lbl = 17
SET LifeExpectancy_lbl = GenPopDeathFemales17_dst
ELSE IF PatientStartAge_lbl = 18
SET LifeExpectancy_lbl = GenPopDeathFemales18_dst
ELSE IF PatientStartAge_lbl = 19
SET LifeExpectancy_lbl = GenPopDeathFemales19_dst
ELSE IF PatientStartAge_lbl = 20
SET LifeExpectancy_lbl = GenPopDeathFemales20_dst
ELSE IF PatientStartAge_lbl = 21
SET LifeExpectancy_lbl = GenPopDeathFemales21_dst
ELSE IF PatientStartAge_lbl = 22
SET LifeExpectancy_lbl = GenPopDeathFemales22_dst
ELSE IF PatientStartAge_lbl = 23
SET LifeExpectancy_lbl = GenPopDeathFemales23_dst
ELSE IF PatientStartAge_lbl = 24
SET LifeExpectancy_lbl = GenPopDeathFemales24_dst
ELSE IF PatientStartAge_lbl = 25
SET LifeExpectancy_lbl = GenPopDeathFemales25_dst
ELSE IF PatientStartAge_lbl = 26
SET LifeExpectancy_lbl = GenPopDeathFemales26_dst
ELSE IF PatientStartAge_lbl = 27
SET LifeExpectancy_lbl = GenPopDeathFemales27_dst
ELSE IF PatientStartAge_lbl = 28
SET LifeExpectancy_lbl = GenPopDeathFemales28_dst
ELSE IF PatientStartAge_lbl = 29
SET LifeExpectancy_lbl = GenPopDeathFemales29_dst
ELSE IF PatientStartAge_lbl = 30
SET LifeExpectancy_lbl = GenPopDeathFemales30_dst
ELSE IF PatientStartAge_lbl = 31
SET LifeExpectancy_lbl = GenPopDeathFemales31_dst
ELSE IF PatientStartAge_lbl = 32
SET LifeExpectancy_lbl = GenPopDeathFemales32_dst
ELSE IF PatientStartAge_lbl = 33
SET LifeExpectancy_lbl = GenPopDeathFemales33_dst
ELSE IF PatientStartAge_lbl = 34
SET LifeExpectancy_lbl = GenPopDeathFemales34_dst
ELSE IF PatientStartAge_lbl = 35
SET LifeExpectancy_lbl = GenPopDeathFemales35_dst
ELSE IF PatientStartAge_lbl = 36
SET LifeExpectancy_lbl = GenPopDeathFemales36_dst
ELSE IF PatientStartAge_lbl = 37
SET LifeExpectancy_lbl = GenPopDeathFemales37_dst
ELSE IF PatientStartAge_lbl = 38
SET LifeExpectancy_lbl = GenPopDeathFemales38_dst
ELSE IF PatientStartAge_lbl = 39
SET LifeExpectancy_lbl = GenPopDeathFemales39_dst
ELSE IF PatientStartAge_lbl = 40
SET LifeExpectancy_lbl = GenPopDeathFemales40_dst
ELSE IF PatientStartAge_lbl = 41
SET LifeExpectancy_lbl = GenPopDeathFemales41_dst
ELSE IF PatientStartAge_lbl = 42
SET LifeExpectancy_lbl = GenPopDeathFemales42_dst
ELSE IF PatientStartAge_lbl = 43
SET LifeExpectancy_lbl = GenPopDeathFemales43_dst
ELSE IF PatientStartAge_lbl = 44
SET LifeExpectancy_lbl = GenPopDeathFemales44_dst
ELSE IF PatientStartAge_lbl = 45
SET LifeExpectancy_lbl = GenPopDeathFemales45_dst
ELSE IF PatientStartAge_lbl = 46
SET LifeExpectancy_lbl = GenPopDeathFemales46_dst
ELSE IF PatientStartAge_lbl = 47
SET LifeExpectancy_lbl = GenPopDeathFemales47_dst
ELSE IF PatientStartAge_lbl = 48
SET LifeExpectancy_lbl = GenPopDeathFemales48_dst
ELSE IF PatientStartAge_lbl = 49
SET LifeExpectancy_lbl = GenPopDeathFemales49_dst
ELSE IF PatientStartAge_lbl = 50
SET LifeExpectancy_lbl = GenPopDeathFemales50_dst
ELSE IF PatientStartAge_lbl = 51
SET LifeExpectancy_lbl = GenPopDeathFemales51_dst
ELSE IF PatientStartAge_lbl = 52
SET LifeExpectancy_lbl = GenPopDeathFemales52_dst
ELSE IF PatientStartAge_lbl = 53
SET LifeExpectancy_lbl = GenPopDeathFemales53_dst
ELSE IF PatientStartAge_lbl = 54
SET LifeExpectancy_lbl = GenPopDeathFemales54_dst
ELSE IF PatientStartAge_lbl = 55
SET LifeExpectancy_lbl = GenPopDeathFemales55_dst
ELSE IF PatientStartAge_lbl = 56
SET LifeExpectancy_lbl = GenPopDeathFemales56_dst
ELSE IF PatientStartAge_lbl = 57
SET LifeExpectancy_lbl = GenPopDeathFemales57_dst
ELSE IF PatientStartAge_lbl = 58
SET LifeExpectancy_lbl = GenPopDeathFemales58_dst
ELSE IF PatientStartAge_lbl = 59
SET LifeExpectancy_lbl = GenPopDeathFemales59_dst
ELSE IF PatientStartAge_lbl = 60
SET LifeExpectancy_lbl = GenPopDeathFemales60_dst
ELSE IF PatientStartAge_lbl = 61
SET LifeExpectancy_lbl = GenPopDeathFemales61_dst
ELSE IF PatientStartAge_lbl = 62
SET LifeExpectancy_lbl = GenPopDeathFemales62_dst
ELSE IF PatientStartAge_lbl = 63
SET LifeExpectancy_lbl = GenPopDeathFemales63_dst
ELSE IF PatientStartAge_lbl = 64
SET LifeExpectancy_lbl = GenPopDeathFemales64_dst
ELSE IF PatientStartAge_lbl = 65
SET LifeExpectancy_lbl = GenPopDeathFemales65_dst
ELSE IF PatientStartAge_lbl = 66
SET LifeExpectancy_lbl = GenPopDeathFemales66_dst
ELSE IF PatientStartAge_lbl = 67
SET LifeExpectancy_lbl = GenPopDeathFemales67_dst
ELSE IF PatientStartAge_lbl = 68
SET LifeExpectancy_lbl = GenPopDeathFemales68_dst
ELSE IF PatientStartAge_lbl = 69
SET LifeExpectancy_lbl = GenPopDeathFemales69_dst
ELSE IF PatientStartAge_lbl = 70
SET LifeExpectancy_lbl = GenPopDeathFemales70_dst
ELSE IF PatientStartAge_lbl = 71
SET LifeExpectancy_lbl = GenPopDeathFemales71_dst
ELSE IF PatientStartAge_lbl = 72
SET LifeExpectancy_lbl = GenPopDeathFemales72_dst
ELSE IF PatientStartAge_lbl = 73
SET LifeExpectancy_lbl = GenPopDeathFemales73_dst
ELSE IF PatientStartAge_lbl = 74
SET LifeExpectancy_lbl = GenPopDeathFemales74_dst
ELSE IF PatientStartAge_lbl = 75
SET LifeExpectancy_lbl = GenPopDeathFemales75_dst
ELSE IF PatientStartAge_lbl = 76
SET LifeExpectancy_lbl = GenPopDeathFemales76_dst
ELSE IF PatientStartAge_lbl = 77
SET LifeExpectancy_lbl = GenPopDeathFemales77_dst
ELSE IF PatientStartAge_lbl = 78
SET LifeExpectancy_lbl = GenPopDeathFemales78_dst
ELSE IF PatientStartAge_lbl = 79
SET LifeExpectancy_lbl = GenPopDeathFemales79_dst
ELSE IF PatientStartAge_lbl = 80
SET LifeExpectancy_lbl = GenPopDeathFemales80_dst
ELSE IF PatientStartAge_lbl = 81
SET LifeExpectancy_lbl = GenPopDeathFemales81_dst
ELSE IF PatientStartAge_lbl = 82
SET LifeExpectancy_lbl = GenPopDeathFemales82_dst
ELSE IF PatientStartAge_lbl = 83
SET LifeExpectancy_lbl = GenPopDeathFemales83_dst
ELSE IF PatientStartAge_lbl = 84
SET LifeExpectancy_lbl = GenPopDeathFemales84_dst
ELSE IF PatientStartAge_lbl = 85
SET LifeExpectancy_lbl = GenPopDeathFemales85_dst
ELSE IF PatientStartAge_lbl = 86
SET LifeExpectancy_lbl = GenPopDeathFemales86_dst
ELSE IF PatientStartAge_lbl = 87
SET LifeExpectancy_lbl = GenPopDeathFemales87_dst
ELSE IF PatientStartAge_lbl = 88
SET LifeExpectancy_lbl = GenPopDeathFemales88_dst
ELSE IF PatientStartAge_lbl = 89
SET LifeExpectancy_lbl = GenPopDeathFemales89_dst
ELSE IF PatientStartAge_lbl = 90
SET LifeExpectancy_lbl = GenPopDeathFemales90_dst
ELSE IF PatientStartAge_lbl = 91
SET LifeExpectancy_lbl = GenPopDeathFemales91_dst
ELSE IF PatientStartAge_lbl = 92
SET LifeExpectancy_lbl = GenPopDeathFemales92_dst
ELSE IF PatientStartAge_lbl = 93
SET LifeExpectancy_lbl = GenPopDeathFemales93_dst
ELSE IF PatientStartAge_lbl = 94
SET LifeExpectancy_lbl = GenPopDeathFemales94_dst
ELSE IF PatientStartAge_lbl = 95
SET LifeExpectancy_lbl = GenPopDeathFemales95_dst
ELSE IF PatientStartAge_lbl = 96
SET LifeExpectancy_lbl = GenPopDeathFemales96_dst
ELSE IF PatientStartAge_lbl = 97
SET LifeExpectancy_lbl = GenPopDeathFemales97_dst
ELSE IF PatientStartAge_lbl = 98
SET LifeExpectancy_lbl = GenPopDeathFemales98_dst
ELSE IF PatientStartAge_lbl = 99
SET LifeExpectancy_lbl = GenPopDeathFemales99_dst
ELSE IF PatientStartAge_lbl = 100
SET LifeExpectancy_lbl = GenPopDeathFemales100_dst
ELSE
IF PatientStartAge_lbl = 0
SET LifeExpectancy_lbl = GenPopDeathMales0_dst
ELSE IF PatientStartAge_lbl = 1
SET LifeExpectancy_lbl = GenPopDeathMales1_dst
ELSE IF PatientStartAge_lbl = 2
SET LifeExpectancy_lbl = GenPopDeathMales2_dst
ELSE IF PatientStartAge_lbl = 3
SET LifeExpectancy_lbl = GenPopDeathMales3_dst
ELSE IF PatientStartAge_lbl = 4
SET LifeExpectancy_lbl = GenPopDeathMales4_dst
ELSE IF PatientStartAge_lbl = 5
SET LifeExpectancy_lbl = GenPopDeathMales5_dst
ELSE IF PatientStartAge_lbl = 6
SET LifeExpectancy_lbl = GenPopDeathMales6_dst
ELSE IF PatientStartAge_lbl = 7
SET LifeExpectancy_lbl = GenPopDeathMales7_dst
ELSE IF PatientStartAge_lbl = 8
SET LifeExpectancy_lbl = GenPopDeathMales8_dst
ELSE IF PatientStartAge_lbl = 9
SET LifeExpectancy_lbl = GenPopDeathMales9_dst
ELSE IF PatientStartAge_lbl = 10
SET LifeExpectancy_lbl = GenPopDeathMales10_dst
ELSE IF PatientStartAge_lbl = 11
SET LifeExpectancy_lbl = GenPopDeathMales11_dst
ELSE IF PatientStartAge_lbl = 12
SET LifeExpectancy_lbl = GenPopDeathMales12_dst
ELSE IF PatientStartAge_lbl = 13
SET LifeExpectancy_lbl = GenPopDeathMales13_dst
ELSE IF PatientStartAge_lbl = 14
SET LifeExpectancy_lbl = GenPopDeathMales14_dst
ELSE IF PatientStartAge_lbl = 15
SET LifeExpectancy_lbl = GenPopDeathMales15_dst
ELSE IF PatientStartAge_lbl = 16
SET LifeExpectancy_lbl = GenPopDeathMales16_dst
ELSE IF PatientStartAge_lbl = 17
SET LifeExpectancy_lbl = GenPopDeathMales17_dst
ELSE IF PatientStartAge_lbl = 18
SET LifeExpectancy_lbl = GenPopDeathMales18_dst
ELSE IF PatientStartAge_lbl = 19
SET LifeExpectancy_lbl = GenPopDeathMales19_dst
ELSE IF PatientStartAge_lbl = 20
SET LifeExpectancy_lbl = GenPopDeathMales20_dst
ELSE IF PatientStartAge_lbl = 21
SET LifeExpectancy_lbl = GenPopDeathMales21_dst
ELSE IF PatientStartAge_lbl = 22
SET LifeExpectancy_lbl = GenPopDeathMales22_dst
ELSE IF PatientStartAge_lbl = 23
SET LifeExpectancy_lbl = GenPopDeathMales23_dst
ELSE IF PatientStartAge_lbl = 24
SET LifeExpectancy_lbl = GenPopDeathMales24_dst
ELSE IF PatientStartAge_lbl = 25
SET LifeExpectancy_lbl = GenPopDeathMales25_dst
ELSE IF PatientStartAge_lbl = 26
SET LifeExpectancy_lbl = GenPopDeathMales26_dst
ELSE IF PatientStartAge_lbl = 27
SET LifeExpectancy_lbl = GenPopDeathMales27_dst
ELSE IF PatientStartAge_lbl = 28
SET LifeExpectancy_lbl = GenPopDeathMales28_dst
ELSE IF PatientStartAge_lbl = 29
SET LifeExpectancy_lbl = GenPopDeathMales29_dst
ELSE IF PatientStartAge_lbl = 30
SET LifeExpectancy_lbl = GenPopDeathMales30_dst
ELSE IF PatientStartAge_lbl = 31
SET LifeExpectancy_lbl = GenPopDeathMales31_dst
ELSE IF PatientStartAge_lbl = 32
SET LifeExpectancy_lbl = GenPopDeathMales32_dst
ELSE IF PatientStartAge_lbl = 33
SET LifeExpectancy_lbl = GenPopDeathMales33_dst
ELSE IF PatientStartAge_lbl = 34
SET LifeExpectancy_lbl = GenPopDeathMales34_dst
ELSE IF PatientStartAge_lbl = 35
SET LifeExpectancy_lbl = GenPopDeathMales35_dst
ELSE IF PatientStartAge_lbl = 36
SET LifeExpectancy_lbl = GenPopDeathMales36_dst
ELSE IF PatientStartAge_lbl = 37
SET LifeExpectancy_lbl = GenPopDeathMales37_dst
ELSE IF PatientStartAge_lbl = 38
SET LifeExpectancy_lbl = GenPopDeathMales38_dst
ELSE IF PatientStartAge_lbl = 39
SET LifeExpectancy_lbl = GenPopDeathMales39_dst
ELSE IF PatientStartAge_lbl = 40
SET LifeExpectancy_lbl = GenPopDeathMales40_dst
ELSE IF PatientStartAge_lbl = 41
SET LifeExpectancy_lbl = GenPopDeathMales41_dst
ELSE IF PatientStartAge_lbl = 42
SET LifeExpectancy_lbl = GenPopDeathMales42_dst
ELSE IF PatientStartAge_lbl = 43
SET LifeExpectancy_lbl = GenPopDeathMales43_dst
ELSE IF PatientStartAge_lbl = 44
SET LifeExpectancy_lbl = GenPopDeathMales44_dst
ELSE IF PatientStartAge_lbl = 45
SET LifeExpectancy_lbl = GenPopDeathMales45_dst
ELSE IF PatientStartAge_lbl = 46
SET LifeExpectancy_lbl = GenPopDeathMales46_dst
ELSE IF PatientStartAge_lbl = 47
SET LifeExpectancy_lbl = GenPopDeathMales47_dst
ELSE IF PatientStartAge_lbl = 48
SET LifeExpectancy_lbl = GenPopDeathMales48_dst
ELSE IF PatientStartAge_lbl = 49
SET LifeExpectancy_lbl = GenPopDeathMales49_dst
ELSE IF PatientStartAge_lbl = 50
SET LifeExpectancy_lbl = GenPopDeathMales50_dst
ELSE IF PatientStartAge_lbl = 51
SET LifeExpectancy_lbl = GenPopDeathMales51_dst
ELSE IF PatientStartAge_lbl = 52
SET LifeExpectancy_lbl = GenPopDeathMales52_dst
ELSE IF PatientStartAge_lbl = 53
SET LifeExpectancy_lbl = GenPopDeathMales53_dst
ELSE IF PatientStartAge_lbl = 54
SET LifeExpectancy_lbl = GenPopDeathMales54_dst
ELSE IF PatientStartAge_lbl = 55
SET LifeExpectancy_lbl = GenPopDeathMales55_dst
ELSE IF PatientStartAge_lbl = 56
SET LifeExpectancy_lbl = GenPopDeathMales56_dst
ELSE IF PatientStartAge_lbl = 57
SET LifeExpectancy_lbl = GenPopDeathMales57_dst
ELSE IF PatientStartAge_lbl = 58
SET LifeExpectancy_lbl = GenPopDeathMales58_dst
ELSE IF PatientStartAge_lbl = 59
SET LifeExpectancy_lbl = GenPopDeathMales59_dst
ELSE IF PatientStartAge_lbl = 60
SET LifeExpectancy_lbl = GenPopDeathMales60_dst
ELSE IF PatientStartAge_lbl = 61
SET LifeExpectancy_lbl = GenPopDeathMales61_dst
ELSE IF PatientStartAge_lbl = 62
SET LifeExpectancy_lbl = GenPopDeathMales62_dst
ELSE IF PatientStartAge_lbl = 63
SET LifeExpectancy_lbl = GenPopDeathMales63_dst
ELSE IF PatientStartAge_lbl = 64
SET LifeExpectancy_lbl = GenPopDeathMales64_dst
ELSE IF PatientStartAge_lbl = 65
SET LifeExpectancy_lbl = GenPopDeathMales65_dst
ELSE IF PatientStartAge_lbl = 66
SET LifeExpectancy_lbl = GenPopDeathMales66_dst
ELSE IF PatientStartAge_lbl = 67
SET LifeExpectancy_lbl = GenPopDeathMales67_dst
ELSE IF PatientStartAge_lbl = 68
SET LifeExpectancy_lbl = GenPopDeathMales68_dst
ELSE IF PatientStartAge_lbl = 69
SET LifeExpectancy_lbl = GenPopDeathMales69_dst
ELSE IF PatientStartAge_lbl = 70
SET LifeExpectancy_lbl = GenPopDeathMales70_dst
ELSE IF PatientStartAge_lbl = 71
SET LifeExpectancy_lbl = GenPopDeathMales71_dst
ELSE IF PatientStartAge_lbl = 72
SET LifeExpectancy_lbl = GenPopDeathMales72_dst
ELSE IF PatientStartAge_lbl = 73
SET LifeExpectancy_lbl = GenPopDeathMales73_dst
ELSE IF PatientStartAge_lbl = 74
SET LifeExpectancy_lbl = GenPopDeathMales74_dst
ELSE IF PatientStartAge_lbl = 75
SET LifeExpectancy_lbl = GenPopDeathMales75_dst
ELSE IF PatientStartAge_lbl = 76
SET LifeExpectancy_lbl = GenPopDeathMales76_dst
ELSE IF PatientStartAge_lbl = 77
SET LifeExpectancy_lbl = GenPopDeathMales77_dst
ELSE IF PatientStartAge_lbl = 78
SET LifeExpectancy_lbl = GenPopDeathMales78_dst
ELSE IF PatientStartAge_lbl = 79
SET LifeExpectancy_lbl = GenPopDeathMales79_dst
ELSE IF PatientStartAge_lbl = 80
SET LifeExpectancy_lbl = GenPopDeathMales80_dst
ELSE IF PatientStartAge_lbl = 81
SET LifeExpectancy_lbl = GenPopDeathMales81_dst
ELSE IF PatientStartAge_lbl = 82
SET LifeExpectancy_lbl = GenPopDeathMales82_dst
ELSE IF PatientStartAge_lbl = 83
SET LifeExpectancy_lbl = GenPopDeathMales83_dst
ELSE IF PatientStartAge_lbl = 84
SET LifeExpectancy_lbl = GenPopDeathMales84_dst
ELSE IF PatientStartAge_lbl = 85
SET LifeExpectancy_lbl = GenPopDeathMales85_dst
ELSE IF PatientStartAge_lbl = 86
SET LifeExpectancy_lbl = GenPopDeathMales86_dst
ELSE IF PatientStartAge_lbl = 87
SET LifeExpectancy_lbl = GenPopDeathMales87_dst
ELSE IF PatientStartAge_lbl = 88
SET LifeExpectancy_lbl = GenPopDeathMales88_dst
ELSE IF PatientStartAge_lbl = 89
SET LifeExpectancy_lbl = GenPopDeathMales89_dst
ELSE IF PatientStartAge_lbl = 90
SET LifeExpectancy_lbl = GenPopDeathMales90_dst
ELSE IF PatientStartAge_lbl = 91
SET LifeExpectancy_lbl = GenPopDeathMales91_dst
ELSE IF PatientStartAge_lbl = 92
SET LifeExpectancy_lbl = GenPopDeathMales92_dst
ELSE IF PatientStartAge_lbl = 93
SET LifeExpectancy_lbl = GenPopDeathMales93_dst
ELSE IF PatientStartAge_lbl = 94
SET LifeExpectancy_lbl = GenPopDeathMales94_dst
ELSE IF PatientStartAge_lbl = 95
SET LifeExpectancy_lbl = GenPopDeathMales95_dst
ELSE IF PatientStartAge_lbl = 96
SET LifeExpectancy_lbl = GenPopDeathMales96_dst
ELSE IF PatientStartAge_lbl = 97
SET LifeExpectancy_lbl = GenPopDeathMales97_dst
ELSE IF PatientStartAge_lbl = 98
SET LifeExpectancy_lbl = GenPopDeathMales98_dst
ELSE IF PatientStartAge_lbl = 99
SET LifeExpectancy_lbl = GenPopDeathMales99_dst
ELSE IF PatientStartAge_lbl = 100
SET LifeExpectancy_lbl = GenPopDeathMales100_dst
‘Set time to first events (note time to preclinical and cancer death are sampled with a multiplier here to ensure monotonically correct ordering)
SET TimeToOCM_lbl = LifeExpectancy_lbl-PatientAge_lbl
SET TimeToProgression_lbl = TPNoAdenomaToAdenoma_dst
IF PatientAge_lbl < MaxCOLage_nbr
SET TimeToNextCOL_lbl = COLInterval1_nbr
ELSE
SET TimeToNextCOL_lbl = LargeN_nbr
SET TimeToCancerDeath_lbl = LargeN_nbr
SET CancerDeathTemp1_lbl = ClinicalCRCSurvival_dst
SET CancerDeathTemp2_lbl = CancerDeathTemp1_lbl*Params_ss[5,SOUR_nbr+SSrowoffset_nbr]
‘Include costs of initial colonoscopy
SET UndiscountedCost_lbl = UndiscountedCost_lbl+Params_ss[18,SSrowoffset_nbr+SOUR_nbr]
SET DiscountedCost_lbl = DiscountedCost_lbl+[Params_ss[18,SSrowoffset_nbr+SOUR_nbr]/[1+DRp_costs_nbr]^[PatientAge_lbl-PatientStartAge_lbl]]
‘Store patient diary (if selected)
IF StorePatientDiary_nbr = 1
SET PatientDiary_ss[1,1+UniqId_lbl] = UniqId_lbl
SET PatientDiary_ss[2,1+UniqId_lbl] = TimeStampModelEntry_lbl
SET PatientDiary_ss[3,1+UniqId_lbl] = PatientStartAge_lbl
SET PatientDiary_ss[4,1+UniqId_lbl] = PatientAge_lbl
SET PatientDiary_ss[5,1+UniqId_lbl] = Sex_lbl
SET PatientDiary_ss[6,1+UniqId_lbl] = histology_lbl
SET PatientDiary_ss[7,1+UniqId_lbl] = LifeExpectancy_lbl
SET PatientDiary_ss[8,1+UniqId_lbl] = TimeToOCM_lbl
SET PatientDiary_ss[9,1+UniqId_lbl] = TimeToProgression_lbl
SET PatientDiary_ss[10,1+UniqId_lbl] = TimeToNextCOL_lbl
SET PatientDiary_ss[11,1+UniqId_lbl] = TimeToCancerDeath_lbl
SET PatientDiary_ss[12,1+UniqId_lbl] = CancerDeathTemp1_lbl
SET PatientDiary_ss[13,1+UniqId_lbl] = CancerDeathTemp2_lbl
‘Set patient run tracker for stability testing
SET StabilityTemp_nbr = UniqId_lbl
Appendix 11 Search strategy for health-utility studies
MEDLINE search strategy
Please note, the MEDLINE search strategies provided were adapted according to each of the databases searched.
-
exp Colorectal Neoplasms/
-
Neoplasms/
-
Carcinoma/
-
Adenocarcinoma/
-
or/2-4
-
Colonic Diseases/
-
Rectal Diseases/
-
exp Colon/
-
exp Rectum/
-
or/6-9
-
5 and 10
-
(carcinoma adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(neoplasia adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(neoplasm$ adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(adenocarcinoma adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(cancer$ adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(tumor$ adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(tumour$ adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
(malignan$ adj3 (colorectal or colon$ or rect$ or intestin$ or bowel)).tw.
-
or/12-19
-
1 or 11 or 20
-
health related quality of life.tw.
-
hrql.tw.
-
hrqol.tw.
-
hql.tw.
-
sf 36.tw.
-
sf thirtysix.tw.
-
sf thirty six.tw.
-
short form 36.tw.
-
short form thirty six.tw.
-
short form thirtysix.tw.
-
shortform 36.tw.
-
shortform thirty six.tw.
-
shortform thirty six.tw.
-
sf36.tw.
-
medical outcomes survey.tw.
-
mos.tw.
-
euroqol.tw.
-
eq 5d.tw.
-
eq5d.tw.
-
qaly$.tw.
-
quality adjusted life years/
-
quality adjusted life year$.tw.
-
hye$.tw.
-
health$ year$ equivalent$.tw.
-
psychological general well being index.tw.
-
psychological general wellbeing index.tw.
-
pgwb$.tw.
-
health utilit$.tw.
-
hui.tw.
-
quality of wellbeing$.tw.
-
quality of well being.tw.
-
qwb$.tw.
-
rosser.tw.
-
trade off$.tw.
-
standard gamble.tw.
-
tto.tw.
-
“Quality of Life”/
-
“Outcome Assessment (Health Care)”/
-
(preference$ or utilit$).tw. and (58 or 59)
-
((preference$ or utilit$) and quality of life).tw.
-
(preference$ adj2 (elicit$ or patient$ or population$ or measure$ or based or cost$)).tw.
-
(utilit$ adj2 (elicit$ or patient$ or population$ or measure$ or based or cost$)).tw.
-
or/22-57,60-63
-
21 and 64
-
limit 65 to yr=“2005-Current”
Databases
The following databases and grey literature sources were searched from inception to present.
-
MEDLINE and MEDLINE in Process & Other Non-Indexed citations (Ovid)
-
EMBASE (Ovid)
-
Cumulative Index to Nursing and Allied Health Literature (EBSCO)
-
BIOSIS previews (WoK)
-
Science Citation Index (Web of Science)
-
Cochrane Database of Systematic Reviews (Cochrane)
-
Cochrane Central Register of Controlled Trials (Cochrane)
-
Database of Abstracts of Reviews of Effects (Cochrane)
-
NHS Health Economic Evaluation Database (Cochrane)
-
Health Technology Assessment database (Cochrane)
-
EconLit (Ovid)
-
Web of Science (WoK)
-
Conference Proceedings index (Web of Science via WoK)
-
ProQuest Dissertations and Theses (ProQuest)
-
Tufts (Cost Effectiveness Analysis Registry).
Limits applied
Results were limited to 2005–current.
Appendix 12 STROBE statement: checklist of items that should be included in reports of cohort studies
The following STROBE table pertains to our workstream, the hospital data set. The screening data set and psychological study involved different, pre-existing study samples.
| Item no. | Recommendation | Reported in chapter/section/page no. | |
|---|---|---|---|
| Title and abstract | 1 | (a) Indicate the study’s design with a commonly used term in the title or the abstract | p. vii |
| (b) Provide in the abstract an informative and balanced summary of what was done and what was found | p. vii, viii | ||
| Introduction | |||
| Background/rationale | 2 | Explain the scientific background and rationale for the investigation being reported | Chapter 1 |
| Objectives | 3 | State specific objectives, including any prespecified hypotheses | Chapter 1, Aims and objectives |
| Methods | |||
| Study design | 4 | Present key elements of study design early in the paper | Chapter 1, Study design and setting |
| Setting | 5 | Describe the setting, locations, and relevant dates, including periods of recruitment, exposure, follow-up and data collection | Chapter 2 |
| Participants | 6 | (a) Give the eligibility criteria, and the sources and methods of selection of participants. Describe methods of follow-up | Chapter 2: Patient eligibility |
| (b) For matched studies, give matching criteria and number of exposed and unexposed | n/a | ||
| Variables | 7 | Clearly define all outcomes, exposures, predictors, potential confounders and effect modifiers. Give diagnostic criteria, if applicable | Chapter 2, Variables |
| Data sources/measurement | 8a | For each variable of interest, give sources of data and details of methods of assessment (measurement). Describe comparability of assessment methods if there is more than one group | Chapter 2: Manual data coding Creating summary values for polyp characteristics Procedure information Defining baseline and surveillance visits |
| Bias | 9 | Describe any efforts to address potential sources of bias | Chapter 2: Patient follow-up (selection bias) Defining baseline and surveillance visits Variables (information bias) |
| Study size | 10 | Explain how the study size was arrived at | Chapter 2, Study size |
| Quantitative variables | 11 | Explain how quantitative variables were handled in the analyses. If applicable, describe which groupings were chosen and why | Chapter 2, Variables |
| Statistical methods | 12 | (a) Describe all statistical methods, including those used to control for confounding | Chapter 2, Statistical methods |
| (b) Describe any methods used to examine subgroups and interactions | Chapter 2, Statistical methods | ||
| (c) Explain how missing data were addressed | Chapter 2: Creating summary values for polyp characteristics Data collection from hospitals |
||
| (d) If applicable, explain how loss to follow-up was addressed | n/a | ||
| (e) Describe any sensitivity analyses | Chapter 2, Statistical methods; Chapter 3, Sensitivity analyses and internal validation | ||
| Results | |||
| Participants | 13a | (a) Report numbers of individuals at each stage of study, e.g. numbers potentially eligible, examined for eligibility, confirmed eligible, included in the study, completing follow-up, and analysed | Chapter 3 |
| (b) Give reasons for non-participation at each stage | n/a | ||
| (c) Consider use of a flow diagram | Chapter 3, Figure 2 | ||
| Descriptive data | 14a | (a) Give characteristics of study participants (e.g. demographic, clinical, social) and information on exposures and potential confounders | Chapter 3, Baseline characteristics of all intermediate-risk patients and those with follow-up |
| (b) Indicate number of participants with missing data for each variable of interest | Chapter 3 | ||
| (c) Summarise follow-up time (e.g., average and total amount) | Chapter 3, Colorectal cancer risk after baseline | ||
| Outcome data | 15a | Report numbers of outcome events or summary measures over time | Chapter 3 |
| Main results | 16 | (a) Give unadjusted estimates and, if applicable, confounder-adjusted estimates and their precision (e.g. 95% CI). Make clear which confounders were adjusted for and why they were included | Chapter 3 |
| (b) Report category boundaries when continuous variables were categorised | n/a | ||
| (c) If relevant, consider translating estimates of relative risk into absolute risk for a meaningful time period | Chapter 3, Long-term cancer risk; Lower- and higher-intermediate-risk groups | ||
| Other analyses | 17 | Report other analyses done, e.g. analyses of subgroups and interactions, and sensitivity analyses | Chapter 3, Lower- and intermediate-risk subgroups; Sensitivity |
| Discussion | |||
| Key results | 18 | Summarise key results with reference to study objectives | Chapter 7, Hospital and screening data sets; Main findings |
| Limitations | 19 | Discuss limitations of the study, taking into account sources of potential bias or imprecision. Discuss both direction and magnitude of any potential bias | Chapter 7, Strengths and limitations |
| Interpretation | 20 | Give a cautious overall interpretation of results considering objectives, limitations, multiplicity of analyses, results from similar studies, and other relevant evidence | Chapter 8 |
| Generalisability | 21 | Discuss the generalisability (external validity) of the study results | Chapter 7, Strengths and limitations Chapter 8 |
| Other information | |||
| Funding | 22 | Give the source of funding and the role of the funders for the present study and, if applicable, for the original study on which the present article is based | Acknowledgements |
List of abbreviations
- AA
- advanced adenoma
- AIC
- Akaike information criterion
- AN
- advanced neoplasia
- ANOVA
- analysis of variance
- BCSP
- Bowel Cancer Screening Programme
- BIC
- Bayesian information criterion
- BSG
- British Society of Gastroenterology
- CEAC
- cost-effectiveness acceptability curve
- CHI
- Community Health Index
- CI
- confidence interval
- CRC
- colorectal cancer
- DH
- Department of Health
- EP
- English Bowel Cancer Screening Pilot
- EPR
- Endoscopy and Pathology Report
- EPRA
- Endoscopy and Pathology Reports Application
- EU
- European Union
- EVPI
- expected value of perfect information
- EVPPI
- expected value of partial perfect information
- FAP
- familial adenomatous polyposis
- FS
- flexible sigmoidoscopy
- FUV1
- follow-up visit 1
- FUV2
- follow-up visit 2
- gFOBT
- guaiac faecal occult blood test
- GHQ
- General Health Questionnaire
- GP
- general practitioner
- HES
- Hospital Episode Statistics
- HGD
- high-grade dysplasia
- HIR
- higher intermediate risk
- HNPCC
- hereditary non-polyposis colorectal cancer
- HR
- hazard ratio
- HRQoL
- health-related quality of life
- HSCIC
- Health and Social Care Information Centre
- HTA
- Health Technology Assessment
- IA
- intermediate adenoma
- IBD
- inflammatory bowel disease
- ICD
- International Statistical Classification of Diseases and Related Health Problems
- ICD-O
- International Classification of Diseases for Oncology
- ICER
- incremental cost-effectiveness ratio
- IQR
- interquartile range
- IR
- intermediate risk
- KP
- Kaiser Permanente Colon Cancer Prevention Program
- LIR
- lower intermediate risk
- LRT
- likelihood ratio test
- LYG
- life-year gained
- MSM
- multistate model
- MVN
- multivariate normal
- NBOCAP
- National Bowel Cancer Audit Programme
- NHSCR
- National Health Service Cancer Registries
- NHSIC
- National Health Service Information Centre
- NICE
- National Institute for Health and Care Excellence
- NIHR
- National Institute for Health Research
- NSS
- National Services Scotland
- ONS
- Office for National Statistics
- OR
- odds ratio
- PAC
- Privacy Advisory Committee
- PAS
- Patient Administration System
- PCQ
- Psychological Consequences of screening Questionnaire
- PI
- principal investigator
- PIAG
- Patient Information Advisory Group
- PSS
- Personal Social Services
- pys
- person-years
- QALY
- quality-adjusted life-year
- R&D
- research and development
- RCT
- randomised controlled trial
- REC
- Research Ethics Committee
- ROC
- receiver operating characteristic
- S
- test sensitivity
- SE
- standard error
- SIR
- standardised incidence ratio
- SNOMED
- Systematized Nomenclature of Medicine
- SNOP
- Systematized Nomenclature of Pathology
- SOP
- standard operating procedure
- TTNE
- time to next event
- UKACR
- UK Association of Cancer Registries
- UKFSST
- UK Flexible Sigmoidoscopy Screening Trial
- VOI
- value of information
- WHO
- World Health Organization
- WTP
- willingness to pay